-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaNuclear Accumulation of Stress Response mRNAs Contributes to the Neurodegeneration Caused by Fragile X Premutation rCGG Repeats
Fragile X–associated tremor/ataxia syndrome (FXTAS) is a neurodegenerative disorder seen in Fragile X premutation carriers. Previous studies found that Fragile X rCGG repeats are sufficient to cause neurodegeneration and that the rCGG repeat-binding proteins Pur α and hnRNP A2/B1 can modulate rCGG–mediated neuronal toxicity. To explore the role of Pur α in rCGG–mediated neurodegeneration further, we took a proteomic approach and identified more than 100 proteins that interact with Pur α. Of particular interest is Rm62, the Drosophila ortholog of p68 RNA helicase, which could modulate rCGG–mediated neurodegeneration. Here we show that rCGG repeats decreased the expression of Rm62 posttranscriptionally, leading to the nuclear accumulation of Hsp70 transcript, as well as additional mRNAs involved in stress and immune responses. Together these findings suggest that abnormal nuclear accumulation of these mRNAs, likely as a result of impaired nuclear export, could contribute to FXTAS pathogenesis.
Published in the journal: . PLoS Genet 7(6): e32767. doi:10.1371/journal.pgen.1002102
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1002102Summary
Fragile X–associated tremor/ataxia syndrome (FXTAS) is a neurodegenerative disorder seen in Fragile X premutation carriers. Previous studies found that Fragile X rCGG repeats are sufficient to cause neurodegeneration and that the rCGG repeat-binding proteins Pur α and hnRNP A2/B1 can modulate rCGG–mediated neuronal toxicity. To explore the role of Pur α in rCGG–mediated neurodegeneration further, we took a proteomic approach and identified more than 100 proteins that interact with Pur α. Of particular interest is Rm62, the Drosophila ortholog of p68 RNA helicase, which could modulate rCGG–mediated neurodegeneration. Here we show that rCGG repeats decreased the expression of Rm62 posttranscriptionally, leading to the nuclear accumulation of Hsp70 transcript, as well as additional mRNAs involved in stress and immune responses. Together these findings suggest that abnormal nuclear accumulation of these mRNAs, likely as a result of impaired nuclear export, could contribute to FXTAS pathogenesis.
Introduction
Fragile X syndrome (FXS) is usually caused by expansion of the CGG trinucleotide repeat in the 5′ untranslated region (5′ UTR) of the Fragile X Mental Retardation 1 (FMR1) gene [1]. Whereas normal individuals generally possess between five and 54 repeats, fully affected individuals have more than 200 CGG repeats on what are referred to as full mutation alleles [2]. Premutation alleles (55–200 CGG repeats) of the FMR1 gene are known to contribute to the Fragile X phenotype through genetic instability, and they can expand into the full mutation during germline transmission [3]. Within the last decade, Fragile X-associated Tremor/Ataxia Syndrome (FXTAS), a late-onset neurodegenerative disorder, has been recognized mainly among many male premutation carriers in or beyond their fifth decade of life [4], and FXTAS is distinct from the neurodevelopmental disorder, FXS.
The most common clinical feature of FXTAS is a progressive action tremor with ataxia. More advanced or severe cases may show a progressive cognitive decline that ranges from executive and memory deficits to dementia [5]. Patients may also present with common psychiatric symptoms, such as increased anxiety, mood liability, and depression [6], [7]. Magnetic resonance imaging (MRI) of adult male patients affected with FXTAS demonstrated mild to moderate global brain atrophy, most common in the fontal and parietal regions, as well as the pons and the cerebellum [8]. Nearly all autopsy studies on the brains of symptomatic premutation carriers show degeneration in the cerebellum, which includes Purkinje neuronal cell loss, Bergman gliosis, spongiosis of the deep cerebellar white matter, and swollen axons [9], [10]. The major neuropathological hallmark and postmortem criterion for definitive FXTAS is eosinophilic, ubiquitin-positive intranuclear inclusions broadly distributed throughout the brain in neurons, astrocytes, and in the spinal column [9].
For the past several years, the hunt has been on to uncover the molecular basis of FXTAS. One unique molecular signature of the fragile X premutation allele is that the level of FMR1 mRNA is significantly elevated, while the FMR1 protein (FMRP) remains relatively unchanged in cells from premutation carriers [11], [12]. Given that the neurodegenerative phenotype of FXTAS is associated specifically with premutation carriers, but not with the full mutation, FMRP deficiency per se is likely not the culprit behind FXTAS [4]. Instead, the neurodegenerative phenotypes associated with FXTAS are suspected of being caused by a gain of function in fragile X premutation rCGG repeat RNAs [3], [13]. The hypothesis is that overproduced rCGG repeats in FXTAS sequester specific RNA-binding proteins and reduce their ability to perform their normal cellular functions, thereby contributing significantly to the pathology of this disorder. The presence of FMR1 mRNA in inclusions found in the brains of FXTAS patients, as well as the formation of similar inclusions upon ectopic expression of rCGG repeats in model systems, have provided strong support for this hypothesis [13]–[16].
Two RNA-binding proteins, Pur α and hnRNP A2/B1, are known to bind rCGG repeats specifically in both mammalian and Drosophila brains [17], [18]. Both Pur α and hnRNP A2/B1 are found to be present in the inclusions of FXTAS brain tissues. Furthermore, overexpression of either Pur α or hnRNP A2/B1 can alleviate neurodegeneration in the fly model of FXTAS [17], [18]. In particular, Pur α knock-out mice appeared normal at birth, but developed severe tremor and spontaneous seizures at two weeks of age due to drastically reduced numbers of neurons in regions of the hippocampus and cerebellum, suggesting that the depletion of Pur α alone could lead to ataxia [19].
In this study, to further investigate the role of Pur α in rCGG-mediated neurodegeneration, we took a proteomic approach to identify the proteins that interact with Pur α. Over 100 proteins, including several known interactors, such as Fmrp, were found to interact with Pur α in vitro. To evaluate the physiological role(s) of Pur α-interacting proteins in rCGG-mediated neuronal toxicity, we further tested their genetic interactions with rCGG repeats using our FXTAS fly model and identified several interactors of Pur α that could genetically modulate the toxicity caused by rCGG repeats. Among these, Rm62, the Drosophila ortholog of the p68 RNA helicase, was of particular interest. Rm62, physically interacting with Pur α, could modulate rCGG-mediated neurodegeneration, and biochemically, fragile X rCGG repeats could decrease the expression of Rm62 posttranscriptionally, leading to the accumulation of Hsp70 transcript, a previously identified target of Rm62, in the nucleus. Further microarray analyses revealed the nuclear accumulation of additional mRNAs involved in stress and immune responses in fragile X premutation flies. These findings suggest an unexpected nuclear accumulation of specific mRNAs caused by fragile X premutation rCGG repeats and point to likely deficits in the nuclear export of specific mRNA as a possible cause of the compromised stress response in neurons expressing rCGG repeats, which would predispose them to neuronal apoptosis.
Results
Identification of a Pur α interactome
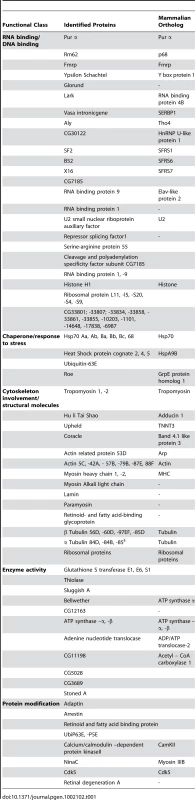
Our previous studies have shown that the neuronal apoptosis caused by fragile X premutation rCGG repeats arose at least in part as a consequence of Pur α sequestration [17]. We therefore hypothesized that genetic modifiers of rCGG-mediated neurodegeneration should be enriched among Pur α-interacting proteins. To test this idea, we took a proteomic approach to identify a comprehensive set of Pur α-interacting proteins (Pur α interactome). GST-Pur α fusion proteins were expressed in Escherichia coli and purified by glutathione-sepharose resins. The resins with the bound proteins were directly used to pack affinity columns. The columns were then loaded with pre-cleared wild-type fly brain extracts, washed extensively, and eluted. The eluted proteins were resolved on an SDS gel. As indicated in Figure 1A, a gel of the indicated size was followed by the analysis of liquid chromatography (LC)-tandem mass spectrometry (LS/MS). GST alone was also used for affinity purification as a negative control (Figure 1A). Through these analyses, we identified over 100 proteins that could interact with Pur α in vitro. The molecular functions of the identified proteins include RNA-binding/DNA-binding, cytoskeleton network, molecular chaperone, and ubiquitin proteasome pathways (Figure 1A and Table 1). Importantly, among these proteins, we also identified Cdk5 and Fmrp, which are known to interact with Pur α biochemically, suggesting the approach we have used to identify the Pur α interactome is reliable.
Fig. 1. Identification of Pur α-interacting proteins. 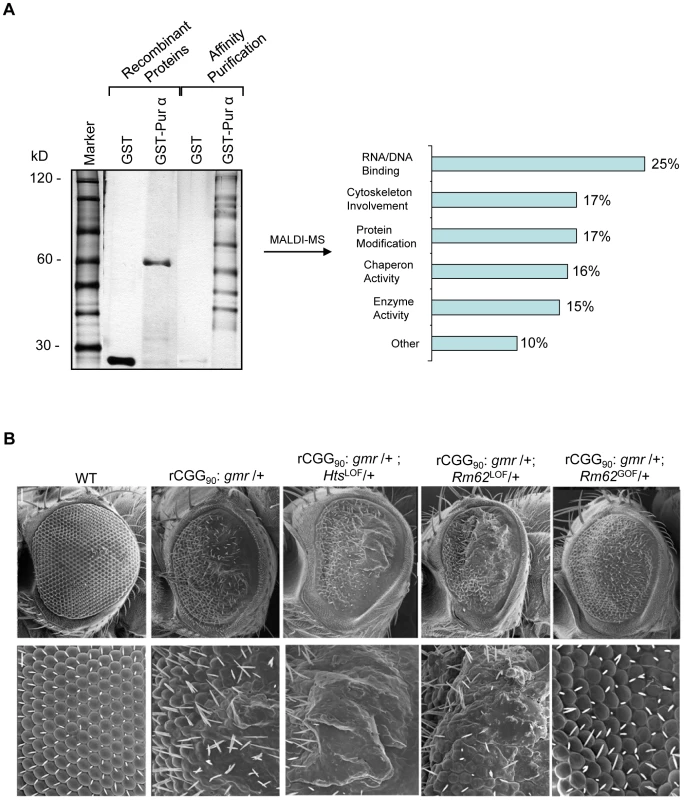
A. Silver-staining gel with distinct bands for recombinant proteins and affinity-purified proteins. The captured proteins were further analyzed by mass spectrometry, and distinct classes of proteins were identified. B. Rm62 and Hts mutants enhance rCGG-mediated neurodegeneration in fly. Column 1: wild-type fly; Column 2: fly expressing (CGG)90-EGFP only; Column 3: fly expressing (CGG)90-EGFP in the heterozygous background of Hts01103 Loss-of-Function (LOF) mutation; Column 4: fly expressing CGG90-EGFP in the heterozygous background of Rm6201084 Loss-of-Function (LOF) mutation; Column 5: fly expressing CGG90-EGFP in the heterozygous background of Rm62(3)3607 overexpression (Gain-of-Function (GOF). SEM eye images are shown. The Pur α–interacting proteins Rm62 and Hts modulate rCGG–mediated neurodegeneration
To determine which Pur α-interacting protein(s) modulates rCGG-mediated neurodegeneration, we conducted a genetic screen based on the fragile X premutation rCGG repeat-mediated neurodegenerative eye phenotype that we observed previously [13]. The screen involved directing the expression of fragile X premutation-length rCGG repeats to the eye with the Gmr-GAL4 driver using the Drosophila GAL4/UAS system. This was followed by crossing Gmr-GAL4, UAS-(CGG)90-EGFP transgenic flies with flies mutant in genes coding the Pur α-interacting proteins. We selected 17 mutant alleles for evaluation based on their availability from several stock centers (Table 2). The progenies were examined for potential suppression or enhancement of the disorganized eye phenotype compared with control rCGG flies. Through this screen, we identified two prominent enhancers that could modulate rCGG-mediated neurodegeneration, l(3)01084 and l(3)01103. P-element insertion l(3)01084 was previously characterized as an insertion within the 5′ region of the Rm62 gene that causes lethality [20] (Figure 1B). Rm62 is a homolog of mammalian p68 RNA helicase, which has been implicated in transcriptional regulation, pre-mRNA splicing, RNA interference, and nucleocytoplasmic shuttling [21]–[26]. The modulation of rCGG-mediated neuronal toxicity by Rm62 was further confirmed using additional loss-of-function alleles of Rm62. Furthermore, overexpression of Rm62 could suppress the neuronal toxicity caused by fragile X premutation rCGG repeats. The other enhancer line, l(3)01103, is a mutant allele of the hu-li tai shao (hts) gene, a homolog of mammalian Adducin, which has been shown to play key roles during early Drosophila oogenesis [27] (Figure 1B).
Tab. 2. Mutant Alleles Corresponding to Pur α-Interacting Proteins Used for Genetic Screen. 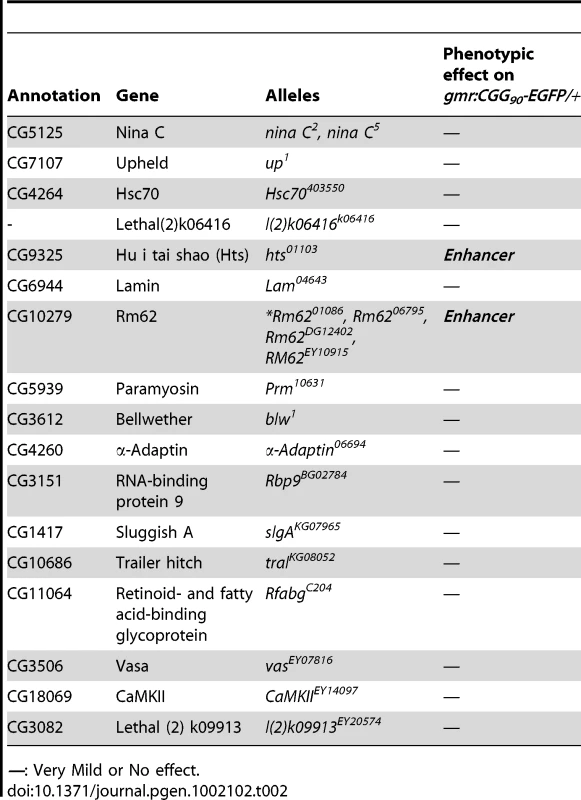
—: Very Mild or No effect. Both Rm62 and Hts directly interact with Pur α
To further determine the nature of biochemical interactions between these two proteins and Pur α we obtained their full-length cDNAs and used them for in vitro translation. Either the radiolabeled Rm62 or Hts protein was incubated with either purified recombinant GST-Pur α or GST alone; radiolabeled luciferase was used as a negative control. Both Rm62 and Hts could directly interact with GST-Pur α, but not with GST alone (Figure 2A). We went on to incubate purified recombinant GST-Pur α or GST with fly S2 cell extract. The captured proteins were gel-separated, followed by a Western blot analysis using specific antibodies against Rm62 protein and Hts. Both endogenous Rm62 and Hts could be captured by GST-Pur α (Figure 2B). To further confirm their interactions, we also performed a reverse pull-down experiment in which we incubated recombinant GST-Rm62 or GST with protein lysate from adult fly heads, which express Pur α at much higher levels than S2 cells. The captured proteins were then used for Western blot analysis with a specific antibody against Pur α protein that we developed previously. As shown in Figure 2C, endogenous Pur α could be captured by GST-Rm62 specifically. Lastly, the addition of RNase did not disrupt the interaction between Pur α and Rm62, suggesting that their interaction is RNA-independent. Given that Pur α is a well-conserved RNA-binding protein between Drosophila and mammals, we further explored the interaction between purified recombinant GST-Pur α and the mouse ortholog of Pur α-interacting proteins. We performed GST pull-down assays using purified recombinant GST-Pur α and wild-type mouse cerebellar lysates. The captured proteins were used for Western blot analysis with the antibody against the mouse ortholog of Rm62, p68 or DDX5 [28]. As shown in Figure 2D, fly Pur α protein could also interact with p68, suggesting that the interaction between Pur α and Rm62 is conserved through evolution.
Fig. 2. Rm62 and Hts directly interact with Pur α and Fragile X premutation rCGG repeats decrease the expression of Rm62 posttranscriptionally. 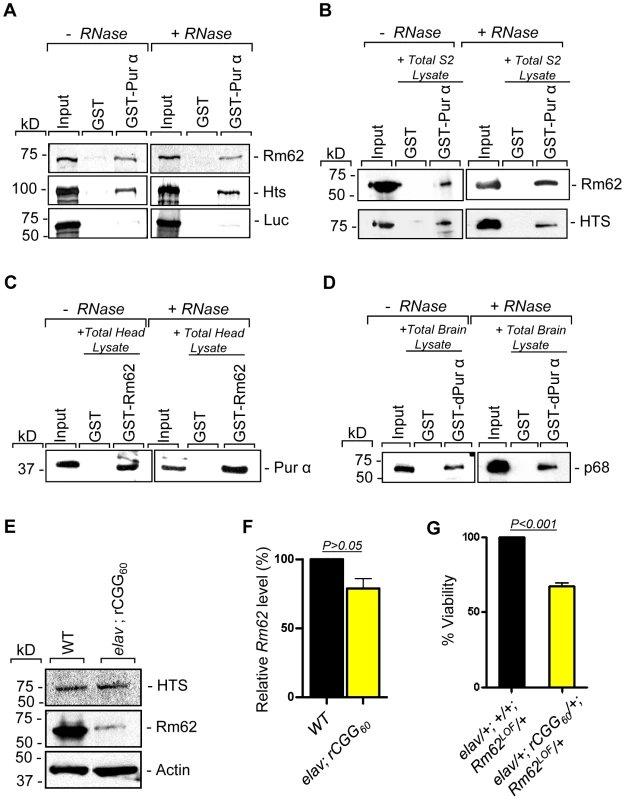
A. Pull-down assay between GST-tagged dPur α and in vitro-translated Rm62 or in vitro-translated Hts or luciferase (negative control). Indicated are samples treated or untreated with RNase prior to binding reaction. In the Input lanes, we loaded 25% of the translation products used in a reaction. Both Rm62 and Hts, but not luciferase, interacts with dPur α. B. Western blot shows the interaction between endogenous Rm62 or endogenous Hts and affinity-purified GST-Pur α. Indicated are samples treated or untreated with RNase prior to binding reaction. C. Western blot shows the interaction between affinity-purified GST-Rm62 and endogenous Pur α in RNase untreated and treated samples. D. Western blot shows the interaction between endogenous mammalian p68 and affinity-purified fly GST-Pur α. E. Protein levels of Rm62 and Hts in wild-type and rCGG-expressing flies. Quantitative analysis of Rm62 and Hts protein levels by Western blot on adult head extracts of the following genotypes: wild-type (WT); elav; rCGG60-expressing flies. Proteins are indicated to the right, corresponding molecular weights to the left. α actin represents a loading control. F. Quantitative analysis of Rm62 mRNA levels by real-time PCR on total RNA obtained from adult heads of wild-type (WT) and rCGG-expressing flies. Quantification is relative to the housekeeping ribosomal protein 32 (Rpl32) mRNA. (E; mean ± SEM n = 3). G. Statistical evaluation of the percent viability displayed by various genotypes: elav/+; +/+; Rm62LOF/+ (Rm62 heterozygous); elav/+; rCGG60-EGFP/+; TM3Sb/+ (Premutation heterozygous), elav/+; +/+; TM3Sb/+ (Internal control); elav/+; rCGG60-EGFP/+; Rm62LOF/+ (interaction). Mean of three data sets was used. Fragile X premutation rCGG repeats decrease the expression of Rm62 posttranscriptionally
Our earlier studies had shown that ectopic expression of rCGG repeats could sequester the soluble pool of Pur α, preventing its normal function and inducing neuronal apoptosis [17]. Given that Rm62 and Hts could directly interact with Pur α and influence rCGG-mediated neuronal toxicity, we examined their expression in flies expressing fragile X premutation rCGG repeats. In rCGG repeat transgenic flies, the severity of their phenotype depends on both dosage and length of the rCGG repeat. Moderate expression of (CGG)90 repeats exclusively in the eyes have an effect on morphology and histology; however, expression of (CGG)90 repeats in the neurons leads to lethality at the embryonic stage, preventing analysis at the adult stage [13]. Therefore, we used a shorter repeat length, r(CGG)60, which allowed us to examine the expression of these proteins in adults. We performed Western blot analysis using whole-head lysates from both wild-type flies and transgenic flies expressing r(CGG)60 repeats in pan-neuronal cells (Elav-GAL4; UAS-(CGG)60-EGFP). Compared with wild-type flies, we observed a significant decrease of Rm62 protein in rCGG repeat-expressing flies, whereas no change was detected with Hts protein (Figure 2E). We further determined the mRNA level of Rm62 by quantitative RT-PCR and detected no significant difference between wild-type and rCGG repeat-expressing flies (Figure 2F). Furthermore, we generated a genetic combination in which rCGG60 repeats were expressed in pan-neuronal cells in the heterozygous background of the Rm62 mutation. The expression of fragile X premutation rCGG repeats significantly reduced the viability of otherwise viable heterozygous Rm62 mutant flies, providing further support for a strong genetic interaction between Rm62 and rCGG repeat-mediated neuronal cell death (Figure 2G). Based on these observations, we chose to focus on the role of Rm62 in the context of rCGG-mediated neurodegeneration.
