-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaBoth α2,3- and α2,6-Linked Sialic Acids on O-Linked Glycoproteins Act as Functional Receptors for Porcine Sapovirus
Although enteropathogenic sapoviruses and noroviruses are leading causes of acute gastroenteritis in both humans and animals, the study of viral pathogenesis and immunity of these ubiquitous pathogens has been hampered due to the lack of a fully permissive cell culture system. Porcine sapovirus Cowden strain provides a suitable system that can be used to identify the molecular mechanisms of viral pathogenesis. Previous studies have shown that carbohydrates and glycolipids play important roles in the attachment of members of the Caliciviridae; histo-blood group antigens (HBGAs) are used by Norovirus genogroups I to IV, as well as members of the Lagovirus, and Recovirus genera, whereas terminal sialic acid is recognized as a receptor for feline calicivirus and murine norovirus. To date, however, the role of carbohydrates in the life cycle of sapoviruses has remained largely unknown. We found that porcine sapovirus binds to susceptible host cells through both α2,3 - and α2,6-linked terminal sialic acids which are attached to O-linked glycoproteins. These efforts, findings and insights will significantly contribute to a better understanding of the sapovirus life cycle.
Published in the journal: . PLoS Pathog 10(6): e32767. doi:10.1371/journal.ppat.1004172
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1004172Summary
Although enteropathogenic sapoviruses and noroviruses are leading causes of acute gastroenteritis in both humans and animals, the study of viral pathogenesis and immunity of these ubiquitous pathogens has been hampered due to the lack of a fully permissive cell culture system. Porcine sapovirus Cowden strain provides a suitable system that can be used to identify the molecular mechanisms of viral pathogenesis. Previous studies have shown that carbohydrates and glycolipids play important roles in the attachment of members of the Caliciviridae; histo-blood group antigens (HBGAs) are used by Norovirus genogroups I to IV, as well as members of the Lagovirus, and Recovirus genera, whereas terminal sialic acid is recognized as a receptor for feline calicivirus and murine norovirus. To date, however, the role of carbohydrates in the life cycle of sapoviruses has remained largely unknown. We found that porcine sapovirus binds to susceptible host cells through both α2,3 - and α2,6-linked terminal sialic acids which are attached to O-linked glycoproteins. These efforts, findings and insights will significantly contribute to a better understanding of the sapovirus life cycle.
Introduction
Caliciviruses (family Caliciviridae) are small (27–40 nm), non-enveloped, icosahedral viruses that possess a single-strand, plus-sense genomic RNA of 7–8 kb [1]. Caliciviruses are important veterinary and human pathogens which are associated with a broad spectrum of diseases in their respective hosts. A member of the genus Lagovirus, rabbit hemorrhagic disease virus (RHDV), is associated with a fatal liver disease in rabbits [2]. Feline calicivirus (FCV), a member of the genus Vesivirus, causes respiratory disease in cats [3], [4]. Caliciviruses in the genera Norovirus and Sapovirus are important acute gastroenteritis pathogens in humans and animals [5], [6]. Each year, human noroviruses cause at least 1.1 million episodes and 218,000 deaths in developing nations as well as approximately 900,000 cases of pediatric gastroenteritis in industrialized nations [7]. Sapoviruses have also been associated with gastroenteritis outbreaks and with disease in pediatric patients [1]. The genus Sapovirus can be divided into five genogroups (GI–GV), among which GI, GII, GIV and GV are known to infect humans, whereas GIII infects porcine species [8]. No fully permissive cell culture system currently exists for the enteric caliciviruses associated with gastroenteritis in humans, hampering the study of viral pathogenesis and immunity of these ubiquitous pathogens [1].
The initial events in a viral infection are induced by binding of the virus to the surface of the host cell, followed by penetration or release of the virus particle into the cytoplasm of the cell. Binding occurs through interactions between the virion and receptors on the plasma membrane of the target cell, and consequently receptors are important determinants of viral tissue tropism and pathogenesis [1]. Among the members of the Calicivirdae family, an attachment factor for RHDV was identified as H-type 2 histo-blood group antigen (HBGA), and this led to further studies identifying factors involved in the attachment of the other members of the family [9]. HBGAs function as the attachment factor of both human and bovine noroviruses [5], [10], while sialic acid linked with gangliosides acts as at least part of the murine norovirus (MNV) receptor [11]. In addition, Tulane virus, the newly discovered rhesus monkey calicivirus, uses HBGA as a receptor [12]. FCV is reported to recognize terminal sialic acid on an N-linked glycoprotein for attachment [4], with junctional adhesion molecule-1 (JAM-1) functioning as a receptor to facilitate FCV penetration and infection of host cells [5], [13]. Although these reports strongly suggest that the recognition of a carbohydrate receptor may be a common feature of many caliciviruses, it is also clear that different caliciviruses recognize different carbohydrate receptors [5]. Importantly, however, virus-like particles of human sapovirus genogroups GI and GV strains are known not to bind salivary HBGAs or synthetic carbohydrates [14].
The close genetic relationship of noroviruses and sapoviruses found in humans and animals has led to concern over the possibility of zoonotic transmission of these viruses [15]. Although animal sapoviruses and noroviruses have not yet been isolated from humans, the detection of antibodies against bovine GIII norovirus and canine GVI norovirus in human serum samples [16], [17], and the detection of human-like GII.4 norovirus in porcine and bovine fecal samples [18] suggest the possible zoonotic transmission of noroviruses. Furthermore, it has been demonstrated that human norovirus GII4-HS66 strain can induce diarrhea and intestinal lesions in the proximal small intestine of gnotobiotic piglets or calves [19]. There is also evidence to suggest that highly conserved receptors across host species can be shared by different viruses, even among different genera or families [10], [20], [21]. Such concerns have prompted us to investigate the ability of porcine sapovirus (PSaV) to recognize carbohydrates present on cultured cells of porcine origin and small intestinal epithelial cells of piglets. In this study, we demonstrate that PSaV binds to both α2,3 - and α2,6-linked sialic acids present on an O-linked glycoprotein.
Results
Attachment and infection of PSaV requires carbohydrate moieties
Although carbohydrate moieties are known to act as receptors or attachment factors for various caliciviruses [5], their role as receptors or attachment factors for members of the Sapovirus genus remains unknown. To determine if PSaV Cowden strain requires carbohydrate moieties for binding and infection, we removed the carbohydrate moieties from permissive porcine LLC-PK cells by treatment with sodium periodate (NaIO4), which is known to cleave carbohydrate groups without altering proteins or membranes [4], [22], [23]. Pretreatment of LLC-PK cells with 1 mM or 5 mM NaIO4 markedly reduced the binding of Alexa 594-labeled PSaV Cowden strain compared to mock treated control (Fig. 1A). To quantify the effect of NaIO4 treatment more accurately, LLC-PK cells were pretreated in a similar manner, and were incubated with radio-labeled PSaV Cowden strain. Cells were washed thoroughly, and virus binding was determined by liquid scintillation counting. Binding of PSaV Cowden strain was reduced to 12% of the levels observed in mock treated cells with 1 mM NaIO4, and to 2% in cells treated with 5 mM NaIO4 (Fig. 1B). The infection rate, as determined by staining cells for the viral antigen VPg, was also significantly reduced; infection rates of 17% and 3% were observed for 1 mM and 5 mM NaIO4, respectively, when compared with mock-treated cells (Fig.1C and 1D). A similar degree of inhibition of binding and infection was observed in FCV F9 strain-infected Crandall-Reese feline kidney (CRFK) cells that were pretreated with NaIO4 (Fig.1B and 1D). However, binding and infection of coxsackievirus B3 (CVB3) Nancy strain, which is known to us decay-accelerating factor as a receptor [24], was not influenced by the pretreatment of HeLa cells with 1 mM or 5 mM NaIO4 (Fig. 1B and 1D, and Fig. S1). In addition, pretreatment of LLC-PK cells with 5 mM NaIO4 had no effect on binding of MNV-1 strain CW1 or vesicular stomatitis virus glycoprotein (VSV-G protein) pseudotyped lentiviruses (Fig. S1). These data strongly indicated that like FCV F9 strain, PSaV Cowden strain utilizes carbohydrate moieties for binding and infection.
Fig. 1. Porcine sapovirus Cowden strain binding and infection requires carbohydrate moieties. 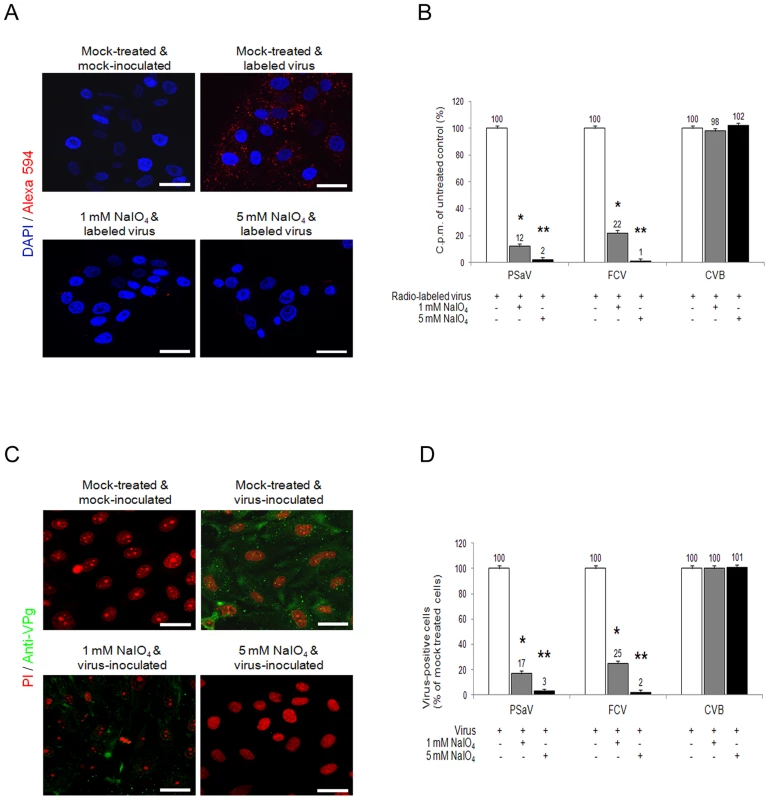
LLC-PK cells were pretreated with 1 mM or 5 mM of NaIO4 to remove carbohydrate moieties. (A) Alexa 594 or Alexa 594-labeled PSaV (MOI of 100 pfu/cell) were bound to pretreated LLC-PK cells, and were subsequently examined for bound virus by confocal microscopy. (B) 35[S]Methionine/Cysteine-labeled PSaV, control FCV or CVB3 (50,000 c.p.m.) were bound to LLC-PK, CRFK or HeLa cells which were pretreated with or without NaIO4. Binding of radio-labeled PSaV, FCV or CVB3 was measured by liquid scintillation counting. (C) PSaV (MOI of 0.1 pfu/cell) was inoculated to NaIO4 pretreated LLC-PK cells, and was subsequently analyzed by immunofluorescence assay to detect the viral antigen VPg, using a rabbit polyclonal antibody 72 h post infection. (D) PSaV, FCV or CVB3 positive cells (%) were quantified in three independent microscope fields. All experiments were performed three independent times, and figures A and C show one representative set of results. The scale bars correspond to 20 µm. Error bars indicate SD from triplicate samples. *p<0.05, **p<0.005. PSaV does not agglutinate any red blood cell (RBC) from various species
HBGAs are complex carbohydrates present on the surfaces of RBCs, and mucosal epithelia, mucin of respiratory, genitourinary, and digestive tract [25]. Among caliciviruses, human and bovine noroviruses, RHDV, and Tulane virus are known to use HBGAs as receptors, resulting in the agglutination of RBCs [5], [12]. To determine if PSaV Cowden strain agglutinated RBCs, hemagglutination assay (HA) was performed by using RBCs that originated from various animal species including pigs, rats, chickens, and humans, which was further classified into ABO and Lewis types. PSaV Cowden strain displayed no hemagglutination activity with RBCs from any species at 4°C or 20°C incubation (Fig. 2). In contrast, influenza A virus PR8 (H1N1) strain agglutinated human, rat, pig and chicken RBCs. P particles of human norovirus VA387 strain and a VP8* of human rotavirus DS-1 strain confirmed binding to corresponding HBGAs [26] (Fig. 2). Whilst positive controls showed differing degrees of HA activities against RBCs from various species, the absence of HA activity by PSaV indicated that PSaV Cowden strain may not recognize HBGAs as receptors for its binding and infection of host cells.
