-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaRFX2 Is a Major Transcriptional Regulator of Spermiogenesis
Failure of spermatogenesis, which is presumed to often result from genetic defects, is a common cause of male sterility. Although numerous genes associated with defects in male spermatogenesis have been identified, numerous cases of genetic male infertility remain unelucidated. We report here that the transcription factor RFX2 is a master regulator of gene expression programs required for progression through the haploid phase of spermatogenesis. Male RFX2-deficient mice are completely sterile. Spermatogenesis progresses through meiosis, but haploid cells undergo a complete block in development just prior to spermatid elongation. Gene expression profiling and ChIP-Seq analysis revealed that RFX2 controls key pathways implicated in cilium/flagellum formation, as well as genes implicated in microtubule and vesicle associated transport. The set of genes activated by RFX2 in spermatids exhibits virtually no overlap with those controlled by other known transcriptional regulators of spermiogenesis, establishing RFX2 as an essential new player in this developmental process. RFX2-deficient mice should therefore represent a valuable new model for deciphering the regulatory networks that direct sperm formation, and thereby contribute to the identification of causes of human male infertility.
Published in the journal: . PLoS Genet 11(7): e32767. doi:10.1371/journal.pgen.1005368
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1005368Summary
Failure of spermatogenesis, which is presumed to often result from genetic defects, is a common cause of male sterility. Although numerous genes associated with defects in male spermatogenesis have been identified, numerous cases of genetic male infertility remain unelucidated. We report here that the transcription factor RFX2 is a master regulator of gene expression programs required for progression through the haploid phase of spermatogenesis. Male RFX2-deficient mice are completely sterile. Spermatogenesis progresses through meiosis, but haploid cells undergo a complete block in development just prior to spermatid elongation. Gene expression profiling and ChIP-Seq analysis revealed that RFX2 controls key pathways implicated in cilium/flagellum formation, as well as genes implicated in microtubule and vesicle associated transport. The set of genes activated by RFX2 in spermatids exhibits virtually no overlap with those controlled by other known transcriptional regulators of spermiogenesis, establishing RFX2 as an essential new player in this developmental process. RFX2-deficient mice should therefore represent a valuable new model for deciphering the regulatory networks that direct sperm formation, and thereby contribute to the identification of causes of human male infertility.
Introduction
Reproductive failure affects 10–15% of couples worldwide, with responsibility distributed about equally between males and females [1,2]. Failure of spermatogenesis is a common cause of male infertility. A large fraction of such cases is believed to result from genetic causes. The advent of forward genetics has led to the identification of over 400 genes associated with male spermatogenic defects [1]. However, the precise etiology of most clinical cases of male infertility remains unknown.
The process of sperm production in the testis is usually described in terms of three phases: the multiplication of diploid spermatogonia, the reduction of chromosome number during meiosis in spermatocytes, and the morphological conversion of round, immotile haploid spermatids into nearly mature sperm by the process of spermiogenesis. The overall process is controlled by master external regulators, including retinoic acid, the pituitary gonadotropins, and testosterone [3]. In contrast, details of the process depend in part on local biochemical communications with closely associated Sertoli cells, and a dynamic gene expression program within the germ cells that involves both transcriptional and post-transcriptional regulation [4,5].
The current study addresses the role of a member of the Regulatory Factor X (RFX) family of transcriptional regulators. Early studies led to description of the X-box as a DNA sequence motif conserved in the promoters of genes encoding MHC class II antigen-presenting proteins [6]. Search for the factor that activates MHC class II genes through this motif led progressively to the identification of a family of RFX transcription factors (TFs) that now numbers 8 members in mammals [7,8] and has its evolutionary origins traced to microorganisms. The most fundamental shared feature is a variant of the winged helix DNA binding domain [9]. In the invertebrate metazoan C. elegans there is single RFX relative (DAF-19), which controls genes important for cilia development and function [10]. In vertebrates several Rfx genes have also been shown to control ciliogenesis (for review see [11,12]). The family can be divided into two groups based on their ability to form homo and heterodimers: RFX 1–4, 6 and 8 are known or predicted to have this property, while RFX5 and 7 do not. This capacity to bind as heterodimers to the same DNA motif makes unambiguous assignment of functional roles to individual RFX proteins difficult when multiple members are present in the same cell.
Mutational analysis has established critical roles for most Rfx genes. Rfx1 deletion in mice was found to be embryonic lethal, in keeping with reports of a wide variety of potential roles [13]. Mouse Rfx3 is critical for cilia development and function, and its absence leads to a variety of severe defects in left/right body patterning, CNS development and differentiation of endocrine cells in the pancreas [14–16]. Disruption of mouse Rfx4 causes failure in the development of dorsal midline brain structures. It is also critical for formation of the subcommissural organ [17–19]. Rfx5 was the object of the original search for the X box regulatory factor, and is strictly required for MHC class II gene expression in humans and mice [20]. Genetic ablation of Rfx6 leads to failure of pancreatic islet development and diabetes in zebra fish and humans [21,22]. Rfx7 deletion was recently shown to affect ciliogenesis in the developing neural tube in Xenopus, where it functions developmentally upstream of Rfx4 [23]. Rfx8 has only been identified by genomic similarity, and functional studies have not been reported (http://www.uniprot.org/uniprot/D3YU81).
In non-mammalian vertebrates Rfx2 is crucial for the differentiation of cells carrying motile cilia and for the development of left/right asymmetry [24–27]. In contrast, the function for Rfx2 in mammals is not well established. Initial expression profiles established in mice demonstrated extremely high transcript levels in testis compared to other organs [28]. This was amply confirmed by multiple genome wide studies (http://germonline.org/). Developmental studies revealed an initial increase in Rfx2 mRNA during the meiotic phase of spermatogenesis [29–31]. This was confirmed at the protein level by immunohistology, showing that RFX2 is restricted to the germ line and not detected in Sertoli cells [32]. RFX2 has also been implicated in the upregulation of several genes in that period [33,34]. Regarding its own regulation, the Rfx2 promoter region contains multiple binding sites for the MYB family of transcriptional regulators [33], of which A-Myb (Mybl1) is critically required for spermatocytes to complete meiosis [35,36]. Because RFX2 expression is greatly reduced in A-Myb deficient mice, it was predicted that Rfx2 is controlled by A-MYB and in turn controls an unknown population of downstream genes important for spermatogenesis [37].
To test this last prediction we inactivated the Rfx2 gene in mice. Rfx2-/- mice develop normally and are healthy, but males are sterile whereas fertility of female mice is not affected. We observed that spermatogenesis proceeded essentially normally through meiosis, but that all spermatids failed to progress past the round cell stage and were shed from the germinal epithelium at approximately step 7 of development. We observed a general defect in Golgi organization and acrosome formation associated with complete failure of axoneme elongation. RNA-seq based transcription-profiling performed with Rfx2-deficient testes at P21 and P30 identified over 100 and 600 genes, respectively, that were downregulated greater than two-fold. ChIP-Seq analysis at P21 identified many of these as being direct RFX2 targets, with a substantial number of them sharing a developmental expression pattern similar to that of Rfx2. Interestingly, this large group of downregulated genes shares almost no overlap with genes regulated by other transcription factors required for spermiogenesis [38–40]. These results clearly place RFX2 among the major transcriptional regulators of the haploid phase of sperm formation.
Results
Generation of Rfx2-/- mice
A null allele was created for Rfx2 by flanking exon 7 with loxP sites, creating targeted ES cells and then chimeric mice that were subsequently bred to obtain germline transmission. Subsequent Cre-induced recombination resulted in the excision of exon 7 (see Materials and Methods and Fig 1). Removal of exon 7 deletes most of the coding region for the DNA binding domain, and is predicted to change the reading frame and introduce premature out-of-frame stop codons if either exon 5 or 6 is spliced to exon 8, 9 or 10. Interbreeding of heterozygous exon 7 deleted mice generated the three expected genotypes for both sexes in the expected Mendelian ratios. Exon 7 was shown to be absent by RT-PCR of testis RNA from Rfx2-/- mice (Fig 1E). RNA-Seq experiments (see below) confirmed the deletion as no reads were obtained for exon 7. RFX2 protein was undetectable by Western blotting (Fig 1E) and immunohistology (Fig 2A and 2B).
Fig. 1. Generation Rfx2-/- mice. 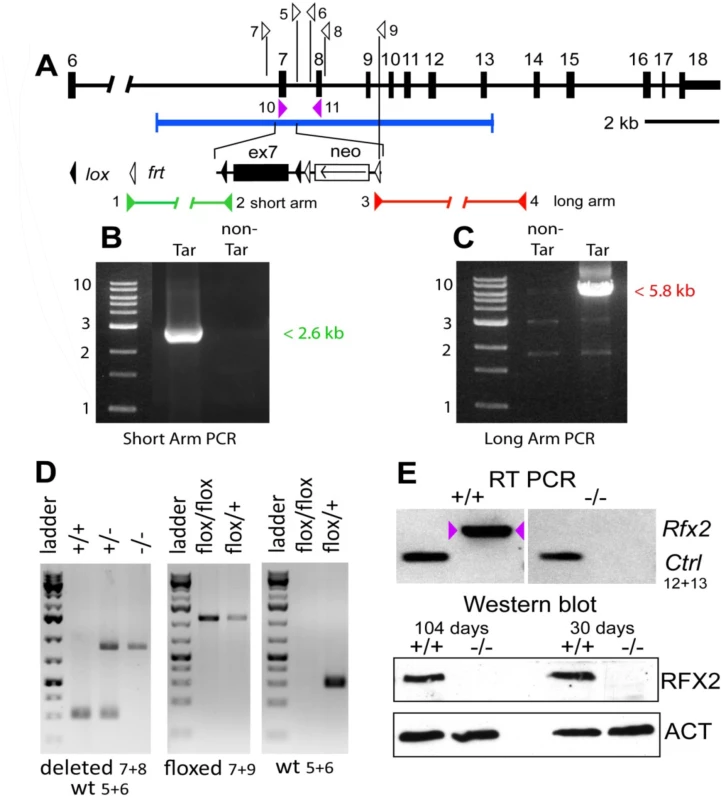
The mouse Rfx2 gene is depicted approximately to scale from exon 6 onward (A). Below is shown the region of homology of the targeting vector, with the details of the floxed exon 7 enlarged. Locations of PCR primers are indicated above and their sequences are provided in S5 Table. ES cell clones were tested for targeted insertions using primer sets with one member lying outside the region of homology either up (B) or downstream (C) of Rfx2 sequences present in the targeting vector. Mice were genotyped by tail biopsy using primer-pairs specific to the WT, floxed or deleted alleles (D). Reverse transcriptase PCR using testis cDNA, and a primer set anchored in exons 7 and 8, confirmed that exon 7 was absent in homozygous Rfx2-/- mice (E). Control amplification (Ctrl) detected a ubiquitously expressed component of the mitochondrial F1 ATP synthase complex (Atp5a1). Western blotting of testis protein extracts from mice of 30 or 104 days of age confirmed that full length RFX2 was not detected in homozygous Rfx2-/- mice (E). Actin (ACT) was used as control. Fig. 2. Spermatogenesis is arrested in Rfx2-/- mice. 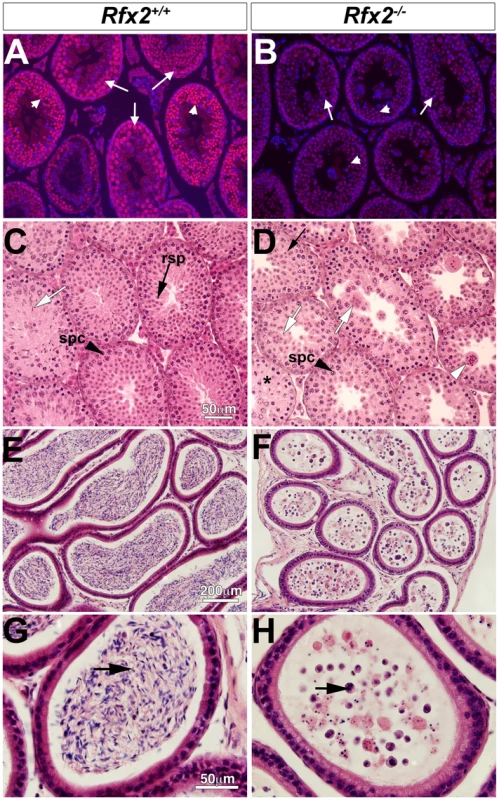
(A-B) Testis sections from Rfx2-/- and Rfx2+/+ mice were stained with anti-RFX2 antibodies. RFX2 is expressed in pachytene spermatocytes (arrows) and early round spermatids (arrowheads). No RFX2 staining is observed in Rfx2-/- sections. (C-D) H&E stained sections from 30 day old Rfx2+/+ and Rfx2-/- littermates are shown. (C) Seminiferous tubules from Rfx2+/+ males show orderly development of germ cells of different stages including spermatocytes (black arrowhead, spc), round spermatids (black arrow, rsp), and late stage of spermatids with condensed nuclei (white arrow). (D) In striking contrast, Rfx2-/- mice exhibit an arrest in differentiation of haploid cells at approximately step 7 of the round spermatid phase (black arrow), without evidence of flagellum formation or condensation of the nucleus. Spermatids start to fuse at this stage (white arrow) leading to the formation of giant multinucleated cells that subsequently develop highly condensed nuclei (white arrowhead). Spermatocytes are normally present (spc, black arrowhead) and undergo meiosis. (E-G) In 3 month-old Rfx2+/+ mice, the cauda region of the epididymis is filled with mature sperm (E, arrow in G). (F and H) The cauda epididymis of a 3-month-old Rfx2-/- mouse contains no mature sperm and only cell remnants (arrow). Spermatogenesis is arrested in Rfx2-/- mice
Rfx2-/- pups grow normally and show no developmental defects. Hence Rfx2-/- mice do not exhibit the ciliopathy hallmarks observed in Rfx3-/- mice or other phenotypes observed for Rfx-deficient mice [13,14,17–19,21]. Rfx2-/- males are sterile, but Rfx2-/- females have no obvious reproductive defects, as Rfx2-/- females (n = 7) produced litters that were comparable to Rfx2+/- females (average litter size was 8 for both genotypes). Initial morphological characterization showed that young adult Rfx2-/- males had slightly smaller testes but that the remainder of the male reproductive tract and accessory glands, such as seminal vesicles, appeared normal. Microscopic examination (Fig 2E–2H) revealed that the epididymis contained no sperm but instead large numbers of degenerating small cells and cell debris. The overall microscopic architecture of the testis was normal, but there was a complete block of spermatogenesis prior to the point where round spermatids should begin to elongate and develop characteristic features of spermatozoa (Fig 2C and 2D). Cells at this stage instead formed multinucleated cells with condensed nuclei, also called symplasts in some studies [41], and were released from the tubules to flow into the epididymis (white arrowhead, Fig 2B).
One of the most readily observed changes between the end of meiosis and the beginning of spermatid elongation is formation of the acrosome, which is characteristically stained by the periodic acid-Schiff (PAS) reagent due to its high glycoprotein content. Light microscopic examination of testes from 30-day-old Rfx2-/- mice, in which the first wave of developing germ cells has only recently encountered the point of arrest, showed that Golgi-derived proacrosomic vesicles appear as expected. However, they frequently fail to attach to the nucleus, and the acrosome becomes increasingly disorganized in appearance (Fig 3A–3H). This can be seen in greater detail using fluorescently tagged peanut agglutinin to label acrosomes (Fig 3I–3P). Almost no spermatids showing a typical cup-shaped acrosome spread over the nucleus, as observed in wild type (WT) step 7 spermatids, can be visualized in Rfx2-/- testis sections. At approximately step 6–7 the mutant spermatids begin to fuse into multinucleate giant cells.
Fig. 3. The characteristic acrosome organelle fails to develop properly in Rfx2-/- mice. 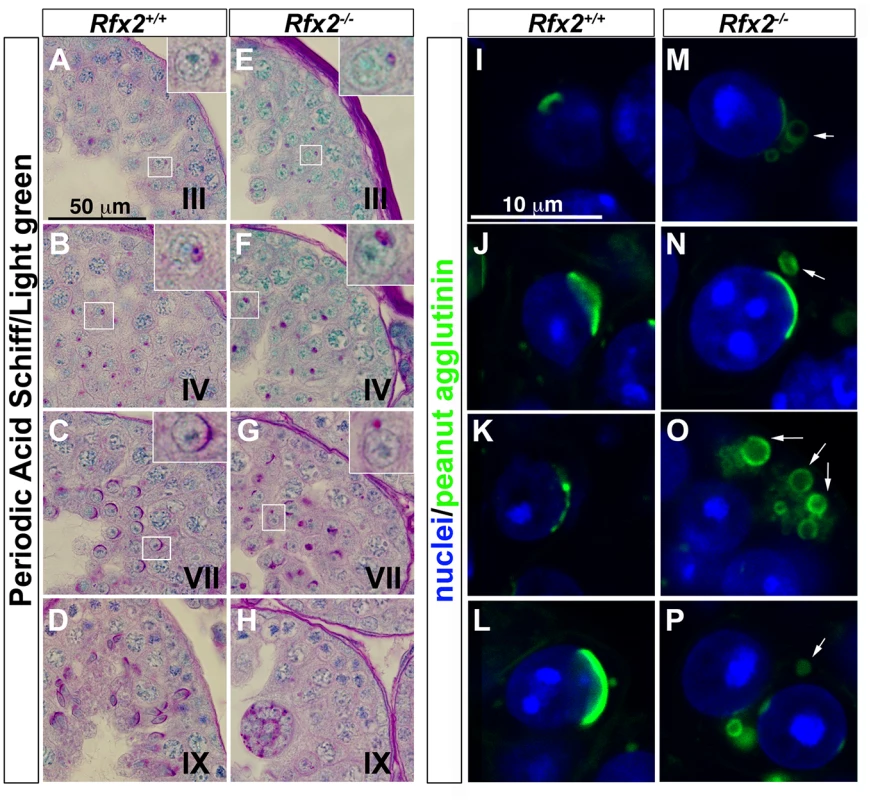
(A-H) Periodic acid shiff (PAS) / light green stained segments of testis sections from Rfx2+/+ mice (A-D) and comparable sections from Rfx2-/- mice (E-H). Developmental stages of the seminiferous tubules are noted on each panel. Insets show a 3-fold magnification of a spermatid from the respective section (box). Insets show in: (A, E) step 2 spermatids, (B, F) step 5 spermatids, (C, G) step 7 spermatids. (I-P) High power views of individual cells with the acrosomes stained by fluorescently-tagged peanut agglutinin. For Rfx2+/+ sections, different step of acrosome development are shown (I: step 2, J-L: step 6–7 spermatids). For Rfx2-/- sections, variations in acrosome morphology are shown for spermatids that are about to arrest (M-P). In the majority of cases the acrosome fails to spread normally over the anterior end of the nucleus and frequently occurs as unattached vesicles (arrows). Arrested spermatid differentiation is not associated with increased apoptosis or accelerated accumulation of transition proteins in the nuclei
WT and Rfx2-/- testis sections were stained by the TUNEL technique to ascertain if the arrest in spermatid development was associated with increased apoptosis. No difference in the distribution or number of TUNEL positive cells was detected (S1A and S1B Fig). Differences in the patterns of phospho-H2AX staining in round spermatids or the condensed nuclei in multi-nucleated bodies were also not observed (S1C Fig). Nuclear condensation in multinucleated cells is thus not due to induction of apoptosis.
We next examined whether the appearance of smaller nuclei among the multi-nucleate bodies could be due to premature or aberrant synthesis of proteins involved in the normal histone to protamine transition. This stepwise process involves multiple proteins including transition proteins TNP1 and TNP2, major players in this process [42]. mRNAs for TNP1 and 2 are not reduced in the knockout mice, as revealed by our RNA-Seq analysis described below. Like many transcripts present at the end of the round spermatid stage, these mRNAs are translationally repressed until the appropriate time. However, TNP1 was not detected by Western blotting at all developmental stages (S2A Fig), whereas TNP2 was present in Rfx2-/- testis even though spermatid development does not reach the stage (steps 9–10) at which it first appears in WT animals (S2A Fig). When sections were stained for TNP2 it was detected largely over the cytoplasm of terminal stage Rfx2-/- spermatids, in contrast with its nuclear localization in Rfx2+/+ mice (S2B and S2C Fig). This suggests that disorganized development may proceed to the point where translational repression of TNP2 mRNA is released, allowing accumulation of TNP2 protein, but that the latter fails to be transported efficiently into the nucleus. These results suggest that premature nuclear accumulation of transition proteins is not likely to be responsible for nuclear condensation in multinucleated cells. We cannot exclude that altered protamine levels could be involved in nuclei condensation, but this would likely be an indirect consequences of earlier nuclear defects as protamines are produced after the spermatid arrest observed in Rfx2-/- testes.
Perturbed proacrosomal vesicle and axoneme formation in Rfx2-/- spermatids
EM analysis was performed to characterize in more detail the cellular defects observed in Rfx2-/- spermatids. Shortly after meiosis in WT spermatids, cell polarity is established such that the Golgi apparatus is oriented with the trans-Golgi facing the nucleus. This favors the transport of budding vesicles to the nuclear surface, where they attach to the nuclear lamina [43,44]. The migration of Golgi vesicles to the nucleus is guided by microtubule tracks and mediated by molecular motors and RAB adapter proteins [45–48]. At the opposite pole of the cell, a single axoneme develops near the plasma membrane from one of the two centrioles and gradually elongates as the basal body migrates inwards toward the nucleus [43]. These processes do not occur correctly in Rfx2-/- mice. The Golgi frequently appears disoriented such that dense-cored proacrosomic vesicles migrate away from the nucleus (Fig 4G–4M). These vesicles fuse to form large dense-cored discs that do not attach to the nucleus (Fig 4G, 4H, 4I and 4J). In other cases, vesicles have fused with the nucleus to form a partial acrosomal cap that only rarely spreads to cover half of the anterior end of the nucleus, as is the case for WT mice (compare Fig 4B–4D with 4H–4J). Occasionally, highly atypical structures are found, such as the development of an acrosome having apparently engulfed multiple vesicles containing cytoplasm (Fig 4I). Nuclei more often show protruding aneurysm-like ruptures in Rfx2-/- cells (Fig 4L). No signs of developing axonemes are found, although these are evident in WT mice (compare Fig 4E and 4F with 4M and 4N and insets). This is in agreement with the absence of acetylated tubulin stained flagella in the lumen of seminiferous tubules (S3 Fig). Interestingly, centriole pairs are found, and can occur in clusters in the giant multinucleated cells (Fig 4N–4R).
Fig. 4. Ultrastructural defects and perturbed Golgi organization in Rfx2-/- testes. 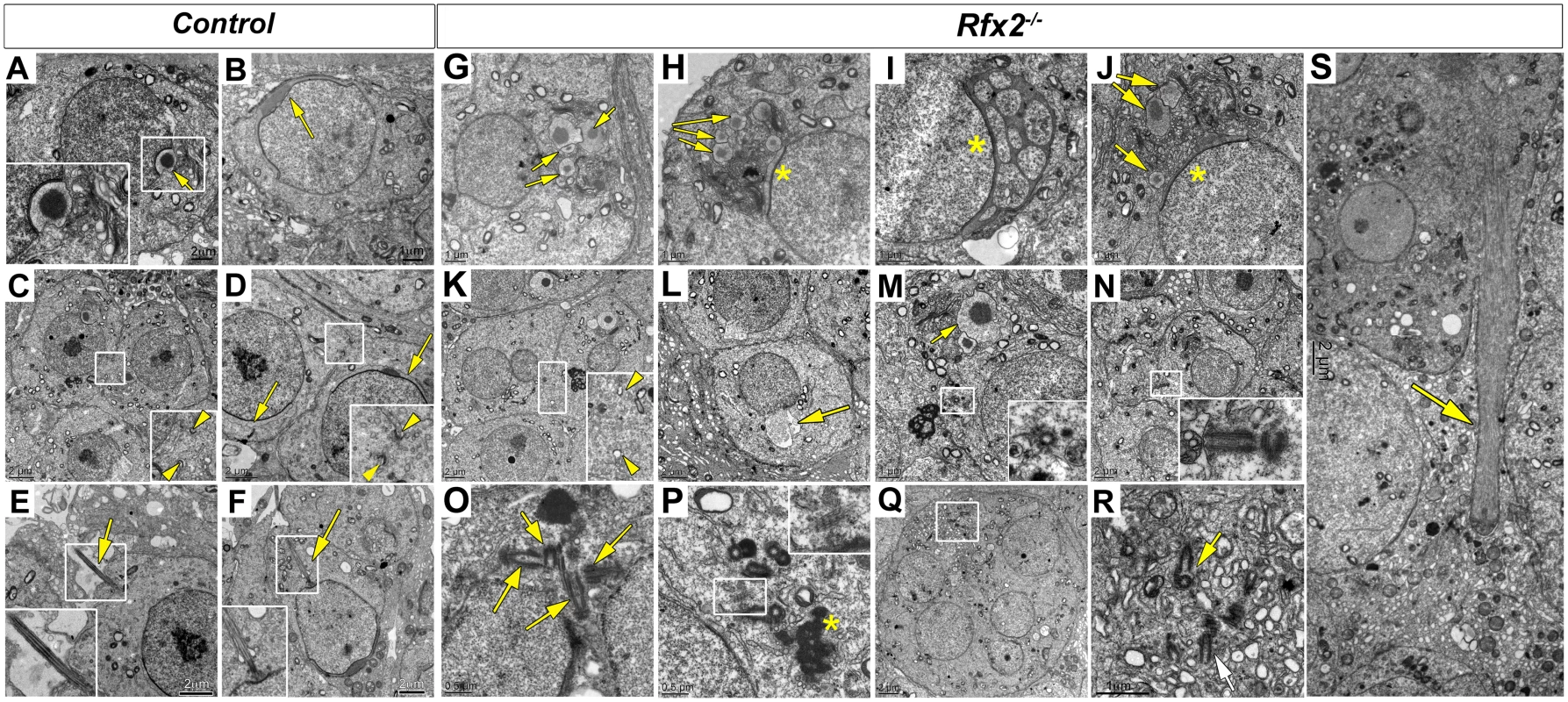
Transmission electron microscopy images of control (A-F) or Rfx2-/- (G-S) spermatids. (A) The acrosome vesicle (arrow, 2-fold magnified inset) has fused with the nucleus and is beginning to flatten at step 4. (B) As development progresses toward the end of the cap phase, acrosomes have flattened and spread over approximately half of the nuclear surface in step 7 spermatids (arrow). (C-D) cytoplasmic bridges (between two arrowheads in insets showing 2-fold enlargements) are visible between two spermatids. The acrosomal cap is marked with arrows. (E) Developing axonemes (arrows, insets show two-fold enlargements of the axonemes) are prevalent among late stage round spermatids and can be seen clearly on an early elongating spermatid of step 9 (F). (G-S) For Rfx2-/- sections various aberrant morphologies are shown for cells at about the point of developmental arrest. (G-J) Examples of aberrant acrosomes observed in Rfx2-/- spermatids. (G) Multiple acrosomic vesicles are present without attachment to the nucleus (arrows) or (H) coexisting with some degree of an attached and flattened acrosome (asterisk). (I) An atypical acrosome (asterisk) has attached and spread but contains multiple cytoplasmic inclusions. (J) Another example of multiple unattached vesicles (arrows). (K) Cytoplasmic bridges (between two arrowheads, 2-fold magnification inset) are apparently normal until spermatids fuse, forming round huge multinucleated cells, and cytoplasmic bridges disappear. (L) Nucleus with a protruding aneurysm-like rupture (arrow). (M, N) Cells contain isolated centriole pairs that have not generated axonemes (inserts). In (M), arrow points to the acrosomal vesicle. (O, P, Q, R) Within multinucleated giant cells, clusters of centrioles occur (arrows), but without associated axonemes. (Q, R) A giant multinucleated cell contains a cluster of centrioles (box enlarged in R, arrows). Ciliary rootlets are visible (2-fold enlarged inset in P), the asterisk indicates the chromatoid body. (S) A large bundle of microtubules that could be an ectopic manchette (arrow). Sizes of scale bars are indicated in individual panels. In WT spermatids, once the acrosome cap has covered slightly more than half the nucleus, the nucleus and associated acrosome move to contact the plasma membrane, which becomes the site of new specialized junctions, the apical ectoplasmic specialization, between the germ cells and Sertoli cells [49,50]. In WT mice, these will control Sertoli cell—germ cell connections until the normal release of mature spermatids at spermiation. In Rfx2-/- spermatids the nucleus/acrosome is not observed to merge with the plasma membrane, suggesting that these junctions do not form. Coincident with this failure, mutant spermatids undergo fusion to form large multinucleate cells. Prior to this point, the mutant spermatids, like all differentiating male germ cells, maintain open cytoplasmic connections via so-called cytoplasmic bridges characterized by well-defined boundaries [51] (Fig 4K). Breakdown of the supporting boundaries of these bridges presumably accompanies the formation of giant cells, but this could not be formally demonstrated from the ultrastructure images, suggesting that once begun this is a rapid process. With the failure to form and maintain the ectoplasmic specialization junctions with Sertoli cells, the spermatids, now largely or entirely in the form of giant cells, are released to pass out of the testis and into the epididymis.
Another prominent ultrastructural feature of the maturing round spermatid is formation of the manchette, a dense arrangement of microtubules attached to the outer edge of the nucleus in a ring just below the border of the acrosome. The manchette is believed to be important for the nuclear shaping that marks the end of the round phase of spermatid differentiation [52,53]. Although no manchette is observed in Rfx2-/- spermatids, dense microtubule clusters are seen in the multinucleated cells, perhaps representing ectopic manchette formation (Fig 4S).
Staining of testis sections for the cis-Golgi marker GM130 revealed a severely altered GM130 distribution in Rfx2-/- mice (Fig 5). Whereas GM130 is mainly concentrated at one pole of WT spermatids, it is also diffusely distributed around nuclei in Rfx2-/- spermatids, illustrating an altered Golgi re-distribution at the onset of the haploid stage.
Fig. 5. Testis sections from P30 Rfx2+/+ or Rfx2-/- mice stained for the cis-Golgi compartment (GM130 antibody, red), the acrosome (peanut agglutinin, green) or nuclei (Dapi, blue). 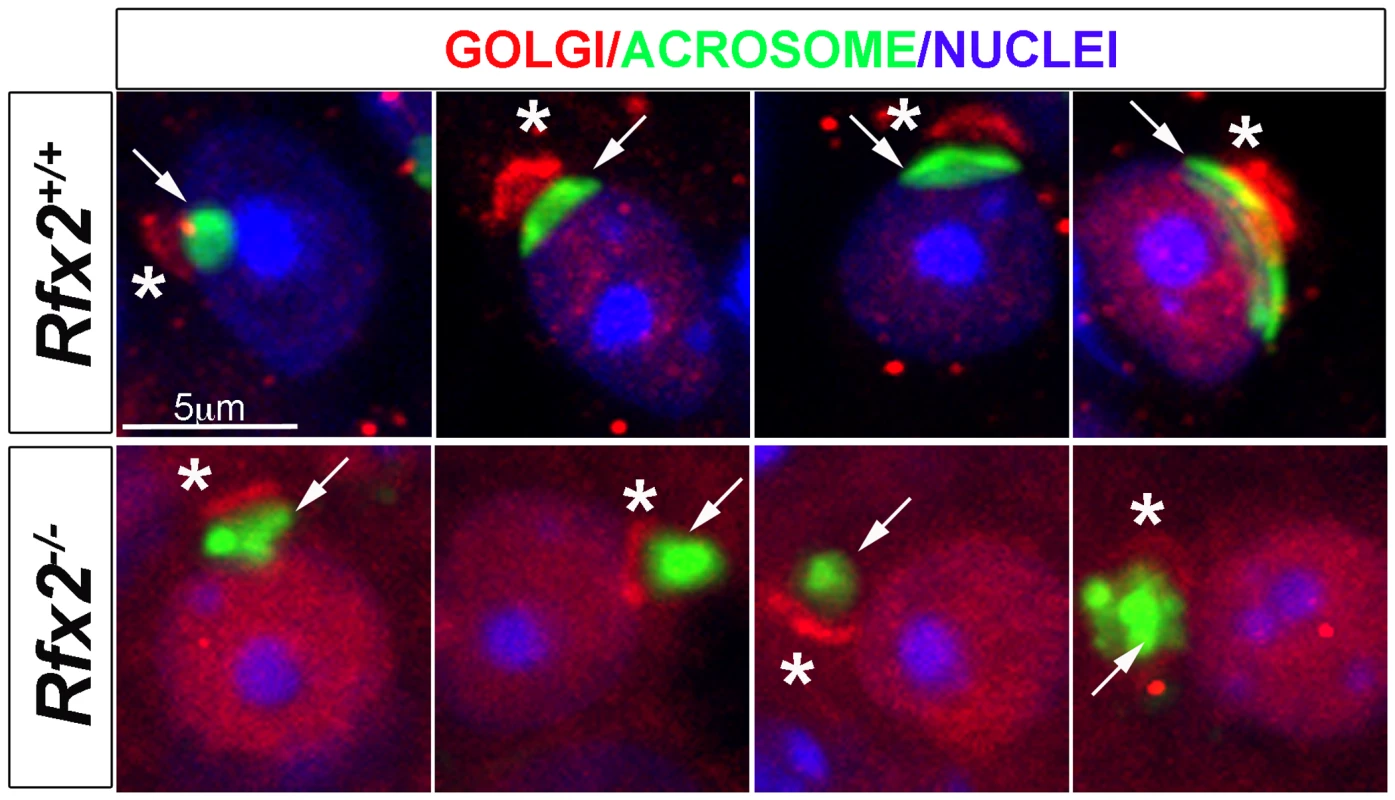
In WT spermatids, the Golgi compartment (asterisks) is concentrated at one pole of the nuclei above the forming acrosome (arrow) on the nucleus. In Rfx2-/- testes, GM130 staining (asterisks) is observed adjacent to the nuclei and acrosome (arrow) and can be correctly orientated in a few situations (left panel), but the overall orientation of the Golgi and acrosome is most frequently disturbed with the cis-Golgi being (from left to right) between the acrosome and the nucleus, apposed to the Golgi and the nucleus, or surrounding the acrosome. A wide range of mRNAs is mis-regulated in Rfx2-/- spermatids
RNA-Seq based transcriptome profiling of testis cells from Rfx2+/+ and Rfx2-/- mice was performed to assess the consequences of the loss of RFX2 function. Animals were chosen at postnatal days 21 (P21) and 29–30, henceforth referred to as (P30) as there are no significant changes in germ cell populations in this time window. At P21, the most advanced tubules contain only very early round spermatids while pachytene spermatocytes are plentiful. At this stage, defects in spermatogenesis are not yet histologically evident in Rfx2-/- mice. At P30, the first wave of germ cells has largely reached the point of arrest observed in Rfx2-/- mice, but potential long term cumulative effects on seminiferous tubules should be minimal. Global representations of RNA-Seq experiments comparing the transcriptomes of WT and Rfx2-/- testes at days P21 and P30 are shown in S4 Fig. At P21, 106 genes were down regulated (p < 0.01) greater than 2-fold, of which 47 were decreased more than 10-fold (Fig 6A, S1 Table). By P30, the number of 2-fold downregulated genes increased to 640, with 151 being decreased 10-fold or more (Fig 6A, S1 Table). Most genes downregulated at P21 were also downregulated at P30 : 95 genes were affected at both time points (Fig 6B and 6C). Markedly fewer genes were upregulated more than 2-fold: 67 genes at P21 and 128 genes at P30 (Fig 6A, S1 Table). Only 9 genes were upregulated significantly at both time points (Fig 6A, S1 Table).
Fig. 6. Perturbed gene expression in Rfx2-/- testis. 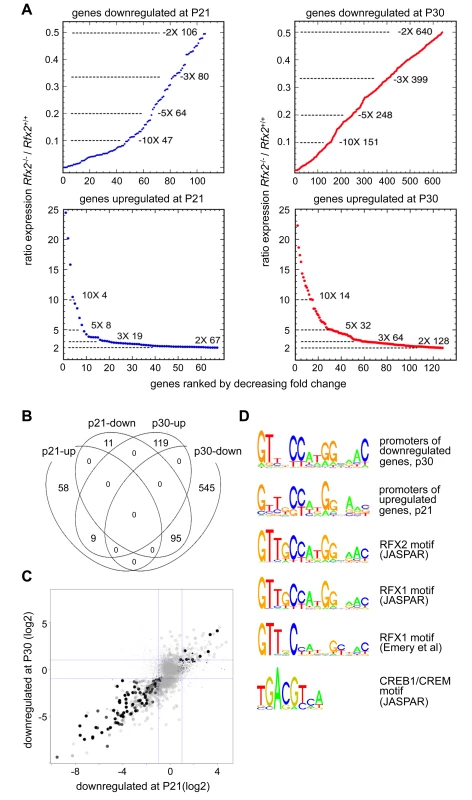
(A) Plots of the cumulative number of downregulated genes as a function of their expression level in Rfx2-/- relative to Rfx2+/+ testis: (top) downregulated genes at P21 and P30; (bottom) genes upregulated at P21 and P30. (B) Venn diagram representing the overlap between each set of differentially expressed genes. (C) Differentially expressed genes were plotted according to their fold-change (log2) at P21 and P30. Dot size is inversely proportional to the p-value at P21. Black to grey scale is inversely proportional to the p-value at P30. (D) Motifs that are significantly enriched in the promoters of P30 downregulated or P21 upregulated genes are compared with the X motifs defined for RFX1 and RFX2 in JASPAR, and the X motif identified experimentally for RFX1 [8]. The motif identified in JASPAR for CREB1/CREM is also reported, to show the absence of homology with RFX binding motifs. To identify genes that are downregulated in Rfx2-/- testis and exhibit a developmental expression program consistent with activation by RFX2 in WT testis, we used the WT expression data described in a recent RNA-Seq analysis of mouse spermatogenesis [29]. Rfx2 expression first increases significantly by day 14, as cells reach early-mid pachytene, and then increases dramatically between days 17 and 21, as the leading cells complete meiosis (Fig 7). Among genes that are downregulated in Rfx2-/- testes and exhibit statistically reliable expression profiles in the data of Laiho et al [29], the majority (47/55 at P21 and 226/281 at P31) exhibits expression profiles during spermatogenesis consistent with activation by Rfx2, (Fig 7, S2 Table).
Fig. 7. Developmental expression profiles of genes downregulated in Rfx2-/- testis. 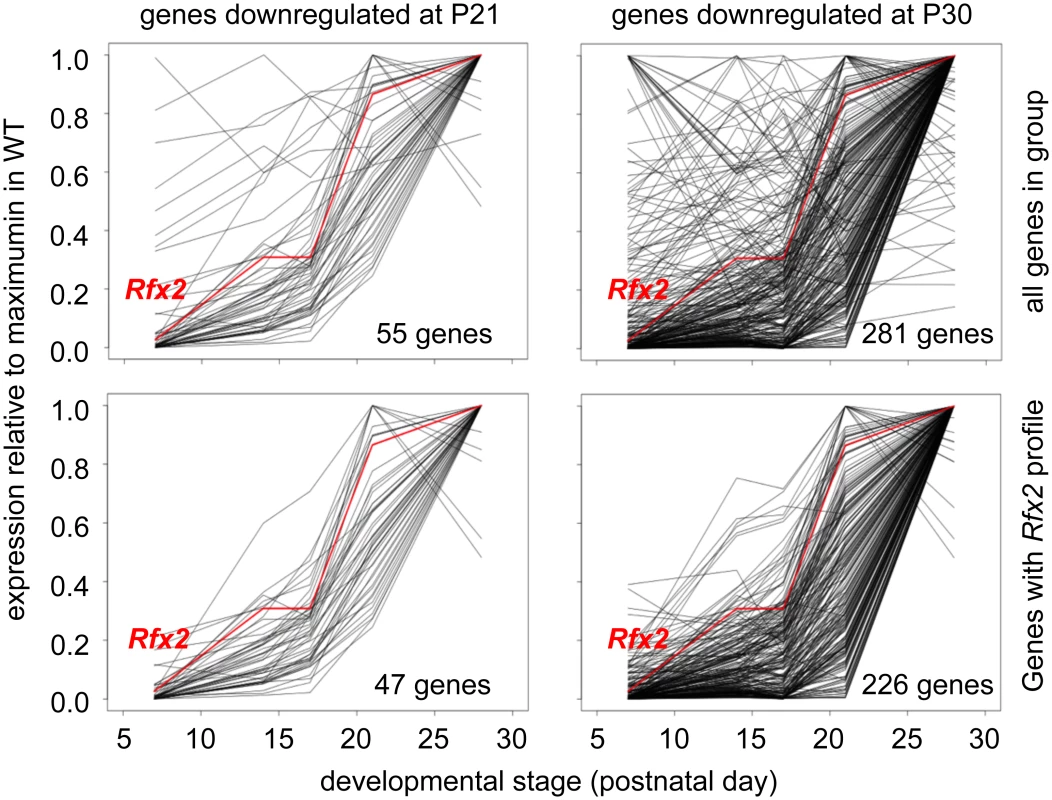
Expression patterns derived from [29], are shown for genes that are downregulated in Rfx2-/- testis at P21 (left plots) or P30 (right plots). The plots show the expression patterns for all genes in the group (top) or for genes exhibiting expression patterns that are the most consistent with activation by RFX2 (lower). Gene totals are less than in Fig 6 because genes exhibiting unreliably low read values in the Laiho et al. data set were not included. Searches for transcription factor binding sites in the promoters of differentially expressed genes, revealed that RFX binding motifs are highly enriched in genes that are upregulated at P21 or downregulated at P21 and/or P30 (S5 Fig). Similarly, an unbiased motif discovery approach identified a 14 bp inverted repeat exhibiting strong homology to the consensus RFX binding site (X box) in the promoter regions (-500 to + 50 bp relative to the predicted transcription start site, TSS) of genes that are upregulated at P21 and downregulated at P30 in Rfx2-/- testis (Fig 6D). Overall, 159 differentially expressed genes contain an X-box in their promoter regions (S1 Table). These observations suggest that RFX2 directly regulates these genes.
Identification of target genes regulated directly by RFX2
To identify genes that are regulated directly by RFX2 in spermatocytes and early spermatids, we carried out chromatin immunoprecipitation followed by high throughput sequencing (ChIP-Seq), using dissociated germ cell preparations from P21 WT mice. We identified nearly 3,000 reproducible and statistically-significant peaks corresponding to binding of RFX2 (Fig 8). Of these peaks, about 1/3 (977) were located within presumptive promoter regions (-500 to - +50), whereas the remaining were distributed in introns, exons and intergenic regions (Fig 8A). The most robust (lowest p-value) peaks were preferentially enriched in promoter regions (Fig 8B). Representative promoter peaks are shown in S6 Fig. The distribution of promoter peaks showed a marked concentration near the TSS (Fig 8C). Genes with promoter peaks will henceforth be designated RFX2 targets.
Fig. 8. ChIP-seq analysis of RFX2 target genes in mouse testis. 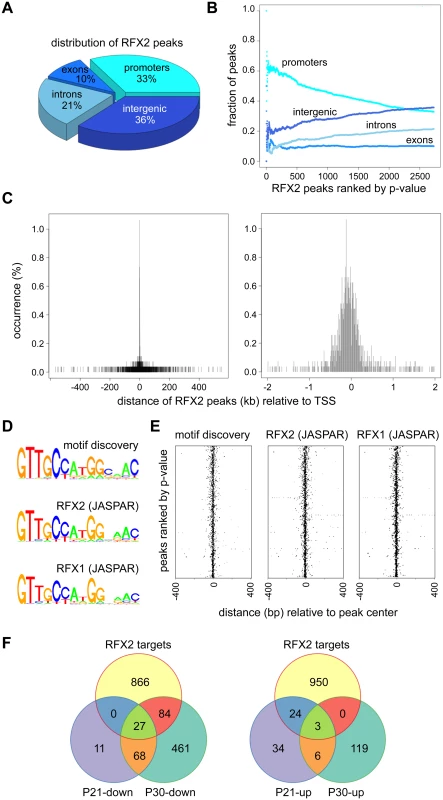
(A) Distribution of RFX2 binding peaks in different genomic regions. (B) Fractions of peaks mapping in the different regions relative to the p-value of the peaks. Peaks are ranked according to increasing p-value. (C) Distribution of peak centers relative to the TSS for genes having peaks within promoter regions. Peaks are mostly centered near the TSS. (D) A motif that is significantly enriched in the promoters of Rfx2 downregulated target genes is compared with the X motifs defined for RFX1 and RFX2 in JASPAR. (E) Position of the de novo identified motif (motif discovery) and predicted RFX1 or RFX2 motifs relative to the TSS of RFX2 target genes. All three motifs cluster near the TSS. (F) Venn diagram showing the overlap between RFX2 targets (identified by ChIP-Seq) and differentially expressed genes at P21 and P30. An unbiased search for overrepresented sequence motifs in the ChIP peaks again identified a 14 nucleotide motif exhibiting an almost perfect match to consensus binding sites for RFX1 and RFX2 (Fig 8D), illustrating that the RFX2 binding motif found in spermatogenesis genes is not different from previously established X-box motifs. As expected, the RFX motifs are mostly located near the center of the ChIP peaks (Fig 8E). In summary, these data clearly show that RFX2 binding sites are located very close to the TSS in the majority of target genes.
Among RFX2 target genes identified by ChIP-seq, 138 were differentially expressed in Rfx2-/- testes (Fig 8F and S1 Table), 111 being downregulated and 27 being upregulated. This pinpoints these genes as primary RFX2 target genes in the mouse testis. A substantial fraction of these target genes exhibit expression patterns during spermatogenesis [29] consistent with activation by RFX2 (compare upper and lower left-hand panels in Fig 9A and 9B). These genes are therefore likely to be directly regulated by RFX2 in the testis.
Fig. 9. Developmental expression profiles of RFX2 target genes. 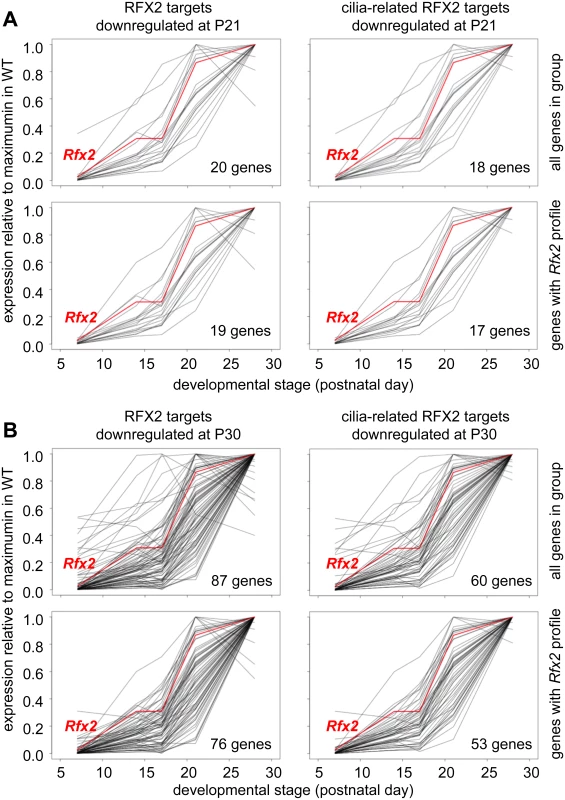
Expression patterns derived from [29], are shown for target genes that are downregulated in Rfx2-/- testis at P21 (A) or P30 (B). The plots show the expression patterns for all genes in the group (top) or for genes exhibiting expression patterns that are the most consistent with activation by RFX2 (lower). Left panels show all genes in the group and right panels only cilia-related genes from each set. RFX2 target genes are involved in cilia assembly and function
Gene ontology analysis (see Supplemental methods) revealed a specific and significant enrichment (p <0.001) in genes associated with GO terms related to ciliogenesis—such as cilium morphogenesis (GO:0060271), cilium organisation (GO:0044782), cilium assembly (GO:0042384) or cell projection GO:0042995—for both the set of direct RFX2 targets identified by ChIP-seq and the set of differentially expressed genes in Rfx2-/- testis (S7 and S8 Figs, S3 Table). We therefore examined the list of downregulated genes for those related to cilia using two reference lists. The first is the database “CilDB” [54]. The second is the “gold standard” list established by the Syscilia consortium [55]. CilDB provides a score corresponding to the number of times a gene is found with low, medium or high confidence in various studies aiming at identifying cilia-associated genes. Among genes that are downregulated in Rfx2-/- testis, 52/106 (49%) at P21 and 231/640 (36%) at P30 are potentially involved in ciliogenesis, as they are found with high confidence in at least one cilia-related study documented in the CilDB database and/or in the Syscilia gold standard list (Fig 10A, S1 Table). Of these, 43/52 at P21 (82%) and 148/231 at P30 (64%) are down regulated more than 3-fold (Fig 10A, S1 Table). Many of these downregulated cilia-related genes are direct targets of RFX2 (Fig 10B, S1 Table). Among genes that are both differentially expressed in Rfx2-/- testis and direct targets of RFX2, 80/138 (57%) are found with high confidence in at least one cilia-related study documented in the CilDB database and/or in the Syscilia gold standard list (S1 Table). Most of these genes exhibit developmental expression profiles [29], similar to Rfx2 (Fig 9A and 9B right-hand panels). Examination of our ChIP-Seq data for a selection of cilia-associated target genes downregulated at P21 and P30 clearly showed peaks in their promoter regions (see S6 Fig for examples). Finally, among 28 cilia-related genes that are upregulated at P21, 11 are direct targets of RFX2 (S9A Fig). These results clearly indicate that genes involved in ciliogenesis are primary targets of RFX2 in the mouse testis.
Fig. 10. Downregulated genes having known or suspected functions related to cilia/flagellum formation, microtubule function or the acrosome. 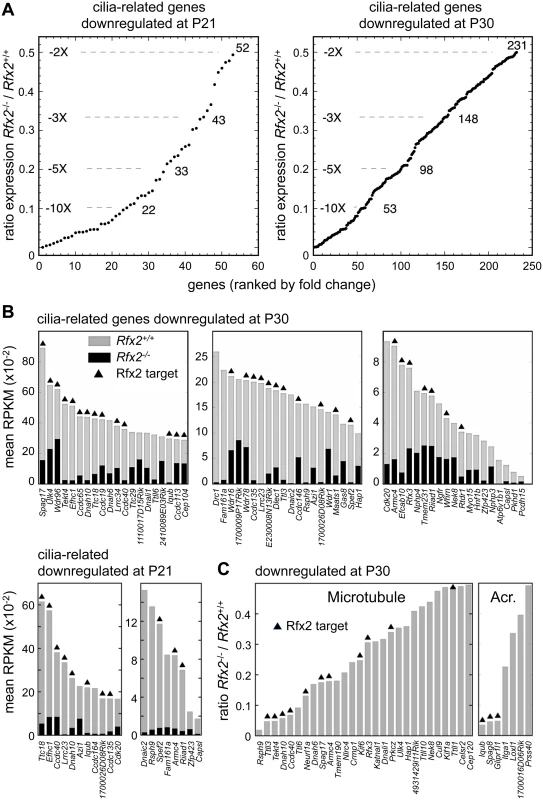
(A) Plot of the cumulative number of cilia-related genes as a function of their expression level in Rfx2-/- relative to Rfx2+/+ testis. Downregulated genes analyzed are included in the Syscilia gold standard list [55] or have at least 1 high-confidence citation in the CilDB database. (B) Bars show the expression levels (RPKM, reads per Kb per million) in Rfx2+/+ (grey) and Rfx2-/- (black) testis for downregulated genes included in the Syscilia Gold or Potential lists. RFX2 targets are indicated above the bars. Genes are ordered according to their expression level in WT mice. (C) Downregulated genes assigned to GO terms related to microtubule or the acrosome (GO function or compartment). Expression in Rfx2-/- testis is expressed relative to Rfx2+/+ testis. RFX2 targets are indicated above the bars. RFX2 regulates genes implicated in cell adhesion and intracellular trafficking
To understand how RFX2 deficiency could lead to specific Golgi-associated and acrosome-formation defects, we examined all GO terms associated with differentially expressed genes at P21 or P30. Many genes deregulated in Rfx2-/- testis were found to match to GO terms associated with intracellular transport, microtubule associated processes or small GTPase regulation, even though these terms were not significantly enriched. For example, of genes that are downregulated in Rfx2-/- testis, 35 belong to gene sets corresponding to the GO terms “microtubule” or “acrosome”, and a substantial fraction of these genes are direct RFX2 targets (Fig 10C). Impaired expression of these genes could contribute to defective cell polarization, proacrosomic vesicle migration and acrosome formation during the first steps of spermatid differentiation.
GO terms that are enriched with a p-value below threshold (0.01<p<0.001), include terms linked to cell-adhesion or cytoskeleton organization. Genes associated with these terms could account for some of the spermatid defects observed in Rfx2-/- mice, in which spermatids lose their contacts with adjacent Sertoli cells and form giant multinucleated cells. These genes include Fndc3c1, which is a paralogue of Fndc3a. The latter is mutated in the mouse sys strain (S4 Table), which has a phenotype strikingly similar to that of Rfx2-/- mice in that it is characterized by the formation of giant multinucleated cells referred to as symplasts [56]. Another interesting candidate is Fascin 1, which is down regulated both at P21 and P30, and is known to be involved in the ectoplasmic specialization required for spermatid-Sertoli cell contacts in the rat testis [57].
Discussion
We demonstrate here that a targeted mutation of the gene encoding RFX2, one of the earliest identified members of the RFX family of transcription factors, leads to a complete block in mouse spermiogenesis. While over 400 mutations have been reported to affect spermatogenesis in the mouse [1], relatively few result in complete arrest of development at the end of the round spermatid stage [58] (S4 Table). Most of these mutations lead to defects in RNA processing or formation of RNA-protein complexes. A number also affect specific cell organelles or cell junctions. Prior to this report, only three of these mutations involved genes encoding TFs. Of these, two (Tbpl1 and Taf7l) [40,59] encode variants of general transcription factors (GTFs), leaving CREM as the only sequence/gene-specific TF established to have a pivotal role in controlling spermiogenesis at the round spermatid stage [60,61]. Our results thus identify RFX2 as a second essential master TF implicated in this stage of spermatogenesis. Of note, none of the transcriptional regulators known to be required for spermiogenesis (see S4 Table) are affected by Rfx2 disruption.
RFX2, CREM and TAF7l control largely non-overlapping gene sets
Genome wide analyses of their effects on transcriptional programs in testis have been performed for mutations in Crem and Taf7l [39,40]. Crem occupies a special place among genes encoding transcriptional regulators of spermatogenesis. In the testis it produces a specific splice variant encoding CREM-tau, a unique isoform of the cAMP-responsive element modulator (CREM), which comprises a family of isoforms that bind to the cAMP response element (CRE). While most of the splice variants produce inhibitory factors, CREM-tau is stimulatory and appears late in meiosis [62]. Crem was one of the earliest targets of engineered mutations [60,61] and proved to be essential for expression of a large number of classic genes expressed in the haploid phase of spermatogenesis. A recent genome wide study identified 627 genes that were downregulated greater than 2-fold in Crem-/- testis [39]. Of these CREM-regulated genes, 277 were shown by ChIP-Seq experiments to be direct targets of CREM in male germ cells [63]. Only 34 of the CREM-regulated genes and 9 of the CREM-occupied genes are downregulated greater than 2-fold in Rfx2-deficient testis (S10A Fig). Furthermore, RFX2 and CREM regulated gene sets exhibit largely distinct patterns of expression during spermatogenesis (S10B Fig). Direct RFX2 target genes tend to parallel the expression pattern of RFX2 itself, starting from little or no significant expression at P7 and rising to half maximal by or before P21. In contrast, direct CREM targets have extremely variable expression at P7 but tend to share large increases after P21 (S10B Fig). Genes that depend directly on RFX2 or CREM thus constitute nearly distinct sets.
Spermatogenesis exhibits a striking dependence on variants of the Pol II GTFs [64], which include TBL1 (TRF2) [59], TAF4b [65], TAF7l [40], and GTF2a1l (ALF) [66]. TAF7l, a variant of TAF7, is a component of TFIID. A targeted mutation of Taf7l led to an arrest in spermatogenesis at the end of the round spermatid stage. Transcriptome analysis of Taf7l-deficient and WT testis [40] identified some 1,440 genes that were down regulated by more than two-fold, and 726 by over three-fold, in the mutant mice. Furthermore TBL1, whose ablation also leads to a developmental arrest of round spermatids [59], was found to co-occupy active promoters with TAF7L, and the two GTFs were suggested to function together on a subset of postmeiotic genes. Remarkably, none of the genes found to be downregulated in Taf7l-deficient testis are also deregulated in RFX2 or CREM deficient testis. This is striking support for the model that there are relatively few master transcriptional regulators for the postmeiotic phase, and that each one controls largely separate groups of genes.
Gtf2a1l (ALF) is yet another GTF expressed during spermatogenesis [66,67]. Targeted mutations have not been reported for this gene. Gtf2a1l was shown to have a binding site for RFX factors [34], although it was not identified by our ChIP-Seq experiment as being a direct RFX2 target. It’s expression is however reduced slightly more than 2-fold in the Rfx2-/- mouse. This reduction may contribute to the broad spectrum of indirectly downregulated genes.
RFX2 regulates a large cohort of cilia related genes in testis
A strikingly large number of Rfx2-regulated genes is associated with the formation and function of cilia and flagella, which are complex organelles that are highly conserved among eukaryotes. In mammals, cilia occur as single non-motile cilia on a wide range of cells, motile monocilia on the embryonic node and multiple motile cilia on various epithelia, whereas flagella are found on sperm cells. The regulation of cilia formation and function by RFX factors traces back to metazoans [10]. As the family expanded through evolution, multiple family members remained associated with this function, including RFX3 and RFX4 in mammals, as well as RFX2 and RFX7 in non-mammalian vertebrates [14,17,18,23,24,26,27]. This study underlines a central role for mouse RFX2 in the regulation of cilia associated genes during spermatogenesis.
RFX proteins control specific subsets of core genes required for cilia formation from C. elegans to mammals (for review see [11,12]). In particular, RFX factors regulate genes encoding intraflagellar transport (IFT) components and Bardet-Biedl syndrome (BBS) proteins required for building cilia. RFX factors were also shown to regulate transition zone components in C. elegans, Drosophila, Xenopus and mice [11,12,68,69]. Transition zone proteins form a specialized structure at the base of the axoneme that connects the distal end of the basal body to the plasma membrane (for review see [70,71]). Whereas many of the IFT genes are indeed bound by RFX2, none are downregulated in Rfx2-/- testes at either time point (S1 Table). Many of the Bbs genes are also bound by RFX2, but none are downregulated more than 2-fold in Rfx2-/- testes. However, expression of some of these genes is significantly decreased by slightly less than the arbitrary 2-fold threshold applied for inclusion in S1 Table. As for IFT and BBS genes, most of the transition zone associated genes are also bound by RFX2 in the testis (Rpgrip1L, B9D1, D2, Tctn1, Tctn2, Tctn3, Tmem216, Tmem138, MKS1, Tmem17, Tmem231, Tmem237, Cep290, Sdccag8). Only two of these are upregulated at P21 (Tcnt1) or downregulated at P30 (Tmem231). Nphp3, Nphp4, and Nek8 are downregulated in Rfx2-/- testis but do not appear to be direct RFX2 targets. Collectively, these results show that RFX2 binds to the promoters of many core genes required for building cilia, but does not seem to play a dominant role in controlling their expression. This implies that other TFs are implicated in regulating genes encoding IFT, BBS and transition zone components in the testis. Other RFX family members expressed in the testis, such as RFX1 or RFX3, are good candidates, as they could function redundantly with RFX2 to regulate core ciliary genes.
Surprisingly, several IFT (IFT74, IFT81) and cilia associated genes are upregulated at P21 in Rfx2-/- testes (S9A Fig, S1 Table). This may imply that RFX2 represses these genes during spermatogenesis. However, since RFX factors can function as homo or heterodimers, an alternative hypothesis is that a controlled balance between different dimeric RFX complexes is required for precise tuning of ciliary gene expression. Perturbing this balance by ablating RFX2 may thus generate new RFX complexes capable of activating these upregulated genes.
In contrast to the core ciliary genes, many genes required for ciliary motility are downregulated in Rfx2-/- testes. Among 32 genes in which defects lead to primary ciliary dyskinesia, six (Dnaic2, Ccdc40, Armc4, Ccdc164/DRC1, Rsph9, Ccdc11) are strongly downregulated in Rfx2-/- testes at both time points, and three others only at P30 (Ccdc65, Zmynd10, Ccno) (for review see [72]). Many genes coding for structural components of flagella or centrioles are also downregulated (S1 Table). RFX2 is thus only critical for the expression of a specific subset of ciliary genes in the testis.
In zebra fish and Xenopus, RFX2 has been implicated in the motile cilia differentiation program required for the establishment of left/right asymmetry during embryonic development and the formation of motile cilia in various epithelia [24,26]. We did not observe any major defects in left/right asymmetry or multiciliated epithelia in Rfx2-/- mice in two different genetic backgrounds. This is unexpected as Rfx2 is expressed strongly in the mouse embryonic node and multiciliated epithelia [25,27]. The absence of aberrant left/right or ciliated-epithelia phenotypes in Rfx2-/- mice could be due to partial redundancy between RFX transcription factors. As Rfx3-/- mice display left/right asymmetry defects, it is possible that RFX3 (or other RFX factors) can fully compensate for the deficiency in RFX2, whereas RFX2 cannot compensate efficiently for the loss of RFX3. The opposite seems to be true in the testes, as the few Rfx3-/- males that survive past birth do not show defects in sperm or flagella production (B. Durand, personal communication), showing that RFX2 and RFX3 do not have redundant functions in this cell type.
In summary, our study implies that RFX TFs have evolved to regulate specific sets of cilia-related genes in different tissues. RFX2 appears to have become specialized for regulating a specific subset of cilia/flagella-related genes required for spermatid development. Double mutants will need to be studied to further understand how different RFX factors exert their respective functions in different mouse tissues. Furthermore, the complex interplay between members of the RFX family, notably Rfx2 and Rfx3, in the regulation of cilia-related genes in different tissues can now be sorted out genetically using the available conditional mutants.
Rfx2-/- mice and mutants defective in RNA processing proteins share a similar arrest in spermatid development
Several mutant mice having defects in RNA biogenesis or small-RNA regulated processes show a developmental arrest in spermatogenesis similar to that of Rfx2-/- mice (S4 Table). The genes mutated in these mouse models are not regulated by RFX2. However, the similar phenotype prompted us to examine whether other genes associated with RNA processing and small RNA pathways could be regulated by RFX2. A recent proteomic analysis identified 88 proteins associated with the Chromatoid Body, which is generally believed to be the site of post-nuclear RNA processing and may also contain translationally suppressed mRNAs [73]. Of these proteins, TEKT4 (tectin 4) is encoded by a gene downregulated in Rfx2-/- testis at P30. Another recent study demonstrated that many genes required for acrosome or flagella function are downregulated in the maelstrom mutant, which carries a mutation in the gene encoding MAEL, a conserved core component of the piRNA pathway [74]. This suggests that RFX2 and MAEL regulated pathways could converge on common genes, and that this might underlie the comparable phenotypes observed in Rfx2-deficient mice and piRNA biogenesis pathway mutants. However, we cannot exclude that RFX2 could regulate the expression of small RNAs that our RNA-seq analysis could not reveal. The transcription factor A-MYB has also been shown to regulate piRNA expression in mouse germ cells and many A-MYB target genes have been identified by ChIP-Seq experiments [75]. The binding motif identified in promoters of A-MYB target genes, including piRNA clusters, does not overlap with the well-defined RFX binding motif [75]. In addition, more than 90% of A-MYB targets identified by ChIP-Seq are unique to A-MYB and do not overlap with RFX2 targets. Thus, although A-MYB could regulate RFX2 [33], the two transcription factors do not regulate the same sets of target genes.