Fragile X premutation rCGG repeats cause the nuclear accumulation of Hsp70 mRNA
Rm62 is the Drosophila ortholog of the p68 RNA helicase. p68 RNA helicase is a prototypical DEAD-box RNA helicase that has been implicated in transcriptional regulation, pre-mRNA splicing, and nucleocytoplasmic shuttling [24], [26], [28]–[30]. In Drosophila, Rm62 is reported to play a novel role in RNA export and gene deactivation, particularly Hsp70 mRNA [31]. Studies from our own and other groups have shown that Hsp70 protein is part of the inclusions induced by fragile X premutation rCGG repeats in both human postmortem brain tissues and FXTAS animal models, including mouse and flies. Like other neurodegenerative disorders, overexpression of Hsp70 can suppress the neuronal cell death caused by rCGG repeats, suggesting an important role for Hsp70 in FXTAS pathogenesis [13].
Because of the role Rm62 plays in the nuclear export of Hsp70 mRNA and the significant reduction of Rm62 in transgenic flies expressing fragile X premutation rCGG repeats, we examined the expression and distribution of Hsp70 mRNA. Total RNA and RNAs from both nuclear and cytoplasmic fractions were isolated from the heads of both wild-type flies and flies expressing rCGG60 repeats in pan-neuronal cells. Proteins from both nuclear and cytoplasmic fractions were further analyzed by Western blots using antibody against histone H3, a protein found exclusively in the nucleus. As shown in Figure 3, histone was present in nuclear fractions with minimum cross-contamination in cytoplasmic fractions prepared from both wild-type and rCGG-expressing fly heads. Furthermore, the cytoplasmic fractions were enriched in tubulin, a protein usually present in cytoplasm. We then determined the levels of Hsp70 mRNAs in total RNAs and RNAs from cytoplasmic and nuclear fractions by quantitative RT-PCR. We observed a three-fold increase in Hsp70 mRNA in the brain total RNAs from flies expressing rCGG60 repeats in pan-neuronal cells compared with wild-type flies (Figure 3A). Interestingly, Hsp70 transcripts were enriched two-fold in the nuclear fraction, while there was no significant change detected in the cytoplasmic fraction from rCGG-expressing flies (Figure 3C). As a negative control, an unrelated transcript, dFmr1, was found to be unaltered in either nuclear or cytoplasmic fractions. Furthermore, the elevated expression of Hsp70 mRNA in flies expressing fragile X premutation rCGG repeats is likely mediated by Rm62, since the partial loss of Rm62 could significantly increase the expression of Hsp70 in rCGG-expressing fly brains.
Fig. 3. Fragile X premutation rCGG repeats cause the nuclear accumulation of Hsp70 mRNA. 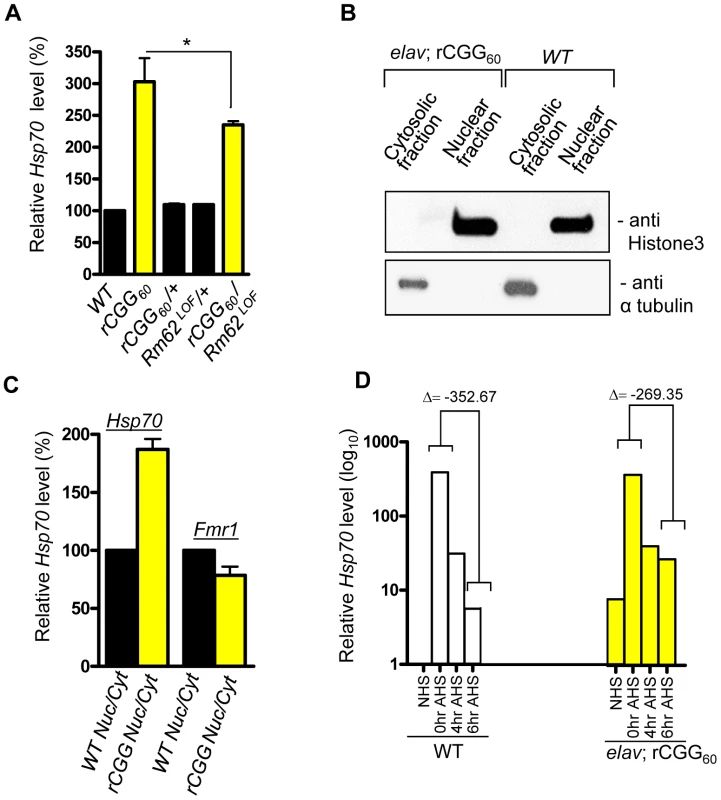
A. Quantitative analysis of Hsp70 mRNA levels by real-time PCR from the adult heads of genotypes: +/+ (wild-type (WT)); elav; rCGG60 (rCGG-expressing homozygous flies); elav/+; rCGG60/+ (rCGG-heterozygous flies); Rm62LOF/+ (Rm62 mutation heterozygous flies); and rCGG60/+; Rm62LOF/+ (interaction). Housekeeping ribosomal protein 32 (Rpl32) mRNA was used as an internal control. *: p<0.05 B. Western blot with anti-histone 3 antibody, and α tubulin. C. Quantitative analysis of Hsp70 mRNA levels by real-time PCR on cytoplasmic and nuclear RNA fractions obtained from adult heads of wild-type (WT) and rCGG-expressing flies. Rpl32 mRNA was used as control. D. Quantitative analysis of Hsp70 mRNA levels in total RNA fractions upon heat shock. Both wild-type (WT) and rCGG-expressing flies were heat shocked for 30 min. No heat shock (NHS) represents non-heat shocked controls. Flies were decapitated at the indicated time after heat shock. Heads were collected and total RNA isolated from them. Both WT and rCGG-expressing flies displayed robust expression of Hsp70 in response to heat shock. After the removal of heat shock, Hsp70 transcripts declined radically in the WT, whereas in rCGG-expressing flies, Hsp70 transcripts show prolonged accumulation. Control samples do not display any overt differences in the timing or expression levels of Hsp70 during the initial response to a short heat shock. Real time against Fmr1 serves as a control on fractionated samples. The data represent mean ± SEM, n = 3. Previous studies have shown that Rm62 functions in RNA export and is involved in the export of Hsp70 mRNA following heat shock [31]. The loss of Rm62 will retard RNA export and increase the time required to shut off the transcription of Hsp70 mRNA [31], so we administered a 30-minute heat shock to both wild-type flies and flies expressing rCGG in pan-neuronal cells (Elav-GAL4; UAS-(CGG)60-EGFP) and allowed them to recover for up to six hours. During the recovery period (0 to 6 h after heat shock (AHS)), timed samples were removed and analyzed for Hsp70 transcript production. The control sample showed high levels of Hsp70 mRNA had accumulated only for a short period following the heat shock, and these levels then began to fall, consistent with previously described kinetics of the heat shock response [32], [33] (Figure 3D). However, in rCGG60 transgenic flies, Hsp70 transcripts persisted for a longer time and could be detected even six hours after the recovery period. This is in contrast to the control samples, where by the end of six hours, Hsp70 transcripts had dropped to near background levels, reflecting the known rapid shutoff and export of Hsp70 mRNA in the hours following a 30-minute heat shock (Figure 3D). This observation is consistent with the earlier finding that the loss of Rm62 could delay the clearance of HS-inducible Hsp70 mRNA. The elevated Hsp70 level in Elav; rCGG transgenic flies prior to heat shock could reflect chronic stress induced by the expression of premutation rCGG repeats. Taken together, these results suggest possible deficits in the nuclear export of Hsp70 mRNA in the presence of Fragile X premutation rCGG repeats, due to reduced Rm62 levels.
