-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaHow Chromatin Is Remodelled during DNA Repair of UV-Induced DNA Damage in
Global genome nucleotide excision repair removes DNA damage from transcriptionally silent regions of the genome. Relatively little is known about the molecular events that initiate and regulate this process in the context of chromatin. We've shown that, in response to UV radiation–induced DNA damage, increased histone H3 acetylation at lysine 9 and 14 correlates with changes in chromatin structure, and these alterations are associated with efficient global genome nucleotide excision repair in yeast. These changes depend on the presence of the Rad16 protein. Remarkably, constitutive hyperacetylation of histone H3 can suppress the requirement for Rad7 and Rad16, two components of a global genome repair complex, during repair. This reveals the connection between histone H3 acetylation and DNA repair. Here, we investigate how chromatin structure is modified following UV irradiation to facilitate DNA repair in yeast. Using a combination of chromatin immunoprecipitation to measure histone acetylation levels, histone acetylase occupancy in chromatin, MNase digestion, or restriction enzyme endonuclease accessibility assays to analyse chromatin structure, and finally nucleotide excision repair assays to examine DNA repair, we demonstrate that global genome nucleotide excision repair drives UV-induced chromatin remodelling by controlling histone H3 acetylation levels in chromatin. The concerted action of the ATPase and C3HC4 RING domains of Rad16 combine to regulate the occupancy of the histone acetyl transferase Gcn5 on chromatin in response to UV damage. We conclude that the global genome repair complex in yeast regulates UV-induced histone H3 acetylation by controlling the accessibility of the histone acetyl transferase Gcn5 in chromatin. The resultant changes in histone H3 acetylation promote chromatin remodelling necessary for efficient repair of DNA damage. Recent evidence suggests that GCN5 plays a role in NER in human cells. Our work provides important insight into how GG-NER operates in chromatin.
Published in the journal: . PLoS Genet 7(6): e32767. doi:10.1371/journal.pgen.1002124
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1002124Summary
Global genome nucleotide excision repair removes DNA damage from transcriptionally silent regions of the genome. Relatively little is known about the molecular events that initiate and regulate this process in the context of chromatin. We've shown that, in response to UV radiation–induced DNA damage, increased histone H3 acetylation at lysine 9 and 14 correlates with changes in chromatin structure, and these alterations are associated with efficient global genome nucleotide excision repair in yeast. These changes depend on the presence of the Rad16 protein. Remarkably, constitutive hyperacetylation of histone H3 can suppress the requirement for Rad7 and Rad16, two components of a global genome repair complex, during repair. This reveals the connection between histone H3 acetylation and DNA repair. Here, we investigate how chromatin structure is modified following UV irradiation to facilitate DNA repair in yeast. Using a combination of chromatin immunoprecipitation to measure histone acetylation levels, histone acetylase occupancy in chromatin, MNase digestion, or restriction enzyme endonuclease accessibility assays to analyse chromatin structure, and finally nucleotide excision repair assays to examine DNA repair, we demonstrate that global genome nucleotide excision repair drives UV-induced chromatin remodelling by controlling histone H3 acetylation levels in chromatin. The concerted action of the ATPase and C3HC4 RING domains of Rad16 combine to regulate the occupancy of the histone acetyl transferase Gcn5 on chromatin in response to UV damage. We conclude that the global genome repair complex in yeast regulates UV-induced histone H3 acetylation by controlling the accessibility of the histone acetyl transferase Gcn5 in chromatin. The resultant changes in histone H3 acetylation promote chromatin remodelling necessary for efficient repair of DNA damage. Recent evidence suggests that GCN5 plays a role in NER in human cells. Our work provides important insight into how GG-NER operates in chromatin.
Introduction
DNA repair is a central facet of DNA metabolism, and nucleotide excision repair (NER) is an important component of a complex cellular response that prevents the loss of genetic information caused by DNA damage. Its importance for the repair of ultraviolet (UV) light induced DNA lesions is dramatically illustrated in humans who suffer from the autosomal recessive disease xeroderma pigmentosum (XP). Defective NER in these individuals severely predisposes them to sunlight-induced skin cancers [1]. The excision of lesions from non-transcribed regions of the human genome involves the global genome nucleotide excision repair (GG-NER) pathway, which in yeast requires the Rad7 and Rad16 GG-NER proteins [1]–[3]. Many of the core enzymatic activities associated with NER have been determined in some detail, but an understanding of how the process functions in relation to chromatin structure is still in its infancy.
DNA in eukaryotic cells is packaged into nucleosomes that form as a result of the wrapping of DNA around histone octamers. Higher-order chromatin structures are formed when nucleosomal arrays are further compacted. Chromatin has a major impact on DNA metabolic processes by controlling the functional interaction of proteins with regulatory and other elements in the DNA [4], [5]. Chromatin remodelling and histone modification are two major mechanisms that contribute to this regulation. Both processes have roles in controlling gene transcription [6], [7] and in NER [8]–[10].
GG-NER in S.cerevisiae requires both the Rad7 and Rad16 proteins [11]–[13]. Rad16 is a member of the SWI/SNF super-family of chromatin remodelling factors [14]. This superfamily of proteins exhibits ATPase activity that is stimulated by DNA or chromatin [15], [16], and all SWI/SNF-like proteins generate superhelical tension in linear DNA fragments via a DNA translocase activity associated with their ATPase function [17], [18]. The generation of superhelicity in DNA is a common mechanism of SWI/SNF-like chromatin remodelling complexes for altering chromatin structure [17]. We recently reported that a Rad7 and Rad16 containing protein complex also has DNA translocase activity. However, it is unable to slide nucleosomes unlike some SWI/SNF superfamily complexes [19]. Although Rad16 is a member of the SWI/SNF super-family, direct evidence of a role in chromatin remodelling is lacking. In this study we have addressed how GG-NER functions during DNA repair in chromatin in yeast cells.
UV irradiation stimulates histone H3 acetylation at lysine 9 and 14 (K9, K14) and chromatin remodelling, both globally and in the MFA2 gene [8], [20]. However, these studies were not able to establish the precise relationship between these two events with respect to their effect on NER, nor did they inform on how these UV induced changes were regulated. Recently we showed that UV induced histone H3 acetylation depends on the Rad16 GG-NER protein. Furthermore, constitutively elevating histone H3 acetylation levels in the MFA2 gene suppresses the requirement for Rad7 and Rad16 during GG-NER [10]. Gene regulation of MFA2 involves the yeast general repressor complex Ssn6-Tup1 [21]. Deletion of TUP1 results in constitutively elevated histone H3 acetylation and modified chromatin structure at the promoter of the MFA2 gene [22]–[24]. Remarkably, Rad7 and Rad16 independent GG-NER occurs in the promoter region of MFA2 in TUP1 deleted cells. This suggested that Rad7 and Rad16 might regulate chromatin structure in response to UV damage during GG-NER via the regulation of histone H3 acetylation levels in chromatin.
In this report we demonstrate that the GG-NER proteins in yeast promote chromatin remodelling necessary for efficient DNA repair, revealing how this processes is regulated in response to DNA damage. We define a series of UV induced, Rad7 and Rad16 dependent events that control histone H3 acetylation which in turn drives chromatin remodelling necessary for efficient GG-NER in yeast. Histone H3 acetylation status at MFA2 is determined by Rad7 and Rad16 controlling the occupancy of the Gcn5 histone acetyl transferase on chromatin in response to UV irradiation. These UV induced histone H3 modifications are required for chromatin remodelling necessary for efficient GG-NER in the region.
Results
UV-induced histone H3 acetylation (K9, K14) requires both Rad7 and Rad16
Acetylation of histone H3 after UV irradiation depends on the presence of Rad16 and this process is necessary for efficient GG-NER [10]. Figure 1A shows that UV induced histone H3 acetylation (K9, K14) at the regulatory region of the MFA2 gene also requires the GG-NER factor Rad7. Therefore Rad7 and Rad16 function in combination to increase histone H3 acetylation levels at MFA2 in response to UV. Since UV induced histone H3 acetylation correlates with efficient GG-NER and elevated levels of histone H3 acetylation at MFA2 suppress the requirement for Rad7 and Rad16 during GG-NER [10], this poses the question as to how Rad7 and Rad16 control histone H3 acetylation.
