-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaThe Analysis of () Mutants Reveals Differences in the Fusigenic Potential among Telomeres
Telomeres are specialized structures that protect chromosome ends from incomplete replication, degradation and end-to-end fusion. Abnormalities in telomere structure or maintenance can promote a variety of human diseases including premature aging and cancer. Although all human telomeres contain the same DNA sequences, they differ from each other in the subtelomeric regions or subtelomeres. Recent work has shown that human subtelomeres control telomere replication and that abnormalities in these structures can lead to localized chromosome instability and disease. However, the relationships between subtelomeres and telomeres are currently poorly understood. Here, we have addressed this problem using the fruit fly Drosophila melanogaster as model system. Drosophila subtelomers are very different from each other as they contain different types of chromatin. We have found that mutations in a gene we called pendolino (peo) cause telomeric fusions (TFs) and that these fusions preferentially involve the telomeres associated with a tightly packed form of chromatin called heterochromatin. Interestingly, none of the 10 mutants with TFs so far described in Drosophila shows the pattern of TFs observed in peo mutants. Thus, our data provide the first demonstration that subtelomeres can affect telomere fusion. We believe that these results will stimulate further studies on the role of subtelomeres in the maintenance of genome stability.
Published in the journal: . PLoS Genet 11(6): e32767. doi:10.1371/journal.pgen.1005260
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1005260Summary
Telomeres are specialized structures that protect chromosome ends from incomplete replication, degradation and end-to-end fusion. Abnormalities in telomere structure or maintenance can promote a variety of human diseases including premature aging and cancer. Although all human telomeres contain the same DNA sequences, they differ from each other in the subtelomeric regions or subtelomeres. Recent work has shown that human subtelomeres control telomere replication and that abnormalities in these structures can lead to localized chromosome instability and disease. However, the relationships between subtelomeres and telomeres are currently poorly understood. Here, we have addressed this problem using the fruit fly Drosophila melanogaster as model system. Drosophila subtelomers are very different from each other as they contain different types of chromatin. We have found that mutations in a gene we called pendolino (peo) cause telomeric fusions (TFs) and that these fusions preferentially involve the telomeres associated with a tightly packed form of chromatin called heterochromatin. Interestingly, none of the 10 mutants with TFs so far described in Drosophila shows the pattern of TFs observed in peo mutants. Thus, our data provide the first demonstration that subtelomeres can affect telomere fusion. We believe that these results will stimulate further studies on the role of subtelomeres in the maintenance of genome stability.
Introduction
Telomeres are nucleoprotein complexes that counterbalance incomplete replication of terminal DNA and protect chromosome ends preventing both activation of cell cycle checkpoints and fusion events. In most organisms, the end replication problem is solved by telomerase, which mediates the addition of short GC-rich repeats to chromosome ends. These repeats specifically bind a discrete number of proteins, which recruit a series of additional factors to form large protein assemblies that ensure proper telomere function and homeostasis (reviewed in [1–4]). Drosophila telomeres are not elongated by telomerase but by targeted transposition of three specialized retroelements called HeT-A, TART and TAHRE (collectively abbreviated with HTT). However, Drosophila telomere formation does not require the HTT arrays; abundant evidence indicates that fly telomeres are epigenetically-determined structures that can assemble at the ends of the chromosomes independently of their terminal DNA sequence (reviewed in [5–7]).
With the exception of budding yeast, in organisms with telomerase telomeres are protected by the conserved shelterin complex. Human shelterin is a six-protein complex (TRF1, TRF2, POT1, TIN2, TPP1 and Rap1) that specifically associates with the telomeric TTAGGG repeats. TRF1, TRF2 directly bind the TTAGGG duplex and POT1 the single stranded overhang. TIN2 and TPP1 do not bind DNA and interconnect TRF1 and TRF2 with POT1. TRF2 interacts with hRap1, a distant homologue of S. cerevisiae Rap1. The shelterin subunits share properties that distinguish them from the non-shelterin telomere-associated proteins: they are specifically enriched at telomeres throughout the cell cycle and appear to function only at telomeres [1]. The human non-shelterin proteins, which are not telomere-specific in localization and function, include the conserved CST complex, HP1a, and proteins involved in DNA repair and/or replication such as the ATM kinase, the Ku70/80 heterodimer, the MRE11/RAD50/NBS1 (MRN) complex, Rad51, the ERCC1/XPF endonuclease, the Apollo exonuclease, the FEN1 nuclease, the RecQ family members WRN and BLM; the RTL1 helicase, RPA70, the Timeless component of the replisome, and the subunits of the conserved ORC prereplication complex. Depletion of any one of the shelterin subunits or the shelterin-associated proteins leads to visible telomere defects ranging from altered packaging of telomeric chromatin (multi telomere signals after FISH), telomere loss and telomere fusion (reviewed in [1–4]; see also [8, 9]).
Most of the Drosophila telomere-capping proteins have been identified by molecular cloning of genes specified by mutations that cause telomeric fusions (TFs) in larval brain cells. Genetic and molecular analyses have thus far identified 11 loci that are required to prevent TF (henceforth they will be designated as TF genes). These are effete (eff; also called UbcD1) that encodes a highly conserved E2 enzyme that mediates protein ubiquitination [10, 11], Su(var)205 that encodes Heterochromatin Protein 1a (HP1a) [12], the Drosophila homologues of the ATM, RAD50, MRE11 and NBS1 DNA repair genes [13–19], without children (woc) that specifies a transcription factor [20]; caravaggio (cav), modigliani (moi), verrocchio (ver) and hiphop that encode the components of the terminin complex [21–25]. HOAP (the cav product HP1/ORC-Associated Protein [21–26]), Moi and HipHop are fast evolving non-conserved proteins that do not share homology with any known telomere-associated protein [22–24]Ver is also a fast evolving protein; however, it contains an OB-fold motif that is structurally homologous to the OB fold of the Stn1 protein of the conserved CST complex [23]. Although the structural characterization of terminin is still incomplete, the extant data suggest that HOAP and HipHop are primarily bound to the DNA duplex while Ver is associated with the single-stranded overhang [6, 22, 23, 26]. In contrast with the other telomere capping proteins (Eff, HP1a, ATM, Mre11, Rad50, Nbs, and Woc) that have multiple localizations and functions, HOAP, HipHop, Moi and Ver localize only at telomeres and appear to function only in telomere maintenance. These properties are similar to the shelterin properties, suggesting that terminin is a functional analog of shelterin [25]. Furthermore, the findings that shelterin subunits are not conserved in flies, that terminin components have no homologues (with the possible exception of Ver) outside Drosophilidae, and that terminin subunits are encoded by fast evolving genes have suggested a hypothesis on terminin evolution. We proposed that the transition between a telomerase-driven and a transposon-driven telomere elongation mechanism generated a divergence in terminal DNA sequences, which exerted a strong selective pressure towards the evolution of sequence-independent telomere binding proteins such as those that comprise terminin. We also hypothesized that non-terminin Drosophila telomere-capping proteins with multiple localizations and functions correspond to ancestral telomere components that did not evolve as rapidly as terminin because of the functional constraints imposed by their participation in diverse cellular processes [23–25].
In addition to their peculiar telomere elongation mechanism, Drosophila telomeres are also characterized by striking variations in their subtelomeric regions. Recent work has shown that subtelomeric regions play important regulatory roles in mammalian telomere behavior. For example, it has been reported that most human telomeres are replicated by forks progressing from subtelomere to telomere [27] and that the timing of telomere replication depends on the type of subtelomeric DNA; the telomeres associated with satellite-like subtelomeric sequences replicate later than telomeres that are not associated with this type of subtelomeric DNA [28]. Furthermore, recent work has shown that chimpanzee telomeres carrying subtelomeric heterochromatin replicate later than telomeres devoid of heterochromatic subtelomeres [29]. However, the fusigenic properties of mammalian telomeres carrying different subtelomeres have never been investigated.
Drosophila is an ideal model organism for investigating the influence of subtelomeric regions on telomere behavior. All Drosophila telomeres terminate with HTT arrays that are capped by terminin; these HTT arrays are juxtaposed to different types of chromatin: canonical constitutive heterochromatin (the Y, XR, and 4L telomeres), clusters of repetitive telomere-associated sequences (designated as TAS, and present at the XL, 2L, 2R, 3L and 3R telomeres), or sequences with both euchromatic and heterochromatic features (4R telomeres). Here we describe a Drosophila gene, pendolino (peo), identified by mutations that preferentially induce TFs between telomeres associated with constitutive heterochromatin. The Peo protein binds terminin but does not have the typical terminin properties, as it is conserved in mammals and associates with several chromosomal sites. In addition, Peo is required for PCNA recruitment and for general DNA replication. However, both the present and previous results strongly suggest that telomere lesions generated by general defects in DNA replication are unable to induce TFs in Drosophila cells. We thus propose that loss of peo function results in specific fusigenic lesions concentrated in heterochromatin-associated telomeres, and that these lesions might be generated during telomere replication.
Results
Isolation and characterization of pendolino (peo)
The pendolino1 (peo1) mutation was isolated by a cytological screen of 120 late lethal mutants mapping to the second chromosome, recovered after I element mobilization by I-R dysgenic crosses (see Materials and Methods). Mitotic cells of DAPI-stained brain preparations from peo1/peo1 larvae displayed very frequent telomeric fusions (TFs; Fig 1A and 1B), often resulting in multicentric linear chromosomes that resemble little “trains” of chromosomes. The pendolino gene was named after this phenotype just as caravaggio, modigliani and verrocchio, which are all names of Italian trains.
Fig. 1. Mutations in peo cause telomeric fusions. 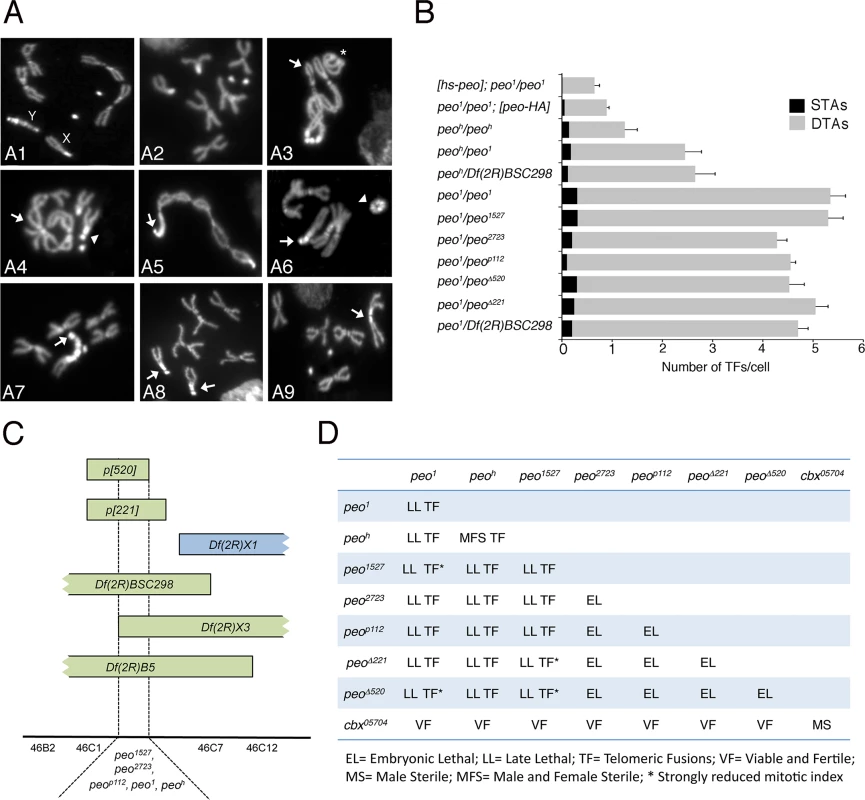
(A) Examples of TFs in peo mutant neuroblasts. (A1-2) Control (Oregon-R) male (A1) and female (A2) metaphases; (A3-A5) peo1/peo1 metaphases showing: (A3) a ring autosome (abbreviated with A; asterisk) and, starting from the arrow, a multicentric chromosome A-XL∙XR-4-4-XR∙XL-A-A; (A4) an XR-4 TF (arrowhead) and an A-A-A-A multicentric chromosome (arrow); (A5) a 4 (arrow)-XR∙XL-A-A-A-A-XL∙XR-4 multicentric chromosome including the entire female complement. (A6-A9) peoh/peoh metaphases showing: (A6) a ring Y chromosome (arrowhead) and a 4 (arrow)-4-XR tricentric chromosome; (A7) a 4 (arrow)-YS∙YL-XR tricentric chromosome; (A8) two 4-XR TFs (arrows); (A9) an XR-XR TF (arrow). (B) Frequencies of TFs in peo mutant combinations. hs-peo and peo-HA are rescued by a construct carrying a wild type copy of the peo gene. At least 250 cells from at least 4 brains were scored for each genotype. (C) Deficiency mapping of peo. (D) Complementation analysis showing the phenotypes of animals heterozygous for the indicated genes/alleles. Recombination analysis with visible markers and deficiency mapping placed peo1 in the 46B-C polytene chromosome interval uncovered by Df(2R)X3 and Df(2R)B5 but not by Df(2R)X1 (Fig 1C). Previous studies mapped to this interval 3 lethal complementation groups [30]. peo1 failed to complement the 1527 and 2723 mutant alleles (henceforth designated as peo1527 and peo2723) of group III for both the lethality and the TF phenotype but complemented representative alleles of groups I and II (Fig 1D). peo1 also failed to complement the P element insertion p112 (henceforth peop112) and the small p221 and p520 deficiencies (henceforth peo∆221 and peo∆520) all generated by the remobilization of the P{w+, ry+}AJN2 insertion [31], originally localized proximally to the Df(2R)X3 breakpoint (Fig 1C and 1D). Finally, we identified another peo mutant allele (peoh) by a cytological screen of a collection of 193 late lethal mutants that arose in the Zucker’s collection of heavily mutagenized viable lines (see Materials and Methods for details). peoh is homozygous viable but male and female sterile, and it is lethal in combination with peo1 (peo1/peoh); the lethal phases of selected combinations of peo mutant alleles and deficiencies are reported in Fig 1D.
To identify the peo gene at the molecular level we exploited the peop112 allele that carries a P{w+, ry+}construct inserted into the gene [30]. Using inverse PCR we found that the P construct is inserted into the 5’ UTR of the longest transcript of the CG10536 gene (Fig 2). CG10536 was originally named ms(2)46C [32] an then renamed crossbronx (cbx) [33]. However, CG10536 does not correspond to ms(2)46C/cbx. The phenotype associated with the ms(2)46C/cbx mutation was attributed to the P{PZ}05704 insertion that maps just proximal to the UTR region of CG10536 (Fig 2). However, this attribution was only tentative because the male sterile phenotype was neither mapped over deficiency nor reverted by P element excision [32]. We found that males homozygous for the P{PZ}05704 insertion are sterile and show the spermatid abnormalities previously described [32]. In contrast, males bearing the same insertion over Df(2R)B5 that uncover the 46B-C interval were fully fertile and displayed normal spermatids (Fig 1D). We thus conclude that the ms(2)46C/cbx mutation maps outside the 46B-C region, and that CG10536 actually corresponds to pendolino.
Fig. 2. Structure of the peo (CG10536) transcripts and available cDNAs. 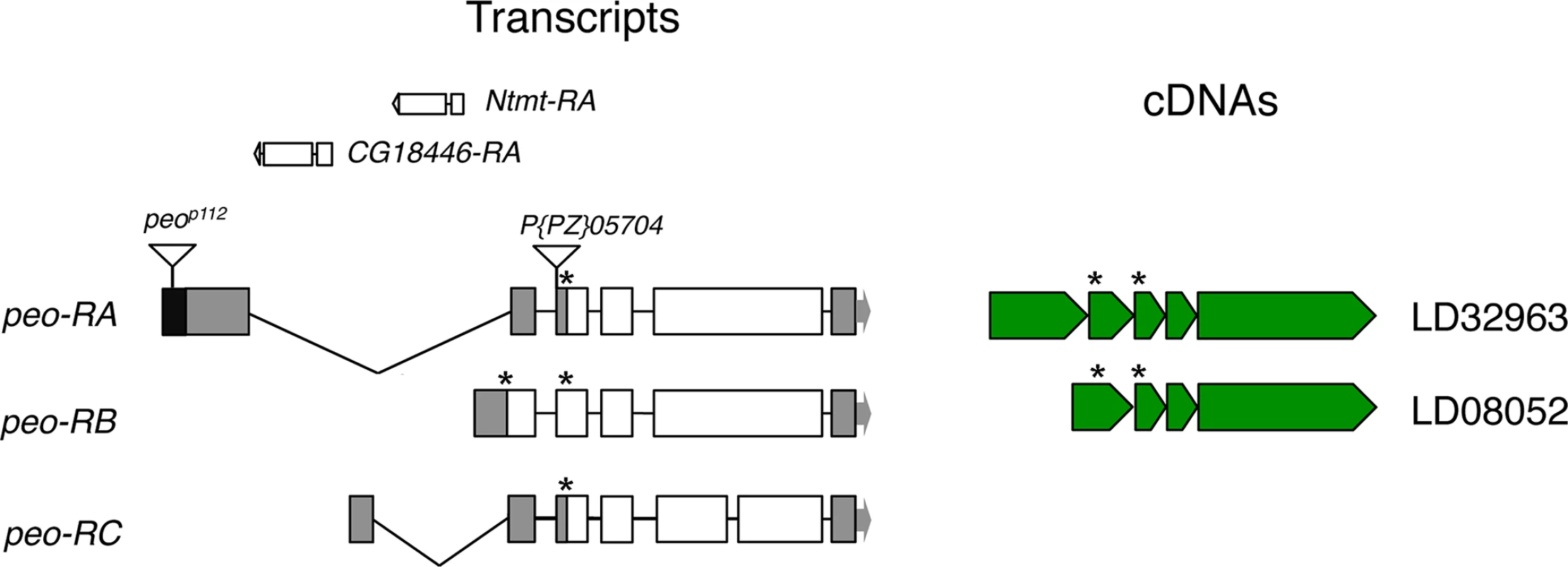
The triangles indicate P element insertions; P[PZ]05704 is not responsible for the crossbronx (cbx) phenotype (see text for detailed explanation). The Ntmt and CG18446 genes are nested into the introns defined by the peo-RA and peo-RC transcripts. The asterisks indicate the positions of ATG start codons. The organization of the peo locus is rather complex. The gene encodes three different transcripts; the long introns of two of these transcripts contain genes (Ntmt and CG18446) with an opposite transcriptional orientation to peo/CG10536 (FlyBase; Fig 2). The three putative peo transcripts encode proteins of 183, 214 and 244 aa and are identified by 2 cDNAs (FlyBase, Fig 2). These proteins share homology with the E2 variant ubiquitin-conjugating enzymes, which are devoid of the catalytic cysteine that mediates ubiquitin transfer [34]. Interestingly, peo has an intronless paralogue (CG16894) that maps to the 56F9 polytene chromosome region. The CG16894 protein shares 35.4 identity and 51.6 similarity with Peo and its function is currently unknown (FlyBase).
Sequencing of peo1 did not revealed nonsense, frameshift, splice-site or missense mutations in the protein-coding sequences of CG10536 with respect to the FlyBase sequences. In addition, sequencing of approximately 2000 bp upstream of the gene ATG and in situ hybridization experiments did not reveal I element insertions. Nonsense, frameshift or splice-site mutations were also absent from the protein coding sequences of peo1527 and peoh mutant genes. All peo mutant alleles displayed several genomic single nucleotide polymorphisms (SNPs) with respect to the Fly Base sequence. However these SNPs were all present in DPGP natural populations (www.dpgp.org/dpgp3), suggesting that they are associated with little if any phenotypic consequences. Collectively, these results suggest that peo1, peo1527 and peoh are regulatory mutations that lower the intracellular amount of the Peo protein (see below). However the molecular lesions in these mutant alleles remain to be identified.
To unambiguously determine the identity of peo we performed rescue experiments using the LD08052 cDNA clone (Fig 2; see also FlyBase). Sequencing confirmed that this cDNA contains the entire coding sequence of the CG10536-RB transcript, which produces the larger protein isoform encoded by the locus (Fig 2). The LD08052 cDNA was fused in frame with the heat shock (hsp70) promoter or used to make a construct containing the tubulin promoter and a 3HA sequence at the 3’ of the gene. Both constructs were then used to make transgenic flies and both rescued the lethality and the TF phenotype of the peo1 mutant flies. In hs-CG10536; peo1/peo1 flies exposed to heat shocks (1h at 37°C every 24 h throughout development), survival of peo1/peo1 individuals with respect their peo1/CyO siblings was 18% of the Mendelian expectation, and the TF frequency dropped to less than 1 per cell from more than 5 per cell (Fig 1B). In the presence of the Tub-CG10536-3HA construct, the survival rate of peo1/peo1 homozygotes with respect to peo1/CyO heterozygotes was 10% and the TF frequency less than 1/cell (Fig 1B). To confirm these rescue data we generated peoh/peoh larvae bearing the Tub-CG10536-3HA construct. The brains of these larvae displayed a 5-fold reduction in TF frequency compared to those of peoh/peoh larvae (0.2 vs 1 TF/ cell). Thus, our results collectively indicate that CG10536 corresponds to peo.
Peo specifically affects heterochromatin-associated telomeres
In most colchicine-treated metaphases from peo1/peo1, peo1/ peo1527, peo1/peo2723, peo1/peop112, peo1/ peo∆221, peo1/peo∆520 and peo1/Df(2R)BSC298 (henceforth Df(2R)BSC298 will be abbreviated with Df), the majority of telomeres were involved in fusions, often forming tangles of chromosomes difficult to resolve (Fig 1A and 1B). However, a careful examination of these tangles led us to estimate the average number of TFs per cell and to determine the relative frequencies of single telomere associations (STAs) and double telomere associations (DTAs). STAs involve a single telomere that fuses with either its sister telomere or another single nonsister telomere. In DTAs, two sister telomeres fuse with another pair of sister telomeres. STAs are likely to be generated during the S-G2 phase, while DTAs are thought to result from the replication of TFs generated during G1 [10, 35]. In peo mutants, DTAs were much more frequent than STAs (Fig 1B) just as in the other TF mutants in which the relative frequencies of STAs and DTAs have been determined, namely eff, Su(var)205, cav, mre11, rad50, nbs, tefu (ATM), woc, moi and ver [10, 12, 15, 16, 21, 20, 23, 24]. This bias towards DTAs may reflect the proximity of Drosophila telomeres during G1 that would be progressively lost as cells proceed through S and G2 [10, 36]. It has been also suggested that telomere fusion occurs primarily during G1, because the DNA repair pathways that join chromosome ends are more active in G1 than in S or G2 [37].
Although peo mutants show a high DTA/STA ratio as the other TF mutants, they exhibit a specific pattern of TFs. The analysis of the peoh mutant that shows ~1 TF/cell allowed a very precise definition of the telomeres involved in fusion events. In peoh homozygous brains of males and females, nearly all TFs involved the telomeres associated with the heterochomatic regions of the chromosomes, namely those of the entirely heterochromatic Y chromosome (YS and YL), the telomere of the right arm of the X chromosome (XR) and the fourth chromosome telomeres (Fig 1A). Of the two telomeres of the fourth chromosome only one is associated with constitutive heterochromatin (4L) but we were not able to distinguish between 4L and 4R, as the DAPI-stained fourth chromosomes appear as brightly fluorescent dots in which the chromosome arms are not discernible. High frequencies of TFs between heterochromatin-associated telomeres (henceforth abbreviated with Ha-telomeres) were also observed in peoh/Df and peoh/peo1 brains (Fig 3). We note that we classified as DTAs all TFs between Ha-telomeres, because the close apposition of the sister chromatids in heterochromatin does not allow a distinction between STAs and DTAs. Thus, the apparent lack of STAs observed in weak peo mutants (Fig 1B) might not reflect a real absence of this type of TFs.
Fig. 3. Involvement of individual telomeres in fusion events in different TF mutants. 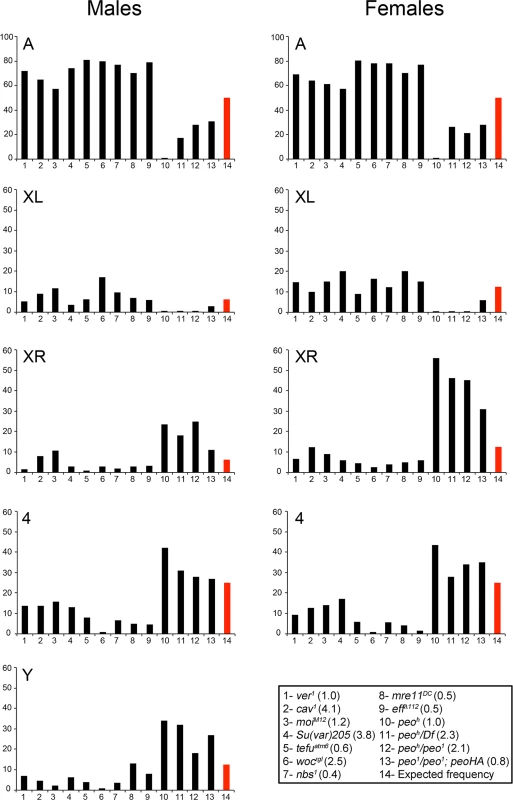
The vertical axes of the graphs show the frequencies (%) with which individual telomeres are involved in TFs. The red columns indicate the expected frequencies assuming a random involvement of telomeres in fusion events. The columns in the graph correspond to numbered genotypes in the box, and the numbers in parentheses next to each genotype indicates the observed TF frequencies. At least 122 fused telomeres from at least 4 brains were scored for each genotype. The Su(var)205 mutant (# 4) is a Su(var)20504/Su(var)20505 heteroallelic combination. The observed number of "heterochromatic telomeres" involved in TFs in each peo mutant combination is significantly higher than would be expected by chance (p < 0.001 in Chi-square test). In contrast, in all other mutants, this number is significantly lower than the expected one (p < 0.001 in Chi-square test). All observed and expected values and the statistical analysis are presented in detail in S1 Table. The high incidence of TFs between Ha-telomeres is not due to an allele-specific effect of peoh, as a high frequency of TFs involving the Ha-telomeres was also observed in peo1/peo1 mutant cells bearing the Tub-peo+-3HA rescue construct (Fig 3). This suggests that the Ha-telomeres are preferentially affected by an impairment of peo function and that this effect is partially masked in strong mutants in which most telomeres are fused. The TF pattern observed in peo mutants is highly specific, as none of the TF mutants we characterized in the past showed a prevalence of TFs between the Ha-telomeres. To precisely compare the patterns of fusions we re-examined all extant Drosophila TF mutants (eff, cav, Su(var)205, mre11, rad50, nbs, tefu, woc, moi and ver). All these mutants displayed an inverse TF pattern compared with peo mutants, with lower than expected frequencies of fusions involving the Y, the XR, and the 4th chromosome telomeres and higher than expected frequencies of TFs between the telomeres of the major autosomes (Fig 3 and S1 Table). We also confirmed that eff mutants do not exhibit a prevalence of fusion between Ha-telomeres (Fig 3) [10]. This finding strongly suggests that the canonical E2 ubiquitin conjugating enzyme encoded by eff and the E2 variant encoded by peo do not function in the same telomere protection pathway.
In interphase nuclei, the heterochromatic regions of the chromosomes aggregate to form an irregular mass of chromatin called chromocenter [38]. Thus, the specific pattern of TFs observed in peo mutants could depend either on an abnormal organization and compaction of the chromocenter or on a specific chromatin composition of heterochromatic telomeres. To discriminate between these possibilities we introduced in a peoh mutant background a Bs w+ y+ Y chromosome that carries a euchromatic fragment marked with Bs w+ and y+ at its YL end [39] (see also FlyBase). We found that in peoh mutant brains the frequency with which the Bs w+ y+ Y chromosome is involved in TFs is more than 5-fold lower than that of a normal Y (Fig 4). Specifically, the formation of Y rings, which in peoh/peoh mutants represents ~90% of the total fusions involving the Y (that together are more than 40% of the observed TFs), was drastically reduced in the presence of a Bs w+ y+ Y. Because the cells bearing a Bs w+ y+ Y chromosome do not exhibit detectable morphological variation in the chromocenter compared to wild type, these results strongly suggest that the preferential involvement of the Ha-telomeres in peo-induced TFs is a consequence of their association with heterochromatin and not of their arrangement within the interphase nucleus.
Fig. 4. Y chromosome ring formation in peo mutants. 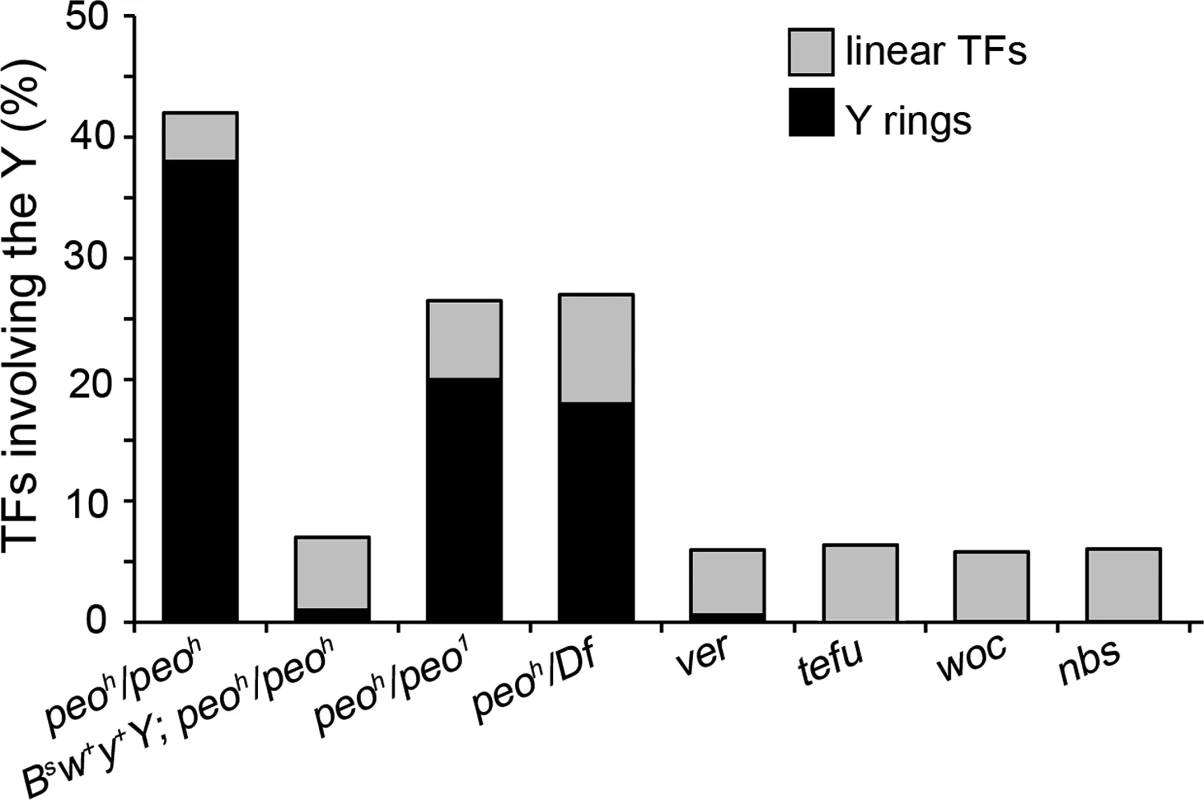
peo mutants brains exhibit an extremely high Y-ring frequency that is not observed in other TF mutants. The frequency of Y rings in peo mutants is much higher than expected assuming a random involvement in TFs of the heterochromatin-associated telomeres (see text) and is strongly reduced when mutants bear a BSw+y+Y that carries a segment of euchromatin appended to the end of its left arm. The particularly high frequency of Y rings in peo mutants is another peculiar feature of their TF pattern. Indeed, the Y rings are rare or virtually absent in mutants in genes such as ver, tefu, woc, and nbs (Fig 4). On the assumption of a random involvement of telomeres in TFs, the expected frequency of Y ring is 1/15 of the total fusions involving the Y chromosome and 1/6 of the fusions involving the Y and either the XR, or the 4th chromosome telomeres. Thus, formation of Y rings in peo mutants is specifically and strikingly frequent. This could reflect a particularly high frequency of fusigenic lesions on the Y telomeres, or the contemporary presence of these lesions on the opposite Y telomeres, or both.
Peo interacts with terminin
We have recently observed that brains from larvae heterozygous for both Su(var)20505 and cav exhibit ~ 0.1 TFs/cells, while in wild type, Su(var)20505/+, cav/+ brains the TF frequency was virtually zero (300 metaphases analyzed in each case). This finding suggests that Su(var)205 and cav genetically interact and that a simultaneous reduction of HP1a and HOAP results in a low level of TFs. We thus asked whether peo exhibits similar genetic interactions with other TF mutants. Double heterozygotes for peo1 and either cav1, moi1, ver2, Su(var)20504 or Su(var)20505 displayed TF frequencies ranging from 0.05 to 0.1 per cell, whereas heterozygotes for peo1 and either effΔ73, wocB111, mre11DC, rad50Δ5.1, nbs1 or tefuatm6 did not exhibit TFs (in all cases, we examined at least 300 metaphases).
We next asked whether the Peo protein physically interacts with the terminin components. A preliminary experiment using the yeast two-hybrid assay suggested that Peo directly interacts with HOAP but not with HP1a (S1 Fig). To confirm this result we performed a GST pulldown assay using bacterially expressed 6His-Peo and GST-tagged HOAP polypeptides of different length. As shown in Fig 5A and 5B, the intact HOAP protein and the HOAP fragments containing the N-terminal region of the protein (aa 1–145) precipitated Peo, while a larger HOAP fragments including the 3 repeated segments of the protein (aa 109–343) failed to bind Peo. We then investigated whether Peo interacts with Moi and Ver. We performed GST pulldown experiments with extracts from human 293T cells expressing Peo-FLAG and either GST alone, GST-Moi, GST-Ver or GST-HOAP. We chose to express Drosophila tagged proteins in human cells because a heterogeneous cellular environment is likely to reduce the probability of indirect interactions among fly proteins. As shown in Fig 5C, Peo-FLAG is precipitated by GST-Moi, GST-Ver and GST HOAP but not GST alone. Collectively, these results provide strong evidence that Peo directly binds HOAP; in addition, they suggest that Peo directly interacts with Moi and Ver.
Fig. 5. Peo directly interacts with HOAP and is also likely to bind Moi and Ver. 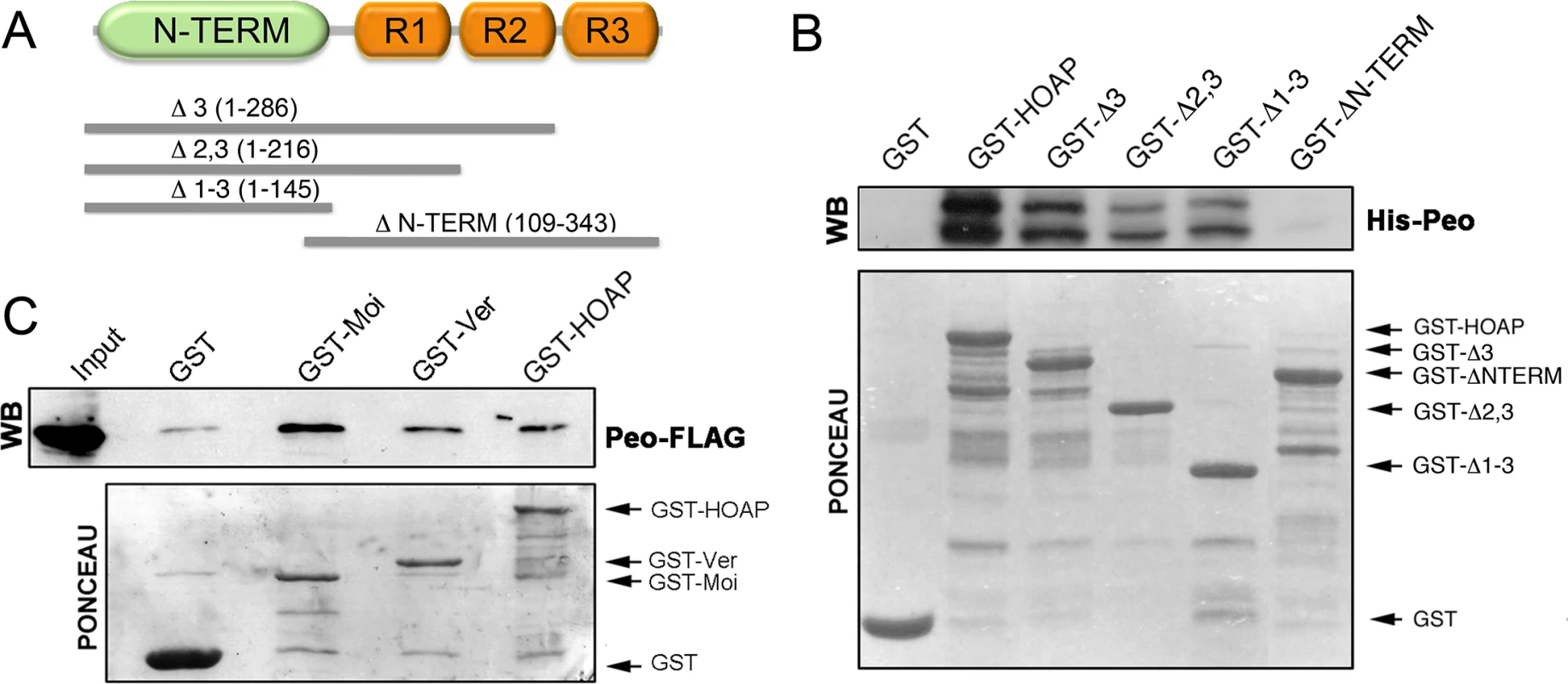
(A) Schematic of HOAP truncations used in GST pulldown experiments; R indicates the repeated segments found in the C-proximal half of the protein. (B) Bacterially purified GST-HOAP segments spanning the N proximal half of the protein precipitate bacterially expressed His-Peo, which is not pulled down by GST alone or the C-proximal half of HOAP. His-Peo was detected using our anti-Peo antibody; the two Peo bands are likely to correspond to different translation products generated in E. coli starting from the two closely spaced ATG codons present on peo cDNA (see Fig 2) (C) GST-HOAP, GST-Moi and GST-Ver precipitate Peo-FLAG from HeLa cell extracts. Peo-FLAG was detected with anti-FLAG antibody. Mapping the Peo sites that interact with the terminin proteins
peo encodes an E2 variant (UEV) enzyme; the UEV proteins are similar to the E2 ubiquitin conjugating enzymes (UBCs) but lack the catalytic cysteine residue that mediates the interaction between ubiquitin and E2 [40]. We elaborated a three-dimensional model of Peo exploiting a series of bionformatic analyses (Fig 6A; see also Material and Methods and S2 Fig). We confirmed that Peo lacks the catalytic cysteine of E2 enzymes. In addition, 8 residues before the catalytic cysteine site, Peo exhibits an HPH tripeptide (S2 Fig) instead of HPN, which is a canonical signature of the E2 superfamily [41]. Peo contains a UEV domain of ~150 amino acids resembling the canonical E2 fold in its hydrophobic core and active site region. This domain consists of three helices packed against a four-stranded antiparallel β-sheet. Next to the UEV domain, Peo contains two C-terminal helices that are present in all E2 proteins but missing in other E2 variant enzymes such as Tsg101 and Mms2. Finally, prediction of potentially disordered regions revealed that Peo also contains a long (50 aa) disordered region at the C-terminus (Figs 6A and S2; see also Materials and Methods).
Fig. 6. Mapping the Peo regions that interact with terminin. 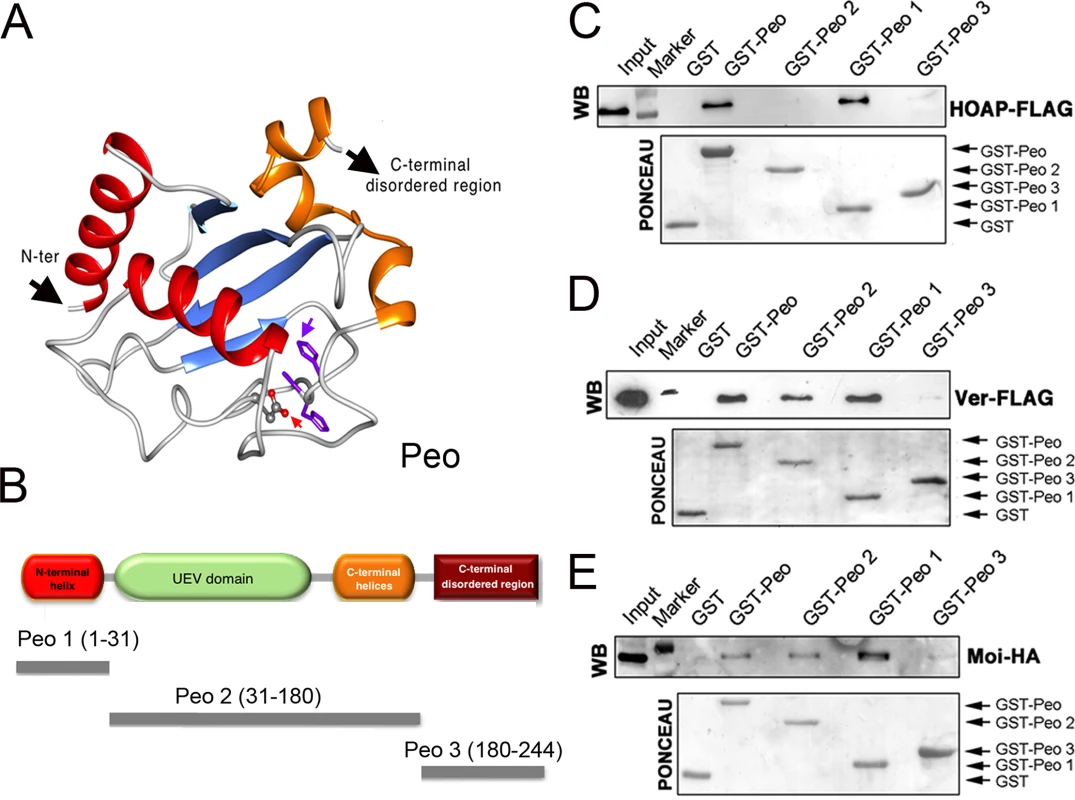
(A) A tridimensional molecular model for Peo. The arrow pointing to “N-ter” indicates the N-terminus of the protein; the arrow pointing to “C-ter” indicates the starting site of the disordered C-terminal region of Peo (not depicted); the variant Asp residue and His-Pro-His motif are represented as sticks and indicated by red and purple arrows, respectively (see Materials and Methods and S2 Fig for construction of the Peo 3D model). (B) Schematic organization of the Peo protein and Peo truncations used for GST pulldown. (C-E) GST-pulldown from S2 cells extracts expressing HOAP-FLAG (C), Ver-FLAG (D) or Moi-HA (E). HOAP-FLAG, Ver-FLAG and Moi-HA were detected with anti-FLAG and anti-HA antibodies. The C-terminal disordered region (included in the Peo 3 fragment) does not interact with any of the terminin components. HOAP specifically interacts with N-terminal region of Peo; in contrast, Moi and Ver interact with both the N terminal and the UEV-containing central regions of the protein. To map the Peo sites that interact with the terminin proteins, we subdivided Peo into 3 GST-tagged fragments: Peo 1 (aa 1–31) that contains the Peo N terminal region which is absent in the short isoform (see Fig 2); Peo 2 (aa 31–180) that includes the central UEV domain of the protein; and Peo 3 (aa180-244) that contains C-terminal disordered region of Peo (Fig 6B). These bacterially expressed GST-tagged Peo fragments were then used to probe Drosophila S2 cell extracts expressing HOAP-HA, Moi-HA or Ver-FLAG (Fig 6C–6E). GST pulldown showed that both Moi and Ver interact with the entire Peo protein (GST-Peo), GST-Peo 1 and GST-Peo 2 but not with GST-Peo 3 or GST alone (Fig 6D and 6E). HOAP did not display the same interaction pattern as Moi and Ver. It was precipitated by GST-Peo and GST-Peo 1 but not by GST-Peo 2 and GST-Peo 3 and thus failed to interact with the Peo UEV domain (Fig 6C).
Peo is not required for terminin recruitment at telomeres
The finding that Peo binds HOAP, Moi and Ver prompted us to ask whether Peo is required for terminin localization at chromosome ends. Although Peo does not appear to interact with HP1a, we also asked whether loss of the wild type function of peo affects HP1a localization at telomeres. Of the latter proteins, only HOAP is clearly detectable at both mitotic and polytene chromosome telomeres; HP1a, Moi and Ver can be easily detected at polytene chromosome ends but not at mitotic telomeres [23, 24, 42]. We thus analyzed HOAP localization in both mitotic and polytene chromosomes of peo1 homozygous mutants; HP1a, Moi and Ver localization was instead studied only in peo1 polytene chromosomes.
An analysis of mitotic chromosomes immunostained for HOAP revealed that mutations in peo do not substantially affect HOAP localization at telomeres, as the frequency of HOAP-stained telomeres in peo mutants and the intensity of the signals were similar to those observed in wild type controls (Fig 7A). Consistent with these results, immunostaining with anti-HOAP and ant-HP1a antibodies showed that peo1/Df mutants exhibit normal concentrations of these proteins at polytene chromosome telomeres (Fig 7B). Because antibodies to Moi or Ver are not currently available, to analyze the localization of these proteins in peo mutants we constructed flies expressing GFP-tagged forms of Moi or Ver in a peo1 mutant background. The analysis of unfixed polytene chromosome nuclei from peo1/peo1 flies expressing either GFP-Moi or Ver-GFP revealed that they exhibit 6 discrete GFP signals (Fig 7C), which we have previously shown to correspond to the 6 euchromatic telomeres of polytene chromosomes [23, 24]. In addition, we found that the intensities of these signals were comparable to those observed in peo1/+ heterozygotes (Fig 7C). Collectively, these results indicate that the wild type function of peo is not required for telomeric localization of HOAP, Moi, Ver and HP1a, and that the strong telomere fusion phenotype observed in peo mutants is not due to the absence of any of these proteins.
Fig. 7. Terminin and HP1a localize normally at the telomeres of peo mutant cells. 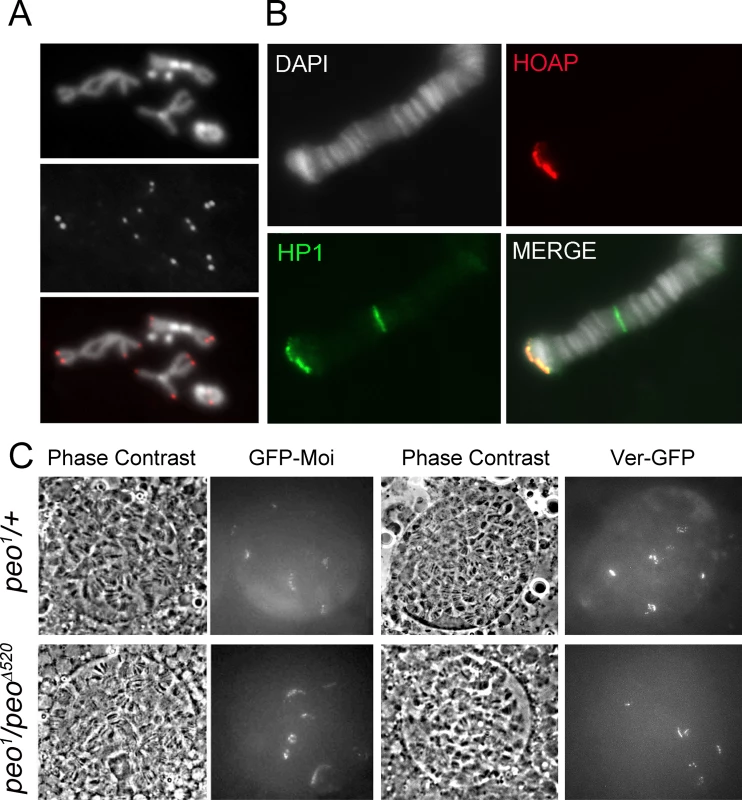
(A) HOAP is normally enriched at the telomeres of brain cell metaphases peo1/peo1 mutants (B). HOAP and HP1a localize normally to the telomere of peo1/peo1 polytene chromosomes (C) Normal localization of Ver-GFP and GFP-Moi in live, unsquashed salivary gland nuclei from peo1/peo∆520. Note that these nuclei display 6 discrete fluorescent signals that are likely to correspond to the telomeres of XL, 2L, 2R, 3L, 3R and 4R. Peo is a non-terminin protein that localizes to multiple chromosome sites
As pointed out in the introduction, terminin subunits are non-conserved fast-evolving proteins that localize and function only at telomeres. Peo is not a terminin-like protein because it does not share any of these properties. Peo is well conserved in Drosophila species (S3 Fig) and has homologues in mouse and humans (Ft1 and AKTIP, respectively). In addition, a three-dimensional model of Drosophila Peo is rather similar to an AKTIP model [43].
To determine the subcellular localization of Peo we raised a rabbit polyclonal antibody against the entirety of Peo and affinity purified it against a GST-Peo fusion protein (see Material and Methods). Western blotting analysis showed that this antibody recognizes 3 bands of ~ 32, ~28 and ~ 25 kDa. In extracts from peo1/Df, and peo1527/Df larval brains, these 3 bands were reduced by approximately 70% compared to +/Df controls (Fig 8A and 8B). Thus, the band with the highest molecular weight is likely to correspond to the longest Peo isoform, while the other two bands might correspond to the shorter isoforms (see Fig 2). In peoh/Df mutant brains, the Peo bands were reduced by approximately 20% with respect to +/Df brains, consistent with the relatively low TF frequency observed in peoh mutants (Fig 1). These results indicate that peo1 and peo1527 are strong hypomorphs compared to the weaker peoh allele.
Fig. 8. Peo expression and localization in polytene chromosomes. 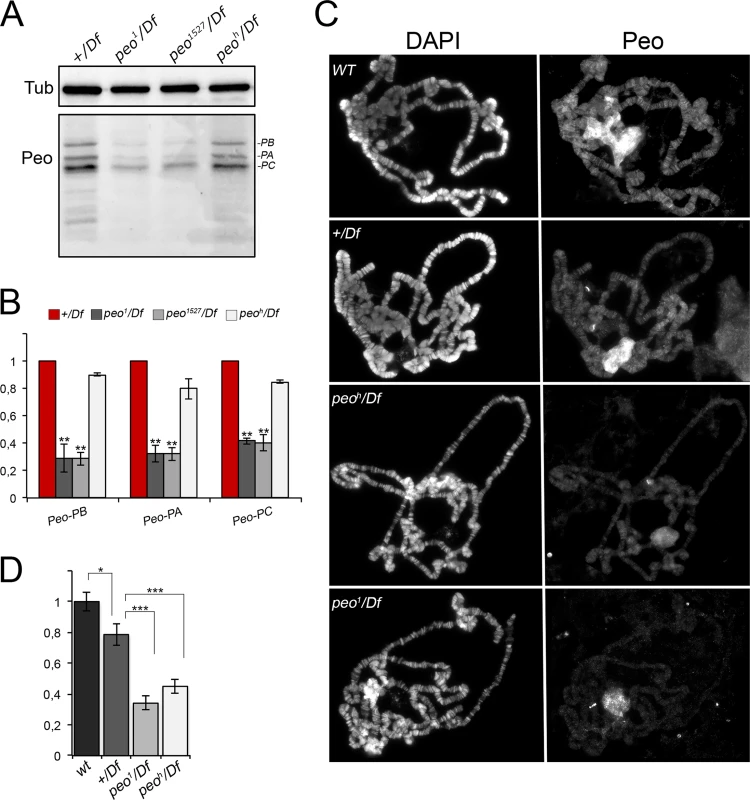
(A) Western blotting showing the levels Peo expression in larval brains. The affinity purified anti-Peo antibody reacts with 3 bands of the apparent molecular weights of 32, 28 and 25 kDa, which are likely to correspond to the 3 Peo isoforms (PA, PB and PC; see Fig 2 and FlyBase). (B) Quantification of the intensities of the 3 bands (see Materials and Methods) after normalization to the tubulin band (Tub) used as loading control; bars show the mean values of 3 experiments ± SEM. In peo1/Df and peo1527/Df brains the 3 bands are reduced by 60–70% compared to +/Df brains used as control; ** significantly different from +/Df with p < 0.01 in the Student's t test. In peoh/Df brains the band intensity reduction is modest, ranging from 10 to 20%. (C) Immunostaining of wild type (WT), +/Df, peoh/Df and peo1/Df polytene chromosomes showing that Peo accumulates in the nucleolus and decorates many chromosome bands. (D) Fluorescence quantification (± SEM) of chromosome arms (but not of the nucleolus; see Materials and Methods) showed that the fluorescence intensities of peoh/Df and peo1/Df chromosomes are both significantly lower than those of either +/Df or wild type chromosomes. * and *** significant with p < 0.05 and p < 0.001 in the Student's t test, respectively. Indirect immunofluorescence experiments on wild type polytene nuclei showed that the anti-Peo antibody stains the nucleolus and many bands along the polytene chromosomes (Fig 8C and 8D). In peoh/Df and peo1/Df nuclei, both the nucleolus and chromosome staining were significantly reduced compared to either +/Df or wild type nuclei, confirming the specificity of the antibody. In addition, the polytene chromosomes of peoh/Df larvae were more intensely stained than those of peo1/Df larvae (Fig 8C and 8D), confirming that the peo1 allele is stronger than peoh.
Although Peo is enriched at numerous polytene bands and interbands, we were not able to detect clear Peo accumulations at chromosome ends. Given that Peo interacts with terminin and it is required to prevent telomere fusion, the most likely explanation for this finding is that Peo is present at the telomere caps in amounts that are not detected by the antibody and the immunostaining technique used here.
Peo is required for DNA replication
We have recently found that AKTIP/Ft1 is required for proper telomeric DNA replication [43]. This finding suggested that peo mutations could also impair DNA replication and specifically affect heterochromatic telomeres, which are likely to replicate at the end of the S phase together with the bulk of heterochromatin (reviewed in [38]). To test this possibility we examined DNA replication in brain cells by analyzing the incorporation of the EdU (5-ethynyl-2’-deoxyuridine) analog of thymidine. Brains were incubated in saline containing 10 mM EdU for 1 h, immediately fixed and then stained with the Click-It Alexa Fluor method to detect EdU (see Methods). In wild type brains, 10% of the nuclei were actively replicating their DNA and incorporated EdU, whereas in peoh/peoh and peo1/peo1 brains the frequency of EdU-labeled nuclei dropped to 7% and 5.5%, respectively (Fig 9A and 9B). In contrast, in woc and ver mutants, the frequency of EdU-positive nuclei was not significantly different from wild type controls, suggesting that the DNA replication defect observed in peo mutants is a specific outcome of the reduced peo activity and not a general consequence of impaired telomere protection.
Fig. 9. Mutations in peo affect DNA replication in brain cell nuclei. 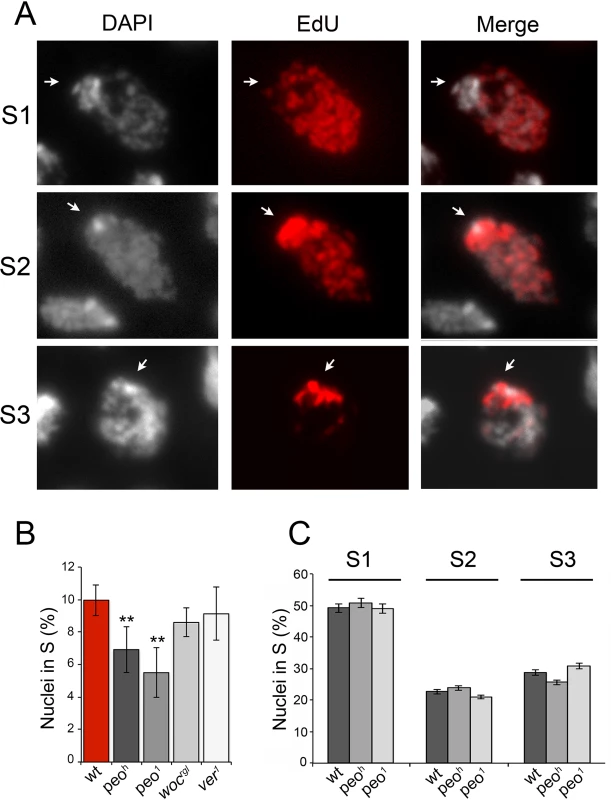
(A) Analysis of EdU-treated brain cell nuclei reveal 3 gross incorporation patterns: S1 nuclei incorporate EdU in most of the nucleus but not in the DAPI stained chromocenter and are presumably in early S; S2 nuclei incorporate EdU in both the chromocenter and the rest of the nucleus and are likely to be in mid-S; S3 nuclei incorporate EdU only in the chromocenter an are thus in late S. (B) peo mutant brains exhibit a significant reduction in the frequency of EdU positive nuclei compared to wild type (wt), ver or woc mutant brains (** significant in Student's t test with p <0.01). The frequencies of EdU positive nuclei in ver and woc brains are not significantly different from the wild type frequency (C) Mutations in peo do not alter the relative frequencies of S1, S2 and S3, nuclei suggesting that Peo is not required for a specific step of the S phase. The frequencies and types of EdU labeled nuclei in each sample (wild type, peoh/peoh and peo1/peo1) were obtained by examining at least 5,000 nuclei from at least 6 brains. EdU staining also allowed subdivision of the S phase according to the incorporation pattern. We distinguished nuclei in early/mid S (S1) in which the nucleus was partially or completely stained with the exception of the heterochromatic chromocenter, nuclei in mid/late S (S2) in which both the chromocenter and the less compact nuclear areas were stained, and nuclei in late S (S3) in which only the chromocenter displayed EdU incorporation (Fig 9). We found that wild type and peo mutant brains do not differ in the relative frequencies of nuclei showing S1, S2 and S3 EdU incorporation patterns. Thus, we conclude that the wild type function of peo is required for general DNA synthesis and not for completion of specific sub-phases of DNA replication.
To gather additional information on the role of Peo in DNA replication we analyzed the distribution of PCNA (proliferating cell nuclear antigen) in peo1/peo1 and peoh/peoh mutant nuclei. PCNA is a processivity factor for DNA polymerases; in nuclei PCNA is either present in a soluble form that can be extracted by detergent treatment or in a detergent-resistant form tightly associated with DNA replication forks (chromatin-bound PCNA) [44]. The analysis of Triton X-extracted brain preparations immunostained for PCNA showed that in wild type 7% of the nuclei were PCNA-positive, while in peo1/peo∆520 and peoh/peoh brains the frequencies of PCNA-stained nuclei were 1.2% and 0.8%, respectively (Fig 10). These results are consistent with those on EdU incorporation and show that in peo mutants the frequency of nuclei with chromatin-bound PCNA is much lower than in control.
Fig. 10. Peo is required for PCNA incorporation into brain cell nuclei. 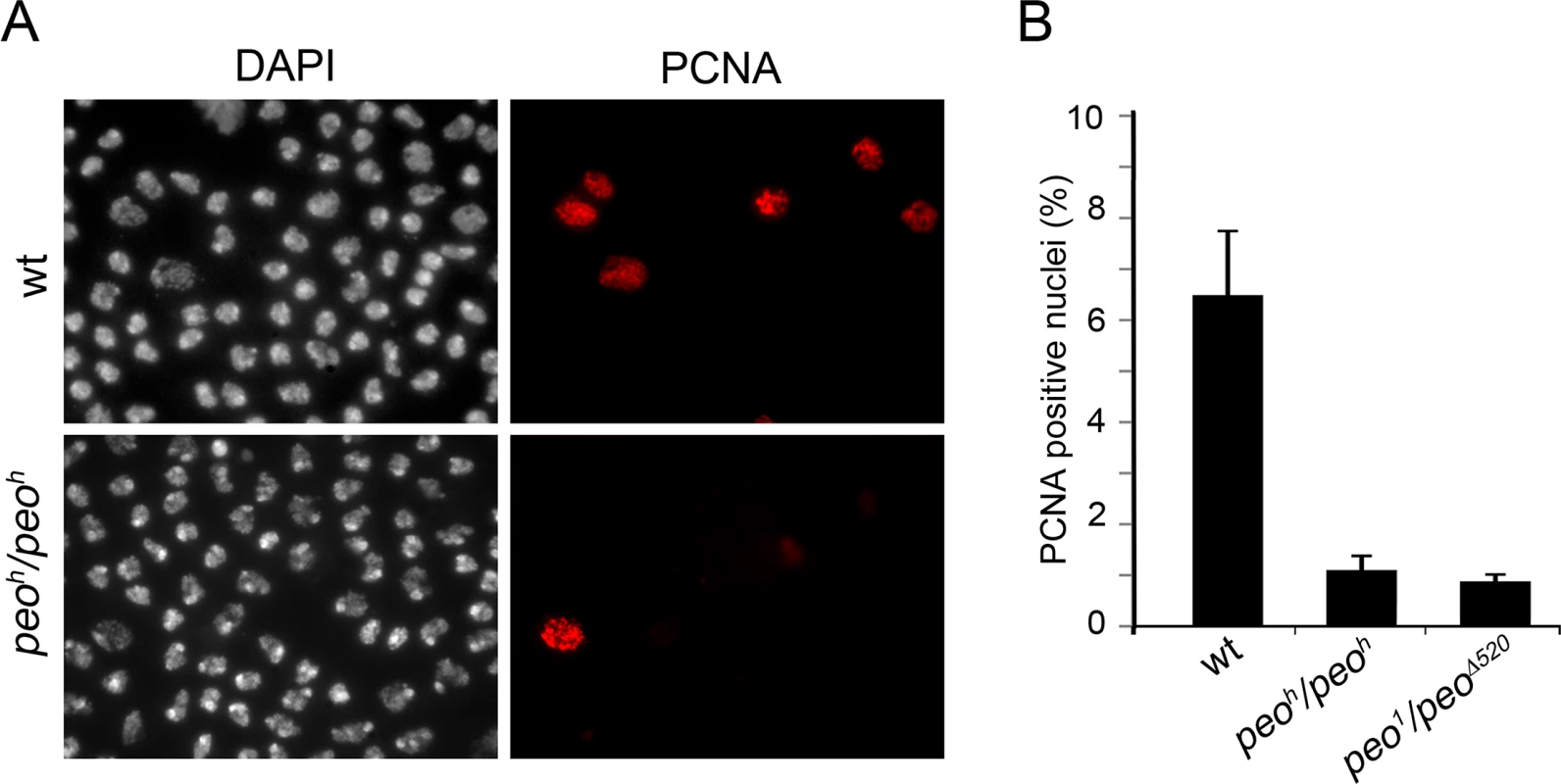
(A) Example of brain cell nuclei stained for PCNA after extraction with Triton X-100. (B) Frequencies of PCNA positive nuclei. The frequencies of PCNA positive nuclei in each sample (wild type, peoh/peoh and peo1/peo∆520) were obtained by examining at least 4000 nuclei from at least 4 brains. A general impairment of DNA replication does not result in TFs
Collectively our results suggest three possibilities: (i) that any impairment of Drosophila telomere replication results in telomere fusion, (ii) that Peo is required for a specific step of telomere replication and that an incorrect execution of this steps results in fusigenic lesions, or (iii) that Peo plays a dual function being independently required for telomere replication and telomere capping. To discriminate between these possibilities we treated wild type and peoh/peoh mutant brains with the DNA polymerase inhibitor aphidicolin (APH), which is known to impair human telomere replication [45–47]. As shown in Fig 11, control and mutant brains were treated in three different ways. Dissected brains were incubated for 1.5 hours in 110 mM APH in NaCl 0.7%. They were then washed, transferred into 3 ml of APH-free saline and fixed 1, 2 or 3 hours after the end of the APH treatment; in all cases, 1 hour before fixation, we added colchicine to the saline to collect metaphases. None of the APH treatments caused TFs in wild type brains or increased the TF frequency in peoh/peoh mutants. We note that the APH treatment was highly effective as it caused a strong reduction in the frequency of mitoses in brains fixed 1 h after the end of the APH treatment (Fig 11B).
Fig. 11. Aphidicolin (APH) does not induce TFs in Drosophila brain cells. 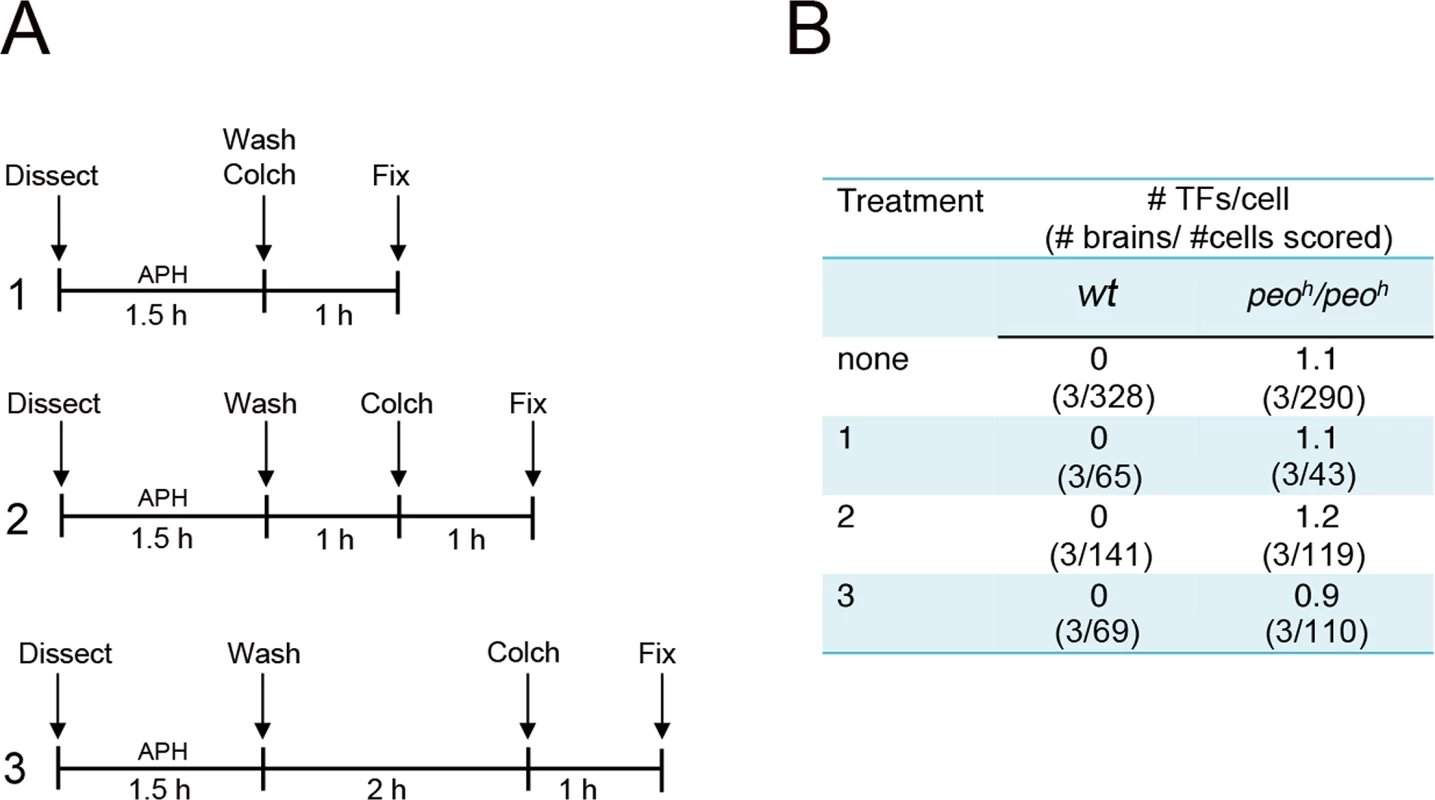
(A) Experimental scheme used. Larval brains were dissected in saline (Dissect), incubated for 1.5 hours in saline plus APH, briefly rinsed in saline (Wash), and then transferred to 3 ml of APH-free Schneider’s medium supplemented with 10% FBS; Colch, addition of colchicine; Fix, fixation. Wild type and peoh/peoh control brains were not treated with APH but processed like the APH-treated brains; they were fixed after 2 hours incubation in the medium following colchicine addition. (B) Results of the experiments outlined in A. Our previous work has shown that mutations in Drosophila timeless2 (tim2) result in chromosome breakage but not in TFs [48]. Tim2 is the Drosophila ortholog of mammalian TIM, a replisome component that facilitates DNA synthesis [49, 50] and is required for telomere replication [8]. These results prompted us to examine the cytological phenotype of brains from mutants in the Blm (formerly mus-309) gene [51], the Drosophila homolog of the Bloom syndrome gene, which encodes a DNA helicase required for mammalian telomere replication [46, 47]. We found that BlmD2/BlmD3 mutant brains exhibit chromosome breaks ranging from 4 to 7% but not TFs (500 metaphases scored from 7 brains). These results suggest that a general impairment of Drosophila telomere replication does not result in TFs.
Discussion
Mutations in peo preferentially affect specific Drosophila telomeres
We have shown that strong peo mutants exhibit an average of 5 TFs/cell; these TFs appear to involve all the telomeres of the Drosophila chromosome complement making difficult a reliable assessment of the relative involvement of individual telomeres in fusion events. However, in weak peo mutants or in strong mutants bearing a peo+ rescue construct, which exhibit ~ 1 TF /cell, the majority of fusions involved the XR, the Y and the 4th chromosome telomeres. These results suggest that these telomeres are preferentially affected by mutations in peo and that this effect is partially masked in strong mutants where most telomeres are fused.
In all the other Drosophila mutants we characterized (eff, Su(var)205, cav, mre11, rad50, nbs, tefu, woc, moi and ver) individual telomeres were engaged in TFs with frequencies that are different from those expected for a random involvement. The telomeres of the major autosomes were involved in TFs more frequently than expected, while participation of the XL telomere in fusions was either slightly lower or conformed to the expected frequency. In contrast, the Y, the XR, and the 4th chromosome telomeres were engaged in TF less than expected (Fig 3 and S1 Table). peo mutants displayed an inverse TF pattern, with higher than expected frequencies of fusions involving the Y, the XR, and the 4th chromosome telomeres (Fig 3 and S1 Table). To the best of our knowledge, this is the first case in which individual telomeres of an organism exhibit different fusigenic capabilities in response to the genetic background. We believe that this finding reflects the peculiar structural differences between the telomeric regions of the different Drosophila chromosomes.
All Drosophila chromosomes of wild type strains terminate with HTT arrays of variable length (made of complete and incomplete HeT-A, TART and TAHRE elements) (reviewed in [5, 52, 53]) and are capped by the multiprotein terminin complex (reviewed in [6]). However Drosophila telomeres differ from each other in both the type of subtelomeric chromatin and in the properties of their HTT arrays (Fig 12). One of the most straightforward features that contradistinguish some of the Drosophila telomeres is their association with constitutive hetrochromatin. Approximately one-third of the Drosophila genome is made of constitutive heterochromatin; the entire Y chromosome, the short arm (XR) and the proximal 40% of the long arm (XL) of the X chromosome, the short arm (4L) and the proximal 70% of the long arm (4R) of the 4th chromosome, and the centric 25% of chromosomes 2 and 3 are heterochromatic [38, 54]. Thus the HTT arrays of YL, YS, XR and 4L are linked to constitutive heterochromatin (Fig 12). The HHT blocks of XL and those of the major autosomes are not directly associated with euchromatin but are instead juxtaposed to divergent clusters of subtelomeric repeats, known as telomere associated sequences (TAS) (reviewed in [55]). The TAS are not only different in sequence but are also occasionally absent from the subtelomeric regions, suggesting that their presence is not essential for proper telomere function [55]. Finally, the 4R telomere is joined to a special type of chromatin that has peculiar features, as well as features shared with both euchromatin and heterochromatin; for example the 4R distal chromatin is enriched in the 4th chromosome-specific Painting of four (Pof) protein and the heterochromatic markers HP1a and histone 3 methylated at lysine 9 (H3K9) (reviewed in [56]) (Fig 12).
Fig. 12. Structure of Drosophila telomeres and subtelomeres. 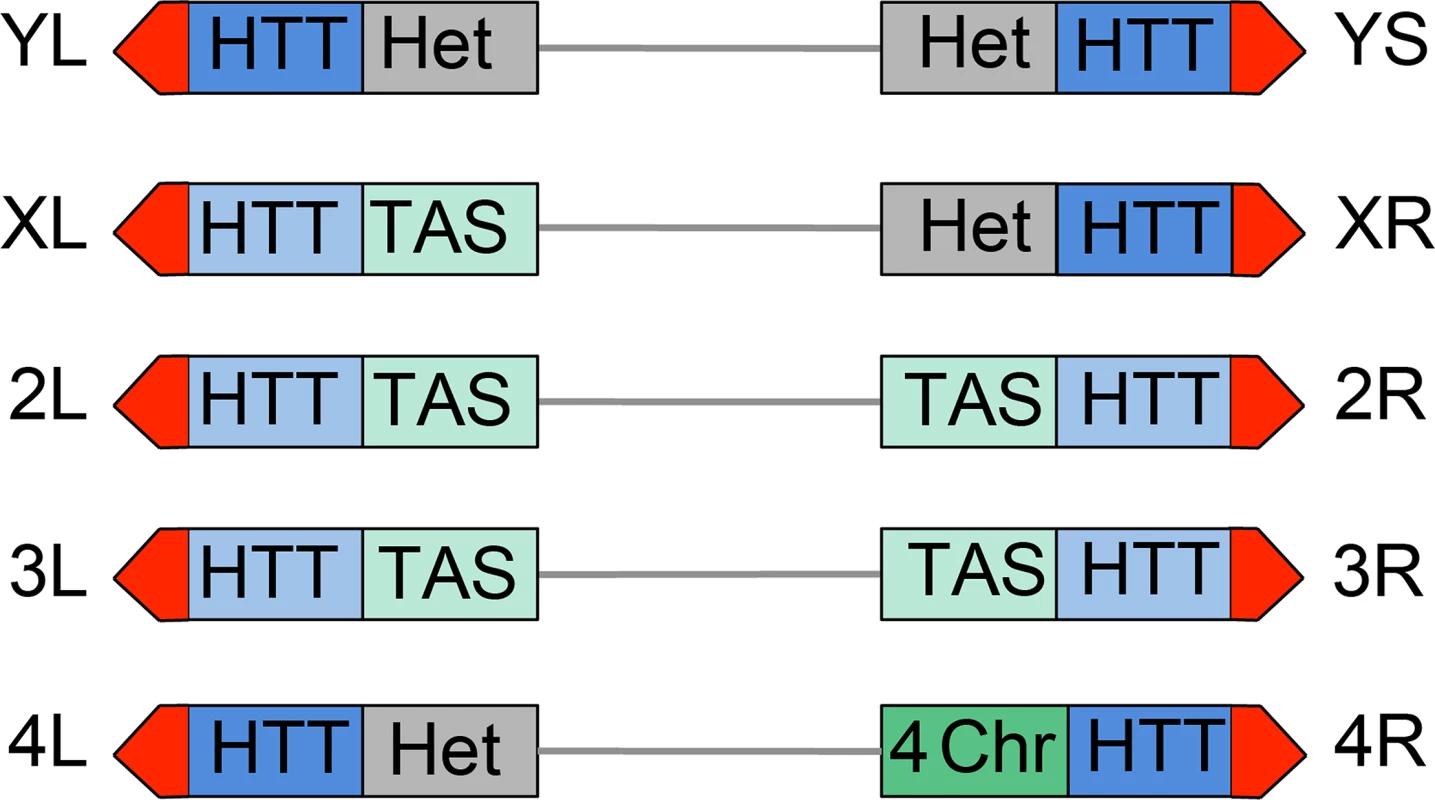
Schematic representation of Drosophila chromosome ends. From a distal to a proximal direction chromosome ends contain the terminin-associated region (red) that may extend for approximately 10 Kb (22), the HTT array that vary in length from 26 to 147 Kb [81] and may either repress (dark blue) or not repress (light blue) the expression of transgenes; the TAS sequences (light green) that comprise about 20 Kb [55, 81], or different types of chromatin: constitutive heterochromatin (Het, grey), or the 4th chromosome chromatin (4-ch, dark green) that has distinctive properties as well as properties shared by euchromatin and heterochromatin. The repressive properties of the YL, XR and 4L HTT have been inferred from those of the YS HTT; the properties of XL and 2L HTT are inferred from those 2L, 3L and 3R HTT. See text for detailed explanation. Additional features that differentiate Drosophila telomeres are the silencing properties of their HTT arrays (Fig 12). Several studies have shown that white+ transgenes inserted within or next to the TAS are partially silenced leading to a variegated eye phenotype (a phenomenon known as telomere position effect or TPE) (reviewed in [5]). However, white+ transgenes inserted into the HTT arrays behave differently depending on the insertion site. Insertions into the HTTs of telomeres not directly joined to heterochromatin such as those of the 2L, 3L and 3R arms do not appear to be subject to TPE [57]. In contrast white+ transgenes inserted within the 4R (only one tested) and YS (6 tested) HTTs lead to a variegated eye phenotype [57, 58]. Furthermore, the white+ transgenes inserted into the HTT array of YS and those embedded into or near the TAS respond differently to genetic modifiers. For example, mutations in Su(var)205 (HP1a), which suppress variegation of genes relocated next to constitutive heterochromatin (position effect variegation or PEV; reviewed in [59, 60]) do not affect the TAS-associated TPE [5, 55] but strongly suppress the TPE of transgenes inserted into the HTT of YS (58). These results indicate that the chromatin of the YS HTT shares some features with constitutive heterochromatin and has different properties from the chromatin that includes the autosomal HTT arrays and the TAS.
These results suggest that the HTT arrays of XR, YL, and 4th chromosome telomeres have properties similar to those of YS, namely they are subject to a “heterochromatinization” process related to their particular location. Numerous studies on PEV have shown that proteins that are typically enriched in the heterochromatin can spread into neighboring euchromatic genes changing their chromatin composition and packaging and downregulating their expression [59, 60]. Thus, although the properties of the HTT arrays of YL, XR and 4th chromosome have not been tested, it is quite likely that they are similar to those of YS. This would suggest that in a peo mutant background the preferential involvement of these telomeres in TF is a consequence of their “heterochromatinization”. This in turn implies that the heterochromatic markers of the HTT regions of the YL, XR and 4h chromosome telomeres extend to the terminin-coated chromosome ends. However, “heterochromatinization” does not extend to the HTT arrays at the end of the left arm end of the BSw+y+Y chromosome, because in peo mutants this marked Y forms much fewer Y rings than a normal Y. Our findings on ring Y formation in peo mutants also suggest that the different “heterochromatic” telomeres respond differently to Peo depletion. The higher than expected frequency of ring Ys observed in peo mutants could be a consequence of the particularly high fusigenic properties of the Y telomeres.
We would like to note that the “heterochromatinization” of the YL, YS, XR and 4th chromosome termini is the natural condition of these telomeres and that we only know that this condition makes them more fusigenic than the other Drosophila telomeres. However, we have no information on the actual properties of these telomeres. Namely, we do not know the properties of their terminin-associated regions. For example, we do not know whether the terminin-coated chromosome ends of YL, YS, XR and 4L have different silencing properties and different responses to PEV and TPE modifiers compared to the terminin-bound regions of the other chromosome ends.
The mechanism underlying TF formation in peo mutants
We have shown that mutations in peo genetically interact with mutation in genes that encode the terminin subunits. Consistent with these results, we have also shown that Peo directly binds terminin and mapped the Peo-HOAP interacting domains. Thus, although Peo does not share the properties of the terminin proteins, it is clearly a component of the Drosophila telomere capping machinery. What is then the role of peo in telomere protection? The finding that mutations in peo do not affect HOAP, Moi and Ver localization at telomeres strongly suggest that loss of peo function does not cause TFs by affecting terminin recruitment at telomeres. Similarly, the normal accumulation of HP1a at the telomeres of peo mutants suggests a telomere fusion mechanism independent of HP1a. More in general, the fact that peo, but not the other TF genes (eff, Su(var)205, cav, mre11, rad50, nbs, woc, moi and ver), preferentially affect “heterochromatic” telomeres suggests that peo might either act upstream to these genes or function in a telomere protection pathway that does not involve them.
The findings that mutations in peo impair DNA replication and preferentially affect late replicating “heterochromatic” telomeres raise the possibility that defective telomere duplication might be fusigenic in Drosophila. However, there are several reasons that lead us to exclude this possibility. Should it be correct, one would expect that impairment of some of the many factors that mediate DNA replication would results in TFs. However, in addition to the aphidicolin treatment and Blm mutations described here, several additional DNA replication factors have been described whose loss fails to induce TFs. Hydroxyurea (HU), which blocks DNA replication by inducing a deoxyribonucleotide triphosphate (dNTP) pool depletion, does not induce TFs in brain cells [48]. In addition, a number of mutations (or RNAi treatments) disrupting different aspect of DNA replication do not results in TFs in Drosophila brain cells or S2 tissue culture cells, although they cause more or less extensive chromosome breakage. These include lesions in the genes/RNAs encoding the origin recognition (ORC) and the minichromosome maintenance (MCM) prereplication complexes, DNA primase, the Cul4 replication licensing factor, the replisome components DNA polymerase alpha, Rpa70, Tim2 and PCNA, as well as the chromatin assembly factor Caf1 that assists in loading the histone tetramer after DNA replication [21, 48, 61–64]. We thus hypothesize that the peo-dependent TFs are generated by a peculiar defect in telomeric DNA replication that creates specific fusigenic lesions. Heterochromatin replication is likely to be different from that of euchromatin, as it requires not only DNA duplication but also reinstallment of specific epigenetic markers such as heterochromatin-associated proteins and histone modifications (reviewed in [65]). It is thus possible that in the presence of weak peo mutations replication of “heterochromatic” telomeres is preferentially affected leading to specific Peo-dependent fusigenic lesions concentrated in the XR, the Y and the 4th chromosome ends. In strong peo mutants, these fusigenic lesions would be generated also in “euchromatic” telomeres resulting into a more general involvement of telomeres in fusion events. However, we cannot exclude the possibility that peo plays a dual function being independently required for DNA replication and telomere capping.
In conclusion, we propose that Peo is a Drosophila telomere-capping protein that preferentially protects chromosome ends associated with heterochromatic markers. Our results also indicate that Peo is required for general DNA replication and most likely also for telomere replication. However, while it is possible that loss of Peo generates specific fusigenic lesions during telomere replication, it is unlikely that a general impairment of Drosophila telomere duplication leads to telomere fusion. In this respect Drosophila telomeres are similar to human telomeres, which fail to fuse following defective replication [8, 46, 47, 66].
Materials and Methods
Drosophila strains
The pendolino1 (peo1) mutation was isolated by a cytological screen of 120 late lethal mutants mapping to the second chromosome, recovered after I element mobilization by I-R dysgenic crosses [67]. The late lethal mutations l(2)1527, l(2)2723, the insertion lines p112, p221 and p520 were described previously [30] and were kindly provided by P. Taghert (Washington University, MO). The peoh allele was isolated from a cytological screen of a collection of 193 late lethal mutants that arose in the Zucker’s collection of heavily mutagenized viable lines [68]. The Df(2R)X3, Df(2R)B5, Df(2R)X1 and Df(2R)BSC298 deletions, the insertion line cbx05704, were obtained from the Bloomington Stock Center and are described in FlyBase. All peo alleles and the deficiencies of 2R were balanced over CyO Tb balancer [69]. The eff, Su(var)205, cav, mre11, rad50, nbs, woc, moi and ver mutations were previously described [10, 12, 15, 16, 21, 23, 24]. The DNA sequences flanking the peop112 insertion were obtained by inverse PCR using standard procedures.
To obtain the rescue constructs, the LD08052 full lenght peo cDNA was cloned into pCASPER4 transformation vectors. Germline transformation was carried out by the BestGene Company. A pCasper4 [w+, peo+] insertion (with a Hsp70 promoter) on the X chromosome was used to establish the [w+, peo+]; peo1/CyO Tb stock. Animals from this stock were heat-shocked for 1h at 37°C every day starting from the embryonic stages; we then examined the brains of non-Tb third instar larvae for the presence and frequency of TFs and the adult flies for the presence of w+, peo+; peo1/peo1 individuals. For the rescue experiments we also employed a pCASPER4 [w+, peo+-HA] insertion (with a Tubulin promoter) on the third chromosomes. We established [w+, peo+-HA]/TM6C; peo1/CyO Tb and [w+, peo+-HA]/TM6C; peoh/CyO Tb stocks and examined them for TFs in non-Tb larvae. The [w+, peo+-HA]/TM6C; peo1/CyO Tb stock was also examined for the presence of peo1/peo1 non Cy flies. Information on the genetic markers and balancers used in this study is available at FlyBase (http://flybase.bio.indiana.edu/). Stocks were maintained and crosses were made on standard Drosophila medium at 25°C.
Anti-Peo antibody production and purification
To generate anti-Peo polyclonal antibodies, rabbits were immunized with bacterially expressed 6His-Peo. Immunization and production of anti-Peo antibodies were carried out by the Agro-Bio Company (France).
To purify the anti-Peo antibody we used a bacterially expressed GST-Peo protein. About 1 mg of this tagged protein was run on a polyacrylamide gel and then blotted onto a nitrocellulose membrane. The membrane strip containing GST-Peo was cut out, washed in 100 mM glycine/HCl pH 2.5 for 5 min, washed for 5 min in TBS, blocked by incubation with 3% BSA for 1 hour, and washed again for 4 min in TBS. The membrane was then cut into small pieces and incubated overnight with rotation at 4°C in 2 ml of serum diluted 1 : 5 in TBS. After centrifugation and removal of supernatant the membrane pieces were washed for 15 min in 50 mM Tris/HCl pH 7.5, 500 mM NaCl, for 5 min in 50 mM Tris/HCl pH 7.5, 100 mM NaCl, and for 10 min in TBS. The membrane was then incubated for 30 min in 1ml of 100 mM glycine/HCl pH 2.5 to elute the antibody. The eluate was then mixed to a proper volume 1M Tris pH 8.8 to bring the final pH to 8.0, and then kept at 4°C before use.
Chromosome cytology and immunostaining
DAPI-stained colchicine-treated larval brain chromosomes were prepared according to [10]. Preparation and immunostaining of mitotic and polytene chromosomes were carried out as described previously [10, 20], with minor modifications.
For anti-PCNA immunostaining, dissected larval brains were incubated in PBS with 0.5% Triton X-100 for 3 min, and then fixed in 3.7% formaldehyde according to [70]. Before immunostaining, brain squash preparations were Triton-X extracted by incubating the slides in 0.1% Triton X-100 containing PBS (PBT), 2 times for 10 min.
The primary antibodies used for immunostaining were: the rabbit anti-Peo described above diluted 1 : 10; rabbit anti-HOAP (1 : 100), mouse anti-HP1a (C1A9; 1 : 10), and mouse anti-PCNA (1 : 20; Abcam ab29). The anti-HP1a antibody C1A9 was obtained from the Developmental Studies Hybridoma Bank, created by the NICHD of the NIH and maintained at The University of Iowa, Department of Biology, Iowa City, IA 52242. After an overnight incubation at 4°C with the primary antibody, slides were washed twice in TBS-Tween 0.1% for 15 min and then incubated for 2 h at room temperature with FITC-conjugated goat anti-mouse (1 : 20; Jackson Laboratories), or AlexaFluor 555-conjugated donkey anti-rabbit (1 : 200; Invitrogen) antibodies. All slides were then mounted in Vectashield medium H-1200 with DAPI to stain DNA. in vivo detection and immunostaining of GFP-tagged proteins on polytene chromosomes were carried out as previously described [24].Chromosome preparations were analyzed using a Zeiss Axioplan epifluorescence microscope (CarlZeiss, Obezkochen, Germany), equipped with a cooled CCD camera (CoolSnap HQ, Photometrics, Woburn, MA). Gray-scale digital images were collected separately, converted to Photoshop format, pseudocolored, and merged.
To quantify the polytene chromosome fluorescence intensity after Peo immunostaining, we used the ImageJ software (National Institute of Mental Health, Bethesda, Maryland, USA). Given that the distribution of fluorescent bands along the chromosomes was rather uniform, for each polytene nucleus we selected 3–4 different chromosome regions of similar length, and measured both their fluorescence and the fluorescence of a close chromosome-free region to correct for background fluorescence. For each genotype (wild type, +/Df, peoh/Df and peo1/Df) shown in Fig 8 we measured at least 30 polytene regions from at least 10 nuclei.
Yeast two-hybrid assay
The coding sequences of Peo, Hp1a and HOAP were PCR-amplified and cloned into pGBKT7 or pGAD-T7 vectors (Clontech). The S. cerevisiae AH109 strain was transformed with the indicated combinations of plasmids and assayed for growth on SD/–His/–Trp/–Leu selection plates supplemented with 20 mM 3-amino-1,2,4-triazole (3-AT), according to the manufacturer’s instructions.
Purification of recombinant proteins
To obtain the GST-Moi, GST-Ver, GST-HOAP and GST-Peo fusion proteins, the corresponding full length cDNAs were cloned in either pGEX-6P1 or pGEX-3X, expressed in bacteria and purified as described previously [24]. The GST-Peo1, GST-Peo2, GST-Peo3, GST-HOAP ∆3, GST-HOAP ∆2, 3, GST-HOAP ∆1–3 and GST-HOAP ∆N-TERM truncated proteins were obtained by cloning the corresponding PCR-generated sequences in pGEX-6P1; bacterially expressed GST fusion proteins were then purified by incubating crude lysates with glutathione sepharose beads (QIAGEN) as recommended by the manufacturer. To generate 6His-Peo, the LD08052 peo full-length cDNA was cloned into the pQE32 expression vector (QIAGEN), and expressed in bacteria; 6His-Peo was affinity purified with a Ni-NTA resin using standard procedures.
Protein extract preparation, GST pulldown and western blot
To obtain extracts for Western Blot analysis, 50 dissected third instar larval brains were lysed in an ice-cold buffer containing 20 mM Hepes KOH pH 7.9, 1.5 mM MgCl2, 10 mM KCl, 420 mM NaCl, 30 mM NaF, 0.2 mM Na3VO4, 25 mM BGP, 0.5 M PMSF, 0.1% NP40, and 1X protease inhibitor cocktail (Roche). To obtain HOAP-FLAG, Ver-FLAG and Peo-FLAG expressing S2 cells, cav, ver or peo cDNAs were cloned in the pAWF vector (DGRC) in frame with the FLAG-coding sequence. For the expression of Moi-HA, the moi full lenght cDNA was fused in frame with the HA-coding sequence and then cloned into a pCASPER4 vector. All constructs were transfected in S2 tissue culture cells using Cellfectin (Invitrogen), and cells were harvested 72 h after transfection. Extracts were lysed in 20 mM Tris pH 8.0, 420 mM NaCl, 1 mM MgCl2, 1 mM DTT, 0.1% NP40, and 1X protease inhibitor cocktail (Roche). For preparation of human cell extracts, HeLa cells expressing Peo-FLAG cloned in the pCDNA vector were harvested after 72hr transfection and lysated in 20 mM Tris pH 8.0, 420 mM NaCl, 1 mM MgCl2, 1 mM DTT, 0.1% NP40 and 1X protease inhibitor cocktail (Roche).
For GST-pulldown assays, protein extracts were incubated with 2 μg of each GST fusion protein bound to sepharose beads in a buffer containing 20 mM Hepes KOH, 20 mM NaF and 0.8% NP40 for 1h at 4°C. Sepharose-bound GST proteins were collected by centrifugation, washed several times with 20 mM Hepes KOH, 20 mM NaF and 1.8% NP40, and resuspended in Laemli buffer in a 30μl final volume for Western Blot analysis. For immunoblotting, protein samples were run into SDS polyacrilammide gels and electro-blotted on a nitrocellulose membrane (Bio-Rad) in a phosphate buffer (390 mM NaH2PO4 /610 mM Na2HPO4). For the detection of HOAP-FLAG, Ver - FLAG, Peo - FLAG, and Moi-HA, membranes were probed with anti-FLAG HRP-conjugated (1 : 1000; Roche), and anti-HA HRP-conjugated (1 : 500; Roche) antibodies; Peo and His-Peo were detected with our rabbit anti-Peo (1 : 100), and Giotto with our anti-Giotto (1 : 5000; [71]). Secondary antibodies were sheep anti-mouse IgG HRP-conjugated (1 : 5000), or donkey anti-rabbit IgG HRP-conjugated (1 : 5000) (both from Amersham Biosciences). The blots were developed using the ECL or ECL Plus method (Amersham Biosciences) and signals were detected with the ChemiDoc scanning system (BioRad). Band intensities were quantified using the image acquisition and analysis Image lab 4.0.1 software (Biorad).
EdU incorporation and staining
EdU labeling was performed as per the manufacturer's instructions (Invitrogen, ClickiT Alexa Fluor 488 Imaging kit). Larval brains were cultured with 10μM EdU in 1 X PBS for 60 min prior to fixation and detection.
Aphidicolin treatment
Brains from third instar larvae were dissected in saline (0.7% NaCl), incubated in saline with 110 μM Aphidicolin (APH) for 1.5 hours, rinsed in saline, and then transferred into a 33 mm Petri dish containing 3 ml of Schneider’s medium (SIGMA) supplemented with 10% fetal bovine serum (FBS, Gibco BRL) for 1, 2 or 3h. Colchicine at a final concentration of 10-5M was added to the medium 1 hour before fixation according to [10]. Control wild type and peoh/peoh brains were incubated in saline without APH for 1.5, and processed like the APH-treated brains; they were fixed after 2 hours incubation n the medium.
Bioinformatic analysis of the Peo structure
As a first step towards the construction of a three-dimensional model of Peo, we used its full-length sequence (Accession code: Q7K4V4) as a query to search the UniProtKB database (http://www.uniprot.org/) using CSI-BLAST [72], with an Expectation (E) value threshold of 10–5. Iterative searches of the database yielded 59 unique sequences. The retrieved sequences were aligned with the CLUSTALW software [73] with default parameters. The multiple sequence alignment (MSA) was next used as a seed to construct a Hidden Markov Model (HMM) of the family.
The HMM was employed to search the Pfam database (http://pfam.sanger.ac.uk/) via the HHpred server [74]. The highest scoring hit was the ubiquitin-conjugating enzyme (UBC) family also known as the E2 enzyme family [34] (probability to be a true positive more than to 99%, E-value equal to 1.2 x10-42). The third scoring hit was the ubiquitin E2 variant (UEV) family (Pfam ID: PF05743) that includes UBC homologs such as Tsg101, Mms2 and UEV1 (probability to be a true positive equal to 96.44%, E-value equal to 1.3 x10-4). Peo belongs to the UEV family because it contains an aspartic acid residue (at position 106, according to SwissProt numbering) in place of the E2 active site cysteine, and it is unable to catalyze ubiquitin transfer as it lacks the cysteine that forms a transient thioester bond with the C-terminus of ubiquitin (Ub).
Prediction of potentially disordered regions using the GeneSilico MetaDisorder server (http://iimcb.genesilico.pl/metadisorder/) revealed that at the C-terminus of Peo there is a stretch of ~ 70 aa (from residue 177 to 244, according SwissProt numbering) that has the tendency to be intrinsically disordered (i.e. lack a unique three dimensional structure at least in the absence of a binding partner), while the region including residues 16–176 shows propensity to form a folded globular domain with a well-defined pattern of secondary structures as revealed by the Quick2d web server analysis [75].
Because no homologous structure with sufficiently high sequence identity with Peo is available, we performed the Peo modeling using the composite approach implemented in I-TASSER server (Iterative Threading ASSEmbly Refinement) [76].The Peo sequence from residue 16 to 176 (predicted to fold in a globular domain) was submitted to the server and the model with the best confidence score (C-score = 0.5) returned by I-TASSER was selected. We added hydrogen atoms in this model using HAAD software [77] and refined it close to the native structure using FG-MD molecular dynamics based algorithm [78].
Our final refined model of Peo was evaluated as a potentially extremely good model (with a predicted LGscore of 2.50) by the PRO-Q model quality assessment program [79]. The QMEAN score [80] was 0.6 (the variability range is 0–1, with 1 being a perfect model). Collectively these parameters indicate that the Peo three-dimensional model is sufficiently accurate for making functional inferences.
Supporting Information
Zdroje
1. Palm W, de Lange T. How shelterin protects mammalian telomeres. Annual Review of Genetics. 2008;42 : 301–34. doi: 10.1146/annurev.genet.41.110306.130350 18680434
2. O'Sullivan RJ, Karlseder J. Telomeres: protecting chromosomes against genome instability. Nature reviews Molecular Cell Biology. 2010;11 : 171–81. doi: 10.1038/nrm2848 20125188
3. Jain D, Cooper JP. Telomeric strategies: means to an end. Annual Review of Genetics. 2010;44 : 243–69. doi: 10.1146/annurev-genet-102108-134841 21047259
4. Stewart JA, Chaiken MF, Wang F, Price CM. Maintaining the end: roles of telomere proteins in end-protection, telomere replication and length regulation. Mutation Research. 2012;730 : 12–9. doi: 10.1016/j.mrfmmm.2011.08.011 21945241
5. Mason JM, Frydrychova RC, Biessmann H. Drosophila telomeres: an exception providing new insights. BioEssays. 2008;30 : 25–37. 18081009
6. Raffa GD, Cenci G, Ciapponi L, Gatti M. Organization and Evolution of Drosophila Terminin: Similarities and Differences between Drosophila and Human Telomeres. Frontiers in Oncology. 2013;3 : 112. doi: 10.3389/fonc.2013.00112 23675571
7. Rong YS. Telomere capping in Drosophila: dealing with chromosome ends that most resemble DNA breaks. Chromosoma. 2008;117 : 235–42. doi: 10.1007/s00412-007-0144-2 18193446
8. Leman AR, Dheekollu J, Deng Z, Lee SW, Das MM, Lieberman PM, et al. Timeless preserves telomere length by promoting efficient DNA replication through human telomeres. Cell Cycle. 2012;11 : 2337–47. doi: 10.4161/cc.20810 22672906
9. Vannier JB, Pavicic-Kaltenbrunner V, Petalcorin MI, Ding H, Boulton SJ. RTEL1 dismantles T loops and counteracts telomeric G4-DNA to maintain telomere integrity. Cell. 2012;149 : 795–806. doi: 10.1016/j.cell.2012.03.030 22579284
10. Cenci G, Rawson RB, Belloni G, Castrillon DH, Tudor M, Petrucci R, et al. UbcD1, a Drosophila ubiquitin-conjugating enzyme required for proper telomere behavior. Genes Dev. 1997;11 : 863–75. 9106658
11. Cipressa F, Romano S, Centonze S, Zur Lage PI, Verni F, Dimitri P, et al. Effete, a Drosophila Chromatin-Associated Ubiquitin-Conjugating Enzyme that Affects Telomeric and Heterochromatic Position Effect Variegation. Genetics. 2013;195 : 147–58 doi: 10.1534/genetics.113.153320 23821599
12. Fanti L, Giovinazzo G, Berloco M, Pimpinelli S. The heterochromatin protein 1 prevents telomere fusions in Drosophila. Mol Cell. 1998;2 : 527–38. 9844626
13. Bi X, Srikanta D, Fanti L, Pimpinelli S, Badugu R, Kellum R, et al. Drosophila ATM and ATR checkpoint kinases control partially redundant pathways for telomere maintenance. Proc Natl Acad Sci U S A. 2005;102 : 15167–72. 16203987
14. Bi X, Wei SC, Rong YS. Telomere protection without a telomerase; the role of ATM and Mre11 in Drosophila telomere maintenance. Curr Biol. 2004;14 : 1348–53. 15296751
15. Ciapponi L, Cenci G, Ducau J, Flores C, Johnson-Schlitz D, Gorski MM, et al. The Drosophila Mre11/Rad50 complex is required to prevent both telomeric fusion and chromosome breakage. Curr Biol. 2004;14 : 1360–6. 15296753
16. Ciapponi L, Cenci G, Gatti M. The Drosophila Nbs protein functions in multiple pathways for the maintenance of genome stability. Genetics. 2006. 173 : 1447–54 16648644
17. Oikemus SR, McGinnis N, Queiroz-Machado J, Tukachinsky H, Takada S, Sunkel CE, et al. Drosophila atm/telomere fusion is required for telomeric localization of HP1 and telomere position effect. Genes Dev. 2004;18 : 1850–61. 15256487
18. Oikemus SR, Queiroz-Machado J, Lai K, McGinnis N, Sunkel C, Brodsky MH. Epigenetic telomere protection by Drosophila DNA damage response pathways. PLoS Genet. 2006;2:e71. 16710445
19. Silva E, Tiong S, Pedersen M, Homola E, Royou A, Fasulo B, et al. ATM is required for telomere maintenance and chromosome stability during Drosophila development. Curr Biol. 2004;14 : 1341–7. 15296750
20. Raffa GD, Cenci G, Siriaco G, Goldberg ML, Gatti M. The putative Drosophila transcription factor woc is required to prevent telomeric fusions. Mol Cell. 2005;20 : 821–31. 16364909
21. Cenci G, Siriaco G, Raffa GD, Kellum R, Gatti M. The Drosophila HOAP protein is required for telomere capping. Nat Cell Biol. 2003;5 : 82–4. 12510197
22. Gao G, Walser JC, Beaucher ML, Morciano P, Wesolowska N, Chen J, et al. HipHop interacts with HOAP and HP1 to protect Drosophila telomeres in a sequence-independent manner. The EMBO Journal. 2010;29 : 819–29. doi: 10.1038/emboj.2009.394 20057353
23. Raffa GD, Raimondo D, Sorino C, Cugusi S, Cenci G, Cacchione S, et al. Verrocchio, a Drosophila OB fold-containing protein, is a component of the terminin telomere-capping complex. Genes Dev. 2010;24 : 1596–601. doi: 10.1101/gad.574810 20679394
24. Raffa GD, Siriaco G, Cugusi S, Ciapponi L, Cenci G, Wojcik E, et al. The Drosophila modigliani (moi) gene encodes a HOAP-interacting protein required for telomere protection. Proc Natl Acad Sci U S A. 2009;106 : 2271–6. doi: 10.1073/pnas.0812702106 19181850
25. Raffa GD, Ciapponi L, Cenci G, Gatti M. Terminin: a protein complex that mediates epigenetic maintenance of Drosophila telomeres. Nucleus. 2011;2 : 383–91. doi: http://dx.doi.org/10.4161/nucl.2.5.17873 21989238
26. Shareef MM, King C, Damaj M, Badagu R, Huang DW, Kellum R. Drosophila heterochromatin protein 1 (HP1)/origin recognition complex (ORC) protein is associated with HP1 and ORC and functions in heterochromatin-induced silencing. Mol Biol Cell. 2001;12 : 1671–85. 11408576
27. Drosopoulos WC, Kosiyatrakul ST, Yan Z, Calderano SG, Schildkraut CL. Human telomeres replicate using chromosome-specific, rather than universal, replication programs. The Journal of Cell Biology. 2012;197 : 253–66. doi: 10.1083/jcb.201112083 22508510
28. Arnoult N, Schluth-Bolard C, Letessier A, Drascovic I, Bouarich-Bourimi R, Campisi J, et al. Replication timing of human telomeres is chromosome arm-specific, influenced by subtelomeric structures and connected to nuclear localization. PLoS Genet. 2010;6:e1000920. doi: 10.1371/journal.pgen.1000920 20421929
29. Novo C, Arnoult N, Bordes WY, Castro-Vega L, Gibaud A, Dutrillaux B, et al. The heterochromatic chromosome caps in great apes impact telomere metabolism. Nucleic acids research. 2013;41 : 4792–801. doi: 10.1093/nar/gkt169 23519615
30. O'Brien MA, Roberts MS, Taghert PH. A genetic and molecular analysis of the 46C chromosomal region surrounding the FMRFamide neuropeptide gene in Drosophila melanogaster. Genetics. 1994;137 : 121–37. 8056304
31. Levis R, Hazelrigg T, Rubin GM. Separable cis-acting control elements for expression of the white gene of Drosophila. The EMBO Journal. 1985;4 : 3489–99. 3004962
32. Castrillon DH, Gonczy P, Alexander S, Rawson R, Eberhart CG, Viswanathan S, et al. Toward a molecular genetic analysis of spermatogenesis in Drosophila melanogaster: characterization of male-sterile mutants generated by single P element mutagenesis. Genetics. 1993;135 : 489–505. 8244010
33. Fabrizio JJ, Hime G, Lemmon SK, Bazinet C. Genetic dissection of sperm individualization in Drosophila melanogaster. Development. 1998;125 : 1833–43. 9550716
34. van Wijk SJ, Timmers HT. The family of ubiquitin-conjugating enzymes (E2s): deciding between life and death of proteins. FASEB Journal. 2010;24 : 981–93. doi: 10.1096/fj.09-136259 19940261
35. de Lange T. Protection of mammalian telomeres. Oncogene. 2002;21 : 532–40. 11850778
36. Csink AK, Henikoff S. Large-scale chromosomal movements during interphase progression in Drosophila. The Journal of Cell Biology. 1998;143 : 13–22. 9763417
37. Konishi A, de Lange T. Cell cycle control of telomere protection and NHEJ revealed by a ts mutation in the DNA-binding domain of TRF2. Genes Dev. 2008;22 : 1221–30. doi: 10.1101/gad.1634008 18451109
38. Gatti M, Pimpinelli S. Functional elements in Drosophila melanogaster heterochromatin. Annual Review of Genetics. 1992;26 : 239–75. 1482113
39. Lindsley DL, Zimm GG. The Genome of Drosophila melanogaster. 1250 Sixth Avenue, San Diego, California 92101–4311: Academic Press, Inc.; 1992.
40. Ye Y, Rape M. Building ubiquitin chains: E2 enzymes at work. Nature Reviews Molecular Cell biology. 2009;10 : 755–64. doi: 10.1038/nrm2780 19851334
41. Cottee PA, Abs ELOYG, Nisbet AJ, Gasser RB. Ubiquitin-conjugating enzyme genes in Oesophagostomum dentatum. Parasitology Research. 2006;99 : 119–25. 16518612
42. Fanti L, Pimpinelli S. HP1: a functionally multifaceted protein. Current opinion in genetics & development. 2008;18 : 169–74.
43. Burla R, Carcuro M, Raffa GD, Galati A, Raimondo D, Rizzo A, et al. AKTIP/Ft1, a new shelterin-interacting factor required for telomere maintenance. PLoS Genet. 2015; 11:e1005167. doi: 10.1371/journal.pgen.1005167
44. Chagin VO, Stear JH, Cardoso MC. Organization of DNA replication. Cold Spring Harbor perspectives in biology. 2010;2:a000737. doi: 10.1101/cshperspect.a000737 20452942
45. Ghosh AK, Rossi ML, Singh DK, Dunn C, Ramamoorthy M, Croteau DL, et al. RECQL4, the protein mutated in Rothmund-Thomson syndrome, functions in telomere maintenance. The Journal of Biological Chemistry. 2012;287 : 196–209. doi: 10.1074/jbc.M111.295063 22039056
46. Barefield C, Karlseder J. The BLM helicase contributes to telomere maintenance through processing of late-replicating intermediate structures. Nucleic Acids Research. 2012;40 : 7358–67. doi: 10.1093/nar/gks407 22576367
47. Sfeir A, Kosiyatrakul ST, Hockemeyer D, MacRae SL, Karlseder J, Schildkraut CL, et al. Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell. 2009;138 : 90–103. doi: 10.1016/j.cell.2009.06.021 19596237
48. Benna C, Bonaccorsi S, Wulbeck C, Helfrich-Forster C, Gatti M, Kyriacou CP, et al. Drosophila timeless2 is required for chromosome stability and circadian photoreception. Curr Biol. 2010;20 : 346–52. doi: 10.1016/j.cub.2009.12.048 20153199
49. Gotter AL, Suppa C, Emanuel BS. Mammalian TIMELESS and Tipin are evolutionarily conserved replication fork-associated factors. Journal of Molecular Biology. 2007;366 : 36–52. 17141802
50. Kondratov RV, Antoch MP. Circadian proteins in the regulation of cell cycle and genotoxic stress responses. Trends in Cell Biology. 2007;17 : 311–7. 17644383
51. Kusano K, Johnson-Schlitz DM, Engels WR. Sterility of Drosophila with mutations in the Bloom syndrome gene—complementation by Ku70. Science. 2001;291 : 2600–2. 11283371
52. Pardue ML, Debaryshe P. Adapting to life at the end of the line: How Drosophila telomeric retrotransposons cope with their job. Mobile Genetic Elements. 2011;1 : 128–34. 22016861
53. Zhang L, Rong YS. Retrotransposons at Drosophila telomeres: host domestication of a selfish element for the maintenance of genome integrity. Biochimica et Biophysica Acta. 2012;1819 : 771–5. doi: 10.1016/j.bbagrm.2012.01.018 22342531
54. Hoskins RA, Carlson JW, Kennedy C, Acevedo D, Evans-Holm M, Frise E, et al. Sequence finishing and mapping of Drosophila melanogaster heterochromatin. Science. 2007;316 : 1625–8. 17569867
55. Mason J, Villasante A. Subtelomeres in Drosophila and Other Diptera. In: Louis EJ, Becker MM, editors. Subtelomeres: Springer Berlin Heidelberg; 2014. p. 211–25.
56. Riddle NC, Shaffer CD, Elgin SC. A lot about a little dot—lessons learned from Drosophila melanogaster chromosome 4. Biochemistry and Cell Biology. 2009;87 : 229–41. doi: 10.1139/O08-119 19234537
57. Biessmann H, Prasad S, Semeshin VF, Andreyeva EN, Nguyen Q, Walter MF, et al. Two distinct domains in Drosophila melanogaster telomeres. Genetics. 2005;171 : 1767–77. 16143601
58. Wang SH, Nan R, Accardo MC, Sentmanat M, Dimitri P, Elgin SC. A distinct type of heterochromatin at the telomeric region of the Drosophila melanogaster Y chromosome. PLoS One. 2014;9:e86451. doi: 10.1371/journal.pone.0086451 24475122
59. Weiler KS, Wakimoto BT. Heterochromatin and gene expression in Drosophila. Annual Review of Genetics. 1995;29 : 577–605. 8825487
60. Elgin SC, Reuter G. Position-effect variegation, heterochromatin formation, and gene silencing in Drosophila. Cold Spring Harbor Perspectives in Biology. 2013;5:a017780. doi: 10.1101/cshperspect.a017780 23906716
61. Gatti M, Baker BS. Genes controlling essential cell-cycle functions in Drosophila melanogaster. Genes Dev. 1989;3 : 438–53. 2498166
62. Loupart ML, Krause SA, Heck MS. Aberrant replication timing induces defective chromosome condensation in Drosophila ORC2 mutants. Curr Biol. 2000;10 : 1547–56. 11137005
63. Somma MP, Ceprani F, Bucciarelli E, Naim V, De Arcangelis V, Piergentili R, et al. Identification of Drosophila mitotic genes by combining co-expression analysis and RNA interference. PLoS Genet. 2008;4:e1000126. doi: 10.1371/journal.pgen.1000126 18797514
64. Balasov M, Huijbregts RP, Chesnokov I. Functional analysis of an Orc6 mutant in Drosophila. Proc Natl Acad Sci U S A. 2009;106 : 10672–7. doi: 10.1073/pnas.0902670106 19541634
65. Wallace JA, Orr-Weaver TL. Replication of heterochromatin: insights into mechanisms of epigenetic inheritance. Chromosoma. 2005;114 : 389–402. 16220346
66. Martinez P, Thanasoula M, Munoz P, Liao C, Tejera A, McNees C, et al. Increased telomere fragility and fusions resulting from TRF1 deficiency lead to degenerative pathologies and increased cancer in mice. Genes Dev. 2009;23 : 2060–75. doi: 10.1101/gad.543509 19679647
67. Dimitri P, Arca B, Berghella L, Mei E. High genetic instability of heterochromatin after transposition of the LINE-like I factor in Drosophila melanogaster. Proc Natl Acad Sci U S A. 1997;94 : 8052–7. 9223313
68. Koundakjian EJ, Cowan DM, Hardy RW, Becker AH. The Zuker collection: a resource for the analysis of autosomal gene function in Drosophila melanogaster. Genetics. 2004;167 : 203–6. 15166147
69. Lattao R, Bonaccorsi S, Guan X, Wasserman SA, Gatti M. Tubby-tagged balancers for the Drosophila X and second chromosomes. Fly. 2011;5 : 369–70. doi: 10.4161/fly.5.4.17283 21785267
70. Bonaccorsi S, Giansanti MG, Gatti M. Spindle assembly in Drosophila neuroblasts and ganglion mother cells. Nat Cell Biol. 2000;2 : 54–6. 10620808
71. Giansanti MG, Bonaccorsi S, Kurek R, Farkas RM, Dimitri P, Fuller MT, et al. The class I PITP giotto is required for Drosophila cytokinesis. Curr Biol. 2006;16 : 195–201. 16431372
72. Biegert A, Soding J. Sequence context-specific profiles for homology searching. Proc Natl Acad Sci U S A. 2009;106 : 3770–5. doi: 10.1073/pnas.0810767106 19234132
73. Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, et al. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Research. 2003;31 : 3497–500. 12824352
74. Soding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Research. 2005;33 (Web Server issue):W244–8. 15980461
75. Biegert A, Mayer C, Remmert M, Soding J, Lupas AN. The MPI Bioinformatics Toolkit for protein sequence analysis. Nucleic Acids Research. 2006;34 (Web Server issue):W335–9. 16845021
76. Roy A, Kucukural A, Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nature Protocols. 2010;5 : 725–38. doi: 10.1038/nprot.2010.5 20360767
77. Li Y, Roy A, Zhang Y. HAAD: A quick algorithm for accurate prediction of hydrogen atoms in protein structures. PloS One. 2009;4:e6701. doi: 10.1371/journal.pone.0006701 19693270
78. Zhang J, Liang Y, Zhang Y. Atomic-level protein structure refinement using fragment-guided molecular dynamics conformation sampling. Structure. 2011;19 : 1784–95. doi: 10.1016/j.str.2011.09.022 22153501
79. Wallner B, Elofsson A. Identification of correct regions in protein models using structural, alignment, and consensus information. Protein Science. 2006;15 : 900–13. 16522791
80. Benkert P, Biasini M, Schwede T. Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics. 2011;27 : 343–50. doi: 10.1093/bioinformatics/btq662 21134891
81. Abad JP, De Pablos B, Osoegawa K, De Jong PJ, Martin-Gallardo A, Villasante A. Genomic analysis of Drosophila melanogaster telomeres: full-length copies of HeT-A and TART elements at telomeres. Molecular Biology and Evolution. 2004;21 : 1613–9. 15163766
Štítky
Genetika Reprodukční medicína
Článek Germline Mutations Confer Susceptibility to Acute Lymphoblastic Leukemia and ThrombocytopeniaČlánek Multiple In Vivo Biological Processes Are Mediated by Functionally Redundant Activities of andČlánek Temporal Expression Profiling Identifies Pathways Mediating Effect of Causal Variant on PhenotypeČlánek Simultaneous DNA and RNA Mapping of Somatic Mitochondrial Mutations across Diverse Human CancersČlánek A Legume Genetic Framework Controls Infection of Nodules by Symbiotic and Endophytic BacteriaČlánek The Eukaryotic-Like Ser/Thr Kinase PrkC Regulates the Essential WalRK Two-Component System inČlánek The Yeast GSK-3 Homologue Mck1 Is a Key Controller of Quiescence Entry and Chronological LifespanČlánek The Role of -Mediated Epigenetic Silencing in the Population Dynamics of Transposable Elements in
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2015 Číslo 6
-
Všechny články tohoto čísla
- Expression of Concern: RNAi-Dependent and Independent Control of LINE1 Accumulation and Mobility in Mouse Embryonic Stem Cells
- Orphan Genes Find a Home: Interspecific Competition and Gene Network Evolution
- Minor Cause—Major Effect: A Novel Mode of Control of Bistable Gene Expression
- Germline Mutations Confer Susceptibility to Acute Lymphoblastic Leukemia and Thrombocytopenia
- Leveraging Identity-by-Descent for Accurate Genotype Inference in Family Sequencing Data
- Is Required for the Expression of Principal Recognition Molecules That Control Axon Targeting in the Retina
- Epigenetic Aging Signatures Are Coherently Modified in Cancer
- Silencing of DNase Colicin E8 Gene Expression by a Complex Nucleoprotein Assembly Ensures Timely Colicin Induction
- A Transposable Element within the Non-canonical Telomerase RNA of Modulates Telomerase in Response to DNA Damage
- The Orphan Gene Regulates Dauer Development and Intraspecific Competition in Nematodes by Copy Number Variation
- 9--13,14-Dihydroretinoic Acid Is an Endogenous Retinoid Acting as RXR Ligand in Mice
- The DnaA Protein Is Not the Limiting Factor for Initiation of Replication in
- FGFR3 Deficiency Causes Multiple Chondroma-like Lesions by Upregulating Hedgehog Signaling
- Multiple Changes of Gene Expression and Function Reveal Genomic and Phenotypic Complexity in SLE-like Disease
- Directed Evolution of RecA Variants with Enhanced Capacity for Conjugational Recombination
- The Regulatory T Cell Lineage Factor Foxp3 Regulates Gene Expression through Several Distinct Mechanisms Mostly Independent of Direct DNA Binding
- MreB-Dependent Inhibition of Cell Elongation during the Escape from Competence in
- DNA Damage Regulates Translation through β-TRCP Targeting of CReP
- Multiple In Vivo Biological Processes Are Mediated by Functionally Redundant Activities of and
- The Analysis of () Mutants Reveals Differences in the Fusigenic Potential among Telomeres
- The Causative Gene in Chanarian Dorfman Syndrome Regulates Lipid Droplet Homeostasis in .
- Temporal Expression Profiling Identifies Pathways Mediating Effect of Causal Variant on Phenotype
- The . Accessory Helicase PcrA Facilitates DNA Replication through Transcription Units
- AKTIP/Ft1, a New Shelterin-Interacting Factor Required for Telomere Maintenance
- Npvf: Hypothalamic Biomarker of Ambient Temperature Independent of Nutritional Status
- Transfer RNAs Mediate the Rapid Adaptation of to Oxidative Stress
- Connecting Circadian Genes to Neurodegenerative Pathways in Fruit Flies
- Response to “Ribosome Rescue and Translation Termination at Non-standard Stop Codons by ICT1 in Mammalian Mitochondria”
- Response to the Formal Letter of Z. Chrzanowska-Lightowlers and R. N. Lightowlers Regarding Our Article “Ribosome Rescue and Translation Termination at Non-Standard Stop Codons by ICT1 in Mammalian Mitochondria”
- Simultaneous DNA and RNA Mapping of Somatic Mitochondrial Mutations across Diverse Human Cancers
- Regulation of Insulin Receptor Trafficking by Bardet Biedl Syndrome Proteins
- Altered Levels of Mitochondrial DNA Are Associated with Female Age, Aneuploidy, and Provide an Independent Measure of Embryonic Implantation Potential
- Non-reciprocal Interspecies Hybridization Barriers in the Capsella Genus Are Established in the Endosperm
- Canine Spontaneous Head and Neck Squamous Cell Carcinomas Represent Their Human Counterparts at the Molecular Level
- Genetic Changes to a Transcriptional Silencer Element Confers Phenotypic Diversity within and between Species
- Functional Assessment of Disease-Associated Regulatory Variants Using a Versatile Dual Colour Transgenesis Strategy in Zebrafish
- Translational Upregulation of an Individual p21 Transcript Variant by GCN2 Regulates Cell Proliferation and Survival under Nutrient Stress
- Independent Neuronal Origin of Seizures and Behavioral Comorbidities in an Animal Model of a Severe Childhood Genetic Epileptic Encephalopathy
- The Human Blood Metabolome-Transcriptome Interface
- A Common Cancer Risk-Associated Allele in the Locus Encodes a Dominant Negative Inhibitor of Telomerase
- A Legume Genetic Framework Controls Infection of Nodules by Symbiotic and Endophytic Bacteria
- The Eukaryotic-Like Ser/Thr Kinase PrkC Regulates the Essential WalRK Two-Component System in
- The Yeast GSK-3 Homologue Mck1 Is a Key Controller of Quiescence Entry and Chronological Lifespan
- Dissection of a Complex Disease Susceptibility Region Using a Bayesian Stochastic Search Approach to Fine Mapping
- Exome Sequencing of Phenotypic Extremes Identifies and as Interacting Modifiers of Chronic Infection in Cystic Fibrosis
- The Role of -Mediated Epigenetic Silencing in the Population Dynamics of Transposable Elements in
- Ancestral Chromatin Configuration Constrains Chromatin Evolution on Differentiating Sex Chromosomes in
- Abnormal Activation of BMP Signaling Causes Myopathy in Null Mice
- Reproductive Mode and the Evolution of Genome Size and Structure in Nematodes
- Replication and Active Partition of Integrative and Conjugative Elements (ICEs) of the SXT/R391 Family: The Line between ICEs and Conjugative Plasmids Is Getting Thinner
- Motor and Sensory Deficits in the Mice Result from Mutation of the ESCRT Component HGS
- Senescence in the Sbds-Deficient Murine Pancreas: Cell-Type Specific Consequences of Translation Insufficiency
- Lipophorin Receptors Recruit the Lipoprotein LTP to the Plasma Membrane to Mediate Lipid Uptake
- Separable Crossover-Promoting and Crossover-Constraining Aspects of Zip1 Activity during Budding Yeast Meiosis
- Context-Dependent Functional Divergence of the Notch Ligands DLL1 and DLL4
- Multilayered Organization of Jasmonate Signalling in the Regulation of Root Growth
- Necrotic Cells Actively Attract Phagocytes through the Collaborative Action of Two Distinct PS-Exposure Mechanisms
- A Novel Feedback Loop That Controls Bimodal Expression of Genetic Competence
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Non-reciprocal Interspecies Hybridization Barriers in the Capsella Genus Are Established in the Endosperm
- Translational Upregulation of an Individual p21 Transcript Variant by GCN2 Regulates Cell Proliferation and Survival under Nutrient Stress
- Exome Sequencing of Phenotypic Extremes Identifies and as Interacting Modifiers of Chronic Infection in Cystic Fibrosis
- The Human Blood Metabolome-Transcriptome Interface
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