Nuclear accumulation of selective mRNAs caused by fragile X premutation rCGG repeats
On the premise that rCGG repeat expression in neurons might influence the RNA export pathway, we sought to systematically identify additional RNAs that are altered in their nuclear-cytoplasmic distribution in the presence of fragile X premutation rCGG repeats. To this end, we carried out microarray studies using total RNAs and RNAs isolated from both nuclear and cytoplasmic fractions. A graphical representation of the experimental design and results is shown in Figure 4A. In the first set, total RNA was isolated from age - and sex-matched heads from wild-type (WT) flies and flies expressing rCGG60 repeats in neurons, and these were classified as wild-type-total and rCGG60-total, respectively. In the second set, RNAs from cytoplasmic fractions were taken from the respective age - and sex-matched heads of wild-type (WT) flies and flies expressing rCGG60 repeats in neurons, and these were classified as wild-type-cytoplasmic and rCGG60-cytoplasmic, respectively. In the third set, RNAs from nuclear fractions were taken from the respective age - and sex-matched heads of wild-type (WT) flies and flies expressing rCGG60 repeats in neurons, and these were classified as wild-type-nuclear and rCGG60-nuclear, respectively. To ensure efficient cell fractionation, all the criteria mentioned above had to be met. Also, we used the same microarray procedures and analyses for each sample. Three biological replicates were analyzed for each class using Affymetrix Drosophila Genome 2.0 Arrays. The signal intensities of each chip were normalized with RMA algorithms. The correlation coefficients for the three biological replicates were greater than 0.93 in all sets, indicating that the gene expression data obtained in this study were highly reproducible. Genes that discriminate among classes were sorted by p-value of the two-sample t-test (with random variance model). In set 1, out of 18,952 probe sets, 1,386 genes were significant at the 0.001 level of the univariate test. In set 2, out of 18,952 probe sets, 3,000 genes were significant at the 0.001 level of the univariate test. Likewise, in set 3, out of 18,952 probe sets, 693 genes were significant at the 0.001 level of the univariate test. For those genes that expressed differentially between the classes within a particular set, we used a fold-change of two or higher as the filtering criterion, as represented in a typical scatter plot of mean log intensities for each sample (Figure 4B). The number of unique RNAs increased in rCGG-total samples, and rCGG-nuclear as well as rCGG-cytoplasmic fractions were displayed in a Venn diagram. Out of 108 genes/probes that were increased in the rCGG-nuclear class, there were overlaps of 36 and 27 genes, with 228 and 237 genes increased in the rCGG-total and rCGG-cytoplasmic class, respectively. Therefore, the remaining 45 genes unique to the rCGG-nuclear class might represent genes that are only altered in their nuclear cytoplasmic distribution, but without a change in the overall transcriptional level in response to rCGG expression (Figure 4C). The identified genes were then annotated with functional assignments and sorted into gene-related categories using a DAVID functional annotation tool [34]. It is noteworthy that genes involved in both immune response and stress response are enriched in our gene list with statistical values of (15%, p value: 2.77E-07), and (10%, p value: 1.34E-05) respectively. We further compared these 45 genes between the classes within a particular set based on the fold enrichment of their intensities of normalized log-transformed gene expressions. As depicted in Figure 4D, genes showing a remarkable fold change were highlighted. For validation, eight genes were selected, including stress response genes, Hsps, Attacin, Turandot, and Cytochrome P450 enzyme. As shown in Figure 4E, quantitative RT PCR on RNA derived from total brain lysates of wild-type and rCGG60 repeat flies indeed showed significant changes in their expression. Hsp70Aa and Fmr1 were used as positive and negative controls, respectively. These findings confirm that nuclear accumulation in the presence of fragile X premutation rCGG repeats is not limited to the Hsp70 transcript and also suggest that the nuclear accumulation of these transcripts could be an important factor in the sequelae associated with FXTAS.
Fig. 4. Identification via microarray analyses of selective mRNAs that accumulate in the nucleus as a result of fragile X premutation rCGG repeats. 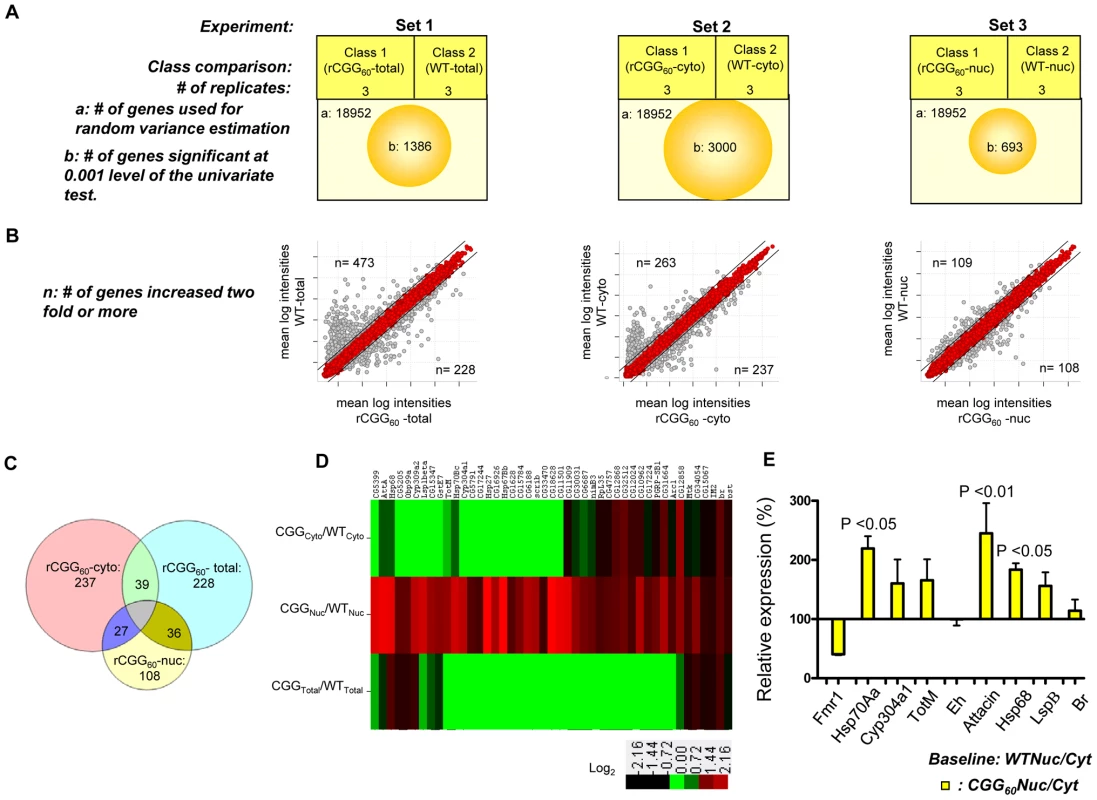
A. The three sets of microarray experiments were carried out in triplicate (three biological replicates) with equal amounts of total RNA obtained from the Drosophila heads of control total, nuclear, and cytoplasmic fractions and the corresponding experimental premutation rCGG repeat samples of total, nuclear, and cytoplasmic fractions. Microarray analyses were carried out by identifying significantly changed genes at the 0.001 level of the univariate test. B. Scatter plot of mean log intensities of each sample demonstrating differentially expressed significant genes with fold-change of 2 or more between the classes within a particular set. C. The numbers of unique differentially expressed genes generated by the different cellular compartments of rCGG sample (comparison of total, cytoplasmic, and nuclear fraction) are displayed in Venn diagrams. D. Fold enrichment depicted by ratios between the intensities of normalized log-transformed gene expressions for 45 genes unique to CGG nuclear fractions in various classes is displayed using the Cluster and TreeView programs for the wild-type and premutation rCGG datasets. The fold of the change is indicated on both sides of the scale bar E. Validation of nuclear enrichment by real-time PCR analysis of the selective genes. Real time against Fmr1 serves as a control. The data represent mean ± SEM, n = 3. rCGG repeats genetically interact with small bristles (sbr), the Drosophila homolog of the human mRNA export factor NXF1
In Drosophila, the small bristles (sbr) and Hel25E genes, homologs of the human mRNA export factor NXF1 and UASP56 RNA helicase, respectively, are essential for exporting the bulk of Drosophila mRNAs, besides heat shock transcripts [35]–[39]. In normal cells, RNA export protein complex shuttles between transcription sites and nuclear pores. Rm62 functions to rapidly remove transcripts from the transcription sites before they begin to move towards nuclear pores. The loss of Rm62 leads to redistribution of export factors within nuclei. Unlike normal cells, where Sbr and other export factors are distributed along the rim of the nucleus and in the nucleoplasm, Rm62 mutations lead to accumulation of Sbr and other export factors in the nucleoplasm. Strong alleles of sbr are lethal, and animals bearing weaker sbr alleles can survive to adulthood, with a few having abnormally thin thoracic bristles, and females being sterile [39]. Rm62 mutations phenotypically resemble mutations in small bristles (sbr) (i.e., lethal and female-sterile) and retain completed mRNAs at their transcription sites [31]. These changes in export factor distribution, as well as the similarities between the Rm62 and sbr mutations, suggest that Rm62 functions upstream or in conjunction with other export factors at sites of gene transcription. If rCGG repeats deplete the Rm62 pool and show biochemical as well as genetic interactions with it, then rCGG repeats might also interact with known RNA export components. To test this hypothesis, male sbr heterozygotes were crossed with elav-GAL4 (X); rCGG60-EGFP. The F1 progenies were collected every 24 hours for four consecutive days or until culture exhausted. A heterozygous combination of sbr/elav-GAL4; rCGG60-EGFP appears to have delayed development and displayed 30–40 times the lethality of its siblings (p = 0.0035). Another cross between sbr/y as well as Elav (X); rCGG60-EGFP separately with wild-type (w1118) did not produce any lethality, suggesting that rCGG repeats influence the Sbr-mediated mRNA export pathway (Figure 5A).
