-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaPARP2 Is the Predominant Poly(ADP-Ribose) Polymerase in Arabidopsis DNA Damage and Immune Responses
All living organisms face constant challenges from environmental factors. Appropriate and rapid responses to external stimuli are crucial for maintenance of genome integrity and cell survival. Poly(ADP-ribosyl)ation is a post-translational modification and contributes to multiple molecular and cellular processes including a prominent role in DNA damage repair. Human PARP1, the founding and most characterized member of the PARP family, accounts for more than 90% of overall molecular and cellular PARP activity in response to DNA damage while PARP2 supplies a minor portion of this PARP activity. Here we show that Arabidopsis PARP2 rather than PARP1 plays the predominant role in poly(ADP-ribosyl)ation and organismal resilience in response to either chemically-induced DNA damage or pathogen infections. We show that the activity and abundance of PARP2 is regulated by both PARP1 and PARG1. We also show that Arabidopsis PARG1 rather than PARG2 is the major contributor to removal poly(ADP-ribose) from acceptor proteins. Core aspects of plant poly(ADP-ribosyl)ation are mediated by substantially different enzymes than in animals, suggesting the likelihood of substantial differences in regulation.
Published in the journal: . PLoS Genet 11(5): e32767. doi:10.1371/journal.pgen.1005200
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1005200Summary
All living organisms face constant challenges from environmental factors. Appropriate and rapid responses to external stimuli are crucial for maintenance of genome integrity and cell survival. Poly(ADP-ribosyl)ation is a post-translational modification and contributes to multiple molecular and cellular processes including a prominent role in DNA damage repair. Human PARP1, the founding and most characterized member of the PARP family, accounts for more than 90% of overall molecular and cellular PARP activity in response to DNA damage while PARP2 supplies a minor portion of this PARP activity. Here we show that Arabidopsis PARP2 rather than PARP1 plays the predominant role in poly(ADP-ribosyl)ation and organismal resilience in response to either chemically-induced DNA damage or pathogen infections. We show that the activity and abundance of PARP2 is regulated by both PARP1 and PARG1. We also show that Arabidopsis PARG1 rather than PARG2 is the major contributor to removal poly(ADP-ribose) from acceptor proteins. Core aspects of plant poly(ADP-ribosyl)ation are mediated by substantially different enzymes than in animals, suggesting the likelihood of substantial differences in regulation.
Introduction
Appropriate and rapid responses to external stimuli can be crucial for maintenance of cellular and organismal viability, especially under stress conditions. Both biotic and abiotic stresses can induce genome DNA damage [1–4]. Maintenance of genome integrity via DNA damage repair then becomes essential, in both germ-line and somatic cells [2, 5, 6].
Poly(ADP-ribosyl)ation is a post-translational modification mediated by poly(ADP-ribose) polymerase (PARP) enzymes, in which negatively charged ADP-ribose units are transferred from donor nicotinamide adenine dinucleotide (NAD+) molecules onto target proteins [7]. PARP enzymes are themselves the most prominent poly(ADP-ribosyl)ation target. Poly(ADP-ribosyl)ation plays a key role in a wide range of cellular responses including DNA repair, chromatin modification, control of transcription and cell death [7–9]. Poly(ADP-ribosyl)ation and PARP proteins have been identified in a wide variety of plants and animals as well as bacteria, fungi and double-stranded DNA viruses [10–12]. In humans, 17 PARP proteins have been identified based on homology to PARP1, the founding member of the PARP family [13]. PARP1 accounts for approximately 90% of the PARP activity in mammalian cells under genotoxic situations, while PARP2 is apparently responsible for the remaining 10% [14–16].
The Arabidopsis genome encodes three PARP proteins that carry a PARP signature motif, as well as RCD1 and five SRO (“Similar to RCD One”) proteins with a variant form of the PARP signature [11, 17–19]. Although the names of plant PARP proteins have in some instances been reversed, the product of the Arabidopsis At2g31320 gene (NCBI NP_850165.1) is herein called PARP1 and the Arabidopsis At4g02390 product (NCBI NP_192148.2) is PARP2, based on their relative similarities to the earlier-named and extensively studied animal homologs (S1 Fig) [11, 20]. PARP2-like proteins are broadly conserved across diverse plant taxa (S2 Fig), while PARP1 is broadly conserved across plants and animals [11, 18]. Arabidopsis PARP3 contains variant active site residues that suggest lack of PARP catalytic function [12], and expression of Arabidopsis PARP3 is restricted to seed tissues [21]. The SROs (including RCD1) are a conserved family of plant-specific proteins that have functions in development and abiotic stress responses [19]. Although a wheat SRO protein that does possesses PARP activity was recently described [22], Arabidopsis SROs contain variant PARP motifs that both bioinformatically and biochemically were found to lack ADP-ribosyl transferase activity [19].
PARPs are widely known for their roles in genotoxic stress, DNA damage repair and programmed cell death in animals [13, 23, 24]. Although additional roles for PARP enzymes are being discovered [9], one of the best-known roles of PARPs is their function as DNA damage sensors. PARP1 in particular binds in its poly(ADP-ribosyl)ated form to ssDNA and dsDNA breaks and initiates events that attract DNA damage repair machinery to the sites of damage [7, 25]. A growing body of evidence indicates that plant PARPs have similar functions. PARP proteins are involved in microhomology mediated back-up non-homologous end joining in Arabidopsis [20]. Arabidopsis PARP1 and PARP2 accumulate rapidly and strongly in response to ionizing radiation, whereas PARP2 is preferentially induced by dehydration and excess cadmium [26]. PARP in soybean cells is differentially involved in responses to mild and intense oxidative stresses, through regulating DNA repair and programmed cell death [27]. Knocking down PARP activities in Arabidopsis and oilseed rape plants by chemical inhibition or gene silencing inhibited cell death and made plants more tolerant to a broad range of abiotic stresses including high light, drought and heat [28, 29]. This is due at least in part to induction of specific abscisic acid signaling pathways [29]. PARP inhibition also enhances Arabidopsis growth by promoting the leaf cell number [30, 31]. In animals, PARP proteins have been implicated in regulation of telomere length, telomere activity and chromosome end protection [32, 33]. However, it appears that Arabidopsis PARP proteins make limited contributions to telomere regulation and maintenance, although telomere dysfunction triggers PARP activation [34].
Poly(ADP-ribosyl)ation is reversible; covalently attached poly(ADP-ribose) can be cleaved from acceptor proteins by poly(ADP-ribose) glycohydrolase (PARG) [7, 8]. Mammalian genomes encode a single PARG gene [35, 36] and mutation of PARG caused enhanced sensitivity to genotoxic stress and elevated accumulation of poly(ADP-ribose), leading to embryonic lethality in mice and Drosophila [37, 38]. However, the human PARG gene undergoes alternative splicing, resulting in multiple PARG protein isoforms that localize to different cellular compartments [39]. Unlike animal models the Arabidopsis genome encodes two PARG genes, PARG1 and PARG2, with 52% amino acid identity (S1 Fig) [11]. PARG1 and PARG2, as well as an inactive pseudogene At2g31860, are all located adjacent to each other on Arabidopsis chromosome 2 [11]. Much less is known about the function of PARGs than PARPs in plant poly(ADP-ribosyl)ation. PARG1 was originally identified to play a role in circadian oscillation in Arabidopsis [40]. The Arabidopsis PARG2 gene is robustly induced by pathogen-associated molecular patterns (PAMPs) and numerous different pathogens, but disruption of PARG1, not PARG2, altered various plant defense responses [17, 41]. Similar to its counterpart, PARP, Arabidopsis PARG1 has also been implicated in drought, osmotic and oxidative stress responses [42].
Despite the above work, the mechanisms by which PARPs and PARGs regulate diverse cellular processes in plants remain largely unknown. The present study used mutational and biochemical approaches to assess the relative contributions of Arabidopsis PARP1/2 and PARG1/2. We present evidence that, unlike in animals, PARP2 rather than PARP1 plays the major role in plant DNA damage and immune responses. We also demonstrate that PARG1 rather than PARG2 is the primary enzyme that counteracts poly(ADP-ribosyl)ation in Arabidopsis. In addition, we discover that PARP1 associates with PARP2, and that PARP1 and PARP2 interact with both PARG1 and PARG2.
Results
Mutant parp2 plants are more sensitive to DNA damage agents than wild-type or parp1 plants
To examine the functional importance of PARP1 and PARP2 in plant poly(ADP-ribosyl)ation and in effective responses to DNA damage agents, we tested sensitivity to genotoxic agents in Arabidopsis parp1 and parp2 single mutants and in parp1parp2 double mutants. Two Arabidopsis T-DNA insertion lines with mutations in PARP1 (At2g31320) and one for PARP2 (At4g02390) were identified: parp1-1 (GABI_380E06), parp1-2 (GABI_382F01) and parp2-1 (GABI_420G03). Double mutant parp1parp2 plants were generated by genetic crosses. The chemical bleomycin is a potent inducer of DNA double-strand breaks (DSB), with a mode of action similar to that of ionizing radiation [43]. Plants were grown on MS plates supplemented with bleomycin and organismal-level sensitivity to DNA damage was scored as the number of plants without true leaves 14 days after germination [44, 45]. As shown in Fig 1A, in bleomycin-free MS plates almost all wild-type and mutant plants produced normal true leaves. In MS plates supplemented with 1.5 μg/ml of bleomycin, over 95% of wild-type plants still produced true leaves, whereas approximately 35% of the parp1-2 plants had no true leaves. Strikingly, more than 50% of the parp2-1 single mutants failed to generate true leaves, similar to parp1-2parp2-1 double mutants (Fig 1A). RT-PCR analysis confirmed that expression of PARP1 or PARP2 was abolished in the respective mutants (S3 Fig). Although contributions of PARP1 cannot be ruled out, ANOVA across three replicate experiments indicated that the growth defects on bleomycin were significantly worse than wild-type only for the parp2 and parp1parp2 double mutants. The experiments suggest that PARP2 is a more substantial contributor than PARP1 to a successful response to bleomycin. Similarly, increased sensitivity to the DNA alkylating agent methyl methane sulfonate (MMS) has been observed in a parp1parp2 double mutant [20, 34].
Fig. 1. Arabidopsis parp mutants are hypersensitive to DNA damage agents. 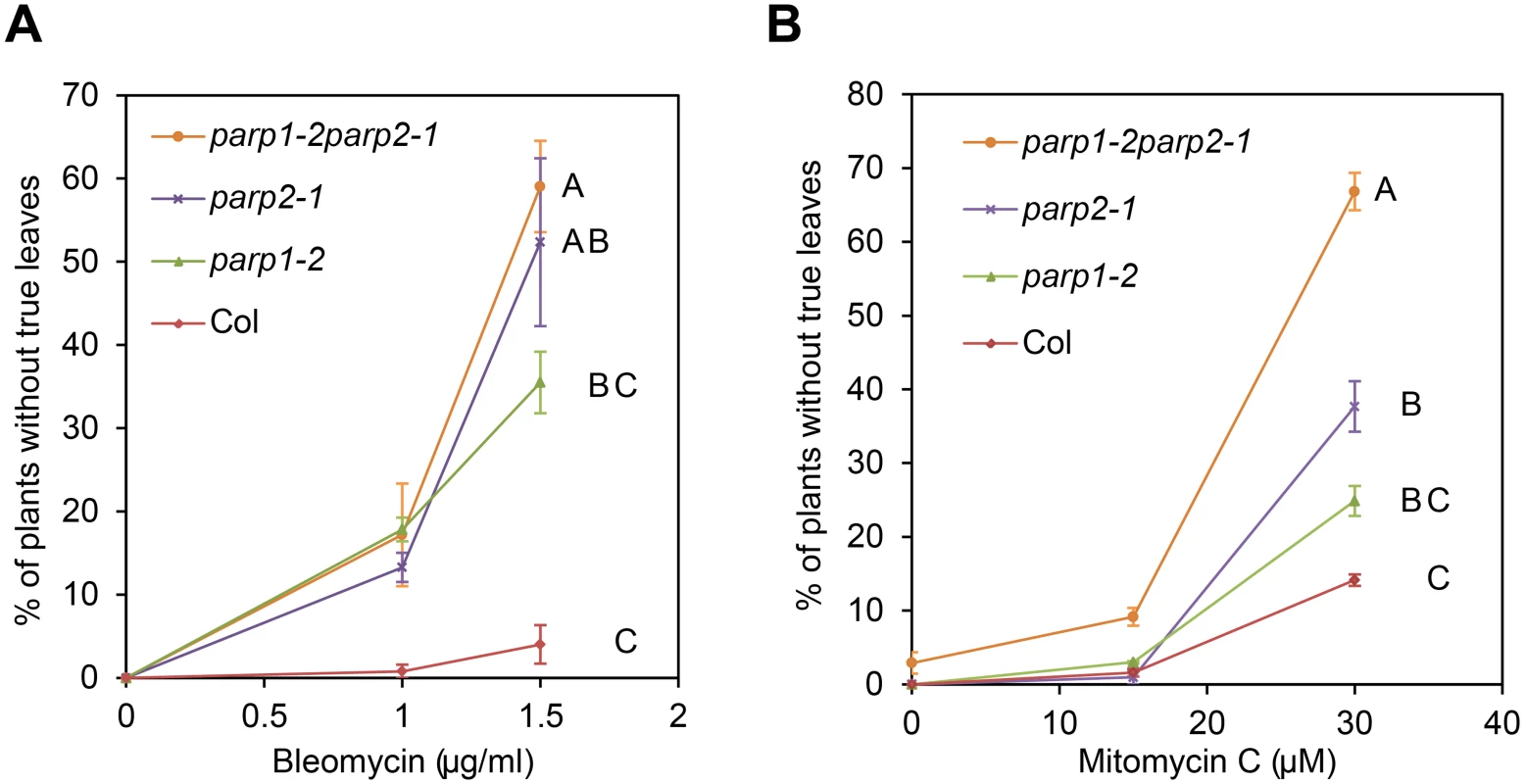
Wild-type Col-0 and parp mutant seeds were grown on MS agar medium supplemented with the genotoxic agents bleomycin (A) or mitomycin C (B). Sensitivity to DNA damage agents was scored as the percentage of plants that had not yet developed true leaves after 14 d. Mean and standard error of the mean are shown for one experiment; experiment was performed three times with similar results. Genotypes not sharing same letter on graph are significantly different at the high concentration (ANOVA Tukey HSD P < 0.05 across three experiments). To further examine the role of PARP1 and PARP2 in DNA damage repair in Arabidopsis we analyzed sensitivity to mitomycin C, an interstrand DNA crosslinking agent [46]. Similar to bleomycin treatment, the parp1-2 and parp2-1 mutants exhibited moderately or markedly increased sensitivity to mitomycin C, respectively (Fig 1B). ANOVA across three replicate experiments again indicated that the growth defects, this time in response to mitomycin C, were significantly worse than wild-type only for the parp2 and parp1parp2 double mutants. However, the P-value for the Col vs. parp1 contrast was 0.054 (very close to P < 0.05 significance), and PARP1 contributions were also indicated because the parp1parp2 double mutant grew significantly less well than the parp2 single mutant. Similar results after mitomycin C treatment also were obtained using a second parp1 mutant allele and parp1/parp2 double mutant line (S4 Fig). These mitomycin C experiments indicate that PARP1 and PARP2 are both required for effective repair of damaged DNA, but that PARP2 plays a stronger role in tolerance of plant DNA damage.
PARP2 accounts for most of the DNA damage-induced PARP activity in Arabidopsis
To investigate whether disruption of the Arabidopsis PARP1 or PARP2 genes disrupts in planta poly(ADP-ribosyl)ation of target proteins, wild-type and parp mutant plants were treated for 18h with increasing concentrations of bleomycin and poly(ADP-ribosyl)ated proteins were monitored on protein immunoblots using an anti-PAR (anti-poly(ADP-ribose)) antibody (Fig 2). Greatly increased levels of poly(ADP-ribosyl)ation were observed in wild-type plants after treatment with bleomycin at concentrations ranging from 2.5 to 10 μg/ml. Substantial amounts of poly(ADP-ribosyl)ated proteins were still detected despite knockout of PARP1 in the parp1-1 or parp1-2 single mutants. This is unexpected given that Arabidopsis PARP1 is the zinc finger-containing homolog of animal PARP1, which has been abundantly demonstrated to make the greatest contribution to poly(ADP-ribosyl)ation in response to DNA damage [7, 10, 14–16]. Knockout of PARP2 rather than PARP1 severely depleted detectable PARP activity in Arabidopsis, with only marginal elevation of poly(ADP-ribosyl)ated proteins detected after bleomycin treatment of parp2-1 mutant plants (Fig 2). Little or no PARP activity was observed in the parp1-1parp2-1 or parp1-2parp2-1 mutants (Fig 2). Under long exposure conditions residual PARP activity in the parp1-2parp2-1 double mutants still remained barely detectable.
Fig. 2. PARP2 plays a dominant role in DNA damage response after bleomycin treatment. 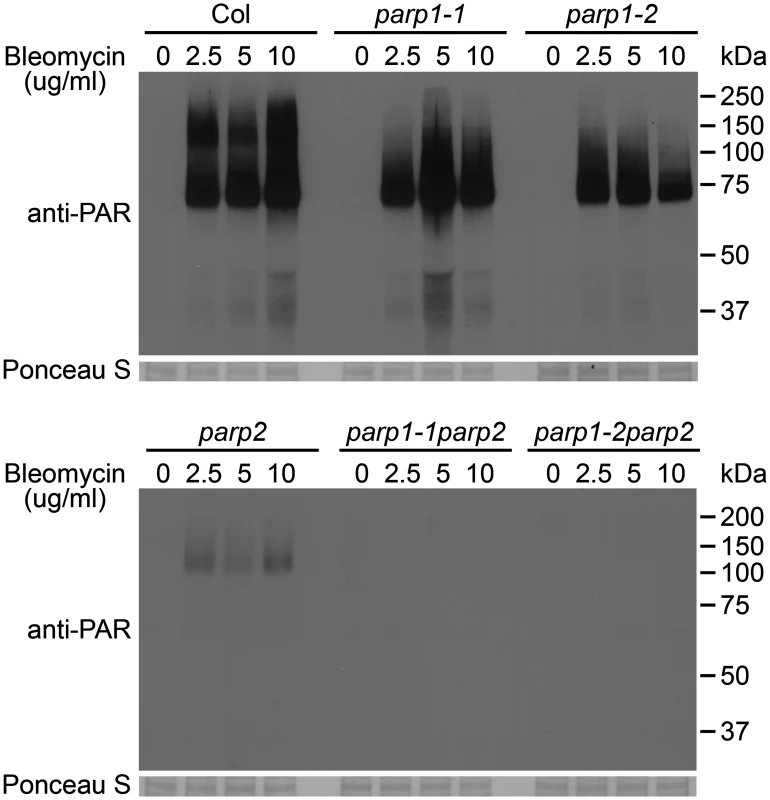
Two-week old Arabidopsis plants were transferred to 0, 2.5, 5 or 10 μg/ml of bleomycin for 18 h. Total proteins were extracted, separated by SDS-PAGE and analyzed by immunoblotting using an anti-PAR antibody. Equivalent loading of lanes was verified using Ponceau S stain. All samples shown and both blots were processed in parallel within the same experiment. Similar results were obtained in three separate experiments. We had obtained RT-PCR evidence that the parp2-1 mutation eliminated PARP2 transcript (Fig 1B) but further tests were then conducted to confirm loss of PARP2 protein production in the parp2-1 mutant, as well as to confirm the specificity of a custom-raised PARP2 polyclonal antibody. In wild-type Col-0 plants treated with bleomycin, PARP2 protein was detected using the anti-PARP2 antibody (S5 Fig). PARP2 protein was still detected in parp1-1 and in parp1-2 mutant plants, but not in the parp2-1 line or in two separate parp1parp2 double mutant lines, indicating that parp2-1 is a null mutant (S5 Fig).
To confirm that the compromised PARP activity in parp2-1 is due to the T-DNA insertion in the PARP2 gene, we complemented the parp2-1 mutant with a construct carrying native Arabidopsis PARP2 promoter sequences driving expression of Arabidopsis PARP2 genomic DNA fused to an HA epitope tag. Five independent T2 transgenic lines with different expression levels of PARP2-HA were chosen for complementation analysis. As shown in S6 Fig, poly(ADP-ribosyl)ation activity was restored in the complemented lines. We further observed that the abundance of poly(ADP-ribosyl)ated proteins present in response to the DNA damaging agent bleomycin correlated with the levels of PARP2 protein in the selected lines (S6 Fig). As a side-matter, the anti-PARP2 antibody was used to detect PARP2 in these complementation experiments. We were not able to detect the PARP2-HA fusion protein using an anti-HA antibody (that worked well with other HA-tagged proteins), possibly due to cleavage or inaccessibility of the C-terminal tag on PARP2. Overall, the above results indicate that PARP1 and PARP2 both contribute to poly(ADP-ribosyl)ation, but that PARP2 is the primary Arabidopsis enzyme responsible for poly(ADP-ribosyl)ation activity in response to DNA damage.
In related experiments we irradiated plants with 150 Gy of γ-radiation, a dose that is sufficient to induce DNA double strand breaks [47], and PARP activity was monitored (Fig 3A). Markedly increased amounts of poly(ADP-ribosyl)ated proteins were detected in wild-type plants from 20 to 60 min after irradiation. This γ-ray-induced PARP activity was substantially reduced in the parp1-2 mutant. An even more complete reduction in poly(ADP-ribosyl)ation was observed in the parp2-1 single mutant, and a similarly complete reduction was observed in the parp1-2parp2-1 double mutant. With the γ-ray-treated plant samples we also measured the level of phosphorylated histone γ-H2AX, a standard indicator of DNA double-strand breaks [48, 49]. Compared to wild-type plants, in the parp1-2 single mutants elevated DNA damage was detected as an increase in the intensity of the γ-H2AX band (Fig 3B). More DNA damage was reproducibly detected in the parp2-1 single mutant than the parp1-2 mutant, and even more significantly increased DNA damage was observed in the parp1-2parp2-1 double mutant (Fig 3B). Hence as in earlier experiments where plant growth in the face of DNA damage was monitored (Fig 1), experiments monitoring abundance of γ-ray-induced DNA double-strand breaks (Fig 3B) detected contributions of PARP1 as well as PARP2. However, these experiments with γ-ray-treated plants again indicated that PARP2 accounts for majority of the cellular PARP activity that is activated in response to DNA damage (Fig 3A).
