-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaPathways of Distinction Analysis: A New Technique for Multi–SNP Analysis of GWAS Data
Genome-wide association studies (GWAS) have become increasingly common due to advances in technology and have permitted the identification of differences in single nucleotide polymorphism (SNP) alleles that are associated with diseases. However, while typical GWAS analysis techniques treat markers individually, complex diseases (cancers, diabetes, and Alzheimers, amongst others) are unlikely to have a single causative gene. Thus, there is a pressing need for multi–SNP analysis methods that can reveal system-level differences in cases and controls. Here, we present a novel multi–SNP GWAS analysis method called Pathways of Distinction Analysis (PoDA). The method uses GWAS data and known pathway–gene and gene–SNP associations to identify pathways that permit, ideally, the distinction of cases from controls. The technique is based upon the hypothesis that, if a pathway is related to disease risk, cases will appear more similar to other cases than to controls (or vice versa) for the SNPs associated with that pathway. By systematically applying the method to all pathways of potential interest, we can identify those for which the hypothesis holds true, i.e., pathways containing SNPs for which the samples exhibit greater within-class similarity than across classes. Importantly, PoDA improves on existing single–SNP and SNP–set enrichment analyses, in that it does not require the SNPs in a pathway to exhibit independent main effects. This permits PoDA to reveal pathways in which epistatic interactions drive risk. In this paper, we detail the PoDA method and apply it to two GWAS: one of breast cancer and the other of liver cancer. The results obtained strongly suggest that there exist pathway-wide genomic differences that contribute to disease susceptibility. PoDA thus provides an analytical tool that is complementary to existing techniques and has the power to enrich our understanding of disease genomics at the systems-level.
Published in the journal: . PLoS Genet 7(6): e32767. doi:10.1371/journal.pgen.1002101
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1002101Summary
Genome-wide association studies (GWAS) have become increasingly common due to advances in technology and have permitted the identification of differences in single nucleotide polymorphism (SNP) alleles that are associated with diseases. However, while typical GWAS analysis techniques treat markers individually, complex diseases (cancers, diabetes, and Alzheimers, amongst others) are unlikely to have a single causative gene. Thus, there is a pressing need for multi–SNP analysis methods that can reveal system-level differences in cases and controls. Here, we present a novel multi–SNP GWAS analysis method called Pathways of Distinction Analysis (PoDA). The method uses GWAS data and known pathway–gene and gene–SNP associations to identify pathways that permit, ideally, the distinction of cases from controls. The technique is based upon the hypothesis that, if a pathway is related to disease risk, cases will appear more similar to other cases than to controls (or vice versa) for the SNPs associated with that pathway. By systematically applying the method to all pathways of potential interest, we can identify those for which the hypothesis holds true, i.e., pathways containing SNPs for which the samples exhibit greater within-class similarity than across classes. Importantly, PoDA improves on existing single–SNP and SNP–set enrichment analyses, in that it does not require the SNPs in a pathway to exhibit independent main effects. This permits PoDA to reveal pathways in which epistatic interactions drive risk. In this paper, we detail the PoDA method and apply it to two GWAS: one of breast cancer and the other of liver cancer. The results obtained strongly suggest that there exist pathway-wide genomic differences that contribute to disease susceptibility. PoDA thus provides an analytical tool that is complementary to existing techniques and has the power to enrich our understanding of disease genomics at the systems-level.
Introduction
Genome-wide association studies (GWAS) have become a powerful and increasingly affordable tool to study the genetic variants associated with disease. Modern GWAS yield information on millions of single nucleotide polymorphism (SNPs) loci distributed across the human genome, and have already yielded insights into the genetic basis of complex diseases [1], [2], including diabetes, inflammatory bowel disease, and several cancers [3]–[7]; a complete list of published GWAS can be found at the National Cancer Institute–National Human Genome Research Institute (NCI-NHGRI) catalog of published genome-wide association studies [8].
Typically, the data produced in GWAS are analyzed by considering each SNP independently, testing the alleles at each locus for association with case status; significant association is indicative of a nearby genetic variation which may play a role in disease susceptibility. Genomic regions of interest may also be subject to haplotype analysis, in which a handful of alleles transmitted together on the same chromosome are tested for association with disease; in this case, the loci which are jointly considered are located within a small genomic region, often confined to the neighborhood of a single gene.
Recently, however, there has been increasing interest in multilocus, systems-based analyses. This interest is motivated by a variety of factors. First, few loci identified in GWAS have large effect sizes (the problem of “missing heritability”) and it is likely that the common–disease, common–variant hypothesis [9], [10] does not hold in the case of complex diseases. Second, single marker associations identified in GWAS often fail to replicate. This phenomenon has been attributed to underlying epistasis [11], and a similar problem in gene expression profiling has been mitigated through the use of gene-set statistics. Most importantly, it is now well understood that because biological systems are driven by complex biomolecular interactions, multi-gene effects will play an important role in mapping genotypes to phenotypes; recent reviews by Moore and coworkers describe this issue well [10], [12]. Additionally, the finding that epistasis and pleiotropy appear to be inherent properties of biomolecular networks [13] rather than isolated occurences motivates the need for systems-level understanding of human genetics.
The impact that biological interaction networks have on our ability to identify genomic causes of complex disease is readily apparent. Consider a biologically crucial mechanism with several potential points of failure, such that an alteration to any will confer disease risk. Because no single alteration is predominant amongst cases, none attain a significant association; indeed, it has long been observed that even in histologically identical tumors, only a fraction may share the same set of mutations (see references in [14] for examples). Additionally, in a robust system, multiple alterations may be necessary to alter the activity of an interaction network; here, healthy individuals may share a subset of the deleterious alleles found in cases, and again these loci will not be detected. This complexity, noted by [10], [12]–[14] and others, has generated considerable interest in multi-locus analysis techniques that take advantage of known interaction information.
Several multi-SNP GWAS analysis approaches have been described in the literature. Thorough reviews are provided in [15], [16], and we briefly describe several here. Building on the well-established Gene Set Enrichment Analysis [17] method initially developed for gene expression data, two articles have proposed an extension of GSEA for SNP data [18], [19]. In these techniques, each SNP is assigned a statistic based on a test of association with the phenotype; a running sum is then used to assess whether large statistics occur more frequently amongst a SNP set of interest than could be expected by chance. While GSEA-type approaches have proven quite useful, their reliance on single-marker statistics means that systematic yet subtle changes in a gene set will be missed if the individual genes do not have a strong marginal association. In the case of a purely epistatic interaction between two SNPs in a set, the set may fail to reach significance altogether.
To address this issue, Yang and colleagues proposed SNPHarvester [20], designed to detect multi-SNP associations even when the marginal effects are weak. To reduce the search space of possible multi-SNP effects, SNPHarvester [20] first removes any SNPs with univarite significance. Using a novel searching algorithm, they identify groups of SNPs that show association with status in a test with degrees of freedom. While this approach can reveal epistatic effects that would be missed by the GSEA-type schemes [18], [19], it has other drawbacks. First, the combinatorial explosion of SNP groups puts a limit on the number of SNPs that may simultaneously be examined. Second, the the arbitrary groupings of SNPs and the exclusion of SNPs with marginal effects can make the biological interpretation of the analysis results difficult.
The notion that cases will more closely resemble other cases than they will controls has motivated a number of interesting distance-based approaches for detecting epistasis. Multi-dimensionality reduction (MDR) has been proposed and applied to SNP data [21]–[23]. In this technique, sets of SNPs are exhaustively searched for combinations that will best partition the samples by examining the cells in that space (corresponding to homozygous minor, heterozygous, or homozygous major alleles for each locus) for overrepresentation of cases. While this method both finds epistatic interactions without requiring marginal effects and can be structured to incorporate expert knowledge, it is limited by the fact the the total number of loci to be combinatorially explored must be restricted to limit computational cost. To address this, an “interleaving” approach in which models are constructed hierarchically has been suggested [22] to reduce the combinatorial search space. A recent and powerful MDR implementation [24] taking advantange of the CUDA parallel computing architecture for graphics processors has made feasible a genome-wide analysis of pairwise SNP interactions. Still, MDR remains computationally challenging, such that expanding the search to other SNP set sizes (rather than restricting to pairwise interactions) can be impeded by combinatorial complexity if an exhaustive search is to be performed.
In order to narrow down the combinatorial complexity of discovering SNP sets using techniques such as MDR, feature selection may be employed. Of particular importance here is the distance-based approach of the Relief family of algorithms [25]–[28]. These are designed to identify features of interest by weighting each feature through a nearest-neighbor approach. The weights are constructed in the following way: for each attribute, one selects samples at random and asks whether the nearest neighbor (across all attributes) from the same class and the nearest neighbor from the other class have the same or different values from the randomly chosen sample. Attributes for which in-class nearest neighbors tend to have the same value are weighted more strongly. Because the distances are computed across all attributes, Relief-type algorithms can identify SNPs that form part of an epistatic group and they provide a means of filtration that does not have the drawbacks of other significance filters.
While these methods have so far been applied to finding small groups of interacting SNPs, one may instead be interested in whether cases and controls exhibit differential distance when considering a large number of genes. A multi-SNP statistic has been proposed in the literature [29]–[31] for determining whether an individual of interest is on average (across a large number of SNPs) “closer” to one population sample than to another. The method, originally proposed by Homer [29], is motivated by the idea that a subtle but systematic variation across a large number of SNPs can produce a discernible difference in the closeness of an individual to one population sample relative to another. While this statistic was originally designed to identify the proband as a member of one of the population samples, it was shown in [30] that out-of-pool cases from a case-control breast cancer study were in general closer (as defined by the statistic presented in [29]) to in-pool cases than they were to in-pool controls, suggesting that the combination of multiple alleles has the potential to distinguish cases from controls.
Building on these ideas, we present a new technique that finds pathway-based SNP-sets that differentiate cases from controls; we call this method Pathways of Distinction Analysis (PoDA). In PoDA, SNP sets are defined based on known relationships (e.g., SNPs in genes sharing a common pathway), and thus incorporate expert knowledge to reduce the search space and provide biological interpretability. Motivated by the differential “closeness” of cases and controls as discussed about and as observed in [30], we hypothesize that if the SNPs come from a pathway that plays a role in disease, there will be greater in-class similarity than across-class similarity in the genotypes for those SNPs; i.e., a case will show greater genetic similarity to other cases than to controls for the SNPs on a disease-related pathway, but will be equidistant for the SNPs on a non-disease-related pathway. Based on this notion, PoDA seeks to identify pathways for which differential heterogeneity is exhibited in cases and controls. In each pathway, PoDA returns a statistic for each sample that quantifies that sample's distance to the remaining cases relative to its distance to the remaining controls for a given pathway's SNPs. PoDA then examines whether the distributions of for the controls differ from those of the cases by computing and testing for significance a Pathway Distinction Score that quantifies the differences in case and control distributions. In this manuscript, we detail the PoDA method and report the results of its application to two data sets.
As we will describe, PoDA improves and complements existing approaches in a number of respects. First, it permits the investigation of arbitrarily large pathways, circumventing the dimensionality issues that are encountered with MDR and SNP-Harvester. Second, it is able to detect pathways that contain an over-abundance of highly-significant markers as well as pathways whose markers have a small but consistent association that would be missed by GSEA-type approaches. Finally, it uses a leave-one-out technique to return for each sample an unsupervised relative distance statistic that can then be used to model disease risk via logistic regression. In addition to providing an effect size for the pathway, this allows the odds of disease for new samples to be obtained by computing its relative distance statistic with respect to the known samples and applying the model.
Methods
Following our conjecture that SNPs associated with the genes in a pathway involved in disease will exhibit more within-group similarity than across-group similarity, we propose Pathways of Distinction Analysis (PoDA), a method designed to address the following questions:
-
Given some set of SNPs, do we find that, on average, cases are “closer” to other cases than to controls (or that controls are “closer” to other controls than to cases)?
-
If we look for these distinctions systematically over all SNP-sets of potential interest, can we use it to single out SNP-sets which may be associated with disease?
In PoDA, a set of SNPs are selected, and for each sample we compute whether it is closer to the pool of remaining cases or controls across that SNP set, using the relative distance statistic described below. Once this is done for every sample, the distribution of the relative distance statistic is compared in the cases and controls using a nonparametric statistic, addressing the first question above. This may be carried out amongst all SNP sets of interest, adjusting the -value for the multiple hypotheses, to find SNP sets for which cases more strongly resemble the population of remaining cases while controls more strongly resemble the population of remaining controls.
We begin with a discussion of how we measure the relative distance of an individual to the other cases vs. other controls. A simple but computationally intensive approach is to represent each sample by a vector in an -dimensional space, where is the number of SNPs in the group of interest. One can then compute, between each sample pair, their distance in this -dimensional space using a Euclidean, Manhattan, or Hamming metric. For each sample, we would define its relative distance statistic as the mean of its distance to other controls minus the mean of its distance to other cases; a sample that is more similar to cases will exhibit a positive statistic, whereas one that is more similar to other controls will exhibit a negative statistic. For the given SNP set, we would then have for each sample a value quantifying its relative distance that was computed without knowledge of that sample's class (i.e., using a leave-one-out scheme) and could then be used in further tests. By doing this for all pathways of interest, one derives a relative distance value for each sample in each pathway.
This brute-force approach, while conceptually clear, has two significant drawbacks. The first is that the distance computation is where is the total number of samples in the study–a considerable undertaking, particularly if many SNP sets are to be analyzed, and one that becomes exceedingly troublesome in the context of permutation tests. The second drawback is that because we are taking the mean of the distances, a sample that is situated squarely within a cluster of cases may have a large case-distance value due to the dispersion of cases around it. Both of these issues are circumvented by instead considering the relative distance to the centroids of the cases and controls in the -dimensional space, a computation that can be performed in for all samples. It is this approach that PoDA employs, as follows:
In [29], [30], the authors consider a measure of individual 's distance to two population samples and at locus ,(1)where and are the minor allele frequencies (MAFs) of SNP in samples and , and is 's genotype at corresponding to homozygous major, heterozygous, and homozygous minor alleles, respectively (i.e., the frequency of minor allele in that individual. The first term quantifies how different 's MAF is from 's for a given locus ; the second term quantifies how different 's MAF is from 's at locus ; and so gives the distance of relative to and at locus . Since the minor allele frequencies and are computed by averaging the genotypes (again, written as ) in samples and respectively, it is clear that is the distance from to the centroid of along the coordinate (and likewise for the term). It can be seen from Eq. 1 that positive implies that is closer to than to , and that negative implies that is closer to than to .
By computing at each locus and taking the standardized mean across the loci, [29] obtain a distance score which quantifies how close is relative to and across all loci under consideration,(2)where denotes the mean of across all loci . That is, provides a means to quantify whether 's MAFs are closer to 's MAFs or 's MAFs on average for the loci under consideration. It is instructive to consider the geometrical interpretation of Eq. 2. Is clear upon inspection that the numerator in Eq. 2 is equal, up to a factor of , to the difference in Manhattan distances between and the (nonstandardized) centroid and and the (nonstandardized) centroid; in this sense, Eq. 2 resembles a nearest-centroid classifier. However, the denominator scales the relative distances by their variance across the SNPs; that is, a sample who is consistently closer to than to for each of the SNPs will obtain a higher than an individual who is variously closer to either across the SNPs under consideration.
By assigning the (non-) controls as and the (non-) cases as , we can compute a statistic quantifying 's distance to other cases relative to 's distance to other controls. If we then apply this systematically to all individuals in the study population (removing that individual, computing the MAF's and for the remaining individuals who comprise and , and then computing in a leave-one-out manner), we can obtain distributions of statistics in cases and controls that may be compared. Here, the null hypothesis is that case and control distributions do not differ, with the alternative hypothesis that the cases have higher values than do controls, i.e., that they are closer (via Eqs. 1–2) to other cases than are controls.
We can use in the following manner to answer the questions posed above by applying it in a leave-one-out manner in each pathway:
-
For a given pathway , select the SNPs associated with that pathway;
-
For every sample , remove from the case or control group as needed, and compute with respect to the remaining cases and controls using the SNPs chosen in step 1.
-
Quantify the differences in distribution of 's for the case samples versus that of the controls and test for significance.
By systematically carrying out the above steps on all pathways of interest, we can identify pathways for which there appears to be differential homogeneity in cases and controls, indicating that the pathway may play a disease-related role. The details of the algorithm are explained below, and summarized in Table 1.
Tab. 1. Procedure for Pathways of Distinction Analysis. 
In [30], we examined Eqs. 1–2 and found that the magnitude of is influenced both by the MAF differences (that is, how distant the centroids of and are) and by correlations between the SNPs under consideration (due to the penalization for variance in provided by the denominator of Eq. 2. These properties are extremely well-suited to the application we propose: pathways with few highly-significant SNPs will yield large differences (due to the influence of ), as will pathways with SNPs that are highly correlated yet have subtle individual MAF differences, corresponding to the concerted action of multiple SNPs.
At the same time, however, we wish to ensure that the pathways we select as having differential are not being influenced large LD blocks covered by the SNPs in the genes on the pathway. That is, we wish to ensure that the SNP correlations which drive are reflective of epistatic effects between different genes rather than recombination events within a gene. To this end, we select a single SNP to represent each gene, based on the desire to detect multi-gene rather than multi-SNP effects.
In practice, SNPs are selected as follows: for each pathway represented in the Pathway Interaction Database [32] (PID, http://www.pid.nci.gov, containing annotations from BioCarta, Reactome, and the NCI/Nature database [32]) and KEGG [33], we select the associated genes. Using dbSNP [34], we retrieve the SNPs associated with the pathway genes that are present in the data, excluding those with missing data or with minor allele frequency in either case of control group. We necessarily exclude pathways for which only one gene is probed by the remaining SNPs. Because we are interested in values that are driven by correlations across genes (and not in individual genes covered by many SNPs with high LD), we select for each gene its most significant SNP in a univariate test of association (Fisher's exact test). It should be noted here that while the SNP chosen for each gene is the most significant of that gene's SNPs, it is not necessarily significantly associated with disease. Our goal here is not to filter based on SNP significance, but rather to select, for each gene, a single marker that is as informative as possible.
Having selected the SNPs of interest, we compute at each locus for every sample by selectively removing it and comparing it to the remaining cases and controls, as described above. For each pathway , we compute for the SNPs that comprise it, yielding a distribution of for cases and another distribution for controls. The difference in the location of the case and control distributions is then quantified nonparametrically by computing the Wilcoxon rank sum statistic, defined as(3)where is the rank of amongst all samples for a given pathway . Eq. 3 thus quantifies, non-parametrically, the degree to which cases are “closer” to other cases and controls “closer” to other controls across a set of SNPs for all individuals in the GWAS.
To illustrate the above, we consider a simulated GWAS of 250 cases and 250 controls and 50 SNPs, shown in Figure 1, and ask whether we are able to detect a 12-SNP pathway in which a subset of SNPs appear to have an epistatic interaction. Alleles were simulated as binomial samples from a source population with MAFs ranging from 0.1 to 0.4 across the 50 SNPs, and case labels were assigned such that a combintion of homozygous minor alleles at SNPs 1 and 2 or 3 (i.e., ) conferred a three-fold relative risk, mimicking an epistatic interaction between SNPs 1 and 2 and SNPs 1 and 3 (Figure 1a). Alone, none of the 50 SNPs showed any association with case status, nor was any SNP significantly out of HWE in either cases or controls. However, the “shared pattern” in SNPs 1–3 is such that a 12 SNP pathway comprising SNPs 1–12 yields greater in cases than in controls as can been seen in Figure 1b, while a random 12 SNP pathway selected from the 50 SNPs (that includes SNP 3, but neither SNP 1 or 2) shows no difference in values as seen in Figure 1c.
Fig. 1. PoDA applied to simulated data. 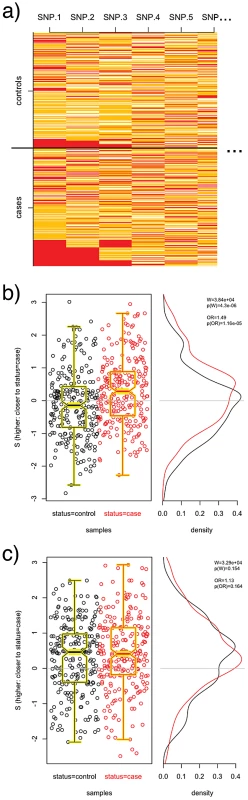
Alleles at 50 loci for 250 cases and 250 controls were simulated such that each SNP was in HWE and not associated with case status, but homozygous minor (red) at both loci 1 and 2 or 1 and 3 yielded a three-fold relative risk (a). A 12-SNP pathway comprising SNPs 1–12 shows differential distributions (b); a random 12-SNP pathway does not (c). Boxplots are overlayed on the scatterplots of for clarity. While the Wilcoxon statistic is normal in the large-sample limit and can be directly compared to a Gaussian, to truly evaluate the significance of for a given pathway , we must address two sources of bias: the number of SNPs per gene, and the size of the pathway. To address these issues, we introduce a normalized Pathway Distinction Score that we test for significance using a resampling procedure.
First, we expect that because we have selected for each gene the single most informative SNP, we are pre-disposed to seeing higher for pathways that contain large genes. Because large genes will be more likely to contain highly-significant SNPs by chance, the concern has been raised that [18], [35] selecting the single most significant SNP as a proxy for the gene (as is done here) will lead to a bias toward pathways that contain an abundance of large genes. To account for this, we follow the approach in [18] and normalize the score via a permutation-based procedure. First, we permute the phenotype labels and in each permutation recalculate as described above, but using the permuted case and control labels. The permuted labels are used both to select the most informative SNP per gene and to compute , , and in Eqns. 1–3). This yields a distribution of under the null hypothesis that the magnitude of is independent of the true case/control classifications. We then normalize the true by the distribution from the permutation procedure, yielding a Distinction Score for pathway that effectively adjusts for different sizes of genes and preserves correlations of SNPs in the same gene:(4)where are the set of obtained for pathway across the permutations. (In practice, 100 permutations are used.) Because the permuted labels are used in the SNP selection, this normalization adjusts for the bias introduced by the fact that large genes have more opportunity to contain significant SNPs by chance. The Pathway Distinction Score thus provides a model-free, gene-size adjusted metric for quantifying the degree to which cases are “closer” to other cases (higher ) than controls.
To test whether is significant, we note that larger pathways may yield high values simply due to the fact that they sample the case anc control differences more thoroughly. Indeed, the question of significance that we wish to address is not simply whether a pathway permits the distinction of cases and controls, but whether it does so better than a random collection of as many SNPs, wherein the SNPs are still selected to be the most informative by gene. To account for the fact that the pathways are of differing sizes, significance of the Distinction Score for a given pathway is assessed through resampling by choosing, at random, the same number of SNPs that are present in that pathway () from the total set of most-informative-SNP-per-gene and recomputing for the random pathway. The value is obtained by counting the fraction of random -SNP sets which give a larger than the true pathway SNPs in resamplings. In this way, we are able to detect pathways that yield large differences of case and control distributions due to their particular SNPs, rather than simply being the result of choosing many SNPs. The value obtained addresses the question of whether the pathway under consideration permits greater separation of cases and controls than would a random collection of most-informative-SNP-per-gene, i.e., whether there exists a more extreme aggregated effect in that pathway than expected by chance.
Results
We applied PoDA to 2287 genotypes obtained from the Cancer Genomic Markers of Susceptibility (CGEMS) breast cancer study. The samples were sourced as described in [4]. Briefly, the samples comprised 1145 breast cancer cases and a comparable number (1142) of matched controls from the participants of the Nurses Health Study. All the participants were American women of European descent. The samples were genotyped against the Illumina 550K arrays, which assays over 550,000 SNPs across the genome.
We also applied it to a smaller liver cancer GWAS [36] comprising 386 hepatocellular carcinoma (HCC) patients and 587 healthy controls from a Korean population. Samples were genotyped against Affymetrix SNP6.0 arrays, which provides SNP information at approximately one million loci.
Breast cancer GWAS results
We begin by applying PoDA to the CGEMS breast cancer GWAS data. Having observed (Figure 1) that PoDA performs as expected for the simulated data, we first turn our attention to a simple test in which we select a SNP set comprising the four SNPs in intron 2 of that were reported to show significant association with case status in [4] (rs11200014, rs2981579, rs1219648, rs2420946). We expect to see a strong difference in the test case and test control distributions, and indeed we do: the cases more frequently have positive than do controls in Figure 2. (The discrete peaks in the distribution are a result of the fact that with four SNPs there exist fewer available values of .) Using a nonparametric Wilcoxon rank sum test with the alternative hypothesis that cases have greater than controls, is obtained, confirming our intuition.
Fig. 2. PoDA applied to four highly-significant SNPs. 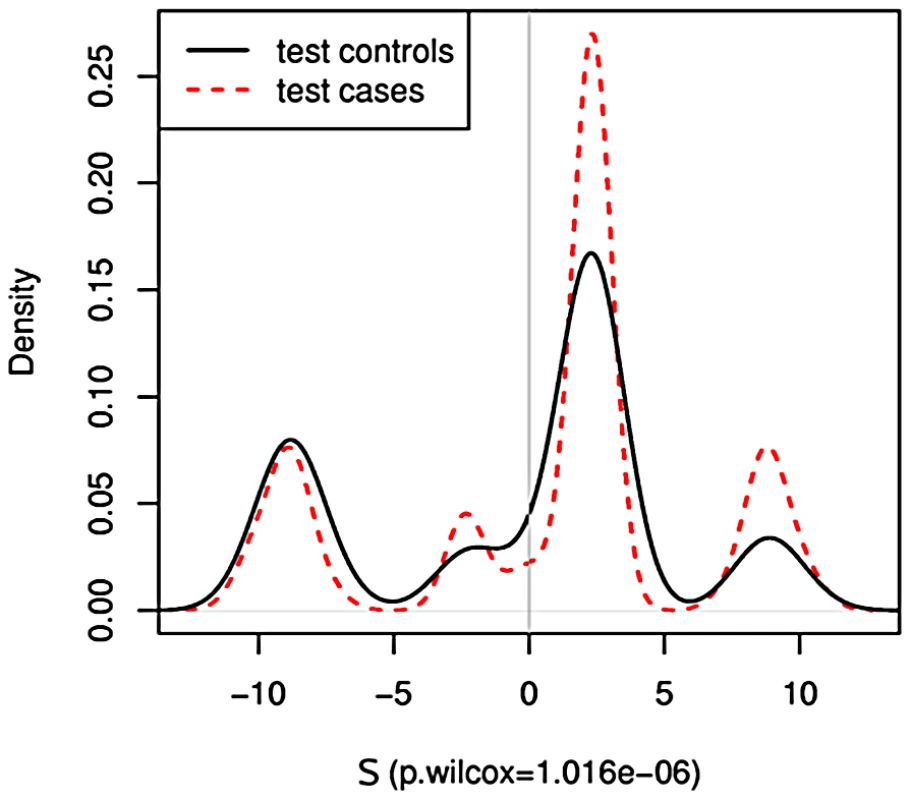
Shown is the distribution of values in CGEMS cases (red) and controls (black) for a SNP-set comprised of four highly-significant SNPs located in the gene [4]. As expected, there is a substantial difference in case and control values, with the cases having higher (i.e., closer to other cases) than controls. The discreteness of the distributions are due to the fact that with four SNPs, a finite number of values are possible. We next applied PoDA systematically to the pathways represented in PID [32] using CGEMS data. Associations between genes and SNPs were made using dbSNP build 129 [34]. 1081 pathways were non-trivially covered in the data set; 69453 SNPs in the data could be associated with at least one of the pathways. Because these 69453 SNPs were associated with 4446 unique genes, 4446 were kept in the analysis (the most significant SNP for each gene of interest). The 1081 pathways ranged from 2 to 229 genes, with a mean of 19. was computed in each pathway for each of the 2287 samples via Eq. 2, and the distinction score (Eq. 4) quantifying differential distributions in cases and controls was computed for each pathway. Significance was assessed as described above, by resampling “dummy” pathways of the same length and computing the fraction of greater scores.
Because PoDA provides for each sample a measure (Eq. 2 of that sample's relative distance from the remaining ones that is obtained without regard to that sample's true class membership, we can use the value as a metric by which to predict the odds of disease. Here, we construct a logistic regression model of case status as a function of to obtain the odds ratio. -values were adjusted for the multiplicity of pathways using FDR adjustment [37], [38].
Pathways with significant and odds ratios are reported in Table 2 and plots of for four of them are illustrated in Figure 3. Although the cases and controls are not crisply separable, a unit increase in over its range from approximately −3 to 3 yields between a 1.5 and 2.0-fold increase in odds. Importantly, given known minor allele frequencies for cases and controls for this set of SNPs, we can model the increase in odds for an unknown individual based on her “closeness” to other cases.
Fig. 3. Four significant pathways in breast cancer data. 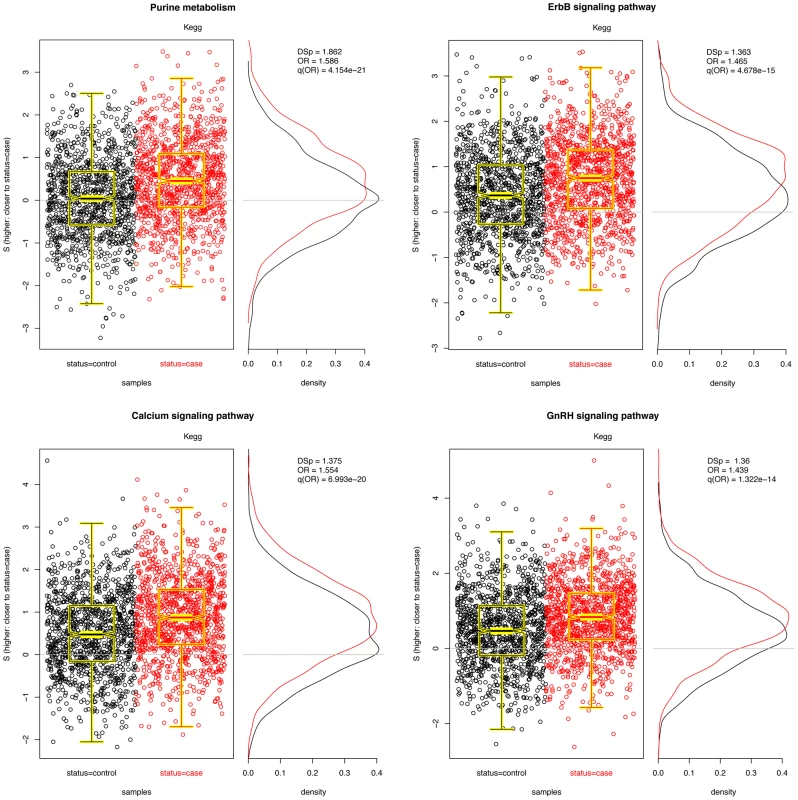
Scatter plots of for each pathway are overlayed with boxplots are given in the left panel; higher values of indicate that the sample is closer to other cases than it is to other controls. Distributions of for cases (red) and controls (black) are given to the right. A significant shift toward higher values is seen in the cases. Odds ratios and FDR-adjusted OR values are given. Tab. 2. PID pathways with significant in the CGEMS breast cancer GWAS. 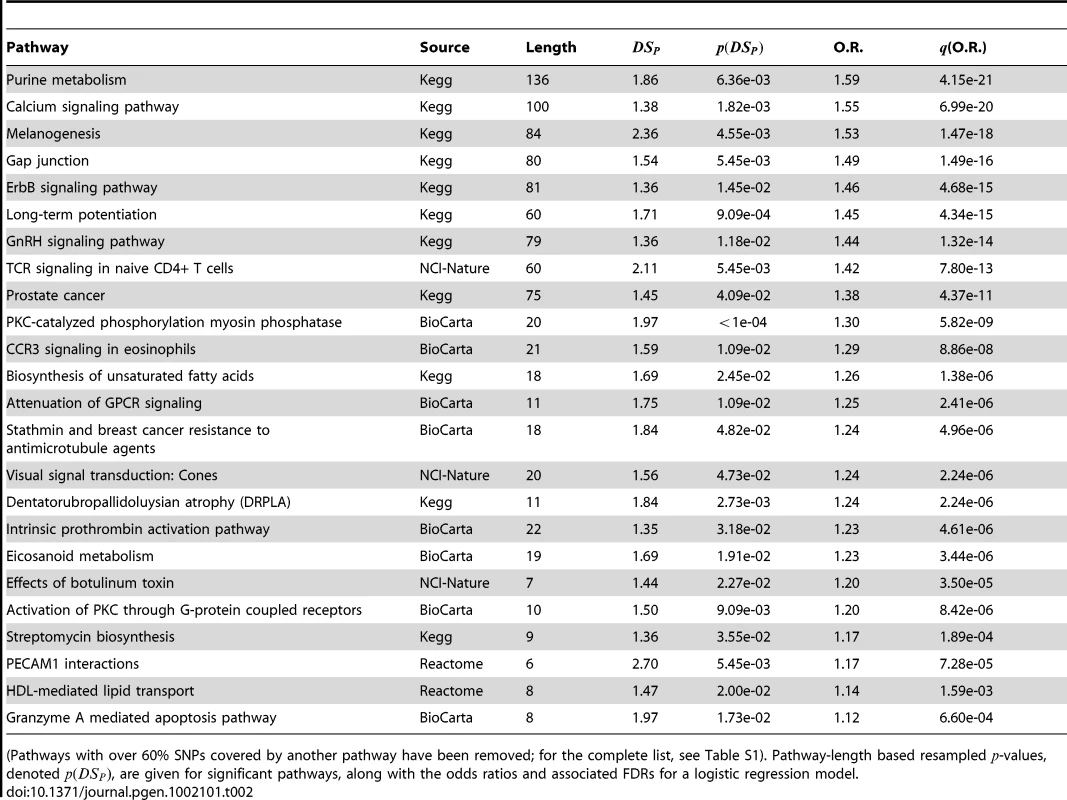
(Pathways with over 60% SNPs covered by another pathway have been removed; for the complete list, see Table S1). Pathway-length based resampled -values, denoted , are given for significant pathways, along with the odds ratios and associated FDRs for a logistic regression model. In order to ensure that the pathways listed were not interrogating the same set of genes, we carried out two checks. First, we computed the SNP overlap between all pairs of significant pathways, sequentially removing pathways that shared in excess of 60% of their genes with another pathway. Because this is done using a greedy algorithm that depends on the order of the pathways input, the culling algorithm was run with different starting orders, and the most frequent output was kept. No pathway remaining in Table 2 shares more than 60% of its SNPs with another pathway. (An un-culled list may be found in Table S1.) Second, we computed the correlation of values between each pair of pathways to assess whether any pathway's statistic was reflecting the same genetic variation as another (i.e., whether samples that had high values for one pathway consistently did so in another). The maximum correlation of values observed between any two pathways in Table 2 was 0.58, suggesting that a different subset of samples is affected in each pathway.
Many of the pathways listed in Table 2 fulfill biological functions that are well known to be cancer-associated, playing a strong role in cell proliferation and migration, processes which are perturbed in malignancies. Purine metabolism–the most significantly associated pathway–has been observed to be altered in cancer cells [39], [40], and the majority of the other significant pathways are directly related to cell migration (e.g., ErbB signaling and gap junction pathways) and cellular signalling (e.g., calcium signaling, PKC-catalyzed phosphorylation of myosin phosphatase, attenuation of GPCR signaling, and activation of PKC through GPCRs) processes that have been implicated in a variety of cancers. In addition, eicosanoids and unsaturated fatty acid metabolism have been associated with breast cancer specifically [41]. In general, the findings in Table 2 suggest that there exist germline genetic differences in these mechanisms that confer a predisposition to disease.
Interestingly, the GnRH (gonadotropin releasing hormone) signaling pathway appears to be significant. GnRH has been linked with HR-positive breast cancer and the use of GnRH analogues in breast cancer treatment has already been proposed [42], [43]. However, a recent large sequencing study found no association of GnRH1 or GnRH receptor gene polymorphisms with breast cancer risk [44], contrary to the author's hypothesis that common, functional polymorphisms of GnRH1 and GnRHR could influence breast cancer risk by modifying hormone production. In contrast to their null findings, our result suggests that there are system-wide variations in GnRH signalling that contribute to risk that are not evident when considering the GnRH1 and GnRHR SNPs independently.
Of the 1081 pathways considered, four–FGF signaling, MAPK signaling, regulation of actin cytoskeleton, and prostate cancer–contained , the gene found to be significantly associated in the initial CGEMS analysis [4]. However, only one–prostate cancer–was significant in comparison to randomly generated pathways of the same length. It may reasonably be asked, then, whether the high significance of the prostate cancer pathway in Table 2 is a result of . To address this, we eliminated the SNP from the prostate cancer pathway; the resampling-based test remained significant ( 8.2e-09), suggesting that the association of the prostate cancer pathway is not driven solely by differences in .
Liver cancer GWAS results
We carried out the same procedure in using data from the liver cancer GWAS described above. Here, 1049 pathways were non-trivially covered in the data set; 53079 SNPs in the data could be associated with at least one of the pathways. Because these 53079 SNPs were associated with 3718 unique genes, 3718 were kept in the analysis (the most significant SNP for each gene of interest). The 1081 pathways ranged from 2 to 193 genes, with a mean of 16. As above, scores for differential distributions in cases and controls were computed for each pathway, resampled values obtained for each pathway size, odds ratios for were obtained, and the multiple hypotheses were corrected using FDR adjustment [37], [38]. Significant pathways are listed in Table 3, and plots of the top three pathways are given in Figure 4a–4d. As in the breast cancer data above, we removed pathways which had over 60% their SNPs covered by another pathway (a complete list, with overlapping pathways, is give in Table S2) and examined the correlation in for all remaining pathways (maximum ).
Fig. 4. Four significant pathways in liver cancer data. 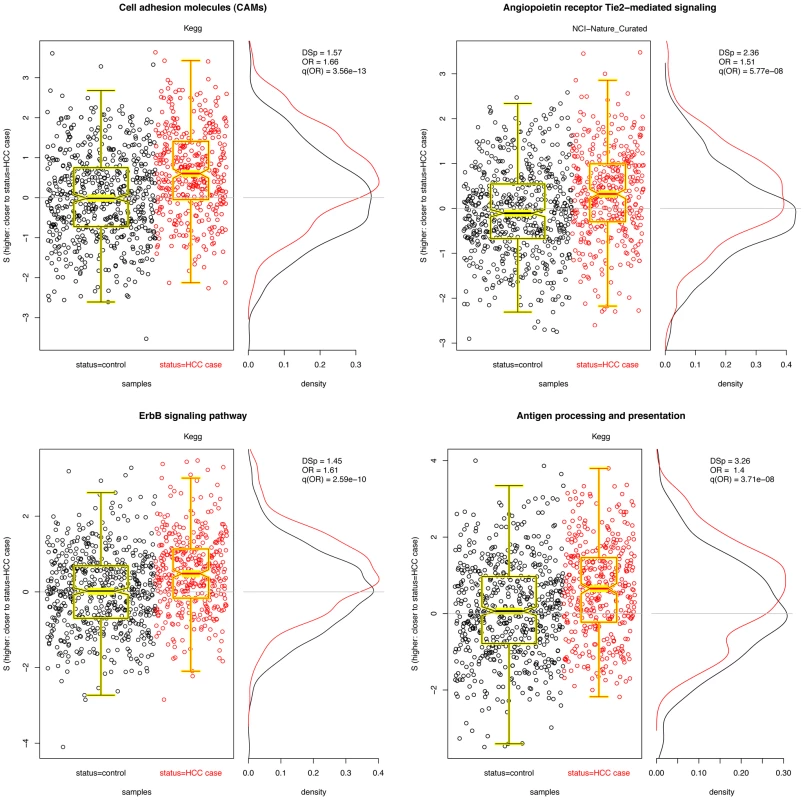
Scatter plots of for each pathway are overlayed with boxplots are given in the left panel; higher values of indicate that the sample is closer to other cases than it is to other controls. Distributions of for cases (red) and controls (black) are given to the right. A significant shift toward higher values is seen in the cases. Odds ratios and FDR-adjusted OR values are given. Tab. 3. PID pathways with significant in the liver cancer GWAS. 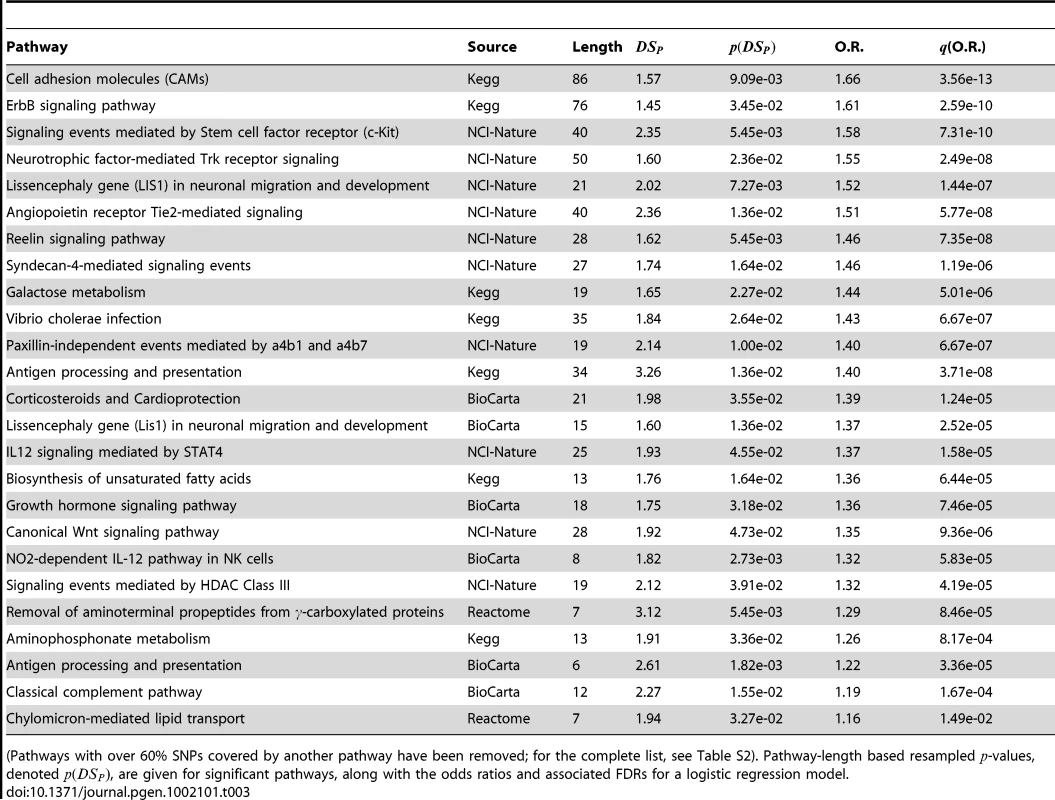
(Pathways with over 60% SNPs covered by another pathway have been removed; for the complete list, see Table S2). Pathway-length based resampled -values, denoted , are given for significant pathways, along with the odds ratios and associated FDRs for a logistic regression model. The results here are interesting. First, we observe that a couple pathways are significant in both the CGEMS breast and liver GWAS with similar effect sizes, namely ErbB signaling and biosynthesis of unsaturated fatty acids. ErbB has a well–established association with cancer; unsaturated fatty acid biosynthesis may link diet to cancer risk, and its appearance may suggest a gene-environment interaction. The commonality of these known cancer-associated pathways across the two studies suggest that there may exist genetic patterns that confer carcinogenesis risk irrespective of the disease site. Along with those shared in the breast cancer data, many of the other significant pathways in the liver cancer data well known to be tumorassociated, including cell adhesion molecules, Wnt signaling, c-Kit receptor, and angiogenesis pathways, further supporting the notion that germline genetic differences in these mechanisms contribute to cancer risk. The appearance of many neuronal pathways is also supported by our understanding of carcinogenesis: thes contain well-known signal transduction molecules including Ras and PKA that may both be driving their conferring increased cancer risk and driving the significance of the pathway [45].
Additionally, six of the 25 significant liver cancer pathways are immune – and inflammation–related, namely, antigen processing and presentation (two, with 60% overlap), classical complement pathway, corticosteroids, IL12 signaling mediated by STAT4, and NO2-dependent IL-12 pathway in NK cells. This is a particularly interesting finding in light of the fact that the original analysis of the liver data [36] suggested that altered T-cell activation plays a direct role in the onset of liver cancer. The involvement of the immune system in liver cancer development has been established in clinical studies and research involving model organisms. Increased activity of helper T-cells, which promote inflammation, is associated with hepatocellular carcinogenesis [46] while activation and proliferation of cytotoxic T-lymphocytes is suppressed in liver cancers [47], [48]. The inflammatory immune response, mediated by interleukins, has also been closely connected to liver cancers in mice [49] and humans [50]–[52]. These findings, coupled with the observation of several significant immune-related pathways in our data, are suggestive of germline polymorphisms in immune response that lead to hepatocellular carcinoma risk.
Combining pathways
In both the breast and liver cancer results, we see observe that even though significant pathways yield between a 1.5 and 2.0-fold increase in odds for each unit increase in (over its typical range of approximately –3 to 3), the cases and controls are not crisply separable based on values. These findings suggest that it may be possible to combine pathways to yield a model that is more predictive than a single pathway alone. However, the values must not simply be put into the regression model because the overlap in pathways will result in some SNPs being double-counted. Rather, we combine pathways by taking the union of their SNPs, and recomputing the statistics. Doing this sequentially for the top pathways in the order as listed in Table 2 and Table 3 yields the values given in Table 4 and Table 5, respectively. Considerably higher ORs are obtained when combining the significant pathways. An illustration of the case and control distributions when using a “superpathway” comprised of the top three pathways in the breast and liver data, respectively, is given in Figure 5. These findings support the notion that the genomic variation contributing to risk is spread over several mechanisms, rather than being concentrated in a single gene.
Fig. 5. Union of top three pathways. 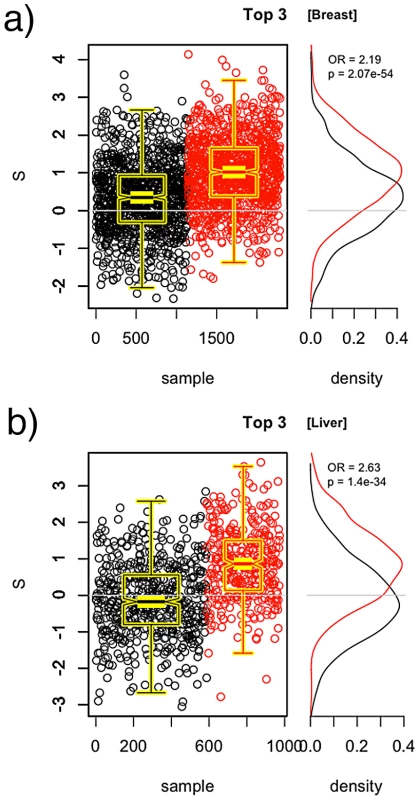
SNPs from the top three pathways are combined to compute for the breast cancer data (a) and the liver cancer data (b). Distributions of for cases (red) and controls (black) are given to the right. A significant shift toward higher values is seen in the cases. Tab. 4. PoDA results for sucessive unions of significant pathways in the CGEMS breast cancer data. 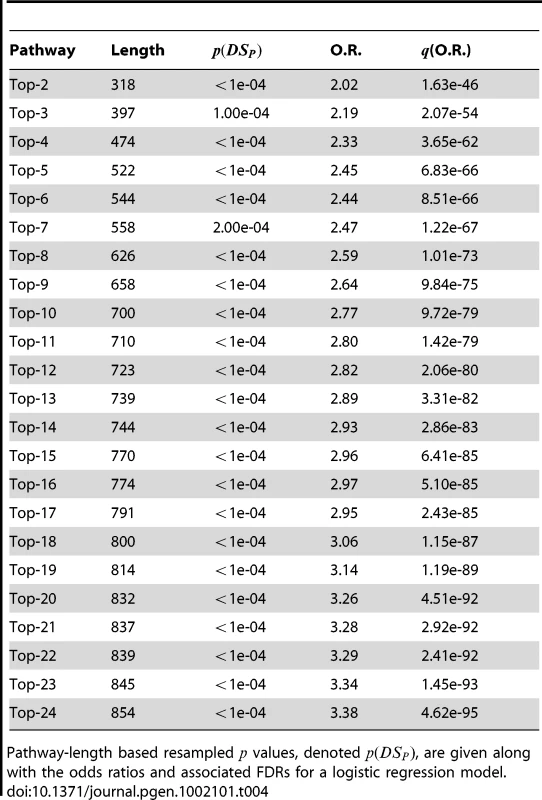
Pathway-length based resampled values, denoted , are given along with the odds ratios and associated FDRs for a logistic regression model. Tab. 5. PoDA results for sucessive unions of significant pathways in the liver cancer data. 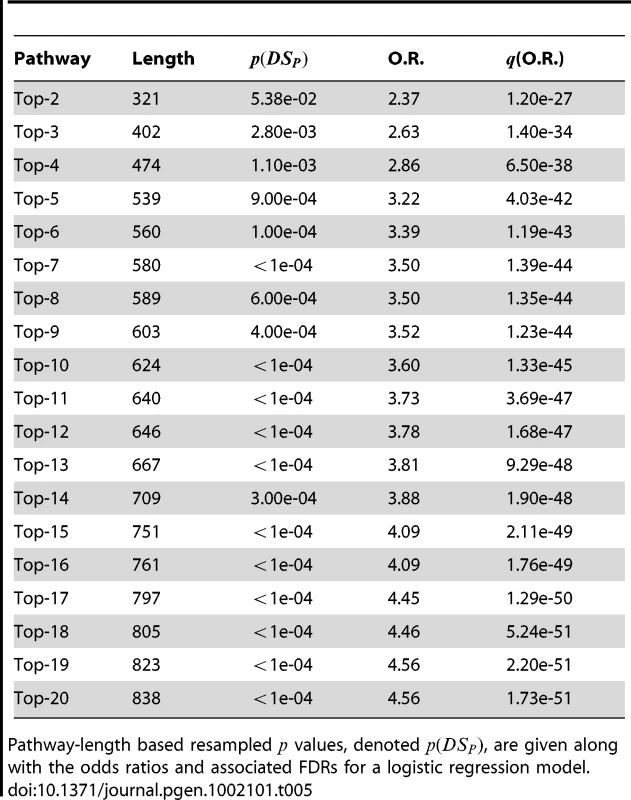
Pathway-length based resampled values, denoted , are given along with the odds ratios and associated FDRs for a logistic regression model. Discussion
We have introduced the Pathways of Distinction analysis method (PoDA) for identifying pathways which can be used to distinguish between phenotype groups. PoDA identifies sets of SNPs in GWAS studies for which cases and controls exhibit differential “closeness” to other cases and controls; that is, it permits one to infer whether cases are more similar to other cases than are controls across a given set of SNPs. Because PoDA is designed to detect the joint effects of multiple SNPs, it presents an approach to GWAS analysis that augments single-SNP or single-gene tests.
We applied PoDA to two GWAS data sets, with highly promising results. In the breast cancer data, we found a number of pathways which are known to play a role in cancers generally and breast cancer specifically, suggesting that differences in these mechanisms which confer disease risk may exist at the germline DNA level. In the liver cancer data, we found an extreme abundance of immune-related pathways, further corroborating the known link between inflammation and hepatocellular carcinoma, and bolstering the observation in [36] that germ-line differences in immune function may play a role in liver carcinogenesis.
PoDA may be used as a complement to other multi-SNP analysis techniques [18]–[21]. Unlike gene-set enrichment type approaches [17]–[19], which search for an overabundance of significant markers in a gene set of interest, PoDA finds both sets containing highly significant markers as well as sets that have a subtle but consistent pattern across all the markers in the set. This permits the detection of pathways in which the joint action of several alterations produce a phenotype and those for which any of several possible alterations, none of them the dominant one, confer predisposition to disease. Indeed, many of the pathways indicated in our analysis of the breast cancer data (Table 2) were not detected using SNP-set enrichment [17]–[19] (data not shown), including the highly significant purine metabolism and GnRH signaling pathways, both of which are biologically relevant (purine metabolism has been implicated in cancers generally due to its role in DNA and RNA synthesis [40], and GnRH has been shown to be clinically important in breast and gynecological cancers [53]). These pathways, along with others that were indicated using PoDA but not enrichment analysis (data not shown), have a statistically significant difference in case and control distributions and remain significant in comparison with randomly-generated pathways of the same length.
Because PoDA effectively measures the closeness of each individual to remaining cases and controls, it bears a conceptual relationship to nearest-neighbor and nearest-centroid classifiers [54], [55], as well as to the distance-based feature selection algorithms like Relief-F and its derivatives [25]–[28]. However, it must be remembered that the goal of PoDA is to indicate mechanisms that may be deleteriously hit at the genomic level even when those hits are heterogeneous, whereas the goal of nearest-centroid classifiers and Relief-F–type feature selection is to derive a minimal set of markers that best classify cases and controls (and thus are the most homogeneously hit). These approaches are complementary, and one can easily envision an application in which (e.g.) Relief-F is run within pathways that are highly significant in the PoDA analysis in order to single out the SNPs driving the effect. In fact, this approach may improve ReliefF's ability to find those genes, since the nearest neighbors from which the Relief SNP weights are calculated would be the nearest-neighbors for that specific pathway, thus discounting heterogeneity introduced by mechanistically unrelated genes. For instance, in the provided example (Figure 1), ReliefF fails to identify the significance of SNPs 1–3 when run using the complete 50-SNP data, but places at least two of SNPs 1, 2 or 3 in the top third of selected features when restricted to SNPs 1–12.
While PoDA has many benefits, it should be noted that when epistasis drives a phenotype with no differences in the minor allele frequencies for the epistatically-interacting genes (as opposed to a slight yet consistent one shown in the example), PoDA as computed via Eqs. 1,2 will miss the pathway. Geometrically, such a situation would mean that the case and control groups have the same centroids while having a different distribution of samples about those centroids. A famous example of this is provided through the non-linearly separable XOR (exclusive or): consider two epistatic loci such that all controls have genotypes in the set and all cases have genotypes in the set (i.e., that a genotype of 1 at either locus can be compensated by a genotype of 1 at the other, but having just one alone–1 at exclusively or –is deleterious). If the loci and each have the same MAF in cases and controls, it is plain to see that the centroids will be in the same location for both groups, and Eq. 1 will yield zero for both cases and controls. If instead of using Eq. 1, we compute pairwise sample-sample distances, we can circumvent this limitation and find such epistatic situations (it is this pairwise approach that permits Relief-F to also uncover nonlinearly interacting SNPs). While we provide the facility for this in the PoDA package, the cost of carrying out the pairwise computation is a considerable increase in computational complexity.
A number of potential avenues exist to extend the application of PoDA further. One possible application is in improving the reproducibility of GWAS results. We note that several of the pathways identified in the breast cancer GWAS data were also implicated in the liver cancer data, which suggests that there may be common features which distinguish individuals to cancer generally. Because different GWA studies–even those of the same phenotypes–often yield different results at the SNP level, it may be possible to find common alterations at the pathway level across disparate GWAS using PoDA.
Extending PoDA further, the scores obtained for each pathway may be examined for over-representation of extreme values in pathways that comprise a particular biological subsystem–one may think of this as a “pathway-set” enrichment analysis (which would be conducted using the a running-sum statistic analogous GSEA [17]), and could use it to answer whether (e.g.) immune-related pathways are hit in liver cancer more often than expected by chance. Alternatively, boosting [56], [57] could be used to find sets of pathways which are more predictive of case status than individual pathways. Either of these approaches would yield a richer, systems-wide view of the connection between genotype and phenotype. Finally, because PID contains topological information regarding pathway connectivity, one may consider sub-networks of pathways, permitting one to find potential chemopreventive and therapeutic targets. Alternatively, Relief-F can be used, as mentioned above, in a pathway–specific manner to yield the subset of SNPs that drive the distinction of cases and controls in high - pathways.
PoDA provides an advantage over existing GWAS analysis methods. Because it does not rely on the significance of individual markers, it has the power to aid in identifying the genomic causes of complex diseases that would not be detected in single-gene tests or enrichment analyses. The size of the SNP set is not limited in PoDA, and since PoDA leverages known biological relationships to find multi-SNP effects, the results are readily interpretable. PoDA may thus be used to augment existing analysis techniques and provide a richer, systems-level understanding of genomics.
Availability
R software to carry out the PoDA computation is available via http://braun.tx0.org/PoDA.
Supporting Information
Zdroje
1. HirschhornJNDalyMJ 2005 Genome-wide association studies for common diseases and complex traits. Nat Rev Genet 6 95 108
2. McCarthyMIAbecasisGRCardonLRGoldsteinDBLittleJ 2008 Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat Rev Genet 9 356 69
3. EastonDFEelesRA 2008 Genome-wide association studies in cancer. Hum Mol Genet 17 R109 15
4. HunterDJKraftPJacobsKBCoxDGYeagerM 2007 A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenopausal breast cancer. Nature Genetics 39 870 874
5. LouHYeagerMLiHBosquetJGHayesRB 2009 Fine mapping and functional analysis of a common variant in MSMB on chromosome 10q11.2 associated with prostate cancer susceptibility. PNAS 106 7933 8
6. LouHYeagerMLiHBosquetJGHayesRB 2009 Fine mapping and functional analysis of a common variant in MSMB on chromosome 10q11.2 associated with prostate cancer susceptibility. PNAS 106 7933 8
7. ThomasGJacobsKBKraftPYeagerMWacholderS 2009 A multistage genome-wide association study in breast cancer identifies two new risk alleles at 1p11.2 and 14q24.1 (RAD51L1). Nat Genet 41 579 84
8. HindorffLASethupathyPJunkinsHARamosEMMehtaJP 2009 Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. PNAS 106 9362 7
9. SchorkNMurraySFrazerKTopolE 2009 Common vs. rare allele hypotheses for complex diseases. Current opinion in genetics & development 19 212 219
10. MooreJAsselbergsFWilliamsS 2010 Bioinformatics challenges for genome-wide association studies. Bioinformatics 26 445
11. GreeneCPenrodNWilliamsSMooreJ 2009 Failure to replicate a genetic association may provide important clues about genetic architecture. PLoS ONE 4 e5639 doi:10.1371/journal.pone.0005639
12. MooreJ 2003 The ubiquitous nature of epistasis in determining susceptibility to common human diseases. Human Heredity 56 73 82
13. TylerAAsselbergsFWilliamsSMooreJ 2009 Shadows of complexity: what biological networks reveal about epistasis and pleiotropy. BioEssays 31 220 227
14. HanahanDWeinbergRA 2000 The hallmarks of cancer. Cell 100 57 70
15. HolmansP 2010 Statistical methods for pathway analysis of genome-wide data for association with complex genetic traits. Advances in genetics 72 141
16. WangKLiMHakonarsonH 2010 Analysing biological pathways in genome-wide association studies. Nature Reviews Genetics 11 843 854
17. SubramanianATamayoPMoothaVKMukherjeeSEbertBL 2005 Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. PNAS 102 15545 50
18. WangKLiMBucanM 2007 Pathway-based approaches for analysis of genomewide association studies. Am J Hum Genet 81 1278
19. HoldenMDengSWojnowskiLKulleB 2008 GSEA-SNP: applying gene set enrichment analysis to SNP data from genome-wide association studies. Bioinformatics 24 2784 5
20. YangCHeZWanXYangQXueH 2009 SNPHarvester: a filtering-based approach for detecting epistatic interactions in genome-wide association studies. Bioinformatics 25 504 11
21. MotsingerARitchieM 2006 Multifactor dimensionality reduction: an analysis strategy for modelling and detecting gene–gene interactions in human genetics and pharmacogenomics studies. Human Genomics 2 318 328
22. MooreJGilbertJTsaiCChiangFHoldenT 2006 A flexible computational framework for detecting, characterizing, and interpreting statistical patterns of epistasis in genetic studies of human disease susceptibility. Journal of theoretical biology 241 252 261
23. CordellH 2009 Detecting gene–gene interactions that underlie human diseases. Nature Reviews Genetics 10 392 404
24. GreeneCSinnott-ArmstrongNHimmelsteinDParkPMooreJ 2010 Multifactor dimensionality reduction for graphics processing units enables genome-wide testing of epistasis in sporadic als. Bioinformatics 26 694
25. KiraKRendellL 1992 A practical approach to feature selection. Proceedings of the Ninth International Workshop on Machine learning 249 256
26. Robnik-ŠikonjaMKononenkoI 1997 An adaptation of relief for attribute estimation in regression. Proc Int Conf on Machine Learning ICML-97 296 304
27. MooreJ 2007 Genome-wide analysis of epistasis using multifactor dimensionality reduction: feature selection and construction in the domain of human genetics. Knowledge Discovery and Data Mining: Challenges and Realities with Real World Data 17 30
28. GreeneCPenrodNKiralisJMooreJ 2009 Spatially Uniform ReliefF (SURF) for computationally-efficient filtering of gene-gene interactions. BioData mining 2 5
29. HomerNSzelingerSRedmanMDugganDTembeW 2008 Resolving individuals contributing trace amounts of DNA to highly complex mixtures using high-density SNP genotyping microarrays. PLoS Genet 4 e1000167 doi:10.1371/journal.pgen.1000167
30. BraunRRoweWSchaeferCZhangJBuetowK 2009 Needles in the haystack: Identifying individuals present in pooled genomic data. PLoS Genet 5 e1000668 doi:10.1371/journal.pgen.1000668
31. VisscherPMHillWG 2009 The limits of individual identification from sample allele frequencies: theory and statistical analysis. PLoS Genet 5 e1000628 doi:10.1371/journal.pgen.1000628
32. SchaeferCFAnthonyKKrupaSBuchoffJDayM 2009 PID: the Pathway Interaction Database. Nucleic Acids Res 37 D674 679
33. KanehisaMArakiMGotoSHattoriMHirakawaM 2008 KEGG for linking genomes to life and the environment. Nucleic Acids Res 36 D480 4
34. SherrySTWardMHKholodovMBakerJPhanL 2001 dbSNP: the NCBI database of genetic variation. Nucleic Acids Res 29 308 311
35. KraftPRaychaudhuriS 2009 Complex diseases, complex genes: keeping pathways on the right track. Epidemiology (Cambridge, Mass) 20 508
36. CliffordRZhangJMeerzamanDLyuMHuY 2010 Genetic variations at loci involved in the immune response are risk factors for hepatocellular carcinoma. Hepatology 52 2034 2043
37. BenjaminiYHochbergY 1995 Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society 57 289 300
38. BenjaminiYYekutieliD 2001 The control of the false discovery rate in multiple testing under dependency. Annals of Statistics 1165 1188
39. WeberG 1977 Enzymology of cancer cells. New England Journal of Medicine 296 541 551
40. WeberG 1983 Enzymes of purine metabolism in cancer. Clinical Biochemistry 16 57 63
41. RoseDConnollyJ 1990 Effects of fatty acids and inhibitors of eicosanoid synthesis on the growth of a human breast cancer cell line in culture. Cancer research 50 7139
42. EidneKFlanaganCHarrisNMillarR 1987 Gonadotropin-releasing hormone (GnRH)-binding sites in human breast cancer cell lines and inhibitory effects of GnRH antagonists. Journal of Clinical Endocrinology & Metabolism 64 425
43. ManniASantenRHarveyHLiptonAMaxD 1986 Treatment of breast cancer with gonadotropin-releasing hormone. Endocrine reviews 7 89
44. CanzianFKaaksRCoxDHendersonKHendersonB 2009 Genetic polymorphisms of the GnRH1 and GNRHR genes and risk of breast cancer in the national cancer institute breast and prostate cancer cohort consortium. BMC cancer 9 257
45. NakagawaraA 2001 Trk receptor tyrosine kinases: a bridge between cancer and neural development. Cancer letters 169 107 114
46. Pentcheva-HoangTCorseEAllisonJP 2009 Negative regulators of T-cell activation: potential targets for therapeutic intervention in cancer, autoimmune disease, and persistent infections. Immunol Rev 229 67 87
47. OrmandyLAHillemannTWedemeyerHMannsMPGretenTF 2005 Increased populations of regulatory T cells in peripheral blood of patients with hepatocellular carcinoma. Cancer Res 65 2457 64
48. UnittERushbrookSMMarshallADaviesSGibbsP 2005 Compromised lymphocytes infiltrate hepatocellular carcinoma: the role of T-regulatory cells. Hepatology 41 722 30
49. NauglerWESakuraiTKimSMaedaSKimK 2007 Gender disparity in liver cancer due to sex differences in MyD88-dependent IL-6 production. Science 317 121 4
50. BudhuASZipserBForguesMYeQHSunZ 2005 The molecular signature of metastases of human hepatocellular carcinoma. Oncology 69 Suppl 1 23 7
51. BudhuAWangXW 2006 The role of cytokines in hepatocellular carcinoma. J Leukoc Biol 80 1197 213
52. BudhuAForguesMYeQHJiaHLHeP 2006 Prediction of venous metastases, recurrence, and prognosis in hepatocellular carcinoma based on a unique immune response signature of the liver microenvironment. Cancer Cell 10 99 111
53. EmonsGGrundkerCGunthertAWestphalenSKavanaghJ 2003 GnRH antagonistsin the treatment of gynecological and breast cancers. Endocrine-related cancer 10 291
54. CoverTHartP 2002 Nearest neighbor pattern classification. IEEE Transactions on Information Theory 13 21 27
55. TibshiraniRHastieTNarasimhanBChuG 2002 Diagnosis of multiple cancer types by shrunken centroids of gene expressionx. PNAS 99 6567 72
56. BuhlmannPHothornT 2007 Boosting algorithms: regularization, prediction and model fitting. Statistical Science 22 477 505
57. MeirRRatschG 2003 An introduction to boosting and leveraging. Lecture Notes in Computer Science 2600 118 183
Štítky
Genetika Reprodukční medicína
Článek Local Absence of Secondary Structure Permits Translation of mRNAs that Lack Ribosome-Binding SitesČlánek Independent Chromatin Binding of ARGONAUTE4 and SPT5L/KTF1 Mediates Transcriptional Gene SilencingČlánek Trade-Off between Bile Resistance and Nutritional Competence Drives Diversification in the Mouse GutČlánek FGF Signaling Regulates the Number of Posterior Taste Papillae by Controlling Progenitor Field SizeČlánek Mammalian BTBD12 (SLX4) Protects against Genomic Instability during Mammalian SpermatogenesisČlánek Identification of Nine Novel Loci Associated with White Blood Cell Subtypes in a Japanese PopulationČlánek Differential Effects of and Risk Variants on Association with Diabetic ESRD in African AmericansČlánek Dynamic Chromatin Localization of Sirt6 Shapes Stress- and Aging-Related Transcriptional Networks
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2011 Číslo 6
-
Všechny články tohoto čísla
- Local Absence of Secondary Structure Permits Translation of mRNAs that Lack Ribosome-Binding Sites
- Statistical Inference on the Mechanisms of Genome Evolution
- Revisiting Heterochromatin in Embryonic Stem Cells
- A Two-Stage Meta-Analysis Identifies Several New Loci for Parkinson's Disease
- Identification of a Sudden Cardiac Death Susceptibility Locus at 2q24.2 through Genome-Wide Association in European Ancestry Individuals
- Genomic Prevalence of Heterochromatic H3K9me2 and Transcription Do Not Discriminate Pluripotent from Terminally Differentiated Cells
- Epistasis between Beneficial Mutations and the Phenotype-to-Fitness Map for a ssDNA Virus
- Recurrent Chromosome 16p13.1 Duplications Are a Risk Factor for Aortic Dissections
- Telomere DNA Deficiency Is Associated with Development of Human Embryonic Aneuploidy
- Genome-Wide Association Study of White Blood Cell Count in 16,388 African Americans: the Continental Origins and Genetic Epidemiology Network (COGENT)
- Unexpected Role for DNA Polymerase I As a Source of Genetic Variability
- Transportin-SR Is Required for Proper Splicing of Genes and Plant Immunity
- How Chromatin Is Remodelled during DNA Repair of UV-Induced DNA Damage in
- Independent Chromatin Binding of ARGONAUTE4 and SPT5L/KTF1 Mediates Transcriptional Gene Silencing
- Two Evolutionary Histories in the Genome of Rice: the Roles of Domestication Genes
- Natural Allelic Variation Defines a Role for : Trichome Cell Fate Determination
- Multiple Common Susceptibility Variants near BMP Pathway Loci , , and Explain Part of the Missing Heritability of Colorectal Cancer
- Pathogenic Mechanism of the FIG4 Mutation Responsible for Charcot-Marie-Tooth Disease CMT4J
- A Functional Variant in Promoter Modulates Its Expression and Confers Disease Risk for Systemic Lupus Erythematosus
- Drift and Genome Complexity Revisited
- Chromosomal Macrodomains and Associated Proteins: Implications for DNA Organization and Replication in Gram Negative Bacteria
- Trade-Off between Bile Resistance and Nutritional Competence Drives Diversification in the Mouse Gut
- Pathways of Distinction Analysis: A New Technique for Multi–SNP Analysis of GWAS Data
- Web-Based Genome-Wide Association Study Identifies Two Novel Loci and a Substantial Genetic Component for Parkinson's Disease
- Chk2 and p53 Are Haploinsufficient with Dependent and Independent Functions to Eliminate Cells after Telomere Loss
- Exome Sequencing Identifies Mutations in High Myopia
- Distinct Functional Constraints Partition Sequence Conservation in a -Regulatory Element
- CorE from Is a Copper-Dependent RNA Polymerase Sigma Factor
- A Single Sex Pheromone Receptor Determines Chemical Response Specificity of Sexual Behavior in the Silkmoth
- FGF Signaling Regulates the Number of Posterior Taste Papillae by Controlling Progenitor Field Size
- Maps of Open Chromatin Guide the Functional Follow-Up of Genome-Wide Association Signals: Application to Hematological Traits
- Increased Susceptibility to Cortical Spreading Depression in the Mouse Model of Familial Hemiplegic Migraine Type 2
- Differential Gene Expression and Epiregulation of Alpha Zein Gene Copies in Maize Haplotypes
- Parallel Adaptive Divergence among Geographically Diverse Human Populations
- Genetic Analysis of Genome-Scale Recombination Rate Evolution in House Mice
- Mechanisms for the Evolution of a Derived Function in the Ancestral Glucocorticoid Receptor
- Mammalian BTBD12 (SLX4) Protects against Genomic Instability during Mammalian Spermatogenesis
- Interferon Regulatory Factor 8 Regulates Pathways for Antigen Presentation in Myeloid Cells and during Tuberculosis
- High-Resolution Analysis of Parent-of-Origin Allelic Expression in the Arabidopsis Endosperm
- Specific SKN-1/Nrf Stress Responses to Perturbations in Translation Elongation and Proteasome Activity
- Graded Nodal/Activin Signaling Titrates Conversion of Quantitative Phospho-Smad2 Levels into Qualitative Embryonic Stem Cell Fate Decisions
- Genome-Wide Analysis Reveals PADI4 Cooperates with Elk-1 to Activate Expression in Breast Cancer Cells
- Trait Variation in Yeast Is Defined by Population History
- Meiosis-Specific Loading of the Centromere-Specific Histone CENH3 in
- A Genome-Wide Survey of Imprinted Genes in Rice Seeds Reveals Imprinting Primarily Occurs in the Endosperm
- Multiple Regulatory Mechanisms to Inhibit Untimely Initiation of DNA Replication Are Important for Stable Genome Maintenance
- SIRT1 Promotes N-Myc Oncogenesis through a Positive Feedback Loop Involving the Effects of MKP3 and ERK on N-Myc Protein Stability
- Bacteriophage Crosstalk: Coordination of Prophage Induction by Trans-Acting Antirepressors
- Role of the Single-Stranded DNA–Binding Protein SsbB in Pneumococcal Transformation: Maintenance of a Reservoir for Genetic Plasticity
- Genomic Convergence among ERRα, PROX1, and BMAL1 in the Control of Metabolic Clock Outputs
- Genome-Wide Association of Bipolar Disorder Suggests an Enrichment of Replicable Associations in Regions near Genes
- Identification of Nine Novel Loci Associated with White Blood Cell Subtypes in a Japanese Population
- DNA Ligase III Promotes Alternative Nonhomologous End-Joining during Chromosomal Translocation Formation
- Differential Effects of and Risk Variants on Association with Diabetic ESRD in African Americans
- Finished Genome of the Fungal Wheat Pathogen Reveals Dispensome Structure, Chromosome Plasticity, and Stealth Pathogenesis
- Dynamic Chromatin Localization of Sirt6 Shapes Stress- and Aging-Related Transcriptional Networks
- Extracellular Matrix Dynamics in Hepatocarcinogenesis: a Comparative Proteomics Study of Transgenic and Null Mouse Models
- Integrating 5-Hydroxymethylcytosine into the Epigenomic Landscape of Human Embryonic Stem Cells
- Vive La Différence: An Interview with Catherine Dulac
- Multiple Loci Are Associated with White Blood Cell Phenotypes
- Nuclear Accumulation of Stress Response mRNAs Contributes to the Neurodegeneration Caused by Fragile X Premutation rCGG Repeats
- A New Mutation Affecting FRQ-Less Rhythms in the Circadian System of
- Cryptic Transcription Mediates Repression of Subtelomeric Metal Homeostasis Genes
- A New Isoform of the Histone Demethylase JMJD2A/KDM4A Is Required for Skeletal Muscle Differentiation
- Genetic Determinants of Lipid Traits in Diverse Populations from the Population Architecture using Genomics and Epidemiology (PAGE) Study
- A Genome-Wide RNAi Screen for Factors Involved in Neuronal Specification in
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Recurrent Chromosome 16p13.1 Duplications Are a Risk Factor for Aortic Dissections
- Statistical Inference on the Mechanisms of Genome Evolution
- Genome-Wide Association Study of White Blood Cell Count in 16,388 African Americans: the Continental Origins and Genetic Epidemiology Network (COGENT)
- Chromosomal Macrodomains and Associated Proteins: Implications for DNA Organization and Replication in Gram Negative Bacteria
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



