-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
Reklamaa GATA Transcription Factor That Directs Disparate Fates in Including Morphogenesis and Siderophore Biosynthesis
Blastomyces dermatitidis belongs to a group of human pathogenic fungi that exhibit thermal dimorphism. At 22°C, these fungi grow as mold that produce conidia or infectious particles, whereas at 37°C they convert to budding yeast. The ability to switch between these forms is essential for virulence in mammals and may enable these organisms to survive in the soil. To identify genes that regulate this phase transition, we used Agrobacterium tumefaciens to mutagenize B. dermatitidis conidia and screened transformants for defects in morphogenesis. We found that the GATA transcription factor SREB governs multiple fates in B. dermatitidis: phase transition from yeast to mold, cell growth at 22°C, and biosynthesis of siderophores under iron-replete conditions. Insertional and null mutants fail to convert to mold, do not accumulate significant biomass at 22°C, and are unable to suppress siderophore biosynthesis under iron-replete conditions. The defect in morphogenesis in the SREB mutant was independent of exogenous iron concentration, suggesting that SREB promotes the phase transition by altering the expression of genes that are unrelated to siderophore biosynthesis. Using bioinformatic and gene expression analyses, we identified candidate genes with upstream GATA sites whose expression is altered in the null mutant that may be direct or indirect targets of SREB and promote the phase transition. We conclude that SREB functions as a transcription factor that promotes morphogenesis and regulates siderophore biosynthesis. To our knowledge, this is the first gene identified that promotes the conversion from yeast to mold in the dimorphic fungi, and may shed light on environmental persistence of these pathogens.
Published in the journal: . PLoS Pathog 6(4): e32767. doi:10.1371/journal.ppat.1000846
Category: Research Article
doi: https://doi.org/10.1371/journal.ppat.1000846Summary
Blastomyces dermatitidis belongs to a group of human pathogenic fungi that exhibit thermal dimorphism. At 22°C, these fungi grow as mold that produce conidia or infectious particles, whereas at 37°C they convert to budding yeast. The ability to switch between these forms is essential for virulence in mammals and may enable these organisms to survive in the soil. To identify genes that regulate this phase transition, we used Agrobacterium tumefaciens to mutagenize B. dermatitidis conidia and screened transformants for defects in morphogenesis. We found that the GATA transcription factor SREB governs multiple fates in B. dermatitidis: phase transition from yeast to mold, cell growth at 22°C, and biosynthesis of siderophores under iron-replete conditions. Insertional and null mutants fail to convert to mold, do not accumulate significant biomass at 22°C, and are unable to suppress siderophore biosynthesis under iron-replete conditions. The defect in morphogenesis in the SREB mutant was independent of exogenous iron concentration, suggesting that SREB promotes the phase transition by altering the expression of genes that are unrelated to siderophore biosynthesis. Using bioinformatic and gene expression analyses, we identified candidate genes with upstream GATA sites whose expression is altered in the null mutant that may be direct or indirect targets of SREB and promote the phase transition. We conclude that SREB functions as a transcription factor that promotes morphogenesis and regulates siderophore biosynthesis. To our knowledge, this is the first gene identified that promotes the conversion from yeast to mold in the dimorphic fungi, and may shed light on environmental persistence of these pathogens.
Introduction
The endemic dimorphic fungi are comprised of seven ascomycetes that include Blastomyces dermatitidis, Histoplasma capsulatum, Coccidioides immitis, Coccidioides posadasii, Paracoccidioides brasiliensis, Sporothrix schenckii, and Penicillium marneffei. These fungi possess the unique ability to switch between two different morphologies, yeast and mold, in response to external stimuli [1]. In nature, they grow as mycelia that produce conidia, which are the infectious particles; when aerosolized spores are inhaled into the warmer lungs of a mammalian host, they convert into pathogenic yeast and cause necrotizing infection [1]. The dimorphic fungi collectively are the most common cause of invasive fungal disease worldwide and account for several million infections each year [2]. Unlike opportunistic fungi, such as Cryptococcus or Aspergillus, the dimorphic fungi can infect both immunocompetent and immunocompromised hosts [3]–[5]. The size of the inhaled inoculum and the integrity of the cell-mediated immune system influence the extent and severity of infection [1],[3]. Clinical manifestations range from asymptomatic infection to symptomatic disease and include pneumonia, acute respiratory distress syndrome, and disseminated disease involving multiple organ systems [1],[3].
The ability of the dimorphic fungi to switch between the two different morphologies is crucial for pathogenesis. Although temperature is postulated to be the major stimulus that induces phase transition, other stimuli, including carbon dioxide tension, steroid hormones, and oxidative stress influence this morphologic switch [1], [6]–[9]. Phase transition is a complex process that involves the coordinated expression and repression of many genes in response to external stimuli, which alters cell wall composition, metabolism, intracellular signaling, and morphology [10]–[13]. The identification of DRK1 (dimorphism-regulating kinase-1) in B. dermatitidis and H. capsulatum offered strong genetic evidence that phase transition is required for pathogenicity [10]. DRK1 functions as a global regulator and has pleotropic effects on the cell, controlling morphogenesis, cell wall composition, sporulation, expression of yeast-phase specific genes, and virulence. DRK1 null mutants remain locked in the mycelial phase, fail to sporulate or express the essential virulence factors BAD1 (Blastomyces adhesin-1 in B. dermatitidis) and CBP1 (Calcium binding protein-1 in H. capsulatum), and are avirulent in a murine model of infection [10]. Three additional genes, RYP1, RYP2, and RYP3, have been described that regulate morphogenesis in H. capsulatum. Silencing the expression of RYP1, 2 or 3 results in hyphal growth at 37°C and inappropriate sporulation [12],[13].
The goal of this study was to identify and characterize additional genes that regulate the phase transition in dimorphic fungi, using B. dermatitidis as a model system. While progress has been made in identifying genes that regulate the morphological transition from mold to yeast, to our knowledge, no genes have been identified that regulate the switch in the other direction in the dimorphic fungi – that is, from the yeast to mold form. The mold form is believed to be required for the growth and survival of the dimorphic fungi in the environment by enabling propagation in soil and transmission to humans through the generation of conidia. Herein, we describe a gene, SREB, identified through insertional mutagenesis, which impacts multiple disparate fates in B. dermatitidis, including the phase transition of yeast to mold, cell growth at 22°C, and the biosynthesis of siderophores.
Results
Insertional mutagenesis and identification of SREB
Agrobacterium tumefaciens-mediated DNA transfer was used to mutagenize haploid, uninucleate conidia of B. dermatitidis strain T53-19. Following selection with hygromycin, 22,000 transformants were visually screened by light microscopy for morphologic alterations including growth as hyphae or pseudohyphae at 37°C or as yeast at 22°C. In this study, one of the mutants identified by the screen, 3-15-1, was characterized in detail. This mutant, unlike the parent strain, was pigmented yellow and failed to complete the conversion from yeast to mold (Figure 1A, 1B). Southern blot hybridization demonstrated a single site of insertion (Figure S1).
Fig. 1. Phenotype of insertional mutant 3-15-1 and conserved motifs in SREB. 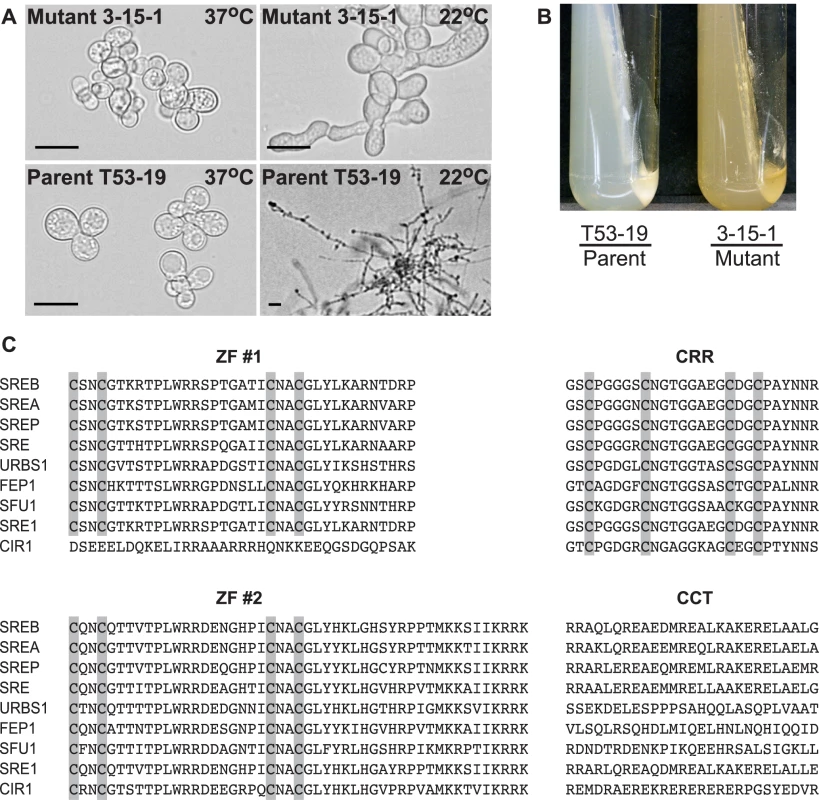
(A) Insertional mutant 3-15-1 failed to convert from yeast to mold after 14 days of incubation at 22°C. Parent strain T53-19 converted from yeast to mycelia within 7 days of shifting the temperature from 37°C to 22°C (Scale bar equals 20 µm). (B) Mutant 3-15-1 developed a yellow pigmentation and discolored the surrounding 7H10 medium, which contains 150 µM FeSO4. In contrast, T53-19 grew as white-colored yeast and did not pigment the medium. (C) The predicted amino acid sequence of B. dermatitidis SREB is aligned with A. nidulans SREA, P. chrysogenum SREP, N. crassa SRE, U. maydis URBS1, S. pombe FEP1, C. albicans SFU1, H. capsulatum SRE1, and C. neoformans CIR1. SREB contained several conserved domains including two zinc fingers (ZF #1, ZF #2) separated by a cysteine-rich region (CRR) and a conserved C-terminus (CCT) with a predicted coiled-coil domain. Conserved cysteine residues in ZF #1, ZF #2, and CRR are highlighted. ClustalW (1.81) was used to align the amino acid sequences. GeneBank accession numbers include AAD25328 (SREA), AAC49628 (SREP), AAC64946 (SRE), AAB05617 (URBS1), AAM29187 (FEP1), XP_723364 (SFU1), ABY66603 (SRE1), and ABG21303 (CIR1). The genomic DNA flanking the insert in 3-15-1 was amplified using adapter PCR, sequenced, and analyzed using a BLASTn search against the genome sequence of B. dermatitidis strain 26199. No rearrangements or deletions were identified in the DNA flanking the insert. Additional BLAST analyses indicated that the insert interrupted a region 692 base-pairs (bp) upstream of a predicted open reading frame with nucleotide homology to Pencillium chrysogenum SREP, which encodes a GATA transcription factor that regulates the biosynthesis of siderophores [14]. We named this homolog SREB (siderophore biosynthesis repressor in Blastomyces) in B. dermatitidis.
SREB sequence analysis
FGENESH analysis of the nucleotide sequence predicted that SREB contained a 1909 nucleotide (nt) coding region interrupted by two short introns (88 and 74 nt). Each intron was located in a zinc-finger coding region and contained the expected 5′-splice donor (GTNNGT) and 3′-splice acceptor (pyrimidine-AG) sequences [15]. The length, location, and number of introns interrupting the open reading frame were conserved among B. dermatitidis SREB, H. capsulatum SRE1, A. nidulans SREA, and N. crassa SRE [16]–[18]. The SREB coding region was predicted to encode a 636 amino acid protein.
The predicted amino acid sequence of SREB had homology to siderophore biosynthesis repressors in other fungi including Aspergillus nidulans SREA, Penicillum chrysogenum SREP, Neurospora crassa SRE, Ustilago maydis URBS1, Schizosaccharomyces pombe FEP1, Candida albicans SFU1, Cryptococcus neoformans CIR1, and Histoplasma capsulatum SRE1 (Figure 1C) [14], [16]–[22]. SREB contained several conserved domains characteristic of GATA transcription factors that regulate iron assimilation, including two zinc finger motifs separated by a cysteine-rich region (CRR) and a C-terminus predicted to have a coiled-coil domain (Figure 1C) [17],[23]. With the exception of C. neoformans CIR1, fungal GATA transcription factors that regulate the acquisition of iron contain two zinc fingers [22]. This zinc finger arrangement is unique because most GATA transcription factors in fungi contain only one zinc finger [17]. The cysteine residues in each zinc finger of SREB were arranged in a conserved class IV motif, Cys-X2-Cys-X17-Cys-X2-Cys [24]. The cysteine-rich region contained four conserved cysteine residues, which have been demonstrated to coordinate the binding of iron in H. capsulatum [16].
Phenotypes of insertional mutant 3-15-1
Mutant 3-15-1 failed to convert from yeast to mycelia or produce conidia following a shift in incubation temperature from 37°C to 22°C (Figure 1A). In contrast, the parent strain T53-19 converted to mycelia when grown at 22°C and produced conidia. Mutant 3-15-1 accumulated little biomass at 22°C, but remained viable (as measured by the exclusion of 0.2% eosin stain), and converted to normal yeast morphology when the incubation temperature was shifted back to 37°C (data not shown).
The yellow-orange pigmentation of mutant 3-15-1 and the predicted amino acid sequence suggested that SREB functioned as a repressor of siderophore biosynthesis. Deletions of SREB homologs in P. chrysogenum (SREP), A. nidulans (SREA), and N. crassa (SRE) produce similar discoloration [14],[17],[18]. To assess for the dysregulation of siderophore biosynthesis in the insertion mutant, we used a colorimetric assay to detect the production of hydroxymate-type sideophores in culture supernatants [25]. Under iron-poor conditions, both T53-19 and 3-15-1 produced an abundance of siderophores as measured by this assay (data not shown). Under iron-replete conditions, mutant 3-15-1 continued to produce siderophores, whereas parent strain T53-19 repressed siderophore biosynthesis (Figure 2A).
Fig. 2. Complementation of insertional mutant 3-15-1. 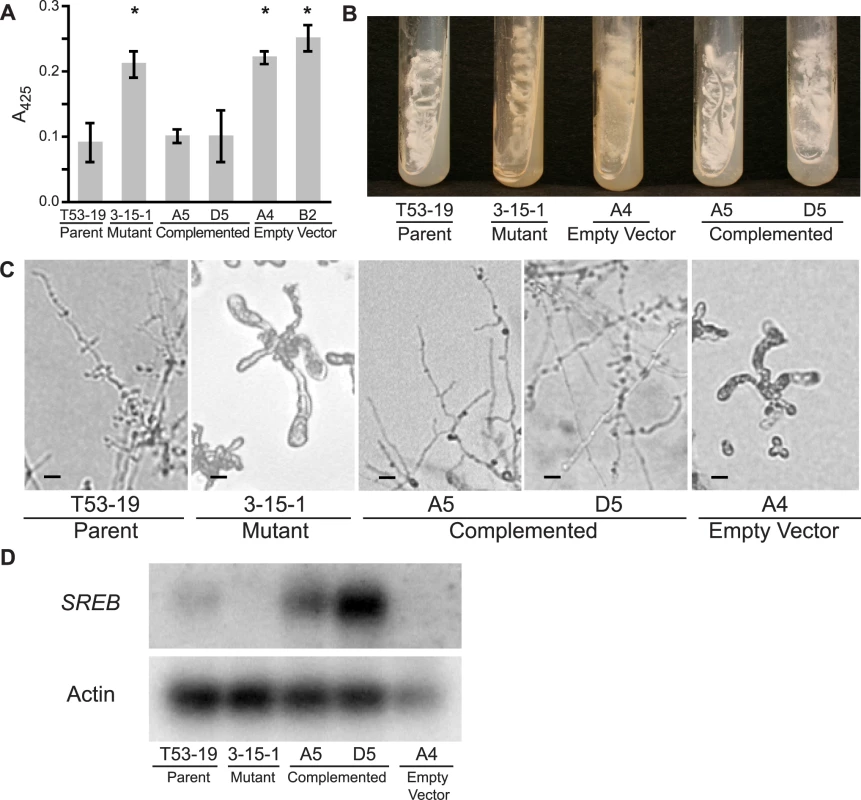
(A) Under iron-replete conditions (10 µM FeSO4), mutant 3-15-1 and empty vector controls fail to suppress siderophore biosynthesis as demonstrated by a 2.5–3.0 fold higher absorbance at 425 nm compared to parental strain T53-19 and complemented strains A5 and D5 (p<0.01 as indicated by asterisk). Data from three independent experiments were analyzed. Siderophore production was measured using the ferric perchlorate assay. (B) Mutant 3-15-1 and empty vector control A4 were pigmented yellow and discolored the surrounding media when grown on 7H10 medium, which contains 150 µM FeSO4. In contrast, parental control T53-19 and complemented strains A5 and D5 were colored white and did not pigment the medium. All strains were incubated for 14 days at 37°C. (C) Mutant 3-15-1 and empty vector control A4 fail to convert to mycelia upon shifting the incubation temperature from 37°C to 22°C. Similar to the wild-type isolate, complemented strains A5 and D5 grew as mycelia at 22°C incubation. (D) Northern blot hybridization demonstrated reduction in transcript abundance in the insertional mutant when compared to the parent strain. Complemented strains A5 and D5 overexpress SREB. Transformation of 3-15-1 with an empty vector failed to restore transcript abundance in strain A4. Complementation of mutant 3-15-1
To determine if the mutant phenotype was from altered expression of SREB, and not due to another mutation incurred during insertional mutagenesis, we set out to complement the mutant phenotype. Insertional mutant 3-15-1 was re-transformed via A. tumefaciens to provide an intact gene copy of SREB and its endogenous promoter. Complemented strains A5 and D5 grew as white colonies that did not discolor the medium, suppressed siderophore production under iron-replete conditions (10 µM FeSO4), and converted fully to mycelia when grown at a temperature of 22°C (Figure 2A-C). Retransformation of 3-15-1 with a vector lacking SREB did not complement the mutant phenotype (empty vector strain) (Figure 2A-C). Whereas Northern analysis demonstrated a reduction in the abundance of SREB transcript in mutant 3-15-1 compared to the parental strain, message levels were overexpressed in both complemented strains (Figure 2D). Thus, complementation reversed the mutant's phenotypic defects, supporting the idea that the insert was responsible for the dysregulation of siderophore biosynthesis and the alteration in morphogenesis.
Disruption of SREB
To confirm that SREB represses the biosynthesis of siderophores and affects morphogenesis in B. dermatitidis, we disrupted this gene in wild-type isolate 26199 using homologous recombination. To minimize the probability that the phenotype observed in mutant 3-15-1 was unique to strain T53-19, we used a different B. dermatitidis strain, 26199, to generate a null mutant. The rate of allelic replacement was 0.04% (1/2670). The null mutant, SREBΔ, grew as yellow-pigmented colonies that discolored the surrounding medium and failed to properly repress siderophore biosynthesis when iron was abundant (Figure 3A, 5B). The intensity of pigmentation was dependant on exogenous iron and independent of temperature (37°C vs. 22°C) (data not shown). In contrast, the parent strain grew as white-colored yeast and repressed the production of siderophores under iron-replete conditions as measured by the ferric perchlorate assay (Figure 3A, 5B). SREBΔ failed to complete the yeast-to-mold phase transition following a shift in temperature from 37°C to 22°C, did not exhibit radial growth, and accumulated little biomass at 22°C (Figure 3A, 3B). The defect in phase transition persisted during prolonged incubation (>14 days) at 22°C; however, a few hyphal strands would develop and could only be observed by light microscopy. Similar to insertional mutant 3-15-1, SREBΔ remained viable at 22°C (as measured by 0.2% eosin exclusion) and converted back to yeast following a shift in temperature from 22°C to 37°C (data not shown). In the yeast form, the SREBΔ mutant grew at the same rate as the parent strain (Figure 3C). The morphologic defect at 22°C was independent of exogenous iron concentrations (data not shown).
Fig. 3. Phenotype of the SREB null mutant. 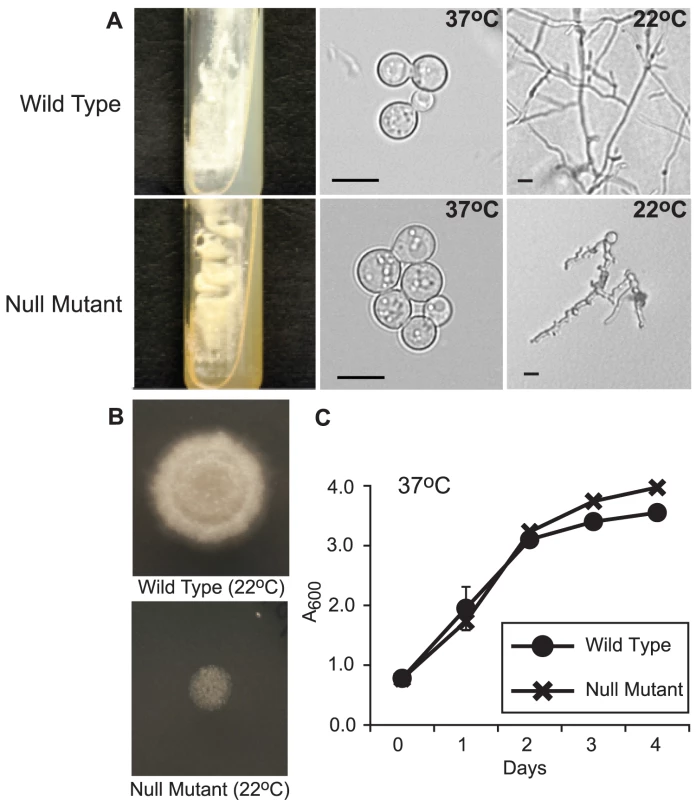
(A) When grown on 7H10 medium containing 150 µM FeSO4, the null mutant (SREBΔ) grew as yellow-orange pigmented colonies that discolored the medium (37°C; 15 days incubation). In contrast the wild-type strain (ATCC 26199) grew as white colored yeast and did not pigment the medium. At 37°C, SREBΔ and wild-type isolates grew as budding yeast. Following a shift in temperature from 37°C to 22°C, SREBΔ failed to complete the conversion from yeast to mycelia (17 days; HMM medium). (B) The null mutant (SREBΔ) does not accumulate significant biomass or expand by radial growth when compared to the wild-type isolate. For each strain, 2.5×104 yeast were spotted on HMM medium and incubated at 22°C for 14 days. (C) The null mutant (SREBΔ) and wild-type isolates have a similar growth rate when they are cultured as yeast at 37°C incubation. Culture density was measured in triplicate at A600. Cultures were grown in liquid HMM supplemented with 10 uM FeSO4. The data were from two independent experiments. Analysis of the null mutant by PCR indicated disruption of SREB and the absence of any deletion or rearrangment of the genomic DNA flanking the transgene (data not shown). Southern blot analyses demonstrated replacement of SREB with a hygromycin resistance cassette and the absence of additional deletions in the genomic DNA flanking the transgene in SREBΔ (Figure 4A-E). Northern analysis demonstrated the loss of SREB transcript in SREBΔ (Figure 4F).
Fig. 4. Southern and Northern blot analyses of the SREB null mutant. 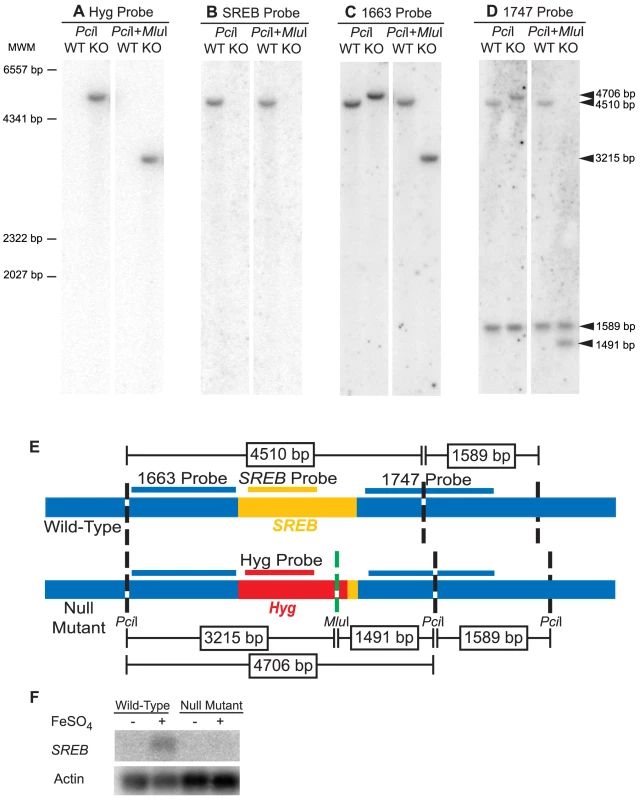
(A-D) Southern analysis of 26199 wild-type (WT) and null mutant SREBΔ (KO). Genomic DNA from WT and KO were digested with PciI alone or in combination with MluI. The hygromycin resistance cassette, but not SREB, contains a MluI restriction site. The blots were probed against hygromycin, Hyg probe (A); SREB, SREB probe (B); the 5′ flank, 1663 probe (C); and 3′ flank, 1747 probe (D). Arrows indicate hybridizing fragments and dashes depict molecular weight markers (MWM). The Hyg and SREB probes failed to hybridize to digested WT or KO DNA, respectively (A, B). The 1663 and 1747 probes, which flank SREB, gave the expected size restriction fragments, which indicated a clean replacement of SREB sequence with the hygromycin resistance cassette (C, D). (E) Schematic illustrating the location of the restriction sites in the wild-type and null mutant (SREBΔ), hybridization sites for probes Hyg, SREB, 1663, and 1747, and expected size of the restriction fragments. Yellow, red, and blue indicate the SREB coding region, hygromycin resistance cassette, and sequence flanking SREB, respectively. (F) Northern analysis of 26199 wild-type and SREB-null mutant when grown under iron-poor (−) and iron-replete (+; 10 µM FeSO4) conditions. SREB transcript is detectable in the wild-type isolate under iron-replete conditions and absent in SREBΔ. Complementation of SREB
To confirm the phenotype in SREBΔ was due to disruption of the siderophore biosynthesis repressor gene, we re-transformed the null mutant using A. tumefaciens to insert a copy of SREB. Complemented strains grew as white-colored colonies and properly suppressed the biosynthesis of siderophores when iron was abundant (Figure 5A, 5B). Following a temperature shift from 37°C to 22°C, complemented yeast strains converted to mold (Figure 5C). This conversion was slower in the complemented strains (14–17 days) when compared to the wild-type isolate (<7 days) (data not shown). The complemented strains underwent radial growth at 22°C; however, colony expansion was less than the wild-type isolate (data not shown). Prolonged incubation did not result in catch-up growth. Analysis of transcript abundance demonstrated restoration of message levels in C#25 and overexpression in C#6 when compared to wild-type and SREBΔ strains (Figure 5D).
Fig. 5. Complementation of the SREB null mutant. 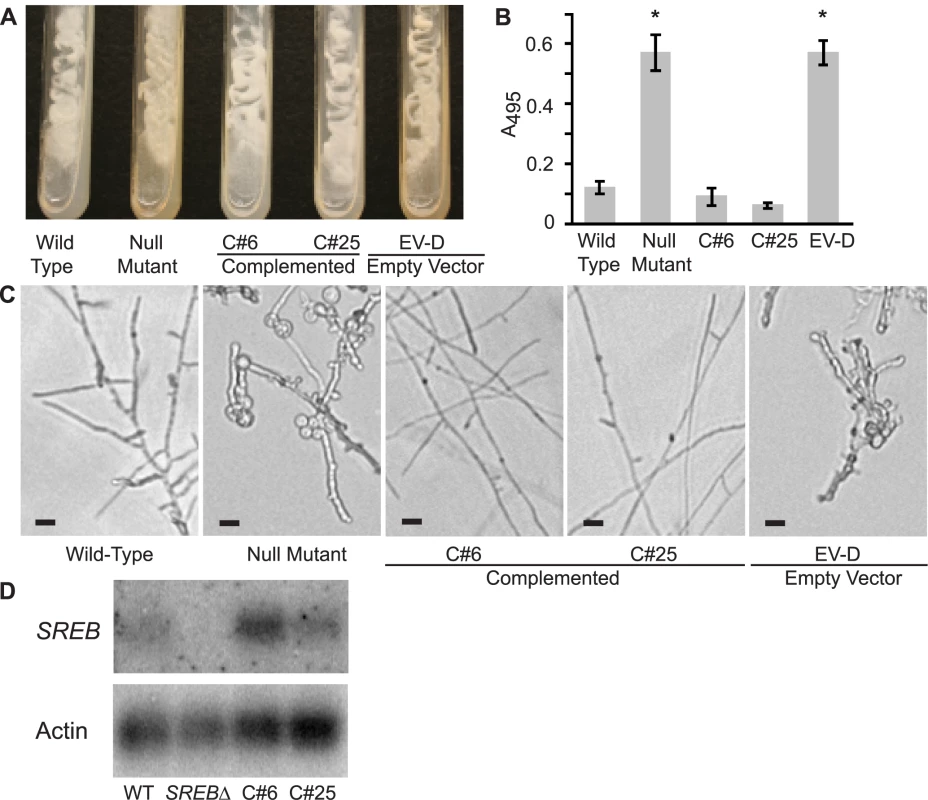
(A) Complemented strains C#6 and C#25 grew as white-colored yeast, similar to the wild-type isolate, on 7H10 slants (150 µM FeSO4). In contrast, the null mutant (SREBΔ) and empty vector strain EV-D grew as yellow-colored yeast and discolored the medium. Cultures were grown at 37°C for 14 days. (B) Complemented strains C#6 and C#25 were able to repress the biosynthesis of siderophores when grown-under iron-replete (10 µM FeSO4) conditions as measured by the ferric perchlorate assay. In contrast, the null mutant and empty vector (EV-D) continued to produce siderophores (p<0.01). Data from three independent experiments were analyzed. (C) Wild-type and complemented strains C#6 and C#25 convert to mycelia within 17 days of incubation at 22°C. In contrast, the null mutant and empty vector EV-D strains fail to complete the conversion to mycelia. (D) Northern blot hybridization demonstrated restoration of SREB transcript abundance in complemented strains C#6 and C#25, when compared to the null mutant. Expression of SREB and regulation of siderophore biosynthesis
To test if the expression of SREB was influenced by the concentration of exogeneous iron, we grew wild-type B. dermatitidis strain 26199 under iron-poor and –replete conditions. Northern blot analysis demonstrated that the expression of SREB was increased during conditions of iron abundance and repressed when iron was limited (Figure 4F).
In fungi, the expression of genes that encode proteins involved with iron assimilation are often co-expressed or -repressed when iron is limited or abundant, respectively. To investigate whether this was also true in B. dermatitidis, we analyzed the expression of several genes in response to exogenous iron. Under iron-poor conditions, B. dermatitidis wild-type strain 26199 induced the expression of genes involved in the biosynthesis of siderophores (SIDA), transport of ornithine from the mitochondria into the cytosol (AMCA), uptake of siderophores (MIRB, MIRC), and a bZIP transcription factor (HAPX) (Figure 6). Conversely, these genes were repressed when iron was abundant (Figure 6). The disruption of SREB de-repressed the expression of each of these genes. Thus, SREB regulates genes involved in siderophore biosynthesis and uptake in B. dermatitidis (Figure 6).
Fig. 6. Northern analysis of candidate genes in the SREB regulon involved with siderophore biosynthesis and uptake. 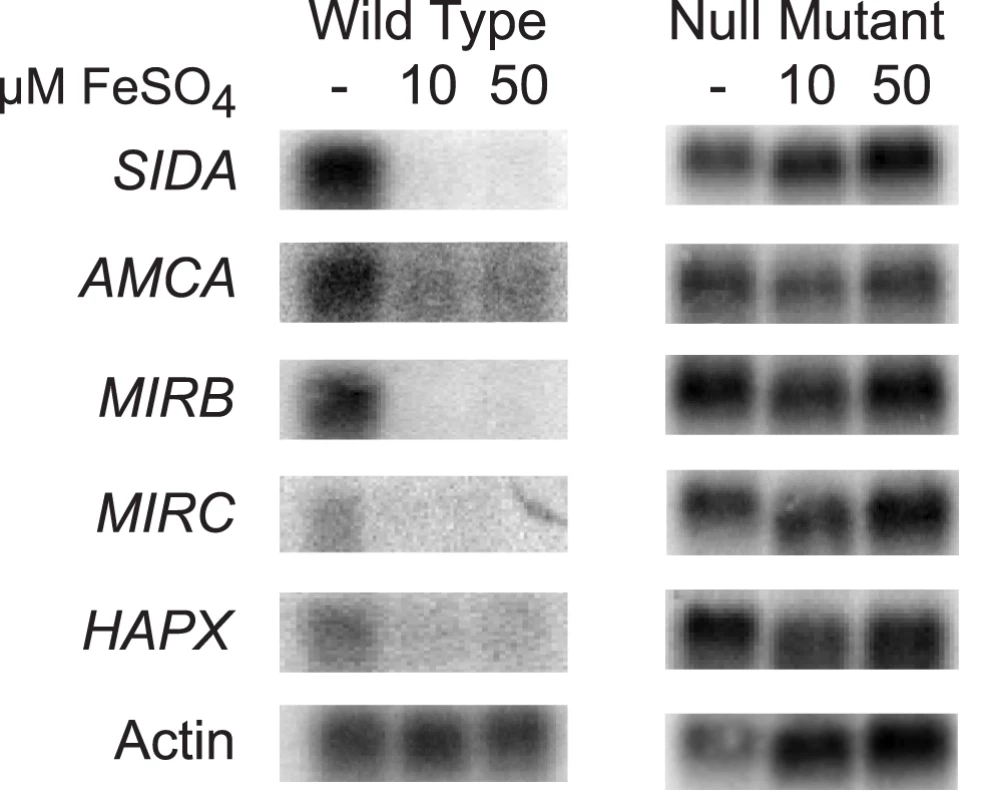
Wild-type B. dermatitidis strain 26199 upregulated the expression of SIDA, AMCA, MIRB, MIRC, and HAPX when grown in iron-poor medium (—); expression of these genes was repressed when iron was abundant (10 and 50 µM FeSO4). Deletion of SREB resulted in de-repression of SIDA, AMCA, MIRB, MIRC, and HAPX under iron-replete conditions. Identification and characterization of B. dermatitidis siderophores
To further characterize the regulatory role of SREB on siderophore biosynthesis, we used LC/MS and reverse-phase HPLC to identify the specific type(s) of siderophores secreted by B. dermatitidis wild-type and null mutant yeast cells. Starting with wild-type cells grown under iron-limited conditions, siderophores from culture supernatant were isolated using column chromatography. Mass spectroscopy of the eluate showed two large peaks at 4.16 and 7.26 minutes with molecular weights of 538.2 and 822.2 that correspond to dimerum acid and coprogen, respectively (Figure 7A-C). Reverse-phase HPLC of the eluate and comparison of retention times to siderophore standards confirmed the identities of these siderophores (Figure 7D). Under iron-replete conditions, wild-type B. dermatitidis repressed the biosynthesis of dimerum acid and coprogen (Figure 7D). In contrast, the null mutant continued to produce both siderophores (Figure 7D).
Fig. 7. Identification and characterization of siderophores in B. dermatitidis. 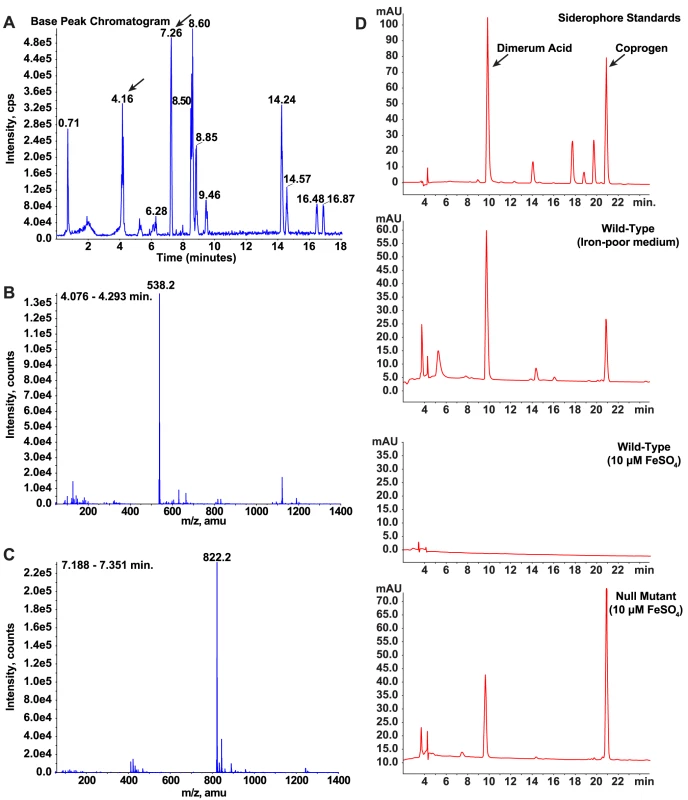
(A-C) Culture supernatant harvested from wild-type B. dermatitidis grown under iron-poor conditions was applied to a column packed with XAD-2 resin. Following a water wash, compounds bound to the resin were eluted with methanol and analyzed by LC/MS. The chromatogram revealed the presence of several compounds in the eluate (A). Analysis of these compounds using mass spectroscopy revealed that two of the peaks (4.16 and 7.26 minutes) had masses consistent with dimerum acid (538.2) and coprogen (822.2), respectively (B, C). (D) Culture supernatants harvested from wild-type B. dermatitidis grown under iron-poor and iron-replete conditions were compared to siderophore standards using HPLC. During conditions of iron-limitation, the wild-type isolate produced and secreted dimerum acid and coprogen. When iron was abundant, no siderophores were detected. In contrast, the null mutant SREBΔ produced and secreted dimerum acid and coprogen under iron-replete conditions. Identification of candidate genes that promote the phase transition
To identify candidate genes regulated by SREB that may promote the phase transition, we first used MAST analysis to search the Blastomyces genome for GATA transcription factor-binding motifs in intergenic regions located ≤2000 bp upstream of predicted genes. Our initial search for the classic GATA transcription factor-binding motif, HGATAR, revealed the presence of this motif upstream of nearly all B. dermatitidis genes. This finding is similar to Schrettl et al., who found widespread distribution of this motif in Aspergillus fumigatus [26].
An extended version of the HGATAR motif, ATC-w-gAta-a, has been recently described and was demonstrated to occur at a 5.4-fold higher frequency in the promoter of genes regulated by A. fumigatus SREA, an SREB homolog, when compared to the entire A. fumigatus genome [26]. We revised our strategy and searched for this extended motif in the promoter of genes in the B. dermatitidis genome. We identified a total of 1,213 genes with at least one of the following motifs located ≤2 kb upstream of the start codon: ATC-(A/T)-GATA-(A/G), ATC-(A/T)-GATA-(T/C), ATC-(A/T)-GATT-A, ATC-(A/T)-GATC-A, ATC-A-GATG-A, ATC-C-GATA-A, and ATC-A-AATA-A. This gene-set included genes involved in siderophore biosynthesis and uptake (i.e. SIDA, MIRB, AMCA). Two or more upstream GATA motifs were present in 232 (19.1%) in the gene-set. Hwang and colleagues identified the motif (G/A)-ATC-(A/T)-GATA-A upstream of siderophore biosynthesis and transport genes regulated by SID1 in H. capsulatum [27]. We found this longer motif upstream of 271 (22.3%) of our 1,213 MAST-identified genes; however, MIRB and MIRC, both involved in siderophore uptake, lacked the motif.
To classify the 1,213 candidate genes into functional categories and facilitate further analysis, we annotated the predicted protein products of these genes as well as the complete B. dermatitidis predicted proteome against the eukaryotic orthologous groups (KOG) database. The results, shown in Table 1, indicate that the KOG-annotated GATA-containing genes fall into many categories of gene function (i.e. transcription, RNA metabolism, signal transduction, cell remodeling and metabolism). The frequency of KOG-annotated genes with upstream GATA motifs within a particular KOG category was compared to the frequency of genes in the same KOG category within all KOG-annotated genes in the B. dermatitidis genome. Three KOG categories were significantly over-represented in the candidate gene-set harboring GATA sites: amino acid transport and metabolism (KOG code E), secondary metabolites biosynthesis, transport and catabolism (KOG code Q), and lipid transport and metabolism (KOG code I) (Table 1 and Table S1). This suggests that these cellular process pathways may be important for SREB regulation, although it does not exclude a role for the GATA-containing genes in other KOG groupings.
Tab. 1. Enrichment of GATA-containing genes in B. dermatitidis according to KOG category*. 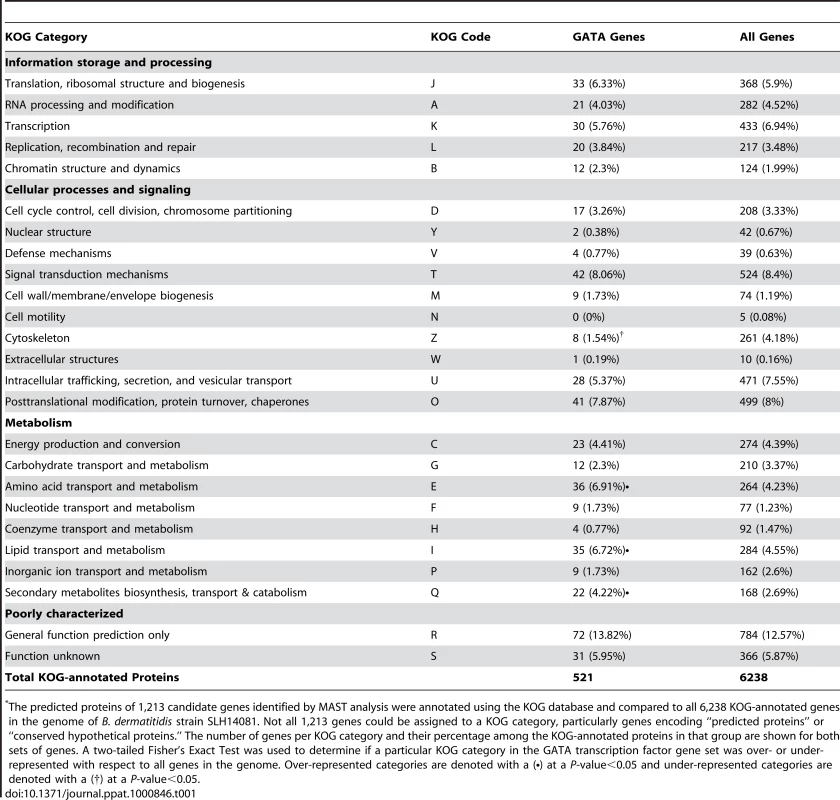
The predicted proteins of 1,213 candidate genes identified by MAST analysis were annotated using the KOG database and compared to all 6,238 KOG-annotated genes in the genome of B. dermatitidis strain SLH14081. Not all 1,213 genes could be assigned to a KOG category, particularly genes encoding “predicted proteins” or “conserved hypothetical proteins.” The number of genes per KOG category and their percentage among the KOG-annotated proteins in that group are shown for both sets of genes. A two-tailed Fisher's Exact Test was used to determine if a particular KOG category in the GATA transcription factor gene set was over- or under-represented with respect to all genes in the genome. Over-represented categories are denoted with a (•) at a P-value<0.05 and under-represented categories are denoted with a (†) at a P-value<0.05. In a complimentary approach to identify genes that may be regulated by SREB, we performed a preliminary microarray analysis. Using an expression array with 70-mer oligonucleotides representing the 10,567 open reading frames of B. dermatitidis strain 26199, we used two-color spotted analysis to compare isogenic wild-type vs. SREBΔ at 37°C and at 22°C 48 hours after the temperature shift downward (data not shown). At least 38 of the genes identified by MAST analysis were differentially expressed (increased or decreased by ≥2-fold), including seven genes classified by KOG to be involved in lipid transport and metabolism. To validate the microarray results, we performed quantitative RT-PCR on a subset of four genes found to be altered in expression; three from the lipid transport and metabolism KOG category, and one from the carbohydrate metabolism category. At 22°C, the null mutant strain failed to upregulate the expression of a lipid transfer protein and acetoacetyl-CoA synthase (Figure 8). Conversely, the expression of a peroxisomal dehydratase was over-expressed at 37°C and 22°C, when compared to the wild-type isolate (Figure 8). We also confirmed the altered expression of a glycosyl hydrolase postulated to be involved in cell-wall remodeling. In the null mutant, this gene is over-expressed at 37°C and 22°C, when compared to the wild-type isolate (Figure 8). Thus, we have begun to identify candidate genes and processes that may be direct or indirect targets of SREB and contribute to the phase transition from yeast to mold.
Fig. 8. Expression of candidate genes in the SREB regulon at 37°C and 22°C. 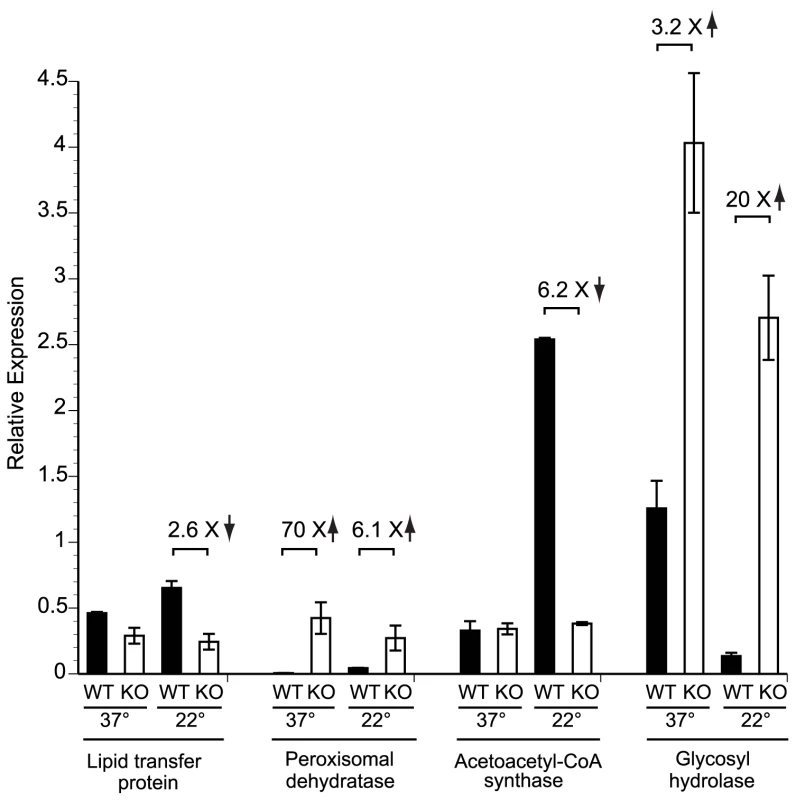
Quantitative RT-PCR (qRT-PCR) was performed to validate the expression of a subset of genes identified by bioinformatic and gene expression analyses. RNA was extracted from the wild-type (WT) strain and null mutant (KO) at 37°C and at 22°C (48 hours following the downward temperature shift). Deletion of SREB resulted in altered expression of genes involved with lipid metabolism and transport (lipid transfer protein, peroxisomal dehydratase, acetoacetyl-CoA synthase) as well as a glycosyl hydrolase that is predicted to have endo-1,3-β-glucanase activity. Data was generated from two biological replicates. qRT-PCR fluorescence was normalized to α-tubulin. Discussion
The use of A. tumefaciens-mediated DNA transfer for insertional mutagenesis has advanced our understanding of the endemic dimorphic fungi at the molecular level [10],[12],[13],[28]. We used this technology to mutate B. dermatitidis conidia and screen for transformants with altered morphology during growth at 22°C and 37°C. Analysis of mutant 3-15-1 uncovered a GATA transcription factor, SREB, which regulates siderophore biosynthesis and affects morphology in B. dermatitidis. GATA transcription factors are zinc-finger proteins that bind conserved motifs to induce or repress gene expression [16],[22],[26],[27],[29]. These genes are found widely in eukaryotes, but they function differently in fungi, plants, and animals [30],[31]. In fungi, GATA transcription factors regulate diverse functions including the response to blue light, switching of mating-type, uptake of nitrogen, pseudohyphal growth during nitrogen starvation, biosynthesis of siderophores, and iron assimilation [17],[22],[29],[32],[33].
Our analysis indicates that SREB has pleotropic effects in B. dermatitidis - it promotes the transition from yeast to mold at environmental temperature and represses the biosynthesis of siderophores. Following a shift of incubation temperature from 37°C to 22°C, the insertional and null mutants were unable to complete the phase transition or accumulate significant biomass when compared to the parent strain. To our knowledge, B. dermatitidis SREB is the first gene identified in the dimorphic fungi that promotes the conversion of yeast to mold. Much of the field's attention has been focused on genes that regulate the phase transition from mold to yeast; only a few genes have been identified that regulate growth or morphology in the dimorphic fungi at environmental temperature (i.e. 22–25°C). In H. capsulatum, the mold-specific gene MS8 regulates mycelial morphology and growth, but not the phase transition [34]. In P. marneffei TupA is required for maintenance of mycelial morphology at 25°C; null mutants convert to mycelia following a temperature shift from 37°C to 25°C, but revert to yeast morphology with prolonged incubation [35].
We hypothesize that B. dermatitidis SREB binds DNA to regulate many genes that, in turn, control such disparate functions as phase transition and the response to abiotic stress, including iron availability. Using MAST analysis we identified a large number of genes with putative GATA transcription factor binding sites. When compared to the entire B. dermatitidis genome, candidate genes involved with the biosynthesis of secondary metabolites as well as amino acid and lipid metabolism were found to be over-represented. Some of these candidate genes were indeed altered in expression in SREBΔ, as detected in preliminary microarray analysis and validated by RT-PCR. The enrichment of genes involved in secondary metabolism and amino acid metabolism were not unexpected, in part, because SREB regulates siderophore biosynthesis, a process that requires the transport and metabolism of amino acids. The abundance of genes containing GATA binding sites involved in lipid transport and metabolism was surprising. To our knowledge, regulation of lipid metabolism and transport in fungi by GATA transcription factors has not been described.
Changes in fatty acid metabolism in the dimorphic fungi are associated with the phase transition and are postulated to impact morphogenesis [36]–[42]. Exposure of H. capsulatum mycelia to unsaturated fatty acids prolongs the mold-to-yeast conversion following a shift in temperature from 25 to 37°C [36]. In contrast, treatment with saturated fatty acids accelerates the phase transition [36]. In C. immitis, exposure to exogenous fatty acids alters the conversion of spherules to mycelia [37]. Reduced expression of the Δ9-desaturase gene, OLE1, in C. albicans, impairs hyphal formation [38]. Differences in the concentration of unsaturated fatty acids (oleic and linoleic acids) and unsaturated sphingolipids (N-2′-hydroxy-(E)-Δ3-octadecenoate) have been described in the yeast and mold forms of H. capsulatum and P. brasiliensis [39]–[42]. In P. brasiliensis, several genes involved in lipid metabolism, have been demonstrated to be phase-regulated [43]. Thus, further investigation of genes involved in fatty acid metabolism may clarify the mechanism by which SREB promotes the phase transition from yeast to mold.
B. dermatitidis insertional and null mutants have multiple alterations in the regulation of iron assimilation, as indicated by their yellow-orange appearance, constitutive production of siderophores, and derepression of iron-regulated genes during conditions of iron abundance.
Iron acquisition must be tightly regulated for proper cellular function and to avoid toxicity due to iron overload [17],[44]. Under iron-replete conditions, SREB represses genes involved in the production (SIDA, AMCA) and uptake (MIRB, MIRC) of siderophores. AMCA encodes a transferase that shuttles ornithine from the mitochondria to the cytosol [44]. The first step in siderophore biosynthesis involves the conversion of ornithine into N5-hydroxy-L-ornithine, which is catalyzed by an L-ornithine-N5-monooxygenase encoded by SIDA [45]. Siderophores secreted into the environment bind iron and then can be taken up by the cell through permeases such as MIRB and MIRC [46]. Analysis of the B. dermatitidis genome did not reveal an ortholog to A. nidulans MIRA, which facilitates the uptake of xenosiderophores, specifically enterobactin [46]. Deletion of SREB resulted in de-repression of SIDA, AMCA, MIRB, and MIRC expression under iron-replete conditions. Similar to P. chrysogenum SREP, N. crassa SRE, and A. nidulans SREA null mutants, disruption of SREB in B. dermatitidis resulted in discoloration of the fungus [14],[18],[45]. In addition, we identified two extracellular siderophores, dimerum acid and coprogen, produced by B. dermatitidis when grown under iron-poor conditions. When iron is abundant, SREB represses the biosynthesis of both these siderophores.
Similar to A. nidulans and H. capsulatum, the expression of B. dermatitidis SREB is upregulated when iron is abundant, and repressed when iron is limited [16],[17]. Repressors of siderophore biosynthesis are not uniformly regulated at the transcriptional level in other fungi, as orthologs of SREB including SRE, URBS1, FEP1, and SFU1 are constitutively expressed regardless of exogenous iron concentrations [18]–[21]. SREB is expressed as a single transcript, similar to SRE and URBS1 [18],[19]. In contrast, SREP, SREA, and FEP1 are expressed as two separate transcripts due to the presence of two transcriptional start sites [14],[17],[20].
B. dermatitidis SREB may participate in a regulatory circuit with the bZIP (basic leucine zipper) transcription factor, HAPX. Computational analysis of the promoter region of HAPX in B. dermatitidis revealed putative GATA binding sites. Moreover, iron-poor conditions induced HAPX expression in wild-type B. dermatitidis, whereas iron abundance reduced its expression. In A. nidulans, HAPX represses SREA as well as genes that encode iron-dependent proteins such as CYCA (cytochrome C), ACOA (aconitase), LYSF (homoaconitase) when iron availability is limited [44]. We found that deletion of SREB resulted in the expression of HAPX under iron-poor and iron-replete conditions.
Our findings support the idea that B. dermatitidis SREB functions as a transcription factor that regulates the biosynthesis of siderophores and promotes the conversion from yeast to mold. We propose that SREB inhibits genes involved with the biosynthesis and uptake of siderophores under conditions of iron abundance. Our findings also suggest that SREB affects phase transition independently of iron assimilation, perhaps, by altering the expression of genes involved with lipid metabolism or cell wall remodeling. The iron-related defects do not explain the failure to convert from yeast to mold since growth under iron-poor conditions had no effect on the defect in morphogenesis. GATA transcription factors in other fungi have been demonstrated to regulate morphogenesis as well as the response to temperature. S. cerevisiae ASH1 encodes a GATA transcription factor that inhibits mating-type switching and induces filamentous growth under conditions of nitrogen limitation [29]. C. neoformans CIR1, an ortholog of B. dermatitidis SREB, regulates genes involved in reductive iron assimilation and siderophore transport, but also genes critical for virulence including those required for thermotolerance, capsule production, and melanin biosynthesis [22].
In summary, we identified and characterized a GATA transcription factor that represses the biosynthesis of siderophores and promotes the phase transition from yeast to mold. To our knowledge, B. dermatitidis SREB is the first gene identified in dimorphic fungi that promotes the conversion of yeast to mycelia. By using bioinformatic and expression analyses we identified several genes whose expression may be directly or indirectly regulated by SREB. We investigated a sample of these genes, including ones in KOG categories for lipid and carbohydrate metabolism, and found that their expression is affected by the deletion of SREB. Future work will strive for a more complete description of how SREB promotes the yeast to mold phase transition. Because growth in the mold form is thought to be essential for the survival of dimorphic fungi in nature and the generation of infectious particles, SREB may be needed for the evolutionary maintenance of this species. The generation of an SREB null mutant provides a unique opportunity to elucidate the SREB regulon and identify genes that govern growth in the mold form, as well as other traits in this human fungal pathogen.
Materials and Methods
Strains and growth conditions
Blastomyces dermatitidis strains used in this study included T53-19 and American Type Culture Collection (ATCC) 26199. T53-19 sporulates, but is weakly virulent in a murine model of infection, and ATCC strain 26199 is highly virulent, but does not sporulate [10],[47]. The genome of strain 26199 has been sequenced by the Genome Sequencing Center at Washington University (http://genome.wustl.edu). B. dermatitidis yeast and mold were grown on Histoplasma macrophage medium (HMM), 3M medium (3M), Potato dextrose agar (PDA), or Middlebrook 7H10 agar medium containing oleic acid-albumin complex (7H10; Becton Dickinson and Company, Franklin Lakes, NJ) [48]–[50]. Agrobacterium tumefaciens strain LBA1100 harboring the Ti helper plasmid pAL1100 (gift from C. van den Hondel; Leiden University, The Netherlands) was maintained on Luria-Bertani (LB) medium supplemented with 0.1% glucose, spectinomycin 100 µg/ml, and kanamycin 100 µg/ml once transformed with a binary vector [28].
Insertional mutagenesis
Conidia from B. dermatitidis strain T53-19 were mutagenized using A. tumefaciens containing pBTS165 [10],[28],[51]. This binary vector contains a resistance cassette, hygromycin phosphotransferase (hph), integrated into the T-DNA that is driven by a glyceraldehyde-3-phosphate dehydrogenase (gpdA) promoter derived from Aspergillus nidulans [10]. Conidia harvested from mycelial cultures by manual disruption were counted using a hemocytometer, suspended in phosphate buffered saline (PBS) to a final concentration of 2×107/ml, and co-cultivated with A. tumefaciens (6×108 cells/ml) on a Biodyne A nylon membrane (Pall Gelman, Ann Arbor, MI) on induction medium containing 200 µM acetosyringone (IMAS medium) [28]. After 72 hours of incubation at 22°C, the biodyne membranes were transferred to 3M medium supplemented with hygromycin 100 µg/ml (AG Scientific Inc., San Diego, CA) and cefotaxime 200 µM (Sigma-Aldrich), and incubated at 37°C or 22°C. Individual transformants were visually screened by light microscopy for altered morphology: growth as hyphae or pseudohyphae at 37°C or yeast at 22°C. Replica plates were used to identify transformants that lost viability upon shifting the incubation temperature from 22°C to 37°C.
Adaptor PCR
Adaptor PCR was used to amplify DNA flanking the pBTS165 insert from insertional mutant 3-15-1 [52]. Following the digestion of genomic DNA by restriction enzymes StuI, HpaI, and XmnI, which do not cut in pBTS165, adaptors were ligated to the restriction fragments using T4 DNA ligase (New England Biolabs, Ipswich, MA). PCR was performed using primers specific for the adaptors and pBTS165. The PCR products were separated by agarose gel electrophoresis and purified using the QIAquick gel extraction kit (Qiagen, Valencia, CA) and sequenced by the DNA Sequencing Laboratory at the University of Wisconsin Biotechnology Center. Sequence flanking the insert was analyzed using GSC (Genome Sequencing Center) BLAST (http://genome.wustl.edu/tools/blast) and National Center for Biotechnology Information (NCBI) tBLASTx (http://blast.ncbi.nlm.nih.gov/Blast.cgi). FGENESH was used to identify predicted exons and introns in the SREB gene (www.softberry.com).
Generation of null mutants
Two vectors, pBTS4-KO1 and pBTS4-KO2, were used to delete SREB in B. dermatitidis strain 26199 by homologous recombination and resulted in two null mutants, T1#23 and T12#16, respectively. Although both null mutants had similar phenotypes, T1#23 contained an additional 2,214 bp deletion in the 5′ untranslated region that was upstream of the disrupted SREB gene. Herein, T12#16, which has no additional deletions, is referred to as SREBΔ. Plasmid pBTS4-KO2 contained 1611 bp of 5′ upstream sequence and 1747 bp of coding and 3′ downstream sequence flanking hph. The 1611 bp and 1747 bp products were amplified from B. dermatitidis 26199 genomic DNA using F and R primers containing SacI, BbsI, SbfI, or ClaI restriction sites (F-1611-SacI 5′-TTTGAGCTCACTTTACTCTTCGGACGGGTTTT; R-1611-BbsI 5′-TTTTCGATTGTCTTCAGCCAAAAGCCCCGTCATTCCTGT; F-1747-SbfI 5′-TT-TCCTGCAGGTTGCAGCGTGAGGCGGAAGA; R-1747-ClaI 5′-TTTATCGATTGACAGGGCAG-GCTACATA). PCR products were separated by agarose gel electrophoresis, purified using QIAquick PCR purification kit (Qiagen, Valencia, CA), sequenced, and ligated into pBTS4 in sequential fashion following restriction digest to flank the hph-resistance cassette [53]. After sequence and restriction digest analyses confirmed integration of the ligated PCR fragments, pBTS4-KO2 was electroporated into A. tumefaciens strain LBA1100 [28]. B. dermatitidis strain 26199 (2×107 yeast/ml) was transformed with A. tumefaciens containing pBTS4-KO2 (6×108 bacteria/ml) on Biodyne A membranes on IMAS medium. After 72 hours of incubation at 22°C, the Biodyne membranes were transferred to HMM medium supplemented with 10–20 µM FeSO4, hygromycin 25 µg/ml, cefotaxime 200 µM, and incubated at 37°C. Transformants were visually screened for yellow pigmentation. The null mutant was cloned to obtain individual colonies and establish a line of cells. SREB gene deletion was confirmed by PCR, and Southern and Northern blot analyses (see below).
Complementation of insertional and null mutants
Insertional mutant 3-15-1 was re-transformed with pBTS47-11+13 using A. tumefaciens-mediated DNA transfer. This plasmid contained the SREB coding region, 1990 bp of 5′ sequence upstream of the start codon, 603 bp of 3′ sequence downstream of the stop codon, and a nourseothricin resistant cassette. Genomic DNA was amplified using primers ggp11-XbaI (5′-TTTCTAGAACAACTACCTCTACATGACACT-GC) and ggp13-SbfI (5′-TTTCCTGCAGGGAGCCTTTTCTTTCTGTCAA). The PCR products were separated by agarose gel electrophoresis, purified using QIAquick PCR gel extraction kit (Qiagen, Valencia, CA), sequenced, and ligated into pBTS47 to generate pBTS47-11+13. The null mutant, SREBΔ, was re-transformed by A. tumefaciens with pBTS47-5331, which contains the SREB coding region, 2655 bp of 5′ sequence upstream of the start codon, 603 bp of 3′ sequence downstream of the stop codon, and a nourseothricin resistant cassette. The protocol for A. tumefaciens-mediated DNA transfer was similar to that described in the previous section. Transformants were screened for white colony pigmentation on HMM medium supplemented with 20 µM FeSO4, nourseothricin 25 ug/ml (Werner Bioagents, Germany), and cefotaxime 200 µM at 37°C incubation.
DNA extraction & Southern blot hybridization
B. dermatitidis was grown to late log phase in liquid HMM at 37°C incubation. Genomic DNA was extracted using the method described by Hogan and Klein [54]. Southern blot hybridization was performed as described [28],[55]. The fate of the transforming DNA in the insertional mutant was determined using probes specific for T-DNA and non-T-DNA sequences. An 822 bp amplicon constructed using primers 5′-CGATG-TAGGAGGGCGTGGATA and 5′-GCTTCTGCGGGCGATTTGTGT was used to probe hph within the T-DNA. An 8 kb BglII restriction fragment generated from pBTS4 was used to probe the non-T-DNA sequence. Deletion of SREB in the null mutant was analyzed using PCR-generated probes specific for the SREB coding region (1303 bp; 5′-CCCGCTCTTTGCTTAACC-CGTATG and 5′-CTGGTGATAAAGAAGGGCTGAA), hph (822 bp; 5′-CGATGTAGGAGGGCG-TGGATA and 5′-GCTTCTGCGGGCGATTTGTGT), 5′ region flanking SREB (1663 bp; 5′-ACTT-TACTCTTCGGACGGGTTTTC and TATCTGCGCTTTTGGTAGTAGGAG), and the 3′ region flanking SREB (1747 bp; 5′-TTGCAGCGTGAGGCGGAAGA and 5′-ACAAATCGTAGCACCAG-TC). All probes were radiolabeled with α-32P dCTP using a Prime-a-Gene labeling system (Promega, Madison, WI). Unincorporated radionucleotides were removed using ProbeQuant G50-micro columns (GE Healthcare, Buckinghamshire, UK). Following hybridization, the blot was washed sequentially with low stringency (0.25 M NaPO4, 2% SDS, 1 mM EDTA) and high stringency (0.04 M NaPO4, 1% SDS, 1 mM EDTA) solutions, exposed to a storage phosphor screen (Molecular Dynamics, Sunnyvale, CA) and scanned using a Storm 660 imaging system (Molecular Dynamics, Sunnyvale, CA).
Ferric perchlorate assay
Ferric perchlorate was used to measure siderophore production semi-quantitatively [25]. B. dermatitidis was grown at 37°C in liquid 3M or HMM under iron-poor or replete (10 µM FeSO4) conditions. Iron-poor media consisted of HMM or 3M prepared with F-12 Ham's nutrient mixture lacking FeSO4, or trace elements lacking FeSO4, respectively. In addition, exogenous iron was not added to these media. As the yeast entered stationary growth (A600 = 3.5−4.0), culture supernatants were collected, filtered (0.2 µM), and added to a ferric perchlorate solution (5 mM Fe(ClO4)3 in 0.1 N HCl). Absorbance was measured at 425 or 495 nm. Plasticware was used whenever possible. Glassware was treated with 2N HCl to remove residual traces of iron [56]. Analysis of variance (ANOVA) was used to analyze the results from the ferric perchlorate assay. Tukey's Honest Significant Difference method was used to adjust the p-values for multiple comparisons.
RNA extraction & Northern blot hybridization
B. dermatitidis was grown to mid-log phase at 37°C in liquid HMM with no added iron (iron-poor medium), 10 µM FeSO4, or 50 µM FeSO4. Total RNA was extracted using the phenol-guanidinium thiocyanate-1-bromo-3-chloropropane extraction method [55]. In brief, yeast were washed with PBS, beaten with beads, and treated with TRI Reagent followed by 1-bromo-3-chloropropane (Molecular Research Center Inc., Cincinnati, OH). RNA was precipitated using a 1∶1 concentration of isopropanol and a high salt solution (Molecular Research Center Inc., Cincinnati, OH), washed with 75% ethanol, and resuspended in water that was pre-treated with diethyl pyrocarbonate (DEPC; Calbiochem, San Diego, CA). Total RNA was further purified using RNeasy kit (Qiagen, Valencia, CA) and enriched for mRNA using oligo(dT)-polystrene chromatography (Sigma-Aldrich). Northern hybridization was performed as described using 2.0-2.3 µg poly(A)+-enriched mRNA per sample [55]. Gene expression was analyzed using probes constructed by PCR against SREB (SreF 5′-CCCGCTCTTTGCTTAACCCGTATG; SreR 5′-CTGGTGATAAAGAAGGGCTGAA) SIDA (SidA-F1 5′-AGACAGTACTCAAGAACGACAA; SidA-R1 5′-GCTGTCATCGCTGGGCTTTAGTGC), MIRB (MirB-F 5′-CTCCTCCTCGTCGCTTTCGCACTA; MirB-R 5′-CCCTGAGGTCCCCGT-AGATGAG), MIRC (MirC-F 5′-TGATGGCATTCTCAACCTCCC; MirC-R 5′-AACCTGCGGTGAT-GAAACCAC), AMCA (AmcA-F 5′-GTCCGCATTACTCATCTG; AmcA-R 5′-CGCCTCATAAATC-GTAA), HAPX (HapX-F 5′-CCGGTACCCCTCAAGCCCACAACT; HapX-R 5′-AAATACTTCAAC-ACGCCCATAACG), and actin (Actin-F 5′-TCGGCCGTCCTCGCCATC and Actin-R 5′-TCCAG-ACTCGTCGTAGTCCTGC).
Real-time PCR
Total RNA was extracted from B. dermatitidis wild-type and SREB null mutant strains grown in HMM at 37°C and 22°C in a similar fashion as described above; modifications included grinding cells frozen in liquid nitrogen in a mortar and pestle. Wild-type and SREB null mutant cells were grown for 48 hours at 22°C prior to RNA extraction. RNA, at 10 ug/sample, was treated with Turbo DNase (Applied Biosystems/Ambion, Austin, Tx) and further purified using RNeasy kit (Qiagen, Valencia, CA). cDNA was generated from 1 ug of DNase-treated RNA using iScript cDNA synthesis kit (Bio-Rad, Inc., Hercules, CA). Real-time PCR reactions were comprised of 1x SSoFast EvagGreen supermix (Bio-Rad), 0.5 mM of each primer, and 1 ul of 10-fold diluted cDNA template in a total volume of 10 ul. All reactions were performed in triplicate for two biological replicates. Real-time PCR was performed using a Bio-Rad iCycler MyiQ. Cycling conditions were 1 cycle at 95°C for 30 seconds followed by 40 cycles of 95°C for 5 seconds and 60°C for 10 seconds. Melting curve analysis was performed following the completion of the PCR. Gene expression was normalized relative to the expression of alpha-tubulin based on R (relative expression) = 2−ΔCt, ΔCt = Cttarget gene–Cttubulin [57]. Primers used to amplify transcripts from the following genes were: Lipid transfer protein (BDBG_03618-1F 5′ - CCATCAATGCTGCCATCAAC; BDBG_03618-1R 5′-GGTCTCACCCTTGTCGTTTG), glycosyl hydrolase (BDBG 03183-1F 5′-GCTCTCCCAAGACATACATCAG, BDBG_03183-1R 5′-CCAT-AGCAAACTTCCCAAAAG), peroxisomal dehydratase (BDBG_00052-1F 5′-CCCATTGTGCTA-ACCTTCAAG, BDBG_00052-1R 5′-AACTCCATCCGTCGCCTC), acetoacetyl-CoA synthase (BDBG_09522-1F 5′-GCTCTCGGCACGCTCATAC, BDBG_09522-1R 5′-GGTGGTGACGG-GAGAAATG) and alpha-tubulin (BDBG_00020-2F 5′-GGTCACTACACCATCGGAAAG-3′, BDBG_00020 2R 5′-CTGGAGGGACGAACAGTTG).
MAST (motif alignment and search tool) analysis and KOG (eukaryotic orthologous groups) annotation
The annotated genome and predicted proteome of B. dermatitidis strain SLH14081 was used for MAST analysis and KOG annotation. The genome of this strain (75.35 Mb; 9,555 genes) has been sequenced and annotated by the Broad Institute (www.broadinstitute.org/annotation/genome/blastomyces_dermatitidis/MultiHome.html and ACBT01000000). The absence of annotation in the sequenced genome of 26199 precluded its use for computational analysis.
MAST/MEME (multiple em for motif elicitation) software in unix (version 4.2.0) was used to identify GATA transcription factor binding motifs in the genome of B. dermatitidis SLH14081 [58]. A fifth-order Markov background model was built for SLH14081 using the MEME utility fasta-get-markov. To find the location of previously identified motifs, MAST was run with a given motif frequency table, the Markov background model (-bfile) and options to produce text output as a ‘hit list’ (–text –hit_list). For a search with the ATCwgAtaa motif [26], a p-value of 0.0005 was used (–mt 0.0005). MAST output and Broad gene coordinates (http://www.broadinstitute.org/annotation/genome/blastomyces_dermatitidis/MultiHome.html) were parsed using a custom perl script to find intergenic motifs <2kb upstream of predicted genes. A total of 84,965 motifs were found in the genome assembly, of which 79,458 were in intergenic regions. Of these, 3,372 copies were found <2 kb upstream of 2,468 genes. Genes with the following motifs were retained: ATC-(A/T)-GATA-(A/G), ATC-(A/T)-GATA-(T/C), ATC-(A/T)-GATT-A, ATC-(A/T)-GATC-A, ATC-A-GATG-A, ATC-C-GATA-A, and ATC-A-AATA-A. These motifs are found upstream of genes regulated by A. fumigatus SREA, an SREB homolog [26].
To discover new motifs using MEME, we identified orthologs in SLH14081 of the iron-upregulated genes from A. fumigatus (BDBG_00046, BDBG_00047, BDBG_00048, BDBG_00050, BDBG_00053, BDBG_00054, BDBG_00055, BDBG_01314, BDBG_02226, BDBG_06775, BDBG_06965, BDBG_08034, BDBG_08208, BDBG_09322) and searched the 1 kb upstream for common motifs; MEME options were set for any number of motifs per region (-mod anr), the above described Markov background model, and a minimum width of 6 (-minw 6). This identified a motif of vATCwGATAA, which is similar to the motif described by Hwang and colleagues [27].
For KOG annotation and analysis, the predicted proteome from B. dermatitidis strain SLH14081 was retrieved from the Broad institute (http://www.broadinstitute.org/annotation/genome/blastomyces_dermatitidis/MultiDownloads.html, accessed: 11/09/2009) and compared against the NCBI KOG database (ftp://ftp.ncbi.nih.gov/pub/mmdb/cdd/, accessed: 11/09/2009)) using RPSBLAST (e-value 1e-05) [59],[60]. Two data sets were generated with the first containing all B. dermatitidis genes encoding proteins that registered a KOG annotation. The second set included B. dermatitidis proteins encoded by the candidate genes with upstream GATA sites. The KOGs for both sets were correlated to their associated categories, and the total number of proteins within each category was tabulated. A two-tailed Fisher's exact test was used to determine if the number of proteins in each category were over - or under-represented when compared to all KOG-annotated proteins in the B. dermatitidis proteome. Categories were considered over-represented if the p-value of the right of the Fisher's exact test was less than 0.05 and over-represented if the left tail was less than 0.05.
Isolation & identification of siderophores
To isolate and identify siderophores produced by B. dermatitidis 26199 wild-type and null mutants, we used column chromatography, liquid chromatography/mass spectroscopy (LC/MS), and reverse-phase high-pressure liquid chromatography (HPLC). Supernatants were harvested from B. dermatitidis grown in liquid HMM at 37°C under iron-poor (no added iron) and iron-replete (10 µM FeSO4) conditions when the cultures entered stationary growth (A600 = 3.5−4.0). Culture supernatants were filtered (0.2 µM), treated with 2% ferric chloride and applied to a column (K 9/30, GE Healthcare) packed with Amberlite XAD-2 resin (Supelco, Bellefonte, PA). The resin and column were prepared according to the manufacturer's recommendations. Following a water wash (7 bed volumes; flow rate of 0.2 ml/min), siderophores were eluted from the resin using methanol (1.7 bed volume; flow rate of 0.1 ml/min), reduced to dryness, and re-suspended in water (100 µl). Colorless supernatants that contained siderophores developed an orange color when treated with ferric chloride. This allowed for visual assessment of binding and elution of siderophores from the resin [61],[62].
The Mass Spectroscopy Facility at the University of Wisconsin Biotechnology Center performed LC/MS analysis of concentrated eluate collected from wild-type B. dermatitidis grown under iron-poor conditions following XAD-2 column chromatography. For HPLC, siderophores were separated on a C18 column (Agilent Eclipse XDB-C18 column; 4.6×150 mm) using a water-acetonitrile gradient containing 0.1% trifluoroacetic acid (Sigma-Aldrich). The gradient of acetonitrile was increased from 5% to 15% over 15 minutes, and 15% to 25% over 35 minutes. The flow rate was 0.5 ml/min and the absorbance was measured at 465 nm. Retention times were compared to siderophore standards (HPLC calibration kit – coprogens and fusarinines; EMC microcollections, Tubingen, Germany).
Accession numbers
The nucleotide sequences for SREB, SIDA, AMCA, MIRB, MIRC, and HAPX from B. dermatitidis strain 26199 were obtained from the Genome Sequencing Center, Washington University, Saint Louis, MO (http://genome.wustl.edu/tools/blast). Although this genome is publically available, it is not annotated. Allelic sequences can be found at the Broad Institute (http://www.broadinstitute.org/annotation/genome/blastomyces_dermatitidis/MultiHome.html) and have the following gene locus identification numbers: SREB (BDBG_01059), SIDA (BDBG_00053), AMCA (BDBG_00128), MIRB (BDBG_05798), MIRC (BDBG_08034), HAPX (BDBG_01314). Additional gene locus numbers include: lipid transfer protein (BDBG_03618), glycosyl hydrolase (BDBG_03183), peroxisomal dehydratase (BDBG_00052), acetoacetyl-CoA synthase (BDBG_09522), and alpha-tubulin (BDBG_00020).
Supporting Information
Zdroje
1. GauthierGM
KleinBS
2008 Insights into fungal morphogenesis and immune evasion. Microbe 3 416 423
2. KleinBS
TebbetsB
2007 Dimorphism and virulence in fungi. Curr Opin Microbiol 10 314 319
3. GauthierGM
SafdarN
KleinBS
AndesDR
2007 Blastomycosis in solid organ transplant recipients. Transpl Infect Dis 9 310 317
4. WalshTJ
RaadI
PattersonTF
ChandrasekarP
DonowitzGR
2007 Treatment of invasive aspergillosis with posaconazole in patients who are refractory to or intolerant of conventional therapy: an externally controlled trial. Clin Infect Dis 44 2 12
5. van der HorstCM
SaagMS
CloudGA
HamillRJ
GraybillJR
1997 Treatment of cryptococcal meningitis with the acquired immunodeficiency syndrome. National Institute of Allergy and Infectious Diseases mycoses study group and AIDS clinical Trials Group. N Engl J Med 337 15 21
6. KlotzSA
DrutzDJ
HuppertM
SunSH
DeMarshPL
1984 The critical role of CO2 in the morphogenesis of Coccidioides immitis in cell-free subcutaneous chambers. J Infect Dis 150 127 134
7. PowellBL
DrutzDJ
HuppertM
SunSH
1983 Relationship of progesterone - and estradiol-binding proteins in Coccidioides immitis to coccidioidal dissemination in pregnancy. Infect Immun 40 478 485
8. RestrepoA
SalazarME
CanoLE
StoverEP
FeldmanD
1984 Estrogens inhibit mycelium-to-yeast transformation in the fungus Paracoccidioides brasiliensis: implications for resistance of females to paracoccidioidomycosis. Infect Immun 46 346 353
9. SugarMA
PicardM
1991 Macrophage - and oxidant-mediated inhibition of the ability of live Blastomyces dermatitidis conidia to transform to the pathogenic yeast phase: implications for the pathogenesis of dimorphic fungal infections. J Infect Dis 163 371 375
10. NemecekJC
WuethrichM
KleinBS
2006 Global control of dimorphism and virulence in fungi. Science 312 583 588
11. SaccoM
MarescaB
KumarBV
KobayashiGS
MedoffG
1981 Temperature - and cyclic nucleotide-induced phase transition of Histoplasma capsulatum. J Bacteriol 146 117 120
12. NguyenVQ
SilA
2008 Temperature-induced switch to the pathogenic yeast form of Histoplasma capsulatum requires RYP1, a conserved transcriptional regulator. Proc Natl Acad Sci U S A 105 4880 4885
13. Hanby WebsterR
SilA
2008 Conserved factors Ryp2 and Ryp3 control cell morphology and infectious spore formation in the fungal pathogen Histoplasma capsulatum. Proc Natl Acad Sci USA 105 14573 14578
14. HaasH
AngermayrK
SotfflerG
1997 Molecular analysis of Penicillium chrysogenum GATA factor encoding gene (sreP) exhibiting significant homology to the Ustilago maydis urbs1 gene. Gene 184 33 37
15. GurrSJ
UnklesSE
KinghornJR
1987 The structure and organization of nuclear genes of filamentous fungi.
KinghornJR
Gene structure of eukaryotic microbes Oxford, England IRL Press 93 139
16. ChaoLY
MarlettaMA
RineJ
2008 Sre1, an iron-modulated GATA DNA-binding protein of iron-uptake genes in the fungal pathogen Histoplasma capsulatum. Biochemistry 47 7274 7283
17. HaasH
ZadraI
StofflerG
AngermayrK
1999 The Aspergillus nidulans GATA factor SREA is involved with regulation of siderophore biosynthesis and control of iron uptake. J Biol Chem 274 4613 4619
18. ZhouLW
HaasH
MarzlufGA
1998 Isolation and characterization of a new gene, sre, which encodes a GATA-type regulatory protein that controls iron transport in Neurospora crassa. Mol Gen Genet 259 532 540
19. VoisardC
WangJ
McEvoyJL
XuP
LeongSA
1993 Urbs1, a gene regulating siderophore biosynthesis in Ustilago maydis, encodes a protein similar to the erythroid transcription factor GATA-1. Mol Cell Biol 13 7091 7100
20. PelletierB
BeaudoinJ
MukaiY
LabbeS
2002 Fep1, an iron sensor regulating iron transporter gene expression in Schizosaccharomyces pombe. J Biol Chem 277 22950 22958
21. LanCY
RodarteG
MurilloLA
JonesT
DavisRW
2004 Regulatory networks affected by iron availability in C. albicans. Mol Microbiol 53 1451 1469
22. JungWH
ShamA
WhiteR
KronstadJW
2006 Iron regulation of the major virulence factors in the AIDS-associated pathogen Cryptococcus neoformans. PLoS Biol 4 e410 doi:10.1371/journal.pbio.0040410
23. WolfE
KimPS
BergerB
1997 MultiCoil: A program for predicting two - and three-stranded coiled coils. Protein Sci 6 1179 1189
24. GronenbornAM
2005 The DNA-binding domain of GATA transcription factors – a prototypical Type IV Cys2-Cys2 zinc finger.
LuchiS
Zinc Finger Proteins: from atomic contact to cellular function New York Kluwer Academic/Plenum Publishers 26 30
25. HolzbergM
ArtisWM
1983 Hydroxymate siderophore production by opportunistic and systemic fungal pathogens. Infect Immun 40 1134 1139
26. SchrettlM
KimHS
EisendleM
KraglC
NiermanWC
2008 SreA-mediated iron regulation in Aspergillus fumigatus. Mol Microbiol 70 27 43
27. HwangLH
MayfieldJA
RineJ
SilA
2008 Histoplasma requires SID1, a member of an iron-regulated siderophore gene cluster, for host colonization. PLoS Pathog 4 e1000044 doi:10.1371/journal.ppat.1000044
28. SullivanTD
RooneyPJ
KleinBS
2002 Agrobacterium tumefaciens integrates transfer DNA into single chromosomal sites of dimorphic fungi and yields homokaryotic progeny from multinucleate yeast. Eukaryot Cell 1 895 905
29. ChandarlapatyS
ErredeB
1998 Ash1, a daughter cell-specific protein, is required for pseudohyphal growth of Saccharomyces cerevisiae. Mol Cell Biol 18 2884 2891
30. ReyesJC
Muro-PastorMI
FlorencioFJ
2004 The GATA family of transcription factors in Arabidopsis and rice. Plant Physiol 134 1718 1732
31. ScazzocchioC
2000 The fungal GATA factors. Curr Opin Microbiol 3 126 31
32. HeQ
ChengP
YangY
WangL
GardnerKH
2002 White collar-1, a DNA binding transcription factor and a light sensor. Science 297 840 843
33. ToddRB
FraserJA
WongKH
DavisMA
HynesMJ
2005 Nuclear accumulation of GATA factor AreA in response to complete nitrogen starvation by regulation of nuclear export. Eukaryotic Cell 4 1646 1653
34. TianX
ShearerGJr
2002 The mold-specific MS8 gene is required for normal hypha formation in the dimorphic pathogenic fungus Histoplasma capsulatum. Eukaryot Cell 1 249 256
35. ToddRB
GreenhalghJR
HynesMJ
AndrianopoulosA
2003 TupA, the Penicillium marneffei Tup1 homologue, represses both yeast and spore development. Mol Microbiol 48 85 94
36. MarescaB
KobayashiGS
1993 Changes in membrane fluidity modulate heat shock gene expression and produced attenuated strains in the dimorphic fungus H. capsulatum. Arch Med Res 24 247 249
37. LonesGW
PeacockCL
McNeyFA
1971 Factors affecting the reversion of Coccidioides immitis spherules to mycelium. Sabouraudia 9 287 296
38. Plaine AKrishnamurthyS
AlbertJ
PrasadT
PrasadJ
2004 Dosage-dependent functions of fatty acid desaturase Ole1p in growth an morphogenesis of Candida albicans. Microbiology 150 1991 2003
39. DomerJE
HamiltonJG
1971 The readily extracted lipids of Histoplasma capsulatum and Blastomyces dermatitidis. Biochim Biophys Acta 231 465 478
40. ManochaMS
1980 Lipid composition of Paracoccidioides brasiliensis: comparison between yeast and mycelial forms. Sabouraudia 18 281 286
41. ToledoMS
LeverySB
SuzukiE
StrausAH
TakahashiHK
2001 Characterization of cerebrosides from the thermally dimorphic mycopathogen Histoplasma capsulatum: expression of 2-hydroxy fatty N-acyl (E)-Δ3-unsaturation correlates with the yeast-mycelium phase transition. Glycobiology 11 113 124
42. ToledoMS
LeverySB
StrausAH
SuzukiE
MomanyM
1999 Characterization of sphingolipids from mycopathogens: Factors correlating with expression of 2-hydroxy fatty acyl (E) - Δ3-unsaturation in cerebrosides of Paracoccidioides brasiliensis and Aspergillus fumigatus. Biochemistry 38 7294 7306
43. MonteiroJP
ClemonsKV
MirelsLF
CollerHA
WuTD
2009 Genomic expression microarray comparison of gene expression patterns in Paracoccidioides brasiliensis mycelia and yeasts in vitro. Microbiology 155 2795 2808
44. HortschanskyP
EisendleM
Al-AbdallahQ
SchmidtAD
BergmannS
2007 Interaction of HapX with the CCAAT-binding complex – a novel mechanism of gene regulation by iron. EMBO J 26 3157 3168
45. ObereggerH
SchoeserM
ZadraI
AbtB
HaasH
2001 SREA is involved in regulation of siderophore biosynthesis, utilization and uptake in Aspergillus nidulans. Mol Microbiol 41 1077 1089
46. HaasH
SchoeserM
LesuisseE
ErnstJF
ParsonW
2003 Characterization of the Aspergillus nidulans transporters for the siderophores enterbactin and triacylfusarinine C. Biochem J 371 505 513
47. BrandhorstTT
WuthrichM
WarnerT
KleinB
1999 Targeted gene disruption reveals an adhesin indispensable for pathogenicity of Blastomyces dermatitidis. J Exp Med 189 1207 1216
48. WorshamPL
GoldmanWE
1988 Quantitative plating of Histoplasma capsulatum without addition of conditioned medium or siderophores. J Med Vet Mycol 26 137 143
49. McVeighI
MortonK
1965 Nutritional studies of Histoplasma capsulatum. Mycopathol Mycol Appl 25 294 308
50. RinaldiMG
1982 Use of potato flake agar in clinical microbiology. J Clin Microbiol 15 1159 1160
51. de GrootMJ
BundockP
HooykaasPJ
BeijersbergenAG
1998 Agrobacterium tumefaciens-mediated transformation of filamentous fungi. Nat Biotechnol 16 839 842
52. SiebertPD
ChenchikA
KelloggDe
LuyanovKA
LukyanovSA
1995 An improved method for walking in uncloned genomic DNA. Nucleic Acid Res 23 1087 1088
53. RooneyPJ
SullivanTD
KleinBS
2001 Selective expression of the virulence factor BAD1 upon morphogenesis to the pathogenic yeast form of Blastomyces dermatitidis: evidence for transcriptional regulation by a conserved mechanism. Mol Microbiol 39 875 889
54. HoganLH
KleinBS
1997 Transforming DNA integrates at multiple sites in the dimorphic fungal pathogen Blastomyces dermatitidis. Gene 186 219 226
55. SambrookJ
RussellDW
2001 Molecular cloning: a laboratory manual, 3rd Ed. Cold Spring Harbor, NY Cold Spring Harbor Laboratory Press 6.33-6.46, 6.50-6.58, 7.9-7.17, 7.31-7.41
56. BothwellTH
ConradME
CookJD
BrosbyWH
FieldingJ
1971 International Committee For Standardization in Hematology. Proposed recommendations for measurement of serum iron in blood. Br J Haemotol 20 451 453
57. LivakKF
SchmittgenTD
2001 Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCt method. Methods 25 402 408
58. BaileyTL
GribskovM
1998 Combining evidence using p-values: application to sequence homology searches. Bioinformatics 14 48 54
59. TatusovRL
FedorovaND
JacksonJD
JacobsAR
KiryutinB
2003 The COG database: an updated version includes eukaryotes. BMC Bioinformatics 4 41
60. Marchler-BauerA
AndersonJB
DerbyshireMK
DeWeese-ScottC
GonzalesNR
2007 CDD: a conserved domain database for interactive domain family analysis. Nucleic Acids Res 35 D237 D240
61. JallalMAF
van der HelmD
1991 in Handbook of Microbial Iron Chelates (Gunther W., ed), pp 235-269, CRC Press, Boca Raton, FL
62. Konetschny-RappS
HuschkaHG
WinkelmannG
JungG
1988 High-performance liquid chromatography of siderophores from fungi. Biometals 1 91 97
Štítky
Hygiena a epidemiologie Infekční lékařství Laboratoř
Článek Suppression of mRNAs Encoding Tegument Tetraspanins from Results in Impaired Tegument TurnoverČlánek Novel Riboswitch Ligand Analogs as Selective Inhibitors of Guanine-Related Metabolic PathwaysČlánek The Physical Relationship between Infectivity and Prion Protein Aggregates Is Strain-DependentČlánek Rhomboid 4 (ROM4) Affects the Processing of Surface Adhesins and Facilitates Host Cell Invasion by
Článek vyšel v časopisePLOS Pathogens
Nejčtenější tento týden
2010 Číslo 4- Jak souvisí postcovidový syndrom s poškozením mozku?
- Měli bychom postcovidový syndrom léčit antidepresivy?
- Farmakovigilanční studie perorálních antivirotik indikovaných v léčbě COVID-19
- 10 bodů k očkování proti COVID-19: stanovisko České společnosti alergologie a klinické imunologie ČLS JEP
-
Všechny články tohoto čísla
- Innate Recognition of Fungal Cell Walls
- Suppression of mRNAs Encoding Tegument Tetraspanins from Results in Impaired Tegument Turnover
- Junín Virus Infection of Human Hematopoietic Progenitors Impairs Proplatelet Formation and Platelet Release via a Bystander Effect Involving Type I IFN Signaling
- The Endosymbiotic Bacterium Induces Resistance to Dengue Virus in
- Natural Regulatory T Cells in Malaria: Host or Parasite Allies?
- Keratinocytes Determine Th1 Immunity during Early Experimental Leishmaniasis
- Spatial and Temporal Association of Outbreaks of H5N1 Influenza Virus Infection in Wild Birds with the 0°C Isotherm
- Novel Riboswitch Ligand Analogs as Selective Inhibitors of Guanine-Related Metabolic Pathways
- RNA Polymerase Activity and Specific RNA Structure Are Required for Efficient HCV Replication in Cultured Cells
- The Physical Relationship between Infectivity and Prion Protein Aggregates Is Strain-Dependent
- Inadequate Clearance of Translocated Bacterial Products in HIV-Infected Humanized Mice
- Topology and Organization of the Type III Secretion Needle Complex Components
- Temperature Modulates Plant Defense Responses through NB-LRR Proteins
- Peptide Inhibitors of Dengue-Virus Entry Target a Late-Stage Fusion Intermediate
- Identification of Host-Dependent Survival Factors for Intracellular through an siRNA Screen
- Exposure to HIV-1 Directly Impairs Mucosal Epithelial Barrier Integrity Allowing Microbial Translocation
- Increased Asymmetric Dimethylarginine in Severe Falciparum Malaria: Association with Impaired Nitric Oxide Bioavailability and Fatal Outcome
- Reconstitution of SARS-Coronavirus mRNA Cap Methylation
- Induces Brain Microvascular Endothelial Cell Detachment from the Matrix and Cleavage of Occludin: A Role for MMP-8
- Two Coregulated Efflux Transporters Modulate Intracellular Heme and Protoporphyrin IX Availability in
- The Type I NADH Dehydrogenase of Counters Phagosomal NOX2 Activity to Inhibit TNF-α-Mediated Host Cell Apoptosis
- Rhomboid 4 (ROM4) Affects the Processing of Surface Adhesins and Facilitates Host Cell Invasion by
- Increased Monocyte Turnover from Bone Marrow Correlates with Severity of SIV Encephalitis and CD163 Levels in Plasma
- The RING-CH Ligase K5 Antagonizes Restriction of KSHV and HIV-1 Particle Release by Mediating Ubiquitin-Dependent Endosomal Degradation of Tetherin
- Molecular Mechanisms of Ethanol-Induced Pathogenesis Revealed by RNA-Sequencing
- Highly Frequent Mutations in Negative Regulators of Multiple Virulence Genes in Group A Streptococcal Toxic Shock Syndrome Isolates
- Emergence and Pathogenicity of Highly Virulent Genotypes in the Northwest United States
- Structural and Functional Analysis of Viral siRNAs
- Prion Shedding from Olfactory Neurons into Nasal Secretions
- a GATA Transcription Factor That Directs Disparate Fates in Including Morphogenesis and Siderophore Biosynthesis
- Three Members of the 6-cys Protein Family of Play a Role in Gamete Fertility
- Complement as an Endogenous Adjuvant for Dendritic Cell-Mediated Induction of Retrovirus-Specific CTLs
- A Genomic Survey of Positive Selection in Provides Insights into the Evolution of Accidental Virulence
- Overcomes Stress of Azole Drugs by Formation of Disomy in Specific Multiple Chromosomes
- Blood Fluke Exploitation of Non-Cognate CD4 T Cell Help to Facilitate Parasite Development
- Antagonism of Tetherin Restriction of HIV-1 Release by Vpu Involves Binding and Sequestration of the Restriction Factor in a Perinuclear Compartment
- The Development of Therapeutic Antibodies That Neutralize Homologous and Heterologous Genotypes of Dengue Virus Type 1
- Deficiencies in Jasmonate-Mediated Plant Defense Reveal Quantitative Variation in Pathogenesis
- Interactions with Bacteria in the Context of Human Health and Disease
- Viral Capsid Is a Pathogen-Associated Molecular Pattern in Adenovirus Keratitis
- Electron Tomography Reveals the Steps in Filovirus Budding
- Selective Condensation Drives Partitioning and Sequential Secretion of Cyst Wall Proteins in Differentiating
- The Effect of Vaccination on the Evolution and Population Dynamics of Avian Paramyxovirus-1
- A Timescale for Evolution, Population Expansion, and Spatial Spread of an Emerging Clone of Methicillin-Resistant
- VacA Toxin/Subunit p34: Targeting of an Anion Channel to the Inner Mitochondrial Membrane
- Production of Extracellular Traps against and in Infected Lung Tissue Is Dependent on Invading Neutrophils and Influenced by Hydrophobin RodA
- A Differential Role for Macropinocytosis in Mediating Entry of the Two Forms of Vaccinia Virus into Dendritic Cells
- Impaired Innate Immunity in Mice but Preserved CD8 T Cell Responses against in -, -, - or -Deficient Mice
- SARS-CoV Pathogenesis Is Regulated by a STAT1 Dependent but a Type I, II and III Interferon Receptor Independent Mechanism
- Proteolysis of Human Thrombin Generates Novel Host Defense Peptides
- Multilayered Mechanism of CD4 Downregulation by HIV-1 Vpu Involving Distinct ER Retention and ERAD Targeting Steps
- PLOS Pathogens
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- The Effect of Vaccination on the Evolution and Population Dynamics of Avian Paramyxovirus-1
- Reconstitution of SARS-Coronavirus mRNA Cap Methylation
- Deficiencies in Jasmonate-Mediated Plant Defense Reveal Quantitative Variation in Pathogenesis
- A Timescale for Evolution, Population Expansion, and Spatial Spread of an Emerging Clone of Methicillin-Resistant
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



