-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaCoping with Stress and the Emergence of Multidrug Resistance in Fungi
article has not abstract
Published in the journal: . PLoS Pathog 11(3): e32767. doi:10.1371/journal.ppat.1004668
Category: Pearls
doi: https://doi.org/10.1371/journal.ppat.1004668Summary
article has not abstract
The incidence of invasive fungal infections poses a serious global health threat, killing nearly 1.4 million people a year—a number comparable to deaths from tuberculosis. Factors that contribute to this disease burden and high mortality include insufficient diagnostics and treatment, and the increasing numbers of individuals with immune system defects who are particularly susceptible to fungal pathogens [1].
Because of the high risk of fungal infections in immunocompromised individuals—e.g., low–birth-weight neonates or patients undergoing organ transplantation or chemotherapy—it has become common medical practice to utilize antifungal prophylaxis in these patients during immunosuppression [2–4]. However, the expanding use of antifungal drugs has been associated with increasing incidence of antifungal drug resistance resulting from inherently less sensitive species and/or acquisition of drug class–specific resistance mechanisms [5]. Most alarming in recent years, multidrug-resistant strains of certain Candida species have emerged that are resistant to azoles and echinocandins, the two most widely used classes of antifungal drugs [6,7].
In this article we explore the notion that frequent and prolonged exposures of fungal cells to antifungal drugs activate fungal stress responses, which both support the short-term cellular adaptation to the drugs [8,9] and promote genetic instability to facilitate the emergence of stable drug resistant mutants refractory to therapy [9,10], including multidrug-resistant (MDR) strains (Fig. 1). Antifungal drug-induced stress has been associated with genetic instability in such distantly related fungi as Candida and Cryptococcus [11,12], suggesting that it is a broadly conserved phenomenon. In this article, we focus on drug resistance in Candida spp. because of its clinical significance, especially with the development of emerging multidrug resistance, and because mechanisms of drug resistance and stress-associated genetic instability are best studied in Candida, both in the clinic and in the lab.
Fig. 1. A model for the formation of multidrug-resistant strains of C. glabrata. 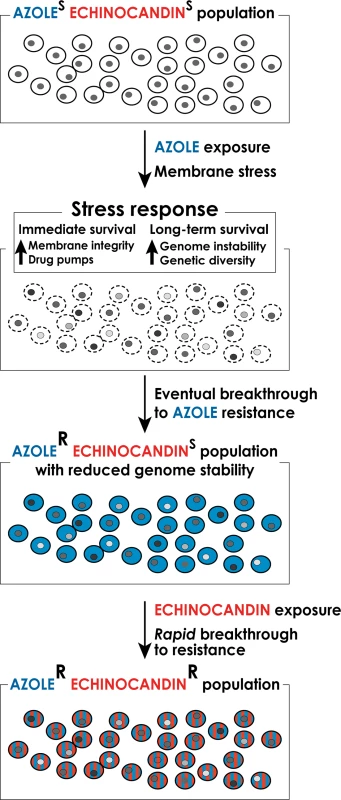
Long-term exposure to antifungal drugs triggers stress responses that both help maintain cellular integrity in the face of acute stress and promote genomic instability and genetic diversity to facilitate the emergence of genetic variants with enhanced intrinsic resistance. In the case of long-term prophylaxis with azole class drugs in Candida glabrata, this process results in formation of azole-resistant mutants with reduced genome stability. These mutants, when challenged with echinocandin class drugs, rapidly develop echinocandin resistance as well, resulting in formation of multidrug-resistant C. glabrata. Genetic Basis of Antifungal Drug Resistance
The most prevalent fungal pathogens belong to the Candida genus, with C. albicans and C. glabrata being the most common and second or third most commonly isolated bloodstream fungal pathogens in the United States [13,14], respectively. For more than three decades, Candida infections have been treated with azole antifungal agents, which target fungal membranes by inhibiting the biosynthesis of ergosterol. However, nearly 20% of C. glabrata strains exhibit intrinsic resistance to azoles, and even susceptible strains rapidly acquire resistance. This inherent resistant property has contributed to C. glabrata being the most frequently isolated Candida species in some high-risk centers [15,16]. To address this issue, echinocandin drugs are now recommended as first-line therapy to treat a range of candidiasis [17], since they do not exhibit cross-resistance to azole-resistant yeasts. A relatively recent addition to the antifungal drug repertoire, echinocandin class compounds target the fungal cell wall by inhibiting the biosynthesis of the key cell wall polymer β-(1,3)D-glucan. Troublingly, however, C. glabrata resistance to echinocandins is on the rise [18], with >10% of isolates now showing resistance in some hospital settings [7]. Many of the echinocandin-resistant C. glabrata isolates are also resistant to azoles, leaving extremely few options to treat patients infected with MDR strains [7].
