-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaAlternative Roles for CRISPR/Cas Systems in Bacterial Pathogenesis
article has not abstract
Published in the journal: . PLoS Pathog 9(10): e32767. doi:10.1371/journal.ppat.1003621
Category: Pearls
doi: https://doi.org/10.1371/journal.ppat.1003621Summary
article has not abstract
CRISPR/Cas Loci Encode Adaptive, RNA-Directed Nucleic Acid Restriction Systems
CRISPR (clustered regularly interspaced short palindromic repeats)/Cas (CRISPR-associated) systems are highly specific bacterial defenses against foreign genetic elements derived from bacteriophages, plasmids, or extracellular chromosomal DNA [1]. These systems consist of a CRISPR array (crRNA array; composed of unique spacer sequences flanked by short repeats) and adjacently encoded Cas proteins [1]. Following transcription, the crRNA array is processed into individual CRISPR RNAs (crRNA) containing a spacer and a partial repeat [2]. The spacers hybridize to complementary nucleic acid targets, triggering their degradation by Cas proteins [1]. In addition, the Cas proteins Cas1 (a dsDNA endonuclease) and Cas2 (a dsDNA and/or ssRNA endonuclease) function to integrate new spacer sequences into the crRNA array, an adaptation phase that allows bacteria to subsequently target foreign genetic elements containing these sequences [3], [4].
There are three main types of CRISPR/Cas systems. All contain Cas1 and Cas2, but are distinguished by specific Cas proteins involved in crRNA maturation, nucleic acid targeting, and cleavage [1]. Specifically, the Type II CRISPR/Cas systems are associated with pathogenic bacteria including Neisseria meningitidis, Campylobacter jejuni, and Streptococcus pyogenes [1], [5]. These systems require a trans-activating CRISPR RNA (tracrRNA) and an endogenous RNase (RNase III) for maturation of crRNAs, as well as two endonuclease domains within Cas9 for cleavage of each strand of the targeted DNA [2], [6], [7]. In addition to their role in defense against foreign nucleic acid, recent work has demonstrated an alternative functionality of Type II CRISPR/Cas systems as being essential to pathogenesis.
CRISPR/Cas Systems Play Critical Roles in Pathogenesis
We recently demonstrated that components of the Type II CRISPR/Cas system in Francisella novicida are necessary for this intracellular bacterial pathogen to evade detection by a host pattern recognition receptor and cause disease [5]. Cas9, in conjunction with tracrRNA and a novel small RNA termed scaRNA (small, CRISPR/Cas-associated RNA), target an endogenous transcript encoding an immunostimulatory bacterial lipoprotein (BLP), leading to mRNA degradation and decreased transcript levels (Figure 1A) [5]. Surprisingly, this action does not rely on any of the crRNAs, but instead is predicted to utilize tracrRNA to target mRNA. In the absence of this regulation, increased BLP levels trigger the activation of a Toll-like Receptor 2 (TLR2)-dependent proinflammatory response, and result in complete attenuation of the bacteria during infection (Figure 1B). These CRISPR/Cas components are therefore critical to the ability of F. novicida to cause disease.
Fig. 1. CRISPR/Cas-mediated evasion of innate immune detection by F. novicida. 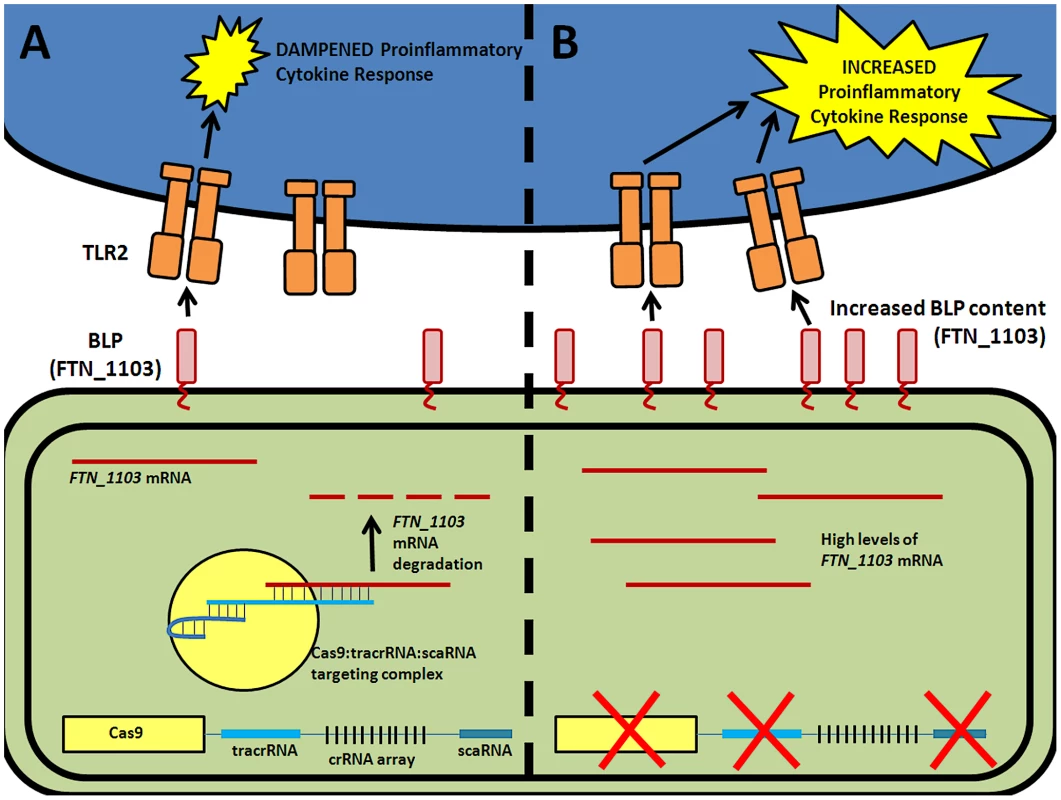
(A) Cas9 (yellow), tracrRNA (blue), and scaRNA (teal) act to target the mRNA of a bacterial lipoprotein (red, BLP), ultimately mediating its repression by degradation. Low levels of BLPs result in a dampened TLR2-dependent proinflammatory cytokine response. (B) In the absence of either of these three CRISPR/Cas components, there is an increase in the abundance of BLP mRNA. This results in increased BLP content, and triggers a robust TLR2-dependent proinflammatory cytokine response. Cas9 is also required for the ability of both Neisseria meningitidis and Campylobacter jejuni to attach to, invade, and replicate within epithelial cells, traits that are essential to their virulence [5], [8]. Currently, the mechanism of Cas9-mediated pathogenesis by these organisms is unknown. It is likely that Cas9, in conjunction with one or more small RNAs, acts to alter the stability of a transcript and that this is important for virulence, similar to its function in F. novicida. Interestingly, the role of Cas9 in C. jejuni virulence correlated with strains producing a sialylated lipooligosaccharide structure in the outer envelope [8]. Coupled with the fact that Cas9 regulates production of a membrane BLP in Francisella, it is tempting to speculate that CRISPR/Cas systems that control mRNA stability may be widely involved in regulation of envelope structure.
In addition to the role of Cas9 in the virulence of bacterial pathogens, Cas2 has been implicated in the ability of Legionella pneumophila to survive within amoebae [9]. Since amoebae are essential to L. pneumophila survival in the environment [10], the role of Cas2 in this process may be critical to environmental persistence and subsequent transmission to other hosts. Exactly how Cas2 mediates Legionella intracellular survival is unknown. Due to its endonuclease activity involved in CRISPR adaptation, Cas2 is hypothesized to also function in either the processing of small RNA regulators during intracellular infection, or to be a direct mediator of mRNA degradation [9]. Interestingly, Cas9 plays no role in the intracellular survival of L. pneumophila in amoebae [9]. Additionally, Cas2 has no currently observed role in F. novicida gene regulation or virulence [5], demonstrating that while Type II CRISPR/Cas systems are similar across species, different organisms may have co-opted distinct components for pathogenesis.
CRISPR/Cas Systems Can Control Bacterial Physiology
Different types of CRISPR/Cas systems have also been observed to contribute to bacterial physiology beyond defense against foreign nucleic acids. The CRISPR/Cas system in Pseudomonas aeruginosa plays a role in modulating biofilm formation [11], [12]. While the precise mechanism is unknown, the data suggest that when P. aeruginosa is lysogenized by a specific bacteriophage, the CRISPR/Cas system interacts with a particular gene in the chromosomally integrated prophage to inhibit the creation of biofilms [11], [12]. It is unclear if the CRISPR/Cas system targets DNA or mRNA, but it is known that the interaction requires the Cas proteins involved in both crRNA maturation and targeting/degradation, as well as a specific targeting crRNA with sequence similarity to the prophage gene [11], [12]. Given the importance of biofilm formation in the pathogenic life cycle of P. aeruginosa [12], it is likely that this intricate CRISPR/Cas regulatory schema plays an important role in infection.
Additionally, Cas1 and the crRNA array in the CRISPR/Cas system of Escherichia coli K-12 (also present in EHEC and UPEC strains [1]) play a role in mediating DNA repair [13]. Given the universality of Cas1 in all known CRISPR/Cas systems, it is intriguing to speculate that it may broadly function in this regard. Further, since DNA damage may occur as a product of host defenses during infection (i.e., production of reactive nitrogen and oxygen species) [14], it is interesting to consider that CRISPR/Cas systems may provide pathogens another layer of redundancy in their ability to resist and repair damage incurred during infection.
Are Self-Targeting CRISPR/Cas Systems Involved in “Autoimmunity” or Gene Regulation?
Another potential example of noncanonical functionality of CRISPR/Cas systems in gene regulation and virulence may involve self-targeting crRNAs with spacer sequences complementary to chromosomally encoded genes [15], [16]. For example, self-targeting crRNAs are predicted to target hypothetical proteins in the pathogens Clostridium botulinum, N. meningitidis, and Yersinia pestis, two sporulation genes and a gene involved in S-layer biosynthesis in Clostridium tetani, and the fdrA gene involved in protein transport in Enterobacter spp. [15]. Since crRNAs are known to target DNA, their specificity for chromosomal genes has been suggested to likely result in detrimental chromosomal cleavage, which can result in large chromosomal deletions, and thus has been termed “autoimmunity” [17]. It has been hypothesized that self-targeting crRNAs can be tolerated if CRISPR/Cas systems in bacteria encoding them are either nonfunctional or are significantly degenerated, thereby preventing “autoimmune” recognition and cleavage of the chromosome [15]. Indeed, CRISPR/Cas systems encoding self-targeting crRNAs often have degenerated Cas proteins [15], [16]. However, this could nonetheless be consistent with a role in gene regulation for at least some self-targeting crRNAs [16]. Inactive Cas1 and Cas2 proteins would not necessarily inhibit self-targeting abilities, but instead prevent acquisition of new crRNAs [3], [4]. Since acquisition of new crRNAs can lead to loss of previously acquired crRNAs [18], degeneration of Cas1 and Cas2 may actually be favored to prevent loss of regulatory crRNAs. Additionally, it has been demonstrated that a catalytically inactive Cas9 is still capable of binding DNA targets and inhibiting transcription, resulting in repression of the targeted gene [19]. Therefore, degenerated Cas proteins could theoretically still participate in gene regulation. Furthermore, the CRISPR/Cas system in P. aeruginosa, capable of targeting a chromosomally integrated element without causing chromosomal degradation, is fully functional against bacteriophage infection, suggesting that chromosomal targeting by an active CRISPR/Cas system does not necessarily lead to “autoimmune” events [11], [12], [20]. Finally, as observed in F. novicida, if mRNA but not DNA is targeted (i.e., FTN_1103) [5], there would be no negative selection against targeting endogenous genes in the chromosome. It is therefore tempting to speculate that self-targeting crRNAs may act as regulatory elements in at least some of the aforementioned and other pathogens.
Perspectives
While well established to play roles in defending bacteria from bacteriophages and other foreign genetic elements, the critical roles that CRISPR/Cas systems play in the ability of pathogenic organisms to evade host defenses and replicate within the host are just now being appreciated. Given that CRISPR/Cas systems are widely distributed among prokaryotes (∼50% of bacteria and 99% of Archaea) and are present in both pathogenic and commensal organisms [1], as well as their specificity and adaptability, it is very likely that more examples of their alternative functions in gene regulation controlling virulence, commensalism, and broader physiology will be revealed. Future work elucidating how CRISPR/Cas systems contribute to bacterial virulence will allow for the identification of novel host defense evasion strategies that bacterial pathogens utilize during infection.
Zdroje
1. MakarovaKS, HaftDH, BarrangouR, BrounsSJ, CharpentierE, et al. (2011) Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol 9 : 467–477.
2. DeltchevaE, ChylinskiK, SharmaCM, GonzalesK, ChaoY, et al. (2011) CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature 471 : 602–607.
3. YosefI, GorenMG, QimronU (2012) Proteins and DNA elements essential for the CRISPR adaptation process in Escherichia coli. Nucleic Acids Res 40 : 5569–5576.
4. DatsenkoKA, PougachK, TikhonovA, WannerBL, SeverinovK, et al. (2012) Molecular memory of prior infections activates the CRISPR/Cas adaptive bacterial immunity system. Nat Commun 3 : 945.
5. SampsonTR, SarojSD, LlewellynAC, TzengYL, WeissDS (2013) A CRISPR/Cas system mediates bacterial innate immune evasion and virulence. Nature 497 : 254–257.
6. JinekM, ChylinskiK, FonfaraI, HauerM, DoudnaJA, et al. (2012) A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337 : 816–821.
7. GasiunasG, BarrangouR, HorvathP, SiksnysV (2012) Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci U S A 109: E2579–2586.
8. LouwenR, Horst-KreftD, de BoerAG, van der GraafL, de KnegtG, et al. (2013) A novel link between Campylobacter jejuni bacteriophage defence, virulence and Guillain-Barré syndrome. Eur J Clin Microbiol Infect Dis 32 : 207–226.
9. GundersonFF, CianciottoNP (2013) The CRISPR-associated gene cas2 of Legionella pneumophila is required for intracellular infection of amoebae. MBio 4: e00074–00013.
10. Abu KwaikY, GaoLY, StoneBJ, VenkataramanC, HarbOS (1998) Invasion of protozoa by Legionella pneumophila and its role in bacterial ecology and pathogenesis. Appl Environ Microbiol 64 : 3127–3133.
11. CadyKC, O'TooleGA (2011) Non-identity-mediated CRISPR-bacteriophage interaction mediated via the Csy and Cas3 proteins. J Bacteriol 193 : 3433–3445.
12. ZegansME, WagnerJC, CadyKC, MurphyDM, HammondJH, et al. (2009) Interaction between bacteriophage DMS3 and host CRISPR region inhibits group behaviors of Pseudomonas aeruginosa. J Bacteriol 191 : 210–219.
13. BabuM, BeloglazovaN, FlickR, GrahamC, SkarinaT, et al. (2011) A dual function of the CRISPR-Cas system in bacterial antivirus immunity and DNA repair. Mol Microbiol 79 : 484–502.
14. SuvarnapunyaAE, LagasseHA, SteinMA (2003) The role of DNA base excision repair in the pathogenesis of Salmonella enterica serovar Typhimurium. Mol Microbiol 48 : 549–559.
15. SternA, KerenL, WurtzelO, AmitaiG, SorekR (2010) Self-targeting by CRISPR: gene regulation or autoimmunity? Trends Genet 26 : 335–340.
16. JorthP, WhiteleyM (2012) An evolutionary link between natural transformation and CRISPR adaptive immunity. MBio 3: e00309–12.
17. VercoeRB, ChangJT, DyRL, TaylorC, GristwoodT, et al. (2013) Cytotoxic chromosomal targeting by CRISPR/Cas systems can reshape bacterial genomes and expel or remodel pathogenicity islands. PLoS Genet 9: e1003454 doi:10.1371/journal.pgen.1003454
18. WestraER, BrounsSJ (2012) The rise and fall of CRISPRs–dynamics of spacer acquisition and loss. Mol Microbiol 85 : 1021–1025.
19. QiLS, LarsonMH, GilbertLA, DoudnaJA, WeissmanJS, et al. (2013) Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152 : 1173–1183.
20. CadyKC, Bondy-DenomyJ, HeusslerGE, DavidsonAR, O'TooleGA (2012) The CRISPR/Cas adaptive immune system of Pseudomonas aeruginosa mediates resistance to naturally occurring and engineered phages. J Bacteriol 194 : 5728–5738.
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2013 Číslo 10- Jak souvisí postcovidový syndrom s poškozením mozku?
- Měli bychom postcovidový syndrom léčit antidepresivy?
- Farmakovigilanční studie perorálních antivirotik indikovaných v léčbě COVID-19
- 10 bodů k očkování proti COVID-19: stanovisko České společnosti alergologie a klinické imunologie ČLS JEP
-
Všechny články tohoto čísla
- Are We There Yet? Recent Progress in the Molecular Diagnosis and Novel Antifungal Targeting of and Invasive Aspergillosis
- Fungal Iron Availability during Deep Seated Candidiasis Is Defined by a Complex Interplay Involving Systemic and Local Events
- Emergence of Azole-Resistant Strains due to Agricultural Azole Use Creates an Increasing Threat to Human Health
- Fungal Adenylyl Cyclase Acts As a Signal Sensor and Integrator and Plays a Central Role in Interaction with Bacteria
- Sensing of the Microbial Neighborhood by
- Antivirulence Therapy for Animal Production: Filling an Arsenal with Novel Weapons for Sustainable Disease Control
- The Cell Biology of : How to Teach Using Animations
- A Structure-Guided Mutation in the Major Capsid Protein Retargets BK Polyomavirus
- RNA Biology in Fungal Phytopathogens
- , , and the Human Mouth: A Sticky Situation
- The Gene Is Essential for Resistance to Human Serum in
- Unisexual Reproduction Drives Evolution of Eukaryotic Microbial Pathogens
- Bacterial Pathogens Activate a Common Inflammatory Pathway through IFNλ Regulation of PDCD4
- Bats and Viruses: Friend or Foe?
- Protein Trafficking through the Endosomal System Prepares Intracellular Parasites for a Home Invasion
- IL-22 Mediates Goblet Cell Hyperplasia and Worm Expulsion in Intestinal Helminth Infection
- B Cells Enhance Antigen-Specific CD4 T Cell Priming and Prevent Bacteria Dissemination following Genital Tract Infection
- Alternative Roles for CRISPR/Cas Systems in Bacterial Pathogenesis
- Chemicals, Climate, and Control: Increasing the Effectiveness of Malaria Vector Control Tools by Considering Relevant Temperatures
- Dengue Vaccines: Strongly Sought but Not a Reality Just Yet
- Feeding Uninvited Guests: mTOR and AMPK Set the Table for Intracellular Pathogens
- Driven Enforced Viral Replication in Dendritic Cells Contributes to Break of Immunological Tolerance in Autoimmune Diabetes
- IL-4Rα-Associated Antigen Processing by B Cells Promotes Immunity in Infection
- A Gammaherpesvirus Uses Alternative Splicing to Regulate Its Tropism and Its Sensitivity to Neutralization
- MicroRNA-155 Promotes Autophagy to Eliminate Intracellular Mycobacteria by Targeting Rheb
- Epigenetic Dominance of Prion Conformers
- MAIT Cells Detect and Efficiently Lyse Bacterially-Infected Epithelial Cells
- The Role of TcdB and TccC Subunits in Secretion of the Tcd Toxin Complex
- A Mechanism for the Inhibition of DNA-PK-Mediated DNA Sensing by a Virus
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Dengue Vaccines: Strongly Sought but Not a Reality Just Yet
- MicroRNA-155 Promotes Autophagy to Eliminate Intracellular Mycobacteria by Targeting Rheb
- Alternative Roles for CRISPR/Cas Systems in Bacterial Pathogenesis
- Feeding Uninvited Guests: mTOR and AMPK Set the Table for Intracellular Pathogens
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



