-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaDeterminants beyond Both Complementarity and Cleavage Govern MicroR159 Efficacy in
In plants, microRNAs (miRNAs) are critical regulators of gene expression. As most validated targets are of high complementarity, whose transcripts are cleaved by the miRNA, both complementarity and cleavage are thought to be the major factors determining the degree to which a target gene is silenced. Here, we explore this principle utilizing the highly conserved miR159-MYB33/MYB65 regulatory module in the model flowering plant Arabidopsis. Firstly, we demonstrate that perfect central complementarity facilitates efficient transcript cleavage but is not required for a strong silencing outcome, as miR159 variants with two central mismatches can recognize and silence MYB33/MYB65 effectively in planta. Driving this silencing is a potent miR159-mediated non-cleavage mechanism that ensures total silencing even when MYB33 transcript levels are very high. Secondly, we demonstrate that the stoichiometric ratio of miRNA to target mRNA is a critical determinant of a silencing outcome, and that ratio becomes increasingly important for inefficient miRNA-target interactions. Finally, we show that nucleotides flanking the miR159 binding site of MYB33 are essential for efficient silencing, demonstrating that the sequence context in which the miRNA target site resides in has a major impact on the silencing outcome. Together, we have shown that although high complementarity underpinned by efficient transcript cleavage may be a prerequisite for a strong silencing outcome, many additional factors that modulate the strength of the miRNA-target interaction are at play. These findings will have ramifications for bioinformatics prediction of miRNA targets and design of artificial miRNAs.
Published in the journal: . PLoS Genet 10(3): e32767. doi:10.1371/journal.pgen.1004232
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004232Summary
In plants, microRNAs (miRNAs) are critical regulators of gene expression. As most validated targets are of high complementarity, whose transcripts are cleaved by the miRNA, both complementarity and cleavage are thought to be the major factors determining the degree to which a target gene is silenced. Here, we explore this principle utilizing the highly conserved miR159-MYB33/MYB65 regulatory module in the model flowering plant Arabidopsis. Firstly, we demonstrate that perfect central complementarity facilitates efficient transcript cleavage but is not required for a strong silencing outcome, as miR159 variants with two central mismatches can recognize and silence MYB33/MYB65 effectively in planta. Driving this silencing is a potent miR159-mediated non-cleavage mechanism that ensures total silencing even when MYB33 transcript levels are very high. Secondly, we demonstrate that the stoichiometric ratio of miRNA to target mRNA is a critical determinant of a silencing outcome, and that ratio becomes increasingly important for inefficient miRNA-target interactions. Finally, we show that nucleotides flanking the miR159 binding site of MYB33 are essential for efficient silencing, demonstrating that the sequence context in which the miRNA target site resides in has a major impact on the silencing outcome. Together, we have shown that although high complementarity underpinned by efficient transcript cleavage may be a prerequisite for a strong silencing outcome, many additional factors that modulate the strength of the miRNA-target interaction are at play. These findings will have ramifications for bioinformatics prediction of miRNA targets and design of artificial miRNAs.
Introduction
Ubiquitously found in plants and animals, microRNAs (miRNAs) are a group of 20–24 nucleotide (nt) small RNAs (sRNAs) that have been demonstrated to be critical regulators of gene expression. In plants, there are hundreds of known miRNAs [1], many of which have been shown to play critical roles in many different developmental and physiological processes [2]. They silence gene expression by guiding the RNA induced silencing complex (RISC) to target mRNAs via base pairing [2]. In plants, the subsequent repression of the target transcript occurs mainly through mRNA cleavage and/or translational inhibition [3].
Plant miRNAs recognise highly complementary binding sites, which enabled accurate bioinformatics prediction of their targets [4]. Moreover, experimentally determined targets were defined by the landmark study of Schwab et al. (2005), formulating the empirical parameters governing miRNA target recognition in plants, which are based purely on complementarity between the miRNA-target mRNA pairs [5]. It was found that there should be no more than one mismatch in the important “seed region” which corresponds to the 5′ end of the miRNA from positions 2 to 12, including no mismatches at the cleavage site (positions 10 and 11), and no more than two contiguous mismatches in the 3′ of the miRNA (positions 13–21) [5], [6]. Although there are examples of miRNA targets that do not fully conform to these sequence parameters [4], [7], generally most experimentally validated miRNA targets satisfy these parameters, and have been verified through degradome sequencing [8], [9]. Consequently, these parameters have been widely accepted and incorporated into many bioinformatic programs that predict miRNA targets [10].
Despite this extensive analysis on the complementary requirements for miRNA target recognition, and the subsequent mechanism of miRNA mediated repression [11], there has been very little analysis on the factors that impact the strength or efficacy of miRNA-mediated gene silencing in plants. It has been clearly shown that alterations in complementarity between the miRNA and its target can impact on the strength of the silencing outcome [12]. Additionally, the abundance of the miRNA can also functionally impact the silencing outcome [13], but generally no other factors have been considered in plants. As most plant miRNA-mRNA duplexes have high complementarity with perfect central pairing, transcript cleavage is generally considered a key silencing mechanism which results in the irreversible destruction of the transcript. This has been hypothesised to clear target transcript, leading to an absolute silencing outcome [14]. However, some plant miRNA targets with near perfect complementarity appear regulated primarily at the translational level [15]–[17], and this mechanism is likely widespread in plants [18]. Unlike cleavage, translational repression is thought not to result in mRNA destruction and thus is potentially reversible [3]. In animals, rather than an absolute silencing or “switch” mechanism, it acts as a tuning mechanism [19], however whether this is the case in plants is unclear. Due to this combinatorial mechanistic complexity, it is generally very difficult to appraise the strength of any miRNA-target interaction. Contributing to this in plants has been the lack of easily scorable systems that can gauge the extent of silencing. Consequently, the efficacy of plant miRNA-mediated regulation has remained largely ignored.
In this paper, we have used the highly conserved Arabidopsis miR159-GAMYB regulatory module to address fundamental questions regarding plant miRNA-mediated gene silencing. The Arabidopsis miR159 family is composed of three genes, MIR159a, MIR159b and MIR159c, although the MIR159c gene appears to be obsolete [20], [21]. Both miR159a and miR159b are predicted to regulate over 20 target genes in Arabidopsis, however regulation of only two targets appears physiologically important; namely MYB33 and MYB65, two redundant genes encoding R2R3 MYB transcription factors [20], [21]. MiR159a and miR159b redundantly silences these two genes in rosettes, and perturbation of their activity enables MYB33/MYB65 expression and results in a decreased rosette size and upwardly curled leaves [20], [22]. This easily scorable phenotype, along with molecular indicators, enables accurate appraisal of miR159 silencing efficiency.
Here, we complement a strong loss-of-function mir159ab double mutant with different artificial miR159 variants to demonstrate that perfect central complementarity is not required for the potent silencing of MYB33 and MYB65. Instead, a strong non-cleavage repression mechanism was evident in transgenic plants accumulating high levels of MYB33 transcripts, and appears to be a major contributing factor behind the strong efficacy of miR159. Finally, we show that factors beyond complementarity are critical for the silencing outcome. Firstly, the stoichiometric ratio of the miRNA:target mRNA strongly impacts the silencing outcome, especially when target cleavage is compromised. Moreover, mutation of sequences flanking the miR159 binding site of MYB33 leads to an attenuation of silencing that is comparable to cleavage site mutations. This reveals an important role for binding site context in plant miRNA-mediated gene regulation, suggesting, in essence, that a miRNA “target site” encompasses sequences that extend beyond the complementary motif to which a miRNA directly binds.
Results
Artificial miR159a variants with up to two central mismatches can fully complement the mir159ab mutant
The mir159ab double mutant presents itself as an ideal tool to examine miRNA-mediated gene silencing, as all mir159ab defects are specifically caused by the deregulation of the redundant GAMYB-like gene pair MYB33/MYB65, and these mutant phenotypes, including leaf curliness and dwarfed stature, are easily scored [20]. Therefore, by complementing mir159ab with different miR159 variants with modified central complementarity to MYB33/MYB65, the impact of central pairing on the silencing outcome of miR159 or their variants can be evaluated.
Since miR159a has the highest complementarity to MYB33/MYB65 and is the most abundant miR159 family member [23], five MIR159a variant constructs were generated by performing site-directed mutagenesis on a MIR159a genomic clone (Figure 1A). The two original mismatches of miR159a to MYB33/MYB65 at positions 7 and 20 were corrected, to keep the miRNA-target pairing free energy of all variants as high as possible (Figure 1B). All five variants were identical except for their number of central mismatches to MYB33/MYB65, which ranged from zero to four. MiR159a0 was included as a positive control to ensure that changes at nucleotide positions 7 and 20 would not abolish target silencing, since these two mismatches are highly conserved between miR159 and the binding site of potential GAMYB target genes from different monocotyledonous and dicotyledonous species (Figure S1). For all constructs, compensatory mutations were made to the miR159a* sequence to maintain the original predicted MIR159a stem-loop secondary structure (Figure S2).
Fig. 1. Artificial miR159a variants with up to two central mismatches are potent silencers of MYB33/MYB65. 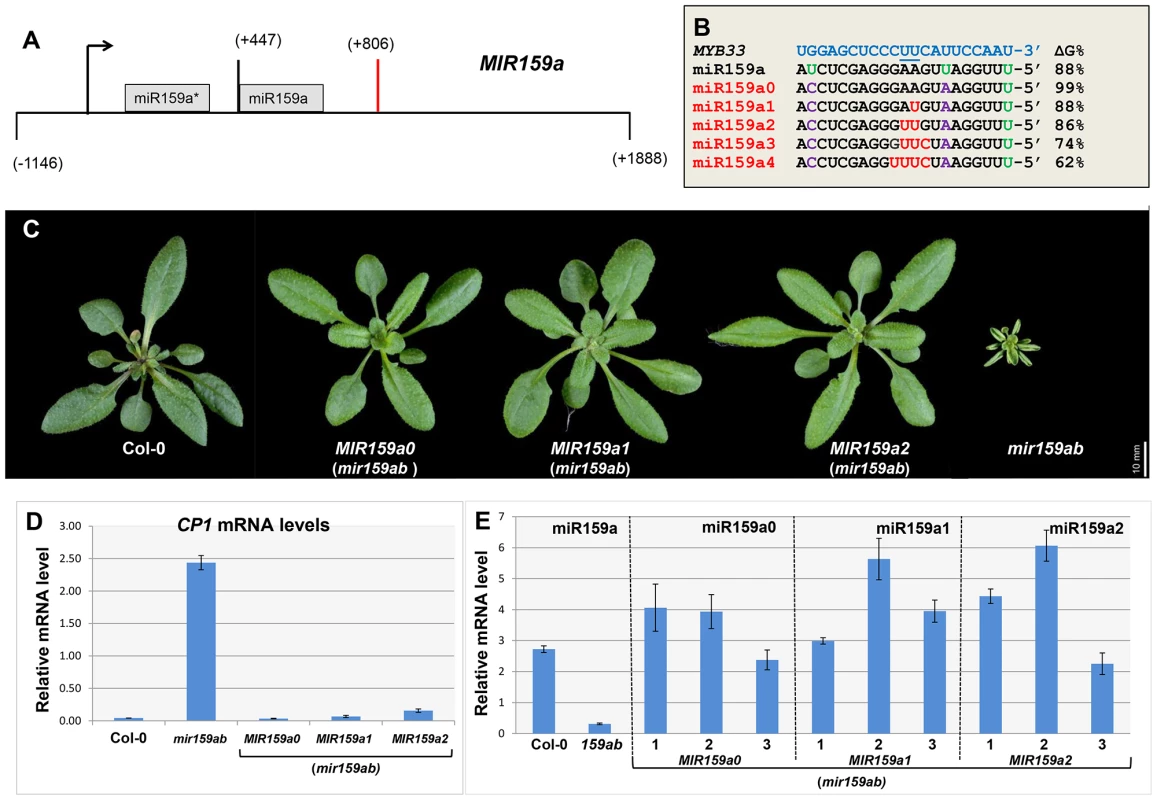
(A) Structure of the MIR159a transgene used to generate miR159a variant constructs. Numbers indicating relative positions to the transcription start site. Black arrow: Transcription start site; Orange bar: Polyadenylation site. (B) The alignment between MYB33 and the miR159a variant sequences used to transform mir159ab. The percentage of free energy pairing with MYB33 compared to a perfect match is listed on the right. Green: original mismatches; Purple: corrected mismatches; Red: introduced mismatches. (C) Aerial view of rosettes from 30-day-old MIR159a0, MIR159a1 and MIR159a2 plants (mir159ab) grown side by side with Col-0 and mir159ab under long day conditions. Scale bar = 10 mm. (D) The mRNA levels of CP1 in the rosettes of mir159ab plants complemented by the MIR159 variants in comparison with wild type and mir159ab. The value for each variant is the average of three independent transgenic lines. All mRNA levels were normalized with CYCLOPHILIN 5. Error bars represent the SEM. (E) Mature miR159a levels in Col-0 and mir159ab, and mature miR159a variant levels in three independent lines of MIR159a0 (mir159ab), MIR159a1 (mir159ab) and MIR159a2 (mir159ab) transformants. All miR159a variant and miR159a levels were normalized with sno101 by comparative quantitation analysis. Error bars represent the SEM. The five MIR159a variant constructs were individually transformed into mir159ab and the phenotypes of multiple primary transformants were scored. Correction of the mismatches at positions 7 and 20 had no influence on the silencing outcome, as all 30 MIR159a0 T1 plants were fully complemented (Figure 1C). Surprisingly, all 44 MIR159a1 and all 46 MIR159a2 primary transformants obtained appeared fully wild type (Figure 1C), suggesting that mismatches at the cleavage site does not prevent silencing. In contrast, all 90 MIR159a3 and 71 MIR159a4 primary transformants appeared indistinguishable from mir159ab (data not shown), indicating that the introduction of more than two central mismatches between miR159 and MYB33/MYB65 prevents complementation of the mir159ab phenotype.
To confirm that these mir159ab plants were fully complemented by the MIR159a1 and MIR159a2 variants, the expression of the GAMYB-like downstream gene CYSTEINE PROTEINASE1 (CP1, AT4G36880) was examined by qRT-PCR, as its mRNA is highly expressed in mir159ab due to the deregulation of MYB33/MYB65 [22]. Consistent with the morphological phenotypes, the mRNA level of CP1 in mir159ab plants complemented by MIR159a0, MIR159a1 or MIR159a2 was close to that of wild-type, indicating that MYB33/MYB65 expression was being suppressed to approximately wild type levels by these variants (Figure 1D). Next, we measured the abundance of the mature miR159a variants in complemented mir159ab plants using customized TaqMan sRNA assays. The levels of mature miR159a0, miR159a1 and miR159a2 in multiple independent transgenic lines were within the same order of magnitude as miR159a in wild type (Figure 1E). This suggests that gross overexpression of the miR159 variants was not an explanation for silencing targets with two central mismatches. Hence, their silencing potency in this regard would not be considered a transgenic artefact.
MiR159a variants with up to two central mismatches can repress mRNA levels through a mechanism that includes transcript cleavage
In both animal and plants, it is a popular notion that miRNA-guided cleavage of target genes is significantly attenuated by central mismatches [24], [25]. Hence, to investigate the mechanism by which the miR159a1 and miR159a2 variants are silencing MYB33 and MYB65, we firstly measured their un-cleaved mRNA levels by using qRT-PCR primers that span the miR159 binding site. Interestingly, MYB33/MYB65 levels were much lower in both MIR159a1 and MIR159a2 lines compared to mir159ab ¸ but were not reduced to wild-type levels. Instead these intermediate MYB mRNA levels appeared dependent on the number of central mismatches, with more mismatches correlating with higher MYB mRNA levels (Figure 2A and B, Figure S4). This demonstrates that miR159a1 and miR159a2 can still reduce MYB33/MYB65 transcript levels, suggesting that the miR159a variants are still mediating cleavage despite the existence of central mismatches.
Fig. 2. MiR159a1 and miR159a2 repress the endogenous MYB33/MYB65 mRNA levels. 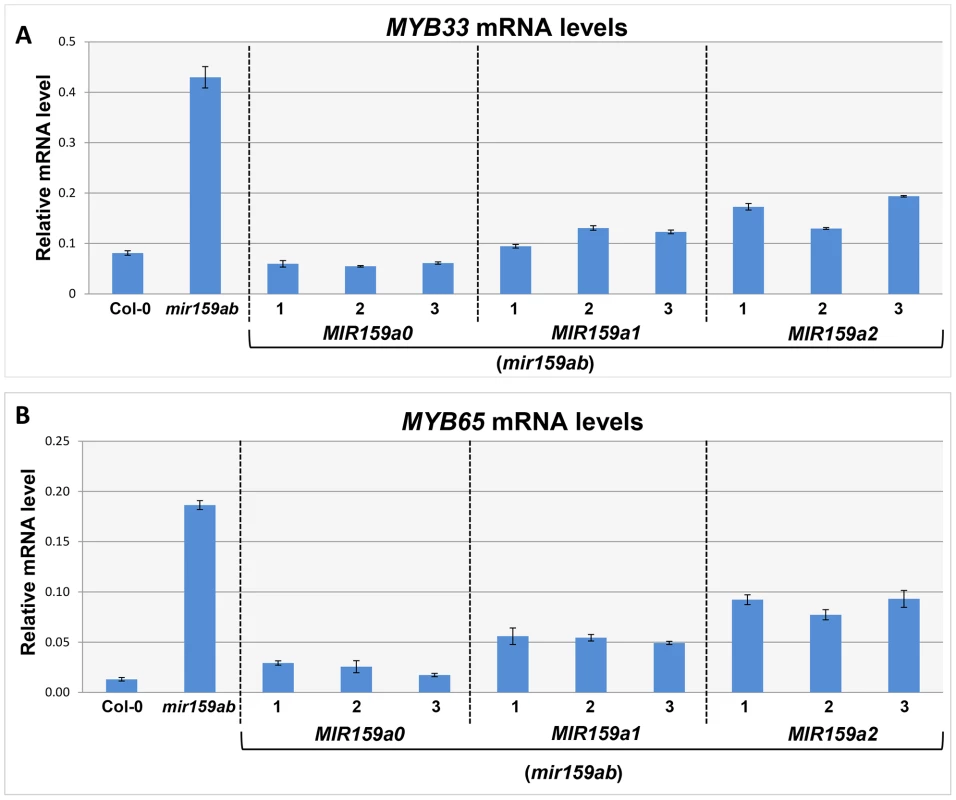
