-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaAdvances and Challenges in Computational Prediction of Effectors from Plant Pathogenic Fungi
article has not abstract
Published in the journal: . PLoS Pathog 11(5): e32767. doi:10.1371/journal.ppat.1004806
Category: Pearls
doi: https://doi.org/10.1371/journal.ppat.1004806Summary
article has not abstract
Fungal Effector Proteins Underpin Diverse Infection Strategies
Fungi occupy diverse environmental niches and many have evolved to live a pathogenic lifestyle, causing devastating diseases in plants and animals. The interface between host and pathogen is complex and constantly evolving. Pathogens secrete effector proteins that manipulate the host to the pathogen’s advantage. Depending on their infection strategy, fungal pathogens may deliver apoplastic effectors into the extracellular spaces and/or cytoplasmic effectors that are taken up by plant cells. Effectors have a broad functional spectrum, ranging from effectors in necrotrophic pathogens with toxic activity that cause plant cell death to avirulence (Avr) effectors in biotrophic pathogens that trigger defense responses and that the plant immune system has evolved to recognize. Molecular studies have revealed over 60 fungal effectors from different species; however, this represents only the tip of the iceberg. For example, only six effectors have thus far been characterized across three rust fungi, while more than 30 Avr specificities have been identified in flax rust and around 50 in each of stem rust, stripe rust, and leaf rust [1]. Similarly, over 40 Avr specificities occur in interactions between Magnaporthe oryzae and rice [2].
With the rising number of sequenced pathogen genomes, computational prediction of effector proteins holds promise as a fast and economical technique to define candidates for subsequent laboratory work. Bacterial effectors delivered to the host via dedicated pathogen-derived delivery mechanisms, such as the type III secretion system, can be predicted using machine learning approaches based on protein sequence information. In oomycetes, consensus sequence motifs implicated in host translocation, such as RXLR, can be exploited for effector prediction. However, computational effector prediction in fungi is challenging due to a lack of known protein features that are common to fungal effectors and the low number of characterized effectors for individual species, which limits the use of machine learning approaches.
Fungal Effector Proteins Generally Lack Sequence Similarity and Conserved Sequence Motifs, but Some Might Share Structural Similarity
Fungal effector prediction is a difficult problem due to the lack of unifying sequence features or structural folds for effectors within and across species. In general, fungal effectors do not share significant sequence similarity with each other, which can be attributed to rapid divergence and host specialization. However, there are some exceptions. The Cladosporium fulvum Ecp6 effector contains LysM domains and has strong sequence similarity to Magnaporthe oryzae Slp1 and other fungal genes [3]. Furthermore, some effector proteins can have a functional annotation that suggests a role in pathogenicity, for example, the chorismate mutase effector in the biotrophic maize pathogen Ustilago maydis [4]. Unlike the oomycete RXLR and Crinkler families of cytoplasmic effectors, no widely conserved sequence-based motifs have thus far been identified for fungal effectors, despite suggestions of RXLR-like sequences in some fungal effectors [5]. There is sporadic evidence of conserved N-terminal sequence motifs in fungal proteins with a secretion signal. For example, effector candidates in the barley powdery mildew fungus, Blumeria graminis f.sp. hordei, share an N-terminal [YFW]xC motif within 30 amino acids of the signal peptide [6]. This motif has also been reported in some effector candidates of rust fungi, but with less positional conservation [7]. In Fusarium, a group of proteins share a conserved [SG]PC[KR]P motif immediately after the signal peptide [8,9]. However, these motifs have not been functionally characterized and can, thus, not be confirmed as fungal effector sequence motifs. [10]. AvrL567 and AvrM from Melampsora lini enter flax cells autonomously mediated by N-terminal uptake domains, however, these do not share conserved motifs or structures [11]. The C-terminal RGD sequence motif in the ToxA effector is required for wheat cell entry [10].
