-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaThe Developmental Intestinal Regulator ELT-2 Controls p38-Dependent Immune Responses in Adult .
C. elegans provides a tractable genetic model to study the regulation of the evolutionarily conserved innate immune system. One of the central signaling modules of innate immunity in all organisms is the p38 pathway, which has been studied extensively in C. elegans. Such studies identified the transcription factors ATF-7 and SKN-1 as proteins mediating downstream effects of the p38 pathway on immune and oxidative stress gene expression. Previous studies in C. elegans also identified ELT-2, a conserved transcription factor important for intestinal development, as a major regulator of immune responses in the adult worm. The current study aimed to characterize the interactions between these two immune regulatory modules. Microarray gene expression analysis in animals with disrupted elt-2 expression revealed two gene subsets that were regulated by elt-2: one that included constitutively regulated genes, and was mostly comprised of digestive enzyme genes, and a second that included genes induced by infection with Pseudomonas aeruginosa. Both subsets were enriched for p38 targets. Genetic analyses and gene expression measurements of elt-2-regulated genes demonstrated that elt-2 cooperates with the p38 pathway and its downstream mediators. These results suggest that ELT-2 functions as a tissue-specific master regulator controlling the contribution of the p38 MAPK pathway to innate immune responses.
Published in the journal: . PLoS Genet 11(5): e32767. doi:10.1371/journal.pgen.1005265
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1005265Summary
C. elegans provides a tractable genetic model to study the regulation of the evolutionarily conserved innate immune system. One of the central signaling modules of innate immunity in all organisms is the p38 pathway, which has been studied extensively in C. elegans. Such studies identified the transcription factors ATF-7 and SKN-1 as proteins mediating downstream effects of the p38 pathway on immune and oxidative stress gene expression. Previous studies in C. elegans also identified ELT-2, a conserved transcription factor important for intestinal development, as a major regulator of immune responses in the adult worm. The current study aimed to characterize the interactions between these two immune regulatory modules. Microarray gene expression analysis in animals with disrupted elt-2 expression revealed two gene subsets that were regulated by elt-2: one that included constitutively regulated genes, and was mostly comprised of digestive enzyme genes, and a second that included genes induced by infection with Pseudomonas aeruginosa. Both subsets were enriched for p38 targets. Genetic analyses and gene expression measurements of elt-2-regulated genes demonstrated that elt-2 cooperates with the p38 pathway and its downstream mediators. These results suggest that ELT-2 functions as a tissue-specific master regulator controlling the contribution of the p38 MAPK pathway to innate immune responses.
Introduction
Induction of local innate immune responses is the first reaction to an invading pathogen, and includes increased expression of antimicrobial effector peptides/proteins, as well as immune modulators. Regulation of these responses depends on signaling modules that are similar in their principles of action from plants to animals, suggesting convergent evolution [1]. Within the animal kingdom these signaling modules often use similar proteins, such as pattern recognition receptors, their downstream signaling cascades, and MAP kinase signaling pathways [2,3]. This conservation warrants the study of innate immune mechanisms in well-characterized invertebrate model organisms, such as Drosophila melanogaster and Caenorhabditis elegans, to better understand their vertebrate counterparts.
Studies of C. elegans immunity have repeatedly converged on the p38 MAPK pathway as a pivotal module in orchestrating immune responses, very similar to its roles in vertebrate innate immune responses [4–7]. The core components of the C. elegans p38 pathway include the NSY-1 MAP3K, the SEK-1 MAP2K, and the PMK-1 MAPK. TIR-1/SARM was shown to serve as an upstream activator during infection [8,9], and VHP-1/DUSP8, as a negative regulator [10]. Downstream to the p38 pathway, several transcription factors have been shown to mediate effects on gene expression: ATF-7, an ATF-2 ortholog, was shown to regulate immune gene expression in the intestine [11]; DAF-19/RFX, was shown to cooperate with ATF-7 in regulating genes involved in neuronal serotonin synthesis, but was also found to contribute to expression of intestinal immune genes [12]; SKN-1/Nrf, better known for regulating oxidative stress responses, was further found to contribute to resistance against bacterial pathogens [13–15]. In addition, ELT-3 was identified as a regulator of epidermal anti-fungal responses, a subset of which was also regulated by the p38 pathway [16].
ELT-3 is one of two C. elegans transcription factors of the GATA family with roles in epithelial development and differentiation, and additional roles in regulating immune responses. ELT-3 is important for epidermal differentiation and epidermis-specific gene expression [17]. The second GATA protein is ELT-2, which is important for terminal development of the intestine and for intestine-specific gene expression [18,19]. Whereas ELT-2 was proposed to be the predominant regulator of all intestinal gene expression, experiments supporting this were performed only in embryos or L1 larvae, leaving the extent of its roles in the adult intestine unresolved [20,21]. We, and others, have shown that ELT-2 regulated specific anti-bacterial responses in the adult intestine [22–24]. Similar roles, both in endodermal development, as well as in adult immune regulation and protection, were described for the Drosophila GATA protein Serpent and for the vertebrate GATA6 [22,25].
Vertebrate GATA transcription factors comprise two homology groups: GATA1-3 are regulators of lymphocyte terminal differentiation and cytokine expression; GATA4-6 are regulators of mesodermal and endodermal differentiation (in the heart, liver, lung, and pancreas), and are considered the orthologs of elt-2 [26,27]. In the adult endoderm, GATA4 and GATA6 were also shown to play key roles in the regulation of stress responses [28,29]. Importantly, MAPK signaling, including signals from the p38 pathway, regulates the activity of GATA4 during stress responses [30]. Thus, it is possible that ELT-2 is similarly regulated during infection.
To better understand the roles of ELT-2 in the adult intestine, particularly its involvement in immune gene regulation, we characterized gene expression following elt-2 knock-down specifically in adults. This identified two gene subsets: one that was constitutively regulated by ELT-2 and included genes involved in digestive degradation of macromolecules; and a second, which was induced in response to infection, and included genes previously implicated in protection from pathogens. Members of the latter demonstrated co-regulation by ELT-2 and the p38 pathway. Subsequent genetic analyses identified genetic interactions between elt-2 and the p38 transcriptional mediator genes atf-7 and skn-1 in regulating C. elegans innate immune responses. Our results suggest a dominant role for elt-2 in the regulation of digestive and metabolic functions of the intestine, and the role of a master regulator for p38-dependent immune responses, cooperating with activated transcription factors to control induced responses.
Results
The constitutive and inducible elt-2 regulon
To identify genes regulated by elt-2, we compared gene expression profiles in animals fed with elt-2 RNAi during the first two days of adulthood (RNAi-ad) to those in control-treated animals, either following a twelve hour infection with Pseudomonas aeruginosa, or exposure to non-pathogenic E. coli (Raw data can be downloaded from GEO, accession no. GSE63846). Adult elt-2 knock-down has been shown to cause a marked decrease in ELT-2 protein levels persisting up to three days after worms were removed from RNAi plates [22]. Successful knock-down is also discernible by eye, as animals present a modest ‘clear’ phenotype, potentially due to reduced fat storage (S1A Fig). Previous work found elt-2(RNAi-ad) animals to be more susceptible to infection, but to have a normal lifespan on dead E. coli, suggesting that effects of post-developmental elt-2 knock-down are largely immune-specific [22].
Microarray analysis identified 429 transcripts, corresponding to 420 genes, which were differentially expressed in elt-2(RNAi) animals compared to control-treated animals (Fig 1A). Prominent clusters of co-regulated genes included a cluster of 187 genes with reduced expression following elt-2 knock-down (‘elt-2-regulated’), suggesting contribution of elt-2 to constitutive expression (Fig 1A and S2 Table); a cluster of 96 genes, that were also suppressed following elt-2 knock-down, and additionally failed to be induced by infection in elt-2(RNAi) animals (‘elt-2-induced’); and a cluster of 43 genes showing elevated expression following elt-2 knock-down, suggesting repression by the transcription factor (‘elt-2-repressed’). qRT-PCR verified elt-2 regulation for three selected ‘elt-2-regulated’, and seven ‘elt-2-induced’ genes (S2A and S2B Fig). Additional measurements for ‘elt-2-induced’ genes in animals exposed to the pathogen for a longer duration (24 hours) similarly showed no infection response in elt-2(RNAi) animals, suggesting that impaired induction represented a complete failure rather than a delay (S2C Fig).
Fig. 1. elt-2 regulated genes. 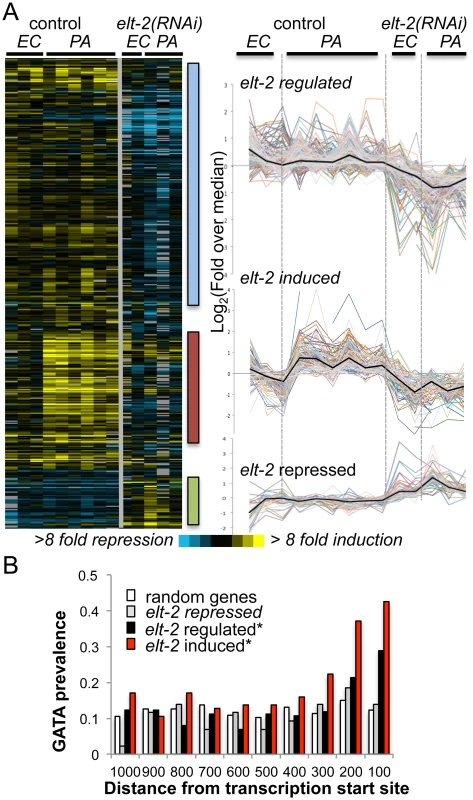
A. microarray analysis. Gene expression profiles for 426 transcripts differentially expressed between elt-2(RNAi) and control-treated wildtype worms, when exposed to P. aeruginosa (PA, 12 hours), compared to E. coli (EC). Left, heatmap of raw values (log2(fold change over reference RNA)), with bars highlighting clusters of interest; right, curves depicting median-centered expression profiles; black curves represent the median. B. GATA motif distribution. Measured for the consensus TGATAA in 1000bp upstream sequences of genes of the designated subsets; shown as #motifs/gene/100bp; asterisks mark significant deviations from random distribution (p<10–8, χ2). To identify potential direct ELT-2 targets in the three subsets, we searched gene promoters for the GATA motif core sequence, TGATAA [20,22]. GATA motifs are prevalent in the genome, as targets for various developmental and tissue-specific transcription factors. However, an examination of GATA motif distribution in upstream sequences of elt-2-dependent genes revealed a statistically-significant enrichment for GATA motifs in proximal promoter regions, in contrast to a uniform distribution in upstream regions of randomly-selected genes (Fig 1B and S3 Table). Focusing on proximal promoter regions (500 bp) to better differentiate between elt-2 targets and non-targets, GATA motifs were identified in 72% of the ‘elt-2 induced’ genes, 50% of ‘elt-2 regulated’ genes, and 47% of ‘elt-2 repressed’, compared to 42% in upstream sequences genome-wide, demonstrating a significant enrichment for GATA motifs in promoters of elt-2-induced and elt-2-regulated genes, but not among ‘elt-2 repressed’ genes (p = 5.6E-10, 0.004 and 0.1, respectively; hypergeometric distribution). Twelve of the GATA-containing genes were among those tested by qRT-PCR (nine of the ‘elt-2-induced’ subset, and three of the ‘elt-2-regulated’ subset) and indeed demonstrated elt-2-dependent expression (S2 Fig). In addition to enrichment of GATA promoter motifs, 55% of the ‘elt-2 induced’, and 32% of the ‘elt-2 regulated’ genes were genes previously reported to be preferentially expressed in the intestine [19,20,31](and Wormbase); only 6/43 (14%) of the ‘elt-2 repressed’ genes were intestinal, while 12/43 were genes shown to be preferentially-expressed in muscle tissue [32]. Together, these analyses suggest that a large fraction of the elt-2 regulated genes, in particular of the ‘elt-2-induced’ genes, are direct ELT-2 targets. Nevertheless, some ‘noise’ is included in these subsets in the form of genes that are indirectly affected by elt-2 knock-down. In the case of ‘elt-2-repressed’ genes it seems that most are affected indirectly, and probably outside of the intestine, suggesting a negligible contribution of elt-2 to direct gene repression.
To learn about potential contributions of putative ELT-2 targets to worm physiology, we next examined their associated GO annotations. Among the ‘elt-2-regulated’ genes enrichment was found for genes involved in innate immunity and defense responses (represented by 13 genes, p = 8.48E-05, Bonferroni corrected), and for genes with hydrolase activity (37 genes, p = 0.038, not corrected)(S4 Table, highlighted in yellow). The former are genes that were previously shown to respond to infection [22], suggesting that they might have been inappropriately assigned as ‘elt-2 regulated’ due to a weak response or noisy measurements, and were more likely to be part of the ‘elt-2 induced’ subset. The more telling members of the ‘elt-2 regulated’ subset appeared to be the hydrolase genes, which mainly included proteases and lipases, and pointed at regulation of these enzymes as an important function of elt-2 in adults. Regulation of three of these enzymes by elt-2 was confirmed by qRT-PCR (S2A Fig). While enrichment for genes annotated as hydrolases is not strictly statistically significant, this may be due to the noise in the ‘elt-2-regulated’ list. Supporting the central role of elt-2 in regulating hydrolytic enzymes in the adult intestine, the overlap between the ‘elt-2-regulated‘ gene list and a previously published list of genes specifically expressed in the adult intestine [20] consisted of fifteen genes, four of which are associated with immune defense functions, seven that encode hydrolytic enzymes, and four unknowns (S4 Table).
In embryos, elt-2 has been shown to contribute significantly to expression of genes encoding structural intestinal proteins [20,33]. However, in agreement with previous results, our microarray data did not reveal effects of elt-2 knock-down in adults on the expression of act-5 (microvilli structure), let-413 (adherens junctions), eps-8 (apical morphogenesis), and ifb-2 (intestinal-specific intermediate filament)[22]. In addition, qRT-PCR analysis found no effect of elt-2 knock-down on the expression of non-hydrolytic genes previously shown to be expressed in the adult intestine: lmp-1 (lysosomal membrane), mrp-5 (membranal transport), and ubl-1 (possibly involved in protein translation)(S3 Fig), whereas hydrolytic enzyme gene expression was reduced in the same RNA samples (as shown in S2A Fig). Together, this indicated that elt-2 was necessary for specific functions in the adult intestine, but not for all.
ELT-2 was previously shown to function synergistically with ELT-7—a co-expressed intestinal GATA transcription factor—in morphological gut differentiation and in larval gut-specific gene expression [21]. It is possible that redundancy between elt-2 and elt-7 masked additional contributions of elt-2 to intestinal gene expression. Nevertheless, the results presented highlight elt-2’s dominant contribution to hydrolytic gene expression.
For the ‘elt-2-induced’ gene subset, all enriched ‘process’ GO annotations were related to defense and innate immune responses (22 genes, p = 1.5E-15)(S4 Table). In addition, ten genes of this subset were annotated with carbohydrate binding, most of which are lectins, which are known to take part in C. elegans innate immune responses, and have been suggested to play roles in pathogen recognition [34]. These enriched annotations support the dominant role previously proposed for elt-2 in regulating intestinal innate immune responses. Interestingly, elt-7 is a member of the ‘elt-2-induced’ subset, suggesting participation in immune responses; however, previous work could not identify any significant contribution of elt-7 to immune protection [22].
Genetic interactions between elt-2 and the p38 pathway
ELT-2 acts as a regulator of intestinal development following activation of its expression. This expression is maintained in adults, possibly through autoregulation [35]. ELT-2 was previously shown to be constitutively nuclear [35]. Therefore, to take part in regulation of induced responses (as demonstrated for ‘elt-2-induced’ genes) its activity must be modulated by some signal transduction pathway(s). A likely candidate is the p38 pathway, which is known to play an important role in regulating C. elegans immune responses [4]. Among genes previously described to be regulated downstream to the MAPKK gene sek-1 or the p38 MAPK gene pmk-1 [36], and included in our filtered dataset, 38% (22/57) and 33% (13/39), respectively, were also regulated by elt-2 (p<4E-8)(Fig 2A). This suggested that elt-2 co-regulated genes with the p38 pathway, potentially downstream to it. To examine this possibility, we knocked down elt-2 in adult sek-1(km4) mutants. While elt-2 knock-down significantly decreased resistance in wildtype animals, its effect on the already compromised resistance of sek-1mutants was marginal (Fig 2B). The fact that overlap between p38 and elt-2 targets was only partial could reflect technical differences between the two studies, resulting in different coverage of the respective datasets; additionally, it may reflect partially aligned regulatory programs, with some contributions to gene expression that are independent of each other. The survival analysis, showing only marginal exacerbation of infection susceptibility of sek-1 mutants by elt-2 RNAi is more consistent with the first possibility.
Fig. 2. elt-2 and the p38 pathway co-regulate immune protection. 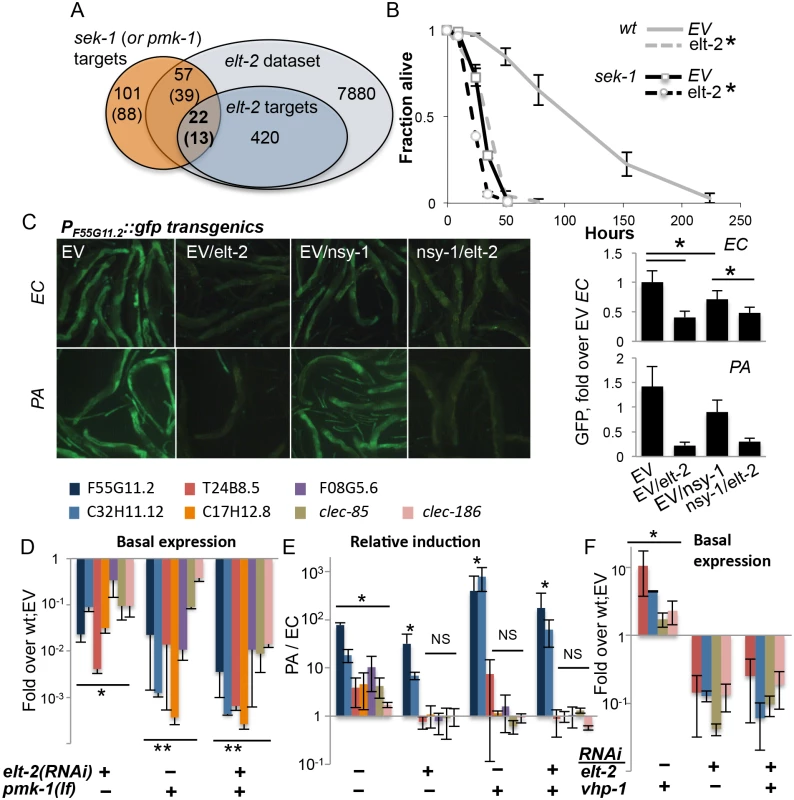
A. Overlaps between sek-1 (or pmk-1) targets (Troemel et al., 2006), and elt-2 targets; gene numbers are shown. B, Survival curves for wildtype and sek-1 animals fed with designated RNAi’s during early adulthood, followed by P. aeruginosa infection; averages ± SDs for three plates (N = 92–151 per group,*p<0.0001 (Logrank test)); shown is a representative of several experiments with similar results. C, PF55G11.2::gfp worms fed with RNAi as designated during adulthood and exposed to P. aeruginosa (PA, 4 hours, N = 10–19 per group) or E. coli (EC, N = 19–25); signal quantification shown on the right, *p<3xE-6, ttest. Shown is a representative experiment of two with similar results. D-F, Gene expression in wildtype or pmk-1(km25) loss-of-function animals fed with designated RNAi’s during larval development. Shown are averages and SDs for two independent experiments D,F, basal expression (values and statistics relative to values in wt;EV (set to 1 and therefore not shown). E, Induction following 12 hours of P. aeruginosa infection, relative to basal expression in similarly-treated worms grown on E. coli. *p<0.01, **p<0.0005 (paired t-test); asterisks mark significant differences for individual gene(s), or for each of the genes in a group designated by a line. We next turned to gene expression, to further examine the relationship between the elt-2 and p38 regulatory modules. We began by examining the expression of a GFP reporter controlled by the promoter of F55G11.2, an early immune response gene regulated by both elt-2 and the p38 pathway [22,36]. RNAi knock-down in adult worms demonstrated that both the p38 MAP3K gene nsy-1, and more so elt-2 were necessary for basal expression from the F55G11.2 promoter (Fig 2C). In response to P. aeruginosa, F55G11.2 induction was apparent within four hours in control-treated animals, but not in elt-2 knock-down animals. Disruption of nsy-1 also reduced immune induction, but not as much as elt-2 disruption. Similar results were observed in pmk-1(km25) mutants, corroborating the co-regulation of F55G11.2 by p38 signaling and elt-2, and the dominant contribution of elt-2 to its expression (S4 Fig).
Using mutants carrying the pmk-1(km25) null allele, we expanded our analysis (and increased its sensitivity) by employing qRT-PCR to follow expression of genes potentially co-regulated by p38 signaling and elt-2. Because p38-dependent responses are more pronounced in younger worms [37], we measured gene expression at the end of larval development. And while knock-down of elt-2 during development has more pronounced effects than during adulthood, giving rise to scrawny worms (S1B Fig), elt-2(RNAi-dev) worms are healthy enough to reach adulthood and lay eggs. Expression was measured for F55G11.2, and for genes that were part of the overlap between elt-2 and p38 targets (Fig 2A): C32H11.12 (‘elt-2-induced’), T24G8.5, clec-85 and clec-186 (all three ‘elt-2-regulated’ according to the microarray analysis, and infection-induced in younger animals according to [22]). Two additional p38 targets were included, C17H12.8, and F08G5.6, the latter of which was previously shown to provide protection from infection [22]. All examined genes included proximal-promoter GATA motifs. qRT-PCR demonstrated that the seven genes were all regulated by both elt-2 and pmk-1. Basal expression was significantly reduced following elt-2 knock-down, compared to age-matched control-treated animals, and was similarly reduced in pmk-1 mutants (Fig 2D). A twelve-hour exposure to P. aeruginosa induced the expression of all seven in wildtype animals, but the regulation of this induction divided the genes into two subsets. Induction of 5/7 genes was abolished by either pmk-1 or elt-2 disruption, indicating dependence on the two factors. However, F55G11.2 and C32H11.12, which depended on pmk-1 or elt-2 for basal expression, were significantly induced above basal levels, even when both pmk-1 and elt-2 were disrupted, suggesting that F55G11.2 and C32H11.12 may be regulated by additional factor(s)(Fig 2E). The relative induction observed in these experiments was not apparent in the GFP reporter strain, presumably due to the increased sensitivity of qRT-PCR compared to fluorescence measurements. Similar experiments were performed with adult worms, which showed significantly lower gene induction during infection, but otherwise, similar contributions of elt-2 and pmk-1 to gene expression (S5 Fig). Lastly, whether elt-2 disruption can exacerbate gene repression in pmk-1 mutants is not clear, since additive effects were observed in two-day adults (S5 Fig), but not in L4 larvae (Fig 2E).
Survival and gene expression analyses in L4 larvae suggested that elt-2 may be epistatic to pmk-1. To examine whether elt-2 knock-down could abrogate pmk-1-dependent gene expression, we knocked down vhp-1, which encodes a phosphatase that dephosphorylates and inactivates PMK-1 [10]. Accordingly, knock-down of vhp-1 caused a significant induction of T24B8.5, C32H11.12, clec-85 and clec-186 (Fig 2F). Simultaneous knock-down of elt-2 abrogated this induction. This was not due to reduced efficiency of vhp-1 RNAi in a double knock-down setting, as vhp-1 knock-down was able to induce gene expression when mixed with another RNAi (see below). Instead, these results suggested that elt-2 was essential for pmk-1 dependent immune gene expression.
Interactions between elt-2 and downstream mediators of the p38 pathway
ATF-7
ATF-7 was reported to regulate gene expression downstream of PMK-1. Normally a repressor of gene expression, its pmk-1-dependent phosphorylation during infection transforms it to an activator [11]. Worms carrying the atf-7(qd22qd130) loss-of-function allele were reported to be impaired for both gene repression and activation. Given the proposed involvement of elt-2 in pmk-1-dependent immune gene expression, it was of interest to examine how elt-2 interacted with atf-7. Survival analysis showed that elt-2 knock-down in developing atf-7 mutants only marginally exacerbated infection susceptibility, as in pmk-1 mutants (Fig 3A); similar results were observed in worms treated with elt-2 RNAi during adulthood (S6A Fig).
Fig. 3. elt-2 is essential for atf-7-dependent immune gene regulation. 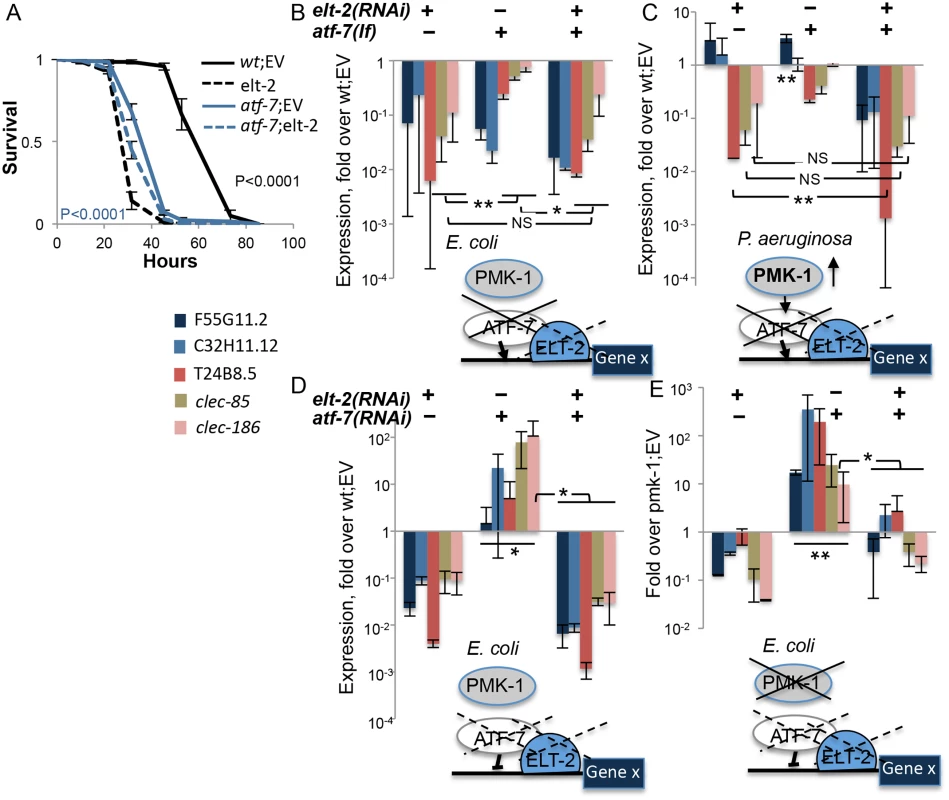
A. Survival curves for wildtype and atf-7(qd22qd130) animals fed with designated RNAi’s during development followed by infection. Averages ± SDs for three plates (N = 129–140 per group) in a representative experiment of several others with similar results. B-E, Gene expression (log scale) in wildtype, pmk-1(km25), and atf-7(qd22qd130) animals, fed with designated RNAi’s during development. Models depict for each panel the mode of disruption, or status, of examined factors (solid-line crosses, loss-of-function mutants; dashed-line crosses, knock-down), placing ELT-2 tentatively at the proximal promoter of immune genes (Gene X) putatively regulated by PMK-1 and ATF-7; atf-7 is depicted as an activator (arrow) or repressor (blunt-ended arrow), based on disruption effects on gene expression. RNA levels were measured in L4/YA worms. Each panel presents averages and SDs for two independent experiments. Asterisks mark inter-group significance with *p<0.05, **p<0.01, and NS, non-significant (paired t-test) for all genes in the group or in underlined subset. Using atf-7(qd22qd130) and pmk-1(km25) mutants, in combination with elt-2 or atf-7 knock-down, qRT-PCR was employed to examine the involvement of elt-2 in pmk-1/atf-7 dependent gene expression. Under normal conditions (growth on E. coli), both elt-2(RNAi) animals and atf-7 mutants showed a strong reduction in immune gene expression compared to wildtype animals (Fig 3B). Similar results were observed in two-day old adults (S6B Fig). Whereas atf-7 is expected to function as a repressor under normal conditions, the results suggested that it was necessary (as was elt-2) for activating gene expression; this is depicted in the model accompanying Fig 3B. Since E. coli strain OP50-1 has been previously reported to be weakly pathogenic [38], it is possible that under basal conditions wildtype ATF-7 functions mostly as an activator. While atf-7 and elt-2 appeared to regulate the same genes, the relationship between them was not immediately apparent: additive contributions of the two were suggested by expression patterns of F55G11.2 and C32H11.12, but dominance of elt-2 was suggested by expression patterns of T24B8.5, clec-85 and clec-186, for which elt-2 knock-down reduced gene expression in wildtype worms or atf-7 mutants to the same extent with no additive effects.
A similar dichotomy in the relationship between atf-7 and elt-2 in regulating target gene expression was observed following infection of wildtype and atf-7 worms with P. aeruginosa, which is known to activate PMK-1 (Fig 3C model), and normally induces the expression of all examined genes (Fig 2E). F55G11.2 and C32H11.12 were modestly induced in response to the pathogen even when elt-2 was knocked down, or in atf-7 mutants (F55G11.2 only)(Fig 3C). Only a double disruption decreased expression of the two genes to levels below those observed in wildtype animals and abolished induction. This result corroborated the roles of atf-7 and elt-2 in positive regulation of immune gene expression, and suggested that for some immune response genes the two factors may provide independent inputs. On the other hand, clec-85, clec-186 and T24B8.5 failed to be induced either in atf-7 mutants or in elt-2(RNAi) animals, and showed in both cases lower RNA levels compared to wildtype control animals, with stronger effects of elt-2 disruption, and mostly with no additive effects of atf-7 disruption (with the exception of T24B8.5). This suggested that in the regulation of other immune genes elt-2 and atf-7 were epistatic.
While experiments in atf-7 loss-of-function mutants pointed at roles in gene activation, atf-7 knock-down experiments in wildtype animals exposed its contributions to gene repression. Knock-down of atf-7 during larval development resulted in derepression, albeit variable, of all examined genes (Fig 3D). This was abolished by elt-2 knock-down. Strong derepression was observed only when atf-7 was knocked down in pmk-1 mutants, when all ATF-7 molecules are expected to be unphosphorylated and therefore in repressive mode (Fig 3E). Again, elt-2 knock-down completely abrogated this derepression, supporting the notion that elt-2 is essential for expression of atf-7-regulated genes.
The results presented in Fig 3 demonstrate that elt-2 is important for atf-7-dependent immune gene expression, basal and induced. In particular, gene expression measurements in pmk-1 mutants suggest that elt-2 is a master regulator without which atf-7-dependent genes cannot be expressed effectively. When ATF-7 was activated, primarily during exposure to P. aeruginosa, but to a lesser degree also on E. coli, it co-regulated genes together with elt-2, demonstrating additive contributions for some genes, but not for others.
SKN-1
While the expression of clec-85, clec-186 and T24B8.5 were fully explained by contributions from elt-2 and atf-7 downstream to the p38 pathway, the expression of F55G11.2 and C32H11.12 was not, and induction, relative to basal expression levels, was still observed when all three were disrupted (Figs 2E and 3C). C32H11.12 was previously shown to be regulated by intestinal SKN-1, and the F55G11.2 promoter is bound by this transcription factor [39,40]. SKN-1 mediates p38-dependent responses to oxidative stress, but was also shown to contribute to immune protection [13–15]. Therefore, we examined whether skn-1 contributed to the expression of the two genes. Both F55G11.2 and C32H11.12 were repressed when any one of elt-2, skn-1, or atf-7 was disrupted (Fig 4A), with accumulating additive effects. However, their infection-induced expression was not significantly reduced until both skn-1 and atf-7 were disrupted (Fig 4B). This suggested that each of the three transcription factors contributed to the expression of F55G11.2 and C32H11.12, and that atf-7 and skn-1 contributed independently to their induction. Whereas skn-1 contributed to the expression of these two p38-dependent genes, it did not affect others. Thus, induction of F55G11.2 and C32H11.12 following p38 activation by vhp-1 knock-down was abolished by skn-1 knock-down, but induction of T24B8.5, clec-85 and clec-186 was not (Fig 4C). Furthermore, not only do elt-2 and skn-1 both contribute to F55G11.2 and C32H11.12 expression, but elt-2 seems to be required for skn-1-dependent regulation, as elt-2 RNAi was able to abolish induction of C32H11.12 following vhp-1 knock-down (Fig 2F).
Fig. 4. skn-1 co-regulates gene expression with atf-7 and elt-2. 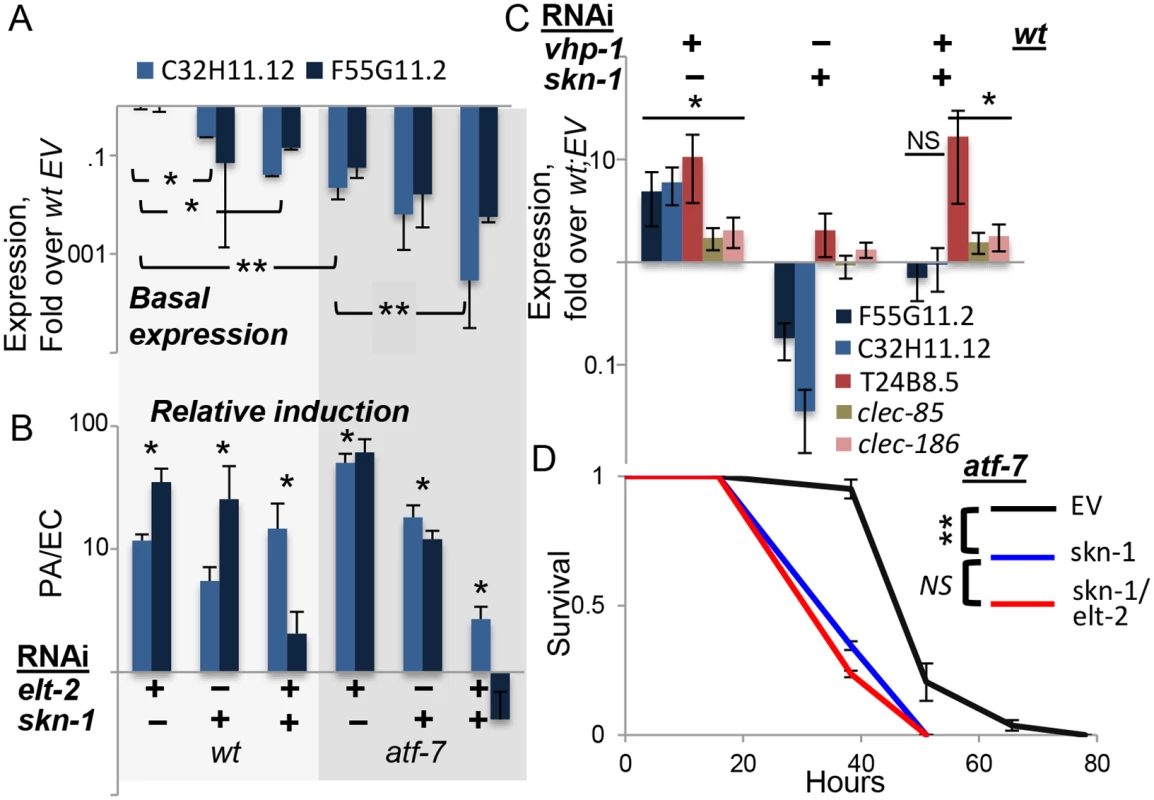
A-C, Gene expression (log scale) in wildtype or atf-7(qd22qd130) animals fed with designated RNAi’s during development. Averages ± SDs for two independent experiments (or three, in C), with *p<0.05 and **p<0.0005 (t-test) for differences between groups joined by line (A), or between marked groups and their respective references (B). A, basal expression following development knock-down, with values and statistics relative to values in wt;EV, not shown. B, similar RNAi treatments as in A, followed by exposure to P. aeruginosa (PA). Responses to PA are shown as fold over basal expression in similarly-treated worms grown on E. coli (EC). C, basal expression following development knock-down; note effective induction of T24B8.5, clec-85 and clec-186 by vhp-1 RNAi in the context of a double knock-down. D, Survival curves for atf-7(qd22qd130) animals fed with designated RNAi’s during development, followed by infection; averages ± SDs for three plates (N = 83–90 per group). In summary, skn-1 seems to be the additional factor needed to explain observed expression patterns of F55G11.2 and C32H11.12. The two genes examined here probably represent a subset of the p38-dependent immune response, regulated not only by atf-7 and elt-2, but also by skn-1. Indeed, survival analysis in atf-7 mutants demonstrated the non-redundant contribution of skn-1 to infection resistance, and further showed no added contribution of elt-2, suggesting that in regulating immune protection elt-2 works with these two regulators but no additional ones (Fig 4D).
Discussion
Our expression analyses in elt-2-disrupted worms define two dominant roles for elt-2 in the adult intestine—regulation of hydrolytic, potentially digestive, enzymes, and regulation of defense/immune genes. Whereas elt-2 has been proposed to regulate all intestinal gene expression, we narrow its role in constitutive intestinal expression by showing that adult elt-2 is important particularly for expression of genes encoding hydrolytic enzymes, but not those that contribute to intestinal structure. Furthermore, we show for the first time that ELT-2 co-regulates induced immune responses together with ATF-7 and SKN-1, functioning as a tissue-specific master regulator controlling the contribution of the p38 pathway to innate immunity.
Regulation of immune responses
ELT-2 was previously shown to be an immune regulator in adult worms, contributing to immune responses and infection resistance [22]. Whereas the vertebrate protein GATA3 activates gene expression following nuclear translocation induced by p38 phosphorylation [41], nuclear localization of the elt-2 ortholog GATA4 was instead shown to be controlled by the kinase GSK3β [42]. In contrast, ELT-2 was proposed to be constitutively localized to the nucleus [35]. Thus, how elt-2 contributed to induced responses was not clear, and if p38 was responsible for infection-induced activation of ELT-2, it was still unclear how this was achieved. While our results cannot rule out ELT-2 phosphorylation by the p38 pathway, they suggest a model in which ELT-2 functions as a master regulator of immune gene expression, cooperating with transcription factors activated by the p38 pathway, namely ATF-7 and SKN-1 (Fig 5). Under normal conditions, ATF-7 functions as a repressor and interferes with elt-2-dependent gene expression; SKN-1 contributes positively to the expression of some genes (of group B, see Fig 5), but not others (group A). Upon exposure to a pathogen, PMK-1 is activated, phosphorylating ATF-7 and transforming it into a transcriptional activator [11]. In this capacity, ATF-7 cooperates with ELT-2 to induce immune gene expression.
Fig. 5. ELT-2-PMK-1-ATF-7-SKN-1 interactions in gene regulation. 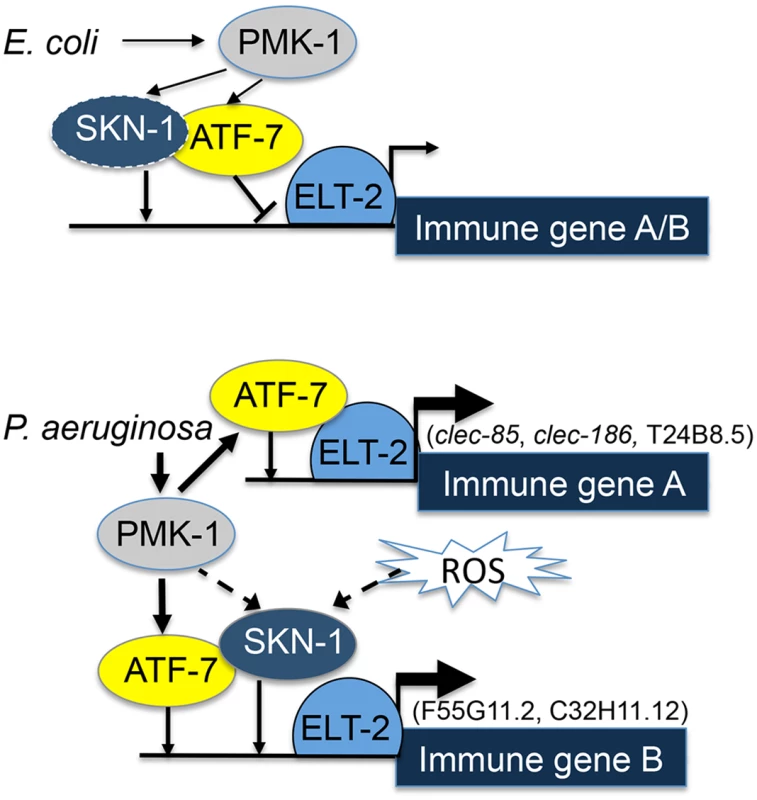
A model. Solid lines represent interactions suggested by results (line thickness is proportional to degree of activation). Dashed lines represent putative alternative options. To better fit this model to the results, it is necessary to consider that under normal conditions activated PMK-1 is present (supported by [43,44]); indeed, “normal” conditions include the presence of E. coli OP50, which is a weak pathogen [38,45]. Thus, by constitutively controlling the interference of ATF-7 with elt-2-dependent expression, PMK-1 plays a role in establishing basal levels of immune gene expression.
Whereas co-regulation by ELT-2 and ATF-7 was sufficient to explain immune responses of group A genes, group B genes additionally depended on SKN-1. Our results support a model in which elt-2 is independently required for atf-7 - and skn-1-dependent gene expression, which could explain the observed additive effects in the contributions of skn-1 and atf-7 to the expression of group B genes. SKN-1 can be directly phosphorylated and activated by the p38 pathway [13], but can alternatively be activated by reactive oxygen species (ROS) generated as part of the protective immune response [14]. Furthermore, alternative sources of ROS (e.g. induced by infecting pathogens [46]) may activate SKN-1 independent of the p38 pathway, as suggested by the reported inability of p38 disruption to completely abolish the induction of oxidative stress response genes during infection [14]. A p38-indepedent SKN-1 activation could explain the results presented in Fig 2, demonstrating induction of F55G11.2 and C32H11.12 in infected pmk-1 mutants. Lastly, a recent report suggested an involvement of the PQM-1 transcription factor in regulating F55G11.2 under normal conditions [47]. pqm-1 affected F55G11.2 expression, but its contribution appears to be small compared to what we have observed with elt-2. While pqm-1 may provide yet another regulatory input to F55G11.2 gene expression, its contribution is not required for explaining F55G11.2’s expression patterns during infection.
Regulation of constitutive intestinal gene expression
As a key regulator of intestinal terminal differentiation, the continued expression of elt-2 in the adult worm has been considered as required for maintenance of intestinal structure and function. Support for this was offered by experiments showing that ectopic elt-2 expression, or elt-2 disruption, during embryogenesis, affected expression of intestinal genes, some of which are expressed in adults [20]. However, with only about 10% overlap between adult and embryonic intestinal gene sets it seems that such experiments might reflect elt-2 contributions in embryos and not necessarily in adults. Differences in elt-2 contributions in different ages have been described. For example, expression of ifb-2, which encodes an intermediate filament protein, is abolished by elt-2 disruption in embryos, but is unaffected in L1 larvae [21]; similarly, it is unaffected in adults [22](and this study), suggesting diminishing regulatory contributions. It was demonstrated that past embryogenesis, elt-2 contributed redundantly to intestinal gene expression with a second intestinal GATA transcription factor, ELT-7 [21]: whereas neither disruption of elt-2, nor elt-7, affected larval ifb-2 expression, disruption of both abolished this expression; this pattern of redundant regulation was shared by several genes, most of which encode intestinal structural proteins. It is quite possible that elt-2, together with elt-7, maintains its contributions to expression of structure-related genes in the adult intestine. However, our results suggest a distinct, and dominant, role for elt-2 in the adult intestine—regulating the expression of hydrolytic enzymes. Such regulation is potentially important for intestinal function (digestion), but also creates a hostile environment for invading pathogens. It is tempting to suggest that the lack of redundancy in regulating these genes (manifested as reduction in gene expression following knock-down of elt-2 alone) is related to the dominant contribution of elt-2 for immune responses.
While hydrolytic enzyme genes are the only ones that we found to be enriched among the ‘elt-2-regulated’ genes, they make up only 20% of this subset. It is possible that additional elt-2-regulated functions are included in this subset, but are obscured by indirectly regulated genes, which our bioinformatic analysis suggests make up a significant part of this gene subset.
In summary, our genome-wide analysis helps distinguish between basal and pathogen-induced elt-2-dependent regulons in the adult worm. Whereas the functional composition of the two appears to be distinct, an overarching theme of anti-bacterial functions is consistent with the idea that bacteria can be both food and pathogens. Additional results further shed light on the largely uncharacterized contribution of elt-2 to induced responses, revealing cooperation with the transcription factors ATF-7 and SKN-1 downstream to the p38 pathway, and suggesting a function of a tissue-specific master regulator. Whereas elt-2 contributions to gene expression during and after development seem to differ both compositionally and mechanistically, it seems that its status as a master regulator is maintained in the adult intestine.
Materials and Methods
Worm strains
They were obtained from the Caenorhabditis Genetics Center and included wild-type N2; sek-1(km4), pmk-1(km25) and atf-7(qd22qd130) signaling mutants; and spe-26(it112) temperature-dependent sterile mutants, which lay unfertilized eggs. PF55G11.2::gfp worms were designed as described below, and further mated to generated PF55G11.2::gfp;pmk-1(km25) worms. Bacterial strains included: E. coli strain OP50-1, Pseudomonas aeruginosa strain PA14, and the latter’s GFP-expressing derivative PA14-GFP [48].
RNAi-mediated knock-down
It was performed with the standard feeding protocol, using bacterial clones from the Ahringer library, with empty RNAi vector (EV) serving as control [22,49]. The exception is atf-7 RNAi, which was from the Open Biosystems library. RNAi feeding was performed for two days, starting at the egg stage (RNAi-dev), or late L4 (RNAi-ad). The protocol used here was previously shown (in worms expressing ELT-2::GFP) to result in a complete knock-down of ELT-2 [22].
Worm growth and infection
All experiments were carried out using synchronized worm populations grown on E. coli at 25°C. Infections were performed using the slow killing protocol, typically at 25°C, or when following survival of sensitive strains, at 20°C [48]. Survival analysis of adult sek-1(km4) mutants was performed with cdc-25.1(RNAi)-sterilized animals [50], to avoid confounding effects of internal egg hatching. Statistical evaluation of differences between survival curves was performed using Kaplan-Meier analysis followed by the Log-rank test.
Microarray experiments
Worms were exposed to RNAi (control or elt-2) beginning at the L4 stage, and following two days were transferred either to E. coli OP50 or to P. aeruginosa PA14-GFP. Following eighteen hours of exposure (control), or twelve hours (elt-2 RNAi), worms were harvested for RNA extraction and microarray analysis. In a previous study we sought to determine the contribution of colonization (and its associated damage), versus specific pathogen recognition, to differential innate immune responses, and what role elt-2 played in regulating these responses. Therefore, worms were separated into those that were conspicuously colonized with the GFP-expressing pathogen, and those that were not visibly colonized. Times of exposure to the pathogen were optimized to maximize colonization variability in the population and were therefore shorter in the more susceptible elt-2(RNAi) worms. In our previous study we focused on immune responses only in control-treated animals and found them to be identical irrespective of colonization status [51]. In the current analysis we focused on the role of elt-2 in innate immune responses as a whole, utilizing data from control-treated animals as a reference for comparison. For this purpose, data from colonized and non-colonized worm groups can be pooled into one group—exposure to pathogen. This results in six independent repeats in control(RNAi) animals exposed to the pathogen, compared to three repeats of similarly-treated animals exposed to E. coli; for the elt-2(RNAi) animals, the exposure to E. coli was performed in duplicate, and to the pathogen—in triplicate. RNA was extracted from worms using Trizol (Invitrogen) (100–700 worms per group), and amplified using the MessageAmp II aRNA Amplification Kit (Ambion), labeled with the ULS aRNA Labeling Kit (Kreatech) and co-hybridized to Epoxy (Corning) microarrays spotted with 60-mer oligonucleotides (Washington University Genome Sequencing Center) with a similarly amplified and labeled reference RNA sample [51]. Filtering for high-quality data resulted in 7,880 genes with expression values >2.5 fold over background in >70% of the microarrays. These gene expression profiles were analyzed with the SAM microarray analysis package [52]; a two-class testing configuration was used to identify genes differentially-expressed during infection in untreated worms compared to elt-2(RNAi) worms, with a false discovery rate of 9%.
PF55G11.2::gfp promoter-reporter strain
A genomic fragment including 1.7 Kb of F55G11.2 upstream region was amplified (annealing: 60°C) using specific primers A-gaagcgcattggtctttga, and B - AGTCGACCTGCAGGCATGCAAGCTttccagcggcggaaact, the latter tailed (capitalized), for subsequent recombinant PCR. This fragment includes part of the F55G11.3 upstream pseudogene, as well as the initial 58 bp of F55G11.2 coding sequence. Recombinant PCR fused this fragment with gfp, as previously described, using the nested primer A* (caatttggacacggcaaact) together with the previously described D* primer [53]. Transgenic animals were generated by microinjecting PCR products, together with the rol-6(su1006) dominant marker, into worms. Genome integration was subsequently achieved by UV irradiation, as described [54]. GFP signal was quantified in worm images using the MetaMorph analysis software (Molecular Devices).
Quantitative (q)RT-PCR
RNA extracted as described above was used as template with primers listed in S1 Table. Gene-specific threshold cycle (Ct) values were normalized to the respective actin values, and presented as fold change over normalized values from control-treated animals exposed to E. coli, or when relative induction was assessed, as fold change in worms exposed to P. aeruginosa over values in worms of similar genetic background/treatment exposed to E. coli. Statistical significance was evaluated with a t-test using actin-normalized Ct values.
Bioinformatics
Management and analysis of gene lists was performed using WormMine (http://www.wormbase.org/tools/wormmine/). Searches for the GATA DNA motif were performed using the MEME suite (http://meme.nbcr.net): FIMO, for analysis of motif distribution; and MAST, for motif prevalence. The DNA motif used for searches was the consensus sequence TGATAA, shared by GATA motifs in different datasets [20,22]. Promoter sequences were retrieved with Worm mart, from Wormbase version WS220. GO analysis was performed with Generic GO Term Finder (http://go.princeton.edu/), using a gene association file downloaded from Wormbase version WS245, and applying Bonferroni correction for p-value calculation (unless otherwise mentioned).
Supporting Information
Zdroje
1. Ausubel FM (2005) Are innate immune signaling pathways in plants and animals conserved? Nat Immunol 6 : 973–979. 16177805
2. Bryant CE, Monie TP (2012) Mice, men and the relatives: cross-species studies underpin innate immunity. Open Biol 2 : 120015. doi: 10.1098/rsob.120015 22724060
3. Fraiture M, Brunner F (2014) Killing two birds with one stone: trans-kingdom suppression of PAMP/MAMP-induced immunity by T3E from enteropathogenic bacteria. Front Microbiol 5 : 320. doi: 10.3389/fmicb.2014.00320 25101059
4. Kim DH, Feinbaum R, Alloing G, Emerson FE, Garsin DA, et al. (2002) A conserved p38 MAP kinase pathway in Caenorhabditis elegans innate immunity. Science 297 : 623–626. 12142542
5. Irazoqui JE, Urbach JM, Ausubel FM (2010) Evolution of host innate defence: insights from Caenorhabditis elegans and primitive invertebrates. Nat Rev Immunol 10 : 47–58. doi: 10.1038/nri2689 20029447
6. Tan MW, Shapira M (2011) Genetic and molecular analysis of nematode-microbe interactions. Cell Microbiol.
7. Arthur JS, Ley SC (2013) Mitogen-activated protein kinases in innate immunity. Nat Rev Immunol 13 : 679–692. doi: 10.1038/nri3495 23954936
8. Couillault C, Pujol N, Reboul J, Sabatier L, Guichou JF, et al. (2004) TLR-independent control of innate immunity in Caenorhabditis elegans by the TIR domain adaptor protein TIR-1, an ortholog of human SARM. Nat Immunol 5 : 488–494. 15048112
9. Liberati NT, Fitzgerald KA, Kim DH, Feinbaum R, Golenbock DT, et al. (2004) Requirement for a conserved Toll/interleukin-1 resistance domain protein in the Caenorhabditis elegans immune response. Proc Natl Acad Sci U S A 101 : 6593–6598. 15123841
10. Kim DH, Liberati NT, Mizuno T, Inoue H, Hisamoto N, et al. (2004) Integration of Caenorhabditis elegans MAPK pathways mediating immunity and stress resistance by MEK-1 MAPK kinase and VHP-1 MAPK phosphatase. Proc Natl Acad Sci U S A 101 : 10990–10994. 15256594
11. Shivers RP, Pagano DJ, Kooistra T, Richardson CE, Reddy KC, et al. (2010) Phosphorylation of the conserved transcription factor ATF-7 by PMK-1 p38 MAPK regulates innate immunity in Caenorhabditis elegans. PLoS Genet 6: e1000892. doi: 10.1371/journal.pgen.1000892 20369020
12. Xie Y, Moussaif M, Choi S, Xu L, Sze JY (2013) RFX transcription factor DAF-19 regulates 5-HT and innate immune responses to pathogenic bacteria in Caenorhabditis elegans. PLoS Genet 9: e1003324. doi: 10.1371/journal.pgen.1003324 23505381
13. Inoue H, Hisamoto N, An JH, Oliveira RP, Nishida E, et al. (2005) The C. elegans p38 MAPK pathway regulates nuclear localization of the transcription factor SKN-1 in oxidative stress response. Genes Dev 19 : 2278–2283. 16166371
14. Hoeven R, McCallum KC, Cruz MR, Garsin DA (2011) Ce-Duox1/BLI-3 generated reactive oxygen species trigger protective SKN-1 activity via p38 MAPK signaling during infection in C. elegans. PLoS Pathog 7: e1002453. doi: 10.1371/journal.ppat.1002453 22216003
15. Papp D, Csermely P, Soti C (2012) A role for SKN-1/Nrf in pathogen resistance and immunosenescence in Caenorhabditis elegans. PLoS Pathog 8: e1002673. doi: 10.1371/journal.ppat.1002673 22577361
16. Pujol N, Zugasti O, Wong D, Couillault C, Kurz CL, et al. (2008) Anti-fungal innate immunity in C. elegans is enhanced by evolutionary diversification of antimicrobial peptides. PLoS Pathog 4: e1000105. doi: 10.1371/journal.ppat.1000105 18636113
17. Gilleard JS, McGhee JD (2001) Activation of hypodermal differentiation in the Caenorhabditis elegans embryo by GATA transcription factors ELT-1 and ELT-3. Mol Cell Biol 21 : 2533–2544. 11259601
18. Fukushige T, Hawkins MG, McGhee JD (1998) The GATA-factor elt-2 is essential for formation of the Caenorhabditis elegans intestine. Dev Biol 198 : 286–302. 9659934
19. McGhee JD, Sleumer MC, Bilenky M, Wong K, McKay SJ, et al. (2007) The ELT-2 GATA-factor and the global regulation of transcription in the C. elegans intestine. Dev Biol 302 : 627–645. 17113066
20. McGhee JD, Fukushige T, Krause MW, Minnema SE, Goszczynski B, et al. (2009) ELT-2 is the predominant transcription factor controlling differentiation and function of the C. elegans intestine, from embryo to adult. Dev Biol 327 : 551–565. doi: 10.1016/j.ydbio.2008.11.034 19111532
21. Sommermann EM, Strohmaier KR, Maduro MF, Rothman JH (2010) Endoderm development in Caenorhabditis elegans: the synergistic action of ELT-2 and -7 mediates the specification—>differentiation transition. Dev Biol 347 : 154–166. doi: 10.1016/j.ydbio.2010.08.020 20807527
22. Shapira M, Hamlin BJ, Rong J, Chen K, Ronen M, et al. (2006) A conserved role for a GATA transcription factor in regulating epithelial innate immune responses. Proc Natl Acad Sci U S A 103 : 14086–14091. 16968778
23. Kerry S, TeKippe M, Gaddis NC, Aballay A (2006) GATA transcription factor required for immunity to bacterial and fungal pathogens. PLoS ONE 1: e77. 17183709
24. Lee SH, Wong RR, Chin CY, Lim TY, Eng SA, et al. (2013) Burkholderia pseudomallei suppresses Caenorhabditis elegans immunity by specific degradation of a GATA transcription factor. Proc Natl Acad Sci U S A 110 : 15067–15072. doi: 10.1073/pnas.1311725110 23980181
25. Petersen UM, Kadalayil L, Rehorn KP, Hoshizaki DK, Reuter R, et al. (1999) Serpent regulates Drosophila immunity genes in the larval fat body through an essential GATA motif. Embo J 18 : 4013–4022. 10406806
26. Molkentin JD (2000) The zinc finger-containing transcription factors GATA-4, -5, and -6. Ubiquitously expressed regulators of tissue-specific gene expression. J Biol Chem 275 : 38949–38952. 11042222
27. Aronson BE, Stapleton KA, Krasinski SD (2014) The role of GATA factors in development, differentiation, and homeostasis of the small intestinal epithelium. Am J Physiol Gastrointest Liver Physiol.
28. Tenhunen O, Sarman B, Kerkela R, Szokodi I, Papp L, et al. (2004) Mitogen-activated protein kinases p38 and ERK 1/2 mediate the wall stress-induced activation of GATA-4 binding in adult heart. J Biol Chem 279 : 24852–24860. 15051723
29. Sartori DJ, Wilbur CJ, Long SY, Rankin MM, Li C, et al. (2014) GATA factors promote ER integrity and beta-cell survival and contribute to type 1 diabetes risk. Mol Endocrinol 28 : 28–39. doi: 10.1210/me.2013-1265 24284823
30. Suzuki YJ (2011) Cell signaling pathways for the regulation of GATA4 transcription factor: Implications for cell growth and apoptosis. Cell Signal 23 : 1094–1099. doi: 10.1016/j.cellsig.2011.02.007 21376121
31. Pauli F, Liu Y, Kim YA, Chen PJ, Kim SK (2006) Chromosomal clustering and GATA transcriptional regulation of intestine-expressed genes in C. elegans. Development 133 : 287–295. 16354718
32. Roy PJ, Stuart JM, Lund J, Kim SK (2002) Chromosomal clustering of muscle-expressed genes in Caenorhabditis elegans. Nature 418 : 975–979. 12214599
33. Bossinger O, Fukushige T, Claeys M, Borgonie G, McGhee JD (2004) The apical disposition of the Caenorhabditis elegans intestinal terminal web is maintained by LET-413. Dev Biol 268 : 448–456. 15063180
34. Schulenburg H, Hoeppner MP, Weiner J 3rd, Bornberg-Bauer E (2008) Specificity of the innate immune system and diversity of C-type lectin domain (CTLD) proteins in the nematode Caenorhabditis elegans. Immunobiology 213 : 237–250. doi: 10.1016/j.imbio.2007.12.004 18406370
35. Fukushige T, Hendzel MJ, Bazett-Jones DP, McGhee JD (1999) Direct visualization of the elt-2 gut-specific GATA factor binding to a target promoter inside the living Caenorhabditis elegans embryo. Proc Natl Acad Sci U S A 96 : 11883–11888. 10518545
36. Troemel ER, Chu SW, Reinke V, Lee SS, Ausubel FM, et al. (2006) p38 MAPK regulates expression of immune response genes and contributes to longevity in C. elegans. PLoS Genet 2: e183. 17096597
37. Youngman MJ, Rogers ZN, Kim DH (2011) A decline in p38 MAPK signaling underlies immunosenescence in Caenorhabditis elegans. PLoS Genet 7: e1002082. doi: 10.1371/journal.pgen.1002082 21625567
38. Hahm JH, Kim S, Paik YK (2011) GPA-9 is a novel regulator of innate immunity against Escherichia coli foods in adult Caenorhabditis elegans. Aging Cell 10 : 208–219. doi: 10.1111/j.1474-9726.2010.00655.x 21108728
39. Bishop NA, Guarente L (2007) Two neurons mediate diet-restriction-induced longevity in C. elegans. Nature 447 : 545–549. 17538612
40. Gerstein MB, Lu ZJ, Van Nostrand EL, Cheng C, Arshinoff BI, et al. (2010) Integrative analysis of the Caenorhabditis elegans genome by the modENCODE project. Science 330 : 1775–1787. doi: 10.1126/science.1196914 21177976
41. Maneechotesuwan K, Xin Y, Ito K, Jazrawi E, Lee KY, et al. (2007) Regulation of Th2 cytokine genes by p38 MAPK-mediated phosphorylation of GATA-3. J Immunol 178 : 2491–2498. 17277157
42. Morisco C, Seta K, Hardt SE, Lee Y, Vatner SF, et al. (2001) Glycogen synthase kinase 3beta regulates GATA4 in cardiac myocytes. J Biol Chem 276 : 28586–28597. 11382772
43. Kawli T, Wu C, Tan MW (2010) Systemic and cell intrinsic roles of Gqalpha signaling in the regulation of innate immunity, oxidative stress, and longevity in Caenorhabditis elegans. Proc Natl Acad Sci U S A 107 : 13788–13793. doi: 10.1073/pnas.0914715107 20647387
44. Twumasi-Boateng K, Wang TW, Tsai L, Lee KH, Salehpour A, et al. (2012) An age-dependent reversal in the protective capacities of JNK signaling shortens C. elegans lifespan. Aging Cell.
45. Garsin DA, Sifri CD, Mylonakis E, Qin X, Singh KV, et al. (2001) A simple model host for identifying Gram-positive virulence factors. Proc Natl Acad Sci U S A 98 : 10892–10897. 11535834
46. Gardner PR (1996) Superoxide production by the mycobacterial and pseudomonad quinoid pigments phthiocol and pyocyanine in human lung cells. Arch Biochem Biophys 333 : 267–274. 8806780
47. Tepper RG, Ashraf J, Kaletsky R, Kleemann G, Murphy CT, et al. (2013) PQM-1 complements DAF-16 as a key transcriptional regulator of DAF-2-mediated development and longevity. Cell 154 : 676–690. doi: 10.1016/j.cell.2013.07.006 23911329
48. Tan MW, Mahajan-Miklos S, Ausubel FM (1999) Killing of Caenorhabditis elegans by Pseudomonas aeruginosa used to model mammalian bacterial pathogenesis. Proc Natl Acad Sci U S A 96 : 715–720. 9892699
49. Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, et al. (2003) Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature 421 : 231–237. 12529635
50. Shapira M, Tan MW (2008) Genetic analysis of Caenorhabditis elegans innate immunity. Methods Mol Biol 415 : 429–442. doi: 10.1007/978-1-59745-570-1_25 18370169
51. Twumasi-Boateng K, Shapira M (2012) Dissociation of Immune Responses from Pathogen Colonization Supports Pattern Recognition in C. elegans. PLoS ONE 7: e35400. doi: 10.1371/journal.pone.0035400 22514739
52. Tusher VG, Tibshirani R, Chu G (2001) Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A 98 : 5116–5121. 11309499
53. Boulin T, Etchberger JF, Hobert O (2006) Reporter gene fusions. WormBook: 1–23.
54. Mariol MC, Walter L, Bellemin S, Gieseler K (2013) A rapid protocol for integrating extrachromosomal arrays with high transmission rate into the C. elegans genome. J Vis Exp: e50773. doi: 10.3791/50773 24379027
Štítky
Genetika Reprodukční medicína
Článek Minor Type IV Collagen α5 Chain Promotes Cancer Progression through Discoidin Domain Receptor-1Článek A Unique Virulence Gene Occupies a Principal Position in Immune Evasion by the Malaria ParasiteČlánek The Centrosomal Linker and Microtubules Provide Dual Levels of Spatial Coordination of CentrosomesČlánek Dissecting the Function and Assembly of Acentriolar Microtubule Organizing Centers in Cells In VivoČlánek MicroRNA-Dependent Transcriptional Silencing of Transposable Elements in Drosophila Follicle CellsČlánek PARP2 Is the Predominant Poly(ADP-Ribose) Polymerase in Arabidopsis DNA Damage and Immune ResponsesČlánek Cooperative Action of Cdk1/cyclin B and SIRT1 Is Required for Mitotic Repression of rRNA SynthesisČlánek Feeding and Fasting Signals Converge on the LKB1-SIK3 Pathway to Regulate Lipid Metabolism in
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2015 Číslo 5
-
Všechny články tohoto čísla
- Genomic Heritability: What Is It?
- Triglyceride-Increasing Alleles Associated with Protection against Type-2 Diabetes
- Epistasis Is a Major Determinant of the Additive Genetic Variance in
- Genetic Regulation of Bone Metabolism in the Chicken: Similarities and Differences to Mammalian Systems
- Minor Type IV Collagen α5 Chain Promotes Cancer Progression through Discoidin Domain Receptor-1
- The Philosophical Approach: An Interview with Ford Doolittle
- Downregulation of the Host Gene by miR-92 Is Essential for Neuroblast Self-Renewal in
- A Unique Virulence Gene Occupies a Principal Position in Immune Evasion by the Malaria Parasite
- Y Fuse? Sex Chromosome Fusions in Fishes and Reptiles
- Regulation of Active DNA Demethylation by a Methyl-CpG-Binding Domain Protein in
- Overlapping Patterns of Rapid Evolution in the Nucleic Acid Sensors cGAS and OAS1 Suggest a Common Mechanism of Pathogen Antagonism and Escape
- Autoselection of Cytoplasmic Yeast Virus Like Elements Encoding Toxin/Antitoxin Systems Involves a Nuclear Barrier for Immunity Gene Expression
- Genetic Architecture of Abdominal Pigmentation in
- Whole Genome DNA Binding Analysis of the Bacterial Replication Initiator and Transcription Factor DnaA
- The Centrosomal Linker and Microtubules Provide Dual Levels of Spatial Coordination of Centrosomes
- Parp3 Negatively Regulates Immunoglobulin Class Switch Recombination
- Burden Analysis of Rare Microdeletions Suggests a Strong Impact of Neurodevelopmental Genes in Genetic Generalised Epilepsies
- Cell Cycle Control by the Master Regulator CtrA in
- Myopathic Lamin Mutations Cause Reductive Stress and Activate the Nrf2/Keap-1 Pathway
- Monoallelic Loss of the Imprinted Gene Promotes Tumor Formation in Irradiated Mice
- Phylum-Level Conservation of Regulatory Information in Nematodes despite Extensive Non-coding Sequence Divergence
- Clustering and Negative Feedback by Endocytosis in Planar Cell Polarity Signaling Is Modulated by Ubiquitinylation of Prickle
- Dissecting the Function and Assembly of Acentriolar Microtubule Organizing Centers in Cells In Vivo
- MicroRNA-Dependent Transcriptional Silencing of Transposable Elements in Drosophila Follicle Cells
- β-Catenin Signaling Biases Multipotent Lingual Epithelial Progenitors to Differentiate and Acquire Specific Taste Cell Fates
- A Simple Auxin Transcriptional Response System Regulates Multiple Morphogenetic Processes in the Liverwort
- Parallel Gene Expression Differences between Low and High Latitude Populations of and .
- The Nutrient-Responsive Hormone CCHamide-2 Controls Growth by Regulating Insulin-like Peptides in the Brain of
- Characterization of TCF21 Downstream Target Regions Identifies a Transcriptional Network Linking Multiple Independent Coronary Artery Disease Loci
- PARP2 Is the Predominant Poly(ADP-Ribose) Polymerase in Arabidopsis DNA Damage and Immune Responses
- Drosophila Spaghetti and Doubletime Link the Circadian Clock and Light to Caspases, Apoptosis and Tauopathy
- Coronary Artery Disease Associated Transcription Factor TCF21 Regulates Smooth Muscle Precursor Cells That Contribute to the Fibrous Cap
- Rescue of DNA-PK Signaling and T-Cell Differentiation by Targeted Genome Editing in a Deficient iPSC Disease Model
- Disruption of Transcriptional Coactivator Sub1 Leads to Genome-Wide Re-distribution of Clustered Mutations Induced by APOBEC in Active Yeast Genes
- Yeast Killer Elements Hold Their Hosts Hostage
- Keeping in Shape the Dogma of Mitochondrial DNA Maternal Inheritance
- Extreme-Depth Re-sequencing of Mitochondrial DNA Finds No Evidence of Paternal Transmission in Humans
- Trading Places—Switching Frataxin Function by a Single Amino Acid Substitution within the [Fe-S] Cluster Assembly Scaffold
- Natural Variation Identifies , a Universal Gene Required for Cell Proliferation and Growth at High Temperatures in
- Mutations in Gene Are Associated with Predisposition to Breast Cancer
- The Whole of a Scientific Career: An Interview with Oliver Smithies
- Cell Specific eQTL Analysis without Sorting Cells
- Cooperative Action of Cdk1/cyclin B and SIRT1 Is Required for Mitotic Repression of rRNA Synthesis
- Systemic Regulation of RAS/MAPK Signaling by the Serotonin Metabolite 5-HIAA
- Reprogramming LCLs to iPSCs Results in Recovery of Donor-Specific Gene Expression Signature
- The Developmental Intestinal Regulator ELT-2 Controls p38-Dependent Immune Responses in Adult .
- Genetic Mechanism of Human Neutrophil Antigen 2 Deficiency and Expression Variations
- Feeding and Fasting Signals Converge on the LKB1-SIK3 Pathway to Regulate Lipid Metabolism in
- Early Lineage Priming by Trisomy of Leads to Myeloproliferation in a Down Syndrome Model
- Turning into a Frataxin-Dependent Organism
- Accounting for Experimental Noise Reveals That mRNA Levels, Amplified by Post-Transcriptional Processes, Largely Determine Steady-State Protein Levels in Yeast
- Biological Significance of Photoreceptor Photocycle Length: VIVID Photocycle Governs the Dynamic VIVID-White Collar Complex Pool Mediating Photo-adaptation and Response to Changes in Light Intensity
- CTXφ Replication Depends on the Histone-Like HU Protein and the UvrD Helicase
- Disruption of miR-29 Leads to Aberrant Differentiation of Smooth Muscle Cells Selectively Associated with Distal Lung Vasculature
- Notch Is Required in Adult Sensory Neurons for Morphological and Functional Plasticity of the Olfactory Circuit
- Post-transcriptional Regulation of Keratinocyte Progenitor Cell Expansion, Differentiation and Hair Follicle Regression by
- PERK Limits Lifespan by Promoting Intestinal Stem Cell Proliferation in Response to ER Stress
- Casein Kinase 1 and Phosphorylation of Cohesin Subunit Rec11 (SA3) Promote Meiotic Recombination through Linear Element Formation
- Fibroblast Growth Factor 9 Regulation by MicroRNAs Controls Lung Development and Links Loss to the Pathogenesis of Pleuropulmonary Blastoma
- Auxin-Mediated Transcriptional System with a Minimal Set of Components Is Critical for Morphogenesis through the Life Cycle in
- The Broad-Spectrum Antiviral Protein ZAP Restricts Human Retrotransposition
- The 4E-BP Caf20p Mediates Both eIF4E-Dependent and Independent Repression of Translation
- Turning into a Frataxin-Independent Organism
- Promotion of Bone Morphogenetic Protein Signaling by Tetraspanins and Glycosphingolipids
- Essential Role of the ESX-5 Secretion System in Outer Membrane Permeability of Pathogenic Mycobacteria
- The Zinc-Finger Antiviral Protein ZAP Inhibits LINE and Alu Retrotransposition
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Drosophila Spaghetti and Doubletime Link the Circadian Clock and Light to Caspases, Apoptosis and Tauopathy
- Autoselection of Cytoplasmic Yeast Virus Like Elements Encoding Toxin/Antitoxin Systems Involves a Nuclear Barrier for Immunity Gene Expression
- Parp3 Negatively Regulates Immunoglobulin Class Switch Recombination
- PERK Limits Lifespan by Promoting Intestinal Stem Cell Proliferation in Response to ER Stress
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



