-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaA Companion Cell–Dominant and Developmentally Regulated H3K4 Demethylase Controls Flowering Time in via the Repression of Expression
Flowering time relies on the integration of intrinsic developmental cues and environmental signals. FLC and its downstream target FT are key players in the floral transition in Arabidopsis. Here, we characterized the expression pattern and function of JMJ18, a novel JmjC domain-containing histone H3K4 demethylase gene in Arabidopsis. JMJ18 was dominantly expressed in companion cells; its temporal expression pattern was negatively and positively correlated with that of FLC and FT, respectively, during vegetative development. Mutations in JMJ18 resulted in a weak late-flowering phenotype, while JMJ18 overexpressors exhibited an obvious early-flowering phenotype. JMJ18 displayed demethylase activity toward H3K4me3 and H3K4me2, and bound FLC chromatin directly. The levels of H3K4me3 and H3K4me2 in chromatins of FLC clade genes and the expression of FLC clade genes were reduced, whereas FT expression was induced and the protein expression of FT increased in JMJ18 overexpressor lines. The early-flowering phenotype caused by the overexpression of JMJ18 was mainly dependent on the functional FT. Our findings suggest that the companion cell–dominant and developmentally regulated JMJ18 binds directly to the FLC locus, reducing the level of H3K4 methylation in FLC chromatin and repressing the expression of FLC, thereby promoting the expression of FT in companion cells to stimulate flowering.
Published in the journal: . PLoS Genet 8(4): e32767. doi:10.1371/journal.pgen.1002664
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1002664Summary
Flowering time relies on the integration of intrinsic developmental cues and environmental signals. FLC and its downstream target FT are key players in the floral transition in Arabidopsis. Here, we characterized the expression pattern and function of JMJ18, a novel JmjC domain-containing histone H3K4 demethylase gene in Arabidopsis. JMJ18 was dominantly expressed in companion cells; its temporal expression pattern was negatively and positively correlated with that of FLC and FT, respectively, during vegetative development. Mutations in JMJ18 resulted in a weak late-flowering phenotype, while JMJ18 overexpressors exhibited an obvious early-flowering phenotype. JMJ18 displayed demethylase activity toward H3K4me3 and H3K4me2, and bound FLC chromatin directly. The levels of H3K4me3 and H3K4me2 in chromatins of FLC clade genes and the expression of FLC clade genes were reduced, whereas FT expression was induced and the protein expression of FT increased in JMJ18 overexpressor lines. The early-flowering phenotype caused by the overexpression of JMJ18 was mainly dependent on the functional FT. Our findings suggest that the companion cell–dominant and developmentally regulated JMJ18 binds directly to the FLC locus, reducing the level of H3K4 methylation in FLC chromatin and repressing the expression of FLC, thereby promoting the expression of FT in companion cells to stimulate flowering.
Introduction
DNA is packaged as chromatin in eukaryotic cells. Nucleosomes, which consist of an octamer of four histones wrapped around 146 base pairs of DNA, are the fundamental unit of chromatin [1]. The flexible N-terminal tails of histones, which protrude from the nucleosome core particle, are subject to several types of covalent modification, including acetylation, methylation, phosphorylation, and ubiquitylation [2]. Each of these modifications is reversible and is required for the dynamic regulation of gene expression [3].
In vivo, the lysine residues in histones display three distinct methylation states: mono - (me1), di - (me2), and tri-methylated (me3). These differences in methylation are important for the recognition of chromatin by chromatin modulators and for the recruitment of other modulators and regulators [4], 5,6,7,8. In addition, lysine methylation at different sites within the histone protein plays distinct roles in gene activation and repression [2], [9]. For example, the methylation of H3K4 and H3K36 is correlated with gene activation, while transcriptional repression has been demonstrated in genomic regions with increased levels of H3K9 and H3K27 methylation [3], [10], [11], [12]. Various developmental processes are regulated by histone methylation in animals and plants. For example, H3K27 methylation mediated by the PRC2 complex is required for embryonic development and stem cell identity in mammals and controls most steps in the development of Arabidopsis [9], [13], [14], [15].
The methylation state of histone proteins is determined by the balance between methylation and demethylation, which is mediated by histone methyltransferases and demethylases, respectively [16], [17], [18]. However, the reversibility of histone methylation in vivo is the latest to be discovered compared to other covalent forms of histone modification. The amine oxidase LSD1 was the first histone demethylase found to demethylate H3K4me2 and H3K4me1 through an FAD-dependent oxidation reaction [18]. LSD1 demethylase family proteins are unable to remove methyl groups from tri-methylated lysines, suggesting the presence of other histone demethylases in eukaryotic cells [18]. More recently, a family of JmjC domain-containing proteins was characterized as histone demethylases which were able to reduce any one of the three histone lysine methylation states at several specific sites in yeast and animals [17], [19], [20], [21]. These histone demethylases are involved in many biological processes in animals, including spermatogenesis, HOX gene regulation, and germ cell development [19], [22], [23].
FLD and LDLs are the homologs of human LSD1 in Arabidopsis. They promote the floral transition by constitutively inhibiting the expression of FLC [24], [25], [26]. The Arabidopsis genome contains 21 JmjC family proteins [27]; however, only five of them have been characterized, and they have been found to be involved in RNA silencing, DNA methylation, flowering time control, circadian clock regulation, BR signaling and shoot regeneration in vitro [28], [29], [30], [31], [32], [33], [34], [35], [36].
Flowering at the appropriate time is the most important factor in achieving reproductive success. To produce the next generation, plants rely on intricate signaling pathways involving a variety of intrinsic factors, including developmental stage and age, and environmental cues such as photoperiod, temperature, and light quantity [37], [38], [39]. Thus, distinct environmental conditions modulate endogenous gene expression to ensure flowering at the correct time [12], [40].
FLOWERING LOCUS C (FLC) encodes a MADS-box transcription factor and floral repressor that regulates flowering time in a dosage-dependent manner by integrating the vernalization, autonomous, PAF1 complex, and H2B ubiquitination pathways [11], [41], [42], [43], [44], [45]. FLC and the functional locus FRIGIDA (FRI) act together to produce winter-annual Arabidopsis accessions, which must be exposed to cold temperature for several weeks to repress FLC expression and promote flowering in the following spring [46], [47]. As endogenous factors, autonomous pathway genes consecutively repress FLC expression [48], [49]. In addition, the FLC antisense transcript affects the expression of sense transcript, thereby influencing flowering time in Arabidopsis [50], [51], [52].
FLC expression is predominant in the shoot apical meristem (SAM); however, it is also expressed in vascular tissue in young leaves and the root tip [44], [53], [54]. FLC expressed in the SAM and leaf vascular tissue contributes to the control of flowering time in Arabidopsis [55]. During vernalization, VIN3 binds FLC chromatin, thereby repressing its expression in the SAM [53]. The components of the autonomous pathway repress, while those of the PAF1 complex activate, FLC expression in the SAM [11], [37], [43]. Thus, although the regulation of FLC expression in the SAM has been extensively studied, little is known about how FLC expression is regulated in leaf vascular tissue.
Histone modification plays crucial roles in the regulation of FLC expression in Arabidopsis. In FLC chromatin, H3K4 hyper-tri-methylation and acetylation are associated with the activation of gene expression [43], [56], [57], [58]. The methylation of H3K27 and H3K9 in FLC chromatin leads to the repression of FLC expression and is required for maintenance of the repression of FLC expression in plants growing in the following spring after vernalization [12], [53], [59]. In addition, H2B monoubiquitination is required for the maintenance of high levels of H3K4me3 and H3K36me2 [11], [45]. A loss of H2B monoubiquitination decreases the level of H3K4me3 and H3K36me2, leading to the repression of FLC transcription and early flowering [11], [45].
FLOWERING LOCUS T (FT), which is a component of the photoperiod pathway, coordinates signals from the vernalization, autonomous, PAF1 complex, and photoperiod pathways to promote flowering in response to increase in day length [60], [61], [62], [63]. FT expression is restricted to leaf phloem companion cells [62]. FT travels from leaves to the SAM, where it interacts with the bZIP transcription factor FD to stimulate floral meristem initiation [64], [65], [66], [67], [68]. As a mobile and systemic signal, FT integrates photoperiod - and FLC-dependent pathways to control flowering time by regulating the expression of floral identity genes. It was shown previously that the companion cells specifically expressed FLC represses FT expression in leaf companion cells and delays the expression of its cofactor FD in the SAM [55]. Thus, FLC systemically blocks the function of FT to repress flowering. However, the regulation of FLC expression in companion cells for the control of floral development has not been characterized.
As mentioned above, several JmjC domain-containing proteins have been reported to be involved in flowering time control [28], [29], . Among them, JMJ14 is a member of JARID family with H3K4 demethylase activity, and is involved in flowering time control through the repression of floral integrators [28], [29], [33], [69]. It is not yet clear whether those floral integrator genes are the direct targets of JMJ14 [28], [29], [69]. ELF6 acts as a repressor in photoperiod pathway [31], and its closest homolog REF6 acts as a FLC repressor in flowering time regulation [31]. Recent results suggest that REF6 is a H3K27 demethylase; thus, FLC is not likely to be the direct target of REF6 [70]. No JmjC domain-containing histone demethylases that target FLC locus have yet been found.
In this paper, we describe the expression pattern and function of JMJ18, a novel JmjC domain-containing histone H3K4 demethylase in Arabidopsis. JMJ18 was predominant expressed in phloem companion cells, and its level of expression increased during vegetative development. JMJ18 was found to promote the floral transition in Arabidopsis by binding FLC chromatin and demethylating H3K4 methylation, leading to the repression of FLC and enhanced expression of the downstream flowering activator FT in companion cells.
Results
JMJ18 demethylates histone H3K4me3 and H3K4me2 in vitro
JMJ18 (At1g30810) belongs to the evolutionarily conserved JARID1 family. Previous studies have shown that JARID1 family proteins demethylate H3K4 methylations in yeast, animal and Arabidosis [28], [29], [33], [69], [71], [72], [73].
To determine whether JMJ18 exhibits histone demethylase activity, we expressed recombinant His-tagged JMJ18 and purified the recombinant protein from insect cells (Figure S1A and S1B). A MALDI-TOF mass spectrometric analysis-based demethylation assay was used to detect the histone demethylation activity of JMJ18. Purified His-JMJ18 was incubated with a variety of histone peptides representing the tri - and di-methylated states of the four lysine residues in histone H3. As shown in Figure 1A, JMJ18 converted H3K4me3 to H3K4me2, but did not alter the H3K4me2 methylation state or the methylation status of any other lysine residue in the peptides (summarized in Figure S1C).
Fig. 1. Mass spectrometric analysis of JMJ18 histone demethylase activity in vitro. 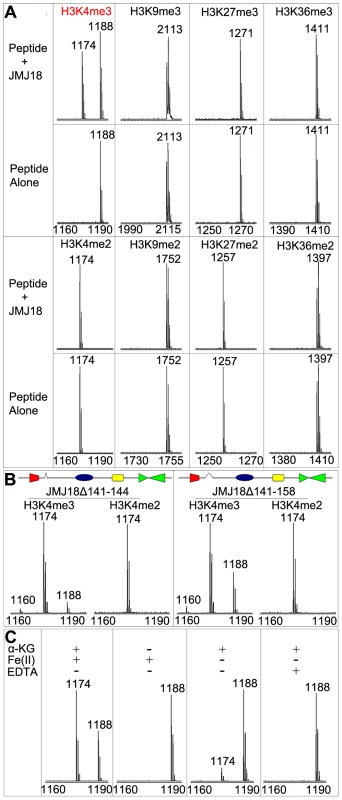
(A) Characterization of JMJ18 histone demethylase site specificity using purified full-length JMJ18. The numbers represent the molecular weights of the peptides. (B) Histone demethylase activity of two truncated forms of JMJ18: His-JMJ18Δ141–144 and His-JMJ18Δ141–158. Tri- and di-methylated H3K4 peptides were used in the assay. Both truncated JMJ18 were able to demethylate tri-methylated H3K4 to the di- and mono-methylated states; however, no demethylase activity was detected if di-methylated H3K4 peptide was used as the substrate. The deleted domain was indicated by an open arrow in the schematic structure of JMJ18. (C) The histone demethylase activity of JMJ18 depends on α-KG and Fe(II). To investigate the demethylase activity of JMJ18 further, we purified two truncated versions of JMJ18, JMJ18Δ141–144 and JMJ18Δ141–158, in which the partial linker sequence between JmjN and JmjC was deleted (Figure 1B), and examined the histone demethylase activity of each using the assay described above. Similar to full-length JMJ18, the truncated proteins were H3K4-specific demethylases (Figure 1B and Figure S1C). Interestingly, both truncated forms of JMJ18 exhibited H3K4me3 and H3K4me2 demethylase activity, and they were able to convert H3K4me3 to H3K4me2 and H3K4me1 in the presence of H3K4me3 peptide (Figure 1B and Figure S1C). This suggests that the linker between JmjN and JmjC blocks the enzyme's demethylase activity toward H3K4me2 in vitro. However, they were unable to demethylate H3K4me2 to H3K3me1 if H3K4me2 peptide was used as the substrate (Figure 1B and Figure S1C).
We further observed that the absence of α-ketoglutarate (α-KG) or presence of EDTA completely abolished the enzyme's demethylase activity (Figure 1C), while a lack of Fe(II) strongly inhibited the enzyme's activity (Figure 1C). Taken together, our results suggest that JMJ18 functions as an H3K4-specific demethylase in vitro, and that its activity is dependent on α-KG and Fe(II).
Characterization of jmj18 mutants
To assess the biological function of JMJ18, we obtained three T-DNA insertion lines from the ABRC [74]: jmj18-1 (SALK_073442), jmj18-2 (GABI_649D05), and jmj18-3 (SAIL_861_F02) (Figure 2A). Reverse transcription (RT)-PCR analysis revealed a lack of full-length JMJ18 mRNA in jmj18-1 and jmj18-2; however, a partial transcript was detected in both lines (Figure 2B). In the third line, the T-DNA was inserted into exon 4 (Figure 2A), and full-length mRNA was detected at a reduced level compared to that in wild-type plants, and the transcript was confirmed by sequencing, suggesting the existence of a knock-down allele (Figure 2B).
Fig. 2. Characterization of three jmj18 mutants. 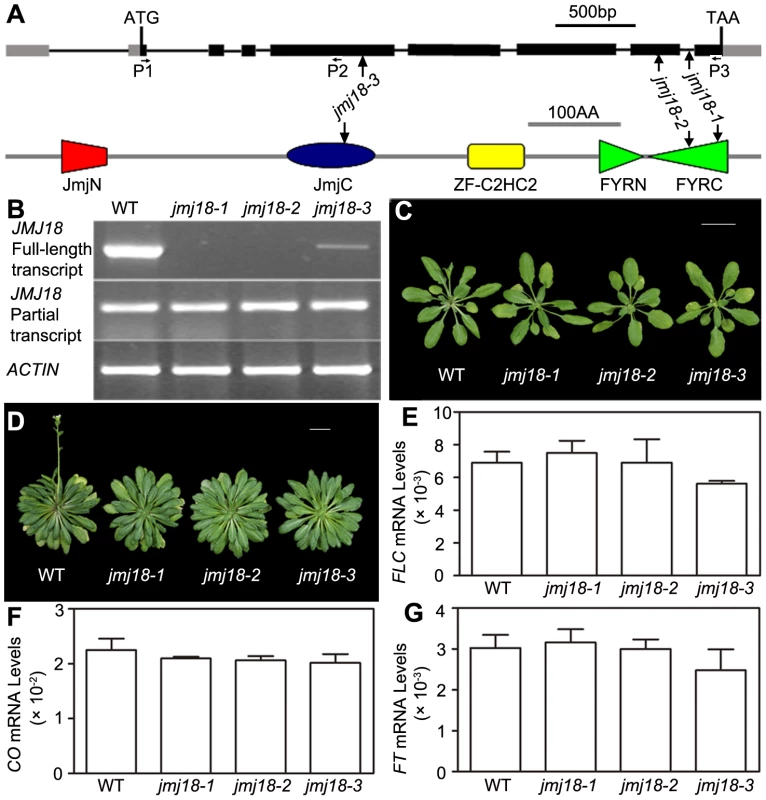
(A) Structures of JMJ18 and its gene product. Arrows indicate the T-DNA insertion sites in the mutants. Introns are represented as lines, exons as filled black boxes, and untranslated regions as filled gray boxes. The primers used for transcript detection (P1–P3) are indicated under the schematic gene structure. (B) Transcript analysis of the jmj18 mutants by RT-PCR. Pairs of P1/P3 and P1/P2 were used for full-length and partial transcripts detection, respectively. The 11-day-old plants of each genotype were collected at dusk for RNA isolation. The jmj18 mutants exhibited a weak late-flowering phenotype under both LD (C) and SD (D) conditions. Bar = 2 cm. Relative expression levels of FLC (E), CO (F), and FT (G) in the jmj18 mutants. The expression level was normalized to that of ACTIN. RNA was isolated from eleven-day-old seedlings. The values are the mean and standard deviation from three independent experiments. There was no significant difference between wild-type and jmj18 mutants for the expression levels of FLC (E), CO (F) or FT (G) tested by Student's t test (P<0.05). All three alleles exhibited a weak but reproducible late-flowering phenotype under long - (LD) and short-day (SD) conditions (Figure 2C, 2D and Table 1). This late flowering phenotype in jmj18-1 was complemented by JMJ18:JMJ18-GFP transformation (Figure S2). Knock-down lines for JMJ18 expression by using both double strand RNA interference (RNAi) and artificial micro-RNA (amiR) were generated, and these lines also exhibit weak late-flowering phenotype under LD conditions (Figure S3 and Table S1). To address the molecular mechanism underlying this phenotype, we examined the expression of three important floral regulators, FLC, CONSTANS (CO), and FT, by quantitative real-time PCR (qRT-PCR). No obvious change in expression compared to wild-type was detected at various vegetative developmental stages, but statistical significant for the expression of FLC and FT between wild-type and jmj18 mutants if any at later vegetative developmental stages (Figure 2E–2G and Figure S4). This suggests that the changes in gene expression are mild, which is consistent with the mild late-flowering phenotype in the jmj18 mutants and knock-down lines.
Tab. 1. jmj18 mutants exhibit late-flowering phenotype. 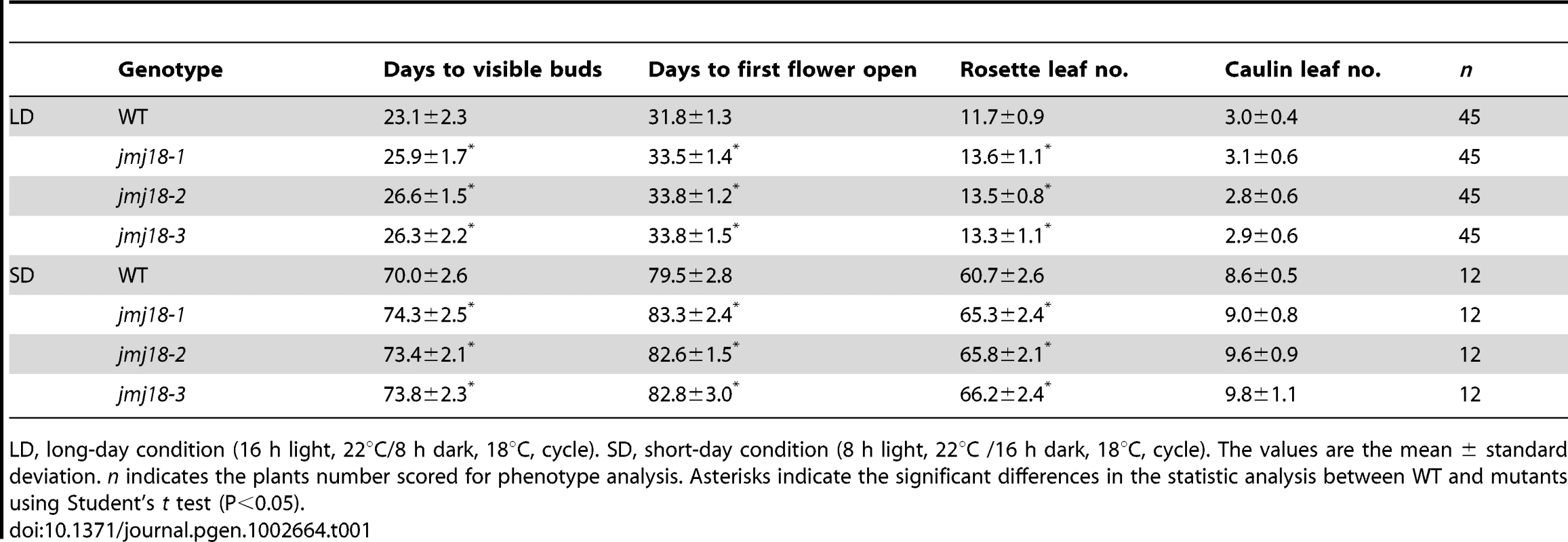
LD, long-day condition (16 h light, 22°C/8 h dark, 18°C, cycle). SD, short-day condition (8 h light, 22°C /16 h dark, 18°C, cycle). The values are the mean ± standard deviation. n indicates the plants number scored for phenotype analysis. Asterisks indicate the significant differences in the statistic analysis between WT and mutants using Student's t test (P<0.05). Spatial expression pattern of JMJ18
To gain a deeper understanding of the function of JMJ18 in Arabidopsis, we examined its expression pattern using wild-type Arabidopsis plants carrying a JMJ18 promoter-fused GUS reporter construct. The whole intergenic region between JMJ18 and its upstream gene was first selected to be the promoter of JMJ18 (Figure S5A). GUS expression was detected in vascular tissue collected from the cotyledons, young leaves, and roots of seedlings (Figure 3A and 3B). Similarly, GUS was detected in the vascular tissue of adult leaves and flowers (Figure 3C and 3D). Similar expression pattern was obtained if the length of promoter was increased from 1.4 to 1.7 kb (Figure S5A–S5D).
Fig. 3. JMJ18 is expressed predominantly in companion cells. 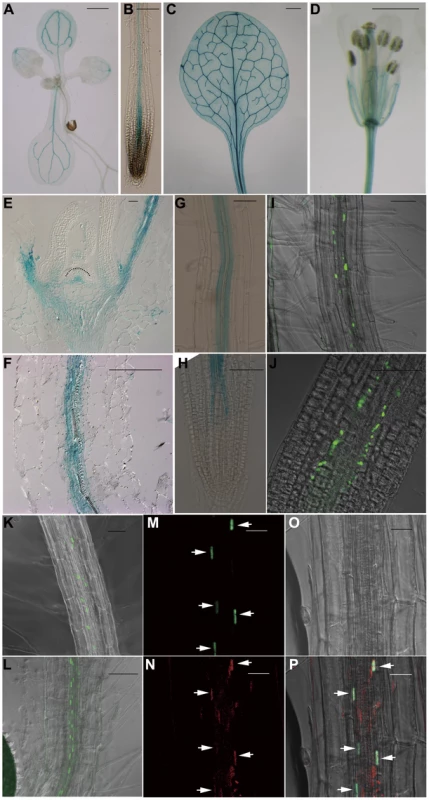
(A–H) Histochemical analysis of JMJ181.4 kb:GUS transgenic plants. Ninety-one percent (22 out of 24 independent lines) of the plants exhibited a similar expression pattern. (A) Nine-day-old seedling. (B) Seven-day-old root. (C) Rosette leaves from a 24-day-old plant. (D) Flower. Bar = 1 mm in (A), (C), and (D), and 0.1 mm in (B). Longitudinal sections of the shoot apex (E) and root (F) from twelve-day-old plants. Bar = 50 µm. GUS staining of mature roots (G) and root tips (H) from seven-day-old seedlings. Bar = 50 µm. (I–K) GFP fluorescence in JMJ181.4 kb:JMJ18-GFP transgenic plants. Eighty-five percent (17 out of 20 independent lines) of the JMJ18:JMJ18-GFP plants displayed a similar expression pattern. Seven-day-old seedlings, mature roots (I) and (K), and root tips (J). Bar = 50 µm. (L) GFP fluorescence in seven-day-old roots of SUC2:JMJ18-GFP transgenic plants. Bar = 50 µm. (M) to (P) JMJ181.4 kb:JMJ18-RFP and SUC2:JMJ18-GFP double-transgenic plants exhibited JMJ18 expression in companion cells. All of the transgenic lines (13 independent lines) observed for the colocalization of JMJ18-GFP and -RFP displayed a similar colocalization pattern. GFP fluorescence (M), RFP fluorescence (N), bright-field image (O), and merged image (P). Bar = 25 µm. Next, JMJ18 expression was examined in detail by microscopy. Longitudinal section analysis showed that JMJ18:GUS was expressed in both the shoot and root phloem, as well as in protophloem, but not in the SAM in seedlings (Figure 3E and 3F). Further, JMJ18 expression was restricted to the cell files of protophloem in the root tip (Figure 3H and Figure S5E–S5J) and phloem companion cells in mature roots (Figure 3F and 3G). These results suggest that JMJ18 is predominant expressed in companion cells.
The phloem cell-dominant expression of JMJ18 was further confirmed in transgenic plants expressing GFP-tagged JMJ18 under the control of the JMJ18 promoter (Figure 3I–3K). JMJ18-GFP was expressed in the protophloem in root tip and phloem in other tissues (Figure 3I–3K and Figure S6A–S6C). We also found that JMJ18-GFP was localized to the nucleus (Figure 3I and Figure S6D), which is consistent with its function as a histone demethylase.
To further verify the expression pattern of JMJ18, the companion cell-specific SUC2 promoter [75], [76] was used to drive JMJ18-GFP expression in wild-type plants. The expression pattern of SUC2:JMJ18-GFP was similar to that of JMJ18:JMJ18-GFP (Figure 3K and 3L). In addition, we generated JMJ18:JMJ18-RFP and SUC2:JMJ18-GFP double-transgenic plants and found that JMJ18-RFP and JMJ18-GFP were colocalized in the nuclei of companion cells (Figure 3M–3P). It was also observed that JMJ18-GUS was also detected in pollen if the anther was stained longer time (Figure S5K–S5N).These results demonstrate that JMJ18 is expressed predominant in phloem companion cells, similar to the floral integrator FT in vegetative developmental stage [62].
The temporal expression pattern of JMJ18 is positively correlated with that of FT
To determine the developmental expression profiles of JMJ18, wild-type seedlings grown under LD conditions were harvested at dusk on days 6, 9, 12, and 15 for RNA isolation. The plants did not form floral meristems; day 15 was the last day before floral meristem formation based on our microscopic observations. JMJ18 was expressed at a low level in young seedlings; however, its expression progressively increased during vegetative development from day 6 to day 15 (Figure 4A). Thus, the expression of JMJ18 is developmentally regulated.
Fig. 4. Temporal expression pattern of JMJ18, JMJ18-induced FT expression, and the floral transition. 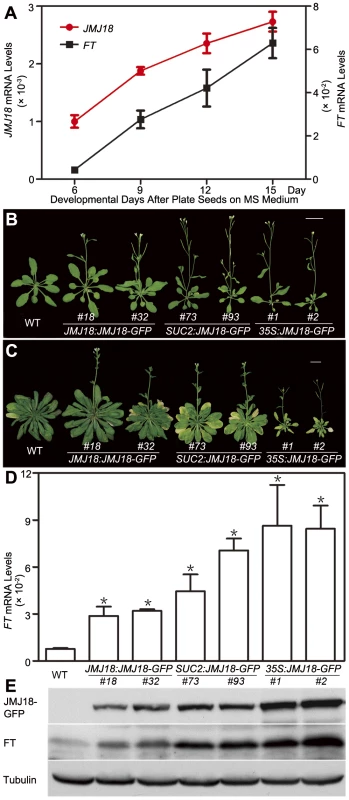
(A) qRT-PCR analysis of JMJ18 and FT expression in plants grown under LD conditions. The expression level was normalized to that of ACTIN. Error bars indicate the standard deviation of three independent biological replicates. (B) JMJ18 overexpression promotes the floral transition under LD conditions. Bar = 2 cm. Twenty-five-day-old plants were analyzed. Twenty-one (23 out of 108), 33 (36 out of 108), and 19 (25 out of 108) percent of the independent lines for JMJ18:JMJ18-GFP, SUC2:JMJ18-GFP, and 35S:JMJ18-GFP, respectively, exhibited earlier flowering compared to wild-type plants. The plants selected for phenotypic analysis were homozygous with one insertion at T4 generation for JMJ18:JMJ18-GFP and SUC2:JMJ18-GFP, and T3 generation for 35S:JMJ18-GFP. Two independent lines per transformation were shown. (C) JMJ18 overexpression promotes the floral transition under SD conditions. Bar = 2 cm. Fifty-eight-day-old plants were analyzed. (D) qRT-PCR analysis of FT mRNA expression in the transgenic plants. The expression level was normalized to that of ACTIN. Error bars indicate the standard deviation of three independent biological replicates. The 11-day-old plants were collected at dusk for the analysis. Asterisks indicate the significant difference between wild-type and overexpression lines using Student's t test (P<0.05). (E) Western blot analysis of JMJ18 and FT expression in the transgenic plants. Tubulin was used as a loading control. The 11-day-old plants were harvested at dusk for the analysis. FT expression was also examined using the same batch of materials. Consistent with previous data [60], [61], FT expression was weak in young seedlings; however, it increased steadily as the plants matured (Figure 4A). There was a significant positive correlation between the expression patterns of JMJ18 and FT during vegetative development, with a correlation coefficient of 0.986 (Figure 4A).
JMJ18 overexpression promotes flowering through FT expression
To further investigate the function of JMJ18, we overexpressed JMJ18 using its endogenous promoter (JMJ18 promoter), the companion cell-specific SUC2 promoter, and the constitutively expressed CaMV35S promoter of which is expressed in and outside of the companion cells in wild-type plants. All three types of overexpression lines exhibited earlier flowering compared to wild-type under inductive LD and non-inductive SD conditions (Figure 4B, 4C and Table 2). Overall, the SUC2:JMJ18-GFP transgenic lines flowered earlier than the JMJ18:JMJ18-GFP transgenic plants; however, the 35S:JMJ18-GFP transgenic lines flowered before either of the other two (Figure 4B, 4C and Table 2). This suggests that the overexpression phenotype was JMJ18 dose-dependent.
Tab. 2. Over-expression of JMJ18 induces floral transition under long-day and short-day conditions. 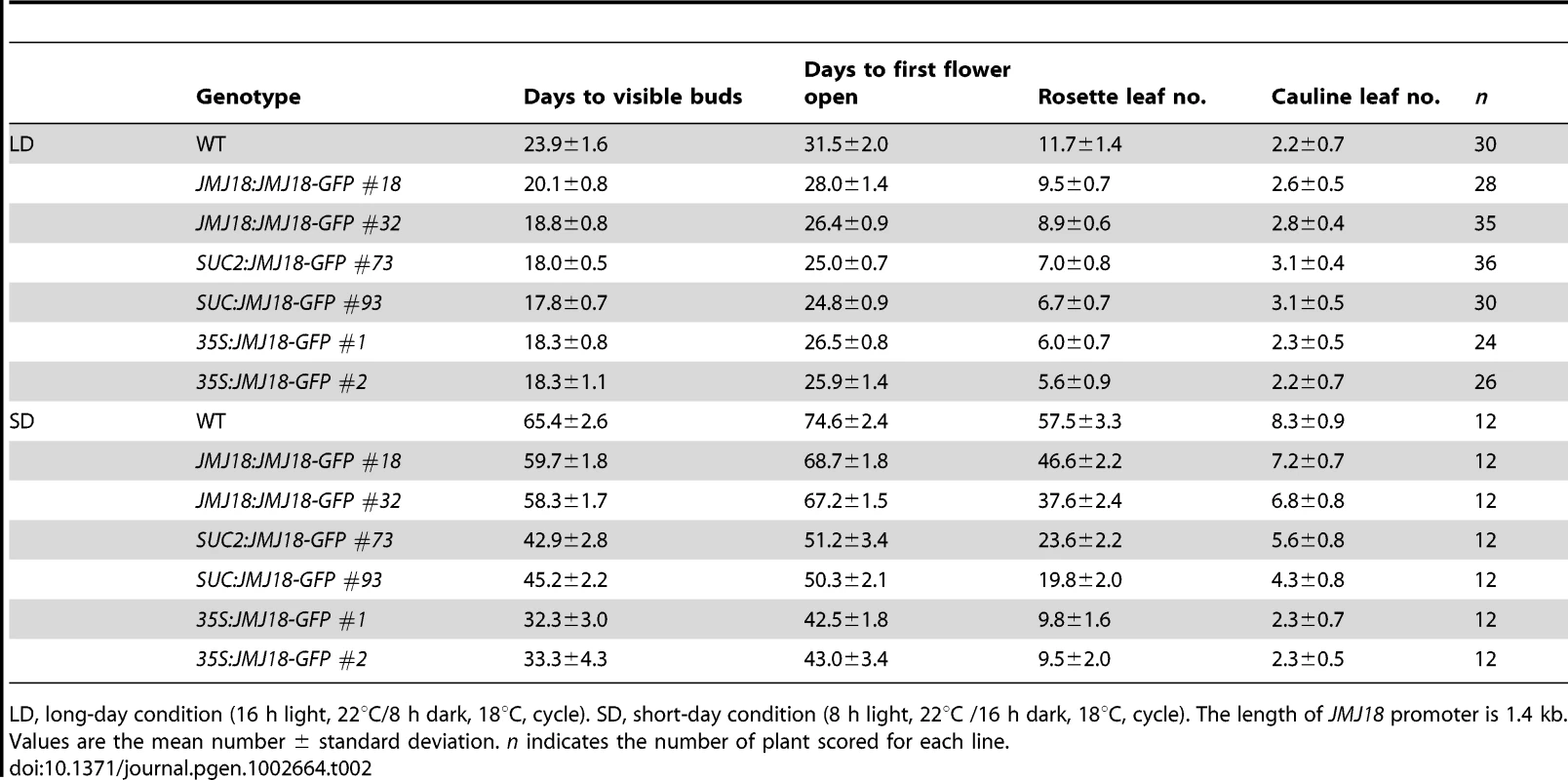
LD, long-day condition (16 h light, 22°C/8 h dark, 18°C, cycle). SD, short-day condition (8 h light, 22°C /16 h dark, 18°C, cycle). The length of JMJ18 promoter is 1.4 kb. Values are the mean number ± standard deviation. n indicates the number of plant scored for each line. To verify this prediction, total protein was extracted from eleven-day-old transgenic plants harvested at dusk, and the abundance of JMJ18 was assessed by Western blotting using anti-JMJ18 antibodies generated against recombinant JMJ18 in rabbits. Consistent with the early-flowering phenotype described above, the expression of JMJ18 in the transgenic plants was as follows (in order from lowest to highest): JMJ18:JMJ18-GFP, SUC2:JMJ18-GFP, and 35S:JMJ18-GFP (Figure 4E and Figure S7). Taking our flowering time and JMJ18 expression data together, the overexpression of JMJ18 promotes flowering in a JMJ18 dose-dependent manner.
The strong positive correlation between the JMJ18 and FT spatio-temporal expression patterns (Figure 3, Figure 4A, and Figure S5) suggests that JMJ18 regulates FT expression in companion cells to control floral development. To investigate this possibility, total RNA was isolated from the same batch of materials used to examine JMJ18 expression. FT expression was increased to varying degrees in all three types of overexpression lines, and the degree of increase in each line was correlated with the abundance of JMJ18 and flowering time (Figure 4D, 4E and Table 2). Similar results were obtained for the expression of TSF, a homolog of FT, in JMJ18 overexpression lines (Figure S8).
It has been shown that FT is one of the most important components of florigen, and its abundance determines flowering behavior [66], [67], [68]. Thus, we next examined the abundance of FT in JMJ18 overexpression lines by Western blotting using anti-FT antibodies generated against recombinant FT in rabbits. The FT level detected in each line was consistent with the mRNA level and abundance of JMJ18 (Figure 4D and 4E). These results indicate that JMJ18 induces FT and its homologs such as TSF transcription and enhances their accumulation to accelerate the vegetative-to-reproductive transition.
The promotion of flowering by JMJ18 mainly depends on FT function
To test whether FT function is genetically necessary for facilitation of the vegetative-to-reproductive transition by JMJ18, ft-11 was crossed with the SUC2:JMJ18-GFP line. The mutation in FT dramatically suppressed the early-flowering phenotype of the SUC2:JMJ18-GFP plants (Figure 5A–5C), suggesting that FT is required for the function of JMJ18 in flowering control.
Fig. 5. Functional FT is necessary for promotion of the floral transition by JMJ18. 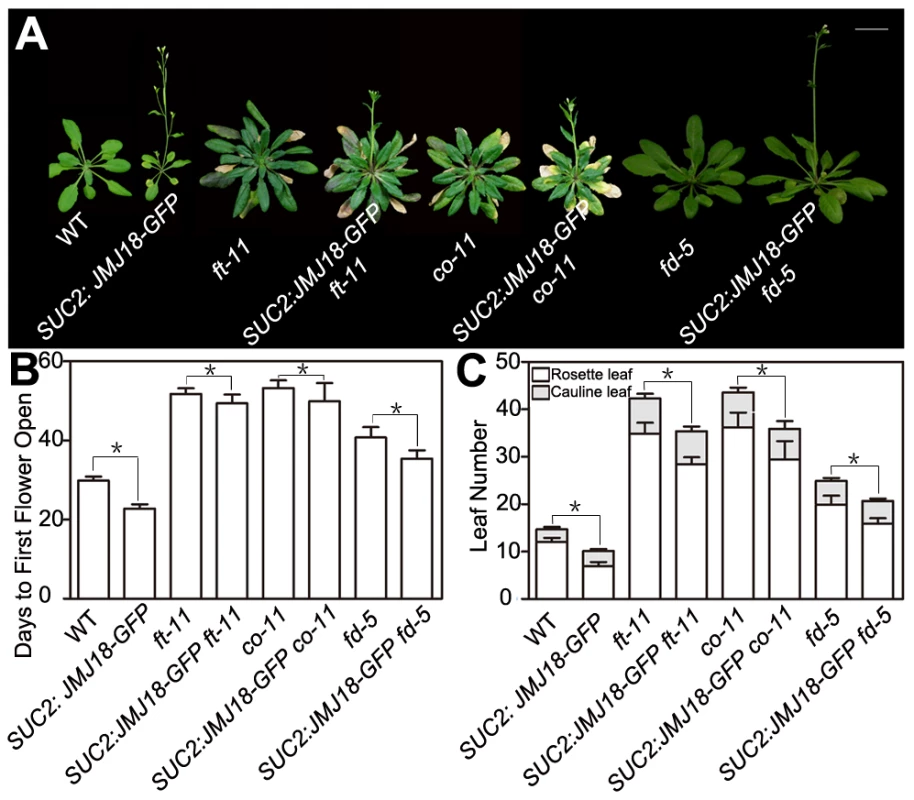
Promotion of the floral transition by JMJ18 is mainly dependent on FT, CO, or FD. (A) WT and SUC2:JMJ18-GFP are 24-day-old plants; ft-11, SUC2:JMJ18-GFP ft-11, co-11, and SUC2:JMJ18-GFP co-11 are 43-day-old plants. fd-5 and SUC2:JMJ18-GFP fd-5 are 35-day-old plants. SUC2:JMJ18-GFP line #73 was used in the experiment. Bar = 2 cm. The values in (B) and (C) are the mean ± standard deviation from at least 20 plants per genotype. Asterisks in (B) and (C) indicate the significant difference between marked plants from the two genotypes by Student's t test (P<0.05). In the photoperiod pathway, CO is responsible for FT induction, and FT expression is obviously delayed in a CO mutant background even under inductive LD conditions [60], [61]. FD is an SAM-specific transcription factor that interacts with FT in the SAM [64]. Both FT and FD are required for floral meristem formation [64], [65]. The mutation of CO or FD also abolished the SUC2:JMJ18-GFP early-flowering phenotype, such as SUC2:JMJ18-GFP co-11 or SUC2:JMJ18-GFP fd-5, flowered at a similar time to co-11 or fd-5 (Figure 5A–5C). In addition, we found that overexpression of JMJ18 in mutant background, such as SUC2:JMJ18-GFP ft-11, SUC2:JMJ18-GFP co-11 and SUC2:JMJ18-GFP fd-5, flowered slightly earlier than the single mutants ft-11, co-11, and fd-5, in terms of both flowering time and rosette leaf number (Figure 5A–5C). These genetic data demonstrate that promotion of the floral transition by JMJ18 mainly depends on functional FT.
The JmjN, JmjC, and zinc-finger domains are required for JMJ18 function
JMJ18 has four conserved domains: the JmjN, JmjC, zinc-finger, and FY-rich domains (Figure 2A). Due to the obvious early-flowering phenotype of JMJ18 overexpressors, we overexpressed truncated JMJ18 (Figure 6A and Figure S9A) in wild-type plants to determine the domain(s) in JMJ18 necessary for its function in planta. The early-flowering phenotype of 35S:JMJ18 (containing the entire JMJ18 CDS) was echoed by that of 35S:JMJ18-GFP (Figure 6B and 6C), suggesting that blocking of the free of C-terminus of JMJ18 by GFP does not affect its function.
Fig. 6. The JmjN, JmjC, and zinc-finger domains are necessary for the promotion of flowering by JMJ18. 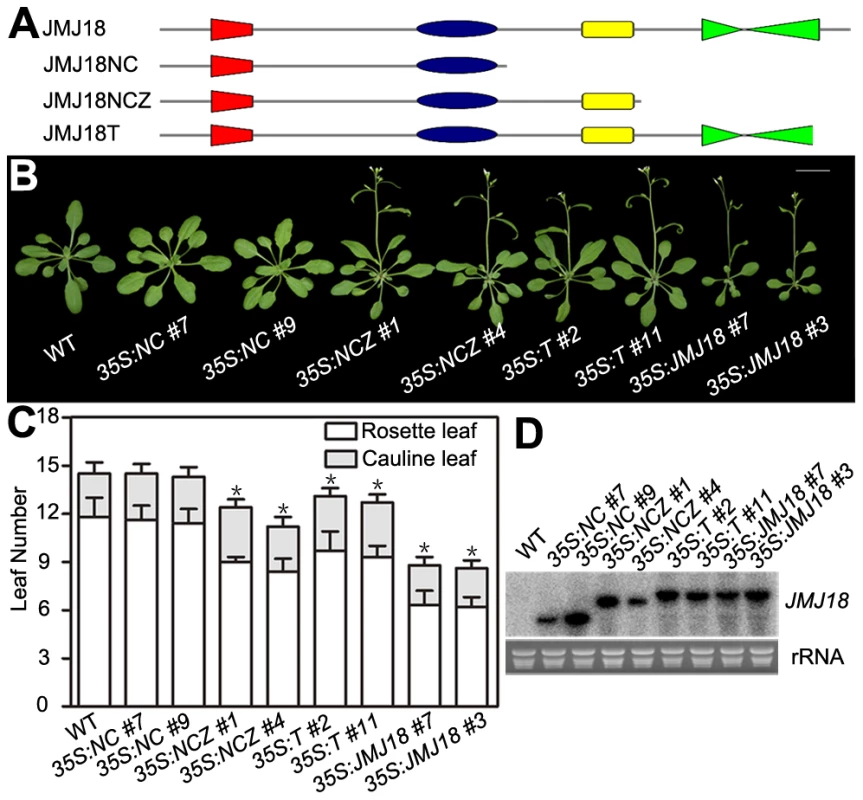
(A) Schemes display the structures of full-length JMJ18 protein as well as truncated versions used for overexpression test. (B) and (C) Flowering time phenotype for the JMJ18 overexpressor lines. All of the plants in (B) were grown for 24 days under LD conditions. All of the 108 independent 35S:NC lines examined flowered at a similar time compared to wild type. Fifteen out of 108 35S:NCZ, 20 out of 108 35S:T, and 27 out of 108 35S:JMJ18 lines displayed an early-flowering phenotype. Two independent lines per transformation were shown. Bar = 2 cm. The values in (C) are the mean ± standard deviation. At least 15 plants were scored for each genotype. Asterisks indicate the significant difference between wild-type and transgenic plants analyzed by Student's t test (P<0.05) using rosette leaf numbers. (D) Northern blot analysis of the expression of truncated JMJ18 in the transgenic lines. 35S:NCZ (containing the JmjN, JmjC, and zinc-finger domains but lacking the FY-rich domain) transgenic plants exhibited a shorter life cycle than wild-type plants; however, the effect was weaker than that observed in 35S:JMJ18 plants (Figure 6B and 6C). Regardless, no early-flowering phenotype was observed in those plants overexpressing truncated JMJ18 with a deletion in both the zinc-finger and FY-rich domains (35S:NC) (Figure 6B and 6C). Northern blot analysis was used to determine whether these truncated transcripts were expressed in the transgenic plants. As shown in Figure 6D, all of the truncated transcripts were expressed. The transgenic plants overexpressing truncated JMJ18 with a deletion in both the JmjN and JmjC domains displayed similar flowering time compared to wild-type plants (Figure S9). Thus, these data suggest that the JmjN, JmjC, and zinc-finger domains are required for the function of JMJ18, while the FY-rich domain is not fully necessary for its function but affects its activity.
The 35S:T (encoding a truncated protein identical to that in jmj18-1 and containing the JmjN, JmjC, and zinc-finger domains) transgenic plants also displayed an obviously earlier floral transition compared to wild-type plants (Figure 6A–6D),which indicates that jmj18-1 and jmj18-2 are weak alleles. And the result is consistent with their weak early-flowering phenotype (Figure 2C, 2D and Table 1).
JMJ18 represses the expression of FLC and MAFs
H3K4 methylation is associated with gene activation; thus, H3K4 demethylases should work as gene repressors, rather than as activators. Therefore, FT is not likely to be the direct target of JMJ18. The target of JMJ18 could be an upstream repressor of FT. If this is the case, the floral repressor FLC is a good candidate. To examine this possibility, the effect of JMJ18 overexpression on FLC expression was examined in JMJ18 overexpressor plants. FLC expression was significantly repressed in all JMJ18 overexpressor lines examined, and the degree of repression was positively correlated with the abundance of JMJ18, but negatively with the level of FT (Figure 4E and Figure 7A), the other members of FLC clade genes (MAF1 to MAF5) were also repressed to different extent in JMJ18 overexpressors (Figure 7C); in comparison, there was no obvious change in the expression of CO, an upstream activator of FT (Figure 7B).
Fig. 7. JMJ18 acts as a repressor of FLC and MAFs. 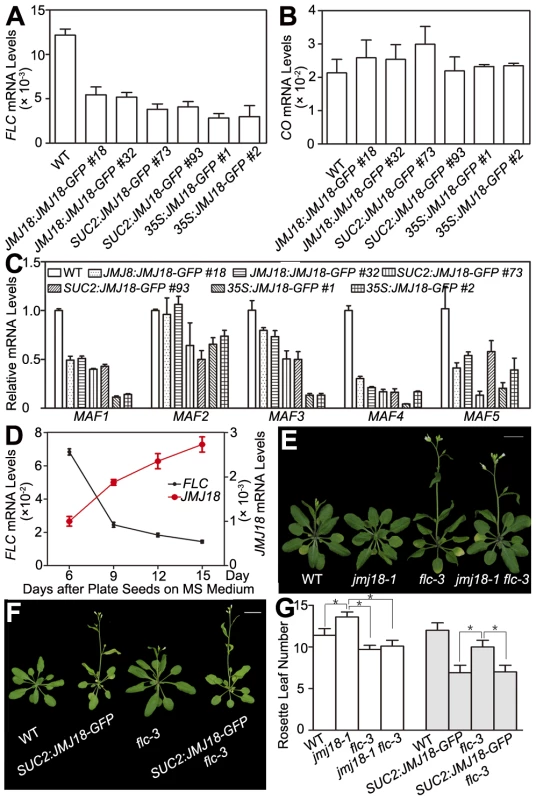
(A) FLC expression was repressed in the JMJ18 overexpressor lines. Asterisks indicate the significant difference between wild-type and transgenic plants analyzed by Student's t test (P<0.05). (B) CO expression was not obviously changed in the JMJ18 overexpressor lines. There was no significant difference between wild-type and transgenic plants analyzed by Student's t test (P<0.05). (C) The expression of MAF1 to MAF5 was repressed in the JMJ18 overexpressor lines. Eleven-day-old plants were used for RNA isolation. Asterisks indicate the significant difference between wild-type and transgenic plants analyzed by Student's t test (P<0.05). The expression level was normalized to that of ACTIN in (A) to (C). (D) Temporal expression patterns of FLC and JMJ18 during vegetative development. The mRNA levels were normalized to that of ACTIN. The error bars indicate the standard deviation from three independent biological replicates. (E) to (G) Genetic analysis of JMJ18 and FLC in flowering time control. The SUC2:JMJ18-GFP #73 was crossed to flc-3. Twenty-eight- (E) and 24-day-old (F) plants were photographed. Bar = 2 cm. Asterisks indicate the significant difference analyzed by Student's t test (P<0.05). To analyze the temporal expression pattern of FLC and its relationship to that of JMJ18 during vegetative development, we measured FLC expression in plants grown under LD conditions on days 6, 9, 12, and 15 as described above. FLC expression was strong at the seedling stage, then decreased during vegetative development, reaching its lowest level before floral meristem formation on day 15 (Figure 7D). There was a strong negative correlation between the developmental expression patterns of JMJ18 and FLC during vegetative development (r = −0.950). Thus, we propose that JMJ18 is a repressor of FLC.
We generated jmj18-1 flc-3 to test whether JMJ18 interacts genetically with FLC. The FLC mutation mainly blocks the late-flowering phenotype of jmj18-1 (Figure 7E, 7G and Table S2). Thus, the mutation in FLC did not enhance the early-flowering phenotype of the JMJ18 overexpressors. In addition, overexpression of JMJ18 in flc, such as SUC2:JMJ18-GFP flc-3, flowered earlier than flc-3 and at almost the same time as SUC2:JMJ18-GFP) (Figure 7F, 7G and Table S2), indicating that endogenous JMJ18 enhances flowering mainly by repressing FLC in wild-type plants, while overexpressing JMJ18 recognized FLC and MAFs as targets. Taken together, those results indicate that JMJ18 and FLC belong to the same genetic pathway in flowering time control, and JMJ18 functions in upstream of FLC.
JMJ18 demethylates the chromatins of FLC and MAFs
Our previous result suggested that JMJ18 functions as an H3K4-specific demethylase in vitro (Figure 1 and Figure S1). To verify the histone demethylase activity of JMJ18 in vivo, nucleoproteins were extracted from jmj18, our JMJ18 overexpressor lines, and wild-type plants. There was no obvious change in the levels of H3K4me3, H3K4me2, and H3K4me1 among the wild-type, jmj18, and JMJ18 overexpressor plants on a global scale (Figure 8A and 8B). A slight decrease in H3K4me3 was detected in the 35S:JMJ18-GFP plants (Figure 8B), but no obvious difference was detected between wild-type and the other JMJ18 overexpressors (Figure 8B). Thus, endogenous JMJ18 may function as a gene-specific H3K4 demethylase in vivo, while overexpressed JMJ18 can target many other loci to demethylate H3K4me3 once it is expressed in many cell types.
Fig. 8. Global H3K4 methylation levels in wild-type, jmj18, and JMJ18 overexpressor lines. 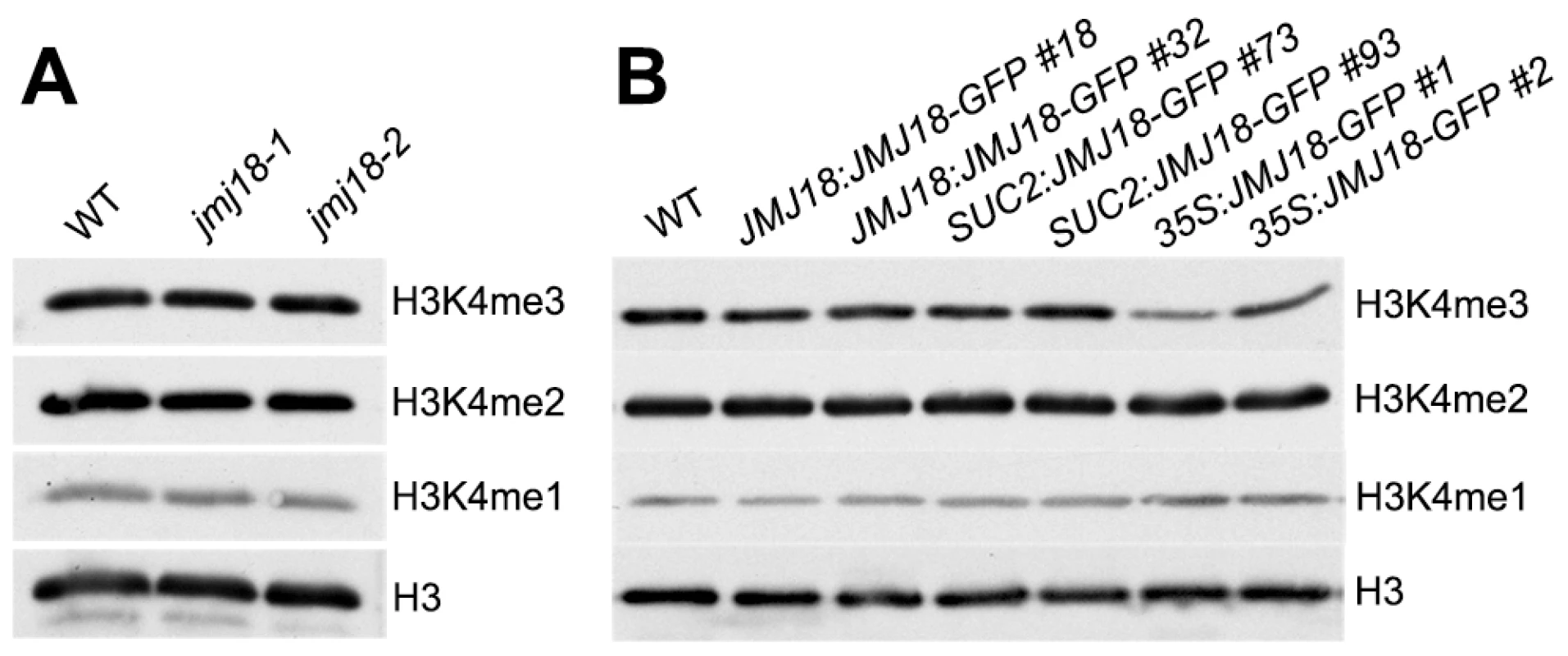
(A) Detection of the global H3K4me3, H3K4me2, and H3K4me1 levels in the jmj18 mutants. (B) Detection of the global H3K4me3, H3K4me2, and H3K4me1 levels in the JMJ18 overexpressor lines. To verify whether JMJ18 demethylates FLC chromatin, chromatin immunoprecipitation (ChIP) was done to detect the level of H3K4 methylation across the entire FLC chromatin region. In wild-type plants, the regions around the transcription start site (regions B4, B5, and B6) had higher levels of H3K4me3 than the other regions (Figure 9A and 9B), which is consistent with previous data [11], [43]. However, the H3K4me3 level was obviously decreased in SUC2:JMJ18-GFP plants across the entire FLC chromatin region compared to that in wild-type plants (Figure 9B). However, the H3K4me3 levels at the house keeping gene ACTIN, which contains high level of H3K4me3, and transposon elements AtMu1 and AtSN1, which are lack of H3K4me3, were not obviously changed in SUC2:JMJ18-GFP plants compared to wild-type plants (Figure S10). The similar results were obtained by using normalizing H3K4me3 level to total H3 and to the input (Figure S11). The level of H3K4me2 in FLC chromatin was also decreased in SUC2:JMJ18-GFP overexpressor plants compared to wild-type plants (Figure 9C). To analyze whether JMJ18 regulates the expression of MAFs through the control of the H3K4 methylation state at their loci, the H3K4me3 and me2 modification levels were detected in SUC2:JMJ18-GFP and wild-type plants. The levels of H3K4me3 at the chromatins of five MAFs were obviously decreased. While, the levels of H3K4me2 were slightly decreased at MAF4, MAF5 but not MAF1, MAF2 and MAF3 (Figure 9D, 9E). These results indicate that JMJ18 demethylates H3K4me3 and H3K4me2 within the FLC and MAFs loci in vivo.