(A) Un-cleaved MYB33 mRNA levels in three independent lines of MIR159a0 (mir159ab), MIR159a1 (mir159ab) and MIR159a2 (mir159ab) transformants; (B) Un-cleaved MYB65 mRNA levels in three independent lines of MIR159a0 (mir159ab), MIR159a1 (mir159ab) and MIR159a2 (mir159ab) transformants. All mRNA levels were normalized with CYCLOPHILIN 5. Measurements are the average of three technical replicates. Error bars represent the SEM. Similar results were obtained with two independent biological replicates. To investigate this possibility, we performed a modified 5′-RACE miRNA cleavage assay [26]–[28]. Instead of gel purifying and cloning PCR products with sizes similar to the expected cleavage product followed by sequencing individual clones, we directly sequenced the entire PCR reaction and analysed the chromatograph peaks to approximate the proportion of degraded MYB33 transcripts that correspond to miR159-guided cleavage products (Figure 3A and S3). For the wild-type control sample, sequencing of the 5′-RACE reaction using a MYB33-specific primer resulted in clean single peaks with very little baseline noise for each MYB33 nucleotide up to the miR159-guided cleavage site, and then into the RNA adaptor sequences (Figure 3B). Such a clean signal indicates that the vast majority of degraded MYB33 transcripts amplified by this assay were miR159-guided 3′-end cleavage products, which is consistent with previous strong degradome signatures of MYB33 [8], [9]. By contrast, the mir159ab sample exhibited overlapping peaks in the portion of the sequence immediately upstream of the miR159 cleavage site (Figure 3C). This demonstrates that the degraded MYB33 transcripts recovered by 5′-RACE in mir159ab were no longer predominantly miR159-guided cleavage products, but mixed populations arising independently of miR159-guided cleavage. By contrast, the chromatographic traces of reactions from MIR159a1 and MIR159a2 plants were indistinguishable from that of wild type (Figure 3D and 3E). This clearly demonstrates that miR159a1 and miR159a2 are capable of mediating cleavage despite the presence of central mismatches. Additionally, as this cleavage is occurring at the canonical MYB33 cleavage site, this suggests that that the intended miR159a1 and miR159a2 variants are being processed from the modified pri-MIR159a transcripts, rather than other possible products arising from imprecise DCL1 processing.
Fig. 3. MiR159a variants with up to two central mismatches can direct target cleavage at the canonical miR159 cleavage site. 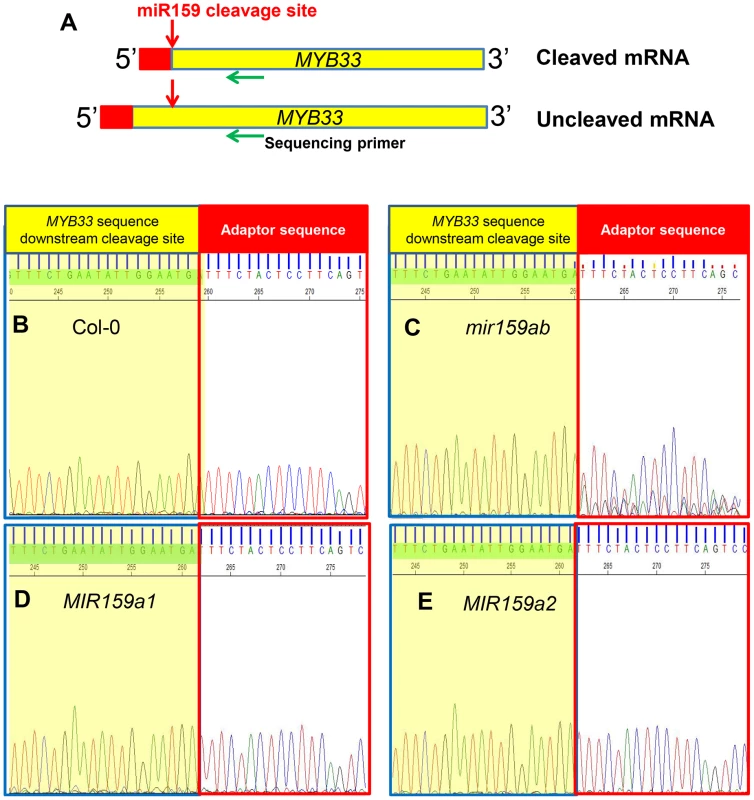
(A) Schematic representation of MYB33 PCR products after a modified 5′-RACE procedure to determine the proportion of degraded MYB33 mRNA that corresponds to miR159-guided cleavage products. Red box: RNA Oligo adaptor; Yellow box: MYB33 sequences; Red arrow: canonical miR159 cleavage site; Green arrow: MYB33 sequencing primer. (B–E) Sequencing chromatographs of the cDNA-adaptor region from 5′-RACE recovered 3′ MYB33 transcripts in: (B) Col-0, (C) mir159ab, (D) MIR159a1 and (E) MIR159a2 plants. Peaks from MYB33 sequence downstream of the miR159 cleavage sites are highlighted in yellow, while those from the adaptor are left white. Three independent transgenic lines were checked for each construct and the results were identical. Although these 5′-RACE assays demonstrate that miR159a1 and miR159a2 can guide cleavage of MYB33, these assays are not quantitative. Therefore, a qRT-PCR assay was developed to quantitate the amount of miR159-guided 3′-end cleavage products of MYB33 mRNA. An equal amount of total RNA from each sample was ligated to a 5′-end RNA adaptor followed by retro-transcription with poly T primers. To specifically amplify miR159-guided cleavage products, a hybrid forward primer was designed that included the last 19 nucleotides of the adaptor followed by five nucleotides of MYB33 sequence immediately downstream of the miR159 cleavage site (Figure S5). The specificity of the assay was confirmed by testing Col-0 and mir159ab samples with two different forward primers (Figure S5). Both un-cleaved MYB33 mRNA and 3′ end cleavage products were measured and the ratio was calculated to obtain an indication of miR159 cleavage efficiency, where the higher this ratio is the more inefficient cleavage is.
