-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaA Functional Variant in Promoter Modulates Its Expression and Confers Disease Risk for Systemic Lupus Erythematosus
Systemic lupus erythematosus (SLE) is a complex autoimmune disease with a strong genetic predisposition, characterized by an upregulated type I interferon pathway. MicroRNAs are important regulators of immune homeostasis, and aberrant microRNA expression has been demonstrated in patients with autoimmune diseases. We recently identified miR-146a as a negative regulator of the interferon pathway and linked the abnormal activation of this pathway to the underexpression of miR-146a in SLE patients. To explore why the expression of miR-146a is reduced in SLE patients, we conducted short parallel sequencing of potentially regulatory regions of miR-146a and identified a novel genetic variant (rs57095329) in the promoter region exhibiting evidence for association with SLE that was replicated independently in 7,182 Asians (Pmeta = 2.74×10−8, odds ratio = 1.29 [1.18–1.40]). The risk-associated G allele was linked to reduced expression of miR-146a in the peripheral blood leukocytes of the controls. Combined functional assays showed that the risk-associated G allele reduced the protein-binding affinity and activity of the promoter compared with those of the promoter containing the protective A allele. Transcription factor Ets-1, encoded by the lupus-susceptibility gene ETS1, identified in recent genome-wide association studies, binds near this variant. The manipulation of Ets-1 levels strongly affected miR-146a promoter activity in vitro; and the knockdown of Ets-1, mimicking its reduced expression in SLE, directly impaired the induction of miR-146a. We also observed additive effects of the risk alleles of miR-146a and ETS1. Our data identified and confirmed an association between a functional promoter variant of miR-146a and SLE. This risk allele had decreased binding to transcription factor Ets-1, contributing to reduced levels of miR-146a in SLE patients.
Published in the journal: . PLoS Genet 7(6): e32767. doi:10.1371/journal.pgen.1002128
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1002128Summary
Systemic lupus erythematosus (SLE) is a complex autoimmune disease with a strong genetic predisposition, characterized by an upregulated type I interferon pathway. MicroRNAs are important regulators of immune homeostasis, and aberrant microRNA expression has been demonstrated in patients with autoimmune diseases. We recently identified miR-146a as a negative regulator of the interferon pathway and linked the abnormal activation of this pathway to the underexpression of miR-146a in SLE patients. To explore why the expression of miR-146a is reduced in SLE patients, we conducted short parallel sequencing of potentially regulatory regions of miR-146a and identified a novel genetic variant (rs57095329) in the promoter region exhibiting evidence for association with SLE that was replicated independently in 7,182 Asians (Pmeta = 2.74×10−8, odds ratio = 1.29 [1.18–1.40]). The risk-associated G allele was linked to reduced expression of miR-146a in the peripheral blood leukocytes of the controls. Combined functional assays showed that the risk-associated G allele reduced the protein-binding affinity and activity of the promoter compared with those of the promoter containing the protective A allele. Transcription factor Ets-1, encoded by the lupus-susceptibility gene ETS1, identified in recent genome-wide association studies, binds near this variant. The manipulation of Ets-1 levels strongly affected miR-146a promoter activity in vitro; and the knockdown of Ets-1, mimicking its reduced expression in SLE, directly impaired the induction of miR-146a. We also observed additive effects of the risk alleles of miR-146a and ETS1. Our data identified and confirmed an association between a functional promoter variant of miR-146a and SLE. This risk allele had decreased binding to transcription factor Ets-1, contributing to reduced levels of miR-146a in SLE patients.
Introduction
Systemic lupus erythematosus (SLE) is a chronic autoimmune disease with a complex etiology and diverse clinical manifestations [1]. The role of genetic factors in the SLE risk has long been established, and demonstrated in familial aggregations, twin studies, and sibling recurrence rates [2]. Recently, high-throughput technologies have facilitated genome-wide association studies (GWASs) across different populations. This approach, accompanied by large-scale replications, has not only confirmed the association of many established susceptibility genes, but has also presented convincing evidence of novel genetic loci involved in SLE [3]–[8]. As members of the Asian Lupus Genetics Consortium, we have also performed a GWAS in Asian populations and have identified variants in ETS1 and WDFY4 that are associated with SLE [9]. A combination of GWAS data from different ethnic groups will clearly provide new insights into the genetics of SLE and further our understanding of the pathogenesis of lupus [10], [11].
To use genomic tools to study the mechanisms of SLE, we and others have independently identified a gene expression signature for lupus patients using microarray profiling [12]–[14], which highlights the pathogenic role of the abnormal activation of the type I interferon (IFN) pathway in human lupus [15]–[17]. Intriguingly, recent investigations suggest a genetic contribution to the variability observed among individuals in the production and signaling of IFN [17], and advances in the genetics of SLE highlight the strong association between the risk of developing lupus and gene variants connected to the production and effects of type I IFN [11], [18].
We recently used a microRNA (miRNA) profiling assay to examine the involvement of miRNAs in SLE, because miRNAs are novel gene expression regulators [19] and important players in shaping the immune response [20]–[22]. This profiling identified a reduction in miR-146a expression in lupus patients, and we showed that the underexpression of miR-146a contributes to lupus pathogenesis by deregulating the activation of the IFN pathway [23]. However, why miR-146a levels are reduced in patients with SLE remains unresolved. miR-146a is encoded at 5q33.3. Interestingly, recent data from GWASs in both European and Asian populations have indicated that this region is a novel susceptibility locus for SLE [3], [7], [9], suggesting a plausible role for a genetic variant around miR-146a in modulating its expression and thus the disease risk.
Several studies have demonstrated unambiguously that genetic variants in miRNA precursors (pre-miRNA) can affect miRNA expression levels by interfering with the miRNA maturation process and are thus associated with disease susceptibility [24]–[26]. We postulate that genetic variants in both the miRNA promoter and the precursor region may alter mature miRNA production. Given the critical regulatory role of miR-146a in the type I IFN pathway and the abovementioned genetic association between this pathway and SLE susceptibility, polymorphisms in the miR-146a gene could also potentially confer a disease risk. To assess whether genetic variants modulate miR-146a expression and thus contribute to the risk of developing SLE, we sequenced the promoter and key regulatory regions of the miR-146a precursor to identify potential functional variants that might be associated with SLE susceptibility. Our subsequent replication and functional studies provide evidence that single-nucleotide polymorphism (SNP) rs57095329 in the miR-146a promoter, which affects its mature level, can confer SLE susceptibility.
Results
Discovery of SLE-associated miR-146a promoter SNPs
miR-146a is located at 5q33.3. The transcription start site (TSS) of its primary transcript (pri-miR-146a) has been identified [27]. To characterize the essential regulatory region for subsequent genetic analysis, we first cloned miR-146a upstream fragments with variable 5′ ends into the pGL3-basic reporter plasmid to analyze its promoter activity. We found that the inclusion of a fragment from nucleotide (nt) −1,091 to nt −611, which contains a known NF-κB-binding site characterized in THP-1 cells [27], was consistently robust to promote luciferase activity in HeLa cells (Figure S1). The inclusion of the more distal region (nt −1,998 to nt −1,091) enhanced neither the basal nor phorbol myristate acetate and ionomycin (hereafter referred to as “PMA+Iono”) -induced activity of the promoter (Figure S1B). Therefore, to look for new genetic variants and to characterize their potential association with SLE, we designed four pairs of primers with which to sequence the upstream region that spans the 1,105-bp promoter (nt −1,091 to nt +14) and the consecutive first exon of pri-miR-146a (Figure S1A), in 360 individuals (180 SLE patients and 180 controls), which served as the discovery panel. We also sequenced the 452-bp region centered on miR-146a precursor or exon 2 of pri-miR-146a, because it potentially affects mature miR-146a production [26], in the same discovery panel. A total of 12 variants were identified, with nine already reported in the dbSNP database Build 130 (Table S1). Five variants had a minor allele frequency (MAF) of >1% (rs17057381, rs73318382, rs57095329, rs6864584, and rs2910164; Figure S1A). Therefore, we extended our sequencing analysis to examine these five SNPs in up to 816 patients and 1,080 controls, who were all Chinese Han individuals living in Shanghai. In this expanded study panel, only rs73318382 and rs57095329 showed an association with SLE (Table S2). These two SNPs are separated by 304 bp and are in strong linkage disequilibrium (LD; r2 = 0.81; Figure S2). When a Bonferroni correction was applied, the association of rs57095329 with SLE remained highly significant (P = 4×10−4). Given that rs57095329 is identified through our candidate region sequencing approach and not included in the HapMap database, it is not surprising that this SNP has not been included in commercial SNP arrays. Because published GWASs in SLE of both Asian and European populations detected association signals at rs2431697 and rs2431099 (Figure S3), 15 kb and 8 kb upstream from miR-146a TSS, respectively, we extended our genotyping of rs2431697 and rs2431099 using 1,896 Shanghai samples. Both SNPs showed significant association with the disease (Table S2), while rs57095329 produced the best association signal among the three SNPs in the same dataset (Table S2). Therefore, we focused on rs57095329 in the subsequent experiments.
Replicated association of rs57095329 with SLE in independent cohorts
We replicated the association between rs57095329 and SLE using a TaqMan genotyping assay in another two panels from Hong Kong, China, and Bangkok, Thailand. We also added 1,536 patients from the central China area to our mainland China cohort, and the newly added patients showed an allele frequency for rs57095329 very similar to that in the discovery panel (MAFs of 20.53% and 20.77%, respectively). This replication provided consistent evidence for the association, revealed by an allelic association analysis (Table 1). When all the samples were included (3,968 patients and 3,214 controls in total) to conduct a meta-analysis, there was strong evidence that the minor G allele of rs57095329 conferred a risk of SLE (Pmeta = 2.74×10−8, odds ratio [OR] = 1.29, 95% confidence interval [CI] = 1.18–1.40; Table 1). There was no significant difference among the ORs for the three independent cohorts (P = 0.33), when the Breslow–Day test installed in PLINK was used [28], although the SNP showed significant allele frequency differences in respective controls. Recessive mode of action seemed to be supported in the Chinese mainland cohort and the cohort from Thailand (OR = 2.47 and 2.11 for the two cohorts, respectively), compared with the allelic OR of 1.35–1.36 for the two cohorts. However, this was not supported by the result for the Hong Kong cohort, where the same OR was observed for both the recessive mode and the allelic test (OR = 1.18), reflecting certain variations among the different cohorts.
Tab. 1. Association between the rs57095329 G allele and SLE in independent cohorts and in the combined sample. 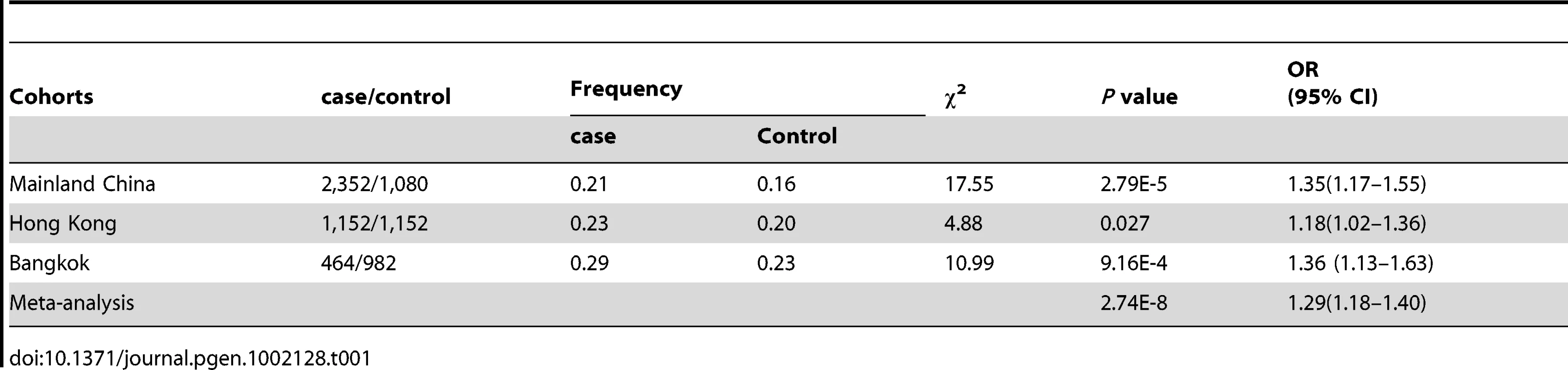
We also examined whether the genetic variant is specifically associated with disease risk in patients with lupus nephritis. Although only the discovery panel in the Chinese mainland cohort showed a significant association in a patient-only analysis, a similar trend was also observed in the Hong Kong and Bangkok cohorts, with a marginal P value of 0.093 and an OR of 1.105 when patients with nephritis were compared with patients without it (Table S3).
Association between rs57095329 and miR-146a expression
We explored the association between rs57095329 and miR-146a expression. Mature miR-146a levels were determined with a TaqMan microRNA assay in 86 healthy controls with known genotypes and available RNA samples. Compared with individuals with the AA genotype, individuals with heterozygous AG genotype for rs57095329 had lower levels of miR-146a (P = 0.0438; Figure 1), while individuals with GG genotype had the lowest miR-146a levels (P = 0.0197; Figure 1). This association indicates that rs57095329, located in the miR-146a promoter, may function by regulating the transcription activity and expression levels of miR-146a.
Fig. 1. Comparison of miR-146a expression levels between groups of healthy individuals with different rs57095329 genotypes. 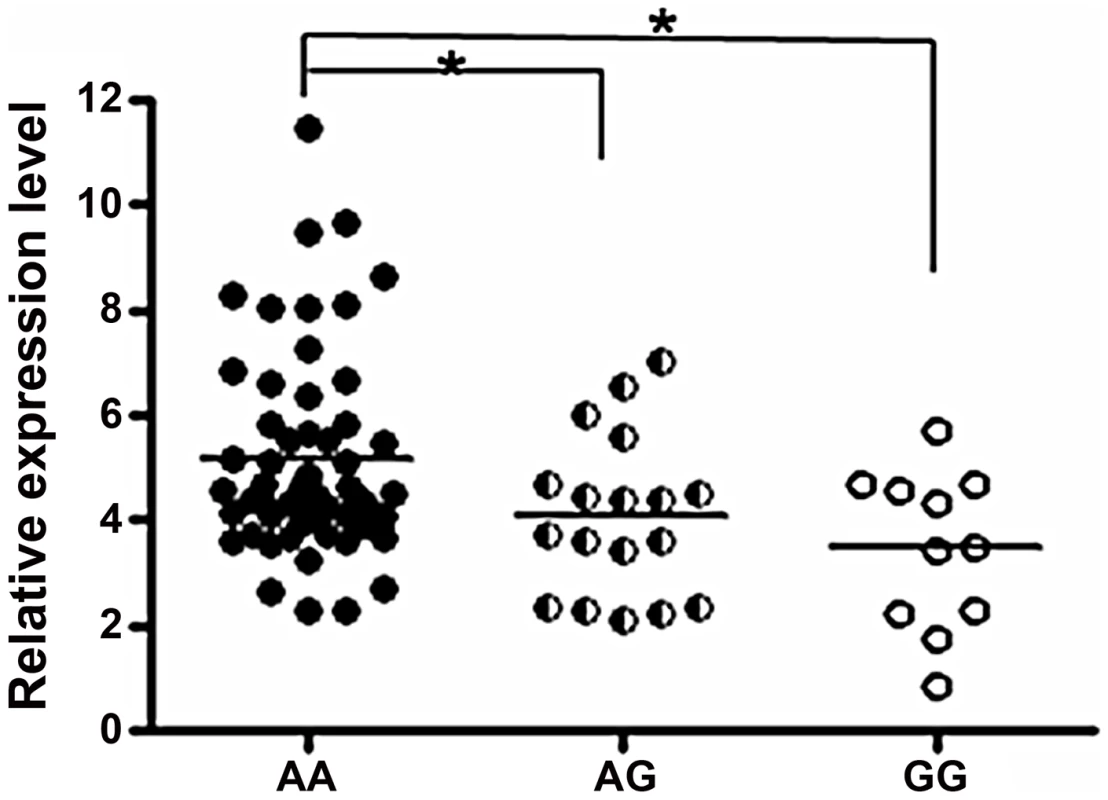
The horizontal line indicates the mean expression level within each group. * indicates P<0.05. Allelic difference of rs57095329 in miR-146a promoter activity
To explore molecular mechanisms of the association between rs57095329 and miR-146a expression, we examined whether the variant is functionally significant by altering the miR-146a promoter activity. We generated reporter gene constructs containing either rs57095329 allele and transfected different cell lines with the reporter plasmids, so that the effect of each allele on the miR-146a promoter activity could be evaluated in the context of the full-length promoter. First, the construct carrying the A allele had higher basal activity in Jurkat T cells than the construct carrying the risk-associated G allele, when a luciferase assay was performed 24 hours after electroporation (Figure 2). This finding is consistent with our previous observation of an association between reduced miR-146a expression and SLE disease. Moreover, when the cells were activated by PMA+Iono or anti-CD3 plus anti-CD28 antibodies after transfection, the induced activity of the promoter with the A allele remained higher (Figure 2). Similarly, an approximately 50% reduction in the activity of the promoter with the risk-associated G allele was observed in HeLa cells under both rested and PMA+Iono-activated conditions (Figure S4A). This difference in promoter activity was also consistently found in steady-state Raji B cells and 293T cells (Figure S4B and S4C). Considering these data together, our reporter gene assay showed that the disease-associated G allele reduced the promoter activity of miR-146a.
