-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
Reklamaβ-Catenin Signaling Biases Multipotent Lingual Epithelial Progenitors to Differentiate and Acquire Specific Taste Cell Fates
Taste is a fundamental sense that helps the body determine whether food can be ingested. Taste dysfunction can be a side effect of cancer therapies, can result from an alteration of the renewal capacities of the taste buds, and is often associated with psychological distress and malnutrition. Thus, understanding how taste cells renew throughout adult life, i.e. how newly born cells replace old cells as they die, is essential to find potential therapeutic targets to improve taste sensitivity in patients suffering taste dysfunction. Here we show that a specific molecular pathway, Wnt/β-catenin signaling, controls renewal of taste cells by regulating separate stages of taste cell turnover. We show that activating this pathway directs the newly born cells to become primarily a specific taste cell type whose role is to support the other taste cells and help them work efficiently.
Published in the journal: . PLoS Genet 11(5): e32767. doi:10.1371/journal.pgen.1005208
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1005208Summary
Taste is a fundamental sense that helps the body determine whether food can be ingested. Taste dysfunction can be a side effect of cancer therapies, can result from an alteration of the renewal capacities of the taste buds, and is often associated with psychological distress and malnutrition. Thus, understanding how taste cells renew throughout adult life, i.e. how newly born cells replace old cells as they die, is essential to find potential therapeutic targets to improve taste sensitivity in patients suffering taste dysfunction. Here we show that a specific molecular pathway, Wnt/β-catenin signaling, controls renewal of taste cells by regulating separate stages of taste cell turnover. We show that activating this pathway directs the newly born cells to become primarily a specific taste cell type whose role is to support the other taste cells and help them work efficiently.
Introduction
The sense of taste is indispensable for feeding behavior. It informs the body whether food is harmful or nutritious, and thus is critical for regulating the intake of essential nutrients. Taste stimuli are detected in the oral cavity by taste buds, which are collections of neuroepithelial cells situated primarily in specialized taste papillae on the tongue surface. In rodents, fungiform papillae (FFP), each housing a single taste bud, are distributed on the anterior two thirds of the tongue, while a single circumvallate papilla (CVP), which contains several hundred taste buds, is situated at the posterior lingual midline. Regardless of location, each taste bud is a heterogeneous collection of ~60–100 elongate cells, which have both neural and epithelial characteristics: neural in that they transduce chemical signals, i.e., salt, sour, sweet, bitter, and umami (savory), into electrochemical signals which are transmitted via sensory afferents to the brain [1]; and epithelial, given their morphology and embryonic origin [2, 3](but see [4]) and the fact that taste cells are continuously renewed throughout life [5].
Taste cells are generated from cytokeratin (Krt) 5 and 14-expressing proliferative basal keratinocytes situated adjacent to taste buds [6]. The Krt14+ progenitor population also produces the non-taste or general epithelium, within which taste buds are embedded, and these cells undergo progressive differentiation, mirroring that of skin, to form the keratinized lingual epithelium [7]. Intriguingly, the pace of renewal of lingual epithelium is rapid, ~5–6 days [8, 9], comparable to that of intestinal epithelium (3–5 days) [10] and epidermis (8–10 days) [11, 12], while taste cells turn over significantly more slowly, on the order of 10–20 days [13–16]. Each taste bud comprises three morphological cell types; Type I cells likely serve a support function, may be salt detectors and are most prevalent within each bud [17]; Type III cells detect sour tastants and are least common; and Type II cells transduce sweet, bitter or umami tastes and occur at intermediate frequency [18–20]. Thus, in order to maintain the sense of taste, the Krt14+ progenitor population must: (1) produce both rapidly renewing, short-lived non-taste epithelium, and more slowly renewing, longer lived taste bud cells; and (2) generate the proper ratio of taste cell types I, II and III within each bud. Currently we have a limited understanding of how cell fate decisions are regulated within the lingual epithelial progenitor population.
Recently, an additional step in the taste bud cell lineage has been defined. Following their terminal cell division, Krt14+ daughters enter the basal compartment of taste buds as ovoid cells, turn on expression of Sonic hedgehog (Shh) [21], and subsequently differentiate into the different mature taste cell types [22]. Importantly, the frequencies with which Shh+ cells differentiate as Type I, II or III taste cells reflect the relative proportions of these cell types resident in the bud, i.e., I > II > III [22]. The point in the taste lineage at which postmitotic Shh+ precursor cell fate is regulated and the underlying molecular mechanisms are unknown.
One candidate is the Wnt/β-catenin pathway. We and others showed previously that Wnt/β-catenin signaling is both sufficient and required for embryonic taste bud development [23, 24]. Moreover, Wnt/β-catenin is a well-known regulator of renewing epithelia and epithelial appendages in adults, including skin, hair follicles and intestine, as well as neuroepithelium [25–30]. In the tongue, LacZ expression driven by the Wnt/β-catenin reporter allele, BATGAL, has revealed that β-catenin signaling is indeed active in cells both in and around adult taste buds, including basal keratinocytes, Shh+ precursor cells and in a subset of each of the 3 differentiated taste bud cell types [31], further strengthening the testable idea that this pathway regulates one or more steps in the taste lineage.
Here, using inducible, taste bud lineage-specific genetic manipulation, we define Wnt/β-catenin function in progressive steps of taste bud cell renewal. We show that activation of β-catenin within Krt14+/Krt5+ progenitors transiently accelerates proliferation, then forces these cells to exit the cell cycle and rapidly differentiate. However, rather than producing the entire spectrum of Krt14+ cell-derived fates, the induced daughter cells are fully diverted from differentiating as general lingual epithelium, and instead become taste cells. Significantly, these daughters differentiate almost exclusively into Type I taste cells—those purported to function primarily as support cells, and to a much smaller extent Type II receptor cells. The third taste cell type, Type III sour detectors, are never induced. Finally, we show that β-catenin activation in postmitotic Shh+ precursor cells within taste buds likewise influences taste cell type differentiation, driving these cells to acquire predominantly a Type I cell fate, but this effect appears to be non-cell autonomous as Shh+ daughter cells with activated β-catenin differentiate into Type II and III cells with a frequency no different from controls. Rather, in FFP taste buds, activated β-catenin appears to function primarily taste bud autonomously to locally affect Type I cell differentiation; while in the posterior CVP, Type I cell fate is also broadly promoted including in taste buds where β-catenin has not been stabilized. We postulate that the different origins of FFP and CV taste buds from ectoderm and endoderm, respectively [32], as well as significant differences in the structure of FF and CV papillae, may underlie these differential effects.
Results
As Krt14+ basal cells give rise to both lingual epithelium and taste bud cells [6], and Krt5 and Krt14 are co-expressed by lingual basal keratinocytes [6, 33], we first confirmed that our doxycycline-mediated Krt5-driven induction system results in Cre-mediated reporter expression in basal keratinocytes, lingual epithelium and taste bud daughter cells. Krt5rtTA;tetOCre;R26RLacZ mice did not express β-galactosidase in the absence of doxycycline (dox), while Xgal+ cells were readily evident in lingual tissue of mice fed dox-supplemented chow (S1A and S1B Fig). Following 4 days of dox feeding of adult mice, the majority of epithelial cells around taste buds were Xgal+ in the posterior CVP and anterior FFP, while only a few Xgal+ cells were detected within taste buds (S1A2 and S1B2 Fig, white arrowheads). The number of Xgal+ cells in taste buds steadily increased with progressively longer dox treatment; after 4 weeks on dox chow, FFP and CVP taste buds were strongly Xgal+ (S1A3–4 and S1B3–4 Fig). Overall, the inducible system resulted in more rapid labeling of CVP taste buds, compared with those of the FFP. To accommodate these differences in efficacy in subsequent experiments, we assessed the CVP after 4 days and the FFP after 7 days of induction, when roughly comparable numbers of Xgal+ cells were evident within buds (compare S1A2 Fig CVP with S1B3 Fig FFP). To further confirm the efficiency of the inducible system, Krt5rtTA;tetOCre;Ctnnb1(Ex3)fl/+ mice were fed dox chow for 4 or 7 days, and tongue tissue examined via β-catenin immunofluorescence. Consistent with R26RLacZ reporter induction, in both FFP and CVP, nuclear β-catenin was elevated in mutant epithelial cells compared to controls (S1C–S1D Fig, white arrowheads). Thus, we established a reliable method for altering β-catenin gene function in adult mouse tongue epithelium.
Activation of β-catenin in basal keratinocytes induces taste bud-like structures at the expense of non-taste epithelium
In mice, Krt14+ basal keratinocytes of the CVP and FFP generate both non-taste lingual epithelium and taste bud cells [6], which express Krt13 [34] and Krt8 [33, 35], respectively. Thus, in the lingual epithelium these 3 keratins (Krt14, Krt13, and Krt8) serve as distinct markers for the 3 cell populations (progenitors, post-mitotic lingual epithelium, and taste buds, respectively), with minimal overlap (Fig 1). In contrast to the distinct, individual taste buds composed of elongate Krt8+ cells present in controls (Fig 1A, Krt8 red, asterisks), after 4 days on dox, the CVP epithelium of Krt5rtTA;tetOCre;Ctnnb1(Ex3)fl/+ mutants was occupied by a large, contiguous field of elongate Krt8+ cells (Fig 1B). When quantified via Krt8 immunofluorescence intensity (see Methods), we found a 2-fold increase in Krt8 signal in mutant CVP compared to controls (188.5 ± 28.2 vs 374.6 ± 21.3 in controls vs mutants; n = 3, p<0.0001, Student’s t-test). By contrast, Krt13+ squamous epithelial cells are found between taste buds in control CVP, but were virtually absent in mutant CVP (Fig 1A and 1B, Krt13 green, white arrowheads in control). In controls, Krt14 is expressed by proliferative keratinocytes situated at the basement membrane (Fig 1, Krt14 cyan, white bent arrows), whereas in mutants with stabilized β-catenin, Krt14+ basal cells were somewhat albeit not significantly diminished; Krt14 immunofluorescence intensity of cells at the basement membrane was decreased by 26% (110.4 ± 14.4 in controls vs 81.8 ± 8 in mutants; n = 3, p = 0.086, Student’s t-test). Instead, in mutants Krt14 immunostaining was evident abnormally in a subset of elongate Krt8+ cells in the expanded taste domain (Fig 1B, yellow arrowheads). Specifically, Krt14 immunofluorescence within the expanded taste epithelium was greatly increased (76.2 ± 15.4 in controls vs 181.9 ± 20.6 in mutants; n = 3, p = 0.0012, Student’s t-test). Overall, we observed a significant shift in the ratio between Krt14 immunofluorescence detected outside of (extragemmal) versus within (intragemmal) the expanded Krt8+ domain (median value = 1.6 in controls vs 0.4 in mutants; p<0.0001, Mann-Whitney test), suggesting the hypothesis that in response to activated β-catenin, progenitors are forced to rapidly differentiate into taste cells. Additionally, Krt14+ progenitors express the Sonic hedgehog (Shh) receptor and target gene, Ptch1 (S2 Fig, control; [36, 37]), while in mutants, Ptch1 expression is lost in the extragemmal compartment of the CVP (S2 Fig, GOF 4 days), further supporting the hypothesis that progenitor cells are reduced by activated β-catenin.
Fig. 1. Stabilized β-catenin depletes progenitors (Krt14+) and causes lingual epithelial cells to differentiate as taste cells (Krt8+) at the expense of non-taste cells (Krt13+). 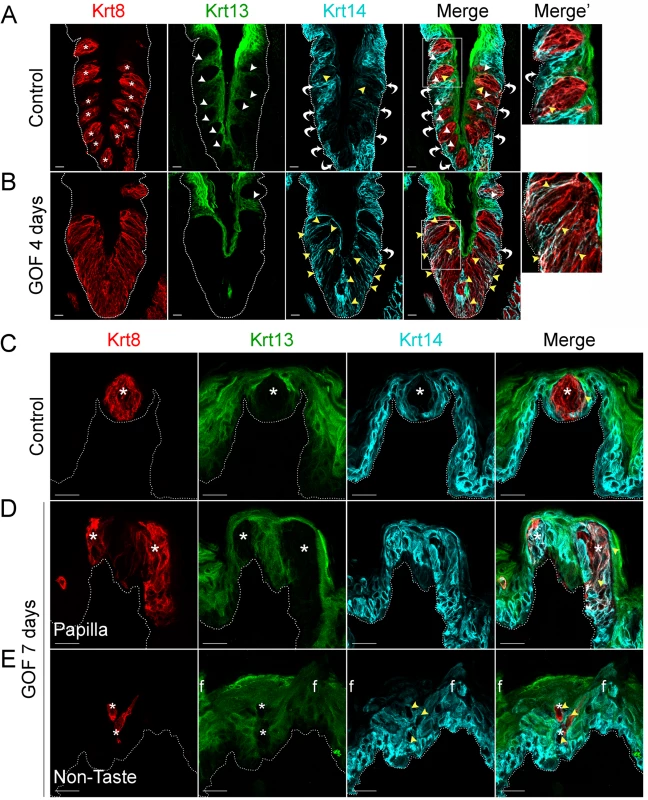
Mutant (Krt5rtTA;tetOCre;Ctnnb1(Ex3)fl/+) and control mice were fed doxycycline-supplemented chow for 4 or 7 days, their tongues harvested, and CVP and anterior tongue sections immunostained for markers of taste cells (Krt8), non-taste squamous epithelium (Krt13), and basal progenitor cells (Krt14). A. In the CVP of control mice, distinct Krt8+ taste buds (red, * indicates a taste bud) are interspersed in the apical epithelium with Krt13+ postmitotic squamous keratinocytes (green, white arrowheads). Ovoid Krt14+ progenitor cells (cyan) are restricted to the basal epithelium (white bent arrows), although occasional elongate Krt8+ cells are also dimly Krt14+ (yellow arrowheads). B. In the CVP of Krt5rtTA;tetOCre;Ctnnb(Ex3)fl/+ mice, Krt8+ cells are greatly expanded, and Krt13+ cells typically found between taste buds are absent (compare white arrowheads in A and B). Ovoid Krt14+ basal cells are also slightly reduced (compare white arrows in A and B, and white arrows in A and B Merge’, while persistent Krt14 staining in the Krt8+ taste field is dramatically increased compared to controls (A and B, yellow arrowheads). C. In the anterior tongue of controls, each FFP houses a single Krt8+ taste bud (red, * indicates a taste bud) surrounded by Krt13+ squamous epithelium (green), while basal cells adjacent to taste buds and throughout the FFP epithelium are Krt14+ (cyan). D. In mutants with stabilized β-catenin, ectopic Krt8+ cell clusters occur in FFP (red), which are devoid of Krt13 (green; white asterisks), while many are Krt14+ (cyan, yellow arrowheads, white signal in merged panel). E. Krt8+ cells (red) are detected ectopically, interspersed among filiform papillae (“f”) of the non-taste lingual epithelium. These Krt8+ cells are Krt13-negative, and frequently Krt14+. Stack images are representative samples from 3 control and 3 mutant mice. Dotted line delineates the basement membrane. Scale bars = 20 μm. Similarly, in the anterior tongue, in contrast to the single Krt8+ taste bud resident in control FFPs (Fig 1C, asterisks), after 7 days of dox, multiple Krt8+ cell clusters were evident within existing FFPs (Fig 1D, asterisks). In mutants, we also detected numerous ectopic Krt8+ cell clusters among the spine-like filiform papillae of the non-taste epithelium (“f” in Fig 1E). Both types of ectopic clusters (in FFP or in non-taste epithelium) comprised elongate Krt8+ cells, which were also Krt13-immunonegative (Fig 1D and 1E, white asterisks), consistent with a taste fate. As in the CVP, Krt14+ basal keratinocytes were disorganized in both FFP and non-taste epithelium of the anterior tongue, and some ectopic Krt8+ cells were also abnormally Krt14+ (Fig 1D and 1E, yellow arrowheads).
To determine if taste cells induced by stabilized β-catenin maintained an organized epithelium, we assessed expression of Claudin4, a tight junction protein, which is associated with epithelial cell polarity and function [38, 39], and is expressed by taste bud cells [40, 41]. In control taste epithelium, Claudin4 is restricted primarily to taste cells, as well as to the squamous layer of the CVP trench and to the apical regions of FFP (Fig 2A and 2B)[40, 41]. Claudin4 expression was expanded, mirroring the expanded taste epithelium of the CVP in mice with stabilized β-catenin (Fig 2A, dotted line). In the anterior tongue, ectopic taste buds situated in the non-taste epithelium and within FFP were also appropriately Claudin4+, as Claudin4 expression was stronger in the apices of ectopic taste buds than in the rest of the epithelium (Fig 2B, arrowheads), indicating that these Krt8+ cells were properly polarized.
Fig. 2. Expression of Claudin4 and evidence of taste pores suggests appropriate cell contacts are induced and maintained in mutant taste buds with stabilized β-catenin. 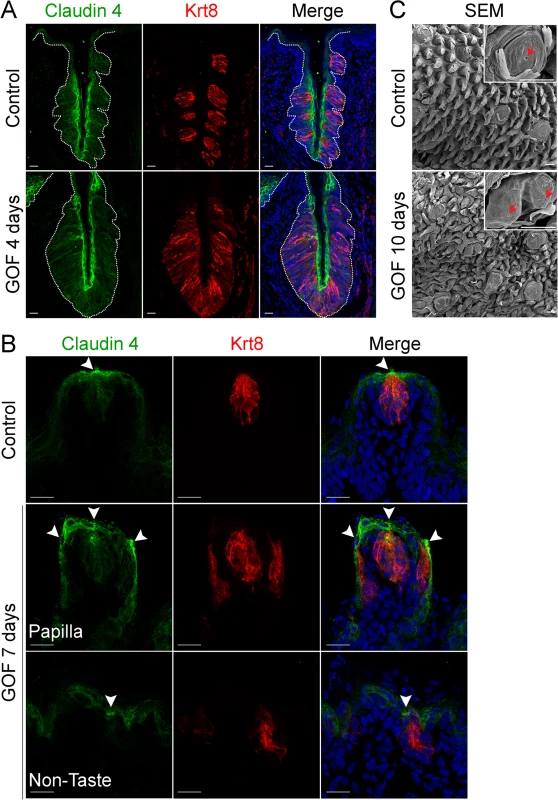
Claudin4 is a tight junctional protein expressed specifically by control taste buds (A,B control). Claudin4 expression was maintained and expanded in mutant CVP taste epithelium (A, GOF 4 days, dotted line), and by ectopic taste buds in the FFP and non-taste epithelium (B, GOF 7 days, arrowheads). FFP in controls have a single taste pore shown via SEM (C, Control, red arrowhead), whereas in mutants treated with dox for 10 days, we encountered FFP with duplicated taste pores (C, GOF 10 days, red arrowhead). NB: red staining in the mesenchyme in A is due to non-specific secondary antibody binding. Nuclei were counterstained with DRAQ5 in blue. Three mice were used in each experimental group. Dotted line delineates the basement membrane. Scale bars = 20 μm in A,B and 30 μm in C. Taste cells within a bud terminate apically in specialized microvilli, which extrude into the taste pore, a small opening which allows the cells access to taste stimuli. This pore is considered the hallmark of a differentiated taste bud [42–45]. As expected in controls, a single taste pore was readily evident within FFP when tongues were examined via SEM (Fig 2C, Control, red arrowhead). However, in mutants treated with dox for 10 days, we found what appeared to be bifurcated taste papillae, each with a taste pore (Fig 2C, GOF 10 days, red arrowheads).
In sum, our data suggest the linked hypotheses that stabilized β-catenin causes Krt14+ epithelial progenitors to produce daughter cells committed to a taste fate (Krt8+) at the expense of a non-taste fate (Krt13+), potentially by forcing precocious differentiation of Krt14+ progenitors, as indicated by loss of Ptch1 and a trend to diminished Krt14 by lingual progenitors, as well as by co-expression of Krt14 and Krt8 in elongate cells.
Stabilized β-catenin alters progenitor proliferation, and accelerates specification of post-mitotic taste precursor cells
Taste bud cell turn-over occurs via proliferation of Krt14+ progenitors adjacent to taste buds, while taste cells within buds, as well as suprabasal Krt13+ epithelial cells, are post-mitotic. To determine if stabilized β-catenin altered proliferation of Krt14+ progenitor cells, we quantified the proliferative index (P.I.) of the basal epithelial compartment of the CVP (P.I. = Ki67+ basal cells/total basal cells; see methods of Nguyen et al., 2012 [46]). Following 2 days of dox, the P.I. of mutants and controls did not differ (Fig 3A, Ki67+ cells were 77.9 ± 2.3% vs. 76.2 ± 2.2% of all basal cells in controls vs. mutants, respectively; n = 3, p = 0.583, Student’s t-test), indicating that the same fraction of Krt14+ progenitors was actively cycling after 2 days of induction. After 4 days, however, proliferation was substantially reduced in the CVP of mutants compared with controls (Fig 3A), and this decrease was not attributable to increased cell death (0 ± 0 vs. 0.06 ± 0.06 TUNEL+ cells in controls vs. mutants after 4 days of dox; n = 3, p = 0.349, Mann-Whitney test).
Fig. 3. Stabilized β-catenin alters proliferation in anterior and posterior taste fields. A, GOF 2 days. 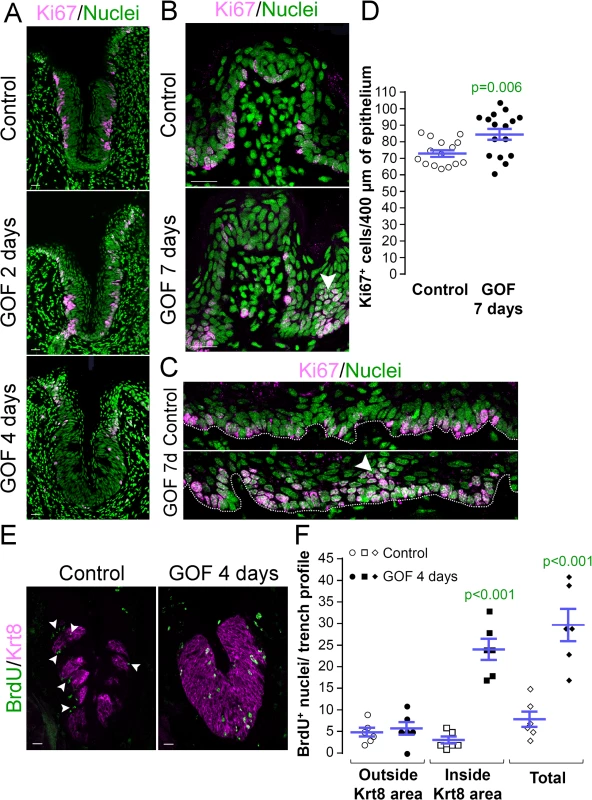
After 2 days of dox, the proportion of Ki67+ basal keratinocytes in mutant CVP epithelium did not differ from controls (77.9 ± 2.3% vs. 76.2 ± 2.2% in controls vs. mutants; n = 3, p = 0.583, Student’s t-test). A, GOF 4 days. By day 4, proliferation in the mutant CVP was virtually abolished. B. In the FFP, the number of Ki67+ cells within the FFP epithelium are comparable in mutants and controls, but in mutants proliferation appears increased in epithelia at the FFP base (GOF 7 days vs control, white arrowheads). C. In controls, only cells in the basal layer of epithelium are Ki67+, while in mutants, multiple layers or clusters of cells were actively proliferating (GOF 7d, arrowhead). D. The number of Ki67+ basal cells per 400μm of epithelium is significantly increased in mutants. E,F. Mutant and control mice were injected with BrdU 48 hours before tongues were harvested after 4 days on dox chow. Postmitotic BrdU+ cells were detected in and around taste buds (Krt8+) in control CVP epithelium (white arrowheads), whereas β-catenin stabilization resulted in increased BrdU+ cells only within the expanded Krt8+ taste field. Nuclei were counterstained with Sytox Green. Stack images comprising 14 compressed 0.75 μm-thick optical sections are representative samples from 3 control and 3 mutant mice. Six CVP trenches and 15–16 sections of non-taste epithelium were analyzed per group. Data are represented as scatter plot (individual symbols), and mean ± SEM (blue bars). Student’s t-test. Dotted line delineates the basement membrane. Scale bars = 20 μm. Progenitor proliferation was also affected in the anterior lingual epithelium. However, in contrast to the diminished proliferative index observed for CVP epithelium, Ki67+ basal cells increased slightly, but significantly in the anterior tongue of mutants compared with controls. Specifically, clusters of Ki67+ cells were observed at the base of the FFP in the mutants (Fig 3B, white arrowhead), suggesting that β-catenin stabilization increased proliferation in specific compartments of the FFP. When examined more broadly, proliferation throughout the non-taste lingual epithelium increased significantly in the mutants, and we detected clusters of suprabasal proliferative cells that are not encountered in controls (Fig 3C and 3D, arrowhead).
To resolve these apparently opposing results, we reexamined progenitor proliferation in the CVP by monitoring the flow of newly born cells into taste epithelium. Mutant and control mice were fed dox for 2 days, then injected with BrdU 48 hours before tongues were harvested after a total of 4 days on dox chow. As expected, in control CVP epithelium, post-mitotic BrdU+ cells were detected in and around Krt8+ taste buds, consistent with the rapid rate of renewal of lingual epithelium [11] and slower turnover of taste bud cells [14–16] (Fig 3E, Control). By comparison, stabilized β-catenin caused a dramatic increase in the number of BrdU+ cells within the CVP epithelium (Fig 3E and 3F) due entirely to an increase in BrdU+ cells inside the expanded Krt8+ taste field (Fig 3E and 3F). Thus, the reduced proliferative index observed for the CVP after 4 days of dox (Fig 3A) is due to earlier accelerated and/or increased proliferation of the progenitor cells followed by en masse differentiation of the progenitors, consistent with the increased proliferation seen in response to activated β-catenin in the anterior tongue epithelium. In sum these data suggest that β-catenin signaling, in addition to causing precocious specification of new taste cells from Krt14+ progenitors, accelerates proliferation of these progenitors, and in the CVP, quickly depletes this population.
Beta-catenin stabilization drives lingual epithelial cells to acquire predominantly a Type I taste cell fate
In mice, mature taste buds are made up of a heterogeneous collection of ~60 differentiated cells, including Type III cells which detect sour, Type II cells which detect sweet, bitter and umami tastes, and Type I cells which are thought to function as support cells and may be salt receptors. As stabilization of β-catenin in lingual progenitor cells results in expanded Krt8+ taste epithelium in CVP, FFP and anterior tongue non-taste epithelium, we next determined if all 3 taste cell types were similarly expanded using specific immunomarkers: SNAP25 for Type III [47], PLCβ2 for Type II [48], and NTPdase2 for Type I cells [49].
