-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaA Multi-layered Protein Network Stabilizes the FtsZ-ring and Modulates Constriction Dynamics
Bacterial cell division is a highly regulated process that must be coordinated with other cellular processes (i.e. DNA replication and chromosome segregation) to promote faithful reproduction. In Escherichia coli, this regulation is most often mediated through the polymerization of the prokaryotic tubulin homolog, FtsZ, which forms a ring-like structure (FtsZ-ring) at midcell. The establishment of the FtsZ-ring marks the site of division and enables the assembly of the macromolecular division machinery (divisome). Here we applied single-molecule based superresolution imaging to reveal the three-dimensional structure of FtsZ in the context of its regulatory proteins: ZapA, ZapB and MatP. We found that these four proteins exist in a multi-layered network that extends from the cell membrane to the chromosome. This layered organization not only helps to stabilize the FtsZ-ring, but also serves to coordinate division with DNA status by influencing constriction rate. Our results not only provide a comprehensive view of the divisome, but also allow new insight to be garnered regarding the structure and function of the divisome.
Published in the journal: . PLoS Genet 11(4): e32767. doi:10.1371/journal.pgen.1005128
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1005128Summary
Bacterial cell division is a highly regulated process that must be coordinated with other cellular processes (i.e. DNA replication and chromosome segregation) to promote faithful reproduction. In Escherichia coli, this regulation is most often mediated through the polymerization of the prokaryotic tubulin homolog, FtsZ, which forms a ring-like structure (FtsZ-ring) at midcell. The establishment of the FtsZ-ring marks the site of division and enables the assembly of the macromolecular division machinery (divisome). Here we applied single-molecule based superresolution imaging to reveal the three-dimensional structure of FtsZ in the context of its regulatory proteins: ZapA, ZapB and MatP. We found that these four proteins exist in a multi-layered network that extends from the cell membrane to the chromosome. This layered organization not only helps to stabilize the FtsZ-ring, but also serves to coordinate division with DNA status by influencing constriction rate. Our results not only provide a comprehensive view of the divisome, but also allow new insight to be garnered regarding the structure and function of the divisome.
Introduction
Prokaryotic cell division is a conserved process that requires the formation of a multi-protein complex (divisome) at midcell [1]. Although many molecular constituents of the divisome have been identified [2], its in-vivo structural organization remains elusive. Understanding divisome architecture will help elucidate the mechanisms by which it achieves cytokinesis and coordinates with other cellular processes.
The central component of the divisome is FtsZ, a highly conserved prokaryotic tubulin homolog that polymerizes at midcell to form a ring-like structure [3]. The FtsZ-ring is not only required to serve as a stable scaffold for the assembly of all other division proteins [4], but may also generate a constrictive force during cytokinesis [5,6]. Based on in vitro polymerization studies, the basic structural units of the E. coli FtsZ-ring are believed to be single-stranded protofilaments that are on average 120 nm long [7]. These protofilaments are attached to the cytoplasmic membrane by binding to two membrane proteins, FtsA and ZipA [8,9].
Recently, high-resolution microscopy studies have revealed that the in vivo FtsZ-ring is a discontinuous structure, comprising a heterogeneous arrangement of FtsZ protofilaments [10–15]. Furthermore, FtsZ molecules within the ring have been shown to dynamically exchange with those in the cytoplasmic pool (half-time τ1/2 ≈ 10 – 30 s) [16,17]. These observations raise the question of how such a disordered, dynamic FtsZ-ring could provide the stable scaffold required for divisome assembly or generate a uniform constrictive force along the septum.
Recent work aimed at answering these questions have focused on a family of FtsZ-ring-associated proteins (Zaps), including: ZapA, ZapB, ZapC, ZapD and ZapE [18–23]. These proteins localize to the midcell in an FtsZ-dependent manner, and promote FtsZ-ring assembly in vivo. While no individual Zap is essential, single deletions lead to elongated cells, abnormal FtsZ-rings and irregular septum morphologies. Double and triple deletions result in synergistic defects [20,22]. These observations suggest that the Zaps carry out important, over-lapping roles in maintaining proper FtsZ-ring structure and function [20–23].
Among the five Zaps, ZapA and ZapB are best understood and are thought to work in concert to stabilize the FtsZ-ring [18,19,24]. We previously showed that in the absence of ZapA and/or ZapB, the FtsZ-ring dissociates into smaller, widely-dispersed FtsZ clusters throughout the midcell region, often leading to incomplete and abnormal septum formation [14]. These results support a model in which ZapA and ZapB stabilize the FtsZ-ring specifically at the division plane. In vitro characterization suggests that ZapA can stabilize the FtsZ-ring by cross-linking FtsZ protofilaments and possibly reducing its GTPase activity [24,25]. ZapB is a 100% coiled-coil protein that self polymerizes into large bundles in vitro [19]. ZapB does not interact directly with FtsZ, but associates indirectly through ZapA [26,27]. Hence, ZapB most likely exerts its stabilizing effect on FtsZ indirectly through ZapA. Interestingly, a recent confocal microscopy study observed that ZapB does not colocalize completely with the FtsZ-ring, but instead resides at the cytoplasmic face of the FtsZ-ring [26]. As such, the ZapB structure may have FtsZ-independent dynamics and functions, suggesting that the structural organization of the entire divisome is not simply dictated by the FtsZ-ring.
Further complicating the picture is the discovery of MatP, a DNA-binding protein involved in condensation and segregation of the terminus (ter) macrodomain of the chromosome [28,29]. MatP was found to interact with ZapB in a bacterial two-hybrid system [29] and recently shown to work with ZapB and ZapA to localize the divisome [30]. Thus, these data suggests a new, attractive model for the structural organization of the divisome—the divisome is extended from the membrane to the chromosome through a three-dimensional, extended protein network formed by these interacting proteins. This large protein network may provide the scaffold function previously attributed to FtsZ alone. Importantly, this network may also coordinate the progression of cell wall constriction with chromosome segregation. To test such a model, in this work we used quantitative superresolution imaging in conjunction with biophysical and genetic investigations to map the spatial organization and characterize the function of the three-dimensional cellular structures formed by FtsZ, ZapA, ZapB and MatP proteins in E. coli cells.
Results
Characterization of ZapA and ZapB structures in live E. coli cells
Using a single-molecule based superresolution imaging method, photoactivated-localization microscopy (PALM) [31], we first characterized the cellular structures formed by FtsZ, ZapA, or ZapB in live E. coli cells with a spatial resolution of ~45 nm [14]. We generated photoactivatable fluorescent fusion proteins mEos2-ZapA and ZapB-mEos2, and used an FtsZ-mEos2 construct described previously [10,14,32]. The mEos2-ZapA and ZapB-mEos2 fusions both rescued the elongated phenotypes of their respective deletion strains and localized to midcell in the absence of the endogenous protein (S1 Fig). To perform live-cell PALM imaging, we expressed these fusions ectopically in wild-type (wt) BW25113 cells and completed imaging in less than 30s. The estimated expression levels of FtsZ-mEos2, mEos2-ZapA and ZapB-mEos2 were ~30%, 45% and 10% of the total cellular concentration of the corresponding wt protein, respectively (Methods).
We found that ZapA and ZapB predominantly formed band-like structures, indicative of rings projected to 2-dimensional (2D) imaging planes (Fig 1, S1 Table). The band-like structure was observed in 41% of cells expressing mEos2-ZapA (ntotal = 229) and 59% of cells expressing ZapB-mEos2 (ntotal = 137), similar to the prevalence observed for FtsZ-mEos2 (51%, ntotal = 201). The remaining cells exhibited a number of different focal or non-planar morphologies, which were also observed previously for FtsZ [10,14] (S2 Fig). These alternate structures appear to precede the band-like structure as they were predominantly observed in shorter cells (S3 Fig, S1 Table). We observed similar morphologies when we applied superresolution imaging [33] to immuno-labeled, native FtsZ and ZapB structures, suggesting that the observed polymorphisms were not artifacts caused by the fused fluorescent proteins (S4 Fig).
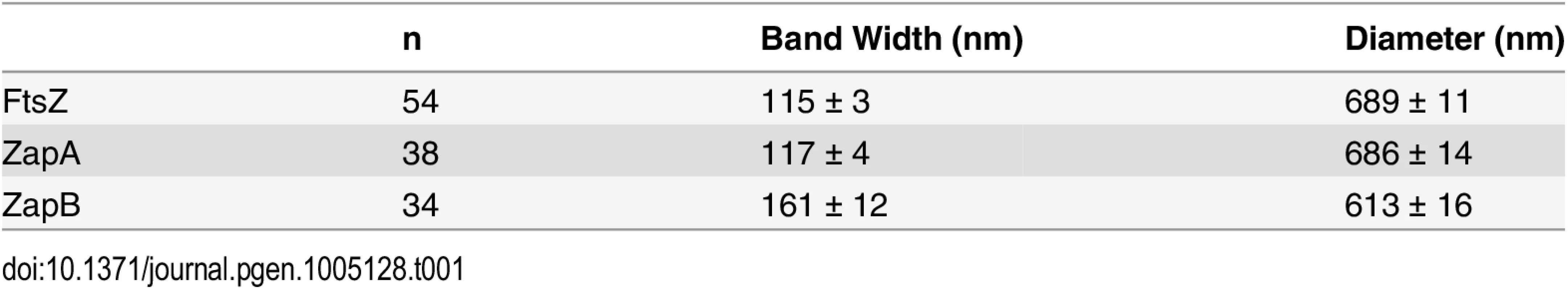
Fig. 1. Live-cell PALM imaging of band-like ZapA, ZapB and FtsZ structures. 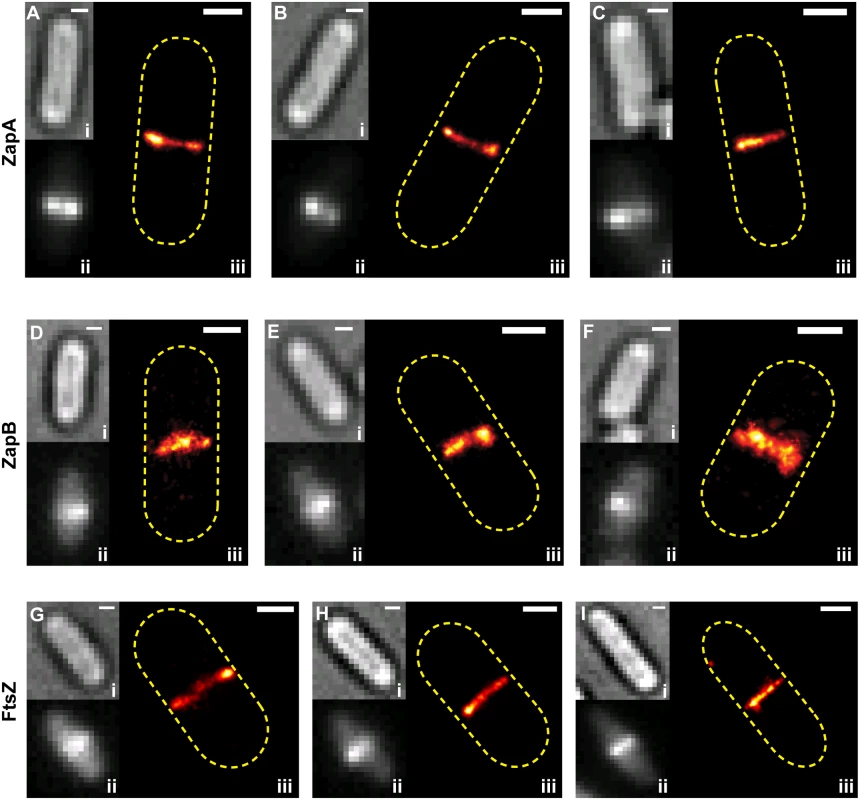
Images of mEos2-ZapA (pJB051, A-C), ZapB-mEos2 (pJB045, D-F) and FtsZ-mEos2 (pJB042, G-I) in wt cells are shown in the order of bright-field image (i), ensemble fluorescence image (ii) and PALM image displayed in pseudocolor (iii). Approximate cell outlines are indicated by yellow dashed lines. Scale Bars, 500 nm. While the three proteins displayed similar structural morphologies, we observed quantitative differences in the width (w) and diameter (d) of the band-like structures formed by each protein (Table 1, Methods). The mEos2-ZapA bands (w = 117 ± 4 nm, d = 686 ± 14 nm, n = 38; x - ± se) were similar (p > 0.3) to those of FtsZ-mEos2 (w = 115 ± 3 nm, d = 689 ± 11 nm, n = 54) (Table 1), but ZapB-mEos2 bands were significantly wider (161 ± 12 nm, p ≈ 1e-4, n = 34) and smaller in diameter (613 ± 16 nm, p < 5e-5). These structural differences are readily apparent in PALM images (Fig 1, iii), but obscured in the corresponding conventional fluorescence images (Fig 1, ii). The higher degree of structural similarity between ZapA and FtsZ relative to that of ZapB and FtsZ likely reflects the fact that ZapA binds FtsZ directly, while ZapB associates with FtsZ indirectly through ZapA [26,27].