More subtle features other than sequence similarity may unify classes of effectors, such as conserved three-dimensional folds. For examples, many oomycete RxLR effectors share a common WY domain fold [12], while the powdery mildew [YFW]xC class effectors are predicted to share a structural fold related to ribonucleases [13]. Similar β-sandwich structures were identified in AvrL567 from Melampsora lini [14], the ToxA effector from Pyrenophora tritici-repentis [15] and in the M. oryzae effector AvrPiz-t [16], suggesting that this fold might be common in fungal effectors. Interestingly, the three-dimensional structure of the M. lini effector AvrM contains a tandem duplicated four-helical motif with similarity to the WY domain of oomycete effectors [12]. Thus, while there may be some structural conservation within certain families of fungal effectors, overall, the lack of conserved structural features suggests difficulty in exploiting these for effector prediction.
Secreted, Small, and Cysteine-Rich: Prediction of Apoplastic Effector Repertoires from Genomes
Given the lack of conserved sequence features, fungal effector prediction approaches have been based on relatively broad criteria, principally the presence of a secretion signal. In addition, most known fungal effectors are small in size and often rich in cysteine residues. Apoplastic effectors, in particular, often contain several disulfide bonds [17] and predicted secretomes of pathogenic fungi contain proteins with elevated levels of cysteines compared to all proteins (Fig 1A). Therefore, the criteria of small and cysteine-rich can be used to mine predicted secretomes for apoplastic effectors and reduce the number of candidates [18,19]. However, not all secreted proteins with small size and high cysteine content will have an effector function and, conversely, not all fungal effectors will be small and cysteine-rich. Many cytoplasmic effectors that are delivered into host cells are low in cysteines and of larger size, which has also been found for several apoplastic effectors (Fig 1B). For example, the AvrLm1 effector from the hemibiotrophic pathogen Leptosphaeria maculans that colonises the apoplast has only one cysteine [20]. Whilst the criteria of small and cysteine-rich are very valuable for screening secretomes for apoplastic effectors, they are not a one-size-fits-all solution for predicting both apoplastic and cytoplasmic effectors, and do not necessarily discriminate between these classes either. For instance, the AvrP4 and AvrP123 effectors of M. lini are small and cysteine-rich, yet are recognised by intracellular immune receptors, suggesting they are delivered to the host cytoplasm [1].
Fig. 1. Cysteine content of predicted fungal secretomes and fungal effector properties. 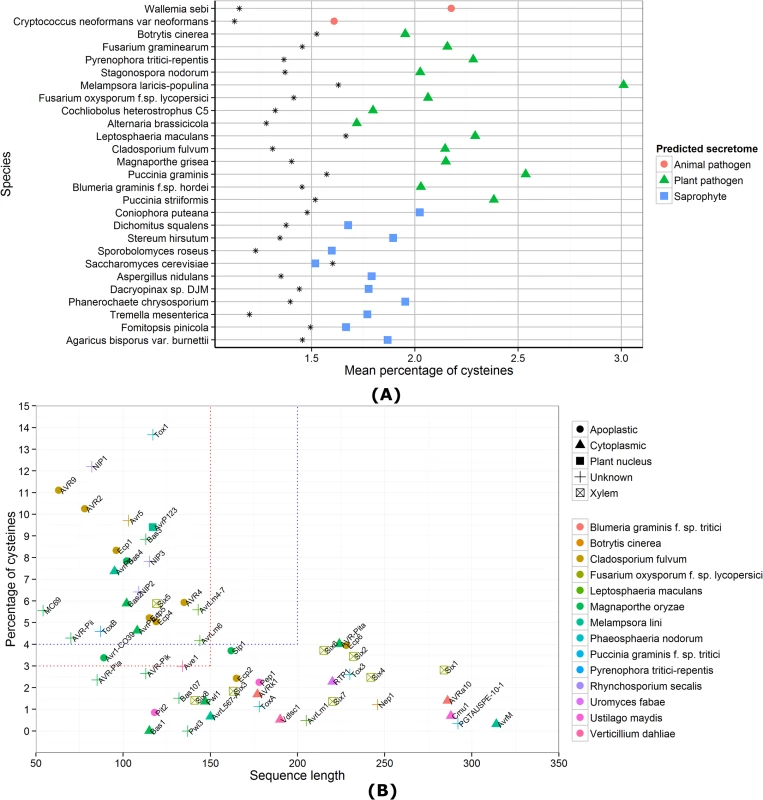
(A) For each species, the mean percentage of cysteines is shown for all predicted genes (as a black star) and the secretome predicted by SignalP 4.1 [31]. Apart from S. cerevisiae, all species have a higher mean percentage of cysteines in their secretomes, compared to the genome-wide mean. (B) Sequence lengths and cysteine content of known fungal effector proteins are shown. The red dotted lines indicate the criteria for small, cysteine-rich defined in Saunders et al. [21] and the blue dotted lines the criteria for small, cysteine-rich defined in Ma et al. [8]. A trend for species-specific conservation of small, cysteine-rich effectors cannot be observed. Even the C. fulvum pathogen that is known to grow extracellularly has two effectors that do not fit under the small, cysteine-rich umbrella defined by commonly used thresholds. Beyond Secreted, Small, and Cysteine-Rich: Dedicated Pipelines for Predicting Apoplastic and Cytoplasmic Effectors
