-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaCRISPR-Cas Immunity against Phages: Its Effects on the Evolution and Survival of Bacterial Pathogens
article has not abstract
Published in the journal: . PLoS Pathog 9(12): e32767. doi:10.1371/journal.ppat.1003765
Category: Pearls
doi: https://doi.org/10.1371/journal.ppat.1003765Summary
article has not abstract
Introduction
Clustered regularly interspaced short palindromic repeats (CRISPR) loci are arrays of short repeats separated by equally short “spacer” sequences [1]–[3]. Along with the CRISPR-associated (cas) genes, they encode an adaptive immune system of archaea and bacteria that protects the cell against viral infection [4]. Remarkably, this system is capable of inserting a short piece of an infecting viral genome as a spacer in the CRISPR array [4], [5] (Figure 1A). The spacer sequence is transcribed and processed to generate a small antisense RNA (the CRISPR RNA or crRNA) (Figure 1B) [6] that is used as a guide for the recognition and destruction of the invader in subsequent infections (Figure 1C) [7]. Thus, spacer acquisition immunizes the bacterium and its progeny against the virus from which it was taken. Because spacers are incorporated in sequential order, CRISPR loci reflect the history of viral infection of the host. Cas proteins participate in all the different steps of this pathway, namely the insertion of spacer sequences into the CRISPR array [8], [9], the biogenesis of crRNAs [10], [11], and the destruction of the infecting viral genome [12], [13].
Fig. 1. The CRISPR immunity pathway. 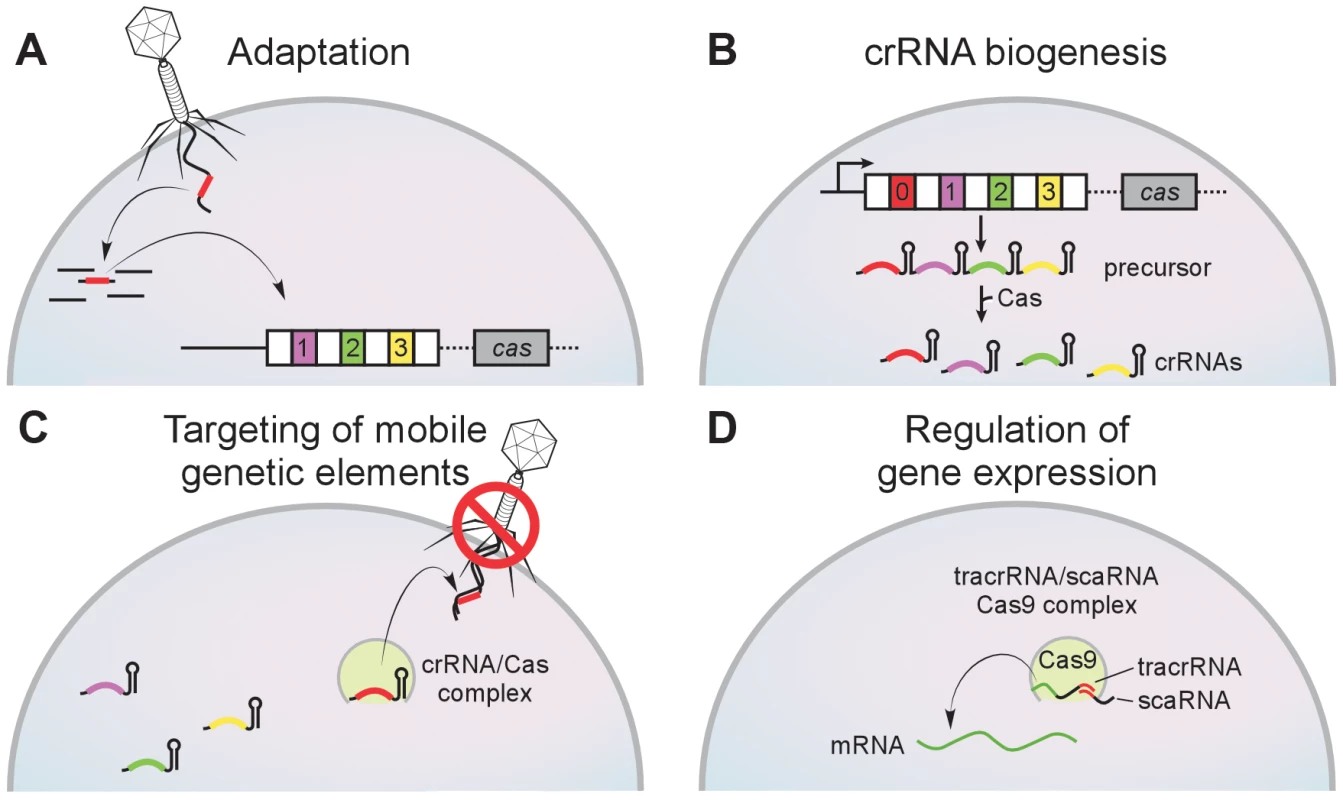
CRISPR loci contain clusters of repeats (white boxes) and spacers (colored boxes) that are flanked CRISPR-associated (cas) genes. (A) During adaptation new spacers derived from the genome of the invading virus are incorporated into the CRISPR array by an unknown mechanism. Repeat duplication is also required. (B) During crRNA biogenesis a CRISPR precursor transcript is processed by Cas endoribonucleases within repeat sequences to generate small crRNAs. (C) During targeting the match between the crRNA spacer and target sequences specifies the nucleolytic cleavage of invading mobile genetic elements such as viruses and plasmids. (D) In the CRISPR-Cas system of F. novicida, the tracrRNA (a small RNA mediated in crRNA biogenesis in this system) contains homology to the BLP (bacterial lipoprotein) transcript. The base-pair interaction between the tracrRNA and the BLP mRNA (mediated also by another small RNA, the scaRNA, and the nuclease Cas9) regulates the expression of this immunomodulatory lipoprotein. Distribution of CRISPR-Cas Loci among Bacterial Pathogens
In spite of the unique role that CRISPR-Cas loci play in antiviral defense, they are not universal. To date, the CRISPR database [14], a webtool that determines the presence of CRISPR arrays in completed genomes, indicates that 119/141 archaeal (84%) and 1012/2113 bacterial (48%) genomes contain CRISPR loci. In bacteria, there are species in which all strains have CRISPR loci, some in which only some strains have these loci, and species without strains having CRISPR loci. Therefore it is not possible to determine unequivocally that lack of CRISPR in certain strains or species is due to loss of these loci. However, because CRISPR sequences are spread thorough horizontal gene transfer [15], [16] and can be easily lost [17]–[20], it has been hypothesized recently that CRISPR are in a constant state of flux and can appear and disappear depending on the selective forces of the environment [20]. The same type of uneven distribution is found when we look at the presence of CRISPR loci in bacterial pathogens in the CRISPR database (http://crispr.u-psud.fr/crispr/).
CRISPR-Cas Systems as a Barrier to Horizontal Gene Transfer
While most of the spacers with matches on GenBank target prokaryotic viruses (phages), there is a still an important fraction that match other targets. A recent study looked at all the spacer hits of archaeal CRISPR loci [21] and reported that 40% of them matched phage sequences. The remaining 60% matched other mobile genetic elements such as conjugative plasmids and transposons (22%), CRISPR-Cas loci (18%), and other genes not associated with mobile elements (hypothetical ORFs and housekeeping genes, 20%). Although an equally extensive study has not been performed with bacterial CRISPR spacers, partial analysis suggests a similar distribution [22], [23]. While the presence of antiphage spacers is key for the defense of the cell, the origin and function of these nonphage targeting spacers is obscure. How are these spacers acquired? One possibility is that these sequences are inserted into CRISPR loci during the transfer of foreign genetic material that commonly occurs between prokaryotes, also known as horizontal gene transfer (HGT) [24]. In this scenario, non-antiphage spacers are acquired during bacteriophage transduction, plasmid conjugation, or upon the uptake of foreign DNA during natural transformation. Alternatively, spacer acquisition only occurs as an adaptive response to phage infection and the nonphage targeting spacers are acquired only from phage transducing particles [25]. Regardless of whether the diversity of the CRISPR spacer repertoire is generated by accident or not, the fact that CRISPR loci can target all sorts of genetic material argues that these loci constitute a barrier against the horizontal transfer of genes and accessory genetic elements. Indeed, CRISPR interference has been shown experimentally to prevent the acquisition of conjugative plasmids [26], integrative conjugative elements [27], and environmental DNA by natural transformation [17], [28]. What is even more puzzling is the function, if any, of these nonphage targeting spacers. Plasmid targeting could eliminate the burden of additional replicating elements inside the cell, and the targeting of housekeeping genes could provide a regulatory function for these spacers. However, plasmids, mobile genetic elements, and foreign genes can provide a fitness advantage or even be essential for survival (e.g., antibiotic resistance genes).
Implications of CRISPR-Mediated Targeting of Mobile Genetic Elements in Bacterial Pathogens
HGT is the major source of genetic diversity for bacterial evolution [24]. In the past century, the introduction of modern antibacterial therapies has accelerated the evolution of pathogens. While it is clear that HGT has played a central role in the spread of virulence factors and antimicrobial resistance genes [29], [30], only a few studies have addressed whether and how CRISPR loci, owing to their potential to regulate HGT, impact the evolution of pathogens. One of these studies investigated the relationship between the CRISPR loci and the prophage content of group A streptococci (GAS, Streptococcus pyogenes), one of the most prevalent human bacterial pathogens. These organisms contain between two to eight prophages, each encoding at least one virulence factor [31]. Bioinformatic analysis revealed that seven of the 13 available GAS genomes contain CRISPR-Cas loci and that there is a mutually exclusive relationship between CRISPR spacer sequences and their prophage targets [32]. This suggests that there is a dynamic relationship between S. pyogenes, its phages, and its CRISPR loci that results in the selection of strains with increased pathogenic adaptations. CRISPR-Cas loci also can impact the spread of antibiotic resistance. Pathogenic staphylococci have acquired resistance to all known antibiotics [33], primarily through the acquisition of conjugative plasmids carrying resistance genes [30]. Staphylococcus epidermidis RP62a is a clinical isolate containing a CRISPR-Cas system with a spacer matching all staphylococcal conjugative plasmids sequenced to date [34]. This spacer provides immunity against the conjugative transfer of these plasmids [26], thereby preventing the acquisition of the antibiotic resistances that they carry. Therefore CRISPR loci could control the dissemination of antibiotic resistance in staphylococci. This does not seem to be the case for Escherichia coli. A study of a collection of 263 natural E. coli isolates from human and animal hosts revealed that CRISPR loci neither match plasmid sequences nor correlate with the presence or absence of plasmids or antibiotic resistance genes [35].
Loss of CRISPR-Cas Loci in Bacterial Pathogens
CRISPR immunity against conjugative plasmids would compromise the survival of S. epidermidis RP62a, and other staphylococci carrying similar CRISPR-Cas systems [36], [37] in hospital or other settings where antibiotics are used. A recent study [20] looked for the transfer of the mupirocin-resistant conjugative plasmid pG0400 into S. epidermidis to determine if a CRISPR-Cas system and its target could coexist to prevent this potentially detrimental antiplasmid activity of CRISPR immunity. Immunity against the plasmid was found to decrease the transfer efficiency by about four orders of magnitude but not absolute. Transconjugants that evaded CRISPR attack were analyzed only to find that in all cases they harbored preexisting CRISPR-Cas mutations that allowed plasmid transfer. Loss of CRISPR-Cas loci upon transfer of antibiotic resistant plasmids also seems to occur in enterococci. A screen of 45 strains of Enterococcus faecalis showed a correlation between the presence of CRISPR-Cas loci and antibiotic resistance genes [38]. Finally, another recent study explored the consequences of CRISPR targeting of Streptococcus pneumoniae capsule genes, essential for pneumococcal infection. During infection, natural transformation of capsule genes allows nonencapsulated, avirulent pneumococci to become encapsulated and kill the mice [39]. A CRISPR-Cas targeting a specific capsule gene was engineered into nonencapsulated S. pneumoniae and used to infect mice in the presence of heat-killed encapsulated pneumococci [17]. Horizontal transfer of capsule genes from heat-killed cells into live, nonencapsulated bacteria was prevented by CRISPR immunity, resulting in the survival of mice. The occasional mice that succumbed to pneumococcal infection, however, contained encapsulated bacteria carrying inactivating mutations in the engineered CRISPR locus. These and other results [18], [19] suggest that CRISPR loci and their targets cannot coexist in the same cell. In the case of strong environmental selection of a targeted gene or mobile element, only CRISPR mutants survive. This is a possible explanation for the lack of CRISPR in S. pneumoniae and S. aureus, two notoriously fast-evolving pathogens, but also in other bacteria and archaea that lack this immune system.
A Direct Role for CRISPR-Cas Systems in Bacterial Pathogenesis
While the reasons for the absence of CRISPR-Cas loci in some fast-evolving pathogens remain a matter of speculation, recent evidence showed that these loci can also promote pathogenesis. A study in Legionella pneumophila showed that cas2, a gene involved in the acquisition of new spacers, is required for the propagation of this pathogen inside amoebae hosts [40], although it is not clear what the function of this gene is during growth. More compelling evidence is found in the intracellular pathogen Francisella novicida. In this bacterium, cas9 is a CRISPR-associated dsDNA nuclease that requires, in addition to the crRNA guide, a tracrRNA (trans-activating crRNA) for cleavage of the invader genome [41], [42]. It was found recently that cas9 is required to repress the production of a bacterial lipoprotein (BLP), a toll-like receptor 2 (TLR2) ligand that induces an innate immune inflammatory response [43]. Repression is independent of the crRNA guides, but requires the tracrRNA and a new small CRISPR-associated RNA (scaRNA) with complementarity to the tracrRNA [44], [45]. The tracrRNA, in turn, contains an ∼85 nt region with partial complementarity to the 3′-end of the BLP messenger, an interaction that leads to the BLP mRNA degradation through an unknown mechanism. This CRISPR-mediated regulation of BLP expression allows F. novicida to evade the host's immune response. A similar mechanism seems to be in place in other pathogens as well: deletion of cas9 in Neisseria meningitidis affected virulence traits such as adherence to and invasion of human epithelial cells [44], and inactivation of cas9 in Campylobacter jejuni resulted in reduced virulence [46]. While the predominance of tracrRNA/scaRNA-mediated regulation remains to be investigated, its existence suggests that CRISPR-Cas loci can be easily converted into regulatory elements that enhance bacterial pathogenesis.
Conclusions
Clearly CRISPR-Cas systems can both prevent the evolution of pathogenesis, and thus be lost or mutated in bacterial pathogens, but also be co-opted by the pathogen to increase virulence. This will depend of a series of factors: whether other antiphage systems can fulfill the function of the lost CRISPR-Cas system, whether the pathogen relies heavily on HGT for survival, and whether the CRISPR-Cas system can be easily converted into a regulator of gene expression. In the face of the lateral transfer of CRISPR systems, the repression of gene expression by CRISPR provides another level of selection for the maintenance of these systems. While the repression of BLP provides a selectable advantage for Francisella, the accidental repression of essential genes (which could be produced by a fortuitous base-pairing of the tracrRNA and an essential transcript) will select against the lateral transfer of some CRISPR-Cas systems into certain hosts. In the future, the application of DNA sequencing technologies to epidemiological studies will allow us to measure correlations between the flux of CRISPR-Cas loci and the acquisition of antibiotic-resistance plasmids and pathogenicity islands or genes, thus allowing us to measure the effect of CRISPR on the emergence of virulence. On the other hand, the importance of CRISPR for pathogenesis provides a new target for antimicrobials with anti-CRISPR activity. Interestingly, phages already found such anti-CRISPR compounds for us: as part of their arms race with bacteria, phages have developed CRISPR inhibitors [47]. The intersection between CRISPR biology and bacterial pathogenesis is a new and exciting research area that is only beginning to be explored.
Zdroje
1. BikardD, MarraffiniLA (2012) Innate and adaptive immunity in bacteria: mechanisms of programmed genetic variation to fight bacteriophages. Curr Opin Immunol 24 : 15–20.
2. WestraER, SwartsDC, StaalsRH, JoreMM, BrounsSJ, et al. (2012) The CRISPRs, they are a-changin': how prokaryotes generate adaptive immunity. Annu Rev Genet 46 : 311–339.
3. WiedenheftB, SternbergSH, DoudnaJA (2012) RNA-guided genetic silencing systems in bacteria and archaea. Nature 482 : 331–338.
4. BarrangouR, FremauxC, DeveauH, RichardsM, BoyavalP, et al. (2007) CRISPR provides acquired resistance against viruses in prokaryotes. Science 315 : 1709–1712.
5. CadyKC, Bondy-DenomyJ, HeusslerGE, DavidsonAR, O'TooleGA (2012) The CRISPR/Cas adaptive immune system of Pseudomonas aeruginosa mediates resistance to naturally occurring and engineered phages. J Bacteriol 194 : 5728–5738.
6. BrounsSJ, JoreMM, LundgrenM, WestraER, SlijkhuisRJ, et al. (2008) Small CRISPR RNAs guide antiviral defense in prokaryotes. Science 321 : 960–964.
7. GarneauJE, DupuisME, VillionM, RomeroDA, BarrangouR, et al. (2010) The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature 468 : 67–71.
8. DatsenkoKA, PougachK, TikhonovA, WannerBL, SeverinovK, et al. (2012) Molecular memory of prior infections activates the CRISPR/Cas adaptive bacterial immunity system. Nat Commun 3 : 945.
9. YosefI, GorenMG, QimronU (2012) Proteins and DNA elements essential for the CRISPR adaptation process in Escherichia coli. Nucleic Acids Res 40 : 5569–5576.
10. CarteJ, WangR, LiH, TernsRM, TernsMP (2008) Cas6 is an endoribonuclease that generates guide RNAs for invader defense in prokaryotes. Genes Dev 22 : 3489–3496.
11. HaurwitzRE, JinekM, WiedenheftB, ZhouK, DoudnaJA (2010) Sequence - and structure-specific RNA processing by a CRISPR endonuclease. Science 329 : 1355–1358.
12. JinekM, ChylinskiK, FonfaraI, HauerM, DoudnaJA, et al. (2012) A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337 : 816–821.
13. WestraER, van ErpPB, KunneT, WongSP, StaalsRH, et al. (2012) CRISPR immunity relies on the consecutive binding and degradation of negatively supercoiled invader DNA by Cascade and Cas3. Mol Cell 46 : 595–605.
14. GrissaI, VergnaudG, PourcelC (2007) The CRISPRdb database and tools to display CRISPRs and to generate dictionaries of spacers and repeats. BMC Bioinformatics 8 : 172.
15. ChakrabortyS, SnijdersAP, ChakravortyR, AhmedM, TarekAM, et al. (2010) Comparative network clustering of direct repeats (DRs) and cas genes confirms the possibility of the horizontal transfer of CRISPR locus among bacteria. Mol Phylogenet Evol 56 : 878–887.
16. GoddeJS, BickertonA (2006) The repetitive DNA elements called CRISPRs and their associated genes: evidence of horizontal transfer among prokaryotes. J Mol Evol 62 : 718–729.
17. BikardD, Hatoum-AslanA, MucidaD, MarraffiniLA (2012) CRISPR interference can prevent natural transformation and virulence acquisition during in vivo bacterial infection. Cell Host Microbe 12 : 177–186.
18. FischerS, MaierLK, StollB, BrendelJ, FischerE, et al. (2012) An archaeal immune system can detect multiple Protospacer Adjacent Motifs (PAMs) to target invader DNA. J Biol Chem 287 : 33351–33363.
19. GudbergsdottirS, DengL, ChenZ, JensenJV, JensenLR, et al. (2011) Dynamic properties of the Sulfolobus CRISPR/Cas and CRISPR/Cmr systems when challenged with vector-borne viral and plasmid genes and protospacers. Mol Microbiol 79 : 35–49.
20. JiangW, ManivI, ArainF, WangY, LevinBR, et al. (2013) Dealing with the evolutionary downside of CRISPR immunity: bacteria and beneficial plasmids. PLoS Genet 9: e1003844 doi:10.1371/journal.pgen.1003844
21. BrodtA, Lurie-WeinbergerMN, GophnaU (2011) CRISPR loci reveal networks of gene exchange in archaea. Biol Direct 6 : 65.
22. HorvathP, Coute-MonvoisinAC, RomeroDA, BoyavalP, FremauxC, et al. (2009) Comparative analysis of CRISPR loci in lactic acid bacteria genomes. Int J Food Microbiol 131 : 62–70.
23. SternA, KerenL, WurtzelO, AmitaiG, SorekR (2010) Self-targeting by CRISPR: gene regulation or autoimmunity? Trends Genet 26 : 335–340.
24. ThomasCM, NielsenKM (2005) Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat Rev Microbiol 3 : 711–721.
25. ErdmannS, GarrettRA (2012) Selective and hyperactive uptake of foreign DNA by adaptive immune systems of an archaeon via two distinct mechanisms. Mol Microbiol 85 : 1044–1056.
26. MarraffiniLA, SontheimerEJ (2008) CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science 322 : 1843–1845.
27. Lopez-SanchezMJ, SauvageE, Da CunhaV, ClermontD, Ratsima HariniainaE, et al. (2012) The highly dynamic CRISPR1 system of Streptococcus agalactiae controls the diversity of its mobilome. Mol Microbiol 85 : 1057–1071.
28. ZhangY, HeidrichN, AmpattuBJ, GundersonCW, SeifertHS, et al. (2013) Processing-Independent CRISPR RNAs Limit Natural Transformation in Neisseria meningitidis. Mol Cell 50 : 488–503.
29. CroucherNJ, HarrisSR, FraserC, QuailMA, BurtonJ, et al. (2011) Rapid pneumococcal evolution in response to clinical interventions. Science 331 : 430–434.
30. WeigelLM, ClewellDB, GillSR, ClarkNC, McDougalLK, et al. (2003) Genetic analysis of a high-level vancomycin-resistant isolate of Staphylococcus aureus. Science 302 : 1569–1571.
31. BisnoAL, BritoMO, CollinsCM (2003) Molecular basis of group A streptococcal virulence. Lancet Infect Dis 3 : 191–200.
32. NozawaT, FurukawaN, AikawaC, WatanabeT, HaobamB, et al. (2011) CRISPR inhibition of prophage acquisition in Streptococcus pyogenes. PLoS ONE 6: e19543 doi:10.1371/journal.pone.0019543
33. FuruyaEY, LowyFD (2006) Antimicrobial-resistant bacteria in the community setting. Nat Rev Microbiol 4 : 36–45.
34. GillSR, FoutsDE, ArcherGL, MongodinEF, DeboyRT, et al. (2005) Insights on evolution of virulence and resistance from the complete genome analysis of an early methicillin-resistant Staphylococcus aureus strain and a biofilm-producing methicillin-resistant Staphylococcus epidermidis strain. J Bacteriol 187 : 2426–2438.
35. TouchonM, CharpentierS, PognardD, PicardB, ArletG, et al. (2012) Antibiotic resistance plasmids spread among natural isolates of Escherichia coli in spite of CRISPR elements. Microbiology 158 : 2997–3004.
36. GoldingGR, BrydenL, LevettPN, McDonaldRR, WongA, et al. (2010) Livestock-associated methicillin-resistant Staphylococcus aureus sequence type 398 in humans, Canada. Emerg Infect Dis 16 : 587–594.
37. HoltDC, HoldenMT, TongSY, Castillo-RamirezS, ClarkeL, et al. (2011) A very early-branching Staphylococcus aureus lineage lacking the carotenoid pigment staphyloxanthin. Genome Biol Evol 3 : 881–895.
38. PalmerKL, GilmoreMS (2010) Multidrug-resistant enterococci lack CRISPR-cas. mBio 1: e00227–00210.
39. GriffithF (1928) The significance of pneumococcal types. J Hyg 27 : 113–159.
40. GundersonFF, CianciottoNP (2013) The CRISPR-associated gene cas2 of Legionella pneumophila is required for intracellular infection of amoebae. mBio 4: e00074–00013.
41. DeltchevaE, ChylinskiK, SharmaCM, GonzalesK, ChaoY, et al. (2011) CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature 471 : 602–607.
42. JiangW, BikardD, CoxD, ZhangF, MarraffiniLA (2013) RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol 31 : 233–239.
43. JonesCL, SampsonTR, NakayaHI, PulendranB, WeissDS (2012) Repression of bacterial lipoprotein production by Francisella novicida facilitates evasion of innate immune recognition. Cell Microbiol 14 : 1531–1543.
44. SampsonTR, SarojSD, LlewellynAC, TzengYL, WeissDS (2013) A CRISPR/Cas system mediates bacterial innate immune evasion and virulence. Nature 497 : 254–257.
45. SampsonTR, SarojSD, LlewellynAC, TzengYL, WeissDS (2013) Corrigendum: a CRISPR/Cas system mediates bacterial innate immune evasion and virulence. Nature 501 : 262.
46. LouwenR, Horst-KreftD, de BoerAG, van der GraafL, de KnegtG, et al. (2013) A novel link between Campylobacter jejuni bacteriophage defence, virulence and Guillain-Barre syndrome. Eur J Clin Microbiol Infect Dis 32 : 207–226.
47. Bondy-DenomyJ, PawlukA, MaxwellKL, DavidsonAR (2013) Bacteriophage genes that inactivate the CRISPR/Cas bacterial immune system. Nature 493 : 429–432.
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek Parental Transfer of the Antimicrobial Protein LBP/BPI Protects Eggs against Oomycete InfectionsČlánek Immune Therapeutic Strategies in Chronic Hepatitis B Virus Infection: Virus or Inflammation Control?Článek Coronaviruses as DNA Wannabes: A New Model for the Regulation of RNA Virus Replication FidelityČlánek The Cyst Wall Protein CST1 Is Critical for Cyst Wall Integrity and Promotes Bradyzoite PersistenceČlánek The Malarial Serine Protease SUB1 Plays an Essential Role in Parasite Liver Stage Development
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2013 Číslo 12- Jak souvisí postcovidový syndrom s poškozením mozku?
- Měli bychom postcovidový syndrom léčit antidepresivy?
- Farmakovigilanční studie perorálních antivirotik indikovaných v léčbě COVID-19
- 10 bodů k očkování proti COVID-19: stanovisko České společnosti alergologie a klinické imunologie ČLS JEP
-
Všechny články tohoto čísla
- Host Susceptibility Factors to Bacterial Infections in Type 2 Diabetes
- LysM Effectors: Secreted Proteins Supporting Fungal Life
- Influence of Mast Cells on Dengue Protective Immunity and Immune Pathology
- Innate Lymphoid Cells: New Players in IL-17-Mediated Antifungal Immunity
- Cytoplasmic Viruses: Rage against the (Cellular RNA Decay) Machine
- Balancing Stability and Flexibility within the Genome of the Pathogen
- The Evolution of Transmissible Prions: The Role of Deformed Templating
- Parental Transfer of the Antimicrobial Protein LBP/BPI Protects Eggs against Oomycete Infections
- Host Defense via Symbiosis in
- Regulatory Circuits That Enable Proliferation of the Fungus in a Mammalian Host
- Immune Therapeutic Strategies in Chronic Hepatitis B Virus Infection: Virus or Inflammation Control?
- Burning Down the House: Cellular Actions during Pyroptosis
- Coronaviruses as DNA Wannabes: A New Model for the Regulation of RNA Virus Replication Fidelity
- CRISPR-Cas Immunity against Phages: Its Effects on the Evolution and Survival of Bacterial Pathogens
- Combining Regulatory T Cell Depletion and Inhibitory Receptor Blockade Improves Reactivation of Exhausted Virus-Specific CD8 T Cells and Efficiently Reduces Chronic Retroviral Loads
- Shaping Up for Battle: Morphological Control Mechanisms in Human Fungal Pathogens
- Identification of the Virulence Landscape Essential for Invasion of the Human Colon
- Nodular Inflammatory Foci Are Sites of T Cell Priming and Control of Murine Cytomegalovirus Infection in the Neonatal Lung
- Hepatitis B Virus Disrupts Mitochondrial Dynamics: Induces Fission and Mitophagy to Attenuate Apoptosis
- Mycobacterial MazG Safeguards Genetic Stability Housecleaning of 5-OH-dCTP
- Systematic MicroRNA Analysis Identifies ATP6V0C as an Essential Host Factor for Human Cytomegalovirus Replication
- Placental Syncytium Forms a Biophysical Barrier against Pathogen Invasion
- The CD8-Derived Chemokine XCL1/Lymphotactin Is a Conformation-Dependent, Broad-Spectrum Inhibitor of HIV-1
- Cyclin A Degradation by Primate Cytomegalovirus Protein pUL21a Counters Its Innate Restriction of Virus Replication
- Genome-Wide RNAi Screen Identifies Novel Host Proteins Required for Alphavirus Entry
- Zinc Sequestration: Arming Phagocyte Defense against Fungal Attack
- The Cyst Wall Protein CST1 Is Critical for Cyst Wall Integrity and Promotes Bradyzoite Persistence
- Biphasic Euchromatin-to-Heterochromatin Transition on the KSHV Genome Following Infection
- The Malarial Serine Protease SUB1 Plays an Essential Role in Parasite Liver Stage Development
- HIV-1 Vpr Accelerates Viral Replication during Acute Infection by Exploitation of Proliferating CD4 T Cells
- A Human Torque Teno Virus Encodes a MicroRNA That Inhibits Interferon Signaling
- The ArlRS Two-Component System Is a Novel Regulator of Agglutination and Pathogenesis
- An In-Depth Comparison of Latent HIV-1 Reactivation in Multiple Cell Model Systems and Resting CD4+ T Cells from Aviremic Patients
- Enterohemorrhagic Hemolysin Employs Outer Membrane Vesicles to Target Mitochondria and Cause Endothelial and Epithelial Apoptosis
- Overcoming Antigenic Diversity by Enhancing the Immunogenicity of Conserved Epitopes on the Malaria Vaccine Candidate Apical Membrane Antigen-1
- The Type-Specific Neutralizing Antibody Response Elicited by a Dengue Vaccine Candidate Is Focused on Two Amino Acids of the Envelope Protein
- Tmprss2 Is Essential for Influenza H1N1 Virus Pathogenesis in Mice
- Signatures of Pleiotropy, Economy and Convergent Evolution in a Domain-Resolved Map of Human–Virus Protein–Protein Interaction Networks
- Interference with the Host Haemostatic System by Schistosomes
- RocA Truncation Underpins Hyper-Encapsulation, Carriage Longevity and Transmissibility of Serotype M18 Group A Streptococci
- Gene Fitness Landscapes of at Important Stages of Its Life Cycle
- Phagocytosis Escape by a Protein That Connects Complement and Coagulation Proteins at the Bacterial Surface
- t Is a Structurally Novel Crohn's Disease-Associated Superantigen
- An Increasing Danger of Zoonotic Orthopoxvirus Infections
- Myeloid Dendritic Cells Induce HIV-1 Latency in Non-proliferating CD4 T Cells
- Transcriptional Analysis of Murine Macrophages Infected with Different Strains Identifies Novel Regulation of Host Signaling Pathways
- Serotonergic Chemosensory Neurons Modify the Immune Response by Regulating G-Protein Signaling in Epithelial Cells
- Genome-Wide Detection of Fitness Genes in Uropathogenic during Systemic Infection
- Induces an Unfolded Protein Response via TcpB That Supports Intracellular Replication in Macrophages
- Intestinal CD103+ Dendritic Cells Are Key Players in the Innate Immune Control of Infection in Neonatal Mice
- Emerging Functions for the RNome
- KSHV MicroRNAs Mediate Cellular Transformation and Tumorigenesis by Redundantly Targeting Cell Growth and Survival Pathways
- HrpA, an RNA Helicase Involved in RNA Processing, Is Required for Mouse Infectivity and Tick Transmission of the Lyme Disease Spirochete
- A Toxin-Antitoxin Module of Promotes Virulence in Mice
- Real-Time Imaging of the Intracellular Glutathione Redox Potential in the Malaria Parasite
- Hypoxia Inducible Factor Signaling Modulates Susceptibility to Mycobacterial Infection via a Nitric Oxide Dependent Mechanism
- Novel Strategies to Enhance Vaccine Immunity against Coccidioidomycosis
- Dual Expression Profile of Type VI Secretion System Immunity Genes Protects Pandemic
- —What Makes the Species a Ubiquitous Human Fungal Pathogen?
- αvβ6- and αvβ8-Integrins Serve As Interchangeable Receptors for HSV gH/gL to Promote Endocytosis and Activation of Membrane Fusion
- -Induced Activation of EGFR Prevents Autophagy Protein-Mediated Killing of the Parasite
- Semen CD4 T Cells and Macrophages Are Productively Infected at All Stages of SIV infection in Macaques
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Influence of Mast Cells on Dengue Protective Immunity and Immune Pathology
- Host Defense via Symbiosis in
- Coronaviruses as DNA Wannabes: A New Model for the Regulation of RNA Virus Replication Fidelity
- Myeloid Dendritic Cells Induce HIV-1 Latency in Non-proliferating CD4 T Cells
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



