-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaReplication and Active Partition of Integrative and Conjugative Elements (ICEs) of the SXT/R391 Family: The Line between ICEs and Conjugative Plasmids Is Getting Thinner
Integrative and conjugative elements (ICEs) constitute a class of mobile genetic elements defined by their ability to integrate into the chromosome of their host cell and to transfer by conjugation. Some of the most studied ICEs belong to the SXT/R391 family, which are major drivers of multidrug resistance dissemination among various pathogenic Gammaproteobacteria. Transfer of SXT/R391 ICEs to a new host first requires its excision from the chromosome as a circular molecule, which may be lost if the cell divides. In silico analyses revealed several putative stabilization systems carried by R391, a prototypical member of the SXT/R391 ICEs family originally isolated from Providencia rettgeri. We discovered that, besides stabilization by integration into the chromosome, stability of SXT/R391 ICEs also depends on toxin/antitoxin systems and plasmid-like features including intracellular replication and active partition. Thus, although it has been known for a long time that ICEs and conjugative plasmids use similar strategies to transfer between bacterial populations, our work reveals additional unforeseen similarities in their mechanisms of maintenance in the host cell.
Published in the journal: . PLoS Genet 11(6): e32767. doi:10.1371/journal.pgen.1005298
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1005298Summary
Integrative and conjugative elements (ICEs) constitute a class of mobile genetic elements defined by their ability to integrate into the chromosome of their host cell and to transfer by conjugation. Some of the most studied ICEs belong to the SXT/R391 family, which are major drivers of multidrug resistance dissemination among various pathogenic Gammaproteobacteria. Transfer of SXT/R391 ICEs to a new host first requires its excision from the chromosome as a circular molecule, which may be lost if the cell divides. In silico analyses revealed several putative stabilization systems carried by R391, a prototypical member of the SXT/R391 ICEs family originally isolated from Providencia rettgeri. We discovered that, besides stabilization by integration into the chromosome, stability of SXT/R391 ICEs also depends on toxin/antitoxin systems and plasmid-like features including intracellular replication and active partition. Thus, although it has been known for a long time that ICEs and conjugative plasmids use similar strategies to transfer between bacterial populations, our work reveals additional unforeseen similarities in their mechanisms of maintenance in the host cell.
Introduction
Integrative and conjugative elements (ICEs) are highly prevalent and widely distributed in bacterial genomes [1–3]. Their ability to self-transfer by conjugation between genetically unrelated bacteria contributes to the emergence of multidrug resistant pathogens in diverse taxonomic groups [4–6]. ICEs usually reside within and replicate with the host cell’s chromosome to be vertically inherited. ICEs eventually excise from the chromosome and form circular covalently closed molecules that serve as the substrate for the conjugative machinery that translocates the ICE DNA to recipient cells [6, 7]. With a few exceptions reported only for ICEs of Actinobacteria, this conjugative machinery usually consists of a relaxase, a type IV coupling protein and a type IV secretion system [1–3, 8].
The SXT/R391 family of ICEs encompasses one of the largest and most diverse set of ICEs studied, including elements that have been found over the past 40 years in clinical and environmental isolates of diverse species of Gammaproteobacteria [9, 10]. ICEs of the SXT/R391 family largely contribute to the spread of antibiotic resistance genes in the seventh-pandemic lineage of Vibrio cholerae, the etiologic agent of cholera, which remains a major cause of mortality and morbidity on a global scale [11]. The ICE SXT is a prototypical member of the SXT/R391 family originally isolated from a 1992 Indian multidrug resistant clinical isolate of V. cholerae O139 [12]. SXT and several variants detected in V. cholerae O139, O1 and non-O1 non-O139 isolates confer resistance to sulfamethoxazole, trimethoprim, streptomycin and chloramphenicol [9]. The second prototypical member of this family is R391, which was originally detected in a 1967 South African isolate of Providencia rettgeri [13]. R391 confers resistance to kanamycin and mercury. Members of the SXT/R391 family all share a common integration site, the 5’ end of prfC, and a highly conserved core of genes and sequences that mediate their regulation, integration/excision and conjugative transfer [10]. Expression of the conjugative function of SXT/R391 ICEs is tightly regulated by SetR, which represses the expression of the master activator genes setC and setD. Their products activate transcription of int, xis and conjugation-associated operons [14]. Repression of setC and setD is alleviated by induction of the bacterial response to DNA damage, which promotes autoproteolysis of SetR [15].
SXT and R391 can exist co-integrated in a tandem fashion in prfC in the same host cell [16, 17]. Such tandem arrays are suitable substrates for frequent homologous recombination events yielding hybrid ICEs that can be easily segregated in exconjugant cells [16, 18]. Interestingly, R391 was reported to be found as a circular extrachromosomal replicative form in a recA recipient strain bearing an integrated copy of R997, another SXT/R391 ICE found in Proteus mirabilis [19]. A similar behavior was also reported for R997 entering a recA recipient bearing an integrated R391. However, no extrachromosomal form of R391 or R997 could be recovered from recA+ hosts. These observations suggest that, at least in specific circumstances, SXT/R391 ICEs are capable of autonomous replication. Autonomous replication was previously suspected for several ICEs and recently well characterized for ICEBs1, an ICE of the Gram-positive bacterium Bacillus subtilis [20–25]. Plasmid-like replication was also shown to be essential for the stability of ICEBs1 [24]. However, whether autonomous replication is relevant to the biology and stability of SXT/R391 ICEs remains to be established.
Breaking with old paradigms about ICEs, we report here that replication is a key step of the lifecycle of SXT/R391 ICEs by using R391 as a model. By monitoring the frequency of excision, the ICE copy number as well as the frequency of loss of a set of mutants, we show that the putative relaxase TraI and the origin of transfer (oriT) are essential for R391 replication and its stability in the progeny of host cells. Furthermore, we demonstrate that, besides diverse non-conserved toxin-antitoxin systems, all SXT/R391 ICEs also encode a conserved plasmid-like type II partitioning system that enhances their stability. Together, these results unravel an unforeseen similarity between the biology of ICEs and conjugative plasmids.
Results
Dynamics of R391: Evidence for autonomous replication
To have a better understanding of SXT/R391 ICEs biology, we evaluated five key factors of R391 lifecycle in Escherichia coli: (i) the dynamics of excision/integration, which will be reported as the frequency of excision in the rest of the manuscript, (ii) the frequency of transfer, (iii) the average copy number per cell in the whole cell population, (iv) the average number of extrachromosomal circular copies per cell, and (v) the ICE stability in the cell population. The frequency of R391 excision was assessed by quantifying by real-time quantitative PCR (qPCR) the relative amount of free integration site (attB) resulting from R391 excision per chromosome as measured by the amount of chromosomal prfC target (Fig 1A). R391 excised at a frequency of 1.90×10-3 ± 0.38×10-3 (Fig 2A), which is about tenfold lower than the excision frequency of SXT (1.76×10-2 ± 0.65×10-2, P = 0.0140, two-tailed Student t-test) in similar conditions. Mating assays showed that R391 transfers at about 5.01×10-4 ± 0.31×10-4 exconjugant/donor (Fig 2B), which is about 20 fold higher than SXT (2.71×10-5 ± 0.55×10-5 exconjugant/donor). Hence there is no correlation between the frequency of excision of these elements measured in the whole cell population and their respective frequency of transfer. This observation indicates that excision is not a factor that limits the rate of dissemination of these two ICEs.
Fig. 1. Genetic determinants of R391 stability. 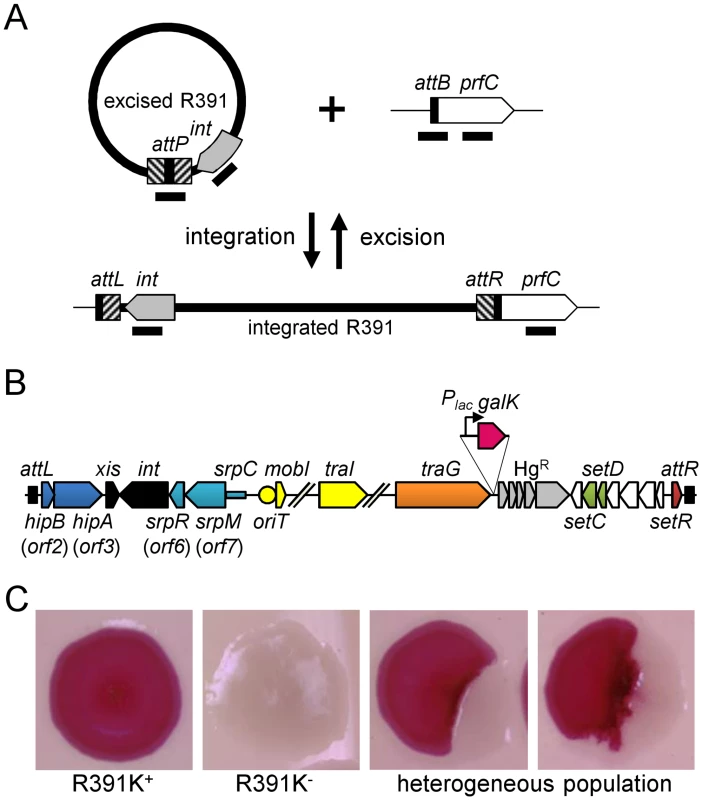
(A) Quantification of R391 integration/excision and replication. The DNA targets (attP, attB, int and prfC) measured by real-time quantitative PCR are shown by black bars. (B) Schematic representation of R391 and R391K structures. The position and orientation of open reading frames (ORFs) are indicated by arrowed boxes. The colors depict the function deduced from functional analyses and BLAST comparisons: dark blue, toxin/antitoxin system; black, site-specific recombination; light blue, active partitioning system; yellow, DNA processing; orange; mating pair formation; light grey, mercury resistance (HgR); green, transcriptional activators; red, transcriptional repressor; white, other or unknown functions. The putative centromere-like region (srpC) and the origin of transfer (oriT) are depicted by a light blue rectangle and a yellow circle, respectively. hipAB and the mercury resistance genes are not conserved features of SXT/R391 ICEs. To construct R391K, the reporter gene galK (in magenta) under the control of the IPTG-inducible Plac promoter was inserted between traG and merR, the first gene of the mercury resistance operon. (C) Colony color-based assay for the detection of R391K-containing colonies on MacConkey D-galactose indicator agar. Colonies bearing galK-tagged R391 (R391K) appeared red on the indicator medium (R391K+) while white colonies lacked the ICE (R391K-). Sectored colonies forming during growth on agar plates are indicative of high instability of R391K mutants. Fig. 2. Effects of the deletion of hipA or abolition of conjugative transfer (ΔtraG) on R391 dynamics. 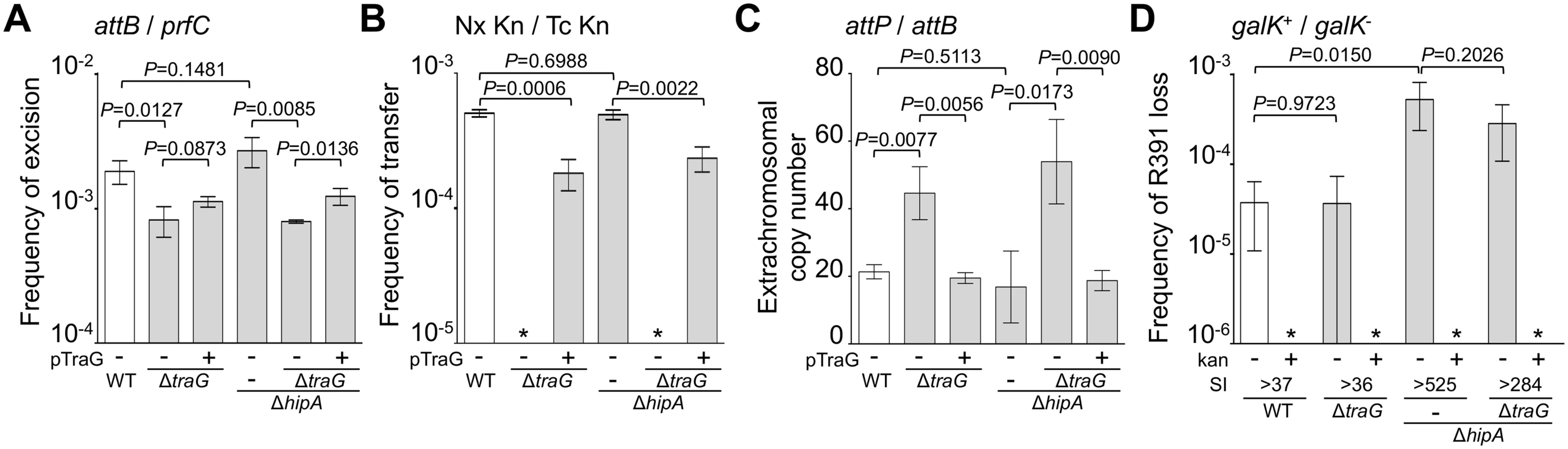
(A) Effect on R391K excision. The frequency of excision of the wild-type R391K ICE (WT) or its ΔhipA and ΔtraG mutants corresponds to the attB/prfC ratio measured by qPCR. (B) Effect on R391K transfer. Conjugation assays were carried out using E. coli VB38 containing R391K (WT) or its ΔhipA and ΔtraG mutants as donors. E. coli MG1655 Nx (VB111) was used as the recipient. Transfer frequencies are expressed as the number of exconjugant per donor CFUs. The asterisk indicates that the frequency of exconjugant formation was below the detection limit (<10-8). (C) Effect on the copy number of excised R391K. Extrachromosomal copy numbers of the wild-type R391 (WT) and its ΔhipA and ΔtraG mutants correspond to attP/attB ratios measured by qPCR. (D) Effect on R391K stability. The frequency of loss of wild-type R391K (WT) or its ΔhipA and ΔtraG mutants was measured as the ratio between the number of R391K- (white colonies) and the number of R391K+ (red colonies) CFUs on MacConkey galactose agar plates after 16 hours of growth in LB medium with (+) or without (-) antibiotic. The stability index (SI) corresponds to the ratio of the frequencies of loss observed with and without antibiotic. The asterisk indicates that the frequency of loss was below the detection limit (<10-6). For panels A, B and C, complementation of the ΔtraG mutation was carried out using traG expressed from an arabinose-inducible promoter (PBAD) provided by pTraG. In all panels, the bars represent the means and standard deviation values obtained from at least three independent experiments. Statistical analyses were carried out using two-tailed Student’s t-tests and the P-value is indicated above the brackets comparing two bars. Using the same approach, we then measured the mean copy number per cell of R391 in the whole cell population as the ratio between the amount of R391-borne int target and the amount of chromosomal prfC target (Fig 1A). This ratio was found to be 0.96 ± 0.04 as expect for a single copy of R391 integrated in the chromosome. We also measured the mean copy number of the extrachromosomal circular form of R391 per cell by establishing the ratio between the amount of attP recombination site resulting from R391 excision and the amount of unoccupied chromosomal attB sites. In theory, each event of R391 excision is expected to yield one unoccupied attB site on the chromosome and one attP site on the circular excised R391 (attP/attB = 1). We observed that this ratio reached 21 ± 2 (Fig 2C), suggesting that R391 is capable of replicating in a small subset of the cell population in which it is excised from the chromosome. This observation is consistent with results previously reported for SXT, for which 4 attP sites on average exist for each unoccupied attB site [16, 26]. New measurements carried out in this study to confirm these reports revealed 3.6 ± 0.2 attP sites per unoccupied attB site for SXT.
We then assessed the stability of R391 by monitoring the number of cells lacking R391K in the cell population after 16 hours of growth (about 20 generations) in LB medium with or without selective pressure. R391K is tagged with the galK reporter gene under the control of the Plac promoter to enable high-level galactokinase activity in a lacI mutant strain such as E. coli VB38 [18], a ΔgalK derivative of E. coli CAG18439 (lacI42::Tn10) (Fig 1B). The frequency of loss was determined as the percentage of white colonies (galK-, devoid of R391K) on MacConkey indicator agar supplemented with 1% galactose (Fig 1C). R391K was found to be inherently stable because it was lost in only 0.0037% of the cell population in the absence of selective pressure (Fig 2D, WT), whereas no detectable loss was observed when cells were grown with kanamycin in liquid culture (detection limit of 0.0001%).
A HipAB-like toxin-antitoxin system stabilizes R391
Stability of many mobile genetic elements relies on a post-segregational killing mechanism, which induces a strong selective disadvantage or even death to cells that have lost them [27–31]. While previous studies have shown that two functional toxin-antitoxin (TA) systems, mosAT and s045-s044, enhance the stability of SXT [32, 33], neither of these TA systems was found in R391. Nevertheless, in silico analysis of the R391 sequence revealed that the two overlapping open reading frames (ORFs) orf02 and orf03, which belong to the variable region I located upstream of xis, encode a putative hipAB-like TA system (Fig 1B) [34, 35]. Indeed, orf03 (hipA) is predicted to encode a HipA-like toxin, while orf02 (hipB) likely codes for the HipB cognate antitoxin, which carries a DNA-binding HTH-XRE (HTH_19) domain. To measure the impact of this putative hipAB-like TA system on R391 stability, we constructed a ΔhipA mutant of R391K. This mutation did not impair the transfer of R391K and had no effect on the excision or extrachromosomal copy number of the element (Fig 2A, 2B and 2C, compare WT and ΔhipA). However, ICE stability was affected as R391K loss increased by 12 fold for the ΔhipA mutant compared to wild-type (Fig 2D). No loss of R391K ΔhipA was detectable in the presence of kanamycin. These results revealed the functionality of the hipAB TA system and its involvement in the stability of R391, as previously demonstrated for mosAT of SXT [32]. However, like the mosAT and s045-s044 loci of SXT, hipAB of R391 is not a conserved feature of SXT/R391 ICEs; therefore hipAB is likely not an inherent and ancestral mechanism used by SXT/R391 ICEs to enhance their stability in their respective hosts. Cell death or growth reduction associated with hipAB after R391 loss was likely to hinder our investigations on R391 stability. To circumvent this issue, the ΔhipA mutant provided us with a useful tool for additional investigations aimed at unraveling other stabilization mechanisms conserved among ICEs of the SXT/R391 family.
Conjugation does not stabilize SXT/R391 ICEs
Conjugation has been shown to be a powerful stabilization mechanism for conjugative plasmids that can reenter in cells having lost them by infectiously spreading in the cell population [28, 36]. While Wozniak and Waldor [32] have shown that conjugation does not promote SXT loss, their assay does not allow to conclude whether conjugation is an efficient mechanism of stabilization of SXT/R391 ICEs. To answer this question, we looked at the frequency of loss of a ΔtraG mutant of R391K. traG codes for an inner-membrane component of the donor cell mating pair formation apparatus that is essential for SXT transfer [37]. As previously reported for SXT, deletion of traG abolished R391K transfer (Fig 2B). However, the inability to transfer did not reduce the stability of R391K, as the frequencies of loss of wild-type R391K and its ΔtraG mutant were nearly identical (Fig 2D). No detectable loss of R391K ΔtraG was observed when kanamycin was added during liquid culture. The frequencies of excision of the ΔtraG and ΔhipA ΔtraG mutants were reduced by ~2.3 fold, while their extrachromosomal copy numbers were more than twice as high as wild-type R391K and its ΔhipA mutant, reaching up to ~54 copies per cells (Fig 2A and 2C). This observation suggests that, once excised from the chromosome, the circular form of R391K accumulates in the cell possibly because a defective mating apparatus caused by the ΔtraG mutation cannot mediate its transfer to a recipient cell.
The putative relaxase TraI is key for transfer, replication and stability of R391
The plasmid-like replication of ICEBs1 was shown to be essential for its stability [24]. Rolling-circle replication of ICEBs1 requires the relaxase NicK and oriT, a cis-acting locus initiating the translocation of DNA through the mating pore.
In a subset of a cell population, R391 seems to be in a multicopy plasmid-like state that may be important for preserving ICE stability when it is excised from the chromosome in actively dividing cells. To test this hypothesis, oriT and traI deletion mutants were constructed in R391K (Fig 1B). traI codes for the putative relaxase of SXT/R391 ICEs that recognize the origin of transfer (oriT) [38]. As expected, the ΔtraI mutation abolished R391K conjugative transfer (Fig 3B). We also observed a ~5-fold reduction of the extrachromosomal copy number of R391K when either traI or oriT were missing compared to wild-type (Fig 3C). The ΔtraI mutation also led to a ~20-fold increase of the frequency of excision and to an 11-fold increase of R391K loss (Fig 3A, 3B and 3D). Combined ΔtraI and ΔhipA mutations led to a 34-fold increase of R391K loss, thereby confirming that traI is important for R391 stability (Fig 3D). Expression of traI in trans from the arabinose-inducible PBAD promoter in pTraI restored and even enhanced the transfer and the stability of both ΔtraI and ΔtraI ΔhipA mutants compared to wild-type (Fig 3B and 3D). Although the copy number of the plasmid-like form of R391K ΔtraI doubled upon complementation with pTraI, it failed to reach the wild-type level (Fig 3C). Interestingly, ΔtraI mutants were so unstable that selective pressure exerted by kanamycin in liquid culture did not, or only slightly, improved R391K stability (Fig 3D). The high instability affecting ΔtraI mutants also led to the formation of sectored colonies on agar plates likely resulting from loss of R391K during colony development (Fig 1C).
Fig. 3. traI and oriT are key factors for R391 replication and stability. 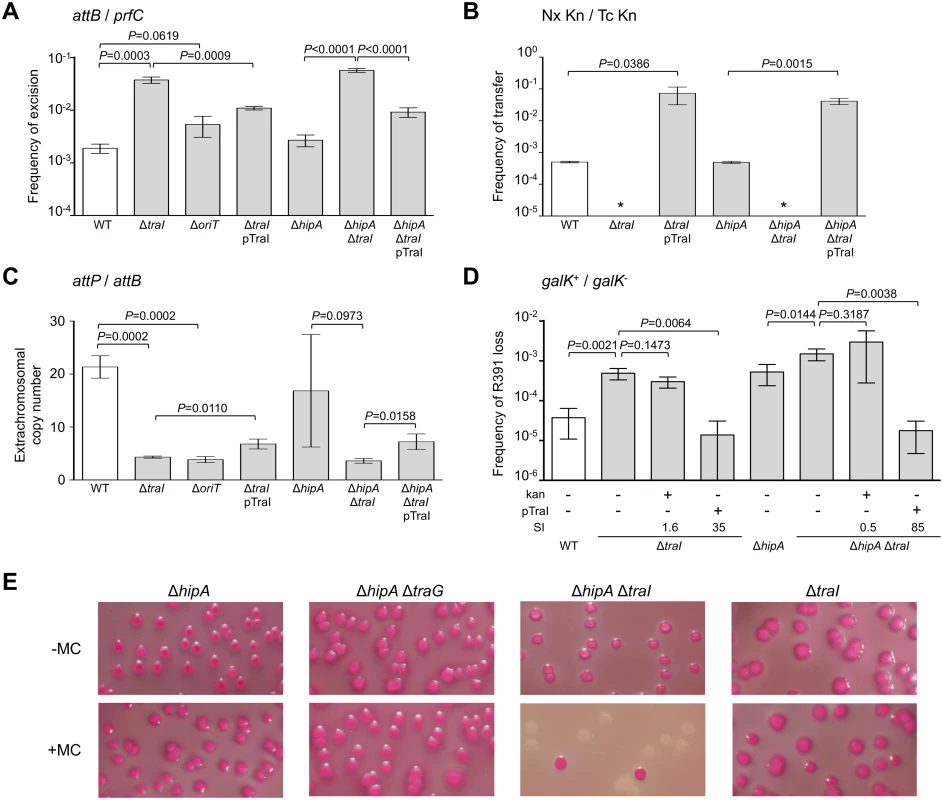
(A) Effect of traI or oriT deletions on R391K excision. (B) Effect of traI deletion on R391K transfer. (C) Effect of traI and oriT deletions on the copy number of excised R391K. (D) Effect of traI deletion on the stability of R391K. For all panels, the experiments were carried out as described in Fig 2. Complementation of the ΔtraI mutation was carried out using traI expressed from an arabinose-inducible promoter (PBAD) provided by pTraI. (E) Effect of mitomycin C on the stability of R391K mutants observed on MacConkey galactose agar after 16-hour growth in LB medium with (+) or without (-) mitomycin C (MC). Since conjugative transfer of SXT/R391 ICEs is known to be stimulated by DNA-damaging agents, we tested the effect of mitomycin C on the stability of the ΔhipA, ΔtraG and ΔtraI mutants of R391K. We observed that the drug did not induce high-frequency loss of the ΔhipA, ΔhipA ΔtraG or ΔtraI mutants of R391K (Fig 3E). In striking contrast, deletion of both ΔhipA and ΔtraI led to a hypersensitivity of R391K to mitomycin C treatment as the ICE was lost in more than 90% of the cell population (Fig 3E). We suspect that ΔtraI mutants are highly unstable; yet in the presence of hipAB, cells that have lost R391K ΔtraI likely have no progeny or strong growth reduction due to the persistence of the HipA toxin, thereby masking this high instability in conditions that strongly induce R391 excision.
SXT/R391 ICEs encode a functional plasmid-like active partition system
In silico analysis of R391 sequence using CD-search on the Conserved Domain Database v3.11 [39, 40] and protein fold recognition server Phyre2 [41] revealed that orf07, the first gene of an operon containing int, codes for a predicted actin-like NTPase structurally related to the ParM plasmid segregation proteins of plasmids R1 and pSK41 (Fig 1B). ParM proteins are a key component of type II ParMRC partitioning systems that mediate plasmid DNA segregation during cell division via a pushing mechanism [42, 43]. ParR adaptor protein connects parC, a cis-acting centromere-like locus, to the ParM filament. ParR proteins have low conservation and their genes are found downstream of the parM gene. The open reading frame orf06, located downstream of orf07, is predicted to code for a small basic protein (pI 9.3) with no recognizable domain (Fig 1B). Hence, orf06 may encode a ParR DNA-binding protein that binds the centromere-like region in partitioning systems. Based on these observations and results described below, orf06 and orf07 were renamed srpR and srpM for SXT/R391 ICEs partitioning proteins R and M, respectively (Fig 1B). By functional analogy with the parMRC partitioning systems carried by the plasmid R1 of E. coli and the staphylococcal plasmid pSK41 [42], the DNA fragment located upstream of srpM likely corresponds to the centromere-like region bound by SrpR and was annotated srpC (Fig 1B). The srpMRC locus is strictly conserved in all SXT/R391 ICEs, suggesting that it may somehow play an important role in their biology.
