-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaIdentification of a Novel Regulatory Mechanism of Nutrient Transport Controlled by TORC1-Npr1-Amu1/Par32
Cells have evolved a variety of mechanisms to control the permeability of the plasma membrane to face environmental perturbations. Transcriptional regulation, endocytosis, gating and activity control of channels and transporters enable global or specific responses to stressful conditions and focused variations in nutrient availability. Emerging data from the yeast model reveal that the conserved TORC1 pathway regulates arrestin-mediated endocytosis of amino-acid transporters. We provide genetic and biochemical evidence for a novel mechanism enabling TORC1 to regulate the inherent activity of transport proteins via the Amu1/Par32 regulator intermediate. This low complexity protein mediates inhibition of specific proteins dedicated to the transport of ammonium, a favored nitrogen source, underscoring that TORC1 selects transporters to be degraded or transiently inactivated and preserved at the cell surface according to the environmental situation. The here-revealed mechanism of transport inhibition by Amu/Par32 is reminiscent to the inhibition of prokaryotic ammonium transport proteins mediated by PII-type proteins, key nitrogen signal transducers widespread in bacteria and Archaea.
Published in the journal: . PLoS Genet 11(7): e32767. doi:10.1371/journal.pgen.1005382
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1005382Summary
Cells have evolved a variety of mechanisms to control the permeability of the plasma membrane to face environmental perturbations. Transcriptional regulation, endocytosis, gating and activity control of channels and transporters enable global or specific responses to stressful conditions and focused variations in nutrient availability. Emerging data from the yeast model reveal that the conserved TORC1 pathway regulates arrestin-mediated endocytosis of amino-acid transporters. We provide genetic and biochemical evidence for a novel mechanism enabling TORC1 to regulate the inherent activity of transport proteins via the Amu1/Par32 regulator intermediate. This low complexity protein mediates inhibition of specific proteins dedicated to the transport of ammonium, a favored nitrogen source, underscoring that TORC1 selects transporters to be degraded or transiently inactivated and preserved at the cell surface according to the environmental situation. The here-revealed mechanism of transport inhibition by Amu/Par32 is reminiscent to the inhibition of prokaryotic ammonium transport proteins mediated by PII-type proteins, key nitrogen signal transducers widespread in bacteria and Archaea.
Introduction
Proteins of the Mep-Amt-Rh superfamily including the human Rhesus factors mediate the transmembrane transport of ammonium from bacteria to mammals [1–4]. Ammonium, hereafter referring to the sum of NH4+ and NH3, is a key nitrogen source for microorganisms and plants whereas it is mainly documented for its role as a blood pH regulator and for the deleterious consequences it has on the central nervous system upon cytotoxic accumulation in mammals for instance [5–7]. Mep-Amt-Rh proteins adopt a trimeric fold, with a proposed conducting pore crossing each of the three monomers [8–13]. The latter are composed of 11 or 12 helices and are prolonged by a cytosolic C-terminal extension showing conserved peculiarities specific to each of the Mep-Amt and Rh subfamilies [14]. In Escherichia coli, the cytosolic extension of each subunit of the AmtB trimers participates to the interaction with a trimeric association of the PII-type protein GlnK, a negative regulator of the ammonium transport activity [15,16]. Yet, PII-type proteins, key transducers of the nitrogen signal, are only found in bacteria, Archaea and a few plant species [17]. Unravelling the mechanisms and the signalling pathways involved in the activity regulation of eukaryotic Mep-Amt-Rh members remains an open field. We recently showed that the activity of Mep2, one of the three Mep-Amt-Rh proteins enabling ammonium transport in the yeast Saccharomyces cerevisiae, is dynamically controlled by the conserved TORC1 (Target Of Rapamycin Complex) pathway [18,19]. Under poor nitrogen supply, TORC1 downregulation relieves inhibition of the Npr1 kinase which enables phosphorylation of an autoinhibitory domain in the C-terminus of Mep2, thereby favouring ammonium transport. In contrast, preferred nitrogen supplementation activates TORC1 and thus inhibits Npr1. In this case, Mep2 is no longer phosphorylated and plasma-membrane phosphatases Psr1 and Psr2 additionally mediate desphosphorylation of preaccumulated phosphorylated Mep2, enabling autoinhibition of ammonium transport activity [18]. The latter mechanism notably contrasted with the known role of TORC1 and of its effector kinase Npr1 in regulating arrestin–mediated endocytosis of transporters but supported previous studies indicating a multiplicity in the mechanisms of Npr1-mediated regulation of Saccharomyces cerevisiae and Candida albicans Mep proteins [20–25]. Seminal works by Grenson and collaborators led to the isolation of mutations suppressing specific defects of Npr1-lacking cells in either amino-acid uptake, including the mut-2, mut-4 and mut-5 mutations, or in ammonium uptake, like the amu1 mutation [25,26]. Of note the mut loci, also known as npi1, npi2 and npi3, were shown to respectively encode variants of the Rsp5 ubiquitin-ligase, the orthologue of mammalian Nedd4, of the Doa4 ubiquitin-hydrolase, and of Bro1, the orthologue of mammalian Alix/Aip-1 [27–31]. While the characterization of the mut suppressor mutations shed light on the mechanism of ubiquitin-mediated endocytosis of permeases and the role of the multivesicular-body pathway in their delivery to the lysosome/vacuole [27–29,32,33], the nature of Amu1 and of the underlying mechanism of ammonium transport control remain unsolved.
Here, we cloned AMU1 by functional complementation identifying YDL173w/PAR32, a gene of unknown function. We show that the phosphorylation status of Amu1/Par32 is dynamically controlled by TORC1-Npr1 in response to the quality of the nitrogen supply. When the function of Npr1 is inhibited, desphosphorylated Amu1/Par32 accumulates at the cell surface and mediates inhibition of specific ammonium transport proteins. This study unravels a novel mode of plasma-membrane permeability tuning, governed by TORC1 and reminiscent of the GlnK-mediated regulation of prokaryotic ammonium transport proteins.
Results
Mep1 and Mep3 are stable at the plasma membrane in Npr1-lacking cells
All three Mep ammonium transport proteins are most produced and active when wild-type cells are grown in the presence of a non-preferred nitrogen source, the MEP2 gene being strongly expressed compared to MEP1 and MEP3 [19]. In these conditions, all three Mep transport activities largely depend on the integrity of the Npr1 kinase [22].
Npr1 is so far reported to protect amino-acid permeases from arrestin-mediated endocytosis and subsequent degradation, while we recently showed that the kinase also regulates the inherent activity of the Mep2 ammonium transport protein by controlling its phosphorylation state [18,20,21,34]. Upon addressing the influence of Npr1 on the protein levels of Mep1 and Mep3, we found that both proteins are not destabilized in the absence of the kinase (Fig 1a and 1b). Mep1 and Mep3 are respectively detected as at least 3 and 2 main forms, both in wild-type and Npr1-lacking cells. In the latter, the relative abundance of the 3 main Mep1 forms is slightly modified, the faster-running one being more predominant in the absence of the kinase. An essential requirement of Npr1 for the presence of one of these immunodetectable Mep1 and Mep3 forms can however not be deduced from these observations. We nevertheless tested whether the different detected Mep1 and Mep3 forms correspond to differentially phosphorylated states of the proteins. Treating protein extracts from wild-type and Npr1-lacking cells with alkaline phosphatase (ALP) led to a removal of the Mep1 and Mep3 slower-running forms, leaving one main intense state with a mobility similar to the faster-running form observed in the absence of ALP (Fig 1b). Hence, Mep1 and Mep3 exist under different phosphorylated states. Fluorescence microscopy revealed that Npr1 is dispensable for proper cell-surface localization of Mep1-GFP and Mep3-GFP as both proteins appeared evenly localized in the presence as in the absence of the kinase (Fig 1c).
Fig. 1. Npr1 is required for Mep1 and Mep3 inherent transport activity. 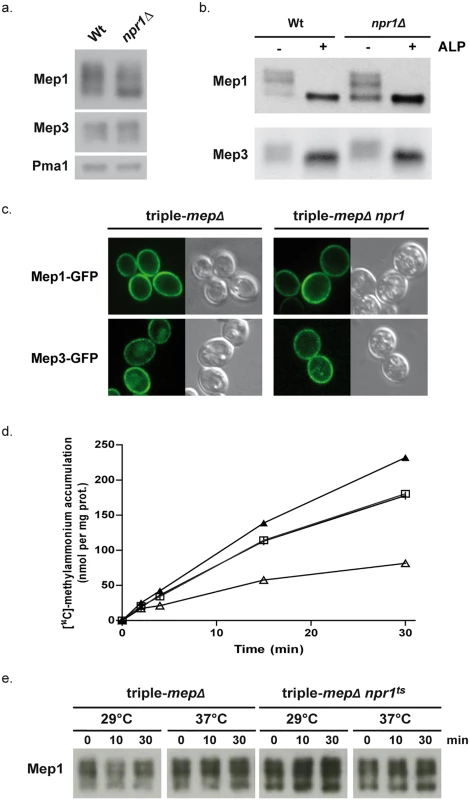
(a) Immunodetection of Mep1 and Mep3 from membrane-enriched cell extracts. Wild-type (23344c) and npr1Δ (30788a) cells were grown in the presence of proline (0.1%) as nitrogen source. The plasma membrane proton ATPase Pma1 was detected as a loading control. (b) Immunodetection of Mep1 and Mep3 from membrane-enriched extracts treated (+) or not (-) with alkaline phosphatase (ALP). Wild-type (23344c) and npr1Δ (30788a) cells were grown in the presence of proline (0.1%) as nitrogen source. (c) Mep1-GFP and Mep3-GFP localization was observed by fluorescence microscopy in triple-mepΔ (31019b) and triple-mepΔ npr1-1 (31052c) cells transformed with the pGAL1Mep1-GFP or pGAL1Mep3-GFP low-copy-number vectors and grown in the presence of proline (0.1%), galactose (3%) and glucose (0.3%). (d) [14C]-methylammonium (0.5 mM) accumulation in proline-grown triple-mepΔ (31019b,│,□) and triple-mepΔ npr1ts (MB063,▲,Δ) cells transformed with YCpMep1 after transfer in a similar medium preheated at 29°C (│,▲) or 37°C (□,Δ). (e) Immunodetection of Mep1 from membrane-enriched extracts of cells collected after the temperature shift as described in Fig 1d. Hence, Npr1 is required for Mep1 and Mep3 inherent transport activity but, in contrast to what has been previously shown for Mep2, the requirement of the kinase does not appear to be simply correlated to the detection of a particular phosphorylated status of the transport proteins.
Npr1 inactivation correlates with Mep1 inactivation
We have previously shown that Npr1 integrity is notably required for the transport activity of Mep1 and Mep3 [22]. We used cells expressing a thermosensitive Npr1 variant [35] to test the consequence of immediate Npr1 inactivation on the Mep-dependent transport activity, focusing on Mep1. In contrast to the low affinity protein Mep3, the Mep1 transport activity can be easily monitored by following accumulation of [14C]-methylammonium (mea), a convenient tracer analogue of ammonium [19,22]. MEP1 was expressed under the control of its own promoter from a low-copy-number centromeric plasmid (YCpMep1) in cells deprived of the three endogenous MEP genes (triple-mepΔ), bearing or not the thermosensitive Npr1 mutation (npr1ts). Inactivation of the Npr1 kinase by shifting npr1ts cells at 37°C clearly reduced the Mep1-dependent accumulation of mea (Fig 1d). Moreover, the initial rate of mea uptake measured just after the shift showed an immediate effect of Npr1 inactivation on the Mep1 activity, revealing a rapid inactivation of Mep1 upon Npr1 inactivation (Triple-mepΔ + YCpMep1 at 29°C: 8.6 nmol min-1 per mg prot; triple-mepΔ + YCpMep1 at 37°C: 6.7 nmol min-1 per mg prot; triple-mepΔ npr1ts + YCpMep1 at 29°C: 8.4 nmol min-1 per mg prot; triple-mepΔ npr1ts +YCpMep1 at 37°C: 1.9 nmol min-1 per mg prot). Immunodetection of Mep1 did not reveal a major impact of Npr1 inactivation on the Mep1 protein level or bands distribution (Fig 1e).
These observations are consistent with Mep1 inherent activity being largely dependent on the maintenance of the Npr1 integrity, and indicate that the activity of plasma-membrane Mep1 is rapidly downregulated upon loss of Npr1 activity.
Mep1 and Mep3 overproduction suppresses the Npr1 requirement
We next evaluated whether overproduction of each of the three Mep proteins could bypass the requirement of Npr1 for their transport function. The Mep functionality was assessed by performing growth tests in the presence of a low ammonium concentration as sole nitrogen source (Fig 2a). Triple-mepΔ cells are unable to grow in the presence of low ammonium concentrations while expression of only one of the three MEP genes enables growth in these conditions [19]. As expected, Mep1 and Mep3 proteins produced from the unique chromosomal MEP1 or MEP3 gene locus were fully dependent on Npr1 for function (Fig 2a) [22]. Expressing the MEP1 and MEP3 genes under their own promoter from a high-copy-number episomal plasmid led to an effective overproduction of the corresponding proteins (Fig 2b). In these conditions, growth on ammonium was observed despite the kinase absence (Fig 2a). MEP2 expression from the episomal plasmid did however not allow Mep2 overproduction (Fig 2b), most likely reflecting the natural high strength of the MEP2 promoter [19]. As expected in this case, the Mep2 function remained totally dependent on Npr1 (Fig 2a).
Fig. 2. Mep1 and Mep3 overproduction suppresses Npr1 requirement. 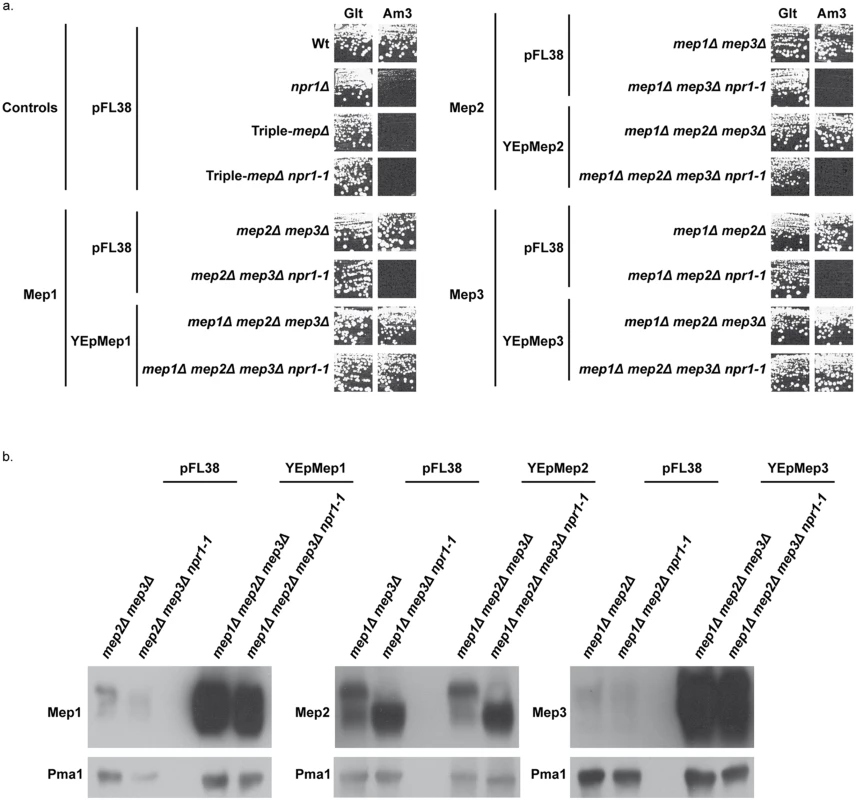
(a) Growth tests on solid medium containing, as the sole nitrogen source, ammonium 3 mM (Am3) or glutamate 0.1% (Glt, positive growth control). Wild-type (23344c), npr1Δ (30788a), mep2Δ mep3Δ (31018b), mep2Δ mep3Δ npr1-1 (31059b), mep1Δ mep3Δ (31022a), mep1Δ mep3Δ npr1-1 (31059d), mep1Δ mep2Δ (31021c) and mep1Δ mep2Δ npr1-1 (31045c) cells were transformed with the empty vector pFL38. Triple-mepΔ (31019b) and triple-mepΔ npr1-1 (31052c) cells were transformed with the empty vector pFL38 and with the high-copy-number plasmids: YEpMep1, YEpMep2 and YEpMep3. (b) Immunodetection of Mep1, Mep2 or Mep3 from membrane-enriched extracts. In the case of Mep2, the cellular extracts were treated with N-glycosidase F. Cells were grown in the presence of proline (0.1%) as nitrogen source. The plasma membrane proton ATPase Pma1 was detected as a loading control. These data reveal that overexpression of the MEP1 and MEP3 genes relieves the Npr1 requirement for the corresponding Mep protein function.
Identification of Amu1 as a negative regulator of ammonium transport
The above findings could be consistent with the existence of a limiting negative factor involved in the Npr1-mediated regulation of Mep1 and Mep3. In 1979, Dubois and Grenson characterized a strain bearing a suppressor mutation in a locus called amu1 (ammonium uptake) enabling yeast growth on low ammonium concentrations despite the lack of a functional Npr1 [25]. Amu1 was proposed to act as a negative regulator of the low-affinity and high-capacity component of the ammonium transport activity of wild-type cells. These data could be consistent with Amu1 regulating Mep1 and/or Mep3, as both proteins possess a lower affinity for their substrate and a higher Vmax compared to Mep2 [19].
Accordingly, while Npr1-lacking cells resist to toxic concentrations of methylammonium, a non-metabolizable ammonium analogue transported via Mep1 and Mep3, cells further bearing a mutated amu1 locus do not grow in these conditions (Fig 3). We used the latter phenotype to clone the AMU1 gene by functional complementation of amu1-1 npr1-1 ura3 cells using a genomic library of the Σ1278b strain and selecting clones able to grow in the presence of a toxic concentration of methylammonium. The selected clones were next controlled for loss of growth on ammonium as sole nitrogen source (Fig 3). Analysis of the plasmidic content of these clones revealed that they all were transformed by a plasmid bearing one common gene, namely the YDL173w ORF.
Fig. 3. The amu1 suppressor mutation restores growth of Npr1-lacking cells on ammonium. 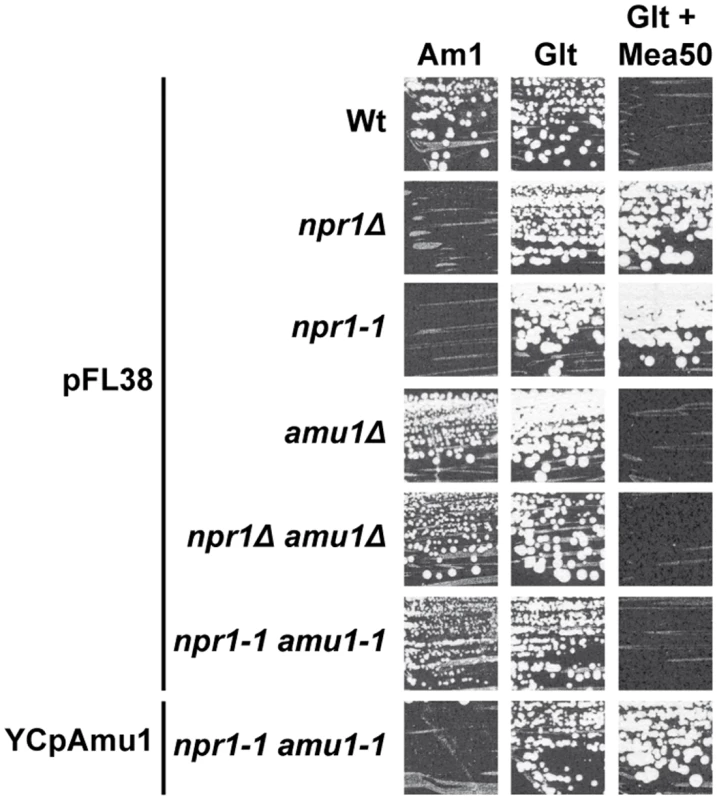
Growth tests on solid medium containing, as the sole nitrogen source, ammonium 1 mM (Am1) or glutamate 0.1% (Glt, positive growth control), supplemented or not with methylammonium 50 mM. Wild-type (23344c), npr1Δ (30788a), npr1-1 (21994b), amu1Δ (MB139) and amu1Δ npr1Δ (36314b) cells were transformed with the empty vector pFL38. amu1-1 npr1-1 (31034c) cells were transformed with the empty vector pFL38 and YCpAmu1. Single expression of YDL173w from a centromeric plasmid complemented the amu1-1 mutation in amu1-1 npr1-1 ura3 cells, conferring resistance to methylammonium and loss of growth on low ammonium (Fig 3). Full deletion of YDL173w in Npr1-lacking cells was sufficient to restore growth on low ammonium and to confer sensitivity to methylammonium.
These findings are consistent with the AMU1 gene corresponding to YDL173w, a gene coding for a protein of unknown function and previously named PAR32 standing for ‘Phosphorylated After Rapamycin, 32kDa’ [36].
The Amu1 protein contains a new repeated motif
The biological function of Amu1/Par32 is unknown. A recent proteomic study identified the cleavage of the N-terminal methionine of Amu1/Par32 and the N-acetylation of the subsequent alanine [37], giving a predicted 294-residue long protein with a calculated molecular weight of 31.75 kDa. The Amu1/Par32 sequence exhibits a biased amino-acid composition with several low-complexity regions. In particular, poly-Asn and poly-Lys stretches are located in the N - and C-terminal region, respectively (Fig 4). The presence of such low-complexity regions often characterizes intrinsically disordered or natively unfolded proteins. Accordingly, disorder prediction algorithms predict an intrinsically unstructured protein based solely on the amino-acid sequence of Amu1/Par32. Of note, the Amu1/Par32 sequence contains an internal repeat ‘GRGGAGN’ present in four copies (Fig 4). No biological function is so far reported to be associated to this particular motif. Sequence similarities are difficult to reveal by simple blast analysis when considering proteins containing large low complexity regions. While one motif was not sufficient in itself, a multi-copy simultaneous search allowed the identification of orthologues, underlying the repetition of the motif as a common feature in the Amu1/Par32 protein family (Fig 4). All identified sequences were found in fungi, with a number of motif repetitions varying between 3 and 6 according to the considered protein.
Fig. 4. Amu1 is only conserved in fungi and contains a four-fold repeated motif. 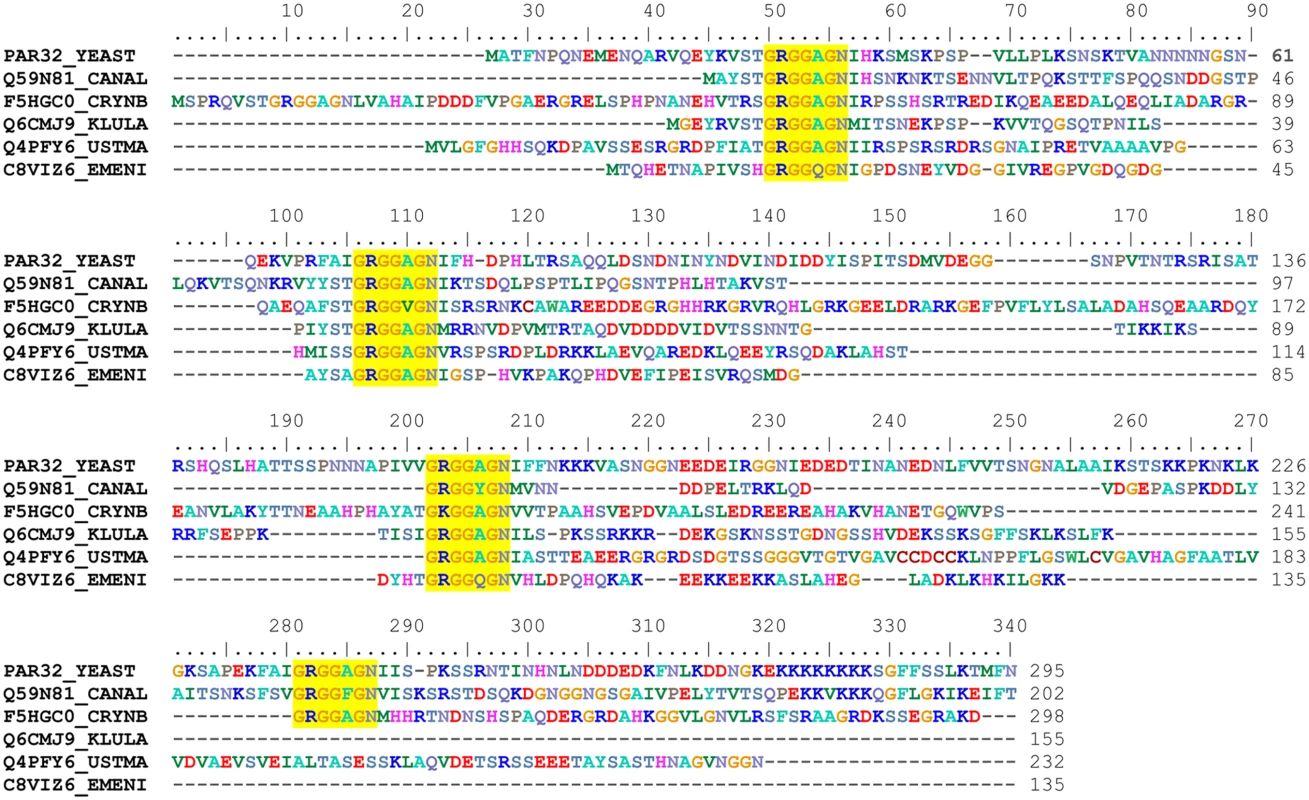
Sequences were retrieved from the UniProt database [38] primarily by a BlastP search [39] and secondly by a FuzzPro search [40] using the following searching pattern, G-R-G-G-X-[AG]-N-X(30,110)-G-R-G-G-X-[AG]-N. A selection of Amu1/Par32 orthologues is displayed in the alignment; the aligned sequences being denoted by their accession numbers: PAR32_YEAST, Saccharomyces cerevisiae, Q59N81_CANAL, Candida albicans, F5HGC0_CRYNB, Crytococcus neoformans, Q6CMJ9_KLULA, Kluyveromyces lactis, Q4PFY6_USTMA, Ustilago maydis and C8VIZ6_EMENI, Aspergillus nidulans. The multiple sequence alignment was automatically generated by ClustalW [41] and manually adjusted using BioEdit [42]. The repeated motif characterizing the Amu1 protein family is highlighted in yellow. Amu1 is required for the Mep1 and Mep3 activity-loss occurring in Npr1-lacking cells
In order to determine which of the three Mep proteins recovers activity in a double npr1 amu1 context, we constructed strains bearing the AMU1 deletion, the npr1 mutation and the deletions of two of the three MEP genes, in all the combinations. Deletion of AMU1 improved growth on low ammonium of npr1 cells expressing specifically MEP1 or MEP3, but not those expressing the MEP2 gene (Fig 5). The simple AMU1 excision had no major impact on growth of cells producing one of the three Mep in the presence of a functional Npr1 kinase.
Fig. 5. The amu1 mutation restores the function of Mep1 and Mep3 in the absence of the Npr1 kinase. 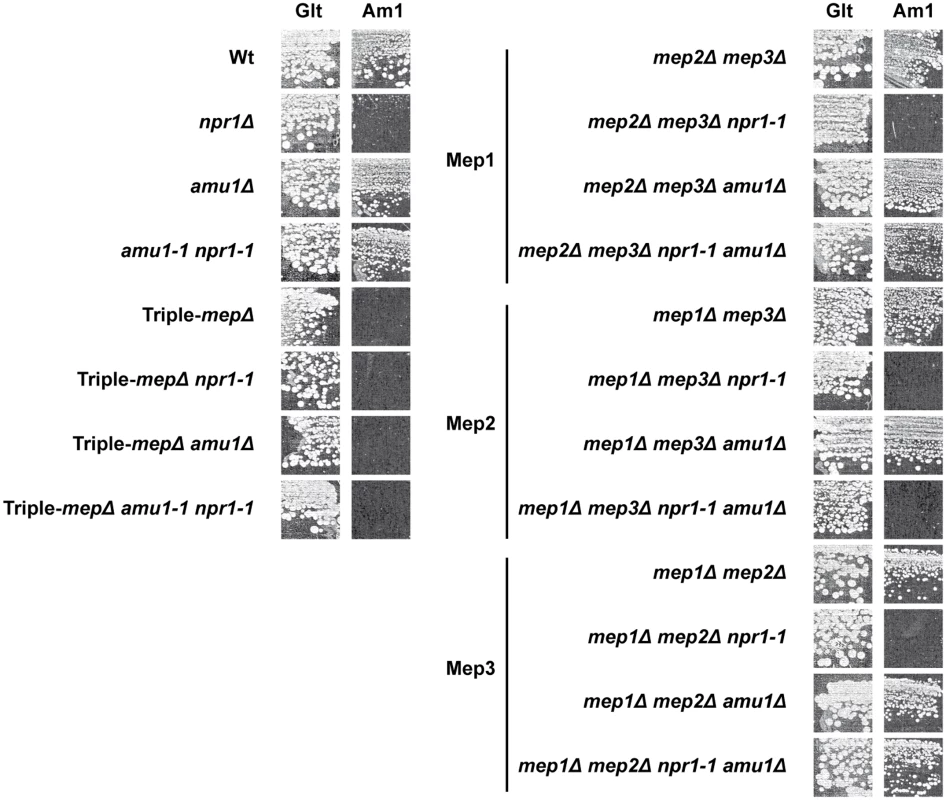
Growth tests on solid medium containing, as the sole nitrogen source, ammonium 1 mM (Am1) or glutamate 0.1% (Glt, positive growth control). Wild-type (23344c), npr1Δ (30788a), amu1Δ (MB139), amu1-1 npr1-1 (31034c), triple-mepΔ (31019b), triple-mepΔ npr1-1 (31052c), triple-mepΔ amu1Δ (AM114), triple-mepΔ amu1Δ npr1-1 (31087c), mep2Δ mep3Δ (31018b), mep2Δ mep3Δ npr1-1 (31059b), mep2Δ mep3Δ amu1Δ (PVV003), mep2Δ mep3Δ amu1Δ npr1-1 (PVV001), mep1Δ mep3Δ (31022a), mep1Δ mep3Δ npr1-1 (31059d), mep1Δ mep3Δ amu1Δ (PVV007), mep1Δ mep3Δ amu1Δ npr1-1 (PVV005), mep1Δ mep2Δ (31021c), mep1Δ mep2Δ npr1-1 (31045c), mep1Δ mep2Δ amu1Δ (PVV011) and mep1Δ mep2Δ amu1Δ npr1-1 (PVV009) cells. Measurements of initial uptake rates of [14C]-methylammonium were consistent with the growth tests (Table 1). An increase in Mep1 activity was observed in Npr1-lacking cells further deleted of AMU1, while Mep2 activity remained low in these conditions, as in npr1 cells.
Tab. 1. Methylammonium uptake activity. 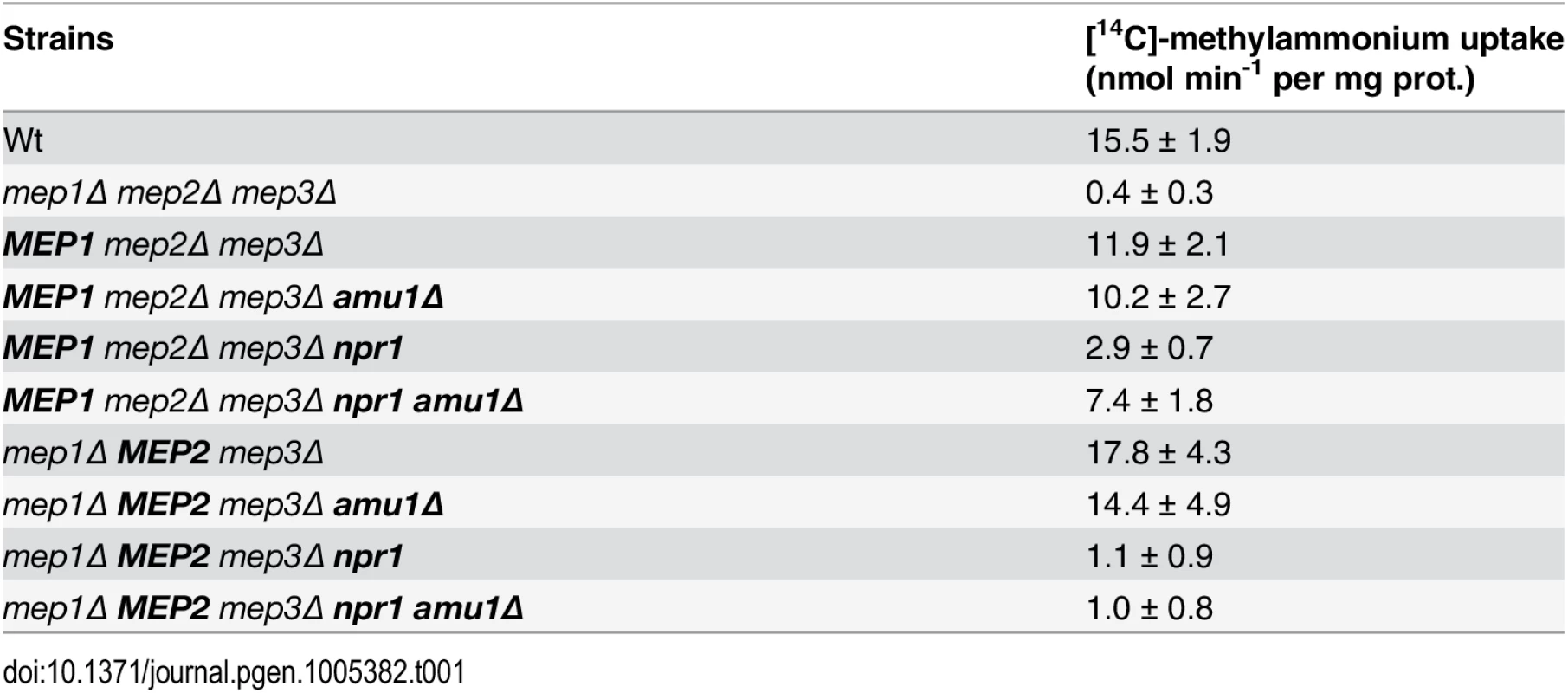
Initial rates of [14C]-methylammonium (0.5 mM) uptake were measured. Wild-type (23344c), triple-mepΔ (31019b), mep2Δ mep3Δ (31018b), mep2Δ mep3Δ npr1-1 (31059b), mep2Δ mep3Δ amu1Δ (PVV003), mep2Δ mep3Δ amu1Δ npr1-1 (PVV001), mep1Δ mep3Δ (31022a), mep1Δ mep3Δ npr1-1 (31059d), mep1Δ mep3Δ amu1Δ (PVV007), and mep1Δ mep3Δ amu1Δ npr1-1 (PVV005) cells were grown in the presence of proline. The standard deviations are indicated for the values corresponding to the averages of two to six independent experiments. Altogether, these observations reveal that loss of AMU1 specifically restores the transport function of Mep1 and Mep3 in Npr1-lacking cells.
Amu1 is phosphorylated in an Npr1-dependent manner
We next tested whether the Npr1 kinase affects the phosphorylation state of the Amu1 protein. In proline-grown Npr1-containing cells, the Amu1-3HA protein produced from the chromosomal tagged-gene was detected as a principal signal of about 63 kDa (Fig 6a). In the latter experiment, overexposure of the autoradiogram also enabled to reveal a signal of about 25 kDa which was however not readily detectable in all the immunoblot experiments. In Npr1-lacking cells, these signals shifted respectively to about 59 and 23 kDa, the faster-running one being hardly detectable. The slower-running Amu1 form could correspond to a multimeric state of the protein or to a complex, both being resistant to denaturing conditions of the SDS-PAGE. Hereafter, when an immunoblot of Amu1-3HA will be discussed, we will mainly focus on the slower-running form.
Fig. 6. Amu1 phosphorylation is controlled by nitrogen supply, the Npr1 kinase and TORC1. 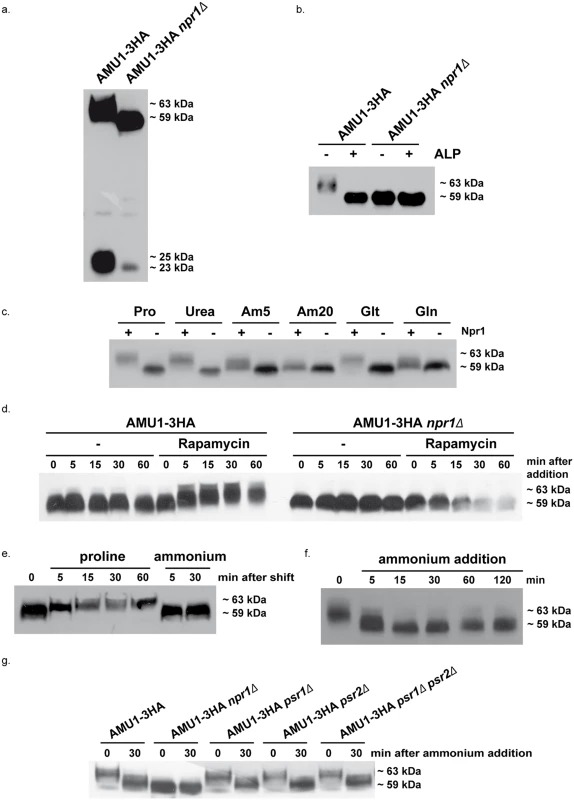
Immunodetection of Amu1-3HA from total cellular extracts. (a-b) AMU1-3HA (MB142) and AMU1-3HA npr1Δ (36307b) cells were grown in the presence of proline (0.1%) as nitrogen source. (b) Total cell extracts were treated (+) or not (-) with alkaline phosphatase (ALP). (c) AMU1-3HA (MB142) and AMU1-3HA npr1Δ (36307b) cells were grown in the presence of proline (0.1%, Pro), urea (0.1%), ammonium (5 or 20 mM, Am), glutamate (0.1%, Glt) or glutamine (0.1%, Gln) as nitrogen sources. (d) AMU1-3HA (MB142) and AMU1-3HA npr1Δ (36307b) cells were grown in the presence of ammonium (20 mM) as nitrogen source. At time t = 0, rapamycin or the drug vehicle alone (-, control) was added to the cell culture. (e) AMU1-3HA (MB142) cells were grown in the presence of ammonium (20 mM) as nitrogen source. At time t = 0, cells were transferred in a proline-medium or in a similar ammonium-medium (control). (f) AMU1-3HA (MB142) cells were grown in the presence of proline (0.1%) as nitrogen source. At time t = 0, ammonium (20 mM) was added to the cell culture. (g) AMU1-3HA (MB142), AMU1-3HA npr1Δ (36307b), AMU1-3HA psr1Δ (PVV152), AMU1-3HA psr2Δ (PVV158), AMU1-3HA psr1Δ psr2Δ (PVV160) cells were grown in the presence of proline (0.1%) as nitrogen source. At time t = 0, ammonium (20 mM) was added to the cell culture. ALP treatment of extracts from Npr1-containing cells was accompanied by a down-shift of the main Amu1-3HA signal from 63 to 59 kDa, thus at the same running-rate observed for the Amu1-3HA signal in Npr1-lacking cells extracts, treated or not with ALP (Fig 6b).
Hence, Amu1-3HA appears phosphorylated in an Npr1-dependent manner.
TORC1 mediates tight nitrogen-dependent control of Amu1 phosphorylation
The Npr1 phosphorylation state and function is regulated by the TORC1 pathway according to the quality of the nitrogen supply [18,43–45]. Immunodetection of Amu1-3HA in Npr1-containing cells grown with nitrogen sources of different quality showed variations in the phosphorylation status of the protein (Fig 6c). The protein was the most phosphorylated during growth on poor nitrogen sources like proline and urea, or intermediate quality nitrogen source like glutamate, namely under conditions where Npr1 is hypophosphorylated and presumed active [44,45]. In contrast, Amu1-3HA appeared to be less phosphorylated during growth with raising concentrations of ammonium or with glutamine, conditions where Npr1 is hyperphosphorylated and inhibited in a TORC1-dependent manner. In Npr1-lacking cells, Amu1-3HA was immunodetected as a fast-running form whatever the nitrogen supply, indicating that the variation in phosphorylation state of Amu1-3HA is triggered by Npr1.
Rapamycin addition, to hinder TORC1 activity in cells growing on high ammonium concentration, was accompanied by a rapid up-shift of the Amu1-3HA signal, not observed in Npr1-lacking cells (Fig 6d). This is consistent with previous findings reporting that Amu1/Par32 is hyperphosphorylated after rapamycin treatment [46]. A similar rapid up-shift was also visible when ammonium-grown cells were transferred to a proline-medium, in keeping with a fast response of the Amu1 phosphorylation status to the quality of the nitrogen source (Fig 6e). Inversely, supplementation of a high ammonium concentration to proline-grown cells was correlated with a rapid down-shift of the Amu1-3HA signal consistent with instant dephosphorylation of Amu1 upon shift from non-preferred to preferred nitrogen supply (Fig 6f). We recently showed that the regulation of the inherent activity of Mep2 involves a dynamic control of the phosphorylation status of S457 in the C-terminal extension of the transport protein [18]. This control is mediated by a balance between Npr1-dependent phosphorylation and Psr1/Psr2-dependent dephosphorylation of Mep2. As the Mep1 and Mep3 negative regulation involves an intermediate protein which is phosphorylated in an Npr1-dependent manner, we tested whether Amu1 could be dephosphorylated via the Psr1 and Psr2 plasma-membrane phosphatases. Our data show that the Amu1-3HA ammonium-induced dephosphorylation was also observed in cells lacking one or both of the Psr phosphatases (Fig 6g). These findings further support a new distinction in the TORC1-Npr1 dependent mechanism of Mep2 regulation compared to Mep1/Mep3.
Altogether, these observations are consistent with TORC1 down-regulation triggering Npr1-dependent phosphorylation of Amu1 and with Amu1 phosphorylation state being tightly and rapidly regulated according to variation of the quality of the nitrogen supply.
Amu1 localization is controlled by TORC1 - and Npr1-dependent phosphorylation
It has been reported that an arrestin-target of Npr1 is specifically hypophosphorylated in Npr1-lacking cells where it accumulates at the plasma membrane [20]. We checked whether Amu1 could respond to the Npr1 regulation through a modified subcellular localization by analyzing the impact of the nitrogen supply and of Npr1 integrity on the localization of Amu1-GFP. In proline-grown cells, Amu1-GFP was most exclusively cytoplasmic in the presence of Npr1, whereas it was detected at the cell surface in Npr1-lacking cells (Fig 7a). Addition of either glutamine or high ammonium concentration to proline-grown Npr1-containing cells triggered an increase of Amu1-GFP at the plasma membrane and a decrease of cytosolic Amu1-GFP revealing a recruitment of the protein at the cell surface (Fig 7b). In all these conditions, Mep1-mCherry expressed from the tagged chromosomal MEP1 locus was principally detected at the plasma membrane. Both Mep1-mCherry and Amu1-GFP labeled the cell surface with discontinuous fluorescence intensity, intense foci of Mep1-mCherry co-localizing with intense Amu1-GFP foci (Fig 7a and 7b). Addition of rapamycin to proline-grown cells prior ammonium supply at least partially prevented the recruitment of Amu1-GFP at the cell surface (Fig 7b), indicating that Amu1-GFP localization can be controlled by TORC1. Accordingly, the Amu1-GFP dephosphorylation induced by ammonium addition was inhibited in cells pretreated with rapamycin (Fig 7c).
Fig. 7. Amu1 localization is controlled by TORC1 and Npr1. 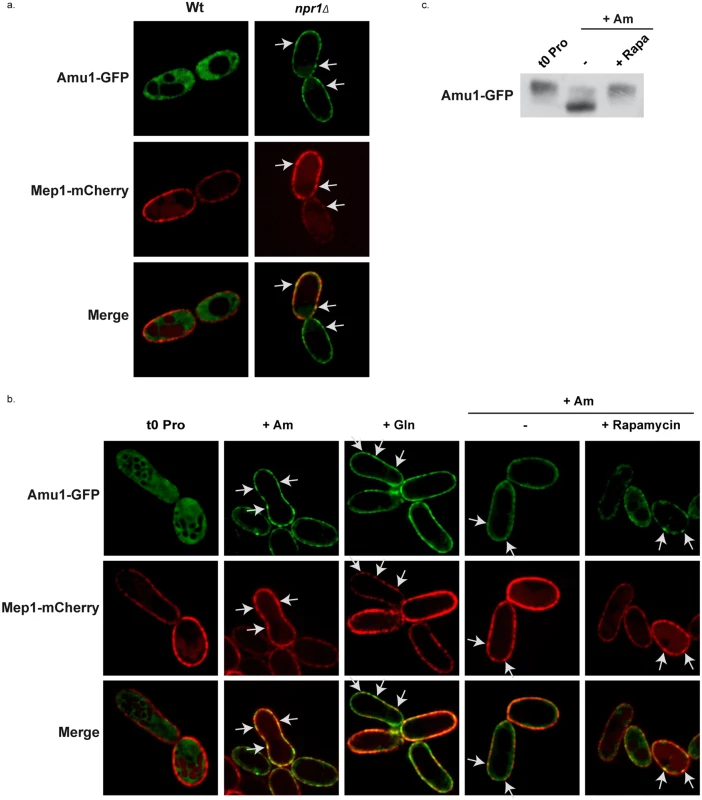
(a-b) Amu1-GFP and Mep1-mCherry localizations were observed by fluorescence microscopy. MEP1-mCherry (MB321) or MEP1-mCherry npr1Δ (MB325) cells were transformed with YCpAmu1-GFP and grown in the presence of proline (0.1%). (b) MEP1-mCherry (MB321) cells transformed with YCpAmu1-GFP were grown in the presence of proline (0.1%) as nitrogen source. At time t = 0, ammonium (Am, 20mM) or glutamine (Gln, 0.1%) was added to the cell culture during 1h (2nd and 3rd columns). Rapamycin or the drug vehicle alone (control, -) was added to the proline-grown cells. 1h later, ammonium (Am, 20 mM) was added to the cell cultures during 1h (4th and 5th columns). (c) Immunodetection of Amu1-GFP from total cellular extracts of amu1Δ (MB139) cells transformed with YCpAmu1-GFP. Rapamycin or the drug vehicle alone (control, -) was added to proline-grown cells. 1h later, ammonium (Am, 20 mM) was added to the cell cultures during 1h. The localization of Amu1 depends on its phosphorylation state
Our data indicate that the kinase prevents plasma-membrane accumulation of Amu1, likely protecting Mep1 and Mep3 from the TORC1-dependent inactivation occurring upon preferred-nitrogen supplementation.
We next wished to address whether Amu1 dephosphorylation could constitute a signal for its cell-surface localization. The Amu1 295-residue-long protein contains no less than 29 serine, 15 threonine and 3 tyrosine residues and, at least 18 putative phosphorylation sites are predicted using the NetPhosYeast 1.0 server (http://www.cbs.dtu.dk/services/NetPhosYeast/). In addition, several independent large-scale phosphoproteomic analyses performed in response to different stimuli, such as rapamycin treatment, cell cycle regulation or general osmotic stress, have revealed numerous phosphorylated sites in Amu1, and also distinct combinations between these phosphorylated sites [36,46–51]. The Amu1 hyperphosphorylation observed upon rapamycin treatment or during growth on proline likely results from the simultaneous phosphorylation of several sites. Taking all these data into consideration, we decided to substitute by alanine 9 serine or threonine residues (S34, S36, S39, S49, S206, S246, S249, S250 and T253) including residues of sites reported to respond to rapamycin treatment [47]. The resulting Amu1-3HA variant, Amu1phos-3HA, was detected as a principal signal of about 59 kDa, showing a down-shift in the migration profile compared to the native Amu1-3HA protein (Fig 8a). The signal was composed of at least two major bands, the faster-running form being detected at a size similar to the Amu1-3HA signal observed in cells lacking the Npr1 kinase. These data indicate that the Npr1-dependent phosphorylation of Amu1phos-3HA is largely reduced.
Fig. 8. The phosphorylation state of Amu1 governs its localization and the integrity of the conserved motif is required for appropriate localization and inhibition function of the protein. 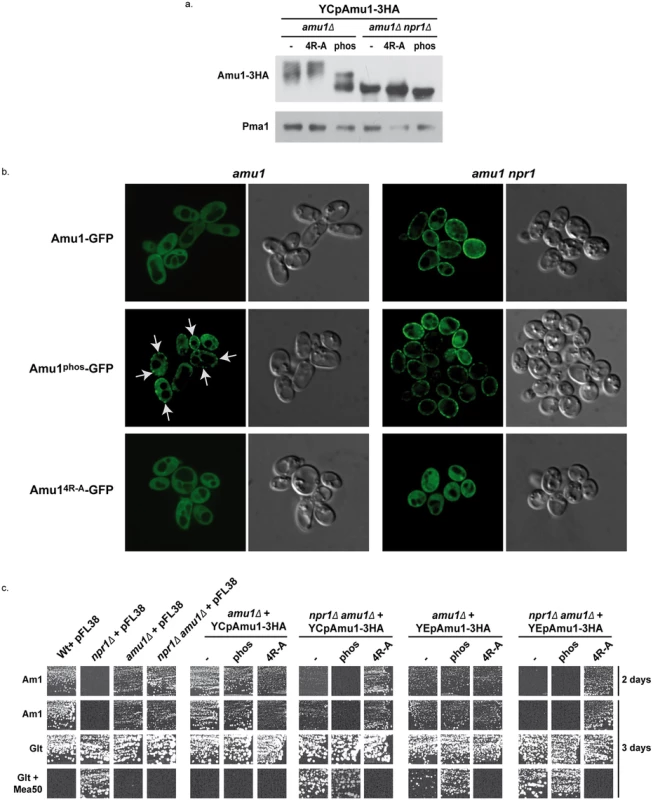
(a) Immunodetection of Amu1-3HA from total extracts of proline (0,1%)-grown cells. amu1Δ (MB139) and amu1Δ npr1Δ (36314b) were transformed with YCpAmu1-3HA, YCpAmu14R-A-3HA, and YCpAmu1phos-3HA. The plasma membrane proton ATPase Pma1 was detected as a loading control. (b) Amu1-GFP, Amu14R-A-GFP and Amu1phos-GFP localization was observed by fluorescence microscopy. amu1Δ (MB139) or amu1 npr1 (31034c or 36314b) cells were transformed with the corresponding YCpAmu1-GFP plasmid and grown in the presence of proline (0.1%). (c) Growth tests on solid medium containing, as the sole nitrogen source, ammonium 1 mM (Am1) or glutamate 0.1% (Glt, positive growth control), supplemented or not with methylammonium 50 mM (Mea50). Wild-type (23344c) and npr1Δ (30788a) were transformed with the empty vector pFL38. amu1Δ (MB139) and amu1Δ npr1Δ (36314b) cells were transformed with the empty vector pFL38, with the low-copy-number plasmids: YCpAmu1-HA, YCpAmu14R-A-HA, YCpAmu1phos-HA, or with the high-copy-number plasmids: YEpAmu1-HA, YEpAmu14R-A-HA, YEpAmu1phos-HA. While Amu1-GFP was mainly cytosolic in proline-grown cells, the Amu1-GFP variant mutated in the 9 potential phosphorylation sites, Amu1phos-GFP, was at least partially directed to the cell surface, despite the presence of the Npr1 kinase (Fig 8b), indicating that the reduction in Amu1 phosphorylation could be linked to its cell-surface localization.
We next addressed the functionality of the Amu1phos-3HA variant by performing growth tests on low ammonium medium as sole nitrogen source and on glutamate medium with a toxic methylammonium concentration (Fig 8c). Double amu1Δ npr1Δ cells expressing a functional Amu1 protein should behave as npr1Δ cells, thus showing no growth on ammonium and resistance to methylammonium, while expression of a non-functional Amu1 protein should not alter the growth of amu1Δ npr1Δ cells on ammonium and the sensitivity to methylammonium. The AMU1-3HA variants were expressed from centromeric (low-copy) and episomal (high-copy) plasmids. Expressing AMU1-3HA from a centromeric plasmid in amu1Δ npr1Δ cells partially inhibited growth on ammonium while it clearly conferred resistance to methylammonium, indicating that the Amu1-tagged protein is partially functional (Fig 8c). Overexpressing AMU1-3HA from an episomal plasmid in amu1Δ npr1Δ cells however fully complemented the AMU1 deletion, suggesting that the partial function alteration of tagged Amu1-3HA can be compensated by overproduction of the protein. As expected, expression of AMU1phos-3HA in amu1Δ npr1Δ cells also complemented the AMU1 deletion, indicating that the mutated protein has conserved its ability to inhibit ammonium transport (Fig 8c). amu1Δ cells producing or overproducing Amu1phos-3HA were able to grow on ammonium as the cells producing the non-mutated Amu1-3HA protein, showing that Amu1phos-3HA was still sensitive to Npr1-mediated inhibition. These data suggest that partial localization of Amu1phos-3HA at the cell surface in cells containing Npr1 is not sufficient to observe a constitutive inhibition of Mep1 and Mep3. However, amu1Δ cells overproducing Amu1phos-3HA appeared more resistant to methylammonium compared to those producing Amu1-3HA, suggesting that the hypophosphorylated variant may have gained a partial ability to inhibit Mep1 and/or Mep3 despite the presence of the Npr1 kinase. Expressing AMU1phos-3HA from a centromeric plasmid in amu1Δ npr1Δ cells appeared to induce a slightly stronger inhibition of growth on ammonium compared to AMU1-3HA, suggesting that the Amu1phos-3HA variant might have an enhanced inhibitory capacity on Mep1 and Mep3 even in the absence of the kinase.
Together, these results highlight the importance of Amu1 phosphorylation for the control of its localization, the hypophosphorylated Amu1phos-3HA variant being at least partially directed to the plasma membrane even if the Npr1 kinase is active. In these conditions, Amu1phos-3HA appears however unable to completely inhibit Mep1 and Mep3, indicating that it is still sensitive to Npr1-mediated inhibition, likely containing additional Npr1-dependent phosphorylation sites.
Amu1 forms a complex with Mep1 and Mep3
Our data indicate that upon preferred nitrogen supply, Mep1 and Mep3 are inactivated in a TORC1 - and Npr1-dependent manner via the Amu1 factor. As Amu1 is targeted to the plasma membrane in this condition, the protein could mediate inactivation of Mep1 and Mep3 via physical interactions.
We performed co-immunoprecipitation assays focusing on potential Amu1-Mep interactions. As Mep1 and Mep3 are poorly expressed proteins, we used an experimental setup enabling to produce Mep-GFP under the control of the strong GAL1 promoter. We show that upon immunoprecipitation of Amu1-3HA, Mep1-GFP and Mep3-GFP were co-immunoprecipitated while Mep2-GFP was not (Fig 9a). It is to note that the co-immunoprecipitation of Mep1-GFP and Mep3-GFP with Amu1-3HA was observed when using proline-grown cells expressing Npr1 and that the addition of glutamine to the proline-grown cells had no significant impact on the efficiency of the co-immunoprecipitation, in the tested experimental conditions.
Fig. 9. Amu1 interacts with Mep1 and Mep3 in vitro. 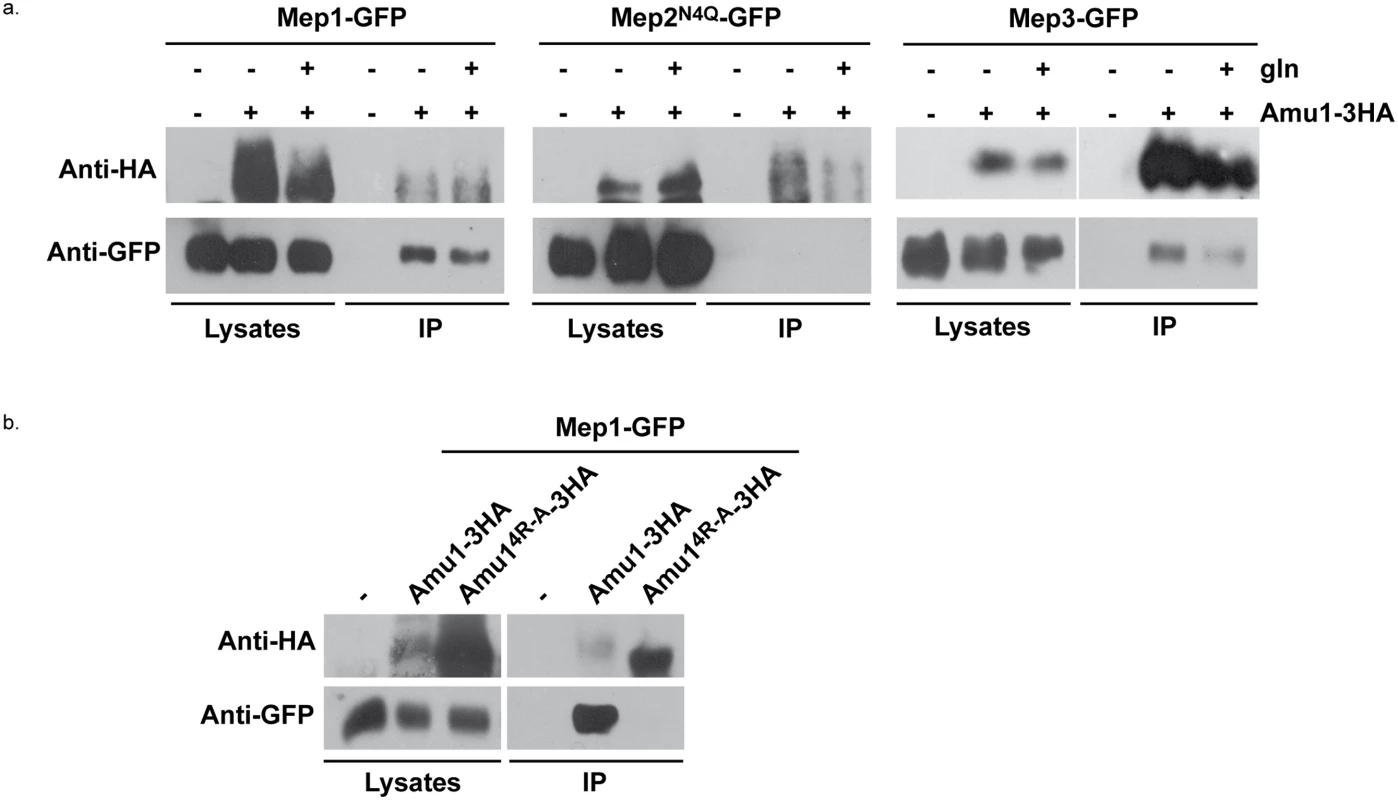
After spheroplasting, the cells were lysed and Amu1-3HA was immunoprecipitated with anti-HA. Immunoblots of the lysates and immunoprecipitates (IP) with anti-HA and anti-GFP antibodies are shown. (a) Wt (23344c, negative control)) and AMU1-3HA (MB142) cells transformed with pGAL1Mep1-GFP, pGAL1Mep2N4Q-GFP or pGAL1Mep3-GFP were grown in the presence of proline (0.1%). Glutamine (gln, 0.1%) was added to AMU1-3HA (MB142) cells transformed with one of the 3 pGAL1Mep-GFP vectors (30 min). (b) Triple-mepΔ (31019b) cells transformed with pGAL1Mep1-GFP (negative control) and, triple-mepΔ amu1Δ (MB310) cells transformed with both pGAL1Mep1-GFP (LEU2) and YEpAmu1-HA (URA3) or with both pGAL1Mep1-GFP (LEU2) and YEpAmu14R-A-HA (URA3) were grown in the presence of proline (0.1%). These results reveal that Mep1-GFP and Mep3-GFP are able to interact in vitro with Amu1-3HA.
The integrity of the conserved and repeated motif of Amu1 is required for its function
We finally assessed the potential functional importance of the conserved repeated ‘GRGGAGN’ motif present in Amu1. We generated a plasmid allowing the expression of tagged Amu1-3HA where the arginine residue of all four motifs was mutated into alanine (Amu14R-A-3HA). Amu14R-A-3HA showed a migration profile similar to native Amu1, with no major alteration in the protein level of the major 63 kDa signal (Fig 8a). It also responded to Npr1 presence or absence similarly to non-mutated Amu1-3HA in terms of apparent phosphorylated and dephosphorylated states. However, Amu14R-A-GFP was cytosolic both in the presence as in the absence of Npr1, revealing a key role of the motif in the cell-surface localization of Amu1 (Fig 8b). The functionality of the Amu14R-A-3HA variant in the process of Mep1 and Mep3 inhibition was assessed by performing growth tests on low ammonium medium or on glutamate medium with a toxic methylammonium concentration (Fig 8c). Consistent with the mislocalization of Amu14R-A-3HA, amu1Δ npr1Δ cells producing or overproducing Amu14R-A-3HA remained able to grow on ammonium and sensitive to methylammonium, showing that this version of Amu1 was unable to inhibit Mep1 and Mep3. In addition, co-immunoprecipitation experiments further revealed that the in vitro interaction between Mep1 and Amu1 was compromised by the mutation of the four repeated motifs of Amu1 (Fig 9b).
These results show a crucial role for the arginine residue of the four repeated motifs in the cell surface localization of Amu1 and in the intrinsic ability of Amu1 to interact and to inhibit the Mep ammonium transport proteins.
Together, our findings are consistent with a direct negative role of Amu1 in the TORC1-Npr1 control of the inherent activity of Mep1 and Mep3.
Discussion
This study reveals a novel mechanism enabling TORC1 and the effector kinase Npr1 to regulate nutrient permeability according to environmental variations. The TORC1-Npr1 pathway is thus able to regulate plasma-membrane transport proteins via three different ways (Fig 10). For instance, the TORC1-Npr1 pathway is so far mainly described for its control of the stability of amino-acid permeases at the plasma membrane [20,21]. Npr1 exerts a phospho-inhibitory control on arrestin-like adaptors thereby preventing recruitment of the Rsp5 ubiquitin-ligase to its permease targets, and consequently protecting them from endocytosis and vacuolar degradation. Seminal studies supported the existence of a different mechanism of Npr1-mediated regulation of yeast Mep ammonium transport proteins and also suggested the existence of a diversity among Npr1-dependent regulatory processes controlling Mep paralogues [22–25]. Consistently, we recently unraveled the molecular mechanism enabling TORC1-Npr1 to fine-tune the inherent activity of the Mep2 ammonium transport protein by dynamically regulating the phosphorylation status of an auto-inhibitory C-terminal domain of the transport protein [18]. Although Npr1 is also required for the activity of the two other Mep1 and Mep3 ammonium transport systems, our data reveal that their regulation indeed involves a different process implicating an inhibitory partner, Amu1/Par32, an ever still functional orphan. On the basis of our findings, we propose a model of Mep1 and Mep3 regulation by TORC1-Npr1 and the nitrogen supply (Fig 10). In the presence of a non-preferred nitrogen source, TORC1 is poorly active and Npr1 is hypophosphorylated and presumably active. In these conditions, Amu1 is phosphorylated in an Npr1-dependent manner and is mainly cytosolic while Mep1 and Mep3 are kept active. The Npr1 kinase might directly phosphorylate Amu1. For instance, a physical interaction between both proteins has been reported by two independent large-scale studies [52,53]. In the presence of a preferred nitrogen source, such as glutamine or a high ammonium concentration, TORC1 is upregulated, Npr1 is hyperphosphorylated and inhibited. In these conditions, Amu1 is dephosphorylated and accumulates at the cell surface. It forms a complex with Mep1 and with Mep3, and mediates inhibition of ammonium transport. Mep1 and Mep3 transport activity could be inhibited upon interaction with Amu1 by physical hindrance of the conducting pore crossing the hydrophobic core of the proteins. Alternatively, physical interaction with Amu1 might prevent C-terminal-dependent activation of transport. The C-terminal extension of several Mep-Amt proteins is indeed reported to regulate transport activation by controlling an allosteric switch [18,54,55]. It is also possible that Amu1 serves as a scaffold for yet to define regulatory protein(s), controlling the activity of Mep1 and Mep3.
Fig. 10. Three different strategies enable the TORC1-pathway to regulate plasma-membrane transport proteins. 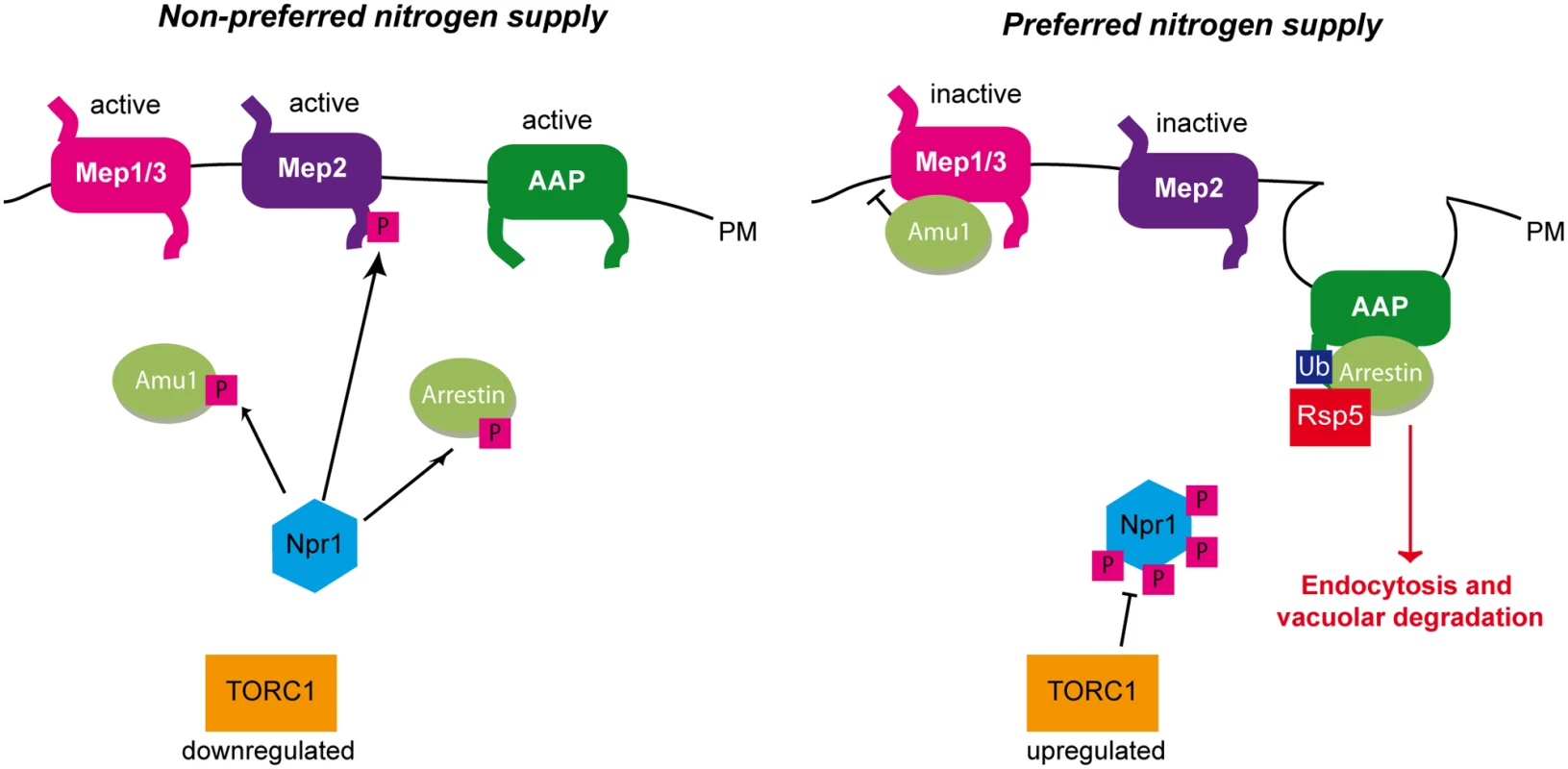
Under non-preferred nitrogen supply, TORC1 is poorly active and Npr1 is hypophosphorylated and able to mediate the phosphorylation of its different targets. Npr1 mediates the phosphorylation of Amu1 which remains cytosolic, while Mep1 and Mep3 are kept active. Npr1 enables C-terminal phosphorylation of the Mep2 ammonium transport protein, thereby silencing an autoinhibitory domain and allowing Mep2 activity. Npr1 mediates phosphorylation of arrestin-like adaptors thereby protecting their amino acid permease (AAP) targets from endocytosis and vacuolar degradation. Under preferred nitrogen supply, TORC1 is upregulated, Npr1 is hyperphosphorylated and inhibited. In these conditions, Amu1 is dephosphorylated, and accumulates at the cell surface, physically interacts with Mep1 and Mep3, and mediates inhibition of ammonium transport. The non-phosphorylated autoinhibitory domain of Mep2 prevents the enhancer domain to activate the transport protein. Dephosphorylated arrestin-like adaptors recruit the Rsp5 ubiquitin-ligase to their AAP targets, which are ubiquitylated, endocytosed and degraded in the vacuole. A regulation by an inhibitory partner is highly reminiscent to the GlnK-mediated control of prokaryotic Mep-Amt proteins. GlnK belongs to the PII protein family of signal transduction proteins widely distributed in bacteria and Archaea [17]. PII proteins play a pivotal role in the control of nitrogen metabolism by regulating the activities of diverse enzymes, transcription factors and membrane transport proteins. Phylogenetic analyses indicate that PII proteins evolved to regulate bacterial AmtB ammonium transport proteins, as evidenced by the early linkage of GLNK with AMTB in operon [56,57]. In E. coli cells, under nitrogen limitation, GlnK is uridylylated and essentially cytoplasmic, while supplementation of high ammonium concentrations is accompanied by deuridylylation of GlnK and interaction with AmtB at the plasma membrane [58]. Crystallisation of the AmtB-GlnK complex reveals that trimeric GlnK regulates trimeric AmtB by insertion of the regulatory deuridylylated T-loop of one GlnK monomers into the pore of the neighboring AmtB monomer in the trimer, and thus by physically blocking the substrate transport [15,16]. GlnK also binds α-ketoglutarate and ATP/ADP and, was recently shown to display an ATPase activity inhibited by α-ketoglutarate [15,16,59,60]. In conditions of nitrogen sufficiency, a drop in cellular α-ketoglutarate leads to a depletion of α-ketoglutarate bound to GlnK, accompanied by a hydrolysis of ATP into ADP. ATP hydrolysis would drive a change in the conformation of the T-loop allowing it to protrude deep into the cytoplasmic exit of the AmtB pore.
Amu1/Par32 is of unknown biochemical function. It is characterized by a four-fold repetition of a new motif ‘GRGGAGN’. The repetition of this motif enables to identify Amu1 orthologues, defining a new family of fungal proteins. Based on prediction algorithms, Amu1 would be classified as an intrinsically disordered protein. These algorithms do however not take into account possible post-translational modifications such as phosphorylation and the impact these could have on folding and function, as recently nicely demonstrated for another intrinsically disordered protein [61]. Although Amu1 is not related in sequence to PII proteins, it is tempting to propose that it could acquire a particular folding/function upon binding to partners, such as the Npr1 kinase or the Mep proteins, and act as a functional analogue of the PII proteins. For instance, we show that the hypophosphorylated Amu1phos variant is at least partially directed to the plasma membrane even in the presence of Npr1. In contrast, the substitution by alanine of the conserved arginine residue in the four motifs in Amu14R-A prevents the cell-surface localization, normally occurring in the absence of Npr1, and this, despite Amu14R-A being dephosphorylated in these conditions. These data suggest that the integrity of the motifs plays a dominant role for the cell-surface localization of dephosphorylated Amu1. Moreover, if Amu14R-A has lost its ability to inhibit Mep1 and Mep3 in vivo, as expected given its mislocalization, Amu14R-A has also lost its proper capacity of interaction with Mep1 in vitro, indicating a key role of the conserved arginine residues. The motifs integrity could thus be required for Amu1 to adopt a particular folding enabling interaction with the Mep proteins. In this model, nutrient-induced dephosphorylation of Amu1 would favor an Amu1 folding process involving the repeated motifs. The dephosphorylation of Amu1 could also be required for the interaction with proteins involved in its cell-surface targeting.
We show that yeast cells have evolved different mechanisms to regulate transport proteins of the same family. While Mep2 shares about 40% of identity with Mep1 and Mep3, the two latter proteins share about 79% identity [19] and likely emerged from the whole-genome duplication event that occurred in the hemiascomycete branch [62]. We previously showed the existence of two functional Mep-Amt subfamilies in fungi distinguishable according to whether the first of two conserved histidines in the conducting pore is preserved, as in yeast Mep2, or replaced by glutamate, as in Mep1 and Mep3 [63]. In fungi, there is usually coexistence of at least one Mep2-type protein with a Mep1/3-type protein. In baker yeast, these two subfamilies notably differ in their kinetic properties and particularly in their optimal pH of transport. We previously proposed that Mep2 and Mep1/Mep3 could transport ammonium via different molecular mechanisms, involving or not a deprotonation step of the recognized NH4+ substrate, leading to opposite direct effects on intracellular pH and to different impacts on physiology. Of note, Mep2-type proteins have a unique status in that they are proposed to play a signalling role in filamentation induction, a dimorphic transition often associated to the virulence of pathogenic fungi [64]. These observations indicate a separation in the evolution history and a functional specialization of proteins of both subfamilies. Although both Mep2 and Mep1/3 regulations are mediated by TORC1-Npr1, evolution of different molecular mechanisms of activity control could enable to discriminate between both subfamily members and to fine-tune each regulatory process in response to specific physiological parameters. Interestingly, in Candida albicans, the CaMep2 activity is abolished in cells lacking CaNpr1 while the CaMep1 activity is only partially compromised, and CaMEP3 appears to be a non-functional gene [24]. Though the potential role of the CaAmu1 protein (Fig 4) in CaMeps regulation is unknown, these data could point to variations to Npr1-dependency of Mep proteins among yeast species. However, it is also conceivable that the distinct functions carried out by Npr1 in Saccharomyces cerevisiae could be supported by distinct Npr1-like, or even distinct kinases, in other species. For instance, in collaboration with the team of Bettina Tudzynski, we recently showed that the expression of at least one of the Fusarium fujikuroi Npr1-like proteins confers growth on ammonium to the Saccharomyces cerevisiae npr1 mutant while being unable to mediate Mep2 phosphorylation [65]. These data suggest that the Fusarium kinase is able to protect yeast Mep1 and/or Mep3 against Amu1-mediated inactivation while being unable to control Mep2 activity.
The TORC1-Npr1 control passing via the Amu1 intermediate affects at least two yeast transport proteins. The latter are dedicated to the transport of a preferred nitrogen source further underlying that TORC1-Npr1 discriminates between transporters to be degraded, transiently inactivated or kept active at the cell surface by controlling at least three different regulatory mechanisms. It will be informative to determine the specter of action of Amu1. Available data already indicate that Amu1 does not control amino-acid permeases like Gap1 and Put4 [25].
Could an Amu1-like regulatory mechanism also apply to mammalian ammonium transport proteins of the Mep/Amt/Rh family? To date the mechanisms and the pathways involved in the activity regulation of Rhesus factors, mammalian counterparts of yeast Mep proteins, are largely unknown. The human RhCG protein shares a similar structure with Mep/Amt and Rh proteins from bacteria and Archaea [8–13]. Recent characterization of a nsSNP variant of human RhCG indicated that the activity regulation of this Rhesus factor could implicate molecular mechanisms similar to those controlling their Mep-Amt homologues [14]. PII proteins are absent from eukaryotes except their presence in plastids of a few plants, and obvious Amu1 homologues are limited to fungi. However, our findings suggest that the molecular mechanism of prokaryotic and fungal Mep-Amt proteins regulation is conserved although the regulatory partners are structurally different. It is tempting to propose that a similar mechanism has evolved to enable the regulation of mammalian Rhesus factors. Determining whether functional analogues of Amu1 and/or PII exist in mammals constitute a challenge for further investigation.
Materials and Methods
Strains, growth conditions
The S. cerevisiae strains used in this study are listed in Table 2. All strains are isogenic with the wild type Σ1278b [66]. Cell transformation and gene deletions were performed as described previously [67,68]. mCherry-tagging of MEP1 at the chromosomal locus was performed by homologous recombination using a PCR fragment amplified from the pFA6a-link-yomCherry-Kan plasmid, a gift from Wendell Lim and Kurt Thorn (Addgene plasmid 44903) [69]. HA-tagging of AMU1 at the chromosomal locus was similarly performed using a PCR fragment amplified from pFA6a-3HA-kanMX6 [70]. Cells were grown in a minimal buffered (pH 6.1) medium with 3% glucose as the carbon source [71]. In experiments in which genes were expressed under the GAL1 promoter, 3% galactose + 0.3% glucose were used for transcriptional induction, and 3% glucose for repression. To this medium, nitrogen sources were added as required by the experiment and as specified in the text. The nitrogen sources used were 0.1% proline, 0.1% urea, 0.1% glutamine, 0.1% glutamate, or (NH4)2SO4 at the specified concentration. When required, the medium was supplemented with 0.0025% uracil to complement the auxotrophy.
Tab. 2. List of strains used in this study. 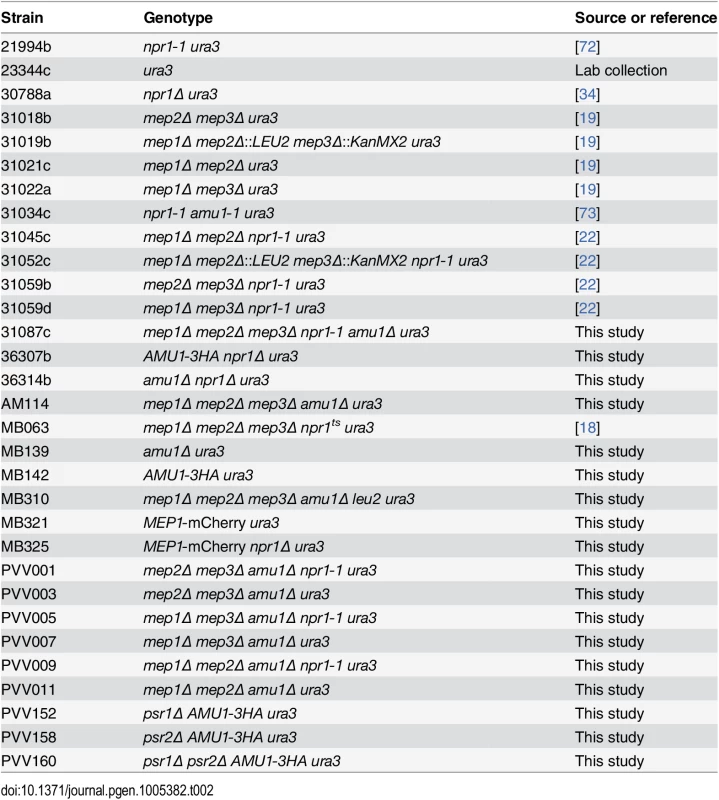
Rapamycin (LC Laboratories) was used at a final concentration of 2 μg/ml from a stock solution prepared in 90% ethanol/10% Tween-20.
Cloning of AMU1
The S. cerevisiae AMU1 gene was cloned by screening a low copy number library [1], representing the genome of Σ1278b strain, for plasmids complementing the amu1-1 mutation in the 31034c recipient strain (npr1-1 amu1-1 ura3). Cells transformed with the plasmid library were plated on a selection minimal medium containing glutamate 0.1% as nitrogen source and 50 mM methylammonium. Among about 125.000 transformants, 10 tested candidates had simultaneously recovered methylammonium resistance and loss of growth on low ammonium (1mM). DNA isolated from these transformants was transformed in E. coli JM109 for purification and amplification. The phenotypes were verified after reintroducing the 10 purified plasmids into 31034c cells. Sequencing of the 10 genomic inserts enabled to identify one single common gene, YDL173w. A HincII-HindIII DNA fragment, containing the single YDL173w ORF with 311 pb upstream and 121 pb downstream sequences, was subcloned into pFL38 [74] to generate YCpAmu1. The latter plasmid was sufficient to complement the amu1-1 mutation of 31034c cells.
Plasmids
Plasmids used in this study are listed in Table 3. Primers used in this study are available upon request. pGAL1Mep1-GFP: the MEP1 gene was amplified by PCR using the YCpMep1 vector [1] as template and then cloned by in vivo recombination in the pGAL1Gap1-GFP vector [34] by replacement of the GAP1 gene. pGAL1Mep3-GFP: the MEP3 gene was amplified by PCR using YCpMep3 vector [19] as template and then similarly used to replace the GAP1 gene in the pGAL1Gap1-GFP vector [34]. YEpMep1, YEpMep2 and YEpMep3 were constructed by transferring the complete insert of YCpMep1 [1], YCpMep2 [19] and YCpMep3 vectors [19] into the pFL44 vector [74] by restriction-ligation. YCpAmu1-GFP: a fragment containing the AMU1 gene and promoter was amplified by PCR using S. cerevisiae (23344c) genomic DNA and then cloned by in vivo recombination in the YCpMep2-GFP vector [18] by replacement of the MEP2 gene and promoter. YCp and YEpAmu1-3HA: a fragment containing a part of AMU1-3HA gene and terminator was amplified by PCR using MB142 (AMU1-3HA) genomic DNA and then cloned by in vivo recombination in the YCp or YEpAmu1 vector. Mutated AMU14R-A and AMU1phos genes were synthesized and cloned in pUC57 vector by GeneCust (Dudelange, Luxembourg). YCp/YEpAmu14R-A-3HA and YCp/YEpAmu1phos-3HA were constructed by transferring the complete insert of the corresponding pUC57-Amu1 into the YCp/YEpAmu1-3HA vector by restriction-ligation. YCpAmu14R-A-GFP and YCpAmu1phos-GFP: a fragment containing a last part of AMU1 gene fused to GFP was amplified by PCR using YCpAmu1-GFP and then cloned by in vivo recombination in the corresponding YCpAmu1-3HA vector by replacement of the 3HA-tag. pGAL1Mep1-GFP (LEU2): the LEU2 gene was amplified by PCR using pFL46 [74] as template and then cloned by in vivo recombination in the pGAL1Mep1-GFP (URA3) vector by replacement of the URA3 gene.
Tab. 3. List of plasmids used in this study. 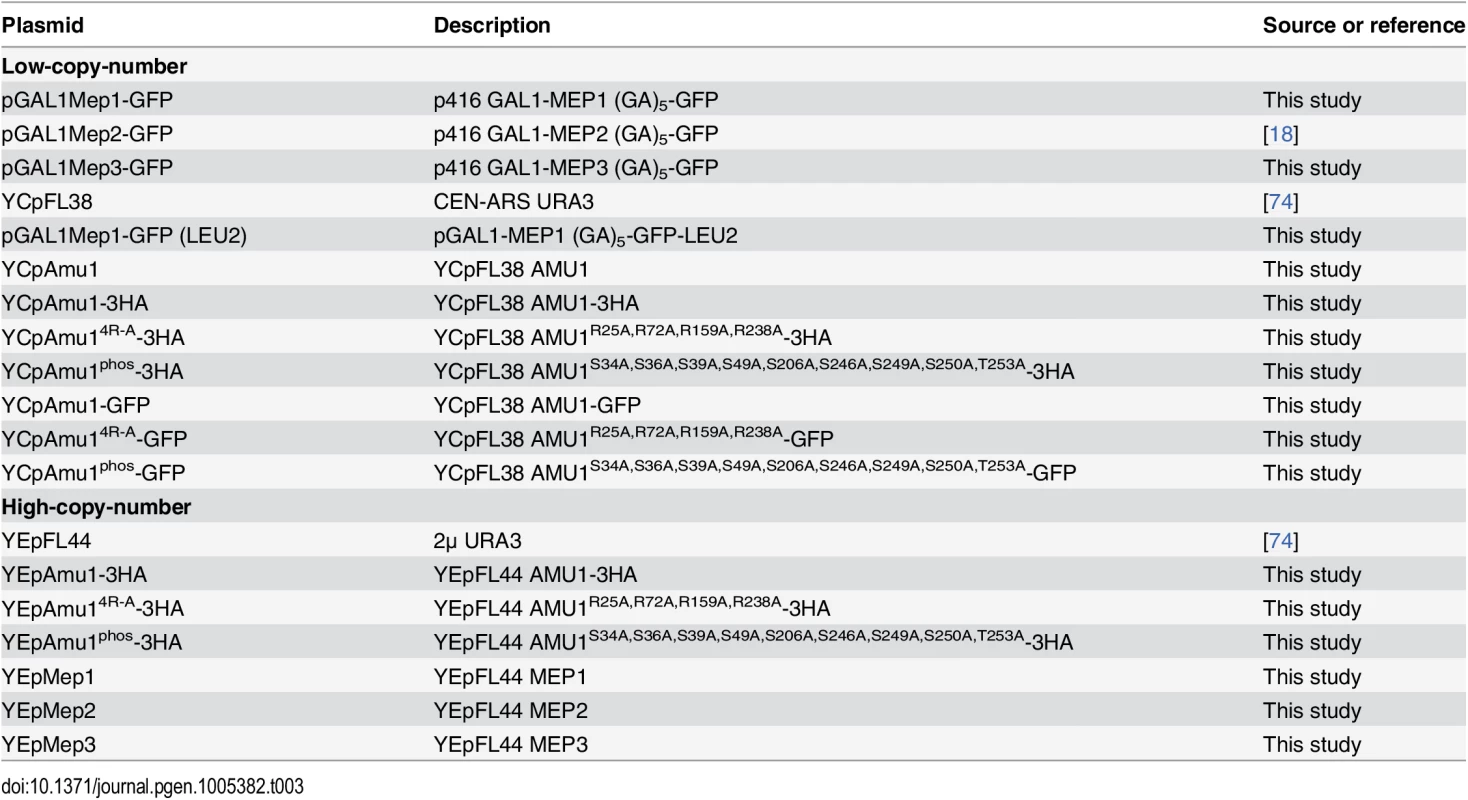
Western immunoblotting
Total protein extracts were performed as described previously [75]. Membrane-enriched cell extracts were prepared as described previously [22]. For blot analysis, equal protein amounts (~20 μg) were loaded onto an 6% to 8% SDS-polyacrylamide gel in a Tricine system [76]. After transfer to a nitrocellulose membrane (Protran, VWR), proteins were probed with a mouse or rabbit antiserum raised against the C-terminal region of Mep1 (1 : 1000)[77], Mep2 (1 : 2000) [77], Mep3 (1 : 1000) [77], HA (1 : 10000) (Roche), GFP (1 : 10000) (Roche) and Pma1 (1 : 10000) [34]. Primary antibodies were detected with horseradish-peroxidase-conjugated anti-rabbit - or anti-mouse-IgG secondary antibodies (GE Healthcare) followed by measurement of chemoluminescence (Lumi-LightPLUS, Roche). For alkaline phosphatase treatment on total cell extracts, the protein pellet collected after TCA precipitation was resuspended in a solution (0.1 M de Tris-HCl pH 6.8, 20% glycérol, 4% SDS, 2% β-mercaptoethanol and 2 mM PMSF) containing proteases inhibitors (Complete Mini, Roche). This extract is then diluted 5 X in a dephosphorylation buffer [CIP 1X (Roche), 50 mM Tris-HCl 1M pH 6.8, 2 mM PMSF] containing proteases inhibitors (Complete Mini, Roche). pH could eventually be adjusted to 7.6 and the extracts were then incubated 2h at 37°C in the presence (or not) of 20 units of calf alkaline phosphatase (Roche). Proteins were precipitated with 10% TCA. For alkaline phosphatase treatment on membrane-enriched cell extracts, the collected membrane pellet was suspended in phosphatase buffer CIP 1X added of 0.1% SDS, 2 mM PMSF and proteinase inhibitors (Complete Mini, Roche). The extracts were then incubated 1h at 37°C in the presence (or not) of 10 units of calf alkaline phosphatase (Roche). Proteins were precipitated with 10% TCA. For N-glycosidase F treatment on membrane-enriched cell extracts, the collected membrane pellet was suspended in buffer (1x PBS, 10mM EDTA pH8, 0.5% octyl-glucopyranoside, 0.2% 2-mercaptoethanol, 3mM PMSF and proteinase inhibitors) and incubated 1 h at 37°C in the presence of 1.5 unit of peptide-N-glycosidase F (PNGase F, Roche).
Co-immunoprecipitation
Co-immunoprecipitation protocol in the presence of a cross-linker was modified from [78]. About 5.108 cells were washed with H2O, and incubated 10 min in prespheroplasting buffer (100 mM Tris pH 9.4, 40 mM β-mercaptoethanol). Spheroplasts were prepared by 30-min incubation in spheroplasting buffer (20 mM HEPES pH 7.4, 0.8 M sorbitol, 0.5X 868 medium, 40 mM β-mercaptoethanol) in the presence of 800 U of lyticase at 20°C. Spheroplasts were washed once in cross-linking buffer (20 mM HEPES pH 7.4, 0.8 M sorbitol, 100 mM potassium acetate) and resuspended in 600 μl of cross-linking buffer added with proteinase inhibitors (Complete Mini, Roche). Cross-linking was conducted with 4 mM dithiobis[succinimidylpropionate] (DSP) (ThermoScientific, 22585) for 30 min at 4°C, and the reaction was quenched by addition of 100 mM Tris pH 7.4 for 15 min at 4°C. Spheroplasts were then lysed in 200 μl of lysis buffer (50 mM Tris pH 7.4, 150 mM NaCl, 1% Nonidet P-40) added with proteinase inhibitors (Complete Mini, Roche), 2 mM phenylmethylsulfonyl fluoride (PMSF), 2 μg/ml leupeptin, 1 μg/ml pepstatin A and 0.5 μg/ml chymostatin; and broken with a glass rod and then by vortex-mixing for 5 min in the presence of glass beads. 700 μl of lysis buffer containing the different proteinase inhibitors were added and the lysates were incubated on a wheel for 30 min to extract membrane proteins. The extracts were centrifuged at 14000 rpm for 10 min. An aliquot of the lysates was collected and proteins were precipitated with 10% TCA. The remaining lysates were incubated for 30 min on a wheel with 25 μl pre-washed Pierce anti-HA magnetic beads (ThermoScientific, 88836). Beads were washed two times with 300 μl lysis buffer and proteins were eluted by incubation for 10 min at 60°C with 50 μl sample buffer (100 mM Tris pH 6.8, 4% SDS, 20% glycerol, 0.02% bromophenol blue). 50 μl of Tris 1M and 2% β-mercaptoethanol were added to the samples. Samples were analyzed by SDS-PAGE, followed by immunoblotting overnight with antibodies against HA (1 : 1000) (Roche) and GFP (1 : 1000) (Roche).
[14C]-Methylammonium uptake assays
Initial rates of [14C]-methylammonium (BioActif) uptake were measured as described for amino acids with cells grown in minimal medium containing proline as nitrogen source. Briefly, 5-ml samples of an exponentially growing culture corresponding to about 0.25 mg protein per ml were put, without any change of the medium, into vessels containing the labelled methylammonium and preheated to 29°C or 37°C in a rotary water bath. One-millilitre samples were then removed at time intervals and poured onto filters (0.45 μM, Millipore) which were immediately washed 5 times with 2 ml iced water before counting.
Fluorescence microscopy
Cells were observed on a Zeiss Axio Observer Z1 microscope, driven by MetaMorph (MDS Analytical Technologies). High-resolution images were captured in the confocal mode using a Yokogawa spindisk head and the HQ2 camera with a laser illuminator from Roper (405 nm 100 mW Vortran, 491 nm 50 mW Cobolt Calypso, and 561 nm 50 mW Cobolt Jive). Images were processed with Adobe Photoshop 8.0 (Adobe Systems, Mountain View, CA) and ImageJ (http://rsb.info.nih.gov/ij).
Zdroje
1. Marini AM, Vissers S, Urrestarazu A, Andre B. Cloning and expression of the MEP1 gene encoding an ammonium transporter in Saccharomyces cerevisiae. EMBO J. 1994;13 : 3456–3463. 8062822
2. Biver S, Belge H, Bourgeois S, Van VP, Nowik M, Scohy S, et al. A role for Rhesus factor Rhcg in renal ammonium excretion and male fertility. Nature. 2008;456 : 339–343. doi: 10.1038/nature07518 19020613
3. Marini AM, Urrestarazu A, Beauwens R, Andre B. The Rh (rhesus) blood group polypeptides are related to NH4+ transporters. Trends Biochem. 1997;22 : 460–461.
4. Huang CH, Peng J. Evolutionary conservation and diversification of Rh family genes and proteins. ProcNatlAcadSciUSA 2005;102 : 15512–15517.
5. Auron A, Brophy PD. Hyperammonemia in review: pathophysiology, diagnosis, and treatment. Pediatr Nephrol. 2012;27 : 207–22. doi: 10.1007/s00467-011-1838-5 21431427
6. Von Wiren N, Merrick M. Regulation and function of ammonium carriers in bacteria, fungi, and plants. Molecular mechanisms controlling transmembrane transport. Topics in Current Genetics 9; 2004. pp. 95–120.
7. Weiner ID, Verlander JW. Molecular physiology of the Rh ammonia transport proteins. Curr Opin Nephrol Hypertens. 2010;19 : 471–7. doi: 10.1097/MNH.0b013e32833bfa4e 20539225
8. Zheng L, Kostrewa D, Berneche S, Winkler FK, Li XD. The mechanism of ammonia transport based on the crystal structure of AmtB of Escherichia coli. ProcNatlAcadSciUSA. 2004;101 : 17090–17095.
9. Khademi S, O’Connell J III, Remis J, Robles-Colmenares Y, Miercke LJ, Stroud RM. Mechanism of ammonia transport by Amt/MEP/Rh: structure of AmtB at 1.35 A. Science 2004;305 : 1587–1594. 15361618
10. Andrade SL, Dickmanns A, Ficner R, Einsle O. Crystal structure of the archaeal ammonium transporter Amt-1 from Archaeoglobus fulgidus. ProcNatlAcadSciUSA. 2005;102 : 14994–14999.
11. Li X, Jayachandran S, Nguyen HH, Chan MK. Structure of the Nitrosomonas europaea Rh protein. ProcNatlAcadSciUSA. 2007;104 : 19279–19284.
12. Lupo D, Li XD, Durand A, Tomizaki T, Cherif-Zahar B, Matassi G, et al. The 1.3-A resolution structure of Nitrosomonas europaea Rh50 and mechanistic implications for NH3 transport by Rhesus family proteins. ProcNatlAcadSciUSA. 2007;104 : 19303–19308.
13. Gruswitz F, Chaudhary S, Ho JD, Schlessinger A, Pezeshki B, Ho C-M, et al. Function of human Rh based on structure of RhCG at 2.1 A. Proc Natl Acad Sci U S A. 2010;107 : 9638–43. doi: 10.1073/pnas.1003587107 20457942
14. Deschuyteneer A, Boeckstaens M, De Mees C, Van Vooren P, Wintjens R, Marini AM. SNPs altering ammonium transport activity of human Rhesus factors characterized by a yeast-based functional assay. PLoS One. 2013;8: e71092. doi: 10.1371/journal.pone.0071092 23967154
15. Gruswitz F, O’Connell J III, Stroud RM. Inhibitory complex of the transmembrane ammonia channel, AmtB, and the cytosolic regulatory protein, GlnK, at 1.96 A. ProcNatlAcadSciUSA. 2007;104 : 42–47.
16. Conroy MJ, Durand A, Lupo D, Li X-D, Bullough P a, Winkler FK, et al. The crystal structure of the Escherichia coli AmtB-GlnK complex reveals how GlnK regulates the ammonia channel. Proc Natl Acad Sci U S A. 2007;104 : 1213–8. 17220269
17. Huergo LF, Chandra G, Merrick M. P(II) signal transduction proteins: nitrogen regulation and beyond. FEMS Microbiol Rev. 2013;37 : 251–83. doi: 10.1111/j.1574-6976.2012.00351.x 22861350
18. Boeckstaens M, Llinares E, Van Vooren P, Marini AM. The TORC1 effector kinase Npr1 fine tunes the inherent activity of the Mep2 ammonium transport protein. Nat Commun. 2014;5 : 3101. doi: 10.1038/ncomms4101 24476960
19. Marini AM, Soussi-Boudekou S, Vissers S, Andre B. A family of ammonium transporters in Saccharomyces cerevisiae. MolCell Biol. 1997;17 : 4282–4293.
20. MacGurn JA, Hsu P-C, Smolka MB, Emr SD. TORC1 Regulates Endocytosis via Npr1-Mediated Phosphoinhibition of a Ubiquitin Ligase Adaptor. Cell. 2011;147 : 1104–1117. doi: 10.1016/j.cell.2011.09.054 22118465
21. Merhi A, André B. Internal Amino Acids Promote Gap1 Permease Ubiquitylation via TORC1/Npr1/14-3-3-Dependent Control of the Bul Arrestin-Like Adaptors. Mol Cell Biol. 2012;32 : 4510–22. doi: 10.1128/MCB.00463-12 22966204
22. Boeckstaens M, Andre B, Marini AM. The yeast ammonium transport protein Mep2 and its positive regulator, the Npr1 kinase, play an important role in normal and pseudohyphal growth on various nitrogen media through retrieval of excreted ammonium. Mol Microbiol. 2007;64 : 534–546. 17493133
23. Rutherford JC, Chua G, Hughes T, Cardenas ME, Heitman J. A Mep2-dependent transcriptional profile links permease function to gene expression during pseudohyphal growth in Saccharomyces cerevisiae. Mol Biol Cell. 2008;19 : 3028–3039. doi: 10.1091/mbc.E08-01-0033 18434596
24. Neuhäuser B, Dunkel N, Satheesh S V, Morschhäuser J. Role of the Npr1 kinase in ammonium transport and signaling by the ammonium permease Mep2 in Candida albicans. Eukaryot Cell. 2011;10 : 332–42. doi: 10.1128/EC.00293-10 21278231
25. Dubois E, Grenson M. Methylamine/ammonia uptake systems in saccharomyces cerevisiae: multiplicity and regulation. MolGenGenet. 1979;175 : 67–76.
26. Grenson M, Acheroy B. Mutations affecting the activity and the regulation of the general amino-acid permease of Saccharomyces cerevisiae. Localisation of the cis-acting dominant pgr regulatory mutation in the structural gene of this permease. Mol Gen Genet. 1982;188 : 261–5. 6759873
27. Hein C, Springael JY, Volland C, Haguenauer-Tsapis R, Andre B. NPl1, an essential yeast gene involved in induced degradation of Gap1 and Fur4 permeases, encodes the Rsp5 ubiquitin-protein ligase. Mol Microbiol. 1995;18 : 77–87. 8596462
28. Springael JY, Galan JM, Haguenauer-Tsapis R, Andre B. NH4+-induced down-regulation of the Saccharomyces cerevisiae Gap1p permease involves its ubiquitination with lysine-63-linked chains. J Cell Sci. 1999;112 : 1375–1383. 10194416
29. Springael JY, Nikko E, Andre B, Marini AM. Yeast Npi3/Bro1 is involved in ubiquitin-dependent control of permease trafficking. FEBS Lett. 2002;517 : 103–109. 12062418
30. Yang B, Kumar S. Nedd4 and Nedd4-2: closely related ubiquitin-protein ligases with distinct physiological functions. Cell Death Differ. 2010;17 : 68–77. doi: 10.1038/cdd.2009.84 19557014
31. Bissig C, Gruenberg J. ALIX and the multivesicular endosome: ALIX in Wonderland. Trends Cell Biol. 2014;24 : 19–25. doi: 10.1016/j.tcb.2013.10.009 24287454
32. Nikko E, Marini A-MM, André B, Andre B. Permease recycling and ubiquitination status reveal a particular role for Bro1 in the multivesicular body pathway. J Biol Chem. 2003;278 : 50732–50743. 14523026
33. MacGurn JA, Hsu P-C, Emr SD. Ubiquitin and membrane protein turnover: from cradle to grave. Annu Rev Biochem. 2012;81 : 231–59. doi: 10.1146/annurev-biochem-060210-093619 22404628
34. De Craene JO, Soetens O, Andre B. The Npr1 kinase controls biosynthetic and endocytic sorting of the yeast Gap1 permease. J Biol Chem. 2001;276 : 43939–43948. 11500493
35. Grenson M. Study of the positive control of the general amino-acid permease and other ammonia-sensitive uptake systems by the product of the NPR1 gene in the yeast Saccharomyces cerevisiae. EurJBiochem. 1983;133 : 141–144.
36. Huber A, Bodenmiller B, Uotila A, Stahl M, Wanka S, Gerrits B, et al. Characterization of the rapamycin-sensitive phosphoproteome reveals that Sch9 is a central coordinator of protein synthesis. Genes Dev. 2009;23 : 1929–43. doi: 10.1101/gad.532109 19684113
37. Van Damme P, Lasa M, Polevoda B, Gazquez C, Elosegui-Artola A, Kim DS, et al. N-terminal acetylome analyses and functional insights of the N-terminal acetyltransferase NatB. Proc Natl Acad Sci U S A. 2012;109 : 12449–54. doi: 10.1073/pnas.1210303109 22814378
38. Ongoing and future developments at the Universal Protein Resource. Nucleic Acids Res. 2011;39: D214–9. doi: 10.1093/nar/gkq1020 21051339
39. Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215 : 403–10. 2231712
40. Rice P, Longden I, Bleasby A. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 2000;16 : 276–7. 10827456
41. Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23 : 2947–8. 17846036
42. Hall T. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. 1999;41 : 95–98.
43. Gander S, Bonenfant D, Altermatt P, Martin DE, Hauri S, Moes S, et al. Identification of the rapamycin-sensitive phosphorylation sites within the Ser / Thr-rich domain of the yeast Npr1 protein kinase. Rapid Commun Mass Spectrom. 2008; 3743–3753. doi: 10.1002/rcm.3790 18980262
44. Neklesa TK, Davis RW. A genome-wide screen for regulators of TORC1 in response to amino acid starvation reveals a conserved Npr2/3 complex. PLoS Genet. 2009;5: e1000515. doi: 10.1371/journal.pgen.1000515 19521502
45. Schmidt A, Beck T, Koller A, Kunz J, Hall MN. The TOR nutrient signalling pathway phosphorylates NPR1 and inhibits turnover of the tryptophan permease. EMBO J. 1998;17 : 6924–6931. 9843498
46. Soulard A, Cremonesi A, Moes S, Schütz F, Jenö P, Hall MN. The rapamycin-sensitive phosphoproteome reveals that TOR controls protein kinase A toward some but not all substrates. Mol Biol Cell. 2010;21 : 3475–86. doi: 10.1091/mbc.E10-03-0182 20702584
47. Iesmantavicius V, Weinert BT, Choudhary C. Convergence of ubiquitylation and phosphorylation signaling in rapamycin-treated yeast cells. Mol Cell Proteomics. 2014;13 : 1979–92. doi: 10.1074/mcp.O113.035683 24961812
48. Soufi B, Kelstrup CD, Stoehr G, Fröhlich F, Walther TC, Olsen J V. Global analysis of the yeast osmotic stress response by quantitative proteomics. Mol Biosyst. 2009;5 : 1337–46. doi: 10.1039/b902256b 19823750
49. Holt LJ, Tuch BB, Villen J, Johnson AD, Gygi SP, Morgan DO. Global Analysis of Cdk1 Substrate Phosphorylation Sites Provides Insights into Evolution. Science (80-). 2009;325 : 1682–1686.
50. Helbig AO, Rosati S, Pijnappel PWWM, van Breukelen B, Timmers MHTH, Mohammed S, et al. Perturbation of the yeast N-acetyltransferase NatB induces elevation of protein phosphorylation levels. BMC Genomics. 2010;11 : 685. doi: 10.1186/1471-2164-11-685 21126336
51. Gnad F, de Godoy LMF, Cox J, Neuhauser N, Ren S, Olsen J V, et al. High-accuracy identification and bioinformatic analysis of in vivo protein phosphorylation sites in yeast. Proteomics. 2009;9 : 4642–52. doi: 10.1002/pmic.200900144 19795423
52. Breitkreutz a., Choi H, Sharom JR, Boucher L, Neduva V, Larsen B, et al. A Global Protein Kinase and Phosphatase Interaction Network in Yeast. Science (80-). 2010;328 : 1043–1046.
53. Fasolo J, Sboner A, Sun MGF, Yu H, Chen R, Sharon D, et al. Diverse protein kinase interactions identified by protein microarrays reveal novel connections between cellular processes. Genes Dev. 2011;25 : 767–78. doi: 10.1101/gad.1998811 21460040
54. Loqué D, Lalonde S, Looger LL, von Wirén N, Frommer WB. A cytosolic trans-activation domain essential for ammonium uptake. Nature. 2007;446 : 195–8. 17293878
55. Yuan L, Gu R, Xuan Y, Smith-Valle E, Loqué D, Frommer WB, et al. Allosteric regulation of transport activity by heterotrimerization of Arabidopsis ammonium transporter complexes in vivo. Plant Cell. 2013;25 : 974–84. doi: 10.1105/tpc.112.108027 23463773
56. Thomas G, Coutts G, Merrick M. The glnKamtB operon. A conserved gene pair in prokaryotes. Trends Genet. 2000;16 : 11–14. 10637624
57. Sant’Anna FH, Trentini DB, de Souto Weber S, Cecagno R, da Silva SC, Schrank IS. The PII superfamily revised: a novel group and evolutionary insights. J Mol Evol. 2009;68 : 322–36. doi: 10.1007/s00239-009-9209-6 19296042
58. Javelle A, Severi E, Thornton J, Merrick M. Ammonium sensing in Escherichia coli. Role of the ammonium transporter AmtB and AmtB-GlnK complex formation. J Biol Chem. 2004;279 : 8530–8538. 14668330
59. Truan D, Huergo LF, Chubatsu LS, Merrick M, Li X-D, Winkler FK. A new P(II) protein structure identifies the 2-oxoglutarate binding site. J Mol Biol. 2010;400 : 531–9. doi: 10.1016/j.jmb.2010.05.036 20493877
60. Radchenko M V, Thornton J, Merrick M. P(II) signal transduction proteins are ATPases whose activity is regulated by 2-oxoglutarate. Proc Natl Acad Sci U S A. 2013;110 : 12948–53. doi: 10.1073/pnas.1304386110 23818625
61. Bah A, Vernon RM, Siddiqui Z, Krzeminski M, Muhandiram R, Zhao C, et al. Folding of an intrinsically disordered protein by phosphorylation as a regulatory switch. Nature. 2014;519 : 106–9. doi: 10.1038/nature13999 25533957
62. Wolfe KH, Shields DC. Molecular evidence for an ancient duplication of the entire yeast genome. Nature. 1997;387 : 708–713. 9192896
63. Boeckstaens M, Andre B, Marini AM. Distinct transport mechanisms in yeast ammonium transport/sensor proteins of the mep/amt/rh family and impact on filamentation. J Biol Chem. 2008;283 : 21362–21370. doi: 10.1074/jbc.M801467200 18508774
64. Lorenz MC, Heitman J. The MEP2 ammonium permease regulates pseudohyphal differentiation in Saccharomyces cerevisiae. EMBO J. 1998;17 : 1236–1247. 9482721
65. Pfannmüller A, Wagner D, Sieber C, Schönig B, Boeckstaens M, Marini AM, et al. The General Amino Acid Permease FfGap1 of Fusarium fujikuroi Is Sorted to the Vacuole in a Nitrogen-Dependent, but Npr1 Kinase-Independent Manner. PLoS One. 2015;10: e0125487. doi: 10.1371/journal.pone.0125487 25909858
66. Bechet J, Grenson M, Wiame JM. Mutations affecting the repressibility of arginine biosynthetic enzymes in Saccharomyces cerevisiae. EurJBiochem. 1970;12 : 31–39.
67. Gietz D, St JA, Woods RA, Schiestl RH. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992;20 : 1425. 1561104
68. Wach A, Brachat A, Pohlmann R, Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;10 : 1793–1808. 7747518
69. Lee S, Lim WA, Thorn KS. Improved blue, green, and red fluorescent protein tagging vectors for S. cerevisiae. PLoS One. 2013;8: e67902. doi: 10.1371/journal.pone.0067902 23844123
70. Longtine MS, McKenzie A, Demarini DJ, Shah NG, Wach A, Brachat A, et al. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14 : 953–61. 9717241
71. Jacobs P, Jauniaux JC, Grenson M. A cis-dominant regulatory mutation linked to the argB-argC gene cluster in Saccharomyces cerevisiae. JMolBiol. 1980;139 : 691–704.
72. Vandenbol M, Jauniaux JC, Vissers S, Grenson M. Isolation of the NPR1 gene responsible for the reactivation of ammonia-sensitive amino-acid permeases in Saccharomyces cerevisiae. RNA analysis and gene dosage effects. EurJBiochem. 1987;164 : 607–612.
73. Feller A, Boeckstaens M, Marini AM, Dubois E. Transduction of the nitrogen signal activating Gln3-mediated transcription is independent of Npr1 kinase and Rsp5-Bul1/2 ubiquitin ligase in Saccharomyces cerevisiae. J Biol Chem. 2006;281 : 28546–54. 16864574
74. Bonneaud N, Ozier-Kalogeropoulos O, Li GY, Labouesse M, Minvielle-Sebastia L, Lacroute F. A family of low and high copy replicative, integrative and single-stranded S. cerevisiae/E. coli shuttle vectors. Yeast. 1991;7 : 609–615. 1767589
75. Volland C, Urban-Grimal D, Geraud G, Haguenauer-Tsapis R, Géraud G, Haguenauer-Tsapis R. Endocytosis and degradation of the yeast uracil permease under adverse conditions. J Biol Chem. 1994;269 : 9833–41. 8144575
76. Schagger H, von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. AnalBiochem. 1987;166 : 368–379.
77. Marini AM, Andre B. In vivo N-glycosylation of the mep2 high-affinity ammonium transporter of Saccharomyces cerevisiae reveals an extracytosolic N-terminus. MolMicrobiol. 2000;38 : 552–564.
78. Copic A, Starr TL, Schekman R. Ent3p and Ent5p exhibit cargo-specific functions in trafficking proteins between the trans-Golgi network and the endosomes in yeast. Mol Biol Cell. 2007;18 : 1803–15. 17344475
Štítky
Genetika Reprodukční medicína
Článek Discovery and Fine-Mapping of Glycaemic and Obesity-Related Trait Loci Using High-Density ImputationČlánek AAA-ATPase FIDGETIN-LIKE 1 and Helicase FANCM Antagonize Meiotic Crossovers by Distinct MechanismsČlánek A Conserved Pattern of Primer-Dependent Transcription Initiation in and Revealed by 5′ RNA-seqČlánek TopBP1 Governs Hematopoietic Stem/Progenitor Cells Survival in Zebrafish Definitive HematopoiesisČlánek Redundant Roles of Rpn10 and Rpn13 in Recognition of Ubiquitinated Proteins and Cellular Homeostasis
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2015 Číslo 7
-
Všechny články tohoto čísla
- LINE-1 Retroelements Get ZAPped!
- /p23: A Small Protein Heating Up Lifespan Regulation
- Hairless Streaks in Cattle Implicate TSR2 in Early Hair Follicle Formation
- Ribosomal Protein Mutations Result in Constitutive p53 Protein Degradation through Impairment of the AKT Pathway
- Molecular Clock of Neutral Mutations in a Fitness-Increasing Evolutionary Process
- Modeling Implicates in Nephropathy: Evidence for Dominant Negative Effects and Epistasis under Anemic Stress
- The Alternative Sigma Factor SigX Controls Bacteriocin Synthesis and Competence, the Two Quorum Sensing Regulated Traits in
- BMP Inhibition in Seminomas Initiates Acquisition of Pluripotency via NODAL Signaling Resulting in Reprogramming to an Embryonal Carcinoma
- Comparative Study of Regulatory Circuits in Two Sea Urchin Species Reveals Tight Control of Timing and High Conservation of Expression Dynamics
- EIN3 and ORE1 Accelerate Degreening during Ethylene-Mediated Leaf Senescence by Directly Activating Chlorophyll Catabolic Genes in
- Genome Wide Binding Site Analysis Reveals Transcriptional Coactivation of Cytokinin-Responsive Genes by DELLA Proteins
- Sensory Neurons Arouse . Locomotion via Both Glutamate and Neuropeptide Release
- A Year of Infection in the Intensive Care Unit: Prospective Whole Genome Sequencing of Bacterial Clinical Isolates Reveals Cryptic Transmissions and Novel Microbiota
- Inference of Low and High-Grade Glioma Gene Regulatory Networks Delineates the Role of Rnd3 in Establishing Multiple Hallmarks of Cancer
- Novel Role for p110β PI 3-Kinase in Male Fertility through Regulation of Androgen Receptor Activity in Sertoli Cells
- A Novel Locus Harbouring a Functional Nonsense Mutation Identified in a Large Danish Family with Nonsyndromic Hearing Impairment
- Checkpoint Activation of an Unconventional DNA Replication Program in
- A Genetic Incompatibility Accelerates Adaptation in Yeast
- The SMC Loader Scc2 Promotes ncRNA Biogenesis and Translational Fidelity
- Blimp1/Prdm1 Functions in Opposition to Irf1 to Maintain Neonatal Tolerance during Postnatal Intestinal Maturation
- Discovery and Fine-Mapping of Glycaemic and Obesity-Related Trait Loci Using High-Density Imputation
- JAK/STAT and Hox Dynamic Interactions in an Organogenetic Gene Cascade
- Emergence, Retention and Selection: A Trilogy of Origination for Functional Proteins from Ancestral LncRNAs in Primates
- MoSET1 (Histone H3K4 Methyltransferase in ) Regulates Global Gene Expression during Infection-Related Morphogenesis
- Arabidopsis PCH2 Mediates Meiotic Chromosome Remodeling and Maturation of Crossovers
- AAA-ATPase FIDGETIN-LIKE 1 and Helicase FANCM Antagonize Meiotic Crossovers by Distinct Mechanisms
- A Conserved Pattern of Primer-Dependent Transcription Initiation in and Revealed by 5′ RNA-seq
- Tempo and Mode of Transposable Element Activity in Drosophila
- The Shelterin TIN2 Subunit Mediates Recruitment of Telomerase to Telomeres
- SAMHD1 Inhibits LINE-1 Retrotransposition by Promoting Stress Granule Formation
- A Genome Scan for Genes Underlying Microgeographic-Scale Local Adaptation in a Wild Species
- TopBP1 Governs Hematopoietic Stem/Progenitor Cells Survival in Zebrafish Definitive Hematopoiesis
- Analysis of the Relationships between DNA Double-Strand Breaks, Synaptonemal Complex and Crossovers Using the Mutant
- Assessing Mitochondrial DNA Variation and Copy Number in Lymphocytes of ~2,000 Sardinians Using Tailored Sequencing Analysis Tools
- Allelic Spectra of Risk SNPs Are Different for Environment/Lifestyle Dependent versus Independent Diseases
- CSB-PGBD3 Mutations Cause Premature Ovarian Failure
- Irrepressible: An Interview with Mark Ptashne
- Genetic Evidence for Function of the bHLH-PAS Protein Gce/Met As a Juvenile Hormone Receptor
- Inactivation of Retinoblastoma Protein (Rb1) in the Oocyte: Evidence That Dysregulated Follicle Growth Drives Ovarian Teratoma Formation in Mice
- Redundant Roles of Rpn10 and Rpn13 in Recognition of Ubiquitinated Proteins and Cellular Homeostasis
- Pyrimidine Pool Disequilibrium Induced by a Cytidine Deaminase Deficiency Inhibits PARP-1 Activity, Leading to the Under Replication of DNA
- Molecular Framework of a Regulatory Circuit Initiating Two-Dimensional Spatial Patterning of Stomatal Lineage
- RFX2 Is a Major Transcriptional Regulator of Spermiogenesis
- A Role for Macro-ER-Phagy in ER Quality Control
- Corp Regulates P53 in via a Negative Feedback Loop
- Common Cell Shape Evolution of Two Nasopharyngeal Pathogens
- Contact- and Protein Transfer-Dependent Stimulation of Assembly of the Gliding Motility Machinery in
- Endothelial Snail Regulates Capillary Branching Morphogenesis via Vascular Endothelial Growth Factor Receptor 3 Expression
- Functional Constraint Profiling of a Viral Protein Reveals Discordance of Evolutionary Conservation and Functionality
- Temporal Coordination of Carbohydrate Metabolism during Mosquito Reproduction
- mTOR Directs Breast Morphogenesis through the PKC-alpha-Rac1 Signaling Axis
- Reversible Oxidation of a Conserved Methionine in the Nuclear Export Sequence Determines Subcellular Distribution and Activity of the Fungal Nitrate Regulator NirA
- Nutritional Control of DNA Replication Initiation through the Proteolysis and Regulated Translation of DnaA
- Cooperation between Paxillin-like Protein Pxl1 and Glucan Synthase Bgs1 Is Essential for Actomyosin Ring Stability and Septum Formation in Fission Yeast
- Encodes a Highly Conserved Protein Important to Neurological Function in Mice and Flies
- Identification of a Novel Regulatory Mechanism of Nutrient Transport Controlled by TORC1-Npr1-Amu1/Par32
- Aurora-A-Dependent Control of TACC3 Influences the Rate of Mitotic Spindle Assembly
- Large-Scale Phenomics Identifies Primary and Fine-Tuning Roles for CRKs in Responses Related to Oxidative Stress
- TFIIS-Dependent Non-coding Transcription Regulates Developmental Genome Rearrangements
- Genome-Wide Reprogramming of Transcript Architecture by Temperature Specifies the Developmental States of the Human Pathogen
- Identification of Chemical Inhibitors of β-Catenin-Driven Liver Tumorigenesis in Zebrafish
- The Catalytic and Non-catalytic Functions of the Chromatin-Remodeling Protein Collaborate to Fine-Tune Circadian Transcription in
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Functional Constraint Profiling of a Viral Protein Reveals Discordance of Evolutionary Conservation and Functionality
- Reversible Oxidation of a Conserved Methionine in the Nuclear Export Sequence Determines Subcellular Distribution and Activity of the Fungal Nitrate Regulator NirA
- Modeling Implicates in Nephropathy: Evidence for Dominant Negative Effects and Epistasis under Anemic Stress
- Nutritional Control of DNA Replication Initiation through the Proteolysis and Regulated Translation of DnaA
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