Clinical resistance to echinocandins resulting in treatment failures is nearly always due to limited mutations in two highly conserved “hot spot regions” of genes encoding subunits of β-glucan synthase: FKS1 for most Candida species and FKS1 and/or FKS2 in C. glabrata [19,20]. Echinocandin resistance is always acquired during therapy, and the resulting amino acid changes in glucan synthase significantly decrease the sensitivity of the enzyme to the drug, resulting in higher MIC values and reduced pharmacodynamic responses [20].
In contrast to the echinocandins, azole drug resistance resulting in clinical failure may be caused by a variety of genetic changes, most of which affect the expression of fungal drug transporters or the structure and/or expression of fungal drug targets [9]. For instance, resistance to azoles can be caused by overexpression of transporters that mediate drug efflux out of the cell [21]. This overexpression can be achieved by amplification of the transporter genes, by large scale chromosomal rearrangement or chromosome gain that result in higher transporter gene copy number, or by point mutations in transcription factors that regulate transporter gene expression [22]. Resistance to azoles can also be mediated by mutations in the gene encoding their cellular target—the Erg11 protein, or by mutations in transcriptional regulators of ERG11 that result in Erg1 overexpression [23,24]. In C. albicans, clinical resistance to azoles frequently occurs in a stepwise manner, with a gradual increase in resistance associated with a sequential accumulation of mutations and genome rearrangements that together result in high-level azole resistance [25]. In contrast, in C. glabrata a single gain-of-function mutation in a transcriptional activator of drug efflux pumps is often sufficient to confer high-level azole resistance [26].
Theoretical and Experimental Support for Stress-Induced Genetic Instability
How do genetic changes that cause drug resistance arise and become fixed in fungal populations? The classical theory of evolution and natural selection posits that rare, random mutations arise in a population (e.g., due to occasional spontaneous errors in DNA replication and repair processes) and that a change in conditions, such as appearance of antifungal drug, would favor mutants that happen to be more fit under the new conditions (i.e., resistant to the drug). However, it seems clear that in thriving populations well adapted to their stable environments, genetic diversity is far less valuable than in maladapted populations, or during times of environmental change. From this point of view, a winning evolutionary strategy for an organism would contain the capacity to enhance its genetic diversity specifically during the times of stress. Indeed, mathematical modeling shows that the ability to generate increased genetic diversity by mutation or recombination specifically during the times of decreased fitness, i.e., stress, is itself a beneficial trait that would be favored by natural selection [27,28]. In other words, an organism with the capacity to modulate its spontaneous mutation and recombination rates—keeping them low during conditions of low stress and increasing them during conditions of high stress—would have a selective advantage over organisms with constant (either constitutively low or high) mutation and recombination rates.
The modeling studies are supported by empirical observations of increased genetic instability in fungi exposed to stresses such as high temperature, starvation, proteotoxic stress, and antifungal drugs. Starving laboratory strains of baker’s yeast (Saccharomyces cerevisiae) induces gross chromosomal rearrangements at rates several-fold higher than non-starving cells [29]. Interestingly, only a fraction of these rearrangements shows increased fitness relative to the parent strain under the starvation conditions, suggesting that rearrangements arise not by the selection of pre-existing genetic variants but by random starvation-induced genetic variability [29]. Furthermore, growth of laboratory yeast strains at high temperature or in the presence of antifungal drugs strongly induces gross chromosomal rearrangements and chromosomal gain or loss [30]. Studies of clinical C. albicans isolates from patients treated with azoles show that these strains frequently exhibit gross chromosomal rearrangements and/or aneuploidy [11]. Likewise, in C. glabrata isolated from patients, chromosomes are frequently reshuffled resulting in new genetic configurations, including appearance of small chromosomes [31,32]. Moreover, passaging C. albicans through a mouse in the absence of drug treatment increases chromosomal rearrangements relative to C. albicans passaged in regular laboratory medium, indicating that the stresses encountered in vivo, such as oxidative stress and elevated temperature promote genetic instability [33].
