-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaFob1 and Fob2 Proteins Are Virulence Determinants of via Facilitating Iron Uptake from Ferrioxamine
Deferoxamine is an iron-chelating agent often used to treat patients with acute iron poisoning, such as seen in dialysis patients with chronic renal failure. These patients are uniquely predisposed to a deadly fungal infection, called mucormycosis, because deferoxamine supplies iron that supports growth of fungi causing this infection. Apart from the important basic knowledge in delineating iron uptake mechanisms in cells, understanding how organisms causing mucormycosis obtain iron from ferrioxamine (deferoxamine bound with iron) is likely to develop strategies to treat mucormycosis infections in patients treated with deferoxamine. In this study we identified two cell surface receptors that bind ferrioxamine and facilitate iron uptake in Rhizopus oryzae, the most causative fungus of mucormycosis. These receptors are required for full virulence of R. oryzae in mice treated with deferoxamine. From genetic and biochemical studies it appears that the fungus binds ferrioxamine via these two receptors then liberates iron through a chemical modification step prior to transporting into the fungal cell without the internalization of deferoxamine.
Published in the journal: . PLoS Pathog 11(5): e32767. doi:10.1371/journal.ppat.1004842
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1004842Summary
Deferoxamine is an iron-chelating agent often used to treat patients with acute iron poisoning, such as seen in dialysis patients with chronic renal failure. These patients are uniquely predisposed to a deadly fungal infection, called mucormycosis, because deferoxamine supplies iron that supports growth of fungi causing this infection. Apart from the important basic knowledge in delineating iron uptake mechanisms in cells, understanding how organisms causing mucormycosis obtain iron from ferrioxamine (deferoxamine bound with iron) is likely to develop strategies to treat mucormycosis infections in patients treated with deferoxamine. In this study we identified two cell surface receptors that bind ferrioxamine and facilitate iron uptake in Rhizopus oryzae, the most causative fungus of mucormycosis. These receptors are required for full virulence of R. oryzae in mice treated with deferoxamine. From genetic and biochemical studies it appears that the fungus binds ferrioxamine via these two receptors then liberates iron through a chemical modification step prior to transporting into the fungal cell without the internalization of deferoxamine.
Introduction
Mucormycoses, caused by fungi in the order Mucorales, are life threatening infections that afflict immunosuppressed patients due to neutropenia, corticosteroids treatment, hyperglycemia, and trauma [1,2]. These relatively uncommon infections, mainly caused by Rhizopus spp., have been steadily increasing in numbers for the last three decades [3,4]. Despite the current aggressive treatment options against mucormycosis which constitutes reversal of immunosuppressive predisposing factors, surgical removal of infected foci (when possible) and antifungal therapy, overall mortality remains at >50% and approaches 100% for patients with brain involvement, prolonged neutropenia and hematogenously disseminated disease [5,6,7,8]. Clearly, novel strategies to prevent and/or treat the disease are critically needed.
Clinical observations strongly link the ability of organisms to obtain iron from the host as an essential virulence factor of Mucorales [9,10]. For example, hyperglycemia, diabetic ketoacidosis and other forms of acidosis predispose the host to mucormycosis because of compromised ability of transferrin to chelate iron, thereby making iron available to invading organisms [9,11,12]. Similarly, dialysis patients with chronic renal failure who suffer from iron overload due to blood transfusion were historically treated with the bacterial siderophore, deferoxamine [13,14,15]. These patients were uniquely susceptible to lethal form of mucormycosis [13,15,16]. Although deferoxamine is an iron-chelator from the perspective of the human host, Rhizopus spp. utilize ferrioxamine (deferoxamine + Fe3+) as a xenosiderophore to obtain previously unavailable iron [17,18]. It was also reported that Rhizopus obtains iron from ferrioxamine by intracellular transport of the reduced iron without deferoxamine internalization [18]. Recently, we found treatment of Rhizopus-infected mice with the iron chelators deferiprone [19] or deferasirox [20] (which are not utilized as xenosiderophores by Rhizopus) markedly improved survival. These results further confirm the unique importance of iron in the pathogenesis of mucormycosis. In the current study, we sought to identify the fungal cell surface protein that binds to ferrioxamine and its role in the pathogenesis of mucormycosis. We provide evidence that the ferrioxamine binding (Fob) cell surface proteins (namely Fob1 and Fob2) are the fungal receptors that mediate attachment to ferrioxamine, thereby facilitating fungal iron uptake from this xenosiderophore via the reductase/high affinity iron permease pathway. Importantly, Fob1 and Fob2 are inducible proteins that are required for full virulence of R. oryzae only in the deferoxamine-treated mouse model of mucormycosis.
Results
Identification of putative ferrioxamine receptors
To isolate putative ferrioxamine receptor(s), R. oryzae protoplasts were allowed to regenerate in the presence or absence of ferrioxamine, which is the iron-rich form of the bacterial siderophore, deferoxamine [21]. SDS-PAGE analysis of proteins demonstrated the presence of a major band at ~ 70 kDa in cell-free supernatants concentrated from R. oryzae regenerating protoplasts in the presence, but not in the absence, of ferrioxamine. Other minor bands were also detected around 40kDa in supernatants from medium with or without ferrioxamine (Fig 1A). For a protein to act as a receptor it must bind to ferrioxamine. Therefore, we incubated the protein preparations collected from these supernatants with radiolabeled ferrioxamine (deferoxamine+55Fe) prior to running on a non-denaturing PAGE followed by autoradiography. A band from proteins concentrated in the supernatant collected from protoplasts regenerated in the presence of ferrioxamine, but not in the absence of ferrioxamine, bound to radiolabeled ferrioxamine (Fig 1B). Because the 70 kDa band was abundant in the ferrioxamine-containing supernatant and absent in the supernatant concentrated from media lacking ferrioxamine, we sequenced this band by MALDITOF MS/MS. The overwhelming indentified protein was predicted to be encoded by RO3G_11000 (352 amino acid), which is annotated as a CBS-domain-containing protein (http://www.broadinstitute.org/, see Discussion for CBS-domain protein definition) in the R. oryzae 99-880 database (also known as R. delemar) (Table 1). We also found that the ORF RO3G_11000 shares ~80% identity with ORF RO3G_05087 (350 amino acid) at the DNA or protein levels. The RO3G_05087 and RO3G_11000 ORFs are predicted to encode proteins with ~ 39 kDa in size and were named FOB1, and FOB2, respectively. These predicted proteins have 4 putative CBS domains and multiple possible N- and O-glycosylation sites (Fig 1C). Interestingly and despite the presence of Fob2 protein in the supernatant of regenerated protoplasts, we did not observe classic features of secreted proteins such as N-terminal signal peptide or C-terminus GPI-anchor sequence. However, using the MEMSAT program it was predicted that Fob1 and Fob2 proteins have one to two transmembrane domains with an extracellular fragment and a cytoplasmic tail (S1 Fig). Further localization analysis using antibody staining confirmed the surface localization of Fob proteins (see below).
Fig. 1. Detection of the ferrioxamine putative receptor in the cell surface material of R. oryzae. 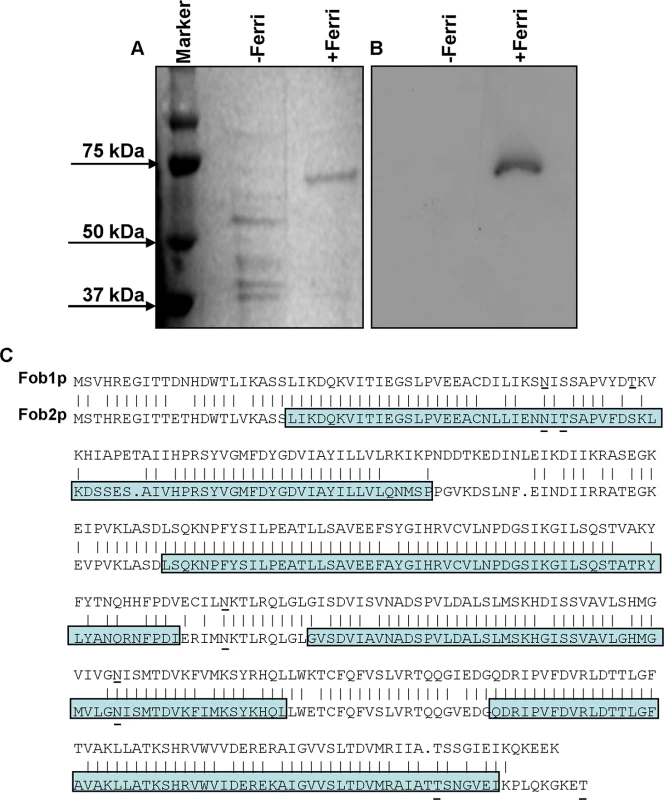
Cell surface material was collected from regenerating R. oryzae protoplasts grown in iron-limited medium with (+Ferri) or without (-Ferri) deferoxamine. Samples were separated on SDS-PAGE and visualized by Coomassie blue (A). Additionally, samples were mixed with [55Fe]ferrioxamine[69] then analyzed by non-denaturing PAGE followed by autoradiography (B). Two putative and closely related ORFs were identified and named FOB1 and FOB2. Highlighted area denotes the four CBS domains while underlined amino acids indicate potential N- and O-glycosylation sites (C). Tab. 1. MALDITOF MS/MS sequence results of the 70 kDa band detected in the supernatant of regenerated protoplasts of R. oryzae grown in the presence of ferrioxamine. 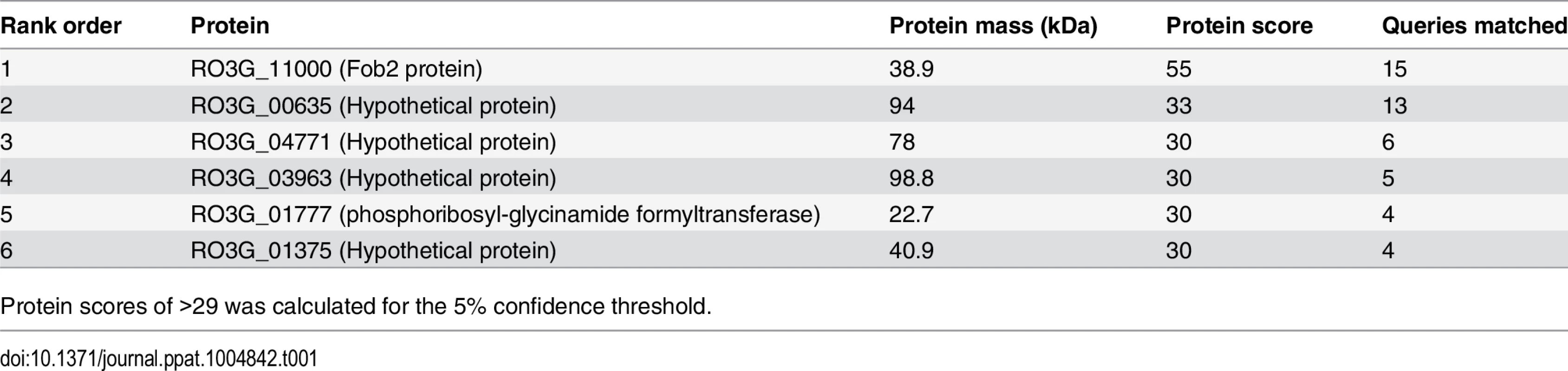
Protein scores of >29 was calculated for the 5% confidence threshold. We also examined whether this family of genes was present in other Mucorales known to cause mucormycosis. Blast search of FOB1 and FOB2 confirmed the presence of orthologs of FOB1/2 in every Mucorales genome published thus far with percent identity at the amino acid levels ranging from 42% for Mortierella verticillata FOB1 ortholog to 78% for Mucor circinelloides f. circinelloides FOB2 (Table 2). Orthologs from other fungi were also found with lesser degree of identity (e.g. ~ 20% for Aspergillus, Saccharomyces cerevisiae, and Candida).
Tab. 2. Sequence homology of FOB1/2 genes from R. oryzae 99-880 to possible Mucorales orthologs. 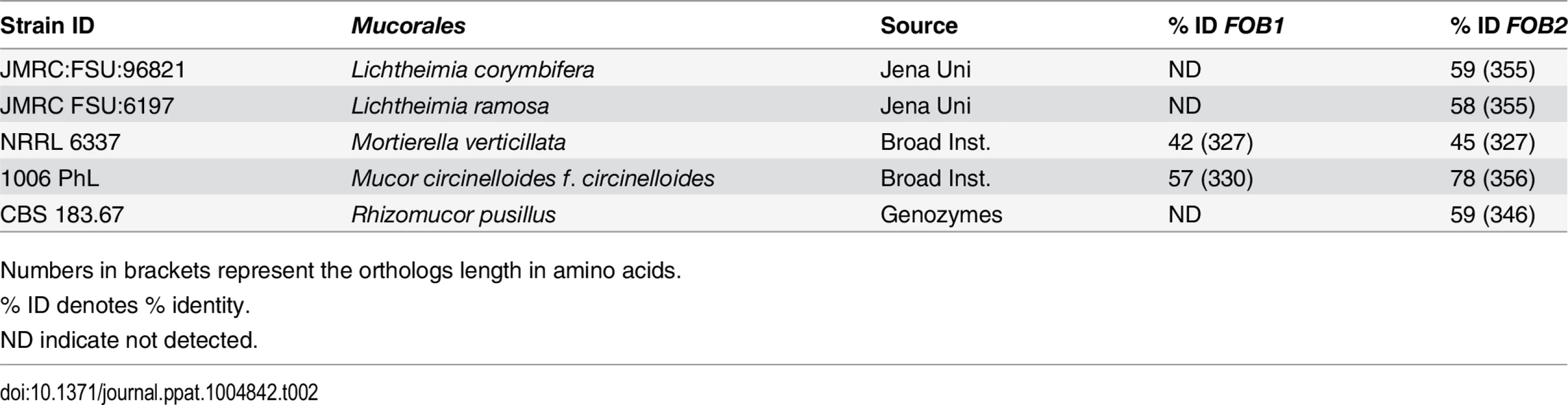
Numbers in brackets represent the orthologs length in amino acids. Putative ferrioxamine receptors are inducible by ferrioxamine, likely to oligomerize, and expressed on the cell surface
Because radiolabeled ferrioxamine bound to a band in protein preparations from supernatants collected from regenerated protoplasts in the presence of ferrioxamine and not in the siderophore’s absence (Fig 1B), we reasoned that the ferrioxamine receptor is likely induced by ferrioxamine. To test if FOB2 and its FOB1 homologue fulfill this criterion, we cultured R. oryzae 99-880 in medium containing ferrioxamine, FeCl3, deferoxamine, rhizoferrin (a siderophore produced by Rhizopus [18,22]), or Fe3+ containing rhizoferrin as the sole source of iron prior to analyzing the expression of FOB1 and FOB2 by qRT-PCR. As can be seen in Fig 2A, only the iron-rich ferrioxamine consistently enhanced the expression of FOB1 and FOB2 by ~ 9 fold vs. any other condition. Neither the iron depleted ferrioxamine (i.e. deferoxamine), nor rhizoferrin with or without iron induced the expression of either FOB1 or FOB2. However, the presence of FeCl3 as a source of iron instead of ferrioxamine resulted in a modest increase in the expression of FOB2.
