-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaExploiting and Subverting Tor Signaling in the Pathogenesis of Fungi, Parasites, and Viruses
article has not abstract
Published in the journal: . PLoS Pathog 7(9): e32767. doi:10.1371/journal.ppat.1002269
Category: Pearls
doi: https://doi.org/10.1371/journal.ppat.1002269Summary
article has not abstract
Tor Signaling Senses Nutrients and Growth Factors to Govern Pathways Involved in the Pathogenesis of Fungi, Parasites, and Viruses
In eukaryotes from yeast to humans, the Tor signaling cascade responds to nutrients and growth factors to orchestrate cell growth and proliferation. The central elements of this signaling cascade are the Tor protein kinases, which are the targets of the potent anti-proliferative and immunosuppressive natural product rapamycin [1]. Most organisms, including mammals, express a single Tor kinase; however, the yeasts Saccharomyces cerevisiae and Schizosaccharomyces pombe contain two Tor homologs [2], and three and four (two classical Tor kinases and two Tor-like kinases) have been identified in the protozoans Leishmania major and Trypanosoma brucei, respectively [3], [4]. The Tor kinases interact with other proteins to form two distinct and ubiquitously conserved complexes known as TORC1 and TORC2 (reviewed in [5]). Under optimal nutrient conditions, TORC2 drives actin polarization, whereas TORC1 promotes ribosome biogenesis and protein translation conducive to cell growth and proliferation while suppressing autophagy. In general, TORC1 is rapamycin sensitive in contrast to TORC2, which is resistant to rapamycin with the exception noted below for T. brucei [4], [6]. This report discusses recent studies uncovering emerging roles for Tor signaling in promoting fungal and protozoan pathogen growth and proliferation tailored to invade and colonize the host. Moreover, protozoans and viruses have also developed strategies to subvert the host Tor signaling cascade and thereby commandeer the translational machinery to evade the immune system and promote viral protein synthesis, respectively.
Tor Regulates Cell–Cell Adhesion in Candida albicans
Candida albicans is the most common opportunistic fungal pathogen of humans, causing skin and mucosal infections as well as potentially fatal systemic infections. Cell–cell adhesion is necessary for C. albicans to form biofilms, an important feature of its pathogenicity repertoire. Interestingly, Tor1 regulates the expression of several cell wall – and hyphal-specific genes, including adhesins and their transcriptional regulators, which elicit biofilm formation in C. albicans [7].
Exposure of C. albicans to rapamycin results in upregulation of the transcriptional activators Bcr1 and Efg1 as well as downregulation of the transcriptional repressors Nrg1 and Tup1, which control adhesin genes [7]. These effects correlate with expression of hyphal-induced genes, including those encoding the adhesins Als1, Als3, and Hwp1 and the gene encoding the cell wall protein Ece1. Importantly, Als1, Als3, and Hwp1 mediate cellular adhesion to a variety of host surfaces and facilitate adhesion during biofilm formation (reviewed in [8]).
Tor1 also regulates morphogenesis and cellular aggregation, which have implications for the pathogenicity and virulence of C. albicans. These effects appear to be at least in part mediated by Mds3, which is a regulator of morphogenetic processes such as the yeast to hyphal transition and biofilm formation [9], [10]. Although at present it is unclear how Mds3 acts, it has been suggested that Mds3 is a negative regulator of Tor1 [11]. Treatment with the Tor inhibitor rapamycin inhibits hyphal growth on solid media and causes extensive cellular aggregation and flocculation. These results are consistent with the model that Tor1 positively controls filamentation and negatively regulates cellular adhesion (Figure 1A) [7].
Fig. 1. Tor signaling governs key processes that dramatically impact pathogenesis of fungi, parasites, and viruses. 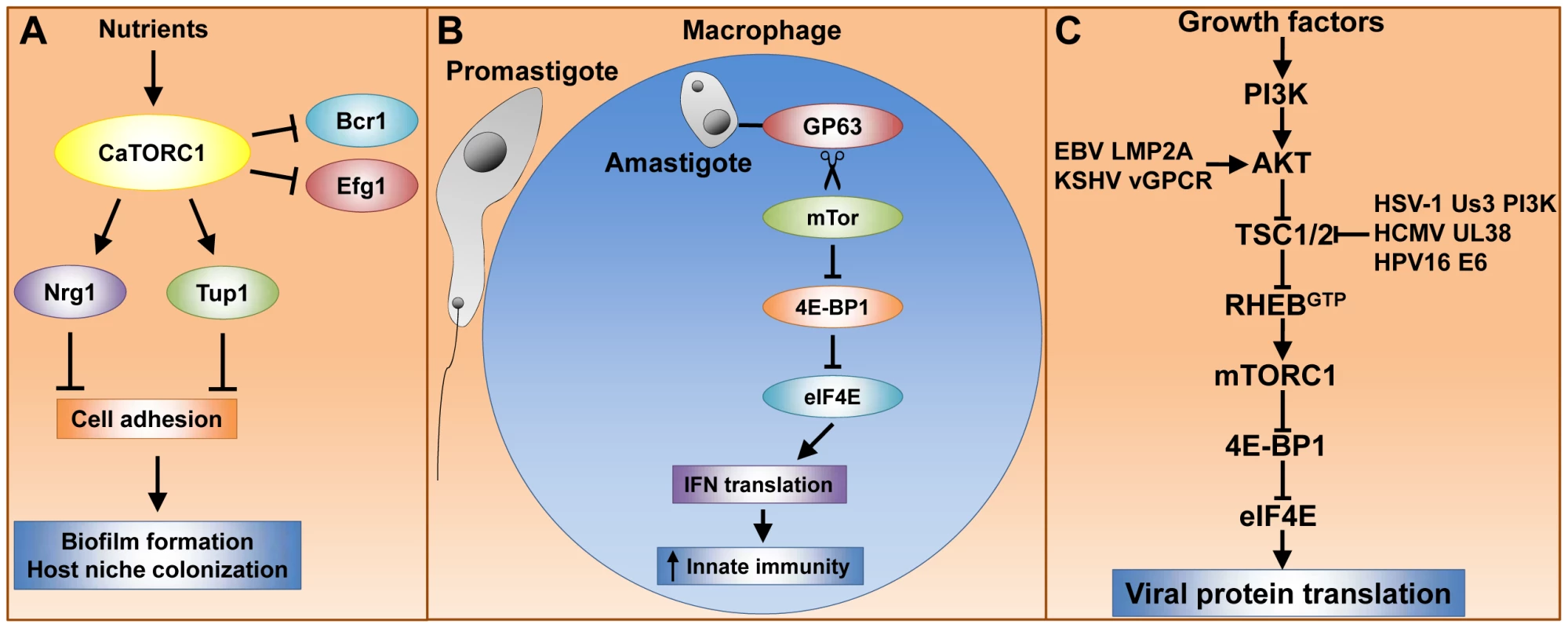
(A) caTor1 controls gene expression of adhesins and their upstream regulators, which elicit cell–cell adhesion, a process required for biofilm formation and host niche colonization. (B) The L. major GP63 cell surface protease promotes mTor cleavage and thereby mTORC1 inactivation. This event prevents mTORC1-dependent IFN type I translation, which dampens the host immune response and results in high parasite cell load. (C) Many human viral pathogens rely upon mTORC1 cap-dependent translation for replication. Growth factors activate mTORC1 via the phosphatidylinositol 3-kinase (PI3K), AKT, TSC1/2, Rheb signaling module (see text for details). Viruses have evolved protein factors capable of stabilizing the RhebGTP active form by either activating AKT or inactivating TSC2. RhebGTP activates mTORC1, enabling cap-dependent translation of early viral transcripts, which leads to viral replication. Rapamycin Potently Inhibits Trypanosome Proliferation by Blocking TORC2 Assembly
T. brucei is a protozoan parasite responsible for causing ∼500,000 annual infections in Africa that result in sleeping sickness, with devastating socioeconomic effects. T. brucei has a complex life cycle that develops in two different hosts (the tsetse fly and vertebrates) and several niches within these hosts. Nutritional stress encountered in the insect triggers the development of the non-infectious procyclic trypomastigote into the infective metacyclic trypomastigote. In addition, nutritional stress also induces adhesion of the parasite to the digestive tract of the insect [12]. Once a trypanosome is delivered into the favorable conditions of vertebrate blood, the epimastigote develops into the bloodstream form of the parasite and rapidly proliferates to establish a successful infection (reviewed in [13]).
Recent studies have shown that Tor signaling profoundly impacts the growth and development of T. brucei. Two classical Tor1 and Tor2 proteins, as well as two Tor-like proteins, have been identified in T. brucei [4]. The tbTor1 kinase associates with TORC1 and controls protein synthesis and cell size while the Tor-like 1 protein regulates autophagy [4], [14], [15]. The tbTor2 kinase populates TORC2 and, in contrast to the paradigm established in other organisms, tbTORC2 is sensitive to rapamycin, whereas tbTORC1 is resistant to rapamycin. tbTORC2 is involved in driving polarized cell growth, endocytosis, and cytokinesis [4].
Trypanosome cells have a highly polarized microtubular network that enables endocytosis and exocytosis to occur at a single site called the flagellar pocket; any perturbation of this network compromises endocytosis with fatal consequences for the organism. Accordingly, either tbTor2 depletion or rapamycin exposure (which prevents tbTORC2 formation) results in aberrant cell morphology with an enlarged flagellar pocket, actin cytoskeleton depolarization, impaired endocytosis, and a block to cytokinesis [4]. Interestingly, rapamycin treatment of the T. brucei bloodstream form cultured in vitro markedly inhibits cell proliferation. Thus, it has been suggested that therapy with less immunosuppressive rapamycin analogs could be highly efficacious against trypanosomiasis [4].
Leishmania major Hijacks the Host Translational Machinery via mTor Proteolysis
The protozoan parasite L. major is the etiologic agent for leishmaniasis and, similar to trypanosomes, develops as promastigotes in the midgut of sandflies and as amastigotes within macrophages of the vertebrate host. L. major contains three Tor homologs, at least one of which (Tor3) is required for macrophage infectivity and virulence [3].
mTOR-dependent translation of type I interferon (IFN) is critical for triggering host innate immune responses to defend against infection, including those caused by parasites [16], [17]. Interestingly, L. major hijacks the host translational machinery via disruption of mTor signaling, thereby enhancing parasite infectivity [18]. The L. major surface glycoprotein GP63, which exhibits Zn-dependent protease activity, is associated with L. major virulence. GP63 directly or indirectly promotes mTor cleavage, resulting in mTORC1 inhibition. This event prevents mTORC1-dependent phosphorylation and inactivation of host 4E-binding protein 1 (4E-BP1). Activated 4E-BP1 forms a physical complex with the mRNA cap binding factor eIF4E (elongation initiation factor 4E), thereby blocking the ability of eIF4E to assemble into the eIF4F complex and cap-dependent translation (reviewed in [19]). In parallel, type I IFN expression, which is deployed by the host immune system to defend against L. major, is decreased, though it is not yet clear what directly causes this phenomenon. Moreover, either rapamycin treatment or 4E-BP1 and 4E-BP2 knockout results in increased type I IFN expression and reduced L. major parasite load in mice. Thus, host mTORC1 is inactivated by L. major GP63 to promote parasite survival within host macrophages (Figure 1B). This model has led to the suggestion that host 4E-BP1/2 could be targeted therapeutically to treat leishmaniasis [18].
Viruses Activate mTor-Dependent Translation to Ensure Infection
Viruses depend on the cellular protein synthesis machinery for viral protein translation. In particular, poxivirus, adenovirus, and human herpes virus mRNA translation is heavily dependent upon mTor to inactivate the repressor 4E-BP1 and thus enable eIF4F cap-dependent translation (reviewed in [19], [20]). Growth factors activate mTor via the AKT-kinase, TSC1/2, Rheb-GTPase signaling module (Figure 1C). In response to growth factors, AKT phosphorylates the tuberous sclerosis heterodimer TSC1/2, thereby inactivating its GTPase activating protein (GAP) activity, which results in the active RhebGTP form and, in turn, mTor activation (reviewed in [21]). Interestingly, herpes and other viruses have developed multiple mechanisms to activate mTor and ensure sustained viral protein translation (Figure 1C). Epstein-Barr virus (EBV) and Kaposi's sarcoma herpes virus (KSHV) encode the proteins LMP2A and G protein–coupled receptor vGPCR, respectively, which activate AKT [22], [23]. The human cytomegalovirus (HCMV) UL38 protein and the human papilloma virus (HPV) E6 oncoprotein antagonize TSC2 [24].
Remarkably, the herpes simplex virus (HSV-1) Us3 kinase acts as an AKT surrogate capable of phosphorylating and inactivating TSC2, resulting in constitutive mTORC1 activation [25]. These results suggest that mTor inhibition should prevent viral replication and, in fact, the mTor kinase catalytic site inhibitor Torin1 potently blocks herpes virus replication, particularly during the early phase of viral infection [26], [27]. The Torin1 studies also revealed that the role of mTor activation during HCMV infection is not confined to inactivating 4E-BP1 or exclusive to the rapamycin-sensitive mTORC1, and additional targets within the mTORC1 and mTORC2 pathway components await elucidation.
Future Directions
The dramatic impact of nutrients as the triggers for morphogenic transitions that promote infection and host colonization in fungi and parasites, as well as the dependence of viral replication on the cellular translational machinery, prompted studies to investigate the roles of the Tor signaling cascade in these diverse infectious processes. Not surprisingly, the exciting results with pathogenic fungi, parasites, and viruses have thus far opened new avenues for further study. Recent studies with the Salmonella effector protein AvraA, which is conserved in Yersinia pathogenic species, suggest that bacteria have also developed strategies to target host signaling cascades (including mTor) to control gene expression and promote virulence [9]. Moreover, there is growing evidence that Tor-regulated processes such as autophagy have dramatic impact on virulence. In particular, it is well established that autophagy can destroy invading pathogens (a process also known as xenophagy) and enhances innate immunity, protecting cells and organisms against infection by a wide range of pathogens, including bacteria, viruses, and parasites (reviewed in [28]). Thus, we can expect that future studies in these arenas will continue to illuminate novel aspects of Tor signaling in microbial pathogenesis that will stimulate the development of new Tor-based therapies to combat infection.
Zdroje
1. HeitmanJMovvaNRHallMN 1991 Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science 253 905 909
2. ShertzCABastidasRJLiWHeitmanJCardenasME 2010 Conservation, duplication, and loss of the Tor signaling pathway in the fungal kingdom. BMC Genomics 11 510
3. Madeira da SilvaLBeverleySM 2010 Expansion of the target of rapamycin (TOR) kinase family and function in Leishmania shows that TOR3 is required for acidocalcisome biogenesis and animal infectivity. Proc Natl Acad Sci U S A 107 11965 11970
4. BarquillaACrespoJLNavarroM 2008 Rapamycin inhibits trypanosome cell growth by preventing TOR complex 2 formation. Proc Natl Acad Sci U S A 105 14579 14584
5. De VirgilioCLoewithR 2006 The TOR signalling network from yeast to man. Int J Biochem Cell Biol 38 1476 1481
6. LoewithRJacintoEWullschlegerSLorbergACrespoJL 2002 Two TOR complexes, only one of which is rapamycin sensitive, have distinct roles in cell growth control. Mol Cell 10 457 468
7. BastidasRJHeitmanJCardenasME 2009 The protein kinase Tor1 regulates adhesin gene expression in Candida albicans. PLoS Pathog 5 e1000294 doi:10.1371/journal.ppat.1000294
8. HoyerLLGreenCBOhSHZhaoX 2008 Discovering the secrets of the Candida albicans agglutinin-like sequence (ALS) gene family–a sticky pursuit. Med Mycol 46 1 15
9. DavisDABrunoVMLozaLFillerSGMitchellAP 2002 Candida albicans Mds3p, a conserved regulator of pH responses and virulence identified through insertional mutagenesis. Genetics 162 1573 1581
10. RichardMLNobileCJBrunoVMMitchellAP 2005 Candida albicans biofilm-defective mutants. Eukaryot Cell 4 1493 1502
11. ZacchiLFGomez-RajaJDavisDA 2010 Mds3 regulates morphogenesis in Candida albicans through the TOR pathway. Mol Cell Biol 30 3695 3710
12. FigueiredoRCRosaDSSoaresMJ 2000 Differentiation of Trypanosoma cruzi epimastigotes: metacyclogenesis and adhesion to substrate are triggered by nutritional stress. J Parasitol 86 1213 1218
13. FennKMatthewsKR 2007 The cell biology of Trypanosoma brucei differentiation. Curr Opin Microbiol 10 539 546
14. BarquillaANavarroM 2009 Trypanosome TOR as a major regulator of cell growth and autophagy. Autophagy 5 256 258
15. BarquillaANavarroM 2009 Trypanosome TOR complex 2 functions in cytokinesis. Cell Cycle 8 697 699
16. CaoWManicassamySTangHKasturiSPPiraniA 2008 Toll-like receptor-mediated induction of type I interferon in plasmacytoid dendritic cells requires the rapamycin-sensitive PI(3)K-mTOR-p70S6K pathway. Nat Immunol 9 1157 1164
17. BogdanCMattnerJSchleicherU 2004 The role of type I interferons in non-viral infections. Immunol Rev 202 33 48
18. JaramilloMGomezMALarssonOShioMTTopisirovicI 2011 Leishmania repression of host translation through mTOR cleavage is required for parasite survival and infection. Cell Host Microbe 9 331 341
19. GingrasACRaughtBSonenbergN 2004 mTOR signaling to translation. Curr Top Microbiol Immunol 279 169 197
20. NormanKLSarnowP 2010 Herpes Simplex Virus is Akt-ing in translational control. Genes Dev 24 2583 2586
21. FingarDCBlenisJ 2004 Target of rapamycin (TOR): an integrator of nutrient and growth factor signals and coordinator of cell growth and cell cycle progression. Oncogene 23 3151 3171
22. MoodyCAScottRSAmirghahariNNathanCOYoungLS 2005 Modulation of the cell growth regulator mTOR by Epstein-Barr virus-encoded LMP2A. J Virol 79 5499 5506
23. SodhiAChaisuparatRHuJRamsdellAKManningBD 2006 The TSC2/mTOR pathway drives endothelial cell transformation induced by the Kaposi's sarcoma-associated herpesvirus G protein-coupled receptor. Cancer Cell 10 133 143
24. MoormanNJCristeaIMTerhuneSSRoutMPChaitBT 2008 Human cytomegalovirus protein UL38 inhibits host cell stress responses by antagonizing the tuberous sclerosis protein complex. Cell Host Microbe 3 253 262
25. ChuluunbaatarURollerRFeldmanMEBrownSShokatKM 2010 Constitutive mTORC1 activation by a herpesvirus Akt surrogate stimulates mRNA translation and viral replication. Genes Dev 24 2627 2639
26. ClippingerAJMaguireTGAlwineJC 2011 The changing role of mTOR kinase in the maintenance of protein synthesis during human cytomegalovirus infection. J Virol 85 3930 3939
27. MoormanNJShenkT 2010 Rapamycin-resistant mTORC1 kinase activity is required for herpesvirus replication. J Virol 84 5260 5269
28. SumpterRJrLevineB 2010 Autophagy and innate immunity: triggering, targeting and tuning. Semin Cell Dev Biol 21 699 711
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek Hostile Takeover by : Reorganization of Parasite and Host Cell Membranes during Liver Stage EgressČlánek A Trigger Enzyme in : Impact of the Glycerophosphodiesterase GlpQ on Virulence and Gene ExpressionČlánek An EGF-like Protein Forms a Complex with PfRh5 and Is Required for Invasion of Human Erythrocytes byČlánek Th2-polarised PrP-specific Transgenic T-cells Confer Partial Protection against Murine ScrapieČlánek Alterations in the Transcriptome during Infection with West Nile, Dengue and Yellow Fever Viruses
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2011 Číslo 9- Jak souvisí postcovidový syndrom s poškozením mozku?
- Měli bychom postcovidový syndrom léčit antidepresivy?
- Farmakovigilanční studie perorálních antivirotik indikovaných v léčbě COVID-19
- 10 bodů k očkování proti COVID-19: stanovisko České společnosti alergologie a klinické imunologie ČLS JEP
-
Všechny články tohoto čísla
- Unconventional Repertoire Profile Is Imprinted during Acute Chikungunya Infection for Natural Killer Cells Polarization toward Cytotoxicity
- Envelope Deglycosylation Enhances Antigenicity of HIV-1 gp41 Epitopes for Both Broad Neutralizing Antibodies and Their Unmutated Ancestor Antibodies
- Co-opts GBF1 and CERT to Acquire Host Sphingomyelin for Distinct Roles during Intracellular Development
- Nrf2, a PPARγ Alternative Pathway to Promote CD36 Expression on Inflammatory Macrophages: Implication for Malaria
- Robust Antigen Specific Th17 T Cell Response to Group A Streptococcus Is Dependent on IL-6 and Intranasal Route of Infection
- Targeting of a Chlamydial Protease Impedes Intracellular Bacterial Growth
- The Protease Cruzain Mediates Immune Evasion
- High-Resolution Phenotypic Profiling Defines Genes Essential for Mycobacterial Growth and Cholesterol Catabolism
- Plague and Climate: Scales Matter
- Exhausted CD8 T Cells Downregulate the IL-18 Receptor and Become Unresponsive to Inflammatory Cytokines and Bacterial Co-infections
- Maturation-Induced Cloaking of Neutralization Epitopes on HIV-1 Particles
- Murine Gamma-herpesvirus Immortalization of Fetal Liver-Derived B Cells Requires both the Viral Cyclin D Homolog and Latency-Associated Nuclear Antigen
- Rapid and Efficient Clearance of Blood-borne Virus by Liver Sinusoidal Endothelium
- Hostile Takeover by : Reorganization of Parasite and Host Cell Membranes during Liver Stage Egress
- A Trigger Enzyme in : Impact of the Glycerophosphodiesterase GlpQ on Virulence and Gene Expression
- Strain Specific Resistance to Murine Scrapie Associated with a Naturally Occurring Human Prion Protein Polymorphism at Residue 171
- Development of a Transformation System for : Restoration of Glycogen Biosynthesis by Acquisition of a Plasmid Shuttle Vector
- Monalysin, a Novel -Pore-Forming Toxin from the Pathogen Contributes to Host Intestinal Damage and Lethality
- Host Phylogeny Determines Viral Persistence and Replication in Novel Hosts
- BC2L-C Is a Super Lectin with Dual Specificity and Proinflammatory Activity
- Expression of the RAE-1 Family of Stimulatory NK-Cell Ligands Requires Activation of the PI3K Pathway during Viral Infection and Transformation
- Structure of the Vesicular Stomatitis Virus N-P Complex
- HSV Infection Induces Production of ROS, which Potentiate Signaling from Pattern Recognition Receptors: Role for S-glutathionylation of TRAF3 and 6
- The Human Papillomavirus E6 Oncogene Represses a Cell Adhesion Pathway and Disrupts Focal Adhesion through Degradation of TAp63β upon Transformation
- Analysis of Behavior and Trafficking of Dendritic Cells within the Brain during Toxoplasmic Encephalitis
- Exposure to the Viral By-Product dsRNA or Coxsackievirus B5 Triggers Pancreatic Beta Cell Apoptosis via a Bim / Mcl-1 Imbalance
- Multidrug Resistant 2009 A/H1N1 Influenza Clinical Isolate with a Neuraminidase I223R Mutation Retains Its Virulence and Transmissibility in Ferrets
- Structure of Herpes Simplex Virus Glycoprotein D Bound to the Human Receptor Nectin-1
- Step-Wise Loss of Bacterial Flagellar Torsion Confers Progressive Phagocytic Evasion
- Complex Recombination Patterns Arising during Geminivirus Coinfections Preserve and Demarcate Biologically Important Intra-Genome Interaction Networks
- An EGF-like Protein Forms a Complex with PfRh5 and Is Required for Invasion of Human Erythrocytes by
- Non-Lytic, Actin-Based Exit of Intracellular Parasites from Intestinal Cells
- The Fecal Viral Flora of Wild Rodents
- The General Transcriptional Repressor Tup1 Is Required for Dimorphism and Virulence in a Fungal Plant Pathogen
- Interferon Regulatory Factor-1 (IRF-1) Shapes Both Innate and CD8 T Cell Immune Responses against West Nile Virus Infection
- A Small Non-Coding RNA Facilitates Bacterial Invasion and Intracellular Replication by Modulating the Expression of Virulence Factors
- Evaluating the Sensitivity of to Biotin Deprivation Using Regulated Gene Expression
- The Motility of a Human Parasite, , Is Regulated by a Novel Lysine Methyltransferase
- Phosphodiesterase-4 Inhibition Alters Gene Expression and Improves Isoniazid – Mediated Clearance of in Rabbit Lungs
- Restoration of IFNγR Subunit Assembly, IFNγ Signaling and Parasite Clearance in Infected Macrophages: Role of Membrane Cholesterol
- Protease ROM1 Is Important for Proper Formation of the Parasitophorous Vacuole
- The Regulated Secretory Pathway in CD4 T cells Contributes to Human Immunodeficiency Virus Type-1 Cell-to-Cell Spread at the Virological Synapse
- Rerouting of Host Lipids by Bacteria: Are You CERTain You Need a Vesicle?
- Transmission Characteristics of the 2009 H1N1 Influenza Pandemic: Comparison of 8 Southern Hemisphere Countries
- Th2-polarised PrP-specific Transgenic T-cells Confer Partial Protection against Murine Scrapie
- Sequential Bottlenecks Drive Viral Evolution in Early Acute Hepatitis C Virus Infection
- Genomic Insights into the Origin of Parasitism in the Emerging Plant Pathogen
- Genomic and Proteomic Analyses of the Fungus Provide Insights into Nematode-Trap Formation
- Influenza Virus Ribonucleoprotein Complexes Gain Preferential Access to Cellular Export Machinery through Chromatin Targeting
- Alterations in the Transcriptome during Infection with West Nile, Dengue and Yellow Fever Viruses
- Protease-Sensitive Conformers in Broad Spectrum of Distinct PrP Structures in Sporadic Creutzfeldt-Jakob Disease Are Indicator of Progression Rate
- Vaccinia Virus Protein C6 Is a Virulence Factor that Binds TBK-1 Adaptor Proteins and Inhibits Activation of IRF3 and IRF7
- c-di-AMP Is a New Second Messenger in with a Role in Controlling Cell Size and Envelope Stress
- Structural and Functional Studies on the Interaction of GspC and GspD in the Type II Secretion System
- APOBEC3A Is a Specific Inhibitor of the Early Phases of HIV-1 Infection in Myeloid Cells
- Impairment of Immunoproteasome Function by β5i/LMP7 Subunit Deficiency Results in Severe Enterovirus Myocarditis
- HTLV-1 Propels Thymic Human T Cell Development in “Human Immune System” Rag2 gamma c Mice
- Tri6 Is a Global Transcription Regulator in the Phytopathogen
- Exploiting and Subverting Tor Signaling in the Pathogenesis of Fungi, Parasites, and Viruses
- The Next Opportunity in Anti-Malaria Drug Discovery: The Liver Stage
- Significant Effects of Antiretroviral Therapy on Global Gene Expression in Brain Tissues of Patients with HIV-1-Associated Neurocognitive Disorders
- Inhibition of Competence Development, Horizontal Gene Transfer and Virulence in by a Modified Competence Stimulating Peptide
- A Novel Metal Transporter Mediating Manganese Export (MntX) Regulates the Mn to Fe Intracellular Ratio and Virulence
- Rhoptry Kinase ROP16 Activates STAT3 and STAT6 Resulting in Cytokine Inhibition and Arginase-1-Dependent Growth Control
- Hsp90 Governs Dispersion and Drug Resistance of Fungal Biofilms
- Secretion of Genome-Free Hepatitis B Virus – Single Strand Blocking Model for Virion Morphogenesis of Para-retrovirus
- A Viral Ubiquitin Ligase Has Substrate Preferential SUMO Targeted Ubiquitin Ligase Activity that Counteracts Intrinsic Antiviral Defence
- Membrane Remodeling by the Double-Barrel Scaffolding Protein of Poxvirus
- A Diverse Population of Molecular Type VGIII in Southern Californian HIV/AIDS Patients
- Disruption of TLR3 Signaling Due to Cleavage of TRIF by the Hepatitis A Virus Protease-Polymerase Processing Intermediate, 3CD
- Quantitative Analyses Reveal Calcium-dependent Phosphorylation Sites and Identifies a Novel Component of the Invasion Motor Complex
- Discovery of the First Insect Nidovirus, a Missing Evolutionary Link in the Emergence of the Largest RNA Virus Genomes
- Old World Arenaviruses Enter the Host Cell via the Multivesicular Body and Depend on the Endosomal Sorting Complex Required for Transport
- Exploits a Unique Repertoire of Type IV Secretion System Components for Pilus Assembly at the Bacteria-Host Cell Interface
- Recurrent Signature Patterns in HIV-1 B Clade Envelope Glycoproteins Associated with either Early or Chronic Infections
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- HTLV-1 Propels Thymic Human T Cell Development in “Human Immune System” Rag2 gamma c Mice
- Hostile Takeover by : Reorganization of Parasite and Host Cell Membranes during Liver Stage Egress
- Exploiting and Subverting Tor Signaling in the Pathogenesis of Fungi, Parasites, and Viruses
- A Viral Ubiquitin Ligase Has Substrate Preferential SUMO Targeted Ubiquitin Ligase Activity that Counteracts Intrinsic Antiviral Defence
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