For wild-type, the steady levels of the miR159-guided 3′end MYB33 cleavage products were detectable, but were approximately 9-fold lower than the un-cleaved MYB33 mRNA levels (Figure 4). Such a low amount could be considered consistent with rapid degradation of these products by exoribonuclease XRN4 [9], [29]. For mir159ab, the levels of 3′ end cleavage products were negligible (Figure 4). Supporting the notion that the miR159a variants are able to guide cleavage, independent transgenic MIR159a1 plants had comparable ratios to wild type, however the ratio values approximately doubled in MIR159a2 plants. Together, these data suggest whilst miR159a2 can still direct cleavage, but it has been attenuated by the introduction of two central mismatches (Figure 4).
Fig. 4. Quantitative comparison of un-cleaved MYB33 mRNA and miR159-guided 3′-end MYB33 cleavage products in MIR159a1 and MIR159a2 variants. 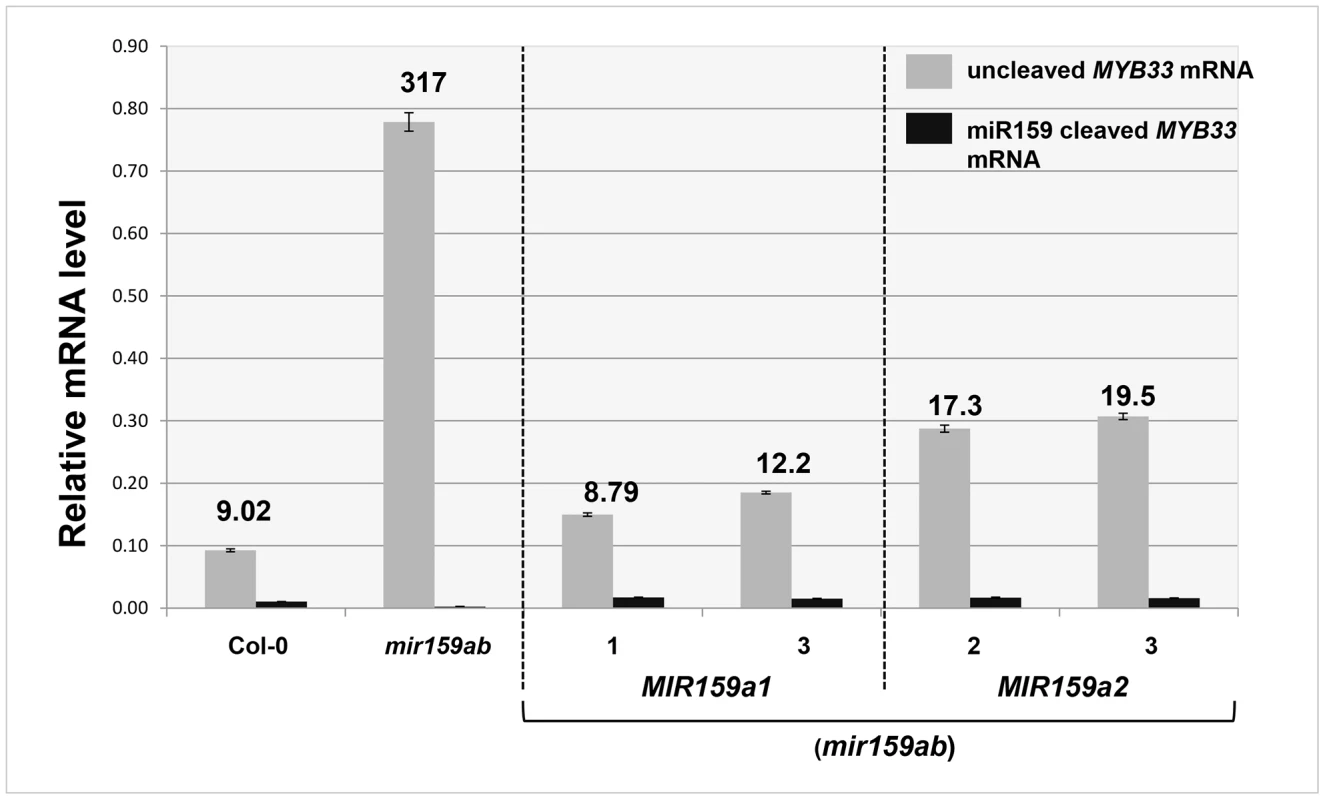
Grey bars represent the un-cleaved MYB33 transcript levels in Col-0, mir159ab, and independent transgenic lines of MIR159a1 (mir159ab) and MIR159a2 (mir159ab). Black bars represent the levels of miR159-guided 3′-end MYB33 cleavage products in the same lines. Numbers show the relative ratio of un-cleaved transcripts to cleavage products. All mRNA levels were normalized with CYCLOPHILIN 5. Measurements are the average of three technical replicates. Similar results were obtained with two independent biological replicates. Error bars represent the SEM. MiR159 can strongly suppress high levels of un-cleaved MYB33 transcripts
Despite the higher levels of un-cleaved MYB33 and MYB65 transcripts, MIR159a1 and MIR159a2 plants appeared morphologically indistinguishable from wild type and the CP1 mRNA remained essentially at wild type levels (Figure 1D). This leads to the question of whether miR159 also recruits a non-cleavage mechanism to achieve complete silencing of MYB33 and MYB65, especially when cleavage is attenuated. To investigate this, we attempted to generate transgenic plants that strongly transcribe MYB33. To do this, MYB33 or mMYB33 (miR159-resistant) constructs were generated (Figure 5B) using a genomic MYB33 clone [30] which contains the endogenous MYB33 promoter. These constructs were transformed into the loss-of-function myb33 mutant, a T-DNA knock-out allele of the MYB33 gene [30]. Myb33 is phenotypically indistinguishable from wild type and the absence of endogenous MYB33 transcript enables accurate quantitation of the MYB33 and mMYB33 transgene mRNA levels.