Deletion of srpR, srpM or both had no measurable effect on the frequency of excision or the extrachromosomal copy number of R391K ΔhipA, and had a slight inhibitory effect (about 4 - to 14-fold reduction) on the frequency of transfer (Fig 4A, 4B and 4C). The lack of impact on the excision frequency confirmed that neither deletion has a polar effect on the expression of the int gene located immediately downstream of srpR (Figs 1B and 4A). Since the deletion of srpM had very little effect on R391 transfer, we used this mutation to further study the phenotype associated with a non-functional srpMRC locus. Cumulative mutations of traG, hipA and srpM did not affect the frequencies of excision or loss of R391K compared to the ΔtraG ΔhipA mutant (Fig 4A and 4D). However, the extrachromosomal copy number of R391K dropped nearly 4 fold when comparing the same mutants (Fig 4C). Furthermore, a ΔtraI ΔhipA ΔsrpM triple mutant exhibited a visible but statistically non-significant 35% reduction of stability compared to the ΔtraI ΔhipA mutant (Fig 4D). However, deletion of srpM led to a 3-fold increase of the stability of R391K ΔtraI ΔhipA after 40 generations (Fig 4D, dark grey bars). Finally, overexpression of traI from pTraI did not completely prevent the loss of R391 ΔtraI ΔhipA ΔsrpM, which was lost about 4 times more frequently than the ΔtraI ΔhipA mutant (Fig 4D). These data revealed that srpM is important for R391 stability when the number of copy of the ICE is low and thus could be a functional active partition system.
Fig. 4. Effect of the SrpMRC active partition system on R391 dynamics. 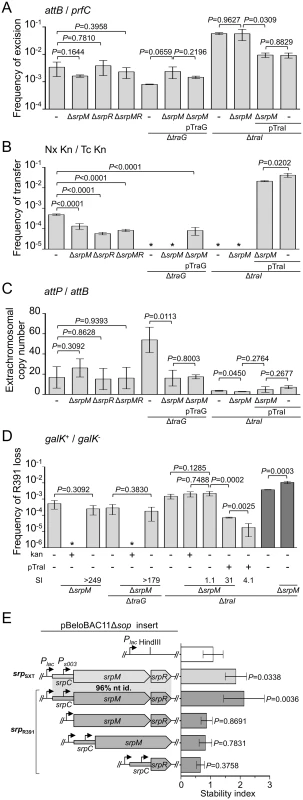
(A) Effect of srpMRC inactivation on R391K excision. (B) Effect of srpMRC inactivation on R391K transfer. (C) Effect of srpMRC inactivation on the copy number of excised R391K. (D) Effect of srpMRC inactivation on R391K stability. For panels A to D, experiments were carried out as described in Fig 2 using R391K ΔhipA derivatives and complementation of the ΔtraI and ΔtraG mutations were carried out using traI and traG expressed from an arabinose-inducible promoter (PBAD) provided by pTraI and pTraG, respectively. For panel D, stability assays carried out over ~40 generations are indicated by dark grey bars. (E) The srpMRC locus stabilizes a partition-deficient single-copy plasmid. The relative stability of the single-copy plasmid pBeloBAC11Δsop and derivatives carrying all or parts of the srpMRC locus under control of the Plac promoter were tested in E. coli TOP10F’. Expression from Plac was induced using 0.02 mM of IPTG and cells were grown for 16 hours in M9 minimal medium. The stability index (SI) corresponds to the ratio of the frequencies of loss observed with and without antibiotic (D) or with and without IPTG (E). The bars represent the mean and standard deviation values obtained from at least three independent experiments. Statistical analyses in panel E were performed using one-way ANOVA with Tukey’s multiple comparison test. P-values indicated above the bars refer to comparison with pBeloBAC11Δsop. To test further whether srpMRC is a functional DNA partitioning system, plasmid stabilization assays were carried out using pBeloBAC11Δsop, an unstable derivative of the single-copy plasmid pBeloBAC11 that lacks its native sopABC partitioning system. In SXT/R391 ICEs, expression of srpM, srpR and int was shown to be driven from Ps003, a promoter exclusively dependent upon activation by the transcriptional activator SetCD (Fig 1B) [14]. To bypass the need for SetCD, the srpMRC loci of R391 and SXT were cloned into pBeloBAC11Δsop downstream of the IPTG-inducible Plac promoter (Fig 4E). Expression of srpMRC loci of SXT (pSrpSXT) or R391 (pSrpR391) upon IPTG induction led to a respective ~1.8 and ~2.1-fold increase of pBeloBAC11Δsop stability, thereby confirming that srpMRC is a functional plasmid stabilization system (Fig 4E and S1 Fig). The absence of srpR, srpM or srpC prevented plasmid stabilization, which was then comparable to the empty vector (Fig 4E and S1 Fig).
SrpR binds the centromere-like sequence srpC of SXT/R391 ICEs
SrpR lacks homologies with known ParR proteins that have been shown to bind parC-centromere-like sequences upstream of parMR genes in plasmids such as R1. To test whether SrpR is capable of binding the srpC locus, we carried out electrophoretic mobility shift essays (EMSA) experiments using purified C-terminally 6xHis-tagged SrpR protein (predicted molecular weight of 10.4 kDa). EMSA assays revealed a specific binding of SrpR to the 615-bp fragment ig(srpM-mobI) which corresponds to the intergenic region between srpM and mobI and likely contains the srpC region (Fig 5A and 5BI). Addition of high concentrations of sonicated salmon sperm DNA (non-specific competitor) did not destabilize SrpR binding to this probe. Further investigation confirmed that SrpR specifically binds the 251-bp srpC region as SrpR binding to srpC was resilient to the addition of the non-specific competitor DNA (Fig 5A and 5BII). The presence of multiple specific shifts suggests that SrpR binds multiple sites or binds as different multimeric forms (Fig 5BII). While SrpR was able to bind to the 298-bp fragment containing oriT, addition of the non-specific competitor DNA destabilized SrpR binding (Fig 5BIII). Non-specific SrpR binding to oriT indicates that SrpR exhibits a significant non-specific affinity for DNA molecules.
Fig. 5. SrpR binds the centromere-like region srpC. 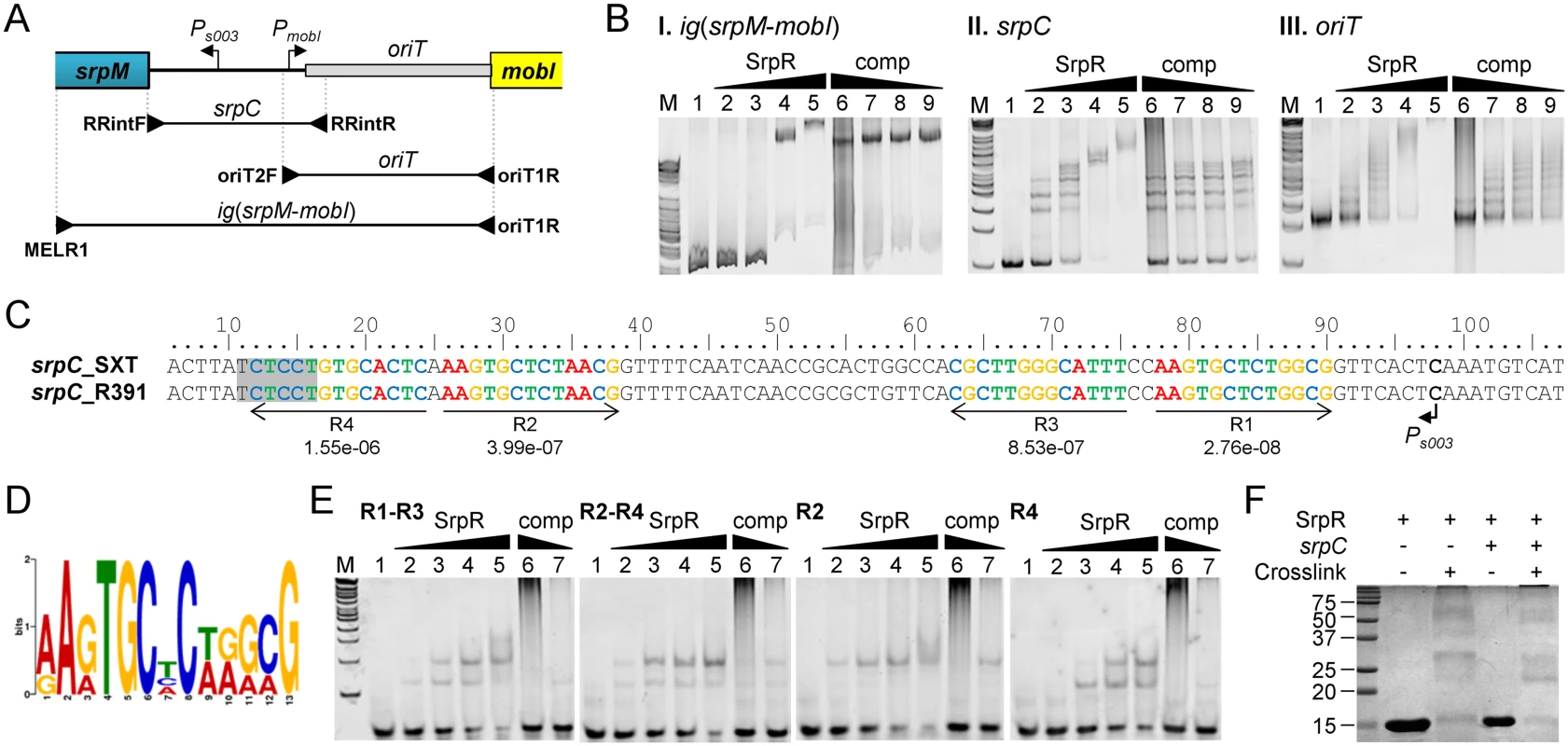
(A) Schematic representation of the intergenic region between the srpMR-int operon and mobI. The positions of oriT and SetCD-dependent promoters Ps003 and PmobI were determined previously [14, 38]. The position of primers used to amplify the probes used in panel B are indicated by arrowheads. (B) Electrophoretic mobility shift assays (EMSA) of DNA fragments of the intergenic region bound by SrpR. Lane M, 2-log DNA ladder; lane 1, 40 ng of DNA probes alone; lanes 2–5, 10, 20, 40, 80 ng of SrpR for ig(srpM-mobI) or 25, 50, 75, 100 ng of SrpR for oriT and srpC; lanes 6–9, 40 ng (ig(srpM-mobI)), 75 ng (oriT) or 50 ng (srpC) of SrpR and 200, 40, 20, 10 pg of sonicated salmon sperm DNA as non-specific competitor DNA (comp). (C) Alignment of the complementary strand of the 5’ UTR upstream of srpM. The repeated sequences identified by MEME are shown with their respective p-values. The shaded area depicts the predicted Shine-Dalgarno sequence of srpM. (D) Logo sequence generated from the four repeated sequences shown in C. (E) EMSA of SrpR on the binding sites R1-R3, R2-R4, R2 and R4. Conditions are the same as those used for srpC with either 200 or 40 pg of sonicated salmon sperm DNA. (F) SrpR multimerization assays. The size in kDa of the Precision Plus Protein Kaleidoscope Standards (BioRad) is indicated on the left side. In silico analysis of the centromere-like srpC region of R391 and SXT using the Multiple Em for Motif Elicitation tool (MEME) [44] revealed four conserved 13-bp direct and inverted repeats that might be recognized by SrpR (Fig 5C and 5D). EMSA results showed that a 40-bp fragment containing either R1-R3 or R2-R4 was bound by SrpR (Fig 5E). Addition of competitor DNA strongly decreased SrpR binding but did not completely alleviate the interaction. Binding of SrpR to the sequences R2 or R4 used as probes was abolished by the addition of the competitor suggesting that half-sites do not produce stable complexes with SrpR (Fig 5E).
Since ParR-like DNA binding proteins have been shown to form multimeric complexes [45], SrpR multimerization assays were carried out using glutaralehyde cross-linking. These assays suggests that, even without any DNA substrate, SrpR seems to be able to assemble as dimeric and tetrameric complexes in solution as shown by the apparition of large bands migrating at compatible molecular weights in a SDS page gel (Fig 5F). Copious amounts of SrpR were also trapped in the well, thereby suggesting that SrpR could be able to assemble in complexes of higher order.
SrpMRC of SXT/R391 ICEs is closely related to a putative partitioning system conserved in IncA/C conjugative plasmids
To assess the relationship of SrpMRC with other type II partitioning systems, we carried out a phylogenetic analysis based on the ParM actin-like homologs found by BlastP. Since the ParR adaptor proteins and parC sequence usually retain low conservation, they were not included in the analysis. Our analyses revealed that as expected, SrpM of R391 clusters with close orthologs encoded by all SXT/R391 (Fig 6A, green branch). SrpM is also closely related to ParM orthologs encoded by a putative type II partitioning system carried by conjugative plasmids of the IncA/C (Fig 6A, red branch) and pAQU groups [46–48]. SrpM and all of these orthologs cluster with more distantly related plasmids such as Rts1 (IncT) [49] and the catabolic plasmids pCAR1/pDK1 (IncP7) [50, 51]. Interestingly, with the exception of Rts1, which seems to lack a gene coding for a ParR protein, the genetic contexts of the orthologous parMRC loci in all these mobile elements are strikingly similar, located between traG - and mobI-like genes, thereby supporting their common ancestry and divergent evolutionary pathways (Fig 6B). This large group of related par loci is distantly related those carried by diverse plasmids broadly distributed among bacterial species of Delta- and Gammaproteobacteria, including the most distantly related parMRC systems carried by the conjugative plasmids R1 and R27 (Fig 6A) [52–54].
Fig. 6. Genetic context and molecular phylogenetic analysis of SrpM. 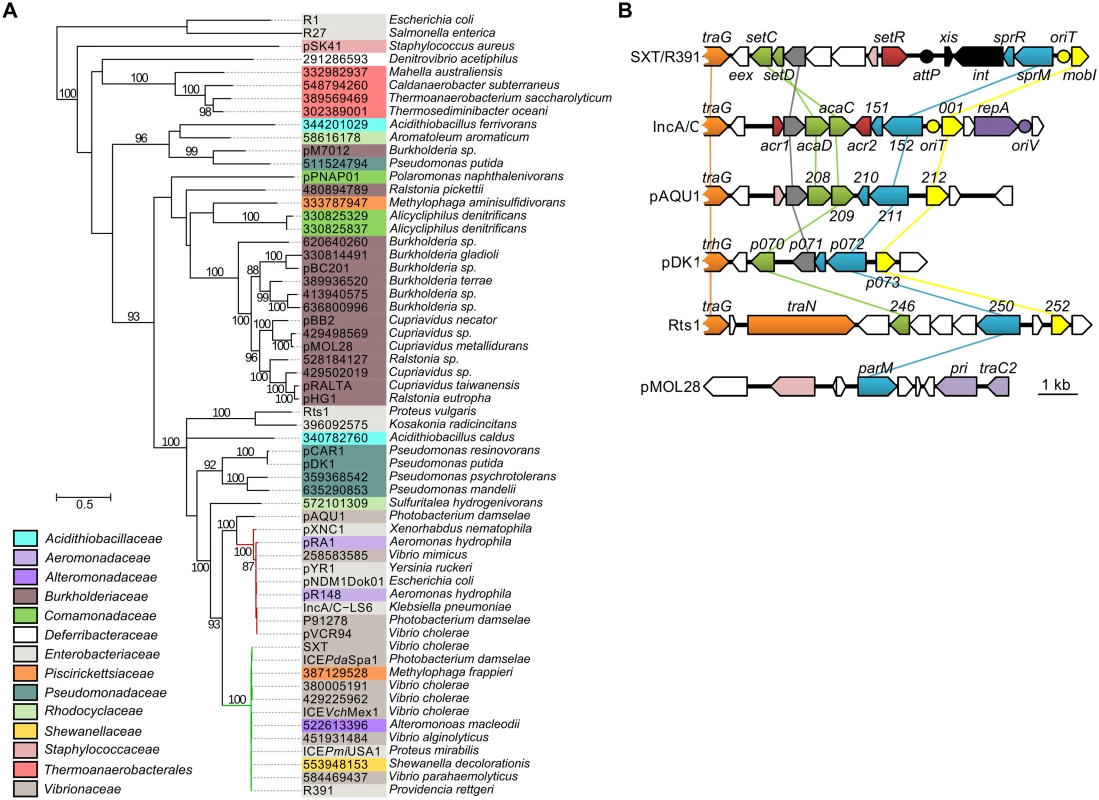
(A) The evolutionary history of SrpM was inferred by using the Maximum Likelihood method based on the Le and Gascuel model [55]. The tree with the highest log likelihood (-15514.1930) is shown. The percentage of trees in which the associated taxa clustered together is shown next to the branches (≥80% cut-off). The background color of each leaf indicates the taxomonic family of original host species from which each element was isolated. SrpM orthologs indicated by their protein sequence GI number are those for which the original element could not be easily identified from data gathered from Genbank and/or for which plasmid naming was inconsistent or not following the rules of Novick et al. [56]. The SXT/R391 ICEs and IncA/C plasmids lineages are shown by green and red branches, respectively. (B) Comparison of the genetic context of genes coding for SrpM orthologs in SXT/R391 ICEs (SXT, AY055428.1), IncA/C plasmids (pVCR94, KF551948), pAQU1 (NC_016983.1), pDK1 (NC_014124.1), Rts1 (NC_003905.1), and pMOL28 (CP000355.2). Arrows of similar color represent genes predicted to have similar functions. Green, transcriptional activator; yellow, MobI-like homologs; red and pink, transcriptional repressor; orange, conjugative transfer; grey, lytic transglycosylase; mauve, replication; black, site-specific recombination; blue, partitioning system; white, other or unknown functions. Yellow circles indicate the position of origins of transfer (oriT). The mauve circle indicates the position of the origin of replication (oriV) of pVCR94. The black circle indicates the position of the attP site for chromosomal integration of SXT by site-specific recombination. Discussion
In our modern world, antibiotics are widespread in most environments, subjecting microorganisms to a strong and constant selective pressure [57, 58]. ICEs circulating among environmental and pathogenic bacteria can take advantage of this selective pressure by collecting and accumulating antibiotic resistance-conferring genes. The selective advantage conferred by antibiotic resistance enhances the stability of ICEs in their hosts as well as their odds to eventually spread into and invade a new bacterial population. However, ICEs likely predate the antibiotic era and have evolved other means to prevent their loss. Indeed, several ICEs are stably maintained despite the lack of genes coding for any obvious selective advantage for their host [59, 60]. One strategy of stabilization consists in a tight control of the excision of the ICE from the chromosome. However, too tight a regulation could prevent its efficient dissemination. For ICEs of the SXT/R391 family, excision and transfer were shown to be coupled with the activation of the host’s SOS response [15]. In bacteria such as E. coli, spontaneous induction of the SOS response in the absence of DNA damaging agents has been shown to occur in 0.3 to 3% of the cell population [61], thereby inherently leading to unscheduled excision that is detrimental to ICE stability. Indeed, cell division occurring after ICE excision can generate ICE-free cell lineages, which likely have a competitive advantage in the absence of selective pressure.
Between attL and xis, R391 bears genes coding for a HipAB-like TA system that enhances the stability to the ICE as inactivation of hipA increased R391K loss by 12-fold (Fig 2D). hipAB is also found at the same position in ICEVchMex1, another member of the SXT/R391 family, which does not seem to confer any heavy metal or antibiotic resistance to its original host [10, 60]. The toxin/antitoxin system mosAT has been shown to strongly improve the stability of SXT [32]. Interestingly, mosAT expression was found to be correlated with activation of SXT excision and conjugative transfer [32]. However, coupling of mosAT expression with SXT excision was later shown to be circumstantial to the activation by SetCD of the expression of the upstream traVA genes [14]. Furthermore, since neither mosAT nor hipAB are conserved in all SXT/R391 ICEs [10], element-specific TA systems located in variable regions should only be considered as auxiliary determinants of stabilization for this family of ICEs. The same holds true for the tad-ata-type TA system s044-s045 carried by SXT in the variable region located between traIDJ and traLEKB [10, 33].
In addition to diverse TA systems encoded by variable DNA, we have shown here that SXT/R391 ICEs rely on specific and conserved strategies to enhance their stability within their host genome. Besides the most obvious one, which is their integration within the host chromosome, our data support the notion that SXT/R391 ICEs are not only capable of replication, but they can also actively segregate the resulting plasmid-like forms.
Conjugation has been shown to be a possible stabilization mechanism for IncP-1 conjugative plasmids in cell populations, allowing the recolonization of plasmid-free cells [36, 62]. However, our results show that conjugation is not a key factor for the stability of SXT/R391 ICEs as a traG mutant that is unable to transfer was no less stable than wild-type R391K (Fig 2D). Interestingly, we found that deletion of traG, which prevents translocation of the ICE DNA to the recipient cell, had unexpected side effects. In such mutants, the frequency of excision decreased while the extrachromosomal copy number increased (Fig 2A and 2C). A plausible explanation for this phenotype is that plasmid-like molecules of R391K can somehow accumulate due to the blocked mating pore. This accumulation of R391K circles would then tend to displace the site-specific recombination reaction from excision toward reintegration, hence the lower excision frequency (Figs 2A and 7). Consistent with this hypothesis, deletion of traI produced the exact opposite effect. We observed that while deletion of traI drastically reduced the number of R391K circles, the frequency of excision was very much increased (Fig 3A and 3C). Impaired replication of R391K seems to displace the site-specific recombination reaction equilibrium toward excision instead of integration (Fig 7).
Fig. 7. General dynamics of SXT/R391 ICEs. 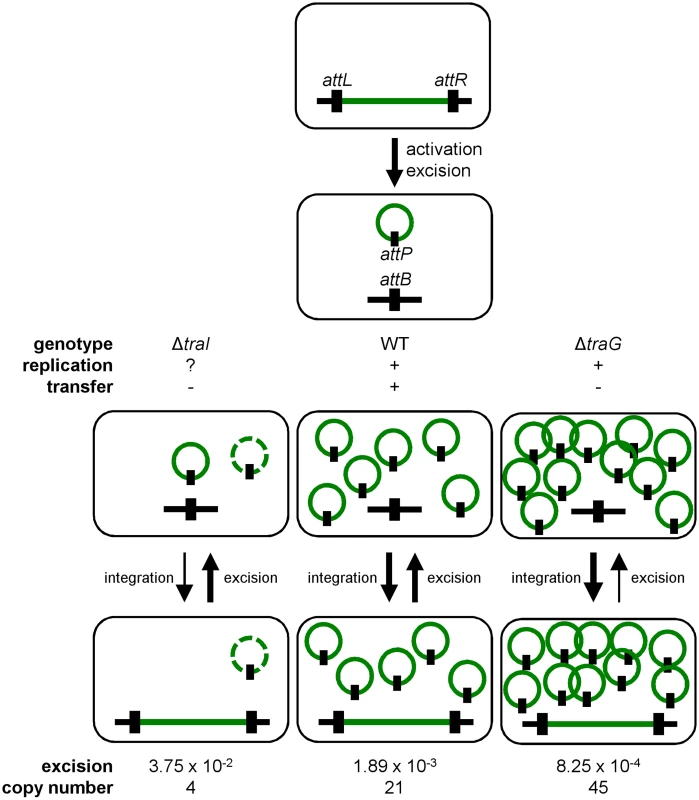
Upon proper stimulation, an integrated ICE is activated and excises from the chromosome. When activated, the ICE is able to replicate and transfer by conjugation. Deletion of traG abolishes ICE conjugative transfer due to the non-functional mating pore, and leads to the accumulation of multiple copies resulting from its replication. Deletion of traI also abolishes ICE conjugative transfer by inhibiting the initiation of transfer at oriT and subsequently impairing rolling-circle replication of the ICE, thereby reducing its copy number. Green line, integrated ICE; green circle, excised ICE, black line, chromosomal integration locus, black rectangle, attachment sites. The dashed green circle represents a putative form of the ICE resulting from low-rate replication by an unknown mechanism. We observed that the frequency of excision does not correlate with the frequency of transfer when comparing SXT and R391. R391 excises at a frequency that is ~10 fold lower than SXT whereas it transfers at a frequency that is ~20 fold higher. We previously reported for SXT that neither its excision from the chromosome of donor cells nor its integration in the chromosome of recipient cells was a step limiting the rate of transfer [14, 26]. This suggests that assembly of the mating apparatus, initiation of transfer or DNA translocation across the cell membranes through the mating pore was the limiting factor. In fact, our data revealed that availability of TraI is a key regulatory element since ovexpression of traI in cells bearing R391K ΔtraI increased the frequency of transfer by 2 logs over wild-type R391K. This observation is supported by RNA-seq data that revealed the relatively low level of expression of traI compared to other tra genes in SXT, R391 and ICEVflInd1, another member of the SXT/R391 family [14]. Therefore, initiation of transfer and/or replication, both depending on TraI and oriT, seem to determine the rate of transfer of SXT/R391 ICEs.
Deletion of traI or oriT drastically reduced the copy number of R391 circles (Fig 3C), which is consistent with a form of replication initiated at oriT by the relaxase TraI. The process of conjugation usually relies on an intercellular rolling-circle replication of conjugative elements, making their intracellular replication also virtually possible [8, 63]. Although ICEs were initially defined as non-replicative elements [6], several recent reports strongly support that single-stranded DNA transferring ICEs can replicate as extrachromosomal plasmid-like molecules, in both Gram-positive and Gram-negative bacteria [20–25]. This replication is initiated at oriT by the relaxase together with other ICE - and host-encoded auxiliary factors [22, 24]. Notably, the transient replication associated with the conjugative transfer of ICEBs1 of B. subtilis, while not required for transfer, plays an important role in the stability [24]. It relies on oriT used as an origin of replication (oriV) and on the conjugative relaxase NicK used as the replication initiator protein. Therefore, the rolling-circle replication module being an intrinsic part of the conjugation module, many ICEs, if not all, might be able to transiently replicate as plasmid-like molecules.
Our work revealed yet another intriguing feature of ICEs of the SXT/R391 family besides replication, which seems to blur the frontier between ICEs and plasmids even more. All SXT/R391 ICEs carry srpMRC, a locus coding for a functional active partition system. Contrary to low-copy plasmids such as F, which must actively segregate in the daughter cells following cell division, SXT/R391 ICEs usually remain quiescent, integrated into the chromosome of their host, and are passively passed on from one generation to another. Active partition of these ICEs would only be required in their transient excised state, even more so if their copy number per cell is low, such as in the traI mutant (Fig 4D). In agreement with this observation, srpMRC is part of the same operon coding for the integrase that catalyzes both the integration and excision of SXT/R391 ICEs, all directly under control of the SetCD activator [14]. Therefore, srpMRC is expressed only prior to excision, replication and transfer of the ICE. We observed that a ΔhipA ΔtraG ΔsrpM R391K mutant has an extrachromosomal copy number similar to the wild-type. The apparent suppression of the effect of the ΔtraG mutation on the extrachromosomal copy number by the loss of srpM suggests a link between conjugation and partition that remains to be elucidated.
Active partition of ICEs could be an overlooked feature that is in fact rather common among ICEs. The ICE PAPI-1 of Pseudomonas aeruginosa encodes the putative active partition system Soj. Deletion of soj leads to high-frequency loss of PAPI-1 [64]. Although the exact mechanism of action of Soj is not well understood, its expression was shown to be stimulated when PAPI-1 excises. ICEs of the pKLC102-ICEclc group, including PAPI-1 and ICEHin1056, were shown to be able to replicate and code for putative partitioning systems [21, 23, 65–67]. Moreover, the core region of Tn4371-like ICEs and the ICE pNOB8 from Sulfolobus codes for ParA and ParB proteins, whose homologs are known to play a role in plasmid partition [68–72]. Finally, ICEA of Mycoplasma agalactiae encodes a ParA homolog that could be part of a partitioning system [73]. All these putative partitioning systems could also be involved in incompatibility with other ICEs and/or plasmids, as well as in transcriptional regulation of ICE - and/or host-borne loci [74–76].
Classification of mobile genetic element is extremely laborious mostly because of their modular structure. Our increasingly precise comprehension of their biology unravels some unexpected features that make them even harder to label [77]. On the one hand, ICEs exhibit phage-like behaviors, such as integration by site-specific recombination and, for some ICEs, regulation controlled by CI - or ImmR-like regulators [15, 37, 59, 78, 79]. On the other hand, ICEs also share several characteristics with plasmids, such as a single-strand DNA intermediate during transfer, their conjugative apparatus and entry exclusion systems (traG/eex) [80, 81]. For instance, the conjugation modules and master activators SetCD and AcaCD of SXT/R391 ICEs and conjugative plasmids of the IncA/C group share a common ancestry [10, 14, 82]. As such SXT/R391 ICEs and IncA/C plasmids offer a dramatic example of divergent evolution from a common ancestor into two different lifestyles. Although SXT/R391 ICEs are capable of transient replication using the relaxase TraI and oriT, this lifestyle does not seem to be sustainable over multiple generations [14]. IncA/C plasmids lack the int and xis genes required for integration and excision, and instead carry a dedicated RepA/C replicon, allowing autonomous, stable and efficient replication in the cell. IncA/C plasmids code for a putative ParMRC-like partitioning system closely related to SrpMRC (vcrx152/vcrx151 in pVCR94) (Fig 6A and 6B). Interestingly, expression of parMRC-like locus of SXT/R391 ICEs and IncA/C plasmids is directly under the control of similar yet distantly related class II transcriptional activator complexes: SetCD for SXT/R391 ICEs and AcaCD for IncA/C plasmids [14, 82, 83] (Fig 6B). Given the pleitropic role of these activators, this mode of regulation directly pairs the expression of DNA segregation functions to expression of conjugative transfer functions. However, although IncA/C plasmids retain a type II parMRC-like partitioning system (actin-type ATPase), they also carry a type I parABC-like partitioning system (Walker-type ATPase) (vcrx031/vcrx032 in pVCR94), which does not seem to be regulated by AcaCD [48, 82, 84, 85]. The exact function and eventual redundancy of each par locus remains to be investigated for IncA/C plasmids. The IncHI1 conjugative plasmid R27 also contains two independent partitioning loci, a type I partitioning system, and a type II partitioning system [54]. The type I partitioning system was shown to be the major stability determinant of R27 whereas type II is the minor stability determinant.
Finally, our results put an end to a long standing question: Do SXT/R391 ICEs behave like plasmids and replicate? R391 and related elements such as R705, R748, R997, and pMERPH were initially reported as R factors belonging to the same J incompatibility group (IncJ) [9, 86]. R391 and R997 were even isolated as circular molecules and physically mapped by restriction analysis [19]. Subsequent identification of SXT as an integrative element, and reports of the site-specific integration of R391 and R997 into the same chromosomal site as SXT highlighted seeming incongruities between otherwise extremely similar mobile genetic elements as revealed by sequence comparison [10, 12, 87–90]. In fact, our results indicate that replication, coupled with partition, is a normal yet transitory step of the lifecycle of SXT/R391 ICEs. The transitory nature of this replication does not allow stable maintenance and inheritance as a plasmid-like form. Therefore, integration into the chromosome remains the main mechanism ensuring stable vertical transmission of SXT/R391 ICEs over multiple generations. In the end, despite using similar strategies for their maintenance in the cell population and transfer between cell populations, ICEs and conjugative plasmids remain distinct entities regarding their respective maintenance by integration or replication.
Materials and Methods
Bacterial strains and media
The bacterial strains and plasmids used in this study are described in Table 1. The strains were routinely grown in lysogeny broth (LB-Miller, EMD) at 37°C in an orbital shaker/incubator and were preserved at -80°C in LB broth containing 15% (vol/vol) glycerol. Antibiotics were used at the following concentrations: ampicillin (Ap), 100 μg/ml; chloramphenicol (Cm), 20 μg/ml; kanamycin (Kn), 50 μg/ml; mitomycin C (MC), 50 ng/ml; nalidixic acid (Nx), 40 μg/ml; rifampicin (Rf), 50 μg/ml; spectinomycin (Sp), 50 μg/ml; sulfamethoxazole (Su), 160 μg/ml; tetracycline (Tc), 12 μg/ml; trimethoprim (Tm), 32 μg/ml. When required, bacterial cultures were supplemented with 0.02 mM of isopropyl β-D-1-thiogalactopyranoside (IPTG) or 0.02% L-arabinose.
Tab. 1. Strains and plasmids used in this study. 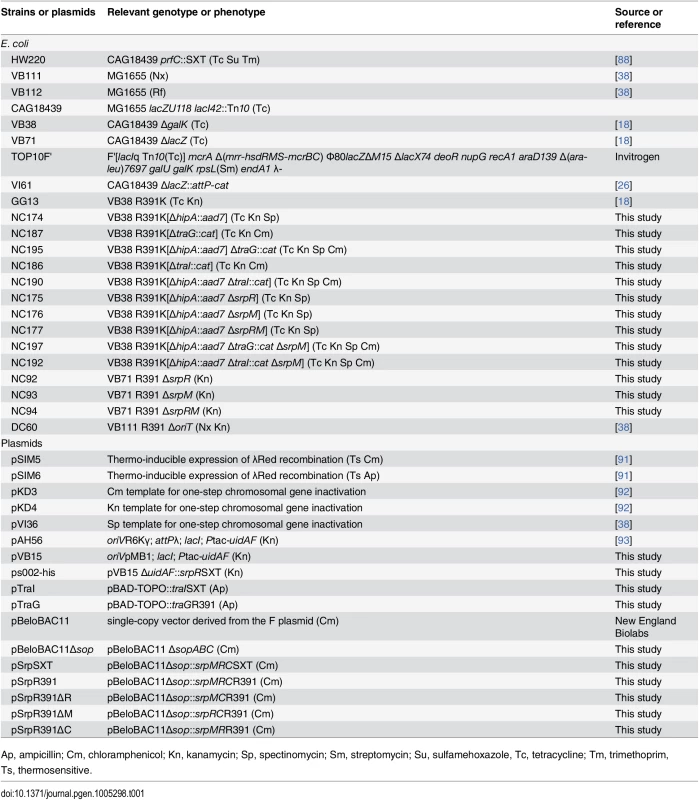
Ap, ampicillin; Cm, chloramphenicol; Kn, kanamycin; Sp, spectinomycin; Sm, streptomycin; Su, sulfamehoxazole, Tc, tetracycline; Tm, trimethoprim, Ts, thermosensitive. Bacterial conjugation assays
Conjugation assays were performed by mixing equal volumes of each donor and recipient strains that were grown overnight at 37°C. The cells were harvested by centrifugation for 3 min at 1200g, washed in 1 volume of LB broth and resuspended in 1/20 volume of LB broth. Mating mixtures were then deposited on LB agar plates and incubated at 37°C for 6 hours. The cells were recovered from the plates in 1 ml of LB broth and serially diluted before plating. Donors, recipients and exconjugants were selected on LB agar plates containing appropriate antibiotics.
Molecular biology methods
Plasmid DNA was prepared using the EZ-10 Spin Column Plasmid DNA Minipreps Kit (Biobasic) according to manufacturer’s instructions. All the enzymes used in this study were purchased from New England BioLabs. PCR assays were performed with the primers described in S1 Table. The PCR conditions were as follows: (i) 3 min at 94°C; (ii) 30 cycles of 30 sec at 94°C, 30 sec at the appropriate annealing temperature, and 1 minute/kb at 68°C; and (iii) 5 min at 68°C. When necessary, PCR products were purified using an EZ-10 Spin Column PCR Products Purification Kit (Biobasic) according to manufacturer’s instructions. E. coli was transformed by electroporation as described by Dower et al. [94] in a BioRad GenePulser Xcell apparatus set at 25 μF, 200 V and 1.8 kV using 1-mm gap electroporation cuvettes. Sequencing reactions were performed by the Plateforme de Séquençage et de Génotypage du Centre de Recherche du CHUL (Québec, QC, Canada).
Plasmid and strain construction
Plasmids and oligonucleotides used in this study are listed in Table 1 and S1 Table. pTraI and pTraG were constructed by cloning traI of SXT and traG of R391 into the TA cloning expression vector pBAD-TOPO (Invitrogen) according to the manufacturer’s instructions. traI was amplified by PCR with its native Shine-Dalgarno sequence using primers pBad-traI_Fw and pBad-traI_Rev and genomic DNA of E. coli HW220 as the template. traG was amplified using the primer pair traGEcoRI.for / traGEcoRI.for and genomic DNA of E. coli GG13 as the template. pVB15 was constructed by amplifying the origin of replication of pUC19 (oriVpMB1) using the primer pair pUC_oriF/pUC_oriR and subsequent cloning into the 5 838-bp fragment of NheI/NotI-digested pAH56 to replace oriVR6K-attPλ and generate the high-copy number expression vector pVB15. ps002-his was then obtained by cloning s002 (srpR) from SXT amplified with the primer pair s002F/s002-hisR into the 4 319-bp fragment of NdeI/BamHI-digested pVB15.
Plasmids used for plasmid stabilization assays were derived from pBeloBAC11Δsop, a pBeloBAC11 vector derivative from which the partitioning system sopABC was deleted by NdeI digestion and re-ligation. The srpMRC locus of SXT (srpMRCSXT) and R391 (srpMRCR391) were amplified by PCR using genomic DNA of strains containing either SXT or R391 as the templates and primers pairs SXTpartHindIIIstop.for/SXTR391partHindIII.rev and R391partHindIIIstop.for/SXTR391partHindIII.rev, respectively. Amplicons were digested by HindIII and cloned into HindIII-digested pBeloBAC11Δsop to generate pSrpSXT and pSrpR391. Subsequent deletions of segments of srpMRCR391 were obtained by high fidelity PCR amplification of the pSrpR391 vector using primer pairs pBeloDelSO02.for/pBeloDelSO02.rev, pBeloDelSO03.for/pBeloDelSO03.rev or pBeloDelparC.for/pBeloDelparC.rev, digestion by NheI and ligation using the T4 DNA ligase. The resulting plasmids were verified by restriction profiling and DNA sequencing.
Deletion mutants of R391::galK (R391K) [18] were constructed using the one-step chromosomal gene inactivation [92] and P1vir transduction [95] techniques. Deletion of hipA, srpR, srpM, srpRM, traI and traG were constructed using primer pairs R391DhipAnoFRT.for/R391DhipAnoFRT.rev, 2SXTR391DSO02.for/2SXTR391DSO02.rev, R391DSO03.for/2SXTR391DSO03.rev, R391DSO03.for/2SXTR391DSO02.rev, R391DtraInoFRT.for/R391DtraInoFRT.rev, R391DtraGnoFRT.for/R391DtraGnoFRT.rev, respectively. Gene resistance cassettes were amplified using the pVI36, pKD3 and pKD4 vectors. The λRed recombination system was expressed using pSIM5 or pSIM6 as described by Datta et al. [91]. If possible, the antibiotic resistance cassette was removed from the resulting construction by Flp-catalyzed excision using the pCP20 vector [96]. All deletions were verified by PCR and antibiotic resistance profiling.
ICE and plasmid stability assays
The stability of R391::galK and derivative mutants was monitored based on the methodology described by Wozniak and Waldor [32]. Cells were grown for 16 hours in 4 ml of LB medium supplemented or not with kanamycin. Serial dilutions were plated on MacConkey agar plates supplemented with 1% D-galactose. Loss of R391 resulted in the formation of white clones (Fig 1C). For each experiment, at least 16 white clones were purified and tested on agar plate for their susceptibility to kanamycin. These clones were also tested by PCR amplification of an internal fragment of R391 using primer pair R391HipBM1.for/R391HipB.rev. The stability of pBeloBAC11Δsop and derivatives containing the srpMRC locus of SXT or R391 was tested for 16 hours in M9 or LB medium using the approach described by Sanchez et al. [97]. Expression of the srp locus from Plac was induced by addition of 0.02 mM IPTG. Relative stability was calculated as the ratio of chloramphenicol resistant colonies in the population in the induced compared to the non-induced conditions. For both ICE and plasmid stability assays, each experiment was carried out at least in biological triplicate.
Determination of ICE dynamics using real-time quantitative PCR
The frequency of excision as well as total copy number in the population and copy number of the excised circular form of the ICE were assessed by real-time quantitative PCR as described elsewhere [20, 26]. Genomic DNA was obtained from cell cultures of E. coli CAG18439 bearing SXT, R391K or its mutants grown for 16 h in LB medium. prfC, attB, attP and int were quantified using primer pairs prfC.qec.F1/prfC.qec.R1, attB.qec.F2/attB.qec.R2, attP.qec.F2/attP.qec.R2 and int.qec.F1/int.qec.R1, respectively (S1 Table). For frequency of excision and copy number determination, E. coli VI61, which contains one chromosomal copy of attB, attP and prfC, was used to simulate 100% of excision and normalize the results. qPCR experiments were performed in triplicate on the RNomics platform of the Laboratoire de Geénomique Fonctionnelle de l’Universiteé de Sherbrooke (http://lgfus.ca) (Sherbrooke, QC, Canada).
Macroscopic observations of colonies
Macroscopic observations were done using a SZX7 zoom stereomicroscope with a DF PLAPO1X-4 objective coupled to a SC30 digital camera via a U-TV1X-2 & U-CMAD3 adaptor (Olympus).
SrpR expression and purification
To express and purify SrpR tagged with a 6×His C-terminal epitope (SrpR6×His), cultures of E. coli BL21 bearing ps002-his were grown overnight, diluted 1 : 500 in fresh 2×YTA broth and incubated at 37°C with agitation. At mid-exponential phase (OD600 of 0.6), protein expression was induced with 0.1 mM IPTG and cultures were incubated for 3 hours. Cells were then harvested by centrifugation at 1500×g for 10 min at 4°C and stored at -20°C. The cell pellet was weighted and re-suspended in Native Purification Buffer (NPB) (50 mM NaH2PO4 pH 8.0, 2.5 M NaCl) containing 0.1% Triton X-100, 1 mM phenylmethanesulfonylfluoride (PMSF), and protease inhibitors at 1 ml / 20g of cell pellet (Protease Inhibitor Cocktail, Sigma). Purification of SrpR6×His was done by Ni-NTA affinity chromatography following the manufacturer’s instructions (Qiagen). Cells were lysed by sonication, cell debris was pelleted by centrifugation, and the supernatant was incubated for 1 h at 4°C with 750 μl of Ni-NTA Agarose resin (QIAGEN) with agitation. The Ni-NTA Agarose resin was then transferred into a column and washed 4 times with 1.25 ml of native wash buffer (NPB with 20 mM imidazole, pH 8.0). SrpR6×His was eluted with native elution buffer (NPB with 250 mM imidazole, pH 8.0) and stored at -20°C. Protein concentration was estimated using a Bradford protein assay (BioRad) and purity was determined by SDS-PAGE analysis.
Electrophoretic mobility shift assays (EMSA)
The linear double-stranded DNA probes srpC (251 bp), oriT (298 bp) and ig(srpM-mobI) (615 bp) used in the EMSA experiments were amplified by PCR using primer pairs RRintF/RRintR, oriT2F/oriT2R and MELR1/oriT1R, respectively, and E. coli HW220 as the template (Table 1 and S1 Table). Probes were purified using an EZ-10 Spin Column PCR Products Purification Kit (Bio Basic) according to the manufacturer's instructions and their concentration was determined using a NanoDrop ND-1000. Probes R3-R1, R4-R2, R2 and R4 were obtained by mixing equimolar concentrations (50 μM) of primers, srpCSXTR3R1F and srpCSXTR3R1R, srpCSXTR4R2F and srpCSXTR4R2R, srpCSXTR2F and srpCSXTR2R, or srpCSXTR4F and srpCSXTR4R, respectively. The primer mixtures were heated at 95°C for 3 min, then annealed by slow cool down overnight.
EMSA assays were carried out using the Electrophoretic Mobility-Shift Assay Kit with SYBR Green & SYPRO Ruby EMSA stains (Life Technologies) according to the manufacturer's instructions. Briefly, a total of 40 ng of DNA probe was used in each reaction. Quantities of SrpR6×His and of the non-specific competitor DNA (sonicated salmon sperm DNA) varied from 10 to 100 ng, and 10 to 200 pg, respectively. The non-specific competitor DNA was mixed with the probe before adding SrpR6×His to maximize competition. All binding reactions were done in a total volume of 10 μl for 15 min at room temperature followed by 10 min incubation on ice. Samples were then loaded on a pre-run (25 min at 100 V) non-denaturing 4% acrylamide gel containing 1× TBE buffer and migration was carried out at 4°C during electrophoresis. SYBR Green staining was done according to the manufacturer's instructions and gel pictures were scanned using a Typhoon FLA 9500 (GE Healthcare Life Sciences) with a LPB filter for SYBR Green I at a 100 μm resolution.
Dimerization assay
Dimerization assays were carried out using 2 μg of purified SrpR6×His. Samples containing srpC were carried out using 1 μg of DNA probe and were incubated prior to the dimerization assay in the same conditions as for the EMSA assays. Samples were incubated with or without 0.6% glutaraldehyde for 30 min at room temperature and 3% of β-mercaptoethanol was added to the samples prior to denaturation at 95°C for 3 min. Samples and ladder (Precision Plus Protein Kaleidoscope Standards, BioRad) were separated by electrophoresis on a 12% SDS-PAGE gel, later stained using Coomassie Brilliant Blue R-250.
Phylogenetic analyses
The molecular phylogenetic analysis of SrpM was conducted in MEGA6 [98] The primary sequence of SrpM encoded by R391 was used to search for homologous sequences in the Genbank Non-redundant protein sequence (nr) database using blastP [99]. Phylogenetic analyses were computed using a protein alignment generated by MUSCLE [100] and poorly aligned regions were removed with the trimAl v1.3 software using the automated heuristic approach [101] prior to phylogenetic analyses. The evolutionary history was inferred by using the Maximum Likelihood method. Initial tree(s) for the heuristic search were obtained automatically by applying Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using a JTT model, and then selecting the topology with superior log likelihood value. A discrete Gamma distribution was used to model evolutionary rate differences among sites (5 categories (+G, parameter = 4.1321)). The rate variation model allowed for some sites to be evolutionarily invariable ([+I], 2.6007% sites). The tree is drawn to scale in iTOL v2 [102], with branch lengths measured by the number of substitutions per site. The analysis involved 60 amino acid sequences with a total of 261 positions in the final dataset.
Supporting Information
Zdroje
1. Guglielmini J, Quintais L, Garcillan-Barcia MP, de la Cruz F, Rocha EP. The Repertoire of ICE in Prokaryotes Underscores the Unity, Diversity, and Ubiquity of Conjugation. PLoS Genet. 2011;7(8):e1002222. Epub 2011/08/31. doi: 10.1371/journal.pgen.1002222 21876676
2. Ghinet MG, Bordeleau E, Beaudin J, Brzezinski R, Roy S, Burrus V. Uncovering the prevalence and diversity of integrating conjugative elements in actinobacteria. PLoS One. 2011;6(11):e27846. Epub 2011/11/25. doi: 10.1371/journal.pone.0027846 22114709
3. Bordeleau E, Ghinet MG, Burrus V. Diversity of integrating conjugative elements in actinobacteria: Coexistence of two mechanistically different DNA-translocation systems. Mob Genet Elements. 2012;2(2):119–24. Epub 2012/08/31. 22934248
4. Wozniak RA, Waldor MK. Integrative and conjugative elements: mosaic mobile genetic elements enabling dynamic lateral gene flow. Nat Rev Microbiol. 2010;8(8):552–63. Epub 2010/07/06. doi: 10.1038/nrmicro2382 20601965
5. Burrus V, Waldor MK. Shaping bacterial genomes with integrative and conjugative elements. Res Microbiol. 2004;155(5):376–86. Epub 2004/06/23. 15207870
6. Burrus V, Pavlovic G, Decaris B, Guedon G. Conjugative transposons: the tip of the iceberg. Mol Microbiol. 2002;46(3):601–10. Epub 2002/11/02. 12410819
7. Carraro N, Burrus V. Biology of Three ICE Families: SXT/R391, ICEBs1, and ICESt1/ICESt3. Microbiology Spectrum. 2014;2(6). doi: 10.1128/microbiolspec.PLAS-0010-2013 25705574
8. de la Cruz F, Frost LS, Meyer RJ, Zechner EL. Conjugative DNA metabolism in Gram-negative bacteria. FEMS Microbiol Rev. 2010;34(1):18–40. Epub 2009/11/19. doi: 10.1111/j.1574-6976.2009.00195.x 19919603
9. Burrus V, Marrero J, Waldor MK. The current ICE age: biology and evolution of SXT-related integrating conjugative elements. Plasmid. 2006;55(3):173–83. Epub 2006/03/15. 16530834
10. Wozniak RA, Fouts DE, Spagnoletti M, Colombo MM, Ceccarelli D, Garriss G, et al. Comparative ICE genomics: insights into the evolution of the SXT/R391 family of ICEs. PLoS Genet. 2009;5(12):e1000786. Epub 2009/12/31. doi: 10.1371/journal.pgen.1000786 20041216
11. Spagnoletti M, Ceccarelli D, Rieux A, Fondi M, Taviani E, Fani R, et al. Acquisition and evolution of SXT-R391 integrative conjugative elements in the seventh-pandemic Vibrio cholerae lineage. MBio. 2014;5(4). Epub 2014/08/21.
12. Waldor MK, Tschape H, Mekalanos JJ. A new type of conjugative transposon encodes resistance to sulfamethoxazole, trimethoprim, and streptomycin in Vibrio cholerae O139. J Bacteriol. 1996;178(14):4157–65. Epub 1996/07/01. 8763944
13. Boltner D, MacMahon C, Pembroke JT, Strike P, Osborn AM. R391: a conjugative integrating mosaic comprised of phage, plasmid, and transposon elements. J Bacteriol. 2002;184(18):5158–69. 12193633
14. Poulin-Laprade D, Matteau D, Jacques PE, Rodrigue S, Burrus V. Transfer activation of SXT/R391 integrative and conjugative elements: unraveling the SetCD regulon. Nucleic Acids Res. 2015;in press. Epub 2015/02/11.
15. Beaber JW, Hochhut B, Waldor MK. SOS response promotes horizontal dissemination of antibiotic resistance genes. Nature. 2004;427(6969):72–4. Epub 2003/12/23. 14688795
16. Burrus V, Waldor MK. Formation of SXT tandem arrays and SXT-R391 hybrids. J Bacteriol. 2004;186(9):2636–45. 15090504
17. Hochhut B, Beaber JW, Woodgate R, Waldor MK. Formation of chromosomal tandem arrays of the SXT element and R391, two conjugative chromosomally integrating elements that share an attachment site. J Bacteriol. 2001;183(4):1124–32. 11157923
18. Garriss G, Waldor MK, Burrus V. Mobile antibiotic resistance encoding elements promote their own diversity. PLoS Genet. 2009;5(12):e1000775. doi: 10.1371/journal.pgen.1000775 20019796
19. Pembroke JT, Murphy DB. Isolation and analysis of a circular form of the IncJ conjugative transposon-like elements, R391 and R997: implications for IncJ incompatibility. FEMS Microbiol Lett. 2000;187(2):133–8. Epub 2000/06/17. 10856646
20. Carraro N, Libante V, Morel C, Decaris B, Charron-Bourgoin F, Leblond P, et al. Differential regulation of two closely related integrative and conjugative elements from Streptococcus thermophilus. BMC Microbiol. 2011;11 : 238. Epub 2011/10/26. doi: 10.1186/1471-2180-11-238 22024428
21. Dimopoulou ID, Russell JE, Mohd-Zain Z, Herbert R, Crook DW. Site-specific recombination with the chromosomal tRNA(Leu) gene by the large conjugative Haemophilus resistance plasmid. Antimicrob Agents Chemother. 2002;46(5):1602–3. Epub 2002/04/18. 11959612
22. Grohmann E. Autonomous plasmid-like replication of Bacillus ICEBs1: a general feature of integrative conjugative elements? Mol Microbiol. 2010;75(2):261–3. Epub 2009/12/01. doi: 10.1111/j.1365-2958.2009.06978.x 19943906
23. Kiewitz C, Larbig K, Klockgether J, Weinel C, Tummler B. Monitoring genome evolution ex vivo: reversible chromosomal integration of a 106 kb plasmid at two tRNA(Lys) gene loci in sequential Pseudomonas aeruginosa airway isolates. Microbiology. 2000;146 (Pt 10):2365–73. 11021913
24. Lee CA, Babic A, Grossman AD. Autonomous plasmid-like replication of a conjugative transposon. Mol Microbiol. 2010;75(2):268–79. Epub 2009/12/01. doi: 10.1111/j.1365-2958.2009.06985.x 19943900
25. Sitkiewicz I, Green NM, Guo N, Mereghetti L, Musser JM. Lateral gene transfer of streptococcal ICE element RD2 (region of difference 2) encoding secreted proteins. BMC Microbiol. 2011;11 : 65. Epub 2011/04/05. doi: 10.1186/1471-2180-11-65 21457552
26. Burrus V, Waldor MK. Control of SXT integration and excision. J Bacteriol. 2003;185(17):5045–54. 12923077
27. Kobayashi I. Behavior of restriction-modification systems as selfish mobile elements and their impact on genome evolution. Nucleic Acids Res. 2001;29(18):3742–56. Epub 2001/09/15. 11557807
28. Bahl MI, Hansen LH, Sorensen SJ. Persistence mechanisms of conjugative plasmids. Methods Mol Biol. 2009;532 : 73–102. Epub 2009/03/10. doi: 10.1007/978-1-60327-853-9_5 19271180
29. Van Melderen L, Saavedra De Bast M. Bacterial toxin-antitoxin systems: more than selfish entities? PLoS Genet. 2009;5(3):e1000437. Epub 2009/03/28. doi: 10.1371/journal.pgen.1000437 19325885
30. Van Melderen L. Toxin-antitoxin systems: why so many, what for? Curr Opin Microbiol. 2010;13(6):781–5. Epub 2010/11/03. doi: 10.1016/j.mib.2010.10.006 21041110
31. Hayes F, Van Melderen L. Toxins-antitoxins: diversity, evolution and function. Crit Rev Biochem Mol Biol. 2011;46(5):386–408. Epub 2011/08/09. doi: 10.3109/10409238.2011.600437 21819231
32. Wozniak RA, Waldor MK. A toxin-antitoxin system promotes the maintenance of an integrative conjugative element. PLoS Genet. 2009;5(3):e1000439. Epub 2009/03/28. doi: 10.1371/journal.pgen.1000439 19325886
33. Dziewit L, Jazurek M, Drewniak L, Baj J, Bartosik D. The SXT conjugative element and linear prophage N15 encode toxin-antitoxin-stabilizing systems homologous to the tad-ata module of the Paracoccus aminophilus plasmid pAMI2. J Bacteriol. 2007;189(5):1983–97. Epub 2006/12/13. 17158670
34. Schumacher MA, Piro KM, Xu W, Hansen S, Lewis K, Brennan RG. Molecular mechanisms of HipA-mediated multidrug tolerance and its neutralization by HipB. Science. 2009;323(5912):396–401. Epub 2009/01/20. doi: 10.1126/science.1163806 19150849
35. Hansen S, Vulic M, Min J, Yen TJ, Schumacher MA, Brennan RG, et al. Regulation of the Escherichia coli HipBA toxin-antitoxin system by proteolysis. PLoS One. 2012;7(6):e39185. Epub 2012/06/22. doi: 10.1371/journal.pone.0039185 22720069
36. Bahl MI, Hansen LH, Sorensen SJ. Impact of conjugal transfer on the stability of IncP-1 plasmid pKJK5 in bacterial populations. FEMS Microbiol Lett. 2007;266(2):250–6. Epub 2006/11/30. 17132149
37. Beaber JW, Hochhut B, Waldor MK. Genomic and functional analyses of SXT, an integrating antibiotic resistance gene transfer element derived from Vibrio cholerae. J Bacteriol. 2002;184(15):4259–69. Epub 2002/07/11. 12107144
38. Ceccarelli D, Daccord A, Rene M, Burrus V. Identification of the origin of transfer (oriT) and a new gene required for mobilization of the SXT/R391 family of integrating conjugative elements. J Bacteriol. 2008;190(15):5328–38. Epub 2008/06/10. doi: 10.1128/JB.00150-08 18539733
39. Marchler-Bauer A, Bryant SH. CD-Search: protein domain annotations on the fly. Nucleic Acids Res. 2004;32(Web Server issue):W327–31. Epub 2004/06/25. 15215404
40. Marchler-Bauer A, Zheng C, Chitsaz F, Derbyshire MK, Geer LY, Geer RC, et al. CDD: conserved domains and protein three-dimensional structure. Nucleic Acids Res. 2013;41(Database issue):D348–52. Epub 2012/12/01. doi: 10.1093/nar/gks1243 23197659
41. Kelley LA, Sternberg MJ. Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc. 2009;4(3):363–71. Epub 2009/02/28. doi: 10.1038/nprot.2009.2 19247286
42. Salje J, Gayathri P, Lowe J. The ParMRC system: molecular mechanisms of plasmid segregation by actin-like filaments. Nat Rev Microbiol. 2010;8(10):683–92. Epub 2010/09/17. doi: 10.1038/nrmicro2425 20844556
43. Moller-Jensen J, Borch J, Dam M, Jensen RB, Roepstorff P, Gerdes K. Bacterial mitosis: ParM of plasmid R1 moves plasmid DNA by an actin-like insertional polymerization mechanism. Mol Cell. 2003;12(6):1477–87. Epub 2003/12/24. 14690601
44. Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, et al. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37(Web Server issue):W202–8. Epub 2009/05/22. doi: 10.1093/nar/gkp335 19458158
45. Schumacher MA. Bacterial plasmid partition machinery: a minimalist approach to survival. Curr Opin Struct Biol. 2012;22(1):72–9. Epub 2011/12/14. doi: 10.1016/j.sbi.2011.11.001 22153351
46. Nonaka L, Maruyama F, Miyamoto M, Miyakoshi M, Kurokawa K, Masuda M. Novel conjugative transferable multiple drug resistance plasmid pAQU1 from Photobacterium damselae subsp. damselae isolated from marine aquaculture environment. Microbes Environ. 2012;27(3):263–72. Epub 2012/03/27. 22446310
47. Johnson TJ, Lang KS. IncA/C plasmids: An emerging threat to human and animal health? Mob Genet Elements. 2012;2(1):55–8. Epub 2012/07/04. 22754754
48. Carraro N, Sauve M, Matteau D, Lauzon G, Rodrigue S, Burrus V. Development of pVCR94DeltaX from Vibrio cholerae, a prototype for studying multidrug resistant IncA/C conjugative plasmids. Front Microbiol. 2014;5 : 44. Epub 2014/02/26. doi: 10.3389/fmicb.2014.00044 24567731
49. Murata T, Ohnishi M, Ara T, Kaneko J, Han CG, Li YF, et al. Complete nucleotide sequence of plasmid Rts1: implications for evolution of large plasmid genomes. J Bacteriol. 2002;184(12):3194–202. Epub 2002/05/25. 12029035
50. Maeda K, Nojiri H, Shintani M, Yoshida T, Habe H, Omori T. Complete nucleotide sequence of carbazole/dioxin-degrading plasmid pCAR1 in Pseudomonas resinovorans strain CA10 indicates its mosaicity and the presence of large catabolic transposon Tn4676. J Mol Biol. 2003;326(1):21–33. 12547188
51. Yano H, Miyakoshi M, Ohshima K, Tabata M, Nagata Y, Hattori M, et al. Complete nucleotide sequence of TOL plasmid pDK1 provides evidence for evolutionary history of IncP-7 catabolic plasmids. J Bacteriol. 2010;192(17):4337–47. Epub 2010/06/29. doi: 10.1128/JB.00359-10 20581207
52. Jensen RB, Gerdes K. Partitioning of plasmid R1. The ParM protein exhibits ATPase activity and interacts with the centromere-like ParR-parC complex. J Mol Biol. 1997;269(4):505–13. Epub 1997/06/20. 9217256
53. Moller-Jensen J, Ringgaard S, Mercogliano CP, Gerdes K, Lowe J. Structural analysis of the ParR/parC plasmid partition complex. EMBO J. 2007;26(20):4413–22. Epub 2007/09/28. 17898804
54. Lawley TD, Taylor DE. Characterization of the double-partitioning modules of R27: correlating plasmid stability with plasmid localization. J Bacteriol. 2003;185(10):3060–7. Epub 2003/05/06. 12730165
55. Le SQ, Gascuel O. An improved general amino acid replacement matrix. Mol Biol Evol. 2008;25(7):1307–20. Epub 2008/03/28. doi: 10.1093/molbev/msn067 18367465
56. Novick RP, Clowes RC, Cohen SN, Curtiss R 3rd, Datta N, Falkow S. Uniform nomenclature for bacterial plasmids: a proposal. Bacteriol Rev. 1976;40(1):168–89. Epub 1976/03/01. 1267736
57. Wellington EM, Boxall AB, Cross P, Feil EJ, Gaze WH, Hawkey PM, et al. The role of the natural environment in the emergence of antibiotic resistance in gram-negative bacteria. Lancet Infect Dis. 2013;13(2):155–65. Epub 2013/01/26. doi: 10.1016/S1473-3099(12)70317-1 23347633
58. Hawkey PM. The growing burden of antimicrobial resistance. J Antimicrob Chemother. 2008;62 Suppl 1:i1–9. Epub 2008/08/14. doi: 10.1093/jac/dkn241 18684701
59. Auchtung JM, Lee CA, Monson RE, Lehman AP, Grossman AD. Regulation of a Bacillus subtilis mobile genetic element by intercellular signaling and the global DNA damage response. Proc Natl Acad Sci U S A. 2005;102(35):12554–9. Epub 2005/08/18. 16105942
60. Burrus V, Quezada-Calvillo R, Marrero J, Waldor MK. SXT-related integrating conjugative element in New World Vibrio cholerae. Appl Environ Microbiol. 2006;72(4):3054–7. Epub 2006/04/07. 16598018
61. McCool JD, Long E, Petrosino JF, Sandler HA, Rosenberg SM, Sandler SJ. Measurement of SOS expression in individual Escherichia coli K-12 cells using fluorescence microscopy. Mol Microbiol. 2004;53(5):1343–57. Epub 2004/09/25. 15387814
62. Bahl MI, Hansen LH, Licht TR, Sorensen SJ. Conjugative transfer facilitates stable maintenance of IncP-1 plasmid pKJK5 in Escherichia coli cells colonizing the gastrointestinal tract of the germfree rat. Appl Environ Microbiol. 2007;73(1):341–3. Epub 2006/11/07. 17085707
63. Llosa M, Gomis-Ruth FX, Coll M, de la Cruz Fd F. Bacterial conjugation: a two-step mechanism for DNA transport. Mol Microbiol. 2002;45(1):1–8. Epub 2002/07/09. 12100543
64. Qiu X, Gurkar AU, Lory S. Interstrain transfer of the large pathogenicity island (PAPI-1) of Pseudomonas aeruginosa. Proc Natl Acad Sci U S A. 2006;103(52):19830–5. Epub 2006/12/21. 17179047
65. Juhas M, Power PM, Harding RM, Ferguson DJ, Dimopoulou ID, Elamin AR, et al. Sequence and functional analyses of Haemophilus spp. genomic islands. Genome Biol. 2007;8(11):R237. Epub 2007/11/13. 17996041
66. Klockgether J, Reva O, Larbig K, Tummler B. Sequence analysis of the mobile genome island pKLC102 of Pseudomonas aeruginosa C. J Bacteriol. 2004;186(2):518–34. Epub 2004/01/02. 14702321
67. Mohd-Zain Z, Turner SL, Cerdeno-Tarraga AM, Lilley AK, Inzana TJ, Duncan AJ, et al. Transferable antibiotic resistance elements in Haemophilus influenzae share a common evolutionary origin with a diverse family of syntenic genomic islands. J Bacteriol. 2004;186(23):8114–22. Epub 2004/11/18. 15547285
68. Ryan MP, Pembroke JT, Adley CC. Novel Tn4371-ICE like element in Ralstonia pickettii and genome mining for comparative elements. BMC Microbiol. 2009;9 : 242. Epub 2009/11/28. doi: 10.1186/1471-2180-9-242 19941653
69. She Q, Phan H, Garrett RA, Albers SV, Stedman KM, Zillig W. Genetic profile of pNOB8 from Sulfolobus: the first conjugative plasmid from an archaeon. Extremophiles. 1998;2(4):417–25. Epub 1998/11/25. 9827331
70. Surtees JA, Funnell BE. Plasmid and chromosome traffic control: how ParA and ParB drive partition. Curr Top Dev Biol. 2003;56 : 145–80. Epub 2003/10/31. 14584729
71. She Q, Brugger K, Chen L. Archaeal integrative genetic elements and their impact on genome evolution. Res Microbiol. 2002;153(6):325–32. Epub 2002/09/18. 12234006
72. She Q, Shen B, Chen L. Archaeal integrases and mechanisms of gene capture. Biochem Soc Trans. 2004;32(Pt 2):222–6. Epub 2004/03/30. 15046576
73. Marenda M, Barbe V, Gourgues G, Mangenot S, Sagne E, Citti C. A new integrative conjugative element occurs in Mycoplasma agalactiae as chromosomal and free circular forms. J Bacteriol. 2006;188(11):4137–41. Epub 2006/05/19. 16707706
74. Miyakoshi M, Shintani M, Inoue K, Terabayashi T, Sai F, Ohkuma M, et al. ParI, an orphan ParA family protein from Pseudomonas putida KT2440-specific genomic island, interferes with the partition system of IncP-7 plasmids. Environ Microbiol. 2012;14(11):2946–59. Epub 2012/08/29. doi: 10.1111/j.1462-2920.2012.02861.x 22925377
75. Baek JH, Rajagopala SV, Chattoraj DK. Chromosome segregation proteins of Vibrio cholerae as transcription regulators. MBio. 2014;5(3):e01061–14. Epub 2014/05/08. doi: 10.1128/mBio.01061-14 24803519
76. Yurimoto H, Hirai R, Matsuno N, Yasueda H, Kato N, Sakai Y. HxlR, a member of the DUF24 protein family, is a DNA-binding protein that acts as a positive regulator of the formaldehyde-inducible hxlAB operon in Bacillus subtilis. Mol Microbiol. 2005;57(2):511–9. Epub 2005/06/28. 15978081
77. Toussaint A, Merlin C. Mobile elements as a combination of functional modules. Plasmid. 2002;47(1):26–35. Epub 2002/01/19. 11798283
78. Bose B, Auchtung JM, Lee CA, Grossman AD. A conserved anti-repressor controls horizontal gene transfer by proteolysis. Mol Microbiol. 2008;70(3):570–82. Epub 2008/09/03. doi: 10.1111/j.1365-2958.2008.06414.x 18761623
79. Bose B, Grossman AD. Regulation of horizontal gene transfer in Bacillus subtilis by activation of a conserved site-specific protease. J Bacteriol. 2011;193(1):22–9. Epub 2010/11/03. doi: 10.1128/JB.01143-10 21036995
80. Audette GF, Manchak J, Beatty P, Klimke WA, Frost LS. Entry exclusion in F-like plasmids requires intact TraG in the donor that recognizes its cognate TraS in the recipient. Microbiology. 2007;153(Pt 2):442–51. Epub 2007/01/30. 17259615
81. Marrero J, Waldor MK. Determinants of entry exclusion within Eex and TraG are cytoplasmic. J Bacteriol. 2007;189(17):6469–73. Epub 2007/06/19. 17573467
82. Carraro N, Matteau D, Luo P, Rodrigue S, Burrus V. The master activator of IncA/C conjugative plasmids stimulates genomic islands and multidrug resistance dissemination. PLoS Genet. 2014;10(10):e1004714. Epub 2014/10/24. doi: 10.1371/journal.pgen.1004714 25340549
83. Carraro N, Matteau D, Burrus V, Rodrigue S. Unraveling the regulatory network of IncA/C plasmid mobilization: when genomic islands hijack conjugative elements. Mob Genet Elements. 2015;in press.
84. Bousquet A, Henquet S, Compain F, Genel N, Arlet G, Decre D. Partition locus-based classification of selected plasmids in Klebsiella pneumoniae, Escherichia coli and Salmonella enterica spp.: An additional tool. J Microbiol Methods. 2015. Epub 2015/01/28.
85. Harmer CJ, Hall RM. The A to Z of A/C plasmids. Plasmid. 2015. Epub 2015/04/26.
86. McGrath BM, O'Halloran JA, Pembroke JT. Pre-exposure to UV irradiation increases the transfer frequency of the IncJ conjugative transposon-like elements R391, R392, R705, R706, R997 and pMERPH and is recA+ dependent. FEMS Microbiol Lett. 2005;243(2):461–5. Epub 2005/02/03. 15686850
87. Beaber JW, Burrus V, Hochhut B, Waldor MK. Comparison of SXT and R391, two conjugative integrating elements: definition of a genetic backbone for the mobilization of resistance determinants. Cell Mol Life Sci. 2002;59(12):2065–70. Epub 2003/02/06. 12568332
88. Hochhut B, Waldor MK. Site-specific integration of the conjugal Vibrio cholerae SXT element into prfC. Mol Microbiol. 1999;32(1):99–110. Epub 1999/04/27. 10216863
89. McGrath BM, Pembroke JT. Detailed analysis of the insertion site of the mobile elements R997, pMERPH, R392, R705 and R391 in E. coli K12. FEMS Microbiol Lett. 2004;237(1):19–26. Epub 2004/07/23. 15268933
90. Murphy DB, Pembroke JT. Monitoring of chromosomal insertions of the IncJ elements R391 and R997 in Escherichia coli K-12. FEMS Microbiol Lett. 1999;174(2):355–61. 10339829
91. Datta S, Costantino N, Court DL. A set of recombineering plasmids for gram-negative bacteria. Gene. 2006;379 : 109–15. Epub 2006/06/06. 16750601
92. Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A. 2000;97(12):6640–5. Epub 2000/06/01. doi: 10.1073/pnas.120163297 10829079
93. Haldimann A, Wanner BL. Conditional-replication, integration, excision, and retrieval plasmid-host systems for gene structure-function studies of bacteria. J Bacteriol. 2001;183(21):6384–93. Epub 2001/10/10. 11591683
94. Dower WJ, Miller JF, Ragsdale CW. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 1988;16(13):6127–45. 3041370
95. Thomason LC, Costantino N, Court DL. E. coli genome manipulation by P1 transduction. Curr Protoc Mol Biol. 2007;Chapter 1:Unit 1 17. Epub 2008/02/12. doi: 10.1002/0471142727.mb0117s79 18265391
96. Cherepanov PP, Wackernagel W. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene. 1995;158(1):9–14. Epub 1995/05/26. 7789817
97. Sanchez A, Rech J, Gasc C, Bouet JY. Insight into centromere-binding properties of ParB proteins: a secondary binding motif is essential for bacterial genome maintenance. Nucleic Acids Res. 2013;41(5):3094–103. Epub 2013/01/25. doi: 10.1093/nar/gkt018 23345617
98. Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013;30(12):2725–9. Epub 2013/10/18. doi: 10.1093/molbev/mst197 24132122
99. Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–402. Epub 1997/09/01. 9254694
100. Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–7. Epub 2004/03/23. 15034147
101. Capella-Gutierrez S, Silla-Martinez JM, Gabaldon T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25(15):1972–3. Epub 2009/06/10. doi: 10.1093/bioinformatics/btp348 19505945
102. Letunic I, Bork P. Interactive Tree Of Life v2: online annotation and display of phylogenetic trees made easy. Nucleic Acids Res. 2011;39(Web Server issue):W475–8. Epub 2011/04/08. doi: 10.1093/nar/gkr201 21470960
Štítky
Genetika Reprodukční medicína
Článek Germline Mutations Confer Susceptibility to Acute Lymphoblastic Leukemia and ThrombocytopeniaČlánek Multiple In Vivo Biological Processes Are Mediated by Functionally Redundant Activities of andČlánek Temporal Expression Profiling Identifies Pathways Mediating Effect of Causal Variant on PhenotypeČlánek Simultaneous DNA and RNA Mapping of Somatic Mitochondrial Mutations across Diverse Human CancersČlánek A Legume Genetic Framework Controls Infection of Nodules by Symbiotic and Endophytic BacteriaČlánek The Eukaryotic-Like Ser/Thr Kinase PrkC Regulates the Essential WalRK Two-Component System inČlánek The Yeast GSK-3 Homologue Mck1 Is a Key Controller of Quiescence Entry and Chronological LifespanČlánek The Role of -Mediated Epigenetic Silencing in the Population Dynamics of Transposable Elements in
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2015 Číslo 6
-
Všechny články tohoto čísla
- Expression of Concern: RNAi-Dependent and Independent Control of LINE1 Accumulation and Mobility in Mouse Embryonic Stem Cells
- Orphan Genes Find a Home: Interspecific Competition and Gene Network Evolution
- Minor Cause—Major Effect: A Novel Mode of Control of Bistable Gene Expression
- Germline Mutations Confer Susceptibility to Acute Lymphoblastic Leukemia and Thrombocytopenia
- Leveraging Identity-by-Descent for Accurate Genotype Inference in Family Sequencing Data
- Is Required for the Expression of Principal Recognition Molecules That Control Axon Targeting in the Retina
- Epigenetic Aging Signatures Are Coherently Modified in Cancer
- Silencing of DNase Colicin E8 Gene Expression by a Complex Nucleoprotein Assembly Ensures Timely Colicin Induction
- A Transposable Element within the Non-canonical Telomerase RNA of Modulates Telomerase in Response to DNA Damage
- The Orphan Gene Regulates Dauer Development and Intraspecific Competition in Nematodes by Copy Number Variation
- 9--13,14-Dihydroretinoic Acid Is an Endogenous Retinoid Acting as RXR Ligand in Mice
- The DnaA Protein Is Not the Limiting Factor for Initiation of Replication in
- FGFR3 Deficiency Causes Multiple Chondroma-like Lesions by Upregulating Hedgehog Signaling
- Multiple Changes of Gene Expression and Function Reveal Genomic and Phenotypic Complexity in SLE-like Disease
- Directed Evolution of RecA Variants with Enhanced Capacity for Conjugational Recombination
- The Regulatory T Cell Lineage Factor Foxp3 Regulates Gene Expression through Several Distinct Mechanisms Mostly Independent of Direct DNA Binding
- MreB-Dependent Inhibition of Cell Elongation during the Escape from Competence in
- DNA Damage Regulates Translation through β-TRCP Targeting of CReP
- Multiple In Vivo Biological Processes Are Mediated by Functionally Redundant Activities of and
- The Analysis of () Mutants Reveals Differences in the Fusigenic Potential among Telomeres
- The Causative Gene in Chanarian Dorfman Syndrome Regulates Lipid Droplet Homeostasis in .
- Temporal Expression Profiling Identifies Pathways Mediating Effect of Causal Variant on Phenotype
- The . Accessory Helicase PcrA Facilitates DNA Replication through Transcription Units
- AKTIP/Ft1, a New Shelterin-Interacting Factor Required for Telomere Maintenance
- Npvf: Hypothalamic Biomarker of Ambient Temperature Independent of Nutritional Status
- Transfer RNAs Mediate the Rapid Adaptation of to Oxidative Stress
- Connecting Circadian Genes to Neurodegenerative Pathways in Fruit Flies
- Response to “Ribosome Rescue and Translation Termination at Non-standard Stop Codons by ICT1 in Mammalian Mitochondria”
- Response to the Formal Letter of Z. Chrzanowska-Lightowlers and R. N. Lightowlers Regarding Our Article “Ribosome Rescue and Translation Termination at Non-Standard Stop Codons by ICT1 in Mammalian Mitochondria”
- Simultaneous DNA and RNA Mapping of Somatic Mitochondrial Mutations across Diverse Human Cancers
- Regulation of Insulin Receptor Trafficking by Bardet Biedl Syndrome Proteins
- Altered Levels of Mitochondrial DNA Are Associated with Female Age, Aneuploidy, and Provide an Independent Measure of Embryonic Implantation Potential
- Non-reciprocal Interspecies Hybridization Barriers in the Capsella Genus Are Established in the Endosperm
- Canine Spontaneous Head and Neck Squamous Cell Carcinomas Represent Their Human Counterparts at the Molecular Level
- Genetic Changes to a Transcriptional Silencer Element Confers Phenotypic Diversity within and between Species
- Functional Assessment of Disease-Associated Regulatory Variants Using a Versatile Dual Colour Transgenesis Strategy in Zebrafish
- Translational Upregulation of an Individual p21 Transcript Variant by GCN2 Regulates Cell Proliferation and Survival under Nutrient Stress
- Independent Neuronal Origin of Seizures and Behavioral Comorbidities in an Animal Model of a Severe Childhood Genetic Epileptic Encephalopathy
- The Human Blood Metabolome-Transcriptome Interface
- A Common Cancer Risk-Associated Allele in the Locus Encodes a Dominant Negative Inhibitor of Telomerase
- A Legume Genetic Framework Controls Infection of Nodules by Symbiotic and Endophytic Bacteria
- The Eukaryotic-Like Ser/Thr Kinase PrkC Regulates the Essential WalRK Two-Component System in
- The Yeast GSK-3 Homologue Mck1 Is a Key Controller of Quiescence Entry and Chronological Lifespan
- Dissection of a Complex Disease Susceptibility Region Using a Bayesian Stochastic Search Approach to Fine Mapping
- Exome Sequencing of Phenotypic Extremes Identifies and as Interacting Modifiers of Chronic Infection in Cystic Fibrosis
- The Role of -Mediated Epigenetic Silencing in the Population Dynamics of Transposable Elements in
- Ancestral Chromatin Configuration Constrains Chromatin Evolution on Differentiating Sex Chromosomes in
- Abnormal Activation of BMP Signaling Causes Myopathy in Null Mice
- Reproductive Mode and the Evolution of Genome Size and Structure in Nematodes
- Replication and Active Partition of Integrative and Conjugative Elements (ICEs) of the SXT/R391 Family: The Line between ICEs and Conjugative Plasmids Is Getting Thinner
- Motor and Sensory Deficits in the Mice Result from Mutation of the ESCRT Component HGS
- Senescence in the Sbds-Deficient Murine Pancreas: Cell-Type Specific Consequences of Translation Insufficiency
- Lipophorin Receptors Recruit the Lipoprotein LTP to the Plasma Membrane to Mediate Lipid Uptake
- Separable Crossover-Promoting and Crossover-Constraining Aspects of Zip1 Activity during Budding Yeast Meiosis
- Context-Dependent Functional Divergence of the Notch Ligands DLL1 and DLL4
- Multilayered Organization of Jasmonate Signalling in the Regulation of Root Growth
- Necrotic Cells Actively Attract Phagocytes through the Collaborative Action of Two Distinct PS-Exposure Mechanisms
- A Novel Feedback Loop That Controls Bimodal Expression of Genetic Competence
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Non-reciprocal Interspecies Hybridization Barriers in the Capsella Genus Are Established in the Endosperm
- Translational Upregulation of an Individual p21 Transcript Variant by GCN2 Regulates Cell Proliferation and Survival under Nutrient Stress
- Exome Sequencing of Phenotypic Extremes Identifies and as Interacting Modifiers of Chronic Infection in Cystic Fibrosis
- The Human Blood Metabolome-Transcriptome Interface
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