Fig. 2. Porcine sapovirus Cowden strain does not agglutinate RBCs from various species. 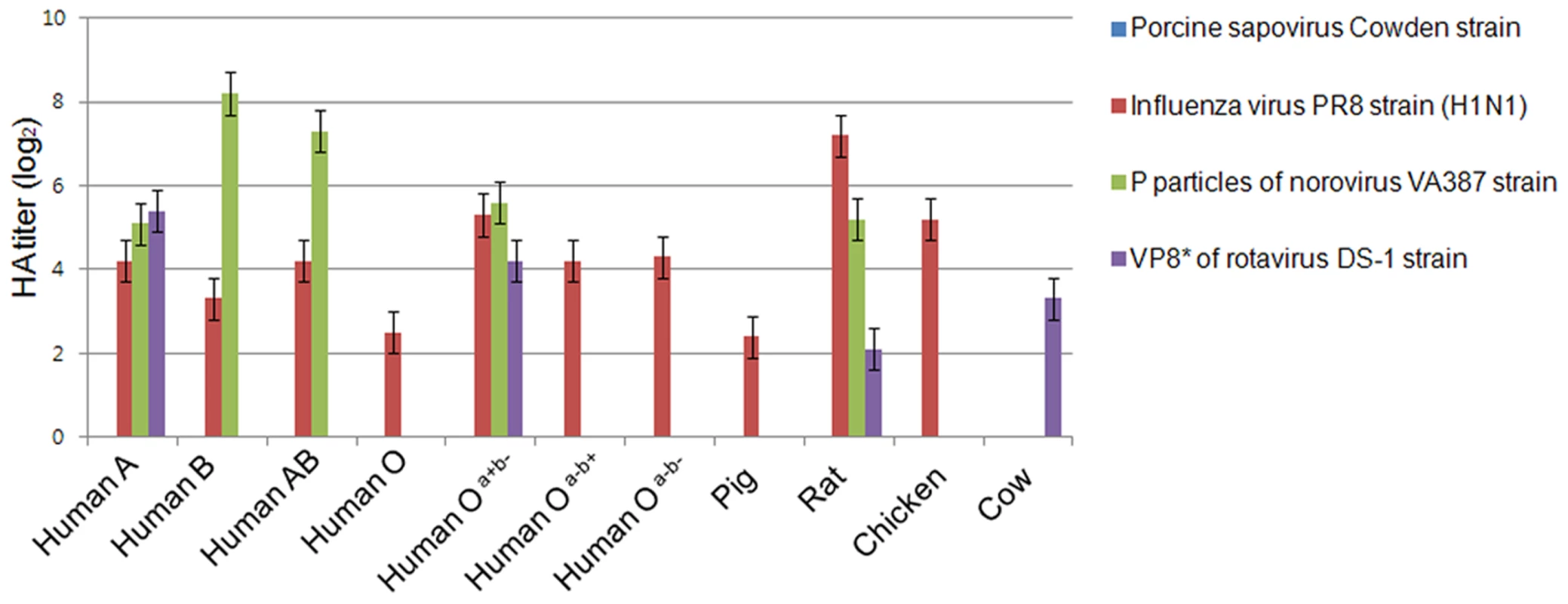
Agglutination of pig, cow, chicken, rat and human (ABO and Lewis types) RBCs by PSaV Cowden strain, human influenza A virus strain A/Puerto Rico/8/34 (H1N1) (PR8 virus), P particles of norovirus VA387 strain, or VP8* of human rotavirus DS-1 strain were determined by hemagglutination assay. Results are shown as average hemagglutination titers (Log2) obtained from triplicate samples in three independent experiments. Error bars represent mean ± SD. PSaV does not interact with synthetic HBGAs
In order to confirm whether PSaV recognizes HBGAs as receptors, a synthetic HBGA binding assay was conducted [26], [27]. The descriptions and structures of all the oligosaccharides tested are provided in Table 1. Consistent with the HA assay, PSaV Cowden strain did not bind to immobilized synthetic oligosaccharides, including the A and H types, both of which are known to be expressed in pigs (Fig. 3) [28]. However, recombinant proteins consisting of the P particles of human norovirus VA387 bound to synthetic oligosaccharides of A type, B type and H type, the P particles of human norovirus VA207 bound to synthetic oligosaccharides Lewis types and H type, and a VP8* of human rotavirus DS-1 bound to synthetic oligosaccharides B type and αGal, respectively (Fig. 3) [5], [26]. Collectively, these results demonstrated that PSaV Cowden strain does not utilize HBGAs as receptors.
Fig. 3. Porcine sapovirus Cowden strain does not bind synthetic histo-blood group antigens (HBGAs). 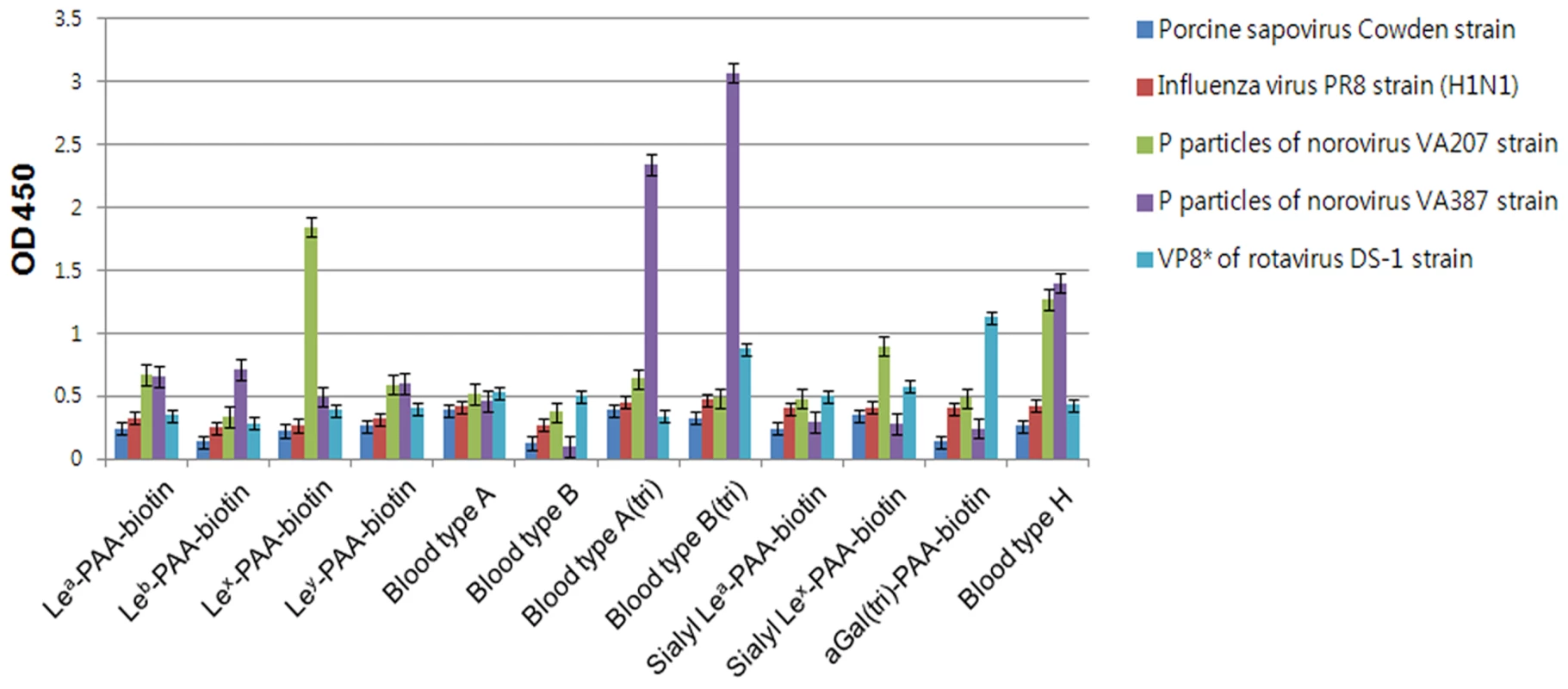
Binding of synthetic HBGAs by PSaV Cowden strain, P particles of norovirus VA387 and VA207 strains, human influenza A virus strain A/Puerto Rico/8/34 (H1N1) (PR8 virus), and VP8* of human rotavirus DS-1 strain were determined using HRP-conjugated-streptavidin. The signal intensities were visualized by TMB at 450 nm in three independent experiments. Error bars represent mean ± SD. Tab. 1. Synthetic oligosaccharide used in this study. 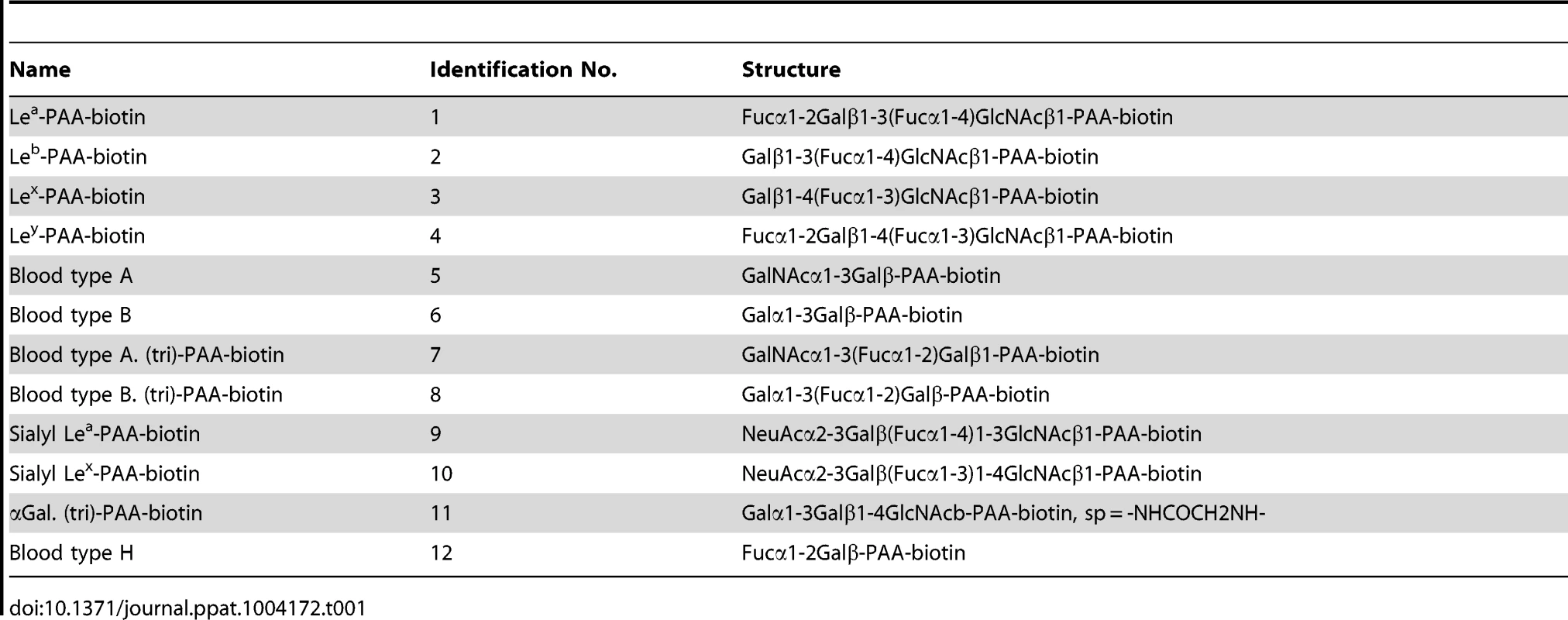
Sialic acid acts as a functional receptor for PSaV
Sialic acid is an abundant carbohydrate moiety on the cell surface [25], which acts as a functional receptor for many viruses, including caliciviruses [4], [11]. To determine if sialic acid is a functional receptor for the PSaV, PSaV Cowden strain was incubated with various concentrations (20–160 mM) of the sialic acid containing molecule, N-acetyl neuraminic acid (NANA), and was then inoculated with LLC-PK cells. NANA at 20 mM significantly reduced the binding activity of Alexa 594-labeled PSaV Cowden strain, and almost completely inhibited binding at 80 mM (Fig. 4A). Binding of radio-labeled PSaV Cowden strain or FCV F9 strain was reduced by NANA in a dose-dependent manner, and was almost completely abolished at 80 mM NANA for PSaV Cowden strain and 40 mM NANA for FCV F9 strain (Fig. 4B). Infection of cells with PSaV Cowden strain was also reduced by incubation with NANA in a dose-dependent manner, and was almost completely inhibited at 80 mM NANA (Fig. 4C and 4D). Similar observations were found with FCV at 40 mM NANA (Fig. 4D). In addition, when plaque reduction assays were performed, PSaV infection was blocked at 80 mM NANA (Fig. S2). Among other monosaccharides and oligosaccharides, N-glycolyl neuraminic acid also inhibited PSaV binding and infection of LLC-PK cells in a dose-dependent manner, but no inhibitory effect of galactose or sialyllactose was found regardless of the concentration (data not shown).
Fig. 4. Porcine sapovirus Cowden strain binding and infection is blocked by N-acetyl neuraminic acid (NANA). 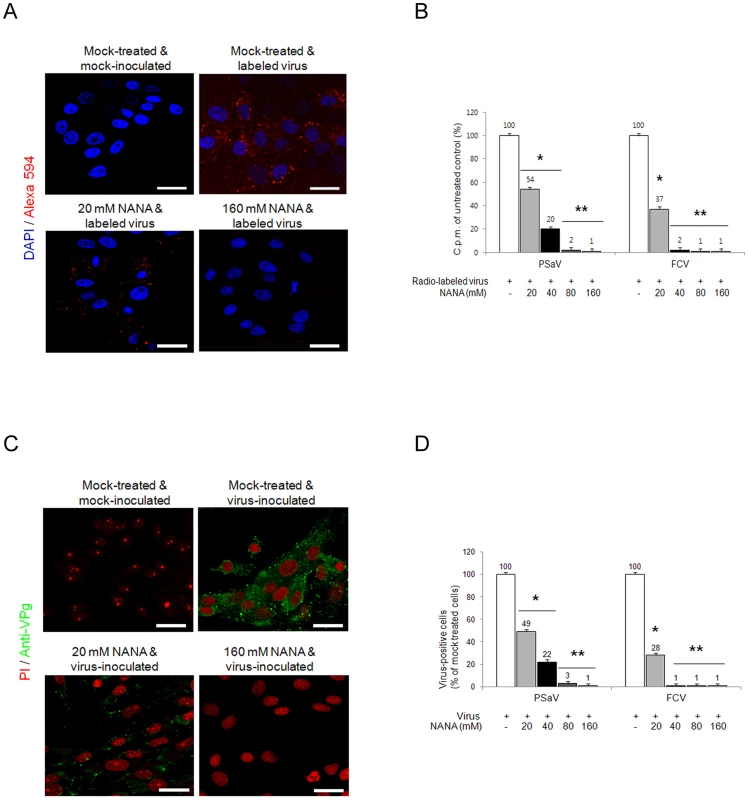
PSaV or control FCV were pre-incubated with a soluble form of NANA at the indicated concentrations to block sialic acid receptors. (A) Alexa 594-labeled PSaV (MOI of 100 pfu/cell) was bound to pretreated LLC-PK cells, and was analyzed for virus binding activity by confocal microscopy. (B) 35[S]Methionine/Cysteine-labeled PSaV or control FCV (50,000 c.p.m.) were bound to pretreated LLC-PK or CRFK cells, and binding of radio-labeled PSaV or FCV was measured by liquid scintillation counting. (C) PSaV (MOI of 0.1 pfu/cell) was incubated with pretreated LLC-PK cells, and was subsequently analyzed by immunofluorescence assay with rabbit polyclonal antibody against PSaV VPg protein 72 h post infection. (D) PSaV or FCV positive cells (%) were enumerated in three independent microscope fields. All experiments were performed in triplicate and figures A and C show representative sets of results. The scale bars correspond to 20 µm. Error bars represent mean ± SD. *p<0.05, **p<0.005. PSaV infects small intestinal epithelial cells, leading to villous atrophy [29]. To confirm whether PSaV binding in small intestinal epithelial cells is also dependent on sialic acid and blocked by NANA, PSaV was incubated with 160 mM of NANA, and was then incubated with porcine small intestinal tissue sections. Duodenal, jejunal and ileal tissue sections incubated with PSaV Cowden strain alone showed a positive signal for PSaV antigens on the villous epithelial cells. The very weak signal observed in the presence of NANA alone is seen only in the duodenal and ileal tissue sections, probably due to increased non-specific binding of the anti-PSaV antibody (Fig. 5). In contrast to these results, intestinal tissue sections incubated with a mixture of PSaV and 160 mM NANA showed markedly decrease of PSaV antigen intensity (Fig. 5). In addition, pretreatment of small intestinal sections with 1 mM NaIO4 (data not shown) or 10 mM NaIO4 markedly reduced the binding activity of PSaV Cowden strain (Fig. 5). To rule out any non-specific effects of NANA addition on the integrity of carbohydrate moieties on the tissue sections the effect of NANA pre-incubation on binding of the P domain of human norovirus VA387, which recognizes HBGAs but not sialic acids as attachment factor, was examined [30]. As expected, a mixture of P domain of VA387 strain and 160 mM NANA had no influence on binding to intestinal epithelial cells (Fig. S3). Moreover, binding of the P domain to intestinal epithelial cells was markedly reduced by the pretreatment of 10 mM NaIO4 but not by 1 mM NaIO4 (Fig. S3). These results fit with previous observations that 1 mM NaIO4 eliminates terminal sialic acids only but 10 mM NaIO4 removes terminal sialic acids as well as HBGAs on carbohydrate moieties [10]. Taken all together, these data strongly indicated that PSaV binds to sialic acid on the cell surface.
Fig. 5. Attachment of porcine sapovirus Cowden strain to porcine intestinal sections is blocked by N-acetyl neuraminic acid (NANA) or pretreatment with NaIO4. 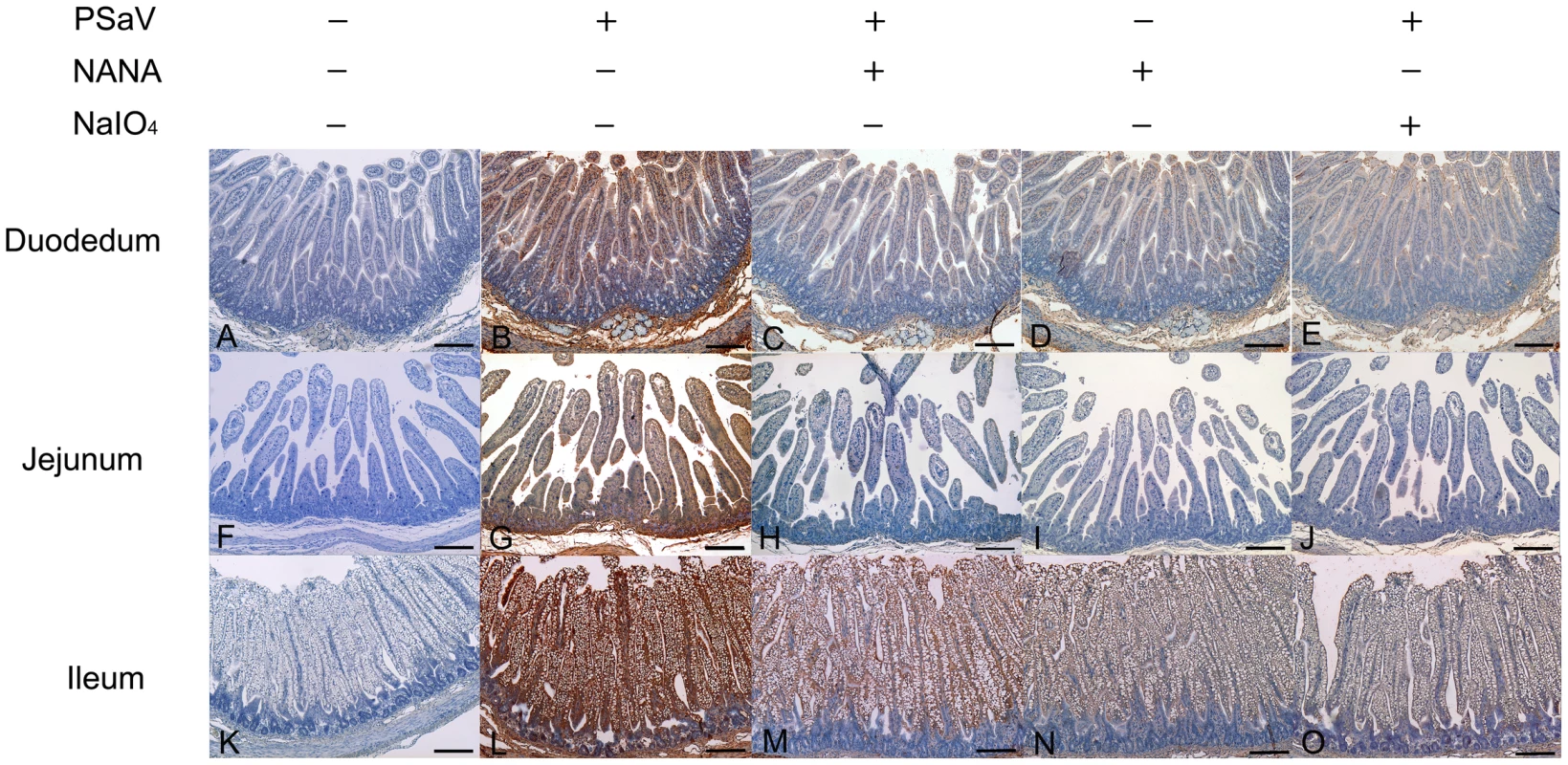
The ability of PSaV to bind to porcine intestinal tissue sections from the duodenum (A–E), jejunum (F–J), and ileum (K–O) was analyzed by immunohistochemistry as described in the Materials and Methods section. In some cases, sections were pretreated with 10 mM NaIO4 prior to the addition of virus (E,J,O) or virus samples were premixed with 160 mM NANA prior to incubating with tissue sections (C,H,M). Binding of PSaV to cells was identified using immunohistochemistry and positive binding is indicated by a red/brown color. Scale bars correspond to 200 µm. This experiment was repeated three independent times and one representative set of results is shown. Both α2,3-linked and α2,6-linked sialic acids are used for PSaV attachment and infection
Sialic acid is attached to glycans via α2,3-, α2,6 - or α2,8-linkages. To determine if PSaV requires these linkages for PSaV binding and infection, LLC-PK cells were pretreated with 200 mU V. cholerae neuraminidase (NA) ml−1, which cleaves α2,3-linked, α2,6-linked and α2,8-linked sialic acids from the underlying glycans [4]. Treated cells were then incubated with either Alexa 594 - or radio-labeled PSaV Cowden strain. Pretreatment with NA markedly reduced the binding of Alexa 594-labeled PSaV (Fig. 6A), and radio-labeled PSaV binding was reduced to 2% of the levels observed in mock treated cells (Fig. 6B). Infection assays, performed by indirect immunofluorescence for the viral antigen VPg, showed almost complete inhibition of PSaV infection at 200 mU NA ml−1 (Fig. 6C and 6D). A similar degree of reduction in binding and infection was observed in the cells infected with α2,6-linked sialic acid-dependent FCV F9 strain (Fig. 6B and 6D) [4] and α2,3-linked sialic acid-dependent influenza virus Kr96 strain (H9N2) (Fig. 6B and 6D, and Fig. S4A and S4B) [31] after pretreatment with NA. In addition, pretreatment of NA had no effect on binding of P domain of human norovirus VA387 strain (Fig. S5).
Fig. 6. Porcine sapovirus binding and infection requires terminal sialic acids. 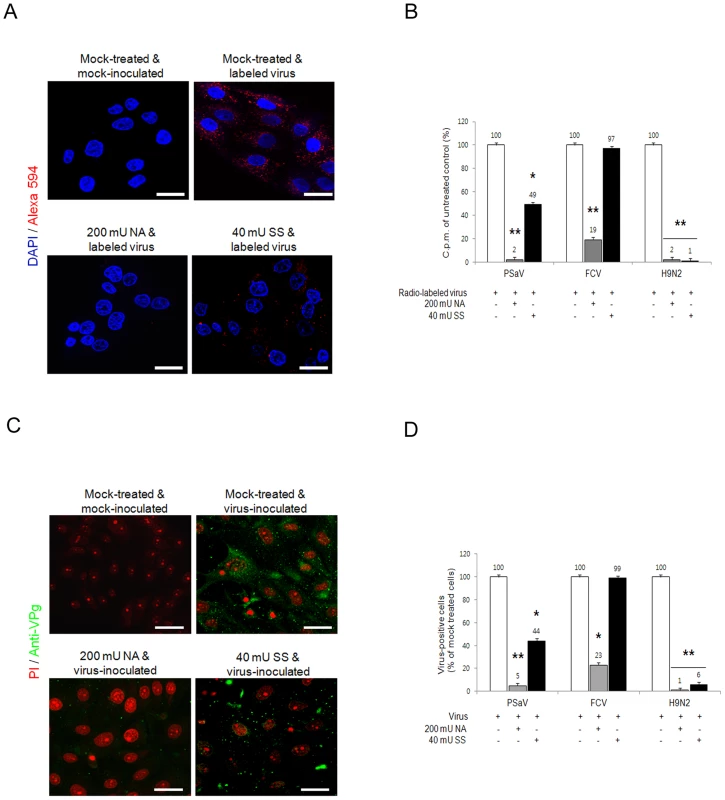
LLC-PK cells were treated with V. cholerae neuraminidase (NA; removes α2,3-, α2,6- and α2,8-linked sialic acid) or sialidase S (SS; removes α2,3-linked sialic acid) from Streptococcus pneumoniae at the indicated concentrations. (A) NA- or SS-treated cells were incubated with either Alex 594 alone or Alexa 594-labeled PSaV (MOI of 100 pfu/cell), and bound virus was detected by confocal microscopy. (B) 35[S]Methionine/Cysteine-labeled PSaV, FCV or influenza virus (H9N2) (50,000 c.p.m.) were bound to LLC-PK, CRFK or MDCK cells, respectively, pretreated with or without SS indicated, and the binding of radio-labeled PSaV, FCV or influenza virus was quantitated by liquid scintillation counting. (C) PSaV (MOI of 0.1 pfu/cell) was incubated with pretreated LLC-PK cells, and was subsequently analyzed by immunofluorescence assay with rabbit polyclonal antibody against PSaV VPg protein 72 h post infection. (D) PSaV, FCV or influenza virus positive cells (%) were enumerated in three independent microscope fields. All experiments were performed independently three times and figures A and C show a single representative set of results. The scale bars correspond to 20 µm. Error bars denote mean ± SD. *p<0.05, **p<0.005. To further identify which specific linkage is used for PSaV binding and infection, LLC-PK cells were initially pretreated with sialidase S (SS) from Streptococcus pneumoniae, which exclusively cleaves α2,3-linked sialic acid from complex carbohydrates and glycoproteins [4]. Pretreatment of LLC-PK cells with 40 mU SS ml−1 reduced PSaV binding to 49% (Fig. 6A and 6B) and decreased PSaV infection to 44% (Fig. 6C and 6D). Importantly, complete inhibition of binding or infection was not achieved, even with increasing doses of SS (data not shown). As expected, pretreatment of 40 mU SS ml−1 significantly blocked binding and infection of α2,3-linked sialic acid-dependent influenza virus Kr96 strain (H9N2) [31] (Fig. 6B and 6D, and Fig. S4A and S4B). In contrast, pretreatment of 40 mU SS ml−1 had no effect on α2,6-linked sialic acid-dependent FCV F9 binding or infection (Fig. 6B and 6D) [4] as well as binding of P domain of HBGAs-dependent human norovirus VA387 strain (Fig. S5) [30]. These data suggested that PSaV Cowden strain may use not only α2,3-linked but also α2,6-linked sialic acids.
