-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaEnhances Protective and Detrimental HLA Class I-Mediated Immunity in Chronic Viral Infection
Killer cell immunoglobulin-like receptors (KIRs) influence both innate and adaptive immunity. But while the role of KIRs in NK-mediated innate immunity is well-documented, the impact of KIRs on the T cell response in human disease is not known. Here we test the hypothesis that an individual's KIR genotype affects the efficiency of their HLA class I-mediated antiviral immune response and the outcome of viral infection. We show that, in two unrelated viral infections, hepatitis C virus and human T lymphotropic virus type 1, possession of the KIR2DL2 gene enhanced both protective and detrimental HLA class I-restricted anti-viral immunity. These results reveal a novel role for inhibitory KIRs. We conclude that inhibitory KIRs, in synergy with T cells, are a major determinant of the outcome of persistent viral infection.
Published in the journal: . PLoS Pathog 7(10): e32767. doi:10.1371/journal.ppat.1002270
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1002270Summary
Killer cell immunoglobulin-like receptors (KIRs) influence both innate and adaptive immunity. But while the role of KIRs in NK-mediated innate immunity is well-documented, the impact of KIRs on the T cell response in human disease is not known. Here we test the hypothesis that an individual's KIR genotype affects the efficiency of their HLA class I-mediated antiviral immune response and the outcome of viral infection. We show that, in two unrelated viral infections, hepatitis C virus and human T lymphotropic virus type 1, possession of the KIR2DL2 gene enhanced both protective and detrimental HLA class I-restricted anti-viral immunity. These results reveal a novel role for inhibitory KIRs. We conclude that inhibitory KIRs, in synergy with T cells, are a major determinant of the outcome of persistent viral infection.
Introduction
Killer cell immunoglobulin-like receptors (KIRs) are a family of transmembrane proteins that are expressed on natural killer (NK) cells and subsets of T cells [1]–[3]. They bind HLA class I molecules and have activatory and inhibitory isoforms [4]. KIRs contribute directly and indirectly to antiviral immunity. Directly, KIRs on NK cells sense the loss of HLA class I molecules from the cell surface and trigger NK-mediated cytolysis. Indirectly, NK cells regulate adaptive immunity via crosstalk with dendritic cells and by the production of chemokines and cytokines [5]–[7].
HLA class I molecules can be grouped into allotypes with similar KIR binding properties [8]. For example, KIR2DL2 binds group C1 HLA-C molecules which have asparagine at residue 80, and, with a weaker affinity, group C2 molecules which have a lysine at position 80 [9].
Early research on KIRs investigated NK-mediated protection by studying disease associations with KIRs in the context of their HLA class I ligands [10]–[11]. There is now compelling evidence that KIRs also regulate adaptive immunity [5]–[7], but it is not known whether this has a significant impact on the response to infection in vivo. Differences between human KIRs and their mouse functional homologues (the Ly49 receptors) and the paucity of KIR allele-specific antibodies have hindered work on the role of KIRs in controlling adaptive immune responses. Here we used immunogenetics to investigate whether KIR genotype modulates HLA-mediated anti-viral protection in vivo. We focussed on HLA class I alleles which have previously been associated with disease outcome and investigated whether these effects were altered by the KIR background. We studied 4 well-documented HLA class I allele-disease associations in two viral infections: human T lymphotropic virus type 1 (HTLV-1) and hepatitis C virus (HCV).
HTLV-1 is a persistent retrovirus that infects 10–20 million people worldwide. Most infected individuals remain lifelong asymptomatic carriers (ACs). However, approximately 10% of infected individuals develop associated diseases including HTLV-1-associated myelopathy/tropical spastic paraparesis (HAM/TSP), an inflammatory disease of the central nervous system that results in progressive paralysis. It is poorly understood why some individuals remain asymptomatic whereas others develop disease, but one strong correlate of disease is the proviral load, which is significantly higher in HAM/TSP patients than in ACs [12]. We have previously shown that HLA-A*02 and C*08 are each associated with both a reduced risk of HAM/TSP and a reduced proviral load in ACs and that HLA-B*54 is associated with an increased prevalence of HAM/TSP and an increased proviral load in HAM/TSP patients [13]–[14].
HCV is among the most widespread viral infections, with 170 million infected people worldwide. As in HTLV-1 infection, the outcome of HCV infection is heterogeneous: the virus persists in approximately 70% of infected individuals while the rest clear the infection spontaneously. Chronic HCV infection can cause serious liver damage including cirrhosis and hepatocellular carcinoma [15]. The origins of this heterogeneity are not completely understood but several genetic determinants have been identified, including HLA-B*57 which is associated with spontaneous clearance in several cohorts [16]–[19].
The aim of this study was to test the hypothesis that KIR genotype determines the efficiency of HLA class I-mediated anti-viral immunity. We tested this hypothesis for 4 HLA class I associations: HLA-C*08, A*02 and B*54 in HTLV-1 infection and B*57 in HCV infection. We show, using multiple independent measures, that for both HCV and HTLV-1, possession of the KIR2DL2 gene enhanced HLA class I-restricted immunity.
Results
Two independent cohorts were studied: HCV-infected individuals (N = 782) and HTLV-1-infected individuals (N = 402). Ten KIR genes were typed; of these, 4 were present at an informative frequency in the HTLV-1 cohort and 9 in the HCV cohort (defined in Methods). The modulation of HLA class I associations by the informative KIRs was analysed; the models included all other known determinants of outcome in the cohorts as covariates.
HTLV-1 and disease status
In the cohort from Southern Japan, HLA-C*08 was associated with a significantly reduced odds of developing HAM/TSP (OR = 0.47, p = 0.03, OR<1 indicates a protective effect while OR>1 indicates a detrimental effect)[14]. We investigated the impact of KIRs on this protective effect by stratifying the cohort by KIR genotype. Of the KIR studied, one particular KIR, KIR2DL2, had a noticeable interaction with C*08 (Table 1 and Figure 1). We found that the C*08 protective effect was weakened and no longer statistically significant in the subset of individuals who were KIR2DL2- (OR = 0.67, p = 0.4) but enhanced in KIR2DL2+ individuals (OR = 0.16, p = 0.02). There were more KIR2DL2 - individuals than KIR2DL2+ individuals so the absence of significance in the KIR2DL2- individuals was not simply due to reduced cohort size. Similarly, HLA-B*54, which is associated with a significantly increased risk of HAM/TSP (OR = 3.11, p = 0.0009), had a weakened impact on disease risk in the absence of KIR2DL2 (OR = 1.70, p = 0.2) but an enhanced impact in the presence of KIR2DL2 (OR = 12.05, p = 0.004). Again, the absence of a significant effect of B*54 in KIR2DL2 - individuals was not attributable to a loss of power. In contrast, although HLA-A*02 was associated with a reduced risk of HAM/TSP, there was no significant additional impact of KIR genotype which could not be attributed to power.
Fig. 1. KIR2DL2 enhances HLA class I mediated antiviral-immunity. 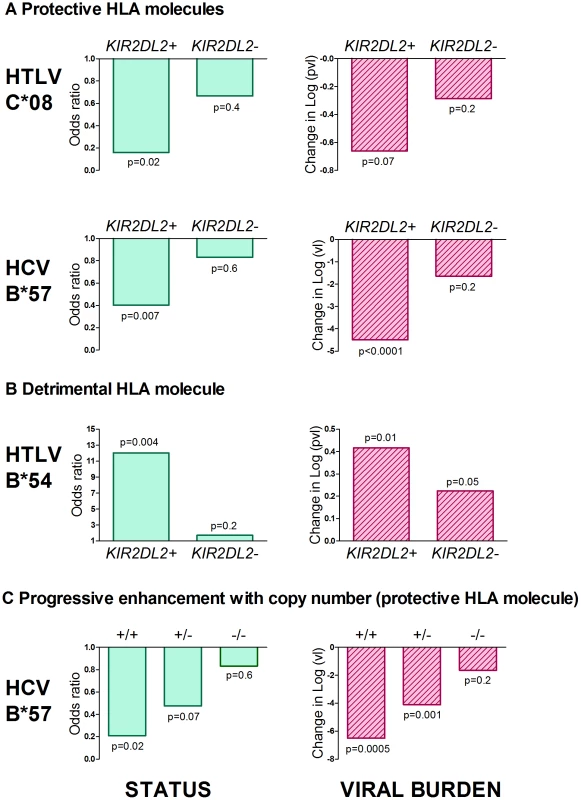
The effect of both protective (Figure 1a) and detrimental (Figure 1b) HLA class I alleles on both status (first column) and viral load (second column) is stronger in KIR2DL2+ individuals than in KIR2DL2- individuals. This effect was seen in both HTLV-1 and HCV infection. We also saw a progressive effect of KIR2DL2 copy number on both status and viral load in HCV (Figure 1c). We could not test for a progressive effect in the HTLV-1 cohort because there are no KIR2DL2 homozygotes. In all cases, the impact on viral burden was considered within (not between) disease status categories and so the observation of an impact on viral burden is independent of the observation of an impact on status. Effect sizes, p values and cohort sizes are provided in Table 1. Tab. 1. KIR2DL2 in HTLV-1 infection: KIR2DL2 enhances the protective effect of C*08 and the detrimental effect of B*54 on HAM/TSP risk and, independently, on proviral load. 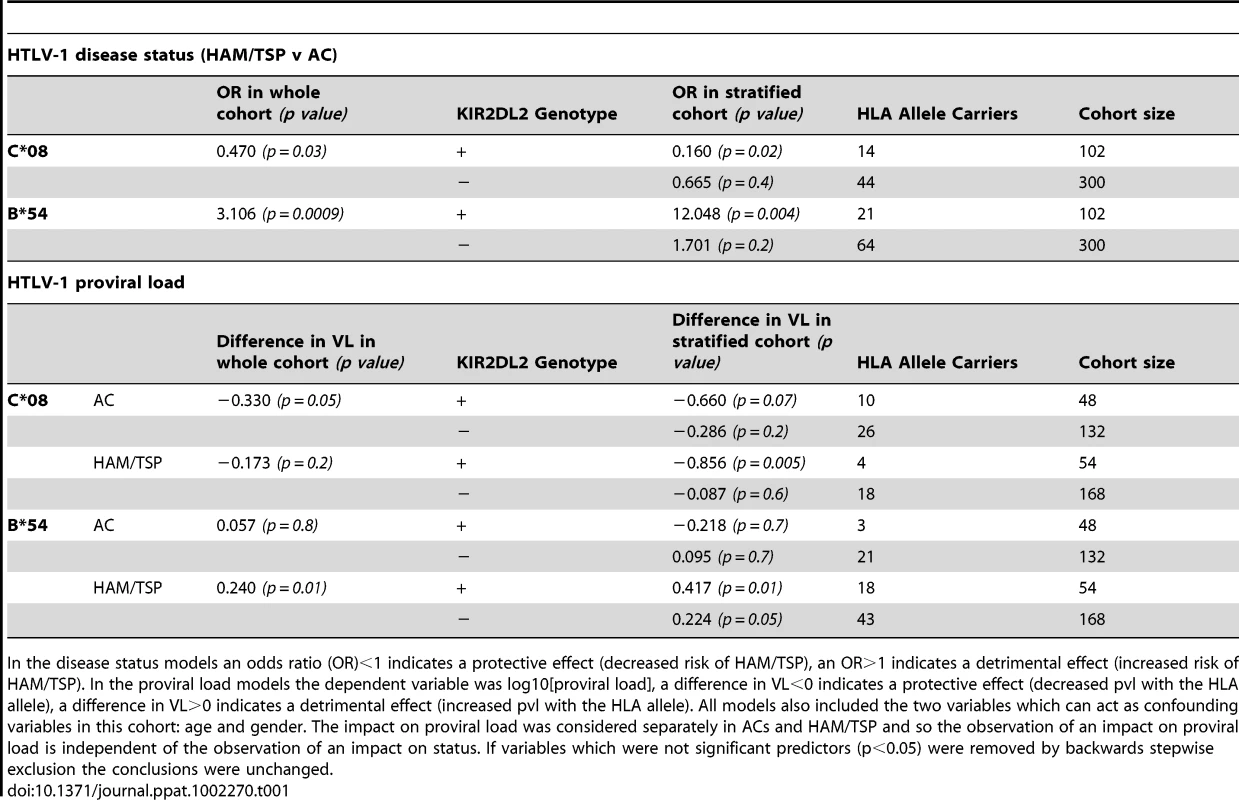
In the disease status models an odds ratio (OR)<1 indicates a protective effect (decreased risk of HAM/TSP), an OR>1 indicates a detrimental effect (increased risk of HAM/TSP). In the proviral load models the dependent variable was log10[proviral load], a difference in VL<0 indicates a protective effect (decreased pvl with the HLA allele), a difference in VL>0 indicates a detrimental effect (increased pvl with the HLA allele). All models also included the two variables which can act as confounding variables in this cohort: age and gender. The impact on proviral load was considered separately in ACs and HAM/TSP and so the observation of an impact on proviral load is independent of the observation of an impact on status. If variables which were not significant predictors (p<0.05) were removed by backwards stepwise exclusion the conclusions were unchanged. HTLV-1 and proviral load
As an independent test of the observation that KIR2DL2 enhanced the effect of both protective and detrimental HLA class I alleles in HTLV-1 infection, we investigated the interaction between HLA class I alleles, KIR2DL2 and HTLV-1 proviral load (pvl). We investigated pvl in ACs and HAM/TSP patients separately, so any observed impact on pvl is independent of the impact on disease status. C*08 has previously been associated with a low pvl in ACs (difference in log10 pvl between C*08+ and C*08- Δ = −0.33, p = 0.05); again, this effect was weakened in KIR2DL2 - individuals (Δ = −0.29 p = 0.2) but enhanced in KIR2DL2+ individuals (Δ = −0.66 p = 0.07); Table 1. Similarly, HLA-B*54, which is associated with a high pvl in HAM/TSP patients (Δ = +0.24 p = 0.01) showed a weakened effect in the absence of KIR2DL2 (Δ = +0.22, p = 0.05) but an enhanced effect in the presence of KIR2DL2 (Δ = +0.42, p = 0.01).
Two previous observations on HTLV-1 immunogenetics have, until now, remained unexplained. Firstly, although C*08 has been associated with a low pvl in ACs it has no detectable impact on pvl in HAM/TSP patients; similarly, B*54, which was associated with a high pvl in HAM/TSP patients, had no impact on pvl in ACs [13]–[14]. Why some HLA class I alleles apparently “cease working” in some populations was unknown. We hypothesised that the lack of the expected C*08 and B*54 effects in HAM/TSP patients and ACs respectively was due to a low frequency of KIR2DL2 in these groups and that the decrease or increase in pvl due to C*08 or B*54 respectively would be manifest only in KIR2DL2+ individuals. Consistent with this hypothesis we found that the frequency of KIR2DL2 carriage in the groups that did not show the expected effect of HLA genotype on pvl was approximately half that of the groups in which HLA-associated effects were observed (prevalence of KIR2DL2+ amongst B*54+ individuals: 12.5% in ACs vs 29.5% in HAM/TSPs; prevalence of KIR2DL2+ amongst C*08+ individuals: 18.2% in HAM/TSPs vs 27.8% in ACs; Table 1). The small numbers of individuals in the stratified cohorts (HAM/TSP KIR2DL2+: C*08+ N = 4, C*08- N = 50. AC KIR2DL2+: B*54+ N = 3, B*54- N = 45) precluded a reliable test for an impact of HLA on pvl in KIR2DL2+ individuals. However, in the larger of these groups there was a significant impact; i.e C*08 was associated with a significant reduction in pvl in KIR2DL2+ individuals (Δ = -0.86, p = 0.005). This provides, for the first time, a plausible explanation for the reported observation [14] that the B*54 effect on pvl was not manifest in ACs and the C*08 effect on pvl was not manifest in HAM/TSP patients.
HTLV-1 and HLA class I specificity
We recently reported that in HTLV-1 infection, HLA class I molecules that bind peptides from the virus protein HBZ are associated with a reduced risk of HAM/TSP and, independently, a reduced pvl [20]. In the same study we showed, using IFNg ELISpot, chromium release and CD107 staining, that HBZ-specific CD8+ T cells were present and functional in fresh PBMC from infected individuals. We therefore investigated the interaction between KIR2DL2 and the protective effect of binding HBZ. We used epitope prediction software [21] to predict the strength of binding of HBZ peptides to an individual's HLA-A and B molecules. We found that ACs had HLA-A and -B molecules that are predicted to bind HBZ significantly more strongly than those in HAM/TSP patients (median difference 12%, p = 0.00005) [20] and that this effect was stronger in KIR2DL2+ individuals (median difference 25%, p = 0.00006) than in KIR2DL2- individuals (median difference 7%, p = 0.06); Figure 2a. We reasoned that this difference in HBZ binding between ACs and HAM/TSP patients was due to HLA-A*02 and B*54, which differ in their HBZ peptide-binding affinities [20] and are associated with different outcomes in HTLV-1 infection. We therefore removed all individuals with A*02 or B*54 from the cohort and repeated the analysis. Surprisingly, we still found the same pattern: possession of HLA molecules that bind HBZ strongly was significantly associated with remaining asymptomatic (median difference 10%, p = 0.04) and this effect was strengthened in KIR2DL2+ individuals (median difference 23%, p = 0.02) but not in KIR2DL2- individuals (median difference 3%, p = 0.2); Figure 2b. This demonstrates that the protective effect of binding HBZ peptides by multiple HLA class I molecules, both A and B, is enhanced by KIR2DL2.
Fig. 2. The protective effect of binding HBZ is enhanced by KIR2DL2. 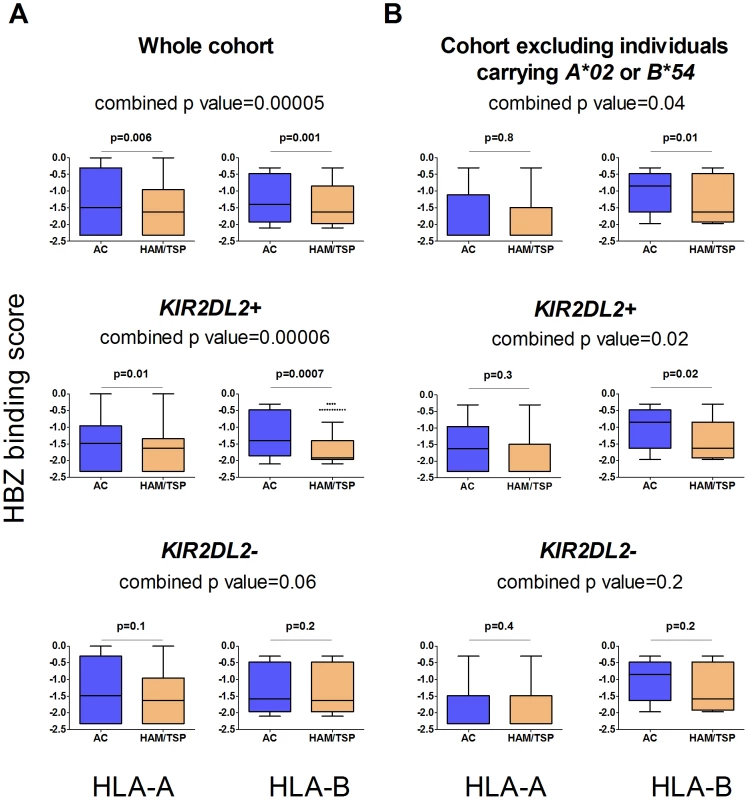
In the whole cohort (Figure 2a) individuals with HLA-A or B molecules which were predicted to bind peptides from HBZ strongly were significantly more likely to be asymptomatic (top row); this effect was stronger in the individuals with KIR2DL2 (middle row) than without (bottom row). When individuals with A*02 or B*54 were removed (Figure 2b) the pattern remained the same. This indicates that the protective effect of binding HBZ peptides by multiple, different HLA class I molecules, both A and B, is enhanced by KIR2DL2.We did not predict HLA-C binding as the relevant algorithms are not available in Metaserver (due to scarcity of HLA C binding data necessary for training the neural networks). HCV and viral clearance
As previously reported, HLA-B*57 was associated with significantly decreased odds of chronic infection (OR = 0.571, p = 0.02). This protective effect was enhanced in the presence of KIR2DL2 (OR = 0.40, p = 0.007) but weakened in the absence of KIR2DL2 (OR = 0.83, p = 0.6) (Table 2). Furthermore, the impact of B*57 was strongest in KIR2DL2 homozygote individuals (OR = 0.21, p = 0.02), weaker in KIR2DL2 heterozygote individuals (OR = 0.48, p = 0.07) and absent in KIR2DL2-negative individuals (OR = 0.83, p = 0.6). KIR2DL2 enhanced the association between B*57 and spontaneous clearance independently and with similar strength in both African Americans and Caucasians (Table S1 in Text S1).
Tab. 2. KIR2DL2 in HCV infection: KIR2DL2 enhances the protective effect of HLA-B*57 on HCV status (spontaneous clearance v chronic infection) and, independently, on HCV viral load. 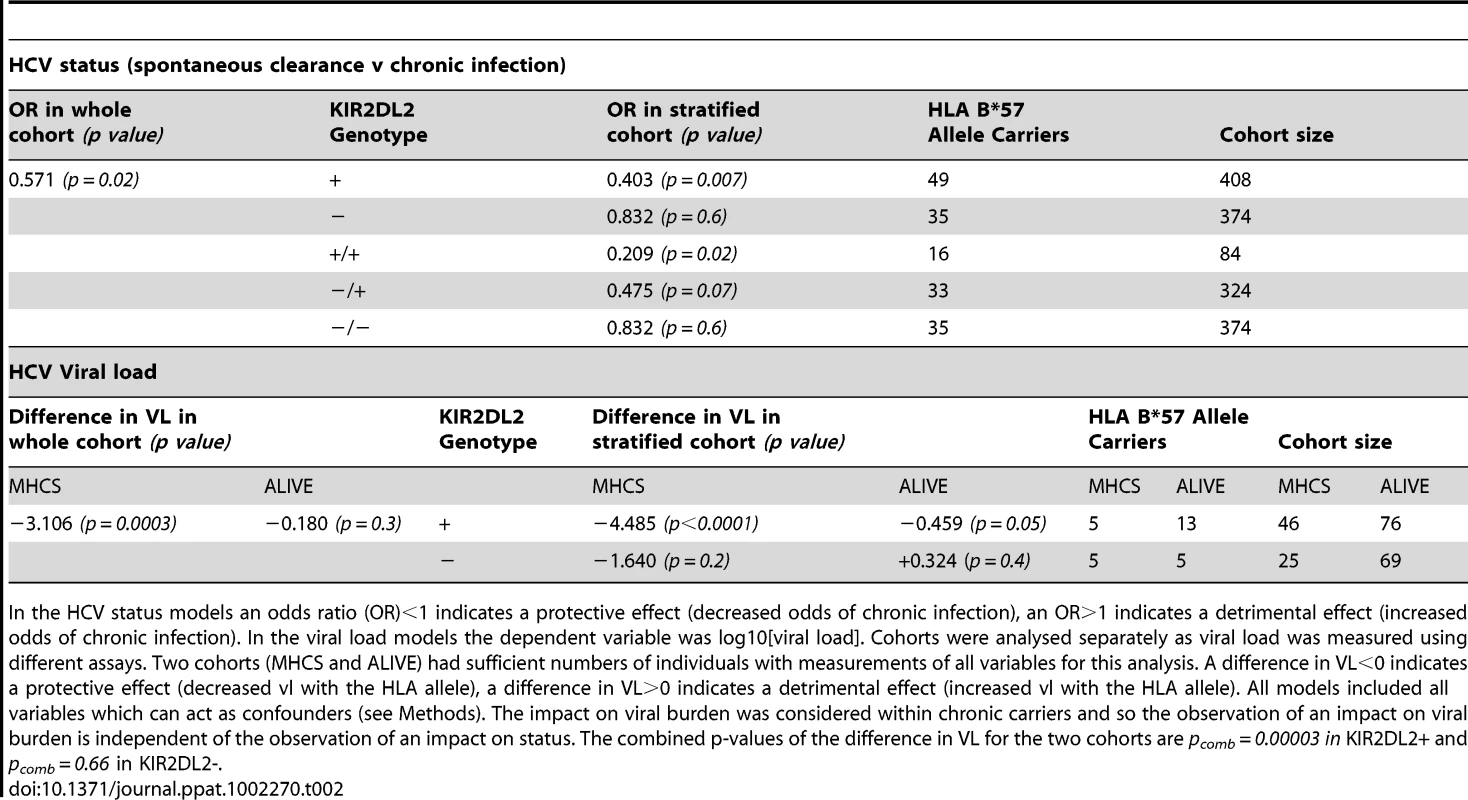
In the HCV status models an odds ratio (OR)<1 indicates a protective effect (decreased odds of chronic infection), an OR>1 indicates a detrimental effect (increased odds of chronic infection). In the viral load models the dependent variable was log10[viral load]. Cohorts were analysed separately as viral load was measured using different assays. Two cohorts (MHCS and ALIVE) had sufficient numbers of individuals with measurements of all variables for this analysis. A difference in VL<0 indicates a protective effect (decreased vl with the HLA allele), a difference in VL>0 indicates a detrimental effect (increased vl with the HLA allele). All models included all variables which can act as confounders (see Methods). The impact on viral burden was considered within chronic carriers and so the observation of an impact on viral burden is independent of the observation of an impact on status. The combined p-values of the difference in VL for the two cohorts are pcomb = 0.00003 in KIR2DL2+ and pcomb = 0.66 in KIR2DL2-. HCV and viral load
Next we investigated the impact of B*57 and KIR2DL2 on HCV viral load. We only considered patients with chronic infection, so any observed impact on viral load is independent of the impact on viral clearance. This analysis was possible in two cohorts: MHCS and ALIVE. We found that B*57 was associated with reduced chronic HCV viral load, particularly in the MHCS cohort (MHCS: difference in log10 VL Δ = −3.1 p = 0.0003; ALIVE: Δ = −0.18 p = 0.3. Combined p = 0.0006). Consistent with our observations in HTLV-1 infection, this reduction was enhanced in the presence of KIR2DL2 (MHCS: Δ = −4.5 p<0.0001, ALIVE: Δ = −0.46 p = 0.05. Combined p = 0.00003) but weakened in the absence of KIR2DL2 (MHCS Δ = −1.64 p = 0.2, ALIVE Δ = +0.32 p = 0.3. Combined p = 0.7); Table 2 and Figure 3. In MHCS (but not ALIVE) we also observed a progressive effect with KIR2DL2 copy number (2 copies: Δ = −6.5, p = 0.0005. 1 copy: Δ = −4.1 p = 0.001. 0 copies Δ = −1.6 p = 0.2); the number of homozygous individuals are too small to draw firm conclusions but this progressive effect is consistent with our other observations.
Fig. 3. The impact of HLA-B*57 and KIR2DL2 on HCV viral load. 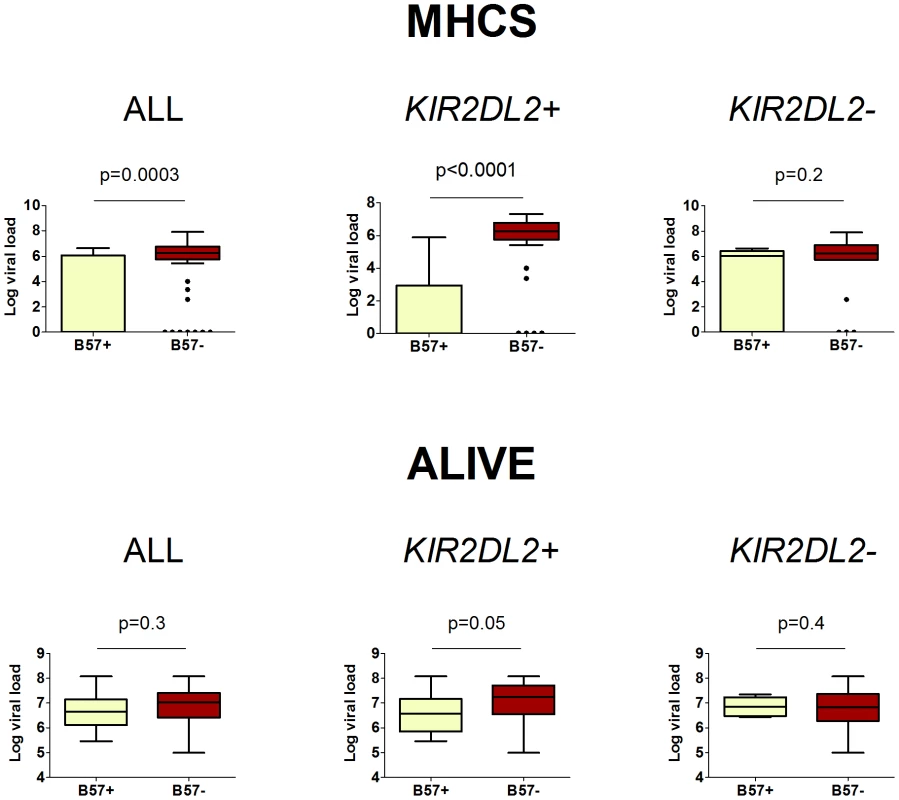
In HCV infection, B*57 is associated with a reduced HCV viral load. This protective effect is enhanced in KIR2DL2+ individuals and reduced or absent in KIR2DL2- individuals. The cohorts were analysed separately as viral load was measured using different assays. Two cohorts, MHCS and ALIVE, had enough individuals with measurements for all factors to perform this analysis. HIV-1 status was a significant determinant of viral load in the ALIVE cohort, including HIV status in the model as a covariate did not change any of our conclusions (ALL p = 0.3; KIR2DL2+ p = 0.05; KIR2DL2- p = 0.4). Summary data from these plots is provided in Table 2. The p-values are derived based on the regression model. These data show that KIR2DL2 enhances both protective and detrimental HLA class I associations. Therefore, KIR2DL2 would not be predicted to have a significant net impact across all HLA class I molecules. That is, possession of KIR2DL2 (alone or with its C1 ligand) without a particular protective or detrimental HLA allele, would not be expected to be significantly protective or detrimental. This prediction was verified (Table S2 in Text S1).
There is strong linkage disequilibrium among the KIR genes and among the HLA class I genes. Analysis of the linked genes indicates that the primary genes driving the observed associations are most likely to be KIR2DL2 in combination with HLA-B*54, C*08 and B*57 rather than individual linked KIR, multiple stimulatory linked KIRs or linked HLA class I genes (sections 3 and 4 in Text S1). We cannot rule out an effect of linkage between KIR2DL2 and neighbouring loci outside the KIR genes. However, there is little evidence of significant linkage between KIRs and even the next closest gene cluster, the LILR [22]. Furthermore, we observed the same effect of KIR2DL2 in three different populations (Japanese, African-American and Caucasian) so a putative linked locus driving the effect would have to be linked to KIR2DL2 in all three populations.
Canonical KIR-HLA binding
Although HLA-C*08, as a group C1 molecule, is expected to bind KIR2DL2, the most frequent subtype in our cohort (Cw*0801, 88%) binds KIR2DL2 very weakly (comparable to background [23]), furthermore HLA-B*54 and HLA-B*57 are not expected to bind KIR2DL2 and the most frequent subtypes in our cohorts (B*5401 and B*5701) have been shown not to bind KIR2DL2 [9], [23]. Finally, KIR2DL2 enhanced the protective effect of binding HBZ peptides by multiple HLA-A and –B molecules. With the exception of B*4601 and B*7301 (which were not responsible for the enhancement, data not shown) KIR2DL2 is not thought to bind HLA-A and B molecules and has been shown not to bind 29 HLA-A and 54 HLA-B allotypes [9]. We therefore hypothesised that the effect of KIR2DL2 on HLA class I-mediated immunity we have observed is not attributable to KIR2DL2 directly binding the HLA molecule whose effect is enhanced. To test this hypothesis we first investigated whether the other group C1 alleles had the same effect as C*08 in HTLV-1 infection. Grouping all the C1 alleles we found no significant association between C1 and decreased risk of HAM/TSP either in the whole cohort or in KIR2DL2+ individuals. Similarly, there was no relation between pvl and C1 in either ACs or HAM/TSP patients. Analysis of the individual C1 alleles confirmed the hypothesis that the C*08 effect we observed was not exhibited by other group C1 alleles (Tables S4 and S5).
HLA-B*54, a group Bw6 HLA allele, is not known to bind any KIR molecule. We therefore tested whether the observed B*54 effect was attributable to C*01, which is in linkage disequilibrium with B*54 and which encodes molecules that bind KIR2DL2. This analysis suggested that B*54, not C*01, was the gene driving the observed detrimental effect on HTLV-1 outcome (section 5.2 in Text S1). This result, and the observation that no other C1 allele shows “B*54-like” behaviour, indicate that, as postulated, the interaction between B*54 and KIR2DL2 cannot be explained by direct KIR-HLA binding.
The most frequent B*57 allele in our cohort is B*5701, which does not bind KIR2DL2 [9]. There are therefore two ways in which the observed interaction between KIR2DL2 and HLA-B*57 could be attributed to “classical” KIR-HLA binding: either KIR2DL2 might bind a class I HLA molecule whose encoding gene is linked to HLA-B*57, or the effect might be due to KIR3DL1/S1, which does bind B*57. Analysis of both these possibilities indicated that they did not explain the KIR2DL2-B*57 effect (section 5.3 and 5.4 in Text S1).
Therefore, as hypothesised, the enhancement of C*08, B*54 and B*57–restricted immunity by KIR2DL2 is not explained by direct binding between the respective HLA molecules and KIR2DL2. Instead, we suggest that KIR2DL2 binds its HLA-C ligands and indirectly modulates C*08, B*54 and B*57–restricted T cells. Consistent with this, we found some evidence that KIR2DL2 enhanced HLA Class I effects more strongly when it's stronger C1 ligands are present (section 6 in Text S1).
Other inhibitory KIR
It seems unlikely that KIR2DL2 behaves fundamentally differently to other inhibitory KIRs. The effect of KIR2DL2 may be most apparent because KIR2DL2 is present at informative frequencies and its C1 and C2 ligands are ubiquitous. We addressed the role of other inhibitory KIRs in 3 ways. We studied the effect of individual inhibitory KIRs (section 4.1 in Text S1), we investigated whether the number of inhibitory KIR:ligands had a cumulative effect (section 7 in Text S1) and we examined the role of the group A KIR haplotypes which are dominated by inhibitory KIRs but do not contain KIR2DL2 (section 8 in Text S1). We found little evidence that the other inhibitory KIRs enhanced HLA class I-mediated immunity but this may be due to small cohort sizes and masking by the dominant KIR2DL2 effect.
Activating KIR
We found no evidence that activating KIR were enhancing HLA class I-restricted immunity. Haplotype B, the more activatory KIR haplotype, enhanced HLA class I associations but this was only true if the haplotype contained KIR2DL2 (section 8 in Text S1). We found no evidence that the cumulative presence of activating KIR enhanced HLA class I restricted immunity (section 4.3 in Text S1). And, as far as it was possible to separate KIR2DL2 and KIR2DS2, which are in tight linkage disequilibrium, the enhancement of HLA class I restricted immunity appeared to be attributable to KIR2DL2 rather than KIR2DS2 (section 4.2 in Text S1).
Discussion
We show that KIR2DL2 enhanced several independent HLA class I-mediated effects in two unrelated viral infections. In HTLV-1 infection, KIR2DL2 enhanced the protective and detrimental effects of HLA-C*08 and B*54 respectively on disease status. KIR2DL2 also enhanced the association between C*08 and low proviral load in ACs and between B*54 and high proviral load in HAM/TSP patients. Additionally, KIR2DL2 enhanced the protective effect of HBZ binding by multiple HLA molecules. Strikingly, on stratifying by KIR2DL2, we observed, for the first time, a protective effect of C*08 on pvl in HAM/TSP patients and explained the lack of impact of B*54 on pvl in ACs. In HCV infection, KIR2DL2 enhanced the protective effect of B*57 on spontaneous clearance and the association between B*57 and low viral load in chronic carriers; for both clearance and viral load a progressive effect with KIR2DL2 copy number was observed. This progressive effect is consistent with reports of an association between KIR gene copy number and the frequency of cell-surface expression of the respective KIR molecule [24]–[25].
There are two mechanisms by which KIR2DL2 could act: it could enhance either NK-mediated or T cell-mediated immunity. That is, NK cell killing of virus-infected cells could be altered by KIR2DL2 expression or, alternatively, the virus-specific CD8+ T cell response could be modified by KIR2DL2 expression (on NK cells or T cells). Two observations indicate that it is the T cell response that is more likely to be enhanced. First, strong binding of HBZ viral peptides via multiple different HLA-A and B molecules was associated with asymptomatic status [20] and this protective effect was enhanced by KIR2DL2. KIR2DL2 is not known to bind HLA-A or–B molecules (with the exception of B*4601 and B*7301) [9], [23], [26] so it is unlikely that the enhancement of the protective effect of binding HBZ by KIR2DL2 is due to direct binding between KIR2DL2 and HBZ peptide in the context of HLA-A and –B molecules. Furthermore, although NK cells exhibit peptide dependence [27], it is hard to reconcile protein-specificity via multiple HLA molecules with an NK cell-mediated mechanism. Second, the KIR2DL2 enhancement could not be explained by binding between KIR2DL2 and any of the 3 HLA class I molecules investigated. One further observation also suggests a T cell-mediated mechanism. Two protective genotypes in HCV infection that are postulated to operate via innate immune mechanisms [28] (namely a SNP upstream of IL28B and KIR2DL3-HLA-C1) had no impact on viral load in chronic infection [10], [29]. The authors hypothesised that this was because innate barriers offer little protection once overcome. In contrast, the KIR2DL2/B*57 effect that we report here had a significant impact on viral load: again, this is perhaps more consistent with adaptive immunity.
Our results indicate that KIR2DL2 enhances HLA class 1-restricted CD8+ T cell-mediated adaptive immunity. KIRs on both NK cells and CD8+ T cells have been reported to shape adaptive immunity [5]-[7]. Of particular interest are reports [30]–[33] that inhibitory KIRs on CD8+ T cells promote the survival of a subset of memory phenotype CD8+ αβ T cells with enhanced cytolytic potential (Tm1 [34]) by reducing activation-induced cell death. Ugolini et al suggested that this phenomenon helps maintain specific CD8+ T lymphocytes during chronic viral infection [30]. Tm1 cells have been described in both HTLV-1 and HCV infections, where they constitute a minority of virus-specific CD8+ T cells but the majority of perforin-bright cells [35]–[36]. Consistent with our findings, these studies showed that the HLA molecule that restricts the T cell whose survival is promoted was independent of the HLA-C molecules that ligated the KIR [30], [34]. We postulate that, in the face of chronic antigen stimulation, protective T cells survive longer if they carry KIR2DL2 and therefore exert stronger protection. Likewise, T cells restricted by HLA alleles associated with increased disease susceptibility also survive for longer in the presence of KIR2DL2 and so are more detrimental (Figure 4). Hence, KIR2DL2 enhances both protective and detrimental HLA class I associations.
Fig. 4. Postulated mechanism: KIR2DL2 reduces clonal exhaustion of CD8+ T cells and is necessary for an effective immune response in the face of chronic antigen stimulation. 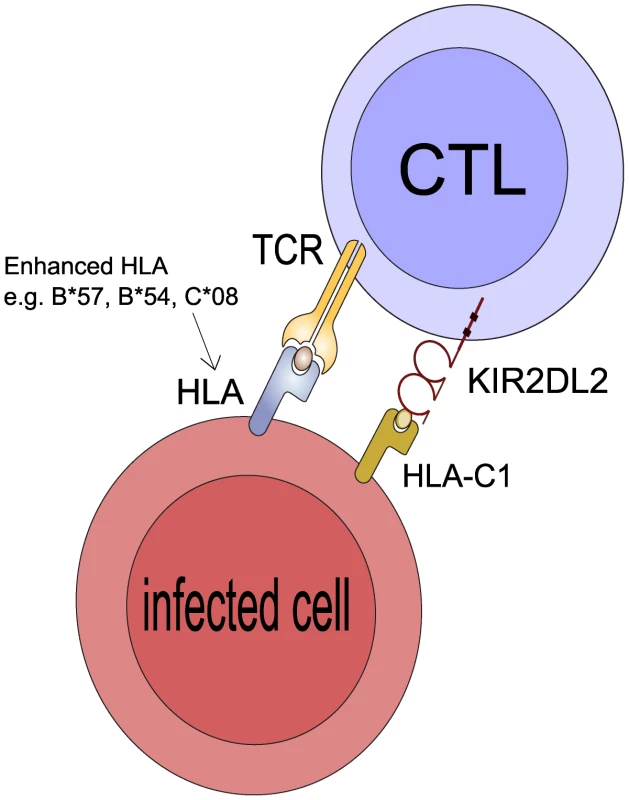
It has been demonstrated that CD8+ T cells that express inhibitory KIR have elevated levels of Bcl-2, are less susceptible to activation induced cell death (AICD) and have a survival advantage leading to their accumulation in vitro and in vivo [30], [31]. We suggest KIR2DL2 binds its HLA-C ligands so that when the CD8+ T cell is activated by engagement of its TCR by the cognate HLA:peptide complex the CD8+ T cell will less likely to undergo AICD. If the CD8+ T cell is restricted by a protective HLA class I molecule (e.g. B*57 in HCV or C*08 in HTLV-1) it will survive for longer than in a person who is KIR2DL2- and its protective effects will be enhanced. Similarly, if the CD8+ T cell is restricted by a detrimental HLA molecule (e.g. B54 in HTLV-1) it will also survive for longer and its detrimental effects will be enhanced. Of note, the HLA molecule whose protective/detrimental effects are enhanced is the HLA molecule that binds the TCR not the HLA that binds KIR2DL2. This postulated mechanism explains 3 striking features of our observations: i) that KIR2DL2, a receptor typically associated with innate immunity, enhances adaptive immunity ii) that the effect cannot be explained by KIR2DL2 binding the enhanced HLA molecules directly iii) that both protective and detrimental HLA effects are enhanced. Alternatively, it is known that NK cells kill activated T cells and that this killing is reduced by inhibitory KIR [37]–[38]. So again, T cells restricted by protective and detrimental HLA class I molecules may survive longer in the presence of inhibitory KIR and thus the protective and detrimental associations would be enhanced.
Ugolini et al proposed that inhibitory KIRs promote T cell survival by increasing the activation threshold of T cells. This may explain why the HLA-A*02 protective effect in HTLV-1 is not significantly enhanced by KIR2DL2. A*02 molecules bind peptides significantly more strongly than other alleles (section 9 in Text S1) and the immunodominant HTLV-1 peptide Tax 11-19 is bound exceptionally strongly. Therefore, even if the T cell activation threshold were increased, the strength of signalling may remain above the threshold and consequently the A*02 protective effect cannot be enhanced.
Why does KIR2DL2 enhance T cell responses whereas the other inhibitory KIRs apparently do not? The effect of KIR2DL2 may be most apparent because KIR2DL2 is present at informative frequencies and its C1 and C2 ligands are ubiquitous; i.e. unlike the other KIR every individual carries a KIR2DL2 ligand.
It will be important to determine whether inhibitory KIRs play a similar role in enhancing CD8+ and possibly CD4+ T cell-mediated immunity to other pathogens and in autoimmune disease. KIR-expressing virus-specific CD8+ T cells have been reported in other chronic infections including HIV-1, CMV and EBV [39]-[41]. Furthermore, in HIV-1 infection, high expression alleles of an inhibitory KIR, KIR3DL1, in the context of HLA-Bw4I have been associated with slow progression to AIDS [11]. In order to explain protection by an inhibitory KIR the authors proposed a model based on NK cell development. Our results suggest an alternative explanation, i.e. that KIR3DL1 enhances protective HLA-B-restricted responses to HIV-1.
In contrast to previous studies of KIR genotype, which investigated the antiviral action of NK cells, we investigated the impact of KIRs on HLA class I-mediated antiviral immunity. We find a clear and consistent effect of KIR2DL2. The effect sizes are striking: KIR2DL2 homozygotes with B*57 are almost 5 times more likely to clear HCV infection spontaneously; if they fail to clear the virus they have a viral load that is reduced by 6.5 logs. Until now, the advantages offered by inhibitory KIRs in virus infections have been unclear. Our data support an alternative role in which inhibitory KIRs enhance both beneficial and detrimental T cell-mediated immunity in persistent viral infection.
Methods
Ethics statement
The HTLV-1 cohort has been approved by the following committees: 1) St. Mary's Local Research Ethics Committee, 1995: title “The immunology and virology of the treatment of HTLV-1-associated inflammatory disease”. Approval reference number: EC3108. 2) Kagoshima University Hospital Clinical Research Ethics Committee: 27th May 1999. Title: “Investigation of HAM pathomechanism: relationship between host genetic background and clinical status of HTLV-1 infection”. Approval reference number: 22. All samples were taken under written informed consent.
Cohorts
The HCV cohort consisted of four sub-cohorts: AIDS Link to Intravenous Experience (ALIVE, N = 262) [42], Multicenter Hemophilia Cohort Study (MHCS, N = 320) [43], Hemophilia Growth and Development Study (HGDS, N = 110) [44] and a UK cohort (N = 341) [10]. 251 individuals were excluded due to incomplete information. The cohort had 257 resolved and 525 chronic patients. HLA class I associations in three of these 4 cohorts (ALIVE, MHCS and HGDS) have previously been reported [18].
The HTLV-1 cohort (N = 431) [14] consists of individuals recruited in Kagoshima Japan. All individuals were of Japanese ethnic origin and resided in Kagoshima Prefecture, Japan. The cohort had 229 HAM/TSP patients and 202 asymptomatic carriers.
HLA genotyping
HLA genotyping of the HCV and HTLV-1 cohorts was performed in previous studies [10], [14], [18].
HCV
Genomic DNA was amplified using locus specific primers as described [10], [18]. The resulting PCR products were blotted onto nitrocellulose membranes and hybridized with sequence specific oligonucleotide probes (SSO). US: alleles were assigned according to the reaction patterns of the SSO probes, ambiguities were resolved by sequencing analysis. UK: PCR products were typed by direct sequencing. HLA types that were not resolved by sequencing or which gave unusual results were also tested by SSO typing using commercial kits (Dynal, RELI SSO, Wirral, UK).
HTLV-1
PCR–sequence-specific primer reactions were performed as described [45].
KIR genotyping
KIR genotyping of the HCV cohort (but not the HTLV-1 cohort) was performed previously [10].
HCV
The presence or absence of ten KIR genes (KIR2DL1, KIR2DL2, KIR2DL3, KIR3DL1, KIR2DS1, KIR2DS2, KIR2DS3, KIR2DS4, KIR2DS5 and KIR3DS1) was determined by PCR using sequence specific primers as described in [46].
HTLV-1
The presence or absence of ten KIR genes (KIR2DL1, KIR2DL2, KIR2DL3, KIR2DL4, KIR3DL1, KIR2DS2, KIR2DS3, KIR2DS4, KIR2DS5 and KIR3DS1) was determined by PCR using sequence specific primers [47]. 30 of the 431 individuals were not KIR genotyped due to insufficient DNA.
This method of KIR typing does not allow the direct determination of KIR2DL2 copy number. Instead we use the allelic nature of KIR2DL2 and KIR2DL3 at the 2DL2/2DL3 locus to infer the number of copies of KIR2DL2.
Viral load
HCV
HCV Cohorts were analysed separately as viral load was measured using different assays. Two cohorts (MHCS and ALIVE) had sufficient numbers of individuals with measurements of all variables for this analysis. For MHCS we used the median of multiple measurements of viral load; for ALIVE, a single measurement was available for each subject. We only analysed viral load in patients with chronic infection, so any observed impact on viral load is independent of an observed impact on viral clearance.
HCV RNA was assessed by branched DNA (Quantiplex HCV RNA 2.0 assay; Chiron Corporation) [ALIVE] or the HCV COBAS AMPLICOR system (COBAS AMPLICOR HCV; Roche Diagnostics) [MHCS].
HTLV-1
The HTLV-1 provirus load in peripheral blood mononuclear cells (PBMC) was measured as described in [48]. Quantitative PCR was performed using an ABI 7700 sequence detector (Perkin–Elmer Applied Biosystems). All DNA standards and samples were amplified in triplicate. A standard curve was generated by using the β-actin gene from HTLV-1-negative PBMC and the Tax gene from TARL-2, a cell line containing a single copy of HTLV-1 proviral DNA. We analysed viral load separately in ACs and HAM/TSP patients, so any observed impact on viral load is independent of any observed impact on disease status.
Statistical analysis
HLA class I
HLA class I alleles which were significantly associated with outcome in our cohorts and that were independently verified (either in a large independent cohort or on an independent outcome) were studied. For HTLV-1 there are three such associations: HLA-A*02 and C*08 which are associated with reduced proviral load in ACs and reduced risk of HAM/TSP and HLA-B*54 which is associated with increased proviral load in HAM/TSP patients and increased risk of HAM/TSP [13]-[14]. For HCV there are two such associations: HLA-B*57 and C*01 which have both been associated with increased odds of viral clearance in two independent cohorts [17], [49]. The C*01-protective effect in HCV infection has been shown to be NK-mediated [10], [28] and therefore we did not consider it here.
KIR
We investigated KIR genes which were present at an informative frequency. We define an informative frequency as sufficient to detect a “moderate” effect size (δ = 0.5, [50]); this is equivalent to at least 32 people carrying the gene and 32 not carrying the gene. Of the KIR genes typed, 4 were present at an informative frequency in the HTLV-I cohort (KIR2DL2, KIR2DS2, KIR2DS3 and KIR3DS1) and 9 in the HCV cohort (KIR2DL2, KIR2DL3, KIR3DL1, KIR2DS1, KIR2DS2, KIR2DS3, KIR2DS4, KIR2DS5 and KIR3DS1).
The effect of individual HLA class I alleles in different KIR genetic background was investigated by stratifying the cohorts for the absence and presence of the informative KIRs and the effect of the already described HLA class I associations were re-evaluated in each stratum. The impact of the HLA alleles on two response variables was studied: 1) status (AC vs. HAM/TSP for HTLV-1 infection and resolved vs. chronic for HCV infection) and 2) viral burden (log10[proviral load] for HTLV-1 infection and log10[viral load] for HCV infection). Multiple logistic regression was used to study the variation in status and multiple linear regression was used for the variation in viral burden. All statistically significant confounding variables were included in the models (HTLV-1: age and gender; HCV: HBV status, mode of infection, SNP rs12979860 and cohort). We focused on the difference between KIR+ and KIR - individuals in the size of the protective/detrimental effects associated with the individual HLA class I molecules (i.e. odds ratios and differences in viral load) rather than p values as the latter comparison is confounded by differences in strata sizes.
We quantified the false discovery rate for our analysis using Monte Carlo methods. For each of the HLA associations studied, we performed 10,000 random stratifications of the relevant (HCV or HTLV-1) cohort with the size of the two strata being the number of KIR2DL2+ and KIR2DL2 - individuals in that cohort and asked how many times we would see odds ratios equal to or more extreme than we observed in the actual cohorts. The probability of making our observations by chance is less than p = 2×10−11.
Linkage disequilibrium (LD) between HLA class I alleles was calculated using the tool available at www.hiv.lanl.gov. The LD between KIRs was calculated based on the Chi-squared test on a 2x2 contingency table. All the reported p values are two-tailed. Where applicable, independent p values were combined using Fisher's combined test. Statistical analysis was performed using R v2.9.2.
Epitope prediction
The binding strength of HLA class I molecules to viral proteins was assessed using epitope prediction software. Prediction of T cell class I epitopes is now highly accurate and algorithms can achieve accuracy of up to 94% [51]. In this study we use the epitope prediction software Metaserver [21] (http://web.bioinformatics.ic.ac.uk/metaserver).
Rank measure for predicted epitopes
We used the rank measure technique [52] in which the strength of an allele's preference for a particular protein is quantified by ranking the strength of binding of the top binding peptide from the protein of interest amongst the strength of binding of peptides from the entire proteome to that allele. Specifically, we split the HTLV-1 proteome into overlapping nonamers offset by a single amino acid and predicted a binding affinity score for each nonomer. For each allele we rank all nonamers from the proteome from the weakest to strongest predicted binding scores. This produces a list of rank values for each protein to that particular allele that quantifies the binding relationship between that allele and the protein. We then invert the rank so the bigger 1/rank, the stronger the preference of the allele for the protein; the logarithm of this measure is plotted on the y axis of Figure 2 as “HBZ binding score”. Each individual therefore contributes up to 4 values (alleles for which no predictive algorithms were available were excluded from the analysis). Binding scores were compared between ACs and HAM/TSP patients using the Wilcoxon rank sum test and reported both as separate p values for HLA-A and B molecules and combined (since we found no evidence to reject the null hypothesis that the HBZ binding score of an individual's A and B molecules was independent, spearman correlation = 0.05 p = 0.5). The median difference in binding score is the median of the difference of average HBZ binding between ACs and HAM/TSP patients expressed as a percent of the AC binding score for HLA-A and –B molecules.
Supporting Information
Zdroje
1. MingariMCMorettaAMorettaL 1998 Regulation of KIR expression in human T cells: a safety mechanism that may impair protective T-cell responses. Immunol Today 19 153 157
2. SpeiserDEPittetMJValmoriDDunbarRRimoldiD 1999 In vivo expression of natural killer cell inhibitory receptors by human melanoma-specific cytolytic T lymphocytes. J Exp Med 190 775 782
3. SpeiserDEValmoriDRimoldiDPittetMJLienardD 1999 CD28-negative cytolytic effector T cells frequently express NK receptors and are present at variable proportions in circulating lymphocytes from healthy donors and melanoma patients. Eur J Immunol 29 1990 1999
4. LongEORajagopalanS 2000 HLA class I recognition by killer cell Ig-like receptors. Semin Immunol 12 101 108
5. GerosaFBaldani-GuerraBNisiiCMarchesiniVCarraG 2002 Reciprocal Activating Interaction between Natural Killer Cells and Dendritic Cells. J Exp Med 195 327 333
6. PiccioliDSbranaSMelandriEValianteNM 2002 Contact-dependent Stimulation and Inhibition of Dendritic Cells by Natural Killer Cells. J Exp Med 195 335 341
7. RauletDH 2004 Interplay of natural killer cells and their receptors with the adaptive immune response. Nat Immunol 5 996 1002
8. WagtmannNRajagopalanSWinterCCPeruzziMLongEO 1995 Killer cell inhibitory receptors specific for HLA-C and HLA-B identified by direct binding and by functional transfer. Immunity 3 801 809
9. MoestaAKNormanPJYawataMYawataNGleimerM 2008 Synergistic polymorphism at two positions distal to the ligand-binding site makes KIR2DL2 a stronger receptor for HLA-C than KIR2DL3. J Immunol 180 3969 3979
10. KhakooSIThioCLMartinMPBrooksCRGaoX 2004 HLA and NK cell inhibitory receptor genes in resolving hepatitis C virus infection. Science 305 872 874
11. MartinMPQiYGaoXYamadaEMartinJN 2007 Innate partnership of HLA-B and KIR3DL1 subtypes against HIV-1. Nat Genet 39 733 740
12. NagaiMUsukuKMatsumotoWKodamaDTakenouchiN 1998 Analysis of HTLV-I proviral load in 202 HAM/TSP patients and 243 asymptomatic HTLV-I carriers: high proviral load strongly predisposes to HAM/TSP. J Neurovirol 4 586 593
13. JefferyKJSiddiquiAABunceMLloydALVineAM 2000 The influence of HLA class I alleles and heterozygosity on the outcome of human T cell lymphotropic virus type I infection. J Immunol 165 7278 7284
14. JefferyKJUsukuKHallSEMatsumotoWTaylorGP 1999 HLA alleles determine human T-lymphotropic virus-I (HTLV-I) proviral load and the risk of HTLV-I-associated myelopathy. Proc Natl Acad Sci U S A 96 3848 3853
15. DustinLBRiceCM 2007 Flying Under the Radar: The Immunobiology of Hepatitis C. Annu Rev Immunol 25 71 99
16. KimAYKuntzenTTimmJNolanBEBacaMA 2011 Spontaneous control of HCV is associated with the expression of HLA-B*57 and preservation of targeted epitopes. Gastroenterology 140 686 696
17. KuniholmMHKovacsAGaoXXueXMartiD 2010 Specific human leukocyte antigen class I and II alleles associated with hepatitis C virus viremia. Hepatology 51 1514 1522
18. ThioCLGaoXGoedertJJVlahovDNelsonKE 2002 HLA-Cw*04 and Hepatitis C Virus Persistence. J Virol 76 4792 4797
19. ChuangWC-MSarkodieFBrownCJOwusu-OforiSBrownJ 2007 Protective effect of HLA-B57 on HCV genotype 2 infection in a West African population. J Med Virol 79 724 733
20. MacNamaraRowanHilburnKadolskyFujiwara 2010 HLA class I binding of HBZ determines outcome in HTLV-1 infection. PLoS Pathog 6 e1001117
21. MacNamaraAKadolskyUBanghamCRAsquithB 2009 T-cell epitope prediction: rescaling can mask biological variation between MHC molecules. PLoS Comput Biol 5 e1000327
22. NormanPJCookMACareyBSCarringtonCVVerityDH 2004 SNP haplotypes and allele frequencies show evidence for disruptive and balancing selection in the human leukocyte receptor complex. Immunogenetics 56 225 237
23. WinterCCGumperzJEParhamPLongEOWagtmannN 1998 Direct binding and functional transfer of NK cell inhibitory receptors reveal novel patterns of HLA-C allotype recognition. J Immunol 161 571 577
24. O'ConnorGMGuinanKJCunninghamRTMiddletonDParhamP 2007 Functional polymorphism of the KIR3DL1/S1 receptor on human NK cells. J Immunol 178 235 241
25. YawataMYawataNDraghiMLittleAMPartheniouF 2006 Roles for HLA and KIR polymorphisms in natural killer cell repertoire selection and modulation of effector function. J Exp Med 203 633 645
26. VilchesCParhamP 2002 KIR: diverse, rapidly evolving receptors of innate and adaptive immunity. Annu Rev Immunol 20 217 251
27. FaddaLBorhisGAhmedPCheentKPageonSV Peptide antagonism as a mechanism for NK cell activation. Proc Natl Acad Sci U S A 107 10160 10165
28. AhlenstielGMartinMPGaoXCarringtonMRehermannB 2008 Distinct KIR/HLA compound genotypes affect the kinetics of human antiviral natural killer cell responses. J Clin Invest 118 1017 1026
29. ThomasDLThioCLMartinMPQiYGeD 2009 Genetic variation in IL28B and spontaneous clearance of hepatitis C virus. Nature 461 798 801
30. UgoliniSArpinCAnfossiNWalzerTCambiaggiA 2001 Involvement of inhibitory NKRs in the survival of a subset of memory-phenotype CD8+ T cells. Nat Immunol 2 430 435
31. YoungNTUhrbergMPhillipsJHLanierLLParhamP 2001 Differential expression of leukocyte receptor complex-encoded Ig-like receptors correlates with the transition from effector to memory CTL. J Immunol 166 3933 3941
32. GatiAGuerraNGaudinCDa RochaSEscudierB 2003 CD158 Receptor Controls Cytotoxic T-Lymphocyte Susceptibility to Tumor-Mediated Activation-Induced Cell Death by Interfering with Fas Signaling. Cancer Res 63 7475 7482
33. YoungNTUhrbergM 2002 KIR expression shapes cytotoxic repertoires: a developmental program of survival. Trends Immunol 23 71 75
34. AnfossiNPascalVVivierEUgoliniS 2001 Biology of T memory type 1 cells. Immunol Rev 181 269 278
35. BieganowskaKHollsbergPBuckleGJLimDGGretenTF 1999 Direct analysis of viral-specific CD8+ T cells with soluble HLA-A2/Tax11-19 tetramer complexes in patients with human T cell lymphotropic virus-associated myelopathy. J Immunol 162 1765 1771
36. BonorinoPLeroyVDufeu-DuchesneTTongiani-DashanSSturmN 2007 Features and distribution of CD8 T cells with human leukocyte antigen class I-specific receptor expression in chronic hepatitis C. Hepatology 46 1375 1386
37. SoderquestKWalzerTZafirovaBKlavinskisLSPolicB Cutting edge: CD8+ T cell priming in the absence of NK cells leads to enhanced memory responses. J Immunol 186 3304 3308
38. TakaoSIshikawaTYamashitaKUchiyamaT The rapid induction of HLA-E is essential for the survival of antigen-activated naive CD4 T cells from attack by NK cells. J Immunol 185 6031 6040
39. AlterGRihnSStreeckHTeigenNPiechocka-TrochaA 2008 Ligand-independent exhaustion of killer immunoglobulin-like receptor-positive CD8+ T cells in human immunodeficiency virus type 1 infection. J Virol 82 9668 9677
40. PoonKMontamat-SicotteDCumberbatchNMcMichaelAJCallanMF 2005 Expression of leukocyte immunoglobulin-like receptors and natural killer receptors on virus-specific CD8+ T cells during the evolution of Epstein-Barr virus-specific immune responses in vivo. Viral Immunol 18 513 522
41. van der VekenLTCampeloMDvan der HoornMAHagedoornRSvan EgmondHM 2009 Functional analysis of killer Ig-like receptor-expressing cytomegalovirus-specific CD8+ T cells. J Immunol 182 92 101
42. VlahovDAnthonyJCMunozAMargolickJNelsonKE 1991 The ALIVE study, a longitudinal study of HIV-1 infection in intravenous drug users: description of methods and characteristics of participants. NIDA Res Monogr 109 75 100
43. GoedertJJKesslerCMAledortLMBiggarRJAndesWA 1989 A prospective study of human immunodeficiency virus type 1 infection and the development of AIDS in subjects with hemophilia. N Engl J Med 321 1141 1148
44. HilgartnerMWDonfieldSMWilloughbyAContantCFEvattBL 1993 Hemophilia growth and development study. Design, methods, and entry data. Am J Pediatr Hematol Oncol 15 208 218
45. BunceMO'NeillCMBarnardoMCNMKrausaPBrowningMJ 1995 Phototyping: comprehensive DNA typing for HLA-A, B, C, DRB1, DRB3, DRB4, DRB5 & DQB1 by PCR with 144 primer mixes utilizing sequence-specific primers (PCR-SSP). Tissue Antigens 46 355 367
46. MartinMPGaoXLeeJ-HNelsonGWDetelsR 2002 Epistatic interaction between KIR3DS1 and HLA-B delays the progression to AIDS. Nat Genet 31 429 434
47. UhrbergMValianteNMShumBPShillingHGLienert-WeidenbachK 1997 Human diversity in killer cell inhibitory receptor genes. Immunity 7 753 763
48. NagaiMUsukuKMatsumotoWKodamaDTakenouchiN 1998 Analysis of HTLV-I proviral load in 202 HAM/TSP patients and 243 asymptomatic HTLV-I carriers: high proviral load strongly predisposes to HAM/TSP. J Virol 4 586 593
49. ThioCLGaoXGoedertJJVlahovDNelsonKE 2002 HLA-Cw*04 and hepatitis C virus persistence. J Virol 76 4792 4797
50. CohenJ 1988 Statistical Power Analysis for the Behavioral Sciences
51. YangXYuX 2009 An introduction to epitope prediction methods and software. Rev Med Virol 19 77 96
52. BorghansJAMMølgaardAde BoerRJKeşmirC 2007 HLA Alleles Associated with Slow Progression to AIDS Truly Prefer to Present HIV-1 p24. PLoS ONE 2 e920 e920
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek Quorum Sensing in Fungi: Q&AČlánek Blood Feeding and Insulin-like Peptide 3 Stimulate Proliferation of Hemocytes in the MosquitoČlánek The DEAD-box RNA Helicase DDX6 is Required for Efficient Encapsidation of a Retroviral GenomeČlánek A Phenome-Based Functional Analysis of Transcription Factors in the Cereal Head Blight Fungus,Článek A Wide Extent of Inter-Strain Diversity in Virulent and Vaccine Strains of AlphaherpesvirusesČlánek The Anti-Sigma Factor TcdC Modulates Hypervirulence in an Epidemic BI/NAP1/027 Clinical Isolate ofČlánek Critical Roles for LIGHT and Its Receptors in Generating T Cell-Mediated Immunity during InfectionČlánek Frequent and Recent Human Acquisition of Simian Foamy Viruses Through Apes' Bites in Central Africa
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2011 Číslo 10- Jak souvisí postcovidový syndrom s poškozením mozku?
- Měli bychom postcovidový syndrom léčit antidepresivy?
- Farmakovigilanční studie perorálních antivirotik indikovaných v léčbě COVID-19
- 10 bodů k očkování proti COVID-19: stanovisko České společnosti alergologie a klinické imunologie ČLS JEP
-
Všechny články tohoto čísla
- Quorum Sensing in Fungi: Q&A
- Discovery of an Ebolavirus-Like Filovirus in Europe
- Toll-like Receptor 7 Controls the Anti-Retroviral Germinal Center Response
- Tubule-Guided Cell-to-Cell Movement of a Plant Virus Requires Class XI Myosin Motors
- Herpesvirus Telomerase RNA (vTR) with a Mutated Template Sequence Abrogates Herpesvirus-Induced Lymphomagenesis
- Mitochondrial Peroxiredoxin Plays a Crucial Peroxidase-Unrelated Role during Infection: Insight into Its Novel Chaperone Activity
- Sustained CD8+ T Cell Memory Inflation after Infection with a Single-Cycle Cytomegalovirus
- Novel Mouse Xenograft Models Reveal a Critical Role of CD4 T Cells in the Proliferation of EBV-Infected T and NK Cells
- Toll-8/Tollo Negatively Regulates Antimicrobial Response in the Respiratory Epithelium
- Exhausted Cytotoxic Control of Epstein-Barr Virus in Human Lupus
- Structural and Functional Analysis of Laninamivir and its Octanoate Prodrug Reveals Group Specific Mechanisms for Influenza NA Inhibition
- Infection Drives IL-17-Mediated Neutrophilic Allergic Airways Disease
- Blood Feeding and Insulin-like Peptide 3 Stimulate Proliferation of Hemocytes in the Mosquito
- HIV-1 Replication in the Central Nervous System Occurs in Two Distinct Cell Types
- Deep Molecular Characterization of HIV-1 Dynamics under Suppressive HAART
- Fitness Landscape of Antibiotic Tolerance in Biofilms
- The DEAD-box RNA Helicase DDX6 is Required for Efficient Encapsidation of a Retroviral Genome
- Preventing Sepsis through the Inhibition of Its Agglutination in Blood
- A Phenome-Based Functional Analysis of Transcription Factors in the Cereal Head Blight Fungus,
- IFITM3 Inhibits Influenza A Virus Infection by Preventing Cytosolic Entry
- Targeting Cattle-Borne Zoonoses and Cattle Pathogens Using a Novel Trypanosomatid-Based Delivery System
- A Wide Extent of Inter-Strain Diversity in Virulent and Vaccine Strains of Alphaherpesviruses
- Coordinated Destruction of Cellular Messages in Translation Complexes by the Gammaherpesvirus Host Shutoff Factor and the Mammalian Exonuclease Xrn1
- Signal Transduction through CsrRS Confers an Invasive Phenotype in Group A
- Biochemical and Structural Insights into the Mechanisms of SARS Coronavirus RNA Ribose 2′-O-Methylation by nsp16/nsp10 Protein Complex
- Histone Deacetylase 8 Is Required for Centrosome Cohesion and Influenza A Virus Entry
- Severe Acute Respiratory Syndrome Coronavirus Envelope Protein Regulates Cell Stress Response and Apoptosis
- Co-opts the FGF2 Signaling Pathway to Enhance Infection
- IRAK-2 Regulates IL-1-Mediated Pathogenic Th17 Cell Development in Helminthic Infection
- Trafficking of Hepatitis C Virus Core Protein during Virus Particle Assembly
- The Anti-interferon Activity of Conserved Viral dUTPase ORF54 is Essential for an Effective MHV-68 Infection
- A Viral Nuclear Noncoding RNA Binds Re-localized Poly(A) Binding Protein and Is Required for Late KSHV Gene Expression
- Suppression of Methylation-Mediated Transcriptional Gene Silencing by βC1-SAHH Protein Interaction during Geminivirus-Betasatellite Infection
- ISG15 Is Critical in the Control of Chikungunya Virus Infection Independent of UbE1L Mediated Conjugation
- Non-Hematopoietic Cells in Lymph Nodes Drive Memory CD8 T Cell Inflation during Murine Cytomegalovirus Infection
- RNA Polymerase II Stalling Promotes Nucleosome Occlusion and pTEFb Recruitment to Drive Immortalization by Epstein-Barr Virus
- Noninfectious Retrovirus Particles Drive the / Dependent Neutralizing Antibody Response
- Endophytic Life Strategies Decoded by Genome and Transcriptome Analyses of the Mutualistic Root Symbiont
- An Integrated Approach to Elucidate the Intra-Viral and Viral-Cellular Protein Interaction Networks of a Gamma-Herpesvirus
- as an Animal Model for the Study of Biofilm Infections
- Homeostatic Proliferation Fails to Efficiently Reactivate HIV-1 Latently Infected Central Memory CD4+ T Cells
- The Anti-Sigma Factor TcdC Modulates Hypervirulence in an Epidemic BI/NAP1/027 Clinical Isolate of
- Enhances Protective and Detrimental HLA Class I-Mediated Immunity in Chronic Viral Infection
- The Mouse IAPE Endogenous Retrovirus Can Infect Cells through Any of the Five GPI-Anchored EphrinA Proteins
- The Urgent Need for Robust Coral Disease Diagnostics
- HacA-Independent Functions of the ER Stress Sensor IreA Synergize with the Canonical UPR to Influence Virulence Traits in
- A Novel Core Genome-Encoded Superantigen Contributes to Lethality of Community-Associated MRSA Necrotizing Pneumonia
- Critical Roles for LIGHT and Its Receptors in Generating T Cell-Mediated Immunity during Infection
- The SARS-Coronavirus-Host Interactome: Identification of Cyclophilins as Target for Pan-Coronavirus Inhibitors
- Frequent and Recent Human Acquisition of Simian Foamy Viruses Through Apes' Bites in Central Africa
- Mechanisms of Trafficking to the Brain
- Defining Emerging Roles for NF-κB in Antivirus Responses: Revisiting the Enhanceosome Paradigm
- The Role of Sialyl Glycan Recognition in Host Tissue Tropism of the Avian Parasite
- Evolutionarily Divergent, Unstable Filamentous Actin Is Essential for Gliding Motility in Apicomplexan Parasites
- The Herpes Simplex Virus-1 Transactivator Infected Cell Protein-4 Drives VEGF-A Dependent Neovascularization
- Distinct Single Amino Acid Replacements in the Control of Virulence Regulator Protein Differentially Impact Streptococcal Pathogenesis
- Soluble Rhesus Lymphocryptovirus gp350 Protects against Infection and Reduces Viral Loads in Animals that Become Infected with Virus after Challenge
- A Genetic Screen Reveals Arabidopsis Stomatal and/or Apoplastic Defenses against pv. DC3000
- Hepatitis C Virus Reveals a Novel Early Control in Acute Immune Response
- Fumarate Reductase Activity Maintains an Energized Membrane in Anaerobic
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Severe Acute Respiratory Syndrome Coronavirus Envelope Protein Regulates Cell Stress Response and Apoptosis
- The SARS-Coronavirus-Host Interactome: Identification of Cyclophilins as Target for Pan-Coronavirus Inhibitors
- Biochemical and Structural Insights into the Mechanisms of SARS Coronavirus RNA Ribose 2′-O-Methylation by nsp16/nsp10 Protein Complex
- Evolutionarily Divergent, Unstable Filamentous Actin Is Essential for Gliding Motility in Apicomplexan Parasites
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



