-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaBdlA, DipA and Induced Dispersion Contribute to Acute Virulence and Chronic Persistence of
Pathogenic bacteria, including the human pathogen Pseudomonas aeruginosa, can cause acute and chronic infections. The difference in these infection modes can be explained by how bacteria grow. Acute infections occur when individual bacteria rapidly replicate, produce high levels of virulence factors, and disseminate from the nidus of infection. Chronic infections occur when bacteria adhere to tissue or implanted medical devices and form multi-cellular, matrix-encased aggregates known as biofilms. The acute-to-chronic infection switch occurs when bacteria transition from planktonic to biofilm growth. However, the contribution of dispersion, the process by which bacteria leave a biofilm to return to planktonic growth, remains unclear. Here, we demonstrate that, while having left a biofilm, dispersed cells are distinct from planktonic cells with respect to gene expression, release of matrix-degrading enzymes, and pathogenicity. We found that a mutant impaired in nutrient-induced dispersion, while enhancing chronic infections, is impaired in mounting acute infections in both plant and mouse hosts. Overall, this work establishes that dispersed cells have a unique virulence phenotype, with nutrient-induced dispersion not only serving as an integral part of both acute and chronic infections but also as a potential mechanism of infection control.
Published in the journal: . PLoS Pathog 10(6): e32767. doi:10.1371/journal.ppat.1004168
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1004168Summary
Pathogenic bacteria, including the human pathogen Pseudomonas aeruginosa, can cause acute and chronic infections. The difference in these infection modes can be explained by how bacteria grow. Acute infections occur when individual bacteria rapidly replicate, produce high levels of virulence factors, and disseminate from the nidus of infection. Chronic infections occur when bacteria adhere to tissue or implanted medical devices and form multi-cellular, matrix-encased aggregates known as biofilms. The acute-to-chronic infection switch occurs when bacteria transition from planktonic to biofilm growth. However, the contribution of dispersion, the process by which bacteria leave a biofilm to return to planktonic growth, remains unclear. Here, we demonstrate that, while having left a biofilm, dispersed cells are distinct from planktonic cells with respect to gene expression, release of matrix-degrading enzymes, and pathogenicity. We found that a mutant impaired in nutrient-induced dispersion, while enhancing chronic infections, is impaired in mounting acute infections in both plant and mouse hosts. Overall, this work establishes that dispersed cells have a unique virulence phenotype, with nutrient-induced dispersion not only serving as an integral part of both acute and chronic infections but also as a potential mechanism of infection control.
Introduction
Pseudomonas aeruginosa is a ubiquitous, Gram-negative opportunistic bacterial pathogen, well known for its remarkable ability to replicate and survive in diverse environments, as well as for causing a variety of acute and chronic human infections. Acute infections are characterized by rapid pathogenic progression, high-level toxin production, and often tissue damage. Chronic infections are characterized by colonization, prolonged persistence, evasion of the host's immune response and tolerance and often resistance to multiple therapeutic agents. The capacity to cause either acute or chronic infections depends to a large extent on the ability of P. aeruginosa to transit from growing planktonically (free living state) to surface-attached communities known as biofilm [1], [2], [3], [4], [5]. While planktonic infections are frequently associated with high virulence and fast growth as observed in sepsis or bacteremia [6], the development of biofilms is considered to be the root cause of chronic infections [1], [2], [3], [7], [8], [9]. Biofilms are composed of microorganisms attached to a solid surface and encased in a hydrated polymeric matrix composed of polysaccharides, protein and DNA. Biofilms form when bacteria adhere to surfaces in moist environments. For example, P. aeruginosa forms what are termed Mode II biofilms [10] within the airways of individuals with cystic fibrosis (CF), with this process playing an important role in CF-associated chronic infections and acting as one of the primary causes of mortality in CF patients [11], [12], [13], [14]. In addition, P. aeruginosa causes a variety of chronic infections in immunocompromised individuals or those suffering from wounds, burns, urinary tract infections, or corneal injury [7], [15], [16], [17], [18], [19].
The transition from a planktonic to surface-attached lifestyle is highly regulated. The two-component regulatory systems (TCS) including the RetS/GacS/GacA/rsmZ signal transduction pathway, as well as the TCS SagS and BfiSR are required for P. aeruginosa to transition to the surface associated mode of growth and to progress from initial attachment to the formation of mature biofilms [1], [2], [3], [4], [5], [20]. Additional regulatory systems required for the formation of mature, three-dimensional biofilms include the TCS BfmSR, MifSR, and SadARS [8], [21], [22]. However, these regulatory systems not only regulate the motile-sessile switch but also the growth-mode dependent expression of virulence factors. For example, genome-wide transcriptional profiling suggested that RetS is required for expression of the Type III secretion system (T3SS) genes (psc and pcr operons), secreted effectors ExoS, ExoT and ExoY [1], [23], [24], and other virulence factors, and for repression of genes responsible for exopolysaccharide components of the P. aeruginosa biofilm matrix [1]. Conversely, inactivation of retS correlated with hyper-adhesion to mammalian cells but loss of cytotoxicity and attenuated virulence in an acute pneumonia model. In addition to RetS and GacS/GacA/rsmZ, regulation of T3SS gene expression also requires SadARS [8]. Considering the number of regulatory systems affecting T3SS expression, it is not surprising that T3SS-mediated cytotoxicity is considered a key virulence mechanism of P. aeruginosa, allowing for the contact-dependent translocation of pathogenicity factors into eukaryotic host cells, with T3SS inactivation attenuating P. aeruginosa virulence in a murine acute infection model [20], [25], [26], [27], [28]. Additional known virulence factors produced by P. aeruginosa include hydrogen cyanide, elastase, phenazines, and rhamnolipids. Hydrogen cyanide is believed to be the primary toxic factor secreted by P. aeruginosa that is responsible for killing Caenorhabditis elegans [29], and has been detected at elevated levels in the infected lungs of patients suffering from CF [30]. Chitinase is expressed at high cell densities and in biofilms and was shown to be able to bind and degrade colloidal chitin [31], while elastase has been shown to cause localized tissue damage [32]. The redox-active pigments phenazines can generate an oxidative stress on the host and are required for “fast killing” of C. elegans [33]. Rhamnolipids interfere with the internalization of attached particles, reducing the level of phagosome-lysosome fusion of internalized targets within macrophages, and inhibiting the response of alveolar macrophages [34], [35].
While the clinical relevance of planktonic and biofilm cells in acute and chronic infections, respectively, has been well established, little is known about the contribution of biofilm dispersion to the virulence phenotype of P. aeruginosa and infections. Dispersion is the last yet a very important step in the development of biofilms that allows bacteria to successfully return from the biofilm to the planktonic growth state to spawn novel communities in new locales [36], [37]. Biofilm dispersion can be induced by exposure to matrix-degrading enzymes and surface protein releasing factors [38], [39], [40]. In P. putida and P. fluorescens, the large adhesive outer-membrane protein LapA mediates attachment to surfaces and to matrix components [41], [42], [43]. Gjermansen et al. [41] demonstrated that in P. putida, release of LapA from the cell surface results in biofilm dispersal and is mediated through the activity of the periplasmic protease LapG. Additional mechanisms linked to dispersion include cell death, with filamentous phage Pf1-mediated cell lysis serving as an important mechanism of differentiation inside microcolonies that facilitates dispersal of a subpopulation of surviving cells [44]. The process of biofilm dispersion in various organisms including P. aeruginosa, P. putida and Schewanella oneidensis is furthermore induced upon sensing a myriad of environmental cues such as variation in oxygen or carbon substrate concentration and sensing the signaling molecule cis-2-decenoic acid [36], [38], [45], [46], [47], [48], [49], [50]. Oxidative or nitrosative stress, induced upon exposure to exogenous or endogenous nitric oxide (NO), has been linked to biofilm dispersion. A role of oxidative or nitrosative stress was further supported by a P. aeruginosa mutant lacking the only enzyme capable of generating metabolic NO through anaerobic respiration (nitrite reductase, ΔnirS) not dispersing [51].
Biofilm dispersion has also been linked to the modulation of the intracellular signaling molecule cyclic di-GMP (c-di-GMP), high levels of which promote sessile growth, while low levels correlate with planktonic existence [52], [53], [54]. Levels of c-di-GMP are enzymatically modulated by diguanylate cyclases (DCG), proteins containing a GGDEF domain, and phosphodiesterases (PDE) harboring either an EAL or HD-GYP domain [52]. In P. aeruginosa, dispersion upon exposure to NO and elevated nutrient concentrations has been linked to the reduction of the cellular c-di-GMP levels, requiring the phosphodiesterases DipA and RbdA [46], [55], [56], [57]. In addition, the membrane-bound phosphodiesterase NbdA was found to be specific to the dispersion response following exposure to NO [48]. The chemotaxis transducer protein BdlA (Biofilm dispersion locus A) appears to play a central role in the dispersion response by P. aeruginosa biofilms, as inactivation of bdlA impaired dispersion by P. aeruginosa biofilms in response to various nutrients, NO, ammonium chloride, and heavy metals [56], [57]. However, BdlA does not directly contribute to the observed reduction of cellular c-di-GMP levels in dispersed cells as BdlA lacks domains required for c-di-GMP modulation. Instead, BdlA harbors two sensory Per Arnt Sim (PAS) domains and a chemoreceptor domain, TarH. The closest known BdlA homolog is the FAD-binding Aer, the redox potential sensor and aerotaxis transducer in Escherichia coli. Alanine replacement mutagenesis of BdlA-PAS domain residues D14A, N23A, W60A, I109A, W182A, that were previously demonstrated to be essential for aerotaxis in Aer, resulted in impaired dispersion, while alanine replacement mutagenesis of residue G31A resulted in the mutant strain transmitting a constant signal-on bias as it rendered P. aeruginosa biofilms hyper-dispersive [58]. The findings suggested BdlA to likely function as a sensory protein [58]. However, for BdlA to contribute to the dispersion response, it must first be activated via unusual, non-processive proteolytic cleavage at a ClpP-protease-like cleavage site located between the PAS sensory domains PASa and PASb within the BdlA protein [45]. Proteolysis of BdlA was stimulated by increased c-di-GMP levels present in biofilms, and dependent on the protease ClpP, the chaperone ClpD, and BdlA phosphorylation at tyrosine-238 [45]. Once activated, BdlA oligomerizes with the phosphodiesterases DipA and RbdA, thus forming a regulatory network that modulates the intracellular c-di-GMP pool to enable dispersion [57], [58].
Considering the link between the motile-sessile transition and the switch in P. aeruginosa virulence towards chronic infections [1], [2], we addressed herein whether induced dispersion, which enables the return to the planktonic mode of growth, is part of an inherent strategy of P. aeruginosa to initiate or contribute to either acute or chronic infections. To address this question, we analyzed virulence factor production and gene expression in cells exhibiting different growth modes by making use of dispersed cells obtained upon induction of dispersion in response to NO or changes in the nutrient glutamate concentration, as well as in a ΔbdlA mutant strain. This mutant was chosen as it is impaired in nutrient - and NO-induced biofilm dispersion [45], [56], [58] while not directly affecting c-di-GMP levels [56], [58]. This was done as intracellular signaling via c-di-GMP has been demonstrated to be an important contributor to virulence in multiple pathogens [59], [60], [61]. To assure, however, that the observed effects on virulence and pathogenicity are not specific to BdlA but instead linked to dispersion, we furthermore made use of mutants inactivated in dipA and rbdA.
Results
Biofilms prior to and post induction of dispersion release fewer degradative proteins than dispersed cells
Several reports have described the process of dispersion to occur from within microcolonies, with dispersing bacteria observed to be motile, followed by them swimming away from the inner portions of the cell cluster through openings in the cluster and entering the bulk liquid [37], [47]. The in vitro observations suggested bacteria within microcolonies and enmeshed by extracellular polymeric substance (EPS) matrix composed of polysaccharides, extracellular DNA, lipids, and proteins (reviewed in [62], [63], [64]) to be able to liberate themselves in order to evacuate from biofilms. We, therefore, hypothesized that dispersion may correlate with the release of enzymes that assist in the degradation of the EPS matrix. To do so, we first determined the concentration of proteins present in culture supernatants produced by 1×109 P. aeruginosa PAO1 bacteria grown planktonically, as biofilms, and following dispersion in response to nitric oxide (NO) or changes in the nutrient concentration, using the Bradford protein assay. NO was chosen as a dispersion inducing cue to mimic the endogenous production of or exogenous exposure to reactive oxygen intermediates to which P. aeruginosa is frequently exposed, both under anaerobic conditions prevalent in the CF lung environment and during infection [48], [51], [57]. Glutamate was used as a dispersion-inducing cue to mimic rapid environmental changes. Analysis of the culture supernatants of P. aeruginosa indicated the presence of supernatant proteins under all growth conditions tested (Fig. 1A). However, the highest amount of proteins was detected in supernatants of dispersed cells followed by supernatants from planktonic cells grown to exponential and stationary phase (Fig. 1A). The lowest concentration was routinely detected in biofilm supernatants (Fig. 1A). Overall, up to 10-times more protein was detected in supernatants of dispersed cells than those of planktonic cells (Fig. 1A). It is of interest to note that no difference in the concentration of supernatant proteins following dispersion in response to NO or changes in the nutrient concentration was noted, indicating protein release to be independent of dispersion inducing conditions (Fig. 1A). As dispersion was induced by NO or glutamate, we also analyzed culture supernatants of planktonic cells exposed to glutamate and NO. No difference in the concentration of protein detected in supernatants of planktonic cells and planktonic cells exposed to glutamate or NO was noted (Fig. 1A). Similar trends were observed upon the analysis of supernatants obtained from the hyper-virulent strain P. aeruginosa PA14 (Fig. S1).
Fig. 1. Dispersion of P. aeruginosa PAO1 biofilms correlates with increased release of proteins into the supernatant. 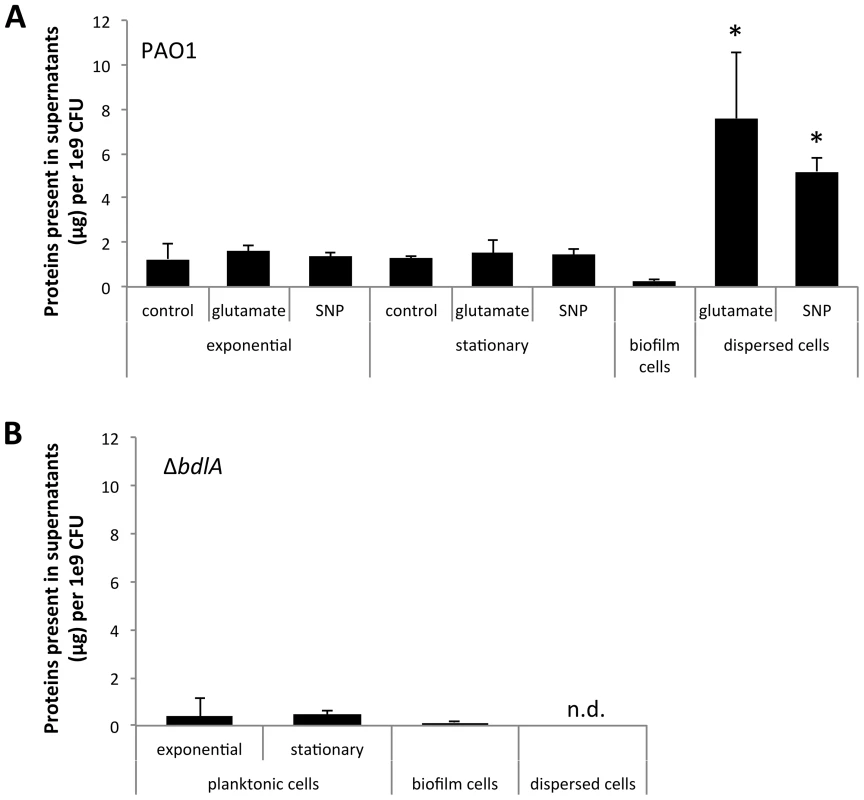
Supernatants were obtained from (A) P. aeruginosa PAO1 and (B) ΔbdlA grown planktonically to exponential and stationary phase, as well as from biofilms and cells dispersed from the biofilm in response to exposure to glutamate (dispersed cells). Dispersed cells were obtained following dispersion in response to glutamate and SNP, which was used as a source of nitric oxide. Likewise, planktonic cells grown to exponential and stationary phase were exposed for 20 min to glutamate or SNP. Planktonic cells not treated with glutamate or nitric oxide are referred to as “control”. Experiments were carried out in triplicate. Error bars indicate standard deviation. The protein concentration of supernatants was determined using the same number of cells (1x109 CFU/ml) regardless of growth conditions. n.d., not determined. Early infection and colonization have been suggested to require the production of a variety of cytotoxic and degradative proteins [1]. To determine whether proteins detected in the supernatants harbored cytotoxic or degradative activity capable of degrading polymeric tissue components or substances that may contribute to the release of bacterial cells from biofilms, we used agar plate-based assays, focusing on the detection of proteolysis, lipid hydrolysis, hemolysis, and Psl degradation. These degradative activities were chosen as they target components of the EPS matrix or are indicators of cytotoxicity. To better quantitate cytotoxic or degradative activities present in the various supernatants, a total of 10 µg of supernatant protein was used for each assay. Supernatants obtained from biofilms, remaining biofilms following nutrient-induced dispersion, dispersed cells following induction of dispersion in response to NO or changes in the medium glutamate concentration, and planktonic cells grown to exponential and stationary phase were used. In addition, supernatants obtained from planktonic cells exposed to NO and glutamate were used as controls. Supernatants obtained from P. aeruginosa PAO1 displayed proteolytic and Psl degradation activities as well as lipid hydrolysis and hemolysis, regardless of the mode of growth tested (Fig. 2). The highest activity with respect to proteolysis, lipid hydrolysis, hemolysis, and Psl degradation was detected in supernatants obtained from planktonic cells grown to exponential and stationary phase (Fig. 2A–B). Exposure of planktonic cells to NO or glutamate did not affect the overall activity present in supernatants (Fig. 2A–B). In contrast, with the exception of lipid hydrolysis, biofilms remaining attached following induction of dispersion demonstrated the least activity (Fig. 2). No significant difference in the cytotoxic and degradative activities was noted in supernatants of dispersed cells obtained following induction of dispersion in response to NO or changes in the nutrient glutamate concentration (Fig. 2C–F). Although supernatants of dispersed cells were characterized by levels approaching 10 times higher protein yield compared to planktonic and biofilm cells (Fig. 1A), the enzyme activity present in supernatants of dispersed cells was intermediate between that of biofilm and planktonic cells (Fig. 2). Overall, the cytotoxic and degradative activities present in 10 µg of supernatant proteins obtained from dispersed cells were, while elevated, more similar to those observed for biofilm supernatants than planktonic supernatants (Fig. 2). Similar results were observed when supernatants obtained from P. aeruginosa PA14 were used (Fig. S2). We furthermore determined DNA hydrolase activity in supernatants. The highest DNA hydrolase activity was found in supernatants of dispersed cells and biofilm cells remaining attached following induction of dispersion (Fig. S3).
Fig. 2. Detection of degradative activity in the extracellular proteome of P. aeruginosa PAO1 is growth mode dependent with P. aeruginosa ΔbdlA impaired in dispersion exhibiting lower degradative activity. 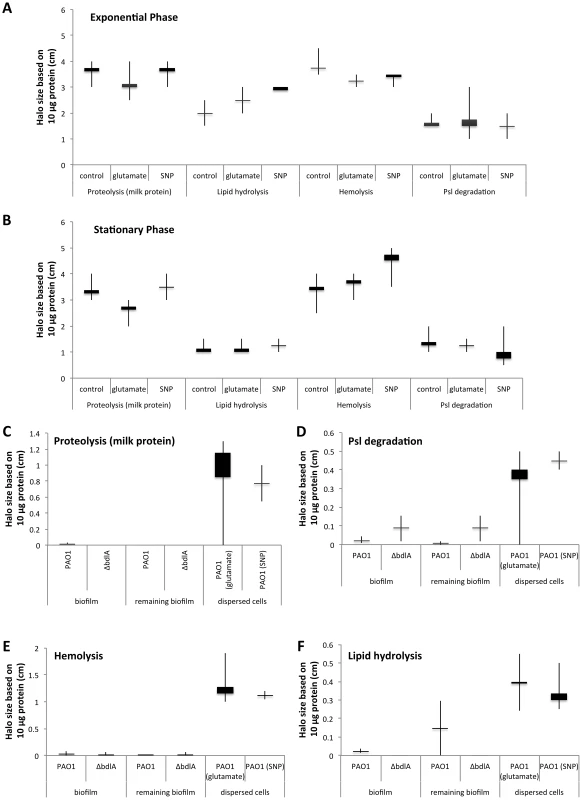
Cytotoxic and degradative activities were determined using 10 µg of supernatant protein in 100 µl of sterile water, followed by measuring the zone of clearance 18 hr post-inoculation of the sterile protein solution into the wells of the respective agar plates. Degradative activity was determined using supernatants obtained from P. aeruginosa grown planktonically to exponential (A) and stationary phase (B). Supernatants of planktonic cells not treated with glutamate or nitric oxide are referred to as “control”. Additionally, supernatants of planktonic cells grown to exponential and stationary phase were exposed for 30 min to glutamate or SNP were used. (C–F) Degradative activities were furthermore determined in supernatants obtained from biofilms, and biofilms post-induction of dispersion with glutamate (remaining biofilm). Dispersed cells were obtained following dispersion in response to glutamate and SNP, which was used as a source of nitric oxide. (C) Proteolytic activity was detected using milk agar plates in supernatants obtained from biofilms, biofilms post-induction of dispersion, and dispersed cells. (D) Lipid hydrolysis was determined using tributyrin containing agar plates. (E) Hemolytic activity was detected using blood agar plates while (F) Psl degradation was detected on agar plates containing Psl extracted from a P. aeruginosa strain overexpressing Psl. Psl degradation was visualized as a zone of clearing following 24 hr incubation and staining the agar plate with iodine. Experiments were carried out at least in triplicate. Error bars indicate standard deviation. Supernatants obtained from the ΔbdlA mutant, impaired in biofilm dispersion, demonstrate reduced enzymatic activities
Our findings suggested that dispersed cells are distinct from planktonic and biofilm cells with respect to cytotoxic or degradative supernatant activities. To determine whether the ability to disperse correlated with the ability to degrade polymeric substances, we made use of a ΔbdlA mutant. Deletion of bdlA was previously demonstrated to render biofilm bacteria deficient in dispersion triggered by multiple environmental cues including various nutrients, NO, ammonium chloride, and heavy metals [45], [56], [58]. Compared to wild type bacteria, ΔbdlA grown planktonically and as biofilms released comparable amounts of proteins into the supernatant (Fig. 1B). Moreover, no difference in cytotoxic and degradative activity was noted for supernatants obtained from ΔbdlA mutant grown planktonically compared to that observed for the wild-type supernatants (not shown). Under biofilm growth conditions, however, very little enzymatic activity was detected in supernatants obtained from ΔbdlA biofilms and biofilms exposed to dispersion-inducing conditions (referred to as remaining biofilms, although no dispersion event occurred) compared to the wild type. In fact, no proteolysis and lipid hydrolysis was detected in supernatants from ΔbdlA biofilms and remaining biofilms, and hemolytic and Psl polysaccharide degradative activities were reduced compared to wild-type biofilms and remaining biofilms (Fig. 2C–F).
Analysis of supernatant proteins of P. aeruginosa PAO1 and ΔbdlA
Our findings not only suggested differences in P. aeruginosa PAO1 and ΔbdlA strains with respect to the release of degradative enzymes but, furthermore, that the inability to disperse likely corresponded with significantly reduced release or absence of degradative enzymes compared to wild-type biofilms. To further confirm differences in proteins released by the two strains, proteins present in supernatants obtained from biofilm cells were analyzed by SDS/PAGE, followed by subsequent protein identification by LC-MS/MS. As expected, marked differences in the extracellular proteins of wild-type and ΔbdlA mutant biofilms were noted (Fig. 3). However, the analysis also revealed the presence of a set of proteins common to the extracellular proteins of both wild type and the ΔbdlA mutant (Fig. 3). These included the outer membrane porins OprD, OprQ, and OprF as well as the flagellin component, FliC, the aminopeptidase PA2939, a component of the ABC transporter PA1342, and the sulfate binding protein precursor PA0283 (Fig. 3, Table 1). The latter two proteins were detected in a growth mode-dependent manner with PA1342 and PA0283 being absent in supernatants obtained from planktonic cells but detectable in supernatants obtained from biofilms (Fig. 3, Table 1). Differences in the extracellular proteome constituted the proteases elastase (LasB) and protease IV (PA3724). While both were detectable in supernatants of P. aeruginosa PAO1 under planktonic and biofilm growth conditions, the proteins were present at elevated levels in supernatants from P. aeruginosa PAO1 grown planktonically to stationary phase. In contrast, no protein bands of respective apparent mass were detected in culture supernatants of the ΔbdlA mutant (Fig. 3, Table 1). The only protease detected in the ΔbdlA mutant was the alkaline metalloproteinase, AprA (Fig. 3, Table 1). The finding of the ΔbdlA strain lacking proteases is consistent with the reduced proteolytic activity in this strain (Fig. 2C). No lipases or hemolysins were detected. Additional differences in the extracellular proteome were noted with respect to exotoxin A. Production of exotoxin A inhibits eukaryotic host cell protein synthesis by interacting with elongation factor-2, resulting in cell death [65]. While exotoxin A was detected in the supernatants of P. aeruginosa PAO1, its presence was limited to biofilms (Fig. 3, see black arrow “F2”, Table 1). In contrast, exotoxin A was detectable in supernatants of the ΔbdlA mutant regardless of growth conditions (Fig. 3, Table 1).
Fig. 3. Analysis of proteins present in supernatants of P. aeruginosa PAO1 and ΔbdlA biofilms. 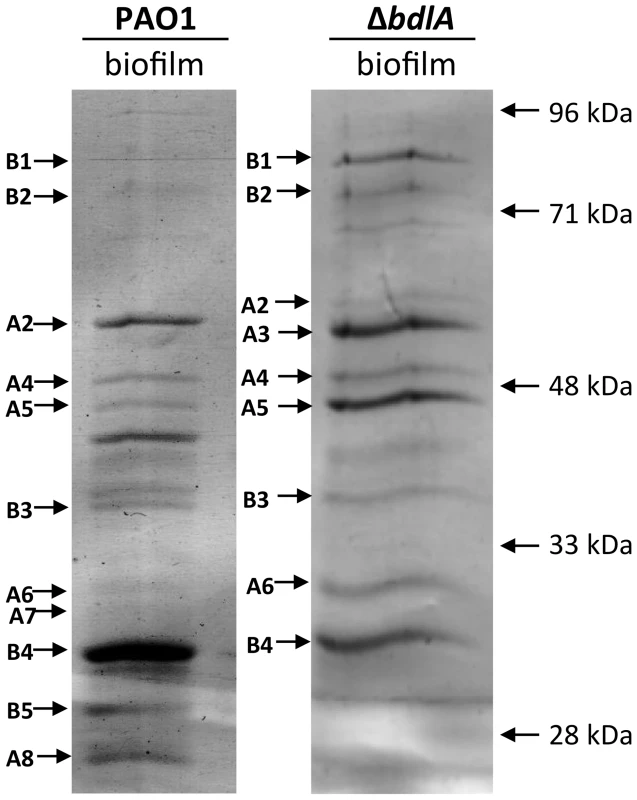
A total of 10 µg supernatant protein obtained from P. aeruginosa PAO1 and ΔbdlA biofilms was loaded per lane. Protein bands indicated by a letter and arrow were selected and subsequently identified by LC-MS/MS (see also Table 1). Experiments were repeated in triplicate and a representative SDS-gel image is shown. Molecular masses are indicated on the right (in kDa). Tab. 1. Identification of proteins present in supernatants. 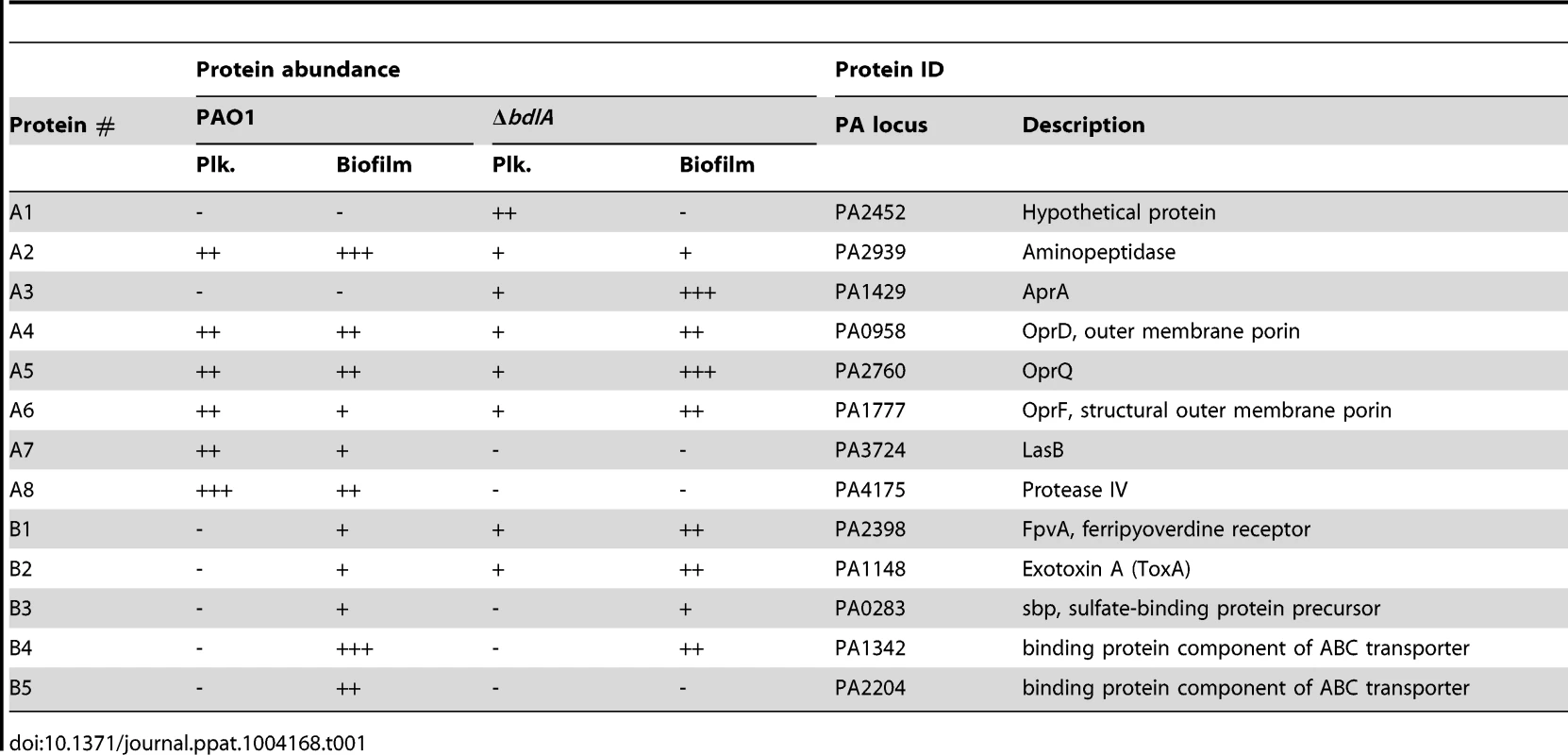
The virulence phenotype of nutrient-induced dispersed cells is distinct from that of planktonic cells
The difference in protease and exotoxin A abundance in the extracellular proteomes obtained from P. aeruginosa wild-type and ΔbdlA biofilms suggested a contribution of dispersion to the production of degradative and virulence factors. To further explore whether dispersion and the return to the planktonic, free-living mode of growth contributes to a switch in virulence gene expression, qRT-PCR was used to quantitatively determine the transcript levels of genes encoding several known virulence factors. In addition to exotoxin A, expression of the virulence factors hydrogen cyanide (hcnA), chitinase (chiC), elastase (lasB), pyocyanin (phz operon), genes encoding T3SS components (pcrV and pscL), and rhamnolipids (rhlA) [66], [67] was assessed. Considering that dispersed cells are exposed to NO or glutamate, we first determined whether expression of virulence genes is affected by the addition of NO or glutamate by analyzing the transcript abundance of virulence genes of interest in exponential phase planktonic cells and exponential phase planktonic cells exposed to glutamate and NO. No significant difference in transcript levels of the tested virulence genes was noted (Fig. 4A) indicating that under the conditions tested, exposure to NO or additional glutamate does not affect the expression of the virulence genes. We therefore compared transcript abundance of virulence genes in biofilms and dispersed cells to untreated exponential phase planktonic cells. Compared to planktonic cells, P. aeruginosa PAO1 grown as biofilms were characterized by significantly reduced expression of chiC, lasB, rhlA, phzB, pscL, and hcnA, but increased pcrV and toxA expression (Fig. 4B, Table S1). Increased toxA transcript levels in P. aeruginosa PAO1 biofilms are in agreement with the biofilm-specific detection of exotoxin A (Fig. 3). Dispersed cells were likewise characterized by reduced expression of virulence genes, regardless of whether NO or changes in the glutamate concentration were used to induce dispersion (Fig. 4B, Table S1). However, all virulence genes tested including pcrV and toxA were significantly reduced in dispersed cells compared to both planktonic or biofilm cells (Fig. 4B, Table S1). Similar changes in transcript levels were observed for dispersed P. aeruginosa PA14 cells compared to planktonic and biofilm cells (Fig. S4) and suggested that dispersion induced upon exposure to environmental cues to be more than merely a transitional episode of cells leaving the biofilm and adapting to the planktonic mode of growth.
Fig. 4. Expression of virulence genes in dispersed cells is distinct from the virulence gene expression profile of planktonic and biofilm cells. 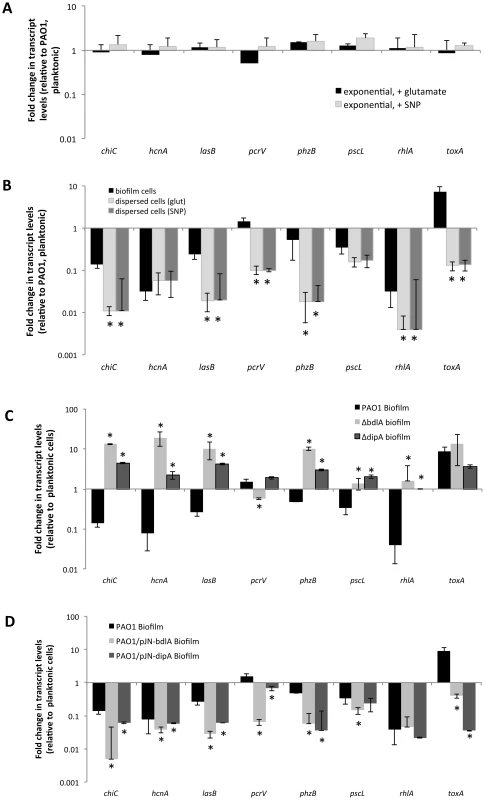
(A) Differential expression of selected virulence genes by P. aeruginosa PAO1 grown planktonically to exponential compared to expontial phase planktonic cells that were exposed for 30 min with 18 mM glutamate or 500 µM SNP, which was used as a source of nitric oxide [51]. (B) Differential expression of selected virulence genes by P. aeruginosa PAO1 grown as biofilms and dispersed cells compared to cells grown planktonically to exponential phase. Differential gene expression was determined by qRT-PCR. Dispersed cells were obtained following induction of dispersion by SNP (nitric oxide) or glutamate. *, significantly different from PAO1 grown as biofilms, P-value <0.01. (C) Differential expression of selected virulence genes by biofilm cells of P. aeruginosa PAO1 wild type and ΔdipA and ΔbdlA mutant strains compared to wild-type cells grown planktonically to exponential phase. *, significantly different from PAO1 grown as biofilms, P-value <0.01. (D) Differential expression of selected virulence genes by P. aeruginosa PAO1, PAO1/pJN-bdlA, and PAO1/pJN-dipA biofilms compared to wild-type cells grown planktonically to exponential phase. Differential gene expression was determined by qRT-PCR. Experiments were carried out at least in triplicate. *, significantly different from PAO1 grown as biofilms, P-value <0.01. Considering the similarity in the trend of virulence gene expression between biofilms and dispersed cells compared to planktonic cells, we hypothesized that dispersion may contribute to virulence gene expression and assessed this hypothesis by testing the dispersion-deficient ΔbdlA mutant strain grown as a biofilm. We anticipated finding transcript levels of virulence genes to be higher in biofilms impaired in dispersion compared to wild-type biofilms. qRT-PCR analysis revealed that transcript levels of chiC, hcnA, lasB, phzB were significantly increased in ΔbdlA biofilms relative to wild-type planktonic cells (and biofilm cells). In contrast, the transcript levels of rhlA and the T3SS genes pscL and pcrV were similar those observed for wild-type planktonic cells (but significantly increased compared to wild-type biofilm cells, Fig. 4C, Table S1). The only transcript present at similar levels in ΔbdlA biofilm and wild-type biofilm cells was toxA. The finding is in agreement with the supernatant protein analysis (Figs. 3, 4C, Table S1). It is of interest to note that similar results were obtained when virulence gene transcript levels of the dispersion-deficient ΔdipA mutant were determined (Fig. 4C).
In contrast, multi-copy expression of bdlA, which renders PAO1/pJN-bdlA biofilms hyper-dispersive [45], [58], resulted in significantly reduced expression of chiC, lasB, rhlA, phzB, pscL, pcrV, and hcnA compared to planktonic cells. Moreover, with the exception of rhlA and toxA, the transcript levels of chiC, lasB, phzB, pscL, pcrV, and hcnA detected in PAO1/pJN-bdlA biofilms were overall reduced compared to wild type biofilms (Fig. 4D, Table S1). Instead, the transcript levels of virulence genes in this hyper-dispersive biofilm were overall similar to those observed for dispersed cells (Fig. 4B-C, Table S1). A similar trend was observed upon overexpression of dipA in biofilms which has previously been demonstrated to also render P. aeruginosa biofilms hyper-dispersive [46]. With the exception of pscl and rhlA, the transcript levels of virulence genes detected in PAO1/pJN-dipA biofilms were significantly different relative to wild type biofilms (Fig. 4D). Our findings indicated non-dispersing and hyper-dispersing biofilms to express virulence genes in a manner distinct from biofilm and planktonic cells, with non-dispersing ΔbdlA and ΔdipA biofilms demonstrating increased expression of virulence genes compared to biofilm and planktonic cells while hyper-dispersing biofilms appeared to express virulence genes at reduced levels compared to planktonic and biofilm cells, with the expression profile overall being more similar to that of dispersed cells. Moreover, our findings further indicate that toxA expression is specific to the biofilm mode of growth.
Inactivation of bdlA and impaired nutrient-induced dispersion capability attenuates the virulence of P. aeruginosa in an Arabidopsis thaliana infection model
Inactivation of bdlA correlated with increased expression of virulence factors but reduced release of cytotoxic and degradative enzymes relative to wild-type cells. In contrast, overexpression of bdlA resulted in significantly reduced transcript levels of virulence genes. We therefore asked whether bdlA inactivation affected the virulence phenotype of this mutant strain using an A. thaliana virulence model. This alternative nonvertebrate, plant host model was chosen as it has been previously demonstrated to result in the identification of bacterial virulence factors and to correlate with virulence outcomes obtained using vertebrate infection models such as the burned mouse pathogenicity model [68], [69]. Compared to the wild type, mutants lacking BdlA were less virulent. While more than 58% of Arabidopsis plants were killed by P. aeruginosa PAO1 within 7 days post infection (Fig. 5A) with the percent of dead plants rising to 70% following 9 days of infection (not shown), the isogenic ΔbdlA mutant was unable to establish infection within 7 days. Only 12% of all plants were killed 7 days post infection (Fig. 5A). In contrast, overexpression of bdlA, which was found to mimic a hyper-biofilm dispersion phenotype in vitro [45], [58], had no additional effect on P. aeruginosa virulence and plant mortality compared to the wild type (Fig. 5A).
Fig. 5. The non-dispersing ΔbdlA mutant and complemented ΔbdlA mutants impaired in biofilm dispersion are avirulent. 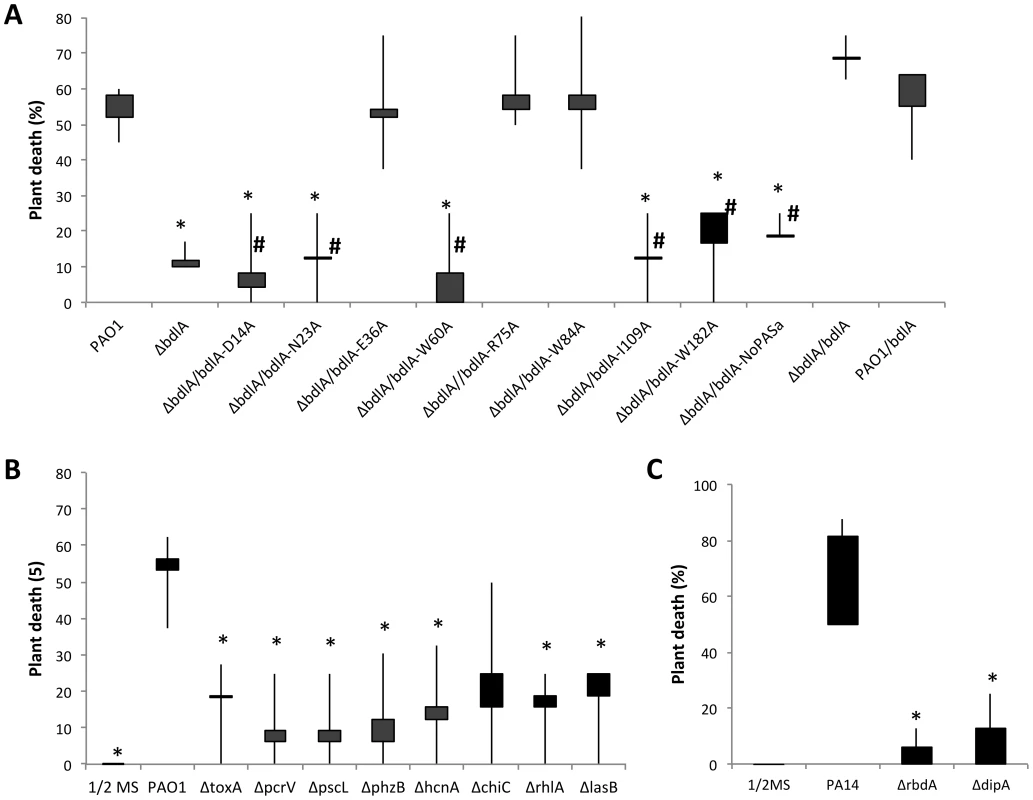
(A) Death of Arabidopsis thaliana 7 days post infection with P. aeruginosa PAO1, the isogenic ΔbdlA mutant and ΔbdlA mutants complemented with bdlA, a truncated BdlA variant (NoPAS-bdlA) and BdlA variants harboring alanine substitutions in various amino acids. #, indicates complemented ΔbdlA strains impaired in nutrient-induced dispersion, see [58]. All other complemented ΔbdlA strains were not impaired in nutrient-induced dispersion. (B) Death of Arabidopsis thaliana 7 days post-infection with P. aeruginosa PAO1 and selected isogenic mutants. Bars indicate average and median plant death rates while vertical lines indicate the highest and lowest plant death rates observed. (C) Death of Arabidopsis thaliana 7 days post infection with P. aeruginosa PA14, and the isogenic ΔdipA and ΔrbdA mutants. Control plants inoculated with ½ MS salts alone showed no symptoms over the course of the experiments. Experiments were carried out in triplicate using 8 plants per strain per replicate. *, significantly different from PAO1, P-value <0.05. To ensure that the factors encoding the tested virulence genes chiC, lasB, rhlA, phzB, pscL, hcnA pcrV and toxA contribute to the pathogenicity of P. aeruginosa in the A. thaliana virulence model, mutants inactivated in these virulence factors were tested. Inactivation of exoA, hcnA, chiC, lasB, phz, and rhlA and T3SS-coding genes pcrV and pcsL resulted in significantly reduced plant death compared to the wild type. While more than 55% of Arabidopsis plants were killed by P. aeruginosa PAO1 within 7 days post infection, infections with mutants inactivated in these virulence genes (with the exception of chiC) resulted in significantly reduced plant mortality, with less than 20% of all plants being killed over the same period of time (Fig. 5B). The findings confirmed a contribution of the respective virulence factors to P. aeruginosa pathogenicity. Considering, however, that the majority of these factors are up-regulated in ΔbdlA biofilms but reduced in PAO1/pJN-bdlA relative to wild-type biofilms, our findings further suggested that the ability to disperse, rather than the differential expression of a particular set of virulence factors, is contributing to the pathogenicity of P. aeruginosa.
To exclude the possibility that the effect on virulence was due to the absence of BdlA rather than linked to the ability to disperse, we made use of recently identified site-directed mutants of BdlA that were unable to restore the ΔbdlA-dispersion phenotype to wild-type levels [58]. These included alanine substitutions of the amino acids D14, N23, W60, I109, and W182 in BdlA. ΔbdlA mutants complemented with any of these BdlA variants were as avirulent as a ΔbdlA mutant (Fig. 5A). Similarly, complementation of ΔbdlA with a truncated BdlA lacking the N-terminal located PAS domain of BdlA, which was previously shown to not restore the ΔbdlA dispersion phenotype to wild-type levels [58], resulted in such a bdlA strain remaining avirulent (Fig. 5A). In contrast, alanine substitution in amino acid positions E36, R75, and W84 that did restore the biofilm-deficient phenotype of ΔbdlA to wild-type levels [58], rendered the complemented bdlA strain as virulent as wild-type bacteria (Fig. 5A). Similarly, a ΔbdlA mutant complemented with intact bdlA was used as positive control and found to be as virulent as the wild type (Fig. 5A). To furthermore support the link between dispersion and virulence, we additionally tested the mutant strains ΔdipA and ΔrbdA, that have been previously demonstrated to be impaired in dispersion of P. aeruginosa biofilms in response to NO and glutamate [46], [55]. Compared to the wild type, mutants lacking DipA and RbdA were significantly less virulent (Fig. 5C). While up to 80% of Arabidopsis plants were killed by the parental P. aeruginosa strain within 7 days post infection (Fig. 5C), the isogenic ΔrbdA and ΔdipA mutants were unable to establish infections. Overall, less than 10% of all plants were killed 7 days post infection (Fig. 5C). Our findings strongly indicate that the process of dispersion is a major contributor to P. aeruginosa pathogenicity, as strains impaired in dispersion are avirulent, while strains capable of the dispersion response demonstrated wild-type virulence levels.
Inactivation of bdlA reduces competitiveness and attenuates the virulence of P. aeruginosa in a murine acute pneumonia model
To determine whether reduced virulence in a plant model of infection correlates with reduced acute infection and attenuation of virulence in vivo, we next examined the ability of the ΔbdlA mutant, which is impaired in dispersion in vitro, to colonize in an acute infection model, using a murine model of acute pneumonia. Wild-type P. aeruginosa is able to colonize the lungs of an infected mouse and grow about 100-fold in the course of 24 hr (Fig. 6A–B). The isogenic ΔbdlA mutant, in comparison, was unable to establish infection; bacterial load was reduced by 10-fold as compared to infection with the wild-type parental strain (Fig. 6A–B). Additionally, the P. aeruginosa ΔbdlA mutant was unable to establish systemic infections and no bacteria were recovered from the liver or spleen (data not shown).
Fig. 6. The non-dispersing P. aeruginosa ΔbdlA and ΔdipA mutant strains are less virulent and competitive compared to P. aeruginosa wild type as determined using an acute murine pneumonia infection model. 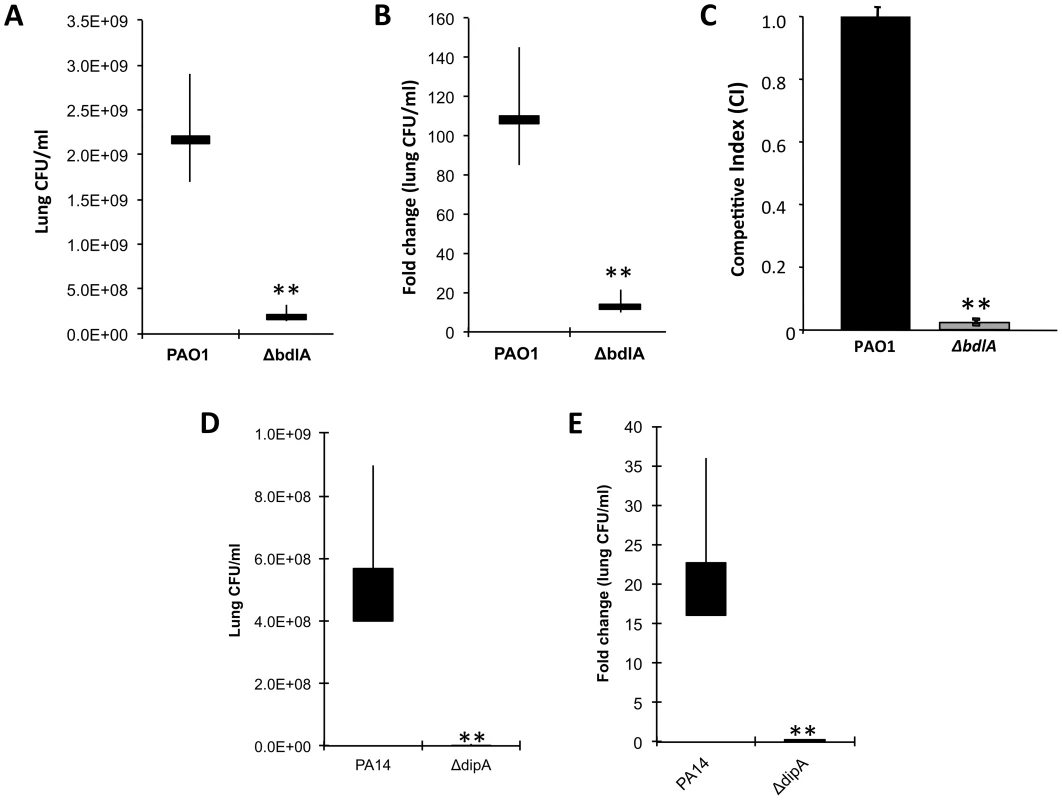
CD1 mice were inoculated (intranasal) with 2×107 CFU of Pseudomonas strains; lungs were harvested 24 h post-inoculation and CFU was determined. Values presented are average, min, max, mean. (A) Bacterial burden in the lung and (B) fold change in lung CFU/ml 24 hr post-infection compared to initial inoculum by P. aeruginosa PAO1 and the isogenic ΔbdlA mutant. Bars indicate average and median lung CFU/ml while vertical lines indicate the highest and lowest lung CFU/ml observed. (C) Competitive index was determined using mixed infection. bdlA is less competitive in an acute murine pneumonia infection model when co-inoculated with P. aeruginosa PAO1. Mice were inoculated intranasally with 1×107 cells (1∶1 ratio) of wild type PA01 and isogenic ΔbdlA mutant. Error bars indicate standard deviation. (D) Bacterial burden in the lung and (E) fold change in lung CFU/ml 24 hr post-infection compared to initial inoculum by P. aeruginosa PA14 and the isogenic ΔdipA mutant. Bars indicate average and median lung CFU/ml while vertical lines indicate the highest and lowest lung CFU/ml observed. A total of 5 mice were used in per study. The values were tested by means of a Fisher test. **, significantly different from wild type (PAO1 or PA14), P-value <0.001. Competitive mixed infection assays have been widely used to assess the fitness of individual P. aeruginosa mutants versus their parental strains during in vivo infection [70], [71]. Wild-type and ΔbdlA mutant bacteria were used to infect adult CD-1 mice (in groups of five) intranasally with 1×107 cells (1∶1 ratio). Following 16 hr, infected lungs were recovered for bacterial load determinations. Remarkably, the ΔbdlA mutant was only 2% as (or 45-fold less) competitive as its parental strain PAO1 (Fig. 6C). To further ensure that the effect on virulence was linked to the ability to disperse, we also examined the ability of the ΔdipA mutant to establish an acute infection. Compared to the parental strain, the isogenic ΔdipA mutant, in comparison, was unable to establish infection, as apparent by the bacterial load being reduced by 80-fold compared to the wild-type strain (Fig. 6D). Moreover, while the bacterial load of wild-type bacteria increased about 20-fold in the course of 24 hr compared to the initial inoculum titer, the ΔdipA mutant load was reduced 4-5-fold (Fig. 6). The results suggested BdlA and DipA, and thus, likely the process of dispersion to play an important role in virulence of P. aeruginosa during acute infection of mouse airways.
Inactivation of bdlA contributes to the persistence of P. aeruginosa in a murine model of chronic pneumonia
To determine the contribution of bdlA and dispersion to the ability of P. aeruginosa to persist at the site of infection in vivo, we next tested this mutant in a murine model of chronic pneumonia. Based on the ΔbdlA dispersion-deficient phenotype in vitro, we expected the P. aeruginosa ΔbdlA mutant to have an advantage in establishing biofilms and/or persistent infection compared to the wild type, due to its reduced virulence phenotype and the reduced number of cells released from the biofilm. We made use of a chronic infection model established by Cash et al. [72] except that we made use of a murine rather than a rat model. The model is based on the intratracheal administration of agarose beads impregnated with P. aeruginosa. This model was chosen because it allows the study of chronic infection, marked by the formation of persistent bacterial biofilm populations at the site of infection that can persist for up to 6 months and mimic biofilm-related infections [70], [71], [73], [74], [75].
Following 14 days of infection, the bacterial load in the lungs of mice infected with ΔbdlA was significantly higher (1.5-log increase) than that of P. aeruginosa PAO1 (Fig. 7). Moreover, compared to the initial inoculum (1.2×106 CFU), a less than a 100-fold reduction was noted for ΔbdlA while a more than 4000-fold reduction was detected between the initial inoculum and the bacterial burden 14 days post infection for the wild type. To ensure that the bacterial load in the lungs coincided with biofilm formation, we also tested P. aeruginosa overexpressing PA2133. PA2133 encodes a phosphodiesterase, and PAO1 strain overexpressing the PA2133 was previously shown by Hickman et al. [76] to be significantly impaired in biofilm formation in vitro, even after 72 h of incubation. Reduced bacterial burden correlated with reduced or impaired biofilm formation as indicated by the reduced detection of PAO1/pJN-PA2133 bacterial cells in the lung. Compared to PAO1/pJN-PA2133, the bacterial burden by P. aeruginosa PAO1 was increased by 1.5 logs, and ΔbdlA showed a 4 log increase (Fig. 7). Decreased burden correlated with a significant reduction in bacterial load compared to the original inoculum. The findings suggested that the bacterial burden at the site of infection depended on the ability to not only form biofilms but also to disperse.
Fig. 7. Impaired dispersion correlates with increased persistence as determined using a chronic murine pneumonia infection model. 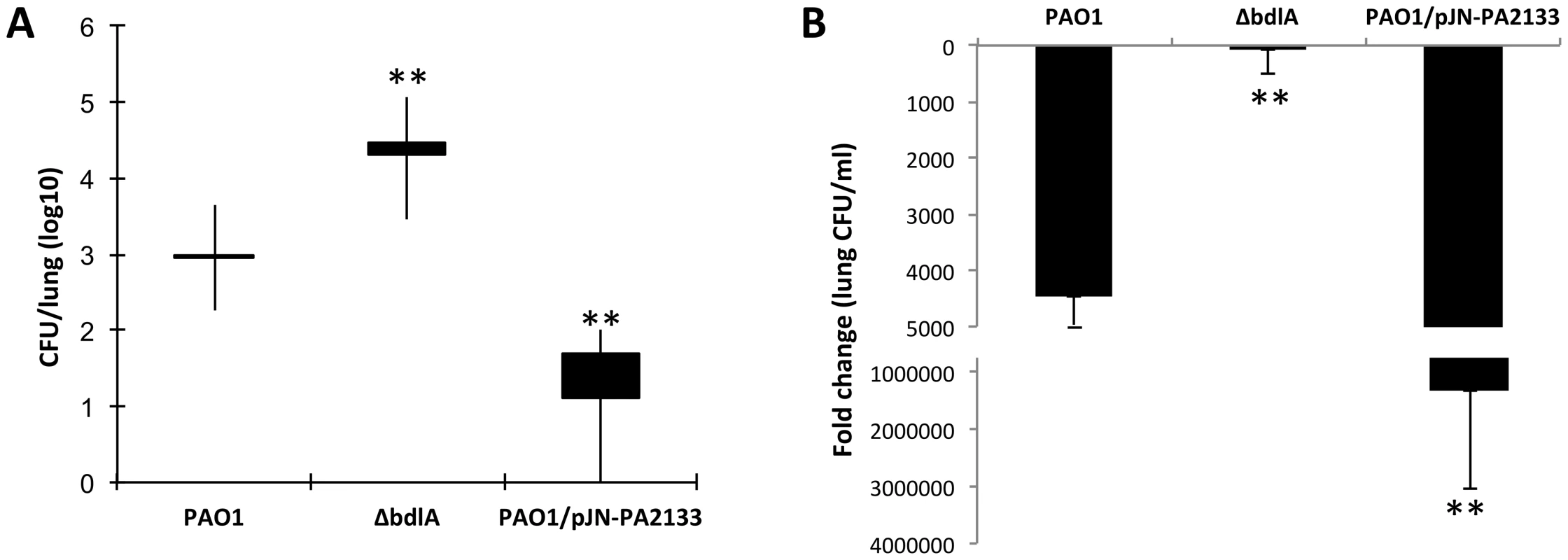
CD1 mice were inoculated intratracheal through oral lavage using feeding needle with 1.2×106 CFU of Pseudomonas strains. Lungs were harvested 14 days post-inoculation and CFU was determined. A total of 10 mice were used in per study. (A) Bacterial burden in the lung. Values presented are average, min, max, mean lung CFU/ml. (B) Fold change in lung CFU/ml 14 days post-infection compared to initial inoculum. Error bars indicate standard deviation. The values were tested by means of a Fisher test. **, significantly different from PAO1, P-value <0.001. Discussion
The opportunistic pathogen P. aeruginosa causes a variety of human diseases, ranging from acute and chronic persisting infections, fatal bacteremia in immunocompromised patients and superficial skin infections to chronic colonization of patients with CF and chronic obstructive pulmonary disease. This remarkable adaptability depends to a large extent on its ability to transition from a planktonic (unattached) to a biofilm mode of growth and to activate the expression of genes required for viability and growth in a particular environment, while simultaneously repressing genes that are unnecessary or even detrimental for survival in that particular niche. Our findings are in support of previous proteomics and genetic studies of P. aeruginosa [1], [2], [3], [8], [9], [77], indicating that planktonic and biofilm cells differ with respect to virulence gene expression with biofilms displaying a biofilm-specific virulence phenotype, and expressing virulence factors up to 30-fold less compared to planktonic cells. The observed down-regulation of virulence factors following biofilm formation likely serves as an additional adaptation for the bacterial communities to avoid recognition and targeting by the immune system. Accordingly, while releasing similar amounts of protein into the supernatant, biofilm cells were found to produce fewer cytotoxic and degradative proteins than planktonic cells (Figs. 1–2). Given that early infection and colonization has been suggested to require the production of a variety of cytotoxic and degradative proteins, some of which function after direct translocation into host cells, the observed difference in cytotoxic and degradative activities is in agreement with planktonic cells causing more considerable host cell damage [1], [78], [79]. In addition to the established acute-to-chronic virulence switch accompanying the motile-to-sessile mode of growth switch, we demonstrate here that dispersion in response to environmental cues is likely another contributing factor to the virulence phenotype of P. aeruginosa. However, while our findings support dispersion to contribute to virulence, dispersion is not simply a reversion of the motile-to-sessile switch. Instead, our findings suggest dispersion to represent a distinct virulence phenotype. For one, dispersed cells were characterized by significantly reduced transcript levels of virulence genes compared to both planktonic and biofilms cells. Moreover, while up to 10-fold higher protein concentrations were detected in supernatants of dispersed cells, the cytotoxic and degradative activity levels were found to be intermediary to those detected in supernatants of planktonic and biofilm cells. Our findings of the distinct virulence gene expression profile of dispersed cells are in agreement with our previous findings of biofilm cells in the dispersion stage and dispersed cells exhibiting a phenotype that is distinct from both planktonic and biofilm bacteria [36], [37], [56], [80].
P. aeruginosa dispersed cells not only display a virulence phenotype distinct from those of planktonic and biofilm cells, but also likely employ mechanisms enabling them to evade the immune response following the loss of protection associated with biofilm growth. This was apparent in the difference of cytotoxic and degradative enzymes present in supernatants compared to planktonic cells. It is likely that the difference represents a rapid initial response at the level of protein activity to liberate the dispersing cells from the enclosing matrix. While direct evidence of dispersion occurring in the lung environment is lacking, dispersion is widely documented in natural systems and is a basic property of biofilms in general. Moreover, it is likely that ability to degrade the biofilm matrix extends to an ability to degrade the surrounding mucus present in the CF lung, which is chemically similar to known biofilm matrix polymers. Additionally, while P. aeruginosa rarely escapes the lung environment, there are indications that dispersion takes place, as dispersed cells are more susceptible to immune function and antimicrobial treatment compared to biofilm cells [47], [56]. For example, Donaldson et al. [81] demonstrated that treatment with aerosolized hypertonic saline causes the vasculature to release water from the blood stream, thereby hydrating the thick mucus, correlated with biofilm bacteria being far more susceptible to the antibiotic amiloride compared to bacteria present in untreated mucus [81]. Differences in virulence factor gene expression may likewise indicate a dispersion-specific adaptation to temporarily avoid the host immune response, enabling dispersed cells to release themselves from the biofilm matrix but remain “immune-masked” until they have either reverted to a planktonic phenotype or have reattached. This was supported by our findings of both ΔbdlA and ΔdipA mutants, which are impaired in dispersion in vitro [45], [46], [56], [58], being attenuated in virulence while demonstrating heightened persistence in vivo (Figs. 5–7). The contribution of BdlA and DipA to virulence and persistence furthermore indicated that both proteins likely contribute to dispersion not only in vitro but also in vivo. The ΔbdlA and ΔdipA virulence phenotypes further suggested the presence of environmental cues in both the plant and the murine lung environment that contribute to dispersion-inducing conditions. This was apparent as the inability to disperse in vitro affected outcomes in both acute and chronic infection models, with dispersion enhancing the virulence of P. aeruginosa during an acute infection, but interfering with the ability to establish chronic infections. The manner in which BdlA, DipA, and dispersion contribute to persistence is distinct from biofilm formation, as inactivation of bdlA or dipA had the opposite effect to overexpression of the phosphodiesterase PA2133, which impairs biofilm formation [76]. Our findings thus likely suggest that in addition to biofilm biomass accumulation, dispersion may also be a contributing factor to P. aeruginosa persistence in the murine lung. This was further supported by the finding of a ΔbdlA mutant complemented with BdlA harboring the D14A, N23A, W60A, I109A, W182A substitutions, that were previously demonstrated to result in a null phenotype for dispersion, being attenuated in virulence. Additional support for a role of dispersion in P. aeruginosa pathogenicity was provided by ΔdipA and ΔrbdA mutants, that are impaired in dispersion in vitro [46], [55], being attenuated in virulence (Figs. 5–6). To our knowledge, this is the first report linking dispersion to virulence of P. aeruginosa in vivo.
It is of interest to note that the ΔbdlA virulence phenotype is similar to that of a ΔretS mutant in the sense that both promote transitions between the planktonic and the sessile lifestyles. While inactivation of retS promotes the motile-sessile transition [1], ΔbdlA mutants are impaired in the sessile-motile transition, with inactivation of both retS or bdlA resulting in decreased initial colonization. While the findings suggest that mutations favoring the biofilm lifestyle (by altering either one of the transitions) would be selected for in vivo, long-term selection in a host would likely drive populations towards a biofilm phenotype rather than favoring a non-dispersing phenotype. Instead, we expect the dispersion phenotype to be a short-term behavioral change in bacteria and to be associated specifically with acute phase infections.
In summary, we demonstrate for the first time that dispersed cells have a unique virulence phenotype, with dispersion and the reversion to the planktonic mode of growth contributing to virulence and persistence of P. aeruginosa in infections. While the ability to disperse in vitro in response to various exogenous cues contributes to virulence of P. aeruginosa in acute infections, dispersion instead reduces the ability of P. aeruginosa to persist at the site of infection. Our observations of dispersion thus reciprocally regulating acute virulence and persistent, chronic infections suggest that dispersion functions as a regulatory switch, mediating the global transition from initial colonization to chronic infections. Thus, our work establishes induced dispersion as not only an integral part of both acute and chronic infections, but also as a potential mechanism of infection control.
Materials and Methods
Ethics statement
The use of animals for this study was reviewed by the IACUC committee at the University of Cincinnati (UC) that is a centralized, campus wide animal care and use program under Laboratory Animal Medical Services (LAMS). LAMS is staffed by three full time veterinarians, six veterinary technicians plus animal care staff. One LAMS veterinarian; one veterinary technician and one LAMS husbandry supervisor is on call after hours. LAMS approved the animal care protocol and use protocol/permit/project license. The approved IACUC protocol number is 12-09-06-01 (“The Molecular Basis of Pseudomonas aeruginosa, Francisella novicida, and Staphylococcus aureus Virulence in Mammalian Hosts”). All animals were handled in strict accordance with good animal practice and animal keeping. UC has an Animal Welfare Assurance on file with the NIH-OLAW (Assurance Number A-3295-01, expires November 30, 2015). UC fully complies with the Guide for the Care and Use of Laboratory Animals (Guide), the Public Health Service Policy on the Humane Care and Use of Laboratory Animals (PHS Policy) and all U.S. Animal Welfare Act Regulations.
Bacterial strains, plasmids, media, and culture conditions
P. aeruginosa strain PAO1 and its isogenic mutant strain bdlA were used in this study. P. aeruginosa strain PA14 was used to validate the generality of the findings. All bacterial strains and plasmids used in this study are listed in Table S2. All planktonic strains were grown in Lennox Broth (LB, BD Biosciences) or minimal medium containing glutamate as the sole carbon source [82] in shake flasks at 220 rpm in the absence or presence of 0.1–1.0% arabinose. Escherichia coli cultures were grown in LB in the absence or presence of 1 mM Isopropyl β-D-1-thiogalactopyranoside (IPTG). Antibiotics for P. aeruginosa were used at the following concentrations: 50–75 µg/mL gentamicin and 200–250 µg/mL carbenicillin.
Planktonic growth conditions
All planktonic cells used in this study were obtained by reinoculating 0.5 ml of overnight grown cells into 50 ml minimal medium. To obtain exponential phase planktonic cells, bacteria were allowed to grow to an optical density (600 nm) of 0.4. To obtain stationary phase planktonic cells, bacteria were allowed to grow for 8 hr at which time they reached an optical density of 1.2. Under the conditions tested, onset of stationary phase was noted following 6–7 hours of growth. Planktonic cells growth to exponential and stationary phase were furthermore exposed to an additional 18 mM glutamate or the addition of 500 µM sodium nitroprusside (SNP, source of NO [51]) 30 min prior to harvesting the planktonic cells. The timing was chosen to mimic the exposure time of dispersed cells to glutamate and NO.
Biofilm formation and dispersion
Biofilms were grown in a continuous flow tube reactor system (1 m long size 14 silicone tubing, Masterflex, Cole Parmer, Inc.) at 22°C for up to 5 days to obtain proteins and RNA. For biofilm dispersion assays, biofilms were cultivated in once-through continuous flow tube reactor system composed of size 13 silicone tubing (Masterflex, Cole Parmer, Inc.) at 22°C for 5 days. After 5 days of biofilm growth, biofilm dispersion was induced by the sudden addition of glutamate (18 mM) to the growth medium as previously described [56]. Moreover, dispersion was induced by 500 µM sodium nitroprusside (SNP) which was used as a source of NO [51]. Dispersion was indicated by an increase in turbidity at 600 nm in the effluent from the silicone tubing.
Analysis of the proteins present in supernatants
Supernatants were obtained from P. aeruginosa PAO1 grown planktonically (exponential and stationary phase), as biofilms, and following dispersion in response to NO or changes in the nutrient glutamate concentration. While the volume of the supernatants varied depending on the growth conditions tested, all supernatants tested represented supernatants produced by 1×109 cells. Briefly, supernatants of cells grown planktonically to exponential and stationary phase and as biofilms were collected by centrifugation. Similarly, supernatants of dispersed cells and biofilms remaining attached to the surface upon induction of dispersion (remaining biofilms) were collected after induction of dispersion of 5 day-old P. aeruginosa biofilms were collected. The resulting supernatant was filter-sterilized, dialyzed, lyophilized, and subsequently resuspended in sterile water. The concentration of proteins present in supernatants was determined using the Bradford assay (Biorad). Cell pellets were collected and sonicated as previously described [37], and cell debris removed by centrifugation. The protein concentration of the resulting total cell extract was determined using the modified method of Lowry [83]. The resulting protein concentration was then used to calculate the number of cells with 1 ug of total protein being equivalent to 3.6*107 CFU [84].
Proteins present in supernatants were visualized by SDS/PAGE analysis using 12% SDS-gels and Coomassie staining. A total of 10 µg of cell extract was loaded onto the gel. Proteins of interest were excised from the gel, tryptic digested, and subsequently identified by LC-MS/MS essentially as previously described using a QStarXL mass spectrometer (Applied Biosystems) [85].
Determination of degradative activities present in the extracellular proteome
Supernatants were tested for the presence of hydrolytic enzymes using agar-plate based biochemical assays. To do so, wells having a 7 mm diameter were punched into the agar. A total of 10 µg of secreted proteins resuspended in a total of 100 µl of water was added to per well. The agar plates were subsequently incubated at 37°C for 18 hr before measurements of the halo surrounding the wells were taken. All measurements were normalized by subtracting the diameter of the well. Lipid hydrolysis was determined using Tributyrin HiVeg Agar Base (BD Bioscience) supplemented with 1% tributyrin while proteolysis was determined using milk agar containing 10% skim milk. Hemolysis was determined using trypticase soy agar supplemented with 5% sheep's blood. Psl degradation was determined using 1.5% agar containing 1% Psl. Psl polysaccharide was extracted using the rapid Psl protocol [86]. Each agar plate contained the Psl equivalent obtained from a Psl overexpressing P. aeruginosa having an OD of 25. Psl degradation was visualized following 24–48 hr of incubation using 1% iodine solution (Grams iodine, Thermo Scientific). DNAse activity was determined using 100 µg of supernatant protein and a colorimetric assay as previously described by Sinicropi et al. [87] using DNA-methyl green as a substrate. A total of 0.2 mg/ml salmon sperm DNA was used and DNA degradation monitored at 620 nm.
Quantitative reverse transcriptase PCR (qRT-PCR)
Isolation of mRNA and cDNA synthesis was carried out as previously described [84], [85], [88], [89]. qRT-PCR was performed using the Eppendorf Mastercycler ep realplex (Eppendorf AG, Hamburg, Germany) and the KAPA SYBR FAST qPCR Kit (KAPABIOSYSTEMS, Woburn, MA), with oligonucleotides listed in Table S3. mreB was used as a control. The stability of mreB levels were verified by 16S RNA abundance using primers HDA1/HDA2 [90]. Relative transcript quantitation was accomplished using the ep realplex software (Eppendorf AG) by first normalizing transcript abundance (based on Ct value) to mreB followed by determining transcript abundance ratios. Melting curve analyses were employed to verify specific single product amplification.
Virulence testing using Arabidopsis thaliana
The role of BdlA in virulence was assessed using the Arabidopsis thaliana infection model which provides a quantitative approach and permits the tracking of bacterial cell proliferation in planta [91]. Following two weeks of growth in 1/2MS (2.2 g/L Murashige and Skoog basal medium), plants were infected with P. aeruginosa PAO1 and its isogenic mutant strains at a final optical density of OD600 = 1.0 in 1/2MS and incubated for a period of 12 days at a 25/22°C, 16 hr-light/8 hr-dark cycle. All plants were inspected daily for signs of infection and/or death as evidenced by wilting, discoloration, and necrosis.
Model of acute pneumonia infection in CD-1 mice
P. aeruginosa strains PAO1 and ΔbdlA were grown to late stationary phase (∼ OD 3.0). Cells were harvested, and washed three times with 10 mM MgSO4. Bacteria were serial diluted in 10 mM MgSO4, mixed in a 1∶1 ratio, and approximately 1×107 cells were intranasally inoculated into the lungs of four CD-1 mice (Charles River, Boston, MA) previously anesthetized with isofluoran. For single infection studies, the mouse lungs were harvested at 24 hr post-infection, homogenized and serially diluted for bacterial burden determination. For competitive index studies, the mouse lungs were harvested at 16 hr post-infection. The bacterial titer is expressed as CFU/g of tissue.
Chronic lung infection model: Bacterial culture, immobilization of P. aeruginosa in agarose beads, and subsequent infection
A chronic infection model established by Cash et al. [72] was used to test whether the dispersion-deficient mutant strains are able to establish a chronic infection. The infection model by Cash was modified by using a murine model instead of a rat model. For the preparation of the agarose beads, P. aeruginosa wild type and mutant strain ΔbdlA, were grown at 32°C in a low phosphate succinate medium [72] to stationary phase and mixed in a 1/10 ratio with 2% agarose in phosphate-buffered saline (PBS, pH 7.4). The mixture was added to heavy mineral oil equilibrated at 55°C, stirred for 6 min at room temperature, and cooled for 10 min. Free bacteria were removed by washing with 0.5% and 0.25% deoxycholic acid sodium salt in PBS once and then in PBS alone three times. The beads were passively filtered through sterile 200 µm diameter nylon mesh and then verified for size (70 - to 150-µm diameter) and uniformity by microscope examination. An aliquot of beads were homogenized, serially diluted, and plated on LB agar plates to enumerate colony-forming units (CFU). A 100 µl inoculum containing 1×106 CFU of viable P. aeruginosa entrapped in agarose beads was then introduced into the lungs of adult CD-1 mice (six week old, groups of 10) via the trachea with nonsurgical methods by a 21-gauge blunt-end needle to the back of the tongue above the tracheal opening. Successful delivery of the beads to the lungs was manifested by choking of the mouse immediately after instillation followed by rapid breathing, and was confirmed by harvesting lungs three minutes post-inoculation and CFU determination of lung homogenates. Mice were sacrificed by asphyxiation in a precharged CO2 chamber, or by transection of the abdominal aorta after an overdose of pentobarbital (2.0 ml per kg body weight). The bacterial load was expressed as CFU/g of tissue.
Statistical analysis
A Student's t-test was performed for pair-wise comparisons of groups, and multivariant analyses were performed using a 1-Way ANOVA followed by a posteriori test using Sigma Stat software. For the animal studies, Fisher exact test was used for statistical analysis (http://www.quantitativeskills.com/sisa/statistics/fishrhlp.htm).
Supporting Information
Zdroje
1. GoodmanAL, KulasekaraB, RietschA, BoydD, SmithRS, et al. (2004) A signaling network reciprocally regulates genes associated with acute infection and chronic persistence in Pseudomonas aeruginosa. Dev Cell 7 : 745–754.
2. VentreI, GoodmanAL, Vallet-GelyI, VasseurP, SosciaC, et al. (2006) Multiple sensors control reciprocal expression of Pseudomonas aeruginosa regulatory RNA and virulence genes. Proc Natl Acad Sci 103 : 171–176.
3. LaskowskiMA, KazmierczakBI (2006) Mutational Analysis of RetS, an Unusual Sensor Kinase-Response Regulator Hybrid Required for Pseudomonas aeruginosa Virulence. Infect Immun 74 : 4462–4473.
4. PetrovaOE, SauerK (2010) The novel two-component regulatory system BfiSR regulates biofilm development by controlling the small RNA rsmZ through CafA. J Bacteriol 192 : 5275–5288.
5. PetrovaOE, SauerK (2011) SagS contributes to the motile-sessile switch and acts in concert with BfiSR to enable Pseudomonas aeruginosa biofilm formation J Bacteriol. 193 : 6614–6628.
6. EmoriTG, GaynesRP (1993) An overview of nosocomial infections, including the role of the microbiology laboratory. Clin Microbiol Rev 6 : 428–442.
7. CostertonJW, StewartPS, GreenbergEP (1999) Bacterial biofilms: a common cause of persistent infections. Science 284 : 1318–1322.
8. KuchmaSL, ConnollyJP, O'TooleGA (2005) A three-component regulatory system regulates biofilm maturation and type III secretion in Pseudomonas aeruginosa. J Bacteriol 187 : 1441–1454.
9. YahrTL, GreenbergEP (2004) The Genetic Basis for the Commitment to Chronic versus Acute Infection in Pseudomonas aeruginosa. Molecular Cell 16 : 497–498.
10. SuS, HassettDJ (2012) Anaerobic Pseudomonas aeruginosa and other obligately anaerobic bacterial biofilms growing in the thick airway mucus of chronically infected cystic fibrosis patients: an emerging paradigm or “Old Hat”? Expert Opin Ther Targets 16 : 859–873.
11. GilliganPH (1991) Microbiology of airway disease in patients with cystic fibrosis. Clin Microbiol Rev 4 : 35–51.
12. GovanJR, DereticV (1996) Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol Rev 60 : 539–574.
13. DonlanRM, CostertonJW (2002) Biofilms: Survival Mechanisms of Clinically Relevant Microorganisms. Clin Microbiol Rev 15 : 167–193.
14. WagnerV, IglewskiB (2008) P. aeruginosa biofilms in CF Infection. Clin Rev Allergy Immunol 35 : 124–134.
15. PruittJBA, McManusAT, KimSH, GoodwinCW (1998) Burn Wound Infections: Current Status. World Journal of Surgery 22 : 135–145.
16. FleiszigSM, EvansDJ (2002) The pathogenesis of bacterial keratitis: studies with Pseudomonas aeruginosa. Clin Exp Optom 85 : 271–278.
17. TakeyamaK, KunishimaY, MatsukawaM, TakahashiS, HiroseT, et al. (2002) Multidrug-resistant Pseudomonas aeruginosa isolated from the urine of patients with urinary tract infection. J Infect Chemother 8 : 58–63.
18. ReinhardtA, KohlerT, WoodP, RohnerP, DumasJ-L, et al. (2007) Development and Persistence of Antimicrobial Resistance in Pseudomonas aeruginosa: a Longitudinal Observation in Mechanically Ventilated Patients. Antimicrob Agents Chemother 51 : 1341–1350.
19. CostertonJW, LewandowskiZ, CaldwellDE, KorberDR, Lappin-ScottHM (1995) Microbial biofilms. Annu Rev Microbiol 49 : 711–745.
20. GooderhamWJ, HancockREW (2009) Regulation of virulence and antibiotic resistance by two-component regulatory systems in Pseudomonas aeruginosa. FEMS Microbiol Rev 33 : 279–294.
21. PetrovaOE, SchurrJR, SchurrMJ, SauerK (2011) The novel Pseudomonas aeruginosa two-component regulator BfmR controls bacteriophage-mediated lysis and DNA release during biofilm development through PhdA. Mol Microbiol 81 : 767–783.
22. PetrovaOE, SchurrJR, SchurrMJ, SauerK (2012) Microcolony formation by the opportunistic pathogen Pseudomonas aeruginosa requires pyruvate and pyruvate fermentation. Mol Microbiol 86 : 819–835.
23. LaskowskiMA, OsbornE, KazmierczakBI (2004) A novel sensor kinase–response regulator hybrid regulates type III secretion and is required for virulence in Pseudomonas aeruginosa. Mol Microbiol 54 : 1090–1103.
24. ZolfagharI, AngusAA, KangPJ, ToA, EvansDJ, et al. (2005) Mutation of retS, encoding a putative hybrid two-component regulatory protein in Pseudomonas aeruginosa, attenuates multiple virulence mechanisms. Microbes Infect 7 : 1305–1316.
25. HolderIA, NeelyAN, FrankDW (2001) Type III secretion/intoxication system important in virulence of Pseudomonas aeruginosa infections in burns. Burns 27 : 129–130.
26. SmithRS, WolfgangMC, LoryS (2004) An adenylate cyclase-controlled signaling network regulates Pseudomonas aeruginosa virulence in a mouse model of acute pneumonia. Infect Immun 72 : 1677–1684.
27. von GotzF, HausslerS, JordanD, SaravanamuthuSS, WehmhonerD, et al. (2004) Expression analysis of a highly adherent and cytotoxic small colony variant of Pseudomonas aeruginosa isolated from a lung of a patient with cystic fibrosis. J Bacteriol 186 : 3837–3847.
28. MikkelsenH, BondNJ, SkindersoeME, GivskovM, LilleyKS, et al. (2009) Biofilms and type III secretion are not mutually exclusive in Pseudomonas aeruginosa. Microbiology 155 : 687–798.
29. GallagherLA, ManoilC (2001) Pseudomonas aeruginosa PAO1 Kills Caenorhabditis elegans by Cyanide Poisoning. J Bacteriol 183 : 6207–6214.
30. CartersonAJ, MoriciLA, JacksonDW, FriskA, LizewskiSE, et al. (2004) The transcriptional regulator AlgR controls cyanide production in Pseudomonas aeruginosa. J Bacteriol 186 : 6837–6844.
31. FoldersJ, AlgraJ, RoelofsMS, van LoonLC, TommassenJ, et al. (2001) Characterization of Pseudomonas aeruginosa Chitinase, a Gradually Secreted Protein. J Bacteriol 183 : 7044–7052.
32. ElsheikhLE, KroneviT, WretlindB, AbaasS, IglewskiBH (1987) Assessment of elastase as a Pseudomonas aeruginosa virulence factor in experimental lung infection in mink. Veterinary Microbiology 13 : 281–289.
33. Mahajan-MiklosS, TanM-W, RahmeLG, AusubelFM (1999) Molecular mechanisms of bacterial virulence elucidated using a Pseudomonas aeruginosa-Caenorhabditis elegans pathogenesis model. Cell 96 : 47–56.
34. McClureCD, SchillerNL (1996) Inhibition of Macrophage Phagocytosis by Pseudomonas aeruginosa Rhamnolipids In Vitro and In Vivo. Current Microbiology 33 : 109–117.
35. JensenPØ, BjarnsholtT, PhippsR, RasmussenTB, CalumH, et al. (2007) Rapid necrotic killing of polymorphonuclear leukocytes is caused by quorum-sensing-controlled production of rhamnolipid by Pseudomonas aeruginosa. Microbiology 153 : 1329–1338.
36. SauerK, CullenMC, RickardAH, ZeefLAH, DaviesDG, et al. (2004) Characterization of nutrient-induced dispersion in Pseudomonas aeruginosa PAO1 biofilm. J Bacteriol 186 : 7312–7326.
37. SauerK, CamperAK, EhrlichGD, CostertonJW, DaviesDG (2002) Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J Bacteriol 184 : 1140–1154.
38. GjermansenM, RagasP, SternbergC, MolinS, Tolker-NielsenT (2005) Characterization of starvation-induced dispersion in Pseudomonas putida biofilms. Environ Microbiol 7 : 894–904.
39. KaplanJB, RagunathC, VelliyagounderK, FineDH, RamasubbuN (2004) Enzymatic detachment of Staphylococcus epidermidis biofilms. Antimicrob Agents Chemother 48 : 2633–2636.
40. LeeSF, LiYH, BowdenGH (1996) Detachment of Streptococcus mutans biofilm cells by an endogenous enzymatic activity. Infection and Immunity 64 : 1035–1038.
41. GjermansenM, NilssonM, YangL, Tolker-NielsenT (2010) Characterization of starvation-induced dispersion in Pseudomonas putida biofilms: genetic elements and molecular mechanisms. Mol Microbiol 75 : 815–826.
42. HinsaSM, Espinosa-UrgelM, RamosJL, O'TooleGA (2003) Transition from reversible to irreversible attachment during biofilm formation by Pseudomonas fluorescens WCS365 requires an ABC transporter and a large secreted protein. Mol Microbiol 49 : 905–918.
43. MondsRD, NewellPD, GrossRH, O'TooleGA (2007) Phosphate-dependent modulation of c-di-GMP levels regulates Pseudomonas fluorescens Pf0-1 biofilm formation by controlling secretion of the adhesin LapA. Mol Microbiol 63 : 656–679.
44. WebbJS, ThompsonLS, JamesS, CharltonT, Tolker-NielsenT, et al. (2003) Cell death in Pseudomonas aeruginosa biofilm development. J Bacteriol 185 : 4585–4592.
45. PetrovaOE, SauerK (2012) Dispersion by Pseudomonas aeruginosa requires an unusual posttranslational modification of BdlA. Proc National Acad Sci 109 : 16690–16695.
46. Basu RoyA, PetrovaOE, SauerK (2012) The phosphodiesterase DipA (PA5017) is essential for Pseudomonas aeruginosa biofilm dispersion. J Bacteriol 194 : 2904–2915.
47. DaviesDG, MarquesCNH (2009) A fatty acid messenger is responsible for inducing dispersion in microbial biofilms. J Bacteriol 191 : 1393–1403.
48. LiY, HeineS, EntianM, SauerK, Frankenberg-DinkelN (2013) NO-induced biofilm dispersion in Pseudomonas aeruginosa is mediated by a MHYT-domain coupled phosphodiesterase. J Bacteriol 195 : 3531–3542.
49. ThormannKM, SavilleRM, ShuklaS, SpormannAM (2005) Induction of rapid detachment in Shewanella oneidensis MR-1 biofilms. J Bacteriol 187 : 1014–1021.
50. ApplegateDH, BryersJD (1991) Effects on carbon and oxygen limitations and calcium concentrations on biofilm removal processes Biotechnol Bioeng. 37 : 17–25.
51. BarraudN, HassettDJ, HwangS-H, RiceSA, KjellebergS, et al. (2006) Involvement of nitric oxide in biofilm dispersal of Pseudomonas aeruginosa. J Bacteriol 188 : 7344–7353.
52. CotterPA, StibitzS (2007) c-di-GMP-mediated regulation of virulence and biofilm formation. Curr Opin Microbiol 10 : 17–23.
53. D'ArgenioDA, MillerSI (2004) Cyclic di-GMP as a bacterial second messenger. Microbiology 150 : 2497–2502.
54. ThormannKM, DuttlerS, SavilleRM, HyodoM, ShuklaS, et al. (2006) Control of formation and cellular detachment from Shewanella oneidensis MR-1 biofilms by cyclic di-GMP. J Bacteriol 188 : 2681–2691.
55. AnS, WuJe, ZhangL-H (2010) Modulation of Pseudomonas aeruginosa biofilm dispersal by a cyclic-di-GMP phosphodiesterase with a putative hypoxia-sensing fomain. Appl Environ Microbiol 76 : 8160–8173.
56. MorganR, KohnS, HwangS-H, HassettDJ, SauerK (2006) BdlA, a chemotaxis regulator essential for biofilm dispersion in Pseudomonas aeruginosa. J Bacteriol 188 : 7335–7343.
57. BarraudN, SchleheckD, KlebensbergerJ, WebbJS, HassettDJ, et al. (2009) Nitric oxide signaling in Pseudomonas aeruginosa biofilms mediates phosphodiesterase activity, decreased cyclic di-GMP levels, and enhanced dispersal. J Bacteriol 191 : 7333–7342.
58. PetrovaOE, SauerK (2012) PAS domain residues and prosthetic group involved in BdlA-dependent dispersion response by Pseudomonas aeruginosa biofilms. J Bacteriol 194 : 5817–5828.
59. DowJM, FouhyY, LuceyJF, RyanRP (2006) The HD-GYP domain, cyclic di-GMP signaling, and bacterial virulence to plants. Mol Plant Microbe Interact 19 : 1378–1384.
60. KulasekaraH, LeeV, BrencicA, LiberatiN, UrbachJ, et al. (2006) Analysis of Pseudomonas aeruginosa diguanylate cyclases and phosphodiesterases reveals a role for bis-(3′-5′)-cyclic-GMP in virulence. Proc Natl Acad Sci 103 : 2839–2844.
61. ChristensenLD, van GennipM, RybtkeMT, WuH, ChiangW-C, et al. (2013) Clearance of Pseudomonas aeruginosa foreign-body biofilm infections through reduction of the cyclic di-GMP level in the bacteria. Infect Immun 81 : 2705–2713.
62. FlemmingH-C, NeuTR, WozniakDJ (2007) The EPS matrix: the “house of biofilm cells”. J Bacteriol 189 : 7945–7947.
63. FlemmingHC, WingenderJ (2010) The biofilm matrix. Nat Rev Microbiol 8 : 623–633.
64. RyderC, ByrdM, WozniakDJ (2007) Role of polysaccharides in Pseudomonas aeruginosa biofilm development. Curr Opin Microbiol 10 : 644–648.
65. ArmstrongS, YatesSP, MerrillAR (2002) Insight into the Catalytic Mechanism of Pseudomonas aeruginosa Exotoxin A. Journal of Biological Chemistry 277 : 46669–46675.
66. WagnerVE, BushnellD, PassadorL, BrooksAI, IglewskiBH (2003) Microarray Analysis of Pseudomonas aeruginosa Quorum-Sensing Regulons: Effects of Growth Phase and Environment. J Bacteriol 185 : 2080–2095.
67. HentzerM, WuH, AndersenJB, RiedelK, RasmussenTB, et al. (2003) Attenuation of Pseudomonas aeruginosa virulence by quorum sensing inhibitors. EMBO J 22 : 3803–3815.
68. RahmeLG, TanM-W, LeL, WongSM, TompkinsRG, et al. (1997) Use of model plant hosts to identify Pseudomonas aeruginosa virulence factors. Proc Natl Acad Sci 94 : 13245–13250.
69. RahmeLG, StevensEJ, WolfortSF, ShaoJ, TompkinsRG, et al. (1995) Common virulence factors for bacterial pathogenicity in plants and animals. Science 268 : 1899–1902.
70. LauGW, RanH, KongF, HassettDJ, MavrodiD (2004) Pseudomonas aeruginosa Pyocyanin Is Critical for Lung Infection in Mice. Infect Immun 72 : 4275–4278.
71. YoonSS, CoakleyR, LauGW, LymarSV, GastonB, et al. (2006) Anaerobic killing of mucoid Pseudomonas aeruginosa by acidified nitrite derivatives under cystic fibrosis airway conditions. J Clin Invest 116 : 436–446.
72. CashHD, WoodsDE, McColloughB, W.GJ, BassJA (1979) A rat model of chronic respiratory infection with Pseudomonas aeruginosa. Am Rev Respir Dis 119 : 453–459.
73. LauGW, BritiganBE, HassettDJ (2005) Pseudomonas aeruginosa OxyR Is Required for Full Virulence in Rodent and Insect Models of Infection and for Resistance to Human Neutrophils. Infect Immun 73 : 2550–2553.
74. Garcia-MedinaR, DunneWM, SinghPK, BrodySL (2005) Pseudomonas aeruginosa acquires biofilm-like properties within airway epithelial cells. Infect Immun 73 : 8298–8305.
75. WoodsDE, CantinA, CooleyJ, KenneyDM, Remold-O'DonnellE (2005) Aerosol treatment with MNEI suppresses bacterial proliferation in a model of chronic Pseudomonas aeruginosa lung infection. Pediatr Pulmonol 39 : 141–149.
76. HickmanJW, TifreaDF, HarwoodCS (2005) A chemosensory system that regulates biofilm formation through modulation of cyclic diguanylate levels. Proc Natl Acad Sci 102 : 14422–14427.
77. CostertonJW, StewartPS, GreenbergEP (1999) Bacterial Biofilms: A Common Cause of Persistent Infections. Science 284 : 1318–1322.
78. PearsonJP, FeldmanM, IglewskiBH, PrinceA (2000) Pseudomonas aeruginosa cell-to-cell signaling is required for virulence in a model of acute pulmonary infection. Infect Immun 68 : 4331–4334.
79. TangH, KaysM, PrinceA (1995) Role of Pseudomonas aeruginosa pili in acute pulmonary infection. Infect Immun 63 : 1278–1285.
80. StoodleyP, SauerK, DaviesDG, CostertonJW (2002) Biofilms as complex differentiated communities. Annu Rev Microbiol 56 : 187–209.
81. DonaldsonSH, BennettWD, ZemanKL, KnowlesMR, TarranR, et al. (2006) Mucus clearance and lung function in cystic fibrosis with hypertonic saline. N Engl J Med 354 : 241–250.
82. SauerK, CamperAK (2001) Characterization of phenotypic changes in Pseudomonas putida in response to surface-associated growth. J Bacteriol 183 : 6579–6589.
83. PetersonGL (1977) A simplification of the protein assay method of Lowry, et al. which is more generally applicable. Anal Biochem 83 : 346–356.
84. Southey-PilligCJ, DaviesDG, SauerK (2005) Characterization of temporal protein production in Pseudomonas aeruginosa biofilms. J Bacteriol 187 : 8114–8126.
85. PetrovaOE, SauerK (2009) A novel signaling network essential for regulating Pseudomonas aeruginosa biofilm development. PLoS Pathog 5: e1000668.
86. ByrdMS, SadovskayaI, VinogradovE, LuH, SprinkleAB, et al. (2009) Genetic and biochemical analyses of the Pseudomonas aeruginosa Psl exopolysaccharide reveal overlapping roles for polysaccharide synthesis enzymes in Psl and LPS production. Mol Microbiol 73 : 622–638.
87. SinicropiD, BakerDL, PrinceWS, ShifferK, ShakS (1994) Colorimetric determination of DNase I activity with a DNA-methyl green substrate. Anal Biochem 222 : 351–358.
88. AllegrucciM, SauerK (2007) Characterization of colony morphology variants isolated from Streptococcus pneumoniae biofilms. J Bacteriol 189 : 2030–2038.
89. AllegrucciM, SauerK (2008) Formation of Streptococcus pneumoniae non-phase-variable colony variants is due to increased mutation frequency present under biofilm growth conditions. J Bacteriol 190 : 6330–6339.
90. McBainAJ, BartoloRG, CatrenichCE, CharbonneauD, LedderRG, et al. (2003) Microbial characterization of biofilms in domestic drains and the establishment of stable biofilm microcosms. Appl Environ Microbiol 69 : 177–185.
91. StarkeyM, RahmeLG (2009) Modeling Pseudomonas aeruginosa pathogenesis in plant hosts. Nat Protocols 4 : 117–124.
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek Recruitment of RED-SMU1 Complex by Influenza A Virus RNA Polymerase to Control Viral mRNA SplicingČlánek Systematic Phenotyping of a Large-Scale Deletion Collection Reveals Novel Antifungal Tolerance GenesČlánek The Contribution of Social Behaviour to the Transmission of Influenza A in a Human Population
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2014 Číslo 6- Jak souvisí postcovidový syndrom s poškozením mozku?
- Měli bychom postcovidový syndrom léčit antidepresivy?
- Farmakovigilanční studie perorálních antivirotik indikovaných v léčbě COVID-19
- 10 bodů k očkování proti COVID-19: stanovisko České společnosti alergologie a klinické imunologie ČLS JEP
-
Všechny články tohoto čísla
- Fungal Nail Infections (Onychomycosis): A Never-Ending Story?
- BdlA, DipA and Induced Dispersion Contribute to Acute Virulence and Chronic Persistence of
- Morphotype Transition and Sexual Reproduction Are Genetically Associated in a Ubiquitous Environmental Pathogen
- A Nucleic-Acid Hydrolyzing Single Chain Antibody Confers Resistance to DNA Virus Infection in HeLa Cells and C57BL/6 Mice
- HopW1 from Disrupts the Actin Cytoskeleton to Promote Virulence in Arabidopsis
- Ly6C Monocytes Become Alternatively Activated Macrophages in Schistosome Granulomas with Help from CD4+ Cells
- Recruitment of RED-SMU1 Complex by Influenza A Virus RNA Polymerase to Control Viral mRNA Splicing
- Contribution of Specific Residues of the β-Solenoid Fold to HET-s Prion Function, Amyloid Structure and Stability
- Antibody Responses to : Role in Pathogenesis and Diagnosis of Encephalitis?
- Discovery of a Novel Compound with Anti-Venezuelan Equine Encephalitis Virus Activity That Targets the Nonstructural Protein 2
- Activation of Focal Adhesion Kinase by Suppresses Autophagy via an Akt/mTOR Signaling Pathway and Promotes Bacterial Survival in Macrophages
- Crossing the Interspecies Barrier: Opening the Door to Zoonotic Pathogens
- Catching Fire: , Macrophages, and Pyroptosis
- IscR Is Essential for Type III Secretion and Virulence
- Selective Chemical Inhibition of Quorum Sensing in Promotes Host Defense with Minimal Impact on Resistance
- The Glycosylated Rv1860 Protein of Inhibits Dendritic Cell Mediated TH1 and TH17 Polarization of T Cells and Abrogates Protective Immunity Conferred by BCG
- A Genome-Wide Tethering Screen Reveals Novel Potential Post-Transcriptional Regulators in
- Structural Insights into SraP-Mediated Adhesion to Host Cells
- Human IGF1 Regulates Midgut Oxidative Stress and Epithelial Homeostasis to Balance Lifespan and resistance in
- Cycling Empirical Antibiotic Therapy in Hospitals: Meta-Analysis and Models
- Rab11 Regulates Trafficking of -sialidase to the Plasma Membrane through the Contractile Vacuole Complex of
- Mitogen and Stress Activated Kinases Act Co-operatively with CREB during the Induction of Human Cytomegalovirus Immediate-Early Gene Expression from Latency
- Profilin Promotes Recruitment of Ly6C CCR2 Inflammatory Monocytes That Can Confer Resistance to Bacterial Infection
- A Central Role for Carbon-Overflow Pathways in the Modulation of Bacterial Cell Death
- An Invertebrate Warburg Effect: A Shrimp Virus Achieves Successful Replication by Altering the Host Metabolome via the PI3K-Akt-mTOR Pathway
- The Highly Conserved Bacterial RNase YbeY Is Essential in , Playing a Critical Role in Virulence, Stress Regulation, and RNA Processing
- A Virulent Strain of Deformed Wing Virus (DWV) of Honeybees () Prevails after -Mediated, or , Transmission
- Systematic Phenotyping of a Large-Scale Deletion Collection Reveals Novel Antifungal Tolerance Genes
- Ubiquitin-Mediated Response to Microsporidia and Virus Infection in
- Preclinical Detection of Variant CJD and BSE Prions in Blood
- Toll-Like Receptor 8 Agonist and Bacteria Trigger Potent Activation of Innate Immune Cells in Human Liver
- Progressive Proximal-to-Distal Reduction in Expression of the Tight Junction Complex in Colonic Epithelium of Virally-Suppressed HIV+ Individuals
- The Triggering Receptor Expressed on Myeloid Cells 2 Inhibits Complement Component 1q Effector Mechanisms and Exerts Detrimental Effects during Pneumococcal Pneumonia
- Differential Activation of Acid Sphingomyelinase and Ceramide Release Determines Invasiveness of into Brain Endothelial Cells
- Forward Genetic Screening Identifies a Small Molecule That Blocks Growth by Inhibiting Both Host- and Parasite-Encoded Kinases
- Defining Immune Engagement Thresholds for Control of Virus-Driven Lymphoproliferation
- Growth Factor and Th2 Cytokine Signaling Pathways Converge at STAT6 to Promote Arginase Expression in Progressive Experimental Visceral Leishmaniasis
- Multimeric Assembly of Host-Pathogen Adhesion Complexes Involved in Apicomplexan Invasion
- Biogenesis of Influenza A Virus Hemagglutinin Cross-Protective Stem Epitopes
- Adequate Th2-Type Response Associates with Restricted Bacterial Growth in Latent Mycobacterial Infection of Zebrafish
- Protective Efficacy of Passive Immunization with Monoclonal Antibodies in Animal Models of H5N1 Highly Pathogenic Avian Influenza Virus Infection
- Fructose-Asparagine Is a Primary Nutrient during Growth of in the Inflamed Intestine
- The Calcium-Dependent Protein Kinase 3 of Influences Basal Calcium Levels and Functions beyond Egress as Revealed by Quantitative Phosphoproteome Analysis
- A Translocated Effector Required for Dissemination from Derma to Blood Safeguards Migratory Host Cells from Damage by Co-translocated Effectors
- Functional Characterization of a Novel Family of Acetylcholine-Gated Chloride Channels in
- Both α2,3- and α2,6-Linked Sialic Acids on O-Linked Glycoproteins Act as Functional Receptors for Porcine Sapovirus
- The Contribution of Social Behaviour to the Transmission of Influenza A in a Human Population
- MicroRNA-146a Provides Feedback Regulation of Lyme Arthritis but Not Carditis during Infection with
- Recombination in Enteroviruses Is a Biphasic Replicative Process Involving the Generation of Greater-than Genome Length ‘Imprecise’ Intermediates
- Cytoplasmic Viral RNA-Dependent RNA Polymerase Disrupts the Intracellular Splicing Machinery by Entering the Nucleus and Interfering with Prp8
- and Are Associated with Murine Susceptibility to Infection and Human Sepsis
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Fungal Nail Infections (Onychomycosis): A Never-Ending Story?
- Profilin Promotes Recruitment of Ly6C CCR2 Inflammatory Monocytes That Can Confer Resistance to Bacterial Infection
- Contribution of Specific Residues of the β-Solenoid Fold to HET-s Prion Function, Amyloid Structure and Stability
- The Highly Conserved Bacterial RNase YbeY Is Essential in , Playing a Critical Role in Virulence, Stress Regulation, and RNA Processing
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



