-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaA Systematic Screen for Tube Morphogenesis and Branching Genes in the Tracheal System
Many signaling proteins and transcription factors that induce and pattern organs have been identified, but relatively few of the downstream effectors that execute morphogenesis programs. Because such morphogenesis genes may function in many organs and developmental processes, mutations in them are expected to be pleiotropic and hence ignored or discarded in most standard genetic screens. Here we describe a systematic screen designed to identify all Drosophila third chromosome genes (∼40% of the genome) that function in development of the tracheal system, a tubular respiratory organ that provides a paradigm for branching morphogenesis. To identify potentially pleiotropic morphogenesis genes, the screen included analysis of marked clones of homozygous mutant tracheal cells in heterozygous animals, plus a secondary screen to exclude mutations in general “house-keeping” genes. From a collection including more than 5,000 lethal mutations, we identified 133 mutations representing ∼70 or more genes that subdivide the tracheal terminal branching program into six genetically separable steps, a previously established cell specification step plus five major morphogenesis and maturation steps: branching, growth, tubulogenesis, gas-filling, and maintenance. Molecular identification of 14 of the 70 genes demonstrates that they include six previously known tracheal genes, each with a novel function revealed by clonal analysis, and two well-known growth suppressors that establish an integral role for cell growth control in branching morphogenesis. The rest are new tracheal genes that function in morphogenesis and maturation, many through cytoskeletal and secretory pathways. The results suggest systematic genetic screens that include clonal analysis can elucidate the full organogenesis program and that over 200 patterning and morphogenesis genes are required to build even a relatively simple organ such as the Drosophila tracheal system.
Published in the journal: . PLoS Genet 7(7): e32767. doi:10.1371/journal.pgen.1002087
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1002087Summary
Many signaling proteins and transcription factors that induce and pattern organs have been identified, but relatively few of the downstream effectors that execute morphogenesis programs. Because such morphogenesis genes may function in many organs and developmental processes, mutations in them are expected to be pleiotropic and hence ignored or discarded in most standard genetic screens. Here we describe a systematic screen designed to identify all Drosophila third chromosome genes (∼40% of the genome) that function in development of the tracheal system, a tubular respiratory organ that provides a paradigm for branching morphogenesis. To identify potentially pleiotropic morphogenesis genes, the screen included analysis of marked clones of homozygous mutant tracheal cells in heterozygous animals, plus a secondary screen to exclude mutations in general “house-keeping” genes. From a collection including more than 5,000 lethal mutations, we identified 133 mutations representing ∼70 or more genes that subdivide the tracheal terminal branching program into six genetically separable steps, a previously established cell specification step plus five major morphogenesis and maturation steps: branching, growth, tubulogenesis, gas-filling, and maintenance. Molecular identification of 14 of the 70 genes demonstrates that they include six previously known tracheal genes, each with a novel function revealed by clonal analysis, and two well-known growth suppressors that establish an integral role for cell growth control in branching morphogenesis. The rest are new tracheal genes that function in morphogenesis and maturation, many through cytoskeletal and secretory pathways. The results suggest systematic genetic screens that include clonal analysis can elucidate the full organogenesis program and that over 200 patterning and morphogenesis genes are required to build even a relatively simple organ such as the Drosophila tracheal system.
Introduction
Elucidating the genetic programs of organ formation and maintenance is a central goal of developmental biology and medicine. Many organogenesis genes have been isolated in systematic genetic screens in model organisms, and many others have been identified by their organ-selective expression patterns and by candidate gene analysis. These approaches have been very successful at discovering the signaling pathways and transcription factors that induce and pattern organs and specify cell fates, but they have been much less successful at identifying the downstream effectors that execute morphogenesis programs, what we call morphogenesis genes [1], [2], [3], [4]. A similar abundance of signaling and transcription factor genes and dearth of morphogenesis genes has obtained from the pioneering genetic dissection of Drosophila body axis formation and other early developmental events [5]. We reasoned that many morphogenesis genes would function in multiple organs and developmental processes, so mutations in these genes would be pleiotropic and hence discarded in most genetic screens. We therefore designed systematic, saturation screens for genes required for Drosophila tracheal system organogenesis that included clonal analysis of gene function in the tracheal system, to identify all tracheal genes including those with pleiotropic phenotypes. The results of a screen of the third chromosome, representing ∼40% of the Drosophila genome [6], are described here, and the results of a first (X) chromosome screen initiated earlier will be described elsewhere ([7]; M. Metzstein and M.A.K., unpublished data).
The Drosophila tracheal (respiratory) system is a branched tubular network that transports oxygen throughout the body [8]. It is one of the most intensively studied and best understood organogenesis programs [9], [10], and it has emerged over the past decade as a paradigm of branching morphogenesis, the developmental process that gives rise to many organs including the lung, vascular system, kidney, and pancreas. Understanding how branching networks are patterned and how cellular tubes are made, shaped, and maintained is of fundamental importance in cell and developmental biology, and in medicine for understanding and treating tubular diseases such as aneurysms and polycystic kidney disease.
The tracheal system develops from 10 pairs of tracheal sacs that arise by invagination of the embryonic ectoderm [8], [11]. Each sac is an epithelial monolayer composed of ∼80 cells. Primary tracheal branches are formed by groups of 3–20 cells that bud from the sacs in different directions and successively sprout secondary and terminal branches. Some specialized primary and secondary branches grow towards and fuse with branches from neighboring sacs to interconnect the tracheal network [12]. The transformation of the simple epithelial sacs into an extensively branched tubular network occurs without cell proliferation, and is mediated by cell migration, rearrangement, and dramatic changes in cell shape [11], [13], [14], [15]. During embryogenesis, the lumens of the developing tracheal branches are filled with a complex and changing matrix, which is cleared and replaced with gas just before the embryo hatches and the tubes become functional in respiration [8], [16]. In the larva, terminal cells ramify extensively to form many new terminal branches (tracheoles), long cytoplasmic extensions that grow toward oxygen-starved cells and then form a cytoplasmic, membrane-bound lumen, creating tiny tubes (<1 um diameter) that supply the targets with oxygen (Figure 1B) [14], [17]. Unlike primary (multicellular) and secondary (unicellular) branches, tubes sealed by intercellular and autocellular junctions (Figure 1B), terminal branches lack cell junctions and resemble the “seamless” endothelial tubes of the mammalian microvasculature [18], [19], [20] and C. elegans excretory system [21].
Fig. 1. Design of tracheal mutant screen. 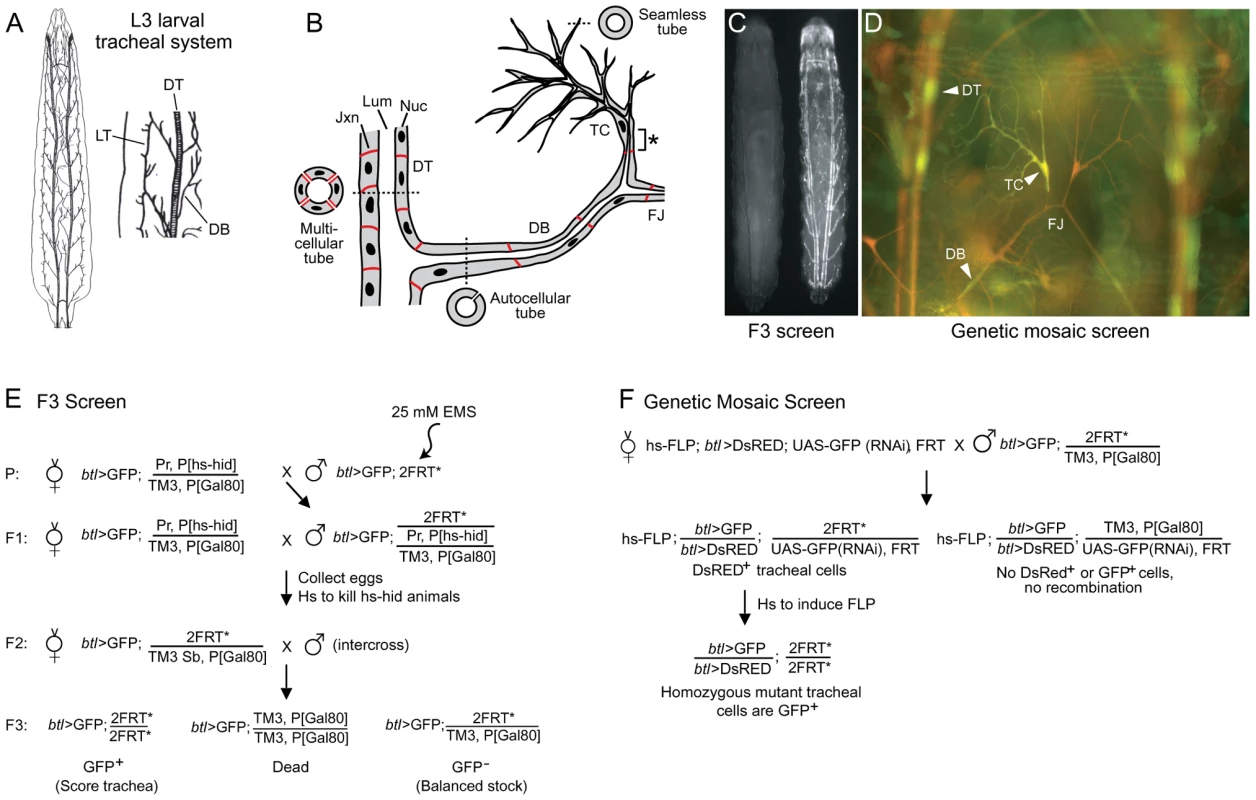
(A) Diagram of Drosophila tracheal system in third instar larva (dorsal view, anterior up unless noted otherwise). A close up of two hemisegments (Tr4 and Tr5) are shown at right, with some primary branches indicated. DT, dorsal trunk; DB, dorsal branch; LT, lateral trunk. (B) Schematic showing cellular structure of dorsal trunk and dorsal branch. Dashed lines indicate plane of section of cross-sections shown. DT is a multicellular tube with multiple cells and intercellular junctions seen in cross-section. DB stalk is an autocellular tube, a single cell wrapped around the luminal space and sealed by an autocellular junction. DB terminal cell (TC) forms multiple terminal branches, each of which is a “seamless” tube lacking junctions. The base of the terminal cell (*), from its junction with a stalk cell to the nucleus, is an autocellular tube. The fusion joint (FJ) is the position where two fusion cells, each of which forms a seamless tube, connect contralateral tracheal hemisegments. Lum, tracheal lumen (black); Jxn, intercellular junctions (red); Nuc, cell nuclei (black). (C) Fluorescence micrograph of two sibling F3 larvae from the F3 screen diagrammed in panel E. The GFP- larva at left is heterozygous for the mutagenized third chromosome; it is nearly invisible because it contains, in trans to the mutagenized chromosome, a Gal80-expressing balancer chromosome that prevents expression of btl-Gal4, UAS-GFP (btl>GFP). The GFP+ larva at right is homozygous for a mutagenized third chromosome; it lacks the Gal80 chromosome, so expresses GFP throughout the tracheal system. (D) Fluorescence micrograph of a segment (Tr5) of the tracheal system from a third instar larva generated by the genetic mosaic strategy shown in panel F. All tracheal cells express btl>DsRED (red); homozygous clones lack the UAS-GFP(RNAi), so express in addition btl>GFP (green). Dorsal trunk (DT), dorsal branch (DB) and terminal cell (TC) clones are marked (arrowheads). Dorsal branch fusion joint (FJ) connecting the left and right hemisegments is indicated. (E) Genetic scheme of F3 screen. EMS, ethyl methanesulfonate; Pr, Prickly; P[hs-hid], heat shock inducible hid transgene; TM3, third chromosome balancer; P[Gal80], transgene with ubiquitous tubulin promoter driving expression of Gal80, a Gal4 inhibitor; 2FRT, two Flp Recombinase Target (FRT) site transgenes (FRT2A on 3L and FRT82B on 3R) flanking the third chromosome centromere; Sb, Stubble; *, mutagenized chromosome. (F) Genetic scheme of the mosaic screen. hs-FLP, heat-inducible FLP recombinase transgene; UAS-GFP(RNAi), Gal4-inducible (Gal4 upstream activating sequence) GFP RNAi transgene. The first important tracheal gene identified was trachealess, isolated in the classical screens for embryonic patterning mutants by the complete and selective absence of the tracheal system [22] and later shown to encode a bHLH-PAS transcription factor, the earliest expressed tracheal-specific gene and a master regulator of tracheal identity [23], [24]. Ten years after the discovery of trachealess, a Drosophila homolog of mammalian FGFRs was isolated and named Breathless because it is selectively expressed in the developing tracheal system and required for branching [25], [26], [27]. Around this time the first systematic screens for tracheal mutants were conducted, screens of P[lacZ] insertions that identified about 50 tracheal genes that subdivided embryonic tracheal development into genetically distinct processes including primary, secondary, and terminal branching, branch fusion, and tube size control [11], [28]. Mapping and molecular characterization of these genes identified many components and modulators of the Breathless FGFR signaling pathway. These include Branchless FGF, which activates Breathless FGFR and plays a central role in controlling and coupling each of these processes by guiding outgrowth of primary branches and inducing expression of key genes encoding transcription factors such as pointed, blistered/pruned, and escargot required, respectively, for secondary and terminal branching and branch fusion [12], [14], [26], [29]. Many other important genes have been identified by their tracheal expression patterns, analysis of candidate genes, and serendipitous discovery of a tracheal function for genes initially studied in other contexts [30], [31], [32], [33]. And, over the past several years, several screens of chemically-induced mutations for tracheal morphogenesis defects in embryos and larval tracheal and air sac primordium clones have been conducted [34], [35], [36] along with more targeted genetic and genomic screens for genes that are expressed or function downstream of some of the key early signaling pathways (branchless, breathless) and transcription factors (trachealess, ribbon) [37], [38], [39], [40]. Together these approaches have implicated ∼100 genes in tracheal development, most of which encode transcription factors or components of signaling pathways (FGF, TGFα/EGF, TGFβ, Wnt, Notch, Slit/Robo, Jak/Stat, and Hedgehog) [1], [2], [4], [9] (Table S1). However, the downstream targets of the signaling pathways and transcription factors, the morphogenesis and maturation genes that create, shape, and stabilize the tubes, have only recently begun to be identified. And, although expected to be a large class, they are substantially under-represented among characterized tracheal genes (Table S1) [4].
We conducted a large-scale screen of chemically-induced mutations to assess the function of nearly all Drosophila third chromosome genes, including early essential genes and genes with pleiotropic phenotypes. We sought to identify most or all of the genetically separable steps in tracheal development; to identify new tracheal genes associated with each step; and to provide an estimate of the total number of genes required to build an organ. We were especially interested in identifying tracheal morphogenesis genes. Our approach involved clonal analysis in the tracheal system of all chemically-induced mutations that did not survive late enough in development as homozygotes to assess their tracheal function, and a secondary screen to exclude general “house-keeping” genes. We isolated mutations representing ∼70 genes, 14 of which we identified molecularly, implicating most of the genes as morphogenesis genes and revealing new cell biological pathways in tracheal development. Many of the mutations affect terminal branch morphogenesis, genetically subdividing this poorly understood process into five major morphogenetic steps including an integral cell growth step.
Results
Screen design
To identify tracheal morphogenesis genes, we screened the third chromosome for EMS-induced mutations that affect larval tracheal morphology. Approximately 4,300 mutagenized third chromosomes were generated, and balanced lines were established for each. Three-quarters (73%) of the lines were homozygous lethal. Assuming a Poisson distribution, there were ∼1.3 lethal mutations per mutagenized third chromosome and a total of 5600 lethal mutations screened. Because there are ∼3600 essential Drosophila genes [6], with roughly 40% (∼1370) on the third chromosome [41], we expected to obtain an average of about four (5600/1370) mutations per gene, with at least one mutation in 97% of all third chromosome genes.
The mutants were screened for tracheal defects in two steps. First, homozygous third instar larvae of the F3 generation (Figure 1E), which carried btl-GAL4 and UAS-GFP transgenes (abbreviated btl>GFP) to label tracheal cells, were scored for tracheal defects by fluorescence microscopy. The balancer chromosome carried a Tub-GAL80 transgene that inhibits Gal4 and blocks expression of UAS-GFP so only homozygous mutant animals expressed GFP, facilitating screening (Figure 1C). For 40% of lines, GFP+ F3 third instar larvae were not recovered, presumably because the homozygous mutations caused early lethality.
These pre-pupal lethal lines were analyzed in a second step of the screen, using a genetic mosaic strategy in which we examined clones of homozygous mutant tracheal cells in otherwise heterozygous larvae (Figure 1F). We devised a variant of the MARCM clone marking strategy [42] employing a UAS-GFP(RNAi) transgene on the homologous chromosome, in trans to the mutation of interest, which allowed us to label all tracheal cells with btl>DsRed and homozygous mutant tracheal cells (lacking UAS-GFP(RNAi)) with btl>GFP (Figure 1D). This facilitated comparison of homozygous mutant cells (DsRed+, GFP+) with surrounding wild type tracheal cells (DsRed+, GFP-), enhancing the sensitivity of the screen and detection of cell non-autonomous effects in the tracheal system.
Overview of screen results
Over 600 mutants with highly penetrant and expressive tracheal defects were identified. However, the vast majority were lethal mutations in genes required cell autonomously for tracheal cell growth and survival, and later found in a secondary screen (see below) to be presumptive housekeeping genes and discarded. 133 tracheal mutations were saved (Table 1 and Table S3), 18 from the F3 screen (alleles with two letter prefix, e.g. PC213) and 115 from the genetic mosaic screen (no prefix).
Tab. 1. Tracheal morphogenesis mutant collection. 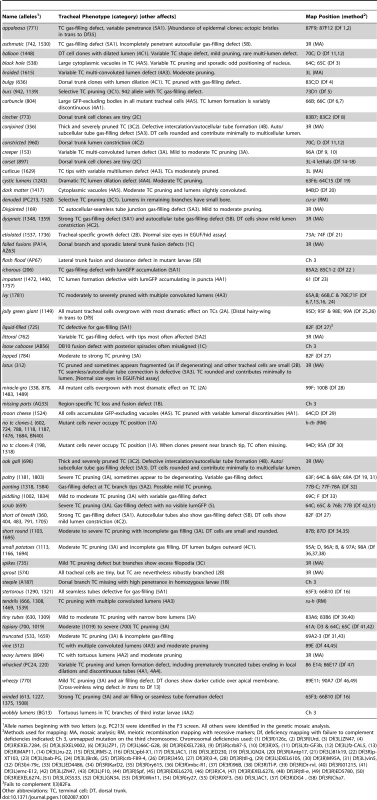
Allele names beginning with two letters (e.g. PC213) were identified in the F3 screen. All others were identified in the genetic mosaic analysis. Most of the mutations could be placed into one of five phenotypic classes (Table 1 and Table S3): (1) cell selection/specification; (2) cell size; (3) branch number, pattern size and shape; (4) tube formation, number, position, and shape; and (5) lumen clearance/gas filling. Within each broad class, phenotypic subgroups were defined and representative mutations in each subgroup were selected and subjected to detailed phenotypic characterization and genetic mapping as detailed below. Some mutations had more than one defect and were placed into more than one subgroup or class. Only a few mutants with cell non-autonomous effects were recovered from the clonal screen (see below), implying that such tracheal mutations are rare.
Genetic complementation groups
Complementation tests allowed assignment of 68 mutations to 24 loci (Table 1). In addition, four mutations that were mapped to specific chromosomal deficiencies were found to be new alleles of extant genes in the mapped intervals (see below). In addition to these 72 definitively assigned mutations in 28 loci, we also characterized and named 30 other mutations with interesting tracheal phenotypes (Table 1). The rest of the saved mutations (Table S3) were not extensively characterized; 12 of these are associated with mapped lethal mutations that complement extant tracheal mutations in the mapped interval, so may represent additional essential tracheal genes.
It is difficult to estimate the number of mutations we obtained in previously known tracheal genes because the mosaic loss of function phenotype is not known for most tracheal genes, and the number of complementation tests necessary to determine this number directly is prohibitive. However, the apparent absence of mutations in two known tracheal genes (stumps and trachealess) whose mosaic phenotype we determined, and the lower than expected allele frequencies (mean 2.6) obtained for the 28 definitively identified loci, indicate that the screen did not achieve the degree of saturation predicted by a Poisson distribution. Nevertheless, the screen was extensive so we think it is likely it identified mutations in most processes and molecular pathways involved in tracheal tube morphogenesis.
Below, we describe each of the major phenotypic classes and subcategories, and representative mutations in each. Most of the mutations are homozygous lethal and all caused highly penetrant and expressive tracheal phenotypes. For ease in presentation, we treat the strongest phenotype in each complementation group as the null phenotype; however, we do not know for most if they truly represent the null condition because it is not readily possible to generate hemizygous (mutant/deficiency) clones for comparison or to exclude partial masking of phenotypes due to perdurance of wild type protein in mutant cells.
Class 1: cell selection/specification mutants
These mutations eliminated specific tracheal cell types or blocked their differentiation.
No mutant terminal cells (1A)
Two complementation groups (no terminal cell clones-3L and no terminal cell clones-3R) gave normal numbers of mutant clones in the mosaic screen but the mutant cells rarely if ever included terminal cells (Figure 2A, 2B). This novel phenotype lead to new insights into the terminal cell selection process (see Discussion).
Fig. 2. Tracheal cell selection/specification mutants. 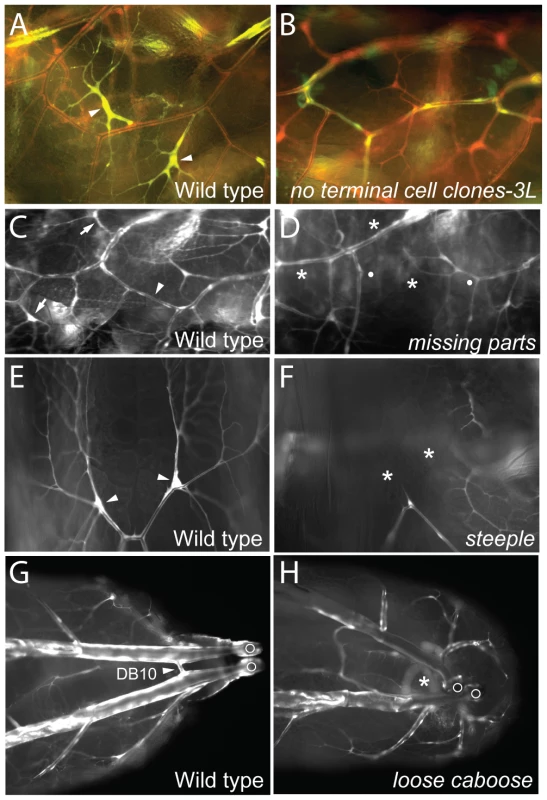
(A, B) Lateral views (anterior left) of a portion of the lateral tracheal trunk (between two transverse connectives) of genetic mosaic third instar larvae with control wild-type clones (A) and homozygous no terminal cell clones-3L clones (B). All tracheal cells express DsRed (red) and tracheal clones also express GFP (green) so appear yellow. Terminal cell clones (arrowheads) are present in A but absent in B. (C–H) Portions of the tracheal system of wild type control and mutant third instar larvae homozygous for the mutations indicated. (C, D) Lateral views (anterior left) of wild type (C) and missing parts mutant (D). Tracheae are labeled with GFP (white). Positions of two normal terminal cells (arrows) and a lateral trunk (LT) fusion joint (arrowhead) are indicated in C. In D, the corresponding terminal cells and LT fusion joint are missing (*), with broken ends of LT indicated by white dots. (E, F) Dorsal views of distal ends of a pair of dorsal branches labeled with GFP (white) in wild type (E) and steeple mutant (F). Note terminal cells (arrowheads in E) are missing (*) in steeple mutant (F). (G, H) Dorsal view of posterior of wild-type (G) and loose caboose mutant (H) with tracheae labeled with GFP (white). Arrowhead, position where contralateral dorsal branches (Tr10) connect to form the DB10 fusion joint (G). DB10 fusion joint is missing (*) in H; in the absence of the fusion joint, the positions of the disconnected parts of the tracheal system are more variable. Open circles, posterior spiracles. Region-specific terminal cell loss (1B)
Two lines lacked terminal cells in specific body regions of homozygous mutant animals. In missing parts (AG33) mutants, dorsal branch, fat body, and CNS terminal cells were frequently missing, but terminal cells in other positions were unaffected (Figure 2C, 2D). steeple (AI87) mutants frequently lacked dorsal branch terminal cells (Figure 2E, 2F), although all other terminal cells were present. These phenotypes demonstrate that there are region - or branch-specific modulators of terminal cell selection, specification, or survival, the existence of which had been suggested by marker expression patterns [11].
Failed branch fusions (1C)
Four lines showed frequent branch fusion failures in homozygous larvae. These included failed fusions (PA14, AZ63) and missing parts (Figure 2D), which showed dorsal branch and lateral trunk fusion defects as well as the terminal cell defects noted above. loose caboose (AB56) mutants had frequent defects in fusion of the most posterior dorsal branches (Figure 2G, 2H).
Class 2: cell size mutants
These mutants had their most profound affects on tracheal cell size. For nearly all mutations, the effect on terminal cell size correlated with branch number: larger cells had more terminal branches and smaller cells had fewer. One exceptional mutant, sprout, is presented below.
General tracheal cell overgrowth (2A)
Two complementation groups showed tracheal cell overgrowth phenotypes in the mosaic screen, and the enlarged terminal cells had more branches (see below). miracle-gro (338,878,1483,1489) mutant cells were several times larger than normal (Figure 3A, 3B), and the lumens of the mutant cells had larger bores and pursued a more tortuous path through the cytoplasm. The latter phenotype was fully penetrant in the seamless tubes of terminal cells and partially penetrant in larger tracheal tubes. jolly green giant (1149) caused a more subtle increase in cell size, most readily detected in terminal cells, and no obvious alteration in lumen morphology. Both miracle-gro and jolly green giant mutations also cause overgrowth of cells outside the tracheal system, because eyes derived from miracle-gro or jolly green giant clones in the EGUF/hid assay [43] described below were enlarged.
Fig. 3. Tracheal cell size mutants. 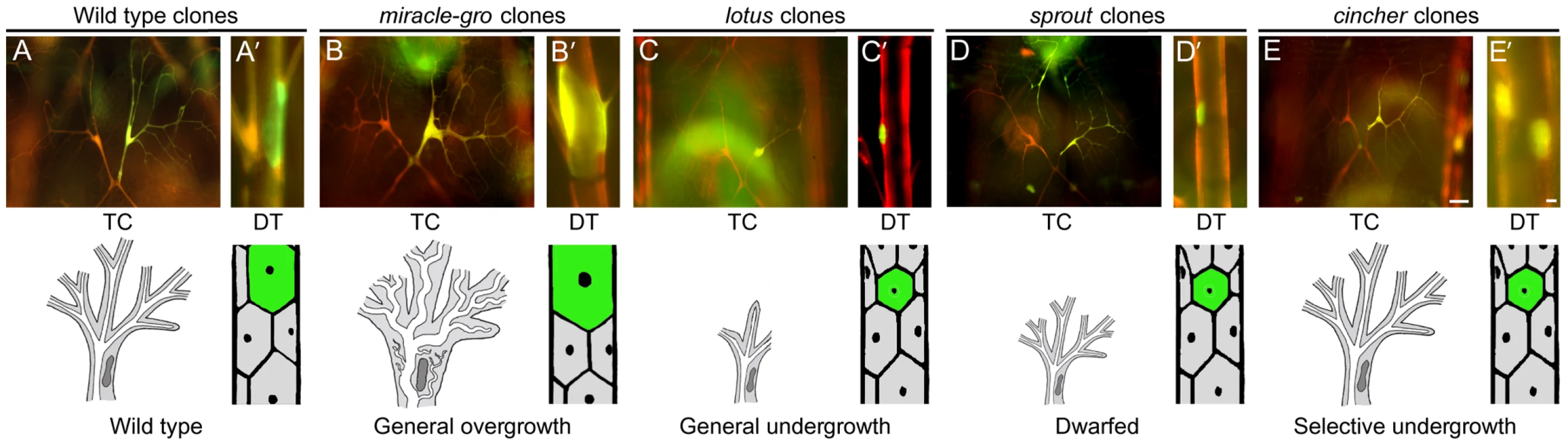
Micrographs (top panels) and schematics (lower panels) of genetic mosaic third instar larva showing terminal cell (TC, A–E) and dorsal trunk (DT, A'–E') clones (GFP+, green; at right) of control wild type (A, A'), miracle-gro338 (B, B'), lotus312 (C, C'), sprout574 (D, D'), and cincher773 (E, E') cells. In A–E, a contralateral control heterozygous terminal cell (DsRED+, red; at left) is included for comparison. The maximal soma cross-sectional area of miracle-gro338 terminal cell clones (0.87±0.05 units in Image J (mean±SEM), n = 10 clones) was four-fold greater than that of wild type control terminal cell clones (0.22±0.03 units). Extra branches in the miraclo-gro clone are highlighted in Figure 4C/4C'. Bar, 50 µm (A–E), 10 µm (A'–E'). General tracheal cell undergrowth (2B)
In roughly 500 lines, homozygous mutant cells in mosaic animals were small or absent, and mutant terminal cells when present were small with few or no branches (Figure 3C). Most of these lines presumably carried mutations in general cell growth (“house-keeping”) genes, but we suspected a subset might carry mutations in genes specifically required for tracheal cell growth. Indeed, we found that clones mutant for the tracheal master regulator trachealess, resembled clones homozygous mutant for the housekeeping gene glutamyl-prolyl-tRNA synthetase (Figure S1). To distinguish tracheal-specific from more general cell growth mutations, we tested the ∼500 tracheal undergrowth mutations in the developing eye. We used the EGUF/hid system [43] in which forced expression of a cell death gene in the eye imaginal disc eliminates wild type cells, so adult eyes develop almost entirely from clones of homozygous mutant cells. Nearly all (>99%) of the tracheal cell undergrowth mutations failed to rescue eye development, resulting in adults lacking eyes or with grossly undersized eyes. This substantiated that these mutations affected more general cell growth genes, and the lines were discarded. However, two tracheal undergrowth mutants, lotus (312) (Figure 3C) and etiolated (1736), formed eyes of normal size and shape, demonstrating that they are tracheal-selective growth mutations. Other aspects of the lotus phenotype are detailed below.
sprout (574) caused a general reduction of tracheal cell size, as well as a growth defect in the EGUF/Hid assay. However, it was distinguished from all the general undergrowth mutations described above by the ability of the small mutant terminal cells to branch extensively, like a bonsai plant (Figure 3D).
Selective tracheal cell undergrowth (2C)
Two mutations, cincher (773) and corset (897), caused a growth defect selective for dorsal trunk cells: homozygous mutant dorsal trunk cells were less than half normal size, whereas mutant terminal cell clones were unaffected (Figure 3E). This selectivity contrasts with that of the ∼500 house-keeping mutants, many of which caused their most pronounced effects on terminal cells, presumably because of the dramatic growth and branching these cells undergo during larval life. Thus, cincher and corset are growth genes required in dorsal trunk but dispensable in terminal cells.
Class 3: branch number, pattern, size, and shape mutants
These mutations affected the number of terminal branches, and in some cases also the position at which new branches bud from the parental branches. Many also affected the shape of the buds and mature branches. Mutations caused different but characteristic spectrums of defects so that, for example, mutations that reduced terminal cell branching to a similar extent could reproducibly give rise to terminal cells of very different morphology, such as short and thick versus elongate and wispy.
Terminal branch pruning (3A)
We identified ∼40 mutants in which terminal cell clones had fewer branches than normal but were distinct from the general cell growth genes described above because mutant dorsal trunk cells were of normal size and morphology. This phenotype is similar to that of blistered (pruned), the canonical terminal branching gene [14],[44]. winded alleles (613,1227,1375,1508) showed severe pruning defects, like blistered null alleles: mutant terminal cells had few branches, and any residual branches were typically thin and wispy and lacked subcellular tubes (Figure 4A, 4B). Other severe pruning mutants were paltry and topiary. Mutations in most other genes of this class showed more modest pruning defects (e.g. lopped, truncated), comparable to blistered partial loss of function alleles.
Fig. 4. Terminal cell branching mutants. 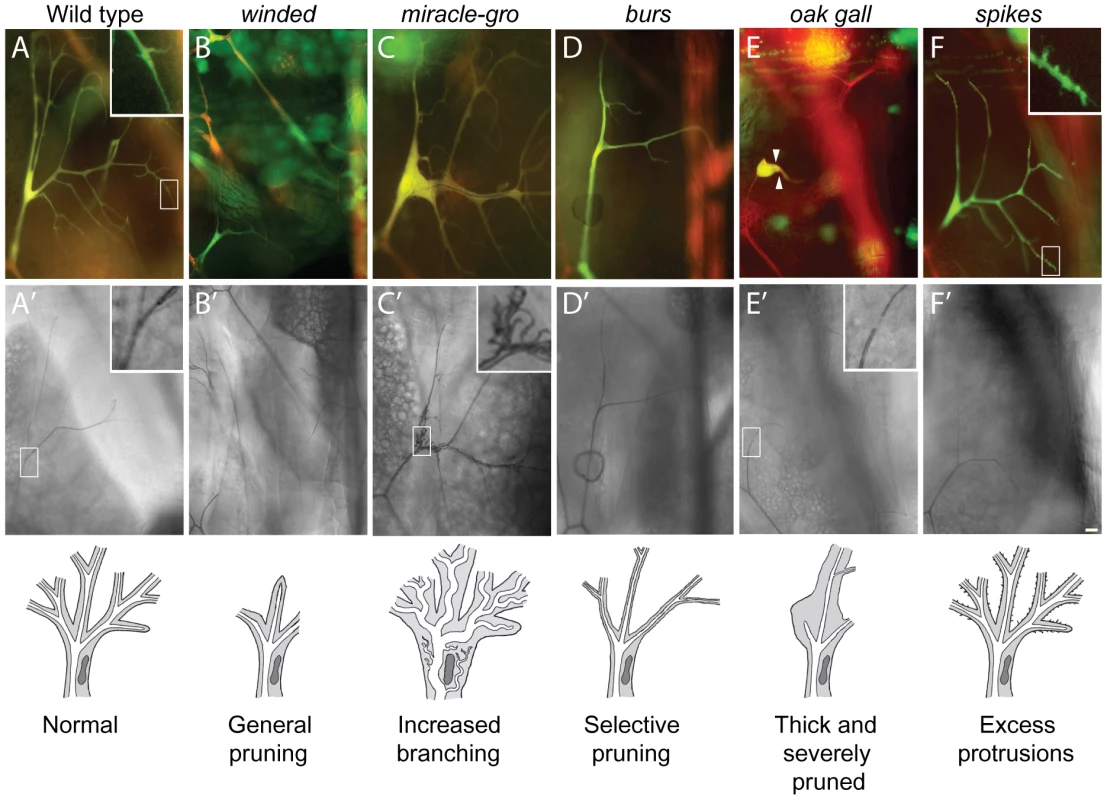
Fluorescence (A–F) and brightfield (A'–F') images of homozygous terminal cell clones (DsRED+, GFP+ so appear yellow in A–F) of the mutations indicated, with schematics of the phenotypes shown below. Open boxes, area enlarged in insets. (A, A') Control wild type clone. There are dozens of terminal branches (A), and each mature branch contains a single, continuous gas-filled lumen (A'). New terminal branches arise from filopodial growth cones (A, inset). (B, B') winded1508 clone. Note absence of terminal branches. (C, C') miracle-gro1483 clone. Note enlarged branches and multiple convoluted seamless tubes in enlarged soma (C', inset). (D, D') burs1139 clone. Note presence of first generation terminal branches but absence of most second and all subsequent generations. (E, E') oak gall696 clone. Note all but one terminal branch is missing, and remaining branch is short and stout (arrowheads). Another phenotype is the tiny gap in the gas-filled lumen at or near the position where autocellular and subcellular tubes connect in terminal cell (E', inset; compare to inset in A'). (F, F') spikes773 clone. Note excess filopodia arising from terminal branches (F, inset) but normal or slightly reduced numbers of mature terminal branches (F'). Bar, 20 µm. Excess branching (3B)
Although many mutations that reduced terminal branching were identified, mutations that increased branching were rare. Indeed, the only mutations that increased branching were the miracle-gro alleles described above, which also increased cell size. Although some extra branches observed in miracle-gro terminal cells might simply be terminal branches that are normally present but bigger than normal and hence easier to detect, there clearly were also extra branches not present in wild type, such as those in proximal positions of miracle-gro terminal cells. In many cases, these extra seamless tubes coursed through the soma of the terminal cell, giving it a striking multiple lumen phenotype (Figure 4C′, inset).
Branch pattern and shape alterations (3C)
Eight mutations affected terminal cell branch pattern or shape and included mutations that caused selective pruning (category 3C1, e.g. burs), thick and severely pruned branches (3C2, e.g. lotus), and excess protrusions (3C3, e.g. spikes), as detailed below.
burs (942, 1139) mutant terminal cells ramified much less extensively than wild type, but the branching defect appears selective for side branches and possibly other later rounds of terminal branching (Figure 4D). Likewise, in the complementation group denuded (PC213, 1520) the first terminal branches appeared largely normal, although sometimes thickened, but subsequent branches were absent or severely compromised: small, unbranched, and with a narrow bore tube. These mutants demonstrate genetic differences between early and later rounds of terminal branching.
Terminal cells mutant for lotus (312), conjoined (356), and oak gall (696) were severely pruned, in the most extreme cases with just a single significant branch, and the remaining branches were unusually thick (Figure 4E). However, lumens within the thickened branches were of normal diameter.
spikes (735) mutant terminal cells had variable numbers of cytoplasmic protrusions emanating along the length of terminal branches (Figure 4F, inset), protrusions that in wild type are typically restricted to the growing tip and sites of lateral sprouting (Figure 4A, inset). Most of the excess protrusions, however, were short and nonproductive as spikes mutant terminal cells had slightly fewer mature terminal branches than normal.
Class 4: tube formation, number, position, and shape mutants
These mutations prevented lumen formation, or altered the number, placement, or shape of the lumens that formed. Most of these mutations did not affect all tracheal tubes, but rather structurally distinct subsets of tubes. We start with a description of mutations that affect the seamless tubes of terminal branches (see Figure 1B; Figure 5A–5F; Figure 6B).
Fig. 5. Tubulogenesis mutants. 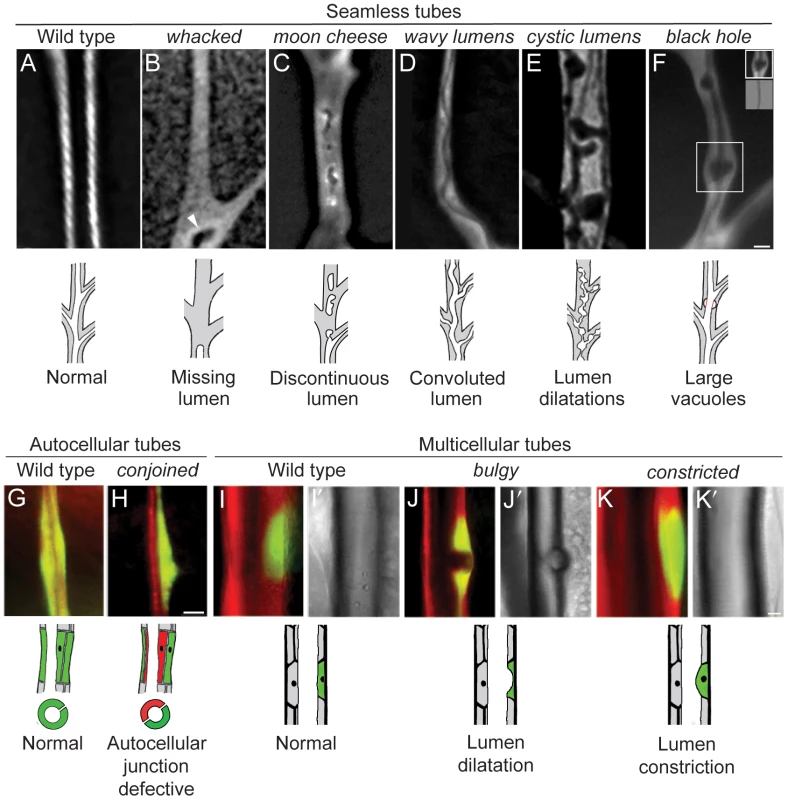
Fluorescence photomicrographs of control wild type (A, G, I) and homozygous mutant (B–F, H, J, K) clones in seamless, autocellular, and multicellular tracheal tubes in third instar larvae. Schematics of the phenotypes are diagrammed below. Clones are marked with GFP (white in A–F, green in G–K) and all tracheal cells with DsRED (red in G–K); brightfield images in I'–K' show air-filled lumens of multicellular tubes. (A) Wild type control clone in seamless tube. (B) whacked220 clone. Note most of the lumen is missing and the terminus of the residual lumen (arrowhead) is dilated and irregularly shaped. (C) moon cheese1524 clone. (D) wavy lumens894 clone. (E) cystic lumens1243 clone. (F) black hole538 clone. The regions where the lumen appears to be dilated (e.g., boxed area, upper inset) are actually regions in which a vacuole, which can be distinguished from the lumen by its accumulation of lumGFP (not shown), intimately surrounds a lumen of normal diameter (lower inset, brightfield view of boxed area). The vacuole is outlined in red in schematic. (G) Wild type control clone in autocellular tube. The single marked cell (GFP+, green) surrounds the lumen, sealed by an autocellular junction. (H) conjoined356 clone. The mutant cell (GFP+, green) does not form an autocellular junction but instead forms the lumen by making intercellular junctions with a heterozygous cell (DsRED+, red). (I) Wild type control clone in dorsal trunk, a multicellular tube. (J) bulgy636 clone. Lumen bulges outward into mutant cell, forming a local dilatation. (K) constricted960 clone. Lumen constricts inward at site of mutant cell by ∼7% relative to the neighboring, fully wild type dorsal trunk segments. Bar, 5 µm (A–F), 10 µm (G,H), 10 µm (I–K). Fig. 6. Lumen clearance and gas-filling mutants. 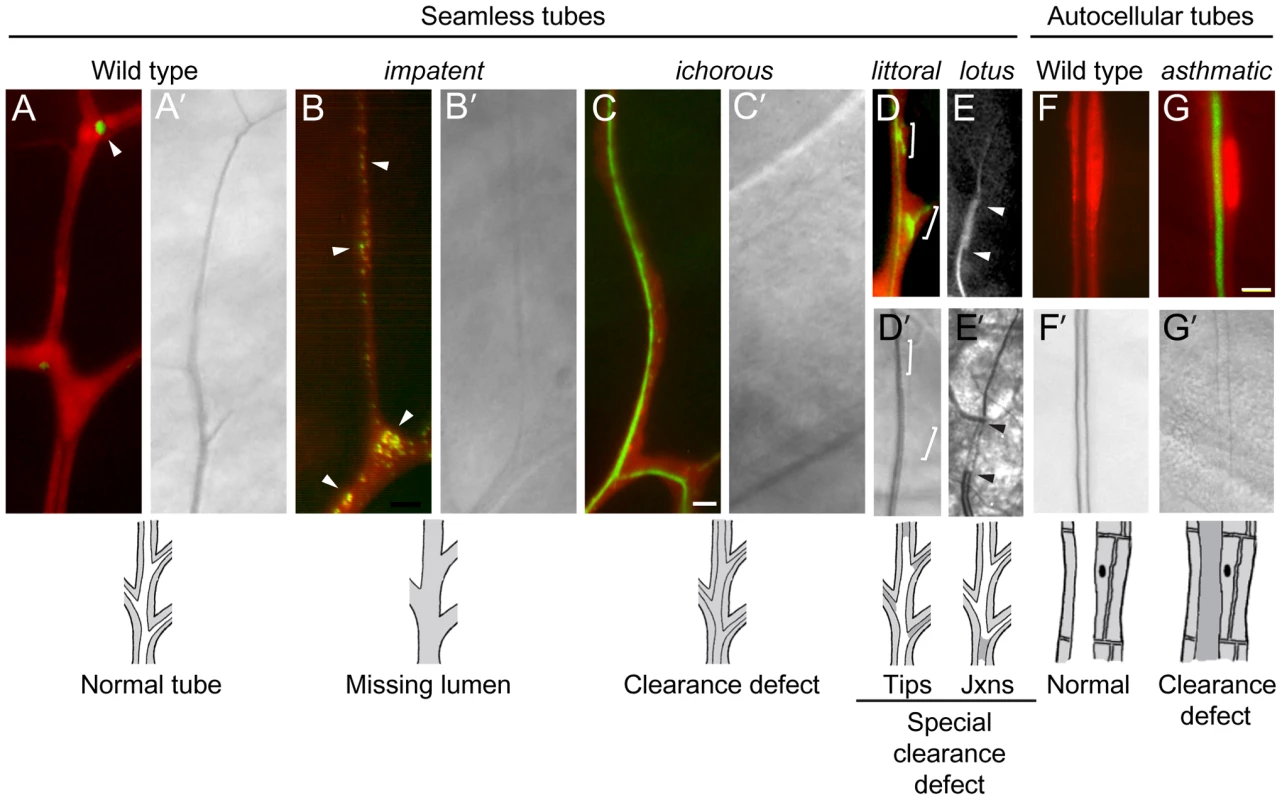
Fluorescence (A–G) and bright field micrographs (A'–G') of control wild type (A, F) and homozygous mutant clones (B–E, G) in seamless and autocellular tracheal tubes as indicated. Clones are labeled with cytoplasmic DsRed (red) and also express lumGFP (green), a secreted form of GFP; the fluorescence micrographs (A–G) are DsRed/lumGFP merged images, except for E, which shows only the lumGFP channel (white). Lumen defects are diagrammed below, with air-filled lumens in white and matrix-filled lumens and tracheal cell cytoplasm in grey. (A, A') Wild type control terminal cell. lumGFP has been cleared from the mature, gas-filled lumen (A'). The only lum-GFP visible is small puncta in the cytoplasm at the tip (A, arrowhead). (B, B') impatent1757 clone. This is a mutant, like those described in Figure 5, in which the seamless lumen is missing (B'): lumGFP is detected only in puncta (B, arrowheads), presumably aberrant intermediates in lumen formation, distributed in the soma and along the lumenless terminal branch. (C, C') ichorous206 clone. Although no mature, gas-filled lumen is detected by brightfield optics (C') as in impatent mutant cells, a lumen has formed–just not cleared–as shown by luminal lumGFP staining (C). (D, D') littoral762 clone. A specialized clearance defect: the central terminal branch forms a normal gas-filled lumen but the tips of growing side branches (brackets) contain a lumen that has not cleared (D') and remains loaded with lumGFP (green, D). (E, E') lotus312 clone. Another specialized clearance defect, restricted to the junction between (arrowheads) the base of the branch (connection with stalk cell) and the seamless tube. The fluorescence signal in E above and below the arrowheads is autofluorescence of the cuticle, not lumGFP. (F, F') Control wild type autocellular tube. (H, H') asthmatic1530 clone. Lumen is difficult to detect (G') because it remains filled with luminal matrix and lumGFP (green, G). Bar, 5 µm (in C, A–E), 10 µm (F,G). Seamless tube defects (4A)
These mutations caused defects in seamless tubes including missing and discontinuous lumens (category 4A1, e.g. impatent), convoluted lumens (4A2, e.g. wavy lumens), multiple convoluted lumens (4A3, e.g. tendrils), lumen dilation and other irregularities (4A4, e.g. cystic lumens), and large vacuoles (4A5, e.g. black hole), as detailed below.
We recovered ∼25 lines in which gas-filled lumens were not detected in terminal cell clones by brightfield microscopy. Some of these mutants are defective in lumen formation as assessed by a secreted GFP reporter (lum-GFP), whereas others form lumens that are difficult to detect by brightfield microscopy because they remain filled with matrix (see below). The three alleles of impatent (1472, 1490, 1757) lacked or had seriously compromised seamless tubes, whereas other aspects of terminal branches appeared normal and lumens of other branches were unaffected. In impatent mutant terminal cells, the lumGFP reporter accumulated in large puncta, up to 1–2 um in diameter, located mostly in the cell body (Figure 6B). These were probably enlarged vesicles representing trapped or aberrant intermediates in lumen formation.
Some of the other mutations in this class displayed a more complex spectrum of defects. whacked mutations (PC24, 220) eliminated the distal portions of seamless tubes, with the lumen typically terminating prematurely in an irregularly-shaped local dilatation (Figure 5B). moon cheese (1524) and carbuncle (804) caused a variable discontinuous lumen phenotype in which regions along the terminal branch were missing lumen but flanked by regions containing blind-ended and irregularly-shaped lumen (Figure 5C). These mutations also caused a fully penetrant large vacuole phenotype (see below).
In wavy lumens (894) and wobbly lumens (BG13), terminal cell lumens formed but followed a convoluted path through the cytoplasm (Figure 5D). Similar convoluted lumens were seen in miracle-gro mutant terminal cells and cells exposed to hypoxia [45]. However, the convoluted lumens of wavy lumens and wobbly lumens were not associated with excessive terminal cell growth and branching as in miracle-gro mutants and under hypoxia: wobbly lumens terminal cells were of normal size and wavy lumens terminal cells were mildly pruned.
miracle-gro mutations caused multiple convoluted lumens in the soma of mutant terminal cells, along with the extra growth and branching described above. Eight additional mutants were identified that had multiple, disorganized lumens but fewer branches than normal. Four compose the complementation group tendrils [46], and vine (512) defines another locus whose phenotype was remarkably similar to tendrils except that it began to manifest a day or so earlier in development. creeper, braided and ivy had similar but less penetrant phenotypes.
Several mutants had irregular but gas-filled tubes. As described above, mutations in whacked had irregularly shaped, and often prematurely terminated tubes. cystic lumens (1243) caused areas of lumen dilation and constriction (Figure 5E) and occasional gas-filling defects. The tube morphology defects in these mutants most closely resemble those previously reported for larger tubes (dorsal trunk) in which the secretion or modification of chitin is affected [47], and more recently by mutation of receptor tyrosine phosphatase activity [48].
Other mutations caused large cytoplasmic vesicles that excluded cytoplasmic GFP in mutant tracheal cells. In black hole (538) mutant cells, the vacuoles were centered over gas-filled lumens, suggesting that transport into the lumenal space is defective (Figure 5F). black hole and dark matter (1417) caused vacuole accumulation specifically in tracheal terminal cells, whereas moon cheese (1524) and carbuncle (804) caused vacuole accumulation in all tracheal cells. For moon cheese and carbuncle, a secreted form of GFP (lum-GFP) accumulated to high levels within mutant cells, particularly within the vacuoles, suggesting a defect in trafficking of secreted proteins. moon cheese and carbuncle also caused a variable discontinuous tube defect described above.
Autocellular tube defects (4B)
lotus (312), conjoined (356), oak gall (696) interfered variably with the formation of tubes sealed by autocellular junctions (Figure 1B): mutant dorsal branch cells appeared either to completely avoid contributing to autocellular tubes, forming exclusively intercellular rather than autocellular junctions (Figure 5G, 5H), or contributed only minimally with most of the cell body protruding basally away from an otherwise smooth epithelial tube. Dorsal trunk clones also protruded basally, although still formed intercellular junctions (not shown). These mutations demonstrate that autocellular junctions and tubes are genetically distinct from intercellular junctions and multicellular tubes (Figure 1B).
Multicellular tube defects (4C)
These mutations caused defects in multicellular tubes, including lumen dilation (category 4C1, e.g. bulgy) and lumen constriction (4C2, e.g. short of breath), as detailed below.
small potatoes (1113, 1166, 1694), bulgy (636), and balloon (1448) caused subtle outward bulges in the lumen of dorsal trunk tubes (Figure 5I, 5J). Bulges occurred at the sites of clones, even single cell clones, hence these mutations identify cell autonomous regulators of multicellular tube diameter.
Eight mutations caused the opposite phenotype: dorsal trunk cells mutant for short of breath (5 alleles), dyspneic (1348, 1359), and constricted (960) caused lumenal constrictions at the sites of mutant cells (Figure 5K), reminiscent of the recently described mutant stenosis [49].
Class 5: lumen clearance/gas-filling mutants
We expected that some mutants from the screen that appeared to lack lumens would instead be lumen clearance and gas-filling mutants in which the lumen was present but difficult to detect by brightfield optics because it remained filled with matrix, which has a similar refractive index as the surrounding cytoplasm. To identify such mutants, lines from the screen that appeared under brightfield optics to lack lumens were subjected to a secondary screen using a transgene expressing a fusion protein containing the signal peptide of p23 [50] linked to GFP. The fusion protein (lumenal-GFP or lumGFP) was designed to transit the secretory pathway, as it does in mammalian cells [50], entering the tracheal lumen and remaining there to mark the lumen of mutants that affect lumenal clearance, such as ichorous and asthmatic (Figure 6C). By contrast, no lumenal accumulation of lumGFP was observed in mutants such as impatent described above that lack or have seriously compromised lumens (Figure 6B), or in wild type control clones because lumGFP is cleared from the lumen during the normal lumenal maturation process (Figure 6A). The only lumGFP that remained in impatent mutant clones was the large puncta already described (Section 4A1), and the only lumGFP detected in control clones was the rare puncta near branch tips or the junctions between branches (Figure 6A). In mutations such as scrub (659), lumGFP was not detected in either matrix-filled lumens or cytoplasmic puncta (data not shown); these mutations might alter lumenal targeting such that lumGFP is secreted from other positions in the cell so does not accumulate intracellularly.
Seamless tube clearance defects (5A)
These mutations caused liquid clearance defects in all seamless tubes (category 5A1, e.g. asthmatic), in the tips of seamless tubes (5A2, e.g. littoral), or at the junction between seamless and autocellular tubes (5A3, e.g. lotus), as detailed below.
Mutants such as asthmatic and ichorous (Figure 6C) described above, and stertorous and liquid-filled, were defective in matrix clearance: mutant terminal cells had normal morphology and formed seamless tubes, but the tubes failed to clear and gas-fill. These cells cannot supply oxygen to their targets, and neighboring wild type cells were frequently found invading the region normally supplied by the mutant cell; under normal conditions, terminal cell domains do not overlap [45], similar to neuronal tiling [51].
Seven mutants (littoral, burs, ivy, panting, 826, 928, 1809) had gas-filling defects that typically affected only new or distal portions of subcellular tubes (Figure 6D). The tip-clearance defects were variable, with some mutant terminal cells more severely affected than others, even within the same mosaic animal. These mutants suggest that the tips of terminal branches have specialized requirements for clearance and gas filling, or that these regions are particularly sensitive to defects in the general machinery.
lotus, oak gall, conjoined, and disjointed mutants showed an exquisitely specific clearance and gas-filling defect: in mutant terminal cells, no gas-filled lumen could be detected connecting the secondary branch tube, which has an autocellular tube, to the terminal branch seamless tubes, which lack junctions and extend throughout the rest of the terminal cell and gas-filled normally (Figure 4E′, Figure 6E and 6E′).
Other branches (5B)
Nine mutants representing four complementation groups showed dramatic defects in liquid clearance/gas-filling of tracheal tubes containing autocellular junctions. Two of the complementation groups, short of breath and dyspneic, were described above, because they also cause defects in dorsal trunk and terminal cells (sections 4C2 and 5A1). asthmatic mutations, which caused a strong terminal cell gas-filling defect as noted above, also caused a partially penetrant autocellular tube gas-filling defect (Figure 6F, 6G). flashflood (AP67) selectively blocked clearance of lateral tracheal trunks, as detected in homozygous third instar larvae.
Mutation mapping and gene identification reveal new cell biological pathways in tracheal development
To begin to define the molecular functions of tracheal genes identified in the screen, we mapped representative mutations and molecularly identified 14 of the genes (Table 2). Six of the identified genes (no tc clones-L, no tc clones-R, short of breath, dyspneic, lopped and failed fusions) were previously implicated in tracheal development. However, new functions were revealed for each by our clonal analysis. Two are allelic to canonical tracheal genes in the branchless/breathless FGF pathway, the breathless FGFR itself [26] and pointed [11], [59], which we showed are differentially required for competition during tip cell selection [13]. Two others, short of breath and dyspneic, which our results implicate in lumen clearance and gas-filling and as cell autonomous promoters of tube expansion, are allelic to krotzkopf verkehrt (kkv) and knickkopf (knk), chitin synthesis pathway genes previously shown to coordinate the behavior of cells in the tracheal epithelium during tube expansion [53], [54], [60], [61]. Our results demonstrate that chitin synthesis genes also have an unexpected, cell autonomous function in lumen clearance and gas filling of autocellular and seamless tubes. lopped784 is allelic to fatiga that encodes Drosophila Hif1 prolyl hydroxylase, and appears to be required in terminal cells for normal branching. Previous studies with hypomorphic fatiga alleles gave opposite results [56], although new studies indicate that early exposure to hypoxia (mimicked by loss of fatiga) result in stunted tracheal development while later exposure stimulates branching [64]. failed fusions is allelic to polychaetoid, which has been implicated in branch fusion and in tracheal cell intercalation, but our mosaic analysis, along with other new data from our lab, suggest another function for polychaetoid in tip cell selection (E. Chao, A.S.G. and M.A.K., unpublished data).
Tab. 2. Molecular identification of tracheal genes. 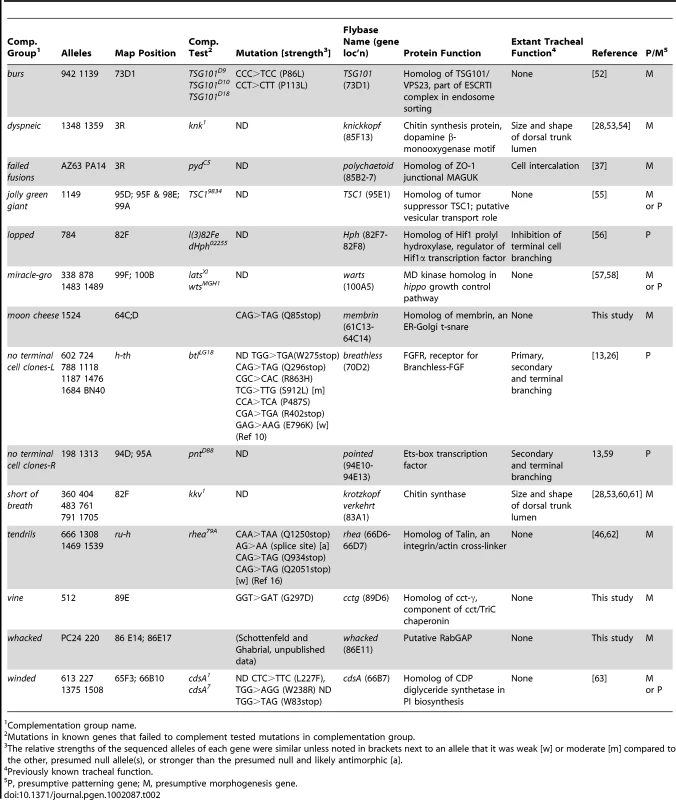
Complementation group name. The other eight molecularly identified genes had not been previously implicated in tracheal development; indeed, three (vine, moon cheese, and whacked) had not been genetically defined (Table 2). All eight identify new cell biological pathways in tracheal development. jolly green giant, which encodes the Drosophila ortholog of TSC1 [55], [65], and miracle-gro (see below) [57], [58] implicate general growth control pathways in tracheal growth and branching. tendrils, which is allelic to rhea and encodes talin [46], and vine, which encodes the Drosophila ortholog of CCTgamma, show that talin-dependent integrin adhesion and a component of the TriChaperonin complex are required for maintenance of terminal branches and lumenal organization.
The four other genes implicate membrane and vesicle trafficking genes in tracheal development. Such genes have been speculated to function in tube morphogenesis but few have been genetically identified. winded, essential for terminal branching, encodes the Drosophila homolog of CdsA [63], an enzyme that converts phosphatidic acid to cds-diacyl glycerol in the production of the membrane lipid, phosphatidyl inositol. moon cheese, another terminal branching gene also implicated in lumen continuity, and burs, a terminal branching gene selectively required for side branches, encode the Drosophila homolog of the ER-Golgi t-SNARE membrin, and TSG101/erupted, a component of the ESCRTI complex that sorts endocytic vesicles to the multivesicular body, respectively [52], [66], [67]. whacked, which promotes the growth and proper shape of terminal cell lumens, encodes a putative RabGAP (A.S.G. and M.A.K., unpublished data). The identification of membrane lipid and vesicle trafficking genes in terminal branching supports the idea that outgrowth of cellular processes and lumen formation require targeting of apical and basolateral membrane components at a distance from the cell soma. It will be important to determine the number of trafficking pathways involved, how the pathways are activated at the appropriate times and places, and how the identified t-SNARE, Rab-GAP, and ESCRTI component function in the pathways.
Thus, all 14 molecularly characterized genes from the screen reveal new cell biological pathways in tracheal development or new functions for established pathways.
Most of the identified genes are morphogenesis genes
The identities of the molecularly characterized genes allowed us to assess the success of the screen in identifying morphogenesis effectors. Although it was not possible to unambiguously classify all 14 genes in this way from their sequence alone, eight very likely function as morphogenesis effectors: the vesicle trafficking genes moon cheese/membrin, burs/TSG101, and whacked/RabGAP; the cell junction and cytoskeletal genes failed fusions/polychaetoid/ZO-1, rhea/tendrils/talin, vine/cctγ; and the chitin synthesis genes short of breath/kkv/chitin synthase and dyspneic/knk. Three others are established patterning genes: the receptor btl/no-terminal cell clones-L, the transcription factor pnt/no terminal cell clones-R, and the transcription factor regulator lopped/fatiga/Hif prolyl hydroxylase. The remaining three are more difficult to categorize because they encode enzymes that likely couple patterning signals to cytoplasmic outgrowth (winded/(CdsA) and cell growth (Tsc1/jolly green giant and miracle-gro), as discussed below. Thus, over three-quarters of the identified genes (11 of 14, 79%) appear to be downstream effectors/morphogenesis genes (8 of 14, 57%) or genes that couple patterning signals to morphogenesis (3 of 14, 21%), supporting our hypothesis that systematic clonal analysis is an effective way of identifying such genes.
A cell growth regulator that also regulates lumen morphogenesis
In addition to identifying new tracheal genes and pathways, the screen suggested new functional connections between pathways. One example came from characterization of the miracle-gro cell overgrowth mutations (Section 2A). In terminal cells, not only was the soma enlarged but there were many ectopic seamless tubes coursing through it (Figure 4C, 4C′ and Figure 7B). This phenotype is nearly unique: it is seen otherwise only upon hyperactivation of the Breathless FGFR pathway (Figure 7C). However, miracle-gro mutations did not map near breathless or any other extant loci in the pathway. Mapping and complementation tests (Table 2) demonstrated that miracle-gro is allelic to warts/lats-1 [57], [58], which encodes a kinase that suppresses cell growth in a well-established general growth control pathway. Thus, loss of a key growth regulator in terminal cells leads not only to excessive cell growth but excessive lumen formation, revealing an unexpected coupling between cell growth control and tubulogenesis.
Fig. 7. Genetic analysis of terminal cell growth control pathway. 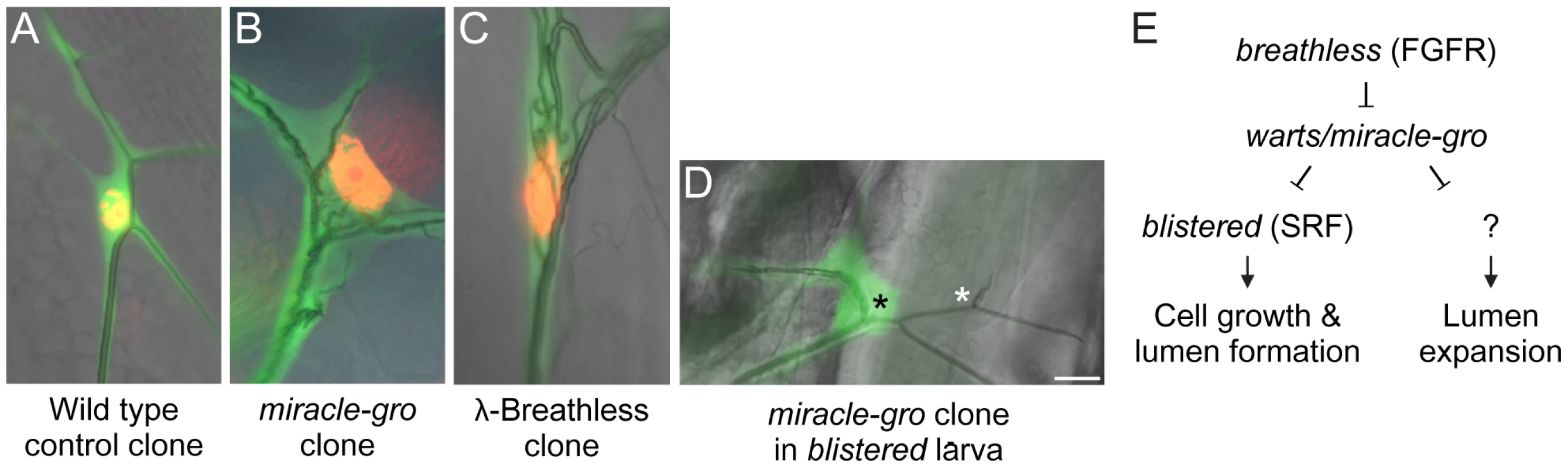
(A–D) Close-ups of the soma of third instar larva terminal cell clones of the indicated genotypes. Terminal cell cytoplasm is marked with GFP (green) and nuclei in A–C are marked with nuclear DsRed2 (red). Note that the cell body and nucleus of the miracle-gro(warts)388/388 clone (B) and the clone expressing λ-Breathless (C), a constitutively active form of Breathless FGFR, are enlarged with ectopic lumens coursing through the soma. By contrast, the soma of the miracle-gro(warts)388/388 clone in a larva homozygous for blistered l(2)3267, a downstream transcription factor in the Breathless pathway (D), is smaller and there are no ectopic lumens (black asterisk). However, the single, truncated lumen of the clone is dilated compared to the truncated lumen of the contralateral control terminal cell (white asterisk). (E) Genetic pathway of terminal cell growth control. Bar, 20 µm. The striking similarity of the warts/miracle-gro loss of function phenotype and the btl pathway gain-of-function phenotype suggested that FGF signaling might stimulate terminal cell growth and tubulogenesis by inactivating Warts function. To test this, we sought to define the genetic epistasis relationship between warts/miracle-gro and breathless-FGFR pathway mutations. Because mutations that disrupt FGF signaling abrogate terminal cell specification [11], [29], it is not possible to generate terminal cells doubly mutant for warts and breathless, so we examined terminal cells doubly mutant for warts/miracle-gro and blistered/pruned/SRF, the downstream transcription factor in the breathless FGFR pathway required for terminal cell growth and branching. Doubly mutant cells were unbranched and small, similar or slightly bigger than blistered mutant terminal cells, and with a single lumen in the soma (Figure 7D). However, the lumen diameter was larger than normal and similar in size to those in warts/miracle-gro mutant terminal cells. Thus, the cell growth and excessive lumen formation seen in warts/miracle-gro mutant terminal cells are dependent on blistered, whereas lumen diameter can be modulated independently of blistered. This supports a model in which Warts/Miracle-gro functions downstream of Breathless FGFR but upstream of Blistered/SRF in the regulation of terminal cell size and lumen number, and upstream of another, as yet unidentified, transcription factor that controls lumen diameter (Figure 7E).
Discussion
Our systematic screen for tracheal mutations on the third chromosome identified new tracheal phenotypes and scores of new tracheal genes as well as new functions for established tracheal genes. Molecular identification of 14 of the genes indicates that most of the isolated genes are downstream effectors/morphogenesis genes, an important category of genes substantially underrepresented in previous screens. Several of the identified genes encode proteins involved in vesicle trafficking, implying that such genes are a major class of morphogenesis genes, at least for terminal branching.
Our screen succeeded in identifying morphogenesis genes because (i) it was systematic and extensive, surveying most third chromosome genes, nearly 40% of the genome; (ii) it included a clonal analysis of mutations using a new cell marking method that allowed facile identification of tracheal functions of pleiotropic mutations such as vesicle trafficking and cytoskeletal genes; and (iii) it employed a secondary screen in another tissue (eye) to exclude mutations in general housekeeping genes. Housekeeping genes are a huge class of genes that would have dominated the results of our clonal screen, as they have in previous clonal screens [35]. Exclusion of housekeeping genes also allowed the identification of tracheal-selective growth regulators, which contributed to our discovery of cell growth control as an important new step in branching (see below).
Many of the mutations we identified, particularly in the clonal analysis, affect terminal branching, a morphogenesis process for which little was known beyond the key signaling pathway and transcription factors that control terminal cell selection. The systematic nature of our screen and the distinct phenotypes of terminal branching mutations we identified provide a comprehensive genetic outline of this morphogenetic process (Figure 8). We propose that terminal branching involves an initial cell selection and specification step plus five major morphogenesis processes: branching, growth, tubulogenesis, gas filling, and maintenance. Each of these processes is associated with one or two defining genes that are required quite generally for the process plus additional genes, mutations in which further subdivide each process into distinct morphogenetic steps or reveal additional levels of regulation. Below we discuss each of these processes and the associated genes, highlighting cell growth regulation because its critical role in branching morphogenesis had not been recognized. We return at the end to discuss implications of the results for the corresponding processes in primary and secondary branching.
Fig. 8. Genetic dissection of terminal branch morphogenesis. 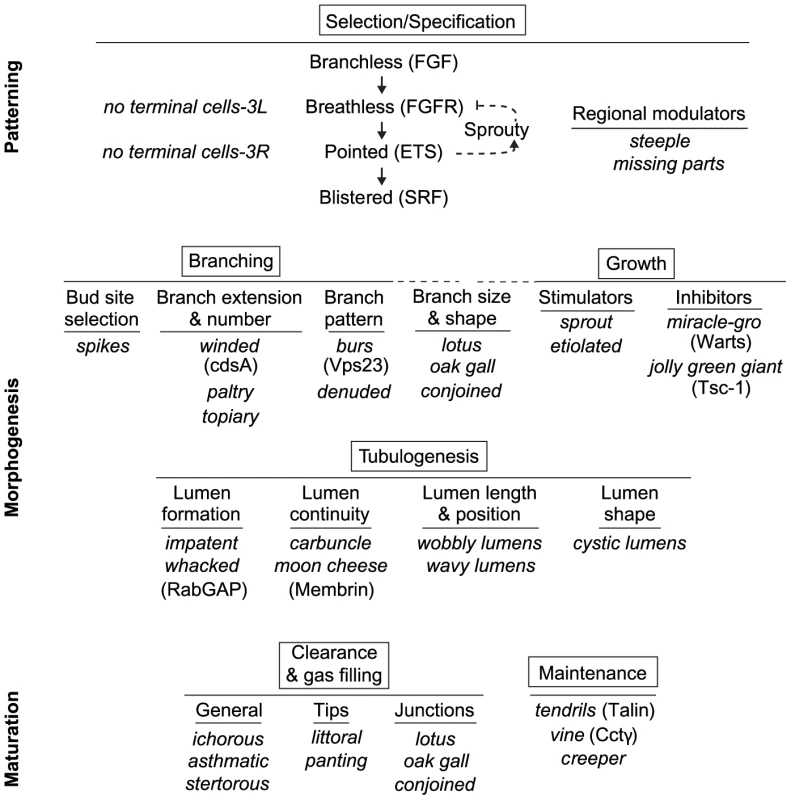
The major, genetically separable processes in the terminal branching program are illustrated, in the order in which they occur, along with representative mutations that disrupt them. There is an initial patterning step (Selection/Specification) that selects and specifies the terminal cell, followed by five morphogenesis (Branching, Growth, Tubulogenesis) and maturation (Clearance/Gas-Filling, Maintenance) steps. The steps can be functionally subdivided further by the more specific phenotypes of the mutants shown. Where the molecular identities of the genes are known, the protein products are given (in parentheses) to indicate some of the molecular functions involved in each step. The SRF transcription factor Blistered (Pruned), a key regulator of terminal branching and the last gene in the Selection/Specification step, presumably controls expression of at least some of the downstream morphogenesis and maturation genes including ones involved in growth and tubulogenesis (Figure 7E). Cell selection and specification
The earliest step of cell selection and specification in terminal branching is a well-characterized process that previous genetic studies have shown is controlled by the Branchless FGF pathway (Bnl/Btl/RAS/Pointed) that induces expression of the Blistered/Pruned SRF transcription factor that selects cells at the ends of budding branches for a terminal branching fate [9], [10]. Re-expression of Bnl FGF later in hypoxic tissues is proposed to initiate terminal branch budding, at least in part by activation of the blistered SRF transcription complex and its downstream effector genes [14], [45].
Our screen identified third chromosome genes previously implicated in the selection process, but revealed an interesting new aspect of the process because of the novel “no terminal cell clones” phenotype. These turned out to be mutations in breathless FGFR and pointed [13], and lead to the discovery there is specialization among cells in a budding branch and only the leaders need to receive the Branchless FGF signal. Cells mutant for Breathless FGFR cannot receive the signal, and are relegated to trailing positions, never to be specified a terminal cell.
We also discovered genes (steeple, missing parts) required for specification of a subset of terminal cells in specific regions or branches. These may encode region-specific enhancers of the Bnl-Btl pathway because sporadic failure of terminal cell formation is seen in animals in which this signaling pathway is partially compromised [11], [13], [34].
Branch budding and extension
Although the Branchless pathway and Blistered SRF transcription complex are key regulators of branch budding and outgrowth [14], [29], little is known of the signal transduction pathway that connects them or of the downstream effectors. winded/cdsA mutations caused a severe, cell autonomous terminal branch pruning phenotype similar to that of blistered/SRF null alleles. winded/cdsA encodes an enzyme (CDP-diacylglycerol synthase) required for phosphoinositide (PI) synthesis, suggesting that a PI-dependent signaling process, presumably like those involved in other receptor tyrosine kinase (RTK) signaling pathways [68], functions downstream of the Btl RTK in the control of Blistered/SRF and branch budding. Some of the other genes with pruned phenotypes (e.g., topiary, paltry, truncated) might encode additional signal transduction components or targets of the SRF transcription factor required for polarized cell growth (see below).
We propose that other branching genes regulate bud site selection and the pattern of branching. spikes encodes a negative regulator of bud site selection because small ectopic buds form in mutant terminal cells. One appealing idea is that spikes restricts the normal budding response of terminal cells to the sites of maximal induction by Branchless FGF. TSG101/erupted/burs and denuded regulate branch pattern by promoting lateral and late rounds of terminal branching, perhaps by catalyzing the local disassembly or reorganization of the cytoskeleton within a maturing terminal branch.
One set of genes (lotus, oak gall, conjoined) affected branch number but also dramatically altered the size and shape of the remaining branches (see below). These were difficult to categorize purely as branching genes or growth regulators, so we place them in a special class at the boundary between those categories because they share features of both. They may function as integrators of branch outgrowth and size control signals.
Growth regulation
A new aspect of the tracheal developmental program highlighted by the mutants is cell size and growth regulation. Outgrowth of terminal branches requires not only chemoattractant signaling to induce and guide migration, but synthesis of cellular and membrane components to support cytoplasmic outgrowth. Terminal cells mutant for glutamyl-prolyl-tRNA synthetase or hundreds of other presumptive house keeping genes failed to form and extend terminal branch buds. Many such growth-promoting genes were presumably among those identified in a previous clonal screen for tracheal mutations [35], but because there are many such mutations and their phenotypes are non-specific (small, sick, or missing cells) and difficult to distinguish from genes simply required for cell viability, it was hard to evaluate their developmental significance.
Three types of data argue that growth control is an integral part of the tracheal developmental program. First, clonal analysis of the master regulator trachealess in terminal cells gave a similar phenotype (Figure S1), implying that terminal cell growth is a process actively regulated by Trachealess. etiolated is a particularly interesting mutant of this class because it resembled trachealess not just in its clonal phenotype in terminal cells, but in its tracheal specificity. Second, we obtained mutations in two canonical growth suppressor genes, warts/miracle-gro and Tsc1/jolly green giant, which gave the opposite phenotype: terminal branches and terminal cells were overgrown with particularly large somas that in warts/miracle-gro mutant cells contained multiple seamless tubes passing through them. This shows that a general growth regulator controls not only cell size but tubulogenesis, an essential step in the tracheal developmental program. Third, the phenotype of warts/miracle-gro mutant terminal cells is very similar to that of activated Btl, and genetic epistasis experiments suggest that warts/miracle-gro functions downstream of, and is negatively regulated by, btl FGFR but upstream of blistered SRF (Figure 7E).
sprout is the most intriguing undergrowth mutant because it was the only one that formed small but normally patterned terminal branches. sprout cleanly decouples branch size from branch budding and outgrowth, so we propose it is a key gene in branch size control.
Three other genes, lotus, oak gall, and conjoined, also function in branch size control, but in a different way. In mutant cells, branches were much thicker and more variable than normal, but the diameter of the seamless tubes that form within them were normal. We propose that these genes function in the size control pathway by regulating the distribution of plasma membrane or other cell constituents among branches. When this process fails, branches become thicker and fewer in number.
Most of the undergrowth mutations and all of the overgrowth mutations affected not only terminal cells but other tracheal cell types and cells outside the tracheal system, implying that the affected genes encode general growth regulators. However, many undergrowth mutations had their most extreme effects on terminal cells, presumably because they are larger and grow more than other cells. But two mutations, cincher and corset, affected the growth of dorsal trunk cells and spared terminal cells. Thus, the growth control programs of these tracheal cell types are genetically separable. Because terminal cell growth appears to be controlled primarily or exclusively by Bnl-Btl signaling and operates selectively under hypoxic conditions, conditions that arrest the growth of most other cell types, cincher and corset might identify specific regulators or components of aerobic growth pathways or other general growth processes dispensable in terminal cells.
Tubulogenesis genes
The striking phenotype of impatent mutant terminal branches, branches that superficially appear normal but lack air-filled tubes and hence are nonfunctional, leads us to propose that impatent encodes a key regulator or component of terminal branch lumen formation. We further propose that lumen shape is governed by cystic lumens, perhaps in conjunction with whacked, mutations in which result in irregularly-shaped lumens, and that lumen length and position are controlled by wobbly lumens and wavy lumens, mutations that cause long and convoluted lumens. Long and convoluted lumens are also seen in terminal cells under hypoxic conditions and other conditions that cause excessive branchless FGF pathway activity and/or terminal cell growth (e.g. warts/miracle-gro), so an appealing model is that these conditions and this signaling pathway inhibit the activity of wobbly lumens and wavy lumens, which themselves function to restrict the length or the position of terminal branch lumens.
Lumen continuity requires carbuncle and membrin/moon cheese, suggesting that lumens of seamless tubes are made piecemeal and these genes promote their connection. We also identified four genes (disjoined, lotus, conjoined, and oak gall) required to make functional connections between seamless tubes and the adjacent, architecturally distinct autocellular tubes that form by wrapping. The short lumenal gap in these mutant terminal cells may result from a failure to connect the tubes or a structural defect that prevents clearance of the connection.
Lumen clearance and gas filling genes
For tracheal branches to become functional, the lumenal matrix must be cleared and replaced by gas, the molecular composition of which is unknown. ichorous and asthmatic are required for clearance and gas filling of most or all terminal branches, and littoral and panting and others are required to clear the tips of terminal branches. Recent studies highlight the importance of secretion into the lumen and subsequent endocytosis of lumenal matrix and liquid during tube expansion and air-filling of large multicellular tracheal tubes [16], [69]; the genes identified in our screen may mediate related processes in seamless tubes.
Maintenance genes
These genes maintain the elaborate shape and structure of terminal branches under mechanical stress such as muscle contraction. In the mutant rhea/tendrils/talin [46], terminal branches begin to form normally but branches break down and their lumens retract as the larva begins to move and the developing branches are subjected to the stress of stretch. The phenotypes of cctgamma/vine, creeper, braided, and ivy are similar to tendrils, suggesting that they function in the same integrin/talin cell adhesion and cytoskeletal support system. For example, the cctgamma/vine chaperonin may facilitate the folding or assembly of talin or some other component in the support system, an appealing hypothesis given that CCT chaperonins have been shown in other systems to mediate the folding of cytoskeletal proteins [70].
These maintenance genes emphasize the importance of analyzing the onset and evolution of a mutant phenotype when elucidating gene function, because similar phenotypes can arise from early developmental aberrations or later defects in maintenance. Other elaborate cell types and organs likely also require maintenance genes. For example, mutations in mouse Dlg5 perturb delivery of adherens complex proteins to the plasma membrane of brain and kidney epithelial tubes, resulting in cyst formation [71]. Many such structural maintenance genes are expected to function late in life so would be missed in typical developmental screens. A major effort should be aimed at their identification and isolation because of their importance in medicine and disease.
Coordination and coupling of morphogenesis processes
How are the genetically separable terminal branch morphogenesis processes described above coordinated and controlled in time and space? An important part of this control and coordination almost certainly involves the Branchless FGF pathway. Expression of both the ligand branchless and the receptor breathless are induced by hypoxia during larval life, and terminal cell outgrowth and branching are stimulated and directed to hypoxic cells by local production of Branchless FGF [56]. One way Bnl-Btl signaling stimulates branching is likely through transcriptional induction of morphogenesis genes via modification and activation of the Blistered /SRF transcription complex. In other systems, the SRF transcription complex has been shown to be regulated by the actin cytoskeleton and to regulate cytoskeletal genes [72], and such genes are almost certainly required for growth of the actin-rich terminal branch buds. It will be important to determine which of the identified morphogenesis genes are regulated by Branchless signaling and SRF, and to identify the full set of downstream targets by transcriptional profiling.
Because Branchless functions as a chemoattractant, it must also provide a spatial cue that guides terminal branch outgrowth. One appealing idea is that the ligand-bound Breathless FGFR at the surface of the terminal cell generates a spatial cue that directs polarized growth of cytoplasmic extensions toward hypoxic, FGF-secreting cells. Such a spatial cue could be used to direct vesicular traffic to the growing ends of terminal cell extensions, both for polarized growth of the new branch and construction of a lumen within it. The spatial cue might be a modified membrane PI because PI signaling functions downstream in many RTK signaling pathways [68] and can regulate vesicle trafficking [73] and tubulogenesis [74], and mutations in the PI synthesis gene cdsA/winded severely abrogated terminal branching.
Although Branchless-Btl signaling likely controls and coordinates many of the events in terminal branch morphogenesis, it is unlikely to be the sole control and coordination mechanism because not all of the morphogenesis events occur at the same time and place. Lumen formation (tubulogenesis) occurs after cytoplasmic outgrowth, and lumen maturation including clearance and gas filling occur even later, in some cases days after the lumen has formed. Likewise, branch and lumen maintenance are late steps in the process. Although there may be delays built into some of the effector pathways downstream of Breathless to stagger its effects, other factors likely also contribute to the timing and spatial organization of the events. For example, ecdysone signaling may gate the timing of lumen clearance and gas filling, and there are presumably cell intrinsic cues that direct transport vesicles carrying integrins and other basolateral markers to the plasma membrane of growing buds, and vesicles carrying apical markers and lumenal components to internal positions.
One surprising finding of our clonal analysis of known tracheal genes was that terminal cell clones of the tracheal master regulator trachealess gave a pruned phenotype. This implies that trachealess is required not only for its well established role in the initiation of tracheal development [23], [24], but also for much later steps in the developmental program such as terminal cell growth and branching. Perhaps it functions in conjunction with SRF and other cell type and stage-specific transcription factors in the program to impart tracheal specificity in the control of downstream effector genes, as shown for the C. elegans pharyngeal master regulator pha-4 [75].
Primary and secondary branch morphogenesis genes
We identified a number of genes required for proper formation of the larger branches of the tracheal system that form earlier than terminal branches and from which they arise. For example, we identified mutants required for proper shape of multicellular tubes, including mutations that cause tracheal dilatations (small potatoes, bulgy, balloon) and others that cause local constriction of multicellular and autocellular tubes (kkv/short of breath, knk/dyspneic, constricted). An especially intriguing set of genes (lotus, conjoined, oak gall) are those required to form autocellular junctions and lumens. These may encode specialized components or regulators of autocellular junctions and tubes, such as proteins required for a cell to wrap on or seal to itself.
Although we identified some primary and secondary branch morphogenesis genes, there was a surprising paucity of such genes relative to the large number of terminal cell branching genes identified; a similarly skewed distribution obtained in a second chromosome screen, if the large number of putative housekeeping genes is excluded [35]). Although it is possible that morphogenesis of these larger branches requires fewer genes, more likely such genes were just not as efficiently identified in our screen. One reason is that some such genes only show a tube phenotype when most or all cells in the branch are mutant, as with breathless (Figure 2B) and grainyhead mutations [76]. Another reason is that perdurance of maternally expressed gene products likely obscures early functions of some genes. Finally, terminal cells have an elaborate structure that may make them more sensitive to mutations and makes phenotypes easier to detect.
The number of genes required to build the tracheal system
Our systematic genetic dissection of an organogenesis process, including a clonal analysis to identify tracheal genes with pleiotropic functions, allows an estimate of the number of genes required to build an organ–an important question not just for developmental biology but for medicine and tissue engineering. Because we identified ∼70 tracheal genes on the third chromosome (Table 1 and Table S3), which represents ∼40% of the genome, the full genome likely contains roughly two hundred genes required to construct the larval tracheal system. This almost certainly represents a lower limit because our screen did not achieve full saturation and, as described above, the screen would miss essential embryonic genes required non-cell autonomously and genes with a significant maternal contribution.
Genomic profiling of developing and mature organs indicates that there are hundreds of differentially expressed genes among different organs, and genetic profiling to identify downstream genes of organ master regulators such as the C. elegans pharyngeal regulator pha-4 [77], the mouse pancreas regulator Pdx1 [78], [79], and the Drosophila tracheal regulator Trachealess (E. Chao and M.A.K. unpublished data), suggests that there are 110–240 genes dependent on the master regulator for expression, at least at certain stages of development. Although it is not known how many of these downstream genes are required for organ morphogenesis, or what fraction of organ morphogenesis genes are both selectively expressed and downstream targets of organ master regulators, these genomic results are in line with the estimate from our genetic studies that organ morphogenesis programs, even ones for relatively simple organs like the Drosophila tracheal system, are likely to involve several hundred genes. The approach used here, involving a clonal analysis in the tracheal system of all mutations that do not survive late enough in development as homozygotes to assess their tracheal function, has begun to be extended to the other major chromosomes to identify the rest of the tracheal morphogenesis program [7], [35] (Metzstein M. and M.A.K., unpublished data); most of the identified mutations fit with the genetic scheme described here, with the exception of a novel set of mutations on the second chromosome that compromise terminal branch mutual avoidance and spacing [35]. The clonal approach could easily be adapted to other organs to systematically dissect additional organogenesis programs.
Materials and Methods
Drosophila strains
D. melanogaster strains used in the screen and meiotic mapping experiments were: (1) btl-GAL4, UAS-GFP; Pr, Hs-hid/TM3Sb, Tub-GAL80, (2) a newly isogenized btl-GAL4, UAS-GFP; FRT2A,FRT82B, (3) y w FLP122; btl-GAL4, UAS-DsRED; FRT82B cu UASi-GFPhp/TM6B, (4) y w FLP122; btl-GAL4, UAS-DsRED; UASi-GFPhp th st FRT2A/MKRS (5) y w ey-FLP; cell-lethal, GMR-hid FRT2A/MKRS, (6) y w ey-FLP; FRT82B, cell-lethal, GMR-hid/MKRS, (7) y w FLP122; breathless-GAL4, UAS-lumGFP, UAS-DsRED; FRT82B TubGal80, (8) y w FLP122; breathless-GAL4, UAS-lumGFP, UAS-DsRED; TubGal80 FRT2A; (9) a newly isogenized ru h th st cu sr e ca; (10) ru h th st cu sr e Pr ca/TM6B. Other strains were: FRT2A, FRT82B (from Trudi Schüpbach); Hs-hid (on chromosome III; from Ruth Lehman), onto which Pr was recombined; TM3Sb, Tub-GAL80 (from Stefan Luschnig); and strains used in complementation tests and deficiency mapping experiments (see Table S3). All other strains, except the mutants isolated here, have been described (http://flybase.bio.indiana.edu) and are available from http://flystocks.bio.indiana.edu.
Vector and transgene construction
UAS-DsRed. This Gal4-dependent DsRed transgene was constructed by inserting a 0.7 kb Kpn I-Xba I restriction fragment containing the DsRed coding sequences from pDsRed (Clontech) between the corresponding sites of the vector pUAST [80]. The resultant plasmid, pUAST-DsRed, was used to establish transgenic lines on the X, second, and third chromosome by P element mediated transformation of w1118 embryos. The second chromosome insertion (line 5A) was recombined with breathless-GAL4 and used here.
pUASTi. This P element vector for generating RNAi transgenes was constructed by PCR amplification (primers Xho+trh-intron F, Kpn+trh-intron B; see Table S2 for primer sequences) and TA cloning of the 221 bp third intron from the trachealess gene into the vector, pCRII-TOPO (Invitrogen). The intron fragment was then excised with Xho I and Kpn I and inserted at those sites in pUAST. Note that the trachealess intron contains an Eco RI site, leaving Bgl II, Not I and Xho I as the only unique restriction sites 5′ of the trachealess intron, and Kpn I and Xba I as the only unique sites 3′ of the trachealess intron. To generate RNAi constructs, a ∼500 bp fragment from the gene of interest is inserted in the forward orientation just upstream of the intron, and in the reverse orientation downstream of the intron, as described below for UAS-GFP(RNAi). Gal4-driven expression of this transgene results in tissue specific transcription of the self-complementary RNA, which is predicted to form a double-stranded “hairpin” conformation that initiates the RNAi response.
UAS-GFP(RNAi). This Gal4-dependent GFP(RNAi) transgene was created by PCR amplification (primers Not-GFP-F, Xho-GFP-R) and insertion of an ∼500 bp fragment of GFP (in the forward, sense orientation) between the Not I and Xho I sites upstream of the intron in pUASTi, and amplification of the same fragment (primers Xba-GFP-F, Kpn-GFP-R) and insertion (in the reverse orientation) between the Kpn I and Xba I sites downstream of the intron, to create plasmid pUASTi-GFP(RNAi). Transgenic flies were generated as above, and insertions were identified on all major chromosomes. For this study, an insertion on 3L (insertion B) was recombined onto ru h th st FRT2A (from Stefan Luschnig) to generate the UASi-GFPhp th st FRT2A chromosome, and an insertion on 3R (insertion 4A) was recombined onto FRT82B cu sr e ca (from S. Luschnig) to generate the FRT82B cu UASi-GFPhp chromosome.
UAS-lumGFP. This Gal4-dependent transgene expressing secreted (lumenal or “lum”) GFP was constructed by inserting an Nhe I-Kpn I restriction fragment with the GFP coding sequence from plum-GFP [50] between the Xba I and Kpn I sites of pBS-KS (Stratagene), and then subcloning the Not I/Kpn I lum-GFP fragment into those same sites in the pUAST vector. Transgenic flies were generated as above, and second and third chromosome insertions were recovered. A second chromosome insertion was recombined onto a breathless-GAL4 bearing second chromosome for use here.
Mutagenesis
A standard F3 EMS mutagenesis screen was performed (Figure 1C). Strains used were homozygous for breathless-GAL4 [81] and UAS-GFP transgenes on chromosome II. Males homozygous for an isogenized FRT2A, FRT82B chromosome III were fed 25 mM EMS as described [82] and mass mated to breathless-GAL4, UAS-GFP; Pr, Hs-hid/TM3, Sb, Tub-GAL80 virgin females. F1 males were each mated to two virgins of the genotype used in the P cross. After five days, parents were removed from the F1 cross and on days five and six F2 larvae were heated to 38°C for 1.5 hours to induce Hs-hid and eliminate animals carrying that chromosome. In the few cases (∼2–3%) where animals carrying Pr, Hs-hid survived, virgins of the appropriate genotype were selected to generate a stock of FRT2A, FRT82B*/TM3, Sb, Tub-GAL80 (*, newly induced mutation). If animals homozygous for the treated third chromosome (non-Sb) were not detected in the F3 or subsequent generations, a lethal mutation was assumed to be present. Two hundred mutagenized chromosomes were assayed in three small-scale pilot screens, and 4100 mutagenized chromosomes were assayed in a final large-scale screen.
F3 screen
Sibling F2 flies (described above) were allowed to mate and were brooded to produce two clutches of F3 individuals, one-quarter of which should be homozygous for the mutagenized third chromosome. The first F3 brood (after five days) was used to maintain the stock and assess for presence of a lethal mutation; the second F3 brood (at 12 days) was screened for tracheal phenotypes (see below). If less than four pairs of flies were obtained in the F2 generation, screening was postponed for a generation. For tracheal phenotype screening, F3 larvae were washed out of their food vials with distilled water and examined under an M2 Zeiss or a Leica fluorescence stereomicroscope. Homozygous third instar larvae were identified by tracheal expression of GFP, and tracheal morphology was analyzed; at least three homozygous larvae were examined for each mutant line. Animals that appeared to have tracheal defects were heat-killed (70–75°C for 3–5 s), mounted in 50% glycerol and examined under a Zeiss Axioplan 2 compound fluorescence microscope. Lines in which a tracheal defect was detected were retested to confirm the phenotype. If no reproducible phenotype was found, the line was discarded. Mutant lines that did not give rise to viable homozygous third instar animals were analyzed in genetic mosaics as follows.
Genetic mosaic screen
Males from the mutant stock established in the F3 screen were crossed to y, w, FLP122; breathless-GAL4, UAS-DsRED; UASi-GFPhp, th, st, FRT2A/MKRS virgin females, and to y, w, FLP122; breathless-GAL4, UAS-DsRED;FRT82B, cu, UASi-GFPhp/ TM6B virgin females, to test mutants on 3L or 3R, respectively. For each arm (3L and 3R) of every mutant stock, a cross with 20–40 pair matings was done. Embryos (0–4 hr old) were heat-treated as above for 0.75–1 hr to induce FLP-mediated recombination; animals 2 hrs old or less at the time of heat treatment typically do not survive. Crosses were maintained at 25°C for five days and then mosaic animals were examined under a fluorescence stereomicroscope. All tracheal cells are marked by expression of UAS-DsRED; mutant tracheal cells also express GFP. This GFP marking of mutant cells was achieved by inducing recombination between a chromosome arm carrying the mutation of interest and the homologous chromosome arm carrying UASi-GFPhp (see above). Daughter cells homozygous for the mutagenized chromosome arm lack the GFP(RNAi) transgene and thus express GFP. Animals of the correct genotype were selected, heat killed, and analyzed for tracheal defects as above. Under these clone induction conditions, ∼60±8 (mean +/ - SEM) tracheal clones were generated per animal (n = 5 animals). Among dorsal branch clones (n = 127 clones), 50% appeared to be composed of a single cell, 37% of two cells, 10% of three cells, and 3% of four or five cells.
EGUF/HID secondary screen
Mutations that caused undergrowth defects in tracheal clones were tested for growth defects in eye development using the EGUF/HID technique that generates eye imaginal discs composed exclusively of mitotic clones of a single genotype [43]. Males from mutant lines were crossed to virgin females of genotype y, w, ey-FLP; *, GMR-hid FRT2A/MKRS, or y, w, ey-FLP; FRT82B, *, GMR-hid/MKRS, where * is the undergrowth mutation. Adult progeny lacking Sb (carried on both of the balancer chromosomes used) were scored for eye size. Mutations unable to support normal eye development were presumed to affect general cell growth and viability genes (“housekeeping” genes) and were discarded.
Lumenal GFP secondary screen
Mutants with no detectable lumen under brightfield optics were tested for presence of liquid-filled or discontinuous lumens using the lumGFP transgene described above, which expresses GFP with a signal peptide that we found accumulates in liquid-filled tracheal lumens but is not detectable in normal, gas-filled lumens. Males from the mutant stocks were crossed to y w FLP122; breathless-GAL4, UAS-lumGFP; TubGal80 FRT2A/MKRS or y w FLP122; breathless-GAL4, UAS-lumGFP; FRT82B TubGal80/MKRS virgin females, and mosaic analysis was carried out as described above in the Genetic Mosaic Screen.
Quantitative analysis of phenotypes
Effect of miracle-gro338 on cell size was determined using ImageJ software to measure the maximal cross-sectional area in a stack of 2D optical sections through the soma of mutant and wild type control terminal cell clones (see Figure 3). Effect on dorsal trunk lumen diameter was determined by comparing lumen diameter between a section of tube containing a single mutant cell and the average diameter of the immediately anterior and posterior regions containing no mutant cells.
Mutation mapping and gene identification
Initial mapping was carried out by complementation tests against a panel of chromosomal deficiencies spanning the third chromosome, and by meiotic recombination mapping using visible recessive markers ru h th st cu sr e ca. Fine scale mapping was carried out using available single nucleotide polymorphisms (SNPs) [83], [84] and new ones specifically identified in this study, in conjunction with complementation tests with chromosomes carrying small, molecularly characterized deletions. The affected gene in the mapped interval was then identified by sequencing candidate genes and comparing their sequences to those in the isogenized parental chromosome to reveal new EMS-induced nonsense mutations or other mutations predicted to compromise gene function, and by complementation tests with extant mutations in the interval.
miracle gro/warts epistasis analysis
To compare the miracle gro/warts terminal cell phenotype to that of activated Breathless FGFR, the MARCM system [42] was used to generate marked (GFP+) clones of tracheal cells expressing λ-Breathless [85], a constitutively dimerized form of the protein, and marked cells at terminal cell positions were examined using a compound fluorescence microscope. To determine the genetic epistasis relationship between miracle-gro/warts and blistered/pruned/SRF, a downstream transcription factor in the Breathless pathway, virgins of the genotype y, w, hsFLP122; bsl(2)3267, btl-Gal4, UAS-GFP/CyO; FRT82B cu, UAS-GFP(RNAi) were crossed to males of the genotype bsl(2)3267, btl-Gal4, UAS-GFP/CyO; FRT82B miracle gro/warts338/MKRS. Mutant animals homozygous for bsl(2)3267 were identified by the strong pruned phenotype of unmarked (GFP-) terminal cells, and marked (GFP+) wts338 mutant terminal cell clones in these animals were examined and scored as above.
Supporting Information
Zdroje
1. AffolterMBellusciSItohNShiloBThieryJP 2003 Tube or not tube: remodeling epithelial tissues by branching morphogenesis. Dev Cell 4 11 18
2. HoganBLKolodziejPA 2002 Organogenesis: molecular mechanisms of tubulogenesis. Nat Rev Genet 3 513 523
3. LubarskyBKrasnowMA 2003 Tube morphogenesis: making and shaping biological tubes. Cell 112 19 28
4. NelsonWJ 2003 Tube morphogenesis: closure, but many openings remain. Trends Cell Biol 13 615 621
5. WieschausEF 1996 From molecular patterns to morphogenesis-The lessons from studies on the fruit fly Drosophila (Nobel Lecture). Angewandte Chemie International Edition in English 35 2188 2194
6. SpradlingACSternDBeatonARhemEJLavertyT 1999 The Berkeley Drosophila Genome Project gene disruption project: Single P-element insertions mutating 25% of vital Drosophila genes. Genetics 153 135 177
7. MetzsteinMMKrasnowMA 2006 Functions of the nonsense-mediated mRNA decay pathway in Drosophila development. PLoS Genet 2 e180
8. ManningGKrasnowMA 1993 Development of the Drosophila tracheal system. BateMMartinez AriasA The Development of Drosophila melanogaster Cold Spring Harbor, NY Cold Spring Harbor Press 609 685
9. GhabrialALuschnigSMetzsteinMMKrasnowMA 2003 Branching morphogenesis of the Drosophila tracheal system. Annu Rev Cell Dev Biol 19 623 647
10. CabernardCNeumannMAffolterM 2004 Cellular and molecular mechanisms involved in branching morphogenesis of the Drosophila tracheal system. J Appl Physiol 97 2347 2353
11. SamakovlisCHacohenNManningGSutherlandDCGuilleminK 1996 Development of the Drosophila tracheal system occurs by a series of morphologically distinct but genetically coupled branching events. Development 122 1395 1407
12. SamakovlisCManningGStenebergPHacohenNCanteraR 1996 Genetic control of epithelial tube fusion during Drosophila tracheal development. Development 122 3531 3536
13. GhabrialASKrasnowMA 2006 Social interactions among epithelial cells during tracheal branching morphogenesis. Nature 441 746 749
14. GuilleminKGroppeJDuckerKTreismanRHafenE 1996 The pruned gene encodes the Drosophila serum response factor and regulates cytoplasmic outgrowth during terminal branching of the tracheal system. Development 122 1353 1362
15. RibeiroCNeumannMAffolterM 2004 Genetic control of cell intercalation during tracheal morphogenesis in Drosophila. Curr Biol 14 2197 2207
16. TsarouhasVSentiKAJayaramSATiklovaKHemphalaJ 2007 Sequential pulses of apical epithelial secretion and endocytosis drive airway maturation in Drosophila. Dev Cell 13 214 225
17. WigglesworthVB 1983 The physiology of insect tracheoles. Adv Insect Physiol 17 88 148
18. BarTGuldnerFHWolffJR 1984 “Seamless” endothelial cells of blood capillaries. Cell Tissue Res 235 99 106
19. KameiMSaundersWBBaylessKJDyeLDavisGE 2006 Endothelial tubes assemble from intracellular vacuoles in vivo. Nature 442 453 456
20. YoshidaYYamadaMWakabayashiKIkutaFKumanishiT 1989 Endothelial basement membrane and seamless-type endothelium in the repair process of cerebral infarction in rats. Virchows Arch A Pathol Anat Histopathol 414 385 392
21. BerryKLBulowHEHallDHHobertO 2003 A C. elegans CLIC-like protein required for intracellular tube formation and maintenance. Science 302 2134 2137
22. JurgensGKludingGNusslein-VolhardCWieschausE 1984 Mutations affecting the pattern of the larval cuticle in Drosophila melanogaster. II. Zygotic loci on the third chromosome. Roux's Archives of Developmental Biology 193 283 295
23. IsaacDDAndrewDJ 1996 Tubulogenesis in Drosophila: a requirement for the trachealess gene product. Genes Dev 10 103 117
24. WilkRWeizmanIShiloBZ 1996 trachealess encodes a bHLH-PAS protein that is an inducer of tracheal cell fates in Drosophila. Genes Dev 10 93 102
25. GlazerLShiloBZ 1991 The Drosophila FGF-R homolog is expressed in the embryonic tracheal system and appears to be required for directed tracheal cell extension. Genes Dev 5 697 705
26. KlambtCGlazerLShiloBZ 1992 breathless, a Drosophila FGF receptor homolog, is essential for migration of tracheal and specific midline glial cells. Genes Dev 6 1668 1678
27. ShishidoEHigashijimaSEmoriYSaigoK 1993 Two FGF-receptor homologues of Drosophila: one is expressed in mesodermal primordium in early embryos. Development 117 751 761
28. BeitelGJKrasnowMA 2000 Genetic control of epithelial tube size in the Drosophila tracheal system. Development 127 3271 3282
29. SutherlandDSamakovlisCKrasnowMA 1996 branchless encodes a Drosophila FGF homolog that controls tracheal cell migration and the pattern of branching. Cell 87 1091 1101
30. IkeyaTHayashiS 1999 Interplay of Notch and FGF signaling restricts cell fate and MAPK activation in the Drosophila trachea. Development 126 4455 4463
31. LinXBuffEMPerrimonNMichelsonAM 1999 Heparan sulfate proteoglycans are essential for FGF receptor signaling during Drosophila embryonic development. Development 126 3715 3723
32. ChiharaTHayashiS 2000 Control of tracheal tubulogenesis by Wingless signaling. Development 127 4433 4442
33. LlimargasM 2000 Wingless and its signalling pathway have common and separable functions during tracheal development. Development 127 4407 4417
34. MyatMMLightfootHWangPAndrewDJ 2005 A molecular link between FGF and Dpp signaling in branch-specific migration of the Drosophila trachea. Dev Biol 281 38 52
35. BaerMMBilsteinALeptinM 2007 A clonal genetic screen for mutants causing defects in larval tracheal morphogenesis in Drosophila. Genetics 176 2279 2291
36. Chanut-DelalandeHJungACLinLBaerMMBilsteinA 2007 A genetic mosaic analysis with a repressible cell marker screen to identify genes involved in tracheal cell migration during Drosophila air sac morphogenesis. Genetics 176 2177 2187
37. JungACRibeiroCMichautLCertaUAffolterM 2006 Polychaetoid/ZO-1 is required for cell specification and rearrangement during Drosophila tracheal morphogenesis. Curr Biol 16 1224 1231
38. LuschnigSBatzTArmbrusterKKrasnowMA 2006 serpentine and vermiform encode matrix proteins with chitin binding and deacetylation domains that limit tracheal tube length in Drosophila. Curr Biol 16 186 194
39. StahlMSchuhRAdryanB 2007 Identification of FGF-dependent genes in the Drosophila tracheal system. Gene Expr Patterns 7 202 209
40. ZhuMYWilsonRLeptinM 2005 A screen for genes that influence fibroblast growth factor signal transduction in Drosophila. Genetics 170 767 777
41. AdamsMDCelnikerSEHoltRAEvansCAGocayneJD 2000 The genome sequence of Drosophila melanogaster. Science 287 2185 2195
42. LeeTLuoL 2001 Mosaic analysis with a repressible cell marker (MARCM) for Drosophila neural development. Trends Neurosci 24 251 254
43. StowersRSSchwarzTL 1999 A genetic method for generating Drosophila eyes composed exclusively of mitotic clones of a single genotype. Genetics 152 1631 1639
44. AffolterMNellenDNussbaumerUBaslerK 1994 Multiple requirements for the receptor serine/threonine kinase thick veins reveal novel functions of TGF beta homologs during Drosophila embryogenesis. Development 120 3105 3117
45. JareckiJJohnsonEKrasnowMA 1999 Oxygen regulation of airway branching in Drosophila is mediated by branchless FGF. Cell 99 211 220
46. LeviBPGhabrialASKrasnowMA 2006 Drosophila talin and integrin genes are required for maintenance of tracheal terminal branches and luminal organization. Development 133 2383 2393
47. SwansonLEBeitelGJ 2006 Tubulogenesis: an inside job. Curr Biol 16 R51 53
48. JeonMZinnK 2009 Receptor tyrosine phosphatases control tracheal tube geometries through negative regulation of Egfr signaling. Development 136 3121 3129
49. ForsterDArmbrusterKLuschnigS 2009 Sec24-dependent secretion drives cell-autonomous expansion of tracheal tubes in Drosophila. Curr Biol 20 62 68
50. BlumRStephensDJSchulzI 2000 Lumenal targeted GFP, used as a marker of soluble cargo, visualises rapid ERGIC to Golgi traffic by a tubulo-vesicular network. J Cell Sci 113 3151-3159
51. GrueberWBYeBMooreAWJanLYJanYN 2003 Dendrites of distinct classes of Drosophila sensory neurons show different capacities for homotypic repulsion. Curr Biol 13 618 626
52. MobergKHSchelbleSBurdickSKHariharanIK 2005 Mutations in erupted, the Drosophila ortholog of mammalian tumor susceptibility gene 101, elicit non-cell-autonomous overgrowth. Dev Cell 9 699 710
53. DevineWPLubarskyBShawKLuschnigSMessinaL 2005 Requirement for chitin biosynthesis in epithelial tube morphogenesis. Proc Natl Acad Sci U S A 102 17014 17019
54. MoussianBTangETonningAHelmsSSchwarzH 2006 Drosophila Knickkopf and Retroactive are needed for epithelial tube growth and cuticle differentiation through their specific requirement for chitin filament organization. Development 133 163 171
55. TaponNItoNDicksonBJTreismanJEHariharanIK 2001 The Drosophila tuberous sclerosis complex gene homologs restrict cell growth and cell proliferation. Cell 105 345 355
56. CentaninLDekantyARomeroNIrisarriMGorrTA 2008 Cell autonomy of HIF effects in Drosophila: tracheal cells sense hypoxia and induce terminal branch sprouting. Dev Cell 14 547 558
57. JusticeRWZilianOWoodsDFNollMBryantPJ 1995 The Drosophila tumor suppressor gene warts encodes a homolog of human myotonic dystrophy kinase and is required for the control of cell shape and proliferation. Genes Dev 9 534 546
58. XuTWangWZhangSStewartRAYuW 1995 Identifying tumor suppressors in genetic mosaics: the Drosophila lats gene encodes a putative protein kinase. Development 121 1053 1063
59. KlambtC 1993 The Drosophila gene pointed encodes two ETS-like proteins which are involved in the development of the midline glial cells. Development 117 163 176
60. AraujoSJAslamHTearGCasanovaJ 2005 mummy/cystic encodes an enzyme required for chitin and glycan synthesis, involved in trachea, embryonic cuticle and CNS development--analysis of its role in Drosophila tracheal morphogenesis. Dev Biol 288 179 193
61. TonningAHemphalaJTangENannmarkUSamakovlisC 2005 A transient luminal chitinous matrix is required to model epithelial tube diameter in the Drosophila trachea. Dev Cell 9 423 430
62. BrownNHGregorySLRickollWLFesslerLIProutM 2002 Talin is essential for integrin function in Drosophila. Dev Cell 3 569 579
63. WuLNiemeyerBColleyNSocolichMZukerCS 1995 Regulation of PLC-mediated signalling in vivo by CDP-diacylglycerol synthase. Nature 373 216 222
64. MortimerNTMobergKH 2009 Regulation of Drosophila embryonic tracheogenesis by dVHL and hypoxia. Dev Biol 329 294 305
65. PotterCJHuangHXuT 2001 Drosophila Tsc1 functions with Tsc2 to antagonize insulin signaling in regulating cell growth, cell proliferation, and organ size. Cell 105 357 368
66. LoweSLPeterFSubramaniamVNWongSHHongW 1997 A SNARE involved in protein transport through the Golgi apparatus. Nature 389 881 884
67. HayJCChaoDSKuoCSSchellerRH 1997 Protein interactions regulating vesicle transport between the endoplasmic reticulum and Golgi apparatus in mammalian cells. Cell 89 149 158
68. GipsSJKandzariDEGoldschmidt-ClermontPJ 1994 Growth factor receptors, phospholipases, phospholipid kinases and actin reorganization. Semin Cell Biol 5 201 208
69. JayaramSASentiKATiklovaKTsarouhasVHemphalaJ 2008 COPI vesicle transport is a common requirement for tube expansion in Drosophila. PLoS One 3 e1964
70. SternlichtHFarrGWSternlichtMLDriscollJKWillisonK 1993 The t-complex polypeptide 1 complex is a chaperonin for tubulin and actin in vivo. Proc Natl Acad Sci U S A 90 9422 9426
71. NechiporukTFernandezTEVasioukhinV 2007 Failure of epithelial tube maintenance causes hydrocephalus and renal cysts in Dlg5-/ - mice. Dev Cell 13 338 350
72. GenesteOCopelandJWTreismanR 2002 LIM kinase and Diaphanous cooperate to regulate serum response factor and actin dynamics. J Cell Biol 157 831 838
73. HauckeV 2005 Phosphoinositide regulation of clathrin-mediated endocytosis. Biochem Soc Trans 33 1285 1289
74. Martin-BelmonteFGassamaADattaAYuWRescherU 2007 PTEN-mediated apical segregation of phosphoinositides controls epithelial morphogenesis through Cdc42. Cell 128 383 397
75. MangoSE 2009 The molecular basis of organ formation: insights from the C. elegans foregut. Annu Rev Cell Dev Biol 25 597 628
76. HemphalaJUvACanteraRBraySSamakovlisC 2003 Grainy head controls apical membrane growth and tube elongation in response to Branchless/FGF signalling. Development 130 249 258
77. GaudetJMuttumuSHornerMMangoSE 2004 Whole-genome analysis of temporal gene expression during foregut development. PLoS Biol 2 e352
78. JonssonJCarlssonLEdlundTEdlundH 1994 Insulin-promoter-factor 1 is required for pancreas development in mice. Nature 371 606 609
79. SvenssonPWilliamsCLundebergJRydenPBergqvistI 2007 Gene array identification of Ipf1/Pdx1-/ - regulated genes in pancreatic progenitor cells. BMC Dev Biol 7 129
80. BrandAHPerrimonN 1993 Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118 401 415
81. ShigaYTanaka-MatakatsuMHayashiS 1996 A nuclear GFP/β-galactosidase fusion protein as a marker for morphogenesis in living Drosophila. Dev Growth Differ 38 99 106
82. AshburnerM 1989 Drosophila: A laboratory handbook. Cold Spring Harbor, NY Cold Spring Harbor Laboratory Press
83. BergerJSuzukiTSentiKAStubbsJSchaffnerG 2001 Genetic mapping with SNP markers in Drosophila. Nat Genet 29 475 481
84. MartinSGDobiKCSt JohnstonD 2001 A rapid method to map mutations in Drosophila. Genome Biol 2 RESEARCH0036
85. LeeTHacohenNKrasnowMMontellDJ 1996 Regulated Breathless receptor tyrosine kinase activity required to pattern cell migration and branching in the Drosophila tracheal system. Genes Dev 10 2912 2921
Štítky
Genetika Reprodukční medicína
Článek Pervasive Sign Epistasis between Conjugative Plasmids and Drug-Resistance Chromosomal MutationsČlánek Stress-Induced PARP Activation Mediates Recruitment of Mi-2 to Promote Heat Shock Gene ExpressionČlánek Histone Crosstalk Directed by H2B Ubiquitination Is Required for Chromatin Boundary IntegrityČlánek A Functional Variant at a Prostate Cancer Predisposition Locus at 8q24 Is Associated with ExpressionČlánek Replication and Explorations of High-Order Epistasis Using a Large Advanced Intercross Line PedigreeČlánek Expression of Tumor Suppressor in Spermatogonia Facilitates Meiotic Progression in Male Germ Cells
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2011 Číslo 7
-
Všechny články tohoto čísla
- Gene-Based Tests of Association
- The Demoiselle of X-Inactivation: 50 Years Old and As Trendy and Mesmerising As Ever
- Variants in and Underlie Natural Variation in Translation Termination Efficiency in
- SHH1, a Homeodomain Protein Required for DNA Methylation, As Well As RDR2, RDM4, and Chromatin Remodeling Factors, Associate with RNA Polymerase IV
- Genome-Wide Association Study Identifies as a Susceptibility Gene for Pediatric Asthma in Asian Populations
- Pervasive Sign Epistasis between Conjugative Plasmids and Drug-Resistance Chromosomal Mutations
- Genetic Anticipation Is Associated with Telomere Shortening in Hereditary Breast Cancer
- Identification of a Mutation Associated with Fatal Foal Immunodeficiency Syndrome in the Fell and Dales Pony
- Stress-Induced PARP Activation Mediates Recruitment of Mi-2 to Promote Heat Shock Gene Expression
- An Epigenetic Switch Involving Overlapping Fur and DNA Methylation Optimizes Expression of a Type VI Secretion Gene Cluster
- Recombination and Population Structure in
- A Rice Plastidial Nucleotide Sugar Epimerase Is Involved in Galactolipid Biosynthesis and Improves Photosynthetic Efficiency
- A Role for Phosphatidic Acid in the Formation of “Supersized” Lipid Droplets
- Colon Stem Cell and Crypt Dynamics Exposed by Cell Lineage Reconstruction
- Loss of the BMP Antagonist, SMOC-1, Causes Ophthalmo-Acromelic (Waardenburg Anophthalmia) Syndrome in Humans and Mice
- Interactions between Glucocorticoid Treatment and Cis-Regulatory Polymorphisms Contribute to Cellular Response Phenotypes
- Translation Reinitiation Relies on the Interaction between eIF3a/TIF32 and Progressively Folded -Acting mRNA Elements Preceding Short uORFs
- DAF-12 Regulates a Connected Network of Genes to Ensure Robust Developmental Decisions
- Adult Circadian Behavior in Requires Developmental Expression of , But Not
- Histone Crosstalk Directed by H2B Ubiquitination Is Required for Chromatin Boundary Integrity
- Proteins in the Nutrient-Sensing and DNA Damage Checkpoint Pathways Cooperate to Restrain Mitotic Progression following DNA Damage
- Complex Evolutionary Events at a Tandem Cluster of Genes Resulting in a Single-Locus Genetic Incompatibility
- () and Its Regulated Homeodomain Gene Mediate Abscisic Acid Response in
- A Functional Variant at a Prostate Cancer Predisposition Locus at 8q24 Is Associated with Expression
- LGI2 Truncation Causes a Remitting Focal Epilepsy in Dogs
- Adaptations to Endosymbiosis in a Cnidarian-Dinoflagellate Association: Differential Gene Expression and Specific Gene Duplications
- The Translation Initiation Factor eIF4E Regulates the Sex-Specific Expression of the Master Switch Gene in
- Somatic Genetics Empowers the Mouse for Modeling and Interrogating Developmental and Disease Processes
- Molecular Mechanisms Generating and Stabilizing Terminal 22q13 Deletions in 44 Subjects with Phelan/McDermid Syndrome
- Replication and Explorations of High-Order Epistasis Using a Large Advanced Intercross Line Pedigree
- Mechanisms of Chromosome Number Evolution in Yeast
- Regulatory Cross-Talk Links Chromosome II Replication and Segregation
- Ancestral Genes Can Control the Ability of Horizontally Acquired Loci to Confer New Traits
- Expression of Tumor Suppressor in Spermatogonia Facilitates Meiotic Progression in Male Germ Cells
- Rare and Common Regulatory Variation in Population-Scale Sequenced Human Genomes
- The Epistatic Relationship between BRCA2 and the Other RAD51 Mediators in Homologous Recombination
- Identification of Novel Genetic Markers Associated with Clinical Phenotypes of Systemic Sclerosis through a Genome-Wide Association Strategy
- NatF Contributes to an Evolutionary Shift in Protein N-Terminal Acetylation and Is Important for Normal Chromosome Segregation
- Araucan and Caupolican Integrate Intrinsic and Signalling Inputs for the Acquisition by Muscle Progenitors of the Lateral Transverse Fate
- Pathologic and Phenotypic Alterations in a Mouse Expressing a Connexin47 Missense Mutation That Causes Pelizaeus-Merzbacher–Like Disease in Humans
- Recombinant Inbred Line Genotypes Reveal Inter-Strain Incompatibility and the Evolution of Recombination
- Epistatic Relationships in the BRCA1-BRCA2 Pathway
- Genome-Wide Association Study Identifies Novel Restless Legs Syndrome Susceptibility Loci on 2p14 and 16q12.1
- Genetic Loci Associated with Plasma Phospholipid n-3 Fatty Acids: A Meta-Analysis of Genome-Wide Association Studies from the CHARGE Consortium
- Fine Mapping of Five Loci Associated with Low-Density Lipoprotein Cholesterol Detects Variants That Double the Explained Heritability
- CHD1 Remodels Chromatin and Influences Transient DNA Methylation at the Clock Gene
- Nonlinear Fitness Landscape of a Molecular Pathway
- Genome-Wide Scan Identifies , , and as Novel Risk Loci for Systemic Sclerosis
- Quantitative and Qualitative Stem Rust Resistance Factors in Barley Are Associated with Transcriptional Suppression of Defense Regulons
- A Systematic Screen for Tube Morphogenesis and Branching Genes in the Tracheal System
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Genome-Wide Association Study Identifies Novel Restless Legs Syndrome Susceptibility Loci on 2p14 and 16q12.1
- Loss of the BMP Antagonist, SMOC-1, Causes Ophthalmo-Acromelic (Waardenburg Anophthalmia) Syndrome in Humans and Mice
- Gene-Based Tests of Association
- Genome-Wide Association Study Identifies as a Susceptibility Gene for Pediatric Asthma in Asian Populations
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



