-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaSelection of Orphan Rhs Toxin Expression in Evolved Serovar Typhimurium
Salmonella Typhimurium is a bacterium that causes intestinal diseases in a number of animals including humans. In mice, this pathogen invades tissues, causing symptoms similar to typhoid fever. In an effort to understand the evolution of this pathogen, we grew S. Typhimurium in either liquid broth or in mice for many generations and examined the resulting “evolved” strains to determine if they were different from the original “parent” culture. We found that many of these evolved strains inhibited the growth of the parent after they were mixed together, and that this growth inhibition requires that the evolved and parental cells are in close contact. Genetic analysis showed that this contact-dependent growth inhibition requires Rhs protein, which has a toxic tip. Salmonella is normally resistant to its Rhs toxin because it also produces an immunity protein that blocks toxin activity. However, evolved cells have undergone a DNA rearrangement that allows them to express a different Rhs toxic tip that inhibits growth of the parental cells, which lack immunity to it. This allows the evolved cells to outgrow the original parental cells. Our work indicates that populations of Salmonella are dynamic, with individuals battling with each other for dominance.
Published in the journal: . PLoS Genet 10(3): e32767. doi:10.1371/journal.pgen.1004255
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004255Summary
Salmonella Typhimurium is a bacterium that causes intestinal diseases in a number of animals including humans. In mice, this pathogen invades tissues, causing symptoms similar to typhoid fever. In an effort to understand the evolution of this pathogen, we grew S. Typhimurium in either liquid broth or in mice for many generations and examined the resulting “evolved” strains to determine if they were different from the original “parent” culture. We found that many of these evolved strains inhibited the growth of the parent after they were mixed together, and that this growth inhibition requires that the evolved and parental cells are in close contact. Genetic analysis showed that this contact-dependent growth inhibition requires Rhs protein, which has a toxic tip. Salmonella is normally resistant to its Rhs toxin because it also produces an immunity protein that blocks toxin activity. However, evolved cells have undergone a DNA rearrangement that allows them to express a different Rhs toxic tip that inhibits growth of the parental cells, which lack immunity to it. This allows the evolved cells to outgrow the original parental cells. Our work indicates that populations of Salmonella are dynamic, with individuals battling with each other for dominance.
Introduction
Bacteria often reside in complex communities such as biofilms in which cells from multiple species touch one another in a three-dimensional network [1]. These environments provide opportunities for cellular interactions, yet the mechanisms underlying contact-dependent competition and cooperation have been largely unexplored until recently. A diverse family of YD-peptide repeat proteins mediates at least two distinct forms of contact-dependent competition in Gram-negative and -positive bacteria [2]. The Rhs (rearrangement hotspot) proteins of Gram-negative enterobacteria [3], [4] are large (∼1,400–1,700 residues) toxic effectors that appear to be exported through the type VI secretion machinery. Related WapA (wall-associated protein A) proteins from Gram-positive bacteria are somewhat larger (∼2,200–3,600 residues) [5] and are likely exported through the general secretory pathway [2]. Rhs and WapA proteins are both characterized by sequence-diverse C-terminal regions (Rhs-CT and WapA-CT) that vary considerably between different strains of the same species. Analysis of several Rhs-CTs and WapA-CTs from Dickeya dadantii 3937 and Bacillus subtilis subspecies revealed that these domains contain the toxin activities responsible for intercellular growth inhibition. All rhs and wapA genes are closely linked to small downstream open reading frames that encode RhsI and WapI immunity proteins, respectively. These immunity proteins are also sequence-diverse and only protect against their cognate Rhs-CT (or WapA-CT) toxins. Thus, Rhs and WapA represent related, yet distinct, delivery platforms for polymorphic toxin domains [2]. Because different strains typically express unique rhs-CT/rhsI (wapA-CT/wapI) alleles, these systems collectively form a complex network of toxin/immunity pairs that are thought to mediate inter-strain competition for environmental resources [2].
The rhs loci of Enterobacteriacae often contain one or more additional rhs-CT/rhsI gene pairs located downstream of the main rhs/rhsI pair. These modules have been termed “orphan” toxin/immunity pairs, because the rhs-CT coding sequences resemble displaced fragments from full-length rhs genes [6]. Orphan rhs-CT genes often contain some coding sequence for portions of the conserved N-terminal regions, but orphan fragments are much smaller than full rhs genes and usually lack translation initiation signals. Therefore, it is unclear whether orphan rhs-CT genes are expressed, raising the question of whether these auxiliary elements are functional. Here, we show that repeated passage of Salmonella enterica serovar Typhimurium LT2 (StLT2) produces “evolved” lineages that deploy the orphan Rhs-CT toxin to inhibit the growth of ancestral cells. We provide evidence that the rhs locus undergoes rearrangement to fuse the rhsmain and rhs-CTorphan genes, thereby providing a mechanism to express and export the Rhs-CTorphan toxin domain. These results indicate that rhs rearrangement provides a selective advantage to a subpopulation of cells, suggesting that rhs plays an important role in clonal selection and bacterial evolution.
Results
In an effort to isolate StLT2 strains with increased fitness, we serially passaged cells for ∼1,000 generations in LB medium [7]. Analysis of six independently evolved cultures revealed that each lineage outcompeted ancestral StLT2 cells in co-culture experiments (Figures 1A & S1A). Remarkably, we observed the same competitive advantage in four of eight StLT2 lineages that were obtained by passage through multiple mouse hosts [8] (Figures 1A & S1B). This competitive advantage was not due to faster growth rate, because four of the evolved lineages grew more slowly than the ancestral strain (Figure S2). To further explore this phenotype, we tested whether evolved lineages inhibit ancestral cells in a contact-dependent manner. We co-cultured evolved and ancestral cells using trans-well culture dishes, in which the two populations are separated by membranes of different porosities [9]. The growth of ancestral cells was inhibited when the populations were separated by a cell-permeable 8.0 µm filter, but not when cell contact was prevented with a 0.4 µm filter (Figure 1B). These results indicate that evolved cells must be in close proximity to target cells in order to inhibit growth. This phenomenon is reminiscent of Rhs-mediated growth inhibition, which we recently characterized for D. dadantii 3937 [2]. StLT2 contains a single rhs locus, which contains a full-length “main” rhs gene (STM0291) and an “orphan” rhs gene fragment (STM0292) (Figure 2). Both rhs genes are closely linked to small open reading frames representing potential rhsI immunity genes (Figure 2), although the predicted rhsImain immunity gene found downstream of rhsmain is not annotated in the genome sequence NC_003197. To determine if the rhs region is responsible for the observed growth inhibition, we tested whether over-expression of either rhsImain or rhsIorphan immunity genes provided protection against evolved StLT2 lineages. Parental StLT2 cells overexpressing rhsImain were still inhibited by the evolved lineages, but overexpression of the rhsIorphan gene fully protected targets from growth inhibition (Figure 1A). These data strongly suggest that evolved StLT2 cells gained the ability to deliver Rhs-CTorphan toxin into neighboring cells.
Fig. 1. Evolved StLT2 cells inhibit the growth of ancestral cells. 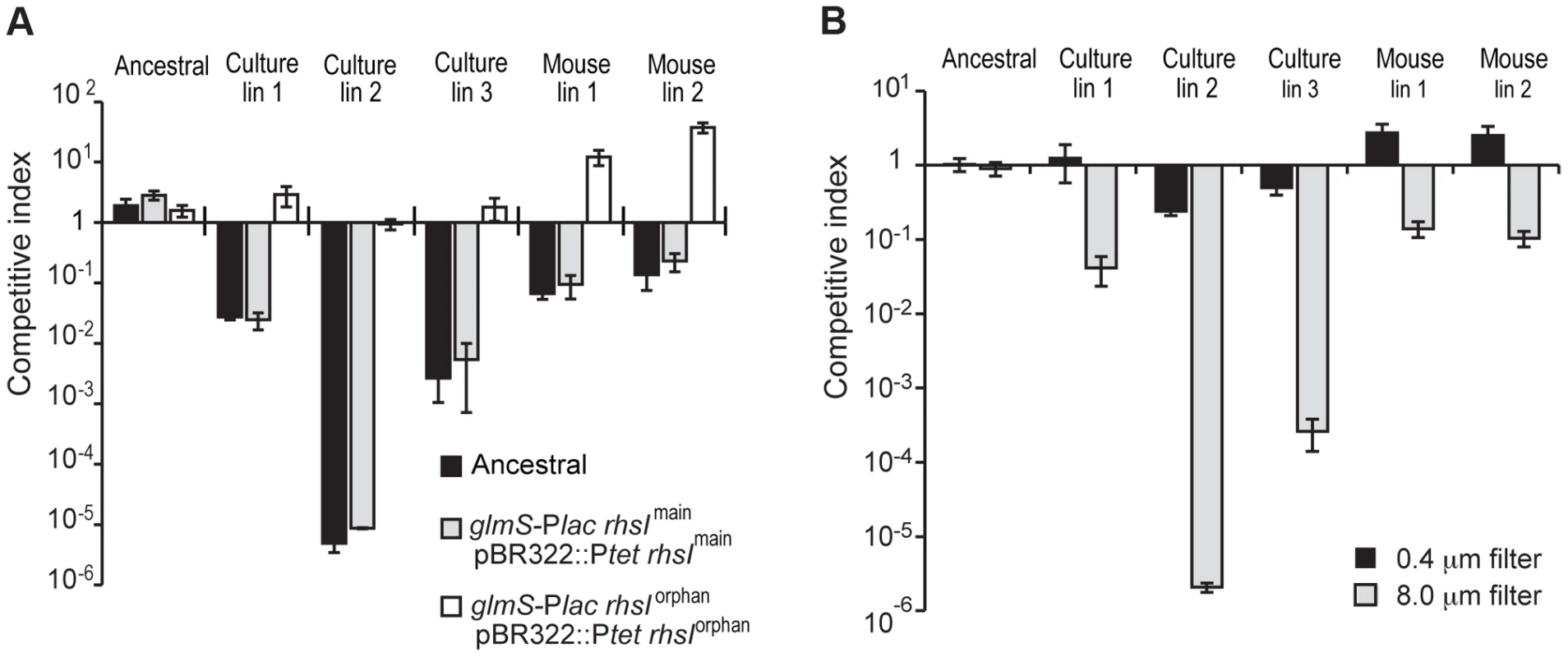
A) The indicated evolved StLT2 lineages were co-cultured with the ancestral strain for 24 h in broth. Viable cell counts for each population were determined as colony forming units and these data were used to calculate the competitive index as described in Methods. Each evolved lineage was competed against ancestral cells (black bars), ancestral cells overexpressing rhsImain (light grey bars) and ancestral cells overexpressing rhsIorphan (white bars). Reported values represent the mean ± SEM for at least three independent experiments. B) The growth inhibition activity of evolved StLT2 requires cell-cell contact. Evolved cells were co-cultured with ancestral cells in adjacent wells of a trans-well incubation chamber. Culture chambers were separated by membranes containing 0.4 µm or 8 µm pores as indicated. Reported competitive indices represent the mean ± SEM for three independent experiments. Fig. 2. Proposed mechanism of rhs locus rearrangement in evolved inhibitor cells. 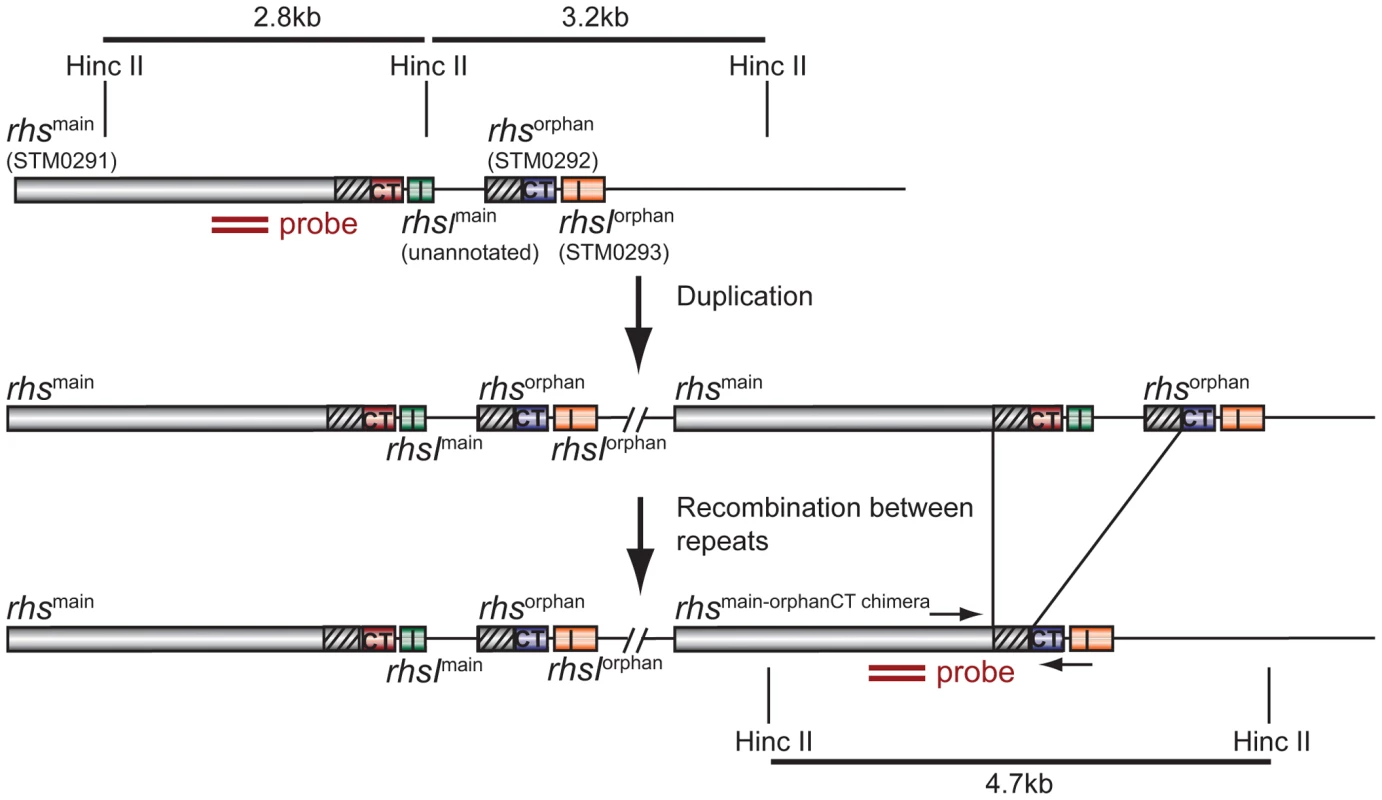
The StLT2 rhs locus contains a full-length rhsmain gene (STM0291) and an rhs-CTorphan gene fragment (STM0292) together with associated rhsI immunity genes. Duplication of the rhs locus would provide the opportunity for subsequent homologous recombination between the 522 bp region of near sequence identity (95%) between rhsmain and rhs-CTorphan (depicted as diagonal hatched regions). The binding site of the Southern blot hybridization probe is indicated by the double ochre bars. Primer binding sites for PCR amplification of the rhsmain/rhs-CTorphan junction are indicated by convergent horizontal arrows. We next tested each rhs/rhsI gene pair to confirm that they encode functional toxin and immunity proteins. Nucleotides 3608 to 4095 of rhsmain and nucleotides 269 to 741 of rhs-CTorphan were cloned under the control of the arabinose-inducible PBAD promoter. The predicted rhsI immunity genes were cloned using a compatible plasmid under control of the IPTG-inducible Ptrc promoter. These plasmids were then introduced into StLT2 cells to evaluate toxin and immunity functions. Induction of either rhs-CTmain or rhs-CTorphan in StLT2 resulted in rapid growth arrest (Figure 3A). In each instance, growth inhibition was neutralized by expression of the cognate rhsI immunity gene. However, co-expression of non-cognate immunity genes did not alleviate growth arrest (Figure 3A), demonstrating that RhsImain and RhsIorphan immunity proteins are specific for their cognate toxins. We obtained essentially identical results upon expressing the rhs main toxin and immunity genes in E. coli cells (Figure 3B). These results indicate that Rhs-CTorphan is capable of inhibiting bacterial growth and support a model in which evolved StLT2 lineages deploy the orphan toxin to inhibit the ancestral strain.
Fig. 3. The StLT2 rhs locus encodes cognate toxin/immunity pairs. 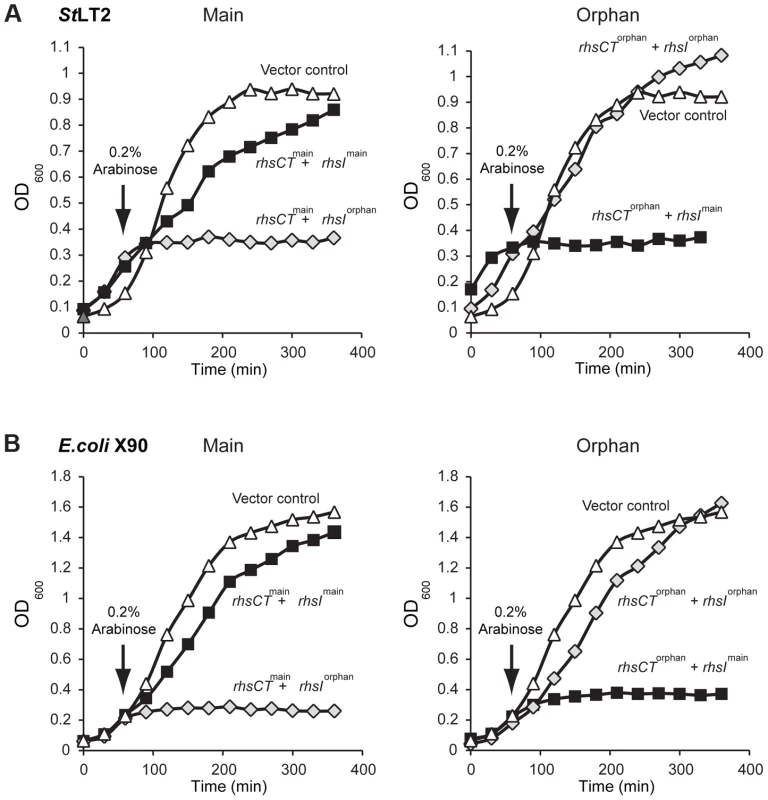
A) Expression of rhs-CTmain or rhs-CTorphan inhibits the growth of StLT2 cells. Expression the plasmid-borne rhs-CT genes was induced by addition of L-arabinose at the indicated time, and cell growth was monitored by measuring the optical density at 600 nm (OD600). The cells also co-expressed either rhsImain (dark squares) or rhsIorphan immunity genes (light grey diamonds) from IPTG-inducible promoters. Growth is compared to control cells that carry the empty vector plasmids (triangles). B) Expression of rhs-CTmain or rhs-CTorphan inhibits the growth of E.coli cells. The rhs-CT and rhsI genes were expressed in E. coli cells from the same plasmids described in panel A, and growth monitored by measuring the OD600 of the cultures. The rhs-CTorphan sequence does not encode a full-length Rhs protein, raising the question of how this toxin is synthesized and exported from evolved cells. The rhsmain and rhs-CTorphan coding regions share 95% sequence identity over 522 base-pairs (Figure 2), raising the possibility that homologous recombination in the evolved lines generates a new full-length rhs gene that encodes the Rhs-CTorphan toxin domain [10]. Bacteria expressing this Rhs chimera would have a growth advantage if rhsIorphan expression is low in ancestral cells. However, the proposed recombination event would also delete the rhsImain gene, rendering the evolved cells sensitive to inhibition by siblings expressing the main Rhs-CT toxin. Therefore, we hypothesized that rhs recombination occurs subsequent to duplication of the locus such that evolved cells retain the rhsImain immunity gene (Figure 2). To test this hypothesis, we analyzed chromosomal DNA from evolved and ancestral lineages by Southern blot. DNA was digested with HincII, which cleaves between the rhsImain and rhs-CTorphan coding sequences, and probed with a labeled DNA fragment that specifically hybridizes to rhsmain (Figure 2). We detected a unique junction fragment representing fusion of rhs-CTorphan to the upstream rhsmain gene in StLT2 lineage 2, which displayed the highest level of growth inhibition of all lineages (Figures 1 & 4A). The wild-type rhs locus was also detected in lineage 2 (Figure 4A), which is consistent with rhs region amplification, but may also indicate distinct populations of recombinant and non-recombinant cells. Orphan rhs recombinants were not detected in the other evolved lineages by Southern blot analysis (Figure 4A). Because the growth inhibition phenotype varied in magnitude between the different evolved strains, it is possible that only a fraction of the evolved StLT2 cells are rhs recombinants. If so, then the proportion of recombined rhs loci in the DNA sample may be below the detection limit of Southern analysis. Therefore we analyzed each evolved lineage with quantitative real-time PCR (qPCR) to measure the relative levels of rhsmain-rhsorphan junction sequences. All five of the evolved lineages contained 10 - to 1,000-fold more rhsmain-rhsorphan junction than ancestral StLT2 (Figure 4B), consistent with the ability of these strains to deploy Rhs-CTorphan toxin.
Fig. 4. Evidence for rhs rearrangement in evolved StLT2. 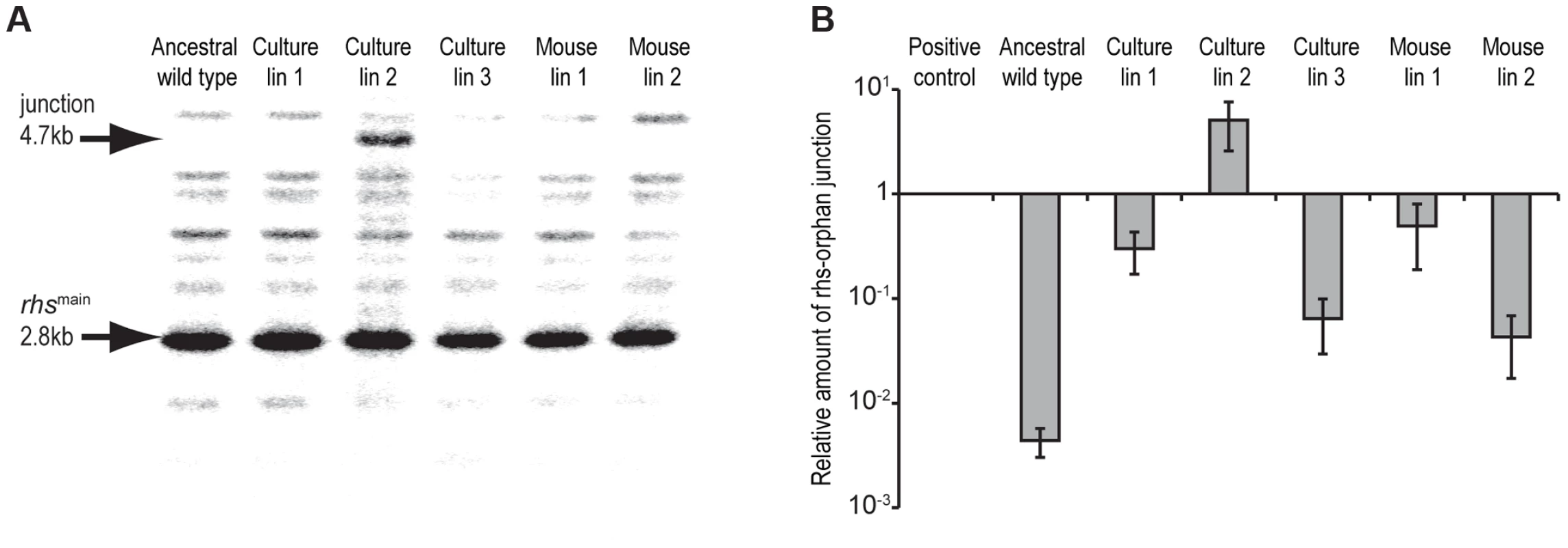
A) Southern blot analysis of evolved lineages. Arrows indicate positions of the 2.8 kbp HincII restriction fragment containing the ancestral rhs region and the 4.7 kbp restriction fragment resulting from recombination between rhsmain and rhs-CTorphan (see Figure 2 for model). B) Real-time qPCR analysis of rhsmain/rhs-CTorphan-CT recombination junctions in evolved StLT2. The levels of rhsmain/rhs-CTorphan junction products are expressed relative to amplified products of a control locus (bamA). Positive control cells are engineered StLT2 that contain a chromosomal deletion fusing rhsmain to rhs-CTorphan. Because only a fraction of the passaged cells appeared to display growth inhibitory activity, we asked whether inhibitor-cell clones could be isolated from each population. As a control, we first isolated colonies from an overnight culture of the ancestral strain and tested these clones for growth inhibition activity. None of the ten ancestral clones tested were inhibitory, suggesting that the proposed rhs rearrangements occur at low frequency. By contrast, approximately 30–90% of the clones isolated from the culture-evolved lineages and ∼20% of the clones from mouse-evolved lineage 1 showed inhibition activity against ancestral cells (Figure 5A). However, no inhibitor clones were isolated from mouse-evolved lineage 2 (Figure 5A). Strikingly, the inhibition activity of these clones varied considerably. For example, competitive index values ranged from 10−1 to 10−5 for competitions between ancestral cells and inhibitory clones isolated from evolved lineage 2 (Figure S3). Although their potencies varied, it appears that each inhibitor-cell clone deployed the Rhs-CTorphan toxin because ancestral cells could be protected through over-expression of rhsIorphan, but not rhsImain (Figure 5B). The presence of DNA fragments corresponding to both ancestral and recombinant rhs loci in lineage 2 (Figure 4A) suggests that either the rhs region was duplicated or there are distinct populations of recombinant and non-recombinant cells. In the latter case, single colonies isolated from the inhibitory lineages would contain only the rhs - rhs-CTorphan junction and not the rhs-CTmain sequence. However, PCR analysis of the single colonies with inhibitory activity in Figure 5B showed that each contained both ancestral and recombinant rhs loci. In addition, sequence analysis of the recombinant PCR product verified that recombination occurred between the regions of homology shared by rhsmain and rhs-CTorphan. Together, these data demonstrate that the evolved populations are heterogeneous with respect to Rhs-CTorphan mediated inhibition activity. Furthermore, these results suggest that the inhibition phenotype of a given culture may be due entirely to a minor subpopulation of potent inhibitor cells.
Fig. 5. A subpopulation of evolved cells has growth inhibition activity. 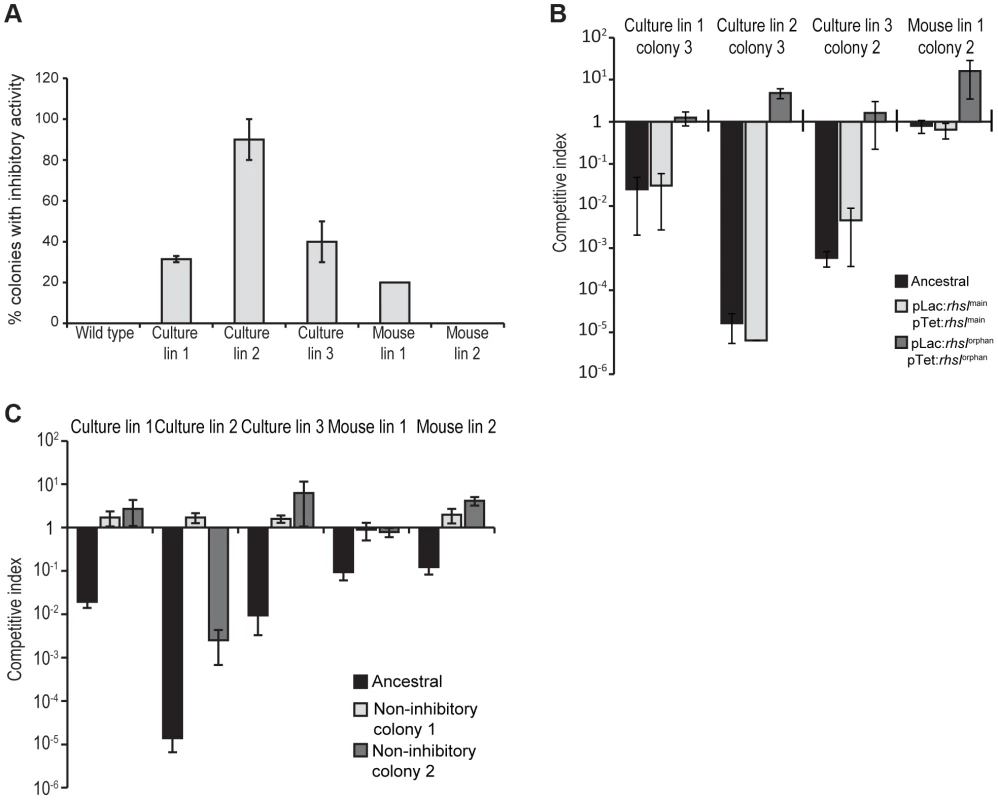
A) Two sets of independent clones were isolated twice from the evolved lineages and tested for growth inhibition activity against ancestral cells. The percentage of evolved clones with inhibition activity is shown. Reported values represent the mean ± SEM for at least two independent experiments. B) Growth inhibition activity of isolated evolved clones. Clones from each evolved lineage were competed against ancestral cells (black bars), ancestral cells overexpressing rhsImain (light grey bars) and ancestral cells overexpressing rhsIorphan (white bars). Reported values represent the mean ± SEM for at least three independent experiments. C) Growth inhibition activity of evolved lineages towards non-inhibitory clones. Evolved lineages were co-cultured with ancestral cells (black bars) and two non-inhibitory clones isolated from the evolved cultures (light grey and white bars). Competitive indices represent the mean ± SEM for at least three independent experiments. Because inhibitor cells represent a subpopulation in the evolved cultures, the other non-recombinant cells in the cohort are presumably resistant to the Rhs-CTorphan toxin. To test this hypothesis, we isolated non-inhibitory clones from each of the evolved cultures and tested them in competition co-cultures against their respective evolved lineages. As predicted, each of the non-inhibitory clones was either fully - or partially-resistant to its cohort lineage (Figure 5C). These cells likely carry uncharacterized resistance mutations that may prevent cell-cell contact, block the delivery of Rhs-CTorphan toxin,, or increase the immunity of these cells to Rhs-CT toxin.
To directly detect Rhs-CTorphan expression in the evolved lineages, we examined cells by immunofluorescence microscopy using polyclonal antibodies against the Rhs-CTorphan toxin. Rhs-CTorphan antigen was detected on the surface of some cells within evolved lineages 1, 2 and 3 as well as mouse-evolved lineages 1 and 2 (Figures 6A & S4). In contrast, the Rhs-CTorphan signal was undetectable on the surface of both ancestral StLT2 cells and cells carrying a deletion of the rhs-CTorphan (Figures 6A & S4). We then quantified the fraction of cells with Rhs-CTorphan antigen on the cell-surface using flow-cytometry. Evolved lineages showed a 2 - to 20-fold increase in the fraction of Rhs-CTorphan-positive cells compared to ancestral StLT2 cells (Figures 6B & S5). Mouse-evolved StLT2 showed very low expression of Rhs-CTorphan antigen on cell surfaces (Figure 6B), consistent with the modest growth inhibition observed for these lineages (Figure 1). Based on Southern blot and RT-qPCR analyses, it seems likely that surface expression of Rhs-CTorphan requires rhs locus rearrangement to generate a chimeric rhs gene. In accord with this conclusion, we also found that over-expression of rhs-CTorphan from a multicopy plasmid does not increase Rhs-CTorphan antigen levels on the cell surface (Figures 6B & S5). Therefore, we sought to detect the predicted Rhs fusion protein using antisera to the Rhs-CTorphan toxin. Western blot analysis revealed an immuno-reactive protein at ∼150 kDa in culture evolved lineages 2 and 3 (Figure 6C). This product corresponds to the expected size of the Rhs fusion protein. Moreover, we were unable to detect the 29 kDa product encoded by rhs-CTorphan in the ancestral and evolved lineages (Figure 6C). Together, these data strongly suggest that the rhs-CTorphan reading frame must recombine with rhsmain to be expressed.
Fig. 6. Expression of Rhs-CT orphan in evolved cells. 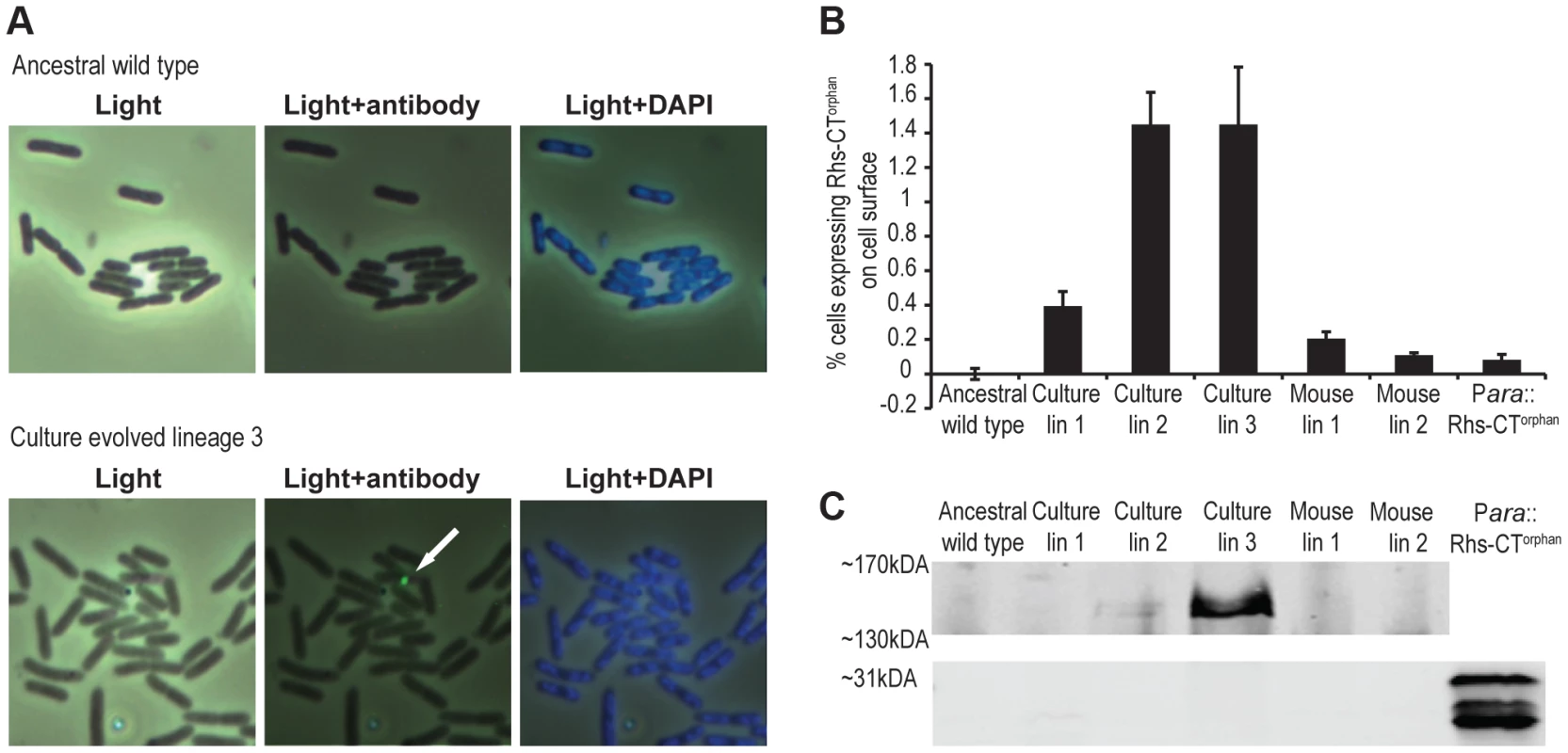
A) Immunofluorescence analysis of ancestral StLT2 and culture-evolved lineage 3 with antibodies against Rhs-CTorphan. Scale is 10 µm×10 µm for each image. B) Quantification of surface-expressed Rhs-CTorphan. Cells were labeled with Rhs-CTorphan antisera and analyzed by flow cytometry as described in Methods. Reported values represent the mean ± SEM for three independent experiments with 50,000 events recorded per sample. C) Immunoblot analysis of Rhs-CTorphan. Proteins were isolated from ancestral and evolved cells and analyzed by immunoblot using antibodies against Rhs-CTorphan. Regions corresponding to predicted migration positions of Rhs-CTorphan (∼31 kDa) and the chimeric fusion protein (∼151 kDa) are shown. Discussion
The results presented here show that serial passage of StLT2, in either laboratory media or within a natural host, leads to enrichment of cells that express Rhs-CTorphan toxin. Analysis of the rhs locus indicates that evolved cells undergo recombination between rhsmain and rhs-CTorphan, forming a gene fusion that allows the Rhs-CTorphan toxin domain to be deployed. Rearranged rhs genes are detected at low levels within the evolved populations, indicating that only a fraction of cells are recombinant inhibitors. A number of observations argue that this subpopulation of cells is responsible for growth inhibition activity. First, the relative competitive advantage of each evolved lineage is correlated with its level of recombinant rhs junctions and surface expression of Rhs-CTorphan antigen. More importantly, ancestral cells are fully protected when they over-express the rhsIorphan immunity gene. Because Rhs immunity proteins are highly specific for their cognate toxins, this latter result demonstrates that Rhs-CTorphan toxin is indeed deployed by the evolved lineages. This result also indicates that ancestral StLT2 cells do not normally express rhsIorphan immunity genes under laboratory conditions. The number of inhibitor cells within each lineage is not known, but can be estimated to be <2% of the population based on flow cytometry measurements of Rhs-CTorphan antigen on cell surfaces. However, we note that this assay may underestimate the actual number of recombinant inhibitor cells because Rhs effectors are likely exported through type VI secretion systems [2], [11]. Although recent studies indicate that the N-terminal PAAR domain found within many Rhs proteins forms the tip of the type VI injection structure [12], other structural studies show that Rhs-peptide repeats form a chamber capable of encapsulating toxin domains [13]. Therefore, much of the Rhs-CTorphan antigen may be inaccessible to antibody until it is delivered to target cells. In accord with this model, we only detect Rhs-CTorphan where two bacteria make contact with one another and never on the surface of individual cells. Regardless of the absolute number of recombinants or rhs expression levels, our results suggest that a small number of inhibitor cells are capable of inhibiting a large excess of ancestral cells. The same phenomenon has been observed during bacterial contact-dependent growth inhibition (CDI), in which each CDI+ cell is able to inhibit 100–1,000 target cells over a few hours [9]. Presumably, the unstructured environment in shaking broth culture promotes a series of transient cell-cell interactions, thereby enabling toxin delivery to multiple ancestral cells.
Chromosomal duplications and amplifications occur frequently in bacteria, typically at rates of about 0.1% per generation for any given locus [14], [15]. However, there is a cost to maintaining amplified regions, and gene duplications are lost during segregation at frequencies up to 10% per generation [16], [17]. Therefore, positive selection is required to retain multiple gene copies. If the amplified region can be stabilized, then the additional gene copy can diverge towards a new function, thus providing a mechanism for evolution [16], [18]. Rearrangement of rhs loci represents a previously unrecognized mechanism for bacteria to exploit chromosomal amplifications for adaptation. We propose that, subsequent to duplication, homologous recombination occurs between rhsmain and rhs-CTorphan to generate a novel chimeric rhs element. This recombination would necessarily delete one copy of the rhsImain, but the other copy would remain and ensure that recombinant cells retain immunity to the Rhsmain toxin should it be deployed by neighboring non-recombinant siblings. This model also predicts that evolved recombinant cells could undergo homologous recombination to restore the original rhs locus (see Figure 2, reverse of the duplication step). Thus, rhs rearrangement could be exploited transiently under conditions where it confers a selective advantage, but rapidly revert back to the ancestral genotype as environmental circumstances dictate.
Analysis of over 150 Salmonella genomes shows that rhs-CT toxin sequences are diverse with at least 57 distinct sequence types (Figure S6A & Table S1). This is a common feature of rhs genes in other bacteria as well and suggests that Rhs mediates inter-strain competition. All Salmonella serovars contain at least one rhs gene, located on pathogenicity islands SPI-6 or SPI-19 [19], [20]. Approximately 50% of these serovars contain at least one predicted rhs orphan sequence, with some strains containing as many as eleven modules. There is generally high conservation of Rhs-CTmain and Rhs-CTorphan. sequences within a given serotype. For example, all sequenced Typhi isolates contain the same Rhs-CTmain and Rhs-CTorphan. sequences, whereas these CT sequence types are only found in one other serotype, thus suggesting that different toxins are linked to serotype and/or the type of infection. However, orphan rhs-CT sequences in one serotype can be present within the main rhs gene of another serotype. For example, the StLT2 Rhs-CTorphan toxin studied in this work is part of the full-length main Rhs in Salmonella enterica serovar Saintpaul SARA23 and some Newport isolates (Figures S6A & S6B). These observations and the association with horizontally transferred elements suggest that rhs genes are exchanged between different serovars and contribute to the evolution of toxin diversity.
Given that Rhs toxins are encoded on pathogenicity islands, it seems likely that these systems also play important roles in Salmonella growth and fitness during pathogenesis. Indeed, StLT2 mutants lacking a chromosomal region containing rhs-CTorphan are outcompeted by wild-type cells in mice [21], and StSL1344 mutants lacking rhs are completely attenuated in pig and cattle models of infection [22]. These observations raise the possibility that rhs locus rearrangement occurs commonly during infections. Intriguingly, StLT2 produces distinct intracellular infection foci, each originating from one or only a few clones [23]. Similarly, analysis of mice orally infected with Yersinia pseudotuberculosis indicates that only a few bacterial clones are able to disseminate from the intestines to the spleen and liver [24]. Clonal invasion has also been reported for Yersinia enterocolitica infections [25], but the mechanisms underlying these apparent dissemination bottlenecks are unknown. Most Yersinia species contain rhs loci with associated orphan gene pairs, raising the possibility that clonal expansion through rhs recombination and growth selection may be a general feature of many enterobacterial infections. Rearrangement could function as a stochastic switch that enables some cells to deploy Rhs-CTorphan and thereby “differentiate” into cells that are specialized for tissue invasion or immune modulation. Although Rhs-mediated inhibition clearly occurs between bacteria, it is also possible that Rhs toxins act directly as virulence factors. The C-terminal region of RhsT from Pseudomonas aeruginosa was recently shown to be delivered into mouse host cells [26]. In the process, the Rhs fragment activates the inflammasome and contributes to pathogenicity.
Materials and Methods
Strains and growth conditions
Bacterial strains were derived from Salmonella enterica serovar Typhimurium LT2 (StLT2) and are listed in Table S2. Bacteria were grown in LB medium [27] supplemented with 50 mM potassium phosphate (pH 7.3). Bacteria were incubated at 37°C with shaking at 200 rpm. Where appropriate, media were supplemented with antibiotics at the following concentrations: ampicillin (Amp), 200 mg/L; chloramphenicol (Cam), 17 mg/L; kanamycin (Kan), 80 mg/L; and tetracycline (Tet), 5 mg/L. Six independent lineages of StLT2 were obtained by serial passage for ∼1,000 generations [7]. Each lineage was passaged daily by dilution of 1.5 µL of overnight culture into 1.5 mL of fresh LB medium. Each evolved lineage was sampled periodically (100–150 generations) and stored at −80°C.
All growth competitions were conducted using ancestral StLT2 marked with the flhC::cat allele, which confers Cam resistance. Non-inhibitory clones isolated from the evolved cultures were transduced with the flhC::cat allele prior to testing for resistance. Ancestral and evolved cells were co-cultured in LB medium supplemented with 50 mM potassium phosphate (pH 7.3) at 37°C with shaking. At time 0 h, ∼106 cfu (1 µL of overnight culture) from evolved and ancestral cultures were suspended in 2 mL of fresh LB (pH ∼7.3) and plated for viable cell counts before shaking incubation for 24 h at 37°C. After 24 h of co-culture, viable cell counts were determined by plating onto LB agar (to enumerate evolved and ancestral cells) and LB agar supplemented with Cam (to enumerate ancestral cells). The competitive index was calculated as the ratio of ancestral∶evolved cells at time 24 h divided by the cell ratio at 0 h. Ancestral StLT2 flhC::cat cells were also supplemented with either rhsImain or rhsIorphan on the chromosome under control of the lac promoter and on plasmid pBR322 under the tet promoter. Chromosomal rhsIorphan and plasmid-borne rhsIorphan individually provided partial protection against the evolved lineages (data not shown), but both copies were required for full immunity. Proximity-dependence of growth inhibition was determined as described previously [9]. Cells were grown to OD600 ∼0.3, then transferred to a trans-well culture plate (BD diagnostics) that separates the two populations with filter containing 0.4 µm (no-contact) or 8.0 µm (contact) pores. Trans-well culture plates were seeded at an evolved∶ancestral cell ratio of 1∶1 and incubated at 37°C with shaking for 24 h. Cultures were then plated onto selective media to determine viable cell counts and to calculate competitive indices.
Construction of plasmids and chromosomal inserts
All oligonucleotides used in this study are presented in Table S3. The rhsImain and rhsIorphan genes were amplified from ancestral StLT2 chromosomal DNA using oligonucleotides 2337/2338 and 2340/2544 (respectively) and ligated to plasmid pBR322 using EcoRV and SalI restriction sites. The immunity genes were also placed under the lac promoter at the glmS locus using bacteriophage λ Red-mediated recombination [28]. Integration constructs containing rhsI genes flanked by a Kan-resistance cassette and glmS-derived homology regions were constructed by overlapping end-PCR as described previously [29]. The following primer pairs were used to amplify: upstream glmS homology (2666/2676), lac promoter (2677/2678 for rhsImain and 2677/2682 for rhsIorphan), rhsImain (2679/2680) or rhsIorphan (2683/2684), Kan-resistance cassette (2618/2619) and downstream glmS homology (2681/2667). The final PCR product was electroporated into StLT2 cells that express Red recombinase proteins, and transformants were selected on LB supplemented with Kan. Integrated immunity genes were verified by PCR analysis using primers 2666/2667 and subsequent DNA sequencing. The flhC::cat and STM0292::kan alleles were generated by PCR using primers 2436/2437 and 2410/2490 to amplify the cat/kan cassettes of plasmids pKD3 and pKD4, respectively. Each PCR product was integrated into the StLT2 chromosome by Red-mediated recombination.
To evaluate toxin activity and the specificity of immunity, individual rhs-CT and rhsI sequences were cloned under the control of inducible promoters on compatible plasmids. The rhs-CTmain and rhs-CTorphan coding sequences were amplified with primers Sty-rhs(E1203)-Nco/Sty-rhs-Xho and Sty-rhs(E1203)-Nco/Sty-orph-rhs-Xho (respectively) and ligated to plasmid pCH450 [30] using NcoI and XhoI restriction sites. The rhsImain and rhsIorphan genes were amplified and ligated to a derivative of plasmid pTrc99A using KpnI and XhoI restriction sites. Rhs-CTorphan was expressed and purified as a non-toxic variant fused to His6-tagged thioredoxin. The his6-trxA sequence was amplified from plasmid pSH21P::trxA [31] using primers pET-Sph and trxA-Bam-TEV-Kpn. The product was digested with SphI/BamHI and ligated to plasmid pET21b to generate plasmid pSH21P::trxA-TEV. The coding sequences for Rhs-CTorphan residues 112–247 and RhsIorphan were amplified using primers Sty-rhs(D1225)-Kpn/Sty-orph-rhsI-Xho) and the His208Ala mutation made by mega-primer PCR using oligonucleotide Sty-CTo1-H208A. The final product was digested with KpnI/XhoI and ligated to plasmid pSH21P::trxA-TEV to generate plasmid pCH10068. The resulting construct was used to overproduce His6-TrxA-Rhs-CT(H208A)orphan fusion protein.
Chromosomal DNA analysis
Chromosomal DNAs were isolated using the Sigma genomic DNA kit and digested with HincII restriction endonuclease. Digested DNAs were resolved by electrophoresis on 0.7% agarose gels at 34V for 15 h and blotted onto nylon membranes by capillary transfer. A probe to nucleotides 2969–3128 of rhsmain was generated by PCR using oligos 2226/2227 and labeled with [32P]-labeled using the Prime-It Random Primer Labeling Kit (Agilent Technologies). Southern blots were visualized by phosphor imaging. Fragment sizes were calculated using a standard curve based on HindIII digested λ ladder (New England Biolabs, USA) run on the same gel. The proportion of rhs recombination junctions was determined by quantitative real-time PCR (qPCR) using oligonucleotides 2226/2231 using the cycle threshold Ct-value method according to the manufacturer (Bio-Rad). Fluorescence was monitored on-line using the MyIQ iCycler real-time PCR system (Bio-Rad). The rhs-rhsorphan junction DNA levels were calculated relative to bamA DNA (oligos 1981/1990) in each sample and normalized to the level of junction DNA in ancestral cells.
Antiserum preparation and immunoblot analysis
His6-TrxA-Rhs-CT(H208A)orphan fusion protein was overproduced in E. coli CH2016 and purified by Ni2+-affinity chromatography as described [32]. The Rhs-CT(H208A)orphan domain was released by TEV protease digestion and used for antiserum production in rabbits (CoCalico Biologicals). Non-specific antibodies were removed by incubation with carbonyl-diimidazole-activated agarose beads linked to soluble protein from E. coli strain CH2016 [33]. Briefly, protein-linked beads were resuspended in 0.5 mL of antiserum (1∶5 dilution) and mixed by rotation for 1 h at room temperature followed by additional incubation for 3 h at 4°C. This process was repeated at least four times with fresh beads.
Evolved lineages were grown to mid-log phase in LB medium supplemented with 50 mM potassium phosphate (pH 7.3) and cells were collected by centrifugation and frozen at −80°C. Cell pellets were resuspended in NuPage LDS-sample buffer (Invitrogen) at 70°C and treated with benzonase to degrade nucleic acids. Cell lysates were run on 3–7% NuPage Tris-acetate gradient gels (Novex) for the detection of the Rhsmain-Rhs-CTorphan chimera, or on 4–10% Precise Tris-glycine gradient gels (Thermo Scientific) to detect Rhs-CTorphan. Gels were electrotransferred to nitrocellulose membranes and the blots incubated with polyclonal antisera against Rhs-CTorphan (1∶1,000 dilution) and secondary anti-rabbit 800CW antiserum (1∶10,000 dilution). Immunoblots were visualized using an Odyssey CLx Infrared Imaging System (LI-COR).
Immunofluorescence microscopy and flow cytometry
Cells were incubated overnight with 4% formaldehyde in 0.15 M phosphate buffered saline (PBS, pH = 7.2). Cells were washed three times with PBS and incubated with polyclonal antibodies to Rhs-CTorphan (1∶50 dilution) for 30 min. Cells were washed with PBS before incubation with secondary anti-rabbit Alexa-Fluor480 antibodies (1∶500 dilution) (Invitrogen) for 30 min on ice. After washing with PBS, cells were applied to poly-D-lysine coated slides, treated with Fluoro-gel II/DAPI (Electron Microscopy Sciences) and visualized by fluorescence microscopy. The fraction of evolved cells expressing Rhs-CTorphan on the surface was determined by flow cytometry. Antibody-labeled cells were analyzed (50,000 events for each sample) with an Accuri C6 flow cytometer with gates set to include bacteria-sized particles. StLT2 Δrhs-CTorphan cells were used to assess non-specific binding of the Rhs-CTorphan antisera. The fraction of cells with surface Rhs-CTorphan antigen was calculated as the ratio of green fluorescent particles in the population after subtracting background fluorescence observed with StLT2 Δrhs-CTorphan cells.
Supporting Information
Zdroje
1. LopezD, VlamakisH, KolterR (2010) Biofilms. Cold Spring Harb Perspect Biol 2: a000398.
2. KoskiniemiS, LamoureuxJG, NikolakakisKC, t'Kint de RoodenbekeC, KaplanMD, et al. (2013) Rhs proteins from diverse bacteria mediate intercellular competition. Proc Natl Acad Sci U S A 110 : 7032–7037.
3. HillCW (1999) Large genomic sequence repetitions in bacteria: lessons from rRNA operons and Rhs elements. Res Microbiol 150 : 665–674.
4. LinRJ, CapageM, HillCW (1984) A repetitive DNA sequence, rhs, responsible for duplications within the Escherichia coli K-12 chromosome. J Mol Biol 177 : 1–18.
5. FosterSJ (1993) Molecular analysis of three major wall-associated proteins of Bacillus subtilis 168: evidence for processing of the product of a gene encoding a 258 kDa precursor two-domain ligand-binding protein. Mol Microbiol 8 : 299–310.
6. PooleSJ, DinerEJ, AokiSK, BraatenBA, t'Kint de RoodenbekeC, et al. (2011) Identification of functional toxin/immunity genes linked to contact-dependent growth inhibition (CDI) and rearrangement hotspot (Rhs) systems. PLoS Genet 7: e1002217.
7. KoskiniemiS, SunS, BergOG, AnderssonDI (2012) Selection-driven gene loss in bacteria. PLoS Genet 8: e1002787.
8. NilssonAI, KugelbergE, BergOG, AnderssonDI (2004) Experimental adaptation of Salmonella typhimurium to mice. Genetics 168 : 1119–1130.
9. AokiSK, PammaR, HerndayAD, BickhamJE, BraatenBA, et al. (2005) Contact-dependent inhibition of growth in Escherichia coli. Science 309 : 1245–1248.
10. LovettST, HurleyRL, SuteraVAJr, AubuchonRH, LebedevaMA (2002) Crossing over between regions of limited homology in Escherichia coli. RecA-dependent and RecA-independent pathways. Genetics 160 : 851–859.
11. SilvermanJM, BrunetYR, CascalesE, MougousJD (2012) Structure and regulation of the type VI secretion system. Annu Rev Microbiol 66 : 453–472.
12. ShneiderMM, ButhSA, HoBT, BaslerM, MekalanosJJ, et al. (2013) PAAR-repeat proteins sharpen and diversify the type VI secretion system spike. Nature 500 : 350–353.
13. BusbyJN, PanjikarS, LandsbergMJ, HurstMR, LottJS (2013) The BC component of ABC toxins is an RHS-repeat-containing protein encapsulation device. Nature 501 : 547–550.
14. ReamsAB, KofoidE, SavageauM, RothJR (2010) Duplication frequency in a population of Salmonella enterica rapidly approaches steady state with or without recombination. Genetics 184 : 1077–1094.
15. AndersonP, RothJ (1981) Spontaneous tandem genetic duplications in Salmonella typhimurium arise by unequal recombination between rRNA (rrn) cistrons. Proc Natl Acad Sci U S A 78 : 3113–3117.
16. BergthorssonU, AnderssonDI, RothJR (2007) Ohno's dilemma: evolution of new genes under continuous selection. Proc Natl Acad Sci U S A 104 : 17004–17009.
17. PetterssonME, SunS, AnderssonDI, BergOG (2009) Evolution of new gene functions: simulation and analysis of the amplification model. Genetica 135 : 309–324.
18. NasvallJ, SunL, RothJR, AnderssonDI (2012) Real-time evolution of new genes by innovation, amplification, and divergence. Science 338 : 384–387.
19. FolkessonA, LofdahlS, NormarkS (2002) The Salmonella enterica subspecies I specific centisome 7 genomic island encodes novel protein families present in bacteria living in close contact with eukaryotic cells. Res Microbiol 153 : 537–545.
20. BlondelCJ, JimenezJC, ContrerasI, SantiviagoCA (2009) Comparative genomic analysis uncovers 3 novel loci encoding type six secretion systems differentially distributed in Salmonella serotypes. BMC Genomics 10 : 354.
21. MulderDT, CooperCA, CoombesBK (2012) Type VI secretion system-associated gene clusters contribute to pathogenesis of Salmonella enterica serovar Typhimurium. Infect Immun 80 : 1996–2007.
22. ChaudhuriRR, MorganE, PetersSE, PleasanceSJ, HudsonDL, et al. (2013) Comprehensive assignment of roles for Salmonella typhimurium genes in intestinal colonization of food-producing animals. PLoS Genet 9: e1003456.
23. SheppardM, WebbC, HeathF, MallowsV, EmilianusR, et al. (2003) Dynamics of bacterial growth and distribution within the liver during Salmonella infection. Cell Microbiol 5 : 593–600.
24. BarnesPD, BergmanMA, MecsasJ, IsbergRR (2006) Yersinia pseudotuberculosis disseminates directly from a replicating bacterial pool in the intestine. J Exp Med 203 : 1591–1601.
25. OellerichMF, JacobiCA, FreundS, NiedungK, BachA, et al. (2007) Yersinia enterocolitica infection of mice reveals clonal invasion and abscess formation. Infect Immun 75 : 3802–3811.
26. KungVL, KhareS, StehlikC, BaconEM, HughesAJ, et al. (2012) An rhs gene of Pseudomonas aeruginosa encodes a virulence protein that activates the inflammasome. Proc Natl Acad Sci U S A 109 : 1275–1280.
27. ScottJR (1968) Genetic studies on bacteriophage P1. Virology 36 : 564–574.
28. DatsenkoKA, WannerBL (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97 : 6640–6645.
29. AiyarA, XiangY, LeisJ (1996) Site-directed mutagenesis using overlap extension PCR. Methods Mol Biol 57 : 177–191.
30. HayesCS, SauerRT (2003) Cleavage of the A site mRNA codon during ribosome pausing provides a mechanism for translational quality control. Mol Cell 12 : 903–911.
31. RuheZC, HayesCS (2010) The N-terminus of GalE induces tmRNA activity in Escherichia coli. PLoS One 5: e15207.
32. DinerEJ, BeckCM, WebbJS, LowDA, HayesCS (2012) Identification of a target cell permissive factor required for contact-dependent growth inhibition (CDI). Genes Dev 26 : 515–525.
33. Garza-SanchezF, GinJG, HayesCS (2008) Amino acid starvation and colicin D treatment induce A-site mRNA cleavage in Escherichia coli. J Mol Biol 378 : 505–519.
Štítky
Genetika Reprodukční medicína
Článek Sex Chromosome Turnover Contributes to Genomic Divergence between Incipient Stickleback SpeciesČlánek Final Pre-40S Maturation Depends on the Functional Integrity of the 60S Subunit Ribosomal Protein L3
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2014 Číslo 3
-
Všechny články tohoto čísla
- , a Gene That Influences the Anterior Chamber Depth, Is Associated with Primary Angle Closure Glaucoma
- Genomic View of Bipolar Disorder Revealed by Whole Genome Sequencing in a Genetic Isolate
- The Rate of Nonallelic Homologous Recombination in Males Is Highly Variable, Correlated between Monozygotic Twins and Independent of Age
- Genetic Determinants Influencing Human Serum Metabolome among African Americans
- Heterozygous and Inherited Mutations in the Smooth Muscle Actin () Gene Underlie Megacystis-Microcolon-Intestinal Hypoperistalsis Syndrome
- Genome-Wide Meta-Analysis of Homocysteine and Methionine Metabolism Identifies Five One Carbon Metabolism Loci and a Novel Association of with Ischemic Stroke
- Cancer Evolution Is Associated with Pervasive Positive Selection on Globally Expressed Genes
- Genetic Diversity in the Interference Selection Limit
- Integrating Multiple Genomic Data to Predict Disease-Causing Nonsynonymous Single Nucleotide Variants in Exome Sequencing Studies
- An Evolutionary Analysis of Antigen Processing and Presentation across Different Timescales Reveals Pervasive Selection
- Cleavage Factor I Links Transcription Termination to DNA Damage Response and Genome Integrity Maintenance in
- DNA Dynamics during Early Double-Strand Break Processing Revealed by Non-Intrusive Imaging of Living Cells
- Genetic Basis of Metabolome Variation in Yeast
- Modeling 3D Facial Shape from DNA
- Dysregulated Estrogen Receptor Signaling in the Hypothalamic-Pituitary-Ovarian Axis Leads to Ovarian Epithelial Tumorigenesis in Mice
- Photoactivated UVR8-COP1 Module Determines Photomorphogenic UV-B Signaling Output in
- Local Evolution of Seed Flotation in Arabidopsis
- Chromatin Targeting Signals, Nucleosome Positioning Mechanism and Non-Coding RNA-Mediated Regulation of the Chromatin Remodeling Complex NoRC
- Nucleosome Acidic Patch Promotes RNF168- and RING1B/BMI1-Dependent H2AX and H2A Ubiquitination and DNA Damage Signaling
- The -Induced Arabidopsis Transcription Factor Attenuates ABA Signaling and Renders Seedlings Sugar Insensitive when Present in the Nucleus
- Changes in Colorectal Carcinoma Genomes under Anti-EGFR Therapy Identified by Whole-Genome Plasma DNA Sequencing
- Selection of Orphan Rhs Toxin Expression in Evolved Serovar Typhimurium
- FAK Acts as a Suppressor of RTK-MAP Kinase Signalling in Epithelia and Human Cancer Cells
- Asymmetry in Family History Implicates Nonstandard Genetic Mechanisms: Application to the Genetics of Breast Cancer
- Co-translational Localization of an LTR-Retrotransposon RNA to the Endoplasmic Reticulum Nucleates Virus-Like Particle Assembly Sites
- Sex Chromosome Turnover Contributes to Genomic Divergence between Incipient Stickleback Species
- DWARF TILLER1, a WUSCHEL-Related Homeobox Transcription Factor, Is Required for Tiller Growth in Rice
- Functional Organization of a Multimodular Bacterial Chemosensory Apparatus
- Genome-Wide Analysis of SREBP1 Activity around the Clock Reveals Its Combined Dependency on Nutrient and Circadian Signals
- The Yeast Sks1p Kinase Signaling Network Regulates Pseudohyphal Growth and Glucose Response
- An Interspecific Fungal Hybrid Reveals Cross-Kingdom Rules for Allopolyploid Gene Expression Patterns
- Temperate Phages Acquire DNA from Defective Prophages by Relaxed Homologous Recombination: The Role of Rad52-Like Recombinases
- Dying Cells Protect Survivors from Radiation-Induced Cell Death in
- Determinants beyond Both Complementarity and Cleavage Govern MicroR159 Efficacy in
- The bHLH Factors Extramacrochaetae and Daughterless Control Cell Cycle in Imaginal Discs through the Transcriptional Regulation of the Phosphatase
- The First Steps of Adaptation of to the Gut Are Dominated by Soft Sweeps
- Bacterial Regulon Evolution: Distinct Responses and Roles for the Identical OmpR Proteins of Typhimurium and in the Acid Stress Response
- Final Pre-40S Maturation Depends on the Functional Integrity of the 60S Subunit Ribosomal Protein L3
- Mitogen-Activated Protein Kinase (MAPK) Pathway Regulates Branching by Remodeling Epithelial Cell Adhesion
- CDP-Diacylglycerol Synthetase Coordinates Cell Growth and Fat Storage through Phosphatidylinositol Metabolism and the Insulin Pathway
- Coronary Heart Disease-Associated Variation in Disrupts a miR-224 Binding Site and miRNA-Mediated Regulation
- TBX3 Regulates Splicing : A Novel Molecular Mechanism for Ulnar-Mammary Syndrome
- Identification of Interphase Functions for the NIMA Kinase Involving Microtubules and the ESCRT Pathway
- Is a Cancer-Specific Fusion Gene Recurrent in High-Grade Serous Ovarian Carcinoma
- LILRB2 Interaction with HLA Class I Correlates with Control of HIV-1 Infection
- Worldwide Patterns of Ancestry, Divergence, and Admixture in Domesticated Cattle
- Parent-of-Origin Effects Implicate Epigenetic Regulation of Experimental Autoimmune Encephalomyelitis and Identify Imprinted as a Novel Risk Gene
- The Phospholipid Flippase dATP8B Is Required for Odorant Receptor Function
- Noise Genetics: Inferring Protein Function by Correlating Phenotype with Protein Levels and Localization in Individual Human Cells
- DUF1220 Dosage Is Linearly Associated with Increasing Severity of the Three Primary Symptoms of Autism
- Sugar and Chromosome Stability: Clastogenic Effects of Sugars in Vitamin B6-Deficient Cells
- Pheromone-Sensing Neurons Expressing the Ion Channel Subunit Stimulate Male Courtship and Female Receptivity
- Gene-Based Sequencing Identifies Lipid-Influencing Variants with Ethnicity-Specific Effects in African Americans
- Telomere Shortening Unrelated to Smoking, Body Weight, Physical Activity, and Alcohol Intake: 4,576 General Population Individuals with Repeat Measurements 10 Years Apart
- A Combination of Activation and Repression by a Colinear Hox Code Controls Forelimb-Restricted Expression of and Reveals Hox Protein Specificity
- An ER Complex of ODR-4 and ODR-8/Ufm1 Specific Protease 2 Promotes GPCR Maturation by a Ufm1-Independent Mechanism
- Epigenetic Control of Effector Gene Expression in the Plant Pathogenic Fungus
- Genetic Dissection of Photoreceptor Subtype Specification by the Zinc Finger Proteins Elbow and No ocelli
- Validation and Genotyping of Multiple Human Polymorphic Inversions Mediated by Inverted Repeats Reveals a High Degree of Recurrence
- CYP6 P450 Enzymes and Duplication Produce Extreme and Multiple Insecticide Resistance in the Malaria Mosquito
- GC-Rich DNA Elements Enable Replication Origin Activity in the Methylotrophic Yeast
- An Epigenetic Signature in Peripheral Blood Associated with the Haplotype on 17q21.31, a Risk Factor for Neurodegenerative Tauopathy
- Lsd1 Restricts the Number of Germline Stem Cells by Regulating Multiple Targets in Escort Cells
- RBPJ, the Major Transcriptional Effector of Notch Signaling, Remains Associated with Chromatin throughout Mitosis, Suggesting a Role in Mitotic Bookmarking
- The Membrane-Associated Transcription Factor NAC089 Controls ER-Stress-Induced Programmed Cell Death in Plants
- The Functional Consequences of Variation in Transcription Factor Binding
- Comparative Genomic Analysis of N-Fixing and Non-N-Fixing spp.: Organization, Evolution and Expression of the Nitrogen Fixation Genes
- An Insulin-to-Insulin Regulatory Network Orchestrates Phenotypic Specificity in Development and Physiology
- Suicidal Autointegration of and Transposons in Eukaryotic Cells
- A Multi-Trait, Meta-analysis for Detecting Pleiotropic Polymorphisms for Stature, Fatness and Reproduction in Beef Cattle
- Genome-Wide DNA Methylation Analysis Predicts an Epigenetic Switch for GATA Factor Expression in Endometriosis
- Genome-Wide DNA Methylation Analysis of Human Pancreatic Islets from Type 2 Diabetic and Non-Diabetic Donors Identifies Candidate Genes That Influence Insulin Secretion
- The and Hybrid Incompatibility Genes Suppress a Broad Range of Heterochromatic Repeats
- The Kil Peptide of Bacteriophage λ Blocks Cytokinesis via ZipA-Dependent Inhibition of FtsZ Assembly
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Worldwide Patterns of Ancestry, Divergence, and Admixture in Domesticated Cattle
- Genome-Wide DNA Methylation Analysis of Human Pancreatic Islets from Type 2 Diabetic and Non-Diabetic Donors Identifies Candidate Genes That Influence Insulin Secretion
- Genetic Dissection of Photoreceptor Subtype Specification by the Zinc Finger Proteins Elbow and No ocelli
- GC-Rich DNA Elements Enable Replication Origin Activity in the Methylotrophic Yeast
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



