-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaComplex Evolutionary Events at a Tandem Cluster of Genes Resulting in a Single-Locus Genetic Incompatibility
Non-additive interactions between genomes have important implications, not only for practical applications such as breeding, but also for understanding evolution. In extreme cases, genes from different genomic backgrounds may be incompatible and compromise normal development or physiology. Of particular interest are non-additive interactions of alleles at the same locus. For example, overdominant behavior of alleles, with respect to plant fitness, has been proposed as an important component of hybrid vigor, while underdominance may lead to reproductive isolation. Despite their importance, only a few cases of genetic over - or underdominance affecting plant growth or fitness are understood at the level of individual genes. Moreover, the relationship between biochemical and fitness effects may be complex: genetic overdominance, that is, increased or novel activity of a gene may lead to evolutionary underdominance expressed as hybrid weakness. Here, we describe a non-additive interaction between alleles at the Arabidopsis thaliana OAK (OUTGROWTH-ASSOCIATED PROTEIN KINASE) gene. OAK alleles from two different accessions interact in F1 hybrids to cause a variety of aberrant growth phenotypes that depend on a recently acquired promoter with a novel expression pattern. The OAK gene, which is located in a highly variable tandem array encoding closely related receptor-like kinases, is found in one third of A. thaliana accessions, but not in the reference accession Col-0. Besides recruitment of exons from nearby genes as promoter sequences, key events in OAK evolution include gene duplication and divergence of a potential ligand-binding domain. OAK kinase activity is required for the aberrant phenotypes, indicating it is not recognition of an aberrant protein, but rather a true gain of function, or overdominance for gene activity, that leads to this underdominance for fitness. Our work provides insights into how tandem arrays, which are particularly prone to frequent, complex rearrangements, can produce genetic novelty.
Published in the journal: . PLoS Genet 7(7): e32767. doi:10.1371/journal.pgen.1002164
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1002164Summary
Non-additive interactions between genomes have important implications, not only for practical applications such as breeding, but also for understanding evolution. In extreme cases, genes from different genomic backgrounds may be incompatible and compromise normal development or physiology. Of particular interest are non-additive interactions of alleles at the same locus. For example, overdominant behavior of alleles, with respect to plant fitness, has been proposed as an important component of hybrid vigor, while underdominance may lead to reproductive isolation. Despite their importance, only a few cases of genetic over - or underdominance affecting plant growth or fitness are understood at the level of individual genes. Moreover, the relationship between biochemical and fitness effects may be complex: genetic overdominance, that is, increased or novel activity of a gene may lead to evolutionary underdominance expressed as hybrid weakness. Here, we describe a non-additive interaction between alleles at the Arabidopsis thaliana OAK (OUTGROWTH-ASSOCIATED PROTEIN KINASE) gene. OAK alleles from two different accessions interact in F1 hybrids to cause a variety of aberrant growth phenotypes that depend on a recently acquired promoter with a novel expression pattern. The OAK gene, which is located in a highly variable tandem array encoding closely related receptor-like kinases, is found in one third of A. thaliana accessions, but not in the reference accession Col-0. Besides recruitment of exons from nearby genes as promoter sequences, key events in OAK evolution include gene duplication and divergence of a potential ligand-binding domain. OAK kinase activity is required for the aberrant phenotypes, indicating it is not recognition of an aberrant protein, but rather a true gain of function, or overdominance for gene activity, that leads to this underdominance for fitness. Our work provides insights into how tandem arrays, which are particularly prone to frequent, complex rearrangements, can produce genetic novelty.
Introduction
Both evolutionary biologists and breeders have long been interested in non-additive interactions among alleles at the same locus. For example, explanations for heterosis or hybrid vigor, a staple of modern agriculture, share many conceptual formalities with models proposed by Bateson, Dobzhansky and Muller to explain how negative heterosis could result from two or more genes that accumulate different changes in separate lineages. The associated phenotypes of hybrid weakness, sterility or lethality in turn may ultimately lead to reproductive isolation and hence speciation ([1]–[3], reviewed in [4], [5]). Hybrid incompatibilities form a continuum from the grey zone of developmental abnormalities through the clearer phenotype of F1 sterility to the severest form, lethality, and it is important to understand the genetic and molecular causes for the entire spectrum of incompatibilities.
F1 incompatibilities have been found in as many as 2% of Arabidopsis thaliana intra-specific hybrids [6]. Several similar cases in A. thaliana and other species involve interactions between alleles of disease resistance genes with other loci in the genome, which cause an autoimmune syndrome known as hybrid necrosis [6]–[8]. That hybrid necrosis is such a relatively common phenomenon is easily explained, since genes involved in plant defense are highly variable between different individuals of the same species [9], [10], and thus make a perfect substrate for causing problems when different genomes are combined. Moreover, several important classes of defense genes, including those encoding nucleotide binding-leucine rich repeat (NB-LRR) proteins and receptor-like kinases (RLKs), commonly occur in tandem arrays, and new alleles are easily created through gene duplication, illegitimate recombination and gene conversion [11]–[19].
In addition to inappropriate activation of the immune system or sterility, aberrant development is often observed in incompatible plant hybrids [20], [21]. Both Triticum and Nicotiana interspecific hybrids frequently suffer from tumor-like tissue proliferation [22], [23]. In Nicotiana hybrids, wounding and physiological stresses enhance tumor formation, and tumors may differentiate into recognizable tissues [24]. Genetically-induced tumors have also been described in hybrids of Brassica, Datura, Solanum and Lilium [24]. Developmental abnormalities in intra-specific moss hybrids have recently been linked to putatively structurally divergent regions [25], similar to the association of hybrid necrosis with structurally diverse disease resistance loci.
While the known cases of F1 hybrid incompatibility are mostly caused by interaction between alleles at unlinked loci, of particular interest are situations of heterozygous advantage (overdominance) or disadvantage (underdominance) due to interaction of divergent alleles at the same locus. Overdominance has been advanced as an important contributor to hybrid vigor, or heterosis [26]–[28]. Conversely, underdominance may underlie hybrid weakness, sterility or lethality, and thus contribute to speciation [20], [21], [29]. It should be noted that cases of heterozygous disadvantage are underdominant with respect to fitness but can be overdominant in the genetic sense: a plant may become less fit due to increased activity of the gene(s) involved.
Although evidence for both single-gene over - and underdominance is easily found in whole-genome expression studies (e.g. [28]), few cases with phenotypic consequences are understood at the molecular level. Schwartz and Laughner [30] reported four decades ago an example in maize, where two partially compromised forms of alcohol dehydrogenase can form a fully active homodimer; a similar case has been described for complementing alleles at the ARF GTPase-encoding GNOM locus of Arabidopsis thaliana [31]. In tomato, a heterozygote for a loss-of-function allele of the SFT gene has increased yield [32]. Finally, a particularly revealing study comes from rice, where sterility ensues when two divergent alleles at the S5 locus are combined [33]. Since this is not observed when either allele is heterozygous with a third, presumably non-functional allele, one can infer that the combination of the two S5 alleles results in gain-of-function activity of the encoded aspartate protease. The S5 interactions thus provide an example of the complex relationship between biochemical and fitness effects, as the underdominant fitness effects are not simply a consequence of reduced gene activity. It also provides a counterpoint to the SFT case, where reduced gene activity has overdominant fitness effects [32].
Here, we report on an intraspecific A. thaliana F1 hybrid, where heterozygosity at a single locus causes a pleiotropic syndrome that includes smaller stature and reduced seed set as well as ectopic outgrowths on leaf petioles. The causal receptor-like kinase (RLK) gene, OUTGROWTH-ASSOCIATED PROTEIN KINASE (OAK), is found in a structurally hypervariable tandem cluster of related RLK genes. During duplication of the ancestral RLK gene, coding sequences were recruited to form a promoter with a new expression domain. Divergence in the extracellular domain of the protein led to evolution of alleles that now interact in the Bla-1/Sha hybrid to produce phenotypes not seen in the parents, making this a case of underdominance for fitness caused by overdominance for gene expression.
Results
Ectopic petiole outgrowths and reduced biomass of Bla-1/Sha hybrids
The aberrant phenotype of Blanes-1 (Bla-1)/Shahdara (Sha) F1 hybrids was identified in a survey of more than 1,300 crosses among over 300 A. thaliana accessions from the world-wide range of the species [6]. Bla-1/Sha F1 plants had a range of phenotypes that were not normally seen in inbred accessions, including the Bla-1 and Sha parents, or in other F1 hybrids: outgrowths on the adaxial surface of the petioles, leaf twisting, leaf lesions, and loss of apical dominance reflected by precocious and increased release of side shoots (Figure 1a–1c). These phenotypes were observed regardless of the direction of the cross. Raising plants in long days at 23°C instead of 16°C restored apical dominance and largely suppressed leaf twisting and lesioning. This partial suppression of the hybrid phenotype at higher temperatures is similar to the suppression of necrosis seen in the Uk-1/Uk-3 and other hybrids with autoimmune defects [6].
Fig. 1. Adaxial outgrowths in Bla-1/Sha hybrids. 
(a) Six-week old plants grown at 16°C, long days, of Sha (left), Bla-1/Sha F1 hybrid (centre) and Bla-1 (right). Arrows indicate de-repressed side shoots in the hybrid. (b) Lesioning is seen in leaves of six-week old F2 hybrid plants grown at 16°C, long days, where the phenotype segregates (present on left leaf, absent on right leaf). (c) Outgrowths on the petioles of Bla-1/Sha F2 plants grown at 23°C, long days. (d–f) Transverse sections of Bla-1 (d) and Bla-1/Sha hybrid (e, f). Outgrowths that are caused by proliferation of parenchyma and/or epidermal cells are visible on the adaxial surface of the petiole. Scale bar = 100 µm. Because the ectopic outgrowth phenotype was particularly striking and reliably observed in all F1 plants, we decided to investigate it in detail. The same phenotype with little variation was seen in approximately 50% of all F2 progeny, compatible with a single-gene, heterozygous genetic basis. The outgrowth phenotype segregated independently of the lesioning in the F2 and subsequent generations.