Fig. 3. PARP2 plays a dominant role in response to ionizing irradiation. 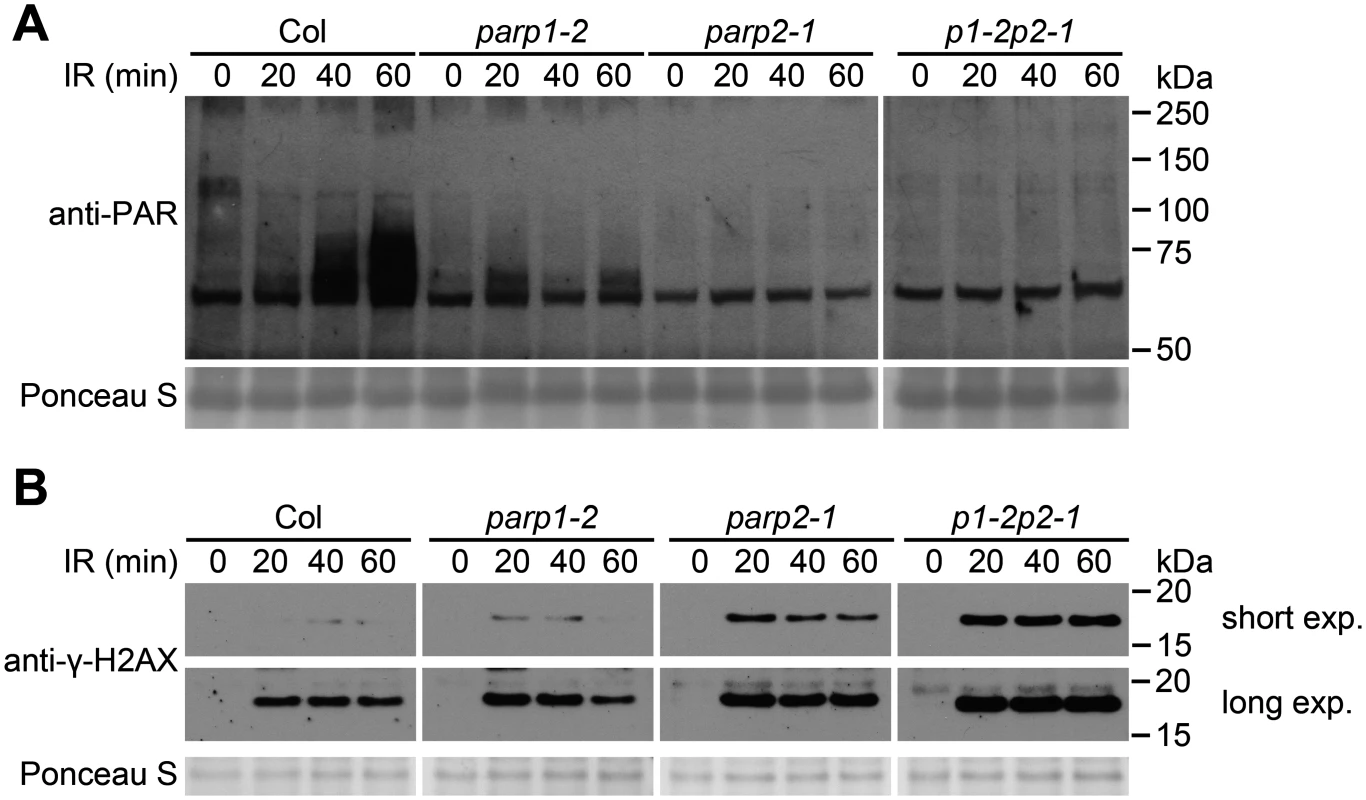
(A) Two-week old Arabidopsis plants grown on MS plates were irradiated with 150 Gy of γ-radiation and then flash-frozen 20, 40 or 60 min after removal from the radiation source. Total proteins were then extracted, separated by SDS-PAGE and analyzed by immunoblotting with an anti-PAR antibody. 0 min sample not exposed to γ-radiation source. All samples shown in (A) were processed in parallel within the same experiment. (B) The level of γ-H2AX was assessed at 20, 40 and 60 min after irradiation as in (A), using an anti-γ-H2AX antibody. Samples all processed in parallel from same experiment. Shorter and longer time exposures of same immunoblot are shown. Equivalent loading of lanes was verified using Ponceau S stain. Experiments were performed three times with similar results. Subcellular localization of PARP1/2 and PARG1/2 proteins
To examine whether Arabidopsis PARPs are targeted to the nucleus as in animals, PARP1 and PARP2 proteins fusions to the C-terminus of green fluorescent protein (GFP) were expressed in leaves of Nicotiana benthamiana by agroinfiltration. Confocal fluorescence microscopy revealed that both PARP1 and PARP2 predominantly accumulate in the nucleus (Fig 4A). To determine the subcellular location of PARPs in Arabidopsis, we expressed PARP1-GFP and PARP2-GFP C-terminal fusion proteins in stable transformants of wild-type Col-0. Fluorescence microscopic examination of transgenic Arabidopsis plants expressing 35S:PARP1-GFP or 35S:PARP2-GFP detected PARP1 and PARP2 only in the nucleus, in both leaves and roots (Fig 4B), consistent with a previous report [50]. Notably, multiple foci were detected throughout the nucleus of root tissues expressing 35S:PARP2-GFP.
Fig. 4. Subcellular localization of Arabidopsis PARP1/2 and PARG1/2. 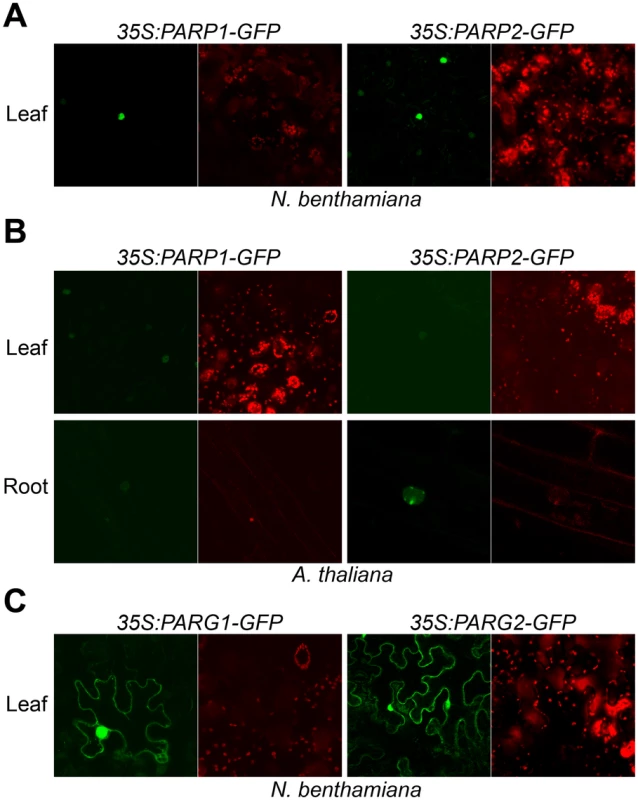
Paired confocal fluorescence microscopy images show same sample; green wavelengths (GFP) on left and red wavelengths (chlorophyll) on right. (A) Arabidopsis PARP1 and PARP2 localized in the nucleus. 35S:AtPARP1-GFP and 35S:AtPARP2-GFP transiently expressed in N. benthamiana epidermal cells within leaves were imaged. (B) PARP1 and PARP2 localized in the nucleus in Arabidopsis. Subcellular localization was carried out in stable transgenic lines carrying 35S:AtPARP1-GFP and 35S:AtPARP2-GFP in the wild-type Col-0 background. (C) Arabidopsis PARG1 and PARG2 localized in the cytoplasm and nucleus. 35S:AtPARG1-GFP and 35S:AtPARG2-GFP were transiently expressed in N. benthamiana and the images were taken 2 d after inoculation. Poly(ADP-ribosyl)ation is impacted by PARP enzymes and also by poly(ADP-ribose) glycohydrolase (PARG) enzymes that remove poly(ADP-ribosyl)ation, so experiments with PARG proteins were also carried out. PARG1-GFP and PARG2-GFP fusion proteins expressed in N. benthamiana were reproducibly observed both in the cytoplasm and nucleus (Fig 4C). PARG2 mRNA abundance was previously shown to be significantly increased in response to virulent or avirulent Pseudomonas syringae pv. tomato (Pst) strains or the PAMPs flg22 or elf18 [17, 41]. In the present study, up-regulation of PARG2 at the protein level was confirmed in transgenic Arabidopsis plants. The abundance of PARG2-GFP protein expressed under control of the PARG2 promoter sequence was substantially increased in leaves within 8 hr after exposure to Pst(avrRpt2) (S7 Fig).
PARG1 (but not PARG2) mediates poly(ADP-ribose) removal in plants responding to bleomycin
Previous work by our group had detected multiple impacts on plant defense for Arabidopsis parg1 mutants, unlike Arabidopsis parg2 mutants [17, 41]. To determine if Arabidopsis PARG1 and/or PARG2 confer detectable poly(ADP-ribose) glycohydrolase activity, parg1-1, parg2-1 and Col-0 plants were treated with 2.5 μg/ml bleomycin and the abundance of poly(ADP-ribosyl)ated proteins was examined. Mutation of PARG1 resulted in significantly elevated presence of poly(ADP-ribosyl)ation in total protein extracts compared with wild-type plants, as might be expected for loss of an active poly(ADP-ribose) glycohydrolase (Fig 5A). Surprisingly but consistent with the Adams-Philips et al. data (2010), disruption of Arabidopsis PARG2 caused little or no increase in poly(ADP-ribosyl)ation (Fig 5A). The data suggest that PARG1 is the primary enzyme that catalyzes the removal of poly(ADP-ribose) from acceptor proteins and that PARG2 makes little or no contribution to this activity. The primary mediator of bleomycin-induced elevation of poly(ADP-ribosyl)ation, PARP2, exhibited increased protein levels in response to bleomycin in all three genetic backgrounds. Notably, the accumulation of PARP2 induced by bleomycin is reduced in the parg1-2 mutant relative to wild-type (Fig 5B), suggesting that PARG1 can influence the levels of PARP2 protein.
Fig. 5. PARG1 is more active than PARG2 in removal of poly(ADP-ribosyl)ation after bleomycin treatment. 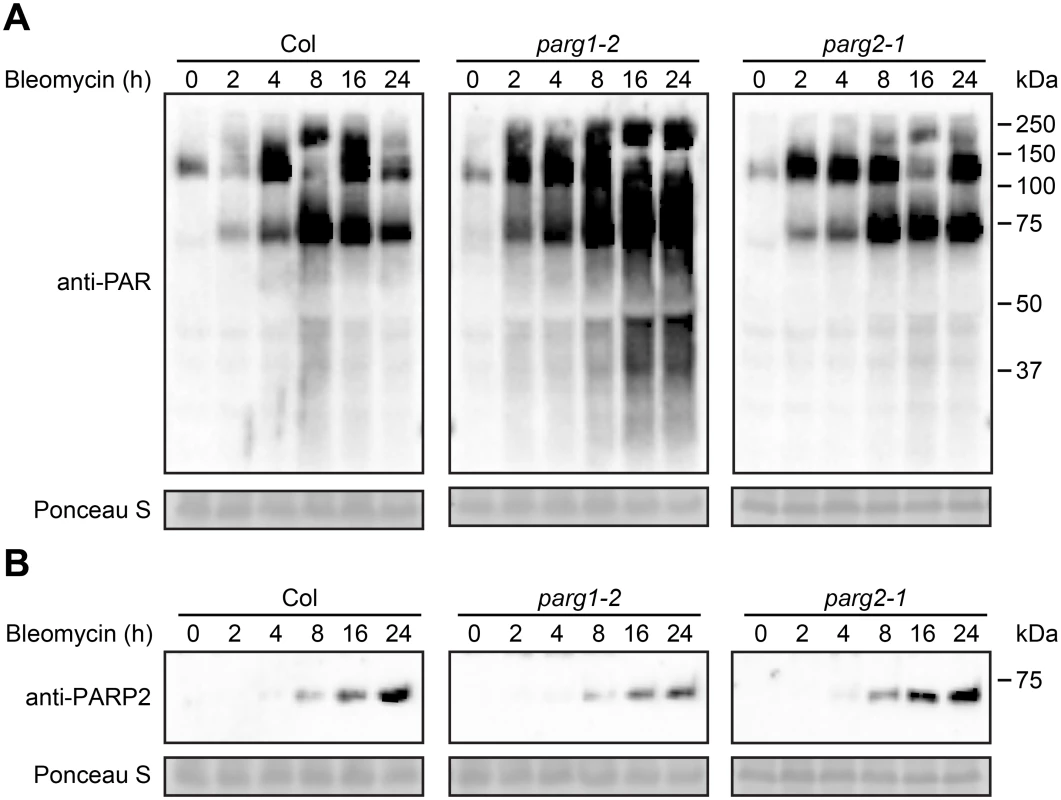
Two-week-old parg1-2, parg2-1 and Col-0 Arabidopsis plants were treated with 2.5 μg/ml bleomycin and samples were collected at indicated times. Total proteins were extracted, separated by SDS-PAGE and analyzed by immunoblotting with anti-PAR (A) or anti-PARP2 (B) antibody. Equivalent loading of lanes was verified using Ponceau S stain. Similar results obtained in two separate experiments. PARP2 activity is regulated by PARP1 in response to DNA alkylating agent mitomycin C
In further work to characterize the role of PARPs in plant DNA damage responses, wild-type and parp mutant plants were treated with mitomycin C to induce DNA cross-linking [51] and the level of poly(ADP-ribosyl)ated proteins was then monitored. As shown in Fig 6, mitomycin C caused increased PARP activity in wild-type plants. As with the bleomycin and γ-ray experiments (Figs 3 and 4), almost no poly(ADP-ribosyl)ation activity was detected after mitomycin C treatment in parp2-1 mutant plants (Fig 6). However, increased rather than decreased abundance of poly(ADP-ribosyl)ation was observed in parp1 mutants, in separate experiments with either the parp1-1 or parp1-2 alleles, in response to mitomycin C (Fig 6). This is unlike the poly(ADP-ribosyl)ation behavior of the same mutants in response to bleomycin or γ-irradiation (Fig 2). The mitomycin C-induced increase in poly(ADP-ribosyl)ation caused by mutation of PARP1 was eliminated if PARP2 was also mutated (parp1-1parp2-1 and parp1-2parp2-1 double mutants, Fig 6). This interesting finding suggests that loss of PARP1 can in some situations lead to elevated PARP2 activity. Said another way, PARP1 may suppress PARP2 activity under certain stress conditions such as after exposure to mitomycin C.
Fig. 6. PARP2 activity is regulated by PARP1 in response to DNA alkylating agent mitomycin C. 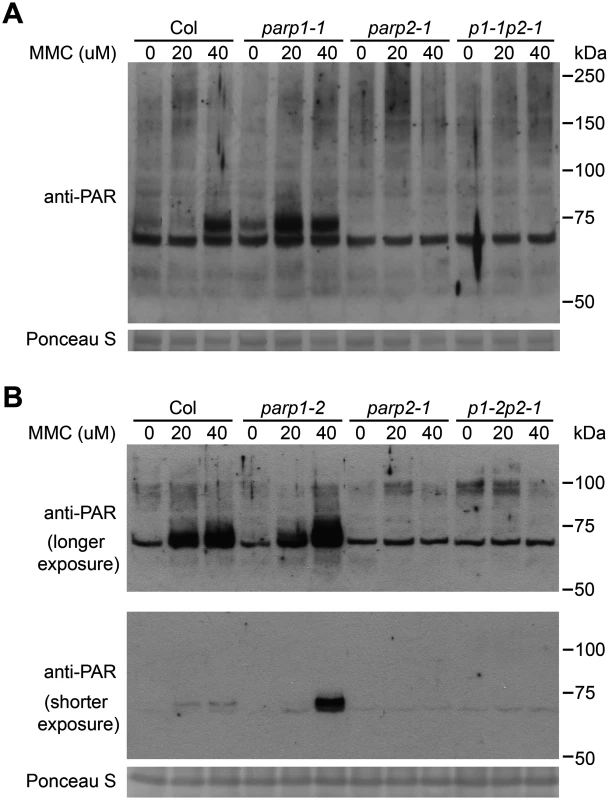
Arabidopsis plants, including (A) parp1-1 or (B) parp1-2 knockout alleles of PARP1, were grown on MS plates supplemented with 0, 20 and 40 μM of mitomycin C (MMC) for two weeks. Total proteins were extracted, separated by SDS-PAGE and analyzed by immunoblotting with anti-PAR antibody. Equivalent loading of lanes was verified using Ponceau S stain. Upper and middle panels of (B) are same blot, showing immunoblot signal after longer and shorter exposure times respectively. Similar results obtained in two separate experiments. PARP2 is required for normal basal resistance responses in Arabidopsis
To examine the role of PARP genes in plant defense responses, the Arabidopsis parp mutants were inoculated with the virulent bacterial pathogen Pseudomonas syringae pv. tomato (Pst) strain DC3000. As shown in Fig 7A, the parp2-1 single and parp1-2parp2-1 double mutants exhibited enhanced susceptibility in comparison to wild-type plants, whereas bacterial growth in the parp1-2 single mutant was similar to that in wild-type. This important result demonstrates an impact of loss of poly(ADP-ribosyl)ation activity on the capacity of Arabidopsis to limit the growth of this virulent bacterial pathogen. The finding also suggests that wild-type PARP2 plays a greater role than PARP1 in basal defense against this pathogen.
Fig. 7. Arabidopsis parp mutants are compromised in basal resistance. 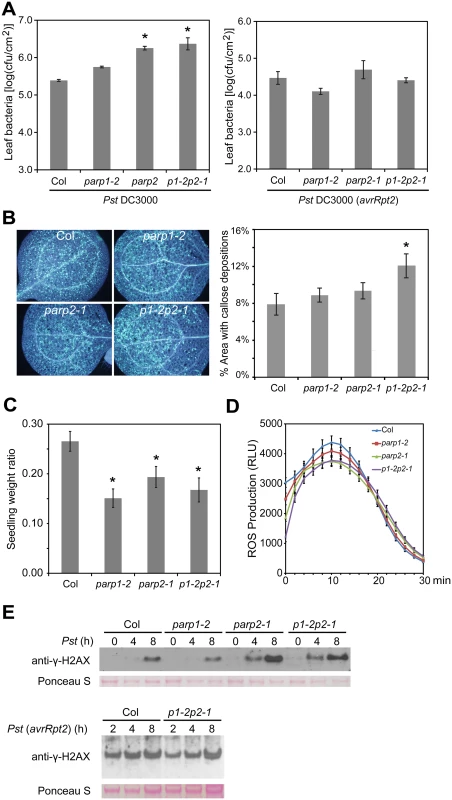
(A) Bacterial population sizes of Pst within leaves. Pst DC3000 strains with or without avrRpt2 were syringe infiltrated into leaf mesophyll at 1×105 cfu/ml and bacterial populations were measured 3 d post-inoculation. Mean ± standard error of mean for one experiment shown. Experiments were performed three times with similar results; * indicates significant difference from Col-0 across the three experiments (ANOVA, Tukey pairwise comparisons, P < 0.05). (B) Flg22-induced callose deposition. Seedlings exposed to 1 μM flg22 for 24 h were fixed and stained with aniline blue to highlight callose deposition. Left panel: representative images of the four genotypes; right panel: data summary for all tested leaves (n = 24 per genotype). * indicates significant difference from Col-0 across the three experiments (ANOVA, Tukey pairwise comparisons, P < 0.05). (C) Seedling growth inhibition due to chronic flg22-induced defense activation. Ratio is weight of individual seedlings grown for 14 d in liquid MS media + 1 μM flg22, divided by mean of seedlings of same genotype grown without flg22 within same experiment (mean ± standard error of mean). * indicates significant difference from Col-0 across the three experiments (ANOVA, Tukey pairwise comparisons, P < 0.05). (D) Flg22-triggered oxidative burst. Reactive oxygen species from leaf discs of the indicated genotype were measured for 30 min after treatment with 1 μM flg22. RLU: relative luminescence units. (E) Pst-induced γ-H2AX accumulation. Arabidopsis plants of the indicated genotype were vacuum-infiltrated with the indicated Pst strain at 1×107 cfu/ml. The level of γ-H2AX in leaf samples from the indicated time points after inoculation was assessed by immunoblot using anti-γ-H2AX antibody. Equivalent loading of lanes was verified using Ponceau S stain. Upper and lower blots are from separate experiments. Similar results obtained in two separate experiments. Experiments with Pst DC3000 expressing the effector protein AvrRpt2 (which triggers RPS2-mediated defense in naturally RPS2+ Arabidopsis Col-0 plants) were also conducted, to test for impacts of poly(ADP-ribosyl)ation on the stronger R gene-mediated defense response (also known as effector-triggered immunity) [52]. No impact of PARP mutations was detected (Fig 7A).
Impacts of PARP genes on PAMP-triggered immunity responses [52–54] were also examined. Callose deposition was not detectably affected in parp1-2 or parp2-1 single mutants, but callose deposition was significantly enhanced in the parp1-2parp2-1 double mutant (Fig 7B). Enhanced callose deposition was also observed in parp1-1parp2-1 double mutant plants (different parp1 allele; S8 Fig). Reinforcing rather than antagonistic roles of PARP and PARG activity were suggested by the related observation that, like parp1parp2 mutants, parg1 mutant plants exhibited elevated callose deposition in response to flg22 (S9 Fig). Seedling growth inhibition assays [55], one of the most sensitive indicators of basal defense activation in response to PAMP treatment, also showed that disruption of PARP activity led to a stronger response to flg22 (Fig 7C). No significant difference in the flg22-induced ROS burst was observed for the parp mutants (Fig 7D).
We recently showed that microbial pathogens including virulent Pst DC3000 induce DNA DSBs in plant host genomes [1]. To monitor DSB induction in Arabidopsis parp mutants in response to Pst, we monitored the accumulation of γ-H2AX. Across replicate experiments, and consistent with the bacterial growth data of Fig 7A, no impact of parp mutations on DSB induction was observed during the strong R gene-mediated responses triggered by Pst DC3000(avrRpt2) (Fig 7E). However, with virulent Pst DC3000 lacking avrRpt2, elevated levels of γ-H2AX were observed 4 and 8 hours after infection in leaves of the parp2-1 and parp1-2parp2-1 mutants as compared to wild-type plants (Fig 7E). No increases of γ-H2AX were observed in parp1-2 plants. These results indicate, as might be predicted [1], that there is a link between plant poly(ADP-ribosyl)ation and prevention or repair of pathogen-induced DNA damage. The results also indicate that in Arabidopsis, PARP2 plays a more significant role than PARP1 in this prevention/repair of pathogen-induced DNA damage.
PARP1 interacts with PARP2; PARG1 interacts with PARG2; PARP1 and PARP2 interact with both PARG1 and PARG2
To determine if plant PARP1 associates with PARP2, we carried out coimmunoprecipitation assays in N. benthamiana. As shown in Fig 8A, myc-tagged PARP1 was coimmunoprecipitated by GFP-tagged PARP2 using an anti-GFP antibody, indicating that at least some PARP1 is present in complexes with PARP2. The interaction was observed without as well as with bleomycin treatment. This readily detectable interaction suggests that PARP2 activity may be regulated in part by physical contact with PARP1. Similarly, PARG1 and PARG2 (the only two PARG proteins in Arabidopsis) also associated with each other (Fig 8B), although that interaction required longer exposures than the PARP1-PARP2 product of Fig 8A to detect a co-IP band of similar intensity. We also investigated whether PARPs interact with PARG1 and/or PARG2. The reproducibly detectable co-immunoprecipitation products in Fig 8C showed that PARP1 interacts with both PARG1 and PARG2, with or without bleomycin treatment. Similarly, PARP2 also complexes with both PARG1 and PARG2 (Fig 8D). Hence the regulation of PARP2 abundance and activity by PARG1 may also be achieved via physical interaction between the two proteins.
Fig. 8. Interactions between PARPs and PARGs. 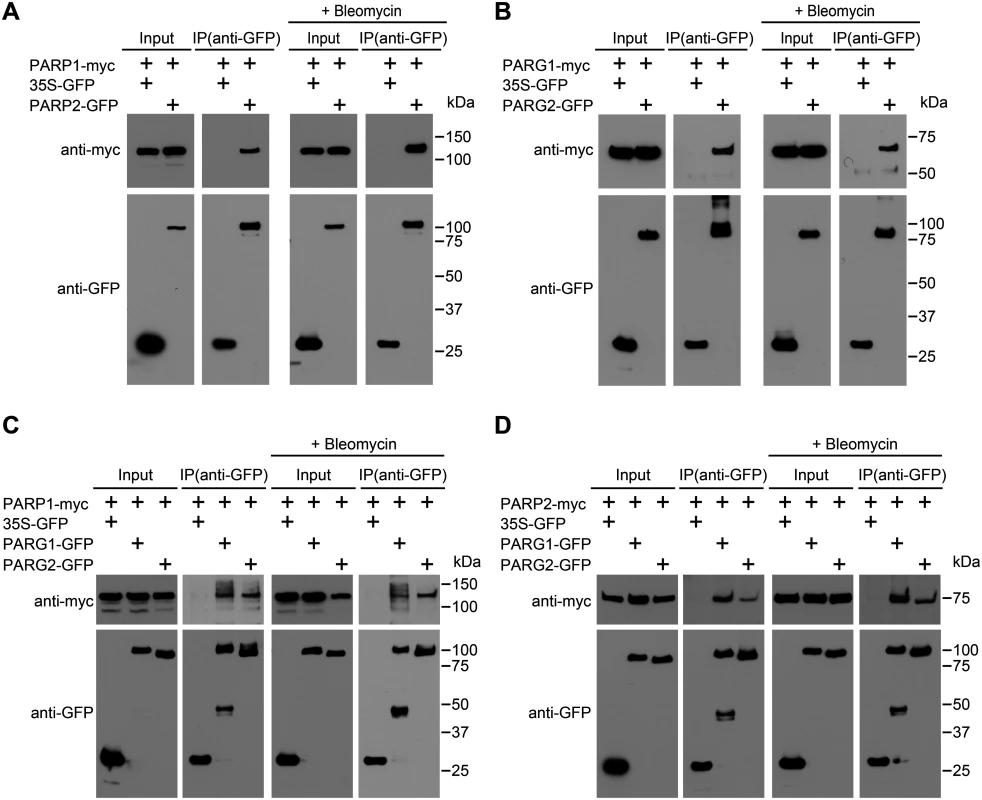
(A) PARP1 associates in vivo with PARP2. (B) PARG1 associates in vivo with PARG2. (C) PARP1 associates in vivo with PARG1 and PARG2. (D) PARP2 associates in vivo with PARG1 and PARG2. The indicated proteins were transiently expressed in N. benthamiana. Input lanes were loaded with total protein extracts, IP lanes were loaded with immunoprecipitation products. Immunoprecipitations were performed with anti-GFP antibodies and immunoblots were analyzed with anti-GFP or anti-myc antibodies, as noted. These experiments were repeated twice with similar results. Discussion
In mammals, PARP1 is by far the predominant contributor to poly(ADP-ribosyl)ation responses to a variety of cellular stresses, and mammalian PARP1 has received the vast majority of poly(ADP-ribose) polymerase research and pharmaceutical industry attention [7–9]. Arabidopsis PARP1 is the poly(ADP-ribose) polymerase with a similar domain structure to mammalian PARP1 (S1 Fig), but the relative roles of the different plant PARPs required investigation [11]. We used genetic and biochemical approaches to elucidate the contributions to Arabidopsis DNA damage and immune responses made by different poly(ADP-ribose) polymerase and poly(ADP-ribose) glycohydrolase gene products. For both groups of enzymes the predicted proteins have significantly divergent domain structures (S1 Fig). In contrast to the paradigm in mammals, we found that PARP2 rather than PARP1 plays the major role biochemically and in organismal-level responses to DNA damage and pathogen infections. Furthermore, we found that PARG1 rather than PARG2 is the primary enzyme that confers poly(ADP-ribose) glycohydrolase activity in Arabidopsis during the tested responses to DNA damage and pathogen infection.
Arabidopsis PARP1 shares a substantially conserved domain structure with human PARP1, whereas Arabidopsis PARP2 is more analogous to human PARP2. In mice, parp2 mutants exhibited some phenotypes similar to parp1 knockouts, despite the dramatic difference between their respective specific enzymatic activities [56–58]. Although neither PARP1 nor PARP2 is required for viability in mice, parp1parp2 double knockouts are embryonic lethal with considerable genomic instability, indicating that the PARP1 and PARP2 gene products together are essential during early embryogenesis and that the deficiency in PARP1 and PARP2 cannot be functionally compensated by other PARP family members [59]. In contrast, when the functions of Arabidopsis PARP1 and PARP2 both were disrupted plants were developmentally normal, demonstrating that this pair of genes is not essential to viability in Arabidopsis
Overall PARP activity in Arabidopsis is significantly decreased when PARP2 is knocked out, suggesting that Arabidopsis PARP2, unlike its counterpart in animals, is responsible for majority of the PARP activity. The predominant role of Arabidopsis PARP2 was detected at more macroscopic levels by the increased sensitivity of parp2 mutant plants to genotoxic stresses, and enhanced susceptibility to virulent Pst growth and to Pst-induced DNA DSB damage. Since poly(ADP-ribosyl)ated proteins were nearly absent in the parp1parp2 double mutant, it is reasonable to propose that other Arabidopsis PARP-domain containing proteins have either low or no PARP activity under the conditions examined in this study. This is consistent with the bioinformatic prediction that Arabidopsis PARP3, RCD1 and SROs lack conserved active catalytic sites in the PARP domains and possibly have lost the ability to bind NAD [12,19]. We had previously reported that P. syringae induces the accumulation of poly(ADP-ribosyl)ated proteins [17] but we know of no studies that have identified the suite of proteins that are poly(ADP-ribosyl)ated during plant-pathogen interactions.
PARP1, the primary PARP that mediates responses such as DNA damage repair in human cells, contains zinc-finger DNA binding domains that are crucial to its function [10, 24] (S1 Fig). Known DNA binding domains are absent from human PARP2. The striking finding that core poly(ADP-ribosyl)ation functions of animal PARP1 have been taken on by PARP2 in plants may be less unusual in light of the fact that plant PARP2 proteins carry N-terminal SAP (SAF-A/B, Acinus, and PIAS) domains (S2 Fig). The SAP domain is a highly conserved sequence-specific or structure-specific DNA binding motif with a four-helix bundle, known to contribute to regulation of chromatin structure and transcription [60, 61]. In contrast to many other DNA recognition protein structures, one end of the helix bundle makes contact with DNA and fits into the minor groove of DNA [62]. One to four distinct SAP domains are present in plant PARP2s, depending on the plant species (S2 Fig). Structural differences in DNA binding domains between Arabidopsis PARP1 and PARP2 likely contribute intriguing differences in the substrate specificities of these two enzymes. Arabidopsis PARP1 and PARP2 also exhibit substantial divergence within their areas of shared domain structure (see Fig 1; for amino acids #130–636 of Arabidopsis PARP2, only 56% are similar (36% identical) to the aligned Arabidopsis PARP1 amino acids #485–982). The substantially divergent structure of plant PARP2 enzymes is likely to be accompanied by substantially divergent mechanisms of regulation.
We found evidence that Arabidopsis PARP2 activity is negatively regulated by PARP1 during responses to mitomycin C, but we did not detect reproducible changes of PARP2 protein abundance in the parp1 mutant under the genotoxic stress conditions we examined. However, it has recently been reported that Arabidopsis PARP1 and PARP2 negatively regulate gene expression of each other [34]. We observed physical interaction (coimmunoprecipitation) of PARP1 and PARP2 in plants responding to bleomycin. The Boltz et al. data and our data point to an intriguing interplay between PARP1 and PARP2, and also suggest that different modes of action of poly(ADP-ribosyl)ation-mediated regulation may exist in plants that are not observed in animals.
In animals, poly(ADP-ribosyl)ation has recently been found to play a significant role during host-pathogen interactions. For example, Helicobactor pylori, a human gastric bacterial pathogen, activates nuclear regulator PARP1 [63]. Several proteins produced by pathogenic viruses have been reported to interact directly with and stimulate animal PARP1 enzymatic activity [64, 65], whereas others prevent PARP1 activation as virulence strategies [66, 67]. In plants, there is also an expanding body of evidence that poly(ADP-ribosyl)ation plays critical roles in pathogenicity as well as in immune responses [17, 41, 68–70]. The present study found that PARP1/2 are positive regulators in plant immune response. The molecular mechanisms by which PARPs regulate plant immunity remain to be fully discovered, but in light of the γ-H2AX findings summarized in Fig 7E, it would appear that one important role of Arabidopsis PARP2 is to minimize the host DNA damage elicited by virulent pathogens such as Pst.
Over the past 30 years, the function of PARPs in maintenance of genome integrity have been extensively characterized [7, 8]. In contrast, study of PARG in poly(ADP-ribosyl)ation has been limited, due in part to its low cellular abundance and high sensitivity to proteases [71], but also to the lethality of knockouts of the sole PARG gene in metazoans [37, 38]. The presence of two PARG genes in plants may offer opportunities for genetic and molecular investigation not available in animal systems. Using Arabidopsis parg null mutants, which are viable, we demonstrated that PARG1 mediates removal of poly(ADP-ribose) whereas PARG2 confers limited PARG activity in the examined conditions. This is in line with the observation that parg1 plants, but not parg2 plants, exhibit increased sensitivity to PAMP treatment and grow less well than wild-type plants in response to DNA-damaging mitomycin C [17]. Because multiple but divergent copies of PARG genes are found in a number of plant species [11], in the future it will be interesting to explore if similar divisions of labor among PARG proteins are common in other species, and if PARG proteins from other plant species differ substantially from those of Arabidopsis in their enzymatic activities and cellular roles.
PARPs and PARGs, enzymes with counteracting enzymatic activities, modify target proteins by addition or removal, respectively, of ADP-ribose polymers. Surprisingly, the present and previous studies showed that parg and parp mutant plants exhibit some unexpectedly similar phenotypes, such as strong seedling growth inhibition and increased callose deposition [17, 41]. This may be attributable to synergistic functions of PARPs and PARGs. There is evidence that in human cells the two enzymes co-localize to target gene promoters and act with a similar rather than antagonistic overall effect to regulate gene expression globally [72]. PARP1 and PARG also function in concert to accelerate single-strand break repair in human cells [73]. Intriguingly, we discovered that the accumulation of PARP2 protein in response to genotoxic agents was reduced relative to wild-type in loss-of-function parg1 mutant plants, consistent with a recent finding made in human HeLa cells that PARP1 transcript and protein expression levels decreased in the parg knockdown. This suggests that PARP1 is regulated by PARG to avoid excess accumulation of poly(ADP-ribose) in a cell [74]. With our detection of plant PARPs and PARGs in the same in vivo complexes, it is plausible to hypothesize that regulation of abundance or activity is achieved in part through physical interactions between PARPs and PARGs [16, 75].
In summary, we have found that although plant PARPs and PARGs have partially overlapping functions Arabidopsis PARP2 and PARG1 play the predominant roles in plant poly(ADP-ribosyl)ation during DNA damage and immune responses. Future studies will further identify the molecular mechanism by which PARPs and PARGs regulate various cellular responses, individually or in a concerted manner, and their functional interplay with each other. Identification of the proteins that are poly(ADP-ribosy)ated is another future research direction that may help to elucidate the regulatory functions of plant poly(ADP-ribosyl)ation, both under normal physiological conditions and in response to the stresses of DNA damage or pathogenic infection.
Materials and Methods
Plant materials and growth conditions
Arabidopsis thaliana plants were grown in Fisons Sunshine Mix #1 soil-less potting mix (Hummert) at 22°C under 9-h light/15-h dark cycles, or MS-grown plants were cultivated on Murashige-Skoog (MS) agar media at 22°C under 16-h light/8-h dark cycles.
The homozygous Arabidopsis T-DNA insertion lines parp1-1 (GABI_380E06), parp1-2 (GABI_382F01), parp2-1 (GABI_420G03), all in the Col-0 background [76], were identified as previously described [77]. Isolation and initial characterization of the parg1 and parg2 T-DNA insertion lines was previously described [17]. Homozygous double-mutant lines were obtained as self-fertilized progeny from crosses of single mutants and were identified using PCR-based allele-specific markers.
Mitomycin C and bleomycin sensitivity assay
Seeds were stratified at 4°C for two d and then approximately 100 seeds were grown for 14 d on each MS agar plate containing different concentrations of mitomycin C (0, 15 and 30 μM) and bleomycin (0, 1, 1.5 μg/ml) (both from Sigma-Aldrich), then the number of plants with and without true leaves was recorded. Sensitivity was scored as the percentage of plants without true leaves, using three or four plates for each concentration (or in a few instances two, due to plate contamination) within each experiment, and the entire experiment was repeated three separate times. Mean and standard error for each chemical concentration are reported from within each experiment.
Seedling growth inhibition assay
To monitor plant defense activation in response to the flagellin epitope flg22 peptide, 6 d old seedlings from MS plates were transferred to 1 ml MS liquid medium + 1 μM flg22 in 24-well plates, and seedling fresh weights were recorded 14 d later for 12 seedlings per treatment, as per [55].
ROS assay
The burst of reactive oxygen species produced in response to the flagellin epitope flg22 peptide was monitored as previously described [78]. Briefly, leaf discs were taken from 5-week-old plants and incubated in 1% dimethyl sulfoxide (DMSO) solution in a 96-well plate overnight, then treated with 1 μM flg22 in 0.1 mg/ml luminol and 0.1 mg/ml horseradish peroxidase immediately prior to 30 min. of luminescence measurement by plate reader (Centro XS3 LB 960, Berthold Technology).
Callose deposition
Arabidopsis seedlings were grown on MS agar plates for five days before being transferred to liquid MS containing 1 μM flg22. After 24 h of treatment, seedlings were fixed in FAA solution (10% formaldehyde, 5% acetic acid and 50% ethanol), cleared in 95% ethanol, and stained with 0.01% aniline blue in 67 mM K2HPO4 with pH adjusted to 12. The stained seedlings were visualized with an Olympus BX60 Epifluorescence Microscope and images of entire cotyledons were captured with an Olympus DP73 camera (with same settings used throughout single experiment). The callose deposits on entire cotyledons were then quantified automatically using ImageJ software by excluding rare wounded leaves and then analyzing the entire area of all images within an experiment after setting ImageJ hue and brightness cutoff levels using images for positive and negative control leaves.
Bacterial growth assay
Leaves of healthy five-week-old Arabidopsis plants were syringe-inoculated with Pst DC3000 or Pst DC3000(avrRpt2) at 1×105 cfu/ml [79]. After 3 d, leaf discs were sampled from inoculated plants and macerated in 10 mM MgCl2. Samples were diluted serially, plated on NYGA plates with rifampicin and kanamycin and the number of colonies was recorded after 2 d incubation at 28°C.
Ionizing radiation and detection of H2AX phosphorylation
For ionizing radiation, Arabidopsis Col-0 plants were irradiated with 150 Gy from a 137Cs source (administered at 2.14 Gy per minute) and tissue samples were collected at the indicated times after removal from the radiation source [47]. To detect pathogen-induced DSBs, 5-week-old plants were infiltrated with a 1×107 cfu/ml solution of Pst bacteria in 10 mM MgCl2 and samples were collected at indicated times. Histone proteins were prepared from leaf tissues as previously described and were subjected to immunoblotting with rabbit anti-human γ-H2AX antibody at 1 : 5000 dilution (Sigma-Aldrich).
Confocal laser scanning microscopy
The full-length cDNAs of PARP1/2 and PARG1/2 were subcloned into the pDONR 207 vector (Invitrogen) and introduced into the destination vector pGWB405, resulting in constructs with C-terminal fusions to GFP under the control of 35S promoter. The sequence-verified constructs were transformed into Agrobacterium tumefaciens strain GV3101(pMP90) and transiently expressed in Nicotiana benthamiana leaves by infiltration. Confocal fluorescence microscopy was carried out at indicated times using a Zeiss 510 Meta confocal laser scanning microscope. Agrobacterium strains carrying 35S:PARP1-GFP and 35S:PARP2-GFP were also used to transform wild-type Col-0 Arabidopsis by floral dip [80]. Stable transgenic T2 lines were selected and the subcellular locations of PARP1 and PARP2 were examined by confocal microscopy.
Complementation of the parp2 mutants
To complement the parp2 mutants, PARP2 genomic DNA fragments including its native promoter was cloned into the pDONR207 (Invitrogen). Using LR reactions, PARP2 was cloned into pGWB3300, a modified vector (Y. Cao and A. Bent) carrying in the multiple cloning site of pCambia3300 (http://www.cambia.org) the Gateway cloning site, C-terminal 3xHA and nos terminator sequences from pGWB13 [82]. This resulted in the generation of PARP2:PARP2-3×HA construct that was then transformed into the parp2-1 mutant background.
Immunoblot analysis
Total proteins were prepared from Arabidopsis plants in extraction buffer (50 mM Tris-HCl (pH7.5), 150 mM NaCl, 5 mM EDTA, 0.5% Triton X-100, 10% glycerol, and Sigma-Aldrich plant protease inhibitor cocktail at 1 : 100). After protein separation by SDS-PAGE, immunoblot analysis was carried out with anti-HA (Roche), anti-poly(ADP-ribose) (anti-PAR) (Trevigen) or anti-PARP2 antibodies. Polyclonal antibodies to Arabidopsis PARP2 protein (custom purchase from Genscript) were raised in rabbit against the synthetic peptide YGKEENDSPVNNDI, which does not share significant homology with PARP1 or other proteins predicted in the Col-0 accession of Arabidopsis. Anti-PARP2 antibodies were purified by peptide affinity chromatography.
Coimmunoprecipitation assays
The cDNAs of PARP1, PARP2, PARG1 and PARG2 were cloned into the GFP-tagged pGWB405 and/or myc-tagged pGWB417 Gateway destination vectors [81] and the resulting constructs were transformed into A. tumefaciens GV3101(pMP90). Leaves of 4–5 week-old N. benthamiana plants were agroinfiltrated with OD600 0.4 of the resulting A. tumefaciens strains, and some samples were then infiltrated two days later with 2 μg/ml of bleomycin solution. Tissues were harvested three days after agroinfiltration and total proteins were prepared in extraction buffer (50 mM Tris-HCl (pH7.5), 150 mM NaCl, 5 mM EDTA, 0.2% Triton X-100, 10% glycerol, and Sigma-Aldrich plant protease inhibitor cocktail at 1 : 100). Immunoprecipitation was carried out with anti-GFP (Abcam) at 4°C overnight followed by incubation with protein A beads (Thermo Scientific) for 1–2 h. The beads were washed three times with extraction buffer without protease inhibitors. The precipitated proteins were eluded with the SDS loading buffer, subjected to SDS-PAGE and immunoblotted with anti-myc (Covance) and anti-GFP (Clontech) antibodies, and detected using Supersignal West Pico or Dura chemiluminescent substrates (Thermo Scientific).
Supporting Information
Zdroje
1. Song J, Bent AF. Microbial pathogens trigger host DNA double-strand breaks whose abundance is reduced by plant defense responses. PLoS Pathog. 2014;10(4):e1004030. doi: 10.1371/journal.ppat.1004030 24699527; PubMed Central PMCID: PMC3974866.
2. Weitzman MD, Weitzman JB. What's the damage? The impact of pathogens on pathways that maintain host genome integrity. Cell Host Microbe. 2014;15(3):283–94. doi: 10.1016/j.chom.2014.02.010 24629335.
3. Gill SS, Tuteja N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol Biochem. 2010;48(12):909–30. doi: 10.1016/j.plaphy.2010.08.016 20870416.
4. Chatzinikolaou G, Karakasilioti I, Garinis GA. DNA damage and innate immunity: links and trade-offs. Trends Immunol. 2014. doi: 10.1016/j.it.2014.06.003 25023467.
5. Papamichos-Chronakis M, Peterson CL. Chromatin and the genome integrity network. Nat Rev Genet. 2013;14(1):62–75. doi: 10.1038/nrg3345 23247436; PubMed Central PMCID: PMC3731064.
6. Ermolaeva MA, Schumacher B. Systemic DNA damage responses: organismal adaptations to genome instability. Trends Genet. 2014;30(3):95–102. doi: 10.1016/j.tig.2013.12.001 24439457.
7. Gibson BA, Kraus WL. New insights into the molecular and cellular functions of poly(ADP-ribose) and PARPs. Nat Rev Mol Cell Biol. 2012;13(7):411–24. doi: 10.1038/nrm3376 22713970.
8. Luo X, Kraus WL. On PAR with PARP: cellular stress signaling through poly(ADP-ribose) and PARP-1. Genes Dev. 2012;26(5):417–32. doi: 10.1101/gad.183509.111 22391446; PubMed Central PMCID: PMC3305980.
9. Burkle A, Virag L. Poly(ADP-ribose): PARadigms and PARadoxes. Mol Aspects Med. 2013;34(6):1046–65. doi: 10.1016/j.mam.2012.12.010 23290998.
10. Hassa PO, Hottiger MO. The diverse biological roles of mammalian PARPS, a small but powerful family of poly-ADP-ribose polymerases. Front Biosci. 2008;13 : 3046–82. 17981777.
11. Briggs AG, Bent AF. Poly(ADP-ribosyl)ation in plants. Trends Plant Sci. 2011;16(7):372–80. doi: 10.1016/j.tplants.2011.03.008 21482174.
12. Lamb RS, Citarelli M, Teotia S. Functions of the poly(ADP-ribose) polymerase superfamily in plants. Cell Mol Life Sci. 2012;69(2):175–89. doi: 10.1007/s00018-011-0793-4 21861184.
13. Schreiber V, Dantzer F, Ame JC, de Murcia G. Poly(ADP-ribose): novel functions for an old molecule. Nat Rev Mol Cell Biol. 2006;7(7):517–28. doi: 10.1038/nrm1963 16829982.
14. Shieh WM, Ame JC, Wilson MV, Wang ZQ, Koh DW, Jacobson MK, et al. Poly(ADP-ribose) polymerase null mouse cells synthesize ADP-ribose polymers. J Biol Chem. 1998;273(46):30069–72. 9804757.
15. Ame JC, Rolli V, Schreiber V, Niedergang C, Apiou F, Decker P, et al. PARP-2, A novel mammalian DNA damage-dependent poly(ADP-ribose) polymerase. J Biol Chem. 1999;274(25):17860–8. 10364231.
16. Schreiber V, Ame JC, Dolle P, Schultz I, Rinaldi B, Fraulob V, et al. Poly(ADP-ribose) polymerase-2 (PARP-2) is required for efficient base excision DNA repair in association with PARP-1 and XRCC1. J Biol Chem. 2002;277(25):23028–36. doi: 10.1074/jbc.M202390200 11948190.
17. Adams-Phillips L, Briggs AG, Bent AF. Disruption of poly(ADP-ribosyl)ation mechanisms alters responses of Arabidopsis to biotic stress. Plant Physiol. 2010;152(1):267–80. doi: 10.1104/pp.109.148049 19889874; PubMed Central PMCID: PMC2799362.
18. Citarelli M, Teotia S, Lamb RS. Evolutionary history of the poly(ADP-ribose) polymerase gene family in eukaryotes. BMC Evol Biol. 2010;10 : 308. doi: 10.1186/1471-2148-10-308 20942953; PubMed Central PMCID: PMC2964712.
19. Jaspers P, Overmyer K, Wrzaczek M, Vainonen JP, Blomster T, Salojarvi J, et al. The RST and PARP-like domain containing SRO protein family: analysis of protein structure, function and conservation in land plants. BMC Genomics. 2010;11 : 170. doi: 10.1186/1471-2164-11-170 20226034; PubMed Central PMCID: PMC2848248.
20. Jia Q, den Dulk-Ras A, Shen H, Hooykaas PJ, de Pater S. Poly(ADP-ribose)polymerases are involved in microhomology mediated back-up non-homologous end joining in Arabidopsis thaliana. Plant Mol Biol. 2013;82(4–5):339–51. doi: 10.1007/s11103-013-0065-9 23625359.
21. Rissel D, Losch J, Peiter E. The nuclear protein Poly(ADP-ribose) polymerase 3 (AtPARP3) is required for seed storability in Arabidopsis thaliana. Plant Biol (Stuttg). 2014. doi: 10.1111/plb.12167 24533577.
22. Liu S, Liu S, Wang M, Wei T, Meng C, Wang M, et al. A wheat SIMILAR TO RCD-ONE gene enhances seedling growth and abiotic stress resistance by modulating redox homeostasis and maintaining genomic integrity. Plant Cell. 2014;26(1):164–80. doi: 10.1105/tpc.113.118687 24443520; PubMed Central PMCID: PMC3963566.
23. Heeres JT, Hergenrother PJ. Poly(ADP-ribose) makes a date with death. Curr Opin Chem Biol. 2007;11(6):644–53. doi: 10.1016/j.cbpa.2007.08.038 17936669.
24. Krishnakumar R, Kraus WL. The PARP side of the nucleus: molecular actions, physiological outcomes, and clinical targets. Mol Cell. 2010;39(1):8–24. doi: 10.1016/j.molcel.2010.06.017 20603072; PubMed Central PMCID: PMC2923840.
25. Huber A, Bai P, de Murcia JM, de Murcia G. PARP-1, PARP-2 and ATM in the DNA damage response: functional synergy in mouse development. DNA Repair (Amst). 2004;3(8–9):1103–8. doi: 10.1016/j.dnarep.2004.06.002 15279798.
26. Doucet-Chabeaud G, Godon C, Brutesco C, de Murcia G, Kazmaier M. Ionising radiation induces the expression of PARP-1 and PARP-2 genes in Arabidopsis. Mol Genet Genomics. 2001;265(6):954–63. 11523787.
27. Amor Y, Babiychuk E, Inze D, Levine A. The involvement of poly(ADP-ribose) polymerase in the oxidative stress responses in plants. FEBS Lett. 1998;440(1–2):1–7. 9862413.