Roles of Cellular Stress Responses in Genetic Instability and Drug Resistance
Cellular stress responses are generally considered in terms of their role in helping cells tolerate and survive acute stress. For instance, echinocandin exposure compromises the cell wall and induces a number of stress signaling cascades, including those dependent on protein kinase C (PKC), Ca2+/calcineurin, HSP90 and high osmolarity glycerol (HOG) kinase, and up-regulation of chitin production to enhance cell wall stability [8,20]. These compensatory mechanisms produce a drug tolerant state where cells appear to resist the fungicidal properties of echinocandin drugs. The enhanced drug tolerance may explain why significant levels of surviving persister C. albicans cells were observed in mice after echinocandin treatment [34]. It is likely that stress response–mediated enhanced drug tolerance allows some cells sufficient time to develop a resistance-conferring mutation in FKS1 or FKS2.
However, the observations of chromosomal instability during stress strongly suggest that another role of cellular stress responses is to help increase genetic diversity during stress, presumably by altering the machinery responsible for maintaining genome integrity [10]. Indeed, several fungal stress response factors have been implicated in promoting genetic instability during stress. For instance, in S. cerevisiae, general stress response transcription factors Msn2 and Msn4 help promote certain types of mutagenesis in response to proteotoxic stress [35]. Furthermore, in both Saccharomyces and Candida, stress-induced aneuploidy depends on the function of a stress-inducible protein chaperone HSP90, which regulates the folding of a number of client proteins that function in chromosome segregation and cell cycle progression [30,36]. Another mechanism of stress-induced aneuploidy has been observed in several Candida species, including C. albicans and C. glabrata, involving mis-regulation of the fungal cell cycle shortly upon exposure to an antifungal agent. Within a few hours of exposure to azoles or echinocandins, Candida cells were observed to uncouple spindle formation and nuclear division from cell division, ultimately leading to the formation of tetraploid cells that underwent unequal division to produce aneuploid progeny [37].
While induction of aneuploidy appears to be an early event following stress, aneuploidy can promote other types of genomic rearrangements and mutagenic lesions [38]. This effect of aneuploidy is attributed to the fact that it alters gene dosage of a subset of the genome, thus disrupting the stoichiometry of complexes involved in chromosome maintenance and DNA repair whose components are encoded by different chromosomes.
Clinical Implications of Stress-Induced Genetic Instability
The potential for stress-induced genetic instability is especially troubling for drugs such as azoles, which are fungistatic—i.e., they do not kill fungal cells but only arrest their growth. As outlined above, arrested cells activate stress responses that promote genome destabilization and genetic diversity. Indeed, data from the clinic indicating rapid development of drug resistance following initiation of therapy [39] support the notion that exposure of C. glabrata to an azole may create fungal cells with highly variable, “evolvable” genomes capable of quick breakthrough of resistance to any other types of drug, such as echinocandins (Fig. 1). Thus, patients encountering fluconazole prophylaxis may show infections with azole resistant C. glabrata followed by rapid breakthrough on echinocandin therapy, resulting in multidrug-resistant strains. In light of this phenomenon, it may be necessary to review the current practice of prophylaxis in some settings, which promotes such genetic instability. Furthermore, there is an urgent need for new fungicidal drugs, for safe combinations of antifungal drugs that would require mutagenesis of more than one target to create resistance (a much more rare event), and for a deeper understanding of mechanisms underlying stress-induced genetic instability so that they could potentially be pharmacologically mitigated during antifungal therapy.
Zdroje
1. Brown GD, Denning DW, Gow NA, Levitz SM, Netea MG, et al. (2012) Hidden killers: human fungal infections. Sci Transl Med 4 : 165rv113. doi: 10.1126/scitranslmed.3004404 23253612
2. Ziakas PD, Kourbeti IS, Mylonakis E (2014) Systemic antifungal prophylaxis after hematopoietic stem cell transplantation: a meta-analysis. Clin Ther 36 : 292–306 e291. doi: 10.1016/j.clinthera.2013.11.010 24439393
3. Greenberg RG, Benjamin DK Jr. (2014) Neonatal candidiasis: Diagnosis, prevention, and treatment. J Infect 69 Suppl 1: S19–22. doi: 10.1016/j.jinf.2014.07.012 25129318
4. Robenshtok E, Gafter-Gvili A, Goldberg E, Weinberger M, Yeshurun M, et al. (2007) Antifungal prophylaxis in cancer patients after chemotherapy or hematopoietic stem-cell transplantation: systematic review and meta-analysis. J Clin Oncol 25 : 5471–5489. 17909198
5. Pfaller MA, Moet GJ, Messer SA, Jones RN, Castanheira M (2011) Geographic variations in species distribution and echinocandin and azole antifungal resistance rates among Candida bloodstream infection isolates: report from the SENTRY Antimicrobial Surveillance Program (2008 to 2009). J Clin Microbiol 49 : 396–399. doi: 10.1128/JCM.01398-10 21068282
6. Pfaller MA, Messer SA, Woosley LN, Jones RN, Castanheira M (2013) Echinocandin and triazole antifungal susceptibility profiles for clinical opportunistic yeast and mold isolates collected from 2010 to 2011: application of new CLSI clinical breakpoints and epidemiological cutoff values for characterization of geographic and temporal trends of antifungal resistance. J Clin Microbiol 51 : 2571–2581. doi: 10.1128/JCM.00308-13 23720791
7. Alexander BD, Johnson MD, Pfeiffer CD, Jimenez-Ortigosa C, Catania J, et al. (2013) Increasing echinocandin resistance in Candida glabrata: clinical failure correlates with presence of FKS mutations and elevated minimum inhibitory concentrations. Clin Infect Dis 56 : 1724–1732. doi: 10.1093/cid/cit136 23487382
8. Walker LA, Gow NA, Munro CA (2010) Fungal echinocandin resistance. Fungal Genet Biol 47 : 117–126. doi: 10.1016/j.fgb.2009.09.003 19770064
9. Cowen LE, Steinbach WJ (2008) Stress, drugs, and evolution: the role of cellular signaling in fungal drug resistance. Eukaryot Cell 7 : 747–764. doi: 10.1128/EC.00041-08 18375617
10. Galhardo RS, Hastings PJ, Rosenberg SM (2007) Mutation as a stress response and the regulation of evolvability. Crit Rev Biochem Mol Biol 42 : 399–435. 17917874
11. Selmecki A, Forche A, Berman J (2006) Aneuploidy and isochromosome formation in drug-resistant Candida albicans. Science 313 : 367–370. 16857942
12. Sionov E, Lee H, Chang YC, Kwon-Chung KJ (2010) Cryptococcus neoformans overcomes stress of azole drugs by formation of disomy in specific multiple chromosomes. PLoS Pathog 6: e1000848. doi: 10.1371/journal.ppat.1000848 20368972
13. Wisplinghoff H, Ebbers J, Geurtz L, Stefanik D, Major Y, et al. (2014) Nosocomial bloodstream infections due to Candida spp. in the USA: species distribution, clinical features and antifungal susceptibilities. Int J Antimicrob Agents 43 : 78–81. doi: 10.1016/j.ijantimicag.2013.09.005 24182454
14. Pfaller MA, Moet GJ, Messer SA, Jones RN, Castanheira M (2011) Candida bloodstream infections: comparison of species distributions and antifungal resistance patterns in community-onset and nosocomial isolates in the SENTRY Antimicrobial Surveillance Program, 2008–2009. Antimicrob Agents Chemother 55 : 561–566. doi: 10.1128/AAC.01079-10 21115790
15. Farmakiotis D, Tarrand JJ, Kontoyiannis DP (2014) Drug-resistant Candida glabrata infection in cancer patients. Emerg Infect Dis 20 : 1833–1840. doi: 10.3201/eid2011.140685 25340258
16. Lortholary O, Desnos-Ollivier M, Sitbon K, Fontanet A, Bretagne S, et al. (2011) Recent exposure to caspofungin or fluconazole influences the epidemiology of candidemia: a prospective multicenter study involving 2,441 patients. Antimicrob Agents Chemother 55 : 532–538. doi: 10.1128/AAC.01128-10 21078946
17. Pappas PG, Kauffman CA, Andes D, Benjamin DK Jr., Calandra TF, et al. (2009) Clinical practice guidelines for the management of candidiasis: 2009 update by the Infectious Diseases Society of America. Clin Infect Dis 48 : 503–535. doi: 10.1086/596757 19191635
18. Pfaller MA, Castanheira M, Lockhart SR, Ahlquist AM, Messer SA, et al. (2012) Frequency of decreased susceptibility and resistance to echinocandins among fluconazole-resistant bloodstream isolates of Candida glabrata. J Clin Microbiol 50 : 1199–1203. doi: 10.1128/JCM.06112-11 22278842
19. Perlin DS (2007) Resistance to echinocandin-class antifungal drugs. Drug Resist Updat 10 : 121–130. 17569573
20. Perlin DS (2014) Echinocandin resistance, susceptibility testing and prophylaxis: implications for patient management. Drugs 74 : 1573–1585. doi: 10.1007/s40265-014-0286-5 25255923
21. White TC, Holleman S, Dy F, Mirels LF, Stevens DA (2002) Resistance mechanisms in clinical isolates of Candida albicans. Antimicrobial Agents and Chemotherapy 46 : 1704–1713. 12019079
22. Schubert S, Barker KS, Znaidi S, Schneider S, Dierolf F, et al. (2011) Regulation of efflux pump expression and drug resistance by the transcription factors Mrr1, Upc2, and Cap1 in Candida albicans. Antimicrob Agents Chemother 55 : 2212–2223. doi: 10.1128/AAC.01343-10 21402859
23. Flowers SA, Barker KS, Berkow EL, Toner G, Chadwick SG, et al. (2012) Gain-of-function mutations in UPC2 are a frequent cause of ERG11 upregulation in azole-resistant clinical isolates of Candida albicans. Eukaryot Cell 11 : 1289–1299. doi: 10.1128/EC.00215-12 22923048
24. Flowers SA, Colon B, Whaley SG, Schuler MA, Rogers PD (2014) The contribution of clinically derived mutations in ERG11 to azole resistance in Candida albicans. Antimicrob Agents Chemother 59 : 450–60. doi: 10.1128/AAC.03470-14 25385095
25. Selmecki AM, Dulmage K, Cowen LE, Anderson JB, Berman J (2009) Acquisition of aneuploidy provides increased fitness during the evolution of antifungal drug resistance. PLoS Genet 5: e1000705. doi: 10.1371/journal.pgen.1000705 19876375
26. Vermitsky J-P, Earhart KD, Smith WL, Homayouni R, Edlind TD, et al. (2006) Pdr1 regulates multidrug resistance in Candida glabrata: gene disruption and genome-wide expression studies. Mol Microbiol 61 : 704–722. 16803598
27. Ram Y, Hadany L (2014) Stress-induced mutagenesis and complex adaptation. Proc Biol Sci 281. pii: 20141025
28. Hadany L, Beker T (2003) On the evolutionary advantage of fitness-associated recombination. Genetics 165 : 2167–2179. 14704195
29. Coyle S, Kroll E (2008) Starvation induces genomic rearrangements and starvation-resilient phenotypes in yeast. Mol Biol Evol 25 : 310–318. 18032404
30. Chen G, Bradford WD, Seidel CW, Li R (2012) Hsp90 stress potentiates rapid cellular adaptation through induction of aneuploidy. Nature 482 : 246–250. doi: 10.1038/nature10795 22286062
31. Shin JH, Chae MJ, Song JW, Jung SI, Cho D, et al. (2007) Changes in karyotype and azole susceptibility of sequential bloodstream isolates from patients with Candida glabrata candidemia. J Clin Microbiol 45 : 2385–2391. 17581937
32. Poláková S, Blume C, Zárate JA, Mentel M, Jørck-Ramberg D, et al. (2009) Formation of new chromosomes as a virulence mechanism in yeast Candida glabrata. Proc Natl Acad Sci USA 106 : 2688–2693. doi: 10.1073/pnas.0809793106 19204294
33. Forche A, Magee PT, Selmecki A, Berman J, May G (2009) Evolution in Candida albicans populations during a single passage through a mouse host. Genetics 182 : 799–811. doi: 10.1534/genetics.109.103325 19414562
34. Slater JL, Howard SJ, Sharp A, Goodwin J, Gregson LM, et al. (2011) Disseminated candidiasis caused by Candida albicans with amino acid substitutions in Fks1 at position Ser645 cannot be successfully treated with micafungin. Antimicrob Agents Chemother 55 : 3075–3083. doi: 10.1128/AAC.01686-10 21502627
35. Shor E, Fox CA, Broach JR (2013) The yeast environmental stress response regulates mutagenesis induced by proteotoxic stress. PLoS Genet 9: e1003680. doi: 10.1371/journal.pgen.1003680 23935537
36. Cowen LE (2009) Hsp90 orchestrates stress response signaling governing fungal drug resistance. PLoS Pathog 5: e1000471. doi: 10.1371/journal.ppat.1000471 19714223
37. Harrison BD, Hashemi J, Bibi M, Pulver R, Bavli D, et al. (2014) A tetraploid intermediate precedes aneuploid formation in yeasts exposed to fluconazole. PLoS Biol 12: e1001815. doi: 10.1371/journal.pbio.1001815 24642609
38. Sheltzer JM, Blank HM, Pfau SJ, Tange Y, George BM, et al. (2011) Aneuploidy drives genomic instability in yeast. Science 333 : 1026–1030. doi: 10.1126/science.1206412 21852501
39. Lewis JS 2nd, Wiederhold NP, Wickes BL, Patterson TF, Jorgensen JH (2013) Rapid emergence of echinocandin resistance in Candida glabrata resulting in clinical and microbiologic failure. Antimicrob Agents Chemother 57 : 4559–4561. doi: 10.1128/AAC.01144-13 23817368
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek A Phospholipase Is Involved in Disruption of the Liver Stage Parasitophorous Vacuole MembraneČlánek Host ESCRT Proteins Are Required for Bromovirus RNA Replication Compartment Assembly and FunctionČlánek Enhanced CD8 T Cell Responses through GITR-Mediated Costimulation Resolve Chronic Viral Infection
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2015 Číslo 3- Jak souvisí postcovidový syndrom s poškozením mozku?
- Měli bychom postcovidový syndrom léčit antidepresivy?
- Farmakovigilanční studie perorálních antivirotik indikovaných v léčbě COVID-19
- 10 bodů k očkování proti COVID-19: stanovisko České společnosti alergologie a klinické imunologie ČLS JEP
-
Všechny články tohoto čísla
- To Be or Not IIb: A Multi-Step Process for Epstein-Barr Virus Latency Establishment and Consequences for B Cell Tumorigenesis
- Is Antigenic Sin Always “Original?” Re-examining the Evidence Regarding Circulation of a Human H1 Influenza Virus Immediately Prior to the 1918 Spanish Flu
- The Great Escape: Pathogen Versus Host
- Coping with Stress and the Emergence of Multidrug Resistance in Fungi
- Catch Me If You Can: The Link between Autophagy and Viruses
- Bacterial Immune Evasion through Manipulation of Host Inhibitory Immune Signaling
- Evidence for Ubiquitin-Regulated Nuclear and Subnuclear Trafficking among Matrix Proteins
- BILBO1 Is a Scaffold Protein of the Flagellar Pocket Collar in the Pathogen
- Production of Anti-LPS IgM by B1a B Cells Depends on IL-1β and Is Protective against Lung Infection with LVS
- Virulence Regulation with Venus Flytrap Domains: Structure and Function of the Periplasmic Moiety of the Sensor-Kinase BvgS
- α-Hemolysin Counteracts the Anti-Virulence Innate Immune Response Triggered by the Rho GTPase Activating Toxin CNF1 during Bacteremia
- Induction of Interferon-Stimulated Genes by IRF3 Promotes Replication of
- Intracellular Growth Is Dependent on Tyrosine Catabolism in the Dimorphic Fungal Pathogen
- HCV Induces the Expression of Rubicon and UVRAG to Temporally Regulate the Maturation of Autophagosomes and Viral Replication
- Spatiotemporal Analysis of Hepatitis C Virus Infection
- Subgingival Microbial Communities in Leukocyte Adhesion Deficiency and Their Relationship with Local Immunopathology
- Interaction between the Type III Effector VopO and GEF-H1 Activates the RhoA-ROCK Pathway
- Attenuation of Tick-Borne Encephalitis Virus Using Large-Scale Random Codon Re-encoding
- Establishment of HSV1 Latency in Immunodeficient Mice Facilitates Efficient Reactivation
- XRN1 Stalling in the 5’ UTR of Hepatitis C Virus and Bovine Viral Diarrhea Virus Is Associated with Dysregulated Host mRNA Stability
- γδ T Cells Confer Protection against Murine Cytomegalovirus (MCMV)
- Rhadinovirus Host Entry by Co-operative Infection
- A Phospholipase Is Involved in Disruption of the Liver Stage Parasitophorous Vacuole Membrane
- Dermal Neutrophil, Macrophage and Dendritic Cell Responses to Transmitted by Fleas
- Elucidation of Sigma Factor-Associated Networks in Reveals a Modular Architecture with Limited and Function-Specific Crosstalk
- A Conserved NS3 Surface Patch Orchestrates NS2 Protease Stimulation, NS5A Hyperphosphorylation and HCV Genome Replication
- Host ESCRT Proteins Are Required for Bromovirus RNA Replication Compartment Assembly and Function
- Disruption of IL-21 Signaling Affects T Cell-B Cell Interactions and Abrogates Protective Humoral Immunity to Malaria
- Compartmentalized Replication of R5 T Cell-Tropic HIV-1 in the Central Nervous System Early in the Course of Infection
- Diminished Reovirus Capsid Stability Alters Disease Pathogenesis and Littermate Transmission
- Characterization of CD8 T Cell Differentiation following SIVΔnef Vaccination by Transcription Factor Expression Profiling
- Visualization of HIV-1 Interactions with Penile and Foreskin Epithelia: Clues for Female-to-Male HIV Transmission
- Sensing Cytosolic RpsL by Macrophages Induces Lysosomal Cell Death and Termination of Bacterial Infection
- PKCη/Rdx-driven Phosphorylation of PDK1: A Novel Mechanism Promoting Cancer Cell Survival and Permissiveness for Parvovirus-induced Lysis
- Metalloprotease NleC Suppresses Host NF-κB/Inflammatory Responses by Cleaving p65 and Interfering with the p65/RPS3 Interaction
- Immune Antibodies and Helminth Products Drive CXCR2-Dependent Macrophage-Myofibroblast Crosstalk to Promote Intestinal Repair
- Adenovirus Entry From the Apical Surface of Polarized Epithelia Is Facilitated by the Host Innate Immune Response
- The RNA Template Channel of the RNA-Dependent RNA Polymerase as a Target for Development of Antiviral Therapy of Multiple Genera within a Virus Family
- Neutrophils: Between Host Defence, Immune Modulation, and Tissue Injury
- CD169-Mediated Trafficking of HIV to Plasma Membrane Invaginations in Dendritic Cells Attenuates Efficacy of Anti-gp120 Broadly Neutralizing Antibodies
- Japanese Encephalitis Virus Nonstructural Protein NS5 Interacts with Mitochondrial Trifunctional Protein and Impairs Fatty Acid β-Oxidation
- Yip1A, a Novel Host Factor for the Activation of the IRE1 Pathway of the Unfolded Protein Response during Infection
- TRIM26 Negatively Regulates Interferon-β Production and Antiviral Response through Polyubiquitination and Degradation of Nuclear IRF3
- Parallel Epigenomic and Transcriptomic Responses to Viral Infection in Honey Bees ()
- A Crystal Structure of the Dengue Virus NS5 Protein Reveals a Novel Inter-domain Interface Essential for Protein Flexibility and Virus Replication
- Enhanced CD8 T Cell Responses through GITR-Mediated Costimulation Resolve Chronic Viral Infection
- Exome and Transcriptome Sequencing of Identifies a Locus That Confers Resistance to and Alters the Immune Response
- The Role of Misshapen NCK-related kinase (MINK), a Novel Ste20 Family Kinase, in the IRES-Mediated Protein Translation of Human Enterovirus 71
- Chitin Recognition via Chitotriosidase Promotes Pathologic Type-2 Helper T Cell Responses to Cryptococcal Infection
- Activates Both IL-1β and IL-1 Receptor Antagonist to Modulate Lung Inflammation during Pneumonic Plague
- Persistence of Transmitted HIV-1 Drug Resistance Mutations Associated with Fitness Costs and Viral Genetic Backgrounds
- An 18 kDa Scaffold Protein Is Critical for Biofilm Formation
- Early Virological and Immunological Events in Asymptomatic Epstein-Barr Virus Infection in African Children
- Human CD8 T-cells Recognizing Peptides from () Presented by HLA-E Have an Unorthodox Th2-like, Multifunctional, Inhibitory Phenotype and Represent a Novel Human T-cell Subset
- Decreased HIV-Specific T-Regulatory Responses Are Associated with Effective DC-Vaccine Induced Immunity
- RSV Vaccine-Enhanced Disease Is Orchestrated by the Combined Actions of Distinct CD4 T Cell Subsets
- Concerted Activity of IgG1 Antibodies and IL-4/IL-25-Dependent Effector Cells Trap Helminth Larvae in the Tissues following Vaccination with Defined Secreted Antigens, Providing Sterile Immunity to Challenge Infection
- Structure of the Low pH Conformation of Chandipura Virus G Reveals Important Features in the Evolution of the Vesiculovirus Glycoprotein
- PPM1A Regulates Antiviral Signaling by Antagonizing TBK1-Mediated STING Phosphorylation and Aggregation
- Lipidomic Analysis Links Mycobactin Synthase K to Iron Uptake and Virulence in .
- Roles and Programming of Arabidopsis ARGONAUTE Proteins during Infection
- Impact of Infection on Host Macrophage Nuclear Physiology and Nucleopore Complex Integrity
- The Impact of Host Diet on Titer in
- Antimicrobial-Induced DNA Damage and Genomic Instability in Microbial Pathogens
- Herpesviral G Protein-Coupled Receptors Activate NFAT to Induce Tumor Formation via Inhibiting the SERCA Calcium ATPase
- The Causes and Consequences of Changes in Virulence following Pathogen Host Shifts
- Small GTPase Rab21 Mediates Fibronectin Induced Actin Reorganization in : Implications in Pathogen Invasion
- Positive Role of Promyelocytic Leukemia Protein in Type I Interferon Response and Its Regulation by Human Cytomegalovirus
- NEDDylation Is Essential for Kaposi’s Sarcoma-Associated Herpesvirus Latency and Lytic Reactivation and Represents a Novel Anti-KSHV Target
- β-HPV 5 and 8 E6 Disrupt Homology Dependent Double Strand Break Repair by Attenuating BRCA1 and BRCA2 Expression and Foci Formation
- An O Antigen Capsule Modulates Bacterial Pathogenesis in
- Variable Processing and Cross-presentation of HIV by Dendritic Cells and Macrophages Shapes CTL Immunodominance and Immune Escape
- Probing the Metabolic Network in Bloodstream-Form Using Untargeted Metabolomics with Stable Isotope Labelled Glucose
- Adhesive Fiber Stratification in Uropathogenic Biofilms Unveils Oxygen-Mediated Control of Type 1 Pili
- Vaccinia Virus Protein Complex F12/E2 Interacts with Kinesin Light Chain Isoform 2 to Engage the Kinesin-1 Motor Complex
- Modulates Host Macrophage Mitochondrial Metabolism by Hijacking the SIRT1-AMPK Axis
- Human T-Cell Leukemia Virus Type 1 (HTLV-1) Tax Requires CADM1/TSLC1 for Inactivation of the NF-κB Inhibitor A20 and Constitutive NF-κB Signaling
- Suppression of RNAi by dsRNA-Degrading RNaseIII Enzymes of Viruses in Animals and Plants
- Spatiotemporal Regulation of a T4SS Substrate by the Metaeffector SidJ
- Antigenic Properties of the Human Immunodeficiency Virus Envelope Glycoprotein Gp120 on Virions Bound to Target Cells
- Dependence of Intracellular and Exosomal microRNAs on Viral Oncogene Expression in HPV-positive Tumor Cells
- Identification of a Peptide-Pheromone that Enhances Escape from Host Cell Vacuoles
- Impaired Systemic Tetrahydrobiopterin Bioavailability and Increased Dihydrobiopterin in Adult Falciparum Malaria: Association with Disease Severity, Impaired Microvascular Function and Increased Endothelial Activation
- Transgenic Expression of the Dicotyledonous Pattern Recognition Receptor EFR in Rice Leads to Ligand-Dependent Activation of Defense Responses
- Comprehensive Antigenic Map of a Cleaved Soluble HIV-1 Envelope Trimer
- Low Doses of Imatinib Induce Myelopoiesis and Enhance Host Anti-microbial Immunity
- Impaired Systemic Tetrahydrobiopterin Bioavailability and Increased Oxidized Biopterins in Pediatric Falciparum Malaria: Association with Disease Severity
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Bacterial Immune Evasion through Manipulation of Host Inhibitory Immune Signaling
- BILBO1 Is a Scaffold Protein of the Flagellar Pocket Collar in the Pathogen
- Antimicrobial-Induced DNA Damage and Genomic Instability in Microbial Pathogens
- Attenuation of Tick-Borne Encephalitis Virus Using Large-Scale Random Codon Re-encoding
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