In order to further identify the sialic acid linkages required for PSaV attachment and infection, LLC-PK cells were pretreated with specific lectins to block each specific isoform of sialic acid: 1) 400 µg Maackia amurensis lectin (MAL) ml−1, which binds preferentially to α2,3-linked sialic acid, and 2) 400 µg Sambucus nigra lectin (SNL) ml−1, which binds preferentially to α2,6-linked sialic acid [4]. Individual pretreatment of both lectins reduced, but did not completely block PSaV binding (Fig. 7A). Quantitation of PSaV binding by using radio-labeled virus also demonstrated reduced binding, with 66% and 62% binding observed, after pretreatment with MAL and SNL, respectively (Fig. 7B). Likewise, infection of LLC-PK cells by PSaV Cowden strain was decreased to 64% by 400 µg MAL ml−1 and 61% by 400 µg ml−1 SNL (Fig. 7C and 7D). In contrast, MAL had no effect on FCV, but SNL significantly reduced binding and infection (Fig. 7B and 7D). To establish whether PSaV recognizes both α2,3-linked and α2,6-linked sialic acids, LLC-PK cells were pretreated with mixtures of both MAL and SNL, and were then infected with PSaV Cowden strain. PSaV binding and infection were decreased, and complete reduction was observed by the treatment of a mixture at 400 µg MAL ml−1 and 400 µg SNL ml−1 (Fig. 7A–7D). Taken together, all findings above confirm that PSaV Cowden strain uses both α2,3-linked and α2,6-linked sialic acids for binding and infection.
Fig. 7. Porcine sapovirus interacts with α2,3- and α2,6-linked sialic acids. 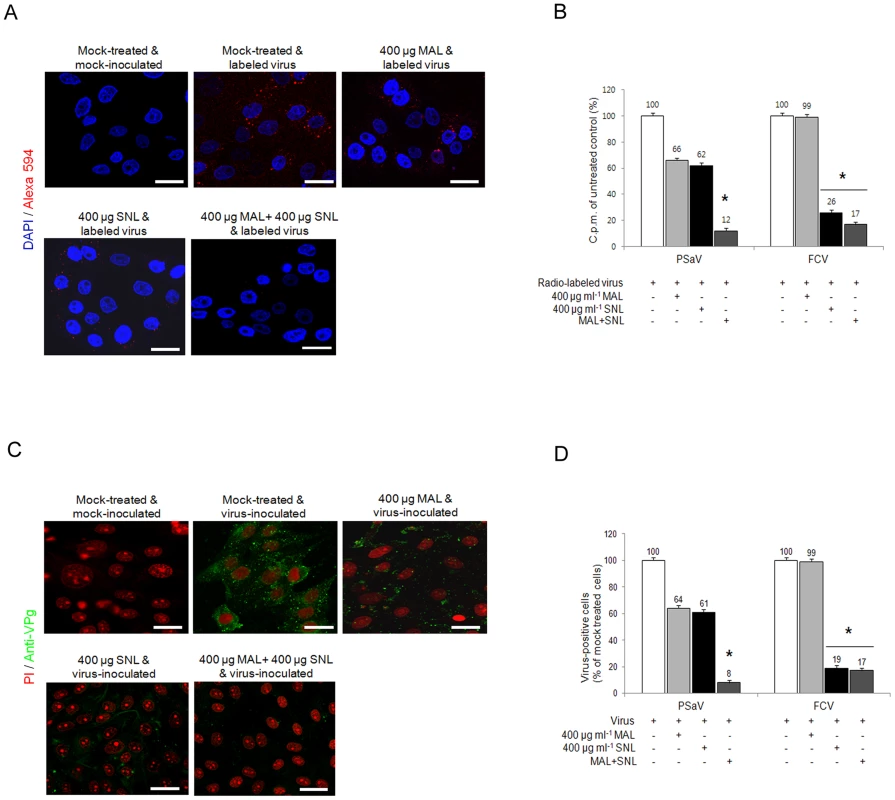
LLC-PK cells were pretreated with M. amurensis lectin (MAL; preferentially binds to α2,3-sialic acid), Sambucus nigra lectin (SNL; preferentially binds to α2,6-linked sialic acid), or combination of both lectins as indicated. (A) Alexa 594 alone or Alexa 594-labeled PSaV (MOI of 100) were bound to pretreated LLC-PK cells, and were observed for their binding activity under confocal microscopy. (B) 35[S]Methionine/Cysteine-labeled PSaV or FCV (50,000 c.p.m.) were bound to LLC-PK or CRFK cells which were pretreated with or without lectins, and binding of radio-labeled PSaV or FCV was measured by liquid scintillation counting. (C) Mock or PSaV (MOI of 0.1) were inoculated to pretreated LLC-PK cells, and were analyzed by immunofluorescence assay with rabbit polyclonal antibody against PSaV VPg protein 72 h post infection. (D) PSaV or FCV positive cells (%) were enumerated in three independent microscope fields. All experiments were performed in triplicate and figures A and C show representative sets of results. The scale bars correspond to 20 µm. The error bars represent the mean ± SD. *p<0.05. Sialic acid for PSaV binding and infection is attached by an O-glycosylation to a glycoprotein
Sialic acids are typically found at the terminal position of N - and O-linked glycans attached to the cell surface and to secreted glycoproteins or glycosphingolipids [32]. To test whether sialic acid moieties used for PSaV binding attach to a glycoprotein, LLC-PK cells were pretreated with either trypsin or chymotrypsin, and were then inoculated with Alexa 594-labeled or radio-labeled PSaV Cowden strain. As shown in Fig. 8A and 8B, pretreatment of 10 µg trypsin ml−1 and 10 µg chymotrypsin ml−1 reduced PSaV attachments to 35% and 41%, respectively, compared to mock treated and PSaV-inoculated control. Furthermore, individual pretreatment of cells with proteases reduced PSaV infection to 25% by trypsin and to 40% by chymotrypsin (Fig. 8C and 8D). Comparatively, FCV binding and infection was reduced by trypsin or chymotrypsin pretreatments, to as low as 12% or 24%, respectively (Fig. 8B and 8D). This inhibition to PSaV binding and infection by trypsin or chymotrypsin pretreatments suggested that like FCV, PSaV Cowden strain binds to sialic acid attached to a glycoprotein.
Fig. 8. Porcine sapovirus Cowden strain interacts with sialic acid on glycoprotein. 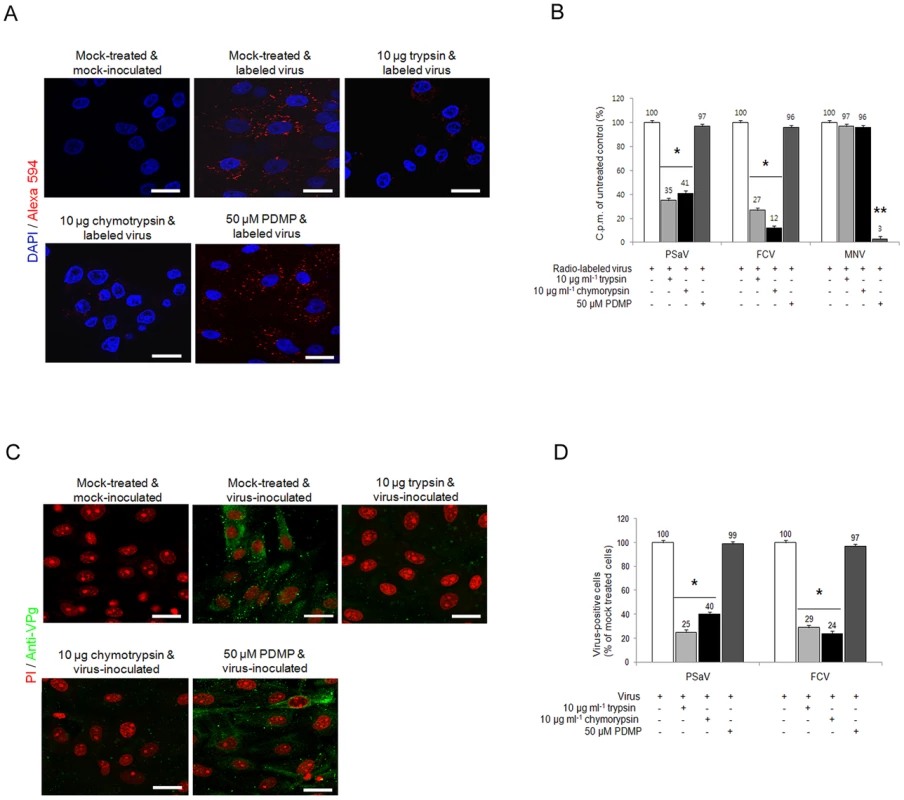
LLC-PK cells were pre-incubated with trypsin (protease), chymotrypsin (protease) or PDMP (lipid metabolic inhibitor) at the indicated concentrations to examine which glycan moieties sialic acid is attached to. (A) Alexa 594 alone or Alexa 594-labeled PSaV (MOI of 100) were bound to pretreated LLC-PK cells, and were observed for their binding activity under confocal microscopy. (B) 35[S]Methionine/Cysteine-labeled PSaV, FCV or MNV (50,000 c.p.m.) were bound to LLC-PK, CRFK or RAW264.7 cells pretreated with or without proteases or PDMP, and binding of radio-labeled PSaV was measured by liquid scintillation counting. (C) Mock or PSaV (MOI of 0.1) were inoculated to pretreated LLC-PK cells, and were analyzed by immunofluorescence assay with rabbit polyclonal antibody against PSaV VPg protein 72 h post infection. (D) PSaV or FCV positive cells (%) were enumerated in three independent microscope fields. All experiments were performed in triplicate and figures A and C show representative sets of results. The scale bars correspond to 20 µm. Error bars show SD in each sample. *p<0.05. To investigate if PSaV also utilizes glycolipid containing sialic acid moieties, LLC-PK cells were pretreated with 50 µM DL-Threo-1-phenyl-2-decanoylamino-3-morpholino-1-propanol (PDMP), a well-known inhibitor of glucosylceramide synthase, and were then infected with PSaV Cowden strain. Regardless of PDMP pretreatment, PSaV bound to and replicated in cells to levels which were identical to those observed in mock treated cells (Fig. 8A–D). Likewise, FCV F9 strain bound to and replicated in CRFK cells despite pretreatment of cells with 50 µM PDMP (Fig. 8B and 8D). As expected, binding of MNV-1 CW1 strain which uses glycolipid as a receptor [11] was significantly inhibited by pretreatment of 50 µM PDMP (Fig. 8 and Fig. S6 and S7). In addition, binding of a VSV-G protein pseudotyped lentivirus, binding of which is also known to at least partially involve glycolipids [33], was also affected by PDMP treatment of porcine LLC-PK cells (Fig. S7). These data confirm that PDMP treatment of LLC-PK cells reduced glycolipid synthesis, therefore confirming that PSaV Cowden strain attaches and infects cells via a glycoprotein containing sialic acid moieties.
As sialic acid moieties may by attached to glycoproteins via both O or N-linkages, the ability of PSaV to bind to LLC-PK pretreated with benzyl 4-O-β-D-galactopyranosyl-β-D-glucopyranoside (benzylGalNAc), an inhibitor of O-linked glycosylation, tunicamycin, an inhibitor of N-linked glycosylation, or PNGase F, which removes N-linked glycans, was examined. Pretreatment of cells with 3 mM benzylGalNAc reduced PSaV attachment to 2% of the levels observed in mock treated cells, whereas 3 µg tunicamycin ml−1 and 200 U PNGase F ml−1 pretreatment had no effect (Fig. 9A and 9B). Infection of cells by PSaV Cowden strain was also reduced by ∼97% by benzylGalNAc treatment, but was unaffected by tunicamycin or PNGase F (Fig. 9C and 9D). As a control, the FCV F9 strain, known to use N-linked sialic acid, was examined. As expected, FCV attachment and infection of CRFK cells was significantly inhibited by pretreatment with tunicamycin and PNGases, but not by benzylGalNAc (Fig. 9B and 9D). These data indicated that PSaV attachment occurs via sialic acid linked by O-linked glycosylation.
Fig. 9. Porcine sapovirus Cowden strain interacts with sialic acid on O-linked glycoprotein. 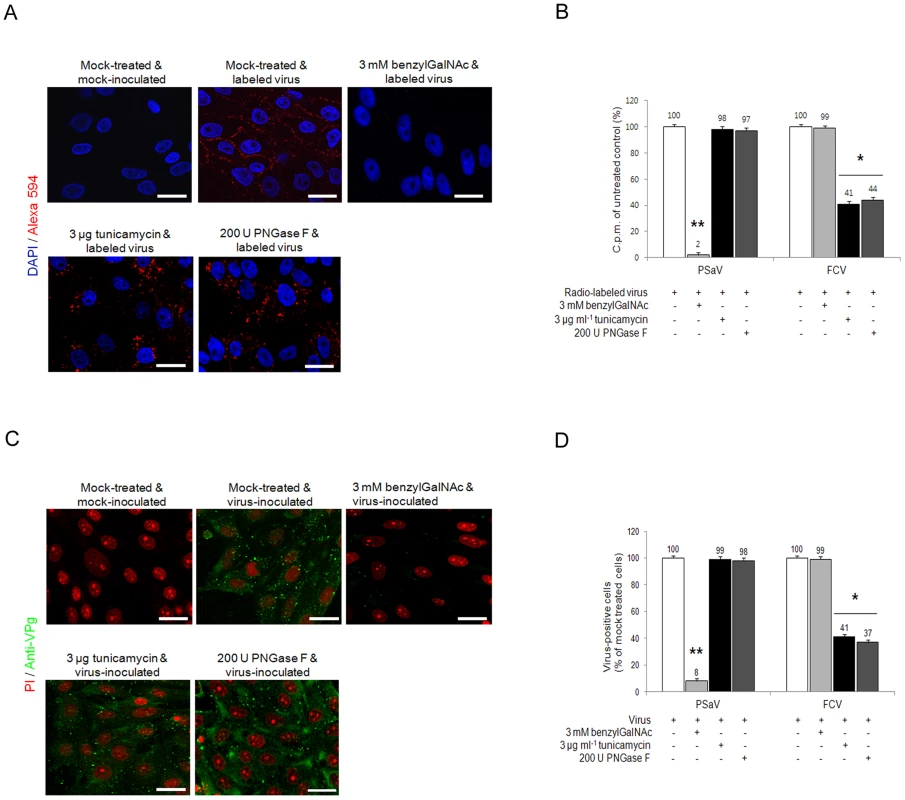
LLC-PK cells were pre-incubated with tunicamycin (N-linked blocker), PNGase F (N-linked blocker) or benzylGalNAc (O-linked blocker) at the indicated concentrations to confirm whether sialic acid is linked to an N or O-glycoprotein. (A) Alexa 594 alone or Alexa 594-labeled PSaV (MOI of 100) were bound to pretreated LLC-PK cells, and were observed for their binding activity under confocal microscopy. (B) 35[S]Methionine/Cysteine-labeled PSaV or FCV (50,000 c.p.m.) were bound to LLC-PK or CRFK cells pretreated with or without tunicamycin, PNGase F or benzylGalNAc, and the binding of radio-labeled PSaV was measured by liquid scintillation counting. (C) Mock or PSaV (MOI of 0.1) were inoculated to pretreated LLC-PK cells, and were analyzed by immunofluorescence assay with rabbit polyclonal antibody against PSaV VPg protein 72 h post infection. (D) PSaV or FCV positive cells (%) were enumerated in three independent microscope fields. All experiments were performed in triplicate and figures A and C show representative sets of results. The scale bars correspond to 20 µm. Error bars show the standard error in each sample. Asterisks indicate significant analysis, p<0.05. Discussion
The lack of an efficient cell culture system of human noroviruses and sapoviruses has hampered the study of virus entry and the molecular mechanisms of virus replication. Among enteric caliciviruses, PSaV Cowden strain is the only cultivable enteric sapovirus and has been shown to replicate in a continuous cell line (LLC-PK), but only in the presence of intestinal contents from gnotobiotic pigs or bile acids as a medium supplement [6]. Although virus-host cell receptor interactions are the first step in the initiation of virus infection, the exact nature of the receptors which are recognized by the Sapovirus genus has not been determined. In the present study, therefore, we used the cell culture adapted PSaV Cowden strain as a model to investigate the entry strategy of an enteric sapovirus in vitro.
Three glycoconjugates with relevance as calicivirus receptors have been described so far; human and bovine noroviruses and Tulane virus utilize HBGAs; MNV uses sialic acid linked to ganglioside or protein in a strain-dependent manner; FCV recognizes terminal sialic acid on an N-linked glycoprotein [4], [5], [10], [11], [12]. Our data would indicate that PSaV does not utilize HBGAs for attachment in agreement with previous work on human sapovirus GI and GV strains that also do not appear to bind HBGAs [14]. Instead, our data Indicate that PSaV, like FCV and some MNV isolates, utilizes sialic acid as a receptor for its binding and infection [4], [11], [34].
Most sialic acids which are recognized as receptors are terminal sialic acids attached to a penultimate galactose by either α2,3-linkage or α2,6-linkage. Previous results have shown that the FCV F9 strain utilizes α2,6-linked sialic acid as a receptor [4], whereas MNV-1 strain CW3 recognizes both α2,3 - and α2,6-linked sialic acids for binding and infection [11]. Our data would indicate that similarly to the MNV-1 CW3 strain [11], PSaV Cowden strain recognizes both α2,3 - and α2,6-linked sialic acids which are attached to glycans as receptors.
The inability of PSaV to form stable agglutinates of RBCs from numerous species, despite the presence of α2,3 and α2,6-linked sialic acid, would suggest that PSaV also forms stabilizing interactions with the specific glycoproteins to which the sialic acids are linked. This is in agreement with our observations that protease treatment reduces virus binding (Fig. 8B) and suggests that this glycoprotein (or glycoproteins) are not expressed on the surface of RBCs. In addition, the conditions under which stable agglutination occurs, i.e., pH, ionic conditions etc., often varies from virus to virus and is dependent on the species of RBCs used [35]. Therefore, it is also possible that the lack of stable agglutination was due to suboptimal conditions used in the HA assay. The identification of specific glycoproteins involved in PSaV binding and the optimal conditions for HA activity of PSaV forms the basis of ongoing work.
The different tissue distribution of α2,3 - and α2,6-linked sialic acids can strongly influence viral tissue tropism and pathogenesis [11], [25], [34], [36]. In pigs, both α2,3 - and α2,6-linked sialic acids are expressed along the epithelial border as well as in goblet cells of the small and large intestines [37]. In addition, both α2,3 - and α2,6-linked sialic acid receptors are distributed extensively in the major organs of pigs, including the trachea, lungs, liver, kidney, spleen, heart, skeletal muscle, and cerebrum [37]. In our previous experiments, we found that pigs orally or intra-venously infected with wild-type virulent PSaV exhibited intestinal pathology as well as viremia [29]. However, systemic infections caused by this strain were not observed, and virus was not isolated from every organ, where both α2,3 - and α2,6-linked sialic acid receptors are well expressed [29]. One explanation for this that the concentration of bile acids which supports the replication of PSaV in cell culture is much higher in the proximal intestine than in the blood and extraintestinal organs [6], [29]. Therefore, it is plausible that the restriction of growth of PSaV within the small intestine is at least partially due to the requirement of a high concentration of bile acids [29]. However, we cannot rule out the possibility that some other potential co-receptors may not be present at extraintestinal sites. It has been suggested that the initial attachment of a virus to a primary receptor enriches the virus at the cell surface and primes the attached virus for interaction with secondary receptor(s) at the cell surface, which is(are) necessary for virus uptake, subsequent uncoating of the nucleic acid, and infection of the target cell [25]. One well characterized example would be the multistep entry of rotavirus into cells, where various cell receptors, including the terminal sialic acid or HBGAs, integrins, and heat shock protein Hsc70 are utilized by the outer most proteins, VP4 and VP7, of rotaviruses [38].
Among caliciviruses, FCV is known to utilize not only sialic acid, but also JAM-1 for virus entry into cells; the latter presumably aiding FCV penetration into the host cells as a co-receptor [4], [13]. In agreement with this hypothesis, cyro-EM reconstruction and biochemical studies of the FCV capsid with JAM-1 indicated that JAM-1 binding results in significant conformational changes in the capsid [39], [40], [41].
Glycosylation produces different types of glycans which are typically attached to cellular proteins and lipids [32], [37]. Different members of caliciviruses seem to utilize different linkage of glycans for their binding and entry, i.e. MNV use sialic acid bearing gangliosides (CW3 like strains) or proteins (CR3 strain) [11], [34], while FCV recognizes sialic acid bearing protein components [4]. In the present study, pretreatment of PDMP had no influence on the binding and infection of PSaV, suggesting that like FCV [4], PSaV does not utilize sialic acid bearing lipids.
Protein glycosylation encompasses N-glycans, O-glycans, and glycosaminoglycans [37]. Different types of glycans which are attached to cellular proteins are utilized by the different viruses, even within the same genus. For example, adeno-associated virus type 5 (AAV5) interacts with sialic acid on N-linked carbohydrates, whereas AAV4 interacts with sialic acid on O-linked carbohydrates, and both viruses require 2,3-linked sialic acid for binding [42]. We found that the sialic acid-bearing glycans used for PSaV binding are attached to cell surface proteins in a similar manner to FCV [4]. Unlike FCV [4], however, carbohydrate moieties linked to terminal sialic acid as a receptor for PSaV are present on an O-linked glycoprotein.
In conclusion, we have demonstrated that unlike noroviruses, PSaV infects cells via both α2,3 - and α2,6-linked sialic acids attached to O-linked glycoproteins. This work has provided new insights into the mechanisms of sapovirus entry, and may provide additional information relevant to the identification of inhibitors of sapovirus pathogenesis.
Materials and Methods
Cells and viruses
LLC-PK cells, Caco2 cells and HeLa cells obtained from the American Type Culture Collection (ATCC, USA) were maintained in Eagle's minimal essential medium (EMEM) containing 10% fetal bovine serum (FBS), 100 U/ml penicillin, and 100 µg/ml streptomycin. CRFK cells and MDCK cells from ATCC was grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 5% FBS, 100 U/ml penicillin, and 100 µg/ml streptomycin. MA-104 cells from ATTCC was grown in α-MEM supplemented with 5% FBS, 100 U/ml penicillin, and 100 µg/ml streptomycin. RAW264.7 cells were grown in RPMI1640 supplemented with 5% FBS, 100 U/ml penicillin, and 100 µg/ml streptomycin. The tissue culture-adapted PSaV Cowden strain was recovered from the full-length infectious clone pCV4A, and was propagated in LLC-PK cells with the supplement of bile acid [6]. The FCV F9 strain, human influenza A virus A/Puerto Rico/8/34 (H1N1) (PR8 virus) strain, human rotavirus DS-1 strain, and CVB3 Nancy strain were purchased from the ATCC. Chicken influenza A virus A/Korea/96/96 (H9N2) (Kr96 virus) strain was obtained from Animal and Plant Quarantine Agency, Korea and MNV-1 CW1 strain was a kind gift of Dr. H.W. Virgin, Washington University School of Medicine, USA. These viruses were propagated in CRFK cells, MA-104 cells, MDCK cells, HeLa cells, or RAW264.7 cells, respectively [4], [11], [43], [44], [45].
Reagents and antibodies
Sodium periodate (Sigma-Aldrich), trypsin (Sigma-Aldrich), chymotrypsin (Sigma-Aldrich), NANA (Fluka, USA), sialyllactose (GeneChem, Korea), galactose (Sigma-Aldrich), MAL (Sigma-Aldrich), and SNL (Sigma-Aldrich) were dissolved in PBS pH 7.2. Tunicamycin (Sigma-Aldrich) and Alexa 594 (Invitrogen, USA) were dissolved in DMSO. PDMP (Calbiochem, USA) and benzylGalNAc (Sigma-Aldrich) were dissolved in ethanol. Other reagents included 35[S]methionine/cysteine (PerkinElmer, USA), NA (Sigma-Aldrich), SS (Prozyme, USA), PNGase F (NEB, UK), anti-PSaV capsid monoclonal antibody [46], anti-PSaV VPg polyclonal antibody, anti-FCV capsid monoclonal antibody (Santa Cruz, USA), anti-CVB3 capsid monoclonal antibody (Millipore, IL, USA), anti-Hu/NoV/GII.4/HS194 virus-like particles (VLPs) polyclonal antibody (kind gift of Dr. L. Saif, The Ohio State University, Ohio, USA), anti-influenza virus nucleoprotein monoclonal antibody (Median Diagnostic, ChunCheon, Korea), anti-rabbit IgG-FITC antibody (Jackson Immuno Research Lab, USA), and anti-mouse IgG-FITC antibody (Santa Cruz).
Cloning, expression and purification of VP8* of rotavirus and P particles of human norovirus
VP8* of human DS-1 rotavirus (RV) strain (P[4] genotype) and P particles of VA387 (GII-4) and VA207 (GII-9) norovirus (NV) strains were cloned, expressed and purified as described previously [26], [27]. Briefly, the cDNAs encoding RV DS-1 strain VP8* and encoding NV VA387 or VA207 P particles with cysteine peptide were cloned into the expression vector pGEX - 4T-1 (glutathione S-transferase [GST]–gene fusion system; GE Healthcare Life Sciences, Piscataway, NJ). After sequence confirmation, the recombinant GST-VP8 and GST-P fusion proteins were expressed in Escherichia coli strain BL21 as described previously (Table 2) [26], [27]. Expression of protein was induced by IPTG (isopropyl-β-D-thiogalactopyranoside; 0.2 mM) at room temperature (22°C) overnight. RV VP8-GST and NV GST-P fusion proteins were purified using the Pierce GST spin purification kit (Pierce, IL, USA) according to the manufacturer's protocol. The P particles of VA387 and VA207 were released from GST by thrombin (Sigma-Aldrich) digestion at room temperature overnight. The concentration of the purified RV VP8* and NV P particles were determined by measuring the absorbance at 280 nm.
Tab. 2. Primers used for amplification of the rotavirus VP8* and norovirus P domain with cysteine-containing tag. 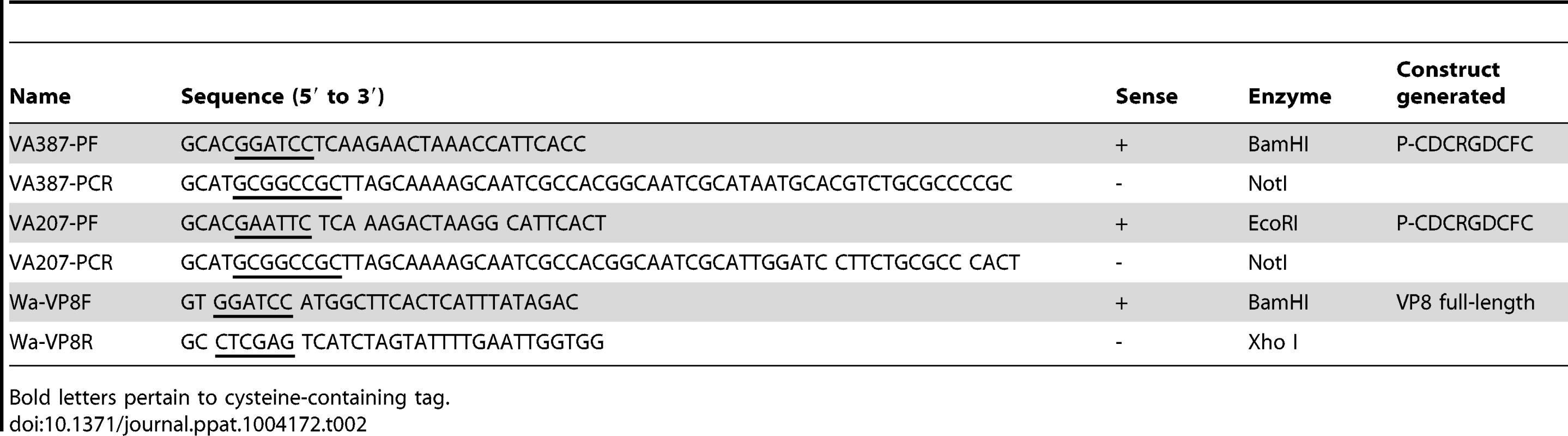
Bold letters pertain to cysteine-containing tag. Hemagglutination assay
Hemagglutination (HA) assay was performed using RBCs from animals and humans. Blood from pig, cow, chicken, and rat were obtained from the College of Veterinary Medicine, Chonnam National University, and human blood was provided by volunteer donors (human ABO, Lewis Oa+b-, Lewis Oa−b+, and Lewis Oa−b− types) at Chonnam National University Hospital. RBCs were packed in PBS pH 7.2 without Ca2+, and were centrifuged at 500×g for 10 min.
PSaV Cowden strain (10 µg/ml), human influenza virus PR8 (H1N1) (5 µg/ml starting dilutions), P particles (10 µg/ml) of norovirus VA387 strain, and VP8* of rotavirus DS-1 strain (10 µg/ml) were diluted serially 2 fold in PBS (0.01 M sodium phosphate, 0.15 M NaCl, pH 5.5) using V-shaped 96-well plates. The HA activity was tested by mixing equal volume of RBCs 1% to the prepared viruses or protein dilutions. The reactions were allowed to proceed for 1 h at 4°C or 20°C, and the agglutination of RBCs was observed and recorded [10]. The HA titer was the reciprocal of the highest virus dilution that allowed sedimentation of the RBCs compared to control wells.
Synthetic oligosaccharides binding assay
The synthetic oligosaccharide-based histo-blood group antigen binding assay was carried out as described elsewhere [47], [48]. Briefly, 96-microtiter plates were coated with PSaV Cowden strain (10 µg/ml), P particles of NV VA207 (10 µg/ml) and VA387 (10 µg/ml) strains, human influenza virus PR8 strain (H1N1) (10 µg/ml), or VP8* of RV DS-1 strain (10 µg/ml) at 4°C overnight. Coated plates were blocked with 5% bovine serum albumin (BSA) for 1 h at room temperature, and each synthetic oligosaccharide-polyacrylamide (PAA)-biotin conjugate (10 µg/ml) was then added and further incubated at 4°C overnight. Oligosaccharides used in this study included Lewis antigens (Lea, Leb, Lex, and Ley), H type 1, H type 2, H type 3, type A disaccharide, type B disaccharide, type A trisaccharide, type B trisaccharide, sLea and sLex tetrasaccharides, which were conjugated with biotin (GlycoTech Co, USA; Table 1). Bound oligosaccharides were detected using HRP-conjugated-streptavidin (Jackson Immuno Research Lab, USA). The signal was visualized by TMB (Komabiotech, Korea) followed by measurement at 450 nm. In each step, the plates were incubated for 1 h at 37°C, after which they were washed five times with PBS-Tween 20.
Labeling of viruses with 35[S] methionine/cysteine
Labeling of PSaV Cowden, FCV F9, CVB3 Nancy, MNV-1 CW1, and influenza virus Kr96 strains with 35[S]methionine/cysteine were carried out as described previously [4]. Briefly, confluent monolayers of permissible cells for above viruses in five 175 cm3 flasks were infected with above each strain, respectively, at a multiplicity of infection (MOI) of 0.1 for 4 h at 37°C. The medium was replaced with RPMI 1640 lacking methionine and cysteine (Sigma-Aldrich) for 2 h. The medium was then supplemented with 1 Mbq 35[S] methionine/cysteine. After 72 h (PSaV Cowden strain), 16 h (FCV F9 strain), 16 h (CVB3 Nancy strain), 16 h (MNV-1 CW1 strain), and 72 h (influenza virus Kr96 strain) post virus inoculation, the cultured virus was frozen and thawed three times. Each virus was pelleted by ultracentrifugation for 10 h at 104,000×g in a Hitachi P28S rotor, and then was purified by sucrose gradient density ultracentrifugation for 10 h at 104,000×g using a Hitachi P28S rotor.
Labeling of viruses with Alexa 594
Labeling of PSaV Cowden, FCV F9, CVB3 Nancy, MNV-1 CW1, P domain of VA387 strain and influenza virus Kr96 strains with Alexa 594 (Invitrogen) was performed following the manufacturer's instruction. Briefly, 1 part of Alexa 594 solution was mixed with 9 parts of a solution containing 1×108 pfu/ml of above each strain or 9 parts of a solution containing 500 µg/ml of P domain of VA387 strain. Each reaction was mixed thoroughly for 30 sec and was then incubated for 1 h at room temperature.
Attachment assay with radiolabeled-viruses
Attachment assays with 35[S]methionine/cysteine-labeled PSaV Cowden, FCV F9, CVB3 Nancy, MNV-1 CW1 or influenza virus Kr96 strains to LLC-PK, CRFK, HeLa, RAW264.7 or MDCK cell lines were performed as described previously with slight modifications [4]. Briefly, subconfluent monolayers of permissible cells grown on 6-well plates were mixed with purified 35[S]methionine/cysteine-labeled above each strain (50,000 c.p.m.), and were then incubated for 45 min on ice. Cells were washed three times with ice-cold PBS, after which they were lysed with 0.1% SDS and 0.1 M NaOH. Total radioactivity in the cell lysate was determined by liquid scintillation counting [4].
Attachment assay with Alexa 594-labeled viruses
To visualize virus attachment to subconfluent monolayers of cells grown on the confocal dish were pretreated with or without inhibitors or enzymes, as described above. Mock - or reagent-treated cells were then inoculated with PSaV Cowden, FCV F9, CVB3 Nancy, MNV-1 CW1, P domain of VA387 strain or influenza virus Kr96 strains labeled with Alexa 594 dye or Alexa 594 dye alone, after which they were incubated for 5 min on ice. Cells were washed 1 time with cold PBS 1% containing fetal bovine serum (PBS-FBS), fixed with 4% formaldehyde in cold PBS for 10 min, and washed 3 times with cold PBS. The cells which were incubated with Alexa 594 dye-labeled viruses or P domain of VA387 strain were stained with 300 nM 4′,6-diamidino-2-phenylindole (DAPI) solution for nucleus staining, mounted with using SlowFade Gold antifade reagent (Invitrogen), and examined using an EZ-C1 confocal microscope and EZ-C1 software (Nikon, Japan). Laser and microscope settings were adjusted according to the manufacturer's instructions. Cells infected with viruses unlabeled with Alexa dye were analyzed by immunofluorescence, as described below.
Attachment assay with RT-qPCR
Attachment assays by RT-qPCR with PSaV Cowden, MNV CW1 or VSV-G protein pesudotyped lentivirus (LV) strains to LLC-PK cell line was performed as described above using Alexa of radio-labeled virus. Briefly, subconfluent monolayers of LLC-PK cells on 24-well plate were pretreated with 50 µM PDMP or 5 mM NaIO4. PDMP-treated cells were inoculated with above each strain (3 TCID50 of PSaV and MNV; 1.25 transducing units per cell of LV), and were then incubated for 45 min on ice. Cells were washed three times with ice-cold PBS, Viral RNA was immediately extracted and analyzed by RT-qPCR with primer specific to each virus. RNA was extracted using GenElute Mammalian Total RNA Miniprep Kit (Sigma). One hundred nanograms were subsequently reverse transcribed using random hexamers. Fragments of ∼200 bp were amplified using the following gene-specific primers: PSaV: 5′-CAACAATGGCACAACAACG-3′ (forward) and 5′-ACAAGCTTCTTCACCCCACA-3′ (reverse); MNV-1 : 5′-TGGACAACGTGGTGAAGGAT-3′ (forward) and 5′-CAAACATCTTTCCCTTGTTC-3′ (reverse), and LV-WPRE: 5′-TCGGCCCTCAATCCAGCGGA-3′ (forward) and 5′-TCGTCTGAGGGCGAAGGCGA-3′ (reverse). Standard curves were generated for all genes quantified. Additional non-template and non-reverse transcriptase samples were routinely analyzed as negative controls. Data were collected using a ViiA 7 Real-time PCR System (Applied Biosystems).
Virus infectivity assay
Infectivity assays of PSaV Cowden, FCV F9, CVB3 Nancy or influenza virus Kr96 strains in the permissible cells, respectively, were carried out as described previously with slight modifications [4]. Briefly, confluent monolayers of each permissible cell were treated with various inhibitors or enzymes as described below. Either mock or treated cells were infected with above each strain at an MOI of 0.1 pfu/cell, and were then incubated at 37°C for 1 h. Cells were washed three times with PBS, and were then replaced with maintenance medium. The cells were incubated for 72 h (PSaV Cowden), 8 h (FCV F9), 9 h (CVB3 Nancy) or 48 h (influenza virus Kr96) at 37°C prior to being fixed with 4% formaldehyde in PBS, and were analyzed by immunofluorescence assay as described below.
Immunofluorescence assay
Immunofluorescence assays was performed as previously described [4]. Briefly, fixed cells in 8-well chamber slides were permeabilized by the addition of 0.2% Triton X-100, incubated for 5 min at room temperature, and then washed with PBS containing 0.1% newborn calf serum (PBS-NCS). Anti-PSaV VPg polyclonal antibody, anti-FCV capsid monoclonal antibody, anti-CVB3 capsid monoclonal antibody or anti-influenza virus nucleoprotein monoclonal antibody was then added at a dilution rate of 1∶100, 1∶200 or 1∶500, respectively. Chamber slides were incubated at 4°C overnight. Cells were washed 3 times with PBS-NCS, and FITC-conjugated secondary antibodies (diluted to 1∶100) were then added. Nuclei were stained with propidium iodide (PI), and cells were examined using confocal microscopy. A total of 700 cells, as indicated by PI or DAPI staining, were counted per condition, and were scored for PSaV VPg protein expression. After image analysis with Metamorph Premier v6.3 software (Molecular Devices, PA), the infected cells were counted as positive for viral antigen if they had a fluorescent intensity which was at least three times the fluorescent intensity of the uninfected controls. The percentage of positive cells was then normalized to that of the untreated control.
Immunohistochemistry analysis
3-day-old piglets obtained from sows by hysterectomy were used to attain the small intestinal segments including the duodenum, jejunum and ileum. Segments were excised after sacrifice, immediately immersed in 10% buffered formalin, processed routinely for paraffin embedment, sectioned, and stained with hematoxylin for histology. For immunohistochemical studies, paraffin-embedded sections were deparaffinized, and were then rehydrated through graded alcohols into 0.1 M PBS, after which they were treated with 0.3% H2O2 to quench endogenous peroxidase. Sections were either incubated in PBS (200 µl), PSaV Cowden strain (1×106 pfu/ml), P domain of VA 387 strain (10 µg/ml), NANA (160 mM, pH 7), PSaV (1×106 pfu/ml) and NANA (160 mM, pH 7) mixture, P domain of VA 387 strain (10 µg/ml) and NANA (160 mM, pH 7) mixture with or without 1 mM or 10 mM NaIO4 pretreatment for 1 h at room temperature. Pretreated sections with NaIO4 were incubated for 30 min prior to the addition of virus. After washing with PBS, a monoclonal antibody against PSaV Cowden strain capsid protein or a polyclonal antibody against Hu/NoV/GII.4/HS194 VLPs was incubated with the sections at 4°C overnight. Sections were then rinsed 3 times with PBS, and incubated with biotinylated secondary antibody (Dako, USA) and peroxidase-conjugated streptavidin (Dako, USA). Reactions were developed with 3-amino-9-ethylcarbazol (AEC; Vector laboratories, USA), and followed by Mayer's hemalum solution (Merck, Germany) for counterstaining.
Plaque reduction assay
Confluent monolayers of LLC-PK cells on 6-well plates were inoculated with PBS (200 µl), PSaV Cowden strain (1×105 pfu/ml), or mixtures of PSaV (1×105 plaque forming unit/ml) and various concentrations of NANA (20–160 mM, pH 7), after which they were incubated in a 5% CO2 incubator. After 2 h of virus adsorption, PBS and virus inocula were thoroughly discarded and washed 3 times with PBS. Overlay medium containing a 1× concentrated MEM, 10% FBS, 1.2% (w/v) avicel (FMC BioPolymer, Belgium), and 200 µM GCDCA was added to each well. Plates were incubated for 96 h in a 5% CO2 incubator. After incubation, inoculated cells were fixed with 20% trichloroacetic acid, and the avicel was then removed. Plaques were visualized by staining with 1% (w/v) crystal violet solution [49], [50].
Treatment of cells with chemicals, metabolic inhibitors and enzymes
Subconfluent monolayers of LLC-PK, CRFK, HeLa, Caco2, RAW264.7 or MDCK cells grown on 6-well plates or 8-well chamber slides for confocal analysis were treated with chemicals, metabolic inhibitors, or enzymes optimized at the specific concentrations, incubation times and temperatures described in Table 3. After pretreatment, cells were washed three times with PBS, and binding and infectivity assays were carried out as described above. Mock and control treatments were performed at the same time.
Tab. 3. Chemicals, metabolic inhibitors, and enzymes used in this study. 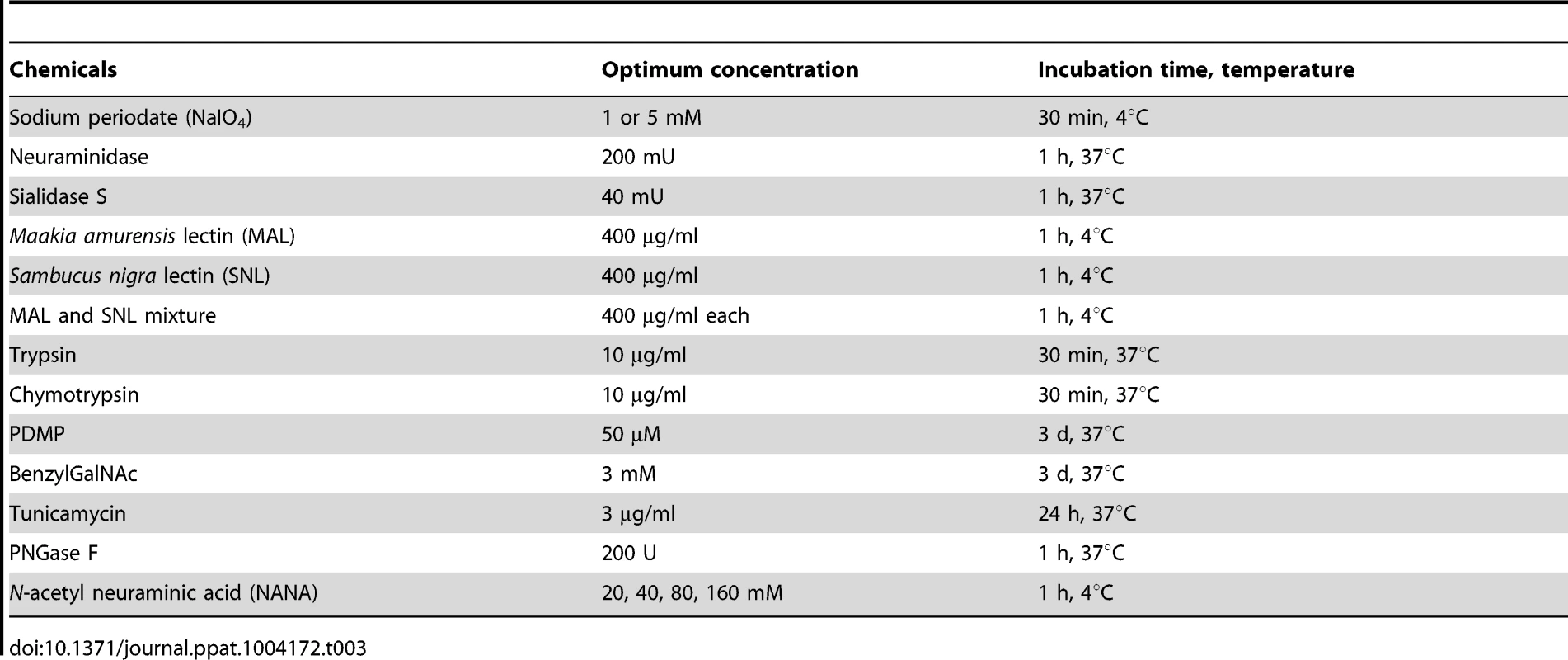
Statistical analysis
Statistical analysis was performed using SPSS version 11.5.1 for window (SPSS, USA). One way analysis of variance (ANOVA) test were used. A P-value <0.05 was considered statistically significant.
Ethics statement
All animals were handled in strict accordance with good animal practices, as described in the NIH Guide for the Care and Use of Laboratory Animals (NIH Publication No. 85–23, 1985, revised 1996). The protocol was approved by the Committee on Ethics of Animal Experiments, CNU with permit number (CNU No. 2012-87). The human blood samples collected with written consent from the patients were handled in strict accordance with human subjects, as described in the Guidance for the Care and Use of Human Samples of the CNU adhered from the WMA Declaration of Helsinki (Ethical Principles for Medical Research Involving Human Subjects). The protocol was approved by the Committee for Research Ethics Concerning Human Subjects, CNU with permit number (CNU IBR No. 1040198-130807-BR-002-01).
Supporting Information
Zdroje
1. Green KY (2007) Caliciviridae: The Noroviruses. In: Knipe DM, Howley PM, eds. Fields Virology, 5th edition. Philadelphia: Lippincott Willisams and Wilkins.
2. NyströmK, Le Gall-ReculéG, GrassiP, AbrantesJ, Ruvoën-ClouetN, et al. (2011) Histo-blood group antigens act as attachment factors of rabbit hemorrhagic disease virus infection in a virus strain-dependent manner. PLoS Pathog 7: e1002188.
3. RadfordAD, CoyneKP, DawsonS, PorterCJ, GaskellRM (2007) Feline calicivirus. Vet Res 38 : 319–335.
4. StuartAD, BrownTD (2007) Alpha2,6-linked sialic acid acts as a receptor for Feline calicivirus. J Gen Virol 88 : 177–186.
5. TanM, JiangX (2010) Norovirus gastroenteritis, carbohydrate receptors, and animal models. PLoS Pathog 6: e1000983.
6. ChangKO, SosnovtsevSV, BelliotG, KimY, SaifLJ, et al. (2004) Bile acids are essential for porcine enteric calicivirus replication in association with down-regulation of signal transducer and activator of transcription 1. Proc Natl Acad Sci USA 101 : 8733–8738.
7. PatelMM, WiddowsonMA, GlassRI, AkazawaK, VinjeJ, et al. (2008) Systematic literature review of role of noroviruses in sporadic gastroenteritis. Emerg Infect Dis 14 : 1224–1231.
8. HansmanGS, OkaT, KatayamaK, TakedaN (2007) Human sapoviruses: genetic diversity, recombination, and classification. Re Med Virol 17 : 133–141.
9. Ruvoen-ClouetN, GaniereJP, Andre-FontaineG, BlanchardD, Le PenduJ (2000) Binding of rabbit hemorrhagic disease virus to antigens of the ABH histo-blood group family. J Virol 74 : 11950–11954.
10. ZakhourM, Ruvoen-ClouetN, CharpilienneA, LangpapB, PoncetD, et al. (2009) The alphaGal epitope of the histo-blood group antigen family is a ligand for bovine norovirus Newbury2 expected to prevent cross-species transmission. PLoS Pathog 5: e1000504.
11. TaubeS, PerryJW, YetmingK, PatelSP, AubleH, et al. (2009) Gangloside-linked terminal sialic acid moieties on murine macrophages function as attachment receptors for murine noroviruses. J Virol 83 : 4092–4101.
12. FarkasT, CrossRW, HargittE3rd, LercheNW, MorrowAL, et al. (2010) Genetic diversity and histo-blood group antigen interactions of rhesus enteric caliciviruses. J Virol 84 : 8617–8625.
13. MakinoA, ShimojimaM, MiyazawaT, KatoK, TohyaY, et al. (2006) Junctional adhesion molecule 1 is a functional receptor for feline calicivirus. J Virol 80 : 4482–4490.
14. Shirato-HorikoshiH, OgawaS, WakitaT, TakedaN (2007) Hansman (2007) Binding activity of norovirus and sapovirus to histo-blood group antigens. Arch Virol 152 : 457–461.
15. Bank-WolfBR, KönigM, ThielHJ (2010) Zoonotic aspects of infections with noroviruses and sapoviruses. Vet Microbiol 140 : 204–212.
16. MesquitaJR, CostantiniVP, CannonJL, LinSC, NascimentoMS, et al. (2013) Presence of antibodies against genogroup VI norovirus in humans. Virol J 10 : 176.
17. WiddowsonM, RockxB, ScheppR, van der PoelWHM, VinjeJ, et al. (2005) Detection of serum antibodies to bovine norovirus in veterinarians and the general population in the Netherlands. J Med Virol 76 : 119–128.
18. MattisonK, ShuklaA, CookA, PollariF, FriendshipR, et al. (2007) Human noroviruses in swine and cattle. Emerg Infect Dis 13 : 1184–1188.
19. SouzaM, AzevedoMS, JungK, CheethamS, SaifLJ (2008) Pathogenesis and immune responses in gnotobiotic calves after infection with the genogroup II.4-HS66 strain of human norovirus. J Virol 82 : 1777–1786.
20. Helenius A (2007) Virus Entry and Uncoating. In: Knipe DM, Howley PM, eds. Fields Virology, 5th edition. Philadelphia: Lippincott Willams and Wilkins.
21. WoolhouseMEJ (2002) Population biology of emerging and re-emerging pathogens. Trends Microbiol 10: S3–S7.
22. Martínez-BarragánJ, AngelRM (2001) Identification of a putative coreceptor on Vero cells that participates in dengue 4 virus infection. J Virol 75 : 7818–7827.
23. WoodwardMP, YoungWWJr, BloodgoodRA (1985) Detection of monoclonal antibodies specific for carbohydrate epitopes using periodate oxidation. J Immunol Methods 78 : 143–153.
24. HafensteinS, BowmanVD, ChipmanPR, KellyCMB, LinF, et al. (2007) Interactionof decay-accelerating factor with coxsakievirus B3. J Virol 81 : 12927–12935.
25. OlofssonS, BergströmT (2005) Glycoconjugateglycans as viral receptors. Ann Med 37 : 154–172.
26. HuangP, XiaM, TanM, ZhongW, WeiC, et al. (2012) Spike protein VP8* of human rotavirus recognizes histo-blood group antigens in a type-specific manner. J Virol 86 : 4833–4843.
27. TanM, ZhongW, SongD, ThorntonS, JiangX (2004) E. coli-expressed recombinant norovirus capsid proteins maintain authentic antigenicity and receptor binding capability. J Med Virol 74 : 641–649.
28. HanagataG, GasaS, SakoF, MakitaA (1990) Human blood group A and H glycolipids in porcine plasma. Evidence for acquisition of the erythrocyte antigens from plasma. FEBS Lett 261 : 312–314.
29. GuoM, HayesJ, ChoKO, ParwaniAV, LucasLM, et al. (2001) Comparative pathogenesis of tissue culture-adapted and wild-type Cowden porcine enteric calicivirus (PEC) in gnotobiotic pigs and induction of diarrhea by intravenous inoculation of wild-type PEC. J Virol 75 : 9239–9251.
30. HuangP, FarkasT, MarionneauS, ZhongW, Ruvoen-ClouetN, et al. (2003) Noroviruses bind to human ABO, lewis, and secretor histo-blood group antigens: Identification of 4 distinct strain-specific patterns. J Infect Dis 188 : 19–31.
31. MatrosovichMN, MatrosovichTY, GrayT, RobertsNA, KlenkHD (2004) Human and avian influenza viruses target different cell types in cultures of human airway epithelium. Proc Natl Acad Sci USA 101 : 4620–4624.
32. VarkiA (2007) Glycan-based interactions involving vertebrate sialic-acid-recognizing proteins. Nature 446 : 1023–1029.
33. SinibaldiL, GoldoniP, SegantiL, SupertiF, TsiangH, OrsiN (1985) Gangliosides in early interactions between vesicular stomatitis virus and CER cells. Microbiologica 8 : 355–365.
34. TaubeS, PerryJW, McGreevyE, YetmingK, PerkinsC, et al. (2012) Murine noroviruses bind glycolipid and glycoprotein attachment receptors in a strain-dependent manner. J Virol 86 : 5584–5593.
35. HorzinekMC (1973) Comparative aspects of togaviruses. J Gen Virol 20 : 87–103.
36. McLeodGM, WigginsHS (1968) Bile-salts in small intestinal contents after ileal resection and in other malabsorption syndromes. Lancet 1 : 873–876.
37. OhtsuboK, MarthJD (2006) Glycosylation in cellular mechanisms of health and disease. Cell 126 : 855–867.
38. LopezS, AriasCF (2006) Early steps in rotavirus cell entry. Curr Top Microbiol Immunol 309 : 39–66.
39. BhellaD, GathererD, ChaudhryY, PinkR, GoodfellowIG (2008) Structural insights into calicivirus attachment and uncoating. J Virol 82(16): 8051–8058.
40. OssiboffRJ, ZhouY, LightfootPJ, PrasadBV, ParkerJS (2010) Conformational changes in the capsid of a calicivirus upon interaction with its functional receptor. J Virol 84(11): 5550–5564.
41. BhellaD, GoodfellowIG (2011) The cryo-electron microscopy structure of feline calicivirus bound to junctional adhesion molecule A at 9-angstrom resolution reveals receptor-induced flexibility and two distinct conformational changes in the capsid protein VP1. J Virol 85(21): 11381–11390.
42. WaltersRW, YiSM, KeshavjeeS, BrownKE, WelshMJ, et al. (2001) Binding of adeno-associated virus type 5 to 2,3-linked sialic acid is required for gene transfer. J Biol Chem 276 : 20610–20616.
43. GenzelY, DietzschC, RappE, SchwarzerJ, ReichlU (2010) MDCK and Vero cells for influenza virus vaccine production: a one-to-one comparison up to lab-scale bioreactor cultivation. Appl Microbiol Biotechnol 88 : 461–475.
44. ParkSH, SaifLJ, JeongC, LimGK, ParkSI, et al. (2006) Molecular characterization of novel G5 bovine rotavirus strains. J Clin Microbiol 44 : 4101–4112.
45. LiX, ShouW, XingX, HuangL, TianL, et al. (2013) Overexpression of 4EBP1, p70S6K, Akt1 or Akt2 differentially promotes Coxsackievirus B3-induced apoptosis in HeLa cells. Cell Death Dis 4: e803–809.
46. RyuMS, JungEH, ChoKO, KangSY (2012) Expression of porcine sapovirus VP1 gene and VP1 specific monoclonal antibody production. Hybridoma 31 : 155–162.
47. HuangP, FarkasT, MarionneauS, ZhongW, Ruvoen-ClouetN, et al. (2003) Noroviruses bind to human ABO, Lewis, and secretor histo-blood group antigens: identification of 4 distinct strain-specific patterns. J Infect Dis 188 : 19–31.
48. HuangP, FarkasT, ZhongW, TanM, ThorntonS, et al. (2005) Norovirus and histo-blood group antigens: demonstration of a wide spectrum of strain specificities and classification of two major binding groups among multiple binding patterns. J Virol 79 : 6714–6722.
49. FukushimaK, TakahashiT, TakaguchiM, UeyamaH, ItoS, et al. (2011) Plaque formation assay for human parainfluenza virus type 1. Biol Pharm Bull 34 : 996–1000.
50. MatrosovichM, MatrosovichT, GartenW, KlenkHD (2006) New low-viscosity overlay medium for viral plaque assays. Virol J 3 : 63.
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek Recruitment of RED-SMU1 Complex by Influenza A Virus RNA Polymerase to Control Viral mRNA SplicingČlánek Systematic Phenotyping of a Large-Scale Deletion Collection Reveals Novel Antifungal Tolerance GenesČlánek The Contribution of Social Behaviour to the Transmission of Influenza A in a Human Population
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2014 Číslo 6- Jak souvisí postcovidový syndrom s poškozením mozku?
- Měli bychom postcovidový syndrom léčit antidepresivy?
- Farmakovigilanční studie perorálních antivirotik indikovaných v léčbě COVID-19
- 10 bodů k očkování proti COVID-19: stanovisko České společnosti alergologie a klinické imunologie ČLS JEP
-
Všechny články tohoto čísla
- Fungal Nail Infections (Onychomycosis): A Never-Ending Story?
- BdlA, DipA and Induced Dispersion Contribute to Acute Virulence and Chronic Persistence of
- Morphotype Transition and Sexual Reproduction Are Genetically Associated in a Ubiquitous Environmental Pathogen
- A Nucleic-Acid Hydrolyzing Single Chain Antibody Confers Resistance to DNA Virus Infection in HeLa Cells and C57BL/6 Mice
- HopW1 from Disrupts the Actin Cytoskeleton to Promote Virulence in Arabidopsis
- Ly6C Monocytes Become Alternatively Activated Macrophages in Schistosome Granulomas with Help from CD4+ Cells
- Recruitment of RED-SMU1 Complex by Influenza A Virus RNA Polymerase to Control Viral mRNA Splicing
- Contribution of Specific Residues of the β-Solenoid Fold to HET-s Prion Function, Amyloid Structure and Stability
- Antibody Responses to : Role in Pathogenesis and Diagnosis of Encephalitis?
- Discovery of a Novel Compound with Anti-Venezuelan Equine Encephalitis Virus Activity That Targets the Nonstructural Protein 2
- Activation of Focal Adhesion Kinase by Suppresses Autophagy via an Akt/mTOR Signaling Pathway and Promotes Bacterial Survival in Macrophages
- Crossing the Interspecies Barrier: Opening the Door to Zoonotic Pathogens
- Catching Fire: , Macrophages, and Pyroptosis
- IscR Is Essential for Type III Secretion and Virulence
- Selective Chemical Inhibition of Quorum Sensing in Promotes Host Defense with Minimal Impact on Resistance
- The Glycosylated Rv1860 Protein of Inhibits Dendritic Cell Mediated TH1 and TH17 Polarization of T Cells and Abrogates Protective Immunity Conferred by BCG
- A Genome-Wide Tethering Screen Reveals Novel Potential Post-Transcriptional Regulators in
- Structural Insights into SraP-Mediated Adhesion to Host Cells
- Human IGF1 Regulates Midgut Oxidative Stress and Epithelial Homeostasis to Balance Lifespan and resistance in
- Cycling Empirical Antibiotic Therapy in Hospitals: Meta-Analysis and Models
- Rab11 Regulates Trafficking of -sialidase to the Plasma Membrane through the Contractile Vacuole Complex of
- Mitogen and Stress Activated Kinases Act Co-operatively with CREB during the Induction of Human Cytomegalovirus Immediate-Early Gene Expression from Latency
- Profilin Promotes Recruitment of Ly6C CCR2 Inflammatory Monocytes That Can Confer Resistance to Bacterial Infection
- A Central Role for Carbon-Overflow Pathways in the Modulation of Bacterial Cell Death
- An Invertebrate Warburg Effect: A Shrimp Virus Achieves Successful Replication by Altering the Host Metabolome via the PI3K-Akt-mTOR Pathway
- The Highly Conserved Bacterial RNase YbeY Is Essential in , Playing a Critical Role in Virulence, Stress Regulation, and RNA Processing
- A Virulent Strain of Deformed Wing Virus (DWV) of Honeybees () Prevails after -Mediated, or , Transmission
- Systematic Phenotyping of a Large-Scale Deletion Collection Reveals Novel Antifungal Tolerance Genes
- Ubiquitin-Mediated Response to Microsporidia and Virus Infection in
- Preclinical Detection of Variant CJD and BSE Prions in Blood
- Toll-Like Receptor 8 Agonist and Bacteria Trigger Potent Activation of Innate Immune Cells in Human Liver
- Progressive Proximal-to-Distal Reduction in Expression of the Tight Junction Complex in Colonic Epithelium of Virally-Suppressed HIV+ Individuals
- The Triggering Receptor Expressed on Myeloid Cells 2 Inhibits Complement Component 1q Effector Mechanisms and Exerts Detrimental Effects during Pneumococcal Pneumonia
- Differential Activation of Acid Sphingomyelinase and Ceramide Release Determines Invasiveness of into Brain Endothelial Cells
- Forward Genetic Screening Identifies a Small Molecule That Blocks Growth by Inhibiting Both Host- and Parasite-Encoded Kinases
- Defining Immune Engagement Thresholds for Control of Virus-Driven Lymphoproliferation
- Growth Factor and Th2 Cytokine Signaling Pathways Converge at STAT6 to Promote Arginase Expression in Progressive Experimental Visceral Leishmaniasis
- Multimeric Assembly of Host-Pathogen Adhesion Complexes Involved in Apicomplexan Invasion
- Biogenesis of Influenza A Virus Hemagglutinin Cross-Protective Stem Epitopes
- Adequate Th2-Type Response Associates with Restricted Bacterial Growth in Latent Mycobacterial Infection of Zebrafish
- Protective Efficacy of Passive Immunization with Monoclonal Antibodies in Animal Models of H5N1 Highly Pathogenic Avian Influenza Virus Infection
- Fructose-Asparagine Is a Primary Nutrient during Growth of in the Inflamed Intestine
- The Calcium-Dependent Protein Kinase 3 of Influences Basal Calcium Levels and Functions beyond Egress as Revealed by Quantitative Phosphoproteome Analysis
- A Translocated Effector Required for Dissemination from Derma to Blood Safeguards Migratory Host Cells from Damage by Co-translocated Effectors
- Functional Characterization of a Novel Family of Acetylcholine-Gated Chloride Channels in
- Both α2,3- and α2,6-Linked Sialic Acids on O-Linked Glycoproteins Act as Functional Receptors for Porcine Sapovirus
- The Contribution of Social Behaviour to the Transmission of Influenza A in a Human Population
- MicroRNA-146a Provides Feedback Regulation of Lyme Arthritis but Not Carditis during Infection with
- Recombination in Enteroviruses Is a Biphasic Replicative Process Involving the Generation of Greater-than Genome Length ‘Imprecise’ Intermediates
- Cytoplasmic Viral RNA-Dependent RNA Polymerase Disrupts the Intracellular Splicing Machinery by Entering the Nucleus and Interfering with Prp8
- and Are Associated with Murine Susceptibility to Infection and Human Sepsis
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Fungal Nail Infections (Onychomycosis): A Never-Ending Story?
- Profilin Promotes Recruitment of Ly6C CCR2 Inflammatory Monocytes That Can Confer Resistance to Bacterial Infection
- Contribution of Specific Residues of the β-Solenoid Fold to HET-s Prion Function, Amyloid Structure and Stability
- The Highly Conserved Bacterial RNase YbeY Is Essential in , Playing a Critical Role in Virulence, Stress Regulation, and RNA Processing
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