In the CVP and FFP, Type I glial cells make up roughly half of the cells within each taste bud, while Type II and III cells each comprise ~10–30% of differentiated taste cells [19, 20]. Thus we reasoned that if β-catenin affected all taste cell fates similarly, then we would see proportionate increases in cell types I, II and III represented in the expanded Krt8+ domains. In the CVP, despite the robust increase in Krt8+ cells, however, the number of SNAP25+ Type III cells in mutant taste epithelium did not differ from controls (Fig 4A and 4B). Stabilization of β-catenin did result in a small increase in the number of PLCβ2+ Type II cells (Fig 4C and 4D), but this minimal gain was insufficient to account for the vast increase in Krt8+ taste cells (see Fig 1A). When we assessed Type I cells, however, we found that stabilized β-catenin induced a dramatic increase in NTPdase2+ epithelium in the CVP, which overlapped the expanded Krt8+ domain (Fig 4E). Because NTPDdase2 localizes to cell membranes, and Type I cells have elaborate sheet-like processes, it is not possible to accurately identify and therefore count individual NTPdase2+ Type I cells [22, 50, 51]. Instead, we used the density of NTPdase2 immunofluorescence as a proxy for the size of the Type I cell population (see Methods). We found a remarkable 2-fold increase in the intensity of NTPdase2 immunofluorescence (Fig 4F), as well as a significant expansion of the area of the CVP epithelium occupied by NTPdase2+ cells (S3A Fig). To verify that corrected fluorescence intensity is a reliable measure of the relative numbers of taste cells within buds, we applied this method to Type II cells, and found a significant correlation between the number and the fluorescence intensity of PLCβ2+ Type II cells (S3B Fig), and that the PLCβ2 fluorescence intensity levels in mutant CVP were slightly, albeit significantly, higher than in controls (S3B Fig), consistent with the small but significant increase in Type II cell number in mutants (see Fig 4D).
Fig. 4. Stabilization of β-catenin in Krt5+ progenitors induces primarily Type I cell differentiation in CVP, FFP and anterior lingual epithelium. 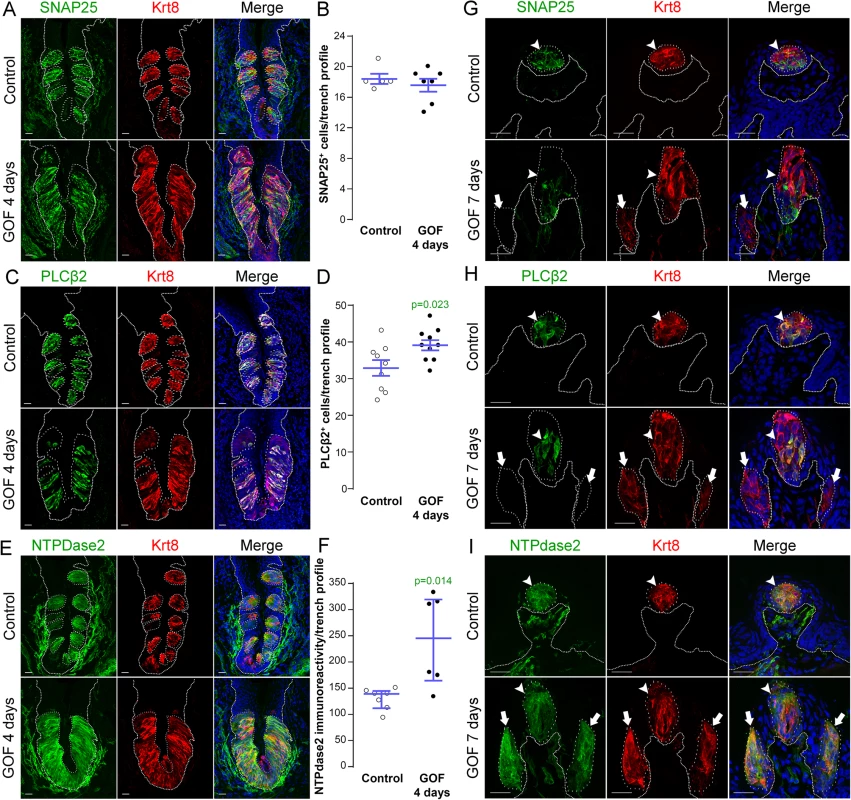
A,B. SNAP25+ Type III taste cells (green) were present in comparable numbers in the CVP of control and mutant mice fed dox chow for 4 days. Note that CVP innervation is also SNAP25+ (green) (Yang et al., 2000). C,D. PLCβ2+ Type II cells (green) were slightly increased in mutant CVP compared to controls. E,F. NTPdase2+ Type I cells (green) cannot be counted (see text), and thus were quantified via measurement of NTPdase2 corrected fluorescence intensity, revealing a 2-fold increase in NTPdase2+ signal in mutant compared to control CVP, accounting for the majority of expanded Krt8+ cells (red in all panels). G. In the anterior tongue of mice fed doxycycline chow for 7 days, endogenous Krt8+ taste buds within FFP (red, white arrowhead) contained SNAP25+ Type III cells (green), while ectopic Krt8+ taste buds (red, white arrow) were devoid of SNAP25+ cells. H. Endogenous FF taste buds house numerous PLCβ2+ Type II taste cells (green, white arrowhead), while ectopic PLCβ2+ cells were not detected in ectopic taste buds (* indicates a PLCβ2-negative ectopic taste bud). I. Ectopic taste buds within FFP of mutants (white arrows), like endogenous FF taste buds (white arrowhead) were always NTPdase2+ (green). Nuclei were counterstained with DRAQ5 in blue. Representative stack images are made of 14 compressed 0.75 μm-thick optical sections. N = 3 control and 3 mutant mice. B,D: Student’s t-test; F: Mann and Whitney test. Data are represented as a scatter plot (individual symbols), and mean ± SEM (blue bars), except F (median with interquartile range). Dotted line delineates the basement membrane. Scale bars = 20 μm. We next determined if Type I cells were also the dominant taste cell fate in ectopic Krt8+ clusters that formed in the anterior tongue in response to activated β-catenin. In the FFP of mice induced with dox for 7 days, SNAP25+ cells were not detected in ectopic Krt8+ clusters (Fig 4G, white arrow). In contrast to the CVP, PLCβ2+ Type II cells were not augmented in the anterior tongue, in that ectopic Krt8+ clusters in FFP were devoid of Type II cells (Fig 4H, white arrows). However, as we observed in the CVP using NTPdase2 to mark Type I cells, ectopic Krt8+ clusters within FFP were strongly NTPDase2+ (Figs 4I and S3C). Likewise, ectopic Krt8+ taste bud-like structures throughout the non-taste lingual epithelium were devoid of Type II and III taste cells after 7 days of dox; rather, ectopic Krt8+ taste buds were made up entirely of NTPdase2+ Type I cells (S4 Fig). Next, we analyzed the anterior tongues of mice induced for 14 days. Interestingly, and consistent with shorter term experimental results in the CVP epithelium, at this time point, 5.8 ± 2% of Krt8+ ectopic clusters housed elongate cells immunopositive for the Type II cell marker PLCβ2 (Fig 5, white arrows); however, in no instance did we observe cells expressing the Type III cell marker SNAP25 in ectopic Krt8 taste cell clusters (356 Krt8+ structures counted, 0 with SNAP25+ cells, n = 3 mice). In sum, stabilized β-catenin within lingual epithelial progenitors drives daughters to rapidly acquire a Type I, to a lesser extent a Type II, but not a Type III cell fate in both the CVP and anterior tongue, albeit over different time spans.
Fig. 5. Prolonged stabilization of β-catenin in Krt5+ progenitors induces differentiation of a small number of Type II cells in ectopic taste bud-like structures in the anterior tongue. 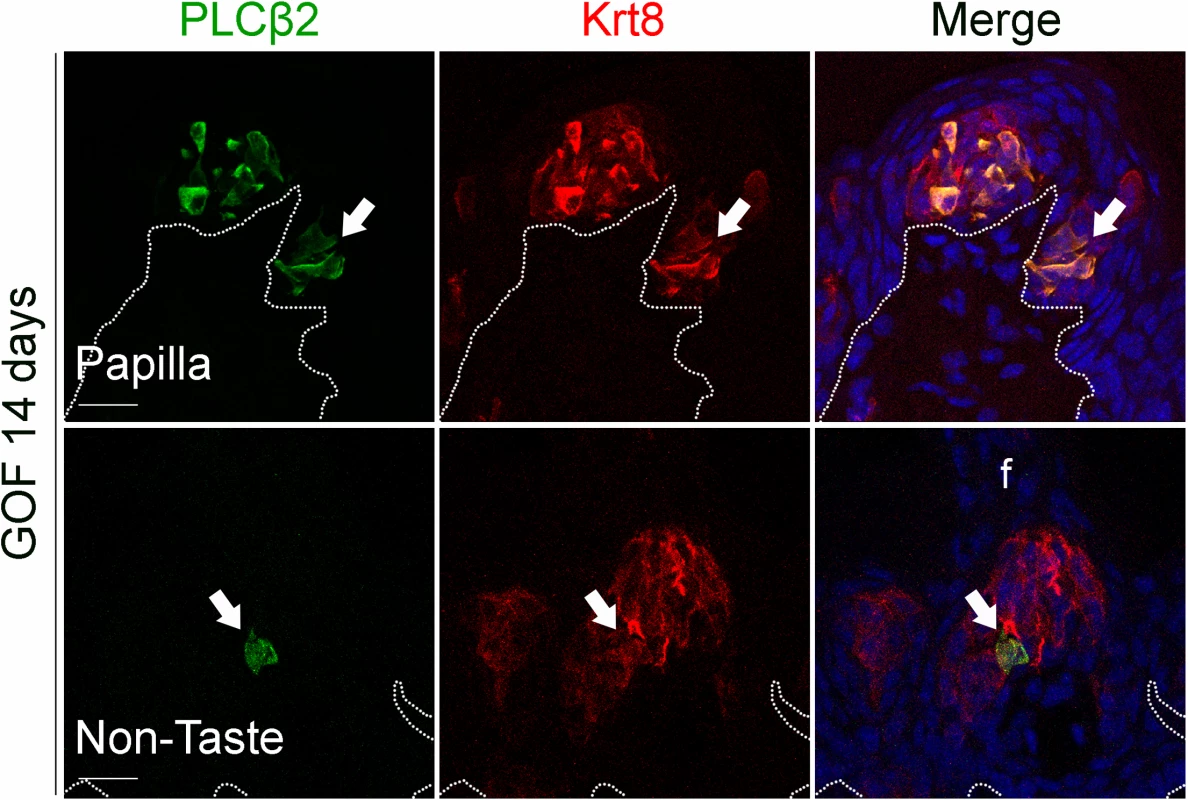
After induction with dox chow for 14 days, 5.8 ± 2% of anterior Krt8+ ectopic structures (total of 485 Krt8+ structures counted, 23 with PLCβ2, n = 3 mice) expressed PLCβ2 (green, white arrows). These ectopic structures never expressed the Type III cell marker SNAP25 (total of 356 Krt8+ structures counted, 0 with SNAP25+ cells, n = 3 mice). Nuclei were counterstained with DRAQ5 in blue. Representative stack images from 3 mutant mice. Dotted line delineates the basement membrane. Scale bars = 20 μm. Beta-catenin stabilization drives epithelial progenitors to generate Type I cells through a normal lineage progression
Shh is expressed specifically by post-mitotic basal cells within buds, and these basal cells are immediate precursors of all three taste cell types [21, 22]. Genetic stabilization of β-catenin in Krt5+ progenitors dramatically increased Shh+ precursor cells in the expanded CVP taste field compared to controls (Fig 6A). Similarly, in the anterior tongue of mutants (Fig 6B–6F), Shh expression was increased in endogenous FF taste buds (Fig 6C–6E, white arrows) and ectopically within the FFP epithelium (Fig 6C–6E, white arrowheads), as well as in ectopic locations throughout the non-gustatory epithelium of the anterior tongue (Fig 6C and 6F, yellow arrowheads). Our data suggest that forced β-catenin stabilization within epithelial progenitors promotes their specification to Shh+ taste bud precursors, which, although capable of giving rise to all 3 taste cell types in controls [22], ultimately biases these cells to differentiate into predominantly Type I taste cells.
Fig. 6. Stabilized β-catenin increases the number of Shh+ precursors of all three taste cell types, but does not impact specific precursors of Type II or III cells. 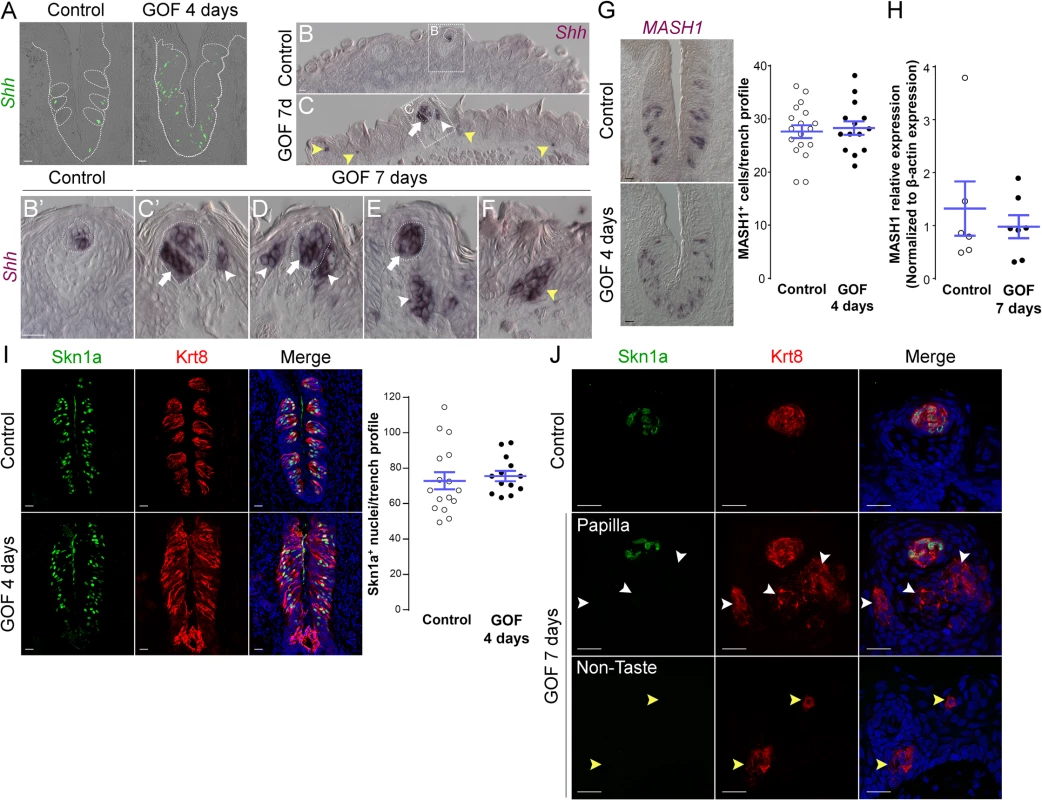
A. In control mice, 1–3 Shh+ basal cells (green) are located in the basal compartment of each taste bud section (dashed circles), whereas in the CVP of mutant mice fed dox chow for 4 days, Shh+ cells (green) are increased and widespread in the expanded taste epithelium (dashed line). B,B’. In the anterior tongue of control mice, Shh+ cells are restricted to FFP taste buds. C-F. In stabilized β-catenin mutants following 7 days on dox, in addition to endogenous Shh+ cells (white arrows), ectopic Shh+ cells are detected in FFP epithelium (white arrowheads in C,C’,D,E), as well as in non-taste epithelium adjacent to filiform papillae (yellow arrowheads in C,F). MASH1 and Skn-1a are expressed in subsets of basal cells within taste buds, and regulate the Type III and Type II cell lineage, respectively. CVP sections of control and mutant mice after 4 days on dox showed that Mash1+ (G) and (I) Skn-1a+ (green) cell numbers were not altered by β-catenin stabilization. Real-time RT-PCR on the anterior tongue epithelium showed no difference in the expression of MASH1 between controls and mutants (H). J. After 7 days on dox chow, Skn-1a+ cells (green) were restricted to endogenous FFP taste buds in control and mutant mice (Krt8+ red, arrow). In mutants, Skn-1a+ cells were not detected in ectopic Krt8+ clusters in FFP epithelium (J, white arrowheads) or non-taste epithelium (J, yellow arrowheads). Nuclei were counterstained with DRAQ5 in blue. Dotted line indicates the basement membrane, and dashed circles indicate taste buds. Representative stack images and data from 3 control and 3 mutant mice. Data are represented as a scatter plot (individual symbols), and mean ± SEM (blue bars). Student’s t-test. Scale bars = 20 μm. A subset of basal cells within adult taste buds expresses Mash1, as well as Shh, and these basal cells have been proposed as precursors of Type III taste cells [52, 53]. We therefore determined if stabilized β-catenin affects specification of Mash1+ precursors. In the CVP, Mash1+ cell number in mutant epithelium did not differ from controls (Fig 6G). In the anterior tongue, while we were unable to detect Mash1 expression via anti-sense RNA or protein in mutants, real-time RT-PCR of peeled anterior tongue epithelium showed no significant difference in Mash1 expression in mutants compared to controls (Fig 6H), consistent with the fact that Type III taste cells are not induced by activated β-catenin (See Figs 4 and S4).
The POU domain transcription factor Skn-1a (POU2f3) is expressed by a sub-population of intragemmal basal cells and by Type II taste cells, and is required for Type II cell fate [54]. Therefore, we explored if stabilized β-catenin regulates the expression of Skn-1a. In contrast to the slight increase in Type II cells (see Figs 4D and 5), the number of Skn-1a+ cells was unchanged in mutant CVP (Fig 6I), suggesting that β-catenin stabilization may increase the number of Type II cells independently of Skn-1a. Consistent with the absence of Type II taste cells in ectopic Krt8+ clusters in the anterior tongue at 7 days, we did not encounter ectopic Skn-1a+ cells in the FFP or non-taste epithelium (Fig 6J).
Beta-catenin stabilization in postmitotic Shh+ precursors biases taste cell fate
Broad activation of β-catenin in Krt5+ keratinocytes drives these progenitors to become Shh+ taste precursor cells, which in turn differentiate primarily into Type I, and infrequently, Type II taste cells (Figs 1 and 3–5). These data establish a role for β-catenin in selecting the fate of epithelial cells derived from Krt5+ progenitors, but left open if β-catenin stabilization within Shh+ taste precursors, once these cells have entered taste buds, affects cell fate selection. Thus, we induced stabilized β-catenin only in Shh+ basal cells in ShhCreERT2;Ctnnb1(Ex3)fl/+;R26R-YFP mice, and compared the relative proportions of Type I, II and III taste cells with those of controls (ShhCreERT2;R26R-YFP).
In the FFP of the anterior tongue, the number of YFP+ cells per labeled taste bud did not differ between mutants and controls (Fig 7A); (although the overall percentage of fungiform taste buds housing YFP+ cells was higher in mutants, S5A Fig). We next asked if the relative proportions of the 3 taste cell types within lineage-labeled taste buds were altered by stabilized β-catenin within Shh+ precursors. We reasoned that if β-catenin activation within Shh+ cells intrinsically biased their differentiation to a Type I fate, then the proportions of Type II and/or Type III taste cells differentiating from Shh+ cells would be reduced, while Type I cells would be increased. As was the case for stabilized β-catenin in the Krt5+ population, NTPdase2-IR of Type I cells was significantly increased in YFP+ taste buds of mutant mice (Fig 7B and 7C). However, the number of Type II (Fig 7D and 7E), and Type III cells (Fig 7F and 7G) resident in YFP+ buds did not differ between mutants and controls. Importantly, Type I cells were significantly changed exclusively in YFP+ taste buds, and not in YFP- taste buds (Fig 7B and 7C), indicating that the control of taste cell differentiation by β-catenin in the FFP is local, i.e., restricted to taste buds housing induced YFP+ mutant cells.
Fig. 7. Stabilization of β-catenin in Shh+ precursor cells biases FFP taste cell fate. 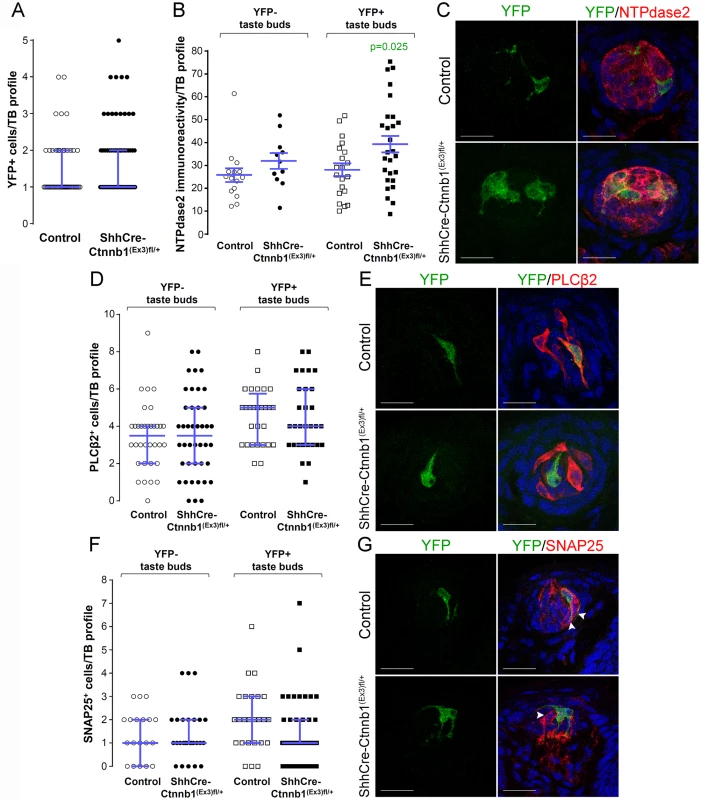
ShhCreERT2;Ctnnb(Ex3)fl/+;R26R-YFP mutant mice and their control counterparts (ShhCreERT2;R26R-YFP) were gavaged with tamoxifen daily for 8 days, and tongues harvested 14 days after the last dose. A. The number of Shh-descendant taste cells labeled with YFP did not differ between mutants and controls. B,C. In mutants, corrected NTPdase2+ immunofluorescence intensity of Type I cells was significantly increased in YFP+ taste buds, but not YFP- taste buds. D, E. Overall the total number of Type II cells did not differ between mutants and controls, nor did the number of Type III cells (F,G). Representative stack images and data from 4–5 control and 4–6 mutant mice. A: 53 vs 72 YFP+ taste bud profiles from 6 control mice vs 6 mutant mice, respectively; B: 15 vs 11 YFP- taste bud profiles and 20 vs 27 YFP+ taste bud profiles from 4 control mice vs 4 mutant mice, respectively; D: 34 vs 42 YFP- taste bud profiles and 28 vs 30 YFP+ taste bud profiles from 4 control mice vs 4 mutant mice, respectively; F: 19 vs 27 YFP- taste bud profiles and 25 vs 42 YFP+ taste bud profiles from 5 control mice vs 6 mutant mice, respectively. Mann & Whitney test, except B (Student’s t-test). Data are represented as scatter plots (individual symbols), and median with interquartile range (blue bars), except B (mean ± SEM). Nuclei are counterstained with DRAQ5 (blue). Scale bars = 20 μm. To determine if this shift in cell fate was due to cell autonomous effects of activated β - catenin within lineage labeled Shh+ cells, we compared the proportions of taste cell types among the YFP+ Shh-descendent cells in mutant versus controls. Due to the contorted morphology of Type I cells, however, we could not identify individual YFP+ Type I cells (see Miura et al., 2014 [22]), and thus could not ascertain if the overall increase in Type I cells (see Fig 7B and 7C) was due to cell-specific activation of β-catenin. However, Shh-descendent Type II and III cells were easily tallied, and we found that cell autonomous activation of β-catenin had no impact on Type II or III cell fate (S1 Table). Thus, our data suggest that activation of β-catenin within Shh+ cells inside taste buds acts by as-yet-to be identified local mechanisms to promote Type I cell differentiation.
We also assessed cell fate in taste buds of the CVP of mice with β-catenin stabilized in Shh+ cells. Unlike the FFP, the number of YFP+ Shh-descendent cells per taste bud was increased in the CVP of mutant mice compared to controls (Fig 8A), whereas similar to anterior taste buds, the proportion of taste buds housing YFP+ cells was minimally albeit significantly increased in mutant CVP versus controls (S5B Fig). In terms of cell lineage, as shown for FFP, the NTPDase2+ Type I cell population in CVP taste buds was increased (Fig 8B and 8C), while the number of Type II and Type III cells in YFP+ taste buds was unchanged in mutant CVP compared to controls (Fig 8D–8G). Further, the fate of Shh-descendent cells with cell autonomous activation of β-catenin did not differ between mutants and controls (S1 Table). Unexpectedly, however, in addition to the increase in NTPdase2-IR Type I cells in mutant YFP+ taste buds, we also found more Type I cells in YFP-negative taste buds in mutants (Fig 8B and 8C), suggesting that in addition to being promoted locally by signals within taste buds, control of Type I cell differentiation in the CVP is also regulated via signals extrinsic to taste buds.
Fig. 8. Beta-catenin stabilization in Shh+ cells biases taste cell fate in the CVP, both taste bud autonomously and indirectly. 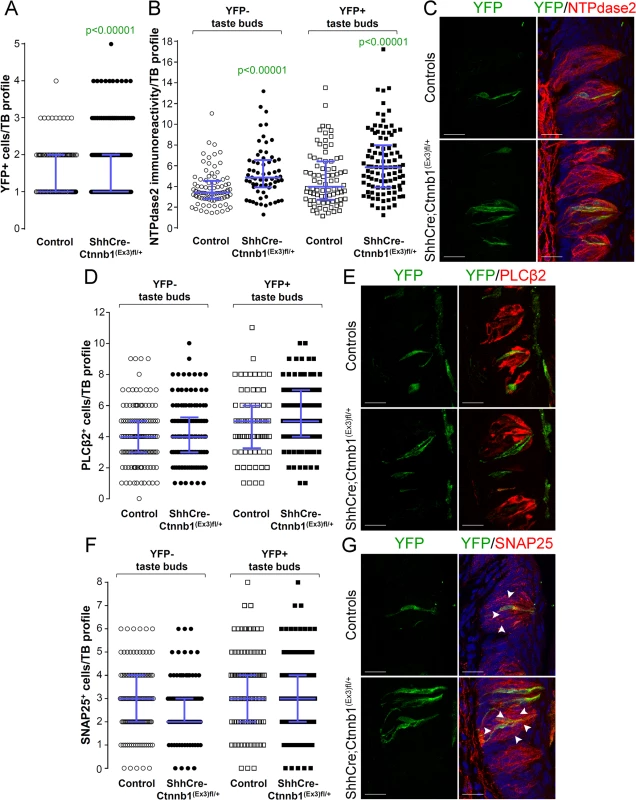
Beta-catenin stabilization in Shh+ precursors significantly increased the number of Shh- descendant taste cells labeled with YFP in mutant mice compared with controls (A). Overall the total number of Type II (D,E) and Type III cells (F,G) did not differ between mutants and controls. NTPdase2 corrected fluorescence intensity of Type I cells was significantly increased in both YFP+ and YFP- taste buds (B,C). Representative stack images and data from 4–5 control and 5–6 mutant mice. A: 178 vs 246 YFP+ taste bud profiles from 6 control mice vs 6 mutant mice, respectively; B: 85 vs 64 YFP- taste bud profiles and 82 vs 100 YFP+ taste bud profiles from 4 control mice vs 4 mutant mice, respectively; D: 133 vs 122 YFP- taste bud profiles and 72 vs 101 YFP+ taste bud profiles from 4 control mice vs 4 mutant mice, respectively; F: 120 vs 93 YFP- taste bud profiles and 106 vs 145 YFP+ taste bud profiles from 5 control mice vs 6 mutant mice, respectively. Mann & Whitney test. Data are represented as scatter plot (individual symbols), and median with interquartile range (blue bars). Nuclei are counterstained with DRAQ5 (blue). Scale bars = 20 μm. Discussion
We showed previously that Wnt/β-catenin signaling is both necessary and sufficient for FFP and early taste bud development during embryogenesis, and moreover, that forced activation of β-catenin induces ectopic and enlarged taste papillae containing enlarged taste buds throughout the lingual epithelium [23]. However, because these embryos died at birth, we were unable to determine if β-catenin also functions in adult taste cell renewal. Here, we show that activation of β-catenin in adult Krt5+ lingual epithelial progenitors results in the formation of ectopic taste buds within FFPs and CVPs, and in non-taste lingual epithelia. These data are consistent with the finding that expression of stabilized β-catenin drives formation of ectopic hair follicles in adult epidermis [55], and show that this pathway can determine lingual epithelial, as well as epidermal, fate decisions in the adult. Beta-catenin signaling is also normally active in lingual filiform papillae [25], but at lower levels than in taste buds. Thus our data suggest that high levels of β-catenin signaling, such as those observed in taste papillae, direct adult lingual cells to a taste fate.
Both within and outside taste papillae, we find that activated β-catenin initially triggers hyperproliferation of Krt5+ lingual progenitors. This finding is consistent with previous data showing that β-catenin signaling is required for normal levels of proliferation of lingual epithelia [25], and suggest that this pathway drives taste bud renewal. Similarly, β-catenin signaling contributes to progenitor cell proliferation in the epidermis, hair follicles, and intestinal epithelium [25, 56, 57]. Interestingly, following an initial bout of proliferation, taste cells expressing stabilized β-catenin are driven to differentiate, predominantly towards a Type I taste cell fate. These data suggest that, in addition to its pro-proliferative role, β-catenin also drives differentiation of taste epithelium.
Our data parallel findings in skin and intestinal epithelia. In each of these tissues, Wnt signaling plays diverse roles, promoting both proliferation and terminal differentiation. For instance, in the small intestine, Wnt signaling is required for progenitor cell proliferation, and also for acquisition of Paneth cell fate [58, 59]. In hair follicles, Wnt signaling is necessary for proliferation of transit amplifying cells in the secondary hair germ and matrix, and also drives matrix cells to terminally differentiate into hair shaft progenitors [25, 60–62]. The mechanisms underlying these seemingly opposing activities are not fully understood, but may depend in part on the level of activity of the signaling pathway. For instance, in adult hair follicle matrix cells and interfollicular epidermis, relatively low levels of signaling are thought to drive proliferation, while high levels of activity cause cells to terminally differentiate [25]. Although active Wnt/β-catenin signaling is evident in all three differentiated taste cell types [31], we find that stabilization of β-catenin in Krt5+ lingual progenitor cells predominantly biases these cells towards a Type I fate, with a lesser induction of Type II taste cell fate, both within and outside existing taste papillae. This differential induction of the different taste cell types may in part reflect differences in their relative proportions in control taste buds, i.e., I > II > III [19, 20], and that Type III cells are longer lived than Type II cells [16], and thus Type III cells would be predicted to be generated least frequently. However, our data more strongly support a model, as in skin epithelia, in which graded levels of β-catenin signaling determine the precise fate of taste cells, with the highest levels promoting Type I fate, while moderate levels of β-catenin are required for Type II differentiation, and both high and mid levels preclude acquisition of Type III fate. Additionally, β-catenin does not bind DNA directly, but rather forms multiprotein complexes that include Lymphoid Enhancer Factor/T Cell Factor (LEF/TCF) family members and cell-type-specific transcription factors [63]. The combination of cooperating factors expressed in any given cell type is thus likely to determine which of β-catenin’s potential target genes are expressed and to contribute to the precise outcome of pathway activation.
The cell autonomous impact of β-catenin on cell fate appears most robust when activated in the Krt5+ progenitor population, while stabilization of β-catenin within Shh+ postmitotic precursors does not alter significantly the fate of these cells. However, increased β-catenin signaling within this lineage-labeled subset does influence cell fate indirectly, as mutant taste buds house more Type I cells in both the FFP and CVP than controls. How this shift in cell fates comes about is not known, but may rely on additional signals emitted from these new taste cells with persistent stabilized β-catenin.
One candidate pathway that may mediate these effects is Sonic hedgehog (Shh) signaling. In the tongues of control mice, Shh expression is restricted to intragemmal basal cells (Type IV) within taste buds [36], which are the immediate precursors of each of the 3 differentiated taste cell types [22]; while Shh target genes Ptch1 and Gli1, are expressed by keratinocytes adjacent to taste buds [36]. This expression pattern suggests that SHH from within buds signals to adjacent progenitors to regulate taste cell renewal [21]. Indeed, we have shown recently that Shh promotes taste bud cell differentiation, as ectopic overexpression of SHH induces the formation of ectopic taste buds throughout the non-taste epithelium [41]. However, distinct from the uniform Type I cells induced by β-catenin, SHH-induced ectopic taste buds possess all 3 differentiated taste cell types [41]. Thus, β-catenin appears to act upstream of Shh as stabilized β-catenin induces excess and ectopic clusters of Shh+ epithelial cells in the anterior and posterior tongue (Fig 6), suggesting that the Shh+ precursor step is obligate for taste cell differentiation in general, yet high β-catenin levels drive Shh+ cells to differentiate predominantly as Type I taste cells.
While generally comparable, elements of β-catenin-mediated regulation of taste cell renewal differ between anterior and posterior tongue. Activated β-catenin drives differentiation of Type I and to a lesser extent Type II cells in both taste fields, but sparse Type II cells are evident in the anterior tongue only after prolonged induction. By contrast, both excess Type I and II cells appear rapidly within 4 days in the CVP epithelium. Additionally, stabilization of β-catenin in Shh+ precursors induced more Type I cells in both YFP+ and YFP- taste buds in the CVP, but in FFP only YFP+ buds were affected. This may be due to differences in the embryonic origins of anterior versus posterior taste buds. The posterior tongue, including the CVP, arises from the foregut endoderm [32], while the anterior tongue is ectodermally derived. Although the oral cavity is lined by a continuous epithelium, and taste buds are thought to be homologous regardless of location [64], these commonalities have arisen by different embryonic histories, which are revealed by differences in BMP4 expression in adult taste buds and FGF functional regulation of taste papilla development [65–67]. Likewise, differences in embryonic origin might cause differential region-specific expression of Wnt pathway components, such as Lgr5 [68, 69], which could in turn contribute to the differential response of anterior and posterior taste epithelia to increased β-catenin signaling [70, 71].
While the impact of β-catenin stabilization in Shh+ precursors was strictly taste bud autonomous in the anterior tongue, i.e., only buds with YFP+ cells were affected, in the posterior CVP, increased β-catenin under the control of the Shh promoter resulted in more Type I cells in taste buds, regardless of whether the taste bud possessed Shh-descendent YFP+ cells or not. This finding implies that stabilized β-catenin in this Shh+ cell population(s) may impact CVP taste cell fate more broadly. The anterior FFP are small, simple structures housing single taste buds, and FFP are typically distributed in the lingual epithelium at low density (~100/cm2) [72]. The single rodent CVP, by contrast contains hundreds of buds, which are packed in close proximity to one another, interspersed with small numbers of Krt14+ cells basolaterally, and Krt13+ cells apicolaterally (see Fig 1A, control). Therefore, increased signaling from Shh+ cells with activated β-catenin may be detectable by adjacent progenitors and taste buds, and thus impact cell fate decisions indirectly. The nature of this signal remains to be explored.
In conclusion, we show that, in parallel with its roles in skin, intestinal and neural epithelia, forced activation of β-catenin signaling promotes acquisition of taste fate, affects both renewal and differentiation of taste buds in adult mice, and does so primarily at the level of the progenitor population. Because radiotherapy targeting head and neck cancers causes taste dysfunction [73], and taste cell renewal is reduced in mice following head and neck radiation [46], our data suggest canonical Wnt signaling as a potential therapeutic target to restore taste sensitivity in these patients. Our data also suggest that, similar to the effects of Shh pathway antagonists [74], systemic cancer therapeutics that block Wnt signaling may cause taste dysfunction, and would need complementary treatment to help restore normal taste function to avoid malnutrition and psychological distress in these patients.
Materials and Methods
Ethics statement
Mice were housed in compliance with the Guide for the Care and Use of Laboratory Animals, Animal Welfare Act and Public Health Service Policy. All procedures were approved by the Institutional Animal Care and Use Committee at the University of Colorado Anschutz Medical Campus.
Animals and procedures
Male and female transgenic mouse lines were all on a mixed background (FVB, 129Sv, C57Bl6). All experimental tissue was generated from adult mice between 7–11 weeks of age. To stabilize β-catenin in epithelial progenitors of taste and non-taste epithelium, trigenic mice were generated: 1) Krt5rtTA—expression of a transcriptional activator rtTA is controlled by the human Krt5 promoter [75]; 2) tetO-Cre tetracycline-sensitive tetO response element controls production of Cre recombinase [76]; 3) Ctnnb1(Ex3)fl—floxed allele of β-catenin with exon 3 flanked by loxP sites [27]. Krt5rtTA;tetOCre;Ctnnb1(Ex3)fl/+ mice were fed the tetracycline analog doxycycline ad libitum in their chow (Bio-Serv, Frenchtown, NJ, 1g/kg) continuously until sacrifice at 2–14 days (longer time points were not possible as mice sickened by 16 days and died around 20 days).
To validate our model, we fed Krt5-rtTA;tetO-Cre;R26RLacZ reporter mice [77] doxycycline chow up to 28 days and then examined lingual tissue via Xgal reaction.
To explore whether β-catenin directly regulates the differentiation of precursors into taste cells, we stabilized β-catenin in Shh-expressing cells by generating trigenic mice: 1) ShhCreERT2—expression of an tamoxifen-inducible Cre recombinase under the Shh promoter [78]; 2) Ctnnb1(Ex3)fl—[27]; 3) R26R-YFP—expression of YFP in the Rosa locus downstream of a LoxP-flanked stop sequence [79]. Mice were gavaged with tamoxifen (100 mg/kgbw; stock solution 10 mg tamoxifen/ml corn oil) once every morning for 8 consecutive days, and were sacrificed 14 days after the last gavage.
Tissue collection
Tongues were dissected from the lower jaw and quickly frozen, fixed via direct immersion, or following transcardial perfusion with Periodate-lysine-paraformaldehyde (PLP) or 4% paraformaldehyde (PFA) in 1× Phosphate Buffer (PB: 29 mM NaH2PO4, 75 mM Na2HPO4). Fixation method is specified in S2 Table for each antiserum used.
Fixed tissue
Mice were anesthetized by i.p. injection of 250 mg/kgbody weight Avertin (2,2,2-Tribromoethanol), and ice cold 0.9% sodium chloride was perfused transcardially to clear blood, followed by Periodate-Lysine-Paraformaldehyde (PLP: 75mM L-lysine monohydrochloride, 1.6% paraformaldehyde, 10 mM sodium periodate) or 4% PFA fixative perfusion. Tongues were then incubated for 3 h in fixative at 4°C, and placed in 20% sucrose (Fisher Scientific, Pittsburgh PA, USA) in 1× PB overnight at 4 °C. Samples were embedded in O.C.T Compound (Tissue-Tek 4583, Sakura Finetek, Torrance CA, USA), frozen on dry ice and stored at -80°C.
Fresh tissue
Mice were euthanized by CO2 inhalation followed by cervical dislocation. The tongues were collected, rinsed in sterile ice-cold 1× Phosphate Buffered Saline (PBS: 29 mM NaH2PO4, 75 mM Na2HPO4, 154 mM NaCl) and embedded in O.C.T Compound, frozen on dry ice and stored at -80°C.
Peeled epithelium
Mice were euthanized by CO2 inhalation followed by cervical dislocation. The tongues were collected, rinsed in ice-cold Normal Tyrode solution (140 mM NaCl, 5 mM KCl, 10 mM HEPES, 4 mM CaCl2, 10 mM glucose, 1 mM MgCl2, 1 mM sodium pyruvate, pH 7.4), and 200 µl of dispase II (3 mg/ml Normal Tyrode solution) was injected underneath the epithelium. Tongues were incubated in calcium-free Tyrode solution at room temperature for 20 min, the epithelium (tip to intermolar eminence) was peeled, immediately placed in a tube and frozen on dry ice. Samples were stored at -80°C.
X-Gal reaction
X-gal reactions were performed on 12 μm cryostat PLP-fixed sections collected on Superfrost Plus Slides. Sections were washed with solution 1 (0.02% Nonidet P40, 2 mM Mgcl2 in 1× PBS pH 7.3), and incubated in reaction solution (5 mM Potassium ferrocyanide, 5mM Potassium ferricyanide, 0.5 mg/ml X-gal in solution 1) at 37°C until desired staining was obtained. Slides were washed in 1× PB, and coverslipped using Fluoromount G (SouthernBiotech, Birmingham AL, USA).
Immunohistochemistry
Immunolabeling was performed on 12 μm cryostat sections collected on Superfrost Plus Slides (Fisher Scientific, Pittsburgh PA, USA) following Gaillard and Barlow (2011) [31]. Sections were rehydrated in 1× PBS prior to staining. Antigen specific protocols are detailed below. Primary and secondary antisera, amplification systems, and dilutions used are listed in S2 Table. Immunoreactivity for each antigen listed was abolished when primary antibodies were omitted. Nuclear counterstain was performed using Sytox Green Nucleic Acid Stain (Invitrogen), or DRAQ5 (Abcam).
β-catenin
Sections were incubated in 10 mM sodium citrate pH 6 + 0.05% tween 20 at 95°C for 15 min, and incubated in blocking solution (2% normal goat serum, 1% bovine serum albumin, 0.3% Triton X100 in 1× PBS, pH 7.3) for 45 min at room temperature. Because the antiserum against β-catenin was produced in mouse, we used the M.O.M. kit from Vector Labs (BMK-2202). First, blocking of non-specific binding sites of the avidin/biotin amplification system was performed by using an Avidin/Biotin kit (Vector Labs SP-2001). Sections were incubated in Avidin solution for 15 min, rinsed, incubated in Biotin solution for 15 min and rinsed again. Slides were dipped in M.O.M. Ig blocking reagent for 1h at room temperature, washed in 1× PBS, incubated in M.O.M. diluent (protein concentrate diluted 1/14 in 1× PBS) for 10 min at room temperature, and incubated in primary antiserum raised against β-catenin (diluted in M.O.M. diluent) overnight at 4°C. Sections were washed, incubated in M.O.M. biotinylated anti-mouse IgG reagent (diluted in M.O.M. diluent) for 1 h at room temperature, washed again, and incubated with Streptavidin Alexa 488 diluted in PBS+0.1% Triton X100 1 h at room temperature. Nuclei were counterstained with DRAQ5 (Abcam, Cambridge, UK) diluted 1/8000 in 1× PB for 30 min at room temperature. Slides were coverslipped using Fluoromount G.
Cell types, keratins, Skn-1a, Mash1, Claudin4, YFP
Fresh tissue or PLP-fixed sections were thawed and post-fixed in 4% PFA for 10 min at room temperature, washed with 1× PBS, incubated for 1.5 h at room temperature in blocking solution (5% normal goat serum, 1% bovine serum albumin, 0.3% Triton X100 in 1× PBS, pH 7.3), and incubated with primary antisera diluted in blocking solution, overnight at 4°C (4 days for MASH1[80]). Sections were washed prior to incubation with secondary antisera diluted in blocking solution for 1 h at room temperature (2 h for MASH1), and washed in 1× PBS. Triple stained fresh tissue sections for Krt8/Krt13/Krt14 were fixed in 4% paraformaldehyde for 10 min at room temperature, and washed in 0.1M PBS. Fixed and fresh sections were counterstained with DRAQ5 as described above, and coverslipped using Fluoromount G.
Ki67
Sections were incubated in 10 mM sodium citrate pH 6 + 0.05% tween 20 at 95°C for 15 min. Once slides reached room temperature, sections were incubated in blocking solution for 1 h at room temperature, then incubated with Ki67 antiserum diluted in blocking solution overnight at 4°C. Sections were rinsed, and blocking of non-specific binding sites of the avidin/biotin amplification system was performed by using an Avidin/Biotin kit (Vector Labs SP-2001), as above. Sections were incubated with the anti-rabbit biotin-conjugated antibody diluted in 1× PBS, 0.1% tween 20, 2.5% normal goat serum for 1 h at room temperature. Samples were then incubated in streptavidin-Alexa 546 diluted in 1% bovine serum albumin, 0.3% Triton X100 in 1× PBS, pH 7.3 for 1 h at room temperature, rinsed, counterstained with SYTOX green (Invitrogen) diluted 1/30000 in 1× PB pH 7.2 for 1 min at room temperature, rinsed again, and slides were mounted with Fluoromount G.
TUNEL assay
To assess cell death, the In Situ Cell Death Detection Kit TMR red (Roche Applied Science, Cat #12156792910) was used. Sections were washed in 0.1M PBS prior to antigen retrieval in 0.1 M Sodium Citrate pH = 6 for 15 min at 90°C. Sections were washed and incubated in a permeabilization solution (0.1% Triton X100 in 0.1% Sodium citrate) for 2 min on ice. Slides were washed and incubated in blocking buffer (50 mM Tris-HCl pH = 7.5, 3% BSA, 20% NGS) 30 min at room temperature. TUNEL reaction was performed according to the manufacturer’s instructions, by mixing 1 volume of Enzyme Solution with 9 volumes of Label Solution, and incubating the sections 60 min at 37°C in humidified atmosphere. Sections were washed, counterstained with Sytox Green, and slides mounted with Fluoromount G. One negative control was included by incubating a slide with the Label Solution only, and one positive control was added by incubating a slide with DNase I (10 U/ml in 50 mM Tris-HCl pH 7.5, 1 mg/ml BSA) 20 min at room temperature prior to executing the TUNEL reaction.
In situ hybridization
Detection of mRNA encoding for Shh was performed as previously described [31]. Antisense RNA probes were synthesized from a linearized plasmid containing a Shh cDNA insert [81], Ptch1 cDNA insert (1318–2362: Genbank MMU46155), or Mash1 cDNA insert (10012: Genbank U68534–783: Genbank M65603, 1276 bp), using FITC-conjugated UTP or digoxigenin-conjugated UTP. Sections were incubated in 4% PFA for 10 min at room temperature, rinsed in 0.1× PBS (14 mM NaCl, 0.3 mM KCl, 0.3 mM Na2HPO4, 0.2 mM KH2PO4), incubated in triethanolamine solution (1.3% triethanolamine, 0.175% HCl 10 N, 0.25% acetic anhydride), rinsed in 0.1× PBS, incubated in hybridization solution (50% formamide, 5× SCC (750 mM NaCl, 75 mM sodium citrate dihydrate), 5× Denhardt’s solution (0.1% Ficoll, 0.1% polyvinylpyrrolidone, and 0.1% bovine serum albumin), 500 μg/ml salmon sperm DNA and 250 μg/ml tRNA) for 2 h at room temperature, then with the RNA probes in hybridization solution overnight at 65°C in a moist chamber. Sections were incubated 90 min at 65°C in 0.2× SSC (30 mM NaCl, 3 mM sodium citrate dihydrate), then in Buffer 2T for 1 h at room temperature, and incubated with peroxidase-coupled anti-digoxigenin antibody diluted 1/600 or alkaline phosphatase-coupled anti-FITC antibody diluted 1/5000 in Buffer 2T overnight in a moist chamber at 4°C. To detect Shh mRNA in the CVP, sections were treated with Streptavidin-Alexa 488 diluted 1/400 (Invitrogen, Carlsbad, CA, USA) for 30 min following a 30 min tyramide-biotin treatment (TSA Biotin Tyramide Reagent, PerkinElmer, Waltham, MA, USA). In the anterior tongue, Shh, Patched1 and MASH1 mRNA transcripts were detected by incubating sections with NBT/BCIP solution (Roche Applied Science, 11681451001) in Buffer 3 (0.1 M Tris-HCl pH 9.5, 0.1 M NaCl, 50 mM MgCl2) at room temperature until desired staining is obtained. Reaction was blocked in Buffer 4 (10 mM Tris-HCl pH 8, 1 mM EDTA) for at least 10 min, and slides were coverslipped with Fluoromount G.
Real-time RT-PCR
Total RNA was extracted from peeled anterior tongue epithelium using the RNeasy Plus Mini kit (Qiagen). Bioanalyzer 2100 (Agilent technologies) was used to assess RNA integrity. cDNA was prepared by Reverse Transcription of 1 μg total RNA using the Omniscript Reverse Transcription kit (Qiagen). Mash1 mRNA levels were normalised to β-actin mRNA levels. cDNA equivalent of 20 ng total RNA, 250 nM of the forward and reverse primers were mixed with the Power SYBR Green PCR Master Mix (Applied Biosystems). Primers sequences were as follows: Mash1 (NM_008553)[82]: forward 5’-GCAACCGGGTCAAGTTGGT-3’, reverse 5’-GTCGTTGGAGTAGTTGGGGG-3’; β-actin (NM_007393)[83]: forward 5’-ACCAACTGGGACGATATGGAGAAGA-3’, reverse 5’-TACGACCAGAGGCATACAGGGACAA-3’. Real-time PCR consisted of forty 95°C/15 s-60°C/60 s cycles. The comparative ΔΔCt method was used for relative quantification of gene expression [84].
Scanning electron microscopy (SEM)
Scanning electron microscope experiments were performed at CDB/CVI Microscopy Core (Perelman School of Medicine, University of Pennsylvania). Tongue samples were washed three times with 1× PBS, fixed overnight in 4% PFA and dehydrated in a graded series of ethanol concentrations through 100% over a period of 1.5 hour. Dehydration in 100% ethanol was done three times. Dehydrated samples were then incubated for 20 min in 50% HMDS in ethanol followed by three changes of 100% HMDS (Sigma-Aldrich Co.) and followed by overnight air-drying as described previously [85]. Then samples were mounted on stubs and sputter coated with gold palladium. Specimens were observed and photographed using a Quanta 250 scanning electron microscope (FEI, Hillsboro, OR, USA) at 10 kV accelerating voltage.
Image acquisition and analysis
Confocal fluorescence images were acquired using a Leica TCS SP5 II laser-scanning confocal microscope and LASAF software. Nomarski images were acquired using a Zeiss Axioplan 2 microscope, camera and software. All sections of the CVP, except the first and last sections which were excluded as they generally contain incomplete trenches, were analyzed. For the anterior tongue, 12 μm serial sections were cut into 6 sets such that sections on each slide were separated by 72 μm. FFP were analyzed in the 3rd through the 12th section, while the 1st and 2nd sections were omitted due to the curved nature of the tongue surface and difficulty in interpreting non-transverse sections through FFP. Thus we analyzed FFP in a region representing 720 μm of the anterior tongue starting ~145 μm from the tongue tip. Proliferative index (P.I.) in the CVP was calculated by dividing the number of Ki67+ basal cells by the number of Sytox Green+ basal cells, i.e., cells residing along the basement membrane, within the portion of the CVP trenches housing taste buds [46]. In the anterior tongue, the number of Ki67+ cells was tallied along 400 μm of non-taste epithelium on the dorsal part of the tongue.
ImageJ (NIH) was used to measure the corrected integrated density of Krt8-, Krt14-, PLCβ2 - and NTPdase2-immunofluorescence signal. NTPdase2 is also expressed by Schwann cells of the CVP innervation (green) [49], but this component of NTPdase2+ signal was excluded from measurements of epithelial signal, as nerve fibers within taste buds are not myelinated [86]. The area, mean gray value and integrated density were measured in the area of interest, and in 4 small areas selected as the background signal. The corrected integrated density was calculated as follows:
Corrected integrated density = Integrated density − (Area selected × Mean value of background) [87].
Immunolabeled cells were tallied by analyzing both 0.75 μm optical sections and compressed z-stacks (14 optical sections). Immunolabeled cells and in situ labeled cells were counted when a nuclear profile was identifiable (nuclear staining, counterstain, or no staining in an elliptical shape within a cytoplasmically stained cell of interest).
Statistical analysis
Statistical analyses were performed using SigmaStat (Systat Software). Normal distribution and equal variances between groups were assessed with a p value set at 5%, to determine whether to run a Mann-Whitney test or a Student’s t-test. Statistical differences were established with a minimum confidence interval of 95%. Non-parametric data are represented as medians, 1st and 3rd quartiles, and error bars represent minimum and maximum values, while parametric data are represented as means ± SEM. Sample sizes for data are presented in the figure legends.
Supporting Information
Zdroje
1. Roper SD. The cell biology of vertebrate taste receptors. Annual review of neuroscience. 1989;12 : 329–53. 2648951
2. Barlow LA, Northcutt RG. Embryonic Origin of Amphibian Taste Buds. Developmental Biology. 1995;169(1):273–85. 7750643
3. Stone LM, Finger TE, Tam PP, Tan SS. Taste receptor cells arise from local epithelium, not neurogenic ectoderm. Proceedings of the National Academy of Sciences. 1995;92(6):1916–20. 7892199
4. Liu H - X, Komatsu Y, Mishina Y, Mistretta CM. Neural crest contribution to lingual mesenchyme, epithelium and developing taste papillae and taste buds. Developmental Biology. 2012;368(2):294–303. doi: 10.1016/j.ydbio.2012.05.028 22659543
5. Feng P, Huang L, Wang H. Taste Bud Homeostasis in Health, Disease, and Aging. Chemical senses. 2014;39(1):3–16. doi: 10.1093/chemse/bjt059 24287552
6. Okubo T, Clark C, Hogan BL. Cell lineage mapping of taste bud cells and keratinocytes in the mouse tongue and soft palate. Stem cells (Dayton, Ohio). 2009;27(2):442–50. doi: 10.1634/stemcells.2008-0611 19038788
7. Presland RB, Dale BA. Epithelial Structural Proteins of the Skin and Oral Cavity: Function in Health and Disease. Critical Reviews in Oral Biology & Medicine. 2000;11(4):383–408. doi: 10.1186/s12967-014-0362-3 25592846
8. Hill MW. Influence of age on the morphology and transit time of murine stratified squamous epithelia. Archives of oral biology. 1988;33(4):221–9. 3165257
9. Potten CS, Booth D, Cragg NJ, O'Shea JA, Tudor GL, Booth C. Cell kinetic studies in murine ventral tongue epithelium: cell cycle progression studies using double labelling techniques. Cell proliferation. 2002;35 : 16–21. 12139704
10. Moore KA, Lemischka IR. Stem Cells and Their Niches. Science. 2006;311(5769):1880–5. 16574858
11. Creamer B, Shorter RG, Bamforth J. The turnover and shedding of epithelial cells. I. The turnover in the gastro-intestinal tract. Gut. 1961;2 : 110–8. 13696345
12. Potten CS, Saffhill R, Maibach HI. Measurement of the transit time for cells through the epidermis and stratum corneum of the mouse and guinea-pig. Cell proliferation. 1987;20(5):461–72. 3450396
13. Beidler LM, Smallman RL. Renewal of cells within taste buds. The Journal of cell biology. 1965;27(2):263–72. 5884625
14. Farbman AI. Renewal of taste bud cells in rat circumvallate papillae. Cell and tissue kinetics. 1980;13(4):349–57. 7428010
15. Hamamichi R, Asano-Miyoshi M, Emori Y. Taste bud contains both short-lived and long-lived cell populations. Neuroscience. 2006;141(4):2129–38. 16843606
16. Perea-Martinez I, Nagai T, Chaudhari N. Functional Cell Types in Taste Buds Have Distinct Longevities. PLoS ONE. 2013;8(1):e53399. doi: 10.1371/journal.pone.0053399 23320081
17. Vandenbeuch A, Clapp TR, Kinnamon SC. Amiloride-sensitive channels in type I fungiform taste cells in mouse. BMC Neurosci. 2008;9 : 1. doi: 10.1186/1471-2202-9-1 18171468
18. Chaudhari N, Roper SD. The cell biology of taste. The Journal of cell biology. 2010;190(3):285–96. doi: 10.1083/jcb.201003144 20696704
19. Ma H, Yang R, Thomas S, Kinnamon J. Qualitative and quantitative differences between taste buds of the rat and mouse. BMC Neuroscience. 2007;8(1):5.
20. Ohtubo Y, Yoshii K. Quantitative analysis of taste bud cell numbers in fungiform and soft palate taste buds of mice. Brain Research. 2011;1367 : 13–21. doi: 10.1016/j.brainres.2010.10.060 20971092
21. Miura H, Kusakabe Y, Harada S. Cell lineage and differentiation in taste buds. Archives of histology and cytology. 2006;69(4):209–25. 17287576
22. Miura H, Scott JK, Harada S, Barlow LA. Sonic hedgehog-expressing basal cells are general post-mitotic precursors of functional taste receptor cells. Developmental Dynamics. 2014;243(10):1286–97. doi: 10.1002/dvdy.24121 24590958
23. Liu F, Thirumangalathu S, Gallant NM, Yang SH, Stoick-Cooper CL, Reddy ST, et al. Wnt-beta-catenin signaling initiates taste papilla development. Nature genetics. 2007;39(1):106–12. 17128274
24. Iwatsuki K, Liu H - X, Gründer A, Singer MA, Lane TF, Grosschedl R, et al. Wnt signaling interacts with Shh to regulate taste papilla development. Proceedings of the National Academy of Sciences. 2007;104(7):2253–8. 17284610
25. Choi Yeon S, Zhang Y, Xu M, Yang Y, Ito M, Peng T, et al. Distinct Functions for Wnt/β-Catenin in Hair Follicle Stem Cell Proliferation and Survival and Interfollicular Epidermal Homeostasis. Cell stem cell. 2013;13(6):720–33. doi: 10.1016/j.stem.2013.10.003 24315444
26. Widelitz RB, Jiang T-X, Lu J, Chuong C-M. β-catenin in Epithelial Morphogenesis: Conversion of Part of Avian Foot Scales into Feather Buds with a Mutated β-Catenin. Developmental Biology. 2000;219(1):98–114. 10677258
27. Harada N, Tamai Y, Ishikawa T-o, Sauer B, Takaku K, Oshima M, et al. Intestinal polyposis in mice with a dominant stable mutation of the [beta]-catenin gene. EMBO J. 1999;18(21):5931–42. 10545105
28. Sangiorgi E, Capecchi MR. Bmi1 is expressed in vivo in intestinal stem cells. Nature genetics. 2008;40(7):915–20. doi: 10.1038/ng.165 18536716
29. Sansom OJ, Reed KR, Hayes AJ, Ireland H, Brinkmann H, Newton IP, et al. Loss of Apc in vivo immediately perturbs Wnt signaling, differentiation, and migration. Genes & Development. 2004;18(12):1385–90.
30. Varela-Nallar L, Inestrosa NC. Wnt signaling in the regulation of adult hippocampal neurogenesis. Frontiers in cellular neuroscience. 2013;7(100):1–11.
31. Gaillard D, Barlow LA. Taste bud cells of adult mice are responsive to Wnt/β-catenin signaling: Implications for the renewal of mature taste cells. Genesis. 2011;49(4):295–306. doi: 10.1002/dvg.20731 21328519
32. Rothova M, Thompson H, Lickert H, Tucker AS. Lineage tracing of the endoderm during oral development. Developmental Dynamics. 2012;241(7):1183–91. doi: 10.1002/dvdy.23804 22581563
33. Asano-Miyoshi M, Hamamichi R, Emori Y. Cytokeratin 14 is expressed in immature cells in rat taste buds. Journal of molecular histology. 2008;39(2):193–9. 17960487
34. Iwasaki S, Aoyagi H, Yoshizawa H. Localization of keratins 13 and 14 in the lingual mucosa of rats during the morphogenesis of circumvallate papillae. Acta histochemica. 2011;113(4):395–401. doi: 10.1016/j.acthis.2010.03.003 20546859
35. Zhang C, Cotter M, Lawton A, Oakley B, Wong L, Zeng Q. Keratin 18 is associated with a subset of older taste cells in the rat. Differentiation; research in biological diversity. 1995;59(3):155–62. 7589899
36. Miura H, Kusakabe Y, Sugiyama C, Kawamatsu M, Ninomiya Y, Motoyama J, et al. Shh and Ptc are associated with taste bud maintenance in the adult mouse. Mech Dev. 2001;106(1–2):143–5. 11472849
37. Liu HX, Ermilov A, Grachtchouk M, Li L, Gumucio DL, Dlugosz AA, et al. Multiple Shh signaling centers participate in fungiform papilla and taste bud formation and maintenance. Developmental Biology. 2013;382(1):82–97. doi: 10.1016/j.ydbio.2013.07.022 23916850
38. Shin K, Fogg VC, Margolis B. Tight Junctions and Cell Polarity. Annual Review of Cell and Developmental Biology. 2006;22(1):207–35.
39. Van Itallie C, Rahner C, Anderson JM. Regulated expression of claudin-4 decreases paracellular conductance through a selective decrease in sodium permeability. J Clin Invest. 2001;107(10):1319–27. 11375422
40. Michlig S, Damak S, Le Coutre J. Claudin-based permeability barriers in taste buds. The Journal of comparative neurology. 2007;502(6):1003–11. 17447253
41. Castillo D, Seidel K, Salcedo E, Ahn C, de Sauvage FJ, Klein OD, et al. Induction of ectopic taste buds by SHH reveals the competency and plasticity of adult lingual epithelium. Development. 2014;141(15):2993–3002. doi: 10.1242/dev.107631 24993944
42. Hosley MA, Oakley B. Postnatal development of the vallate papilla and taste buds in rats. The Anatomical record. 1987;218(2):216–22. 3619089
43. Bradley RM, Stern IB. The development of the human taste bud during the foetal period. Journal of anatomy. 1967;101(Pt 4):743–52.
44. Belecky TL, Smith DV. Postnatal development of palatal and laryngeal taste buds in the hamster. The Journal of comparative neurology. 1990;293(4):646–54. 2329198
45. Mbiene JP, Farbman AI. Evidence for stimulus access to taste cells and nerves during development: an electron microscopic study. Microscopy research and technique. 1993;26(2):94–105. 8241557
46. Nguyen HM, Reyland ME, Barlow LA. Mechanisms of taste bud cell loss after head and neck irradiation. J Neurosci. 2012;32(10):3474–84. doi: 10.1523/JNEUROSCI.4167-11.2012 22399770
47. Yang R, Crowley HH, Rock ME, Kinnamon JC. Taste cells with synapses in rat circumvallate papillae display SNAP-25-like immunoreactivity. The Journal of comparative neurology. 2000;424(2):205–15. 10906698
48. Clapp TR, Yang R, Stoick CL, Kinnamon SC, Kinnamon JC. Morphologic characterization of rat taste receptor cells that express components of the phospholipase C signaling pathway. The Journal of comparative neurology. 2004;468(3):311–21. 14681927
49. Bartel DL, Sullivan SL, Lavoie EG, Sevigny J, Finger TE. Nucleoside triphosphate diphosphohydrolase-2 is the ecto-ATPase of type I cells in taste buds. The Journal of comparative neurology. 2006;497(1):1–12. 16680780
50. Lawton DM, Furness DN, Lindemann B, Hackney CM. Localization of the glutamate-aspartate transporter, GLAST, in rat taste buds. The European journal of neuroscience. 2000;12(9):3163–71. 10998100
51. Pumplin DW, Yu C, Smith DV. Light and dark cells of rat vallate taste buds are morphologically distinct cell types. The Journal of comparative neurology. 1997;378(3):389–410. 9034899
52. Seta Y, Oda M, Kataoka S, Toyono T, Toyoshima K. Mash1 is required for the differentiation of AADC-positive type III cells in mouse taste buds. Dev Dyn. 2011;240(4):775–84. doi: 10.1002/dvdy.22576 21322090
53. Kito-Shingaki A, Seta Y, Toyono T, Kataoka S, Kakinoki Y, Yanagawa Y, et al. Expression of GAD67 and Dlx5 in the Taste Buds of Mice Genetically Lacking Mash1. Chemical senses. 2014;39(5):403–14. doi: 10.1093/chemse/bju010 24682237
54. Matsumoto I, Ohmoto M, Narukawa M, Yoshihara Y, Abe K. Skn-1a (Pou2f3) specifies taste receptor cell lineage. Nature neuroscience. 2011;14(6):685–7. doi: 10.1038/nn.2820 21572433
55. Gat U, DasGupta R, Degenstein L, Fuchs E. De Novo hair follicle morphogenesis and hair tumors in mice expressing a truncated beta-catenin in skin. Cell. 1998;95(5):605–14. 9845363
56. Lim X, Tan SH, Koh WLC, Chau RMW, Yan KS, Kuo CJ, et al. Interfollicular Epidermal Stem Cells Self-Renew via Autocrine Wnt Signaling. Science. 2013;342(6163):1226–30. doi: 10.1126/science.1239730 24311688
57. Lien W-H, Fuchs E. Wnt some lose some: transcriptional governance of stem cells by Wnt/β-catenin signaling. Genes & Development. 2014;28(14):1517–32.
58. Clevers H. The Intestinal Crypt, A Prototype Stem Cell Compartment. Cell. 2013;154(2):274–84. doi: 10.1016/j.cell.2013.07.004 23870119
59. Andreu P, Peignon G, Slomianny C, Taketo MM, Colnot S, Robine S, et al. A genetic study of the role of the Wnt/β-catenin signalling in Paneth cell differentiation. Developmental Biology. 2008;324(2):288–96. doi: 10.1016/j.ydbio.2008.09.027 18948094
60. Lowry WE, Blanpain C, Nowak JA, Guasch G, Lewis L, Fuchs E. Defining the impact of beta-catenin/Tcf transactivation on epithelial stem cells. Genes Dev. 2005;19(13):1596–611. 15961525
61. Zhang Y, Andl T, Yang SH, Teta M, Liu F, Seykora JT, et al. Activation of beta-catenin signaling programs embryonic epidermis to hair follicle fate. Development. 2008;135(12):2161–72. doi: 10.1242/dev.017459 18480165
62. Lien W - H, Polak L, Lin M, Lay K, Zheng D, Fuchs E. In vivo transcriptional governance of hair follicle stem cells by canonical Wnt regulators. Nature cell biology. 2014;16(2):179–90. doi: 10.1038/ncb2903 24463605
63. Clevers H. Wnt/[beta]-Catenin Signaling in Development and Disease. Cell. 2006;127(3):469–80. 17081971
64. Northcutt RG. Taste Buds: Development and Evolution. Brain, Behavior and Evolution. 2004;64(3):198–206. 15353910
65. Nguyen HM, Barlow LA. BMP4 expression differs in anterior fungiform versus posterior circumvallate taste buds of mice. BMC Neurosci. 2010;11(1):129.
66. Petersen CI, Jheon AH, Mostowfi P, Charles C, Ching S, Thirumangalathu S, et al. FGF Signaling Regulates the Number of Posterior Taste Papillae by Controlling Progenitor Field Size. PLoS genetics. 2011;7(6):e1002098. doi: 10.1371/journal.pgen.1002098 21655085
67. Kist R, Watson M, Crosier M, Robinson M, Fuchs J, Reichelt J, et al. The formation of endoderm-derived taste sensory organs requires a pax9-dependent expansion of embryonic taste bud progenitor cells. PLoS genetics. 2014;10(10):e1004709. doi: 10.1371/journal.pgen.1004709 25299669
68. Chai R, Xia A, Wang T, Jan TA, Hayashi T, Bermingham-McDonogh O, et al. Dynamic Expression of Lgr5, a Wnt Target Gene, in the Developing and Mature Mouse Cochlea. JARO. 2011;12(4):455–69. doi: 10.1007/s10162-011-0267-2 21472479
69. Yamamoto S, Nakase H, Matsuura M, Honzawa Y, Matsumura K, Uza N, et al. Heparan sulfate on intestinal epithelial cells plays a critical role in intestinal crypt homeostasis via Wnt/beta-catenin signaling. Am J Physiol Gastrointest Liver Physiol. 2013;305(3):G241–9. doi: 10.1152/ajpgi.00480.2012 23744737
70. Takeda N, Jain R, Li D, Li L, Lu MM, Epstein JA. Lgr5 Identifies Progenitor Cells Capable of Taste Bud Regeneration after Injury. PLoS ONE. 2013;8(6):e66314. 23824276
71. Yee KK, Li Y, Redding KM, Iwatsuki K, Margolskee RF, Jiang P. Lgr5-EGFP Marks Taste Bud Stem/Progenitor Cells in Posterior Tongue. Stem cells (Dayton, Ohio). 2013:N/A-N/A.
72. Zhang GH, Zhang HY, Deng SP, Qin YM. Regional differences in taste bud distribution and alpha-gustducin expression patterns in the mouse fungiform papilla. Chemical senses. 2008;33(4):357–62. doi: 10.1093/chemse/bjm093 18296428
73. Sandow PL, Hejrat-Yazdi M, Heft MW. Taste Loss and Recovery Following Radiation Therapy. Journal of dental research. 2006;85(7):608–11. 16798859
74. Ruat M, Hoch L, Faure H, Rognan D. Targeting of Smoothened for therapeutic gain. Trends in Pharmacological Sciences. 2014;35(5):237–46. doi: 10.1016/j.tips.2014.03.002 24703627
75. Diamond I, Owolabi T, Marco M, Lam C, Glick A. Conditional Gene Expression in the Epidermis of Transgenic Mice Using the Tetracycline-Regulated Transactivators tTA and rTA Linked to the Keratin 5 Promoter. J Invest Dermatol. 2000;115(5):788–94. 11069615
76. Perl A - KT, Wert SE, Nagy A, Lobe CG, Whitsett JA. Early restriction of peripheral and proximal cell lineages during formation of the lung. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(16):10482–7. 12145322
77. Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nature genetics. 1999;21(1):70–1. 9916792
78. Harfe BD, Scherz PJ, Nissim S, Tian H, McMahon AP, Tabin CJ. Evidence for an Expansion-Based Temporal Shh Gradient in Specifying Vertebrate Digit Identities. Cell. 2004;118(4):517–28. 15315763
79. Srinivas S, Watanabe T, Lin C-S, William C, Tanabe Y, Jessell T, et al. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Developmental Biology. 2001;1(1):4.
80. Kim EJ, Battiste J, Nakagawa Y, Johnson JE. Ascl1 (Mash1) lineage cells contribute to discrete cell populations in CNS architecture. Molecular and Cellular Neuroscience. 2008;38(4):595–606. doi: 10.1016/j.mcn.2008.05.008 18585058
81. Kitamura K, Miura H, Yanazawa M, Miyashita T, Kato K. Expression patterns of Brx1 (Rieg gene), Sonic hedgehog, Nkx2.2, Dlx1 and Arx during zona limitans intrathalamica and embryonic ventral lateral geniculate nuclear formation. Mech Dev. 1997;67(1):83–96. 9347917
82. Kokovay E, Wang Y, Kusek G, Wurster R, Lederman P, Lowry N, et al. VCAM1 Is Essential to Maintain the Structure of the SVZ Niche and Acts as an Environmental Sensor to Regulate SVZ Lineage Progression. Cell stem cell. 2012;11(2):220–30. doi: 10.1016/j.stem.2012.06.016 22862947
83. Ceradini DJ, Kulkarni AR, Callaghan MJ, Tepper OM, Bastidas N, Kleinman ME, et al. Progenitor cell trafficking is regulated by hypoxic gradients through HIF-1 induction of SDF-1. Nat Med. 2004;10(8):858–64. 15235597
84. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods (San Diego, Calif. 2001;25(4):402–8. 11846609
85. Braet F, De Zanger R, Wisse E. Drying cells for SEM, AFM and TEM by hexamethyldisilazane: a study on hepatic endothelial cells. Journal of microscopy. 1997;186(Pt 1):84–7.
86. Yoshie S, Wakasugi C, Teraki Y, Fujita T. Fine structure of the taste bud in guinea pigs. I. Cell characterization and innervation patterns. Archives of histology and cytology. 1990;53(1):103–19. 2364007
87. Gavet O, Pines J. Progressive Activation of CyclinB1-Cdk1 Coordinates Entry to Mitosis. Developmental Cell. 2010;18(4):533–43. doi: 10.1016/j.devcel.2010.02.013 20412769
Štítky
Genetika Reprodukční medicína
Článek Minor Type IV Collagen α5 Chain Promotes Cancer Progression through Discoidin Domain Receptor-1Článek A Unique Virulence Gene Occupies a Principal Position in Immune Evasion by the Malaria ParasiteČlánek The Centrosomal Linker and Microtubules Provide Dual Levels of Spatial Coordination of CentrosomesČlánek Dissecting the Function and Assembly of Acentriolar Microtubule Organizing Centers in Cells In VivoČlánek MicroRNA-Dependent Transcriptional Silencing of Transposable Elements in Drosophila Follicle CellsČlánek PARP2 Is the Predominant Poly(ADP-Ribose) Polymerase in Arabidopsis DNA Damage and Immune ResponsesČlánek Cooperative Action of Cdk1/cyclin B and SIRT1 Is Required for Mitotic Repression of rRNA SynthesisČlánek The Developmental Intestinal Regulator ELT-2 Controls p38-Dependent Immune Responses in Adult .Článek Feeding and Fasting Signals Converge on the LKB1-SIK3 Pathway to Regulate Lipid Metabolism in
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2015 Číslo 5
-
Všechny články tohoto čísla
- Genomic Heritability: What Is It?
- Triglyceride-Increasing Alleles Associated with Protection against Type-2 Diabetes
- Epistasis Is a Major Determinant of the Additive Genetic Variance in
- Genetic Regulation of Bone Metabolism in the Chicken: Similarities and Differences to Mammalian Systems
- Minor Type IV Collagen α5 Chain Promotes Cancer Progression through Discoidin Domain Receptor-1
- The Philosophical Approach: An Interview with Ford Doolittle
- Downregulation of the Host Gene by miR-92 Is Essential for Neuroblast Self-Renewal in
- A Unique Virulence Gene Occupies a Principal Position in Immune Evasion by the Malaria Parasite
- Y Fuse? Sex Chromosome Fusions in Fishes and Reptiles
- Regulation of Active DNA Demethylation by a Methyl-CpG-Binding Domain Protein in
- Overlapping Patterns of Rapid Evolution in the Nucleic Acid Sensors cGAS and OAS1 Suggest a Common Mechanism of Pathogen Antagonism and Escape
- Autoselection of Cytoplasmic Yeast Virus Like Elements Encoding Toxin/Antitoxin Systems Involves a Nuclear Barrier for Immunity Gene Expression
- Genetic Architecture of Abdominal Pigmentation in
- Whole Genome DNA Binding Analysis of the Bacterial Replication Initiator and Transcription Factor DnaA
- The Centrosomal Linker and Microtubules Provide Dual Levels of Spatial Coordination of Centrosomes
- Parp3 Negatively Regulates Immunoglobulin Class Switch Recombination
- Burden Analysis of Rare Microdeletions Suggests a Strong Impact of Neurodevelopmental Genes in Genetic Generalised Epilepsies
- Cell Cycle Control by the Master Regulator CtrA in
- Myopathic Lamin Mutations Cause Reductive Stress and Activate the Nrf2/Keap-1 Pathway
- Monoallelic Loss of the Imprinted Gene Promotes Tumor Formation in Irradiated Mice
- Phylum-Level Conservation of Regulatory Information in Nematodes despite Extensive Non-coding Sequence Divergence
- Clustering and Negative Feedback by Endocytosis in Planar Cell Polarity Signaling Is Modulated by Ubiquitinylation of Prickle
- Dissecting the Function and Assembly of Acentriolar Microtubule Organizing Centers in Cells In Vivo
- MicroRNA-Dependent Transcriptional Silencing of Transposable Elements in Drosophila Follicle Cells
- β-Catenin Signaling Biases Multipotent Lingual Epithelial Progenitors to Differentiate and Acquire Specific Taste Cell Fates
- A Simple Auxin Transcriptional Response System Regulates Multiple Morphogenetic Processes in the Liverwort
- Parallel Gene Expression Differences between Low and High Latitude Populations of and .
- The Nutrient-Responsive Hormone CCHamide-2 Controls Growth by Regulating Insulin-like Peptides in the Brain of
- Characterization of TCF21 Downstream Target Regions Identifies a Transcriptional Network Linking Multiple Independent Coronary Artery Disease Loci
- PARP2 Is the Predominant Poly(ADP-Ribose) Polymerase in Arabidopsis DNA Damage and Immune Responses
- Drosophila Spaghetti and Doubletime Link the Circadian Clock and Light to Caspases, Apoptosis and Tauopathy
- Coronary Artery Disease Associated Transcription Factor TCF21 Regulates Smooth Muscle Precursor Cells That Contribute to the Fibrous Cap
- Rescue of DNA-PK Signaling and T-Cell Differentiation by Targeted Genome Editing in a Deficient iPSC Disease Model
- Disruption of Transcriptional Coactivator Sub1 Leads to Genome-Wide Re-distribution of Clustered Mutations Induced by APOBEC in Active Yeast Genes
- Yeast Killer Elements Hold Their Hosts Hostage
- Keeping in Shape the Dogma of Mitochondrial DNA Maternal Inheritance
- Extreme-Depth Re-sequencing of Mitochondrial DNA Finds No Evidence of Paternal Transmission in Humans
- Trading Places—Switching Frataxin Function by a Single Amino Acid Substitution within the [Fe-S] Cluster Assembly Scaffold
- Natural Variation Identifies , a Universal Gene Required for Cell Proliferation and Growth at High Temperatures in
- Mutations in Gene Are Associated with Predisposition to Breast Cancer
- The Whole of a Scientific Career: An Interview with Oliver Smithies
- Cell Specific eQTL Analysis without Sorting Cells
- Cooperative Action of Cdk1/cyclin B and SIRT1 Is Required for Mitotic Repression of rRNA Synthesis
- Systemic Regulation of RAS/MAPK Signaling by the Serotonin Metabolite 5-HIAA
- Reprogramming LCLs to iPSCs Results in Recovery of Donor-Specific Gene Expression Signature
- The Developmental Intestinal Regulator ELT-2 Controls p38-Dependent Immune Responses in Adult .
- Genetic Mechanism of Human Neutrophil Antigen 2 Deficiency and Expression Variations
- Feeding and Fasting Signals Converge on the LKB1-SIK3 Pathway to Regulate Lipid Metabolism in
- Early Lineage Priming by Trisomy of Leads to Myeloproliferation in a Down Syndrome Model
- Turning into a Frataxin-Dependent Organism
- Accounting for Experimental Noise Reveals That mRNA Levels, Amplified by Post-Transcriptional Processes, Largely Determine Steady-State Protein Levels in Yeast
- Biological Significance of Photoreceptor Photocycle Length: VIVID Photocycle Governs the Dynamic VIVID-White Collar Complex Pool Mediating Photo-adaptation and Response to Changes in Light Intensity
- CTXφ Replication Depends on the Histone-Like HU Protein and the UvrD Helicase
- Disruption of miR-29 Leads to Aberrant Differentiation of Smooth Muscle Cells Selectively Associated with Distal Lung Vasculature
- Notch Is Required in Adult Sensory Neurons for Morphological and Functional Plasticity of the Olfactory Circuit
- Post-transcriptional Regulation of Keratinocyte Progenitor Cell Expansion, Differentiation and Hair Follicle Regression by
- PERK Limits Lifespan by Promoting Intestinal Stem Cell Proliferation in Response to ER Stress
- Casein Kinase 1 and Phosphorylation of Cohesin Subunit Rec11 (SA3) Promote Meiotic Recombination through Linear Element Formation
- Fibroblast Growth Factor 9 Regulation by MicroRNAs Controls Lung Development and Links Loss to the Pathogenesis of Pleuropulmonary Blastoma
- Auxin-Mediated Transcriptional System with a Minimal Set of Components Is Critical for Morphogenesis through the Life Cycle in
- The Broad-Spectrum Antiviral Protein ZAP Restricts Human Retrotransposition
- The 4E-BP Caf20p Mediates Both eIF4E-Dependent and Independent Repression of Translation
- Turning into a Frataxin-Independent Organism
- Promotion of Bone Morphogenetic Protein Signaling by Tetraspanins and Glycosphingolipids
- Essential Role of the ESX-5 Secretion System in Outer Membrane Permeability of Pathogenic Mycobacteria
- The Zinc-Finger Antiviral Protein ZAP Inhibits LINE and Alu Retrotransposition
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Drosophila Spaghetti and Doubletime Link the Circadian Clock and Light to Caspases, Apoptosis and Tauopathy
- Autoselection of Cytoplasmic Yeast Virus Like Elements Encoding Toxin/Antitoxin Systems Involves a Nuclear Barrier for Immunity Gene Expression
- Parp3 Negatively Regulates Immunoglobulin Class Switch Recombination
- PERK Limits Lifespan by Promoting Intestinal Stem Cell Proliferation in Response to ER Stress
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



