-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaHell's BELs: acterial 3 igases That Exploit the Eukaryotic Ubiquitin Machinery
article has not abstract
Published in the journal: . PLoS Pathog 10(8): e32767. doi:10.1371/journal.ppat.1004255
Category: Pearls
doi: https://doi.org/10.1371/journal.ppat.1004255Summary
article has not abstract
How Do E3s Work in Eukaryotic Cells?
A common post-translational modification in eukaryotes is the covalent attachment of ubiquitin, a 76 amino acid protein, to specific proteins. Most commonly, ubiquitin is conjugated to ε–amino groups of lysine residues, and in unusual cases it can be conjugated to serine and cysteine residues or the terminal amino group of a protein. This process, referred to as ubiquitination (or ubiquitylation), can result in a variety of outcomes for a protein, depending upon how many Ub molecules are attached, whether a polyubiquitin chain is formed, and the nature of the chain. Mono-ubiquitination can result in relocalization of proteins, while most polyubiquitin chains (e.g., K48 and K11-linked chains) direct proteins for proteasomal degradation. Linkages of Ub formed using Lys 63 or by end-to-end linkages (also known as Met Ub) [1] are not directed to the proteasome and can mediate protein trafficking, scaffolding of protein complexes, or enzyme activation. Ub chains are also used for targeting invading microbes for clearance via xenophagy [2].
Ubiquitination is carried out by a series of enzymes. First, a ubiquitin activating enzyme (E1) forms a thioester with the C-terminus of Ub. The activated Ub is then transferred to one of many (∼40 human) ubiquitin conjugating enzymes (E2s). Finally, the E3 enzymes (perhaps over 500 human E3s) direct the transfer of Ub to specific substrates. In eukaryotic cells there are two general classes of E3 ubiquitin ligases. The HECT (Homologous to E6-AP Carboxyl Terminus) and Ring Between Ring (RBR) domain E3s possess an invariant catalytic Cys residue that accepts Ub from a charged E2 before catalyzing transfer of Ub to substrates. Other E3s contain a Really Interesting New Gene (RING) or RING-like domain (U-box) that recruits a charged E2, as well as a domain that recruits substrates. Ub is then transferred from the E2 to the substrate, with the E3 serving primarily as a scaffold. Some RING E3s are single polypeptides, while the cullin-RING Ligases (CRLs) are modular multisubunit complexes [3]. Mammalian CRLs are nucleated by one of seven cullin family members, with a RING domain protein that binds to its C-terminus. The N-terminal region of the cullin binds specific cullin adaptor proteins that engage substrate receptor proteins; the most studied class of substrate receptors are the F-Box proteins. CRLs are subject to an additional level of control by a ubiquitin-like modifier protein, Nedd8.
E3 enzymes have two important roles. First, they recognize substrates and position them for ubiquitination. Second, E3s dictate the nature of the Ub linkage(s), which will determine the substrate's fate. For HECT E3s, the Ub chain type is dictated by the C-terminal lobe of the HECT domain [4]. By contrast, RING-type E3s direct Ub chain type based upon the charged E2 that they recruit [5].
Successful pathogens use proteins that interfere with host cell function that are delivered into eukaryotic host cells via specialized secretion systems and collectively referred to as “effectors.” Remarkably, although ubiquitin is restricted to eukaryotic cells, the past decade has revealed that both bacterial and viral pathogens use effectors to interfere with or manipulate the ubiquitination system [6]. This involves a large number of Bacterially encoded E3 ubiquitin Ligases (BELs). There are multiple RING-type BELs, HECT-like BELs, and even BELs that bear no resemblance to known eukaryotic ubiquitin ligases (Figure 1A).
Fig. 1. Bacteria encode E3 ubiquitin ligases of many classes. 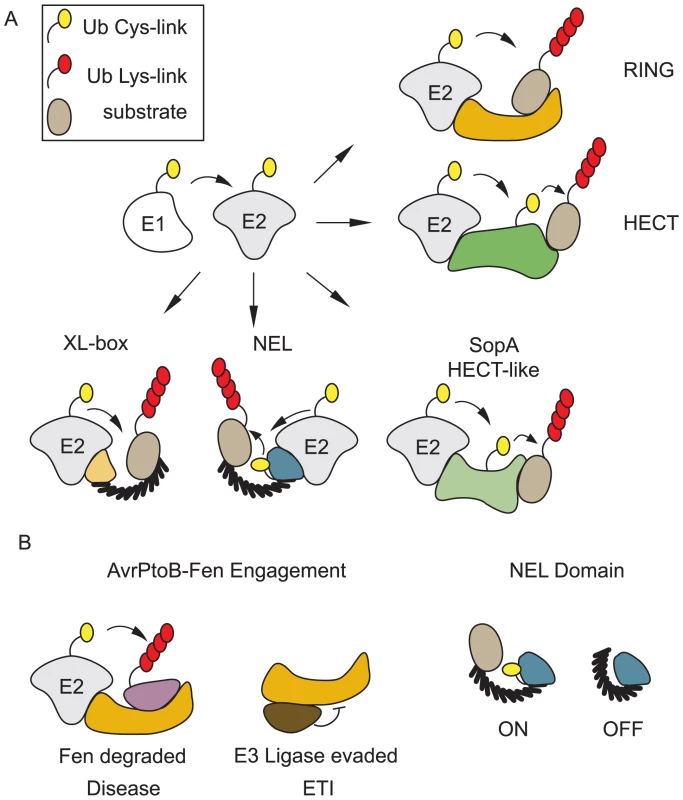
BELs share little or no sequence homology with eukaryotic E3s; however, structural similarities exist between BELs and eukaryotic E3s. Some BELs display entirely novel interactions with their cognate E2s. A. There are examples of bacterial E3 ligases that function similar to RING-type or HECT E3s in eukaryotic cells. Some BELs have novel mechanisms of ubiquitin transfer and interact with E2s in unique ways. B. Evidence exists for regulation of BEL activity inside eukaryotic cells. NEL domain enzymes are autoinhibited until they engage their substrates. AvrPtoB, a RING-type E3, can promote disease by ubiquitinating and causing degradation of the R proteins Pto and Fen (purple). Pto (brown) can also associate with AvrPtoB through an alternate domain allowing it to evade ubiquitination and initiating an ETI. BELs That RING
The first identification of a bacterially encoded E3 ubiquitin ligase was AvrPtoB from the plant pathogen Pseudomonas syringae pv. tomato (Pst) [7]. Plants use resistance proteins (R proteins) that recognize effectors from would-be pathogens. When R proteins engage bacterial effectors, they initiate an Effector Triggered Immune (ETI) response that prevents systemic disease [8]. AvrPtoB effectors from some P. syringae strains are recognized by the R protein Fen that initiates ETI. Although Pst AvrPtoB shared no sequence similarity to proteins of known function, the crystal structure revealed striking structural conservation with RING E3s [9]. AvrPtoB was shown to possess E3 ligase activity in vitro and to suppress ETI, demonstrating a role in pathogenesis [7]. AvrPtoB disrupts ETI by targeting Fen and Pto kinases for ubiquitination [10].
Bioinformatic approaches revealed that Legionella pneumophila encodes a number of F-box containing proteins [11]. Studies have demonstrated that these F-Box proteins function in infected cells as components of CRLs. In this case, BEL activity generates free amino acids that serve as fuel for Legionella growth inside infected cells [12]. Legionella encodes a BEL, LubX, that contains multiple U-boxes [13]. LubX targets another Legionella effector, the kinase SidH, for destruction, establishing LubX as a “metaeffector” that may act to coordinate spatiotemporal control of the effector repertoire within the host cell [14]. Thus, the targets of BELs should not necessarily be assumed to be host-encoded proteins.
One abundant class of RING-type E3s is the NleG family, encoded in enterohaemorrhagic Escherichia coli and Citrobacter rodentium [15]. NleGs were identified using NMR structural studies. Although no sequence similarity exists, the NleGs show close structural similarity to RING-type U-box proteins. As yet, no phenotypes have been associated with nleG mutants, but their large number points toward an important role.
HECT-Like BELs
Discovery of HECT-like BELs followed a similar path as did AvrPtoB and NleG, driven in part by structural biology. The closely related effectors SopA, from Salmonella enterica, and NleL, from enterpathogenic E. coli, were both known to play a role in dampening host inflammation upon infection. While these effectors share no sequence homology to proteins of known function, Zhou and coworkers demonstrated that SopA functioned as a HECT-like enzyme [16]. The mechanisms of ubiquitin transfer had an absolute requirement for a catalytic Cys residue, and SopA was shown to form a Cys∼Ub thioester intermediate. These data suggest a mechanism of Ub transfer similar in mechanism to that of HECT enzymes, but it is clear that there will be significant differences as well. First, the proposed substrate-binding domain is adjacent to the E2 binding site, in contrast to what has been proposed for eukaryotic HECTs [17]. Secondly, SopA and NleL interact with the same region of Ubc8 as do mammalian HECT or RING domain E3s, but differ in the precise E2 residues that are required for BEL activity [18].
BELs That Are NELs—And More…
Shigella spp. use T3SS effectors to cause shigellosis. The most abundant effectors produced by Shigella upon contact with human cells are the IpaH proteins, a class of proteins that exist in many gram-negative pathogens of animals and plants [19]. Their N-terminal domain consists of a series of leucine rich repeat (LRR) domains that share high similarity with YopM from Yersinia spp. [20]. The C-terminal domain is highly conserved among IpaH family members. Using yeast as a surrogate genetic system, IpaH9.8 was shown to possess E3 ubiquitin ligase activity and destroy the MAPKK Ste7 in a proteasome-dependent manner [19]. The IpaH family member SspH1 from Salmonella was shown to also be an E3 ligase that could ubiquitinate a known mammalian interacting protein, PKN1, in vitro [19]. Earlier studies had already shown that substrate specificity was dictated by the LRR domains [21]. The ability to ubiquitinate substrates was shown to rely on a Cys residue that is invariant among the more than 50 proteins that comprise the IpaH family [19]. Again, the IpaH family members shared no sequence similarity to proteins of known function. The crystal structure was solved independently by three groups and revealed that the catalytic domain was entirely alpha helical, had no resemblance to other E3 enzymes, and was coined the NEL domain (novel E3 ligase) [22]–[24]. Similar to HECTs, NEL domains use the invariant Cys residue to form a thioester linkage with Ub [22], [23]. Mutations in E2 enzymes that ablate activity towards RING or HECT domain E3s were shown to be fully functional for Ub transfer to NELs, suggesting a novel E2–E3 interaction [22]. Elegant structural studies showed that NEL domain BELs recognize E2∼Ub conjugates (activated E2s) and use a region of E2s that had not before been implicated in Ub transfer. Based on these studies, a radically new “see-saw” mechanism for Ub transfer was proposed for NEL-domain BELs [25].
The most recent addition to the BEL family is the XL-box domain BEL, XopL, from Xanthomonas campestris pv. vesicatoria (Xcv), a pathogen of tomatoes and peppers [26]. The general architecture of XopL is similar to NEL domain E3s in that it contains an N-terminal domain of LRRs with homology to those of NEL LRRs. The C-terminal domain is a novel E3 ubiquitin ligase domain known as the XL-box. The LRR domain alone is required for suppressing ETI; however, the catalytic domain is required to cause disease in the plant [26]. The XL domain lacks Cys residues, suggesting they function in a mechanism similar to RING-type E3s. Some E2 residues that are absolutely required for RING and HECT function were also required for XopL function, but others were not [26].
Is BEL Activity Regulated?
The activity of NEL domain E3s is negatively regulated by the LRR domain in the absence of their substrates, presumably to prevent premature autoubiquitination until they can productively engage their substrates [22], [23], [27]. The first NEL domain enzyme–substrate structure has been characterized [28]. The LRR of SspH1 binds the HR1b coiled-coil subdomain of PKN1. This report provides the first direct evidence that substrate engagement activates the catalytic activity of NELs. In this case, a straightforward “displacement model” between a linear PKN1 motif and the inhibitory residues within the NEL domain compete for binding to residues within the LRR [28]. As LRR domains are remarkably diverse scaffolds for protein–protein interactions, it remains to be tested if the SspH1–PKN1 paradigm will emerge as a universal mechanism for effector–substrate recognition and activation for BELs. Precise BEL–substrate interactions were recently shown to effect distinct outcomes in the Pst system. The R proteins Fen and Pto interact near the Ring domain of AvrPtoB, resulting in their ubiquitination and degradation [10]. Pto can also interact with an AvrPtoB domain distal to the Ring domain [29]. Binding at the distal domain allows Pto to evade ubiquitination by the AvrPtoB E3 ligase and to activate an ETI response.
Though they do not have a ubiquitin system, bacteria encode a wide variety of E3 ubiquitin ligases that are delivered into the host cells that they infect using specialized secretion systems. A trend among BELs is that while they possess little to no sequence homology with eukaryotic E3s, they often share structural similarity. Notable exceptions are NEL domain and XL-box BELs, suggesting the idea that there are structurally related NEL domain enzymes encoded by eukaryotes. Determining the spectrum of BEL substrates in their respective hosts is an achievable goal. The identification of the eukaryotic proteins that BELs target will increase our understanding of immune functions and provide insights to help combat infection.
Zdroje
1. BehrendsC, HarperJW (2011) Constructing and decoding unconventional ubiquitin chains. Nat Struct Mol Biol 18 : 520–528.
2. FujitaN, YoshimoriT (2011) Ubiquitination-mediated autophagy against invading bacteria. Curr Opin Cell Biol 23 : 492–497.
3. PetroskiMD, DeshaiesRJ (2005) Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol 6 : 9–20.
4. KimHC, HuibregtseJM (2009) Polyubiquitination by HECT E3s and the determinants of chain type specificity. Mol Cell Biol 29 : 3307–3318.
5. WilliamsonA, BanerjeeS, ZhuX, PhilippI, IavaroneAT, et al. (2011) Regulation of ubiquitin chain initiation to control the timing of substrate degradation. Mol Cell 42 : 744–757.
6. AndersonDM, FrankDW (2012) Five mechanisms of manipulation by bacterial effectors: a ubiquitous theme. PLoS Pathog 8: e1002823.
7. AbramovitchRB, JanjusevicR, StebbinsCE, MartinGB (2006) Type III effector AvrPtoB requires intrinsic E3 ubiquitin ligase activity to suppress plant cell death and immunity. Proc Natl Acad Sci U S A 103 : 2851–2856.
8. OhCS, MartinGB (2011) Effector-triggered immunity mediated by the Pto kinase. Trends Plant Sci 16 : 132–140.
9. JanjusevicR, AbramovitchRB, MartinGB, StebbinsCE (2006) A bacterial inhibitor of host programmed cell death defenses is an E3 ubiquitin ligase. Science 311 : 222–226.
10. RosebrockTR, ZengL, BradyJJ, AbramovitchRB, XiaoF, et al. (2007) A bacterial E3 ubiquitin ligase targets a host protein kinase to disrupt plant immunity. Nature 448 : 370–374.
11. PriceCT, Al-KhodorS, Al-QuadanT, SanticM, HabyarimanaF, et al. (2009) Molecular mimicry by an F-box effector of Legionella pneumophila hijacks a conserved polyubiquitination machinery within macrophages and protozoa. PLoS Pathog 5: e1000704.
12. PriceCT, Al-QuadanT, SanticM, RosenshineI, Abu KwaikY (2011) Host proteasomal degradation generates amino acids essential for intracellular bacterial growth. Science 334 : 1553–1557.
13. KuboriT, HyakutakeA, NagaiH (2008) Legionella translocates an E3 ubiquitin ligase that has multiple U-boxes with distinct functions. Mol Microbiol 67 : 1307–1319.
14. KuboriT, ShinzawaN, KanukaH, NagaiH (2010) Legionella metaeffector exploits host proteasome to temporally regulate cognate effector. PLoS Pathog 6: e1001216.
15. WuB, SkarinaT, YeeA, JobinMC, DileoR, et al. (2010) NleG Type 3 effectors from enterohaemorrhagic Escherichia coli are U-Box E3 ubiquitin ligases. PLoS Pathog 6: e1000960.
16. ZhangY, HigashideWM, McCormickBA, ChenJ, ZhouD (2006) The inflammation-associated Salmonella SopA is a HECT-like E3 ubiquitin ligase. Mol Microbiol 62 : 786–793.
17. DiaoJ, ZhangY, HuibregtseJM, ZhouD, ChenJ (2008) Crystal structure of SopA, a Salmonella effector protein mimicking a eukaryotic ubiquitin ligase. Nat Struct Mol Biol 15 : 65–70.
18. LinDY, DiaoJ, ChenJ (2012) Crystal structures of two bacterial HECT-like E3 ligases in complex with a human E2 reveal atomic details of pathogen-host interactions. Proc Natl Acad Sci U S A 109 : 1925–1930.
19. RohdeJR, BreitkreutzA, ChenalA, SansonettiPJ, ParsotC (2007) Type III secretion effectors of the IpaH family are E3 ubiquitin ligases. Cell Host Microbe 1 : 77–83.
20. HaragaA, MillerSI (2003) A Salmonella enterica serovar typhimurium translocated leucine-rich repeat effector protein inhibits NF-kappa B-dependent gene expression. Infect Immun 71 : 4052–4058.
21. HaragaA, MillerSI (2006) A Salmonella type III secretion effector interacts with the mammalian serine/threonine protein kinase PKN1. Cell Microbiol 8 : 837–846.
22. SingerAU, RohdeJR, LamR, SkarinaT, KaganO, et al. (2008) Structure of the Shigella T3SS effector IpaH defines a new class of E3 ubiquitin ligases. Nat Struct Mol Biol 15 : 1293–1301.
23. ZhuY, LiH, HuL, WangJ, ZhouY, et al. (2008) Structure of a Shigella effector reveals a new class of ubiquitin ligases. Nat Struct Mol Biol 15 : 1302–1308.
24. QuezadaCM, HicksSW, GalanJE, StebbinsCE (2009) A family of Salmonella virulence factors functions as a distinct class of autoregulated E3 ubiquitin ligases. Proc Natl Acad Sci U S A 106 : 4864–4869.
25. LevinI, EakinC, BlancMP, KlevitRE, MillerSI, et al. (2010) Identification of an unconventional E3 binding surface on the UbcH5 ∼ Ub conjugate recognized by a pathogenic bacterial E3 ligase. Proc Natl Acad Sci U S A 107 : 2848–2853.
26. SingerAU, SchulzeS, SkarinaT, XuX, CuiH, et al. (2013) A pathogen type III effector with a novel E3 ubiquitin ligase architecture. PLoS Pathog 9: e1003121.
27. ChouYC, KeszeiAF, RohdeJR, TyersM, SicheriF (2012) Conserved structural mechanisms for autoinhibition in IpaH ubiquitin ligases. J Biol Chem 287 : 268–275.
28. KeszeiAF, TangX, McCormickC, ZeqirajE, RohdeJR, et al. (2014) Structure of an SspH1-PKN1 complex reveals the basis for host substrate recognition and mechanism of activation for a bacterial E3 ubiquitin ligase. Mol Cell Biol 34 : 362–373.
29. MathieuJ, SchwizerS, MartinGB (2014) Pto Kinase Binds Two Domains of AvrPtoB and its Proximity to the Effector E3 Ligase Determines if It Evades Degradation and Activates Plant Immunity. PLoS Pathog In Press.
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek Disruption of Fas-Fas Ligand Signaling, Apoptosis, and Innate Immunity by Bacterial PathogensČlánek A Tick Gut Protein with Fibronectin III Domains Aids Congregation to the Gut during TransmissionČlánek The Vi Capsular Polysaccharide Enables Serovar Typhi to Evade Microbe-Guided Neutrophil ChemotaxisČlánek Structure of CfaA Suggests a New Family of Chaperones Essential for Assembly of Class 5 Fimbriae
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2014 Číslo 8- Jak souvisí postcovidový syndrom s poškozením mozku?
- Měli bychom postcovidový syndrom léčit antidepresivy?
- Farmakovigilanční studie perorálních antivirotik indikovaných v léčbě COVID-19
- 10 bodů k očkování proti COVID-19: stanovisko České společnosti alergologie a klinické imunologie ČLS JEP
-
Všechny články tohoto čísla
- Regulatory RNAs Involved in Bacterial Antibiotic Resistance
- From Dandruff to Deep-Sea Vents: -like Fungi Are Ecologically Hyper-diverse
- Pathogenicity and Epithelial Immunity
- Mother–Infant HIV Transmission: Do Maternal HIV-Specific Antibodies Protect the Infant?
- Hell's BELs: acterial 3 igases That Exploit the Eukaryotic Ubiquitin Machinery
- Disruption of Fas-Fas Ligand Signaling, Apoptosis, and Innate Immunity by Bacterial Pathogens
- Primary Seronegative but Molecularly Evident Hepadnaviral Infection Engages Liver and Induces Hepatocarcinoma in the Woodchuck Model of Hepatitis B
- TLR2 Signaling Decreases Transmission of by Limiting Bacterial Shedding in an Infant Mouse Influenza A Co-infection Model
- Production of an Attenuated Phenol-Soluble Modulin Variant Unique to the MRSA Clonal Complex 30 Increases Severity of Bloodstream Infection
- Inhibition of the TRAIL Death Receptor by CMV Reveals Its Importance in NK Cell-Mediated Antiviral Defense
- Early Mucosal Sensing of SIV Infection by Paneth Cells Induces IL-1β Production and Initiates Gut Epithelial Disruption
- Limited HIV Infection of Central Memory and Stem Cell Memory CD4+ T Cells Is Associated with Lack of Progression in Viremic Individuals
- Virus-Specific Regulatory T Cells Ameliorate Encephalitis by Repressing Effector T Cell Functions from Priming to Effector Stages
- A Tick Gut Protein with Fibronectin III Domains Aids Congregation to the Gut during Transmission
- The HIV-1 Envelope Transmembrane Domain Binds TLR2 through a Distinct Dimerization Motif and Inhibits TLR2-Mediated Responses
- Infection with MERS-CoV Causes Lethal Pneumonia in the Common Marmoset
- VGIII Isolates Causing Infections in HIV/AIDS Patients in Southern California: Identification of the Local Environmental Source as Arboreal
- Diverse Host-Seeking Behaviors of Skin-Penetrating Nematodes
- Capsid Protein VP4 of Human Rhinovirus Induces Membrane Permeability by the Formation of a Size-Selective Multimeric Pore
- The Murine Gammaherpesvirus Immediate-Early Rta Synergizes with IRF4, Targeting Expression of the Viral M1 Superantigen to Plasma Cells
- Characterization of an Insecticidal Toxin and Pathogenicity of against Insects
- The Vi Capsular Polysaccharide Enables Serovar Typhi to Evade Microbe-Guided Neutrophil Chemotaxis
- Histone Deacetylase Inhibitors Impair the Elimination of HIV-Infected Cells by Cytotoxic T-Lymphocytes
- A Locus Encompassing the Epstein-Barr Virus Kinase Regulates Expression of Genes Encoding Viral Structural Proteins
- Distinct APC Subtypes Drive Spatially Segregated CD4 and CD8 T-Cell Effector Activity during Skin Infection with HSV-1
- Structure of CfaA Suggests a New Family of Chaperones Essential for Assembly of Class 5 Fimbriae
- Adoptive Transfer of EBV Specific CD8 T Cell Clones Can Transiently Control EBV Infection in Humanized Mice
- Schistosome Feeding and Regurgitation
- EVM005: An Ectromelia-Encoded Protein with Dual Roles in NF-κB Inhibition and Virulence
- Rabies Virus Hijacks and Accelerates the p75NTR Retrograde Axonal Transport Machinery
- Why HIV Virions Have Low Numbers of Envelope Spikes: Implications for Vaccine Development
- Identification of Anti-virulence Compounds That Disrupt Quorum-Sensing Regulated Acute and Persistent Pathogenicity
- HIV-1 Receptor Binding Site-Directed Antibodies Using a VH1-2 Gene Segment Orthologue Are Activated by Env Trimer Immunization
- Cooperation between Epstein-Barr Virus Immune Evasion Proteins Spreads Protection from CD8 T Cell Recognition across All Three Phases of the Lytic Cycle
- Parasite Extracellular Vesicles: Mediators of Intercellular Communication
- RC1339/APRc from Is a Novel Aspartic Protease with Properties of Retropepsin-Like Enzymes
- Cyclic di-GMP-dependent Signaling Pathways in the Pathogenic Firmicute
- Non-random Escape Pathways from a Broadly Neutralizing Human Monoclonal Antibody Map to a Highly Conserved Region on the Hepatitis C Virus E2 Glycoprotein Encompassing Amino Acids 412–423
- Neutrophil Elastase Causes Tissue Damage That Decreases Host Tolerance to Lung Infection with Species
- Ly6C Monocyte Recruitment Is Responsible for Th2 Associated Host-Protective Macrophage Accumulation in Liver Inflammation due to Schistosomiasis
- SGNH Hydrolase-Like Proteins AlgJ and AlgX Have Similar Topology but Separate and Distinct Roles in Alginate Acetylation
- Why Sexually Transmitted Infections Tend to Cause Infertility: An Evolutionary Hypothesis
- Late Engagement of CD86 after Influenza Virus Clearance Promotes Recovery in a FoxP3 Regulatory T Cell Dependent Manner
- Determinants of Influenza Transmission in South East Asia: Insights from a Household Cohort Study in Vietnam
- A Novel Signal Transduction Pathway that Modulates Quorum Sensing and Bacterial Virulence in
- Host Responses to Group A Streptococcus: Cell Death and Inflammation
- A Cysteine Protease Inhibitor of Is Essential for Exo-erythrocytic Development
- EBNA3C Augments Pim-1 Mediated Phosphorylation and Degradation of p21 to Promote B-Cell Proliferation
- On the Front Line: Quantitative Virus Dynamics in Honeybee ( L.) Colonies along a New Expansion Front of the Parasite
- Assembly and Architecture of the EBV B Cell Entry Triggering Complex
- NLR-Associating Transcription Factor bHLH84 and Its Paralogs Function Redundantly in Plant Immunity
- The PDZ-Binding Motif of Severe Acute Respiratory Syndrome Coronavirus Envelope Protein Is a Determinant of Viral Pathogenesis
- Strain-Specific Properties and T Cells Regulate the Susceptibility to Papilloma Induction by Papillomavirus 1
- Human Cytomegalovirus pUL79 Is an Elongation Factor of RNA Polymerase II for Viral Gene Transcription
- The GAP Activity of Type III Effector YopE Triggers Killing of in Macrophages
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Disruption of Fas-Fas Ligand Signaling, Apoptosis, and Innate Immunity by Bacterial Pathogens
- Ly6C Monocyte Recruitment Is Responsible for Th2 Associated Host-Protective Macrophage Accumulation in Liver Inflammation due to Schistosomiasis
- Host Responses to Group A Streptococcus: Cell Death and Inflammation
- Pathogenicity and Epithelial Immunity
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