Rfx2-/- mice display polarization defects in post meiotic germ cells
Early in normal spermatid development, a crucial polarity develops such that the acrosome will form and define the anterior end of the sperm, while at the opposite cellular pole, the centriole pair will move to the plasma membrane and initiate growth of the axoneme, which will eventually grow to form the interior elements of the flagellum [43,44]. Failure of normal acrosome cap formation is a characteristic of polarity loss. The mechanisms that set up this polarity are not well understood in spermatids. The PAR3-PAR6-aPKC complex was shown to be required to establish cell polarity [76], but we did not observed any defects of the distribution of these markers (S3 Fig), suggesting that only downstream actors of this complex are likely to be affected in Rfx2-/- mice. Cell polarity in many systems is set up by cell junctions with surrounding tissue and involves a set of highly conserved genes. Spermatids are connected to Sertoli cells by adherens-like junctions at this stage [50]. Mutation of junctional adhesion molecule-C (JAM-C) resulted in general polarization failure of spermatids and acrosomic failure similar to that observed in Rfx2-/- mice [77]. Around 20 genes required for cell adhesion or cell junctions are downregulated in Rfx2-/- testes, which could account for the observed spermatid defect.
Microtubules and associated motors responsible for movement of vesicles and macromolecules are effectors of cell polarization, and required for Golgi organization and acrosome formation in spermatids [47,48,78,79]. 24 genes mapping to Golgi GO terms are downregulated in Rfx2-/- testes (S9B Fig) and this may contribute to disruption of overall Golgi organization and acrosome formation in Rfx2-/- spermatids. For example, RAB27a/b is known to function as an adapter for acrosome-bound vesicles [46], and Rab27b is down regulated ~3-fold. 20 other downregulated genes also have GTPase regulator activity. Furthermore, some 28 microtubule-related genes are downregulated in Rfx2-/- testis, which could also contribute to overall polarity and vesicular transport failure (Fig 10C). Hence, defective spermatogenesis in Rfx2-/- mice is likely to result from alterations in several different cellular pathways.
Concluding remarks
Our results demonstrate that RFX2 is a crucially important TF for orchestrating the postmeiotic phase of sperm development. The morphology of the mutant and characterization of RFX2-regulated pathways may provide a useful basis for assigning genetic causes to human infertility, and ultimately for deciphering the regulatory networks that direct sperm formation.
Materials and Methods
Generation of Rfx2-deficient mice
The conditional targeting vector (Fig 1A) carried loxP sites inserted into the introns flanking exon 7, which encodes a large segment of the DNA binding domain. Deletion of exon 7 is predicted to alter the reading frame if either exon 5 or 6 is spliced to exon 8 (exon 6 is variably included in the mature transcript). The vector also contained a FRT-flanked neomycin selection cassette. The vector was electroporated into R1 ES cells (strain 129/Sv) by the Mouse Biology Program at University of California, Davis. Targeted clones were identified using PCR and primer sets with outside members located both upstream and downstream of the homology region of the vector (Fig 1B and 1C). Correctly targeted clones were introduced into C57BL/6 blastocysts to generate chimeras. These were bred to C57Bl/6 mice and offspring tested for transmission by PCR analysis of tail biopsy extracts (Fig 1D) using primers described in S5 Table. For initial experiments exon 7 was removed by breeding to a universal Cre-expressing strain, FVB/N-Tg(ACTB-cre)2Mrt/3, and deletion of exon 7 in offspring was verified by PCR (Fig 1D). Heterozygous mice of either gender were fertile and were mated together to generate Rfx2-/-neo+ mice, which were maintained as a mixed background line. Age-matched littermates were used whenever possible. A second line, without the neo cassette, was developed by breeding to a flipase expressing strain, B6;SJL-Tg(ACTFLPe)9205Dym/J [80]. Offspring were selected for loss of the neo cassette and flipase transgene. This strain was backcrossed onto a C57BL/6 genetic background. The phenotype of Rfx2-/- animals appears identical with respect to testis morphology for animals from either the C57BL/6 or mixed background lines.
Mice were treated in accordance with the American Veterinary Medicine Association (AVMA) Guidelines for Euthanasia of Laboratory Animals and with French Institutional guidelines in compliance with French and European animal welfare regulations (authorization n° 562, French Ministry of Education and Research). All animal studies were approved by the Institutional Animal Care and Use Committee (IACUC) of the University of South Carolina (AUP No 1818) and by the Ethics Committee of the University of Lyon-1 (BH 2008–01).
Histology
Animals were sacrificed by CO2 anesthesia. Testes were fixed in Bouin’s solution and epididymis in PBS containing 4% paraformaldehyde and processed to produce paraffin sections. Dewaxed sections were stained with hematoxylin and eosin or by PAS using Light Green SF Yellowish as counterstain.
Western blotting
SDS extraction of testes and Western blotting were carried out as previously described [81]. Separation was in an 8% (RFX2) or 15% gel (TNP1, TNP2). Primary antisera to TNP1 (1 : 5000) and TNP2 (1 : 5000) [82], RFX2 (1 : 250, C-15, Santa Cruz Biotechnology, Santa Cruz, CA) and actin (1 : 1000, A-2066, Sigma-Aldrich, St Louis, MO) were detected with HRP-conjugated donkey anti-rabbit IgG and anti-goat IgG (1 : 1000, Jackson ImmunoResearch Laboratories, West Grove, PA) and chemiluminescence [83].
Reverse transcriptase PCR
For semi-quantitative RT-PCR, total RNA (1 μg) was reverse transcribed using the Superscript III First Strand Synthesis Kit (Invitrogen/Life Technologies) and random hexamer primers according to the manufacturer’s directions in a final volume of 20 μl. After a 1 to 10 dilution, 1 μl was used for a 50 μl PCR reaction. Control reactions lacking reverse transcriptase were done in parallel.
Electron microscopy
Mice (P25 to P40) were anesthetized by i.p. injection of sodium pentobarbital (0.1 ml/100g b.w.—CEVA santé Animale, France) and fixed by intra-cardiac injection (adapted from [84]) with 2% glutaraldehyde and 0.5% paraformaldehyde in 0.1 M Sodium cacodylate, pH 7.35. Testes were cut into cubes of 1 mm3 in fresh fixative solution and left for 2 days at 4°C. After extensive washing in 0.15 M sodium cacodylate (4 x 1h at RT and o/n at 4°C) samples were postfixed in 0.15M sodium cacodylate containing 1% OsO4 for 1 hour at RT, briefly rinsed in distilled water and dehydrated through a graded series of ethanol solutions (from 30° to 100°) and two baths of propylene oxide for 15 min each. After substitution and impregnation, small blocks were embedded in epoxy resin in flat silicon molds and polymerized at 56°C for 48h. Ultrathin sections were cut with a UC7 Leica ultramicrotome. Ultrathin sections were contrasted in aqueous uranyl-acetate and lead citrate solutions using a Leica ultrostainer. Sections were observed with a Philips CM120 transmission electron microscope at 120Kv.
Immunohistochemical analysis
Testes were isolated from Rfx2+/+ and Rfx2-/- mice (P30 to P40), fixed o/n in 4% PFA, embedded and frozen in 15% sucrose, 7.5% gelatin. Immunofluorescence and apoptosis experiments were performed with 14 μm cryosections. Briefly, samples were permeabilized in 0.1% Triton for 5 min, and nonspecific antibody binding was blocked with 3% BSA, 10% FBS. Primary antibodies were incubated o/n at 4°C. Secondary antibodies (anti-rabbit Alexa 488 or anti-mouse Alexa 594 or Alexa 555, Invitrogen, both at 1 : 500) were incubated for 2 h at RT. Nuclei were counterstained with DRAQ5. Fluorescence was observed under a LSM 510 Confocal microscope. Primary antibodies were: anti mouse H2AX (1/500, Millipore), Acetylated-tubulin (1/1000, T6793 Sigma), Beta-catenin (1/50, C2206 Sigma), ZO-1 (1/25, PA5-28858 Zymed), PAR3 (1/100, 07–330 Millipore), PAR6 (1/50, sc-67393 Santa Cruz Biotechnology), aPKC (1/400, sc-216 Santa Cruz Biotechnology). PAR6 and aPKC antibodies were kind gifts from M. Montcouquiol.
Acrosomes
Testes were fixed in 4% paraformaldehyde in PBS overnight, incubated in 30% sucrose, embedded in sucrose/gelatin and cut with a cryostat. Frozen sections (20 μm) were washed in PBS and incubated 2 hours at RT in PBS with Fluorescein isothiocyanate-conjugated peanut agglutinin (FITC-PNA, Sigma L7381) at a final concentration of 20 μg/ml. Nuclei were counterstained with Hoechst. Sections were washed twice in PBS and mounted in Dako fluorescence mounting medium (Dako S3023) and observed with a 63X/1.4 oil objective on a Leica SP5 spectral confocal.
Apoptosis
Apoptotic cells were visualized according to the TUNEL assay protocol from Roche Diagnostics (Roche 11684795910). 20 μm thick frozen sections (as prepared above) were washed in PBS, permeabilized 2 min at 4°C with 0.1% Triton X-100 in 0.1% sodium citrate freshly prepared, washed twice in PBS 1X and incubated in the TUNEL label solution for 60 min at 37°C, washed twice in PBS and mounted and observed as described above.
TNP2
Decapsulated testes were fixed in 4% paraformaldehyde in PBS overnight at 4°C and embedded in paraffin. Sections were mounted on Superfrost plus slides (Fisher Thermo Scientific), rehydrated and subjected to antigen retrieval by microwave heating as described previously [81]. Sections were blocked in PBS containing 0.1% Triton and 10% donkey serum for 30 min at room temp and then exposed to the primary antibody overnight at 4°C. After washing 3X with PBS, 0.1% TritonX-100 slides were exposed to the secondary antibody for 30–45 min at room temp (biotinylated IgG, 1 : 200, Jackson ImmunoResearch), washed briefly, exposed to Cy2 or Cy3 conjugated streptavidin (1 : 1000, Jackson) for 15 min at RT, washed, and cover-slipped using Vectashield containing DAPI (1 μg/ml). Slides were examined using an Olympus B41 microscope and digital images recorded with an Olympus DP71 camera. Images were processed using Photoshop.
RNA Seq and bioinformatics
See supplementary material.
Supporting Information
Zdroje
1. Matzuk MM, Lamb DJ (2008) The biology of infertility: research advances and clinical challenges. Nat Med 14 : 1197–1213. doi: 10.1038/nm.f.1895 18989307
2. Visser L, Repping S (2010) Unravelling the genetics of spermatogenic failure. Reproduction 139 : 303–307. doi: 10.1530/REP-09-0229 19776097
3. O'Donnell L, Meachem SJ, Stanton PG, McLaughlan RI (2005) Endocrine Regulation of Spermatogenesis. In: de Kretser DM, editor. Knobil and Neill's Physiology of Reproduction. 3rd ed. New York: Academic Press. pp. 1017–1070.
4. Bettegowda A, Wilkinson MF (2010) Transcription and post-transcriptional regulation of spermatogenesis. Philos Trans R Soc Lond, B, Biol Sci 365 : 1637–1651. doi: 10.1098/rstb.2009.0196 20403875
5. Kerr JB, Loveland KL, O'Bryan MK, de Kretser DM (2005) Cytology of the Testis and Intrinsic Control Mechanisms. In: de Kretser DM, editor. Knobil and Neill's Physiology of Reproduction. 3rd ed. New York: Academic Press. pp. 827–948.
6. Dorn A, Durand B, Marfing C, Le Meur M, Benoist C, et al. (1987) Conserved major histocompatibility complex class II boxes—X and Y—are transcriptional control elements and specifically bind nuclear proteins. Proc Natl Acad Sci U S A 84 : 6249–6253. 3114745
7. Aftab S, Semenec L, Chu JS, Chen N (2008) Identification and characterization of novel human tissue-specific RFX transcription factors. BMC Evol Biol 8 : 226. doi: 10.1186/1471-2148-8-226 18673564
8. Emery P, Durand B, Mach B, Reith W (1996) RFX proteins, a novel family of DNA binding proteins conserved in the eukaryotic kingdom. Nucleic Acids Res 24 : 803–807. 8600444
9. Gajiwala KS, Chen H, Cornille F, Roques BP, Reith W, et al. (2000) Structure of the winged-helix protein hRFX1 reveals a new mode of DNA binding. Nature 403 : 916–921. 10706293
10. Swoboda P, Adler HT, Thomas JH (2000) The RFX-type transcription factor DAF-19 regulates sensory neuron cilium formation in C. elegans. Mol Cell 5 : 411–421. 10882127
11. Thomas J, Morlé L, Soulavie F, Laurençon A, Sagnol S, et al. (2010) Transcriptional control of genes involved in ciliogenesis: a first step in making cilia. Biol Cell 102 : 499–513. doi: 10.1042/BC20100035 20690903
12. Choksi SP, Lauter G, Swoboda P, Roy S (2014) Switching on cilia: transcriptional networks regulating ciliogenesis. Development 141 : 1427–1441. doi: 10.1242/dev.074666 24644260
13. Feng C, Xu W, Zuo Z (2009) Knockout of the regulatory factor X1 gene leads to early embryonic lethality. Biochem Biophys Res Commun 386 : 715–717. doi: 10.1016/j.bbrc.2009.06.111 19559676
14. Bonnafe E, Touka M, AitLounis A, Baas D, Barras E, et al. (2004) The transcription factor RFX3 directs nodal cilium development and left-right asymmetry specification. Mol Cell Biol 24 : 4417–4427. 15121860
15. Benadiba C, Magnani D, Niquille M, Morlé L, Valloton D, et al. (2012) The ciliogenic transcription factor RFX3 regulates early midline distribution of guidepost neurons required for corpus callosum development. PLoS Genet 8: e1002606. doi: 10.1371/journal.pgen.1002606 22479201
16. Ait-Lounis A, Baas D, Barras E, Benadiba C, Charollais A, et al. (2007) Novel function of the ciliogenic transcription fator RFX3 in development of the endocrine pancreas. Diabetes 56 : 950–959. 17229940
17. Blackshear PJ, Graves JP, Stumpo DJ, Cobos I, Rubenstein JL, et al. (2003) Graded phenotypic response to partial and complete deficiency of a brain-specific transcript variant of the winged helix transcription factor RFX4. Development 130 : 4539–4552. 12925582
18. Zarbalis K, May SR, Shen Y, Ekker M, Rubenstein JL, et al. (2004) A focused and efficient genetic Screening Strategy in the mouse: Identification of mutations that disrupt cortial development. Plos Biology 2 : 1177–1187.
19. Zhang D, Zeldin DC, Blackshear PJ (2007) Regulatory factor X4 variant 3: a transcription factor involved in brain development and disease. J Neurosci Res 85 : 3515–3522. 17510980
20. Steimle V, Durand B, Barras E, Zufferey M, Hadam MR, et al. (1995) A novel DNA-binding regulatory factor is mutated in primary MHC class II deficiency (bare lymphocyte syndrome). Genes Dev 9 : 1021–1032. 7744245
21. Smith SB, Qu H-Q, Taleb N, Kishimoto NY, Scheel DW, et al. (2010) Rfx6 directs islet formation and insulin production in mice and humans. Nature 463 : 775–780. doi: 10.1038/nature08748 20148032
22. Soyer J, Flasse L, Raffelsberger W, Beucher A, Orvain C, et al. (2010) Rfx6 is an Ngn3-dependent winged helix transcription factor required for pancreatic islet cell development. Development 137 : 203–212. doi: 10.1242/dev.041673 20040487
23. Manojlovic Z, Earwood R, Kato A, Stefanovic B, Kato Y (2014) RFX7 is required for the formation of cilia in the neural tube. Mech Dev 132 : 28–37. doi: 10.1016/j.mod.2014.02.001 24530844
24. Bisgrove BW, Makova S, Yost HJ, Brueckner M (2012) RFX2 is essential in the ciliated organ of asymmetry and an RFX2 transgene identifies a population of ciliated cells sufficient for fluid flow. Dev Biol 363 : 166–178. doi: 10.1016/j.ydbio.2011.12.030 22233545
25. Chung M-I, Kwon T, Tu F, Brooks ER, Gupta R, et al. (2014) Coordinated genomic control of ciliogenesis and cell movement by RFX2. Elife 3: e01439. doi: 10.7554/eLife.01439 24424412
26. Chung M-I, Peyrot SM, Leboeuf S, Park TJ, Mcgary KL, et al. (2012) RFX2 is broadly required for ciliogenesis during vertebrate development. Dev Biol 363 : 155–165. doi: 10.1016/j.ydbio.2011.12.029 22227339
27. Liu Y, Pathak N, Kramer-Zucker A, Drummond IA (2007) Notch signaling controls the differentiation of transporting epithelia and multiciliated cells in the zebrafish pronephros. Development 134 : 1111–1122. 17287248
28. Reith W, Ucla C, Barras E, Gaud A, Durand B, et al. (1994) RFX1, a transactivator of hepatitis B virus enhancer I, belongs to a novel family of homodimeric and heterodimeric DNA-binding proteins. Mol Cell Biol 14 : 1230–1244. 8289803
29. Laiho A, Kotaja N, Gyenesei A, Sironen A (2013) Transcriptome profiling of the murine testis during the first wave of spermatogenesis. PLoS ONE 8: e61558. doi: 10.1371/journal.pone.0061558 23613874
30. Schultz N, Hamra FK, Garbers DL (2003) A multitude of genes expressed solely in meiotic or spermatogenic cells offers a myriad of contraceptive targets. Proc Natl Acad Sci U S A 100 : 12201–12206. 14526100
31. Shima JE, McLean DJ, McCarrey JR, Griswold MD (2004) The murine testicular transcriptome: characterizing gene expression in the testis during the progression of spermatogenesis. Biol Reprod 71 : 319–330. 15028632
32. Horvath GC, Kistler WS, Kistler MK (2004) RFX2 is a potential transcriptional regulatory factor for histone H1t and other genes expressed during the meiotic phase of spermatogenesis. Biol Reprod 71 : 1551–1559. 15229132
33. Horvath GC, Kistler MK, Kistler WS (2009) RFX2 is a candidate downstream amplifier of A-MYB regulation in mouse spermatogenesis. BMC Dev Biol 9 : 63. doi: 10.1186/1471-213X-9-63 20003220
34. Kim MJ, Li D, Cui Y, Mueller K, Chears WC, et al. (2006) Regulatory factor interactions and somatic silencing of the germ cell-specific ALF gene. J Biol Chem 281 : 34288–34298. 16966320
35. Toscani A, Mettus RV, Coupland R, Simpkins H, Litvin J, et al. (1997) Arrest of spermatogenesis and defective breast development in mice lacking A-myb. Nature 386 : 713–717. 9109487
36. Bolcun-Filas E, Bannister LA, Barash A, Schimenti KJ, Hartford SA, et al. (2011) A-MYB (MYBL1) transcription factor is a master regulator of male meiosis. Development 138 : 3319–3330. doi: 10.1242/dev.067645 21750041
37. Kistler WS, Horvath GC, Dasgupta A, Kistler MK (2009) Differential expression of Rfx1-4 during mouse spermatogenesis. Gene Expr Patterns 9 : 515–519. doi: 10.1016/j.gep.2009.07.004 19596083
38. Hogeveen KN, Sassone-Corsi P (2006) Regulation of gene expression in post-meiotic male germ cells: CREM-signalling pathways and male fertility. Hum Fertil (Camb) 9 : 73–79.
39. Kosir R, Juvan P, Perse M, Budefeld T, Majdic G, et al. (2012) Novel Insights into the Downstream Pathways and Targets Controlled by Transcription Factors CREM in the Testis. PLoS ONE 7: e31798. doi: 10.1371/journal.pone.0031798 22384077
40. Zhou H, Grubisic I, Zheng K, He Y, Wang PJ, et al. (2013) Taf7l cooperates with Trf2 to regulate spermiogenesis. Proc Natl Acad Sci USA 110 : 16886–16891. doi: 10.1073/pnas.1317034110 24082143
41. MacGregor GR, Russell LD, Van Beek MEAB, Hanten GR, Kovac MJ, et al. (1990) Symplastic spermatids (sys): A recessive insertional mutation in mice causing a defect in spermatogenesis. Proc Natl Acad Sci USA 87 : 5016–5020. 2164218
42. Meistrich ML, Mohapatra B, Shirley CR, Zhao M (2003) Roles of transition nuclear proteins in spermiogenesis. Chromosoma 111 : 483–488. 12743712
43. Clermont Y, Oko R, Hermo L (1993) Cell Biology of Mammalian Spermatogenesis. In: Desjardins C, Ewing LL, editors. Cell and Molecular Biology of the Testis. New York: Oxford University Press. pp. 332–376.
44. Russell LD, Ettlin RA, Sinha Hikim AP, Clegg ED (1990) Histological and Histopathological Evaluation of the Testis. Clearwater: Cache River Press.
45. Berruti G, Paiardi C (2011) Acrosome biogenesis: Revisiting old questions to yield new insights. Spermatogenesis 1 : 95–98. 22319656
46. Kierszenbaum AL, Tres LL, Rivkin E, Kang-Decker N, van Deursen JMA (2004) The acroplaxome is the docking site of Golgi-derived myosin Va/Rab27a/b - containing proacrosomal vesicles in wild-type and Hrb mutant mouse spermatids. Biol Reprod 70 : 1400–1410. 14724135
47. Moreno RD, Palomino J, Schatten G (2006) Assembly of spermatid acrosome depends on microtubule organization during mammalian spermiogenesis. Dev Biol 293 : 218–227. 16540102
48. Sperry AO (2012) The dynamic cytoskeleton of the developing male germ cell. Biol Cell 104 : 297–305. doi: 10.1111/boc.201100102 22276751
49. Lee NP, Cheng CY (2004) Ectoplasmic specialization, a testis-specific actin-based adherens junction type: is this a potential target for male contraceptive development? Hum Reprod Update 10 : 349–369. 15192055
50. Russell LD, Tallon-Doran M, Weber JE, Wong V, Peterson RN (1983) Three-dimensional reconstruction of a rat stage V Sertoli Cell: III. A study of specific cellular relationships. Am J Anat 167 : 181–192. 6613903
51. Dym M, Fawcett DW (1971) Further observations on the numbers of spermatogonia, spermatocytes, and spermatids connected by intercellular bridges in the mammalian testis. Biol Reprod 4 : 195–215. 4107186
52. Kierszenbaum AL, Tres LL (2004) The acrosome-acroplaxome-manchette complex and the shaping of the spermatid head. Arch Histol Cytol 67 : 271–284. 15700535
53. Meistrich ML (1993) Nuclear Morphogenesis during Spermiogenesis. In: de Kretser DM, editor. Molecular Biology of the Male Reproductive System. New York: Academic Press. pp. 67–97.
54. Arnaiz O, Malinowska A, Kolotz C, L S, Dadlez M, et al. (2009) Cildb: a knowledgebase for centrosomes and cilia. Database (Oxford) 2009: bap022.
55. van Dam TJ, Wheway G, Slaats GG, Group SS, Huynen MA, et al. (2013) The SYSCILIA gold standard (SCGSv1) of known ciliary components and its applications within a systems biology consortium. Cilia 2 : 7. doi: 10.1186/2046-2530-2-7 23725226
56. Obholz KL, Akopyan A, Waymire KG, MacGregor GR (2006) FNDC3A is required for adhesion between spermatids and Sertoli cells. Dev Biol 298 : 498–513. 16904100
57. Gungor-Ordueri NE, Celik-Ozenci C, Cheng CY (2014) Fascin 1 is an actin filament-bundling protein that regulates ectoplasmic specialization dynamics in the rat testis. Am J Physiol Endocrinol Metab 307: E738–753. doi: 10.1152/ajpendo.00113.2014 25159326
58. Yan W (2009) Male infertility caused by spermiogenic defects: lessons from gene knockouts. Molec Cell Endocrinol 306 : 24–32. doi: 10.1016/j.mce.2009.03.003 19481682
59. Martianov I, Fimia GM, Dierich A, Parvinen M, Sassone-Corsi P, et al. (2001) Late arrest of spermiogenesis and germ cell apoptosis in mice lacking the TBP-like TLF/TRF2 gene. Mol Cell 7 : 509–515. 11463376
60. Blendy JA, Kaestner KH, Weinbauer GF, Nieschlag E, Schütz G (1996) Severe impairment of spermatogenesis in mice lacking the CREM gene. Nature 380 : 162–165. 8600391
61. Nantel F, Monaco L, Foulkes NS, Masquilier D, LeMeur M, et al. (1996) Spermiogenesis deficiency and germ-cell apoptosis in CREM-mutant mice. Nature 380 : 159–162. 8600390
62. Nantel F, Sassone-Corsi P (1996) A transcriptional master switch during the spermatogenesis differentiation program. Front Biosci 1: d266–269. 9159233
63. Martianov I, Choukrallah MA, Krebs A, Ye T, Legras S, et al. (2010) Cell-specific occupancy of an extended repertoire of CREM and CREB binding loci in male germ cells. BMC Genomics 11 : 530. doi: 10.1186/1471-2164-11-530 20920259
64. Kolthur-Seethaaram U, Martianov I, Davidson I (2008) Specialization of the general transcriptional machinery in male germ cells. Cell Cycle 7 : 3493–3498. 19001848
65. Falender AE, Freiman RN, Geles KG, Lo KC, Hwang K, et al. (2005) Maintenance of spermatogenesis requires TAF4b, a gonad-specific subunit of TFIID. Genes Dev 19 : 794–803. 15774719
66. Upadhyaya AB, Lee SH, DeJong J (1999) Identification of a general transcription factor TFIIAa/b homolog selectively expressed in testis. J Biol Chem 274 : 18040–18048. 10364255
67. White-Cooper H, Davidson I (2011) Unique aspects of transcription regulation in male germ cells. Cold Spring Harb Perspect Biol 3: a002626. doi: 10.1101/cshperspect.a002626 21555408
68. Laurencon A, Dubruille R, Efimenko E, Grenier G, Bissett R, et al. (2007) Identification of novel regulatory factor X (RFX) target genes by comparative genomics in Drosophila species. Genome Biol 8: R195. 17875208
69. Piasecki BP, Burghoorn J, Swoboda P (2010) Regulatory Factor X (RFX)-mediated transcriptional rewiring of ciliary genes in animals. Proc Natl Acad Sci U S A 107 : 12969–12974. doi: 10.1073/pnas.0914241107 20615967
70. Garcia-Gonzalo FR, Reiter JF (2012) Scoring a backstage pass: mechanisms of ciliogenesis and ciliary access. J Cell Biol 197 : 697–709. doi: 10.1083/jcb.201111146 22689651
71. Reiter JF, Blacque OE, Leroux MR (2012) The base of the cilium: roles for transition fibres and the transition zone in ciliary formation, maintenance and compartmentalization. EMBO Rep 13 : 608–618. doi: 10.1038/embor.2012.73 22653444
72. Lobo LJ, Zariwala MA, Noone PG (2014) Primary ciliary dyskinesia. QJM 107 : 691–699. doi: 10.1093/qjmed/hcu063 24652656
73. Meikar O, Vagin VV, Chalmel F, Sõstar K, Lardenois A, et al. (2014) An atlas of chromatoid body components. RNA 20 : 483–495. doi: 10.1261/rna.043729.113 24554440
74. Castañeda J, Genzor P, van der Heijden GW, Sarkeshik A, Yates JR 3rd, et al. (2014) Reduced pachytene piRNAs and translation underlie spermiogenic arrest in Maelstrom mutant mice. EMBO J.
75. Li XZ, Roy CK, Dong X, Bolcun-Filas E, Wang J, et al. (2013) An ancient transcription factor initiates the burst of piRNA production during early meiosis in mouse testes. Mol Cell 50 : 67–81. doi: 10.1016/j.molcel.2013.02.016 23523368
76. Wong EW, Cheng CY (2009) Polarity proteins and cell-cell interactions in the testis. Int Rev Cell Mol Biol 278 : 309–353. doi: 10.1016/S1937-6448(09)78007-4 19815182
77. Gliki G, Ebnet K, Aurrand-Lions M, Imhof BA, Adams RH (2004) Spermatid differentiation requires the assembly of a cell polarity complex downstream of junctional adhesion molecule-C. Nature 431 : 320–324. 15372036
78. O'Donnell L, O'Bryan MK (2014) Microtubules and spermatogenesis. Semin Cell Dev Biol.
79. Sugioka K, Sawa H (2012) Formation and functions of asymmetric microtubule organization in polarized cells. Curr Opin Cell Biol 24 : 517–525. doi: 10.1016/j.ceb.2012.05.007 22704717
80. Rodriguez CI, Buchholz F, Galloway J, Sequerra R, Kasper J, et al. (2000) High-efficiency deleter mice show that FLPe is an alternative to Cre-loxP. Nat Genet 25 : 139–140. 10835623
81. Ma W, Horvath GC, Kistler MK, Kistler WS (2008) Expression patterns of SP1 and SP3 during mouse spermatogenesis: SP1 down-regulation correlates with two successive promoter changes and translationally compromised transcripts. Biol Reprod 79 : 289–300. doi: 10.1095/biolreprod.107.067082 18417714
82. Oko RJ, Jando V, Wagner CL, Kistler WS, Hermo LS (1996) Chromatin reorganization in rat spermatids during the disappearance of testis-specific histone, H1t, and the appearance of transition proteins TP1 and TP2. Biol Reprod 54 : 1141–1157. 8722637
83. Hann C, Behrmann I (2007) A cost effective non-commercial ECL-solution for Western blot detections yielding strong signals and low background. J Immunol Methods 318 : 11–19. 17141265
84. Gage GJ, Kipke DR, Shain W (2012) Whole animal perfusion fixation for rodents. J Vis Exp.
Štítky
Genetika Reprodukční medicína
Článek Discovery and Fine-Mapping of Glycaemic and Obesity-Related Trait Loci Using High-Density ImputationČlánek AAA-ATPase FIDGETIN-LIKE 1 and Helicase FANCM Antagonize Meiotic Crossovers by Distinct MechanismsČlánek A Conserved Pattern of Primer-Dependent Transcription Initiation in and Revealed by 5′ RNA-seqČlánek TopBP1 Governs Hematopoietic Stem/Progenitor Cells Survival in Zebrafish Definitive HematopoiesisČlánek Redundant Roles of Rpn10 and Rpn13 in Recognition of Ubiquitinated Proteins and Cellular Homeostasis
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2015 Číslo 7
-
Všechny články tohoto čísla
- LINE-1 Retroelements Get ZAPped!
- /p23: A Small Protein Heating Up Lifespan Regulation
- Hairless Streaks in Cattle Implicate TSR2 in Early Hair Follicle Formation
- Ribosomal Protein Mutations Result in Constitutive p53 Protein Degradation through Impairment of the AKT Pathway
- Molecular Clock of Neutral Mutations in a Fitness-Increasing Evolutionary Process
- Modeling Implicates in Nephropathy: Evidence for Dominant Negative Effects and Epistasis under Anemic Stress
- The Alternative Sigma Factor SigX Controls Bacteriocin Synthesis and Competence, the Two Quorum Sensing Regulated Traits in
- BMP Inhibition in Seminomas Initiates Acquisition of Pluripotency via NODAL Signaling Resulting in Reprogramming to an Embryonal Carcinoma
- Comparative Study of Regulatory Circuits in Two Sea Urchin Species Reveals Tight Control of Timing and High Conservation of Expression Dynamics
- EIN3 and ORE1 Accelerate Degreening during Ethylene-Mediated Leaf Senescence by Directly Activating Chlorophyll Catabolic Genes in
- Genome Wide Binding Site Analysis Reveals Transcriptional Coactivation of Cytokinin-Responsive Genes by DELLA Proteins
- Sensory Neurons Arouse . Locomotion via Both Glutamate and Neuropeptide Release
- A Year of Infection in the Intensive Care Unit: Prospective Whole Genome Sequencing of Bacterial Clinical Isolates Reveals Cryptic Transmissions and Novel Microbiota
- Inference of Low and High-Grade Glioma Gene Regulatory Networks Delineates the Role of Rnd3 in Establishing Multiple Hallmarks of Cancer
- Novel Role for p110β PI 3-Kinase in Male Fertility through Regulation of Androgen Receptor Activity in Sertoli Cells
- A Novel Locus Harbouring a Functional Nonsense Mutation Identified in a Large Danish Family with Nonsyndromic Hearing Impairment
- Checkpoint Activation of an Unconventional DNA Replication Program in
- A Genetic Incompatibility Accelerates Adaptation in Yeast
- The SMC Loader Scc2 Promotes ncRNA Biogenesis and Translational Fidelity
- Blimp1/Prdm1 Functions in Opposition to Irf1 to Maintain Neonatal Tolerance during Postnatal Intestinal Maturation
- Discovery and Fine-Mapping of Glycaemic and Obesity-Related Trait Loci Using High-Density Imputation
- JAK/STAT and Hox Dynamic Interactions in an Organogenetic Gene Cascade
- Emergence, Retention and Selection: A Trilogy of Origination for Functional Proteins from Ancestral LncRNAs in Primates
- MoSET1 (Histone H3K4 Methyltransferase in ) Regulates Global Gene Expression during Infection-Related Morphogenesis
- Arabidopsis PCH2 Mediates Meiotic Chromosome Remodeling and Maturation of Crossovers
- AAA-ATPase FIDGETIN-LIKE 1 and Helicase FANCM Antagonize Meiotic Crossovers by Distinct Mechanisms
- A Conserved Pattern of Primer-Dependent Transcription Initiation in and Revealed by 5′ RNA-seq
- Tempo and Mode of Transposable Element Activity in Drosophila
- The Shelterin TIN2 Subunit Mediates Recruitment of Telomerase to Telomeres
- SAMHD1 Inhibits LINE-1 Retrotransposition by Promoting Stress Granule Formation
- A Genome Scan for Genes Underlying Microgeographic-Scale Local Adaptation in a Wild Species
- TopBP1 Governs Hematopoietic Stem/Progenitor Cells Survival in Zebrafish Definitive Hematopoiesis
- Analysis of the Relationships between DNA Double-Strand Breaks, Synaptonemal Complex and Crossovers Using the Mutant
- Assessing Mitochondrial DNA Variation and Copy Number in Lymphocytes of ~2,000 Sardinians Using Tailored Sequencing Analysis Tools
- Allelic Spectra of Risk SNPs Are Different for Environment/Lifestyle Dependent versus Independent Diseases
- CSB-PGBD3 Mutations Cause Premature Ovarian Failure
- Irrepressible: An Interview with Mark Ptashne
- Genetic Evidence for Function of the bHLH-PAS Protein Gce/Met As a Juvenile Hormone Receptor
- Inactivation of Retinoblastoma Protein (Rb1) in the Oocyte: Evidence That Dysregulated Follicle Growth Drives Ovarian Teratoma Formation in Mice
- Redundant Roles of Rpn10 and Rpn13 in Recognition of Ubiquitinated Proteins and Cellular Homeostasis
- Pyrimidine Pool Disequilibrium Induced by a Cytidine Deaminase Deficiency Inhibits PARP-1 Activity, Leading to the Under Replication of DNA
- Molecular Framework of a Regulatory Circuit Initiating Two-Dimensional Spatial Patterning of Stomatal Lineage
- RFX2 Is a Major Transcriptional Regulator of Spermiogenesis
- A Role for Macro-ER-Phagy in ER Quality Control
- Corp Regulates P53 in via a Negative Feedback Loop
- Common Cell Shape Evolution of Two Nasopharyngeal Pathogens
- Contact- and Protein Transfer-Dependent Stimulation of Assembly of the Gliding Motility Machinery in
- Endothelial Snail Regulates Capillary Branching Morphogenesis via Vascular Endothelial Growth Factor Receptor 3 Expression
- Functional Constraint Profiling of a Viral Protein Reveals Discordance of Evolutionary Conservation and Functionality
- Temporal Coordination of Carbohydrate Metabolism during Mosquito Reproduction
- mTOR Directs Breast Morphogenesis through the PKC-alpha-Rac1 Signaling Axis
- Reversible Oxidation of a Conserved Methionine in the Nuclear Export Sequence Determines Subcellular Distribution and Activity of the Fungal Nitrate Regulator NirA
- Nutritional Control of DNA Replication Initiation through the Proteolysis and Regulated Translation of DnaA
- Cooperation between Paxillin-like Protein Pxl1 and Glucan Synthase Bgs1 Is Essential for Actomyosin Ring Stability and Septum Formation in Fission Yeast
- Encodes a Highly Conserved Protein Important to Neurological Function in Mice and Flies
- Identification of a Novel Regulatory Mechanism of Nutrient Transport Controlled by TORC1-Npr1-Amu1/Par32
- Aurora-A-Dependent Control of TACC3 Influences the Rate of Mitotic Spindle Assembly
- Large-Scale Phenomics Identifies Primary and Fine-Tuning Roles for CRKs in Responses Related to Oxidative Stress
- TFIIS-Dependent Non-coding Transcription Regulates Developmental Genome Rearrangements
- Genome-Wide Reprogramming of Transcript Architecture by Temperature Specifies the Developmental States of the Human Pathogen
- Identification of Chemical Inhibitors of β-Catenin-Driven Liver Tumorigenesis in Zebrafish
- The Catalytic and Non-catalytic Functions of the Chromatin-Remodeling Protein Collaborate to Fine-Tune Circadian Transcription in
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Functional Constraint Profiling of a Viral Protein Reveals Discordance of Evolutionary Conservation and Functionality
- Reversible Oxidation of a Conserved Methionine in the Nuclear Export Sequence Determines Subcellular Distribution and Activity of the Fungal Nitrate Regulator NirA
- Modeling Implicates in Nephropathy: Evidence for Dominant Negative Effects and Epistasis under Anemic Stress
- Nutritional Control of DNA Replication Initiation through the Proteolysis and Regulated Translation of DnaA
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