28. De Block M, Verduyn C, De Brouwer D, Cornelissen M. Poly(ADP-ribose) polymerase in plants affects energy homeostasis, cell death and stress tolerance. Plant J. 2005;41(1):95–106. doi: 10.1111/j.1365-313X.2004.02277.x 15610352.
29. Vanderauwera S, De Block M, Van de Steene N, van de Cotte B, Metzlaff M, Van Breusegem F. Silencing of poly(ADP-ribose) polymerase in plants alters abiotic stress signal transduction. Proc Natl Acad Sci U S A. 2007;104(38):15150–5. doi: 10.1073/pnas.0706668104 17823244; PubMed Central PMCID: PMC1986628.
30. Schulz P, Neukermans J, Van der Kelen K, Muhlenbock P, Van Breusegem F, Noctor G, et al. Chemical PARP inhibition enhances growth of Arabidopsis and reduces anthocyanin accumulation and the activation of stress protective mechanisms. PLoS One. 2012;7(5):e37287. doi: 10.1371/journal.pone.0037287 22662141; PubMed Central PMCID: PMC3360695.
31. Schulz P, Jansseune K, Degenkolbe T, Meret M, Claeys H, Skirycz A, et al. Poly(ADP-ribose)polymerase activity controls plant growth by promoting leaf cell number. PLoS One. 2014;9(2):e90322. doi: 10.1371/journal.pone.0090322 24587323; PubMed Central PMCID: PMC3938684.
32. De Vos M, Schreiber V, Dantzer F. The diverse roles and clinical relevance of PARPs in DNA damage repair: current state of the art. Biochem Pharmacol. 2012;84(2):137–46. doi: 10.1016/j.bcp.2012.03.018 22469522.
33. Abd Elmageed ZY, Naura AS, Errami Y, Zerfaoui M. The poly(ADP-ribose) polymerases (PARPs): new roles in intracellular transport. Cell Signal. 2012;24(1):1–8. doi: 10.1016/j.cellsig.2011.07.019 21840394.
34. Boltz KA, Jasti M, Townley JM, Shippen DE. Analysis of poly(ADP-Ribose) polymerases in Arabidopsis telomere biology. PLoS One. 2014;9(2):e88872. doi: 10.1371/journal.pone.0088872 24551184; PubMed Central PMCID: PMC3923816.
35. Lin W, Ame JC, Aboul-Ela N, Jacobson EL, Jacobson MK. Isolation and characterization of the cDNA encoding bovine poly(ADP-ribose) glycohydrolase. J Biol Chem. 1997;272(18):11895–901. 9115250.
36. Ame JC, Apiou F, Jacobson EL, Jacobson MK. Assignment of the poly(ADP-ribose) glycohydrolase gene (PARG) to human chromosome 10q11.23 and mouse chromosome 14B by in situ hybridization. Cytogenet Cell Genet. 1999;85(3–4):269–70. doi: 15310 10449915.
37. Hanai S, Kanai M, Ohashi S, Okamoto K, Yamada M, Takahashi H, et al. Loss of poly(ADP-ribose) glycohydrolase causes progressive neurodegeneration in Drosophila melanogaster. Proc Natl Acad Sci U S A. 2004;101(1):82–6. doi: 10.1073/pnas.2237114100 14676324; PubMed Central PMCID: PMC314142.
38. Koh DW, Lawler AM, Poitras MF, Sasaki M, Wattler S, Nehls MC, et al. Failure to degrade poly(ADP-ribose) causes increased sensitivity to cytotoxicity and early embryonic lethality. Proc Natl Acad Sci U S A. 2004;101(51):17699–704. doi: 10.1073/pnas.0406182101 15591342; PubMed Central PMCID: PMC539714.
39. Meyer-Ficca ML, Meyer RG, Coyle DL, Jacobson EL, Jacobson MK. Human poly(ADP-ribose) glycohydrolase is expressed in alternative splice variants yielding isoforms that localize to different cell compartments. Exp Cell Res. 2004;297(2):521–32. doi: 10.1016/j.yexcr.2004.03.050 15212953.
40. Panda S, Poirier GG, Kay SA. tej defines a role for poly(ADP-ribosyl)ation in establishing period length of the arabidopsis circadian oscillator. Dev Cell. 2002;3(1):51–61. 12110167.
41. Adams-Phillips L, Wan J, Tan X, Dunning FM, Meyers BC, Michelmore RW, et al. Discovery of ADP-ribosylation and other plant defense pathway elements through expression profiling of four different Arabidopsis-Pseudomonas R-avr interactions. Mol Plant Microbe Interact. 2008;21(5):646–57. Epub 2008/04/09. doi: 10.1094/MPMI-21-5-0646 18393624.
42. Li G, Nasar V, Yang Y, Li W, Liu B, Sun L, et al. Arabidopsis poly(ADP-ribose) glycohydrolase 1 is required for drought, osmotic and oxidative stress responses. Plant Sci. 2011;180(2):283–91. doi: 10.1016/j.plantsci.2010.09.002 21421372.
43. Povirk LF. DNA damage and mutagenesis by radiomimetic DNA-cleaving agents: bleomycin, neocarzinostatin and other enediynes. Mutat Res. 1996;355(1–2):71–89. 8781578.
44. Durrant WE, Wang S, Dong XN. Arabidopsis SNI1 and RAD51D regulate both gene transcription and DNA recombination during the defense response. Proc Natl Acad Sci U S A. 2007;104(10):4223–7. doi: 10.1073/pnas.0609357104 ISI:000244972400098.
45. Song J, Durrant WE, Wang S, Yan S, Tan EH, Dong X. DNA repair proteins are directly involved in regulation of gene expression during plant immune response. Cell Host Microbe. 2011;9(2):115–24. doi: 10.1016/j.chom.2011.01.011 21320694.
46. Dronkert ML, Kanaar R. Repair of DNA interstrand cross-links. Mutat Res. 2001;486(4):217–47. 11516927.
47. Friesner JD, Liu B, Culligan K, Britt AB. Ionizing radiation-dependent gamma-H2AX focus formation requires ataxia telangiectasia mutated and ataxia telangiectasia mutated and Rad3-related. Mol Biol Cell. 2005;16(5):2566–76. doi: 10.1091/mbc.E04-10-0890 15772150; PubMed Central PMCID: PMC1087258.
48. Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem. 1998;273(10):5858–68. Epub 1998/04/16. 9488723.
49. Kinner A, Wu W, Staudt C, Iliakis G. Gamma-H2AX in recognition and signaling of DNA double-strand breaks in the context of chromatin. Nucleic Acids Res. 2008;36(17):5678–94. doi: 10.1093/nar/gkn550 18772227; PubMed Central PMCID: PMC2553572.
50. Babiychuk E, Van Montagu M, Kushnir S. N-terminal domains of plant poly(ADP-ribose) polymerases define their association with mitotic chromosomes. Plant J. 2001;28(3):245–55. 11722768.
51. Tomasz M, Lipman R, Chowdary D, Pawlak J, Verdine GL, Nakanishi K. Isolation and structure of a covalent cross-link adduct between mitomycin C and DNA. Science. 1987;235(4793):1204–8. 3103215.
52. Jones JD, Dangl JL. The plant immune system. Nature. 2006;444(7117):323–9. 17108957.
53. Dodds PN, Rathjen JP. Plant immunity: towards an integrated view of plant-pathogen interactions. Nat Rev Genet. 2010;11(8):539–48. doi: 10.1038/nrg2812 20585331.
54. Boller T, Felix G. A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu Rev Plant Biol. 2009;60 : 379–406. doi: 10.1146/annurev.arplant.57.032905.105346 19400727.
55. Gomez-Gomez L, Boller T. FLS2: an LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol Cell. 2000;5(6):1003–11. 10911994.
56. Wang ZQ, Stingl L, Morrison C, Jantsch M, Los M, SchulzeOsthoff K, et al. PARP is important for genomic stability but dispensable in apoptosis. Genes Dev. 1997;11(18):2347–58. doi: 10.1101/gad.11.18.2347 WOS:A1997XX74800004.
57. de Murcia JM, Niedergang C, Trucco C, Ricoul M, Dutrillaux B, Mark M, et al. Requirement of poly(ADP-ribose) polymerase in recovery from DNA damage in mice and in cells. Proc Natl Acad Sci U S A. 1997;94(14):7303–7. 9207086; PubMed Central PMCID: PMC23816.
58. Masutani M, Nozaki T, Nakamoto K, Nakagama H, Suzuki H, Kusuoka O, et al. The response of Parp knockout mice against DNA damaging agents. Mutat Res. 2000;462(2–3):159–66. 10767627.
59. Menissier de Murcia J, Ricoul M, Tartier L, Niedergang C, Huber A, Dantzer F, et al. Functional interaction between PARP-1 and PARP-2 in chromosome stability and embryonic development in mouse. EMBO J. 2003;22(9):2255–63. doi: 10.1093/emboj/cdg206 12727891; PubMed Central PMCID: PMC156078.
60. Aravind L, Koonin EV. SAP—a putative DNA-binding motif involved in chromosomal organization. Trends Biochem Sci. 2000;25(3):112–4. 10694879.
61. Sahara S, Aoto M, Eguchi Y, Imamoto N, Yoneda Y, Tsujimoto Y. Acinus is a caspase-3-activated protein required for apoptotic chromatin condensation. Nature. 1999;401(6749):168–73. doi: 10.1038/43678 10490026.
62. Okubo S, Hara F, Tsuchida Y, Shimotakahara S, Suzuki S, Hatanaka H, et al. NMR structure of the N-terminal domain of SUMO ligase PIAS1 and its interaction with tumor suppressor p53 and A/T-rich DNA oligomers. J Biol Chem. 2004;279(30):31455–61. doi: 10.1074/jbc.M403561200 15133049.
63. Nossa CW, Jain P, Tamilselvam B, Gupta VR, Chen LF, Schreiber V, et al. Activation of the abundant nuclear factor poly(ADP-ribose) polymerase-1 by Helicobacter pylori. Proc Natl Acad Sci U S A. 2009;106(47):19998–20003. doi: 10.1073/pnas.0906753106 19897724; PubMed Central PMCID: PMC2785281.
64. Gordon-Shaag A, Yosef Y, Abd El-Latif M, Oppenheim A. The abundant nuclear enzyme PARP participates in the life cycle of simian virus 40 and is stimulated by minor capsid protein VP3. J Virol. 2003;77(7):4273–82. 12634384; PubMed Central PMCID: PMC150672.
65. Carbone M, Reale A, Di Sauro A, Sthandier O, Garcia MI, Maione R, et al. PARP-1 interaction with VP1 capsid protein regulates polyomavirus early gene expression. J Mol Biol. 2006;363(4):773–85. doi: 10.1016/j.jmb.2006.05.077 16979186.
66. Muthumani K, Choo AY, Zong WX, Madesh M, Hwang DS, Premkumar A, et al. The HIV-1 Vpr and glucocorticoid receptor complex is a gain-of-function interaction that prevents the nuclear localization of PARP-1. Nat Cell Biol. 2006;8(2):170–9. doi: 10.1038/ncb1352 16429131; PubMed Central PMCID: PMC3142937.
67. Zhou J, Huang JD, Poon VK, Chen DQ, Chan CC, Ng F, et al. Functional dissection of an IFN-alpha/beta receptor 1 promoter variant that confers higher risk to chronic hepatitis B virus infection. J Hepatol. 2009;51(2):322–32. doi: 10.1016/j.jhep.2009.03.020 19501422.
68. Bartsch M, Gobbato E, Bednarek P, Debey S, Schultze JL, Bautor J, et al. Salicylic acid-independent ENHANCED DISEASE SUSCEPTIBILITY1 signaling in Arabidopsis immunity and cell death is regulated by the monooxygenase FMO1 and the Nudix hydrolase NUDT7. Plant Cell. 2006;18(4):1038–51. doi: 10.1105/tpc.105.039982 16531493; PubMed Central PMCID: PMC1425861.
69. Ge X, Li GJ, Wang SB, Zhu H, Zhu T, Wang X, et al. AtNUDT7, a negative regulator of basal immunity in Arabidopsis, modulates two distinct defense response pathways and is involved in maintaining redox homeostasis. Plant Physiol. 2007;145(1):204–15. doi: 10.1104/pp.107.103374 17660350; PubMed Central PMCID: PMC1976571.
70. Jambunathan N, Mahalingam R. Analysis of Arabidopsis growth factor gene 1 (GFG1) encoding a nudix hydrolase during oxidative signaling. Planta. 2006;224(1):1–11. doi: 10.1007/s00425-005-0183-y 16328543.
71. Bonicalzi ME, Haince JF, Droit A, Poirier GG. Regulation of poly(ADP-ribose) metabolism by poly(ADP-ribose) glycohydrolase: where and when? Cell Mol Life Sci. 2005;62(7–8):739–50. doi: 10.1007/s00018-004-4505-1 15868399.
72. Frizzell KM, Gamble MJ, Berrocal JG, Zhang T, Krishnakumar R, Cen Y, et al. Global analysis of transcriptional regulation by poly(ADP-ribose) polymerase-1 and poly(ADP-ribose) glycohydrolase in MCF-7 human breast cancer cells. J Biol Chem. 2009;284(49):33926–38. doi: 10.1074/jbc.M109.023879 19812418; PubMed Central PMCID: PMC2797163.
73. Fisher AE, Hochegger H, Takeda S, Caldecott KW. Poly(ADP-ribose) polymerase 1 accelerates single-strand break repair in concert with poly(ADP-ribose) glycohydrolase. Mol Cell Biol. 2007;27(15):5597–605. doi: 10.1128/MCB.02248-06 17548475; PubMed Central PMCID: PMC1952076.
74. Uchiumi F, Watanabe T, Ohta R, Abe H, Tanuma S. PARP1 gene expression is downregulated by knockdown of PARG gene. Oncol Rep. 2013;29(5):1683–8. doi: 10.3892/or.2013.2321 23467693; PubMed Central PMCID: PMC3658849.
75. Keil C, Grobe T, Oei SL. MNNG-induced cell death is controlled by interactions between PARP-1, poly(ADP-ribose) glycohydrolase, and XRCC1. J Biol Chem. 2006;281(45):34394–405. doi: 10.1074/jbc.M606470200 16963444.
76. Kleinboelting N, Huep G, Kloetgen A, Viehoever P, Weisshaar B. GABI-Kat SimpleSearch: new features of the Arabidopsis thaliana T-DNA mutant database. Nucleic Acids Res. 2012;40(Database issue):D1211–5. doi: 10.1093/nar/gkr1047 22080561; PubMed Central PMCID: PMC3245140.
77. Enns LC, Kanaoka MM, Torii KU, Comai L, Okada K, Cleland RE. Two callose synthases, GSL1 and GSL5, play an essential and redundant role in plant and pollen development and in fertility. Plant Mol Biol. 2005;58(3):333–49. doi: 10.1007/s11103-005-4526-7 16021399.
78. Gomez-Gomez L, Felix G, Boller T. A single locus determines sensitivity to bacterial flagellin in Arabidopsis thaliana. Plant J. 1999;18(3):277–84. 10377993.
79. Whalen MC, Innes RW, Bent AF, Staskawicz BJ. Identification of Pseudomonas syringae pathogens of Arabidopsis and a bacterial locus determining avirulence on both Arabidopsis and soybean. Plant Cell. 1991;3 : 49–59. 1824334
80. Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16(6):735–43. 10069079.
81. Nakagawa T, Suzuki T, Murata S, Nakamura S, Hino T, Maeo K, et al. Improved Gateway binary vectors: high-performance vectors for creation of fusion constructs in transgenic analysis of plants. Biosci Biotechnol Biochem. 2007;71(8):2095–100. doi: 10.1271/bbb.70216 17690442.
82. Nakagawa T, Kurose T, Hino T, Tanaka K, Kawamukai M, Niwa Y, et al. Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J Biosci Bioeng. 2007;104(1):34–41. doi: 10.1263/jbb.104.34 17697981.
Štítky
Genetika Reprodukční medicína
Článek Minor Type IV Collagen α5 Chain Promotes Cancer Progression through Discoidin Domain Receptor-1Článek A Unique Virulence Gene Occupies a Principal Position in Immune Evasion by the Malaria ParasiteČlánek The Centrosomal Linker and Microtubules Provide Dual Levels of Spatial Coordination of CentrosomesČlánek Dissecting the Function and Assembly of Acentriolar Microtubule Organizing Centers in Cells In VivoČlánek MicroRNA-Dependent Transcriptional Silencing of Transposable Elements in Drosophila Follicle CellsČlánek Cooperative Action of Cdk1/cyclin B and SIRT1 Is Required for Mitotic Repression of rRNA SynthesisČlánek The Developmental Intestinal Regulator ELT-2 Controls p38-Dependent Immune Responses in Adult .Článek Feeding and Fasting Signals Converge on the LKB1-SIK3 Pathway to Regulate Lipid Metabolism in
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2015 Číslo 5
-
Všechny články tohoto čísla
- Genomic Heritability: What Is It?
- Triglyceride-Increasing Alleles Associated with Protection against Type-2 Diabetes
- Epistasis Is a Major Determinant of the Additive Genetic Variance in
- Genetic Regulation of Bone Metabolism in the Chicken: Similarities and Differences to Mammalian Systems
- Minor Type IV Collagen α5 Chain Promotes Cancer Progression through Discoidin Domain Receptor-1
- The Philosophical Approach: An Interview with Ford Doolittle
- Downregulation of the Host Gene by miR-92 Is Essential for Neuroblast Self-Renewal in
- A Unique Virulence Gene Occupies a Principal Position in Immune Evasion by the Malaria Parasite
- Y Fuse? Sex Chromosome Fusions in Fishes and Reptiles
- Regulation of Active DNA Demethylation by a Methyl-CpG-Binding Domain Protein in
- Overlapping Patterns of Rapid Evolution in the Nucleic Acid Sensors cGAS and OAS1 Suggest a Common Mechanism of Pathogen Antagonism and Escape
- Autoselection of Cytoplasmic Yeast Virus Like Elements Encoding Toxin/Antitoxin Systems Involves a Nuclear Barrier for Immunity Gene Expression
- Genetic Architecture of Abdominal Pigmentation in
- Whole Genome DNA Binding Analysis of the Bacterial Replication Initiator and Transcription Factor DnaA
- The Centrosomal Linker and Microtubules Provide Dual Levels of Spatial Coordination of Centrosomes
- Parp3 Negatively Regulates Immunoglobulin Class Switch Recombination
- Burden Analysis of Rare Microdeletions Suggests a Strong Impact of Neurodevelopmental Genes in Genetic Generalised Epilepsies
- Cell Cycle Control by the Master Regulator CtrA in
- Myopathic Lamin Mutations Cause Reductive Stress and Activate the Nrf2/Keap-1 Pathway
- Monoallelic Loss of the Imprinted Gene Promotes Tumor Formation in Irradiated Mice
- Phylum-Level Conservation of Regulatory Information in Nematodes despite Extensive Non-coding Sequence Divergence
- Clustering and Negative Feedback by Endocytosis in Planar Cell Polarity Signaling Is Modulated by Ubiquitinylation of Prickle
- Dissecting the Function and Assembly of Acentriolar Microtubule Organizing Centers in Cells In Vivo
- MicroRNA-Dependent Transcriptional Silencing of Transposable Elements in Drosophila Follicle Cells
- β-Catenin Signaling Biases Multipotent Lingual Epithelial Progenitors to Differentiate and Acquire Specific Taste Cell Fates
- A Simple Auxin Transcriptional Response System Regulates Multiple Morphogenetic Processes in the Liverwort
- Parallel Gene Expression Differences between Low and High Latitude Populations of and .
- The Nutrient-Responsive Hormone CCHamide-2 Controls Growth by Regulating Insulin-like Peptides in the Brain of
- Characterization of TCF21 Downstream Target Regions Identifies a Transcriptional Network Linking Multiple Independent Coronary Artery Disease Loci
- PARP2 Is the Predominant Poly(ADP-Ribose) Polymerase in Arabidopsis DNA Damage and Immune Responses
- Drosophila Spaghetti and Doubletime Link the Circadian Clock and Light to Caspases, Apoptosis and Tauopathy
- Coronary Artery Disease Associated Transcription Factor TCF21 Regulates Smooth Muscle Precursor Cells That Contribute to the Fibrous Cap
- Rescue of DNA-PK Signaling and T-Cell Differentiation by Targeted Genome Editing in a Deficient iPSC Disease Model
- Disruption of Transcriptional Coactivator Sub1 Leads to Genome-Wide Re-distribution of Clustered Mutations Induced by APOBEC in Active Yeast Genes
- Yeast Killer Elements Hold Their Hosts Hostage
- Keeping in Shape the Dogma of Mitochondrial DNA Maternal Inheritance
- Extreme-Depth Re-sequencing of Mitochondrial DNA Finds No Evidence of Paternal Transmission in Humans
- Trading Places—Switching Frataxin Function by a Single Amino Acid Substitution within the [Fe-S] Cluster Assembly Scaffold
- Natural Variation Identifies , a Universal Gene Required for Cell Proliferation and Growth at High Temperatures in
- Mutations in Gene Are Associated with Predisposition to Breast Cancer
- The Whole of a Scientific Career: An Interview with Oliver Smithies
- Cell Specific eQTL Analysis without Sorting Cells
- Cooperative Action of Cdk1/cyclin B and SIRT1 Is Required for Mitotic Repression of rRNA Synthesis
- Systemic Regulation of RAS/MAPK Signaling by the Serotonin Metabolite 5-HIAA
- Reprogramming LCLs to iPSCs Results in Recovery of Donor-Specific Gene Expression Signature
- The Developmental Intestinal Regulator ELT-2 Controls p38-Dependent Immune Responses in Adult .
- Genetic Mechanism of Human Neutrophil Antigen 2 Deficiency and Expression Variations
- Feeding and Fasting Signals Converge on the LKB1-SIK3 Pathway to Regulate Lipid Metabolism in
- Early Lineage Priming by Trisomy of Leads to Myeloproliferation in a Down Syndrome Model
- Turning into a Frataxin-Dependent Organism
- Accounting for Experimental Noise Reveals That mRNA Levels, Amplified by Post-Transcriptional Processes, Largely Determine Steady-State Protein Levels in Yeast
- Biological Significance of Photoreceptor Photocycle Length: VIVID Photocycle Governs the Dynamic VIVID-White Collar Complex Pool Mediating Photo-adaptation and Response to Changes in Light Intensity
- CTXφ Replication Depends on the Histone-Like HU Protein and the UvrD Helicase
- Disruption of miR-29 Leads to Aberrant Differentiation of Smooth Muscle Cells Selectively Associated with Distal Lung Vasculature
- Notch Is Required in Adult Sensory Neurons for Morphological and Functional Plasticity of the Olfactory Circuit
- Post-transcriptional Regulation of Keratinocyte Progenitor Cell Expansion, Differentiation and Hair Follicle Regression by
- PERK Limits Lifespan by Promoting Intestinal Stem Cell Proliferation in Response to ER Stress
- Casein Kinase 1 and Phosphorylation of Cohesin Subunit Rec11 (SA3) Promote Meiotic Recombination through Linear Element Formation
- Fibroblast Growth Factor 9 Regulation by MicroRNAs Controls Lung Development and Links Loss to the Pathogenesis of Pleuropulmonary Blastoma
- Auxin-Mediated Transcriptional System with a Minimal Set of Components Is Critical for Morphogenesis through the Life Cycle in
- The Broad-Spectrum Antiviral Protein ZAP Restricts Human Retrotransposition
- The 4E-BP Caf20p Mediates Both eIF4E-Dependent and Independent Repression of Translation
- Turning into a Frataxin-Independent Organism
- Promotion of Bone Morphogenetic Protein Signaling by Tetraspanins and Glycosphingolipids
- Essential Role of the ESX-5 Secretion System in Outer Membrane Permeability of Pathogenic Mycobacteria
- The Zinc-Finger Antiviral Protein ZAP Inhibits LINE and Alu Retrotransposition
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Drosophila Spaghetti and Doubletime Link the Circadian Clock and Light to Caspases, Apoptosis and Tauopathy
- Autoselection of Cytoplasmic Yeast Virus Like Elements Encoding Toxin/Antitoxin Systems Involves a Nuclear Barrier for Immunity Gene Expression
- Parp3 Negatively Regulates Immunoglobulin Class Switch Recombination
- PERK Limits Lifespan by Promoting Intestinal Stem Cell Proliferation in Response to ER Stress
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



