-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaVariants in and Underlie Natural Variation in Translation Termination Efficiency in
Translation termination is a highly controlled process in the cell. In Saccharomyces cerevisiae, various regulatory factors employ genetic and epigenetic mechanisms to control this process. We used a quantitative dual luciferase reporter assay to demonstrate a difference in translation termination efficiency between two different yeast strains, BY4724 and RM11-1a. We then used a recently developed linkage mapping technique, extreme QTL mapping (X-QTL), to show that this difference is largely explained by a coding polymorphism in TRM10 (which encodes a tRNA–methylating enzyme) and a regulatory polymorphism in SUP45 (which encodes one of the yeast translation termination factors). BY and RM carry variants of TRM10 and SUP45 with opposite effects on translation termination efficiency. These variants are common among 63 diverse S. cerevisiae strains and are in strong linkage disequilibrium with each other. This observation suggests that selection may have favored allelic combinations of the two genes that maintain an intermediate level of translation termination efficiency. Our results also provide genetic evidence for a new role of Trm10p in translation termination efficiency.
Published in the journal: . PLoS Genet 7(7): e32767. doi:10.1371/journal.pgen.1002211
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1002211Summary
Translation termination is a highly controlled process in the cell. In Saccharomyces cerevisiae, various regulatory factors employ genetic and epigenetic mechanisms to control this process. We used a quantitative dual luciferase reporter assay to demonstrate a difference in translation termination efficiency between two different yeast strains, BY4724 and RM11-1a. We then used a recently developed linkage mapping technique, extreme QTL mapping (X-QTL), to show that this difference is largely explained by a coding polymorphism in TRM10 (which encodes a tRNA–methylating enzyme) and a regulatory polymorphism in SUP45 (which encodes one of the yeast translation termination factors). BY and RM carry variants of TRM10 and SUP45 with opposite effects on translation termination efficiency. These variants are common among 63 diverse S. cerevisiae strains and are in strong linkage disequilibrium with each other. This observation suggests that selection may have favored allelic combinations of the two genes that maintain an intermediate level of translation termination efficiency. Our results also provide genetic evidence for a new role of Trm10p in translation termination efficiency.
Introduction
Translational fidelity is essential for functional integrity of the cell. Efficient termination is an important aspect of translational fidelity, and a multitude of mechanisms participate in this highly regulated process. Translation termination in eukaryotes is mediated by two termination factors, eRF1 and eRF3 [1]. eRF1, encoded by SUP45 in Saccharomyces cerevisiae, recognizes all three stop codons (UAG, UAA, and UGA) and facilitates release of the nascent polypeptide chain from the translational machinery [2], [3]. GTPase activity of eRF3, encoded by SUP35 in S. cerevisiae, is required to couple the recognition of translation termination signals by eRF1 to efficient polypeptide chain release [4].
In a successful translation termination event, termination factors efficiently recognize stop codons. However, in certain instances, transfer RNAs (tRNAs) outcompete termination factors in stop codon recognition. The resulting misincorporation of an amino acid into the nascent peptide is known as translational readthrough. Therefore, during translation, any event that directly or indirectly makes a tRNA more likely to bind to a stop codon increases readthrough.
It is widely accepted that the efficiency of translation termination is modulated by both cis - and trans-acting factors [5]. In S. cerevisiae, the sequence surrounding the stop codon has been shown to play a major role in translation termination efficiency [6], [7]. Several trans factors have also been shown to affect translation termination, either directly through contacts with release factors or indirectly, as demonstrated by genetic experiments (reviewed in [8]). Moreover, recent studies of translation termination in S. cerevisiae have revealed genetic and epigenetic regulatory mechanisms that may enable controlled readthrough of stop codons, which can have significant effects on cellular processes such as mRNA degradation and, in some cases, can confer a beneficial phenotype to the cell [9]. The most studied example of such a mechanism is [PSI+], the prion conformation of the Sup35 protein, which can have pleiotropic effects on growth that vary among different yeast strains [10].
Although our knowledge of translation termination has grown in the past few decades, one can envision that many factors that modulate this complex process remain to be discovered. Natural genetic variation provides a framework for finding such factors. Linkage analysis has been successfully used to find the genetic basis of complex phenotypes in yeast at the cellular level, including growth in different chemical environments [11], sporulation efficiency [12] and growth at high temperatures [13], as well as phenotypes at the molecular level, such as genome-wide mRNA expression levels [14], [15].
Here, we employed linkage analysis to study translation termination efficiency. We used extreme QTL mapping (X-QTL) [16] to find the genetic basis for the observed difference in readthrough between two S. cerevisiae strains, RM11-1a (a wine strain hereafter referred to as RM) and BY4724 (a laboratory strain hereafter referred to as BY). We show that a coding polymorphism in TRM10, which encodes a tRNA-methylating enzyme with an unknown physiological role in the cell [17], affects readthrough in yeast. Moreover, we show that cis-regulatory variation that alters the expression level of SUP45 is another factor involved in translation termination efficiency variation between BY and RM. These two yeast strains carry alleles of TRM10 and SUP45 with opposing effects on readthrough. The BY and RM alleles of both TRM10 and SUP45 are common in a diverse collection of S. cerevisiae strains [18] and are in significant linkage disequilibrium (LD), suggesting that readthrough may be subject to stabilizing selection.
Results
Dual luciferase assay reveals readthrough difference between BY and RM
In order to measure readthrough in the two parent strains, we took advantage of a dual luciferase reporter system [19]. This system uses tandem Renilla and firefly luciferase genes that are separated by a single in-frame stop codon. The activity of the firefly luciferase, encoded by the distal open reading frame, provides a quantitative measure of the readthrough of the stop codon that separates the two open reading frames. The activity of the Renilla luciferase, encoded by the proximal open reading frame, serves as an internal control for mRNA abundance. Thus, the relative abundance of these light-emitting proteins measures the efficiency of translation termination. Here, we used two separate reporters; one with UGA (stop codon) and one with CGA (sense codon) separating the Renilla and firefly open reading frames. For each strain, we calculated the readthrough as the ratio of firefly to Renilla luciferase activity in the presence of the stop codon, normalized by the observed ratio for the sense codon constructs (Table 1). We found that the readthrough in RM is higher than in BY.
Tab. 1. Readthrough measured with the dual luciferase reporter system. 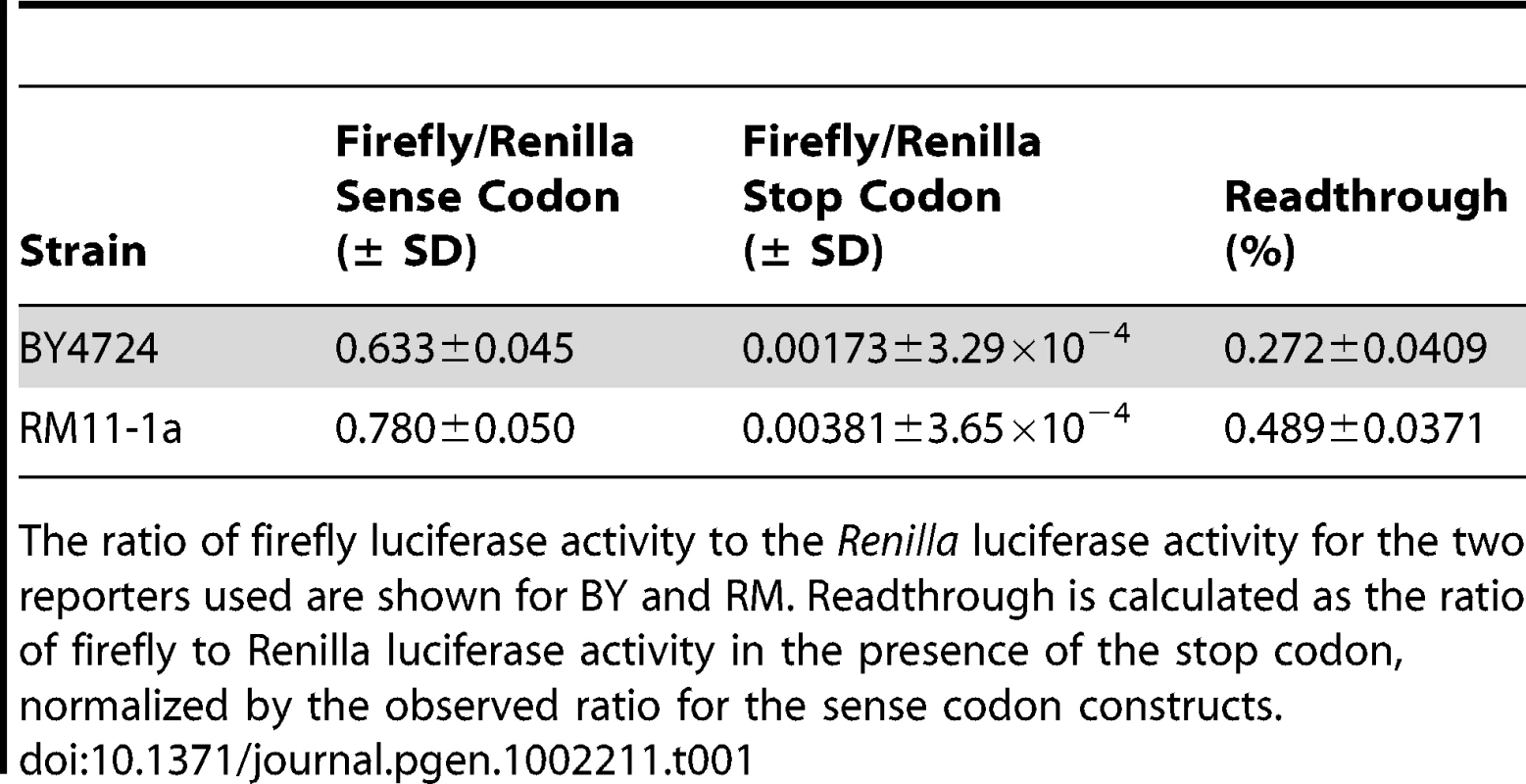
The ratio of firefly luciferase activity to the Renilla luciferase activity for the two reporters used are shown for BY and RM. Readthrough is calculated as the ratio of firefly to Renilla luciferase activity in the presence of the stop codon, normalized by the observed ratio for the sense codon constructs. Missense change in TRM10 affects readthrough
We used X-QTL to examine the genetic basis of the readthrough difference between BY and RM in a large pool of segregants from a cross between these strains. In order to be able to select those segregants in the tails of the distribution of readthrough, we constructed a GFP reporter with a UGA stop codon inserted at the beginning of the GFP coding sequence and integrated it into the genomes of BY and RM (Materials and Methods). We also integrated an intact GFP reporter (without the stop codon) in the same position. Then, we transferred these reporters into BY and RM strains with suitable markers for X-QTL (Materials and Methods). For each strain, we calculated readthrough as the ratio of the GFP signal in the presence of the stop codon to the GFP signal in the absence of the stop codon. We showed that readthrough measured using the GFP reporter is in agreement with readthrough measured using the dual luciferase assay for both BY and RM (Table S1).
To map the genetic basis of the readthrough difference, we harvested a MATa pool from a sporulation culture of BY×RM diploid hybrids containing the GFP reporter and sorted out segregants from the two extremes of the readthrough distribution by fluorescence-activated cell sorting (FACS). We selected the top 1% of the segregating population (high GFP signal) and the bottom 1% of the segregating population (low GFP signal). Samples from both selected pools as well as a sample of the whole (unselected) population were subsequently genotyped as previously described [16].
Comparisons of the high and low segregant pools to the whole population showed allele frequency differences on chromosome XV, at the same position but in opposite directions in the two selected tails (Figure 1A). The directions of the skew suggested that carrying a BY allele in this region results in increased readthrough, whereas carrying the RM allele results in a decrease in readthrough. Based on functional annotations available in the Saccharomyces Genome Database and sequence comparison between BY and RM for the genes in this region (Figure S1), we selected TRM10 as a candidate for further investigation. TRM10 encodes a tRNA modifying enzyme, which methylates the N-1 position of guanosine-9 in ten tRNAs in yeast [17]. Comparison of the coding sequence of TRM10 between BY and RM showed eight single nucleotide polymorphisms (SNPs) between the two yeast strains, among which five are nonsynonymous substitutions. In order to test the causality of TRM10 polymorphisms for the observed peak on chromosome XV, we made allele replacement strains in both BY and RM (replacing TRM10 with the version from the other strain) and measured readthrough in the newly made strains using the dual luciferase assay. Results of this experiment showed that replacing TRM10 in each strain with the alternative allele of this gene changed readthrough in the direction predicted from the X-QTL results. RM-TRM10BY showed higher readthrough than RM. BY-TRM10RM showed lower readthrough than BY (Figure 1B). These results showed that the effect of the TRM10 coding polymorphism on readthrough is in the opposite direction from the difference observed in the parent strains; swapping TRM10 increased the difference in readthrough between BY and RM. This observation suggested the presence of other polymorphic factor(s) that influence translation termination efficiency.
Fig. 1. TRM10 role in readthrough. 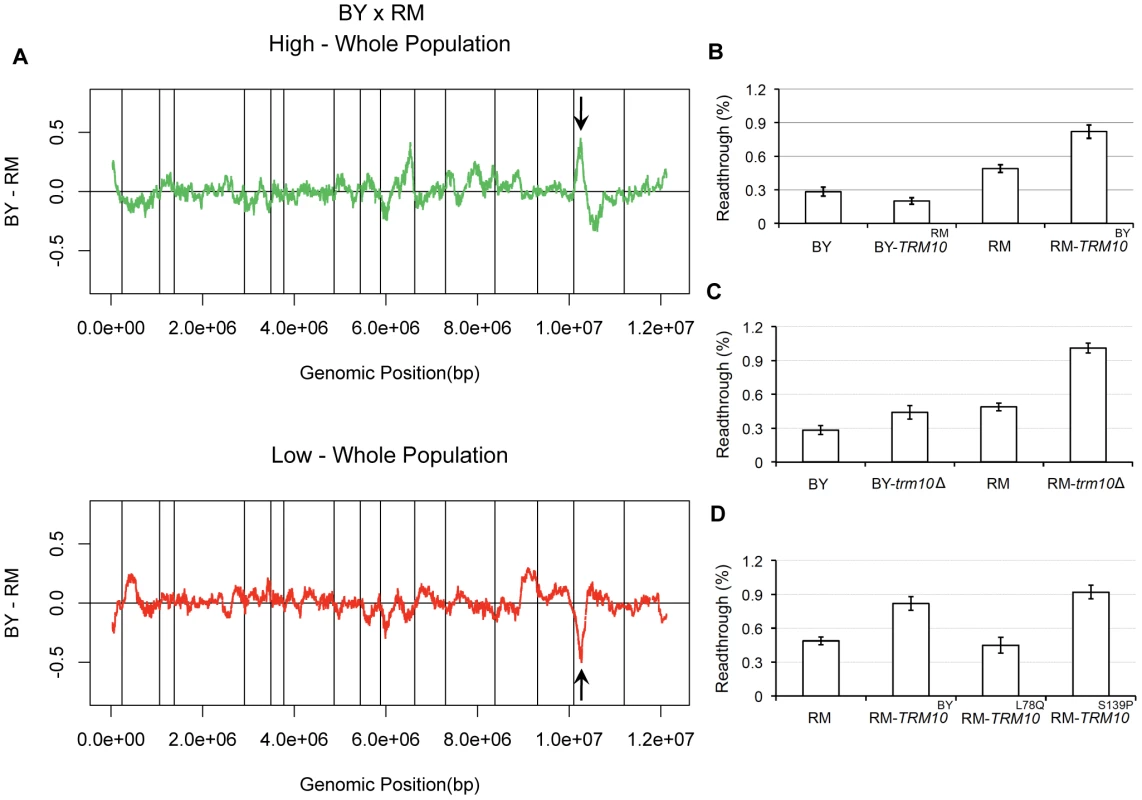
A) X-QTL results for mapping readthrough in the BY×RM cross. The comparisons of the selected top 1% of the segregating population and the bottom 1% of the segregating population to the whole population are shown, with enrichment of the BY allele indicated by deviations above zero and enrichment of the RM allele indicated by deviations below zero. Sliding window averages (40 kb) are plotted. The black arrows show allele frequency skew on chromosome XV. B) Readthrough measured by dual luciferase assay for the parent strains and the two TRM10 swapped strains are shown. C) Readthrough for the parent strains and TRM10 knocked out strains are shown. D) Readthrough is shown for the RM parent strain, RM parent strain with the BY allele of TRM10 (RM-TRM10BY) and two RM strains made by site-directed mutagenesis RM-TRM10L78Q and RM-TRM10S139P. To further analyze the relationship between the Trm10p tRNA methylation activity and translation termination efficiency, we made complete TRM10 deletions in both genetic backgrounds. Readthrough measurements using the dual luciferase assay showed that deleting TRM10 in both BY and RM increases readthrough, which provides further evidence for the role of Trm10p tRNA modification in translation termination efficiency (Figure 1C). Moreover, these results suggest that BY carries a partial loss of function allele of TRM10, because the BY allele of TRM10 is associated with higher readthrough. To identify the causal polymorphism, we also made strains with TRM10L78Q and TRM10S139P single amino acid changes in the RM background using site-directed mutagenesis. These two polymorphisms were chosen based on fungal protein sequence alignment from the Saccharomyces Genome Database, which showed that these residues are highly conserved in different yeast species. Readthrough measurements in RM-TRM10L78Q and RM-TRM10S139P identified the serine to proline substitution at position 139 as the causal polymorphism (Figure 1D).
To determine whether TRM10 is the sole factor explaining the observed allele frequency skew on chromosome XV, we carried out X-QTL with segregants from a cross between BY and RM - TRM10BY (i.e. TRM10 was no longer polymorphic, with both parent strains carrying the BY allele). The pool selected from the high tail of the readthrough distribution showed higher average GFP signal relative to the high tail in the original BY×RM cross (data not shown), and no allele frequency skew was observed in the TRM10 region on chromosome XV in either selected pool (Figure 2A).
Fig. 2. SUP45 role in readthrough. 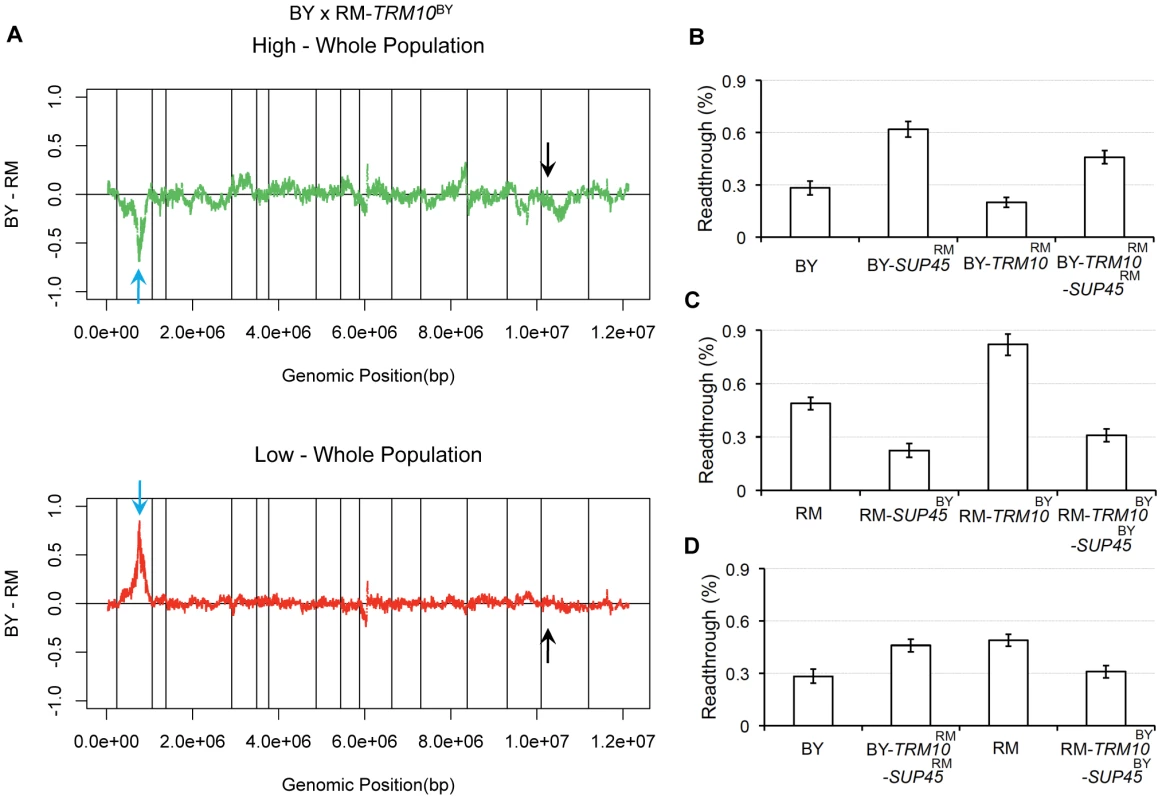
A) X-QTL results for mapping readthrough in the TRM10-fixed population. The comparisons of the selected segregating population with high GFP signal and the selected segregating population with low GFP signal to the whole population are shown. Sliding window averages (40 kb) are plotted. Black arrows show TRM10 locus on chromosome XV and blue arrows show the allele frequency skew on chromosome II. B and C) Effect of swapping SUP45 coding and upstream sequence on readthrough is shown in B) BY and C) RM backgrounds. D) Readthrough for the parent strains, BY and RM, are compared to strains swapped for TRM10 (coding sequence) and SUP45 (coding and upstream sequence), BY-TRM10RM-SUP45RM and RM-TRM10BY-SUP45BY. SUP45 expression level polymorphism contributes to readthrough difference between BY and RM
X-QTL results from the cross with both parent strains carrying the BY allele of TRM10 showed a new region of allele frequency skew on chromosome II (Figure 2A). Our ability to detect this locus was improved by the overall increase in the GFP signal resulting from the increased readthrough conferred by the BY allele of TRM10. The direction of the skew on chromosome II was in the direction expected from the difference between the parent strains: the RM allele at this locus was enriched in the high-readthrough pool and depleted from the low-readthrough pool. This locus (Figure S2) contains SUP45, which encodes the yeast translation termination factor responsible for stop codon recognition. Sequence comparison between BY and RM showed six synonymous SNPs in the coding sequence and four nucleotide substitutions and two indels in the 400-base pair upstream noncoding region. We previously showed that expression level of SUP45 is lower in RM (relative expression level = 0.00333±0.0406) than in BY (relative expression level = 0.315±0.0415) [20]. This study also showed, using an independent panel of 109 segregants, that the expression level difference mapped to the location of the SUP45 gene. These findings strongly suggested that the observed allele frequency skew on chromosome II is due to cis-regulatory polymorphism that alters the expression level of SUP45 between BY and RM.
To test this hypothesis, we made SUP45 allele replacement strains in BY and RM, in both TRM10-wild type and TRM10-swapped backgrounds. In the first set of replacements, we swapped only the SUP45 coding sequence. In the second set, we replaced the SUP45 coding sequence as well as the 400-base pair upstream region. We used the dual luciferase assay to measure readthrough in the newly made strains. In both TRM10-wild type and TRM10-swapped strains, replacing the SUP45 coding sequence along with the upstream region had a significant effect on readthrough (Figure 2B and 2C), whereas replacing just the coding sequence of SUP45 did not have a significant effect on readthrough (Figure S3). These results support the role of SUP45 expression level in translation termination efficiency.
When we swapped both TRM10 (coding sequence) and SUP45 (coding and upstream sequence) in each parent strain with the alleles of these genes from the other (donor) strain, translation termination efficiency changed to the level of the donor strain (Figure 2D). These data demonstrate that the polymorphisms in these two genes explain the difference in translation termination efficiency between BY and RM. We also performed X-QTL in BY×RM-SUP45BY (coding and upstream regions), as well as in BY×RM-TRM10BY (coding region)-SUP45BY (coding and upstream regions). X-QTL with the segregant pool fixed for the BY allele of SUP45 showed the expected allele frequency skew in the region of chromosome XV containing TRM10 (Figure 3A). X-QTL with the segregant pool fixed for the BY alleles of both TRM10 and SUP45 showed no significant allele frequency skews anywhere in the genome (Figure 3B), providing further evidence that these two genes explain most of the difference in translation termination efficiency between BY and RM.