Fig. 5. Fragile X premutation rCGG repeats display genetic interaction with the nuclear export factor, small bristles, and the expression of molecular chaperone Hsp70 suppresses rCGG repeat-mediated neurodegeneration in vivo. 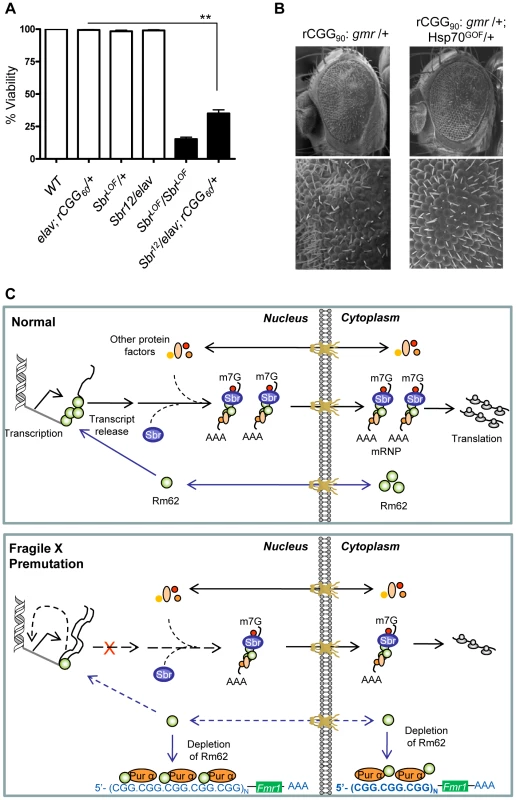
A. Statistical evaluation of the percent viability displayed by Sbr homozygous and heterozygous flies or premutation heterozygous flies, and their respective interaction experiments Sbr12LOF/elav; rCGG60-EGFP/+. Genotypes: +/+ (wild-type); Sbr12LOF/Sbr12LOF; Sbr12LOF/+; elav/+; rCGG60-EGFP/+ and Sbr12LOF/elav; rCGG60-EGFP/+ (interaction). Mean of three data sets was used. Error bars indicate SEM. **: p<0.001. B. Shown are SEM pictures of the eyes of adult flies expressing gmr: (CGG)90-EGFP/+ only (left), in comparison to Gmr: (CGG)90-EGFP/+; Hsp70GOF/+. C. Model representing various interactions involving fragile X premutation rCGG repeats and its potential impact on mRNA nuclear export. Discussion
Fragile X-associated tremor/ataxia syndrome is a neurodegenerative disorder seen in adult fragile X syndrome premutation carriers. Although several studies, including our own, have proposed a model of RNA-mediated sequestration, in which the sequestration of several RNA-binding proteins (RBP) by rCGG repeat-binding causes cell death and neurodegeneration, the precise mechanisms by which expanded rCGG repeats exert their toxic effects are less understood. Here we report an unexpected nuclear accumulation of specific mRNAs caused by fragile X premutation rCGG repeats, which indicates that a potential deficit in the nuclear export of these mRNAs could contribute to the pathogenesis of FXTAS.
Our earlier studies demonstrated that Pur α and hnRNP A2/B1 are rCGG-repeat-binding proteins and that they could modulate rCGG-mediated neuronal toxicity [17], [18]. In particular, the loss of Pur α in mice is known to cause neurological phenotypes reminiscent of those observed in FXTAS patients [19]. Therefore, to better understand the set of molecular interactions that are dysregulated as a consequence of the sequestration of rCGG repeat-binding proteins (RBPs), we used proteomic analyses to identify the Pur α interactome, which includes proteins involved in different biological pathways. However, whether these interacting proteins could be involved in rCGG repeat-mediated neuronal toxicity was unclear. To address this question, we further tested the roles of Pur α-interacting proteins with the existing fly mutants in rCGG-mediated neurodegeneration. Interestingly, two proteins, Rm62 and Hts, the Drosophila orthologs of the p68 RNA helicase and Adducin, respectively, were found to prominently modulate rCGG-mediated neuronal toxicity. These findings suggest that, despite Pur α being involved in multiple biological pathways, only selective pathway(s) altered by fragile X premutation rCGG repeats and the sequestration of Pur α are involved in rCGG-mediated neurodegeneration. Our methods demonstrate the utility and power of combining a proteomic approach with genetic-interaction tests to identify key biological pathway(s) relevant to human diseases.
Rm62/Dmp68 is a member of the DEAD-box family, which includes a large number of proteins that play important roles in almost all aspects of RNA metabolism [28], [29]. Specifically, Rm62/Dmp68 is found to be a multifunctional protein involved in gene transcription, pre-mRNA, pre-rRNA, and pre-miRNA processing, RNAi, and RNA export [21]-[23],[25],[26],[30],[31]. The belief now is that Rm62/Dmp68 may be associated with mRNP export, promoting gene silencing by removing transcripts from their active site of synthesis to promote an inactive heterochromatin configuration [31]. The mammalian ortholog of Rm62/Dmp68 is found to shuttle between cytoplasm and the nucleus [30]. In flies, disruption of its action alters the nuclear distribution of known RNA export components, producing defects similar to those seen in sbr mutants, a known component of the RNA export machinery [31]. Given the importance of Rm62/Dmp68 as part of an mRNP complex involved in mRNA transport, it is likely that its depletion or sequestration caused by fragile X premutation rCGG repeats could lead to dysfunction of mRNA transport in the cell and eventual cell death and neuronal toxicity. In line with our observation, other studies have shown that premutation rCGG repeats can biochemically associate with several other DEAD-box RNA helicases, like DDX3, DDX5/p68, and DDX17, as well as colocalize with CGG60 RNA aggregates, resulting in the formation of inclusions in cells [40]; this suggests sequestration as a potential mechanism for the Rm62 posttranscriptional reduction.
In our FXTAS fly model, total steady-state levels of Hsp70 transcripts are increased, which could reflect an initial cellular response to the stress caused by fragile X premutation rCGG repeats. However, we found that most of the increased Hsp70 mRNAs are trapped in the nucleus, instead of being transported out of the nucleus to produce Hsp70 protein and respond to cell stress. Interestingly, this altered distribution of Hsp70 transcripts and their delay in turnover is similar to what was seen with Hsp70 transcripts in the Rm62/p68 mutation. Therefore, fragile X premutation rCGG repeats could likely produce deficits in the nuclear export of selective transcripts mediated by Rm62, such as Hsp70 mRNA. This nuclear accumulation will interfere with the neurons' ability to produce sufficient proteins to respond to cellular stress (Figure 5C). To identify additional accumulated mRNAs in the nucleus of neurons expressing fragile X premutation rCGG repeats, we performed gene expression profiling using total RNAs and RNAs from both nuclear and cytoplasmic fractions. Through these analyses, we identified transcripts that were uniquely enriched/restricted in the nuclear fraction. The gene functions of these nuclear-accumulated transcripts cover a variety of biological processes. For example, gene functions like oxidoreductases indicate the involvement of oxidative stress. Likewise, chaperones indicate maintenance of the conformational homeostasis of cellular proteins and RNAs. The cytokine-like interleukin-6 (IL-6) and immune-induced molecules (IM2) regulate part of the immune system. Discovering that these nuclear-enriched transcripts fell into stress, apoptotic, and immunity/defense pathways rather than other biological processes implies that they are either a causative event, or are a consequence of FXTAS pathogenesis. There is a growing body of evidence from both animal and human studies to suggest that stress can intensify inflammation and increase a person's risk for developing various neurological disorders, like Parkinson's disease, multiple sclerosis (MS), and other inflammatory diseases [41], [42]. These studies, along with our findings presented here, suggest that in the presence of toxic RNAs, such as fragile X premutation rCGG repeats, a continuous build up of stress might allow the inflammatory process to run amok and may compromise homeostasis integrity, triggering neuronal cell death/neurodegeneration. Significantly with regard to the data provided above, overexpression of Hsp70 suppresses toxicity of premutation rCGG repeats (Figure 5B). This paradox could be explained by that the nucleus becomes saturated due to induced Hsp70 transcripts that are forced out of the nucleus to respond to cell stress. While our study provides insights into potential roles of stress response pathways in FXTAS neurodegeneration, in the future, a detailed analysis of accumulated mRNAs will be important to understand their implications in the pathogenesis of FXTAS.
In summary, through systematic proteomic, genetic, and microarray analyses, here we show that the nuclear accumulation of select mRNAs caused by fragile X premutation rCGG repeats may contribute to FXTAS pathogenesis, and the mechanism could be via impaired nuclear export due to the decreased levels of Rm62 seen upon fragile X premutation rCGG expression.
Materials and Methods
Drosophila strains and genetics
The wild-type strain used was W118. All insertions, Elav-Gal4 (C115) and Gmr-Gal4, were from Bloomington Stock Centre (Bloomington, Indiana, USA). Transgenic flies expressing rCGG90 and rCGG60 repeats were previously generated in the lab (Peng Jin, Emory University School of Medicine, Atlanta, USA). A stable line expressing rCGG60 repeats under the control of Elav-Gal4 was established. Rm6201084 and Rm62CB02119 fly lines were kindly provided by Allan Spradling. Rm62(3)3607, the Rm62 overexpression fly line, was kindly provided by Mani Ramaswami. Other P-element lesions as depicted in Table 2 were from Bloomington. All crosses were grown on standard medium at 25°C, except that the Elav; rCCG60 line was maintained at room temperature.
Construction, expression, and purification of GST-Pur α fusion protein
Drosophila melanogaster Pur α cDNAs were cloned into vector pGEX-2TK using the respective pairs of primers 5′-ATGTGGATCCATGGAAGATCTCCTCGTG-3′ and 5′ - ATGTGAATTCTTAGGACGTGCCATTGAC-3′ with the restriction sites (underlined). Rm62 coding sequence containing a site for ‘Gateway entry clone’ was obtained from cDNA, RE11923, using the respective pairs of primers 5′ CACCATGCTTAAGCTTGTGCAATACATAG and GTCGAAGCGCGAGTGTCT with the entry site (underlined). Rm62 CDS was subsequently cloned in pDEST24 Gateway expression vector. The open reading frame of both fusion genes was confirmed by sequencing. Both pGEX-Pur α and pGEX-2TK were individually transformed into the E. coli expression strain BL21 (DE3). Overexpression of pGEX-Pur α or pGEX-2TK alone was induced by 16 µM IPTG for 7 h at room temperature. Overexpression of pDEST-Rm62 or pDEST24 alone was induced by 0.2% L-arabinose for 8 h at room temperature. Bacteria expressing corresponding proteins were lysed in B-PER Protein Extraction Reagents (Pierce, Thermo Scientific). Purification of GST-tagged Pur α and GST alone was carried out by affinity chromatography using glutathione immobilized to a matrix sepharose, which were treated with both DNase and RNase before being used for capture experiment.
Capture of the interacting proteins with dPur α
The GST-fusion protein affinity purification was performed based on a previously reported procedure [43]. Total protein lysates were obtained from adult wild-type fly heads using 50 mM Hepes, pH 7.2, 0.6 M NaCl, 15% glycerol, 20 mM CHAPS, 0.1% Triton, 1 mM DTT, 1 mM EGTA, 1 mM EDTA, and protease inhibitors (Complete Mini, Roche). Ten milliliters of each extract (10 mg/ml) were dialyzed against 1L of dialysis buffer (20 mM Hepes, pH 7.2, 0.1 M NaCl, 5% glycerol, 1 mM DTT, 1 mM EGTA, 1 mM EDTA) twice to reduce the concentration of salt and detergent. After dialysis, 1 mM of DTT and protease inhibitors were added and centrifuged at 500,000 g for 2 h (60K on SW 60 Ti rotor). Supernatant was collected and run through columns of glutathione beads (200 µl) previously equilibrated with the purified GST-Pur α or GST alone. After washing, the sequential elution of bound proteins was performed with the following buffers: Buffer 1 (20 mM Hepes, pH 7.2, 0.2 M to NaCl, 15% glycerol, 0.1% Triton, 1 mM DTT, 1 mM EGTA, 1 mM EDTA); Buffer 2 (20 mM Hepes, pH 7.2, 0.6 M NaCl, 15% glycerol, 0.5% Triton, 1 mM DTT, 1 mM EGTA, 1 mM EDTA); and Buffer 3 (20 mM Hepes, pH 7.2, 2 M NaCl, 15% glycerol, 2% Triton, 1 mM DTT, 1 mM EGTA, 1 mM EDTA), respectively. Eluants were combined, precipitated with cold acetone, washed, and resuspended in protein loading buffer (2% SDS, 60 mM Tris-HCl pH 6.8, 100 mM DTT, 10% glycerol, 0.01% bromophenol blue) for mass spectrometric sequencing analyses.
Liquid chromatography-tandem mass spectrometry (LC-MS/MS)
Protein identification was performed on an optimized LC-MS/MS platform as described previously [44]. The captured proteins by affinity-purified GST-Pur α and GST alone were separated on a 10% SDS gel, and each lane was cut into multiple pieces, followed by in-gel tryptic digestion. Peptide samples were dissolved and analyzed by nanoscale reverse phase chromatography coupled with an LTQ-Orbitrap mass spectrometer (Thermo Finnigan). All MS/MS spectra were searched against a composite database containing the NCBI mouse or fly reference database and a decoy database. The dataset was filtered to reduce the protein false-discovery rate to less than 1% [45]. To further eliminate false identifications, proteins matched by a single peptide were manually validated.
GST pull-down assay
Full-length cDNA of Rm62 (RE11923) and Hts (SD02552) were obtained from DGRC, Indiana, USA. Both cDNAs were separately used for in vitro transcription–translation in rabbit reticulocyte lysate in the presence of [35S] methionine, according to the manufacturer's instructions (Promega). In vitro-translated proteins were mixed with purified GST-Pur α or GST alone. The pull-down assay was performed in the presence of 150 mM NaCl. Total cell lysate obtained from S2 cells or from adult fly heads or from mouse cerebellum was incubated overnight with Pur α-GST or Rm62-GST or GST only. Beads were extensively washed in lysis buffer, directly boiled in SDS-PAGE loading buffer, and subjected to SDS-PAGE analysis. 1X proteinase inhibitor was maintained at each step. For the RNAse treatment, a potent RNase Cocktail, a mixture of RNase A (500 U/ml) and RNase T1 (20,000 U/ml) (Ambion, Applied Biosystems) was used in a dilution of 2.5 µl/50 µl of sample.
Protein and RNA isolation
Heads from adult flies expressing either fragile X premutation rCGG repeats (Elav-GAL4; UAS-(CGG)60-EGFP) or WT were collected and homogenized in 200 µL of ice-cold lysis buffer (10 mM Tris, pH 7.4, 150 mM NaCl, 30 mM EDTA, 0.5% Triton X-100) with 2X complete protease inhibitors (Roche) and kept on ice for 30 min. Supernatant of 12,000 g was collected, and the amount of total protein was determined by Bradford assay.
S2 cells were cultured in Schneider cell medium (Gibco BRL/Invitrogen) + 10% FBS. Total cell extracts were prepared by lysing S2 cells in buffer (150 mM NaCl, 20 mM Tris-Hcl pH 7.4, 5 mM MgCl2, 0.4% Triton X-100, protease inhibitor (complete mini, Roche)), followed by incubation on ice for 30 min. The supernatant of 12,000 g was collected, and the amount of total protein was determined by Bradford assay.
For the RNA isolation, adult heads of the required genotype were mashed in Trizol (Gibco BRL Life Technologies) using plastic kontes. After precipitation, RNA was further cleaned with RNeasy (QIAGEN).
Western blot analysis
We used standard Western blotting techniques with Tris Buffer, pH 8.3. For the detection of Rm62, transfer was carried in CAPS buffer, pH 11. Antibodies were used at the following concentrations: rat anti-Rm62 (1∶800; kind gift from Elissa Lei, Bethesda, MD, USA); Hts supernatant (1∶5; Hybridoma); rabbit anti-Pur α (generated previously in the lab; mMad1 (kind gift from Frances Fuller-Pace); histone H3 (1∶5000, Abcam);α tubulin (1∶1000, Abcam); goat anti-mouse HRP (1∶5000); Sigma); goat anti-rabbit HRP (1∶5000; Sigma); and goat anti-rat HRP (1∶5000; The Jackson Laboratory).
RT-PCR and quantitative RT-PCRs
For RT-PCR, RNA was reverse-transcribed with random primers using the high-capacity cDNA reverse transcription kit (Applied Biosystems) according to the manufacturer's instructions. Real-time PCR was performed with gene-specific primers and Power SYBR Green PCR Master Mix (Applied Biosystems) using 7500 Fast Real-Time PCR system (Applied Biosystems). Primers were designed using Primer Express software (Applied Biosystems) or the commercially available QuantiTect primers from Qiagen. For Rm62 the following primer combination was used: CGAGAAGCTAATCAGGAAATCAATC and ACCGTCGTAGCGCGAGTT. For Hsp70 the following primer combination was used: CTCGTCGGCGGATCCA and GGAAGAACTCCTGCAGCAGACT. For Fmr1 the following primer combination was used: CTCGTCGGCCCAGCA and GGAAGAACTTCGAGCAGACT. For the endogenous control, QuantiTect Dm_RpL32_1_SG (Qiagen) was used according to the manufacturer's instructions.
Preparation of nuclear and cytoplasmic fractionations
Flies of the required genotypes were snap-frozen in dry ice and then decapitated. Heads were collected and crushed to powder using motorized plastic kontes in dry ice. Powdered heads were resuspended in cell fractionation buffer from the PARIS kit (Ambion) and kept on ice for 30 min to 1 h, followed by low-speed centrifugation of 500 g to pellet nuclei together with debris. Carefully, supernatant was collected as the cytoplasmic fraction, and the pellet as nuclear fraction. The pellet/nuclear fraction was washed again with cell fractionation buffer to remove any lingering supernatant. Protein and RNA were isolated from the same homogenate using the PARIS kit (Ambion) according to the manufacturer's instructions. RNA was quantified, and RT-PCR was performed using ABI (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions.
Microscopy and immunohistochemistry
Genetic interactions based on the rough eye enhancement or suppression were initially assessed under the dissection microscope for more than 50 flies. To confirm suppression and to document phenotypes, we compared 6-10 SEM images of eyes from experimental animals with those of “average” (rCGG)90. For scanning electron microscopy (SEM) images, whole flies were serially dehydrated in ethanol, dried with hexamethyldisilazane (Sigma-Aldrich), and analyzed with an ISI DS-130 LaB6 SEM/STEM microscope.
Microarray analyses
cRNA amplification and fluorescence labeling was performed according to the supplier's instructions using the Affymetrix 3′IVT kit (Affymetrix Technologies). The labeled target was combined and allowed to hybridize to probes on the Drosophila genome 2.0 array according to instructions. The arrays were washed using the Midi _euk2v3 fluidics protocol and scanned using the Microarray Scanner laser-based detection system. All normalizations were performed using default settings.
Image data were quantified using the Affymetrix expression console. All analysis was performed using Bayesian infinite mixture models as implemented in the BBR software, version 3.8.1 (http://linus.nci.nih.gov/BRB-ArrayTools.html), an integrated package for the visualization and statistical analysis of DNA microarray gene expression data. Gene expression data were normalized using the robust multi-array average (RMA) statistical algorithms built in BRB. All filtering parameters were turned off. Class comparison and cluster analysis was performed using the Bayesian infinite mixture models as implemented in the BBR software. Heat map was generated by clustering genes and arrays with complete linkage uncentered correlation using Cluster 3 and Java TreeView [46].
Significantly differentially expressed genes were annotated with functional assignments to help determine which gene categories were enriched with differentially expressed genes. Genes were annotated and biological processes analyzed using the Database for Annotation, Visualization and Integrated Discovery (DAVID) (http://david.abcc.ncifcrf.gov/) [34].
Statistical method
Statistical analysis was performed using ANOVA with post-hoc t-tests (two samples assuming equal variances) using GraphPad Prism software to determine significance, and indicated p values. All data are shown as mean with standard error of mean (mean ± SEM).
Zdroje
1. WarrenSTShermanSL 2001 The fragile X syndrome. ScriverCRBeaudetALValleDChildsBKinzlerKWVogelsteinB The Metabolic & Molecular Bases of Inherited Disease New York McGraw-Hill Companies 1257 1290
2. ShermanS 2002 Epidemiology. HagermanRJ Fragile X Syndrome: Diagnosis, Treatment and Research Baltimore, MD The Johns Hopkins University Press 136 168
3. HagermanRJHagermanPJ 2002 The fragile X premutation: into the phenotypic fold. Curr Opin Genet Dev 12 278 283
4. HagermanPJHagermanRJ 2004 The fragile-X premutation: a maturing perspective. Am J Hum Genet 74 805 816
5. GrigsbyJBregaAGJacquemontSLoeschDZLeeheyMA 2006 Impairment in the cognitive functioning of men with fragile X-associated tremor/ataxia syndrome (FXTAS). J Neurol Sci 248 227 233
6. BacalmanSFarzinFBourgeoisJACogswellJGoodlin-JonesBL 2006 Psychiatric phenotype of the fragile X-associated tremor/ataxia syndrome (FXTAS) in males: newly described fronto-subcortical dementia. J Clin Psychiatry 67 87 94
7. HesslDTassoneFLoeschDZBerry-KravisELeeheyMA 2005 Abnormal elevation of FMR1 mRNA is associated with psychological symptoms in individuals with the fragile X premutation. Am J Med Genet B Neuropsychiatr Genet 139 115 121
8. JacquemontSFarzinFHallDLeeheyMTassoneF 2004 Aging in individuals with the FMR1 mutation. Am J Ment Retard 109 154 164
9. GrecoCMBermanRFMartinRMTassoneFSchwartzPH 2006 Neuropathology of fragile X-associated tremor/ataxia syndrome (FXTAS). Brain 129 243 255
10. GrecoCMHagermanRJTassoneFChudleyAEDel BigioMR 2002 Neuronal intranuclear inclusions in a new cerebellar tremor/ataxia syndrome among fragile X carriers. Brain 125 1760 1771
11. TassoneFHagermanRJTaylorAKGaneLWGodfreyTE 2000 Elevated levels of FMR1 mRNA in carrier males: a new mechanism of involvement in the fragile-X syndrome. Am J Hum Genet 66 6 15
12. KennesonAZhangFHagedornCHWarrenST 2001 Reduced FMRP and increased FMR1 transcription is proportionally associated with CGG repeat number in intermediate-length and premutation carriers. Hum Mol Genet 10 1449 1454
13. JinPZarnescuDCZhangFPearsonCELucchesiJC 2003 RNA-mediated neurodegeneration caused by the fragile X premutation rCGG repeats in Drosophila. Neuron 39 739 747
14. WillemsenRHoogeveen-WesterveldMReisSHolstegeJSeverijnenLA 2003 The FMR1 CGG repeat mouse displays ubiquitin-positive intranuclear neuronal inclusions; implications for the cerebellar tremor/ataxia syndrome. Hum Mol Genet 12 949 959
15. TassoneFIwahashiCHagermanPJ 2004 FMR1 RNA within the intranuclear inclusions of fragile X-associated tremor/ataxia syndrome (FXTAS). RNA Biology 1 103 105
16. ArocenaDGIwahashiCKWonNBeilinaALudwigAL 2005 Induction of inclusion formation and disruption of lamin A/C structure by premutation CGG-repeat RNA in human cultured neural cells. Hum Mol Genet 14 3661 3671
17. JinPDuanRQurashiAQinYTianD 2007 Pur alpha binds to rCGG repeats and modulates repeat-mediated neurodegeneration in a Drosophila model of fragile X tremor/ataxia syndrome. Neuron 55 556 564
18. SofolaOAJinPQinYDuanRLiuH 2007 RNA-binding proteins hnRNP A2/B1 and CUGBP1 suppress fragile X CGG premutation repeat-induced neurodegeneration in a Drosophila model of FXTAS. Neuron 55 565 571
19. KhaliliKDel ValleLMuralidharanVGaultWJDarbinianN 2003 Puralpha is essential for postnatal brain development and developmentally coupled cellular proliferation as revealed by genetic inactivation in the mouse. Mol Cell Biol 23 6857 6875
20. SpradlingACSternDMKissIRooteJLavertyT 1995 Gene disruptions using P transposable elements: an integral component of the Drosophila genome project. Proc Natl Acad Sci U S A 92 10824 10830
21. BondATMangusDAHeFJacobsonA 2001 Absence of Dbp2p alters both nonsense-mediated mRNA decay and rRNA processing. Mol Cell Biol 21 7366 7379
22. EndohHMaruyamaKMasuhiroYKobayashiYGotoM 1999 Purification and identification of p68 RNA helicase acting as a transcriptional coactivator specific for the activation function 1 of human estrogen receptor alpha. Mol Cell Biol 19 5363 5372
23. IshizukaASiomiMCSiomiH 2002 A Drosophila fragile X protein interacts with components of RNAi and ribosomal proteins. Genes Dev 16 2497 2508
24. LinCYangLYangJJHuangYLiuZR 2005 ATPase/helicase activities of p68 RNA helicase are required for pre-mRNA splicing but not for assembly of the spliceosome. Mol Cell Biol 25 7484 7493
25. LiuZR 2002 p68 RNA helicase is an essential human splicing factor that acts at the U1 snRNA-5′ splice site duplex. Mol Cell Biol 22 5443 5450
26. WilsonBJBatesGJNicolSMGregoryDJPerkinsND 2004 The p68 and p72 DEAD box RNA helicases interact with HDAC1 and repress transcription in a promoter-specific manner. BMC Mol Biol 5 11
27. YueLSpradlingAC 1992 hu-li tai shao, a gene required for ring canal formation during Drosophila oogenesis, encodes a homolog of adducin. Genes Dev 6 2443 2454
28. Fuller-PaceFV 2006 DExD/H box RNA helicases: multifunctional proteins with important roles in transcriptional regulation. Nucleic Acids Res 34 4206 4215
29. Janknecht R Multi-talented DEAD-box proteins and potential tumor promoters: p68 RNA helicase (DDX5) and its paralog, p72 RNA helicase (DDX17). Am J Transl Res 2 223 234
30. WangHGaoXHuangYYangJLiuZR 2009 P68 RNA helicase is a nucleocytoplasmic shuttling protein. Cell Res 19 1388 1400
31. BuszczakMSpradlingAC 2006 The Drosophila P68 RNA helicase regulates transcriptional deactivation by promoting RNA release from chromatin. Genes Dev 20 977 989
32. DiDomenicoBJBugaiskyGELindquistS 1982 The heat shock response is self-regulated at both the transcriptional and posttranscriptional levels. Cell 31 593 603
33. DiDomenicoBJBugaiskyGELindquistS 1982 Heat shock and recovery are mediated by different translational mechanisms. Proc Natl Acad Sci U S A 79 6181 6185
34. DennisGJrShermanBTHosackDAYangJGaoW 2003 DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol 4 P3
35. GatfieldDLe HirHSchmittCBraunICKocherT 2001 The DExH/D box protein HEL/UAP56 is essential for mRNA nuclear export in Drosophila. Curr Biol 11 1716 1721
36. HeroldAKlymenkoTIzaurraldeE 2001 NXF1/p15 heterodimers are essential for mRNA nuclear export in Drosophila. Rna 7 1768 1780
37. HeroldATeixeiraLIzaurraldeE 2003 Genome-wide analysis of nuclear mRNA export pathways in Drosophila. Embo J 22 2472 2483
38. JimenoSRondonAGLunaRAguileraA 2002 The yeast THO complex and mRNA export factors link RNA metabolism with transcription and genome instability. Embo J 21 3526 3535
39. WilkieGSZimyaninVKirbyRKoreyCFrancis-LangH 2001 Small bristles, the Drosophila ortholog of NXF-1, is essential for mRNA export throughout development. Rna 7 1781 1792
40. SellierCRauFLiuYTassoneFHukemaRK 2010 Sam68 sequestration and partial loss of function are associated with splicing alterations in FXTAS patients. EMBO J 29 1248 1261
41. BarnhamKJMastersCLBushAI 2004 Neurodegenerative diseases and oxidative stress. Nat Rev Drug Discov 3 205 214
42. Savvateeva-PopovaEMedvedevaAPopovAEvgen'evM 2008 Role of non-coding RNAs in neurodegeneration and stress response in Drosophila. Biotechnol J 3 1010 1021
43. ChoeEALiaoLZhouJYChengDDuongDM 2007 Neuronal morphogenesis is regulated by the interplay between cyclin-dependent kinase 5 and the ubiquitin ligase mind bomb 1. J Neurosci 27 9503 9512
44. XuPDuongDMPengJ 2009 Systematical optimization of reverse-phase chromatography for shotgun proteomics. J Proteome Res 8 3944 3950
45. PengJEliasJEThoreenCCLickliderLJGygiSP 2003 Evaluation of multidimensional chromatography coupled with tandem mass spectrometry (LC/LC-MS/MS) for large-scale protein analysis: the yeast proteome. J Proteome Res 2 43 50
46. EisenMBSpellmanPTBrownPOBotsteinD 1998 Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A 95 14863 14868
Štítky
Genetika Reprodukční medicína
Článek Local Absence of Secondary Structure Permits Translation of mRNAs that Lack Ribosome-Binding SitesČlánek Independent Chromatin Binding of ARGONAUTE4 and SPT5L/KTF1 Mediates Transcriptional Gene SilencingČlánek Trade-Off between Bile Resistance and Nutritional Competence Drives Diversification in the Mouse GutČlánek FGF Signaling Regulates the Number of Posterior Taste Papillae by Controlling Progenitor Field SizeČlánek Mammalian BTBD12 (SLX4) Protects against Genomic Instability during Mammalian SpermatogenesisČlánek Identification of Nine Novel Loci Associated with White Blood Cell Subtypes in a Japanese PopulationČlánek Differential Effects of and Risk Variants on Association with Diabetic ESRD in African AmericansČlánek Dynamic Chromatin Localization of Sirt6 Shapes Stress- and Aging-Related Transcriptional Networks
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2011 Číslo 6
-
Všechny články tohoto čísla
- Local Absence of Secondary Structure Permits Translation of mRNAs that Lack Ribosome-Binding Sites
- Statistical Inference on the Mechanisms of Genome Evolution
- Revisiting Heterochromatin in Embryonic Stem Cells
- A Two-Stage Meta-Analysis Identifies Several New Loci for Parkinson's Disease
- Identification of a Sudden Cardiac Death Susceptibility Locus at 2q24.2 through Genome-Wide Association in European Ancestry Individuals
- Genomic Prevalence of Heterochromatic H3K9me2 and Transcription Do Not Discriminate Pluripotent from Terminally Differentiated Cells
- Epistasis between Beneficial Mutations and the Phenotype-to-Fitness Map for a ssDNA Virus
- Recurrent Chromosome 16p13.1 Duplications Are a Risk Factor for Aortic Dissections
- Telomere DNA Deficiency Is Associated with Development of Human Embryonic Aneuploidy
- Genome-Wide Association Study of White Blood Cell Count in 16,388 African Americans: the Continental Origins and Genetic Epidemiology Network (COGENT)
- Unexpected Role for DNA Polymerase I As a Source of Genetic Variability
- Transportin-SR Is Required for Proper Splicing of Genes and Plant Immunity
- How Chromatin Is Remodelled during DNA Repair of UV-Induced DNA Damage in
- Independent Chromatin Binding of ARGONAUTE4 and SPT5L/KTF1 Mediates Transcriptional Gene Silencing
- Two Evolutionary Histories in the Genome of Rice: the Roles of Domestication Genes
- Natural Allelic Variation Defines a Role for : Trichome Cell Fate Determination
- Multiple Common Susceptibility Variants near BMP Pathway Loci , , and Explain Part of the Missing Heritability of Colorectal Cancer
- Pathogenic Mechanism of the FIG4 Mutation Responsible for Charcot-Marie-Tooth Disease CMT4J
- A Functional Variant in Promoter Modulates Its Expression and Confers Disease Risk for Systemic Lupus Erythematosus
- Drift and Genome Complexity Revisited
- Chromosomal Macrodomains and Associated Proteins: Implications for DNA Organization and Replication in Gram Negative Bacteria
- Trade-Off between Bile Resistance and Nutritional Competence Drives Diversification in the Mouse Gut
- Pathways of Distinction Analysis: A New Technique for Multi–SNP Analysis of GWAS Data
- Web-Based Genome-Wide Association Study Identifies Two Novel Loci and a Substantial Genetic Component for Parkinson's Disease
- Chk2 and p53 Are Haploinsufficient with Dependent and Independent Functions to Eliminate Cells after Telomere Loss
- Exome Sequencing Identifies Mutations in High Myopia
- Distinct Functional Constraints Partition Sequence Conservation in a -Regulatory Element
- CorE from Is a Copper-Dependent RNA Polymerase Sigma Factor
- A Single Sex Pheromone Receptor Determines Chemical Response Specificity of Sexual Behavior in the Silkmoth
- FGF Signaling Regulates the Number of Posterior Taste Papillae by Controlling Progenitor Field Size
- Maps of Open Chromatin Guide the Functional Follow-Up of Genome-Wide Association Signals: Application to Hematological Traits
- Increased Susceptibility to Cortical Spreading Depression in the Mouse Model of Familial Hemiplegic Migraine Type 2
- Differential Gene Expression and Epiregulation of Alpha Zein Gene Copies in Maize Haplotypes
- Parallel Adaptive Divergence among Geographically Diverse Human Populations
- Genetic Analysis of Genome-Scale Recombination Rate Evolution in House Mice
- Mechanisms for the Evolution of a Derived Function in the Ancestral Glucocorticoid Receptor
- Mammalian BTBD12 (SLX4) Protects against Genomic Instability during Mammalian Spermatogenesis
- Interferon Regulatory Factor 8 Regulates Pathways for Antigen Presentation in Myeloid Cells and during Tuberculosis
- High-Resolution Analysis of Parent-of-Origin Allelic Expression in the Arabidopsis Endosperm
- Specific SKN-1/Nrf Stress Responses to Perturbations in Translation Elongation and Proteasome Activity
- Graded Nodal/Activin Signaling Titrates Conversion of Quantitative Phospho-Smad2 Levels into Qualitative Embryonic Stem Cell Fate Decisions
- Genome-Wide Analysis Reveals PADI4 Cooperates with Elk-1 to Activate Expression in Breast Cancer Cells
- Trait Variation in Yeast Is Defined by Population History
- Meiosis-Specific Loading of the Centromere-Specific Histone CENH3 in
- A Genome-Wide Survey of Imprinted Genes in Rice Seeds Reveals Imprinting Primarily Occurs in the Endosperm
- Multiple Regulatory Mechanisms to Inhibit Untimely Initiation of DNA Replication Are Important for Stable Genome Maintenance
- SIRT1 Promotes N-Myc Oncogenesis through a Positive Feedback Loop Involving the Effects of MKP3 and ERK on N-Myc Protein Stability
- Bacteriophage Crosstalk: Coordination of Prophage Induction by Trans-Acting Antirepressors
- Role of the Single-Stranded DNA–Binding Protein SsbB in Pneumococcal Transformation: Maintenance of a Reservoir for Genetic Plasticity
- Genomic Convergence among ERRα, PROX1, and BMAL1 in the Control of Metabolic Clock Outputs
- Genome-Wide Association of Bipolar Disorder Suggests an Enrichment of Replicable Associations in Regions near Genes
- Identification of Nine Novel Loci Associated with White Blood Cell Subtypes in a Japanese Population
- DNA Ligase III Promotes Alternative Nonhomologous End-Joining during Chromosomal Translocation Formation
- Differential Effects of and Risk Variants on Association with Diabetic ESRD in African Americans
- Finished Genome of the Fungal Wheat Pathogen Reveals Dispensome Structure, Chromosome Plasticity, and Stealth Pathogenesis
- Dynamic Chromatin Localization of Sirt6 Shapes Stress- and Aging-Related Transcriptional Networks
- Extracellular Matrix Dynamics in Hepatocarcinogenesis: a Comparative Proteomics Study of Transgenic and Null Mouse Models
- Integrating 5-Hydroxymethylcytosine into the Epigenomic Landscape of Human Embryonic Stem Cells
- Vive La Différence: An Interview with Catherine Dulac
- Multiple Loci Are Associated with White Blood Cell Phenotypes
- Nuclear Accumulation of Stress Response mRNAs Contributes to the Neurodegeneration Caused by Fragile X Premutation rCGG Repeats
- A New Mutation Affecting FRQ-Less Rhythms in the Circadian System of
- Cryptic Transcription Mediates Repression of Subtelomeric Metal Homeostasis Genes
- A New Isoform of the Histone Demethylase JMJD2A/KDM4A Is Required for Skeletal Muscle Differentiation
- Genetic Determinants of Lipid Traits in Diverse Populations from the Population Architecture using Genomics and Epidemiology (PAGE) Study
- A Genome-Wide RNAi Screen for Factors Involved in Neuronal Specification in
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Recurrent Chromosome 16p13.1 Duplications Are a Risk Factor for Aortic Dissections
- Statistical Inference on the Mechanisms of Genome Evolution
- Genome-Wide Association Study of White Blood Cell Count in 16,388 African Americans: the Continental Origins and Genetic Epidemiology Network (COGENT)
- Chromosomal Macrodomains and Associated Proteins: Implications for DNA Organization and Replication in Gram Negative Bacteria
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání