-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
Reklama/p23: A Small Protein Heating Up Lifespan Regulation
article has not abstract
Published in the journal: . PLoS Genet 11(7): e32767. doi:10.1371/journal.pgen.1005188
Category: Perspective
doi: https://doi.org/10.1371/journal.pgen.1005188Summary
article has not abstract
Dauer and Life Span
The life span of non-renewing organisms is determined by the potential of their individual cells to maintain their functions while aging. Nematodes, like Caenorhabditis elegans with their 20 days of adult life, have proven to be excellent model systems to study organismal lifespan, its variability, and its regulation [1–3]. Early on, the life span could be linked to environmental conditions, like growth temperature and food intake [1,4]. In general, in these experiments, organisms develop and age slower and live longer at lower growth temperatures. This is evident from a clear relationship between temperature and life span ranging from 35 days to 9 days upon temperature changes from 10 to 25.5 degrees [1]. On the higher end of this temperature range, C. elegans can enter the dauer state, which is also found in response to starvation or the presence of dauer pheromone [5–7]. The formation of this stress-resistant state, which enables survival of the organism for longer than 3 months, requires morphological changes to the cuticule and inhibition of further development. Interestingly, it is entirely reversible without effects on the later adult life span [8]. This decision has been analyzed genetically in detail, identifying genes that promote dauer entry (DAF-c) and those that prevent dauer entry (DAF-d). These studies unravel the pathways, which cooperate in the decision whether to enter the path to the dauer state instead of normal development. The most prominent of those are the homologs of the insulin-like receptor DAF-2, the FOXO-transcription factor DAF-16, and the steroid hormone receptor DAF-12, amongst others [9–11]. The decision making requires the sensing of environmental factors and alteration of developmental programs in different tissues. Thus the number of genes influencing this decision is considerable.
Interestingly, several genes that control the entry into the stress - and starvation-resistant dauer state also exert control over the normal life span of the nematode [12,13]. Early aging markers include disorganization of muscular structure and reduction of pharyngeal activity and motility [14,15]. In this context, lower temperature, like some aging-related mutations, delays these early aging markers and likewise postpones later aging markers, like swallowing difficulties and general loss of motility. Despite knowing the individual function of many dauer-influencing genes, the reconstruction of regulated cellular pathways is complicated. This also originates from the fact that different cells are participating in the pathways as well as a contribution of humoral controls, implying that several individual cellular decisions culminate to regulate these pathways [16,17].
daf-41/p23 Reprograms Temperature-Dependence in Aging
For many years it has been known that the cellular chaperone network is also contributing to these phenomena. In this context, the daf-21 allele p673, representing a mutation in the heat shock protein 90, was known to cause constitutive dauer entry [18]. Moreover, the general regulator of the heat shock response HSF-1 was known to influence the life span in cooperation with other dauer genes and its depletion causes early onset of aging [19,20].
In this issue of PLOS Genetics, Horikawa and coworkers address critical questions at the crossroads of stress-resistance, longevity, and chaperone involvement by investigating a deletion mutant of the cochaperone p23, an effector protein of Hsp90. They first determine that the deletion strain constitutively enters dauer and name the gene daf-41. They find that the usual temperature-dependence of the lifespan is altered in this strain with a much smaller temperature-influence than known for the wild-type background (Fig 1). This makes the deletion strain short lived at low temperatures and long lived at high temperatures. While it has been thought up until recently that the slow development and aging at low temperatures reflects the slower turnover of metabolites and the slower rate of all biochemical processes based on plain physical principles, being able to influence this effect by genetic means implies the existence of a biological control. Recent reports had suggested that such programs may exist [21]. Also, temperature-sensitive neurons had been reported to influence the aging process, similar to the findings reported here [22]. In general, these studies show that development does not necessarily has to be slow at low temperatures and fast at high temperatures, and, importantly, with daf-41 a regulator is uncovered that influences this program.
Fig. 1. Nematode lifespan is regulated by temperature amongst other influences. 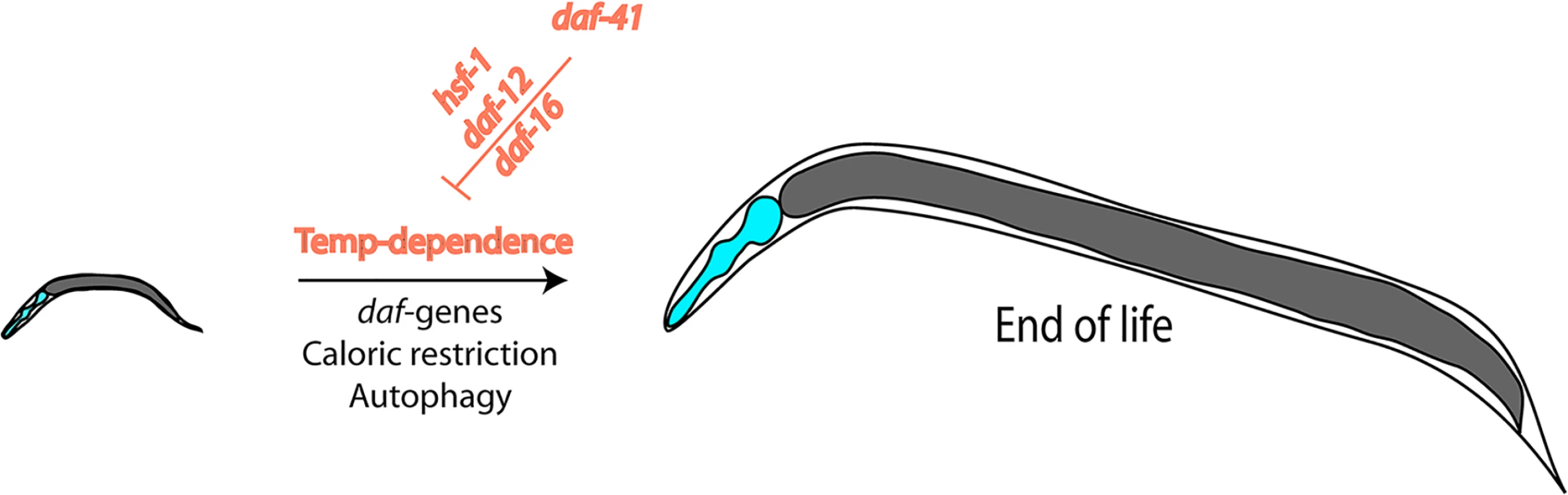
This temperature dependence is the result of transcriptional networks and DAF-41/p23, which influences the transcriptional activities of HSF-1, DAF-12, and DAF-16. Horikawa and coworkers also provide information on the mechanism—how such a response may be regulated by addressing the transcriptional networks influenced by the daf-41 mutation. They show that the deletion of p23 influences several transcriptional outputs. At low temperatures, the influence on the steroid hormone receptor DAF-12 and on DAF-16 is especially relevant. At high temperatures, instead, the lifespan regulation originates from influences on the activity of the heat-shock factor HSF-1, making the stress response stronger in the absence of the Hsp90 cofactor. Thus, p23 appears to balance the transcriptional responses of these three transcription factors relevant for dauer formation, longevity, and aging, and in this way enables control over these processes at different temperatures.
DAF-41/p23: How Can it Work?
Temperature-dependent growth is observed in all organisms, from bacteria to metazoa, and generally has been attributed to differences in metabolic rates. The recent observations now cast in doubt the general belief that plain physics controls the temperature-dependent effects on life span, as at least metazoa appear to have developed a program that controls these growth rates based on transcriptional networks. It is thus very exciting to see that another potentially deterministic program controls the progression of C. elegans through its larval stages to adulthood and during aging and adjusts the growth rate to the environmental conditions.
While the molecular details at the basis of this regulation are still elusive, it is worthwhile to look at the known biochemical functions of p23 and its interaction partner Hsp90. Hsp90 is known to regulate transcriptional outputs by influencing the activity of dozens of transcription factors via their cellular stability. p23, likewise, has been shown to influence the activity of transcription factors, including steroid hormone receptors and HSF-1 [23]. In several cases p23 and Hsp90 cooperate, but also individual activities have been observed for p23 [24]. The biochemical and structural aspects of their interaction are well studied, including by structural characterization of the Hsp90-p23 complex [25]. p23 binds to a conformational state of Hsp90 adopted during its ATPase cycle. Specifically, it recognizes an ATP-induced N-terminal dimerized conformation, which is populated just prior to ATP hydrolysis and remains bound to Hsp90 during the hydrolysis reaction. It controls the hydrolysis rate and enables stable complexes between Hsp90 and its protein clients. Processed client proteins are released after ATP hydrolysis. Essentially, this characteristic behavior contributed to the identification of the protein p23 more than 20 years ago, when it was uncovered as part of the protein assemblies involved in steroid hormone receptor maturation in mammals [26,27]. The influence of Hsp90 and p23 on their clients is not fully understood and rarely has a combinatorial effect on several clients being studied. However, tinkering with Hsp90 inhibition leads to many different phenotypic traits in flies [28], showing that in other model systems, several signaling networks are influenced simultaneously.
With the study of Horikawa and coworkers, this chaperone machinery now also moves into the center of life span regulation. Previously Hsp90, like p23, was found to regulate Hsf1 activity [29], and its depletion strongly induces the heat-shock response [30]. The chaperones’ involvement in the cellular folding process of transcription factors was, until now, seen as a contribution to the regulation of individual transcription factors. The new picture emerging is that by regulating simultaneously the activity of several transcriptional outputs, p23 enables complex developmental decision making. The present study thus integrates the cellular chaperone network surrounding the molecular chaperone Hsp90/DAF-21 into the complex decision making to enter the dauer state and to determine the organismal life span.
Zdroje
1. Klass MR (1977) Aging in the nematode Caenorhabditis elegans: major biological and environmental factors influencing life span. Mech Aging Dev 6 : 413–429. 926867
2. Gershon D (1970) Studies on aging in Nematodes. I. The nematode as a model organism for aging research. Exp Gerontol 5 : 7–12. 5446386
3. Cassada RC, Russell RL (1975) The dauerlarva, a post-embryonic developmental variant of the nematode Caenorhabditis elegans. Dev Biol 46 : 326–342. 1183723
4. Klass MR (1983) A method for the isolation of longevity mutants in the nematode Caenorhabditis elegans and initial results. Mech Aging Dev 22 : 279–286. 6632998
5. Swanson MM, Riddle DL (1981) Critical periods in the development of the Caenorhabditis elegans dauer larva. Dev Biol 84 : 27–40. 7250500
6. Golden JW, Riddle DL (1982) A pheromone influences larval development in the nematode Caenorhabditis elegans. Science 218 : 578–580. 6896933
7. Butcher RA, Ragains JR, Kim E, Clardy J (2008) A potent dauer pheromone component in Caenorhabditis elegans that acts synergistically with other components. Proc Natl Acad Sci U S A 105 : 14288–14292. doi: 10.1073/pnas.0806676105 18791072
8. Klass M, Hirsh D (1976) Non-aging developmental variant of Caenorhabditis elegans. Nature 260 : 523–525. 1264206
9. Antebi A, Yeh WH, Tait D, Hedgecock EM, Riddle DL (2000) daf-12 encodes a nuclear receptor that regulates the dauer diapause and developmental age in C. elegans. Genes Dev 14 : 1512–1527. 10859169
10. Larsen PL, Albert PS, Riddle DL (1995) Genes that regulate both development and longevity in Caenorhabditis elegans. Genetics 139 : 1567–1583. 7789761
11. Fielenbach N, Antebi A (2008) C. elegans dauer formation and the molecular basis of plasticity. Genes Dev 22 : 2149–2165. doi: 10.1101/gad.1701508 18708575
12. Kenyon C, Chang J, Gensch E, Rudner A, Tabtiang R (1993) A C. elegans mutant that lives twice as long as wild type. Nature 366 : 461–464. 8247153
13. Tissenbaum HA (2012) Genetics, life span, health span, and the aging process in Caenorhabditis elegans. J Gerontol A Biol Sci Med Sci 67 : 503–510. doi: 10.1093/gerona/gls088 22499764
14. Huang C, Xiong C, Kornfeld K (2004) Measurements of age-related changes of physiological processes that predict lifespan of Caenorhabditis elegans. Proc Natl Acad Sci U S A 101 : 8084–8089. 15141086
15. Herndon LA, Schmeissner PJ, Dudaronek JM, Brown PA, Listner KM, et al. (2002) Stochastic and genetic factors influence tissue-specific decline in aging C. elegans. Nature 419 : 808–814. 12397350
16. Gottlieb S, Ruvkun G (1994) daf-2, daf-16 and daf-23: genetically interacting genes controlling Dauer formation in Caenorhabditis elegans. Genetics 137 : 107–120. 8056303
17. Riddle DL, Swanson MM, Albert PS (1981) Interacting genes in nematode dauer larva formation. Nature 290 : 668–671. 7219552
18. Birnby DA, Link EM, Vowels JJ, Tian H, Colacurcio PL, et al. (2000) A transmembrane guanylyl cyclase (DAF-11) and Hsp90 (DAF-21) regulate a common set of chemosensory behaviors in caenorhabditis elegans. Genetics 155 : 85–104. 10790386
19. Hsu AL, Murphy CT, Kenyon C (2003) Regulation of aging and age-related disease by DAF-16 and heat-shock factor. Science 300 : 1142–1145. 12750521
20. Morley JF, Morimoto RI (2004) Regulation of longevity in Caenorhabditis elegans by heat shock factor and molecular chaperones. Mol Biol Cell 15 : 657–664. 14668486
21. Xiao R, Zhang B, Dong Y, Gong J, Xu T, et al. (2013) A genetic program promotes C. elegans longevity at cold temperatures via a thermosensitive TRP channel. Cell 152 : 806–817. doi: 10.1016/j.cell.2013.01.020 23415228
22. Lee SJ, Kenyon C (2009) Regulation of the longevity response to temperature by thermosensory neurons in Caenorhabditis elegans. Curr Biol 19 : 715–722. doi: 10.1016/j.cub.2009.03.041 19375320
23. Zelin E, Zhang Y, Toogun OA, Zhong S, Freeman BC (2012) The p23 molecular chaperone and GCN5 acetylase jointly modulate protein-DNA dynamics and open chromatin status. Mol Cell 48 : 459–470. doi: 10.1016/j.molcel.2012.08.026 23022381
24. Echtenkamp FJ, Zelin E, Oxelmark E, Woo JI, Andrews BJ, et al. (2011) Global functional map of the p23 molecular chaperone reveals an extensive cellular network. Mol Cell 43 : 229–241. doi: 10.1016/j.molcel.2011.05.029 21777812
25. Ali MM, Roe SM, Vaughan CK, Meyer P, Panaretou B, et al. (2006) Crystal structure of an Hsp90-nucleotide-p23/Sba1 closed chaperone complex. Nature 440 : 1013–1017. 16625188
26. Hutchison KA, Stancato LF, Owens-Grillo JK, Johnson JL, Krishna P, et al. (1995) The 23-kDa acidic protein in reticulocyte lysate is the weakly bound component of the hsp foldosome that is required for assembly of the glucocorticoid receptor into a functional heterocomplex with hsp90. J Biol Chem 270 : 18841–18847. 7642537
27. Johnson JL, Beito TG, Krco CJ, Toft DO (1994) Characterization of a novel 23-kilodalton protein of unactive progesterone receptor complexes. Mol Cell Biol 14 : 1956–1963. 8114727
28. Rutherford SL, Lindquist S (1998) Hsp90 as a capacitor for morphological evolution. Nature 396 : 336–342. 9845070
29. Zou J, Guo Y, Guettouche T, Smith DF, Voellmy R (1998) Repression of heat shock transcription factor HSF1 activation by HSP90 (HSP90 complex) that forms a stress-sensitive complex with HSF1. Cell 94 : 471–480. 9727490
30. Guisbert E, Czyz DM, Richter K, McMullen PD, Morimoto RI (2013) Identification of a tissue-selective heat shock response regulatory network. PLoS Genet 9: e1003466. doi: 10.1371/journal.pgen.1003466 23637632
Štítky
Genetika Reprodukční medicína
Článek Discovery and Fine-Mapping of Glycaemic and Obesity-Related Trait Loci Using High-Density ImputationČlánek AAA-ATPase FIDGETIN-LIKE 1 and Helicase FANCM Antagonize Meiotic Crossovers by Distinct MechanismsČlánek A Conserved Pattern of Primer-Dependent Transcription Initiation in and Revealed by 5′ RNA-seqČlánek TopBP1 Governs Hematopoietic Stem/Progenitor Cells Survival in Zebrafish Definitive HematopoiesisČlánek Redundant Roles of Rpn10 and Rpn13 in Recognition of Ubiquitinated Proteins and Cellular Homeostasis
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2015 Číslo 7
-
Všechny články tohoto čísla
- LINE-1 Retroelements Get ZAPped!