Fig. 3. Polymorphisms in TRM10 and SUP45 explain readthrough difference between BY and RM. 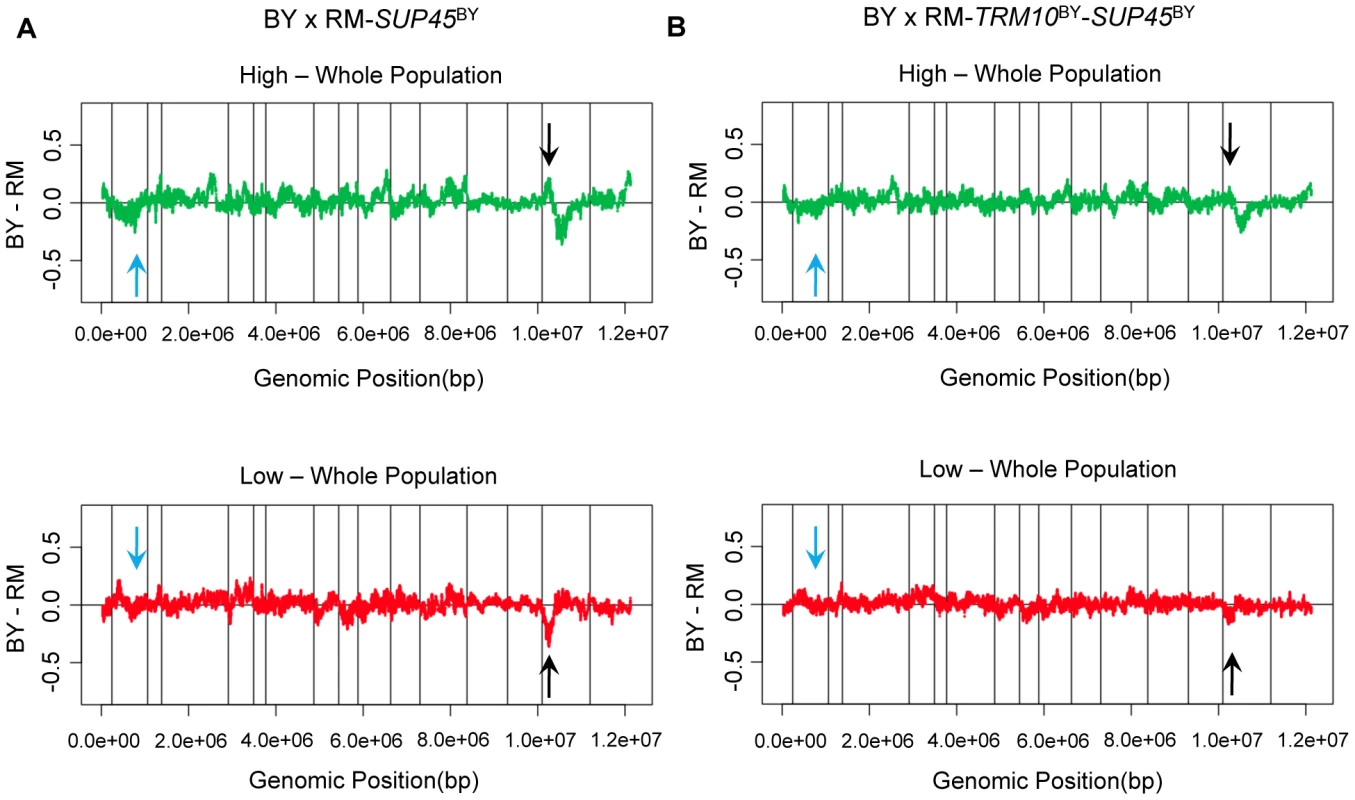
X-QTL results for mapping readthrough in A) SUP45-fixed population and B) TRM10 and SUP45-fixed population. The comparisons of the selected segregating population with high GFP signal and the selected segregating population with low GFP signal to the whole population are shown. Sliding window averages (40 kb) are plotted. Black arrows show TRM10 locus on chromosome XV and blue arrows show the SUP45 locus on chromosome II. Linkage disequilibrium between TRM10 and SUP45
We used a previously published polymorphism survey [18] to determine the frequencies of the BY and RM alleles of TRM10 and SUP45 among 63 diverse S. cerevisiae strains isolated from a broad range of sources. We found that both alleles of both genes are common among these S. cerevisiae strains, with allele frequencies of 0.46 and 0.44 for the BY alleles of TRM10 and SUP45, respectively. This observation shows that these variants are not restricted to strains adapted to the laboratory environment, but rather represent naturally occurring polymorphisms in the species. Strains which carry the BY allele of TRM10 include laboratory strains, Sake strains, clinical isolates and oak strains. Most of these strains also carry the BY allele of SUP45 (Table S2). The SUP45BY-TRM10BY and SUP45RM-TRM10RM haplotypes are present in S. cerevisiae strains more frequently than expected based on random association (76% vs. 50%). These data suggested that TRM10 and SUP45 might be in linkage disequilibrium (LD).
We confirmed the presence of strong LD between these genes by calculating D' and r2 between SUP45 and TRM10 and obtained values of 0.537 and 0.266, respectively. LD could arise in part due to the population structure present in S. cerevisiae [18]. To test whether the observed LD is significantly higher than what is expected solely due to structure, we used polymorphisms from 63 diverse S. cerevisiae [18] strains and calculated LD between 1000 randomly chosen SNP pairs from different chromosomes. We observed nine pairs out of 1000 with higher r2 than the TRM10-SUP45 pair (Figure S4), which showed that LD between TRM10 and SUP45 SNPs is significantly higher than what is expected due to structure alone (p-value = 0.009). We obtained similar results for D' (p-value = 0.011).
This non-random association of alleles suggests a functional association between the two genes. The two overrepresented haplotypes carry alleles with opposing effects on readthrough. The BY allele of TRM10 results in higher readthrough, whereas the BY allele of SUP45 results in lower readthrough; conversely, the RM allele of TRM10 results in lower readthrough whereas the RM allele of SUP45 results in higher readthrough. These two combinations keep readthrough within the range ∼0.3–0.5%, above that of the SUP45BY-TRM10RM combination (∼0.2%) and below that of the SUP45RM-TRM10BY combination (∼0.8%). The fact that the two allelic combinations with intermediate readthrough are overrepresented suggests that readthrough may be subject to stabilizing selection.
Discussion
We have shown that a difference in translation termination efficiency between two yeast strains is explained by polymorphisms in two genes, TRM10 and SUP45. TRM10 encodes a tRNA-modifying enzyme, which methylates the N-1 position of guanosine-9 in some tRNAs [17]. In the S. cerevisiae, m1G is found at position 9 in 10 out of 34 tRNAs. Despite the evolutionary conservation of this modification and the conservation of the protein [17], a cellular role for m1G9 modification has not been reported. Previous studies have not found an obvious growth defect in yeast cells lacking Trm10p under standard growth conditions in rich or minimal media. Recently, a temperature-sensitive phenotype was reported in yeast cells lacking Trm10p in the presence of 5 - fluorouracil [21]. This study showed that 5 - fluorouracil targets tRNA-modifying enzymes and therefore reduces a number of tRNA modifications. Loss of most of the tRNA modifications in the presence of 5 - fluorouracil, in combination with loss of the m1G9 modification in the absence of Trm10p, resulted in destabilization of hypomodified tRNAs, which explained the growth defect and suggested a role for m1G9 modification in tRNA stability. Here, we provide genetic evidence supporting a role for m1G9 modification in translation termination efficiency. We showed that a serine to proline substitution at position 139 of Trm10p contributes to the readthrough difference between BY and RM. One of the Trm10p substrates is the tryptophan tRNA, which decodes UGG, one of the closest codons to UGA, the opal stop codon in the dual luciferase and GFP reporters used here. According to the Pfam domain annotation, the Trm10p tRNA methylating domain spans residues 104 to 276. A serine to proline substitution at position 139 may decrease Trm10p tRNA methylation activity. Loss of this modification may increase the ability of near cognate tRNAs (such as the tryptophan tRNA) to outcompete SUP45p in UGA stop codon recognition, resulting in increased readthrough. These data support a role of tRNA methylation by Trm10p in efficient translation termination.
We also showed that the other factor involved in the translation termination efficiency difference between BY and RM is a regulatory polymorphism that alters the expression level of SUP45. Sup45p is the yeast translation termination factor responsible for stop codon recognition. It was previously reported that reducing the cellular level of Sup45p decreases the efficiency of translation termination [22]. When the cellular level of Sup45p is low, for example as a result of a lower expression level of SUP45, near cognate tRNAs are more likely to outcompete Sup45p in stop codon recognition, which in turn increases readthrough.
BY alleles of both TRM10 and SUP45 are common among S. cerevisiae strains. This observation shows that these alleles are not restricted to strains adapted to the laboratory environment, but rather represent naturally occurring polymorphisms. Despite the fact that these two genes reside on different chromosomes and are thus not physically linked, we found strong and significant linkage disequilibrium (LD) between these two genes. This non-random allelic association provides evidence that natural selection has favored the SUP45BY-TRM10BY and SUP45RM-TRM10RM haplotypes over the others. The preferred haplotypes consist of alleles with opposing effects on readthrough, and thus strains that carry them should exhibit intermediate readthrough relative to the other two haplotypes. This observation suggests that readthrough may be subject to stabilizing selection. High readthrough may be disadvantageous due to production of too many inappropriately extended proteins. On the other hand, keeping readthrough from dropping too low may protect yeast mRNAs whose translation requires the suppression of leaky stop codons [23].
Materials and Methods
Strains, media, and plasmids
Cultures were grown in minimal medium containing 0.67% (w/v) yeast nitrogen base without amino acids (Difco) containing 2% (w/v) glucose (SMD) or 2% galactose (SMGal) or 4% raffinose (SMRaf), as specified. Additional nutritional supplements or drugs were added as required. YPD plates were made as described [24]. For sporulation, SPO++ was used (http://www.genomics.princeton.edu/dunham/sporulationdissection.htm). To make a readthrough GFP reporter, a 78-nucleotide region from the ade1-14 allele [25] was inserted in-frame with GFP coding region into HindIII site (6th codon) of pGAL-GFP plasmid (kindly provided by James Broach). The insertion was confirmed by sequencing. To include a selectable marker, NATMX cassette was inserted into a NotI site downstream of GFP coding sequence in pGAL-GFP-ade1-14 plasmid. To integrate this GFP reporter in yeast genome, the region containing pGAL-GFP-ade1-14-NATMX was amplified with primers with 40 base pairs of homology to regions upstream and downstream of YDL242W (an open-reading frame unlikely to encode a protein, based on available experimental and comparative sequence data from http://www.yeastgenome.org/). This PCR product was then used in transformation of BY4724 [26] and RM11-1a [14]. Insertion of the GFP reporter inside YDL242W was then confirmed by PCR. We then transferred this GFP reporter into strains with suitable genetic markers. To do so, we crossed these new BY and RM strains with GFP reporter into BY MATα can1Δ::STE2pr-SpHIS5 lyp1Δ his3Δ1 and RM MATα AMN1BY his3Δ0::NatMX ho::HphMX [16], respectively. After sporulating the obtained diploids and genotyping the dissected tetrads, we selected BY Matα his3Δ1 lyp1Δ can1Δ::STE2prSpHIS5 YDL242W::pGAL-GFP-ade1-14-NATMX and RM Mata AMN1BY his3Δ::NATMX, YDL242W::pGAL-GFP-ade1-14-NATMX .
TRM10 and SUP45 replacement strains were generated by a two-step replacement method [27]. TRM10 was replaced with URA3-KanMX cassette from pCORE in BY4724 and RM11-1a generating trm10Δ::URA3-KanMX knockout strains. TRM10 alleles from the donor strains were amplified by PCR with approximately 200-bp overlapping sequence and introduced into recipient strains to replace URA3-KanMX cassette. For SUP45 replacement URA3-KanMX cassette from pCORE was inserted downstream of SUP45 coding sequence in recipient strains. pCORE cassette was then replaced by the PCR-amplified SUP45 allele from donor strains. Allele replacements were confirmed by sequencing.
Sequencing
The RM TRM10 and SUP45 sequences were obtained from the whole genome-sequencing project at the Broad Institute (http://www.broad.mit.edu/annotation/genome/saccharomyces_cerevisiae/Home.html).
All sequencing was done using standard dideoxy methods.
Dual luciferase assay
Dual luciferase assay was performed as explained before [19]. Dual luciferase reporter plasmids were kindly provided by David Bedwell. Plasmids with the stop codon (pDB691) or the sense codon (pDB690) were transformed into the indicated yeast strains, and transformants were selected on SMD drop-out plates lacking uracil. Transformed strains were grown in liquid SMD medium to a cell density of 0.5–0.7 A600 units/mL as measured using Synergy 2 Multi-Mode Microplate Reader (BioTek Instruments). The luciferase assay was performed using the Dual-Luciferase Reporter Assay System (Promega; E1910). Approximately 104 yeast cells from each strain expressing the indicated dual luciferase reporter were lysed using 100 µL of Passive Lysis Buffer in a 96-well plate (Costar; 3370). Two microliters of the lysate were added to 10 µL of the Luciferase Assay Reagent II in an opaque 96-well plate (Costar; 3614). Relative luminescence units (RLUs) produced by firefly luciferase activity were then measured for 10 seconds using Synergy 2 Multi-Mode Microplate Reader (BioTek Instruments). 10 µL of Stop&Glo buffer was then added to quench the firefly activity and activate the Renilla luciferase activity. RLUs were again measured for 10 seconds to determine the Renilla luciferase activity. Negative controls that contained all the reaction components except cell lysates were used to determine the background for each luciferase reaction and were subtracted from the experimental values obtained. Percent readthrough is expressed as the mean ± the standard deviation of values obtained from at least eight independent dual luciferase assay including at least four biological replicates.
X-QTL
All X-QTL experiments were done in duplicates. MATa haploid segregants from the indicated cross were selected as explained before [16]. To create the segregating pool, a single colony of the diploid progenitor was inoculated into 5 mL YPD and grown to stationary phase. The diploid culture was spun down and the supernatant was decanted. The diploid pellet was then resuspended in 50 mL SPO++ sporulation medium. The sporulation was kept at room temperature (∼22°C) with shaking and monitored for the fraction of diploids that had sporulated. Once more than 50% of the diploids had sporulated, 10 mL of the sporulation were spun down and then the supernatant was decanted. The pellet was resuspended in 2 mL water. 600 µL β-glucoronidase (Sigma; G7770) were added to the preparation, and the mixture was incubated at 30°C for one hour. Water was added to the sample so that the total volume was 20 mL. The spore preparation was spread onto SMD + canavanine/thialysine plates (Sigma; C9758 for canavanine (L-canavanine sulphate salt); A2636 for thialysine (S-(2-aminoethyl) - L-cysteine hydrochloride)), with 100 µL of sample going onto each plate. The plates were incubated at 30°C for two days. Then 10 mL of water were poured onto each plate and a sterile spreader was used to remove the segregants from the plate. The cell mixtures from each plate were then pipetted off the plates into a container. The pool was spun down and the water decanted. Haploid segregants were then inoculated into liquid SMRaf plus canavanine medium at a concentration of ∼1×107 cells mL−1. The cells were grown for approximately two generations to a density of ∼2×107 cells mL−1. To induce the GAL promoter, cells were spun down and then were transferred to liquid SMGal plus canavanine with a density of ∼2×106 cells mL−1and were incubated in 30°C while shaking on a rotary shaker at 200 rpm for four hours. Cells were then sorted using BD FACS Vantage SE, collecting 20,000 cells from top 1% (High GFP) and bottom 1% (Low GFP) of the whole population. High GFP, Low GFP and a sample of the whole population were then plated on YPD plates and were incubated at 30°C for two days. DNA was extracted from the grown cells using Genomic-tip 100/G columns (Qiagen; 10243). DNA was labeled using the BioPrime Array CGH Genomic Labeling Module (Invitrogen; 18095-012) with the sample being labeled with Cy3 dUTP and the reference being labeled with Cy5 dUTP. We used a BY/RM diploid as the reference for all hybridizations. Labeled samples were then hybridized onto the allele-specific genotyping microarray with isothermal probes that assays ∼18,000 single nucleotide polymorphisms (SNPs) between BY and RM [16]. Hybridization intensities were extracted and normalized using the rank invariant method in the Agilent Feature Extraction software package. For a given SNP, the difference in the log10 ratios of BY and RM-specific probes on a single array (or log10 intensity difference) was computed. Background allele frequency changes that occur during pool construction were removed from the data for top 1% (High GFP) and bottom 1% (Low GFP) selection by subtracting the log10 intensity difference obtained for the whole (unselected) population from the log10 intensity difference observed in the High and Low GFP selections. These steps were conducted in R (http://www.r-project.org/).
Supporting Information
Zdroje
1. ZhouravlevaGFrolovaLLe GoffXLe GuellecRInge-VechtomovS 1995 Termination of translation in eukaryotes is governed by two interacting polypeptide chain release factors, eRF1 and eRF3. EMBO J 14 4065 4072
2. FrolovaLLe GoffXRasmussenHHChepereginSDrugeonG 1994 A highly conserved eukaryotic protein family possessing properties of polypeptide chain release factor. Nature 372 701 703
3. StansfieldIAkhmalokaTuiteMF 1995 A mutant allele of the SUP45 (SAL4) gene of Saccharomyces cerevisiae shows temperature-dependent allosuppressor and omnipotent suppressor phenotypes. Curr Genet 27 417 426
4. Salas-MarcoJBedwellDM 2004 GTP hydrolysis by eRF3 facilitates stop codon decoding during eukaryotic translation termination. Mol Cell Biol 24 7769 7778
5. JanzenDMGeballeAP 2001 Modulation of translation termination mechanisms by cis - and trans-acting factors. Cold Spring Harbor symposia on quantitative biology 66 459 467
6. NamyOHatinIRoussetJP 2001 Impact of the six nucleotides downstream of the stop codon on translation termination. EMBO Rep 2 787 793
7. TorkSHatinIRoussetJPFabretC 2004 The major 5′ determinant in stop codon read-through involves two adjacent adenines. Nucleic Acids Res 32 415 421
8. RospertSRakwalskaMDubaquieY 2005 Polypeptide chain termination and stop codon readthrough on eukaryotic ribosomes. Rev Physiol Biochem Pharmacol 155 1 30
9. von der HaarTTuiteMF 2007 Regulated translational bypass of stop codons in yeast. Trends Microbiol 15 78 86
10. ShorterJLindquistS 2005 Prions as adaptive conduits of memory and inheritance. Nat Rev Genet 6 435 450
11. PerlsteinEORuderferDMRobertsDCSchreiberSLKruglyakL 2007 Genetic basis of individual differences in the response to small-molecule drugs in yeast. Nat Genet 39 496 502
12. DeutschbauerAMDavisRW 2005 Quantitative trait loci mapped to single-nucleotide resolution in yeast. Nat Genet 37 1333 1340
13. SteinmetzLMSinhaHRichardsDRSpiegelmanJIOefnerPJ 2002 Dissecting the architecture of a quantitative trait locus in yeast. Nature 416 326 330
14. BremRBYvertGClintonRKruglyakL 2002 Genetic dissection of transcriptional regulation in budding yeast. Science 296 752 755
15. EhrenreichIMGerkeJPKruglyakL 2009 Genetic dissection of complex traits in yeast: insights from studies of gene expression and other phenotypes in the BYxRM cross. Cold Spring Harb Symp Quant Biol 74 145 153
16. EhrenreichIMTorabiNJiaYKentJMartisS 2010 Dissection of genetically complex traits with extremely large pools of yeast segregants. Nature 464 1039 1042
17. JackmanJEMontangeRKMalikHSPhizickyEM 2003 Identification of the yeast gene encoding the tRNA m1G methyltransferase responsible for modification at position 9. RNA 9 574 585
18. SchachererJShapiroJARuderferDMKruglyakL 2009 Comprehensive polymorphism survey elucidates population structure of Saccharomyces cerevisiae. Nature 458 342 345
19. KeelingKMLanierJDuMSalas-MarcoJGaoL 2004 Leaky termination at premature stop codons antagonizes nonsense-mediated mRNA decay in S. cerevisiae. RNA 10 691 703
20. SmithENKruglyakL 2008 Gene-environment interaction in yeast gene expression. PLoS Biol 6 e83 doi:10.1371/journal.pbio.0060083
21. GustavssonMRonneH 2008 Evidence that tRNA modifying enzymes are important in vivo targets for 5-fluorouracil in yeast. RNA 14 666 674
22. StansfieldIEurwilaichitrLAkhmalokaTuiteMF 1996 Depletion in the levels of the release factor eRF1 causes a reduction in the efficiency of translation termination in yeast. Mol Microbiol 20 1135 1143
23. NamyODuchateau-NguyenGHatinIHermann-Le DenmatSTermierM 2003 Identification of stop codon readthrough genes in Saccharomyces cerevisiae. Nucleic Acids Res 31 2289 2296
24. BurkeDDawsonDStearnsT Cold Spring Harbor Laboratory 2000 Methods in yeast genetics : a Cold Spring Harbor Laboratory course manual. Plainview N.Y.: Cold Spring Harbor Laboratory Press xvii, 205 p.
25. ManogaranALKirklandKTLiebmanSW 2006 An engineered nonsense URA3 allele provides a versatile system to detect the presence, absence and appearance of the [PSI+] prion in Saccharomyces cerevisiae. Yeast 23 141 147
26. BrachmannCBDaviesACostGJCaputoELiJ 1998 Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14 115 132
27. StoriciFLewisLKResnickMA 2001 In vivo site-directed mutagenesis using oligonucleotides. Nat Biotechnol 19 773 776
Štítky
Genetika Reprodukční medicína
Článek Pervasive Sign Epistasis between Conjugative Plasmids and Drug-Resistance Chromosomal MutationsČlánek Stress-Induced PARP Activation Mediates Recruitment of Mi-2 to Promote Heat Shock Gene ExpressionČlánek Histone Crosstalk Directed by H2B Ubiquitination Is Required for Chromatin Boundary IntegrityČlánek A Functional Variant at a Prostate Cancer Predisposition Locus at 8q24 Is Associated with ExpressionČlánek Replication and Explorations of High-Order Epistasis Using a Large Advanced Intercross Line PedigreeČlánek Expression of Tumor Suppressor in Spermatogonia Facilitates Meiotic Progression in Male Germ Cells
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2011 Číslo 7
-
Všechny články tohoto čísla
- Gene-Based Tests of Association
- The Demoiselle of X-Inactivation: 50 Years Old and As Trendy and Mesmerising As Ever
- Variants in and Underlie Natural Variation in Translation Termination Efficiency in
- SHH1, a Homeodomain Protein Required for DNA Methylation, As Well As RDR2, RDM4, and Chromatin Remodeling Factors, Associate with RNA Polymerase IV
- Genome-Wide Association Study Identifies as a Susceptibility Gene for Pediatric Asthma in Asian Populations
- Pervasive Sign Epistasis between Conjugative Plasmids and Drug-Resistance Chromosomal Mutations
- Genetic Anticipation Is Associated with Telomere Shortening in Hereditary Breast Cancer
- Identification of a Mutation Associated with Fatal Foal Immunodeficiency Syndrome in the Fell and Dales Pony
- Stress-Induced PARP Activation Mediates Recruitment of Mi-2 to Promote Heat Shock Gene Expression
- An Epigenetic Switch Involving Overlapping Fur and DNA Methylation Optimizes Expression of a Type VI Secretion Gene Cluster
- Recombination and Population Structure in
- A Rice Plastidial Nucleotide Sugar Epimerase Is Involved in Galactolipid Biosynthesis and Improves Photosynthetic Efficiency
- A Role for Phosphatidic Acid in the Formation of “Supersized” Lipid Droplets
- Colon Stem Cell and Crypt Dynamics Exposed by Cell Lineage Reconstruction
- Loss of the BMP Antagonist, SMOC-1, Causes Ophthalmo-Acromelic (Waardenburg Anophthalmia) Syndrome in Humans and Mice
- Interactions between Glucocorticoid Treatment and Cis-Regulatory Polymorphisms Contribute to Cellular Response Phenotypes
- Translation Reinitiation Relies on the Interaction between eIF3a/TIF32 and Progressively Folded -Acting mRNA Elements Preceding Short uORFs
- DAF-12 Regulates a Connected Network of Genes to Ensure Robust Developmental Decisions
- Adult Circadian Behavior in Requires Developmental Expression of , But Not
- Histone Crosstalk Directed by H2B Ubiquitination Is Required for Chromatin Boundary Integrity
- Proteins in the Nutrient-Sensing and DNA Damage Checkpoint Pathways Cooperate to Restrain Mitotic Progression following DNA Damage
- Complex Evolutionary Events at a Tandem Cluster of Genes Resulting in a Single-Locus Genetic Incompatibility
- () and Its Regulated Homeodomain Gene Mediate Abscisic Acid Response in
- A Functional Variant at a Prostate Cancer Predisposition Locus at 8q24 Is Associated with Expression
- LGI2 Truncation Causes a Remitting Focal Epilepsy in Dogs
- Adaptations to Endosymbiosis in a Cnidarian-Dinoflagellate Association: Differential Gene Expression and Specific Gene Duplications
- The Translation Initiation Factor eIF4E Regulates the Sex-Specific Expression of the Master Switch Gene in
- Somatic Genetics Empowers the Mouse for Modeling and Interrogating Developmental and Disease Processes
- Molecular Mechanisms Generating and Stabilizing Terminal 22q13 Deletions in 44 Subjects with Phelan/McDermid Syndrome
- Replication and Explorations of High-Order Epistasis Using a Large Advanced Intercross Line Pedigree
- Mechanisms of Chromosome Number Evolution in Yeast
- Regulatory Cross-Talk Links Chromosome II Replication and Segregation
- Ancestral Genes Can Control the Ability of Horizontally Acquired Loci to Confer New Traits
- Expression of Tumor Suppressor in Spermatogonia Facilitates Meiotic Progression in Male Germ Cells
- Rare and Common Regulatory Variation in Population-Scale Sequenced Human Genomes
- The Epistatic Relationship between BRCA2 and the Other RAD51 Mediators in Homologous Recombination
- Identification of Novel Genetic Markers Associated with Clinical Phenotypes of Systemic Sclerosis through a Genome-Wide Association Strategy
- NatF Contributes to an Evolutionary Shift in Protein N-Terminal Acetylation and Is Important for Normal Chromosome Segregation
- Araucan and Caupolican Integrate Intrinsic and Signalling Inputs for the Acquisition by Muscle Progenitors of the Lateral Transverse Fate
- Pathologic and Phenotypic Alterations in a Mouse Expressing a Connexin47 Missense Mutation That Causes Pelizaeus-Merzbacher–Like Disease in Humans
- Recombinant Inbred Line Genotypes Reveal Inter-Strain Incompatibility and the Evolution of Recombination
- Epistatic Relationships in the BRCA1-BRCA2 Pathway
- Genome-Wide Association Study Identifies Novel Restless Legs Syndrome Susceptibility Loci on 2p14 and 16q12.1
- Genetic Loci Associated with Plasma Phospholipid n-3 Fatty Acids: A Meta-Analysis of Genome-Wide Association Studies from the CHARGE Consortium
- Fine Mapping of Five Loci Associated with Low-Density Lipoprotein Cholesterol Detects Variants That Double the Explained Heritability
- CHD1 Remodels Chromatin and Influences Transient DNA Methylation at the Clock Gene
- Nonlinear Fitness Landscape of a Molecular Pathway
- Genome-Wide Scan Identifies , , and as Novel Risk Loci for Systemic Sclerosis
- Quantitative and Qualitative Stem Rust Resistance Factors in Barley Are Associated with Transcriptional Suppression of Defense Regulons
- A Systematic Screen for Tube Morphogenesis and Branching Genes in the Tracheal System
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Genome-Wide Association Study Identifies Novel Restless Legs Syndrome Susceptibility Loci on 2p14 and 16q12.1
- Loss of the BMP Antagonist, SMOC-1, Causes Ophthalmo-Acromelic (Waardenburg Anophthalmia) Syndrome in Humans and Mice
- Gene-Based Tests of Association
- Genome-Wide Association Study Identifies as a Susceptibility Gene for Pediatric Asthma in Asian Populations
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