Two-color PALM imaging reveals structural deviations between FtsZ, ZapA and ZapB
Analysis of the single-color PALM images above indicated that ZapB structures could deviate from those of FtsZ and ZapA in vivo. Thus, to directly compare their spatial arrangements, we performed two-color PALM imaging in live E. coli cells. We expressed either Dronpa-ZapA or ZapB-Dronpa together with FtsZ-PAmCherry1 in the same cell. The functionality and expression level of these fusions were similar to those of the mEos2 constructs described above (S5 Fig).
We found that structures formed by Dronpa-ZapA and FtsZ-PAmCherry1 largely overlapped, and adopted similar structural morphologies in ~80% of cells, consistent with incorporation of ZapA into the FtsZ-ring (Fig 2Ai-iii, ntotal = 96). In the remaining cells, however, the two structures showed deviations significantly larger than our spatial resolution in two-color PALM imaging (~50 nm) (Fig 2Aiv-vi). On several occasions (n = 7), Dronpa-ZapA structures were sandwiched in between FtsZ-PAmCherry1 structures (Fig 2Av-vi), suggesting a possible bridging function for ZapA. We observed more substantial deviations between ZapB-Dronpa and FtsZ-PAmCherry1 (Fig 2B). In ~70% of cells (ntotal = 88), ZapB-Dronpa structures appeared to be encompassed by FtsZ-PAmCherry1 and located toward the inner surface of FtsZ-ring (Fig 2B).
Fig. 2. Two color-PALM imaging of ZapA-FtsZ, ZapB-FtsZ and ZapA-ZapA pairs. 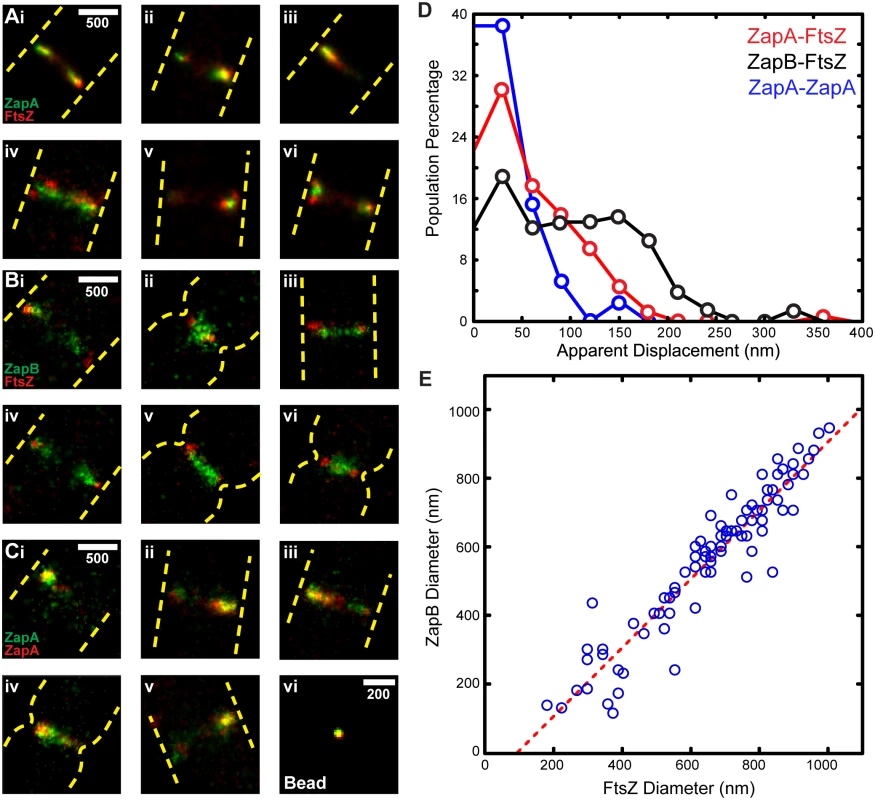
Cropped PALM images of cells expressing Dronpa-ZapA and FtsZ-PAmCherry1 (Ai-vi), ZapB-Dronpa and FtsZ-PAmCherry1 (Bi-vi), or Dronpa-ZapA and PAmCherry1-ZapA (Ci-v). Approximate cell outlines are indicated in yellow dashed lines. (Cvi) 2C-PALM image of a 100 nm TetraSpeck bead. (D) Histograms of the apparent displacement between the given protein pairs in individual cells. The mean values for the ZapA-FtsZ, ZapB-FtsZ and ZapA-ZapA pairs are 55 ± 50 nm (n = 158), 97 ± 70 nm (n = 132) and 33 ± 33 nm (n = 39), respectively (x- ± sd). (E) The diameters for ZapB-Dronpa and FtsZ-PAmCherry1 structures visualized simultaneously in the same cell (blue circles) were best fit to a line (red) where y = 1.0x - 95. The R2 of the fitted line to the data was 0.89. Scale Bars, as labeled in nm. The structural deviations we observed between the FtsZ-ZapA and FtsZ-ZapB pairs were not observed in a control strain where Dronpa-ZapA was co-expressed with PAmCherry1-ZapA and imaged under the same condition (Fig 2Ci-v). Multi-color fluorescent beads imaged in both color channels also showed complete overlap within our spatial resolution (Fig 2Cvi). These results suggest that the observed structural deviations were not imaging artifacts, nor caused by the fusion protein dynamics or photoproperties of Dronpa and PAmCherry1.
To quantify the degree of colocalization for the ZapA-FtsZ, ZapB-FtsZ and ZapA-ZapA pairs, we performed a coordinate-based cross-correlation analysis [34–36]. In each cell, we determined the cross-correlation value between two different species at midcell as a function of their displacement along the short axis of the cell. The displacement value at maximal cross-correlation value was assigned as the 'apparent displacement' for each cell (Fig 2D). We found that the average apparent displacement between Dronpa-ZapA and FtsZ-PAmCherry1 molecules was 55 ± 50 nm (n = 158; x - ± sd). Molecules of ZapB-Dronpa were displaced farther away from FtsZ-PAmCherry1 molecules with an average of 97 ± 70 nm (n = 132). Both of these displacements are significantly larger than that of the control ZapA-ZapA pair (33 ± 33 nm, n = 39) (Fig 2D). The latter reflects our spatial resolution in resolving two cellular structures. These results further support our qualitative observations that ZapA can appreciably deviate from the FtsZ-ring, and that ZapB assembles into a discrete structure internal to the FtsZ-ring.
Separation between FtsZ and ZapB ring-like structures remains constant throughout constriction
In cells showing visible constrictions, we often observed that both FtsZ and ZapB formed ring-like structures. To investigate whether the relative spatial arrangement between FtsZ and ZapB changes during cell constriction, we measured the corresponding diameters of these ring-like structures in each individual cell. We found that the diameters of the FtsZ-ring and ZapB-rings were linearly correlated with each other, and the scatter plot can be best fit by a line with a slope of 1.0 and x-intercept at 95 nm (Fig 2E). This observation indicates that the radial separation (95/2 = 47.5 nm) between FtsZ and ZapB structures is maintained throughout constriction. These results suggest that there may be a specific mechanism to maintain the relative spatial separation between FtsZ and ZapB, and such a separation may be important for successful cell wall constriction.
iPALM reveals relative spatial arrangements of FtsZ, ZapA and ZapB with respect to the inner membrane
While two-color PALM imaging allowed us to directly visualize the relative arrangement between two protein species in the same cell, the nature of 2D imaging of three-dimensional (3D) structures prevented us from accurately quantifying the relative spatial arrangement of FtsZ, ZapA and ZapB along the radial axis. Therefore, we performed 3D superresolution imaging using interferometric PALM (iPALM). iPALM employs the same principles as PALM imaging, but also utilizes the interference of light from the same molecule in two different paths to determine its z-position with high precision [37,38]. Under our imaging conditions we achieved a, x-, y-, and z-resolution of ~20 nm.
iPALM images of FtsZ-mEos2 and mEos2-ZapA showed similar, irregular punctate structures that curved along the cell periphery (Fig 3A). ZapB-mEos2, however, appeared much more cohesive, exhibiting large, contiguous structures that often lacked curvature. This observation is consistent with our previous observations that ZapB structures significantly deviate from FtsZ and ZapA.
Fig. 3. iPALM imaging and z-position measurements of FtsZ, ZapA, and ZapB. 
(A) Cross-sectional views of the midcell region of representative wt cells expressing FtsZ-mEos2, mEos2-ZapA, ZapB-mEos2 or mEos2-MTSBs (three cells for each strain) are displayed in respective rows. All images are to scale and share the same z-dimensions as labeled on the left. The dashed rectangle box (gray) in the bottom right-most image is representative of the user-defined box we used to calculate mean z-positions. (B) Histograms (20 nm bins) of the average measured z-positions for each protein structure in individual cells illustrate the internal nature of ZapB. Scale Bar, 250 nm. To measure the relative radial displacement of FtsZ, ZapA and ZapB molecules, we compared their mean z-positions to that of the coverslip surface coated with Alexa Fluor 568 [38]. To restrict our analysis to midcell regions close to the coverslip, we excluded cells that showed visible indentation at midcell in corresponding DIC (Differential Interference Contrast) images, and only used molecules inside a user-defined region at the bottom of each non-constricting cell (Fig 3A). The mean z-position of these molecules was then calculated for each individual cell, and the corresponding distribution for each protein in different cells was plotted in Fig 3B. The distributions of the three proteins show substantial overlap, but clearly indicate that ZapB molecules are displaced internal to the FtsZ-ring. Quantifying the mean z-positions reveals that FtsZ and ZapA structures reside close to each other (z = 80 ± 2 nm, n = 125 for FtsZ, and 74 ± 1 nm, n = 250 for ZapA, respectively; x - ± se), while the ZapB structures are displaced significantly internal to the FtsZ-ring (z = 117 ± 2 nm, n = 226). We note that the ~40 nm difference between FtsZ and ZapB observed by iPALM is in a similar range with what can be estimated from single-color, 2D PALM measurements (343FtsZ radius – 306ZapB radius = 37 nm), or from the correlation between the diameters of ZapB and FtsZ band-like structures in two-color, 2D PALM imaging (x-intercept / 2 = 47.5 nm, Fig 2E).