- /p23: A Small Protein Heating Up Lifespan Regulation
- Hairless Streaks in Cattle Implicate TSR2 in Early Hair Follicle Formation
- Ribosomal Protein Mutations Result in Constitutive p53 Protein Degradation through Impairment of the AKT Pathway
- Molecular Clock of Neutral Mutations in a Fitness-Increasing Evolutionary Process
- Modeling Implicates in Nephropathy: Evidence for Dominant Negative Effects and Epistasis under Anemic Stress
- The Alternative Sigma Factor SigX Controls Bacteriocin Synthesis and Competence, the Two Quorum Sensing Regulated Traits in
- BMP Inhibition in Seminomas Initiates Acquisition of Pluripotency via NODAL Signaling Resulting in Reprogramming to an Embryonal Carcinoma
- Comparative Study of Regulatory Circuits in Two Sea Urchin Species Reveals Tight Control of Timing and High Conservation of Expression Dynamics
- EIN3 and ORE1 Accelerate Degreening during Ethylene-Mediated Leaf Senescence by Directly Activating Chlorophyll Catabolic Genes in
- Genome Wide Binding Site Analysis Reveals Transcriptional Coactivation of Cytokinin-Responsive Genes by DELLA Proteins
- Sensory Neurons Arouse . Locomotion via Both Glutamate and Neuropeptide Release
- A Year of Infection in the Intensive Care Unit: Prospective Whole Genome Sequencing of Bacterial Clinical Isolates Reveals Cryptic Transmissions and Novel Microbiota
- Inference of Low and High-Grade Glioma Gene Regulatory Networks Delineates the Role of Rnd3 in Establishing Multiple Hallmarks of Cancer
- Novel Role for p110β PI 3-Kinase in Male Fertility through Regulation of Androgen Receptor Activity in Sertoli Cells
- A Novel Locus Harbouring a Functional Nonsense Mutation Identified in a Large Danish Family with Nonsyndromic Hearing Impairment
- Checkpoint Activation of an Unconventional DNA Replication Program in
- A Genetic Incompatibility Accelerates Adaptation in Yeast
- The SMC Loader Scc2 Promotes ncRNA Biogenesis and Translational Fidelity
- Blimp1/Prdm1 Functions in Opposition to Irf1 to Maintain Neonatal Tolerance during Postnatal Intestinal Maturation
- Discovery and Fine-Mapping of Glycaemic and Obesity-Related Trait Loci Using High-Density Imputation
- JAK/STAT and Hox Dynamic Interactions in an Organogenetic Gene Cascade
- Emergence, Retention and Selection: A Trilogy of Origination for Functional Proteins from Ancestral LncRNAs in Primates
- MoSET1 (Histone H3K4 Methyltransferase in ) Regulates Global Gene Expression during Infection-Related Morphogenesis
- Arabidopsis PCH2 Mediates Meiotic Chromosome Remodeling and Maturation of Crossovers
- AAA-ATPase FIDGETIN-LIKE 1 and Helicase FANCM Antagonize Meiotic Crossovers by Distinct Mechanisms
- A Conserved Pattern of Primer-Dependent Transcription Initiation in and Revealed by 5′ RNA-seq
- Tempo and Mode of Transposable Element Activity in Drosophila
- The Shelterin TIN2 Subunit Mediates Recruitment of Telomerase to Telomeres
- SAMHD1 Inhibits LINE-1 Retrotransposition by Promoting Stress Granule Formation
- A Genome Scan for Genes Underlying Microgeographic-Scale Local Adaptation in a Wild Species
- TopBP1 Governs Hematopoietic Stem/Progenitor Cells Survival in Zebrafish Definitive Hematopoiesis
- Analysis of the Relationships between DNA Double-Strand Breaks, Synaptonemal Complex and Crossovers Using the Mutant
- Assessing Mitochondrial DNA Variation and Copy Number in Lymphocytes of ~2,000 Sardinians Using Tailored Sequencing Analysis Tools
- Allelic Spectra of Risk SNPs Are Different for Environment/Lifestyle Dependent versus Independent Diseases
- CSB-PGBD3 Mutations Cause Premature Ovarian Failure
- Irrepressible: An Interview with Mark Ptashne
- Genetic Evidence for Function of the bHLH-PAS Protein Gce/Met As a Juvenile Hormone Receptor
- Inactivation of Retinoblastoma Protein (Rb1) in the Oocyte: Evidence That Dysregulated Follicle Growth Drives Ovarian Teratoma Formation in Mice
- Redundant Roles of Rpn10 and Rpn13 in Recognition of Ubiquitinated Proteins and Cellular Homeostasis
- Pyrimidine Pool Disequilibrium Induced by a Cytidine Deaminase Deficiency Inhibits PARP-1 Activity, Leading to the Under Replication of DNA
- Molecular Framework of a Regulatory Circuit Initiating Two-Dimensional Spatial Patterning of Stomatal Lineage
- RFX2 Is a Major Transcriptional Regulator of Spermiogenesis
- A Role for Macro-ER-Phagy in ER Quality Control
- Corp Regulates P53 in via a Negative Feedback Loop
- Common Cell Shape Evolution of Two Nasopharyngeal Pathogens
- Contact- and Protein Transfer-Dependent Stimulation of Assembly of the Gliding Motility Machinery in
- Endothelial Snail Regulates Capillary Branching Morphogenesis via Vascular Endothelial Growth Factor Receptor 3 Expression
- Functional Constraint Profiling of a Viral Protein Reveals Discordance of Evolutionary Conservation and Functionality
- Temporal Coordination of Carbohydrate Metabolism during Mosquito Reproduction
- mTOR Directs Breast Morphogenesis through the PKC-alpha-Rac1 Signaling Axis
- Reversible Oxidation of a Conserved Methionine in the Nuclear Export Sequence Determines Subcellular Distribution and Activity of the Fungal Nitrate Regulator NirA
- Nutritional Control of DNA Replication Initiation through the Proteolysis and Regulated Translation of DnaA
- Cooperation between Paxillin-like Protein Pxl1 and Glucan Synthase Bgs1 Is Essential for Actomyosin Ring Stability and Septum Formation in Fission Yeast
- Encodes a Highly Conserved Protein Important to Neurological Function in Mice and Flies
- Identification of a Novel Regulatory Mechanism of Nutrient Transport Controlled by TORC1-Npr1-Amu1/Par32
- Aurora-A-Dependent Control of TACC3 Influences the Rate of Mitotic Spindle Assembly
- Large-Scale Phenomics Identifies Primary and Fine-Tuning Roles for CRKs in Responses Related to Oxidative Stress
- TFIIS-Dependent Non-coding Transcription Regulates Developmental Genome Rearrangements
- Genome-Wide Reprogramming of Transcript Architecture by Temperature Specifies the Developmental States of the Human Pathogen
- Identification of Chemical Inhibitors of β-Catenin-Driven Liver Tumorigenesis in Zebrafish
- The Catalytic and Non-catalytic Functions of the Chromatin-Remodeling Protein Collaborate to Fine-Tune Circadian Transcription in
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Functional Constraint Profiling of a Viral Protein Reveals Discordance of Evolutionary Conservation and Functionality
- Reversible Oxidation of a Conserved Methionine in the Nuclear Export Sequence Determines Subcellular Distribution and Activity of the Fungal Nitrate Regulator NirA
- Modeling Implicates in Nephropathy: Evidence for Dominant Negative Effects and Epistasis under Anemic Stress
- Nutritional Control of DNA Replication Initiation through the Proteolysis and Regulated Translation of DnaA
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