Fig. 1. Histone H3 acetylation and occupancy of Gcn5 at the MFA2 promoter. 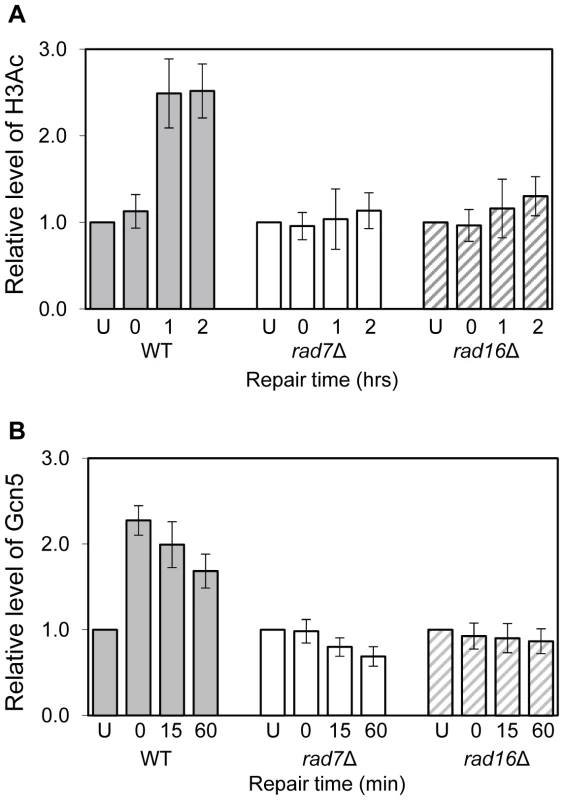
(A) ChIP analysis of Histone H3 acetylation (H3Ac) at the MFA2 promoter using H3Ac (Lys 9 and Lys 14) antibody was performed in wild type (WT), rad7Δ and rad16Δ cells. U: untreated samples; 0: cells received 100 J/m2 of ultraviolet without repair; 1 and 2: cells were irradiated with ultraviolet and then allowed to repair in YPD medium for one or two hours respectively. Acetylation level shown is the fold change relative to unirradiated cells. Data are the average of at least three independent experiments ± SD. (B) ChIP with anti-myc antibody was performed in wild type, rad7Δ and rad16Δ cells. Gcn5 binding is presented as the fold change relative to untreated cells. Data are the average of at least three independent experiments ± SD. Rad7 and Rad16 control histone H3 acetylation status by regulating Gcn5 occupancy at MFA2
Rad7 and Rad16 control UV induced histone H3 acetylation at MFA2 and these proteins are not required for GG-NER when histone H3 acetylation is constitutively elevated in the region. We speculated that during GG-NER Rad7 and Rad16 mediate changes in histone H3 acetylation after UV by controlling the accessibility of the histone acetyl transferase Gcn5, which regulates histone H3 acetylation at MFA2. To test this we performed Gcn5 chromatin immunoprecipitation (ChIP) experiments in the promoter of the MFA2 gene. Figure 1B shows the relative levels of Gcn5 binding at the repressed MFA2 promoter in the absence of UV (U) or following UV irradiation at the times indicated (0, 15 and 60 minutes) in wild type, rad7Δ and rad16Δ strains. In the absence of UV irradiation, background levels of Gcn5 occupancy are detected in all three strains. However, after UV, a rapid increase in Gcn5 occupancy is observed in the wild type, but not in the rad7Δ or rad16Δ strains. In wild type cells, decreasing levels of Gcn5 occupancy at MFA2 were observed with increasing time after UV irradiation and as repair occurred. Therefore in wild type cells Gcn5 occupies the promoter of the MFA2 gene at low levels, resulting in background levels of histone H3 acetylation at MFA2 in the absence of UV. Following UV, a Rad7 and Rad16 dependent increase in Gcn5 occupancy (Figure 1B) and histone H3 acetylation (Figure 1A) is observed at MFA2.
Histone H3 acetylation regulates chromatin structure at the promoter of MFA2
We measured chromatin changes at MFA2 in TUP1 deleted α-cells where histone H3 is hyperacetylated and where the requirement for Rad7 and Rad16 during GG-NER is abrogated. Tup1 is a component of a repressor complex that regulates gene expression at MFA2. In α mating type cells where the chromatin is repressed, the deletion of TUP1 correlates with altered chromatin structure in MFA2 and other TUP1 regulated genes [10], [23], [25]. To confirm this we compared the MNase sensitive sites in naked DNA and chromatin from wild type and tup1Δ α-cells on both DNA strands of the MFA2 promoter region (Figure 2A and 2B, Figure S1, and Text S1). Figure 2A and 2B reveal that MNase digestion is almost identical between tup1Δ α-cell chromatin and naked DNA, whereas chromatin from wild type α-cells exhibits significantly reduced MNase digestion due to protection by the positioned nucleosomes designated N-1 and N-2. Autoradiograms are shown in Figure S1. Therefore chromatin structure is altered in TUP1 deleted α-cells. To further explore the effect of histone acetylation on chromatin structure we examined the accessibility of the restriction enzyme RsaI to nucleosomal core DNA. Chromatin was treated with RsaI restriction enzyme and purified DNA was digested using HaeIII. Restriction with HaeIII generated a 599 bp DNA fragment (Figure 2C). A double restriction digest with RsaI and HaeIII of naked DNA generated a smaller fragment of 419 bp (Figure 2C). In wild type α-cells MFA2 is repressed by positioned nucleosomes and RsaI has only limited access to the DNA at its restriction site located within nucleosome N-2. RsaI digests only 8.7±1.9% of the total MFA2 fragments (Figure 2D, Lane 2). However, in wild type a-cells and tup1Δ α-cells (Figure 2D, Lanes 1 and 5) where MFA2 is derepressed, RsaI cuts in both strains to the extent of 60.3±1.0% and 74.5±2.2% of the total HaeIII fragments, respectively. Therefore, restriction enzyme sites are masked in chromatin from wild type α-cells, but are accessible in chromatin from wild type a-cells and tup1Δ α-cells.
Fig. 2. Densitometric scan of MNase sensitive regions of the MFA2 promoter. 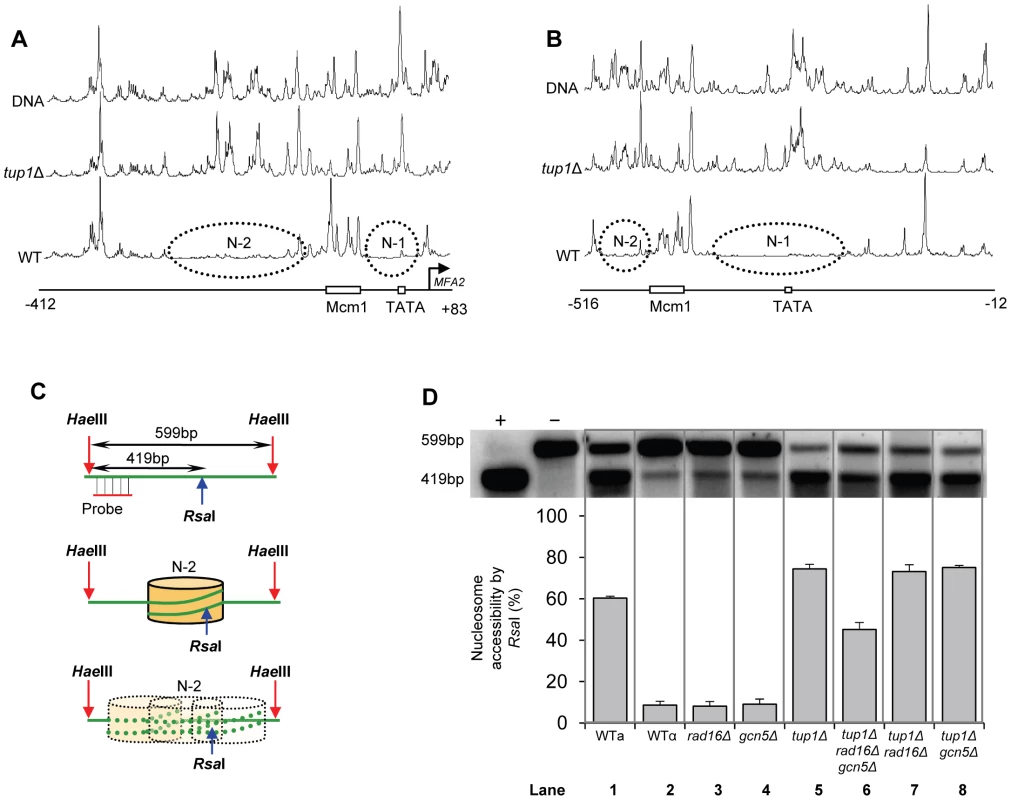
(A and B) Relative MNase sensitivity is expressed graphically from scans of the gels shown in Figure S1. Trace A: transcribed strand (TS); Trace B: non-transcribed strand (NTS). The positioned nucleosomes observed in wild type cells are represented by ellipses N-1 and N-2. (C) Schematic representation of the assay in D. The middle of nucleosome N-2 of MFA2 promoter has a single RsaI restriction site within the HaeIII restriction fragment. The probe shown detects either the full-length 599 bp of HaeIII fragment or 419 bp of RsaI and HaeIII double digested fragment. The protection rendered by nucleosome N-2 limits the accessibility of RsaI to the site. (D) Southern blot analysis of RsaI accessibility to the MFA2 promoter N-2 site. Lane −: naked DNA digested by HaeIII only; lane +: naked DNA digested by both HaeIII and RsaI. Lanes 1–8 represent HaeIII degisted DNA purified from RsaI digested chromatin samples from the strains listed. The lower panel shows the data graphically. Increased histone H3 acetylation levels at MFA2 in TUP1 deleted α cells is dependent on Gcn5 and Rad16
The relationship between chromatin accessibility and histone H3 acetylation status was examined by measuring the histone H3 acetylation levels in the MFA2 promoter in the absence of and following UV irradiation. In Figure 1A, and in Figure 3, a three fold increased UV induced histone H3 acetylation is observed in wild type α-cells. In the tup1Δα strain an eight-fold elevation in constitutive histone H3 acetylation is observed and no further increase in H3 acetylation is seen following UV irradiation. A similar result was noted in tup1Δrad16Δ α-cells. Intriguingly, in tup1Δgcn5Δ α-cells histone H3 acetylation remains constitutively high, despite the loss of the Gcn5 histone acetyl transferase in this strain.
