-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaRedundant Roles of Rpn10 and Rpn13 in Recognition of Ubiquitinated Proteins and Cellular Homeostasis
At least two major ubiquitin receptor subunits that directly capture ubiquitin chains have been identified in the proteasome: Rpn10 and Rpn13. Analyses in Saccharomyces cerevisiae have suggested only a modest role of Rpn10 and Rpn13 in the recruitment of ubiquitinated proteins, as double deletion of Rpn10 and Rpn13 causes very mild phenotypes. Considering that ubiquitin recognition is an essential process for protein degradation by the proteasome and that failure in degradation of ubiquitinated proteins leads to human diseases such as neurodegeneration, it is important to evaluate the role of Rpn10 and Rpn13 in mammals. Liver-specific deletion of either Rpn10 or Rpn13 showed modest impairment, but simultaneous loss of both Rpn10 and Rpn13 caused severe liver injury accompanied by massive accumulation of ubiquitin conjugates and failure in recruiting mHR23B and ubiquilin/Plic-1 and -4 proteins, which deliver ubiquitinated proteins to the proteasome. Our findings indicate that the largely redundant roles of Rpn10 and Rpn13 in ubiquitin recognition and recruitment of mHR23B and ubiquilin/Plic-1 and -4 are essential for cellular homeostasis in mammals and should provide information for understanding the mechanism of ubiquitin recognition by the 26S proteasome in mammals and for development of therapeutic agents targeting protein degradation.
Published in the journal: . PLoS Genet 11(7): e32767. doi:10.1371/journal.pgen.1005401
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1005401Summary
At least two major ubiquitin receptor subunits that directly capture ubiquitin chains have been identified in the proteasome: Rpn10 and Rpn13. Analyses in Saccharomyces cerevisiae have suggested only a modest role of Rpn10 and Rpn13 in the recruitment of ubiquitinated proteins, as double deletion of Rpn10 and Rpn13 causes very mild phenotypes. Considering that ubiquitin recognition is an essential process for protein degradation by the proteasome and that failure in degradation of ubiquitinated proteins leads to human diseases such as neurodegeneration, it is important to evaluate the role of Rpn10 and Rpn13 in mammals. Liver-specific deletion of either Rpn10 or Rpn13 showed modest impairment, but simultaneous loss of both Rpn10 and Rpn13 caused severe liver injury accompanied by massive accumulation of ubiquitin conjugates and failure in recruiting mHR23B and ubiquilin/Plic-1 and -4 proteins, which deliver ubiquitinated proteins to the proteasome. Our findings indicate that the largely redundant roles of Rpn10 and Rpn13 in ubiquitin recognition and recruitment of mHR23B and ubiquilin/Plic-1 and -4 are essential for cellular homeostasis in mammals and should provide information for understanding the mechanism of ubiquitin recognition by the 26S proteasome in mammals and for development of therapeutic agents targeting protein degradation.
Introduction
The ubiquitin-proteasome system is the main non-lysosomal proteolytic pathway through which regulatory proteins and misfolded proteins are degraded in eukaryotic cells [1,2]. Ubiquitin chains are covalently attached to target proteins through the coordinated effort of an enzymatic cascade. Ubiquitinated proteins are then recognized and degraded by the 26S proteasome in an ATP-dependent manner. The 26S proteasome is composed of one proteolytically active 20S core particle (CP) and 19S regulatory particles (RP) attached to one or both ends of the CP [3].
The RP plays an essential role in the degradation of ubiquitinated proteins by recognizing ubiquitin chains, deubiquitinating and unfolding substrate proteins, opening the gate of the CP, and translocating the substrates into the CP. The RP can be divided into two subcomplexes; the base and the lid [2]. The base contains six ATPase subunits Rpt1–Rpt6 and two large non-ATPase subunits Rpn1 (Q8VDM4) and Rpn2 (Q3TXS7), which function as scaffolds for molecules that modulate proteasome functions, such as Rpn13 (encoded by Adrm1 [56436] in mice), Uch37 (Q9WUP7), and Usp14 (Q9JMA1) [4–8]. The lid has been shown to be essential for the degradation of ubiquitinated proteins through the function of Rpn11 (O35593), which cleaves ubiquitin (P0CG50) chains from substrates prior to degradation [9,10].
The RP has two major ubiquitin receptor subunits, Rpn10 (P38886) and Rpn13 (O13563), which directly bind to ubiquitin chains [11–13]. Rpn10 and Rpn13 can also receive ubiquitinated proteins from extraproteasomal UBL-UBA proteins, such as HR23 (P54728), ubiquilin (also called Plic) (Q8R317, Q99NB8), and Ddi1 (Q9DAF3), which have been reported to bind to either Rpn1, Rpn10, or Rpn13 via ubiquitin-like (UBL) domains and to ubiquitin chains via ubiquitin-associated (UBA) domains [14,15].
Rpn10 is composed of an N-terminal von Willebrand factor A (VWA) domain and a C-terminal ubiquitin interacting motif (UIM). While S. cerevisiae Rpn10 has a single UIM that preferentially binds to K48-linked ubiquitin chains [16], human Rpn10 has two UIMs and binds to both K48-and K63-linked ubiquitin chains with equally high affinities by using the two UIMs in a cooperative manner [17–20]. Previously, we demonstrated that mice (Mus musculus) lacking the UIMs of the Rpn10 (Rpn10ΔUIM) exhibited embryonic lethality, suggesting the importance of UIMs in Rpn10-mediated recognition of ubiquitinated proteins [21]. Rpn13 has an N-terminal pleckstrin-like receptor of ubiquitin (PRU) domain, which binds preferentially to the proximal ubiquitin of K48-linked diubiquitin chains [13]. Rpn10 and Rpn13 can bind simultaneously to a K48-linked diubiquitin; Rpn13 binds to the proximal ubiquitin while Rpn10 binds to the distal one [20]. However, it is unclear whether such coordination of Rpn10 and Rpn13 occurs in vivo.
The recognition pathways for ubiquitinated substrates appear to have diverged in different species. For example, neither Rpn10 nor Rpn13 is essential in S. cerevisiae [12,22]. However, Rpn10 is essential in mice and Drosophila melanogaster [21,23]. In addition, Rpn13-null mice carrying a gene trap mutation were smaller at birth and infertile due to defective gametogenesis [24]. Similarly, the UBL-UBA proteins are not essential for S. cerevisiae cell growth, while some have been shown to be essential in mouse development [12,25–27]. Although both Rpn10 and Rpn13 are considered major receptors for direct recognition of ubiquitinated substrates by the 26S proteasome, the biological significance of Rpn13 and detailed mechanisms of recognition of ubiquitinated proteins by these two receptors are still not fully understood [14,28].
In this study, to examine the recognition pathway for ubiquitinated substrates in mice, we generated Rpn13-null mice and liver-specific Rpn13-deficient mice. Rpn13-null mice died soon after birth. We also revealed that the deletion of both Rpn10-UIM and Rpn13 in the liver caused significant accumulation of ubiquitinated proteins due to impaired recognition of ubiquitinated proteins and defects in recruitment of mHR23B and ubiquilin/Plic-1 and -4 to the proteasome. Our results indicate that the largely, if not entirely, redundant roles of Rpn10 and Rpn13 in ubiquitin recognition and recruitment of mHR23B and ubiquilin/Plic-1 and -4 are essential for cellular homeostasis in mammals.
Results
Loss of Rpn13 causes neonatal lethality in mice
Rpn13 is encoded by Adrm1 in the mouse genome. A targeting vector was designed to modify the Adrm1 gene by homologous recombination in ES cells so that exon 3 and exon 4 of the gene were flanked by loxP sites (S1 Fig). The floxed allele was confirmed by Southern blot analysis (S1 Fig). Deletion of exon 3 and exon 4 of the gene by expressing DNA recombinase Cre causes a frame shift and emergence of a stop codon in exon 5. Therefore, even if splicing occurred between exon 2 and exon 5, the function of Rpn13 would be disrupted. To obtain mice lacking Rpn13 (Rpn13KO), Adrm1F/F mice were crossed with mice expressing Cre recombinase throughout the body under the control of the adenovirus-derived EIIa promoter [29]. Mice heterozygous for Adrm1 (Adrm1+/-) were healthy, fertile, and did not show any obvious phenotypes. Intercrosses of Adrm1+/- mice revealed that Rpn13KO mice were delivered at lower frequency than the expected Mendelian frequency (Fig 1A). The body weight of Adrm1-/- neonates (0.86 ± 0.13 g, n = 9) was significantly smaller than that of Adrm1+/+ and Adrm1+/- neonates (1.07 ± 0.11 g, n = 47; p < 0.01), and all Adrm1-/- neonates were runted, cyanotic, did not move spontaneously, and died within 1 day after birth (Fig 1B). Eventually, no Adrm1-/- mice were obtained at postnatal day 28 (P28) (Fig 1A). These results indicate that a Rpn13 homozygous mutation causes postnatal lethality.
Fig. 1. Loss of Rpn13 causes neonatal lethality in mice. 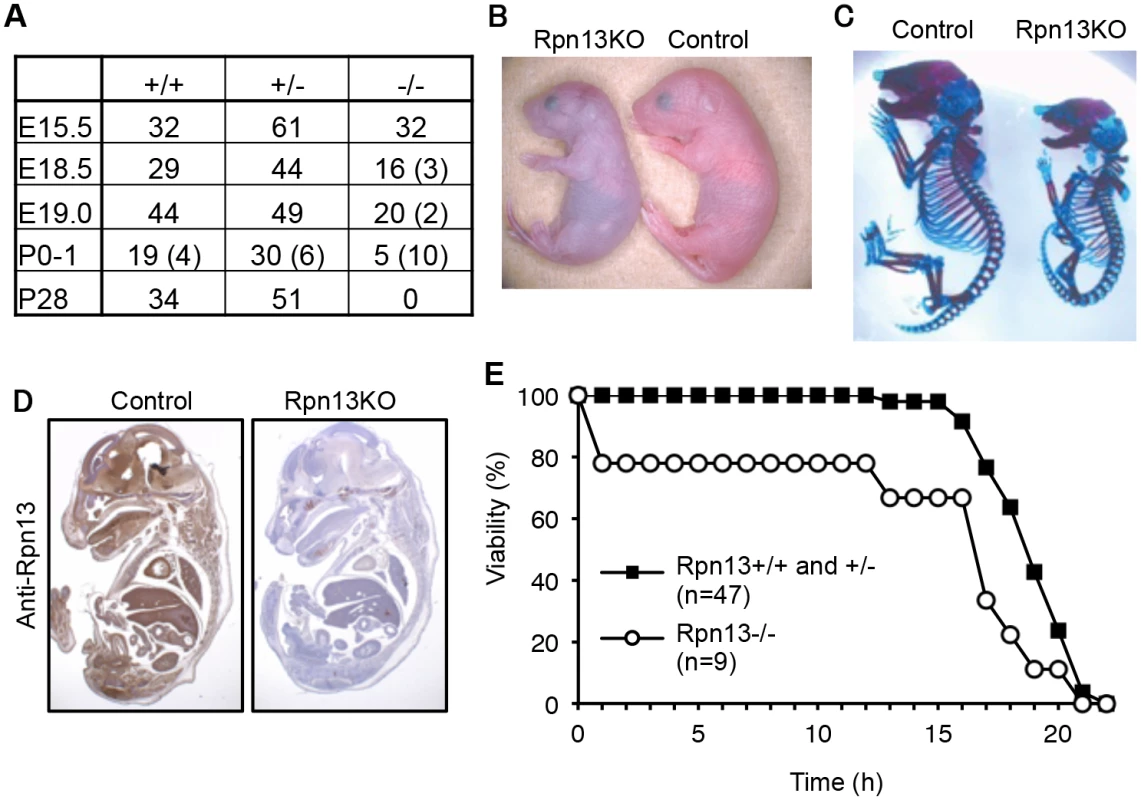
(A) Genotype frequencies of embryos produced from Adrm1+/- (Rpn13+/-) mouse intercrosses. Numbers in parenthesis indicate resorbed fetuses or dead newborns. E: Embryonic day, P: Postnatal day. (B) Gross appearance of control and Rpn13KO littermates shortly after birth. (C) Skeletal analysis of E18.5 littermates by Alzarin red (bone) and Alcian blue (cartilage) staining. (D) Immunohistochemical analysis of sagittally sectioned E18.5 littermates by Rpn13 antibody. (E) Survival curves of newborn mice. Control and Rpn13KO mice were delivered by cesarean section. To understand the cause of postnatal death, we examined the skeletal structure of P0 mice (Fig 1C), whole body sections of embryonic day 18.5 (E18.5) embryos (Fig 1D), histology of the placenta of E18.5 and E15.5 embryos (S2 Fig), and the heart of P0 neonates (S2 Fig). We confirmed loss of Rpn13 protein in a whole body section (Fig 1D). However, we were not able to detect apparent morphological and histological defects in organs of Rpn13KO mice other than their small sizes. Since newborn Rpn13KO mice were cyanotic and did not show an abnormality in the heart that would cause congestion (S2 Fig), we performed histological analysis of the lung. At E18.5, there was no difference between control and Rpn13KO mice (S2 Fig). However, pulmonary alveoli of Rpn13KO newborns did not expand, while those of control littermates did (S2 Fig). This result indicates that Rpn13KO mice failed to breathe after birth. Because Rpn13KO neonates appeared to be breathing in response to mechanical skin stimulation, we tested whether cesarean delivery followed by stimulation for breathing would rescue the early death of Rpn13KO neonates. Most Rpn13KO neonates delivered by cesarean section survived more than ten hours, which is longer than the survival time of Rpn13KO neonates obtained by natural delivery, yet shorter than that of control littermates (Fig 1E). Histologically, these stimulated Rpn13KO neonates showed expanded alveoli, similar to control littermates (S2 Fig). These results suggest that one of the major causes of the early neonatal death of Rpn13KO mice is spontaneous breathing failure.
Rpn13 deficiency in the liver impairs degradation of ubiquitinated proteins
To examine the biochemical basis of the significance of Rpn13, we generated liver-specific Rpn13 knockout (Rpn13LKO) mice by crossing Adrm1-floxed mice with transgenic mice that expressed Cre recombinase in postnatal hepatocytes under the control of the albumin (Alb) promoter [30]. Rpn13LKO mice were born without any developmental defects. We confirmed loss of Rpn13 protein in the 8-week-old Rpn13LKO liver (Fig 2A). Loss of Rpn13 caused concurrent loss of Uch37, a deubiquitinating enzyme bound to Rpn13, consistent with our previous work [4]. In addition, we observed increases in protein levels of the CP subunits α3 (Q9R1P0) and β2 (P70195) and the RP subunits Rpn8 (A1L3B8) and Rpt6 (P62196) in the Rpn13LKO liver, compared to the control liver (Fig 2A).
Fig. 2. Rpn13 deficiency in the liver impairs degradation of ubiquitinated proteins. 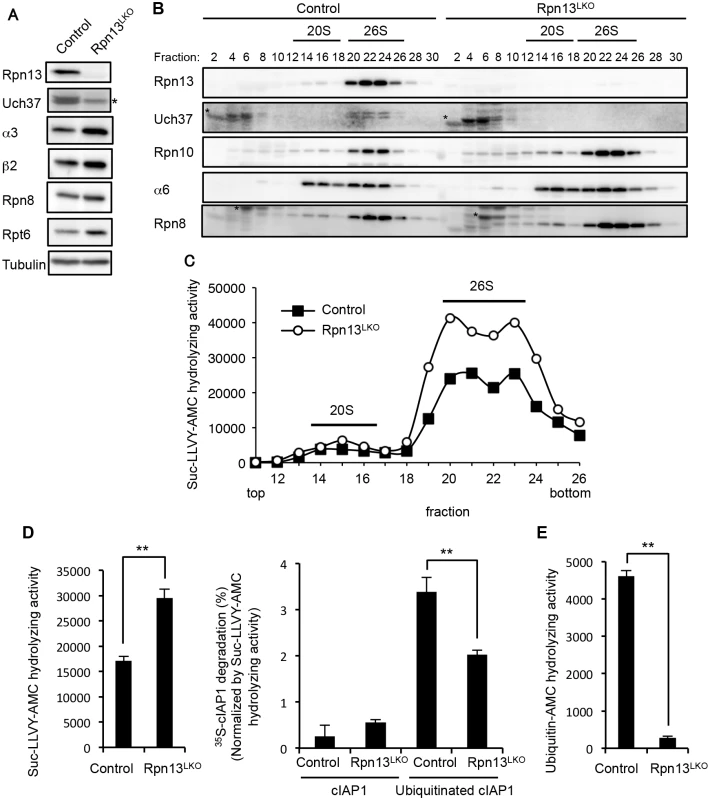
(A) Immunoblot analysis of liver lysates from 8-week-old control and Rpn13LKO mice with antibodies against the indicated proteins. Asterisk indicates a nonspecific band. (B) Lysates from control and Rpn13LKO livers were fractionated by glycerol gradient centrifugation (8 to 32% glycerol from fraction 1 to 30) and an equal amount of each fraction was used for immunoblot analysis using antibodies against the indicated proteins. Asterisks indicate nonspecific bands. (C) Each fraction of (B) was assayed for chymotrypsin-like activity using Suc-LLVY-AMC as a substrate. (D) The 26S proteasome fractions of (C) (fractions 20–23) were subjected to the assay of chymotrypsin-like activity (left panel), and degradation of 35S-labeled cIAP1 with or without ubiquitination was measured and normalized by chymotrypsin-like activity (right panel). Data are mean ±standard deviations from triplicate experiments. **p < 0.01. (E) The deubiquitinating activities of 26S proteasome fractions of (C) were measured using ubiquitin-AMC as a substrate. Data are mean ± standard deviations from triplicate experiments. **p < 0.01 We next fractionated liver lysates by glycerol gradient centrifugation, followed by immunoblot analysis and measurement of peptidase activity of each fraction. The absence of Rpn13 and Uch37 did not affect the assembly of the 26S proteasome in Rpn13LKO liver as shown by the normal distribution of Rpn10, α6 (Q3TS44) (CP), and Rpn8 (RP). As suggested by the results in Fig 2A, the amount of the 26S proteasome increased in Rpn13LKO liver (Fig 2B). This observation was confirmed by increased peptidase activity in the 26S proteasome fractions of the Rpn13LKO liver, compared with the control liver (Fig 2C). Consistent with this, mRNA levels of proteasome subunits in the Rpn13LKO liver were increased (S3 Fig). These results suggest that absence of Rpn13 attenuates degradation of ubiquitinated proteins and induces a feedback increase in the expression of proteasome subunits [31–34].
To confirm this view, we employed the 26S proteasome fraction (fraction 20–23 in Fig 2C) for in vitro degradation of ubiquitinated cIAP1 (Q62210) protein [35]. The 26S fraction of Rpn13LKO liver lysates contained a larger amount of the 26S proteasome than that of control liver lysates, and hence exhibited higher peptidase activity (Fig 2B, 2C and 2D, left panel). Therefore, the degradation rate of cIAP1 was normalized by the peptidase activity. This revealed that the 26S proteasome without Rpn13 had a moderate defect in degrading ubiquitinated cIAP (Fig 2D, right panel). Also, the 26S proteasome lacking Rpn13 showed low deubiquitinating activity compared to the control 26S proteasome, most likely due to loss of Uch37 (Fig 2E). These results indicate that Rpn13 plays an important role in degradation of ubiquitinated proteins in the mouse liver.
Simultaneous deletion of Rpn10-UIM and Rpn13 causes severe liver injury
We previously reported that mice lacking the UIMs of Rpn10 (Rpn10ΔUIM) are embryonic lethal and that liver-specific ablation of Rpn10-UIM (Rpn10LΔUIM) caused accumulation of ubiquitinated proteins in the liver [21]. Since Δrpn10 and Δrpn13 leads to synthetic defects in proteasome function in S. cerevisiae [12,36], we next examined whether absence of Rpn10-UIM and Rpn13 also exhibit a synthetic effect in mouse liver.
Liver-specific double knockout for Rpn10-UIM and Rpn13 (DKO) caused an aberrantly icteric liver at two weeks of age, compared to controls (Fig 3A). Histological analysis of these livers as well as each single knockout liver revealed that whereas either Rpn10LΔUIM or Rpn13LKO livers had almost normal liver histology indistinguishable from the control liver, DKO livers had disorganized architecture (Fig 3B). Immunohistochemistry using antibodies against the Rpn10 C-terminus (corresponding to the UIM domain) and Rpn13 proteins confirmed the absence of both Rpn10-UIM and Rpn13 proteins in the DKO liver (Fig 3C). The DKO liver exhibited multiple areas of hepatocyte loss (Fig 3D) and hypertrophic hepatocytes (S4 Fig). Immunostaining with an anti-ubiquitin antibody detected strong ubiquitin-positive signals in the cytosol of DKO hepatocytes (Fig 3E). We also observed vacuolated nuclei in the DKO hepatocytes (S4 Fig). At 5 weeks of age, the expression of the extracellular matrix genes Col1a1 (12842) and Epcam (17075) were upregulated in DKO livers (Fig 3F, left panel), and as a consequence DKO liver displayed fibrosis as detected by Azan staining (Fig 3F, right panel); chronic hepatitis and cholestasis are known to lead to liver fibrosis [37]. At 7 weeks of age, focal necrosis was apparent in the DKO liver (S4 Fig).
Fig. 3. Simultaneous deletion of Rpn10-UIM and Rpn13 causes severe liver injury. 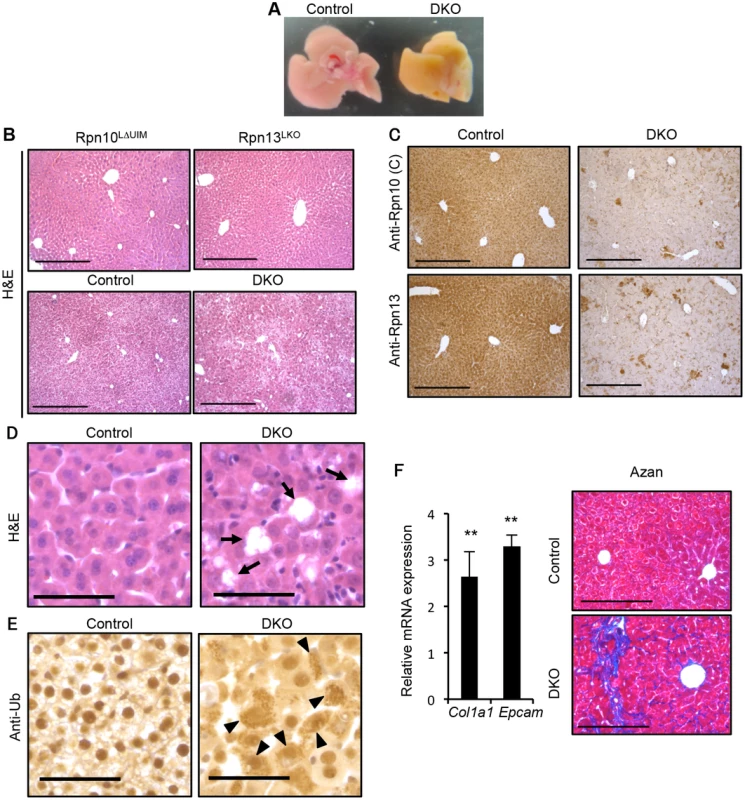
(A) Representative macroscopic images of 2-week-old control and DKO livers. (B) H&E staining of 4-week-old Rpn10LΔUIM, Rpn13LKO and 2-week-old control and DKO livers. All scale bars (black lines), 300 μm. (C) Immunohistochemical analysis on representative liver paraffin sections from 2-week-old mice by using Rpn10 (C) and Rpn13 antibodies. Scale bars, 300 μm. (D and E) Representative H&E staining (D) and immunohistochemical analysis of ubiquitin (E) on liver sections from 2-week-old mice. Arrows in (D) indicate regions of sloughing hepatocytes. Arrowheads in (E) indicate hepatocytes with high accumulation of ubiquitin in cytosol. All scale bars (black lines), 50 μm. (F) Azan staining of liver sections from 5-week-old control and DKO mice (right panels). All scale bars (black lines), 200 μm. Real-time RT-PCR was used to measure the expression of transcripts encoding fibrosis markers (Col1a1 and Epcam) in the livers of 3–6-week-old control and DKO mice (left panels). Data represent levels of transcripts in each genotype liver relative to those in control liver and are expressed as means; error bars denote SEM. **p < 0.01 (n = 4 each genotype). Consistent with the icteric appearance of the DKO liver, serum markers of cholestasis such as alkaline phosphatase (ALP), γ-glutamyltranspeptidase (γ-GTP), cholesterol, bilirubin, and bile acid, as well as markers of liver injury (alanine and asparatate aminotransferases; ALT, AST) were significantly increased in 4-week-old DKO mice (S4 Fig), suggesting that either impairment of the biliary system or hepatocyte death is the cause of the liver pathology. We employed quantitative RT-PCR analysis to assess expression of genes involved in the biliary system. Reduction in Slc10a1 (20493), which mediates bile acid uptake into hepatocytes, and increase in Abcc4 (239273), which promotes efflux of bile acid from hepatocytes, suggests compensatory regulation to reduce the high concentration of bile acid in the DKO liver. This is consistent with the expression pattern observed in liver injury (S4 Fig) [38]. Furthermore, the expression of genes involved in bile acid synthesis (Cyp8b1 [13124] and Cyp7a1 [13122]) was not increased in DKO liver (S4 Fig). These results suggest that the principal defect resulting from simultaneous loss of Rpn10 and Rpn13 is hepatocyte injury.
Redundant roles of Rpn10 and Rpn13 in degradation of ubiquitinated proteins
We next examined how deficiency of Rpn10-UIM and Rpn13 affects proteasome function. We confirmed the loss of both the full-length Rpn10 and Rpn13 proteins in the 3-week-old DKO liver (Fig 4A). In addition, increases in the CP and RP subunits were observed in the DKO liver, suggesting that expression of proteasome subunits was induced in the DKO liver by the feedback mechanism (Fig 4A). Indeed, we confirmed increased mRNA expression of CP and RP subunits in the DKO liver (Fig 4B). The assembly of the 26S proteasome was normal in DKO livers, as revealed by normal distribution of the CP subunit α6 and the RP subunit Rpn8 (Fig 4C). The truncated form of Rpn10 (Rpn10ΔUIM) was incorporated correctly into 26S proteasomes, as we previously reported [21]. Loss of Uch37 proteins was also observed in DKO livers (Fig 4C). Consistent with the increase in assembled proteasomes, the proteasome peptidase activity of the DKO liver was nearly twice than that of the control liver (Fig 4D).
Fig. 4. Redundant roles of Rpn10 and Rpn13 in degradation of ubiquitinated proteins. 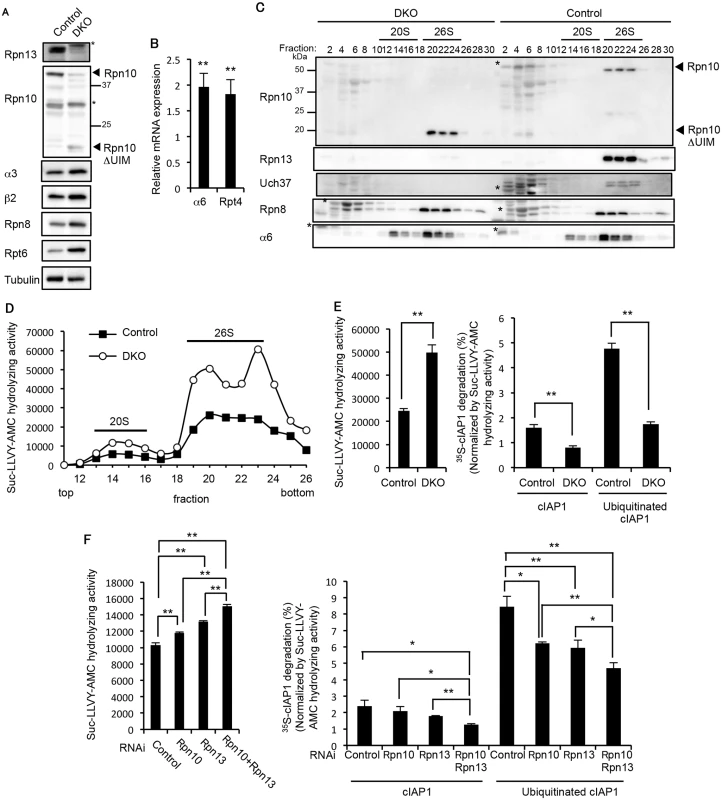
(A) Immunoblot analysis of liver lysates from 3-week-old control and DKO mice with antibodies against the indicated proteins. (B) Real-time RT-PCR was performed to measure the mRNA expressions of the proteasome subunits α6 and Rpt4 in the liver of 2–4-week-old control and DKO mice. Data represent transcript levels in DKO livers relative to those in control livers and are expressed as means; error bars denote SEM. **p < 0.01 (n = 7 for each genotype). (C) Lysates from control and DKO livers were fractionated by glycerol gradient centrifugation (8 to 32% glycerol from fraction 1 to 30) and an equal amount of each fraction was used for immunoblot analysis using antibodies against the indicated proteins. Asterisks indicate nonspecific bands. (D) Each fraction of (C) was assayed for chymotrypsin-like activity using Suc-LLVY-AMC as a substrate. (E) The 26S proteasome fractions of (D) (fractions 20–23) were subjected to the assay of chymotrypsin-like activity (left panel). Degradation rates of 35S-labeled cIAP1 with or without ubiquitination were measured and normalized by chymotrypsin-like activity (right panel). Data are mean ± standard deviations from triplicate experiments. **p < 0.01. (F) Lysates from HeLa cells transfected with indicated siRNAs were assayed for Suc-LLVY-AMC hydrolyzing activity (left panel) and degradation of 35S-labeled cIAP1 with or without ubiquitination. cIAP degradation rates are normalized by Suc-LLVY-AMC hydrolyzing activity (right panel). Data are mean ± standard deviations from three experiments. *p < 0.05; **p < 0.01. To assess the ubiquitin-mediated protein degrading activity of DKO proteasomes, the 26S proteasome fractions (fraction 20–23 in Fig 4D) were subjected to an in vitro degradation assay of ubiquitinated cIAP1, and activity was normalized by the respective peptidase activities (Fig 4E, left panel). The degradation rate of ubiquitinated cIAP1 was markedly reduced in the DKO proteasome compared to the control proteasome (Fig 4E, right panel), suggesting severe defects in degradation of ubiquitinated proteins in the absence of both Rpn10 and Rpn13. Intriguingly, degradation of unmodified cIAP1 was also reduced in the DKO proteasome compared to the control proteasome (Fig 4E, right panel). Although the mechanism of ubiquitin-independent protein degradation by the 26S proteasome is not fully understood, some structural changes caused by loss of Rpn13 and the UIM domain of Rpn10 might affect the degradation mechanism. Similar results were obtained using lysates of HeLa cells treated with siRNAs against Rpn10, Rpn13, or combinations of these. Rpn10/Rpn13-double knockdown caused significant increase in peptidase activity compared to single knockdown of either Rpn10 or Rpn13 (Fig 4F, left panel). The degradation rate of ubiquitinated cIAP1 was significantly reduced in Rpn10/Rpn13-double knockdown compared to single knockdown of either Rpn10 or Rpn13 (Fig 4F, right panel). These results indicate that the Rpn10 loss and Rpn13 loss synthetically affect degradation of ubiquitinated proteins.
Defective binding of ubiquitinated proteins and the UBL-UBA proteins mHR23B and ubiquilins to Rpn10ΔUIM/ΔRpn13 proteasomes
Since both Rpn10-UIM and Rpn13 can recognize ubiquitin chains, we examined whether binding of ubiquitinated proteins to the 26S proteasome was affected in the Rpn10LΔUIM, Rpn13LKO, and DKO livers. The 26S proteasome was immunoprecipitated from each liver lysate and subjected to immunoblot analysis. While the amount of ubiquitinated proteins was increased in Rpn10LΔUIM, the amount of ubiquitinated proteins coprecipitated with the 26S proteasome was decreased in Rpn10LΔUIM livers compared to control (Fig 5A, left panel). The amount of ubiquitinated proteins in Rpn13LKO livers was similar to that in control livers, but ubiquitinated proteins that coprecipitated with the 26S proteasome was decreased in Rpn13LKO livers compared to control (Fig 5A, middle panel). These results indicate that either loss of Rpn10 or Rpn13 causes failure in recruiting ubiquitinated proteins to the proteasome.
Fig. 5. Defective binding of ubiquitinated and UBL-UBA proteins to Rpn10ΔUIM/ΔRpn13 proteasomes. 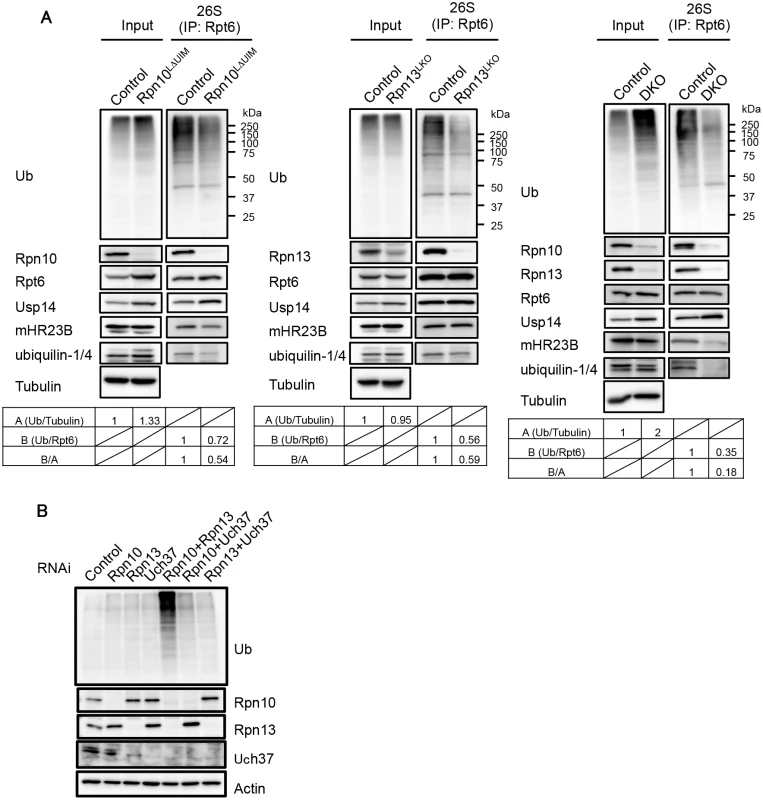
(A) Homogenates from mouse livers were immunoprecipitated with an anti-Rpt6 antibody and subjected to immunoblotting with the indicated antibodies. Values for the relative band intensities of ubiquitin normalized by tubulin (input) or Rpt6 (IP) are shown as A and B, with the control being set to one. Values for B/A indicate the relative amount of bound ubiquitinated proteins to the amount of input ubiquitinated proteins. (B) HEK293T cells were transfected with siRNA against Rpn10, Rpn13, or Uch37. Where indicated, cells were transfected with a mixture of siRNAs. After 96h, cell extracts were subjected to SDS-PAGE, followed by immunoblotting with the indicated antibodies. In DKO liver, the accumulation of ubiquitinated proteins was augmented compared to each single knockout liver, yet the amount of co-precipitated ubiquitinated proteins was much smaller than the control liver, especially in terms of the relative amount of bound ubiquitinated proteins to that of input ubiquitinated proteins (Fig 5A, right panel). We also observed an increase in Usp14 bound to the Rpn10LΔUIM and DKO proteasomes (Fig 5A, left and right panels), consistent with previous observations that ubiquitin stress enhances binding of Ubp6 (P43593), a Usp14 ortholog, with the proteasome [7,21,39]. The reduction in proteasome-bound ubiquitinated proteins strongly suggests that recruitment of ubiquitinated proteins is severely impaired in proteasomes lacking Rpn10-UIM or Rpn13 and that simultaneous deletion of Rpn10-UIM and Rpn13 enhances this effect.
Ubiquitinated proteins are either directly recognized by Rpn10 and Rpn13 or bound and delivered to the proteasome by UBL-UBA proteins [14]. To examine whether the association of UBL-UBA proteins with the proteasome was affected in the absence of Rpn10-UIM and Rpn13, the immunoprecipitates were also probed for mHR23B and ubiquilin-1/4 (the antibody detects both proteins). mHR23B bound to Rpn10LΔUIM proteasomes was reduced compared to that bound to the control proteasomes (Fig 5A, left panel), consistent with our previous observations [21]. The amount of ubiquilin-1/4 bound to Rpn10LΔUIM proteasomes was also decreased (Fig 5A, left panel). On the other hand, the amount of mHR23B and ubiquilin-1/4 in Rpn13LKO proteasomes were comparable to and slightly decreased compared to control proteasomes, respectively (Fig 5A, middle panel). However, mHR23B and ubiquilin-1/4 were only faintly detected in DKO proteasomes (Fig 5A, right panel). To date, Rpn1 has been considered the major subunit for recruitment of UBL-UBA proteins in S. cerevisiae [40–42]. These results demonstrate that both Rpn10 and Rpn13 are the major subunits for recruitment of mHR23B and ubiquilin-1/4 in mammals, where Rpn10 and Rpn13 play partially redundant roles. These results also suggest that accumulation of ubiquitinated proteins is due not only to failure in direct recognition of ubiquitin chains by Rpn10 and Rpn13, but also impairment in delivery of ubiquitinated proteins by mHR23B and ubiquilin-1/4.
Since deletion of Rpn13 leads to concurrent loss of Uch37, the possibility remains that loss of Uch37 rather than Rpn13 is responsible for the massive accumulation of ubiquitinated proteins in the absence of Rpn13. To exclude this possibility, we compared the levels of ubiquitinated proteins in HEK293T cells treated with siRNAs against Rpn10, Rpn13, Uch37, or combinations of these (Fig 5B). Consistent with the results in mice, Rpn10/Rpn13-double knockdown caused profound accumulation of ubiquitinated proteins compared to single knockdowns of either Rpn10 or Rpn13. Notably, Rpn10/Uch37-double knockdown did not cause such accumulation (Fig 5B), indicating that the synthetic effect of Rpn13 deletion with Rpn10 deletion is not due to loss of Uch37 but due to loss of Rpn13 itself. These results also show that the effects of Rpn10 loss and Rpn13 loss work synthetically. Taken together with the histological analysis (Fig 3), these results suggest that a large part of ubiquitinated proteins can be recognized either through Rpn10 or Rpn13, whether directly or indirectly, and that the roles of Rpn10 and Rpn13 are largely, if not entirely, redundant.
Synthetic effect of Rpn10 and Rpn13 deletions on cellular stress
To further test redundant roles of Rpn10-UIM and Rpn13 in degradation of ubiquitinated proteins in the liver, we examined the protein levels of Nrf1 (Q61985) and β-catenin (Q02248), both of which are constitutively ubiquitinated and constantly degraded by the proteasome [34,43]. The protein levels of Nrf1 and β-catenin in Rpn10LΔUIM and Rpn13LKO single knockout livers were comparable to those in the control liver. However, both proteins were remarkably accumulated in the DKO liver, although the extent varied between litters (Fig 6A and S5 Fig). These increases are likely due to a defect in protein turnover, because mRNAs of Nrf1 (Nfe2l1) (18023) and β-catenin (Ctnnb1) (12387) were not increased in the DKO liver (Fig 6B). These results support the view that Rpn10 and Rpn13 act redundantly to degrade ubiquitinated proteins in general.
Fig. 6. Synthetic effect of Rpn10-UIM and Rpn13 deletion on degradation of ubiquitinated proteins and cellular stress. 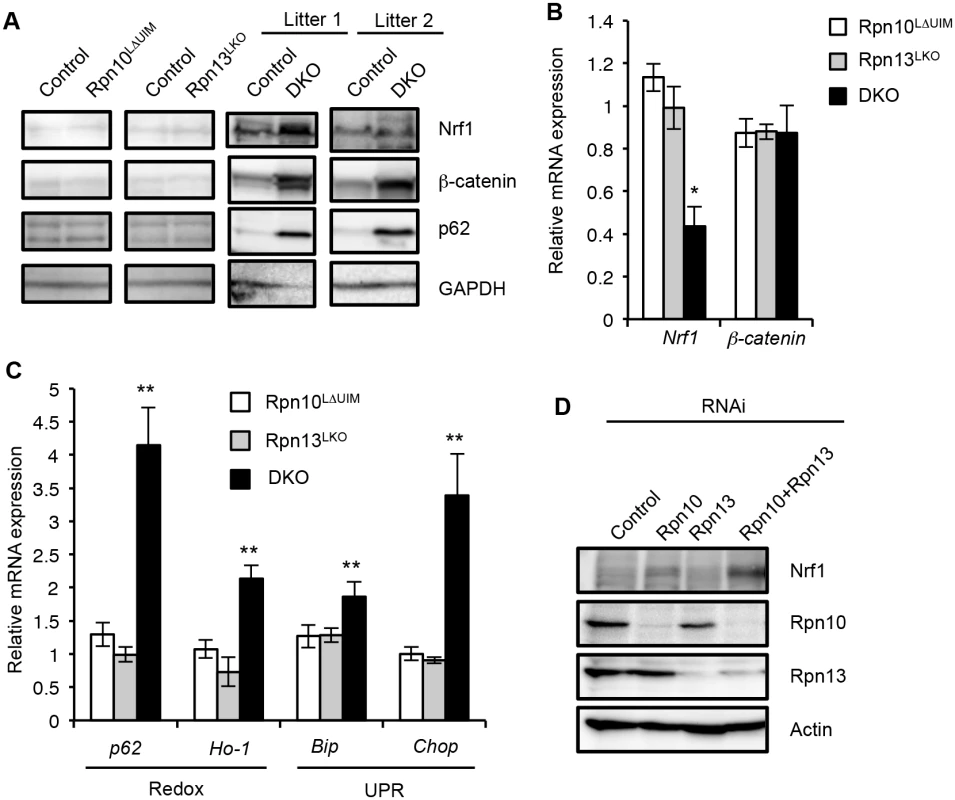
(A) Immunoblot analysis of whole-cell extracts of livers from indicated genotypes of mice (2-month-old for Rpn10LΔUIM and Rpn13LKO, and 2–4-week-old DKO) with antibodies against indicated proteins. (B and C) Real-time RT-PCR was used to measure the expression of transcripts encoding Nrf1, β-catenin, redox pathway (p62 [18412] and Ho-1 [15368]), and UPR pathway (Bip [14828] and Chop [13198]) genes in the livers of 3–6-week-old control and DKO mice. Data represent levels of transcripts in each genotype liver relative to those in control liver and are expressed as means; error bars denote SEM. *p < 0.05; **p < 0.01 (n = 3 for Rpn10LΔUIM and Rpn13LKO, and n = 4 for DKO). (D) HeLa cells were transfected with siRNA against Rpn10 and Rpn13. Where indicated, cells were transfected with a mixture of siRNAs. After 96h, whole-cell extracts were subjected to SDS-PAGE, followed by immunoblotting with the indicated antibodies. It has been reported that inhibition of the proteasome causes oxidative stress and ER stress, which then induce expressions of redox genes and the unfolded protein response (UPR), respectively [44,45]. We therefore examined whether induction of such genes is also exaggerated when both Rpn10-UIM and Rpn13 are deficient. Quantitative RT-PCR analysis showed a remarkable increase in mRNA for redox genes (p62 and Ho-1) and UPR genes (Bip and Chop) in DKO livers, compared to the single knockout mice (Fig 6C). The increase in p62 proteins is likely due to upregulation of p62 expression (Fig 6A). We also confirmed accumulation of Nrf1 proteins in Rpn10/Rpn13-double knockdown HeLa cells (Fig 6D). These results indicate that simultaneous loss of Rpn10-UIM and Rpn13 induces cellular stress more severe than loss of either Rpn10-UIM or Rpn13 and further supports the view that Rpn10 and Rpn13 act redundantly for cellular homeostasis.
Liver injury in DKO liver induce regeneration accompanied by fibrosis
Despite severe liver injury at 2 weeks of age (Fig 3A–3E), DKO mice survived as long as 50 weeks. We examined the expression of pro-apoptotic genes (Noxa [58801] and Puma [170770]) and proliferation marker genes (Gpc3 [14734] and Afp [11576]) in 3–6-week-old mice (Fig 7A). This analysis revealed that both pro-apoptotic and proliferation-related genes were upregulated in the DKO liver. Indeed, immunostaining of liver sections for the proliferation marker Ki-67 confirmed hepatocyte proliferation in DKO mice, especially at 8 weeks of age (Fig 7B). These results suggest that liver regeneration following loss of hepatic tissue caused by liver injury occurs in DKO mice.
Fig. 7. Spontaneous liver injury, fibrosis and regeneration in DKO mice. 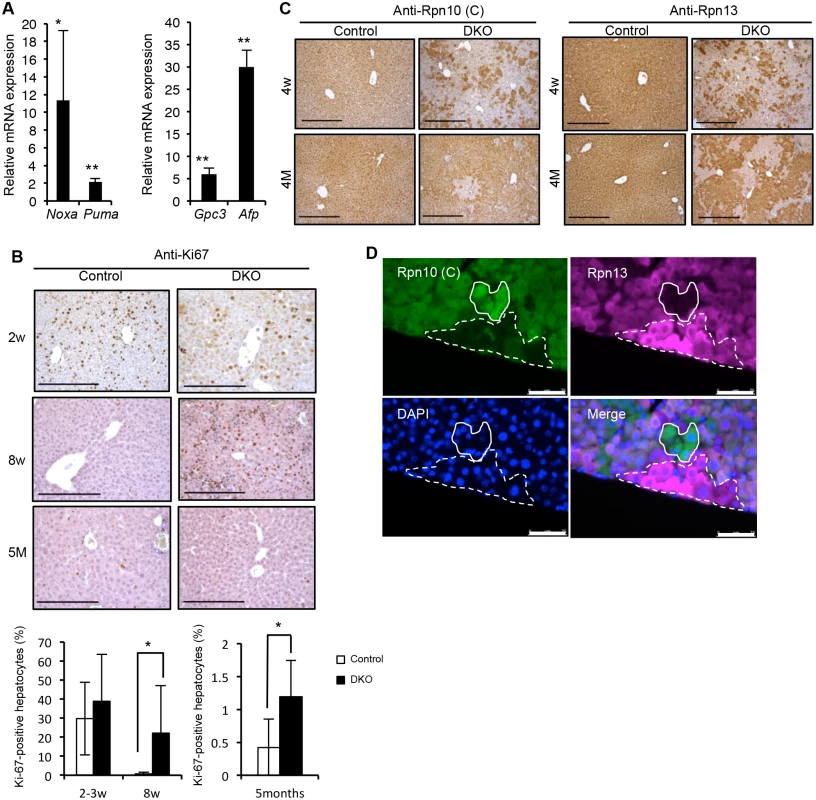
(A) Real-time RT-PCR was used to measure the expressions of p53 target pro-apoptotic related genes Noxa and Puma (left panel) and proliferation marker genes Gpc3 and Afp (right panel) in the livers of 3–6-week-old control and DKO mice. Data represent levels of transcripts in each genotype liver relative to those in control liver and are expressed as means; error bars denote SEM. *p < 0.05; **p < 0.01 (n = 4 each genotype). (B) Representative liver sections of 2- (top panels), 8-week (middle panels), and 5-month-old (bottom panels) control and DKO mice were stained for Ki-67 followed by quantification of the percentage of Ki-67 positive proliferating hepatocytes calculated from three high-power-fields (HPF) analysis. Results are shown as means, and error bars indicate SEM (n = 3–7 mice for each genotype). All scale bars (black lines), 100 μm. (C) Immunohistochemical analyses on representative liver paraffin sections from 4-week-old and 4-month-old mice by using Rpn10 C-terminal antibodies, refer to Rpn10 (C), and Rpn13 antibodies. All scale bars (black lines), 300 μm. (D) Immunofluorescent analysis of liver sections from 4-week-old DKO mice by using Rpn10 (C) and Rpn13 antibodies. 4’, 6-Diamidino-2-phenylindole (DAPI) was used for nuclear counterstaining. The dashed line region represents Rpn13 (+)/Rpn10 (-) hepatocytes, while the lined region represents Rpn13 (-)/Rpn10 (+) hepatocytes. All scale bars (white lines), 75 μm. We then immunostained the liver sections for the expression of Rpn10-UIM and Rpn13. At 4 weeks of age, the majority of hepatocytes expressed neither Rpn10-UIM nor Rpn13, but compared to 2 weeks of age, some Rpn10-UIM - or Rpn13-positive areas appeared (Figs 3C and 7C). At 4 months of age, the majority of hepatocytes were Rpn10-UIM - or Rpn13-positive (Fig 7C). These results demonstrate that liver regeneration by hepatocytes expressing either Rpn10 or Rpn13 occurred in adult DKO liver.
We observed clusters of cells strongly expressing either Rpn10 or Rpn13 in DKO liver, although the mechanism by which these cells recovered expression of Rpn10 and Rpn13 is unclear (Fig 7D). These results indicate that recovery of either Rpn10 or Rpn13 is sufficient to regenerate hepatocytes, further demonstrating that hepatocytes lacking both Rpn10 and Rpn13 are unable to survive and proliferate in mammals.
Discussion
In this study, we showed that Rpn13 plays a redundant role with Rpn10 in recognition and degradation of ubiquitinated proteins in mouse livers. Hepatocyte-specific ablation of either Rpn10-UIM or Rpn13 did not cause obvious defects and showed only mild accumulation of ubiquitinated proteins. However, simultaneous loss of both Rpn13 and Rpn10-UIM caused failure in degradation of ubiquitinated proteins both in vivo and in vitro and led to intense accumulation of ubiquitintead proteins in hepatocytes. An NMR study showed that Rpn10 and Rpn13 can bind simultaneously to a single diubiquitin, where Rpn13 and Rpn10 preferably bind to the proximal and distal ubiquitin, respectively [20]. In addition, proteasome structures obtained by electron microscopy showed that the distance between Rpn10 and Rpn13 is long enough to determine the minimal length for a ubiquitin chain to be recognized by the proteasome, and that simultaneous binding of Rpn10 and Rpn13 to a single ubiquitin chain can orient the chain for deubiquitination, which might promote degradation of ubiquitinated proteins [46,47]. However, our results suggest that most proteins are efficiently degraded when either Rpn10 or Rpn13 is intact, as shown by accumulation of ubiquitinated proteins (Fig 5A and 5B), accumulation of Nrf1 and β-catenin (Fig 6A and 6D), and liver injury (Figs 3 and 6C). On the other hand, some proteins accumulated in the absence of Rpn10-UIM or Rpn13 could be specific substrates for either Rpn10 or Rpn13. Considering that both Rpn10 - and Rpn13-deficient mice are embryonic lethal, there are also non-redundant roles for these receptors, and defects in degradation of each receptor-specific substrates might be the cause of lethality in Rpn10 - and Rpn13-deficient mice. The specificity may be determined by the relative position and orientation between the ubiquitinated site and the degradation initiation site of the substrate [48].
An unexpected finding is that the UBL-UBA proteins mHR23B and ubiquilin-1 and -4 were only faintly detected in 26S proteasomes lacking both Rpn10-UIM and Rpn13 (Fig 5A). In S. cerevisiae, Rpn1 has been shown to contribute to recruitment of UBL-UBA proteins [14,40]. However, our results clearly indicate that Rpn10 and Rpn13 are major acceptors of mHR23B and ubiquilin-1 and -4 in mice. S. cerevisiae Rpn10 has only one UIM, whereas mammalian Rpn10 has two UIMs. It has been reported that the first UIM binds to ubiquitin chains while the second UIM binds to the UBL domain of UBL-UBA proteins in mammalian Rpn10 [49]. It is possible that the second UIM of Rpn10, which is found in metazoa [47], has taken over the role of Rpn1 as an acceptor of UBL-UBA proteins. These results also suggest that recognition of ubiquitinated proteins by the mammalian proteasome is not the same as that by the S. cerevisiae proteasome.
Despite lack of the two ubiquitin receptor subunits and binding of mHR23B and ubiquilin-1/4, DKO proteasomes still bound ubiquitinated proteins (Fig 5A). This suggests the presence of other ubiquitin receptor(s) on the 26S proteasome. The existence of such additional ubiquitin receptor(s) had been proposed in S. cerevisiae [12,36]. Indeed, Dss1/Sem1/Rpn15 (O14140) were recently identified as additional ubiquitin receptor subunits of the proteasome in Saccharomyces pombe [50].
Rpn13 was found to be present in a substoichiometric amount in the proteasomes of S. cerevisiae, D. melanobaster, and human [51–53]. A recent study demonstrated that the 26S proteasome contains only one Rpn13, which defines the asymmetry of the 26S proteasome [54]. In addition, ubiquitination of Rpn13 decreases proteasome ability to bind and degrade ubiquitinated proteins [55]. Considering that the phenotype of Rpn13 mice is less severe that of Rpn10ΔUIM mice, which are lethal at embryonic day 8–9 [21], contribution of Rpn13 in degradation of ubiquitinated proteins seems to be smaller than Rpn10. Therefore, we suggest that Rpn13 plays an auxiliary regulatory role in protein degradation by the proteasome under certain conditions such as proteotoxic stress and cell proliferation, especially in tumor cells. Consistent with this, an Rpn13 inhibitor was recently reported to be toxic to tumor cells but tolerated by normal cells [56]. It has been suggested that Rpn10, Rpn13, and other ubiquitin receptor(s) may have distinct substrate specificity to cover a broad range of ubiquitinated substrates [57]. Thus, further analysis is required to reveal the complicated mechanisms of ubiquitin recognition and degradation by the 26S proteasome, which would provide useful information for elucidating physiological functions of each ubiquitin receptor and for designing drugs targeting these receptors.
Materials and Methods
Gene targeting of Adrm1 (Rpn13)
A targeting vector for Adrm1 (Rpn13) conditional deletion was constructed by inserting loxP sequences into intron 2 and intron 4 so that exon 3 and 4 was deleted by the expression of Cre recombinase. A neomycin cassette flanked by FRT sites was inserted into intron 2. RENKA embryonic stem cells were screened as described previously [58]. For Southern blot analysis, genomic DNA was digested with BamHI and was hybridized with the probes shown in S1 Fig. EIIa-Cre and Alb-Cre were purchased from the Jackson Laboratory. PCR primers used for mouse genotyping are listed in S1 Table. All animal experiments were performed under the guidelines of Science Council of Japan, with approval of procedures by the Institutional Animal Care Committee of Graduate School of Pharmaceutical Sciences, the University of Tokyo (approval number M25-19). Mice were housed in pathogen-free facilities.
Cesarean delivery
Cesarean section was performed at 19 days postcoitus, and pups were placed in a humidified thermostat-controlled chamber.
Histological examination
Tissues were dissected, fixed in 4% paraformaldehyde, embedded in paraffin, and sectioned. Sections were stained with Mayer’s hematoxylin, followed by eosin staining. For immunohistochemical analysis, all tissue sections were subjected to antigen retrieval using the microwave method in 0.01 M citrate buffer for 10 min. After blocking, sections were incubated with primary antibodies overnight at 4°C. Sections were incubated with biotinylated secondary antibodies that were detected using VECTASTAIN ABC kits (Vector) and the DAB substrate (Sigma). For immunofluorescent analysis, sections were blocked in 0.5% goat serum in TBST and then incubated with primary antibodies, followed by Alexa 488 or 633-labeled secondary antibody (Life Technologies). Paraffin-embedded tissue sections were used for AZAN staining. Staining of skeleton was performed with Alizarin red and Alcian blue. Serum tests were performed by a commercial laboratory (Oriental Yeast Co., Ltd.).
RNA isolation, reverse transcription, and real-time PCR
For real-time PCR analysis, total RNAs were isolated from the livers of 5-week-old mice by using an RNAspin kit (Roche), reverse transcribed to cDNA using a SuperScript VILO cDNA Synthesis Kit (Life Technologies), and subjected to real-time PCR using the Light cycler 480 system (Roche). PCR primers and universal probes (Roche), which are listed in S2 Table, were designed according to the Universal Probe Assay Design Center. Glucuronidase beta (Gusβ) was used for normalization. Real-time PCR data were analyzed by the ΔΔCT method.
Immunological analysis
Mouse livers were homogenized and subjects to immunoblotting and immunoprecipitation as described previously [21]. Whole-cell extracts from the livers and HeLa cells were homogenized and lysed in buffer containing 50 mM Tris-HCl (pH 7.5), 1% SDS, 5 mM EDTA, and 10 mM 2-ME, followed by sonication. The antibodies for α3, α6, β2, Rpt6, Rpn8, Rpn10 (N), Rpn10 (C), Rpn13, Uch37, mHR23B, Usp14, and polyubiquitin were described previously [21, 59]. A polyclonal antibody that recognizes both ubiquilin-1 and ubiquilin-4 was raised by immunizing rabbits with recombinant full-length ubiquilin-4 proteins. For immunohistochemistry, monoclonal antibodies against Rpn13 were raised by immunizing rats with recombinant full-length Rpn13 proteins. The antibodies for Tubulin (sc-5286; Santa Cruz Biotechnology), Ki-67 (RM-9106; Thermo Scientific), Nrf1 (sc-13031; Santa Cruz Biotechnology), β-catenin (610153; BD biosciences), p62 (PM045; MBL), Actin (MAB1501R; Millipore), and GAPDH (MCA4739; AbD Serotec) were purchased. For immunoprecipitation, liver homogenates were immunoprecipitated with an anti-Rpt6 antibody as described previously [21]. Band intensities were quantified using Fusion software (M&S Instruments Inc.).
Glycerol gradient analysis
Liver homogenates were clarified by centrifugation at 20,000 x g and subjected to 8 to 32% (vol/vol) linear glycerol gradient centrifugation (22 h, 83,000 x g) as described previously [59].
Assay of proteasome activity
The assays of proteasome chymotryptic peptidase activity, deubiquitination assay, and degradation of polyubiquitinated 35S-labeled cIAP1 protein have been described previously [4].
RNAi experiments
siRNA-mediated knockdown was performed as described previously. The targeted sequences are as follows: Rpn10, 5’-GGAGCAGAGUUUGGCCAGGCGGAAU-3’; Rpn13, 5’-GGAGGGUCUACGUGCUGAAGUUCAAA-3’; Uch37, 5’-ACCGAGCTCATTAAAGGATTCGGTT-3’. Cells were harvested 96 h after transfection of siRNAs [4].
Supporting Information
Zdroje
1. Glickman MH, Ciechanover A. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol Rev. 2002;82 : 373–428. 11917093
2. Glickman MH, Rubin DM, Coux O, Wefes I, Pfeifer G, Cjeka Z, et al. A subcomplex of the proteasome regulatory particle required for ubiquitin-conjugate degradation and related to the COP9-signalosome and elF3. Cell. 1998;94 : 615–623. 9741626
3. Baumeister W, Walz J, Zühl F, Seemüller E. The proteasome: Paradigm of a self-compartmentalizing protease. Cell. 1998. pp. 367–380. 9476896
4. Hamazaki J, Iemura S-I, Natsume T, Yashiroda H, Tanaka K, Murata S. A novel proteasome interacting protein recruits the deubiquitinating enzyme UCH37 to 26S proteasomes. EMBO J. 2006;25 : 4524–4536. 16990800
5. Qiu X-B, Ouyang S-Y, Li C-J, Miao S, Wang L, Goldberg AL. hRpn13/ADRM1/GP110 is a novel proteasome subunit that binds the deubiquitinating enzyme, UCH37. EMBO J. 2006;25 : 5742–5753. 17139257
6. Yao T, Song L, Xu W, DeMartino GN, Florens L, Swanson SK, et al. Proteasome recruitment and activation of the Uch37 deubiquitinating enzyme by Adrm1. Nat Cell Biol. 2006;8 : 994–1002. 16906146
7. Leggett DS, Hanna J, Borodovsky A, Crosas B, Schmidt M, Baker RT, et al. Multiple associated proteins regulate proteasome structure and function. Mol Cell. 2002;10 : 495–507. 12408819
8. Verma R, Chen S, Feldman R, Schieltz D, Yates J, Dohmen J, et al. Proteasomal proteomics: identification of nucleotide-sensitive proteasome-interacting proteins by mass spectrometric analysis of affinity-purified proteasomes. Mol Biol Cell. 2000;11 : 3425–3439. 11029046
9. Verma R, Aravind L, Oania R, McDonald WH, Yates JR, Koonin E V, et al. Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science. 2002;298 : 611–615. 12183636
10. Yao T, Cohen RE. A cryptic protease couples deubiquitination and degradation by the proteasome. Nature. 2002;419 : 403–407. 12353037
11. Deveraux Q, Ustrell V, Pickart C, Rechsteiner M. A 26 S protease subunit that binds ubiquitin conjugates. J Biol Chem. 1994;269 : 7059–7061. 8125911
12. Husnjak K, Elsasser S, Zhang N, Chen X, Randles L, Shi Y, et al. Proteasome subunit Rpn13 is a novel ubiquitin receptor. Nature. 2008;453 : 481–488. doi: 10.1038/nature06926 18497817
13. Schreiner P, Chen X, Husnjak K, Randles L, Zhang N, Elsasser S, et al. Ubiquitin docking at the proteasome through a novel pleckstrin-homology domain interaction. Nature. 2008;453 : 548–552. doi: 10.1038/nature06924 18497827
14. Finley D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu Rev Biochem. 2009;78 : 477–513. doi: 10.1146/annurev.biochem.78.081507.101607 19489727
15. Hiyama H, Yokoi M, Masutani C, Sugasawa K, Maekawa T, Tanaka K, et al. Interaction of hHR23 with S5a. The ubiquitin-like domain of hHR23 mediates interaction with S5a subunit of 26 S proteasome. J Biol Chem. 1999;274 : 28019–28025. 10488153
16. Fatimababy AS, Lin YL, Usharani R, Radjacommare R, Wang HT, Tsai HL, et al. Cross-species divergence of the major recognition pathways of ubiquitylated substrates for ubiquitin/26S proteasome-mediated proteolysis. FEBS J. 2010;277 : 796–816. doi: 10.1111/j.1742-4658.2009.07531.x 20059542
17. Hofmann RM, Pickart CM. In Vitro Assembly and Recognition of Lys-63 Polyubiquitin Chains. J Biol Chem. 2001;276 : 27936–27943. 11369780
18. Hyoung TK, Kwang PK, Lledias F, Kisselev AF, Scaglione KM, Skowyra D, et al. Certain pairs of ubiquitin-conjugating enzymes (E2s) and ubiquitin-protein ligases (E3s) synthesize nondegradable forked ubiquitin chains containing all possible isopeptide linkages. J Biol Chem. 2007;282 : 17375–17386. 17426036
19. Saeki Y, Kudo T, Sone T, Kikuchi Y, Yokosawa H, Toh-e A, et al. Lysine 63-linked polyubiquitin chain may serve as a targeting signal for the 26S proteasome. EMBO J. 2009;28 : 359–371. doi: 10.1038/emboj.2008.305 19153599
20. Zhang N, Wang Q, Ehlinger A, Randles L, Lary JW, Kang Y, et al. Structure of the S5a:K48-Linked Diubiquitin Complex and Its Interactions with Rpn13. Mol Cell. 2009;35 : 280–290. doi: 10.1016/j.molcel.2009.06.010 19683493
21. Hamazaki J, Sasaki K, Kawahara H, Hisanaga S-I, Tanaka K, Murata S. Rpn10-mediated degradation of ubiquitinated proteins is essential for mouse development. Mol Cell Biol. 2007;27 : 6629–6638. 17646385
22. Van Nocker S, Sadis S, Rubin DM, Glickman M, Fu H, Coux O, et al. The multiubiquitin-chain-binding protein Mcb1 is a component of the 26S proteasome in Saccharomyces cerevisiae and plays a nonessential, substrate-specific role in protein turnover. Mol Cell Biol. 1996;16 : 6020–6028. 8887631
23. Szlanka T, Haracska L, Kiss I, Deák P, Kurucz E, Andó I, et al. Deletion of proteasomal subunit S5a/Rpn10/p54 causes lethality, multiple mitotic defects and overexpression of proteasomal genes in Drosophila melanogaster. J Cell Sci. 2003;116 : 1023–1033. 12584246
24. Al-Shami A, Jhaver KG, Vogel P, Wilkins C, Humphries J, Davis JJ, et al. Regulators of the proteasome pathway, Uch37 and Rpn13, play distinct roles in mouse development. PLoS One. 2010;5.
25. Díaz-Martínez LA, Kang Y, Walters KJ, Clarke DJ. Yeast UBL-UBA proteins have partially redundant functions in cell cycle control. Cell Div. 2006;1 : 28. 17144915
26. Kim I, Mi K, Rao H. Multiple interactions of rad23 suggest a mechanism for ubiquitylated substrate delivery important in proteolysis. Mol Biol Cell. 2004;15 : 3357–3365. 15121879
27. Saeki Y, Saitoh A, Toh-e A, Yokosawa H. Ubiquitin-like proteins and Rpn10 play cooperative roles in ubiquitin-dependent proteolysis. Biochem Biophys Res Commun. 2002;293 : 986–992. 12051757
28. Fu H, Lin YL, Fatimababy AS. Proteasomal recognition of ubiquitylated substrates. Trends in Plant Science. 2010. pp. 375–386. doi: 10.1016/j.tplants.2010.03.004 20399133
29. Lakso M, Pichel JG, Gorman JR, Sauer B, Okamoto Y, Lee E, et al. Efficient in vivo manipulation of mouse genomic sequences at the zygote stage. Proc Natl Acad Sci U S A. 1996;93 : 5860–5865. 8650183
30. Postic C, Shiota M, Niswender KD, Jetton TL, Chen Y, Moates JM, et al. Dual roles for glucokinase in glucose homeostasis as determined by liver and pancreatic?? cell-specific gene knock-outs using Cre recombinase. J Biol Chem. 1999;274 : 305–315. 9867845
31. Lundgren J, Masson P, Mirzaei Z, Young P. Identification and characterization of a Drosophila proteasome regulatory network. Mol Cell Biol. 2005;25 : 4662–4675. 15899868
32. Meiners S, Heyken D, Weller A, Ludwig A, Stangl K, Kloetzel PM, et al. Inhibition of proteasome activity induces concerted expression of proteasome genes and de novo formation of mammalian proteasomes. J Biol Chem. 2003;278 : 21517–21525. 12676932
33. Radhakrishnan SK, Lee CS, Young P, Beskow A, Chan JY, Deshaies RJ. Transcription Factor Nrf1 Mediates the Proteasome Recovery Pathway after Proteasome Inhibition in Mammalian Cells. Mol Cell. 2010;38 : 17–28. doi: 10.1016/j.molcel.2010.02.029 20385086
34. Steffen J, Seeger M, Koch A, Krüger E. Proteasomal degradation is transcriptionally controlled by TCF11 via an ERAD-dependent feedback loop. Mol Cell. 2010;40 : 147–158. doi: 10.1016/j.molcel.2010.09.012 20932482
35. Silke J, Kratina T, Chu D, Ekert PG, Day CL, Pakusch M, et al. Determination of cell survival by RING-mediated regulation of inhibitor of apoptosis (IAP) protein abundance. Proc Natl Acad Sci U S A. 2005;102 : 16182–16187. 16263936
36. Peth A, Uchiki T, Goldberg AL. ATP-Dependent steps in the binding of ubiquitin conjugates to the 26s proteasome that commit to degradation. Mol Cell. 2010;40 : 671–681. doi: 10.1016/j.molcel.2010.11.002 21095592
37. Bataller R, Brenner DA. Liver fibrosis. J Clin Invest. 2005;115 : 209–218. 15690074
38. Geier A, Wagner M, Dietrich CG, Trauner M. Principles of hepatic organic anion transporter regulation during cholestasis, inflammation and liver regeneration. Biochimica et Biophysica Acta—Molecular Cell Research. 2007. pp. 283–308.
39. Hanna J, Meides A, Zhang DP, Finley D. A Ubiquitin Stress Response Induces Altered Proteasome Composition. Cell. 2007;129 : 747–759. 17512408
40. Elsasser S, Gali RR, Schwickart M, Larsen CN, Leggett DS, Müller B, et al. Proteasome subunit Rpn1 binds ubiquitin-like protein domains. Nat Cell Biol. 2002;4 : 725–730. 12198498
41. Gomez TA, Kolawa N, Gee M, Sweredoski MJ, Deshaies RJ. Identification of a functional docking site in the Rpn1 LRR domain for the UBA-UBL domain protein Ddi1. BMC Biol. 2011;9 : 33. doi: 10.1186/1741-7007-9-33 21627799
42. Rosenzweig R, Bronner V, Zhang D, Fushman D, Glickman MH. Rpn1 and Rpn2 coordinate ubiquitin processing factors at proteasome. J Biol Chem. 2012;287 : 14659–14671. doi: 10.1074/jbc.M111.316323 22318722
43. Tauriello DVF, Maurice MM. The various roles of ubiquitin in Wnt pathway regulation. Cell Cycle. 2010. pp. 3700–3709. doi: 10.4161/cc.9.18.13204 20930545
44. Fribley A, Wang C-Y. Proteasome inhibitor induces apoptosis through induction of endoplasmic reticulum stress. Cancer Biol Ther. 2006;5 : 745–748. 16861900
45. Sasaki K, Hamazaki J, Koike M, Hirano Y, Komatsu M, Uchiyama Y, et al. PAC1 gene knockout reveals an essential role of chaperone-mediated 20S proteasome biogenesis and latent 20S proteasomes in cellular homeostasis. Mol Cell Biol. 2010;30 : 3864–3874. 0 doi: 10.1128/MCB.00216-10 20498273
46. Lasker K, Förster F, Bohn S, Walzthoeni T, Villa E, Unverdorben P, et al. Molecular architecture of the 26S proteasome holocomplex determined by an integrative approach. Proc Natl Acad Sci U S A. 2012;109 : 1380–7. doi: 10.1073/pnas.1120559109 22307589
47. Sakata E, Bohn S, Mihalache O, Kiss P, Beck F, Nagy I, et al. Localization of the proteasomal ubiquitin receptors Rpn10 and Rpn13 by electron cryomicroscopy. Proceedings of the National Academy of Sciences. 2012. pp. 1479–1484.
48. Bhattacharyya S, Yu H, Mim C, Matouschek A. Regulated protein turnover: snapshots of the proteasome in action. Nat Rev Mol Cell Biol. 2014;15 : 122–133. doi: 10.1038/nrm3741 24452470
49. Kang Y, Chen X, Lary JW, Cole JL, Walters KJ. Defining how ubiquitin receptors hHR23a and S5a bind polyubiquitin. J Mol Biol. 2007;369 : 168–76. 17408689
50. Paraskevopoulos K, Kriegenburg F, Tatham MH, Rösner HI, Medina B, Larsen IB, et al. Dss1 Is a 26S Proteasome Ubiquitin Receptor. Mol Cell. 2014;56 : 453–61. doi: 10.1016/j.molcel.2014.09.008 25306921
51. Bohn S, Beck F, Sakata E, Walzthoeni T, Beck M, Aebersold R, et al. Structure of the 26S proteasome from Schizosaccharomyces pombe at subnanometer resolution. Proc Natl Acad Sci U S A. 2010;107 : 20992–20997. doi: 10.1073/pnas.1015530107 21098295
52. Fabre B, Lambour T, Garrigues L, Ducoux-Petit M, Amalric F, Monsarrat B, et al. Label-free quantitative proteomics reveals the dynamics of proteasome complexes composition and stoichiometry in a wide range of human cell lines. J Proteome Res. 2014;13 : 3027–3037. doi: 10.1021/pr500193k 24804812
53. Wang X, Huang L. Identifying dynamic interactors of protein complexes by quantitative mass spectrometry. Mol Cell Proteomics. 2008;7 : 46–57. 17934176
54. Berko D, Herkon O, Braunstein I, Isakov E, David Y, Ziv T, et al. Inherent asymmetry in the 26S proteasome is defined by the ubiquitin receptor RPN13. J Biol Chem. 2014;289 : 5609–5618. doi: 10.1074/jbc.M113.509380 24429290
55. Besche HC, Sha Z, Kukushkin N V., Peth A, Hock EM, Kim W, et al. Autoubiquitination of the 26S Proteasome on Rpn13 Regulates Breakdown of Ubiquitin Conjugates. EMBO J. 2014;33 : 1159–1176. doi: 10.1002/embj.201386906 24811749
56. Anchoori R, Karanam B, Peng S, Wang J, Jiang R, Tanno T, et al. A bis-Benzylidine Piperidone Targeting Proteasome Ubiquitin Receptor RPN13/ADRM1 as a Therapy for Cancer. Cancer Cell. 2013;24 : 791–805. doi: 10.1016/j.ccr.2013.11.001 24332045
57. Mayor T, Graumann J, Bryan J, MacCoss MJ, Deshaies RJ. Quantitative profiling of ubiquitylated proteins reveals proteasome substrates and the substrate repertoire influenced by the Rpn10 receptor pathway. Mol Cell Proteomics. 2007;6 : 1885–1895. 17644757
58. Mishina M, Sakimura K. Conditional gene targeting on the pure C57BL/6 genetic background. Neurosci Res. 2007;58 : 105–112. 17298852
59. Hirano Y, Hendil KB, Yashiroda H, Iemura S, Nagane R, Hioki Y, et al. A heterodimeric complex that promotes the assembly of mammalian 20S proteasomes. Nature. 2005;437 : 1381–1385. 16251969
Štítky
Genetika Reprodukční medicína
Článek Discovery and Fine-Mapping of Glycaemic and Obesity-Related Trait Loci Using High-Density ImputationČlánek AAA-ATPase FIDGETIN-LIKE 1 and Helicase FANCM Antagonize Meiotic Crossovers by Distinct MechanismsČlánek A Conserved Pattern of Primer-Dependent Transcription Initiation in and Revealed by 5′ RNA-seqČlánek TopBP1 Governs Hematopoietic Stem/Progenitor Cells Survival in Zebrafish Definitive Hematopoiesis
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2015 Číslo 7
-
Všechny články tohoto čísla
- LINE-1 Retroelements Get ZAPped!
- /p23: A Small Protein Heating Up Lifespan Regulation
- Hairless Streaks in Cattle Implicate TSR2 in Early Hair Follicle Formation
- Ribosomal Protein Mutations Result in Constitutive p53 Protein Degradation through Impairment of the AKT Pathway
- Molecular Clock of Neutral Mutations in a Fitness-Increasing Evolutionary Process
- Modeling Implicates in Nephropathy: Evidence for Dominant Negative Effects and Epistasis under Anemic Stress
- The Alternative Sigma Factor SigX Controls Bacteriocin Synthesis and Competence, the Two Quorum Sensing Regulated Traits in
- BMP Inhibition in Seminomas Initiates Acquisition of Pluripotency via NODAL Signaling Resulting in Reprogramming to an Embryonal Carcinoma
- Comparative Study of Regulatory Circuits in Two Sea Urchin Species Reveals Tight Control of Timing and High Conservation of Expression Dynamics
- EIN3 and ORE1 Accelerate Degreening during Ethylene-Mediated Leaf Senescence by Directly Activating Chlorophyll Catabolic Genes in
- Genome Wide Binding Site Analysis Reveals Transcriptional Coactivation of Cytokinin-Responsive Genes by DELLA Proteins
- Sensory Neurons Arouse . Locomotion via Both Glutamate and Neuropeptide Release
- A Year of Infection in the Intensive Care Unit: Prospective Whole Genome Sequencing of Bacterial Clinical Isolates Reveals Cryptic Transmissions and Novel Microbiota
- Inference of Low and High-Grade Glioma Gene Regulatory Networks Delineates the Role of Rnd3 in Establishing Multiple Hallmarks of Cancer
- Novel Role for p110β PI 3-Kinase in Male Fertility through Regulation of Androgen Receptor Activity in Sertoli Cells
- A Novel Locus Harbouring a Functional Nonsense Mutation Identified in a Large Danish Family with Nonsyndromic Hearing Impairment
- Checkpoint Activation of an Unconventional DNA Replication Program in
- A Genetic Incompatibility Accelerates Adaptation in Yeast
- The SMC Loader Scc2 Promotes ncRNA Biogenesis and Translational Fidelity
- Blimp1/Prdm1 Functions in Opposition to Irf1 to Maintain Neonatal Tolerance during Postnatal Intestinal Maturation
- Discovery and Fine-Mapping of Glycaemic and Obesity-Related Trait Loci Using High-Density Imputation
- JAK/STAT and Hox Dynamic Interactions in an Organogenetic Gene Cascade
- Emergence, Retention and Selection: A Trilogy of Origination for Functional Proteins from Ancestral LncRNAs in Primates
- MoSET1 (Histone H3K4 Methyltransferase in ) Regulates Global Gene Expression during Infection-Related Morphogenesis
- Arabidopsis PCH2 Mediates Meiotic Chromosome Remodeling and Maturation of Crossovers
- AAA-ATPase FIDGETIN-LIKE 1 and Helicase FANCM Antagonize Meiotic Crossovers by Distinct Mechanisms
- A Conserved Pattern of Primer-Dependent Transcription Initiation in and Revealed by 5′ RNA-seq
- Tempo and Mode of Transposable Element Activity in Drosophila
- The Shelterin TIN2 Subunit Mediates Recruitment of Telomerase to Telomeres
- SAMHD1 Inhibits LINE-1 Retrotransposition by Promoting Stress Granule Formation
- A Genome Scan for Genes Underlying Microgeographic-Scale Local Adaptation in a Wild Species
- TopBP1 Governs Hematopoietic Stem/Progenitor Cells Survival in Zebrafish Definitive Hematopoiesis
- Analysis of the Relationships between DNA Double-Strand Breaks, Synaptonemal Complex and Crossovers Using the Mutant
- Assessing Mitochondrial DNA Variation and Copy Number in Lymphocytes of ~2,000 Sardinians Using Tailored Sequencing Analysis Tools
- Allelic Spectra of Risk SNPs Are Different for Environment/Lifestyle Dependent versus Independent Diseases
- CSB-PGBD3 Mutations Cause Premature Ovarian Failure
- Irrepressible: An Interview with Mark Ptashne
- Genetic Evidence for Function of the bHLH-PAS Protein Gce/Met As a Juvenile Hormone Receptor
- Inactivation of Retinoblastoma Protein (Rb1) in the Oocyte: Evidence That Dysregulated Follicle Growth Drives Ovarian Teratoma Formation in Mice
- Redundant Roles of Rpn10 and Rpn13 in Recognition of Ubiquitinated Proteins and Cellular Homeostasis
- Pyrimidine Pool Disequilibrium Induced by a Cytidine Deaminase Deficiency Inhibits PARP-1 Activity, Leading to the Under Replication of DNA
- Molecular Framework of a Regulatory Circuit Initiating Two-Dimensional Spatial Patterning of Stomatal Lineage
- RFX2 Is a Major Transcriptional Regulator of Spermiogenesis
- A Role for Macro-ER-Phagy in ER Quality Control
- Corp Regulates P53 in via a Negative Feedback Loop
- Common Cell Shape Evolution of Two Nasopharyngeal Pathogens
- Contact- and Protein Transfer-Dependent Stimulation of Assembly of the Gliding Motility Machinery in
- Endothelial Snail Regulates Capillary Branching Morphogenesis via Vascular Endothelial Growth Factor Receptor 3 Expression
- Functional Constraint Profiling of a Viral Protein Reveals Discordance of Evolutionary Conservation and Functionality
- Temporal Coordination of Carbohydrate Metabolism during Mosquito Reproduction
- mTOR Directs Breast Morphogenesis through the PKC-alpha-Rac1 Signaling Axis
- Reversible Oxidation of a Conserved Methionine in the Nuclear Export Sequence Determines Subcellular Distribution and Activity of the Fungal Nitrate Regulator NirA
- Nutritional Control of DNA Replication Initiation through the Proteolysis and Regulated Translation of DnaA
- Cooperation between Paxillin-like Protein Pxl1 and Glucan Synthase Bgs1 Is Essential for Actomyosin Ring Stability and Septum Formation in Fission Yeast
- Encodes a Highly Conserved Protein Important to Neurological Function in Mice and Flies
- Identification of a Novel Regulatory Mechanism of Nutrient Transport Controlled by TORC1-Npr1-Amu1/Par32
- Aurora-A-Dependent Control of TACC3 Influences the Rate of Mitotic Spindle Assembly
- Large-Scale Phenomics Identifies Primary and Fine-Tuning Roles for CRKs in Responses Related to Oxidative Stress
- TFIIS-Dependent Non-coding Transcription Regulates Developmental Genome Rearrangements
- Genome-Wide Reprogramming of Transcript Architecture by Temperature Specifies the Developmental States of the Human Pathogen
- Identification of Chemical Inhibitors of β-Catenin-Driven Liver Tumorigenesis in Zebrafish
- The Catalytic and Non-catalytic Functions of the Chromatin-Remodeling Protein Collaborate to Fine-Tune Circadian Transcription in
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Functional Constraint Profiling of a Viral Protein Reveals Discordance of Evolutionary Conservation and Functionality
- Reversible Oxidation of a Conserved Methionine in the Nuclear Export Sequence Determines Subcellular Distribution and Activity of the Fungal Nitrate Regulator NirA
- Modeling Implicates in Nephropathy: Evidence for Dominant Negative Effects and Epistasis under Anemic Stress
- Nutritional Control of DNA Replication Initiation through the Proteolysis and Regulated Translation of DnaA
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



