-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaMultiple In Vivo Biological Processes Are Mediated by Functionally Redundant Activities of and
Amongst the small number of miRNA knockouts that exhibit substantially overt phenotypes, mutants of Drosophila mir-279 are notable. Previous studies have uncovered its essential requirements in a range of developmental and behavioral assays. Surprisingly, we find that the phenotypes attributed to mir-279 deletions depend on the unanticipated loss of expression of the downstream locus mir-996, whose genomic locus is retained in extant mir-279 mutants. These miRNAs share their seed regions but are divergent elsewhere in the mature sequences. We use precise genetic engineering to show that a single endogenous copy of either mir-279 or mir-996 can fully rescue viability, olfactory neuron, and circadian rhythm defects of double deletion animals. These data and genetic reagents set a new foundation for developmental and behavioral studies of this critical miRNA locus. More generally, these data demonstrate that multiple loss-of-function phenotypes can be rescued by endogenous expression of divergent seed family members, highlighting the importance and potentially sufficiency of this region for in vivo function.
Published in the journal: . PLoS Genet 11(6): e32767. doi:10.1371/journal.pgen.1005245
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1005245Summary
Amongst the small number of miRNA knockouts that exhibit substantially overt phenotypes, mutants of Drosophila mir-279 are notable. Previous studies have uncovered its essential requirements in a range of developmental and behavioral assays. Surprisingly, we find that the phenotypes attributed to mir-279 deletions depend on the unanticipated loss of expression of the downstream locus mir-996, whose genomic locus is retained in extant mir-279 mutants. These miRNAs share their seed regions but are divergent elsewhere in the mature sequences. We use precise genetic engineering to show that a single endogenous copy of either mir-279 or mir-996 can fully rescue viability, olfactory neuron, and circadian rhythm defects of double deletion animals. These data and genetic reagents set a new foundation for developmental and behavioral studies of this critical miRNA locus. More generally, these data demonstrate that multiple loss-of-function phenotypes can be rescued by endogenous expression of divergent seed family members, highlighting the importance and potentially sufficiency of this region for in vivo function.
Introduction
microRNAs (miRNAs) are ~22 nucleotide (nt) regulatory RNAs derived from hairpin precursors [1], and there are 100s ~ 1000 miRNA loci in well-studied animal genomes [2]. As animal miRNAs regulate targets exhibiting as little as 7 nt of complementarity to their 5' regions (principally nts 2–8, known as the "seed region"), they coordinate large regulatory networks [3]. Collectively, the developmental and physiological impacts of miRNA-mediated regulation are extensive and substantial [4,5].
The first miRNAs discovered, nematode lin-4 and let-7, exhibit strong developmental defects and have key individual targets that mediate substantial aspects of their phenotype [6–8]. As well, gain-of-function neural phenotypes associated with loss of 3' UTR elements from Notch target genes identified the functional logic of miRNA binding sites and highlighted additional key targets of miRNAs [9–11]. On the other hand, it is now well-appreciated that knockouts of individual miRNA genes frequently lack substantial phenotypes [12,13], and that the typical range of miRNA-mediated repression is modest [14,15]. Such findings have been interpreted to reflect that miRNAs are usually for "fine-tuning" or "robustness" of gene expression [5,16], but perhaps dispensable for major aspects of development, metabolism and behavior.
Nevertheless, a handful of animal miRNA mutants exhibit dramatic phenotypes in one or more settings. A particularly compelling example is miR-279 [17,18]. This miRNA is one of just a few loci across all animals, including nematode lin-4 [19], let-7 [8], lsy-6 [20], and mouse mir-96 [21,22], to have emerged from forward loss-of-function genetics, attesting to the strength of its mutant phenotype. By comparison, virtually every other miRNA studied in intact animals originated from gain-of-function screening or a directed knockout (although clearly in some cases these proved to have substantial effects).
A mutant of mir-279 initially emerged from a genetic screen for altered patterning of olfactory neurons, yielding a line with ectopic CO2-sensing neurons in the maxillary palp [18]. This was associated with a transposon insertion near mir-279, which was phenocopied by multiple mir-279 deletion alleles. In this setting, the transcription factors encoded by nerfin-1 and escargot are critical miR-279 targets [18,23]. Subsequent studies defined additional functions of miR-279, including to mediate normal circadian activity [24] and for specification and migration of border cells [25]. Curiously, these other settings were associated with deregulation of JAK-STAT signaling, although via different mechanisms. miR-279 restricts the JAK-STAT ligand unpaired in circadian pacemaker cells [24], whereas in ovarian border cells it represses the transcription factor STAT [25]. Altogether, these studies highlight diverse requirements for miR-279 in development and behavior.
The mir-996 locus was later identified in the vicinity of mir-279 and shown to encode a similar seed, but they otherwise have distinct mature sequences and were originally suggested to derive from separate genes [26,27]. Notably, miR-996 has not been implicated in any biological processes, since available mir-279 deletion alleles do not affect the mir-996 locus, and mir-279 mutant phenotypes can be rescued by a genomic transgene that contains only mir-279 and lacks mir-996 sequence [18]. Indeed, the deep conservation of divergent non-seed regions of miR-279 and miR-996, and the observation that mir-279 is ancestral and that mir-996 emerged more recently during arthropod evolution [28], suggest that miR-996 may have neofunctionalized from miR-279 to acquire some distinct activity.
In this study, we generated single and double mutants of mir-279 and mir-996, and cursory examination suggested that miR-996 was dispensable for overt development and behavior, while miR-279 was essential. However, our studies unexpectedly reveal that all mir-279 single deletion mutants affect the expression of miR-996. In essence, then, all studies of mir-279 mutants to date [18,23–25] have effectively been of double mutants. To remedy this, we engineered a defined set of backgrounds, using recombineered knockin/knockout transgenes introduced into the double deletion, to distinguish individual miRNA sequence requirements from dosage effects. We find that miR-996 contributes essential function in all known biological settings of miR-279 activity, such that a single genomic dose of either mir-996 or mir-279 provides nearly wildtype rescue to double deletion mutants in all characterized neural and non-neural settings. These data strongly support the notion that the seed region is the major determinant of in vivo miRNA function in animals.
Results
Generation of single and double deletion alleles of the mir-279 and mir-996 loci
The mir-996 hairpin is located ~1.5 kb downstream of the mir-279 hairpin (Fig 1A). While they share seed regions, the rest of their mature regions are distinct and well-conserved (Fig 1B). When we initially annotated mir-996, its hairpin was embedded in the annotated gene CG31044 (FlyBase Release 5.12), which was predicted to encode a short, non-conserved, protein. At the time, we hypothesized that CG31044 might represent the primary transcript for mir-996, distinct from mir-279 [26]. A similar inference of separate transcription units for these miRNAs was reported in a concurrent study [27]. Phylogenomic tracing indicates that mir-279 is ancestral and that mir-996 has adopted a derived sequence [28]. A third seed member, mir-286, is genomically unlinked from the mir-279/996 cluster and moreover deployed in a spatially and temporally distinct manner, being essentially restricted to early embryogenesis [26,29,30] (S1 Fig). Nevertheless, mature miR-279 is more similar to miR-286 than it is to miR-996 (Fig 1A). This suggests that miR-279 and miR-996 are selected for distinct sequences, presumably related to some separable functions.
Fig. 1. The mir-279/996 locus and phenotypes of single and double deletion alleles. 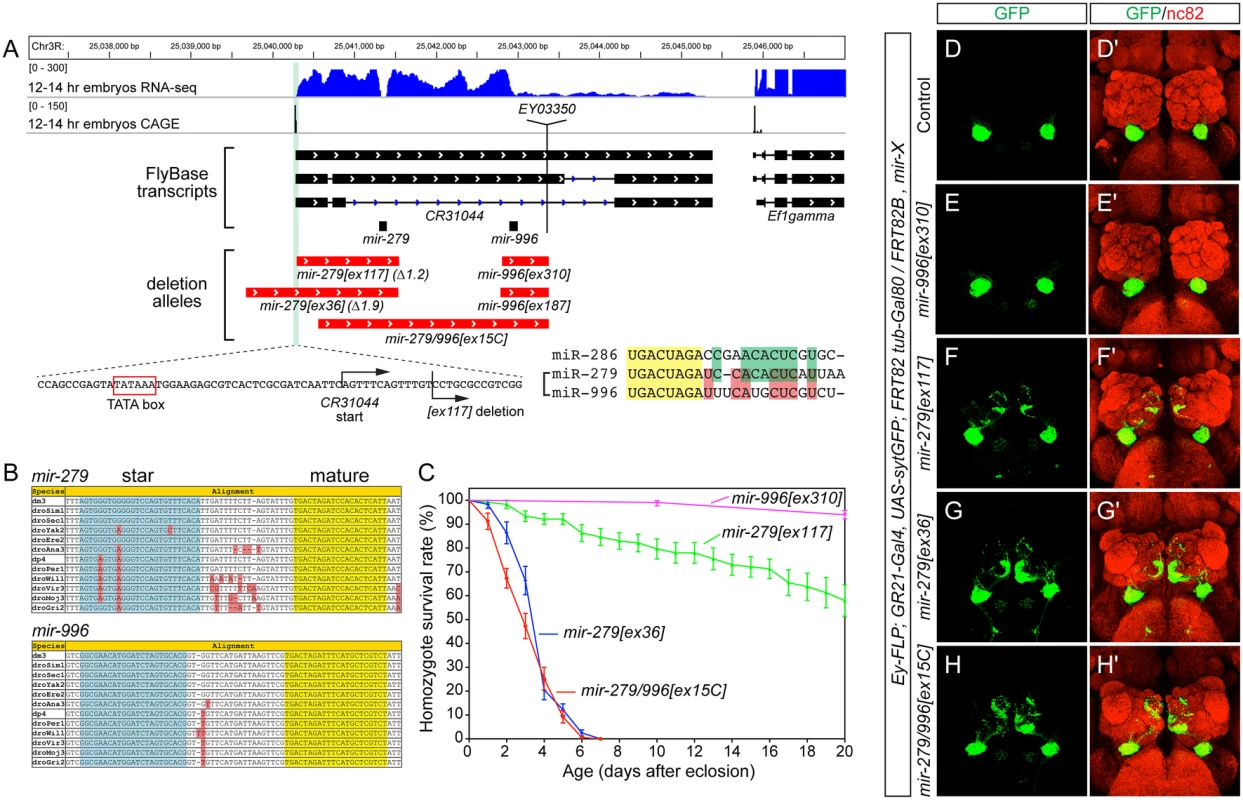
(A) Annotation of the mir-279/996 region in the Drosophila R5/dm3 version. RNA-Seq data (in blue) reveals characteristic "dropoffs" in read density at the Drosha cleavage sites for mir-279 and mir-996. CAGE data (in black) reveals a single peak in the mir-279/996 region located downstream of a TATA box, likely representing the start of a shared primary mir-279/996 transcript (i.e., CR31044). The genomic regions deleted in five mir-279/996 alleles are shown in red. Zoom-in of the 5' breakpoint of the mir-279[ex117] deletion shows it removes sequence just downstream of the CR31044 TSS, leaving the putative TATA box intact. The mature sequences of three members of the miR-279 seed family are shown; note that miR-286 is encoded elsewhere in the genome but is more related to miR-279 than is miR-996. (B) Alignments of pri-mir-279 and pri-mir-996 demonstrate their distinct mature miRNAs are perfectly conserved across across 12 Drosophilids. (C) Distinct lifespan of various mir-279/996 deletion alleles. n = 100 for mir-996[ex310] mutants, n>150 were assayed for the different mir-279 mutant alleles; equal numbers of males and females were included. Error bars represent SEM. (D-H) MARCM analysis of GR21+ neurons CO2-sensing neurons. The tester stock genotype is shown, where "X" refers to the mutations as labeled. (D, D') The normal projection pattern of GR21+ neurons to ventral glomeruli in control clones. (E, E') Deletion of mir-996 does not perturb CO2-sensing neurons. (F, F') mir-279[ex117] clones exhibit substantial ectopic projections to medial glomeruli, whereas mir-279[ex36] clones (G, G') and mir-279/996[ex15C] clones (H, H') exhibit more extensive misprojections. Our previous efforts yielded two alleles in this region, [ex117] and [ex36], that delete mir-279 but spare the mir-996 locus (Fig 1A). As the phenotypes of these mutants were rescued by a ~3kb genomic transgene bearing only mir-279, and lacked mir-996 sequence [18], mir-279 appeared to be causal. Nevertheless, we were interested to assess whether miR-996 contributes to any biological settings known to depend on miR-279. We screened excisions of a P element inserted downstream of mir-996, and recovered two small deletions ([ex187] and [ex310]) that selectively remove this locus. We also recovered a longer deletion ([ex15C]) that removes both mir-279 and mir-996 loci, thus establishing an apparent allelic series of single and double mutants of these miRNAs (Fig 1A).
Both mir-996 single deletions were homozygous viable and lacked obvious morphological or behavioral defects. We measured the lifespan of mir-996[ex310] mutants and this was normal (Fig 1C). We also analyzed the projections of GR21+ olfactory neurons. In wildtype, these CO2-sensing neurons are present only in the antenna and they project to ventral glomeruli. We visualized these in control ey-FLP; FRT82B MARCM clones generated in the GR21-Gal4, UAS-synaptotagmin-GFP background, and stained for GFP-labeled projections to brains that were counterstained with nc82 (Fig 1D). The GR21+ projections of mir-996 deletion MARCM clones were identical to wildtype (Fig 1E). In contrast, mir-279 deletions induced ectopic medial projections (Fig 1F and 1G), as described [18], and reflected the generation of ectopic CO2-sensing neurons in the maxillary palp.
Our newly generated mir-279/996 double deletion mutant [ex15C] exhibited similar gross phenotypes as mir-279[ex36], with respect to lifespan (Fig 1C) and ectopic GR21+ projections (Fig 1H). As well, the pharate lethality of homozygous [ex15C] mutants was well-rescued by the mir-279-only genomic transgene. Altogether, these findings were consistent with an interpretation that miR-279 is primarily responsible for essential genetic requirements of this two-miRNA locus.
Unanticipated effects of mir-279 deletions on mir-996 expression
While the initial genetic data were reasonably explained by phenotypic dominance of miR-279, certain other observations remained difficult to account for. Perhaps most germane was the fact that the mir-279[ex117] and mir-279[ex36] single deletions, both null for mir-279, exhibited distinct viability. While mir-279[ex36] is lethal within a few days of eclosion, mir-279[ex117] adults can eclose and survive for weeks with optimal care, despite their locomotor difficulties (Fig 1C). The discrepancy of these alleles was exploited in the circadian rhythm studies of Sehgal and colleagues; such behavioral studies require adult viability of at least one week [24]. Their analyses utilized a stock of mir-279[ex117] that had been outcrossed to remove potential second-site aberrations.
A plausible model was that mir-279[ex36] bears an unlinked mutation responsible for its stronger defects. However, we were unable to recover homozygous mir-279[ex36] stocks that survived longer, even after extensive outcrossing of this mutant chromosome. In addition, flies carrying mir-279[ex36] in trans to a deficiency of the region showed the same gross phenotypes. Finally, we could rescue the viability and locomotor behavior of this mutant using the 3kb mir-279-only genomic transgene. Taken together, these findings suggested that phenotypic differences between mir-279[ex117] and mir-279[ex36] must reside extremely close to the mir-279 hairpin. For example, there might theoretically be another non-coding function of the primary mir-279 transcript, or even perhaps a peptide encoded by this region.
However, another scenario we considered was that miR-996 might be affected by available miR-279 alleles. Northern analysis of small RNAs in different mir-279 and mir-996 alleles confirmed this hypothesis. The mir-996[ex310] homozygous mutant lacked mature miR-996, validating its nature as a null allele and demonstrating specificity of the miR-996 probe; this mutant expressed miR-279 normally (Fig 2A). Both mir-279 "single" alleles and the mir-279/996 double deletion failed to express mature miR-279, as expected, but all of these mutants also proved deficient for miR-996. mir-279[ex117] expressed <10% the normal level of miR-996, and mir-279[ex36] did not detectably express miR-996 (Fig 2B). We obtained similar results by examining different female tissues (Fig 2) as well as male tissues (S2 Fig). Therefore, we conclude that the available mir-279 "single" mutants are unexpectedly also strong or nearly null alleles of mir-996.
Fig. 2. Severe loss of mature miR-996 expression in mir-279 deletion alleles. 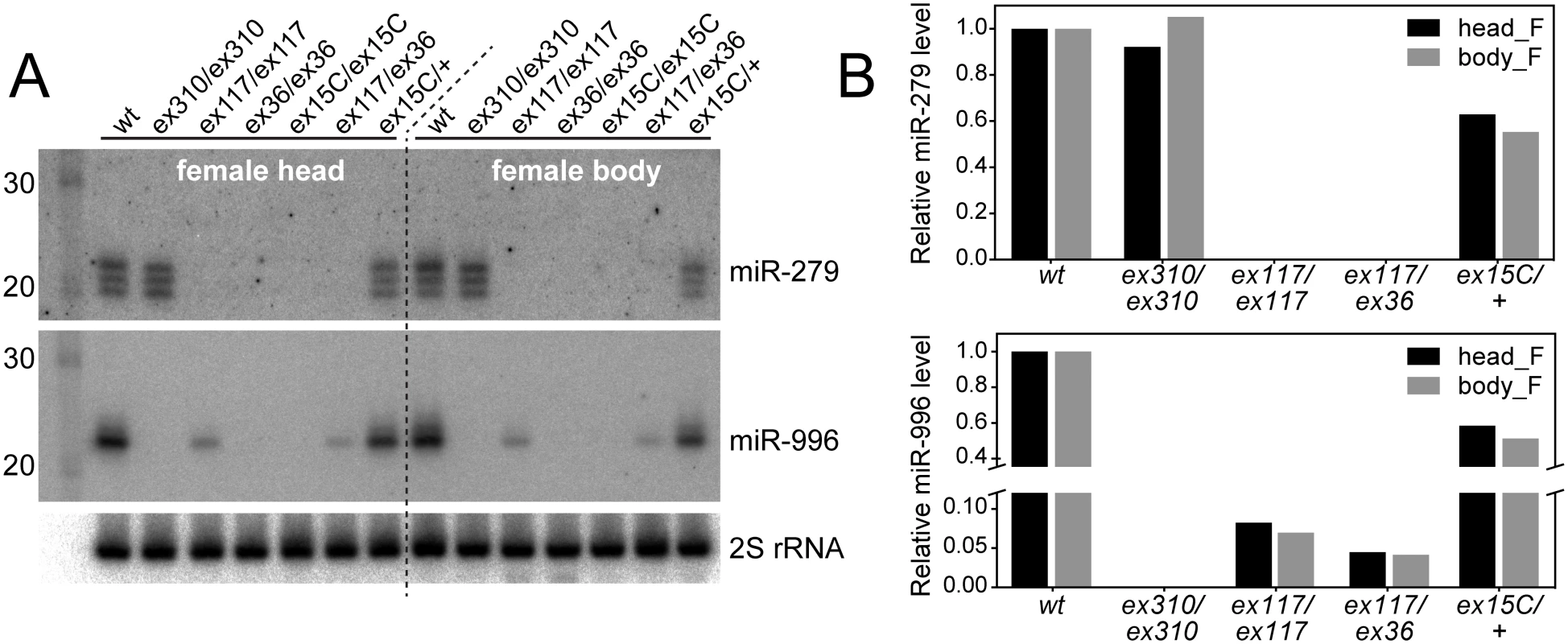
(A) Northern blots of miR-279 and miR-996 in various mir-279 and mir-996 homozygous or trans-heterozygous allele combinations. In mir-279 alleles [ex117] and [ex36] that retain the mir-996 genomic DNA, the levels of mature miR-996 are strongly diminished ([ex117]) or nearly undetectable ([ex36]). mir-996[ex310] is a deletion of the mir-996 region that does not affect mir-279 and mir-279/996[ex15C] deletes both miRNAs. (B) Quantifications of mature miR-279 and miR-996 levels. Homozygous [ex117] mutants expressed ~10% of the wild type level of miR-996 and [ex117/ex36] transheterozygous mutants expressed ~5% of miR-996. Note that the expression level for both miRNAs is copy-number dependent, since heterozygous mir-279/996[ex15C] flies expressed roughly half of both miRNAs compared to wild type animals. Consideration of current modENCODE transcriptomic data at the mir-279/996 region proved informative (Fig 1). Our small RNA analyses showed that miR-279 and miR-996 belong to the same expression cluster across diverse Drosophila tissue and cell line small RNA libraries [31], indicating their coordinate deployment. Inspection of companion transcriptome data [32] revealed relatively continuous, although graded, levels of RNA-seq reads across the entire locus, consistent with the notion of a single primary mir-279/996 transcript. It is commonly observed in Drosophila that 3' fragments of Drosha-cleaved primary transcripts are less stable than 5' Drosha fragments [33,34], and evidently at the mir-279/996 locus, the 3'-most fragment of its primary transcript is least stable of them all (Fig 1A). On the basis of such transcriptome data, the provenance of CR31044 was expanded in the most recent FlyBase release (5.47), such that it now includes both mir-279 and mir-996 (Fig 1A).
Analysis of capped analysis of gene expression (CAGE) data revealed a 5' transcription start site ~1kb upstream of the mir-279 hairpin. This lies <30nt downstream of a typical TATA box sequence (GTATATAAA), suggesting that as the promoter for the mir-279/996 transcription unit. The deletion extents of the "mir-279" alleles, relative to the transcription start, were notable. mir-279[ex36] removes sequence upstream of the promoter, presumably explaining why this allele strongly compromises expression of the downstream miRNA. On the other hand, mir-279[ex117] deletes to within 14 nt of the transcription start site (Fig 1A), which does not abolish, but apparently debilitates expression and/or processing of the intact mir-996 hairpin. Altogether, these molecular observations are consistent with our genetic inference that both miRNAs may contribute to mutant phenotypes uncovered by chromosomal aberrations of the region.
Generation of genetically defined miR-279 and miR-996 single and double mutants
Since the available allelic series did not permit assessment of phenotypes caused by specific loss of miR-279, we sought an alternative strategy to analyze "clean" mir-279 and mir-996 mutant backgrounds. To do so, we recombineered a genomic transgene that includes the full 16.6kb intergenic region between the upstream (CG14508) and downstream (Ef1gamma) protein-coding genes (Fig 3A), and thus may be expected to confer full miRNA rescue. To permit direct comparison with variant forms, we used the phiC31 system to integrate transgenes into a common genomic site. The wildtype 16.6 kb transgene restored accumulation of mature miR-279 and miR-996 (Fig 3B) and fully rescued viability of mir-279/996[ex15C] double deletion homozygotes (Fig 3C). Using this genomic fragment, we then generated a series of mutant transgenes in which we specifically deleted 100bp covering either the mir-279 or mir-996 hairpins (1x-mir-279 and 1x-mir-996, which essentially serve as mir-996-KO and mir-279-KO transgenes, respectively) or replaced either miRNA with the non-cognate hairpin (2x-mir-279 and 2x-mir-996) (Fig 3A).
Fig. 3. Modified genomic transgenes to assess individual miR-279/996 functions. 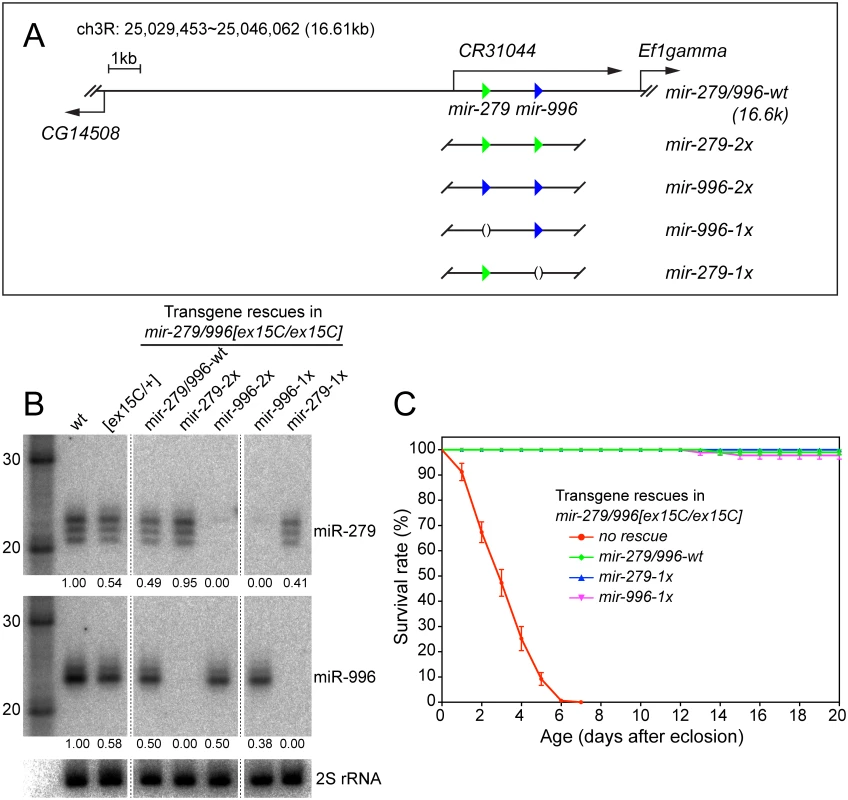
(A) In the 16.6kb mir-279/996 rescue transgene, the 5' end extends to cover a portion of the upstream CG14508 gene and 3' end extends into the downstream Ef1gamma gene. Green and blue triangles represent mir-279 and mir-996 hairpins, respectively. The wildtype genomic fragment was modified to replace the mir-279 and mir-996 hairpins with either a deletion or the non-cognate miRNA. (B) Northern blots verify the expression of miR-279 and miR-996 from different genomic transgenes that were introduced into mir-279/996[ex15C] double mutant homozygous animals. RNA samples were extracted from whole flies carrying one copy of individual transgenes. Intensities of miR-279 and miR-996 expression were quantified by normalization to homozygous wild type and marked below each lane. mir-996-2x expressed only comparable amount of miR-996 as mir-279/996-wt and mir-996-1x transgenes. (C) Rescue of the lifespan defect in mir-279/996[ex15C] double mutants by 16.6kb transgenes. The mir-279/996[ex15C] data shown here are the same as plotted in Fig 1C. For each genotype, 100 flies including equal number of males and females were assayed; error bars represent SEM. We placed one copy of each transgene into mir-279/996[ex15C] homozygotes, and performed Northern blotting for the two miRNAs from adult females. These tests demonstrated specific expression of mature miR-279 and miR-996 in the designated genotypes (Fig 1A). This confirmed their status as a bona fide panel of single mutants of the mir-279/996 locus, an allelic series that was not functionally fulfilled by corresponding single genomic deletions. For reasons that are not apparent, the amount of mature miR-996 from the transgenic copies, especially the reprogrammed allele, was not as robust as the endogenous chromosomal locus (Fig 3B). Nevertheless, each of the four transgenes fully rescued adult viability of mir-279/996 double deletion backgrounds. This provided an initial view into the complicated genetics of this locus. Strikingly, endogenous expression of only a single copy of either 1x-mir-279 or 1x-mir-996 transgenes in the double mutant, thus recapitulating full knockout of either miRNA on top of heterozygosity for the other, was sufficient to rescue adult viability (Fig 3C). This indicates that there is no essential requirement for the unique miR-279 sequence, and that one allele of either mir-279 or mir-996 supports normal viability of Drosophila.
We proceeded to subject these engineered miRNA backgrounds to detailed phenotypic study, to ascertain the extent to which defects previously attributed to miR-279 might actually depend on the joint function of miR-279 and miR-996.
Both miR-279 and miR-996 mediate specification of CO2-sensing neurons
Under MARCM clonal conditions, mir-996 single hairpin deletions exhibit normal specification of CO2-sensing neurons within the antenna, and these project to ventral glomeruli in the central brain (Fig 1D and 1E). In contrast, mir-279 single hairpin deletions, which we now recognize as deficient for mature miR-996, exhibit ectopic CO2-sensing neurons in the palp, and these project to medial glomeruli. In particular, mir-279[ex117] (which retains some expression of miR-996) exhibits a weaker GR21 phenotype than mir-279[ex36] or mir-279/996[ex15C] (which are nearly null or definitively null for both miRNAs, respectively) under clonal conditions (Fig 1F–1H).
These observations strongly hinted that endogenous miR-996 contributes to suppression of CO2-sensing neurons in the maxillary palp. We sought to test this under non-clonal conditions, which are expected to be more sensitive than clonal conditions, which may potentially be rendered less potent by perdurance. It is more difficult to obtain mir-279/996[ex15C] homozygotes compared to MARCM mutant adults, but these proved to exhibit strong and fully penetrant GR21 projection phenotypes (Fig 4A). In fact, under these non-clonal conditions, this genuine double deletion mutant exhibited slightly stronger phenotypes than either of the shorter deletions that physically remove only mir-279 but compromise mir-996 expression (Fig 4G).
Fig. 4. miR-996 restores proper CO2-sensing neurons to mir-279/996 double mutants. 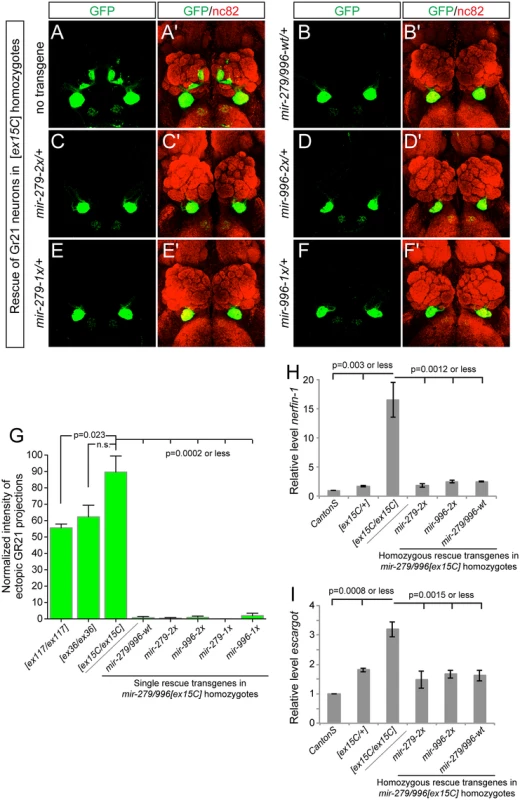
(A-F) Representative images of GR21 projections in brains from mir-279/996[ex15C] whole animal mutants and rescued backgrounds bearing one copy of each 16.6 kb genomic transgene. Genotype for the mutant is: Gr21-Gal4, UAS-syt-GFP/+; mir-279/996[ex15C], and rescued genotypes are: Gr21-Gal4, UAS-syt-GFP/mir[rescue]; mir-279/996[ex15C]. (A, A') Double deletion mir-279/996[ex15C] homozygote. (B-F) Rescue of mir-279/996[ex15C] homozygotes by a copy of the wildtype transgene (B, B'), by a single mir-279-only transgene (C, C'), by a single mir-996-only transgene (D, D'), by a two-copy mir-279-only transgene (E, E'), and by a two-copy mir-996-only transgene (F, F'). (G) Quantification of ectopic Gr21 axon projection normalized to the normal ventral projection. 4–6 brains were analyzed for each genotype, error bars represent SEM, and p-values calculated by Student's two-tailed T-test, equal variance. (H-I) qPCR assays show elevated nerfin-1 (H) and escargot (I) transcript levels in mir-279/996[ex15C] homozygous heads. These defects are comparably rescued by mir-279-2x or mir-996-2x transgenes as they are by wildtype mir-279/996 genomic transgene, to target levels that are slightly higher than in Canton S heads but similar to [ex15C]/+ heads. Error bars represent SD, p-values calculated using unpaired T-tests from the combined data of independent, triplicate qPCR assays. The mir-279/996[ex15C/ex15C] phenotype was fully rescued by a single insertion of the wildtype 16.6kb mir-279/996 transgene, validating its status as a fully functional genomic fragment (Fig 4B). Moreover, single insertions of either "2x" transgene, in which mir-279 was substituted for mir-996, and vice versa, also provided complete rescue of the ectopic CO2-sensing neurons (Fig 4C and 4D). In more stringent assays, we then showed that single insertions of either "1x" transgenes, recapitulating mir-279-knockout and mir-996-knockout conditions, similarly provided essentially full suppression of the double deletion phenotype (Fig 4E and 4F). We quantified these rescues, expressed as the relative amounts of ectopic GFP+ projections in the brain, in Fig 4G.
The ectopic CO2 neuron phenotype was shown to be driven by derepression of specific miR-279 targets, namely the transcription factors encoded by nerfin-1 and escargot [18,23]. Although CO2-sensing neurons comprise only a small number of cells in the nervous system, we were able to detect massive derepression of nerfin-1 (Fig 4H) and substantial upregulation of escargot (Fig 4I) transcripts in whole mir-279/996[ex15C/ex15C] adult heads, relative to Canton S control heads. Based on the degree of misregulation, we infer that miR-279/996 must regulate nerfin-1 outside of the CO2-sensing apparatus. In mutant heads bearing either wildtype mir-279/996 genomic transgene, or mir-279-only or mir-996-only transgenes, we observed comparable restoration of nerfin-1 and escargot transcript levels by the various miRNAs (Fig 4H and 4I). The rescued levels were consistently slightly greater than Canton S but were actually similar to mir-279/996[ex15C/+] heterozygotes in all cases. This might reflect marginally incomplete rescue by the transgenes, or alternatively, some genetic background variation between this control and the miRNA deletion genotypes. In either case, we conclude that this dramatic, fully-penetrant, neural cell specification phenotype requires the joint activity of the miR-279 and miR-996 miRNAs, and that either miRNA suffices to direct normal development of these neurons via joint repression of shared critical target genes.
Both miR-279 and miR-996 mediate normal adult rhythmic behavior
We next tested the potential involvement of miR-996 in maintenance of circadian behavior. We initially studied the hypomorphic mir-279[ex117] homozygous condition, and confirmed previous observations [24] that a majority of mir-279[ex117] mutant flies displayed arrhythmic locomotor activity in constant darkness (Fig 5A and 5B). Unexpectedly, however, we further observed that 30% of mutant individuals were weakly rhythmic (Fig 5C). The quantification of percentage of rhythmic animals, their circadian period, and their power of rhythmicity are shown in Table 1. Restoration of either miR-279 or miR-996 on the [ex117] background, in either two doses or in a single dose, fully recovered behavioral rhythmicity (Fig 5D–5F and Table 1). The normalized activity profiles of all the different transgene combinations in the [ex117] background are provided in S3 Fig.
Tab. 1. Both miR-279 and miR-996 contribute to circadian rhythm. 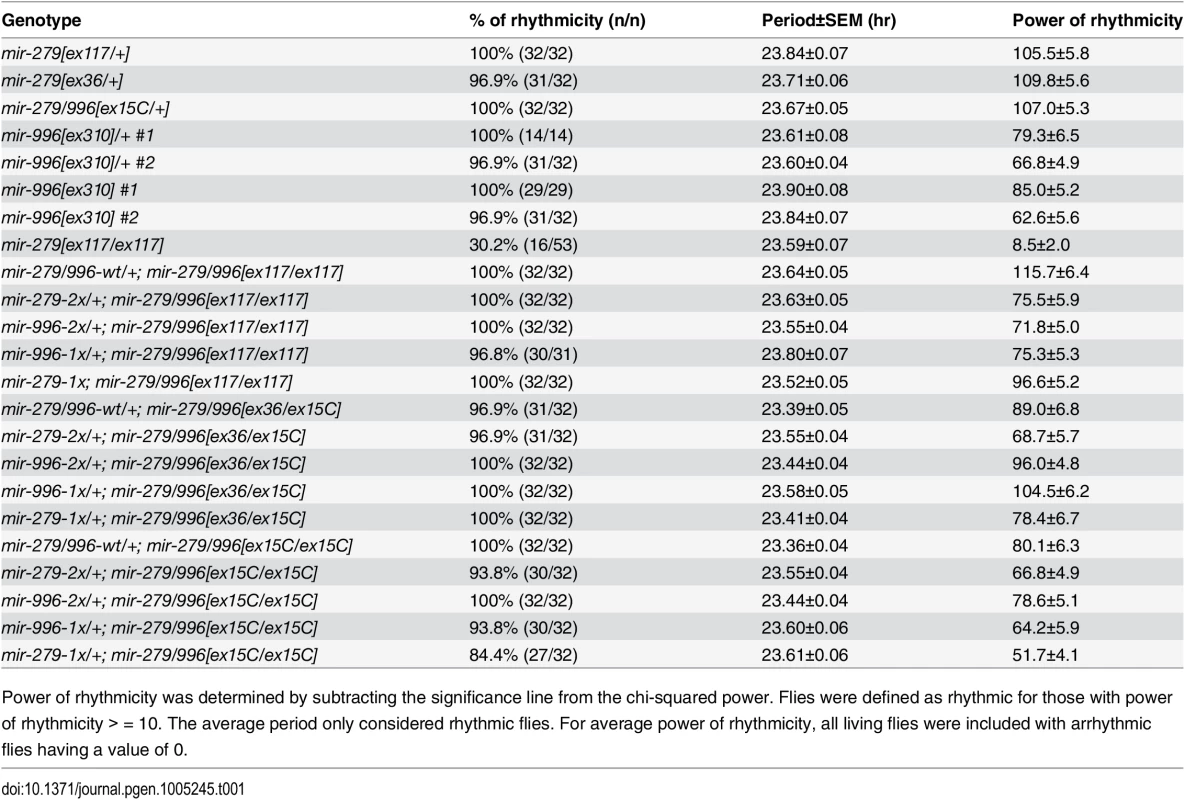
Power of rhythmicity was determined by subtracting the significance line from the chi-squared power. Flies were defined as rhythmic for those with power of rhythmicity > = 10. The average period only considered rhythmic flies. For average power of rhythmicity, all living flies were included with arrhythmic flies having a value of 0. Fig. 5. Both miR-279 and miR-996 contribute to maintenance of circadian rhythm. 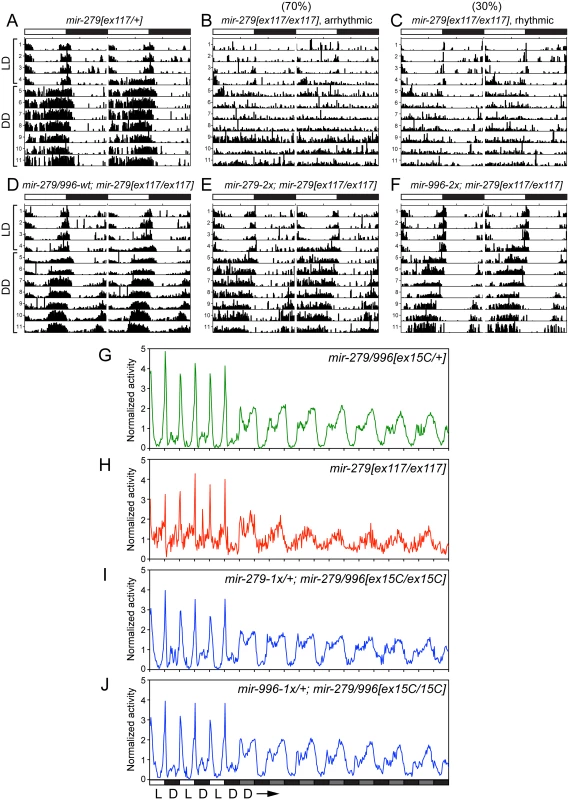
(A-F) Typical activity profiles of individual flies of various mir-279/996 genotypes. Animals were entrained in 12hr-light/12hr-dark (LD) cycles for four days, and then kept in constant darkness (DD) for seven more days. In LD cycles, white bars represent the light phase (day) and black bars represent the dark phase (night). During DD cycles, grey and black bars represent the subjective day and night time, respectively. (A) mir-279[ex117] heterozygotes behave normally in that they maintain circadian activity in the dark, although the strong morning and evening activity peaks and mid-day siesta are not as well maintained. (B-C) In mir-279[ex117] homozygotes, the majority of animals gradually lost behavioral rhythmicity after transferring to constant darkness (B), but about 1/3 of animals could maintain circadian activity in constant darkness (C). Note that all mir-279[ex117/ex117] animals exhibited generally less activity than heterozygotes. The activity and circadian defects in mir-279[ex117/ex117] animals were rescued by single copies of the wild-type 16.6kb mir-279/996 transgene (D) or the 2x-mir-279-only (E) or 2x-mir-996-only (F) transgenes. (G-J) Averaged activity profiles of various mir-279/996 genotypes. (G) mir-279/996[ex15C] heterozygotes exhibit robust behavioral rhythmicity after transferring to constant darkness, but mir-279[ex117] homozygotes do not. In the mir-279/996[ex15C] homozygous background (which is normally mostly lethal by ~4 days), expression of only a single 1x-mir-279 (I) or single 1x-mir-996 (J) transgene can restore normal rhythmic behavior in constant darkness. n = ~32 for each genotype; the number of flies assayed for each genotype are indicated in Table 1. We performed more stringent tests by asking if either miRNA could rescue the coordinate absence of both miR-279 and miR-996. Circadian rhythm assays require flies to be mobile for at least a week. Since homozygous mir-279[ex36] and mir-279/996[ex15C] adult flies are poorly able to stand or walk, and die within a few days (Fig 1C), these genotypes cannot be assayed for circadian behavior. Heterozygotes of the double deletion [ex15C] exhibit normal rhythmic behavior (Fig 5G); for comparison, the normalized activity pattern of surviving [ex117] homozygotes is shown in Fig 5H. When we assayed our panel of recombineered transgenes in mir-279/996[ex36/ex15C] transheterozygotes, which should be close to a null condition for the locus, we observed normal rhythmic behavior in all cases (Table 1). Finally, we performed the strictest test by assaying rescues of [ex15C] homozygotes. Strikingly, all transgene isoforms, including single mir-279 and mir-996 versions, fully restored the normal circadian clock in the mutant flies (Fig 5I and 5J and Table 1). Altogether, these data indicate that intact mir-996 expression fully complements loss of mir-279 in both nervous system development and adult neurophysiology.
Similar and distinct capacities of miR-279/996 in gain-of-function analyses
The evidence gathered indicates that miR-279 and miR-996 play surprisingly similar roles in diverse developmental and behavioral settings. Nevertheless, their distinct conserved 3' sequences (Fig 1B) suggests that these miRNAs have diversified in some way during evolution. As we showed in the head, endogenous miR-279 and miR-996 exhibit comparable activity to restrict the accumulation of nerfin-1 and escargot transcripts (Fig 4H). To probe this further, we compared the gain-of-function activities of these miRNAs using luciferase sensor assays in S2 cells. We tested 3' UTR sensors for nine genes bearing conserved miR-279 family target sites, including all of those identified in previous in vivo studies. Transfection of ub-Gal4 and UAS-dsRed-miRNA expression constructs led to significant repression of all sensors (Fig 6A). As a negative control, the Hairless 3' UTR does not contain any miR-279/996 binding site and the sensor was not repressed upon miRNA transfection (Fig 6A). We verified comparable ectopic expression of both miR-279 and miR-996 in these transfection experiments (Fig 6B). Therefore, the capacities of miR-279 and miR-996 in S2 cells are similar.
Fig. 6. Comparison of gain-of-function activities of miR-279 and miR-996. 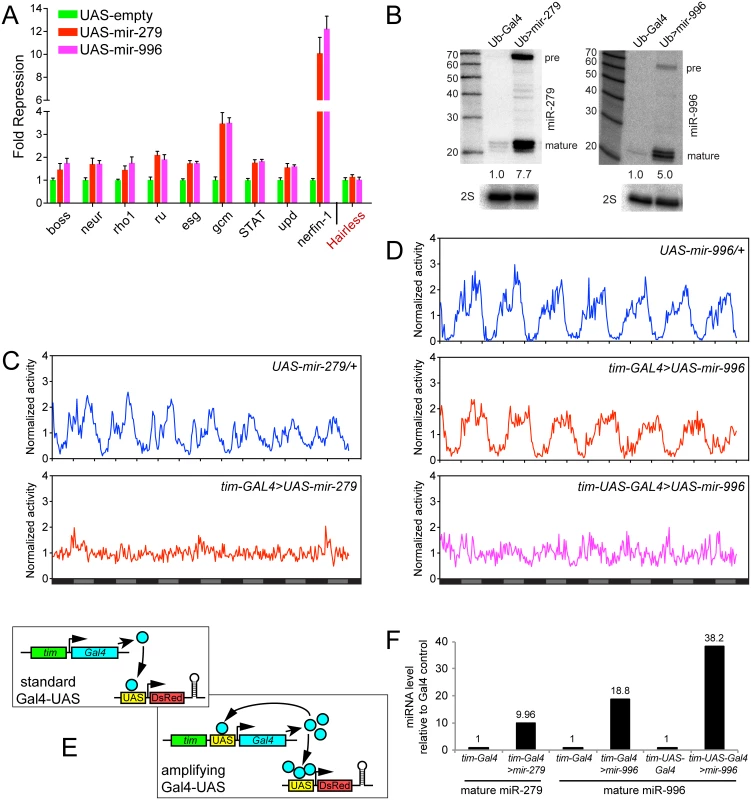
(A) Luciferase sensor assays in S2 cells indicated that 3' UTRs of multiple miR-279 targets are all additionally responsive to miR-996. The control Hairless 3' UTR has no miR-279/996 seed match and was not repressed by these miRNAs. Error bars represent standard deviation from quadruplicate assays. (B) Northern confirmation of ectopic miR-279 and miR-996 in S2 cell experiments. pre = pre-miRNA hairpin, mature = mature miRNA product. Overexpressed miRNAs were calculated relative to endogenous mature miRNAs, normalized to 2S loading control. (C-D) Averaged activity profiles for control and miRNA overexpressing flies for 7 days in constant darkness since the second day after transferring from LD to DD. Some of these experiments utilized the amplifier driver tim-UAS-Gal4, as schematized in (E). (C) Overexpression of miR-279 by tim-Gal4 induced strong arrhythmia. (D) Ectopic expression of miR-996 by tim-Gal4 had no significant affect on circadian behavior, but further induction by tim-UAS-Gal4 led to a complete arrhythmia. n = ~32 for each genotype; the number of flies assayed for each genotype is indicated in Table 2. (F) Validation that higher levels of mature miR-996 are induced by tim-UAS-Gal4, compared to tim-Gal4. Overexpressed miRNAs were quantified as in (B) using Northern blotting, and normalized to 2S loading control. These tests also confirm that tim-Gal4>UAS-mir-996 flies effectively misexpressed miR-996, even though they lacked circadian defects. To explore their ectopic activities in vivo, we overexpressed the miRNAs in the circadian neurons with tim-Gal4. In accordance with previous studies [24], overexpression of miR-279 in circadian tissues strongly disrupted the adult behavioral rhythm. In contrast to the unactivated transgene background, tim>mir-279 animals quickly became arrhythmic following their transfer to constant darkness (Fig 6C). However, ectopic miR-996 only weakly affected circadian rhythm, with 13% arrhythmic flies observed with only one of the two insertions (Fig 6D). Quantitative data for these genotypes is shown in Table 2. This was not due to inability to produce miR-996, since the degree of accumulation of ectopic miR-996 induced by tim-Gal4 was greater than for miR-279 (Fig 6F). Such phenotypic differences suggested that miR-279 has stronger capacity to influence circadian cell activity, even though endogenous mir-996 is completely able to compensate for the absence of mir-279.
Tab. 2. Circadian rhythm defects upon overexpression of miR-279 family miRNAs. 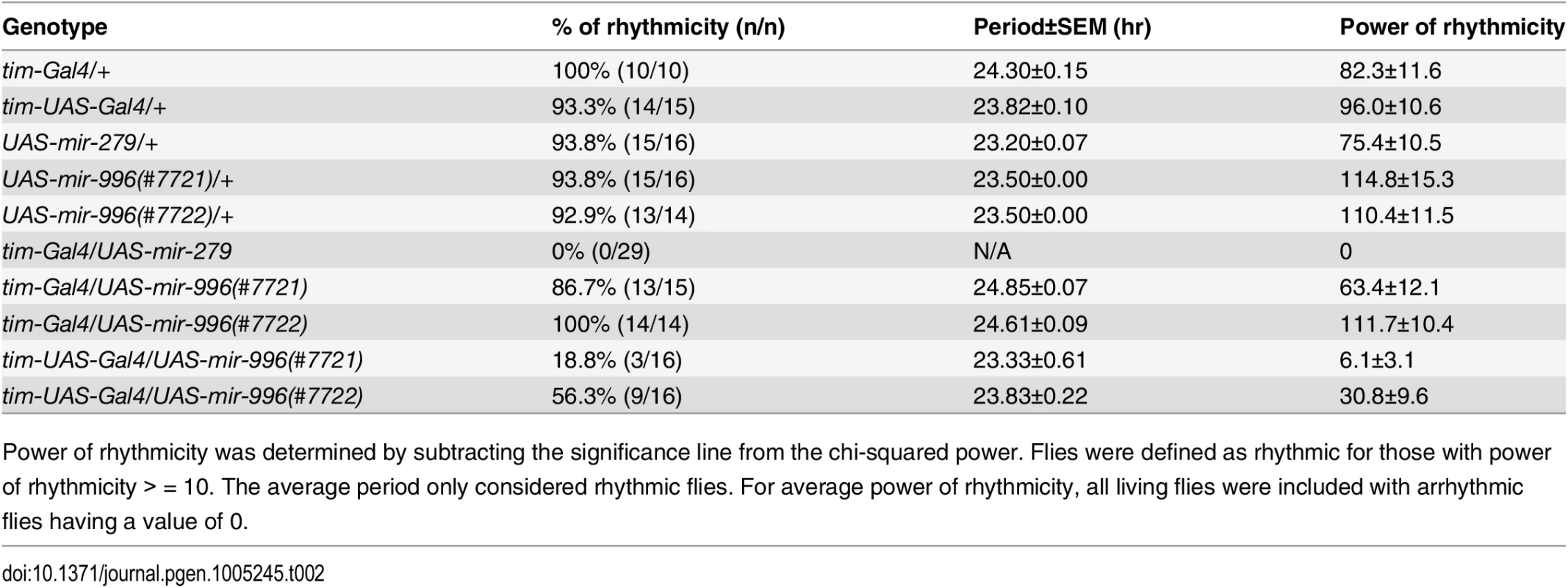
Power of rhythmicity was determined by subtracting the significance line from the chi-squared power. Flies were defined as rhythmic for those with power of rhythmicity > = 10. The average period only considered rhythmic flies. For average power of rhythmicity, all living flies were included with arrhythmic flies having a value of 0. Nevertheless, given the substantial overlapping capacities of these miRNAs for target regulation (Fig 6A), we asked whether increased levels of mir-996 could influence circadian rhythm. For these tests, we utilized tim-UAS-Gal4, which auto-potentiates Gal4 expression in tim-expressing neurons (Fig 6E). We used Northern blotting to verify that more mature miR-996 was generated in the latter condition (Fig 6F). Interestingly, in this sensitized overexpression background, miR-996 induced substantial behavioral arrhythmia (81% and 44% of the independent UAS-mir-996 insertions exhibited arrhythmia, Fig 6D and Table 2). In summary, these gain-of-function experiments reveal intrinsic differences between miR-279 and miR-996, which otherwise exhibit surprising genetic redundancy under carefully controlled endogenous conditions. This is perhaps counter to normal expectation, in which overexpressed seed family miRNAs more typically exhibit similar properties even on the transcriptome level [35,36], but may instead display functional distinctions under physiological settings.
Discussion
miRNAs with diverse and profound endogenous phenotypic requirements
Despite extensive experimental and computational evidence for the pervasive nature of animal miRNA target networks, genetic studies have not generally supported the notion that animals rely upon miRNAs to the same degree, as say, transcription factors and signaling pathways [37]. This was strikingly evident with the systematic knockout of C. elegans miRNAs, which revealed barely any developmental or behavioral phenotypes [12]. Similarly, a genomewide collection of D. melanogaster miRNA knockouts reveals a variety of phenotypes, but these are generally quantitative in nature and include few documented developmental defects [38]. While this might partly be due to functional overlap amongst members of similar miRNA families, compound knockouts of C. elegans miRNA families revealed overt consequences only for a minority of families [39,40]. Only upon further sensitization, by reducing the activity of other gene broad regulators, did additional miRNA knockouts exhibit phenotypes [41]. Together with studies of dozens of miRNAs that mostly exhibit phenotypes under sensitized conditions [42–44], an emerging concept is that miRNAs mostly act as robustness factors [5,16].
Nevertheless, select miRNAs have proven to be essential for certain development or physiological processes. Our current studies affirm and extend the broad impact of the mir-279/mir-996 locus, which generates phenotypically critical miRNAs of profound impact. Together, these miRNAs are fully essential for organismal viability, for normal cell specification of olfactory neuron subtypes, and for rhythmic behavior via circadian pacemaker cells. An unexpected conclusion of this work was to uncover that these two miRNAs, previously inferred to derive from separate transcription units [26], actually provide highly overlapping in vivo activities. Although the stringent evolutionary conservation of these miRNAs is de facto evidence that they are not truly "redundant", we demonstrate using precise genetic engineering that single genomic copies of either mir-279 or mir-996 can fully compensate for the deletion of all four miRNA alleles in diverse developmental and physiological settings.
A notable feature of the function of this miRNA family operon is that they mediate their effects through multiple key, setting-specific, targets. For example, the transcription factors encoded by nerfin-1 and escargot are the critical miR-279/996 targets whose de-repression induces ectopic CO2-sensing neurons, and whose heterozygosity confers substantial rescue of ectopic CO2-sensing neurons in flies that lack these miRNAs [18,23]. On the other hand, miR-279/996 have substantial effects on different aspects of the JAK-STAT signaling pathway in circadian pacemaker cells and ovarian border cells, by targeting the ligand unpaired [24] and the transcription factor STAT [25], respectively.
While manipulation of these various targets can substantially rescue setting-specific phenotypes caused by loss of miR-279 and miR-996, none of them rescue the extremely abbreviated lifespan of the double mutant. It remains to be seen whether this marked phenotype is due to combined de-repression of multiple characterized targets, or to a different pathway or target network. Our newly characterized mir-279/996 alleles and rescue backgrounds comprise valuable reagents for future study of these miRNAs.
Dominance of seed sequences for in vivo miRNA functions
Early genetic studies revealed the principle of miRNA seed targeting [10,11], many years before the complementary miRNAs were identified [9,45]. Since then, a wealth of experimental studies have shown that ~7 nt seed matches are sufficient to confer substantial regulation by miRNAs, not only in culture cells [46] but also in the animal [47,48]. Genomewide studies show that seed-matching is the dominant mode of conserved target recognition [49–51]. In addition, overexpression tests clearly demonstrate that the dominant transcriptome signature induced by ectopic miRNAs is seed-based, and maintained even upon substitution of the remainder of the miRNA sequence [35,36].
Nevertheless, considerable debate continues about the contribution of "non-seed" target sites to miRNA networks. Notably, some of the earliest miRNA targets found, which definitively mediate regulatory interactions critical for development, lack continuous seed matching [7,8]. In the classic example of let-7:lin-41 pairing, the atypical architecture of a bulged seed supplemented by extended 3' pairing is proposed to permit specific recognition that cannot be achieved by other let-7 family members in C. elegans [40,52]. A number of subsequent directed studies define functional and/or conserved miRNA:target pairing configurations, characterized by 3' compensatory pairing, distributive complementarity, or centered pairing [47,53,54]. The existence of such sites has been attractive from the point of view that they might help explain strong evolutionary constraints on entire miRNA sequences, which are not satisfactorily explained by seed regions alone. However, it remains to be seen whether mutations of non-seed miRNA sequences are of consequence to in vivo miRNA-mediated phenotypes.
On the other hand, available genetic studies suggest limits on the contribution of non-seed regions to in vivo miRNA phenotypes. For example, all known miRNA point mutant alleles, other than ones that broadly affect biogenesis, invariably prove to alter seed regions [6,8,21,22,55,56]. In addition, Horvitz and colleagues showed that embryo/larval lethality caused by compound mutations of the 8-member mir-35 family or the 6-member mir-51 family could both be rescued by individual family members [39]. Our studies of miR-279 and miR-996 provide a new testbed for this, since this locus is responsible for several of the most overt developmental and behavioral phenotypes ascribed to animal miRNAs. While we provide functional evidence that these miRNAs are not identical in regulatory capacity and/or processing, it is still striking from the biological viewpoint that multiple essential in vivo phenotypes can be fully satisfied by either of these non-seed divergent miRNAs.
It should be informative to conduct similar studies for some of the other miRNAs that exhibit overt phenotypic requirements, potentially by systematic mutation of 3' regions, to assess their relative contribution to organismal phenotypes. With the advent of CRISPR/Cas9 genome engineering, it is now feasible to address these questions with respect to miRNA genes and miRNA sites in the genomes of intact organisms [57,58].
Materials and Methods
Generation of new mir-279/996 deletion alleles
The mir-996 single deletion and mir-279/996 double deletion alleles were generated by imprecise excision of P{EPgy2}CR31044[EY03350], which is inserted 370bp downstream of the mir-996 hairpin. We crossed these to the TMS, Δ2-3/TM6B jumpstarter and induced transposition in P{EPgy2}CR31044[EY03350]/TMS, Δ2–3 animals. Following the segregation of TMS, Δ2–3, we screened ~500 candidate excision chromosomes for deletions in the mir-279/mir-996 region using the the following PCR amplicons: mir279F excision CAAGAAACCACCCCGAGAAGAAGAAG mir279R excision AGCAGGTGTTACAGTTACACTCAAACG.
The mir-996[ex310] deletion contains a 568 deletion with 9bp of P-element sequence left, while the mir-996[ex187] deletion contains a 584 bp deletion with 154bp of P-element sequence left. The mir-279/996[ex15C] allele bears a 2825bp deletion that removes both miRNA hairpins and retains 84bp of P-element sequence.
Generation of mir-279/996 genomic rescue transgenes
The 3.0kb genomic sequence containing only mir-279 was cloned from the genome and inserted into the pBDP vector [59]. For the large rescue transgenes, we retrieved 16.6kb extending into both upstream and downstream protein-coding genes of the mir-279/996 locus from the BAC CH322-35G11 (BACPAC Resources) and cloned it into the attB-P[acman]-AmpR vector by recombineering as described [60]. The mir-279 or mir-996 hairpins were targeted with an rpsL-neo cassette (Gene Bridges), which was flanked by the ~50bp left and right homology arms for the miRNA and carried two BbvCI restriction sites between the rpsL-neo cassette and the homology arms. We then deleted the rpsL-neo cassette from the targeted construct by BbvCI digestion and the remaining vector was re-ligated to generate the mir-279-1x or mir-996-1x construct.
To generate the mir-279-2x construct, genomic fragments 13.5kb upstream and 3.0kb downstream of the mir-996 hairpin were retrieved from the CH322-35G11 BAC and cloned between the AscI and NotI sites of the attB-P[acman]-AmpR vector. The 5’ end of the upstream and 3' end of the downstream fragment were identical to the ends of the 16.6kb wild type genomic fragment. The mir-279 hairpin was PCR cloned and inserted to the 5' end of the mir-996 downstream fragment, then the resultant 3.1kb piece was digested out and ligated with the 13.5kb mir-996 upstream sequence to generate the mir-279-2x construct. Similar procedures were followed to generate the mir-996-2x construct. Such mir-279-2x and mir-996-2x constructs carried a NotI site at the 5' side and an AscI site at the 3' side of the ectopic hairpin. Sequences of the primers used are listed in S1 Table. Transgenes were generated using the phiC31 system (BestGene Inc).
Other Drosophila mutants and transgenes
The mir-279[ex117] (also known as Δ1.2) and mir-279[ex36] (also known as Δ1.9) alleles were generated in the lab previously [18], but outcrossed stocks were obtained from Amita Sehgal [24] and used in this study. Other previously-described stocks utilized in this study include UAS-luc-mir-279 and UAS-DsRed-mir-996 [61], tim-Gal4 and tim-UAS-Gal4 [62], and the MARCM tester stock eyflp; Gr21-Gal4, UAS-sytGFP; FRT82, tubGal80 [18,63].
Small RNA northern blot
Total RNAs were extracted using Trizol LS (Life Technologies) following the manufacturer’s protocol. RNA samples were separated on 12% polyacrylamide denaturing gels (National Diagnostics), transferred to the GeneScreen Plus (Perkin Elmer) membrane, crosslinked with UV light and hybridized with γ-32P-labeled LNA (Exiqon) antisense probes for miR-279 and miR-996 at 45°C overnight. Signals were exposed to an Imaging Plate (Fujifilm) for 2~3 days for appropriate signal intensity. 2S RNA was hybridized with DNA probe and exposed for 30 min as the loading control. Signal quantifications were performed in the Image Gauge software and levels of miR-279 and miR-996 in different genotypes were normalized to 2S rRNA.
Lifespan assay
Late stage pupae of mir-279[ex117], mir-279[ex36] and mir-279/996[ex15C] mutants were transferred from culture bottles to clean petri dishes humidified with wet Kimwipe papers. This was necessary because of the overall poor vigor of the mutant stocks. For flies carrying rescue transgenes, no extra care was needed as all wildtype and modified constructs fully restored normal robustness of the mutant backgrounds. Adult flies of all genotypes were collected within 24 hours of eclosion into normal food vials and maintained in room temperature (22°C) in low density (10 flies per vial). Flies were transferred to fresh vials every day for mutants and every 3 to 5 days for rescued genotypes and scored for survivors across a time frame of 20 days. Again, the additional care was necessary to extend the lifespan of the mutants, which would normally succumb much more prematurely due to becoming trapped in the food.
Immunohistochemistry
Adult heads were fixed on ice for 2~3 hours in the fixative solution containing 4% paraformaldehyde (PFA) and 0.2% Triton-X-100 in PBS (0.2% PBST). Heads were then rinsed with 0.2% PBST for 3 times and brains were dissected in the blocking solution (5% normal goat serum in 0.2% PBST). Both primary and secondary antibodies were incubated overnight at room temperature. Brains were washed 3 times with 0.2% PBST before and after the secondary antibody incubation and mounted in the Vectashield mounting medium with DAPI (H-1200, Vector Labs). Antibodies were used as follows: mouse-anti-nc82 (1 : 20, Developmental Studies Hybridoma Bank), rabbit-anti-GFP (1 : 1000, Invitrogen), and Alexa Fluor-488, -568 secondary antibodies (1 : 500, Molecular Probes).
Circadian behavior assay
Flies were entrained in 12-hour light/12-hour dark (LD) cycles at 25°C for 5 days before transferred into constant darkness (DD) during the dark phase of the LD cycle. Locomotor activities of individual flies were recorded with the Drosophila Activity Monitor System (TriKinetics) every 1 min, data were then binned at 30 min with the DAMFileScan and the circadian period was calculated using ClockLab (Actimetrics) from data collected for 7 days in DD conditions. The power of rhythmicity was calculated as the chi-squared power above the significance line [64].
Luciferase assay
The 3' UTRs of Hairless and predicted miR-279 target genes were cloned between the XhoI and NotI sites of a modified psiCHECK-2 vector [65]. Sensor plasmid and Ub-Gal4 were cotransfected with UAS-dsRed-miRNA [61] or empty pUAST vector into S2-R+ cells with the Effectene (Qiagene) reagents. Luciferase levels were measured using the Dual-Glo Luciferase Assay System (Promega). Primer sequences for 3' UTR cloning are listed in S1 Table.
Gene expression analysis
We prepared cDNA from Trizol-extracted RNA that was treated with DNase and reverse transcribed using QuantiTect Reverse Transcription Kit (Qiagen). qPCR reactions were performed using SYBR select master mix (Life Technologies). Data were normalized to Rpl32 amplification. Primer sequences for qPCR amplicons are listed in S1 Table.
Supporting Information
Zdroje
1. Yang JS, Lai EC (2011) Alternative miRNA biogenesis pathways and the interpretation of core miRNA pathway mutants. Molecular cell 43 : 892–903. doi: 10.1016/j.molcel.2011.07.024 21925378
2. Kozomara A, Griffiths-Jones S (2011) miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic acids research 39: D152–157. doi: 10.1093/nar/gkq1027 21037258
3. Bartel DP (2009) MicroRNAs: target recognition and regulatory functions. Cell 136 : 215–233. doi: 10.1016/j.cell.2009.01.002 19167326
4. Sun K, Lai EC (2013) Adult-specific functions of animal microRNAs. Nature reviews Genetics 14 : 535–548. doi: 10.1038/nrg3471 23817310
5. Mendell JT, Olson EN (2012) MicroRNAs in stress signaling and human disease. Cell 148 : 1172–1187. doi: 10.1016/j.cell.2012.02.005 22424228
6. Lee RC, Feinbaum RL, Ambros V (1993) The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75 : 843–854. 8252621
7. Wightman B, Ha I, Ruvkun G (1993) Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 75 : 855–862. 8252622
8. Reinhart BJ, Slack F, Basson M, Pasquinelli A, Bettinger J, et al. (2000) The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403 : 901–906. 10706289
9. Lai EC (2002) microRNAs are complementary to 3' UTR sequence motifs that mediate negative post-transcriptional regulation. Nature genetics 30 : 363–364. 11896390
10. Lai EC, Burks C, Posakony JW (1998) The K box, a conserved 3' UTR sequence motif, negatively regulates accumulation of Enhancer of split Complex transcripts. Development 125 : 4077–4088. 9735368
11. Lai EC, Posakony JW (1997) The Bearded box, a novel 3' UTR sequence motif, mediates negative post-transcriptional regulation of Bearded and Enhancer of split Complex gene expression. Development 124 : 4847–4856. 9428421
12. Miska EA, Alvarez-Saavedra E, Abbott AL, Lau NC, Hellman AB, et al. (2007) Most Caenorhabditis elegans microRNAs Are Individually Not Essential for Development or Viability. PLoS genetics 3: e215. 18085825
13. Smibert P, Lai EC (2008) Lessons from microRNA mutants in worms, flies and mice. Cell cycle 7 : 2500–2508. 18719388
14. Baek D, Villen J, Shin C, Camargo FD, Gygi SP, et al. (2008) The impact of microRNAs on protein output. Nature 455 : 64–71. doi: 10.1038/nature07242 18668037
15. Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, et al. (2008) Widespread changes in protein synthesis induced by microRNAs. Nature 455 : 58–63. doi: 10.1038/nature07228 18668040
16. Ebert MS, Sharp PA (2012) Roles for microRNAs in conferring robustness to biological processes. Cell 149 : 515–524. doi: 10.1016/j.cell.2012.04.005 22541426
17. Lai EC, Tomancak P, Williams RW, Rubin GM (2003) Computational identification of Drosophila microRNA genes. Genome biology 4: R42.41–R42.20.
18. Cayirlioglu P, Kadow IG, Zhan X, Okamura K, Suh GS, et al. (2008) Hybrid neurons in a microRNA mutant are putative evolutionary intermediates in insect CO2 sensory systems. Science 319 : 1256–1260. doi: 10.1126/science.1149483 18309086
19. Chalfie M, Horvitz HR, Sulston JE (1981) Mutations that lead to reiterations in the cell lineages of C. elegans. Cell 24 : 59–69. 7237544
20. Johnston RJ, Hobert O (2003) A microRNA controlling left/right neuronal asymmetry in Caenorhabditis elegans. Nature 426 : 845–849. 14685240
21. Lewis MA, Quint E, Glazier AM, Fuchs H, De Angelis MH, et al. (2009) An ENU-induced mutation of miR-96 associated with progressive hearing loss in mice. Nature genetics 41 : 614–618. doi: 10.1038/ng.369 19363478
22. Mencia A, Modamio-Hoybjor S, Redshaw N, Morin M, Mayo-Merino F, et al. (2009) Mutations in the seed region of human miR-96 are responsible for nonsyndromic progressive hearing loss. Nature genetics 41 : 609–613. doi: 10.1038/ng.355 19363479
23. Hartl M, Loschek LF, Stephan D, Siju KP, Knappmeyer C, et al. (2011) A New Prospero and microRNA-279 Pathway Restricts CO2 Receptor Neuron Formation. The Journal of neuroscience: the official journal of the Society for Neuroscience 31 : 15660–15673. doi: 10.1523/JNEUROSCI.2592-11.2011 22049409
24. Luo W, Sehgal A (2012) Regulation of Circadian Behavioral Output via a MicroRNA-JAK/STAT Circuit. Cell 148 : 765–779. doi: 10.1016/j.cell.2011.12.024 22305007
25. Yoon WH, Meinhardt H, Montell DJ (2011) miRNA-mediated feedback inhibition of JAK/STAT morphogen signalling establishes a cell fate threshold. Nature cell biology 13 : 1062–1069. doi: 10.1038/ncb2316 21857668
26. Ruby JG, Stark A, Johnston WK, Kellis M, Bartel DP, et al. (2007) Evolution, biogenesis, expression, and target predictions of a substantially expanded set of Drosophila microRNAs. Genome research 17 : 1850–1864. 17989254
27. Stark A, Kheradpour P, Parts L, Brennecke J, Hodges E, et al. (2007) Systematic discovery and characterization of fly microRNAs using 12 Drosophila genomes. Genome research 17 : 1865–1879. 17989255
28. Mohammed J, Siepel A, Lai EC (2014) Diverse modes of evolutionary emergence and flux of conserved microRNA clusters. RNA in press.
29. Bushati N, Stark A, Brennecke J, Cohen SM (2008) Temporal Reciprocity of miRNAs and Their Targets during the Maternal-to-Zygotic Transition in Drosophila. Curr Biol 18 : 501–506. doi: 10.1016/j.cub.2008.02.081 18394895
30. Aboobaker AA, Tomancak P, Patel N, Rubin GM, Lai EC (2005) Drosophila microRNAs exhibit diverse spatial expression patterns during embryonic development. Proceedings of the National Academy of Sciences of the United States of America 102 : 18017–18022. 16330759
31. Wen J, Mohammed J, Bortolamiol-Becet D, Tsai H, Robine N, et al. (2014) Diversity of miRNAs, siRNAs and piRNAs across 25 Drosophila cell lines. Genome research 24 : 1236–1250. doi: 10.1101/gr.161554.113 24985917
32. Brown JB, Boley N, Eisman R, May G, Stoiber M, et al. (2014) Diversity and dynamics of the Drosophila transcriptome. Nature 512 : 393–399. 24670639
33. Graveley BR, Brooks AN, Carlson JW, Duff MO, Landolin JM, et al. (2011) The developmental transcriptome of Drosophila melanogaster. Nature 471 : 473–479. doi: 10.1038/nature09715 21179090
34. Manak JR, Dike S, Sementchenko V, Kapranov P, Biemar F, et al. (2006) Biological function of unannotated transcription during the early development of Drosophila melanogaster. Nature genetics 38 : 1151–1158. 16951679
35. Linsley PS, Schelter J, Burchard J, Kibukawa M, Martin MM, et al. (2007) Transcripts targeted by the microRNA-16 family cooperatively regulate cell cycle progression. Molecular and cellular biology 27 : 2240–2252. 17242205
36. Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, et al. (2005) Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature 433 : 769–773. 15685193
37. Lai EC (2015) Two decades of miRNA biology: lessons and challenges. RNA 21 : 675–677. doi: 10.1261/rna.051193.115 25780186
38. Chen YW, Song S, Weng R, Verma P, Kugler JM, et al. (2014) Systematic Study of Drosophila MicroRNA Functions Using a Collection of Targeted Knockout Mutations. Developmental cell 31 : 784–800. doi: 10.1016/j.devcel.2014.11.029 25535920
39. Alvarez-Saavedra E, Horvitz HR (2010) Many families of C. elegans microRNAs are not essential for development or viability. Curr Biol 20 : 367–373. doi: 10.1016/j.cub.2009.12.051 20096582
40. Abbott AL, Alvarez-Saavedra E, Miska EA, Lau NC, Bartel DP, et al. (2005) The let-7 MicroRNA family members mir-48, mir-84, and mir-241 function together to regulate developmental timing in Caenorhabditis elegans. Developmental cell 9 : 403–414. 16139228
41. Brenner JL, Jasiewicz KL, Fahley AF, Kemp BJ, Abbott AL (2010) Loss of individual microRNAs causes mutant phenotypes in sensitized genetic backgrounds in C. elegans. Curr Biol 20 : 1321–1325. doi: 10.1016/j.cub.2010.05.062 20579881
42. Smibert P, Lai EC (2010) A view from Drosophila: multiple biological functions for individual microRNAs. Seminars in cell & developmental biology 21 : 745–753.
43. Small EM, Olson EN (2011) Pervasive roles of microRNAs in cardiovascular biology. Nature 469 : 336–342. doi: 10.1038/nature09783 21248840
44. Li X, Cassidy JJ, Reinke CA, Fischboeck S, Carthew RW (2009) A microRNA imparts robustness against environmental fluctuation during development. Cell 137 : 273–282. doi: 10.1016/j.cell.2009.01.058 19379693
45. Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T (2001) Identification of novel genes coding for small expressed RNAs. Science 294 : 853–858. 11679670
46. Doench JG, Sharp PA (2004) Specificity of microRNA target selection in translational repression. Genes & development 18 : 504–511.
47. Brennecke J, Stark A, Russell RB, Cohen SM (2005) Principles of MicroRNA-Target Recognition. PLoS biology 3: e85. 15723116
48. Lai EC, Tam B, Rubin GM (2005) Pervasive regulation of Drosophila Notch target genes by GY-box-, Brd-box-, and K-box-class microRNAs. Genes & development 19 : 1067–1080.
49. Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB (2003) Prediction of mammalian microRNA targets. Cell 115 : 787–798. 14697198
50. Stark A, Brennecke J, Russell RB, Cohen SM (2003) Identification of Drosophila MicroRNA Targets. PLoS biology 1: E60. 14691535
51. Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, et al. (2005) Combinatorial microRNA target predictions. Nature genetics 37 : 495–500. 15806104
52. Vella MC, Choi EY, Lin SY, Reinert K, Slack FJ (2004) The C. elegans microRNA let-7 binds to imperfect let-7 complementary sites from the lin-41 3'UTR. Genes & development 18 : 132–137.
53. Lal A, Navarro F, Maher CA, Maliszewski LE, Yan N, et al. (2009) miR-24 Inhibits cell proliferation by targeting E2F2, MYC, and other cell-cycle genes via binding to "seedless" 3'UTR microRNA recognition elements. Molecular cell 35 : 610–625. doi: 10.1016/j.molcel.2009.08.020 19748357
54. Shin C, Nam JW, Farh KK, Chiang HR, Shkumatava A, et al. (2010) Expanding the MicroRNA Targeting Code: Functional Sites with Centered Pairing. Molecular cell 38 : 789–802. doi: 10.1016/j.molcel.2010.06.005 20620952
55. Sarin S, O'Meara MM, Flowers EB, Antonio C, Poole RJ, et al. (2007) Genetic screens for Caenorhabditis elegans mutants defective in left/right asymmetric neuronal fate specification. Genetics 176 : 2109–2130. 17717195
56. Nairz K, Rottig C, Rintelen F, Zdobnov E, Moser M, et al. (2006) Overgrowth caused by misexpression of a microRNA with dispensable wild-type function. Developmental biology 291 : 314–324. 16443211
57. Bassett AR, Azzam G, Wheatley L, Tibbit C, Rajakumar T, et al. (2014) Understanding functional miRNA-target interactions in vivo by site-specific genome engineering. Nature communications 5 : 4640. doi: 10.1038/ncomms5640 25135198
58. Ecsedi M, Rausch M, Grosshans H (2015) The let-7 microRNA Directs Vulval Development through a Single Target. Developmental cell 32 : 335–344. doi: 10.1016/j.devcel.2014.12.018 25669883
59. Pfeiffer BD, Jenett A, Hammonds AS, Ngo TT, Misra S, et al. (2008) Tools for neuroanatomy and neurogenetics in Drosophila. Proceedings of the National Academy of Sciences of the United States of America 105 : 9715–9720. doi: 10.1073/pnas.0803697105 18621688
60. Venken KJ, He Y, Hoskins RA, Bellen HJ (2006) P[acman]: a BAC transgenic platform for targeted insertion of large DNA fragments in D. melanogaster. Science 314 : 1747–1751. 17138868
61. Bejarano F, Bortolamiol-Becet D, Dai Q, Sun K, Saj A, et al. (2012) A genome-wide transgenic resource for conditional expression of Drosophila microRNAs. Development 139 : 2821–2831. doi: 10.1242/dev.079939 22745315
62. Blau J, Young MW (1999) Cycling vrille expression is required for a functional Drosophila clock. Cell 99 : 661–671. 10612401
63. Jones WD, Cayirlioglu P, Kadow IG, Vosshall LB (2007) Two chemosensory receptors together mediate carbon dioxide detection in Drosophila. Nature 445 : 86–90. 17167414
64. Pfeiffenberger C, Lear BC, Keegan KP, Allada R (2010) Processing circadian data collected from the Drosophila Activity Monitoring (DAM) System. Cold Spring Harbor protocols 2010: pdb prot5519. doi: 10.1101/pdb.prot5519 21041392
65. Okamura K, Hagen JW, Duan H, Tyler DM, Lai EC (2007) The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell 130 : 89–100. 17599402
Štítky
Genetika Reprodukční medicína
Článek Germline Mutations Confer Susceptibility to Acute Lymphoblastic Leukemia and ThrombocytopeniaČlánek Temporal Expression Profiling Identifies Pathways Mediating Effect of Causal Variant on PhenotypeČlánek Simultaneous DNA and RNA Mapping of Somatic Mitochondrial Mutations across Diverse Human CancersČlánek A Legume Genetic Framework Controls Infection of Nodules by Symbiotic and Endophytic BacteriaČlánek The Eukaryotic-Like Ser/Thr Kinase PrkC Regulates the Essential WalRK Two-Component System inČlánek The Yeast GSK-3 Homologue Mck1 Is a Key Controller of Quiescence Entry and Chronological LifespanČlánek The Role of -Mediated Epigenetic Silencing in the Population Dynamics of Transposable Elements in
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2015 Číslo 6
-
Všechny články tohoto čísla
- Expression of Concern: RNAi-Dependent and Independent Control of LINE1 Accumulation and Mobility in Mouse Embryonic Stem Cells
- Orphan Genes Find a Home: Interspecific Competition and Gene Network Evolution
- Minor Cause—Major Effect: A Novel Mode of Control of Bistable Gene Expression
- Germline Mutations Confer Susceptibility to Acute Lymphoblastic Leukemia and Thrombocytopenia
- Leveraging Identity-by-Descent for Accurate Genotype Inference in Family Sequencing Data
- Is Required for the Expression of Principal Recognition Molecules That Control Axon Targeting in the Retina
- Epigenetic Aging Signatures Are Coherently Modified in Cancer
- Silencing of DNase Colicin E8 Gene Expression by a Complex Nucleoprotein Assembly Ensures Timely Colicin Induction
- A Transposable Element within the Non-canonical Telomerase RNA of Modulates Telomerase in Response to DNA Damage
- The Orphan Gene Regulates Dauer Development and Intraspecific Competition in Nematodes by Copy Number Variation
- 9--13,14-Dihydroretinoic Acid Is an Endogenous Retinoid Acting as RXR Ligand in Mice
- The DnaA Protein Is Not the Limiting Factor for Initiation of Replication in
- FGFR3 Deficiency Causes Multiple Chondroma-like Lesions by Upregulating Hedgehog Signaling
- Multiple Changes of Gene Expression and Function Reveal Genomic and Phenotypic Complexity in SLE-like Disease
- Directed Evolution of RecA Variants with Enhanced Capacity for Conjugational Recombination
- The Regulatory T Cell Lineage Factor Foxp3 Regulates Gene Expression through Several Distinct Mechanisms Mostly Independent of Direct DNA Binding
- MreB-Dependent Inhibition of Cell Elongation during the Escape from Competence in
- DNA Damage Regulates Translation through β-TRCP Targeting of CReP
- Multiple In Vivo Biological Processes Are Mediated by Functionally Redundant Activities of and
- The Analysis of () Mutants Reveals Differences in the Fusigenic Potential among Telomeres
- The Causative Gene in Chanarian Dorfman Syndrome Regulates Lipid Droplet Homeostasis in .
- Temporal Expression Profiling Identifies Pathways Mediating Effect of Causal Variant on Phenotype
- The . Accessory Helicase PcrA Facilitates DNA Replication through Transcription Units
- AKTIP/Ft1, a New Shelterin-Interacting Factor Required for Telomere Maintenance
- Npvf: Hypothalamic Biomarker of Ambient Temperature Independent of Nutritional Status
- Transfer RNAs Mediate the Rapid Adaptation of to Oxidative Stress
- Connecting Circadian Genes to Neurodegenerative Pathways in Fruit Flies
- Response to “Ribosome Rescue and Translation Termination at Non-standard Stop Codons by ICT1 in Mammalian Mitochondria”
- Response to the Formal Letter of Z. Chrzanowska-Lightowlers and R. N. Lightowlers Regarding Our Article “Ribosome Rescue and Translation Termination at Non-Standard Stop Codons by ICT1 in Mammalian Mitochondria”
- Simultaneous DNA and RNA Mapping of Somatic Mitochondrial Mutations across Diverse Human Cancers
- Regulation of Insulin Receptor Trafficking by Bardet Biedl Syndrome Proteins
- Altered Levels of Mitochondrial DNA Are Associated with Female Age, Aneuploidy, and Provide an Independent Measure of Embryonic Implantation Potential
- Non-reciprocal Interspecies Hybridization Barriers in the Capsella Genus Are Established in the Endosperm
- Canine Spontaneous Head and Neck Squamous Cell Carcinomas Represent Their Human Counterparts at the Molecular Level
- Genetic Changes to a Transcriptional Silencer Element Confers Phenotypic Diversity within and between Species
- Functional Assessment of Disease-Associated Regulatory Variants Using a Versatile Dual Colour Transgenesis Strategy in Zebrafish
- Translational Upregulation of an Individual p21 Transcript Variant by GCN2 Regulates Cell Proliferation and Survival under Nutrient Stress
- Independent Neuronal Origin of Seizures and Behavioral Comorbidities in an Animal Model of a Severe Childhood Genetic Epileptic Encephalopathy
- The Human Blood Metabolome-Transcriptome Interface
- A Common Cancer Risk-Associated Allele in the Locus Encodes a Dominant Negative Inhibitor of Telomerase
- A Legume Genetic Framework Controls Infection of Nodules by Symbiotic and Endophytic Bacteria
- The Eukaryotic-Like Ser/Thr Kinase PrkC Regulates the Essential WalRK Two-Component System in
- The Yeast GSK-3 Homologue Mck1 Is a Key Controller of Quiescence Entry and Chronological Lifespan
- Dissection of a Complex Disease Susceptibility Region Using a Bayesian Stochastic Search Approach to Fine Mapping
- Exome Sequencing of Phenotypic Extremes Identifies and as Interacting Modifiers of Chronic Infection in Cystic Fibrosis
- The Role of -Mediated Epigenetic Silencing in the Population Dynamics of Transposable Elements in
- Ancestral Chromatin Configuration Constrains Chromatin Evolution on Differentiating Sex Chromosomes in
- Abnormal Activation of BMP Signaling Causes Myopathy in Null Mice
- Reproductive Mode and the Evolution of Genome Size and Structure in Nematodes
- Replication and Active Partition of Integrative and Conjugative Elements (ICEs) of the SXT/R391 Family: The Line between ICEs and Conjugative Plasmids Is Getting Thinner
- Motor and Sensory Deficits in the Mice Result from Mutation of the ESCRT Component HGS
- Senescence in the Sbds-Deficient Murine Pancreas: Cell-Type Specific Consequences of Translation Insufficiency
- Lipophorin Receptors Recruit the Lipoprotein LTP to the Plasma Membrane to Mediate Lipid Uptake
- Separable Crossover-Promoting and Crossover-Constraining Aspects of Zip1 Activity during Budding Yeast Meiosis
- Context-Dependent Functional Divergence of the Notch Ligands DLL1 and DLL4
- Multilayered Organization of Jasmonate Signalling in the Regulation of Root Growth
- Necrotic Cells Actively Attract Phagocytes through the Collaborative Action of Two Distinct PS-Exposure Mechanisms
- A Novel Feedback Loop That Controls Bimodal Expression of Genetic Competence
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Non-reciprocal Interspecies Hybridization Barriers in the Capsella Genus Are Established in the Endosperm
- Translational Upregulation of an Individual p21 Transcript Variant by GCN2 Regulates Cell Proliferation and Survival under Nutrient Stress
- Exome Sequencing of Phenotypic Extremes Identifies and as Interacting Modifiers of Chronic Infection in Cystic Fibrosis
- The Human Blood Metabolome-Transcriptome Interface
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



