-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaThe Gene Is Essential for Resistance to Human Serum in
Trypanosoma brucei gambiense causes 97% of all cases of African sleeping sickness, a fatal disease of sub-Saharan Africa. Most species of trypanosome, such as T. b. brucei, are unable to infect humans due to the trypanolytic serum protein apolipoprotein-L1 (APOL1) delivered via two trypanosome lytic factors (TLF-1 and TLF-2). Understanding how T. b. gambiense overcomes these factors and infects humans is of major importance in the fight against this disease. Previous work indicated that a failure to take up TLF-1 in T. b. gambiense contributes to resistance to TLF-1, although another mechanism is required to overcome TLF-2. Here, we have examined a T. b. gambiense specific gene, TgsGP, which had previously been suggested, but not shown, to be involved in serum resistance. We show that TgsGP is essential for resistance to lysis as deletion of TgsGP in T. b. gambiense renders the parasites sensitive to human serum and recombinant APOL1. Deletion of TgsGP in T. b. gambiense modified to uptake TLF-1 showed sensitivity to TLF-1, APOL1 and human serum. Reintroducing TgsGP into knockout parasite lines restored resistance. We conclude that TgsGP is essential for human serum resistance in T. b. gambiense.
Published in the journal: . PLoS Pathog 9(10): e32767. doi:10.1371/journal.ppat.1003686
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1003686Summary
Trypanosoma brucei gambiense causes 97% of all cases of African sleeping sickness, a fatal disease of sub-Saharan Africa. Most species of trypanosome, such as T. b. brucei, are unable to infect humans due to the trypanolytic serum protein apolipoprotein-L1 (APOL1) delivered via two trypanosome lytic factors (TLF-1 and TLF-2). Understanding how T. b. gambiense overcomes these factors and infects humans is of major importance in the fight against this disease. Previous work indicated that a failure to take up TLF-1 in T. b. gambiense contributes to resistance to TLF-1, although another mechanism is required to overcome TLF-2. Here, we have examined a T. b. gambiense specific gene, TgsGP, which had previously been suggested, but not shown, to be involved in serum resistance. We show that TgsGP is essential for resistance to lysis as deletion of TgsGP in T. b. gambiense renders the parasites sensitive to human serum and recombinant APOL1. Deletion of TgsGP in T. b. gambiense modified to uptake TLF-1 showed sensitivity to TLF-1, APOL1 and human serum. Reintroducing TgsGP into knockout parasite lines restored resistance. We conclude that TgsGP is essential for human serum resistance in T. b. gambiense.
Introduction
Throughout their evolution in sub-Saharan Africa, humans have been under assault by a range of different pathogens. One defining challenge is that posed by African trypanosomes, a species complex of blood-borne protozoan parasites transmitted by tsetse flies [1]. The principle pathogenic species in Africa are Trypanosoma brucei, T. congolense and T. vivax, although only Trypanosoma brucei sub-species are able to infect humans. A key feature of these parasites is the ability to undergo antigenic variation by modifying the variant specific glycoprotein (VSG) enveloping the cell that renders the mammalian adaptive immune system largely ineffective [2]. Components of the innate immune system therefore contribute significantly to defence against these organisms [3]. Critical to these defences is the serum protein apolipoprotein L1 (APOL1) found in some catarrhine primates, including humans [4], [5]. The protein is able to kill the majority of trypanosome species in a dose-dependent manner [5]. APOL1 is delivered to parasites in two fractions of the high-density lipoprotein (HDL) component of serum, termed trypanolytic factor 1 and 2 (TLF-1 and TLF-2) [6]. TLF-1 binds to the parasite through an interaction between the haptoglobin-related protein (HPR) surrounding the TLF-1 particle and the haptoglobin haemoglobin receptor (HpHbR) in the flagellar pocket of the parasite [7]–[9]. Under the acidic conditions found in the lysosome, APOL1 changes conformation and embeds in the lysosomal membrane, forming pores in the organelle, leading to cell death [5], [10]. A proportion of TLF-2 similarly enters trypanosomes via HpHbR, although an alternate route also contributes to uptake [11].
Although TLF-1 and 2 kill the majority of trypanosome species, two sub-species of T. brucei have evolved to overcome this innate immunity. T. b. rhodesiense and T. b. gambiense are both resistant to lysis by APOL1 and establish bloodstream infections in humans [1]. T. b. rhodesiense causes an acute form of the disease and is found in East Africa whereas T. b. gambiense is found in West and Central Africa. T. b. gambiense causes a more chronic form of the disease and is responsible for 97% of all human cases of trypanosomiasis [12]. The mechanism of human serum resistance for T. b. rhodesiense involves the expression of a truncated VSG, termed serum resistance associated (SRA) protein [13], [14]. SRA binds to APOL1 in the lysosome, preventing lysis [14]. However, the SRA gene is absent from T. b. gambiense, the more prevalent human infective sub-species [15]. The T. b. gambiense subspecies consists of two sub-groups (1 and 2) that differ in phenotype, including their associated pathology. Group 1 T. b. gambiense parasites are the most prevalent of the human infective trypanosomes and are responsible for the vast majority of cases [16]. Group 1 T. b. gambiense can be distinguished by both their reduced efficacy of HpHbR for binding TLF-1, due to a conserved single nucleotide polymorphism [17]–[19] and also by the presence of a specific truncated VSG, TgsGP [20]. The TgsGP gene is present in all group 1 isolates examined to date but not in T. b. brucei, T. b. rhodesiense or group 2 T. b. gambiense [20]–[23]. The specificity of TgsGP to group 1 T. b. gambiense and its resemblance to SRA, in that it is a truncated VSG gene, led to a suggestion that this gene may confer human serum resistance to group 1 T. b. gambiense [20]. The gene was transfected into T. b. brucei where it did not confer increased resistance to human serum. It was hypothesized that if TgsGP was involved in human serum resistance other factors would also be required to confer the phenotype in T. b. brucei [20]. Efforts to delete the gene from T. b. gambiense were unsuccessful and the function of TgsGP remained unknown [20]. Here we have successfully deleted the TgsGP gene from T. b. gambiense and demonstrated that it is essential for human serum resistance and requires a T. b. gambiense genetic background in order to function.
Results
Deletion of TgsGP in wild-type group 1 T. b. gambiense
To assess whether TgsGP is involved in human serum resistance in T. b. gambiense, the gene was deleted from the genome of a group 1 T. b. gambiense strain. All strains of T. b. gambiense investigated so far are hemizygous for TgsGP, allowing a complete knockout with just one round of transfection [20]–[22]. Although it was postulated that TgsGP was an essential gene and could not be deleted [20], several TgsGP−/0 clones were generated in this study. One of the clones was selected for analysis and used for subsequent assays. The deletion of TgsGP from the clone was confirmed by PCR (Figure 1A). The TgsGP−/0 T. b. gambiense clones was unable to survive in the presence of normal human serum (Figure 2) or recombinant APOL1 (Figure 2), with significantly fewer surviving cells compared to wild-type T. b. gambiense (human serum t-test p = 0.001, APOL1 t-test p<0.001). The clone grew in the presence of non-lytic serum in a similar manner to wild-type T. b. gambiense (t-test p = 0.145). This indicates that TgsGP is involved in protecting against the trypanolytic protein APOL1.
Fig. 1. PCR amplification of TgsGP and RT-PCR of HpHbR in wild-type and transfected lines. 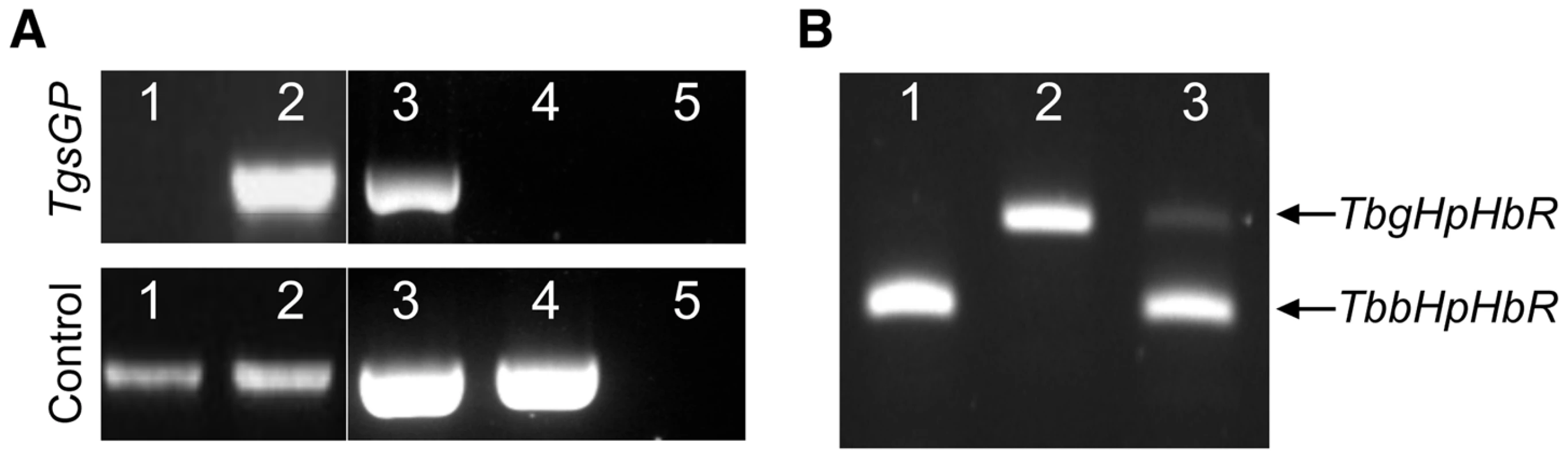
(A) Amplification of TgsGP and a control gene (cathepsin L) by PCR in [1] wild-type T. b. brucei [2], TgsGP−/+ T. b. brucei [3], wild-type T. b. gambiense, [4] TgsGP−/0 T. b. gambiense [5] and negative control. (B) RT-PCR amplification of HpHbR followed by HpyCH4V restriction digestion of [1] wild-type T. b. brucei, [2] wild-type T. b. gambiense and [3] TbbHbHpR−/+TgsGP−/0 T. b. gambiense. Fig. 2. TgsGP is essential for resistance to human serum in T. b. gambiense. 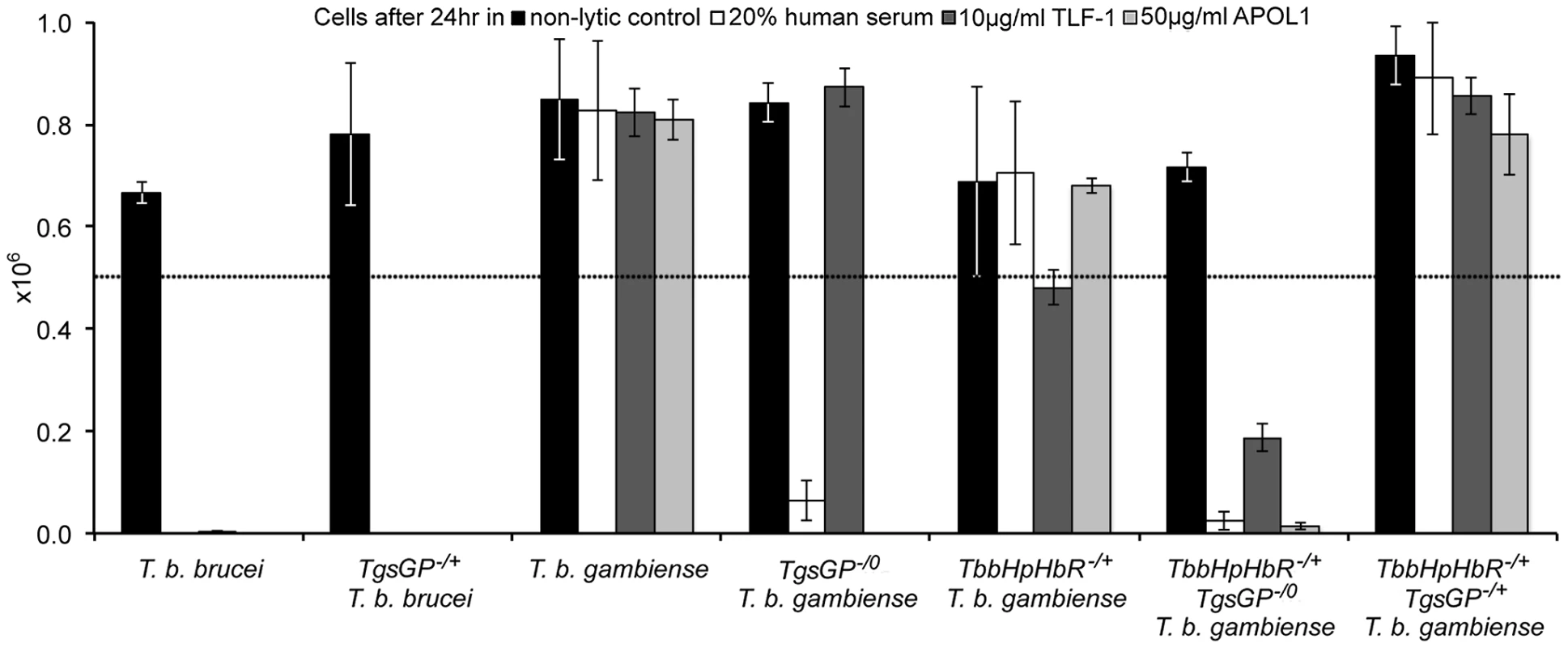
The number of surviving cells after 24% human serum (open box), 10 µg/ml TLF-1 (dark grey box), 50 µg/ml recombinant APOL1 (light grey box) or a non-lytic 20% FBS control (black box). The dotted line indicates the starting concentration of 5×105 cells. The cell lines assayed were wild-type T. b. brucei; TgsGP−/+ T. b. brucei; wild-type T. b. gambiense; TgsGP−/0 T. b. gambiense; TbbHbHpR−/+ T. b. gambiense; TbbHbHpR−/+ TgsGP−/0 T. b. gambiense and TbbHbHpR−/+ TgsGP−/+ T. b. gambiense. Standard error is shown, n = 4 for each data point. The clone was able to grow in the presence of TLF-1 and the number of cells after 24 hours does not differ significantly from that of the wild-type