Fig. 2. FOB1 and FOB2 expression is induced by ferrioxamine and to a lesser extent by ferric iron but not by deferoxamine. 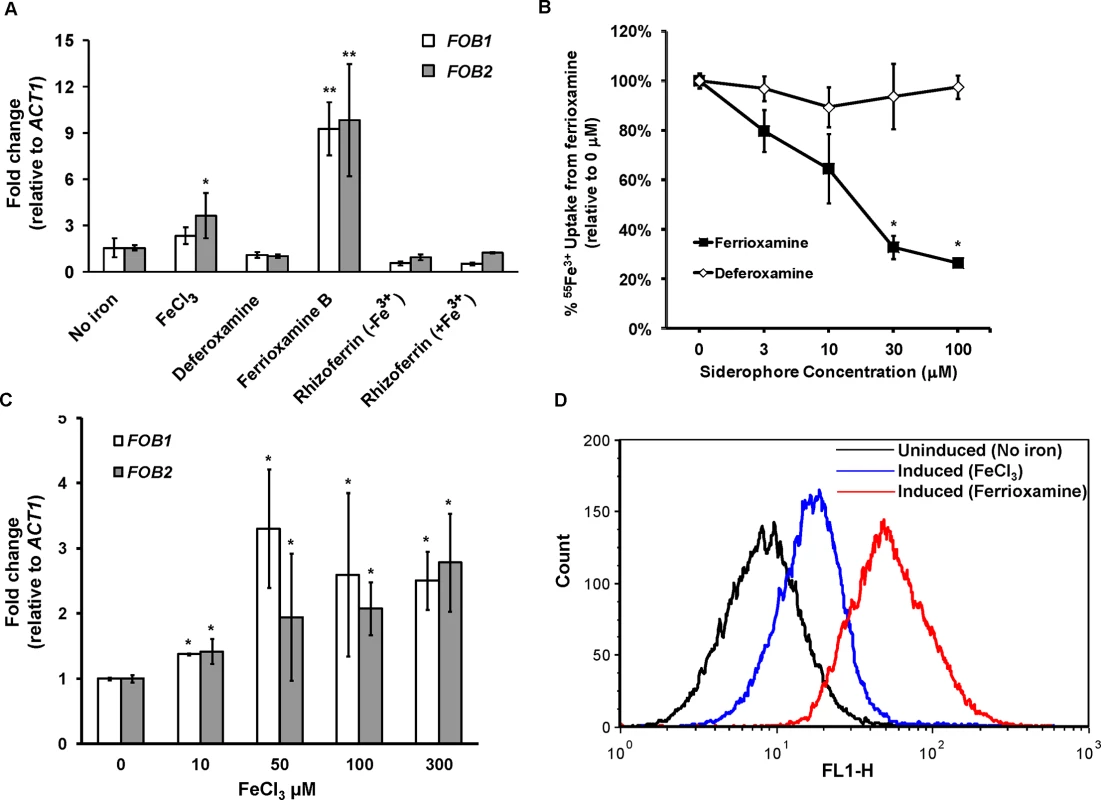
The expression of FOB1 and FOB2 was studied in response to iron sources in vitro. (A) qRT-PCR was used to study the expression of FOB1 and FOB2 in media without added iron, 10 μM of ferrioxamine, deferoxamine, rhizoferrin without Fe3+, rhizoferrin with Fe3+ or 100 μM FeCl3. (B) Cold ferrioxamine but not deferoxamine inhibits 55Fe3+ uptake from ferrioxamine (introduced at a concentration of 10 μM). (C) Compared to ferrioxamine, FeCl3 demonstrated modest and non-linear increase in FOB1 and FOB2 expression as determined by qRT-PCR. (D) Flow cytometry using antiFob2p polyclonal antibodies demonstrated the cell surface expression of Fob1 and Fob2 proteins in medium containing ferrioxamine but not in medium without iron supplement. Error bars represent the standard deviation of the mean from at least two independent assays (n> 6 per group). *P<0.05 vs. no iron, while **P<0.001 vs. no iron or FeCl3. If FOB2 and/or FOB1 indeed encode a ferrioxamine receptor, the lack of their expression by deferoxamine indicated that iron uptake from radiolabeled ferrioxamine should not be inhibited by deferoxamine but rather by cold ferrioxamine. To test this possibility, iron uptake of 55Fe3+ from ferrioxamine by R. oryzae was carried out at increased concentrations of cold ferrioxamine or deferoxamine. Cold ferrioxamine, but not deferoxamine, inhibited 55Fe3+ uptake by >70% when introduced at 3–10 fold higher concentrations than the radiolabeled ferrioxamine (Fig 2B).
Because FeCl3 had a modest effect on the expression of FOB1 and FOB2 when compared to ferrioxamine, while deferoxamine had no effect on their expression, this implicated ferric iron as a critical signal for triggering the expression of these two genes. We studied the expression of FOB1 and FOB2 in response to varying concentrations of FeCl3. As can be seen in Fig 2C, FeCl3, modestly enhanced the expression of both genes in a non-linear manner. Collectively, these results show that the expression of FOB1 and FOB2 is maximal only when the siderophore is iron–rich (i.e. ferrioxamine), the iron-depleted form (i.e. deferoxamine) does not induce nor it inhibits iron uptake from ferrioxamine, and the native siderophore of rhizoferrin does not induce the expression of these two genes.
To study if enhancement in FOB1 and FOB2 gene expression resulted in enhanced protein expression on R. oryzae cell surface, we raised antibodies against an E. coli-produced Fob2 protein (rFob2p) by vaccinating mice and used these polyclonal antibodies to detect the expression of Fob proteins on R. oryzae cell surface by FACS analysis. Sera collected from mice vaccinated with rFob2p + adjuvant had an antibody titer of 1 : 32,000 vs. a titer of 1 : 1600 of sera collected from mice vaccinated with the adjuvant alone. Additionally, sera collected from rFob2p vaccinated mice reacted to the E. coli produced Fob2 protein as well as a protein band isolated from whole cell protein extract of R. oryzae wild-type hyphae only when grown in the presence of ferrioxamine. These two bands were detected at ~40 kDa with slight difference in size which is likely attributed to the presence of His-tag on the rFob2p. Also the antibodies did not detect any bands from whole cell extracts of R. oryzae strain in which both FOB1 and FOB2 were down regulated regardless of the presence or absence of ferrioxamine in the growth medium (see below for FOB1/FOB2 inhibition by RNA-i) (Fig 3A for Western blot and S2A Fig for SDS-PAGE gel picture). Analysis of the recognized bands by nano-LC ESI MS/MS confirmed the identity of the proteins to Fob2p (Table 3). Interestingly, we could not detect the protein in cell-free supernatants collected from media containing R. oryzae wild-type or FOB1/FOB2 inhibition mutant grown in the presence or absence of ferrioxamine (Fig 3B for Western blot, and S2B Fig for SDS-PAGE gel picture). Furthermore, radiolabeled ferrioxamine reacted to crude cell protein extracts or proteins immunoprecipitated by anti-Fob2 antibodies from R. oryzae wild-type whole cell extracts only when grown in a medium containing ferrioxamine (Fig 3C). Finally, the immunoprecipitated band from the whole cell extract of R. oryzae wild-type cells as well as the immunoprecipitated band from rFob2p had sizes of around 40 kDa (Fig 3D for Western blot and S2C Fig for SDS-PAGE gel picture) similar to what noticed in Fig 3A. These two immunoprecipitated bands were confirmed to be Fob2 protein by nano-LC ESI MS/MS (Table 3). Because immunoprecipitation of whole cell extracts of R. oryzae wild-type cells resulted in almost no apparent band on SDS-PAGE gel, we reasoned that the majority of the protein was retained bound to the antibody coated beads. Therefore, we ran the beads on a SDS-PAGE gel after boiling them. Coomassie Brilliant Blue stain showed two major bands at ~40 kDa as well as ~70 kDa (S2D Fig). Because the 70 kDa band corresponded to the size of the identified Fob2 protein from concentrated supernatant of regenerated protoplasts grown in ferrioxamine-containing medium (Fig 1A), we sequenced this band as well as the 40 kDa band by nano-LC ESI MS/MS. The major protein present in these two bands was identified as Fob2p (Table 3). Collectively, these results demonstrated the specificity of the antibodies to Fob proteins (the antibodies are likely to recognize Fob1 and Fob2 proteins due to the predicted similar size and the 80% identity), strongly indicate that Fob proteins are cell-associated rather than secreted, and likely Fob2 proteins oligomerize under certain conditions to form larger protein size.
Fig. 3. Western blot analysis demonstrating that Fob proteins are cell-associated, not secreted, and bind to radiolabeled ferrioxamine. 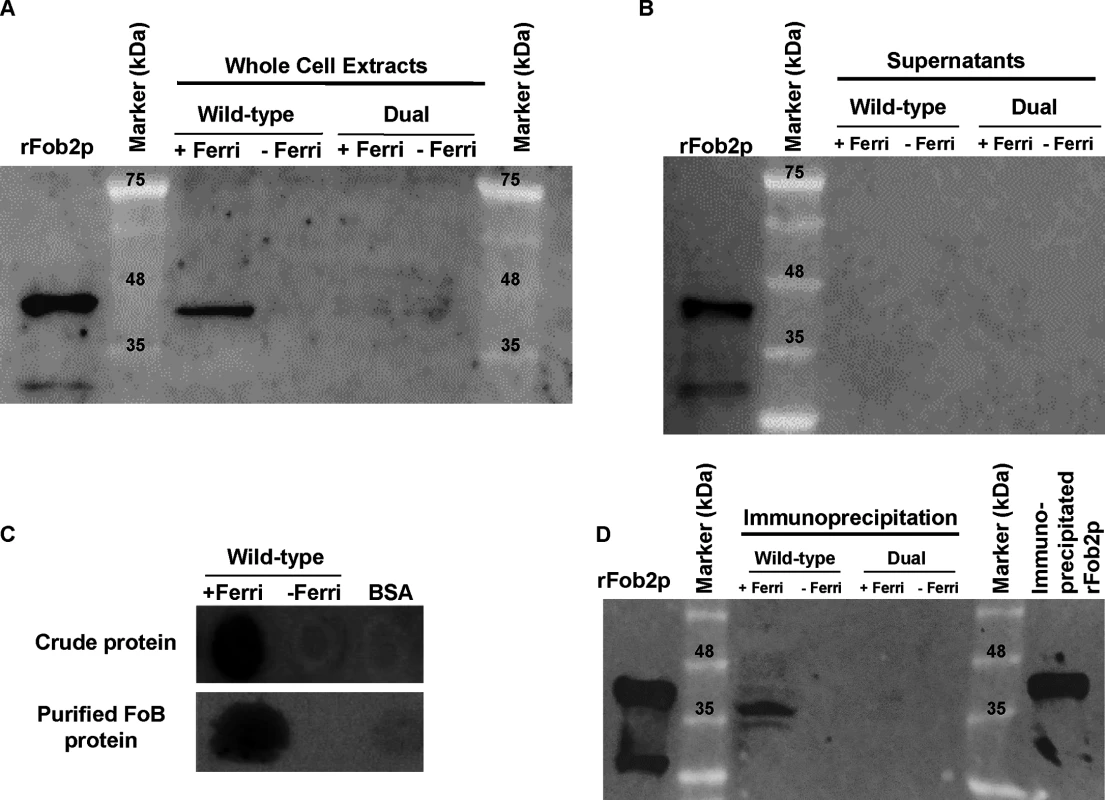
Antibodies were raised against E. coli-produced Fob2 protein and used to study the expression pattern of Fob proteins. (A) Western blot analysis of whole cell proteins extracted from R. oryzae wild-type or R. oryzae mutant with dual inhibition of FOB1/ FOB2 by RNA-i (see Fig 6 for generation of this mutant). (B) Western blot analysis of supernatant (cell-free) collected from R. oryzae wild-type hyphae or R. oryzae mutant with dual inhibition of FOB1/ FOB2 growing for overnight in media with or without ferrioxamine. (C) Whole cell proteins from R. oryzae wild-type grown in medium with or without ferrioxamine were loaded on membrane under non-denaturing conditions prior to mixing with [55Fe]ferrioxamine[69] then analyzed with autoradiography. Also, Fob proteins from the same crude cell wall extracts were separated by immunoprecipitation using anti-IgG Fob2 antibodies coupled to protein G beads column prior to incubating with [55Fe]ferrioxamine under non-denaturing conditions. The beads were washed extensively with washing buffer prior to spotting them on membrane followed by autoradiography. (D) Western blot analysis of whole cell proteins extracted from R. oryzae wild-type or R. oryzae mutant with dual inhibition of FOB1/ FOB2 following immunoprecipitation using anti-IgG Fob2 antibodies as in C. rFob2p was included in A, B, and D as a control showing the expected band size of ~40 kDa (bands in A, B and D below 35 kDa from rFob2p lane represent degradation of rFob2p. Note, that the marker is overlaid on each of the western blots to determine band sizes. Tab. 3. nano-LC ESI MS/MS sequence data of 40 or 70 kDa bands from whole cell extracts of R. oryzae wild-type before and after immunoprecipitation. 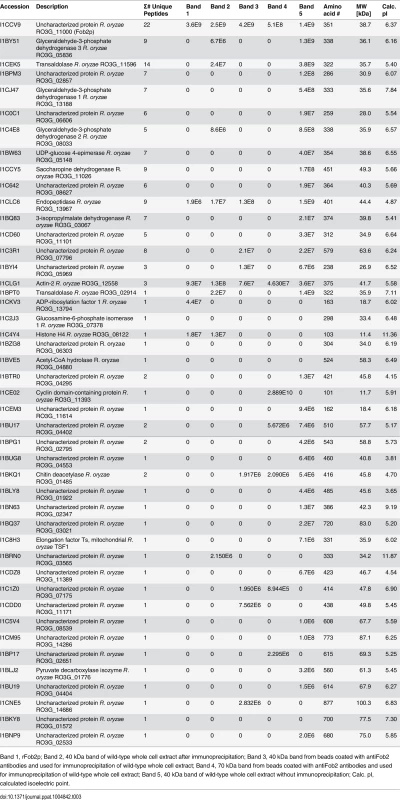
Band 1, rFob2p; Band 2, 40 kDa band of wild-type whole cell extract after immunoprecipitation; Band 3, 40 kDa band from beads coated with antiFob2 antibodies and used for immunoprecipitation of wild-type whole cell extract; Band 4, 70 kDa band from beads coated with antiFob2 antibodies and used for immunoprecipitation of wild-type whole cell extract; Band 5, 40 kDa band of wild-type whole cell extract without immunoprecipitation; Calc. pI, calculated isoelectric point. To discern if the proteins are expressed on the cell surface or intracellularly, we used the anti-Fob2p antibodies to stain R. oryzae germlings without permeabilization and studied the staining patterns by flow cytometry and confocal imaging. We noticed enhanced fluorescence when R. oryzae wild-type cells were incubated with ferrioxamine and to a lesser extent when FeCl3 was used as the sole source of iron compared to fungal cells grown in the absence of iron source (Fig 2D). Confocal images using these antibodies also confirmed the cell surface localization of Fob proteins when the fungal germlings were incubated with ferrioxamine (S3 Fig). Collectively, these results show that Fob proteins are expressed on the cell surface of germinated R. oryzae when ferrioxamine is present.
FOB1 or FOB2 gene silencing by RNA-i modestly reduces iron uptake and slows growth of R. oryzae on medium supplemented with ferrioxamine
To determine the role of these putative receptors in the uptake of iron from ferrioxamine, we down regulated the expression of either of FOB1 or FOB2 (Fig 4A). As expected RNA-i constructs targeting either of the 2 ORFs resulted in ~90% inhibition of the expression of the intended target (i.e., FOB1 or FOB2). However, there was no inhibition in the expression of the other gene (Fig 4B and 4C). In contrast, gene silencing of one FOB appeared to induce the expression of the other gene (e.g., a plasmid targeting FOB1 blocked expression of FOB1 expression, while FOB2 expression increased compared to cells transformed with empty plasmid and grown in the presence of ferrioxamine) (Fig 4B and 4C). These findings were further confirmed by immunostaining and FACS analysis. Cell surface expression of R. oryzae transformed with RNA-i constructs targeting either FOB1 or FOB2 had reduced levels compared to R. oryzae cells transformed with empty plasmid and grown in the presence of ferrioxamine. However, this reduction in Fob protein expression did not reach the low levels of R. oryzae transformed with the empty plasmid and grown in the absence of ferrioxamine (Fig 4D). These results indicate that targeting one FOB gene results in a gene expression compensatory effect of the other.
Fig. 4. RNA-i targeting a single putative ferrioxamine receptor leads to compensatory effect with the other receptor. 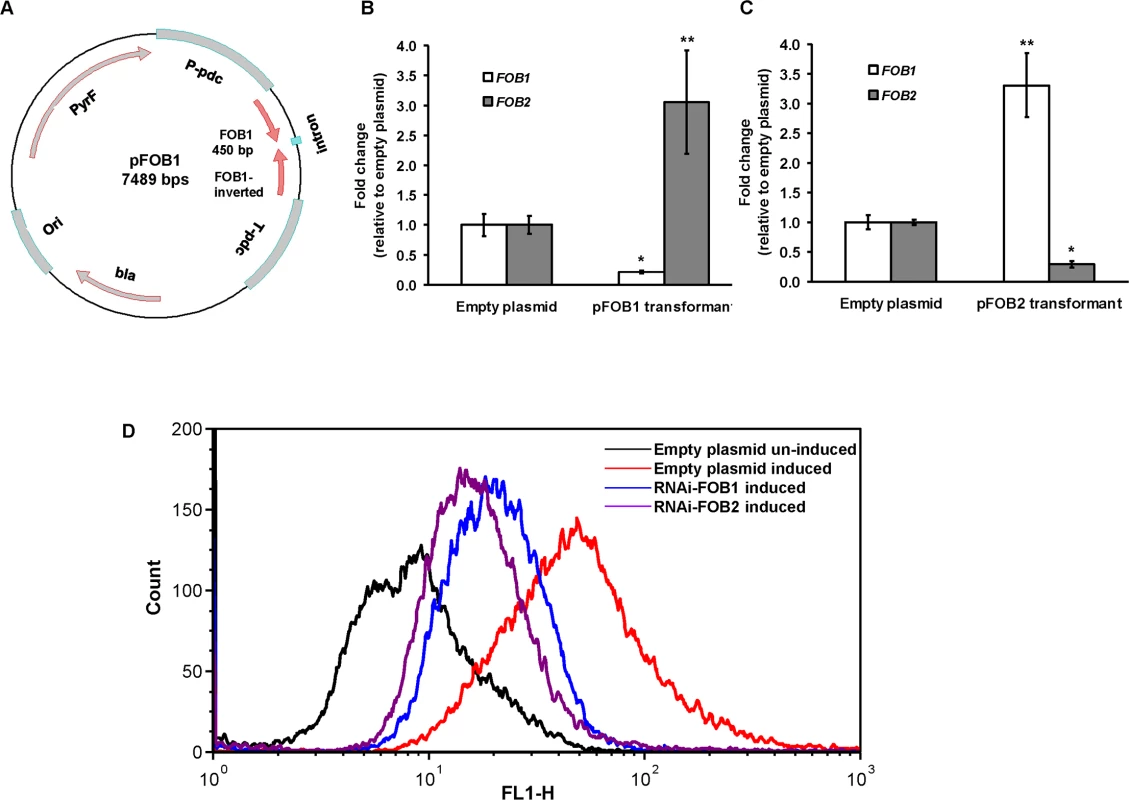
Plasmid pFOB1 (or pFOB2) targeting inhibition of a single putative ferrioxamine receptor (A). Plasmid pFOB1 (B) or pFOB2 (C) successfully inhibited the expression of FOB1 and FOB2, respectively as determined by qRT-PCR. However, a compensatory gene expression was observed in these transformants from the untargeted gene. Results (n = 6 per group) were expressed as fold change in gene expression relative to gene expression in empty plasmid transformant. Error bars represent the standard deviation of the mean from two independent assays. * and **P<0.05 compared to FOB1 and FOB2 expression in empty plasmid transformant, respectively. Flow cytometry using antiFob2 polyclonal antibodies confirmed cell surface expression and the compensatory effect of the untargeted gene. Induction and un-induction conditions were carried out using YNB with or without 10 μM ferrioxamine (D). Having generated the two R. oryzae strains with reduced expression in FOB1 or FOB2, we compared the FOB1 or FOB2 silencing effect on the germination and growth of R. oryzae in a medium supplemented with ferrioxamine. Silencing FOB1 or FOB2 did not have an effect on the ability of R. oryzae to germinate in a medium supplemented with ferrioxamine (Fig 5A). In contrast, R. oryzae transformed with RNA-i constructs targeting FOB1 or FOB2 gene expression resulted in 20–25% inhibition in growth on medium supplemented with ferrioxamine as a sole source of iron when compared to R. oryzae wild-type or R. oryzae strain transformed with the empty plasmid (Fig 5B). This reduction in growth is likely due to the compromised ability of R. oryzae transformed with RNA-i constructs, targeting FOB1 or FOB2, to take up iron from ferrioxamine since attenuation of either FOB1 or FOB2 reduced the ability of R. oryzae to take up iron from radiolabeled ferrioxamine (Fig 5C). Although initial incubation periods of 1–2 hrs showed 60–80% reduction in 55Fe3+ uptake from ferrioxamine among R. oryzae transformed with either RNA-i constructs targeting FOB1 or FOB2 compared to wild-type or R. oryzae transformed with empty plasmid, only 45% inhibition was detected after 4 hrs. This modest reduction in iron uptake is likely due to the compensatory expression of the other FOB gene.
Fig. 5. Single gene inhibition of FOB1 or FOB2 slightly reduced germination, growth and iron uptake of R. oryzae when ferrioxamine is used as a sole source of iron. 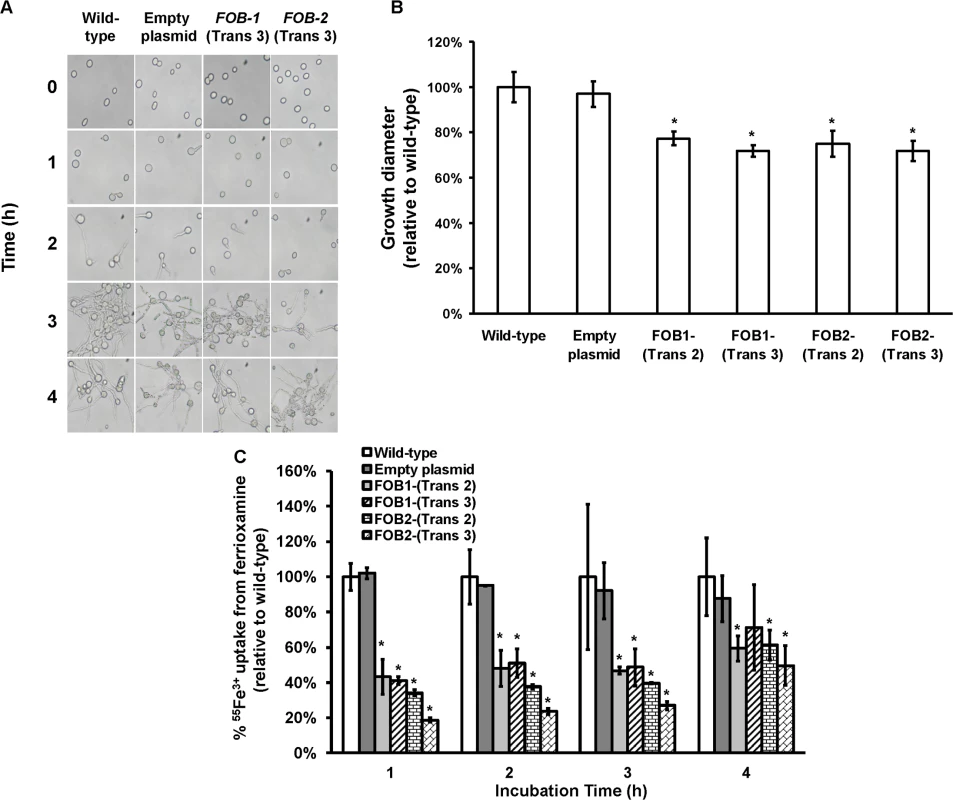
(A) To determine effect of gene inhibition on R. oryzae germination, cells were pregerminated in YNB+CSM-URA medium supplemented with 10 μM ferrioxamine as a sole source of iron. At selected time intervals samples were taken from the medium and examined by light microscopy. (B) To determine effect of gene inhibition on growth of R. oryzae, 105 spores of each strain were plated in the middle of the YNB+CSM-URA agar plates supplemented with 10 μM ferrioxamine as a sole source of iron and the colony diameter (mm) was measured. Two independent transformants from each construct were tested in triplicate (n = 9). *P <0.05 vs. wild-type and empty plasmid strains. (C) Single gene inhibition attenuated 55Fe3+ from ferrioxamine. Two independent transformants from each construct were tested in triplicate (n = 9). *P <0.05 vs. Wild-type and empty plasmid strains. In B and C, error bars represent the standard deviation of the mean from three independent assays. Gene silencing of both FOB1 and FOB2 affects germination and drastically reduces growth and iron uptake from ferrioxamine
Because gene silencing of one of the two FOB genes resulted in a modest effect on growth and iron uptake from ferrioxamine, we employed a strategy of silencing both genes in a single construct driven by a single pdc promoter (i.e., pFOB1/2) (Fig 6A). Upon transforming R. oryzae with this construct we were able to reduce the expression of FOB1 and FOB2 by >90% (Fig 6B). Additionally, anti-Fob2p antibodies could not detect any bands from whole cell protein extract of R. oryzae transformed with dual inhibition construct and grown in the presence or absence of ferrioxamine (Fig 3A). Moreover, the inhibition of FOB1/FOB2 translated to near complete blocking of cell surface expression to the level of R. oryzae transformed with the empty plasmid and grown in the absence of ferrioxamine (Figs 6C and S3). Furthermore, the dual inhibition of FOB1 and FOB2 resulted in severe retardation of R. oryzae germination (Fig 7A) and more than 40–50% inhibition in growth on medium supplemented with ferrioxamine (Fig 7B) when compared to wild-type strain or R. oryzae transformed with empty plasmid. Finally, this retardation in germination and growth on ferrioxamine supplemented medium was accompanied by up to 85% inhibition of iron uptake from ferrioxamine (Fig 7C). Notably, there was no difference in growth of any of the three strains on FeCl3 supplemented YNB medium (S4 Fig). Collectively, these results clearly implicate Fob1 and Fob2 proteins in the uptake of iron from ferrioxamine.
Fig. 6. Dual gene inhibition strategy completely abrogates FOB1 and FOB2 expression. 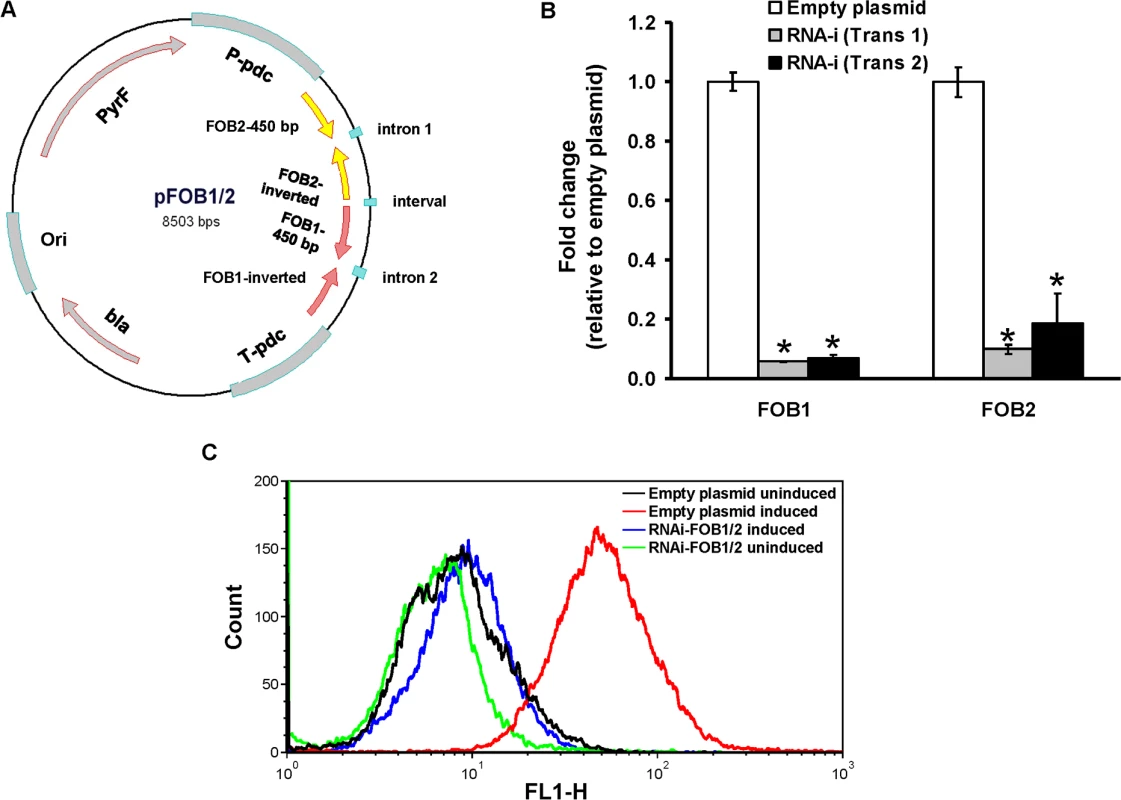
Plasmid pFOB1/FOB2 targeting inhibition of the dual putative ferrioxamine receptors (A). Plasmid pFOB1/FOB2 almost completely blocked FOB1 and FOB2, respectively as determined by qRT-PCR (B). Results were expressed as fold change in gene expression relative to empty plasmid transformant. Error bars represent the standard deviation of the mean from two independent assays. *P<0.005 compared to empty plasmid transformant. Flow cytometry using antiFob2 polyclonal antibodies confirmed the lack of expression of either Fob1 or Fob2 proteins on the cell surface of R. oryzae transformed with pFOB1/FOB2 relative to cells transformed with empty plasmid (C). Induction and un-induction conditions were carried out using YNB with or without 10 μM ferrioxamine. Fig. 7. Dual gene inhibition significantly attenuated the ability of R. oryzae to germinate, grow and take up iron from ferrioxamine. 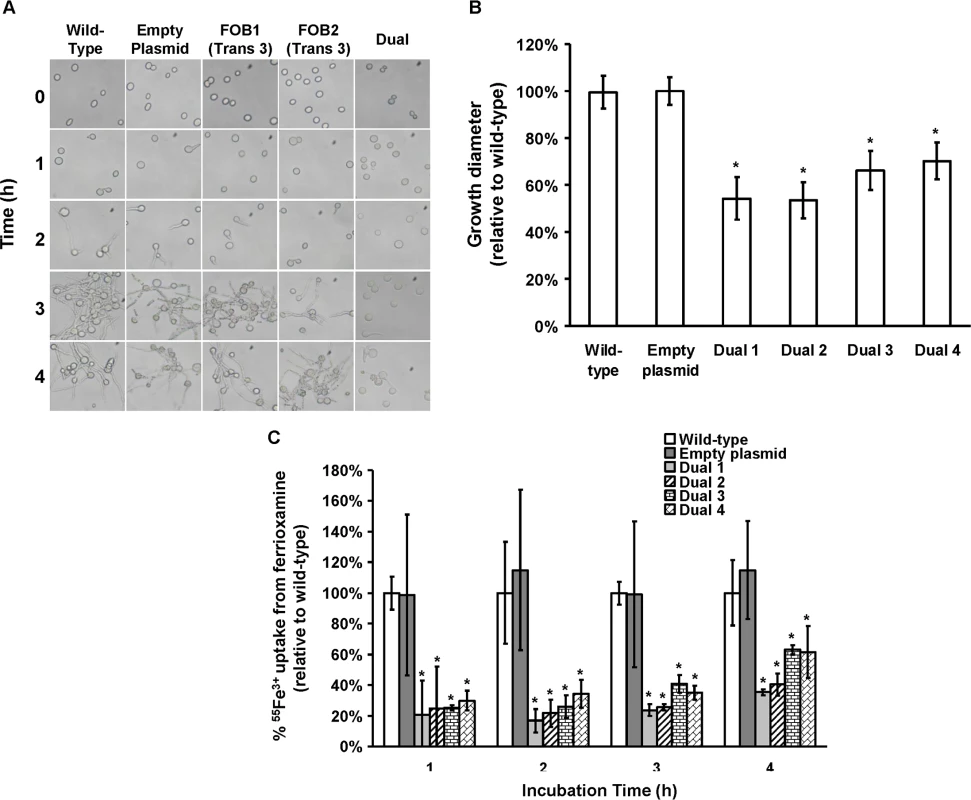
(A) To determine effect of dual gene inhibition on R. oryzae germination, cells were pregerminated in YNB+CSM-URA medium supplemented with 10 μM ferrioxamine as a sole source of iron. At selected time intervals samples were taken from the medium and examined by light microscopy. (B) To determine effect of dual gene inhibition on growth of R. oryzae, 105 spores of each strain were plated in the middle of the YNB+CSM-URA agar plates supplemented with 10 μM ferrioxamine as a sole source of iron and the colony diameter (mm) was measured. Four independent transformants were tested in triplicate (n = 9). *P <0.01 vs. wild-type and empty plasmid strains. (C) Dual gene inhibition attenuated 55Fe3+ from ferrioxamine. Four independent transformants were tested in triplicate (n = 9). *P <0.05 vs. wild-type or empty plasmid strains. In B and C, error bars represent the standard deviation of the mean from three independent assays. FOB1 and FOB2 are required for full virulence of R. oryzae in the deferoxamine-treated but not the diabetic ketoacidotic mouse models
Because FOB1 and FOB2 are involved in iron uptake from ferrioxamine and since iron acquisition from the host is a critical virulence factor for mucormycosis [9,10,23], we hypothesized that these two genes are critical determinants of virulence especially in a mouse model of deferoxamine treatment. To test this hypothesis, we compared the virulence of R. oryzae with reduced cell surface expression of Fob1 and Fob2 proteins to wild-type or to R. oryzae transformed with empty plasmid using a deferoxamine-treated mouse model infected with a high inocula of 105 spores or a lower dose of 103 spores. At the higher inoculum, R. oryzae cells harboring the empty plasmid was as virulent as wild-type R. oryzae (median survival time of 3 days of the wild-type and the empty plasmid strains infected mice P = 0.33). In contrast, mice infected with one of two independently generated dual RNA-i transformants had attenuated virulence with > 21 day median survival time and 1/2 of the mice surviving the lethal infection (P<0.001). Further, at the lower inoculum of 103 spores, the two RNA-i transformants were avirulent with 100% of the mice surviving the infection and appeared healthy at the termination of the experiment, while mice infected with R. oryzae transformed with the empty plasmid had 80% mortality as early as 5 days post infection (P<0.001) (Fig 8A). In support of these data, mice infected with the RNA-i transformant had ~ 2 log reduction in fungal burden in the brains and kidneys (primary and secondary target organs) when compared to same organs recovered from mice infected with wild-type cells or those infected with the empty plasmid transformant (P <0.0001) (Fig 8B). To compare the severity of infection, we conducted histopathological examination on mice organs infected with R. oryzae transformed with the empty plasmid vs. those infected with R. oryzae transformed with RNA-i plasmid targeting both FOB1 and FOB2. Brains and kidneys harvested from mice infected with R. oryzae transformed with RNA-i construct had minimal inflammation, edema and minimal to no presence of fungal elements. In contrast, organs taken from mice infected with R. oryzae transformed with the empty plasmid had abundance of fungal abscesses characterized by phagocyte infiltration and substantial edema (Fig 8C).
Fig. 8. FOB1 and FOB2 are required for the full virulence of R. oryzae in the deferoxamine-treated and not the DKA mouse models. 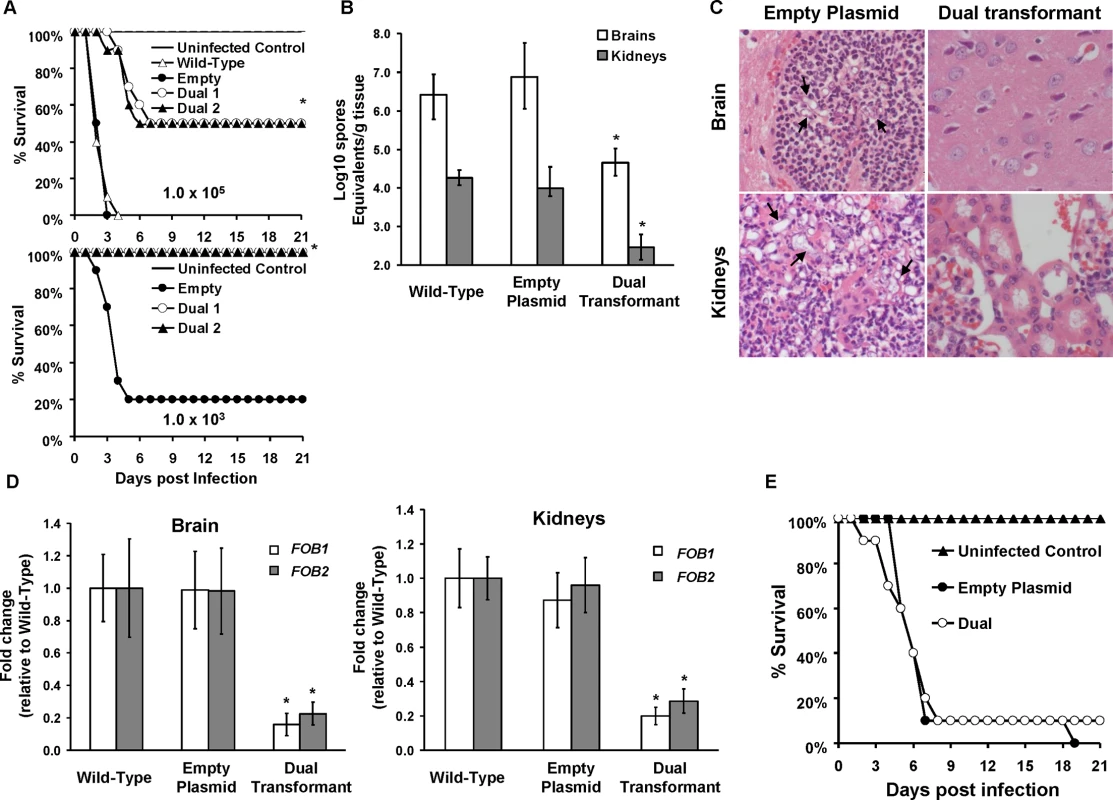
(A) Survival of mice (n = 10 per arm) treated with three doses of deferoxamine (100 mg/kg/dose) via i.p. injection on day -1, 0, and +1 relative to intravenous infection with R. oryzae strains. For the targeted inoculum of 1 x 105, mice were infected with wild-type (6.5 x 104 spores), R. oryzae transformed with empty plasmid (8.6 x 104 spores) or R. oryzae with dual attenuated expression of FOB1 and FOB2 (Dual 1 at 4.6 x 104 spores or Dual 2 at 3.6 x 104 spores). For the targeted inoculum of 1 x 103, mice were infected with R. oryzae transformed with the empty plasmid (9.5 x 102 spores) or R. oryzae with dual attenuated expression of FOB1 and FOB2 (Dual 1 at 7.9 x 102 spores or Dual 2 at 4.9 x 102 spores). *P<0.001 compared to mice infected with wild-type or R. oryzae transformed with empty plasmid. (B) Tissue fungal burden (determined by qPCR) of target organs (brains and kidneys) harvested from mice (n = 10 per arm) infected with wild-type (7 x 104 spores), R. oryzae transformed with empty plasmid (5.7 x 104 spores) or R. oryzae with dual attenuated expression of FOB1 and FOB2 (4.8 x 104 spores). Data are expressed as median ± interquartile range. y axis value represents the lower limit of detection in the assay. *P<0.05 compared to tissues harvested from mice infected with wild-type or empty plasmid transformant. (C) Histopathological examination (H&E stain) of target organs revealed minimal inflammation and fungal elements in brains and kidneys of mice infected with RNA-i transformant, while organs collected from the empty plasmid transformant infected mice had abscesses with abundant hyphae (arrows) and neutrophil infiltration. Magnification of 400 x. (D) In vivo expression of FOB1 and FOB2 genes in brain and kidneys harvested from mice infected with wild-type, empty plasmid or RNA-i dual transformant as determined by qRT-PCR using specific primers to each of the FOB genes. Data are expressed as mean ± SD. * P<0.001 vs. wild-type or empty plasmid. Error bars represent the standard deviation of the mean from 10 mice per arm. (E) DKA mice (n = 10 per arm) were infected with R. oryzae transformed with empty plasmid (2.8 x 103 spores) or R. oryzae with dual attenuated expression of FOB1 and FOB2 (2.4 x 103 spores). To confirm that the RNA-i construct inhibited FOB1 and FOB2, we assessed the pattern of in vivo expression of these genes in fungi recovered from the same mouse organs that were processed for tissue fungal burden. Relative to the fungal actin house keeping gene (ACT1), FOB1 and FOB2 were expressed in organs collected from mice infected with the wild-type, or the empty plasmid transformant. Importantly, fungal cells recovered from brain and kidneys of mice infected with R. oryzae transformed with the RNA-i construct had ~80% reduction in FOB1 or FOB2 gene expression (Fig 8D). These results indicate that FOB1 and FOB2 are expressed in vivo and the reduced virulence in mice infected with R. oryzae transformed with the RNA-i is due to attenuation of expression of both FOB genes. Finally, R. oryzae transformed with the dual construct of pFOB1/FOB2 had identical virulence to R. oryzae transformed with the empty plasmid in the diabetic ketoacidotic mice (Fig 8E). Collectively, these data show that FOB1 and FOB2 are required for maximal virulence of R. oryzae only in the deferoxamine-treated mice.
Iron uptake from ferrioxamine is dependent on the reductase/permease pathway
Having established the importance of FOB1 and FOB2 in the uptake of iron from ferrioxamine in vivo, we wanted to investigate the mechanism by which R. oryzae obtains iron from ferrioxamine. Since one of the most common pathways of obtaining iron in fungi and R. oryzae is the reductase/permease pathway [9], we sought to investigate if this pathway plays a role in iron uptake from ferrioxamine. R. oryzae wild-type cells growing in medium containing ferrioxamine as a sole source of iron demonstrated ~ a 3 fold increase in reductase activity when compared to cells growing on medium containing iron in the form of FeCl3 (Fig 9A). Importantly, the extracellular, membrane-impermeable ferrous chelator bathophenanthroline disulfonate (BPS) inhibited growth of R. oryzae when ferrioxamine is used as a sole source of iron (Fig 9B). This inhibition was due to chelating ferrous since increased concentrations of FeCl3 reversed the inhibitory effect of BPS (Fig 9C) and the addition of BPS blocked 55Fe3+ uptake from ferrioxamine (Fig 9D). Similar results were obtained with the membrane-permeable ferrous chelator bipyridyl (S5 Fig). Since ferrioxamine chelates ferric iron, this inhibition implies the reduction of the ferric iron to ferrous prior to transportation to the fungal cell. Previously, we have shown that targeted gene disruption of FTR1 in the multinucleated R. oryzae results in a mutant with reduced copies of the high affinity iron permease (FTR1) rather than a completely ftr1 null mutant [24]. This mutant with reduced copies of FTR1 had a retarded growth on medium containing ferrioxamine B as a sole source of iron which implies a role for Ftr1p in iron transport from ferrioxamine [24]. To determine if Ftr1p is required to transport the released ferrous to the fungal cell wall, we compared the ability of this R. oryzae strain with reduced FTR1 copy number to wild-type or PyrF-complemented strain in their ability to take up 55Fe3+ from ferrioxamine. After 6 hours of incubation with radiolabeled ferrioxamine, R. oryzae with reduced FTR1 copy number (i.e., putative ftr1 mutant) had significant decrease in taking up 55Fe3+ compared to wild-type cells (P<0.01). Moreover, after 8 hours of incubation with ferrioxamine, R. oryzae with reduced FTR1 copy number had significant reduction in 55Fe3+ uptake when compared to wild-type or PyrF-complemented strains (P<0.005) (Fig 10A).
Fig. 9. R. oryzae iron uptake from ferrioxamine is mediated by reductase activity. 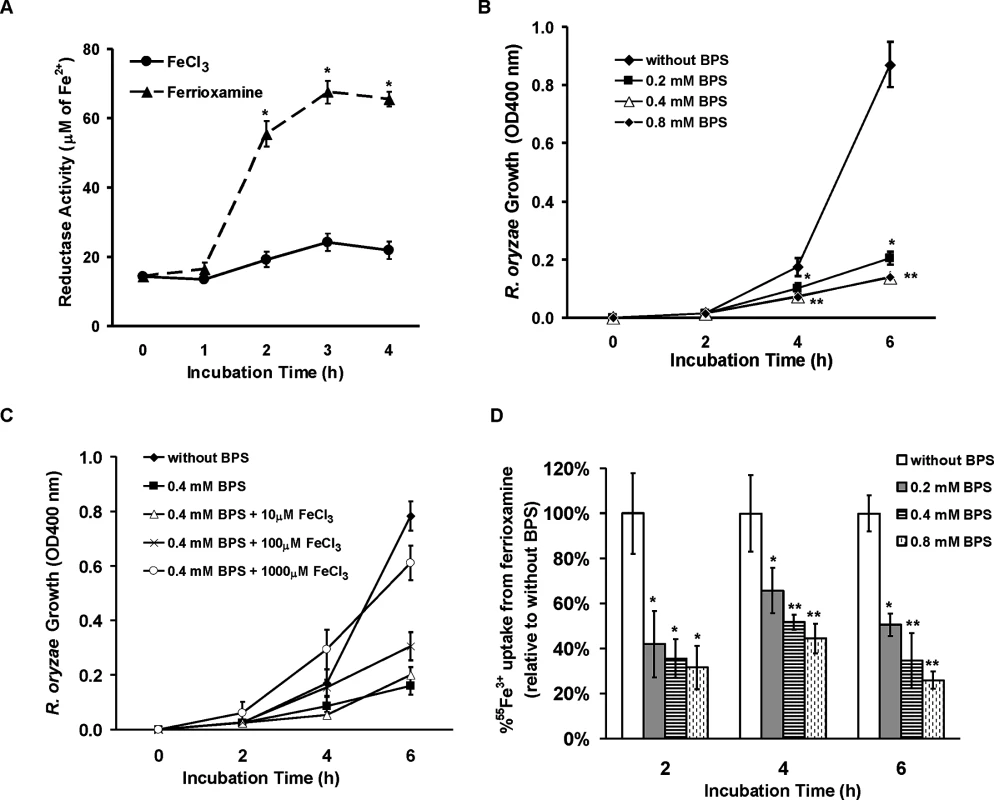
(A) Iron uptake from ferrioxamine is associated with increased reductase activity as compared to iron uptake from FeCl3 (n = 4 per each time point). *P<0.03 vs. reductase activity of FeCl3 at the corresponding time point. (B) The ferrous chelator BPS inhibits growth of R. oryzae on medium supplemented with ferrioxamine as a sole source of iron (n = 6 per group and per time point). *P <0.006 vs. without BPS, while **P <0.006 vs. without BPS or 0.2 mM BPS. (C) FeCl3 reverses growth inhibition mediated by BPS (n = 4 per group and per time point). (D) BPS inhibits 55Fe3+ uptake from ferrioxamine (n = 8 per group and per time point). * P<0.04 vs. without BPS and **P <0.003 vs. without BPS or with 0.2 mM BPS. Error bars represent the standard deviation of the mean from two independent assays. Fig. 10. Reduction of FTR1 expression decreases the ability of R. oryzae to take up iron from ferrioxamine in vitro and attenuates R. oryzae virulence in the deferoxamine-treated mice. 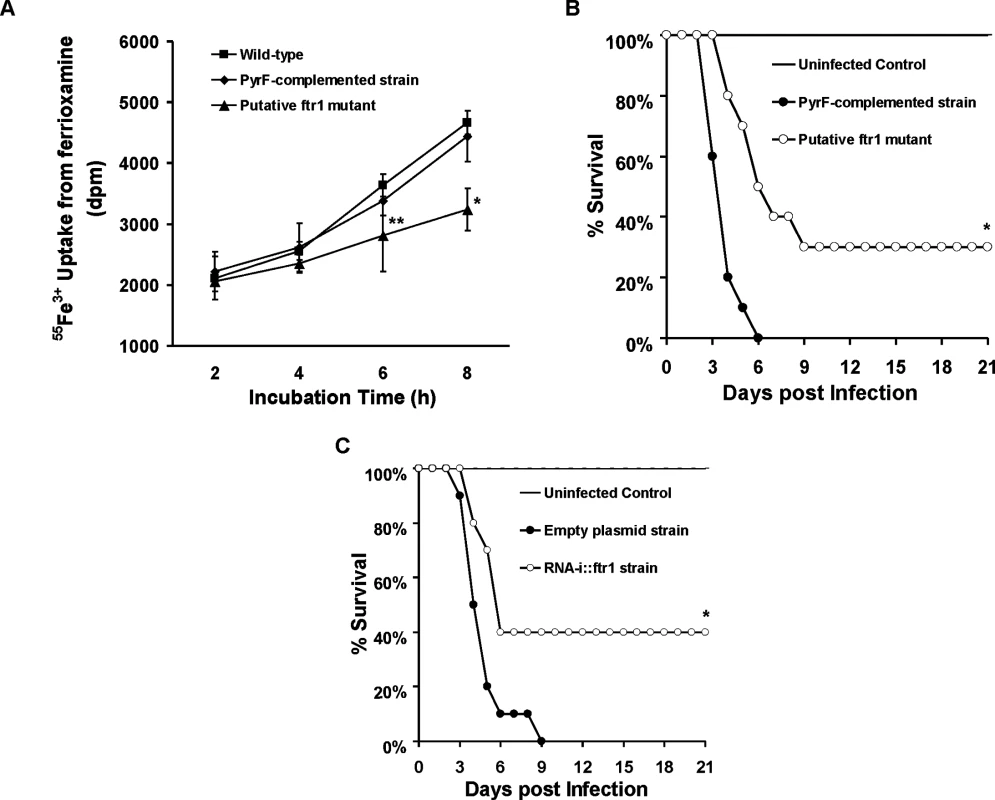
(A) 55Fe3+ uptake from ferrioxamine by wild-type, PyrF-complemented, or putative ftr1 strains (a strain with reduced copies of FTR1 [24], n = 6 per arm from two independent experiments). *P<0.006 vs. wild-type or PyrF-complemented strains. **P<0.01 vs. wild-type strain. (B) Survival of deferoxamine-treated mice (n = 10 per arm) infected with R. oryzae PyrF-complemented strain (9.4 x 102 spores), or R. oryzae putative ftr1 null (6.5 x 102). *P = 0.001 compared to mice infected with R. oryzae PyrF-complemented strain. (C) Survival of deferoxamine-treated mice (n = 10 per arm) infected with R. oryzae transformed with the empty plasmid (9.1 x 102 spores), or R. oryzae transformed with RNA-i construct targeting FTR1 (RNAi::ftr1) (9.5 x 102) [24]. *P = 0.013 compared to mice infected R. oryzae transformed with empty plasmid. To confirm these in vitro findings, we compared the virulence of two R. oryzae with attenuated expression of FTR1 to their corresponding control strains in the deferoxamine-treated mouse model. R. oryzae strain with reduced FTR1 copy number (Fig 10B) and R. oryzae transformed with RNA-i construct targeting FTR1 expression [24] (Fig 10C) had attenuated virulence in the deferoxamine-treated mouse model when compared to PyrF-complemented strain or R. oryzae transformed with the RNA-i empty plasmid, respectively. Specifically, at day 21 post infection, 30% of mice infected with reduced FTR1 copy number strain survived the infection vs. 0% for mice infected with the PyrF-complemented strain (P = 0.001) and 40% of mice infected with R. oryzae transformed with RNA-i construct targeting FTR1 survived the infection vs. 0% survival for mice infected with R. oryzae transformed with the empty plasmid (P = 0.013). Collectively, these results clearly show that iron transport from ferrioxamine relies, at least in part, on the reductase/permease system.
Potential role of other putative transporters in iron-uptake from ferrioxamine
Because the ftr1 mutants were not completely avirulent in deferoxamine-treated mice, we investigated the possibility of the involvement of other mechanisms in transporting iron from ferrioxamine. All identified eukaryotic siderophore transporters belong to the siderophore–iron transporters (SIT) family, a subfamily of the major facilitator superfamily [25,26,27,28]. We have conducted homology searches of the S. cerevisiae SIT genes [25,29,30], including ARN1, ARN2, ARN3/SIT1, and ARN4 with the published R. oryzae genome data base. Although the overall amino acid identity was low (i.e. 20–23%), we identified 9 ORFs that share homology with the S. cerevisiae SIT encoding genes and therefore potentially encode Sit proteins in R. oryzae (S1 Table). We compared the pattern of expression of four of these genes (SIT1, 4, 6, and 9) with the highest homology to S. cerevisiae SIT genes to FOB1, FOB2, and FTR1 over time in the presence of deferoxamine or ferrioxamine as iron-poor and iron-rich media, respectively. In iron-rich ferrioxamine medium, FOB2 expression was induced as early as 30 min and reached a maximum expression of 9 fold increase by 24 hrs. Interestingly, FOB1 expression was significantly increased only after 24 h incubation (Fig 11A). This enhancement of FOB gene expression was accompanied by gradual reduction in iron starvation genes (i.e. FTR1 and SIT) that reached >99% decrease after 24 h (FTR1 and SIT9 had almost complete reduction in expression after 30 min of incubation) when compared to 10 min data set (Fig 11B). In contrast and as expected, none of the FOB gene expression was enhanced in the presence of deferoxamine (Fig 11C). Finally, FTR1 and putative SIT gene level of expression did not considerably change in deferoxamine containing medium over time (level of expression fluctuated between 0.3–1.4 fold relative to ACT1) (Fig 11D). These results confirm the induction of FOB genes in response to ferrioxamine and that this induction changes the conditions of the fungal cell from iron-deplete to iron-replete conditions as shown by the reduction in the iron-starvation gene expression. Further, the similar pattern of SIT genes expression to the FTR1 expression in ferrioxamine containing medium suggests that these genes might play a role in iron uptake from this siderophore.
Fig. 11. FOB genes are induced in ferrioxamine containing medium, while iron transporter genes such as FTR1 and SIT-like genes are transiently expressed and tightly regulated. 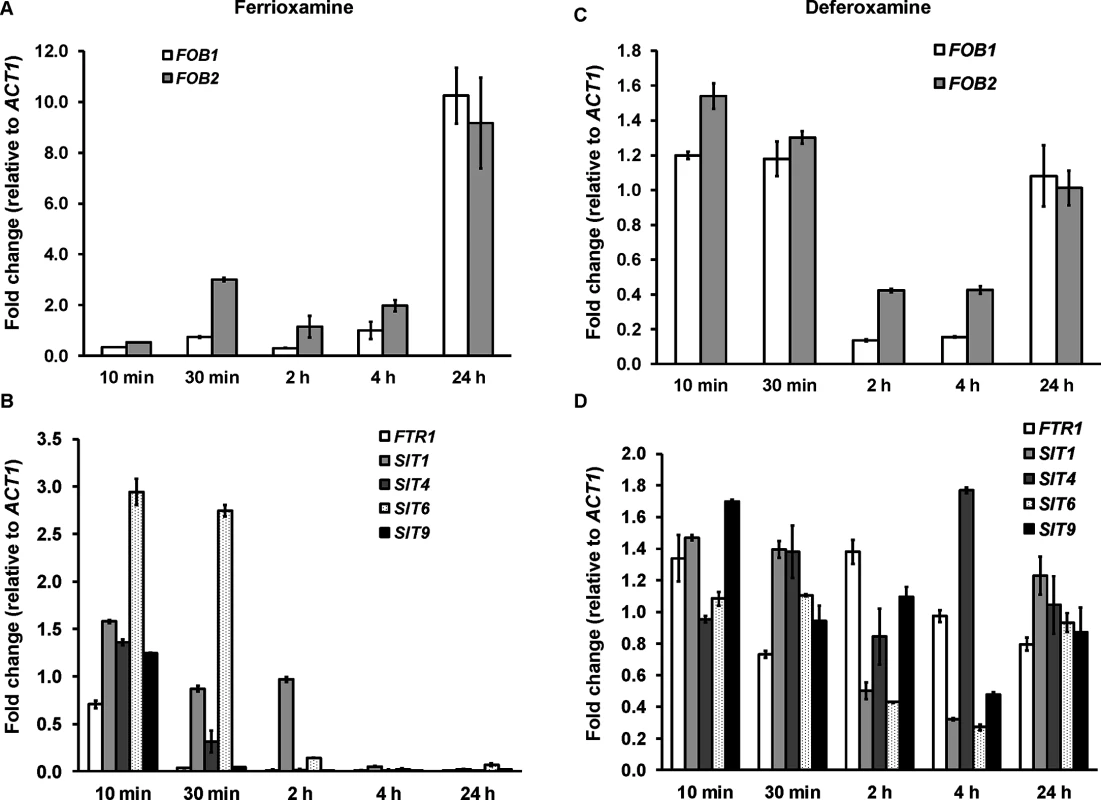
qRT-PCR was used to study the expression of FOB1, FOB2, FTR1, or SIT-like genes in media supplemented with 10 μM ferrioxamine (A, B) or deferoxamine (C, D). Only ferrioxamine strongly induced the expression of FOB genes (A). Iron starvation genes (FTR1 and SIT-like genes) were transiently expressed in ferrioxamine-containing medium over time (B) and expressed with minimal variation in deferoxamine-containing medium (D). Error bars represent the standard deviation of the mean from two independent assays (n = 6 per group). Discussion
The unique predisposition of dialysis patients receiving deferoxamine for treating iron overload to mucormycosis has been clinically known for decades [13,15,16]. Previous data demonstrated that Rhizopus species can utilize iron from ferrioxamine as a xenosiderophore to obtain iron from the host [17,31,32]. This process was found to be energy dependent and possibly doesn’t require internalization of the siderophore [18]. In this study, we have identified two ORFs that encode Fob1 and Fob2 proteins which are required for iron uptake from ferrioxamine. We introduce evidence that these proteins are receptors to which ferrioxamine binds to during the process of obtaining iron by R. oryzae. First, Fob proteins are mainly induced by ferrioxamine and detected in the supernatants of regenerating R. oryzae protoplasts before they are covalently incorporated into the nascent cell surface [33]. Second, these two proteins were found to be expressed on the cell surface of R. oryzae, a criterion required for cell receptors. Third, whole cell extracts from R. oryzae grown in the presence, but not the absence, of ferrioxamine bound to radiolabeled ferrioxamine under non-denaturing conditions. Fourth, Fob2 protein isolated from whole cell extracts using anti-Fob2 antibodies also bound to radiolabeled ferrioxamine. Finally, a mutant of R. oryzae with attenuated FOB1 and FOB2 expression bound much less, and had retardation in, iron uptake from radiolabeled ferrioxamine versus the wild-type strain. Collectively, these features indicate that Fob1 and Fob2 proteins are ferrioxamine induced, surface exposed, and directly interact with the siderophore.
The identified Fob proteins were found to belong to the CBS domain family. CBS domain is a conserved protein sequence region, which is found in all kingdoms of life [34]. It was first described in Cystathionine β Synthase, hence the name CBS. Other proteins belonging to this family include inosine monophosphate dehydrogenase, AMP kinase, and chloride channels. Therefore, CBS-containing proteins functions are very diverse and could range from affecting metabolism, multimerization and sorting of proteins, channel gating, to ligand binding. Based on their function, CBS domain proteins can be present in the cytoplasm or associated with the cell membrane [34]. For example, despite the lack of classical features of secreted proteins such as N-terminal signal peptide sequence, CBS domain proteins were reported to be associated with cell membrane in eukaryotic cells functioning as CLC chloride channels with outer membrane sequence, transmembrane domain, and a cytoplasmic tail [35]. Concordant with this report is our finding that Fob1p and Fob2p are predicted to be surface associated by using the MEMSAT program (S1 Fig). In most cases, however, CBS-domain proteins bind to ligands related to adenosine, including S-adenosylmethionine (SAM), AMP, ADP [36], as well as recently reported DNA and RNA fragments [37,38]. They also have been reported to bind to metallic ions such as Mg2+ [39]. To our knowledge, we show for the first time that a ferrioxamine receptor belongs to the CBS domain family of proteins. Given the diverse function of these proteins and the previous report of binding metallic ions, it is possible that Fob1 and Fob2 proteins on R. oryzae cell surface bind ferrioxamine via Fe3+. This assumption is supported by our results that only ferrioxamine, but not deferoxamine, induces the expression of FOB1 and FOB2 and by the fact that deferoxamine was not able to competitively inhibit the uptake of radiolabeled ferrioxamine by R. oryzae Importantly, ferric-rich siderophores are known to induce a conformational change of the secondary structure of their receptors and likely explain the ability of ferrioxamine and inability of deferoxamine to induce and bind to Fob proteins [40,41].
An apparent discrepancy in our results is the actual size of the Fob2p of ~40 kDa as indicated by the predicted sequence, the size of the rFob2p and recognized band from whole cell extracts vs. the originally isolated 70 kDa band. We initially assumed that the presence of multiple possible N - and O-glycosylation sites might account for this increase in size of the protein due to post-translational modification. However, the isolation of Fob proteins from whole R. oryzae extracts at the predicted size of 40 kDa argues against a post-translational modification of the protein. A more likely explanation is the possible oligomerization of the protein when the sample is present in concentrated form mixed with SDS. First, the 70 kDa band was detected in concentrated supernatants from regenerating R. oryzae protoplasts in SDS-PAGE. Second, when we immunoprecipitated whole cell extracts from wild-type cells, the majority of the sample remained bound to the antiFob2p antibody-coated beads and that sample contained Fob proteins at the ~40 and ~70 kDa bands when separated on SDS-PAGE. It is prudent to mention that cell membrane proteins are notorious for oligomerization even in denaturing conditions when mixed with SDS [42,43]. In fact, a study found that SDS enhances the dimerization of β-amyloid proteins from human cortical tissues and the level of this unnatural dimerization increases with the increased concentration of SDS in the sample [44]. Alternatively, CBS domain proteins were reported to oligomerize (e.g. Streptococcus pyogenes IMPDH protein [34]), therefore it is possible that Fob proteins are present as homo - or heterodimers. Our sequence data which identified Fob2p in two independent samples detected at ~70 kDa without the identification of Fob1p suggest that Fob2p is the major form expressed. However, it is possible that Fob1p is expressed in low quantities and is required for the receptor activity by forming a heterodimer with Fob1p. Investigations into the possibility of homodimer vs. heterodimer formation of Fob2p and Fob1p or the possibility of induction of unnatural oligomerization during sample processing are currently underway.
Deferoxamine is a siderophore belonging to the hydroxamate family that is mainly isolated from bacteria including Gram-positives (e.g., Streptomyces and Nocardia) as well as Gram negatives Enterobacteriaceae [45,46,47]. Up to now, deferoxamine has not been isolated from fungi but many studies indicated the utilization of this siderophore by fungi in obtaining iron including Mucorales, Aspergillus, Cryptococcus [17] and S. cerevisiae [25,27,48]. In addition to utilizing deferoxamine as xenosiderophores, Mucorales are known for secreting their own siderophore, rhizoferrin [22,49]. However, rhizoferrin belongs to the carboxylate family which is structurally distinct from the hydroxamate siderophores [22,50]. Interestingly, rhizoferrin (with or without iron) did not induce the expression of FOB1 or FOB2 indicating that these genes encode receptors specific to iron uptake from ferrioxamine. However, the ability of Mucorales to utilize iron from structurally distinct siderophores points to the critical role iron plays in the survival of this group of fungi.
Our in vitro data with down regulation of a single FOB gene demonstrated a modest effect on germination, growth and iron uptake from medium supplemented with ferrioxamine. These results can be explained by the fact that the untargeted gene was overexpressed and had a compensatory effect. This compensatory mechanism by related genes is well documented when other genes of the family are nullified [51]. However, this compensatory gene expression effect did not translate into full restoration of cell surface protein to the level of R. oryzae transformed with empty plasmid and grown in ferrioxamine containing medium (Fig 4D). The reduction of fluorescence in a single inhibition strain could be due to the possibility that the enhancement of non-targeted gene expression did not rise to the level that would entirely compensate for inhibition of the other gene. It is also possible that the gene expression of the non-targeted gene does not localize entirely to the cell surface.
Our strategy of targeting both genes with a single construct successfully drastically attenuated the expression of FOB1 and FOB2, resulting in a significant retardation in germination, growth and iron uptake from medium supplemented with ferrioxamine as a sole source of iron. However, the inhibition of growth was not complete. Further, the inhibition of iron uptake from ferrioxamine by R. oryzae with abrogated FOB1/FOB2 expression was marginally reduced at later time points (Fig 7C, compare 4 h with earlier time points). These two phenomena can be explained by the fact that old or heat-damaged ferrioxamine (due to incubation at 37oC) is unstable and undergoes an autoreduction step to release ferrous iron [52], that is taken up by the fungus. This assumption is corroborated by the yielding of a pink-colored complex in the presence of ferrozine in our experiment. It is also probable that secondary mechanisms of obtaining iron from ferrioxamine which are independent from FOB1/FOB2 are operative. Our results with pattern of expression of SIT-like genes in medium supplemented with ferrioxamine or deferoxamine over time supports the model of SIT-like genes being involved in iron uptake from ferrioxamine. Specifically and similar to FTR1 gene, SIT-like genes were transiently expressed as early as 10 min then the expression faded over time which indicates change of the conditions of the fungal cell from iron-deplete to iron-replete conditions. This transient expression and tight regulation is critical for the organism to avoid the toxic carnage of excess iron [53,54].
Importantly, a R. oryzae mutant with dual attenuation of FOB1/FOB2 expression had severe virulence retardation in the deferoxamine-treated mice vs. mice infected with wild-type or empty plasmid strains with 50% of the mice infected with R. oryzae transformed with RNA-i targeting FOB1/FOB2 surviving the infection and looking healthy. It is imperative to point out that this difference in virulence was not detected in the DKA mouse model which is consistent with the fact that Fob1 and Fob2 are proteins induced by ferrioxamine. Fob1 and Fob2 are likely dispensable in the DKA model since Fe3+ is released from transferrin by the action of acidosis and hyperglycemia [9,11] and transported into the fungal cell via the Ftr1p [24].
In microorganisms, the mechanism of iron uptake from ferrioxamine involves either the uptake of the entire siderophore complex, reduction of the Fe3+ to Fe2+ to release it from the siderophore followed by uptake of iron via oxidase/permease system, or both. For example, in S. cerevisiae it has been reported that both mechanisms are operative [30,55]. A previous study demonstrated that iron uptake from ferrioxamine by R. oryzae is likely energy dependent, require a reductive step and doesn’t involve the uptake of the ferrioxamine complex [18]. In contrast, by using a fluorescent derivative of deferoxamine, a recent study showed that the entire siderophore was taken up by the R. oryzae cells [56]. Our results mainly support the model by which R. oryzae obtains iron from ferrioxamine via the reductase/permease system. First, the reductase activity of R. oryzae when incubated with ferrioxamine far exceeds the reductase activity when fungal cells were incubated with FeCl3. Second, the addition of the ferrous chelator BPS inhibited growth of R. oryzae in medium with ferrioxamine as a sole source of iron and this inhibition was due to attenuation of iron uptake from ferrioxamine. Finally, we previously reported that a R. oryzae mutant with reduced copy number of the high affinity iron permease (FTR1) had reduced growth on medium supplemented with ferrioxamine [24]. In this study we show that this growth inhibition is likely due to the diminished ability of this mutant to take iron from ferrioxamine. More significantly, we demonstrated reduced virulence in the deferoxamine-treated mouse model of mucormycosis, even though the R. oryzae mutant with reduced FTR1 copy number or inhibited expression of FTR1 do not have full abrogation of the function of the permease activity [24]. However, a minor mechanism reliant on internalization of the entire siderophore cannot be completely ruled out because the effect of BPS in vitro and the attenuated virulence with ftr1 mutants were not complete. It is possible that the reductase/permease pathway represent a major uptake pathway for iron acquisition from ferrioxamine, while the uptake of the siderophore by shuttle mechanism[56] represents a secondary mechanism. This secondary mechanism of iron uptake from ferrioxamine is supported by the pattern of transient and tightly regulated SIT-like gene expression in ferrioxamine containing medium (Fig 11). Also the assumption that ferrioxamine is taken by a Sit-like shuttle transporter is a secondary mechanism is enforced by the fact that the patchy distribution of fluorescent ferrioxamine was observed only at the later time point of 24 hours of incubation and not earlier [56]. Consequently, we propose that R. oryzae overexpresses Fob1 and Fob2 in response to ferrioxamine, which accumulates in a host being treated with deferoxamine to eliminate iron overload toxicity. These receptors bind ferrioxamine and through fungal cell surface reductase activity, Fe3+ is released from the siderophore as Fe2+ prior to being transported across the fungal cell membrane by the oxidase/permease complex (i.e., Fet3/Ftr1) to support fungal growth and virulence (Fig 12A). In the absence of Fob1/Fob2 or the Ftr1p, this process is compromised and the fungal virulence is reduced (Fig 12B). Also in this model, transport of the entire ferrioxamine siderophore by Sit-like transporter is likely to be operative as a secondary pathway.
Fig. 12. Proposed mechanism of iron uptake by R. oryzae from ferrioxamine. 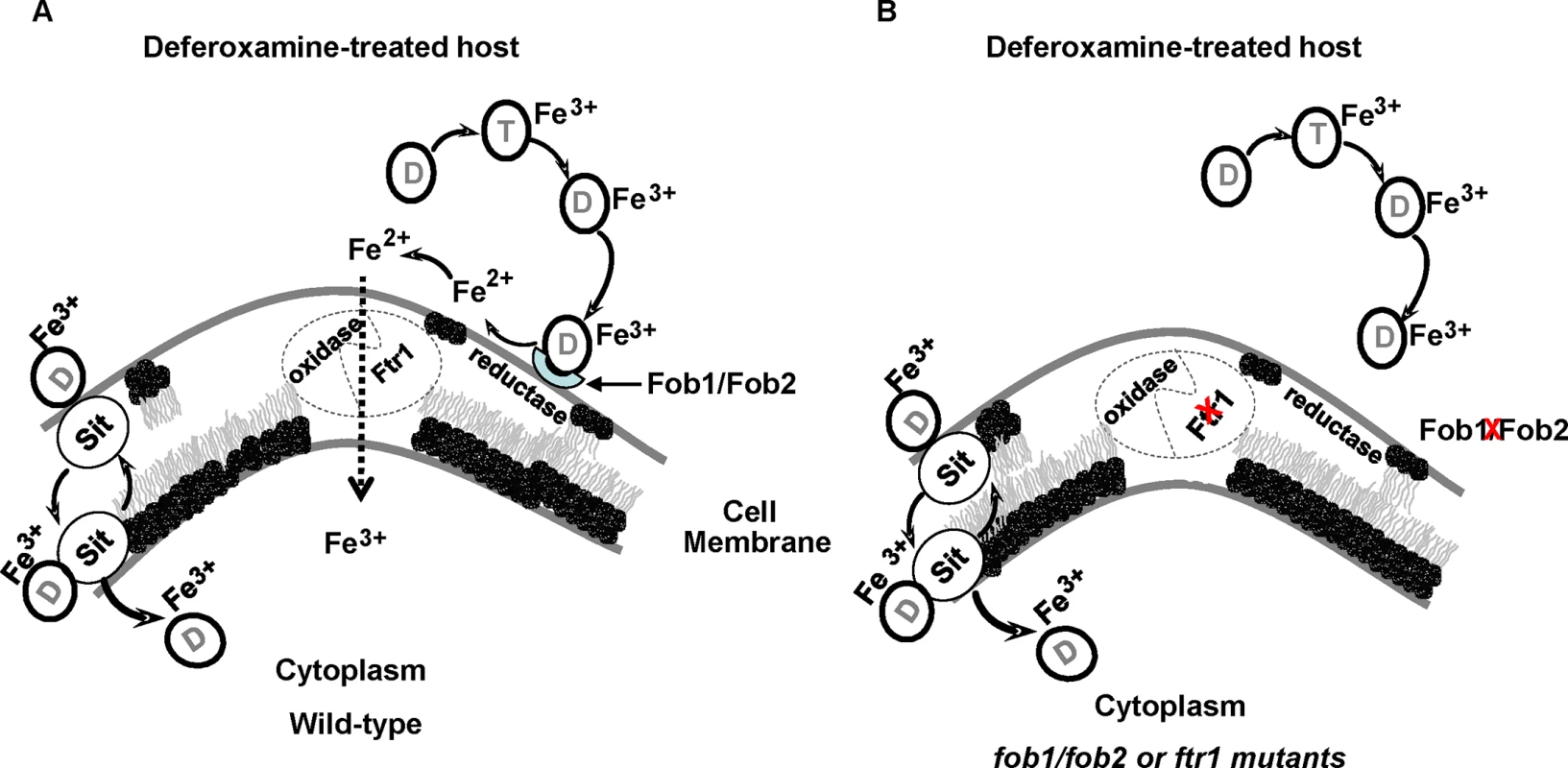
Deferoxamine (D) directly chelates iron from transferrin (T), resulting in ferrioxamine which binds to Fob1 and/or Fob2 proteins on the cell surface. R. oryzae then liberates Fe2+ iron by the actions of cell surface reductases then transported into the cell by oxidase/Ftr1 complex (A). In the absence of Fob1/Fob2 or Ftr1 proteins, the ability of R. oryzae to obtain iron from ferrioxamine is compromised and the virulence is abrogated in deferoxamine-treated mouse model (B). Sit-mediated shuttle mechanism of ferrioxamine transportation probably acts as a secondary mechanism of iron uptake from ferrioxamine. In summary, we have identified Fob1 and Fob2 proteins as ferrioxamine receptors in R. oryzae. These cell surface proteins are induced by ferrioxamine and are required for mucormycosis pathogenesis in the deferoxamine-treated mouse model. The mechanism of iron uptake from ferrioxamine is reliant on binding of the iron-rich siderophore to its receptor, followed by transport of iron to the fungal cell via the reductase/permease system without internalization of the siderophore-iron complex. These two receptors appear to be conserved in at least another five Mucorales and can be the subject of future novel therapy to maintain the use of deferoxamine for treating iron-overload.
Materials and Methods
Organisms and culture conditions
All organisms used in this study are listed in S2 Table. R. oryzae 99-880 is a clinical strain isolated from a patient with a brain abscess and obtained from the Fungus Testing Laboratory of
University of Texas Health Science Center at San Antonio. The organism was routinely grown on potato dextrose agar (PDA) plates (BD) for 4-5 days at 37°C. R. oryzae M16 is a pyrF null mutant that is derived from R. oryzae 99-880 and is unable to synthesize its own uracil [57], was grown on YPD medium (MP Biomedicals) supplemented with 100 μg/ml uracil. R. oryzae PyrF-complemented strain, R. oryzae with reduced FTR1 copy number, and R. oryzae transformed with RNA-i targeting FTR1 expression were all derived from strain 99-880 and M16 and previously described in detail [24]. In RNA-i experiments, a chemically-defined synthetic medium containing yeast nitrogen base (YNB) supplemented with complete supplemental mixture without uracil (CSM-URA) (MP Biomedicals) (i.e., YNB+CSM-URA) (formulation/liter, 17 g yeast nitrogen base without amino acids (YNB) (BD), 200 g dextrose, and 7.7 g complete supplemental mixture minus uracil (CSM-URA)) was used. To count and isolate single colonies all media above were supplemented with 0.1% Triton X-100 (Sigma). Spores were collected in endotoxin free PBS containing 0.01% Tween 80, washed twice with PBS, and counted with a hemacytometer for inoculum preparation.
Isolation of ferrioxamine putative receptors
To identify the R. oryzae cell surface protein(s) that interacts with ferrioxamine, freshly harvested R. oryzae 99-880 spores (2.5x108) were added to 50 ml YPD and pregerminated with shaking at 37°C. After 4 hours, the spores were pelleted by centrifugation and washed twice with 0.5 M sorbitol buffer, and suspended in 50 ml protoplasting solution which consists of 10 mM sodium phosphate, pH 6.4, 0.5 M sorbitol, 250 μg/ml lysing enzyme (Sigma), 150 μg/ml chitinase (Sigma) and 150 μg/ml chitosanase (US Biologicals). The suspension was incubated for ~ 2 h at 30°C with gentle shaking (100 rpm) until majority of the spores (> 90%) formed protoplasts as determined by light microscopy examination and their sensitivity to water. Protoplasts were then collected by centrifugation at low speed (100 rpm), washed twice with 0.5 M sorbitol, and then resuspended in 10 ml YNB with amino acids containing 0.5 M sorbitol in the presence or absence of 10 μM ferrioxamine (Sigma). The protoplasts were allowed to regenerate for 2 h by incubating at 37°C with gentle shaking at 100 rpm. During the early stages of this regeneration process, many surface protein precursors are shed into the extracellular medium but not yet covalently incorporated into the nascent cell surface, thereby enabling their easy isolation and identification [33]. Thus, supernatants containing cell wall materials from regenerated protoplasts were isolated from fungal cells by centrifugation at 1000 rpm, followed by filtration through a 0.25 μm pore-size membrane after the addition of Halt Protease Inhibitor Cocktails (Thermo Scientific). The filtrate was then concentrated using iConcentrator tube with molecular weight cutoff = 9.0 kDa (Thermo Scientific) and protein concentration was measured with the Bradford method (Bio-Rad). Proteins from supernatant of regenerated protoplasts in the presence or absence of ferrioxamine were separated on 10% SDS-polyacrylamide gel (Bio-Rad) and stained with Coomassie dye R-250 (Pierce). To investigate if the proteins from supernatant of regenerated protoplasts bind to ferrioxamine B, 25 μg of protein extract was mixed with 55Fe-labeled ferrioxamine (for preparation, see below), and incubated for 1 h at room temperature. The mixture was then mixed with native protein sample buffer and separated by running a native gel (Bio-Rad) in the absence of SDS and exposed to X-ray film. The candidate band from SDS-PAGE was cut and microsequenced using MALDITOF MS/MS (UCLA Molecular Instrumentation Center).
Preparation of radiolabeled ferrioxamine
55Fe3+ labeled ferrioxamine was prepared as previously described [58]. Briefly, 55FeCl3 (3 μl of 3.5 Ci/mmol, Perkin Elmer) was mixed with iron-free deferoxamine B (Sigma) in a molar ratio of 0.9 : 1 in a volume of 100 μl, and incubated for 2 h at room temperature. Free iron was then removed by adding sodium phosphate (pH 7.4) followed by centrifugation at 20,000 g for 2 min. Supernatant was diluted to 3.0 ml with ultra-pure water with a resistivity of 18.2 megohm-cm (Barnstead International, Dubuque, Iowa) and treated with 150 mg Chelex 100 resin (Bio-Rad) twice to get rid of any residual free iron.
FOB1, FOB2, and SIT transporter expression in vitro
To quantify gene expression of putative ferrioxamine receptors and other SIT genes of R. oryzae in vitro, total RNA was isolated from mycelia that have been grown in YNB with amino acids containing 10 μM of siderophore (ferrioxamine, deferoxamine, rhizoferrin with or without ferric iron [EMC Microcollections GmbH])) or varying concentrations of FeCl3 overnight with shaking at 37°C. The RNA was isolated using RNeasy Plant Mini Kit (Qiagen) after grinding the mycelia in liquid nitrogen. cDNA was synthesized from 2 μg of total RNA from each sample after treating with Turbo DNA-free DNase I (Life Technologies) to remove any contaminating DNA. After removing DNase I with DNase inactivation Reagent (Life Technologies), cDNA was synthesized using RETROscript kit (Life technologies). qPCR was carried out using the Power SYBR Green method in the StepOne Real-Time PCR System (Life Technologies) with a thermal-cycling program as follows: initial denaturing step for 10 min at 95°C, followed by 40 cycles of denaturing at 95°C for 15 s, and annealing/elongation at 60°C for 1 min. R. oryzae actin gene (ACT1) was used as a reference control, putative ferrioxamine receptor gene-specific primers are listed in S3 Table. The comparative Ct method was used for analysis [59].
Attenuation of FOB1 and FOB2 expression by RNAi
RNA-i was used to silence FOB1 or FOB2 expression individually or collectively. For individual silencing, two reverse-complemented fragments (~450 bp) were PCR-amplified and cloned into plasmid pRNAi-pdc-intron in an opposite direction and separated by a 100 bp-intron sequence, hence the total insert is ~1 kb [24]. Two constructs, termed pRNAi-FOB1 and pRNAi-FOB2, targeting FOB1 and FOB2 expression were respectively generated. To silence both genes at the same time, the 1 kb insert from pRNAi-FOB1 was cut and re-cloned downstream of pRNAi-FOB2, separated by a 50-bp interval of random sequence to yield pFOB1/2. All constructs were transformed into R. oryzae M16 (pyrF null mutant) using a biolistic delivery system (Bio-Rad) as previously described [60] and transformants were selected on YNB+CSM-URA plates supplemented with 0.1% triton. Primers used for this work are listed in S3 Table.
Growth rate determination
To compare growth rate among R. oryzae isolates on medium with ferrioxamine as a sole source of iron, 105 spores of each strain were plated in the middle of the YNB+CSM-URA agar plates which have been treated with 1 mM of ascorbic acid (Sigma) and 1 mM ferrozine (Sigma) to chelate iron and then supplemented with 10 μM ferrioxamine. The plates were incubated at 37°C and the diameter of the colony was calculated after 24 or 48 h.
To determine the effect of the ferrous chelators BPS or bipyridyl on the growth of R. oryzae in the presence of ferrioxamine, R. oryzae wild-type spores (5x105/ml) were added to YNB+CSM-URA medium supplemented with 10 μM ferrioxamine and one of the two ferrous chelators (0.2–0.8 mM of BPS or 0.2–0.4 mM bipyridyl). In some experiments, FeCl3 was added in increased concentrations (10–1000 μM) to investigate if iron can reverse the effect of the ferrous chelator. Cultures were incubated at 37°C with shaking. At selected time intervals, 1 ml sample was taken from the culture and the optical density measured at 400 nm [17].
Germination studies
Spores from R. oryzae wild-type, transformants transformed with empty plasmid or RNA-i constructs targeting FOB1, FOB2, or FOB1/FOB2 were harvested from their plates as above and inoculated into YNB+CSM-URA medium supplemented with 10 μM ferrioxamine as a sole source of iron at 5 x 106 spores/ml. The cultures were incubated at 37°C with shaking (200 rpm). At selected time periods, a 10 μl sample was collected from each culture, and examined with a phase contrast microscope to determine if there is any effect of FOB gene silencing on germination. Pictures were taken with a Nikon E5400 camera.
Iron uptake
To determine the ability of different strains of R. oryzae to take up iron from ferrioxamine, we used our previously described methods [24]. Briefly, R. oryzae spores (5 x 106/ml) were pre-germinated for 3 h in YNB medium supplemented with 1 mM ferrozine and 1 mM ascorbic acid at 37°C with shaking. Cells were harvested by centrifugation, washed twice with assay buffer (YNB + 10 mM 4-morpholinepropanesulfonic acid + 1 mM ferrozine), and resuspended in 7 ml of assay buffer supplemented with 10 μM of 55Fe-labeled ferrioxamine at a concentration of 5 x 106 spores/ml, and incubated at 37°C with shaking. In some experiments, to determine if iron uptake from ferrioxamine required the reduction of Fe3+ to Fe2+, the ferrous chelator BPS (0.2–0.8 mM) or bipyridyl (0.2–0.4 mM) were added to the assay buffer. At different time points, 1 ml of sample was collected by Whatman GF/C glass filters (GE Healthcare) through a vacuum filtration manifold (Millipore) and washed twice with cold water. Cell associated 55Fe was counted in a liquid scintillation counter (Packard Instrument, Downers Grove, IL).
For competition studies using cold deferoxamine or ferrioxamine, 55Fe3+ uptake studies were conducted as above while including increasing concentrations of cold deferoxamine or ferrioxamine. Iron uptake was determined after 2 h incubation period and the results expressed as % inhibition relative to samples run without the inclusion of cold siderophores.
Reductase activity
To determine the reductase activity of R. oryzae wild-type in the presence of ferrioxamine or FeCl3, we used the method of Dancis et al. [61]. Briefly 5x107 of R. oryzae spores were cultured in 10 ml of YNB+CSM-URA medium supplemented with 100 μM FeCl3 or 10 μM ferrioxamine. The culture was incubated at 37°C with shaking. At time intervals, 1 ml of the culture was collected, centrifuged at 3000 rpm and the spores washed twice with cold distilled water. The spores were suspended in 1 ml of assay buffer (0.05 M sodium citrate (pH 6.5) supplemented with 5% glucose (w/v)). The culture was incubated at 30°C for 15 min before adding BPS and FeCl3 to a final concentration of 1 mM for both compounds. The mixture is vortexed and then incubated for additional 20 min, after which spores were removed by centrifugation and the optical density of the supernatant determined at 535 nm using a blank processed similarly but without any added spores. The amount of generated Fe2+ was estimated with a standard curve constructed from solutions of known Fe2+ concentrations using FeSO4.
Fob2 protein purification and antibody production
To determine the cell surface localization of the putative ferrioxamine receptors, we sought to raise polyclonal antibodies against one of the two proteins (i.e., Fob1p or Fob2p). Since Fob1 and Fob2 predicted proteins are ~ 80% identical at the amino acid level, we chose to express FOB2 (RO3G_11000). To express FOB2, we first PCR-amplified R. oryzae FOB2 gene (S3 Table), the PCR product was gel-purified and digested with BamHI/PstI before cloning into E. coli expression vector pQE32 (Qiagen) to yield pQE32/FOB2. This resulted in a construct with 6xhis tag on the N-terminus of Fob2 protein. Plasmid pQE32/FOB2 was transformed into E. coli XL-10 gold (Agilent Technologies) and transformants isolated on Luria-Bertani (LB) agar plates supplemented with ampicillin (100 μg/ml). Gene cloning was confirmed by DNA sequencing. To purify Fob2 protein, a single colony from the transformed E. coli was grown overnight. On the second day, the culture was diluted 200 times in fresh medium containing ampicillin, and incubated with shaking at 37°C until reaching an OD = 0.6. At this stage, 1 mM IPTG was added to the culture which was then incubated at 37°C with shaking for additional four hours. The bacterial cells were then harvested by centrifugation, and 6xhis tagged Fob2p was purified to >90 homogeneity with HisPur Cobalt Purification Kit according to the instructional manual (Pierce). The purified recombinant protein was extensively dialyzed in phosphate buffered saline (PBS) (Mediatech, VA), and the purity and size confirmed by SDS-PAGE analysis followed by MALDI-TOF–mass spectrometry/mass spectrometry analysis.
To generate anti-Fob2p antibodies, normal BALB/c mice were immunized by subcutaneous injection of 20 μg recombinant Fob2p mixed in equal amounts with complete freund’s adjuvant (CFA, Sigma-Aldrich). The mice were boosted with a subcutaneous administration of a similar dose of the protein mixed with incomplete freund’s adjuvant (IFA) three weeks following the initial immunization. Mice vaccinated with the diluent (i.e., PBS) mixed with CFA/IFA without rFob2p using the same regimen served as control mice. Twelve days after the boost, blood samples were collected from mice, and anti-Fob2p Ab titers were determined by using ELISA plates coated with Fob2p as we previously described [62]. The ELISA titer was taken as the reciprocal of the last serum dilution that gave a positive OD reading (i.e., more than the mean OD of negative control samples plus 2 standard deviations).
Localization of Fob1 and Fob2 to the cell surface of R. oryzae
To prove Fob proteins are R. oryzae cell surface proteins, freshly harvested spores grown on YNB+CSM-URA were pregerminted for 3–4 hours in the presence or absence of 10 μM deferoxamine. The pregerminated spores were incubated with the anti-Fob2p IgG at 1 : 100 for 1 hour on ice after a blocking step using 1.5% goat serum in PBS for 1 hour. Cells were washed 3 times with Tris-buffered saline (TBS; 0.01 M Tris HCl [pH 7.4], 0.15 M NaCl) containing 0.05% Tween 20, then counterstained with alexa 488-conjugated anti-mouse secondary at 1 : 100 for 1 hour on ice. The stained cells were imaged with Leica confocal microscope using an excitation wavelength of 488 nm.
To quantify the expression of Fob proteins on R. oryzae cell surface, the pregerminated spores were stained as above, then 1 ml samples were analyzed using a FACSCalibur (Becton Dickinson) instrument equipped with an argon laser emitting at 488 nm. Fluorescence emission was read with a 515/40 bandpass filter. Fluorescence data were collected with logarithmic amplifiers. The mean fluorescence intensities of 104 events were calculated using CELLQUEST software [20,63].
Whole cell extraction, secreted proteins, western blotting and ferrioxamine binding studies
R. oryzae (5 x 107 cells) were grown overnight in YNB+CSM-URA with or without 10 μM ferrioxamine. Mycelia were collected by filtration, washed briefly with PBS, and then ground thoroughly in liquid nitrogen using pestle and mortar for 3 min. The ground powder was immediately transferred to microfuge tube containing 500 μl extraction buffer which consisted of 50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 10 mM MgCl2. The extraction buffer was supplemented with 1X Halt Protease Inhibitor Cocktails (Thermo Scientific). The sample was vortexed vigorously for 1 min, then centrifuged for 5 min at 21000 g at 4°C. The supernatant was transferred to a new tube and the protein concentration determined using Bradford method.
For detecting protein secretion, supernatants from R. oryzae cells growing in YNB+CSM-URA with or without 10 μM ferrioxamine for overnight, were collected after filtration of the hyphal mate using 0.22 μM membrane units (Millipore). The cell-free supernatants were concentrated by 200 x and the total proteins measured as above.
For Western blotting, 5 μg of each sample was used to separate proteins on an SDS-PAGE. Separated proteins were transferred to PVDF membranes (GE Water & Process Technologies), and treated with Western blocking reagent (Roche) for 1 h at room temperature. PVDF membranes were then probed with mouse anti-Fob2p antibodies (1 : 100 diluted). After washing 3 times with TBS-T buffer (Tris-buffered saline + 0.05% Tween 20), the membranes were incubated with HRP-conjugated sheep anti-mouse IgG (1 : 10,000 dilution) (Sigma-Aldrich). Fob bands were visualized by adding the HRP substrate (SuperSignal West Dura Extended Duration Substrate, Thermo Scientific), and the chemiluminescent signal was detected using a Sony CCD camera. In-gel tryptic digest followed by nano-liquid chromatography–tandem mass spectrometry analysis was used to confirm the identity of the band.
Dot blots were used to determine the direct binding of radiolabeled ferrioxamine to crude protein extracts or Fob proteins purified by immunoprecipitation using anti-Fob2p antibodies. Briefly, anti-Fob2p antibodies (1 : 10 dilution) were coupled to protein G column according to the manufacturer’s recommendation (Thermo Scientific). Protein extracts (100 μl), or bovine serum albumin (BSA) were added to the column at room temperature for 1 h after which the flow through is discarded followed by washing the column 3 times with PBS. 55Fe3+ labeled ferrioxamine was then added to the anti-Fob2 antibodies-coupled protein G column and incubated for another 1 h at room temperature. Next, the flow through is discarded and the resin was washed three times with PBS before blotting beads containing antibody/antigen complex on PVDF membrane followed by exposure to X-ray film.
In vivo virulence studies
The contribution of FOB to R. oryzae virulence was determined using both a deferoxamine-treated and DKA mouse models. In both models, ICR mice (≥20 g) (Taconic, USA) were used. For the deferoxamine-treated mouse model we adapted the method of Abe et al [64]. Briefly, mice were injected i.p. with three doses of deferoxamine (100 mg/kg/per dose) given on days -1, 0, and +1 relative to infection with R. oryzae. For the DKA mouse model, mice were rendered diabetic in slight ketoacidosis with a single i.p. injection of 210 mg/kg streptozotocin in 0.2 ml citrate buffer 10 days prior to fungal challenge as we previously described [20,65,66,67]. Glycosuria and ketonuria were confirmed in all mice 7 days after streptozotocin treatment. In both models, mice were infected with 105 spores by intravenous injection in the tail vein. Additionally, in the deferoxamine-treated model, a lower inoculum of 103 was tested. The primary efficacy endpoint was time to moribundity. In some experiments, as a secondary endpoint, fungal burden in the brains and kidneys (primary and secondary target organs) was determined 48 hours after infection by quantitative PCR assay. Briefly, mouse organs were disrupted/homogenized in Lysing Matrix tubes (MP Biomedicals) using FastPrep FP120 Homogenizer (Thermo Electron Corporation), total DNA was then isolated according to the instructional manual of DNeasy Tissue Kit (Qiagen). qPCR was then carried out as previously described [68], using R. oryzae-specific 18S rRNA primers (S3 Table). Values were expressed as log10 spore equivalent/g tissue. Histopathological examination was carried out on sections of the harvested organs after fixing in 10% zinc formalin. The fixed organs were embedded in paraffin, and 5-mm sections were stained with H&E to detect R. oryzae hyphae [20].
For in vivo expression of FOB genes, brains and kidneys were collected from mice 48 hours after wild-type, empty plasmid–transformed, or RNA-i transformed R. oryzae infection were flash frozen in liquid nitrogen, disrupted/homogenized as above, and then processed for RNA extraction using a Tri Reagent solution (Ambion). Reverse transcription was performed with RETROscript (Ambion) using primers listed in S3 Table. For quantitative RT-PCR, SYBR green assays were performed. Constitutively expressed ACT1 was used as a control for all reactions. Calculations and statistical analyses were performed using StepOne Real-Time PCR System (Applied Biosystems).
Also, the contribution of the reductase/permease pathway to the pathogenesis of R. oryzae in the deferoxamine-treated mouse model was determined by comparing the virulence of R. oryzae with reduced copy number of FTR1 or with reduced expression of FTR1 due to RNA-i targeting FTR1 [24], to their corresponding control strains of PyrF-complemented or R. oryzae transformed with the empty plasmid, respectively. Infection was carried out as above with a targeted inoculum of 1 x 103 spores. The primary efficacy endpoint was time to moribundity.
Ethics statement
All procedures involving mice were approved by the IACUC of the Los Angeles Biomedical Research Institute at Harbor-UCLA Medical Center (protocol # 11671-08), according to the NIH guidelines for animal housing and care.
Statistical analysis
Differences in FOB expression, growth rates, germination, reductase activity, iron uptake, and tissue fungal burden were compared by the nonparametric Wilcoxon rank-sum test. The nonparametric log-rank test was used to determine differences in survival times. A P value <0.05 was considered significant.
Supporting Information
Zdroje
1. Ibrahim AS, Edwards JE Jr., Filler SG, Spellberg B (2011) Mucormycosis and Entomophtoramycosis (Zygomycosis). In: Kauffman CA PP, Sobel JD, Dismukes WE, 2nd Ed., editor. Essentials of Clinical Mycology. New York: Springer. pp. 265–280.
2. Kontoyiannis DP, Lewis RE (2006) Invasive zygomycosis: update on pathogenesis, clinical manifestations, and management. Infect Dis Clin North Am 20 : 581–607, vi. 16984870
3. Bitar D, Van Cauteren D, Lanternier F, Dannaoui E, Che D, et al. (2009) Increasing incidence of zygomycosis (mucormycosis), France, 1997–2006. Emerg Infect Dis 15 : 1395–1401. doi: 10.3201/eid1509.090334 19788806
4. Chakrabarti A, Chatterjee SS, Das A, Panda N, Shivaprakash MR, et al. (2009) Invasive zygomycosis in India: experience in a tertiary care hospital. Postgrad Med J 85 : 573–581. doi: 10.1136/pgmj.2008.076463 19892892
5. Gleissner B, Schilling A, Anagnostopolous I, Siehl I, Thiel E (2004) Improved outcome of zygomycosis in patients with hematological diseases? Leuk Lymphoma 45 : 1351–1360. 15359632
6. Ibrahim AS (2011) Host cell invasion in mucormycosis: role of iron. Curr Opin Microbiol 14 : 406–411. doi: 10.1016/j.mib.2011.07.004 21807554
7. Kauffman CA (2004) Zygomycosis: reemergence of an old pathogen. Clin Infect Dis 39 : 588–590. 15356828
8. Kontoyiannis DP, Wessel VC, Bodey GP, Rolston KV (2000) Zygomycosis in the 1990s in a tertiary-care cancer center. Clin Infect Dis 30 : 851–856. 10852735
9. Ibrahim AS, Spellberg B, Edwards J Jr. (2008) Iron acquisition: a novel perspective on mucormycosis pathogenesis and treatment. Curr Opin Infect Dis 21 : 620–625. doi: 10.1097/QCO.0b013e3283165fd1 18978530
10. Sugar AM (2005) Agents of Mucormycosis and Related Species. In: Mandell GL, Bennett JE, Dolin R, editors. Principles and Practice of Infectious Diseases. 6th ed. Philadelphia, PA: Elsevier. pp. 2979.
11. Artis WM, Fountain JA, Delcher HK, Jones HE (1982) A mechanism of susceptibility to mucormycosis in diabetic ketoacidosis: transferrin and iron availability. Diabetes 31 : 1109–1114. 6816646
12. Walsh TJ, Bloom BE, Kontoyiannis DP (2012) Meeting the challenges of an emerging pathogen: the Henry Schueler 41&9 Foundation International Forum on Mucormycosis. Clin Infect Dis 54 Suppl 1: S1–4. doi: 10.1093/cid/cir862 22247440
13. Boelaert JR (1994) Mucormycosis (zygomycosis): is there news for the clinician? Journal of Infection 28 Suppl 1 : 1–6. 8077686
14. Boelaert JR, Fenves AZ, Coburn JW (1989) Registry on mucormycosis in dialysis patients [letter]. Journal of Infectious Diseases 160 : 914. 2809271
15. Boelaert JR, van Roost GF, Vergauwe PL, Verbanck JJ, de Vroey C, et al. (1988) The role of desferrioxamine in dialysis-associated mucormycosis: report of three cases and review of the literature. Clinical Nephrology 29 : 261–266. 3293856
16. Boelaert JR, Vergauwe PL, Vandepitte JM (1987) Mucormycosis infection in dialysis patients [letter]. Annals of Internal Medicine 107 : 782–783. 3662297
17. Boelaert JR, de Locht M, Van Cutsem J, Kerrels V, Cantinieaux B, et al. (1993) Mucormycosis during deferoxamine therapy is a siderophore-mediated infection. In vitro and in vivo animal studies. Journal of Clinical Investigation 91 : 1979–1986. 8486769
18. de Locht M, Boelaert JR, Schneider YJ (1994) Iron uptake from ferrioxamine and from ferrirhizoferrin by germinating spores of Rhizopus microsporus. Biochemical Pharmacology 47 : 1843–1850. 8204101
19. Ibrahim AS, Edwards JE Jr., Fu Y, Spellberg B (2006) Deferiprone iron chelation as a novel therapy for experimental mucormycosis. J Antimicrob Chemother 58 : 1070–1073. 16928702
20. Ibrahim AS, Gebermariam T, Fu Y, Lin L, Husseiny MI, et al. (2007) The iron chelator deferasirox protects mice from mucormycosis through iron starvation. J Clin Invest 117 : 2649–2657. 17786247
21. Maruyama HB, Azuma H, Koto Y, Suhara Y (1975) Desferrioxamine B, an inhibitor of Escherichia coli motility, reversing the stimulating effect of cyclic adenosine 3',5'-monophosphate. Antimicrob Agents Chemother 7 : 377–380. 166612
22. Thieken A, Winkelmann G (1992) Rhizoferrin: a complexone type siderophore of the Mucorales and entomophthorales (Zygomycetes). FEMS Microbiol Lett 73 : 37–41. 1387861
23. Kwon-Chung KJ, Bennett JE (1992) Mucormycosis. Medical Mycology. Philadelphia: Lea & Febiger. pp. 524–559.
24. Ibrahim AS, Gebremariam T, Lin L, Luo G, Husseiny MI, et al. (2010) The high affinity iron permease is a key virulence factor required for Rhizopus oryzae pathogenesis. Mol Microbiol 77 : 587–604. doi: 10.1111/j.1365-2958.2010.07234.x 20545847
25. Yun CW, Ferea T, Rashford J, Ardon O, Brown PO, et al. (2000) Desferrioxamine-mediated iron uptake in Saccharomyces cerevisiae. Evidence for two pathways of iron uptake. J Biol Chem 275 : 10709–10715. 10744769
26. Yun CW, Tiedeman JS, Moore RE, Philpott CC (2000) Siderophore-iron uptake in Saccharomyces cerevisiae. Identification of ferrichrome and fusarinine transporters. J Biol Chem 275 : 16354–16359. 10748025
27. Lesuisse E, Simon-Casteras M, Labbe P (1998) Siderophore-mediated iron uptake in Saccharomyces cerevisiae: the SIT1 gene encodes a ferrioxamine B permease that belongs to the major facilitator superfamily. Microbiology 144 (Pt 12): 3455–3462. 9884238
28. Heymann P, Gerads M, Schaller M, Dromer F, Winkelmann G, et al. (2002) The siderophore iron transporter of Candida albicans (Sit1p/Arn1p) mediates uptake of ferrichrome-type siderophores and is required for epithelial invasion. Infect Immun 70 : 5246–5255. 12183576
29. Moore RE, Kim Y, Philpott CC (2003) The mechanism of ferrichrome transport through Arn1p and its metabolism in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A 100 : 5664–5669. 12721368
30. Philpott CC, Protchenko O (2008) Response to iron deprivation in Saccharomyces cerevisiae. Eukaryot Cell 7 : 20–27. 17993568
31. Boelaert JR, Fenves AZ, Coburn JW (1989) Mucormycosis among patients on dialysis [letter]. New England Journal of Medicine 321 : 190–191. 2747752
32. Boelaert JR, Van Cutsem J, de Locht M, Schneider YJ, Crichton RR (1994) Deferoxamine augments growth and pathogenicity of Rhizopus, while hydroxypyridinone chelators have no effect. Kidney International 45 : 667–671. 8196268
33. Pitarch A, Nombela C, Gil C (2008) Collection of proteins secreted from yeast protoplasts in active cell wall regeneration. Methods Mol Biol 425 : 241–263. doi: 10.1007/978-1-60327-210-0_20 18369901
34. Ignoul S, Eggermont J (2005) CBS domains: structure, function, and pathology in human proteins. Am J Physiol Cell Physiol 289: C1369–1378. 16275737
35. Jentsch TJ, Neagoe I, Scheel O (2005) CLC chloride channels and transporters. Curr Opin Neurobiol 15 : 319–325. 15913981
36. Kemp BE (2004) Bateman domains and adenosine derivatives form a binding contract. J Clin Invest 113 : 182–184. 14722609
37. McLean JE, Hamaguchi N, Belenky P, Mortimer SE, Stanton M, et al. (2004) Inosine 5'-monophosphate dehydrogenase binds nucleic acids in vitro and in vivo. Biochem J 379 : 243–251. 14766016
38. Aguado-Llera D, Oyenarte I, Martinez-Cruz LA, Neira JL (2010) The CBS domain protein MJ0729 of Methanocaldococcus jannaschii binds DNA. FEBS Lett 584 : 4485–4489. doi: 10.1016/j.febslet.2010.10.006 20934423
39. Ishitani R, Sugita Y, Dohmae N, Furuya N, Hattori M, et al. (2008) Mg2+-sensing mechanism of Mg2+ transporter MgtE probed by molecular dynamics study. Proc Natl Acad Sci U S A 105 : 15393–15398. doi: 10.1073/pnas.0802991105 18832160
40. Stintzi A, Barnes C, Xu J, Raymond KN (2000) Microbial iron transport via a siderophore shuttle: a membrane ion transport paradigm. Proc Natl Acad Sci U S A 97 : 10691–10696. 10995480
41. Braun V, Hantke K, Koster W (1998) Bacterial iron transport: mechanisms, genetics, and regulation. Met Ions Biol Syst 35 : 67–145. 9444760
42. Cole G, Simonetti K, Ademi I, Sharpe S (2012) Dimerization of the transmembrane domain of human tetherin in membrane mimetic environments. Biochemistry 51 : 5033–5040. doi: 10.1021/bi201747t 22667354
43. Columbus L, Lipfert J, Klock H, Millett I, Doniach S, et al. (2006) Expression, purification, and characterization of Thermotoga maritima membrane proteins for structure determination. Protein Sci 15 : 961–975. 16597824
44. Watt AD, Perez KA, Rembach A, Sherrat NA, Hung LW, et al. (2013) Oligomers, fact or artefact? SDS-PAGE induces dimerization of beta-amyloid in human brain samples. Acta Neuropathol 125 : 549–564. doi: 10.1007/s00401-013-1083-z 23354835
45. Lisiecki P, Wysocki P, Mikucki J (1999) [Hydroxamate siderophores in enterococci]. Med Dosw Mikrobiol 51 : 249–257. 10803254
46. Lisiecki P, Wysocki P, Mikucki J (2000) Occurrence of siderophores in enterococci. Zentralbl Bakteriol 289 : 807–815. 10705612
47. Muller G, Raymond KN (1984) Specificity and mechanism of ferrioxamine-mediated iron transport in Streptomyces pilosus. J Bacteriol 160 : 304–312. 6480557
48. Protchenko O, Ferea T, Rashford J, Tiedeman J, Brown PO, et al. (2001) Three cell wall mannoproteins facilitate the uptake of iron in Saccharomyces cerevisiae. J Biol Chem 276 : 49244–49250. 11673473
49. de Locht M, Boelaert JR, Schneider YJ (1994) Iron uptake from ferrioxamine and from ferrirhizoferrin by germinating spores of Rhizopus microsporus. Biochem Pharmacol 47 : 1843–1850. 8204101
50. Kuhn S, Braun V, Koster W (1996) Ferric rhizoferrin uptake into Morganella morganii: characterization of genes involved in the uptake of a polyhydroxycarboxylate siderophore. J Bacteriol 178 : 496–504. 8550472
51. Schaller M, Korting HC, Schafer W, Bastert J, Chen W, et al. (1999) Secreted aspartic proteinase (Sap) activity contributes to tissue damage in a model of human oral candidosis. Mol Microbiol 34 : 169–180. 10540295
52. Gutteridge JM, Quinlan GJ, Swain J, Cox J (1994) Ferrous ion formation by ferrioxamine prepared from aged desferrioxamine: a potential prooxidant property. Free Radic Biol Med 16 : 733–739. 8070676
53. Fu Y, Lee H, Collins M, Tsai HF, Spellberg B, et al. (2004) Cloning and functional characterization of the Rhizopus oryzae high affinity iron permease (rFTR1) gene. FEMS Microbiol Lett 235 : 169–176. 15158278
54. Howard DH (1999) Acquisition, transport, and storage of iron by pathogenic fungi. Clin Microbiol Rev 12 : 394–404. 10398672
55. Lesuisse E, Labbe P (1989) Reductive and non-reductive mechanisms of iron assimilation by the yeast Saccharomyces cerevisiae. J Gen Microbiol 135 : 257–263. 11699493
56. Larcher G, Dias M, Razafimandimby B, Bomal D, Bouchara JP (2013) Siderophore production by pathogenic mucorales and uptake of deferoxamine B. Mycopathologia 176 : 319–328. doi: 10.1007/s11046-013-9693-5 23982284
57. Skory CD, Ibrahim AS (2007) Native and modified lactate dehydrogenase expression in a fumaric acid producing isolate Rhizopus oryzae 99-880. Curr Genet 52 : 23–33. 17551728
58. Dertz EA, Stintzi A, Raymond KN (2006) Siderophore-mediated iron transport in Bacillus subtilis and Corynebacterium glutamicum. J Biol Inorg Chem 11 : 1087–1097. 16912897
59. Schmittgen TD, Livak KJ (2008) Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 3 : 1101–1108. 18546601
60. Skory CD (2003) Induction of Rhizopus oryzae pyruvate decarboxylase genes. Curr Microbiol 47 : 59–64. 12783195
61. Dancis A, Klausner RD, Hinnebusch AG, Barriocanal JG (1990) Genetic evidence that ferric reductase is required for iron uptake in Saccharomyces cerevisiae. Mol Cell Biol 10 : 2294–2301. 2183029
62. Ibrahim AS, Spellberg BJ, Avenissian V, Fu Y, Filler SG, et al. (2005) Vaccination with recombinant N-terminal domain of Als1p improves survival during murine disseminated candidiasis by enhancing cell-mediated, not humoral, immunity. Infect Immun 73 : 999–1005. 15664943
63. Fu Y, Ibrahim AS, Sheppard DC, Chen YC, French SW, et al. (2002) Candida albicans Als1p: an adhesin that is a downstream effector of the EFG1 filamentation pathway. Mol Microbiol 44 : 61–72. 11967069
64. Abe F, Inaba H, Katoh T, Hotchi M (1990) Effects of iron and desferrioxamine on Rhizopus infection. Mycopathologia 110 : 87–91. 2142253
65. Gebremariam T, Liu M, Luo G, Bruno V, Phan QT, et al. (2014) CotH3 mediates fungal invasion of host cells during mucormycosis. J Clin Invest 124 : 237–250. doi: 10.1172/JCI71349 24355926
66. Ibrahim AS, Avanessian V, Spellberg B, Edwards JE Jr. (2003) Liposomal amphotericin B, and not amphotericin B deoxycholate, improves survival of diabetic mice infected with Rhizopus oryzae. Antimicrob Agents Chemother 47 : 3343–3344. 14506054
67. Liu M, Spellberg B, Phan QT, Fu Y, Lee AS, et al. (2010) The endothelial cell receptor GRP78 is required for mucormycosis pathogenesis in diabetic mice. J Clin Invest 120 : 1914–1924. doi: 10.1172/JCI42164 20484814
68. Ibrahim AS, Bowman JC, Avanessian V, Brown K, Spellberg B, et al. (2005) Caspofungin inhibits Rhizopus oryzae 1,3-beta-D-glucan synthase, lowers burden in brain measured by quantitative PCR, and improves survival at a low but not a high dose during murine disseminated zygomycosis. Antimicrob Agents Chemother 49 : 721–727. 15673756
69. Aso H, Miyoshi S, Nakao H, Okamoto K, Yamamoto S (2002) Induction of an outer membrane protein of 78 kDa in Vibrio vulnificus cultured in the presence of desferrioxamine B under iron-limiting conditions. FEMS Microbiol Lett 212 : 65–70. 12076789
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek Neutrophil-Derived MMP-8 Drives AMPK-Dependent Matrix Destruction in Human Pulmonary TuberculosisČlánek Circumventing . Virulence by Early Recruitment of Neutrophils to the Lungs during Pneumonic PlagueČlánek Admixture in Humans of Two Divergent Populations Associated with Different Macaque Host SpeciesČlánek Human and Murine Clonal CD8+ T Cell Expansions Arise during Tuberculosis Because of TCR SelectionČlánek Selective Recruitment of Nuclear Factors to Productively Replicating Herpes Simplex Virus GenomesČlánek Remembering MumpsČlánek Human Cytomegalovirus miR-UL112-3p Targets TLR2 and Modulates the TLR2/IRAK1/NFκB Signaling PathwayČlánek Induces the Premature Death of Human Neutrophils through the Action of Its Lipopolysaccharide
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2015 Číslo 5- Jak souvisí postcovidový syndrom s poškozením mozku?
- Měli bychom postcovidový syndrom léčit antidepresivy?
- Farmakovigilanční studie perorálních antivirotik indikovaných v léčbě COVID-19
- 10 bodů k očkování proti COVID-19: stanovisko České společnosti alergologie a klinické imunologie ČLS JEP
-
Všechny články tohoto čísla
- Parasites and Their Heterophagic Appetite for Disease
- The Elusive Role of the Prion Protein and the Mechanism of Toxicity in Prion Disease
- Intestinal Colonization Dynamics of
- Activation of Typhi-Specific Regulatory T Cells in Typhoid Disease in a Wild-Type . Typhi Challenge Model
- The Engineering of a Novel Ligand in gH Confers to HSV an Expanded Tropism Independent of gD Activation by Its Receptors
- Neutrophil-Derived MMP-8 Drives AMPK-Dependent Matrix Destruction in Human Pulmonary Tuberculosis
- Group Selection and Contribution of Minority Variants during Virus Adaptation Determines Virus Fitness and Phenotype
- Phosphatidic Acid Produced by Phospholipase D Promotes RNA Replication of a Plant RNA Virus
- A Ribonucleoprotein Complex Protects the Interleukin-6 mRNA from Degradation by Distinct Herpesviral Endonucleases
- Characterization of Transcriptional Responses to Different Aphid Species Reveals Genes that Contribute to Host Susceptibility and Non-host Resistance
- Circumventing . Virulence by Early Recruitment of Neutrophils to the Lungs during Pneumonic Plague
- Natural Killer Cell Sensing of Infected Cells Compensates for MyD88 Deficiency but Not IFN-I Activity in Resistance to Mouse Cytomegalovirus
- Manipulation of the Xanthophyll Cycle Increases Plant Susceptibility to
- Ly6C Monocytes Regulate Parasite-Induced Liver Inflammation by Inducing the Differentiation of Pathogenic Ly6C Monocytes into Macrophages
- Admixture in Humans of Two Divergent Populations Associated with Different Macaque Host Species
- Expression in the Fat Body Is Required in the Defense Against Parasitic Wasps in
- Experimental Evolution of an RNA Virus in Wild Birds: Evidence for Host-Dependent Impacts on Population Structure and Competitive Fitness
- Inhibition and Reversal of Microbial Attachment by an Antibody with Parasteric Activity against the FimH Adhesin of Uropathogenic .
- The EBNA-2 N-Terminal Transactivation Domain Folds into a Dimeric Structure Required for Target Gene Activation
- Human and Murine Clonal CD8+ T Cell Expansions Arise during Tuberculosis Because of TCR Selection
- The NLRP3 Inflammasome Is a Pathogen Sensor for Invasive via Activation of α5β1 Integrin at the Macrophage-Amebae Intercellular Junction
- Sequential Conformational Changes in the Morbillivirus Attachment Protein Initiate the Membrane Fusion Process
- A Two-Component DNA-Prime/Protein-Boost Vaccination Strategy for Eliciting Long-Term, Protective T Cell Immunity against
- cAMP-Signalling Regulates Gametocyte-Infected Erythrocyte Deformability Required for Malaria Parasite Transmission
- Response Regulator VxrB Controls Colonization and Regulates the Type VI Secretion System
- Evidence for a Novel Mechanism of Influenza Virus-Induced Type I Interferon Expression by a Defective RNA-Encoded Protein
- Dust Devil: The Life and Times of the Fungus That Causes Valley Fever
- TNF-α Induced by Hepatitis C Virus via TLR7 and TLR8 in Hepatocytes Supports Interferon Signaling via an Autocrine Mechanism
- The Recent Evolution of a Maternally-Inherited Endosymbiont of Ticks Led to the Emergence of the Q Fever Pathogen,
- L-Rhamnosylation of Wall Teichoic Acids Promotes Resistance to Antimicrobial Peptides by Delaying Interaction with the Membrane
- Rapid Sequestration of by Neutrophils Contributes to the Development of Chronic Lesion
- Selective Recruitment of Nuclear Factors to Productively Replicating Herpes Simplex Virus Genomes
- The Expression of Functional Vpx during Pathogenic SIVmac Infections of Rhesus Macaques Suppresses SAMHD1 in CD4 Memory T Cells
- Fob1 and Fob2 Proteins Are Virulence Determinants of via Facilitating Iron Uptake from Ferrioxamine
- TRAF1 Coordinates Polyubiquitin Signaling to Enhance Epstein-Barr Virus LMP1-Mediated Growth and Survival Pathway Activation
- Vaccine-Elicited Tier 2 HIV-1 Neutralizing Antibodies Bind to Quaternary Epitopes Involving Glycan-Deficient Patches Proximal to the CD4 Binding Site
- Remembering Mumps
- The Role of Horizontal Gene Transfer in the Evolution of the Oomycetes
- Advances and Challenges in Computational Prediction of Effectors from Plant Pathogenic Fungi
- Investigating Fungal Outbreaks in the 21st Century
- Systems Biology for Biologists
- How Does the Dinoflagellate Parasite Outsmart the Immune System of Its Crustacean Hosts?
- FCRL5 Delineates Functionally Impaired Memory B Cells Associated with Exposure
- Phospholipase D1 Couples CD4 T Cell Activation to c-Myc-Dependent Deoxyribonucleotide Pool Expansion and HIV-1 Replication
- Influenza A Virus on Oceanic Islands: Host and Viral Diversity in Seabirds in the Western Indian Ocean
- Geometric Constraints Dominate the Antigenic Evolution of Influenza H3N2 Hemagglutinin
- Widespread Recombination, Reassortment, and Transmission of Unbalanced Compound Viral Genotypes in Natural Arenavirus Infections
- Gammaherpesvirus Co-infection with Malaria Suppresses Anti-parasitic Humoral Immunity
- A Single Protein S-acyl Transferase Acts through Diverse Substrates to Determine Cryptococcal Morphology, Stress Tolerance, and Pathogenic Outcome
- Survives with a Minimal Peptidoglycan Synthesis Machine but Sacrifices Virulence and Antibiotic Resistance
- Mechanisms of Stage-Transcending Protection Following Immunization of Mice with Late Liver Stage-Arresting Genetically Attenuated Malaria Parasites
- The Myelin and Lymphocyte Protein MAL Is Required for Binding and Activity of ε-Toxin
- Genome-Wide Identification of the Target Genes of AP2-O, a AP2-Family Transcription Factor
- An Atypical Mitochondrial Carrier That Mediates Drug Action in
- Human Cytomegalovirus miR-UL112-3p Targets TLR2 and Modulates the TLR2/IRAK1/NFκB Signaling Pathway
- Helminth Infection and Commensal Microbiota Drive Early IL-10 Production in the Skin by CD4 T Cells That Are Functionally Suppressive
- Circulating Pneumolysin Is a Potent Inducer of Cardiac Injury during Pneumococcal Infection
- ExoT Induces Atypical Anoikis Apoptosis in Target Host Cells by Transforming Crk Adaptor Protein into a Cytotoxin
- Discovery of a Small Non-AUG-Initiated ORF in Poleroviruses and Luteoviruses That Is Required for Long-Distance Movement
- Induces the Premature Death of Human Neutrophils through the Action of Its Lipopolysaccharide
- Varicella Viruses Inhibit Interferon-Stimulated JAK-STAT Signaling through Multiple Mechanisms
- Paradoxical Immune Responses in Non-HIV Cryptococcal Meningitis
- Recovery of Recombinant Crimean Congo Hemorrhagic Fever Virus Reveals a Function for Non-structural Glycoproteins Cleavage by Furin
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Human Cytomegalovirus miR-UL112-3p Targets TLR2 and Modulates the TLR2/IRAK1/NFκB Signaling Pathway
- Paradoxical Immune Responses in Non-HIV Cryptococcal Meningitis
- Expression in the Fat Body Is Required in the Defense Against Parasitic Wasps in
- Survives with a Minimal Peptidoglycan Synthesis Machine but Sacrifices Virulence and Antibiotic Resistance
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