Fig. 9. JMJ18 binds FLC chromatin and mediates the level of H3K4 methylation in FLC chromatin. 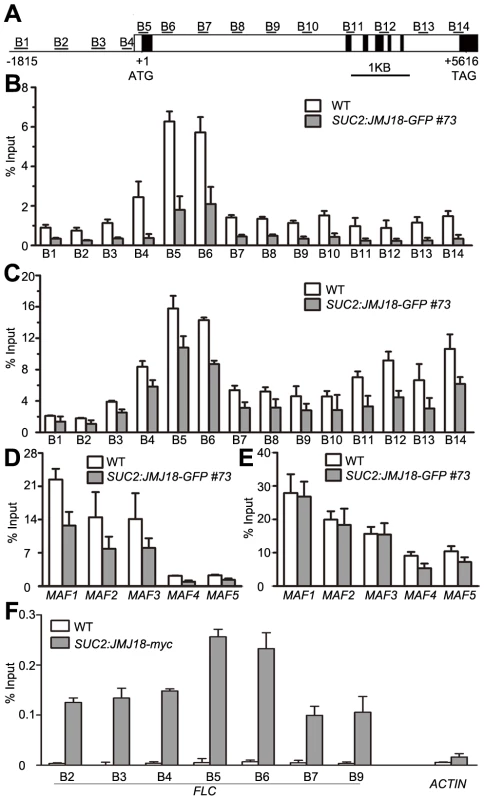
(A) Schematic genomic structure of the FLC locus, and the probes used to measure the histone methylation level. Open boxes represent introns, while filled boxes represent exons. B1–B14 indicate the FLC regions recognized by the probes during the analysis of the H3K4 methylation level by ChIP. (B) ChIP analysis of the H3K4me3 level across FLC in wild-type and JMJ18 overexpressor plants. (C) ChIP analysis of the H3K4me2 level across FLC in wild-type and JMJ18 overexpressor plants. (D) and (E) ChIP analysis of the H3K4me3 and H3K4me2 levels for other members of FLC clade genes in wild-type and JMJ18 overexpressor plants. (F) ChIP analysis of the binding of JMJ18 to FLC chromatin in eleven-day-old plants. The enrichments were normalized to the input. ACTIN was used as a negative control. The values are the mean ± standard deviation from three biological replicates. JMJ18 directly associates with FLC chromatin
JMJ18 controls FLC expression by modulating the H3K4 methylation level at FLC chromatin, suggesting that JMJ18 associates with the FLC locus and mediates the methylation of FLC chromatin directly. To investigate this possibility, ChIP was used to detect the binding of JMJ18 to the FLC locus. Due to the increased level of nonspecific binding of GFP to Arabidopsis chromatin under our experimental conditions, we used SUC2:JMJ18-myc transgenic plants instead of SUC2:JMJ18-GFP plants in our assay. The SUC2:JMJ18-myc transgenic plants exhibited similar phenotypes to the SUC2:JMJ18-GFP plants, including an early-flowering phenotype, altered FLC and FT expression, and reduced H3K4me3 and H3K4me2 levels across FLC chromatin (Figure S12). A c-myc-specific antibody was used to precipitate chromatin from wild-type and SUC2:JMJ18-myc plants. Compared to the wild-type plants, the FLC chromatin of the transgenic plants was significantly enriched with JMJ18 protein, with varying levels of occupancy across the entire FLC chromatin region, while, there was no obvious binding of JMJ18 to the chromatin of ACTIN, which was a negative control, compared with its binding to FLC chromatin (Figure 9F). Regions B5 and B6 exhibited the highest binding ability (Figure 9F), whereas the H3K4 methylation level was higher than in the other regions (Figure 9B and 9C), demonstrating that JMJ18 associates with FLC chromatin and that the binding of JMJ18 to FLC is necessary for the dynamic of H3K4 methylation in FLC chromatin.
JMJ18 overexpression suppresses the FRI late-flowering phenotype
FRI is the major determinant of ecotype differences in flowering time in Arabidopsis [46], [47]. To examine the influence of JMJ18 on flowering time promotion, JMJ18 was overexpressed in FRI plants by the introduction of FRI into JMJ18:JMJ18-GFP, SUC2:JMJ18-GFP, and 35S:JMJ18-GFP transgenic lines via crossing, respectively. The overexpression of JMJ18 significantly suppressed the late-flowering phenotype of FRI in a JMJ18 dose-dependent manner (Figure 10A and Table 3).
Fig. 10. JMJ18 overexpression suppresses the FRI late-flowering phenotype. 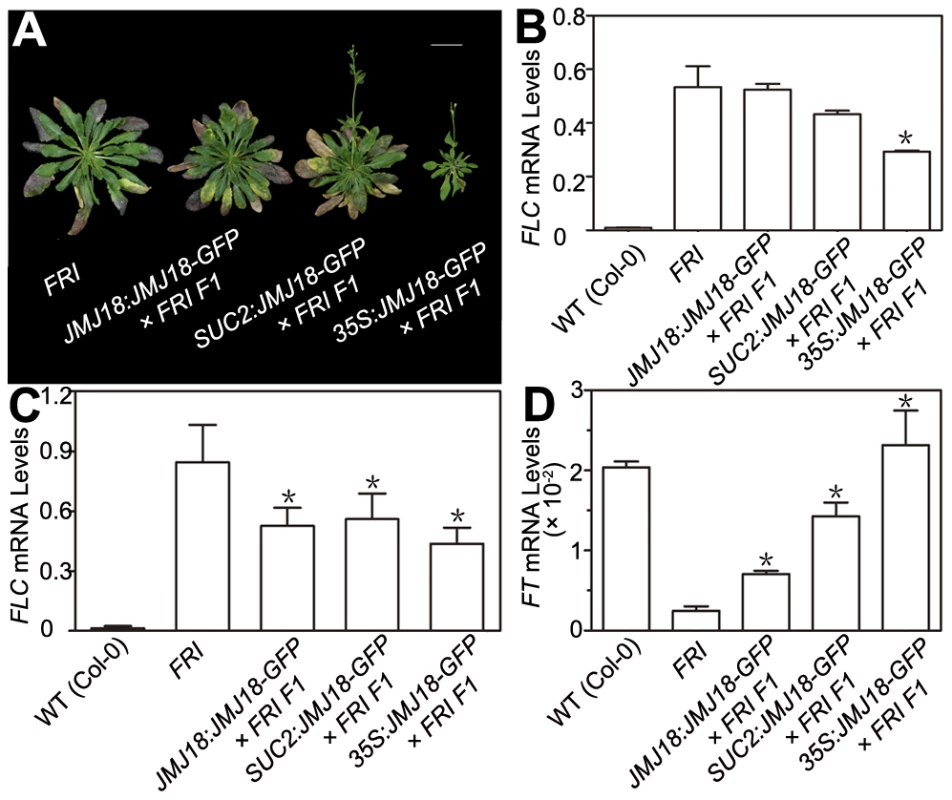
(A) Flowering time in FRI wild-type and F1 plants from a cross between FRI wild-type and JMJ18 overexpressor plants. All of the plants were 48 days old at analysis except for 35:JMJ18-GFP #1×FRI F1, which was 38 days old. All plants were grown under LD conditions. Bar = 2 cm. Expression levels of FLC in eleven-day-old whole seedlings (B) and 26-day-old rosette leaves (C). (D) FT expression in eleven-day-old seedlings. The expression level was normalized to that of ACTIN. The values are the mean and standard deviation from three independent biological replicates. The lines crossed with FRI were: JMJ18:JMJ18-GFP #34, SUC2:JMJ18-GFP #73, and 35S:JMJ18-GFP #1. Asterisks in (B), (C) and (D) indicate the significant difference for the expression levels of FLC and FT between FRI and other genotypes analyzed by Student's t test (P<0.05). Tab. 3. JMJ18 overexpression suppresses the FRI late-flowering phenotype. 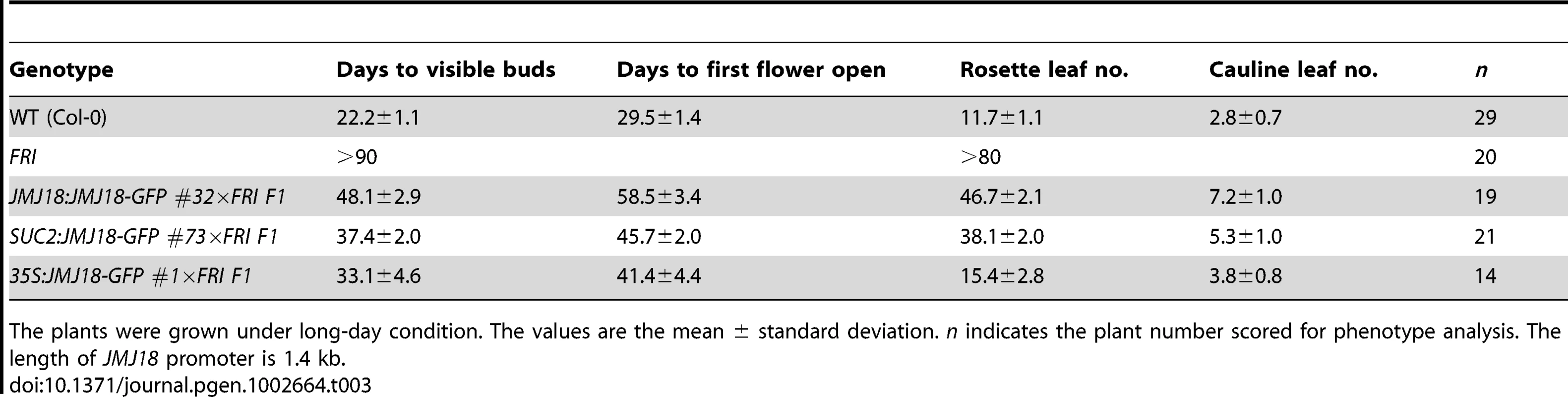
The plants were grown under long-day condition. The values are the mean ± standard deviation. n indicates the plant number scored for phenotype analysis. The length of JMJ18 promoter is 1.4 kb. FRI represses the floral transition by activating FLC expression [46], [77]. To determine whether JMJ18 suppresses the FRI late-flowering phenotype by decreasing FLC expression, FLC transcription in FRI wild-type and JMJ18 overexpressors was measured. FLC expression was decreased in the transgenic lines compared to that in the FRI background, with a more pronounced effect in leaves than in complete seedlings except for 35S:JMJ18-GFP (Figure 10B and 10C), which is consistent with the expression pattern of FLC and JMJ18 in our transgenic lines. This is because FLC was expressed in both the SAM and companion cells, with predominant expression in the SAM, while both the JMJ18 and SUC2 promoters were specifically expressed in companion cells. Thus, this result supports the notions that JMJ18 promotes flowering in FRI by repressing FLC expression, and that FLC expressed in both the SAM and companion cells contributes to flowering time control. Consistent with our previous results (Figure 4D and Figure S12D) and early-flowering phenotype (Figure 10A and Table 3), the expression of FT in these transgenic lines was obviously activated (Figure 10D), indicating that JMJ18 represses FLC expression and induces FT expression in an FRI background as well.
Discussion
JMJ18 is a histone H3K4 demethylase in Arabidopsis
Several JARID1 family proteins have been characterized as H3K4 demethylases in various organisms [9], [28], [29], [33], [69], [71], [72], [73]. There are 6 members of JARID1 proteins in Arabidopsis, JMJ14 (at4g20400), JMJ15 (at2g34880), JMJ16 (at1g08620), JMJ17 (at1g63490), JMJ18 (at1g30810), and JMJ19 (at2g38950). To examine whether Arabidopsis JMJ18 is a real histone demethylase, we characterized its histone demethylase activity in vitro and in vivo. We examined the histone demethylase activity of JMJ18 in vitro by purifying recombinant JMJ18 from insect cells and analyzing it by MALDI-TOF mass spectrometry. In that analysis, JMJ18 exhibited histone H3K4-specific demethylase activity (Figure 1A, 1B, and Figure S1), which was dependent on Fe(II) and α-KG (Figure 1C). Additionally, JMJ18 was able to demethylate histone H3K4me3 to H3K4me2 and H3K4me1 (Figure 1A, 1B, and Figures S1C). This observation was confirmed in vivo using FLC and MAFs chromatins (Figure 9B–9E, Figures S11A–11B and S12E–S12F), but not in a genome-wide analysis of histone H3 in jmj18 mutants or in overexpressing lines driven by JMJ18- or SUC2 - promoters (Figure 8), and the H3K4me3 modification was not obviously changed at ACTIN, AtMu1 and AtSN1 loci neither (Figures S10 and S11C), suggesting that JMJ18 may function as a gene-specific H3K4 demethylase in Arabidopsis. However, it is obvious that global H3K4me3 in 35S:JMJ18-GFP is reduced (Figure 8B), indicating that JMJ18 can target many other loci to demethylate H3K4me3 but not H3K4me2 once it is expressed in many cell types. Thus, the target specificity of JMJ18 can be fulfilled by gene specific targeting mechanisms or expression restriction or both. It was observed that the reduction in H3K4me2 is less than that H3K4me3 in the chromatins FLC and MAFs by JMJ18 overexpression (Figure 9B–9E and –S12F), and full-length JMJ18 lacks demethylase activity towards H3K4me2 to H3K4me1 (Figure 1A and Figure S1C), indicating that JMJ18 favors H3K4me3 to H3K4me2 demethylation. Our results also indicate that JARID1 family proteins are conserved histone H3K4 demethylases from human to plants.
Interestingly, full-length JMJ18 only demethylated H3K4me3 to H3K4me2 (Figure 1A). However, truncated JMJ18 lacking the partial linker between the JmjN and JmjC domains exhibited demethylase activity toward both H3K4me3 and H3K4me2 (Figure 1B and Figure S1C). Taken together (Figure 9B–9E and Figure S12E–S12F), our results suggest that the linker between the JmjN and JmjC domains in JMJ18 blocks its H3K4me2 demethylase activity in vitro. This linker could not block the H3K4me2 demethylase activity of JMJ18 in vivo by functioning as a protein-interaction domain; however, this requires further study. At any rate, this observation raises an interesting question as to the structural function of the linker sequence in JMJ18 itself and within the JMJ18 complex.
JMJ18 is a companion cell–predominant, developmentally regulated gene
Cell - or tissue-specific expression is a significant part of enabling a gene to achieve its function in plant cell fate determination and development [64], [65], [78]. Since the transgenic plants carrying JMJ18:JMJ18-GFP transformation could complement the late-flowering phenotype of jmj18-1 (Figure S2), it was possible to characterize the expression pattern of JMJ18 in Arabidopsis based on JMJ18 promoter-reporter expression (Figure 3, Figures S5 and S6). We found that JMJ18:GUS and JMJ18:GFP were expressed in the vascular tissue of cotyledons, young leaves, adult leaves, hypocotyl and flowers (Figure 3A, 3C, 3D; Figures S5 and S6), not expressed in the SAM where FLC was expressed dominantly (Figure 3E). In the roots of the transgenic plants, signals for GUS and GFP were predominantly observed in protophloem at the root tip, and in phloem in other regions of the root (Figure 3B, 3F–3K; Figures S5 and S6D). In addition, different length of promoter-driven reporter exhibits similar expression pattern (Figure 3, Figures S5 and S6), indicating that JMJ18 is predominantly expressed in the phloem of vascular tissue during vegetative development. Recently, Hong et al. (2009) reported that the expression of JMJ18 (Atjmj8 in their study) is expressed broader in root and leaf [79]. We constructed a same version of JMJ18 promoter-GUS fusion as Hong et al (2009) and transformed it into the wild-type plants. We observed that all 21 independent lines transformed with 1.7 kb length promoter-GUS we examined exhibits similar expression pattern, and the two versions of promoter (1.4 and 1.7 kb in length) exhibit very similar expression patterns (Figure 3, Figures S5 and S6). One possibility for the discrepancy between the two studies is due to the over-staining of GUS of tissues in study of Hong et al. (2009) [79].
Phloem is composed of sieve elements and companion cells. An analysis of the cell-specific expression pattern of JMJ18:GFP in phloem indicated that JMJ18 is predominantly expressed in phloem companion cells (Figure 3I, 3K, and Figure S5). This result was confirmed by examining the cellular localization of tagged JMJ18 in the phloem using Arabidopsis coexpressing JMJ18:JMJ18-RFP and SUC2:JMJ18-GFP. SUC2 is a companion cell-specific gene in Arabidopsis [76], [80]. JMJ18-RFP and -GFP were colocalized in the same cell in the roots of the transgenic plants (Figure 3M–3P), indicating that JMJ18 is predominantly expressed in companion cells. Thus, our results reveal that JMJ18 is a companion cell-dominant histone H3K4 demethylase in Arabidopsis.
In addition, the expression of JMJ18 increased during vegetative development, reaching its high level before the formation of the floral meristem (Figure 4A). This suggests that JMJ18 encodes a companion cell-dominant, developmentally-regulated histone demethylase in Arabidopsis.
JMJ18 regulates flowering time
To determine the role of JMJ18 in Arabidopsis development, we characterized three jmj18 T-DNA insertion mutants. All three mutants displayed a late-flowering phenotype under LD and SD conditions (Figure 2C, 2D and Table 1). The knock-down lines for JMJ18 using both RNAi and amiR also exhibit delayed flowering (Figure S3 and Table S1). The late-flowering phenotype in jmj18 is complemented by transforming JMJ18:JMJ18-GFP to jmj18 (Figure S2). However, the flowering phenotype was weak, and the changes in the expression of flowering marker genes in jmj18 were very mild (Figure 2E–2G, Figures S3 and S4).
To determine the reason for the weak phenotype of jmj18, we further characterized the three jmj18 alleles. We found that the T-DNA was inserted at the 3′-end of JMJ18, behind the JmjN, JmjC, and zinc-finger domains, in both jmj18-1 and jmj18-2 (Figure 2A). Partial transcripts of JMJ18 were detected in jmj18-1 and jmj18-2 (Figure 2B). In addition, the truncated JMJ18 in jmj18-1 and jmj18-2 was functional in planta (Figure 6B and 6C). For the third allele, although the T-DNA was inserted in the fourth exon of JMJ18, full-length JMJ18 mRNA was detected at reduced levels in jmj18-3 (Figure 2B). These results indicate that the three jmj18 alleles were weak mutants. Unfortunately, no other allele for jmj18 is currently available.
However, the increase in JMJ18 activity caused by the overexpression of JMJ18 significantly enhanced flowering in Arabidopsis (Figure 4B, 4C; Figure 6B, 6C; Figure 10A; Table 2 and Table 3; and ). This observation was true for all JMJ18 overexpressors driven by the constitutive CaMV35S promoter, companion cell-specific SUC2 promoter, and endogenous JMJ18 promoter (Figure 4B, 4C; Figure 6B, 6C; Figure 10A; Table 2 and Table 3; and Figure S12A, S12B). The above data suggest that JMJ18 is a flowering time regulator in Arabidopsis.
JMJ18 directly represses FLC expression and indirectly induces FT expression in companion cells to control flowering time in Arabidopsis
FLC is a central repressor of flowering in Arabidopsis that works in part through the repression of a companion cell-specific flowering activator, FT [55]. FLC is expressed in both the SAM and companion cells, and the expressed FLC in both tissues contributes to flowering time control in Arabidopsis [55]. The regulation of FLC expression in the SAM has been extensively studied [11], [24], [43], [53]; however, little is known about the regulation of FLC expression in companion cells.
In the present work, we found that JMJ18 was predominantly expressed in companion cells in vegetative tissue (Figure 3, Figures S5 and S6). In addition, we confirmed that the expression pattern of JMJ18 was strongly negatively correlated with that of FLC, but strongly positively correlated with that of FT, during vegetative development in Arabidopsis (Figure 4A and Figure 7D). These results suggest that JMJ18 works as an endogenous developmental signal to regulate the expression of FLC and FT in the control of flowering time.
We further found that JMJ18 binds FLC chromatin (Figure 9F), and decreases the levels of H3K4me3 and H3K4m2 in FLC chromatin (Figure 9B, 9C; Figures S11A and S12E–S12F). In addition, an increase in JMJ18 abundance obviously decreased the mRNA levels of FLC and MAFs (Figure 4E; Figure 7A, 7C, 7D; Figure 10B, 10C; and Figure S12C), and obviously increased the expression of FT at both the mRNA and protein levels (Figure 4D, 4E; Figure 10D; Figure S12D). Furthermore, the promotion of flowering time by JMJ18 was mainly dependent on functional FT (Figure 5A–5C), but was not enhanced by mutation in FLC (Figure 7F, 7G, and Table 2). These results indicate that JMJ18 works as an endogenous, companion cell-dominant developmental signal that directly represses FLC expression and indirectly induces FT expression in companion cells to control flowering time during vegetative development in Arabidopsis. It was noted that overexpressed JMJ18-GFP in companion cells reduced both levels of H3K4me3 and H3K4me2 on FLC, but only H3K4me3 on MAFs (Figure 9B–9E and Figure S12E–S12F). The difference of the decrease in the levels of H3K4 methylation between FLC and MAFs in JMJ18 overexpression line may suggest that JMJ18 is more important for the regulation of FLC than MAFs in Arabidopsis.
In Arabidopsis, the flowering transition is a complicated process. It needs to integrate the environmental cues and internal signals; in addition, these signals will be sensed and regulated in different tissues or organs in the plant [81], [82]. There are two main tissues involved in the flowering control: the vascular tissue in the leaf and the SAM [37], [83]. The character of the apical meristem is determined not only by the processes occurring in apical meristem, but also by the signals transmitted from vascular tissue. Thus, different flowering regulators should be functional and regulated in distinct tissues or steps, integrated in the SAM for the precise control of flowering time. As flowering regulators, the JmjC domain-containing demethylases are functional in both vascular tissue and SAM, and target different genes. Since REF6 is characterized as a H3K27 demethylase and represses FLC in both SAM and leaves, FLC can not be the direct target of REF6 [31], [70]. However, its homolog ELF6, an H3K4 demethylase, is expressed in leaves and targets FT to repress flowering [28]. Another H3K4 demethylase, JMJ14, is restricted in leaves to repress flowering by decreasing the expression of the floral integrators including FT, SOC1, LFY and AP1 [28], [29], [69]. In this report, we demonstrated that JMJ18 is a H3K4 demethylase and specially expressed in the vascular tissue (Figure 1, Figure 3, Figure 9; Figures S5, S6, S11, and S12E–S12F). The expression of JMJ18 is developmental-regulated (Figure 4A). JMJ18 directly represses the expression of FLC and releases the expression of FT in vascular tissue (Figure 4, Figure 5, Figure 7, Figure 9, Figure 10, and Figure S12), then the released FT is transmitted from the vascular tissue to the SAM to stimulating flowering. These results suggest that plants may have evolved to the point where they use a family of proteins with different expression patterns which target various genes to integrate environmental cues and internal signals to precisely control flowering time. Thus, this study provides novel insight into the regulation of FLC expression in companion cells and the epigenetic control of the floral transition in Arabidopsis.
Methods
Plant materials and growth conditions
The Arabidopsis plants used in our experiments were of the Columbia-0 ecotype except for FRI. The seeds were first sterilized with 2.25% bleach, then washed three times with water, stratified for three days at 4°C, and then put on Murashige and Skoog (MS) medium (Sigma-Aldrich) containing 1% sucrose and 0.3% phytagel (Sigma-Aldrich). Following ten days of growth under LD (16 h of light, 22°C/8 h of dark, 18°C) or SD (8 h of light, 22°C /16 h of dark, 18°C) conditions in a growth chamber (Percival CU36L5) under a cool white fluorescent light (160 µmolm−2 s−1), the plants were transplanted to soil and grown in a growth room under LD (16 h of light, 22°C/8 h of dark, 18°C) or SD (8 h of light, 22°C/6 h of dark, 18°C) conditions at 50% relative humidity.
jmj18-1 (SALK_073442), jmj18-2 (GABI-649D05), jmj18-3 (SAIL-861-F02), ft-11 (GABI-290E08), and co-11 (SAIL_24_H04) were obtained from the ABRC. flc-3, fd-5, and FRI-Col were described previously [44], [64], [84]. All mutations were confirmed by PCR and sequencing.
Plant transformation
To construct JMJ18:GUS, 1,361 and 1667-bp genomic sequence upstream of the JMJ18 ATG were PCR-amplified and fused with GUS. The primers used for the two versions of promoter were forward for 1,361 bp 5′-CTGCAGACAAGACAGAAGAAGCAGGAAAC-3′, 1667 bp 5′-CTGCAGTGCGAGTTCTTCTTTCAATGTG-3′ and reverse for both, 5′-TCTAGAATGAATTGAAAAATCAATACTACTCAA-3′. The SUC2 promoter was cloned as described previously [76], [80] using the primers SUC2 promoter forward 5′-CTGCAGAAAATCTGGTTTCATATTAATTTCA-3′, and reverse 5′-TCTAGAATTTGACAAACCAAGAAAGTAAGA-3′.
We used the following primers to amplify the full-length JMJ18 CDS: JMJ18 forward 5′-TCTAGAATGGAAAATCCTCCATTAGAATC-3′, and reverse, 5′-GGATCCCCATCAAATCTACTCCGAAAAGTT-3′. GFP, RFP, or 6×c-myc was fused to the C-terminus of the JMJ18 CDS to produce tagged JMJ18. To construct 35S:NC, 35S:NCZ, 35S:T, 35S:JMJ18 and 35S:ZY, the following primers were used to amplify the truncated or full-length JMJ18 CDS: JMJ18 forward 5′-TCTAGAATGGAAAATCCTCCATTAGAATC-3′; JMJ18 NC reverse 5′-GAGCTCTTAATATAGTTCCACAGCGTTCTGTC-3′; JMJ18 NCZ reverse GAGCTCTTAACCTTCCTTTAACTTCTTCTCCTC-3′; JMJ18 T reverse, 5′-GAGCTCTTAAACTATAGAGGGTGAGAGGAAACC-3′; JMJ ZY forward, 5′-TCTAGAATGCAGAACGCTGTGGAACTATATAG-3′ and JMJ18 CDS reverse 5′-GAGCTCTTACATCAAATCTACTCCGAAAAGTT-3′.
For plant transformation, the constructs were transformed into Agrobacterium tumefaciens strain GV3101. Plants were transformed using the floral dip method [85]. Transformants were selected on MS medium containing kanamycin or hygromycin. Single insertion lines were selected based on the segregation of antibiotic resistance.
JMJ18 purification and histone demethylase activity assay
Full-length and truncated (JMJ18Δ420–432 bp and JMJ18Δ420–474 bp) versions of JMJ18 were generated by PCR amplification from Arabidopsis cDNA and cloned into the pFastBac expression vector (Invitrogen). Baculoviruses expressing the various proteins were generated by transfecting the appropriate vector into High5 insect cells. The proteins were purified using Ni-NTA affinity resin (Qiagen). Select fractions were concentrated and purified by size-exclusion chromatography.
The histone demethylase assay and MALDI-TOF mass spectrometry were performed as described previously [21]. About 2 µg of purified full-length or truncated recombinant JMJ18 were incubated with 10 µM peptides at 37°C for 3 h in histone demethylase reaction buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 50 µM [NH4]2Fe[SO4]2, 2 mM ascorbic acid, and 1 mM α-KG). The reaction was stopped by freezing, and the mixture was diluted for mass spectrometry. The peptides used were: H3K4me3 ARTK(me3)QTARKS, H3K4me2 ARTK(me2)QTARKS, H3K36me3 ATGGVK(me3)KPHRYR, H3K36me2 ATGGVK(me2)KPHRYR, H3K9me3 ARTKQTARK(me3)STGGKAPRAQ, H3K9me2 ARTKQTARK(me2)STGGKAY, H3K27me3 ATKAARK(me3)SAPATG, and H3K27me2 ATKAARK(me2)SAPATG.
Histological and cytological analyses of JMJ18 expression
GUS staining was performed as described previously [86], [87]. Twenty-four independent JMJ18:GUS T2 lines were used for histochemical analysis, with at least ten individual plants observed for each line. Pictures were taken using a stereomicroscope (Leica MZ16) or light microscope (Zeiss Imager M1).
For the observation of GFP fluorescence, roots were cut from seedlings grown on MS medium under LD conditions for seven days. For JMJ18:JMJ18-GFP, 20 independent lines were observed, with at least ten individual plants analyzed per line. Images were collected using a Zeiss LSM 510 Meta confocal laser scanning microscope as described previously [80].
Observation of the colocalization of JMJ18-GFP and -RFP
JMJ18:JMJ18-RFP was constructed to pCAMBIA2300 and introduced into SUC2:JMJ18-GFP (in pCAMBIA1300) transgenic plants. The double transformants were selected on MS medium containing kanamycin and hygromycin. Transgenic lines expressing JMJ18-GFP and -RFP were selected and photographed using a Zeiss LSM 510 Meta confocal laser scanning microscope.
Gene expression analysis
Total RNA was isolated from the plant materials indicated in the text using RNAiso plus (Takara) and treated with RNA-free DNase (Promega) to remove all remaining DNA. Three micrograms of total RNA were used to synthesize first-strand cDNA with a reverse transcription kit (Fermentas). The cDNAs were diluted to 60 µl with sterilized water. One microliter of diluted cDNA was used for real-time PCR amplification with SYBR Premix Ex Taq (Takara). Three biological replicates were performed to calculate the mRNA abundance. Standard deviations were calculated from the replicates. The primers used were listed in Table S3.
ChIP assay
Plant tissue (1.5 g) was collected after growth under LD conditions for eleven days. ChIP was performed as described previously with three biological replicates [88], [89]. The results are shown as absolute enrichment compared to the input or to total H3. The antibodies used were: anti-H3K4me3 (Upstate 04-745), -H3K4me2 (Upstate 07-030), and -c-myc (Sigma-Aldrich M4439). The primers used to measure the amount of DNA from the ChIP products were listed in Table S4.
Preparation of anti-JMJ18 and -FT antibodies
Purified recombinant JMJ18 from insect cells and FT from E. coli were used as antigens to immunize rabbits. Each rabbit was immunized once every fourteen days with 1 mg of protein. After four successive immunizations, the anti-serum was examined by ELISA and Western blotting using total protein from the mutant and overexpressor lines.
Western blot analysis
For immunoblotting, Arabidopsis seedlings were ground to a powder in liquid nitrogen then homogenized in extraction buffer (10 mM Tris-HCl [pH 7.5], 150 mM NaCl, 10 mM MgCl2, 1% Nonidet P-40, complete protease inhibitor [Roche], and 1 mM phenylmethylsulfonyl fluoride). The extracts were then centrifuged, the pellet removed, and the supernatant boiled in 6× SDS sample buffer. The proteins in the samples were separated by 8% SDS-PAGE, transferred to polyvinylidene difluoride membranes (Millipore), and detected using different antibodies. The antibody used for Tubulin detection was anti-α-Tubulin (Sigma-Aldrich T5168).
Double-strand RNA and amiRNA interferences
To knock-down the expression of JMJ18, a double strand RNA and artificial microRNA interference approaches were used. For a double strand RNA RNAi, a unique 616 bp JMJ18 sequence was amplified by PCR. The gene-specific primers were: forward 5′-GGGGTACCTCTAGAGGTGAAGGCTCTTTGGGAACTC-3′, reverse 5′-CGGGATCCGAGCTCCTCGACCGATACACCAAGGTTC-3′. The self-complementary hairpin RNA was constructed to pTCK303 as described by Wesley et al. (2001) [90].
Three primer pairs were designed through the web (http://wmd2.weigelworld.org/cgi-bin/mirnatools.pl?page=1) for artificial microRNA interference assay. The primers are: amiR-s, 5′-GATGAGTCTTTAAAATGCAGGGCTCTCTCTTTTGTATTCC-3′, amiR-a, 5′-GAGCCCTGCATTTTAAAGACTCATCAAAGAGAATCAATGA-3′, amiR*s, 5′-GAGCACTGCATTTTATAGACTCTTCACAGGTCGTGATATG-3′, amiR*a, 5′-GAAGAGTCTATAAAATGCAGTGCTCTACATATATATTCCT-3′. The constructs for amiRNAi were established by following the protocols from Schwab et al.(Schwab et al., 2006). The resulting three constructs were delivered to wild-type through agrobacterium–mediated transformation to generate JMJ18 amiRNA interference lines.
Accession numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative database under the following accession numbers: ACTIN (At5g09810), CO (At5g15840), FLC (At5g10140), FT (At1g65480), FRI (At4g00650), JMJ18 (At1g30810), MAF1 (At1g77080), MAF2 (At5g65050), MAF3 (At5g65060), MAF4 (At5g65070), MAF5 (At5g65080), SUC2 (At1g22710) and TSF (At4g20370).
Supporting Information
Zdroje
1. LugerKMaderAWRichmondRKSargentDFRichmondTJ 1997 Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389 251 260
2. KouzaridesT 2007 Chromatin modifications and their function. Cell 128 693 705
3. LiBCareyMWorkmanJL 2007 The role of chromatin during transcription. Cell 128 707 719
4. BannisterAJZegermanPPartridgeJFMiskaEAThomasJO 2001 Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410 120 124
5. KimTBuratowskiS 2009 Dimethylation of H3K4 by Set1 recruits the Set3 histone deacetylase complex to 5′ transcribed regions. Cell 137 259 272
6. ZhangXGermannSBlusBJKhorasanizadehSGaudinV 2007 The Arabidopsis LHP1 protein colocalizes with histone H3 Lys27 trimethylation. Nat Struct Mol Biol 14 869 871
7. TurckFRoudierFFarronaSMartin-MagnietteMLGuillaumeE 2007 Arabidopsis TFL2/LHP1 specifically associates with genes marked by trimethylation of histone H3 lysine 27. PLoS Genet 3 e86 doi:10.1371/journal.pgen.0030086
8. WangGGSongJWangZDormannHLCasadioF 2009 Haematopoietic malignancies caused by dysregulation of a chromatin-binding PHD finger. Nature 459 847 851
9. LiuCLuFCuiXCaoX 2010 Histone methylation in higher plants. Annu Rev Plant Biol 61 395 420
10. ZhaoZYuYMeyerDWuCShenWH 2005 Prevention of early flowering by expression of FLOWERING LOCUS C requires methylation of histone H3 K36. Nat Cell Biol 7 1256 1260
11. CaoYDaiYCuiSMaL 2008 Histone H2B monoubiquitination in the chromatin of FLOWERING LOCUS C regulates flowering time in Arabidopsis. Plant Cell 20 2586 2602
12. BastowRMylneJSListerCLippmanZMartienssenRA 2004 Vernalization requires epigenetic silencing of FLC by histone methylation. Nature 427 164 167
13. JiangDWangYHeY 2008 Repression of FLOWERING LOCUS C and FLOWERING LOCUS T by the Arabidopsis Polycomb repressive complex 2 components. PLoS ONE 3 e3404 doi:10.1371/journal.pone.0003404
14. PasiniDBrackenAPJensenMRLazzerini DenchiEHelinK 2004 Suz12 is essential for mouse development and for EZH2 histone methyltransferase activity. EMBO J 23 4061 4071
15. RingroseLParoR 2004 Epigenetic regulation of cellular memory by the Polycomb and Trithorax group proteins. Annu Rev Genet 38 413 443
16. KloseRJKallinEMZhangY 2006 JmjC-domain-containing proteins and histone demethylation. Nat Rev Genet 7 715 727
17. KloseRJYamaneKBaeYZhangDErdjument-BromageH 2006 The transcriptional repressor JHDM3A demethylates trimethyl histone H3 lysine 9 and lysine 36. Nature 442 312 316
18. ShiYLanFMatsonCMulliganPWhetstineJR 2004 Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 119 941 953
19. AggerKCloosPAChristensenJPasiniDRoseS 2007 UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature 449 731 734
20. CloosPAChristensenJAggerKMaiolicaARappsilberJ 2006 The putative oncogene GASC1 demethylates tri - and dimethylated lysine 9 on histone H3. Nature 442 307 311
21. WhetstineJRNottkeALanFHuarteMSmolikovS 2006 Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell 125 467 481
22. YamaneKToumazouCTsukadaYErdjument-BromageHTempstP 2006 JHDM2A, a JmjC-containing H3K9 demethylase, facilitates transcription activation by androgen receptor. Cell 125 483 495
23. OkadaYScottGRayMKMishinaYZhangY 2007 Histone demethylase JHDM2A is critical for Tnp1 and Prm1 transcription and spermatogenesis. Nature 450 119 123
24. HeYMichaelsSDAmasinoRM 2003 Regulation of flowering time by histone acetylation in Arabidopsis. Science 302 1751 1754
25. JiangDYangWHeYAmasinoRM 2007 Arabidopsis relatives of the human lysine-specific Demethylase1 repress the expression of FWA and FLOWERING LOCUS C and thus promote the floral transition. Plant Cell 19 2975 2987
26. LiuFQuesadaVCrevillenPBaurleISwiezewskiS 2007 The Arabidopsis RNA-binding protein FCA requires a lysine-specific demethylase 1 homolog to downregulate FLC. Mol Cell 28 398 407
27. LuFLiGCuiXLiuCWangXJ 2008 Comparative analysis of JmjC domain-containing proteins reveals the potential histone demethylases in Arabidopsis and rice. J Integr Plant Biol 50 886 896
28. JeongJHSongHRKoJHJeongYMKwonYE 2009 Repression of FLOWERING LOCUS T chromatin by functionally redundant histone H3 lysine 4 demethylases in Arabidopsis. PLoS ONE 4 e8033 doi:10.1371/journal.pone.0008033
29. LuFCuiXZhangSLiuCCaoX 2010 JMJ14 is an H3K4 demethylase regulating flowering time in Arabidopsis. Cell Res 20 387 390
30. MiuraANakamuraMInagakiSKobayashiASazeH 2009 An Arabidopsis jmjC domain protein protects transcribed genes from DNA methylation at CHG sites. EMBO J 28 1078 1086
31. NohBLeeSHKimHJYiGShinEA 2004 Divergent roles of a pair of homologous jumonji/zinc-finger-class transcription factor proteins in the regulation of Arabidopsis flowering time. Plant Cell 16 2601 2613
32. SazeHShiraishiAMiuraAKakutaniT 2008 Control of genic DNA methylation by a jmjC domain-containing protein in Arabidopsis thaliana. Science 319 462 465
33. SearleIRPontesOMelnykCWSmithLMBaulcombeDC 2010 JMJ14, a JmjC domain protein, is required for RNA silencing and cell-to-cell movement of an RNA silencing signal in Arabidopsis. Genes Dev 24 986 991
34. YuXLiLGuoMChoryJYinY 2008 Modulation of brassinosteroid-regulated gene expression by Jumonji domain-containing proteins ELF6 and REF6 in Arabidopsis. Proc Natl Acad Sci U S A 105 7618 7623
35. LiWLiuHChengZJSuYHHanHN 2011 DNA Methylation and Histone Modifications Regulate De Novo Shoot Regeneration in Arabidopsis by Modulating WUSCHEL Expression and Auxin Signaling. PLoS Genet 7 e1002243 doi:10.1371/journal.pgen.1002243
36. LuSXKnowlesSMWebbCJCelayaRBChaC 2010 The JmjC domain-containing protein JMJ30 regulates period length in the Arabidopsis circadian clock. Plant Physiol
37. BaurleIDeanC 2006 The timing of developmental transitions in plants. Cell 125 655 664
38. WangJWCzechBWeigelD 2009 miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell 138 738 749
39. WuGParkMYConwaySRWangJWWeigelD 2009 The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 138 750 759
40. HayamaRCouplandG 2003 Shedding light on the circadian clock and the photoperiodic control of flowering. Curr Opin Plant Biol 6 13 19
41. AusinIAlonso-BlancoCJarilloJARuiz-GarciaLMartinez-ZapaterJM 2004 Regulation of flowering time by FVE, a retinoblastoma-associated protein. Nat Genet 36 162 166
42. BaurleIDeanC 2008 Differential interactions of the autonomous pathway RRM proteins and chromatin regulators in the silencing of Arabidopsis targets. PLoS ONE 3 e2733 doi:10.1371/journal.pone.0002733
43. HeYDoyleMRAmasinoRM 2004 PAF1-complex-mediated histone methylation of FLOWERING LOCUS C chromatin is required for the vernalization-responsive, winter-annual habit in Arabidopsis. Genes Dev 18 2774 2784
44. MichaelsSDAmasinoRM 1999 FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11 949 956
45. SchmitzRJTamadaYDoyleMRZhangXAmasinoRM 2009 Histone H2B deubiquitination is required for transcriptional activation of FLOWERING LOCUS C and for proper control of flowering in Arabidopsis. Plant Physiol 149 1196 1204
46. JohansonUWestJListerCMichaelsSAmasinoR 2000 Molecular analysis of FRIGIDA, a major determinant of natural variation in Arabidopsis flowering time. Science 290 344 347
47. LempeJBalasubramanianSSureshkumarSSinghASchmidM 2005 Diversity of flowering responses in wild Arabidopsis thaliana strains. PLoS Genet 1 e6 doi:10.1371/journal.pgen.0010006
48. LimMHKimJKimYSChungKSSeoYH 2004 A new Arabidopsis gene, FLK, encodes an RNA binding protein with K homology motifs and regulates flowering time via FLOWERING LOCUS C. Plant Cell 16 731 740
49. SimpsonGGDijkwelPPQuesadaVHendersonIDeanC 2003 FY is an RNA 3′ end-processing factor that interacts with FCA to control the Arabidopsis floral transition. Cell 113 777 787
50. HornyikCTerziLCSimpsonGG 2010 The spen family protein FPA controls alternative cleavage and polyadenylation of RNA. Dev Cell 18 203 213
51. LiuFMarquardtSListerCSwiezewskiSDeanC 2010 Targeted 3′ processing of antisense transcripts triggers Arabidopsis FLC chromatin silencing. Science 327 94 97
52. SwiezewskiSLiuFMagusinADeanC 2009 Cold-induced silencing by long antisense transcripts of an Arabidopsis Polycomb target. Nature 462 799 802
53. SungSAmasinoRM 2004 Vernalization in Arabidopsis thaliana is mediated by the PHD finger protein VIN3. Nature 427 159 164
54. SheldonCCConnABDennisESPeacockWJ 2002 Different regulatory regions are required for the vernalization-induced repression of FLOWERING LOCUS C and for the epigenetic maintenance of repression. Plant Cell 14 2527 2537
55. SearleIHeYTurckFVincentCFornaraF 2006 The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis. Genes Dev 20 898 912
56. JiangDGuXHeY 2009 Establishment of the winter-annual growth habit via FRIGIDA-mediated histone methylation at FLOWERING LOCUS C in Arabidopsis. Plant Cell 21 1733 1746
57. PienSFleuryDMylneJSCrevillenPInzeD 2008 ARABIDOPSIS TRITHORAX1 dynamically regulates FLOWERING LOCUS C activation via histone 3 lysine 4 trimethylation. Plant Cell 20 580 588
58. TamadaYYunJYWooSCAmasinoRM 2009 ARABIDOPSIS TRITHORAX-RELATED7 is required for methylation of lysine 4 of histone H3 and for transcriptional activation of FLOWERING LOCUS C. Plant Cell 21 3257 3269
59. SungSHeYEshooTWTamadaYJohnsonL 2006 Epigenetic maintenance of the vernalized state in Arabidopsis thaliana requires LIKE HETEROCHROMATIN PROTEIN 1. Nat Genet 38 706 710
60. KardailskyIShuklaVKAhnJHDagenaisNChristensenSK 1999 Activation tagging of the floral inducer FT. Science 286 1962 1965
61. KobayashiYKayaHGotoKIwabuchiMArakiT 1999 A pair of related genes with antagonistic roles in mediating flowering signals. Science 286 1960 1962
62. TakadaSGotoK 2003 Terminal flower2, an Arabidopsis homolog of heterochromatin protein1, counteracts the activation of flowering locus T by constans in the vascular tissues of leaves to regulate flowering time. Plant Cell 15 2856 2865
63. HeYAmasinoRM 2005 Role of chromatin modification in flowering-time control. Trends Plant Sci 10 30 35
64. AbeMKobayashiYYamamotoSDaimonYYamaguchiA 2005 FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309 1052 1056
65. WiggePAKimMCJaegerKEBuschWSchmidM 2005 Integration of spatial and temporal information during floral induction in Arabidopsis. Science 309 1056 1059
66. CorbesierLVincentCJangSFornaraFFanQ 2007 FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316 1030 1033
67. JaegerKEWiggePA 2007 FT protein acts as a long-range signal in Arabidopsis. Curr Biol 17 1050 1054
68. MathieuJWarthmannNKuttnerFSchmidM 2007 Export of FT protein from phloem companion cells is sufficient for floral induction in Arabidopsis. Curr Biol 17 1055 1060
69. YangWJiangDJiangJHeY 2010 A plant-specific histone H3 lysine 4 demethylase represses the floral transition in Arabidopsis. Plant J 62 663 673
70. LuFCuiXZhangSJenuweinTCaoX 2011 Arabidopsis REF6 is a histone H3 lysine 27 demethylase. Nat Genet 43 715 719
71. ChristensenJAggerKCloosPAPasiniDRoseS 2007 RBP2 belongs to a family of demethylases, specific for tri-and dimethylated lysine 4 on histone 3. Cell 128 1063 1076
72. IwaseSLanFBaylissPde la Torre-UbietaLHuarteM 2007 The X-linked mental retardation gene SMCX/JARID1C defines a family of histone H3 lysine 4 demethylases. Cell 128 1077 1088
73. LiFHuarteMZaratieguiMVaughnMWShiY 2008 Lid2 is required for coordinating H3K4 and H3K9 methylation of heterochromatin and euchromatin. Cell 135 272 283
74. AlonsoJMStepanovaANLeisseTJKimCJChenH 2003 Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653 657
75. BonkeMThitamadeeSMahonenAPHauserMTHelariuttaY 2003 APL regulates vascular tissue identity in Arabidopsis. Nature 426 181 186
76. ImlauATruernitESauerN 1999 Cell-to-cell and long-distance trafficking of the green fluorescent protein in the phloem and symplastic unloading of the protein into sink tissues. Plant Cell 11 309 322
77. ClarkeJHDeanC 1994 Mapping FRI, a locus controlling flowering time and vernalization response in Arabidopsis thaliana. Mol Gen Genet 242 81 89
78. FletcherJCBrandURunningMPSimonRMeyerowitzEM 1999 Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science 283 1911 1914
79. HongEHJeongYMRyuJYAmasinoRMNohB 2009 Temporal and spatial expression patterns of nine Arabidopsis genes encoding Jumonji C-domain proteins. Mol Cells 27 481 490
80. AnHRoussotCSuarez-LopezPCorbesierLVincentC 2004 CONSTANS acts in the phloem to regulate a systemic signal that induces photoperiodic flowering of Arabidopsis. Development 131 3615 3626
81. YantLMathieuJSchmidM 2009 Just say no: floral repressors help Arabidopsis bide the time. Curr Opin Plant Biol 12 580 586
82. TurckFFornaraFCouplandG 2008 Regulation and identity of florigen: FLOWERING LOCUS T moves center stage. Annu Rev Plant Biol 59 573 594
83. KobayashiYWeigelD 2007 Move on up, it's time for change–mobile signals controlling photoperiod-dependent flowering. Genes Dev 21 2371 2384
84. LeeIAmasinoRM 1995 Effect of Vernalization, Photoperiod, and Light Quality on the Flowering Phenotype of Arabidopsis Plants Containing the FRIGIDA Gene. Plant Physiol 108 157 162
85. CloughSJBentAF 1998 Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16 735 743
86. HemerlyASFerreiraPde Almeida EnglerJVan MontaguMEnglerG 1993 cdc2a expression in Arabidopsis is linked with competence for cell division. Plant Cell 5 1711 1723
87. JeffersonRAKavanaghTABevanMW 1987 GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6 3901 3907
88. BowlerCBenvenutoGLaflammePMolinoDProbstAV 2004 Chromatin techniques for plant cells. Plant J 39 776 789
89. SalehAAlvarez-VenegasRAvramovaZ 2008 An efficient chromatin immunoprecipitation (ChIP) protocol for studying histone modifications in Arabidopsis plants. Nat Protoc 3 1018 1025
90. WesleySVHelliwellCASmithNAWangMBRouseDT 2001 Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J 27 581 590
Štítky
Genetika Reprodukční medicína
Článek A Genome-Wide Screen for Genetic Variants That Modify the Recruitment of REST to Its Target GenesČlánek Population Structure of Hispanics in the United States: The Multi-Ethnic Study of AtherosclerosisČlánek Differing Requirements for RAD51 and DMC1 in Meiotic Pairing of Centromeres and Chromosome Arms inČlánek Transcriptional Regulation of Rod Photoreceptor Homeostasis Revealed by NRL Targetome AnalysisČlánek Cell Contact–Dependent Outer Membrane Exchange in Myxobacteria: Genetic Determinants and MechanismČlánek Formation of Rigid, Non-Flight Forewings (Elytra) of a Beetle Requires Two Major Cuticular Proteins
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2012 Číslo 4
-
Všechny články tohoto čísla
- Runs of Homozygosity Implicate Autozygosity as a Schizophrenia Risk Factor
- Modifier Genes and the Plasticity of Genetic Networks in Mice
- The DSIF Subunits Spt4 and Spt5 Have Distinct Roles at Various Phases of Immunoglobulin Class Switch Recombination
- A Genome-Wide Screen for Genetic Variants That Modify the Recruitment of REST to Its Target Genes
- Population Structure of Hispanics in the United States: The Multi-Ethnic Study of Atherosclerosis
- Deep Sequencing of Plant and Animal DNA Contained within Traditional Chinese Medicines Reveals Legality Issues and Health Safety Concerns
- Differing Requirements for RAD51 and DMC1 in Meiotic Pairing of Centromeres and Chromosome Arms in
- Insulin Signaling Mediates Sexual Attractiveness in
- Progressive Telomere Dysfunction Causes Cytokinesis Failure and Leads to the Accumulation of Polyploid Cells
- Long-Range Chromosome Organization in : A Site-Specific System Isolates the Ter Macrodomain
- Regulation of Budding Yeast Mating-Type Switching Donor Preference by the FHA Domain of Fkh1
- Polyglutamine Toxicity Is Controlled by Prion Composition and Gene Dosage in Yeast
- Patterns of Regulatory Variation in Diverse Human Populations
- Sequence-Specific Targeting of Dosage Compensation in Favors an Active Chromatin Context
- Whole-Exome Sequencing and Homozygosity Analysis Implicate Depolarization-Regulated Neuronal Genes in Autism
- Replication Fork Reversal after Replication–Transcription Collision
- Common Variants at 9p21 and 8q22 Are Associated with Increased Susceptibility to Optic Nerve Degeneration in Glaucoma
- Coordinate Regulation of Lipid Metabolism by Novel Nuclear Receptor Partnerships
- Epigenome-Wide Scans Identify Differentially Methylated Regions for Age and Age-Related Phenotypes in a Healthy Ageing Population
- A Coordinated Interdependent Protein Circuitry Stabilizes the Kinetochore Ensemble to Protect CENP-A in the Human Pathogenic Yeast
- Budding Yeast Dma Proteins Control Septin Dynamics and the Spindle Position Checkpoint by Promoting the Recruitment of the Elm1 Kinase to the Bud Neck
- , a Homolog of a Deaf-Blindness Gene, Regulates Circadian Output and Slowpoke Channels
- Transcriptional Regulation of Rod Photoreceptor Homeostasis Revealed by NRL Targetome Analysis
- Cell Contact–Dependent Outer Membrane Exchange in Myxobacteria: Genetic Determinants and Mechanism
- Defective Membrane Remodeling in Neuromuscular Diseases: Insights from Animal Models
- Formation of Rigid, Non-Flight Forewings (Elytra) of a Beetle Requires Two Major Cuticular Proteins
- SPE-44 Implements Sperm Cell Fate
- A Shared Role for RBF1 and dCAP-D3 in the Regulation of Transcription with Consequences for Innate Immunity
- A Companion Cell–Dominant and Developmentally Regulated H3K4 Demethylase Controls Flowering Time in via the Repression of Expression
- The HEN1 Ortholog, HENN-1, Methylates and Stabilizes Select Subclasses of Germline Small RNAs
- Improved Statistics for Genome-Wide Interaction Analysis
- The Probability of a Gene Tree Topology within a Phylogenetic Network with Applications to Hybridization Detection
- Context-Dependent Dual Role of SKI8 Homologs in mRNA Synthesis and Turnover
- Mu Insertions Are Repaired by the Double-Strand Break Repair Pathway of
- Competition between Replicative and Translesion Polymerases during Homologous Recombination Repair in Drosophila
- An Unbiased Assessment of the Role of Imprinted Genes in an Intergenerational Model of Developmental Programming
- Type 2 Diabetes Risk Alleles Demonstrate Extreme Directional Differentiation among Human Populations, Compared to Other Diseases
- Mutations in and Cause “Splashed White” and Other White Spotting Phenotypes in Horses
- Fine-Scale Mapping of Natural Variation in Fly Fecundity Identifies Neuronal Domain of Expression and Function of an Aquaporin
- Dynamics of Brassinosteroid Response Modulated by Negative Regulator LIC in Rice
- Genetic Inhibition of Solute-Linked Carrier 39 Family Transporter 1 Ameliorates Aβ Pathology in a Model of Alzheimer's Disease
- The Functions of Mediator in Support a Role in Shaping Species-Specific Gene Expression
- Patterns of Ancestry, Signatures of Natural Selection, and Genetic Association with Stature in Western African Pygmies
- Dissection of Pol II Trigger Loop Function and Pol II Activity–Dependent Control of Start Site Selection
- PIWI Associated siRNAs and piRNAs Specifically Require the HEN1 Ortholog
- Genome-Wide Patterns of Gene Expression in Nature
- Hypoxia Disruption of Vertebrate CNS Pathfinding through EphrinB2 Is Rescued by Magnesium
- A New Role for Translation Initiation Factor 2 in Maintaining Genome Integrity
- Sex Reversal in C57BL/6J XY Mice Caused by Increased Expression of Ovarian Genes and Insufficient Activation of the Testis Determining Pathway
- The Rac GTP Exchange Factor TIAM-1 Acts with CDC-42 and the Guidance Receptor UNC-40/DCC in Neuronal Protrusion and Axon Guidance
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- A Coordinated Interdependent Protein Circuitry Stabilizes the Kinetochore Ensemble to Protect CENP-A in the Human Pathogenic Yeast
- Coordinate Regulation of Lipid Metabolism by Novel Nuclear Receptor Partnerships
- Defective Membrane Remodeling in Neuromuscular Diseases: Insights from Animal Models
- Formation of Rigid, Non-Flight Forewings (Elytra) of a Beetle Requires Two Major Cuticular Proteins
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



