-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaArabidopsis PCH2 Mediates Meiotic Chromosome Remodeling and Maturation of Crossovers
In the reproductive cells of many eukaryotes, a process called meiosis generates haploid gametes. During meiosis, homologous parental chromosomes (homologs) recombine forming crossovers (CO) that provide genetic variation. CO formation generates physical links called chiasmata, which are essential for accurate homolog segregation. CO control designates a sub-set of recombination precursors that will mature to form at least one chiasma between each homolog pair. Recombination is accompanied by extensive chromosome reorganization. Formation of a proteinaceous axis organizes the pairs of sister chromatids of each homolog into conjoined linear looped chromatin arrays. Pairs of homologs then align and synapse becoming closely associated along their length by a protein structure, the synaptonemal complex (SC). The SC is disassembled at the end of prophase I and recombination is completed. We have investigated the link between recombination and chromosome remodelling by analysing the role of a protein, PCH2, which we show is required for remodelling of the chromosome axis during SC formation. In wild type, immunolocalization reveals depletion of the axis-associated signal of the axis component, ASY1, along synapsed regions of the chromosomes. In the absence of PCH2, the ASY1 signal is not depleted from the chromosome axis and the SC does not form normally. Although this defect in chromosome remodelling has no obvious effect on CO designation, CO maturation is perturbed such that the formation of at least one CO per homolog pair no longer occurs.
Published in the journal: . PLoS Genet 11(7): e32767. doi:10.1371/journal.pgen.1005372
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1005372Summary
In the reproductive cells of many eukaryotes, a process called meiosis generates haploid gametes. During meiosis, homologous parental chromosomes (homologs) recombine forming crossovers (CO) that provide genetic variation. CO formation generates physical links called chiasmata, which are essential for accurate homolog segregation. CO control designates a sub-set of recombination precursors that will mature to form at least one chiasma between each homolog pair. Recombination is accompanied by extensive chromosome reorganization. Formation of a proteinaceous axis organizes the pairs of sister chromatids of each homolog into conjoined linear looped chromatin arrays. Pairs of homologs then align and synapse becoming closely associated along their length by a protein structure, the synaptonemal complex (SC). The SC is disassembled at the end of prophase I and recombination is completed. We have investigated the link between recombination and chromosome remodelling by analysing the role of a protein, PCH2, which we show is required for remodelling of the chromosome axis during SC formation. In wild type, immunolocalization reveals depletion of the axis-associated signal of the axis component, ASY1, along synapsed regions of the chromosomes. In the absence of PCH2, the ASY1 signal is not depleted from the chromosome axis and the SC does not form normally. Although this defect in chromosome remodelling has no obvious effect on CO designation, CO maturation is perturbed such that the formation of at least one CO per homolog pair no longer occurs.
Introduction
During meiosis genetic crossovers (COs), the products of homologous recombination, in conjunction with sister chromatid cohesion establish physical links, referred to cytologically as chiasmata, between homologous chromosome pairs (homologs) to ensure accurate chromosome segregation at the first nuclear division that follows prophase I. In the absence of crossing over the homologs segregate randomly. This leads to the formation of aneuploid gametes following the second meiotic division [1]. Recombination is initiated by the programmed formation of DNA double-strand breaks (DSBs), catalysed by the topoisomerase type II related protein Spo11 [2,3]. In Saccharomyces cerevisiae (budding yeast) around 40% of DSBs are repaired as non-CO (NCO) products with the remainder progressing to form COs [4]. In Arabidopsis thaliana and other multicellular organisms the proportion of COs is substantially less, typically 5–10% [5]. Importantly, the CO/NCO balance is highly controlled. This control is manifested in several ways. First, each pair of homologs acquires at least one CO. Second, CO interference ensures that multiple COs are well spaced along the chromosomes. Finally, CO homeostasis maintains CO numbers in the face of perturbations that may affect the number of earlier recombinational interactions [6–10]. It is hypothesized that a CO patterning phenomenon, that can be simulated by the beam-film model, underlies these three features of CO control [11,12].
In budding yeast, DSBs form in early leptotene coincident with the elaboration of a proteinaceous chromosome axis that organizes each pair of sister chromatids into linear looped chromatin arrays conjoined by a shared axis. DSBs occur in the context of the chromosome axis [13–15]. At the transition from leptotene to zygotene, formation of the synaptonemal complex (SC), a tripartite structure comprising the chromosome axes linked by overlapping transverse filaments (TFs), is initiated at multiple synapsis initiation sites [1,16,17]. Synapsis continues throughout zygotene bringing the axes into close apposition and is completed at the onset of pachytene when the SC is fully formed. This programmed morphogenesis of the chromosome axes and SC is critical for the coordination of recombination, playing important roles in the meiosis-specific bias that favours inter-homolog recombination and the maturation of CO designated recombination intermediates [18–26].
In budding yeast mutation of the PCH2 gene, which encodes a member of the conserved AAA+ ATPase protein family, disrupts remodelling of the chromosome axis during prophase I of meiosis [27,28]. In wild type cells the chromosome axis protein, Hop1, and the SC transverse filament protein, Zip1, appear to load uniformly at a basal level along the chromosomes. Superimposed on this, each forms a series of non-overlapping, alternating hyper-abundant domains. In a pch2 mutant this domainal loading is disrupted to give a uniform overlapping signal for each protein along the chromosomes [27,28]. Pch2 may modulate inter-homolog bias by remodelling the chromosome structure in the vicinity of DSBs and have a role in a recombination checkpoint [29,30]. Loss of the protein also affects CO formation. In one study, a pch2∆ mutant had increased COs on larger chromosomes, while CO frequency on the small chromosome III was unaffected [31]. Genetic data suggested the mutant also exhibited a defect in CO interference. A link with CO interference was also established in a parallel study, although in this instance no effect on overall CO number was observed [28]. However, further analysis based on the distribution of foci of the E3 ligase Zip3, which arise at CO designated intermediates and so provide an early marker for CO interference, reported that interference is not affected in a pch2 deletion mutant [32].
Orthologs of PCH2 have been identified in a variety of organisms. In mouse, analysis of a weak hypomorphic allele of TRIP13 (PCH2) indicated that the protein was required for the efficient repair of DSBs that enter the NCO pathway but not COdesignated intermediates, which were processed normally. Despite the presence of unrepaired DSBs synapsis was normal in these mice [33]. Subsequently, a study of a more severe Trip13 mutant reported a defect in CO formation and synapsis [34]. Similar to Pch2 in budding yeast, TRIP13 is required for the depletion of the Hop1 orthologs HORMAD1 and HORMAD2 along synapsed regions of the chromosome axes [35]. In Drosophila, PCH2 acts in a checkpoint to monitor defects in recombination and chromosome structure [36]. In Caenorhabditis elegans it is reported to maintain the fidelity of recombination and synapsis during prophase I by acting to constrain these processes [37]. A PCH2 ortholog, referred to as CRC1 (CENTRAL REGION COMPONENT1) has also been identified in rice (Oryza sativa) [38]. The CRC1 protein is 43.8% identical to TRIP13 and 23.1% identical to Pch2 from budding yeast. The crc1 mutant is completely asynaptic and forms univalents at metaphase I due to a failure to make DSBs [39].
Here we describe the identification and analysis of the PCH2 orthologs from Brassica oleracea and its close relative Arabidopsis thaliana. Using super-resolution structured illumination microscopy (SIM) we reveal dynamic changes in localization of PCH2 in relation to chromosome axis and SC morphogenesis during meiotic prophase I. Analysis of Arabidopsis mutants lacking PCH2 reveals a meiotic role that is markedly different to that reported for the rice CRC1 protein. Loss of PCH2 results in a failure to deplete ASY1 from the chromosome axes during zygotene coupled with a synaptic defect. Although recombination initiation and CO designation appears to occur normally during early prophase I, the defects in remodelling of the chromosome axes which influence SC formation are associated with a disruption of the patterned formation of COs along the homologous chromosomes.
Results
PCH2 and ASY1 co-immunoprecipitate in a meiotic protein complex
Protein complexes were immunoprecipitated from Brassica oleracea var. alboglabra A12DH pollen mother cells (PMCs) in meiotic prophase I using an anti-ASY1 antibody as previously described [40]. Co-precipitating proteins were analysed by mass-spectrometry and identified using the Brassica rapa sequence [41]. Up to 10 unique peptides corresponding to 25% sequence coverage (124/490 amino acids) of the Bra013827 predicted gene product were detected in three independent experiments and were absent from control samples (S1A Fig). The protein was identified as a P-loop containing nucleoside triphosphate hydrolase superfamily member. BLAST searches revealed that the protein is 87% identical to the Arabidopsis At4g24710 predicted gene product. ClustalW2 analysis (http://www.ebi.ac.uk) showed that Bra013827 and At4g24710 are members of a sub-family of the AAA+ ATPase super-family that contains the budding yeast PCH2 and mouse TRIP13 genes (S1B Fig).
Atpch2 mutants exhibit a reduced fertility phenotype
To determine whether At4g24710 encodes a functional ortholog of Pch2/TRIP13 we obtained three T-DNA insertion lines: SAIL_1187_C06, SALK_031449 and SALK_130138, hereafter referred to as Atpch2-1, Atpch2-2 and Atpch2-3 respectively. For all lines, the T-DNA insertion site was confirmed by DNA sequencing and the absence of a full-length AtPCH2 transcript confirmed by RT-PCR (S2 and S3 Figs). The vegetative phenotype of each line was indistinguishable from wild type Arabidopsis, Col-0, but their fertility was reduced (S4A and S4B Fig). Quantification of the fertility defect in Atpch2-1 revealed a slight, yet significant reduction in mean silique length from 1.66 ± 0.05 cm in wild type to 1.41 ± 0.06 cm (n = 50; P<0.05) in Atpch2-1 (n = 50). This was accompanied by numerous gaps between the seeds within the siliques such that overall the mean seed-set was significantly reduced from 67.8 per silique in wild type to 34.6 in Atpch2-1 (n = 50; P< 0.01). Analysis of Atpch2-2 and Atpch2-3 revealed very similar fertility defects (S4C Fig).
Loss of AtPCH2 results in a reduction of chiasmata
The Atpch2 reduced fertility phenotype suggested a meiotic defect. Cytogenetic analysis of DAPI stained chromosome spreads from Atpch2-1 PMCs at leptotene revealed the threadlike chromosomes with no obvious differences to the wild type controls (Fig 1A and 1B). In wild type PMCs, the homologs achieved full synapsis at pachytene with the threadlike signals visibly paired along their lengths, giving a thicker appearance than at leptotene (Fig 1C). However, in Atpch2-1 pachytene stage cells were not observed, instead the majority of the chromosomes remained as single threadlike signals with some limited regions where paired axes were visible (Fig 1D). During diplotene both Atpch2-1 and wild type chromosomes desynapsed and began to condense, such that by diakinesis chiasmata linking the homologs were visible. At metaphase I, following further condensation, distinct bivalents were observed. Five bivalents were invariably present in wild type, but some Atpch2-1 nuclei contained a mixture of bivalent and univalent chromosomes (Fig 1E and 1F). To quantify this we counted chiasmata in Atpch2-1 in metaphase I chromosome spreads using fluorescent in situ hybridization (FISH) with 45S and 5S rDNA probes to identify individual chromosomes [42] (Fig 1G and 1H). This revealed a significant reduction in the mean chiasma frequency in Atpch2-1 compared to wild type (6.9; n = 50 versus 9.6; n = 50; P < 0.001). No univalents were observed in the wild type sample, whereas they were present at a frequency of 10.0% in Atpch2-1 with all chromosomes affected. Similar results were obtained for Atpch2-2 (6.9; n = 37; P < 0.001; univalent frequency 14.6%) and Atpch2-3 (6.2; n = 26; P < 0.001; univalent frequency 7.7%). As a consequence of this, in contrast to wild type, mis-segregation of the chromosomes was observed at the first meiotic division in Atpch2-1 (Fig 1I and 1J) leading to unbalanced tetrads (Fig 1K and 1L). No precocious sister chromatid separation was observed suggesting that there was no cohesion defect. Analyses of Atpch2-2 and Atpch2-3 revealed that the meiotic defect in the three mutants is essentially identical (S5A–S5H Fig). To confirm that the observed phenotype was due to a loss of AtPCH2 function, an allelism test was conducted by crossing Atpch2-1 with Atpch2-2. Phenotypic and cytological analysis of the Atpch2-1/Atpch2-2 progeny revealed the same defects as in the parental lines confirming that these arose due to the loss of AtPCH2 function (S5I–S5M Fig).
Fig. 1. Meiotic stages from wild type Arabidopsis and Atpch2-1 pollen mother cells. 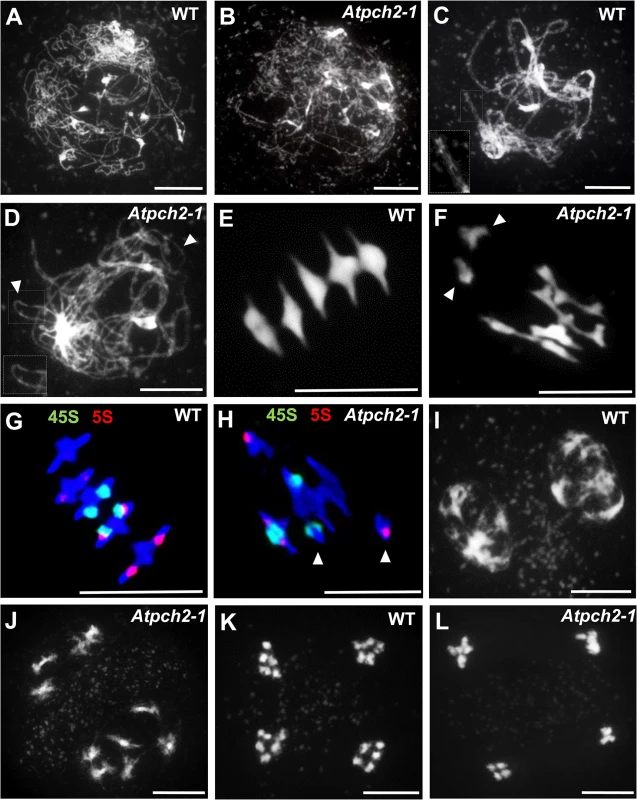
Chromosome spread preparations from wild type (A,C,E,G,I,K) and Atpch2-1 (B,D,F,H,J,L) PMCs. (A,B) leptotene; (C,D) pachytene (note in Atpch2-1 cell is at late prophase I normal pachytene stage was not observed). Arrowheads mark unsynapsed regions. (E,F) metaphase I. Arrowheads mark univalent chromosomes; (G,H) metaphase I stage labelled with 5S (red) and 45S (green) rDNA probes to identify the individual chromosomes. Arrowheads mark univalent chromosomes; (I,J) dyad; (K-L) tetrad. DNA is stained with DAPI. Bar = 10 μm. We next investigated if the reduction in chiasmata in Atpch2-1 reflected a defect in CO formation. Approximately 85% of COs in Arabidopsis exhibit CO interference [43]. Formation of these, so-called Class I COs, require a group of proteins known as ZMMs (Zip1, Zip2, Zip3/Hei10, Zip4, Mer3, Msh4 and Msh5) [5,18]. The remainder (Class II) are insensitive to CO interference and dependent on the structure-specific endonuclease AtMUS81 [44]. To determine if the loss of AtPCH2 affected one or both classes of COs we generated an Atmsh5-1/Atpch2-1 double mutant. In Atmsh5-1, the number of chiasmata per PMC ranged between 0 and 4 with a mean chiasma frequency of 1.2 (n = 50) (S6B Fig). In comparison, the mean chiasma frequency in the Atmsh5-1/Atpch2-1 double mutant was significantly reduced to 0.3 (n = 50; P < 0.001), with the number of chiasmata per nucleus ranging between 0 and 2 (S6C Fig).
Thus, overall the cytological analysis suggests that the reduction in chiasmata in Atpch2-1 arises from a recombination defect that impacts on both interference sensitive and insensitive CO formation, rather than through an effect on sister chromatid cohesion.
Chromosome axis remodelling is disrupted in Atpch2-1
The failure to observe pachytene stage PMCs in Atpch2-1 suggested a defect in formation of the SC. To investigate further, we examined chromosome axis reorganization during early to mid-prophase I using immunocytochemistry combined with fluorescence light microscopy and SIM. At leptotene in wild type Arabidopsis, the HORMA domain protein ASY1 is detected in chromosome spreads of PMCs as a linear axis-associated signal. This appears to be comprised of a series of alternating regions of higher and lower signal intensity, suggestive of a domainal organization of ASY1 abundance along the chromosome axis [19,45] (Fig 2A and 2C). Analysis of Atpch2-1 PMCs at leptotene did not reveal any obvious differences, with localization of ASY1 appearing normal (Fig 2B and 2D). Similarly, the cohesin complex protein SYN1 [46,47] (S7A and S7B Fig) and the chromosome axis protein ASY3 [19] (S7C and S7D Fig) appeared unaffected in Atpch2-1, with both forming a linear axis-associated signal from leptotene through to mid-prophase I. That SYN1 localization was normal supported the cytological observation that there was no evidence of a sister chromatid cohesion defect. Consistent with these observations, comparison of the mean total axis length per PMC at leptotene was not significantly different to wild type (Atpch2-1: 229 μm versus wt: 220 μm, n = 10, P = 0.53).
Fig. 2. Immunolocalization of ASY1 and ZYP1 in wild type and Atpch2-1 during prophase I. 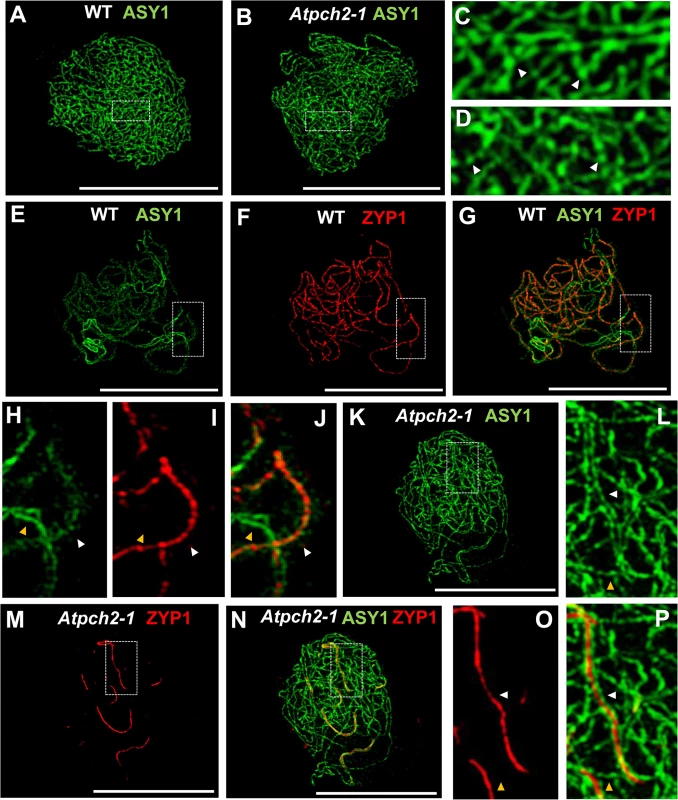
(A-D) Immunolocalization of ASY1 (green) on chromosome spread preparations from wild type (A,C) and Atpch2-1 (B,D) nuclei at leptotene. Panels C and D show magnified sections of axes from A and B respectively. White arrowheads mark the regions of ASY1 with higher signal intensity. (E-J) Immunolocalization of ASY1 (green) (E,G,H,J) and ZYP1 (red) (F,G,I,J) and merge images (G,J) on chromosome spreads from wild type at zygotene. Figs H, I and J show magnified sections of axes from E, F and G respectively. (K-P) Immunolocalization of ASY1 (green) (K,L,N,P) and ZYP1 (red) (M-P) and merge images (N,P) on chromosome spread preparations from Atpch2-1 at mid/late-prophase I. Figs L, O and P show magnified sections of axes from K, M and N respectively. White arrowheads represent synapsed regions while yellow arrowheads represent unsynapsed regions. DNA is stained with DAPI (blue). Bar = 10 cm. Previous immunolocalization studies show that the Arabidopsis SC TF protein ZYP1 begins to polymerize between the aligned homologous chromosomes from multiple sites of synapsis initiation at the onset of zygotene. Polymerization continues throughout zygotene until completion of SC formation at pachytene [20]. Dual-localization of ZYP1 and ASY1 in wild type Arabidopsis revealed that SC formation is accompanied by a reduction in the intensity of ASY1 signal which appeared less continuous and appeared to be associated with the chromatin loops rather than the axis along synapsed regions (Fig 2E–2J: compare synapsed segment with unsynapsed region in 2G and 2J; S8A–S8C Fig). Quantification of the relative intensity of the ASY1 signal (S9 Fig) indicated a significant reduction of 67.0% (n = 23; P = <0.001) on the synapsed region compared to the unsynapsed axes (S9C and S9D Fig; S1 Table). Analysis of Atpch2-1 PMCs at mid/late-prophase I suggested that unlike wild type, the ASY1 signal intensity along the synapsed compared to unsynapsed regions remained unchanged (n = 22; P = 0.25) (S9E and S9F Fig; S1 Table). However, the differentiation of the ASY1 signal into putative domains of high and low intensity appeared enhanced in the mutant, possibly a consequence of the delayed synapsis and increased axis compaction relative to leptotene (S9F Fig). SC polymerization was compromised in the mutant (Fig 2M–2P). Stretches of ZYP1 were detected but varied in number and length from cell to cell. On average the SC length at late prophase I in Atpch2-1 was 32% that of wild type (57 μM, n = 16 versus 179 μM, n = 8), although this ranged from 13% to 57%.
PCH2 distribution during prophase I
To gain further insight into the relationship between PCH2 and the components of the chromosome axes, we conducted immunolocalization studies using an anti-PCH2 antibody on chromosome spreads of wild type PMCs from Arabidopsis and B. oleracea (Figs 3 and 4). Analysis of Arabidopsis using SIM revealed numerous chromatin-associated PCH2 foci (mean 165; n = 10) in G2 coinciding with the appearance of foci and short stretches of ASY1 (Fig 3A). Most PCH2 foci remained distinct from the ASY1 signal (Fig 3A, inset 3C). As the chromosome axis formed in leptotene, the ASY1 signal became more linear. At this stage the proteins appeared associated, with 51.2% (n = 12) of the PCH2 foci overlapping the ASY1 signal to some extent (Fig 3B, inset 3D), possibly a consequence of the chromosome reorganization that occurs at leptotene. As the SC formed during zygotene PCH2 distribution changed. ASY1 associated foci were no longer apparent. Instead PCH2 now tracked the depleted ASY1 signal along the synapsed region, forming a linear array of foci that tended to coalesce (Fig 3E and 3I). Dual-immunolocalization of PCH2 and ZYP1 confirmed that the PCH2 signal localized to the regions where SC nucleates and was present as foci along the SC during zygotene through pachytene (Fig 3J–3P and S10A–S10C Fig). Analysis in Atasy1 and Atasy3 mutants where SC formation is severely compromised, such that only short stretches or accumulations of ZYP1 are formed [19,23], also revealed colocalization of the PCH2 and ZYP1 signals (S10D–S10I Fig). No PCH2 signal was detected in any of the three Atpch2 mutant lines (S11 Fig).
Fig. 3. Immunolocalization of PCH2 in wild type Arabidopsis. 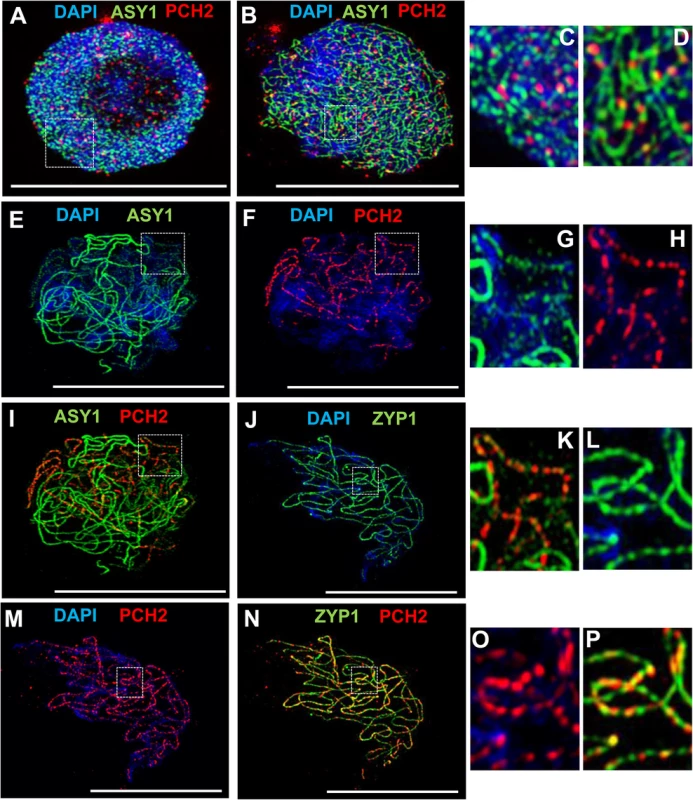
(A-D) Dual localization of ASY1 (green) and PCH2 (red) on chromosome spread preparations from wild type PMCs at G2 (A) and leptotene (B). Panels (C) and (D) show magnified sections of (A) and (B) respectively. (E-I,K) Immunolocalization of ASY1 (green) and PCH2 (red) and merge (I,K) in wild type at mid-prophase I. Panels G, H and K show magnified sections of axes from images E, F and I respectively. (J,L-P) Immunolocalization of ZYP1 (green) and PCH2 (red) and merge (N,P) in a wild type nucleus at mid-prophase I. Panels L, O and P represent magnified sections of axes from J, M and N respectively. DNA is stained with DAPI. Bar = 10 cm. Fig. 4. Immunolocalization of PCH2 in B. oleracea at the leptotene/zygotene transition. 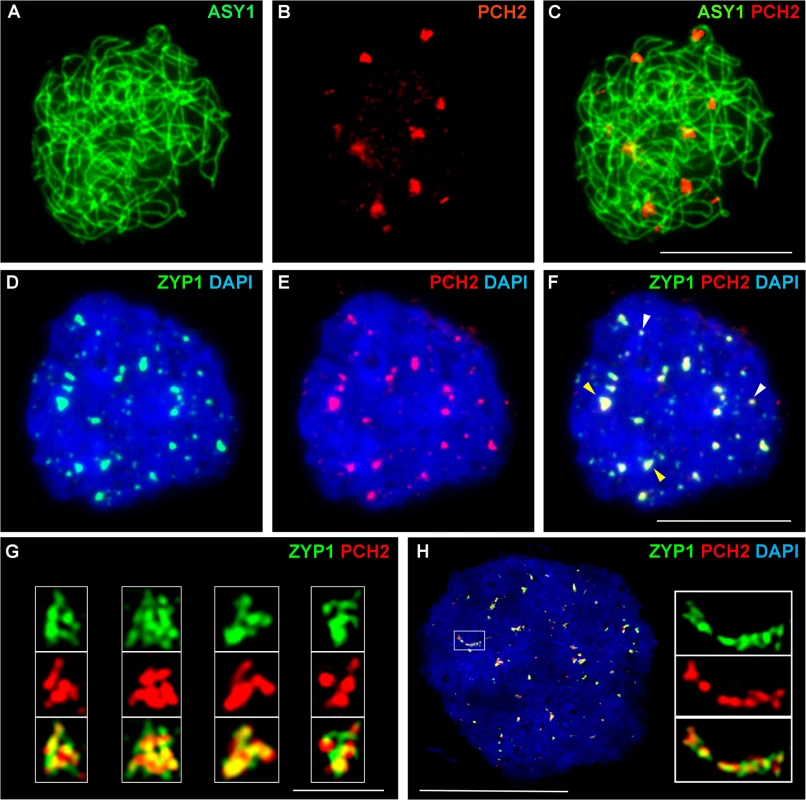
(A-C) Dual localization of ASY1 (green) and PCH2 (red) on chromosome spread preparations from B. oleracea PMCs at the leptotene/zygotene transition. (D-F) Dual localization of ZYP1 (green) and PCH2 (red) at SC nucleation sites. Yellow arrows indicate examples of large ‘arrowhead’ shaped foci and white arrows indicate the smaller SC nucleation sites. (G) SIM images of arrowhead SC nucleation sites stained with ZYP1 (green) and PCH2 (red). (H) SIM image of dual localization of ZYP1 (green) and PCH2 (red) on a nascent stretch of SC. DNA is stained with DAPI (blue). Bar = 1 μm in (G) and 10 μm in all other images. Immunolocalization in B. oleracea PMCs revealed that similar to Arabidopsis, numerous PCH2 foci were detected in late G2/early leptotene (S12A and S12B Fig). At late leptotene/early zygotene PCH2 formed fewer, large foci (mean number per nucleus = 14.2; range = 10–20; n = 18) (Fig 4A–4C). Dual localization of ZYP1 and PCH2 at this stage indicated that these foci correspond to sites of SC nucleation at the leptotene/zygotene transition (Fig 4D–4F) and SIM analysis revealed that the ZYP1 signal at the nucleation site often appeared to form a ‘arrowhead-like’ shape to which PCH2 co-localized (Fig 4G). From the SIM images the arrowhead-like foci were estimated to have a mean length of 602 nm (range = 560–640 nm; n = 40) and a mean maximum width of 419 nm (range 400–480 nm; n = 40) and seemed quite consistent in number (mean 15 per nucleus; range = 12–19; n = 5). In a larger sample, analysed using fluorescence microscopy, a mean of 12.2 foci per nucleus was observed (n = 50). Although the range (5–22) was greater than in the SIM sample, most nuclei (76.0%) contained 10 or more foci. The slight variation in the number of PCH2 foci observed in these experiments probably reflected the dynamics of the process and the increased resolution afforded by SIM relative to fluorescence microscopy. In addition to the large foci, slightly more numerous smaller ZYP1 foci were also observed at early zygotene (mean number per nucleus = 17.0; n = 50). These also co-localized with PCH2 (Fig 4D–4F). As the SC began to extend, SIM revealed extensive overlap between ZYP1 and PCH2 signals each appearing to be comprised of multiple smaller foci (Fig 4H). At zygotene, the ASY1 signal appeared to be reduced along synapsed regions of the chromosomes (S12C Fig) (64.3% reduction relative to unsynapsed axes; n = 14), which were decorated with numerous small PCH2 foci. At pachytene, PCH2 foci were still detected along the entire length of the ZYP1-stained SC as well as in the surrounding chromatin (S12D Fig).
Prophase I progression is delayed in Atpch2-1
In budding yeast deletion of PCH2 results in an accumulation of nuclei in pachytene and a delay in progression through meiosis I [27]. We therefore investigated if the protein has a role in prophase I progression in Arabidopsis. 5-ethynyl-2’-deoxyuridine (EdU) was used to pulse-label Atpch2-1 PMCs during meiotic S-phase [48]. Progression through meiosis was then monitored (S13A Fig). In wild type and Atpch2-1, EdU labelled leptotene nuclei were detected 10h post S-phase. By 25h all labelled wild type PMCs were at zygotene or pachytene and at zygotene in Atpch2-1. At 32h the wild type PMCs had exited pachytene and were at diplotene/diakinesis and by 36h were at the dyad stage, whereas Atpch2-1 PMCs were still at zygotene suggesting a delay of 5-8h (S13A and S13B Fig).
Fluorescent tetrad analysis supports a CO defect in Atpch2-1
The cytological analysis (see earlier) suggested a defect in CO formation in Atpch2 mutants. To further examine the recombination phenotype of Atpch2-1 we used the fluorescent-tagged-line (FTL) system [49,50] which relies on the segregation of three genetically linked transgenic markers, each encoding a distinct pollen-specific fluorescent protein expressed post-meiotically. The FTLs are in a qrt1-2 mutant background which prevents the separation of the gametes and facilitates the visualisation of the meiotic recombination events that have occurred between the transgenic markers in the tetrad pollen [51,52]. Three pairs of adjacent genetic intervals, one on each arm of chromosome 5 and another on chromosome 2 were examined (S14 Fig). This revealed that the genetic map distance determined using the Perkins mapping equation [53] in the adjacent intervals I5c and I5d was not significantly affected by the Atpch2-1 mutation (I5c wild type 6.1 cM v Atpch2-1 6.8 cM; P = 0.17; I5d wild type 5.5 cM v Atpch2-1 6.0 cM; P = 0.28) (Fig 5A). However, interval I5a showed a significant decrease in recombination frequency in the presence of Atpch2-1 (15.1 cM) compared to wild type (27.7 cM; P < 0.001), whereas the map distance in interval I5b exhibited a significant increase from 17.3 cM in wild type to 22.3 cM in the mutant (P < 0.001) (Fig 5A). A significant increase in map distance was observed in intervals l2f and l2g in the presence of the Atpch2-1 mutation (l2f wild type 6.1 cM / Atpch2-1 8.0 cM P < 0.001; l2g wild type 5.1 cM / Atpch2-1 7.1 cM P < 0.001) (Fig 5A).
Fig. 5. Recombination frequency and chiasma distribution in Atpch2-1. 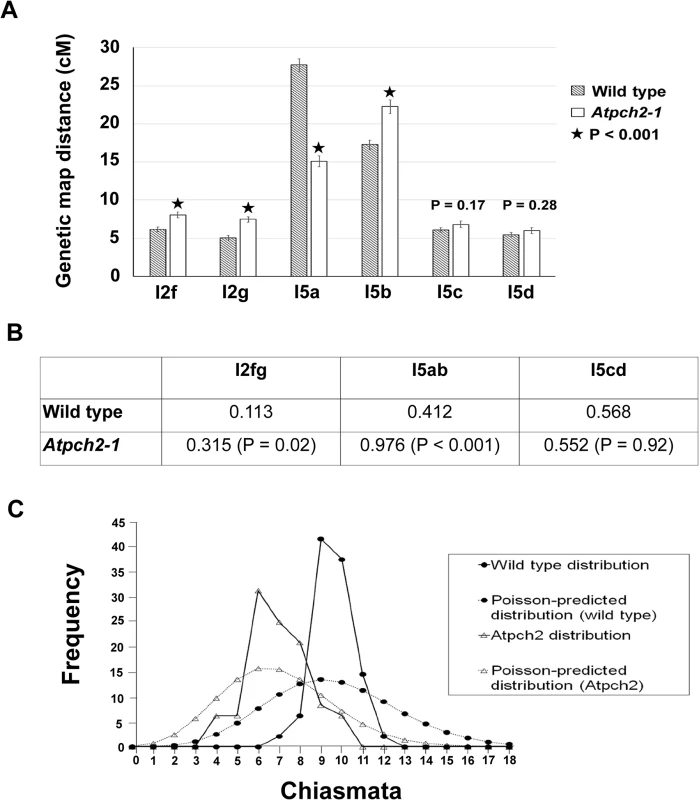
(A) Genetic map distance of four distinct intervals on chromosome 5 and two intervals on chromosome 2 in wild type Arabidopsis and Atpch2-1 mutant. Black stars represent a statistical difference in the genetic map distance between wild type and mutant. P value is indicated on the graph when the genetic map distance of an interval is not statistically different between wild type and mutant. Error bars represent the standard error of the mean. (B) Three pairs of adjacent intervals l2fg, I5ab and I5cd were used to estimate genetic CO interference for wild type and Atpch2-1 mutant. CO interference ratio for each pair of intervals is indicated in the table and P value is shown in parentheses. (C) Observed (solid line) and Poisson-predicted (dotted line) distributions of chiasma numbers per cell for wild type (black circle) and Atpch2-1 mutant (white triangle). We used the FTL data to obtain a genetic estimate for CO interference in adjacent intervals by calculating the Interference Ratio (IR). This method, developed by Malkova et al. [54], uses the ratio of the genetic map distance in an interval with and without the presence of a CO in an adjacent interval to provide an estimate of the strength of CO interference. When COs in adjacent intervals are entirely independent of each other the IR is 1, indicating no interference. Values less than 1 indicate increasing levels of (positive) interference with a value of 0 indicating complete interference. IR ratios greater than 1 are indicative of negative interference. The CO interference ratio was 0.412 for I5ab in wild type. In Atpch2-1, the genetic map distance of I5a was similar with and without the presence of a CO in the interval I5b (14.8 cM with a CO in interval I5b vs 15.2 cM without a CO in interval I5b). The CO interference ratio is 0.976 and is statistically higher than wild type (Z-score = 5.40; P < 0.001) (Fig 5B). This suggests that CO interference is reduced in the interval I5ab in Atpch2-1. In contrast, the CO interference ratio of I5cd is similar in wild type (0.568) and in Atpch2-1 (0.552; Z-score = 0.01; P = 0.92) (Fig 5B). The interference ratio for interval l2fg is also increased in the Atpch2-1 mutant. In wild type the ratio is 0.113 whereas in Atpch2-1 it is 0.315 (P = 0.021).
We also used the FTL data to estimate the coefficient of coincidence (CoC) for the three pairs of intervals. The CoC is calculated by dividing the observed frequency of double COs in two adjacent intervals by the expected frequency assuming no interference [55]. When interference is absent the CoC is 1 and where it is complete the CoC is 0. The overall result was similar to that obtained for the IR (S2 Table). For l5a/b interference appeared reduced (CoC wild type = 0.46 v CoC Atpch2-1 = 0.99), for l5c/d it was unchanged (CoC wild type = 0.60 v CoC Atpch2-1 = 0.60) and for l2fg there was an apparent decrease (CoC wild type = 0.13 v CoC Atpch2-1 = 0.37).
Numerical distribution of chiasmata in Atpch2-1
In the absence of CO control the numerical distribution of chiasmata between cells is predicted to fit a Poisson distribution [56]. This expectation is borne out in ZMM mutants such as Atmsh4 and Atmer3, whereas in wild type the distribution is non-Poissonian. [43,57,58]. We analysed the chiasma distribution in the sample of Atpch2-1 cells described above. The number of chiasmata per nucleus ranged between 4 and 10 in Atpch2-1 and between 7 and 12 in wild type. Further inspection revealed that the proportion of Atpch2-1 PMCs with a chiasma frequency close to the mean of 6.9 was over-represented in the sample analysed, with 74% having between 6 and 8 chiasmata per cell (vs 42.8% if the numerical distribution of chiasmata was random) (Fig 5C). Over-distribution of chiasma around the mean is also a feature of wild type [43]. The chiasma distribution in Atpch2-1 differed significantly from a Poisson distribution (X(11)2 = 45.2; P < 0.001). This was also confirmed in Atpch2-2 (X(11)2 = 37.2; P < 0.001) and Atpch2-3 (X(11)2 = 40.00; P < 0.001).
Later stages of recombination are aberrant in Atpch2-1
We investigated the basis for the reduction in chiasmata in Atpch2-1 using immunolocalization of recombination pathway proteins on prophase I chromosome spreads from Atpch2-1 PMCs. Immunolocalization of the strand-exchange proteins RAD51 and DMC1 which are recruited to DSBs at leptotene was used to monitor early recombination and immunolocalization of the ZMM protein AtMSH4 was used to detect later recombination progress [43,59,60]. There were no significant differences between wild type (Fig 6A, 6C and 6E) and Atpch2-1 (Fig 6B, 6D and 6F) PMCs. At mid-leptotene the mean number of RAD51 foci in Atpch2-1 was 144 versus 146 in wild type (n = 12; P = 0.37) (Fig 6A and 6B). For DMC1 the corresponding values were 167 versus 173 (n = 12, P = 0.56) (Fig 6C and 6D). In PMCs at the leptotene/zygotene transition the mean number of MSH4 foci was 150 in Atpch2-1 versus 152 in wild type (n = 12; P = 0.63) (Fig 6E and 6F).
Fig. 6. Dual localization of ASY1 and recombination pathway proteins in wild type and Atpch2-1 meiotic nuclei at early prophase I. 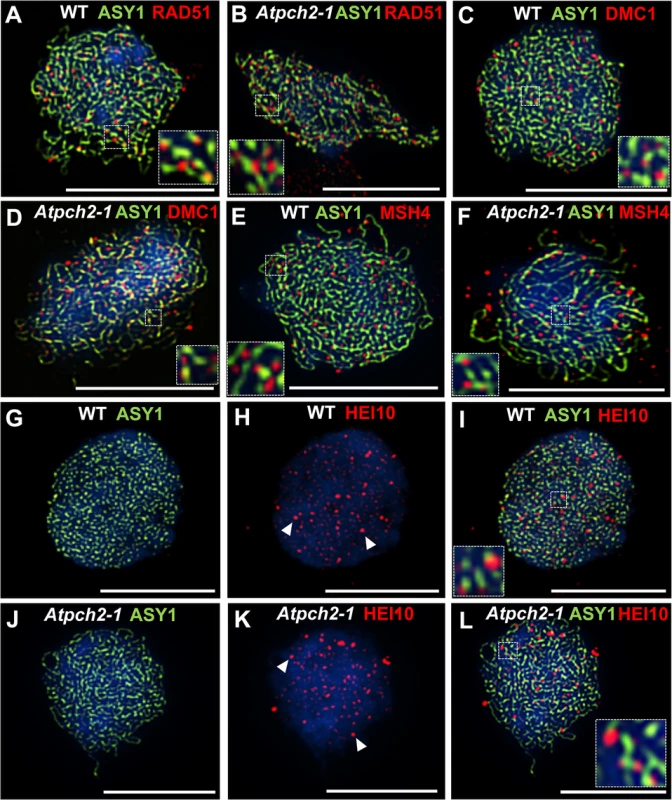
Dual localization of ASY1 (green) and RAD51 (red) on wild type (A) and Atpch2-1 (B) PMCs at mid-leptotene; ASY1 (green) and DMC1 (red) on wild type (C) and Atpch2-1 (D) PMCs at mid-leptotene; ASY1 (green) and MSH4 (red) on wild type (E) and Atpch2-1 (F) PMCs at leptotene/zygotene transition; ASY1 (green) and HEI10 (red) on wild type (G-I) and Atpch2-1 (J-L) PMCs at late-leptotene. DNA is stained with DAPI (blue). Bar = 10 μm. HEI10 (Human enhancer of invasion-10) is a member of the Zip3/Hei10 family of proteins which are thought to possess SUMO/ubiquitin E3 ligase activity [61]. Studies reveal that Zip3/Hei10 marks the sites of future type I COs [61,62]. In Sordaria macrospora Hei10 foci that mark COs are ~300 nm in size and emerge from a much larger population of small axis-associated foci during early/mid-prophase I [63]. Dual localisation of ASY1 and HEI10 on chromosome spreads of Arabidopsis wild type and Atpch2-1 PMCs at late leptotene showed that in both cases HEI10 formed a very similar large number of foci (166 versus 165 respectively, n = 10; P = 0.81) along the chromosome axes (Fig 6G–6L). As prophase I progressed the foci decreased in number and disappeared by pachytene. Most foci were small (~175 nm) but in addition, a number of larger (>250 nm) HEI10 foci were observed in both sets of PMCs. In wild type at the leptotene/zygotene transition we observed 7 to 15 large HEI10 foci (mean 10.6, n = 33). This remained constant through late pachytene (mean 9.9 range 9–12, n = 14) (Fig 7A, 7C and 7I). During early prophase I the number and distribution of large HEI10 foci in Atpch2-1 PMCs was not significantly different to wild type (mean 10.6 versus 10.2; P = 0.20; n = 21). However, at mid/late prophase the mean number of large HEI10 foci was significantly reduced to 6.9 (n = 27) compared to wild type nuclei (P < 0.001) (Fig 7B, 7D and 7I). HEI10 foci were mostly found as singletons on the stretches of SC in Atpch2-1 (83.4% n = 185) (Fig 7B and 7D), with two or three HEI10 foci observed in 14.6% and 2.0% of cases respectively.
Fig. 7. Dual localization of ZYP1 and recombination pathway proteins in wild type Arabidopsis and Atpch2-1 meiotic nuclei at mid/late prophase. 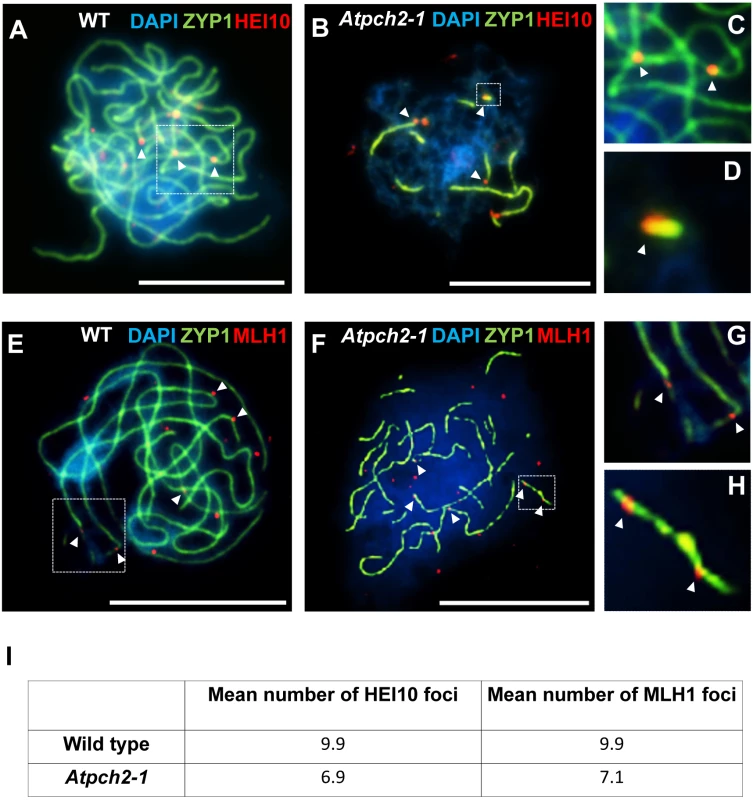
(A-D) Dual localization of ZYP1 (green) and HEI10 (red) on wild type (A,C) and Atpch2-1 (B,D) PMCs. Panels C and D show magnified sections of SC from A and B respectively. (E-H) Dual localization of ZYP1 (green) and MLH1 (red) on wild-type (E,G) and Atpch2-1 (F,H) PMCs. Panels G and H show magnified sections of SC from E and F respectively. DNA is stained with DAPI (blue). Bar = 10 μm. (I) Table showing the mean number of HEI10 and MLH1 foci in wild type and Atpch2-1. Dual localization of HEI10 and ZYP1 in B. oleracea PMCs at the leptotene/zygotene transition revealed that most of the large ZYP1 foci at SC nucleation sites that had been shown to co-localize with PCH2 at this stage (see earlier), also co-localized with HEI10 (86.0% foci; n = 30 nuclei) (S15 Fig).
To confirm that the reduction in large HEI10 foci in Atpch2-1 reflects a reduced number of mature CO intermediates we analysed the distribution of the late recombination protein MLH1 which marks the sites of Type I COs/chiasmata [64]. Dual immunolocalization of MLH1 and ZYP1 (N-terminus Ab, see Materials and Methods) on chromosome spreads of wild type PMCs, showed the number of MLH1 foci per nucleus varied between 9 and 11 with a mean count of 9.9 (n = 12) at pachytene (Fig 7E, 7G and 7I). In Atpch2-1 the mean number of MLH1 foci per nucleus was 7.1 (n = 12), a significant reduction compared to wild type (P < 0.002) (Fig 7F, 7H and 7I). We noted that in both cases the MLH1 foci were often adjacent to the ZYP1 signal rather than directly over the SC central region. Similar to the distribution of HEI10 foci, MLH1 foci were mostly observed as singletons on stretches of SC in Atpch2-1 PMCs (61.2% n = 85) with two or three foci occurring in 28.2% and 10.6% cases respectively (Fig 7F and 7H).
Discussion
Chromosome axis remodeling at the leptotene/zygotene transition is defective in the absence of PCH2
Formation of the chromosome axis in early prophase I appears unaffected by loss of PCH2 based on immunolocalization of the axis proteins and axis length measurements. This differs from rice where the PCH2 ortholog, CRC1, is required for recruitment of the ASY1 ortholog PAIR2 onto the chromosome axes at leptotene [38]. This difference between the two plant species is perhaps surprising but it is not the first example where the phenotype of a rice meiotic mutant is different to that in other plants. For instance, loss of ZYP1 in Arabidopsis and barley results in a reduction of CO formation whereas mutation of the corresponding rice gene, ZEP1, leads to increased COs [20,65,66].
At mid-prophase I in Arabidopsis and B. oleracea, PCH2 forms foci along the SC which correlate with regions of ASY1 signal depletion on the axes. The overall distribution of PCH2, together with the fact that ASY1 signal intensity is not reduced along the synapsed axes in Atpch2 mutants, suggests that PCH2 participates in the depletion of ASY1 from the axis at the leptotene/zygotene transition. This could be a direct effect since biochemical studies in budding yeast show that Pch2 can bind to Hop1 in vitro and binding is strongly enhanced if its ATP hydrolysis activity is blocked [67]. In addition, Pch2 was shown to displace Hop1 from double-stranded DNA. Direct interaction in vivo has not been established as it is argued that this would be transient in the presence of ATP [67]. Based on the number of peptides recovered, PCH2 is found as an abundant component of a complex that is co-precipitated with ASY1 from Brassica PMCs. Although this could reflect a direct interaction, PCH2 may be co-precipitated as part of a larger chromosome axis-protein complex. Thus, an alternative possibility is that the reduction in the ASY1 signal is an indirect consequence of PCH2-dependent reorganization of the chromosome axis at the onset of zygotene.
PCH2 is important for synapsis but does not appear to be an integral SC component
In rice, loss of the PCH2 ortholog, CRC1 leads to a failure to form SC. This is unsurprising given that DSBs are not formed in a crc1 mutant [38]. Nevertheless, studies indicate that CRC1 localizes to the central region of the SC at pachytene and interacts with the SC transverse filament protein ZEP1 in a yeast two-hybrid assay, suggesting it is a component of the SC [38]. Analysis of the Atpch2 mutants indicates that PCH2 plays a critical role in formation of the SC, since loss of the protein results in a substantial defect in polymerization of the SC transverse filament protein ZYP1. An average reduction in SC length of 68% was observed but this was quite variable ranging from 43% to 87%. Similar to rice, co-localization between PCH2 and ZYP1 is also observed from the beginning of zygotene through pachytene. In Atasy1 and Atasy3 mutants, where SC polymerization is compromised, PCH2 is associated with the residual ZYP1 signal. Association of ZYP1 and PCH2 is also supported by SIM analysis of the B. oleracea SC as it begins to extend, although this suggests that they are not forming a homogeneous complex. This could reflect that any interaction between the proteins is transient. Since ASY1 appears to be the target for PCH2, it is conceivable that ZYP1 or another component of the SC central region acts to couple/guide the PCH2/ASY1 interaction. A precedent for this is seen in the bacterial P1 plasmid partitioning system in which the ParA ATPase is functionally coupled by the ParB protein to move its plasmid DNA cargo via a diffusion-rachet mechanism [68]. Furthermore the interaction between the C. elegans PCH2 ortholog, PCH-2, and the HORMAD spindle checkpoint protein Mad2, has been shown to involve an adaptor protein, p31 [69].
The SC nucleations seen in B. oleracea were consistent in size and their arrowhead-like shape is likely a consequence of the convergence of the homolog axes at the synaptic site. It is noteworthy that in most nuclei examined the number of large foci is broadly similar to the chiasma frequency (13–15) in B. oleracea [70,71]. Moreover most of the arrowhead-like ZYP1 foci (86.0%) co-localized with HEI10, a further indication that they occur at designated CO sites. Nuclei with fewer large foci may have been at a slightly earlier stage and reflect the dynamic nature of the initial appearance of foci. The smaller, slightly more numerous ZYP1 foci that were also present at early zygotene are likely to be additional synapsis initiation sites. The apparent existence of two classes of SC nucleation structures is reminiscent of observations in S. macrospora [72]. These have revealed distinct types of designations, one defining SC nucleation sites that correspond to CO designated recombination events and another that defines a similar number of sites where SC nucleation alone occurs. Importantly, the distribution of both classes exhibit interference and fits the prediction of the ‘beam-film’ model [11,12]. This posits that mechanical stress arises within a chromatin-axis meshwork as a result of global chromatin expansion during leptotene. Subsequent bi-directional relief of this stress results in a set of CO designations (and SC nucleations) that are spatially separated along the chromosomes. Further studies will be required to establish if the observed SC nucleations in B. oleracea also reflect a corresponding underlying interference-dependent distribution.
In other species loss of PCH2 orthologs leads to a variety of different effects on synapsis. In mouse, mutation of the Pch2 ortholog TRIP13 also results in a synaptic defect, albeit less severe than in Arabidopsis, with the unsynapsed regions accounting for just under 30% of the total axis length [34]. Loss of Pch2 in budding yeast does not appear to affect SC formation [27]. However, budding yeast forms high levels of COs and each designated CO site is thought to nucleate SC formation [73]. Hence, loss of Pch2 may not impact on SC installation to the degree observed in Arabidopsis where the relative CO rates are far lower. PCH2 also impacts on SC formation in C. elegans but in this case SC formation occurs more quickly than in wild type [37]. Interestingly, this defect was suppressed at lower temperatures. Thus loss of Pch2 has differing effects on the extent of SC polymerization in different species but in each case is associated with a recombination defect. Together, these observations suggest that PCH2 is not an integral structural component of the SC and more likely, regulates the coordination of synapsis with the controlled formation of COs.
Early recombination pathway events appear normal and DSBs are repaired in Atpch2-1
The controlled formation of COs via homologous recombination is an essential feature of meiosis. Studies of PCH2 in several species have linked loss of the protein to a variety of recombination defects. In the most severe case, loss of the rice PCH2 ortholog, CRC1, is reported to result in a failure to form DSBs [38]. In budding yeast, a minor role for Pch2 in DSB formation has been reported [74]. It is also involved in processing of early occurring, low abundance DSBs and loss of the protein leads to a coordinate delay in the repair of DSBs to form both CO and NCO products [27,28].
In mouse, studies suggest that a severe reduction in TRIP13 expression does not compromise DSB formation but loading of RAD51 onto the resected DSBs is reduced [34]. In Arabidopsis, immunolocalization of RAD51 and DMC1 in Atpch2-1 PMCs indicated that early stages in recombination occur normally. As there is no evidence of chromosome fragmentation, it seems DSBs are also repaired, albeit with a reduction in CO formation, but progression through prophase I is delayed by 5-8h. This is reminiscent of that seen in some meiotic mutants and is indicative of an underlying defect in the recombination pathway [20,43,64]. Since a significant reduction in CO frequency was observed in an Atpch2/Atmsh5 double mutant relative to an Atmsh5 mutant it appears that loss of PCH2 impacts on the formation of both Class I and Class II COs.
CO interference is likely established in Atpch2-1 but maturation of designated COs appears defective
Studies in different species have reported a CO interference defect associated with mutation of Pch2/TRIP13. Genetic analysis in budding yeast using intervals across a range of chromosomes of different sizes has revealed an increased frequency of closely spaced double-CO events in the absence of Pch2 [28,31]. In a Trip13 hypomorphic mutant mouse, despite an overall reduction in MLH1 foci at pachytene, a small, yet significant, reduction in the mean inter-focus distance between pairs of foci was observed. This implies that although the COs remain subject to interference, there has been some weakening in its effect, although a subtle change in the positioning of the DSB complexes cannot be excluded [34]. Despite these observations recent evidence from budding yeast has found that inter-focus distance of Zip3 foci that mark future COs is not affected by loss of Pch2, indicating CO interference is normal [32]. Why the discrepancy? Zip3 foci are the earliest known marker of CO designation, appearing in late leptotene. However, maturation of designated intermediates to form COs is dependent on additional later events during the remainder of prophase I [5]. Other analyses of pch2 mutants have used genetic markers or MLH1 foci, which mark mature CO sites. Hence it is conceivable that while loss of Pch2/Trip13 affects the final CO patterning, CO designation occurs and hence interference is initially established. Analysis of Atpch2-1 is consistent with this possibility. The localization of HEI10 foci in wild type and Atpch2-1 at early prophase I was identical. Numerous small axis-associated foci were observed together with around 10 large (~250 nm) foci. At present, it is not technically possible to measure inter-focus distance at early prophase I in Arabidopsis, nevertheless inspection of the nuclei reveals these large HEI10 foci are usually spatially well separated. These are still observed at mid/late prophase I when a similar number of MLH1 foci, which mark interference sensitive CO sites, are also observed. By analogy with budding yeast and S. macrospora where the appearance of Zip3/Hei10 foci are indicative of CO designation in early prophase I, it seems likely this is also the case in Arabidopsis as the number of HEI10 foci at the leptotene/zygotene transition appeared normal in the absence of PCH2. However, the maturation of the CO designated intermediates is compromised by the defect in remodelling of the chromosomes axes in Atpch2-1, leading to a deficit in COs. Overall, our data imply that in Arabidopsis, as in budding yeast and S. macrospora, CO designation and interference arise, and are complete, during zygotene.
We noted that in both wild type and Atpch2-1 some MLH1 foci appeared adjacent to the ZYP1 signal rather than directly over it. This has not been previously recorded in Arabidopsis. It may be a consequence of the spreading procedure but it is worth noting that in this study, the anti-ZYP1 antibody was raised to the N-terminus of the protein which is predicted to mark the central region of the SC. This could suggest the MLH1 containing complexes are not in direct contact with the SC central region. However the basis and significance of this remains unclear.
Other features of the CO distribution in the mutants indicate that CO interference is established. A predicted outcome of CO interference is that the numerical distribution of interference-sensitive COs between nuclei does not fit a Poisson distribution, whereas the converse applies for non-interfering COs [56]. The distribution of COs in Atpch2-1 does not fit a Poisson distribution, suggesting that COs remain subject to spatial patterning and do not arise by the random maturation of a proportion of the recombination initiations into COs. Also, there was a strong tendency for any HEI10 or MLH1 foci that were found associated with stretches of SC in Atpch2-1 to occur as single foci.
Although the data indicate that CO designation occurs normally, it seems that precursor maturation to form CO products is perturbed in Atpch2. This is manifested in several ways. Most obviously, the mean chiasma frequency in Atpch2 mutants is ~7, a reduction of around 30% relative to wild type. This is accompanied by the presence of univalents at metaphase I at a frequency of ~10%. A global reduction in CO formation has also been reported in TRIP13/Pch2-deficient mice and PCH2-deficient C. elegans [34,37]. In budding yeast, an increase in COs has been reported for some genetic intervals whereas in others wild type levels were recorded [31]. Also, the distribution of MLH1 foci in the mouse Trip13 mutants suggests there are chromosomal regions which show an increase in CO frequency [34]. This could suggest variation between different species but it is worth noting that despite the global reduction in COs in the absence of PCH2 an increase in recombination frequency was observed in 3 out of 6 intervals (l2f, I2g and 15b) in the Arabidopsis FTL lines used in this study. However, this comes with the caveat that this approach scores only viable tetrads which could influence the analysis. Estimation of genetic CO interference using the FTL lines suggested its effect may be diminished in at least some chromosomal regions, since a reduction in strength was detected over regions of chromosome 2 and chromosome 5. This apparent contradiction with the cytological evidence can perhaps be reconciled by data from a study of CO patterning in S. macrospora applying the beam-film model to experimental data. This showed that under some circumstances a normal interference signal is established and remains, yet CO interference as measured using CoC as a metric appears to be reduced [7,72].
An altered pattern of COs combined with a synaptic defect and a reduction in genetic interference has been reported in a kinesin mutant, Atpss1, and Ataxr1, a mutant in the E1 enzyme Arabidopsis neddylation complex [55,75]. Both mutants are strongly defective in synapsis with univalents observed at metaphase I. In each case HEI10 and MLH1 foci are observed in late prophase I in approximately wild type numbers but, in contrast to Atpch2-1, often clustered along the limited stretches of SC that have formed. An effect on the distribution of MLH1 foci has also been reported in as1, an asynaptic mutant of tomato [76]. The genetic basis of the as1 mutation is unknown but it is associated with changes in compaction of the chromosome axes. Relative to wild type, the average SC length in as1 was reduced by 81% with MLH1 inter-focus distance decreased by 71%. However the median number of MLH1 foci was unchanged, although the range was more variable. It is hypothesized that the tendency of these plant mutants to maintain CO numbers may reflect a homeostatic mechanism [76]. This is not so obvious in Atpch2-1 but it was notable that the mean reduction in CO frequency (~30%) was not-coordinate with that in SC length (~68%).
This study demonstrates that in the absence of PCH2, remodelling of the chromosome axis at zygotene and the normal patterned maturation of CO designated intermediates in Arabidopsis are aberrant. This further emphasises the functional inter-relationship between the chromosome axis and the controlled formation of COs.
Materials and Methods
Plant material and nucleic acid extraction
A. thaliana ecotype Columbia (0) was used for wild type analysis. T-DNA insertion lines Atpch2-1: SAIL_1187_C06, Atpch2-2: SALK_031449 and Atpch2-3: SALK_130138 were obtained from NASC for mutant analysis. Plants were grown, material harvested and nucleic acid extractions were performed as previously described by Higgins et al. [43].
Proteomic analysis of Brassica PMCs
AtPCH2 peptides were identified by mass spectrometry in protein extracts from Brassica oleracea var. alboglabra A12DHd PMCs following co-immunoprecipitation with affinity purified anti-ASY1 antibody as previously described [40].
T-DNA insertion site mapping
The T-DNA insertion site of the mutant lines was confirmed as previously described [43]. Details of the primers used are presented in S3 Table.
RNA extraction and RT-PCR
RNA extraction and RT-PCR was carried out as previously described [43]. Details of the primers are given in S3 Table.
Nucleic acid sequencing
Nucleotide sequencing was carried out by the Genomics and Proteomics Unit, School of Biosciences, University of Birmingham, UK.
Antibody production
An anti-PCH2 antibody was raised in rabbit against a 15-residue peptide from the C-terminus of Arabidopsis PCH2 (Abmart Inc., Shanghai, China). Due to the high level of sequence identity between the PCH2 proteins in Arabidopsis and Brassica the antibody was also effective for immunolocalization in Brassica.
Cytological procedures
Cytological studies were carried out as previously described [43]. The following antibodies were used: anti-AtPCH2 (rat 1/200 dilution), anti-AtASY3 (rabbit, 1/200 dilution) [19], anti-AtASY1 (rabbit/rat, 1/1000 dilution) [45], anti-AtMSH4 (rabbit, 1/500 dilution) [43], anti-AtZYP1 (N-terminus Ab aa residues 1–415; C-terminus Ab aa residues 422–845; rabbit/rat, 1/500 dilution), anti-AtRAD51 (rabbit 1/500 dilution), anti-AtSYN1 (rabbit 1/500 dilution), anti-AtDMC1 (rabbit 1/500 dilution) [20,23], anti-AtMLH1 (rabbit/rat, 1/200 dilution) [64], anti-AtHEI10 (rabbit 1/500 dilution) and anti-γH2AX (ser 139, catalog no. 07–164 Upstate Biotechnology; rabbit, 1/100 dilution). Microscopy was carried out using a Nikon 90i Fluorescence Microscope (Tokyo, Japan). Image capture, image analysis and processing were conducted using NIS-Elements-F software (Nikon, Tokyo, Japan) as previously described [19]. Image deconvolution was carried out using the function “Mexican hat”. This allows better discrimination of the signals. This function performs filtration on the intensity component (or on every selected component—when working with multichannel images) of an image using convolution with 5x5 kernel. Mexican Hat kernel is defined as a combination of Laplacian kernel and Gaussian kernel it marks edges and also reduces noise. SIM was carried out using the OMX facility at the University of Dundee (http://microscopy.lifesci.dundee.ac.uk/omx/).
In Arabidopsis, ASY1 intensity analysis was conducted on chromosome spread preparations stained with anti-ASY1 antibody (rat, 1 in 5000 dilution) and anti-ZYP1 (rabbit, 1 in 500 dilution). 5μl of 6μm, 0.3% relative intensity InSpeck Red microspheres (Life Technologies), were added to slides before coverslips. PMCs and microspheres were imaged using specific exposure times. Randomly selected, non-overlapping sections of axis, ~2–4μm in length, were defined as regions of interest and were analysed for mean signal intensity using Nikon NIS-elements software. Intensities were normalised based on mean intensity of the microspheres. Intensity raw data is shown in grey-scale values. For B. oleracea, ASY1 intensity was determined in on PMC chromosome spreads at zygotene comparing non-overlapping segments of unsynapsed and synapsed sections of axis ~2–4μm in length.
Chiasma counts were carried out as previously described [42]. Chromosome spread preparations from PMCs at metaphase I were examined by light microscopy after fluorescence in situ hybridization (FISH) using 45S and 5S rDNA probes. The use of FISH enabled the identification of individual chromosomes. The overall shape of individual bivalents allowed the number and position of individual chiasmata to be determined and this was also informed by the position of the FISH signals.
The time course of progress through prophase I in wild type and Atpch2-1 was determined as previously described [48] except that 5-ethynyl-2’-deoxyuridine (EdU) was used to label the PMCs which were analyzed at 5 h intervals from 0–30 h and at 2 h intervals thereafter up to 36 h.
Fluorescent tetrad analysis was carried out as described Berchowitz and Copenhaver [49] using genetic intervals I2f and I2g on chromosome 2 (FTL coordinates for the I2fg interval: FTL#800 18286716 bp DsRed2; FTL#3411 18957093 bp YFP; FTL#3263 19373634 bp AmCyan) and I5a, I5b, I5c, and I5d on chromosome 5 as described [49]. Pollen was scored through eCFP, eYFP and DsRed2 filters using an Olympus BX-61 epifluorescence microscope. The Stahl Lab Online Tools (http://molbio.uoregon.edu/~fstahl/) was used for statistical analyses of the data.
Statistical procedures
The statistical procedures were carried out as described previously [43]. Chi-squared (Χ2) tests were used to determine agreement between the observed chiasma counts and those expected from a Poisson distribution. Numbers of foci in wild type and mutant PMCs were compared using the Wilcoxon signed-rank test. Mean intensities between synapsed and unsynapsed sections of axis/SC were analysed using a 2 tail paired T-test.
Supporting Information
Zdroje
1. Zickler D, Kleckner N (1998) The leptotene-zygotene transition of meiosis. Annual Review of Genetics 32 : 619–697. 9928494
2. Keeney S, Giroux CN, Kleckner N (1997) Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell 88 : 375–384. 9039264
3. Bergerat A, deMassy B, Gadelle D, Varoutas PC, Nicolas A, et al. (1997) An atypical topoisomerase II from archaea with implications for meiotic recombination. Nature 386 : 414–417. 9121560
4. Mancera E, Bourgon R, Brozzi A, Huber W, Steinmetz LM (2008) High-resolution mapping of meiotic crossovers and non-crossovers in yeast. Nature 454 : 479–U471. doi: 10.1038/nature07135 18615017
5. Osman K, Higgins JD, Sanchez-Moran E, Armstrong SJ, Franklin FCH (2011) Pathways to meiotic recombination in Arabidopsis thaliana. New Phytologist 190 : 523–544. doi: 10.1111/j.1469-8137.2011.03665.x 21366595
6. Jones GH, Franklin FC (2006) Meiotic crossing-over: obligation and interference. Cell 126 : 246–248. 16873056
7. Wang S, Zickler D, Kleckner N, Zhang L (2015) Meiotic crossover patterns: Obligatory crossover, interference and homeostasis in a single process. Cell cycle (Georgetown, Tex) 14 : 305–314.
8. Copenhaver GP, Housworth EA, Stahl FW (2002) Crossover interference in Arabidopsis. Genetics 160 : 1631–1639. 11973316
9. Shinohara M, Oh SD, Hunter N, Shinohara A (2008) Crossover assurance and crossover interference are distinctly regulated by the ZMM proteins during yeast meiosis. Nature Genetics 40 : 299–309. doi: 10.1038/ng.83 18297071
10. Sturtevant AH (1915) The behavior of chromosomes as studied through linkage. Z Induct Abstammungs-Vererbungsl, 13 : 234–287.
11. Kleckner N, Zickler D, Jones GH, Dekker J, Padmore R, et al. (2004) A mechanical basis for chromosome function. Proceedings of the National Academy of Sciences of the United States of America 101 : 12592–12597. 15299144
12. Zhang L, Liang Z, Hutchinson J, Kleckner N (2014) Crossover Patterning by the Beam-Film Model: Analysis and Implications. Plos Genetics 10.
13. Kleckner N (2006) Chiasma formation: chromatin/axis interplay and the role(s) of the synaptonemal complex. Chromosoma 115 : 175–194. 16555016
14. Blat Y, Protacio RU, Hunter N, Kleckner N (2002) Physical and Functional Interactions among Basic Chromosome Organizational Features Govern Early Steps of Meiotic Chiasma Formation. Cell 111 : 791–802. 12526806
15. Panizza S, Mendoza MA, Berlinger M, Huang L, Nicolas A, et al. (2011) Spo11-Accessory Proteins Link Double-Strand Break Sites to the Chromosome Axis in Early Meiotic Recombination. Cell 146 : 372–383. doi: 10.1016/j.cell.2011.07.003 21816273
16. Page SL, Hawley RS (2004) The genetics and molecular biology of the synaptonemal complex. Annual Review of Cell and Developmental Biology 20 : 525–558. 15473851
17. Fung JC, Rockmill B, Odell M, Roeder GS (2004) Imposition of crossover interference through the nonrandom distribution of synapsis initiation complexes. Cell 116 : 795–802. 15035982
18. Boerner GV, Kleckner N, Hunter N (2004) Crossover/noncrossover differentiation, synaptonemal complex formation, and regulatory surveillance at the leptotene/zygotene transition of meiosis. Cell 117 : 29–45. 15066280
19. Ferdous M, Higgins JD, Osman K, Lambing C, Roitinger E, et al. (2012) Inter-Homolog Crossing-Over and Synapsis in Arabidopsis Meiosis Are Dependent on the Chromosome Axis Protein AtASY3. Plos Genetics 8.
20. Higgins JD, Sanchez-Moran E, Armstrong SJ, Jones GH, Franklin FCH (2005) The Arabidopsis synaptonemal complex protein ZYP1 is required for chromosome synapsis and normal fidelity of crossing over. Genes Dev 19 : 2488–2500. 16230536
21. Kim KP, Weiner BM, Zhang LR, Jordan A, Dekker J, et al. (2010) Sister Cohesion and Structural Axis Components Mediate Homolog Bias of Meiotic Recombination. Cell 143 : 924–937. doi: 10.1016/j.cell.2010.11.015 21145459
22. Mao-Draayer Y, Galbraith AM, Pittman DL, Cool M, Malone RE (1996) Analysis of meiotic recombination pathways in the yeast Saccharomyces cerevisiae. Genetics 144 : 71–86. 8878674
23. Sanchez-Moran E, Santos JL, Jones GH, Franklin FCH (2007) ASY1 mediates AtDMC1-dependent interhomolog recombination during meiosis in Arabidopsis. Genes & Development 21 : 2220–2233.
24. Schwacha A, Kleckner N (1997) Interhomolog bias during meiotic recombination: Meiotic functions promote a highly differentiated interhomolog-only pathway. Cell 90 : 1123–1135. 9323140
25. Xu LH, Weiner BM, Kleckner N (1997) Meiotic cells monitor the status of the interhomolog recombination complex. Genes & Development 11 : 106–118.
26. Storlazzi A, Tesse S, Ruprich-Robert G, Gargano S, Poeggeler S, et al. (2008) Coupling meiotic chromosome axis integrity to recombination. Genes & Development 22 : 796–809.
27. Boerner GV, Barot A, Kleckner N (2008) Yeast Pch2 promotes domainal axis organization, timely recombination progression, and arrest of defective recombinosomes during meiosis. Proceedings of the National Academy of Sciences of the United States of America 105 : 3327–3332. doi: 10.1073/pnas.0711864105 18305165
28. Joshi N, Barot A, Jamison C, Boerner GV (2009) Pch2 Links Chromosome Axis Remodeling at Future Crossover Sites and Crossover Distribution during Yeast Meiosis. Plos Genetics 5.
29. Zanders S, Brown MS, Chen C, Alani E (2011) Pch2 Modulates Chromatid Partner Choice During Meiotic Double-Strand Break Repair in Saccharomyces cerevisiae. Genetics 188 : 511–521. doi: 10.1534/genetics.111.129031 21515575
30. Ho H-C, Burgess SM (2011) Pch2 Acts through Xrs2 and Tel1/ATM to Modulate Interhomolog Bias and Checkpoint Function during Meiosis. Plos Genetics 7.
31. Zanders S, Alani E (2009) The pch2 Delta Mutation in Baker's Yeast Alters Meiotic Crossover Levels and Confers a Defect in Crossover Interference. Plos Genetics 5.
32. Zhang L, Wang S, Yin S, Hong S, Kim KP, et al. (2014) Topoisomerase II mediates meiotic crossover interference. Nature 511 : 551–556. doi: 10.1038/nature13442 25043020
33. Li X, Schimenti JC (2007) Mouse pachytene checkpoint 2 (Trip13) is required for completing meiotic recombination but not Synapsis. Plos Genetics 3 : 1365–1376.
34. Roig I, Dowdle JA, Toth A, de Rooij DG, Jasin M, et al. (2010) Mouse TRIP13/PCH2 Is Required for Recombination and Normal Higher-Order Chromosome Structure during Meiosis. Plos Genetics 6.
35. Wojtasz L, Daniel K, Roig I, Bolcun-Filas E, Xu HL, et al. (2009) Mouse HORMAD1 and HORMAD2, Two Conserved Meiotic Chromosomal Proteins, Are Depleted from Synapsed Chromosome Axes with the Help of TRIP13 AAA-ATPase. Plos Genetics 5.
36. Joyce EF, McKim KS (2009) Drosophila PCH2 Is Required for a Pachytene Checkpoint That Monitors Double-Strand-Break-Independent Events Leading to Meiotic Crossover Formation. Genetics 181 : 39–51. doi: 10.1534/genetics.108.093112 18957704
37. Deshong AJ, Ye AL, Lamelza P, Bhalla N (2014) A Quality Control Mechanism Coordinates Meiotic Prophase Events to Promote Crossover Assurance. Plos Genetics 10.
38. Miao C, Tang D, Zhang H, Wang M, Li Y, et al. (2013) CENTRAL REGION COMPONENT1, a Novel Synaptonemal Complex Component, Is Essential for Meiotic Recombination Initiation in Rice. Plant Cell 25 : 2998–3009. doi: 10.1105/tpc.113.113175 23943860
39. Nonomura KI, Nakano M, Murata K, Miyoshi K, Eiguchi M, et al. (2004) An insertional mutation in the rice PAIR2 gene, the ortholog of Arabidopsis ASY1, results in a defect in homologous chromosome pairing during meiosis. Molecular Genetics and Genomics 271 : 121–129. 14758540
40. Osman K, Roitinger E, Yang J, Armstrong S, Mechtler K, et al. (2013) Analysis of meiotic protein complexes from Arabidopsis and Brassica using affinity-based proteomics. Methods in molecular biology (Clifton, NJ) 990 : 215–226.
41. Wang X, Wang H, Wang J, Sun R, Wu J, et al. (2011) The genome of the mesopolyploid crop species Brassica rapa. Nature Genetics 43 : 1035–U1157. doi: 10.1038/ng.919 21873998
42. Sanchez-Moran E, Armstrong SJ, Santos JL, Franklin FCH, Jones GH (2002) Variation in chiasma frequency among eight accessions of Arabidopsis thaliana. Genetics 162 : 1415–1422. 12454084
43. Higgins JD, Armstrong SJ, Franklin FCH, Jones GH (2004) The Arabidopsis MutS homolog AtMSH4 functions at an early step in recombination: evidence for two classes of recombination in Arabidopsis. Genes & Development 18 : 2557–2570.
44. Higgins JD, Buckling EF, Franklin FCH, Jones GH (2008) Expression and functional analysis of AtMUS81 in Arabidopsis meiosis reveals a role in the second pathway of crossing-over. Plant Journal 54 : 152–162. doi: 10.1111/j.1365-313X.2008.03403.x 18182028
45. Armstrong SJ, Caryl AP, Jones GH, Franklin FC (2002) Asy1, a protein required for meiotic chromosome synapsis, localizes to axis-associated chromatin in Arabidopsis and Brassica. J Cell Sci 115 : 3645–3655. 12186950
46. Bai XF, Peirson BN, Dong FG, Xue C, Makaroff CA (1999) Isolation and characterization of SYN1, a RAD21-like gene essential for meiosis in Arabidopsis. Plant Cell 11 : 417–430. 10072401
47. Cai X, Dong FG, Edelmann RE, Makaroff CA (2003) The Arabidopsis SYN1 cohesin protein is required for sister chromatid arm cohesion and homologous chromosome pairing. Journal of Cell Science 116 : 2999–3007. 12783989
48. Armstrong SJ, Franklin FCH, Jones GH (2003) A meiotic time-course for Arabidopsis thaliana. Sexual Plant Reproduction 16 : 141–149.
49. Berchowitz LE, Copenhaver GP (2008) Fluorescent Arabidopsis tetrads: a visual assay for quickly developing large crossover and crossover interference data sets. Nature Protocols 3 : 41–50. doi: 10.1038/nprot.2007.491 18193020
50. Francis KE, Lam SY, Harrison BD, Bey AL, Berchowitz LE, et al. (2007) Pollen tetrad-based visual assay for meiotic recombination in Arabidopsis. Proceedings of the National Academy of Sciences of the United States of America 104 : 3913–3918. 17360452
51. Francis KE, Lam SY, Copenhaver GP (2006) Separation of Arabidopsis pollen tetrads is regulated by QUARTET1, a pectin methylesterase gene. Plant Physiology 142 : 1004–1013. 16980565
52. Preuss D, Rhee SY, Davis RW (1994) Tetrad analysis is possible in Arabidopsis with mutation in the QUARTET (QRT) genes. Science 264 : 1458–1460. 8197459
53. Perkins DD (1949) Biochemical mutants in the smut fungus Ustilago maydis Genetics 34 : 607–626. 17247336
54. Malkova A, Swanson J, German M, McCusker JH, Housworth EA, et al. (2004) Gene conversion and crossing over along the 405-kb left arm of Saccharomyces cerevisiae chromosome VII. Genetics 168 : 49–63. 15454526
55. Jahns MT, Vezon D, Chambon A, Pereira L, Falque M, et al. (2014) Crossover Localisation Is Regulated by the Neddylation Posttranslational Regulatory Pathway. Plos Biology 12.
56. Jones G (1987) Chiasmata. Meiosis: 213–244.
57. Chen CB, Zhang W, Timofejeva L, Gerardin Y, Ma H (2005) The Arabidopsis ROCK-N-ROLLERS gene encodes a homolog of the yeast ATP-dependent DNA helicase MER3 and is required for normal meiotic crossover formation. Plant Journal 43 : 321–334. 16045469
58. Mercier R, Jolivet S, Vezon D, Huppe E, Chelysheva L, et al. (2005) Two meiotic crossover classes cohabit in Arabidopsis: One is dependent on MER3, whereas the other one is not. Current Biology 15 : 692–701. 15854901
59. Klimyuk VI, Jones JDG (1997) AtDMC1, the Arabidopsis homologue of the yeast DMC1 gene: Characterization, transposon-induced allelic variation and meiosis-associated expression. Plant Journal 11 : 1–14. 9025299
60. Li WX, Chen CB, Markmann-Mulisch U, Timofejeva L, Schmelzer E, et al. (2004) The Arabidopsis AtRAD51 gene is dispensable for vegetative development but required for meiosis. Proceedings of the National Academy of Sciences of the United States of America 101 : 10596–10601. 15249667
61. Chelysheva L, Vezon D, Chambon A, Gendrot G, Pereira L, et al. (2012) The Arabidopsis HEI10 Is a New ZMM Protein Related to Zip3. Plos Genetics 8.
62. Wang K, Wang M, Tang D, Shen Y, Miao C, et al. (2012) The Role of Rice HEI10 in the Formation of Meiotic Crossovers. Plos Genetics 8.
63. De Muyt A, Zhang L, Piolot T, Kleckner N, Espagne E, et al. (2014) E3 ligase Hei10: a multifaceted structure-based signaling molecule with roles within and beyond meiosis. Genes & Development 28 : 1111–1123.
64. Jackson N, Sanchez-Moran E, Buckling E, Armstrong SJ, Jones GH, et al. (2006) Reduced meiotic crossovers and delayed prophase I progression in AtMLH3-deficient Arabidopsis. Embo Journal 25 : 1315–1323. 16467846
65. Barakate A, Higgins JD, Vivera S, Stephens J, Perry RM, et al. (2014) The synaptonemal complex protein ZYP1 is required for imposition of meiotic crossovers in barley. The Plant Cell.
66. Wang M, Wang KJ, Tang D, Wei CX, Li M, et al. (2010) The Central Element Protein ZEP1 of the Synaptonemal Complex Regulates the Number of Crossovers during Meiosis in Rice. Plant Cell 22 : 417–430. doi: 10.1105/tpc.109.070789 20154151
67. Chen C, Jomaa A, Ortega J, Alani EE (2014) Pch2 is a hexameric ring ATPase that remodels the chromosome axis protein Hop1. Proceedings of the National Academy of Sciences of the United States of America 111: E44–E53. doi: 10.1073/pnas.1310755111 24367111
68. Hwang LC, Vecchiarelli AG, Han Y - W, Mizuuchi M, Harada Y, et al. (2013) ParA-mediated plasmid partition driven by protein pattern self-organization. Embo Journal 32 : 1238–1249. doi: 10.1038/emboj.2013.34 23443047
69. Ye Q, Rosenberg, S.C., Moeller, A., Speir, J.A., Su, T.Y., Corbett, K.D. (2015) TRIP13 is a protein-remodelling AAA+ATPase that catalyses MAD2 confromation switching. eLife doi: 10.7554/eLife.07367
70. Nilsson NO, Sall T, Bengtsson BO (1993) Chiasma and recombination data in plants-are they compatible? Trends in Genetics 9 : 344–348. 8068067
71. Wills AB (1966) Meiotic behaviour in Brassiceae. Caryologia 19 : 103–&.
72. Zhang L, Espagne E, de Muyt A, Zickler D, Kleckner NE (2014) Interference-mediated synaptonemal complex formation with embedded crossover designation. Proceedings of the National Academy of Sciences of the United States of America 111: E5059–E5068. doi: 10.1073/pnas.1416411111 25380597
73. Henderson KA, Keeney S (2004) Tying synaptonemal complex initiation to the formation and programmed repair of DNA double-strand breaks. Proceedings of the National Academy of Sciences of the United States of America 101 : 4519–4524. 15070750
74. Farmer S, Hong E-JE, Leung W-K, Argunhan B, Terentyev Y, et al. (2012) Budding Yeast Pch2, a Widely Conserved Meiotic Protein, Is Involved in the Initiation of Meiotic Recombination. Plos One 7.
75. Duroc Y, Lemhemdi A, Larcheveque C, Hurel A, Cuacos M, et al. (2014) The Kinesin AtPSS1 Promotes Synapsis and is Required for Proper Crossover Distribution in Meiosis. Plos Genetics 10.
76. Qiao H, Offenberg HH, Anderson LK (2012) Altered distribution of MLH1 foci is associated with changes in cohesins and chromosome axis compaction in an asynaptic mutant of tomato. Chromosoma 121 : 291–305. doi: 10.1007/s00412-012-0363-z 22350750
Štítky
Genetika Reprodukční medicína
Článek Discovery and Fine-Mapping of Glycaemic and Obesity-Related Trait Loci Using High-Density ImputationČlánek AAA-ATPase FIDGETIN-LIKE 1 and Helicase FANCM Antagonize Meiotic Crossovers by Distinct MechanismsČlánek A Conserved Pattern of Primer-Dependent Transcription Initiation in and Revealed by 5′ RNA-seqČlánek TopBP1 Governs Hematopoietic Stem/Progenitor Cells Survival in Zebrafish Definitive HematopoiesisČlánek Redundant Roles of Rpn10 and Rpn13 in Recognition of Ubiquitinated Proteins and Cellular Homeostasis
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2015 Číslo 7
-
Všechny články tohoto čísla
- LINE-1 Retroelements Get ZAPped!
- /p23: A Small Protein Heating Up Lifespan Regulation
- Hairless Streaks in Cattle Implicate TSR2 in Early Hair Follicle Formation
- Ribosomal Protein Mutations Result in Constitutive p53 Protein Degradation through Impairment of the AKT Pathway
- Molecular Clock of Neutral Mutations in a Fitness-Increasing Evolutionary Process
- Modeling Implicates in Nephropathy: Evidence for Dominant Negative Effects and Epistasis under Anemic Stress
- The Alternative Sigma Factor SigX Controls Bacteriocin Synthesis and Competence, the Two Quorum Sensing Regulated Traits in
- BMP Inhibition in Seminomas Initiates Acquisition of Pluripotency via NODAL Signaling Resulting in Reprogramming to an Embryonal Carcinoma
- Comparative Study of Regulatory Circuits in Two Sea Urchin Species Reveals Tight Control of Timing and High Conservation of Expression Dynamics
- EIN3 and ORE1 Accelerate Degreening during Ethylene-Mediated Leaf Senescence by Directly Activating Chlorophyll Catabolic Genes in
- Genome Wide Binding Site Analysis Reveals Transcriptional Coactivation of Cytokinin-Responsive Genes by DELLA Proteins
- Sensory Neurons Arouse . Locomotion via Both Glutamate and Neuropeptide Release
- A Year of Infection in the Intensive Care Unit: Prospective Whole Genome Sequencing of Bacterial Clinical Isolates Reveals Cryptic Transmissions and Novel Microbiota
- Inference of Low and High-Grade Glioma Gene Regulatory Networks Delineates the Role of Rnd3 in Establishing Multiple Hallmarks of Cancer
- Novel Role for p110β PI 3-Kinase in Male Fertility through Regulation of Androgen Receptor Activity in Sertoli Cells
- A Novel Locus Harbouring a Functional Nonsense Mutation Identified in a Large Danish Family with Nonsyndromic Hearing Impairment
- Checkpoint Activation of an Unconventional DNA Replication Program in
- A Genetic Incompatibility Accelerates Adaptation in Yeast
- The SMC Loader Scc2 Promotes ncRNA Biogenesis and Translational Fidelity
- Blimp1/Prdm1 Functions in Opposition to Irf1 to Maintain Neonatal Tolerance during Postnatal Intestinal Maturation
- Discovery and Fine-Mapping of Glycaemic and Obesity-Related Trait Loci Using High-Density Imputation
- JAK/STAT and Hox Dynamic Interactions in an Organogenetic Gene Cascade
- Emergence, Retention and Selection: A Trilogy of Origination for Functional Proteins from Ancestral LncRNAs in Primates
- MoSET1 (Histone H3K4 Methyltransferase in ) Regulates Global Gene Expression during Infection-Related Morphogenesis
- Arabidopsis PCH2 Mediates Meiotic Chromosome Remodeling and Maturation of Crossovers
- AAA-ATPase FIDGETIN-LIKE 1 and Helicase FANCM Antagonize Meiotic Crossovers by Distinct Mechanisms
- A Conserved Pattern of Primer-Dependent Transcription Initiation in and Revealed by 5′ RNA-seq
- Tempo and Mode of Transposable Element Activity in Drosophila
- The Shelterin TIN2 Subunit Mediates Recruitment of Telomerase to Telomeres
- SAMHD1 Inhibits LINE-1 Retrotransposition by Promoting Stress Granule Formation
- A Genome Scan for Genes Underlying Microgeographic-Scale Local Adaptation in a Wild Species
- TopBP1 Governs Hematopoietic Stem/Progenitor Cells Survival in Zebrafish Definitive Hematopoiesis
- Analysis of the Relationships between DNA Double-Strand Breaks, Synaptonemal Complex and Crossovers Using the Mutant
- Assessing Mitochondrial DNA Variation and Copy Number in Lymphocytes of ~2,000 Sardinians Using Tailored Sequencing Analysis Tools
- Allelic Spectra of Risk SNPs Are Different for Environment/Lifestyle Dependent versus Independent Diseases
- CSB-PGBD3 Mutations Cause Premature Ovarian Failure
- Irrepressible: An Interview with Mark Ptashne
- Genetic Evidence for Function of the bHLH-PAS Protein Gce/Met As a Juvenile Hormone Receptor
- Inactivation of Retinoblastoma Protein (Rb1) in the Oocyte: Evidence That Dysregulated Follicle Growth Drives Ovarian Teratoma Formation in Mice
- Redundant Roles of Rpn10 and Rpn13 in Recognition of Ubiquitinated Proteins and Cellular Homeostasis
- Pyrimidine Pool Disequilibrium Induced by a Cytidine Deaminase Deficiency Inhibits PARP-1 Activity, Leading to the Under Replication of DNA
- Molecular Framework of a Regulatory Circuit Initiating Two-Dimensional Spatial Patterning of Stomatal Lineage
- RFX2 Is a Major Transcriptional Regulator of Spermiogenesis
- A Role for Macro-ER-Phagy in ER Quality Control
- Corp Regulates P53 in via a Negative Feedback Loop
- Common Cell Shape Evolution of Two Nasopharyngeal Pathogens
- Contact- and Protein Transfer-Dependent Stimulation of Assembly of the Gliding Motility Machinery in
- Endothelial Snail Regulates Capillary Branching Morphogenesis via Vascular Endothelial Growth Factor Receptor 3 Expression
- Functional Constraint Profiling of a Viral Protein Reveals Discordance of Evolutionary Conservation and Functionality
- Temporal Coordination of Carbohydrate Metabolism during Mosquito Reproduction
- mTOR Directs Breast Morphogenesis through the PKC-alpha-Rac1 Signaling Axis
- Reversible Oxidation of a Conserved Methionine in the Nuclear Export Sequence Determines Subcellular Distribution and Activity of the Fungal Nitrate Regulator NirA
- Nutritional Control of DNA Replication Initiation through the Proteolysis and Regulated Translation of DnaA
- Cooperation between Paxillin-like Protein Pxl1 and Glucan Synthase Bgs1 Is Essential for Actomyosin Ring Stability and Septum Formation in Fission Yeast
- Encodes a Highly Conserved Protein Important to Neurological Function in Mice and Flies
- Identification of a Novel Regulatory Mechanism of Nutrient Transport Controlled by TORC1-Npr1-Amu1/Par32
- Aurora-A-Dependent Control of TACC3 Influences the Rate of Mitotic Spindle Assembly
- Large-Scale Phenomics Identifies Primary and Fine-Tuning Roles for CRKs in Responses Related to Oxidative Stress
- TFIIS-Dependent Non-coding Transcription Regulates Developmental Genome Rearrangements
- Genome-Wide Reprogramming of Transcript Architecture by Temperature Specifies the Developmental States of the Human Pathogen
- Identification of Chemical Inhibitors of β-Catenin-Driven Liver Tumorigenesis in Zebrafish
- The Catalytic and Non-catalytic Functions of the Chromatin-Remodeling Protein Collaborate to Fine-Tune Circadian Transcription in
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Functional Constraint Profiling of a Viral Protein Reveals Discordance of Evolutionary Conservation and Functionality
- Reversible Oxidation of a Conserved Methionine in the Nuclear Export Sequence Determines Subcellular Distribution and Activity of the Fungal Nitrate Regulator NirA
- Modeling Implicates in Nephropathy: Evidence for Dominant Negative Effects and Epistasis under Anemic Stress
- Nutritional Control of DNA Replication Initiation through the Proteolysis and Regulated Translation of DnaA
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