Fig. 2. Allelic difference of rs57095329 in miR-146a promoter activity. 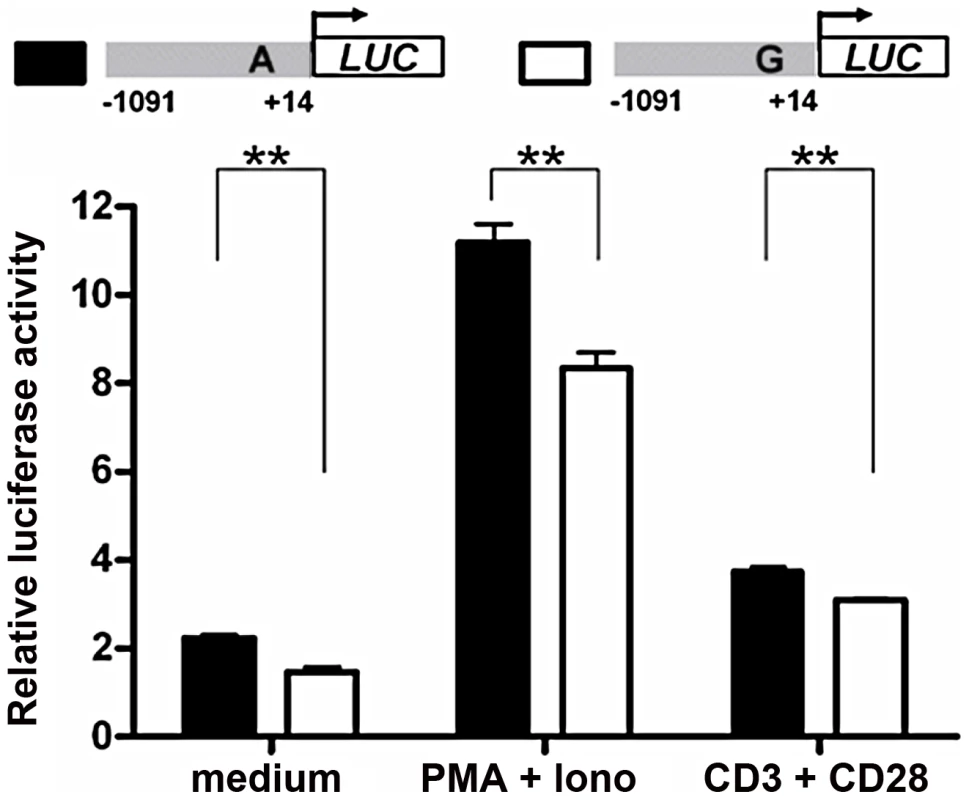
Shown is a schematic representation of reporter gene constructs driven by the full-length miR-146a promoter containing one or other of the rs57095329 alleles (upper) and the relative luciferase activity of the two constructs in Jurkat cells in both the steady (medium) and activated states (lower). For activation, the cells were stimulated with PMA+Iono or with anti-CD3+anti-CD28 antibodies for 6 hours (see Materials and Methods). The data shown are means ± SEM and are representative of three independent experiments performed in triplicate. ** indicates P<0.01. Allelic difference of rs57095329 in nuclear protein–binding activity
To examine whether allelic difference in promoter activity may be attributable to their different binding capacities for nuclear factors, two probes corresponding to the 24-bp miR-146a promoter region, centered on rs57095329, were synthesized and biotin-labeled for an electrophoretic mobility shift assay (EMSA), and unlabeled oligonucleotides were used as the “competitors”. Nuclear extracts were then prepared from resting and anti-CD3+anti-CD28-activated Jurkat cells. As shown in Figure 3, probe A formed much more DNA–protein complexes with the nuclear extracts from resting Jurkat cells than did probe G (lane 2 versus lane 7), indicating that the promoter carrying the A allele of rs57095329 binds more robustly to nuclear proteins. Once the cells were activated, both probes were able to bind more nuclear proteins. Similarly, in this case, probe A exhibited much stronger binding than probe G (Figure 3: lane 4 versus lane 9). All the DNA–protein complexes were reduced or abolished by the addition of excessive corresponding “competitor” oligonucleotides, demonstrating the binding specificity. Similar results were observed with PMA+Iono-stimulated Jurkat and HeLa cells (Figure S5).
Fig. 3. Allelic difference of rs57095329 in binding affinity to nuclear proteins. 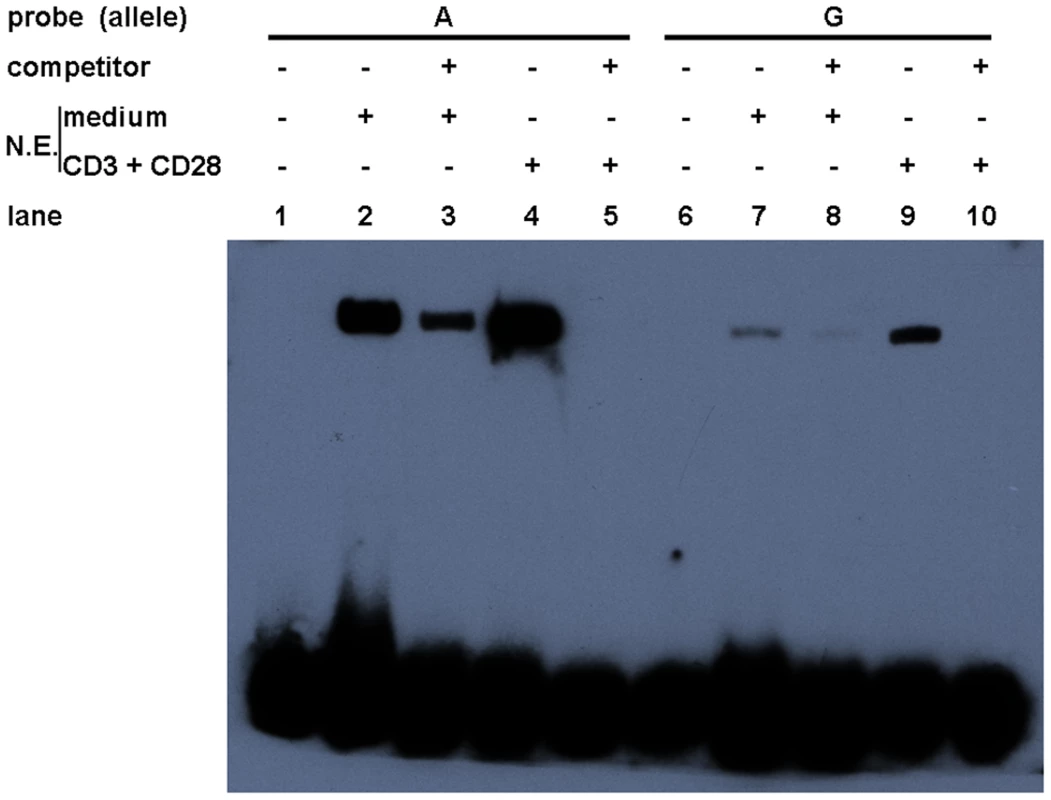
Shown is a gel-shift assay of the allelic probes (A or G) with nuclear extracts (N.E.) prepared from either rested (medium) or anti-CD3+anti-CD28-activated Jurkat cells. Also shown are the results of a competition assay performed with the addition of 100-fold unlabeled competitor oligonucleotides. The experiment was repeated three times. Alteration of Ets-1 binding to the miR-146a promoter by rs57095329
The findings described above illustrated that rs57095329 alleles conferred differential binding affinity of nuclear extracts to the miR-146a promoter. To identify which proteins bind at or near this SNP to regulate the expression levels of miR-146a, we performed a bioinformatics search. The Genomatix online tool suggested that the multipotent transcription factor Ets-1 binds to the rs57095329 region (Figure S6). Interestingly, it has been shown that mutation of this predicted Ets-1-binding site resulted in a great reduction in the activity of an miR-146a promoter–reporter gene [29]. Here, we performed the following assays and further confirmed the pivotal role of Ets-1 in regulating miR-146a expression: the transient expression of Ets-1 greatly enhanced the reporter gene activity from the full-length miR-146a promoter in Jurkat cells, compared with that of another transcription factor, PBX1 (Figure 4A); knockdown of Ets-1 by small interfering RNA (siRNA) in Jurkat cells directly impaired the induction of pri-miR-146a upon T-cell activation (Figure 4B); and the overexpression of Ets-1 dramatically enhanced, whereas the knockdown of Ets-1 consistently reduced, the miR-146a promoter–reporter gene activity in HeLa cells (Figure S7A and S7B).
Fig. 4. Ets-1 regulates miR-146a expression and accounts for the different regulatory activities of the rs57095329 alleles. 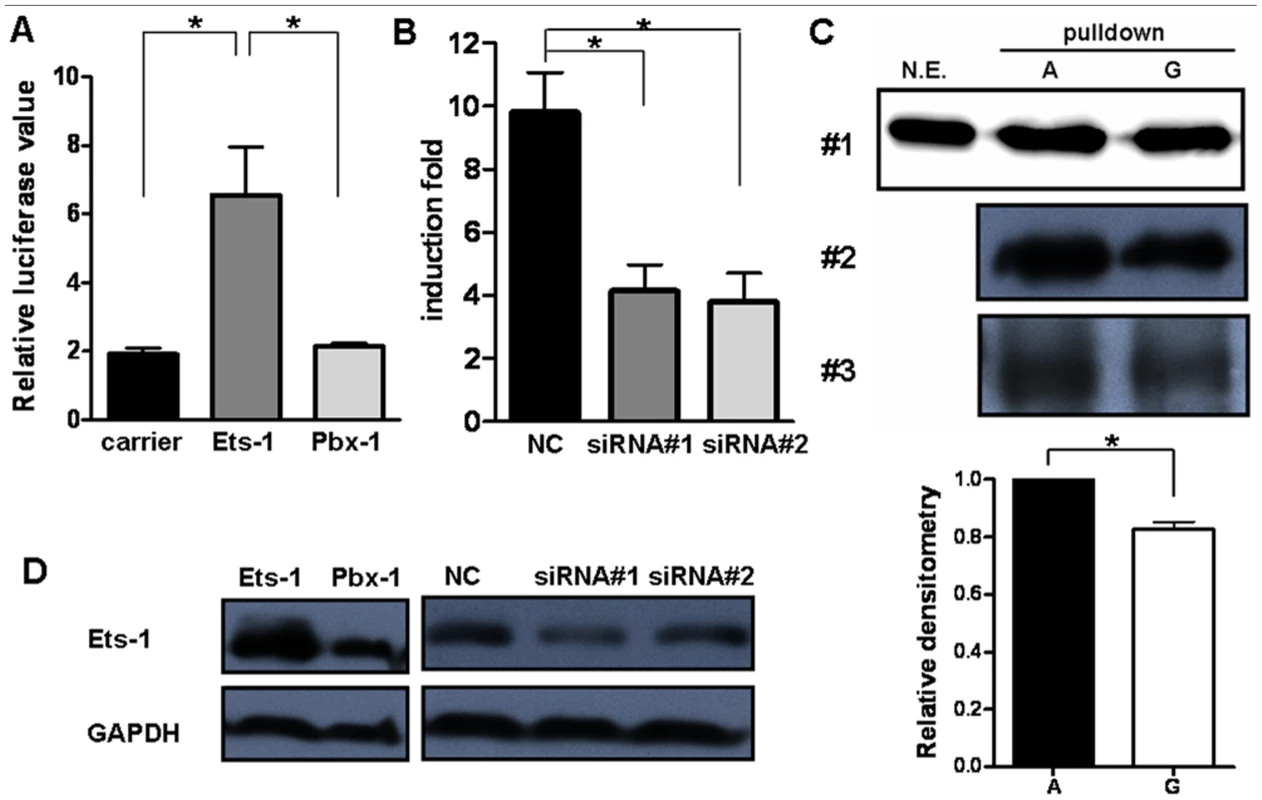
(A) Comparison of the miR-146a promoter–reporter gene activities in Jurkat cells after cotransfection with an equal amount of irrelevant carrier vector or ETS1- or PBX1-expressing vector. The data shown are means ± SEM and are representative of three independent experiments. * indicates P<0.05. (B) Real-time PCR analysis of the fold induction of pri-miR-146a in Jurkat cells after the transfection of ETS1 siRNA (#1 and #2) or a negative control (NC), followed by cell activation with anti-CD3+anti-CD28 antibodies for 9 hours. The data shown are means ± SEM and are representative of three independent experiments. * indicates P<0.05. (C) Streptavidin–agarose pulldown assay of transcription factors bound to allelic probes of rs57095329. Biotinylated A or G probes were incubated with nuclear extracts from anti-CD3+anti-CD28-antibody-activated Jurkat cells in the presence of streptavidin–agarose beads. The precipitated proteins were analyzed by western blotting with an anti-Ets-1 antibody (upper). The assay was repeated three times. For experiment #1, the nuclear extracts (N.E.) that were used as input for the pulldown assay were also directly blotted as the control. Also shown is a comparison of the relative densitometry values for the blotted bands corresponding to the allelic probes (lower). * indicates P<0.05. (D) Western blot analysis of Ets-1 levels in Jurkat cells after transfection of the same amounts of the indicated expression vectors or siRNA as used in A or B, respectively. The cells were collected 24 hours or 72 hours after transfection for overexpression or for siRNA-mediated knockdown assays, respectively. GAPDH was used as the loading control. We assessed whether the allelic difference of rs57095329 in regulatory activity is attributable to different binding affinity for Ets-1. We cotransfected different amounts of Ets-1 with the reporter gene construct containing either the A or G allele of rs57095329 into HeLa cells. Increasing the protein levels of Ets-1 greatly enhanced the promoter activity of both constructs. However, the activity ratios of the two constructs (G/A) gradually decreased (Figure S8), suggesting that the inferior ability of the G-allele-containing sequence to bind Ets-1 could be compensated by increasing the levels of Ets-1. To examine the binding affinity more directly, we performed a promoter pulldown assay using streptavidin-conjugated agarose beads. When incubated with nuclear extracts from anti-CD3+anti-CD28-activated Jurkat cells, the biotin-labeled A probe bound more Ets-1 protein than the G probe, as demonstrated by western blotting analysis of total agarose beads precipitated proteins with an anti-Ets-1 antibody (Figure 4C). Taken together, these results demonstrate that rs57095329 alters Ets-1 binding, and the risk-associated G allele is less competent than the A allele in the regulation of miR-146a expression.
Interaction analysis of rs57095329 with ETS1 SNP rs1128334
It is intriguing that our previous GWAS and that of others identified an association between a functional variant of ETS1 and SLE [8], [9]. Therefore, we investigated whether there is an interaction between the risk variants of the two genes, rs1128334 in ETS1 and rs57095329 in miR-146a. No epistatic effect was detected between the two variants (P = 0.46), when analyzed with a conditional logistic regression test, with the interaction between the two variants treated as a covariate using PLINK [28]. However, we observed additive effects of the risk alleles of miR-146a and ETS1, suggesting that individuals carrying two or more of these alleles are at greater risk than those carrying only one allele (Figure 5). This additive effect between ETS1 and miR-146a SNPs is also supported by a consistent increase in OR values in analysis of the enlarged samples containing 4,302 individuals with known genotypes for both SNPs (Table S4).
Fig. 5. Joint effects of the risk-associated alleles of ETS1 and miR-146a. 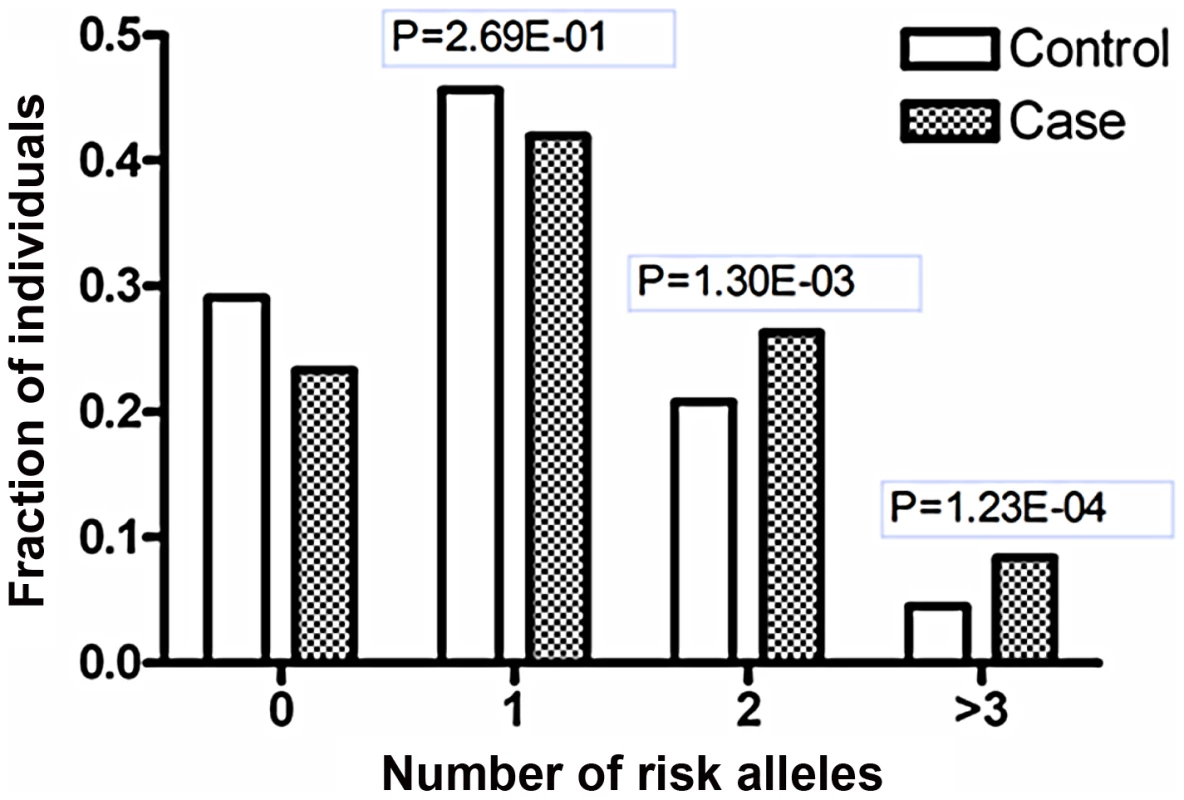
Shanghai individuals (703 patients and 983 controls) with known genotypes for both variants (rs1128334 in ETS1 and rs57095329 in miR-146a) were analyzed for the distributions of the risk-associated alleles for SLE; fraction of each group with different numbers of risk alleles was shown. The P values were calculated with the χ2 test for the differences in the allele counts between the patients and the controls in the groups with two or more risk-associated alleles, compared with the groups with zero risk-associated alleles. Discussion
miRNAs have been shown to play an essential role in immune homeostasis, and aberrations in the miRNA-mediated regulation of immune-cell development and function has been linked to autoimmune diseases [30]. In an miRNA profiling study, we recently identified a significant reduction in miR-146a expression in lupus patients [23]. Here, we extended this study to determine why the expression of miR-146a is reduced in SLE patients. Prompted by the genetic association between the type I IFN pathway and the risk of SLE and by evidence that polymorphic variants in miRNA precursors can modulate miRNA biogenesis and disease risk, we sequenced key regions of pri-miR-146a and identified an SLE-associated SNP, rs57095329, within the promoter of miR-146a, which functionally affects miR-146a expression levels and thus contributes to the risk of SLE. The association of this variant with SLE was consistent in three independent cohorts from mainland China, Hong Kong, and Bangkok (Thailand). Individuals carrying the risk-associated G allele tended to express lower levels of miR-146a. To the best of our knowledge, this is the first report of an association between a functional genetic variant in an miRNA promoter and a human disease. It will be interesting to investigate the association between rs57095329 and SLE in other ethnic groups. Another functional variant located in the miR-146a precursor, rs2910164, has been associated with cancer development [26], [31], [32], but showed no significant association with SLE in our initial sequencing experiments.
Among the multiple immunological aberrations present in lupus patients, the type I IFN system is thought to play a crucial role in its pathogenesis [15]–[17]. Intriguingly, a number of genes involved in IFN signaling have already been associated with various autoimmune diseases, including SLE [33]. Functional variants in genes encoding key components of the IFN pathway, such as TYK2, IRF5, and STAT4, have been identified and characterized, and their association with SLE has been extensively replicated [34]–[39]. Our recent work characterized the role of miR-146a as a negative regulator of the type I IFN pathway by targeting key signaling proteins [23]. Here, the delineation of an SLE-susceptible variant of the miR-146a promoter further supports the notion that polymorphic variants linked to IFN pathway molecules contribute to the pathogenesis of lupus. miR-146a is embedded in a non-coding RNA with a previously unknown function, so our findings highlight the importance of exploring genetic variants in such regions, which have been more or less ignored in previous genetic studies.
Our findings underline the regulatory role of Ets-1 in miR-146a expression, and attribute the allelic difference of rs57095329 to different affinity for Ets-1. Rs57095329 is not located at the core sequence of the Ets-1-binding site (Figure S6), so it only causes an affinity difference, whereas Ets-1 recognition is still well preserved. Nevertheless, the risk-associated G allele of rs57095329 does affect the strongly conserved A residue near the Ets-1 core motif (Figure S6), highlighting the relevance of this SNP. Besides, this is a germ-line regulatory polymorphism and thus potentially functions in each cell type, as reflected in our consistent observation of the reduced activity of a reporter gene carrying the risk-associated G allele in various cell lines (Figure 2 and Figure S4). The attenuation of the promoter activity by the risk-associated G allele of rs57095329 thus accounts, at least partly, for the underexpression of miR-146a in lupus patients. Intriguingly, ETS1 has been characterized as a susceptibility gene for SLE in GWAS results from others and our group [8], [9]. The reduced expression of ETS1 was shown to be associated with the risk-associated allele of rs1128334 compared with the protective allele, identified in an allelic expression assay [9]. The finding that Ets-1 knockdown led to an inability to induce miR-146a expression in vitro (Figure 4B) was consistent with reduced miR-146a expression in patients with SLE who have reduced Ets-1 levels. It seems that both rs1128334 in ETS1 and rs57095329 in miR-146a may reduce the expression of miR-146a, through the reduced availability of Ets-1 and a reduced binding affinity for Ets-1, respectively. However, we did not detect interaction between the two variants. This may not be surprising because both of these variants only have a quantitative effect on their respective functions. Therefore, an additive effect is observed between the two variants (Figure 5 and Table S4) rather than a strong interaction, which would be the case if both of them totally abolished a function. We propose a working model for the genetic link between ETS1 and miR-146a to illustrate the genetic contribution to the reduced expression of miR-146a in SLE patients (Figure 6). We fully appreciate that Ets-1 can modulate a large collection of genes for their expression, and are not trying to limit the contribution of its 3′UTR SNP to SLE solely to affecting miR-146a expression. Yet our functional study, the genotype-expression data, and the additive effect of the two SNPs together provide an interesting connection between these two SLE susceptibility genes.
Fig. 6. Diagram of the genetic link between ETS1 and miR-146a and genetic contribution to miR-146a expression. 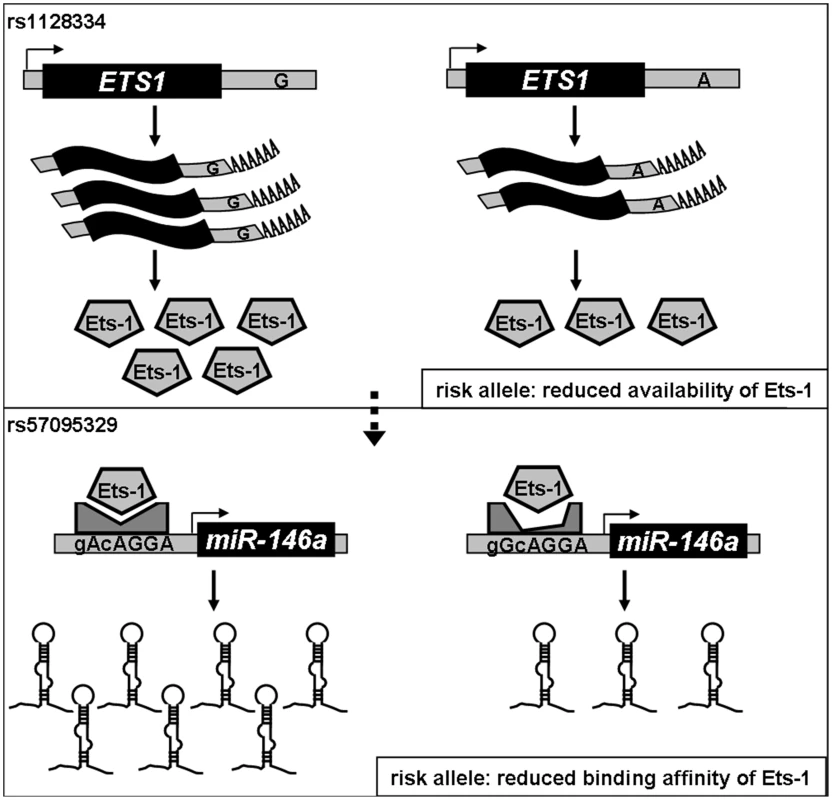
Expression of cellular miR-146a depends on the binding of Ets-1 to the miR-146a promoter, so both the availability of and its binding affinity for Ets-1 could affect miR-146a expression. We previously found that the A allele of rs1128334 located in the ETS1 3′ untranslated region is associated with SLE by reducing Ets-1 expression (upper panel). In this study, we show that the binding affinity for Ets-1 is affected by the risk-associated G allele of rs57095329, which causes an incompetent compositional change (lower panel), and therefore represents another genetic factor that contributes to the reduced expression of miR-146a in SLE patients. Recent SLE GWASs identified disease association of two SNPs (rs2431697 and rs2431099) that are upstream of miR-146a gene region [3], [7], [9] and our genotyping confirmed their association on our Chinese samples (Table S2). We therefore performed the following analysis to clarify the genetic signals of association across this region: 1) Imputation of rs57095329 into our Asian GWAS dataset, using the individuals of Asian ancestry from the 1000 genome project as the reference panel, suggested that this SNP represented an independent signal (conditional P value of 0.90); 2) rs2431099 and rs2431697 was in intermediate LD with each other while they were not in LD with rs57095329 (Figure S2); 3) Conditional analysis indicated that the association of rs57095329 with SLE was independent of those detected at rs2431697 and rs2431099, while the association of rs2431099 with SLE could be attributed to rs2431697 (Table S5); 4) Haplotype analysis showed that rs57095329 and rs2431697 were two independent SLE-associated loci, while rs57095329 had a stronger association in Chinese (Table S6); and 5) There was no correlation between miR-146a expression levels and the genotypes of rs2431697 or rs2431099 (Figure S9). Therefore we have newly identified a relevant SNP (rs57095329) by direct sequencing that the genotyping arrays in GWASs missed due to incomplete coverage, and these SNPs may confer a disease risk through different and independent mechanisms.
In conclusion, our findings add an miRNA gene, miR-146a, to the list of SLE-susceptible genes. A genetic variant of the miR-146a promoter, rs57095329, is functionally significant in modulating the expression of miR-146a by altering its binding affinity for Ets-1.
Materials and Methods
Ethics statement
This study was conducted according to the principles expressed in the Declaration of Helsinki. Informed consent was obtained from all subjects. The Shanghai study was approved by the Institutional Review Board of Renji Hospital. The studies of the Hong Kong, Anhui, and Thai samples were approved by the Institutional Review Board of the University of Hong Kong and Hospital Authority, Hong Kong West Cluster, New Territory West Cluster, and Hong Kong East Cluster; the Research Ethics Committee of Anhui Medical University; and the Ethics Committee of the Faculty of Medicine, Chulalongkorn University, respectively.
Subjects
We recruited 816 SLE patients and 1,080 sex - and age-matched controls, all of whom were from the Chinese Han population in Shanghai, China. Other Chinese mainland samples consisted of 1,536 SLE patients living in central China, collected by collaborators in Anhui province. For the independent replications, samples collected by collaborators in Hong Kong (case vs control: 1,152 vs 1,152) and Bangkok, Thailand (464 vs 982, respectively) were included. All SLE patients fulfilled the American College of Rheumatology (ACR) classification criteria for SLE, and 1,254 patients met the ACR criteria for lupus nephritis.
DNA sequencing and genotyping
Consecutive overlapping amplicons corresponding to the miR-146a promoter region were amplified from genomic DNA extracted from peripheral blood leukocytes. The products were purified and directly sequenced on a 3730 automated sequencer (Applied Biosystems). The 452-bp DNA region around the miR-146a precursor was also amplified and sequenced. The primer pairs used are shown in Table S7.
In the replication stage, SNP rs57095329 was genotyped with the specified TaqMan SNP genotyping probes (Applied Biosystems). The assay was run on a 7900HT sequence detection system (Applied Biosystems) and the data were analyzed with the affiliated SDS software, version 2.3. The genotypes of rs57095329 were found to be in Hardy–Weinberg equilibrium (P>0.01) in the controls of all three cohorts. The average call rate for all samples was 92%.
Real-time PCR
Total RNA was extracted from peripheral blood leukocytes or cultured cells using TRIzol (Invitrogen), followed by reverse transcription using a reverse transcriptase kit obtained from Takara. To determine the quantity of pri-miR-146a, the cDNA was amplified by real-time PCR with SYBR Green RT–PCR kit (Takara), and the expression of RPL13A was used as the internal control. The primers used are shown in Table S7. To determine the quantity of mature miR-146a, the specific TaqMan MicroRNA Assay kit (Applied Biosystems) was used, and the expression levels were normalized to snRNA U6. The assays were performed on a 7900HT real-time instrument (Applied Biosystems). Relative expression levels were calculated using the 2−ΔΔCt method.
Constructs
To create the miR-146a promoter–luciferase reporter constructs, three fragments of variable lengths, corresponding to the upstream region of the TSS of pri-miR-146a, were amplified and cloned into the pGL3-basic luciferase vector (Promega). To compare the activities of miR-146a promoters containing the different rs57095329 alleles, the full-length 1105-bp fragment was amplified from individual homozygous templates. The ETS1 overexpression vector was a kind gift from Dr Gang Pei, and the PBX1 overexpression plasmid was created by replacing the inserted ETS1 sequence with the PBX1 coding sequence. The primers used are shown in Table S7. All constructs were verified by sequencing.
Cell culture, transfection, and stimulation
Jurkat and Raji cells were grown in RPMI 1640 medium supplemented with 10% fetal bovine serum. These two cell lines were electroporated with 2 µg of the indicated luciferase reporter vector and 0.2 µg of a modified pRL-TK vector, using a nucleofector device (Amaxa). Alternatively, the reporter gene vectors were electroporated in combination with 1.5 µg of an ETS1 - or PBX1-expressing vector. For the knockdown of ETS1, 3 µg of ETS1 siRNA or negative control oligonucleotides (all from GenePharma, Shanghai) were transfected. HeLa and 293T cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum. These two cell lines were transfected using Lipofectamine 2000 (Invitrogen), with the ETS1 - or PBX1-expressing vector or ETS1 siRNA alone, or in combination with 50 ng of the indicated luciferase reporter vector and 5 ng of a modified pRL-TK plasmid. Where indicated, an irrelevant “carrier” vector was added to ensure that equal total amounts of plasmid DNA were transfected among the groups. For cell activation, Jurkat and HeLa cells were stimulated with PMA (100 ng/mL; Sigma) and ionomycin (1 µM; Sigma) for the indicated times. Alternatively, Jurkat cells were activated with plate-bound anti-CD3 antibody (coating solution: 5 µg/mL; eBioscience) and soluble anti-CD28 antibody (2 µg/mL; eBioscience).
Reporter gene assay
Cells were cultured for 24 hours or 48 hours after transfection with the reporter gene vectors together with the ETS1 expression vector or siRNA, respectively. The cells were then maintained resting or activated for 6 hours and lysed. Their luciferase activity was measured on a luminometer (LB960; Berthold) using the Dual-Luciferase Reporter Assay System (Promega). The ratio of firefly luciferase to Renilla luciferase was calculated for each well.
EMSA
Jurkat and HeLa cells (1×107) were activated or left to rest for 2 hours, and then their nuclear proteins were extracted with a Nuclear Extract Kit (Active Motif), according to the manufacturer's protocol. The protein concentrations were determined with the DC Protein Assay Kit (Bio-Rad). Double-stranded allelic probes were synthesized and labeled with biotin by Takara (the sequence is shown in Figure S6). EMSA was carried out with a gel-shift kit purchased from Active Motif. The competition assay was performed by adding cognate unlabeled oligonucleotides. After incubation, the protein–DNA complexes were separated on a nondenaturing 6% polyacrylamide gel and then transferred to a nitrocellulose membrane (Millipore). The signals were detected using a luminoimage analyzer.
