-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaThe Role of -Mediated Epigenetic Silencing in the Population Dynamics of Transposable Elements in
The piwi-interacting RNAs (piRNAs) are small RNAs that can suppress the expression of selfish transposable elements (TEs) in many animal genomes. One mechanism by which piRNAs silence TEs is through the formation of heterochromatin, which is condensed chromatin and generally associated with repressed gene expression. Several functional studies have demonstrated that piRNA-mediated heterochromatin of TEs can spread to adjacent genes. We hypothesized that this spread of TE-induced heterochromatin influences the function of adjacent genes, ultimately resulting in selection against individual TEs. Consistent with our hypothesis, we found that sequences and genes adjacent to TEs are enriched in heterochromatic marks. We determine that this TE-induced variation in epigenetic status is probably piRNA-dependent and that this change in chromatin state influences the expression levels of adjacent genes. Importantly, TEs that lead to higher heterochromatin enrichment of adjacent genes are more strongly selected against, demonstrating the evolutionary consequences of TE-induced epigenetic silencing. In contrast to previously studied deleterious impacts of TEs, which depend on TEs’ physical disruptions of DNAs, our proposed functional effect of TEs is mediated through their epigenetic influence. Our study suggests that the piRNA-dependent epigenetic impact of TEs may play an important role in the evolutionary dynamics of TEs.
Published in the journal: . PLoS Genet 11(6): e32767. doi:10.1371/journal.pgen.1005269
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1005269Summary
The piwi-interacting RNAs (piRNAs) are small RNAs that can suppress the expression of selfish transposable elements (TEs) in many animal genomes. One mechanism by which piRNAs silence TEs is through the formation of heterochromatin, which is condensed chromatin and generally associated with repressed gene expression. Several functional studies have demonstrated that piRNA-mediated heterochromatin of TEs can spread to adjacent genes. We hypothesized that this spread of TE-induced heterochromatin influences the function of adjacent genes, ultimately resulting in selection against individual TEs. Consistent with our hypothesis, we found that sequences and genes adjacent to TEs are enriched in heterochromatic marks. We determine that this TE-induced variation in epigenetic status is probably piRNA-dependent and that this change in chromatin state influences the expression levels of adjacent genes. Importantly, TEs that lead to higher heterochromatin enrichment of adjacent genes are more strongly selected against, demonstrating the evolutionary consequences of TE-induced epigenetic silencing. In contrast to previously studied deleterious impacts of TEs, which depend on TEs’ physical disruptions of DNAs, our proposed functional effect of TEs is mediated through their epigenetic influence. Our study suggests that the piRNA-dependent epigenetic impact of TEs may play an important role in the evolutionary dynamics of TEs.
Introduction
Transposable elements (TEs) are genetic elements that increase their copy number in the host genome by copying themselves to new genomic locations. Despite reported incidences of potentially adaptive TEs [1–6], the majority of TEs are considered deleterious to their host and widely viewed as “genomic parasites”. TE insertions can disrupt sequence and function of host genetic elements [7]. Additionally, ectopic recombination between nonhomologous TE copies leads to potentially deleterious chromosomal rearrangements [8–12]. Although deleterious impacts of TE insertions are broadly appreciated, our picture for the functional and evolutionary mechanisms of TEs containment in natural populations is still incomplete.
Because of the replicative nature of most transposition mechanisms of TEs, it is critical to understand the evolutionary forces counterbalancing the constant increase of TEs. Theoretical models demonstrate that the TE copy number in an outbreeding population reaches an equilibrium when the increase in TE copy number via transposition is counterbalanced by the removal of TEs [13,14]. Regulating the transposition rate such that it equals the excision rate of TEs is one possible mechanism of achieving equilibrium. This, however, is not supported by empirical observations (reviewed in [15]). In addition, theoretical studies have shown that transposition rate has only a minimal impact on the predicted frequency spectrum of TEs [13,14]. Regulation of transposition rate is unlikely to account for the observed low population frequency of most TEs in outbreeding populations such as Drosophila ([4,16–19], reviewed in [15]). Instead, both theoretical analyses and empirical results support that selection against deleterious impacts of TEs plays a primary role in the evolutionary dynamics of TEs [15,20,21]. Without regulated TE transposition, selection alone can result in an equilibrium of TE copy number and lead to the skewed frequency spectrum of TEs in natural populations.
Most previous work on the deleterious effects of TEs has centered around consequences of TE-mediated physical disruption of genomic DNA, such as TEs’ insertion into functional elements and ectopic recombination between nonhomologous TEs. The exploration of the potential functional and evolutionary consequences of TEs’ epigenetic impacts have been limited [22]. In Drosophila, a class of small interfering RNA, piwi-interacting RNAs (piRNAs), are enriched for TE sequences and post-transcriptionally regulate TE transposition in the germline [23–26]. In addition, piRNAs can suppress TE transposition by inducing heterochromatic formation of euchromatic TEs in the germline [27–31] as well as in larval and adult somatic tissues [32,33]. Constitutive pericentric and telomeric heterochromatin “spreads” and usually results in stochastic silencing of the nearby genes, a phenomenon known as “Position-effect variegation” (PEV [34], reviewed in [35–37]). Similarly, piRNA-mediated heterochromatin of euchromatic TEs was shown to spread into adjacent host genes by using reporter constructs [32]. The resulting perturbation of host gene expression due to the spread of heterochromatin from adjacent TEs probably has deleterious consequences. These TEs are expected to be removed by selection even if they do not generate physical disruptions of the DNA, leading to a skewed frequency spectrum of TEs [20].
Despite detailed functional studies investigating the epigenetic influence of TEs on surrounding sequences, the influence of naturally occurring TEs on the chromatin states and functions of nearby genes has not been explored on a local or genomic scale in Drosophila. The connection between TE-induced epigenetic changes and the evolutionary genomic consequences of TE insertions is also lacking. In this study, we tested our hypothesis that the spread of piRNA-mediated heterochromatin of TEs to adjacent genes is deleterious and represents an important force shaping the population dynamics of TEs (Fig 1).
Fig. 1. Proposed model for the evolutionary impacts of piRNA-mediated epigenetic silencing of TEs. 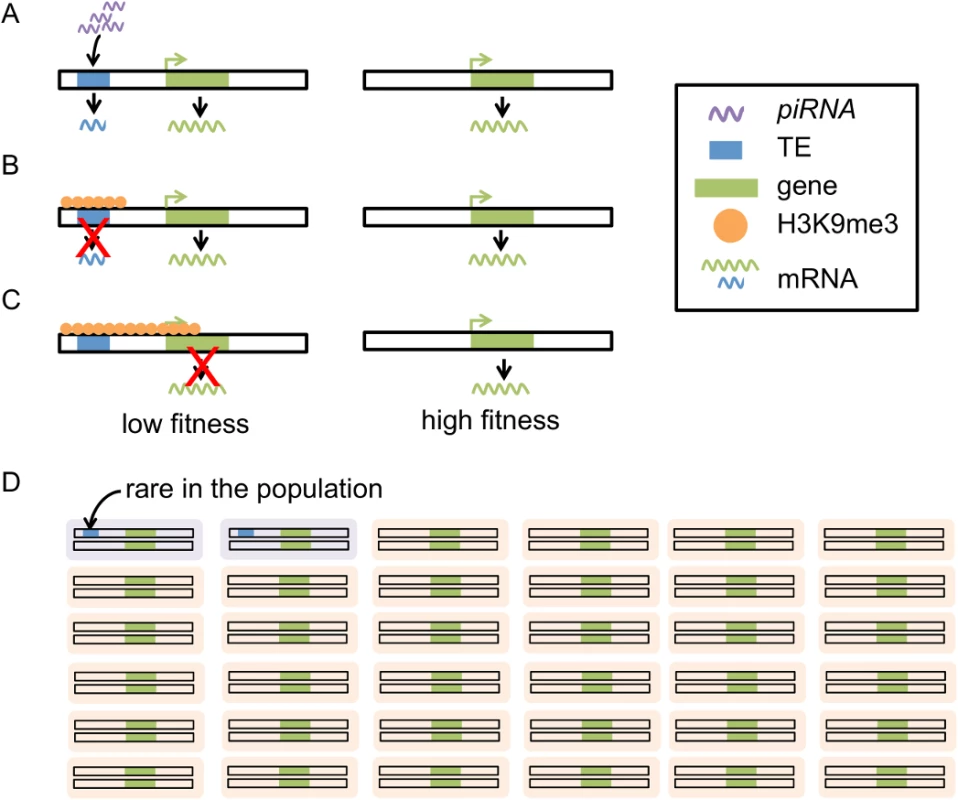
piRNAs target TEs that have complementary sequences (A), leading to installation of heterochromatic marks (i.e. H3K9me) on TE sequences (B). The boundaries of heterochromatin are generally dynamic and heterochromatin induced by TEs can “spread” into adjacent sequence. The spread of heterochromatin from TEs into genes could lead to lowered gene expression, impairing the function of genes (C). Individuals with these TE insertions will have lower fitness (purple in D) than individuals without (orange in D). TEs with this mutational impact are expected to be selected against, remaining rare in the population (D). Throughout our study, we used the distribution of histone modifications as an index for the chromatin state of a region. Within the nucleus, DNA is wrapped around core histones (H2A, H2B, H3, and H4) to form nucleosomes, the fundamental unit of chromatin. Modifications of histones, such as methylation and acetylation, at different positions of the core histones have been associated with biological consequences, particularly gene expression states (reviewed in [38]). For instance, tri-methylation of histone H3 lysine 4 (H3-K4me3) is enriched at transcription start sites and found to correlate positively with gene expression levels [39,40]. The methylation of histone H3 lysine 9 (H3K9me), particularly di - and tri - methylations, is generally regarded as “repressive” mark of the chromatin and is found enriched in the heterochromatic regions of the Drosophila genome [38–41]. We focused on the genomic variation in the level of H3K9me, which allows us to relate TE-composition to the heterochromatic states of sequences.