Sophisticated approaches for predicting apoplastic and cytoplasmic effector candidates have emerged that do not solely rely on rules, such as a predicted secretion signal, small size, and cysteine content, but also include other lines of evidence associated with fungal effectors and are potentially powerful for predicting effector candidates without making a priori assumptions on their properties.
For haustorially delivered rust fungi effectors, Saunders et al. [21] developed a ranking method according to criteria associated with experimentally verified effectors (details given in Fig 2). First, secretomes are clustered into tribes based on sequence similarity scores. Second, tribes are ranked according to the likelihood of obtaining at least the same number of proteins with the given effector property by chance. Whilst high-scoring tribes were predicted that contain likely effector families, the pipeline failed to recognize the Puccinia graminis f. sp. tritici effector PGTAUSPE-10-1 [22] as a candidate. The same pipeline was also applied to M. lini with thresholds informed by the known rust effectors, which returned 200 high priority tribes of candidate effectors [23], and to P. striiformis f. sp. tritici [24] combined with evidence of sequence polymorphisms and in planta expression. The combination of additional lines of evidence is very useful to reduce the set of high-priority candidates. For example, Sperschneider et al. [25] combined evidence for diversifying selection; conservation, predominantly in fungal pathogen genomes; and induction in planta and in haustoria to identify a list of 42 haustorially delivered effector candidates in P. graminis f. sp. tritici and successfully recovered PGTAUSPE-10-1 as the top candidate [22].
Fig. 2. Lines of evidence that have been used for predicting fungal effector proteins and examples for fungal effector prediction pipelines. 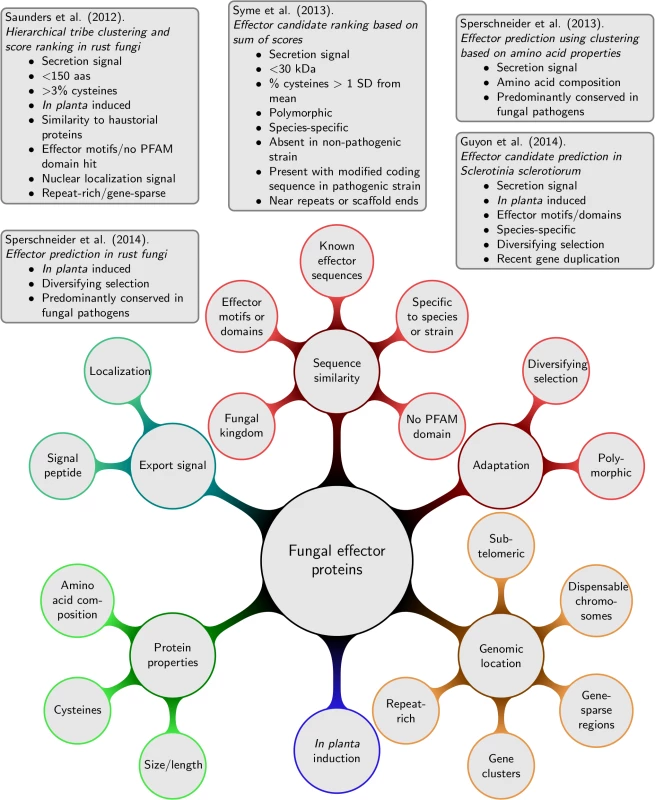
For necrotrophic pathogens, Guyon et al. [26] returned 78 effector candidates from Sclerotinia sclerotiorum, again, using several independent lines of evidence as shown in Fig 2. Syme et al. [27] used the sum of effector evidence scores (details in Fig 2) to rank Stagonospora nodorum effector candidates that are not found or that are highly divergent in a re-sequenced, non-pathogenic strain. An unsupervised exploration of fungal effector properties in cereal pathogens was performed in Sperschneider et al. [9]. Proteins that were predominantly conserved across fungal pathogens were clustered based on their amino acid properties and other sequence-derived features. This revealed putative effector clusters with enrichment in secretion signals for several fungal pathogens infecting cereals. Interestingly, some of these protein clusters are enriched in secreted proteins that have a high content of small amino acids and cysteines, whereas others are enriched in features not commonly associated with fungal effectors. This supports the view that our current knowledge of fungal effectors is still incomplete.
Fungal Effector Prediction from Genomic Sequences: A Unified Way Forward
Whilst the full scope of fungal effectors remains a mystery, in particular for animal pathogens, characterized plant pathogen effectors have been found to be extremely versatile, targeting diverse host cell compartments and elements of the plant immune system [28]. Despite increasing insight into effector functions through molecular and structural studies, the only universal features thus far identified of fungal effectors are that they are secreted and differentially expressed during in planta infection. However, they are not necessarily computationally predicted to be secreted, as exemplified by fungal effectors that lack a predicted signal peptide and must instead use an unconventional secretion pathway [29]. Approaches for predicting fungal effectors from genomic sequences must be able to look beyond sequence-similarity-based methods and should not rely purely on selecting small and cysteine-rich proteins from the secretome as effector candidates. Classifiers that integrate other evidence for effector function, such as in planta expression data, signatures of diversifying selection, genomic features, or taxonomic information, are equally powerful and do not make a priori assumptions on effector protein properties. Future studies will be required to determine if there are structural folds or other molecular features common to fungal effectors targeting the same host cell machinery. It will be interesting to apply concepts from effector prediction in fungal plant pathogens (Fig 2) to the prediction of effectors in fungal animal pathogens to explore possible similarities [30]. Finally, an increase in the number of identified fungal effectors might enable machine learning approaches for unbiased prediction, which could lead to the discovery of protein properties common to fungal effectors.
Zdroje
1. Garnica DP, Nemri A, Upadhyaya NM, Rathjen JP, Dodds PN (2014) The ins and outs of rust haustoria. PLoS Pathog 10: e1004329. doi: 10.1371/journal.ppat.1004329 25211126
2. Zhang S, Xu JR (2014) Effectors and effector delivery in Magnaporthe oryzae. PLoS Pathog 10: e1003826. doi: 10.1371/journal.ppat.1003826 24391496
3. Bolton MD, van Esse HP, Vossen JH, de Jonge R, Stergiopoulos I, et al. (2008) The novel Cladosporium fulvum lysin motif effector Ecp6 is a virulence factor with orthologues in other fungal species. Mol Microbiol 69 : 119–136. doi: 10.1111/j.1365-2958.2008.06270.x 18452583
4. Djamei A, Schipper K, Rabe F, Ghosh A, Vincon V, Kahnt J, et al. (2011) Metabolic priming by a secreted fungal effector. Nature 478 : 395–398. doi: 10.1038/nature10454 21976020
5. Kale SD, Gu B, Capelluto DG, Dou D, Feldman E, Rumore A, et al. (2010) External lipid PI3P mediates entry of eukaryotic pathogen effectors into plant and animal host cells. Cell 142 : 284–295. doi: 10.1016/j.cell.2010.06.008 20655469
6. Godfrey D, Bohlenius H, Pedersen C, Zhang Z, Emmersen J, Thordal-Christensen H. (2010) Powdery mildew fungal effector candidates share N-terminal Y/F/WxC-motif. BMC Genomics 11 : 317. doi: 10.1186/1471-2164-11-317 20487537
7. Duplessis S, Cuomo CA, Lin YC, Aerts A, Tisserant E, Veneault-Fourrey C, et al. (2011) Obligate biotrophy features unraveled by the genomic analysis of rust fungi. Proc Natl Acad Sci U S A 108 : 9166–9171. doi: 10.1073/pnas.1019315108 21536894
8. Ma LJ, van der Does HC, Borkovich KA, Coleman JJ, Daboussi MJ, Di Pietro A, et al. (2010) Comparative genomics reveals mobile pathogenicity chromosomes in Fusarium. Nature 464 : 367–373. doi: 10.1038/nature08850 20237561
9. Sperschneider J, Gardiner DM, Taylor JM, Hane JK, Singh KB, Manners JM. (2013) A comparative hidden Markov model analysis pipeline identifies proteins characteristic of cereal-infecting fungi. BMC Genomics 14 : 807. doi: 10.1186/1471-2164-14-807 24252298
10. Manning VA, Hamilton SM, Karplus PA, Ciuffetti LM (2008) The Arg-Gly-Asp-containing, solvent-exposed loop of Ptr ToxA is required for internalization. Mol Plant Microbe Interact 21 : 315–325. doi: 10.1094/MPMI-21-3-0315 18257681
11. Rafiqi M, Gan PH, Ravensdale M, Lawrence GJ, Ellis JG, Jones D, et al. (2010) Internalization of flax rust avirulence proteins into flax and tobacco cells can occur in the absence of the pathogen. Plant Cell 22 : 2017–2032. doi: 10.1105/tpc.109.072983 20525849
12. Win J, Krasileva KV, Kamoun S, Shirasu K, Staskawicz BJ, Banfield M. (2012) Sequence divergent RXLR effectors share a structural fold conserved across plant pathogenic oomycete species. PLoS Pathog 8: e1002400. doi: 10.1371/journal.ppat.1002400 22253591
13. Pedersen C, Ver Loren van Themaat E, McGuffin LJ, Abbott JC, Burgis TA, Barton G, et al. (2012) Structure and evolution of barley powdery mildew effector candidates. BMC Genomics 13 : 694. doi: 10.1186/1471-2164-13-694 23231440
14. Wang CI, Guncar G, Forwood JK, Teh T, Catanzariti AM, Lawrencec G, et al. (2007) Crystal structures of flax rust avirulence proteins AvrL567-A and-D reveal details of the structural basis for flax disease resistance specificity. Plant Cell 19 : 2898–2912. 17873095
15. Sarma GN, Manning VA, Ciuffetti LM, Karplus PA (2005) Structure of Ptr ToxA: an RGD-containing host-selective toxin from Pyrenophora tritici-repentis. Plant Cell 17 : 3190–3202. 16214901
16. Zhang ZM, Zhang X, Zhou ZR, Hu HY, Liu M, Zhou B, et al. (2013) Solution structure of the Magnaporthe oryzae avirulence protein AvrPiz-t. J Biomol NMR 55 : 219–223. doi: 10.1007/s10858-012-9695-5 23334361
17. Stergiopoulos I, de Wit PJ (2009) Fungal effector proteins. Annu Rev Phytopathol 47 : 233–263. doi: 10.1146/annurev.phyto.112408.132637 19400631
18. de Jonge R (2012) In silico identification and characterization of effector catalogs. Methods Mol Biol 835 : 415–425. doi: 10.1007/978-1-61779-501-5_25 22183668
19. Hacquard S, Joly DL, Lin YC, Tisserant E, Feau N, Delaruelle C, et al. (2012) A comprehensive analysis of genes encoding small secreted proteins identifies candidate effectors in Melampsora larici-populina (poplar leaf rust). Mol Plant Microbe Interact 25 : 279–293. doi: 10.1094/MPMI-09-11-0238 22046958
20. Gout L, Fudal I, Kuhn ML, Blaise F, Eckert M, Cattolico L, et al. (2006) Lost in the middle of nowhere: the AvrLm1 avirulence gene of the Dothideomycete Leptosphaeria maculans. Mol Microbiol 60 : 67–80. 16556221
21. Saunders DG, Win J, Cano LM, Szabo LJ, Kamoun S, Raffaele S. (2012) Using hierarchical clustering of secreted protein families to classify and rank candidate effectors of rust fungi. PLoS One 7: e29847. doi: 10.1371/journal.pone.0029847 22238666
22. Upadhyaya NM, Mago R, Staskawicz BJ, Ayliffe M, Ellis J, Dodds PN. (2013) A Bacterial Type III Secretion Assay for Delivery of Fungal Effector Proteins into Wheat. Mol Plant Microbe Interact.
23. Nemri A, Saunders DG, Anderson C, Upadhyaya NM, Win J, Lawrence GJ, et al. (2014) The genome sequence and effector complement of the flax rust pathogen Melampsora lini. Front Plant Sci 5 : 98. doi: 10.3389/fpls.2014.00098 24715894
24. Cantu D, Segovia V, MacLean D, Bayles R, Chen X, Kamoun S, et al. (2013) Genome analyses of the wheat yellow (stripe) rust pathogen Puccinia striiformis f. sp. tritici reveal polymorphic and haustorial expressed secreted proteins as candidate effectors. BMC Genomics 14 : 270. doi: 10.1186/1471-2164-14-270 23607900
25. Sperschneider J, Ying H, Dodds PN, Gardiner DM, Upadhyaya NM, Singh KB, et al. (2014) Diversifying selection in the wheat stem rust fungus acts predominantly on pathogen-associated gene families and reveals candidate effectors. Front Plant Sci 5 : 372. doi: 10.3389/fpls.2014.00372 25225496
26. Guyon K, Balague C, Roby D, Raffaele S (2014) Secretome analysis reveals effector candidates associated with broad host range necrotrophy in the fungal plant pathogen Sclerotinia sclerotiorum. BMC Genomics 15 : 336. doi: 10.1186/1471-2164-15-336 24886033
27. Syme RA, Hane JK, Friesen TL, Oliver RP (2013) Resequencing and comparative genomics of Stagonospora nodorum: sectional gene absence and effector discovery. G3 (Bethesda) 3 : 959–969. doi: 10.1534/g3.112.004994 23589517
28. Rovenich H, Boshoven JC, Thomma BP (2014) Filamentous pathogen effector functions: of pathogens, hosts and microbiomes. Curr Opin Plant Biol 20 : 96–103. doi: 10.1016/j.pbi.2014.05.001 24879450
29. Liu T, Song T, Zhang X, Yuan H, Su L, Li W, et al. (2014) Unconventionally secreted effectors of two filamentous pathogens target plant salicylate biosynthesis. Nat Commun 5 : 4686. doi: 10.1038/ncomms5686 25156390
30. Sharon A, Shlezinger N (2013) Fungi infecting plants and animals: killers, non-killers, and cell death. PLoS Pathog 9: e1003517. doi: 10.1371/journal.ppat.1003517 24009499
31. Petersen TN, Brunak S, von Heijne G, Nielsen H (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8 : 785–786. doi: 10.1038/nmeth.1701 21959131
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek Neutrophil-Derived MMP-8 Drives AMPK-Dependent Matrix Destruction in Human Pulmonary TuberculosisČlánek Circumventing . Virulence by Early Recruitment of Neutrophils to the Lungs during Pneumonic PlagueČlánek Admixture in Humans of Two Divergent Populations Associated with Different Macaque Host SpeciesČlánek Human and Murine Clonal CD8+ T Cell Expansions Arise during Tuberculosis Because of TCR SelectionČlánek Selective Recruitment of Nuclear Factors to Productively Replicating Herpes Simplex Virus GenomesČlánek Fob1 and Fob2 Proteins Are Virulence Determinants of via Facilitating Iron Uptake from FerrioxamineČlánek Remembering MumpsČlánek Human Cytomegalovirus miR-UL112-3p Targets TLR2 and Modulates the TLR2/IRAK1/NFκB Signaling PathwayČlánek Induces the Premature Death of Human Neutrophils through the Action of Its Lipopolysaccharide
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2015 Číslo 5- Jak souvisí postcovidový syndrom s poškozením mozku?
- Měli bychom postcovidový syndrom léčit antidepresivy?
- Farmakovigilanční studie perorálních antivirotik indikovaných v léčbě COVID-19
- 10 bodů k očkování proti COVID-19: stanovisko České společnosti alergologie a klinické imunologie ČLS JEP
-
Všechny články tohoto čísla
- Parasites and Their Heterophagic Appetite for Disease
- The Elusive Role of the Prion Protein and the Mechanism of Toxicity in Prion Disease
- Intestinal Colonization Dynamics of
- Activation of Typhi-Specific Regulatory T Cells in Typhoid Disease in a Wild-Type . Typhi Challenge Model
- The Engineering of a Novel Ligand in gH Confers to HSV an Expanded Tropism Independent of gD Activation by Its Receptors
- Neutrophil-Derived MMP-8 Drives AMPK-Dependent Matrix Destruction in Human Pulmonary Tuberculosis
- Group Selection and Contribution of Minority Variants during Virus Adaptation Determines Virus Fitness and Phenotype
- Phosphatidic Acid Produced by Phospholipase D Promotes RNA Replication of a Plant RNA Virus
- A Ribonucleoprotein Complex Protects the Interleukin-6 mRNA from Degradation by Distinct Herpesviral Endonucleases
- Characterization of Transcriptional Responses to Different Aphid Species Reveals Genes that Contribute to Host Susceptibility and Non-host Resistance
- Circumventing . Virulence by Early Recruitment of Neutrophils to the Lungs during Pneumonic Plague
- Natural Killer Cell Sensing of Infected Cells Compensates for MyD88 Deficiency but Not IFN-I Activity in Resistance to Mouse Cytomegalovirus
- Manipulation of the Xanthophyll Cycle Increases Plant Susceptibility to
- Ly6C Monocytes Regulate Parasite-Induced Liver Inflammation by Inducing the Differentiation of Pathogenic Ly6C Monocytes into Macrophages
- Admixture in Humans of Two Divergent Populations Associated with Different Macaque Host Species
- Expression in the Fat Body Is Required in the Defense Against Parasitic Wasps in
- Experimental Evolution of an RNA Virus in Wild Birds: Evidence for Host-Dependent Impacts on Population Structure and Competitive Fitness
- Inhibition and Reversal of Microbial Attachment by an Antibody with Parasteric Activity against the FimH Adhesin of Uropathogenic .
- The EBNA-2 N-Terminal Transactivation Domain Folds into a Dimeric Structure Required for Target Gene Activation
- Human and Murine Clonal CD8+ T Cell Expansions Arise during Tuberculosis Because of TCR Selection
- The NLRP3 Inflammasome Is a Pathogen Sensor for Invasive via Activation of α5β1 Integrin at the Macrophage-Amebae Intercellular Junction
- Sequential Conformational Changes in the Morbillivirus Attachment Protein Initiate the Membrane Fusion Process
- A Two-Component DNA-Prime/Protein-Boost Vaccination Strategy for Eliciting Long-Term, Protective T Cell Immunity against
- cAMP-Signalling Regulates Gametocyte-Infected Erythrocyte Deformability Required for Malaria Parasite Transmission
- Response Regulator VxrB Controls Colonization and Regulates the Type VI Secretion System
- Evidence for a Novel Mechanism of Influenza Virus-Induced Type I Interferon Expression by a Defective RNA-Encoded Protein
- Dust Devil: The Life and Times of the Fungus That Causes Valley Fever
- TNF-α Induced by Hepatitis C Virus via TLR7 and TLR8 in Hepatocytes Supports Interferon Signaling via an Autocrine Mechanism
- The Recent Evolution of a Maternally-Inherited Endosymbiont of Ticks Led to the Emergence of the Q Fever Pathogen,
- L-Rhamnosylation of Wall Teichoic Acids Promotes Resistance to Antimicrobial Peptides by Delaying Interaction with the Membrane
- Rapid Sequestration of by Neutrophils Contributes to the Development of Chronic Lesion
- Selective Recruitment of Nuclear Factors to Productively Replicating Herpes Simplex Virus Genomes
- The Expression of Functional Vpx during Pathogenic SIVmac Infections of Rhesus Macaques Suppresses SAMHD1 in CD4 Memory T Cells
- Fob1 and Fob2 Proteins Are Virulence Determinants of via Facilitating Iron Uptake from Ferrioxamine
- TRAF1 Coordinates Polyubiquitin Signaling to Enhance Epstein-Barr Virus LMP1-Mediated Growth and Survival Pathway Activation
- Vaccine-Elicited Tier 2 HIV-1 Neutralizing Antibodies Bind to Quaternary Epitopes Involving Glycan-Deficient Patches Proximal to the CD4 Binding Site
- Remembering Mumps
- The Role of Horizontal Gene Transfer in the Evolution of the Oomycetes
- Advances and Challenges in Computational Prediction of Effectors from Plant Pathogenic Fungi
- Investigating Fungal Outbreaks in the 21st Century
- Systems Biology for Biologists
- How Does the Dinoflagellate Parasite Outsmart the Immune System of Its Crustacean Hosts?
- FCRL5 Delineates Functionally Impaired Memory B Cells Associated with Exposure
- Phospholipase D1 Couples CD4 T Cell Activation to c-Myc-Dependent Deoxyribonucleotide Pool Expansion and HIV-1 Replication
- Influenza A Virus on Oceanic Islands: Host and Viral Diversity in Seabirds in the Western Indian Ocean
- Geometric Constraints Dominate the Antigenic Evolution of Influenza H3N2 Hemagglutinin
- Widespread Recombination, Reassortment, and Transmission of Unbalanced Compound Viral Genotypes in Natural Arenavirus Infections
- Gammaherpesvirus Co-infection with Malaria Suppresses Anti-parasitic Humoral Immunity
- A Single Protein S-acyl Transferase Acts through Diverse Substrates to Determine Cryptococcal Morphology, Stress Tolerance, and Pathogenic Outcome
- Survives with a Minimal Peptidoglycan Synthesis Machine but Sacrifices Virulence and Antibiotic Resistance
- Mechanisms of Stage-Transcending Protection Following Immunization of Mice with Late Liver Stage-Arresting Genetically Attenuated Malaria Parasites
- The Myelin and Lymphocyte Protein MAL Is Required for Binding and Activity of ε-Toxin
- Genome-Wide Identification of the Target Genes of AP2-O, a AP2-Family Transcription Factor
- An Atypical Mitochondrial Carrier That Mediates Drug Action in
- Human Cytomegalovirus miR-UL112-3p Targets TLR2 and Modulates the TLR2/IRAK1/NFκB Signaling Pathway
- Helminth Infection and Commensal Microbiota Drive Early IL-10 Production in the Skin by CD4 T Cells That Are Functionally Suppressive
- Circulating Pneumolysin Is a Potent Inducer of Cardiac Injury during Pneumococcal Infection
- ExoT Induces Atypical Anoikis Apoptosis in Target Host Cells by Transforming Crk Adaptor Protein into a Cytotoxin
- Discovery of a Small Non-AUG-Initiated ORF in Poleroviruses and Luteoviruses That Is Required for Long-Distance Movement
- Induces the Premature Death of Human Neutrophils through the Action of Its Lipopolysaccharide
- Varicella Viruses Inhibit Interferon-Stimulated JAK-STAT Signaling through Multiple Mechanisms
- Paradoxical Immune Responses in Non-HIV Cryptococcal Meningitis
- Recovery of Recombinant Crimean Congo Hemorrhagic Fever Virus Reveals a Function for Non-structural Glycoproteins Cleavage by Furin
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Human Cytomegalovirus miR-UL112-3p Targets TLR2 and Modulates the TLR2/IRAK1/NFκB Signaling Pathway
- Paradoxical Immune Responses in Non-HIV Cryptococcal Meningitis
- Expression in the Fat Body Is Required in the Defense Against Parasitic Wasps in
- Survives with a Minimal Peptidoglycan Synthesis Machine but Sacrifices Virulence and Antibiotic Resistance
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