Fig. 3. Histone H3 acetylation at the MFA2. 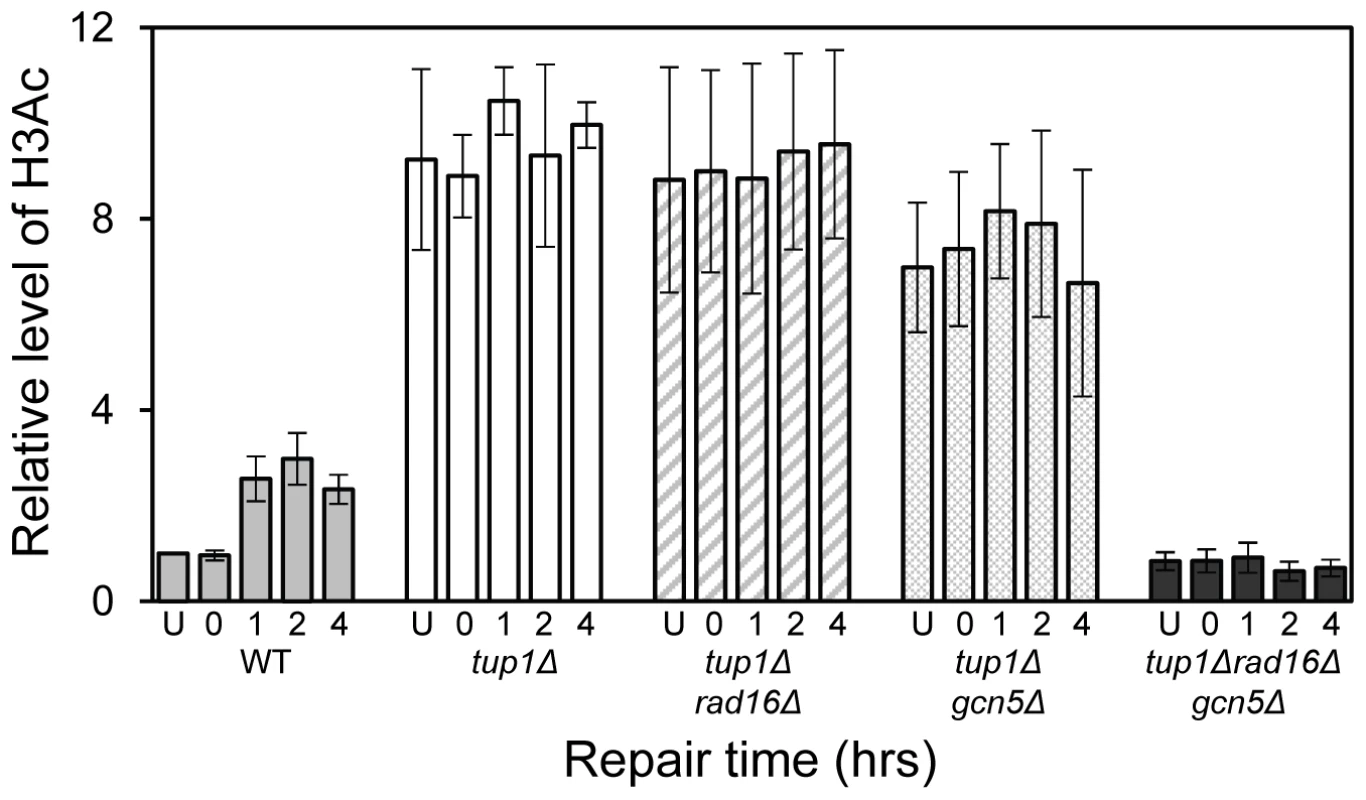
ChIP analysis of Histone H3 acetylation (H3Ac) was performed using H3Ac (Lys 9 and Lys 14) antibodies. U: untreated samples; 0: cells received 100 J/m2 of ultraviolet without repair; 1, 2 or 4: cells were irradiated with ultraviolet and then were allowed to repair in YPD for the number of hours indicated. Acetylation level is presented as the fold change relative to unirradiated wild type cells. Data are the average of at least three independent experiments ± SD. Increased chromatin accessibility at MFA2 in TUP1 deleted α cells depends on Rad16 and Gcn5
Figure 2D lanes 3 and 4 demonstrate that in RAD16 or GCN5 deleted α-cells chromatin structure remains closed as evidenced by low-level RsaI cutting observed (8.2%±2.3% and 9.0%±2.6% respectively), similar to levels seen in wild type α cells (Figure 2D, Lane 2). In tup1Δrad16Δ double mutant α-cells, open chromatin structure is retained as high levels of restriction enzyme cutting are observed (73.1%±3.4%) (Figure 2D, Lane 7), similar to levels seen in tup1Δ α-cells (Figure 2D, Lane 5). An open chromatin structure was also seen in tup1Δgcn5Δ α-cells shown in lane 8 (75.1%±1.0% RsaI enzyme cutting). This was unexpected, since Gcn5 is deleted in this strain. But the result is consistent with the constitutively high histone H3 acetylation level detected (Figure 3), explaining the increased chromatin accessibility observed in this strain (Figure 2D, lane 8). Note that deleting RAD16 in tup1Δgcn5Δ α-cells to create a tup1Δrad16Δgcn5Δα triple mutant strain results in significantly reduced restriction enzyme cutting indicating the presence of a more repressive chromatin structure at the site (45.2%±3.4% RsaI enzyme cutting) (Figure 2D, lane 6).
Increased histone H3 acetylation levels and open chromatin structure are required for Rad7 - and Rad16-independent GG-NER
Rad7 and Rad16 independent GG-NER occurs in genomic regions where constitutively elevated levels of histone H3 acetylation are observed, such as the promoter of MFA2 in tup1Δ α-cells (Figure 4, Figure S2, and Text S1) [10]. The absence of CPD repair at MFA2 in the tup1Δ,rad14Δ mutant proves that repair in the tup1Δ,rad16Δ α-cells occurs unequivocally via Rad7 and Rad16 independent GG-NER [10]. This suggested that Rad7 and Rad16 mediated UV induced histone H3 acetylation is necessary for efficient GG-NER. We examined this by measuring repair of CPDs in the promoter of MFA2 in tup1Δrad16Δ α-cells and in tup1Δrad16Δgcn5Δ α-cells, where the histone acetyl transferase gene GCN5 is deleted (Figure S2). Figure 4 shows the time taken to remove 50% of the CPDs (T50%) from the nontranscribed strand at the positions indicated. As seen previously, GG-NER in tup1Δrad16Δ α-cells, or tup1Δrad7Δα is restored to near wild type levels compared to the lack of repair seen in the rad16Δ α single mutant cells (Figure 4). Therefore Rad7 and Rad16 are not required for GG-NER at MFA2 when histone H3 acetylation levels are elevated creating an open chromatin structure (Figure 3 and Figure 2D, lane 5). To determine the significance of UV induced histone H3 acetylation levels and chromatin structure on efficient GG-NER we examined repair in tup1Δrad16Δgcn5Δ α-cells. Figure 4 reveals that loss of hisotne H3 acetylation which causes reduced chromatin accessibility [See Figure 3 and Figure 2D, lane 6] in this triple mutant strain, results in significantly reduced GG-NER in the region of nucleosomes N-1 and N-2 (see Figure 2A and 2B) upstream of the transcriptional start site (Figure 4: open diamonds). Repair in a small region in the vicinity of the transcriptional start site is unaffected. Therefore, the Rad7 and Rad16 independent GG-NER observed at MFA2 in TUP1 deleted cells is primarily due to the constitutively elevated levels of histone H3 acetylation and open chromatin structure in the region. We observed only background levels of histone H3 acetylation in the triple mutated strain and this results in a less accessible chromatin structure and reduced NER activity. This might imply that histone acetylation is not solely responsible for chromatin remodeling necessary for NER, because in the absence of detectable histone H3 acetylation, chromatin remains partially ‘open’. However our observations reveal that histone H3 acetylation does play a significant role in chromatin remodeling necessary for efficient NER. We also noted that in the absence of Gcn5, histone H3 acetylation at K9 and K14 can still be detected and this acetylation is dependent on Rad16, since acetylation is lost in the triple mutated strain (Figure 3). This underscores the significance of Rad16 in controlling histone acetylation status in the region, and demonstrates that redundancy exists with respect to the histone acetyl transferase that can be recruited to the chromatin. These observations are considered in more detail in the Discussion section.
Fig. 4. Repair of CPDs at the MFA2 promoter. 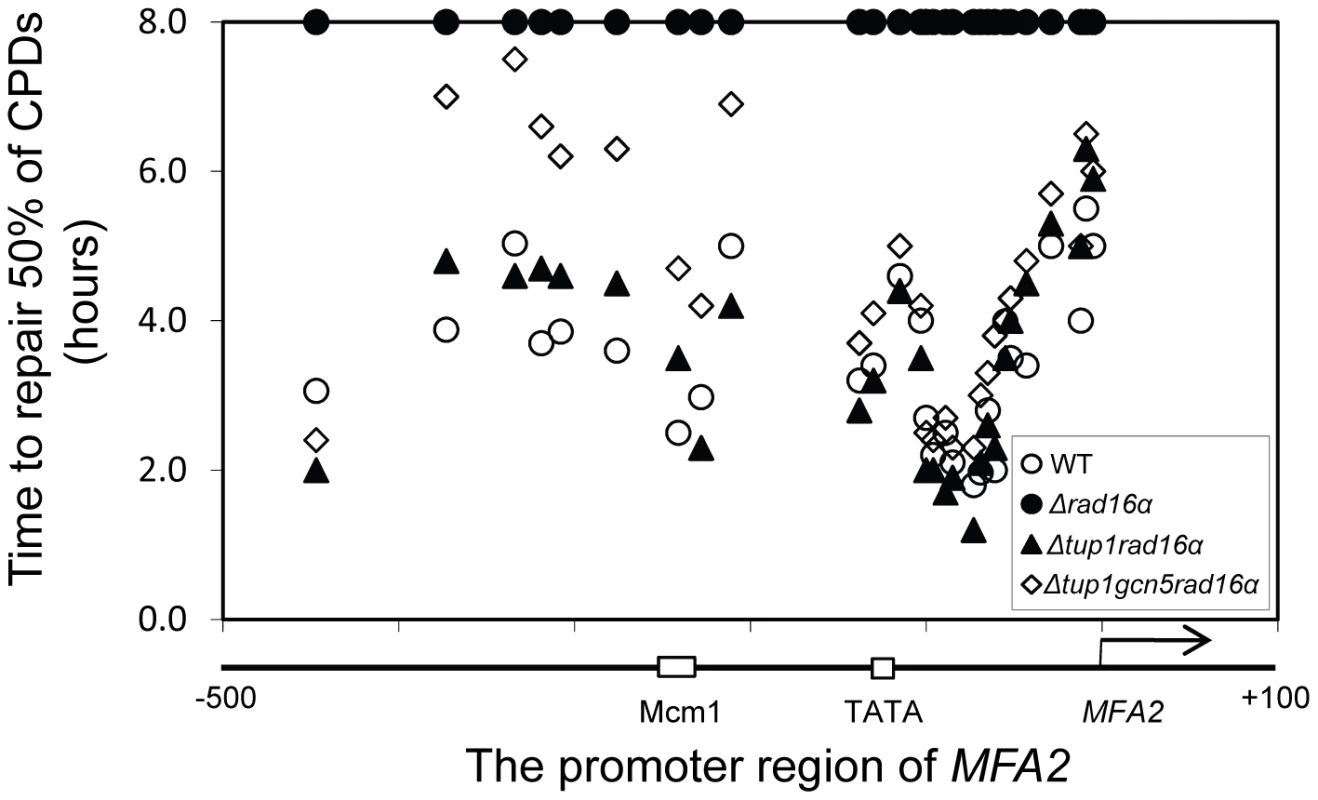
Time to remove 50% of the initial CPDs (T50%) at the sites indicated. T50% of a single CPD or a clustered group of CPDs with similar repair rates was calculated as described previously (Teng et al, 2002) [20]. The T50% of unrepaired CPDs (T50%≥8 h) were represented at the 8 h level on the graph. See also Figure S2. The ATPase and RING domains of Rad16 contribute to efficient UV survival
Rad16 has two known catalytic functions: a DNA translocase activity associated with the ATPase domain [19], and an E3 ubiquitin ligase activity associated with the C3HC4 RING domain embedded within the ATPase domain [see Figure 5A] [26], [27]. We introduced point mutations into each of the catalytic domains of Rad16 to examine their effect on GG-NER. The ATPase activity was tested by mutating the conserved Walker A box catalytic residue lysine 216 to alanine (K216A). This mutation creates an ATPase null mutant [27]. We call this the RAD16 ATPase mutant. We also mutated the RING domain of Rad16 to test the role of the E3 ligase activity. RING domains have conserved cysteine and histidine residues that coordinate two zinc atoms. A conserved hydrophobic residue is also essential for the interaction between the RING domain and specific E2 ubiquitin conjugating enzymes. We made two point mutations in conserved cysteine and histidine residues; cysteine 552 to alanine and histidine 554 to alanine (C552A,H554A). We call this the RAD16 RING mutant. Finally, we tested the effect of mutating both the ATPase and RING domains of Rad16 by introducing these mutations (K216A,C552A,H554A) into a single strain. Figure 5B compares the UV sensitivity in each of these strains compared to the parental wild type, and Rad16 deleted strains. The individual RAD16 ATPase and RING mutant strains show intermediate UV sensitivity. Whereas the double mutant strain is as sensitive as the Rad16 deleted strain. These observations confirm previous findings that both the ATPase and RING E3 ligase catalytic activities contribute independently to efficient GG-NER and UV survival [27].
Fig. 5. The effect of mutating specific domains in Rad16. 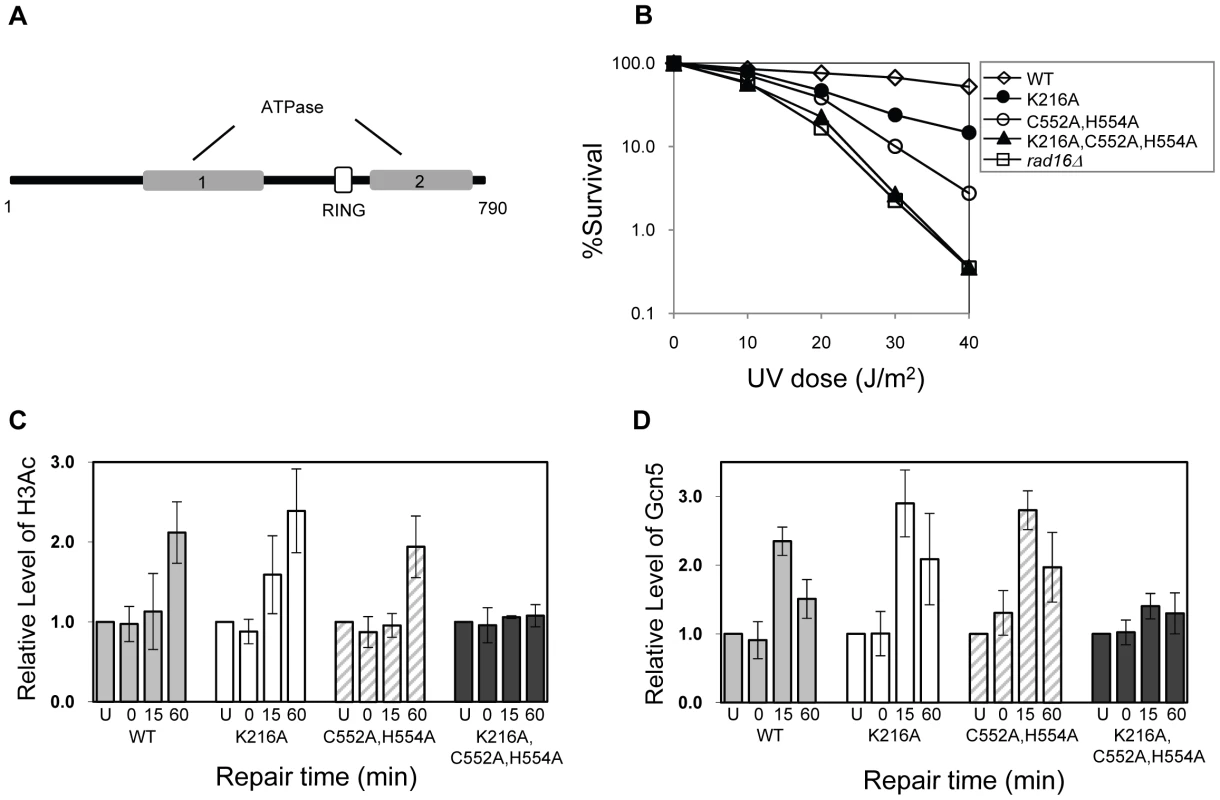
(A) The domain structure of Rad16. (B) UV survival curves of the strains indicated. The result shows the average of three independent experiments. (C) Histone H3 acetylation at the MFA2 promoter. ChIP analysis of Histone H3 acetylation (H3Ac) was performed using H3Ac (Lys 9 and Lys 14) antibody. U: untreated samples; 0: cells received 100 J/m2 of ultraviolet without repair; 15 and 60: cells were irradiated with UV and then were allowed to repair in medium for the times indicated. Acetylation level shown as the fold-change relative to unirradiated cells. Data are the average of at least three independent experiments ± SD. (D). The occupancy of Gcn5 at the MFA2 promoter ChIP was performed with anti-myc antibody. Gcn5 binding is presented as the fold change relative to untreated cells. Data are represented as average of at least three independent experiments ± SD. The ATPase and RING domains of Rad16 are required for UV-induced Gcn5 occupancy and histone H3 acetylation
We examined the effect of the RAD16 point mutations on the level of histone H3 acetylation and Gcn5 occupancy at MFA2. We performed histone H3 acetylation (K9, K14) ChIP experiments in the promoter of MFA2. Figure 5C shows the relative levels of acetylated histone H3 at the repressed MFA2 in the absence of UV (U) or after UV irradiation at the times indicated (0, 15 and 60 minutes). In the absence of UV irradiation, background levels of histone H3 acetylation are detected in all four strains. However, following UV, a rapid increase in histone H3 acetylation is observed in the wild type strain and in the single RAD16 ATPase and RING mutated strains, but not in the RAD16 ATPase, RING double mutant strain, where UV induced histone H3 acetylation is abolished. Similar results were obtained when Gcn5 occupancy was examined in these strains, Figure 5D.
The ATPase and RING domains of Rad16 are required for efficient GG-NER
Finally, we examined the repair of CPDs at MFA2 in wild type and each of the point mutated strains described above (Figure 6A, Figure S4, and Text S1). A typical autoradiogram is shown in Figure S3. In Figure 6A repair was expressed as the time taken to remove 50% of the CPDs (T50%) from the nontranscribed strand at the nucleotide positions indicated. As seen previously, GG-NER in the nontranscribed strand of MFA2 proceeds efficiently in wild type cells (Figure 6A). Mutating either the ATPase domain or the RING domain of RAD16 individually impairs UV lesion removal, but GG-NER continues less efficiently (Figure S4 and Text S1). This correlates with the near wild type levels of histone H3 acetylation, Gcn5 occupancy (Figure 5C and 5D), and intermediate UV sensitivity (Figure 5B) of these strains. GG-NER in the ATPase, RING domain double mutated strain is abolished over almost the whole of the MFA2 promoter region and occurs at a level seen in the RAD16 deleted strain (Figure 6A and Figure 4). This correlates with the lack of UV induced histone H3 acetylation and Gcn5 occupancy in the region (Figure 5C and 5D), and the high level of UV sensitivity (Figure 5B) observed in this strain. These observations demonstrate that the ATPase and RING domains of Rad16 function in combination to regulate UV induced Gcn5 occupancy and histone H3 acetylation status, which ultimately controls chromatin structure at MFA2 in response to DNA damage. In Figure 6B we demonstrate the lack of UV induced chromatin remodelling observed in the ATPase, RING double mutated strain compared to the remodelling observed in the wild type strain using the restriction enzyme accessibility assay described earlier in Figure 2C and 2D. This confirms the importance of chromatin remodelling to the GG-NER process.
Fig. 6. Repair of CPDs at the MFA2 promoter. 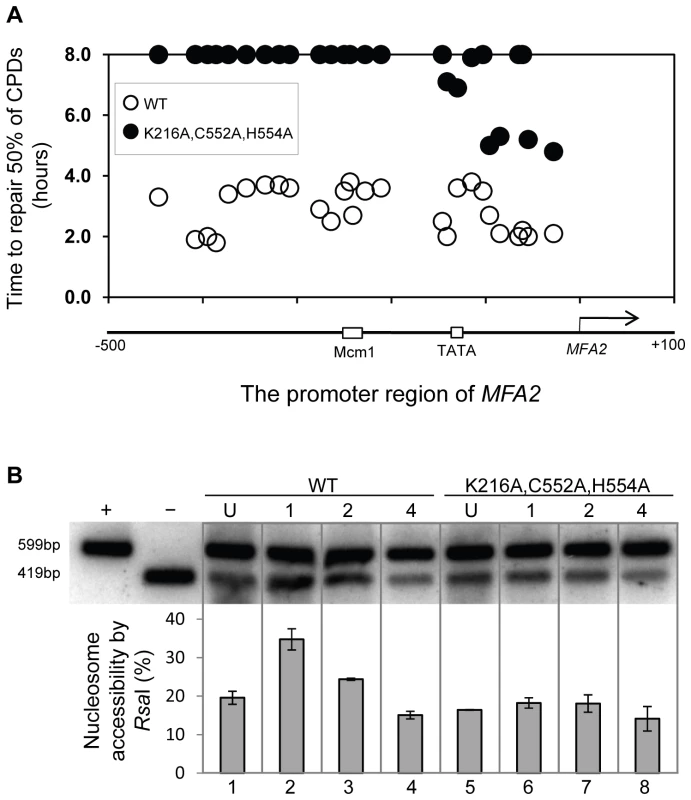
(A) Time to remove 50% of the initial CPDs (T50%) at given sites. T50% of a single CPD or a clustered group of CPDs with a similar repair rate was calculated as described previously (Teng et al, 2002) [20]. The T50% of unrepaired CPDs (T50%≥8 h) were represented at the 8 hour level on the graph. See also Figure S3. (B) Southern blot analysis of RsaI accessibility to the MFA2 promoter N-2 nucleosomal DNA, as described in legend to Figure 2D. Discussion
We've shown that Rad7 and Rad16 proteins are required for UV induced histone H3 acetylation at MFA2. These GG-NER factors regulate the acetylation status by controlling the occupancy of the histone acetyl transferase Gcn5 at this locus. In unirradiated wild type cells only background levels of Gcn5 are detected at MFA2, whereas increased Gcn5 occupancy is seen following UV irradiation. This correlates with increased acetylation of histone H3 observed in wild type cells in response to UV. In Rad7 and Rad16 deleted cells no increased Gcn5 occupancy or increased histone H3 acetylation is observed at MFA2 in response to UV. This indicates that both events are Rad7 and Rad16 dependent in wild type cells. Increased histone acetylation levels have long been associated with changes in chromatin structure, particularly with respect to generating an open chromatin structure needed for gene transcription [8]. To address the impact of histone H3 acetylation on chromatin structure at MFA2 in response to UV, we employed two methods: a nucleosome mapping assay, and a restriction enzyme accessibility assay. We examined these events in TUP1 deleted cells since Tup1 is a component of the Ssn6-Tup1 general repressor complex, which regulates gene expression in a range of genes including MFA2. In α mating type yeast cells MFA2 is repressed, but in TUP1 deleted α-cells histone H3 levels at MFA2 are constitutively elevated which results in an open chromatin structure at MFA2. We found that cells with elevated levels of histone H3 acetylation as is the case when TUP1 alone, TUP1,RAD16 and TUP1,GCN5 are deleted in α-cells also have an open chromatin structure as demonstrated in the restriction enzyme accessibility assay in Figure 2D. We were surprised to detect elevated levels of histone H3 acetylation, and open chromatin structure in TUP1,GCN5 deleted cells since the histone acetyl transferase Gcn5 known to function at MFA2 in wild-type cells is absent in this strain [8]. We speculate that in GCN5 deleted cells, an alternative histone acetyl transferase can substitute for GCN5. Significantly, this redundancy is dependent on Rad16, since in tup1Δrad16Δgcn5Δ triple mutant cells, histone H3 acetylation is reduced to background levels and the open chromatin structure is altered to a more repressed state. We suggest that these observations underscore the significance of Rad16 in regulating the histone acetylation status of chromatin in the region, and indicate that Rad16 determines histone acetyl transferase recruitment to the chromatin. We examined the significance of histone H3 acetylation at MFA2 on lesion removal during GG-NER by measuring repair in TUP1 deleted cells. To determine whether the elevated levels of histone H3 acetylation and open chromatin structure observed in TUP1,RAD16 deleted cells promotes the repair observed in these cells, we examined repair in the tup1Δrad16Δgcn5Δ triple mutant strain where histone H3 levels are diminished to background levels, and chromatin accessibility is significantly reduced. We found that the near wild type level of repair observed in the TUP1,RAD16 deleted cells was significantly reduced in the tup1Δrad16Δgcn5Δ mutant cells indicating the importance of histone H3 acetylation and chromatin structure to the repair observed in the region. Despite detecting only background levels of histone H3 acetylation in the triple mutant strain, we still detect a partially open chromatin structure, which results in a reduced but not totally defective NER efficiency. Our findings demonstrate that UV induced histone H3 acetylation is playing an important role in chromatin remodelling during NER, but recognise that other factors might also be influencing the process.
Finally, we investigated whether either of the known activities associated with Rad16 was responsible for controlling this series of events. Strains carrying point mutations in the ATPase or the C3HC4 RING domain of Rad16, or a double mutant carrying both these mutations were examined. Previous studies showed that the Rad16 ATPase mutant has no detectable ATPase function [27], and the Rad16 RING mutant has no E3 ligase activity. UV survival experiments showed an intermediate UV sensitivity for the Rad16 ATPase and RING domain single mutants, while the double domain mutant showed higher UV sensitivity, similar to that observed in the Rad16 deleted strain (Figure 5). Therefore both the ATPase and E3 ligase functions of Rad16 are required for efficient GG-NER, in agreement with previous studies [26], [27]. This observation suggests that a UV induced ubiquitination event, possibly involving a histone or alternatively another NER factor, is likely important in initiating the chromatin remodelling process. It is established that UV induced histone ubiquitination is observed in human cells and is necessary for efficient NER [28]. We showed that both Rad16 domains contribute to efficient GG-NER. Figure S4 shows reduced levels of CPD removal from the nontranscribed strand of the MFA2 promoter in each of the single domain mutant strains, and defective lesion removal only in the double domain mutant strain (Figure 6A). This observation correlates with the level of Gcn5 occupancy and histone H3 acetylation levels observed in these strains (Figure 5C and 5D). Loss of UV induced Gcn5 occupancy and histone H3 acetylation is only observed in the double mutant strain suggesting that the ATPase and RING domains of Rad16 are both required for efficient chromatin remodelling during GG-NER. Figure 6B confirms that efficient GG-NER observed in the wild type strain is dependent on UV induced chromatin remodelling since failure to remodel chromatin in the ATPase, RING double mutant strain results in defective repair. Collectively our results demonstrate that during GG-NER the Rad7 and Rad16 proteins promote efficient repair by regulating histone acetyl transferase occupancy on chromatin in response to UV. This explains how histone H3 acetylation status and chromatin structure is controlled in response to DNA damage, and that this process is necessary for efficient GG-NER. Our results are consistent with a model for UV induced chromatin remodelling in yeast cells described in Figure 7 (See Text S1 for further discussion).
Fig. 7. Model for UV-induced chromatin remodeling during GG-NER. 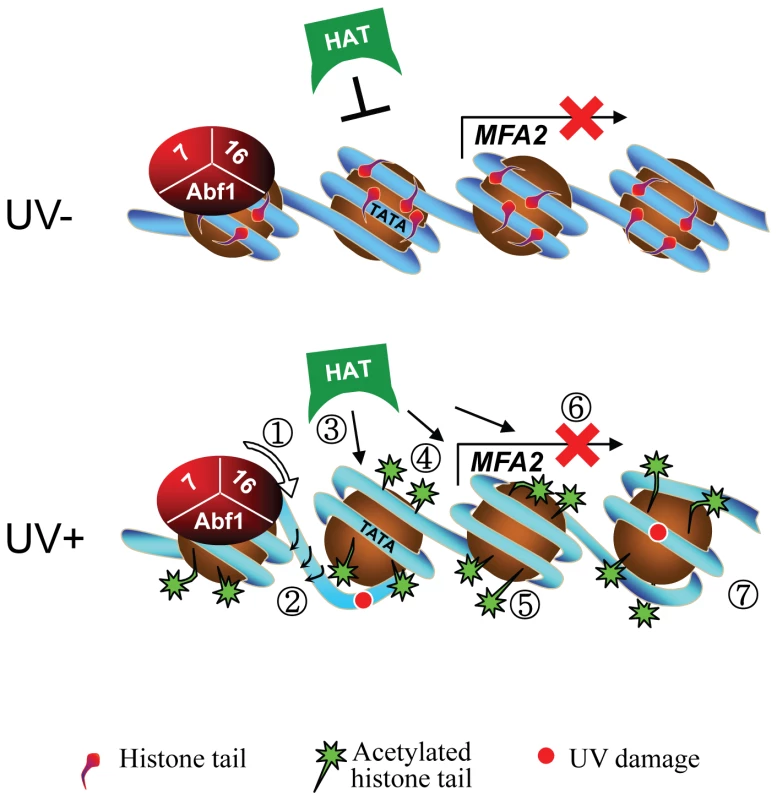
Top panel. In the absence of UV, basal levels of histone acetyl transferase occupancy are detected on the chromatin of the MFA2 promoter. The absence of histone acetyl transferase occupancy is marked by the presence of an inhibitory link. Consequently, histone H3 tails remain unacetylated and chromatin remains repressed. Lower Panel. Following UV the DNA translocase (1) and E3 ligase (2) activities of Rad16 in the GG-NER complex promote increased histone acetyl transferase occupancy on chromatin as indicated by the presence of arrows (3) and histone H3 acetylation (4) that drives chromatin remodeling as shown by a more open chromatin structure around the nucleosomes (5). Failure of the GG-NER complex to slide nucleosomes may prevent transcription factor binding explaining the continued repression of MFA2 transcription (6) despite chromatin remodeling. GG-NER dependent chromatin remodeling promotes efficient lesion removal (7). It was recently reported that Gcn5 is recruited to sites of UV induced DNA damage in human cells [29]. However, its role in chromatin remodelling was not determined. Our studies provide important insight into how chromatin is remodelled to facilitate efficient DNA repair following UV induced DNA damage in human cells.
Materials and Methods
Plasmids and yeast strains
The details of plasmids and yeast strains used in this study can be found in Text S1 and in Table S1.
UV survival assays
Cells were grown in synthetic complete medium with leucine dropout (SC-leu−) to mid-log phase (around 2×107 cells/ml). Following mild sonication, cells were plated on SC-leu− agar plates, then irradiated with the germicidal UV lamp at the indicated UV doses. Following irradiation, plates were immediately wrapped in foil and incubated for 3 days at 30°C. Survival was derived from the number of colonies relative to that in the unirradiated control. Experiments were performed in triplicate.
Chromatin immunoprecipitation (ChIP)
This was performed as in Yu et al, [8] with modifications. In brief, proteins were cross-linked to DNA by addition of formaldehyde to 100 ml yeast cells (about 2×109 cells) to a final concentration of 1% for 20 min at room temperature. 5.5 ml of Glycine (2.5 M) was added to stop cross-linking. Cells were lysed by the addition of 0.5 ml of glass beads (Sigma), and vortexed for 30 min on a Disruptor Genie at 4°C. The cell lysate was sonicated to generate DNA fragments ranging from 200–500 bps in length. Sonication was carried out using the Bioruptor (Diagenode) following the manufacturer's instruction at 4°C, power position “H”, 20 seconds on and 40 seconds off for 6 cycles. 50 µl of pre-washed pan mouse or anti-rabbit IgG Dynabeads was incubated with 2.5 µg of mouse anti-Myc (9E11, Abcam) antibody, or 2.5 µl of rabbit anti-acetyl histone H3 (at K9 and K14, Upstate Biotechnology) at 30°C for 30 min, then the antibody bound Dynabeads were subsequently incubated with 100 µl sheared chromatin solution equivalent to 108 cells in a total volume of 0.5 ml for 3 hours at 21°C. After elution with pronase buffer (125 mM Tris pH 7.5, 25 mM EDTA, 2.5% SDS) from Dynabeads beads, formaldehyde cross-linking was reversed by incubating the eluate at 65°C overnight in the presence of 125 µg of pronase. Finally, DNA was purified with PCR purification kit (QIAGEN). 50 µl of chromatin solution was taken as input control for each sample. Quantitative PCR was performed in real time using iQ SYBR Green Supermix (Bio-Rad) and diluted DNA in the Bio-Rad MyiQ. PCR was performed in triplicate for each sample, and melting curves were executed to ensure single PCR products. Primers for amplifying nucleosome N-2 in the promoter region of MFA2 are:
-
primer 1, AAAGCAGCATGTTTTCATTTGAAACA;
-
primer 2, TATGGGCGTCCTATGCATGCAC.
Chromatin preparation, MNase digestion, and the high-resolution nucleosome mapping
These were carried out as described previously [30]
Restriction enzyme accessibility
Chromatin was prepared as described in Teng et al, [30] with modifications. In brief, cells from 200 ml YPD (2–4×109 cells) were pelleted, washed in cold PBS and 1 M Sorbitol, and spheroplasted in 1 m lysis solution (1 M Sorbitol, 5 mM 2-mercatoethanol) containing 20 mg of Zymolyase-20T per 1 g of cells for 20 min at 30°C. Spheroplasts were washed with cold 1 M Sorbitol, and lysed in 7 ml Ficoll solution (18% Ficoll, 20 mM KH2PO4, pH 6.8, 1 mM MgCl2, 0.25 mM EGTA, 0.25 mM EDTA) per 1 g cell. Collecting nuclei by centrifugation, and washing the pellet with RsaI restriction enzyme reaction buffer, chromatin from 4×108 cells was incubated with 300 units of RsaI for 3 hours at 37°C. Purified DNA from the digest was subjected to a secondary digestion by HaeIII and then resolved on 1.5% agarose gel in 1×TAE buffer. Southern transfer of DNA to GeneScreen Plus Hybridization Transfer Membrane (Perkin Elmer) preparation was described previously [20].
Preparation of radioactive probes for Southern blot analysis
These were undertaken as described in Teng et al, [20]. Details are available in Text S1.
UV treatment of yeast cells, DNA isolation, and high-resolution mapping of CPD sites
These were undertaken as described by Reed et al, [31] and Teng et al, [32]. Details are available in Text S1.
Supporting Information
Zdroje
1. FriedbergEWalkerGCSiedeWWoodRDSchultzRAEllenbergerT 2005 DNA Repair And Mutagenesis ASM Press
2. VerhageRZeemanAMde GrootNGleigFBangDD 1994 The RAD7 and RAD16 genes, which are essential for pyrimidine dimer removal from the silent mating type loci, are also required for repair of the nontranscribed strand of an active gene in Saccharomyces cerevisiae. Mol Cell Biol 14 6135 6142
3. SugasawaKShimizuYIwaiSHanaokaF 2002 A molecular mechanism for DNA damage recognition by the xeroderma pigmentosum group C protein complex. DNA Repair (Amst) 1 95 107
4. WolffeA 2000 Chromatin Structure and Function: Academic Press, San Diego, CA
5. AtaianYKrebsJE 2006 Five repair pathways in one context: chromatin modification during DNA repair. Biochem Cell Biol 84 490 504
6. WuJGrunsteinM 2000 25 years after the nucleosome model: chromatin modifications. Trends Biochem Sci 25 619 623
7. WaterborgJH 2002 Dynamics of histone acetylation in vivo. A function for acetylation turnover? Biochem Cell Biol 80 363 378
8. YuYTengYLiuHReedSHWatersR 2005 UV irradiation stimulates histone acetylation and chromatin remodeling at a repressed yeast locus. Proc Natl Acad Sci U S A 102 8650 8655
9. GongFFahyDSmerdonMJ 2006 Rad4–Rad23 interaction with SWI/SNF links ATP-dependent chromatin remodeling with nucleotide excision repair. Nat Struct Mol Biol 13 902 907
10. TengYLiuHGillHWYuYWatersR 2008 Saccharomyces cerevisiae Rad16 mediates ultraviolet-dependent histone H3 acetylation required for efficient global genome nucleotide-excision repair. EMBO Rep 9 97 102
11. LiSSmerdonMJ 2004 Dissecting transcription-coupled and global genomic repair in the chromatin of yeast GAL1-10 genes. J Biol Chem 279 14418 14426
12. ReedSHAkiyamaMStillmanBFriedbergEC 1999 Yeast autonomously replicating sequence binding factor is involved in nucleotide excision repair. Genes Dev 13 3052 3058
13. VerhageRAvan GoolAJde GrootNHoeijmakersJHvan de PutteP 1996 Double mutants of Saccharomyces cerevisiae with alterations in global genome and transcription-coupled repair. Mol Cell Biol 16 496 502
14. BangDDVerhageRGoosenNBrouwerJvan de PutteP 1992 Molecular cloning of RAD16, a gene involved in differential repair in Saccharomyces cerevisiae. Nucleic Acids Res 20 3925 3931
15. EisenJASwederKSHanawaltPC 1995 Evolution of the SNF2 family of proteins: subfamilies with distinct sequences and functions. Nucleic Acids Res 23 2715 2723
16. WhitehouseIFlausACairnsBRWhiteMFWorkmanJL 1999 Nucleosome mobilization catalysed by the yeast SWI/SNF complex. Nature 400 784 787
17. HavasKFlausAPhelanMKingstonRWadePA 2000 Generation of superhelical torsion by ATP-dependent chromatin remodeling activities. Cell 103 1133 1142
18. Van KomenSPetukhovaGSigurdssonSStrattonSSungP 2000 Superhelicity-driven homologous DNA pairing by yeast recombination factors Rad51 and Rad54. Mol Cell 6 563 572
19. YuSSmirnovaJBFriedbergECStillmanBAkiyamaM 2009 ABF1-binding Sites Promote Efficient Global Genome Nucleotide Excision Repair. J Biol Chem 284 966 973
20. TengYYuYWatersR 2002 The Saccharomyces cerevisiae histone acetyltransferase Gcn5 has a role in the photoreactivation and nucleotide excision repair of UV-induced cyclobutane pyrimidine dimers in the MFA2 gene. J Mol Biol 316 489 499
21. KeleherCAReddMJSchultzJCarlsonMJohnsonAD 1992 Ssn6-Tup1 is a general repressor of transcription in yeast. Cell 68 709 719
22. BoneJRRothSY 2001 Recruitment of the yeast Tup1p-Ssn6p repressor is associated with localized decreases in histone acetylation. J Biol Chem 276 1808 1813
23. CooperJPRothSYSimpsonRT 1994 The global transcriptional regulators, SSN6 and TUP1, play distinct roles in the establishment of a repressive chromatin structure. Genes Dev 8 1400 1410
24. MalaveTMDentSY 2006 Transcriptional repression by Tup1-Ssn6. Biochem Cell Biol 84 437 443
25. DuckerCESimpsonRT 2000 The organized chromatin domain of the repressed yeast a cell-specific gene STE6 contains two molecules of the corepressor Tup1p per nucleosome. EMBO J 19 400 409
26. GilletteTGYuSZhouZWatersRJohnstonSA 2006 Distinct functions of the ubiquitin-proteasome pathway influence nucleotide excision repair. EMBO J 25 2529 2538
27. RamseyKLSmithJJDasguptaAMaqaniNGrantP 2004 The NEF4 complex regulates Rad4 levels and utilizes Snf2/Swi2-related ATPase activity for nucleotide excision repair. Mol Cell Biol 24 6362 6378
28. KapetanakiMGGuerrero-SantoroJBisiDCHsiehCLRapic-OtrinV 2006 The DDB1-CUL4ADDB2 ubiquitin ligase is deficient in xeroderma pigmentosum group E and targets histone H2A at UV-damaged DNA sites. Proc Natl Acad Sci U S A 103 2588 2593
29. GuoRChenJMitchellDLJohnsonDG. GCN5 and E2F1 stimulate nucleotide excision repair by promoting H3K9 acetylation at sites of damage. Nucleic Acids Res 39 1390 1397
30. TengYYuSWatersR 2001 The mapping of nucleosomes and regulatory protein binding sites at the Saccharomyces cerevisiae MFA2 gene: a high resolution approach. Nucleic Acids Res 29 E64 64
31. ReedSHBoiteuxSWatersR 1996 UV-induced endonuclease III-sensitive sites at the mating type loci in Saccharomyces cerevisiae are repaired by nucleotide excision repair: RAD7 and RAD16 are not required for their removal from HML alpha. Mol Gen Genet 250 505 514
32. TengYLiSWatersRReedSH 1997 Excision repair at the level of the nucleotide in the Saccharomyces cerevisiae MFA2 gene: mapping of where enhanced repair in the transcribed strand begins or ends and identification of only a partial rad16 requisite for repairing upstream control sequences. J Mol Biol 267 324 337
Štítky
Genetika Reprodukční medicína
Článek Local Absence of Secondary Structure Permits Translation of mRNAs that Lack Ribosome-Binding SitesČlánek Independent Chromatin Binding of ARGONAUTE4 and SPT5L/KTF1 Mediates Transcriptional Gene SilencingČlánek Trade-Off between Bile Resistance and Nutritional Competence Drives Diversification in the Mouse GutČlánek FGF Signaling Regulates the Number of Posterior Taste Papillae by Controlling Progenitor Field SizeČlánek Mammalian BTBD12 (SLX4) Protects against Genomic Instability during Mammalian SpermatogenesisČlánek Identification of Nine Novel Loci Associated with White Blood Cell Subtypes in a Japanese PopulationČlánek Differential Effects of and Risk Variants on Association with Diabetic ESRD in African AmericansČlánek Dynamic Chromatin Localization of Sirt6 Shapes Stress- and Aging-Related Transcriptional Networks
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2011 Číslo 6
-
Všechny články tohoto čísla
- Local Absence of Secondary Structure Permits Translation of mRNAs that Lack Ribosome-Binding Sites
- Statistical Inference on the Mechanisms of Genome Evolution
- Revisiting Heterochromatin in Embryonic Stem Cells
- A Two-Stage Meta-Analysis Identifies Several New Loci for Parkinson's Disease
- Identification of a Sudden Cardiac Death Susceptibility Locus at 2q24.2 through Genome-Wide Association in European Ancestry Individuals
- Genomic Prevalence of Heterochromatic H3K9me2 and Transcription Do Not Discriminate Pluripotent from Terminally Differentiated Cells
- Epistasis between Beneficial Mutations and the Phenotype-to-Fitness Map for a ssDNA Virus
- Recurrent Chromosome 16p13.1 Duplications Are a Risk Factor for Aortic Dissections
- Telomere DNA Deficiency Is Associated with Development of Human Embryonic Aneuploidy
- Genome-Wide Association Study of White Blood Cell Count in 16,388 African Americans: the Continental Origins and Genetic Epidemiology Network (COGENT)
- Unexpected Role for DNA Polymerase I As a Source of Genetic Variability
- Transportin-SR Is Required for Proper Splicing of Genes and Plant Immunity
- How Chromatin Is Remodelled during DNA Repair of UV-Induced DNA Damage in
- Independent Chromatin Binding of ARGONAUTE4 and SPT5L/KTF1 Mediates Transcriptional Gene Silencing
- Two Evolutionary Histories in the Genome of Rice: the Roles of Domestication Genes
- Natural Allelic Variation Defines a Role for : Trichome Cell Fate Determination
- Multiple Common Susceptibility Variants near BMP Pathway Loci , , and Explain Part of the Missing Heritability of Colorectal Cancer
- Pathogenic Mechanism of the FIG4 Mutation Responsible for Charcot-Marie-Tooth Disease CMT4J
- A Functional Variant in Promoter Modulates Its Expression and Confers Disease Risk for Systemic Lupus Erythematosus
- Drift and Genome Complexity Revisited
- Chromosomal Macrodomains and Associated Proteins: Implications for DNA Organization and Replication in Gram Negative Bacteria
- Trade-Off between Bile Resistance and Nutritional Competence Drives Diversification in the Mouse Gut
- Pathways of Distinction Analysis: A New Technique for Multi–SNP Analysis of GWAS Data
- Web-Based Genome-Wide Association Study Identifies Two Novel Loci and a Substantial Genetic Component for Parkinson's Disease
- Chk2 and p53 Are Haploinsufficient with Dependent and Independent Functions to Eliminate Cells after Telomere Loss
- Exome Sequencing Identifies Mutations in High Myopia
- Distinct Functional Constraints Partition Sequence Conservation in a -Regulatory Element
- CorE from Is a Copper-Dependent RNA Polymerase Sigma Factor
- A Single Sex Pheromone Receptor Determines Chemical Response Specificity of Sexual Behavior in the Silkmoth
- FGF Signaling Regulates the Number of Posterior Taste Papillae by Controlling Progenitor Field Size
- Maps of Open Chromatin Guide the Functional Follow-Up of Genome-Wide Association Signals: Application to Hematological Traits
- Increased Susceptibility to Cortical Spreading Depression in the Mouse Model of Familial Hemiplegic Migraine Type 2
- Differential Gene Expression and Epiregulation of Alpha Zein Gene Copies in Maize Haplotypes
- Parallel Adaptive Divergence among Geographically Diverse Human Populations
- Genetic Analysis of Genome-Scale Recombination Rate Evolution in House Mice
- Mechanisms for the Evolution of a Derived Function in the Ancestral Glucocorticoid Receptor
- Mammalian BTBD12 (SLX4) Protects against Genomic Instability during Mammalian Spermatogenesis
- Interferon Regulatory Factor 8 Regulates Pathways for Antigen Presentation in Myeloid Cells and during Tuberculosis
- High-Resolution Analysis of Parent-of-Origin Allelic Expression in the Arabidopsis Endosperm
- Specific SKN-1/Nrf Stress Responses to Perturbations in Translation Elongation and Proteasome Activity
- Graded Nodal/Activin Signaling Titrates Conversion of Quantitative Phospho-Smad2 Levels into Qualitative Embryonic Stem Cell Fate Decisions
- Genome-Wide Analysis Reveals PADI4 Cooperates with Elk-1 to Activate Expression in Breast Cancer Cells
- Trait Variation in Yeast Is Defined by Population History
- Meiosis-Specific Loading of the Centromere-Specific Histone CENH3 in
- A Genome-Wide Survey of Imprinted Genes in Rice Seeds Reveals Imprinting Primarily Occurs in the Endosperm
- Multiple Regulatory Mechanisms to Inhibit Untimely Initiation of DNA Replication Are Important for Stable Genome Maintenance
- SIRT1 Promotes N-Myc Oncogenesis through a Positive Feedback Loop Involving the Effects of MKP3 and ERK on N-Myc Protein Stability
- Bacteriophage Crosstalk: Coordination of Prophage Induction by Trans-Acting Antirepressors
- Role of the Single-Stranded DNA–Binding Protein SsbB in Pneumococcal Transformation: Maintenance of a Reservoir for Genetic Plasticity
- Genomic Convergence among ERRα, PROX1, and BMAL1 in the Control of Metabolic Clock Outputs
- Genome-Wide Association of Bipolar Disorder Suggests an Enrichment of Replicable Associations in Regions near Genes
- Identification of Nine Novel Loci Associated with White Blood Cell Subtypes in a Japanese Population
- DNA Ligase III Promotes Alternative Nonhomologous End-Joining during Chromosomal Translocation Formation
- Differential Effects of and Risk Variants on Association with Diabetic ESRD in African Americans
- Finished Genome of the Fungal Wheat Pathogen Reveals Dispensome Structure, Chromosome Plasticity, and Stealth Pathogenesis
- Dynamic Chromatin Localization of Sirt6 Shapes Stress- and Aging-Related Transcriptional Networks
- Extracellular Matrix Dynamics in Hepatocarcinogenesis: a Comparative Proteomics Study of Transgenic and Null Mouse Models
- Integrating 5-Hydroxymethylcytosine into the Epigenomic Landscape of Human Embryonic Stem Cells
- Vive La Différence: An Interview with Catherine Dulac
- Multiple Loci Are Associated with White Blood Cell Phenotypes
- Nuclear Accumulation of Stress Response mRNAs Contributes to the Neurodegeneration Caused by Fragile X Premutation rCGG Repeats
- A New Mutation Affecting FRQ-Less Rhythms in the Circadian System of
- Cryptic Transcription Mediates Repression of Subtelomeric Metal Homeostasis Genes
- A New Isoform of the Histone Demethylase JMJD2A/KDM4A Is Required for Skeletal Muscle Differentiation
- Genetic Determinants of Lipid Traits in Diverse Populations from the Population Architecture using Genomics and Epidemiology (PAGE) Study
- A Genome-Wide RNAi Screen for Factors Involved in Neuronal Specification in
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Recurrent Chromosome 16p13.1 Duplications Are a Risk Factor for Aortic Dissections
- Statistical Inference on the Mechanisms of Genome Evolution
- Genome-Wide Association Study of White Blood Cell Count in 16,388 African Americans: the Continental Origins and Genetic Epidemiology Network (COGENT)
- Chromosomal Macrodomains and Associated Proteins: Implications for DNA Organization and Replication in Gram Negative Bacteria
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