Streptavidin–agarose pulldown and western blotting
The pulldown assay was performed following a protocol described elsewhere [40], with slight modification. Biotin-labeled allelic probes were incubated with equal amounts of nuclear extract from activated Jurkat cells for 2 hours at room temperature, in the presence of streptavidin–agarose beads (GE Healthcare) and protein inhibitors. The precipitated protein–DNA complex was dissociated from the agarose beads by suspending the pellet in Laemmli sample buffer (Bio-Rad) and heating it. The supernatants were then subjected to SDS–PAGE. The proteins were transferred onto a PVDF membrane (Bio-Rad), blotted with an anti-Ets-1 antibody, and detected with ECL solution (Pierce). To evaluate the Ets-1 protein levels after the transfection of the overexpression vectors or siRNAs, the Jurkat and HeLa cells were lysed in RIPA buffer (Thermo Scientific), and the supernatants were similarly used for immunoblotting. Anti-ETS1, anti-GAPDH, and horseradish-peroxidase-conjugated secondary antibodies were all obtained from Santa Cruz Biotechnology.
Data analysis
For single SNP analysis, PLINK was used for the basic allelic test and other tests in the patients and the controls [28]. LD patterns were analyzed and displayed with HaploView [41]. Review manager was used to perform meta-analysis. IMPUTE version 2 was used to perform imputation. Other data were analyzed with GraphPad Prism 4 software, version 4.03. The nonparametric Mann–Whitney test was used to compare miR-146a expression between the genotype groups, and an unpaired t test was used to compare reporter gene activities. Two-tailed P values<0.05 were considered to be statistically significant.
Supporting Information
Zdroje
1. D'CruzDPKhamashtaMAHughesGR 2007 Systemic lupus erythematosus. Lancet 369 587 596
2. RahmanAIsenbergDA 2008 Systemic lupus erythematosus. N Engl J Med 358 929 939
3. HarleyJBAlarcon-RiquelmeMECriswellLAJacobCOKimberlyRP 2008 Genome-wide association scan in women with systemic lupus erythematosus identifies susceptibility variants in ITGAM, PXK, KIAA1542 and other loci. Nat Genet 40 204 210
4. HomGGrahamRRModrekBTaylorKEOrtmannW 2008 Association of systemic lupus erythematosus with C8orf13-BLK and ITGAM-ITGAX. N Engl J Med 358 900 909
5. GrahamRRCotsapasCDaviesLHackettRLessardCJ 2008 Genetic variants near TNFAIP3 on 6q23 are associated with systemic lupus erythematosus. Nat Genet 40 1059 1061
6. KozyrevSVAbelsonAKWojcikJZaghloolALinga ReddyMV 2008 Functional variants in the B-cell gene BANK1 are associated with systemic lupus erythematosus. Nat Genet 40 211 216
7. GatevaVSandlingJKHomGTaylorKEChungSA 2009 A large-scale replication study identifies TNIP1, PRDM1, JAZF1, UHRF1BP1 and IL10 as risk loci for systemic lupus erythematosus. Nat Genet 41 1228 1233
8. HanJWZhengHFCuiYSunLDYeDQ 2009 Genome-wide association study in a Chinese Han population identifies nine new susceptibility loci for systemic lupus erythematosus. Nat Genet 41 1234 1237
9. YangWShenNYeDQLiuQZhangY 2010 Genome-wide association study in Asian populations identifies variants in ETS1 and WDFY4 associated with systemic lupus erythematosus. PLoS Genet 6 e1000841 doi:10.1371/journal.pgen.1000841
10. CrowMK 2008 Collaboration, genetic associations, and lupus erythematosus. N Engl J Med 358 956 961
11. FlesherDLSunXBehrensTWGrahamRRCriswellLA 2010 Recent advances in the genetics of systemic lupus erythematosus. Expert Rev Clin Immunol 6 461 479
12. BennettLPaluckaAKArceECantrellVBorvakJ 2003 Interferon and granulopoiesis signatures in systemic lupus erythematosus blood. J Exp Med 197 711 723
13. HanGMChenSLShenNYeSBaoCD 2003 Analysis of gene expression profiles in human systemic lupus erythematosus using oligonucleotide microarray. Genes Immun 4 177 186
14. BaechlerECBatliwallaFMKarypisGGaffneyPMOrtmannWA 2003 Interferon-inducible gene expression signature in peripheral blood cells of patients with severe lupus. Proc Natl Acad Sci U S A 100 2610 2615
15. PascualVFarkasLBanchereauJ 2006 Systemic lupus erythematosus: all roads lead to type I interferons. Curr Opin Immunol 18 676 682
16. RonnblomLElorantaMLAlmGV 2006 The type I interferon system in systemic lupus erythematosus. Arthritis Rheum 54 408 420
17. CrowMK 2007 Type I interferon in systemic lupus erythematosus. Curr Top Microbiol Immunol 316 359 386
18. RonnblomLAlmGVElorantaML 2009 Type I interferon and lupus. Curr Opin Rheumatol 21 471 477
19. BartelDP 2004 MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116 281 297
20. BaltimoreDBoldinMPO'ConnellRMRaoDSTaganovKD 2008 MicroRNAs: new regulators of immune cell development and function. Nat Immunol 9 839 845
21. LodishHFZhouBLiuGChenCZ 2008 Micromanagement of the immune system by microRNAs. Nat Rev Immunol 8 120 130
22. XiaoCRajewskyK 2009 MicroRNA control in the immune system: basic principles. Cell 136 26 36
23. TangYLuoXCuiHNiXYuanM 2009 MicroRNA-146A contributes to abnormal activation of the type I interferon pathway in human lupus by targeting the key signaling proteins. Arthritis Rheum 60 1065 1075
24. DuanRPakCJinP 2007 Single nucleotide polymorphism associated with mature miR-125a alters the processing of pri-miRNA. Hum Mol Genet 16 1124 1131
25. MenciaAModamio-HoybjorSRedshawNMorinMMayo-MerinoF 2009 Mutations in the seed region of human miR-96 are responsible for nonsyndromic progressive hearing loss. Nat Genet 41 609 613
26. JazdzewskiKMurrayELFranssilaKJarzabBSchoenbergDR 2008 Common SNP in pre-miR-146a decreases mature miR expression and predisposes to papillary thyroid carcinoma. Proc Natl Acad Sci U S A 105 7269 7274
27. TaganovKDBoldinMPChangKJBaltimoreD 2006 NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci U S A 103 12481 12486
28. PurcellSNealeBTodd-BrownKThomasLFerreiraMA 2007 PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81 559 575
29. CurtaleGCitarellaFCarissimiCGoldoniMCarucciN 2010 An emerging player in the adaptive immune response: microRNA-146a is a modulator of IL-2 expression and activation-induced cell death in T lymphocytes. Blood 115 265 273
30. LuoXTsaiLMShenNYuD 2010 Evidence for microRNA-mediated regulation in rheumatic diseases. Ann Rheum Dis 69 Suppl 1 i30 36
31. XuTZhuYWeiQKYuanYZhouF 2008 A functional polymorphism in the miR-146a gene is associated with the risk for hepatocellular carcinoma. Carcinogenesis 29 2126 2131
32. XuBFengNHLiPCTaoJWuD 2010 A functional polymorphism in Pre-miR-146a gene is associated with prostate cancer risk and mature miR-146a expression in vivo. Prostate 70 467 472
33. Delgado-VegaAMAlarcon-RiquelmeMEKozyrevSV 2010 Genetic associations in type I interferon related pathways with autoimmunity. Arthritis Res Ther 12 Suppl 1 S2
34. SigurdssonSNordmarkGGoringHHLindroosKWimanAC 2005 Polymorphisms in the tyrosine kinase 2 and interferon regulatory factor 5 genes are associated with systemic lupus erythematosus. Am J Hum Genet 76 528 537
35. GrahamRRKozyrevSVBaechlerECReddyMVPlengeRM 2006 A common haplotype of interferon regulatory factor 5 (IRF5) regulates splicing and expression and is associated with increased risk of systemic lupus erythematosus. Nat Genet 38 550 555
36. GrahamRRKyogokuCSigurdssonSVlasovaIADaviesLR 2007 Three functional variants of IFN regulatory factor 5 (IRF5) define risk and protective haplotypes for human lupus. Proc Natl Acad Sci U S A 104 6758 6763
37. RemmersEFPlengeRMLeeATGrahamRRHomG 2007 STAT4 and the risk of rheumatoid arthritis and systemic lupus erythematosus. N Engl J Med 357 977 986
38. KariukiSNKirouKAMacDermottEJBarillas-AriasLCrowMK 2009 Cutting edge: autoimmune disease risk variant of STAT4 confers increased sensitivity to IFN-alpha in lupus patients in vivo. J Immunol 182 34 38
39. RulloOJWooJMWuHHoftmanADMaranianP 2010 Association of IRF5 polymorphisms with activation of the interferon alpha pathway. Ann Rheum Dis 69 611 617
40. WuKK 2006 Analysis of protein-DNA binding by streptavidin-agarose pulldown. Methods Mol Biol 338 281 290
41. BarrettJCFryBMallerJDalyMJ 2005 Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21 263 265
Štítky
Genetika Reprodukční medicína
Článek Local Absence of Secondary Structure Permits Translation of mRNAs that Lack Ribosome-Binding SitesČlánek Independent Chromatin Binding of ARGONAUTE4 and SPT5L/KTF1 Mediates Transcriptional Gene SilencingČlánek Trade-Off between Bile Resistance and Nutritional Competence Drives Diversification in the Mouse GutČlánek FGF Signaling Regulates the Number of Posterior Taste Papillae by Controlling Progenitor Field SizeČlánek Mammalian BTBD12 (SLX4) Protects against Genomic Instability during Mammalian SpermatogenesisČlánek Identification of Nine Novel Loci Associated with White Blood Cell Subtypes in a Japanese PopulationČlánek Differential Effects of and Risk Variants on Association with Diabetic ESRD in African AmericansČlánek Dynamic Chromatin Localization of Sirt6 Shapes Stress- and Aging-Related Transcriptional Networks
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2011 Číslo 6
-
Všechny články tohoto čísla
- Local Absence of Secondary Structure Permits Translation of mRNAs that Lack Ribosome-Binding Sites
- Statistical Inference on the Mechanisms of Genome Evolution
- Revisiting Heterochromatin in Embryonic Stem Cells
- A Two-Stage Meta-Analysis Identifies Several New Loci for Parkinson's Disease
- Identification of a Sudden Cardiac Death Susceptibility Locus at 2q24.2 through Genome-Wide Association in European Ancestry Individuals
- Genomic Prevalence of Heterochromatic H3K9me2 and Transcription Do Not Discriminate Pluripotent from Terminally Differentiated Cells
- Epistasis between Beneficial Mutations and the Phenotype-to-Fitness Map for a ssDNA Virus
- Recurrent Chromosome 16p13.1 Duplications Are a Risk Factor for Aortic Dissections
- Telomere DNA Deficiency Is Associated with Development of Human Embryonic Aneuploidy
- Genome-Wide Association Study of White Blood Cell Count in 16,388 African Americans: the Continental Origins and Genetic Epidemiology Network (COGENT)
- Unexpected Role for DNA Polymerase I As a Source of Genetic Variability
- Transportin-SR Is Required for Proper Splicing of Genes and Plant Immunity
- How Chromatin Is Remodelled during DNA Repair of UV-Induced DNA Damage in
- Independent Chromatin Binding of ARGONAUTE4 and SPT5L/KTF1 Mediates Transcriptional Gene Silencing
- Two Evolutionary Histories in the Genome of Rice: the Roles of Domestication Genes
- Natural Allelic Variation Defines a Role for : Trichome Cell Fate Determination
- Multiple Common Susceptibility Variants near BMP Pathway Loci , , and Explain Part of the Missing Heritability of Colorectal Cancer
- Pathogenic Mechanism of the FIG4 Mutation Responsible for Charcot-Marie-Tooth Disease CMT4J
- A Functional Variant in Promoter Modulates Its Expression and Confers Disease Risk for Systemic Lupus Erythematosus
- Drift and Genome Complexity Revisited
- Chromosomal Macrodomains and Associated Proteins: Implications for DNA Organization and Replication in Gram Negative Bacteria
- Trade-Off between Bile Resistance and Nutritional Competence Drives Diversification in the Mouse Gut
- Pathways of Distinction Analysis: A New Technique for Multi–SNP Analysis of GWAS Data
- Web-Based Genome-Wide Association Study Identifies Two Novel Loci and a Substantial Genetic Component for Parkinson's Disease
- Chk2 and p53 Are Haploinsufficient with Dependent and Independent Functions to Eliminate Cells after Telomere Loss
- Exome Sequencing Identifies Mutations in High Myopia
- Distinct Functional Constraints Partition Sequence Conservation in a -Regulatory Element
- CorE from Is a Copper-Dependent RNA Polymerase Sigma Factor
- A Single Sex Pheromone Receptor Determines Chemical Response Specificity of Sexual Behavior in the Silkmoth
- FGF Signaling Regulates the Number of Posterior Taste Papillae by Controlling Progenitor Field Size
- Maps of Open Chromatin Guide the Functional Follow-Up of Genome-Wide Association Signals: Application to Hematological Traits
- Increased Susceptibility to Cortical Spreading Depression in the Mouse Model of Familial Hemiplegic Migraine Type 2
- Differential Gene Expression and Epiregulation of Alpha Zein Gene Copies in Maize Haplotypes
- Parallel Adaptive Divergence among Geographically Diverse Human Populations
- Genetic Analysis of Genome-Scale Recombination Rate Evolution in House Mice
- Mechanisms for the Evolution of a Derived Function in the Ancestral Glucocorticoid Receptor
- Mammalian BTBD12 (SLX4) Protects against Genomic Instability during Mammalian Spermatogenesis
- Interferon Regulatory Factor 8 Regulates Pathways for Antigen Presentation in Myeloid Cells and during Tuberculosis
- High-Resolution Analysis of Parent-of-Origin Allelic Expression in the Arabidopsis Endosperm
- Specific SKN-1/Nrf Stress Responses to Perturbations in Translation Elongation and Proteasome Activity
- Graded Nodal/Activin Signaling Titrates Conversion of Quantitative Phospho-Smad2 Levels into Qualitative Embryonic Stem Cell Fate Decisions
- Genome-Wide Analysis Reveals PADI4 Cooperates with Elk-1 to Activate Expression in Breast Cancer Cells
- Trait Variation in Yeast Is Defined by Population History
- Meiosis-Specific Loading of the Centromere-Specific Histone CENH3 in
- A Genome-Wide Survey of Imprinted Genes in Rice Seeds Reveals Imprinting Primarily Occurs in the Endosperm
- Multiple Regulatory Mechanisms to Inhibit Untimely Initiation of DNA Replication Are Important for Stable Genome Maintenance
- SIRT1 Promotes N-Myc Oncogenesis through a Positive Feedback Loop Involving the Effects of MKP3 and ERK on N-Myc Protein Stability
- Bacteriophage Crosstalk: Coordination of Prophage Induction by Trans-Acting Antirepressors
- Role of the Single-Stranded DNA–Binding Protein SsbB in Pneumococcal Transformation: Maintenance of a Reservoir for Genetic Plasticity
- Genomic Convergence among ERRα, PROX1, and BMAL1 in the Control of Metabolic Clock Outputs
- Genome-Wide Association of Bipolar Disorder Suggests an Enrichment of Replicable Associations in Regions near Genes
- Identification of Nine Novel Loci Associated with White Blood Cell Subtypes in a Japanese Population
- DNA Ligase III Promotes Alternative Nonhomologous End-Joining during Chromosomal Translocation Formation
- Differential Effects of and Risk Variants on Association with Diabetic ESRD in African Americans
- Finished Genome of the Fungal Wheat Pathogen Reveals Dispensome Structure, Chromosome Plasticity, and Stealth Pathogenesis
- Dynamic Chromatin Localization of Sirt6 Shapes Stress- and Aging-Related Transcriptional Networks
- Extracellular Matrix Dynamics in Hepatocarcinogenesis: a Comparative Proteomics Study of Transgenic and Null Mouse Models
- Integrating 5-Hydroxymethylcytosine into the Epigenomic Landscape of Human Embryonic Stem Cells
- Vive La Différence: An Interview with Catherine Dulac
- Multiple Loci Are Associated with White Blood Cell Phenotypes
- Nuclear Accumulation of Stress Response mRNAs Contributes to the Neurodegeneration Caused by Fragile X Premutation rCGG Repeats
- A New Mutation Affecting FRQ-Less Rhythms in the Circadian System of
- Cryptic Transcription Mediates Repression of Subtelomeric Metal Homeostasis Genes
- A New Isoform of the Histone Demethylase JMJD2A/KDM4A Is Required for Skeletal Muscle Differentiation
- Genetic Determinants of Lipid Traits in Diverse Populations from the Population Architecture using Genomics and Epidemiology (PAGE) Study
- A Genome-Wide RNAi Screen for Factors Involved in Neuronal Specification in
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Recurrent Chromosome 16p13.1 Duplications Are a Risk Factor for Aortic Dissections
- Statistical Inference on the Mechanisms of Genome Evolution
- Genome-Wide Association Study of White Blood Cell Count in 16,388 African Americans: the Continental Origins and Genetic Epidemiology Network (COGENT)
- Chromosomal Macrodomains and Associated Proteins: Implications for DNA Organization and Replication in Gram Negative Bacteria
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



