-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaNLR-Associating Transcription Factor bHLH84 and Its Paralogs Function Redundantly in Plant Immunity
In plants and animals, NLR immune receptors are utilized to detect pathogen-derived molecules and activate immunity. However, the mechanisms of plant NLR activation remain unclear. Here, we report on bHLH84, which functions as a transcriptional activator. Simultaneously knocking out three closely related bHLH paralogs partially suppresses the autoimmunity of snc1 and compromises RPS4-mediated defense, while overexpression of these close paralogs renders strong autoimmunity, suggesting functional redundancy in the gene family. In planta co-immunoprecipitation revealed interactions between not only bHLH84 and SNC1, but also bHLH84 and RPS4. Therefore bHLH84 family transcription factors associate with these NLRs to activate defense responses, enabling potentially faster and more robust transcriptional reprogramming upon pathogen recognition.
Published in the journal: . PLoS Pathog 10(8): e32767. doi:10.1371/journal.ppat.1004312
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1004312Summary
In plants and animals, NLR immune receptors are utilized to detect pathogen-derived molecules and activate immunity. However, the mechanisms of plant NLR activation remain unclear. Here, we report on bHLH84, which functions as a transcriptional activator. Simultaneously knocking out three closely related bHLH paralogs partially suppresses the autoimmunity of snc1 and compromises RPS4-mediated defense, while overexpression of these close paralogs renders strong autoimmunity, suggesting functional redundancy in the gene family. In planta co-immunoprecipitation revealed interactions between not only bHLH84 and SNC1, but also bHLH84 and RPS4. Therefore bHLH84 family transcription factors associate with these NLRs to activate defense responses, enabling potentially faster and more robust transcriptional reprogramming upon pathogen recognition.
Introduction
Plants have evolved a sophisticated immune system to fight against invading microbial pathogens that threaten their normal growth and development. Plant immunity is in part mediated by resistance (R) proteins that recognize pathogen proteins known as effectors [1]–[3]. The majority of R proteins are NLR receptors that contain leucine-rich repeats (LRRs) at the C-terminus, a central nucleotide-binding site (NBS) and either a Toll/Interleukin-1 receptor (TIR) or a coiled-coil (CC) domain at the N-terminus [4]. In Arabidopsis, genetically downstream of the R proteins are the EDS1 (ENHANCED DISEASE SUSCEPTIBILITY 1)/PAD4 (PHYTOALEXIN DEFICIENT 4)/SAG101 (SENESCENCE-ASSOCIATED GENE101) complex and NDR1 (NON-RACE-RESISTANCE 1), which mainly mediate TIR-NB-LRR or CC-NB-LRR triggered defense responses, respectively [5]–[8].
While the mechanisms underlying effector recognition by R proteins have been intensively studied, little is known about the post-recognition events leading to defense activation. Recently, it has been shown that the nuclear pool of certain R proteins, including MLA10 (MILDEW A LOCUS 10) in barley, N in tobacco, Pb1 (Panicle blast 1) in rice, and RPS4 (RESISTANT TO P.SYRINGAE 4), RRS1 (RESISTANT TO RALSTONIA SOLANACEARUM 1) and SNC1 (SUPPRESSOR OF NPR1-1, CONSTITUTIVE1) in Arabidopsis, is important for the activation of defense responses [9]–[14]. The latest discoveries on the interactions between some of these R proteins and their associating transcription factors (TFs) further shed light on the activation mechanism of nuclear R proteins. For example, MLA10 interacts with WRKY TFs to de-repress PAMP (PATHOGEN-ASSOCIATED MOLECULAR PATTERN) triggered basal defense [9]. The active state of MLA10 can also release MYB6 (MYB DOMAIN PROTEIN 6) from WRKY suppression and promote its binding to cis-elements to initiate defense responses [15]. CC-type NLR Pb1 in rice interacts with WRKY45 and this interaction is believed to protect the TF from proteasomal degradation in the nucleus [16]. In addition, SNC1 associates with transcriptional co-repressor TPR1 (TOPLESS RELATED 1) to negatively regulate the expression of known defense suppressors, thereby activating plant immunity [17]. Lately, studies on N in tobacco showed that it is able to associate with the TF SPL6 (SQUAMOSA PROMOTER BINDING PROTEIN-LIKE 6) upon effector recognition [18]. From these data, it has been hypothesized that some NLRs associate with TFs inside the nucleus to directly participate in transcriptional reprogramming to regulate downstream defense responses.
In Arabidopsis, the gain-of-function NLR mutant snc1 constitutively expresses PATHOGENESIS RELATED (PR) defense marker genes and exhibits enhanced disease resistance against virulent bacteria Pseudomonas syringae pv. maculicola (P.s.m.) ES4326 and oomycete Hyaloperonospora arabidopsidis (H.a.) Noco2 [19], [20]. As snc1 displays strong autoimmune phenotypes while remaining fully fertile, it has become a useful tool for dissecting NLR mediated resistance. Forward genetic screens designed to isolate positive regulators of immunity were conducted in the snc1 background and over a dozen Modifier of snc1 (MOS) genes have been identified. Characterizations of the MOS genes and their encoded protein products have revealed complicated regulatory events surrounding snc1 mediated autoimmunity, which include nucleocytoplasmic trafficking, RNA processing, protein modification and transcriptional regulation [21], [22]. However, genetic redundancy and lethality may have prevented some essential positive regulators from being discovered through forward genetic approaches. Here, we employed a targeted reverse genetic screen to search for candidate TFs participating in the regulation of snc1-mediated defense. One basic Helix-loop-Helix (bHLH) type TF, which is a putative transcriptional activator, was isolated from the screen and found to be able to associate with NLRs to activate immunity.
Results
A targeted reverse genetic screen
Previously, SNC1 was found to participate directly in transcriptional reprogramming with TPR/MOS10 repressor proteins that do not directly bind DNA [17]. We did not find a DNA-binding TF that functions together with SNC1 from the MOS forward genetic screens, suggesting that multiple TFs may function redundantly in snc1-mediated immunity. To search for novel TFs regulating plant immunity, a reverse genetic screen was employed. As UV irradiation has been shown to induce resistance to pathogens and to induce transcription of defense related genes [23]–[25], we selected 36 putative TFs which show >1.7-fold enhanced expression level upon UV treatment based on publically available microarray data from The Arabidopsis Information Resource (Table S1). The genomic sequences of these genes were cloned into a binary vector pCambia1305 containing C-terminus GFP and HA double tags. Using the floral dip method [26], overexpression transgenic plants in snc1 and Col-0 backgrounds were generated. From the primary screen, we searched for transformants either suppressing or enhancing the dwarf morphology of snc1 or causing dwarfism in Col-0 background. Transgenic plants exhibiting heritable altered morphology were subject to a secondary screen, where altered resistance was examined using a Hyaloperonospora arabidopsidis (H.a.) Noco2 infection assay. Screening data for these candidate TFs are summarized in Table S1.
From the screen, we identified several TFs that displayed phenotypes in only snc1 or Col-0 background, but not in both when overexpressed (Table S1). However, overexpression of three TFs, At2g31230, At2g14760 or At5g61590, resulted in stunted growth in both the snc1 and Col-0 backgrounds (Table S1, Figure 1A and 1B). We selected two TFs with the strongest phenotypes for further analysis. At2g14760 encodes bHLH84, a predicted basic helix-loop-helix TF, while At5g61590 encodes ERF107, which belongs to the ethylene-response-factor (ERF) TF family.
Fig. 1. Characterization of bHLH84 and ERF107 overexpression (OX) lines. 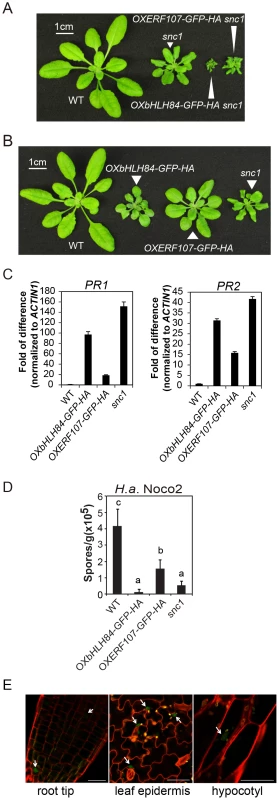
A. Morphology of wild type (WT), snc1 and representative transgenic lines of OXbHLH84-GFP-HA and OXERF107-GFP-HA in snc1 background. For both A and B, plants were grown on soil for four weeks before the pictures were taken. B. Morphology of WT, OXbHLH84-GFP-HA, OXERF107-GFP-HA and snc1 plants. Genes were overexpressed in Col-0 WT background. C. Relative PR1 and PR2 gene expression in WT, OXbHLH84-GFP-HA, and OXERF107-GFP-HA plants as determined by real-time PCR. Total RNA samples were extracted from 12-day-old plants grown on solid MS medium and reverse transcribed to cDNA using Superscript II reverse transcriptase. All genotypes of plants were grown simultaneously on the same large petri plate. The expression of PR1 and PR2 was normalized to that of ACTIN1, and the value of each genotype was compared to that of WT. D. Quantification of H.a. Noco2 sporulation on WT, OXbHLH84-GFP-HA, OXERF107-GFP-HA and snc1 plants. 2.5-week-old plants were inoculated with H.a. Noco2 at a concentration of 105 spores/mL water. Oomycete spores on the leaf surface were quantified seven days after inoculation. Bars represent means of four replicates ± SD. Variant letters represent statistical differences among the indicated genotypes as analyzed by StatsDirect software (p<0.05). E. Detection of GFP green fluorescence in OXbHLH84-GFP-HA seedlings using confocal fluorescence microscopy. The pictures were taken from root tip and leaf epidermis of 10-day-old plants and hypocotyl of 5-day-old seedlings. Cell walls were visualized in red using propidium iodide (PI) staining. Arrows point to the nuclei with GFP signal. Scale bar = 20 µm. Characterization of the OXbHLH84-GFP-HA and OXERF107-GFP-HA lines
To further explore the functions of bHLH84 and ERF107 in plant immunity, we isolated homozygous overexpression transgenic lines in Col-0 background. As shown in Figure 1B, both OXbHLH84-GFP-HA and OXERF107-GFP-HA plants exhibited dwarf morphology compared with WT plants. We further examined defense marker PR gene expression in these transgenic plants using real-time PCR. As shown in Figure 1C, the expression of both PR1 and PR2 was significantly up-regulated, with about 100 - and 35 - fold changes, respectively, in OXbHLH84-GFP-HA, indicating that the defense responses were constitutively activated. In OXERF107-GFP-HA transgenic plants, both PR1 and PR2 were around 15-fold up-regulated. Consistent with PR gene expression, resistance against virulent pathogen H.a. Noco2 was enhanced in both OXbHLH84-GFP-HA and OXERF107-GFP-HA plants (Figure 1D). As OXbHLH84-GFP-HA plants displayed more severe immune phenotypes than OXERF107-GFP-HA plants, we chose to focus solely on the functional study of bHLH84. Consistent with its predicted TF function, bHLH84-GFP-HA fluorescence was detected in the nuclei when the OXbHLH84-GFP-HA seedlings were examined by confocal fluorescence microscopy (Figure 1E).
bHLH84 functions as a transcriptional activator
To further investigate how bHLH84 regulates plant immunity, we tested whether it is a bona fide transcription factor by conducting a previously established protoplast transcription activity transient assay [27]. In this assay, the β-glucuronidase (GUS) reporter gene is driven by 2×Gal4 DNA-binding sites (DBS). Co-transformation of bHLH84 fused with the Gal4 DNA-binding domain (DBD) together with the reporter constructs in Arabidopsis mesophyll protoplasts resulted in drastically enhanced GUS expression (Figure 2A) compared to the control transfection, suggesting that bHLH84 functions as a transcriptional activator.
Fig. 2. Mutant analysis of transcriptional activator bHLH84 and its paralogs. 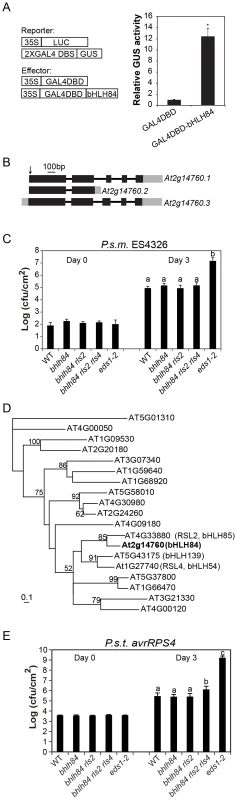
A. Relative GUS activities were assayed using Arabidopsis mesophyll protoplasts cotransfected with the reporter and the indicated effector constructs. The 35S-driven luciferase (LUC) report construct served as internal transfection control. * indicates statistical significance analyzed by unpaired student's t-test (p<0.01). B. Gene structure of bHLH84 (At2g14760). There are three different splice variants of bHLH84. Boxes indicate exons and lines indicate introns. Grey regions show the UTR regions. The position of the T-DNA insertion in bhlh84 is indicated with an arrow. C. Bacterial growth of P.s.m. ES4326 on four-week-old plants of the indicated genotypes at 0 (Day 0) and 3 days (Day 3) post-inoculation, with bacterial inoculum of OD600 = 0.0001. eds1-2 plants served as enhanced disease susceptibility (EDS) control. Bars represent means of five replicates ±SD. Statistical difference among the indicated genotypes were analyzed with a StatsDirect software (p<0.001). D. A phylogenetic tree of bHLH84 and its close paralogs in Arabidopsis. The amino acid sequences of bHLH84 and its paralogs in Arabidopsis were used to generate the tree, using a method as previously described [61]. E. Bacterial growth of the avirulent pathogen P.s.t. avrRPS4 on plants of the indicated genotype. Four-week-old plants were infiltrated with a bacterial suspension at OD600 = 0.002. Leaf discs within the infected area were taken at day 0 and day 3 to measure the bacterial growth. Bars represent means of five replicates ±SD. Statistical differences were analyzed by one-way analysis using StatsDirect software. Variant letters represent statistical difference among the indicated genotype. (p<0.05). For each experiment, five plants with two leaves per plant were inoculated. Two leaf discs from each plant were assayed as one replicate. Similar results were observed in four independent trials. Knocking out bHLH84 and its two close paralogs does not compromise basal immunity while attenuating RPS4-mediated defense response
bHLH TFs constitute one of the largest TF families in Arabidopsis, with 147 members including bHLH84 [28]. bHLH84 has three alternatively spliced variants according to available expressed sequence tag (EST) data (Figure 2B). Based on sequence analysis, At2g14760.2 encodes a truncated protein without the C-terminal bHLH DNA binding domain, while the other two variants encode full-length proteins [28]. However, when the coding region of bHLH84 was amplified from cDNA of WT plants and sequenced, only At2g14760.1 was observed, suggesting that At2g14760.1 is the dominantly expressed version.
To further investigate the contribution of bHLH84 in plant immunity, knock-out analysis of bHLH84 was carried out. A T-DNA allele of bHLH84 (SALK_064296) was obtained from the Arabidopsis Biological Resource Centre (ABRC). As shown in Figure 2B, the T-DNA inserts in the first exon of At2g14760.1. As a consequence, the expression of bHLH84 was abolished (Figure S1A). SALK_064296 was thus assigned as bhlh84. When bhlh84 leaves were challenged with virulent bacterial pathogen Pseudomonas syringae pv maculicola (P.s.m.) ES4326, they exhibited similar bacterial growth as WT (Figure 2C), indicating that the immune response is not compromised in the knock-out mutant.
To investigate whether genetic redundancy masks the function of bHLH84, we carried out a phylogenetic analysis of bHLH84 and its paralogs. As RSL2 (ROOT HAIR DEFECTIVE 6-LIKE 2) is the closest paralog of bHLH84 (Figure 2D; [29]), a T-DNA knock-out line for this gene, SALK_048849, was obtained from ABRC. As shown in Figure S1B, no expression of RSL2 was detectable in SALK_048849, which was named as rsl2. Double mutant bhlh84 rsl2 was created and subjected to pathogen infection experiments. As shown in Figure 2C, the bhlh84 rsl2 double mutant did not exhibit resistance defects either. As RSL4 (ROOT HAIR DEFECTIVE 6-LIKE 4) is functionally redundant with RSL2 in regulating root hair growth [29], we further created the triple mutant by crossing bhlh84 rsl2 with rsl4 rsl2, which was characterized by Yi et al., 2010 [29]. The triple mutant bhlh84 rsl2 rsl4 still did not exhibit obvious defects upon infection with P.s.m. ES4326 compared to WT plants (Figure 2C), indicating that knocking out bHLH84 and its two paralogs does not compromise basal defense responses. Since no good T-DNA mutant line was available for bHLH139, we were not able to test higher level of redundancy using knockout approach.
To further examine the contribution of these TFs in specific R protein mediated immunity, we challenged single, double and triple mutant plants with Pseudomonas syringae pv tomato (P.s.t.) carrying either avrRPS4 or hopA1, which are effectors recognized by TIR-NB-LRR proteins RPS4 and RPS6, respectively. As shown in Figure 2E, significantly more P.s.t. avrRPS4 growth was observed in bhlh84 rsl2rsl4 triple mutant plant, while no detectable difference was observed when the TF mutants were challenged with P.s.t. hopA1 (Figure S2), suggesting that these bHLH TFs contribute redundantly to RPS4-mediated immunity.
Simultaneously knocking out bHLH84, RSL2 and RSL4 partially suppresses the autoimmunity of snc1
To investigate the biological function of bHLH84 and its paralogs in snc1-mediated immunity, we crossed bhlh84 rsl2 with snc1 and isolated triple mutant snc1 bhlh84 rsl2. The dwarf phenotype of snc1 was not suppressed in the triple mutant (Figure 3A). We further crossed snc1 bhlh84 rsl2 with rsl4 rsl2 [29] and isolated quadruple mutant snc1 bhlh84 rsl2 rsl4 from the F2 generation by genotyping bhlh84, rsl4 and snc1 loci. The quadruple mutant plants were significantly larger than those of snc1 (Figure 3A). Consistent with the morphological suppression, the expression of PR1 and PR2 in the quadruple mutant was significantly decreased compared to snc1 plants while only slight reduction was observed in the triple mutant (Figure 3B). In addition, when the quadruple mutant seedlings were challenged with H.a. Noco2 and P.s.m. ES4326, more pathogen growth was observed compared to snc1, although the resistance was not restored to wild type levels (Figure 3C and 3D). Taken together, the bhlh84 rsl2 rsl4 triple mutant partially suppresses snc1, suggesting that bHLH84 and its paralogs are functionally redundant and required for the autoimmunity of snc1.
