-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaWorldwide Patterns of Ancestry, Divergence, and Admixture in Domesticated Cattle
The DNA of domesticated plants and animals contains information about how species were domesticated, exported, and bred by early farmers. Modern breeds were developed by lengthy and complex processes; however, our use of 134 breeds and new analytical models enabled us to reveal some of the processes that created modern cattle diversity. In Asia, Africa, North and South America, humpless (Bos t. taurus or taurine) and humped (Bos t. indicus or indicine) cattle were crossbred to produce hybrids adapted to the environment and local production systems. The history of Asian cattle involves the domestication and admixture of several species whereas African taurines arose through the introduction of domesticated Fertile Crescent taurines and their hybridization with wild African aurochs. African taurine genetic background is commonly observed among European Mediterranean breeds. The absence of indicine introgression within most European taurine breeds, but presence within three Italian breeds is consistent with at least two separate migration waves of cattle to Europe, one from the Middle East which captured taurines in which indicine introgression had already occurred and the second from western Africa into Spain with no indicine introgression. This second group seems to have radiated from Spain into the Mediterranean resulting in a cline of African taurine introgression into European taurines.
Published in the journal: . PLoS Genet 10(3): e32767. doi:10.1371/journal.pgen.1004254
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004254Summary
The DNA of domesticated plants and animals contains information about how species were domesticated, exported, and bred by early farmers. Modern breeds were developed by lengthy and complex processes; however, our use of 134 breeds and new analytical models enabled us to reveal some of the processes that created modern cattle diversity. In Asia, Africa, North and South America, humpless (Bos t. taurus or taurine) and humped (Bos t. indicus or indicine) cattle were crossbred to produce hybrids adapted to the environment and local production systems. The history of Asian cattle involves the domestication and admixture of several species whereas African taurines arose through the introduction of domesticated Fertile Crescent taurines and their hybridization with wild African aurochs. African taurine genetic background is commonly observed among European Mediterranean breeds. The absence of indicine introgression within most European taurine breeds, but presence within three Italian breeds is consistent with at least two separate migration waves of cattle to Europe, one from the Middle East which captured taurines in which indicine introgression had already occurred and the second from western Africa into Spain with no indicine introgression. This second group seems to have radiated from Spain into the Mediterranean resulting in a cline of African taurine introgression into European taurines.
Introduction
High-throughput genotyping assays have allowed population geneticists to use genome-wide marker sets to analyze the histories of many species, including human [1], cattle [2]–[4], sheep [5], dog [6], horse [7], yeast [8], mouse [9], [10], rice [11], [12], maize [13]–[16], grape [17], and wheat [18]. We previously described the phylogeny of domesticated bovine populations using their genetic variation inferred from a sample of 40,843 single-nucleotide polymorphisms (SNPs) [3]. Although we had sampled 48 cattle breeds, we did not have samples from key geographic regions including China and Southeast Asia, Anatolia, the Baltic States, southern and eastern Africa, and the Iberian Peninsula. As a consequence of those gaps in geographic sampling, we were unable to address the origins of cattle in these regions and the extent to which these cattle influenced the population structure of regions such as the New World.
We have now assembled a genomic data set which represents the largest population sampling of any mammalian species. This allows for an extremely detailed description of the population structure of domesticated cattle worldwide. Using this data set, we accurately establish the patterns of exportation, divergence, and admixture for domesticated cattle.
Results and Discussion
Worldwide patterns
We used principal component analysis (PCA) [19], ancestry graphs implemented in TreeMix [20], and ancestry models implemented in ADMIXTURE [21] to analyze the relationships between 134 breeds of domesticated bovids (Table S1). These breeds arose from three domesticated (sub)species: Bos javanicus, Bos taurus indicus and Bos taurus taurus (we use the terms breed and population interchangeably, due to the different definitions of breed worldwide). The principal source of SNP genotype variation was between B. t. taurus and B. t. indicus breeds (Figure 1). This split corresponds to the cattle which originated from the two separate major centers of domestication in the Fertile Crescent and Indus Valley [22]. Although Bos javanicus has a more distant common ancestor compared with Bos t. indicus and Bos t. taurus [3], the uneven sample sizes and ascertainment of SNPs common in Bos t. taurus in the design of the BovineSNP50 assay [23] caused the Bos t. indicus/Bos t. taurus split to be the main source of variation in these data. The second principal component split African taurine cattle from Eurasian taurine, indicine, and Bali cattle.
Fig. 1. Principal component analysis of 1,543 animals genotyped with 43,043 SNPs. 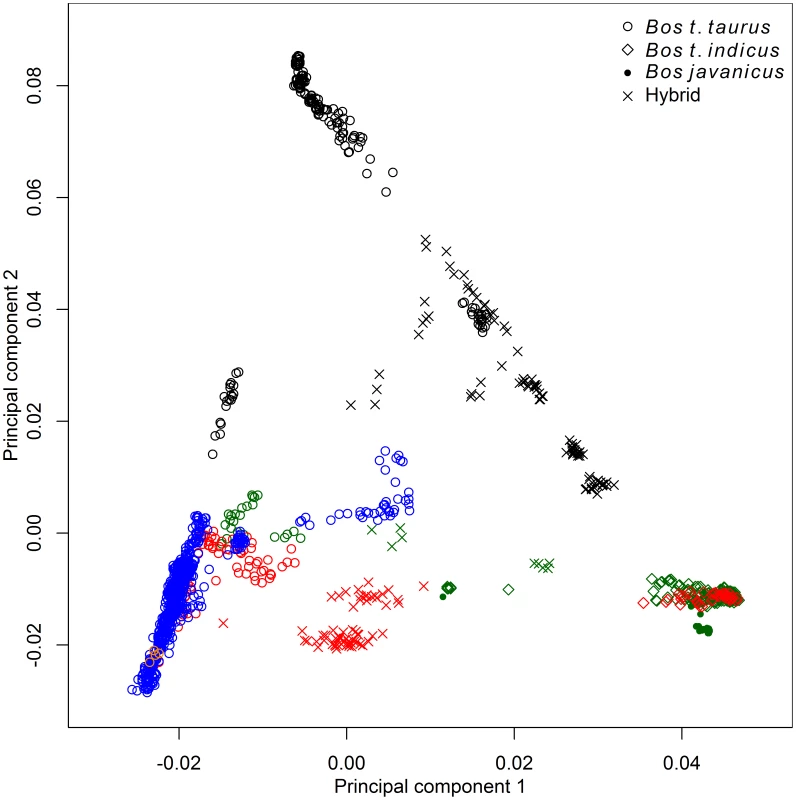
Points were colored according to geographic origin of breed; black: Africa, green: Asia, red: North and South America, orange: Australia, and blue: Europe. Early farmers were able to expand their habitat range because of the availability of a reliable supply of food and likely displaced indigenous hunter-gatherer populations by introducing new diseases [24]. The genomes of modern cattle reflect the history of animal movements by migratory farmers out of the ancient centers of cattle domestication. We first ran TreeMix with all 134 populations to identify patterns of divergence (Figure 2). We next ran TreeMix with 74 representative populations (Figure 3, residuals presented in Figure S1) and began to add migration edges to the phylogenetic model (Figure 4, residuals presented in Figure S2, see Methods for an explanation of TreeMix). The proportion of the variance in relatedness between populations explained by the model began to asymptote at 0.998 (a value also obtained by simulations [20]) when 17 migration edges were fit (Figure S3). The consistency of these migration edges was evaluated using 5 independent runs of TreeMix with 17 migration edges (Figure S4). In addition to the migratory routes previously described from the Fertile Crescent to Europe [3], we now find strong evidence of exportations from the Indian subcontinent to China and southeast Asia, India to Africa, Africa to the Iberian Peninsula and Mediterranean Europe, India to the Americas, and Europe to the Americas (Figures 4 and 5, discussed in detail in the following subsections). Subsequent to these initial exportations, there have been countless exportations and importations of cattle worldwide. When domesticated cattle were present and new germplasm was imported, the introduced cattle were frequently crossed with the local cattle resulting in an admixed population. Admixed populations were most readily identified when Bos t. indicus and Bos t. taurus animals were hybridized, which occurred in China, Africa, and the Americas (crosses in Figure 1).
Fig. 2. Phylogram of the inferred relationships between 134 cattle breeds. 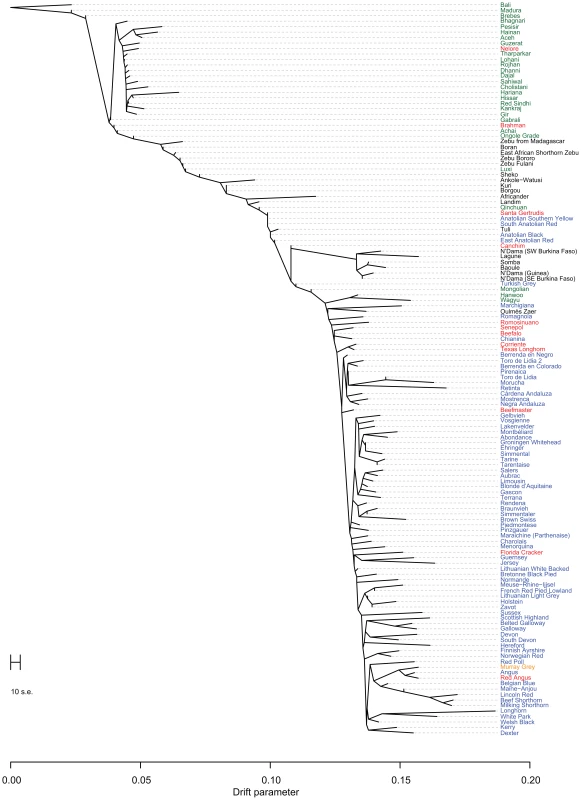
Breeds were colored according to their geographic origin; black: Africa, green: Asia, red: North and South America, orange: Australia, and blue: Europe. Scale bar shows 10 times the average standard error of the estimated entries in the sample covariance matrix (See [20]). Fig. 3. Phylogram of the inferred relationships between 74 cattle breeds. 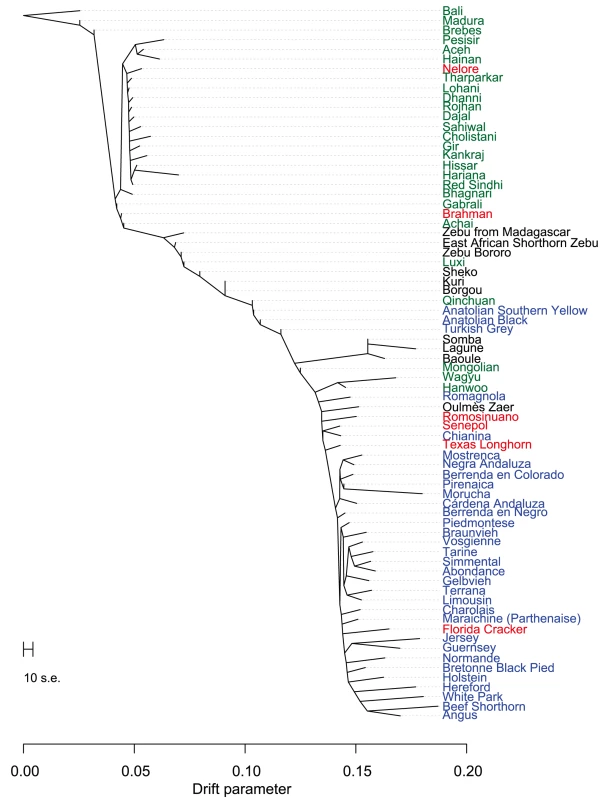
Breeds were colored according to their geographic origin; black: Africa, green: Asia, red: North and South America, orange: Australia, and blue: Europe. Scale bar shows 10 times the average standard error of the estimated entries in the sample covariance matrix. Fig. 4. Phylogenetic network of the inferred relationships between 74 cattle breeds. 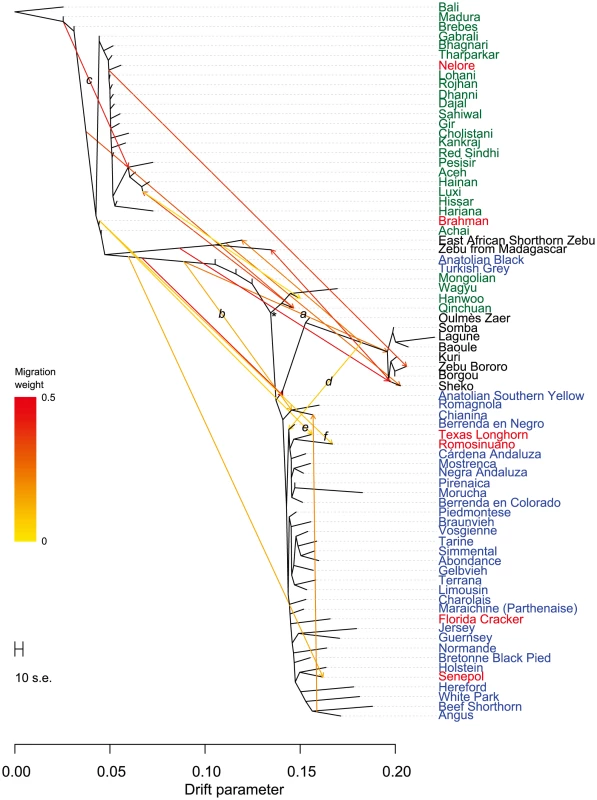
Breeds were colored according to their geographic origin; black: Africa, green: Asia, red: North and South America, orange: Australia, and blue: Europe. Scale bar shows 10 times the average standard error of the estimated entries in the sample covariance matrix. Common ancestor of domesticated taurines is indicated by an asterisk. Migration edges were colored according to percent ancestry received from the donor population. Migration edge a is hypothesized to be from wild African auroch into domesticates from the Fertile Crescent. Migration edge b is hypothesized to be introgression from hybrid African cattle. Migration edge c is hypothesized to be introgression from Bali/indicine hybrids into other Indonesian cattle. Migration edge d signals introgression of African taurine into Iberia. Migration edges e and f represent introgression from Brahman into American Criollo. Fig. 5. Worldwide map with country averages of ancestry proportions with 3 ancestral populations (K = 3). 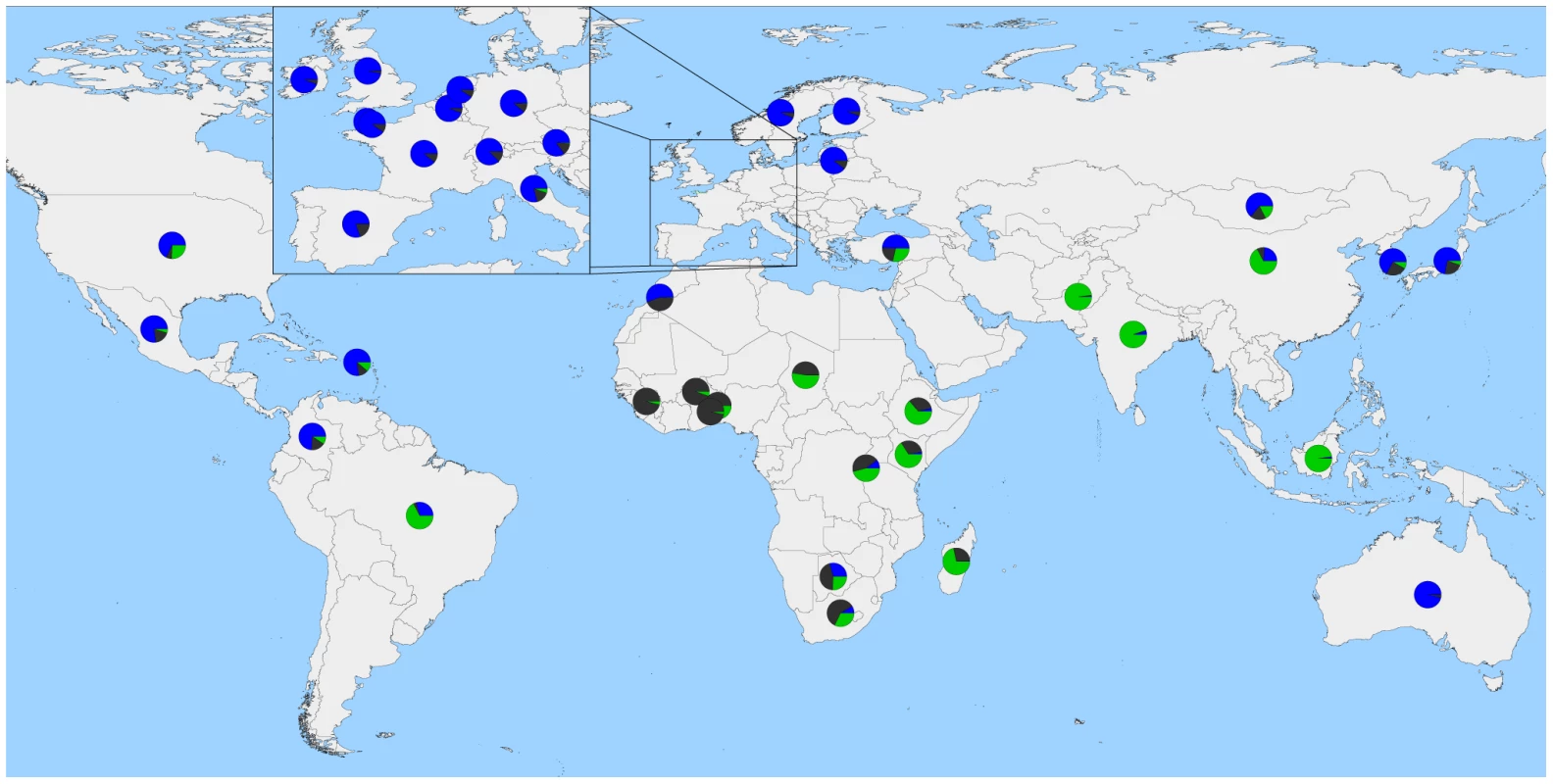
Blue represents Eurasian Bos t. taurus ancestry, green represents Bos javanicus and Bos t. indicus ancestry, and dark grey represents African Bos. t. taurus ancestry. Please note, averages do not represent the entire populations of each country, as we do not have a geographically random sample. In the late 18th and 19th centuries, European cattlemen began forming closed herds which they developed into breeds [25]. Because breeds are typically reproductively isolated with little or no interbreeding, we found that the cross-validation error estimates continued to decrease as we increased the number of ancestral populations K modeled in the admixture analysis (Table S2). This reflects the large differences in allele frequencies that exist between breeds resulting from separate domestication events, geographic dispersal and isolation, breed formation, and the use of artificial insemination. The method of Evanno et al. [26], which evaluates the second order rate of change of the likelihood function with respect to K (ΔK), identified K = 2 as the optimum level of K (Figure S5). This method was overwhelmed by the early divergence between indicine and taurine cattle, and was not sensitive to the hierarchical relationships of populations and breeds [27]. As we increased the value of K, we recapitulated the increasingly fine structure represented in the branches of the phylogeny (Figures 6, S6, S7, S8, S9, S10).
Fig. 6. Ancestry models with 3 ancestral populations (K = 3). 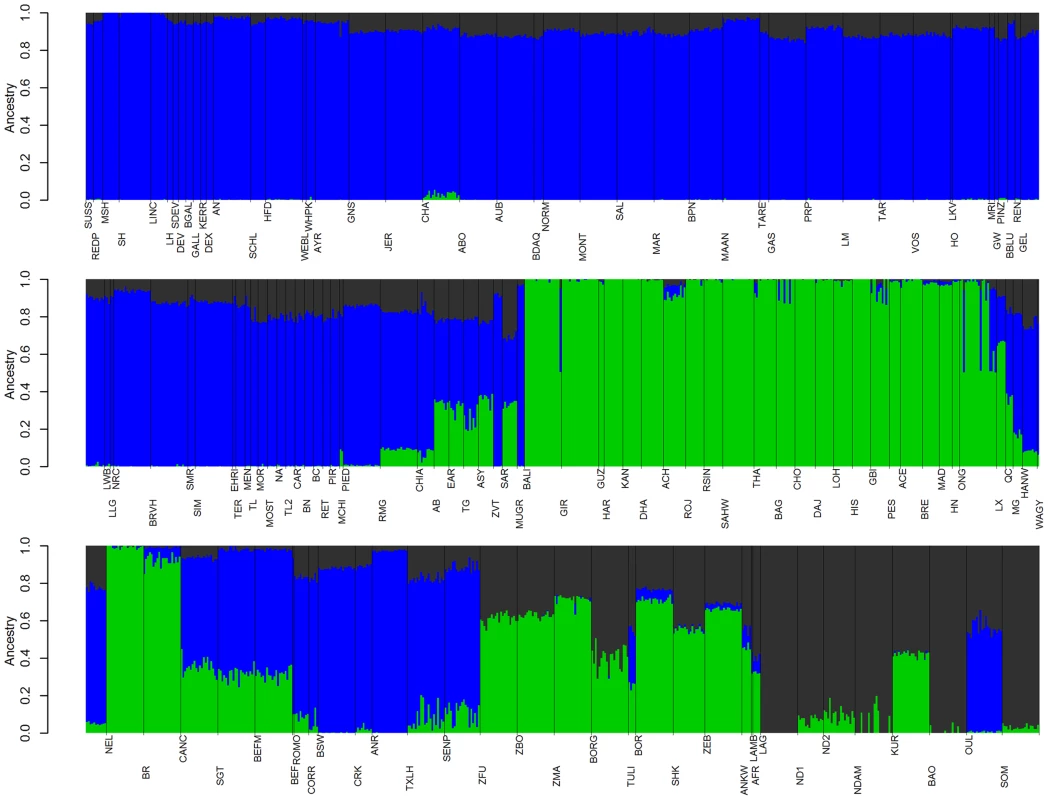
Blue represents Eurasian Bos t. taurus ancestry, green represents Bos javanicus and Bos t. indicus ancestry, and dark grey represents African Bos. t. taurus ancestry. See Supplementary Figures S5, S6, S7, S8, S9, S10 for other values of K. Modern Anatolian cattle are not representatives of early domesticated cattle
Anatolian breeds (AB, EAR, TG, ASY, and SAR) are admixed between blue Fertile Crescent, grey African-like, and green indicine-like cattle (Figures 5 and 6), and we infer that they do not represent the taurine populations originally domesticated in this region due to a history of admixture. Zavot (ZVT), a crossbred breed [25], has a different history with a large portion of ancestry similar to Holsteins (Figures 2 and S8, S9, S10). The placement of Anatolian breeds along principal components 1 and 2 in Figure 1 [23], the ancestry estimates in Figure 6, their extremely short branch lengths in Figures 2–4, and significant f3 statistics confirm that modern Anatolian breeds are admixed (see Methods for explanation of f-statistics). For example, the Anatolian Southern Yellow (ASY) has 3,003 significant f3 tests, the most extreme of which has Vosgienne (VOS, a taurine breed) and Achai (ACH, an indicine breed) as sister groups with a Z-score of −43.69. Our results support previous work using microsatellite loci [28] which inferred Anatolian cattle to possess indicine introgression. We further demonstrate that Anatolian breeds have introgression from African taurine. We calculated f4 statistics with East Anatolian Red, Anatolian Southern Yellow, and Anatolian Black as sister, and N'Dama, Somba, Lagune, Baole, Simmental, Holstein, Hereford, and Shorthorn as the opposing sister group. From Figure 2, we would expect these relationships to be tree-like. But 45 of the possible 84 f4 tests indicated significant levels of admixture. The most significant was f4(East Anatolian Red, Anatolian Southern Yellow; Somba, Shorthorn) = −0.0026±0.0003 (Z-score = −8.10, alternative trees have Z-scores of 9.88 and 5.20).
Divergence within the taurine lineage
If African and Asian taurines were both exported from the Fertile Crescent in similar numbers at about the same time, we would expect them to be approximately equally diverged from European taurines. However, African taurines were consistently revealed to be more diverged from European and Asian taurines (Figures 1, 2, 3, and 5, Anatolian breeds are not considered in this comparison because of their admixed history). Two factors appear to influence this divergence. First, European cattle were exported into Asia and admixed with Asian taurines. In the admixture models in which K = 15 or 20 (Figures S9 and S10), there was evidence of European taurine admixture in the Mongolian (MG), Hanwoo (HANW), and Wagyu (WAGY) breeds. We ran TreeMix with 14 representative populations and estimated Wagyu to have 0.188±0.069 (p-value = 0.003) of their genome originating from northwestern European ancestry (Figure 7). We also see some runs of TreeMix placing a migration edge from Chianina cattle to Asian taurines (Figure S4). We ran f4 tests with Mongolian, Hanwoo, Wagyu, Tharparkar (THA), or Kankraj (KAN) as sister populations, and Piedmontese (PIED), Simmental (SIM), Brown Swiss (BSW), Braunvieh (BRVH), Devon (DEV), Angus (AN), Shorthorn (SH), or Holstein (HO) as the opposing pair of sister groups. From previous research [3] and Figures 2 and 3, these relationships should be tree-like if there were no admixture. For 53 of the possible 280 tests, the Z-score was more extreme than ±2.575829. The most extreme test statistics were f4(Wagyu, Mongolian; Simmental, Shorthorn) = −0.003 (Z-score = −5.21, other rearrangements of these groups had Z-scores of 7.32 and 16.55) and f4(Hanwoo, Wagyu; Piedmontese, Shorthorn) = 0.002 (Z-score = 4.90, other rearrangements of these groups had Z-scores of 21.79 and 27.77). When K = 20, Hanwoo appear to have a Mediterranean influence, whereas Wagyu have a northwestern European, including British, influence (Figure S10). We conclude that there were two waves of European introgression into Far East Asian cattle, first with Mediterranean cattle (which carried African taurine and indicine alleles) brought along the Silk Road [29] and later from 1868 to 1918 when Japanese cattle were crossed with British and Northwest European cattle [25].
Fig. 7. Phylogenetic network of the inferred relationships between 14 cattle breeds. 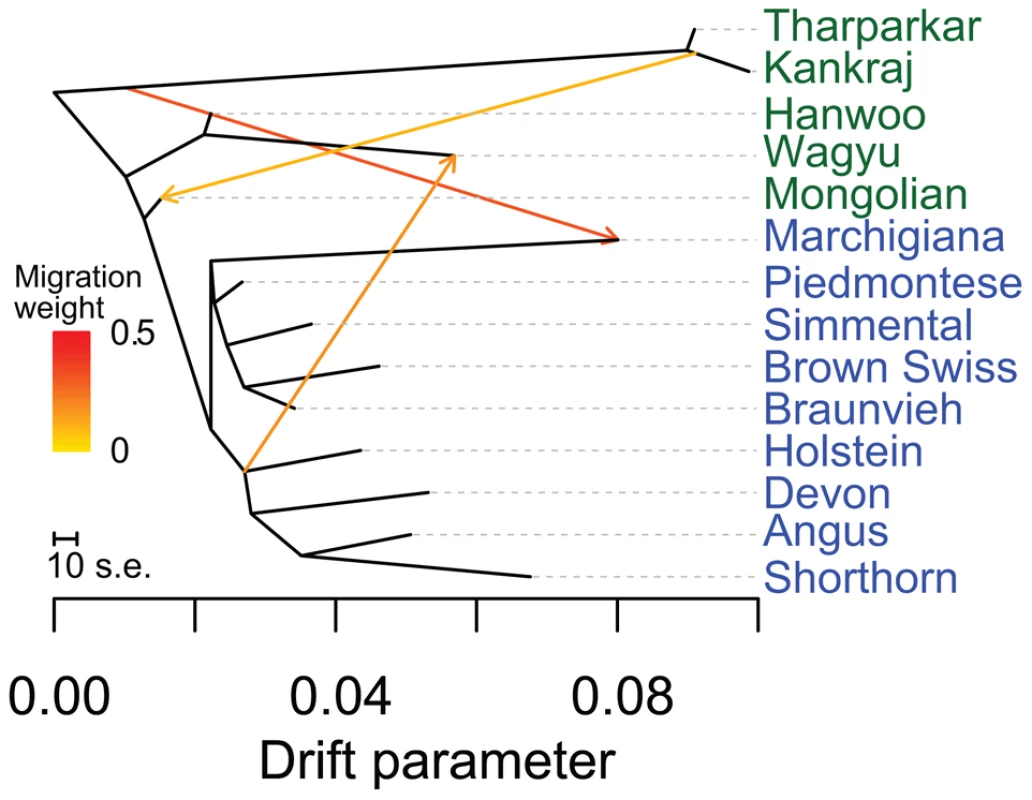
Breeds were colored according to their geographic origin; green: Asia, and blue: Europe. Scale bar shows 10 times the average standard error of the estimated entries in the sample covariance matrix. Migration edges were colored according to percent ancestry received from the donor population. Migration edges show indicine introgression into Mongolian cattle, African taurine and indicine ancestry in Marchigiana, and a northern European influence on Wagyu. The second factor that we believe underlies the divergence of African taurine is a high level of wild African auroch [30], [31] introgression. Principal component (Figure 1), phylogenetic trees (Figures 2 and 3), and admixture (Figure 6) analyses all reveal the African taurines as being the most diverged of the taurine populations. Because of this divergence, it has been hypothesized that there was a third domestication of cattle in Africa [32]–[36]. If there was a third domestication, African taurine would be sister to the European and Asian clade. When no migration events were fit in the TreeMix analyses, African cattle were the most diverged of the taurine populations (Figures 2 and 3), but when admixture was modeled to include 17 migrations, all African cattle, except for East African Shorthorn Zebu and Zebu from Madagascar which have high indicine ancestry, were sister to European cattle and were less diverged than Asian or Anatolian cattle (Figure 4), thus ruling out a separate domestication. Our phylogenetic network (Figure 4) shows that there was not a third domestication process, rather there was a single origin of domesticated taurine (Asian, African, and European all share a recent common ancestor denoted by an asterisk in Figure 4, with Asian cattle sister to the rest of the taurine lineage), followed by admixture with an ancestral population in Africa (migration edge a in Figure 4, which is consistent across 6 separate TreeMix runs, Figure S4). This ancestral population (origin of migration edge a in Figure 4) was approximately halfway between the common ancestor of indicine and the common ancestor of taurine. We conclude that African taurines received as much as 26% (estimated as 0.263 in the network, p-value<2.2e-308) of their ancestry from admixture with wild African auroch, with the rest being Fertile Crescent domesticate in origin. Although three other migration edges originate from the branch between indicine and taurine (such as edge b), all of the receiving populations show indicine ancestry in the ADMIXTURE models. But African auroch are extinct and samples were not available for the ADMIXTURE model, thus the admixed auroch ancestry of African taurines cannot specifically be discovered by this model [27], [37] and African taurine, especially Lagune, are depicted as having a single ancestry without indicine influence (Figures 5 and 6, see f3 and f4 statistics reported later). Unlike ADMIXTURE, TreeMix can model admixture from an unsampled population by placing a migration edge more basal along a branch of the phylogeny, in this case African auroch.
Others have observed distinct patterns of linkage disequilibrium in African taurines, resulting in larger estimates of ancestral effective population size than for either Bos t. taurus or Bos t. indicus breeds [2] consistent with greater levels of admixture from wild aurochs. Just as Near Eastern domesticated pig mitochondrial lineages were replaced by mitochondria from indigenous wild populations [38], we infer that the divergent T1d African mitochondrial subgroup [39] previously observed originated either from Fertile Crescent domesticates or admixture with wild African auroch. Similar patterns of admixture from wild forebears have been observed in other species [38], such as pig [40]–[42], chicken [43], and corn [14], and this conclusion represents the most parsimonious explanation of our results. We hypothesize that the auroch introgression in Africa may have been driven by trypanosomiasis resistance in African auroch which may be the source of resistance in modern African taurine populations [44]. Admixture with distant relatives has had an important impact on the immune system of other species, such as human [45] and possibly chicken [46]. More sophisticated demographic models and unbiased whole-genome sequence data will be needed to further test these hypotheses.
Indicine admixture in Africa
African cattle also demonstrate a geographical gradient of indicine ancestry [47]. Taurine cattle in western Africa possess from 0% to 19.9% indicine ancestry (Figures 5 and 6, LAG, ND1, ND2, NDAM, BAO, OUL, SOM), with an average of 3.3%. Moving from west to east and from south to central Africa, the percent of indicine ancestry increases from 22.7% to 74.1% (Figures 5 and 6, ZFU, ZBO, ZMA, BORG, TULI, BOR, SHK, ZEB, ANKW, LAMB, an AFR), with an average of 56.9%. As we increased values of K to 10, 15, and 20 (Figures S8, S9, S10), we revealed two clusters of indicine ancestry possibly resulting from the previously suggested two waves of indicine importation into Africa, the first occurring in the second millennium BC and the second during and after the Islamic conquests [25], [34], [48]. The presence of two separate clades of African cattle in Figure 4 also supports the idea of two waves of indicine introgression.
Admixture in Asia
Asian cattle breeds were derived from cattle domesticated in the Indian subcontinent or imported from the Fertile Crescent and Europe. Cattle in the north and northeast are primarily of Bos t. taurus ancestry (Figures 5 and 6; HANW, WAGY, and MG), but Hanwoo and Mongolian also have Bos t. indicus ancestry (Figures 5, 6, S9, and S10). Cattle in Pakistan, India, southern China and Indonesia are predominantly Bos t. indicus (Figures 5 and 6; ONG, MAD, BRE, HN, ACE, PES, ACH, HAR, BAG, GUZ, SAHW, GBI, CHO, GIR, KAN, THA, RSIN, HIS, LOH, ROJ, DHA, and DAJ). Cattle located between these two geographical regions are Bos t. taurus×Bos t. indicus hybrids (Figures 1, 4, 5, and 6; QC and LX). Our results suggest an additional source for increased indicine diversity—admixture with domesticated cattle from other species. In addition to cattle domesticated from aurochs (Bos primigenius), bovids were also domesticated from water buffalo (Bubalus bubalis), yak (Bos grunniens), gaur (Bos gaurus), and banteng (Bos javanicus), represented in our sample by the Bali breed [25], [49]. We find that the Indonesian Brebes (BRE) and Madura (MAD) breeds have significant Bos javanicus (BALI) ancestry demonstrated by the short branch lengths in Figures 2–4, shared ancestry with Bali in ADMIXTURE analyses (light green in Figures S8, S9, S10), and significant f3 statistics (Table S3). The Indonesian Pesisir and Aceh and the Chinese Hainan and Luxi breeds also have Bali ancestry (migration edge c in Figure 4, migration edges in Figure S4, and light green in Figures S8 and S9).
Admixture in Europe
Cattle were imported into Europe from the southeast to the northwest. The descendants of Durham Shorthorns (the ancestral Shorthorn breed [25]) were the most distinct group of European cattle as they clustered at the extremes of principal component 2 (lower left hand corner of Figure 1), and they formed a distinct cluster in the ADMIXTURE analyses whenever K was greater than 4 (Figures S6, S7, S8, S9, S10). As shown in Figures S6 through S10, f3 statistics in Table S4, and from their breed histories [25], many breeds share ancestry with Shorthorn cattle, including Milking Shorthorn, Beef Shorthorn, Lincoln Red, Maine-Anjou, Belgian Blue, Santa Gertrudis, and Beefmaster.
From the previous placement of the American Criollo breeds including Romosinuano, Texas Longhorn, and Corriente, it has been posited that Iberian cattle became admixed as a result of an introgression of cattle from Africa into the local European cattle [3], [50], [51]. Our genotyping of individuals from 11 Spanish breeds supported, but clarified, this hypothesis. On average, Spanish cattle had 19.3% of African ancestry when K = 3, with a minimum of 8.8% and a maximum of 23.4%, which supports previous analyses of mitochondrial DNA [52], [53]. Migration edge d in the phylogenetic network (Figure 4, and consistently seen in Figure S4) estimates that Iberian cattle, Texas Longhorn, and Romosinuano derive 7.5% of their ancestry from African taurine introgression, similar to the ancestry estimates from the models with larger K values (Figures S8, S9, S10). The Oulmès Zaer (OUL) breed from Morocco also shows that cattle were transported from Iberia and France to Africa (tan and red in Figure S10, and short branch length in Figure 4). However, the 11 Spanish breeds had no more indicine ancestry than all other European taurine breeds (essentially none for the majority of breeds, see Figures 5 and 6). Maraichine (MAR), Gascon (GAS), Limousin (LIM), and other breeds from France, and Piedmontese cattle (PIED) from northwest Italy have a similar ancestry. These data indicate that the reason that the American Criollo breeds were found to be sister to European cattle in our previous work [3] was because of their higher proportion of indicine ancestry. The 5 sampled American Criollo breeds had, on average, 14.7% African ancestry (minimum of 6.2% and maximum of 20.4%) and 8.0% indicine ancestry (minimum of 0.6% and maximum of 20.3%).
Other Italian breeds (MCHI, CHIA, and RMG) share ancestry with both African taurine and indicine cattle (Figures 6, S6, S7, S8). This introgression may have come from Anatolian or East African cattle that carried both African taurine and indicine ancestry, which is modeled as migration edge b in Figure 4. The placement of Italian breeds is not consistent across independent TreeMix runs (Figure S4), likely due to their complicated history of admixture.
We also used f-statistics to explore the evidence for African taurine introgression into Spain and Italy. We did not see any significant f3 statistics, but this test may be underpowered because of the low-level of introgression. With Italian and Spanish breeds as a sister group and African breeds, including Oulmès Zaer, as the other sister group, we see 321 significant tests out of 1,911 possible tests. Of these 321 significant tests, 218 contained Oulmès Zaer. We also calculated f4 statistics with the Spanish breeds as sister and the African taurine breeds as sister (excluding Oulmès Zaer). With this setup, out of the possible 675 tests we saw only 1 significant test, f4(Berrenda en Negro, Pirenaica;Lagune, N'Dama (ND2)) = 0.0007, Z-score = 3.064. With Italian cattle as sister and African taurine as sister (excluding Oulmès Zaer), we saw 17 significant tests out of the 90 possible. Patterson et al. [54] defined the f4-ratio as f4(A, O; X, C)/f4(A, O; B, C), where A and B are a sister group, C is sister to (A,B), X is a mixture of B and C, and O is the outgroup. This ratio estimates the ancestry from B, denoted as , and the ancestry from C, as . We calculated this ratio using Shorthorn as A, Montbeliard as B, Lagune as C, Morucha as X, and Hariana as O. We choose Shorthorn, Montbeliard, Lagune, and Hariana as they appeared the least admixed in the ADMIXTURE analyses. We choose Morucha because it appears as solid red with African ancestry in Figure S10. This statistic estimated that Morucha is 91.23% European ( = 0.0180993/0.0198386) and 8.77% African, which is similar to the proportion estimated by TreeMix. The multiple f4 statistics with Italian breeds as sister and African breeds as the opposing sister support African admixture into Italy. The f4-ratio test with Morucha also supports our conclusion of African admixture into Spain.
Preservation of pure taurine in Africa and lack of widespread indicine ancestry in Europe
It has recently been concluded that indicine ancestry is a common feature of European cattle genomes [55]. However, our data refute this conclusion. McTavish et al. relied on the Evanno test to arrive at an optimal number of ancestral populations of K = 2, which masks the fact that there are cattle breeds in Africa with 100% African taurine ancestry (Figure 6). Although our K = 2 ADMIXTURE results suggested that most African breeds had at least 20% indicine ancestry (Figure S5), when we increased K to 3, Lagune (LAG) revealed no indicine ancestry, and Baoule (BAO) and N'Dama (NDAM) possess very little indicine ancestry. If the K = 2 model was correct, we would expect to see numerous significant f3 and f4 tests with Eurasian taurine and indicine as sister groups. Whereas, if the K = 3 model more accurately reflected the heritage of European and African taurines, we would not observe any significant f3 or f4 tests showing admixture of taurine and indicine in the ancestry of African taurine. For the Lagune, Baoule and N'Dama (NDAM and ND2) breeds we found no significant f3 statistics. Among the 225 f4 statistics calculated with NDAM, LAG, BAO, ND2, SH, and MONT as sisters and BALI, GIR, HAR, SAHW, PES, and ACE as the opposing sister group, only 36 were significantly different from 0 (Table 1). When ND2 was excluded from the results, only 4 tests were significant (Table 1), and we have no evidence that the Lagune breed harbors indicine alleles. Thus, we conclude that contrary to the assumptions and conclusions of [55] cattle with pure taurine ancestry do exist in Africa. Further, we conclude that indicine ancestry in European taurine cattle is extremely rare, and that some breeds, especially those prevalent near the Mediterranean, possess African taurine introgression—but with the exception of the Charolais, Marchigiana, Chianina and Romagnola breeds—not African hybrid or African indicine introgression. We concur that Texas Longhorn and other American Criollo breeds possess indicine ancestry, but infer that this introgression occurred after the arrival of Spanish cattle in the New World and likely originated from Brahman cattle (migration edges e and f in Figure 4). In TreeMix replicates, Texas Longhorn and Romosinuano are either sister to admixed Anatolian breeds or they receive a migration edge that originates near Brahman (Figure S4). To reiterate, Iberian cattle do not have indicine ancestry, American Criollo breeds originated from exportations from Iberia, Brahman cattle were developed in the United States in the 1880's [25], American Criollo breeds carry indicine ancestry, and the introgression likely occurred from Brahman cattle.
Tab. 1. Significant f4 statistics for African taurine breeds and populations.1 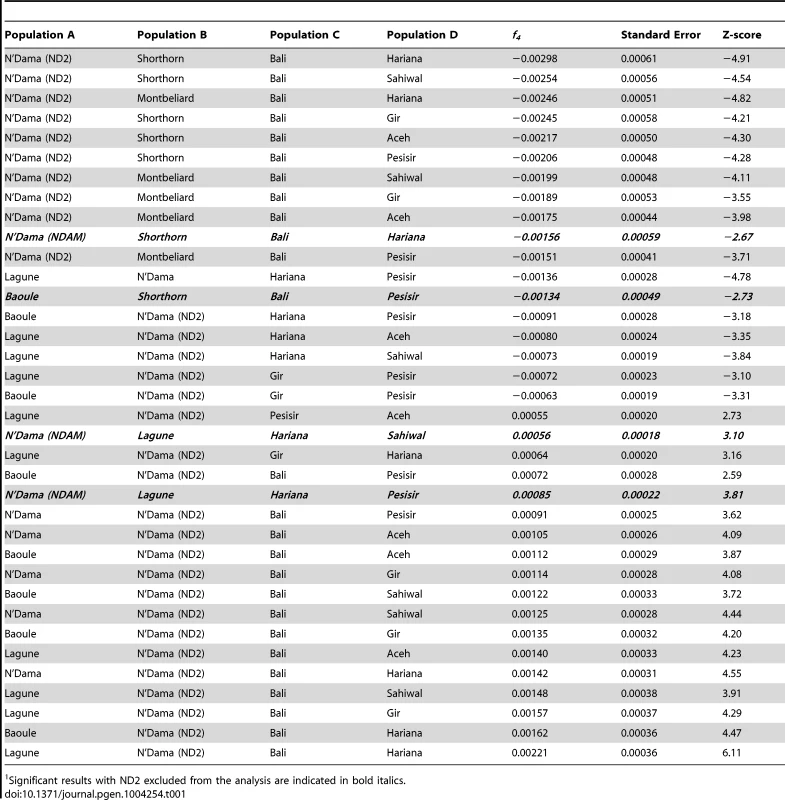
Significant results with ND2 excluded from the analysis are indicated in bold italics. Domestication, exportation, admixture, and breed formation have had tremendous impacts on the variation present within and between cattle breeds. In Asia, Africa, North and South America, cattle breeders have crossbred Bos t. taurus and Bos t. indicus cattle to produce hybrids which were well suited to the environment and endemic production systems. In this study, we clarify the relationships between breeds of cattle worldwide, and present the most accurate cattle “Tree of Life” to date in Figure 4. We elucidate the complicated history of Asian cattle involving the domestication and subsequent admixture of several bovid species. We provide evidence for admixture between domesticated Fertile Crescent taurine and wild African auroch in Africa to form the extant African taurine breeds. We also observe African taurine content within the genomes of European Mediterranean taurine breeds. The absence of indicine content within the majority of European taurine breeds, but the presence of indicine within three Italian breeds is consistent with two separate introductions, one from the Middle East potentially by the Romans which captured African taurines in which indicine introgression had already occurred and the second from western Africa into Spain which included African taurines with no indicine introgression. It was this second group of cattle which likely radiated from Spain into Southern France and the Alps. The prevalence of admixture further convolutes the cryptic history of cattle domestication.
Materials and Methods
Sample selection
We used 1,543 samples in total, including 234 samples from [3] and 425 samples from [4], see Table S1. We selected samples that had fewer than 10% missing genotypes, and for breeds with fewer than 20 genotyped samples, we used all available samples which passed the missing genotype data threshold. When pedigree data were absent for a breed, the 20 samples with the highest genotype call rates were selected. For breeds which had pedigree information, we filtered any animals whose sire or dam was also genotyped. For identified half-siblings, we sampled only the sibling with the highest genotype call rate. After removing genotyped animals known to be closely related, we selected the 20 animals with the highest genotype call rate to represent the breed. All DNA samples were collected in an ethical manner under University of Missouri ACUC approved protocol 7505.
Genotyping
Samples were genotyped with the Illumina BovineSNP50 BeadChip [56]. Autosomal SNPs and a single pseudo-autosomal SNP were analyzed, because the data set from Gautier et al. [4] excluded SNPs located exclusively on the X chromosome. We also filtered all SNPs which mapped to “chromosome unknown” of the UMD3.1 assembly [57]. In PLINK [58], [59], we removed SNPs with greater than 10% missing genotypes and with minor allele frequencies less than 0.0005 (1/[2*Number of Samples] = 0.000324, thus the minor allele had to be observed at least once in our data set). The average total genotype call rate in the remaining individuals was 0.993. Genotype data were deposited at DRYAD (doi:10.5061/dryad.th092) [60].
Principal component analysis
The sample genotype covariance matrix was decomposed using SMARTPCA, part of EIGENSOFT 4.2 [19]. To limit the effects of linkage disequilibrium on the estimation of principal components, for each SNP the residual of a regression on the previous two SNPs was input to the principal component analysis (see EIGENSOFT POPGEN README).
TreeMix analysis
TreeMix [20] models the genetic drift at genome-wide polymorphisms to infer relationships between populations. It first estimates a dendrogram of the relationships between sampled populations. Next it compares the covariance structure modeled by this dendrogram to the observed covariance between populations. When populations are more closely related than modeled by a bifurcating tree it suggests that there has been admixture in the history of those populations. TreeMix then adds an edge to the phylogeny, now making it a phylogenetic network. The position and direction of these edges are informative; if an edge originates more basally in the phylogenetic network it indicates that this admixture occurred earlier in time or from a more diverged population.
TreeMix was used to create a maximum likelihood phylogeny of the 134 breeds. Because TreeMix was slow to add migration events (modeled as “edges”) to the complete data set of 134 breeds, we also analyzed subsets of the data containing considerably fewer breeds. For these subsets, breeds with fewer than 4 samples were removed. To speed up the analysis, we iteratively used the previous graph with m-1 migrations as the starting graph and added one migration edge for a total of m migrations. We rooted the graphs with Bali cattle, used blocks of 1000 SNPs, and used the -se option to calculate standard errors of migration proportions. Migration edges were added until 99.8% of the variance in ancestry between populations was explained by the model. We also ensured that the incorporated migration edges were statistically significant. To further evaluate the consistency of migration edges, we ran TreeMix five separate times with -m set to 17.
Admixture analysis
ADMIXTURE 1.21 was used to evaluate ancestry proportions for K ancestral populations [21]. We ran ADMIXTURE with cross-validation for values of K from 1 through 20 to examine patterns of ancestry and admixture in our data set. Map figure was generated in R using rworldmap (http://cran.r-project.org/web/packages/rworldmap/index.html).
f3 and f4 statistics
The f3 and f4 statistics are used to detect correlations in allele frequencies that are not compatible with population evolution following a bifurcating tree; these statistics provide support for admixture in the history of the tested populations [54], [61]. The THREEPOP program from TreeMix was used to calculate f3 statistics [54] for all possible triplets from the 134 breeds. The FOURPOP program of TreeMix was used to calculate f4 statistics for subsets of the breeds.
Supporting Information
Zdroje
1. NovembreJ, RamachandranS (2011) Perspectives on human population structure at the cusp of the sequencing era. Annu Rev Genomics Hum Genet 12 : 245–274 Available: http://www.ncbi.nlm.nih.gov/pubmed/21801023.
2. GibbsRA, TaylorJF, Van TassellCP, BarendseW, EversoleKA, et al. (2009) Genome-wide survey of SNP variation uncovers the genetic structure of cattlebreeds. Science (80-) 324 : 528–532 Available: http://www.ncbi.nlm.nih.gov/pubmed/19390050.
3. DeckerJE, PiresJC, ConantGC, McKaySD, HeatonMP, et al. (2009) Resolving the evolution of extant and extinct ruminants with high-throughput phylogenomics. Proc Natl Acad Sci U S A 106 : 18644–18649 Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2765454&tool=pmcentrez&rendertype=abstract. Accessed 13 August 2013.
4. GautierM, LaloëD, Moazami-GoudarziK (2010) Insights into the genetic history of French cattle from dense SNP data on 47 worldwide breeds. PLoS One 5: e13038 Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2948016&tool=pmcentrez&rendertype=abstract. Accessed 13 September 2013.
5. KijasJW, TownleyD, DalrympleBP, HeatonMP, MaddoxJF, et al. (2009) A genome wide survey of SNP variation reveals the genetic structure of sheep breeds. PLoS One 4: e4668 Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2652362&tool=pmcentrez&rendertype=abstract. Accessed 29 February 2012.
6. VonholdtBM, PollingerJP, LohmuellerKE, HanE, ParkerHG, et al. (2010) Genome-wide SNP and haplotype analyses reveal a rich history underlying dog domestication. Nature 464 : 898–902 Available: http://www.ncbi.nlm.nih.gov/pubmed/20237475. Accessed 2 March 2012.
7. McCueME, BannaschDL, PetersenJL, GurrJ, BaileyE, et al. (2012) A high density SNP array for the domestic horse and extant Perissodactyla: Utility for association mapping, genetic diversity, and phylogeny studies. PLoS Genet 8: e1002451 Available: http://dx.plos.org/10.1371/journal.pgen.1002451. Accessed 16 January 2012.
8. SchachererJ, ShapiroJA, RuderferDM, KruglyakL (2009) Comprehensive polymorphism survey elucidates population structure of Saccharomyces cerevisiae. Nature 458 : 342–345 Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2782482&tool=pmcentrez&rendertype=abstract. Accessed 31 July 2013.
9. YangH, WangJR, DidionJP, BuusRJ, BellTA, et al. (2011) Subspecific origin and haplotype diversity in the laboratory mouse. Nat Genet 43 : 648–655 Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3125408&tool=pmcentrez&rendertype=abstract.
10. StaubachF, LorencA, MesserPW, TangK, PetrovDA, et al. (2012) Genome patterns of selection and introgression of haplotypes in natural populations of the house mouse (Mus musculus). PLoS Genet 8: e1002891 Available: http://dx.plos.org/10.1371/journal.pgen.1002891. Accessed 31 July 2013.
11. McNallyKL, ChildsKL, BohnertR, DavidsonRM, ZhaoK, et al. (2009) Genomewide SNP variation reveals relationships among landraces and modern varieties of rice. Proc Natl Acad Sci U S A 106 : 12273–12278 Available: http://www.pnas.org/cgi/content/long/106/30/12273. Accessed 31 July 2013.
12. ZhaoK, WrightM, KimballJ, EizengaG, McClungA, et al. (2010) Genomic diversity and introgression in O. sativa reveal the impact of domestication and breeding on the rice genome. PLoS One 5: e10780 Available: http://dx.plos.org/10.1371/journal.pone.0010780. Accessed 31 July 2013.
13. YanJ, ShahT, WarburtonML, BucklerES, McMullenMD, et al. (2009) Genetic characterization and linkage disequilibrium estimation of a global maize collection using SNP markers. PLoS One 4: e8451 Available: http://dx.plos.org/10.1371/journal.pone.0008451. Accessed 31 July 2013.
14. Van HeerwaardenJ, DoebleyJ, BriggsWH, GlaubitzJC, GoodmanMM, et al. (2011) Genetic signals of origin, spread, and introgression in a large sample of maize landraces. Proc Natl Acad Sci U S A 108 : 1088–1092 Available: http://www.pnas.org/cgi/content/long/1013011108v1. Accessed 4 December 2012.
15. Van HeerwaardenJ, HuffordMB, Ross-IbarraJ (2012) Historical genomics of North American maize. Proc Natl Acad Sci U S A 109 : 12420–12425 Available: http://www.pnas.org/cgi/content/long/109/31/12420. Accessed 31 July 2013.
16. HuffordMB, LubinksyP, PyhäjärviT, DevengenzoMT, EllstrandNC, et al. (2013) The genomic signature of crop-wild introgression in maize. PLoS Genet 9: e1003477 Available: http://dx.plos.org/10.1371/journal.pgen.1003477. Accessed 31 July 2013.
17. MylesS, BoykoAR, OwensCL, BrownPJ, GrassiF, et al. (2011) Genetic structure and domestication history of the grape. Proc Natl Acad Sci U S A 108 : 3530–3535 Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3048109&tool=pmcentrez&rendertype=abstract. Accessed 14 March 2012.
18. CavanaghCR, ChaoS, WangS, HuangBE, StephenS, et al. (2013) Genome-wide comparative diversity uncovers multiple targets of selection for improvement in hexaploid wheat landraces and cultivars. Proc Natl Acad Sci U S A 110 : 8057–8062 Available: http://www.pnas.org/cgi/content/long/110/20/8057. Accessed 31 July 2013.
19. PattersonNJ, PriceAL, ReichD (2006) Population structure and eigenanalysis. PLoS Genet 2: e190 Available: http://www.ncbi.nlm.nih.gov/pubmed/17194218. Accessed 12 March 2012.
20. PickrellJK, PritchardJK (2012) Inference of population splits and mixtures from genome-wide allele frequency data. PLoS Genet 8: e1002967 Available: http://dx.plos.org/10.1371/journal.pgen.1002967. Accessed 16 November 2012.
21. AlexanderDH, NovembreJ, LangeK (2009) Fast model-based estimation of ancestry in unrelated individuals. Genome Res 19 : 1655–1664 Available: http://www.ncbi.nlm.nih.gov/pubmed/19648217.
22. LoftusRT, MacHughDE, BradleyDG, SharpPM, CunninghamP (1994) Evidence for two independent domestications of cattle. Proc Natl Acad Sci U S A 91 : 2757–2761 Available: http://www.ncbi.nlm.nih.gov/pubmed/8146187.
23. McVeanG (2009) A genealogical interpretation of principal components analysis. PLoS Genet 5: e1000686 Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2757795&tool=pmcentrez&rendertype=abstract. Accessed 11 July 2011.
24. DiamondJ (2002) Evolution, consequences and future of plant and animal domestication. Nature 418 : 700–707 Available: http://www.ncbi.nlm.nih.gov/pubmed/12167878.
25. Felius M (1995) Cattle breeds - an encyclopedia. Doetinchem, Netherlands: Misset.
26. EvannoG, RegnautS, GoudetJ (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14 : 2611–2620 Available: http://www.ncbi.nlm.nih.gov/pubmed/15969739. Accessed 3 October 2012.
27. ReichD, ThangarajK, PattersonNJ, PriceAL, SinghL (2009) Reconstructing Indian population history. Nature 461 : 489–494 Available: http://www.ncbi.nlm.nih.gov/pubmed/19779445.
28. LoftusRT, ErtugrulO, HarbaAH, El-BarodyMA, MacHughDE, et al. (1999) A microsatellite survey of cattle from a centre of origin: the Near East. Mol Ecol 8 : 2015–2022 Available: http://www.ncbi.nlm.nih.gov/pubmed/10632853. Accessed 5 September 2013.
29. ChristianD (2000) Silk roads or steppe roads? The silk roads in world history. J World Hist 11 : 1–26 Available: http://muse.jhu.edu/journals/jwh/summary/v011/11.1christian.html. Accessed 6 January 2014.
30. StockF, Gifford-GonzalezD (2013) Genetics and African Cattle Domestication. African Archaeol Rev 30 : 51–72 Available: http://link.springer.com/10.1007/s10437-013-9131-6.
31. LinseeleV (2004) Size and size change of the African aurochs during the Pleistocene and Holocene. J African Archaeol 2 : 165–185 Available: http://www.african-archaeology.de/index.php?page_id=154&journal_id=6&pdf_id=96. Accessed 10 December 2013.
32. TroyCS, MacHughDE, BaileyJF, MageeDA, LoftusRT, et al. (2001) Genetic evidence for Near-Eastern origins of European cattle. Nature 410 : 1088–1091 Available: http://www.ncbi.nlm.nih.gov/pubmed/11323670.
33. BradleyDG, MacHughDE, CunninghamP, LoftusRT (1996) Mitochondrial diversity and the origins of African and European cattle. Proc Natl Acad Sci U S A 93 : 5131–5135 Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=39419&tool=pmcentrez&rendertype=abstract.
34. HanotteO, BradleyDG, OchiengJW, VerjeeY, HillEW, et al. (2002) African pastoralism: genetic imprints of origins and migrations. Science (80-) 296 : 336–339 Available: http://www.ncbi.nlm.nih.gov/pubmed/11951043.
35. MarshallF, WeissbrodL (2011) Domestication Processes and Morphological Change. Curr Anthropol 52: S397–S413 Available: http://www.jstor.org/stable/info/10.1086/658389. Accessed 24 August 2013.
36. Pérez-PardalL, RoyoLJ, Beja-Pereiraa, ChenS, CantetRJC, et al. (2010) Multiple paternal origins of domestic cattle revealed by Y-specific interspersed multilocus microsatellites. Heredity (Edinb) 105 : 511–519 Available: http://www.ncbi.nlm.nih.gov/pubmed/20332805. Accessed 11 September 2013.
37. FalushD, StephensM, PritchardJK (2003) Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164 : 1567–1587 Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=1462648&tool=pmcentrez&rendertype=abstract.
38. LarsonG, BurgerJ (2013) A population genetics view of animal domestication. Trends Genet 29 : 197–205 Available: http://www.ncbi.nlm.nih.gov/pubmed/23415592. Accessed 22 May 2013.
39. BonfiglioS, GinjaC, De GaetanoA, AchilliA, OlivieriA, et al. (2012) Origin and spread of Bos taurus: new clues from mitochondrial genomes belonging to haplogroup T1. PLoS One 7: e38601 Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3369859&tool=pmcentrez&rendertype=abstract. Accessed 9 August 2012.
40. LarsonG, AlbarellaU, DobneyK, Rowley-ConwyP, SchiblerJ, et al. (2007) Ancient DNA, pig domestication, and the spread of the Neolithic into Europe. Proc Natl Acad Sci U S A 104 : 15276–15281 Available: http://www.pnas.org/content/104/39/15276.short. Accessed 28 August 2013.
41. OttoniC, FlinkLG, EvinA, GeörgC, De CupereB, et al. (2013) Pig domestication and human-mediated dispersal in western Eurasia revealed through ancient DNA and geometric morphometrics. Mol Biol Evol 30 : 824–832 Available: http://www.ncbi.nlm.nih.gov/pubmed/23180578. Accessed 13 September 2013.
42. LarsonG, CucchiT, FujitaM, Matisoo-SmithE, RobinsJ, et al. (2007) Phylogeny and ancient DNA of Sus provides insights into neolithic expansion in Island Southeast Asia and Oceania. Proc Natl Acad Sci U S A 104 : 4834–4839 Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=1829225&tool=pmcentrez&rendertype=abstract.
43. ErikssonJ, LarsonG, GunnarssonU, Bed'homB, Tixier-BoichardM, et al. (2008) Identification of the yellow skin gene reveals a hybrid origin of the domestic chicken. PLoS Genet 4: e1000010 Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2265484&tool=pmcentrez&rendertype=abstract. Accessed 22 May 2013.
44. StewartJL (1951) The West African Shorthorn cattle. Their value to Africa as trypanosomiasis-resistant animals. Vet Rec 63 : 454.
45. Abi-RachedL, JobinMJ, KulkarniS, McWhinnieA, DalvaK, et al. (2011) The shaping of modern human immune systems by multiregional admixture with archaic humans. Science 334 : 89–94 Available: http://www.ncbi.nlm.nih.gov/pubmed/21868630. Accessed 6 August 2013.
46. DowningT, LynnDJ, ConnellS, LloydAT, Bhuiyan aK, et al. (2009) Evidence of balanced diversity at the chicken interleukin 4 receptor alpha chain locus. BMC Evol Biol 9 : 136 Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3224688&tool=pmcentrez&rendertype=abstract. Accessed 6 September 2013.
47. MacHughDE, ShriverMD, LoftusRT, CunninghamP, BradleyDG (1997) Microsatellite DNA variation and the evolution, domestication and phylogeography of taurine and zebu cattle (Bos taurus and Bos indicus). Genetics 146 : 1071–1086 Available: http://www.genetics.org/cgi/content/abstract/146/3/1071.
48. Ajmone-MarsanP, GarciaJF, LenstraJA (2010) On the origin of cattle: How aurochs became cattle and colonized the world. Evol Anthropol Issues, News, Rev 19 : 148–157 Available: http://doi.wiley.com/10.1002/evan.20267. Accessed 5 September 2013.
49. Cockrill WR (1974) The husbandry and health of the domestic buffalo. Cockrill WR, editor Rome: Food and Agriculture Organization of the United Nations.
50. CymbronT, LoftusRT, MalheiroMI, BradleyDG (1999) Mitochondrial sequence variation suggests an African influence in Portuguese cattle. Proc Biol Sci 266 : 597–603 Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=1689806&tool=pmcentrez&rendertype=abstract. Accessed 23 July 2013.
51. CymbronT, FreemanAR, Isabel MalheiroM, VigneJ-D, BradleyDG (2005) Microsatellite diversity suggests different histories for Mediterranean and Northern European cattle populations. Proc Biol Sci 272 : 1837–1843 Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=1559860&tool=pmcentrez&rendertype=abstract. Accessed 31 December 2012.
52. MirolPM, GiovambattistaG, LirónJP, DuloutFN (2003) African and European mitochondrial haplotypes in South American Creole cattle. Heredity (Edinb) 91 : 248–254 Available: http://www.ncbi.nlm.nih.gov/pubmed/12939625.
53. LirónJP, BraviCM, MirolPM, Peral-GarcíaP, GiovambattistaG (2006) African matrilineages in American Creole cattle: evidence of two independent continental sources. Anim Genet 37 : 379–382 Available: http://www.ncbi.nlm.nih.gov/pubmed/16879351. Accessed 5 September 2013.
54. PattersonN, MoorjaniP, LuoY, MallickS, RohlandN, et al. (2012) Ancient admixture in human history. Genetics 192 : 1065–1093 Available: http://www.ncbi.nlm.nih.gov/pubmed/22960212. Accessed 13 September 2013.
55. McTavishEJ, DeckerJE, SchnabelRD, TaylorJF, HillisDM (2013) New World cattle show ancestry from multiple independent domestication events. Proc Natl Acad Sci U S A 110: E1398–406 Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3625352&tool=pmcentrez&rendertype=abstract. Accessed 21 May 2013.
56. MatukumalliLK, LawleyCT, SchnabelRD, TaylorJF, AllanMF, et al. (2009) Development and characterization of a high density SNP genotyping assay for cattle. PLoS One 4: e5350 Available: http://www.ncbi.nlm.nih.gov/pubmed/19390634.
57. ZiminAV, DelcherAL, FloreaL, KelleyDR, SchatzMC, et al. (2009) A whole-genome assembly of the domestic cow, Bos taurus. Genome Biol 10: R42 Available: http://www.ncbi.nlm.nih.gov/pubmed/19393038. Accessed 5 March 2012.
58. PurcellSM, NealeB, Todd-BrownK, ThomasL, FerreiraMAR, et al. (2007) PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81 : 559–575 Available: http://pngu.mgh.harvard.edu/purcell/plink/. Accessed 1 March 2012.
59. Purcell SM (2009) PLINK. Available: http://pngu.mgh.harvard.edu/purcell/plink/.
60. DeckerJE, McKaySD, RolfMM, KimJ, AlcaláAM, et al. (2013) Data from: Worldwide Patterns of Ancestry, Divergence, and Admixture in Domesticated Cattle. Dryad Digit Repos doi:10.5061/dryad.th092
61. ReichD, ThangarajK, PattersonN, PriceAL, SinghL (2009) Reconstructing Indian population history. Nature 461 : 489–494 Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2842210&tool=pmcentrez&rendertype=abstract. Accessed 2 March 2013.
Štítky
Genetika Reprodukční medicína
Článek Sex Chromosome Turnover Contributes to Genomic Divergence between Incipient Stickleback SpeciesČlánek Final Pre-40S Maturation Depends on the Functional Integrity of the 60S Subunit Ribosomal Protein L3
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2014 Číslo 3
-
Všechny články tohoto čísla
- , a Gene That Influences the Anterior Chamber Depth, Is Associated with Primary Angle Closure Glaucoma
- Genomic View of Bipolar Disorder Revealed by Whole Genome Sequencing in a Genetic Isolate
- The Rate of Nonallelic Homologous Recombination in Males Is Highly Variable, Correlated between Monozygotic Twins and Independent of Age
- Genetic Determinants Influencing Human Serum Metabolome among African Americans
- Heterozygous and Inherited Mutations in the Smooth Muscle Actin () Gene Underlie Megacystis-Microcolon-Intestinal Hypoperistalsis Syndrome
- Genome-Wide Meta-Analysis of Homocysteine and Methionine Metabolism Identifies Five One Carbon Metabolism Loci and a Novel Association of with Ischemic Stroke
- Cancer Evolution Is Associated with Pervasive Positive Selection on Globally Expressed Genes
- Genetic Diversity in the Interference Selection Limit
- Integrating Multiple Genomic Data to Predict Disease-Causing Nonsynonymous Single Nucleotide Variants in Exome Sequencing Studies
- An Evolutionary Analysis of Antigen Processing and Presentation across Different Timescales Reveals Pervasive Selection
- Cleavage Factor I Links Transcription Termination to DNA Damage Response and Genome Integrity Maintenance in
- DNA Dynamics during Early Double-Strand Break Processing Revealed by Non-Intrusive Imaging of Living Cells
- Genetic Basis of Metabolome Variation in Yeast
- Modeling 3D Facial Shape from DNA
- Dysregulated Estrogen Receptor Signaling in the Hypothalamic-Pituitary-Ovarian Axis Leads to Ovarian Epithelial Tumorigenesis in Mice
- Photoactivated UVR8-COP1 Module Determines Photomorphogenic UV-B Signaling Output in
- Local Evolution of Seed Flotation in Arabidopsis
- Chromatin Targeting Signals, Nucleosome Positioning Mechanism and Non-Coding RNA-Mediated Regulation of the Chromatin Remodeling Complex NoRC
- Nucleosome Acidic Patch Promotes RNF168- and RING1B/BMI1-Dependent H2AX and H2A Ubiquitination and DNA Damage Signaling
- The -Induced Arabidopsis Transcription Factor Attenuates ABA Signaling and Renders Seedlings Sugar Insensitive when Present in the Nucleus
- Changes in Colorectal Carcinoma Genomes under Anti-EGFR Therapy Identified by Whole-Genome Plasma DNA Sequencing
- Selection of Orphan Rhs Toxin Expression in Evolved Serovar Typhimurium
- FAK Acts as a Suppressor of RTK-MAP Kinase Signalling in Epithelia and Human Cancer Cells
- Asymmetry in Family History Implicates Nonstandard Genetic Mechanisms: Application to the Genetics of Breast Cancer
- Co-translational Localization of an LTR-Retrotransposon RNA to the Endoplasmic Reticulum Nucleates Virus-Like Particle Assembly Sites
- Sex Chromosome Turnover Contributes to Genomic Divergence between Incipient Stickleback Species
- DWARF TILLER1, a WUSCHEL-Related Homeobox Transcription Factor, Is Required for Tiller Growth in Rice
- Functional Organization of a Multimodular Bacterial Chemosensory Apparatus
- Genome-Wide Analysis of SREBP1 Activity around the Clock Reveals Its Combined Dependency on Nutrient and Circadian Signals
- The Yeast Sks1p Kinase Signaling Network Regulates Pseudohyphal Growth and Glucose Response
- An Interspecific Fungal Hybrid Reveals Cross-Kingdom Rules for Allopolyploid Gene Expression Patterns
- Temperate Phages Acquire DNA from Defective Prophages by Relaxed Homologous Recombination: The Role of Rad52-Like Recombinases
- Dying Cells Protect Survivors from Radiation-Induced Cell Death in
- Determinants beyond Both Complementarity and Cleavage Govern MicroR159 Efficacy in
- The bHLH Factors Extramacrochaetae and Daughterless Control Cell Cycle in Imaginal Discs through the Transcriptional Regulation of the Phosphatase
- The First Steps of Adaptation of to the Gut Are Dominated by Soft Sweeps
- Bacterial Regulon Evolution: Distinct Responses and Roles for the Identical OmpR Proteins of Typhimurium and in the Acid Stress Response
- Final Pre-40S Maturation Depends on the Functional Integrity of the 60S Subunit Ribosomal Protein L3
- Mitogen-Activated Protein Kinase (MAPK) Pathway Regulates Branching by Remodeling Epithelial Cell Adhesion
- CDP-Diacylglycerol Synthetase Coordinates Cell Growth and Fat Storage through Phosphatidylinositol Metabolism and the Insulin Pathway
- Coronary Heart Disease-Associated Variation in Disrupts a miR-224 Binding Site and miRNA-Mediated Regulation
- TBX3 Regulates Splicing : A Novel Molecular Mechanism for Ulnar-Mammary Syndrome
- Identification of Interphase Functions for the NIMA Kinase Involving Microtubules and the ESCRT Pathway
- Is a Cancer-Specific Fusion Gene Recurrent in High-Grade Serous Ovarian Carcinoma
- LILRB2 Interaction with HLA Class I Correlates with Control of HIV-1 Infection
- Worldwide Patterns of Ancestry, Divergence, and Admixture in Domesticated Cattle
- Parent-of-Origin Effects Implicate Epigenetic Regulation of Experimental Autoimmune Encephalomyelitis and Identify Imprinted as a Novel Risk Gene
- The Phospholipid Flippase dATP8B Is Required for Odorant Receptor Function
- Noise Genetics: Inferring Protein Function by Correlating Phenotype with Protein Levels and Localization in Individual Human Cells
- DUF1220 Dosage Is Linearly Associated with Increasing Severity of the Three Primary Symptoms of Autism
- Sugar and Chromosome Stability: Clastogenic Effects of Sugars in Vitamin B6-Deficient Cells
- Pheromone-Sensing Neurons Expressing the Ion Channel Subunit Stimulate Male Courtship and Female Receptivity
- Gene-Based Sequencing Identifies Lipid-Influencing Variants with Ethnicity-Specific Effects in African Americans
- Telomere Shortening Unrelated to Smoking, Body Weight, Physical Activity, and Alcohol Intake: 4,576 General Population Individuals with Repeat Measurements 10 Years Apart
- A Combination of Activation and Repression by a Colinear Hox Code Controls Forelimb-Restricted Expression of and Reveals Hox Protein Specificity
- An ER Complex of ODR-4 and ODR-8/Ufm1 Specific Protease 2 Promotes GPCR Maturation by a Ufm1-Independent Mechanism
- Epigenetic Control of Effector Gene Expression in the Plant Pathogenic Fungus
- Genetic Dissection of Photoreceptor Subtype Specification by the Zinc Finger Proteins Elbow and No ocelli
- Validation and Genotyping of Multiple Human Polymorphic Inversions Mediated by Inverted Repeats Reveals a High Degree of Recurrence
- CYP6 P450 Enzymes and Duplication Produce Extreme and Multiple Insecticide Resistance in the Malaria Mosquito
- GC-Rich DNA Elements Enable Replication Origin Activity in the Methylotrophic Yeast
- An Epigenetic Signature in Peripheral Blood Associated with the Haplotype on 17q21.31, a Risk Factor for Neurodegenerative Tauopathy
- Lsd1 Restricts the Number of Germline Stem Cells by Regulating Multiple Targets in Escort Cells
- RBPJ, the Major Transcriptional Effector of Notch Signaling, Remains Associated with Chromatin throughout Mitosis, Suggesting a Role in Mitotic Bookmarking
- The Membrane-Associated Transcription Factor NAC089 Controls ER-Stress-Induced Programmed Cell Death in Plants
- The Functional Consequences of Variation in Transcription Factor Binding
- Comparative Genomic Analysis of N-Fixing and Non-N-Fixing spp.: Organization, Evolution and Expression of the Nitrogen Fixation Genes
- An Insulin-to-Insulin Regulatory Network Orchestrates Phenotypic Specificity in Development and Physiology
- Suicidal Autointegration of and Transposons in Eukaryotic Cells
- A Multi-Trait, Meta-analysis for Detecting Pleiotropic Polymorphisms for Stature, Fatness and Reproduction in Beef Cattle
- Genome-Wide DNA Methylation Analysis Predicts an Epigenetic Switch for GATA Factor Expression in Endometriosis
- Genome-Wide DNA Methylation Analysis of Human Pancreatic Islets from Type 2 Diabetic and Non-Diabetic Donors Identifies Candidate Genes That Influence Insulin Secretion
- The and Hybrid Incompatibility Genes Suppress a Broad Range of Heterochromatic Repeats
- The Kil Peptide of Bacteriophage λ Blocks Cytokinesis via ZipA-Dependent Inhibition of FtsZ Assembly
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Worldwide Patterns of Ancestry, Divergence, and Admixture in Domesticated Cattle
- Genome-Wide DNA Methylation Analysis of Human Pancreatic Islets from Type 2 Diabetic and Non-Diabetic Donors Identifies Candidate Genes That Influence Insulin Secretion
- Genetic Dissection of Photoreceptor Subtype Specification by the Zinc Finger Proteins Elbow and No ocelli
- GC-Rich DNA Elements Enable Replication Origin Activity in the Methylotrophic Yeast
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