T. b. gambiense strain (t-test p = 0.511). Wild-type T. b. gambiense is resistant to lysis by TLF-1 due to reduced efficacy of their HpHbR for binding TLF-1. Thus lethal amounts of the lytic particle are not internalised by the parasites [18], [19]. It is likely that TgsGP−/0 T. b. gambiense clones are able to grow in the presence of TLF-1 because it possesses the T. b. gambiense HpHbR allele that is less efficient at binding TLF-1.
Deletion and reintroduction of TgsGP in TbbHpHbR−/+ T. b. gambiense
As previously detailed, group 1 T. b. gambiense is characterised by a non-functional HpHbR which results in a reduced uptake of TLF-1 and to a lesser extent TLF-2 [17]–[19], [24]. To investigate the effect of the loss of TgsGP in combination with TLF-1 uptake, a T. b. gambiense strain expressing a functional T. b. brucei HpHbR (TbbHpHbR) and lacking TgsGP was created (termed TbbHpHbR−/+ TgsGP−/0). Expression of both wild-type and ectopic TbbHpHbR alleles was confirmed by RT-PCR (Figure 1B). An allele-specific HpyCh4V restriction site present in the open reading frame of TbbHpHbR, but absent in TbgHpHbR, was used to distinguish between the alleles (Figure 1B) and demonstrated that both alleles were expressed, although the TbgHpHbR allele exhibits lower expression relative to the TbbHpHbR allele. The strain expresses a fully functional HpHbR and hence takes up TLF-1 to a degree similar to T. b. brucei, confirmed by fluorescence microscopy (Figure 3). TbbHpHbR−/+ TgsGP−/0 T. b. gambiense clones were killed in the presence of normal human serum, recombinant APOL1 or, unlike TgsGP−/0 clones, physiological levels of TLF-1 (Figure 2). The number of remaining cells at 24 hours was significantly lower than wild-type T. b. gambiense (human serum t-test p = 0.001, TLF-1 t-test p<0.001, APOL1 t-test p<0.001). However, the cells were able to grow in the presence of non-lytic serum in a similar manner to wild-type T. b. gambiense (t-test = 0.690). A T. b. gambiense clone with TgsGP and the functional TbbHpHbR was able to grow in the presence of human serum and APOL1 (Figure 2) with cell number not significantly differing from wild-type T. b. gambiense (human serum t-test p = 0.936, APOL1 t-test p = 0.465) or in the presence of non-lytic serum (t-test p = 0.972). However, the clone displayed a trypanostatic growth effect in physiological levels of purified TLF-1 with significantly fewer surviving cells compared to wild type (Figure 2) (t-test p = 0.001).
Fig. 3. Uptake of TLF-1 across strains. 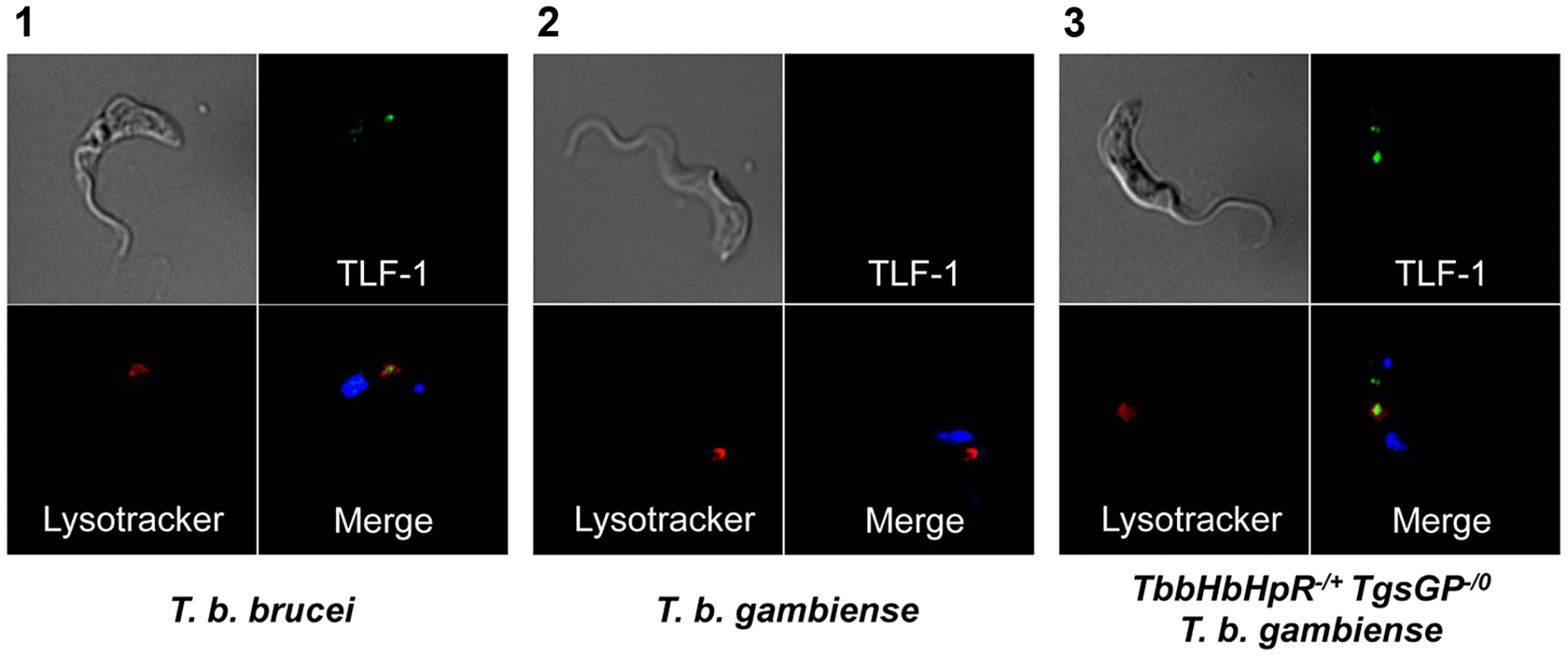
Uptake of TLF-1 after one hour in [1] wild-type T. b. brucei [2] wild-type T. b. gambiense and [3] TbbHbHpR−/+ TgsGP−/0 T. b. gambiense by co-localization of fluorescently tagged TLF-1 (green) with the lysosomal marker Lysotracker (red). The kinetoplast and nucleus were also stained using DAPI (blue). To confirm that the loss of resistance to human serum, APOL1 and TLF-1 in TbbHpHbR−/+ TgsGP−/0 T. b. gambiense was due to the loss of TgsGP, the gene was re-introduced into this background. Resistance to human serum, TLF-1 and APOL1 was rescued by the re-introduction of TgsGP, confirming that this gene is essential for resistance to lysis (Figure 2). When the same TgsGP add-back construct was transfected into a human serum sensitive T. b. brucei, it did not confer resistance to any lytic component (Figure 2), confirming earlier work [20].
Localisation of TgsGP
Previous work has shown that TgsGP localises to the flagellar pocket in T. b. gambiense and this is likely to be the site of interaction between TLF and TgsGP [20]. A possible hypothesis for the observation that when TgsGP is transfected into in TbbHpHbR−/+ TgsGP−/0 T. b. gambiense background it restores human serum resistance but does not confer resistance in T. b. brucei [20] (figure 2) is that the protein is not trafficked correctly to the flagellar pocket. In order to verify localisation, TgsGP was transfected into wild-type T. b. brucei with the addition of a TY tag into a HindIII restriction site at position 1130 of the TgsGP ORF, upstream of the predicted GPI anchor sequence [25], [26]. Immunofluorescence with anti-TY antibodies shows clear localisation of TY-TgsGP adjacent to the kinetoplast and co-localization with fluorescent Concanavalin A, which acts as a marker for the flagellar pocket [27], (Figure 4). However, these cells were killed in human serum, TLF-1 or APOL1 (Figure S1). A similar localisation is observed when the TY-tagged TgsGP protein is expressed in TbbHpHbR−/+ TgsGP−/0 T. b. gambiense (Figure 4), with strong signal close to the kinetoplast and a more diffuse signal closer to the nucleus. In this case, the capacity to grow in human serum, TLF-1 and APOL1 was restored by the reintroduction of the TY-tagged TgsGP (Figure S1). As an identical construct was used in both transfections, it is probable that group 1 T. b. gambiense possess a protein or mechanism complementing TgsGP that is absent in T. b. brucei.
Fig. 4. Localisation of TY-TgsGP. 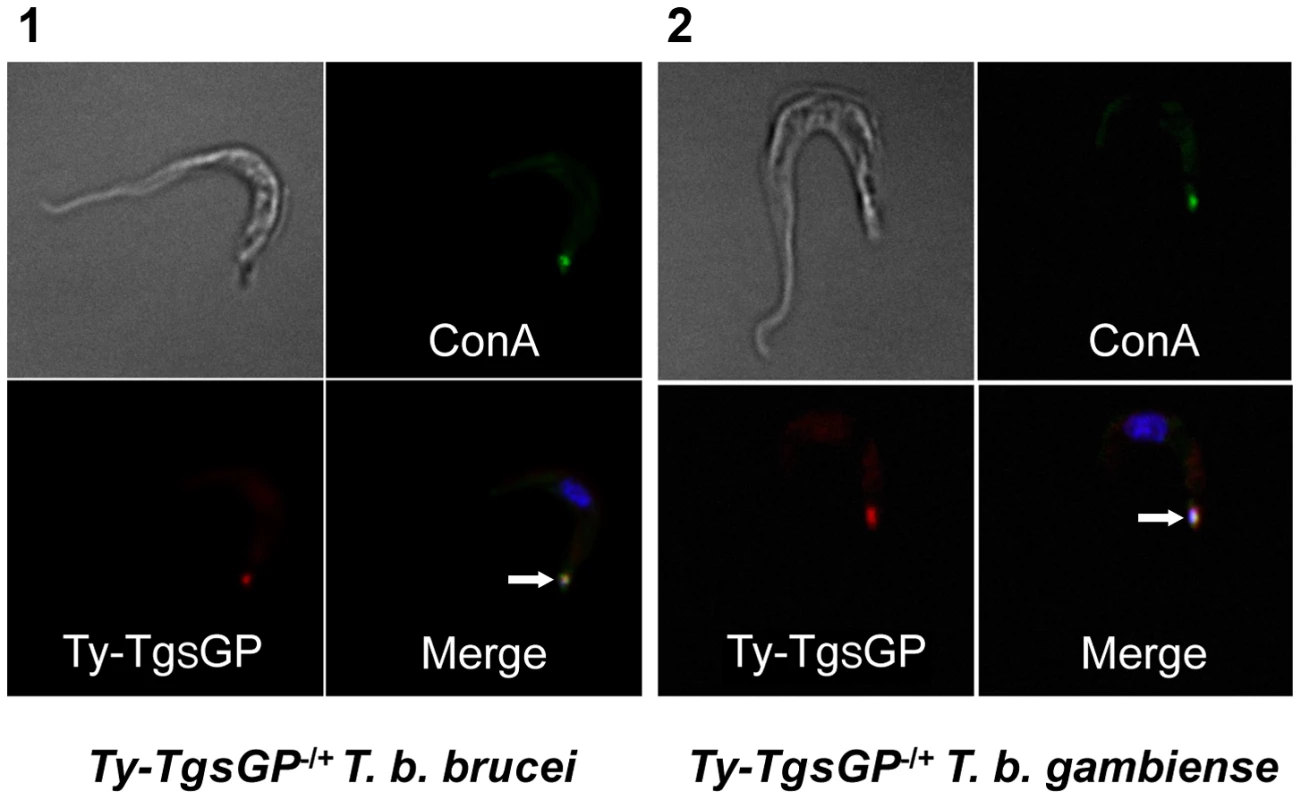
Localisation of TY-tagged TgsGP (red) relative to un-endocytosed FITC-labeled Concanavalin A bound to glycoproteins in the flagellar pocket (green) and DAPI stained nucleus and kinetoplast (blue) in [1] TY-TgsGP−/+ T. b. brucei and [2] TbbHbHpR−/+ TY-TgsGP−/+ T. b. gambiense. The flagellar pocket (revealed by Concanavalin A and kinetoplast position) is indicated with a white arrow. Discussion
This study demonstrates that the TgsGP gene is essential for resistance to human serum in the most clinically important T. brucei sub-species, group 1 T. b. gambiense. Previous work has shown that TgsGP did not confer resistance to human serum when ectopically expressed in T. b. brucei [20], which was confirmed here. As originally hypothesized [20], it appears likely that this is due to other factor(s) or mechanism(s) that works in concert with TgsGP, which are absent in T. b. brucei. By removing TgsGP from T. b. gambiense itself, we have demonstrated that the gene is necessary for resistance to human serum. Elucidation of a gene essential to human serum resistance in group 1 T. b gambiense unlocks new avenues for future treatment of human African sleeping sickness. These include peptide screens that neutralise the TgsGP protein, targeted antibodies or the possibility of using TgsGP as a vaccine candidate, as expression is required for parasite survival in humans. Additionally, there exists the potential that variants of APOL1 may offer protection against T. b. gambiense. Sera from individuals possessing certain APOL1 alleles has been shown to affect the growth of T. b. rhodesiense and it has been suggested that these alleles may be protective against T. b. rhodesiense [28], [29]. However, this has yet to be confirmed in a case control study. Nevertheless, it is likely that there are variant APOL1 alleles that protect against group 1 T. b. gambiense in resistant individuals, such as the reportedly resistant Bambuti people of the Mbomo region in the Democratic Republic of the Congo [30] or recently described asymptomatic and self-cured cases from Côte d'Ivoire [31].
One other benefit of our study is the trypanosome research community now possesses a representative group 1 T. b. gambiense strain that is easily cultured, is no longer human serum resistant, yet only differs from the wild-type by a single gene. This is a powerful biological resource that could replace T. b. brucei as the common laboratory model for the human disease, which maybe useful, particularly as several drugs display different efficacies between sub-species [1]. As such, identifying TgsGP as a gene essential for resistance to human serum in group 1 T. b. gambiense will likely be important to future control of the disease.
Materials and Methods
Trypanosomes strains and maintenance
Bloodstream form T. b. brucei Lister 427 (MITat 1.2) was grown at 37°C under 5% CO2 in HMI9 medium supplemented with 20% foetal bovine serum (Sigma-Aldrich) and 20% Serum-Plus (Sigma-Aldrich). The bloodstream form group 1 T. b. gambiense strain ELIANE (MHOM/CI/52/ELIANE) was isolated from a patient infected while in Côte d'Ivoire [22]. It was cultured in modified HMI9 [32] supplemented with 20% serum plus (SAFC Biosciences Ltd.). Similar to other group 1 T. b. gambiense strains, ELIANE is consistently resistant to lysis by human serum, despite repeated passage.
Transfection of T. b. brucei and group 1 T. b. gambiense
T. b. gambiense and T. brucei strains were transfected using the protocols outlined in [33]. For ectopic expression of TgsGP in T. b. brucei and reinsertion into the TgsGP−/0 T. b. gambiense strains, the TgsGP ORF was inserted into the pURAN vector [34] using G418 for selection. Ectopic expression of TbbHbHpR in T. b. gambiense was achieved using the tubulin-targeting TbbHbHpR pTub-phelo construct, using phleomycin for selection [17]. For deletion of TgsGP from the genome of T. b. gambiense and TbbHbHpR−/+T. b. gambiense, 500 base pairs from both the upstream and downstream regions of TgsGP (sequence AM237444.1, http://www.genedb.org) were inserted into a vector containing a hygromycin resistance cassette. Insertion of TY-tagged TgsGP into the deletion strain T. b. gambiense and T. b. brucei was performed by inserting a TY tag into a HindIII restriction site at position 1130 of the TgsGP ORF. This sequence was ligated into the pURAN vector [34], [35] and transfectants were screened using a G418 selection marker. This insertion site is upstream of the predicted GPI anchor site identified using the big-PI software package [25] and a GPI prediction protocol validated for trypanosomes [26]. Correct integration for constructs was assessed by PCR and/or RT-PCR. All primers used in the studies and their targets, are listed in Table S1.
RT-PCR of expressed TbbHpHbR in group 1 T. b. gambiense
Total RNA was isolated from cells using RNeasy kit (Qiagen) according to manufacturers' instruction, with additional DNase steps. 2 µg RNA was subject to a second round of DNase treatment (Invitrogen) prior to cDNA synthesis using Superscript III (Invitrogen), according to manufacturers' instructions. RT-PCR was performed using Taq DNA polymerase and the primers are described in Table S1. For RFLP analysis of HpHpR, the amplified product was cleaned using GeneJet PCR purification column, digested with HpyCh4V and the digested products separated on a 2% agarose gel.
TLF-1 purification
TLF-1 purification, labeling and survival assays were performed as previously described [17], [36].
Generation of recombinant APOL1
APOL1 synthesis and purification was performed as previously described [36]. Protein purity was estimated using a Nanodrop spectrometer (Nanodrop) and SDS-PAGE. A Western blot using an antibody raised against an APOL1 peptide (Sigma-Aldrich) was used to verify that the bands present were APOL1.
Lysis survival assays
To assess survival in human serum, trypanosomes were diluted to 5×105 per ml in HMI9 and incubated for 24 hours with 20% human serum or 20% non-lytic foetal bovine serum (FBS). The number of surviving trypanosomes in each well was recorded after 24 hours using a haemocytometer. To assess survival in TLF-1 and APOL1, trypanosomes were diluted to 5×105 per ml in HMI9 with FBS. Cells were incubated with a physiological amount of TLF-1 (10 µg/ml). For the recombinant APOL1 assays, a concentration of 50 µg−1 ml was used as this had previously been determined to kill 100% of T. b. brucei cells in a 24-hour assay [36]. The number of cells in each well was counted with a haemocytometer at 24 hours. There were four replicates for each data point. The number of surviving cells for each treatment were compared between each of the T. b. gambiense clones and wild-type T. b. gambiense using the unpaired 2-tailed t-test function of the Minitab 14 Statistics Package (Minitab).
Immunofluorescence assays
TLF-1 immunofluorescence assays were performed as previously described [17], [36]. Immunofluorescence localisation of TY-TgsGP was performed with approximately 106 bloodstream-cultured parasites in mid-log phase. Cells were incubated with 5 mg/ml FITC conjugated Concanavalin A in serum-free HMI9 for 20 minutes at 4°C. The Concanavalin A binds to glycoproteins in the flagellar pocket but is not endocytosed due to the reduced temperature, thus labeling the flagellar pocket [27]. Cells were then fixed by immersion in chilled methanol for 30 minutes. Slides were incubated for 1 hour with 1∶500 primary mouse anti-TY antibody (Iain Johnston, University of Glasgow), washed with PBS and then incubated with 1∶1000 of AlexaFluor568 anti-mouse secondary (Invitrogen). The slides were mounted using 50% glycerol, 0.1% DAPI and 2.5% DABCO. Parasites were imaged using a Deltavision Core system and SoftWorx package (Applied Precision). Images were composited using the ImageJ software package [37].
Supporting Information
Zdroje
1. BarrettMP, BurchmoreRJS, StichA, LazzariJO, FraschAC, et al. (2003) The trypanosomiases. Lancet 362 : 1469–1480 doi:10.1016/S0140-6736(03)14694-6
2. VincendeauP, BouteilleB (2006) Immunology and immunopathology of African trypanosomiasis. An Acad Bras Cienc 78 : 645–665.
3. PaysE, VanhollebekeB (2009) Human innate immunity against African trypanosomes. Current Opinion in Immunology 21 : 493–498.
4. PoelvoordeP, VanhammeL, AbbeeleJVD, SwitzerWM, PaysE (2004) Distribution of apolipoprotein L-I and trypanosome lytic activity among primate sera. Mol Biochem Parasitol 134 : 155–157 doi:10.1016/j.molbiopara.2003.11.006
5. VanhammeL, Paturiaux-HanocqF, PoelvoordeP, NolanDP, LinsL, et al. (2003) Apolipoprotein L-I is the trypanosome lytic factor of human serum. Nature 422 : 83–87 doi:10.1038/nature01461
6. RaperJ, PortelaMPM, LugliE, FrevertU, TomlinsonS (2001) Trypanosome lytic factors: novel mediators of human innate immunity. Current Opinion in Microbiology 4 : 402–408.
7. VanhollebekeB, De MuylderG, NielsenMJ, PaysA, TebabiP, et al. (2008) A Haptoglobin-Hemoglobin Receptor Conveys Innate Immunity to Trypanosoma brucei in Humans. Science 320 : 677–681 doi:10.1126/science.1156296
8. ShiflettAM, BishopJR, PahwaA, HajdukSL (2005) Human high density lipoproteins are platforms for the assembly of multi-component innate immune complexes. J Biol Chem 280 : 32578–32585.
9. ShiflettA, FaulknerSD, CotlinLF, WidenerJ, StephensN, et al. (2007) African trypanosomes: intracellular trafficking of host defense molecules. J Eukaryot Microbiol 54 : 18–21 doi:10.1111/j.1550-7408.2006.00228.x
10. CampilloN, CarringtonM (2003) The origin of the serum resistance associated (SRA) gene and a model of the structure of the SRA polypeptide from Trypanosoma brucei rhodesiense. Mol Biochem Parasitol 127 : 79–84 doi:10.1016/S0166-6851(02)00306-7
11. BullardW, KieftR, CapewellP, VeitchNJ, MacleodA, et al. (2012) Haptoglobin-hemoglobin receptor independent killing of African trypanosomes by human serum. Virulence 3 : 72–76.
12. SimarroPP, DiarraA, PostigoJAR, FrancoJR, JanninJG (2011) The Human African Trypanosomiasis Control and Surveillance Programme of the World Health Organization 2000–2009: The Way Forward. PLoS Neglected Tropical Diseases 5: e1007 doi:10.1371/journal.pntd.0001007
13. De GreefC, HamersR (1994) The serum resistance-associated (SRA) gene of Trypanosoma brucei rhodesiense encodes a variant surface glycoprotein-like protein. Mol Biochem Parasitol 68 : 277–284.
14. XongHV, VanhammeL, ChamekhM, ChimfwembeCE, Van Den AbbeeleJ, et al. (1998) A VSG expression site-associated gene confers resistance to human serum in Trypanosoma rhodesiense. Cell 95 : 839–846.
15. De GreefC, ImberechtsH, MatthyssensG, Van MeirvenneN, HamersR (1989) A gene expressed only in serum-resistant variants of Trypanosoma brucei rhodesiense. Mol Biochem Parasitol 36 : 169–176 doi:10.1016/0166-6851(89)90189-8
16. GibsonW (1986) Will the real Trypanosoma b. gambiense please stand up. Parasitol Today 2 : 255–257.
17. KieftR, CapewellP, TurnerCMR, VeitchNJ, MacleodA, et al. (2010) Mechanism of Trypanosoma brucei gambiense (group 1) resistance to human trypanosome lytic factor. Proceedings of the National Academy of Sciences 107 : 16137–16141.
18. DeJesusE, KieftR, AlbrightB, StephensNA, HajdukSL (2013) A Single Amino Acid Substitution in the Trypanosoma brucei gambiense Haptoglobin-Hemoglobin Receptor Abolishes TLF-1 Binding. PLoS Pathogens e1003317 doi:10.1371/journal.ppat.1003317
19. HigginsMK, TkachenkoO, BrownA (2013) Structure of the trypanosome haptoglobin–hemoglobin receptor and implications for nutrient uptake and innate immunity. Proceedings of the National Academy of Sciences 110 (5) 1905–10 doi:10.1073/pnas.1214943110/-/DCSupplemental
20. BerberofM, Pérez-MorgaD, PaysE (2001) A receptor-like flagellar pocket glycoprotein specific to Trypanosoma brucei gambiense. Mol Biochem Parasitol 113 : 127–138.
21. GibsonW, NemetschkeL, Ndung'uJ (2010) Conserved sequence of the TgsGP gene in Group 1 Trypanosoma brucei gambiense. Infection, Genetics and Evolution 10 : 453–458.
22. RadwanskaM, ClaesF, MagezS, MagnusE, Pérez-MorgaD, et al. (2002) Novel primer sequences for polymerase chain reaction-based detection of Trypanosoma brucei gambiense. Am J Trop Med Hyg 67 : 289–295.
23. CapewellP, CooperA, DuffyCW, TaitA, TurnerCM, et al. (2013) Human and animal trypanosomes in Côte d'Ivoire form a single breeding population. PLoS ONE doi:10.1371/journal.pone.0067852
24. SymulaRE, BeadellJS, SistromM (2012) Trypanosoma brucei gambiense Group 1 Is Distinguished by a Unique Amino Acid Substitution in the HpHb Receptor Implicated in Human Serum Resistance. PLoS Neglected Tropical Diseases 6: e1728.
25. EisenhaberB, BorkP, EisenhaberF (1999) Prediction of Potential GPI-modification Sites in Proprotein Sequences. Journal of Molecular Biology 292 : 741–758 doi:10.1006/jmbi.1999.3069
26. BöhmeU, CrossGAM (2002) Mutational analysis of the variant surface glycoprotein GPI-anchor signal sequence in Trypanosoma brucei. Journal of Cell Science 805–816.
27. BalberAE, FrommelTO (1988) Trypanosoma brucei gambiense and T. b. rhodesiense: Concanavalin A Binding to the Membrane and Flagellar Pocket of Bloodstream and Procyclic Forms. J Eukaryot Microbiol 35 : 214–219 doi:10.1111/j.1550-7408.1988.tb04326.x
28. LecordierL, VanhollebekeB, PoelvoordeP, TebabiP, Paturiaux-HanocqF, et al. (2009) C-Terminal Mutants of Apolipoprotein L-I Efficiently Kill Both Trypanosoma brucei brucei and Trypanosoma brucei rhodesiense. PLoS Pathogens 5: e1000685 doi:10.1371/journal.ppat.1000685
29. GenoveseG, FriedmanDJ, RossMD, LecordierL, UzureauP, et al. (2010) Association of Trypanolytic ApoL1 Variants with Kidney Disease in African Americans. Science 329 : 841–845 doi:10.1126/science.1193032
30. Frezil JL (1983) Human trypanosomiasis in the Congo. Paris: ORSTOM.
31. JamonneauV, IlboudoH, KaboréJ, KabaD, KoffiM, et al. (2012) Untreated Human Infections by Trypanosoma brucei gambiense Are Not 100% Fatal. PLoS Neglected Tropical Diseases 6: e1691 doi:10.1371/journal.pntd.0001691
32. HirumiH, HirumiK (1989) Continuous cultivation of Trypanosoma brucei blood stream forms in a medium containing a low concentration of serum protein without feeder cell layers. J Parasitol 75 : 985–989.
33. GiroudC, OttonesF, CoustouV, DacheuxD, BiteauN, et al. (2009) Murine Models for Trypanosoma brucei gambiense Disease Progression—From Silent to Chronic Infections and Early Brain Tropism. PLoS Neglected Tropical Diseases 3: e509 doi:10.1371/journal.pntd.0000509.t003
34. LigtenbergMJ, BitterW, KieftR, SteverdingD, JanssensH, et al. (1994) Reconstitution of a surface transferrin binding complex in insect form Trypanosoma brucei. The EMBO Journal 13 : 2565.
35. BastinP, BagherzadehZ, MatthewsKR, GullK (1996) A novel epitope tag system to study protein targeting and organelle biogenesis in Trypanosoma brucei. Mol Biochem Parasitol 77 : 235–239.
36. CapewellP, VeitchNJ, TurnerCMR, RaperJ, BerrimanM, et al. (2011) Differences between Trypanosoma brucei gambiense groups 1 and 2 in their resistance to killing by trypanolytic factor 1. PLoS Neglected Tropical Diseases 5: e1287.
37. AbràmoffMD, MagalhãesPJ, RamSJ (2004) Image processing with ImageJ. Biophotonics international 11 : 36–42.
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2013 Číslo 10- Jak souvisí postcovidový syndrom s poškozením mozku?
- Měli bychom postcovidový syndrom léčit antidepresivy?
- Farmakovigilanční studie perorálních antivirotik indikovaných v léčbě COVID-19
- 10 bodů k očkování proti COVID-19: stanovisko České společnosti alergologie a klinické imunologie ČLS JEP
-
Všechny články tohoto čísla
- Are We There Yet? Recent Progress in the Molecular Diagnosis and Novel Antifungal Targeting of and Invasive Aspergillosis
- Fungal Iron Availability during Deep Seated Candidiasis Is Defined by a Complex Interplay Involving Systemic and Local Events
- Emergence of Azole-Resistant Strains due to Agricultural Azole Use Creates an Increasing Threat to Human Health
- Fungal Adenylyl Cyclase Acts As a Signal Sensor and Integrator and Plays a Central Role in Interaction with Bacteria
- Sensing of the Microbial Neighborhood by
- Antivirulence Therapy for Animal Production: Filling an Arsenal with Novel Weapons for Sustainable Disease Control
- The Cell Biology of : How to Teach Using Animations
- A Structure-Guided Mutation in the Major Capsid Protein Retargets BK Polyomavirus
- RNA Biology in Fungal Phytopathogens
- , , and the Human Mouth: A Sticky Situation
- The Gene Is Essential for Resistance to Human Serum in
- Unisexual Reproduction Drives Evolution of Eukaryotic Microbial Pathogens
- Bacterial Pathogens Activate a Common Inflammatory Pathway through IFNλ Regulation of PDCD4
- Bats and Viruses: Friend or Foe?
- Protein Trafficking through the Endosomal System Prepares Intracellular Parasites for a Home Invasion
- IL-22 Mediates Goblet Cell Hyperplasia and Worm Expulsion in Intestinal Helminth Infection
- B Cells Enhance Antigen-Specific CD4 T Cell Priming and Prevent Bacteria Dissemination following Genital Tract Infection
- Alternative Roles for CRISPR/Cas Systems in Bacterial Pathogenesis
- Chemicals, Climate, and Control: Increasing the Effectiveness of Malaria Vector Control Tools by Considering Relevant Temperatures
- Dengue Vaccines: Strongly Sought but Not a Reality Just Yet
- Feeding Uninvited Guests: mTOR and AMPK Set the Table for Intracellular Pathogens
- Driven Enforced Viral Replication in Dendritic Cells Contributes to Break of Immunological Tolerance in Autoimmune Diabetes
- IL-4Rα-Associated Antigen Processing by B Cells Promotes Immunity in Infection
- A Gammaherpesvirus Uses Alternative Splicing to Regulate Its Tropism and Its Sensitivity to Neutralization
- MicroRNA-155 Promotes Autophagy to Eliminate Intracellular Mycobacteria by Targeting Rheb
- Epigenetic Dominance of Prion Conformers
- MAIT Cells Detect and Efficiently Lyse Bacterially-Infected Epithelial Cells
- The Role of TcdB and TccC Subunits in Secretion of the Tcd Toxin Complex
- A Mechanism for the Inhibition of DNA-PK-Mediated DNA Sensing by a Virus
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Dengue Vaccines: Strongly Sought but Not a Reality Just Yet
- MicroRNA-155 Promotes Autophagy to Eliminate Intracellular Mycobacteria by Targeting Rheb
- Alternative Roles for CRISPR/Cas Systems in Bacterial Pathogenesis
- Feeding Uninvited Guests: mTOR and AMPK Set the Table for Intracellular Pathogens
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



