-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaDual Expression Profile of Type VI Secretion System Immunity Genes Protects Pandemic
The Vibrio cholerae type VI secretion system (T6SS) assembles as a molecular syringe that injects toxic protein effectors into both eukaryotic and prokaryotic cells. We previously reported that the V. cholerae O37 serogroup strain V52 maintains a constitutively active T6SS to kill other Gram-negative bacteria while being immune to attack by kin bacteria. The pandemic O1 El Tor V. cholerae strain C6706 is T6SS-silent under laboratory conditions as it does not produce T6SS structural components and effectors, and fails to kill Escherichia coli prey. Yet, C6706 exhibits full resistance when approached by T6SS-active V52. These findings suggested that an active T6SS is not required for immunity against T6SS-mediated virulence. Here, we describe a dual expression profile of the T6SS immunity protein-encoding genes tsiV1, tsiV2, and tsiV3 that provides pandemic V. cholerae strains with T6SS immunity and allows T6SS-silent strains to maintain immunity against attacks by T6SS-active bacterial neighbors. The dual expression profile allows transcription of the three genes encoding immunity proteins independently of other T6SS proteins encoded within the same operon. One of these immunity proteins, TsiV2, protects against the T6SS effector VasX which is encoded immediately upstream of tsiV2. VasX is a secreted, lipid-binding protein that we previously characterized with respect to T6SS-mediated virulence towards the social amoeba Dictyostelium discoideum. Our data suggest the presence of an internal promoter in the open reading frame of vasX that drives expression of the downstream gene tsiV2. Furthermore, VasX is shown to act in conjunction with VasW, an accessory protein to VasX, to compromise the inner membrane of prokaryotic target cells. The dual regulatory profile of the T6SS immunity protein-encoding genes tsiV1, tsiV2, and tsiV3 permits V. cholerae to tightly control T6SS gene expression while maintaining immunity to T6SS activity.
Published in the journal: . PLoS Pathog 9(12): e32767. doi:10.1371/journal.ppat.1003752
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1003752Summary
The Vibrio cholerae type VI secretion system (T6SS) assembles as a molecular syringe that injects toxic protein effectors into both eukaryotic and prokaryotic cells. We previously reported that the V. cholerae O37 serogroup strain V52 maintains a constitutively active T6SS to kill other Gram-negative bacteria while being immune to attack by kin bacteria. The pandemic O1 El Tor V. cholerae strain C6706 is T6SS-silent under laboratory conditions as it does not produce T6SS structural components and effectors, and fails to kill Escherichia coli prey. Yet, C6706 exhibits full resistance when approached by T6SS-active V52. These findings suggested that an active T6SS is not required for immunity against T6SS-mediated virulence. Here, we describe a dual expression profile of the T6SS immunity protein-encoding genes tsiV1, tsiV2, and tsiV3 that provides pandemic V. cholerae strains with T6SS immunity and allows T6SS-silent strains to maintain immunity against attacks by T6SS-active bacterial neighbors. The dual expression profile allows transcription of the three genes encoding immunity proteins independently of other T6SS proteins encoded within the same operon. One of these immunity proteins, TsiV2, protects against the T6SS effector VasX which is encoded immediately upstream of tsiV2. VasX is a secreted, lipid-binding protein that we previously characterized with respect to T6SS-mediated virulence towards the social amoeba Dictyostelium discoideum. Our data suggest the presence of an internal promoter in the open reading frame of vasX that drives expression of the downstream gene tsiV2. Furthermore, VasX is shown to act in conjunction with VasW, an accessory protein to VasX, to compromise the inner membrane of prokaryotic target cells. The dual regulatory profile of the T6SS immunity protein-encoding genes tsiV1, tsiV2, and tsiV3 permits V. cholerae to tightly control T6SS gene expression while maintaining immunity to T6SS activity.
Introduction
Vibrio cholerae is the etiological agent of the diarrheal disease cholera. This pathogen utilizes a wide array of virulence factors during host infection including the well-characterized cholera toxin (CT) and toxin-coregulated pilus (TCP). In addition, V. cholerae possesses numerous other virulence factors including the type VI secretion system (T6SS), a recently described mechanism used by numerous Gram-negative bacteria to export effectors across their cell envelopes. In contrast to TCP and CT, whose presence is restricted to a subset of virulent V. cholerae strains, the V. cholerae T6SS is highly conserved and is present in strains of all serogroups sequenced to date. Structurally, the T6SS resembles an inverted bacteriophage; it assembles in the V. cholerae cytoplasm and docks onto a baseplate complex situated in the bacterial envelope. Two alleles on the small and large chromosome encode hemolysin-coregulated protein (Hcp) [1], which polymerizes [2] and forms the inner tube of the T6SS injectosome and acts as a chaperone for T6SS effectors [3]. Contraction of the outer sheath (formed by VipA and VipB) around the formed Hcp nanotube leads to the ejection of the Hcp tube decorated with a VgrG trimer consisting of three different VgrG proteins: VgrG-1, VgrG-2, and VgrG-3 [4]–[10]. VgrG-1 and VgrG-3 carry enzymatic C-terminal extensions that crosslink actin or degrade the peptidoglycan layer, respectively, upon translocation into target cells [4], [6], [10]–[12]. ATP hydrolysis by the inner membrane protein VasK provides the energy for Hcp secretion [13]. A recent report suggests that proteins from the PAAR (proline-alanine-alanine-arginine) repeat superfamily form a sharp conical extension on the VgrG cap. This extension is able to load additional T6SS effectors besides the VgrGs, which are then delivered simultaneously into target cells in a single contraction-driven translocation event [14].
The O37 serogroup V. cholerae strain V52 [15] constitutively synthesizes T6SS structural proteins and effectors and actively engages in T6SS-mediated virulence under standard laboratory conditions [4], [5], [16]. Conversely, the pandemic-causing O1 serogroup strain C6706 possesses a full complement of T6SS-encoding genes, but does not express T6SS genes encoding structural apparatus components and effectors under laboratory conditions. Null-mutations in the genes encoding the quorum sensing regulator LuxO and the global regulator TsrA lead to production and secretion of the T6SS protein Hcp, and T6SS-mediated virulence in V. cholerae strain C6706, demonstrating that this pandemic strain employs a tightly controlled, fully active T6SS [17].
The V. cholerae T6SS is encoded by three separate gene clusters [5] (Figure 1A). One source of T6SS regulation, VasH (encoded by VCA0117), is encoded in all V. cholerae genomes sequenced so far and acts as an activator of the alternate sigma factor 54. A regulatory role for VasH in the expression of genes within the two auxiliary T6SS gene clusters encoding (i) Hcp-1 and VgrG-1, and (ii) VgrG-2 and VasX has been established in V. cholerae El Tor and classical O1, and non-O1/non-O139 strains [18]–[20].
Fig. 1. Dual regulatory profile of T6SS immunity protein-encoding genes. 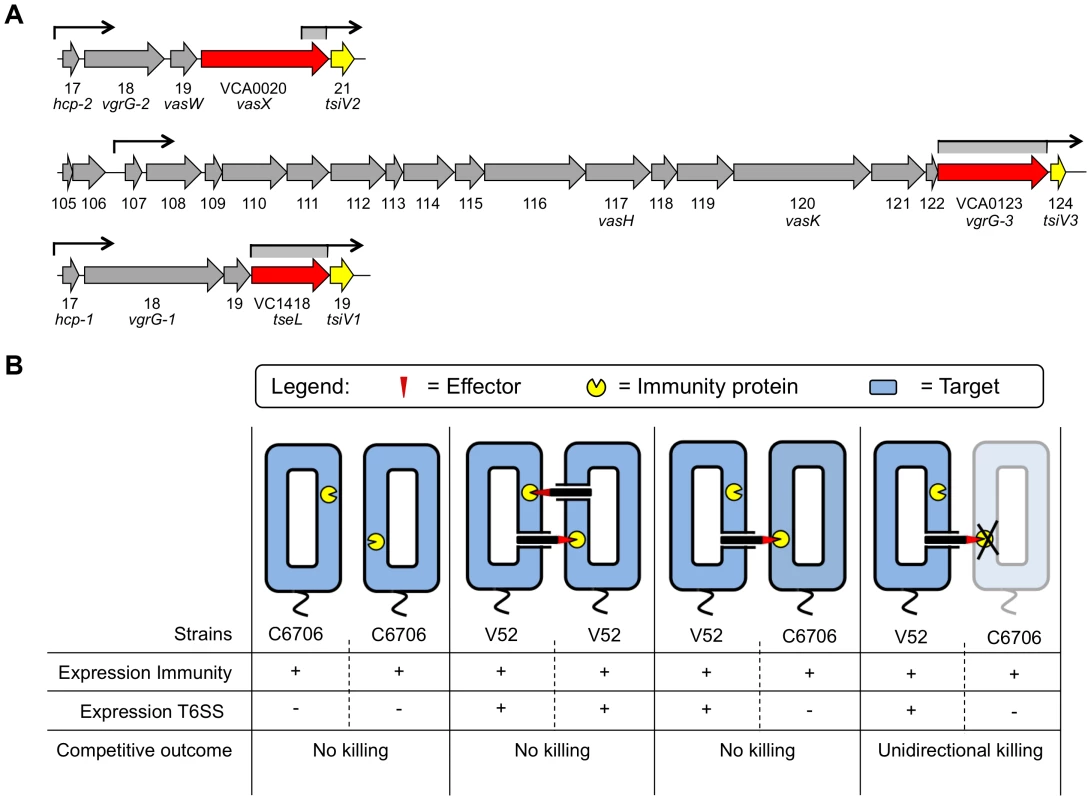
(A) Schematic representation of the V. cholerae T6SS gene clusters. Toxin-encoding genes are shown in red and immunity protein-encoding genes in yellow. Promoters upstream of the individual T6SS clusters are indicated by arrows. Regions with promoter activity that drive expression of immunity protein-encoding genes are indicated in grey below arrows. (B) Two V. cholerae strains, C6706 and V52, exemplify the dual regulatory profile of the T6SS system. In the T6SS-off state, C6706 expresses only the T6SS immunity proteins (first panel). In contrast, V52 expresses structural, effector, and immunity T6SS proteins, the latter of which provide protection from T6SS-active kin bacteria (second panel). When T6SS-active V52 come in contact with C6706 in the T6SS-off state, C6706 is protected from a T6SS-mediated attack without engaging in T6SS-mediated virulence (third panel). C6706 succumbs to killing by V52 when immunity genes are removed or not expressed (fourth panel). T6SSs mediate toxicity towards both eukaryotes and prokaryotes [4], [5], [16], [21]–[32]. The V. cholerae T6SS mediates virulence towards murine macrophages and the amoeboid host Dictyostelium discoideum [4], [5], [16], [18], [21] as well as a variety of Gram-negative bacteria including Escherichia coli, Salmonella Typhimurium, and Citrobacter rodentium [16], [33]. Importantly, V52 bacteria do not kill each other, and some bacteria such as V. cholerae C6706 are resistant to killing by V52 [16]. Although all sequenced V. cholerae strains possess a full complement of T6SS-encoding genes, only some strains are known to express these genes under laboratory conditions. V52 has a constitutively active T6SS and produces hallmark T6SS proteins such as Hcp and VgrGs when grown in liquid culture [5]. In contrast, C6706 maintains a T6SS-silent state and does not produce Hcp and VgrGs under standard laboratory conditions [34]; yet C6706 is immune to killing by V52 [11], [16].
T6SS-mediated killing of prokaryotes has been reported in other bacterial species including Pseudomonas aeruginosa, Serratia marcescens, and Burkholderia thailandensis [23], [25], [26], [35], [36]. The P. aeruginosa T6SS encodes effectors as part of a toxin/antitoxin (TA) gene system in which the effector (toxin) is injected into the target cell where it degrades the peptidoglycan in the target cell unless inhibited by an immunity protein (antitoxin) [35].