Outgrowths were occasionally noted in the Bla-1 parent, but with incomplete penetrance that varied greatly between experiments (Table S1). Onset of outgrowth formation in Bla-1, when it occurred, was much later than in the F1 hybrids. Crosses of each parental line to the reference accession Col-0 did not produce any progeny with outgrowths, but they were, as expected, seen in about one quarter of progeny after Col-0/Bla-1 and Sha/Col-0 F1 hybrids were crossed to each other.
Analysis of transverse sections revealed that outgrowths originated from proliferating parenchyma and/or epidermal cells on the adaxial surface of the petiole (Figure 1d–1f). The vascular system of the petioles appeared normal. Because of their determinate nature, we concluded that the outgrowths did not constitute undifferentiated callus.
We also asked whether the gene(s) causing the hybrid phenotypes of outgrowth and lesioning might affect overall plant performance. In a segregating F2 population of five-week old plants, we found that outgrowths alone were correlated with a 29% reduction in rosette weight, while lesioning or lesioning plus outgrowths reduced growth by over 50% (Table S2; 2-way ANOVA outgrowths p = 0.0003, lesioning p<0.0001). In addition, we assessed seed set as a proxy for lifetime fitness. Due to confounding factors such as differential flowering times in Sha and Bla-1, we measured seed set after the incompatibility was reconstituted in the Col-0 reference background (see below for further details). Seed set was reduced by 90% in F1 hybrids that were phenotypically comparable to the natural hybrids (two-tailed, unequal variance t-test: p<<0.001; Figure S1). In two other independent crosses that resulted in a more severe incompatibility phenotype, all the hybrids died within two months, and thus did not produce any seeds at all. This indicates that the Bla-1/Sha OAK incompatibility greatly reduces lifetime fitness.
Because wounding and physiological stresses enhance the formation of tumors in Nicotiana, where these may differentiate into recognizable tissues [24], we examined the effects of wounding, by pricking the petioles of Bla-1/Sha F1 plants with a fine needle. Outgrowth formation was not enhanced, but we found that increased humidity suppressed outgrowth formation (Figure S2). This is reminiscent of the suppression of constitutive activation of disease resistance in the ssi4 mutant by high humidity [34].
Compared to normal tissue, induction of callus from Nicotiana hybrid tumors requires less auxin [35]. Some A. thaliana tumor forming lines also produce callus tissue that can continue to proliferate on hormone-free media [36]. To test auxin response in our system, transverse sections of leaf and petiole tissue were induced to form callus. Although the Bla-1 parent had a relatively higher auxin requirement for callus formation, there was no difference between the Sha parent and the Bla-1/Sha hybrids (Figure S3). Thus, the outgrowths are probably genetically distinct from the Nicotiana tumors.
Genome-wide expression studies
Microarray analysis with triplicate Affymetrix ATH1 arrays using RNA extracted from three-week-old aerial tissue identified 356 genes differentially expressed in the hybrids compared to the parents. There was no significant up - or down-regulation of any particular known pathways or reactions based on the SkyPainter tool [37], but several, often overlapping, Gene Ontology (GO) categories were enriched among the differentially expressed genes, most notably several related to pathogen response (Table S3; [38]). Whether this reflects a link to disease resistance remains unclear, since some well-known markers for pathogen response, such as PR1 or the defensin gene PDF1.2(b), were down-regulated in the hybrids (Tables S4 and S5). In any case, as with the morphological phenotype, there was no overwhelming connection to the hybrid necrosis syndrome as seen in many other incompatible A. thaliana F1 hybrids [21].
Ectopic outgrowths caused by a hypervariable protein kinase gene cluster
Using F2 and F3 progeny, we mapped the outgrowth phenotype to a single genomic region on chromosome 5 containing 17 genes in the reference accession Col-0 (At5g59560 to At5g59700; Figure S4). A tandem array of four genes that encode a distinct clade of closely related receptor-like kinases (RLKs; At5g59650 to At5g59680) [17] were of particular interest, because RLKs are one of the most variable gene families in the A. thaliana genome [9].
We recovered the genomic regions from At5g59616 (encoding a protein kinase-related protein) to At5g59690 (histone H4) by long-range PCR from Bla-1 and Sha, and found the RLK cluster to be highly variable (Figure 2a). In Col-0 only, there are two transposons and a pseudogene upstream of the RLK genes. In Sha, the first RLK gene in the cluster, At5g59650, is missing and the upstream gene At5g59616 is only partially present. In both Bla-1 and Sha, a 150 bp remnant of the second RLK gene, At5g59660, indicates that a deletion likely occurred in the Bla-1/Sha lineage. Also in both Bla-1 and Sha, the third RLK gene of the cluster, At5g59670, has been duplicated to give rise to At5g59670a and At5g59670b (Table S6). In addition to Bla-1 and Sha, the At5g59670 duplication was detected by PCR analysis of the OAK promoter in 36 of 87 diverse A. thaliana accessions (Table S7), while a Col-0 like promoter was found in 45 accessions. Assays for both promoter types were positive in two accessions, indicating either illegitimate recombination or a different duplication event. The PCR assays failed in the remaining four accessions.
Fig. 2. OAK kinase cluster architecture. 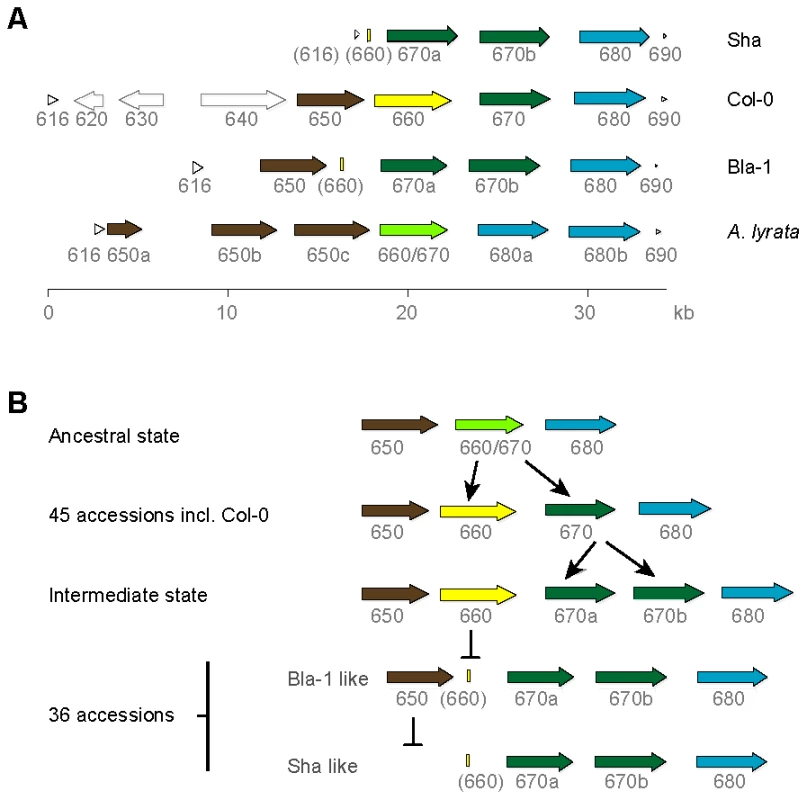
(a) Last three digits of At5g59XXX gene identifier given. Truncated genes are indicated by brackets around the gene identifier. CACTA transposons and the pseudogene in Col-0 are indicated by light grey, unfilled arrows. (b) Hypothesized events in the evolution of the OAK kinase cluster in Bla-1 and Sha. Reconstruction of the ancestral state of the tandem array, by comparison with the close relative A. lyrata [39], suggested the presence of three tandem RLK genes in the last common ancestor of A. thaliana and A. lyrata. The central gene was duplicated in the A. thaliana lineage to produce At5g59660 and At5g59670, whereas in A. lyrata, there have been subsequent duplications of the two flanking RLK genes, resulting in a cluster with six genes. Given the presence of a remnant of At5g59660 in Bla-1 and Sha and that the Col-0-like At5g59670 is found in over half the accessions tested, the ancestral state of this cluster in A. thaliana is likely to have been a cluster of four RLK genes as found in Col-0 (Figure 2b).
Two alleles of a single RLK cause novel growth phenotypes
To determine whether any of the RLK genes contribute to the outgrowth phenotype, a genomic copy of each gene from Bla-1 and Sha was individually introduced into the Bla-1, Sha and Col-0 backgrounds. Only plants transformed with At5g59670b from Bla-1 or Sha developed outgrowths (Figure 3a). Unexpectedly, while At5g59670b from Bla-1 induced outgrowths most effectively in Sha, and At5g59670b from Sha in Bla-1, outgrowths were also seen, albeit at lower frequency, upon transformation of either gene into the recurrent parent or into Col-0. This suggests a dosage effect, perhaps due to elimination of negative regulatory elements or epigenetic marks in the transgene that normally suppress expression of the endogenous locus, such that the transgenic proteins are present at an elevated level compared to native OAKSha or OAKBla-1. This is supported by some transgenic lines in which we saw a 3∶1 ratio of normal to affected plants in the T2 generation, such that a hemizygous state gives a wild-type phenotype while homozygosity for the transgene leads to a Bla-1/Sha-like phenotype. A similar increase in incompatibility severity after transgenic reconstitution was also observed for DM1 in the case of Uk-1/Uk-3 [6].
Fig. 3. Identification of At5g59670b homologs as sufficient and necessary for outgrowths. 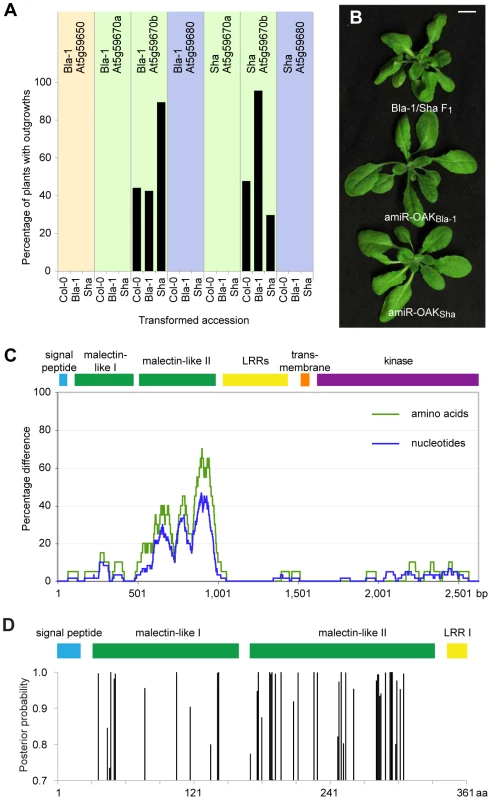
(a) Fraction of T1 plants (n≥90, except for Bla-1 transformed with Sha At5g59680 where n = 56) with outgrowths. (b) Suppression of outgrowths with amiRNAs against OAK (At5g59670b) from Bla-1 or Sha. (c) Divergence between OAK (At5g59670b) alleles from Bla-1 and Sha (sliding windows of 60 bp and 20 amino acids, respectively). (d) Identification of individual sites in the N-terminal part of OAK protein under positive selection (as determined by Bayesian Posterior Probability) across 34 accessions using PAML [43]. The second malectin-like domain is enriched for such sites. To determine whether the RLKs were not only sufficient, but also necessary for the outgrowths, artificial miRNAs (amiRNAs) were designed against individual RLKs [40]. Only Bla-1/Sha plants with an amiRNA directed against At5g59670b showed a suppression of the hybrid phenotype (outgrowths, leaf twisting and apical dominance; Figure 2b and Figure S5). We therefore refer to At5g59670b as OUTGROWTH-ASSOCIATED PROTEIN KINASE (OAK).
Comparison of Bla-1 and Sha OAK alleles
The Bla-1 and Sha OAK primary transcripts are each 3.9 kb long, with 13 exons, and a 5′ untranslated region of 92 nt (expressed in Bla-1 and Sha petioles) or up to 123 nt (expressed in Sha pedicels and peduncles), as determined by 5′ RACE-PCR. Both OAK alleles encode proteins of 873 amino acids, with 9% of residues being different. The majority of polymorphisms are located in a 152 amino acid region, between positions 180 and 331, where 55 residues differ (Figure 3c). Among the remaining 721 residues, there are only 19 replacements.
Like many other plant RLKs, the OAK proteins include a signal peptide, potential leucine-rich repeats (LRRs; in OAK, four to five), a transmembrane domain, and a cytoplasmic kinase domain (Michael Hothorn, personal communication; Figure 3d and Figure S6). In addition, two related regions with similarity to a carbohydrate-binding domain in ER-localized malectin proteins from animals [41] are found between the signal peptide and the LRRs (http://toolkit.tuebingen.mpg.de/hhpred/; Michael Hothorn, personal communication). Interestingly, the region that is very different between the Bla-1 and Sha proteins, from residue 180 to 331, coincides almost perfectly with the second predicted malectin-like domain, from residue 169 to 331. An analysis of OAK and its homologs (OAKSha, OAKBla-1, At5g59670aSha, At5g59670aBla-1 and At5g59670Col-0), using the Codeml program of PAML to assess dN/dS ratios, did not provide evidence for directional or diversifying selection across the entire protein [42], [43]. However, an Bayesian Posterior Probability analysis of positive selection at individual residues, using At5g59670Col-0 as a reference, suggested that several codons in the second malectin-like domain are under positive selection [44]. A broader analysis of 34 accessions from which OAK sequences could be recovered supported these conclusions (Figure 3d).
To determine if the second malectin-like region in OAK homologs is generally hypervariable, we performed a sliding window analysis of all eleven RLKs in the Col-0, Bla-1 and Sha clusters (Figure S7). Most highly conserved are the LRR and kinase domains. We also examined in detail the duplicated genes encoding the At5g59670 proteins. At5g59670aSha and OAKSha stood out, because they are identical across the first 598 amino acids of the protein. At the nucleotide level, the two genes include an identical 2.7 kb fragment, which most likely reflects a recent gene conversion event that extends from 13 bp upstream of the translational start site to the first 60 bp of the kinase encoding sequences. In conclusion, the divergence between the second malectin-like domain of OAKBla-1 and OAKSha is not representative of the variation between RLKs encoded by orthologs and paralogs in this cluster.
Role of divergent promoter sequences in causing the OAK hybrid phenotype
To determine the contribution of non-coding and coding sequences of OAK to the outgrowth phenotype, we performed a series of domain swaps between OAKBla-1, OAKSha, and At5g59670Col-0 (Figure 4a). Similar to plants transformed with the non-chimeric fragments, T1 transformants frequently showed more severe phenotypes than were observed in the F1 hybrids. This indicated that divergent OAK alleles have the potential to cause even stronger incompatibilities than seen between the accessions Bla-1 and Sha.
Fig. 4. Contribution of both the OAK promoter and extracellular domain to outgrowths. 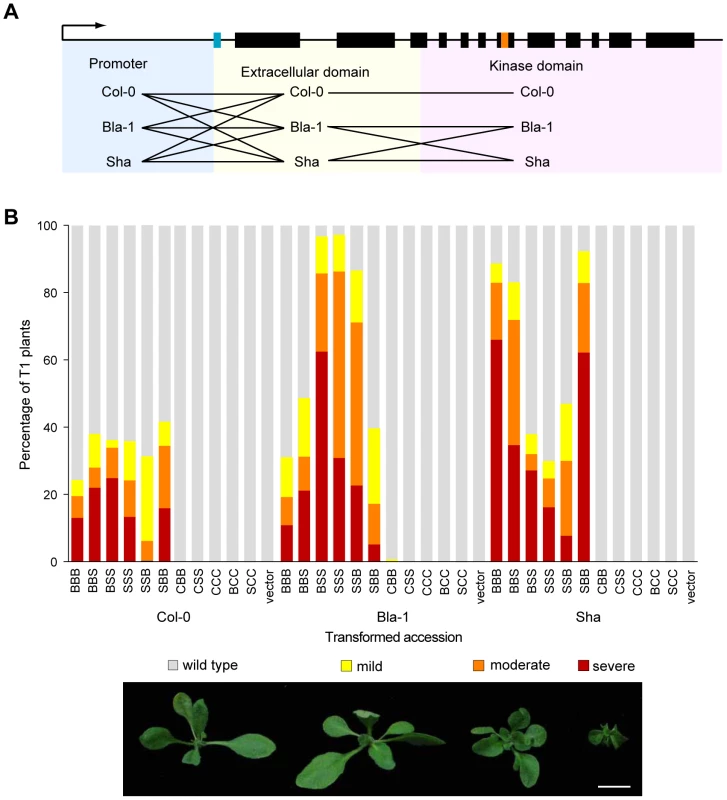
(a) Overview of domain swaps. (b) Phenotypic distribution of T1 plants (n≥90). Three-letter code indicates composition of chimeras. E.g., BBS, promoter and extracellular domain from Bla-1, kinase domain from Sha. Examples of phenotypic classes are shown at the bottom: mild (outgrowths, but otherwise normal leaves), moderate (outgrowths, shortened petioles, mild leaf twisting, normal lamina size) or severe (stunted plants, petioles almost absent, reduced lamina surface, seed rarely obtained). Scale bar = 1 cm. The first major conclusion from the experiments with the chimeric transgenes was that the promoter region contributed to the outgrowth phenotype, because outgrowths were only observed when a particular recombinant protein was expressed from either the OAKBla-1 or OAKSha promoter, but never with the At5g59670Col-0 promoter (Figure 4b). GUS reporter experiments demonstrated that the OAK promoters from Bla-1 and Sha were active in the vascular system of the petioles, in a pattern consistent with the location of the outgrowths (Figure 5). In contrast, the At5g59670Col-0 promoter drove expression in the leaf lamina, explaining why it could not cause petiole outgrowths. The activity domain of the At5g59670aBla-1 promoter was similar to that of the At5g59670Col-0 promoter, but with additional expression in the lamina of the cotyledons. Finally, the At5g59670aSha promoter was active in all seedling tissues, but in isolated patches that differed from plant to plant. Thus, despite the encoded proteins being closely related, the promoters conditioned a surprisingly wide spectrum of expression patterns, with differences both between duplicates within an accession and among orthologs from different accessions.
Fig. 5. Activity domains of OAK homolog promoters. 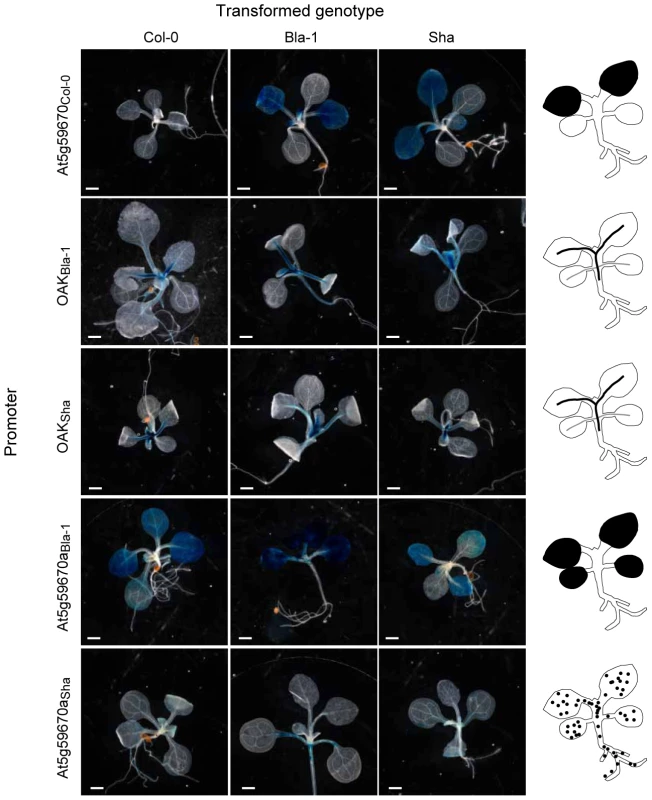
A representative T1 plant for each promoter∶GUS construct transformed into Col-0, Bla-1 and Sha is shown, with diagrams of the expression domain on the far right. Scale bar = 100 µm. Diversity and origin of promoters in the OAK cluster
The OAKBla-1 and OAKSha promoters are more similar to each other than are the coding regions, being 97% identical in the 1,238 bp upstream of the start codon. OAK promoter sequences could be recovered from a further 32 accessions. Pairwise identity for all 34 accessions including Bla-1 and Sha was between 97 and 100%. Given the high similarity of the promoter region, the duplication of At5g59670 to form OAK is unlikely to have occurred more than once. Therefore while the change in expression domain has determined how the incompatibility is expressed, the causative changes for the incompatibility are not within the promoter region. In comparison, over the first 1,077 bp of the coding region, the pairwise identity for the 34 accessions ranged from 87 to 100%, with a mean of 94%. One accession that was identical to Sha throughout both the promoter and coding region was Kondara, which we found to be incompatible with Bla-1 as well. Across the entire RLK cluster, there were only two nucleotide differences in 17.5 kb, and both were in non-coding sequences. Kondara was therefore not considered separately in any of the sequence analyses. Further crosses of Bla-1 and Sha to other accessions with the OAK gene revealed that while most accessions are compatible, a similar incompatibility phenotype is seen in Sha x Bak-2, Sha x Leo-1, Mer-6 x Bla-1 and Leb-3 x Bla-1 hybrids (all incompatibilities between Bla-1-like and Sha-like haplotype groups based on the second malectin domain; Figure S8). Less severe incompatibilities with a late onset of outgrowth formation were found in crosses of Bla-1 to a number of accessions with a second malectin domain that fell into a different haplotype group (ICE91, ICE92, ICE152, ICE153, Vash-1 and Valsi-1).
Using NeighborNet implemented in SplitsTree [45], we examined the relationship between the RLKs from the 34 accessions based on the promoter sequences and the extracellular domains (amino acids 1 to 360; Figure 6a, 6b). Similarity in the coding region was not always reflected in promoter similarity, and vice versa, suggesting a history of recombination or gene conversion events. The SplitsTree analysis suggested four major haplotypes at the OAK locus. Analysis with STRUCTURE [46], where we treated polymorphisms in the OAK locus as linked markers on a chromosome, confirmed that there are four major haplotype groups, with half of the accessions studied showing contributions from more than one haplotype (Figure 6c). Within-locus switching between haplotype groups was confirmed by visual inspection of sequence alignments between individual accessions. This likely reflects high levels of gene conversion or recombination within the OAK gene.
Fig. 6. Phylogenetic analysis of OAK from multiple accessions. 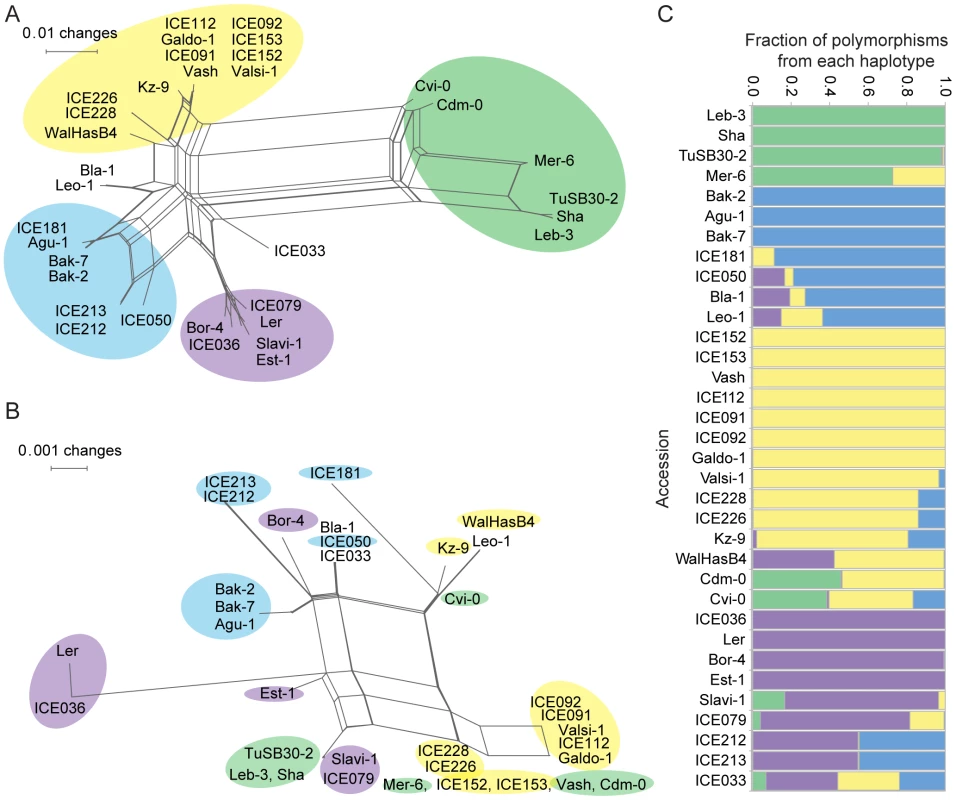
Splitstree [45] was used to examine the phylogenetic relationship of OAK from 34 accessions based on (a) 1,540 bp coding sequences downstream of the translational start site or (b) 1,196 bp promoter sequence. Color code in (b) reflects cluster membership in (a), highlighting variable correlation between promoter and coding region similarity. (c) STRUCTURE analysis [46] of haplotype contributions to each accession based on promoter and coding regions. A search of the Col-0 reference genome for the possible origin of the OAK promoter revealed that most of it probably arose from the coding region of one of the RLK genes, spanning intron 2 to exon 7 (encoding amino acids 207 to 383 of At5g59670). Although these regions are only 60 to 70% identical to the OAK promoter (BLASTN v2.2.25, E-value 1×10−61), they present the best matches in the Col-0 genome (second best hit is to LRR-RLK gene At3g46330, E-value 3×10−13) indicating that this is the most likely origin of the OAK promoter. While the promoter includes potential coding sequences, there are several in-frame stop codons upstream of the predicted OAK translation start. The OAKBla-1 and OAKSha promoters show similar levels of identity with RLK coding sequences across the cluster, but it seems most likely that the duplication of the At5g59670 gene involved an additional duplication that led to conversion of the region coding largely for the second malectin-like domain into a promoter. Interestingly, this is also the portion of the coding sequence that is most different between Bla-1 and Sha. The 260 bp promoter region immediately upstream of the start codon of OAK is most similar to sequences found in triplicate in the At5g59670Col-0 promoter (Figure S9).
Role of the protein and kinase activity in causing the OAK hybrid phenotype
A second conclusion of the chimeric transgene experiments was that in addition to the promoter, the protein, and the extracellular domain in particular, contributed to the outgrowth phenotype (Figure 4a, 4b). The At5g59670Col-0 protein did not cause an incompatibility phenotype even when expressed under the OAKBla-1 or OAKSha promoters. Swapping the extracellular and cytoplasmic domains between the OAKBla-1 and OAKSha proteins showed that the cytoplasmic domains were broadly equivalent. However, introduction of the extracellular domain of OAKBla-1 into the Sha genotype, or vice versa, greatly increased the proportion of affected T1 plants. This result is supported by the incompatibility between Leo-1 and Sha, where Leo-1 has an extracellular domain identical to Bla-1, but only two amino acid differences in the cytoplasmic domain compared to Sha (Figure S10). Further attempts to narrow down the causal region within the extracellular domain with additional chimeras were not successful.
We tested the hypothesis that the outgrowth phenotype resulted from ectopic activation of a kinase-dependent signaling pathway by mutating key residues in the kinase catalytic domain [47]. Double mutants of D693N and K695R should lack all kinase activity. In the Sha background, over 80% of T1 plants carrying the Bla-1 kinase-active construct had a moderate or severe phenotype, while only one third of T1 plants transformed with the Bla-1 kinase-dead construct had any phenotype, and this was always mild. When the Sha kinase-dead construct was transformed back into the Sha accession, all T1 transformants were wild type in appearance, which contrasts with 30% of T1 plants expressing the Sha kinase-active construct having a mild to severe phenotype (Figure 7a). Results were comparable with Bla-1 transformants, although in this case some plants with a moderate phenotype were observed after transformation with the Sha kinase-dead construct.
Fig. 7. Requirement of OAK kinase activity and expression domain for hybrid phenotype. 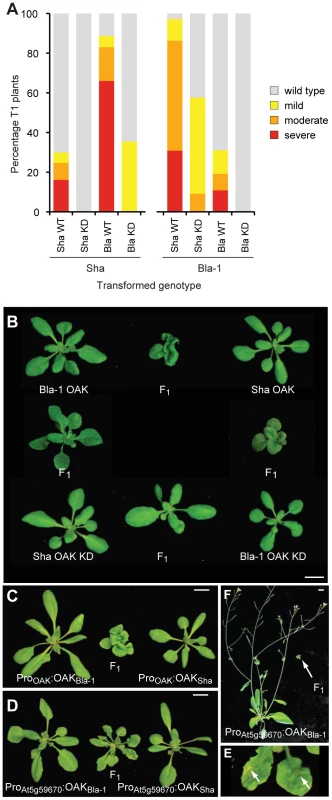
(a) Phenotypic distribution of T1 plants (n≥90) expressing kinase dead (KD) or wild-type (WT) versions of OAK. (b) Crosses of Col-0 plants carrying Bla-1/Sha POAK∶OAK KD constructs. Representative F1 plants from crosses among five pairs of independent, phenotypically normal T1 plants are shown with alongside the parental lines. Scale bar = 1 cm. (c) Crosses of five pairs of phenotypically normal Col-0 plants transformed with POAK∶OAKSha and POAK∶OAKBla-1, or (d, e) with PAt5g59670∶OAKSha and PAt5g59670∶OAKBla-1. Plants in (b-d) are 4-weeks old, in (e) 6-weeks old. Arrows in (f) indicate regions of cell death visible to the naked eye on a close-up of the F1 plant in (d). Because RLKs can form homo - and heterodimers [48], we tested the effects of combining Bla-1 and Sha kinase-dead versions in the neutral Col-0 reference background. We transformed both kinase-active and -dead versions individually into Col-0 and then generated the four possible combinations by crossing (Figure 7b, 7c). The F1 hybrids in which only one of the transgenes expressed a kinase-active version had a less severe phenotype than those carrying both Bla-1 and Sha kinase-active versions. All F1 progeny from five crosses using OAK kinase-dead forms of both Bla-1 and Sha were wild type in appearance. This finding not only confirmed that kinase activity of OAK is required for its function, but also suggested that OAK can act as a heteroallelic dimer or multimer, because a kinase active version of one OAK allele can at least partially complement a kinase-dead version of the other OAK allele. In addition, these data indicated that other RLKs present at the OAK cluster in Col-0 are unlikely to be involved in the outgrowth phenotype.
Further circumstantial evidence suggesting that OAK proteins form dimers or multimers was obtained by expressing only the extracellular domain of OAKBla-1 or OAKSha in hybrid plants. Expression under the native promoter in particular suppressed the outgrowth phenotype in many OAKBla-1/OAKSha heterozygous plants (Figure S11). We propose that by binding to OAK proteins, the extracellular domains reduce the number of active OAKBla-1 or OAKSha heterodimers.
The OAK kinase can couple to the salicylic acid pathway
Curiosity led us to examine the consequences of mis-expressing the incompatible OAK alleles from the Col-0 promoter in the putative ancestral domain of the leaf lamina. We introduced ProAt5g59670-Col∶OAKBla and ProAt5g59670-Col∶OAKSha chimeric transgenes into the Col-0 reference background, and crossed the transformants, which were wild type in appearance, to each other. As described above, performing this experiment with the OAK wild-type alleles from Bla-1 and Sha reproduced the Bla-1/Sha hybrid phenotype with petiole outgrowths. Co-expressing the Bla-1 and Sha OAK proteins from the Col-0 promoter resulted in a new incompatibility phenotype, ranging from patches of cell death visible to the naked eye on the leaf lamina and abbreviated inflorescences, to severely stunted plants (Figure 7d–7f). It is striking that the altered expression domain leads essentially to a diametrically opposite phenotype, ectopic cell death instead of ectopic cell proliferation.
Tissue necrosis and ectopic cell death are typical responses to pathogen infection that rely on salicylic acid signaling [49]. To determine whether the cell death we observed was associated with increased activity of this pathway, we used a transgene that drives constitutive expression of a bacterial salicylate hydroxylase, nahG, which converts salicylic acid to catechol [50]. The Pro35S∶nahG transgene suppressed the cell death phenotype caused by co-expression of OAKBla-1 and OAKSha proteins from the Col-0 promoter, but had no effect on the ectopic outgrowths and other phenotypes seen when the proteins were expressed from their own promoters in Col-0 (Figure S12). This not only indicated that OAK proteins can couple to alternative downstream signaling pathways (as is known for the BAK1 RLK [51]), but also that the ancestral function might have involved detection of microbes, a known function of different RLKs [52]–[54]. Mutation of other key genes in disease resistance pathways (PAD4, EDS1, and NDR1) [49] had no effect on the aberrant phenotypes caused by co-expression of the OAK alleles under either the OAK or the Col-0 At5g59670 promoter.
Discussion
We have identified a case of a single-gene incompatibility interaction that leads to multiple aberrant phenotypes in hybrids between A. thaliana accessions Bla-1 and Sha. The phenotypes include reduced stature, leaf twisting, a loss of apical dominance and ectopic outgrowths on the petioles in addition to a decrease in lifetime fitness as measured by seed set. In the genetic sense, the Bla-1 and Sha OAK alleles can be thought of behaving in an overdominant fashion, since the action of either allele (which can cause milder versions of the hybrid phenotype in a foreign background on their own) is enhanced by the other allele. However, considering that the phenotypes are not normally seen in the parents or in other hybrids, and that one of them is reduced growth, the alleles behave in an underdominant fashion when it comes to fitness, as measured by seed set under laboratory conditions.
The causal gene for the Bla-1/Sha incompatibility, OAK, is an RLK that is part of a highly variable tandem array, with evidence of gene conversion, duplications and deletions in the recent evolutionary past. OAK was formed by a whole-gene duplication event in a common ancestor of Bla-1 and Sha, with the additional duplication of a segment of coding DNA that now forms most of the OAK promoter. This gene duplication is present in approximately one third of A. thaliana accessions sampled, but the Bla-1 and Sha alleles themselves are rare. The new promoter changed the OAK expression domain from the leaf lamina to the leaf petiole. Although this change is expression domain is required for manifestation of the OAK incompatibility, it is not in itself causal as the new promoter probably arose only once, and most accessions carrying the OAK gene are compatible with Bla-1 and Sha. Notably, the coding sequences that became part of the promoter include those coding for the second malectin-like domain, which has diverged between Bla-1, Sha and other accessions after the initial duplication. Changes in cis-regulatory sequences are an important source of interspecific variation [55], but such drastic intraspecific shifts in expression domains as we have observed are rare.
A function for OAK in disease resistance or development?
The A. thaliana genome encodes over 600 RLKs. Approximately two thirds of A. thaliana RLKs are predicted to contain structurally diverse extracellular domains [15], which often include LRRs [56]. These extracellular domains are involved in perceiving a wide range of ligands, including small proteins, steroids, and carbohydrates. The function and ligands of most plant RLKs are unknown, but known activities of LRR-RLKs include both control of plant development (e.g., BRI1 in brassinosteroid response [57], CLV1 in meristem maintenance [58] and ERECTA in pleiotropic patterning processes [59]) and microbe detection (e.g., Xa21, FLS2 and GmNARK [52]–[54]). The RLK genes constitute one of the most variable gene families in A. thaliana, which has been interpreted as many RLKs evolving in response to pathogen pressure [9]. Local and genome-wide duplications, along with gene conversion, have contributed to the expansion and diversification of RLKs in plants [12], and RLK genes are overrepresented in tandem arrays [15], [60], although those with known roles in plant development are generally not located in tandem arrays [17].
Circumstantial evidence that might point to an interaction of OAK-like RLKs with microbes include the microarray results and the high variability of the OAK gene cluster. OAK does not appear to be required for normal development, since amiRNA-mediated knockdown of OAK activity has no obvious adverse effects. However, it is also possible that OAK acts redundantly in plant development given that the incompatibility phenotype manifests itself primarily as morphological abnormalities. In addition, the mis-expression experiments using the Col-0 promoter revealed that OAKs can trigger typical salicylic-acid dependent cell death as is often seen in response to pathogen attack, although OAK coupling to downstream signaling pathways may be dependent on the expression pattern of alternative interactors. Following the BAK1 paradigm [51], it is conceivable that the availability of OAK interaction partners determine its activity in plant development versus microbe-interactions. The similarity of the OAK extracellular domains to the carbohydrate-binding protein malectin [41] might indicate that OAK-like RLKs interact with carbohydrates found on the surface of microbes. Alternatively, their function might be detection of damaged self, according to the concept of indirect recognition of pathogens through damage-associated molecular patterns (DAMPs) [61]. A role for OAK in plant immunity through perception of self damage would be reminiscent of previously reported cases of hybrid incompatibility that involve disease resistance genes [6]–[8], [62].
Causes for increased OAK activity in hybrids
Some RLKs function as hetero - or homodimers, with auto - and trans-phosphorylation required for function of the complex. For example, BAK1 and BRI1 form heteromultimers, and a multi-step pathway involving auto - and trans-phosphorylation events activates downstream signaling [63]. Our experiments with kinase-dead versions demonstrated that kinase activity is important for OAK function. The limited effects of the kinase-dead Sha allele in the Bla-1 background, and vice versa, indicate partial complementation by the opposite kinase-active allele, which is suggestive of heteroallelic dimer or multimer formation. In addition, the suppression of the hybrid phenotypes by expression of the Bla-1 or Sha OAK extracellular domain alone provides further support for this scenario.
We do not know whether the change in expression pattern associated with the acquisition of a new promoter by the Bla-1 and Sha OAK alleles subsequently became subject to positive selection, or whether these alleles lack a beneficial function all together. However, the fact that the unusually high divergence in sequence between the two alleles is largely restricted to the second malectin-like domains suggests positive selection or a gene conversion event. We speculate that these sequence changes also altered the affinity for potential ligands. The fact that the Bla-1 and Sha proteins on their own can cause a hybrid-like phenotype, albeit less effectively than when they are combined, suggests that each protein on its own can interact with this potential, unknown ligand. We speculate that OAK heterodimers have increased affinity for such a ligand, leading to ectopic activation of the downstream signaling pathway and aberrant development.
Evolution of incompatible OAK alleles
Several incompatibilities in F1 and F2 hybrids have recently been linked to disease resistance (R) genes. At least one of the A. thaliana factors, and likely another in A. thaliana and rice each, appears to be encoded in a highly polymorphic cluster of NB-LRR genes, the most common class of R genes, and at the same time the most polymorphic gene family in plants [6], [8], [9], [62], [64], [65]. Indeed, more broadly, copy number variation is a recurring factor in reproductive isolation [66]. It has been proposed that the occurrence of disease resistance genes in clusters is critical for generating diversity of resistance specificities, because the tandem arrays support high rates of gene conversion and illegitimate recombination [67]. Indeed, complex histories of transposon insertions, translocations, and gene duplications and rearrangements have also contributed to the formation of NB-LRR gene clusters [11], [13], [16], [18], [19]. RLK genes share with NB-LRR genes the frequent occurrence in tandem arrays and extreme diversity [9], [12], [15]. The complex evolutionary history of the OAK cluster is thus not atypical for this gene family.
Most hybrid incompatibilities described so far involve multiple loci and as such are classical examples of the Bateson, Dobzhansky and Muller model where derived alleles of two or more genes interact to produce underdominant fitness outcomes (e.g.[8], [21], [62], [68]). In contrast, the incompatibility we describe here is due to interaction of two different alleles at a single locus. Due to the high level of polymorphisms, it is difficult to know what the ancestral allele at the OAK locus looked like immediately after duplication. The incompatible OAK alleles may have evolved through mutations within both the Sha and Bla-1 lineages, with the current alleles remaining compatible with the ancestral allele. Alternatively, all important mutation and gene conversion events may have occurred in only one lineage, through multiple intermediate allelic forms that were never incompatible with the immediately ancestral allele [69]. Either way, evolution of the current situation would not require that plants passed through a fitness valley with heterozygosity for the two incompatible OAK alleles.
Conclusions
Not many cases of single-gene hybrid incompatibility have been described in plants: in rice, incompatible alleles of the S5 locus cause most hybrids between the japonica and indica varieties to be female sterile [33]. It is not inconceivable that heterodimers are involved, similar to what appears to be the case for OAK, and dimer formation may be an important pre-condition for evolution of single-gene incompatibilities. We note that passage through a fitness valley is not required so long as the genetic changes causing incompatibility evolve in multiple steps within separate genetic backgrounds. In this way, two alleles could cause underdominance for fitness and reduce or abolish gene flow, but only upon crossing of lines that have diverged independently from a common ancestor. If there were strong positive selection for two different alleles that caused underdominance or sterility in hybrids, then they could eventually contribute to a speciation event.
In animals single-gene single-generation speciation occurs in snails, where shell chirality is maternally determined, with opposite chirality forming a strong pre-mating barrier [70], [71]. Extenuating factors that could allow rapid speciation based on a single locus, even after one generation, include transient silencing of genes, for example, by parental imprinting, or incomplete sterility of the hybrid. If an incompatible allele arises, but is silenced for one generation, this would allow for the production of multiple offspring that are pre-or post-zygotically incompatible with individuals carrying the ancestral allele. Offspring with the new allele can self or interbreed to establish a subpopulation before this allele is lost again by genetic drift. Similarly, if the heteroallelic combination is sublethal, then F2 offspring homozygous for the new allele can be produced. If, in turn, the homozygous form is subject to positive selection, the allele may become established in the population [70]. Such as scenario is particularly applicable to self-fertilizing species such as Arabidopsis thaliana.
Whether the sort of developmental abnormalities we have observed in Bla-1/Sha F1 hybrids can contribute to cumulative reproductive isolation is of course not known. Nevertheless, that OAK has the potential to greatly reduce reproductive success can be inferred from the severe phenotypes in some plants transformed with active OAK constructs, the necrosis seen when incompatible OAKs are co-expressed from the Col-0 promoter, and the decrease in lifetime fitness as measured via seed set. All together, we propose that the occurrence of genes in variable tandem repeats, such as NB-LRR genes in several hybrid necrosis cases [6], [8], [62], or RLKs as in the present case, predisposes them to being sources for the creation of novel hybrid phenotypes. Whether, as with other mutations, these are normally disadvantageous or not, will require further systematic analyses of hybrid incompatibilities in a broad range of taxa.
Materials and Methods
Plant material
Bla-1 (N28079) and Sha (N28735) were obtained from the European Arabidopsis Stock Centre. Plants were grown at 16°C with 16 hours light, or 23°C with 8 or 16 hours of light, as indicated. Transgenic seedlings were selected on soil by BASTA resistance, and at least 90 T1 plants phenotyped, unless otherwise indicated.
Transgenic plants
Genomic constructs spanned sequences from immediately downstream of the translational stop codon of the preceding gene to 200 bp downstream of the predicted translational stop. AmiRNAs were designed using WMD3 (http://wmd3.weigelworld.org/). Constructs were transformed into plants by the Agrobacterium tumefaciens floral-dip method [72] using strain GV3101 pMP90RK or ASE. For reporter gene analysis, the promoter region between the stop codon of the previous gene and the translational start codon of the OAK homolog was inserted into pGWB433 using Gateway LR clonase (Invitrogen, Darmstadt, Germany).
Seed set
Independent ProOAK∶OAKBla-1 and ProOAK∶OAKSha T1 plants in Col-0 that did not show any morphological defects were crossed to each other to create F1 populations, which were raised in randomly distributed individual pots without selection for the transgenes. Plants were genotyped, and seeds collected from each plant after three months of growth and weighed. The weight of individual seeds was determined by weighing 500 seeds for each of three plants per genotype, and total and individual seed weight were used to calculate total seed number per plant.
Humidity assay
Plants were grown in 23°C (long days) at 65% ambient humidity; or under mild drought-stress with minimal watering (but equal ambient humidity); or in saturated humidity with water surrounding the pots and the tray covered.
Histology
Bla-1 and Bla-1/Sha petioles were fixed in 3.7% formaldehyde, 5% acetic acid, 50% ethanol, embedded in an ASP300 (Leica, Nussloch, Germany) tissue processor in paraffin. Transverse sections of 8 µm thickness, stained with 0.02% Toluidine Blue after dewaxing, were examined with a Zeiss Axioplan 2 microscope.
Callus assay
Seeds were stratified for one week on ½ strength MS plates. Seedlings were grown in Percival LE Intellus chambers (Perry, IA, USA) under 23°C long days until the 4-6 leaf stage. At least 40 transverse sections per genotype of leaves (1 mm thick) and petioles (2 mm thick) were placed on callus induction medium (3.1 g/L Gamborg's B5 salts, 2% glucose, 2.6 mM MES, pH 5.7, 0.8% agar) with 2.2 µM to 22 nM 2,4-dichlorophenoxyacetic acid (2,4-D) and 200 nM to 200 pM kinetin. Callus formation was assessed after 12 days.
Expression analysis
RNA was extracted from leaves of individual plants using the Qiagen (Hilden, Germany) Plant RNeasy Mini kit. One µg of RNA was DNaseI treated, and cDNA synthesized with hexamer primers (Fermentas RevertAid kit, St. Leon-Rot, Germany). qRT-PCR was performed with Invitrogen (St. Louis, MO, USA) SYBR Green PCR Mastermix and the MJR Opticon Continuous Fluorescence Detection System (Bio-Rad, Hercules, CA, USA). Two technical replicates were performed per sample. Expression was normalized to β-TUBULIN-2 (At5g62690) and an amplification efficiency of 2.0 per cycle was used in the calculations. The average across three biological replicates is shown with standard deviation. The 5′ untranslated regions of OAK were identified by 5′ RACE (GeneRacer, Invitrogen, Darmstadt, Germany) on RNA from petioles (Bla-1 and Sha) or pedicels and peduncles (Sha).
GUS staining
Twelve-day old seedlings grown on ½ strength MS plates with kanamycin selection were fixed in 90% acetone on ice for 20 minutes. X-gluc stained tissue [72] was examined with a Leica MZFLIII microscope.
Microarrays
Affymetrix (Santa Clara, CA, USA) ATH1 microarrays were probed as described [73].
Genetic mapping
Coarse mapping was performed with the Sequenom (San Diego, CA, USA) MassARRAY platform. For high-resolution mapping, approximately 750 F2 and F3 plants were genotyped with microsatellite and CAPS markers [72].
Phylogenetic and statistical analyses
For the sliding window analysis of divergence, amino acid sequences were aligned with MUSCLE (http://www.ebi.ac.uk/Tools/muscle/) and nucleotide sequences with BlastX (http://blast.ncbi.nlm.nih.gov/Blast.cgi).
For analysis of population structure, nucleotide sequences were aligned with Lasergene SeqMan. Networks were calculated with SplitsTree [45] using the default parameter settings for NeighborNet. For analysis of haplotypes and recombination, STRUCTURE (version 2.3.2.1) [46] was used with 200,000 iterations for the burnin and 800,000 iterations for the final analysis. A k value of 4 was used based on the SplitsTree results, with all other parameters as default.
Analyses of potential positive selection was performed with the Codeml programme implemented in PAML (version 3.15), using default settings [74]. A likelihood ratio test was used to identify residues under positive selection with Bayesian posterior probability calculated through the Bayes Empirical Bayes (BEB) tool [44]. Sites with dN/dS>1 and a high probability (>95%) are likely to be under positive selection.
A 2-way ANOVA analysis for interaction of lesioning, outgrowth formation and biomass was performed using a web service (http://faculty.vassar.edu/lowry/anova2×2.html).
Supporting Information
Zdroje
1. DobzhanskyT 1937 Genetics and the Origin of Species. New York Columbia University Press 404 420
2. BatesonW 1909 V. Heredity and variation in modern lights. SewardAC Darwin and modern science Cambridge Cambridge University Press 85 101
3. MullerHJ 1942 Isolating mechanisms, evolution and temperature. Biol Symp 6 71 125
4. CoyneJ 1992 Genetics and speciation. Nature 355 511 515
5. OrrH 1996 Dobzhansky, Bateson, and the genetics of speciation. Genetics 144 1331 1335
6. BombliesKLempeJEpplePWarthmannNLanzC 2007 Autoimmunity as a mechanism for hybrid necrosis, a genetic incompatibility syndrome in plants. PLoS Biol 5 e236 doi:10.1371/journal.pbio.0050236
7. KrügerJThomasCMGolsteinCDixonMSSmokerM 2002 A tomato cysteine protease required for Cf-2-dependent disease resistance and suppression of autonecrosis. Science 296 744 747
8. YamamotoETakashiTMorinakaYLinSWuJ 2010 Gain of deleterious function causes an autoimmune response and Bateson-Dobzhansky-Muller incompatibility in rice. Mol Genet Genomics 1 11
9. ClarkRMSchweikertGToomajianCOssowskiSZellerG 2007 Common sequence polymorphisms shaping genetic diversity in Arabidopsis thaliana. Science 317 338 342
10. JorgensenTHEmersonBC 2008 Functional variation in a disease resistance gene in populations of Arabidopsis thaliana. Mol Ecol 17 4912 4923
11. MichelmoreRWMeyersBC 1998 Clusters of resistance genes in plants evolve by divergent selection and a birth-and-death process. Genome Res 8 1113 1130
12. ShiuSHBleeckerAB 2001 Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc Natl Acad Sci USA 98 10763 10768
13. RichlyEKurthJLeisterD 2002 Mode of amplification and reorganization of resistance genes during recent Arabidopsis thaliana evolution. Mol Biol Evol 19 76 84
14. MeyersBCKozikAGriegoAKuangHMichelmoreRW 2003 Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis. Plant Cell 15 809 834
15. ShiuSHBleeckerAB 2003 Expansion of the receptor-like kinase/Pelle gene family and receptor-like proteins in Arabidopsis. Plant Physiol 132 530 543
16. BaumgartenACannonSSpanglerRMayG 2003 Genome-level evolution of resistance genes in Arabidopsis thaliana. Genetics 165 309 319
17. ShiuSHKarlowskiWMPanRTzengYHMayerKFX 2004 Comparative analysis of the receptor-like kinase family in Arabidopsis and rice. Plant Cell 16 1220 1234
18. KuangHWooSSMeyersBCNevoEMichelmoreRW 2004 Multiple genetic processes result in heterogeneous rates of evolution within the major cluster disease resistance genes in lettuce. Plant Cell 16 2870 2894
19. LeisterD 2004 Tandem and segmental gene duplication and recombination in the evolution of plant disease resistance gene. Trends Genet 20 116 122
20. RiesebergLHWillisJH 2007 Plant speciation. Science 317 910 914
21. BombliesKWeigelD 2007 Hybrid necrosis: autoimmunity as a potential gene-flow barrier in plant species. Nat Rev Genet 8 382 393
22. JoshiMG 1972 Occurrence of genetic tumours in Triticum interspecies hybrids. Theor Appl Genet 42 227 228
23. SmithHH 1988 The inheritance of genetic tumors in Nicotiana hybrids. J Hered 79 277 283
24. AhujaMR 1965 Genetic control of tumor formation in higher plants. Quart Rev Biol 40 329 340
25. McDanielSFWillisJHShawAJ 2008 The genetic basis of developmental abnormalities in interpopulation hybrids of the moss Ceratodon purpureus. Genetics 179 1425 1435
26. EastEM 1936 Heterosis. Genetics 21 375 397
27. CrowJF 1948 Alternative hypotheses of hybrid vigor. Genetics 33 477 487
28. BirchlerJYaoHChudalayandiSVaimanDVeitiaR 2010 Heterosis. Plant Cell 22 2105 2112
29. CoyneJAOrrHA 2004 Speciation. Sunderland, MA Sinauer 557
30. SchwartzDLaughnerWJ 1969 A molecular basis for heterosis. Science 166 626 627
31. BuschMMayerUJürgensG 1996 Molecular analysis of the Arabidopsis pattern formation of gene GNOM: gene structure and intragenic complementation. Mol Gen Genet 250 681 691
32. KriegerULippmanZBZamirD 2010 The flowering gene SINGLE FLOWER TRUSS drives heterosis for yield in tomato. Nat Genet 42 459 463
33. ChenJDingJOuyangYDuHYangJ 2008 A triallelic system of S5 is a major regulator of the reproductive barrier and compatibility of indica-japonica hybrids in rice. Proc Natl Acad Sci USA 105 11436 11441
34. ZhouFMenkeFLHYoshiokaKModerWShiranoY 2004 High humidity suppresses ssi4-mediated cell death and disease resistance upstream of MAP kinase activation, H2O2 production and defense gene expression. Plant J 39 920 932
35. BayerMHAhujaMR 1968 Tumor formation in Nicotiana: auxin levels and auxin inhibitors in normal and tumor-prone genotypes. Planta 79 292 298
36. CampellBTownC 1991 Physiology of hormone autonomous tissue lines derived from radiation-induced tumors of Arabidopsis thaliana. Plant Physiol 97 1166 1173
37. TsesmetzisNCouchmanMHigginsJSmithADoonanJH 2008 Arabidopsis reactome: a foundation knowledgebase for plant systems biology. Plant Cell 20 1426 1436
38. CarbonSIrelandAMungallCShuSMarshallB 2009 AmiGO: online access to ontology and annotation data. Bioinformatics 25 288 289
39. HuTTPattynPBakkerEGCaoJChengJ-F 2011 The Arabidopsis lyrata genome sequence and the basis of rapid genome size change. Nat Genet in press
40. OssowskiSSchwabRWeigelD 2008 Gene silencing in plants using artificial microRNAs and other small RNAs. Plant J 53 674 690
41. SchallusTJaeckhCFehérKPalmaASLiuY 2008 Malectin: a novel carbohydrate-binding protein of the endoplasmic reticulum and a candidate player in the early steps of protein N-glycosylation. Mol Biol Cell 19 3404 3414
42. NeiMGojoboriT 1986 Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol 3 418 426
43. YangZ 2007 PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol 24 1586 1591
44. YangZWongWNielsenR 2005 Bayes empirical bayes inference of amino acid sites under positive selection. Mol Biol Evol 22 1107 1118
45. HusonDHBryantD 2006 Application of phylogenetic networks in evolutionary studies. Mol Biol Evol 23 254 267
46. FalushDStephensMPritchardJK 2003 Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164 1567 1587
47. KnightonDZhengJTen EyckLAshfordVXuongN 1991 Crystal structure of the catalytic subunit of cyclic adenosine monophosphate-dependent protein kinase. Science 253 407 414
48. BecraftP 2002 Receptor kinase signaling in plant development. Annu Rev Cell Dev Biol 18 163 192
49. GlazebrookJ 2001 Genes controlling expression of defense responses in Arabidopsis - 2001 status. Curr Opin Plant Biol 4 301 308
50. GaffneyTFriedrichLVernooijBNegrottoDNyeG 1993 Requirement of salicylic acid for the induction of systemic acquired resistance. Science 261 754 756
51. ShanLHePLiJHeeseAPeckSC 2008 Bacterial effectors target the common signaling partner BAK1 to disrupt multiple MAMP receptor-signaling complexes and impede plant immunity. Cell Host Microbe 4 17 27
52. LeeSWHanSWSririyanumMParkCJSeoYS 2009 A type I-secreted, sulfated peptide triggers XA21-mediated innate immunity. Science 326 850 853
53. Gomez-GomezLBollerT 2000 FLS2: an LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol Cell 5 1003 1011
54. SearleIMenALaniyaTBuzasDIturbe-OrmaetxeI 2003 Long-distance signaling in nodulation directed by a CLAVATA1-like receptor kinase. Science 299 109 112
55. SternDOrgogozoV 2009 Is genetic evolution predictable? Science 323 746 751
56. GouXPHeKYangHYuanTLinHH 2010 Genome-wide cloning and sequence analysis of leucine-rich repeat receptor-like protein kinase genes in Arabidopsis thaliana. BMC Genomics 11 19
57. KinoshitaTCaño-DelgadoASetoHHiranumaSFujiokaS 2005 Binding of brassinosteroids to the extracellular domain of plant receptor kinase BRI1. Nature 433 167 171
58. OgawaMShinoharaHSakagamiYMatsubayashiY 2008 Arabidopsis CLV3 peptide directly binds CLV1 ectodomain. Science 319 294
59. ShpakEDMcAbeeJMPillitteriLJToriiKU 2005 Stomatal patterning and differentiation by synergistic interactions of receptor kinases. Science 309 290 293
60. ZhangLGautBS 2003 Does recombination shape the distribution and evolution of tandemly arrayed genes (TAGs) in the Arabidopsis thaliana genome? Genome Res 13 2533 2540
61. BollerTFelixG 2009 A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu Rev Plant Biol 60 379 406
62. AlcázarRGarciaAVParkerJEReymondM 2009 Incremental steps toward incompatibility revealed by Arabidopsis epistatic interactions modulating salicylic acid pathway activation. Proc Natl Acad Sci USA 106 334 339
63. WangXKotaUHeKBlackburnKLiJ 2008 Sequential transphosphorylation of the BRI1/BAK1 receptor kinase complex impacts early events in brassinosteroid signaling. Dev Cell 15 220 235
64. DanglJLJonesJD 2001 Plant pathogens and integrated defence responses to infection. Nature 411 826 833
65. McNallyKLChildsKLBohnertRDavidsonRMZhaoK 2009 Genomewide SNP variation reveals relationships among landraces and modern varieties of rice. Proc Natl Acad Sci USA 106 12273 12278
66. RiesebergLBlackmanB 2010 Speciation genes in plants. Ann Bot 106 439 455
67. HulbertSWebbCSmithSSunQ 2001 Resistance gene complexes: evolution and utilization. Annu Rev Phytopathol 39 285 312
68. LeeHChouJCheongLChangNYangS 2008 Incompatibility of nuclear and mitochondrial genomes causes hybrid sterility between two yeast species. Cell 135 1065 1073
69. PhillipsPC 2008 Epistasis--the essential role of gene interactions in the structure and evolution of genetic systems. Nat Rev Genet 9 855 867
70. OrrH 1991 Is single-gene speciation possible? Evolution 45 764 769
71. UeshimaRAsamiT 2003 Single-gene speciation by left-right reversal. Nature 425 679
72. WeigelDGlazebrookJ 2002 Arabidopsis: A Laboratory Manual. Cold Spring Harbor, NY Cold Spring Harbor Laboratory Press 354
73. LaubingerSZellerGHenzSRSachsenbergTWidmerCK 2008 At-TAX: a whole genome tiling array resource for developmental expression analysis and transcript identification in Arabidopsis thaliana. Genome Biol 9 R112
74. YangZWongWSNielsenR 2005 Bayes empirical bayes inference of amino acid sites under positive selection. Mol Biol Evol 22 1107 1118
Štítky
Genetika Reprodukční medicína
Článek Pervasive Sign Epistasis between Conjugative Plasmids and Drug-Resistance Chromosomal MutationsČlánek Stress-Induced PARP Activation Mediates Recruitment of Mi-2 to Promote Heat Shock Gene ExpressionČlánek Histone Crosstalk Directed by H2B Ubiquitination Is Required for Chromatin Boundary IntegrityČlánek A Functional Variant at a Prostate Cancer Predisposition Locus at 8q24 Is Associated with ExpressionČlánek Replication and Explorations of High-Order Epistasis Using a Large Advanced Intercross Line PedigreeČlánek Expression of Tumor Suppressor in Spermatogonia Facilitates Meiotic Progression in Male Germ Cells
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2011 Číslo 7
-
Všechny články tohoto čísla
- Gene-Based Tests of Association
- The Demoiselle of X-Inactivation: 50 Years Old and As Trendy and Mesmerising As Ever
- Variants in and Underlie Natural Variation in Translation Termination Efficiency in
- SHH1, a Homeodomain Protein Required for DNA Methylation, As Well As RDR2, RDM4, and Chromatin Remodeling Factors, Associate with RNA Polymerase IV
- Genome-Wide Association Study Identifies as a Susceptibility Gene for Pediatric Asthma in Asian Populations
- Pervasive Sign Epistasis between Conjugative Plasmids and Drug-Resistance Chromosomal Mutations
- Genetic Anticipation Is Associated with Telomere Shortening in Hereditary Breast Cancer
- Identification of a Mutation Associated with Fatal Foal Immunodeficiency Syndrome in the Fell and Dales Pony
- Stress-Induced PARP Activation Mediates Recruitment of Mi-2 to Promote Heat Shock Gene Expression
- An Epigenetic Switch Involving Overlapping Fur and DNA Methylation Optimizes Expression of a Type VI Secretion Gene Cluster
- Recombination and Population Structure in
- A Rice Plastidial Nucleotide Sugar Epimerase Is Involved in Galactolipid Biosynthesis and Improves Photosynthetic Efficiency
- A Role for Phosphatidic Acid in the Formation of “Supersized” Lipid Droplets
- Colon Stem Cell and Crypt Dynamics Exposed by Cell Lineage Reconstruction
- Loss of the BMP Antagonist, SMOC-1, Causes Ophthalmo-Acromelic (Waardenburg Anophthalmia) Syndrome in Humans and Mice
- Interactions between Glucocorticoid Treatment and Cis-Regulatory Polymorphisms Contribute to Cellular Response Phenotypes
- Translation Reinitiation Relies on the Interaction between eIF3a/TIF32 and Progressively Folded -Acting mRNA Elements Preceding Short uORFs
- DAF-12 Regulates a Connected Network of Genes to Ensure Robust Developmental Decisions
- Adult Circadian Behavior in Requires Developmental Expression of , But Not
- Histone Crosstalk Directed by H2B Ubiquitination Is Required for Chromatin Boundary Integrity
- Proteins in the Nutrient-Sensing and DNA Damage Checkpoint Pathways Cooperate to Restrain Mitotic Progression following DNA Damage
- Complex Evolutionary Events at a Tandem Cluster of Genes Resulting in a Single-Locus Genetic Incompatibility
- () and Its Regulated Homeodomain Gene Mediate Abscisic Acid Response in
- A Functional Variant at a Prostate Cancer Predisposition Locus at 8q24 Is Associated with Expression
- LGI2 Truncation Causes a Remitting Focal Epilepsy in Dogs
- Adaptations to Endosymbiosis in a Cnidarian-Dinoflagellate Association: Differential Gene Expression and Specific Gene Duplications
- The Translation Initiation Factor eIF4E Regulates the Sex-Specific Expression of the Master Switch Gene in
- Somatic Genetics Empowers the Mouse for Modeling and Interrogating Developmental and Disease Processes
- Molecular Mechanisms Generating and Stabilizing Terminal 22q13 Deletions in 44 Subjects with Phelan/McDermid Syndrome
- Replication and Explorations of High-Order Epistasis Using a Large Advanced Intercross Line Pedigree
- Mechanisms of Chromosome Number Evolution in Yeast
- Regulatory Cross-Talk Links Chromosome II Replication and Segregation
- Ancestral Genes Can Control the Ability of Horizontally Acquired Loci to Confer New Traits
- Expression of Tumor Suppressor in Spermatogonia Facilitates Meiotic Progression in Male Germ Cells
- Rare and Common Regulatory Variation in Population-Scale Sequenced Human Genomes
- The Epistatic Relationship between BRCA2 and the Other RAD51 Mediators in Homologous Recombination
- Identification of Novel Genetic Markers Associated with Clinical Phenotypes of Systemic Sclerosis through a Genome-Wide Association Strategy
- NatF Contributes to an Evolutionary Shift in Protein N-Terminal Acetylation and Is Important for Normal Chromosome Segregation
- Araucan and Caupolican Integrate Intrinsic and Signalling Inputs for the Acquisition by Muscle Progenitors of the Lateral Transverse Fate
- Pathologic and Phenotypic Alterations in a Mouse Expressing a Connexin47 Missense Mutation That Causes Pelizaeus-Merzbacher–Like Disease in Humans
- Recombinant Inbred Line Genotypes Reveal Inter-Strain Incompatibility and the Evolution of Recombination
- Epistatic Relationships in the BRCA1-BRCA2 Pathway
- Genome-Wide Association Study Identifies Novel Restless Legs Syndrome Susceptibility Loci on 2p14 and 16q12.1
- Genetic Loci Associated with Plasma Phospholipid n-3 Fatty Acids: A Meta-Analysis of Genome-Wide Association Studies from the CHARGE Consortium
- Fine Mapping of Five Loci Associated with Low-Density Lipoprotein Cholesterol Detects Variants That Double the Explained Heritability
- CHD1 Remodels Chromatin and Influences Transient DNA Methylation at the Clock Gene
- Nonlinear Fitness Landscape of a Molecular Pathway
- Genome-Wide Scan Identifies , , and as Novel Risk Loci for Systemic Sclerosis
- Quantitative and Qualitative Stem Rust Resistance Factors in Barley Are Associated with Transcriptional Suppression of Defense Regulons
- A Systematic Screen for Tube Morphogenesis and Branching Genes in the Tracheal System
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Genome-Wide Association Study Identifies Novel Restless Legs Syndrome Susceptibility Loci on 2p14 and 16q12.1
- Loss of the BMP Antagonist, SMOC-1, Causes Ophthalmo-Acromelic (Waardenburg Anophthalmia) Syndrome in Humans and Mice
- Gene-Based Tests of Association
- Genome-Wide Association Study Identifies as a Susceptibility Gene for Pediatric Asthma in Asian Populations
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