To orient these structures in the context of the inner membrane (IM), we determined the z-position of mEos2 fused to the membrane-targeting sequence of MinD (V199-S219) from Bacillus subtilis (mEos2-MTSBs) (Fig 3A). mEos2-MTSBs distributed randomly and sparsely on the membrane; the mean z-position from the coverslip surface was measured at z = 67 ± 1 nm (n = 315), consistent with the expected value when the thickness of cell envelope was considered [39,40]. Thus, we used the z-position of mEos2-MTSBs as a proxy for the cytoplasmic face of the inner membrane and estimated that FtsZ is displaced ~13 nm away from the IM on average.
We note that our estimates of apparent displacement do not take into account the molecular size of the mEos2 label (~4 nm) [41]. To determine the true displacement between two mEos2-labeled proteins, one must know how mEos2 is oriented with respect to each labeled protein. Without this information, however, we can still estimate the error associated with our apparent displacement measurements by treating the size of mEos2 as an uncertainty (42+42=5.7 nm). Comparing this uncertainty to the measured apparent displacements of FtsZ-ZapA (6 nm), FtsZ-ZapB (37 nm) and FtsZ-IM (13 nm), we conclude that FtsZ and ZapA reside on a similar radial plane, and that ZapB is significantly displaced into the cytoplasm relative to FtsZ. The 13 nm distance between FtsZ and the IM requires more careful consideration because the 5.7 nm uncertainty level is relatively large. Nevertheless, by taking into account the size of mEos2 and how mEos2 is attached to FtsZ, the displacement between FtsZ and the IM measured from iPALM can be estimated (S1 Text). This estimate will allow us to assess the amount of force FtsZ protofilaments could exert on the membrane through specific force generation mechanisms.
MatP enhances FtsZ-ring stability
Next, we investigated the role of MatP in maintaining the layered FtsZ-ZapA-ZapB structure. We previously showed that deletion of MatP resulted in dispersed, mislocalized FtsZ structures under fast growth conditions [14]. This effect can be explained by an indirect mechanism, in which the abnormal nucleoid architecture caused by deletion of matP [28] results in aberrant distributions of the nucleoid-occlusion factor SlmA [42], which consequently disrupts FtsZ-ring assembly at the midcell. Our previous observation of SlmA mislocalization in ΔmatP cells supported this mechanism [14]. To further test whether MatP also has a direct role in maintianing FtsZ-ring at the midcell through its interaction with ZapB and ZapA, we constructed a double deletion strain (ΔmatPΔslmA), which allowed us to observe the effect of ΔmatP independent of the nucleoid-occlusion effect caused by SlmA. We reasoned that if abnormally-distributed SlmA is solely responsible for mislocalizing the FtsZ-ring in ΔmatP cells, then deletion of slmA should revert cells to normal FtsZ-ring localization, as deletion of slmA alone does not show any detectable defect in FtsZ-ring localization (S6 Fig) [42]. In contrast to this expectation, we found that ΔmatPΔslmA cells displayed a similar FtsZ-ring mislocalization phenotype as ΔmatP, albeit with a slightly reduced frequency (S6 Fig). This observation indicates that MatP has a SlmA-independent role in stabilizing the FtsZ-ring at midcell, possibly mediated by the physical connection between FtsZ and MatP through ZapA and ZapB. This is consistent with a recent report in which similar FtsZ-ring positioning defect was observed in related mutant backgrounds [30].
MatP is located ~30 nm internal to ZapB
To determine where MatP is positioned inside the cell with respect to ZapB, we performed live-cell PALM imaging on wt and ΔmatP cells ectopically expressing a MatP-mEos2 fusion protein (Fig 4Ai-iv). Consistent with previous studies [28,29], MatP-mEos2 typically appeared as one or two large clusters in both strains, suggesting that MatP-mEos2 localizes correctly. The MatP-mEos2 clusters we observed were on average 100 ± 58 nm in diameter (x - ± sd, n = 613, S7 Fig). Given our resolution, this size is much larger than a single molecule of MatP-mEos2 would appear, suggesting that the clusters likely comprise multiple closely-associated MatP molecules. This finding is consistent with the ability of MatP dimers to associate with a series of matS sites on the chromosome and further tetramerize to condense the ter macrodomain [43].
Fig. 4. PALM imaging of MatP and z-position estimation. 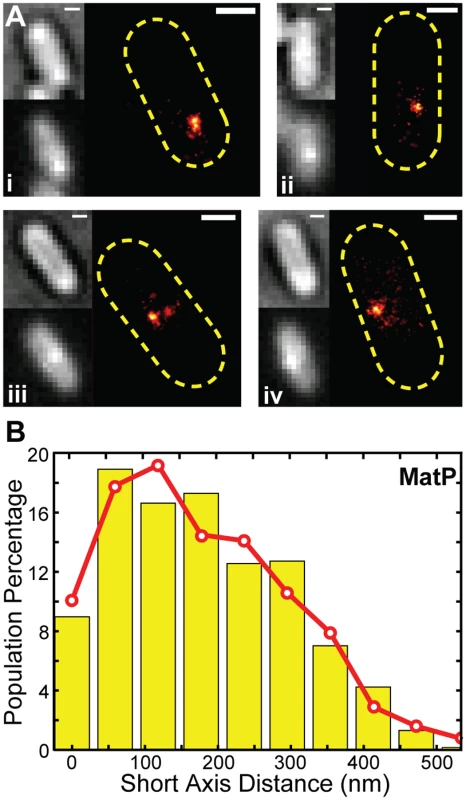
(A) Four representative PALM images for cells expressing MatP-mEos2 in the order of bright-field, ensemble fluorescence, and PALM image. Scale Bars, 500 nm. (B) Distribution of short-axis displacements of MatP-mEos2 clusters from the middle of the cell (yellow bars) fitted with a model (red line) in which a normally-distributed radial displacement is randomly projected to a 2D imaging plane (S1 Text). The fitted radial displacement is 285 ± 120 nm (red line, x- ± sd). We measured the displacement of each MatP-mEos2 cluster from the cell center along the long and short axes of the cell (S8A Fig). We found that the long axis displacement exhibited a cell-length dependent decrease (S8B Fig), as expected from the directed movement of the ter macrodomain toward midcell during the cell cycle [28]. The short axis displacement, however, was independent of cell length (S8C Fig). The short axis displacement is the 2D projection of the radial displacement of each cluster from the cell center (S8A Fig). Assuming that MatP-mEos2 clusters are randomly distributed angularly and projected to the 2D imaging plane, the observed short axis displacement distribution of MatP-mEos2 clusters can be best described by a model in which MatP clusters are on average 280 ± 120 nm (x - ± sd, n = 613) radially away from the cell center (S1 Text, Figs 5B, S8–S9). Comparing this measurement with the typical diameter of ZapB band-like structures, we estimate that MatP resides ~30 nm (607ZapB radius ÷ 2–280) internal to ZapB on average. Thus, MatP forms another distinct layer internal to the FtsZ-ring. Given the wide distributions of MatP and ZapB z-positions (Fig 3B, 4B), our data suggest that MatP and ZapB are appropriately positioned to enable direct interaction, and that this interaction serves to stabilize the FtsZ-ring. We note, however, that we cannot exclude the possibility that other proteins may bridge the ZapB-MatP interaction.
Deletion of matP increases the turnover rates of FtsZ, ZapA and ZapB
As we have shown, MatP plays a direct role in stabilizing the FtsZ-ring through its interaction with ZapB. To further understand how MatP exerts this influence, we probed the turnover dynamics of FtsZ, ZapA, and ZapB in the presence and absence of MatP. Previous fluorescence recovery after photobleaching (FRAP) measurements showed that the FtsZ-ring and its associated proteins exchange dynamically with their cytoplasmic pools with a half-time of 10–30 s depending on growth conditions and strain background [16,17,25,44]. We reason that one way for MatP to stabilize the FtsZ-ring could be by promoting the stable formation of large, polymerized ZapB assemblies at midcell, which consequently stabilizes ZapA and FtsZ, and could be reflected by a reduction in the turnover rate of ZapB.
We ectopically expressed FtsZ-GFP, GFP-ZapA or GFP-ZapB in wt and ΔmatP cells, and used FRAP to measure their turnover half-time under our slow growth conditions. Interestingly, we found that in the wt background, the FRAP half-time for FtsZ-GFP and GFP-ZapA were comparable to each other at 12.3 ± 0.2 (n = 58, x - ± se) and 14.2 ± 1.1 s (n = 51), respectively, but that of GFP-ZapB was significantly longer (19.8 ± 1.0 s, n = 59, p < 1e-10) (Fig 5A, Table 2). The slower turnover rate of GFP-ZapB is consistent with the highly polymeric nature of ZapB assemblies observed in vitro [19,27], and also agrees with our in vivo observations that ZapB structures are generally larger and more cohesive compared to those of FtsZ and ZapA (Fig 3A). In the ΔmatP mutant, the turnover half-times of both GFP-ZapB and GFP-ZapA were significantly reduced by ~50% to 11.6 ± 0.4 s (n = 56, p < 0.001) and 5.7 ± 0.2 s (n = 59, p< 0.001), respectively, supporting an influential role for MatP on their turnover dynamics (Fig 5B, Table 2). The half-time of FtsZ-GFP turnover was also significantly reduced in the ΔmatP mutant (10.5 ± 0.3 s, n = 59, p< 0.001), but to a lesser degree (~15%). This smaller effect may be due to the fact that MatP’s interaction with FtsZ is more indirect than its interactions with ZapA or ZapB, and/or that FtsZ’s self-polymerization properties and interactions with other proteins may have a greater influence on its turnover rate than MatP does.
Fig. 5. FRAP measurements of FtsZ, ZapA and ZapB turnover rates in BW25113 and ΔmatP cells. 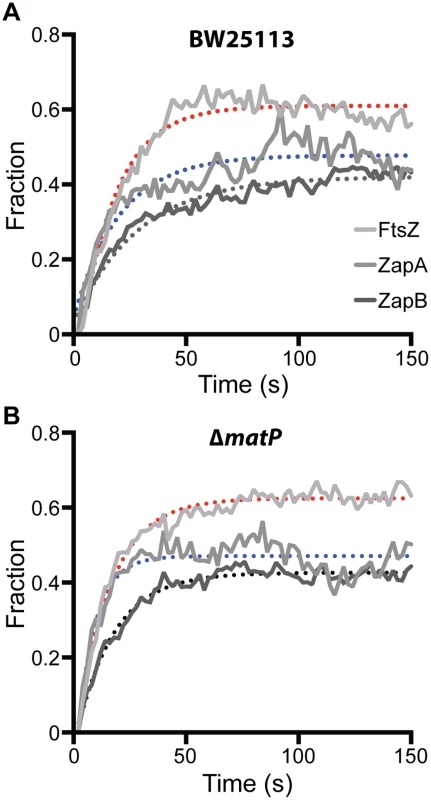
Average fluorescence recovery trajectories for FtsZ (light grey), ZapA (grey) and ZapB (dark grey) are displayed for wt (A) and ΔmatP (B) cells as a fraction of pre-bleached intensity. Trajectories were fit with single exponentials illustrated as dotted lines (FtsZ, red; ZapA, blue; ZapB, black). The respective half-times are listed in Table 2. Tab. 2. Half-time for fluorescence recovery of FtsZ, ZapA and ZapB. 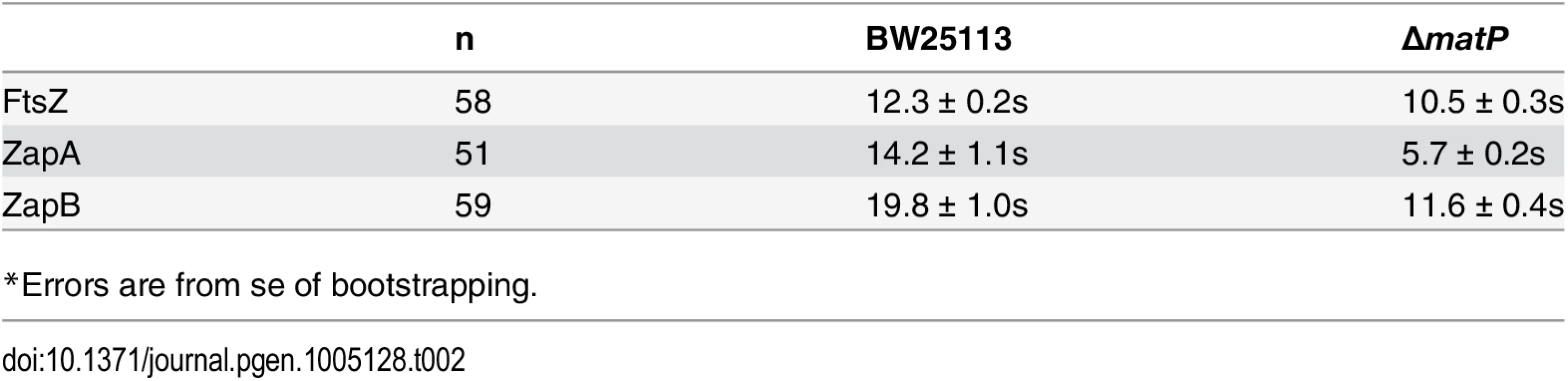
*Errors are from se of bootstrapping. We note that these FRAP measurements were obtained under slow growth conditions, where the loss of MatP has little to no observable effect on the localization of FtsZ [14]. Nevertheless, here we observed significant effects on the turnover dynamics for ZapA and ZapB structures. These results suggest that MatP modulates the dynamic turnover of all three protein structures even in the absence of obvious mislocalization phenotypes.
ΔmatP cells have shorter constriction periods
The FtsZ-ring has long been regarded as not only the critical structural component of the divisome, but also a primary driving force for cell constriction [6,45]. If structural stability of the FtsZ-ring is important for active force generation, constriction may be slowed in cells where the structural stability of the FtsZ-ring is compromised by disruption of the FtsZ-ZapA-ZapB-MatP network. To test this idea we measured the constriction period, τc, in wt and ΔmatP cells ectopically expressing similar low levels of FtsZ-GFP under our slow growth condition (S10 Fig) using time-lapse fluorescence microscopy (Methods). We defined constriction initiation as the time when an indentation of cell wall was first visible in bright-field images, and the end of constriction as the time when FtsZ-GFP fluorescence completely disappeared from, and did not return to, the midcell (S11 Fig). Surprisingly, we found that the constriction periods of ΔmatP (40.3 ± 2.6 min, n = 37; x - ± se) and ΔmatPΔslmA cells (34.3 ± 2.2 min, n = 50) were significantly shorter than that of the wt cells (48.8 ± 3.0 min, n = 44, p < 0.04), while the doubling time remained similar (~200 min, Table 3). These results are contrary to our expectation that FtsZ-ring stability promotes constriction progress. As we will discuss below, two possible mechanisms coordinating the rate of constriction and nucleoid segregation could contribute to this observed phenomenon.
Tab. 3. Time-lapse analysis of FtsZ-GFP in mutant strains. 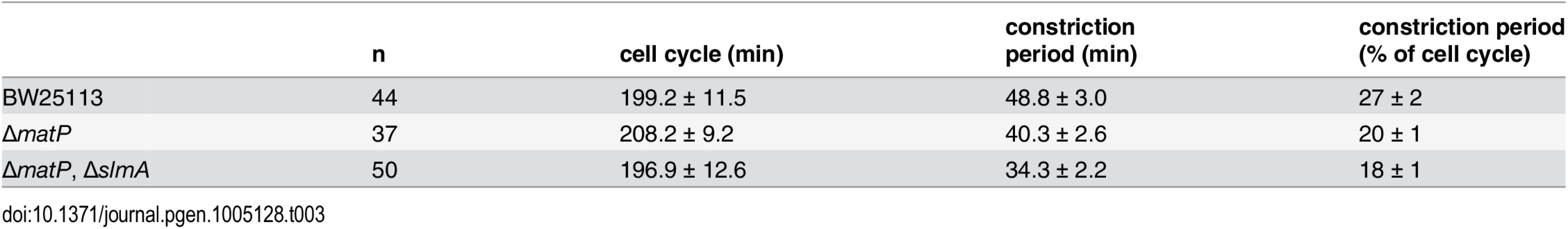
Discussion
Structural and spatial arrangement of the FtsZ-ZapA-ZapB-MatP network
In this work, we provided a quantitative characterization of the spatial organization and function of the FtsZ-ZapA-ZapB-MatP network in E. coli cells. By taking advantage of the superior spatial resolution and detection sensitivity offered by single-molecule based superresolution imaging, we quantified the spatial arrangement of each protein and showed that they are positioned to form a large, multi-layered network extending from the membrane to nucleoid at the midcell. We found that the FtsZ-ring comprises a heterogeneous arrangement of FtsZ clusters and is displaced on average 13 nm away from the cytoplasmic face of the inner membrane. ZapA adopts a similar heterogeneous clustered arrangement to that of FtsZ and resides at a similar radial plane. Interestingly, ~20% of ZapA structures deviated from FtsZ structures, and in a few cases ZapA clusters appeared to be sandwiched in between FtsZ clusters. ZapB forms wider, larger and more cohesive structures that are displaced on average ~40 nm internal to FtsZ and ZapA. Finally, MatP forms puncta with an average diameter of ~100 nm and is located on average ~30 nm internal to ZapB, or ~280 nm away from the center of the cell. As MatP binds to the ter region of the chromosome, it is reasonable to expect that its position also reflects that of the ter region of the nucleoid at midcell. From these measurements a three-dimensional architecture of a multi-layered protein network begins to emerge (Fig 6). As we will discuss below, these quantitative measurements provide an important physical framework upon which mechanisms regarding constrictive force generation, divisome assembly, FtsZ-ring function, and the interplay between cell wall constriction and chromosome segregation should be considered.
Fig. 6. The multi-layered organization of the E. coli divisome. 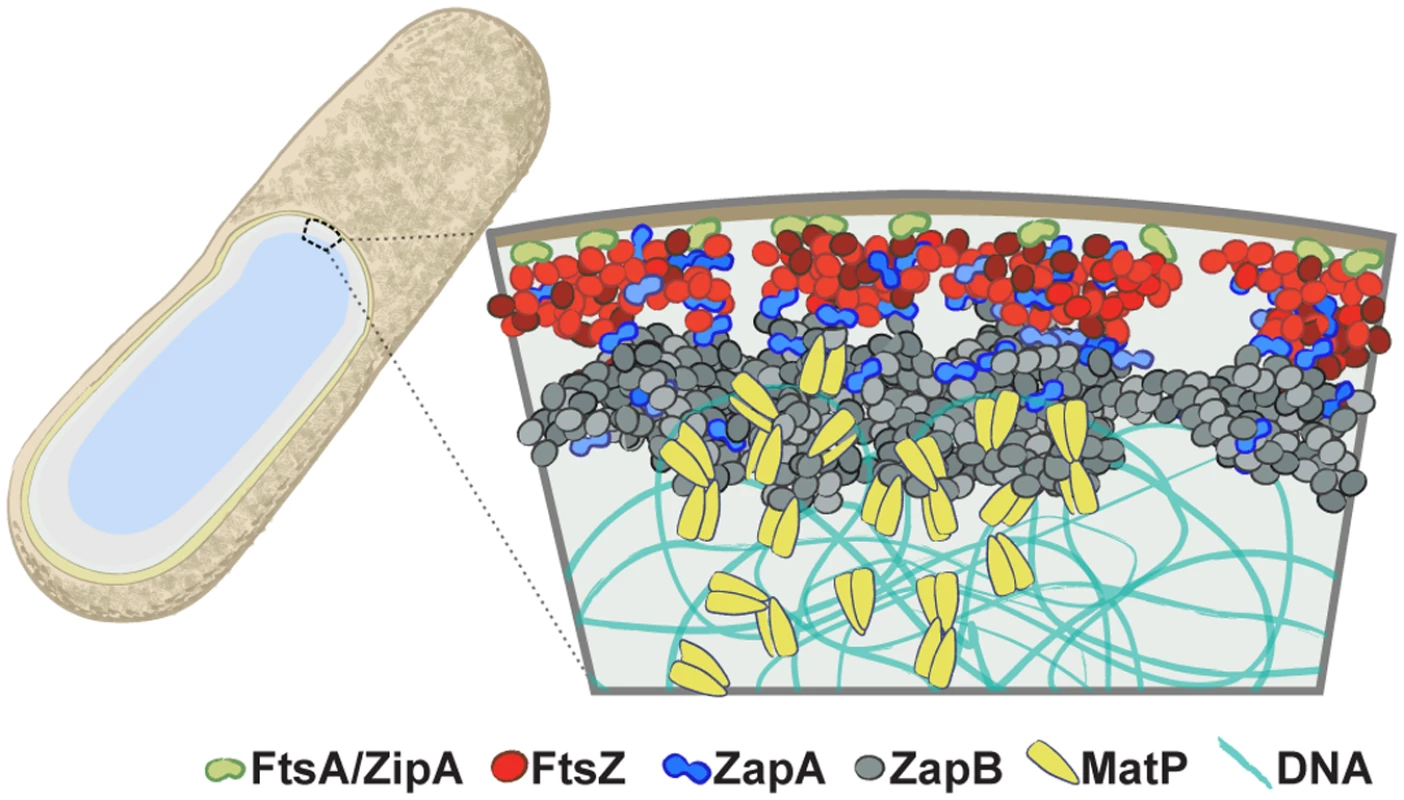
A schematic illustrating the relative radial arrangement of FtsA/ZipA-FtsZ-ZapA-ZapB-MatP. FtsA (green) and ZipA (orange) tether the punctuate FtsZ (red) structure to the inner membrane. ZapA (blue) mimics FtsZ but can deviate, possibly by interacting with ZapB or a number of membrane proteins [50,51]. The large, internal ZapB (grey) structure indirectly associates with FtsZ through ZapA, and is anchored on the chromosome through its associations with MatP (yellow). Structural deviations of ZapA and ZapB from FtsZ
Historically, the FtsZ-ring has been regarded as the main structural component of the divisome, serving as a scaffold for the assembly of all other division proteins at the constriction site [46]. Hence, its structural organization was thought to determine that of the other divisome components through a complex interaction network. We and others previously showed that the FtsZ-ring is not a smooth, uniformly organized structure, but rather a heterogeneously arranged assembly of FtsZ clusters [10–15]. Here, we show that ZapA also displays a heterogeneous organization that morphologically mimics the FtsZ-ring. This heterogeneity was in contrast to the largely uniform appearance of ZapB-mEos2, indicating that the heterogeneous nature of FtsZ and ZapA is specific to their assembly.
We additionally show that the structures of both ZapA and ZapB can significantly deviate from those of FtsZ using two-color PALM. These observations suggest that, although the midcell localization of ZapA and ZapB is dependent on FtsZ, their structures do not replicate that of FtsZ, and there is unlikely a strictly-defined protein complex with fixed stoichiometry between FtsZ, ZapA and ZapB. Note that in a recent superresolution study, structural deviations of FtsA and ZipA from FtsZ were also observed [47], suggesting that this may be a common feature of divisome assembly. Although no defined complex appears to predominate, we find it interesting that the radial separation between FtsZ and ZapB is conserved throughout constriction (Fig 2E). We reason that maintaining the relative spatial arrangement between FtsZ and ZapB structures may aid the efficiency of constriction.
What leads to the apparent structural deviations of ZapA and ZapB from FtsZ? It is likely that the inherent oligomerization properties of these proteins coupled with their interactions with other division proteins play a large role. In vitro biochemical studies have shown that FtsZ polymers exhibit remarkable polymorphism, assembling into single-stranded protofilaments, sheets, bundles, helices and toroids [48]. ZapA itself does not extensively polymerize but exists in a dimer-tetramer equilibrium [49]; ZapB, on the contrary, readily forms large, bundled polymers [19]. This self-polymerization capability of ZapB likely contributes to its ability to substantially deviate from FtsZ structures.
The protein interactions exhibited by FtsZ, ZapA, and ZapB are also consistent with their locations relative to FtsZ. Bacterial two-hybrid and in vivo FRET have shown that FtsZ mainly interacts with FtsA, ZipA, ZapA, and FtsK, while ZapA associates with a large number of inner membrane proteins that do not interact with FtsZ, including: FtsQ, FtsL, FtsB, FtsW and FtsN [50–52]. Notably, these latter proteins are involved in septum synthesis during cell wall constriction. The membrane-proximal location of ZapA is conducive to interactions between ZapA and these proteins, and these interactions may facilitate the constriction progress. It will be interesting to apply the tools developed in this study to investigate the degree of colocalization of other divisome proteins with FtsZ and ZapA. Different colocalization patterns may reflect specific roles for FtsZ and ZapA in supporting these proteins’ functions in cell wall constriction, further elucidating the relationship between the structural organization and function of the divisome.
ZapB does not exhibit the interaction promiscuity of ZapA, and has only been shown to interact with ZapA and MatP [26,29]. These limited protein-protein interactions are consistent with the fact that, compared to ZapA, ZapB is displaced an additional ~40 nm into the cytoplasm, near MatP. It is likely that the morphology and localization of FtsZ, ZapA and ZapB structures are greatly influenced by their respective interacting partners, and that these interactions are not uniformly distributed across the structures.
Role of the FtsZ-ZapA-ZapB-MatP network in positioning the division plane
Correctly positioning the FtsZ-ring at the midcell in E. coli is largely attributed to two negative regulatory systems: MinCDE and SlmA [53]. MinCDE is a three-component system that oscillates from pole to pole, preventing polar FtsZ-ring formation [54]. SlmA is a DNA-activated FtsZ antagonist that prevents FtsZ-ring formation over the bulk nucleoid regions except the ter macrodomain [55]. Together, these two systems create a midcell zone where the FtsZ-ring can stably polymerize. Here we show that a third system, the ZapA-ZapB-MatP network, also contributes to the midcell positioning of the FtsZ-ring by providing a physical tether to the ter region of the chromosome. Disabling this linkage by deleting any one of the three proteins leads to dispersed FtsZ clusters in a large region around the midcell [14]. Importantly, the effect in the absence of MatP is not solely mediated by abnormally distributed SlmA, as FtsZ mislocalization persists in a ΔmatPΔslmA strain. We postulate that as the ter region moves toward the midcell at the beginning of cell cycle and resides there until the end of DNA replication [28], it provides a convenient positive positioning system to reinforce the midcell localization of the divisome—MatP promotes the localization of ZapB to midcell through its direct interaction with ZapB, which in turn influences the localization of ZapA and further FtsZ. This mechanism is supported by recent work from the Männik and Sherratt groups, who showed that the FtsZ-ring colocalizes with the MatP-bound ter macrodomain at the center of nucleoid in cells depleted of both MinC and SlmA [30]. Interestingly, this colocalization was diminished, but not completely abolished, in the absence of all the three systems, suggesting the presence of other positioning systems. The variety and redundancy of positioning systems highlights the importance of the FtsZ-ring and the robust nature of its highly-evolved regulatory system.
Role of the FtsZ-ZapA-ZapB-MatP linkage in coordinating division with nucleoid segregation
One unexpected finding of this study is that the deletion of matP leads to a faster cell wall constriction rate. This finding is counter-intuitive, because we have shown that the presence of MatP and the associated protein network helps to position and maintain the FtsZ-ring at the midcell, and the deletion of MatP leads to mislocalized FtsZ-ring and faster turnover dynamics of ZapA and ZapB. If the stability of the FtsZ-ring is indeed essential for efficient cell wall constriction, we should expect the deletion of MatP to slow down constriction, instead of speeding it up.
One possible explanation for this apparent paradox is that the cell wall constriction machinery has the ability to proceed much faster than nucleoid segregation. However, under normal conditions, cell wall constriction does not proceed at its maximum speed because some divisome constituents may exist to impede, or slow down cell wall constriction to allow time for nucleoid segregation. If the rates of the two processes are not balanced, a septum could form over an unsegregated nucleoid, which is detrimental for cell division.
How would MatP exert its influence on cell wall constriction rate? This influence could be transmitted through the network of FtsZ-ZapA-ZapB-MatP, or by the altered distribution of SlmA on the nucleoid. We showed in this study that the effects of ΔmatP on FtsZ mislocalization and constriction rate remained in the ΔmatPΔslmA double deletion strain, suggesting that the physical linkage of FtsZ-ZapA-ZapB-MatP indeed plays a direct role. We propose that there could be two different mechanisms mediated by the linkage to explain the influence of MatP on cell wall constriction rate.
In the first mechanism, the physical linkage between the membrane and nucleoid by the FtsA-FtsZ-ZapA-ZapB-MatP protein network could act as a steric hindrance to prevent cell wall constriction from proceeding too fast. As both ends of the linkage are anchored, this protein network effectively couples cell wall and nucleoid segregation mechanically so that cell wall constriction can only complete when the ter macrodomains are resolved at the end of DNA replication and moved outside of the midcell. If this linkage is disabled, unchecked cell wall constriction may not always allow sufficient time for complete nucleoid segregation, perhaps explaining the occasional nucleoid segregation defects observed in cells lacking MatP [28]. This mechanism is similar to one previously proposed for FtsK, which can interact with divisome proteins (FtsZ, FtsL, FtsQ) and the chromosome [56]. A recent study found that the ordered segregation of sister chromosomes by FtsK requires the presence of MatP, suggesting that these two proteins may coordinate with each other [57].
An alternative mechanism could be that the additional stability provided by the presence of the ZapA-ZapB-MatP network actually inhibits the ability of FtsZ to exert an active force to drive cell wall constriction. The inhibitory effect of an overly-stable FtsZ-ring is supported by the lethality of FtsZ overexpression [58]. In this mechanism, MatP facilitates the midcell localization of ZapA by interacting with ZapB, which further promotes the inhibitory bundling effect of ZapA on FtsZ. In vitro it has been shown that ZapA promotes FtsZ polymerization [18,24]. We and others showed that ZapA and ZapB promote FtsZ-ring assembly in vivo by aligning and corralling FtsZ polymers at the midcell. It may be possible that a less bundled, highly dynamic FtsZ-ring could be more active in directing/driving cell constriction.
One possible way to differentiate the above two mechanisms would be to examine the constriction rate in a ΔzapA or ΔzapB strain. If the physical, steric hindrance mechanism has a larger role, disabling the linkage by deleting zapA or zapB should have the same effect as deleting matP. If the activity inhibition mechanism has a larger role, deleting zapA or zapB should have a larger impact on cell wall constriction rate. This is because ZapA and ZapB can still localize to the midcell to inhibit FtsZ activity in the absence of MatP, albeit less efficiently. Previously we have observed that a subpopulation of ΔzapA and ΔzapB cells had much faster cell cycle time than wt cells, which we attributed to the ability of rapid reinitiation of previously primed division sites. We were unable to quantitatively verify whether the constriction rates of these cells were indeed faster than that in ΔmatP cells due to the highly dynamic nature of FtsZ-ring and abnormal septum formation in ΔzapA and ΔzapB cells. It would be interesting to design further experiments to examine these hypotheses.
Materials and Methods
Bacterial strains, growth conditions and materials
Bacterial strains and plasmids are indicated in S2 Table. Construction of strains and plasmids is detailed in the S1 Text and primers used are listed in S3 Table. Prior to imaging, all cells were grown from a single colony in LB media overnight at 37°C. For our default slow growth condition, cells were then diluted in M9 minimal media supplemented with 0.4% Glucose, MEM Vitamins and MEM Amino Acids (M9+), and grown at room temperature (RT) for at least 20 hrs. For our fast growth condition, cells were diluted in EZ Rich Defined Media (Teknova) supplemented with 0.4% Glucose and incubated at 37°C. When appropriate, we added 150 μg ml-1 chloramphenicol, 50 μg ml-1 kanamycin, 50 μg ml-1 carbenicillin or 100 μg ml-1 spectinomycin. Expression of FtsZ-mEos2 (pJB042) and mEos2-ZapA (pJB051) was induced with 20 μM IPTG for 2 hrs. Expression of ZapB-mEos2 (pJB045) and MatP-mEos2 (pJB128) was induced with 5–10 or 33 μM IPTG for 1hr, respectively. The dual-labeled Dronpa-ZapA—FtsZ-PAmCherry1 (pJB089) and Dronpa-ZapA—PAmCherry1-ZapA (pJB090) strains were induced with 20 μM IPTG for 2 hrs. For the ZapB-FtsZ 2C-PALM sample, FtsZ-PAmCherry1 (pJB066) was induced with 0.2% Arabinose for 30 min, while ZapB-Dronpa (pJB073) was uninduced. For all constructs, induction was followed by a washing step and a 2–3 hr outgrowth at RT without inducer. Fixation for iPALM samples was performed using 4% (v/v) formaldehyde in PBS (pH 7.4) for 45 min at RT.
Fluorescence imaging
All ensemble and PALM image acquisition and sample preparation were performed as described previously [10,59]. All PALM images were constructed from 3,000 frames acquired at a frame rate of ~100 s-1 with a constant 405 nm activation (~5 W cm-2). Single-molecule identification and image reconstruction were described previously [59]. Images are displayed in pseudo-color ('Red Hot') via ImageJ with a pixel size of 15 nm. Measurements of cell length and band width were performed using custom MATLAB (The MathWorks, Inc., Natick, MA) software and are described elsewhere [10,59]. Diameters of band-like structures were determined by first projecting the band intensity along the short axis of the cell. Peak intensities were then identified using the 'findpeaks' MATLAB function and the distance between the two most distal peaks was calculated. Immuno-based superresolution (STORM)[60] imaging of FtsZ was performed with Alexa Fluor 568 Goat Anti-Rabbit IgG (Invitrogen, GAR-568), as described previously with α-FtsZ (a gift from H. Erickson). STORM imaging of ZapB was performed similarly with α-ZapB (a gift from K. Gerdes) and GAR-568 applied at 1 : 500 and 1 : 1,000, respectively.
Two-color PALM imaging utilized the OptoSplit II (Andor) device, which simultaneously projected the two emission signals onto separate halves of the CCD chip. We obtained 1,500 frames at a rate of 100 s-1 using 405-nm activation at ~500 mW cm-2 followed by a second 1,500-frame acquisition at ~5 W cm-2. Both acquisitions employed a constant 488-nm and 561-nm excitation at a power density of ~1 kW cm-2.
Image overlay of the two-color images employed a transformation step that was achieved by using the multi-colored emission spectrum of 100 nm TetraSpeck beads (Life Technologies, Inc.), as described previously [61]. Briefly, we acquired hundreds of snapshots of single TetraSpeck beads simultaneously in both channels at various positions across the imaging region, generating a large dataset of control points. We then calculated the transformation of two-color data using these control points and the 'cp2tform' function in MATLAB [62]. This type of global transformations resulted in ~18-nm registration error in our microscope setup.
iPALM imaging and sample preparation was performed as described previously [37,38] with the following exceptions. Gold-embedded coverslips were coated with 0.1% Poly-L-Lysine (Ted Pella) for 40 min, washed with PBS (pH 7.4), and dried with purified air. Alexa Fluor 568 carboxylic acid (Life Technologies, Inc.) was diluted (~2e-9) in PBS (pH 7.4), applied to the coverslip for 15 min, and then washed and dried as above. Fixed cellular samples were subsequently applied in a similar fashion. Each image was produced from 45,000-100,000 frames. The average spatial resolution obtained for the x-, y - and z-axes was 21 nm, 23 nm, and 17 nm, respectively.
We obtained at least three biological replicates for each fusion protein on different days. iPALM data were processed via the PeakSelector v9.3 software [37] to extract molecular coordinates and fitting errors. Further analysis was performed by custom MATLAB software. Specifically, for each iPALM image, we first determined the z-position of the coated coverslip surface by fitting the Alexa Fluor 568 signal to a Gaussian function. This fit was typically characterized by a FWHM of ~10 nm. We then drew a user-defined box centered at the bottom of the cell, which in general had a z-depth of 150 nm and width of 200 nm to avoid molecules along the curved regions (Fig 3). The z-positions of all molecules in this box were then averaged and the relative distance to the coverslip was taken as the mean z-position of the protein in the cell.
Estimating mEos2 fusion concentration
We used the fluorescence intensity of mEos2 after excitation with a 488-nm laser to quantify the relative expression levels of FtsZ-mEos2, mEos2-ZapA and ZapB-mEos2. We used custom MATLAB software to measure the integrated cellular intensity (A.U.) of each fusion protein: FtsZ = 46,000 ± 2,000, ZapA = 94,000 ± 5,000, and ZapB = 40,000 ± 3,000 (x - ± se). Taking into account the previously determined expression level of FtsZ-mEos2 under the same growth conditions (30% of FtsZtotal) [14] and the previously determined endogenous concentrations of the three proteins (FtsZ ≈ 5,000, ZapA ≈ 5,000 and ZapB ≈ 15,000 molecules cell-1) [10,14,18,19,63], we estimate that the fusions are present at 30%, 45% and 10% of the total (wt + fusion) protein concentration.
Cross-correlation analysis of 2C-PALM images
We used a previously published coordinate-based cross-correlation analysis to identify the apparent displacement between different protein pairs in 2C-PALM images [34,35]. Briefly, regions around the midcell of each 2C-PALM image were cropped and the cross-correlation between the two proteins of interest was calculated as a function of displacement along the short axis of the cell. Apparent displacement between a given protein pair in a single cell was defined as the displacement value that exhibited maximal cross-correlation. Fig 2D shows the distributions of this value for FtsZ-ZapA, FtsZ-ZapB and ZapA-ZapA pairs. The mean apparent displacement value for ZapA-ZapA (33 nm) reflects our resolution in determining the displacement between two protein structures.
Fluorescence recovery after photobleaching
Fluorescence recovery after photobleaching (FRAP) was carried out on wild-type BW25113 cells or ΔmatP mutants (JW0939) ectopically expressing FtsZ-GFP (pXY027), GFP-ZapA (pJB154) or GFP-ZapB (pJB150) in the absence of inducer. Imaging was conducted on cells immobilized on 3% Agarose gel pad (M9+ without MEM Vitamins) using an Olympus IX-71 inverted microscope with excitation from a 488-nm laser (OBIS, Coherent, Santa Clara, CA). The excitation laser was passed through a linear polarizer (Thorlabs, Newton, NJ), expanded to a 10mm diameter, and split with a polarizing beamsplitting cube (Thorlabs, Newton, NJ) to generate two excitation beams. The reflected beam was then focused with an achromatic doublet lens (f = 750.0 mm, Ø2", Thorlabs, Newton, NJ) to allow epi-fluorescence illumination and recombined with the transmitted beam by another polarizing beamsplitting cube. Both beams illuminated the sample through a 100×, 1.45 NA TIRFM objective. The transmitted beam was focused by the objective to a ~300 nm radius spot for photobleaching and controlled by an independent shutter. The epi-fluorescence imaging beam (reflected beam) was focused to a radius of ~50 μm. The intensity of the photobleaching beam and imaging beam were 2 kW/cm2 and 5 W/cm2, respectively.
For imaging, the focus of the photobleaching beam was positioned to the midcell periphery of an individual cell. Timelapse fluorescence images were then acquired with the imaging beam at a rate of 30 min-1 for 4 min. The photobleaching step occurred during the second timelapse acquisition, all other frames were acquired using epi-fluorescence illumination. The exposure time for all acquisitions was 50 ms.
For FRAP analysis, we cropped the bleached area and whole midcell region and plotted their average intensity in each frame against time: IBleach(N),IMidcell(N), where N is the frame number. We calculated the bleaching ratio as (IBleach(1)-IBleach(3))/IBleach(3))IBleach(1), and only used trajectories where this value was greater than 40%. We normalized individual trajectories to [0,1], with the first acquisition post-photobleach (IBleach (3)) set as 0. We typically observed large fluctuations at the tail end of the bleached area (IBleach(N)) trajectories, whereas trajectories of the whole midcell region (IMidcell(N)) were much more stable. Consequently, we used the average intensity of last 60 frames of the whole midcell region (< IMidcell(61:120) >) as the maximum to normalize each bleach trajectory (IBleach,Nor(N)).
We averaged the normalized trajectories of each dataset across all cells from at least two days of imaging and estimated their recovery rates by fitting the data to single exponential functions. These normalized trajectories were then resampled 3000 times by bootstrapping. The resampled trajectories were then averaged and fit to obtain the recovery half-time. We then calculated the standard error from the distribution of bootstrapped recovery half-time.
Time-lapse fluorescence imaging and analysis for constriction periods
Imaging of FtsZ-GFP in BW25113, ΔmatP and ΔmatPΔslmA was performed using plasmid JW0093 [64]. Each strain was first inoculated from a fresh colony into LB, grown overnight at 37°C to saturation, then diluted 1 : 100 into M9+ media and grown overnight at 30°C to saturation. This saturated culture was then diluted 1 : 200 into fresh M9+ media and grown at RT until mid-log phase (0.2–0.4 OD600). This 3-step culture preparation ensured similar expression levels of FtsZ-GFP among all three strains (see S10 Fig). The midlog culture was deposited onto a 3% agarose gel pad made with M9+ lacking MEM vitamins. Bright-field and green fluorescence images were acquired at 5-minute intervals. Constriction onset was determined from bright-field images as the first frame with visible cell wall indentation (S11 Fig). To limit interference from adjacent cells, only cells located at the outer edge of the growing microcolony were analyzed. The end of constriction, and thus the end of the mother cell cycle, and beginning of daughter cell cycles, was determined by the complete and unrecovered loss of GFP fluorescence from midcell (S11 Fig). Measurement bias in identification of constriction onset and completion may lead to underestimation of constriction times because detection of cell wall indentation is diffraction-limited [65] and detection of fluorescence loss at constriction completion is limited by signal sensitivity. However, comparison of relative constriction times between the three strains remains valid as the same criteria were applied consistently between all three strains.
Supporting Information
Zdroje
1. Egan AJF, Vollmer W (2012) The physiology of bacterial cell division. Annals of the New York Academy of Sciences 1277 : 8–28. doi: 10.1111/j.1749-6632.2012.06818.x 23215820
2. Harry E, Monahan L, Thompson L (2006) Bacterial Cell Division: The Mechanism and Its Precison. International Review of Cytology. Elsevier, Vol. 253. pp. 27–94. doi: 10.1016/S0074-7696(06)53002-5
3. Bi EF, Lutkenhaus (1991) FtsZ ring structure associated with division in Escherichia coli. Nature 354 : 161–164. doi: 10.1038/354161a0 1944597
4. Addinall SG, Bi E, Lutkenhaus (1996) FtsZ ring formation in fts mutants. Journal of Bacteriology 178 : 3877–3884. 8682793
5. Osawa M, Anderson DE, Erickson HP (2009) Curved FtsZ protofilaments generate bending forces on liposome membranes. The EMBO Journal 28 : 3476–3484. doi: 10.1038/emboj.2009.277 19779463
6. Osawa M, Anderson DE, Erickson HP (2008) Reconstitution of Contractile FtsZ Rings in Liposomes. Science 320 : 792–794. doi: 10.1126/science.1154520 18420899
7. Chen Y (2005) Rapid in Vitro Assembly Dynamics and Subunit Turnover of FtsZ Demonstrated by Fluorescence Resonance Energy Transfer. Journal of Biological Chemistry 280 : 22549–22554. doi: 10.1074/jbc.M500895200 15826938
8. Hale CA, de Boer PA (1997) Direct binding of FtsZ to ZipA, an essential component of the septal ring structure that mediates cell division in E. coli. Cell 88 : 175–185. 9008158
9. Pichoff S, Lutkenhaus (2005) Tethering the Z ring to the membrane through a conserved membrane targeting sequence in FtsA. Mol Microbiol 55 : 1722–1734. doi: 10.1111/j.1365-2958.2005.04522.x 15752196
10. Fu G, Huang T, Buss JA, Coltharp C, Hensel Z, et al. (2010) In Vivo Structure of the E. coli FtsZ-ring Revealed by Photoactivated Localization Microscopy (PALM). PLoS ONE 5: e12680. doi: 10.1371/journal.pone.0012680 20856929
11. Jennings PC, Cox G, Monahan LG, Harry E (2011) Super-resolution imaging of the bacterial cytokinetic protein FtsZ. Micron 42 : 336–341. doi: 10.1016/j.micron.2010.09.003 20933427
12. Biteen JS, Goley ED, Shapiro L, Moerner WE (2012) Three-Dimensional Super-Resolution Imaging of the Midplane Protein FtsZ in Live Caulobacter crescentusCells Using Astigmatism. ChemPhysChem 13 : 1007–1012. doi: 10.1002/cphc.201100686 22262316
13. Strauss MP, Liew ATF, Turnbull L, Whitchurch CB, Monahan LG, et al. (2012) 3D-SIM Super Resolution Microscopy Reveals a Bead-Like Arrangement for FtsZ and the Division Machinery: Implications for Triggering Cytokinesis. Plos Biol 10: e1001389. doi: 10.1371/journal.pbio.1001389 22984350
14. Buss JA, Coltharp C, Huang T, Pohlmeyer C, Wang S-C, et al. (2013) In vivo organization of the FtsZ-ring by ZapA and ZapB revealed by quantitative super-resolution microscopy. Mol Microbiol 89 : 1099–1120. doi: 10.1111/mmi.12331 23859153
15. Holden SJ, Pengo T, Meibom KL, Fernandez Fernandez C, Collier J, et al. (2014) High throughput 3D super-resolution microscopy reveals Caulobacter crescentus in vivo Z-ring organization. Proc Natl Acad Sci USA 111 : 4566–4571. doi: 10.1073/pnas.1313368111 24616530
16. Stricker J, Maddox P, Salmon ED, Erickson HP (2002) Rapid assembly dynamics of the Escherichia coli FtsZ-ring demonstrated by fluorescence recovery after photobleaching. Proceedings of the National Academy of Sciences 99 : 3171–3175. doi: 10.1073/pnas.052595099 11854462
17. Anderson DE, Gueiros-Filho FJ, Erickson HP (2004) Assembly Dynamics of FtsZ Rings in Bacillus subtilis and Escherichia coli and Effects of FtsZ-Regulating Proteins. Journal of Bacteriology 186 : 5775–5781. doi: 10.1128/JB.186.17.5775-5781.2004 15317782
18. Gueiros-Filho FJ (2002) A widely conserved bacterial cell division protein that promotes assembly of the tubulin-like protein FtsZ. Genes & Development 16 : 2544–2556. doi: 10.1101/gad.1014102
19. Ebersbach G, Galli E, Møller-Jensen J, Löwe J, Gerdes K (2008) Novel coiled-coil cell division factor ZapB stimulates Z ring assembly and cell division. Mol Microbiol 68 : 720–735. doi: 10.1111/j.1365-2958.2008.06190.x 18394147
20. Durand-Heredia J, Yu HH, De Carlo S, Lesser CF, Janakiraman A (2011) Identification and Characterization of ZapC, a Stabilizer of the FtsZ Ring in Escherichia coli. Journal of Bacteriology 193 : 1405–1413. doi: 10.1128/JB.01258-10 21216995
21. Hale CA, Shiomi D, Liu B, Bernhardt T, Margolin W, et al. (2011) Identification of Escherichia coli ZapC (YcbW) as a Component of the Division Apparatus That Binds and Bundles FtsZ Polymers. Journal of Bacteriology 193 : 1393–1404. doi: 10.1128/JB.01245-10 21216997
22. Durand-Heredia J, Rivkin E, Fan G, Morales J, Janakiraman A (2012) Identification of ZapD as a Cell Division Factor That Promotes the Assembly of FtsZ in Escherichia coli. Journal of Bacteriology 194 : 3189–3198. doi: 10.1128/JB.00176-12 22505682
23. Marteyn BS, Karimova G, Fenton AK, Gazi AD, West N, et al. (2014) ZapE is a novel cell division protein interacting with FtsZ and modulating the Z-ring dynamics. mBio 5: e00022–14. doi: 10.1128/mBio.00022-14 24595368
24. Small E, Marrington R, Rodger A, Scott DJ, Sloan K, et al. (2007) FtsZ Polymer-bundling by the Escherichia coli ZapA Orthologue, YgfE, Involves a Conformational Change in Bound GTP. Journal of Molecular Biology 369 : 210–221. doi: 10.1016/j.jmb.2007.03.025 17428494
25. Dajkovic A, Pichoff S, Lutkenhaus, Wirtz D (2010) Cross-linking FtsZ polymers into coherent Z rings. Mol Microbiol 78 : 651–668. doi: 10.1111/j.1365-2958.2010.07352.x 20969647
26. Galli E, Gerdes K (2010) Spatial resolution of two bacterial cell division proteins: ZapA recruits ZapB to the inner face of the Z-ring. Mol Microbiol 76 : 1514–1526. doi: 10.1111/j.1365-2958.2010.07183.x 20487275
27. Galli E, Gerdes K (2011) FtsZ-ZapA-ZapB Interactome of Escherichia coli. Journal of Bacteriology 194 : 292–302. doi: 10.1128/JB.05821-11 22056926
28. Mercier R, Petit M-A, Schbath S, Robin S, Karoui El M, et al. (2008) The MatP/matS site-specific system organizes the terminus region of the E. coli chromosome into a macrodomain. Cell 135 : 475–485. doi: 10.1016/j.cell.2008.08.031 18984159
29. Espeli O, Borne R, Dupaigne P, Thiel A, Gigant E, et al. (2012) A MatP-divisome interaction coordinates chromosome segregation with cell division in E. coli. The EMBO Journal 31 : 3198–3211. doi: 10.1038/emboj.2012.128 22580828
30. Bailey MW, Bisicchia P, Warren BT, Sherratt DJ, Männik J (2014) Evidence for divisome localization mechanisms independent of the Min system and SlmA in Escherichia coli. PLoS Genet 10: e1004504. doi: 10.1371/journal.pgen.1004504 25101671
31. Betzig E, Patterson GH, Sougrat R, Lindwasser OW, Olenych S, et al. (2006) Imaging Intracellular Fluorescent Proteins at Nanometer Resolution. Science 313 : 1642–1645. doi: 10.1126/science.1127344 16902090
32. Coltharp C, Kessler RP, Xiao J (2012) Accurate Construction of Photoactivated Localization Microscopy (PALM) Images for Quantitative Measurements. PLoS ONE 7: e51725. doi: 10.1371/journal.pone.0051725 23251611
33. Rust MJ, Bates M, Zhuang X (2006) Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM). Nature Methods 3 : 793–796. doi: 10.1038/nmeth929 16896339
34. Veatch SL, Machta BB, Shelby SA, Chiang EN, Holowka DA, et al. (2012) Correlation Functions Quantify Super-Resolution Images and Estimate Apparent Clustering Due to Over-Counting. PLoS ONE 7: e31457. doi: 10.1371/journal.pone.0031457 22384026
35. Sengupta P, Jovanovic-Talisman T, Lippincott-Schwartz J (2013) Quantifying spatial organization in point-localization superresolution images using pair correlation analysis. Nat Protoc 8 : 345–354. doi: 10.1038/nprot.2013.005 23348362
36. Coltharp C, Yang X, Xiao J (2014) Quantitative analysis of single-molecule superresolution images. Curr Opin Struct Biol 28C: 112–121. doi: 10.1016/j.sbi.2014.08.008
37. Shtengel G, Galbraith JA, Galbraith CG, Lippincott-Schwartz J, Gillette JM, et al. (2009) Interferometric fluorescent super-resolution microscopy resolves 3D cellular ultrastructure. Proceedings of the National Academy of Sciences 106 : 3125–3130. doi: 10.1073/pnas.0813131106 19202073
38. Kanchanawong P, Shtengel G, Pasapera AM, Ramko EB, Davidson MW, et al. (2010) Nanoscale architecture of integrin-based cell adhesions. Nature 468 : 580–584. doi: 10.1038/nature09621 21107430
39. Matias VRF, Beveridge TJ (2005) Cryo-electron microscopy reveals native polymeric cell wall structure in Bacillus subtilis 168 and the existence of a periplasmic space. Mol Microbiol 56 : 240–251. doi: 10.1111/j.1365-2958.2005.04535.x 15773993
40. Sleytr UB, Schuster B, Egelseer E-M, Pum D (2014) S-layers: principles and applications. FEMS Microbiology Reviews 38 : 823–864. doi: 10.1111/1574-6976.12063 24483139
41. Zhang M, Chang H, Zhang Y, Yu J, Wu L, et al. (2012) Rational design of true monomeric and bright photoactivatable fluorescent proteins. Nature Methods: 1–6. doi: 10.1038/nmeth.2021 22312634
42. Bernhardt T, de Boer PA (2005) SlmA, a Nucleoid-Associated, FtsZ Binding Protein Required for Blocking Septal Ring Assembly over Chromosomes in. Molecular Cell 18 : 555–564. doi: 10.1016/j.molcel.2005.04.012 15916962
43. Dupaigne P, Tonthat NK, Espeli O, Whitfill T, Boccard FEDER, et al. (2012) Molecular Basis for a Protein-Mediated DNA-Bridging Mechanism that Functions in Condensation of the E. coli Chromosome. Molecular Cell 48 : 560–571. doi: 10.1016/j.molcel.2012.09.009 23084832
44. Geissler B, Shiomi D, Margolin W (2007) The ftsA* gain-of-function allele of Escherichia coli and its effects on the stability and dynamics of the Z ring. Microbiology 153 : 814–825. doi: 10.1099/mic.0.2006/001834-0 17322202
45. Erickson HP, Anderson DE, Osawa M (2010) FtsZ in Bacterial Cytokinesis: Cytoskeleton and Force Generator All in One. Microbiology and Molecular Biology Reviews 74 : 504–528. doi: 10.1128/MMBR.00021-10 21119015
46. Weiss DS (2004) Bacterial cell division and the septal ring. Mol Microbiol 54 : 588–597. doi: 10.1111/j.1365-2958.2004.04283.x 15491352
47. Rowlett VW, Margolin W (2014) 3D-SIM super-resolution of FtsZ and its membrane tethers in Escherichia coli cells. Biophysical Journal 107: L17–L20. doi: 10.1016/j.bpj.2014.08.024 25418183
48. Popp D, Iwasa M, Narita A, Erickson HP, Maéda Y (2009) FtsZ condensates: An in vitro electron microscopy study. Biopolymers 91 : 340–350. doi: 10.1002/bip.21136 19137575
49. Low HH, Moncrieffe MC, Löwe J (2004) The Crystal Structure of ZapA and its Modulation of FtsZ Polymerisation. Journal of Molecular Biology 341 : 839–852. doi: 10.1016/j.jmb.2004.05.031 15288790
50. Maggi S, Massidda O, Luzi G, Fadda D, Paolozzi L, et al. (2008) Division protein interaction web: identification of a phylogenetically conserved common interactome between Streptococcus pneumoniae and Escherichia coli. Microbiology 154 : 3042–3052. doi: 10.1099/mic.0.2008/018697-0 18832310
51. Alexeeva S, Gadella TWJ Jr, Verheul J, Verhoeven GS, Blaauwen den T (2010) Direct interactions of early and late assembling division proteins in Escherichia coli cells resolved by FRET. Mol Microbiol 77 : 384–398. doi: 10.1111/j.1365-2958.2010.07211.x 20497333
52. Pazos M, Natale P, Margolin W, Vicente M (2013) Interactions among the early Escherichia coli divisome proteins revealed by bimolecular fluorescence complementation. Environ Microbiol 15 : 3282–3291. doi: 10.1111/1462-2920.12225 23957637
53. Monahan LG, Liew ATF, Bottomley AL, Harry E (2014) Division site positioning in bacteria: one size does not fit all. Front Microbio 5 : 19. doi: 10.3389/fmicb.2014.00019
54. de Boer PA, Crossley RE, Rothfield LI (1989) A division inhibitor and a topological specificity factor coded for by the minicell locus determine proper placement of the division septum in E. coli. Cell 56 : 641–649. 2645057
55. Cho H, McManus HR, Dove SL, Bernhardt T (2011) Nucleoid occlusion factor SlmA is a DNA-activated FtsZ polymerization antagonist. Proc Natl Acad Sci USA 108 : 3773–3778. doi: 10.1073/pnas.1018674108 21321206
56. Grenga L, Luzi G, Paolozzi L, Ghelardini P (2008) The Escherichia coli FtsK functional domains involved in its interaction with its divisome protein partners. FEMS Microbiology Letters 287 : 163–167. doi: 10.1111/j.1574-6968.2008.01317.x 18759781
57. Stouf M, Meile J-C, Cornet F (2013) FtsK actively segregates sister chromosomes in Escherichia coli. Proc Natl Acad Sci USA 110 : 11157–11162. doi: 10.1073/pnas.1304080110 23781109
58. Dai K, Lutkenhaus (1992) The proper ratio of FtsZ to FtsA is required for cell division to occur in Escherichia coli. Journal of Bacteriology 174 : 6145–6151. 1400163
