-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaA New Mutation Affecting FRQ-Less Rhythms in the Circadian System of
We are using the fungus Neurospora crassa as a model organism to study the circadian system of eukaryotes. Although the FRQ/WCC feedback loop is said to be central to the circadian system in Neurospora, rhythms can still be seen under many conditions in FRQ-less (frq knockout) strains. To try to identify components of the FRQ-less oscillator (FLO), we carried out a mutagenesis screen in a FRQ-less strain and selected colonies with altered conidiation (spore-formation) rhythms. A mutation we named UV90 affects rhythmicity in both FRQ-less and FRQ-sufficient strains. The UV90 mutation affects FRQ-less rhythms in two conditions: the free-running long-period rhythm in choline-depleted chol-1 strains becomes arrhythmic, and the heat-entrained rhythm in the frq10 knockout is severely altered. In a FRQ-sufficient background, the UV90 mutation causes damping of the free-running conidiation rhythm, reduction of the amplitude of the FRQ protein rhythm, and increased phase-resetting responses to both light and heat pulses, consistent with a decreased amplitude of the circadian oscillator. The UV90 mutation also has small but significant effects on the period of the conidiation rhythm and on growth rate. The wild-type UV90 gene product appears to be required for a functional FLO and for sustained, high-amplitude rhythms in FRQ-sufficient conditions. The UV90 gene product may therefore be a good candidate for a component of the FRQ-less oscillator. These results support a model of the Neurospora circadian system in which the FRQ/WCC feedback loop mutually interacts with a single FLO in an integrated circadian system.
Published in the journal: . PLoS Genet 7(6): e32767. doi:10.1371/journal.pgen.1002151
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1002151Summary
We are using the fungus Neurospora crassa as a model organism to study the circadian system of eukaryotes. Although the FRQ/WCC feedback loop is said to be central to the circadian system in Neurospora, rhythms can still be seen under many conditions in FRQ-less (frq knockout) strains. To try to identify components of the FRQ-less oscillator (FLO), we carried out a mutagenesis screen in a FRQ-less strain and selected colonies with altered conidiation (spore-formation) rhythms. A mutation we named UV90 affects rhythmicity in both FRQ-less and FRQ-sufficient strains. The UV90 mutation affects FRQ-less rhythms in two conditions: the free-running long-period rhythm in choline-depleted chol-1 strains becomes arrhythmic, and the heat-entrained rhythm in the frq10 knockout is severely altered. In a FRQ-sufficient background, the UV90 mutation causes damping of the free-running conidiation rhythm, reduction of the amplitude of the FRQ protein rhythm, and increased phase-resetting responses to both light and heat pulses, consistent with a decreased amplitude of the circadian oscillator. The UV90 mutation also has small but significant effects on the period of the conidiation rhythm and on growth rate. The wild-type UV90 gene product appears to be required for a functional FLO and for sustained, high-amplitude rhythms in FRQ-sufficient conditions. The UV90 gene product may therefore be a good candidate for a component of the FRQ-less oscillator. These results support a model of the Neurospora circadian system in which the FRQ/WCC feedback loop mutually interacts with a single FLO in an integrated circadian system.
Introduction
Circadian rhythms are approximately 24 h cycles of behavior, physiology, etc. that are driven by an endogenous biological clock. They are found at all levels of eukaryotic life, and in some prokaryotes. Molecular models for the endogenous oscillators that drive these rhythms in eukaryotes are based on rhythmic transcription of a small number of “clock genes”, translation of the rhythmic transcripts into rhythmic levels of clock proteins, and negative regulation of the clock genes by their own proteins. These models have been called transcription/translation feedback loops (TTFL).
Although a great deal of evidence has accumulated to support the concept of negative and positive feedback loops regulating the expression of these clock genes, there are many indications that the TTFL model is inadequate as a complete clock mechanism (reviewed in [1]). In recent mammalian examples, it was found that cycling of mammalian clock proteins CLOCK and CRY is not required for rhythmicity in fibroblasts [2]; large reductions in overall transcription rate and levels of clock proteins do not eliminate circadian oscillations in mouse fibroblasts [3]; and rhythmic cAMP signaling is required to sustain rhythmic transcription in mammalian cells [4]. These and other findings are not compatible with the canonical TTFL models.
In the prokaryotic cyanobacterium Synechococcus elongatus, a post-translational oscillator has been shown to be the core pacemaker. It can operate in vitro in the absence of any transcription or translation, but in vivo a TTFL is coupled to it as a slave oscillator [5], [6]. The architecture of the cyanobacterial clock provides certain advantages that may be relevant to eukaryotic clock systems as well [6]. Progress in understanding circadian oscillators in eukaryotes will depend on identifying clock components outside of the TTFL [7], [8].
The filamentous fungus Neurospora crassa is a model organism that has provided many insights into the molecular basis of circadian rhythmicity [9]–[11]. Asexual spore formation (conidiation) is controlled by the circadian clock in Neurospora, and rhythmic spore formation can be easily monitored during growth as a pattern of thick conidiation “bands” alternating with thin growth in “interbands” as the fungal mycelium advances across a solid agar surface. The current model for the Neurospora circadian oscillator (called the FRQ/WCC TTFL) consists of interlocked negative and positive feedback loops involving the clock genes frq, wc-1 and wc-2 [9], in which a complex of the WC-1 and WC-2 proteins (WCC) activates transcription of frq and is in turn negatively regulated by FRQ protein.
However, there have been many reports of rhythmicity with periods in the circadian range that can be seen in strains with null mutations in frq or wc genes. These include conidiation rhythms [12]–[22] and molecular/biochemical rhythms [23]–[26]. These rhythms are said by some to be driven by many different “FRQ-less oscillators” (FLOs) that may be completely separate from the FRQ/WCC feedback loop [26]–[28]. An alternative view sees the FRQ/WCC TTFL as a component of a larger circadian system in which the FRQ/WCC TTFL interacts with a single FLO [29]. Neurospora provides a unique eukaryotic system in which we can directly access clock components outside of the TTFL by assaying rhythmicity in FRQ-less strains in which the TTFL is not functioning. We are using this system to identify components of FLO by searching for mutations affecting rhythmicity in FRQ-less strains. Such mutations can also provide insight into the interactions between the FRQ/WCC TTFL and the FLO. The new mutation we report here disrupts two FRQ-less rhythms and also affects the FRQ/WCC TTFL, suggesting that a single FLO may interact with the FRQ/WCC TTFL in a circadian architecture similar to that of cyanobacteria.
Results
Isolation and initial characterization of a mutation affecting FRQ-less rhythmicity
To isolate mutations affecting the FLO, we conducted a mutagenesis screen to identify mutations affecting conidiation rhythms in a frq knockout background. We used UV light to mutagenize uninucleate microconidial spores of a strain carrying both the frq10 knockout allele and chol-1. Strains carrying the chol-1 mutation express conidiation rhythms in FRQ-less strains on choline-deficient media, and these rhythms are therefore an example of a FLO [16]. On high choline, chol-1 strains are identical to chol+ [16]. The parent strain for mutagenesis was both frq10 and chol-1 and was therefore rhythmic without choline but arrhythmic on high choline (Figure 1A, upper 4 tubes). We analyzed 600 colonies derived from mutagenized spores on individual race tubes and found one mutant that was arrhythmic both with and without choline (Figure 1A, lower 4 tubes). The mutation was named UV90, as it was the 90th spore analyzed after UV mutagenesis.
Fig. 1. Phenotypes of UV90 mutants. 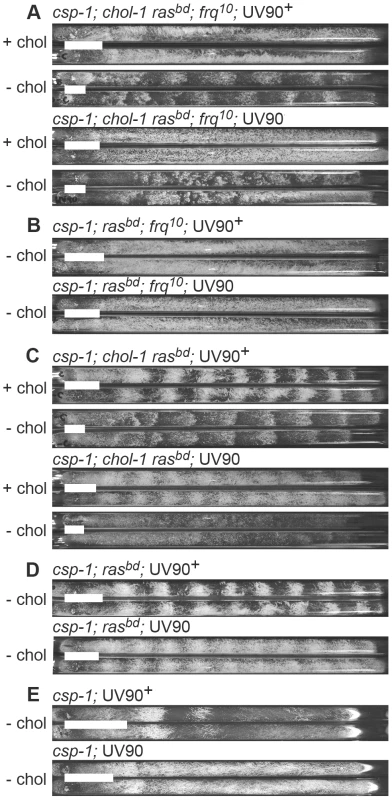
Cultures were grown at 22°C in DD in race tubes on MA medium with (+ chol) or without (−chol) 100 µM choline. Growth was from left to right. White bars represent an average distance for 24 h of growth. Two sibling strains from a cross are shown in pairs. All strains carry the csp-1 mutation. A–D: Progeny are from the backcross of csp-1; chol-1 rasbd; frq10; UV90 to rasbd. A. All strains carry the ras-1bd, frq10 and chol-1 mutations and were grown with or without choline. B. All strains carry the ras-1bd and frq10 mutations and were grown without choline. C. All strains carry the ras-1bd and chol-1 mutations and were grown with or without choline. D. All strains carry the ras-1bd mutation and were grown without choline. E. Progeny are from the cross of csp-1; rasbd; UV90 to Mauriceville. All strains were grown without choline. This strain, genotype csp-1; chol-1 rasbd; frq10; UV90, was backcrossed to rasbd and the phenotypes of the progeny were analyzed. The chol-1 phenotype was assayed by comparing growth rates on choline-supplemented and choline-deficient media. The conidiation banding phenotypes were easily identified among the progeny and these phenotypes were consistently identified by two individuals using blind-coded tubes or photographs. Periods and growth rates of all strains described below are listed in Table 1, and conidiation phenotypes are presented in Figure 1.
Tab. 1. Periods and growth rates of UV90 wild-type and mutant strains. 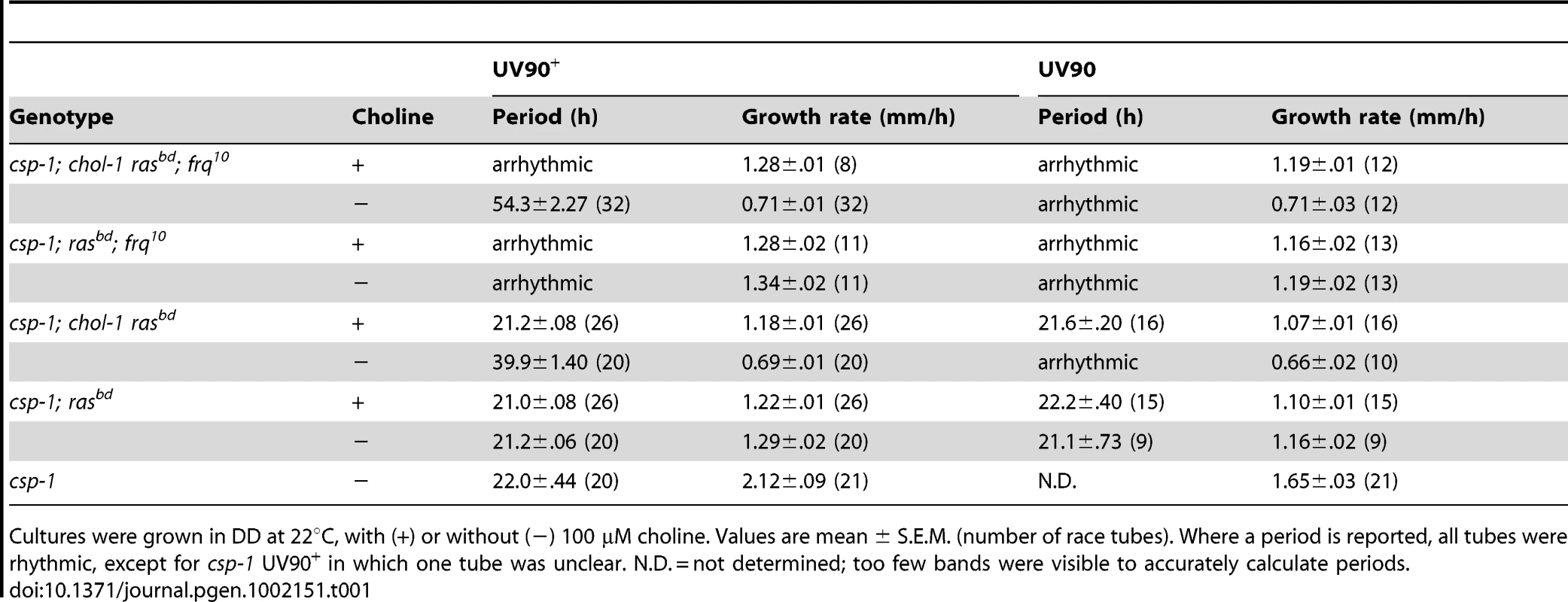
Cultures were grown in DD at 22°C, with (+) or without (−) 100 µM choline. Values are mean ± S.E.M. (number of race tubes). Where a period is reported, all tubes were rhythmic, except for csp-1 UV90+ in which one tube was unclear. N.D. = not determined; too few bands were visible to accurately calculate periods. The chol-1 progeny were identified by slow growth on low choline medium and rapid growth on high choline (Table 1). Among the chol-1 progeny, the frq10; UV90 double mutant phenotype was identified by its similarity to the mutant parent: short aerial hyphae with very even conidiation on high choline (Figure 1A, tubes 5 and 6) and heavier conidiation and disrupted rhythmicity on low choline (Figure 1A, tubes 7 and 8). Densitometry traces of individual chol-1; frq10 progeny are presented in Figure 2. The chol+ progeny were identified by rapid growth on both high and low choline (Table 1). Among the chol+ progeny, the frq10; UV90 double mutants (Figure 1B) were identified as having a phenotype on choline-free medium that was identical to the chol-1; frq10; UV90 parent strain on high choline (Figure 1A, tubes 5 and 6).
Fig. 2. Densitometry of chol-1; frq10 progeny. 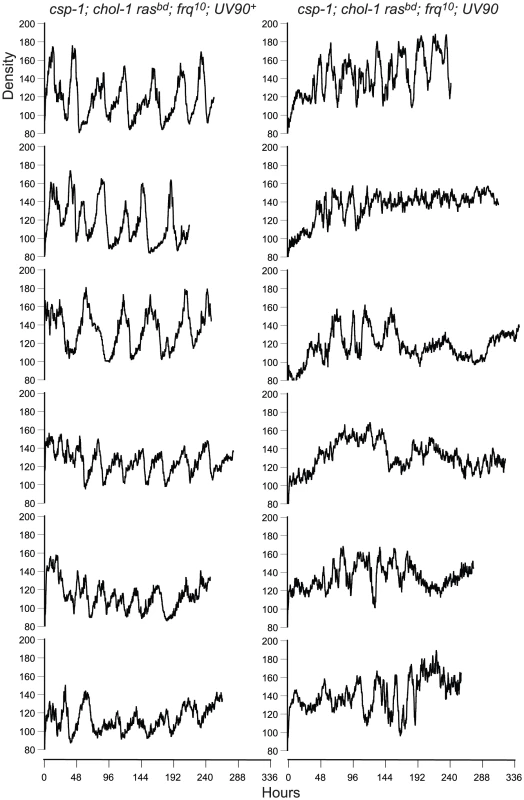
Race tube cultures of csp-1; chol-1 rasbd; frq10 progeny from the backcross of csp-1; chol-1 rasbd; frq10; UV90 to rasbd were analyzed by densitometry. Six progeny from each class were randomly chosen for analysis. Left column: UV90+ progeny. Right column: UV90 progeny. All cultures were grown without choline. Density is in arbitrary units. Time is in hours after transfer from LL (constant light) to DD. When the frq10 mutation was crossed out of the original UV90 strain, the effects of the UV90 mutation on the circadian system when the FRQ/WCC oscillator is functional could be observed. The chol-1; frq+ progeny (Figure 1C) were identified by the presence of rhythmicity on high choline (Figure 1C, tubes 1, 2, 5 and 6). Progeny in which the long-period rhythm on low choline was abolished (Figure 1C, tubes 7 and 8) were identified as UV90. Densitometry traces of individual chol-1; frq+ progeny are presented in Figure 3. These progeny also showed a low-amplitude rhythm on high choline (Figure 1C, tubes 5 and 6).
Fig. 3. Densitometry of chol-1; frq+ progeny. 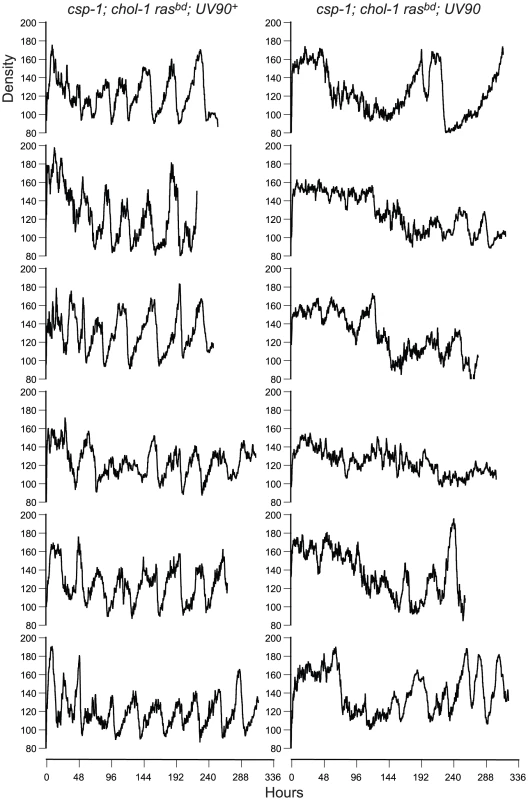
Race tube cultures of csp-1; chol-1 rasbd progeny from the backcross of csp-1; chol-1 rasbd; frq10; UV90 to rasbd were analyzed by densitometry. All other conditions as in Figure 2. The chol+; frq+ progeny (Figure 1D) were identified as indistinfguishable on choline-deficient medium from the phenotypes of the chol-1 progeny on high choline (Figure 1C, tubes 1, 2, 5 and 6). The UV90 progeny were identified by the damped conidiation rhythm (Figure 1D, tubes 3 and 4). The low amplitude rhythm of frq+ UV90 strains was due to an increase in average levels of conidiation, as shown by densitometry (Figure 4A).
Fig. 4. Densitometry of chol+; frq+ progeny. 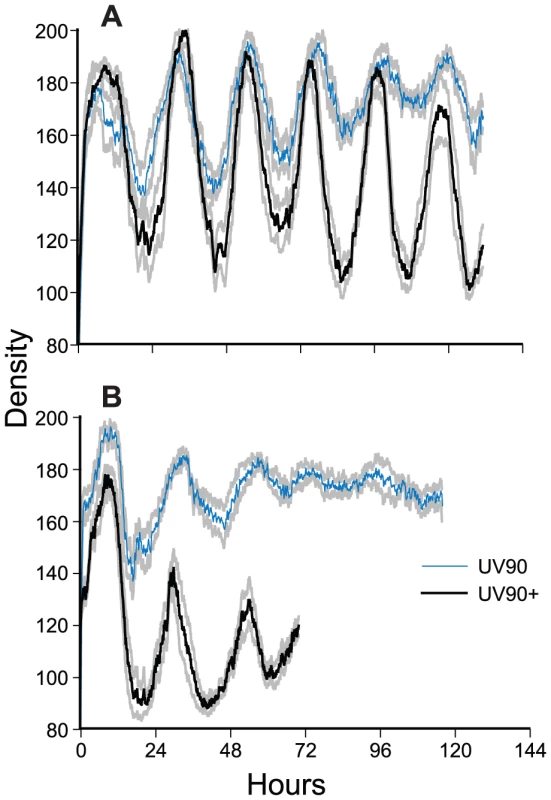
Race tube cultures of progeny from the cross of csp-1; rasbd; UV90 to Mauriceville were analyzed by densitometry. Six csp-1 progeny from each class were randomly chosen for analysis. Densitometry traces were averaged for each class and plotted ± one S.E.M. A. rasbd progeny. B. ras+ progeny. Thick black lines: UV90+ progeny. Thin blue lines: UV90 progeny. Gray lines: ± one S.E.M. All cultures were grown without choline. Density is in arbitrary units. Time is in hours after transfer from LL to DD. One of the csp-1; rasbd; UV90 progeny was backcrossed to the Mauriceville wild-type to remove the rasbd mutation. The ras+ progeny were identified by growth rates more rapid than the rasbd strains (Table 1). Although these progeny (Figure 1E) produced very poor banding rhythms, as expected, a UV90 phenotype was noticeable, increasing conidiation levels and producing nearly constant conidiation (Figure 1E, tubes 3 and 4, and Figure 4B).
We conclude that the UV90 mutation severely compromises the function of the FLO in chol-1, completely disrupting conidiation rhythmicity in the frq10 knockout background in choline-deficient conditions. UV90 also damps the amplitude of the conidiation rhythm in frq+ strains in the presence of a functional FRQ/WCC TTFL.
The UV90 phenotype was found to segregate in backcrosses with the expected 1∶1 ratio for segregation of a single-gene mutation. In the backcross of the original csp-1; chol-1 rasbd; frq10; UV90 mutant to rasbd, out of 88 csp-1; rasbd progeny assayed, 44 were classified as UV90 and 44 as UV90+ based on banding phenotypes on high and low choline. In the cross to Mauriceville of a putative csp-1; rasbd; UV90 strain, out of 40 csp-1; rasbd progeny assayed, 21 were classified as UV90 and 19 were classified as UV90+ based on banding phenotype.
Among the chol-1 progeny of the backcross of the original mutant to rasbd, the two UV90 banding phenotypes segregated together. Out of 45 csp-1; chol-1 rasbd progeny tested, 39 gave clear phenotypes on choline-deficient medium and 6 were unclear. Of those 39 clear phenotypes, 20 produced bands on low choline similar to UV90+ and all had high-amplitude banding rhythms on high choline; 19 were arrhythmic on low choline and all produced low-amplitude bands on high choline similar to UV90. The UV90 mutation slightly decreased the growth rates in most cases (Table 1). This property also co-segregated with the UV90 banding phenotypes. Figure 5 presents growth rate data for frq+ progeny grown on high and low choline media, and it can be seen that progeny with a UV90+ banding phenotype, both chol+ and chol-1, clustered together at higher growth rates than UV90 progeny.
Fig. 5. Segregation of growth rates. 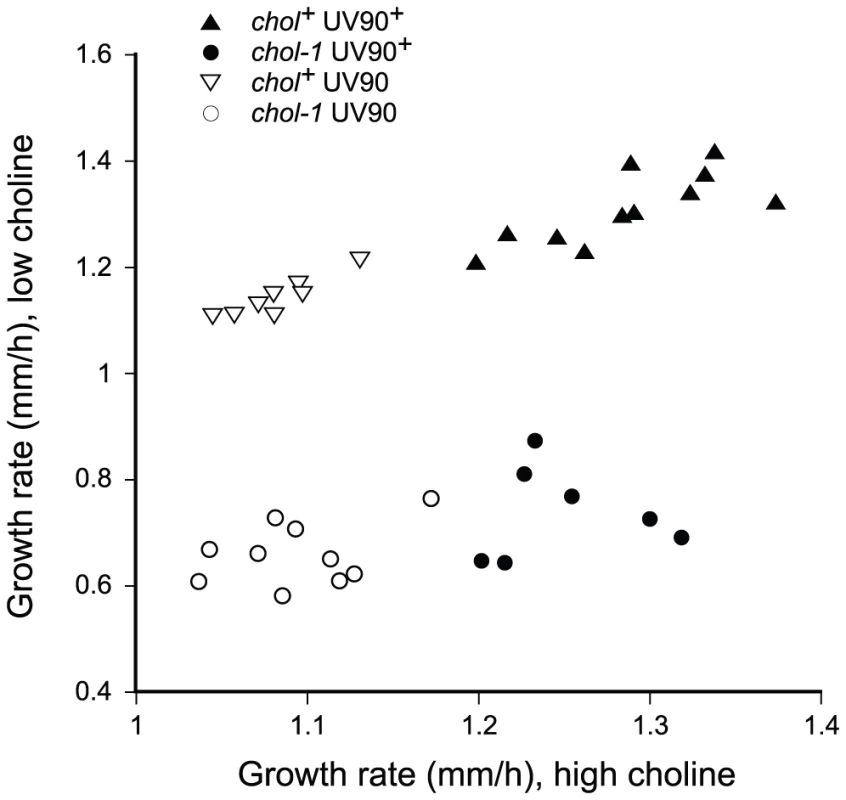
Progeny from the backcross of csp-1; chol-1 rasbd; frq10; UV90 to rasbd were grown at 22°C in DD in race tubes on MA medium with 100 µM choline (high choline) or without added choline (low choline) and growth rates were determined. All progeny carry the csp-1, frq+ and rasbd alleles. Note the change in scales between the X and Y axes. The UV90 mutation mapped to the right arm of linkage group (LG) VI. In the backcross of the original UV90 strain to rasbd, the UV90 phenotype was unlinked to either frq (LG VIIR) or chol-1 (LG IVR): out of 88 progeny tested, 50.0% were recombinant with frq and 48.9% were recombinant with chol-1. In a cross to the multiply-marked alcoy tester strain [30], recombination frequencies were: 51.9% with cot-1 (LG IVL/VR), 40.7% with al-1 (LG IR/IIL) and 16.7% with ylo-1 (LG IIIR/VIR). These results suggested that UV90 is linked to either LG III or LG VI. The original UV90 strain was crossed to the Mauriceville wild-type and mapped using cleaved amplified polymorphic sequence (CAPS) markers and bulked segregant analysis according to the method of Jin et al. [31]. The UV90 phenotype was unlinked to a CAPS marker on LG III (3–52-EcoRI in Jin et al.) and was linked to CAPS markers 6–39-HaeII on the left of the centromere on LG VI, and 6–68-MspI on the right of the centromere. Recombination rates between UV90 and 6–39-HaeII were roughly 5–10% (estimated from the relative band intensities of the cleaved PCR fragments from the bulked segregants) and recombination between UV90 and 6–68-MspI was roughly 0–5%, suggesting that UV90 maps to the right arm of LG VI, in a gene-rich region of the chromosome.
Effects of UV90 on growth rate and temperature compensation of period
One of the defining characteristics of a circadian rhythm is that the period of the rhythm does not change appreciably when the organism is maintained at different constant temperatures; this property of “temperature compensation” distinguishes circadian clocks from simple chemical reactions that increase in rate with an increase in temperature. We assayed the conidiation rhythm of our new mutant strain at different constant growth temperatures to determine the effects of the UV90 mutation on period and temperature compensation of the rhythm. All strains were wild-type for frq. Both chol+ and chol-1 strains were assayed on high choline to repair the defect in chol-1. As shown in Figure 6A, the UV90 mutation significantly increased the period by several hours at some temperatures, and altered the response of the period to temperature in comparison to the UV90+ strains. The UV90 mutation also decreased the growth rate by small but significant amounts at temperatures above 19°C (Figure 6B). These results suggest that the UV90 mutation may have minor effects on metabolism affecting growth, and minor but significant effects on the temperature compensation of the circadian clock in the presence of a functional TTFL.
Fig. 6. Effects of UV90 mutation on growth rate and temperature compensation of period. 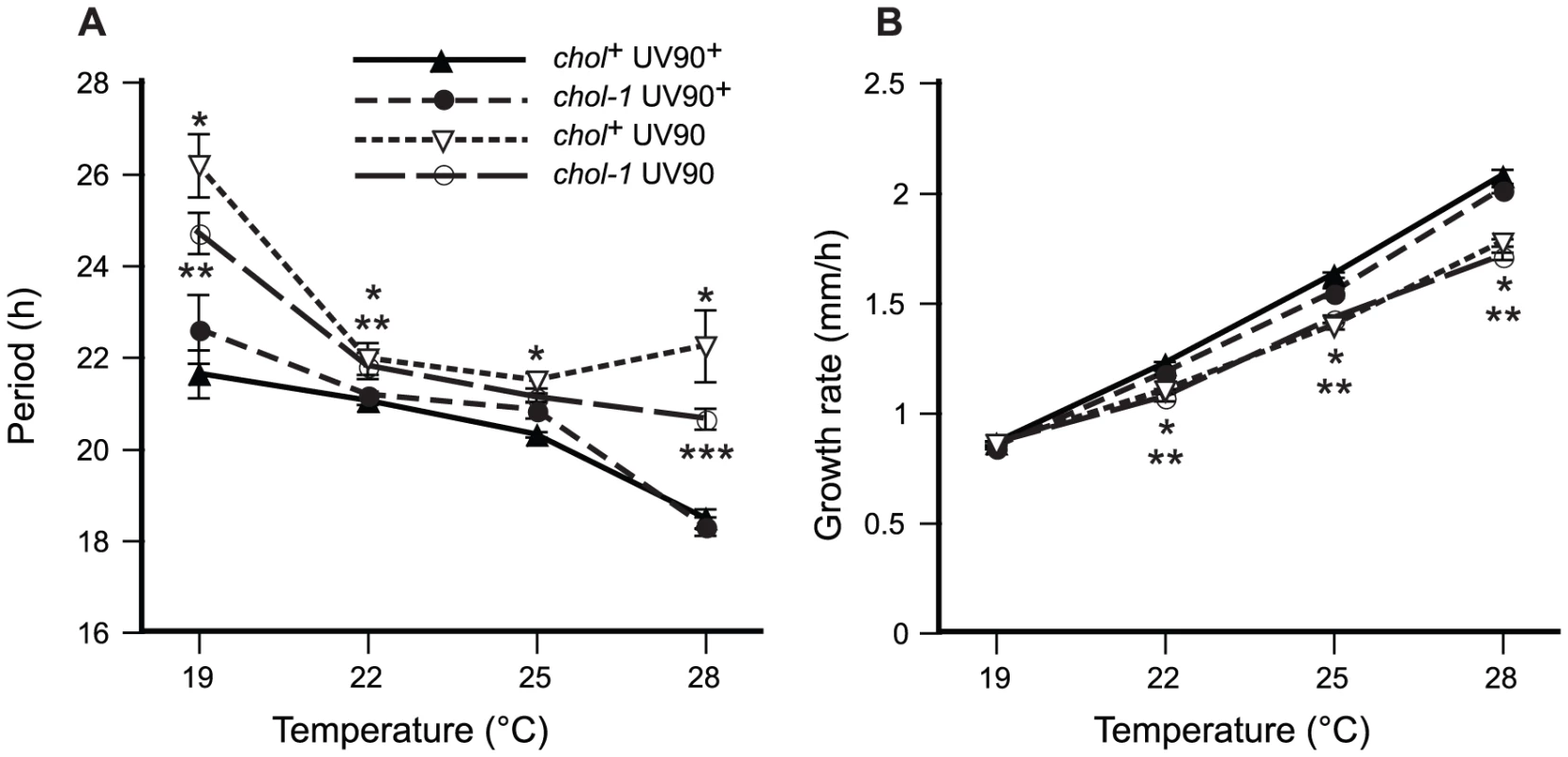
The period of the conidiation rhythm and the growth rate were assayed at different constant growth temperatures on medium containing 100 µM choline. Means are plotted ± one S.E.M., N = 6–26. Closed symbols: UV90+ strains. Open symbols: UV90 strains. Triangles: chol-1+ strains. Circles: chol-1 strains. All strains carry the rasbd and csp-1 mutations and are wild-type for frq. A. Period of the conidiation rhythm, in hours. *: chol+ UV90 significantly different from chol+ UV90+, p<0.01. **: chol-1 UV90 significantly different from chol-1 UV90+, p<0.05. ***: chol-1 UV90 significantly different from chol-1 UV90+, p<0.01. B. Growth rate, in mm/h. Note that most S.E.M.s are smaller than the plot symbols. *: chol+ UV90 significantly different from chol+ UV90+, p<0.01. **: chol-1 UV90 significantly different from chol-1 UV90+, p<0.01. Effects of UV90 on entrainment to heat pulses
It has been shown that FRQ-less strains can be entrained to repeated pulses of high temperature and behave as if they contain a functional heat-entrainable oscillator [15], . This heat-entrainable oscillator is a second example of a FLO. We used 2-hour pulses of 32°C on cultures growing at 22°C to entrain the conidiation rhythm to various T-cycles (where T = total number of hours in the cycle) (Figure 7). Changes in peak timing and peak shape in different T-cycles are indicative of entrainment of an underlying oscillator [19], [32]. In the presence of wild-type frq (Figure 7A–7D), the UV90 mutation had small but significant effects on the shape of the entrained peaks. In the FRQ-less background (Figure 7E–7H), the UV90 mutation had a dramatic effect on the peak timing, shifting the major peak to a much earlier time. We conclude that the UV90 mutation has a greater effect on entrainment behavior in the absence of functional FRQ (in the frq10 strain) than in the presence of functional FRQ, and therefore is likely to primarily affect the FLO rather than the FRQ/WCC feedback loop.
Fig. 7. Effects of UV90 mutation on entrainment to high temperature pulses. 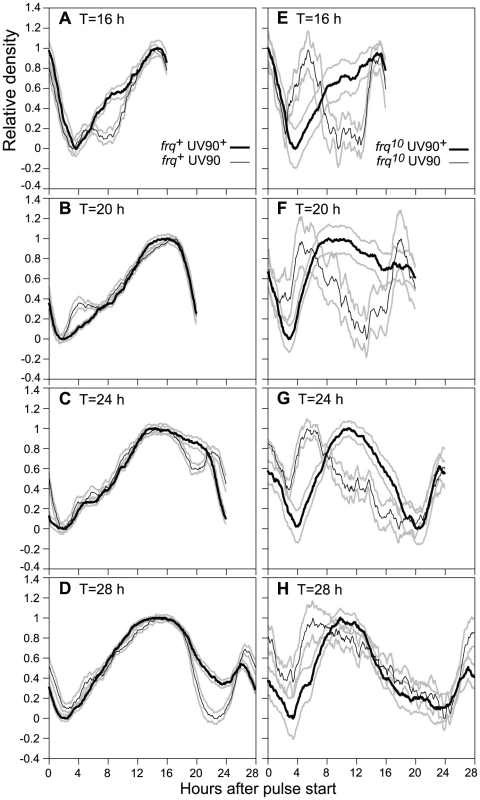
All strains carry the ras-1bd, csp-1 and chol-1 mutations and were grown on high choline to repair the chol-1 defect. Cultures were grown in DD at 22°C and were subjected to 2-h 32°C pulses at repeated intervals of T hours (16, 20, 24 or 28 h), in which T is the length of one cycle from one pulse start to the next pulse start. Culture density was determined and the last two complete cycles on each race tube were averaged for 6 race tubes. Density traces were normalized and are plotted ± one S.E.M., N = 12, against hours after the beginning of the 2-h heat pulse. Thick black lines: wild-type for UV90; thin black lines: UV90 mutant; grey lines: ± S.E.M. A–D. Comparison of UV90 wild-type with UV90 mutant in the frq+ background. E–H. Comparison of UV90 wild-type with UV90 mutant in the frq10 background. Effects of UV90 on phase-resetting
The damping of the amplitude of the conidiation rhythm seen in UV90 mutant strains with functional FRQ (Figure 1D) could be due to either an effect on the output from the circadian oscillator, such as continuous activation of the conidiation developmental pathway, or an effect on the amplitude of the circadian oscillator itself. To distinguish between these two possibilities, the amplitude of the oscillator was probed with phase-resetting stimuli. According to oscillator theory [33], [34], an oscillator with a small amplitude will respond to a particular stimulus with a larger phase resetting response than will an oscillator with a large amplitude. A similar effect should be seen with any type of stimulus, regardless of the input pathway it uses [33].
We used both light pulses and high temperature pulses to probe the amplitude of the oscillator in the UV90 mutant strains in the frq+ chol+ background. Data were plotted in both the traditional phase response curve (PRC) format, plotting phase shifts against the circadian time of the pulse, and in the phase transition curve (PTC) format [35], plotting new phase against old phase. The PTC format is preferred, as it does not introduce artifactual breaks in the data where large phase delays meet large phase advances [36]. As seen in Figure 8, the PRCs for the UV90 mutant showed larger phase shifts than the wild-type for both light and heat stimuli. The PTCs indicate that under these conditions, the wild-type produced type 1 PTCs and the UV90 mutant produced type 0 PTCs. A type 1 PTC is indicative of a weaker response than a type 0 PTC [34]–[36]. Because the same stimuli produced a weak response from wild-type and a stronger response from the UV90 mutant, these results are consistent with a smaller oscillator amplitude for the UV90 mutant [36].
Fig. 8. Effects of UV90 mutation on phase-resetting. 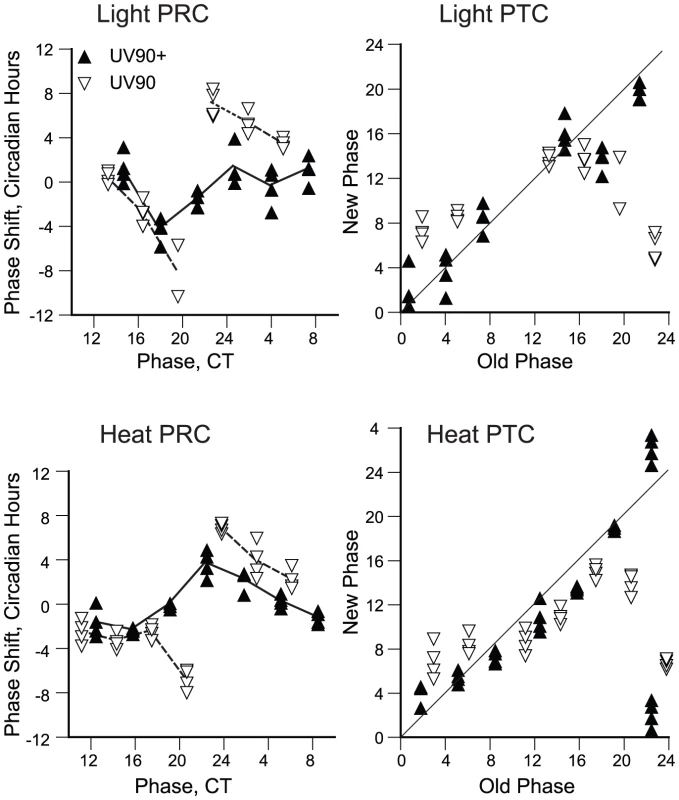
All strains carry the ras-1bd and csp-1 mutations. Cultures were grown in DD at 22°C. Solid triangles: UV90+. Open triangles: UV90. Each point represents an individual race tube. Top row: Resetting by 2 min light pulses. Bottom row: Resetting by 15 min heat pulses. Left column: Phase response curves, plotting the change in phase (phase shift in circadian hours) against the circadian time (CT) of the pulse. Average phase shifts are connected by lines. Right column: Phase transition curves, plotting the new phase after the pulse in circadian hours against the circadian time of the pulse (“old phase”). Diagonal lines indicate the locus of “no effect” where new phase equals old phase. Note that the UV90+ data for old phase 22.5 in the heat PTC have been double-plotted to show the continuity of the data. Effects of UV90 on expression of FRQ protein
We assayed the relative levels of FRQ protein in the UV90 mutant strain and the UV90 wild-type to determine whether the UV90 mutation affects the expression of FRQ. Three circadian cycles were assayed every three hours, from 3 to 72 h in DD (constant darkness), and levels of FRQ protein were directly compared between the two strains. The pattern of FRQ expression in the wild-type (Figure 9A) was very similar to the pattern we previously reported for cultures on solid agar medium across two cycles from 24 to 69 h in DD [21]. The phosphorylation pattern of FRQ was similar in mutant and wild-type (Figure 9B). Expression of FRQ in the UV90 mutant was lower than wild-type in the first cycle and rapidly damped out such that expression was much lower in the second and third cycles (Figure 9A). Mean expression levels from three replicate experiments indicated that the level of FRQ expression in UV90 was significantly lower than in UV90+ (Figure 9C). We conclude that the UV90 mutation affects the functioning of the TTFL by damping the rhythm of FRQ protein. It is interesting to note that, although the levels of FRQ protein in the UV90 mutant were significantly lower than UV90 wild-type (Figure 9), there was very little effect on the period of the conidiation rhythm at the temperature used in this experiment (22°C, Figure 6; see also Table 1).
Fig. 9. Effects of UV90 mutation on expression of FRQ protein. 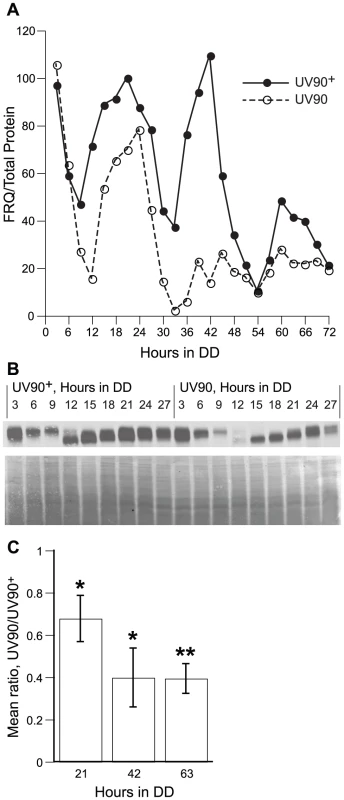
Both strains carry the ras-1bd, csp-1 and chol-1 mutations and were grown on high choline to repair the chol-1 defect. Cultures were grown in DD for varying lengths of time and protein extracts were assayed for FRQ protein by immunoblotting. A. Relative level of FRQ, normalized to total protein by Coomassie staining. Solid circles: UV90 wild-type; open circles: UV90 mutant. Points are from a single series of experiments; similar results were obtained in a replicate experiment. B. Upper panel: Immunoblotting of FRQ protein for one cycle, 3–27 h in DD, corresponding to the time-series in A. UV90 wild-type samples are on the left half and UV90 mutant samples on the right half of the blot. Lower panel: Coomassie stained membrane of samples in upper panel. C. Ratios of relative FRQ levels in UV90 vs. UV90+. Ratios were calculated at three peak time points (21, 42 and 63 hours in DD) in three independent experiments and mean ratios are plotted ± one S.E.M. *: significantly less than 1.00, p<0.05. **: significantly less than 1.00, p<0.01. Discussion
The UV90 mutation was found to affect two FRQ-less rhythms: the free-running rhythm under low-choline conditions (Figure 1A and 1C) and the heat-entrained rhythm in choline-sufficient conditions (Figure 7E–7H). Some prd mutations also affect more than one FRQ-less rhythm: The prd-1 mutation affects FRQ-less rhythms in the cel [37] and chol-1 [29] mutant backgrounds, in geraniol-supplemented cultures [17], and under heat entrained conditions [29], and prd-2 affects the last three rhythms but has not been assayed for effects on cel. As with UV90, it is not known whether these prd mutations affect other FRQ-less rhythms that have not yet been assayed. Each of the products of the UV90, prd-1 and prd-2 genes may have multiple molecular targets and multiple functions, affecting several independent FLOs. Alternatively, if we assume the simplest model, that each gene product has one major function and one major target, then these results are consistent with the hypothesis that all FRQ-less rhythms are driven by a single FRQ-less oscillator, or FLO [29].
The various FRQ-less rhythms that have been reported appear in some cases to have different characteristics, and this may be seen to support the hypothesis of multiple independent FLOs. For example, temperature compensation is reported to be defective in some cases [14], [18], and in others the period is compensated within a temperature range but not outside of that range [16], [22]. This is similar to the observations of differences in temperature compensation between period-affecting frq mutants and other clock-affecting mutants in the presence of a functional FRQ/WCC TTFL, in which the genetic background can affect temperature compensation in frq+ strains [38]. FRQ-less rhythms have been reported from several different laboratories, and the assay conditions, media, and genetic backgrounds of the strains differ. It is possible that under the widely varying conditions and genetic backgrounds used to assay the various FRQ-less rhythms, the properties observed may be affected by those conditions. The same suite of properties has not been assayed for all FRQ-less rhythms, and it is often impossible to directly compare them. For example, the molecular rhythm in DAG [25] has not been characterized beyond the observation that it persists in frq-less strains; the nitrate reductase rhythm [23] is known to persist in frq and wc null mutants and in both constant dark and constant light; but in neither case has temperature compensation been assayed nor is it known what molecular mechanisms drive these rhythms. In the absence of further details about the characteristics of the various FRQ-less rhythms, the most parsimonious assumption is that a single oscillator can drive all observed FRQ-less rhythms and this single FLO is affected by multiple gene products such as UV90, prd-1 and prd-2. In the absence of a functional FRQ/WCC TTFL, the FLO may continue to oscillate with a low amplitude and with alterations of properties such as temperature compensation and period stability; the various conditions under which FRQ-less rhythms can be observed may act on different parameters of the FLO to increase its amplitude and allow FRQ-less rhythms with differing properties to be expressed.
Two lines of evidence suggest that the UV90 mutation primarily affects the FLO rather than the FRQ/WCC feedback loop. First, the free-running conidiation rhythm in low-choline frq10 cultures is completely abolished by UV90 while a low-amplitude rhythm persists in the presence of functional frq (Figure 1). Second, entrainment to heat pulses is affected more strongly in the frq10 background than in frq+ (Figure 7). However, the UV90 mutation does have significant effects in the presence of functional frq. The amplitude of the free-running conidiation rhythm is dampened (Figure 1D), the temperature compensation of the free-running conidiation rhythm is affected (Figure 6), the amplitude of the oscillator as assayed by phase resetting is reduced (Figure 8) and the expression of FRQ protein is dampened (Figure 9). The UV90 gene product may have several molecular functions and may affect the FRQ/WCC TTFL independently of its effects on the FLO, but we prefer the simpler explanation stated previously that UV90 has a single major target. In this case, these findings lend support to the hypothesis that the FRQ/WCC feedback loop and the FLO interact with each other.
The long-period conidiation rhythm in the chol-1 strain grown on choline-deficient medium persists in the absence of the FRQ/WCC TTFL, in either frq knockout or wc mutant strains [16]. This defines it as a FRQ-less rhythm driven by a FLO, but the long-period conidiation rhythm is also seen in frq+ strains. This raises the question: What is happening to the FRQ/WCC TTFL when frq+ cultures on low choline are producing long-period conidiation rhythms driven by the FLO? Shi et al. [28] found short-period rhythms of frq promoter activity in chol-1 cultures with long-period conidiation rhythms, and concluded that the FLO in chol-1 is “in no way connected to the circadian system” [28]. These authors did not use mathematical techniques to look for relative coordination or frequency demultiplication between the short-period molecular rhythm and the long-period conidiation rhythm and so might have missed evidence of interactions. We have previously reported evidence for an influence of the FRQ/WCC TTFL on the FLO in chol-1: a short-period frq1 mutation [39] and a wc-2 mutation [16] can significantly alter the long period in chol-1, and introducing the frq10 mutation into chol-1 can lengthen the long period (Table 1 and [29]).
If our suggestion that the UV90 mutation primarily affects the FLO is correct, then our results now provide evidence for an influence of the FLO on the FRQ/WCC TTFL. The UV90 gene product is required for normal functioning of the FLO and to sustain the amplitude of the FRQ/WCC TTFL. Evidence that prd mutations can affect rhythmicity in both frq+ and frq10 backgrounds [29] suggests interactions between these two oscillators. The complete system maybe more complex, with multiple targets for the UV90 and prd gene products, and multiple oscillators, but the current data set can be explained with this two-oscillator model. Further experiments will be needed clarify the functions of the gene products in question, identify their targets, and describe the mechanism of the FLO(s).
We conclude that the simplest model for the architecture of the circadian system of Neurospora [29] is a single FLO that mutually interacts with, and is required to support, the FRQ/WCC TTFL. The fungal circadian system may thus be conceptually similar to the cyanobacterial system, in which a post-translational core oscillator interacts with a TTFL [6]. The coupling of a TTFL with a post-transcriptional oscillator is an emerging theme in circadian biology [40], with examples coming from plants [41], [42] and animals [8], [43] as well as the cyanobacteria. Mathematical modeling demonstrates that such a system of coupled oscillators could be more robust to noise and changes in growth rate than either oscillator alone [6], [44]. Neurospora has long been a fruitful model system for elucidating mechanisms of circadian rhythmicity common to many organisms, and therefore our results should encourage the search for circadian system components outside of the TTFL loops in other eukaryotic organisms.
Materials and Methods
Strains and growth conditions
All strains carried the ras-1bd (formerly bd) and csp-1 mutations, as previously described [15], [29]. The frq10 mutation is a knockout created by targeted gene disruption [12]. The chol-1 mutation requires choline for normal growth as previously described [16], [45]. Multiple mutant strains were created by standard crossing methods, and were backcrossed to the rasbd strain to verify genotypes by segregation of the expected phenotypes. Cultures were grown on maltose/arginine (MA) medium containing Vogel's salts, 0.5% maltose, 0.01% arginine, 2% agar, and either high choline (100 µM) or no choline supplementation. On 100 µM choline, the chol-1 strains are not distinguishable from chol+ strains [39]. Cultures were initially grown in constant light on agar plates, and small plugs of mycelium were transferred to race tubes (for rhythm assays) or to cellophane-covered Petri plates (for biochemical assays) as previously described [21], [29] before transfer to constant darkness (DD) at 22°C.
Mutagenesis
The parent strain for mutagenesis was genotype csp-1; chol-1 rasbd; frq10. Uninucleate microconidia were prepared by the method of Maheshwari [46], [47], using iodoacetate to induce microconidiation. Microconidia were suspended in water and exposed to ultraviolet light. Survival rates (compared to untreated microconidia) ranged between 10% to 80% in various experiments. Treated microconidia were plated on a sorbose-containing medium to induce colonial growth and individual colonies were tested for rhythmic phenotypes on race tubes.
Temperature compensation
All strains carried the wild-type frq+ allele (in addition to ras-1bd and csp-1) and either chol+ or chol-1, and UV90+ or UV90. The four genotypes used were: (1) csp-1; rasbd, (2) csp-1; rasbd; UV90, (3) csp-1; chol-1 rasbd, and (4) csp-1; chol-1 rasbd; UV90. Cultures were grown on race tubes containing MA medium with 100 µM choline. The period of the conidiation rhythm and the growth rate were assayed for each tube by linear regression as previously described [29], [39]. At 19°C the UV90 strains produced only two or three conidiation bands before the rhythm damped out, therefore only the second period on each tube was used for all strains at this temperature. The number of tubes averaged for means ranged between 6 and 12. Means were compared using the two-tailed Student's t-test for samples with equal variances. UV90 strains were compared to the corresponding UV90+ strains, i.e., chol+ UV90 was compared to chol+ UV90+, and chol-1 UV90 was compared to chol-1 UV90+.
Entrainment to high temperature pulses
All strains carried chol-1 (in addition to ras-1bd and csp-1) and either frq+ or frq10, and UV90+ or UV90. The four genotypes used were: (1) csp-1; chol-1 rasbd, (2) csp-1; chol-1 rasbd; UV90, (3) csp-1; chol-1 rasbd; frq10, and (4) csp-1; chol-1 rasbd; frq10; UV90. Cultures were grown in DD at 22°C on race tubes containing MA medium with 100 µM choline to repair the chol-1 defect. 2-hour heat pulses to 32°C were delivered as previously described [15], [29] and density traces were collected and analysed as previously described [15], [29]. Average density traces were calculated from the last two complete cycles in each experiment, averaging two cycles per tube and six tubes per set for N = 12. Standard errors were calculated for each average pixel value. Average density profiles were normalized by setting the minimum and maximum pixel values to 0 and 1, and confidence intervals were plotted as ± one S.E.M.
Phase resetting assays
All strains carried the wild-type alleles of chol+ and frq+ (in addition to ras-1bd and csp-1). The two genotypes used were: (1) csp-1; rasbd, and (2) csp-1; rasbd; UV90. Cultures were grown in DD at 22°C on race tubes containing MA medium. For light resetting, groups of 4 tubes were exposed to a 2 min pulse of cool-white fluorescent light at 20–24 µmol/m2/s at 3 h intervals between 24 to 39 h in DD. For heat pulse resetting, groups were exposed to a 15 min pulse of 37°C at 3 h intervals between 22 to 40 h in DD. After growth had finished at 22°C, the positions of the bands that formed after the pulses were used to calculate the phase (in circadian time) of each race tube relative to the average phase of the un-pulsed control group. Circadian time (CT) was calculated using the periods determined from the un-pulsed controls, defining one period (approx. 22 h) as equal to 24 circadian hours. One circadian hour is therefore 1/24th of a period. CT 12 is defined as the light-to-dark transition. Phase shifts were calculated as the difference in circadian hours between the pulsed tubes and the average phase of the un-pulsed controls, and were plotted against the CT of the pulses to generate phase response curves. To generate phase transition curves, the new phases of the pulsed tubes (in CT) were plotted against the CT of the pulses, defined as the phases (in CT) of the controls (“old phase”) at the time of the pulses.
Western blotting for FRQ expression
The method was essentially as previously described [21]. Cultures of the rasbd; csp-1; chol-1 and rasbd; csp-1; chol-1 UV90 strains were grown at 22°C on top of cellophane on MA medium with 100 µM choline in 150 mm Petri plates. Cultures were initially grown in constant light, then transferred to DD, and harvested after 72 hours of total growth. Circadian phase (reported as hours in darkness) was varied by varying the time at which cultures were transferred from light to dark. Plates were transferred every three hours, and samples were collected from 3 to 72 h in DD. Cultures were harvested, protein was extracted, and FRQ was detected as previously described [21]. The primary antibodies were generously supplied by M. Merrow [48] and M. Brunner. Samples were divided into three sets for electrophoresis and blotting: 3–27, 27–48, and 48–72 h in DD. Both UV90+ and UV90 samples were run on the same gel to directly compare expression levels between strains. FRQ was normalized against total protein by staining blots with Coomassie Blue after immunodetection. To plot one complete time series from 3 to 72 h, normalized values were adjusted to equalize the repeated samples between sets (27 and 48 h). To calculate mean ratios, three time points were chosen near the peak values in each cycle. Three independent experiments were carried out at each time point, running UV90+ and UV90 samples on the same gel for direct comparison of FRQ levels. The relative level of FRQ in UV90 was divided by the relative level in UV90+ to calculate the ratio. A ratio of 1.00 indicates that FRQ levels are the same in the UV90+ and UV90 samples. The means and S.E.M.s of the three independent ratios were calculated. One-tailed one sample t-tests were used to test the null hypothesis that the means are not less than 1.00, that is, that UV90 FRQ levels are not lower than UV90+ levels.
Zdroje
1. Lakin-ThomasPL 2006 Transcriptional feedback oscillators: Maybe, maybe not. J Biol Rhythms 21 83 92
2. FanYZHidaAAndersonDAIzumoMJohnsonCH 2007 Cycling of CRYPTOCHROME proteins is not necessary for circadian clock function in mammalian fibroblasts. Curr Biol 17 1091 1100
3. DibnerCSageDUnserMBauerCD'EysmondT 2009 Circadian gene expression is resilient to large fluctuations in overall transcription rates. EMBO J 28 123 134
4. O'NeillJSMaywoodESCheshamJETakahashiJSHastingsMH 2008 cAMP-dependent signaling as a core component of the mammalian circadian pacemaker. Science 320 949 953
5. DongGGoldenSS 2008 How a cyanobacterium tells time. Curr Opin Microbiol 11 541 546
6. QinXByrneMXuYMoriTJohnsonCH 2010 Coupling of a core post-translational pacemaker to a slave transcription/translation feedback loop in a circadian system. PLoS Biol 8 e1000394 doi:10.1371/journal.pbio.1000394
7. ZhangEELiuACHirotaTMiragliaLJWelchG 2009 A genome-wide RNAi screen for modifiers of the circadian clock in human cells. Cell 139 199 210
8. HastingsMHMaywoodESO'NeillJS 2008 Cellular circadian pacemaking and the role of cytosolic rhythms. Curr Biol 18 R805 R815
9. DunlapJCLorosJJ 2006 How fungi keep time: circadian system in Neurospora and other fungi. Curr Opin Microbiol 9 579 587
10. Lakin-ThomasPL 2006 New models for circadian systems in microorganisms. FEMS Microbiol Lett 259 1 6
11. LiuYBell-PedersenD 2006 Circadian rhythms in Neurospora crassa and other filamentous fungi. Eukaryotic Cell 5 1184 1193
12. AronsonBDJohnsonKADunlapJC 1994 Circadian clock locus Frequency: Protein encoded by a single open reading frame defines period length and temperature compensation. Proc Natl Acad Sci U S A 91 7683 7687
13. DragovicZTanYGörlMRoennebergTMerrowM 2002 Light reception and circadian behavior in “blind” and “clock-less” mutants of Neurospora crassa. EMBO J 21 3643 3651
14. GranshawTTsukamotoMBrodyS 2003 Circadian rhythms in Neurospora crassa: Farnesol or geraniol allow expression of rhythmicity in the otherwise arrhythmic strains frq10, wc-1, and wc-2. J Biol Rhythms 18 287 296
15. Lakin-ThomasPL 2006 Circadian clock genes frequency and white collar-1 are not essential for entrainment to temperature cycles in Neurospora crassa. Proc Natl Acad Sci U S A 103 4469 4474
16. Lakin-ThomasPLBrodyS 2000 Circadian rhythms in Neurospora crassa: Lipid deficiencies restore robust rhythmicity to null frequency and white-collar mutants. Proc Natl Acad Sci U S A 97 256 261
17. LombardiLSchneiderKTsukamotoMBrodyS 2007 Circadian rhythms in Neurospora crassa: Clock mutant effects in the absence of a frq-based oscillator. Genetics 175 1175 1183
18. LorosJJFeldmanJF 1986 Loss of temperature compensation of circadian period length in the frq-9 mutant of Neurospora crassa. J Biol Rhythms 1 187 198
19. MerrowMBrunnerMRoennebergT 1999 Assignment of circadian function for the Neurospora clock gene frequency. Nature 399 584 586
20. YoshidaYMaedaTLeeBHasunumaK 2008 Conidiation rhythm and light entrainment in superoxide dismutase mutant in Neurospora crassa. Molecular Genetics and Genomics 279 193 202
21. SchneiderKPerrinoSOelhafenKLiSZatsepinA 2009 Rhythmic conidiation in constant light in Vivid mutants of Neurospora crassa. Genetics 181 917 931
22. BrodySOelhafenKSchneiderKPerrinoSGoetzA 2010 Circadian rhythms in Neurospora crassa: Downstream effectors. Fungal Genet Biol 47 159 168
23. ChristensenMKFalkeidGLorosJJDunlapJCLilloC 2004 A nitrate induced frq-less oscillator in Neurospora crassa. J Biol Rhythms 19 280 286
24. CorreaALewisAZGreeneAVMarchIJGomerRH 2003 Multiple oscillators regulate circadian gene expression in Neurospora. Proc Natl Acad Sci U S A 100 13597 13602
25. RamsdaleMLakin-ThomasPL 2000 sn-1,2-diacylglycerol levels in the fungus Neurospora crassa display circadian rhythmicity. J Biol Chem 275 27541 27550
26. de PaulaRMLewisZAGreeneAVSeoKSMorganLW 2006 Two circadian timing circuits in Neurospora crassa cells share components and regulate distinct rhythmic processes. J Biol Rhythms 21 159 168
27. IwasakiHDunlapJC 2000 Microbial circadian oscillatory systems in Neurospora and Synechococcus: models for cellular clocks. Curr Opin Microbiol 3 189 196
28. ShiMLarrondoLFLorosJJDunlapJC 2007 A developmental cycle masks output from the circadian oscillator under conditions of choline deficiency in Neurospora. Proc Natl Acad Sci U S A 104 20102 20107
29. LiSLakin-ThomasPL 2010 Effects of prd circadian clock mutations on FRQ-less rhythms in Neurospora. J Biol Rhythms 25 71 80
30. PerkinsDD 1991 Neurospora alcoy linkage tester stocks with group VII marked, and their use for mapping translocations. Fungal Genet News 38 83
31. JinYAllanSBaberLBhattaraiEKLambTM 2007 Rapid genetic mapping in Neurospora crassa. Fungal Genet Biol 44 455 465
32. RoennebergTDragovicZMerrowM 2005 Demasking biological oscillators: Properties and principles of entrainment exemplified by the Neurospora circadian clock. Proc Natl Acad Sci U S A 102 7742 7747
33. Lakin-ThomasPLBrodySCoteGG 1991 Amplitude model for the effects of mutations and temperature on period and phase resetting of the Neurospora circadian oscillator. J Biol Rhythms 6 281 297
34. JohnsonCHElliotJAFosterRG 2003 Entrainment of circadian programs. Chronobiol Int 20 741 774
35. WinfreeAT 2001 The Geometry of Biological Time New York Springer 777
36. Lakin-ThomasPL 1995 A beginners guide to limit cycles, their uses and abuses. Biol Rhythm Res 26 216 232
37. Lakin-ThomasPLBrodyS 1985 Circadian rhythms in Neurospora crassa: interactions between clock mutations. Genetics 109 49 66
38. GardnerGFFeldmanJF 1981 Temperature compensation of circadian period length in clock mutants of Neurospora crassa. Plant Physiol 68 1244 1248
39. Lakin-ThomasPL 1998 Choline depletion, frq mutations, and temperature compensation of the circadian rhythm in Neurospora crassa. J Biol Rhythms 13 268 277
40. MerrowMRoennebergT 2007 Circadian clock: time for a phase shift of ideas? Curr Biol 17 R636 R638
41. MergenhagenDSchweigerHG 1975 The effect of different inhibitors of transcription and translation on the expression and control of circadian rhythm in individual cells of Acetabularia. Exp Cell Res 94 321 326
42. O'NeillJSvan OoijenGDixonLETroeinCCorellouF 2011 Circadian rhythms persist without transcription in a eukaryote. Nature 469 554 558
43. O'NeillJSReddyAB 2011 Circadian clocks in human red blood cells. Nature 469 498 504
44. ZwickerDLubenskyDKTen WoldePR 2010 Robust circadian clocks from coupled protein-modification and transcription-translation cycles. Proc Natl Acad Sci U S A 107 22540 22545
45. Lakin-ThomasPL 1996 Effects of choline depletion on the circadian rhythm in Neurospora crassa. Biol Rhythm Res 27 12 30
46. MaheshwariR 1999 Microconidia of Neurospora crassa. Fungal Genet Biol 26 1 18
47. PanditAMaheshwariR 1993 A simple method of obtaining pure microconidia in Neurospora crassa. Fungal Genet News 40 64 65
48. GörlMMerrowMHuttnerBJohnsonJRoennebergT 2001 A PEST-like element in FREQUENCY determines the length of the circadian period in Neurospora crassa. EMBO J 20 7074 7084
Štítky
Genetika Reprodukční medicína
Článek Local Absence of Secondary Structure Permits Translation of mRNAs that Lack Ribosome-Binding SitesČlánek Independent Chromatin Binding of ARGONAUTE4 and SPT5L/KTF1 Mediates Transcriptional Gene SilencingČlánek Trade-Off between Bile Resistance and Nutritional Competence Drives Diversification in the Mouse GutČlánek FGF Signaling Regulates the Number of Posterior Taste Papillae by Controlling Progenitor Field SizeČlánek Mammalian BTBD12 (SLX4) Protects against Genomic Instability during Mammalian SpermatogenesisČlánek Identification of Nine Novel Loci Associated with White Blood Cell Subtypes in a Japanese PopulationČlánek Differential Effects of and Risk Variants on Association with Diabetic ESRD in African AmericansČlánek Dynamic Chromatin Localization of Sirt6 Shapes Stress- and Aging-Related Transcriptional Networks
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2011 Číslo 6
-
Všechny články tohoto čísla
- Local Absence of Secondary Structure Permits Translation of mRNAs that Lack Ribosome-Binding Sites
- Statistical Inference on the Mechanisms of Genome Evolution
- Revisiting Heterochromatin in Embryonic Stem Cells
- A Two-Stage Meta-Analysis Identifies Several New Loci for Parkinson's Disease
- Identification of a Sudden Cardiac Death Susceptibility Locus at 2q24.2 through Genome-Wide Association in European Ancestry Individuals
- Genomic Prevalence of Heterochromatic H3K9me2 and Transcription Do Not Discriminate Pluripotent from Terminally Differentiated Cells
- Epistasis between Beneficial Mutations and the Phenotype-to-Fitness Map for a ssDNA Virus
- Recurrent Chromosome 16p13.1 Duplications Are a Risk Factor for Aortic Dissections
- Telomere DNA Deficiency Is Associated with Development of Human Embryonic Aneuploidy
- Genome-Wide Association Study of White Blood Cell Count in 16,388 African Americans: the Continental Origins and Genetic Epidemiology Network (COGENT)
- Unexpected Role for DNA Polymerase I As a Source of Genetic Variability
- Transportin-SR Is Required for Proper Splicing of Genes and Plant Immunity
- How Chromatin Is Remodelled during DNA Repair of UV-Induced DNA Damage in
- Independent Chromatin Binding of ARGONAUTE4 and SPT5L/KTF1 Mediates Transcriptional Gene Silencing
- Two Evolutionary Histories in the Genome of Rice: the Roles of Domestication Genes
- Natural Allelic Variation Defines a Role for : Trichome Cell Fate Determination
- Multiple Common Susceptibility Variants near BMP Pathway Loci , , and Explain Part of the Missing Heritability of Colorectal Cancer
- Pathogenic Mechanism of the FIG4 Mutation Responsible for Charcot-Marie-Tooth Disease CMT4J
- A Functional Variant in Promoter Modulates Its Expression and Confers Disease Risk for Systemic Lupus Erythematosus
- Drift and Genome Complexity Revisited
- Chromosomal Macrodomains and Associated Proteins: Implications for DNA Organization and Replication in Gram Negative Bacteria
- Trade-Off between Bile Resistance and Nutritional Competence Drives Diversification in the Mouse Gut
- Pathways of Distinction Analysis: A New Technique for Multi–SNP Analysis of GWAS Data
- Web-Based Genome-Wide Association Study Identifies Two Novel Loci and a Substantial Genetic Component for Parkinson's Disease
- Chk2 and p53 Are Haploinsufficient with Dependent and Independent Functions to Eliminate Cells after Telomere Loss
- Exome Sequencing Identifies Mutations in High Myopia
- Distinct Functional Constraints Partition Sequence Conservation in a -Regulatory Element
- CorE from Is a Copper-Dependent RNA Polymerase Sigma Factor
- A Single Sex Pheromone Receptor Determines Chemical Response Specificity of Sexual Behavior in the Silkmoth
- FGF Signaling Regulates the Number of Posterior Taste Papillae by Controlling Progenitor Field Size
- Maps of Open Chromatin Guide the Functional Follow-Up of Genome-Wide Association Signals: Application to Hematological Traits
- Increased Susceptibility to Cortical Spreading Depression in the Mouse Model of Familial Hemiplegic Migraine Type 2
- Differential Gene Expression and Epiregulation of Alpha Zein Gene Copies in Maize Haplotypes
- Parallel Adaptive Divergence among Geographically Diverse Human Populations
- Genetic Analysis of Genome-Scale Recombination Rate Evolution in House Mice
- Mechanisms for the Evolution of a Derived Function in the Ancestral Glucocorticoid Receptor
- Mammalian BTBD12 (SLX4) Protects against Genomic Instability during Mammalian Spermatogenesis
- Interferon Regulatory Factor 8 Regulates Pathways for Antigen Presentation in Myeloid Cells and during Tuberculosis
- High-Resolution Analysis of Parent-of-Origin Allelic Expression in the Arabidopsis Endosperm
- Specific SKN-1/Nrf Stress Responses to Perturbations in Translation Elongation and Proteasome Activity
- Graded Nodal/Activin Signaling Titrates Conversion of Quantitative Phospho-Smad2 Levels into Qualitative Embryonic Stem Cell Fate Decisions
- Genome-Wide Analysis Reveals PADI4 Cooperates with Elk-1 to Activate Expression in Breast Cancer Cells
- Trait Variation in Yeast Is Defined by Population History
- Meiosis-Specific Loading of the Centromere-Specific Histone CENH3 in
- A Genome-Wide Survey of Imprinted Genes in Rice Seeds Reveals Imprinting Primarily Occurs in the Endosperm
- Multiple Regulatory Mechanisms to Inhibit Untimely Initiation of DNA Replication Are Important for Stable Genome Maintenance
- SIRT1 Promotes N-Myc Oncogenesis through a Positive Feedback Loop Involving the Effects of MKP3 and ERK on N-Myc Protein Stability
- Bacteriophage Crosstalk: Coordination of Prophage Induction by Trans-Acting Antirepressors
- Role of the Single-Stranded DNA–Binding Protein SsbB in Pneumococcal Transformation: Maintenance of a Reservoir for Genetic Plasticity
- Genomic Convergence among ERRα, PROX1, and BMAL1 in the Control of Metabolic Clock Outputs
- Genome-Wide Association of Bipolar Disorder Suggests an Enrichment of Replicable Associations in Regions near Genes
- Identification of Nine Novel Loci Associated with White Blood Cell Subtypes in a Japanese Population
- DNA Ligase III Promotes Alternative Nonhomologous End-Joining during Chromosomal Translocation Formation
- Differential Effects of and Risk Variants on Association with Diabetic ESRD in African Americans
- Finished Genome of the Fungal Wheat Pathogen Reveals Dispensome Structure, Chromosome Plasticity, and Stealth Pathogenesis
- Dynamic Chromatin Localization of Sirt6 Shapes Stress- and Aging-Related Transcriptional Networks
- Extracellular Matrix Dynamics in Hepatocarcinogenesis: a Comparative Proteomics Study of Transgenic and Null Mouse Models
- Integrating 5-Hydroxymethylcytosine into the Epigenomic Landscape of Human Embryonic Stem Cells
- Vive La Différence: An Interview with Catherine Dulac
- Multiple Loci Are Associated with White Blood Cell Phenotypes
- Nuclear Accumulation of Stress Response mRNAs Contributes to the Neurodegeneration Caused by Fragile X Premutation rCGG Repeats
- A New Mutation Affecting FRQ-Less Rhythms in the Circadian System of
- Cryptic Transcription Mediates Repression of Subtelomeric Metal Homeostasis Genes
- A New Isoform of the Histone Demethylase JMJD2A/KDM4A Is Required for Skeletal Muscle Differentiation
- Genetic Determinants of Lipid Traits in Diverse Populations from the Population Architecture using Genomics and Epidemiology (PAGE) Study
- A Genome-Wide RNAi Screen for Factors Involved in Neuronal Specification in
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Recurrent Chromosome 16p13.1 Duplications Are a Risk Factor for Aortic Dissections
- Statistical Inference on the Mechanisms of Genome Evolution
- Genome-Wide Association Study of White Blood Cell Count in 16,388 African Americans: the Continental Origins and Genetic Epidemiology Network (COGENT)
- Chromosomal Macrodomains and Associated Proteins: Implications for DNA Organization and Replication in Gram Negative Bacteria
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



