-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaThe UPR Branch IRE1- in Plants Plays an Essential Role in Viral Infection and Is Complementary to the Only UPR Pathway in Yeast
The unfolded protein response (UPR) is crucial to life as it regulates gene expression in response to stress in the endoplasmic reticulum (ER). There are two functionally overlapping UPR branches in plants, e.g., IRE1-bZIP60 and S1P/S2P-bZIP17/bZIP28, but only one, IRE1p-HAC1, in yeast. Despite recent significant progress in understanding UPR, a functional connectivity of IRE1 and its splicing target has not been established in multicellular eukaryotes. It is unknown if a single UPR branch in plants has any unique biological functions. Given that all eukaryotes are equipped with the IRE1-mediated pathway, are IRE1 downstream signaling effectors conserved among kingdoms and at what degree? Here, we show that IRE1 and its substrate bZIP60 function as a matched enzyme-substrate pair to mediate virus-host interactions in plants. We further provide evidence that a single UPR branch, IRE1-bZIP60, rather than S1P/S2P-bZIP17/bZIP28, determines viral pathogenesis, indicating that the two UPR arms may have distinct functions in plants. Finally, we demonstrate that the spliced form bZIP60 and HAC1p, rather than IRE1, are functionally replaceable to cope with abiotic stress in yeast, suggesting that the downstream signaling of the IRE1-mediated splicing is evolutionary conserved in plants and yeast. These data shed new lights into UPR in multicellular eukaryotes.
Published in the journal: . PLoS Genet 11(4): e32767. doi:10.1371/journal.pgen.1005164
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1005164Summary
The unfolded protein response (UPR) is crucial to life as it regulates gene expression in response to stress in the endoplasmic reticulum (ER). There are two functionally overlapping UPR branches in plants, e.g., IRE1-bZIP60 and S1P/S2P-bZIP17/bZIP28, but only one, IRE1p-HAC1, in yeast. Despite recent significant progress in understanding UPR, a functional connectivity of IRE1 and its splicing target has not been established in multicellular eukaryotes. It is unknown if a single UPR branch in plants has any unique biological functions. Given that all eukaryotes are equipped with the IRE1-mediated pathway, are IRE1 downstream signaling effectors conserved among kingdoms and at what degree? Here, we show that IRE1 and its substrate bZIP60 function as a matched enzyme-substrate pair to mediate virus-host interactions in plants. We further provide evidence that a single UPR branch, IRE1-bZIP60, rather than S1P/S2P-bZIP17/bZIP28, determines viral pathogenesis, indicating that the two UPR arms may have distinct functions in plants. Finally, we demonstrate that the spliced form bZIP60 and HAC1p, rather than IRE1, are functionally replaceable to cope with abiotic stress in yeast, suggesting that the downstream signaling of the IRE1-mediated splicing is evolutionary conserved in plants and yeast. These data shed new lights into UPR in multicellular eukaryotes.
Introduction
The accumulation of unfolded protein in the endoplasmic reticulum (ER) results in ER stress that triggers the unfolded protein response (UPR), a complex signal transduction cascade that orchestrates adaptation to ER stress or induces apoptosis if ER stress remains unmitigated [1–6]. In mammalian cells, the UPR is mediated by three classes of membrane-associated sensor transducers including inositol-requiring transmembrane kinase/endonuclease (IRE1), protein kinase RNA (PKR)-like ER kinase (PERK), and activating transcription factor 6 (ATF6) [2,3]. In contrast to animals, the UPR in yeast is controlled by only the IRE1p-mediated signaling pathway, which triggers the expression of over 5% genes mainly encoding chaperones and ER-associated protein degradation components in the genome [7]. The IRE1 lumenal domain at the N-terminus serves as a UPR sensor domain and the C-terminal cytoplasmic portion encompassing serine/threonine protein kinase and endoribonuclease domains functions as an effector domain [3]. Upon sensing ER stress, IRE1 dimerizes or oligomerizes in the plane of the ER membrane through the binding of unfolded proteins to its UPR sensor domain or the release of oligomerization-repressing chaperones, or both, allowing for trans-autophosphorylation of juxtaposed kinase domains [8–10]. Based on the crystal structure of the dual catalytic region of IRE1, the trans-autophosphorylation of the kinase domain of IRE1 is suggested to permit unfettered binding to nucleotides (nt), which in turn promotes dimerization of IRE1 to compose the active ribonuclease site, thus unmasking the dormant endoribonucleolytic activity [3,11].
The allosteric activation of IRE1 entails the sequence-specific cleavage of a single known messenger RNA encoding a basic leucine zipper (bZIP) transcription factor—ATF/CREB1 (HAC1) in yeast [12] or X-box binding protein-1 (XBP1) in metazoans [13]. The IRE1-dependent mRNA cleavage is an unconventional splicing, which occurs predominantly in the cytoplasm in a spliceosome-independent manner [14]. While the unconventional splicing mediated by IRE1 does not comply with Chambon’s rule (GU-AG rule) at the exon-intron border, it requires the existence of a pair of characteristic stem–loop structures in mRNA, which drive the projected splicing sites close to the ribonuclease catalytic sites in the cytosolic domain of IRE1 [15,16]. IRE1 catalyzes the cleavage at the conserved sites in both of the 7-nt loops of the mRNA precursors unspliced HAC1 (HAC1 U) and XBP1 (XBP1 U), excising a 252 - and 26-bp intervening intron to produce the spliced form of HAC1 (HAC1 S) and XBP1 (XBP1 S), respectively [17], leading to a frame-shift and introduction of a new termination codon in both coding sequences. Owing to the frame-shift, the encoded HAC1 S and XBP1 S proteins both gain a transcriptional activation domain (AD) at their C-termini, which is necessary for the transcription of downstream genes [18–20]. In yeast, the translation of HAC1 U mRNA is hampered due to the presence of a translational inhibitor in the intron, and relief of this repression via producing HAC1 S is the key activating event for the yeast UPR [20]. By contrast, in metazoans both XBP1 U and XBP1 S are translated [13,19]. Nevertheless, XBP1 U complexes with the XBP1 S protein, which is more stable and transcriptionally active for UPR target genes, and exports it to the cytoplasm for proteasome-dependent degradation because a nuclear export signal and a degradation domain are present in the C-terminus of XBP1 U [13,19]. As a result, XBP1 S action is shut down during the later phase of ER stress, and XBP1 U is thus regarded as an inhibitor of the UPR in higher eukaryotes [13,19,21].
In plants, two UPR pathways have recently been identified, one mediated by IRE1-bZIP60, and the other by site-1/site-2 proteases (S1P/S2P)-bZIP17/bZIP28, which is analogous to the animal ATF6 pathway [15,22–25]. Although two genes, IRE1A and IRE1B in the genome of Arabidopsis (Arabidopsis thaliana), were found to encode IRE1 homologs a decade ago [26,27], their involvement in the plant UPR remained undetermined until most recently when bZIP60 mRNA was identified as the RNA target of IRE1A/IRE1B for unconventional splicing [1,15,22,28–30]. bZIP60 does not share a high sequence identity with HAC1 and XBP1 at both nucleotide and protein levels. However, like HAC1 and XBP1 mRNA, bZIP60 mRNA can fold into an IRE1 recognition site composed of two stem loops, each containing the bases at three positions remarkably conserved from yeast to mammalians [15,22,31,32]. Although the second loop from bZIP60 mRNA consists of 8 instead of 7 nt, the “kissing” stem loops in which the two stems are capable of base-pairing with each other are still formed to protrude the cleavage sites to the catalytic sites of IRE1, which is essential for IRE1-dependent splicing of HAC1 and XBP1 mRNAs [11,15,22,31,33]. In response to pathogen infection (i.e., Pseudomonas syringae), heat or salicylic acid stimuli, as well as ER stress agents, such as tunicamycin (Tm) and dithiothreitol (DTT), bZIP60 mRNA is spliced to remove a 23-bp fragment in Arabidopsis [15,22,30]. As a result, a translational frame-shift occurs downstream of the splicing sites and eliminates a single transmembrane domain (TMD) encoded by unspliced bZIP60 (bZIP60 U) to produce bZIP60 S, which is an active transcription factor that up-regulates the UPR target genes, such as BiP (coding for lumenal binding proteins), CAM (calmodulin), CRT (calreticulin) and PDI (protein disulphide isomerase) [15,22,30,33]. Although the IRE1-mediated mRNA splicing apparently is a conserved strategy for the IRE1 signaling across eukaryotes [34], functional inter-kingdom equivalence of bZIP60 with HAC1 and XBP1 is yet to be demonstrated.
Analyses of three independent homozygous transfer DNA (T-DNA) insertion lines of IRE1A (ire1a-2, SALK_018112; ire1a-3, WiscDsLox420D09, ire1a-4, SAIL_1256_F04) and a knockout mutant of IRE1B (ire1b-4; SAIL_238_F07) showed that IRE1A has little effect on the bZIP60 mRNA splicing in Arabidopsis seedlings in response to DTT or Tm treatment, whereas IRE1B plays a major role in the bZIP60 mRNA processing [15,30]. However, the findings obtained in a different ire1b mutant (ire1b-1, GABI_638B07) demonstrated that the single IRE1B mutation does not affect the bZIP60 splicing caused by Tm treatment, whereas stress-induced bZIP60 splicing is eliminated in the ire1a-2 ire1b-1 or ire1a-3 ire1b-4 double mutant [22,30]. Thus, IRE1A and IRE1B seem functionally redundant through splicing bZIP60 mRNA. In addition, the ire1a-3 ire1b-4 double mutant, but not the single mutant lines, develops a short-root phenotype as a result of a disorder in cell elongation in the transition zone/elongation zone [28], whereas the mutation in their RNA target bZIP60 does not lead to the similar short-root phenotype [35]. Therefore, the function of IRE1A and IRE1B with respect to the bZIP60 mRNA splicing in specific biological event(s) still remains mysterious. To make it more complex, several recent studies have also demonstrated that the two arms of the UPR in plants functionally overlap in abiotic stress [24,25,32,36,37], which motivates us to ask whether the single branch of the two UPR arms has any unique functions.
Here, we report that IRE1A and IRE1B are fully functionally redundant for the production of bZIP60 S to determine the extent of plant diseases caused by Turnip mosaic virus (TuMV) infection. Moreover, the IRE1-bZIP60 mRNA pair mediating viral infection is independent of the S1P/S2P-bZIP17/bZIP28 pathway. To the best of our knowledge, this is the first report showing that a biological process in plants is regulated by a single UPR pathway, in which IRE1 and bZIP60 mRNA function in a projected linear manner. We further show that bZIP60 and HAC1, rather than IRE1, are functionally replaceable to cope with abiotic stress response in yeast. Taken together, our data unravel an evolutionarily conserved role of the IRE1-bZIP60 pathway in regulation of abiotic and biotic stresses, shedding new lights on the complex UPR signaling pathways.
Results
bZIP60 Is Spliced in Response to TuMV Infection
Recent studies have shown that viral infection may trigger UPR in plants [38,39]. To investigate how the UPR is implicated in viral infection, we first examined whether the IRE1-bZIP60 pathway is activated under viral attack. A pair of primers that specifically captures bZIP60 S transcripts was designed to detect bZIP60 splicing by RT-PCR (S1A Fig, S1 Table). In Arabidopsis local leaves inoculated with TuMV-GFP, a recombinant TuMV tagged by green fluorescence protein (GFP), the bZIP60 S greatly accumulated at 2.5 and 5.5 days post-infection (dpi), compared with the controls (Fig 1A). The absence of the 23-bp intron in the amplified products was confirmed by colony diagnostic testing and DNA sequencing (S1B and S1C Fig). Quantitative analyses demonstrated that the level of bZIP60 S in TuMV-infected plants was significantly higher than that in controls at both time points (Fig 1B). Although bZIP60 U was also significantly increased at 2.5 dpi in response to TuMV challenge, it returned to the level not significantly different from that in the buffer or agrobacterium-inoculated controls at 5.5 dpi (Fig 1A and 1B). To investigate if the IRE1-bZIP60 pathway is also activated in systemically infected leave, the bZIP60 S was monitored following an approach recently developed by Moreno et al. [30] (Fig 1C). Result demonstrated that a unique cDNA fragment corresponding to the spliced form of bZIP60 mRNA was clearly evident in the newly emerging leaves of Arabidopsis seedlings inoculated with TuMV, but barely detectable in the corresponding leaves of control plants rubbed without or with buffer (Fig 1C and 1D). Taken together, these data indicated that TuMV infection induces bZIP60 mRNA splicing in both locally and systemically infected levels.
Fig. 1. The splicing of bZIP60 is induced in response to TuMV infection. 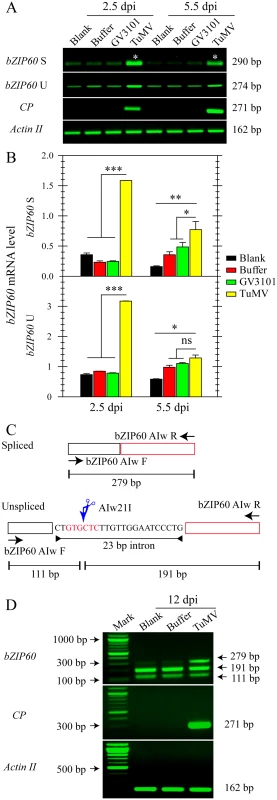
(A) Semi-quantitative RT-PCR analysis of bZIP60 U, bZIP60 S and coat protein (CP) expression at 2.5 and 5.5 dpi in the local rosette leaves from the indicated treatments. Actin II was analyzed as a loading control. Note that we loaded 4 times the amount of control for bZIP60 S since its level is much lower in vivo less than bZIP60 U [22]. PCR product sizes are indicated at right. Note that colony diagnostic test and sequencing showed that the two bands marked with asterisks do not contain the 23-bp intron (see S1 Fig). (B) The mRNA level of bZIP60 S and bZIP60 U was determined by qRT-PCR. Actin II was used as an internal control for qRT-PCR. Data represent means with SD of three biological replicates. * P<0.05, ** P<0.01, *** P<0.001, unpaired two-tailed Student’s test. ns, non-significant. (C) and (D) Flanking assay for detecting bZIP60 S in systemically infected leaves. (C) Schematic representation of the flanking approach used in this study [30]. The primer sets flanking the 23-bp intron amplify the bZIP60 S (top) and bZIP60 U (bottom). Due to the absence of Alw21I restriction site in spliced fragments, only un-spliced fragments are digested by Alw21I restriction enzyme, thus producing two smaller fragments. (D) A unique fragment corresponding to spliced forms of bZIP60 could only be detected under TuMV attack, whereas the other two smaller fragments corresponding to the Alw21I digested products were detectable in all groups. Actin II and CP were analyzed as a control and an indicator of TuMV infection. PCR product sizes are indicated at right. Since the splicing of bZIP60 mRNA is initiated by TuMV infection, we explored its downstream signaling by determining the expression of ER stress marker genes such as BiP, CRT, and PDI, which have been shown as the targets of bZIP60 [39]. In local inoculation leaves, the expression of BiP3, BiP1/2 and PDI was remarkably increased at 3, 6 and 9 dpi in response to TuMV infection, compared to the controls (S2B and S2C, S2E Fig). The expression of the ER marker gene CRT was also increased at 3 dpi under TuMV attack, even though it was barely detectable during the late phases of infection (S2E Fig). Overall, the expression of these UPR marker genes was also up-regulated at 6 and 9 dpi in systemically infected leaves (S2D and S2F Fig). These results suggested that the bZIP60 UPR signaling pathway is indeed activated in both local and systemic leaves in response to TuMV infection.
The Viral Membrane Protein 6K2 Is an Inducer of bZIP60 Splicing
To determine which TuMV protein(s) is responsible for inducing the splicing of bZIP60, transient expression assays were performed in Nicotiana benthamiana (N. benthamiana). Like other potyviruses, TuMV encodes a total of 11 mature proteins, i.e., P1, HcPro, P3, P3N-PIPO, 6K1, CI, 6K2, NIaVPg, NIaPro, NIb and CP [40]. Plant expression vectors encoding each of the 11 viral proteins fused with GFP (viral factor-GFP fusion) or their reciprocal form (GFP-viral factor fusion) were created and transiently expressed in N. benthamiana via agroinfiltration (S3 Fig). Here, the sequence of NtbZIP60 was used for the splicing assay in N. benthamiana [39]. NtbZIP60 is homologous to Arabidopsis bZIP60. NtbZIP60 mRNA was predicted to fold a conserved twin hairpin loop based on the RNA structure prediction program M-Fold [41] and the sequence in the double hairpin loop region is nearly identical in at least 20 bZIP60 homologues in plants (Figs 2A, S4 and S5), suggesting that NtbZIP60 is a potential target of unconventional splicing in N. benthamiana. Using a primer set specific for spliced NtbZIP60 (NtbZIP60 S), quantitative RT-PCR (qRT-PCR) revealed that TuMV infection triggered NtbZIP60 splicing in N. benthamiana as expected (Fig 2B, S1 Table). Of the 11 viral factors, the viral membrane protein 6K2 (either in the form of 6K2-GFP or GFP-6K2) strongly induced the accumulation of NtbZIP60 S (Fig 2B). These data suggested that the viral membrane protein 6K2 is an inducer of bZIP60 splicing.
Fig. 2. TuMV 6K2 is an inducer of bZIP60 splicing. 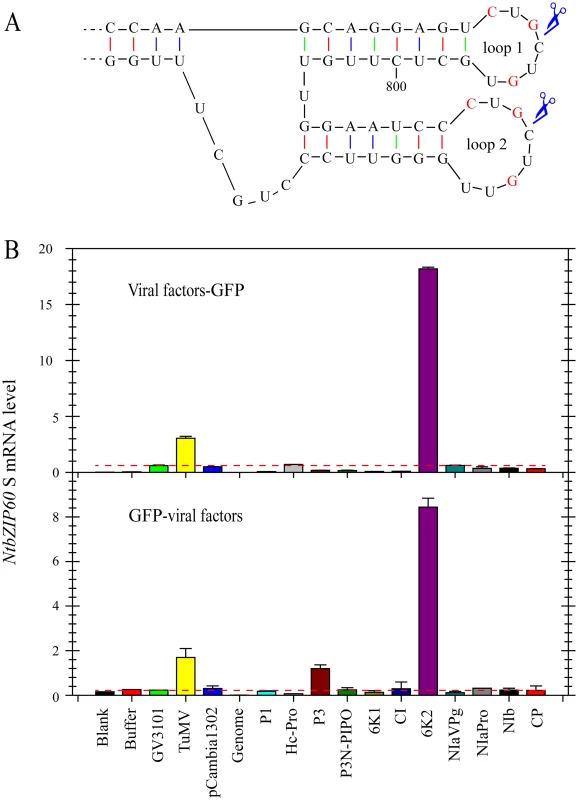
(A) Twin hairpin loop structure in NtbZIP60 mRNA, which is the magnification of red boxed area in S4 Fig. Each of the two loops contains three conserved bases (red). Scissors indicate predicted cleavage sites. (B) qRT-PCR analysis of NtbZIP60 S level in N. benthamiana. 11 TuMV factors fused with a C-terminal (top) or N-terminal (bottom) GFP were transiently expressed alone via agroinflitration. At 2.5 dpi, the transient expression of each construct was verified under confocal by observing GFP (see S3 Fig), and RNA was extracted from the agroinflitrated leaves. The RNAs from un-infiltrated leaves (Blank) and the leaves infiltrated with buffer, GV3101, GV3101 containing TuMV infectious clone and pCabmibia1302 were used as controls. Genomic DNA was also analyzed as a control by qRT-PCR. 18S RNA was used as an internal control. Note that the lines drawn according to the value of NtbZIP60 S caused by GV3101 were used to highlight that only TuMV 6K2 in both types of constructs could strongly induce NtbZIP60 splicing. The Data represent means with SD of three biological replicates. Viral Accumulation Is Reduced in bzip60-2 Mutant but Not in bzip60-1 Mutant
Since TuMV and TuMV 6K2 induced the splicing of bZIP60 mRNA in plants, the role of bZIP60 S in viral pathogenesis was examined. Here, two independent T-DNA insertion mutants in bZIP60 (bzip60-1, SALK_050203; bzip60-2, SAIL_283_B03) were used [15,42]. Seedlings of the wild type Arabidopsis and the bzip60-1 mutant inoculated with TuMV developed typical TuMV symptoms, including mosaic, leaf yellowing and stunted growth, compared to the mock-inoculated control (Fig 3A). However, the viral symptoms in the bzip60-2 mutant were much milder than the wild type and the bzip60-1 mutant, although the stature of TuMV-infected bzip60-2 mutant was smaller than that of mock-inoculated plants (Fig 3A). Quantitative analyses indicated that TuMV accumulated to similar levels in the wild type and bzip60-1 plants (Fig 3B), but in the bzip60-2 mutant, the level of the virus was much lower than that in the wild type and bzip60-1 mutant (Fig 3B, P<0.01). These data clearly indicated that viral pathogenesis is alleviated in the bzip60-2 mutant. Intriguingly, in comparison with that in mock-inoculated plants, bZIP60 S transcripts were significantly accumulated in the systemically infected leaves of both wild type and bzip60-1 plants inoculated with TuMV (Fig 3C, P<0.01), and the bZIP60 S level in the wild type was significantly higher than that in the bzip60-1 mutant (Fig 3C, P<0.05). In contrast, no bZIP60 S was detectable in the bzip60-2 mutant either under or without TuMV attack (Fig 3C). Taken together, these data suggested an association of TuMV accumulation and viral pathogenesis with the bZIP60 S.
Fig. 3. Viral pathogenesis is alleviated in the bzip60-2 mutant. 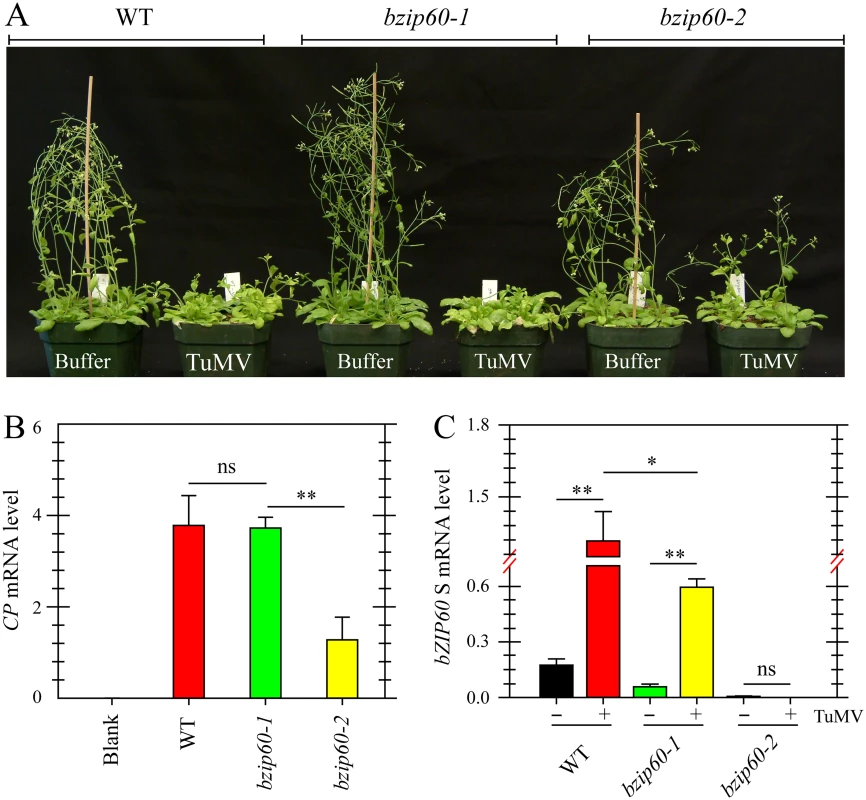
(A) Phenotypes of the wild type, bzip60-1 and bzip60-2 mutants at 18 dpi after inoculation with buffer or TuMV. Note that the bzip60-2 mutant, but not the bzip60-1 mutant, alleviated TuMV symptoms, compared to the wild type. (B) and (C) qRT-PCR analysis of TuMV CP (B) and bZIP60 S (C) in the wild type, bzip60-1 and bzip60-2 mutants. At 18 dpi after inoculation with buffer or TuMV, RNA was extracted from the systemically infected leaves, and qRT-PCR was carried out. For detecting CP level, the RNAs were collected from the wild type, bzip60-1 and bzip60-2 mutants under buffer treatment as a blank control. Actin II was used as an internal control. Data represent means with SD of three biological replicates. * P<0.05, ** P<0.01, unpaired two-tailed Student’s test. ns, non-significant. bzip60-1 and bzip60-2 Are Non-RNA Null Mutants and Transcribe Incomplete bZIP60 ORF
The finding that the bzip60-1 and bzip60-2 mutants develop contrasting viral symptoms promoted us to re-examine the molecular characterization of the two mutants. By sequencing the T-DNA flanking regions, we mapped the T-DNA insertion at position 41 and 1116 nt downstream from the translation initiation codon (ATG1) of bZIP60 genomic DNA in bzip60-1 and bzip60-2 (Figs 4A and S6A and S6B), respectively, indicating that the genomic DNA structure in the two mutants was disrupted. Genomic PCR analyses indicated that both bzip60 mutants are homozygous mutant lines (Fig 4A and 4B). RT-PCR with three specific pairs of primers (NF + NR, MF + MR, and CF + CR) was carried out to further determine whether the bzip60-1 and bzip60-2 mutants represent RNA-null mutants (Fig 4A, S1 Table). Although no bZIP60 amplicon was detectable in the bzip60-1 mutant using the primers flanking the insertion site (NF + NR), bZIP60 transcripts were present in the bzip60-1 mutant using the downstream primer sets (MF + MR and CF + CR) (Fig 4A and 4C). In contrast, bZIP60 amplicon could not be detected in the bzip60-2 mutant by the downstream primer set (CF + CR), while other two amplicons could be seen using the upstream primer sets (NF + NR and MF + MR) (Fig 4A and 4C). These results suggested that neither bzip60-1 nor bzip60-2 is an RNA-null mutant, although both of them indeed do not express full-length bZIP60 transcripts.
Fig. 4. Molecular characterization of the bzip60 mutants. 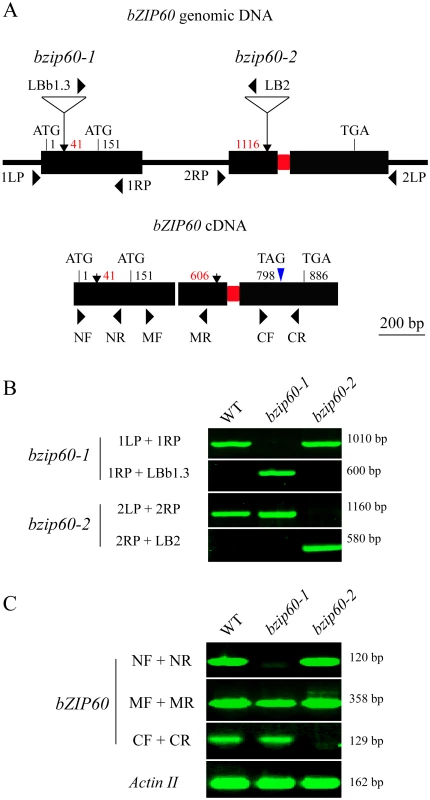
(A) Schematic representation of bZIP60 genomic DNA (top) and cDNA (bottom) structure with intron and exons. The positions of the T-DNA insertion in the two bzip60 mutants are indicated by arrows in genomic DNA, and corresponding positions in the cDNA structure are also shown. The start and stop codons of two in-frame ORFs are labeled (ATG1 for bZIP60 and ATG151 for bZIP60ΔN). Note that a 23-bp sequence within the second exon to be spliced through unconventional splicing is represented by a red rectangle. The new termination codon (TAG798) due to a frame-shift caused by splicing is indicated by a blue triangle. (B) and (C) PCR with genomic DNA (B) and cDNA (C) from the wild type and the bzip60 mutants to test the homozygosis and the presence of non-full length bZIP60 mRNA fragments in the mutant lines. PCR product sizes are indicated at right. The primer locations are shown in (A), and primer sequences are given in S1 Table. In (C), Actin II served as a control. Analyses of the bZIP60 cDNA sequence revealed that there is a potential in-frame start codon at position 151 relative to the start codon of the previously identified bZIP60 ORF (Figs 4A, S6A and S7A). Thus, the bzip60-1 mutant might produce a shorter version of bZIP60 ORF (bZIP60ΔN). To prove this assumption, 5′ rapid amplification of cDNA ends (5′ RACE) was used to determine the 5′ end of bZIP60 mRNA in this mutant. DNA sequencing results indicated that all selected cDNAs from the bzip60-1 mutant after different treatments contain the intact ORF from the alternative start codon, although the 5′ end of bZIP60ΔN varied in length (S7A Fig). In addition, analyses of the bzip60-2 mutant revealed that T-DNA insertion in the bZIP60 genome disrupts the 3′ end of bZIP60 mRNA, which causes the premature termination of bZIP60 translation at nt 52 of the T-DNA sequence (S7B Fig, Stop ΔC). Taken together, these findings showed that the bzip60-2 mutant transcribes incomplete bZIP60 mRNA (bZIP60ΔC) coding for a truncated bZIP60 without the C-terminus, whereas the bzip60-1 mutant produces bZIP60ΔN transcripts encoding a shorter version of bZIP60 lacking the N-terminal 50 amino acids (aa) (S8A Fig).
bzip60-1 Is a Knockdown Mutant and bzip60-2 a bZIP60 Splicing Knockout Mutant
To characterize the two types of shorter bZIP60 mRNAs of bzip60-1 and bzip60-2 mutants, we further analyzed bZIP60 mRNA expression and splicing in the wild type and mutants. Since the IRE1-mediated unconventional splicing could be detected in Arabidopsis flowers under unstressed conditions [35], we isolated RNAs from opened and unopened flowers, according to a previous report [43]. The expression levels of bZIP60 in the wild type and bZIP60ΔN in the bzip60-1 mutant as well as the bZIP60ΔC in the bzip60-2 mutant were quantitatively determined using a primer set sitting between MF and MR (Fig 4A, S1 Table). It was found that, in both types of flowers, the bZIP60 level in the wild type was significantly higher than the bZIP60ΔN level in the bzip60-1 mutant (S9A Fig, P<0.01). The bZIP60ΔC transcript in the bzip60-2 mutant accumulated to a level similar to bZIP60 mRNA in the wild type (S9A Fig). Therefore, T-DNA insertion did not affect bZIP60ΔC expression in the bzip60-2 mutant, but greatly inhibited the bZIP60ΔN expression in the bzip60-1 mutant. It should be pointed out that bZIP60 mRNA in the wild type, bZIP60ΔN mRNA in the bzip60-1 mutant, and bZIP60ΔC mRNA in the bzip60-2 mutant were all up-regulated in opened flowers in comparison with unopened flowers (S9A Fig).
Analysis of bZIP60 mRNA splicing revealed that bZIP60 S was not detected in both types of flowers in the bzip60-2 mutant (S9B Fig). As for the bzip60-1 mutant, the bZIP60ΔN S level of the unopened flowers was significantly lower than the bZIP60 S level in the wild type (S9B Fig, P<0.001). However, in the wild type and bzip60-1 plants, the level of bZIP60 S (bZIP60ΔN S for bzip60-1) was significantly higher in unopened flowers than in opened ones (S9B Fig, P<0.001 in wild type and P<0.01 in bizp60-1), suggesting that the splicing of bZIP60 mRNA might be part of the scheduled organelle development programs.
Previous publications have shown that treatment of Arabidopsis with ER stress agents, such as DTT and Tm, induces bZIP60 mRNA splicing [15,22,30]. We thus also analyzed bZIP60 mRNA splicing in the bzip60-1 and the bzip60-2 seedlings treated with DTT or Tm. After treatment with ER stress agents DTT or Tm, the bzip60-1 seedlings rather than the bzip60-2 seedlings produced the detectable spliced form of bZIP60 mRNA, although the level of bZIP60ΔN S in the bzi60-1 mutant was lower than that of bZIP60 S in the wild type (S9C Fig). Clearly, bZIP60 splicing is eliminated in the bzip60-2 mutant but only reduced in the bzip60-1 mutant.
It has been reported that a pair of kissing hairpin loops with three conserved bases in each loop is the recognition sites of IRE1 (S5 and S10A Figs) [15]. Since the spliced form of bZIP60 could be detected in the bzip60-1 mutant, it is possible that bZIP60ΔN mRNA could still be able to fold into twin kissing loops. To test this idea, we predicted the secondary structure of bZIP60 mRNA using M-Fold [41]. Among 29 predicted different free energy forms of RNA structures, 89.6% of bZIP60ΔN contains the conserved double loops, similar to the full-length bZIP60 mRNA (S10B Fig). Interestingly, the predicted conserved loops are also present in one of bZIP60ΔN mRNA species resulting from 5’ RACE which even contains a short batch of nt from the T-DNA (S10B Fig). These data supported the assumption that the truncated bZIP60 mRNA in the bzip60-1 mutant is spliced in the same manner as bZIP60 mRNA in wild type. Taken together these data suggested that the bzip60-1 mutant is a bZIP60 expression knockdown mutant whose bZIP60ΔN transcripts can be spliced and the bzip60-2 mutant is a bZIP60 splicing knockout mutant.
bZIP60ΔN S Is Targeted to the Nucleus as bZIP60 S
In a recent study, Deng et al. have shown that as an active transcriptional factor, bZIP60 S, rather than bZIP60 U, is located in the nucleus [15]. Since bZIP60ΔN S in the bzip60-1 mutant supports virus infection like full-length bZIP60 S (Fig 3A and 3B), it should at least have capability to enter the nucleus. To prove this assumption, we used NucPred program to position the nucleus localization signal (NLS) in bZIP60 [44]. It was found that both bZIP60 S and bZIP60ΔN S contain five NLS consensus motifs (K/RR/KxR/K) (Figs 5A and S8A) [45]. Among the five predicted NLS motifs, NLS2 and NLS1 are produced due to the splicing-mediated frame-shift, and other three NLSs (NLS3, NLS4 and NLS5) are also present in bZIP60 U and bZIP60ΔN U (Figs 5A and S8A). Further analyses showed that NLS1, NLS3 and NLS4 are highly conserved in at least 20 plant bZIP60 homologues (S11 Fig). Although the unspliced forms of bZIP60 and bZIP60ΔN also contain another conserved NLS motif (NLS6) downstream of the TMD motif, it is absent in the spliced forms of bZIP60 and bZIP60ΔN and thus was not included in this study (Figs 5A, S8A and S11). Mutation analyses showed that none of mutated NLS3, NLS2 or NLS1 alone prevented bZIP60 S from entry into the nucleus (Fig 5B). Also, introduction of a double mutation into NLS3 and NLS2 or NLS2 and NLS1 of the bZIP60 S did not affect its nucleus-targeting (Fig 5B). However, either a double mutation of NLS3 and NLS1 or the deletion of NLS3 plus mutation of NLS1 of the bZIP60 S compromised its nucleus-targeting, leading to co-localizing with an ER labeling marker (Fig 5B). Moreover, a triple mutation of NLS4, NLS5 and NLS2 did not arrest the localization of bZIP60 S into the nucleus (Fig 5B). These data indicated that either NLS3 or NLS1 can direct the targeting of bZIP60 S protein into the nucleus, and that none of NLS4, NLS5 or NLS2 is a functional nucleus-targeting signal motif. We therefore concluded that bZIP60 S and bZIP60ΔN S both have functional NLSs (Figs 5A and S8A).
Fig. 5. Identification of NLSs of bZIP60 S. 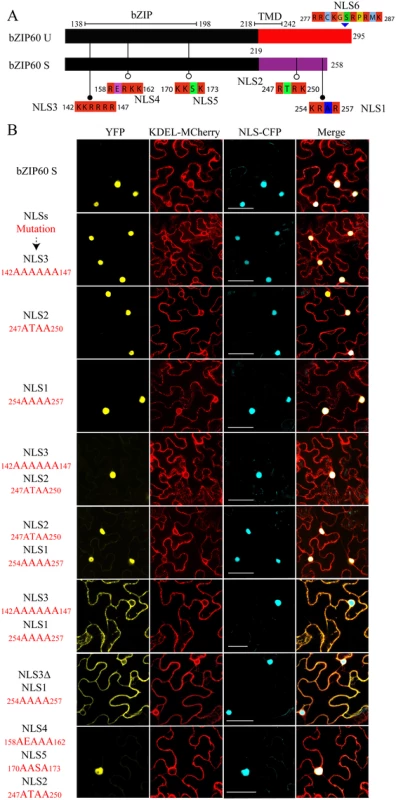
(A) The predicted NLS motifs distributed in bZIP60 U and bZIP60 S (see S8 Fig). Note that only bZIP60 U contains the TMD, and the NLSs located at the C-terminus of bZIP60 S have been mentioned previously [33] and thus named as NLS1 and NLS2. (B) Sub-cellular localization of YFP-bZIP60 S following mutation or deletion (for NLS3) of the predicted NLS(s) of bZIP60 S. KDEL-MCherry and NLS-CFP were used as the ER marker and the nucleus reporter, respectively. Experiments were repeated three times with similar results Bars = 25 μm. Homodimerization of bZIP60 S and bZIP60ΔN S
It is well known that bZIP proteins form homodimers and/or heterodimers to regulate gene transcription [23,46]. Although bZIP60 without C-terminus (aa 218–258) can heterodimerize with bZIP28 [23], it remains unconfirmed whether bZIP60 S homodimerizes. In addition, given that bZIP60ΔN S contains functional NLSs, and the bzip60-1 mutant develops typical viral symptoms like the wild type (Figs 3A, 3B, 5 and S8), the bZIP60ΔN S was surmised to behave the same as the full-length bZIP60 S. To verify this assumption, we first examined if bZIP60 S or bZIP60ΔN S self-interacts using the gold yeast two-hybrid (Y2H) system. In yeast, bZIP60 S and bZIP60ΔN S were shown to have no auto-activation and toxicity effects (Fig 6A). When bZIP60 S and bZIP60ΔN S served as both bait and prey, bZIP60 S and bZIP60ΔN S both indeed showed a strong tendency to homodimerize even under high-stringent selection conditions (QDO medium plus a high concentration of AbA) (Figs 6A and S12A).
Fig. 6. Homodimerization of bZIP60 S and bZIP60ΔN S. 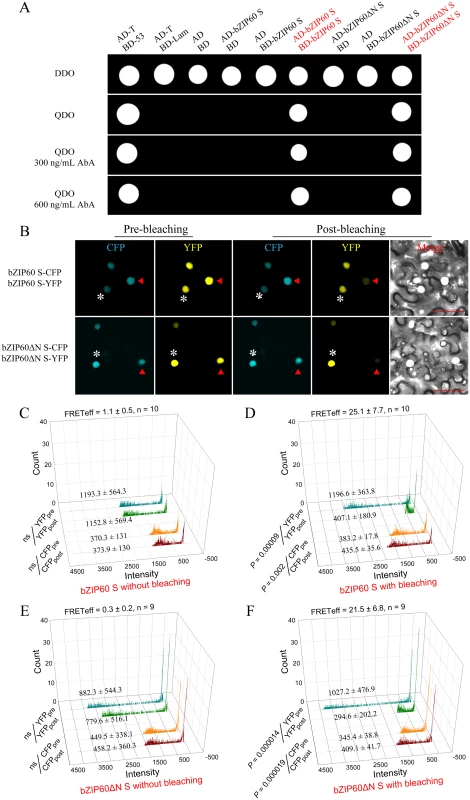
(A) Self-interactions of bZIP60 S and bZIP60ΔN S in the Y2H assay. Homodimerziation was examined by yeast growth on a QDO medium in the presence of AbA. Note that both bZIP60 S and bZIP60ΔN S showed a strong homo-interaction, indicated by yeast growth even under higher concentration AbA (also see S12A Fig). Data are representative of three repeats. (B) Localization and self-interactions of bZIP60 S and bZIP60ΔN S in vivo by photo-bleaching assay. Images were captured from CFP and YFP channels before and after YFP-fusion proteins photo-bleaching using a 514 nm beam at 100% output power. Note that the donor fluorescence was increased following photo-bleaching the acceptor (red triangles), whereas it kept constant without the acceptor photo-bleaching (white asterisks). Bars = 50 μm. (C), (D), (E) and (F) Emission of donor and acceptor images without (C and E) or with (D and F) photo-bleaching FRET. The average fluorescence intensity per photon of pre- and post-bleaching images from donor and acceptor was presented, and the FRET efficiency was calculated based on at least 9 independent photo-bleaching results. Next, we employed fluorescence resonance energy transfer (FRET) to further determine the self-interactions of bZIP60 S or bZIP60ΔN S in living cells. As expected, bZIP60 S and bZIP60ΔN S both were located in the nucleus, co-localized with an NLS-tagged reporter (S12B Fig). However, no FRET signal was evident in cells co-transformed with bZIP60 S-CFP and NLS-YFP or the reciprocal combination (S12B Fig). The same results were also obtained from co-transformation with bZIP60ΔN S-CFP and NLS-YFP or the reciprocal combination (S12B Fig). Nevertheless, a strong FRET emission was observed in cells co-expressing bZIP60 S-CFP and bZIP60 S-YFP proteins or co-expressing bZIP60ΔN S-CFP and bZIP60ΔN S-YFP proteins (S12B Fig). The results were further corroborated by the FRET acceptor photo-bleaching assay. After photo-destruction of bZIP60 S-YFP energy acceptor, a significant increase in the fluorescent intensity of bZIP60 S-CFP was observed, indicating energy transfer between bZIP60 S-CFP and bZIP60 S-YFP (Fig 6B and 6D, red triangles, P = 0.002). Similarly, the energy transfer between bZIP60ΔN S-CFP and bZIP60ΔN S-YFP was also detectable with a FRET efficiency similar to that found in the combination of bZIP60 S-CFP and bZIP60 S-YFP (Fig 6B, 6D and 6F, red triangles). In control cells without photo-destruction of bZIP60 S-YFP and bZIP60ΔN S-YFP, no significant change in fluorescence intensity of bZIP60 S-CFP and bZIP60ΔN S-CFP could be observed (Fig 6B, 6C and 6E, white asterisks). These results indicated that bZIP60 S and bZIP60ΔN S both homodimerize.
Either bZIP60 S or bZIP60ΔN S Can Rescue the Virus Suppression Phenotype
To prove that the mild viral symptoms phenotype of the bzip60-2 mutant is caused by the loss-of-function of bZIP60 S, and bZIP60ΔN S fulfills the same function as bZIP60 S in TuMV infection, we complemented the bzip60-2 mutant using bZIP60 S and bZIP60ΔN S under the control of the native promoter (pbZIP60-bZIP60 S and pbZIP60-bZIP60ΔN S). We found that, in response to TuMV, the transgenic lines with bZIP60ΔN S or bZIP60 S developed the same typical viral symptoms as the wild type (Fig 7A). Quantitative analyses of virus accumulation revealed that only the bzip60-2 mutant produced much fewer viruses than the wild type (Fig 7B, P<0.01), and no difference in virus accumulation could be detected among the wild type and the transgenic lines carrying either bZIP60ΔN S or bZIP60 S (Fig 7B). These data unambiguously proved that either bZIP60ΔN S or bZIP60 S rescues the virus suppression phenotype of the bzip60-2 mutant. We therefore concluded that the biotic stress-resistance phenotype of the bzip60-2 mutant arise from the loss-of-function mutation in bZIP60 S, and bZIP60ΔN S has the same function as bZIP60 S in TuMV infection in plants.
Fig. 7. Either bZIP60 S or bZIP60ΔN S can rescue the virus suppression phenotype of the bzip60-2 mutant. 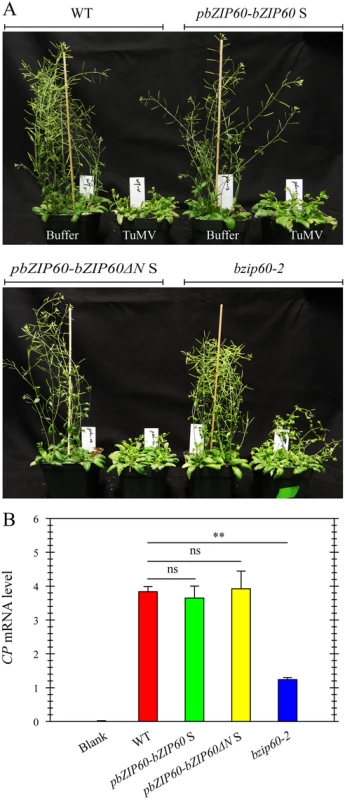
(A) Phenotypes of the wild-type, the transgenic lines of pbZIP60-bZIP60 S and pbZIP60-bZIP60ΔN S and the bzip60-2 mutant at 18 dpi after inoculation with buffer or TuMV. Note that both transformants developed typical TuMV symptoms, compared to the wild type and bzip60-2 mutant. (B) qRT-PCR analysis of TuMV CP mRNA levels. At 18 dpi after inoculation with buffer or TuMV, RNA was extracted from systemically infected leaves, and qRT-PCR was carried out. Actin II was used as an internal control. Data represent means with SD of three biological replicates. ** P<0.05, unpaired two-tailed Student’s test. ns, non-significant. Loss-of-Function of IRE1A and IRE1B Inhibits Viral Infection
To definitively establish the role of IRE1-bZIP60 S signaling pathway in the development of viral symptoms, two ire1a ire1b double mutants (ire1a-2 ire1b-4 and ire1a-3 ire1b-4) were inoculated with TuMV since both IRE1A and IRE1B have been suggested to process bZIP60 mRNA [15,28,30]. Under TuMV attack, virus-induced symptom development in these double mutants was markedly delayed and viral accumulation was also significantly inhibited, compared with the wild type (Fig 8A and 8F, P<0.001), whereas the three single mutants displayed typical viral symptoms like the wild type (S13 Fig). In contrast to an increased level of bZIP60 S mRNA in TuMV-infected wild type plants, no bZIP60 S mRNA was detectable in the two ire1a ire1b double mutants (Fig 8E). The vanished bZIP60 S mRNA in the ire1a-3 ire1b-4 double mutant was largely restored following transformation with IRE1A under the control of its native promoter or with IRE1B under the control of a DEX-inducible promoter in the presence of DEX (Fig 8E). In parallel, TuMV infection was also rescued by complementation of the ire1a-3 ire1b-4 double mutant with IRE1A or IRE1B alone (Figs 8B, 6C and 8F). Importantly, the virus suppression phenotype of the ire1a-3 ire1b-4 double mutant could also be rescued by introducing bZIP60 S or bZIP60ΔN S into the double mutant (Fig 8D and 8F). Therefore, it was concluded that the IRE1A or IRE1B mediated bZIP60 splicing is essential for TuMV infection.
Fig. 8. Loss-of-function of IRE1A and IRE1B inhibits viral infection. 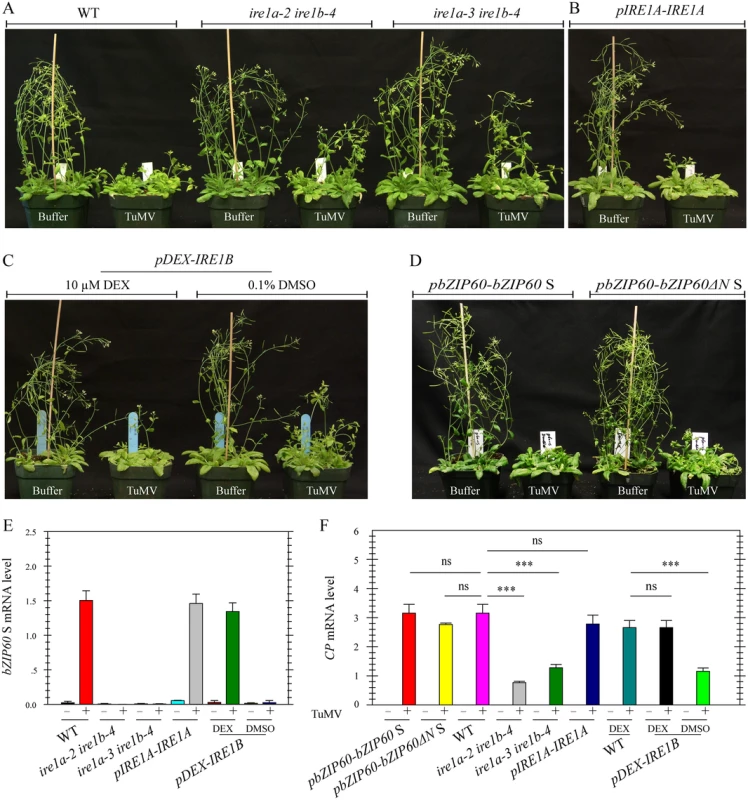
(A) Phenotypes of the wild type and the two different ire1a ire1b double mutants (ire1a-2 ire1b-4 and ire1a-3 ire1b-4) at 18 dpi after inoculation with buffer or TuMV. Note that both double mutants developed slight TuMV symptoms, compared to the wild type and the single mutants (see S13 Fig). (B), (C) and (D) Complementation of ire1a-3 ire1b-4 by IRE1A under the control of its native promoter (B), IRE1B under the control of a DEX-inducible promoter (C), or bZIP60 S and bZIP60ΔN S under the control of bZIP60 promoter (D) rescued the virus suppression phenotype of ire1a-3 ire1b-4. For dexamethasone (DEX) treatment, 0.1% DMSO (the vehicle of DEX) and DEX (10 μM) were spared onto seedlings 3 d prior to TuMV infection and every 3 d during the whole infection period. (E) and (F) qRT-PCR analyses of bZIP60 S (E) and TuMV CP (F) in the wild type, two double mutants and transgenic lines. At 18 dpi after inoculation with buffer or TuMV, RNA was extracted from systemically infected leaves, and qRT-PCR was carried out. Actin II was used as an internal control. Data represent means with SD of three biological replicates. *** P<0.01, unpaired two-tailed Student’s test. ns, non-significant. bZIP60 S and bZIP60ΔN S Are Functionally Complementary with HAC1p S in Yeast
Since bZIP60ΔN S and bZIP60 S have equivalent functions in TuMV infection, a biotic stress (Figs 3, 7 and 8), it is very tempting to assume that they have similar function in abiotic stress. In order to prove this, we used a yeast complementation system developed based on HAC1-deficient yeast strains (CRY1 Δhac1p::TRP) [10,47]. Both HAC1p and bZIP60 contain a conserved DNA binding domain (BD or bZIP domain) (Figs 9A and S8). Moreover, like HAC1p, bZIP60 S and bZIP60ΔN S have a functional NLS [45] that is also located immediate upstream of the BD domain (Figs 5 and S8, NLS3), i.e., RKRAKTK in HAC1p (Fig 9A, shaded in pink) [48] and KKRRRR in bZIP60 (Fig 9A, shaded in black). cDNAs encoding HAC1p U, HAC1p S, bZIP60 U, bZIP60 S, bZIP60ΔN U and bZIP60ΔN S were cloned into a yeast expression vector in frame fused to the C-terminus of YFP under the control of a GAL1 inducible promoter. In yeast cells, in the presence of 2% galactose and 2 mM DTT, the cells expressing YFP-HAC1 U and YFP-HAC1 S displayed nuclear fluorescence, indicated by 4’, 6-Diamidino-2-phenylindole (DAPI), whereas YFP alone from the empty vector distributed throughout the cytoplasm (Fig 9B). Both bZIP60 S and bZIP60ΔN S tagged with YFP at their N-termini were efficiently targeted to the nucleus (Fig 9B), whereas the fusion proteins YFP-bZIP60 U and YFP-bZIP60ΔN U were found in the cytoplasm (Fig 9B). This was likely due to that the unspliced forms of bZIP60 contain a TMD that is absent in the spliced forms of bZIP60 (S8 and S11 Figs). In addition, consistent with the results obtained in N. benthamiana, the bZIP60 S protein with mutated NLS3 and NLS1 was found in the cytoplasm in yeast (Figs 5B and 9B).
Fig. 9. Both bZIP60 S and bZIP60ΔN S are localized to the nucleus in yeast. 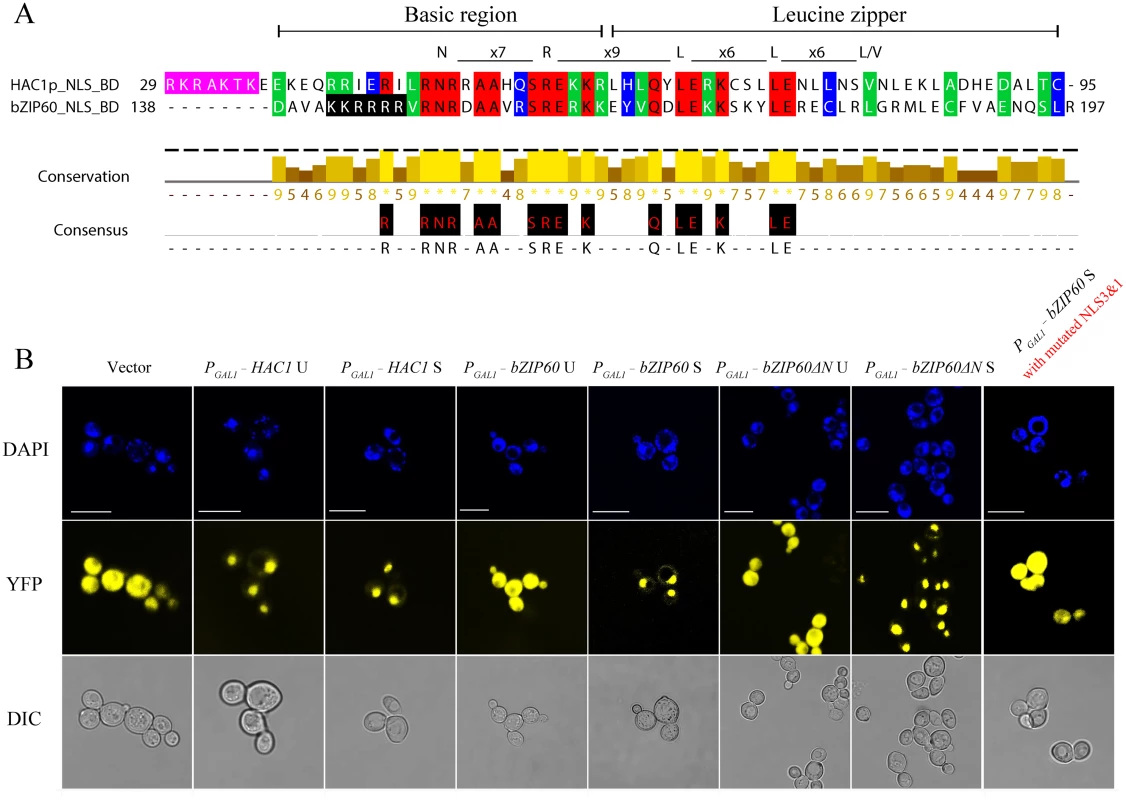
(A) The amino acid sequence of the bZIP domain is highly conserved between HAC1p and bZIP60. The identical residues are highlighted with red, and conserved residues with a conservation threshold of 9 and 8 are shaded with green and blue, respectively. A consensus sequence is given below with a histogram. A schematic of the bZIP domain consensus is shown above by extremely conserved residues and distance [54]. The typical bZIP domain contains basic region and leucine zipper. Note that the NLS shaded in pink in HAC1p, whereas the NLS shaded in black in bZIP60. (B) Detection of subcellular localization of HAC1p and bZIP60 by confocal imaging of CRY1 Δhac1::TRP strains. The transformed yeast cells with 2-micron plasmids were induced by galactose in the presence of 2 mM DTT. At 8 h post-induction, the yeast cells were stained with DAPI for 1 h and processed for confocal observation. Note that HAC1p U, HAC1p S, bZIP60 S and bZIP60ΔN S localized to the nucleus indicated by DAPI staining, whereas bZIP60 U, bZIP60ΔN U as well as bZIP60 S with mutated NLS3 and NLS1 displayed YFP signal throughout the cytoplasm, like the empty vector control. It is worth mentioning that DAPI also stains the mitochondria. Bars = 10 μm. It has been reported that the last 18 aa (residues 221–238) in HAC1p S is a domain for transcription activation (AD), which results from the unconventional splicing to remove 252 nt (S8 Fig) [18]. In Arabidopsis, the transcriptional activation activity of bZIP60 is located to aa 41–80 (S8 Fig) [22,49]. Comparison of the two ADs revealed a highly conserved motif corresponding to aa 59–76 of bZIP60 S, which is also present in bZIP60ΔN S (Figs 10A and S8). Since both bZIP60 S and bZIP60ΔN S contain NLS, AD and BD domains as HAC1p, functional complementation was performed to test whether bZIP60 S and bZIP60ΔN S could execute the functions of HAC1p in yeast. Considering that bZIP60ΔC from the bzip60-2 mutant also contains AD and BD domains as well as a functional NLS (S8 Fig, NLS3), it was therefore included in the functional complementation assay. Here, we designed two types of bZIP60ΔC, i.e., bZIP60ΔC1 and bZIP60ΔC2 (S8A and S14 Figs). cDNAs of HAC1p and bZIP60 were expressed in the Δhac1::TRP cells using a CEN-ARS plasmid containing a GAL1 inducible promoter. Compared to the control yeast grown in SGal-TRP with 0.1% DMSO, the Δhac1p::TRP cells displayed an obvious growth defect under 0.2 μg/mL Tm treatment (Fig 10B). Intriguingly, bZIP60 S and bZIP60ΔN S, rather than bZIP60 U, bZIP60ΔN U and bZIP60ΔCs, successfully rescued the ER stress phenotype like HAC1p U and HAC1p S (Figs 10B and S14B). As expected, the bZIP60 S protein with mutated NLS3 and NLS1 failed to rescue the Tm-sensitive phenotype (S14B Fig). Previous studies have shown that constitutive expression of HAC1p S has an adverse effect on yeast growth under normal conditions [12,50]. Consistent with these results, the yeast cells expressing bZIP60 S or bZIP60ΔN S, but not bZIP60 U, bZIP60ΔN U nor bZIP60ΔCs, also exhibited limited growth in the presence of 2% galactose (Figs 11C and S14A). As expected, the yeast cells displayed normal growth in non-inducible medium (Figs 11A, 11B and S14B). Together, the results suggested that bZIP60 S or bZIP60ΔN S functions like HAC1p to rescue the ER-stress sensitive phenotype in yeast.
Fig. 10. Either bZIP60 S or bZIP60ΔN S can rescue HAC1-deficient yeast under ER stress. 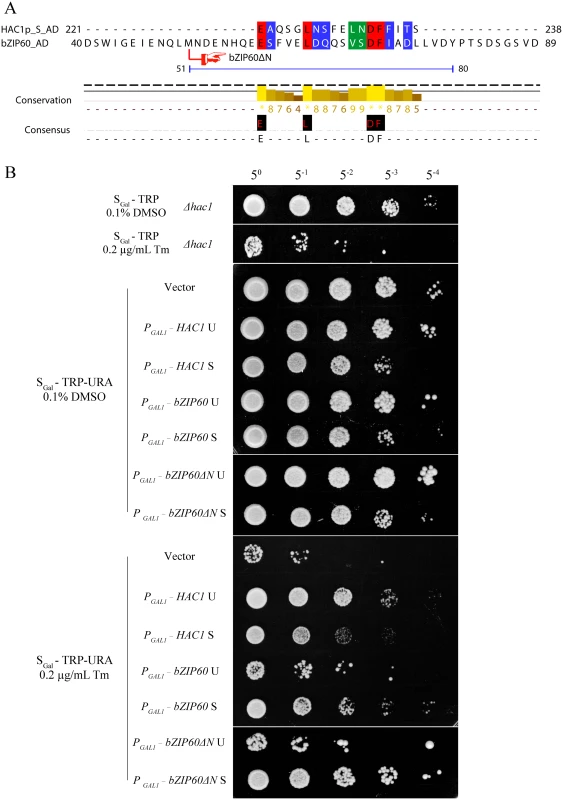
(A) Alignment of the amino acid sequence of the ADs between HAC1p S and bZIP60. A region corresponding to amino acids 59–76 in the AD of bZIP60 was found to be highly identical to the AD of HAC1p S. The identical residues are highlighted with red, and conserved residues with a conservation threshold of 9 and 8 are shaded with green and blue, respectively. A consensus sequence is given below with a histogram. Note that both bZIP60 S and bZIP60ΔN S contain the conserved AD region (see S8 Fig). (B) Functional complementation in CRY1 Δhac1::TRP strains with HAC1p and bZIP60. The untransformed or transformed yeast cells with CEN-ARS plasmids, which were grown in the raffinose-containing medium for 8 h, were switched to galactose-containing medium for induction from OD600 = 0.3. At 10 h post-induction, the cells were normalized to an OD600 = 1.0 and 5-fold serial dilutions were spotted on galactose-containing plates in the presence of 0.1% DMSO (control) or 0.2 μg/mL Tm. The plates were kept at 30°C for 48 h. Note that the expression of bZIP60 S and bZIP60ΔN S, not bZIP60 U or bZIP60ΔN U, inhibits yeast growth under normal condition but increases ER stress tolerance, and the induction of both HAC1 U and HAC1 S leads to retarded growth and enhanced ER tolerance (see Fig 11C). Fig. 11. Effects of constitutive expression of HAC1 and bZIP60 on yeast growth. 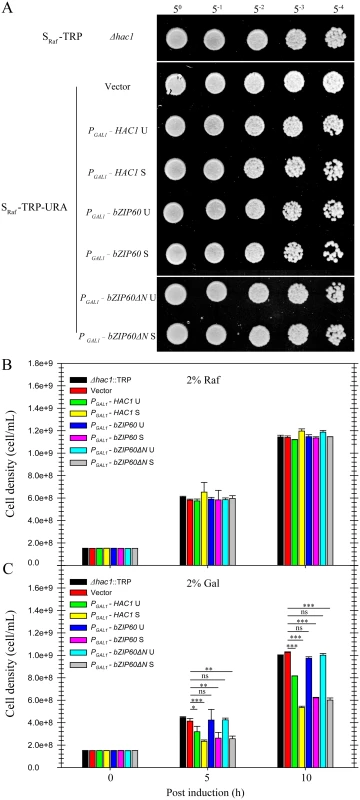
(A) to (C) The untransformed or transformed CRY1 Δhac1::TRP cells, which were grown in the raffinose-containing medium for 8 h, were switched to galactose- or raffinose-containing medium for culture from OD600 = 0.3. (A) After 10 h culture in the presence of raffinose, the cells were normalized to an OD600 = 1.0 and 5-fold serial dilutions were spotted on raffinose-containing plates. The plates were kept at 30°C for 48 h. (B) and (C) Quantitative measurement of the effects of constitutive expression of HAC1p and bZIP60 on yeast growth in the presence of raffinose (B) and galactose (C). After 5 and 10 h culture, the yeast cell density was determined by measuring the OD600 (1 OD600 = 5e+8). Note that the expression of bZIP60 S and bZIP60ΔN S, not bZIP60 U or bZIP60ΔN U, inhibits yeast growth, whereas the induction of both HAC1 U and HAC1 S leads to retarded growth (see Fig 10B). Data represent means with SD of three experiments. * P<0.05, ** P<0.01, *** P<0.001, unpaired two-tailed Student’s test. ns, non-significant. Discussion
bZIP60 S of Arabidopsis Is a Functional Homolog of Yeast HAC1p
In this work, we found that the bzip60-2 mutant displayed an anti-biotic stress phenotype, i.e., inhibition of viral infection, whereas the other independent mutant bzip60-1 exhibited the same susceptible phenotype as wild type plants (Fig 3). Our experimental evidence revealed that both bzip60-1 and bzip60-2 mutants are non-RNA null mutants and produce an N-terminal truncated mRNA (bZIP60ΔN) and a C-terminal truncated mRNA (bZIP60ΔC), respectively (Figs 4, S6, S7 and S8A). Our data also showed that the bzip60-2 mutant is a splicing knockout mutant, whereas the bzip60-1 mutant is an expression knockdown mutant and bZIP60ΔN mRNA can undergo splicing (Figs 3, S7, S8A, S9 and S10). As bZIP60ΔN mRNAs contain an in-frame start codon 150 nt downstream of AUG of wild type bZIP60 ORF (Figs 4A and S7), the bZIP60ΔN S protein in the bzip60-1 mutant, like bZIP60 S, bears functional AD and BD domains as well as the intact C-terminus (Figs 9, 10 and S8A) [22,49]. Moreover, bZIP60 S and bZIP60ΔN S both contain two functional NLSs (Figs 5, 6B, S8A, S11 and S12B), i.e., NLS1 at their C-terminal regions, which is generated from the splicing-mediated frame-shift, and NLS3 located upstream of their BD domains (S8A Fig). Like bZIP60 S, bZIP60ΔN S self-interacts to homodimerize (Figs 6 and S12). Taken together, we concluded that bZIP60ΔN S executes the equivalent function as the wild type bZIP60 S. This conclusion is strengthened by two further lines of evidence obtained in complementation assays in plants under biotic stress and in yeast under abiotic stress (Figs 7, 10 and 11; see discussion below).
The bZIP60ΔN S-producing bzip60-1 mutant has long been considered as an RNA-null mutant to ascertain the behavior of the UPR, but many confusing findings have been generated. First, it has been reported that a low level of truncated bZIP60 mRNA is present in the bzip60-1 mutant [42,51] and the expression of several UPR marker genes is not significantly affected in this mutant [42]. Second, in response to ER stress, the expression of the UPR marker genes, including BiP1, BiP2, and BiP3, is up-regulated in the bzip60-1 mutant, like the wild type [15,52], and the bzip60-1 mutant can activate the ER stress response [53]. Third, under ER stress, the bzip60-1 mutant develops only a modest ER stress phenotype similar to the wild type controls, whereas the double mutant of IRE1A and IRE1B displays a marked stress injury [23,28]. Finally, the gametes bearing the triple mutation of IRE1A, IRE1B and bZIP28 are lethal, but the double bzip60-1 bzip28-2 mutant not [35]. Therefore, the finding of bZIP60ΔN S as a functional derivative of the full-length bZIP60 S presented in this work unambiguously helps to clarify these long-standing confusions.
As HAC1 is the splicing target of IRE1 in yeast, the identification of bZIP60 mRNA as the substrate of IRE1 in Arabidopsis raises the question if bZIP60 S and bZIP60ΔN S are the functional homolog of yeast HAC1p [15]. To answer this question, we first determined if bZIP60 protein derivatives localize to the nucleus of yeast. We found that, like yeast HAC1p U and HAC1p S, bZIP60 S and bZIP60ΔN S were predominantly concentrated in the nucleus in yeast under DTT treatment (Fig 9B). However, the bZIP60 S with mutated NLS3 and NLS1 accumulated in the cytoplasm and failed to enter the nucleus in plant cells and in yeast (Figs 5 and 9B), suggesting that the mechanisms of nuclear import used for bZIP proteins may be highly conserved across kingdoms. This notion is also in agreement with the finding that both bZIP60 and HAC1p carry a functional NLS (NLS3 for bZIP60) at the similar N-terminal region of their BD domains, and this NLS is highly conserved among bZIP60 homologs from at least 20 plant species (Figs 5, 9 and S11). It should be pointed out that bZIP60 U and bZIP60ΔN U were located in the cytoplasm in yeast, even though they contain a functional NLS (NLS3) (Figs 9B and S8). The exclusion of bZIP60 U and bZIP60ΔN U from the nucleus may be attributed to the TMD that anchors the bZIP60 U and bZIP60ΔN U proteins to the ER membrane in yeast (Figs 9B and S8), consistent with the findings obtained in Arabidopsis and tobacco suspension cells [15,52]. The removal of the small 23-bp intron of bZIP60 U and bZIP60ΔN U leads to a frame-shift that eliminates the TMD and acquires another functional NLS (NLS1), enabling bZIP60 S and bZIP60ΔN S to be targeted to the nucleus (S5 and S11 Figs) [15]. Therefore, we concluded that yeast IRE1 cannot splice bZIP60 U into bZIP60 S which is translated into the nucleus-targeting bZIP60 S. Given that the plant IRE1 could not splice the yeast HAC1 mRNA either in Arabidopsis protoplasts [26] or in yeast itself (S15 Fig), we speculate that it is the secondary structure difference between bZIP60 and HAC1 mRNAs that does not allow for splicing in the heterologous systems.
The complementation assay conducted in a Δhac1 yeast strain demonstrated that, like HAC1p, both bZIP60 S and bZIP60ΔN S, but not bZIP60 U nor bZIP60ΔN U, rescued the ER stress phenotype of HAC1-deficient strain (Fig 10B), indicating the two spliced forms are a functional homolog of yeast HACp in abiotic stress tolerance. As bZIP signature-bearing proteins, HAC1p acquires the C-terminal 18-aa AD after IRE1-mediated splicing [18], whereas bZIP60 has the N-terminal, splicing-independent AD (S8 Fig) [49]. In spite of this positional difference, the two transcriptional activation domains share high sequence similarity (Fig 10A). The identification of bZIP60 S and bZIP60ΔN S as a homolog of HAC1p was further corroborated by the yeast growth assay showing that, under normal condition, constitutive expression of either bZIP60 S or bZIP60ΔN S rather than their unspliced forms slowed down yeast growth (Fig 11), a phenotype resulting from the constitutive expression of HAC1p [12,50]. Based on these findings, we propose that although the sequences of the transcription factors, i.e., bZIP60 and HAC1p, and the IRE1-mediated splicing mechanisms have diverged from the last eukaryotic ancestor, the downstream ER stress signaling mechanisms have evolved to functionally converge at least in yeast and plants.
It should be mentioned that although bZIP60ΔC in the bzip60-2 mutant has all functional NLSs as well as AD and BD domains, it failed to rescue the Tm-sensitive phenotype of HAC1-deficient yeast and did not inhibit yeast growth under normal conditions (S8 and S14 Figs). This is most likely due to the fact that it lacks the intact C-terminus (aa 202–258) of the wild type bZIP60 S (S8 Fig). According to a previous report, a truncated bZIP60 without the large C-terminal region (aa 218–258) is not able to homodimerize (S8 Fig), although it can still heterodiemrize with a same truncated bZIP28 [23]. Therefore, the IRE1-mediated bZIP60 splicing not only leads to the elimination of the TMD and acquisition of a functionally redundant NLS, but also produces a new C-terminal sequence that is essential for bZIP60 S homodimerization to carry out transcriptional regulation. Alternatively, the C-terminal sequence, which is immediately proximal to the BD domain, might be important for the proper formation of the superimposing coiled-coil structure that binds targeted DNAs [54]. Based on these results and analyses, it is reasonable to conclude that bZIP60ΔC in the bzip60-2 mutant is non-functional.
IRE1-dependent bZIP60 Splicing Plays an Important Role in Viral Pathogenesis
As the primary UPR arm, the role of IRE1 and its mRNA substrate in viral infection has been intensively explored in mammalian cells [55,56]. In human hepatoma cells expressing hepatitis C virus (HCV) sub-genomic replicons, IRE1 is activated as indicated by the enhanced XBP1 S mRNA level, but the transcriptional regulation activity of XBP1 S is inhibited [56,57]. Similar to the case of HCV, infection with murine coronavirus mouse hepatitis virus also causes a progressive increase in XBP1 S mRNA with very little XBP1 S protein [58]. Therefore, the activation of IRE1 in parallel with the concomitant inhibition of XBP1 S (in either mRNA or protein level or both) has been viewed as an effective strategy utilized by mammalian viruses to cope with the IRE1-XBP1 branch-mediated antiviral responses [58,59].
In this work, we found that, in response to TuMV infection, the IRE1-bZIP60 arm of the UPR was activated in both locally and systemically infected leaves, indicated by the accumulation of bZIP60 S (Figs 1, S1 and S2). We also found that upon TuMV infection, the bzip60-1 mutant with up-regulated bZIP60ΔN S, albeit at a lower level, developed typical viral symptoms and allowed viruses to accumulate at the same level as the wild type (Fig 3). In contrast, the other independent mutant bzip60-2 without detectable bZIP60 S significantly inhibited the viral accumulation and remarkably suppressed the development of disease symptoms (Fig 3). We further provided genetic evidence that either bZIP60 S or bZIP60ΔN S could rescue the virus suppression phenotype of the bzip60-2 mutant (Fig 7). These data clearly indicated that the virus suppression phenotype in the bzip60-2 mutant arise from the loss-of-function of bZIP60 S. Consistently, two different double mutants of IRE1A and IRE1B, in which bZIP60 U splicing was blocked, displayed reduced levels of viral RNA accumulation and suppressed viral symptom development like the bzip60-2 mutant (Figs 3 and 8), suggesting that bZIP60 S, not bZIP60 U, plays a crucial function in favoring virus infection. The absence of bZIP60 S and suppression of virus infection in the double mutants of IRE1A and IRE1B were rescued by complementation with either IRE1A or IRE1B alone (Fig 8). Moreover, the virus suppression phenotype in the ire1a-3 ire1b-4 double mutant could also be recovered by the introduction of bZIP60 S or bZIP60ΔN (Fig 8). These results directly demonstrated that IRE1 and its processed bZIP60 S function as a linear pair in promoting virus infection (Figs 3, 7 and 8). To our knowledge, this is the first report showing that that the projected cognate system of IRE1 and bZIP60 behaves like a host factor in viral infection in plants. It should be mentioned that although IRE1 and bZIP60 were both involved as a matched system in viral pathogenesis (Figs 3, 7 and 8), neither the double mutant of IRE1A and IRE1B nor the bzip60-2 mutant could completely prevent viral infection (Figs 3 and 8). This result is in accordance with the established conception of the UPR functioning as a buffer or a homeostat to cope with diverse ER stresses [47,60,61].
Nevertheless, the role of IRE1 and bZIP60 S in virus infection in plants apparently contradicts with the function of their counterparts as an resistance mechanism to some viruses in mammalian cells and to a bacterial pathogen in plants [30,58,59]. This is probably due to the complexity of UPR signaling pathways and speciality of virus-host interactions. Indeed, several recent studies have shown that the UPR could be hijacked by virus to favor viral infection. In lung epithelial cell, influenza A virus activates the IRE1 pathway, with little or no concomitant activation of PERK and ATF6 pathways, and inhibition of IRE1 activity leads to reduced viral replication [62]. In N. benthamiana, silencing NtbZIP60 suppresses the expression of the UPR marker genes and reduces Potato virus X (PVX) accumulation [38,39]. It is possible that during viral infection, the IRE1-bZIP60 pathway-mediated UPR may aim to alleviate cytotoxicity by up-regulating ER molecular chaperons since membrane-associated virus replication or accumulation of large amounts of viral proteins can break the homeostatic cellular environment. This reasoning is supported by our observation that TuMV infection induced the accumulation of ER chaperones including BiP3, BiP1/2, CRT and PDI (S2 Fig) as well as by several earlier reports that virus infection up-regulated the expression of bZIP60 and ER marker genes in plants [63–65].
The increased expression of ER-resident chaperones may further facilitate virus infection through direct involvement in virus infection process [66]. In the single-celled yeast, the host Ssa1/2p molecular chaperone (yeast homologue of HSP70) is required for the assembly of the tombusvirus replicase to enhance viral RNA replication [67,68]. In plants, HSP70 induced by potyvirus infection is also regulated by the cytoplasmic UPR pathway [69,70]. As a component of a membrane-associated viral ribonucleoprotein complex, HSP70 has a role, together with its co-chaperone CPIP, in preventing the potyviral coat protein from interfering with viral gene expression [66], and the Hsp70-15-deficient mutant is more tolerant to virus infection [71]. In agreement with these findings, we also found that activation of the UPR in N. benthamiana through treatment with pharmacological small molecular chaperones did promote TuMV infection (S16 Fig). It is worth pointing out that although BiP2 was induced by TuMV infection, the bip2-2 mutant developed normal disease symptoms (S2 and S13 Figs). This is likely due to that molecular chaperones could complement each other or that bip2-2 might not be a clean knockout mutant [72].
In this work, we found that among 11 viral factors of TuMV, 6K2 significantly induced the splicing of NtbZIP60 mRNA in N. benthamiana (Fig 2B). The potyviral 6K2 protein is an integral membrane protein and elicits the formation of ER-derived virus replication factories at ER exit sites [40]. The finding presented here is consistent with previous reports that virus-encoded ER targeting proteins induce the UPR. For instance, among seven proteins encoded by simian virus 5, only the HN glycoprotein that is inserted into the ER is capable of stimulating UPR response [73]. This also holds true for the ER-resident proteins encoded by flaviviruses or retroviruses such as human immunodeficiency virus [57,74]. In Arabidopsis and N. benthamiana, the PVX viral movement protein TGBp3 that also resides in the ER is sufficient to elicit the UPR [39,75]. Therefore, the potyviral 6K2 protein is a new member in the group of virus-encoded ER-targeting and UPR-inducing proteins. However, the mechanism of 6K2 triggering the UPR is yet to be determined. In an attempt to test if 6K2 interacts with IRE1 to activate the UPR, we found no interactions between 6K2 and IRE1 (S17 Fig). It is possible that 6K2 induces the UPR through its physical interaction with the ER or subsequent ER remodelling. Elucidation of the molecular mechanisms by which the viral protein(s) triggers the UPR will certainly advance our understanding of the UPR itself as well as virus-host interactions in general.
IRE1 and Its Splicing Substrate As a Linear Pair Is Evolutionarily Conserved in Higher Eukaryotes
In yeast, Δhac1 and Δire1 mutants exhibit indistinguishable growth phenotypes and share highly correlated gene-expression profiles [7]. Search for additional mRNA substrates of IRE1p using three independent genome-scale methods did not identify any other mRNA except HAC1 mRNA [76]. Therefore, it is suggested that IRE1p and HAC1 mRNA have evolved as a matched enzyme-substrate pair to carry out the signal transduction between the ER and nucleus of the UPR [76]. In this report, we showed that IRE1 and bZIP60 mRNA as a cognate system to determine the viral pathogenesis in Arabidopsis (Figs 3, 7 and 8), unambiguously demonstrating that the ancient pair also plays an important role in biotic stress in plants.
Given that the IRE1p and HAC1 mRNA pathway functions in the simple one-celled eukaryotic organism of yeast as a linear pair during development or stress responses and the IRE1 and bZIP60 mRNA duet manipulates the virus-host interactions in plants, it is reasonable to propose that the coupling of IRE1 and its splicing substrate is evolutionarily conserved in higher eukaryotes. In Caenorhabditis elegans, deletion of either IRE1 or its splicing target XBP1 is synthetically lethal with deletion of either ATF-6 or PEK-1, due to a developmental arrest at larval stage 2 [77]. In mammalians, IRE1-/- or XBP1-/- mouse embryo perishes at a similar early stage of gestation (between days 9.5 and 11.5 of gestation), indicating the linear pair of IRE1 and XBP1 is essential for individual development [13,78]. The conserved property of the linear enzyme-substrate pair may be also reflected by the specificity and uniqueness of IRE1 splicing mRNA substrate. To date, bZIP60 mRNA is the only known substrate of IRE1A/IRE1B in Arabidopsis [22,30,35].
In view of the fundamental roles played by the IRE1p and HAC1 mRNA pair in yeast, we suggest that the corresponding counterpart pairs in other higher eukaryotes may function in diverse biological processes. However, the existence of functionally redundant genes and overlapping pathways hampers the identification and further elucidation of functions mediated by the IRE1-mRNA substrate pathway. In Arabidopsis, the pollen viability of the single UPR pathway mutants is similar to that of wild type plants, whereas the triple mutant ire1a-2 ire1b-4 bzip28-2 is lethal, caused by a severe defect in male gametophyte [35]. These observations suggest that the UPR pathways could functionally complement each other for plant development. Similar results have been observed in C. elegans, in which ATF-6 acts synergistically with PEK-1 to complement the developmental requirement for IRE1-XBP1 [77]. In this study, we proved that the virus suppression phenotype resulting from dysfunction of the IRE1-bZIP60 pathway is independent of the S1P/S2P-bZIP17/bZIP28 arm (S13 Fig), directly showing a single branch of the UPR determines a biological process. To our knowledge, this is the first evidence that the single UPR arm functions alone in plants.
It is worth stressing that IRE1-mediated UPR response in higher multicellular organisms is apparently more complex and diverse than that in yeast. In Arabidopsis, a double mutant of IRE1A and IRE1B showed a short-root phenotype [28], but both the bZIP60 slicing knockout mutant (bzip60-2) (S18 Fig) and the bZIP60 expression knockdown mutant (bzip60-1) [35] displayed normal root growth, suggesting that IRE1 may activate other cellular component(s) to execute the regulatory function of IRE1 in root growth. In mammalian cells, IRE1 not only splices XBP1 mRNA but also activates Jun N-terminal kinase, a serine-directed protein kinase, in response to ER stress in embryonic fibroblasts, and directly interacts with pro-apoptotic factors, such as BAX and BAK, to contribute to apoptosis in ER-stressed cells [78,79]. Most recently, IRE1 has been shown to selectively degrade microRNAs (miRs -17, -34a, -96, and -125b) [80]. These findings suggest that IRE1 may regulate the UPR signaling through protein–protein interaction, mRNA splicing, microRNA degradation and other unknown mechanisms in a multicellular context, even in the single-celled yeast [34,81].
Materials and Methods
Plant Materials
Arabidopsis thaliana used in this study is in Columbia-0 (Col-0) background, with the exception of ire1a-3 in Col background as well as of bzip60-2 and ire1b-4 in Col-3 qrt background. The mutants bzip60-1 (SALK_050203), bzip60-2 (SAIL_283_B03), bzip17 (SALK_104326), bzip28-2 (SALK_132285), bip2-2 (SALK_047956), ire1a-2 (SALK_018112) and ire1b-4 (SAIL_238_F07) were obtained from the Arabidopsis Biological Resource Center. The mutant ire1a-3 (WiscDsLox420D09) and two different double mutants (ire1a-3 ire1b-4 and ire1a-2 ireb-4) were described previously [28,30].
Unless stated otherwise, all Arabidopsis plants were grown in a growth chamber with a 14 h photoperiod (100 μmol photons m-2 s-1) and a relative humidity of 75% at 23/21°C (light/dark). The N. benthamiana plants were grown in a growth room with a 16 h photoperiod (80–100 μmol photons m-2 s-1) and a relative humidity of 75% at 22°C.
Identification of Homozygous Mutants
The homozygous line containing the T-DNA insertion in the gene of interest was screened essentially as described previously [82]. The genotyping primers were listed in S1 Table.
Virus Infection Assay
TuMV was introduced into plants via either agro-infiltration or mechanical inoculation according to a previous report [82]. For mechanical inoculation, TuMV-infected N. benthamiana leaves were used as an inoculum.
RNA Extraction, PCR, RT-PCR and Real-Time RT-PCR
RNA extraction, PCR, RT-PCR and real-time RT-PCR analyses were performed essentially as described previously [28,82]. The primer sets used in this study were listed in S1 Table. The RNAs from yeast CRY1 treated with or without 2 mM DTT were exacted using NucleoSpin RNA II (Clontech) to obtain cDNAs of IRE1, HAC1 U and HAC1 S.
Entry Vector Construction
Unless stated otherwise, Phusion High-Fidelity DNA Polymerase (NEB, USA) was used to amplify all DNA sequences using the primer sets listed in S1 Table, and Gateway technology (Invitrogen, USA) was employed to generate plasmids. Coding sequences of HAC1p U, HAC1p S and IRE1p were amplified using yeast cDNA as described above. Coding sequences of bZIP60 U, bZIP60 S, bZIP60ΔN U, bZIP60ΔN S, bZIP60ΔC1, bZIP60ΔC2, IRE1A and IRE1B were amplified using Arabidopsis cDNA (cDNA from DTT-treated seedlings was used for amplification of bZIP60 S and bZIP60ΔN S, whereas cDNA from bzip60-2 seedlings for amplification of bZIP60ΔC2). Coding regions of P1, HC-Pro, P3, 6K1, CI, 6K2, NIaVPg, NIaPro, NIb and CP of TuMV were amplified from the TuMV infectious clone [40]. With the exception of pENTRTM 1A Dual Selection vector (A10462, Invitrogen) used for IRE1B, all amplified coding sequences were recombined into pDONR221 via the BP reaction (Invitrogen, USA). The entry vector containing P3N-PIPO was described in our previous work [40]. To highlight the nucleus and to produce donor - and acceptor-only samples (used in FRET assays), constructs bearing 35S::NLS-CFP and 35S::NLS-YFP were created following the BP and LR reactions using the primers listed in S1 Table.
Transient Expression of Viral Factors in N. benthamiana
The linearized products of the entry vectors containing virus single factor sequences were recombined into the binary destination vector pEarleygate103 for expression of fusion proteins containing viral factors-GFP or into the vector pMDC43 for expression of GFP-viral factor fusions. The resulting binary destination vectors were electroporated into Agrobacterium tumefaciens (GV3101). The GV3101 cells harbouring relevant expression constructs were re-suspended with the infiltration buffer to OD600 = 0.3, and then infiltrated into the leaves of 3-week-old N. benthamiana seedling. At 2.5 dpi, the transient expression of each construct was verified under an inverted confocal microscope (TCS SP2, Leica, Germany) by observing GFP at an excitation wavelength of 488 nm and an emission 510–550 nm, and the agroinfiltrated leaves were harvested for RNA extraction. The experiments were repeated three times, and each treatment contains at least three independent plants.
Mutation of NLSs in bZIP60 S and Sub-cellular Localization
The mutation of the putative NLSs of bZIP60 S was conducted on the entry vector bearing the cDNA of bZIP60 S using the primers listed in S1 Table, based on the QuikChange Lightning Site-Directed Mutagenesis Kit (210519, Agilent). The mutated vectors were recombined into the pEarlyGate104 vector via the LR reaction (Invitrogen, USA), to generate constructs with YFP fused to the N-terminus of bZIP60 S under the control of the CaMV 35S promoter (35S::YFP-bZIP60 S with mutated NLSs). 3-week-old N. benthamiana leaves were co-transformed via agroinfiltration to express these YFP-fusion proteins as well the nucleus indicator (35S::NLS-CFP) and a ER marker KDEL-MCherry [40]. Two days after transformation, their subcellular localizations were observed under the confocal microscope using a sequential scanning model. Three band-pass (BP) filters (BP 465–520 nm, BP 565–585 nm, and BP 590–630 nm) were used for CFP, YFP and MCherry signal collection, which were excited at 458 nm, 514 nm, and 543 nm respectively.
Y2H Assay
The entry vectors with cDNAs of bZIP60 S and bZIP60ΔN S were recombined into pGBKT7-GW (bait) and pGADT7-GW (prey) vectors using the LR reaction (Invitrogen, USA). Sets of constructs were co-transformed into Y2H Gold yeast strain (Clontech). The AD-T and BD-53 combination was utilized as positive control, whereas the AD-T and BD-Lam set as well as the empty pGBKT7 (BD) and pGADT7 (AD) were used as negative controls. Yeast transformants were selected on synthetic minimal double dropout medium deficient in TRP and LEU (DDO). Protein interactions were assessed on quadruple dropout medium deficient in HIS, TRP, LEU and adenine (QDO) in the presence of different concentrations of aureobasidin A (AbA).
FRET Assay
To test homodimerization of bZIP60 S or bZIP60ΔN S in living cells, the cDNAs of bZIP60 S and bZIP60ΔN S were cloned into pEarlyGate101 and pEarlyGate102 to generate YFP - and CFP-fusion proteins, respectively (Invitrogen, USA). 3-week-old N. benthamiana leaves were co-transformed with the indicated sets of constructs. Two days after transformation, sensitized emission FRET was determined under an inverted confocal microscope (TCS SP2, Leica, Germany). Images in donor (excitation 458 nm; emission 465 to 505 nm), acceptor (excitation 514 nm; emission 525 to 600 nm), and FRET (emission 525 to 600 nm) channels were captured. For acceptor photo-bleaching FRET, the fluorescence of the CFP and YFP channels was scanned as for sensitized emission FRET before and after photo-bleaching. Bleaching of the acceptor fluorescence signal was performed using a 514-nm beam at maximum intensity for 10 frames. The energy transfer efficiency between the paired proteins was quantified according to the change in fluorescence intensity of the acceptor and the donor before and after photo-bleaching.
Complementation Test in Arabidopsis
To create complementation constructs, we cloned the bZIP60 promoter containing a 3356 bp region immediately upstream of the ATG into the pMDC43 Gateway vector-substituting 2 × 35 S promoter to generate pMDC43-pbZIP60 destination vector using bZIP60HindIII-F and bZIP60KpnI-R primers (S1 Table). The entry vectors bearing the cDNAs of bZIP60 S and bZIP60ΔN S were recombined into the pMDC43-pbZIP60 destination vector via the LR reaction, to generate the vectors pbZIP60-bZIP60 S and pbZIP60-bZIP60ΔN S. The resulting constructs were introduced into GV3101 by electroporation. The bzip60-2 mutant and ire1a-3 ire1b-4 double mutant plants were transformed by the floral-dip method [83], and transformants were selected on solid half-strength MS medium supplemented with hygromycin (20 μg/mL) and confirmed by RT-PCR. The resulting homozygous transgenic lines (T2 generation) were used for phenotypic analyses.
For complementation assays by IRE1A or IRE1B, transgenic lines were made previously [28].
Complementation Test in Yeast
To observe the subcellular localization of HAC1p and bZIP60 and to test their functional complementation, the relevant entry vectors were recombined via the LR reaction with a Gateway destination vector pAG423GAL-EYFP-ccdB (Plasmid 14341, Addgene) or a CEN-ARS Gateway destination vector pAG416GAL-ccdB-HA (Plasmid 14243, Addgene). The resulting destination vectors and the empty vectors were transferred into the CRY1 Δhac1::TRP strains with the Quick & Easy Yeast Transformation Mix (631851, Clontech). The CRY1 Δhac1::TRP yeast cells transformed with appropriate vectors were grown at 28°C in synthetic media lacking TRP and HIS and containing 2% raffinose (2 x SRaf-TRP-HIS). At exponential growth, the yeast cultures were spun down, washed and re-suspended to an OD600 = 0.3 with synthetic media lacking TRP and HIS and containing 2% galactose (2 x SGal-TRP-HIS) to induce the expression of the fusion constructs at 28°C. After 8 h induction, the yeast cells were then incubated with 2 μg/mL DAPI for 1 h and processed for microscopy to visualize nuclei. DAPI signal were visualized with excitation at 405 nm and emission at 450–500 nm, and YFP signal was captured in another detection channel using a 514 nm excitation light and a 525–550 nm band-pass filter.
For functional complementation assays, the CRY1 Δhac1::TRP cells transformed with appropriate vectors were grown overnight to mid-log phase at 28°C in synthetic media lacking TRP and URA containing 2% glucose (2 x SGlu-TRP-URA). The yeast cells were then spun down, washed, and cultured in 2 x SRaf-TRP-URA media for 8 h to relief the glucose repression of Gal1 promoter. The cells were pelleted, washed and re-suspended to an OD600 = 0.3 in 2 x SGal-TRP-URA media or in 2 x SRaf-TRP-URA media (as controls) to induce the expression of HAC1p and bZIP60. At 5 h and 10 h post-induction, the cell density was measured by a spectrophotometer (SmartSpecTm plus, Bio-Rad) to determine yeast growth. At 10 h post-induction, the induced and non-induced yeast cells were pelleted, washed and diluted to an OD600 = 1.0 with sterile water. 5-fold serial dilutions of the cells were spotted on the 2 x SGal-TRP-URA plates in the presence of 0.1% DMSO or 0.2 μg/mL Tm and incubated for 48 h at 30°C. The non-induced cells were also spotted on 2 x SRaf-TRP-URA plates in the presence of 0.1% DMSO.
Sequence Analyses
Multiple sequence alignment was generated by ClustalW [84]. Domain and NLS motif were predicted using SMART (http://smart.embl-heidelberg.de/) and NucPred (http://www.sbc.su.se/~maccallr/nucpred/), respectively. Figures were created by SigmaPlot 12.5.
Accession Numbers
Gene sequences used in this study can be retrieved under the following accession numbers: AT1G42990 (bZIP60), AT2G40950 (bZIP17), AT3G10800 (bZIP28), AT2G17520 (IRE1A), AT5G24360 (IRE1B), AT5G42020 (BiP2), AT1G09080 (BiP3), AT1G21750 (PDI), AT1G56340 (CRT), AT3G18780 (Actin II), AB281271 (NtbZIP60), AJ236016 (Nt18s RNA), D26506 (HAC1), NM_001179209 (IRE1) and EF028235 (TuMV).
Supporting Information
Zdroje
1. Chen Y, Brandizzi F (2013) IRE1: ER stress sensor and cell fate executor. Trends Cell Biol 23 : 547–555. doi: 10.1016/j.tcb.2013.06.005 23880584
2. Hetz C (2012) The unfolded protein response: controlling cell fate decisions under ER stress and beyond. Nat Rev Mol Cell Biol 13 : 89–102. doi: 10.1038/nrm3270 22251901
3. Walter P, Ron D (2011) The unfolded protein response: from stress pathway to homeostatic regulation. Science 334 : 1081–1086. doi: 10.1126/science.1209038 22116877
4. Smith JA (2014) A new paradigm: innate immune sensing of viruses via the unfolded protein response. Front Microbiol 5 : 222. doi: 10.3389/fmicb.2014.00222 24904537
5. Williams B, Verchot J, Dickman M (2014) When supply does not meet demand-ER stress and plant programmed cell death. Front Plant Sci 5 : 211. doi: 10.3389/fpls.2014.00211 24926295
6. Wang J, Pareja KA, Kaiser CA, Sevier CS (2014) Redox signaling via the molecular chaperone BiP protects cells against endoplasmic reticulum-derived oxidative stress. eLIFE 3: e03496. doi: 10.7554/eLife.03496 25053742
7. Travers KJ, Patil CK, Wodicka L, Lockhart DJ, Weissman JS, et al. (2000) Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell 101 : 249–258.
8. Li H, Korennykh AV, Behrman SL, Walter P (2010) Mammalian endoplasmic reticulum stress sensor IRE1 signals by dynamic clustering. Proc Natl Acad Sci USA 107 : 16113–16118. doi: 10.1073/pnas.1010580107 20798350
9. Shamu CE, Walter P (1996) Oligomerization and phosphorylation of the Ire1p kinase during intracellular signaling from the endoplasmic reticulum to the nucleus. EMBO J 15 : 3028–3039. 8670804
10. Gardner BM, Walter P (2011) Unfolded proteins are Ire1-activating ligands that directly induce the unfolded protein response. Science 333 : 1891–1894.
11. Lee KP, Dey M, Neculai D, Cao C, Dever TE, et al. (2008) Structure of the dual enzyme Ire1 reveals the basis for catalysis and regulation in nonconventional RNA splicing. Cell 132 : 89–100. doi: 10.1016/j.cell.2007.10.057 18191223
12. Cox JS, Walter P (1996) A novel mechanism for regulating activity of a transcription factor that controls the unfolded protein response. Cell 87 : 391–404.
13. Calfon M, Zeng H, Urano F, Till JH, Hubbard SR, et al. (2002) IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP1 mRNA. Nature 415 : 92–96. 11780124
14. Uemura A, Oku M, Mori K, Yoshida H (2009) Unconventional splicing of XBP1 mRNA occurs in the cytoplasm during the mammalian unfolded protein response. J Cell Sci 122 : 2877–2886. doi: 10.1242/jcs.040584 19622636
15. Deng Y, Humbert S, Liu JX, Srivastava R, Rothstein SJ, et al. (2011) Heat induces the splicing by IRE1 of a mRNA encoding a transcription factor involved in the unfolded protein response in Arabidopsis. Proc Natl Acad Sci USA 108 : 7247–7252. doi: 10.1073/pnas.1102117108 21482766
16. Zhang L, Wang A (2012) Virus-induced ER stress and the unfolded protein response. Front Plant Sci 3 : 293. doi: 10.3389/fpls.2012.00293 23293645
17. Hetz C, Martinon F, Rodriguez D, Glimcher LH (2011) The unfolded protein response: integrating stress signals through the stress sensor IRE1α. Physiol Rev 91 : 1219–1243. doi: 10.1152/physrev.00001.2011 22013210
18. Mori K, Ogawa N, Kawahara T, Yanagi H, Yura T (2000) mRNA splicing-mediated C-terminal replacement of transcription factor Hac1p is required for efficient activation of the unfolded protein response. Proc Natl Acad Sci USA 97 : 4660–4665. 10781071
19. Yoshida H, Oku M, Suzuki M, Mori K (2006) pXBP1 (U) encoded in XBP1 pre-mRNA negatively regulates unfolded protein response activator pXBP1 (S) in mammalian ER stress response. J Cell Biol 172 : 565–575.
20. Rüegsegger U, Leber JH, Walter P (2001) Block of HAC1 mRNA translation by long-range base pairing is released by cytoplasmic splicing upon induction of the unfolded protein response. Cell 107 : 103–114.
21. Lin JH, Li H, Yasumura D, Cohen HR, Zhang C, et al. (2007) IRE1 signaling affects cell fate during the unfolded protein response. Science 318 : 944–949. 17991856
22. Nagashima Y, Mishiba K-i, Suzuki E, Shimada Y, Iwata Y, et al. (2011) Arabidopsis IRE1 catalyses unconventional splicing of bZIP60 mRNA to produce the active transcription factor. Sci Rep 1 : 29.
23. Liu JX, Howell SH (2010) bZIP28 and NF-Y transcription factors are activated by ER stress and assemble into a transcriptional complex to regulate stress response genes in Arabidopsis. Plant Cell 22 : 782–796. doi: 10.1105/tpc.109.072173 20207753
24. Gao H, Brandizzi F, Benning C, Larkin RM (2008) A membrane-tethered transcription factor defines a branch of the heat stress response in Arabidopsis thaliana. Proc Natl Acad Sci USA 105 : 16398–16403. doi: 10.1073/pnas.0808463105 18849477
25. Liu JX, Srivastava R, Che P, Howell SH (2007) Salt stress responses in Arabidopsis utilize a signal transduction pathway related to endoplasmic reticulum stress signaling. Plant J 51 : 897–909.
26. Noh S-J, Kwon CS, Chung W (2002) Characterization of two homologs of Ire1p, a kinase/endoribonuclease in yeast, in Arabidopsis thaliana. Biochim Biophys Acta (BBA)-Gene Structure and Expression 1575 : 130–134. 12020828
27. Koizumi N, Martinez IM, Kimata Y, Kohno K, Sano H, et al. (2001) Molecular characterization of two Arabidopsis Ire1 homologs, endoplasmic reticulum-located transmembrane protein kinases. Plant Physiol 127 : 949–962. 11706177
28. Chen Y, Brandizzi F (2012) AtIRE1A/AtIRE1B and AGB1 independently control two essential unfolded protein response pathways in Arabidopsis. Plant J 69 : 266–277. doi: 10.1111/j.1365-313X.2011.04788.x 21914012
29. Deng Y, Srivastava R, Howell SH (2013) Endoplasmic reticulum (ER) stress response and its physiological roles in plants. Int J Mol Sci 14 : 8188–8212. doi: 10.3390/ijms14048188 23591838
30. Moreno AA, Mukhtar MS, Blanco F, Boatwright JL, Moreno I, et al. (2012) IRE1/bZIP60-mediated unfolded protein response plays distinct roles in plant immunity and abiotic stress responses. PLoS ONE 7: e31944. doi: 10.1371/journal.pone.0031944 22359644
31. Oikawa D, Tokuda M, Hosoda A, Iwawaki T (2010) Identification of a consensus element recognized and cleaved by IRE1α. Nucleic Acids Res 38 : 6265–6273. doi: 10.1093/nar/gkq452 20507909
32. Gonzalez TN, Sidrauski C, Dörfler S, Walter P (1999) Mechanism of non-spliceosomal mRNA splicing in the unfolded protein response pathway. EMBO J 18 : 3119–3132. 10357823
33. Howell SH (2013) Endoplasmic reticulum stress responses in plants. Annu Rev Plant Biol 64 : 477–499. doi: 10.1146/annurev-arplant-050312-120053 23330794
34. Kimmig P, Diaz M, Zheng J, Williams CC, Lang A, et al. (2012) The unfolded protein response in fission yeast modulates stability of select mRNAs to maintain protein homeostasis. eLIFE 1: e00048. doi: 10.7554/eLife.00048 23066505
35. Deng Y, Srivastava R, Howell SH (2013) Protein kinase and ribonuclease domains of IRE1 confer stress tolerance, vegetative growth, and reproductive development in Arabidopsis. Proc Natl Acad Sci USA 110 : 19633–19638. doi: 10.1073/pnas.1314749110 24145452
36. Fujita M, Mizukado S, Fujita Y, Ichikawa T, Nakazawa M, et al. (2007) Identification of stress-tolerance-related transcription-factor genes via mini-scale Full-length cDNA Over-eXpressor (FOX) gene hunting system. Biochem Bioph Res Co 364 : 250–257.
37. Liu JX, Srivastava R, Che P, Howell SH (2007) An endoplasmic reticulum stress response in Arabidopsis Is mediated by proteolytic processing and nuclear relocation of a membrane-associated transcription factor, bZIP28. Plant Cell 19 : 4111–4119. 18156219
38. Ye CM, Chen S, Payton M, Dickman MB, Verchot J (2013) TGBp3 triggers the unfolded protein response and SKP1-dependent programmed cell death. Mol Plant Pathol 14 : 241–255. doi: 10.1111/mpp.12000 23458484
39. Ye C, Dickman MB, Whitham SA, Payton M, Verchot J (2011) The unfolded protein response is triggered by a plant viral movement protein. Plant Physiol 156 : 741–755. doi: 10.1104/pp.111.174110 21474436
40. Wei T, Zhang C, Hong J, Xiong R, Kasschau KD, et al. (2010) Formation of complexes at plasmodesmata for potyvirus intercellular movement is mediated by the viral protein P3N-PIPO. PLoS Pathog 6: e1000962.
41. Zuker M (2003) Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31 : 3406–3415. 12824337
42. Lu DP, Christopher DA (2008) Endoplasmic reticulum stress activates the expression of a sub-group of protein disulfide isomerase genes and AtbZIP60 modulates the response in Arabidopsis thaliana. Mol Genet Genomics 280 : 199–210. doi: 10.1007/s00438-008-0356-z 18574595
43. Francis KE, Lam SY, Copenhaver GP (2006) Separation of Arabidopsis pollen tetrads is regulated by QUARTET1, a pectin methylesterase gene. Plant Physiol 142 : 1004–1013. 16980565
44. Brameier M, Krings A, MacCallum RM (2007) NucPred—predicting nuclear localization of proteins. Bioinformatics 23 : 1159–1160. 17332022
45. Fontes MR, Teh T, Kobe B (2000) Structural basis of recognition of monopartite and bipartite nuclear localization sequences by mammalian importin-α. J Mol Biol 297 : 1183–1194. 10764582
46. Vinson C, Myakishev M, Acharya A, Mir AA, Moll JR, et al. (2002) Classification of human B-ZIP proteins based on dimerization properties. Mol Cell Biol 22 : 6321–6335. 12192032
47. Pincus D, Chevalier MW, Aragón T, Van Anken E, Vidal SE, et al. (2010) BiP binding to the ER-stress sensor Ire1 tunes the homeostatic behavior of the unfolded protein response. PLoS Biol 8: e1000415. doi: 10.1371/journal.pbio.1000415 20625545
48. Pal B, Chan NC, Helfenbaum L, Tan K, Tansey WP, et al. (2007) SCFCdc4-mediated degradation of the Hac1p transcription factor regulates the unfolded protein response in Saccharomyces cerevisiae. Mol Biol Cell 18 : 426–440. 17108329
49. Iwata Y, Yoneda M, Yanagawa Y, Koizumi N (2009) Characteristics of the nuclear form of the Arabidopsis transcription factor AtbZIP60 during the endoplasmic reticulum stress response. Biosci Biotech Bioch 73 : 865–869.
50. Valkonen M, Penttilä M, Saloheimo M (2003) Effects of inactivation and constitutive expression of the unfolded-protein response pathway on protein production in the yeast Saccharomyces cerevisiae. Appl Environ Microbiol 69 : 2065–2072. 12676684
51. Humbert S, Zhong S, Deng Y, Howell SH, Rothstein SJ (2012) Alteration of the bZIP60/IRE1 pathway affects plant response to ER stress in Arabidopsis thaliana. PloS one 7: e39023. doi: 10.1371/journal.pone.0039023 22701744
52. Iwata Y, Fedoroff NV, Koizumi N (2008) Arabidopsis bZIP60 is a proteolysis-activated transcription factor involved in the endoplasmic reticulum stress response. Plant Cell 20 : 3107–3121. doi: 10.1105/tpc.108.061002 19017746
53. Tajima H, Iwata Y, Iwano M, Takayama S, Koizumi N (2008) Identification of an Arabidopsis transmembrane bZIP transcription factor involved in the endoplasmic reticulum stress response. Biochem Biophys Res Commun 374 : 242–247. doi: 10.1016/j.bbrc.2008.07.021 18634751
54. Jakoby M, Weisshaar B, Dröge-Laser W, Vicente-Carbajosa J, Tiedemann J, et al. (2002) bZIP transcription factors in Arabidopsis. Trends Plant Sci 7 : 106–111. 11906833
55. Netherton CL, Parsley JC, Wileman T (2004) African swine fever virus inhibits induction of the stress-induced proapoptotic transcription factor CHOP/GADD153. J Virol 78 : 10825–10828. 15367650
56. Tardif KD, Waris G, Siddiqui A (2005) Hepatitis C virus, ER stress, and oxidative stress. Trends Microbiol 13 : 159–163. 15817385
57. Tardif KD, Mori K, Kaufman RJ, Siddiqui A (2004) Hepatitis C virus suppresses the IRE1-XBP1 pathway of the unfolded protein response. J Biol Chem 279 : 17158–17164. 14960590
58. Bechill J, Chen Z, Brewer JW, Baker SC (2008) Coronavirus infection modulates the unfolded protein response and mediates sustained translational repression. J Virol 82 : 4492–4501. doi: 10.1128/JVI.00017-08 18305036
59. Medigeshi GR, Lancaster AM, Hirsch AJ, Briese T, Lipkin WI, et al. (2007) West Nile virus infection activates the unfolded protein response, leading to CHOP induction and apoptosis. J Virol 81 : 10849–10860. 17686866
60. DeSalvo J, Kuznetsov JN, Du J, Leclerc GM, Leclerc GJ, et al. (2012) Inhibition of Akt potentiates 2-DG–Induced apoptosis via downregulation of UPR in acute lymphoblastic leukemia. Mol Cancer Res 10 : 969–978. doi: 10.1158/1541-7786.MCR-12-0125 22692960
61. Adolph TE, Tomczak MF, Niederreiter L, Ko H-J, Böck J, et al. (2013) Paneth cells as a site of origin for intestinal inflammation. Nature 503 : 272–276. doi: 10.1038/nature12599 24089213
62. Hassan IH, Zhang MS, Powers LS, Shao JQ, Baltrusaitis J, et al. (2012) Influenza A viral replication is blocked by inhibition of the inositol-requiring enzyme 1 (IRE1) stress pathway. J Biol Chem 287 : 4679–4689. doi: 10.1074/jbc.M111.284695 22194594
63. Yang C, Guo R, Jie F, Nettleton D, Peng J, et al. (2007) Spatial analysis of Arabidopsis thaliana gene expression in response to Turnip mosaic virus infection. Mol Plant Microbe Interact 20 : 358–370. 17427806
64. García-Marcos A, Pacheco R, Martiáñez J, González-Jara P, Díaz-Ruíz JR, et al. (2009) Transcriptional changes and oxidative stress associated with the synergistic interaction between Potato virus X and Potato virus Y and their relationship with symptom expression. Mol Plant Microbe Interact 22 : 1431–1444. doi: 10.1094/MPMI-22-11-1431 19810812
65. Whitham SA, Quan S, Chang HS, Cooper B, Estes B, et al. (2003) Diverse RNA viruses elicit the expression of common sets of genes in susceptible Arabidopsis thaliana plants. Plant J 33 : 271–283.
66. Hafrén A, Hofius D, Rönnholm G, Sonnewald U, Mäkinen K (2010) HSP70 and its cochaperone CPIP promote potyvirus infection in Nicotiana benthamiana by regulating viral coat protein functions. Plant Cell 22 : 523–535. doi: 10.1105/tpc.109.072413 20154150
67. Pogany J, Stork J, Li Z, Nagy PD (2008) In vitro assembly of the Tomato bushy stunt virus replicase requires the host Heat shock protein 70. Proc Natl Acad Sci USA 105 : 19956–19961. doi: 10.1073/pnas.0810851105 19060219
68. Serva S, Nagy PD (2006) Proteomics analysis of the tombusvirus replicase: Hsp70 molecular chaperone is associated with the replicase and enhances viral RNA replication. J Virol 80 : 2162–2169. 16474124
69. Sugio A, Dreos R, Aparicio F, Maule AJ (2009) The cytosolic protein response as a subcomponent of the wider heat shock response in Arabidopsis. Plant Cell 21 : 642–654.
70. Aparicio F, Thomas CL, Lederer C, Niu Y, Wang D, et al. (2005) Virus induction of heat shock protein 70 reflects a general response to protein accumulation in the plant cytosol. Plant Physiol 138 : 529–536. 15805473
71. Jungkunz I, Link K, Vogel F, Voll LM, Sonnewald S, et al. (2011) AtHsp70-15-deficient Arabidopsis plants are characterized by reduced growth, a constitutive cytosolic protein response and enhanced resistance to TuMV. Plant J 66 : 983–995. doi: 10.1111/j.1365-313X.2011.04558.x 21418353
72. Maruyama D, Endo T, Nishikawa S-i (2010) BiP-mediated polar nuclei fusion is essential for the regulation of endosperm nuclei proliferation in Arabidopsis thaliana. Proc Natl Acad Sci USA 107 : 1684–1689. doi: 10.1073/pnas.0905795107 20080634
73. Watowich SS, Morimoto RI, Lamb RA (1991) Flux of the paramyxovirus hemagglutinin-neuraminidase glycoprotein through the endoplasmic reticulum activates transcription of the GRP78-BiP gene. J Virol 65 : 3590–3597. 2041085
74. Sung SC, Chao CY, Jeng KS, Yang JY, Lai M (2009) The 8ab protein of SARS-CoV is a luminal ER membrane-associated protein and induces the activation of ATF6. Virology 387 : 402–413. doi: 10.1016/j.virol.2009.02.021 19304306
75. Bamunusinghe D, Hemenway CL, Nelson RS, Sanderfoot AA, Ye CM, et al. (2009) Analysis of potato virus X replicase and TGBp3 subcellular locations. Virology 393 : 272–285. doi: 10.1016/j.virol.2009.08.002 19729179
76. Niwa M, Patil CK, DeRisi J, Walter P (2005) Genome-scale approaches for discovering novel nonconventional splicing substrates of the Ire1 nuclease. Genome Biol 6: R3.
77. Shen X, Ellis RE, Sakaki K, Kaufman RJ (2005) Genetic interactions due to constitutive and inducible gene regulation mediated by the unfolded protein response in C. elegans. PLoS Genet 1: e37. 16184190
78. Urano F, Wang XZ, Bertolotti A, Zhang Y, Chung P, et al. (2000) Coupling of stress in the ER to activation of JNK protein kinases by transmembrane protein kinase IRE1. Science 287 : 664–666. 10650002
79. Hetz C, Bernasconi P, Fisher J, Lee AH, Bassik MC, et al. (2006) Proapoptotic BAX and BAK modulate the unfolded protein response by a direct interaction with IRE1α. Science 312 : 572–576. 16645094
80. Upton J-P, Wang L, Han D, Wang ES, Huskey NE, et al. (2012) IRE1α cleaves select microRNAs during ER stress to derepress translation of proapoptotic caspase-2. Science 338 : 818–822. doi: 10.1126/science.1226191 23042294
81. Cross BC, Ron D (2012) New twists in the unfolded protein response. eLIFE 1: e00243. doi: 10.7554/eLife.00243 23066509
82. Huang TS, Wei T, Laliberte JF, Wang A (2010) A host RNA helicase-like protein, AtRH8, interacts with the potyviral genome-linked protein, VPg, associates with the virus accumulation complex, and is essential for infection. Plant Physiol 152 : 255–266. doi: 10.1104/pp.109.147983 19880609
83. Clough SJ, Bent AF (1998) Floral dip: a simplified method forAgrobacterium-mediated transformation ofArabidopsis thaliana. Plant J 16 : 735–743. 10069079
84. Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ (2009) Jalview Version 2-a multiple sequence alignment editor and analysis workbench. Bioinformatics 25 : 1189–1191. doi: 10.1093/bioinformatics/btp033 19151095
Štítky
Genetika Reprodukční medicína
Článek Retraction: Astakine 2—the Dark Knight Linking Melatonin to Circadian Regulation in CrustaceansČlánek Adventures in WonderlandČlánek Genomic Location of the Major Ribosomal Protein Gene Locus Determines Global Growth and InfectivityČlánek Spatial Fluctuations in Expression of the Heterocyst Differentiation Regulatory Gene in FilamentsČlánek Genome-Wide Negative Feedback Drives Transgenerational DNA Methylation Dynamics in ArabidopsisČlánek Systematic Dissection of the Sequence Determinants of Gene 3’ End Mediated Expression ControlČlánek The Chromatin Remodeler CHD8 Is Required for Activation of Progesterone Receptor-Dependent EnhancersČlánek Selection against Heteroplasmy Explains the Evolution of Uniparental Inheritance of MitochondriaČlánek The DNA Helicase Recql4 Is Required for Normal Osteoblast Expansion and Osteosarcoma FormationČlánek Dual-Specificity Anti-sigma Factor Reinforces Control of Cell-Type Specific Gene Expression in
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2015 Číslo 4
-
Všechny články tohoto čísla
- Retraction: Astakine 2—the Dark Knight Linking Melatonin to Circadian Regulation in Crustaceans
- Adventures in Wonderland
- Experimental Swap of 's Assortative Mating Preferences Demonstrates Key Role of X-Chromosome Divergence Island in Incipient Sympatric Speciation
- Chromosome Replacement and Deletion Lead to Clonal Polymorphism of Berry Color in Grapevine
- The Protein Quality Control Machinery Regulates Its Misassembled Proteasome Subunits
- Genome-Wide Association Study Identifies as a Critical Gene for Susceptibility to Noise-Induced Hearing Loss
- Genomic Location of the Major Ribosomal Protein Gene Locus Determines Global Growth and Infectivity
- Viable Neuronopathic Gaucher Disease Model in Medaka () Displays Axonal Accumulation of Alpha-Synuclein
- Multi-locus Analysis of Genomic Time Series Data from Experimental Evolution
- The Genetic Legacy of the Expansion of Turkic-Speaking Nomads across Eurasia
- Lack of GDAP1 Induces Neuronal Calcium and Mitochondrial Defects in a Knockout Mouse Model of Charcot-Marie-Tooth Neuropathy
- The Pif1 Helicase, a Negative Regulator of Telomerase, Acts Preferentially at Long Telomeres
- Inhibiting K63 Polyubiquitination Abolishes No-Go Type Stalled Translation Surveillance in
- SYD-1C, UNC-40 (DCC) and SAX-3 (Robo) Function Interdependently to Promote Axon Guidance by Regulating the MIG-2 GTPase
- Spatial Fluctuations in Expression of the Heterocyst Differentiation Regulatory Gene in Filaments
- Synergistic and Independent Actions of Multiple Terminal Nucleotidyl Transferases in the 3’ Tailing of Small RNAs in Arabidopsis
- Host Genetic Variation Influences Gene Expression Response to Rhinovirus Infection
- Contribution of Large Region Joint Associations to Complex Traits Genetics
- Volatility of Mutator Phenotypes at Single Cell Resolution
- Proteolysis of Virulence Regulator ToxR Is Associated with Entry of into a Dormant State
- Genome-Wide Negative Feedback Drives Transgenerational DNA Methylation Dynamics in Arabidopsis
- A Multi-layered Protein Network Stabilizes the FtsZ-ring and Modulates Constriction Dynamics
- Systematic Dissection of the Sequence Determinants of Gene 3’ End Mediated Expression Control
- Genome Sequencing of the Perciform Fish Provides Insights into Molecular and Genetic Mechanisms of Stress Adaptation
- Natural Variant E610G Is a Semi-dominant Suppressor of IAP-Induced RNA Processing Defects
- The Alkaline Response Pathway: Identification of a Novel Rim Pathway Activator
- Transgenerational Inheritance of Diet-Induced Genome Rearrangements in Drosophila
- A Single Nucleotide Polymorphism Uncovers a Novel Function for the Transcription Factor Ace2 during Hyphal Development
- DNA Damage Response and Spindle Assembly Checkpoint Function throughout the Cell Cycle to Ensure Genomic Integrity
- The Functional Interplay Between the t(9;22)-Associated Fusion Proteins BCR/ABL and ABL/BCR in Philadelphia Chromosome-Positive Acute Lymphatic Leukemia
- Extreme Recombination Frequencies Shape Genome Variation and Evolution in the Honeybee,
- Beyond Glycolysis: GAPDHs Are Multi-functional Enzymes Involved in Regulation of ROS, Autophagy, and Plant Immune Responses
- Comprehensive Profiling of Amino Acid Response Uncovers Unique Methionine-Deprived Response Dependent on Intact Creatine Biosynthesis
- Windpipe Controls Intestinal Homeostasis by Regulating JAK/STAT Pathway via Promoting Receptor Endocytosis and Lysosomal Degradation
- Ataxin-2 Regulates Translation in a New BAC-SCA2 Transgenic Mouse Model
- Cross-Population Joint Analysis of eQTLs: Fine Mapping and Functional Annotation
- The Power of Gene-Based Rare Variant Methods to Detect Disease-Associated Variation and Test Hypotheses About Complex Disease
- The Chromatin Remodeler CHD8 Is Required for Activation of Progesterone Receptor-Dependent Enhancers
- Competition between VanU Repressor and VanR Activator Leads to Rheostatic Control of Vancomycin Resistance Operon Expression
- A Missense Change in the Gene Links Aberrant Autophagy to a Neurodegenerative Vacuolar Storage Disease
- Simultaneous Discovery, Estimation and Prediction Analysis of Complex Traits Using a Bayesian Mixture Model
- Selection against Heteroplasmy Explains the Evolution of Uniparental Inheritance of Mitochondria
- Genome-Destabilizing Effects Associated with Top1 Loss or Accumulation of Top1 Cleavage Complexes in Yeast
- Imputation-Based Population Genetics Analysis of Malaria Parasites
- Heterozygosity for a Hypomorphic Polβ Mutation Reduces the Expansion Frequency in a Mouse Model of the Fragile X-Related Disorders
- Neto-Mediated Intracellular Interactions Shape Postsynaptic Composition at the Neuromuscular Junction
- Ndd1 Turnover by SCF Is Inhibited by the DNA Damage Checkpoint in
- Frameshift Variant Associated with Novel Hoof Specific Phenotype in Connemara Ponies
- The DNA Helicase Recql4 Is Required for Normal Osteoblast Expansion and Osteosarcoma Formation
- Spastin Binds to Lipid Droplets and Affects Lipid Metabolism
- Maintenance of Glia in the Optic Lamina Is Mediated by EGFR Signaling by Photoreceptors in Adult Drosophila
- Auxin Influx Carriers Control Vascular Patterning and Xylem Differentiation in
- Dual-Specificity Anti-sigma Factor Reinforces Control of Cell-Type Specific Gene Expression in
- The Lowe Syndrome Protein OCRL1 Is Required for Endocytosis in the Zebrafish Pronephric Tubule
- Postnatal Loss of Hap1 Reduces Hippocampal Neurogenesis and Causes Adult Depressive-Like Behavior in Mice
- CAPER Is Vital for Energy and Redox Homeostasis by Integrating Glucose-Induced Mitochondrial Functions via ERR-α-Gabpa and Stress-Induced Adaptive Responses via NF-κB-cMYC
- Distinct and Cooperative Activities of HESO1 and URT1 Nucleotidyl Transferases in MicroRNA Turnover in
- The Evolutionary Origination and Diversification of a Dimorphic Gene Regulatory Network through Parallel Innovations in and
- MAPK Signaling Pathway Alters Expression of Midgut ALP and ABCC Genes and Causes Resistance to Cry1Ac Toxin in Diamondback Moth
- Spatio-temporal Remodeling of Functional Membrane Microdomains Organizes the Signaling Networks of a Bacterium
- Asymmetric Transcript Discovery by RNA-seq in . Blastomeres Identifies , a Gene Important for Anterior Morphogenesis
- A Stress-Induced Small RNA Modulates Alpha-Rhizobial Cell Cycle Progression
- Systematic Profiling of Poly(A)+ Transcripts Modulated by Core 3’ End Processing and Splicing Factors Reveals Regulatory Rules of Alternative Cleavage and Polyadenylation
- The UPR Branch IRE1- in Plants Plays an Essential Role in Viral Infection and Is Complementary to the Only UPR Pathway in Yeast
- A Non-canonical RNA Silencing Pathway Promotes mRNA Degradation in Basal Fungi
- Co-chaperone p23 Regulates . Lifespan in Response to Temperature
- Re-replication of a Centromere Induces Chromosomal Instability and Aneuploidy
- Shade Avoidance Components and Pathways in Adult Plants Revealed by Phenotypic Profiling
- Lipid-Induced Epigenomic Changes in Human Macrophages Identify a Coronary Artery Disease-Associated Variant that Regulates Expression through Altered C/EBP-Beta Binding
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Lack of GDAP1 Induces Neuronal Calcium and Mitochondrial Defects in a Knockout Mouse Model of Charcot-Marie-Tooth Neuropathy
- Proteolysis of Virulence Regulator ToxR Is Associated with Entry of into a Dormant State
- Frameshift Variant Associated with Novel Hoof Specific Phenotype in Connemara Ponies
- Ataxin-2 Regulates Translation in a New BAC-SCA2 Transgenic Mouse Model
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