Results
Distance from intergenic TE insertion correlates negatively with H3K9me3 enrichment
Previous local studies indicated that the density of heterochromatic marks is elevated in sequences adjacent to TEs [32], which our hypothesis relies on (Fig 1B and 1C). To confirm this finding with naturally occurring TEs on a genomic scale, we used the genome annotation of the D. melanogaster reference genome (the Drosophila genome with the best annotated TEs), and the modEncode H3K9 tri-methylation (H3K9me3) data of the same strain at nine developmental time stages (six embryonic stages, two larval stages, and one pupal stage, [40]). These experiments were performed using whole animals, which consist of heterogeneous tissues and cell types. Histone modifications could significantly vary across cell types [42] and it is difficult to interpret the modifications of samples consisting of heterogeneous tissues and/or cell types as binary states [43,44]. Accordingly, we analyzed H3K9me3 data quantitatively, using the average read density of H3K9me3 for our following analyses (see Materials and Methods).
To investigate whether the TE-induced heterochromatin spreads beyond TEs, we first examined the enrichment of heterochromatic marks adjacent to TEs. We did this by estimating the H3K9me3 density at sequences that are 10kb upstream and downstream of euchromatic TEs. We only considered 10kb sequences that are entirely within intergenic regions to avoid the potential influences from functional elements (genes and regulatory sequences), which are generally depleted of “silent” marks (such as H3K9me) and enriched with other “active” histone modifications ([39,40], reviewed in [38]). We observed that the H3K9me3 density of 1kb nonoverlapping windows decreases with increasing distance from TEs (Fig 2A and 2B). The windows closest to a TE (0-1kb and 1-2kb) have significantly higher H3K9me3 density than the most distant window analyzed (9-10kb, Mann-Whitney U test, p < 0.011; S1 Table) for all developmental stages. For some embryonic stages, the significant difference in H3K9me3 enrichment is still observable for windows that are even farther from TEs (S1 Table).
Fig. 2. The decay of H3K9me3 density of intergenic sequences adjacent to TEs. 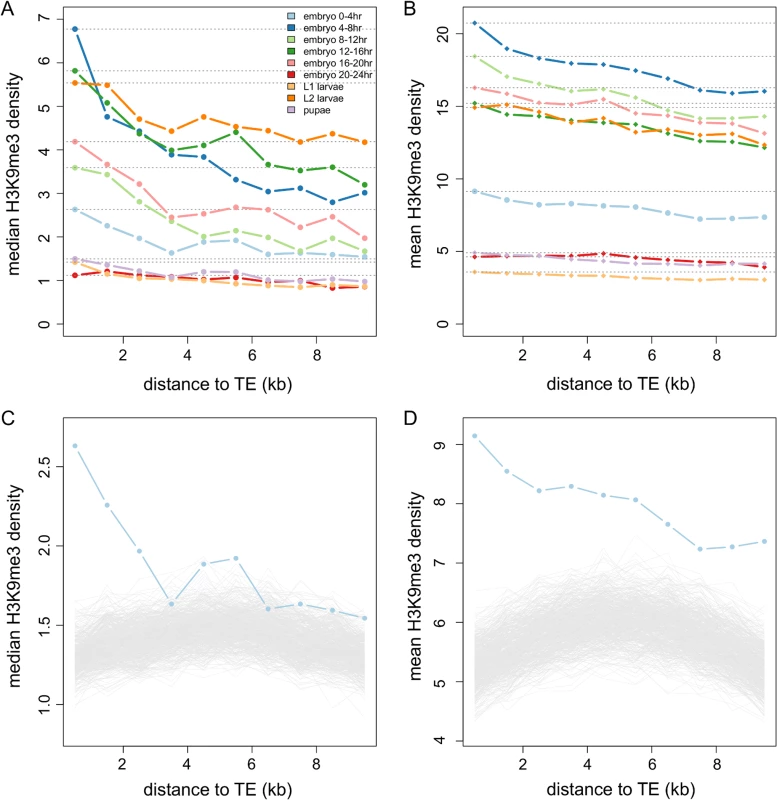
The median (A) and mean (B) of H3K9me3 density in nonoverlapping 1kb windows of intergenic sequences decreases as the windows are farther from TEs. Different colors are for different developmental stages: embryo 0–4 hr (light blue), embryo 4–8 hr (blue), embryo 8–12 hr (light green), embryo 12–16 hr (green), embryo 16–20 hr (pink), embryo 20–24 hr (red), L1 larvae (yellow), L2 larvae (orange), and pupae (purple). Dashed lines are the median/mean of the H3K9me3 density in the window closest to TEs. The observed median (C) and mean (D) H3K9me3 densities of windows adjacent to TEs are higher than those adjacent to 1,000 sets of randomly chosen TE-size sequences (gray lines), particularly for windows that are closer to TEs. Results of 0–4 hr embryos are shown here. Note that the scales of y-axis are different between (A) and (B) and between (C) and (D). We compared our observations to null genomic expectations by randomly choosing genomic segments that are of the same sizes and the same large-scale chromosome locations as those of TEs and investigating the decay in H3K9me3 enrichment near these “TE-size” sequences (see Materials and Methods). The median (Fig 2C) and mean (Fig 2D) H3K9me3 of windows adjacent to TEs are indeed higher than those of random sequences for most developmental stages (see S1 and S2 Figs for all developmental stages). Our genomic observations support predictions from functional data [32] that the presence of TEs influences the chromatin status of adjacent sequences.
Genes adjacent to TEs have higher H3K9me3 than other genes
In order to support our hypothesis, it is critical to demonstrate that the spreading of TE-induced heterochromatin also influences the chromatin states of neighboring genes in addition to intergenic sequences (Fig 1C). Using the same H3K9me3 data of the reference strain at nine developmental stages, we contrasted the enrichment of H3K9me3 marks in euchromatic genes that have TEs inside their introns (in gene), in 1kb, 1-2kb, 2-5kb, 5-10kb upstream/downstream of the gene to those of genes that have no TEs within 10kb upstream/downstream. Similar to our observation for intergenic sequences, we found genes with TEs nearby have significantly higher H3K9me3 density than genes without TEs nearby. H3K9me3 enrichment decreases as the windows move further away from TEs (Fig 3A for 0–4 embryo, see S3 Fig for all developmental stages). Consistently, we found significant negative correlations between a gene’s H3K9me3 density and its distance from the nearest TE (Spearman rank ρ = -0.189 ~ -0.040, p < 0.05 for L1 and L2 larvae and p < 10–3 for all other developmental stages, S2 Table). In addition, the number of TEs in bins further away from genes is less correlated with a gene’s H3K9me3 density (Fig 3B). These observations suggest that genic H3K9me3 enrichment correlates with a gene’s surrounding TE composition, both in terms of TE number and distance between genes and TEs.
Fig. 3. The associations between H3K9me3 density of genes and their neighborhood TE content. 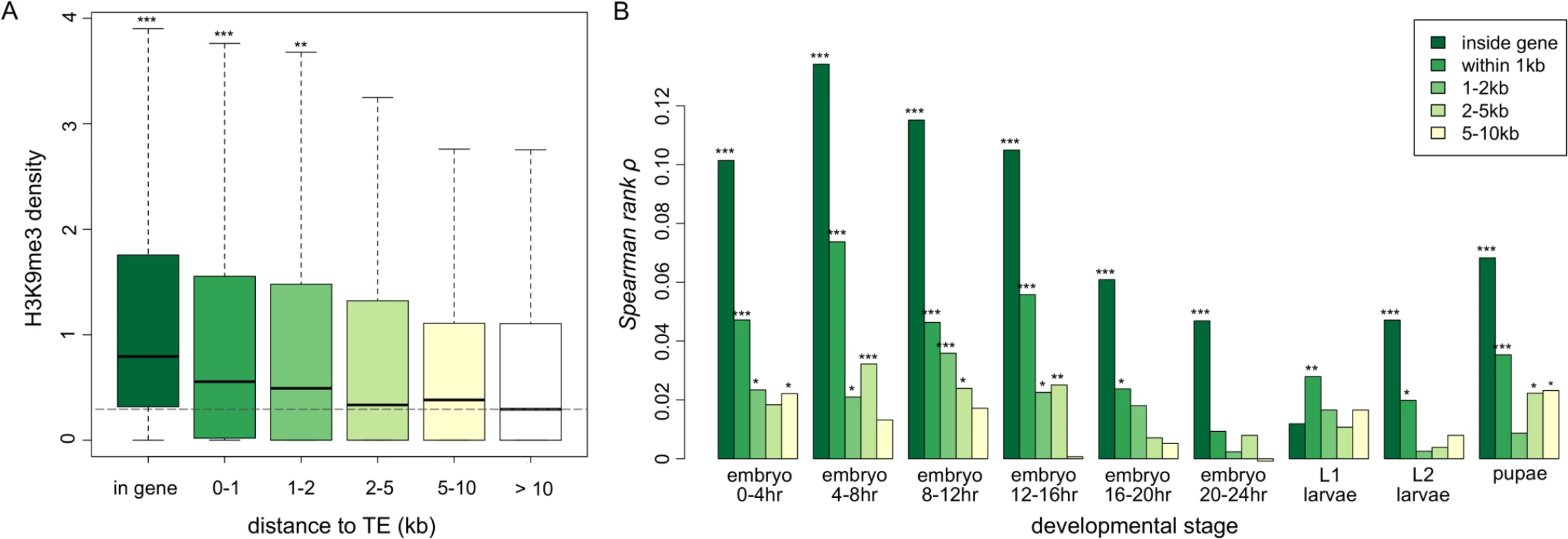
(A) Boxplots for the H3K9me3 density of genes that are of different distance from TEs. The H3K9me3 density of genes that have at least one TE in the introns of the gene (in gene), within 1kb, 1-2kb, 2-5kb, and 5-10kb upstream/downstream from the gene are significantly lower than those of genes that have no TE 10kb upstream/downstream (whose median of H3K9me3 density is shown as a dashed line). Numbers of genes in each category are 588 (in gene), 579 (within 1kb), 323 (1-2kb), 859 (2-5kb), 1162 (5-10kb), and 8072 (no TE in 10kb). All comparisons are against genes without TEs in 10kb upsteam/downstream. Result of 0–4 hr embryo is shown. (B) The spearman rank correlation coefficients between H3K9me3 density of a gene and the number of adjacent TEs within a specific window decrease as the distance between genes and TEs increases. The median, mean, and maximum number of TEs in specific windows are (1) in gene: 0 (median), 0.0523 (mean) and 8 (maximum), (2) 0-1kb: 0 (median), 0.0555 (mean), and 4 (maximum), (3) 1-2kb: 0 (median), 0.0341 (mean), 2 (maximum), (4) 2-5kb: 0 (median), 0.101 (mean), and 5 (maximum), and (5) 5-10kb: 0 (median), 0.158 (mean), and 7 (maximum). Notations for p-values are * (p < 0.05), ** (p < 0.01), and *** (p < 0.001). Our observations are not merely driven by genes that are in highly heterochromatinized genomic regions. Excluding genes in H3K9me3 enriched regions in another strain of the modEncode project (at embryonic or larval stage of Oregon-R strain) or in genomic regions that are classified as state 7 (highly enriched with H3K9me2/3) or state 8 (moderately enriched with H3K9me2/3) in either S2 or BG3 cell lines [39] gave similar results (S4–S6 Figs for comparisons of H3K9me3 density of genes that are of different distance from TEs; S7–S9 Figs for correlations between H3K9me3 density of genes and the nearby number of TEs).