Fig. 3. bhlh84 rsl2 rsl4 partially suppresses the autoimmunity of snc1. 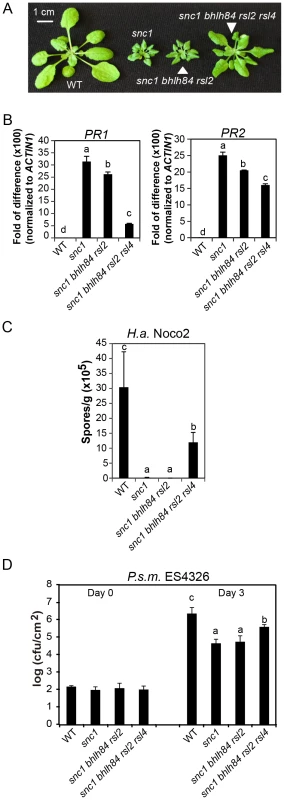
A. Morphology of WT, snc1, snc1 bhlh84 rsl2, and snc1 bhlh84 rsl2 rsl4 quadruple mutant. B. Relative PR1 and PR2 gene expression in the indicated genotypes was determined by real-time PCR. The experiment was carried out as in Figure 1C except that the plants used were two-week-old. Statistical differences among the indicated genotypes were analyzed by a StatsDirect software, which are indicated using different letters (p<0.05). C. Growth of H.a. Noco2 on the indicated genotypes was measured and analyzed using a similar method as used in Figure 1D, except that spores were collected at 8 days post inoculation. D. Bacterial growth of P.s m. ES4326 on four-week-old plants of the indicated genotypes at 0 and 3 days post-inoculation with bacterial inoculum of OD600 = 0.001. Bars represent means of five replicates ±SD. Statistically different groups were analyzed by StatsDirect and labelled by different letters (p<0.005). When we further isolated snc1 rsl2 rsl4 (Figure S3A and S3B), the triple mutant was slightly larger than snc1. Since snc1 bhlh84 rsl2 plants were indistinguishable from snc1 in size, it can thus be concluded that these three TFs are not equally redundant; RSL4 seems to play a slightly larger role than bHLH84 in snc1-mediated autoimmunity.
Overexpression of bHLH84, RSL2, or RSL4 exhibits extreme dwarfism likely due to autoimmunity
To further test the redundant roles of bHLH84 and its paralogs, we overexpressed bHLH84, RSL4 or RSL2 in Col-0 by transforming plants with the coding sequence of each gene without any epitope tags under the control of the 35S promoter. When screening T1 populations, multiple plants with extremely dwarf morphology were observed for each genotype (Figure 4). Intriguingly, plants of intermediate sizes were observed in the transgenic lines overexpressing bHLH84, while the majority of the plants overexpressing RSL4 or RSL2 were tiny and gradually perished, presumably as a result of extreme autoimmunity. The phenotypic similarity in these overexpression progeny further supports the functional redundancy among these three TFs in regulating plant immunity.
Fig. 4. bHLH84, RSL2 and RSL4 all exhibit dwarfism when overexpressed. 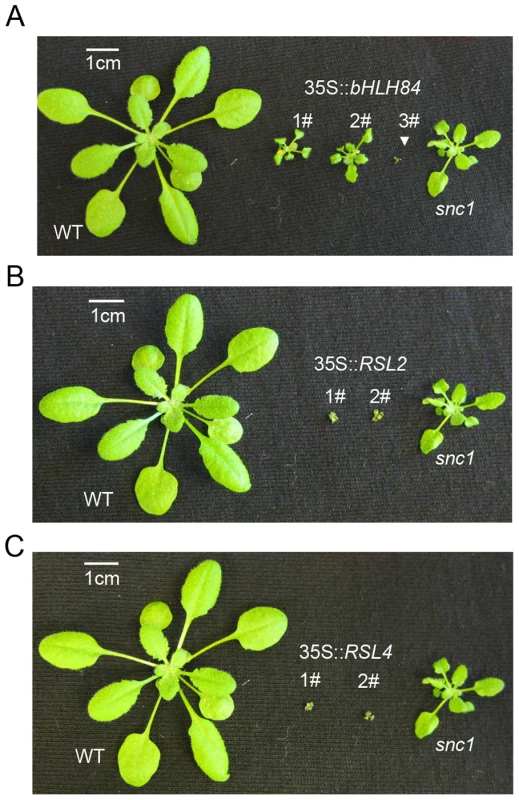
A. Morphology of WT, three representative T1 transgenic plants of 35S:bHLH84 in Col-0, and snc1. B. Morphology of WT, two representative T1 transgenic plants of 35S:RSL2 in Col-0, and snc1. C. Morphology of WT, two representative T1 transgenic plants of 35S:RSL4 in Col-0, and snc1. All pictures were taken from four-week-old soil-grown plants. SNC1 contributes to the constitutive activation of defense responses in OXbHLH84-GFP-HA transgenic plants
As with snc1, the dwarf morphology of OXbHLH84-GFP-HA plants was largely suppressed when grown at 28°C (Figure S4) [30]. This observation led us to ask whether SNC1 is required for the autoimmunity of OXbHLH84-GFP-HA. As shown in Figure 5A, the snc1-r1 allele (a loss-of-function allele of SNC1 in which 8 bp of the first exon of SNC1 is deleted from fast neutron mutagenesis; [20]) could largely suppress the dwarf morphology of OXbHLH84-GFP-HA. Consistent with the observed morphological suppression, defense response phenotypes conferred by OXbHLH84-GFP-HA, including up-regulation of PR gene expression and resistance to P.s.m. ES4326 and H.a. Noco2, were significantly suppressed by snc1-r1 (Figures 5B, 5C and 5D), indicating that a functional SNC1 is indispensable for the effects of bHLH84 overexpression. As CPR1 (CONSTITUTIVE EXRPRESSER OF PR GENES 1) targets SNC1 for degradation [31], we crossed OXbHLH84-GFP-HA with plants overexpressing CPR1 (OXCPR1). The dwarf morphology and enhanced resistance of OXbHLH84-GFP-HA were largely suppressed (Figure 5), providing further support that SNC1 contributes to the autoimmune phenotypes associated with OXbHLH84-GFP-HA. In addition, the bHLH84-GFP-HA protein level in snc1-r1 or OXCPR1 background was not changed (Figure S5), suggesting that SNC1 does not affect bHLH84 protein accumulation.
Fig. 5. Epistasis analysis between OXbHLH84-GFP-HA and snc1-r1, eds1-2, sid2-2, OXCPR1, and ndr1-1. 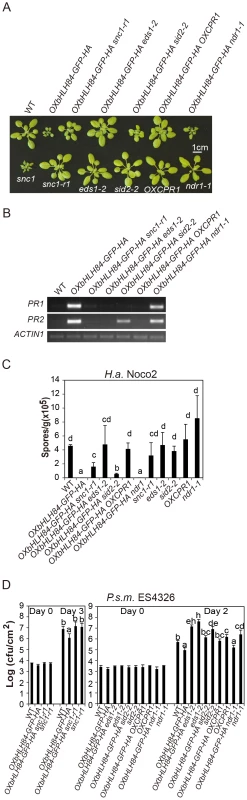
A. Morphology of four-week-old soil-grown plants of the indicated genotypes. B. PR1 and PR2 gene expression of the indicated genotypes as determined by RT-PCR. The experiment was carried out as in Figure 1C except that gene expression was determined by RT- PCR. C. Quantification of H.a. Noco2 sporulation on the indicated genotypes using the same method as in Figure 1D (p<0.05). D. Bacterial growth of P.s.m. ES4326 on four-week-old plants of the indicated genotypes at 0 and 2/3 days post-inoculation with bacterial inoculum of OD600 = 0.001. Bars represent means of five replicates ±SD. The same statistical analysis method as described in Figure 3D was used (p<0.05). The experiment was repeated three times with similar results. Epistasis analysis reveals that constitutive activation of defense responses in OXbHLH84-GFP-HA is EDS1- and SID2- dependent and NDR1- independent
To further dissect the function of bHLH84 in plant defense pathways, OXbHLH84-GFP-HA was crossed with various mutants of key components in plant immunity, including eds1-2, sid2-2, and ndr1-1 [8], [32], [33]. As shown in Figure 5A, eds1-2 and sid2-2 could fully and partially suppress the morphology of OXbHLH84-GFP-HA in terms of leaf shape and plant size, respectively, while ndr1-1 had little effect. The enhanced PR gene expression and resistance to H.a. Noco2 and P.s.m. ES4326 were fully suppressed by eds1-2 and partially by sid2-2 (Figure 5B, 5C and 5D), indicating that EDS1 and SA are required for the autoimmunity in OXbHLH84-GFP-HA. In contrast, ndr1-1 was not able to suppress the enhanced PR gene expression, H.a. Noco2 and P.s.m. ES4326 resistance conferred by OXbHLH84-GFP-HA, indicating that the constitutive activation of defense responses in OXbHLH84-GFP-HA is NDR1-independent.
bHLH84 does not directly regulate SNC1 transcription
As SNC1 is required for the constitutive activation of the defense responses of OXbHLH84-GFP-HA plants, we asked whether bHLH84 could directly regulate SNC1 transcription. We observed that the transcription and protein levels of SNC1 in OXbHLH84-GFP-HA plants were slightly higher than in WT (Figure S6). However, this up-regulation of SNC1 is probably due to the positive feed-back effect resulting from the high SA in the autoimmune transgenic plants [34]. To avoid interference from the feed-back up-regulation of SNC1, we used OXbHLH84-GFP-HA eds1-2 plants to examine SNC1 transcription level. Real-time PCR showed that no significant change in SNC1 transcription was detected in OXbHLH84-GFP-HA eds1-2 compared to eds1-2 control plants (Figure 6A). As a consequence, the SNC1 protein level in OXbHLH84-GFP-HA eds1-2 was similar to that of eds1-2 (Figure 6B). In addition, we tested the transcript levels of selected R genes including RPS6, RPS4, RPP2, RPP4, RPS2, RPS5, and RPM1 in the OXbHLH84-GFP-HA eds1-2 background. Similar to SNC1, none of the tested R genes showed over 1.2-fold transcriptional changes when compared to eds1-2 (Figure S7A). In addition, no significant up-regulation of R genes was observed in OXbHLH84-GFP-HA snc1-r1 double mutant compared to snc1-r1 control plants (Figure S7B). Taken together, bHLH84 does not seem to participate in the direct transcriptional regulation of SNC1 or other tested R genes, unless bHLH84 recruits both EDS1 and SNC1 for this regulation.
Fig. 6. bHLH84 does not regulate SNC1 transcription. 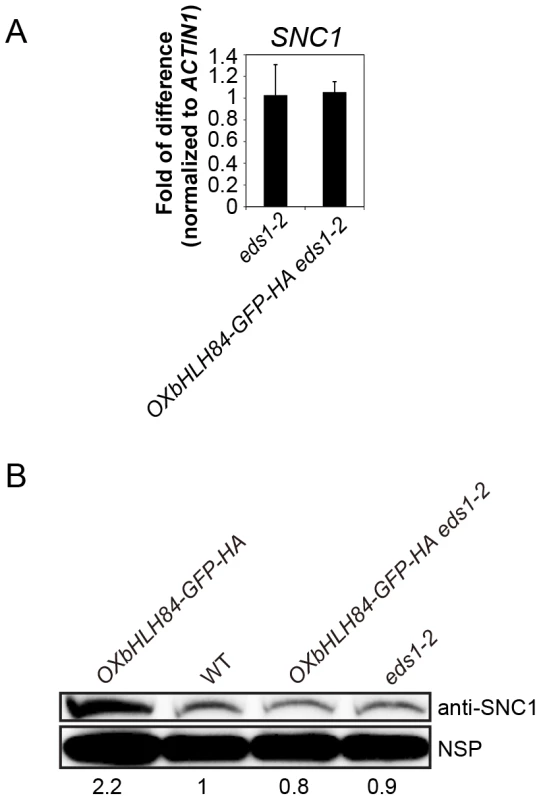
A. SNC1 expression level in eds1-2 and OXbHLH84-GFP-HA eds1-2 plants as determined by real-time PCR. SNC1 transcripts were amplified from cDNA, with primers specific to SNC1 cDNA by real-time PCR. The value of SNC1 expression for each genotype was normalized to that of ACTIN1. The value of each genotype was normalized to that of eds1-2. Bars represent means of three replicates ±SD. Statistical differences among the indicated genotypes were analyzed by StatsDirect software, which are represented using different letters (p<0.05). B. SNC1 protein levels in OXbHLH84-GFP-HA, WT, OXbHLH84-GFP-HA eds1-2 and eds1-2 plants. Total protein was extracted from leaves of four-week-old soil-grown plants. SNC1 protein levels were examined by immunoblot using an anti-SNC1 antibody [62]. NSP, a non-specific protein band that was used as internal loading control. Image J was used to quantify the band intensities of SNC1 and NSP. The band intensity of SNC1 relative to NSP was calculated for each genotype and normalized to the value of WT. The amount of SNC1 relative to WT is shown at the bottom of each genotype. bHLH84 interacts with SNC1 and RPS4 in planta
As the dependence of OXbHLH84-GFP-HA on a functional SNC1 and the partial suppression of snc1 by bhlh84 rsl2 rsl4 resembles the genetic interactions between SNC1 and TPR1/MOS10, and SNC1 interacts with TPR1 [17], we further tested whether bHLH84 associates with SNC1. We attempted a nuclear co-immunoprecipitation (co-IP) experiment using OXbHLH84-GFP-HA transgenic plants, which carry C-terminal GFP and HA double tags. Unfortunately, we were unable to detect the bait after immunoprecipitation in the elution, while all the proteins were found in the flow-through fraction (Figure S8A). As an alternate approach, we transformed Arabidopsis plants with a construct expressing bHLH84 under its native promoter and containing an N-terminal GFP tag. The protein produced was functional, as the transgenic plants resembled the original OXbHLH84-GFP-HA plants (Figure S8B). However, when they were used for co-IP with anti-GFP beads, the bait still could not be pulled down (Figure S8C). The inability of bHLH84 to be pulled down using immunoprecipitation could be due to unknown structural complexity of the protein. Since we were not able to carry out a co-IP experiment with bHLH84 as bait using epitope-tagged bHLH84 transgenic plants, we decided to examine the interaction between SNC1 and bHLH84 using the Nicotiana benthamiana transient expression system [35]. Interestingly, when both proteins were expressed in N. benthamiana leaves, we consistently observed a faster hypersensitive response (HR), which was obvious a few hours earlier compared to when SNC1-FLAG was expressed with the control vector (Figure S9A and S9B). This was further confirmed by the ion leakage analysis of the infiltrated leaves (Figure 7A). Both proteins were expressed efficiently in N. benthamiana (Figure 7B). When co-immunoprecipitation was carried out, SNC1-FLAG could specifically pull down bHLH84-HA, but not an unrelated nuclear protein MAC5A-HA (Figure 7C, [36]), indicating that bHLH84 can interact with SNC1 in planta.
Fig. 7. bHLH84 interacts with SNC1 or RPS4 in planta. 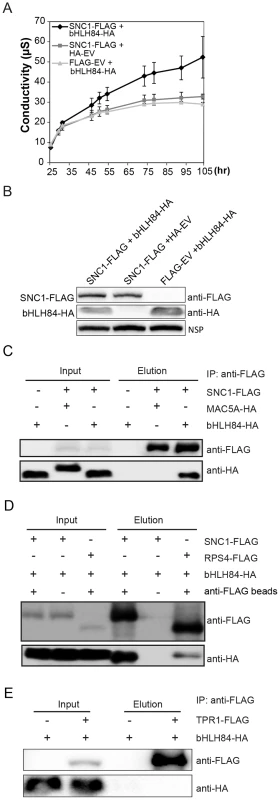
A. Ion leakage as measured in N. benthamiana leaves infiltrated by Agrobacterium containing the indicated constructs. Agrobacterium containing pCambia1300-35S -FLAG or pCambia1300-35S- HA empty vectors (EV) served as control. Leaf disc samples were collected at different time points post infiltration. Bars represent means of three replicates ±SD. For each replicate, 12 leaf discs were used. B. Protein expression of bHLH84-HA and SNC1-FLAG in N.benthamiana leaves with the indicated infiltration. NSP, signals from a non-specific protein band that served as loading control. C. bHLH84-HA co-immunoprecipitates with SNC1-FLAG when co-expressed in N. benthamiana leaves. Four-week-old N. benthamiana leaves were co-infiltrated with Agrobacterium containing pCambia1300-35S-bHLH84-HA and pCambia1300-35S-SNC1-FLAG at OD600 = 0.2 for each strain. N. benthamiana leaves co-infiltrated with Agrobacterium containing pCambia1300-35S-FLAG EV and pCambia1300-35S-bHLH84-HA or pCambia1300-35S-SNC1-FLAG and pGreen229-MAC5A-HA [36] were used as negative controls. 1.5 g of N. benthamiana leaf tissues for each infiltration were collected 36 hours post-inoculation and the protein extracts from the leaves were subjected to IP using anti-FLAG beads. Input indicates protein sample before IP. Elution indicates protein sample competitively eluted from the beads by 3×FLAG peptides. D. bHLH84-HA co-immunoprecipitates with RPS4-FLAG when co-expressed in N. benthamiana leaves. A similar experimental procedure was carried out as in Figure 7C. Protein sample of SNC1-FLAG and bHLH84-HA incubated with anti-FLAG beads served as positive control while proteins incubated with protein A agarose beads without the conjugated anti-FLAG antibody served as negative control. E. bHLH84-HA did not co-immunoprecipitate with TPR1-FLAG when co-expressed in N. benthamiana leaves. A similar experimental procedure was carried out as in Figure 7C except that pCambia1305-ProTPR1-TPR1-FLAG was used instead of pCambia1300-35S-SNC1-FLAG. As bHLH84 is able to interact with SNC1 in planta, we further examined the interaction specificity between bHLH84-HA and other R proteins by conducting co-IP of bHLH84-HA with either RPS4-FLAG, RPS2-FLAG or RPS6-FLAG. As shown in Figure 7D, RPS4-FLAG could also immunoprecipitate bHLH84-HA, although not as efficiently as SNC1-FLAG. However, RPS2-FLAG or RPS6-FLAG could not pull down bHLH84-HA (Figure S10). Taken together, bHLH84-HA can specifically interact with SNC1-FLAG or RPS4-FLAG in planta.
SNC1 was previously shown to interact with transcriptional co-repressor TPR1, which does not contain a DNA binding domain [17]. Additionally, the SNC1-dependent phenotypes observed upon overexpressing bHLH84 are similar to those observed when TPR1 is overexpressed. We therefore asked whether bHLH84 interacts with TPR1. As shown in Figure 7E, bHLH84-HA could not be pulled down by TPR1-FLAG, indicating that bHLH84 does not interact with TPR1 in planta. In addition, when we co-expressed SNC1-FLAG, bHLH84-HA and TPR1-HA in N. benthamiana, SNC1-FLAG was able to pull down both TPR1-HA and bHLH84-HA (Figure S11). The IP efficiency of TPR1-HA by SNC1-FLAG with all three proteins expressed was comparable to that with only TPR1-HA and SNC1-FLAG expressed. On the other hand, the IP efficiency of bHLH84-HA by SNC1-FLAG varied from trial to trial. Taken together, these data suggest that the interactions of SNC1-bHLH84 and SNC1-TPR1 in planta are independent, although whether there is competition between bHLH84 and TPR1 in associating with SNC1 is unclear.
To further investigate whether bHLH84 is able to directly interact with SNC1, we carried out yeast-two-hybrid experiment by co-transforming bHLH84 fused with AD and SNC1 fused with BD. Since we failed in making a full-length SNC1 construct, we made truncated SNC1 segments. As shown in Figure S12, yeast cells transformed with bHLH84-AD and different truncated SNC1 fused with BD were not able to grow on the selection plates, suggesting that bHLH84 does not directly interact with the truncated SNC1 segments in yeast. Moreover, the interaction between bHLH84 and SNC1 probably demands a properly folded full-length SNC1 or an intermediate partner. As EDS1 is required for the function of bHLH84 and EDS1 was shown to interact with SNC1 [37], we asked whether EDS1 or its interacting protein PAD4 [7] might be the intermediate partner. However, we did not detect interaction between bHLH84 and EDS1or bHLH84 and PAD4 (Figure S13), suggesting that EDS1or PAD4 is not likely mediating the interaction between SNC1 and bHLH84.
Discussion
From a targeted reverse genetic screen, we have identified a group of TFs, bHLH84 and its paralogs RSL2 and RSL4, which serve as transcriptional regulators for plant immunity. bHLH84 constitutively activates defense responses when overexpressed, and this activation is SNC1-dependent. bHLH84 was further demonstrated to be a transcriptional activator. In addition, the autoimmune phenotypes of snc1 can be partially suppressed by bhlh84 rsl2 rsl4 triple mutant, suggesting that bHLH84 and SNC1 are mutually dependent. bHLH84 does not seem to directly regulate the transcription of SNC1 or other tested R genes. However, the specific interaction between bHLH84 and NLRs including SNC1 and RPS4 in planta suggests that it associates with nuclear NLRs to mediate downstream transcriptional reprogramming. As we failed to observe association between bHLH84 and the repressor protein TPR1 which also interacts with SNC1, we propose that bHLH84 activates defense responses by forming a complex with SNC1 that functions in parallel with the SNC1-TPR1 complex to activate downstream positive regulators (Figure S14).
The targeted reverse genetic screen is a useful approach to identify new players in biological pathways
Previous work on MLA, N, RRS1 and SNC1 suggests that the interactions between some nuclear R proteins and their associating TFs are essential in regulating defense responses [9], [11], [12], [15], [17], [18]. Different approaches have been utilized to isolate TFs that are able to interact with nuclear R proteins. TPR1, which associates with SNC1 to repress negative regulators of immunity, was isolated from a forward genetic screen for suppressors of snc1 [17]. Yeast-two-hybrid screens have been successfully used to identify TFs in plant immunity. For example, SPL6 was initially identified from a yeast-two-hybrid screen and was further confirmed to interact with N in tobacco [18]. In addition, identified from yeast-two-hybrid screens, MYB6 and WRKY1 were shown to interact with MLA in barley to initiate disease resistance signaling in an antagonistic manner [15]. In this study, we used an alternative reverse genetic screen and successfully identified a group of novel TFs that play critical roles in plant immunity.
Our targeted reverse genetic approach has several advantages. Since plant defense to UV radiation is regulated by many of the same factors as pathogen resistance [23]–[25], while UV treatment datasets exclude a large number of genes that are manipulated by pathogen effectors which are not directly related to defense responses [38], the number of target genes we chose from the UV-induced database is more manageable for a reverse genetics screen. All the selected TFs were overexpressed in both Col-0 and snc1 backgrounds, facilitating rapid identification of both defense enhancers and suppressors (Table S1). Furthermore, the functional redundancy predicament often encountered in forward genetic screens can be effectively avoided by using the overexpression approach. Finally, our approach evades self-activation problems that are often associated with yeast-two-hybrid screens for transcriptional activators. Specifically, bHLH84 exhibits strong self-activation when fused with GAL4 binding domain in yeast (data not shown), thus cannot be identified from a yeast-two-hybrid screen. However, our screen does rely on the availability of high-quality microarray data, which may still overlook TFs with relatively low expression level changes.
bHLH84 functions as a transcriptional activator that is able to bind N1 - or N2-boxes
As bHLH84 was shown to be a transcriptional activator, we attempted chromatin immunoprecipitation (ChIP) to identify target genes of bHLH84. However, as with our co-IP experiments (Figure S8), the bHLH84-GFP-HA protein could not be pulled down when subjected to ChIP (Figure S15). Thus we were unable to identify the target DNA of bHLH84 in planta. Using yeast-one-hybrid assay as an alternative approach, we attempted to identify the DNA-binding sequences of bHLH84. Many bHLH type TFs were shown to bind sequences containing a consensus core element E-box (5′-CANNTG-3′), with the palindromic G-box (5′-CACGTG-3′) being the most typical form [39]. Some bHLH proteins bind to non-E-box sequences (N-box), such as 5′-CACGc/aG-3′ and 5′-CGCGTG-3′ [40], [41]. As shown in Figure S16A and Figure S16B, compared with the bHLH84 alone or cis-element alone negative controls, the most enhanced yeast growth was observed on SD-Leu-Trp-His media when AD-bHLH84 was co-transformed with pHIS2-N1-box, while considerably enhanced growth was observed when AD-bHLH84 was co-transformed with pHIS2-N2-box. No enhanced yeast growth was observed in G-box or N3 box co-transformations. These data suggest that bHLH84 is able to bind N1 - and N2-boxes, but not N3 - or G-boxes. These data are consistent with the prediction that TFs in this bHLH subfamily are non E-box binders [42]. Although the potential binding sites of bHLH84 have been revealed, it is still difficult to predict its target genes. More sophisticated ChIP experiments designed in the future may be able to solve this problem.
bHLH84 and its paralogs are implicated in plant immunity
The bHLH-containing proteins constitute a large conserved TF family in eukaryotes [43], [44]. They have been studied intensively in yeast and humans, providing evidence for their regulatory functions in cell proliferation and cellular differentiation pathways [45]–[48].
While only a few bHLH proteins have been studied in detail in plants, they have been shown to serve regulatory functions in multiple biological pathways. For example, a group of bHLH TFs in Zea mays regulate the production of the purple anthocyanin pigments by interacting with R2R3-MYB TFs [49]. In Arabidopsis, GL3 (GLABRA3) regulates trichome development through its interaction with MYB-like TF GL1(GLABRA1) [50]. Another small subfamily of bHLH TFs, referred to as phytochrome-interacting factors (PIFs), have been shown to play diverse functions including regulating light signaling pathways, seed germination, seedling photomorphogenesis, and shade avoidance responses via their interactions with phytochromes [51]–[56]. In addition, JAM1 (ABA-INDUCIBLE BHLH-TYPE TRANSCRIPTION FACTOR/JA-ASSOCIATED MYC2-LIKE), acts as a transcriptional repressor and negatively regulates JA signaling [57]. bHLH84 and its paralogs have previously been shown to regulate root hair elongation [29], [58]. However, they are the first few bHLH TFs found to be involved in plant immunity. Since bHLH TFs form one of the largest TF families in plants, it is difficult to imagine that these three TFs are the only bHLHs involved in immune regulation. Lethality of the knockout mutants or redundancy could be the factors prohibiting others from being discovered. Future novel methods, such as our overexpression approach, may facilitate the functional studies of more TFs in large families.
bHLH84 and its paralogs function redundantly in NLR-mediated immunity
As one of the largest TF families in Arabidopsis with 147 members, bHLH TFs are further subdivided into 12 major subfamilies based on sequence similarity. bHLH84 and its paralogs belong to the VIIIc subgroup [28]. In this study, we have experimentally shown that bHLH84, RSL2 and RSL4 redundantly regulate defense responses. Overexpression of any of these proteins results in constitutive activation of defense responses (Figure 4). Their redundancy was further demonstrated using the triple mutant of bhlh84 rsl4 rsl2, which is able to partially suppress the autoimmune phenotypes of snc1 (Figure 3), and compromise RPS4-mediated defense responses (Figure 2E). It is possible that additional members of the VIIIc subfamily are also functionally redundant with bHLH84. Future construction of higher order bhlh mutants may provide insight into the additional redundant relationships among these family members.
Typically, the bHLH domain contains approximately 60 amino acids and is comprised of a stretch of hydrophilic and basic residues at the N terminus, followed by two amphipathic alpha-helices connected by an intervening loop [44]. The helix-loop-helix and the basic region of the bHLH are required for DNA-binding, whereas the helix-loop-helix region alone often enables homo - or heterodimerization with other bHLH proteins. Since the single mutants of bhlh84, rsl2 and rsl4 do not exhibit obvious phenotypes, we speculate that if dimerization occurs, it would most likely be homodimerization rather than heterodimerization. The dimerized bHLH84 or its paralogs may bind to the same DNA region, thus regulating immunity in a similar manner. In addition, bHLH TFs often associate with other types of TFs, including MYBs and bZIPs for transcriptional reprogramming [49], [56], thus we cannot exclude the possibility that there are more unknown TFs that are also involved in the bHLH84-SNC1 complex.
As the expression level of SNC1 is comparable in eds1-2 and OXbHLH84-GFP-HA eds1-2 backgrounds (Figure 6), bHLH84 does not seem to regulate SNC1 expression. In addition, we did not observe transcriptional up-regulation of tested R genes in OXbHLH84-GFP-HA snc1-r1 or OXbHLH84-GFP-HA eds1-2 plants (Figure S7), suggesting that bHLH84 does not directly regulate the transcription of R genes.
As we also detected attenuated immunity against P.s.t. avrRps4 in bhlh84 rsl2 rsl4 triple mutant (Figure 2E), and interaction between RPS4 and bHLH84 in N.benthamina (Figure 7D), bHLH84 and its paralogs seem to be not just specific to SNC1. As both RPS4's and SNC1's nuclear localizations are critical to their defense activation [10], [14], we speculate that these bHLH TFs may work together with selective nuclear TIR-NB-LRRs to trigger downstream immunity. More in-depth investigations on the interactions of other nuclear TIR-NB-LRR proteins with these TFs might reveal more R proteins working together with these bHLH proteins.
bHLH84 and TPR1 function in parallel to regulate SNC1-mediated resistance
Overexpression of either bHLH84 or TPR1 results in SNC1-dependent autoimmunity, indicating that both bHLH84 and TPR1 positively regulate SNC1-mediated defense responses. Both bHLH84 and TPR1 were shown to associate with SNC1, although no interaction was detected between bHLH84 and TPR1, suggesting that bHLH84-SNC1 and TPR1-SNC1 probably function in distinct complexes (Figure 7, S11 and S14). Their downstream target genes are probably different, as bHLH84 is a transcriptional activator while TPRs are repressors. Defense activation induced by SNC1 is likely achieved through a combination of activation of positive regulators and repression of negative regulators.
Materials and Methods
Construction of plasmids
The genomic sequences of selected TFs, excluding the stop codon and including approximately 1.5 kb sequence upstream of the start codon, were amplified by PCR with two different restriction enzyme sites separately introduced at the two primer ends. The chosen restriction enzyme sites were KpnI, SalI, SacI, XbaI or PstI. The amplified fragments were then digested and ligated to modified pCambia1305 vectors harboring C-terminal GFP and HA tags. These constructs were transformed into snc1 and Col-0 using the floral dip method [26].
For overexpression of bHLH84, RSL2 and RSL4, coding sequences of the genes were amplified by PCR with two different restriction enzyme sites separately introduced at the two primer ends. The primer sequences can be found in Table S2. The fragments were then digested and ligated to the pG229HAN vector with a 35S promoter.
For the pCambia1300-35S-SNC1-FLAG, pCambia1300-35S-RPS4-FLAG and pCambia1300-35S-RPS6-FLAG constructs used in the transient expression in N. benthamiana, the genomic region of SNC1, RPS4 or RPS6 without the stop codon, was cloned into the pCambia1300 vector with a 35S promoter and a C-terminus FLAG tag. For other pCambia1300 constructs used in the transient expression, the CDS regions of the genes were cloned into the corresponding vectors. The primer sequences can be found in Table S2
Transgenic screening
Approximately 0.4 g of T1 transgenic seeds for each construct were first plated on solid MS medium containing 30 µg/ml Hygromycin B. 48 one-week-old transformant seedlings per genotype were selected and subsequently transplanted on soil. Col-0 and snc1 seeds were planted on solid MS medium without any selection and transplanted on soil at the same time to serve as controls. Among the transgenic plants of each genotype, the transformants which showed varied sizes were kept, and T2 seeds from these plants were planted on Hygromycin B plates to analyze transgene copy number, check for the presence of the transgene and validate the background using primers specific to the SNC1 locus [20]. The transgenic plants with heritable phenotypes and with the correct backgrounds were then subjected to H.a. Noco2 infection to examine whether their altered morphology is correlated with altered resistance. Resistance was scored based on the degree of deviation from that observed in the control plants. More specifically, transgenic plants in Col-0 background showing similar sporulation as Col-0 were scored as no change (NC). Plants showing less sporulation than Col-0 were scored as showing enhanced resistance phenotype with “+”. Plants exhibiting a little sporulation were scored as having more enhanced resistance phenotype with “++”, while the ones showing no sporulation were scored as the most enhanced resistance phenotype as “+++”. For transgenic plants in the snc1 background, plants showing more sporulation than snc1 were scored as suppressing phenotype with “−”, while the ones showing less sporulation than snc1 were scored as enhancing phenotype with “+”.
Confocal microscopy
Leaves from one-week-old seedlings were soaked in 1 mg/mL (1∶1 [g/v]) propidium iodide (PI) for 3 minutes and rinsed briefly with water before visualization. Root tissues were submerged in 1 µg/ml (1∶1 [g/v]) PI for 10 seconds and mounted in water. For GFP and PI visualization, a Nikon ECLIPSE 80i Confocal microscope was used under 488 nm and 543 nm filter sets.
Transient protein expression and co-immunoprecipitation in N. benthamiana
Transient protein expression in N. benthamiana was carried out as previously described [35]. The IP protocol was modified from [59]. Briefly, Agrobacteria containing the binary vector pCambia1300 constructed with the target genes and tags were cultured in LB media with kanamycin selection at 28°C overnight. The bacteria were inoculated into a new culture media (10.5 g/L K2HPO4, 4.5 g/L KH2PO4, 1.0 g/L (NH4)2SO4, 0.5 g/L NaCitrate, 1 mM MgSO4, 0.2% glucose, 0.5% glycerol, 50 µM acetosyringone, and 10 mM N-morpholino-ethanesulfonic acid (MES) (pH 5.6), 50 µg/mL Kanamycin) by 1∶50 dilution and cultured for a further 8–12 hours. The bacteria were then harvested by centrifugation at 4000 rpm for 10 minutes and resuspended in MS buffer (4.4 g/L MS, 10 mM MES, 150 µM acetosyringone) to a final concentration of OD600 = 0.2 for infiltration into four-week-old N. benthamiana leaves.
For co-immunoprecipitation, 3 g of N. benthamiana leaves were collected at 36 hours post-infiltration and ground into fine powder in liquid nitrogen using a cold mortar and pestle. The powder was mixed with 6 ml extraction buffer (10% glycerol, 25 mM Tris pH 7.5, 1 mM EDTA, 150 mM NaCl, 10 mM DTT, 2% w/v PVPP, protease inhibitor cocktail) and homogenized by further grinding. All the following steps were carried out at 4°C. The samples were centrifuged at 15000 g for 10 minutes and the supernatants were transferred to new tubes. These two steps were repeated twice before NP40 (Nonidet P-40 Substitute) was added into each supernatant to a final concentration of 0.15%. 30 µl pre-washed protein A or protein G agarose beads were added into each supernatant and incubated for 30 minutes. The mixtures were centrifuged at 4000 rpm for 2 minutes to remove the beads. Each supernatant was incubated with 30 µl anti-FLAG beads or protein A agarose beads for 3 hours, and the beads were pelleted down by centrifuging at 8000 rpm for 1 minute and washed 8 times using extraction buffer containing 0.15% NP40. Proteins specifically bound to the beads were competitively eluted using 100 µl 250 µg/ml 3×FLAG peptides. All the samples were boiled in SDS loading buffer for 5 minutes before running on SDS-PAGE gel.
Arabidopsis protoplast transient assay for transcriptional activity
The isolation and transfection of Arabidopsis protoplasts and the reporter gene assay were previously described in [27]. Briefly, the Arabidopsis protoplasts were transfected with the reporter construct, the effector construct and the internal control construct as illustrated in Figure 2A. GUS expression was determined using MUG assay (Acros Organics from Fisher Scientific). Fluorescence was measured using a fluorescence spectrophotometer (360/460 nm). The internal LUC expression was examined using a Dual-Luciferase reporter assay system (Promega, E1910).
Ion leakage assay
The ion leakage assay was performed as previously described [60], with a few modifications. Briefly, twelve leaf discs (7 mm in diameter) per measurement were punched from the infiltrated area at 23 hr post infiltration and placed in a 60 mm petri dish containing 10 ml of ddH2O. After 30 minutes, the water was removed and another 10 ml of ddH2O was added into the petri dish containing the leaf discs. Conductivity was measured using a 545 Conductivity Multi-purpose Cell (VWR Scientific) at the indicated time points.
Yeast-one-hybrid and yeast-two-hybrid assays
For yeast-one-hybrid assay, the pHIS2 derivatives (harboring the N1-, N2-, N3 - and G-box cis-elements) were co-transformed with the construct of pAD-bHLH84 into the yeast strain Y187. For each co-transformation of pAD-bHLH84 and pHIS2 derivatives, yeast cells co-transformed with pHIS2 empty vector (EV) and pAD-bHLH84 as well as yeast cells cotransformed with pAD EV and the pHIS2 derivatives were used as negative controls. The positive transformants were isolated from SD-Trp-Leu medium. The transformants were then analyzed on the SD-Trp-Leu-His medium supplemented with 60 mM and 100 mM 3-Amino-1,2,4-Triazole (3AT).
For yeast-two-hybrid assays, the pGBKT7 derivatives containing various truncated SNC1 fragments were co-transformed with pAD-bHLH84 into yeast strain Y1347. pGBKT7 EV cotransformed with pAD-bHLH84 was used as a negative control. The positive transformants were isolated from SD-Trp-Leu medium. The transformants were then analyzed on SD-Trp-Leu-His medium supplemented with 3 mM 3AT.
Supporting Information
Zdroje
1. JonesJD, DanglJL (2006) The plant immune system. Nature 444 : 323–329.
2. Hammond-KosackKE, JonesJD (1996) Resistance gene-dependent plant defense responses. Plant Cell 8 : 1773–1791.
3. ChisholmST, CoakerG, DayB, StaskawiczBJ (2006) Host-microbe interactions: shaping the evolution of the plant immune response. Cell 124 : 803–814.
4. MaekawaT, KuferTA, Schulze-LefertP (2011) NLR functions in plant and animal immune systems: so far and yet so close. Nat Immunol 12 : 817–826.
5. AartsN, MetzM, HolubE, StaskawiczBJ, DanielsMJ, et al. (1998) Different requirements for EDS1 and NDR1 by disease resistance genes define at least two R gene-mediated signaling pathways in Arabidopsis. Proc Natl Acad Sci U S A 95 : 10306–10311.
6. FeysBJ, WiermerM, BhatRA, MoisanLJ, Medina-EscobarN, et al. (2005) Arabidopsis SENESCENCE-ASSOCIATED GENE101 stabilizes and signals within an ENHANCED DISEASE SUSCEPTIBILITY1 complex in plant innate immunity. Plant Cell 17 : 2601–2613.
7. FeysBJ, MoisanLJ, NewmanMA, ParkerJE (2001) Direct interaction between the Arabidopsis disease resistance signaling proteins, EDS1 and PAD4. Embo j 20 : 5400–5411.
8. CenturyKS, HolubEB, StaskawiczBJ (1995) NDR1, a locus of Arabidopsis thaliana that is required for disease resistance to both a bacterial and a fungal pathogen. Proc Natl Acad Sci U S A 92 : 6597–6601.
9. ShenQH, SaijoY, MauchS, BiskupC, BieriS, et al. (2007) Nuclear activity of MLA immune receptors links isolate-specific and basal disease-resistance responses. Science 315 : 1098–1103.
10. WirthmuellerL, ZhangY, JonesJD, ParkerJE (2007) Nuclear accumulation of the Arabidopsis immune receptor RPS4 is necessary for triggering EDS1-dependent defense. Curr Biol 17 : 2023–2029.
11. DeslandesL, OlivierJ, TheulieresF, HirschJ, FengDX, et al. (2002) Resistance to Ralstonia solanacearum in Arabidopsis thaliana is conferred by the recessive RRS1-R gene, a member of a novel family of resistance genes. Proc Natl Acad Sci U S A 99 : 2404–2409.
12. DeslandesL, OlivierJ, PeetersN, FengDX, KhounlothamM, et al. (2003) Physical interaction between RRS1-R, a protein conferring resistance to bacterial wilt, and PopP2, a type III effector targeted to the plant nucleus. Proc Natl Acad Sci U S A 100 : 8024–8029.
13. Burch-SmithTM, SchiffM, CaplanJL, TsaoJ, CzymmekK, et al. (2007) A novel role for the TIR domain in association with pathogen-derived elicitors. PLoS Biol 5: e68.
14. ChengYT, GermainH, WiermerM, BiD, XuF, et al. (2009) Nuclear pore complex component MOS7/Nup88 is required for innate immunity and nuclear accumulation of defense regulators in Arabidopsis. Plant Cell 21 : 2503–2516.
15. ChangC, YuD, JiaoJ, JingS, Schulze-LefertP, et al. (2013) Barley MLA immune receptors directly interfere with antagonistically acting transcription factors to initiate disease resistance signaling. Plant Cell 25 : 1158–1173.
16. InoueH, HayashiN, MatsushitaA, XinqiongL, NakayamaA, et al. (2013) Blast resistance of CC-NB-LRR protein Pb1 is mediated by WRKY45 through protein-protein interaction. Proc Natl Acad Sci U S A 110 : 9577–9582.
17. ZhuZ, XuF, ZhangY, ChengYT, WiermerM, et al. (2010) Arabidopsis resistance protein SNC1 activates immune responses through association with a transcriptional corepressor. Proc Natl Acad Sci U S A 107 : 13960–13965.
18. PadmanabhanMS, MaS, Burch-SmithTM, CzymmekK, HuijserP, et al. (2013) Novel positive regulatory role for the SPL6 transcription factor in the N TIR-NB-LRR receptor-mediated plant innate immunity. PLoS Pathog 9: e1003235.
19. LiX, ClarkeJD, ZhangY, DongX (2001) Activation of an EDS1-mediated R-gene pathway in the snc1 mutant leads to constitutive, NPR1-independent pathogen resistance. Mol Plant Microbe Interact 14 : 1131–1139.
20. ZhangY, GoritschnigS, DongX, LiX (2003) A gain-of-function mutation in a plant disease resistance gene leads to constitutive activation of downstream signal transduction pathways in suppressor of npr1-1, constitutive 1. Plant Cell 15 : 2636–2646.
21. JohnsonKC, DongOX, HuangY, LiX (2012) A Rolling Stone Gathers No Moss, but Resistant Plants Must Gather Their MOSes. Cold Spring Harb Symp Quant Biol 77 : 259–268.
22. XiaS, ChengYT, HuangS, WinJ, SoardsA, et al. (2013) Regulation of Transcription of Nucleotide-Binding Leucine-Rich Repeat-Encoding Genes SNC1 and RPP4 via H3K4 Trimethylation. Plant Physiol 162 : 1694–1705.
23. KunzBA, DandoPK, GriceDM, MohrPG, SchenkPM, et al. (2008) UV-induced DNA damage promotes resistance to the biotrophic pathogen Hyaloperonospora parasitica in Arabidopsis. Plant Physiol 148 : 1021–1031.
24. KliebensteinDJ, LimJE, LandryLG, LastRL (2002) Arabidopsis UVR8 regulates ultraviolet-B signal transduction and tolerance and contains sequence similarity to human regulator of chromatin condensation 1. Plant Physiol 130 : 234–243.
25. KunzBA, CahillDM, MohrPG, OsmondMJ, VonarxEJ (2006) Plant responses to UV radiation and links to pathogen resistance. Int Rev Cytol 255 : 1–40.
26. CloughSJ, BentAF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16 : 735–743.
27. TiwariS, WangS, HagenG, GuilfoyleTJ (2006) Transfection assays with protoplasts containing integrated reporter genes. Methods Mol Biol 323 : 237–244.
28. HeimMA, JakobyM, WerberM, MartinC, WeisshaarB, et al. (2003) The basic helix-loop-helix transcription factor family in plants: a genome-wide study of protein structure and functional diversity. Mol Biol Evol 20 : 735–747.
29. YiK, MenandB, BellE, DolanL (2010) A basic helix-loop-helix transcription factor controls cell growth and size in root hairs. Nat Genet 42 : 264–267.
30. ZhuY, QianW, HuaJ (2010) Temperature modulates plant defense responses through NB-LRR proteins. PLoS Pathog 6: e1000844.
31. ChengYT, LiY, HuangS, HuangY, DongX, et al. (2011) Stability of plant immune-receptor resistance proteins is controlled by SKP1-CULLIN1-F-box (SCF)-mediated protein degradation. Proc Natl Acad Sci U S A 108 : 14694–14699.
32. ParkerJE, HolubEB, FrostLN, FalkA, GunnND, et al. (1996) Characterization of eds1, a mutation in Arabidopsis suppressing resistance to Peronospora parasitica specified by several different RPP genes. Plant Cell 8 : 2033–2046.
33. NawrathC, MetrauxJP (1999) Salicylic acid induction-deficient mutants of Arabidopsis express PR-2 and PR-5 and accumulate high levels of camalexin after pathogen inoculation. Plant Cell 11 : 1393–1404.
34. YangS, HuaJ (2004) A haplotype-specific Resistance gene regulated by BONZAI1 mediates temperature-dependent growth control in Arabidopsis. Plant Cell 16 : 1060–1071.
35. Van den AckervekenG, MaroisE, BonasU (1996) Recognition of the bacterial avirulence protein AvrBs3 occurs inside the host plant cell. Cell 87 : 1307–1316.
36. MonaghanJ, XuF, XuS, ZhangY, LiX (2010) Two putative RNA-binding proteins function with unequal genetic redundancy in the MOS4-associated complex. Plant Physiol 154 : 1783–1793.
37. BhattacharjeeS, HalaneMK, KimSH, GassmannW (2011) Pathogen effectors target Arabidopsis EDS1 and alter its interactions with immune regulators. Science 334 : 1405–1408.
38. KayS, BonasU (2009) How Xanthomonas type III effectors manipulate the host plant. Curr Opin Microbiol 12 : 37–43.
39. Toledo-OrtizG, HuqE, QuailPH (2003) The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell 15 : 1749–1770.
40. GroszmannM, BylstraY, LampugnaniER, SmythDR (2010) Regulation of tissue-specific expression of SPATULA, a bHLH gene involved in carpel development, seedling germination, and lateral organ growth in Arabidopsis. J Exp Bot 61 : 1495–1508.
41. OhsakoS, HyerJ, PanganibanG, OliverI, CaudyM (1994) Hairy function as a DNA-binding helix-loop-helix repressor of Drosophila sensory organ formation. Genes Dev 8 : 2743–2755.
42. Carretero-PauletL, GalstyanA, Roig-VillanovaI, Martinez-GarciaJF, Bilbao-CastroJR, et al. (2010) Genome-wide classification and evolutionary analysis of the bHLH family of transcription factors in Arabidopsis, poplar, rice, moss, and algae. Plant Physiol 153 : 1398–1412.
43. MurreC, McCawPS, VaessinH, CaudyM, JanLY, et al. (1989) Interactions between heterologous helix-loop-helix proteins generate complexes that bind specifically to a common DNA sequence. Cell 58 : 537–544.
44. Cornelis MurreGB, van DijkMarc A, EngelIsaac, FurnariBeth A, MassariMark E, MatthewsJames R, QuongMelanie W, RiveraRichard R, StuiverMaarten H (1994) Structure and function of helix-loop-helix proteins. Biochimica et Biophysica Acta (BBA) - Gene Structure and Expression 1218 : 129–135.
45. StevensJD, RoalsonEH, SkinnerMK (2008) Phylogenetic and expression analysis of the basic helix-loop-helix transcription factor gene family: genomic approach to cellular differentiation. Differentiation 76 : 1006–1022.
46. CrewsST (1998) Control of cell lineage-specific development and transcription by bHLH-PAS proteins. Genes Dev 12 : 607–620.
47. IshibashiJ, PerryRL, AsakuraA, RudnickiMA (2005) MyoD induces myogenic differentiation through cooperation of its NH2 - and COOH-terminal regions. J Cell Biol 171 : 471–482.
48. UittenbogaardM, PeavyDR, ChiaramelloA (1999) Expression of the bHLH gene NSCL-1 suggests a role in regulating cerebellar granule cell growth and differentiation. J Neurosci Res 57 : 770–781.
49. LudwigSR, HaberaLF, DellaportaSL, WesslerSR (1989) Lc, a member of the maize R gene family responsible for tissue-specific anthocyanin production, encodes a protein similar to transcriptional activators and contains the myc-homology region. Proc Natl Acad Sci U S A 86 : 7092–7096.
50. PayneCT, ZhangF, LloydAM (2000) GL3 encodes a bHLH protein that regulates trichome development in arabidopsis through interaction with GL1 and TTG1. Genetics 156 : 1349–1362.
51. CastillonA, ShenH, HuqE (2007) Phytochrome Interacting Factors: central players in phytochrome-mediated light signaling networks. Trends Plant Sci 12 : 514–521.
52. ShinJ, KimK, KangH, ZulfugarovIS, BaeG, et al. (2009) Phytochromes promote seedling light responses by inhibiting four negatively-acting phytochrome-interacting factors. Proc Natl Acad Sci U S A 106 : 7660–7665.
53. StephensonPG, FankhauserC, TerryMJ (2009) PIF3 is a repressor of chloroplast development. Proc Natl Acad Sci U S A 106 : 7654–7659.
54. LeivarP, QuailPH (2011) PIFs: pivotal components in a cellular signaling hub. Trends Plant Sci 16 : 19–28.
55. LeivarP, MonteE, OkaY, LiuT, CarleC, et al. (2008) Multiple phytochrome-interacting bHLH transcription factors repress premature seedling photomorphogenesis in darkness. Curr Biol 18 : 1815–1823.
56. ChenD, XuG, TangW, JingY, JiQ, et al. (2013) Antagonistic Basic Helix-Loop-Helix/bZIP Transcription Factors Form Transcriptional Modules That Integrate Light and Reactive Oxygen Species Signaling in Arabidopsis. Plant Cell 25 : 1657–1673.
57. NakataM, MitsudaN, HerdeM, KooAJ, MorenoJE, et al. (2013) A bHLH-Type Transcription Factor, ABA-INDUCIBLE BHLH-TYPE TRANSCRIPTION FACTOR/JA-ASSOCIATED MYC2-LIKE1, Acts as a Repressor to Negatively Regulate Jasmonate Signaling in Arabidopsis. Plant Cell 25 : 1641–1656.
58. BruexA, KainkaryamRM, WieckowskiY, KangYH, BernhardtC, et al. (2012) A gene regulatory network for root epidermis cell differentiation in Arabidopsis. PLoS Genet 8: e1002446.
59. MoffettP, FarnhamG, PeartJ, BaulcombeDC (2002) Interaction between domains of a plant NBS-LRR protein in disease resistance-related cell death. Embo j 21 : 4511–4519.
60. NomuraH, KomoriT, UemuraS, KandaY, ShimotaniK, et al. (2012) Chloroplast-mediated activation of plant immune signalling in Arabidopsis. Nat Commun 3 : 926.
61. GaoM, WangX, WangD, XuF, DingX, et al. (2009) Regulation of cell death and innate immunity by two receptor-like kinases in Arabidopsis. Cell Host Microbe 6 : 34–44.
62. LiY, LiS, BiD, ChengYT, LiX, et al. (2010) SRFR1 negatively regulates plant NB-LRR resistance protein accumulation to prevent autoimmunity. PLoS Pathog 6: e1001111.
63. XuF, XuS, WiermerM, ZhangY, LiX (2012) The cyclin L homolog MOS12 and the MOS4-associated complex are required for the proper splicing of plant resistance genes. Plant J 70 : 916–928.
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek Disruption of Fas-Fas Ligand Signaling, Apoptosis, and Innate Immunity by Bacterial PathogensČlánek A Tick Gut Protein with Fibronectin III Domains Aids Congregation to the Gut during TransmissionČlánek The Vi Capsular Polysaccharide Enables Serovar Typhi to Evade Microbe-Guided Neutrophil Chemotaxis
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2014 Číslo 8- Jak souvisí postcovidový syndrom s poškozením mozku?
- Měli bychom postcovidový syndrom léčit antidepresivy?
- Farmakovigilanční studie perorálních antivirotik indikovaných v léčbě COVID-19
- 10 bodů k očkování proti COVID-19: stanovisko České společnosti alergologie a klinické imunologie ČLS JEP
-
Všechny články tohoto čísla
- Regulatory RNAs Involved in Bacterial Antibiotic Resistance
- From Dandruff to Deep-Sea Vents: -like Fungi Are Ecologically Hyper-diverse
- Pathogenicity and Epithelial Immunity
- Mother–Infant HIV Transmission: Do Maternal HIV-Specific Antibodies Protect the Infant?
- Hell's BELs: acterial 3 igases That Exploit the Eukaryotic Ubiquitin Machinery
- Disruption of Fas-Fas Ligand Signaling, Apoptosis, and Innate Immunity by Bacterial Pathogens
- Primary Seronegative but Molecularly Evident Hepadnaviral Infection Engages Liver and Induces Hepatocarcinoma in the Woodchuck Model of Hepatitis B
- TLR2 Signaling Decreases Transmission of by Limiting Bacterial Shedding in an Infant Mouse Influenza A Co-infection Model
- Production of an Attenuated Phenol-Soluble Modulin Variant Unique to the MRSA Clonal Complex 30 Increases Severity of Bloodstream Infection
- Inhibition of the TRAIL Death Receptor by CMV Reveals Its Importance in NK Cell-Mediated Antiviral Defense
- Early Mucosal Sensing of SIV Infection by Paneth Cells Induces IL-1β Production and Initiates Gut Epithelial Disruption
- Limited HIV Infection of Central Memory and Stem Cell Memory CD4+ T Cells Is Associated with Lack of Progression in Viremic Individuals
- Virus-Specific Regulatory T Cells Ameliorate Encephalitis by Repressing Effector T Cell Functions from Priming to Effector Stages
- A Tick Gut Protein with Fibronectin III Domains Aids Congregation to the Gut during Transmission
- The HIV-1 Envelope Transmembrane Domain Binds TLR2 through a Distinct Dimerization Motif and Inhibits TLR2-Mediated Responses
- Infection with MERS-CoV Causes Lethal Pneumonia in the Common Marmoset
- VGIII Isolates Causing Infections in HIV/AIDS Patients in Southern California: Identification of the Local Environmental Source as Arboreal
- Diverse Host-Seeking Behaviors of Skin-Penetrating Nematodes
- Capsid Protein VP4 of Human Rhinovirus Induces Membrane Permeability by the Formation of a Size-Selective Multimeric Pore
- The Murine Gammaherpesvirus Immediate-Early Rta Synergizes with IRF4, Targeting Expression of the Viral M1 Superantigen to Plasma Cells
- Characterization of an Insecticidal Toxin and Pathogenicity of against Insects
- The Vi Capsular Polysaccharide Enables Serovar Typhi to Evade Microbe-Guided Neutrophil Chemotaxis
- Histone Deacetylase Inhibitors Impair the Elimination of HIV-Infected Cells by Cytotoxic T-Lymphocytes
- A Locus Encompassing the Epstein-Barr Virus Kinase Regulates Expression of Genes Encoding Viral Structural Proteins
- Distinct APC Subtypes Drive Spatially Segregated CD4 and CD8 T-Cell Effector Activity during Skin Infection with HSV-1
- Structure of CfaA Suggests a New Family of Chaperones Essential for Assembly of Class 5 Fimbriae
- Adoptive Transfer of EBV Specific CD8 T Cell Clones Can Transiently Control EBV Infection in Humanized Mice
- Schistosome Feeding and Regurgitation
- EVM005: An Ectromelia-Encoded Protein with Dual Roles in NF-κB Inhibition and Virulence
- Rabies Virus Hijacks and Accelerates the p75NTR Retrograde Axonal Transport Machinery
- Why HIV Virions Have Low Numbers of Envelope Spikes: Implications for Vaccine Development
- Identification of Anti-virulence Compounds That Disrupt Quorum-Sensing Regulated Acute and Persistent Pathogenicity
- HIV-1 Receptor Binding Site-Directed Antibodies Using a VH1-2 Gene Segment Orthologue Are Activated by Env Trimer Immunization
- Cooperation between Epstein-Barr Virus Immune Evasion Proteins Spreads Protection from CD8 T Cell Recognition across All Three Phases of the Lytic Cycle
- Parasite Extracellular Vesicles: Mediators of Intercellular Communication
- RC1339/APRc from Is a Novel Aspartic Protease with Properties of Retropepsin-Like Enzymes
- Cyclic di-GMP-dependent Signaling Pathways in the Pathogenic Firmicute
- Non-random Escape Pathways from a Broadly Neutralizing Human Monoclonal Antibody Map to a Highly Conserved Region on the Hepatitis C Virus E2 Glycoprotein Encompassing Amino Acids 412–423
- Neutrophil Elastase Causes Tissue Damage That Decreases Host Tolerance to Lung Infection with Species
- Ly6C Monocyte Recruitment Is Responsible for Th2 Associated Host-Protective Macrophage Accumulation in Liver Inflammation due to Schistosomiasis
- SGNH Hydrolase-Like Proteins AlgJ and AlgX Have Similar Topology but Separate and Distinct Roles in Alginate Acetylation
- Why Sexually Transmitted Infections Tend to Cause Infertility: An Evolutionary Hypothesis
- Late Engagement of CD86 after Influenza Virus Clearance Promotes Recovery in a FoxP3 Regulatory T Cell Dependent Manner
- Determinants of Influenza Transmission in South East Asia: Insights from a Household Cohort Study in Vietnam
- A Novel Signal Transduction Pathway that Modulates Quorum Sensing and Bacterial Virulence in
- Host Responses to Group A Streptococcus: Cell Death and Inflammation
- A Cysteine Protease Inhibitor of Is Essential for Exo-erythrocytic Development
- EBNA3C Augments Pim-1 Mediated Phosphorylation and Degradation of p21 to Promote B-Cell Proliferation
- On the Front Line: Quantitative Virus Dynamics in Honeybee ( L.) Colonies along a New Expansion Front of the Parasite
- Assembly and Architecture of the EBV B Cell Entry Triggering Complex
- NLR-Associating Transcription Factor bHLH84 and Its Paralogs Function Redundantly in Plant Immunity
- The PDZ-Binding Motif of Severe Acute Respiratory Syndrome Coronavirus Envelope Protein Is a Determinant of Viral Pathogenesis
- Strain-Specific Properties and T Cells Regulate the Susceptibility to Papilloma Induction by Papillomavirus 1
- Human Cytomegalovirus pUL79 Is an Elongation Factor of RNA Polymerase II for Viral Gene Transcription
- The GAP Activity of Type III Effector YopE Triggers Killing of in Macrophages
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Disruption of Fas-Fas Ligand Signaling, Apoptosis, and Innate Immunity by Bacterial Pathogens
- Ly6C Monocyte Recruitment Is Responsible for Th2 Associated Host-Protective Macrophage Accumulation in Liver Inflammation due to Schistosomiasis
- Host Responses to Group A Streptococcus: Cell Death and Inflammation
- Pathogenicity and Epithelial Immunity
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



