-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaPolyglutamine Toxicity Is Controlled by Prion Composition and Gene Dosage in Yeast
Polyglutamine expansion causes diseases in humans and other mammals. One example is Huntington's disease. Fragments of human huntingtin protein having an expanded polyglutamine stretch form aggregates and cause cytotoxicity in yeast cells bearing endogenous QN-rich proteins in the aggregated (prion) form. Attachment of the proline(P)-rich region targets polyglutamines to the large perinuclear deposit (aggresome). Aggresome formation ameliorates polyglutamine cytotoxicity in cells containing only the prion form of Rnq1 protein. Here we show that expanded polyglutamines both with (poly-QP) or without (poly-Q) a P-rich stretch remain toxic in the presence of the prion form of translation termination (release) factor Sup35 (eRF3). A Sup35 derivative that lacks the QN-rich domain and is unable to be incorporated into aggregates counteracts cytotoxicity, suggesting that toxicity is due to Sup35 sequestration. Increase in the levels of another release factor, Sup45 (eRF1), due to either disomy by chromosome II containing the SUP45 gene or to introduction of the SUP45-bearing plasmid counteracts poly-Q or poly-QP toxicity in the presence of the Sup35 prion. Protein analysis confirms that polyglutamines alter aggregation patterns of Sup35 and promote aggregation of Sup45, while excess Sup45 counteracts these effects. Our data show that one and the same mode of polyglutamine aggregation could be cytoprotective or cytotoxic, depending on the composition of other aggregates in a eukaryotic cell, and demonstrate that other aggregates expand the range of proteins that are susceptible to sequestration by polyglutamines.
Published in the journal: . PLoS Genet 8(4): e32767. doi:10.1371/journal.pgen.1002634
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1002634Summary
Polyglutamine expansion causes diseases in humans and other mammals. One example is Huntington's disease. Fragments of human huntingtin protein having an expanded polyglutamine stretch form aggregates and cause cytotoxicity in yeast cells bearing endogenous QN-rich proteins in the aggregated (prion) form. Attachment of the proline(P)-rich region targets polyglutamines to the large perinuclear deposit (aggresome). Aggresome formation ameliorates polyglutamine cytotoxicity in cells containing only the prion form of Rnq1 protein. Here we show that expanded polyglutamines both with (poly-QP) or without (poly-Q) a P-rich stretch remain toxic in the presence of the prion form of translation termination (release) factor Sup35 (eRF3). A Sup35 derivative that lacks the QN-rich domain and is unable to be incorporated into aggregates counteracts cytotoxicity, suggesting that toxicity is due to Sup35 sequestration. Increase in the levels of another release factor, Sup45 (eRF1), due to either disomy by chromosome II containing the SUP45 gene or to introduction of the SUP45-bearing plasmid counteracts poly-Q or poly-QP toxicity in the presence of the Sup35 prion. Protein analysis confirms that polyglutamines alter aggregation patterns of Sup35 and promote aggregation of Sup45, while excess Sup45 counteracts these effects. Our data show that one and the same mode of polyglutamine aggregation could be cytoprotective or cytotoxic, depending on the composition of other aggregates in a eukaryotic cell, and demonstrate that other aggregates expand the range of proteins that are susceptible to sequestration by polyglutamines.
Introduction
A variety of human neurodegenerative disorders are associated with expansions of polyglutamine (poly-Q) repeats in certain proteins [1], [2]. One well known example is Huntington's disease (HD), which is caused by an expansion of the poly-Q stretch, located within the N-terminal stretch of the essential protein called huntingtin (Htt) [3]. Poly-Q expansion promotes formation of aggregates by the proteolytic Htt fragments containing an expanded poly-Q stretch [4], [5]. As the poly-Q-expanded N-terminal region of Htt is shown to aggregate and produce HD-like neurodegeneration in the mouse model, it is clear that this region is sufficient for reproducing the characteristic features of poly-Q aggregation and toxicity [6], [7]. Poly-Q associated pathologies can not be explained solely by the loss of the cellular function of a respective protein, e. g. Htt (for review, see [1]). Sequestration of other essential proteins by poly-Q aggregates was proposed to be a possible mechanism for toxicity [1], [8]. However, different experimental models suggested different candidates for sequestration [9]–[12], which decreased enthusiasm for the sequestration model.
To complicate matters further, expanded poly-Q proteins form various types of aggregates in mammalian cells [13], [14]. In the case of Htt, both nuclear and cytoplasmic aggregates were found [4], [15], [16]. Their contributions to poly-Q pathogenicity remain a topic of intense discussion [17], [18]. At least, most researchers agree that one type of cytoplasmic aggregated structure, so-called “aggresome”, plays a cytoprotective role via assembling poly-Q expanded Htt at one site and possibly promoting its autophagy-dependent clearance [19]–[22]. The aggresome is located perinuclearly, associated with the centrosome, and assembled with participation of the microtubular cytoskeleton. Other misfolded proteins can also be sequestered into an aggresome, indicating that this structure serves as a universal quality control depot for aggregating proteins [19], [23]–[27].
Experimental assays for studying the molecular mechanism of poly-Q aggregation and toxicity have been developed in the yeast Saccharomyces cerevisiae [28]–[34]. It has been shown that cytoplasmic aggregation and toxicity of the chimeric protein, generated by a fusion of the expanded poly-Q stretch of Htt to the green fluorescent protein (GFP), is facilitated by the presence of the endogenous yeast prions, [PIN+] and/or [PSI+] [30], [35]. In the absence of a prion, aggregates of this construct were rare, and no significant cytotoxicity was detected. However, in the presence of a prion, multiple peripherally located aggregates were formed, and cytotoxicity was observed [30]. The prions [PIN+] and [PSI+] are self-perpetuating aggregates of the endogenous yeast proteins Rnq1 (unknown function) and Sup35 (translation termination, or release factor, also called eRF1), respectively (for review, see [36]). Both of these proteins contain QN-rich prion domains (PrDs) that are responsible for aggregation properties (for review, see [37], [38]). It is likely that pre-existing prion aggregates nucleate aggregation of poly-Q expanded huntingtin. In the case of the Rnq1 prion, it was shown that poly-Q aggregates sequester some cytoskeletal components and inhibit endocytosis, which apparently contributes to cytotoxicity [39]. Inhibition of endocytosis was also detected in mammalian cells expressing poly-Q [40]. As mammalian Htt has been proposed to play a role in vesicle trafficking [41], these results are likely relevant to human HD.
Flanking sequences modulate poly-Q toxicity [32], [42]. In yeast strains containing the Rnq1 prion, cytotoxicity was eliminated by using a longer Htt fragment, which includes a proline (P)-rich stretch in addition to poly-Q. This P-rich stretch was shown to target aggregated poly-Q protein into a single perinuclear microtubule-dependent deposit, co-localized with the spindle body (yeast counterpart of a centrosome) and therefore resembling a mammalian aggresome [42]. The cytoprotective role of the aggresome, as opposed to cytotoxicity of some other types of aggregates, recapitulates the situation previously observed in mammalian cells [20], [21], [23].
While the prion form of Sup35 protein ([PSI+]) also promotes poly-Q toxicity in the yeast assay [35], the mechanism for this toxicity has not been studied in detail previously. In our current work, we demonstrate that [PSI+]-dependent poly-Q toxicity is not counteracted by aggresome formation, but is ameliorated by an increased dosage of some components of the translational termination machinery. These data show that targets of poly-Q toxicity and the cytoprotective potential of the aggresome depend on the composition of endogenous aggregated proteins in a eukaryotic cell.
Results
Aggresome formation does not ameliorate polyglutamine toxicity in the presence of Sup35 prion
To distinguish between the different patterns of poly-Q aggregation in yeast, we have employed the previously described constructs (Figure 1A) that produce the N-proximal region of Htt, fused to the FLAG epitope at the N-terminus and the green fluorescent protein (GFP) at the C-terminus. The N-terminal Htt region included the poly-Q stretch, which is either followed (poly-QP) or not followed (poly-Q) by the P-rich region. The poly-Q expanded versions (103Q and 103QP) contained a stretch of 103 glutamine residues, which corresponds to a severe form of Huntington's disease, while control non-aggregating versions (25Q and 25QP) contained 25 glutamine residues. As there was no difference in the effects of 25Q and 25QP, some of the figures show only the 25Q control. As described previously [30], the 103Q construct was toxic to yeast strains containing either [PIN+] (Rnq1 protein in a prion form) or [PSI+] (Sup35 protein in a prion form), with a combination of both prions showing an additive effect (Figure 1B). Also in agreement with previous observations [42], the 103QP construct was not toxic to the strains containing only Rnq1 prion. Surprisingly, the 103QP construct was toxic to the strains containing the Sup35 prion, independently in the presence or absence of Rnq1 prion (Figure 1B). Fluorescence microscopy confirmed that 103QP preferentially formed a single perinuclear aggregate deposit (aggresome) in the cells containing Rnq1 and/or Sup35 prions, while 103Q produced multiple peripheral aggregates (Figure 1C). Therefore, the ability of poly-QP to form an aggresome was not affected by the Sup35 prion, however, amelioration of toxicity by the aggresome was impaired. These data show that the mechanism of polyglutamine toxicity, promoted by the Sup35 prion, is different from the mechanism of polyglutamine toxicity promoted by the Rnq1 prion. Unless stated otherwise, all further experiments were performed in the strains containing the [PIN+] prion, thereby comparing the [PSI+] and [psi−] derivatives so that we could distinguish the effects attributed specifically to the [PSI+] prion.
Fig. 1. Polyglutamine toxicity and aggregation in the yeast strains with various prion compositions. 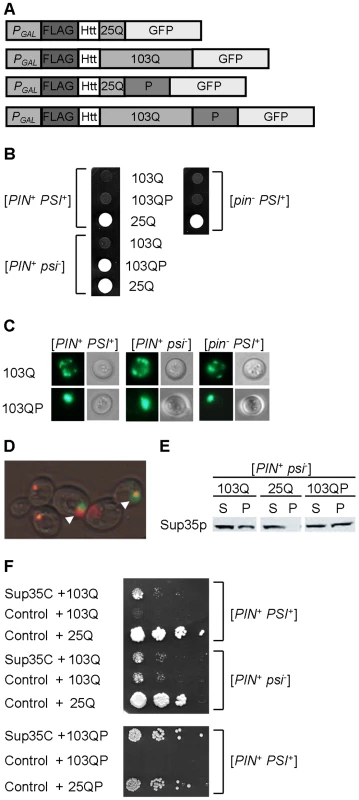
A – Polyglutamine constructs used in this work. All constructs were under the control of the galactose-inducible promoter (PGAL), and contained the FLAG epitope, N-terminal 17 amino acid residues and poly-Q stretch of human Htt, and were fused to the gene coding for green fluorescent protein (GFP) at C-terminus. Numbers indicate length of poly-Q stretch. Poly-QP constructs also contained the proline-rich region of Htt (designated as P), immediately following the poly-Q stretch. B – Expanded poly-Q without a P-rich region (103Q), expressed under the PGAL promoter on -Ura/Gal medium, is toxic in the presence of either [PIN+] or [PSI+] (or both), with two prions showing an additive effect. In contrast, expanded poly-Q with a P-rich region (103QP) is toxic only in the presence of [PSI+]. The 25Q construct, not exhibiting toxicity under these conditions, is shown as a control. The 25QP construct (not shown) behaved in the same way as 25Q. C – 103Q and 103QP form multiple peripheral aggregates and single aggregate (aggresome), respectively, in cells containing either or both prions ([PIN+] and/or [PSI+]), as visualized by fluorescence microscopy. Perinuclear location of aggresome (not shown) was confirmed by DAPI staining as described previously [42]. D – Overexpressed Sup35NM-DsRed (red) forms large clumps in the [PSI+] cells, that overlap with the 103QP-GFP aggresome (green), as pointed by arrows. E - Expression of 103Q or 103QP promotes aggregation of Sup35 in the [psi−] strain as seen by an increase of pellet (P) versus supernatant (S) fraction, in comparison to the respective strain expressing 25Q. Centrifugation analysis was followed by Western blotting and immunostaining with the Sup35 antibody. F - Expression of the Sup35 derivative, lacking the prion and middle domains (Sup35C), decreases 103Q and 103QP toxicity in the [PIN+ PSI+] strain but does not influence 103Q toxicity in the [PIN+ psi−] strain. SUP35C gene was under control of the endogenous SUP35 promoter. Serial decimal dilutions were spotted onto -Ura/Gal medium. Polyglutamine accumulation leads to Sup35 sequestration
Notably, when Sup35NM, tagged with DsRed, and 103QP-GFP are co-overproduced in the [PSI+] strain, most of the Sup35NM-DsRed is eventually assembled into one large deposit, that is either partially or completely overlapping with the 103QP aggresome (Figure 1D). This indicates a possibility of sequestration of Sup35 by the aggresome, and agrees with the previous observation [43], confirmed by us (Figure 1E) that polyglutamines promote aggregation of a fraction of Sup35, even in a [psi−] strain. Notably, 103QP toxicity in the [PSI+] cells was ameliorated by introducing the Sup35 derivative (designated Sup35C) that lacks the N-terminal (prion) and middle domains and therefore, is functional but unable to be incorporated into the prion aggregates (Figure 1F). Expression of Sup35C also decreased toxicity of 103Q in the [PIN+ PSI+] strain down to the levels observed in the [PIN+ psi−] strain. However, Sup35C did not affect toxicity of 103Q in [PIN+ psi−]. These data confirm that in the [PSI+] strain (but not in the [PIN+ psi−] strain), sequestration of Sup35 contributes to polyglutamine toxicity.
Isolation of the anti-polyQ-toxic (AQT) mutants
Next, we looked for other genetic factors influencing the [PSI+]-dependent polyglutamine toxicity. As the ubiquitin-proteasome system (UPS) is known to influence poly-Q effects in mammalian cells, we have studied poly-Q toxicity in the yeast strain with a deletion of the UBC4 gene, coding for one of the major yeast ubiquitin-conjugating enzymes [44]. Ubc4Δ did not improve growth in the presence of polyglutamines (Figure 2A) however the ubc4Δ 103Q strain produced spontaneously arising fast-growing papillae (Figure 2B). Three independent papillae were analyzed further, and each was confirmed to stably reproduce the anti-toxic phenotype (Figure 2B and 2C), and also to ameliorate toxicity of 103QP (Figure 2D). These derivatives were named AQT for Anti-polyQ Toxicity, with respective phenotype designated as Aqt+. Specific effect of AQT on toxicity caused by expanded polyglutamines was especially pronounced after longer periods of incubation (Figure 2C). All AQT derivatives retained the [PIN+] and [PSI+] prions (data not shown). In each derivative, the Aqt+ phenotype was dominant (see Figure 2E as an example) and segregated in a Mendelian fashion in meiosis (Table S1). All pairwise genetic crosses between three independent AQT derivatives produced 4 AQT: 0 wild-type pattern of segregation in the vast majority of tetrads (Table S2), indicating that all AQT derivatives are formally confined to a single genetic locus. Reintroduction of the wild-type UBC4 gene into the AQT strain decreased but did not completely eliminate amelioration of toxicity, indicating that ubc4Δ strengthens the Aqt+ phenotype but is not required for its manifestation (Figure 2F).
Fig. 2. Isolation and characterization of anti-polyQ toxicity (AQT) derivatives. 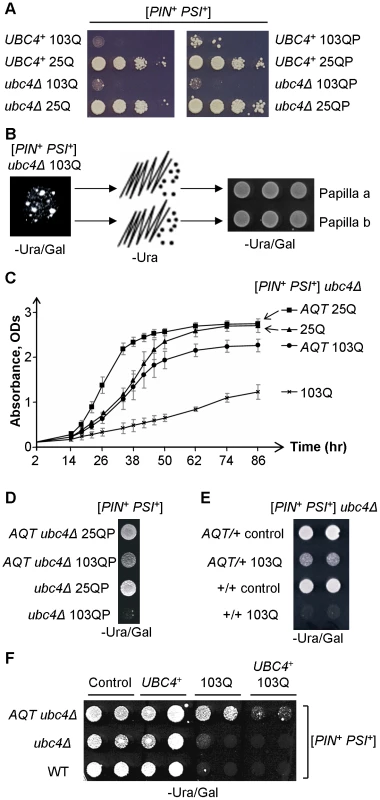
A – Ubc4Δ has no significant effect on toxicity of 103Q or 103QP in the [PIN+ PSI+] background. Serial decimal dilutions were spotted onto -Ura/Gal medium. B – Papillae arise spontaneously in the ubc4Δ [PIN+ PSI+] strain expressing 103Q, and are able to stably maintain the anti-polyQ-toxic phenotype after colony purification. These papillae were designated as AQT. C – Comparison of the growth curves of [PIN+ PSI+] ubc4Δ strains that differ by polyglutamine constructs and by the presence or absence of AQT. Growth was measured by optical density at 600 nm in the liquid –Ura medium with galactose and raffinose instead of glucose. At least 3 independent cultures were characterized per each combination. Error bars represent standard deviations. D – AQT ameliorates 103QP toxicity. E – AQT is dominant (all strains are [PIN+ PSI+] and ubc4Δ homozygotes). F – Reintroduction of the UBC4 gene under galactose-inducible promoter on a multicopy plasmid partly suppresses but does not completely eliminate anti-toxic effect of AQT. -Ura/Gal plates are scored on panels D, E and F. Despite their anti-toxic effect, AQT derivatives retained the typical mode of cytologically detectable aggregation for both 103Q (multiple peripheral aggregates, Figure 3A) and 103QP (single perinuclear aggregate, Figure 3B), indicating that amelioration of toxicity is not due to a lack of aggregation. Overproduction of Sup35 protein or its prion domain, Sup35N, is known to inhibit growth of the [PSI+] strains [37], [38]. This effect was ameliorated in AQT derivatives (Figure 3C). The AQT strains also exhibited additional phenotypes that were not directly related to amelioration of toxicity, such as compensation of the ubc4Δ-mediated temperature sensitivity and loss of the invasive growth capability (Figure S1). AQT also slightly increased the growth of the [PIN+ psi−] ubc4Δ strain expressing 103Q, especially after shorter incubation periods (Figure S1D), possibly due to increased robustness of the AQT ubc4Δ strain in the stressful conditions.
Fig. 3. Effects of AQT on polyglutamine aggregation and Sup35 toxicity. 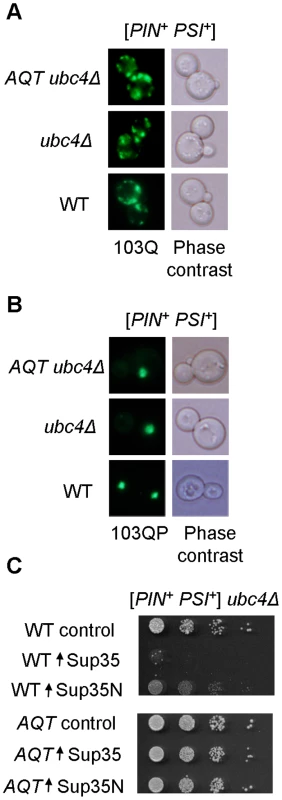
A and B – Typical aggregation patterns of 103Q (multiple dots, A) and 103QP (single clump, B) are not affected by AQT, as confirmed by fluorescence microscopy. C – AQT mutant ameliorates toxicity of excess Sup35 or Sup35N in the [PSI+] strain. Sup35 and Sup35N proteins were expressed from centromeric plasmid under control of the galactose-inducible promoter. Cells were grown on the -Ura/glucose medium selective for the plasmid for 1 day. Serial decimal dilutions were plated onto -Ura/Gal medium. AQT derivatives are disomics of chromosome II
Genetic crosses aimed at characterizing the inheritance of AQT revealed that AQT is centromere-linked (Figure S2A). Moreover, when the original AQT derivative, obtained in the strain bearing the ubc4Δ::HIS3 disruption was mated to the wild-type strain bearing the ubc4Δ::KanMX disruption (causing resistance to the antibiotic G418), both AQT and KanMX markers segregated 2∶2 in meiosis as expected, while majority of tetrads did show 3∶1 or 4∶0 segregation for the His+ phenotype, indicative of the presence of two copies of the HIS3 allele in the cross (Figure 4A). Notably, all AQT spores (76 total) obtained from tetrads with a 2∶2 ratio for KanMX were His+. The simplest scenario compatible with these ratios is that the AQT derivative is a disomic of chromosome II, where the ubc4Δ:: HIS3 allele is located. As chromosome segregation is controlled by the centromere, this would also explain the centromere linkage of AQT. Indeed, fractionation of the yeast chromosomes via CHEF (Contour-clamped Homogeneous Electric Field) gel electrophoresis confirmed that each of the three independent AQT derivatives contains an additional copy of the chromosome II band (Figure S2B). Extra-copy of chromosome II also co-segregated with AQT in tetrad analysis (data not shown). Microarray-based analysis of genomic DNA of the three original AQT derivatives and two AQT meiotic segregants confirmed that each of these strains contains an extra copy of every piece of the coding material in chromosome II (Figure 4B). Overall, our data demonstrate that AQT is associated with an extra copy of chromosome II.
Fig. 4. AQT derivatives are disomic for chromosome II, and extra-copy of SUP45 is responsible for antitoxicity. 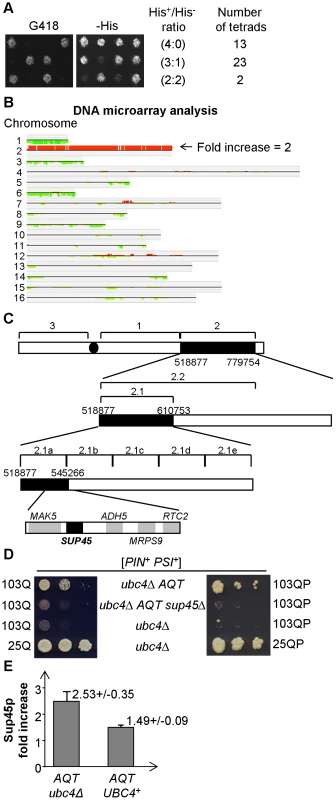
A – Tetrad analysis of a diploid obtained from mating of the AQT strain bearing the ubc4Δ::HIS3 transplacement, to the strain bearing the ubc4Δ::KanMX transplacement, demonstrates presence of at least 2 copies of the HIS3 gene versus one copy of the KanMX gene. This can be concluded from the fact that majority of tetrads produce more than 2 His+ spores, in contrast to the typically 2∶2 segregation by G418 resistance caused by KanMX. All AQT spores in this cross were His+ (not shown). B – Hybridization of total DNA to a complete DNA microarray of the S. cerevisiae genome confirms that all the coding material of chromosome II is duplicated in the AQT strain. Comparison is performed according to CLAC (CLuster Along Chromosome) consensus plot. For procedure, see Text S1. C – Sequential deletion mapping of the chromosome II extra copy in the AQT strain. The AQT#7 derivative (see Figure S2B) was used in these experiments. Each numbered region corresponds to a respective deletion. Deletions eliminating the antitoxicity phenotype in the [PSI+] background are shown as boxes filled in black. All deletions were verified by PCR. Five ORFs located within region 2.1a were each deleted individually; among those deletions, only deletion of SUP45 eliminated AQT as shown on panels B and C. D – Elimination of the antitoxic effect on 103Q and 103QP by the sup45 deletion in AQT strain. Serial decimal dilutions were spotted onto -Ura/Gal medium. E – Sup45 protein levels are elevated in the AQT strain, more profoundly in ubc4Δ background than in the presence of wild type UBC4 gene (UBC4+). Sup45p level is shown relative to the isogenic monosomic (non-AQT) control in each case. Ade2 protein was used as the loading control. At least 3 measurements with independent cultures were performed in each case. Error bars correspond to standard deviations. In each case the difference in Sup45 levels between the AQT and non-AQT strain is statistically significant as confidence limits do not overlap, and differences between the UBC4+ and ubc4Δ strains are statistically significant according to t-criterium (PHo<0.01). Extra copy of the SUP45 gene is responsible for the amelioration of [PSI+]-dependent polyglutamine toxicity in the AQT derivatives
Sequential deletion analysis of the extra copy of chromosome II in the AQT strain confined the gene responsible for amelioration of toxicity to the region of the right arm, located between positions 528161 and 537490 (Figure 4C). This region contains 5 ORFs, including the essential gene SUP45, that codes for a translation termination factor Sup45, or eRF1, working together with Sup35 (eRF3) [45]. We have disrupted the copy of the SUP45 gene, located on the duplicated chromosome II in the AQT strain, and have shown that this disruption eliminates the anti-toxicity effect on both 103Q and 103QP (Figure 4D). Notably, other phenotypes, associated with chromosome II disomy but not related to amelioration of polyglutamine toxicity in the [PSI+] strain, including slightly increased growth of the [PIN+ psi−] strain in the presence of 103Q, were not affected by sup45Δ (Figure S1B, S1C and S1E).
Western blot analysis confirmed that the AQT derivative contains more Sup45 protein, compared to the isogenic wild type strain (Figure 4E). This increase was more profound in the ubc4Δ than in the UBC4+ background. This explains why the AQT effect was better seen in ubc4Δ. Thus, an increase in Sup45 levels due to the presence of an extra-copy of the SUP45 gene is responsible for the anti-toxic (Aqt+) phenotype in the [PSI+] background.
Plasmid-mediated overproduction of a release factor also ameliorates polyglutamine toxicity
Next, we checked if an increase in the Sup45 levels, produced by means other than duplication of chromosome II, would also ameliorate the [PSI+]-dependent polyglutamine toxicity. Indeed, introduction of the centromeric plasmid bearing the SUP45 gene under its own (Figure 5A) or galactose-inducible (Figure 5B) promoter (in the latter case, under inducing conditions) ameliorated toxicity of both 103Q and 103QP. Anti-toxic effect of plasmid-borne SUP45 was clearly detected in both ubc4Δ and UBC4+ backgrounds, indicating that it is less sensitive to the presence of Ubc4 protein, compared to the chromosomal extra copy. For both plasmids, Sup45 overproduction was confirmed by protein analysis (Figure 5C and 5D). Ability of the extra-copy of SUP45 to ameliorate polyglutamine toxicity was abolished by a deletion of 19 C-terminal amino acids (Figure 5E), that impairs Sup45 function in translation and interaction with Sup35 [46], or by the missense mutation sup45-103, T62C (Figure 5F) that also impairs Sup45 function in translation termination [47]. Thus, Sup45 ability to ameliorate toxicity depends on the same sequence elements that control its function in translational machinery.
Fig. 5. Modulation of polyglutamine toxicity by the plasmid-borne release factor genes. 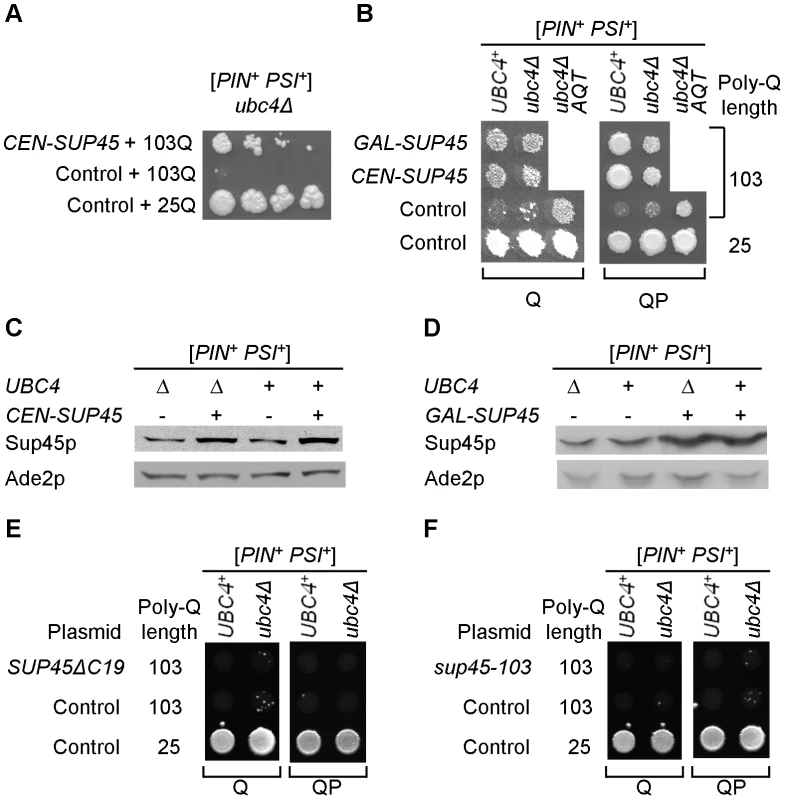
A - An extra copy of SUP45 gene, located on the centromeric plasmid under endogenous promoter, ameliorates toxicity of 103Q in the ubc4Δ [PSI+] strain, as seen from serial decimal dilutions plated onto the galactose medium selective for both poly-Q and SUP45 (or control) plasmids. B – Amelioration of [PSI+]-dependent polyglutamine toxicity by a plasmid-borne extra copy of SUP45 gene is detected for both endogenous (CEN-SUP45) galactose-inducible (GAL-SUP45) promoters, for both 103Q and 103QP constructs, and in both ubc4Δ and UBC4+ strains. Antitoxic effect of the plasmid-borne SUP45 gene in the ubc4Δ strain is comparable to antitoxic effect of AQT. Toxicity was scored on the galactose medium selective for both poly-Q and SUP45 (or control) plasmids. C and D – Centromeric plasmids with SUP45 gene under endogenous (C) or galactose-inducible PGAL (D) promoters increase levels of Sup45 protein (Sup45p) both [UBC4+] and ubc4Δ strains. Cultures were grown in liquid -Ura -Leu glucose (C) or -Ura -Leu galactose/raffinose (D) medium. Ade2 (Ade2p) protein is shown as a loading control. E and F – Plasmids, expressing the SUP45 alleles with either C-terminal deletion, SUP45ΔC19 (that abolishes Sup45 function and interaction with Sup35) (E) or missense mutation sup45-103, T62C (that impairs Sup45 function) (F) from the endogenous SUP45 promoter, do not ameliorate 103Q and 103QP toxicity, as scored on the galactose medium selective for both plasmids. Aggregation patterns of polyQ and release factors in the wild-type and AQT strains
As both polyglutamines and prion form of Sup35 form SDS-resistant polymers in the yeast cells, we have checked if patterns of their aggregation are influenced by the presence of an extra copy of SUP45. Both 103Q and 103QP proteins exhibit a broad range of distribution of the SDS-resistant polymers by size, as demonstrated by semi-denaturing agarose gel electrophoresis (SDD-AGE), with 103QP containing more protein in the higher molecular weight (MW) fraction (Figure 6A). This result confirms that the aggresome, formed by 103QP, contains insoluble protein aggregates, in contrast to the juxtanuclear quality control compartment (JUNQ) observed in the yeast cells with a defect of the ubiquitin-proteasome system [48]. Neither 103Q nor 103QP polymer distribution was significantly affected by AQT (Figure 6A). In the wild type [PSI+] cells containing non-expanded polyglutamines (25Q), Sup35 prion polymers were distributed within a relatively narrow range of sizes (Figure 6B). However, in the presence of either 103Q or 103QP, size range of the Sup35 polymers was increased and higher molecular weight (MW) polymers were accumulated, suggesting that some Sup35 could be associated with 103Q (or 103QP) polymers, therefore partly following their distribution. Notably, the Sup35 polymer size range became narrower in the presence of AQT, and the high MW fraction, which depends on 103Q/QP, disappeared (Figure 6B). This suggests that the extra dosage of Sup45 somewhat counteracts the increase in size of the Sup35 polymers and possibly, their interaction with polyglutamines.
Fig. 6. Effects of AQT on aggregation of release factors. 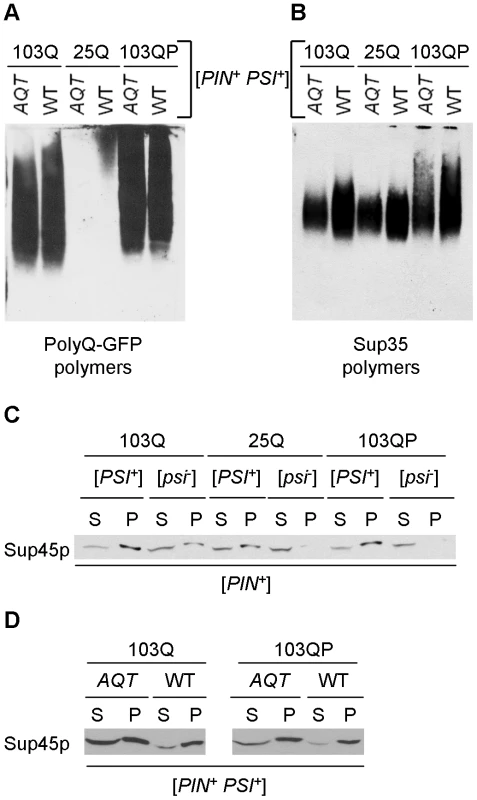
A and B – Fractionation of the polyQ/QP-GFP (A) and Sup35 (B) polymers by sizes in the ubc4Δ [PSI+] strains either with (AQT) or without (WT) AQT. Polymers were separated by semi-denaturing agarose gel electrophoresis (SDD-AGE, see Text S1). Filter obtained from one and the same gel was reacted to either GFP (A) or Sup35C (B) antibodies. Polyglutamines alter distribution of Sup35 polymers, and this effect is counteracted by AQT. Experiment has been repeated with 3 independent cultures per each combination, each time with the same result. C – Expression of 103Q promotes aggregation of Sup45 protein (Sup45p) in the ubc4Δ [PIN+ psi−] strain, and expression of either 103Q or 103QP increases aggregate-associated fraction of Sup45 in the ubc4Δ [PSI+] strain, as detected by an increase in the pellet (P) versus supernatant (S) fraction in comparison to the respective strain expressing 25Q. Centrifugation was followed by Western blotting and reaction to the Sup45 antibody. D – Proportion of soluble (supernatant, S) versus aggregate-associated (pellet, P) Sup45 protein is significantly increased in AQT ubc4Δ [PSI+] strain, compared to the identical non-AQT (WT) strain, as determined by centrifugation analysis, followed by Western blotting and reaction to the Sup45 antibody. We have also checked effects of polyglutamines and gene dosage on patterns of Sup45 aggregation. Sequestration of Sup45 by the Sup35 prion aggregates is known to contribute to cytotoxicity of overproduced Sup35 in [PSI+] strains [49]. We could not detect aggregate-associated Sup45 by SDD-AGE (data not shown), apparently because it is not converted into an amyloid form and is therefore released after SDS treatment. However, centrifugation analysis demonstrated that presence of either [PSI+] prion or 103Q protein resulted in the shift of a fraction of Sup45 protein to the pelletable (aggregate-associated) form, with both prion and 103Q together having an additive effect (Figure 6C). 103QP did not exhibit any observable effect on Sup45 aggregation in the [psi−] strain, however it further increased Sup45 aggregation in the presence of [PSI+]. Remarkably, proportion of the pelletable versus soluble Sup45 was decreased in the AQT (disomic) [PSI+] strain expressing 103Q or 103QP, compared to the identical strain not possessing disomy (Figure 6D). This showed that an increase in Sup45 levels counteracted its sequestration by aggregates.
Overall, our data indicate that both release factors, Sup35 and Sup45, are sequestered by polyglutamine aggregates in the [PSI+] cells, and that excess Sup45 not only restores supply of functional Sup45 but also changes the mode of Sup35 aggregation.
Polyglutamines and translational readthrough
As our data point to sequestration of release factors as a mechanism of polyglutamine toxicity, we have checked if polyglutamines increase translational readthrough of stop codons. For this purpose, the chimeric constructs bearing a stop codon between the PGK1 and lacZ ORFs have been employed. Surprisingly, no increase in translational readthrough (measured by β-galactosidase activity) has been detected in the presence of 103Q (Table S3). One possible explanation of these data is that damage to translational machinery, caused by the aggregation and sequestration of release factors in the presence of polyglutamines, is so severe that translation is arrested and not proceeding beyond the stop codon. Another (but not mutually exclusive) possibility is that cytotoxicity is related to non-translational functions of Sup35/45. Indeed, it has been reported that the immediate consequence of the severe shortage of a release factor in yeast is not translational defect per se, but cytoskeleton damage leading to cell death [50].
Discussion
Prion role in polyglutamine toxicity
Our data demonstrate that mechanism of polyglutamine toxicity depends on the prion composition of the cell. In fact, it appears that polyglutamine protein is not the toxicity agent itself, but rather amplifies the effects of the endogenous prion aggregates by sequestering them and making them more rigid. In case of Rnq1 prion, sequestration of Rnq1 by polyglutamines also leads to sequestration of the cytoskeletal proteins, interacting with Rnq1, and subsequent impairment of endocytosis [39], [40]. In this case, toxicity is relieved by re-localization of polyglutamines to an aggresome that removes polyQ (and possibly Rnq1) from the endocytic sites [42]. However, in case of Sup35 prion, relocalization is not sufficient for amelioration of toxicity. This could be due to the fact that Sup35 itself is an essential protein, and/or due to its normal distribution all over the cytoplasm, making it impossible to define specific toxicity sites. Additive action of Rnq1 and Sup35 prions on 103Q toxicity in the absence of the P-rich region also confirms that their cytotoxic effects are at least partly independent of each other.
Remarkably, our data show for the first time that the aggresome is not always cytoprotective. Moreover, it is possible that formation of the aggresome in the cell containing an essential protein in the form of self-perpetuating amyloid (prion) is itself cytotoxic due to sequestration of this essential protein. However, it remains uncertain whether toxicity is primarily driven by sequestration of Sup35 into an aggresome, or by its sequestration into the smaller polyglutamine aggregates remaining in the cytoplasm. While some Sup35 is definitely detected in the aggresome (Figure 1D), we don't know if the functional fraction of Sup35 is sequestered there. Moreover, while it is obvious that some fraction of Sup35 should retain function in the [PSI+] strain, as elimination of Sup35 is lethal [51], it remains unknown whether this functional component of Sup35 is represented by residual non-aggregated Sup35, smaller oligomers, or both. However, it is more likely that a fraction of oligomeric Sup35 remains functional, as amount of monomeric Sup35 retained by the strong [PSI+] strains, that is essentially at the limit of detection, seems too low for maintaining viability. Indeed, Sup35C regions are not included in the amyloid core and may stay enzymatically active, so that only size, composition and/or location of the aggregate would modulate its functionality in the cellular context. Changes in the distribution of Sup35 polymers by size in the presence of polyglutamines (Figure 6B), clearly show that certain alterations of Sup35 aggregation patterns, making them more similar to poly-Q aggregation patterns, coincide with toxicity.
Modulation of polyglutamine toxicity by Sup45 dosage
Amelioration of [PSI+]-dependent cytotoxicity by extra-dosage of the Sup35 functional partner, Sup45, confirms that toxicity results from sequestration of release factor(s) by polyglutamine aggregates. Possibly Sup35, containing a QN-rich domain, is sequestered directly, while Sup45 is sequestered via its interaction with Sup35. Indeed, ability of polyglutamines to facilitate aggregation of endogenous QN-rich proteins even in a non-prion strain has been reported previously [43], [52] and confirmed by us (Figure 1E and Figure 6D), and it was shown that Sup35 prion aggregates produced at high levels cause toxicity via sequestering Sup45 [49]. In agreement with these data, AQT (i. e., extra copy of SUP45) ameliorates both polyglutamine toxicity (Figure 2) and toxicity of excess Sup35 (Figure 3C) in the [PSI+] cells. There is probably a competition for the Sup35/Sup45 complex between polyglutamine aggregates and functional sites (ribosome etc.) at which the Sup35/Sup45 complex is supposed to act. Therefore, an increased abundance of Sup45 not only increases a proportion of non-sequestered Sup45 but also partly counteracts sequestration of Sup35, that can be seen as a change in size distribution of the Sup35 polymers (Figure 6B). Hence, the antitoxic effect of excess Sup45.
Role of ubc4 deletion in AQT detection
The AQT derivatives were originally detected in the strain, lacking the major ubiquitin-conjugating enzyme Ubc4. One obvious reason for this is that amelioration of toxicity by AQT is more pronounced in the absence of Ubc4 (Figure 2F), making detection of the anti-toxic papillae easier. It is possible that Ubc4 promotes ubiquitination and subsequent degradation of a fraction of excess Sup45, that agrees with a more profound increase in Sup45 protein levels, which was detected in the ubc4Δ strain bearing an extra-copy of SUP45, in comparison to the isogenic UBC4+ strain (Figure 4E). In addition, ubc4Δ may influence patterns of polyglutamine aggregation and/or ability of polyglutamines to sequester other proteins. Indeed, defects of the ubiquitin system are known to promote aggresome formation in mammalian cells [20], and ubc4Δ influences formation and aggregation of the [PSI+] prion yeast, as well as levels of some Hsps and patterns of their interactions with prion aggregates [53].
Another, although not necessarily exclusive explanation of increased AQT appearance in ubc4Δ cells is that a lack of Ubc4 may affect chromosome segregation and/or recombination, therefore increasing the frequency of chromosome non-disjunction. Ubiquitination and ubiquitin-dependent protein degradation are involved in regulation of DNA repair and chromosome segregation [54], [55]. Ubc4 is implicated in ubiquitination of histones [56], and ubc4Δ is shown to affect proper segregation of some yeast plasmids [57]. Persisting variations of the chromosome II size in AQT strains (for example, see Figure S2B) and occasional appearance of weak additional bands on the CHEF gels of the ubc4Δ strains (data not shown) speak in favor of a detrimental effect of ubc4Δ on chromosome stability. It is possible that the presence of the foreign DNA (KanMX insertion) on chromosome II of the ubc4Δ strain aids in destabilization of this specific chromosome. Irrespective of the mechanism of the ubc4Δ effect, this deletion is required for neither maintenance of the chromosome II extra copy nor toxicity amelioration. The UBC4+ strains with an extra copy of chromosome II were obtained by genetic cross and dissection and continued to maintain a disomy (data not shown), and amelioration of toxicity by excess Sup45 was still detected in the UBC4+ strain, at a lower level in case of chromosome II extra copy (Figure 2F) and at more profound level for the plasmid-mediated excess Sup45 that is less sensitive to the presence of Ubc4 (Figure 5).
Relevance of yeast data to human polyglutamine disorders
Involvement of translational machinery in HD has been suspected from some results in mammalian systems [58]. It remains unknown if polyglutamines can sequester the human homologs of Sup35 and Sup45 (respectively, eRF3 and eRF1), as mammalian ortholog of Sup35 does not have a QN-rich domain. However, our results could be relevant to mammalian systems in a more general way. About 40% of the variation in the age of HD onset, in cases where the polyglutamine repeat is of the same length, is due to DNA variation [59]. Our work provides a potential explanation for such a variation by demonstrating that changes in the abundance of the sequestered protein(s), occurring via alteration of either gene dosage or gene expression, can modulate polyglutamine toxicity. A non-DNA component of variations in polyglutamine toxicity can be explained by differences in the composition of other aggregated proteins (e. g. endogenous self-perpetuating aggregates or prions) present in the cell. Our results show that prion composition of the cell not only drives polyglutamine toxicity but also determines a pathway via which polyglutamines influence cell physiology, as proteins already associated with the other aggregates are more likely to be sequestered by polyglutamines. Mammalian cells contain a variety of proteins with the prion-like QN-rich domains, and machinery for propagation of the QN-rich protein aggregates exists in mammals [60]. Protein aggregation can also be induced by oxidative damage and other stresses. It was reported that artificially generated β-rich aggregates may sequester other proteins [61]. It is therefore entirely possible that organisms or tissues (or both) differ by the aggregate composition, and this in turn influences their susceptibility to polyglutamine disorders. Composition of endogenous aggregates may also modulate which proteins are sequestered by polyglutamines, as proteins associated with other aggregates interacting with polyglutamines are more likely to be sequestered, like Sup45 in the cells containing the Sup35 prion. This could explain why different groups are coming out with different conclusions in regard to both mechanisms of polyglutamine toxicity and contributions of different types of polyglutamine aggregates.
Materials and Methods
Yeast strains and growth conditions
The Saccharomyces cerevisiae strains, used in this study and listed in Table S4, are derivatives of GT81 series [62] of the prototype haploid genotype ade1 his3 leu2 lys2 trp1 ura3, with different mating types and various prion compositions. The individual gene deletions were made by using PCR-mediated transplacement with the cassette bearing either Schizosaccharomyces pombe HIS5 gene, an ortholog of S. cerevisiae HIS3 gene (thus designated in this paper as HIS3), or bacterial kanr gene, which causes resistance to G418 in yeast [63]. Spontaneous AQT mutants were initially obtained in the strain GT349 (MATa ubc4Δ::HIS3 [PIN+ PSI+]), as described in Results.
Standard yeast media, procedures (including transformation, phenotype scoring, velveteen replica plating, mating and sporulation), and growth conditions were used [64]. Yeast cultures were grown at 30°C except for the temperature-sensitivity assays (employing 39°C). Tetrad dissection was performed by using the MSM System 300 micromanipulator from Singer Instrument Co. Ltd. Analysis of yeast chromosomes by CHEF (Contour-clamped Homogeneous Electric Field) is described in Text S1.
Polyglutamine toxicity was detected as growth inhibition on the synthetic dropout medium with galactose instead of glucose where polyglutamine constructs were selectively maintained and induced. As most of our polyglutamine constructs were expressed from plasmids bearing the URA3 marker, the plasmid-selective galactose medium (-Ura/Gal) was used, unless stated otherwise. Polyglutamine toxicity becomes more evident after longer incubation periods, as also confirmed by growth curves (see Figure 2C). Typically, velveteened plates were scanned following 5–10 days of incubation after a second passage on galactose medium, while spotted from solution (without dilutions) plates were scanned after 3–5 days, and serial decimal dilutions spotted onto galactose medium were scanned after 2–3 weeks.
Plasmids
Major plasmids used in this study are described in Text S1. A list of plasmids is available in Table S5.
Construction of chromosomal deletions
Strategy of making chromosomal deletions is described in Text S1.
Fluorescence microscopy
Fluorescence microscopy was performed according to standard techniques, as described in Text S1.
Protein analysis
Protein isolation and electrophoresis are described in Text S1. Semi-Denaturing Detergent-Agarose Gel Electrophoresis (SDD-AGE), used to fractionate the SDS-resistant protein polymers according to their sizes, was performed according to the standard protocol [65] with slight modifications. Proteins were diluted in 2% SDS, incubated for 5 min at room temperature before loading, run in the 1.5% agarose gel with 0.1% SDS in 1X TAE buffer containing 0.1% SDS, transferred to nitrocellulose membrane (Whatman) by capillary blotting, and reacted to appropriate antibody. Assay for β-galactosidase activity was performed according to the standard protocol [66], except that cell debris was removed by centrifugation to avoid light scattering before the OD reading at 420 nm was taken.
Antibodies
Description of antibodies used in this study can be found in Text S1.
DNA microarray analysis
Gene copy number was determined by hybridization to the complete DNA microarray of the Saccharomyces cerevisiae genome, as described previously [67]. Detailed information can be found in the Text S1.
Supporting Information
Zdroje
1. ShaoJDiamondMI 2007 Polyglutamine diseases: emerging concepts in pathogenesis and therapy. Hum Mol Genet 16 R115 123
2. La SpadaARTaylorJP 2010 Repeat expansion disease: progress and puzzles in disease pathogenesis. Nat Rev Genet 11 247 258
3. The Huntington's Disease Collaborative Research Group 1993 A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington's disease chromosomes. Cell 72 971 983
4. DiFigliaMSappEChaseKODaviesSWBatesGP 1997 Aggregation of huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain. Science 277 1990 1993
5. LunkesALindenbergKSBen-HaiemLWeberCDevysD 2002 Proteases acting on mutant huntingtin generate cleaved products that differentially build up cytoplasmic and nuclear inclusions. Mol Cell 10 259 269
6. MangiariniLSathasivamKSellerMCozensBHarperA 1996 Exon 1 of the HD gene with an expanded CAG repeat is sufficient to cause a progressive neurological phenotype in transgenic mice. Cell 87 493 506
7. StackECKubilusJKSmithKCormierKDel SignoreSJ 2005 Chronology of behavioral symptoms and neuropathological sequela in R6/2 Huntington's disease transgenic mice. J Comp Neurol 490 354 370
8. ZoghbiHYOrrHT 1999 Polyglutamine diseases: protein cleavage and aggregation. Curr Opin Neurobiol 9 566 570
9. SteffanJSKazantsevASpasic-BoskovicOGreenwaldMZhuYZ 2000 The Huntington's disease protein interacts with p53 and CREB-binding protein and represses transcription. Proc Natl Acad Sci USA 97 6763 6768
10. HayDGSathasivamKTobabenSStahlBMarberM 2004 Progressive decrease in chaperone protein levels in a mouse model of Huntington's disease and induction of stress proteins as a therapeutic approach. Hum Mol Genet 13 1389 1405
11. RavikumarBVacherCBergerZDaviesJELuoS 2004 Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nat Genet 36 585 595
12. YamanakaTTosakiAMiyazakiHKurosawaMFurukawaY 2010 Mutant huntingtin fragment selectively suppresses Brn-2 POU domain transcription factor to mediate hypothalamic cell dysfunction. Hum Mol Genet 19 2099 2112
13. RossCAPoirierMA 2004 Protein aggregation and neurodegenerative disease. Nat Med 10 Suppl S10 17
14. HandsSLWyttenbachA 2010 Neurotoxic protein oligomerisation associated with polyglutamine diseases. Acta Neuropathol 120 419 437
15. DaviesSWTurmaineMCozensBADiFigliaMSharpAH 1997 Formation of neuronal intranuclear inclusions underlies the neurological dysfunction in mice transgenic for the HD mutation. Cell 90 537 548
16. ScherzingerELurzRTurmaineMMangiariniLHollenbachB 1997 Huntingtin-encoded polyglutamine expansions form amyloid-like protein aggregates in vitro and in vivo. Cell 90 549 558
17. RossCAWoodJDSchillingGPetersMFNuciforaFCJr 1999 Polyglutamine pathogenesis. Philos Trans R Soc Lond B Biol Sci 354 1005 1011
18. RossCATabriziSJ 2011 Huntington's disease: from molecular pathogenesis to clinical treatment. Lancet Neurol 10 83 98
19. JohnstonJAWardCLKopitoRR 1998 Aggresomes: a cellular response to misfolded proteins. J Cell Biol 143 1883 1898
20. WaelterSBoeddrichALurzRScherzingerELuederG 2001 Accumulation of mutant huntingtin fragments in aggresome-like inclusion bodies as a result of insufficient protein degradation. Mol Biol Cell 12 1393 1407
21. TaylorJPTanakaFRobitschekJSandovalCMTayeA 2003 Aggresomes protect cells by enhancing the degradation of toxic polyglutamine-containing protein. Hum Mol Genet 12 749 757
22. OlzmannJALiLChinLS 2008 Aggresome formation and neurodegenerative diseases: therapeutic implications. Curr Med Chem 15 47 60
23. KopitoRR 2000 Aggresomes, inclusion bodies and protein aggregation. Trends Cell Biol 10 524 530
24. Garcia-MataRGaoYSSztulE 2002 Hassles with taking out the garbage: aggravating aggresomes. Traffic 3 388 396
25. OlzmannJALiLChudaevMVChenJPerezFA 2007 Parkin-mediated K63-linked polyubiquitination targets misfolded DJ-1 to aggresomes via binding to HDAC6. J Cell Biol 178 1025 1038
26. ChinLSOlzmannJALiL 2010 Parkin-mediated ubiquitin signalling in aggresome formation and autophagy. Biochem Soc Trans 38 144 149
27. BondziCBrunnerAMMunyikwaMRConnorCDSimmonsAN 2011 Recruitment of the oncoprotein v-ErbA to aggresomes. Mol Cell Endocrinol 332 196 212
28. KrobitschSLindquistS 2000 Aggregation of huntingtin in yeast varies with the length of the polyglutamine expansion and the expression of chaperone proteins. Proc Natl Acad Sci USA 97 1589 1594
29. MuchowskiPJSchaffarGSittlerAWankerEEHayer-HartlMK 2000 Hsp70 and Hsp40 chaperones can inhibit self-assembly of polyglutamine proteins into amyloid-like fibrils. Proc Natl Acad Sci USA 97 7841 7846
30. MeriinABZhangXHeXNewnamGPChernoffYO 2002 Huntington toxicity in yeast model depends on polyglutamine aggregation mediated by a prion-like protein Rnq1. J Cell Biol 157 997 1004
31. DuennwaldMLJagadishSGiorginiFMuchowskiPJLindquistS 2006 A network of protein interactions determines polyglutamine toxicity. Proc Natl Acad Sci USA 103 11051 11056
32. DuennwaldMLJagadishSMuchowskiPJLindquistS 2006 Flanking sequences profoundly alter polyglutamine toxicity in yeast. Proc Natl Acad Sci USA 103 11045 11050
33. DuennwaldML 2011 Polyglutamine misfolding in yeast: toxic and protective aggregation. Prion 5 285 290
34. MasonRPGiorginiF 2011 Modeling Huntington disease in yeast: perspectives and future directions. Prion 5 269 276
35. GokhaleKCNewnamGPShermanMYChernoffYO 2005 Modulation of prion-dependent polyglutamine aggregation and toxicity by chaperone proteins in the yeast model. J Biol Chem 280 22809 22818
36. WicknerRBEdskesHKShewmakerFNakayashikiTEngelA 2007 Yeast prions: evolution of the prion concept. Prion 1 94 100
37. TuiteMFCoxBS 2003 Propagation of yeast prions. Nat Rev Mol Cell Biol 4 878 890
38. Inge-VechtomovSGZhouravlevaGAChernoffYO 2007 Biological roles of prion domains. Prion 1 228 235
39. MeriinABZhangXMiliarasNBKazantsevAChernoffYO 2003 Aggregation of expanded polyglutamine domain in yeast leads to defects in endocytosis. Mol Cell Biol 23 7554 7565
40. MeriinABZhangXAlexandrovIMSalnikovaABTer-AvanesianMD 2007 Endocytosis machinery is involved in aggregation of proteins with expanded polyglutamine domains. FASEB J 21 1915 1925
41. CavistonJPHolzbaurEL 2009 Huntingtin as an essential integrator of intracellular vesicular trafficking. Trends Cell Biol 19 147 155
42. WangYMeriinABZaarurNRomanovaNVChernoffYO 2009 Abnormal proteins can form aggresome in yeast: aggresome-targeting signals and components of the machinery. FASEB J 23 451 463
43. UrakovVNVishnevskayaABAlexandrovIMKushnirovVVSmirnovVN 2010 Interdependence of amyloid formation in yeast: implications for polyglutamine disorders and biological functions. Prion 4 45 52
44. SeufertWJentschS 1990 Ubiquitin-conjugating enzymes UBC4 and UBC5 mediate selective degradation of short-lived and abnormal proteins. EMBO J 9 543 550
45. StansfieldIJonesKMKushnirovVVDagkesamanskayaARPoznyakovskiAI 1995 The products of the SUP45 (eRF1) and SUP35 genes interact to mediate translation termination in Saccharomyces cerevisiae. EMBO J 14 4365 4373
46. KallmeyerAKKeelingKMBedwellDM 2006 Eukaryotic release factor 1 phosphorylation by CK2 protein kinase is dynamic but has little effect on the efficiency of translation termination in Saccharomyces cerevisiae. Eukaryot Cell 5 1378 1387
47. MoskalenkoSEZhuravlevaGASoomMShabel'skaiaSVVolkovKV 2004 Characterization of missense mutations in the SUP45 gene of Saccharomyces cerevisiae encoding translation termination factor eRF1. Genetika 40 599 606
48. KaganovichDKopitoRFrydmanJ 2008 Misfolded proteins partition between two distinct quality control compartments. Nature 454 1088 1095
49. VishveshwaraNBradleyMELiebmanSW 2009 Sequestration of essential proteins causes prion associated toxicity in yeast. Mol Microbiol 73 1101 1114
50. ValouevIAKushnirovVVTer-AvanesyanMD 2002 Yeast polypeptide chain release factors eRF1 and eRF3 are involved in cytoskeleton organization and cell cycle regulation. Cell Motil Cytoskeleton 52 161 173
51. DagkesamanskayaARTer-AvanesyanMD 1991 Interaction of the yeast omnipotent suppressors SUP1(SUP45) and SUP2(SUP35) with non-mendelian factors. Genetics 128 513 520
52. DerkatchILUptainSMOuteiroTFKrishnanRLindquistSL 2004 Effects of Q/N-rich, polyQ, and non-polyQ amyloids on the de novo formation of the [PSI+] prion in yeast and aggregation of Sup35 in vitro. Proc Natl Acad Sci U S A 101 12934 12939
53. AllenKDChernovaTATennantEPWilkinsonKDChernoffYO 2007 Effects of ubiquitin system alterations on the formation and loss of a yeast prion. J Biol Chem 282 3004 3013
54. PaganoM 1997 Cell cycle regulation by the ubiquitin pathway. FASEB J 11 1067 1075
55. Al-HakimAEscribano-DiazCLandryMCO'DonnellLPanierS 2010 The ubiquitous role of ubiquitin in the DNA damage response. DNA Repair 9 1229 1240
56. SinghRKKabbajMHPaikJGunjanA 2009 Histone levels are regulated by phosphorylation and ubiquitylation-dependent proteolysis. Nat Cell Biol 11 925 933
57. SleepDFinnisCTurnerAEvansL 2001 Yeast 2 microm plasmid copy number is elevated by a mutation in the nuclear gene UBC4. Yeast 18 403 421
58. KingMAHandsSHafizFMizushimaNTolkovskyAM 2008 Rapamycin inhibits polyglutamine aggregation independently of autophagy by reducing protein synthesis. Mol Pharmacol 73 1052 1063
59. WexlerNSLorimerJPorterJGomezFMoskowitzC 2004 Venezuelan kindreds reveal that genetic and environmental factors modulate Huntington's disease age of onset. Proc Natl Acad Sci USA 101 3498 3503
60. KrammerCKryndushkinDSuhreMHKremmerEHofmannA 2009 The yeast Sup35NM domain propagates as a prion in mammalian cells. Proc Natl Acad Sci USA 106 462 467
61. OlzschaHSchermannSMWoernerACPinkertSHechtMH 2011 Amyloid-like aggregates sequester numerous metastable proteins with essential cellular functions. Cell 144 67 78
62. ChernoffYOGalkinAPLewitinEChernovaTANewnamGP 2000 Evolutionary conservation of prion-forming abilities of the yeast Sup35 protein. Mol Microbiol 35 865 876
63. LongtineMSMcKenzieA3rdDemariniDJShahNGWachA 1998 Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14 953 961
64. ShermanF 2002 Getting started with yeast. Methods Enzymol 350 3 41
65. HalfmannRLindquistS 2008 Screening for amyloid aggregation by Semi-Denaturing Detergent-Agarose Gel Electrophoresis. J Vis Exp doi 10.3791/838
66. MillerJH 1972 Experiments in Molecular Genetics Cold Spring Harbor, NY Cold Spring Harbor Laboratory
67. LemoineFJDegtyarevaNPLobachevKPetesTD 2005 Chromosomal translocations in yeast induced by low levels of DNA polymerase a model for chromosome fragile sites. Cell 120 587 598
68. Le GoffCZemlyankoOMoskalenkoSBerkovaNInge-VechtomovS 2002 Mouse GSPT2, but not GSPT1, can substitute for yeast eRF3 in vivo. Genes Cells 7 1043 1057
69. Ter-AvanesyanMDKushnirovVVDagkesamanskayaARDidichenkoSAChernoffYO 1993 Deletion analysis of the SUP35 gene of the yeast Saccharomyces cerevisiae reveals two non-overlapping functional regions in the encoded protein. Mol Microbiol 7 683 692
70. SerioTRCashikarAGMoslehiJJKowalASLindquistSL 1999 Yeast prion [psi+] and its determinant, Sup35p. Methods Enzymol 309 649 673
71. LaneyJDHochstrasserM 2003 Ubiquitin-dependent degradation of the yeast Matα2 repressor enables a switch in developmental state. Genes Dev 17 2259 2270
72. FiroozanMGrantCMDuarteJATuiteMF 1991 Quantitation of readthrough of termination codons in yeast using a novel gene fusion assay. Yeast 7 173 183
73. StoriciFResnickMA 2006 The delitto perfetto approach to in vivo site-directed mutagenesis and chromosome rearrangements with synthetic oligonucleotides in yeast. Methods Enzymol 409 329 345
74. SikorskiRSHieterP 1989 A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122 19 27
75. NewnamGPBirchmoreJLChernoffYO 2011 Destabilization and recovery of a yeast prion after mild heat shock. J Mol Biol 408 432 448
76. AllenKDWegrzynRDChernovaTAMèullerSNewnamGP 2005 Hsp70 chaperones as modulators of prion life cycle: novel effects of Ssa and Ssb on the Saccharomyces cerevisiae prion [PSI+]. Genetics 169 1227 1242
77. NarayananVMieczkowskiPAKimHMPetesTDLobachevKS 2006 The pattern of gene amplification is determined by the chromosomal location of hairpin-capped breaks. Cell 125 1283 1296
78. ArguesoJLWestmorelandJMieczkowskiPAGawelMPetesTD 2008 Double-strand breaks associated with repetitive DNA can reshape the genome. Proc Natl Acad Sci USA 105 11845 11850
79. DeRisiJLIyerVRBrownPO 1997 Exploring the metabolic and genetic control of gene expression on a genomic scale. Science 278 680 686
80. WangPKimYPollackJNarasimhanBTibshiraniR 2005 A method for calling gains and losses in array CGH data. Biostatistics 6 45 58
Štítky
Genetika Reprodukční medicína
Článek A Genome-Wide Screen for Genetic Variants That Modify the Recruitment of REST to Its Target GenesČlánek Population Structure of Hispanics in the United States: The Multi-Ethnic Study of AtherosclerosisČlánek Differing Requirements for RAD51 and DMC1 in Meiotic Pairing of Centromeres and Chromosome Arms inČlánek Transcriptional Regulation of Rod Photoreceptor Homeostasis Revealed by NRL Targetome AnalysisČlánek Cell Contact–Dependent Outer Membrane Exchange in Myxobacteria: Genetic Determinants and MechanismČlánek Formation of Rigid, Non-Flight Forewings (Elytra) of a Beetle Requires Two Major Cuticular Proteins
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2012 Číslo 4
-
Všechny články tohoto čísla
- Runs of Homozygosity Implicate Autozygosity as a Schizophrenia Risk Factor
- Modifier Genes and the Plasticity of Genetic Networks in Mice
- The DSIF Subunits Spt4 and Spt5 Have Distinct Roles at Various Phases of Immunoglobulin Class Switch Recombination
- A Genome-Wide Screen for Genetic Variants That Modify the Recruitment of REST to Its Target Genes
- Population Structure of Hispanics in the United States: The Multi-Ethnic Study of Atherosclerosis
- Deep Sequencing of Plant and Animal DNA Contained within Traditional Chinese Medicines Reveals Legality Issues and Health Safety Concerns
- Differing Requirements for RAD51 and DMC1 in Meiotic Pairing of Centromeres and Chromosome Arms in
- Insulin Signaling Mediates Sexual Attractiveness in
- Progressive Telomere Dysfunction Causes Cytokinesis Failure and Leads to the Accumulation of Polyploid Cells
- Long-Range Chromosome Organization in : A Site-Specific System Isolates the Ter Macrodomain
- Regulation of Budding Yeast Mating-Type Switching Donor Preference by the FHA Domain of Fkh1
- Polyglutamine Toxicity Is Controlled by Prion Composition and Gene Dosage in Yeast
- Patterns of Regulatory Variation in Diverse Human Populations
- Sequence-Specific Targeting of Dosage Compensation in Favors an Active Chromatin Context
- Whole-Exome Sequencing and Homozygosity Analysis Implicate Depolarization-Regulated Neuronal Genes in Autism
- Replication Fork Reversal after Replication–Transcription Collision
- Common Variants at 9p21 and 8q22 Are Associated with Increased Susceptibility to Optic Nerve Degeneration in Glaucoma
- Coordinate Regulation of Lipid Metabolism by Novel Nuclear Receptor Partnerships
- Epigenome-Wide Scans Identify Differentially Methylated Regions for Age and Age-Related Phenotypes in a Healthy Ageing Population
- A Coordinated Interdependent Protein Circuitry Stabilizes the Kinetochore Ensemble to Protect CENP-A in the Human Pathogenic Yeast
- Budding Yeast Dma Proteins Control Septin Dynamics and the Spindle Position Checkpoint by Promoting the Recruitment of the Elm1 Kinase to the Bud Neck
- , a Homolog of a Deaf-Blindness Gene, Regulates Circadian Output and Slowpoke Channels
- Transcriptional Regulation of Rod Photoreceptor Homeostasis Revealed by NRL Targetome Analysis
- Cell Contact–Dependent Outer Membrane Exchange in Myxobacteria: Genetic Determinants and Mechanism
- Defective Membrane Remodeling in Neuromuscular Diseases: Insights from Animal Models
- Formation of Rigid, Non-Flight Forewings (Elytra) of a Beetle Requires Two Major Cuticular Proteins
- SPE-44 Implements Sperm Cell Fate
- A Shared Role for RBF1 and dCAP-D3 in the Regulation of Transcription with Consequences for Innate Immunity
- A Companion Cell–Dominant and Developmentally Regulated H3K4 Demethylase Controls Flowering Time in via the Repression of Expression
- The HEN1 Ortholog, HENN-1, Methylates and Stabilizes Select Subclasses of Germline Small RNAs
- Improved Statistics for Genome-Wide Interaction Analysis
- The Probability of a Gene Tree Topology within a Phylogenetic Network with Applications to Hybridization Detection
- Context-Dependent Dual Role of SKI8 Homologs in mRNA Synthesis and Turnover
- Mu Insertions Are Repaired by the Double-Strand Break Repair Pathway of
- Competition between Replicative and Translesion Polymerases during Homologous Recombination Repair in Drosophila
- An Unbiased Assessment of the Role of Imprinted Genes in an Intergenerational Model of Developmental Programming
- Type 2 Diabetes Risk Alleles Demonstrate Extreme Directional Differentiation among Human Populations, Compared to Other Diseases
- Mutations in and Cause “Splashed White” and Other White Spotting Phenotypes in Horses
- Fine-Scale Mapping of Natural Variation in Fly Fecundity Identifies Neuronal Domain of Expression and Function of an Aquaporin
- Dynamics of Brassinosteroid Response Modulated by Negative Regulator LIC in Rice
- Genetic Inhibition of Solute-Linked Carrier 39 Family Transporter 1 Ameliorates Aβ Pathology in a Model of Alzheimer's Disease
- The Functions of Mediator in Support a Role in Shaping Species-Specific Gene Expression
- Patterns of Ancestry, Signatures of Natural Selection, and Genetic Association with Stature in Western African Pygmies
- Dissection of Pol II Trigger Loop Function and Pol II Activity–Dependent Control of Start Site Selection
- PIWI Associated siRNAs and piRNAs Specifically Require the HEN1 Ortholog
- Genome-Wide Patterns of Gene Expression in Nature
- Hypoxia Disruption of Vertebrate CNS Pathfinding through EphrinB2 Is Rescued by Magnesium
- A New Role for Translation Initiation Factor 2 in Maintaining Genome Integrity
- Sex Reversal in C57BL/6J XY Mice Caused by Increased Expression of Ovarian Genes and Insufficient Activation of the Testis Determining Pathway
- The Rac GTP Exchange Factor TIAM-1 Acts with CDC-42 and the Guidance Receptor UNC-40/DCC in Neuronal Protrusion and Axon Guidance
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- A Coordinated Interdependent Protein Circuitry Stabilizes the Kinetochore Ensemble to Protect CENP-A in the Human Pathogenic Yeast
- Coordinate Regulation of Lipid Metabolism by Novel Nuclear Receptor Partnerships
- Defective Membrane Remodeling in Neuromuscular Diseases: Insights from Animal Models
- Formation of Rigid, Non-Flight Forewings (Elytra) of a Beetle Requires Two Major Cuticular Proteins
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



