-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaTmprss2 Is Essential for Influenza H1N1 Virus Pathogenesis in Mice
Annual influenza epidemics and occasional pandemics pose a severe threat to human health. Host cell factors required for viral spread but not for cellular survival are attractive targets for novel approaches to antiviral intervention. The cleavage activation of the influenza virus hemagglutinin (HA) by host cell proteases is essential for viral infectivity. However, it is unknown which proteases activate influenza viruses in mammals. Several candidates have been identified in cell culture studies, leading to the concept that influenza viruses can employ multiple enzymes to ensure their cleavage activation in the host. Here, we show that deletion of a single HA-activating protease gene, Tmprss2, in mice inhibits spread of mono-basic H1N1 influenza viruses, including the pandemic 2009 swine influenza virus. Lung pathology was strongly reduced and mutant mice were protected from weight loss, death and impairment of lung function. Also, after infection with mono-basic H3N2 influenza A virus body weight loss and survival was less severe in Tmprss2 mutant compared to wild type mice. As expected, Tmprss2-deficient mice were not protected from viral spread and pathology after infection with multi-basic H7N7 influenza A virus. In conclusion, these results identify TMPRSS2 as a host cell factor essential for viral spread and pathogenesis of mono-basic H1N1 and H3N2 influenza A viruses.
Published in the journal: . PLoS Pathog 9(12): e32767. doi:10.1371/journal.ppat.1003774
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1003774Summary
Annual influenza epidemics and occasional pandemics pose a severe threat to human health. Host cell factors required for viral spread but not for cellular survival are attractive targets for novel approaches to antiviral intervention. The cleavage activation of the influenza virus hemagglutinin (HA) by host cell proteases is essential for viral infectivity. However, it is unknown which proteases activate influenza viruses in mammals. Several candidates have been identified in cell culture studies, leading to the concept that influenza viruses can employ multiple enzymes to ensure their cleavage activation in the host. Here, we show that deletion of a single HA-activating protease gene, Tmprss2, in mice inhibits spread of mono-basic H1N1 influenza viruses, including the pandemic 2009 swine influenza virus. Lung pathology was strongly reduced and mutant mice were protected from weight loss, death and impairment of lung function. Also, after infection with mono-basic H3N2 influenza A virus body weight loss and survival was less severe in Tmprss2 mutant compared to wild type mice. As expected, Tmprss2-deficient mice were not protected from viral spread and pathology after infection with multi-basic H7N7 influenza A virus. In conclusion, these results identify TMPRSS2 as a host cell factor essential for viral spread and pathogenesis of mono-basic H1N1 and H3N2 influenza A viruses.
Introduction
Annual influenza epidemics and unpredictable pandemics pose a severe threat to human health, exemplified by the estimated 30–50 million deaths caused by the 1918 pandemic. Current therapy targets viral proteins, neuraminidase and M2, but is hampered by development of resistance [1], due to the high mutation rate of the virus. Novel antiviral strategies are urgently required and invariable host cell factors essential for viral spread are attractive targets.
The cleavage of the influenza virus hemagglutinin (HA) by host cell proteases is essential for viral infectivity [2], [3]. The HA proteins of highly pathogenic avian influenza viruses harbor multiple basic amino acids at their cleavage site and are activated by furin [4]. In contrast, low pathogenic avian and human influenza viruses contain a mono-basic cleavage site in their HA proteins. Several studies showed that multiple secreted proteases can activate human influenza viruses for infection of cell lines (see [5], [6] for examples and [7] for a review). However, the analysis of cultured human respiratory epithelium demonstrated that influenza virus cleavage activation is a cell-associated process and no evidence for a role of secreted proteases was obtained [8]. Subsequently, the type II transmembrane serine protease (TTSP) family member TMPRSS2, a membrane associated protease, was shown to activate HA proteins of diverse human influenza viruses in cell culture [9], [10], [11], [12]. In addition, TMPRSS2 was found to be expressed in human respiratory epithelium positive for alpha 2,6-linked sialic acid [13]. However, the role of TMPRSS2 in influenza virus spread and pathogenesis in the infected host has not yet been studied.
Therefore, we investigated if TMPRSS2 contributes to influenza virus replication and pathogenesis in experimentally infected mice. We focused our analysis on viruses of the H1N1 (including the pandemic 2009 influenza virus) and H3N2 subtypes, since viruses of these subtypes are presently circulating in the human population. Our study shows that deletion of Tmprss2 in knock-out mice strongly limits virus spread and lung pathology after H1N1 influenza A virus infection. The deletion of Tmprss2 also reduces body weight loss and mortality after H3N2 infection but to a much lower degree than for H1N1 infected mice.
Results
Tmprss2 is essential for spread and pathogenesis of H1N1 influenza viruses in mice
To assess the role of TMPRSS2 during influenza virus infection in vivo, we used mice carrying a deletion of the Tmprss2 gene [14]. Non-infected Tmprss2 knock-out mice did not show a phenotype in the absence of infection, as described previously [14] and RT-PCR analysis of kidney tissue confirmed the absence of full length Tmprss2 transcripts. Upon intranasal infection of mice with mouse-adapted PR8M (A/PuertoRico/8/34 H1N1 Münster variant, [15]), wild type mice lost weight significantly after infection and 50% of infected mice died, whereas Tmprss2 knock-out mice did not exhibit body weight loss and showed no signs of disease (Figs. 1A; Fig. S1). The same results were obtained after infection with a human isolate of the pandemic HA4 (A/Hamburg/4/2009 H1N1) virus (Figure 1B). Also, after infection with a lethal dose of the more virulent PR8F virus isolate (A/PuertoRico/8/34 H1N1 Freiburg variant) all wild-type mice died within seven days post infection whereas no knock-out mice showed symptoms of disease (Figure 1C). Similar results were observed for blood oxygen saturation levels which provide a measurement for lung function. Whereas wild type mice exhibited a significant drop in peripheral blood oxygen saturation that peaked at day 8 post infection (p.i.) with PR8M virus, Tmprss2−/− mutant mice showed only a very mild change (Figure 2).
Fig. 1. Tmprss2 is essential for spread and pathogenesis of H1N1 influenza viruses. 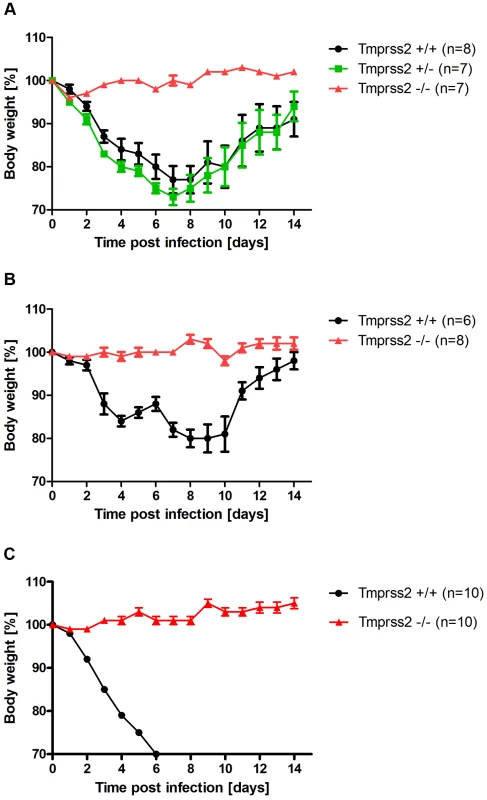
Eight to eleven weeks old female mice were infected with 2×105 FFU mouse-adapted PR8M (H1N1; A), 2×105 FFU HA4 (pH1N1, B), 2×103 FFU mouse-adapted PR8F (H1N1; C). Body weight loss was monitored until day 14 p.i. Mice with a weight loss of more than 30% of the starting bodyweight were euthanized and recorded as dead. Weight loss data represent mean values +/− SEM. Note that only data of surviving mice are presented (e.g. about 50% of infected mice died after infection with PR8M, see Fig. S1). Body weight loss was significantly different between wild type and homozygous mutant mice at day 6 p.i. (p<0.0001 for PR8M infected mice, and p<0.0001 for HA4 infected mice, using the non-parametric Mann Whitney U test) and between heterozygous and homozygous mutant mice (PR8M infected mice, p<0.0001 using the Mann Whitney U test). Fig. 2. Tmprss2−/− mutant mice do not exhibit drop in blood oxygen saturation. 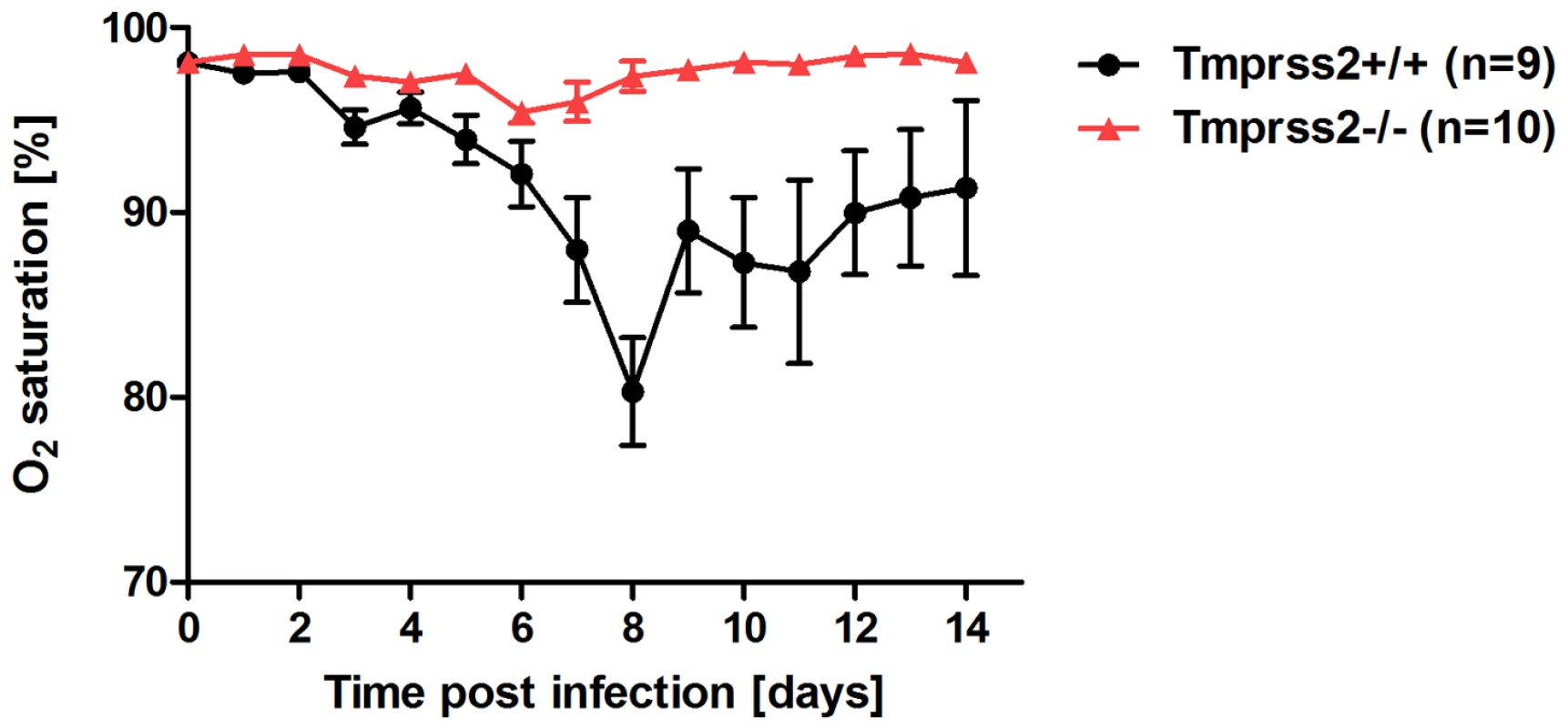
Tmprss2−/− and wild type mice were infected with 2×105 FFU PR8M virus and oxygen saturation in the peripheral blood was measured until day 14 p.i. Tmprss2−/− mice showed only a very mild drop in oxygen saturation whereas wild type mice exhibited a significant decrease that peaked at day 8 p.i. Histological analyses of infected lungs revealed a similar onset of the influenza infection on day 1 post infection in Tmprss2−/− and wild type mice with infected epithelial cells in the bronchiole (Figure 3A,B,E,F). However, at day 3 p.i. virus was spreading into the alveolar regions of wild type mice whereas it was only found in bronchioles in Tmprss2−/− mice (Figure 3C,D and G,H, respectively). Furthermore, infected wild type mice showed a strong increase in lung infiltrates and also in the number of infected cells whereas in Tmprss2 knock-out mice a much lower number of infiltrating cells and infected cells were observed (Figure 3 K,L and O,P, respectively). Thus, the absence of TMPRSS2 largely protects animals from virus spread and virus induced pathogenesis.
Fig. 3. Mild lung pathology and reduced viral spread is observed in Tmprss2−/− mutant mice. 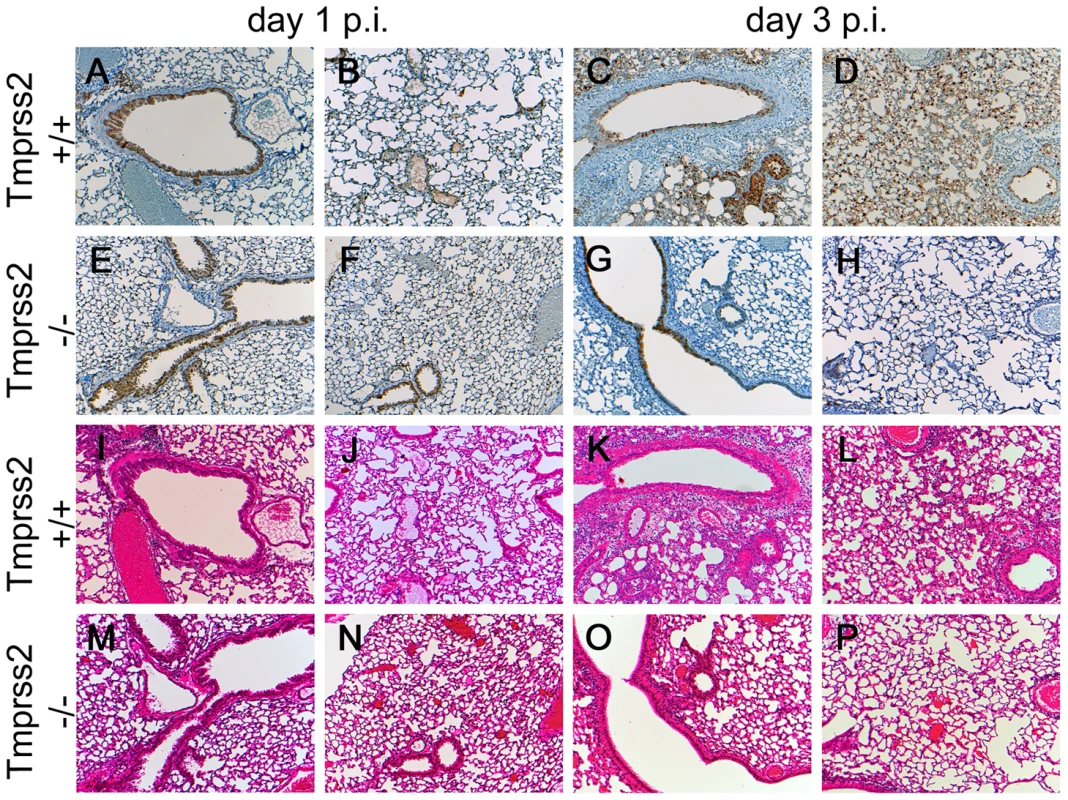
Eight to twelve weeks old mice were infected intra-nasally with 2×105 FFU of PR8M virus. Serial lung sections were stained at day 1 and day 3 p.i. with anti-influenza antibody and haematoxylin (A–H) or with haematoxylin/eosin (I–P). The overall lung tissues were more densely consolidated with larger numbers of infiltrating immune cells in wild type (I–L) compared to Tmprss2−/− mice (M–P). In addition, the airways of Tmprss2+/+ mice were surrounded by higher numbers neutrophils and macrophages (I–L) whereas airways of Tmprss2−/− mice showed lower numbers of immune cell infiltrations (M–P). Virus-infected cells at day 1 p.i. were observed mainly in bronchiolar regions in the lungs of both mice (A, B, E, F). Both wild type and mutant mice showed influenza-positive cells at day 3 p.i. (C, G). However, the overall number of infected cells was lower in Tmprss2−/− compared to wild type mice. Furthermore, infected cells were mostly limited to bronchiolar regions in Tmprss2−/− whereas in wild type mice the virus also spread significantly into the alveolar regions (D, H). Next, we assessed if protection from pathogenesis was due to reduced viral spread. After infection with PR8M, we could detect infectious viral particles in the lung in both homozygous Tmprss2−/− and wild type mice. However, the number of infectious particles was close to background at day 1 p.i. in Tmprss2−/− mice and was markedly reduced at days 2 and 3 p.i. compared to wild type mice (Figure 4). Influenza-specific antibodies were readily detectable in sera of Tmprss2 knock-out mice (Figure S2) after infection with PR8M and PR8F virus demonstrating that the inoculated virus was able to infect lung cells and elicit a humoral immune response. In conclusion, these results show that TMPRSS2 is critical for efficient spread and pathogenesis of epidemic and pandemic H1N1 influenza viruses in vivo.
Fig. 4. Viral load in the lungs of Tmprss2−/− mice after infection with H1N1 (PR8M) influenza A virus. 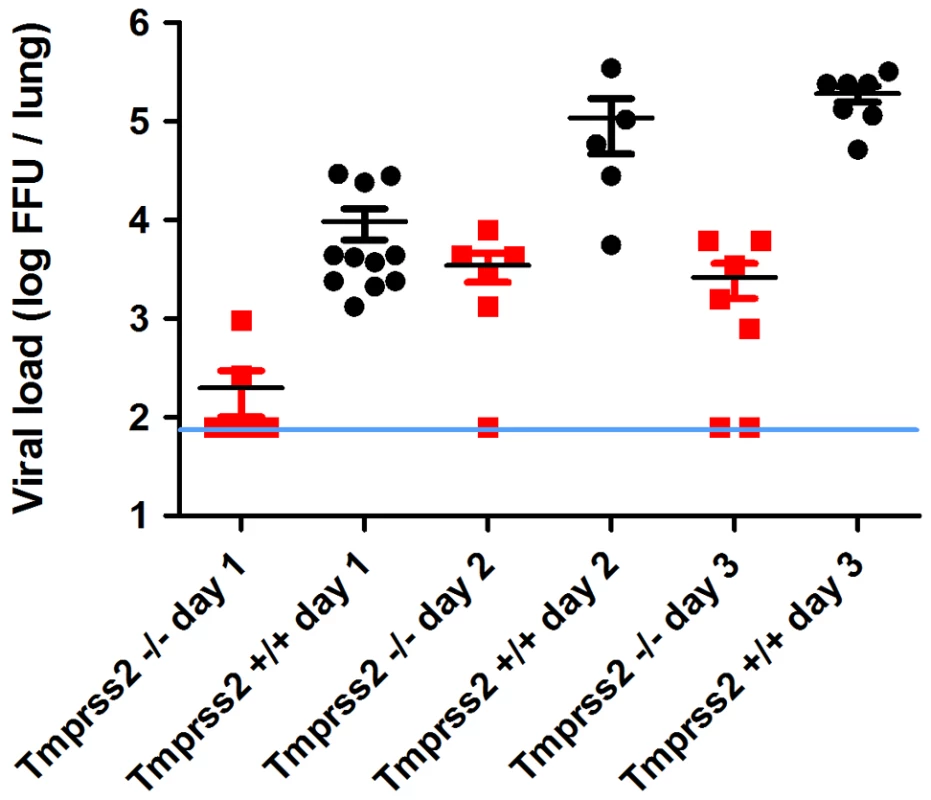
Eight to eleven weeks old female mice were infected with 2×105 FFU of the PR8M virus. Infectious virus particles were determined in lung homogenates. Viral load was higher in infected wild type mice compared to infected homozygous mutant mice at days 1, 2 and 3 p.i. Individual values, mean and SEM are presented. Detection limit of the assay is at 80 infectious particles per lung indicated by the blue line. Day 1 p.i. n = 9 for Tmprss2−/−, n = 11 for Tmprss2+/+, day 2 p.i. n = 6 for Tmprss2−/−, n = 5 for Tmprss2+/+, day 3 p.i. n = 7 for Tmprss2−/−, n = 7 for Tmprss2+/+. Tmprss2 is required for HA cleavage in mouse lungs
Next, we sought to obtain direct evidence for the lack of proteolytic cleavage of HA in Tmprss2 deficient mice. For this, broncho-alveolar lavages (BAL) of infected mice were collected and the proteolytic processing of the HA precursor protein HA0 was analyzed. After infection with a high dose of PR8M virus, processed HA1 as well as non-processed HA0 protein were detected in infected wild type mice whereas only HA0 protein was found in homozygous Tmprss2 mutant mice (Figure 5). These results demonstrate that TMPRSS2 is essential for efficient HA cleavage activation in mice.
Fig. 5. The hemagglutinin of H1N1 PR8M influenza virus is not processed in Tmprss2 knock-out mice. 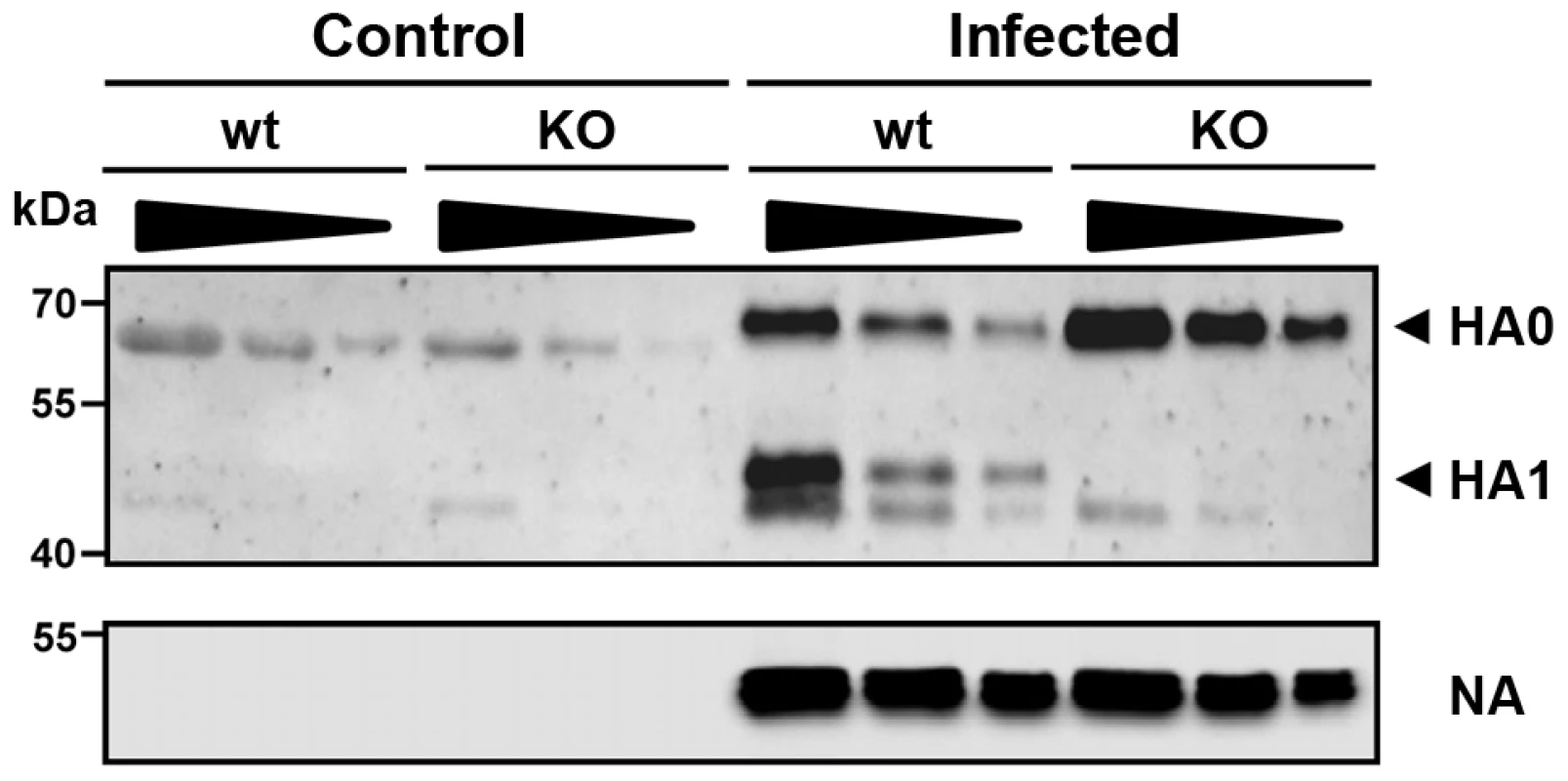
BAL from infected wild type and Tmprss2−/− male mice was harvested at day 1 after infection with 2×105 FFU PR8M and viral particles were concentrated by centrifugation through a sucrose cushion. As control, BAL from non-infected wild type and Tmprss2−/− was analyzed. Each sample was loaded undiluted (first lane), and in two dilutions (second lane 1∶1.33, third lane 1∶2). The virus-containing pellets were then analyzed for HA cleavage by Western blots. As loading control, the stripped membranes were incubated with anti-influenza A virus antibody confirming that equal amounts of proteins were loaded for respective undiluted and diluted samples. Tmprss2-deficient mice were also protected from pathogenesis after infection with H3N2 virus but to a lesser extent
At present, influenza viruses of the HA subtypes H1 and H3 are circulating in humans. Therefore, we investigated if a H3 virus was also dependent on expression of a functional Tmprss2 gene. After infection with a low dose (101 Focus Forming Units (FFU)) of a mouse-adapted H3N2 virus (A/HK/01/68 [16]), body weight loss is less severe and survival is increased in Tmprss2 knock-out compared to wild type mice (Figure 6A, B). After infection with a higher dose (2×103 FFU) of H3N2 virus, mortality was significantly lower in Tmprss2−/− mice compared to wild type mice (Figure 7A). However, no significant differences were observed in the amount of infectious particles at day 1 to 3 p.i. (Figure 7B). Thus, activity of TMPRSS2 is required for the processing of both H1N1 and H3N2 but H3N2 viruses may be cleaved by other proteases in addition to TMPRSS2.
Fig. 6. Tmprss2 knock-out mice show reduced body weight loss and mortality after infection with low dose H3N2 influenza A virus infections. 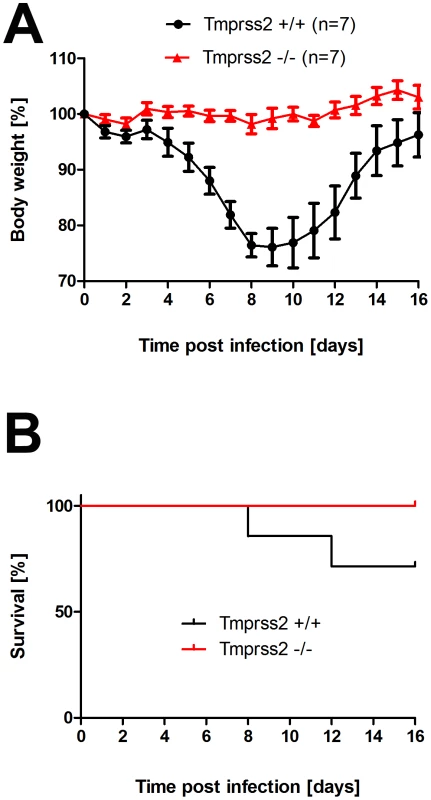
Eight to eleven weeks old female mice were infected with 101 FFU mouse-adapted H3N2 influenza virus by intra-nasal application and bodyweight (A) and survival (B) was monitored until day 14 p.i. In addition to mice that were found dead, mice with a weight loss of more than 30% of the starting body weight were euthanized and recorded as dead. Homozygous Tmprss2 knock-out mice lost significantly less weight than wild type (e.g. p = 0,0006 at day 7, and p = 0,0006 at day 8, using MWU test) mice and showed reduced mortality compared to wild type mice, although this difference was not significant (using the log rank test). Fig. 7. Tmprss2 knock-out mice show reduced mortality after infection with high dose H3N2 influenza A virus infections. 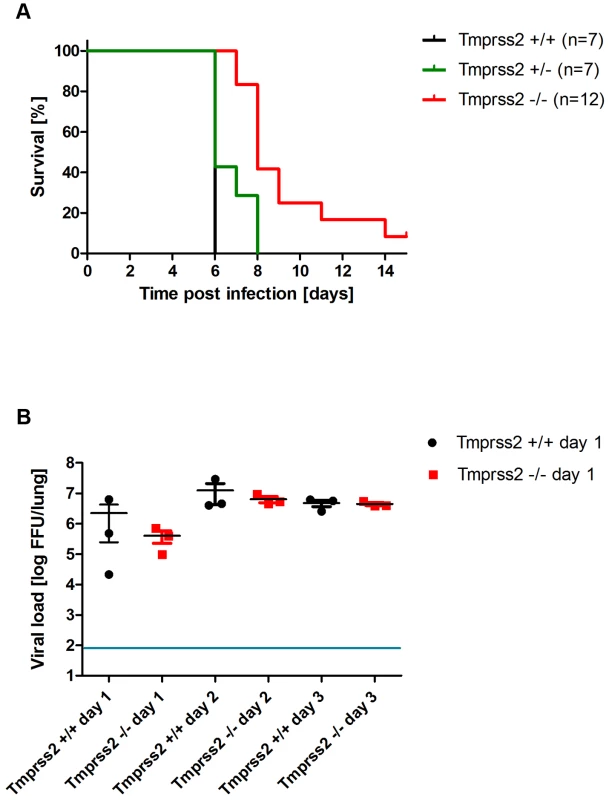
Eight to eleven weeks old female mice were infected with 2×103 FFU mouse-adapted H3N2 influenza virus by intra-nasal application and survival (A) was monitored until day 14 p.i. In addition to mice that were found dead, mice with a weight loss of more than 30% of the starting body weight were euthanized and recorded as dead. Infectious viral particles were determined in lung homogenates (B). Individual values, mean and SEM are presented. Detection limit of the assay is at 80 infectious particles per lung indicated by the blue line. Homozygous Tmprss2 knock-out mice showed significantly reduced mortality compared to wild type and heterozygote mice (p<0.0001 and p = 0.0032, respectively, using the log rank test). Viral load was not significantly different in infected wild type mice compared to infected homozygous mutant mice at days 1 to 3 p.i. Tmprss2 is not required for spread and pathogenesis of a virus with a multi-basic cleavage site
In cell culture, TMPRSS2 is dispensable for cleavage activation of viruses with a multi-basic cleavage site [10]. Thus, if the resistance of Tmprss2 knock-out mice to H1N1 infection was indeed due to lack of HA processing, a virus with a multi-basic cleavage site should spread efficiently and cause disease. To investigate this, we infected mice with SC35M (mouse-adapted A/Seal/Massachusetts/1/80, H7N7) influenza virus which contains a multi-basic HA cleavage site. Mortality and body weight loss in infected Tmprss2−/− mice were not significantly different compared to wild type and Tmprss2+/− infected mice (Figure 8), suggesting that the presence of a multi-basic cleavage site renders viral spread and pathogenesis independent of TMPRSS2 expression.
Fig. 8. Murine Tmprss2 is not required for spread of H7N7 influenza A virus. 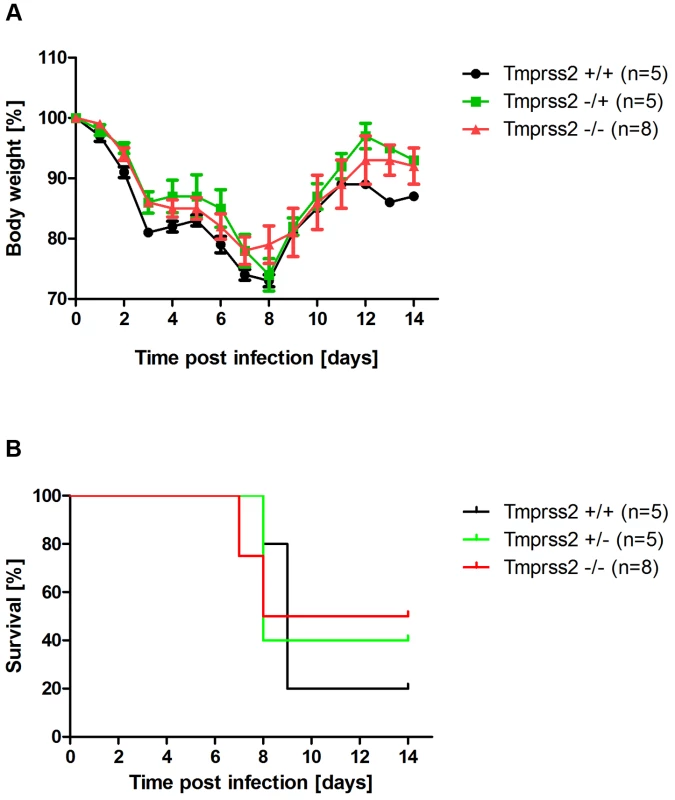
Eight to eleven weeks old female mice were infected with 2×104 FFU mouse-adapted SC35M (H7N7) influenza virus by intra-nasal application and bodyweight (A) and survival (B) was monitored until day 14 p.i. In addition to mice that were found dead, mice with a weight loss of more than 30% of the starting bodyweight were euthanized and recorded as dead. No significant differences were observed in survival between heterozygous and homozygous mutant mice after H7N7 infections (using the log rank test). Cleavage-activation of diverse influenza viruses by murine Tmprss2 in vitro
Finally, we investigated if murine Tmprss2 was able to activate HA proteins of H1N1 and H3N2 viruses. The co-expression of protease and the respective HA proteins of PR8M, HA4, and H3N2 viruses in transfected cells facilitated HA cleavage of all viruses (Figure S3A). Additionally, expression of TMPRSS2 in this cell culture system allowed the spread of PR8M and HA4 viruses in a trypsin-independent fashion (Figure S3B). In contrast, spread of mouse-adapted SC35M virus, containing a multi-basic cleavage site did not depend on TMPRSS2 expression [10]. Thus, murine Tmprss2, like its human homologue [11], [17] can activate HA.
Discussion
Cleavage activation of influenza virus HA by host cell proteases is essential for viral infectivity [2], [3]. However, the nature of the proteases required for the cleavage activation of viruses with a mono-basic HA cleavage site in the infected host organism remains unclear. At least eight candidate enzymes from different protease families have been suggested based on cell culture studies [18], leading to the concept that redundant proteolytic enzymes activate influenza viruses in the host. Here, we show for the first time that the deletion of a single protease, TMPRSS2, in mice largely abrogates viral spread and protects animals from severe pathology and death after H1N1 and, to a lower extent, H3N2 influenza virus infection.
After infection of Tmprss2−/− mice with H1N1 virus, no processing of the HA precursor protein HA0 was observed in BAL. However, an initial increase in viral titers was measured from day 1 to day 2 p.i. whereas at later times p.i. viral titers rapidly decreased. This initial increase in viral titers is expected because the virus used for infections had been produced in embryonated chicken eggs. It therefore carries an activated HA allowing it to enter cells and replicate [19], [20]. In addition, it is conceivable that other proteases besides TMPRSS2 may facilitate low levels of H1 cleavage and allow limited viral spread which is rapidly cleared once the antiviral immune responses have been activated. However, the cleavage activation by such alternative enzymes must be very inefficient since viral titers in H1N1 infected Tmprss2−/− mice were markedly reduced compared to wild type animals and no weight loss was observed.
Surprisingly, H3N2 virus which also carries a mono-basic cleavage site in the HA, was able to replicate in Tmprss2−/− mice. However, body weight loss and mortality were significantly reduced in knock-out mice compared to wild type mice in a dose-dependent manner. On the other hand, viral load was not significantly lower in mutant mice after infection with 2×103 FFU. The amino acid sequence in the HA loop which is recognized by proteases differs between HA subtypes H1 and H3 (Figure 9). A recent study demonstrated that different proteases, including TMPRSS2, TMPRSS11d (HAT) and even trypsin, cleave HA from different subtypes and variants with varying efficiency [21]. In addition, ST14 (matriptase) has the capability to cleave HA of particular H1 subtype strains but only minimal cleavage was observed for H2 and H3 [22]. Furthermore, KLK5 and 12 (kallikrein related-peptidase 5 and 12) have been described as host proteases that are capable of cleavage activation of viral H1, H2 and H3 HA in vitro [23]. Thus, H3N2 appears to be processed to some extent by TMPRSS2, resulting in the reduced pathology in Tmprss2 knock-out mice but also other proteases are able to cleave H3 hemagglutinin in vivo. Finally, it is noteworthy that Tmprss2−/− mutant mice were not protected from infection with an H7N7 virus which contains a multi-basic HA cleavage site and can be activated by ubiquitously expressed proteases [4].
Fig. 9. Alignment of amino acid sequences of the protease loop region from H1N1 and H3N2 influenza A viruses. 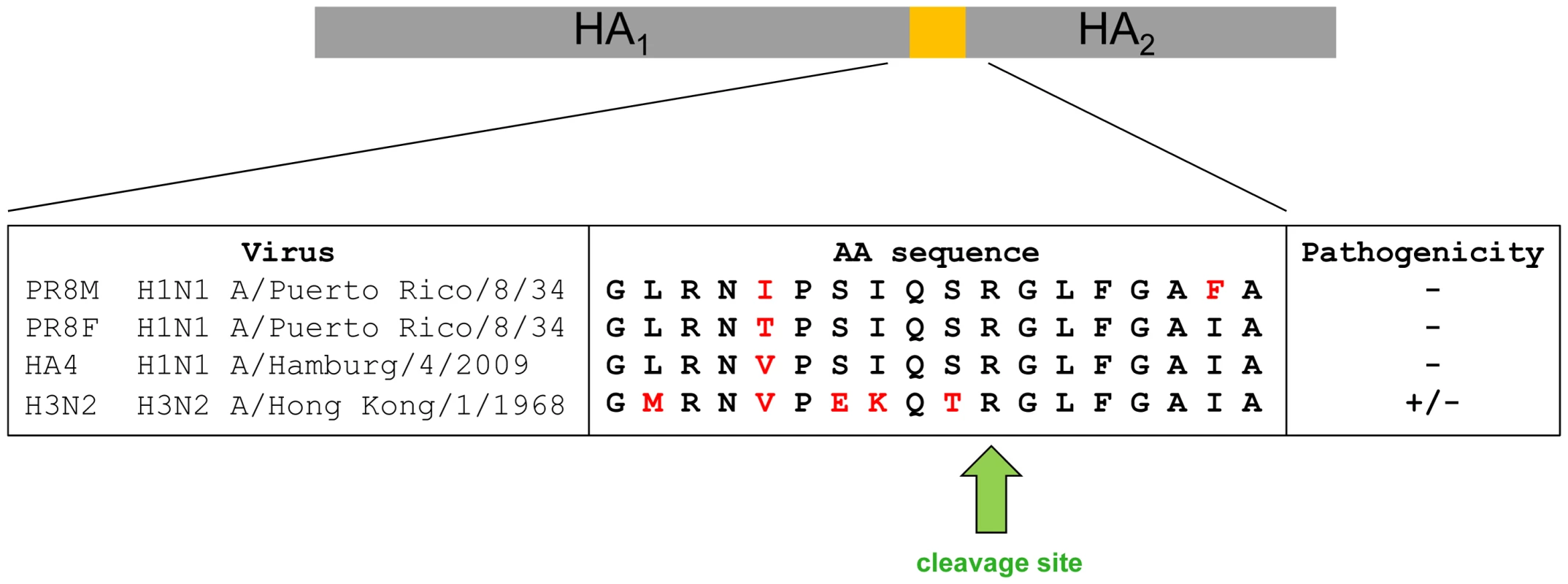
Pathology is strongly reduced in Tmprss2−/− mice after infection with H1N1 virus and diminished after infection with H3N2 virus. Our findings may have potential for the development of future influenza virus therapeutics. Broad spectrum protease inhibitors have been shown to inhibit influenza virus in cell culture and in vivo [24], [25], [26], [27], [28] but unwanted side effects are a major concern. The results reported here suggest that targeting a single protease, TMPRSS2, may be sufficient to achieve a notable therapeutic benefit against H1N1 influenza viruses and possibly other subtypes. Blocking of TMPRSS2 may not be associated with severe unwanted side effects, since Tmprss2−/− mice are healthy and do not show any phenotypic alterations in the absence of an infection [14]. Furthermore, TMPRSS2 inhibitors might exert activity against diverse respiratory infections. For example, human metapneumovirus [29], the emerging MERS-coronavirus [30], [31] and SARS-coronavirus [32], [33], [34] can also be activated by TMPRSS2 in cell culture and might use this protease to support their spread in the infected host. It should, however, be noted that our findings in the mouse model system require validation in humans.
Materials and Methods
Ethics statement
All experiments in mice were approved by an external committee according to the national guidelines of the animal welfare law in Germany (‘Tierschutzgesetz in der Fassung der Bekanntmachung vom 18. Mai 2006 (BGBl. I S. 1206, 1313), das zuletzt durch Artikel 20 des Gesetzes vom 9. Dezember 2010 (BGBl. I S. 1934) geändert worden ist.’). The protocol used in these experiments has been reviewed by an ethics committee and approved by the ‘Niedersächsisches Landesamt für Verbraucherschutz und Lebensmittelsicherheit, Oldenburg, Germany’ (Permit Number: 33.9.42502-04-051/09).
Virus, mice and plasmids
Original stocks of viruses were obtained from Stefan Ludwig, University of Münster (PR8M, A/PuertoRico/8/34 H1N1, Münster variant), from Peter Stäheli, University of Freiburg (PR8F, A/PuertoRico/8/34 H1N1, Freiburg variant and SC35M [35]), from Otto Haller, University of Freiburg (H3N2, [16]) and from Thorsten Wolff, Robert-Koch-Institute, Berlin (HA4). The different virulence of PR8M and PR8F virus isolates has been described before [15]. SC35M (H7N7) was originally derived from the seal, adapted to the mouse by serial passages in the mouse lung [35], and the laboratory strain which was used here was generated by reverse genetics using the plasmid rescue system [36]. Virus stocks of PR8 were prepared by infection of 10-day-old embryonated chicken eggs and for HA4 on MDCK cells as described [37]. Mutant Tmprss2−/− mice were on a mixed C57BL/6J – 129 background [14]. Animals were maintained under specific pathogen free conditions at the animal facility of the HZI in Braunschweig. Heterozygous mutant mice were interbred and wild type, heterozygous and homozygous mutant mice were genotyped by PCR analysis and then used for infections. Genotyping of Tmprss2 alleles was carried out using a three primer strategy (P1 5′TGTGCCCTTGGACAGATGACTC3′, P2 5′GGACTACAGATATGAGGTGTTC3′, P3 5′AGGCCAGAGGCCACTTGTGTAG3′) that allows to distinguish between wild type (yielding a 540 bp product) and knock-out (yielding a 400 bp product) alleles, respectively. Expression plasmids encoding human and mouse proteases TMPRSS2 were published earlier [10], [13]. Previously described expression plasmids for PR8M [38] were used as templates for amplification of the respective coding regions using oligonucleotides PR8-HA-5Acc: GGGGGTACCACCATGAAGGCAAACCTACTGGTCCTG, PR8-HA-3Nhe: GGGCGCTAGCTCAGATGCATATTCTGCACTG. The resulting PCR products were inserted into plasmid pCAGGS using the Acc65I and NheI sites. For cloning of HA4 HA protein, expression plasmid kindly provided by Prof. Klenk [39] was used as templates for amplification of the coding region using oligonucleotides swi09-HA-5Eco: GGGAATTCACCATGAAGGCAATACTAGTAGTTCTGC and swi09-HA-3Xho: GGGCTCGAGTTAAATACATATTCTACACTGTAGAG. The resulting PCR products were inserted into plasmid pCAGGS using EcoRI and XhoI.
Infection of mice and measurement of body weight loss and survival
For infection experiments, female mice at the age of 8–11 weeks were anesthetized by intra-peritoneal injection of Ketamin-Xylazine solution in sterile NaCl (50 mg/ml Ketamine, Invesa Arzneimittel GmbH, Freiburg; 2% Xylazine, Bayer Health-Care, Leverkusen) with a dose adjusted to the individual body weight. Infection was performed by intranasal application of virus solution in 20 µl of sterile phosphate-buffered saline. Subsequently survival and body weight loss were monitored until day 14 p.i. In addition to mice that were found dead, mice with a weight loss of more than 30% of the starting body weight were euthanized and recorded as dead.
Determining infectious viral particles
Viral load in infected lungs was determined on MDCK II (Madin-Darby Canine Kidney II) cells using the FFU assay as described [15]. Detection limit of the assay is at 80 infectious particles/lung. Thus, for samples where no virus was detected, the data points were set to 80 FFU/lung.
Analysis of processing of HA from infected lungs
For analysis of HA in viral particles, Broncho-alveolar lavages (BALs) of wild type and Tmprss2 knock-out mice infected with 2×105 FFU PR8M were harvested at day 1 p.i., centrifuged at full speed and supernatant were loaded on a 20% sucrose cushion for concentration of viral particles for 2–3 h at full speed and 4°C. The pelleted viral particles were lysed in 2×SDS loading buffer. For immuno-blotting, the lysates were separated by SDS gel electrophoresis, blotted on a nitrocellulose membrane and HA was detected by staining with a rabbit anti-PR8HA antibody (Sino biological) at a dilution of 1∶500, followed by incubation with a horseradish peroxidase (HRP)-coupled anti-rabbit antibody (Dianova) at a dilution of 1∶10.000. As loading control, membranes were stripped (Stripping Buffer: 1M Tris-HCl (pH 6,8), 10% SDS, 100 mM ß-Mercaptoethanol) for 30 min at 50°C, washed 1 h with dH2O and stained for NA by using an anti-influenza A virus goat serum (Millipore) at a dilution of 1∶500 followed by incubation with a horseradish peroxidase (HRP)-coupled anti-goat antibody (Dianova) at a dilution of 1∶10.000. Bands were visualized by using a commercially available kit ECL Prime Western Blotting Detection Reagents (Amersham).
Pulsoxymetric determination of oxygen saturation
Eight to twelve weeks old mice were infected intranasally with 2×105 FFU PR8M and the amount of oxygen saturation was determined by the MouseOx® system (STARR Life Science Corp.) over a period of 14 days. For oxygen measurements mice were anesthetized using an isoflurane inhalator.
Histological and immunohistochemical analyses
Lungs were prepared and immersion-fixed for 24 hours in 4% buffered formaldehyde solution (pH 7.4), dehydrated in a series of graded ethanol and embedded in paraffin. Sections (0.5 µm) were cut from five evenly distributed levels of the paraffin blocks and stained with haematoxylin and eosin. For immunohistochemical studies, sections were stained with a polyclonal primary antibody (against influenza A H1N1 virions; Virostat) overnight at 4°C and subsequently tissue sections were incubated for 30 min with secondary antibody (rabbit anti-goat-biotin; KPL; Gaithersburg, Madison, USA) and counterstained with haematoxylin.
Influenza virus cleavage activation by TMPRSS2 in cell culture
293T cells were transiently transfected with expression plasmids encoding human or mouse TMPRSS2 or control transfected with empty plasmid. At 24 h post transfection, cells were infected with either of the influenza viruses PR8M, HA4 or SC35M at a multiplicity of infection (MOI) of 1. After 1 h incubation at 37°C, virus was removed and fresh MEM medium (supplemented with 0.2% BSA and 1 µg/ml TPCK-trypsin or PBS) was added to the cells. At 48 h p.i., supernatants were harvested, cleared from debris by centrifugation for 5 min at 3.500 rpm and stored at −80°C until quantification of infectious virus particles by focus formation assay (FFU) as described previously [15].
Supporting Information
Zdroje
1. HurtAC, ChotpitayasunondhT, CoxNJ, DanielsR, FryAM, et al. (2012) Antiviral resistance during the 2009 influenza A H1N1 pandemic: public health, laboratory, and clinical perspectives. Lancet Infect Dis 12 : 240–248.
2. KlenkHD, RottR, OrlichM, BlodornJ (1975) Activation of influenza A viruses by trypsin treatment. Virology 68 : 426–439.
3. LazarowitzSG, ChoppinPW (1975) Enhancement of the infectivity of influenza A and B viruses by proteolytic cleavage of the hemagglutinin polypeptide. Virology 68 : 440–454.
4. Stieneke-GroberA, VeyM, AnglikerH, ShawE, ThomasG, et al. (1992) Influenza virus hemagglutinin with multibasic cleavage site is activated by furin, a subtilisin-like endoprotease. EMBO J 11 : 2407–2414.
5. KidoH, YokogoshiY, SakaiK, TashiroM, KishinoY, et al. (1992) Isolation and characterization of a novel trypsin-like protease found in rat bronchiolar epithelial Clara cells. A possible activator of the viral fusion glycoprotein. J Biol Chem 267 : 13573–13579.
6. MurakamiM, TowatariT, OhuchiM, ShiotaM, AkaoM, et al. (2001) Mini-plasmin found in the epithelial cells of bronchioles triggers infection by broad-spectrum influenza A viruses and Sendai virus. Eur J Biochem 268 : 2847–2855.
7. BertramS, GlowackaI, SteffenI, KuhlA, PohlmannS (2010) Novel insights into proteolytic cleavage of influenza virus hemagglutinin. Rev Med Virol 20 : 298–310.
8. ZhirnovOP, IkizlerMR, WrightPF (2002) Cleavage of influenza a virus hemagglutinin in human respiratory epithelium is cell associated and sensitive to exogenous antiproteases. J Virol 76 : 8682–8689.
9. Böttcher-FriebertshauserE, SteinDA, KlenkHD, GartenW (2011) Inhibition of influenza virus infection in human airway cell cultures by an antisense peptide-conjugated morpholino oligomer targeting the hemagglutinin-activating protease TMPRSS2. J Virol 85 : 1554–1562.
10. BertramS, GlowackaI, BlazejewskaP, SoilleuxE, AllenP, et al. (2010) TMPRSS2 and TMPRSS4 facilitate trypsin-independent spread of influenza virus in Caco-2 cells. J Virol 84 : 10016–10025.
11. BöttcherE, MatrosovichT, BeyerleM, KlenkHD, GartenW, et al. (2006) Proteolytic activation of influenza viruses by serine proteases TMPRSS2 and HAT from human airway epithelium. J Virol 80 : 9896–9898.
12. WangW, XieH, YeZ, VassellR, WeissCD (2010) Characterization of lentiviral pseudotypes with influenza H5N1 hemagglutinin and their performance in neutralization assays. J Virol Methods 165 : 305–310.
13. BertramS, HeurichA, LavenderH, GiererS, DanischS, et al. (2012) Influenza and SARS-Coronavirus Activating Proteases TMPRSS2 and HAT Are Expressed at Multiple Sites in Human Respiratory and Gastrointestinal Tracts. PLoS ONE 7: e35876.
14. KimTS, HeinleinC, HackmanRC, NelsonPS (2006) Phenotypic analysis of mice lacking the Tmprss2-encoded protease. Mol Cell Biol 26 : 965–975.
15. BlazejewskaP, KoscinskiL, ViegasN, AnhlanD, LudwigS, et al. (2011) Pathogenicity of different PR8 influenza A virus variants in mice is determined by both viral and host factors. Virology 412 : 36–45.
16. HallerO, ArnheiterH, LindenmannJ (1979) Natural, genetically determined resistance toward influenza virus in hemopoietic mouse chimeras. Role of mononuclear phagocytes. J Exp Med 150 : 117–126.
17. ChaipanC, KobasaD, BertramS, GlowackaI, SteffenI, et al. (2009) Proteolytic activation of the 1918 influenza virus hemagglutinin. J Virol 83 : 3200–3211.
18. KidoH, OkumuraY, YamadaH, LeTQ, YanoM (2007) Proteases essential for human influenza virus entry into cells and their inhibitors as potential therapeutic agents. Curr Pharm Des 13 : 405–414.
19. GotohB, OgasawaraT, ToyodaT, InocencioNM, HamaguchiM, et al. (1990) An endoprotease homologous to the blood clotting factor X as a determinant of viral tropism in chick embryo. EMBO J 9 : 4189–4195.
20. GotohB, YamauchiF, OgasawaraT, NagaiY (1992) Isolation of factor Xa from chick embryo as the amniotic endoprotease responsible for paramyxovirus activation. FEBS Lett 296 : 274–278.
21. GallowaySE, ReedML, RussellCJ, SteinhauerDA (2013) Influenza HA subtypes demonstrate divergent phenotypes for cleavage activation and pH of fusion: Implications for host range and adaptation. PLoS Pathog 9: e1003151.
22. HamiltonBS, GludishDW, WhittakerGR (2012) Cleavage Activation of the Human-Adapted Influenza Virus Subtypes by Matriptase Reveals Both Subtype - and Strain-Specificity. J Virol 86 : 10579–86.
23. HamiltonBS, WhittakerGR (2013) Cleavage activation of human-adapted influenza virus subtypes by kallikrein-related peptidases 5 and 12. J Biol Chem 288 : 17399–17407.
24. ZhirnovOP, KlenkHD, WrightPF (2011) Aprotinin and similar protease inhibitors as drugs against influenza. Antiviral Res 92 : 27–36.
25. BahgatMM, BlazejewskaP, SchughartK (2011) Inhibition of lung serine proteases in mice: a potentially new approach to control influenza infection. Virol J 8 : 27.
26. ZhirnovOP, OvcharenkoAV, BukrinskayaAG (1984) Suppression of influenza virus replication in infected mice by protease inhibitors. J Gen Virol 65 (Pt 1): 191–196.
27. ZhirnovOP (1987) High protection of animals lethally infected with influenza virus by aprotinin-rimantadine combination. J Med Virol 21 : 161–167.
28. OvcharenkoAV, ZhirnovOP (1994) Aprotinin aerosol treatment of influenza and paramyxovirus bronchopneumonia of mice. Antiviral Res 23 : 107–118.
29. ShiroganeY, TakedaM, IwasakiM, IshiguroN, TakeuchiH, et al. (2008) Efficient multiplication of human metapneumovirus in Vero cells expressing the transmembrane serine protease TMPRSS2. J Virol 82 : 8942–8946.
30. GiererS, BertramS, KaupF, WrenschF, HeurichA, et al. (2013) The spike protein of the emerging betacoronavirus EMC uses a novel coronavirus receptor for entry, can be activated by TMPRSS2, and is targeted by neutralizing antibodies. J Virol 87 : 5502–5511.
31. ShiratoK, KawaseM, MatsuyamaS (2013) Middle East Respiratory Syndrome Coronavirus (MERS-CoV) Infection Mediated by the Transmembrane Serine Protease TMPRSS2. J Virol doi: 10.1128/JVI.01890-13
32. GlowackaI, BertramS, MullerMA, AllenP, SoilleuxE, et al. (2011) Evidence that TMPRSS2 activates the severe acute respiratory syndrome coronavirus spike protein for membrane fusion and reduces viral control by the humoral immune response. J Virol 85 : 4122–4134.
33. ShullaA, Heald-SargentT, SubramanyaG, ZhaoJ, PerlmanS, et al. (2011) A transmembrane serine protease is linked to the severe acute respiratory syndrome coronavirus receptor and activates virus entry. J Virol 85 : 873–882.
34. MatsuyamaS, NagataN, ShiratoK, KawaseM, TakedaM, et al. (2010) Efficient activation of the severe acute respiratory syndrome coronavirus spike protein by the transmembrane protease TMPRSS2. J Virol 84 : 12658–12664.
35. GabrielG, DauberB, WolffT, PlanzO, KlenkHD, et al. (2005) The viral polymerase mediates adaptation of an avian influenza virus to a mammalian host. Proc Natl Acad Sci U S A 102 : 18590–18595.
36. HoffmannE, StechJ, LenevaI, KraussS, ScholtissekC, et al. (2000) Characterization of the Influenza a Virus Gene Pool in Avian Species in Southern China: Was H6N1 a Derivative or a Precursor of H5N1? Journal of Virology 74 : 6309–6315.
37. WilkE, SchughartK (2012) The mouse as model system to study host-pathogen interactions in influenza A infections. Curr Protoc Mouse Biol 2 : 177–205.
38. GrimmD, StaeheliP, HufbauerM, KoernerI, Martínez-SobridoL, et al. (2007) Replication fitness determines high virulence of influenza A virus in mice carrying functional Mx1 resistance gene. PNAS 104 : 6806–6811.
39. GerlachT, KuhlingL, UhlendorffJ, LaukemperV, MatrosovichT, et al. (2012) Characterization of the neuraminidase of the H1N1/09 pandemic influenza virus. Vaccine 30 : 7348–52.
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek Parental Transfer of the Antimicrobial Protein LBP/BPI Protects Eggs against Oomycete InfectionsČlánek Immune Therapeutic Strategies in Chronic Hepatitis B Virus Infection: Virus or Inflammation Control?Článek Coronaviruses as DNA Wannabes: A New Model for the Regulation of RNA Virus Replication FidelityČlánek CRISPR-Cas Immunity against Phages: Its Effects on the Evolution and Survival of Bacterial PathogensČlánek The Cyst Wall Protein CST1 Is Critical for Cyst Wall Integrity and Promotes Bradyzoite PersistenceČlánek The Malarial Serine Protease SUB1 Plays an Essential Role in Parasite Liver Stage Development
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2013 Číslo 12- Jak souvisí postcovidový syndrom s poškozením mozku?
- Měli bychom postcovidový syndrom léčit antidepresivy?
- Farmakovigilanční studie perorálních antivirotik indikovaných v léčbě COVID-19
- 10 bodů k očkování proti COVID-19: stanovisko České společnosti alergologie a klinické imunologie ČLS JEP
-
Všechny články tohoto čísla
- Host Susceptibility Factors to Bacterial Infections in Type 2 Diabetes
- LysM Effectors: Secreted Proteins Supporting Fungal Life
- Influence of Mast Cells on Dengue Protective Immunity and Immune Pathology
- Innate Lymphoid Cells: New Players in IL-17-Mediated Antifungal Immunity
- Cytoplasmic Viruses: Rage against the (Cellular RNA Decay) Machine
- Balancing Stability and Flexibility within the Genome of the Pathogen
- The Evolution of Transmissible Prions: The Role of Deformed Templating
- Parental Transfer of the Antimicrobial Protein LBP/BPI Protects Eggs against Oomycete Infections
- Host Defense via Symbiosis in
- Regulatory Circuits That Enable Proliferation of the Fungus in a Mammalian Host
- Immune Therapeutic Strategies in Chronic Hepatitis B Virus Infection: Virus or Inflammation Control?
- Burning Down the House: Cellular Actions during Pyroptosis
- Coronaviruses as DNA Wannabes: A New Model for the Regulation of RNA Virus Replication Fidelity
- CRISPR-Cas Immunity against Phages: Its Effects on the Evolution and Survival of Bacterial Pathogens
- Combining Regulatory T Cell Depletion and Inhibitory Receptor Blockade Improves Reactivation of Exhausted Virus-Specific CD8 T Cells and Efficiently Reduces Chronic Retroviral Loads
- Shaping Up for Battle: Morphological Control Mechanisms in Human Fungal Pathogens
- Identification of the Virulence Landscape Essential for Invasion of the Human Colon
- Nodular Inflammatory Foci Are Sites of T Cell Priming and Control of Murine Cytomegalovirus Infection in the Neonatal Lung
- Hepatitis B Virus Disrupts Mitochondrial Dynamics: Induces Fission and Mitophagy to Attenuate Apoptosis
- Mycobacterial MazG Safeguards Genetic Stability Housecleaning of 5-OH-dCTP
- Systematic MicroRNA Analysis Identifies ATP6V0C as an Essential Host Factor for Human Cytomegalovirus Replication
- Placental Syncytium Forms a Biophysical Barrier against Pathogen Invasion
- The CD8-Derived Chemokine XCL1/Lymphotactin Is a Conformation-Dependent, Broad-Spectrum Inhibitor of HIV-1
- Cyclin A Degradation by Primate Cytomegalovirus Protein pUL21a Counters Its Innate Restriction of Virus Replication
- Genome-Wide RNAi Screen Identifies Novel Host Proteins Required for Alphavirus Entry
- Zinc Sequestration: Arming Phagocyte Defense against Fungal Attack
- The Cyst Wall Protein CST1 Is Critical for Cyst Wall Integrity and Promotes Bradyzoite Persistence
- Biphasic Euchromatin-to-Heterochromatin Transition on the KSHV Genome Following Infection
- The Malarial Serine Protease SUB1 Plays an Essential Role in Parasite Liver Stage Development
- HIV-1 Vpr Accelerates Viral Replication during Acute Infection by Exploitation of Proliferating CD4 T Cells
- A Human Torque Teno Virus Encodes a MicroRNA That Inhibits Interferon Signaling
- The ArlRS Two-Component System Is a Novel Regulator of Agglutination and Pathogenesis
- An In-Depth Comparison of Latent HIV-1 Reactivation in Multiple Cell Model Systems and Resting CD4+ T Cells from Aviremic Patients
- Enterohemorrhagic Hemolysin Employs Outer Membrane Vesicles to Target Mitochondria and Cause Endothelial and Epithelial Apoptosis
- Overcoming Antigenic Diversity by Enhancing the Immunogenicity of Conserved Epitopes on the Malaria Vaccine Candidate Apical Membrane Antigen-1
- The Type-Specific Neutralizing Antibody Response Elicited by a Dengue Vaccine Candidate Is Focused on Two Amino Acids of the Envelope Protein
- Tmprss2 Is Essential for Influenza H1N1 Virus Pathogenesis in Mice
- Signatures of Pleiotropy, Economy and Convergent Evolution in a Domain-Resolved Map of Human–Virus Protein–Protein Interaction Networks
- Interference with the Host Haemostatic System by Schistosomes
- RocA Truncation Underpins Hyper-Encapsulation, Carriage Longevity and Transmissibility of Serotype M18 Group A Streptococci
- Gene Fitness Landscapes of at Important Stages of Its Life Cycle
- Phagocytosis Escape by a Protein That Connects Complement and Coagulation Proteins at the Bacterial Surface
- t Is a Structurally Novel Crohn's Disease-Associated Superantigen
- An Increasing Danger of Zoonotic Orthopoxvirus Infections
- Myeloid Dendritic Cells Induce HIV-1 Latency in Non-proliferating CD4 T Cells
- Transcriptional Analysis of Murine Macrophages Infected with Different Strains Identifies Novel Regulation of Host Signaling Pathways
- Serotonergic Chemosensory Neurons Modify the Immune Response by Regulating G-Protein Signaling in Epithelial Cells
- Genome-Wide Detection of Fitness Genes in Uropathogenic during Systemic Infection
- Induces an Unfolded Protein Response via TcpB That Supports Intracellular Replication in Macrophages
- Intestinal CD103+ Dendritic Cells Are Key Players in the Innate Immune Control of Infection in Neonatal Mice
- Emerging Functions for the RNome
- KSHV MicroRNAs Mediate Cellular Transformation and Tumorigenesis by Redundantly Targeting Cell Growth and Survival Pathways
- HrpA, an RNA Helicase Involved in RNA Processing, Is Required for Mouse Infectivity and Tick Transmission of the Lyme Disease Spirochete
- A Toxin-Antitoxin Module of Promotes Virulence in Mice
- Real-Time Imaging of the Intracellular Glutathione Redox Potential in the Malaria Parasite
- Hypoxia Inducible Factor Signaling Modulates Susceptibility to Mycobacterial Infection via a Nitric Oxide Dependent Mechanism
- Novel Strategies to Enhance Vaccine Immunity against Coccidioidomycosis
- Dual Expression Profile of Type VI Secretion System Immunity Genes Protects Pandemic
- —What Makes the Species a Ubiquitous Human Fungal Pathogen?
- αvβ6- and αvβ8-Integrins Serve As Interchangeable Receptors for HSV gH/gL to Promote Endocytosis and Activation of Membrane Fusion
- -Induced Activation of EGFR Prevents Autophagy Protein-Mediated Killing of the Parasite
- Semen CD4 T Cells and Macrophages Are Productively Infected at All Stages of SIV infection in Macaques
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Influence of Mast Cells on Dengue Protective Immunity and Immune Pathology
- Host Defense via Symbiosis in
- Coronaviruses as DNA Wannabes: A New Model for the Regulation of RNA Virus Replication Fidelity
- Myeloid Dendritic Cells Induce HIV-1 Latency in Non-proliferating CD4 T Cells
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



