-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaGenesis of Mammalian Prions: From Non-infectious Amyloid Fibrils to a Transmissible Prion Disease
The transmissible agent of prion disease consists of a prion protein in its abnormal, β-sheet rich state (PrPSc), which is capable of replicating itself according to the template-assisted mechanism. This mechanism postulates that the folding pattern of a newly recruited polypeptide chain accurately reproduces that of a PrPSc template. Here we report that authentic PrPSc and transmissible prion disease can be generated de novo in wild type animals by recombinant PrP (rPrP) amyloid fibrils, which are structurally different from PrPSc and lack any detectable PrPSc particles. When induced by rPrP fibrils, a long silent stage that involved two serial passages preceded development of the clinical disease. Once emerged, the prion disease was characterized by unique clinical, neuropathological, and biochemical features. The long silent stage to the disease was accompanied by significant transformation in neuropathological properties and biochemical features of the proteinase K-resistant PrP material (PrPres) before authentic PrPSc evolved. The current work illustrates that transmissible prion diseases can be induced by PrP structures different from that of authentic PrPSc and suggests that a new mechanism different from the classical templating exists. This new mechanism designated as “deformed templating” postulates that a change in the PrP folding pattern from the one present in rPrP fibrils to an alternative specific for PrPSc can occur. The current work provides important new insight into the mechanisms underlying genesis of the transmissible protein states and has numerous implications for understanding the etiology of neurodegenerative diseases.
Published in the journal: . PLoS Pathog 7(12): e32767. doi:10.1371/journal.ppat.1002419
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1002419Summary
The transmissible agent of prion disease consists of a prion protein in its abnormal, β-sheet rich state (PrPSc), which is capable of replicating itself according to the template-assisted mechanism. This mechanism postulates that the folding pattern of a newly recruited polypeptide chain accurately reproduces that of a PrPSc template. Here we report that authentic PrPSc and transmissible prion disease can be generated de novo in wild type animals by recombinant PrP (rPrP) amyloid fibrils, which are structurally different from PrPSc and lack any detectable PrPSc particles. When induced by rPrP fibrils, a long silent stage that involved two serial passages preceded development of the clinical disease. Once emerged, the prion disease was characterized by unique clinical, neuropathological, and biochemical features. The long silent stage to the disease was accompanied by significant transformation in neuropathological properties and biochemical features of the proteinase K-resistant PrP material (PrPres) before authentic PrPSc evolved. The current work illustrates that transmissible prion diseases can be induced by PrP structures different from that of authentic PrPSc and suggests that a new mechanism different from the classical templating exists. This new mechanism designated as “deformed templating” postulates that a change in the PrP folding pattern from the one present in rPrP fibrils to an alternative specific for PrPSc can occur. The current work provides important new insight into the mechanisms underlying genesis of the transmissible protein states and has numerous implications for understanding the etiology of neurodegenerative diseases.
Introduction
Prion diseases, or transmissible spongiform encephalopathies (TSEs), are fatal neurodegenerative disorders that can be sporadic, inherited, or infectious in origin. Misfolding and aggregation of the normal, cellular form of the prion protein (PrPC) into an abnormal β-sheet rich state underlies the pathogenic mechanisms of the prion diseases for all three origins [1]. The “protein only” hypothesis of prion propagation postulates that the transmissible agent of prion diseases consists of a prion protein in its abnormal, β-sheet rich conformation (PrPSc), which is capable of propagating itself in an autocatalytic manner by recruiting and converting PrPC [2], [3].
Recent years have witnessed a number of studies where transmissible prion diseases were generated in animals de novo by inoculating prion infectious material produced in vitro [4]–[10]. All studies on generating prion infectivity can be divided into two large groups, where the materials for inoculating animals were produced using either (i) Protein Misfolding Cyclic Amplification (PMCA) [8]–[10], or (ii) in vitro fibrillation protocols that utilized recombinant PrP (rPrP) [4]–[7]. In the studies that employed the first approach, application of serial PMCA (sPMCA) accomplished two purposes: (i) generating PrPSc particles de novo and (ii) amplification of newly formed PrPSc to amounts that can effectively produce clinical disease in wild type animals within a short incubation time and 100% success rate [8]–[10]. Because de novo generation of PrPSc in sPMCA followed stochastic behavior, it remains unclear whether a large amount of infectivity produced after multiple rounds of sPMCA was a result of amplification of a few or even a single PrPSc particle. In the second approach, rPrP was converted into amyloid fibrils in vitro without application of PMCA [4]–[7]. In these studies, transmissible diseases were generated either in transgenic animals with high levels of PrPC expression and after relatively long incubation time [4], [5], [7], or in wild type animals, but with a less than 100% success rate and after a long silent stage [6]. Critical concerns that rPrP amyloid fibrils did not induce the disease de novo but only accelerated an on-going pathogenic process have been raised regarding the studies performed on transgenic mice [11]–[13]. For the experiments conducted on wild type animals, a long silent stage and less than 100% success rate led to the general assumption that the preparations of amyloid fibrils contain minuscule amounts of PrPSc or particles with a structure of PrPSc, and that this tiny sub-fraction is responsible for the disease.
In the current study we report that no PrPSc can be detected in preparations of rPrP amyloid fibrils using a sPMCA format that could detect as little as a single PrPSc particle. Nevertheless, despite lack of PrPSc in the preparations of rPrP fibrils, transmissible prion disease was produced in wild type animals. When induced by rPrP fibrils, which are known to be structurally different from PrPSc [14]–[17], the clinical disease developed only at the third serial passage. The long silent stage to the disease that involved two serial passages was found to be accompanied by significant changes in biochemical and neuropathological properties of the proteinase K (PK) -resistant PrP (PrPres). These changes involved dramatic transformation of a PK-resistant profile, changes in the PrPres deposition pattern and expansion of brain areas targeted by PrPres. Clinical, neuropathological, and biochemical features of the new prion disease were different from those produced in rodents by previously known, hamster-adapted prion strains or strains generated by sPMCA [8]–[10], consistent with the idea that the new strain originated from a unique amyloid structure. The current work introduces a new hypothesis that transmissible prion diseases can be induced by cross-β PrP structures substantially different from that of authentic PrPSc. This hypothesis suggests that a new templating mechanism (which will be referred to as “deformed templating”) different from the classical templating might exist. The deformed templating involves a switch from one PrP folding pattern present in amyloid fibrils to an alternative folding pattern present in PrPSc. The current work provides important new insight into the mechanisms underlying genesis and evolution of the transmissible states of the prion protein and has numerous implications for understanding the etiology of prion and other neurodegenerative diseases.
Results
Serial transmission of rPrP amyloid fibrils
Full-length Syrian hamster rPrP was converted into amyloid fibrils in vitro and subjected to an annealing procedure (brief heating to 80°C) in the presence of BSA prior to inoculation as previously described [6], [18]. All experiments on expression and purification of rPrP, its conversion into fibrils, annealing and preparation of inocula were performed in a facility and with equipment that have never been exposed to TSEs. Annealed rPrP fibrils were inoculated intracerebrally into Syrian hamsters. While none of the animals developed any signs of prion disease for up to 661 days post inoculation, one out of seven animals showed atypical, PK-resistant, low molecular weight (LMW) bands of 21, 16 and 13 kDa ([6] and Figure 1A). These PK-resistant material (PrPres) could be detected by the antibodies against the C-terminal PrP region R1 (epitope 225–231) or SAF-84 (epitope 160–170), but not by 3F4 (epitope 109–112) (Figure 1A and B). Interestingly, when used as seed in sPMCA reaction with beads (sPMCAb) [19] , the brain material with atypical PrPres gave rise to PK-resistant products with a band-shift typical for authentic PrPSc that could be detected by 3F4 and the C-terminal antibodies (Figure 1C). These PK-resistant products will be referred to as typical or standard PrPres. We do not know whether it was atypical PrPres that gave rise to typical PrPres in sPMCAb or small, undetectable amounts of typical PrPres that might have already been present in brain homogenates (BH).
Fig. 1. Serial passages of rPrP fibrils and de novo generation of PrPSc in Syrian hamsters. 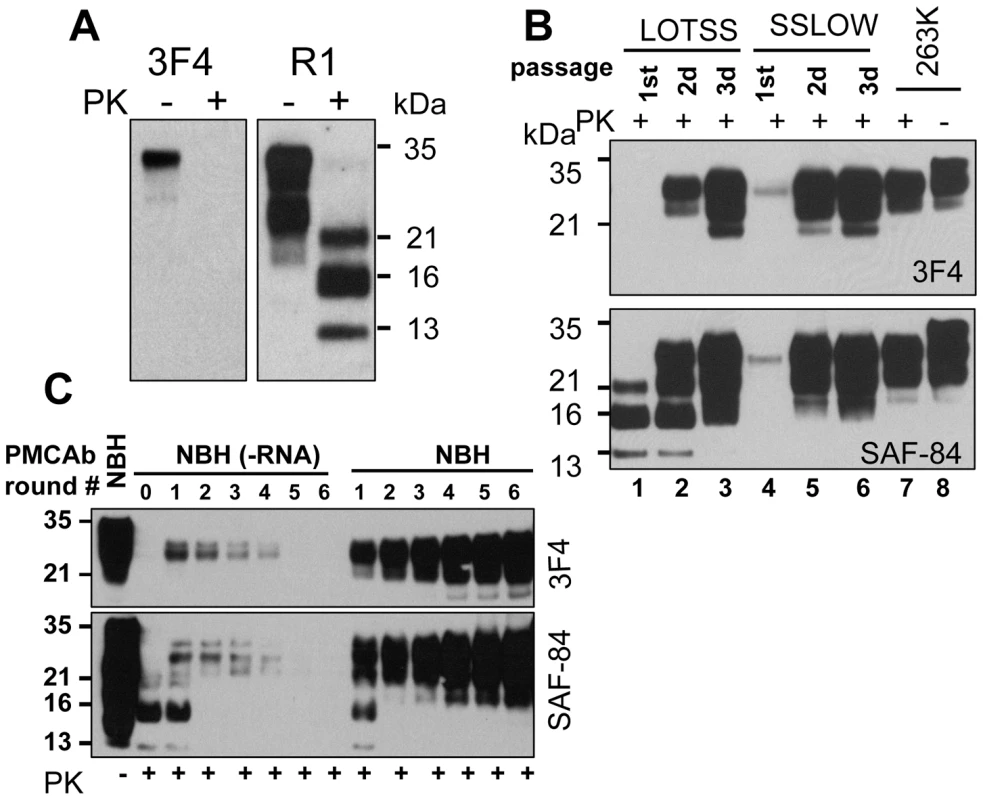
(A) Western blotting of brain homogenate (BH) from the animal inoculated with BSA-annealed rPrP amyloid fibrils. Western blots were stained with 3F4 (left panel) or R1 (right panel) antibody. Undigested samples were loaded at 1/10th the amount of the digested samples. (B) Western blotting of BHs of animals from the 1st, 2nd, or 3rd passages of BSA-annealed rPrP fibrils (lanes 1, 2 and 3, respectively) stained with 3F4 (top panel) or SAF-84 (bottom panel) antibody. The new prion strain produced as a result of inoculation of BSA-annealed rPrP fibrils will be designated as LOTSS. The 1st, 2nd, or 3rd passages of SSLOW, the strain that was previously produced as a result of inoculation of normal BH (NBH)-annealed rPrP fibrils (lanes 4, 5 and 6 respectively) [6], or 263 K are provided as references. (C) sPMCAb of brain material from the 1st passage of LOTSS. 10% BH from the animal inoculated with BSA-annealed rPrP amyloid fibrils was diluted 10-fold into 10% RNA-depleted (NBH(-RNA)) or NBH and subjected to sPMCAb. Each PMCAb round consisted of 48 cycles, 30 min each; 10-fold dilutions were used for each subsequent sPMCAb round. Western blots were stained with 3F4 (top panel) or SAF-84 (bottom panel) antibody. Undigested 10% NBH is provided as a reference. BHs from the remaining six animals that lacked any PrPres material on Western blot were tested in PMCAb. After a single PMCAb round, BH from one more animal showed PrPres products detectible by 3F4 antibody (Figure S1A). By the sixth sPMCAb round, BH from an additional animal showed PrPres positive by 3F4 antibody (Figure S1B). Ten non-seeded sPMCAb reactions were all negative after six sPMCAb rounds (Figure S1C) arguing against possibility of cross-contamination or generation of PrPSc or PrPres de novo in the sPMCAb format employed here. As we reported previously, sPMCAb reactions seeded with normal BHs (NBHs) from old animals were also all negative after six sPMCAb rounds [19]. Therefore, three out of seven animals inoculated with rPrP fibrils showed signs of infection as judged by appearance of PMCAb-active PrPres material in their brains, although the amounts of PrPres material was highly variable. As previously described, no clinical signs or PrPres material were found in animals inoculated with any of the controls or in uninoculated age-matched controls ([6] and Table S1).
To perform a second passage, 10% BH containing atypical PrPres was inoculated into a new group. Surprisingly, none of the animals developed clinical signs of the disease before they were euthanized at 661 days (Table S1). However, six out of seven hamsters showed large amounts of standard and atypical PrPres (Figures 1B and S2). The remaining animal showed predominantly atypical, LMW PrPres detectible by SAF-84 antibody and miniscule amounts of standard PrPres (Figure S2, lane 6). After a single PMCAb round, BH from this animal showed PrPres products detectible by 3F4 antibody (Figure S2, lane 9). These results illustrated that all animals were infected in a second passage. The variation in the ratio of standard to atypical PrPres in animals from the 2nd passage (Figure S2) suggested that transformation in PrPres structure and PK-resistance pattern occurred at a variable rate within the same group of animals. Considering that no clinical signs were observed despite substantial amounts of PrPres, the new synthetic strain was designated as LOTSS (LOw Toxicity Synthetic Strain).
The amount of PrPres in LOTSS-infected animals from the 2nd passage was similar to those found at the terminal stage of 263 K-inoculated hamsters (Figure 1B). While no signs of clinical disease were observed, LOTSS PrPres from the 2nd passage showed biochemical attributes of authentic PrPSc. LOTSS standard PrPres could be amplified in sPMCAb and displayed an amplification rate similar to that of 263 K PrPSc (Figure S3A). The resistance of LOTSS standard PrPres to PK digestion was similar to that of 263 K (Figure S3B).
Histopathological studies of animals from the 2d passage of LOTSS revealed characteristic signs of TSE infection including spongiform degeneration, neuronal loss, reactive astrogliosis and deposition of disease-associated PrP in the brains and spinal cords (Figures 2 and 3). No inflammatory changes were observed in the brains and no immunoreactivity for disease-associated PrP was found in the spleens (data not shown). Spongiform change, neuronal loss, and reactive astrogliosis were predominantly found in the thalamus and the brainstem followed by caudate-putamen and spinal cord, however, they were minor in the frontal cortex, hippocampus or cerebellum (Figure 3A and C). Similar to the animals infected with SSLOW, a synthetic strain that was previously produced as a result of inoculation of NBH-annealed rPrP fibrils [6], LOTSS-infected brains from the 2nd passage displayed prominent, large plaques and amorphous deposits in the subpial, periventricular and periaqueductal subependymal regions (Figures 2 and 3). Only mild synaptic PrP immunoreactivity was found in the thalamus, brainstem, caudate-putamen and spinal cord, while no noticeable synaptic PrP deposition was observed in frontal cortex, hippocampus, or cerebellum (Figures 2 and 3C). Previously, synaptic PrP immunoreactivity was defined as a type of disease-associated PrP that co-localizes with synaptophysin [20]. Overall, the lesion profile and the pattern of PrP immunostaining was reminiscent of those observed in SSLOW-inoculated animals [6], but significantly different from those in 263 K-infected brains.
Fig. 2. Analysis of PrP deposition in LOTSS-inoculated animals from the 2d passage. 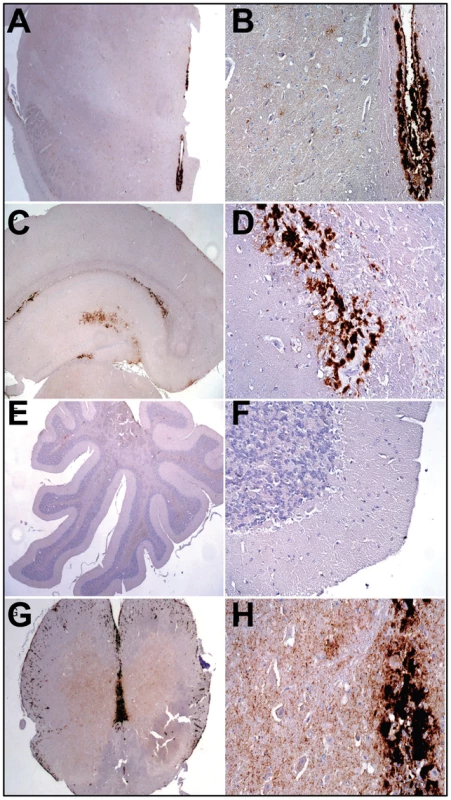
(A, B) PrP plaques in the subependymal regions and minor synaptic immunoreactivity in the basal ganglia and thalamus. (C, D) PrP plaques in the subependymal regions in hippocampus and frontal cortex. (E, F) Very minor focal PrP deposition in cerebellum. (G, H) Plaques around the aqueductus and synaptic PrP immunoreactivity in the posterior and anterior horns of spinal cord. Fig. 3. Histopathological analysis of brains of LOTSS-inoculated animals. 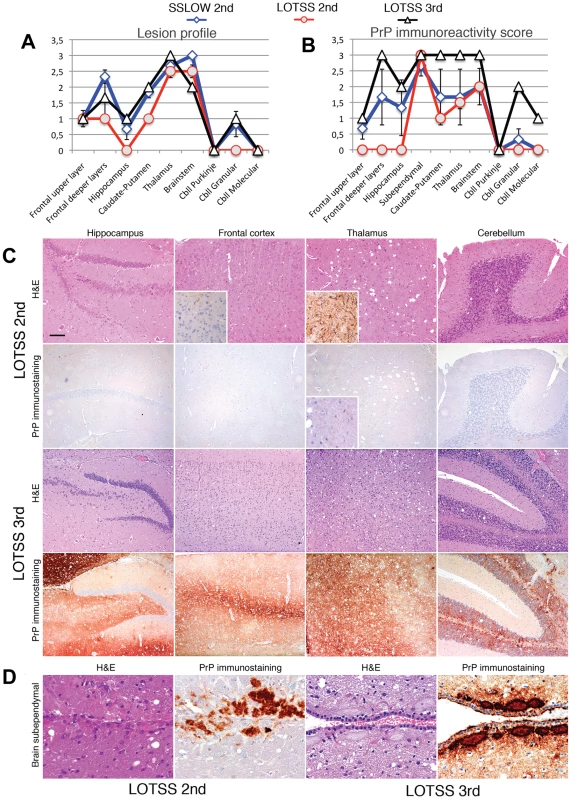
Lesion profile (A) and PrP immunopositivity score (B) for hamsters from the 2nd (o, red lines) or 3rd passages (Δ, black lines) of LOTSS. The histopathological profiles for SSLOW-inoculated hamsters (2nd passage, ◊ blue line) were previously described in [6] and are provided as a reference. The lesion profile was obtained by averaging the scores for spongiform change, neuronal loss and gliosis for three animals within each group. (C) Comparison of spongiform changes in the hippocampus, frontal cortex, thalamus, and cerebellum stained with Hematoxylin and Eosin (H&E; upper panels) or anti-PrP 3F4 antibody (lower panels) in hamsters from 2nd or 3rd passages of LOTSS, as indicated. Insets in H&E stained images of the frontal cortex and thalamus from LOTSS 2nd animals represent GFAP immunostaining, and enlargement of the PrP immunostaining. (D) Brain subependymal region stained with H&E and 3F4 antibody in LOTSS-inoculated animals from 2nd (left panel) or 3rd (right panel) passages. Scale bar = 00 µm in C and = 60 µm in D. Differences between LOTSS and SSLOW
In previous studies, we showed that serial passaging of rPrP fibrils annealed in NBH in Syrian hamster produced a synthetic strain (designated as SSLOW) with highly unique clinical and neuropathological features [6]. To test whether LOTSS is different from SSLOW or 263 K, we employed a conformational stability assay that monitors GdnHCl-induced denaturation using 3F4, D18 and SAF-84 antibodies that recognize PrP regions 109–112, 133–157 and 160–170, respectively (Figure 4). Consistent with previous study [21], the denaturation profiles monitored by three antibodies were superimposable for 263 K (Figure 4A). This result suggests that 263 K follows apparent two-state unfolding without notable stable intermediates. In contrast to 263 K, SSLOW showed a dissociation of the denaturation profile monitored by SAF-84 from those monitored by 3F4 or D18 (Figure 4B), while LOTSS exhibited dissociation of the profiles monitored by D18 and SAF-84 from that of 3F4 (Figure 4C). This experiment revealed that (i) three strains showed three individual strain-specific unfolding patterns; (ii) SSLOW and LOTTS exhibited more complex unfolding behavior than 263 K, (iii) in LOTSS, the central and the C-terminal domain were found to be more stable than the N-terminal domain. Furthermore, the region tracked by D18 antibody was found to be considerably more stable in LOTSS than in SSLOW or 263 K. An alternative experimental format of the conformational stability assay that employs PK-digestion and Western blot instead of dot blot confirmed that three strains were conformationally different (Figure S4).
Fig. 4. Analysis of epitope-specific conformational stability. 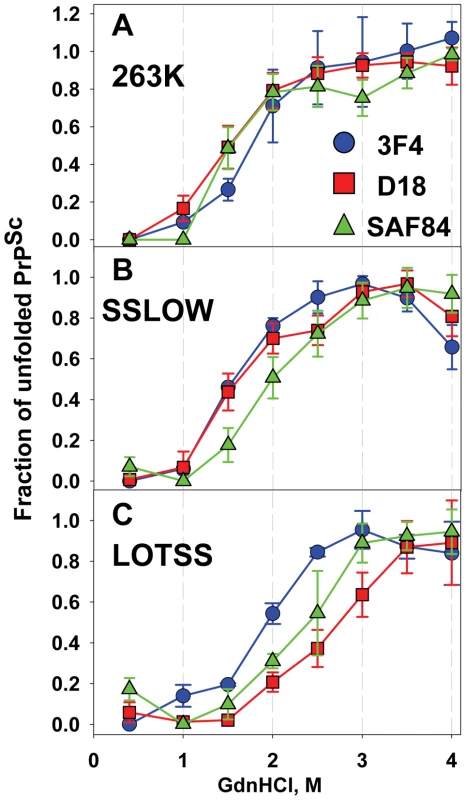
Conformational stability profiles for 263 K (A), SSLOW (B), or LOTSS (C) monitored in GdnHCl-induced denaturation assay using 3F4 (•, blue), D18 (▪, red) or SAF-84 (▴, green) antibodies. Data represent average ± SD of four denaturation experiments. Brain materials from SSLOW- or LOTSS-inoculated animals from the 2nd passages were used. 96-well dot blot was used instead of Western blot for quantitative analysis of multiple replicas in a single blot assay. In addition to the strain-specific differences in unfolding pattern, LOTSS was found to be different from SSLOW or 263 K with respect to the ratio of intensities of SAF-84 to 3F4 immunostaining. Regardless of whether BHs were treated with PK or not and regardless of whether Western blot or dot were used for analysis, LOTSS-infected BHs consistently showed substantially higher ratios of SAF-84 to 3F4 intensities than SSLOW or 263 K (Figures S5 and 1B).
RNA-dependency emerged during the first passage
Previous studies established that RNA serves as a cofactor in replication of hamster prion strains [22], [23]. Furthermore, formation of authentic mammalian PrPSc in vitro de novo from purified PrPC or rPrP was also shown to require RNA [9], [10]. Because the current study employed the fibrillation protocols that lacked RNA or any other cellular components, rPrP amyloid fibrils did not require RNA for forming or self-propagating their individual cross-β structures [18]. Therefore, we were interested in testing whether RNA-dependency evolved with a transformation of rPrP amyloid structure into a structure of PrPres. To address this question, sPMCAb reactions were seeded with BH that contained atypical PKres products from the 1st passage, and amplification was carried out using normal or RNA-depleted BHs as a source of PrPC. In the presence of RNA, a gradual increase in the amounts of standard, 3F4-positive PrPres material was observed with each subsequent round (Figure 1C). This result illustrates that in the presence of RNA, LOTSS PrPres amplification rate exceeded 10-fold per round. However, in RNA-depleted BH, the amounts of PrPres decreased with each subsequent round and disappeared below detectable levels by the 5th round suggesting that the amplification rate was less than 10-fold per round (Figure 1C). In our experience, this amplification rate was similar to the rate displayed by 263 K in RNA-depleted NBH (data not shown). Therefore, in contrast to amyloid fibrillation reactions, the amplification of LOTSS PrPres showed a strong RNA-dependency, just like amplification of other hamster strains [23]. Notably, an increase in PK-resistant material during PMCAb was entirely attributed to the amplification of standard PrPres, whereas no detectible amplification of the atypical PrPres that produces 13 - or 16-kDa PK-resistant bands was observed regardless of the presence or absence of RNA. Overall, this experiment revealed that the transformation from the RNA-independent structure of amyloid fibrils to an RNA-dependent structure of LOTSS PrPres occurred during the first passage.
Evolution of LOTSS neuropathological and biochemical features during serial transmission
Despite substantial amounts of PrPres in the brains and the fact that LOTSS PrPres exhibits key physical attributes of authentic PrPSc, LOTSS-infected animals remained healthy in the second passage. There is a possibility that the clinical signs associated with LOTSS were not clearly visible and only involved impairment of cognitive functions. Lack of clinical signs typical for TSEs might indicate that LOTSS is an intrinsically silent strain, i.e. it is characterized by transmissible formation of PrPres that displays minimal neurotoxicity. Alternatively, LOTSS might not be silent but the second passage represents a silent stage in evolution of LOTSS pathological features. To distinguish between these alternatives, a third passage was conducted. The very first clinical signs of the disease including startle response to sound and touch were noticed starting from 10th to 12th months post inoculation. Similarly to SSLOW-inoculated animals, the progression of disease in LOTSS-infected animals was very slow. After the first signs, 5 to 8 months were required for the disease to progress to the terminal stage (Table S1). Overall, the set of clinical symptoms was similar to that of SSLOW strains (severe obesity, dry hair and hair loss, difficulty in righting, substantially reduced activity) but different from previously described hamster strains.
Three animals were analyzed at 307 days postinoculation, at which two animals were yet asymptomatic, while the third one began to show the first signs. While the PrPres amount varied between these three animals, with the largest amount observed in the symptomatic brain, the relative ratio of atypical PrPres (positive for SAF-84) to standard PrPres (positive for 3F4 and SAF-84) remained approximately the same and was similar to those observed in majority of animals at the end of the 2nd passage (Figure 5A). Because of the complexity in the PKres profile and, specifically, the overlap between the upper 21 kDa band of atypical PrPres with the middle band of standard PrPres, the dynamics in PrPres profile is shown in parallel as a schematic diagram (Figure 5B). The variation in the total PrPres amounts by 307 days was similar to the range seen at the end of life on the 2nd passage (Figure 5A).
Fig. 5. Evolution of PrPres during LOTSS serial transmission. 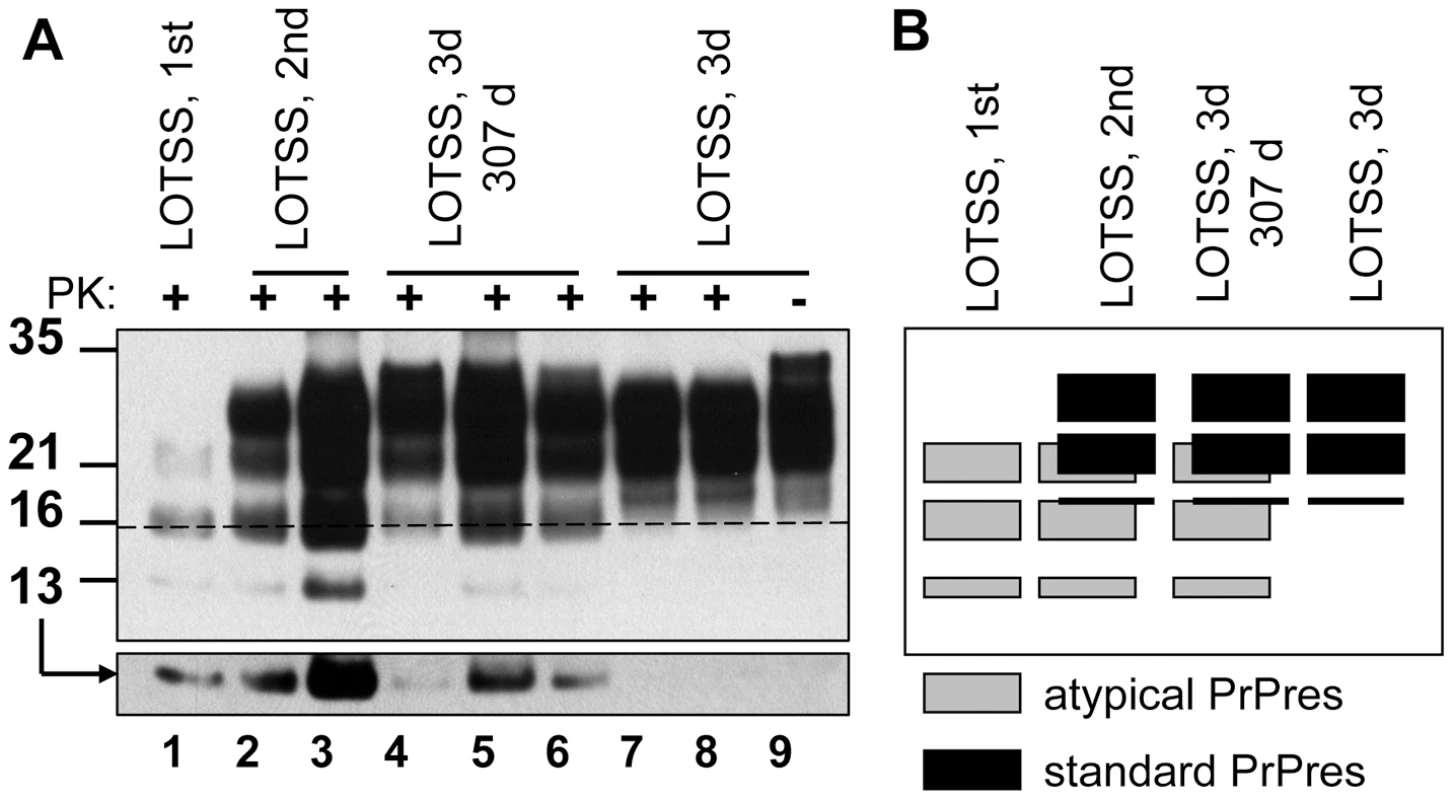
(A) Western blotting of BHs of animals from the 1st, 2nd, or 3rd passages of LOTSS stained with SAF-84 antibodies. Animals from the 1st and 2nd were euthanized at 661 days postinoculation in the absence of clinical disease, whereas animals from the 3rd passage were euthanized at the beginning of first clinical signs, i.e. 307 days postinoculation (lanes 4, 5 and 6), or at the end of the terminal stage (lanes 7, 8). Overexposed Western blot with 13 kDa atypical PrPres fragment is shown in bottom panel. Dashed line marks the central position of the atypical 16 kDa band. Undigested 10% NBH is provided as a reference. (B) Schematic representation of the dynamics in PrPres profile during serial transmission, where gray boxes represent atypical PrPres, whereas black boxes represent standard PrPres. The remaining animals of the 3rd passage were euthanized between 485 and 568 days postinoculation, as they approached the terminal stage (Table S1). Only a very modest if any increase in the total amount of PrPres was observed during the progression of clinical disease (Figure 5A). Surprisingly, the relative ratio of atypical to standard PrPres changed dramatically within the clinical stage (Figure 5A). As judged from the disappearance of the 16 and 13 kDa bands, atypical PrPres was fully replaced by standard PrPres as the clinical disease progressed to the terminal stage.
Overall, the atypical PrPres was observed during three serial passages, however, its amount diminished to undetectable levels during the 3rd passage clinical stage (Figures 1B and 5). While the atypical PrPres was capable of replicating in a brain, this isoform was not amplifiable under standard sPMCAb conditions (Figure 1C). This first signs of standard PrPres appeared in the first passage, although at the levels detectible only by PMCAb, but not Western blotting (Figure 1A and C). While the accumulation rate of standard PrPres varied between animals within the same group (Figure S2), the amount of standard PrPres continued to increase gradually during the 2nd and 3rd passages until it fully replaced the atypical PrPres by the end of the 3rd passage.
Neuropathological analysis of animals euthanized at 307 days postinoculation in a 3rd passage revealed plaques and granular PrP deposits in the subpial, periventricular, and subependymal areas, as well as fine perineuronal and synaptic PrP immunoreactivity in thalamus and basal ganglia (Figure 6). These features were similar to those observed in LOTSS-inoculated animals from the 2nd passage (Figure 2). However, in contrast to the animals from the 2nd passage, the frontal cortex showed prominent diffuse synaptic PrP immunoreactivity and perineuronal PrP deposition (Figure 6).
Fig. 6. Comparison of PrP deposition at the early and late stages of the disease during the 3rd passage. 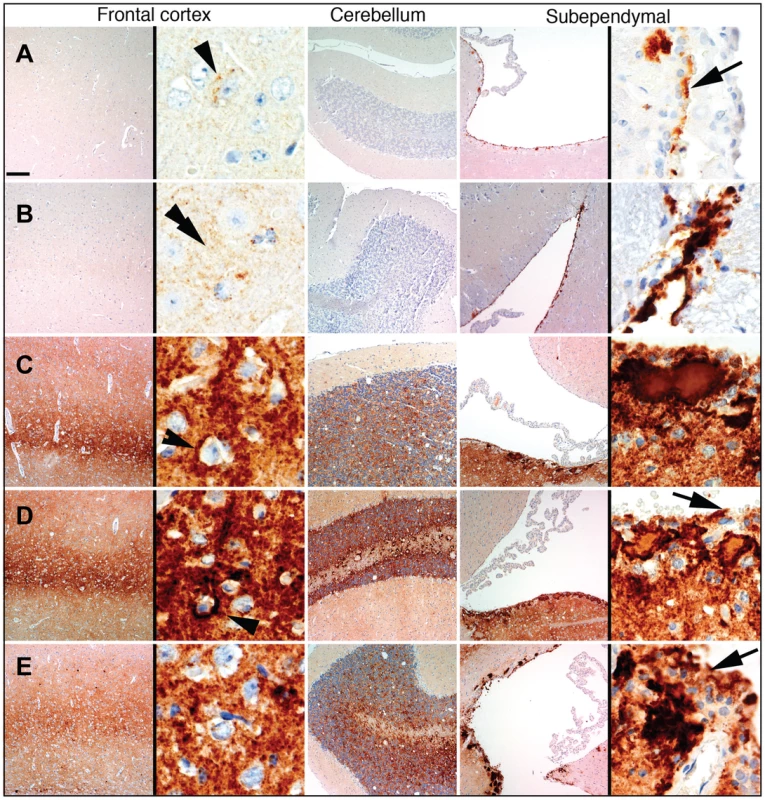
PrP deposition in frontal cortex, cerebellum or subependymal areas in two animals euthanized at 307 days postinoculation (A, B), and three animals euthanized at the terminal stage of the disease at 494, 521, and 494 days postinoculation (C, D, and E, respectively). Scale bar upper left represents 40 µm for all except for the right panels of the frontal cortex and subependymal images where it represents 5 µm. Arrowheads indicate perineuronal deposits, double arrowhead indicates fine synaptic immunoreactivity and arrows indicate granular immunoreactivity around ependymal cells. Upon development of clinical disease in animals from the 3rd passage, a dramatic increase in synaptic PrP immunoreactivity was observed in all examined regions, including the hippocampus, frontal cortex, thalamus, and granular layer of the cerebellum (Figures 3C and 6). In the animals from the 2nd LOTSS passage, only a few brain regions showed weak synaptic PrP immunoreactivity indicating that this pathological hallmark had been evolving with serial transmission (Figures 2 and 4C). Furthermore, the progression of clinical disease was accompanied by accumulation of prominent perineuronal PrP deposition in the deeper layers of the frontal cortex (Figure 6) and thalamus. During the clinical course, the smaller plaques or granular deposits in the subependymal areas evolved into large plaques (Figure 6). In addition, at the terminal stage, perivascular miniplaques appeared in the cerebellum and brainstem (Figure 6 C, D and E).
Overall, LOTSS 3rd passage presented with a quantitative and qualitative change in the neuropathological phenotype. While subependymal plaques were observed in animals from both 2nd and 3rd passages, the animals of the 3rd passage were characterized by substantially more pronounced PrP immunoreactivity including immunopositivity in the regions that did not show any PrP deposition in LOTSS 2nd. In addition, prominent diffuse synaptic and perineuronal PrP deposition appeared as a correlate for the clinical symptoms in LOTSS 3rd indicating that the lack of clinical disease in 2nd passage could be attributed to the limited synaptic deposition. These results suggest that LOTSS pathological features evolve over substantial time period, which might take several serial passages. Comparisons of PrP immunoreactivity score shows that new brain regions including frontal layers, hippocampus and cerebellum became involved during the 3rd passage (Figure 3B). The lesion profile obtained by averaging the spongiform changes, neuronal loss and gliosis was found to be very similar in LOTSS - and SSLOW-inoculated animals but different from the profiles in other hamster strains suggesting that LOTSS and SSLOW belong to the same family of strains (Figure 3A).
Preparation of rPrP amyloid fibrils lacks any detectible PrPSc
Two alternative mechanisms can be put forward to explain the low success rate in triggering transmissible prion disease by rPrP amyloid fibrils. According to one mechanism, the preparations of rPrP amyloid fibrils contain very small amounts of PrPSc or particles with the structure of authentic PrPSc. If this is true, the low success rate and long silent stage of the disease are attributed to the long time required for amplification of this extremely small amount of PrPSc. An alternative mechanism postulates that there are no PrPSc particles in the preparations of amyloid fibrils. Instead, amyloid fibrils, while structurally different from PrPSc [14]–[17], nevertheless are capable of triggering formation of PrPSc when inoculated into animals. In this case, the low success rate and silent stage of the disease are attributed to inefficient seeding of PrPres by recombinant PrP amyloids and subsequent evolution of PrPres structural and biological features, a process that eventually give rise to PrPSc.
To distinguish between two mechanisms, we employed sPMCAb that can detect miniscule amounts of PrPSc. In the first series of experiments, we tested whether miniscule amounts of LOTSS PrPSc could be efficiently detected by sPMCAb. 10% BHs prepared from the 2nd passage of LOTSS were diluted in ten fold serial steps, then aliquots from each dilution were used to seed sPMCAb reactions (Figure 7A). In 100 µl of PMCAb reaction volume, 109 - and 1010-fold diluted LOTSS brain material was detected with 100% and 83% success rate, respectively (Figure 7B). The reactions seeded with brain material diluted to 1011-fold or higher were all negative. Six sPMCAb rounds were sufficient for amplification of the highest dilutions of LOTSS brain material to the level detectible by Western blot. Non-seeded normal BH subjected to six sPMCAb rounds were all negative illustrating that positive signals were not due to spontaneous formation or cross-contamination (Figure S1C). As judged from the fitting of experimental data on sPMCAb amplification of serially diluted LOTSS brain material (Figure 7B), a PMCAb50 titer was found to be 1010.26 (Figure 7B). This value represents a dilution fold at which there is a 50% chance of finding at least one PMCAb-active PrPSc particle capable of initiating PMCAb amplification per 100 µl of the reaction volume. This experiment demonstrated that the sPMCAb format employed here can effectively detect miniscule amounts and, presumably, as little as a single LOTSS PrPSc particle.
Fig. 7. Preparations of rPrP fibrils have no detectible PrPSc. 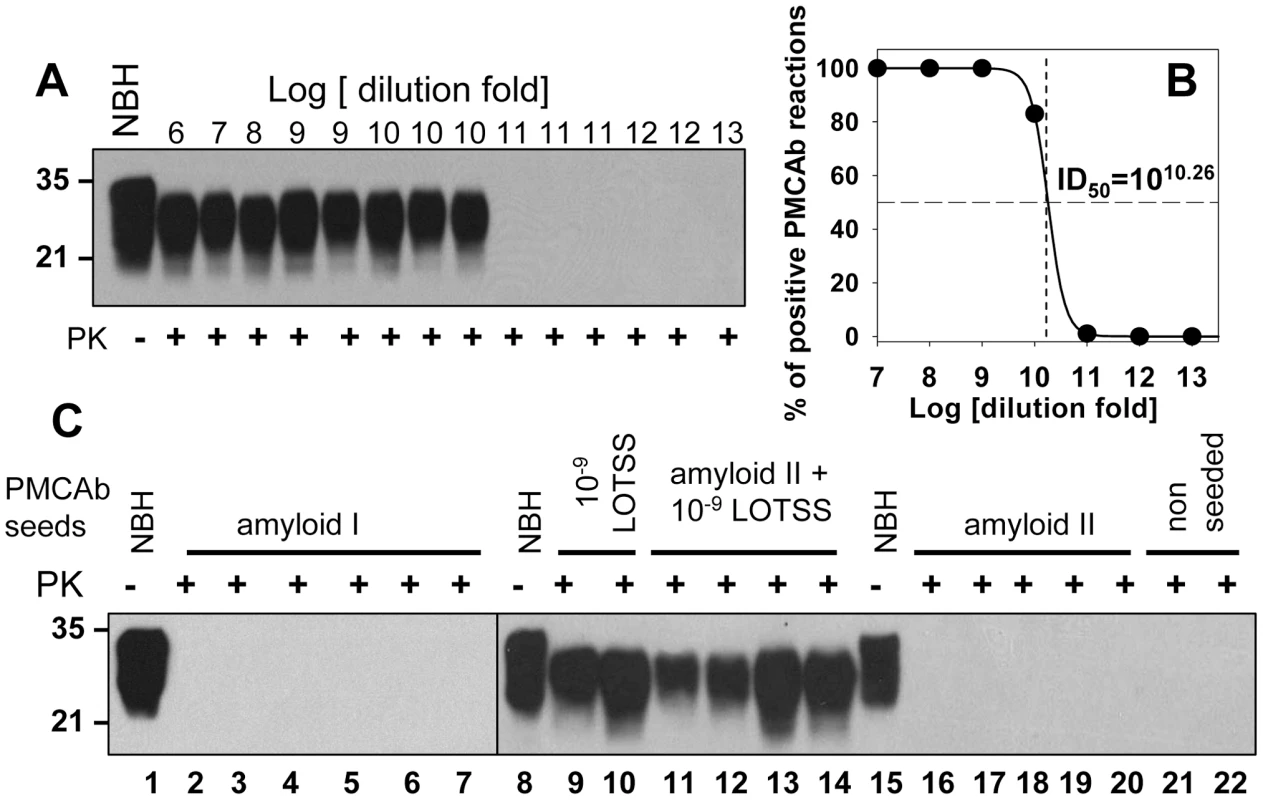
PMCAb titration of LOTSS brain material (A and B). (A) LOTSS brain material was serially diluted to up to 1013-fold and each dilution was subjected to six rounds of sPMCAb. Representative results are shown on Western blot stained with 3F4 antibody. Undigested 10% NBH is provided as a reference. (B) For each dilution of brain material, the fractions of sPMCAb reactions with a positive signal on Western blot were plotted against the logarithm of dilution (•) and non-linear least-square regression to a sigmoidal function (black solid line, R = 0.9999) was used to calculate PMCAb50 titer. (C) sPMCAb reactions were seeded with two independent preparations of BSA-annealed rPrP amyloid fibrils (lanes 2–7 with amyloid I or lanes 16–20 with amyloid II), 109-fold diluted LOTSS brain material (lanes 9, 10), or with amyloid fibrils II and 109-fold diluted LOTSS brain material (lanes 11–14), then six rounds of sPMCAb were conducted for each condition. The final concentraton of rPrP amyloid fibrils in sPMCAb reaction was 10 µg/ml for amyloid I or 5 µg/ml for amyloid II. Two non-seeded reactions are shown as negative control. Brains from LOTSS-inoculated animals from the 2nd passages were used for all experiments. Undigested 10% NBH is provided as a reference. Western blots were stained with 3F4. In the next series of experiments, sPMCAb was employed for detecting PrPSc in the preparations of rPrP amyloid fibrils subjected to annealing in the presence of BSA. Eleven sPMCAb reactions seeded with rPrP amyloid fibrils from two independent amyloid preparations were conducted. No positive signals were detected in any reactions after six sPMCAb rounds (Figure 7C). As judged from the previous experiment on PMCAb titration, six sPMCAb rounds were sufficient for detecting LOTSS PrPSc at the level of a single particle in 100 µl of the reaction volume.
To rule out the possibility that the negative result in detecting PrPSc in the preparations of amyloid fibrils was due to the inhibitory effect of fibrils on amplification of PrPSc, sPMCAb reactions were seeded with rPrP amyloid fibrils mixed with 109-fold diluted LOTSS brain material. 109-fold diluted brain material represents the highest dilution at which 100% sPMCAb reactions were positive in the PMCAb titration experiment. Four independent sPMCAb reactions were conducted and all gave a positive signal on Western blot (Figure 7C). This experiment confirmed that the presence of rPrP fibrils did not diminish the sensitivity of detection of LOTSS PrPSc by sPMCAb. Taken together, these experiments revealed that the preparations of rPrP amyloid fibrils do not contain any PrPSc particles that could be detected by sPMCAb providing strong support for the second mechanism. In our experience and consistent with the previously published data [24], sPMCAb is ∼400 to 4000 fold more sensitive than bioassay.
Transition from rPrP fibrils to LOTSS PrPSc
During serial transmission of LOTSS, we observed a dynamic transformation of the PK-resistant band pattern. PrPres consisted of atypical C-terminal fragments at the 1st passage, and then atypical PrPres shifted to a predominantly 21–30 kDa PK-resistant core typical for PrPSc at the 3rd passage (Figures 1B and 5). To determine whether the atypical PrPres encompasses the same region as the PK-resistant core of rPrP fibrils, BH from the animal of the 1st passage was treated with PNGase F to remove carbohydrates. The PK-resistant core of annealed rPrP fibrils was previously found to consists of two major fragments of ∼138–231 and 152/153–231 [18], [25]. Upon treatment with PNGase F, the three PK-resistant bands of 21, 16 and 13 kDa merged into a single band of ∼13 kDa illustrating that these three bands represent di-, mono - and unglycosylated forms of a single PK-resistant fragment (Figure 8). This C-terminal fragment reacted with R1 (epitope 225–231) and SAF-84 (epitope 160–170), but not with 3F4 (epitope 109–112) or D18 (epitope 133–157) antibodies (Figures 1A, and 8A and B). Therefore, the atypical PrPres fragments matched the lower PK-resistant band of the fibrillar core that encompassed resides 152/153–231. The difference between PAGE-mobility of the rPrP 152/153–231 fragment and 13 kDa atypical PrPres fragment was likely due to a GPI-anchor in the last one. This result supports the hypothesis that the structure of LOTSS PrPres originated from rPrP fibrils.
Fig. 8. Analysis of the C-terminal PrPres fragments. 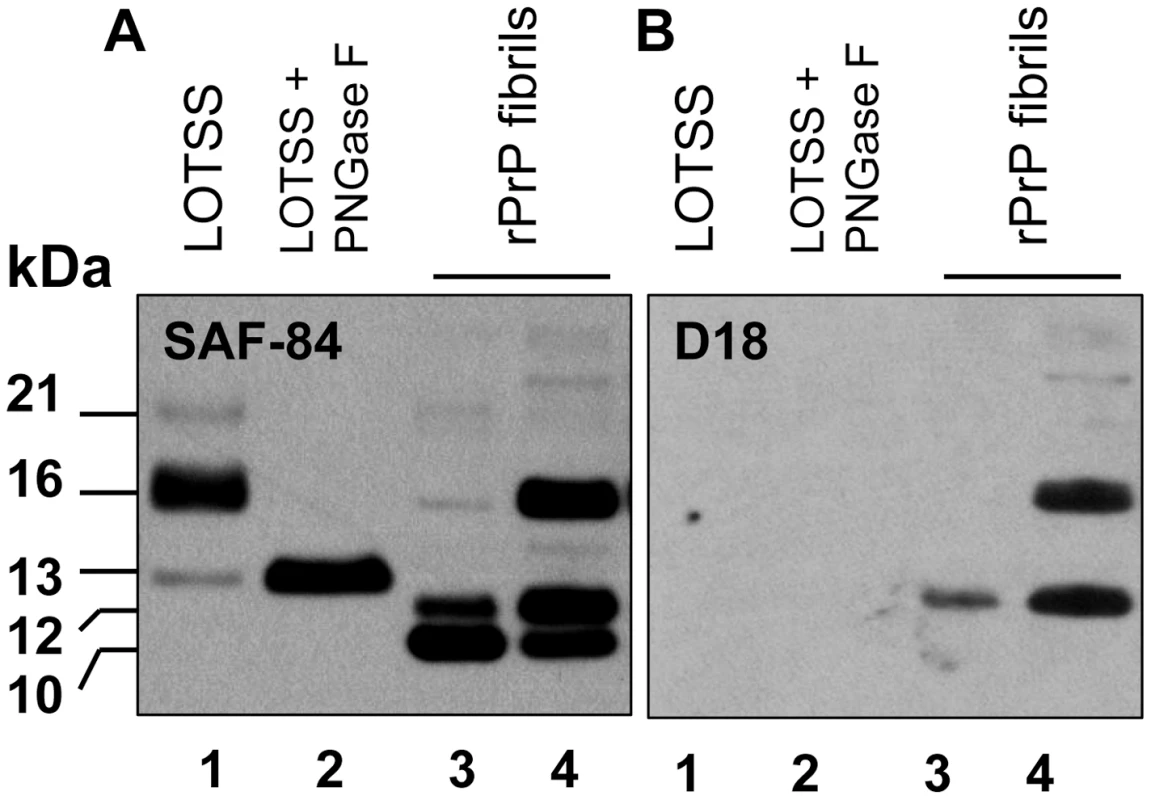
Western blot of PK-digested BH from the animal inoculated with BSA-annealed rPrP amyloid fibrils (lanes 1, 2) or BSA-annealed rPrP amyloid fibrils (lanes 3 and 4) stained with SAF-84 (panel A) or D18 (panel B) antibodies. BH in lane 2 was treated with PNGase F. Before PK digestion, rPrP fibrils were mixed with 10% NBH to balance the condition for PK digestion. Samples in lanes 1, 2 and 3 were treated with 20 µg/ml of PK, and the sample in lane 4 – with 10 µg/ml of PK. Western blot was stained with SAF-84. Together with previous data (Figure 1), the results on epitope mapping illustrate that a dynamic transformation of LOTSS PrPres occurred during three serial passages. This transformation involved an increase in the size of the PK-resistant core from the atypical PrPres to the one typical for authentic PrPSc. Notably, the change in size was accompanied by a significant shift in the ratio of three PrP glycoforms recruited by PrPres. While atypical PrPres preferred monoglycosylated PrP (16 kDa band, Figure 1B), standard PrPres at the 3rd passage favored a diglycosylated PrP (Figure 1B and D), a feature common for all previously known hamster strains.
Discussion
In the current work, transmissible prion disease was generated in wild type animals upon inoculation of rPrP amyloid fibrils produced in vitro in the absence of a cellular environment or co-factors. When triggered by recombinant fibrils, only a small fraction of animals showed signs of infection. Furthermore, it took two additional serial passages for a fully developed disease with a distinct set of symptoms to evolve. Two alternative models could be put forward to explain the low success rate in infecting the animals by rPrP fibrils (Figure 9). According to the first model, the preparations of rPrP amyloid fibrils contain very small amounts of PrPSc or particles with a structure of authentic PrPSc (Figure 9A). The second model proposes that the formation of PrPSc and emergence of transmissible prion diseases could be triggered in wild type animals by seeding material that lacks PrPSc (Figure 9B). This model takes into consideration previous data that showed that the structures of PrPSc and rPrP fibrils are fundamentally different as evident from X-ray diffraction analysis [14], [17], FTIR spectroscopy [26], [27], AFM and EM imaging and seeding-specificity assay [16]. Because of the structural differences, rPrP fibril-induced seeding of PrPSc is not efficient, which could explain the low rate of infection in the first passage. For the same reasons, transformation of the rPrP amyloid structure into PrPSc structure might involve several steps before fully infectious authentic PrPSc emerges, yielding the long silent stage to clinical disease.
Fig. 9. Schematic representation of two mechanisms responsible for generating transmissible prion diseases de novo. 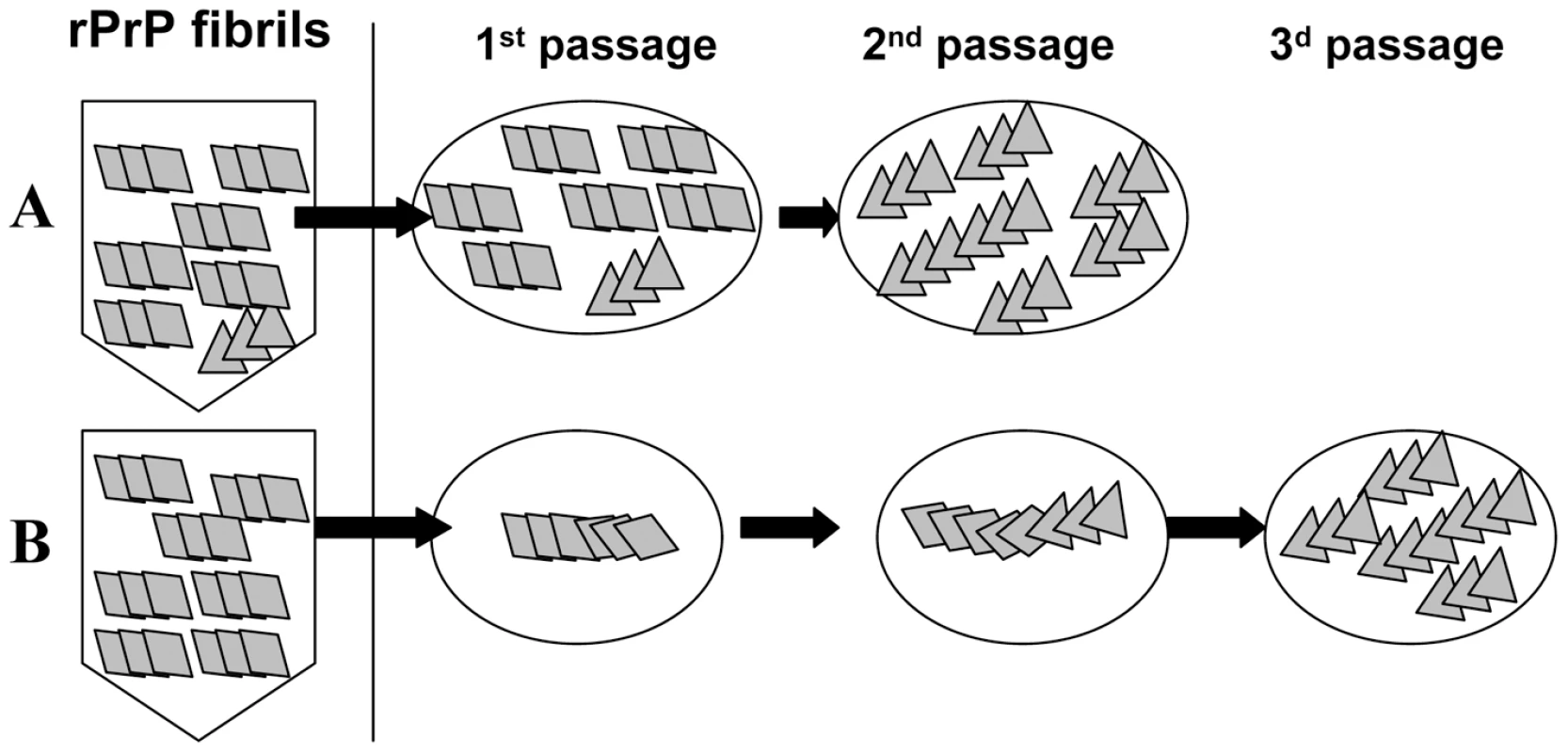
According to the first mechanism (A), the preparations of rPrP amyloid fibrils contain very small amounts of PrPSc. The silent stage of the disease is attributed to the long time required for amplification of this extremely small amount of PrPSc. A second mechanism referred to as deformed templating postulates that there are no PrPSc particles in the preparations of amyloid fibrils (B). Instead, when inoculated into animals, amyloid fibrils can seed conversion of PrPC into PrPres, although with a low efficiency. PrPres undergoes slow transformation, a process that might require long silent stage, before authentic PrPSc emerges. The results of the current and previously published studies strongly support the second model. First, no PrPSc was found in the preparations of rPrP amyloid in an assay format that detects miniscule amounts of prions. One could argue that the efficiency of prion amplification and the sensitivity of detection in sPMCAb are strain-specific. We found that the efficiency of LOTSS PrPSc amplification in sPMCAb was similar to that of 263 K PrPSc (Figure 3SA). 100% and 83% of sPMCAb reactions seeded with asymptomatic LOTSS brains serially diluted 109 or 1010-fold, respectively, were positive, whereas all reactions seeded with 1011-fold and higher dilutions were negative (Figure 7). This result suggests that the limiting dilution was reached in the experiment on sPMCAb titration. It also illustrates that 1010-fold diluted LOTSS brain material has only one or very few PMCAb-active PrPSc particles per 100 µl of the reaction volume, and that these quantities of LOTSS PrPSc could be effectively detected in our assay format. If one assumes that the first model is correct, the amount of infectivity per 50 µl of inoculum should be equivalent to approximately 0.5–1 infectious dose to account for the low infection rate in the first passage (3 out of 7 animals). This amount of infectivity is equivalent to ∼10,000–100,000 PrP molecules or to ∼100–1000 PrPSc particles, assuming that an average PrPSc particle consists of ∼100 PrP molecules [24]. This amount of PrPSc is well above the detection limits of sPMCAb, and should have been easily detected if present in preparations of rPrP fibrils.
Second, the experimental protocol used for producing rPrP amyloid fibrils employs denaturants (a mixture of 1M GdnHCl and 3M urea) - solvent conditions under which PrPSc undergoes denaturation. Because rPrP fibrils are much more conformationally stable than PrPSc [6], [21], [28], rPrP fibrils can be formed under solvent conditions where PrPSc is largely denatured. Furthermore, conversion of PrPC into authentic PrPSc in vitro requires RNA and lipids [9], [10], whereas rPrP amyloid fibrils were formed in the absence of any cellular co-factors. Therefore, it is highly unlikely that PrPSc particles with authentic structures could be formed during the preparation of rPrP fibrils conducted in the absence of co-factors essential for authentic PrPSc structures and under solvent conditions that promote PrPSc denaturation.
Third, when transmissible prion disease is triggered by rPrP amyloid fibrils, a decrease in PrPSc conformational stability was observed during serial passages of synthetic prions [6], [29]. Similar dynamics in PrPSc conformational stability was observed regardless of whether transgenic mice or Syrian hamsters were used for inoculating rPrP fibrils suggesting that a common pathway in the genesis and evolution of transmissible prion protein structures might exist [6], [29]. Observed changes in the physical properties illustrate that PrPSc structure undergoes transformation during serial passages providing a direct support for the second model.
Fourth, the first model has difficulties in explaining two silent passages. While the first silent passage could be attributed to the low titer of infectious material in the preparation of rPrP fibrils, it is challenging to explain the second silent passage without assuming that the neurotoxic properties of LOTSS PrPSc are evolving slowly. Together with a dynamic change in PK-resistant profile, the second silent passage is indicative of transformation in PrPres structure and its neurotoxic features, which is consistent with the second model.
Fifth, significant quantitative and qualitative changes in PrP deposition were observed during LOTSS serial transmission. In animals from the 2nd passage, LOTSS-specific PrP was predominantly found in the form of subpial and subependymal plaques, whereas animals from the 3rd passage showed a dramatic increase in diffuse synaptic immunoreactivity and perineuronal deposition in addition to plaques. Furthermore, involvement of new brain regions positive for PrP immunostaining was observed during the serial passage of LOTSS, pointing out possible changes in strain neurotropism, which again is consistent with a dynamic transformation in PrPres biological and physical features. Previously, a change in the pattern of PrP accumulation was found to accompany strain adaptation during serial transmission in a new host [30]. The changes in neurotropism or distribution pattern could not be attributed solely to an increase in the total amounts of LOTSS PrPres, because certain brain regions including frontal deeper layers, hippocampus and granular cerebellum displayed a disproportionally high increase in the amounts of LOTSS-specific PrP deposition in the 3rd passage despite a shorter life span.
Sixth, in recent studies on synthetic prions, a strong correlation between conformational stability of rPrP amyloid fibrils, the stability of PrPSc produced in animals upon inoculating rPrP fibrils and the incubation time to disease was described [5]. If a miniscule fraction in the preparation of rPrP fibrils is responsible for the disease, the correlation between stability of rPrP amyloid, which is a bulk property of fibril preparation, and PrPSc would be challenging to explain. Again, these results provide a strong support for the second model.
Seventh, as judged from clinical and neuropathological features, both synthetic strains generated de novo upon inoculating Syrian hamsters with rPrP fibrils (LOTSS and SSLOW) were remarkably different from all previously known rodent strains including those generated in sPMCA [8]–[10]. Among the most distinguishable LOTSS and SSLOW features were the long incubation time to and slow progression of clinical disease, accumulation of large plaques in subpial and subependymal areas, distinctive lesion and PrP immunoreactivity profiles and unusual clinical phenotype (obesity, hair loss). The fact that SSLOW and LOTSS can be separated into a distinct class of prion strains remarkably different from strains produced in sPMCA or isolated from animals is consistent with the hypothesis that SSLOW or LOTSS PrPSc originated from unique structures or rPrP fibrils.
Eighth, observation of the atypical, C-terminal, low molecular weight PK-resistant products in the first passage strongly supports the last conclusion. Both, atypical PrPres and rPrP fibrils had very similar if not identical PK-resistant cores that encompassed epitopes for SAF-84 and R1, but not 3F4 antibodies [18], [25]. This result supports the idea that the structure of fibrillar rPrP gave origin to LOTSS PrPres.
Ninth, the transformation in LOTSS PrPres structure is demonstrated by an increase in the size of the PK-resistant core from atypical to standard and the change in glycosylation preferences. This transformation provides direct support for the second model. While atypical PrPres predominantly recruited monoglycylated PrP, the LOTSS PrPSc preferred diglycosylated PrP, a feature which is common for all currently known hamster-adapted prion strains.
All the data on synthetic prions accumulated to date support the hypothesis that transmissible prion diseases can be triggered by PrP structures substantially different from that of authentic PrPSc and in the absence of PrPSc in preparation of rPrP fibrils (Figure 9 B). This hypothesis assumes that only a partial overlap or distant similarities in structures of fibrillar rPrP and PrPSc are sufficient for triggering transmissible prion diseases in animals. While the precise mechanistic details for inducing PrPSc formation by conformationally different PrP structures remain to be elucidated, the model of conformational switching provides one possible explanation of how such transformation occurs [31], [32]. According to this model, the global folding pattern of PrP molecules within amyloid fibrils and PrPSc are different, nevertheless, they share a common structural motif. For instance, a common β-strand that can link two structures provides opportunity for limited templating. This model is consistent with experimental observations that the global structures of PrPSc and rPrP fibrils are different [14], [15], [17], and explains that there is a correlation between conformational stability of two structures [5]. Because of only partial overlap between the two structures, the seeding of PrPSc by rPrP fibrils is inefficient, which explains the low infection rate in the first passage. Previous studies on molecular imaging of single amyloid fibrils provided a proof of principle that the conformational switching between two alternative PrP folding patterns can occur within an individual PrP fibril or particle [31].
The current study proposes that a new mechanism designated as ‘deformed templating’ that is different from the classical templating might exist. In classical templating, the folding pattern of a newly recruited polypeptide chain accurately reproduces that of the template. In deformed templating, while the template provides limited seeding, a newly recruited polypeptide chain acquires a new folding pattern which only partially overlaps with the folding pattern of the template. Two glycosyl groups and a GPI anchor present in PrPC might impose steric constraints on the spectrum of folding patterns available to PrPC, therefore providing a driving force behind switching PrP folding patterns from rPrP fibril-specific to PrPS-specific. In fact, the evolution of PrPSc structure might involve at least two steps. The first step involves a transformation of the fibrillar rPrP structure to the structure of atypical PrPres, which is characterized by a short PK-resistant core, a preference for monoglycosylated PrP, and inactivity in PMCAb. We do not know how long it took for the transition from fibrillar rPrP to atypical PrPres to occur in animals. The second step is a transformation from atypical PrPres to LOTSS PrPSc that displays a standard PK-resistant core, favors diglycosylated PrP, is PMCAb-active and shows RNA-dependency in sPMCAb amplification.
If the hypothesis that a fibril-specific PrP folding pattern can template a distinct folding pattern within PrPSc is correct, one can assume that the opposite reaction, i.e. the seeding of rPrP fibrils by PrPSc is also possible. Indeed, several assays exploited the phenomenon of PrPSc-seeded conversion of α-rPrP into amyloid fibrils for detecting miniscule amounts of PrPSc [33], [34]. Interestingly, the structure of rPrP fibrils produced as a result of seeding by PrPSc only distantly resembled those of PrPSc structure and had limited infectivity [35].
Although a number of unique features warrant classification of SSLOW and LOTSS into a separate class of prion strains, PrPSc of both synthetic strains was predominantly diglycosylated, and PMCAb amplification of both strains showed strong RNA dependency, features that were common with other hamster strains [22], [23]. Interestingly, the RNA dependency in LOTSS amplification emerged during the first passage (Figure 1C), whereas the first clinical signs were observed only during the 3d passage. These results suggest that the two stages in the evolution of PrPSc structure – (1) transformation of fibrillar structure, which is RNA independent, into a structure that relies on RNA in self-replication and (ii) acquiring neurotoxic features essential for clinical disease, are separated. Dramatic increase in diffuse synaptic PrP immunoreactivity at the 3rd passage suggests that the clinical expression of the disease might require synaptic PrP deposition. The mechanism underlying significant changes in the PrP deposition pattern during LOTSS serial transmission and possible connections between changes in PrPSc structure and its deposition pattern remain to be explored in future studies.
Inter-species prion transmission to new hosts including human is known to involve a long clinically silent stage that accompanies an adaptation to a new host [36], [37]. In the case of inter-species transmission, the long silent stage, which is referred to as a transmission barrier, is attributed at least in part to the differences in amino acid sequence of host PrPC and donor PrPSc. Moreover, the cross-species barrier could also be due to change in preferences for species-specific cofactors in prion replication [23]. Despite identity in the amino acid sequence of the rPrP inoculum and host PrPC, the silent stage in the current studies was found to be very long and consisted of two passages. Such a long silent stage is presumably attributable to the evolution of PrPSc physical and biological features and appears to be more prominent than those associated with inter-species transmission despite the fact that no change in the same amino acid sequence was involved. Consistent with this view, FTIR and X-ray diffraction analysis revealed that PrPSc isolated from different species display very similar folding patterns, which appear to be fundamentally different from the PrP folding pattern within rPrP fibrils [17], [26], [27], [38].
In other studies on inter-species transmission including the work on transmission of BSE to wild type mice or transmission of the hamster-adapted strain 263 K to transgenic mice carrying murine PRNP gene with a mutation P101L [39], [40], the clinical disease was observed in the first passage but in the absence of detectable amounts of PrPres. In the current studies, the second passage of LOTSS produced large amounts of PrPres, however, without any detectible clinical symptoms. Both sets of data were consistent with the emerging hypothesis that infectious and neurotoxic PrP species might be two different entities [41].
Much of the public health risk from prion diseases derives from their long asymptomatic incubation times [42]. During the asymptomatic period humans pose a real risk of spreading the infection through invasive medical procedures or tissue or blood donation [43]. The current work raises the importance of using antibodies against several PrP regions including the C-terminal region for effective diagnosis of prion diseases. Noteworthy, C-terminal PK-resistant fragments with a length similar to that of atypical PrPres described here were found in a majority of individuals with sporadic CJD [44]. If asymptomatic stages are accompanied by accumulation of predominantly atypical PrPres, while the procedures that rely only on N-terminal antibodies such as 3F4 are employed, the pathogenic process would remain undetected. This study raises a great concern that transmissible prion diseases can be triggered by protein structures which are considered to be not-infectious. Furthermore, spread of transmissible, disease-related prion structures can occur undetected during a silent stage of prion evolution when no clinical expression of the disease can be observed.
The hypothesis that transmissible prion diseases can be triggered by cross-β PrP structures substantially different from that of authentic PrPSc has large implications for understanding the ethiology of prion and, possibly, other neurodegenerative diseases. Several recent studies documented that amyloid forms of several proteins linked to neurodegenerative diseases were capable of seeding their own aggregation in a prion-like manner in a cell and spreading from cell to cell through the nervous system (reviewed in [45]–[47]). The recent study provided the first illustration that pathological changes associated with non-prion neurodegenerative diseases could be induced or transmitted through inoculation of the aggregated forms of non-prion proteins such as Aβ [48]. Furthermore, a growing amount of evidence illustrates that amyloids can template structures different from their own [31]. It is generally assumed that self-perpetuating aggregation requires identity in amino acid sequence between seeds and substrate. An increasing number of studies, however, suggest the possibility of cross-talk between non-related amyloidogenic proteins [49]–[51]. In vivo, amyloidosis of one protein was found to be triggered by fibrils of an unrelated protein in a manner similar to cross-seeded polymerization [49]–[51]. Cross-talk between several yeast prion proteins provides another example of how direct interactions between newly forming and pre-existing heterologous fibrils might take place in a cell [52]–[54]. Moreover, pathological studies revealed that protein aggregates produced from two different proteins or peptides, including PrP, Aβ, α-synuclein, immunoglobulin light chain λ, and β2 microglobulin often co-localize within the same amyloid plaques in a variety of organs or tissues [55]–[59]. The promiscuous nature of the propagating activity of amyloid structures can lead to devastating consequences. For instance, cross-talk between non-related amyloidogenic proteins may offer a possible explanation for development of the age-related conformational disorders that are considered to be sporadic. It would be interesting in future studies to define the spectrum of structures and sequences capable of triggering PrPC to PrPSc conversion and induce transmissible prion diseases.
Materials and Methods
Ethics statement
This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocol was approved by the Institutional Animal Care and Use Committee of the University of Maryland, Baltimore (Assurance Number A32000–01; Permit Number: 0309001).
Expression and purification of rPrP and formation of rPrP fibrils
Syrian hamster full-length recombinant PrP encompassing residues 23–231 (rPrP) was expressed and purified according to the previously described procedure [60] with minor modifications [6]. Lyophilized rPrP was dissolved in 6 M GdnHCl to prepare a 3 mg/ml stock solution of rPrP. To form fibrils, the rPrP stock solution was diluted to a final protein concentration of 0.5 mg/ml and incubated at 37°C in 20 mM sodium acetate (pH 5.0), 1 M GdnHCl, 3 M Urea, 150 mM NaCl, 10 mM EDTA for 4 days with continuous horizontal shaking at 600 rpm. Amyloid formation was confirmed by increased Thioflavin T fluorescence and electron microscopy [60]. Fibrils were dialyzed into 10 mM sodium acetate, pH 5.0.
To perform annealing, rPrP fibrils at 0.1 mg/ml were heat-treated in PBS, pH 7.4, in the presence of 5 mg/ml BSA (Sigma, catalog #A3294) and 0.1% Triton X-100 using the following procedure: 50 µl aliquots in thin-wall PCR tubes with domed caps were placed into the PCR machine and subjected to 5 cycles of 1 min incubation at 80°C followed by 1 min incubation at 37°C. All aliquots were pooled together and the combined volume was immediately transferred to the animal facility for inoculation.
Bioassay
For the first passage, weanling Syrian hamsters were inoculated intracerebrally with the preparation of rPrP fibrils described above. Each animal was anesthetized with interperitoneal pentobarbital before receiving 50 µl of inoculum. Hamsters were observed daily for disease starting from the third month postinoculation. They were euthanized at 661 days post inoculation without any signs of clinical disease, their brains removed aseptically and saved for subsequent passage and analysis.
For the second and third passages, 10% BHs prepared by sonication in PBS, pH 7.4 (see below), were dispersed by an additional 30 sec of sonication immediately before inoculation. Each hamster received 50 µl of inoculum intracerebrally under general anesthesia (2% O2/4 MAC isoflurane). After inoculation, hamsters were observed daily for disease.
Proteinase K digestion
Brains were collected aseptically and cut in half with disposable scalpels. One half was used to prepare 10% BHs, while the second half was stored at −80°C for future analysis or fixed in formalin for histopathology. Homogenization was performed on ice in PBS, pH 7.4, with a Sonics vibra cell VCX750 (Newtown, CT) tip sonicatior fitted with a stepped microtip by 2 pulses of 30 sec sonication with 30 sec cooling on ice between the pulses. For the Proteinase K digestion in sarcosyl, an aliquot of 10% brain homogenate was mixed with an equal volume of 4% sarcosyl in PBS, supplemented with 50 mM Tris, pH 7.5, and digested with 20 µg/ml PK for 30 min at 37°C with 1000 rpm shaking (Eppendorf Thermomixer). Alternatively, 1% brain homogenate in conversion buffer was supplemented with 0.25% SDS and 50 µg/ml PK and digested at 37°C for 1 hour. Proteinase K digestion was stopped by adding SDS sample buffer and heating the samples for 10 min in a boiling water bath. After loading onto NuPAGE 12% BisTris gels and transfer to PVDF membrane, PrP was detected with 3F4 (epitope 109–112), D18 (epitope 133–157), SAF-84 (epitope 160–170), or R1 (epitope 225–231) antibody, as indicated. For PK digestion of recombinant fibrils annealed in BSA, fibrils were diluted to 10 µg/ml in 1% NBH supplemented with 0.25% SDS and digested with 10 or 20 µg/ml PK for 1 hour at 37°C.
Deglycosylation of PrPres
Removal of N-linked glycans was performed as previously described [61], using glycerol free PNGase F and supplied buffers (New England BioLabs). 5% BH in sarcosyl was digested with 20 µg/ml Proteinase K as described above. After termination of PK digestion with 2 mM PMSF, the resulting 36 µl of digest was supplemented with 4 µl of 10x glycoprotein buffer and heated at 95°C for 10 min. After heat-treatment, 5 µl of 10xG7 reaction buffer, 5 µl of 10% NP-40, and 3 µl of PNGase F enzyme were added, and the samples were incubated overnight at 37°C. The next day, the samples were boiled for 10 min in SDS sample buffer (Invitrogen) and loaded onto NuPAGE 12% BisTris gels. After transfer to PVDF, PrP was detected with SAF-84 or D18 antibody as indicated.
Conformational stability assay
10% brain homogenates were sonicated for 30 sec at 50% power efficiency within Misonix-4000 microplate horn filled with 350 ml water. Then the samples were diluted to 1% BH with conversion buffer and incubated with various concentrations of GdnHCl within 96 Deep Well 1 ml plate (Fisher #12–566–120) for 1 hour at room temperature. After incubation, all samples were diluted with PBS to A final concentration of GdnHCl of 0.4 M and incubated for an additional 1 h at room temperature. The incubation was followed by loading 50 µl of each denaturation point onto a nitrocellulose membrane using a BioDot microfiltration apparatus (Bio-Rad) according to the manufacturer' manual. After two washes with PBS, the membrane was removed from the apparatus, immediately blocked with 2.5% non-fat milk in PBST for 30 min, incubated with antibody overnight at +4°C, and developed according to the common immunodetection protocol.
To compare relative exposure of 3F4 and SAF-84 epitopes in LOTSS, SSLOW and 263 K, equal amounts of brain homogenates were denatured with 5 M GdnHCl and loaded onto nitrocellulose membrane with a BioDot apparatus. To eliminate the possibility of PrPC influencing the data, the relative exposure of epitopes was also analyzed after mild denaturation with 1 M GdnHCl and PK digestion. Chemiluminescent signals were measured by a Typhoon 9200 variable mode imager.
Protein misfolding cyclic amplification
Healthy hamsters were euthanized and immediately perfused with PBS, pH 7.4, supplemented with 5 mM EDTA. Brains were dissected, 10% brain homogenate (w/v) was prepared using ice-cold conversion buffer and glass/Teflon tissue grinders cooled on ice and attached to a constant torque homogenizer (Heidolph RZR2020). The brains were ground at low speed until homogeneous, then 5 additional strokes completed the homogenization. The composition of conversion buffer was as previously described [62]: Ca2+-free and Mg2+-free PBS, pH 7.5, supplemented with 0.15 M NaCl, 1.0% Triton, and 1 tablet of Complete protease inhibitors cocktail (Roche, Cat# 1836145) per 50 ml of conversion buffer. The resulting 10% NBH in conversion buffer was precleared by 2 min centrifugation at 500 g and the resulting supernatant was used as the substrate in PMCA with beads (PMCAb) reactions [19]. 100 µl samples in 0.2 ml thin-wall PCR tubes containing 3 Teflon 3/32”beads (McMaster-Carr #9660 K12) were placed in a floating rack inside Misonix-4000 microplate horn sonicator, filled with 350 ml water. Two coils of rubber tubing attached to a circulating water bath were installed for maintaining 37°C inside the sonicator chamber. The standard sonication program consisted of 30 sec sonication pulses delivered at 50% power efficiency applied every 30 min during a 24 hours period, therefore, each round consisted of 48 cycles. For each subsequent round, 10 µl of the reaction from previous round were added to the 90 µl of a fresh substrate. To produce RNA-depleted BH, precleared NBH was incubated with 100 µg/ml RNase A (Sigma, Cat. # R-4875) for 1 hour at 37°C prior to its use as a substrate in PMCAb. Complete digestion was confirmed using RNA content analysis in 1.2% Agarose gel and ethidium bromide staining.
For the titration experiment, 100 µl aliquots of 10% BH were sonicated with 3 Teflon 3/32” beads for 30 sec in PMCA conditions before serial 10-fold dilutions into conversion buffer. sPMCAb titration was done for two animals from the 2nd passage of LOTSS. To determine PMCAb50 values, nonlinear least-square regression analysis of the data to the following equation was performed in Sigma Plot:where F is percent of positive PMCAb reactions, x is the logarithm of the dilution fold, and A and B are two fitting parameters that define the position of a limiting dilution transition on the x axis and the slope of the transition, respectively. PMCAb50 was calculated according to the equation:
To analyze production of PK-resistant PrP material in PMCA, 10 µl of the sample were supplemented with 5 µl SDS and 5 µl PK, to a final concentration of SDS and PK of 0.25% and 50 µg/ml respectively, followed by incubation at 37°C for 1 hour. The digestion was terminated by addition of SDS-sample buffer and heating the samples for 10 min in boiling water bath. Samples were loaded onto NuPAGE 12% BisTris gels, transferred to PVDF membrane, and probed with 3F4 or SAF-84 antibodies.
Histopathological studies
Formalin fixed brain halves divided at the midline (right hemisphere), spinal cord, and spleen, were processed for hematoxylin-eosin stain as well as for immunohistochemistry for PrP, using the mouse monoclonal anti-PrP antibody 3F4 (1∶1000, Covance, Berkeley, CA, USA) and anti-glial fibrillar acidic protein (GFAP; 1∶3000, Dako, Glostrup, Denmark). Blocks were treated in formic acid (96%) prior to embedding in paraffin. For detection of disease-associated PrP, we applied a pretreatment of 30 minutes hydrated autoclaving at 121°C followed by 5 minutes in 96% formic acid. We evaluated all tissues for the presence of inflammation and PrP immunoreactivity, and the brain for the presence of spongiform change and degree of gliosis. Degree of spongiform change, neuronal loss and gliosis, and intensity of PrP immunostaining were semiquantitatively evaluated (0: none; 1: mild; 2: moderate; 3 severe) in the following anatomical regions as previously described: frontal cortex, hippocampus, caudate-putamen, thalamus, brainstem and cerebellum [6]. Lesion profiles were obtained by averaging scores of spongiform change, neuronal loss, and gliosis for each anatomical region and animal group.
Supporting Information
Zdroje
1. PrusinerSB 1996 Prion diseases. NathansonNAhmedRGonzalez-ScaranoFGriffinDHolmesK Viral Pathogenesis New York Raven Press 855 911
2. PrusinerSB 1982 Novel proteinaceous infectious particles cause scrapie. Science 216 136 144
3. GriffithJS 1967 Self-replication and scrapie. Nature 215 1043 1044
4. LegnameGBaskakovIVNguyenH-OBRiesnerDCohenFE 2004 Synthetic mammalian prions. Science 305 673 676
5. ColbyDWGilesKLegnameGWilleHBaskakovIV 2009 Design and construction of diverse mammalian prion strains. Proc Acad Natl Sci U S A 106 20417 20422
6. MakaravaNKovacsGGBocharovaOVSavtchenkoRAlexeevaI 2010 Recombinant prion protein induces a new transmissible prion disease in wild type animals. Acta Neuropathol 119 177 187
7. ColbyDWWainRBaskakovIVLegnameGPAlmerCG 2010 Protease-sensitive synthetic prions. PLoS Pathog 6 e1000736
8. BarriaMAMukherjeeAGonzalez-RomeroDMoralesRSotoC 2009 De Novo Generation of Infectious Prions In Vitro Produces a New Disease Phenotype. PLOS Pathog 5 e1000421
9. DeleaultNRHarrisBTReesJRSupattaponeS 2007 Formation of native prions from minimal components in vitro. Proc Acad Natl Sci U S A 104 9741 9746
10. WangFWangXYuanC-GMaJ 2010 Generating a Prion Bacterially Expressed Recombinant Prion Protein. Science 327 1132 1135
11. CaugheyBBaronGSChesebroBJeffreyM 2009 Getting a grip on prions: oligomers, amyloids, and pathological membrane interactions. Annu Rev Biochem 78 177 204
12. CaugheyBBaronGS 2006 Prions and their partners in crime. Nature 443 803 810
13. SotoC 2011 Prion hypothesis: the end of the controversy? Trends Biochem Sci 36 151 158
14. WilleHBianWMcDonaldMKendallAColbyDW 2009 Natural and synthetic prion structure from X-ray fiber diffraction. Proc Acad Natl Sci U S A 106 16990 16995
15. WilleHZhangG-FBaldwinMACohenFEPrusinerSB 1996 Separation of scrapie prion infectivity from PrP amyloid polymers. J Mol Biol 259 608 621
16. PiroJRWangFWalshDJReesJRMaJ 2011 Seeding Specificity and Ultrastructural Characteristics of Infectious Recombinant Prions. Biochemistry 50 7111 7116
17. OstapchenkoVGSawayaMRMakaravaNSavtchenkoRNilssonKP 2010 Two amyloid states of the prion protein display significantly different folding patterns. J Mol Biol 400 908 921
18. BocharovaOVMakaravaNBreydoLAndersonMSalnikovVV 2006 Annealing PrP amyloid firbils at high temperature results in extension of a proteinase K resistant core. J Biol Chem 281 2373 2379
19. Gonzalez-MontalbanNMakaravaNOstapchenkoVGSavtchenkoRAlexeevaI 2011 Highly Efficient Protein Misfolding Cyclic Amplification. PLoS Pathog 7 e1001277
20. KovacsGGPreusserMStrohschneiderMBudkaH 2005 Subcellular Localization of Disease-Associated Prion Protein in the Human Brain. Am J Pathol 166 287 294
21. PeretzDScottMGrothDWilliamsonABurtonD 2001 Strain-specified relative conformational stability of the scrapie prion protein. Protein Sci 10 854 863
22. DeleaultNRLucassenRWSupattaponeS 2003 RNA molecules stimulate prion protein conversion. Nature 425 717 720
23. DeleaultNRKascsakRGeogheganJCSupattaponeS 2010 Species-dependent differences in cofactor utilization for formation of the protease-resistant prion protein in vitro. Biochemistry 49 3928 3934
24. SaaPCastillaJSotoC 2006 Ultra-efficient Replication of Infectious Prions by Automated Protein Misfolding Cyclic Amplification. J Biol Chem 281 35245 35252
25. BocharovaOVBreydoLSalnikovVVGillACBaskakovIV 2005 Synthetic prions generated in vitro are similar to a newly identified subpopulation of PrPSc from sporadic Creutzfeldt-Jakob Disease PrPSc . Prot Science 14 1222 1232
26. SpassovSBeekesMNaumannD 2006 Structural differences between TSEs strains investigated by FT-IR spectroscopy. Biochim Biophys Acta 1760 1138 1149
27. MakaravaNBaskakovIV 2008 The same primary structure of the prion protein yields two distinct self-propagating states. J Biol Chem 283 15988 15996
28. SunYBreydoLMakaravaNYangQBocharovaOV 2007 Site-specific conformational studies of PrP amyloid fibrils revealed two cooperative folding domain within amyloid structure. J Biol Chem 282 9090 9097
29. LegnameGNguyenH-OBBaskakovIVCohenFEDeArmondSJ 2005 Strain-specified characteristics of mouse synthetic prions. Proc Natl Aca Sci U S A 102 2168 2173
30. KimuraKKuboMYokoyamaT 2000 Characteristics of prion protein (PrP(Sc)) in the brains of hamsters inoculated serially with a mouse-passaged scrapie strain. J Comp Pathol 122 123 130
31. MakaravaNOstapchenkoVGSavtchenkoRBaskakovIV 2009 Conformational switching within individual amyloid fibrils. J Biol Chem 284 14386 14395
32. BaskakovIV 2009 Switching in amyloid structure within individual fibrils: implication for strain adaptation, species barrier and strain classification. FEBS Lett 583 2618 2622
33. ColbyDWZhangQWangSGrothDLegnameG 2007 Prion detection by an amyloid seeding assay. Proc Acad Natl Sci U S A 104 20914 20919
34. AtarashiRMooreRASimVLHughsonAGDorwardDW 2007 Ultrasensitive detection of scrapie prion protein using seeded conversion of recombinant prion protein. Nature Methods 4 645 650
35. KimJICaliISurewiczKKongQRaymondGJ 2010 Mammalian prions generated from bacterially expressed prion protein in the absence of any mammalian cofactors. J Biol Chem 285 14083 14087
36. HillAFJoinerSLinehanJDesbruslaisMLantosPL 2000 Species-barrier-independent prion replication in apparently resistant species. Proc Natl Acad Sci U S A 97 10248 10253
37. RaceRRainseARaymondGCaugheyBChesebroB 2001 Long-Term Subclinical Carrier State Preceedes Scrapie Replication and Adaptation in a Resistant Species: Analogies to Bovine Spongiform Encephalopathy and Varian Creutzfeldt-Jakob Disease in Humans. J Virol 2001 10106 10112
38. WilleHMcDonaldMBianWKendallABorovinskiyA 2008 X-ray fiber diffraction reveals major structural differences between brain-derived prions and recombinant prion protein amyloid. Abstracts, International Conference "Prion-2008"; 8-10 October 2008; Madrid, Spain
39. LasmézasCIDeslysJ-PRobainOJaeglyABeringueV 1997 Transmission of the BSE agent to mice in the absence of detectable abnormal prion protein. Science 275 402 405
40. BarronRMCampbellSLKingDBellonAChapmanKE 2007 High titer of transmissible spongiform encephalopathy infectivity associated with extremely low levels of PrPSc in vitro. J Biol Chem 35878 35886
41. SandbergMKAl-DoujailyHSharpsBClarkeARCollingeJ 2011 Prion propagation and toxicity in vitro occur in two distinct mechanistic phases. Nature 470 540 542
42. ComoyEECasaloneCLescoutra-EtchegarayNZanussoGFreireS 2008 Atypical BSE (BASE) transmitted from asymptomatic aging cattle to a primate. Plos ONE 3 e3017
43. PedenAHHeadMWRitchieDLBellJEIronsideJW 2004 Preclinical vCJD after blood transfusion in a PRNP codon 129 heterozygous patient. Lancet 364 527 529
44. ZouWQCapellariSParchiPSyMSGambettiP 2003 Identification of Novel Proteinase K-resistant C-terminal Fragments of PrP in Creutzfeldt-Jakob Disease. J Biol Chem 278 40429 40436
45. MillerG 2009 Could they all be prion diseases? Science 326 1337 1339
46. FrostBDiamondMI 2010 Prion-like mechanisms on neurodegenerative diseases. Nat Rev Neuroscience 11 155 159
47. AguzziARajendranL 2009 The transcellular spread of cytosolic amyloids, prions, and prionoids. Neuron 64 783 790
48. EiseleYSObermullerUHeilbronnerGBaumannFKaeserSA 2010 Peripherally applied Abeta-containing inoculates induce cerebral beta-amyloidosis. Science 330 980 982
49. JeanLThomasBTahiri-AlaouiAShawMVauxDJ 2007 Heterologous amyloid seeding: revisiting the role of acetylcholinesterase in Alzheimer' disease. Plos ONE 2 e652
50. YanJFuXGeFZhangBYaoJ 2007 Cross-seeding and cross-competition in mouse apolipoprotein A-II amyloid fibrils and protein A amyloid fibrils. Am J Pathol 171 172 180
51. MoralesREstradaLDDiaz-EspinozaRMorales-ScheihingDJaraMC 2010 Molecular cross talk between misfolded proteins in animal models of Alzheimer's and prion diseases. J Neurosci 30 4528 4535
52. DerkatchILBradleyMEZhouPChernoffYOLiebmanSW 1997 Genetic and environmental factors affecting the de novo appearance of the [PSI+] prion in Saccharomyces cerevisiae. Genetics 147 507 519
53. DerkatchILBradleyMEHongJYLiebmanSW 2001 Prions affect the appearance of other prions: the story of [PIN(+)]. Cell 106 171 182
54. DerkatchILUptainSMQuteiroTFKrishnanRLindquistSL 2004 Effects of Q/N-rich, polyQ, and non-polyQ, amyloids on the de novo formation of the [PSI+] prion in yeast and aggregation of Sup35 in vitro. Proc Acad Natl Sci U S A 101 12934 12939
55. HaikSPrivatNAdjouKTSazdovitchVDormontD 2002 Alpha-synuclein-immunoreactive deposits in human and animal prion diseases. Acta Neuropath (Berlin) 103 516 520
56. AdjouKTAllixSOuidjaMOBackerSCouquetC 2007 Alpha-synuclein accumulates in the brain of scrapie-affected sheep and goats. J Comp Pathol 137 78 81
57. TakahashiMHoshiiYKawanoHGondoTIshiharaT 1996 Ulstrastructural evidence for collocalization of kappa light chain - and beta 2-microglobulin -derived amyloid using double labelling immunogold electron microscopy. Virchows Arch 429 383 388
58. MiyazonoMKitamotoTDoh-uraKIwakiTTateishiJ 1992 Creutzfeldt-Jakob disease with codon 129 polymorphism (Valine): a comparative study of patients with codon 102 point mutation or without mutations. Acta Neuropathol 84 349 354
59. GaluskeRADrachLMNichtweissMMarquardtGFranzK 2004 Colocalization of different types of amyloid in the walls of cerebral blood vessels of patients from cerebral amyloid angiopathy and spontaneous intracranial hemorrhage: a report of 5 cases. Clin Neuropathol 23 113 119
60. BocharovaOVBreydoLParfenovASSalnikovVVBaskakovIV 2005 In vitro conversion of full length mammalian prion protein produces amyloid form with physical property of PrPSc. J Mol Biol 346 645 659
61. DeleaultAMDeleaultNRHarrisBTReesJRSupattaponeS 2008 The effects of prion protein proteolysis and disaggregation on the strain properties of hamster scrapie. J Gen Virol 89 2642 2650
62. CastillaJSaaPHetzCSotoC 2005 In vitro generation of infectious scrapie prions. Cell 121 195 206
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek Role of Permissive Neuraminidase Mutations in Influenza A/Brisbane/59/2007-like (H1N1) VirusesČlánek Allelic Variation on Murine Chromosome 11 Modifies Host Inflammatory Responses and Resistance toČlánek Multifaceted Regulation of Translational Readthrough by RNA Replication Elements in a TombusvirusČlánek Latent KSHV Infection of Endothelial Cells Induces Integrin Beta3 to Activate Angiogenic PhenotypesČlánek Controlling Viral Immuno-Inflammatory Lesions by Modulating Aryl Hydrocarbon Receptor Signaling
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2011 Číslo 12- Jak souvisí postcovidový syndrom s poškozením mozku?
- Měli bychom postcovidový syndrom léčit antidepresivy?
- Farmakovigilanční studie perorálních antivirotik indikovaných v léčbě COVID-19
- 10 bodů k očkování proti COVID-19: stanovisko České společnosti alergologie a klinické imunologie ČLS JEP
-
Všechny články tohoto čísla
- Inhibition of Apoptosis and NF-κB Activation by Vaccinia Protein N1 Occur via Distinct Binding Surfaces and Make Different Contributions to Virulence
- Genesis of Mammalian Prions: From Non-infectious Amyloid Fibrils to a Transmissible Prion Disease
- Kaposi's Sarcoma Herpesvirus microRNAs Target Caspase 3 and Regulate Apoptosis
- Nutritional Immunology: A Multi-Dimensional Approach
- Role of Permissive Neuraminidase Mutations in Influenza A/Brisbane/59/2007-like (H1N1) Viruses
- Vaccinomics and Personalized Vaccinology: Is Science Leading Us Toward a New Path of Directed Vaccine Development and Discovery?
- Symbiont Infections Induce Strong Cytoplasmic Incompatibility in the Tsetse Fly
- Allelic Variation on Murine Chromosome 11 Modifies Host Inflammatory Responses and Resistance to
- Computational and Biochemical Analysis of the Effector AvrBs2 and Its Role in the Modulation of Type Three Effector Delivery
- Granzyme B Inhibits Vaccinia Virus Production through Proteolytic Cleavage of Eukaryotic Initiation Factor 4 Gamma 3
- Association of Activating KIR Copy Number Variation of NK Cells with Containment of SIV Replication in Rhesus Monkeys
- Fungal Virulence and Development Is Regulated by Alternative Pre-mRNA 3′End Processing in
- versus the Host: Remodeling of the Bacterial Outer Membrane Is Required for Survival in the Gastric Mucosa
- Follicular Dendritic Cell-Specific Prion Protein (PrP) Expression Alone Is Sufficient to Sustain Prion Infection in the Spleen
- Autophagy Protein Atg3 is Essential for Maintaining Mitochondrial Integrity and for Normal Intracellular Development of Tachyzoites
- Longevity and Composition of Cellular Immune Responses Following Experimental Malaria Infection in Humans
- Sequential Adaptive Mutations Enhance Efficient Vector Switching by Chikungunya Virus and Its Epidemic Emergence
- Acquisition of Pneumococci Specific Effector and Regulatory Cd4 T Cells Localising within Human Upper Respiratory-Tract Mucosal Lymphoid Tissue
- The Meaning of Death: Evolution and Ecology of Apoptosis in Protozoan Parasites
- Deficiency of a Niemann-Pick, Type C1-related Protein in Is Associated with Multiple Lipidoses and Increased Pathogenicity
- Feeding Cells Induced by Phytoparasitic Nematodes Require γ-Tubulin Ring Complex for Microtubule Reorganization
- Eight RGS and RGS-like Proteins Orchestrate Growth, Differentiation, and Pathogenicity of
- Prion Uptake in the Gut: Identification of the First Uptake and Replication Sites
- Nef Decreases HIV-1 Sensitivity to Neutralizing Antibodies that Target the Membrane-proximal External Region of TMgp41
- Multifaceted Regulation of Translational Readthrough by RNA Replication Elements in a Tombusvirus
- A Temporal Role Of Type I Interferon Signaling in CD8 T Cell Maturation during Acute West Nile Virus Infection
- The Membrane Fusion Step of Vaccinia Virus Entry Is Cooperatively Mediated by Multiple Viral Proteins and Host Cell Components
- HIV-1 Capsid-Cyclophilin Interactions Determine Nuclear Import Pathway, Integration Targeting and Replication Efficiency
- Neonatal CD8 T-cell Hierarchy Is Distinct from Adults and Is Influenced by Intrinsic T cell Properties in Respiratory Syncytial Virus Infected Mice
- Two Novel Transcriptional Regulators Are Essential for Infection-related Morphogenesis and Pathogenicity of the Rice Blast Fungus
- Five Questions about Non-Mevalonate Isoprenoid Biosynthesis
- The Human Cytomegalovirus UL11 Protein Interacts with the Receptor Tyrosine Phosphatase CD45, Resulting in Functional Paralysis of T Cells
- Wall Teichoic Acids of Limit Recognition by the Drosophila Peptidoglycan Recognition Protein-SA to Promote Pathogenicity
- A Novel Role for the NLRC4 Inflammasome in Mucosal Defenses against the Fungal Pathogen
- Inflammasome-dependent Pyroptosis and IL-18 Protect against Lung Infection while IL-1β Is Deleterious
- CNS Recruitment of CD8+ T Lymphocytes Specific for a Peripheral Virus Infection Triggers Neuropathogenesis during Polymicrobial Challenge
- Latent KSHV Infection of Endothelial Cells Induces Integrin Beta3 to Activate Angiogenic Phenotypes
- A Receptor-based Switch that Regulates Anthrax Toxin Pore Formation
- Targeting of Heparin-Binding Hemagglutinin to Mitochondria in Macrophages
- Chikungunya Virus Neutralization Antigens and Direct Cell-to-Cell Transmission Are Revealed by Human Antibody-Escape Mutants
- Ce-Duox1/BLI-3 Generated Reactive Oxygen Species Trigger Protective SKN-1 Activity via p38 MAPK Signaling during Infection in
- Structural Elucidation and Functional Characterization of the Effector Protein ATR13
- Controlling Viral Immuno-Inflammatory Lesions by Modulating Aryl Hydrocarbon Receptor Signaling
- SAMHD1-Deficient CD14+ Cells from Individuals with Aicardi-Goutières Syndrome Are Highly Susceptible to HIV-1 Infection
- Acid Stability of the Hemagglutinin Protein Regulates H5N1 Influenza Virus Pathogenicity
- Cryo Electron Tomography of Herpes Simplex Virus during Axonal Transport and Secondary Envelopment in Primary Neurons
- A Novel Human Cytomegalovirus Locus Modulates Cell Type-Specific Outcomes of Infection
- Juxtamembrane Shedding of AMA1 Is Sequence Independent and Essential, and Helps Evade Invasion-Inhibitory Antibodies
- Pathogenesis and Host Response in Syrian Hamsters following Intranasal Infection with Andes Virus
- IRGM Is a Common Target of RNA Viruses that Subvert the Autophagy Network
- Epstein-Barr Virus Evades CD4 T Cell Responses in Lytic Cycle through BZLF1-mediated Downregulation of CD74 and the Cooperation of vBcl-2
- Quantitative Multicolor Super-Resolution Microscopy Reveals Tetherin HIV-1 Interaction
- Late Repression of NF-κB Activity by Invasive but Not Non-Invasive Meningococcal Isolates Is Required to Display Apoptosis of Epithelial Cells
- Polar Flagellar Biosynthesis and a Regulator of Flagellar Number Influence Spatial Parameters of Cell Division in
- Epstein-Barr Virus Nuclear Antigen 3C Stabilizes Gemin3 to Block p53-mediated Apoptosis
- The Enteropathogenic (EPEC) Tir Effector Inhibits NF-κB Activity by Targeting TNFα Receptor-Associated Factors
- Toward an Integrated Model of Capsule Regulation in
- A Systematic Screen to Discover and Analyze Apicoplast Proteins Identifies a Conserved and Essential Protein Import Factor
- A Host Small GTP-binding Protein ARL8 Plays Crucial Roles in Tobamovirus RNA Replication
- Comparative Pathobiology of Fungal Pathogens of Plants and Animals
- Synergistic Roles of Eukaryotic Translation Elongation Factors 1Bγ and 1A in Stimulation of Tombusvirus Minus-Strand Synthesis
- Engineered Immunity to Infection
- Inflammatory Monocytes and Neutrophils Are Licensed to Kill during Memory Responses
- Sialidases Affect the Host Cell Adherence and Epsilon Toxin-Induced Cytotoxicity of Type D Strain CN3718
- Eurasian-Origin Gene Segments Contribute to the Transmissibility, Aerosol Release, and Morphology of the 2009 Pandemic H1N1 Influenza Virus
- SARS Coronavirus nsp1 Protein Induces Template-Dependent Endonucleolytic Cleavage of mRNAs: Viral mRNAs Are Resistant to nsp1-Induced RNA Cleavage
- Identification and Characterization of a Novel Non-Structural Protein of Bluetongue Virus
- Functional Analysis of the Kinome of the Wheat Scab Fungus
- Norovirus Regulation of the Innate Immune Response and Apoptosis Occurs via the Product of the Alternative Open Reading Frame 4
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Controlling Viral Immuno-Inflammatory Lesions by Modulating Aryl Hydrocarbon Receptor Signaling
- Fungal Virulence and Development Is Regulated by Alternative Pre-mRNA 3′End Processing in
- Epstein-Barr Virus Nuclear Antigen 3C Stabilizes Gemin3 to Block p53-mediated Apoptosis
- Engineered Immunity to Infection
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