59. Buss JA, Coltharp C, Xiao J (2013) Super-resolution Imaging of the Bacterial Division Machinery. JoVE. doi: 10.3791/50048
60. Huang B, Wang W, Bates M, Zhuang X (2008) Three-dimensional super-resolution imaging by stochastic optical reconstruction microscopy. Science 319 : 810–813. doi: 10.1126/science.1153529 18174397
61. Hensel Z, Weng X, Lagda AC, Xiao J (2013) Transcription-Factor-Mediated DNA Looping Probed by High-Resolution, Single-Molecule Imaging in Live E. coli Cells. Plos Biol 11: e1001591. doi: 10.1371/journal.pbio.1001591 23853547
62. Churchman LS, Spudich JA (2012) Colocalization of Fluorescent Probes: Accurate and Precise Registration with Nanometer Resolution. Cold Spring Harbor Protocols 2012: pdb.top067918–pdb.top067918. doi: 10.1101/pdb.top067918 22301660
63. Mohammadi T, Ploeger GEJ, Verheul J, Comvalius AD, Martos A, et al. (2009) The GTPase activity of Escherichia coli FtsZ determines the magnitude of the FtsZ polymer bundling by ZapA in vitro. Biochemistry 48 : 11056–11066. doi: 10.1021/bi901461p 19842714
64. Kitagawa M, Ara T, Arifuzzaman M, Ioka-Nakamichi T, Inamoto E, et al. (2006) Complete set of ORF clones of Escherichia coli ASKA library (A Complete Set of E. coli K-12 ORF Archive): Unique Resources for Biological Research. DNA Research 12 : 291–299. doi: 10.1093/dnares/dsi012
65. Reshes G, Vanounou S, Fishov I, Feingold M (2008) Timing the start of division in E. coli: a single-cell study. Phys Biol 5 : 046001. doi: 10.1088/1478-3975/5/4/046001 18997273
66. Li Y, Hsin J, Zhao L, Cheng Y, Shang W, et al. (2013) FtsZ Protofilaments Use a Hinge-Opening Mechanism for Constrictive Force Generation. Science 341 : 392–395. doi: 10.1126/science.1239248 23888039
Štítky
Genetika Reprodukční medicína
Článek Retraction: Astakine 2—the Dark Knight Linking Melatonin to Circadian Regulation in CrustaceansČlánek Adventures in WonderlandČlánek Genomic Location of the Major Ribosomal Protein Gene Locus Determines Global Growth and InfectivityČlánek Spatial Fluctuations in Expression of the Heterocyst Differentiation Regulatory Gene in FilamentsČlánek Genome-Wide Negative Feedback Drives Transgenerational DNA Methylation Dynamics in ArabidopsisČlánek Systematic Dissection of the Sequence Determinants of Gene 3’ End Mediated Expression ControlČlánek The Chromatin Remodeler CHD8 Is Required for Activation of Progesterone Receptor-Dependent EnhancersČlánek Selection against Heteroplasmy Explains the Evolution of Uniparental Inheritance of MitochondriaČlánek The DNA Helicase Recql4 Is Required for Normal Osteoblast Expansion and Osteosarcoma FormationČlánek Dual-Specificity Anti-sigma Factor Reinforces Control of Cell-Type Specific Gene Expression in
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2015 Číslo 4
-
Všechny články tohoto čísla
- Retraction: Astakine 2—the Dark Knight Linking Melatonin to Circadian Regulation in Crustaceans
- Adventures in Wonderland
- Experimental Swap of 's Assortative Mating Preferences Demonstrates Key Role of X-Chromosome Divergence Island in Incipient Sympatric Speciation
- Chromosome Replacement and Deletion Lead to Clonal Polymorphism of Berry Color in Grapevine
- The Protein Quality Control Machinery Regulates Its Misassembled Proteasome Subunits
- Genome-Wide Association Study Identifies as a Critical Gene for Susceptibility to Noise-Induced Hearing Loss
- Genomic Location of the Major Ribosomal Protein Gene Locus Determines Global Growth and Infectivity
- Viable Neuronopathic Gaucher Disease Model in Medaka () Displays Axonal Accumulation of Alpha-Synuclein
- Multi-locus Analysis of Genomic Time Series Data from Experimental Evolution
- The Genetic Legacy of the Expansion of Turkic-Speaking Nomads across Eurasia
- Lack of GDAP1 Induces Neuronal Calcium and Mitochondrial Defects in a Knockout Mouse Model of Charcot-Marie-Tooth Neuropathy
- The Pif1 Helicase, a Negative Regulator of Telomerase, Acts Preferentially at Long Telomeres
- Inhibiting K63 Polyubiquitination Abolishes No-Go Type Stalled Translation Surveillance in
- SYD-1C, UNC-40 (DCC) and SAX-3 (Robo) Function Interdependently to Promote Axon Guidance by Regulating the MIG-2 GTPase
- Spatial Fluctuations in Expression of the Heterocyst Differentiation Regulatory Gene in Filaments
- Synergistic and Independent Actions of Multiple Terminal Nucleotidyl Transferases in the 3’ Tailing of Small RNAs in Arabidopsis
- Host Genetic Variation Influences Gene Expression Response to Rhinovirus Infection
- Contribution of Large Region Joint Associations to Complex Traits Genetics
- Volatility of Mutator Phenotypes at Single Cell Resolution
- Proteolysis of Virulence Regulator ToxR Is Associated with Entry of into a Dormant State
- Genome-Wide Negative Feedback Drives Transgenerational DNA Methylation Dynamics in Arabidopsis
- A Multi-layered Protein Network Stabilizes the FtsZ-ring and Modulates Constriction Dynamics
- Systematic Dissection of the Sequence Determinants of Gene 3’ End Mediated Expression Control
- Genome Sequencing of the Perciform Fish Provides Insights into Molecular and Genetic Mechanisms of Stress Adaptation
- Natural Variant E610G Is a Semi-dominant Suppressor of IAP-Induced RNA Processing Defects
- The Alkaline Response Pathway: Identification of a Novel Rim Pathway Activator
- Transgenerational Inheritance of Diet-Induced Genome Rearrangements in Drosophila
- A Single Nucleotide Polymorphism Uncovers a Novel Function for the Transcription Factor Ace2 during Hyphal Development
- DNA Damage Response and Spindle Assembly Checkpoint Function throughout the Cell Cycle to Ensure Genomic Integrity
- The Functional Interplay Between the t(9;22)-Associated Fusion Proteins BCR/ABL and ABL/BCR in Philadelphia Chromosome-Positive Acute Lymphatic Leukemia
- Extreme Recombination Frequencies Shape Genome Variation and Evolution in the Honeybee,
- Beyond Glycolysis: GAPDHs Are Multi-functional Enzymes Involved in Regulation of ROS, Autophagy, and Plant Immune Responses
- Comprehensive Profiling of Amino Acid Response Uncovers Unique Methionine-Deprived Response Dependent on Intact Creatine Biosynthesis
- Windpipe Controls Intestinal Homeostasis by Regulating JAK/STAT Pathway via Promoting Receptor Endocytosis and Lysosomal Degradation
- Ataxin-2 Regulates Translation in a New BAC-SCA2 Transgenic Mouse Model
- Cross-Population Joint Analysis of eQTLs: Fine Mapping and Functional Annotation
- The Power of Gene-Based Rare Variant Methods to Detect Disease-Associated Variation and Test Hypotheses About Complex Disease
- The Chromatin Remodeler CHD8 Is Required for Activation of Progesterone Receptor-Dependent Enhancers
- Competition between VanU Repressor and VanR Activator Leads to Rheostatic Control of Vancomycin Resistance Operon Expression
- A Missense Change in the Gene Links Aberrant Autophagy to a Neurodegenerative Vacuolar Storage Disease
- Simultaneous Discovery, Estimation and Prediction Analysis of Complex Traits Using a Bayesian Mixture Model
- Selection against Heteroplasmy Explains the Evolution of Uniparental Inheritance of Mitochondria
- Genome-Destabilizing Effects Associated with Top1 Loss or Accumulation of Top1 Cleavage Complexes in Yeast
- Imputation-Based Population Genetics Analysis of Malaria Parasites
- Heterozygosity for a Hypomorphic Polβ Mutation Reduces the Expansion Frequency in a Mouse Model of the Fragile X-Related Disorders
- Neto-Mediated Intracellular Interactions Shape Postsynaptic Composition at the Neuromuscular Junction
- Ndd1 Turnover by SCF Is Inhibited by the DNA Damage Checkpoint in
- Frameshift Variant Associated with Novel Hoof Specific Phenotype in Connemara Ponies
- The DNA Helicase Recql4 Is Required for Normal Osteoblast Expansion and Osteosarcoma Formation
- Spastin Binds to Lipid Droplets and Affects Lipid Metabolism
- Maintenance of Glia in the Optic Lamina Is Mediated by EGFR Signaling by Photoreceptors in Adult Drosophila
- Auxin Influx Carriers Control Vascular Patterning and Xylem Differentiation in
- Dual-Specificity Anti-sigma Factor Reinforces Control of Cell-Type Specific Gene Expression in
- The Lowe Syndrome Protein OCRL1 Is Required for Endocytosis in the Zebrafish Pronephric Tubule
- Postnatal Loss of Hap1 Reduces Hippocampal Neurogenesis and Causes Adult Depressive-Like Behavior in Mice
- CAPER Is Vital for Energy and Redox Homeostasis by Integrating Glucose-Induced Mitochondrial Functions via ERR-α-Gabpa and Stress-Induced Adaptive Responses via NF-κB-cMYC
- Distinct and Cooperative Activities of HESO1 and URT1 Nucleotidyl Transferases in MicroRNA Turnover in
- The Evolutionary Origination and Diversification of a Dimorphic Gene Regulatory Network through Parallel Innovations in and
- MAPK Signaling Pathway Alters Expression of Midgut ALP and ABCC Genes and Causes Resistance to Cry1Ac Toxin in Diamondback Moth
- Spatio-temporal Remodeling of Functional Membrane Microdomains Organizes the Signaling Networks of a Bacterium
- Asymmetric Transcript Discovery by RNA-seq in . Blastomeres Identifies , a Gene Important for Anterior Morphogenesis
- A Stress-Induced Small RNA Modulates Alpha-Rhizobial Cell Cycle Progression
- Systematic Profiling of Poly(A)+ Transcripts Modulated by Core 3’ End Processing and Splicing Factors Reveals Regulatory Rules of Alternative Cleavage and Polyadenylation
- The UPR Branch IRE1- in Plants Plays an Essential Role in Viral Infection and Is Complementary to the Only UPR Pathway in Yeast
- A Non-canonical RNA Silencing Pathway Promotes mRNA Degradation in Basal Fungi
- Co-chaperone p23 Regulates . Lifespan in Response to Temperature
- Re-replication of a Centromere Induces Chromosomal Instability and Aneuploidy
- Shade Avoidance Components and Pathways in Adult Plants Revealed by Phenotypic Profiling
- Lipid-Induced Epigenomic Changes in Human Macrophages Identify a Coronary Artery Disease-Associated Variant that Regulates Expression through Altered C/EBP-Beta Binding
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Lack of GDAP1 Induces Neuronal Calcium and Mitochondrial Defects in a Knockout Mouse Model of Charcot-Marie-Tooth Neuropathy
- Proteolysis of Virulence Regulator ToxR Is Associated with Entry of into a Dormant State
- Frameshift Variant Associated with Novel Hoof Specific Phenotype in Connemara Ponies
- Ataxin-2 Regulates Translation in a New BAC-SCA2 Transgenic Mouse Model
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání