-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaRibosomal Protein Mutations Result in Constitutive p53 Protein Degradation through Impairment of the AKT Pathway
The p53 tumor suppressor is the most commonly mutated gene in human cancers. However, cancer cells exploit multiple mechanisms to silence the p53 pathway in addition to inactivation of the p53 gene. We previously reported that one of these mechanisms is found in tumor cells with ribosomal protein (RP) gene mutations. These cells transcribe wild type p53 mRNA yet do not stabilize p53 protein when exposed to DNA damaging agents. In this work we demonstrate that this loss of p53 protein is due to its constitutive degradation. This degradation is due to impairment of the AKT pathway, which normal signals for p53 to stabilize when the DNA is damaged. By re-activating the AKT pathway in RP-mutant cells we are able to restore p53 stabilization and activity, which may hold clinical significance for cancer treatment.
Published in the journal: . PLoS Genet 11(7): e32767. doi:10.1371/journal.pgen.1005326
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1005326Summary
The p53 tumor suppressor is the most commonly mutated gene in human cancers. However, cancer cells exploit multiple mechanisms to silence the p53 pathway in addition to inactivation of the p53 gene. We previously reported that one of these mechanisms is found in tumor cells with ribosomal protein (RP) gene mutations. These cells transcribe wild type p53 mRNA yet do not stabilize p53 protein when exposed to DNA damaging agents. In this work we demonstrate that this loss of p53 protein is due to its constitutive degradation. This degradation is due to impairment of the AKT pathway, which normal signals for p53 to stabilize when the DNA is damaged. By re-activating the AKT pathway in RP-mutant cells we are able to restore p53 stabilization and activity, which may hold clinical significance for cancer treatment.
Introduction
The stabilization of the p53 tumor suppressor is a pivotal event in the programmed cell death response. Levels of p53 protein are normally kept very low through its physical association with the MDM2 protein, an E3 ubiquitin ligase that constitutively ubiquitinates p53 and targets it for proteasomal degradation [1]. Many kinds of cellular stress, including DNA damage and oncogene presence, activate different signaling pathways that result in the dissociation of p53 and MDM2. p53 then stabilizes and translocates to the nucleus where it targets genes that arrest the cell cycle and turn on DNA repair, or genes that induce apoptotic cell death if the damage is deemed irreparable [2].
The stabilization of p53 has been reported to trigger human bone marrow failures such as dyskeratosis congenita and Fanconi anemia [3,4]. While Fanconi anemia is predominantly linked to mutations in DNA repair enzymes, several genes found mutated in dyskeratosis congenita patients have a known role in the rRNA maturation steps of early ribosome biogenesis. The mutation of these latter genes in zebrafish stabilizes p53, as does the mutation of several other genes important for the processing of rRNA [5–7]. In human bone marrow failures syndromes linked to RP haploinsufficiency such as Diamond-Blackfan anemia (DBA) and 5q-myelodysplastic syndrome, the loss of hematopoietic progenitor CD34+ cells by p53-induced apoptosis is believed by some to be the major cause of cytopenia [8]. However, the contribution of p53-induced apoptosis specifically to the cytopenia phenotype remains controversial. Recent studies demonstrated that patient CD34+ hematopoietic progenitor cells carrying mutations in the most commonly mutated gene linked to DBA (RPS19) do not reveal any hallmarks of apoptosis as they are induced to differentiate into erythrocytes [9]. This work also showed that while the co-depletion of p53 with RPL11 in CD34+ cells reduced some apoptotic effects, it did not restore the proliferation capacity lost upon RPL11 depletion alone. Therefore the contribution of p53 stabilization to the loss of erythrocytes in DBA patients is possibly less significant than previously thought.
In addition to being important for erythrocyte production, there also exist several reports indicating a role for RPs as tumor suppressors. Human patients with 5q-MDS or DBA are more predisposed to developing malignancies, both acute myeloid leukemia (AML) and solid tumors, respectively [10,11]. Importantly, the advent of exome sequencing has unveiled a surprising number of somatic RP gene mutations in an array of human cancers. These recent exome sequencing reports have identified mutations in RPL5 and RPL10 in T-cell acute lymphoblastic leukemia, mutations in RPL5 in gliomas, and mutations in RPL22 in human endometrioid endometrial cancer and colorectal cancer [12–14].
Embryonic zebrafish mutants and morphants are popular models of DBA and RP loss. In mutant lines where RP genes are disrupted by murine virus integrations, the homozygous embryos reveal a progressive reduction of the RP over the first 3 days post fertilization (dpf) coupled to a failure of hemoglobin-expressing cells to develop [15,16]. In contrast, haploinsufficient RP embryos reveal no cytopenia or any other conspicuous phenotype except for a mild growth defect that does not affect their development into adulthood [17]. Once reaching adulthood however, many of the haploinsufficient mutant lines reveal the formation of malignant peripheral nerve sheath tumors (MPNSTs), a tumor type rarely observed in laboratory strains of zebrafish [18]. Interestingly, this exact tumor type arises with the same frequency in zebrafish carrying homozygous mutations of p53 in a highly conserved residue within one of the DNA-binding domains (M214K) [19]. Further study into this revealed that while wild type p53 mRNA was normally transcribed in the RP-mutant tumor cells, p53 protein was unable to be detected [20]. This observation was despite the application of ionizing radiation and/or proteasome inhibition with MG132, two conditions that were found to stabilize p53 in zebrafish tumor cells carrying wild type RP genes.
Actinomycin D is a drug that disrupts ribosome biogenesis by inhibiting rRNA polymerase and results in the stabilization of p53 [21]. It was recently reported that this induction of p53 requires the activity of the survival factor AKT/PKB [22], a kinase that responds to the stimulation of many growth factor receptors, including insulin receptors [23]. While activated AKT has many functions, one major event of insulin stimulation directly downstream of AKT is the phosphorylation and inhibition of glycogen synthase kinase-3 (GSK-3) [24]. Under normal conditions GSK-3 phosphorylates and activates MDM2 in a way that promotes p53 degradation [25]. However upon DNA damage by ionizing radiation, the phosphorylation of AKT results in the inactivation of GSK-3 [26]. This AKT-mediated inactivation of GSK-3 in response to ionizing radiation begins with the DNA-dependent protein kinase (DNA-PK), a protein that recognizes the double-stranded breaks on DNA and signals through a cascade that ultimately results in p53 stabilization and programmed cell death [27]. Thus one mechanism of the p53 stabilization in response to ionizing radiation is through the activation of DNA-PK and AKT inhibiting the downstream activity of GSK-3 and MDM2.
We previously reported that the loss of RP genes in mammalian cells and in zebrafish embryos results in a loss of AKT activity that could be overcome by the addition of insulin [16]. This observation led us to consider the possibility that the AKT pathway was involved in the regulation of p53 in RP mutant cells, and that the impairment of the DNA damage pathway through AKT may have a role in the predisposition to malignancy caused by RP gene mutations.
Results
The hematopoietic phenotype of zebrafish with RP loss is not dependent on p53
The RP mutant zebrafish lines we used for this study were generated by viral insertions in the introns of RP genes, two of which (rpS7 and rpL11) have homologues found mutated in DBA patients [28,29]. We find that at 2 dpf these embryos display normal expression of the scl transcription factor required for the genesis of hematopoietic stem cells in both primitive and definitive hematopoiesis, a result that was also observed in zebrafish embryos with deletions in the rpS19 gene (S1 Fig) [30–33]. However, RP mutants reveal a marked decrease in the expression of the βE1-globin gene, which (at this stage of development) normally becomes up regulated as cells commit to the erythrocyte lineage (S1 Fig) [34]. Zebrafish embryos carry maternal stores of RPs such that these mutants reveal a progressive loss of RP expression. The rpS7 mutants express about 50% of wild type rpS7 levels at 1 dpf while this percent reduction is not observed in rpL11 mutants until 3 dpf [16,35]. This likely explains why despite the fact that embryos from both mutant lines reveal hematopoietic and developmental phenotypes, these are more severe in the rpS7 mutants [16,35]. Because these phenotypes at 1 dpf are much more pronounced in rpS7 embryos, we selected them for the following analysis of apoptosis. S2 Fig provides images of the mutants compared to wild types and shows the morphological features that we use to initially select the mutants, such as the smaller head and eye, inflated hindbrain, and the dent in the mid-hind brain barrier.
We measured overall levels of cell death in developing embryos carrying the rpS7 mutation with acridine orange (AO), a stain commonly used to detect cells undergoing apoptosis in zebrafish embryos [36]. Fig 1A and 1B show that at 1 dpf, rpS7 mutant embryos reveal a significantly larger number of apoptotic cells compared to wild type controls. Closer visualization of an rpS7 mutant in Fig 1C reveals that these AO-stained cells are found clustered in the brain area and evenly distributed on the entire surface of the tail. The injection of a translation-blocking morpholino that we have previously demonstrated is specific to zebrafish p53 (p53 MO) but not a missense control morpholino (mis MO), completely rescued the number of cells undergoing apoptosis in the rpS7 mutants, reinforcing a role for p53 in cell death as a result of RP loss (Fig 1A and 1B) [37]. The p53 MO also results in the rescue of morphological phenotypes commonly observed in ribosome biogenesis mutants such as the inflation of the hindbrain vesicle and pericardial edemas, a rescue effect that has been previously demonstrated in other zebrafish models of RP loss (S2 Fig) [15,38]. To determine if the p53 MO was sufficient to rescue the defective hematopoiesis of the mutants we used o-dianisidine, which stains hemoglobin-expressing cells evident at 2 dpf. Staining with o-dianisidine revealed that despite the rescue of apoptotic cells by p53 depletion observed at 1 dpf in the rpS7 mutants, the p53 MO was not able to rescue hematopoietic development to any appreciable degree (Fig 1D).
Fig. 1. Early cell death of RP mutant cells requires p53. 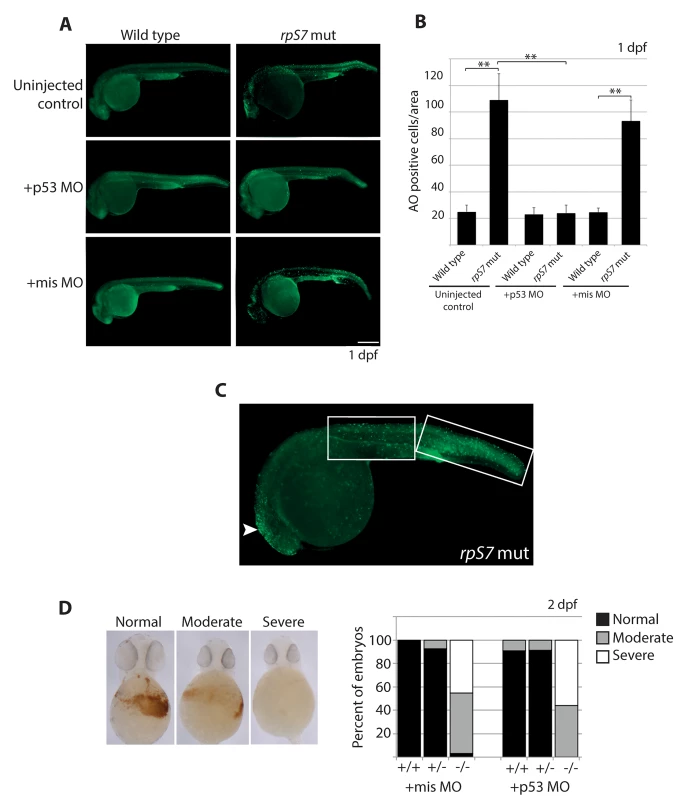
A) Acridine orange staining allowing visualization of dying cells in wild type or rpS7 mutants at 1 dpf that are uninjected, injected with the p53 MO, or injected with the missense MO. Size bar = 0.25mm. B) Quantification of (A). **p<0.01. C) Acridine orange staining of a rpS7 mutant is observed predominantly in the brain region (arrowhead) or distributed over the surface of the tail (white boxes). D) Scoring results from o-dianisidine staining allowing visualization of hemoglobin-expressing cells in clutches of either 111 embryos (+mis MO) or 177 embryos (+p53 MO) from an rpS7 pairing. The apoptotic response to DNA damage is caspase-independent in RP mutants
The levels of caspase-induced apoptosis in human CD34+ cells with RP loss vary depending on the RP [9]. To determine if the loss of RPs triggers caspase-induced apoptosis in zebrafish embryos we measured both the basal levels of activated caspase 2 or 3/7 and their levels in response to ionizing radiation, comparing wild type embryos with those carrying mutations in rpS7 and rpL11, as well as rpS3 and rpL36 genes. Fig 2A and 2B indicate that in 2 dpf embryos the caspase 2 and 3/7 basal levels in the mutants is equivalent to wild types, and the caspase response to DNA damage is severely impaired in all the mutants. In fact, the RP mutant lines show the same suppressed caspase 2 and 3/7 response to ionizing radiation as the homozygous p53M214K/M214K mutant line (Fig 2A and 2B), which is severely impaired in its ability to induce apoptosis [19]. To determine if other hallmarks of apoptosis such as DNA fragmentation are present in RP mutants we performed TUNEL assays on rpS7 embryos at 2 dpf. Fig 2C and 2D show that while there is no appreciable difference in the levels of TUNEL-positive cells between the rpS7 mutant and wild type embryos, the exposure of mutants to ionizing radiation results in a similar significant increase of TUNEL-positive cells as observed in the wild types. Closer visualization of these DNA-damaged embryos reveals a localization of TUNEL-positive cells in the brain area similar to what is seen with the AO staining, however the localization of TUNEL-positive cells in the tail region is found much more restricted to the dorsal area above the notochord (Fig 2E). This may be due to the limits of penetration of the AO stain, or may suggest that the cells in this dorsal area are especially sensitive to DNA damage-induced apoptosis, as are the rapidly proliferating cells in the brain.
Fig. 2. Caspase 3/7 activation is increasingly impaired in RP mutants. 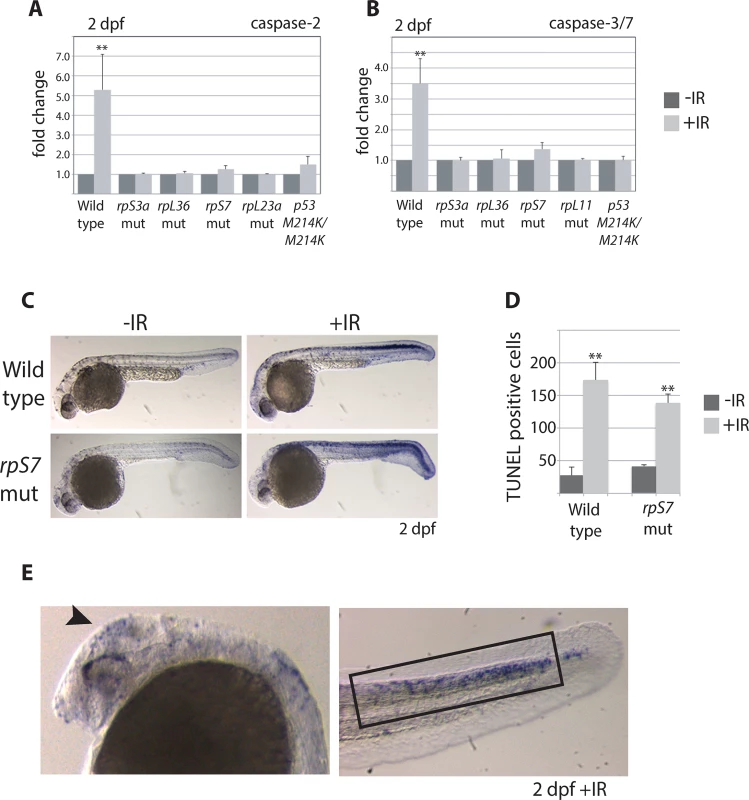
A) Caspase 2 and B) 3/7 activity is measured using a proluminescent caspase-3/7 DEVD-aminoluciferin substrate in 2 dpf embryos either untreated or exposed to 25 Gy ionizing radiation (IR). C) TUNEL analysis of 2 dpf wild type or rpS7 embryos reveals cells with DNA fragmentation when embryos are exposed to 25 Gy IR. D) The number of TUNEL-positive cells in (C) are quantified. E) TUNEL-positive cells in both wild type and rpS7 mutants are observed predominantly in the brain region (arrowhead) or in the tail tissue dorsal to the notochord (black box). *p<0.05, **p<0.01. p53 stabilization but not transcription is impaired in RP mutants
The increased transcription of the zebrafish p53 gene and its p53Δ113 isoform (a target gene of stabilized p53) has been described in several models of RP loss and likely reflects the early response of p53 that triggers an up regulation of its own transcription and the transcription of p53Δ113 [15,35,38–41]. In line with these results, we found using real-time quantitative PCR analysis with primers that amplify both full-length p53 and p53Δ113 that p53 mRNA levels were significantly higher in rpS7 and rpL11 mutants at 1 and 2 dpf both in the presence and absence of ionizing radiation compared to untreated wild type embryos (Fig 3A and 3B). Semi-quantitative PCR analysis of p53 mRNA levels in several other RP-mutant embryos (rpS3a, rpL23a, and rpL36) at 2 dpf similarly revealed equivalent levels of p53 transcription in the mutants compared to wild types (S3 Fig). However, when we performed western blotting analysis using a zebrafish p53-specific antibody, we were unable to detect any appreciable amount of p53 protein in the rpS7 or rpL11 mutants in either the presence or absence of ionizing radiation at either 1 or 2 dpf (Fig 3C). This is the case in all the mutant RP lines we tested including in rpS3a, rpL23a, and rpL36 (S3 Fig). We often observe what may be a p53-specific isoform such as p53Δ113 on the western blots, but this may also be a p53 degradation product and in this work we cannot be certain of its exact identity. Taken together, the results suggest that although the p53 response to the RP mutation on a transcriptional level may function normally, an additional level of p53 post-translational regulation exists in the presence of RP mutations that serves to reduce p53 protein.
Fig. 3. p53 protein stabilization is impaired independently of p53 mRNA levels. 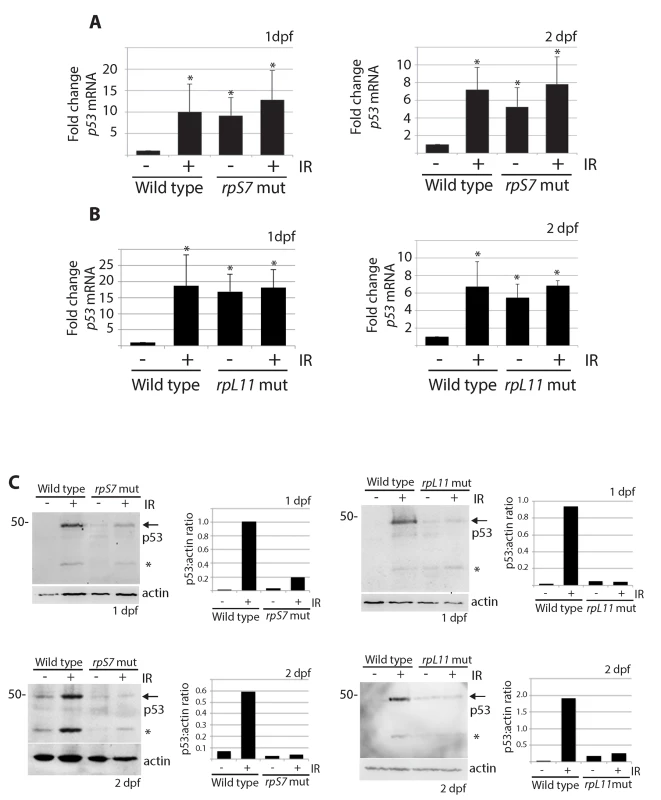
A) qPCR analysis measuring levels of p53 mRNA in wild type or rpS7 mutants at 1 or 2 dpf either untreated or exposed to 25 Gy ionizing radiation. B) qPCR analysis of p53 mRNA levels in wild type or rpL11 mutants at 1 or 2 dpf either untreated or exposed to 25 Gy ionizing radiation. *p<0.05. C) Western blot analysis of p53 protein levels and the quantification of the p53:actin ratio of in rpS7 or rpL11 mutants at 1 or 2 dpf either untreated or exposed to 25 Gy ionizing radiation. * indicates either a p53-specific isoform or a degradation product. p53 protein is degraded by the proteasome and rescued by insulin in RP mutants
We recently demonstrated that AKT phosphorylation activity is impaired in zebrafish embryos carrying mutations in rpS7, rpL11, and rpS3a [16]. Since the loss of AKT activity would theoretically lead to an increase of p53 proteasomal degradation due to an increase of GSK-3 phosphorylation of MDM2, we used the proteasome inhibitor MG132 to determine if we could restore p53 expression. Fig 4A illustrates that MG132 is able to stabilize p53 in wild type embryo cells as expected, and moreover it is able to stabilize p53 in the rpS7 mutants. This supports the notion that the loss of p53 in the RP mutants is the result of excessive proteasomal degradation. We therefore hypothesized that stimulating AKT in the presence of ionizing radiation may rescue the p53 response to DNA damage. Fig 4B (lanes 1–4) shows p53 stabilization approximately 6 hours after exposure of wild type embryos to ionizing radiation when the embryos are treated with the anti-oxidant Trolox, insulin, or both. Fig 4B (lanes 5–8) illustrates that the addition of insulin, but not Trolox, immediately following the ionizing radiation results in a rescue of the stabilization of p53 in rpS7 mutants. These data suggest that overcoming the RP mutation-induced inhibition of AKT with insulin, which would stimulate AKT more directly than Trolox, is able to rescue the p53 stabilization response to ionizing radiation. A diagram illustrating how the impairment of the AKT pathway can lead to p53 protein degradation through GSK-3 is shown in Fig 4C.
Fig. 4. p53 protein is degraded by the proteasome and rescued by insulin in RP mutants. 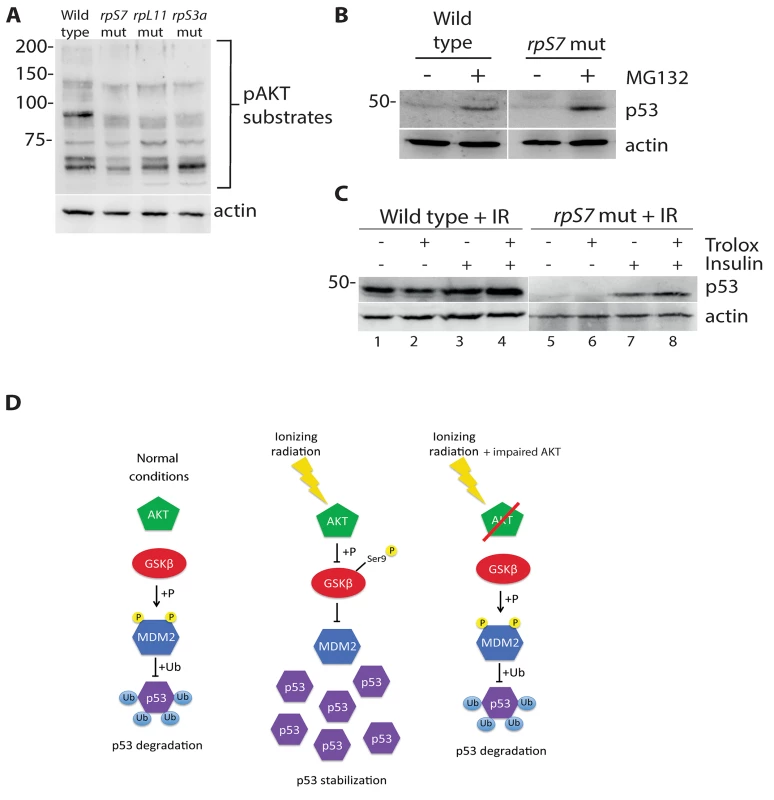
A) Western blot analysis of p53 levels in 2 dpf wild type or rpS7 mutant cells untreated or exposed to 20μM of the proteasome inhibitor MG132. B) Western blot analysis of p53 protein levels in either wild type or rpS7 mutants exposed to 25 Gy ionizing radiation followed by the addition of 350nM insulin and/or 10mM Trolox. C) Model illustrating how impaired AKT activity by RP mutations may result in the failure of p53 to stabilize in response to ionizing radiation. GSK-3 inhibition restores p53 in RP mutant cells
We hypothesized that the impairment of AKT activity observed in cells with RP mutations could cause constitutive activation of GSK-3, phosphorylation of MDM2, and ultimately resulting in the constitutive degradation of p53. Therefore we reasoned that the inhibition of GSK-3 with lithium chloride (LiCl) in cells with RP mutations could restore p53 stabilization. Fig 5A and 5B show a partial rescue of p53 stabilization in rpL11 and rpS7 mutant embryos exposed to ionizing radiation followed by 6 hours of LiCl treatment. Finally, p53 expression is completely restored in rpS7 haploinsufficient MPNST tumor cells when the cells are plated overnight with different dosages of LiCl (Fig 5C). This figure also shows LiCl induces the expression of what may be different isoforms of p53 such as p53Δ113 resulting from restored transcriptional activity of p53 in the tumor cells, for LiCl treatment of MPNST tumor cells expressing the transcriptionally impaired p53M214K/M214K mutant does not result in the appearance of same bands (Fig 5D). However we are still unable to state for certain what the exact identity of these bands are, although they appear to be specific to p53 activation. These data strongly implicate the impairment of AKT, in particular the loss of AKT inhibition of GSK-3, as a driving force behind the high levels of constitutive degradation of p53 observed in tumor cells with RP gene haploinsufficiency.
Fig. 5. Lithium chloride restores p53 stabilization in RP mutant embryos and tumor cells. 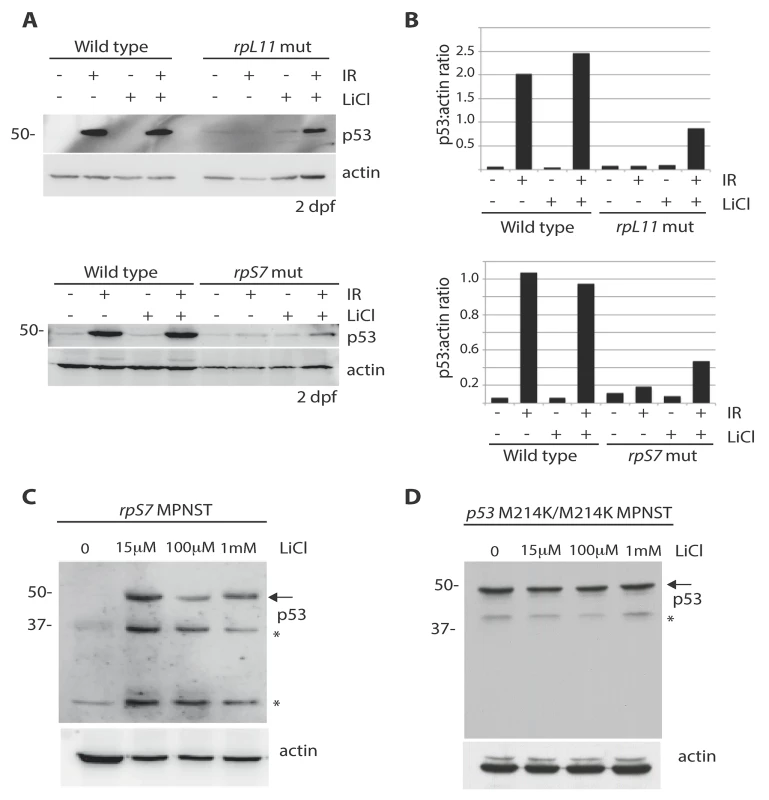
A) Western blot analysis of p53 levels in 2 dpf wild type, rpS7, or rpL11 mutant embryos either untreated or exposed to 25 Gy ionizing radiation followed by the addition of 100μM LiCl. Note the partial rescue of p53 stabilization in the RP mutant embryos when exposed to LiCl after IR. B) Quantification of the p53:actin ratio of the western blots from (A). C) Western blot analysis of p53 levels in rpS7 MPNST tumor cells untreated or incubated with varying concentrations of LiCl. D) Western blot analysis of p53 levels in p53 M214K/M214K MPNST tumor cells untreated or incubated with varying concentrations of LiCl. * indicates either a p53-specific isoform or a degradation product. Discussion
While stabilization of the p53 tumor suppressor has long been held the culprit for the cytopenia phenotype observed in human diseases linked to RP gene mutations, it has been a decade-long mystery as to why no p53 protein is detectable in RP haploinsufficient tumor cells. In the context of the RP mutant zebrafish embryos, we show that a similar loss of p53 protein is evident as the embryos age to 2 dpf, the maternal stores of RPs are depleted, and the RP deficiency resulting from the mutation becomes more severe. We do not believe that this loss of p53 in RP mutants is simply due to the inability of the cells to make protein per se, for in other zebrafish models of ribosome biogenesis deficiency such as nucleostemin, gnl2, and nop10 mutants we are able to visualize robust p53 stabilization by western blot analysis even in the absence of ionizing radiation up until 4 dpf [5,6]. Interestingly, the nop10, gnl2, and nucleostemin genes all code for proteins with important roles in early ribosome biogenesis and the processing of rRNA. While it has been shown that there are indeed detectable defects in rRNA processing in DBA patient CD34+ cells with RP mutations, these are by and large more subtle than the defects we observe upon the loss of nop10, gnl2, or nucleostemin [5,6,42]. This suggests that, as with actinomycin D, defective rRNA processing is a critical mediator of p53 stabilization and may in fact be a causative agent in the cytopenia phenotype of human diseases such as dyskeratosis congenita. However, our data suggest the molecular pathology underlying the anemia in diseases linked to RP mutations is likely to implicate mechanisms that go beyond stabilization of the p53 protein.
The fact that others and we observe no difference in early hematopoietic stem cell expression in RP mutant zebrafish embryos coupled to the anemia failing to be rescued upon p53 loss suggests that the anemia phenotype cannot be explained by the organism’s general p53 response to the ribosome biogenesis defects induced by RP loss [30]. Clearly the embryos younger than 1 dpf are experiencing some p53-induced apoptosis otherwise we would see no AO staining rescue upon injection of the p53 MO and no partial rescue of the morphological phenotypes. We therefore propose that p53 has such an early effect in the developing RP mutant embryo that by the time of 1 dpf (approximately 30 hours post fertilization is when the experiments would begin) all that we are able to observe by laboratory techniques are the apoptotic cells in the wake of a brief p53 activation. The remaining live cells at 1 dpf appear devoid of p53 protein, and if the apoptotic cells do still carry p53 protein we cannot detect it by western blotting. We were unable to detect p53 by western blotting in any embryo younger than 1 dpf, but this is likely a technical issue reflecting the abundance of yolk protein still present at this early stage. The apoptotic cells rescued by p53 loss that we are able to observe at 1 dpf also do not appear to be important for red cell development, as we detect no rescue of the anemia phenotype upon p53 loss.
The results of TUNEL assay suggests that some DNA fragmentation upon acute DNA damage is still possible in the 2 dpf RP mutant cells despite no evidence of p53 stabilization or caspase activation. Other studies have reported similar findings of caspase-independent DNA fragmentation in cancer epithelial cells in response to diverse toxins, and the authors of this work suggest that the DNA damage induced by these toxins may lead to noncaspase-mediated proteolytic activation of DNases [43]. Interestingly, studies in Drosophila have shown that ionizing radiation leads to cells acquiring RP haploinsufficiency, and that these cells undergo apoptosis that is also p53-independent [44]. Taken all together, our work supports a model where the impairment of mature red blood cell formation in organisms with RP deficiency is due to cell loss that is not dependent on either p53 stabilization or caspase activation. However, it should be pointed out here that the work in this study is entirely based on zebrafish models of DBA, which carry homozygous RP mutations (as noted in the introduction, the haploinsufficient mutants have no embryonic phenotype except a slight growth defect). Therefore it may be that p53 has a more prominent role in cells with true RP haploinsufficiency, and this remains an issue that must be kept in mind when interpreting data generated from zebrafish models of DBA.
While other studies have suggested that the p53 MO or p53M214K/M214K mutant background rescues both general apoptosis and the number of hemoglobin-expressing cells in embryos deficient for RPs [15,38], we believe our genetic models are more consistent than using morphants and that our approach to quantifying the mutant phenotypes are both more robust (we use sample sizes N > 100) and reliable (we genotype the embryos after the blind scoring to resolutely identify the homozygotes). Other studies support us by demonstrating that p53 independent pathways are contributing to the anemia phenotype of RP-mutant zebrafish embryos, and that the loss of p53 rescues the morphological abnormalities but not the anemia phenotype of embryos with reduced RP expression [45–47].
It remains an open question as to what the major pathways beyond p53 and caspase activation contribute to the death of RP deficient cells and the anemia phenotype in zebrafish models of DBA. Interestingly, a recent study has shown that the mutation of rpS19 in zebrafish embryo erythrocytes specifically reduces the translation, but not the transcription of the hemoglobin gene hbbe3 [30]. We also recently reported that the up regulation of autophagy, the cellular process of self-digestion that is tightly regulated during hematopoiesis, is observed in both zebrafish models of DBA and in human DBA cells with RP haploinsufficiency [16]. It may therefore be that RP loss in maturing erythrocytes derails their proper differentiation by failing to translate critical mRNAs, or by the constitutive activation of autophagy resulting in the erythrocyte progenitor cells essentially eating themselves before they are able to fully differentiate. The former possibility is supported by several findings suggesting that L-leucine, an amino acid that increases translation by activating the mTOR pathway, is able to partially rescue the anemia phenotype of zebrafish RP morphants and increases the number of erythroid cells in red cell culture assays where CD34+ cells are infected with shRNAs against RPS19 or RPS14 [47,48]. The latter possibility is enticing for one would not expect constitutively up regulated autophagy to elicit an apoptotic response. Or the cells may be undergoing an as of yet unknown p53 - and caspase-independent mechanism of cell death that awaits identification.
We had previously reported that the addition of MG132 to the MPNST cells with RP gene haploinsufficiency was not able to restore p53 stabilization [20]. At the time this led us to believe that the block in p53 expression was at the level of protein synthesis, since MG132 should be able to stabilize any protein undergoing ubiquitination and proteasomal degradation. However our present study suggests otherwise, that in fact the MG132 treatment is not sufficient to restore p53 stabilization in RP mutant MPNST cells. There may be additional factors at the tumor level contributing to the degradation of p53 that are as of yet not known, or it may be that the robustness of the degradation in the tumors is much stronger than in the embryos. While our present study is not able to delineate between these possibilities, it may be that more powerful proteasome inhibitors are capable of restoring p53 to the same levels as we observe with the LiCl treatment of tumor cells, and these studies will be of great future interest. It should also be pointed out here that given the very wide range of effects that LiCl has on many cellular signaling pathways, there may be other effects beyond the AKT pathway that are contributing to the restoration of p53 that we observe.
Very recent work suggests that a mechanism for malignant transformation in RP-haploinsufficient cells involves cells acquiring oncogenic mutations that allow for the bypass of a 60S ribosomal subunit integrity checkpoint [49]. We propose that these mutations (which have yet to be identified) coupled to excessive degradation of wild type p53 would be sufficient to promote malignant transformation. Interestingly, the T-ALL cells with RPL5 and RPL10 mutations, similar to the RP-haploinsufficient zebrafish tumor cells, the p53 gene remains wild type (personal communication with Dr. De Keersmaecker). We thus posit that cells with RP deficiencies are able to exert selective pressure to overcome the p53 activity not just by acquiring p53 gene mutations but also by suppressing AKT activity.
DBA is a rare disease with a prevalence that is approximately 7 in 1 million live births [50]. In addition, since the majority of DBA patients die early in life from bone marrow failure or from complications stemming from chronic blood transfusions, the number of DBA patients who ultimately experience malignant transformation is very low (it would be next to impossible for us to obtain DBA patient-derived tumor tissue for p53 analysis). In the absence of a mammalian RP mutant cancer model, the zebrafish RP mutants to date remain the best possible option for molecular studies of RP mutation-driven tumors. That said, we believe our results may still have some important implications for human leukemia. For example, acute myeloid leukemia (AML) has very low rates of p53 genetic inactivation (similar to the RP mutant MPNSTs) compared to very high rates in many other tumor types [51,52]. Inhibition of GSK-3 with small molecules in AML cells leads to increased differentiation, impaired growth and proliferation, and the induction of apoptosis [53]. Interestingly, the immunomodulating agent lenalidomide, used to treat multiple myeloma, results in the phosphorylation of GSK-3 at the same serine residues that inhibit GSK-3 phosphorylation of MDM2 [54]. This drug has also been reported in 5q-MDS patients to result in increased survival and a reduced risk of transformation to AML [55], raising the possibility that one mechanism of action of lenalidomide is to inhibit GSK-3 phosphorylation of MDM2 and restore a normal p53 response to the acquisition of oncogenic mutations. In sum, we suggest the new mechanisms driving p53 loss that we report here may be useful pathways to target in some cancers.
Materials and Methods
Zebrafish strains and maintenance
Zebrafish mutants were created and maintained as described [19,28,56]. Animal experiments were conducted in accordance with the Dutch guidelines for the care and use of laboratory animals, with the approval of the Animal Experimentation Committee (Dier Experimenten Commissie) of the Royal Netherlands Academy of Arts and Sciences (Koninklijke Nederlandse Akademie van Wetenschappen [KNAW] (Protocol # 08.2001).
In situ hybridizations
Performed as previously described using probes against scl [31] and βE1-globin [57]. At least three independent stains were performed with clutches > 60 embryos. Clutches were stained simultaneously and mutants confirmed afterwards using Sanger sequencing, as previously described [58].
Morphology
Embryos were sorted by gross phenotype and photographed with a Leica MZ FLIII microscope. At least three independent clutches were analyzed and the expected Mendelian ratio of mutants was always observed. Mutants were confirmed afterwards using Sanger sequencing, as previously described [58].
Morpholino injections
Embryos were microinjected with ~1ng MO at the one-cell stage. The p53 MO (Gene Tools, Inc. Philomath, OR, USA) is 5’-GCGCCATTGCTTTGCAAGAATTG-3’ and has been previously described [59]. The missense MO sequence is 5'-CATGTTCAACTATGTGTTAGCTTCA-3' (Gene Tools, Inc. Philomath, OR, USA)
Acridine orange staining and cell counting
Live 1 dpf embryos were incubated in E3-embryo medium + 10μg/mL AO stain (Sigma) in the dark for 30 min. Photos were obtained using a Leica MZ FLIII microscope and cells were blindly counted within a defined area that included the tail starting at the dorsal end of the yolk extension using Image J v1.44 software. At least 8 animals per condition were used for counting. Embryos were genotyped afterwards using Sanger sequencing, as previously described [58].
O-dianisidine staining
At least 100 embryos per clutch of a mating between two heterozygous hi1034b fish were injected at the one-cell stage with MOs as described above. At 2 dpf they were stained with o-dianisidine (Sigma) as previously described [16]. Scoring of the phenotype severity was done blindly. Embryos were genotyped afterwards using Sanger sequencing as previously described [58].
TUNEL assays
2 dpf embryos were untreated or exposed to 25 Gy of ionizing radiation. Six hours after irradiation TUNEL assays were performed as previously described and TUNEL-positive cells counted as for AO staining [6].
Caspase assays
Embryos at 2 dpf were either untreated or subjected to 25 Gy ionizing radiation, and mechanically lysed 6 hours later with a P200 pipette (each sample = 3 embryos per well of a 96-well plate) using 100μL of the western blotting lysis buffer described above. The Caspase-Glo® 2 or 3/7 assay (Promega, Madison, WI, USA) was then performed per the manufacturer’s instructions.
cDNA synthesis and quantitative PCR
5 embryos per sample were added to 100μL Trizol (Life Technologies, Carlsbad, CA, USA), RNA was isolated and used to make cDNA with the iScript cDNA Synthesis Kit (Bio-Rad, Hercules, CA, USA) per the manufacturer’s instructions. Primers used were as follows: p53 forward 5’ - GCTTGTCACAGGGGTCATTT-3’, p53 reverse 5’-ACAAAGGTCCCAGTGGAGTG-3’, GAPDH forward 5’-GGATCTGACAGTCCGTCTTGAGAA-3’, GAPDH reverse 5’ - CCATTGAAGTCAGTGGACACAACC, actin forward 5’-GCCCATCTATGAGGGTTACG-3’, actin reverse 5’-GCAAGATTCCATACCCAGGA-3’. Quantitative PCR (qPCR) was performed using iQ SYBR Green Super Mix and a MyiQ Single-Color PCR thermal cycler (Biorad, Hercules, CA, USA). Each experiment was performed in biological triplicate. p53 mRNA expression in mutants relative to wild types was normalized to GAPDH and calculated according to the Cτ method. Semi-quantitative analysis was performed with a standard PCR method using either p53 or actin primers, the products run on a 1% agarose gel. Statistics were performed with a Student’s t-test.
Western blotting
Embryos were subject to 25 Gy ionizing radiation and then lysed 6 hours later. 350nM insulin, 10mM Trolox or 100μM LiCl (all from Sigma) was added to the E3 media immediately following the ionizing radiation. For the MG132 (Sigma) experiment, embryo cells were dissociated mechanically with a 200μL pipette tip and added to DMEM media +10% FCS with or without 20μM MG132 for 6 hours at 28°C before lysing the cells. The zebrafish specific p53 antibody and western blotting of zebrafish embryos and MPNST cells has been previously described [20]. For MPNST cells the tumor was dissected, cells mechanically dissociated with a P200 pipette, and plated in a 6 well dish with the indicated concentration of LiCl at 28°C in DMEM + 10% fetal calf serum (Life Technologies, Carlsbad, CA, USA) media overnight. 50μg of total protein from the MPNST cells was used for each sample, measured by Bradford assay (BioRad, Hercules, CA, USA). Other antibodies used at 1 : 1000 dilution included anti-phospho-AKT (Cell Signaling #9275) and anti-actin (Santa Cruz Biotech, #sc-1616, Santa Cruz, CA, USA). Antibodies used at 1 : 5000 included donkey anti-goat IgG-HRP (Santa Cruz Biotech, #sc-2020, Santa Cruz, CA, USA) and sheep anti-mouse IgG-HRP (GE Healthcare, #NA931, Little Chalfont, Buckinghamshire, United Kingdom). Quantifications of western blots were performed using Image J v1.44 software. Unless otherwise stated, all embryos subject to western blotting were 2 dpf.
Supporting Information
Zdroje
1. Lane DP (2005) Exploiting the p53 pathway for the diagnosis and therapy of human cancer. Cold Spring Harb Symp Quant Biol 70 : 489–497. 16869788
2. Horn HF, Vousden KH (2007) Coping with stress: multiple ways to activate p53. Oncogene 26 : 1306–1316. 17322916
3. Carrillo J, Gonzalez A, Manguan-Garcia C, Pintado-Berninches L, Perona R (2014) p53 pathway activation by telomere attrition in X-DC primary fibroblasts occurs in the absence of ribosome biogenesis failure and as a consequence of DNA damage. Clin Transl Oncol 16 : 529–538. doi: 10.1007/s12094-013-1112-3 24065372
4. Ceccaldi R, Parmar K, Mouly E, Delord M, Kim JM, et al. (2012) Bone marrow failure in Fanconi anemia is triggered by an exacerbated p53/p21 DNA damage response that impairs hematopoietic stem and progenitor cells. Cell Stem Cell 11 : 36–49. doi: 10.1016/j.stem.2012.05.013 22683204
5. Essers PB, Pereboom TC, Goos YJ, Paridaen JT, Macinnes AW (2014) A comparative study of nucleostemin family members in zebrafish reveals specific roles in ribosome biogenesis. Dev Biol 385 : 304–315. doi: 10.1016/j.ydbio.2013.10.029 24211311
6. Pereboom TC, van Weele LJ, Bondt A, MacInnes AW (2011) A zebrafish model of dyskeratosis congenita reveals hematopoietic stem cell formation failure resulting from ribosomal protein-mediated p53 stabilization. Blood 118 : 5458–5465. doi: 10.1182/blood-2011-04-351460 21921046
7. Azuma M, Toyama R, Laver E, Dawid IB (2006) Perturbation of rRNA synthesis in the bap28 mutation leads to apoptosis mediated by p53 in the zebrafish central nervous system. J Biol Chem 281 : 13309–13316. 16531401
8. Dutt S, Narla A, Lin K, Mullally A, Abayasekara N, et al. (2011) Haploinsufficiency for ribosomal protein genes causes selective activation of p53 in human erythroid progenitor cells. Blood 117 : 2567–2576. doi: 10.1182/blood-2010-07-295238 21068437
9. Moniz H, Gastou M, Leblanc T, Hurtaud C, Cretien A, et al. (2012) Primary hematopoietic cells from DBA patients with mutations in RPL11 and RPS19 genes exhibit distinct erythroid phenotype in vitro. Cell Death Dis 3: e356. doi: 10.1038/cddis.2012.88 22833095
10. Vlachos A, Rosenberg PS, Atsidaftos E, Alter BP, Lipton JM (2012) Incidence of neoplasia in Diamond Blackfan anemia: a report from the Diamond Blackfan Anemia Registry. Blood 119 : 3815–3819. doi: 10.1182/blood-2011-08-375972 22362038
11. Boultwood J, Lewis S, Wainscoat JS (1994) The 5q-syndrome. Blood 84 : 3253–3260. 7949083
12. De Keersmaecker K, Atak ZK, Li N, Vicente C, Patchett S, et al. (2013) Exome sequencing identifies mutation in CNOT3 and ribosomal genes RPL5 and RPL10 in T-cell acute lymphoblastic leukemia. Nature genetics 45 : 186–190. doi: 10.1038/ng.2508 23263491
13. Lawrence MS, Stojanov P, Mermel CH, Robinson JT, Garraway LA, et al. (2014) Discovery and saturation analysis of cancer genes across 21 tumour types. Nature 505 : 495–501. doi: 10.1038/nature12912 24390350
14. Novetsky AP, Zighelboim I, Thompson DM Jr., Powell MA, Mutch DG, et al. (2013) Frequent mutations in the RPL22 gene and its clinical and functional implications. Gynecol Oncol 128 : 470–474. doi: 10.1016/j.ygyno.2012.10.026 23127973
15. Chakraborty A, Uechi T, Higa S, Torihara H, Kenmochi N (2009) Loss of ribosomal protein L11 affects zebrafish embryonic development through a p53-dependent apoptotic response. PLoS One 4: e4152. doi: 10.1371/journal.pone.0004152 19129914
16. Heijnen HF, van Wijk R, Pereboom TC, Goos YJ, Seinen CW, et al. (2014) Ribosomal protein mutations induce autophagy through S6 kinase inhibition of the insulin pathway. PLoS Genet 10: e1004371. doi: 10.1371/journal.pgen.1004371 24875531
17. Lai K, Amsterdam A, Farrington S, Bronson RT, Hopkins N, et al. (2009) Many ribosomal protein mutations are associated with growth impairment and tumor predisposition in zebrafish. Dev Dyn 238 : 76–85. doi: 10.1002/dvdy.21815 19097187
18. Amsterdam A, Sadler KC, Lai K, Farrington S, Bronson RT, et al. (2004) Many ribosomal protein genes are cancer genes in zebrafish. PLoS Biol 2: E139. 15138505
19. Berghmans S, Murphey RD, Wienholds E, Neuberg D, Kutok JL, et al. (2005) tp53 mutant zebrafish develop malignant peripheral nerve sheath tumors. Proc Natl Acad Sci U S A 102 : 407–412. 15630097
20. MacInnes AW, Amsterdam A, Whittaker CA, Hopkins N, Lees JA (2008) Loss of p53 synthesis in zebrafish tumors with ribosomal protein gene mutations. Proceedings of the National Academy of Sciences of the United States of America 105 : 10408–10413. doi: 10.1073/pnas.0805036105 18641120
21. Choong ML, Yang H, Lee MA, Lane DP (2009) Specific activation of the p53 pathway by low dose actinomycin D: a new route to p53 based cyclotherapy. Cell Cycle 8 : 2810–2818. 19657224
22. Chen CS, Ho DR, Chen FY, Chen CR, Ke YD, et al. (2014) AKT mediates actinomycin D-induced p53 expression. Oncotarget 5 : 693–703. 24525337
23. Alessi DR, Andjelkovic M, Caudwell B, Cron P, Morrice N, et al. (1996) Mechanism of activation of protein kinase B by insulin and IGF-1. The EMBO journal 15 : 6541–6551. 8978681
24. Cross DA, Alessi DR, Cohen P, Andjelkovich M, Hemmings BA (1995) Inhibition of glycogen synthase kinase-3 by insulin mediated by protein kinase B. Nature 378 : 785–789. 8524413
25. Kulikov R, Boehme KA, Blattner C (2005) Glycogen synthase kinase 3-dependent phosphorylation of Mdm2 regulates p53 abundance. Molecular and cellular biology 25 : 7170–7180. 16055726
26. Kwon O, Kim KA, He L, Kim SO, Kim MS, et al. (2008) Ionizing radiation can induce GSK-3beta phosphorylation and NF-kappaB transcriptional transactivation in ATM-deficient fibroblasts. Cell Signal 20 : 602–612. doi: 10.1016/j.cellsig.2007.10.022 18243662
27. Boehme KA, Kulikov R, Blattner C (2008) p53 stabilization in response to DNA damage requires Akt/PKB and DNA-PK. Proceedings of the National Academy of Sciences of the United States of America 105 : 7785–7790.
28. Golling G, Amsterdam A, Sun Z, Antonelli M, Maldonado E, et al. (2002) Insertional mutagenesis in zebrafish rapidly identifies genes essential for early vertebrate development. Nature genetics 31 : 135–140. 12006978
29. Boria I, Garelli E, Gazda HT, Aspesi A, Quarello P, et al. (2010) The ribosomal basis of Diamond-Blackfan Anemia: mutation and database update. Human mutation 31 : 1269–1279. doi: 10.1002/humu.21383 20960466
30. Zhang Y, Ear J, Yang Z, Morimoto K, Zhang B, et al. (2014) Defects of protein production in erythroid cells revealed in a zebrafish Diamond-Blackfan anemia model for mutation in RPS19. Cell Death Dis 5: e1352. doi: 10.1038/cddis.2014.318 25058426
31. Juarez MA, Su F, Chun S, Kiel MJ, Lyons SE (2005) Distinct roles for SCL in erythroid specification and maturation in zebrafish. J Biol Chem 280 : 41636–41644. 16210319
32. Davidson AJ, Zon LI (2004) The 'definitive' (and 'primitive') guide to zebrafish hematopoiesis. Oncogene 23 : 7233–7246. 15378083
33. Mikkola HK, Klintman J, Yang H, Hock H, Schlaeger TM, et al. (2003) Haematopoietic stem cells retain long-term repopulating activity and multipotency in the absence of stem-cell leukaemia SCL/tal-1 gene. Nature 421 : 547–551. 12540851
34. Tsiftsoglou AS, Vizirianakis IS, Strouboulis J (2009) Erythropoiesis: model systems, molecular regulators, and developmental programs. IUBMB Life 61 : 800–830. doi: 10.1002/iub.226 19621348
35. Danilova N, Sakamoto KM, Lin S (2011) Ribosomal protein L11 mutation in zebrafish leads to haematopoietic and metabolic defects. British journal of haematology 152 : 217–228. doi: 10.1111/j.1365-2141.2010.08396.x 21114664
36. Tucker B, Lardelli M (2007) A rapid apoptosis assay measuring relative acridine orange fluorescence in zebrafish embryos. Zebrafish 4 : 113–116. 18041929
37. MacInnes AW, Amsterdam A, Whittaker CA, Hopkins N, Lees JA (2008) Loss of p53 synthesis in zebrafish tumors with ribosomal protein gene mutations. Proc Natl Acad Sci U S A 105 : 10408–10413. doi: 10.1073/pnas.0805036105 18641120
38. Taylor AM, Humphries JM, White RM, Murphey RD, Burns CE, et al. (2012) Hematopoietic defects in rps29 mutant zebrafish depend upon p53 activation. Exp Hematol 40 : 228–237 e225. doi: 10.1016/j.exphem.2011.11.007 22120640
39. Danilova N, Sakamoto KM, Lin S (2008) Ribosomal protein S19 deficiency in zebrafish leads to developmental abnormalities and defective erythropoiesis through activation of p53 protein family. Blood 112 : 5228–5237. doi: 10.1182/blood-2008-01-132290 18515656
40. Duan J, Ba Q, Wang Z, Hao M, Li X, et al. (2011) Knockdown of ribosomal protein S7 causes developmental abnormalities via p53 dependent and independent pathways in zebrafish. The international journal of biochemistry & cell biology 43 : 1218–1227.
41. Chen J, Ng SM, Chang C, Zhang Z, Bourdon JC, et al. (2009) p53 isoform delta113p53 is a p53 target gene that antagonizes p53 apoptotic activity via BclxL activation in zebrafish. Genes Dev 23 : 278–290. doi: 10.1101/gad.1761609 19204115
42. Flygare J, Aspesi A, Bailey JC, Miyake K, Caffrey JM, et al. (2007) Human RPS19, the gene mutated in Diamond-Blackfan anemia, encodes a ribosomal protein required for the maturation of 40S ribosomal subunits. Blood 109 : 980–986. 16990592
43. Cummings BS, Kinsey GR, Bolchoz LJ, Schnellmann RG (2004) Identification of caspase-independent apoptosis in epithelial and cancer cells. J Pharmacol Exp Ther 310 : 126–134. 15028782
44. McNamee LM, Brodsky MH (2009) p53-independent apoptosis limits DNA damage-induced aneuploidy. Genetics 182 : 423–435. doi: 10.1534/genetics.109.102327 19364807
45. Jia Q, Zhang Q, Zhang Z, Wang Y, Zhang W, et al. (2013) Correction: Transcriptome Analysis of the Zebrafish Model of Diamond-Blackfan Anemia from RPS19 Deficiency via p53-Dependent and-Independent Pathways. PLoS One 8.
46. Torihara H, Uechi T, Chakraborty A, Shinya M, Sakai N, et al. (2011) Erythropoiesis failure due to RPS19 deficiency is independent of an activated Tp53 response in a zebrafish model of Diamond-Blackfan anaemia. Br J Haematol 152 : 648–654. doi: 10.1111/j.1365-2141.2010.08535.x 21223253
47. Yadav GV, Chakraborty A, Uechi T, Kenmochi N (2014) Ribosomal protein deficiency causes Tp53-independent erythropoiesis failure in zebrafish. Int J Biochem Cell Biol 49 : 1–7. doi: 10.1016/j.biocel.2014.01.006 24417973
48. Payne EM, Virgilio M, Narla A, Sun H, Levine M, et al. (2012) L-Leucine improves the anemia and developmental defects associated with Diamond-Blackfan anemia and del(5q) MDS by activating the mTOR pathway. Blood 120 : 2214–2224. doi: 10.1182/blood-2011-10-382986 22734070
49. Sulima SO, Patchett S, Advani VM, De Keersmaecker K, Johnson AW, et al. (2014) Bypass of the pre-60S ribosomal quality control as a pathway to oncogenesis. Proc Natl Acad Sci U S A 111 : 5640–5645. doi: 10.1073/pnas.1400247111 24706786
50. Vlachos A, Ball S, Dahl N, Alter BP, Sheth S, et al. (2008) Diagnosing and treating Diamond Blackfan anaemia: results of an international clinical consensus conference. British journal of haematology 142 : 859–876. doi: 10.1111/j.1365-2141.2008.07269.x 18671700
51. Fenaux P, Preudhomme C, Quiquandon I, Jonveaux P, Lai JL, et al. (1992) Mutations of the P53 gene in acute myeloid leukaemia. British journal of haematology 80 : 178–183. 1550773
52. Vogelstein B, Lane D, Levine AJ (2000) Surfing the p53 network. Nature 408 : 307–310. 11099028
53. Banerji V, Frumm SM, Ross KN, Li LS, Schinzel AC, et al. (2012) The intersection of genetic and chemical genomic screens identifies GSK-3alpha as a target in human acute myeloid leukemia. J Clin Invest 122 : 935–947. doi: 10.1172/JCI46465 22326953
54. Bjorklund CC, Ma W, Wang ZQ, Davis RE, Kuhn DJ, et al. (2011) Evidence of a role for activation of Wnt/beta-catenin signaling in the resistance of plasma cells to lenalidomide. The Journal of biological chemistry 286 : 11009–11020. doi: 10.1074/jbc.M110.180208 21189262
55. List AF, Bennett JM, Sekeres MA, Skikne B, Fu T, et al. (2014) Extended survival and reduced risk of AML progression in erythroid-responsive lenalidomide-treated patients with lower-risk del(5q) MDS. Leukemia 28 : 1033–1040. doi: 10.1038/leu.2013.305 24150217
56. Westerfield M (2000) The zebrafish book. A guide for the laboratory use of zebrafish (Danio rerio). Eugene, OR: Univ. of Oregon Press.
57. Burns CE, Traver D, Mayhall E, Shepard JL, Zon LI (2005) Hematopoietic stem cell fate is established by the Notch-Runx pathway. Genes Dev 19 : 2331–2342. 16166372
58. van Rooijen E, Voest EE, Logister I, Korving J, Schwerte T, et al. (2009) Zebrafish mutants in the von Hippel-Lindau tumor suppressor display a hypoxic response and recapitulate key aspects of Chuvash polycythemia. Blood 113 : 6449–6460. doi: 10.1182/blood-2008-07-167890 19304954
59. Langheinrich U, Hennen E, Stott G, Vacun G (2002) Zebrafish as a model organism for the identification and characterization of drugs and genes affecting p53 signaling. Curr Biol 12 : 2023–2028. 12477391
Štítky
Genetika Reprodukční medicína
Článek Discovery and Fine-Mapping of Glycaemic and Obesity-Related Trait Loci Using High-Density ImputationČlánek AAA-ATPase FIDGETIN-LIKE 1 and Helicase FANCM Antagonize Meiotic Crossovers by Distinct MechanismsČlánek A Conserved Pattern of Primer-Dependent Transcription Initiation in and Revealed by 5′ RNA-seqČlánek TopBP1 Governs Hematopoietic Stem/Progenitor Cells Survival in Zebrafish Definitive HematopoiesisČlánek Redundant Roles of Rpn10 and Rpn13 in Recognition of Ubiquitinated Proteins and Cellular Homeostasis
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2015 Číslo 7
-
Všechny články tohoto čísla
- LINE-1 Retroelements Get ZAPped!
- /p23: A Small Protein Heating Up Lifespan Regulation
- Hairless Streaks in Cattle Implicate TSR2 in Early Hair Follicle Formation
- Ribosomal Protein Mutations Result in Constitutive p53 Protein Degradation through Impairment of the AKT Pathway
- Molecular Clock of Neutral Mutations in a Fitness-Increasing Evolutionary Process
- Modeling Implicates in Nephropathy: Evidence for Dominant Negative Effects and Epistasis under Anemic Stress
- The Alternative Sigma Factor SigX Controls Bacteriocin Synthesis and Competence, the Two Quorum Sensing Regulated Traits in
- BMP Inhibition in Seminomas Initiates Acquisition of Pluripotency via NODAL Signaling Resulting in Reprogramming to an Embryonal Carcinoma
- Comparative Study of Regulatory Circuits in Two Sea Urchin Species Reveals Tight Control of Timing and High Conservation of Expression Dynamics
- EIN3 and ORE1 Accelerate Degreening during Ethylene-Mediated Leaf Senescence by Directly Activating Chlorophyll Catabolic Genes in
- Genome Wide Binding Site Analysis Reveals Transcriptional Coactivation of Cytokinin-Responsive Genes by DELLA Proteins
- Sensory Neurons Arouse . Locomotion via Both Glutamate and Neuropeptide Release
- A Year of Infection in the Intensive Care Unit: Prospective Whole Genome Sequencing of Bacterial Clinical Isolates Reveals Cryptic Transmissions and Novel Microbiota
- Inference of Low and High-Grade Glioma Gene Regulatory Networks Delineates the Role of Rnd3 in Establishing Multiple Hallmarks of Cancer
- Novel Role for p110β PI 3-Kinase in Male Fertility through Regulation of Androgen Receptor Activity in Sertoli Cells
- A Novel Locus Harbouring a Functional Nonsense Mutation Identified in a Large Danish Family with Nonsyndromic Hearing Impairment
- Checkpoint Activation of an Unconventional DNA Replication Program in
- A Genetic Incompatibility Accelerates Adaptation in Yeast
- The SMC Loader Scc2 Promotes ncRNA Biogenesis and Translational Fidelity
- Blimp1/Prdm1 Functions in Opposition to Irf1 to Maintain Neonatal Tolerance during Postnatal Intestinal Maturation
- Discovery and Fine-Mapping of Glycaemic and Obesity-Related Trait Loci Using High-Density Imputation
- JAK/STAT and Hox Dynamic Interactions in an Organogenetic Gene Cascade
- Emergence, Retention and Selection: A Trilogy of Origination for Functional Proteins from Ancestral LncRNAs in Primates
- MoSET1 (Histone H3K4 Methyltransferase in ) Regulates Global Gene Expression during Infection-Related Morphogenesis
- Arabidopsis PCH2 Mediates Meiotic Chromosome Remodeling and Maturation of Crossovers
- AAA-ATPase FIDGETIN-LIKE 1 and Helicase FANCM Antagonize Meiotic Crossovers by Distinct Mechanisms
- A Conserved Pattern of Primer-Dependent Transcription Initiation in and Revealed by 5′ RNA-seq
- Tempo and Mode of Transposable Element Activity in Drosophila
- The Shelterin TIN2 Subunit Mediates Recruitment of Telomerase to Telomeres
- SAMHD1 Inhibits LINE-1 Retrotransposition by Promoting Stress Granule Formation
- A Genome Scan for Genes Underlying Microgeographic-Scale Local Adaptation in a Wild Species
- TopBP1 Governs Hematopoietic Stem/Progenitor Cells Survival in Zebrafish Definitive Hematopoiesis
- Analysis of the Relationships between DNA Double-Strand Breaks, Synaptonemal Complex and Crossovers Using the Mutant
- Assessing Mitochondrial DNA Variation and Copy Number in Lymphocytes of ~2,000 Sardinians Using Tailored Sequencing Analysis Tools
- Allelic Spectra of Risk SNPs Are Different for Environment/Lifestyle Dependent versus Independent Diseases
- CSB-PGBD3 Mutations Cause Premature Ovarian Failure
- Irrepressible: An Interview with Mark Ptashne
- Genetic Evidence for Function of the bHLH-PAS Protein Gce/Met As a Juvenile Hormone Receptor
- Inactivation of Retinoblastoma Protein (Rb1) in the Oocyte: Evidence That Dysregulated Follicle Growth Drives Ovarian Teratoma Formation in Mice
- Redundant Roles of Rpn10 and Rpn13 in Recognition of Ubiquitinated Proteins and Cellular Homeostasis
- Pyrimidine Pool Disequilibrium Induced by a Cytidine Deaminase Deficiency Inhibits PARP-1 Activity, Leading to the Under Replication of DNA
- Molecular Framework of a Regulatory Circuit Initiating Two-Dimensional Spatial Patterning of Stomatal Lineage
- RFX2 Is a Major Transcriptional Regulator of Spermiogenesis
- A Role for Macro-ER-Phagy in ER Quality Control
- Corp Regulates P53 in via a Negative Feedback Loop
- Common Cell Shape Evolution of Two Nasopharyngeal Pathogens
- Contact- and Protein Transfer-Dependent Stimulation of Assembly of the Gliding Motility Machinery in
- Endothelial Snail Regulates Capillary Branching Morphogenesis via Vascular Endothelial Growth Factor Receptor 3 Expression
- Functional Constraint Profiling of a Viral Protein Reveals Discordance of Evolutionary Conservation and Functionality
- Temporal Coordination of Carbohydrate Metabolism during Mosquito Reproduction
- mTOR Directs Breast Morphogenesis through the PKC-alpha-Rac1 Signaling Axis
- Reversible Oxidation of a Conserved Methionine in the Nuclear Export Sequence Determines Subcellular Distribution and Activity of the Fungal Nitrate Regulator NirA
- Nutritional Control of DNA Replication Initiation through the Proteolysis and Regulated Translation of DnaA
- Cooperation between Paxillin-like Protein Pxl1 and Glucan Synthase Bgs1 Is Essential for Actomyosin Ring Stability and Septum Formation in Fission Yeast
- Encodes a Highly Conserved Protein Important to Neurological Function in Mice and Flies
- Identification of a Novel Regulatory Mechanism of Nutrient Transport Controlled by TORC1-Npr1-Amu1/Par32
- Aurora-A-Dependent Control of TACC3 Influences the Rate of Mitotic Spindle Assembly
- Large-Scale Phenomics Identifies Primary and Fine-Tuning Roles for CRKs in Responses Related to Oxidative Stress
- TFIIS-Dependent Non-coding Transcription Regulates Developmental Genome Rearrangements
- Genome-Wide Reprogramming of Transcript Architecture by Temperature Specifies the Developmental States of the Human Pathogen
- Identification of Chemical Inhibitors of β-Catenin-Driven Liver Tumorigenesis in Zebrafish
- The Catalytic and Non-catalytic Functions of the Chromatin-Remodeling Protein Collaborate to Fine-Tune Circadian Transcription in
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Functional Constraint Profiling of a Viral Protein Reveals Discordance of Evolutionary Conservation and Functionality
- Reversible Oxidation of a Conserved Methionine in the Nuclear Export Sequence Determines Subcellular Distribution and Activity of the Fungal Nitrate Regulator NirA
- Modeling Implicates in Nephropathy: Evidence for Dominant Negative Effects and Epistasis under Anemic Stress
- Nutritional Control of DNA Replication Initiation through the Proteolysis and Regulated Translation of DnaA
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