TA systems are not restricted to T6SS gene clusters, and one other notable TA system subset are colicins—small secreted peptides produced by and toxic to certain strains of E. coli [37]. Colicins employ a diverse range of mechanisms to kill target cells, including degradation of RNA and DNA [38]–[44], inhibition of murein synthesis [45], and pore-formation in the inner membrane [46]–[49]. In the latter case, the pore in the inner membrane forms a voltage-dependent ion channel that dissipates membrane potential and halts cellular respiration [48]–[55]. Pore-formation in the inner membrane can occur only if colicin is presented to the target cell from the periplasmic face [56], [57]. Consequently, cytoplasmic production of colicins is not toxic to the producing cell [57].
We previously identified the protein VasX and characterized this T6SS effector with respect to its role in T6SS-mediated killing of D. discoideum [21]. In this study, we characterize the mechanism by which VasX acts as a bacterial effector. We also present evidence that VasW, the product of the gene directly upstream of vasX, is necessary for VasX-mediated bacterial killing. By screening a C6706 T6SS transposon library [58], we identified the VasX immunity protein-encoding gene VCA0021 located directly downstream of vasX, and two other T6SS-encoding genes, VCA0124 and VC1419, that encode additional immunity proteins. We demonstrate that all three immunity protein-encoding genes are controlled in a dual regulatory fashion, ensuring that even T6SS-silent strains such as C6706 are protected from T6SS-mediated killing by neighboring bacterial competitors (Figure 1B).
Materials and Methods
Bacterial strains and growth conditions
The strains and plasmids used in this study are listed in Table 1 and Table 2, respectively. A streptomycin-resistant V. cholerae strain V52 (O37 serogroup) lacking hapA, rtxA, and hlyA genes was used as a T6SS-active strain in all experiments in this study unless otherwise noted in the figure legend. A streptomycin - and rifampicin-resistant V. cholerae strain C6706 (O1 serogroup) was used as a T6SS-silent strain in all experiments. DH5α λpir and SM10 λpir were used for cloning, and mating of pWM91-based plasmids, respectively. Unless stated otherwise, bacteria were grown in Luria-Bertani (LB) broth at 37°C with shaking (200 rpm). Antibiotic concentrations used were 100 µg·mL−1 ampicillin, 10 µg·mL−1 chloramphenicol, 100 µg·mL−1 streptomycin, 50 µg·mL−1 kanamycin, and 50 µg·mL−1 rifampicin. Arabinose was added at a final concentration of 0.1% to induce expression from the PBAD promoter.
In-frame deletions and plasmid construction
In-frame deletion of T6SS-encoding genes was performed as described previously [59]. Construction of the vasX, vasH, and vasK knockout constructs was also previously described [5], [18], [21]. VasW, vgrG-3, tseL, tsiV1, tsiV2, and tsiV3 knockout constructs were created using the primers listed in Table 3. PCR products resulting from primer combinations A/B and C/D were stitched together by overlapping PCR. The resulting knockout construct was digested with BamHI and cloned into the suicide plasmid pWM91 [59]. V52ΔvgrG-3 [5] was used as the parent strain in which vasW was deleted to create the V52ΔvgrG-3ΔvasW double mutant. V52ΔvasH was used as the parent strain in which vasX was deleted to create the V52ΔvasHΔvasX double mutant.
Tab. 3. Oligonucleotide sequences. 
F, forward; R, reverse; Construction of plasmid pBAD24-vasX was described previously [21]. VasX::FLAG and vasW::FLAG were PCR-amplified using the primers listed in Table 3 with Accuprime Pfx DNA polymerase (Invitrogen) and cloned into pCR2.1 TOPO TA (Invitrogen) following 10 minute incubation at 72°C with TopTaq polymerase (Qiagen) to add 3′ adenine overhangs. VasX::FLAG was excised using KpnI and XbaI and subcloned into pBAD24. VasW::FLAG was excised using EcoRI and XbaI and subcloned into pBAD24 [60].
The VasX immunity protein-encoding gene tsiV2 was PCR amplified with a C-terminal 6×His tag using Phusion Taq (Thermo Fisher) and ligated into pJET1.2/blunt (Thermo Fisher). The fragment was excised with KpnI and XbaI restriction enzymes, subcloned into pBAD33 [60], and transformed into E. coli Top10 (Invitrogen). A triparental mating technique [61] was used to introduce pBAD33-tsiV2::6×His into V. parahaemolyticus RIMD2210633. Top10/pBAD33-tsiV2::6×His (CmR), V. parahaemolyticus (RifR), and DH5α/pRK2013 (helper plasmid containing tra and mob genes) were mixed at a 1∶1∶1 ratio, spotted onto nonselective LB plates, and incubated for 6 hours at 37°C. The spot was harvested into 1 mL LB and pelleted at 13,000 rpm for 5 minutes. Cells were resuspended in 100 µL LB and plated on thiosulfate-citrate-bile salts-sucrose (TCBS) agar plates containing 50 µg·mL−1 rifampicin and 10 µg·mL−1 chloramphenicol to select for V. parahaemolyticus/pBAD33-tsiV2::6×His. Plates were incubated overnight at 37°C. Resulting colonies were restreaked onto TCBSrif50chlor10 to exclude false-positive transformants.
Construction of the periplasmic expression vectors pBAD24-LS and pBAD24-SecP::core was described previously [12]. The vasX gene was cloned in-frame with the sequence encoding a sec signal peptide (SecP) using XbaI and HindIII restriction sites. The resulting construct was transformed into E. coli Top10 (Invitrogen) for expression analysis and then into the indicated Vibrio strains.
Plasmid pAH6 [62] was a generous gift from Dr. Jun Zhu (University of Pennsylvania School of Medicine) and contains a promoter-less lacZ gene. VasX (4-3258, 4-1345, 1575-3258, 2208-3258), tseL, vgrG-3, and Phcp were amplified using Phusion Taq (Thermo Fisher), cloned into the blunt-ended cloning vector pJET1.2/blunt (Thermo Fisher), and transformed into E. coli Top10 (Invitrogen). All vasX constructs, Phcp, and tseL were excised using XbaI and HindIII restriction sites and subcloned into pAH6. VgrG-3 was excised using the XbaI restriction site and subcloned into pAH6. The resulting constructs were transformed into E. coli Top10, followed by V. cholerae V52, V52ΔvasH, and C6706.
Plasmids pBAD24-tsiV1::FLAG, pBAD24-tsiV2::FLAG, and pBAD24-tsiV3::FLAG were created using the Gateway recombination system. Briefly, the LR recombination site, including the ccdB negative selection sequence, was amplified out of pBAD-DEST-49 using the primers listed in Table 2 (forward primer adds a KpnI site, reverse primer adds a stop codon, XbaI site, and FLAG epitope tag sequence). The PCR product was digested with XbaI and KpnI and ligated into the corresponding sites of pBAD24 to create pBAD24gw::FLAG. The Gateway recombination reaction between pDONR221-tsiV2 (Harvard Institute of Proteomics) and pBAD24gw::FLAG was performed according to the manufacturer's protocol (Invitrogen, Carlsbad, CA). The resulting plasmids were transformed into E. coli Top10, followed by transformation into V. cholerae C6706ΔtsiV1, C6706ΔtsiV2, C6706ΔtsiV3, C6706ΔvasX, C6706ΔtseL, or C6706ΔvgrG-3.
Bioinformatics and statistical analyses
Bioinformatics were performed using BLASTn (nucleotide) and BLASTp (protein) algorithms (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Statistical significance was determined using the Student's one-tailed, paired t-test with a significance cut-off of p<0.05.
Bacterial killing assay
Killing assays were performed as described previously [16]. Briefly, bacterial strains were grown overnight on selective LB plates and resuspended in LB broth (or LB 3% NaCl for V. parahaemolyticus). Predator and prey were mixed at a 10∶1 ratio and spotted onto prewarmed LB agar plates and incubated at 37°C for 4 hours. Spots were harvested, serially diluted, and 10 µl of each dilution was spotted onto a selective LB plate. Plates were incubated overnight at 37°C and surviving prey (CFU·mL−1) were enumerated. The competitive index was calculated by dividing prey CFUs after exposure to V52 by prey CFUs following exposure to V52ΔvasK.
Cytoplasmic production of VasX in C6706ΔtsiV2
An overnight culture of C6706ΔtsiV2/pBAD24-vasX was back-diluted 1∶100 in selective LB broth in the presence or absence of 0.1% arabinose to induce expression of vasX. Strains were grown in a 96 well plate on a Heidolph Titromax 1000 vibrating shaker at 900 rpm. OD600 readings were taken every 30 min for 8 h using a BioRad XMark microplate spectrophotometer. At the 7-hour time point, a cell lysate sample was collected from each sample, mixed with sodium dodecyl sulfate (SDS) protein sample buffer, and boiled for 10 min. The protein samples were subjected to SDS-PAGE followed by western blotting to detect VasX and DnaK.
β-galactosidase assay
The OD600 was measured of overnight liquid cultures diluted 1/10 in Z buffer (0.06 M Na2HPO4, 0.04 M NaH2PO4, 0.01 M KCl, 0.001 M MgSO4, pH 7.0) containing 2.7% β-mercaptoethanol. To lyse bacteria, 10 µL of 0.1% SDS and 20 µL of chloroform were added and the mixture was vortexed for 5 sec. Tubes were incubated in a 28°C waterbath for 10 min, followed by addition of 200 µL ortho-nitrophenyl-β-galactoside (ONPG) buffer (4 mg·mL−1 ONPG dissolved in Z buffer using a sonicating waterbath). The time required for development of a yellow color was recorded and the reaction was halted by the addition of 500 µL of 1 M Na2CO3. The OD420 of each sample was measured using a BioRad XMark microplate spectrophotometer. Miller units were calculated based on the equation (OD420/(OD600·time·volume)×103).
Strains V52, C6706, and V52ΔvasH used in this assay each possess a chromosomal copy of lacZ. Strains O395w, NIH41w, MAK757w, C6709w, and N16961w contain a disrupted lacZ gene. Mutation of lacZ was accomplished using the suicide vector pJL1. E. coli SM10 λpir/pJL1 was mixed at a ∼1∶1 ratio with the recipient V. cholerae strains and incubated on a prewarmed LB agar plate for 6 hours at 37°C. Bacterial mixtures were harvested and resuspended in 1 mL LB and subjected to 4 serial dilutions; 200 µL of diluted sample were spread onto selective LB agar plates. Transconjugants were restreaked onto LB containing ampicillin and 40 µg·mL−1 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside (x-gal) to ensure disruption of lacZ resulted in white colony formation.
CFU recovery time course
Strains grown overnight were back-diluted 1∶100 in selective LB broth with 0.1% arabinose. At 0, 2, 4, 6, and 8 h, a 200 µL sample was taken and serially diluted. 10 µL of each dilution was spotted onto selective LB plates and incubated overnight at 37°C.
Membrane potential assay and flow cytometry
The BacLight Bacterial Membrane Potential Kit (Molecular Probes, Invitrogen) was used to determine whether VasX, when provided with a Sec signal peptide (SecP) to export SecP::VasX to the periplasm, caused dissipation of the membrane potential. Overnight cultures of the indicated strains were back-diluted 1∶30 in selective LB with arabinose (0.1%) and grown for 2 h at 37°C with shaking (225 rpm). Cells were diluted to ∼106 cells·mL−1 in filtered PBS. Carbonyl cyanide m-chlorophenyl hydrazone (CCCP, depolarizing control) and 3,3-diethyloxacarbocyanine iodide (DiOC2(3)) stain were added (where appropriate) to final concentrations of 5 µM and 15 µM, respectively, and incubated for 30 min in the dark. Stained cells were analyzed using a BD LSR II flow cytometer with a 488 nm laser and PE-Texas Red (601 long pass and 616/23 band pass filters) and FITC (502 long pass and 525/50 band pass filters) detectors. Forward and side scatter and fluorescence data were collected with logarithmic signal amplification. Red/green ratios were calculated based on double positive (Texas Red and FITC) cells by collecting 50,000 events per strain tested.
Flow cytometry analysis of cells incubated with propidium iodide
Overnight bacterial cultures were diluted 1∶100 in LB containing appropriate antibiotics in the presence of 0.1% arabinose. Cultures were grown under inducing conditions for 2 hours at 37°C with shaking. As a positive control for propidium iodide staining, dead cells were prepared by ethanol treatment: 1 mL of bacterial culture was centrifuged at 13,000 rpm for 3 minutes. The supernatant was aspirated and the pellet was resuspended in 1 mL of 70% ethanol and incubated for 20 minutes at room temperature. Cells were pelleted by centrifugation at 13,000 rpm for 3 minutes and the pellet was resuspended in 1 mL filtered PBS. ∼106 cells (living or dead) were diluted into 1 mL of filtered PBS. 1.5 µL of 20 mM PI was added to each sample followed by incubation for 15 minutes in the dark.
Stained cells were analyzed using a BD LSRFortessa Cell Analyzer with a 488 nm laser, and a PE (550 long pass and 575/26 band pass filters) detector. Forward and side scatter and fluorescence were collected with logarithmic signal amplification. The percentage of cells permeable to PI was recorded by collecting 50,000 events per strain tested.
Secretion profiles
Overnight bacterial cultures were diluted 1∶100 in LB (+/−0.1% arabinose) and grown to late-logarithmic phase. Bacterial pellet samples were resuspended in SDS protein sample buffer and boiled for 10 min. Culture supernatants were isolated, filter sterilized, and concentrated with 20% trichloroacetic acid. Supernatant proteins were resuspended in SDS protein sample buffer and boiled for 10 minutes. Samples were subjected to SDS-PAGE followed by western blotting with α-Hcp [5] and α-DnaK (loading and lysis controls, obtained from Stressgen) antibodies.
Results
Identification of three V. cholerae genes encoding T6SS-immunity proteins
V. cholerae V52 uses its constitutively active T6SS to kill other Gram-negative bacteria such as E. coli strain MG1655 [16]. Interestingly, two 7th pandemic El Tor strains C6706 and N16961 (with silent T6SSs), as well as V52 are resistant to this killing phenotype. C6706 and N16961 contain a full complement of T6SS-encoding genes; however, when spotted on nutrient agar plates, these strains do not produce structural components (e.g., Hcp and VgrGs) required for the assembly of a functional T6SS. Therefore, C6706 and N16961 do not engage in T6SS-mediated killing of prokaryotes or eukaryotes under laboratory conditions [18], [34], [63]. We hypothesized that C6706 and N16961 (and other V. cholerae strains with silent T6SSs) encode immunity proteins within theT6SS gene clusters that are produced in a silent (T6SS-off) state. Dong et al. applied a high-throughput parallel sequencing technique to address this problem and identified the T6SS immunity protein-encoding genes, tsiV1 (VC1419), tsiV2 (VCA0021), and tsiV3 (VCA0124), along with their corresponding T6SS effector genes tseL (VC1418), vasX (VCA0020), and vgrG-3 (VCA0123), respectively [11]. We independently identified the same immunity genes by screening a subset of a transposon library of C6706 [58] to identify insertions within the three T6SS gene clusters that result in the loss of T6SS immunity against V52. Mutation of VCA0021 (tsiV2), VCA0124 (tsiV3), and VC1419 (tsiV1) [isolated as a polar mutation in VC1418 (tseL)] resulted in sensitivity to killing by V52 (Figure S1). We noted that mutants with transposons in VCA0114, VCA0115, and VCA0121 also became susceptible to killing by V52. However, we decided not to analyze these three genes further because deletion of these genes in strain V52 [64], which employs a constitutively active T6SS, does not interfere with the protection from killing by V52 kin bacteria. Episomal expression of VCA0021 (tsiV2) and VCA0124 (tsiV3) protects the respective C6706 mutants from T6SS-mediated killing by V52 (Figures 2A and 2B). Complementation of the VC1418 null-mutation did not restore immunity (Figure S2). However, episomal expression of VC1419 in C6706ΔVC1418 restored immunity to killing by V52 (Figure S2), implying that VC1419 is the third immunity protein-encoding gene and confirming the findings by Dong et al. [11]. Subsequently, in-frame deletion of VC1419 rendered C6706 susceptible to killing by V52 and this could be complemented by expression of VC1419 in-trans (Figure 2B).
Fig. 2. Episomal expression of VCA0021, VCA0124, and VC1419 protects respective mutants from T6SS killing. 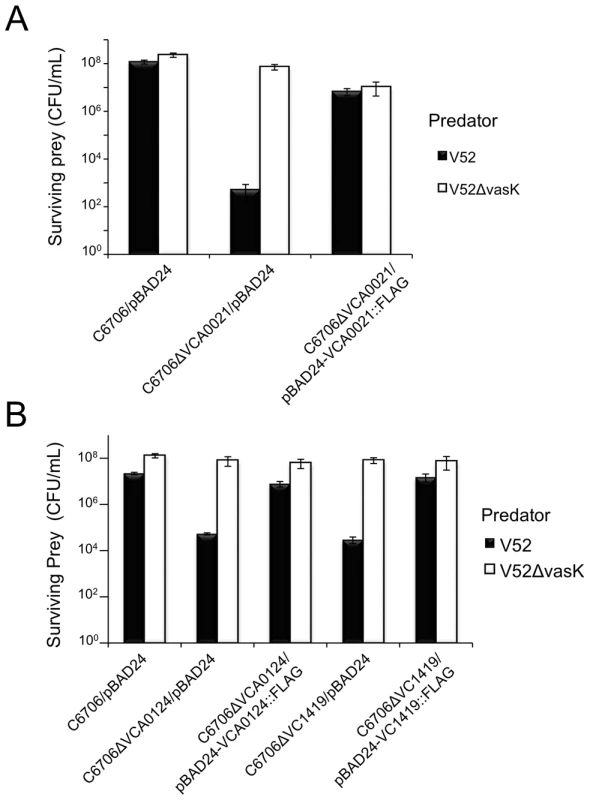
(A) Survival of rifampicin-resistant prey strains C6706 or C6706ΔVCA0021 harboring empty vector (pBAD24) or pBAD24-VCA0021::FLAG was determined by measuring CFU following exposure to the rifampicin-sensitive predator listed in the legend. V52ΔvasK is a T6SS-null strain that serves as a negative control for T6SS-mediated bacterial killing. Arabinose was included in all samples to drive expression from the PBAD promoter. These data are representative of three independent experiments, each performed in technical duplicate. Error bars indicate the standard deviation. (B) Survival of rifampicin-resistant prey strains C6706, C6706ΔVCA0124, or C6706ΔVC1419 harboring empty vector (pBAD24), pBAD24-VCA0124::FLAG, or pBAD24-VC1419::FLAG was determined by measuring CFU following exposure to a rifampicin-sensitive predator (listed in the legend) – wild-type V52 or the T6SS-null strain V52ΔvasK (negative control). Arabinose was included in all samples to drive expression from the PBAD promoter. These data are representative of two independent experiments, each performed in technical duplicate. Error bars indicate the standard deviation. The bacterial killing mechanism of VasX has not been characterized. Thus, we focused on the T6SS gene cluster encoding vasX and tsiV2 to understand the mode-of-action of VasX and the ability of C6706 to employ TsiV2 to achieve immunity to VasX-mediated toxicity in the T6SS-off state.
VasX is required for T6SS-mediated bacterial killing
Secondary structure predictions indicated that VasX has homology to colicins [64]. Because we previously observed that V52 kills E. coli MG1655 in a T6SS-dependent manner, we first tested whether VasX was required for this killing phenotype. As shown in Figure 3A, V52 with a disabled T6SS due to a vasK null-mutation (V52ΔvasK) was unable to kill E. coli; however, V52 with a native vasK but lacking vasX retained the ability to kill E. coli to the same extent as wild-type V52. Thus, we concluded that VasX is dispensable for V52's ability to kill E. coli, probably because the other two T6SS effectors, VgrG-3 and TseL, compensate for the absence of VasX. To test whether VasX is important for killing E. coli in the absence of VgrG-3 and TseL, we performed a killing assay with a V52 predator that does not produce TseL and VgrG3, and relies on VasX for T6SS-mediated virulence. We observed that this predator strain was able to reduce the number of viable E. coli by ∼10-fold compared to fully attenuated V52ΔvasK or V52 missing all three T6SS effector genes (Figure S3). Although predator strains using VasX or VgrG-3 as the only T6SS toxin exhibited a similar Hcp secretion defect, their killing ability differed significantly (Figure S3), suggesting that VasX contributes to killing of E. coli. In conclusion, these data suggest that VasX is sufficient to kill E. coli when it is the sole effector utilized by V52, but VgrG-3 and TseL can compensate for the lack of VasX toxicity.
Fig. 3. VasX is a bacterial toxin that is lethal when presented from the periplasm. 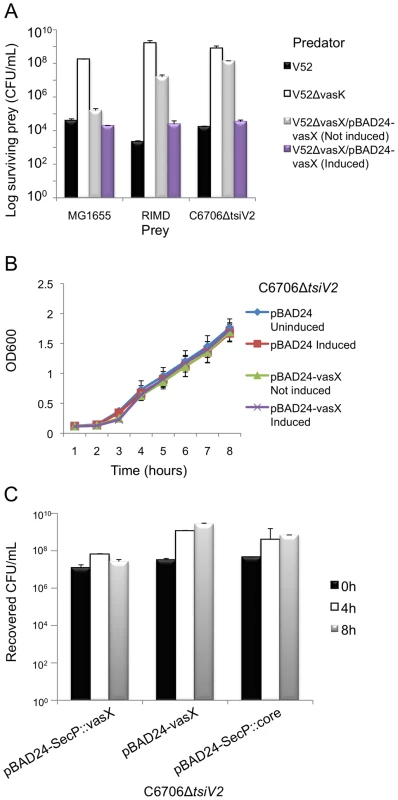
(A) VasX is required for killing V. parahaemolyticus RIMD and C6706ΔtsiV2, but not E. coli MG1655. Survival of rifampicin-resistant prey (listed on the x-axis) was determined by counting CFU following exposure to the indicated rifampicin-sensitive predator (legend). V52ΔvasK is a T6SS-null strain and was used as a negative control. Arabinose was included where indicated (“induced”) to drive expression from the PBAD promoter. These data are representative of three independent experiments, each performed in technical duplicate. Error bars indicate the standard deviation. (B) Episomal expression of vasX does not affect bacterial growth. C6706ΔtsiV2 harboring the plasmids indicated in the legend were grown in liquid culture and the OD600 was measured at the time points indicated on the x-axis. Arabinose was included where indicated (“induced”) to drive expression from the PBAD promoter. These data represent two independent experiments performed in technical triplicate. Error bars indicate the standard deviation. (C) Delivery of VasX to the periplasm is toxic to the producing cell. C6706ΔtsiV2 harboring the plasmids indicated on the x-axis were grown for 8 hours in liquid culture. The recovered CFU/mL were enumerated after harvesting samples at the time points indicated in the legend. Arabinose was included in all samples to drive expression from the PBAD promoter. These data represent three independent experiments performed in technical duplicate. Error bars indicate the standard deviation. We determined that V. cholerae C6706 lacking tsiV2, the gene encoding the immunity protein for VasX, became susceptible to killing by V52 (Figure 3A). This killing occurred in a VasX-dependent manner such that TsiV2 conferred immunity specifically to VasX (Figure 3A). We also tested whether VasX was involved in killing of other Vibrio species, namely Vibrio parahaemolyticus strain RIMD2210633 (hereafter referred to as V. parahaemolyticus RIMD). V. parahaemolyticus RIMD encodes a functional T6SS [65], [66]; however, the ability of this strain to engage in T6SS-dependent killing of bacterial neighbors has yet to be determined. As shown in Figure 3A, VasX is required for maximal killing of V. parahaemolyticus RIMD as a vasX mutant of V52, still capable of employing the T6SS effectors TseL and VgrG-3, is attenuated with regard to the killing of V. parahaemolyticus RIMD. As expected, episomal expression of vasX complemented the killing phenotype. Other Vibrio species including Vibrio alginolyticus and Vibrio fischeri were also susceptible to VasX-mediated killing (data not shown). Episomal expression of the immunity protein-encoding gene tsiV2 in V. parahaemolyticus RIMD provided intermediate protection against killing by V52 (Figure S4). In contrast, production of TsiV2 in MG1655 did not provide protection against VasX-mediated killing (data not shown).
Taken together, these data indicate that although VasX is not required for V52 to kill E. coli MG1655 due to the compensatory effects of VgrG-3 and TseL, VasX is important for killing other Vibrio species.
VasX targets the inner membrane of prey bacteria
The V. cholerae VasX protein is important for T6SS-dependent killing of other bacteria [11] (Figure 3A) and has homology to colicins [64]. Colicins can kill target cells by degrading DNA and RNA, by forming pores in the inner membrane, or by inhibiting murein synthesis. Previous bioinformatic analysis suggested that VasX has three transmembrane domains in its C-terminus [21]; therefore, we hypothesized that the toxic activity of VasX involves damage to the inner membrane of target cells. To be toxic, pore-forming colicins must be presented to the target cell from the periplasmic face [56], [57]. Fittingly, we observed that VasX expression was not toxic towards V. cholerae C6706 lacking tsiV2 (Figure 3B). To ensure that VasX was produced, we collected a lysate sample from each sample presented in Figure 3B and performed a western blot with antibodies against VasX and DnaK (loading control) (Figure S5). As we detected VasX under inducing conditions in live bacteria, we reasoned that VasX is not toxic to the producing cell, because to be toxic it has to be presented to the inner membrane of target cells from the periplasmic space similar to pore-forming colicins.
We then determined whether VasX is toxic when presented to bacteria from the periplasmic face. As shown in Figure 3C, providing VasX with a SecP secretion signal to target SecP::VasX to the periplasm reduced the growth rate of C6706 lacking tsiV2. The observed toxicity caused by SecP::VasX was not due to increased export of proteins to the periplasm as VgrG-3 (missing the peptidoglycan binding domain) [12] targeted to the periplasm (denoted as “SecP::core”) was not toxic to the producing cell. We concluded from this data that VasX targeted to the periplasm of the producing cell is toxic to that cell.
Knowing that periplasmic localization of VasX resulted in autotoxicity, we postulated that VasX inserts into the inner membrane of target cells similar to pore-forming colicins that share homology with VasX. To test whether production of SecP::VasX compromised membrane integrity of the producing cell, we performed an SDS lysis assay [67] and determined that production of SecP::VasX increased the sensitivity of a tsiV2 mutant of C6706 to SDS compared to the control strains (data not shown). This finding suggests that periplasmic VasX compromises membrane integrity.
If VasX resembles a pore-forming colicin, insertion into the inner membrane would result in increased cellular permeability, ion leakage, and dissipation of the bacterial membrane potential. To test whether production of SecP::VasX dissipates the membrane potential in the producing cell, we used the BacLight Bacterial Membrane Potential Kit (Molecular Probes, Invitrogen). In this experiment, the fluorescent dye DiOC2(3) was used to stain all cells green. If the cell is actively respiring, the dye accumulates within the cell and shifts towards red emission. Stained cells were analyzed by flow cytometry using Texas Red and FITC filters and the red/green ratio was indicative of the strength of the membrane potential in analyzed cells. CCCP, which uncouples the proton gradient, served as a positive control for dissipation of membrane potential. Production of SecP::VasX in a tsiV2 mutant of C6706 resulted in a lower red fluorescence compared to cells producing cytoplasmic VasX, SecP-tagged VgrG-3 core (SecP::core), or empty vector control (Figure S6). When CCCP was added, the red fluorescence intensity decreased to levels comparable to cells producing SecP::VasX, implying that SecP::VasX disrupts the membrane potential to an extent similar to CCCP. Calculation of the red/green ratios indicated that the membrane potential for cells producing SecP::VasX is significantly reduced compared to controls (Figure 4A). Furthermore, SecP::VasX uncoupled the proton gradient to the same extent as the positive control CCCP (Figure 4A). Thus, only when VasX was presented from the periplasmic face, did it compromise the integrity of the inner membrane and dissipate the membrane potential (Figure 4A).
Fig. 4. VasX compromises the integrity of the inner membrane in target cells. 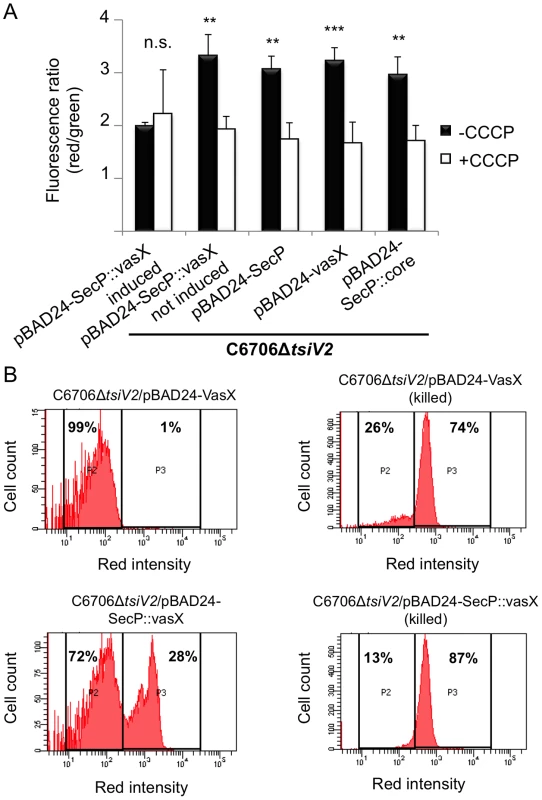
(A) VasX dissipates the target cell's membrane potential. C6706ΔtsiV2 harboring the plasmids indicated on the x-axis were analyzed using the BacLight Membrane Potential Kit and flow cytometry. The red/green fluorescence ratio was calculated for each condition. Carbonyl cyanide m-chlorophenyl hydrazone (CCCP) is a chemical that uncouples the proton gradient and was used as a positive control for dissipation of membrane potential in this experiment. Arabinose was included in all samples (except in the sample noted as “not induced”) to drive expression from the PBAD promoter. These data are representative of three independent experiments. Error bars indicate standard deviation. *** = p<0.001, ** = p<0.005 relative to SecP::vasX (induced, -CCCP). p-values were calculated using the Student's one-tailed, paired t-test. (B) Cells producing periplasmic VasX are permeable to propidium iodide (PI). The strain indicated at the top of each histogram (living or ethanol-killed) was incubated in the presence of PI and analyzed by flow cytometry. P2 represents cells not permeable to PI and P3 represents cells permeable to PI. The percentage of cells represented in P2 and P3 populations is indicated. These data represent three independent experiments. Next, we tested whether SecP::VasX made the tsiV2 null-mutant of C6706 permeable to propidium iodide (PI) – an intercalating agent bound to a fluorescent molecule that binds DNA. Normally, PI is used to assess bacterial cell viability because it is membrane impermeant and does not penetrate healthy cells. However, in the case of dead cells, or those with damaged membranes, PI enters the cell and binds DNA. We hypothesized that cells producing SecP::VasX would be permeable to PI because they have damaged membranes. The tsiV2-mutant of C6706 producing SecP::VasX or VasX was grown in liquid culture in the presence of arabinose and then incubated with PI. As a positive control for PI uptake, both strains were killed by incubation in ethanol following incubation with PI. Cells were analyzed by flow cytometry to assess the permeability of these cells at a population level. Both strains that were killed in ethanol exhibited increased permeability to PI (74% of the total population for cells producing VasX and 87% of the cell population for those producing SecP::VasX) (Figure 4B). ∼30% of cells producing SecP::VasX (without ethanol incubation) showed greater uptake of PI compared to those producing VasX (Figure 4B). This indicates that production of SecP::VasX increases cellular permeability to PI. We thus conclude that VasX is a bacterial colicin-like effector that disrupts the inner membrane of target cells.
VasW is an accessory protein of VasX
The V. cholerae vasX gene is contained within a satellite T6SS-cluster along with hcp-2, vgrG-2, tsiV2, and vasW (Figure 1A). Speculating that the gene products act as a cooperative unit, we set out to determine whether VasW (encoded immediately upstream of vasX) was important for bacterial killing. We deleted vasW from the V52 chromosome and tested this mutant's ability to kill V. parahaemolyticus RIMD. As shown in Figure 5A, deletion of vasW resulted in an attenuated phenotype towards V. parahaemolyticus RIMD, similar to the result observed when the V52 strain lacking vasX was used as the predator (Figure 5A). Episomal expression of vasX in the vasW null-mutant of V52 did not restore killing, demonstrating that the vasW mutation did not display a polar effect on vasX. Episomal expression of vasW restored killing of V. parahaemolyticus RIMD comparable to wild-type levels (Figure 5A). Based on these results, we hypothesized that VasW is important for VasX function.
Fig. 5. VasW plays an accessory role in VasX-mediated bacterial killing. 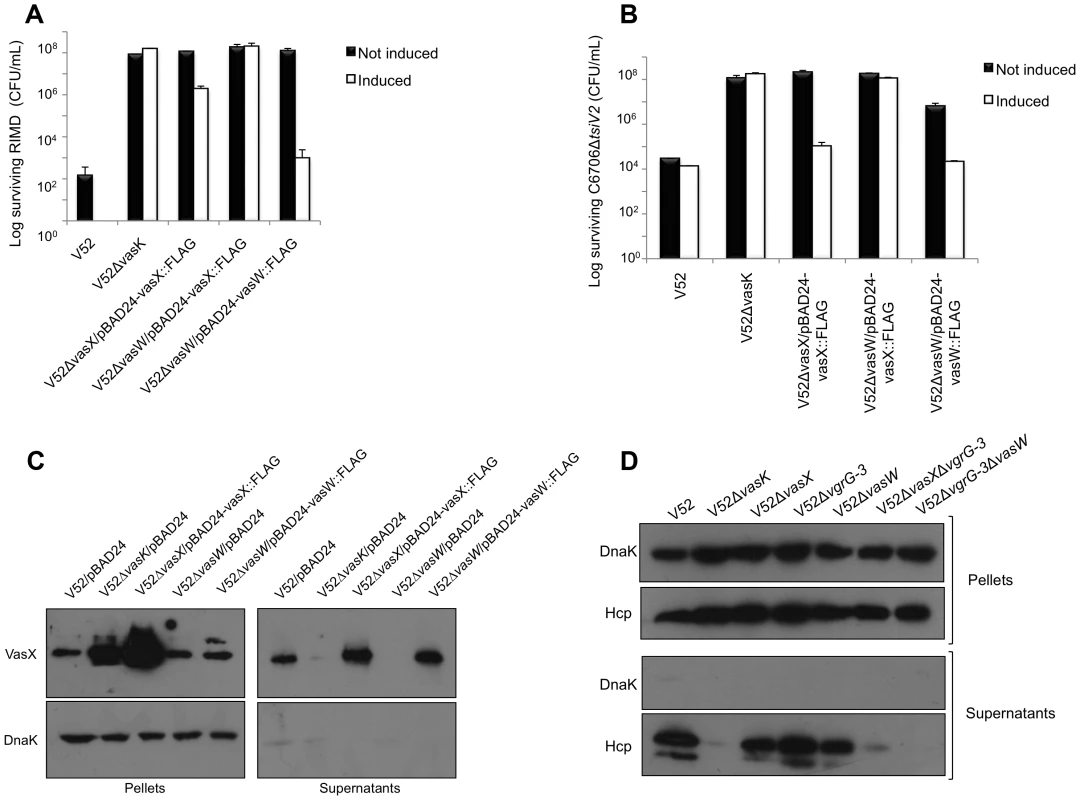
(A) VasW is required for V52 to kill Vibrio parahaemolyticus RIMD. Survival of rifampicin-resistant RIMD was determined by measuring CFU following exposure to the indicated rifampicin-sensitive predator listed on the x-axis. Arabinose was included where indicated (“induced”) to drive expression from the PBAD promoter. These data represent three independent experiments. Error bars indicate the standard deviation. (B) VasW is required for V52 to kill C6706ΔtsiV2. Survival of rifampicin-resistant C6706ΔtsiV2 was determined by measuring CFU following exposure to the indicated rifampicin-sensitive predator listed on the x-axis. Arabinose was included where indicated (“induced”) to drive expression from the PBAD promoter. These data represent three independent experiments. Error bars indicate the standard deviation. (C) V52ΔvgrG-3 with deletions in vasW or vasX does not secrete Hcp. Bacterial pellet and supernatant samples from the strains indicated at the top of the blot were subjected to western blotting with α-Hcp and α-DnaK (loading and lysis control) antibodies. (D) V52ΔvasW does not secrete VasX. Bacterial pellet and supernatant samples from the strains indicated at the top of the blot were subjected to western blotting with α-VasX and α-DnaK (loading and lysis control) antibodies. To determine whether the lack of bacterial killing by the vasW null-mutant of V52 resulted from impairment of VasX function, we used the vasW null-mutant V52ΔvasW as predator in a killing assay against a C6706 mutant that lacked the immunity protein-encoding gene tsiV2 and that had been shown to be sensitive to VasX-mediated killing by V52 [11]. As shown in Figure 5B, the killing ability of V52ΔvasW towards C6706ΔtsiV2 was attenuated and this phenotype could be complemented by expression of vasW from the plasmid pBAD24 (Figure 5B). Therefore, we conclude that V. cholerae VasW assists in VasX-mediated killing of other bacteria and this defect in killing could result from improper localization of VasX or the inability of VasX to be secreted.
We previously determined that VasX depends on other T6SS proteins such as VgrG-2 and Hcp for secretion into culture supernatants [21]. Given that deletion of vasW results in similar phenotypes compared to V52ΔvasX, we asked whether VasW was required for V52 to secrete VasX. As shown in Figure 5C, the vasW null-mutant of V52 was unable to secrete VasX into culture supernatants, and episomal expression of vasW was able to restore VasX secretion. Because VasX secretion requires Hcp translocation [21], we tested whether Hcp could be found in culture supernatants of the vasW mutant. As expected, wild-type V52 secreted Hcp into culture supernatants, while V52 lacking vasK did not (Figure 5D). Hcp secretion was not affected by deletion of vasW in V52 (Figure 5D). We and others previously observed that V52 lacking both vasX and vgrG-3 is devoid of Hcp secretion, whereas V52 lacking either vgrG-3 or vasX retains the ability to secrete Hcp (Figure 5D and [11]). Because VasW appears to be important for VasX function, we tested whether a V52 double mutant lacking vgrG-3 and vasW was unable to secrete Hcp, similar to what we observed for a V52 mutant lacking vasX and vgrG-3. We observed that the absence of vgrG-3 and vasW abolished Hcp secretion (Figure 5D), suggesting that VasX and VasW act in a coordinated fashion. Taken together, these data indicate that VasW plays a crucial role in VasX secretion and in its function as a bacterial effector.
VasX possesses an internal promoter for tsiV2
Given that C6706 does not have an active T6SS under laboratory conditions (i.e., does not produce VasX or Hcp), we hypothesized that expression of the immunity protein-encoding gene tsiV2 does not require the T6SS regulator VasH, an alternate sigma factor σ54 that works at the operon promoter (located in the 400 base pairs upstream of hcp-2) to drive expression of hcp-2, vgrG-2, vasX, and likely tsiV2 [18], [19]. This operon promoter is not active in C6706 as evidenced by a lack of Hcp production under standard laboratory conditions [34]. We speculated that a promoter exists within vasX capable of driving transcription of the downstream gene tsiV2, which would explain why wild-type C6706 cells are immune to killing by V52. To test this, we made transcriptional fusions of full-length vasX (4-3258, lacking the start codon) and different vasX truncations (nucleotides 4-1345, 1575-3258, and 2208-3258) to promoterless lacZ in the plasmid pAH6, as outlined in Figure 6A. The hcp-2 promoter region (Phcp, basepairs −1 to −400 upstream of hcp-2) was also fused to lacZ as a control. Plasmids were transformed into V52, V52 lacking vasH, and C6706. The level of lacZ expression within cells was then determined by performing β-galactosidase assays.
Fig. 6. Dual expression profile of the immunity protein-encoding gene tsiV2. 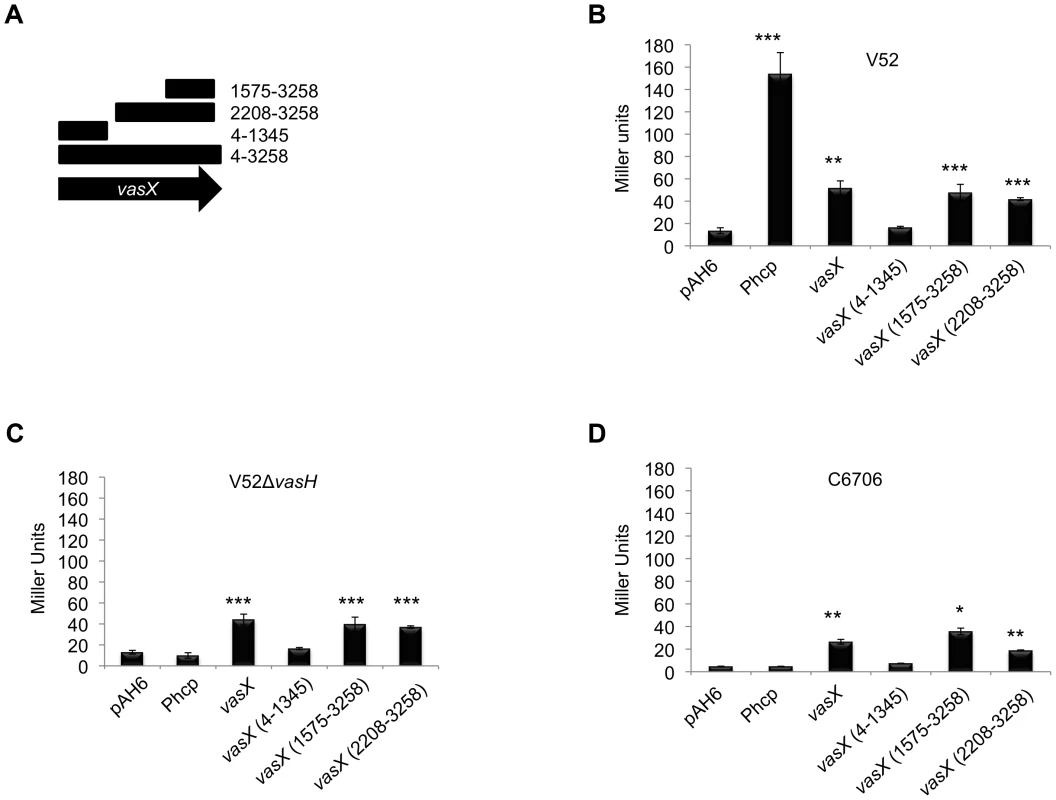
(A) Schematic representation of vasX fragments cloned upstream of lacZ in the plasmid pAH6. (B, C, D) A promoter exists within vasX. β-galactosidase assays were performed using the strains indicated at the top of the graph. Fragments of vasX, or the hcp-2 promoter present in pAH6 are indicated on the x-axis. Data are representative of two independent experiments performed in triplicate and the error bars indicate the standard deviation. *** = p<0.001, ** = p<0.005, * = p<0.01 relative to the empty vector control. p-values were calculated based on the Student's one-tailed, paired t-test. V52 permits constitutive expression of T6SS-encoding genes from the promoter upstream of hcp-2 as indicated by the significant amount of LacZ production driven by the operon promoter Phcp. Full length vasX, vasX (1575-3258), and vasX (2208-3258) drove expression of lacZ to levels significantly greater than the empty vector control by ∼5-fold, but less than Phcp (Figure 6B). Deletion of vasH in V52 prohibited transcription from the hcp-2 promoter, but not transcription from within full-length vasX (4-3258) and truncated vasX (1575-3258 and 2208-3258) (Figure 6C). When the reporter plasmids were transformed into the T6SS-silent strain C6706, we observed significant LacZ production in the presence of full-length vasX, vasX (1575-3258), and vasX (2208-3258), but not in the presence of Phcp or vasX (4-1345) (Figure 6D). These data suggest the existence of a promoter within the 3′-end of vasX that can drive expression of tsiV2 independently of VasH. Additional promoter dissection experiments demonstrated that the 3′-terminal 253-bp region of vasX (3006-3258) is sufficient to drive expression of a lacZ reporter (Figure S7).
To test the biological role of the promoter within vasX, we performed two independent killing assays. First, we challenged V52 lacking vasH, vasX or both with wild-type V52 and isogenic mutants lacking vasK or vasX. When challenged with a wild-type V52 predator, V52 prey and V52 prey lacking vasH were resistant to killing; however, V52 prey that lacked vasX were more susceptible to V52 predation than V52 prey that lacked vasH (Figure 7A). The V52 ΔvasXΔvasH double mutant prey exhibited greater sensitivity to killing by V52 than V52 lacking either vasX or vasH (Figure 7A). Furthermore, a V52 predator lacking vasX was able to kill E. coli MG1655 but failed to kill V52 prey regardless of their respective mutations, suggesting that null mutations of vasX and vasH impaired immunity to VasX.
Fig. 7. VasX contains a promoter to drive expression of tsiV2 in several V. cholerae strain backgrounds. 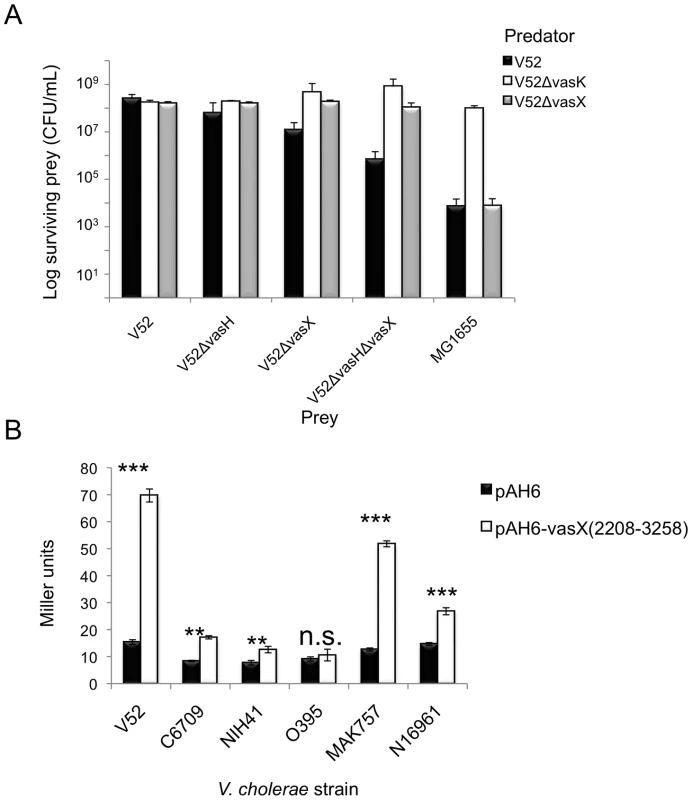
(A) V52 becomes susceptible to killing following deletion of vasH and vasX. Rifampicin-sensitive V52 derivatives (predator) were mixed with rifampicin-resistant V52, V52ΔvasH, V52ΔvasX, V52ΔvasHΔvasX, and E. coli MG1655 (prey). Surviving prey were enumerated by selection on LB agar containing rifampicin and the results were plotted. Data are representative of three independent experiments. Error bars indicate the standard deviation. (B) The internal tsiV2 promoter is recognized in several strains. V. cholerae strains indicated on the x-axis were transformed with pAH6-vasX(2208-3258) or plasmid control. Transformed strains were subjected to β-galactosidase assays and the Miller units were calculated and plotted. Data represent two independent experiments performed in triplicate; error bars indicate the standard deviation. *** = p<0.001, ** = p<0.005, * = p<0.01 relative to the empty vector control. n.s; not significant. p-values were calculated based on the Student's one-tailed, paired t-test. In the second series of killing assays, we constructed a C6706 derivative containing an in-frame deletion mutation of vasX, and used this strain as prey in a killing assay with a V52 predator. We reasoned that in this strain, deletion of the putative tsiV2 promoter would render C6706 lacking vasX susceptible to killing by V52. C6706ΔvasX (with empty vector pBAD24) were killed by V52 (Figure S8). Importantly, trans-expression of vasX was unable to restore immunity to killing by V52 because expression from pBAD24 does not restore promoter function to drive expression of tsiV2 on the C6706 chromosome (Figure S8). Expression of tsiV2 from pBAD24 protected C6706ΔvasX from being killed by V52 (Figure S8). Taken together, these data indicate that the promoter within vasX can drive transcription of tsiV2 in a VasH-independent manner.
To determine whether the promoter in vasX was functional specifically in V52 or in a variety of strains, we also transformed a pAH6 empty vector or pAH6-vasX (2208-3258) into V. cholerae O1 serogroup strains O395w, C6709w, NIH41w, MAK 757w, and N16961w. As shown in Figure 7B, V52, C6709w, NIH41w, MAK 757w, and N16961w had significantly higher LacZ activities compared to their empty vector controls. In contrast, O395w did not have higher levels of LacZ with pAH6-vasX (2208-3258) compared to the empty vector control (Figure 7B). These data indicate that the promoter found within vasX can elicit transcription of downstream genes in a variety of V. cholerae strains and that this phenomenon is not specific to V52.
TseL and vgrG-3 possess internal promoters for tsiV1 and tsiV3, respectively
Based on the data presented in Figures 6 and 7, we tested whether tseL and vgrG-3 also contain internal promoters to drive VasH-independent transcription of tsiV1 and tsiV3, respectively. We made transcriptional fusions of tseL and vgrG-3 to promoterless lacZ in pAH6 and transformed the plasmids into V52, V52 lacking vasH, and C6706. Similar to what we observed with vasX, both tseL and vgrG-3 resulted in statistically significant production of β-galactosidase compared to the empty vector control. Production of β-galactosidase was independent of VasH (Figure S9), and the amount of β-galactosidase activity measured was higher for vgrG-3 than for either vasX or tseL (Figures 6 and S9).
Last, we performed killing assays using C6706 lacking tseL or vgrG-3 as prey to determine if deletion of the effector gene, and thus the putative promoter for the downstream immunity protein-encoding genes, rendered these strains susceptible to killing by V52. As shown in Figure S2, C6706 lacking tseL or vgrG-3 (with empty vector pBAD24) were killed by V52. Episomal expression of tseL and vgrG-3 in their respective mutants did not restore immunity (Figure S2). However, immunity to V52 could be restored by complementing tseL and vgrG-3 null-mutations in C6706 with tsiV1 and tsiV3, respectively (Figure S2). Taken together, these data indicate that internal promoters located within tseL and vgrG-3 can drive transcription of tsiV1 and tsiV3 in a VasH-independent manner.
Discussion
TA systems are commonly employed by bacteria to ward off bacterial competitors [23], [33], [35], [37], [68]–[71]. In some bacteria, the presence of an antitoxin or immunity protein prevents self-intoxication and killing by toxins produced by sister cells. Similarly, it appears that T6SS immunity proteins protect against an oncoming attack by neighboring T6SS-active bacteria. By screening a C6706 T6SS transposon library for susceptibility to killing by V. cholerae V52 (Figure S1), we identified three T6SS immunity protein-encoding genes, namely tsiV1, tsiV2, and tsiV3 (Figure 1A). These data agree with recent findings of Dong et al. who also identified tsiV1, tsiV2, and tsiV3 as V. cholerae T6SS immunity protein-encoding genes using a Tn-seq approach [11].
Episomal expression of tsiV2 in the Vibrio species V. parahaemolyticus RIMD provided significant protection against killing by V52 (Figure S4). In contrast, episomal expression of tsiV2 in E. coli MG1655 did not provide protection against T6SS-mediated killing – even when VasX was the only bacterial effector employed (data not shown). Importantly, V. parahaemolyticus RIMD has a functional T6SS but does not possess homologs to VasX or TsiV2 according to BLASTn and BLASTp analyses (data not shown). MG1655 also does not possess genes similar to vasX and tsiV2 (data not shown). Therefore, we postulate that even though V. parahaemolyticus RIMD contains T6SS gene clusters, putative immunity proteins are not cross-protective against V52 effectors. Furthermore, we suggest that TsiV2 provides (intermediate) protection in V. parahaemolyticus RIMD, but not in E. coli MG1655, because TsiV2 requires other T6SS proteins or cofactors for proper function and/or localization within the prey cell that are produced by V. parahaemolyticus but not E. coli.
In all TA systems, including those in which the toxin is a colicin, the presence of an immunity protein prevents toxicity. Commonly, the gene encoding the immunity protein is located directly adjacent to the effector gene [56], [72], [73]. Using β-galactosidase assays, we demonstrated that a promoter exists within the 1,051 nucleotides of the 3′end of vasX to drive expression of lacZ in V52, V52ΔvasH, C6706, C6709w, NIH41w, MAK 757w, and N16961w (Figures 6 and 7B). The promoter was determined to be within the 3′-terminal 253-bp region of vasX in a subsequent experiment with strain C6706 (Figure S7). Expression driven by promoters within vasX, vgrG-3, and tseL in the strains tested implies that C6706 produces immunity proteins under laboratory conditions to mediate protection against a T6SS-onslaught by bacterial neighbors (Figures 6, 7, S2, S8, and S9). We are currently conducting experiments to identify the exact sequence that encodes these promoters.
Constitutive expression of T6SS immunity protein-encoding genes could be advantageous for several reasons. The most obvious reason for maintaining basal expression of genes encoding immunity proteins is for protection against an attack by sister cells. Bacteria with activated T6SS gene clusters could readily kill sister cells that live within close quarters (i.e., biofilms) and lack expression of immunity protein-encoding genes. Constitutive expression of immunity protein-encoding genes, independent of other genes encoding T6SS components, would also allow the bacterium to conserve energy as the T6SS secretion apparatus is a large, dynamic structure [7], [8], [33], the production of which likely requires vast energy expenditure. Previously it was shown that infections of infant mice with V. cholerae V52 unable to shut down its T6SS caused inflammation and actin cross-linking in the mouse gut [74]. By employing strict regulatory mechanisms for T6SS proteins while maintaining continuous expression of immunity protein-encoding genes (such is the case with C6706), V. cholerae could prevent identification by the host immune system and avoid a loss of viability at the time of T6SS activation [75].
Outside the human host, the T6SS-off state can be advantageous in mixed bacterial populations. Immunity protein-encoding genes only protect against T6SS effectors of the same species, sensitizing V. cholerae to T6SS effectors of other organisms including P. aeruginosa [33]. In a mixed population of V. cholerae and P. aeruginosa, P. aeruginosa employed its T6SS to kill V. cholerae only if V. cholerae used its T6SS to attack first. V. cholerae with an inactive T6SS did not induce a counter-attack from neighboring P. aeruginosa and the two bacterial species maintained a peaceful coexistence [33]. We speculate that maintaining constitutive expression of genes encoding immunity proteins independently from other T6SS-encoding genes provides the bacterium with a fitness advantage in the event it engages in T6SS dueling with another T6SS-active kin bacterium.
The three immunity protein-encoding genes tsiV1, tsiV2, and tsiV3 are located directly downstream of their corresponding effector genes tseL, vasX, and vgrG-3, respectively. We previously demonstrated that VgrG-3 degrades the peptidoglycan of target cells [12], a phenotype that has also been described for T6SS bacterial effectors produced by P. aeruginosa [35]. TseL is a predicted class III lipase [11], however the role of TseL within the context of the V. cholerae T6SS remains to be determined. Here we showed that VasX is an effector that targets prokaryotes lacking the VasX immunity protein-encoding gene tsiV2 and that the accessory protein VasW is required for secretion of VasX.
We previously demonstrated that V. cholerae V52 uses its T6SS to kill E. coli [16], and data presented here indicate that all three effectors – TseL, VgrG-3, and VasX are active against E. coli; however, the presence of TseL and VgrG-3 can compensate for the lack of VasX (Figures 3A and S3). A similar conclusion was reached by Dong et al. such that VasX is sufficient but not required for V52 to kill E. coli [11]. Interestingly, when other Vibrio species such as V. parahaemolyticus RIMD or a mutant of V. cholerae strain C6706 lacking tsiV2 were used as prey, VasX had a much stronger phenotype (Figure 3A). In this case, VasX was a more effective effector against V. parahaemolyticus RIMD and C6706, but less effective for killing E. coli. This could be due to factors such as a requirement for specific receptors on target cells or varying degrees of susceptibility to T6SS effectors.
We determined that VasW, a protein whose gene lies directly upstream of vasX, is required for secretion of VasX and that V52 lacking vasW has a phenotype similar to V52 lacking vasX. We and others [11] demonstrated that a double mutant lacking vasX and vgrG-3 fails to secrete the T6SS substrate Hcp into culture supernatants. Dong et al. suggested that VgrG-3 and VasX interact to form part of the T6SS secretion apparatus [11]. Because V52 lacking both vgrG-3 and vasW also does not secrete Hcp (Figure 5D), we propose that VasW acts as an accessory protein responsible for mediating the interaction between VasX and structural proteins of the secretion apparatus. In light of the recently proposed mechanism for T6SS effector translocation [14], VasW could bind to the PAAR protein located at tip of the injectosome to prepare VasX for T6SS-mediated translocation. VasW-mediated recruitment of VasX to the T6SS injectosome could then function as a checkpoint for ejection of the inner Hcp tube decorated with a VgrG-3-containing tip.
Previously, VasX was reported to have homology to colicins [64], secreted proteins produced by and toxic to certain strains of E. coli [37]. The data presented here suggest that similar to pore-forming colicins, VasX perturbs the inner membrane of target cells. Although we cannot conclude that VasX acts via pore-formation, our data indicate that VasX dissipates the target cell's membrane potential and increases cellular permeability – both characteristics of pore-forming toxins [70], [76]–[78].
The observation that periplasmic localization of VasX is toxic to the producer cell also indicates similarities between VasX and pore-forming colicins. Cytoplasmic production of pore-forming colicins does not result in toxicity, because pore formation can occur only when the colicin is presented to the cell from the periplasmic face [56], [57], [67]. We also observed that production of VasX in C6706 that lacks the immunity protein-encoding gene tsiV2 was not toxic to the producing cell (Figure 3B), but providing VasX with the Sec signal peptide resulted in autotoxicity (Figure 3C). Along these lines, we previously demonstrated that VasX is a secreted protein that is present in the cytoplasm and membrane fractions of V52; however, VasX is undetectable in the periplasmic fraction by western blot analysis [21]. It appears that VasX either bypasses the periplasm, or is present transiently in small quantities en route out of the cell, as its presence in the periplasm could be toxic.
Interestingly, cells producing SecP::VasX initially increased in cell number, but then began to die. After 8 hours of induction the bacterial concentration was similar to the starting bacterial concentration (Figure 3C). It is unclear why we did not observe a more significant reduction in the number of surviving bacteria; however, a similar growth/toxicity phenotype referred to as “quasilysis” has been described for colicin lysis proteins that perforate colicin-producing cells and release colicin molecules into the extracellular milieu [56], [79]–[82]. We propose a model whereby VasX is injected into the periplasm of target cells along with T6SS structural proteins and other putative effectors. Upon arrival in the periplasm, VasX inserts into the inner membrane permeabilizing the target cell – an activity similar to quasilysis
We previously determined that VasX has a role in T6SS-mediated virulence towards the amoeba D. discoideum and that VasX binds eukaryotic lipids [19]. The fact that VasX has the ability to target both prokaryotic and eukaryotic cells suggests that VasX targets a cellular structure common to both cell types such as the cytoplasmic membrane. A similar model has been proposed by the Mougous group regarding T6SS lipase effectors that target cytoplasmic membranes of both prokaryotes and eukaryotes [79]. In the case of D. discoideum, we propose that perturbation of the plasma membrane is responsible for the VasX-mediated phenotype observed in this eukaryotic host. Targeting the membrane rather than a single protein/receptor is an evolutionarily advantageous mechanism employed by bacterial effectors because developing resistance to this toxic mechanism would prove challenging for target cells.
Supporting Information
Zdroje
1. WilliamsSG, VarcoeLT, AttridgeSR, ManningPA (1996) Vibrio cholerae Hcp, a secreted protein coregulated with HlyA. Infect Immun 64 : 283–289.
2. BallisterER, LaiAH, ZuckermannRN, ChengY, MougousJD (2008) In vitro self-assembly of tailorable nanotubes from a simple protein building block. Proc Natl Acad Sci U S A 105 : 3733–3738.
3. SilvermanJM, AgnelloDM, ZhengH, AndrewsBT, LiM, et al. (2013) Haemolysin coregulated protein is an exported receptor and chaperone of type VI secretion substrates. Mol Cell 51 : 584–593.
4. PukatzkiS, MaAT, RevelAT, SturtevantD, MekalanosJJ (2007) Type VI secretion system translocates a phage tail spike-like protein into target cells where it cross-links actin. Proc Natl Acad Sci U S A 104 : 15508–15513.
5. PukatzkiS, MaAT, SturtevantD, KrastinsB, SarracinoD, et al. (2006) Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc Natl Acad Sci U S A 103 : 1528–1533.
6. LeimanPG, BaslerM, RamagopalUA, BonannoJB, SauderJM, et al. (2009) Type VI secretion apparatus and phage tail-associated protein complexes share a common evolutionary origin. Proc Natl Acad Sci U S A 106 : 4154–4159.
7. BaslerM, MekalanosJJ (2012) Type 6 secretion dynamics within and between bacterial cells. Science 337 : 815.
8. BaslerM, PilhoferM, HendersonGP, JensenGJ, MekalanosJJ (2012) Type VI secretion requires a dynamic contractile phage tail-like structure. Nature 483 : 182–186.
9. BonemannG, PietrosiukA, DiemandA, ZentgrafH, MogkA (2009) Remodelling of VipA/VipB tubules by ClpV-mediated threading is crucial for type VI protein secretion. EMBO J 28 : 315–325.
10. MaAT, McAuleyS, PukatzkiS, MekalanosJJ (2009) Translocation of a Vibrio cholerae type VI secretion effector requires bacterial endocytosis by host cells. Cell Host Microbe 5 : 234–243.
11. DongTG, HoBT, Yoder-HimesDR, MekalanosJJ (2013) Identification of T6SS-dependent effector and immunity proteins by Tn-seq in Vibrio cholerae. Proc Natl Acad Sci U S A 110 : 2623–2628.
12. BrooksTM, UnterwegerD, BachmannV, KostiukB, PukatzkiS (2013) Lytic activity of the Vibrio cholerae type VI secretion toxin VgrG-3 is inhibited by the antitoxin TsaB. J Biol Chem 288 : 7618–7625.
13. MaLS, LinJS, LaiEM (2009) An IcmF family protein, ImpLM, is an integral inner membrane protein interacting with ImpKL, and its walker a motif is required for type VI secretion system-mediated Hcp secretion in Agrobacterium tumefaciens. J Bacteriol 191 : 4316–4329.
14. ShneiderMM, ButhSA, HoBT, BaslerM, MekalanosJJ, et al. (2013) PAAR-repeat proteins sharpen and diversify the type VI secretion system spike. Nature 500 : 350–353.
15. ZinnakaY, CarpenterCCJr (1972) An enterotoxin produced by noncholera Vibrios. Johns Hopkins Med J 131 : 403–411.
16. MacIntyreDL, MiyataST, KitaokaM, PukatzkiS (2010) The Vibrio cholerae type VI secretion system displays antimicrobial properties. Proc Natl Acad Sci U S A 107 : 19520–19524.
17. ZhengJ, ShinOS, CameronDE, MekalanosJJ (2010) Quorum sensing and a global regulator TsrA control expression of type VI secretion and virulence in Vibrio cholerae. Proc Natl Acad Sci U S A 107 : 21128–21133.
18. KitaokaM, MiyataST, BrooksTM, UnterwegerD, PukatzkiS (2011) VasH is a transcriptional regulator of the type VI secretion system functional in endemic and pandemic Vibrio cholerae. J Bacteriol 193 : 6471–6482.
19. BernardCS, BrunetYR, GavioliM, LloubesR, CascalesE (2011) Regulation of type VI secretion gene clusters by sigma54 and cognate enhancer binding proteins. J Bacteriol 193 : 2158–2167.
20. DongTG, MekalanosJJ (2012) Characterization of the RpoN regulon reveals differential regulation of T6SS and new flagellar operons in Vibrio cholerae O37 strain V52. Nucleic Acids Res
21. MiyataST, KitaokaM, BrooksTM, McAuleySB, PukatzkiS (2011) Vibrio cholerae requires the type VI secretion system virulence factor VasX to kill Dictyostelium discoideum. Infect Immun 79 : 2941–2949.
22. UnterwegerD, KitaokaM, MiyataST, BachmannV, BrooksTM, et al. (2012) Constitutive type VI secretion system expression gives Vibrio cholerae intra - and interspecific competitive advantages. PLoS One 7: e48320.
23. HoodRD, SinghP, HsuF, GuvenerT, CarlMA, et al. (2010) A type VI secretion system of Pseudomonas aeruginosa targets a toxin to bacteria. Cell Host Microbe 7 : 25–37.
24. MougousJD, CuffME, RaunserS, ShenA, ZhouM, et al. (2006) A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science 312 : 1526–1530.
25. SchwarzS, WestTE, BoyerF, ChiangWC, CarlMA, et al. (2010) Burkholderia type VI secretion systems have distinct roles in eukaryotic and bacterial cell interactions. PLoS Pathog 6(8): e1001068.
26. MurdochSL, TrunkK, EnglishG, FritschMJ, PourkarimiE, et al. (2011) The opportunistic pathogen Serratia marcescens utilizes type VI secretion to target bacterial competitors. J Bacteriol 193 : 6057–6069.
27. AubertDF, FlannaganRS, ValvanoMA (2008) A novel sensor kinase-response regulator hybrid controls biofilm formation and type VI secretion system activity in Burkholderia cenocepacia. Infect Immun 76 : 1979–1991.
28. BurtnickMN, BrettPJ, HardingSV, NgugiSA, RibotWJ, et al. (2011) The cluster 1 type VI secretion system is a major virulence determinant in Burkholderia pseudomallei. Infect Immun 79 : 1512–1525.
29. BurtnickMN, DeShazerD, NairV, GherardiniFC, BrettPJ (2010) Burkholderia mallei cluster 1 type VI secretion mutants exhibit growth and actin polymerization defects in RAW 264.7 murine macrophages. Infect Immun 78 : 88–99.
30. SchellMA, UlrichRL, RibotWJ, BrueggemannEE, HinesHB, et al. (2007) Type VI secretion is a major virulence determinant in Burkholderia mallei. Mol Microbiol 64 : 1466–1485.
31. WangX, WangQ, XiaoJ, LiuQ, WuH, et al. (2009) Edwardsiella tarda T6SS component evpP is regulated by esrB and iron, and plays essential roles in the invasion of fish. Fish Shellfish Immunol 27 : 469–477.
32. SuarezG, SierraJC, ShaJ, WangS, ErovaTE, et al. (2008) Molecular characterization of a functional type VI secretion system from a clinical isolate of Aeromonas hydrophila. Microb Pathog 44 : 344–361.
33. BaslerM, HoBT, MekalanosJJ (2013) Tit-for-tat: type VI secretion system counterattack during bacterial cell-cell interactions. Cell 152 : 884–894.
34. IshikawaT, SabharwalD, BromsJ, MiltonDL, SjostedtA, et al. (2012) Pathoadaptive conditional regulation of the type VI secretion system in Vibrio cholerae O1 strains. Infect Immun 80 : 575–584.
35. RussellAB, HoodRD, BuiNK, LeRouxM, VollmerW, et al. (2011) Type VI secretion delivers bacteriolytic effectors to target cells. Nature 475 : 343–347.
36. RussellAB, SinghP, BrittnacherM, BuiNK, HoodRD, et al. (2012) A widespread bacterial type VI secretion effector superfamily identified using a heuristic approach. Cell Host Microbe 11 : 538–549.
37. GratiaA (1925) Sur un remarquable exemple d'antagonism entre deux souches de colibacille. Comptes Rendus des Seances et Memoires de la Societe de Biologie 93 : 1040–1041.
38. ChakKF, KuoWS, LuFM, JamesR (1991) Cloning and characterization of the ColE7 plasmid. J Gen Microbiol 137 : 91–100.
39. CooperPC, JamesR (1984) Two new E colicins, E8 and E9, produced by a strain of Escherichia coli. J Gen Microbiol 130 : 209–215.
40. SchallerK, NomuraM (1976) Colicin E2 is DNA endonuclease. Proc Natl Acad Sci U S A 73 : 3989–3993.
41. SeniorBW, HollandIB (1971) Effect of colicin E3 upon the 30S ribosomal subunit of Escherichia coli. Proc Natl Acad Sci U S A 68 : 959–963.
42. BowmanCM, DahlbergJE, IkemuraT, KoniskyJ, NomuraM (1971) Specific inactivation of 16S ribosomal RNA induced by colicin E3 in vivo. Proc Natl Acad Sci U S A 68 : 964–968.
43. BowmanCM, SidikaroJ, NomuraM (1971) Specific inactivation of ribosomes by colicin E3 in vitro and mechanism of immunity in colicinogenic cells. Nat New Biol 234 : 133–137.
44. BoonT (1972) Inactivation of ribosomes in vitro by colicin E 3 and its mechanism of action. Proc Natl Acad Sci U S A 69 : 549–552.
45. SchallerK, HoltjeJV, BraunV (1982) Colicin M is an inhibitor of murein biosynthesis. J Bacteriol 152 : 994–1000.
46. ElkinsP, BunkerA, CramerWA, StauffacherCV (1997) A mechanism for toxin insertion into membranes is suggested by the crystal structure of the channel-forming domain of colicin E1. Structure 5 : 443–458.
47. VetterIR, ParkerMW, TuckerAD, LakeyJH, PattusF, et al. (1998) Crystal structure of a colicin N fragment suggests a model for toxicity. Structure 6 : 863–874.
48. ReevesP (1968) Mode of action of colicins of types E1, E2, E3, and K. J Bacteriol 96 : 1700–1703.
49. LuriaSE (1964) On the Mechanisms of Action of Colicins. Ann Inst Pasteur (Paris) 107: SUPPL: 67–73.
50. ZakharovSD, CramerWA (2002) Insertion intermediates of pore-forming colicins in membrane two-dimensional space. Biochimie 84 : 465–475.
51. ZakharovSD, KotovaEA, AntonenkoYN, CramerWA (2004) On the role of lipid in colicin pore formation. Biochim Biophys Acta 1666 : 239–249.
52. ScheinSJ, KaganBL, FinkelsteinA (1978) Colicin K acts by forming voltage-dependent channels in phospholipid bilayer membranes. Nature 276 : 159–163.
53. FieldsKL, LuriaSE (1969) Effects of colicins E1 and K on cellular metabolism. J Bacteriol 97 : 64–77.
54. FieldsKL, LuriaSE (1969) Effects of colicins E1 and K on transport systems. J Bacteriol 97 : 57–63.
55. PhillipsSK, CramerWA (1973) Properties of the fluorescence probe response associated with the transmission mechanism of colicin E1. Biochemistry 12 : 1170–1176.
56. CascalesE, BuchananSK, DucheD, KleanthousC, LloubesR, et al. (2007) Colicin biology. Microbiol Mol Biol Rev 71 : 158–229.
57. EspessetD, DucheD, BatyD, GeliV (1996) The channel domain of colicin A is inhibited by its immunity protein through direct interaction in the Escherichia coli inner membrane. EMBO J 15 : 2356–2364.
58. CameronDE, UrbachJM, MekalanosJJ (2008) A defined transposon mutant library and its use in identifying motility genes in Vibrio cholerae. Proc Natl Acad Sci U S A 105 : 8736–8741.
59. MetcalfWW, JiangW, DanielsLL, KimSK, HaldimannA, et al. (1996) Conditionally replicative and conjugative plasmids carrying lacZ alpha for cloning, mutagenesis, and allele replacement in bacteria. Plasmid 35 : 1–13.
60. GuzmanLM, BelinD, CarsonMJ, BeckwithJ (1995) Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol 177 : 4121–4130.
61. WhiteAP, Allen-VercoeE, JonesBW, DeVinneyR, KayWW, et al. (2007) An efficient system for markerless gene replacement applicable in a wide variety of enterobacterial species. Can J Microbiol 53 : 56–62.
62. HsiaoA, LiuZ, JoelssonA, ZhuJ (2006) Vibrio cholerae virulence regulator-coordinated evasion of host immunity. Proc Natl Acad Sci U S A 103 : 14542–14547.
63. MiyataST, KitaokaM, WieteskaL, FrechC, ChenN, et al. (2010) The Vibrio cholerae type VI secretion system: evaluating its role in the human disease cholera. Front Microbiol 1 : 117.
64. ZhengJ, HoB, MekalanosJJ (2011) Genetic analysis of anti-amoebae and anti-bacterial activities of the type VI secretion system in Vibrio cholerae. PLoS One 6: e23876.
65. Gode-PotratzCJ, McCarterLL (2011) Quorum sensing and silencing in Vibrio parahaemolyticus. J Bacteriol 193 : 4224–4237.
66. MaL, ZhangY, YanX, GuoL, WangL, et al. (2012) Expression of the type VI secretion system 1 component Hcp1 is indirectly repressed by OpaR in Vibrio parahaemolyticus. ScientificWorldJournal 2012 : 982140.
67. EspessetD, CordaY, CunninghamK, BenedettiH, LloubesR, et al. (1994) The colicin A pore-forming domain fused to mitochondrial intermembrane space sorting signals can be functionally inserted into the Escherichia coli plasma membrane by a mechanism that bypasses the Tol proteins. Mol Microbiol 13 : 1121–1131.
68. AokiSK, PammaR, HerndayAD, BickhamJE, BraatenBA, et al. (2005) Contact-dependent inhibition of growth in Escherichia coli. Science 309 : 1245–1248.
69. RakinA, BoolgakowaE, HeesemannJ (1996) Structural and functional organization of the Yersinia pestis bacteriocin pesticin gene cluster. Microbiology 142(Pt 12): 3415–3424.
70. AokiSK, WebbJS, BraatenBA, LowDA (2009) Contact-dependent growth inhibition causes reversible metabolic downregulation in Escherichia coli. J Bacteriol 191 : 1777–1786.
71. HanKD, AhnDH, LeeSA, MinYH, KwonAR, et al. (2013) Identification of chromosomal HP0892-HP0893 toxin-antitoxin proteins in Helicobacter pylori and structural elucidation of their protein-protein interaction. J Biol Chem 288(8): 6004–13.
72. DouganG, SaulM, WarrenG, SherrattD (1978) A Functional Map of Plasmid ColE1. Molecular and General Genetics 158 : 325–327.
73. AndreoliPM, OverbeekeN, VeltkampE, van EmbdenJD, NijkampHJ (1978) Genetic map of the bacteriocinogenic plasmid CLO DF13 derived by insertion of the transposon Tn901. Mol Gen Genet 160 : 1–11.
74. MaAT, MekalanosJJ (2010) In vivo actin cross-linking induced by Vibrio cholerae type VI secretion system is associated with intestinal inflammation. Proc Natl Acad Sci U S A 107 : 4365–4370.
75. MiyataST, BachmannV, PukatzkiS (2013) Type VI secretion system regulation as a consequence of evolutionary pressure. Journal of Medical Microbiology 62(Pt 5): 663–76.
76. YangCC, KoniskyJ (1984) Colicin V-treated Escherichia coli does not generate membrane potential. J Bacteriol 158 : 757–759.
77. DankertJR, UrataniY, GrabauC, CramerWA, HermodsonM (1982) On a domain structure of colicin E1. A COOH-terminal peptide fragment active in membrane depolarization. J Biol Chem 257 : 3857–3863.
78. UrataniY, HoshinoT (1984) Pyocin R1 inhibits active transport in Pseudomonas aeruginosa and depolarizes membrane potential. J Bacteriol 157 : 632–636.
79. JacobF, SiminovitchL, WollmanE (1952) Sur la biosynthese d'une colicine et sur son mode d'action. Annales de l'Institut Pasteur (Paris) 83 : 295–315.
80. JakesKS, ModelP (1979) Mechanism of export of colicin E1 and colicin E3. J Bacteriol 138 : 770–778.
81. CavardD, HowardSP, LloubesR, LazdunskiC (1989) High-level expression of the colicin A lysis protein. Mol Gen Genet 217 : 511–519.
82. LuirinkJ, de GraafFK, OudegaB (1987) Uncoupling of synthesis and release of cloacin DF13 and its immunity protein by Escherichia coli. Mol Gen Genet 206 : 126–132.
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek Parental Transfer of the Antimicrobial Protein LBP/BPI Protects Eggs against Oomycete InfectionsČlánek Immune Therapeutic Strategies in Chronic Hepatitis B Virus Infection: Virus or Inflammation Control?Článek Coronaviruses as DNA Wannabes: A New Model for the Regulation of RNA Virus Replication FidelityČlánek CRISPR-Cas Immunity against Phages: Its Effects on the Evolution and Survival of Bacterial PathogensČlánek The Cyst Wall Protein CST1 Is Critical for Cyst Wall Integrity and Promotes Bradyzoite PersistenceČlánek The Malarial Serine Protease SUB1 Plays an Essential Role in Parasite Liver Stage Development
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2013 Číslo 12- Jak souvisí postcovidový syndrom s poškozením mozku?
- Měli bychom postcovidový syndrom léčit antidepresivy?
- Farmakovigilanční studie perorálních antivirotik indikovaných v léčbě COVID-19
- 10 bodů k očkování proti COVID-19: stanovisko České společnosti alergologie a klinické imunologie ČLS JEP
-
Všechny články tohoto čísla
- Host Susceptibility Factors to Bacterial Infections in Type 2 Diabetes
- LysM Effectors: Secreted Proteins Supporting Fungal Life
- Influence of Mast Cells on Dengue Protective Immunity and Immune Pathology
- Innate Lymphoid Cells: New Players in IL-17-Mediated Antifungal Immunity
- Cytoplasmic Viruses: Rage against the (Cellular RNA Decay) Machine
- Balancing Stability and Flexibility within the Genome of the Pathogen
- The Evolution of Transmissible Prions: The Role of Deformed Templating
- Parental Transfer of the Antimicrobial Protein LBP/BPI Protects Eggs against Oomycete Infections
- Host Defense via Symbiosis in
- Regulatory Circuits That Enable Proliferation of the Fungus in a Mammalian Host
- Immune Therapeutic Strategies in Chronic Hepatitis B Virus Infection: Virus or Inflammation Control?
- Burning Down the House: Cellular Actions during Pyroptosis
- Coronaviruses as DNA Wannabes: A New Model for the Regulation of RNA Virus Replication Fidelity
- CRISPR-Cas Immunity against Phages: Its Effects on the Evolution and Survival of Bacterial Pathogens
- Combining Regulatory T Cell Depletion and Inhibitory Receptor Blockade Improves Reactivation of Exhausted Virus-Specific CD8 T Cells and Efficiently Reduces Chronic Retroviral Loads
- Shaping Up for Battle: Morphological Control Mechanisms in Human Fungal Pathogens
- Identification of the Virulence Landscape Essential for Invasion of the Human Colon
- Nodular Inflammatory Foci Are Sites of T Cell Priming and Control of Murine Cytomegalovirus Infection in the Neonatal Lung
- Hepatitis B Virus Disrupts Mitochondrial Dynamics: Induces Fission and Mitophagy to Attenuate Apoptosis
- Mycobacterial MazG Safeguards Genetic Stability Housecleaning of 5-OH-dCTP
- Systematic MicroRNA Analysis Identifies ATP6V0C as an Essential Host Factor for Human Cytomegalovirus Replication
- Placental Syncytium Forms a Biophysical Barrier against Pathogen Invasion
- The CD8-Derived Chemokine XCL1/Lymphotactin Is a Conformation-Dependent, Broad-Spectrum Inhibitor of HIV-1
- Cyclin A Degradation by Primate Cytomegalovirus Protein pUL21a Counters Its Innate Restriction of Virus Replication
- Genome-Wide RNAi Screen Identifies Novel Host Proteins Required for Alphavirus Entry
- Zinc Sequestration: Arming Phagocyte Defense against Fungal Attack
- The Cyst Wall Protein CST1 Is Critical for Cyst Wall Integrity and Promotes Bradyzoite Persistence
- Biphasic Euchromatin-to-Heterochromatin Transition on the KSHV Genome Following Infection
- The Malarial Serine Protease SUB1 Plays an Essential Role in Parasite Liver Stage Development
- HIV-1 Vpr Accelerates Viral Replication during Acute Infection by Exploitation of Proliferating CD4 T Cells
- A Human Torque Teno Virus Encodes a MicroRNA That Inhibits Interferon Signaling
- The ArlRS Two-Component System Is a Novel Regulator of Agglutination and Pathogenesis
- An In-Depth Comparison of Latent HIV-1 Reactivation in Multiple Cell Model Systems and Resting CD4+ T Cells from Aviremic Patients
- Enterohemorrhagic Hemolysin Employs Outer Membrane Vesicles to Target Mitochondria and Cause Endothelial and Epithelial Apoptosis
- Overcoming Antigenic Diversity by Enhancing the Immunogenicity of Conserved Epitopes on the Malaria Vaccine Candidate Apical Membrane Antigen-1
- The Type-Specific Neutralizing Antibody Response Elicited by a Dengue Vaccine Candidate Is Focused on Two Amino Acids of the Envelope Protein
- Tmprss2 Is Essential for Influenza H1N1 Virus Pathogenesis in Mice
- Signatures of Pleiotropy, Economy and Convergent Evolution in a Domain-Resolved Map of Human–Virus Protein–Protein Interaction Networks
- Interference with the Host Haemostatic System by Schistosomes
- RocA Truncation Underpins Hyper-Encapsulation, Carriage Longevity and Transmissibility of Serotype M18 Group A Streptococci
- Gene Fitness Landscapes of at Important Stages of Its Life Cycle
- Phagocytosis Escape by a Protein That Connects Complement and Coagulation Proteins at the Bacterial Surface
- t Is a Structurally Novel Crohn's Disease-Associated Superantigen
- An Increasing Danger of Zoonotic Orthopoxvirus Infections
- Myeloid Dendritic Cells Induce HIV-1 Latency in Non-proliferating CD4 T Cells
- Transcriptional Analysis of Murine Macrophages Infected with Different Strains Identifies Novel Regulation of Host Signaling Pathways
- Serotonergic Chemosensory Neurons Modify the Immune Response by Regulating G-Protein Signaling in Epithelial Cells
- Genome-Wide Detection of Fitness Genes in Uropathogenic during Systemic Infection
- Induces an Unfolded Protein Response via TcpB That Supports Intracellular Replication in Macrophages
- Intestinal CD103+ Dendritic Cells Are Key Players in the Innate Immune Control of Infection in Neonatal Mice
- Emerging Functions for the RNome
- KSHV MicroRNAs Mediate Cellular Transformation and Tumorigenesis by Redundantly Targeting Cell Growth and Survival Pathways
- HrpA, an RNA Helicase Involved in RNA Processing, Is Required for Mouse Infectivity and Tick Transmission of the Lyme Disease Spirochete
- A Toxin-Antitoxin Module of Promotes Virulence in Mice
- Real-Time Imaging of the Intracellular Glutathione Redox Potential in the Malaria Parasite
- Hypoxia Inducible Factor Signaling Modulates Susceptibility to Mycobacterial Infection via a Nitric Oxide Dependent Mechanism
- Novel Strategies to Enhance Vaccine Immunity against Coccidioidomycosis
- Dual Expression Profile of Type VI Secretion System Immunity Genes Protects Pandemic
- —What Makes the Species a Ubiquitous Human Fungal Pathogen?
- αvβ6- and αvβ8-Integrins Serve As Interchangeable Receptors for HSV gH/gL to Promote Endocytosis and Activation of Membrane Fusion
- -Induced Activation of EGFR Prevents Autophagy Protein-Mediated Killing of the Parasite
- Semen CD4 T Cells and Macrophages Are Productively Infected at All Stages of SIV infection in Macaques
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Influence of Mast Cells on Dengue Protective Immunity and Immune Pathology
- Host Defense via Symbiosis in
- Coronaviruses as DNA Wannabes: A New Model for the Regulation of RNA Virus Replication Fidelity
- Myeloid Dendritic Cells Induce HIV-1 Latency in Non-proliferating CD4 T Cells
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání