-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaLysM Effectors: Secreted Proteins Supporting Fungal Life
article has not abstract
Published in the journal: . PLoS Pathog 9(12): e32767. doi:10.1371/journal.ppat.1003769
Category: Pearls
doi: https://doi.org/10.1371/journal.ppat.1003769Summary
article has not abstract
Introduction
Fungi occupy a plethora of niches and play essential roles in diverse environments through decomposition of organic material as saprophytes or through establishment of symbiotic relationships with plants and animals that range from mutually beneficial to pathogenic. During colonization of their niches, fungi secrete proteins that include carbohydrate-degrading enzymes to feed on complex molecules and effectors that mediate the establishment of interactions with host organisms [1]. Although effectors are typically thought to be species - or even lineage-specific, some effectors are widespread among pathogens, such as the necrosis - and ethylene-inducing-like proteins (NLPs) that are widely spread in bacteria, fungi, and oomycetes [2], [3]. Several studies have shown that NLPs contribute to pathogen virulence through phytotoxic activity, but more recent work has revealed that some NLPs act in processes other than pathogenicity, such as fungal growth and sporulation [4]. A more recently identified class of conserved effectors are LysM effectors: fungal effectors that carry no recognizable protein domains other than lysin motifs (LysMs) [5]. Intriguingly, like NLPs, LysM effectors occur in both pathogenic and in nonpathogenic fungi.
Plant Pathogen LysM Effectors: Virulence Factors through Interactions with Chitin
Microbial pathogens carry conserved structures, termed microbe-associated molecular patterns (MAMPs), that are recognized by host cell surface receptors and trigger an immune response [6], [7]. Chitin, a major constituent of fungal cell walls, is a well-described MAMP, and several plasma membrane–localized chitin receptors have been identified in plants that all contain extracellular LysMs, well-known carbohydrate-binding protein domains [8]–[10]. To overcome host immunity, genuine pathogens secrete effector molecules that manipulate host physiology, including immune responses, to support host colonization [2], [7]. Likely, also other microbes that establish intimate relationships with host plants, such as mutualistic symbiotic microbes and endophytes, secrete effectors to bring about their association.
The fungal tomato leaf mould pathogen Cladosporium fulvum secretes the LysM-containing effector Ecp6 that binds chitin with high specificity [11], [12]. Ecp6 does not protect fungal hyphae against the hydrolytic activity of tomato chitinases, a function that was previously assigned to C. fulvum effector Avr4 that contains an invertebrate chitin-binding domain [13], [14]. Consequently, it was speculated that Ecp6 interferes with chitin detection by the host [5]. Indeed, Ecp6 was demonstrated to perturb chitin-induced immunity, and it was proposed that Ecp6 functions by sequestration of cell wall–derived chitin fragments that would otherwise be perceived by host immune receptors [12] (Figure 1). The crystal structure of Ecp6 showed that two LysM domains (LysM1 and LysM3) collectively bind a single chitin molecule [15] (Figure 1). This ligand-induced composite binding groove is deeply buried in the effector and displays ultra-high (picomolar) chitin-binding affinity, which is significantly higher than that of plant immune receptors [15]. Through analysis of a crystal structure of the Arabidopsis chitin elicitor receptor kinase (AtCERK1) it was previously demonstrated that only one of the three LysM domains in this immune receptor binds chitin [16]. Moreover, the structural orientation of the three LysM domains in AtCERK1 does not permit intramolecular LysM dimerization as observed in Ecp6 [15], [16]. Interestingly, the singular LysM domain of Ecp6 that is not involved in the intramolecular composite binding site (LysM2) also contains a functional chitin-binding site (Figure 1), and has the capacity to perturb chitin-induced immunity [12], [15]. Since the chitin-binding affinity of this singular LysM domain is significantly lower than that of the composite binding site, it is unlikely to deregulate chitin-induced immunity merely by chitin oligosaccharide sequestration. As it has been suggested that chitin-induced immune receptor dimerization is required for the activation of immune signalling, LysM2 may perturb chitin-induced immunity through interference with this dimerization [15], [16] (Figure 1, Figure 2). Since LysM effectors produced by the wheat blotch fungus Mycosphaerella graminicola and the rice blast pathogen Magnaporthe oryzae, Mg3LysM and Slp1 respectively, similarly suppress chitin-triggered immunity, it seems that deregulation of chitin-triggered immunity is an important function of LysM effectors [17], [18]. Nevertheless, functional analysis of M. graminicola LysM effectors has revealed that they may have additional functions during host colonization [10], [17]. Fungal cell wall chitin is a target of plant chitinases that act in fungal immunity; exochitinases release chitin oligosaccharide MAMPs from fungal cell walls that can induce host immune responses, which include the secretion of endochitinases that cause hyphal lysis [8], [19]. Interestingly, M. graminicola Mg1LysM and Mg3LysM prevent hyphal lysis by plant chitinases, whereas Ecp6 and Slp1 do not have this capacity [12], [17], [18] (Figure 2). Thus, functional diversification of LysM effectors during host colonization has occurred in plant pathogens.
Fig. 1. Three-dimensional structure of the Cladosporium fulvum LysM effector Ecp6. 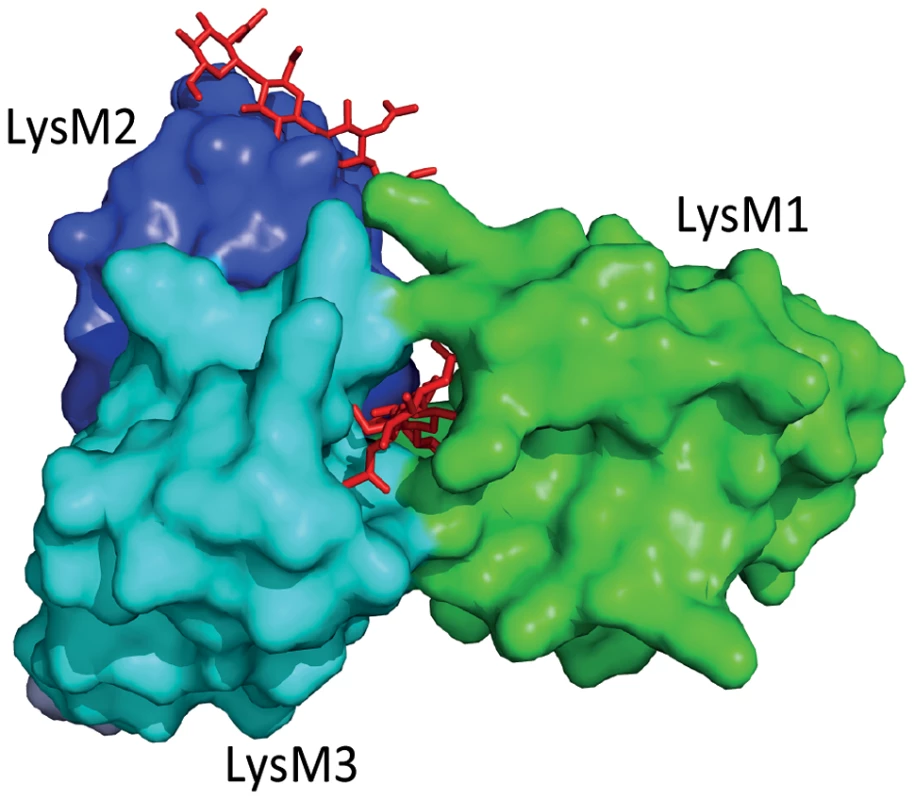
Two LysM domains of Ecp6 (LysM1 and LysM3) cooperate to form a binding groove that binds a single chitin oligosaccharide molecule (chitin tetramer oligosaccharide in red) with picomolar affinity. The remaining, singular LysM domain (LysM2) also has a functional chitin-binding site, although its affinity for chitin binding is significantly lower than that of the composite binding site. Fig. 2. Overview of the diverse roles that fungal LysM effectors may play in fungal physiology. 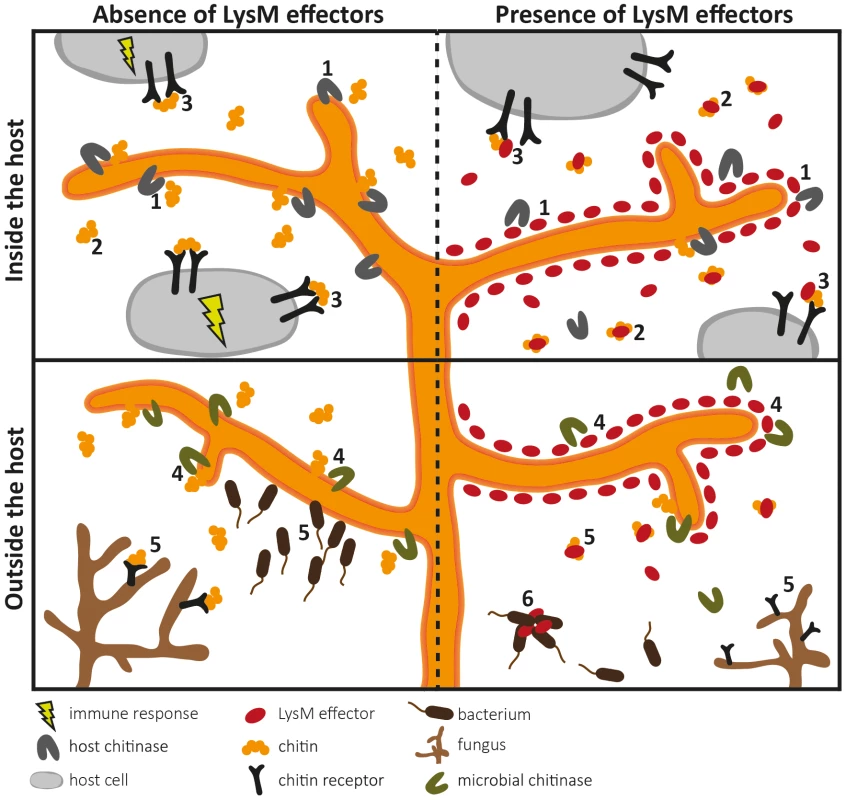
LysM effectors may act during host colonization (upper panels) and outside the host (lower panels). Pathogen LysM effectors have been implicated in two different pathogenicity-related processes (upper panels). Firstly, LysM effectors may protect fungal hyphae against degradation by hydrolytic enzymes secreted by the host (1). Secondly, LysM effectors may secure fungal cell wall–derived chitin fragments so that chitin cannot stimulate an immune response because LysM effectors efficiently scavenge chitin fragments (2), or interfere with host receptor activation by preventing ligand-induced dimerization (3). As LysM effectors also occur in nonpathogenic fungi (lower panels), they may protect fungal hyphae against hydrolytic enzymes secreted by mycoparasites (4). In addition, chitin sequestration might prevent attraction of such microbes (5). Some LysM effectors may recognize chitin-related carbohydrates such as peptidoglycan and immobilize bacterial competitors (6). LysM Effectors as Virulence Factors of Mammalian Pathogens?
Genome mining revealed that LysM effectors are not confined to plant pathogens as, for instance, genomes of most (opportunistic) fungal pathogens of mammals contain LysM effector genes as well [5]. For instance, in the dermatophyte Trichophyton rubrum, causal agent of athlete's foot, as well as in related dermatophyte species, the gene family encoding LysM effectors appears to be expanded [20]. Similar to plants, mammals do not synthesize chitin but can respond to chitin with an immune response, which includes the production of chitinases [21]. These observations tempt speculation that fungal pathogens of mammals secrete LysM effectors to deal with host immunity in a similar fashion as plant pathogens [10]. Furthermore, allergies such as asthma are associated with fungal infections, although the underlying mechanisms presently remain unclear [22]. Emerging evidence suggests an important role for host chitinases that might mediate host responses to chitin and its derivatives [22], which may again be influenced by fungal LysM effectors. However, the fact that chitin is not universally recognized as a MAMP in mammalian systems argues against the hypothesis that fungal mammalian pathogens secrete LysM effectors to establish infection [23]. Furthermore, the genome of the human pathogenic yeast Candida albicans, as well as of most other Candida spp. that occur as opportunistic human pathogens, appears to lack LysM effector genes [5]. Similarly, in the genome of the skin-associated fungus Malassezia globosa that is responsible for the onset of dandruff and other skin disorders, and the fungus Pneumocystis jirovecii that causes pneumonia among immunocompromised hosts, no LysM effector genes are found [24], [25]. Since many mammalian pathogens show a low degree of host adaptation and lack host specificity, it has been suggested that infection by mammalian fungal pathogens does not require effector activity [1]. In contrast to plant pathogens, most fungal pathogens of mammals spend a considerable amount of their life cycle free-living in the environment and only infect mammalian hosts in an opportunistic manner. Thus, mammalian fungal pathogens may use their LysM effector homologues in processes other than host colonization, such as survival in the environment. The aforementioned absence of LysM effector genes in Candida albicans, Malassezia globosa, and Pneumocystis jirovecii, which are among the few fungal species that are commensals of humans and animals and that do not occur free-living in the environment, seems to support this hypothesis [5], [24], [25].
LysM Effectors of Saprophytes: Diverse Possibilities
Considering that LysM effectors are ubiquitous in fungi, it could be argued that they might act in general physiological processes, such as cell wall modification. Fungi secrete lytic enzymes that break chitin polymers and in this manner maintain cell wall flexibility to allow hyphal growth, branching, morphogenesis, and spore germination. Recently, a Trichoderma atroviride LysM effector was found to be coexpressed with an adjacent chitinase gene [26]. Since addition of the purified LysM effector to T. atroviride inhibited spore germination in vitro, a role in hyphal growth was proposed for this LysM effector. However, further experimental evidence that includes targeted deletion of the LysM effector gene in T. atroviride is required to support such a role. Their occurrence in saprophytes may furthermore suggest that LysM effectors contribute to growth in any fungal niche, as likely other microbes are encountered that compete for the same niche or may act as mycoparasites. In this respect, several hypotheses can be envisaged. Extrapolating the findings for LysM effectors of plant pathogens, LysM effectors may protect fungi against chitinases and other hydrolytic enzymes produced by mycoparasites. Moreover, sequestration of cell wall–derived chitin oligosaccharides may be relevant if mycoparasites would be attracted by gradients of such fragments (Figure 2). One step further, LysM effectors may also have functions that are not associated with chitin binding. Originally, LysMs were identified in bacterial lysozymes (hence the name of the domain) that bind and hydrolyse peptidoglycan, a chitin-related glycan and a major component of bacterial cell walls [27]. LysMs occur in various peptidoglycan-binding proteins, and thus it is conceivable that some LysM effectors bind peptidoglycan as well. Such LysM effectors may help fungi to affect bacterial competitors in their niches, for instance because they immobilize them in a similar fashion as antibodies do [28] (Figure 2).
Concluding Remarks
Fungal LysM effectors are versatile proteins that occur in fungal species with extremely divergent lifestyles. Conceivably, LysM effectors function in various ecological niches. In addition, even LysM effectors of plant pathogens that function in the same niche (the plant host) and that bind the same substrate (chitin) were demonstrated to have distinct roles in promoting fungal virulence [10], [17]. Furthermore, pathogens interact with other microbes, both in the free-living stage and during colonization of their hosts where they may encounter opportunistic pathogens, commensals, and endophytes. In this respect it is interesting to note that strains of the vascular wilt pathogen Verticillium dahliae have a significantly expanded LysM effector family of six to seven members [29], [30]. However, functional analysis has revealed that only one of these LysM effectors is induced in planta and contributes to pathogenicity, while the role of the others still remains obscure [30]. V. dahliae is known to survive as a resting structure in the soil for decades in the absence of suitable host plants, and it is tempting to speculate that its LysM effectors contribute to persistence of these structures through protection against microbial activity. Therefore, the study of LysM effectors of fungi that thrive in a variety of niches will reveal additional LysM effector functions that are relevant for pathogenic fungi as well.
Zdroje
1. LoweRGT, HowlettBJ (2012) Indifferent, affectionate, or deceitful: lifestyles and secretomes of fungi. PLoS Pathog 8: e1002515 doi:10.1371/journal.ppat.1002515
2. de JongeR, BoltonMD, ThommaBPHJ (2011) How filamentous pathogens co-opt plants: the ins and outs of fungal effectors. Curr Opin Plant Biol 14 : 1–7.
3. GijzenM, NürnbergerT (2006) Nep1-like proteins from plant pathogens: recruitment and diversification of the NPP1 domain across taxa. Phytochemistry 67 : 1800–1807.
4. SanthanamP, van EsseHP, AlbertI, FainoL, NürnbergerT, et al. (2013) Evidence for functional diversification within a fungal NEP1-like protein family. Mol Plant Microbe Interact 26 : 278–286.
5. de JongeR, ThommaBPHJ (2009) Fungal LysM effectors – extinguishers of host immunity? Trends Microbiol 17 : 151–157.
6. NürnbergerT, BrunnerF (2002) Innate immunity in plants and animals: emerging parallels between the recognition of general elicitors and pathogen-associated molecular patterns. Curr Opin Plant Biol 5 : 318–324.
7. ThommaBPHJ, NürnbergerT, JoostenMHAJ (2011) Of PAMPs and effectors: the blurred PTI-ETI dichotomy. Plant Cell 23 : 4–15.
8. FelixG, RegenassM, BollerT (1993) Specific perception of subnanomolar concentrations of chitin fragments by tomato cells: induction of extracellular alkalinization, changes in protein phosphorylation, and establishment of a refractory state. Plant J 4 : 307–316.
9. KakuH, NishizawaY, Ishii-MinamiN, Akimoto-TomiyamaC, DohmaeN, et al. (2006) Plant cells recognize chitin fragments for defence signalling through a plasma membrane receptor. Proc Natl Acad Sci U S A 103 : 11086–11091.
10. KombrinkA, Sánchez-ValletA, ThommaBPHJ (2011) The role of chitin detection in plant-pathogen interactions. Microbes Infect 13 : 1168–1176.
11. BoltonMD, van EsseHP, VossenJH, de JongeR, StergiopoulosI, et al. (2008) The novel Cladosporium fulvum lysin motif effector Ecp6 is a virulence factor with orthologues in other fungal species. Mol Microbiol 69 : 119–136.
12. de JongeR, van EsseHP, KombrinkA, ShinyaT, DesakiY, et al. (2010) Conserved fungal LysM effector Ecp6 prevents chitin-triggered immunity in plants. Science 329 : 953–955.
13. van den BurgHA, HarrisonSJ, JoostenMHAJ, VervoortJ, de WitPJGM (2006) Cladosporium fulvum Avr4 protects fungal cell wall against hydrolysis by plant chitinases accumulating during infection. Mol Plant Microbe Interact 19 : 1420–1430.
14. van EsseHP, BoltonMD, StergiopoulosI, de WitPJGM, ThommaBPHJ (2007) The chitin-binding Cladosporium fulvum effector protein Avr4 is a virulence factor. Mol Plant Microbe Interact 20 : 1092–1101.
15. Sánchez-ValletA, Saleem-BatchaR, KombrinkA, HansenG, ValkenburgD-J, et al. (2013) Fungal effector Ecp6 outcompetes host immune receptor for chitin binding through intrachain LysM dimerization. eLife 2: e00790.
16. LiuT, LiuZ, SongC, HuY, HanZ, et al. (2012) Chitin-induced dimerization activates a plant immune receptor. Science 336 : 1160–1164.
17. MarshallR, KombrinkA, MotteramJ, Loza-ReyesE, LucasJ, et al. (2011) Analysis of two in planta expressed LysM effector homologues from the fungus Mycosphaerella graminicola reveals novel functional properties and varying contributions to virulence on wheat. Plant Physiol 156 : 756–769.
18. MentlakTA, KombrinkA, ShinyaT, RyderLS, OtomoI, et al. (2012) Effector-mediated suppression of chitin-triggered immunity by Magnaporthe oryzae is necessary for rice blast disease. Plant Cell 24 : 322–335.
19. SchlumbaumA, MauchF, VogeliU, BollerT (1986) Plant chitinases are potent inhibitors of fungal growth. Nature 324 : 365–367.
20. MartinezDA, OliverBG, GräserY, GoldbergJM, LiW, et al. (2013) Comparative genome analysis of Trichophyton rubrum and related dermatophytes reveals candidate genes involved in infection. MBio 3: e00259–12.
21. LeeCG, Da SilvaCA, LeeJ-Y, HartlD, EliasJA (2008) Chitin regulation of immune responses: an old molecule with new roles. Curr Opin Immunol 20 : 684–689.
22. GoldmanDL, VicencioAG (2012) The chitin connection. MBio 3: e00056–12.
23. Mora-MontesHM, NeteaMG, FerwerdaG, LenardonMD, BrownGD, et al. (2011) Recognition and blocking of innate immunity cells by Candida albicans chitin. Infect Immun 79 : 1961–1970.
24. XuJ, SaundersCW, HuP, GrantRA, BoekhoutT, et al. (2007) Dandruff-associated Malassezia genomes reveal convergent and divergent virulence traits shared with plant and human fungal pathogens. Proc Natl Acad Sci U S A 104 : 18730–18735.
25. CisséOH, PagniM, HauserPM (2012) De novo assembly of the Pneumocystis jirovecii genome from a single bronchoalveolar lavage fluid specimen from a patient. MBio 4: e00428–12.
26. Seidl-SeibothV, ZachS, FrischmannA, SpadiutO, DietschC, et al. (2013) Spore germination of Trichoderma atroviride is inhibited by its LysM protein TAL6. FEBS J 280 : 1226–1236.
27. BuistG, SteenA, KokJ, KuipersOP (2008) LysM, a widely distributed protein motif for binding to (peptido)glycans. Mol Microbiol 68 : 838–847.
28. YangJ, WangW, WeiX, QiuL, WangL, et al. (2010) Peptidoglycan recognition protein of Chlamys farreri (CfPGRP-S1) mediates immune defenses against bacterial infection. Dev Comp Immunol 34 : 1300–1307.
29. KlostermannSJ, SubbaraoKV, KangS, VeroneseP, GoldSE, et al. (2011) Comparative genomics yields insights into niche adaptation of plant vascular wilt pathogens. PLoS Pathog 7: e1002137 doi:10.1371/journal.ppat.1002137
30. de JongeR, BoltonMD, KombrinkA, van den BergGCM, YadetaKA, et al. (2013) Extensive chromosomal reshuffling drives evolution of virulence in an asexual pathogen. Genome Res 23 : 1271–1282.
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek Parental Transfer of the Antimicrobial Protein LBP/BPI Protects Eggs against Oomycete InfectionsČlánek Immune Therapeutic Strategies in Chronic Hepatitis B Virus Infection: Virus or Inflammation Control?Článek Coronaviruses as DNA Wannabes: A New Model for the Regulation of RNA Virus Replication FidelityČlánek CRISPR-Cas Immunity against Phages: Its Effects on the Evolution and Survival of Bacterial PathogensČlánek The Cyst Wall Protein CST1 Is Critical for Cyst Wall Integrity and Promotes Bradyzoite PersistenceČlánek The Malarial Serine Protease SUB1 Plays an Essential Role in Parasite Liver Stage Development
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2013 Číslo 12- Jak souvisí postcovidový syndrom s poškozením mozku?
- Měli bychom postcovidový syndrom léčit antidepresivy?
- Farmakovigilanční studie perorálních antivirotik indikovaných v léčbě COVID-19
- 10 bodů k očkování proti COVID-19: stanovisko České společnosti alergologie a klinické imunologie ČLS JEP
-
Všechny články tohoto čísla
- Host Susceptibility Factors to Bacterial Infections in Type 2 Diabetes
- LysM Effectors: Secreted Proteins Supporting Fungal Life
- Influence of Mast Cells on Dengue Protective Immunity and Immune Pathology
- Innate Lymphoid Cells: New Players in IL-17-Mediated Antifungal Immunity
- Cytoplasmic Viruses: Rage against the (Cellular RNA Decay) Machine
- Balancing Stability and Flexibility within the Genome of the Pathogen
- The Evolution of Transmissible Prions: The Role of Deformed Templating
- Parental Transfer of the Antimicrobial Protein LBP/BPI Protects Eggs against Oomycete Infections
- Host Defense via Symbiosis in
- Regulatory Circuits That Enable Proliferation of the Fungus in a Mammalian Host
- Immune Therapeutic Strategies in Chronic Hepatitis B Virus Infection: Virus or Inflammation Control?
- Burning Down the House: Cellular Actions during Pyroptosis
- Coronaviruses as DNA Wannabes: A New Model for the Regulation of RNA Virus Replication Fidelity
- CRISPR-Cas Immunity against Phages: Its Effects on the Evolution and Survival of Bacterial Pathogens
- Combining Regulatory T Cell Depletion and Inhibitory Receptor Blockade Improves Reactivation of Exhausted Virus-Specific CD8 T Cells and Efficiently Reduces Chronic Retroviral Loads
- Shaping Up for Battle: Morphological Control Mechanisms in Human Fungal Pathogens
- Identification of the Virulence Landscape Essential for Invasion of the Human Colon
- Nodular Inflammatory Foci Are Sites of T Cell Priming and Control of Murine Cytomegalovirus Infection in the Neonatal Lung
- Hepatitis B Virus Disrupts Mitochondrial Dynamics: Induces Fission and Mitophagy to Attenuate Apoptosis
- Mycobacterial MazG Safeguards Genetic Stability Housecleaning of 5-OH-dCTP
- Systematic MicroRNA Analysis Identifies ATP6V0C as an Essential Host Factor for Human Cytomegalovirus Replication
- Placental Syncytium Forms a Biophysical Barrier against Pathogen Invasion
- The CD8-Derived Chemokine XCL1/Lymphotactin Is a Conformation-Dependent, Broad-Spectrum Inhibitor of HIV-1
- Cyclin A Degradation by Primate Cytomegalovirus Protein pUL21a Counters Its Innate Restriction of Virus Replication
- Genome-Wide RNAi Screen Identifies Novel Host Proteins Required for Alphavirus Entry
- Zinc Sequestration: Arming Phagocyte Defense against Fungal Attack
- The Cyst Wall Protein CST1 Is Critical for Cyst Wall Integrity and Promotes Bradyzoite Persistence
- Biphasic Euchromatin-to-Heterochromatin Transition on the KSHV Genome Following Infection
- The Malarial Serine Protease SUB1 Plays an Essential Role in Parasite Liver Stage Development
- HIV-1 Vpr Accelerates Viral Replication during Acute Infection by Exploitation of Proliferating CD4 T Cells
- A Human Torque Teno Virus Encodes a MicroRNA That Inhibits Interferon Signaling
- The ArlRS Two-Component System Is a Novel Regulator of Agglutination and Pathogenesis
- An In-Depth Comparison of Latent HIV-1 Reactivation in Multiple Cell Model Systems and Resting CD4+ T Cells from Aviremic Patients
- Enterohemorrhagic Hemolysin Employs Outer Membrane Vesicles to Target Mitochondria and Cause Endothelial and Epithelial Apoptosis
- Overcoming Antigenic Diversity by Enhancing the Immunogenicity of Conserved Epitopes on the Malaria Vaccine Candidate Apical Membrane Antigen-1
- The Type-Specific Neutralizing Antibody Response Elicited by a Dengue Vaccine Candidate Is Focused on Two Amino Acids of the Envelope Protein
- Tmprss2 Is Essential for Influenza H1N1 Virus Pathogenesis in Mice
- Signatures of Pleiotropy, Economy and Convergent Evolution in a Domain-Resolved Map of Human–Virus Protein–Protein Interaction Networks
- Interference with the Host Haemostatic System by Schistosomes
- RocA Truncation Underpins Hyper-Encapsulation, Carriage Longevity and Transmissibility of Serotype M18 Group A Streptococci
- Gene Fitness Landscapes of at Important Stages of Its Life Cycle
- Phagocytosis Escape by a Protein That Connects Complement and Coagulation Proteins at the Bacterial Surface
- t Is a Structurally Novel Crohn's Disease-Associated Superantigen
- An Increasing Danger of Zoonotic Orthopoxvirus Infections
- Myeloid Dendritic Cells Induce HIV-1 Latency in Non-proliferating CD4 T Cells
- Transcriptional Analysis of Murine Macrophages Infected with Different Strains Identifies Novel Regulation of Host Signaling Pathways
- Serotonergic Chemosensory Neurons Modify the Immune Response by Regulating G-Protein Signaling in Epithelial Cells
- Genome-Wide Detection of Fitness Genes in Uropathogenic during Systemic Infection
- Induces an Unfolded Protein Response via TcpB That Supports Intracellular Replication in Macrophages
- Intestinal CD103+ Dendritic Cells Are Key Players in the Innate Immune Control of Infection in Neonatal Mice
- Emerging Functions for the RNome
- KSHV MicroRNAs Mediate Cellular Transformation and Tumorigenesis by Redundantly Targeting Cell Growth and Survival Pathways
- HrpA, an RNA Helicase Involved in RNA Processing, Is Required for Mouse Infectivity and Tick Transmission of the Lyme Disease Spirochete
- A Toxin-Antitoxin Module of Promotes Virulence in Mice
- Real-Time Imaging of the Intracellular Glutathione Redox Potential in the Malaria Parasite
- Hypoxia Inducible Factor Signaling Modulates Susceptibility to Mycobacterial Infection via a Nitric Oxide Dependent Mechanism
- Novel Strategies to Enhance Vaccine Immunity against Coccidioidomycosis
- Dual Expression Profile of Type VI Secretion System Immunity Genes Protects Pandemic
- —What Makes the Species a Ubiquitous Human Fungal Pathogen?
- αvβ6- and αvβ8-Integrins Serve As Interchangeable Receptors for HSV gH/gL to Promote Endocytosis and Activation of Membrane Fusion
- -Induced Activation of EGFR Prevents Autophagy Protein-Mediated Killing of the Parasite
- Semen CD4 T Cells and Macrophages Are Productively Infected at All Stages of SIV infection in Macaques
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Influence of Mast Cells on Dengue Protective Immunity and Immune Pathology
- Host Defense via Symbiosis in
- Coronaviruses as DNA Wannabes: A New Model for the Regulation of RNA Virus Replication Fidelity
- Myeloid Dendritic Cells Induce HIV-1 Latency in Non-proliferating CD4 T Cells
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