It is important to examine whether the observed associations between the H3K9me3 enrichment of a gene and its neighborhood TE content can be attributed to other confounding factors. In D. melanogaster, TEs are known to accumulate in genomic regions with low recombination rate [16,18,45–48] and potentially regions with low gene density (although see [47]). We found that genic H3K9me3 density is negatively correlated with local gene density (Spearman rank ρ = -0.217 ~ -0.097, p < 10–16 for all developmental stages) and weakly correlated with its local recombination rate (Spearman rank ρ = -0.040 ~ -0.021, p < 0.05 for all but one developmental stage). However, we still observed significant positive correlations between a gene’s H3K9me3 density and its adjacent TE numbers using partial correlation analyses (S10 Fig for controlling for gene density and S11 Fig for controlling for local recombination rate), suggesting that neither the effects of recombination rate nor gene density can solely account for our observations.
Genic H3K9me3 enrichment is positively correlated with piRNA targeting of adjacent TEs
Heterochromatin formation of TEs in somatic tissues is observed to be piRNA-dependent [32]. We hypothesized that TEs targeted by large number of piRNAs are likely to be transcriptionally silenced, and, consequently, more likely to influence the chromatin states of nearby genes. We estimated the piRNA density of TEs [the average (per bp) number of piRNAs mapped to TEs; see materials and methods] in the reference genome using ovarian piRNA sequence of the reference strain (generated by Shpiz et al. [49]). It is worth noting that the H3K9me3 and piRNA data were collected using different types of tissues [embryos, larvae, and pupae (H3K9me3) vs ovary (piRNA)]. Nevertheless, embryonic piRNAs are known to be maternally deposited [50,51].
Consistent with our hypothesis that TEs targeted by piRNAs are more likely to influence the chromatin states of adjacent genes, we observed significant positive correlations between the H3K9me3 density of a gene at an early embryonic stage (0–4 hour embryo) and the ovarian piRNA density of its nearest TE [Spearman rank ρ = 0.043, p = 0.016 (sense piRNA vs H3K9me3); Spearman rank ρ = 0.084, p < 10–5 (antisense piRNA vs H3K9me3)]. As expected, these correlations are stronger for gene-TE pairs that are closer to each other than those that are farther apart [sense TE piRNA vs. genic H3K9me3: Spearman rank ρ = 0.081 (p = 0.008) for short distances between a gene and its nearest TE; antisense TE piRNA vs. genic H3K9me3: Spearman rank ρ = 0.131 (p < 10–4) for short, 0.063 (p = 0.038) for intermediate, and 0.057 (p = 0.061) for long distances between a gene and its nearest TE; see S12 Fig]. It is worth noting that, although significant, the observed correlations between embryonic H3K9me3 density and ovarian piRNA density of nearest TEs are rather weak. This might be due to the differences between ovarian and embryonic piRNAs. Future studies using embryonic piRNAs could further address this issue. Similar to previous studies [49,52,53], we identified a small fraction of piRNAs that target host genes (S3 Table). However, we did not observe similar positive correlations between H3K9me3 density and ovarian piRNA density of a gene (S3 Table), suggesting that our observed higher H3K9me3 of genes with TE nearby is more attributable to the spreading of H3K9me3 from transcriptionally silenced TE instead of by piRNAs’ transcriptional silencing directly at the host genes.
TE insertions lead to reduced expression of adjacent genes
H3K9me has been widely associated with the silencing of gene expression (reviewed in [38]). Indeed, we observed significant negative correlations between the H3K9me3 density of a gene and its expression level at the corresponding developmental stage (Spearman rank ρ = -0.248 ~ -0.029, p < 0.05 for all genes or only genes having TE within 10kb, S4 Table). Given our observation that genes with adjacent TEs have higher H3K9me3 density than genes without, transcriptional output from the former genes should be relatively lower. Indeed, in the majority of developmental stages, genes with TEs nearby have significantly lower expression levels than genes without TEs (S5 Table).
The analysis reported above cannot distinguish the causal relationship between a gene’s expression level and its nearby TE content. Our observations are consistent with either TE insertions leading to reduced expression of adjacent genes or TEs preferentially inserting near genes with low expression. To genomically assess these alternative scenarios, we compared the expression of alternative “alleles” (with or without TEs) of a gene, using TE polymorphism [19] and adult expression data [54] from a North American population. We compared the average expression rank of alleles with at least one TE within a window (“with TE”) to those of “without TE” alleles and performed permutation to assess significance. There is an excess of genes that have significantly (permutation p-value < 0.05) lower expression of “with TE” alleles than those of “without TE” alleles for all window sizes chosen and for both adult female and adult male (Fisher’s Exact, p < 0.05, odds ratio = 1.46~2.60, Table 1; see S6 and S7 Tables for results of all genes). Because female and male individuals from the same strain have the same type of allele, it is expected that genes having differential expression between “with TE” and “without TE” alleles in one sex should also show the same pattern in another sex. Indeed, we found an excess of genes that are differentially expressed between “with TE” and “without TE” alleles in both female and male (S8 Table). In short, alleles with nearby TEs are more likely to have lower expressions than their homologs without TEs. It is worth noting that the disruption of regulatory sequences by TE insertion could also contribute to this observation [55].
Tab. 1. Differences in expression levels between alleles with and without TEs. 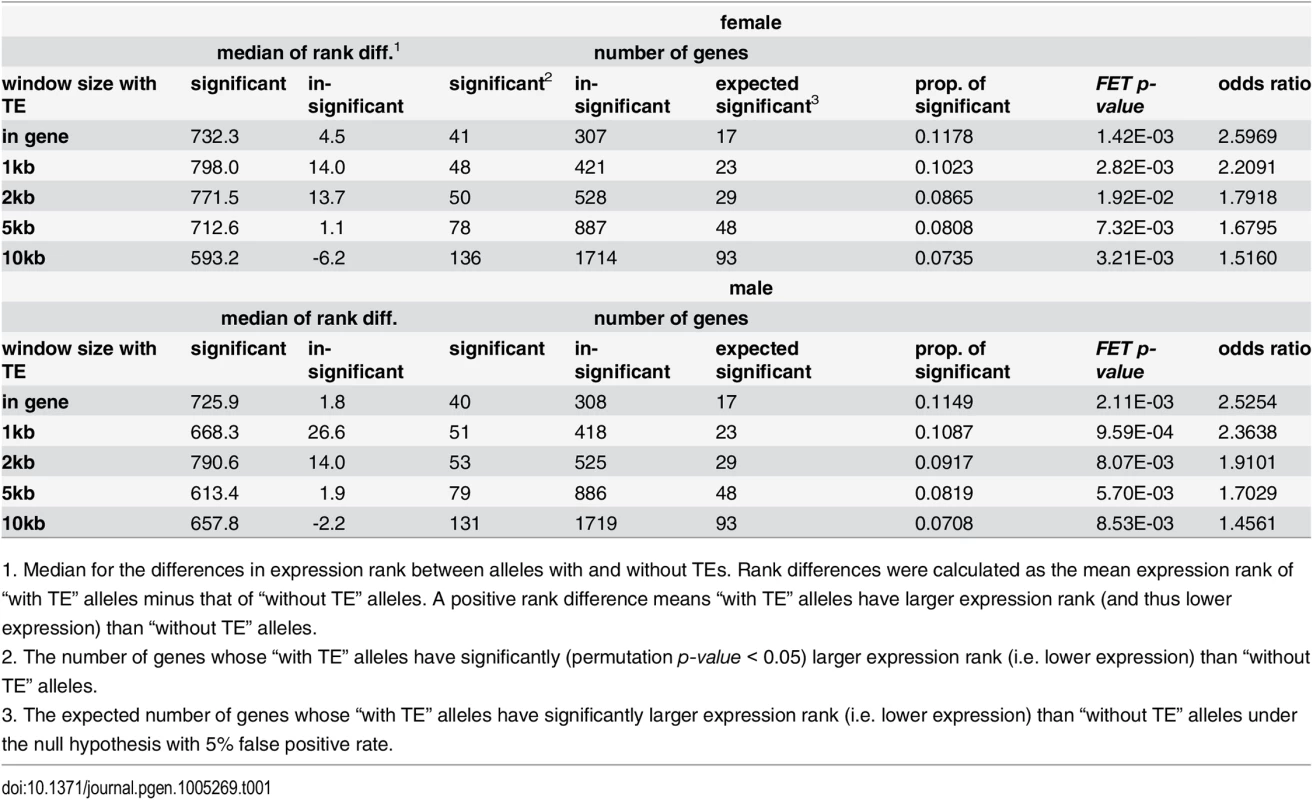
1. Median for the differences in expression rank between alleles with and without TEs. Rank differences were calculated as the mean expression rank of “with TE” alleles minus that of “without TE” alleles. A positive rank difference means “with TE” alleles have larger expression rank (and thus lower expression) than “without TE” alleles. TEs adjacent to genes with high H3K9me3 have lower population frequencies
Most TE insertions in the Drosophila population are polymorphic [15,18,19,48] and individual TE insertions present at lower population frequencies are expected to have experienced stronger selection removing them. Our hypothesis predicts that TE insertions inducing higher heterochromatic mark enrichment of neighboring genes should have larger deleterious impacts, are more likely removed from the population by selection, and as a result should segregate at lower population frequencies than other TEs. We classified reference TEs into those that are observed (high-frequency TEs) and not observed (low-frequency TEs) in a North American population [19]. In the reference genome, genes whose nearest TE occurs at low frequency (not observed in the North American population) have higher median H3K9me3 density than those near high-frequency TEs (Mann-Whitney U test, p < 0.01 for all developmental stages except for L1 larvae and pupae, Fig 4). Furthermore, we found significant negative correlations between a gene’s H3K9me3 density and the population frequency of its nearest TE in the North American population (Spearman rank ρ = -0.152 ~ -0.083, p < 0.01 for all developmental stages except for L1 larvae and pupae, S9 Table). Unless multiple independent insertions at the same site happen frequently, the alternative hypothesis that H3K9me3 enrichment inhibits insertions cannot explain our observed negative associations between genic H3K9me3 enrichment and population frequencies of their nearest TEs. Indeed, even though insertion site preference has been reported for several TE families [56–58], multiple independent insertions from the same TE family at the same genomic location have not been documented in Drosophila.
Fig. 4. Comparisons of the H3K9me3 density for genes whose nearest TEs have different population frequencies. 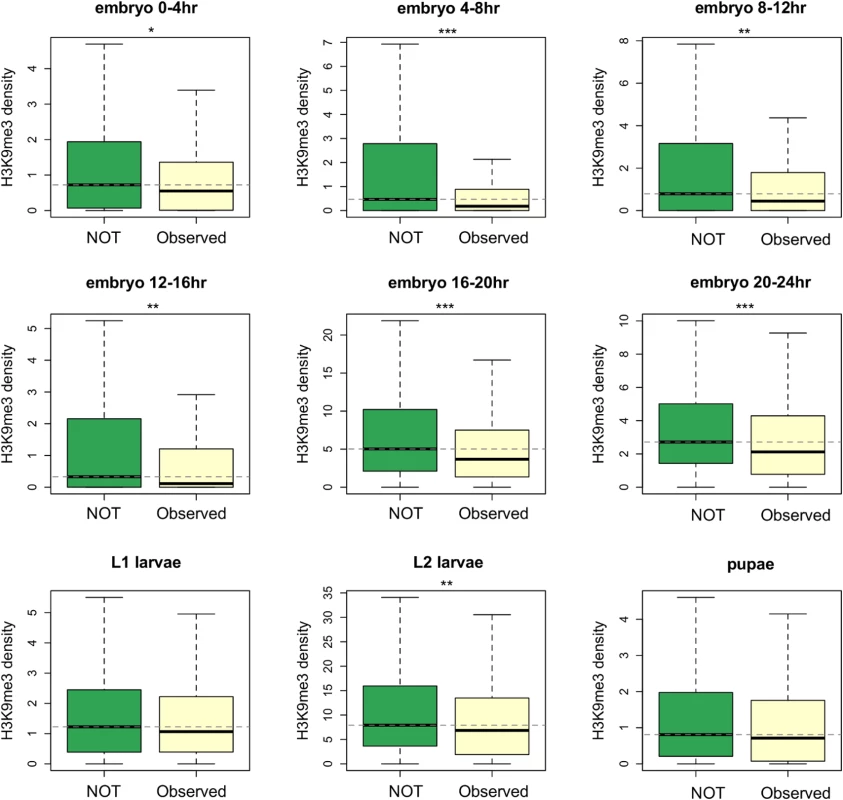
TEs that are observed in the North American D. melanogaster population (Observed) have higher population frequencies than those that are not observed in the population (NOT). Genes whose nearest TEs are observed in the population have significantly lower H3K9me3 density. Dashed lines show the median of H3K9me3 density for genes whose nearest TE are not observed in the population. Numbers of genes included in the analysis are 297 (for Observed) and 491 (for NOT). Notations for p-values are * (p < 0.05), ** (p < 0.01), and *** (p < 0.001). Another prediction of our model is that TEs inducing heterochromatin spreading at more developmental stages are expected to have larger cumulative deleterious impacts. Indeed, we found a significant negative correlation between the numbers of developmental stages during which a gene is enriched for H3K9me3 (top 10% genome-wide) and the population frequency of its nearest TE (Spearman rank ρ = -0.159, p < 10–5). This association is not merely driven by genes that have low (below first quantile) H3K9me3 density in all developmental stages, because a significant negative correlation was still observed after removing these genes (Spearman rank ρ = -0.160, p < 10–5). Removing genes with high H3K9me3 enrichment in another strain or cell lines (see Materials and Methods) and using a different threshold to categorize “high H3K9me3 genes” both gave consistent results [Spearman rank ρ = -0.148, p< 10–3 (exclude genes with high H3K9me3 in Oregon-R), ρ = -0.166, p< 10–4 (exclude genes in 7 or 8 chromatin state in cell lines), and -0.112, p = 0.002 (use top 25% genome-wide threshold)].
Compared to young and actively transposing TE families, old TE families were found to have lower piRNA density [59]. According to our model, we expect old TE families to be less likely be epigenetically silenced and influence the chromatin states of their adjacent genes. Members of old TE families are usually observed at higher population frequencies due to the age of their insertion events. This has the potential to confound our observations. We performed ANOVA that jointly considers the effect of the population frequency and family identity of nearest TEs on genic H3K9me3 enrichment. Our analysis still found that whether a gene’s nearest TE is observed in the North American population or not significantly contributes to the variation of genic H3K9me3 density in the majorities of developmental stages (S10 Table), suggesting that variation in TE age across families is unlikely the sole factor driving our observation.
Our observed negative associations between H3K9me3 of a gene and the frequency of the nearest TE could be confounded by the longer length of TEs adjacent to genes enriched with H3K9me3 (Spearman rank ρ between a gene’s H3K9me3 and the length of nearest TE, 0.036 ~ 0.118, p < 0.05 for all developmental stages). Longer TEs are expected to undergo ectopic recombination more frequently, more likely to be selected against, and thus be at lower population frequencies, a theoretical result which has been supported by empirical data [9,10,48,60]. However, given equal piRNA density, longer TEs represent larger piRNA targets and are more likely to be silenced as well as to have deleterious epigenetic impacts on adjacent genes. Furthermore, we found that there is a large positive correlation between piRNA density and TE length [Spearman rank ρ = 0.684 (sense piRNA) and 0.797 (anti-sense piRNA), p < 10–16 for both comparisons]. The positive correlations between H3K9me3 density of a gene and the length of its nearest TE also depend on the distance between genes and TEs [For 0-4hr embryo, Spearman rank ρ = 0.151 (short distance between a gene and its nearest TE), 0.120 (intermediate), and 0.083 (long), p < 0.01 for all; see S13 Fig for results of all developmental stages], suggesting that the high H3K9me3 of genes near long TEs is probably due to the spread of TE heterochromatin.
Another potential confounding factor comes from the fact that TEs in low recombination regions of the genome are present at higher population frequencies [16,18,48,61]. This observation is generally attributed to lower probability of ectopic recombination [9,10] and/or less effective purifying selection against TEs because of selective interference [62]. Accordingly, we performed logistic regression analyses to investigate the association between genes’ H3K9me3 signature and the population frequencies of nearest TEs while accounting for the effect of recombination rate (see Materials and Methods). We still found that the H3K9me3 density of a gene and the number of developmental stages in which a gene is enriched with H3K9me3 are significant negative predictors for whether the nearest TE is observed in the North American population or not (S11 Table).
Discussion
Selection removes deleterious TE insertions from the population and plays an important role in the containment of TEs. TE-induced physical disruptions of the genetic elements, such as insertions into functional elements or ectopic recombination between nonhomologous TE insertions, have been viewed as the primary source of the negative fitness impacts of TEs. Empirical investigation and theoretical discussion of TEs’ deleterious impact via epigenetic mechanisms has been limited [22]. In this study, we hypothesized that the piRNA-mediated epigenetic silencing of TEs perturbs transcription of adjacent genes and shapes the population dynamics of TEs (Fig 1). Using a genomic approach, we discovered an elevated density of repressive chromatin marks, H3K9me3, in sequences and genes up to several kb away from TEs (Fig 2 and Fig 3), which supports an important component of our hypothesis (Fig 1B and 1C). The H3K9me3 density of a gene heavily depends on its neighboring TE content (number and distance) and the strongest associations were observed for genes that are within 2kb from TEs. These genes account for 10.86% of the euchromatic genes, suggesting that the spread of TEs’ heterochromatin could influence the chromatin status and function of an appreciable number of genes in the genome. In accordance with our hypothesis (Fig 1C), the presence of nearby TEs is also associated with reduced gene expression (Table 1 and S5 Table). Our comparisons of with-TE and without-TE alleles of the same gene clearly demonstrated that the transcriptional consequence of TE insertions is not due to other confounding factors and further extend previous observations on the mutational impacts of TEs on gene expression [55]. Importantly, our observations vary with distance between genes and TEs, where larger distance between genes and TEs are associated with weaker effects. This is consistent with an important aspect of our model that the mutagenic effect of TEs is through the spread of heterochromatin from TEs. Future empirical studies investigating the chromatin states of with-TE and without-TE alleles of the same gene will help further distinguish the causal relationship of these observed associations, which could be attributed to either the spread of TE-induced epigenetic silencing or the preferential insertions of TEs near genes with high heterochromatic marks.
Supporting our hypothesis that the epigenetic silencing of TEs can have deleterious fitness impacts (Fig 1D), we found a negative association between H3K9me3 density of a gene and the population frequency of its nearest TE. This observation might be confounded by the fact that TEs adjacent to genes enriched with H3K9me3 tend to be longer, which is expected to increase the rate of ectopic recombination. However, ends of chromosomes that are highly heterochromatic are known to have reduced rates of crossing over. If rates of crossing over are also suppressed at TE-induced heterochromatin in the euchromatic regions, TEs with high H3K9me3 enrichment would be less likely to undergo ectopic recombination, and as a result would be removed by selection less frequently. Under this scenario, we would expect an opposite pattern from our observation—a positive correlation between H3K9me3 density of a gene and the population frequency of its nearest TE. In addition, TEs adjacent to genes with high H3K9me3 in multiple tissues also have lower population frequencies. This phenomenon is unlikely to be explained by ectopic recombination removing TEs, but is consistent with the prediction of our hypothesis that TEs epigenetically influencing adjacent genes in more developmental stages have larger overall deleterious impacts. Importantly, the preferential insertion of TEs near genes enriched with heterochromatic marks cannot account for either observation, suggesting that our observed associations between a gene’s H3K9me3 enrichment and its neighborhood TE content should be more attributable to the spread of TE-induced heterochromatin.
We observed that the H3K9me3 density of genes is positively correlated with the piRNA density of their nearest TEs, suggesting that our observed patterns depend on the piRNA-pathway. Even though the heterochromatin establishment by the piRNA-pathway occurs primarily in early embryos [63], the piRNA-dependent heterochromatin formed at this stage was found to have a lasting effect and significantly influence the chromatin states in adults [32,63]. Indeed, most of our observations are supported by H3K9me3 data from early embryos through pupae. Accordingly, the epigenetic silencing of TEs during early embryonic development, which depends on maternally deposited piRNAs, can influence the chromatin states of adjacent genes that are expressed at different developmental stages and have large cumulative mutational impacts.
Interestingly, the strength and statistical significance of our observed associations between genic H3K9me3 and the properties of their adjacent TEs (including distance from, number, and population frequency) varies across developmental stages. Earlier embryonic stages consistently showed the strongest associations, while larval and pupal stages generally showed the weakest or even statistically insignificant patterns. The effect of TE-induced heterochromatin spreading might be suppressed by other mechanisms of heterochromatin regulation differently at different developmental stages. Intriguingly, flies at later developmental stages, which usually showed weaker associations between genic H3K9me3 density and neighboring TE composition, consist of a greater diversity of differentiated tissues and cell types. It will be important in future work to investigate the tissue specificity of the epigenetic impact of TEs. This can elucidate the temporal and spatial variation in TE-induced heterochromatin spreading and will enable precise identification of the functional effects and evolutionary consequences of TE insertions.
It is worth mentioning that even though our observed associations between genic H3K9me3 density and the neighboring TE content are significant, they are not particularly strong. The paucity of strong correlations might be an issue of power, however. TEs are generally scarce around functional elements [19,64,65] and most TEs appear as singletons in natural populations [4,15–19]. The majority of genes included in our analyses have few adjacent TEs and their nearest TEs are absent in the North American population, providing us limited variation to estimate correlations. In addition, other biological processes can also influence the chromatin state of genes. By comparing chromatin states across a single genome (which was the focus of most of our analyses), we were unable to distinguish the epigenetic effects of TEs from those of other biological processes. Investigation of variation in chromatin states between “with TE” and “without TE” alleles, which presumably only differ with respect to their neighboring TE composition, can help further address the relative importance of the epigenetic effects of TEs on the chromatin states of genes and the evolutionary dynamics of TEs.
It was previously reported that the epigenetic silencing of TEs via methylation could influence the expression of adjacent genes in Arabidopsis thaliana [22]. Epigenetically silenced (methylated) TEs, but not unmethylated TEs, were found to be associated with lower expression of nearby genes and are present in lower population frequencies. Even though DNA methylation is rare in the D. melanogaster genome and definite evidence for methylated TEs is still missing [66,67], we observed a similar epigenetic impact of TEs on the expression of neighboring genes with a different mechanism (histone modifications). We provided complementary observations by investigating the associations between the heterochromatic mark enrichment of genes and their neighborhood TE composition. Importantly, combining our observed elevated H3K9me3 density surrounding TEs and the PEV phenomenon that has been known for decades ([34], reviewed in [35–37]), we are able to provide a mechanistic explanation for the observed deleterious impacts of TE’s epigenetic silencing.
There are several properties that make the piRNA-mediated epigenetic silencing of TEs an especially attractive mechanism for containment of TE copy number in natural populations. The generation of piRNAs only depends on the transcription of TE sequences and is thus general to virtually all classes and families of TEs [20]. Importantly, unlike other small RNAs, piRNAs are generated and amplified through a feed-forward ping-pong cycle (i.e. a positive feedback loop) [23,24]. The TE transcripts are targeted by anti-sense piRNAs and processed into sense-piRNAs, which are involved in generating additional antisense piRNAs. Despite previous suggestions that antisense piRNAs are mostly generated from piRNA clusters in heterochromatic regions [24], a recent functional study [49] and statistical analysis [59] both showed that euchromatic TE copy number is a major determinant of piRNA amount. It is expected that the probability that the DNA sequence of a TE is targeted by piRNAs increases with piRNA amount and, accordingly, the number of TEs that are epigenetically silenced will depend both on the amount of piRNAs (which is positively correlated with TE copy number of a family [59]) and TE copy number. Consequently, the number of epigenetically silenced TEs and the resulting mutational impacts due to TE-heterochromatin spreading might depend quadratically on TE copy number. This can potentially provide the required synergistic epistasis of TEs’ deleterious impacts for stable containment of TE copy number [13].
Our study demonstrates that the spread of piRNA-mediated heterochromatin of TEs is another important, though previously unexplored, mechanism leading to removal of TEs. Given piRNA’s wide phylogenetic distribution in animals [68], we expect this selective mechanism against TEs also plays an important role in the containment of TEs in other organisms. Further investigations of the relative roles of different selective mechanisms in the containment of TEs, particularly the potential interference between our proposed TE-induced indirect epigenetic effects and the widely empirically supported ectopic recombination between TEs, will help piece together our picture of TE dynamics in natural populations.
Materials and Methods
Genome annotations
Our analyses used D. melanogaster reference genome annotation 5.21 for genes, TEs, and other functional sequences. We only included genes and TEs that are in the euchromatic regions of the genome, using the heterochromatin-euchromatin boundaries defined in [69]. Genes and TEs on the 4th chromosome were excluded from the analyses as well.
Processing H3K9me3 ChIP-seq data
We used H3K9me3 ChIP-seq data generated by modEncode [40], which used samples from nine developmental stages of the reference D. melanogaster strain (0-4hr, 4-8hr, 8-12hr, 12-16hr, 16-20hr, and 20-24hr embryos, and L1 larvae, L2 larvae, and pupae). We used the modEncode processed wiggle files, which are background subtracted H3K9me3 read density (normalized read counts of uniquely mapped reads of H3K9me3 experiments subtracting the re-scaled read counts of uniquely mapped reads of the input experiments, [70]). Windows that have negative background-subtracted read density (no enrichment of H3K9me3) were assigned to zero.
The effect of TEs on the H3K9me3 density of adjacent intergenic sequences
The average H3K9me3 read density of the upstream/downstream 10kb sequences of TEs was estimated in 1kb nonoverlapping windows. We excluded 10kb sequences that overlap with any annotations other than “intergenic” and analyzed the right side and left side of the sequences separately. To generate a null expectation for the H3K9me3 density decay near TEs, we randomly chose sequences that have the same size and are on the same chromosome as TEs included in the analyses. To account for the observed large-scale variation in histone modifications across D. melanogaster genome [39,40], we divided each chromosomal arm into 4Mb bins (5–7 bins per chromosome) and randomly selected “TE-size” segments within the same bins as the reference TEs. The number of randomly chosen sequences is the same as the number of TEs included in the analysis. For each randomly selected segment, we estimated the H3K9me3 density of adjacent sequences using the same methods as for TEs. This procedure was repeated 1,000 times.
The effect of TEs on the H3K9me3 density of adjacent genes
The H3K9me3 density of genes was estimated as weighted averages across all exons of the longest isoforms. We excluded genes that have TEs inside their coding or noncoding exons. We further removed genes whose 10kb upstream/downstream sequences overlaps with the euchromatin-heterochromatin boundaries, because of how we categorized genes according to their neighboring TE content (see below). Genes are categorized into nonoverlapping groups according to their distance from the nearest TE (having TEs in introns, in 1kb, in 1-2kb, in 2-5kb, and in 5-10kb upstream/downstream of the gene, or have no TEs 10kb upstream/downstream of the gene). These genes account for 3.66% (in gene), 4.65% (in 1kb), 2.55% (1-2kb), 6.83% (2-5kb), and 9.26% (5-10kb) of the analyzed euchromatic genes (12,204 genes). We also counted the number of TEs that are inside introns, less than 1kb, 1-2kb, 2-5kb, and 5-10kb away from each gene. For TEs that span over multiple windows, we classified them to windows that are closest to genes. For analyses considering the properties of nearest TEs, genes that have more than one TEs of equal distance were excluded, resulting in 3,193 gene-TE pairs. Gene-TE pairs were further categorized into three equal-size bins according to the distance between them (short: 0-1451bp, intermediate: 1452-5184bp, and long: > 5184bp). Recombination rate were interpolated for the mid-point of genes or TEs using [71]. Gene density was estimated as the number of genes in a 100kb window centered on the focused gene. S12 Table includes estimated H3K9me3 density, TE states, and other genic attributes of analyzed genes.
Estimation of piRNA density
The ovarian piRNA of reference strain [49] were processed and mapped without mismatches (using BWA [72]) to reference genome release 5 following methods in [73]. We estimated the density (average per bp) of piRNAs mapped to sense (sense piRNA) and antisense (antisense piRNA) strands of genes/TEs. For piRNAs mapped to multiple genomic locations, each of the n mapped positions in the genome is counted as having 1/n read mapped. Analyses using all mapped piRNAs (uniquely mapped and multiply mapped) or only uniquely mapped piRNAs gave consistent results. We presented the results based on all mapped piRNAs because this represents the full probability that a TE is targeted by the piRNA-pathway.
Testing the expressional impacts of TEs
To investigate the impact of H3K9me3 on gene expression, we used modEncode developmental stage expression data that were generated also using reference D. melanogaster strain [74]. The normalized and standardized expression level (RPKM) was downloaded from FlyBase. No (zero) expression of genes could be due to the presence of nearby TE insertions or simply the absence of expression at a particular developmental stage. Because we could not distinguish these alternatives, we excluded genes that have zero expression in the analysis of the particular developmental stage.
There are 21 strains of a North American population that have both microarray-based gene expression data [54] and TE calls [19]. We used these strains to investigate the expressional impacts of TEs within population. We downloaded processed gene expression data from the supplementary data of [54]. To avoid the systematic difference in expression level across microarray experiments and/or individuals, we used expressional rank (from highest to lowest) within each samples. In [19], for each strain, each TE insertion site was annotated as “present” (with TE), “absent” (no TE), or “no call” (missing data due to low sequencing coverage). For each gene, the “with TE” alleles are those that have one or more TE insertion sites called as “present” within introns, in 1kb, 2kb, 5kb, or 10kb windows from the gene. An allele is categorized as “without TE” if all the known TE insertion sites near it were called as “absent”. An allele is treated as missing data when any of the known TE insertion sites near it was called as “no call” and none of them were called as “present” (see S13 Table for TE states of all alleles). For genes that have at least two “with TE” and at least two “without TE” alleles, we calculated the difference in mean expression rank between “with TE” alleles and “without TE” alleles. For each gene, we used all possible permutations with respect to TE labels to find the null distribution of rank differences between “with TE” and “without TE” alleles (“with TE” alleles minus “without TE” alleles). Positive differences suggest that the “with TE” alleles have larger expressional rank, or lower expression, than “without TE” alleles. One tail p-values were calculated as the proportion of permuted combinations that have differences greater than or equal to the observed differences. To ensure that a gene could potentially have a significant (p < 0.05) p-value, we further restricted our analysis to genes that have more than 20 possible permuted combinations. We also randomly chose the same number of genes that have no TE alleles (“without TE genes”) as the number of “genes with TE alleles”. The alleles of these “without TE genes” were randomly partitioned into two groups accordingly to the observed allele frequencies for “genes with TE alleles”, and we used these “without TE genes” to assess the false positive rate of above procedures. This process was repeated 100 times for each window size and separately for each sex. We found that the false positive rate of our approach is slightly smaller than the expected 5% (S14 Table).
Variation in selective pressure on TEs
We used the same TE polymorphism data of the North American population [19] to investigate the evolutionary impacts of heterochromatin spreads from TEs. It is worth noting that this part of the analyses included all 131 genomes whose TEs has been annotated in the North American population [19], compared to 21 in the above expressional impact analysis. Only TEs observed in the reference genome were included in our analysis. For TEs that are only observed in the reference genome, their population frequencies in the North American population is zero.
We performed two-way Analysis of Variance analysis (ANOVA) to test whether the nearest TE is observed in the North American population (binary variable), the family of nearest TE (categorical variable, 52 TE families), and the interaction between these two variables contribute to the variation in H3K9me3 density of genes (model: H3K9me3 density ~ observed/not + TE family + observed/not * TE family). The H3K9me3 density of genes has an overall exponential distribution and a large number of genes with zero H3K9me3 density, which made us unable to identify appropriate transformation for the response variable (H3K9me3 density). Accordingly, we restricted our analysis to genes that have positive H3K9me3 density and log-transformed the H3K9me3 density of genes. We also performed mixed linear model analysis to investigate the influence of TE frequency on genic H3K9me3 while treating the influence of TE family as random. Mixed linear model regression was performed using R package nlme version 3.1.
We performed logistic regressions, using whether a reference TE is observed (one) or not (zero) in the North American population as response variable and the H3K9me3 status of its nearest gene and TE’s local recombination rate as predictors. We first performed regression analysis that included only one predictor variable at a time to determine the regression model (linear, quadratic, or logarithmic). We then included all predictor variables and performed backward model selection based on AIC to determine the regression model. Full regression models used are:
where logit p is the log odds of whether a reference TE is observed in the North American population (one) or not (zero) and “no. dev. stages” is the number of developmental stages a gene has top 10% H3K9me3 density genome-wide. All statistical analyses were performed using R.Supporting Information
Zdroje
1. Daborn PJ, Yen JL, Bogwitz MR, Le Goff G, Feil E, Jeffers S, et al. A Single P450 Allele Associated with Insecticide Resistance in Drosophila. Science. 2002;297 : 2253 –2256. doi: 10.1126/science.1074170 12351787
2. Schlenke TA, Begun DJ. Strong selective sweep associated with a transposon insertion in Drosophila simulans. Proc Natl Acad Sci U S A. 2004;101 : 1626–1631. doi: 10.1073/pnas.0303793101 14745026
3. Aminetzach YT, Macpherson JM, Petrov DA. Pesticide Resistance via Transposition-Mediated Adaptive Gene Truncation in Drosophila. Science. 2005;309 : 764 –767. doi: 10.1126/science.1112699 16051794
4. González J, Lenkov K, Lipatov M, Macpherson JM, Petrov DA. High rate of recent transposable element-induced adaptation in Drosophila melanogaster. PLoS Biol. 2008;6: e251. doi: 10.1371/journal.pbio.0060251 18942889
5. Schmidt JM, Good RT, Appleton B, Sherrard J, Raymant GC, Bogwitz MR, et al. Copy number variation and transposable elements feature in recent, ongoing adaptation at the Cyp6g1 locus. PLoS Genet. 2010;6: e1000998. doi: 10.1371/journal.pgen.1000998 20585622
6. Maside X, Bartolomé C, Charlesworth B. S-element insertions are associated with the evolution of the Hsp70 genes in Drosophila melanogaster. Curr Biol CB. 2002;12 : 1686–1691. 12361573
7. Finnegan DJ. Transposable elements. Curr Opin Genet Dev. 1992;2 : 861–867. 1335807
8. Montgomery E, Charlesworth B, Langley CH. A test for the role of natural selection in the stabilization of transposable element copy number in a population of Drosophila melanogaster. Genet Res. 1987;49 : 31–41. 3032743
9. Montgomery EA, Huang SM, Langley CH, Judd BH. Chromosome rearrangement by ectopic recombination in Drosophila melanogaster: genome structure and evolution. Genetics. 1991;129 : 1085–1098. 1783293
10. Langley CH, Montgomery E, Hudson R, Kaplan N, Charlesworth B. On the role of unequal exchange in the containment of transposable element copy number. Genet Res. 1988;52 : 223–235. 2854088
11. Batzer MA, Deininger PL. Alu repeats and human genomic diversity. Nat Rev Genet. 2002;3 : 370–379. doi: 10.1038/nrg798 11988762
12. Mieczkowski PA, Lemoine FJ, Petes TD. Recombination between retrotransposons as a source of chromosome rearrangements in the yeast Saccharomyces cerevisiae. DNA Repair. 2006;5 : 1010–1020. doi: 10.1016/j.dnarep.2006.05.027 16798113
13. Charlesworth B, Charlesworth D. The Population Dynamics of Transposable Elements. Genet Res. 1983;42 : 1–27. doi: 10.1017/S0016672300021455
14. Langley CH, Brookfield JF, Kaplan N. Transposable elements in mendelian populations. I. A theory. Genetics. 1983;104 : 457–471. 17246142
15. Charlesworth B, Langley CH. The population genetics of Drosophila transposable elements. Annu Rev Genet. 1989;23 : 251–287. doi: 10.1146/annurev.ge.23.120189.001343 2559652
16. Charlesworth B, Lapid A. A study of ten families of transposable elements on X chromosomes from a population of Drosophila melanogaster. Genet Res. 1989;54 : 113–125. 2558961
17. Charlesworth B, Lapid A, Canada D. The distribution of transposable elements within and between chromosomes in a population of Drosophila melanogaster. II. Inferences on the nature of selection against elements. Genet Res. 1992;60 : 115–130. 1334900
18. Kofler R, Betancourt AJ, Schlötterer C. Sequencing of Pooled DNA Samples (Pool-Seq) Uncovers Complex Dynamics of Transposable Element Insertions in Drosophila melanogaster. PLoS Genet. 2012;8: e1002487. doi: 10.1371/journal.pgen.1002487 22291611
19. Cridland JM, Macdonald SJ, Long AD, Thornton KR. Abundance and Distribution of Transposable Elements in Two Drosophila QTL Mapping Resources. Mol Biol Evol. 2013;30 : 2311–2327. doi: 10.1093/molbev/mst129 23883524
20. Lee YCG, Langley CH. Transposable elements in natural populations of Drosophila melanogaster. Philos Trans R Soc Lond B Biol Sci. 2010;365 : 1219–1228. doi: 10.1098/rstb.2009.0318 20308097
21. Blumenstiel JP, Chen X, He M, Bergman CM. An Age-of-Allele Test of Neutrality for Transposable Element Insertions. Genetics. 2014;196 : 523–538. doi: 10.1534/genetics.113.158147 24336751
22. Hollister JD, Gaut BS. Epigenetic silencing of transposable elements: A trade-off between reduced transposition and deleterious effects on neighboring gene expression. Genome Res. 2009;19 : 1419–1428. doi: 10.1101/gr.091678.109 19478138
23. Gunawardane LS, Saito K, Nishida KM, Miyoshi K, Kawamura Y, Nagami T, et al. A slicer-mediated mechanism for repeat-associated siRNA 5’ end formation in Drosophila. Science. 2007;315 : 1587–1590. doi: 10.1126/science.1140494 17322028
24. Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, et al. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128 : 1089–1103. doi: 10.1016/j.cell.2007.01.043 17346786
25. Klattenhoff C, Theurkauf W. Biogenesis and germline functions of piRNAs. Dev Camb Engl. 2008;135 : 3–9. doi: 10.1242/dev.006486
26. Senti K - A, Brennecke J. The piRNA pathway: a fly’s perspective on the guardian of the genome. Trends Genet. 2010;26 : 499–509. doi: 10.1016/j.tig.2010.08.007 20934772
27. Grewal SIS, Elgin SCR. Transcription and RNA interference in the formation of heterochromatin. Nature. 2007;447 : 399–406. doi: 10.1038/nature05914 17522672
28. Klenov MS, Lavrov SA, Stolyarenko AD, Ryazansky SS, Aravin AA, Tuschl T, et al. Repeat-associated siRNAs cause chromatin silencing of retrotransposons in the Drosophila melanogaster germline. Nucleic Acids Res. 2007;35 : 5430–5438. doi: 10.1093/nar/gkm576 17702759
29. Klenov MS, Sokolova OA, Yakushev EY, Stolyarenko AD, Mikhaleva EA, Lavrov SA, et al. Separation of stem cell maintenance and transposon silencing functions of Piwi protein. Proc Natl Acad Sci U S A. 2011;108 : 18760–18765. doi: 10.1073/pnas.1106676108 22065765
30. Wang SH, Elgin SCR. Drosophila Piwi functions downstream of piRNA production mediating a chromatin-based transposon silencing mechanism in female germ line. Proc Natl Acad Sci U S A. 2011;108 : 21164–21169. doi: 10.1073/pnas.1107892109 22160707
31. Thomas AL, Rogers AK, Webster A, Marinov GK, Liao SE, Perkins EM, et al. Piwi induces piRNA-guided transcriptional silencing and establishment of a repressive chromatin state. Genes Dev. 2013;27 : 390–399. doi: 10.1101/gad.209841.112 23392610
32. Sentmanat MF, Elgin SCR. Ectopic assembly of heterochromatin in Drosophila melanogaster triggered by transposable elements. Proc Natl Acad Sci. 2012;109 : 14104–14109. doi: 10.1073/pnas.1207036109 22891327
33. Sentmanat M, Wang SH, Elgin SCR. Targeting heterochromatin formation to transposable elements in Drosophila: Potential roles of the piRNA system. Biochem Mosc. 2013;78 : 562–571. doi: 10.1134/S0006297913060023 23980883
34. Muller HJ. Types of visible variations induced by X-rays inDrosophila. J Genet. 1930;22 : 299–334. doi: 10.1007/BF02984195
35. Talbert PB, Henikoff S. Spreading of silent chromatin: inaction at a distance. Nat Rev Genet. 2006;7 : 793–803. doi: 10.1038/nrg1920 16983375
36. Girton JR, Johansen KM. Chromatin structure and the regulation of gene expression: the lessons of PEV in Drosophila. Adv Genet. 2008;61 : 1–43. doi: 10.1016/S0065-2660(07)00001-6 18282501
37. Elgin SCR, Reuter G. Position-effect variegation, heterochromatin formation, and gene silencing in Drosophila. Cold Spring Harb Perspect Biol. 2013;5: a017780. doi: 10.1101/cshperspect.a017780 23906716
38. Kouzarides T. Chromatin Modifications and Their Function. Cell. 2007;128 : 693–705. doi: 10.1016/j.cell.2007.02.005 17320507
39. Kharchenko PV, Alekseyenko AA, Schwartz YB, Minoda A, Riddle NC, Ernst J, et al. Comprehensive analysis of the chromatin landscape in Drosophila melanogaster. Nature. 2011;471 : 480–485. doi: 10.1038/nature09725 21179089
40. Nègre N, Brown CD, Ma L, Bristow CA, Miller SW, Wagner U, et al. A cis-regulatory map of the Drosophila genome. Nature. 2011;471 : 527–531. doi: 10.1038/nature09990 21430782
41. Riddle NC, Minoda A, Kharchenko PV, Alekseyenko AA, Schwartz YB, Tolstorukov MY, et al. Plasticity in patterns of histone modifications and chromosomal proteins in Drosophila heterochromatin. Genome Res. 2011;21 : 147–163. doi: 10.1101/gr.110098.110 21177972
42. Henry GL, Davis FP, Picard S, Eddy SR. Cell type–specific genomics of Drosophila neurons. Nucleic Acids Res. 2012;40 : 9691–9704. doi: 10.1093/nar/gks671 22855560
43. Consortium TEP. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489 : 57–74. doi: 10.1038/nature11247 22955616
44. Arthur RK, Ma L, Slattery M, Spokony RF, Ostapenko A, Nègre N, et al. Evolution of H3K27me3-marked chromatin is linked to gene expression evolution and to patterns of gene duplication and diversification. Genome Res. 2014;24 : 1115–1124. doi: 10.1101/gr.162008.113 24985914
45. Eanes WF, Wesley C, Charlesworth B. Accumulation of P elements in minority inversions in natural populations of Drosophila melanogaster. Genet Res. 1992;59 : 1–9. 1315298
46. Sniegowski PD, Charlesworth B. Transposable element numbers in cosmopolitan inversions from a natural population of Drosophila melanogaster. Genetics. 1994;137 : 815–827. 8088526
47. Bartolomé C, Maside X, Charlesworth B. On the abundance and distribution of transposable elements in the genome of Drosophila melanogaster. Mol Biol Evol. 2002;19 : 926–937. 12032249
48. Petrov DA, Fiston-Lavier A-S, Lipatov M, Lenkov K, González J. Population genomics of transposable elements in Drosophila melanogaster. Mol Biol Evol. 2011;28 : 1633–1644. doi: 10.1093/molbev/msq337 21172826
49. Shpiz S, Ryazansky S, Olovnikov I, Abramov Y, Kalmykova A. Euchromatic Transposon Insertions Trigger Production of Novel Pi - and Endo-siRNAs at the Target Sites in the Drosophila Germline. PLoS Genet. 2014;10: e1004138. doi: 10.1371/journal.pgen.1004138 24516406
50. Blumenstiel JP, Hartl DL. Evidence for maternally transmitted small interfering RNA in the repression of transposition in Drosophila virilis. Proc Natl Acad Sci U S A. 2005;102 : 15965–15970. doi: 10.1073/pnas.0508192102 16247000
51. Brennecke J, Malone CD, Aravin AA, Sachidanandam R, Stark A, Hannon GJ. An epigenetic role for maternally inherited piRNAs in transposon silencing. Science. 2008;322 : 1387–1392. doi: 10.1126/science.1165171 19039138
52. Saito K, Inagaki S, Mituyama T, Kawamura Y, Ono Y, Sakota E, et al. A regulatory circuit for piwi by the large Maf gene traffic jam in Drosophila. Nature. 2009;461 : 1296–1299. doi: 10.1038/nature08501 19812547
53. Robine N, Lau NC, Balla S, Jin Z, Okamura K, Kuramochi-Miyagawa S, et al. A broadly conserved pathway generates 3’UTR-directed primary piRNAs. Curr Biol CB. 2009;19 : 2066–2076. doi: 10.1016/j.cub.2009.11.064 20022248
54. Ayroles JF, Carbone MA, Stone EA, Jordan KW, Lyman RF, Magwire MM, et al. Systems genetics of complex traits in Drosophila melanogaster. Nat Genet. 2009;41 : 299–307. doi: 10.1038/ng.332 19234471
55. Cridland JM, Thornton KR, Long AD. Gene Expression Variation in Drosophila melanogaster Due to Rare Transposable Element Insertion Alleles of Large Effect. Genetics. 2015;199 : 85–93. doi: 10.1534/genetics.114.170837 25335504
56. Linheiro RS, Bergman CM. Whole Genome Resequencing Reveals Natural Target Site Preferences of Transposable Elements in Drosophila melanogaster. PLoS ONE. 2012;7: e30008. doi: 10.1371/journal.pone.0030008 22347367
57. Liao G, Rehm EJ, Rubin GM. Insertion site preferences of the P transposable element in Drosophila melanogaster. Proc Natl Acad Sci U S A. 2000;97 : 3347–3351. 10716700
58. Spradling AC, Bellen HJ, Hoskins RA. Drosophila P elements preferentially transpose to replication origins. Proc Natl Acad Sci. 2011; doi: 10.1073/pnas.1112960108
59. Kelleher ES, Barbash DA. Analysis of piRNA-Mediated Silencing of Active TEs in Drosophila melanogaster Suggests Limits on the Evolution of Host Genome Defense. Mol Biol Evol. 2013;30 : 1816–1829. doi: 10.1093/molbev/mst081 23625890
60. Petrov DA, Aminetzach YT, Davis JC, Bensasson D, Hirsh AE. Size Matters: Non-LTR Retrotransposable Elements and Ectopic Recombination in Drosophila. Mol Biol Evol. 2003;20 : 880–892. doi: 10.1093/molbev/msg102 12716993
61. Bartolomé C, Maside X. The lack of recombination drives the fixation of transposable elements on the fourth chromosome of Drosophila melanogaster. Genet Res. 2004;83 : 91–100. 15219154
62. Hill WG, Robertson A. The effect of linkage on limits to artificial selection. Genet Res. 1966;8 : 269–294. 5980116
63. Gu T, Elgin SCR. Maternal Depletion of Piwi, a Component of the RNAi System, Impacts Heterochromatin Formation in Drosophila. PLoS Genet. 2013;9: e1003780. doi: 10.1371/journal.pgen.1003780 24068954
64. Kaminker JS, Bergman CM, Kronmiller B, Carlson J, Svirskas R, Patel S, et al. The transposable elements of the Drosophila melanogaster euchromatin: a genomics perspective. Genome Biol. 2002;3: RESEARCH0084.
65. Quesneville H, Bergman CM, Andrieu O, Autard D, Nouaud D, Ashburner M, et al. Combined Evidence Annotation of Transposable Elements in Genome Sequences. PLoS Comput Biol. 2005;1. doi: 10.1371/journal.pcbi.0010022
66. Schaefer M, Lyko F. Lack of evidence for DNA methylation of Invader4 retroelements in Drosophila and implications for Dnmt2-mediated epigenetic regulation. Nat Genet. 2010;42 : 920–921. doi: 10.1038/ng1110-920 20980983
67. Takayama S, Dhahbi J, Roberts A, Mao G, Heo S-J, Pachter L, et al. Genome methylation in D. melanogaster is found at specific short motifs and is independent of DNMT2 activity. Genome Res. 2014;24 : 821–830. doi: 10.1101/gr.162412.113 24558263
68. Grimson A, Srivastava M, Fahey B, Woodcroft BJ, Chiang HR, King N, et al. Early origins and evolution of microRNAs and Piwi-interacting RNAs in animals. Nature. 2008;455 : 1193–1197. doi: 10.1038/nature07415 18830242
69. Begun DJ, Holloway AK, Stevens K, Hillier LW, Poh Y-P, Hahn MW, et al. Population genomics: whole-genome analysis of polymorphism and divergence in Drosophila simulans. PLoS Biol. 2007;5: e310. doi: 10.1371/journal.pbio.0050310 17988176
70. Kharchenko PV, Tolstorukov MY, Park PJ. Design and analysis of ChIP-seq experiments for DNA-binding proteins. Nat Biotechnol. 2008;26 : 1351–1359. doi: 10.1038/nbt.1508 19029915
71. Comeron JM, Ratnappan R, Bailin S. The Many Landscapes of Recombination in Drosophila melanogaster. PLoS Genet. 2012;8: e1002905. doi: 10.1371/journal.pgen.1002905 23071443
72. Li H, Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009;25 : 1754–1760. doi: 10.1093/bioinformatics/btp324 19451168
73. Olovnikov I, Ryazansky S, Shpiz S, Lavrov S, Abramov Y, Vaury C, et al. De novo piRNA cluster formation in the Drosophila germ line triggered by transgenes containing a transcribed transposon fragment. Nucleic Acids Res. 2013;41 : 5757–5768. doi: 10.1093/nar/gkt310 23620285
74. Graveley BR, Brooks AN, Carlson JW, Duff MO, Landolin JM, Yang L, et al. The developmental transcriptome of Drosophila melanogaster. Nature. 2011;471 : 473–479. doi: 10.1038/nature09715 21179090
Štítky
Genetika Reprodukční medicína
Článek Germline Mutations Confer Susceptibility to Acute Lymphoblastic Leukemia and ThrombocytopeniaČlánek Multiple In Vivo Biological Processes Are Mediated by Functionally Redundant Activities of andČlánek Temporal Expression Profiling Identifies Pathways Mediating Effect of Causal Variant on PhenotypeČlánek Simultaneous DNA and RNA Mapping of Somatic Mitochondrial Mutations across Diverse Human CancersČlánek A Legume Genetic Framework Controls Infection of Nodules by Symbiotic and Endophytic BacteriaČlánek The Eukaryotic-Like Ser/Thr Kinase PrkC Regulates the Essential WalRK Two-Component System inČlánek The Yeast GSK-3 Homologue Mck1 Is a Key Controller of Quiescence Entry and Chronological Lifespan
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2015 Číslo 6
-
Všechny články tohoto čísla
- Expression of Concern: RNAi-Dependent and Independent Control of LINE1 Accumulation and Mobility in Mouse Embryonic Stem Cells
- Orphan Genes Find a Home: Interspecific Competition and Gene Network Evolution
- Minor Cause—Major Effect: A Novel Mode of Control of Bistable Gene Expression
- Germline Mutations Confer Susceptibility to Acute Lymphoblastic Leukemia and Thrombocytopenia
- Leveraging Identity-by-Descent for Accurate Genotype Inference in Family Sequencing Data
- Is Required for the Expression of Principal Recognition Molecules That Control Axon Targeting in the Retina
- Epigenetic Aging Signatures Are Coherently Modified in Cancer
- Silencing of DNase Colicin E8 Gene Expression by a Complex Nucleoprotein Assembly Ensures Timely Colicin Induction
- A Transposable Element within the Non-canonical Telomerase RNA of Modulates Telomerase in Response to DNA Damage
- The Orphan Gene Regulates Dauer Development and Intraspecific Competition in Nematodes by Copy Number Variation
- 9--13,14-Dihydroretinoic Acid Is an Endogenous Retinoid Acting as RXR Ligand in Mice
- The DnaA Protein Is Not the Limiting Factor for Initiation of Replication in
- FGFR3 Deficiency Causes Multiple Chondroma-like Lesions by Upregulating Hedgehog Signaling
- Multiple Changes of Gene Expression and Function Reveal Genomic and Phenotypic Complexity in SLE-like Disease
- Directed Evolution of RecA Variants with Enhanced Capacity for Conjugational Recombination
- The Regulatory T Cell Lineage Factor Foxp3 Regulates Gene Expression through Several Distinct Mechanisms Mostly Independent of Direct DNA Binding
- MreB-Dependent Inhibition of Cell Elongation during the Escape from Competence in
- DNA Damage Regulates Translation through β-TRCP Targeting of CReP
- Multiple In Vivo Biological Processes Are Mediated by Functionally Redundant Activities of and
- The Analysis of () Mutants Reveals Differences in the Fusigenic Potential among Telomeres
- The Causative Gene in Chanarian Dorfman Syndrome Regulates Lipid Droplet Homeostasis in .
- Temporal Expression Profiling Identifies Pathways Mediating Effect of Causal Variant on Phenotype
- The . Accessory Helicase PcrA Facilitates DNA Replication through Transcription Units
- AKTIP/Ft1, a New Shelterin-Interacting Factor Required for Telomere Maintenance
- Npvf: Hypothalamic Biomarker of Ambient Temperature Independent of Nutritional Status
- Transfer RNAs Mediate the Rapid Adaptation of to Oxidative Stress
- Connecting Circadian Genes to Neurodegenerative Pathways in Fruit Flies
- Response to “Ribosome Rescue and Translation Termination at Non-standard Stop Codons by ICT1 in Mammalian Mitochondria”
- Response to the Formal Letter of Z. Chrzanowska-Lightowlers and R. N. Lightowlers Regarding Our Article “Ribosome Rescue and Translation Termination at Non-Standard Stop Codons by ICT1 in Mammalian Mitochondria”
- Simultaneous DNA and RNA Mapping of Somatic Mitochondrial Mutations across Diverse Human Cancers
- Regulation of Insulin Receptor Trafficking by Bardet Biedl Syndrome Proteins
- Altered Levels of Mitochondrial DNA Are Associated with Female Age, Aneuploidy, and Provide an Independent Measure of Embryonic Implantation Potential
- Non-reciprocal Interspecies Hybridization Barriers in the Capsella Genus Are Established in the Endosperm
- Canine Spontaneous Head and Neck Squamous Cell Carcinomas Represent Their Human Counterparts at the Molecular Level
- Genetic Changes to a Transcriptional Silencer Element Confers Phenotypic Diversity within and between Species
- Functional Assessment of Disease-Associated Regulatory Variants Using a Versatile Dual Colour Transgenesis Strategy in Zebrafish
- Translational Upregulation of an Individual p21 Transcript Variant by GCN2 Regulates Cell Proliferation and Survival under Nutrient Stress
- Independent Neuronal Origin of Seizures and Behavioral Comorbidities in an Animal Model of a Severe Childhood Genetic Epileptic Encephalopathy
- The Human Blood Metabolome-Transcriptome Interface
- A Common Cancer Risk-Associated Allele in the Locus Encodes a Dominant Negative Inhibitor of Telomerase
- A Legume Genetic Framework Controls Infection of Nodules by Symbiotic and Endophytic Bacteria
- The Eukaryotic-Like Ser/Thr Kinase PrkC Regulates the Essential WalRK Two-Component System in
- The Yeast GSK-3 Homologue Mck1 Is a Key Controller of Quiescence Entry and Chronological Lifespan
- Dissection of a Complex Disease Susceptibility Region Using a Bayesian Stochastic Search Approach to Fine Mapping
- Exome Sequencing of Phenotypic Extremes Identifies and as Interacting Modifiers of Chronic Infection in Cystic Fibrosis
- The Role of -Mediated Epigenetic Silencing in the Population Dynamics of Transposable Elements in
- Ancestral Chromatin Configuration Constrains Chromatin Evolution on Differentiating Sex Chromosomes in
- Abnormal Activation of BMP Signaling Causes Myopathy in Null Mice
- Reproductive Mode and the Evolution of Genome Size and Structure in Nematodes
- Replication and Active Partition of Integrative and Conjugative Elements (ICEs) of the SXT/R391 Family: The Line between ICEs and Conjugative Plasmids Is Getting Thinner
- Motor and Sensory Deficits in the Mice Result from Mutation of the ESCRT Component HGS
- Senescence in the Sbds-Deficient Murine Pancreas: Cell-Type Specific Consequences of Translation Insufficiency
- Lipophorin Receptors Recruit the Lipoprotein LTP to the Plasma Membrane to Mediate Lipid Uptake
- Separable Crossover-Promoting and Crossover-Constraining Aspects of Zip1 Activity during Budding Yeast Meiosis
- Context-Dependent Functional Divergence of the Notch Ligands DLL1 and DLL4
- Multilayered Organization of Jasmonate Signalling in the Regulation of Root Growth
- Necrotic Cells Actively Attract Phagocytes through the Collaborative Action of Two Distinct PS-Exposure Mechanisms
- A Novel Feedback Loop That Controls Bimodal Expression of Genetic Competence
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Non-reciprocal Interspecies Hybridization Barriers in the Capsella Genus Are Established in the Endosperm
- Translational Upregulation of an Individual p21 Transcript Variant by GCN2 Regulates Cell Proliferation and Survival under Nutrient Stress
- Exome Sequencing of Phenotypic Extremes Identifies and as Interacting Modifiers of Chronic Infection in Cystic Fibrosis
- The Human Blood Metabolome-Transcriptome Interface
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání





