-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaDrift and Genome Complexity Revisited
article has not abstract
Published in the journal: . PLoS Genet 7(6): e32767. doi:10.1371/journal.pgen.1002092
Category: Viewpoints
doi: https://doi.org/10.1371/journal.pgen.1002092Summary
article has not abstract
Introduction
Recently, Whitney and Garland [1] (hereafter “WG”) reanalyzed a dataset presented in Lynch and Conery [2] (hereafter “LC”) using phylogenetic statistical techniques. Contrary to LC, WG found little support for the idea that Neu (the product of effective population size and the mutation rate) is statistically related to genome size or six other genomic attributes. Lynch [3] has responded with criticisms of the WG approach and interpretations. Below we carefully consider these criticisms, present additional analyses, and conclude that the WG analyses are robust. In addition, we explore the consistency of some predictions of the mutational-hazard (MH) hypothesis [3] and provide some guidance regarding future tests.
Given that both analyses used the same dataset, the heart of the issue is the choice of analysis techniques and interpretation of results. Below, we use the terms “phylogenetic” and “nonphylogenetic” to describe the techniques employed by WG and LC, respectively. “Nonphylogenetic” remains in quotes because, in fact, species-level regression or correlation analyses that do not explicitly incorporate phylogenetic history do assume a particular phylogeny—a star phylogeny (polytomy) in which all species are equally related and all branches have equal lengths [4], [5] .
The Appropriateness of Phylogenetic Analyses
Lynch [3] argues that both Neu and measures of genome complexity (e.g., genome size) are so evolutionarily labile that analyses incorporating a hierarchical phylogenetic tree are unnecessary and potentially misleading (but see [6]). The issue can be empirically addressed [7], [8]. The key test of whether a phylogenetic or “nonphylogenetic” regression analysis is more appropriate examines the regression residuals for phylogenetic signal [8], [9]. Phylogenetic signal in the residuals is evidence that the evolutionary response of the dependent variable to the independent variable was not so rapid as to make phylogeny unimportant in regression analyses. This was the agnostic approach taken in WG, letting the statistics indicate the best-fit model. The phylogenetic models had better fit (see Table 1 in [1]), indicating significant phylogenetic signal in the residuals. These models did not support the hypothesis that Neu explains a significant fraction of the variation in genomic attributes such as genome size.
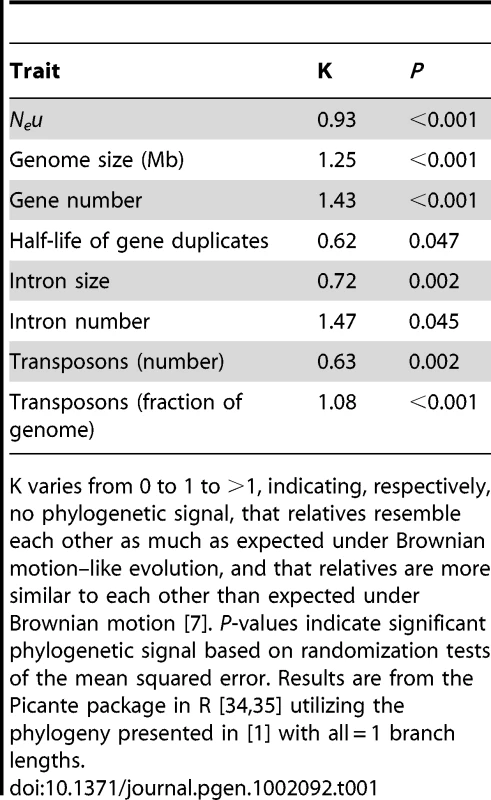
Although the key insight regarding trait lability is determined from the phylogenetic signal of the regression residuals, it can also be instructive to examine phylogenetic signal for particular traits. Table 1 presents estimates of phylogenetic signal (K) for the dataset under discussion; all traits show significant (and often extremely strong) phylogenetic signal, indicating that species cannot be considered statistically independent entities for any of these traits [7]. Such strong phylogenetic signal may be counterintuitive for Neu, which is a population-level trait as opposed to a “standard” individual-level morphological trait. However, Ne can be construed as an emergent trait that reflects several other traits (e.g., mating system, dispersal ability, social group size, body size) that generally do show phylogenetic signal (e.g., [7]). In any case, the empirical data do not support Lynch's contention that Neu (as estimated by πs, the average nucleotide heterozygosity at silent sites) is so labile as to “hav[e] no shared phylogenetic history” across the species in the dataset.
Next, Lynch argues that phylogenetic techniques are inappropriate for the current dataset because “. . . phylogenetic inertia is overshadowed by other evolutionary effects. For example, for the two most closely related species . . . mouse and human . . . numerous shared features of genome architecture are a consequence of convergent evolution, not shared ancestry.” He observes that genome sizes in different species may be determined by the abundances of different transposable element (TE) families. Although it is certainly true that genome architecture can be superficially similar because of convergent evolution, and that such convergence can evolve via different underlying components (e.g., different TEs in the case of genome size), these observations do not automatically override the necessity for phylogenetic analyses. Phylogenetic nonindependence must be accounted for if it exists, no matter how it arises. Phylogenetic signal in the residuals of the regression of genome size on Neu (see WG and Table 2 of the current article) indicates that related species could share similar values of other traits (aside from Neu) that influence genome size. We posit that traits influencing the proliferation of TEs (e.g., mating system, methylation propensity, RNAi-mediated interference) show phylogenetic signal and are partly responsible for the nonindependence observed among residual genome sizes of closely related species. Another non-mutually-exclusive hypothesis is that related taxa share physiological traits that partly determine the environments in which they can live (e.g., [10], [11]), and that the resulting shared environmental conditions have caused selection favoring similar-sized genomes. Regardless of one's ability to identify the lower-level traits involved, phylogenetic nonindependence of residuals is present in the current dataset (WG and Table 2 of the current article), and ignoring it can lead to incorrect inferences about associations between traits.
Finally, Lynch makes two general criticisms of phylogenetic methods. First, he asserts “it can be shown” that the phylogenetically independent contrast method inflates the sampling variance of the independent variable and decreases r2 values by ≈30%. No justification or citation is given for this assertion, and we know of no such bias. Moreover, r2 values are generally not directly comparable across “nonphylogenetic” and phylogenetic regression models [9]. Second, citing [12], Lynch states that ordinary least-squares (OLS) correlations are “on average, unbiased” and that similar correlations are expected “whether or not shared phylogenetic history is accounted for.” Indeed, empirically, parameter estimates from the two types of analyses are often similar (see also [5], [13]). However, this average outcome across studies does not prevent phylogenetic versus “nonphylogenetic” analyses from giving very different answers for a particular dataset, which is clearly the case here. Thus, any conclusion that a “nonphylogenetic” analysis will always provide the correct inference is not warranted.
Estimation of Neu
Lynch identifies three issues relating to Neu and to estimating Neu via πs : 1) estimates of πs are associated with high sampling variance; 2) because of constraints on Ne and u, many prokaryote species will have similar Neu values; and 3) πs in unicellular species is subject to downward bias resulting from selection on silent sites, perhaps causing prokaryotic Neu estimates to be off by more than an order of magnitude. These issues are properly viewed as criticisms of the dataset itself, not the chosen analysis. They are equally applicable to the OLS analysis of LC and have no bearing on whether a phylogenetic versus “nonphylogenetic” analysis is more appropriate.
We note that error in the independent variable can be incorporated into both phylogenetic and “nonphylogenetic” regression analyses using special techniques (e.g., [14]). However, such techniques require that the error be quantified. For the current dataset, error in πs is not quantified, and thus neither we nor Lynch have the opportunity to apply such techniques.
Tree Topologies and Branch Lengths
Lynch argues that potential uncertainties associated with tree topology and branch lengths weaken the conclusions of WG. We agree that errors in topologies and branch lengths can influence the outcomes of phylogenetically based statistical analyses [4], [5], [15]. However, the key point is that a “nonphylogenetic” analysis (e.g., the OLS regression performed in LC) is not phylogeny-free. Regression analyses assume that residuals in the dependent (Y) variable are independent and identically distributed. Under Brownian-motion-like evolution, the only phylogenetic tree that generates the appropriate variance–covariance matrix (an identity matrix) is a star phylogeny, in which each taxon is equally related to all other taxa and branch lengths are equal [4], [5]. In effect, the LC analysis assumes that humans are no more closely related to mice than to bacteria. Clearly, if there are critical errors in tree topology (and branch lengths) that undermine the conclusions of the alternate analyses under discussion here, then they are found in the star phylogeny assumed by LC.
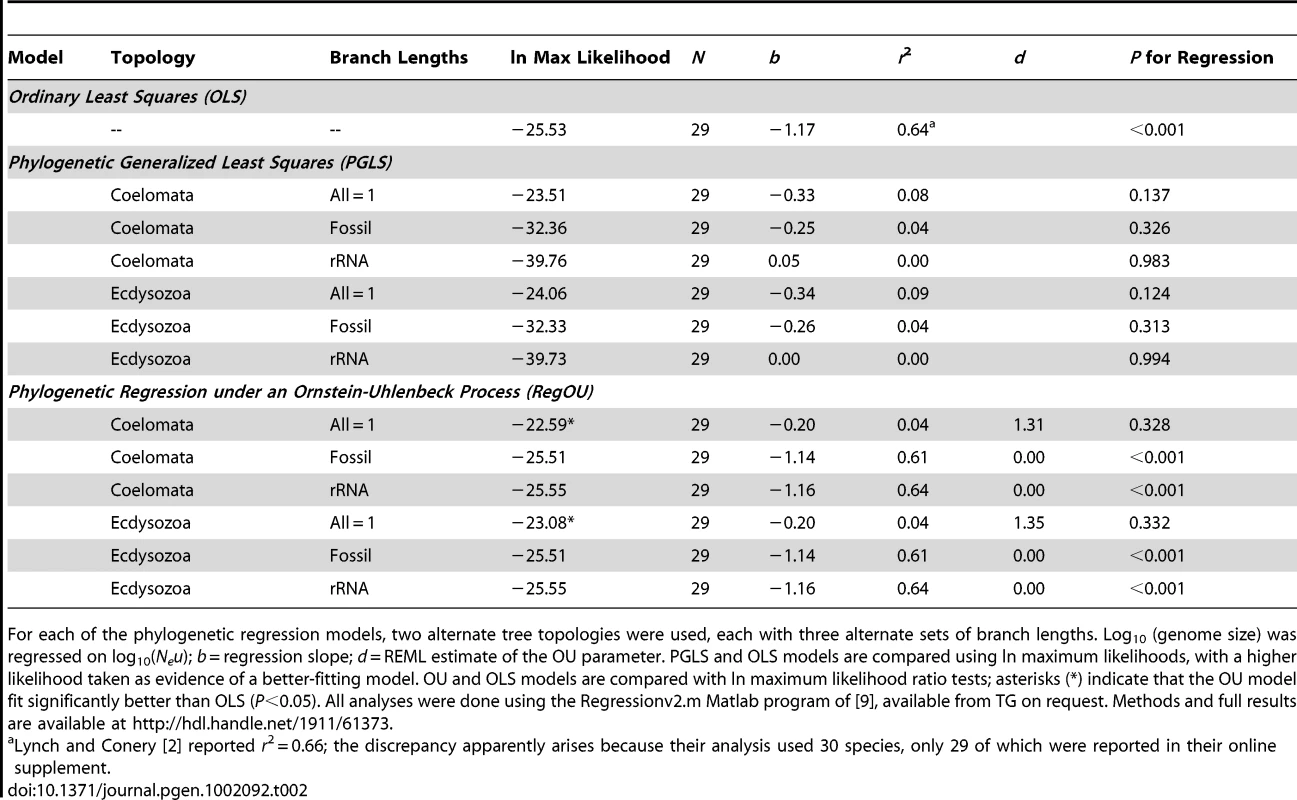
The sensitivity of a phylogenetic comparative analysis is often assessed by examining alternative topologies and/or branch lengths (e.g., [16]). To assess the robustness of the WG results, we have investigated a second topology suggested by Lynch [3] and two additional sets of branch lengths. The WG topology followed the “Coelomata hypothesis,” whereas the alternate topology reflects the “Ecdysozoa hypothesis” and unites nematodes and arthropods in a monophyletic group [17]. We did not investigate a third topology suggested by Lynch, as it is not supported in recent analyses [18]–[20]. Three sets of branch lengths were calculated for the two trees: arbitrary lengths (all = 1) as in WG, lengths derived from fossil-based divergence times, and lengths based on ribosomal RNA substitutions. Full methodological details are available as supplementary material from the Rice Digital Scholarship Archive at http://hdl.handle.net/1911/61373. Consistent with the WG results, none of the six phylogenetic generalized least-squares (PGLS) analyses found statistically significant relationships between Neu and genome size, and the models using all = 1 branch lengths best fit the data (had the highest likelihoods) regardless of the topology (Table 2). Thus, the conclusion of no relationship between Neu and genome size appears robust to substantial variation in topologies and branch lengths.
The analyses of topologies and branch lengths described above (including the star topology assumed by OLS) all assume a Brownian motion–like model of residual trait evolution. If residual evolution has not been Brownian motion–like, then both PGLS and OLS analyses may be suspect. This is why WG explored an additional model—the Ornstein-Uhlenbeck (OU) model, which is based on a diffusion process in which a particle wanders via a random walk, but is bounded by a restraining force whose power increases with distance from the starting point [7], [21]. Felsenstein ([21], p. 464) argued that the OU process is a good model for “the motion of a population which is wandering back and forth on a selective peak under the influence of genetic drift” or for “the wanderings of an adaptive peak in the phenotype space.” WG verified that a regression model with residuals modeled as an OU process (RegOU; [9]) fit significantly better than OLS, and found that it also did not support a relationship between Neu and genome size. We have expanded those results by examining RegOU models for the full set of topologies and branch lengths (Table 2). Again, the best-fitting models for both topologies had starter branch lengths of 1.0 and did not support a significant relationship between Neu and genome size (Table 2).
Thresholds
Lynch [3] states that the MH hypothesis predicts threshold (nonlinear) relationships on a log scale between Neu and measures of genome complexity, including genome size. Therefore, he argues that the WG analyses of linear relationships are inherently flawed. We find this argument inconsistent, given that a central analysis of LC examines the relationship between log Neu and log genome size and reports a highly significant linear relationship (r2 = 0.66; their Figure 1b). Furthermore, neither LC nor [22] discuss thresholds or nonlinearity in the Neu / genome size relationship, nor is there obvious visual evidence of thresholds in the data (Figure 1b of [2]; Figure 4.8 of [22]; Figure 3a of [1]). As with genome size, three of the remaining six attributes analyzed in WG (gene number, the half-life of gene duplicates, and intron size) are clearly not associated with thresholds in LC, given that they are presented as linear relationships or, in the case of gene number, a slightly curvilinear relationship (see Figures 1–3 of [2]).
WG did perhaps err in conducting linear analyses of Neu against three other genomic attributes associated with thresholds in LC: intron number, transposon number, and transposon fraction. However, Lynch's argument that a “substantial reduction in the correlation of [Neu with] genomic attributes” does not contradict the MH hypothesis but instead follows from WG's use of phylogenetic techniques is not correct: the problem is not that WG used PGLS, but that within PGLS, they chose to model linear rather than threshold relationships for these particular attributes. PGLS is capable of modeling any relationship possible with OLS [23], including linear, polynomial, and break-point relationships (e.g., segmented regression [24]).
A simple approach to test for threshold effects of Neu is via the PGLS equivalent of ANCOVA [9] on two groups separated into low versus high Neu. Of the 15 species with Neu and intron number data in the LC dataset, only two fall into the “high” Neu class (Neu>0.015); similarly, of the 18 species with transposon number (or fraction) data, only three fall into the “high” Neu class (Neu>0.0128). These highly unbalanced designs do not allow confidence in analysis via either regular or phylogenetic ANCOVA. Therefore, the LC dataset does not permit robust conclusions about the responses of introns and transposons to Neu thresholds, regardless of whether one utilizes phylogenetic or “nonphylogenetic” techniques.
Lessons from Other Studies
Lynch takes issue with WG's interpretations of two other studies. In both cases, he argues that the metric used to estimate the strength of drift/selection (allozyme-derived Ne [25]; Ka/Ks [26]) is inappropriate for investigating relationships between drift and genome complexity. We argue below that allozyme-derived Ne is in fact informative for the dataset in [25]. The merits of Ka/Ks have been discussed elsewhere [26]–[28] and will not be treated further here. Despite concerns about the Ka/Ks metric, Lynch [3] nonetheless views the results in bacteria [26] as “compelling support” for the MH hypothesis.
Whitney et al. [25] examined allozyme-based estimates of Ne and genome size for 205 species of seed plants; using phylogenetically independent contrasts, no significant relationship was detected. (OLS analysis found a significant negative relationship, apparently the basis of Lynch's characterization of the results as “consistent” with the MH hypothesis.) Lynch argues first that allozyme data are not useful for estimating Neu, because allozymes are products of protein-sequence variation and thus are less reliable surrogates of neutral variation than silent sites. We agree that there are likely constraints on allozyme H that limit the maximum Neu that can be estimated; however, it does not follow that the signal of Neu is completely erased. In fact, as discussed in [25], a significant positive correlation exists between allozyme-based and sequence-based Neu estimates in a subset of the plant dataset. Furthermore, for a subset of the LC dataset for which allozyme data were available, allozyme-based Neu was as strongly related to genome size as was sequence-based Neu [25]. Lynch also argues that regressions in [25] should have used Neu rather than Ne. In that analysis, Ne was calculated from heterozygosity H via Ne = ((1–H)−2–1)/(8u), assuming a constant u of 10−5. That assumption means that, computationally, it makes absolutely no difference whether Neu or Ne were used; neither had a significant relationship with genome size in phylogenetic analyses.
Kuo et al. [26] analyzed 42 paired bacterial genomes, using the efficacy of purifying selection in coding regions (as estimated by Ka/Ks) to quantify genetic drift. Bacterial taxa experiencing greater levels of genetic drift—implying a smaller evolutionary Ne—had smaller genomes. Lynch [3] argues that these results support the MH hypothesis because “the theory predicts that with increasing power of random genetic drift, effectively neutral genomic features will evolve in the direction of mutation bias” and because “there is a deletion bias in bacteria” in contrast to an insertion bias in eukaryotes. Thus, the predicted Neu and genome size/complexity relationship is positive for prokaryotes and negative for eukaryotes. These statements appear to represent a revision of the MH hypothesis, which in previous treatments [2], [22] had assumed an insertion bias in both groups and a continuous, negative Neu versus genome size relationship across prokaryotes and eukaryotes.
The assertion that mutation bias differs in direction for prokaryotes and eukaryotes is difficult to evaluate. We note that studies examining mutation bias typically find a deletion bias in both groups (e.g., [29] and references therein). More importantly, most of these studies use sequence data from diverged lineages to estimate the ratio of insertions to deletions. In previous discussions, Lynch has argued [22], [30] that such studies do not accurately estimate the quantity of interest (de novo mutation bias), in contrast to lab mutation accumulation studies involving relaxation of selection. We agree: indels in sequence data from naturally diverged lineages reflect not only mutation but also subsequent selection and drift and thus may not represent the de novo mutation spectrum. However, lab mutation accumulation studies [31], [32] are simply too few to allow generalizations about mutation biases in prokaryotes versus eukaryotes. The lack of hard data on de novo mutation bias means that any nonzero correlation between Neu and genome size can be judged “consistent” with the MH hypothesis simply by claiming the appropriate mutation bias.
Regardless, the new prediction for decreasing prokaryotic genome size with decreasing Neu is not supported by the LC dataset, whether analyzed using “nonphylogenetic” or phylogenetic methods. We regressed genome size on Neu using both OLS and PGLS for just the seven bacterial species and found no statistical relationship in either analysis (b = −0.19 and −0.11, P = 0.47 and 0.49, respectively). Although the sample size is small, we note the trends are for genome size and Neu to move in opposite directions, counter to the prediction if a deletion bias in bacteria is assumed.
In summary, the datasets of Whitney et al. [25] and of LC do not support the MH hypothesis regardless of the assumed direction of mutation bias. The Kuo et al. data [26] contradict the MH hypothesis, assuming a universal insertion bias, but support it under an assumption of a deletion bias in prokaryotes. We conclude, as did WG, that current comparative datasets examining drift and genome size provide little support for the MH hypothesis.
Conclusions
We agree with Lynch [3] that the MH hypothesis should not be rejected based on the difficulty of performing formal hypothesis tests. We note, however, that such difficulty does not in turn justify acceptance based on inappropriate statistical models. We find the theoretical population genetic basis of the original LC argument sound: smaller effective population size should result in an increasing role for drift relative to selection and an increasing probability of fixation of slightly deleterious mutations that alter genome size and complexity. Our focus, however, is not whether effective population size plays a role, but how important it might be relative to numerous other factors that might influence genome size and complexity. Does Neu explain 66% of the variation in genome size across the tree of life, 6%, or 0.6%? The WG analysis and those presented herein suggest that, given the demonstrated phylogenetic nonindependence of the data at hand, the 66% estimate claimed by LC is far too high; in fact, any influence of Neu on genome size is not statistically detectable in better-fitting phylogenetic regression models (Table 2). Finally, we question whether simple regression models (regardless of whether they are phylogenetic or “nonphylogenetic”) can ever provide unequivocal support for the MH hypothesis. One of the major criticisms expressed in WG and in [33] is that Neu is highly correlated with other aspects of organismal biology, including body size, mating system, developmental rate, and metabolic rate. Thus, comparative analyses using only Neu as a predictor variable may be uninformative about the actual mechanisms driving genome size and complexity; multivariate analyses are needed.
Zdroje
1. WhitneyKDGarlandTJr
2010
Did genetic drift drive increases in genome
complexity?
PLoS Genet
6
e1001080
doi:10.1371/journal.pgen.1001080
2. LynchMConeryJS
2003
The origins of genome complexity.
Science
302
1401
1404
3. LynchM
2011
Statistical inference on the mechanisms of genome
evolution.
PLoS Genet
7
e1001389. doi: 10.1371/journal.pgen.1001389
4. GarlandTJrBennettAFRezendeEL
2005
Phylogenetic approaches in comparative
physiology.
J Exp Biol
208
3015
3035
5. GarlandTMidfordPEIvesAR
1999
An introduction to phylogenetically based statistical methods,
with a new method for confidence intervals on ancestral
values.
Am Zool
39
374
388
6. LynchMConeryJS
2004b
Testing genome complexity - response.
Science
304
390
7. BlombergSPGarlandTIvesAR
2003
Testing for phylogenetic signal in comparative data: Behavioral
traits are more labile.
Evolution
57
717
745
8. FreckletonRPHarveyPHPagelM
2002
Phylogenetic analysis and comparative data: A test and review of
evidence.
Am Nat
160
712
726
9. LavinSRKarasovWHIvesARMiddletonKMGarlandT
2008
Morphometrics of the avian small intestine compared with that of
nonflying mammals: A phylogenetic approach.
Physiol Zool
81
526
550
10. HueyRBDeutschCATewksburyJJVittLJHertzPE
2009
Why tropical forest lizards are vulnerable to climate
warming.
Proc Biol Sci
276
1939
1948
11. SwansonDLGarlandT
2009
The evolution of high summit metabolism and cold tolerance in
birds and its impact on present-day distributions.
Evolution
63
184
194
12. RicklefsREStarckJM
1996
Applications of phylogenetically independent contrasts: A mixed
progress report.
Oikos
77
167
172
13. RohlfFJ
2006
A comment on phylogenetic correction.
Evolution
60
1509
1515
14. IvesARMidfordPEGarlandT
2007
Within-species variation and measurement error in phylogenetic
comparative methods.
Syst Biol
56
252
270
15. Diaz-UriarteRGarlandT
1998
Effects of branch length errors on the performance of
phylogenetically independent contrasts.
Syst Biol
47
654
672
16. HutcheonJMGarlandT
2004
Are megabats big?
J Mammal Evol
11
257
276
17. AdoutteABalavoineGLartillotNLespinetOPrud'hommeB
2000
The new animal phylogeny: Reliability and
implications.
Proc Natl Acad Sci U S A
97
4453
4456
18. DelsucFBrinkmannHChourroutDPhilippeH
2006
Tunicates and not cephalochordates are the closest living
relatives of vertebrates.
Nature
439
965
968
19. DunnCWHejnolAMatusDQPangKBrowneWE
2008
Broad phylogenomic sampling improves resolution of the animal
tree of life.
Nature
452
745-U745
20. PhilippeHDerelleRLopezPPickKBorchielliniC
2009
Phylogenomics revives traditional views on deep animal
relationships.
Curr Biol
19
706
712
21. FelsensteinJ
1988
Phylogenies and quantitative characters.
Annu Rev Ecol Syst
19
445
471
22. LynchM
2007
The origins of genome architecture.
Sunderland (Massachusetts)
Sinauer Associates
23. GarlandTIvesAR
2000
Using the past to predict the present: Confidence intervals for
regression equations in phylogenetic comparative methods.
Am Nat
155
346
364
24. ChappellR
1989
Fitting bent lines to data, with applications to
allometry.
J Theor Biol
138
235
256
25. WhitneyKDBaackEJHamrickJLGodtMJWBarringerBC
2010
A role for nonadaptive processes in plant genome size
evolution?
Evolution
64
2097
2109
26. KuoCHMoranNAOchmanH
2009
The consequences of genetic drift for bacterial genome
complexity.
Genome Res
19
1450
1454
27. DaubinVMoranNA
2004
Comment on "The origins of genome complexity".
Science
306
978a
28. YangZHBielawskiJP
2000
Statistical methods for detecting molecular
adaptation.
Trends Ecol Evol
15
496
503
29. KuoCHOchmanH
2009
Deletional bias across the three domains of life.
Genome Biol Evol
1
145
152
30. LynchMConeryJS
2004a
Response to comment on "The origins of genome
complexity".
Science
306
978a
31. DenverDRMorrisKLynchMThomasWK
2004
High mutation rate and predominance of insertions in the
Caenorhabditis elegans nuclear genome.
Nature
430
679
682
32. NilssonAIKoskiniemiSErikssonSKugelbergEHintonJCD
2005
Bacterial genome size reduction by experimental
evolution.
Proc Natl Acad Sci U S A
102
12112
12116
33. CharlesworthBBartonN
2004
Genome size: Does bigger mean worse?
Curr Biol
14
R233
R235
34. KembelSWCowanPDHelmusMRCornwellWKMorlonH
2010
Picante: R tools for integrating phylogenies and
ecology.
Bioinformatics
26
1463
1464
35. R Development Core Team
2010
R: A language and environment for statistical
computing.
Version 2.11.1
Vienna
R Foundation for Statistical Computing
Štítky
Genetika Reprodukční medicína
Článek Local Absence of Secondary Structure Permits Translation of mRNAs that Lack Ribosome-Binding SitesČlánek Independent Chromatin Binding of ARGONAUTE4 and SPT5L/KTF1 Mediates Transcriptional Gene SilencingČlánek Trade-Off between Bile Resistance and Nutritional Competence Drives Diversification in the Mouse GutČlánek FGF Signaling Regulates the Number of Posterior Taste Papillae by Controlling Progenitor Field SizeČlánek Mammalian BTBD12 (SLX4) Protects against Genomic Instability during Mammalian SpermatogenesisČlánek Identification of Nine Novel Loci Associated with White Blood Cell Subtypes in a Japanese PopulationČlánek Differential Effects of and Risk Variants on Association with Diabetic ESRD in African AmericansČlánek Dynamic Chromatin Localization of Sirt6 Shapes Stress- and Aging-Related Transcriptional Networks
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2011 Číslo 6
-
Všechny články tohoto čísla
- Local Absence of Secondary Structure Permits Translation of mRNAs that Lack Ribosome-Binding Sites
- Statistical Inference on the Mechanisms of Genome Evolution
- Revisiting Heterochromatin in Embryonic Stem Cells
- A Two-Stage Meta-Analysis Identifies Several New Loci for Parkinson's Disease
- Identification of a Sudden Cardiac Death Susceptibility Locus at 2q24.2 through Genome-Wide Association in European Ancestry Individuals
- Genomic Prevalence of Heterochromatic H3K9me2 and Transcription Do Not Discriminate Pluripotent from Terminally Differentiated Cells
- Epistasis between Beneficial Mutations and the Phenotype-to-Fitness Map for a ssDNA Virus
- Recurrent Chromosome 16p13.1 Duplications Are a Risk Factor for Aortic Dissections
- Telomere DNA Deficiency Is Associated with Development of Human Embryonic Aneuploidy
- Genome-Wide Association Study of White Blood Cell Count in 16,388 African Americans: the Continental Origins and Genetic Epidemiology Network (COGENT)
- Unexpected Role for DNA Polymerase I As a Source of Genetic Variability
- Transportin-SR Is Required for Proper Splicing of Genes and Plant Immunity
- How Chromatin Is Remodelled during DNA Repair of UV-Induced DNA Damage in
- Independent Chromatin Binding of ARGONAUTE4 and SPT5L/KTF1 Mediates Transcriptional Gene Silencing
- Two Evolutionary Histories in the Genome of Rice: the Roles of Domestication Genes
- Natural Allelic Variation Defines a Role for : Trichome Cell Fate Determination
- Multiple Common Susceptibility Variants near BMP Pathway Loci , , and Explain Part of the Missing Heritability of Colorectal Cancer
- Pathogenic Mechanism of the FIG4 Mutation Responsible for Charcot-Marie-Tooth Disease CMT4J
- A Functional Variant in Promoter Modulates Its Expression and Confers Disease Risk for Systemic Lupus Erythematosus
- Drift and Genome Complexity Revisited
- Chromosomal Macrodomains and Associated Proteins: Implications for DNA Organization and Replication in Gram Negative Bacteria
- Trade-Off between Bile Resistance and Nutritional Competence Drives Diversification in the Mouse Gut
- Pathways of Distinction Analysis: A New Technique for Multi–SNP Analysis of GWAS Data
- Web-Based Genome-Wide Association Study Identifies Two Novel Loci and a Substantial Genetic Component for Parkinson's Disease
- Chk2 and p53 Are Haploinsufficient with Dependent and Independent Functions to Eliminate Cells after Telomere Loss
- Exome Sequencing Identifies Mutations in High Myopia
- Distinct Functional Constraints Partition Sequence Conservation in a -Regulatory Element
- CorE from Is a Copper-Dependent RNA Polymerase Sigma Factor
- A Single Sex Pheromone Receptor Determines Chemical Response Specificity of Sexual Behavior in the Silkmoth
- FGF Signaling Regulates the Number of Posterior Taste Papillae by Controlling Progenitor Field Size
- Maps of Open Chromatin Guide the Functional Follow-Up of Genome-Wide Association Signals: Application to Hematological Traits
- Increased Susceptibility to Cortical Spreading Depression in the Mouse Model of Familial Hemiplegic Migraine Type 2
- Differential Gene Expression and Epiregulation of Alpha Zein Gene Copies in Maize Haplotypes
- Parallel Adaptive Divergence among Geographically Diverse Human Populations
- Genetic Analysis of Genome-Scale Recombination Rate Evolution in House Mice
- Mechanisms for the Evolution of a Derived Function in the Ancestral Glucocorticoid Receptor
- Mammalian BTBD12 (SLX4) Protects against Genomic Instability during Mammalian Spermatogenesis
- Interferon Regulatory Factor 8 Regulates Pathways for Antigen Presentation in Myeloid Cells and during Tuberculosis
- High-Resolution Analysis of Parent-of-Origin Allelic Expression in the Arabidopsis Endosperm
- Specific SKN-1/Nrf Stress Responses to Perturbations in Translation Elongation and Proteasome Activity
- Graded Nodal/Activin Signaling Titrates Conversion of Quantitative Phospho-Smad2 Levels into Qualitative Embryonic Stem Cell Fate Decisions
- Genome-Wide Analysis Reveals PADI4 Cooperates with Elk-1 to Activate Expression in Breast Cancer Cells
- Trait Variation in Yeast Is Defined by Population History
- Meiosis-Specific Loading of the Centromere-Specific Histone CENH3 in
- A Genome-Wide Survey of Imprinted Genes in Rice Seeds Reveals Imprinting Primarily Occurs in the Endosperm
- Multiple Regulatory Mechanisms to Inhibit Untimely Initiation of DNA Replication Are Important for Stable Genome Maintenance
- SIRT1 Promotes N-Myc Oncogenesis through a Positive Feedback Loop Involving the Effects of MKP3 and ERK on N-Myc Protein Stability
- Bacteriophage Crosstalk: Coordination of Prophage Induction by Trans-Acting Antirepressors
- Role of the Single-Stranded DNA–Binding Protein SsbB in Pneumococcal Transformation: Maintenance of a Reservoir for Genetic Plasticity
- Genomic Convergence among ERRα, PROX1, and BMAL1 in the Control of Metabolic Clock Outputs
- Genome-Wide Association of Bipolar Disorder Suggests an Enrichment of Replicable Associations in Regions near Genes
- Identification of Nine Novel Loci Associated with White Blood Cell Subtypes in a Japanese Population
- DNA Ligase III Promotes Alternative Nonhomologous End-Joining during Chromosomal Translocation Formation
- Differential Effects of and Risk Variants on Association with Diabetic ESRD in African Americans
- Finished Genome of the Fungal Wheat Pathogen Reveals Dispensome Structure, Chromosome Plasticity, and Stealth Pathogenesis
- Dynamic Chromatin Localization of Sirt6 Shapes Stress- and Aging-Related Transcriptional Networks
- Extracellular Matrix Dynamics in Hepatocarcinogenesis: a Comparative Proteomics Study of Transgenic and Null Mouse Models
- Integrating 5-Hydroxymethylcytosine into the Epigenomic Landscape of Human Embryonic Stem Cells
- Vive La Différence: An Interview with Catherine Dulac
- Multiple Loci Are Associated with White Blood Cell Phenotypes
- Nuclear Accumulation of Stress Response mRNAs Contributes to the Neurodegeneration Caused by Fragile X Premutation rCGG Repeats
- A New Mutation Affecting FRQ-Less Rhythms in the Circadian System of
- Cryptic Transcription Mediates Repression of Subtelomeric Metal Homeostasis Genes
- A New Isoform of the Histone Demethylase JMJD2A/KDM4A Is Required for Skeletal Muscle Differentiation
- Genetic Determinants of Lipid Traits in Diverse Populations from the Population Architecture using Genomics and Epidemiology (PAGE) Study
- A Genome-Wide RNAi Screen for Factors Involved in Neuronal Specification in
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Recurrent Chromosome 16p13.1 Duplications Are a Risk Factor for Aortic Dissections
- Statistical Inference on the Mechanisms of Genome Evolution
- Genome-Wide Association Study of White Blood Cell Count in 16,388 African Americans: the Continental Origins and Genetic Epidemiology Network (COGENT)
- Chromosomal Macrodomains and Associated Proteins: Implications for DNA Organization and Replication in Gram Negative Bacteria
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání