-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaAn Interaction between KSHV ORF57 and UIF Provides mRNA-Adaptor Redundancy in Herpesvirus Intronless mRNA Export
The hTREX complex mediates cellular bulk mRNA nuclear export by recruiting the nuclear export factor, TAP, via a direct interaction with the export adaptor, Aly. Intriguingly however, depletion of Aly only leads to a modest reduction in cellular mRNA nuclear export, suggesting the existence of additional mRNA nuclear export adaptor proteins. In order to efficiently export Kaposi's sarcoma-associated herpesvirus (KSHV) intronless mRNAs from the nucleus, the KSHV ORF57 protein recruits hTREX onto viral intronless mRNAs allowing access to the TAP-mediated export pathway. Similarly however, depletion of Aly only leads to a modest reduction in the nuclear export of KSHV intronless mRNAs. Herein, we identify a novel interaction between ORF57 and the cellular protein, UIF. We provide the first evidence that the ORF57-UIF interaction enables the recruitment of hTREX and TAP to KSHV intronless mRNAs in Aly-depleted cells. Strikingly, depletion of both Aly and UIF inhibits the formation of an ORF57-mediated nuclear export competent ribonucleoprotein particle and consequently prevents ORF57-mediated mRNA nuclear export and KSHV protein production. Importantly, these findings highlight that redundancy exists in the eukaryotic system for certain hTREX components involved in the mRNA nuclear export of intronless KSHV mRNAs.
Published in the journal: . PLoS Pathog 7(7): e32767. doi:10.1371/journal.ppat.1002138
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1002138Summary
The hTREX complex mediates cellular bulk mRNA nuclear export by recruiting the nuclear export factor, TAP, via a direct interaction with the export adaptor, Aly. Intriguingly however, depletion of Aly only leads to a modest reduction in cellular mRNA nuclear export, suggesting the existence of additional mRNA nuclear export adaptor proteins. In order to efficiently export Kaposi's sarcoma-associated herpesvirus (KSHV) intronless mRNAs from the nucleus, the KSHV ORF57 protein recruits hTREX onto viral intronless mRNAs allowing access to the TAP-mediated export pathway. Similarly however, depletion of Aly only leads to a modest reduction in the nuclear export of KSHV intronless mRNAs. Herein, we identify a novel interaction between ORF57 and the cellular protein, UIF. We provide the first evidence that the ORF57-UIF interaction enables the recruitment of hTREX and TAP to KSHV intronless mRNAs in Aly-depleted cells. Strikingly, depletion of both Aly and UIF inhibits the formation of an ORF57-mediated nuclear export competent ribonucleoprotein particle and consequently prevents ORF57-mediated mRNA nuclear export and KSHV protein production. Importantly, these findings highlight that redundancy exists in the eukaryotic system for certain hTREX components involved in the mRNA nuclear export of intronless KSHV mRNAs.
Introduction
Post-transcriptional events which regulate mRNA biogenesis are fundamental to the control of gene expression [1]. A nascent mRNA is therefore steered through multimeric RNA-protein complexes that mediate its capping, splicing, polyadenylation, nuclear export and ultimately its translation [2], [3]. A key aspect of these post-transcriptional events is that they are intrinsically linked [4]. For example, the act of splicing is coupled to the deposition of two distinct multiple protein complexes onto the mRNA which are involved in further processing events, namely the human transcription and export complex (hTREX) [5]–[7] and the exon-exon junction complex (EJC) [8]. The hTREX complex associates with the 5′end of the first exon by virtue of interactions with the cap-binding complex, and facilitates the nuclear export of the bulk mRNA through the TAP-mediated pathway [6]. In contrast, the EJC is deposited 20–24 nucleotides upstream of each exon-exon boundary and plays a role in mRNA surveillance [9] and translation enhancement [10]–.
The TREX complex is conserved from yeast to metazoans [3], [13], [14]. The human TREX complex comprises several core components: Aly (a NXF/TAP adaptor protein); UAP56 (a DEAD-box helicase); Tex1 (a protein of unknown function) and the stable multi-protein hTHO complex (hHpr1, hTho2, fSAP79, fSAP35 and fSAP24) [3]. Moreover, recent proteomic analysis has identified CIP29/Tho1 as a hTREX component that is conserved in both yeast and metazoans [15]. The precise mechanism of how hTREX is assembled onto the mRNA is not fully understood or characterised. UAP56 is thought to associate with mRNA at an early stage during the assembly of the spliceosome and functions to mediate the recruitment of Aly, CIP and the THO complex in an ATP-dependent manner to form hTREX [15], [16]. This involvement of the spliceosome in hTREX assembly reflects the splicing-dependent nature of mRNA nuclear export [16]–[18]. In addition to splicing, a functional 7-methylguanosine 5′ cap is also essential for hTREX recruitment, due to an interaction between Aly and the cap-binding complex protein, CBP80 [6]. Such cap-dependent recruitment of the export complex affords the mRNA polarity upon exiting the nuclear pore. Once assembled onto the mRNA, hTREX then instigates the recruitment of the nuclear export factor TAP, and its heterodimeric partner, p15, at the nuclear periphery, via a direct interaction with Aly [18]–[20]. TAP binding then elicits a RNA handover mechanism which results in the remodelling of the protein-mRNA interactions within the ribonucleoprotein complex [21]. Subsequently, TAP associates with the nucleoporins through central and carboxy-terminal domains, directing the ribonucleoprotein though the nuclear pore complex into the cytoplasm [22].
Surprisingly, considering the central role played by Aly in TAP recruitment, gene knockdown experiments performed in Drosophila melanogaster and Caenorhabditis elegans have shown that only UAP56, in contrast to Aly and THO-complex proteins, is required for bulk mRNA nuclear export [23]–[25]. Moreover, a genome-wide RNAi study in D. melanogaster reported that the conserved THO-complex was only required by a subset of transcripts for nuclear export [26], [27]. This data indicates a degree of redundancy is present in these pathways and suggests the existence of additional export adaptor proteins which are involved in bulk mRNA nuclear export. In support of this idea, a novel mRNA export adaptor protein has recently been identified that utilises the UAP56/TAP-mediated pathway. UAP56-interacting factor (UIF) was initially identified in silica, by virtue of sequence similarity to the characterised UAP56-binding domain found in Aly [28]. Notably, cellular expression levels of UIF appear to be linked in vivo to the relative expression of Aly, as miRNA-mediated depletion of Aly led to a dramatic increase in UIF expression. Importantly, simultaneous depletion of both Aly and UIF leads to a dramatic nuclear accumulation of bulk mRNA [28]. Therefore, it is believed that Aly and UIF bind independently to the same mRNA providing multiple export adaptor proteins to recruit multiple TAP molecules to ensure efficient mRNA nuclear export. Moreover, the observation that UIF expression increases in Aly-depleted cells is believed to be a redundancy mechanism that ensures cellular survival should Aly expression be compromised.
Given the importance of the formation of multimeric mRNA-protein complexes in mRNA biogenesis, it is not surprising that viruses manipulate and exploit these pathways. This is particularly important for herpesviruses which replicate in the host-cell nucleus and express numerous lytic intronless mRNAs. Due to the reliance of herpesviruses on the host cell machinery for efficient processing of their mRNAs, an immediate issue arises concerning the mechanism by which the viral intronless mRNAs are efficiently exported from the nucleus, given that the majority of cellular bulk mRNA nuclear export is intimately linked, and dependent upon, splicing [29]. We have investigated this potential roadblock to herpesvirus gene expression and replication in the gamma-2 herpesvirus, Kaposi's sarcoma-associated herpesvirus (KSHV) [30], which is associated with the AIDS-related malignancies Kaposi's sarcoma (KS), primary effusion lymphoma (PEL) and multicentric Castleman's disease [31]–[33]. To circumvent the roadblock of efficient intronless viral mRNA nuclear export, KSHV encodes a multi-functional protein termed ORF57/Mta. KSHV ORF57 is a functionally conserved protein found in all herpesviruses that plays a pivotal role in enhancing viral gene expression at a post-transcriptional level [34], [35]. ORF57 has been implicated in multiple steps of RNA biogenesis, including enhancing viral splicing, protecting viral RNAs from degradation to enhancing viral mRNA nuclear export and translation [36]–[39].
We have demonstrated that KSHV ORF57 promotes the nuclear export of intronless viral mRNAs via the TAP-mediated pathway, by directly interacting with the hTREX export adaptor, Aly [37]. Moreover, we investigated the composition and assembly of these export-competent intronless KSHV ribonucleoprotein particles (vRNP) and showed that ORF57 functions to recruit the complete hTREX complex to intronless viral mRNA, an event that is essential for viral intronless mRNA export and KSHV replication [37]. Furthermore, these properties are also conserved in other gamma-2 herpesvirus ORF57 homologues, such as the Herpesvirus saimiri (HVS) ORF57 protein [40], [41]. These data suggest that Aly is essential for ORF57-mediated KSHV intronless mRNA export, as well as playing an important role in mRNA nuclear export in other herpesviruses. However, experiments involving siRNA-mediated depletion of Aly report only a modest effect on ORF57-mediated KSHV intronless mRNA export, although only partial depletion of Aly was achieved [42]. This data correlates with depletion-related studies on the role of Aly in mRNA export in higher eukaryotes where, surprisingly, Aly has been shown to be dispensable in mRNA export [23], [24]. Similar stories are also evident for other herpesviruses mRNA export proteins. For example, an observed interaction between ICP27 (the HSV-1 ORF57 homologue) and Aly was initially reported as important for HSV-1 mRNA export [43]. However, subsequent functional studies using siRNA-mediated depletion of Aly led to the authors suggesting that Aly is not essential for ICP27-mediated HSV-1 mRNA export [44]. This suggests that additional cellular mRNA export proteins play important roles in herpesvirus intronless mRNA export. Indeed, recently it has been demonstrated that the SR proteins, SRp20 and 9G8, can contribute to efficient export of herpes simplex virus 1 mRNAs [45].
Herein we report a novel interaction between the KSHV ORF57 protein and the recently identified mRNA export adaptor protein, UIF. Moreover, we provide data to suggest that ORF57 may preferentially bind Aly compared to UIF. Furthermore, we investigate whether the linked expression of UIF and Aly plays a role in the apparent redundancy of Aly in herpesvirus intronless mRNA nuclear export. We provide the first evidence that the ORF57-UIF interaction enables the recruitment of the complete hTREX and the nuclear export factor, TAP, to KSHV intronless mRNA in Aly-depleted cells. Strikingly, we demonstrate that depletion of both Aly and UIF inhibit the formation of an ORF57-mediated nuclear export competent ribonucleoprotein particle and consequently prevent ORF57-mediated nuclear export of intronless viral mRNAs and KSHV protein production. Importantly, these findings highlight that redundancy exists in the eukaryotic system for certain hTREX components involved in the mRNA nuclear export of intronless KSHV mRNAs.
Results
KSHV ORF57 interacts with the UAP56 interacting protein, UIF
KSHV ORF57 interacts directly with the cellular export adaptor protein Aly to recruit cellular hTREX, comprising UAP56 and the hTHO complex, onto a viral intronless mRNA to form an export competent ribonucleoprotein particle [37]. However, ORF57 and homologues can mediate nuclear export of an intronless viral mRNA in Aly-depleted cells [42], suggesting that alternative export pathways may be targeted by the ORF57 protein. Therefore, to determine whether ORF57 interacts with alternative export adaptor proteins, GST-pulldown and co-immunoprecipitations assays were performed to assess if ORF57 interacted with the recently identified UAP56 interacting protein, UIF. Initially, recombinant GST-, GST-UAP56 or GST-ORF57 fusion proteins were produced and used in GST-pulldown assays. It must be noted however, that although full length GST-ORF57 is produced, a large proportion of the product is degraded as previously observed [37]. GST-pulldown experiments were therefore performed using equal amounts of total protein from each GST construct immobilised to beads followed by incubation with 293T cell lysates transfected with pUIF-Flag. Analysis showed that UIF interacted with both UAP56 and KSHV ORF57 (Figure 1A). To confirm these results co-immunoprecipitation experiments were also performed. 293T cells were transfected with either pEGFP, pUAP56-myc or pORF57GFP in the presence of pUIF-Flag and used in co-immunoprecipitation experiments with GFP or UAP56-specific antibodies. Results confirmed the interaction between UIF and KSHV ORF57 (Figure 1B).
Fig. 1. KSHV ORF57 interacts directly with UIF. 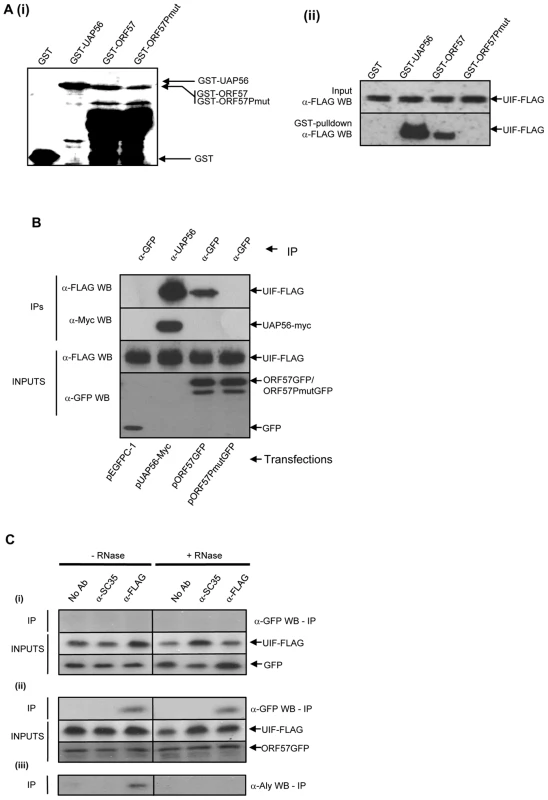
(A) (i) Bacterially expressed GST-, GST-UAP56-, GST-ORF57- and GST-ORF57Pmut-bound to glutathione agarose beads and separated by SDS-PAGE, proteins were visualised by coomassie staining. (ii) GST Pull-down assays were performed using pUIF-FLAG transfected cell lysates. Precipitated UIF protein was detected by western blot analysis using a FLAG-specific antibody. UIF-FLAG transfected cell lysate served as a loading control (Input). (B) Immunoprecipitations using GFP- or UAP56-specific antibodies were performed using cell lysates cotransfected with UIF-FLAG in the presence of either pEGFP, pUAP56-Myc, pORF57GFP or pORF57PmutGFP. Precipitated UIF-FLAG, UAP56-myc, GFP, ORF57GFP and ORF57PmutGFP were detected by western blot analysis using antibodies specific to FLAG, myc or GFP. Transfected cell lysates served as a loading control (Inputs). (C) Immunoprecipitations were performed in the presence or absence of RNase, using a no antibody control, (SC35)- or FLAG-specific antibodies on cell lysates cotransfected with pUIF-FLAG in the presence of either (i) pGFP and (ii) pORF57GFP. Precipitated proteins were detected by western blotting using GFP- or FLAG-specific antibodies. (iii) ORF57-transfected co-immunoprecipitations were also immunoblotted with an Aly-specific antibody to confirm the activity of the RNase digestion to abolish the RNA-dependent interaction between Aly and UIF. We have previously identified an ORF57 mutant protein, ORF57PmutGFP, which is unable to interact with Aly and therefore recruit the remainder of the hTREX complex onto viral intronless mRNAs [37]. Moreover, we demonstrated that this mutant is unable to efficiently export viral intronless mRNA from the nucleus suggesting that the recruitment of a complete hTREX complex is required for ORF57-mediated nuclear export. ORF57PmutGFP contains site-directed alterations of two proline residues within a PxxP poly-proline motif, situated in the previously identified minimal Aly-binding domain encompassing residues 181–215. We have previously demonstrated that although ORF57PmutGFP is unable to bind Aly, it still retains features of the wild type ORF57 protein, namely localising to nuclear speckles, the ability to homodimerise, bind KSHV RTA and bind intronless viral mRNA [37]. To assess whether this mutant could interact with UIF, GST-pulldown experiments and co-immunoprecipitation experiments were performed as described above using GST-ORF57Pmut and pORF57PmutGFP, respectively. In both cases the mutant ORF57 protein, which fails to bind Aly, also lacks the ability to interact with UIF (Figure 1A and 1B). Importantly, this suggests that the failure of ORF57PmutGFP to recruit hTREX and efficiently export intronless viral mRNAs from the nucleus may be due to the inability to bind either Aly or UIF.
To determine if the interaction between ORF57 and UIF depended on RNA bridging, co-immunoprecipitation experiments were repeated in the absence and presence of RNase. 293T cells were transfected with either pEGFP or pORF57GFP in the presence of pUIF-Flag and co-immunoprecipitation assays were performed using a polyclonal Flag-specific antibody. In addition, no antibody and a negative control antibody (α-SC-35) were also used in the analysis. ORF57 was readily precipitated using the Flag-specific antibody in contrast to negative controls. Moreover, the observed interaction was still detected in the presence of RNase suggesting the interactions are not due to RNA bridging (Figure 1C). To ensure the RNase treatment was effective the immunoprecipitations were also blotted with an Aly-specific antibody. Results show that the UIF-Aly interaction is RNA dependent as previously described [28], [36].
In order to address potential overexpression artefacts of the above co-immunoprecipitation experiments and also determine whether ORF57 interacts with UIF during KSHV lytic replication, latently-infected BCBL-1 cells remained uninduced or reactivated using the phorbol ester, TPA. Lytic expression was confirmed by the detection of ORF57 using Western blot analysis in the reactivated samples (Figure 2). Uninduced and reactivated cell lysates were then incubated with no antibody control, ORF57 - or UIF-specific antibodies. Reciprocal western blot analysis using the antibodies in reverse demonstrated that ORF57 interacts with UIF during KSHV lytic replication (Figure 2). Therefore, these data provide the first evidence of a viral protein associating with UIF.
Fig. 2. ORF57 interacts directly with UIF during KSHV lytic replication. 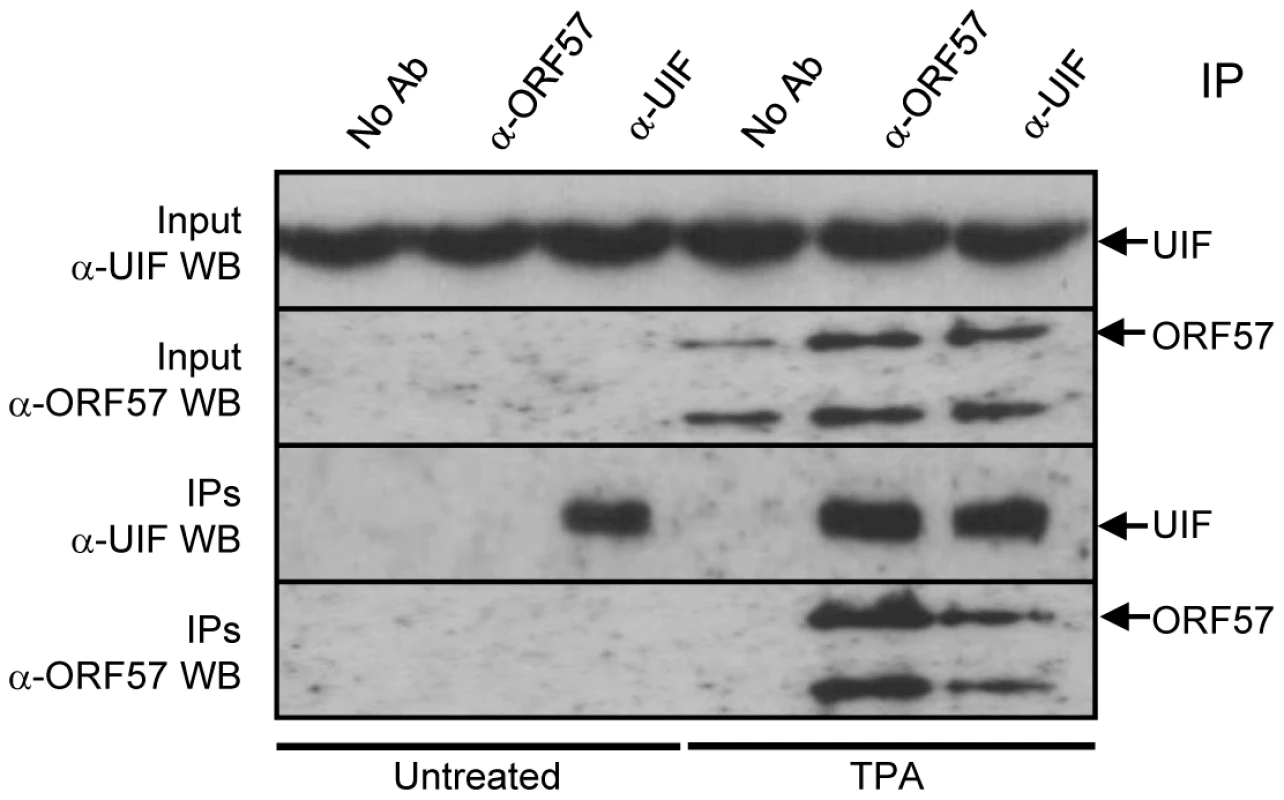
Immunoprecipitations were performed on unreactivated or reactivated BCBL-1 cells using no antibody control, ORF57- or UIF-specific antibodies. Precipitated proteins were detected by western blot using UIF- and ORF57-specific antibodies. BCBL-1 cells were reactivated by the addition of TPA (20 ng/ml). ORF57 is a nucleocytoplasmic protein that is predominately observed in the nucleus, specifically colocalising with nuclear speckle and nucleoli-associated proteins [42], [46]. Therefore, we were interested to determine whether ORF57 colocalises with UIF in either of these subnuclear domains. To this end, 293T cells were cultured on poly-L lysine coated coverslips and transfected with either pORF57-mCherry or pUIF-GFP alone or in combination. The subcellular localisation of ORF57 and UIF were observed via direct fluorescence, in addition indirect-immunofluorescence was performed to identify nuclear speckles and the nucleolus using SC35 - (Figure 3Bii) and B23 - (Figure 3Dii) specific antibodies, respectively. As previously observed ORF57 colocalises with both subnuclear domain markers (Figure 3Bii and 3Dii). Moreover, UIF was also found to localise with these subnuclear structures and also colocalises with the ORF57 protein (Figure 3B and 3D). However, results demonstrate that the majority of ORF57 and UIF colocalise in the nucleolus whereas only a proportion of ORF57 and UIF colocalise with the nuclear speckle marker, SC35.
Fig. 3. KSHV ORF57 colocalises with UIF in the nucleus and nucleolus. 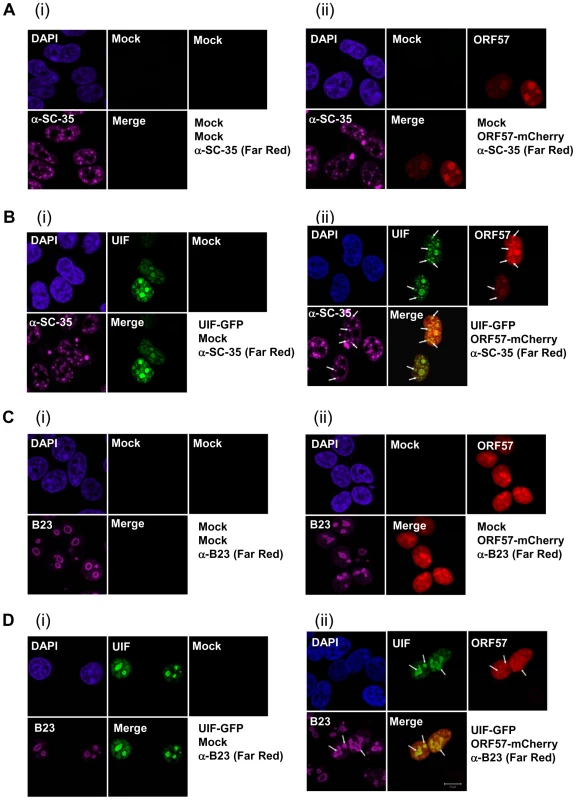
293T cells were either mock (Ai, Ci), or transfected with pORF57-mCherry (Aii, Cii) or pUIF-GFP (Bi, Di) and in combination (Bii, Dii), incubated for 24 h, fixed and immunofluoresence staining performed. ORF57 and UIF were visualised by direct fluorescence of Cherry and GFP, respectively. Subcellular localisation within the nuclear speckles or nucleolus was confirmed using antibodies specific to SC-35 (A, B) or B23 (C, D), respectively. A merge of the mCherry/GFP channels is also included for all images. White arrows indicate nuclear speckles (Bii) or nucleolar localisation (Dii). ORF57 facilitates the loading of UIF onto KSHV intronless mRNAs
One major difference between the mRNA export adaptor proteins Aly and UIF is the mechanism they utilise to be loaded onto mRNA. Aly has been shown to associate with mRNA in a UAP56 and splicing-dependent manner [47], in contrast UIF is loaded onto mRNA via the histone chaperone FACT [28]. We have previously demonstrated that ORF57 is required for the recruitment of Aly and the remainder of the hTREX complex onto viral intronless mRNAs, therefore we were intrigued to determine if UIF could associate with intronless viral mRNAs in an ORF57-independent manner using RNA-immunoprecipitation (RNA-IP) assays. A vector expressing KSHV ORF47 (a late structural intronless gene) was transfected into 293T cells with either pEGFP or pORF57GFP. Total cell lysates were then used in immunoprecipitations performed with either No, Y14 - (negative control), UIF - or GFP-specific antibodies and the amount of ORF47 precipitated was measured by qRT-PCR. RNA-IPs performed on cell extracts transfected with pORF47 and pEGFP failed to show an interaction between UIF and the viral intronless ORF47 mRNA (Figure 4). In contrast, extracts from cells transfected with both pORF47 and pORF57GFP displayed a clear interaction between UIF, ORF57GFP and the intronless viral ORF47 mRNA (Figure 4). These data show that although UIF can associate with cellular spliced and unspliced single exon cellular mRNAs, ORF57 is required for the recruitment of UIF onto intronless viral mRNA.
Fig. 4. ORF57 is required for the recruitment of UIF to a KSHV intronless mRNA. 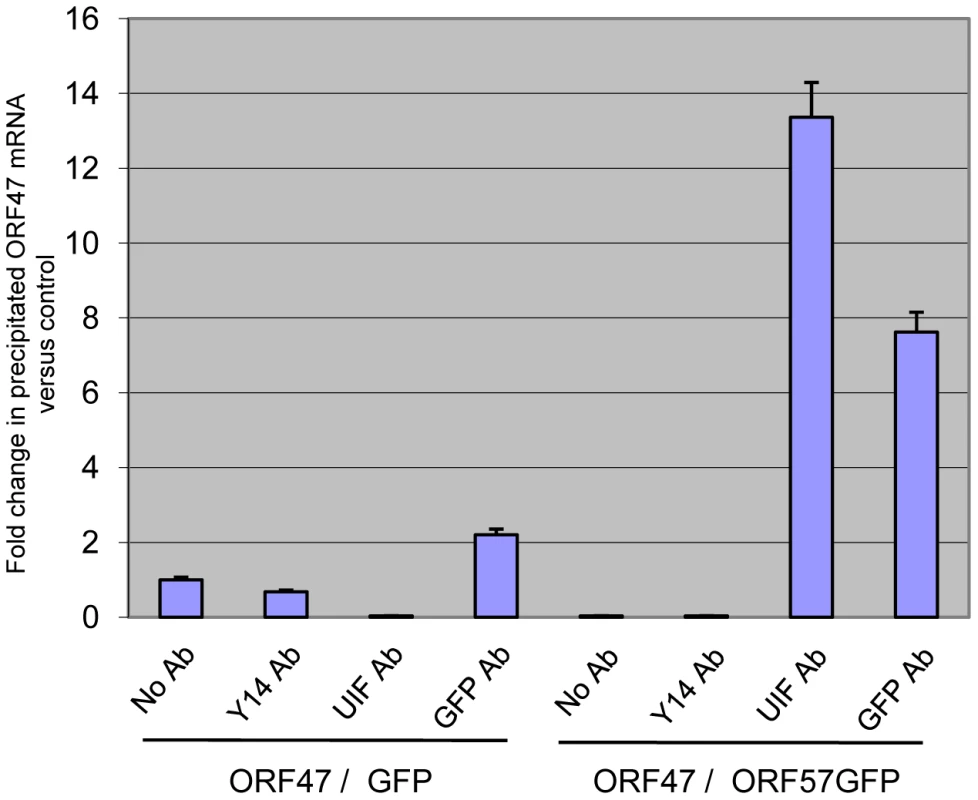
RNA immunoprecipitations were performed in 293T cells cotransfected with KSHV pORF47 in the presence of either pEGFP or pORF57GFP. After UV crosslinking cell lysates were immunoprecipitated using GFP- or UIF-specific antibodies. In addition, no antibody and a negative control Y14 antibody were also used as controls. Protein was then digested, and immunoprecipitated RNA was analysed by qRT-PCR. UIF links ORF57 to the hTREX component, UAP56
We have previously demonstrated that the nuclear export adaptor protein, Aly, is recruited to viral intronless mRNAs in a splicing-independent manner by directly interacting with ORF57. Once bound it then leads to the recruitment of the remaining components of hTREX, in turn leading to efficient nuclear export of these viral intronless mRNAs via a TAP-mediated pathway [37]. We therefore next sought to determine if UIF can perform a similar function by linking ORF57 to hTREX components such as UAP56. Initially, we determined whether ORF57 interacted with UIF directly using GST-pulldown assays. Recombinant GST - and GST-ORF57 proteins were immobilised to beads and incubated with purified recombinant UIF-6xHis or a negative control purified recombinant HVS ORF73-6xHis protein [48]. UIF-6xHis was precipitated by GST-ORF57 but not the negative GST control, moreover ORF73-6xHis failed to interact with either GST or GST-ORF57 (Figure 5A). These data provide further support for the direct interaction between ORF57 and UIF.
Fig. 5. ORF57 is linked to the hTREX complex by UIF; however, ORF57 preferentially interacts with Aly over UIF. 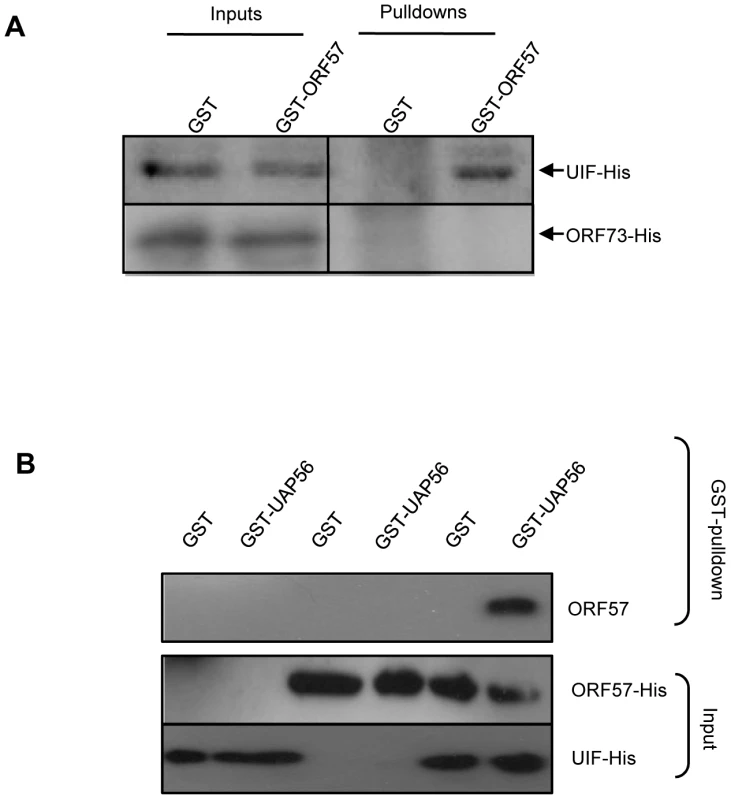
(A) Recombinant GST, and GST-ORF57 were bound to glutathione agarose beads and GST Pull-down assays performed using purified recombinant UIF-His or control ORF73-His proteins. Precipitated proteins and inputs were analysed by western blotting using a His-specific antibody. (B) Reconstitutive GST pulldowns were performed using GST or GST-UAP56 bound to glutathione agarose beads and incubated with recombinant purified ORF57-His or Aly-His alone or in combination. Precipitated proteins and inputs were analysed by western blotting using a His-specific antibody. Given the fact that ORF57 and UIF interact directly, we next determined whether UIF can bridge the interaction between ORF57 and hTREX components, such as UAP56, which we have previously shown fails to interact with ORF57 directly [37]. Reconstitutive GST-pulldowns were therefore performed using recombinant GST - and GST-UAP56 proteins immobilised to beads and incubating with either purified recombinant ORF57-6xHis or purified recombinant UIF-6xHis alone or in combination. No interaction with GST or GST-UAP56 was observed in the presence of ORF57-6xHis alone. In contrast, an interaction between GST-UAP56 and ORF57 was observed in the presence of purified UIF-6xHis protein (Figure 5B), suggesting that UIF can facilitate the formation of the ORF57-hTREX complex. This provides the first evidence to demonstrate that UIF could function to assemble the hTREX complex on viral intronless mRNAs.
ORF57 may preferentially bind Aly over UIF
The above data demonstrate that UIF interacts directly with ORF57 and suggest that it can function to bridge an interaction between ORF57 and the remaining hTREX components. This mechanism is similar to our previous observations regarding the functional significance of the Aly-ORF57 interaction, and therefore leads to the intriguing question of whether ORF57 has a preference for Aly binding over UIF or vice versa. To address this question we performed competitive GST pulldown assays. Recombinant GST-ORF57 protein was immobilised to beads and incubated with a constant amount (1 µg) of purified recombinant Aly-6xHis protein, in addition the pulldown was spiked with increasing amounts of purified recombinant UIF-6xHis protein (0, 0.5, 1, 2, 3 µg). Western blot analysis was then performed using an Aly-specific antibody. Results demonstrate that the binding of Aly to GST-ORF57 is only slightly reduced in the presence of increasing amount of UIF (Figure 6Ai), suggesting that UIF cannot out-compete Aly for ORF57 binding. Similar spiked experiments were performed using a constant amount of UIF and increasing amounts of Aly. In contrast, results showed that even low quantities of Aly led to a dramatic loss of UIF binding to the ORF57 protein (Figure 6Aii). These results reveal that Aly can out-compete UIF for ORF57 binding, suggesting that ORF57 may preferentially bind Aly to form an export competent ribonucleoprotein particle.
Fig. 6. KSHV ORF57 may preferentially bind Aly over UIF. 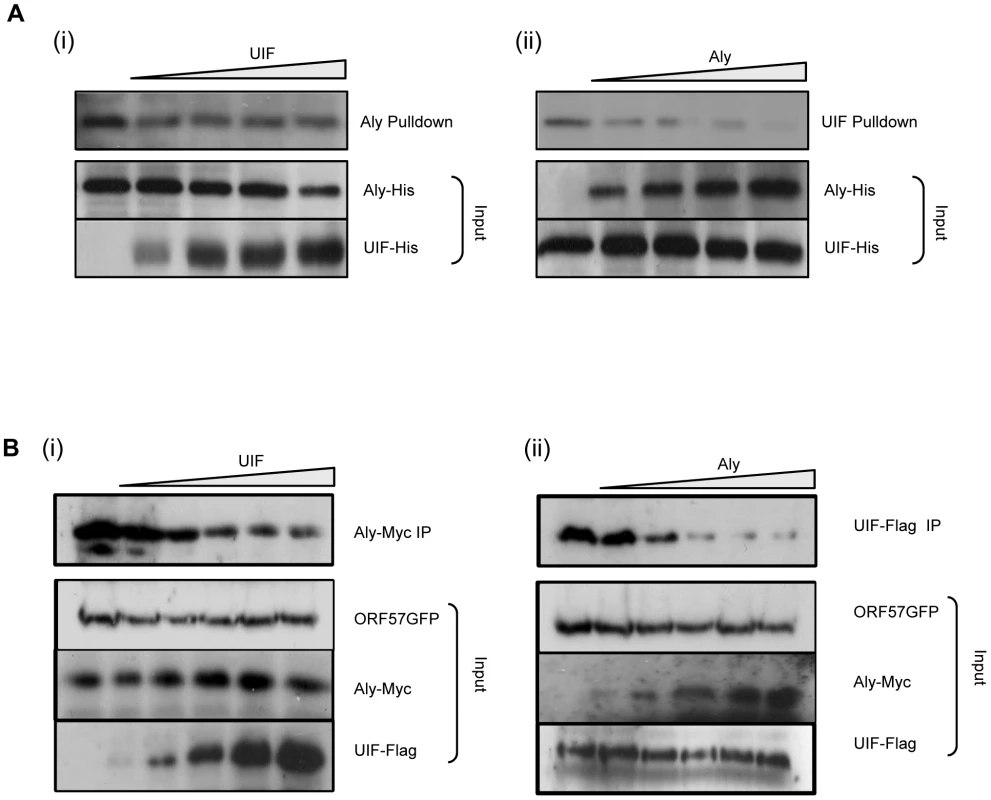
(A) Competition assays were performed using recombinant GST-ORF57 bound to glutathione agarose beads and incubated with (i) a constant amount of purified Aly-His (1 µg) and increasing amounts of purified UIF-His (0, 0.5, 1, 2, 3 µg), (ii) a constant amount of purified UIF-His (1 µg) and increasing amounts of purified Aly-His (0, 0.5, 1, 2, 3 µg), Precipitated protein and inputs were analysed by western blotting using a His-specific antibody. (B) Dose-dependent co-immunoprecipitations were performed using 293T cells cotransfected with (i) 0.5 ug of pORF57GFP and 0.5 ug of pAly-myc, in addition to increasing amounts (0, 0.1, 0.5, 0.8, 1.2 ug) of pUIF-Flag or (ii) 0.5 ug of pORF57GFP and 0.5 ug of pUIF-Flag, in addition to increasing amounts (0, 0.1, 0.5, 0.8, 1.2 ug) of pAly-myc, empty vector was also added to ensure a similar amount of DNA was transfected in each sample. After 24 hours, cell lysates were incubated with GFP-TRAP-Affinity agarose beads and the amount of precipitated (i) Aly or (ii) UIF was identified by immunoblotting with Myc- or Flag-specific antibodies, respectively. Western blots for input loading are also shown for ORF57, Aly and UIF constructs. However as shown in Figure 1A, although bacterial expression of full length GST-ORF57 results in a full length ORF57 protein, a large proportion of degraded products are also produced. Therefore, to further assess the possibility that ORF57 may interact with Aly preferentially over UIF, dose-dependent coimmunoprecipitation assays were performed. To this end, 293T cells were cotransfected with 0.5 ug of pORF57GFP and 0.5 ug of pAly-myc, in addition to increasing amounts of pUIF-Flag (0, 0.1, 0.5, 0.8, 1.2 ug). After 24 hours, cell lysates were incubated with GFP-TRAP-Affinity agarose beads and the amount of precipitated Aly was identified by immunoblotting with a Myc-specific antibody. Results again show that the binding of Aly is only slightly reduced in the presence of increasing amounts of UIF (Figure 6Bi). Moreover, reciprocal dose-dependent coimmunoprecipitations were performed using 0.5 ug of pORF57GFP and 0.5 ug of pUIF-Flag, in addition to increasing amounts of pAly-myc (0, 0.1, 0.5, 0.8, 1.2 ug). In contrast, results suggest that higher concentrations of Aly can significantly reduce the amount of precipitated UIF (Figure 6Bii). These results corroborate the above GST pulldown assays and suggest that ORF57 may preferentially bind Aly over UIF to form an export competent ribonucleoprotein particle.
Depletion of both Aly and UIF is required to inhibit ORF57-mediated virus ribonucleoprotein particle formation
Having established that both Aly and UIF can bridge an interaction between ORF57 and hTREX components, such as UAP56, we next sought to determine the effect of depleting Aly and UIF either singularly, or in combination, on the ability of ORF57 to form an export competent ribonucleoprotein particle containing the complete hTREX complex and the nuclear export factor TAP. To this end, we have utilised doxycycline inducible 293 cell lines expressing miRNAs targeting Aly, UIF or both Aly and UIF [28]. Effective depletion of Aly, UIF or both proteins can be observed after 72 hours post doxycycline induction (Figure 7A). However, a caveat of this type of experiment is that depletion of multiple mRNA export factors in combination may firstly be toxic to the host cell and second inhibit the expression of ORF57 itself as recently reported [49]. Characterisation of the cell viability and growth of the cells depleted with both Aly and UIF has previously been performed and results show they are viable and grow for 4 days post knockdown prior to cell death at day 6 [28]. Therefore all experiments using these cell lines were performed in this 4 day window. Moreover, to ensure ORF57 protein production, cells were transfected at 48 hours prior to complete Aly or UIF depletion at 72 hours.
Fig. 7. Knockdown of both Aly and UIF impairs the ability of ORF57 form an export competent viral ribonucleoprotein particle. 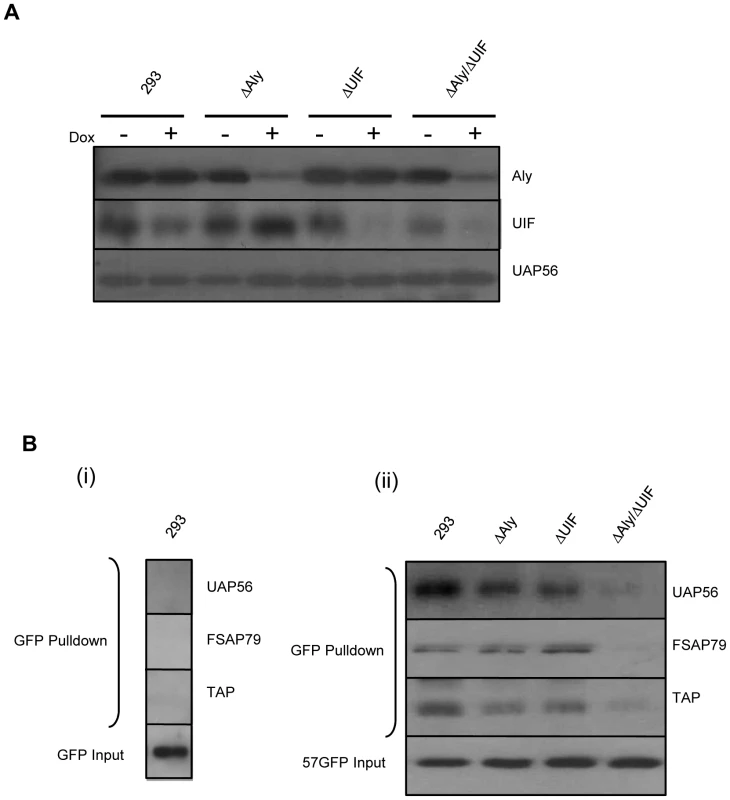
(A) 293, 293ΔAly, 293ΔUIF and 293ΔAlyΔUIF cells were mock treated or treated with 2 µg/ml doxycyclin grown for 72 h. Cell lysates were analysed by western blotting using Aly-, UIF- and UAP56-specific antibodies. (B) (i) 293 cells were transfected with pEGFP and grown for a further 24 h. Cell lysates were incubated with GFP-TRAP-Affinity beads and after washing, the precipitated proteins were detected by western blotting using GFP-, UAP56-, FSAP79- and TAP-specific antibodies. (ii) 293, 293ΔAly, 293ΔUIF and 293ΔAlyΔUIF cells were treated with 2 µg/ml doxycyclin and grown for 48 h before being transfected with pGFP-ORF57 and grown for a further 24 h. Cell lysates were incubated with GFP-TRAP-Affinity beads and after washing, the precipitated proteins were detected by western blotting using GFP-, UAP56-, FSAP79- and TAP-specific antibodies. Transfected cell lysates were used as an input control. To assess viral ribonucleoprotein particle formation, wild type 293 cells and each miRNA-targeted cell line were induced with doxycycline to deplete the respective proteins and after 48 hours' induction, each cell line was transfected with pORF57GFP. After a further 24 hours when maximum Aly and UIF depletion has occurred, cell lysates were used in co-immunoprecipitation experiments using GFP-TRAP-Affinity agarose beads. Western blot analysis was then performed using UAP56-, FSAP79 - (a hTHO complex component) and TAP-specific antibodies. As a negative control, GFP was also transfected into the wild type 293 cell line and co-immunoprecipitations performed using GFP-TRAP-Affinity agarose beads, no interactions were observed with any of the hTREX components or TAP. However, results showed that expression of ORF57 in the wild type 293 cell line led to the precipitation of UAP56, FSAP79 and TAP suggesting that ORF57 expression leads to the formation of an export competent ribonucleoprotein particle (Figure 7B). Similar complex formation was observed in cell lines depleted singularly for Aly and UIF, where ORF57 can precipitate UAP56, FSAP79 and TAP (Figure 7B). In contrast, depletion of Aly and UIF in combination significantly reduced the interaction between ORF57 and the hTREX components and the nuclear export factor TAP. Importantly, these data demonstrate that either Aly or UIF are required for the formation of an ORF57-mediated nuclear export competent ribonucleoprotein particle.
Either Aly or UIF is required for ORF57-mediated mRNA nuclear export and KSHV protein production
The above data suggest that ORF57 must interact with either export adaptor protein, Aly or UIF, to recruit hTREX and the nuclear export protein TAP, to form an export competent ribonucleoprotein particle. Therefore, we next determined whether both UIF and Aly were required for efficient ORF57-mediated nuclear export of viral intronless mRNAs. To this end, we assessed the ability of ORF57 to enhance the nuclear export of the KSHV intronless ORF47 mRNA, using a previously described assay to compare the accumulation of ORF47 mRNA in the cytoplasm [46]. Essentially, cells are transfected with a plasmid expressing the intronless KSHV ORF47 gene in addition to either GFP or wild type ORF57 constructs. After 24 hours, RNA is extracted from total and cytoplasmic fractions and RNA levels quantified using qRT-PCR. Total RNA levels are assessed to ensure similar expression levels of the ORF47 mRNA in each sample, where an increase in cytoplasmic levels of ORF47 mRNA signifies an increase in ORF57-mediated mRNA export levels. Therefore, to assess the ability of ORF57 to export ORF47 mRNA from the nucleus in the absence of either UIF or Aly or both, wild type 293 cells and each miRNA-targeted cell line were induced with doxycycline to deplete the respective proteins and after 48 hours induction, each cell line was transfected with pORF57GFP and pORF47. Again, this allowed sufficient time to express ORF57 prior to optimal export adaptor protein depletion. After a further 24 hours, RNA was extracted from total and cytoplasmic fractions and ORF47 levels assessed by qRT-PCR. Results demonstrated that ORF47 mRNA levels from total cell fractions are similar in wild type and the depleted cell lines. Moreover, in the control 293 cell line ORF47 mRNA accumulates in the cytoplasm in the presence of ORF57 as previously described [46]. Similarly, mRNA can accumulate in the cytoplasm of cells depleted singularly for Aly and UIF, however, a reduction in export efficiency was observed of approximately 40% and 23% of wild type levels, respectively (Figure 8A). In contrast, depletion of both Aly and UIF together led to a dramatic reduction of ORF47 mRNA accumulation in the cytoplasm with an 80% decrease compared to wild type levels (Figure 8A).
Fig. 8. Knockdown of both Aly and UIF impairs the ability of ORF57 to export KSHV intronless mRNA from the nucleus and reduces KSHV late protein production. 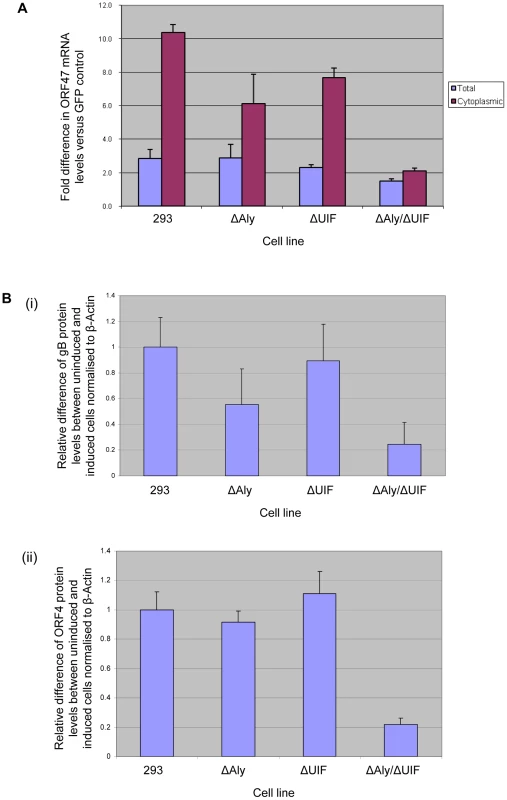
(A) 293, 293ΔAly, 293ΔUIF and 293ΔAlyΔUIF cells were treated with 2 µg/ml doxycyclin and grown for 48 h before being transfected with pORF47 in the presence of either pEGFP or pGFP-ORF57. 24 h post-transfection RNA was isolated from total and cytoplasmic fractions and relative levels were analysed by qRT-PCR using GAPDH as a reference. Fold increase was determined by ΔΔcT and statistical significance by a non-paired t-test. Data from 3 independent experiments is presented as fold increase versus pGFP-transfected controls. (B) 293, 293ΔAly, 293ΔUIF and 293ΔAlyΔUIF cells were mock treated or treated with 2 µg/ml doxycyclin and grown for 48 h before infection with KSHV at a MOI = 1. At 48 h post-infection cells lysates were analysed by western blot using antibodies specific to KSHV (i) gB and (ii) ORF4 proteins. Results are shown of densitometry analysis of the western blots from 3 independent experiments carried out using the ImageJ software and is shown as expression of gB or ORF4 between uninduced and induced cell lines relative to β-actin. We next tested whether the observed reduction in the ability of ORF57 to export intronless mRNAs from the nucleus in cell lines depleted for Aly and UIF had any effect on KSHV protein production. To this end, the wild type 293 cells and each miRNA-targeted cell line were induced with doxycycline to deplete the respective proteins and after 48 hours induction, each cell line was infected with recombinant KSHV at a MOI = 1. This time point was used to allow sufficient time to express ORF57 prior to optimal export adaptor protein depletion. After a further 48 hours, the cell lysates were analysed by immunoblotting using KSHV glycoprotein B - and ORF4-specific antibodies. Results showed that gB protein expression in cell lines singularly depleted for either Aly or UIF was reduced by ∼42% and ∼10%, respectively, whereas little or no reduction was observed for ORF4 protein levels in the singularly depleted cells. Strikingly, however depletion of both Aly and UIF led to a dramatic reduction in both gB and ORF4 expression levels of 78% and 79%, respectively (Figure 8B). These results suggest that depletion of UIF has limited if any effect of virus replication, however, depletion of UIF together with Aly had a dramatic negative effect on KSHV protein production. However, it must be noted that this reduction in protein levels may also stem from altered levels of one or more key cellular proteins involved in KSHV lytic protein production.
Taken together, our data suggest that either one of the cellular nuclear export adaptor proteins, Aly or UIF, is required for the formation of an ORF57-mediated nuclear export competent ribonucleoprotein particle which is essential for KSHV protein production.
Discussion
The nuclear export of bulk mRNA is mediated by the conserved heterodimeric export receptor, TAP/p15 [3]. Cellular mRNAs gain access to TAP/p15 via interaction with a group of RNA-binding proteins termed export adaptors. The first mRNA export adaptor to be identified in higher systems was Aly/REF, and subsequent work from a number of groups led to the current model where Aly is recruited to the 5′ cap of spliced mRNA along with several other proteins to form a multimeric protein complex termed hTREX [6]. The hTREX complex facilitates the association of bound mRNAs with TAP/p15 thus licensing nuclear export. In addition to Aly, several other hTREX components have been identified including the DEAD-box helicase UAP56, hTex1, the multi-protein THO complex and recently, CIP29 [15]. While the underlying mechanism of hTREX-mediated mRNA export is loosely understood, the individual functions of the hTREX components remain elusive.
Perhaps the greatest enigma surrounding TAP/p15-mediated mRNA export is the apparent redundancy that exists for certain hTREX proteins. This is particularly true for Aly, where a number of different studies have shown that the metazoan homologue, REF1, is not required for the bulk export of mRNA [23], [24]. These studies suggest that additional mRNA export adaptors must exist which can function to link nascent mRNA to the TAP/p15 heterodimer. Moreover, this raises the intriguing possibility that, via the use of numerous different mRNA export adaptor proteins, a further layer of control may exist to regulate gene expression. Indeed, several recent reports have highlighted that differences exist within component members of mRNA export complexes associated with different classes of mRNAs. For example, HSP70 mRNA only requires Aly and the co-adaptor Thoc5 to mediate TAP recruitment [50]. Moreover, an alternative mRNA export (AREX) complex, distinct to hTREX has recently been identified which comprises the related RNA helicase URH49, instead of UAP56 [51]. Interestingly, each helicase regulates a specific set of mRNAs associated with distinct subsets of key mitotic regulators. In addition, members of the SPEN family of proteins, RBM15 and OTT3 are functionally similar, in that they can bind RNA and physically interact with TAP. However, the association of OTT3 with TAP is attenuated compared to RBM15, leading to speculation that strong and weak variants exist that may function during developmental or tissue specific mRNA processing events [52]. These data galvanise the hypothesis that ultimately it is the recruitment of TAP/p15 that is required for nuclear export, and that one function of the export adaptor proteins is to provide selectivity to this system. Such a hypothesis is consistent with, and offers an explanation to, conflicting data regarding the nuclear export of KSHV intronless mRNAs.
Herpesviruses hijack the TAP/p15-mediated mRNA export pathway in order to enhance the nuclear export of viral intronless mRNA. We have previously shown that during KSHV replication the virus-encoded ORF57 protein procures the hTREX complex (and subsequently TAP/p15) via a direct interaction with Aly, facilitating the efficient export of KSHV intronless mRNAs [37]. We proposed therefore, that as the ORF57-Aly interaction provides the link between the virus mRNA and hTREX, it was likely that Aly would be essential for KSHV mRNA export. This hypothesis was supported by data showing that an ORF57 mutant, ORF57PmutGFP, unable to bind Aly was no longer functional in virus mRNA export. However, similarly to previous studies in D. melanogaster and C. elegans, siRNA-mediated depletion of Aly did not translate to a decrease in ORF57-mediated nuclear export of KSHV intronless mRNA, although only partial knockdown of Aly was observed [42]. Correspondingly, the HSV homologue of ORF57, ICP27, was shown to directly interact with Aly. Moreover, studies in Xenopus laevis oocytes showed ICP27 dramatically stimulated the export of intronless viral mRNAs, and a mutant ICP27 protein that failed to interact with REF is inactive in viral mRNA export [43]. Again however, siRNA-mediated depletion of Aly has been shown not to affect HSV-1 mRNA export [44].
Herein, we demonstrate that redundancy exists in the eukaryotic system for certain hTREX components involved in the mRNA nuclear export of intronless KSHV mRNAs. Evidence for such redundancy in export adapter proteins was recently provided by the identification of a second mRNA export adaptor protein, UIF [28]. Importantly, cellular expression levels of UIF appear to be linked in vivo to the relative expression of Aly, as depletion of Aly leads to a dramatic increase in UIF expression. This would therefore account for the modest reduction in mRNA nuclear export in Aly-depleted cells. Indeed, as shown in Figure 1 and 5, ORF57 interacts directly with UIF and thus is able to recruit hTREX/TAP/p15 allowing efficient intronless virus mRNA nuclear export in Aly-depleted cells (Figure 8).
Recent analysis has also suggested that additional mechanisms exist to ensure the nuclear export of viral transcripts in other herpesviruses. For example, ICP27 can bind directly to TAP, suggesting ICP27 can bypass nuclear export adapter proteins [53]. However, although analysis of ICP27 mutants unable to interact with TAP export showed greatly reduced intronless viral mRNA export, it was not completely abolished suggesting other cellular proteins may have a role. Indeed, recent analysis has shown that nuclear accumulation of HSV-1 mRNA is reduced when cells were treated with siRNAs specific for the SR proteins, SRp20 and 9G8, confirming that other cellular export proteins, such as SR proteins, can contribute to HSV-1 mRNA nuclear export [45]. Similarly, the EBV ORF57 homologue, SM/EB2, can interact with SRp20, although to date, this interaction has been implicated in alternative splicing mechanisms [54]. However, EBV SM/EB2 has been previously shown to interact with alternative cellular export factors, such as CRM-1 [55]. An alternative approach may be employed by the hCMV ORF57 homologue, UL69, which interacts with other hTREX proteins required for bulk mRNA nuclear export, such as UAP56 [56]. However, current work is ongoing to determine if these homologues interact with UIF. Moreover, the role of UIF may also have wider implications in the field of virology. Influenza A virus produces capped and polyadenylated mRNAs in the nucleus of infected cells that resemble mature cellular mRNAs, which require export by the TAP-mediated pathway [57]. Depletion of Aly had little effect on viral mRNA export, but reduction of UAP56 levels strongly inhibited trafficking and/or translation of influenza mRNAs [58]. It will now be interesting to determine whether UIF also substitutes for Aly function in this viral system.
There are however, some important mechanistic differences between Aly and UIF which have implications for KSHV intronless mRNA nuclear export. The hTREX component, CIP29, bridges the Aly-UAP56 interaction to form a trimeric complex that is assembled in an ATP-dependent manner [15]. Importantly, the recruitment of Aly to the mRNA requires an interaction with the 5′ cap and is dependent on splicing [6]. However, UIF appears to be co-transcriptionally loaded onto burgeoning mRNAs via an interaction with the histone chaperone, FACT [28]. It appears therefore that Aly and UIF are deposited onto the same mRNA separately and independently, a hypothesis supported by ribonuclease-treated co-immunoprecipitation experiments, which show that the interaction between Aly and UIF is facilitated by RNA-bridging [28], [36]. These data suggest that there are two distinct cellular mechanisms that can each recruit TAP to an mRNA. This raises a number of interesting questions with regards to how ORF57 orchestrates the recruitment of hTREX (and ultimately TAP/p15) via UIF. As seen in Figure 4, UIF is recruited to KSHV intronless mRNA only in the presence of ORF57, this is in stark contrast to the mechanism by which UIF is loaded onto cellular mRNA. Why UIF is not loaded onto KSHV intronless transcripts via FACT is unclear. One possible explanation is that FACT does not interact with RNA polymerase II during the transcription of ORF47 mRNA in this assay, possibly due to incomplete chromatinisation of vector DNA. Alternatively, recruitment of UIF to both spliced and unspliced mRNA maybe partially dependent on UAP56 and we have previously shown that UAP56 recruitment to KSHV mRNA is dependent on the ORF57 protein [37].
As mentioned above, Aly and UIF are loaded separately onto the same cellular mRNA via different mechanisms and both function to ultimately recruit TAP/p15 to the mRNA via interactions with hTREX. Intriguingly, we show in Figure 6, that ORF57 may preferentially bind to Aly over UIF, using both competitive GST-pulldown and dose-dependent coimmunoprecipitation assays. Why KSHV ORF57 would evolve to preferentially bind Aly over UIF is at present uncertain. One possibility is that Aly is the major export adaptor protein and UIF forms a backup or default pathway. This is not without precedent as proteins expression levels suggest that Aly is more abundantly expressed than UIF and UIF protein levels significantly increase in Aly-depleted cells [28]. Alternatively, it is possible that ORF57 may have a higher affinity for Aly due to important functional differences in how the Aly export adaptor recruits the remaining hTREX components to virus mRNA, compared with UIF. Alternatively, Aly and UIF could recruit different components of the hTREX complex to a KSHV mRNA, highlighted by the Aly-specific recruitment of CIP29, and that the export of KSHV intronless mRNA is more reliant on these Aly-recruited hTREX proteins.
As discussed earlier, a number of siRNA-mediated studies have proposed that Aly is not essential for KSHV intronless mRNA export. However, we have previously described an ORF57 mutant protein, ORF57Pmut, which is unable to interact with Aly and failed to export viral intronless mRNAs [37]. The region mutated in ORF57Pmut maps to a PxxP motif in the N-terminal region of the protein. It is not known whether the PxxP motif mutated in ORF57Pmut is a direct interaction site for Aly, or if this mutant confers some structural change of ORF57 in the Aly binding region. Importantly, herein we have shown that this mutant is also unable to interact with UIF, suggesting that ORF57Pmut is ‘dead’ with regards to export adaptor interaction. This explains therefore why this mutant is unable to export viral intronless mRNAs, as it is unable to bind to either Aly or UIF (Figure 1). This result is also confirmed by depletion of both these export adaptors which lead to a block in KSHV mRNA nuclear export. Importantly, Aly depletion in these and previous studies have shown that UIF expression is increased and therefore UIF probably replaces Aly as the dominant export adaptor protein. It is tempting to speculate that the link between increased UIF expression in Aly-depleted cells is a redundancy mechanism that ensures cellular survival should Aly expression be compromised.
The fact that ORF57Pmut is unable to interact with both Aly and UIF would suggest that the PxxP motif is either the complete ORF57 interacting motif, or part of the interacting motif, for Aly and UIF binding, and that the binding sites for the two proteins are either identical or overlap to some degree. Alternatively, the PxxP motif may cause a loss of interaction of both Aly and UIF by altering the structure of each of the binding sites. Importantly, our competition assays demonstrate that ORF57 may preferentially bind to Aly over UIF. These observations suggest that Aly and UIF may compete for a binding site on ORF57, and further studies are now required to determine if this is the case. Interestingly, we have recently identified the key residues that interact directly with Aly in both HSV-1 ICP27 and herpesvirus saimiri (HVS) ORF57 using solution-state NMR and mapped this interaction to a WRV/A motif [59]. Due to the sequence differences between ORF57 homologues this motif does not appear in KSHV ORF57, although the region of KSHV ORF57 that interacts with Aly has been mapped to the N terminus (aa 1–215). We are currently investigating the interacting residues for both Aly and UIF within this N-terminal region of KSHV ORF57 using solution-state NMR.
In summary, our results demonstrate the first known interaction between a viral protein and the newly described export adaptor protein, UIF. Importantly, the ORF57-UIF interaction is sufficient to recruit the hTREX complex onto viral intronless mRNAs and highlights that redundancy exists in the eukaryotic system for certain hTREX components involved in the mRNA nuclear export of intronless KSHV mRNAs. It now seems clear that the events which lead up to TAP/p15 recruitment to the mRNA are not linear. Indeed, it appears that multiple pathways exist by which an mRNA can bind TAP/p15 and be licensed for nuclear export. The existence of numerous export adaptor proteins may partly be explained in terms of redundancy but there is strong evidence to suggest that this also generates specificity within the system.
Materials and Methods
Plasmid and antibody details
Details of oligonucleotides used for qRT-PCR have been described previously [37], [46]. KSHV, hTREX and UIF-related plasmid constructs have been described previously [6], [28], [37]. KSHV ORF57 - and ORF4 - specific antibodies were a kind gift from Gary Hayward (Johns Hopkins, Baltimore) and Brad Spiller (Cardiff University), respectively. Antibodies against SC-35, Flag, Myc and Aly (Sigma), GFP and mCherry (Clontech), B23 (Santa Cruz), KSHV gB (Abcam) and GAPDH (Abcam) were purchased from their respective suppliers. Western blot analysis was carried out using specific antibodies at 1∶1000 dilution, except for UIF-specific antibody (1∶250) and GFP-specific antibody (1∶5000). Antibodies used for immunofluorescence studies were at a dilution of 1∶250.
Cell culture, viruses and transfection
293 inducible cells lines which specifically deplete Aly, UIF and both Aly and UIF have been previously reported [28]. They were produced using the FLP-In T-REX 293 cells (Invitrogen) system to express miRNAs to each specific export adapter protein, miRNA sequences are detailed in Hautbergue et al., 2009. HEK-293T cells, HEK-293T BAC36 cells harbouring a recombinant KSHV BAC36 genome and FLP-In T-REX 293 cells were cultured in Dulbecco's modified Eagle medium (DMEM, Invitrogen) supplemented with glutamine, 10% foetal calf serum (FCS, Invitrogen) and penicillin-streptomycin. 293T BAC36 cells were reactivated using TPA (20 ng/ml) for the designated time. miRNA expression from FLP-IN T-REX 293 cells was induced with 2 µg/ml doxycyclin (Sigma) for the designated time. Plasmid transfections were carried out using Lipofectamine 2000 (Invitrogen) or GeneJuice (Novagen) and were carried out as per the manufacturer's instructions. rKSHV.219 (KSHV) was produced from the latently infected Vero line [60]. This virus specifies red fluorescent protein (RFP) from the KSHV lytic PAN promoter, green fluorescent protein (GFP) from the EF-1α promoter, and encodes a puromycin resistance gene. Vero cells stably infected with rKSHV.219 were maintained in MEM Eagles medium, 2.2 g/L NaHCO3, 10% fetal calf serum, puromycin (5 ug/ml) (Sigma-Aldrich, Poole, UK) and penicillin and streptomycin (Invitrogen). To induce KSHV lytic replication in these cells, they were infected with BacK50, a baculovirus construct encoding the lytic switch RTA protein, and treated with 1.25 mM sodium butyrate (Sigma). 48 h after KSHV reactivation, the supernatant was harvested, centrifuged (500g, 15 mins) to remove cell debris, and the virions concentrated by centrifugation (65,000g, 4 h). The virion pellet was resuspended overnight in EBM2 medium (Lonza, Clonetics). The rKSHV.219 titre was determined on 293 cells, quantifying GFP-positive cells by fluorescence microscopy. 293 and 293 derived cells were infected with KSHV. To this end, cells were plated at 1.25×105 cells per well in 24-well plates for infection and cultured overnight. The culture medium was then removed and replaced with virus diluted in EBM2 basal media after 24 hrs. Cells were then centrifuged for 30 min at 420× g at room temperature. Cells were transferred to a 37°C incubator (5% CO2, humidified) for 90 min. Virus supernatant was removed and cells were washed once in cell culture media and incubated for 48 hrs before being harvested.
Expression and purification of recombinant proteins
Recombinant GST, GST-ORF57, GST-ORF57pmut, GST-UAP56 and UIF-His, Aly-His and ORF73-His were expressed and purified as previously described [36],[37],[61]. Purification of Baculovirus recombinant ORF57-6xHis was as per the manufacturer's instructions (Invitrogen) using the pFASTBac protocol.
In vitro pull-down assays and immunoprecipitation assays
GST pull-down experiments and co-immunoprecipitations were performed as described previously [62], [63]. GFP-TRAP-Affinity (Chromotek) experiments were performed as per the manufacturer's instructions. RNA immunoprecipitation experiments were carried out as follows: 1×107 adherent 293T cells were transiently transfected with appropriate GFP-containing plasmid DNA. After the appropriate amount of time cells were washed in ice-cold PBS and UV irradiated (900 mJ/cm2) using a Stratalinker 2400 (Stratagene) to crosslink protein and RNA. Cells were then scraped, transferred to an RNA-free tube and pelleted at 300× g for 3 min. Cell pellets were then resuspended in 2 ml lysis buffer [Dulbecco's PBS, 1% Nonidet P-40 (v/v), 1 µl/ml RNaseOUT (Invitrogen), 1× Complete EDTA-free Protease inhibitor (Roche)]. Cells were left on ice for 30 min before being centrifuged for 10 min at 15,000× g. The clear lysate was then transferred to a clean RNA-free tube. 1 ml of the cleared lysate was added to 30 µl pre-washed GFP-TRAP-Affinity agarose beads (Chromotek) per IP and immunoprecipitated at 4°C with end-over-end mixing for 4 hrs. Beads were washed 3 times in ice-cold PBS containing 1× Complete EDTA-free protease inhibitor (Roche) followed by a further 2 times in PBS. Beads where then incubated in protease buffer (Dulbecco's PBS, 1% Nonidet P-40 (v/v), 0.1% SDS (w/v), 0.5 mg/ml Proteinase K) for 30 min at 50°C. RNA was extracted using TRIzol reagent (Invitrogen) as per the manufacturer's directions. cDNA was then produced from 10 µl of RNA using Superscript II RT (Invitrogen) and qPCR performed to analyse the relative levels of cDNA. RT-ve samples were used as controls.
GST pull-down competition assays
Bacterially expressed GST-tagged ORF57 was immobilised to GST beads and used for GST pulldown competition assays. Recombinant His-tagged Aly or UIF was expressed and purified as previously described [36], [37]. Equal amounts of Aly-His (1 µg) were used in the pull-downs with increasing amounts of UIF-His (0, 0.5, 1, 2, 3 µg). The converse experiments were also performed with equal amounts of UIF-His (1 µg) and increasing amounts of Aly-His (0, 0.5, 1, 2, 3 µg).
Real-time qRT-PCR
To assess ORF57-mediated ORF47 mRNA export efficiency, 293T and inducible cells were cotransfected with ORF47 and ORF57 expression constructs. After 24 hours RNA was extracted from total and cytoplasmic fractions using TRIzol (Invitrogen) as described by the manufacturer. Cytoplasmic fractions were produced by lysis of cells in 200 µl of PBS 1% Triton-X 100(v/v) containing 40 U of RNaseOUT (Invitrogen), prior to TRIzol purification. RNA was DNase treated using the Ambion DNase-free kit, as per the manufacturer's instructions, and RNA (1 µg) from each fraction was reverse transcribed with SuperScript II (Invitrogen), as per the manufacturer's instructions, using oligo(dT) primers (Promega). 10 ng of cDNA was used as template in SensiMixPlus SYBR qPCR reactions (Quantace), as per manufacturer's instructions, using a Rotor-Gene Q 5plex HRM Platform (Qiagen), with a standard 3-step melt program (95°C for 15 seconds, 60°C for 30 seconds, 72°C for 20 seconds). With GAPDH as internal control mRNA, quantitative analysis was performed using the comparative CT method as previously described [46].
Immunofluorescence
Immunofluorescence staining and visualisation by microscopy was carried out as previously described [64]. Visualisation was performed on an LSM 510 Meta confocal microscope (Zeiss) and images were analysed using the LSM imaging software (Zeiss).
Zdroje
1. MooreMJ 2005 From birth to death: the complex lives of eukaryotic mRNAs. Science 309 1514 1518
2. BentleyDL 2005 Rules of engagement: co-transcriptional recruitment of pre-mRNA processing factors. Curr Opin Cell Biol 17 251 256
3. KohlerAHurtE 2007 Exporting RNA from the nucleus to the cytoplasm. Nat Rev Mol Cell Biol 8 761 773
4. MooreMJProudfootNJ 2009 Pre-mRNA processing reaches back to transcription and ahead to translation. Cell 136 688 700
5. ReedRChengH 2005 TREX, SR proteins and export of mRNA. Curr Opin Cell Biol 17 269 273
6. ChengHDufuKLeeCSHsuJLDiasA 2006 Human mRNA export machinery recruited to the 5′ end of mRNA. Cell 127 1389 1400
7. MasudaSDasRChengHHurtEDormanN 2005 Recruitment of the human TREX complex to mRNA during splicing. Genes Dev 19 1512 1517
8. Le HirHIzaurraldeEMaquatLEMooreMJ 2000 The spliceosome deposits multiple proteins 20–24 nucleotides upstream of mRNA exon-exon junctions. EMBO J 19 6860 6869
9. ChangYFImamJSWilkinsonMF 2007 The nonsense-mediated decay RNA surveillance pathway. Annu Rev Biochem 76 51 74
10. DiemMDChanCCYounisIDreyfussG 2007 PYM binds the cytoplasmic exon-junction complex and ribosomes to enhance translation of spliced mRNAs. Nat Struct Mol Biol 14 1173 1179
11. MaXMYoonSORichardsonCJJulichKBlenisJ 2008 SKAR links pre-mRNA splicing to mTOR/S6K1-mediated enhanced translation efficiency of spliced mRNAs. Cell 133 303 313
12. NottALe HirHMooreMJ 2004 Splicing enhances translation in mammalian cells: an additional function of the exon junction complex. Genes Dev 18 210 222
13. ReedRHurtE 2002 A conserved mRNA export machinery coupled to pre-mRNA splicing. Cell 108 523 531
14. StrasserKMasudaSMasonPPfannstielJOppizziM 2002 TREX is a conserved complex coupling transcription with messenger RNA export. Nature 417 304 308
15. DufuKLivingstoneMJSeebacherJGygiSPWilsonSA 2010 ATP is required for interactions between UAP56 and two conserved mRNA export proteins, Aly and CIP29, to assemble the TREX complex. Genes Dev 24 2043 2053
16. LuoMJReedR 1999 Splicing is required for rapid and efficient mRNA export in metazoans. Proc Natl Acad Sci U S A 96 14937 14942
17. ValenciaPDiasAPReedR 2008 Splicing promotes rapid and efficient mRNA export in mammalian cells. Proc Natl Acad Sci U S A 105 3386 3391
18. ZhouZLuoMJStraesserKKatahiraJHurtE 2000 The protein Aly links pre-messenger-RNA splicing to nuclear export in metazoans. Nature 407 401 405
19. KatahiraJStrasserKPodtelejnikovAMannMJungJU 1999 The Mex67p-mediated nuclear mRNA export pathway is conserved from yeast to human. EMBO J 18 2593 2609
20. StutzFBachiADoerksTBraunICSeraphinB 2000 REF, an evolutionary conserved family of hnRNP-like proteins, interacts with TAP/Mex67p and participates in mRNA nuclear export. RNA 6 638 650
21. HautbergueGMHungMLGolovanovAPLianLYWilsonSA 2008 Mutually exclusive interactions drive handover of mRNA from export adaptors to TAP. Proc Natl Acad Sci U S A 105 5154 5159
22. FribourgSBraunICIzaurraldeEContiE 2001 Structural basis for the recognition of a nucleoporin FG repeat by the NTF2-like domain of the TAP/p15 mRNA nuclear export factor. Mol Cell 8 645 656
23. GatfieldDIzaurraldeE 2002 REF1/Aly and the additional exon junction complex proteins are dispensable for nuclear mRNA export. J Cell Biol 159 579 588
24. LongmanDJohnstoneILCaceresJF 2003 The Ref/Aly proteins are dispensable for mRNA export and development in Caenorhabditis elegans. RNA 9 881 891
25. HeroldAKlymenkoTIzaurraldeE 2001 NXF1/p15 heterodimers are essential for mRNA nuclear export in Drosophila. RNA 7 1768 1780
26. RehwinkelJHeroldAGariKKocherTRodeM 2004 Genome-wide analysis of mRNAs regulated by the THO complex in Drosophila melanogaster. Nat Struct Mol Biol 11 558 566
27. FarnyNGHurtJASilverPA 2008 Definition of global and transcript-specific mRNA export pathways in metazoans. Genes Dev 22 66 78
28. HautbergueGMHungMLWalshMJSnijdersAPChangCT 2009 UIF, a New mRNA export adaptor that works together with REF/ALY, requires FACT for recruitment to mRNA. Curr Biol 19 1918 1924
29. BoyneJRWhitehouseA 2006 gamma-2 Herpes virus post-transcriptional gene regulation. Clin Microbiol Infect 12 110 117
30. BoyneJRJacksonBRWhitehouseA 2010 ORF57: Master regulator of KSHV mRNA biogenesis. Cell Cycle 9 2702 2703
31. ChangYMoorePS 1996 Kaposi's sarcoma (KS)-associated herpesvirus and its role in KS. Infect Agents Dis 5 215 222
32. BoshoffCChangY 2001 Kaposi's sarcoma - Associated herpesvirus: A new DNA tumor virus. Ann Rev Med 52 453 470
33. GanemD 2006 KSHV infection and the pathogenesis of Kaposi's sarcoma. Annu Rev Pathol 1 273 296
34. MajerciakVZhengZM 2009 Kaposi's sarcoma-associated herpesvirus ORF57 in viral RNA processing. Front Biosci 14 1516 1528
35. SwaminathanS 2005 Post-transcriptional gene regulation by gamma herpesviruses. J Cell Biochem 95 698 711
36. BoyneJRJacksonBTaylorAMacnabSWhitehouseA 2010 KSHV ORF57 interacts with PYM to enhance the translation of viral intronless mRNAs. EMBO J 29 1851 1864
37. BoyneJRColganKJWhitehouseA 2008 Recruitment of the complete hTREX complex is required for Kaposi's sarcoma-associated herpesvirus intronless mRNA nuclear export and virus replication. PLoS Pathog 4 e1000194
38. MajerciakVYamanegiKAllemandEKruhlakMKrainerAR 2008 Kaposi's sarcoma-associated herpesvirus ORF57 functions as a viral splicing factor and promotes expression of intron-containing viral lytic genes in spliceosome-mediated RNA splicing. J Virol 82 2792 2801
39. SahinBBPatelDConradNK 2010 Kaposi's sarcoma-associated herpesvirus ORF57 protein binds and protects a nuclear noncoding RNA from cellular RNA decay pathways. PLoS Pathog 6 e1000799
40. ColganKJBoyneJRWhitehouseA 2009 Uncoupling of hTREX demonstrates that UAP56 and hTHO-complex recruitment onto herpesvirus saimiri intronless transcripts is required for replication. J Gen Virol 90 1455 1460
41. BoyneJRColganKJWhitehouseA 2008 Herpesvirus saimiri ORF57: a post-transcriptional regulatory protein. Front Biosci 13 2928 2938
42. MajerciakVYamanegiKNieSHZhengZM 2006 Structural and functional analyses of Kaposi sarcoma-associated herpesvirus ORF57 nuclear localization signals in living cells. J Biol Chem 281 28365 28378
43. KoffaMDClementsJBIzaurraldeEWaddSWilsonSA 2001 Herpes simplex virus ICP27 protein provides viral mRNAs with access to the cellular mRNA export pathway. EMBO J 20 5769 5778
44. JohnsonLALiLSandri-GoldinRM 2009 The cellular RNA export receptor TAP/NXF1 is required for ICP27-mediated export of herpes simplex virus 1 RNA, but the TREX complex adaptor protein Aly/REF appears to be dispensable. J Virol 83 6335 6346
45. Escudero-PaunettoLLiLHernandezFPSandri-GoldinRM 2010 SR proteins SRp20 and 9G8 contribute to efficient export of herpes simplex virus 1 mRNAs. Virology 401 155 164
46. BoyneJRWhitehouseA 2009 Nucleolar disruption impairs Kaposi's sarcoma-associated herpesvirus ORF57-mediated nuclear export of intronless viral mRNAs. FEBS Letters 583 3549 3556
47. LuoMLZhouZMagniKChristoforidesCRappsilberJ 2001 Pre-mRNA splicing and mRNA export linked by direct interactions between UAP56 and Aly. Nature 413 644 647
48. GriffithsRWhitehouseA 2007 Herpesvirus saimiri episomal persistence is maintained via interaction between open reading frame 73 and the cellular chromosome-associated protein MeCP2. J Virol 81 4021 4032
49. MajerciakVDengMZhengZM 2010 Requirement of UAP56, URH49, RBM15, and OTT3 in the expression of Kaposi sarcoma-associated herpesvirus ORF57. Virology 407 206 212
50. KatahiraJInoueHHurtEYonedaY 2009 Adaptor Aly and co-adaptor Thoc5 function in the Tap-p15-mediated nuclear export of HSP70 mRNA. EMBO J 28 556 567
51. YamazakiTFujiwaraNYukinagaHEbisuyaMShikiT 2010 The closely related RNA helicases, UAP56 and URH49, preferentially form distinct mRNA export machineries and coordinately regulate mitotic progression. Mol Biol Cell 21 2953 2965
52. UranishiHZolotukhinASLindtnerSWarmingSZhangGM 2009 The RNA-binding motif protein 15B (RBM15B/OTT3) acts as cofactor of the nuclear export receptor NXF1. J Biol Chem 284 26106 26116
53. ChenIHLiLSilvaLSandri-GoldinRM 2005 ICP27 recruits Aly/REF but not TAP/NXF1 to herpes simplex virus type 1 transcription sites although TAP/NXF1 is required for ICP27 export. J Virol 79 3949 3961
54. VermaDBaisSGaillardMSwaminathanS 2010 Epstein-Barr Virus SM protein utilizes cellular splicing factor SRp20 to mediate alternative splicing. J Virol 84 11781 11789
55. BoyleSMRuvoloVGuptaAKSwaminathanS 1999 Association with the cellular export receptor CRM 1 mediates function and intracellular localization of Epstein-Barr virus SM protein, a regulator of gene expression. J Virol 73 6872 6881
56. LischkaPTothZThomasMMuellerRStammingerT 2006 The UL69 transactivator protein of human cytomegalovirus interacts with DEXD/H-Box RNA helicase UAP56 to promote cytoplasmic accumulation of unspliced RNA. Mol Cell Biol 26 1631 1643
57. HaoLSakuraiAWatanabeTSorensenENidomCA 2008 Drosophila RNAi screen identifies host genes important for influenza virus replication. Nature 454 890 893
58. ReadEKDigardP 2010 Individual influenza A virus mRNAs show differential dependence on cellular NXF1/TAP for their nuclear export. J Gen Virol 91 1290 1301
59. TunnicliffeRBHautbergueGMKalraPJacksonBRWhitehouseA 2011 Structural basis for the recognition of cellular mRNA export factor REF by herpes viral proteins HSV-1 ICP27 and HVS ORF57. PLoS Pathog 7 e1001244
60. VieiraJO'HearnPM 2004 Use of the red fluorescent protein as a marker of Kaposi's sarcoma-associated herpesvirus lytic gene expression. Virology 325 225 240
61. CalderwoodMAHallKTMatthewsDAWhitehouseA 2004 The herpesvirus saimiri ORF73 gene product interacts with host-cell mitotic chromosomes and self-associates via its C terminus. J Gen Virol 85 147 153
62. GouldFHarrisonSMHewittEWWhitehouseA 2009 Kaposi's sarcoma-associated herpesvirus RTA promotes degradation of the Hey1 repressor protein through the ubiquitin proteasome pathway. J Virol 83 6727 6738
63. HallKTGilesMSCalderwoodMAGoodwinDJMatthewsDA 2002 The herpesvirus saimiri open reading frame 73 gene product interacts with the cellular protein p32. J Virol 76 11612 11622
64. BoyneJRWhitehouseA 2006 Nucleolar trafficking is essential for nuclear export of intronless herpesvirus mRNA. Proc Natl Acad Sci U S A 103 15190 15195
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek A Typhimurium-Typhi Genomic Chimera: A Model to Study Vi Polysaccharide Capsule Function In VivoČlánek SUMO Pathway Dependent Recruitment of Cellular Repressors to Herpes Simplex Virus Type 1 GenomesČlánek A Structural Model for Binding of the Serine-Rich Repeat Adhesin GspB to Host Carbohydrate ReceptorsČlánek Dynamic Evolution of Pathogenicity Revealed by Sequencing and Comparative Genomics of 19 IsolatesČlánek Widespread Endogenization of Genome Sequences of Non-Retroviral RNA Viruses into Plant GenomesČlánek The Cost of Virulence: Retarded Growth of Typhimurium Cells Expressing Type III Secretion System 1Článek A Role for the Chemokine RANTES in Regulating CD8 T Cell Responses during Chronic Viral Infection
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2011 Číslo 7- Jak souvisí postcovidový syndrom s poškozením mozku?
- Měli bychom postcovidový syndrom léčit antidepresivy?
- Farmakovigilanční studie perorálních antivirotik indikovaných v léčbě COVID-19
- 10 bodů k očkování proti COVID-19: stanovisko České společnosti alergologie a klinické imunologie ČLS JEP
-
Všechny články tohoto čísla
- What Do We Really Know about How CD4 T Cells Control ?
- “Persisters”: Survival at the Cellular Level
- E6 and E7 from Beta Hpv38 Cooperate with Ultraviolet Light in the Development of Actinic Keratosis-Like Lesions and Squamous Cell Carcinoma in Mice
- Selection of Resistant Bacteria at Very Low Antibiotic Concentrations
- The Extracytoplasmic Domain of the Ser/Thr Kinase PknB Binds Specific Muropeptides and Is Required for PknB Localization
- CD39/Adenosine Pathway Is Involved in AIDS Progression
- Hypoxia and a Fungal Alcohol Dehydrogenase Influence the Pathogenesis of Invasive Pulmonary Aspergillosis
- One Is Enough: Effective Population Size Is Dose-Dependent for a Plant RNA Virus
- Effects of Interferon-α/β on HBV Replication Determined by Viral Load
- A Typhimurium-Typhi Genomic Chimera: A Model to Study Vi Polysaccharide Capsule Function In Vivo
- Dual Chaperone Role of the C-Terminal Propeptide in Folding and Oligomerization of the Pore-Forming Toxin Aerolysin
- Rotavirus Stimulates Release of Serotonin (5-HT) from Human Enterochromaffin Cells and Activates Brain Structures Involved in Nausea and Vomiting
- Dissociation of Infectivity from Seeding Ability in Prions with Alternate Docking Mechanism
- The Impact of Recombination on dN/dS within Recently Emerged Bacterial Clones
- The Regulation of Sulfur Metabolism in
- Illumination of Parainfluenza Virus Infection and Transmission in Living Animals Reveals a Tissue-Specific Dichotomy
- A Permeable Cuticle Is Associated with the Release of Reactive Oxygen Species and Induction of Innate Immunity
- A Concerted Action of Hepatitis C Virus P7 and Nonstructural Protein 2 Regulates Core Localization at the Endoplasmic Reticulum and Virus Assembly
- SUMO Pathway Dependent Recruitment of Cellular Repressors to Herpes Simplex Virus Type 1 Genomes
- Re-localization of Cellular Protein SRp20 during Poliovirus Infection: Bridging a Viral IRES to the Host Cell Translation Apparatus
- Divergent Effects of Human Cytomegalovirus and Herpes Simplex Virus-1 on Cellular Metabolism
- A Structural Model for Binding of the Serine-Rich Repeat Adhesin GspB to Host Carbohydrate Receptors
- Transformation of Natural Genetic Variation into Genomes
- EBV Latency Types Adopt Alternative Chromatin Conformations
- Global mRNA Degradation during Lytic Gammaherpesvirus Infection Contributes to Establishment of Viral Latency
- Dynamic Evolution of Pathogenicity Revealed by Sequencing and Comparative Genomics of 19 Isolates
- Microbial Virulence as an Emergent Property: Consequences and Opportunities
- Widespread Endogenization of Genome Sequences of Non-Retroviral RNA Viruses into Plant Genomes
- Structural Basis of Chemokine Sequestration by CrmD, a Poxvirus-Encoded Tumor Necrosis Factor Receptor
- Cross-Species Transmission of a Novel Adenovirus Associated with a Fulminant Pneumonia Outbreak in a New World Monkey Colony
- An Interaction between KSHV ORF57 and UIF Provides mRNA-Adaptor Redundancy in Herpesvirus Intronless mRNA Export
- Elevated 17β-Estradiol Protects Females from Influenza A Virus Pathogenesis by Suppressing Inflammatory Responses
- The Role of IL-15 Deficiency in the Pathogenesis of Virus-Induced Asthma Exacerbations
- Fluorescence Lifetime Imaging Unravels Metabolism and Its Crosstalk with the Host Cell
- Programmed Death (PD)-1-Deficient Mice Are Extremely Sensitive to Murine Hepatitis Virus Strain-3 (MHV-3) Infection
- Hemoglobin Promotes Nasal Colonization
- Crystallography of a Lewis-Binding Norovirus, Elucidation of Strain-Specificity to the Polymorphic Human Histo-Blood Group Antigens
- The Cost of Virulence: Retarded Growth of Typhimurium Cells Expressing Type III Secretion System 1
- A Genome-Wide Approach to Discovery of Small RNAs Involved in Regulation of Virulence in
- Requires Glycerol for Maximum Fitness During The Tick Phase of the Enzootic Cycle
- C Metabolic Flux Analysis Identifies an Unusual Route for Pyruvate Dissimilation in Mycobacteria which Requires Isocitrate Lyase and Carbon Dioxide Fixation
- A Role for the Chemokine RANTES in Regulating CD8 T Cell Responses during Chronic Viral Infection
- Glycosaminoglycans and Sialylated Glycans Sequentially Facilitate Merkel Cell Polyomavirus Infectious Entry
- Regulation of Stomatal Tropism and Infection by Light in : Evidence for Coordinated Host/Pathogen Responses to Photoperiod?
- Multiple Translocation of the Effector Gene among Chromosomes of the Rice Blast Fungus and Related Species
- Comparative Genomics Yields Insights into Niche Adaptation of Plant Vascular Wilt Pathogens
- Unique Cell Adhesion and Invasion Properties of O:3, the Most Frequent Cause of Human Yersiniosis
- C-Terminal Region of EBNA-2 Determines the Superior Transforming Ability of Type 1 Epstein-Barr Virus by Enhanced Gene Regulation of LMP-1 and CXCR7
- Novel Chikungunya Vaccine Candidate with an IRES-Based Attenuation and Host Range Alteration Mechanism
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Requires Glycerol for Maximum Fitness During The Tick Phase of the Enzootic Cycle
- Comparative Genomics Yields Insights into Niche Adaptation of Plant Vascular Wilt Pathogens
- The Role of IL-15 Deficiency in the Pathogenesis of Virus-Induced Asthma Exacerbations
- “Persisters”: Survival at the Cellular Level
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