Fig. 5. High levels of un-cleaved MYB33 transcripts remain strongly repressed. 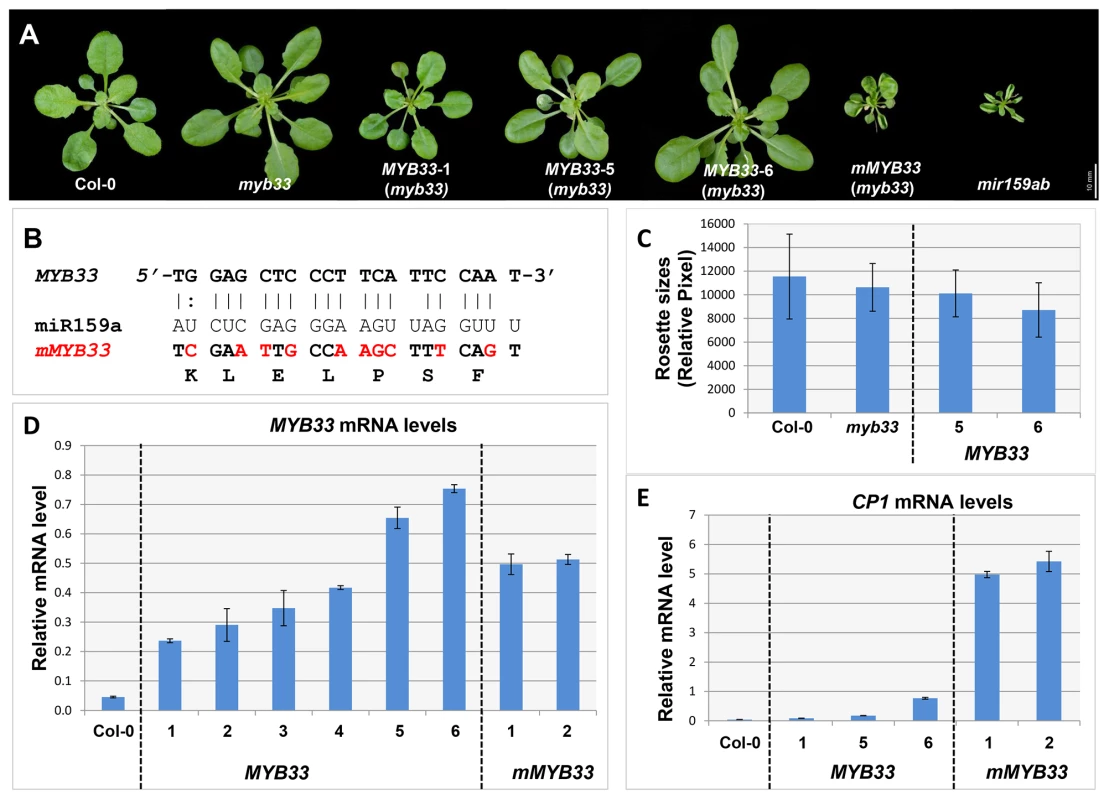
(A) Aerial view of 29-day-old rosettes from MYB33 and mMYB33 transgenic plants (myb33 genotype) grown under short day conditions. Scale bar = 10 mm. (B) Alignments between miR159a and the wild type and mutated miR159 target sites in MYB33 and mMYB33 constructs used to transform Arabidopsis. Nucleotide changes made in mMYB33 are highlighted in red. Both MYB33 and mMYB33 encode the same protein. (C) Average rosette sizes of 29-day-old T2 transgenic plants from MYB33 lines grown side by side with Col-0 and myb33. Unit of measurement shown is relative pixel. Multiple plants for each line were grown on soil side by side with Col-0 and myb33. More than fifteen transgenic plants in each line were analysed. Error bars represent SD. (D) The steady state mRNA levels of un-cleaved MYB33 in the rosettes of multiple MYB33 (lines 1–6) and mMYB33 (lines 1–2) T2 transgenic lines. (E) Relative mRNA levels of CP1 in the rosettes of the same MYB33 and mMYB33 lines. All mRNA levels were normalized with CYCLOPHILIN 5. All measurements are the average of three technical replicates with error bars representing the SEM. Consistent with previous experiments [30], [31], expression of mMYB33 caused pleiotropic developmental defects in myb33, as all 20 primary transformants had upwardly curled leaves and dwarfed stature resembling mir159ab (Figure 5A). In stark contrast, none of the 30 MYB33 primary transformants recovered had any conspicuous defects (Figure 5A). QRT-PCR was then performed on rosettes to quantitate the steady state level of un-cleaved MYB33 transcripts. As would be predicted, the MYB33 transcript level was elevated in mMYB33 lines (1 and 2), being approximately ten fold higher compared to wild type (Figure 5D). Surprisingly, all six MYB33 transgenic lines examined also accumulated un-cleaved MYB33 transcripts to a much greater level than wild type, where 5–15 fold increases were observed (Figure 5D; lines 1–6). This suggests that miR159 is unable to reduce MYB33 mRNA to a wild type level through a transcript cleavage mechanism. Moreover, in MYB33 lines 5 and 6, the un-cleaved MYB33 mRNA levels were even higher than those in the mMYB33 lines. The expression of these transcripts appeared to be strongly repressed by miR159, as the corresponding transgenic plants exhibited no curly leaves or reduction in rosette size (Figure 5A and 5C). In addition, whereas the CP1 mRNA level was dramatically elevated in mMYB33 plants (∼120 times higher than wild type), such increases were not seen in any MYB33 plants (Figure 5E). These data together support the previous notion that miR159 suppresses MYB33 not only by transcript cleavage, but also by a non-cleavage mechanism(s) [22]. Therefore, in this case, the steady state mRNA levels are a poor indicator of silencing.
To investigate the reason why miR159 fails to repress MYB33 mRNA to wild type levels in MYB33 plants, we quantified 3′-end MYB33 cleavage products and un-cleaved MYB33 transcript levels as previously described. Interestingly, in the MYB33 samples (lines 1 and 2), the 3′ end cleavage product appeared to increase proportionally with the levels of uncleaved MYB33 mRNA, while no 3′ end cleavage product was apparent in the mMYB33 sample (Figure 6). This indicates that cleavage is unlikely to be attenuated by the high MYB33 transcript level, and the amount of cleavage products present appears dependent on the amount of MYB33 transcripts present.
Fig. 6. Quantitative comparison of un-cleaved MYB33 mRNA and miR159-guided 3′-end MYB33 cleavage products in MYB33 and mMYB33 plants. 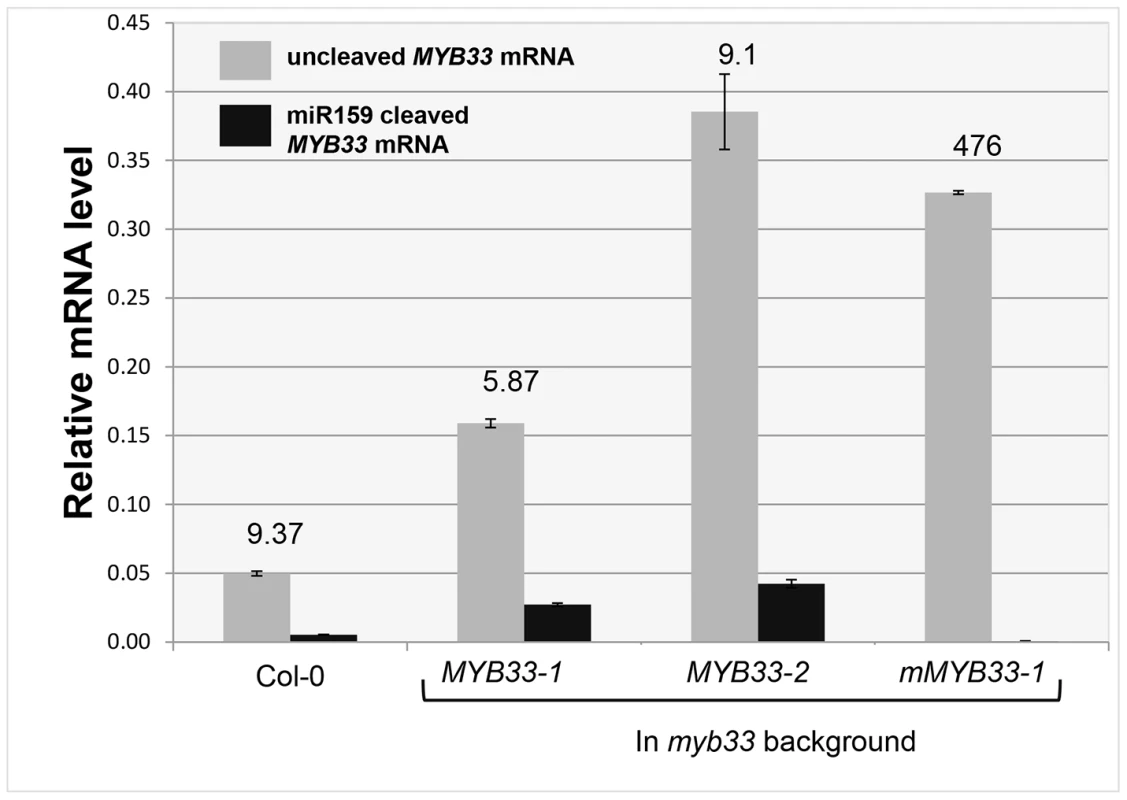
Grey bars represent the un-cleaved MYB33 transcript levels in Col-0, MYB33 and mMYB33 transgenic lines. Black bars represent the levels of miR159-guided 3′-end MYB33 cleavage products in the same lines. Numbers show the relative ratio of un-cleaved transcripts to cleavage products. All mRNA levels were normalized with CYCLOPHILIN 5. Measurements are the average of three technical replicates. Similar results were obtained with independent biological replicates. Error bars represent the SEM. MYB33 transgenes with central mismatches cannot be fully silenced by endogenous miR159
The miR159a variants result could lead to a surprising conclusion that central matches are not important for a strong silencing outcome. To further investigate this, three MYB33 variants were made, all of which were identical except that the number of mismatches to miR159 at positions 10 and 11 ranged from zero to two (Figure 7B). To keep the overall complementarity and free energy close to the original value when pairing to miR159a, the original mismatches at positions 7 and 20 were corrected (Figure 7B). Therefore, the positions of mismatches of MYB33-0 cm, MYB33-1 cm and MYB33-2 cm to miR159a mirror those of miR159a0, miR159a1 and miR159a2 to the endogenous MYB33/MYB65 genes respectively. Although these sequence alterations result in one to two amino acid changes in the MYB33 protein, all substitutions made were conservative changes to minimize possible changes to the biochemical properties of the protein. Primary transformants for each construct were generated in the myb33 background.
Fig. 7. MYB33 variants with central mismatches are not fully silenced. 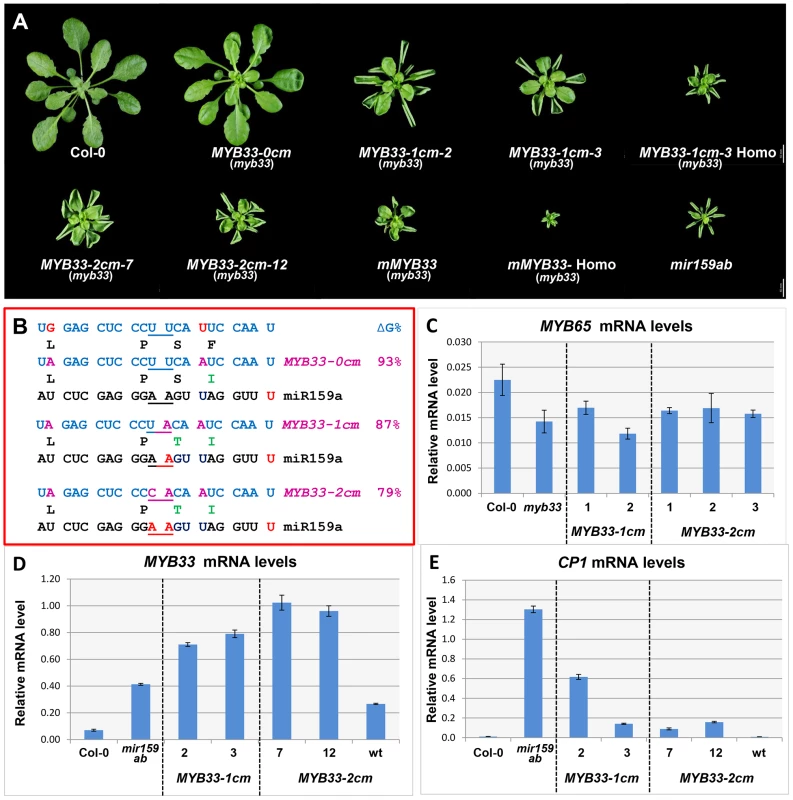
(A) Aerial view of rosettes from 35-day-old MYB33-1 cm and MYB33-2 cm plants (myb33 genotype) grown side by side with Col-0, mir159ab and mMYB33 under short day conditions. Homo stands for homozygous plants for the transgene. Scale bar = 10 mm. (B) The alignment between the different MYB33 variants and miR159a. The sequences of the amino acids encoded by the miR159 target site in different MYB33 transgenes are shown below. The percentage of free energy when pairing to miR159a compared to the perfect match is shown on the right. Red: mismatches; Purple: nucleotides mutated in the MYB33 miR159 target site. Amino acid changes compared to the original MYB33 protein are highlighted with green. (C) The steady state MYB65 mRNA level in the rosette of selected MYB33-1 cm and MYB33-2 cm transgenic lines with mutant phenotypes. Levels are not statistically different between the variants and parental myb33 plants (D) The steady-state MYB33 mRNA levels in rosettes of the transgenic lines shown in (A). (E) CP1 mRNA levels in the rosette of the transgenic lines shown in (A). All mRNA levels were normalized with CYCLOPHILIN 5. Measurements are the average of three technical replicates. Error bars represent the SEM. Consistent with the result that mir159ab can be fully complemented by miR159a0, all 52 MYB33-0 cm transgenic plants generated were indistinguishable from wild type (Figure 5A). However, counter to miR159a1 and miR159a2, all 46 MYB33-1 cm and 41 out of 42 MYB33-2 cm T1 plants recovered had developmental defects, exhibiting phenotypes characteristic of mir159ab, including curled leaves and a smaller rosette (Figure 7A). This demonstrates that the MYB33-1 cm and MYB33-2 cm transgenes are not efficiently silenced by endogenous miR159. Molecular and phenotypic analyses were carried out to investigate this. Firstly, to dismiss that MYB33-1 cm and MYB33-2 cm are acting as decoys to suppress endogenous miR159 activity [32], we measured MYB65 transcript levels in the MYB33 variants and control plants. As the mRNA levels were found to be similar in all plants (Figure 7C), this demonstrates that MYB65 has not been deregulated in MYB33 variants, meaning that miR159 activity has not been perturbed. Therefore, the abnormal rosette phenotypes were resulting from failure of miR159 to silence the MYB33 variants. To phenotypically assess the severity of the developmental phenotypes, multiple MYB33-1 cm and MYB33-2 cm T2 transgenic lines were grown side by side with Col-0, mir159ab and mMYB33 plants. Although most transgenic MYB33-1 cm and MYB33-2 cm lines displayed traits characteristic of mir159ab, the extent of leaf curl and the reduction in rosette size of most plants were less severe compared to either mir159ab or mMYB33 plants (Figure 7A). Therefore, the repression of MYB33-1 cm or MYB33-2 cm by miR159 does not appear completely abolished.
To examine this, total RNA was extracted from individual transgenic lines using rosettes of multiple plants exhibiting similar phenotypes (Figure 7A). QRT-PCR analysis found that the un-cleaved MYB33 mRNA levels were 10–15 fold higher in MYB33-1 cm and MYB33-2 cm lines compared to wild type, while only a six-fold increase was observed in mir159ab (Figure 7D). Even considering the fact that MYB65 is also de-regulated in mir159ab, these very high MYB33-1 cm/2 cm mRNA levels would have been expected to result in phenotypic severities more akin to that of mir159ab, if they were completely devoid of miR159 regulation. Moreover, transcript levels of CP1, which reflects the abundance of MYB protein, were found to be much lower in all MYB33-1 cm/2 cm lines examined than in mir159ab (Figure 7E). Together, these data suggest that the high levels of MYB33-1 cm and MYB33-2 cm transcripts are still being partially repressed by miR159, which again suggests a non-cleavage mechanism in operation.
5′-RACE was also performed to assay the degraded 3′-end MYB33 mRNA population as described before (Figure S3). A MYB33 transgenic line with a very high MYB33 mRNA level was included as control (line 5 from Figure 7), and its sequencing chromatograph was identical to that of wild type (Figure 8C). This demonstrates that the vast majority of 3′ degraded MYB33 transcripts amplified by this assay are miR159-guided 3′ end cleavage products, regardless of how high the transcript level is. In contrast, no signals from the adaptor sequence but those from the transgene were recovered for mMYB33, which is completely resistant to cleavage (Figure 8D). For MYB33-1 cm, past the cleavage site, the signal became a mixture of adaptor and MYB33-1 cm sequence, indicating that although some miR159-guided cleavage products were present, it was a mixed population (Figure 8A). For MYB33-2 cm, the signal from the adapter sequence corresponding to the canonical miR159 cleavage site was negligible, where the signal was dominated by MYB33-2 cm sequences (Figure 8B). This demonstrates that 3′ end miR159-guided cleavage products are poorly represented in the reactions from the MYB33-1 cm and MYB33-2 cm samples when compared to the MYB33 sample, arguing that miR159-guided cleavage of MYB33-1 cm and MYB33-2 cm is not as efficient as MYB33, which is likely due to the introduction of central mismatches.
Fig. 8. MiR159-guided cleavage is attenuated in MYB33-1 cm and MYB33-2 cm plants. 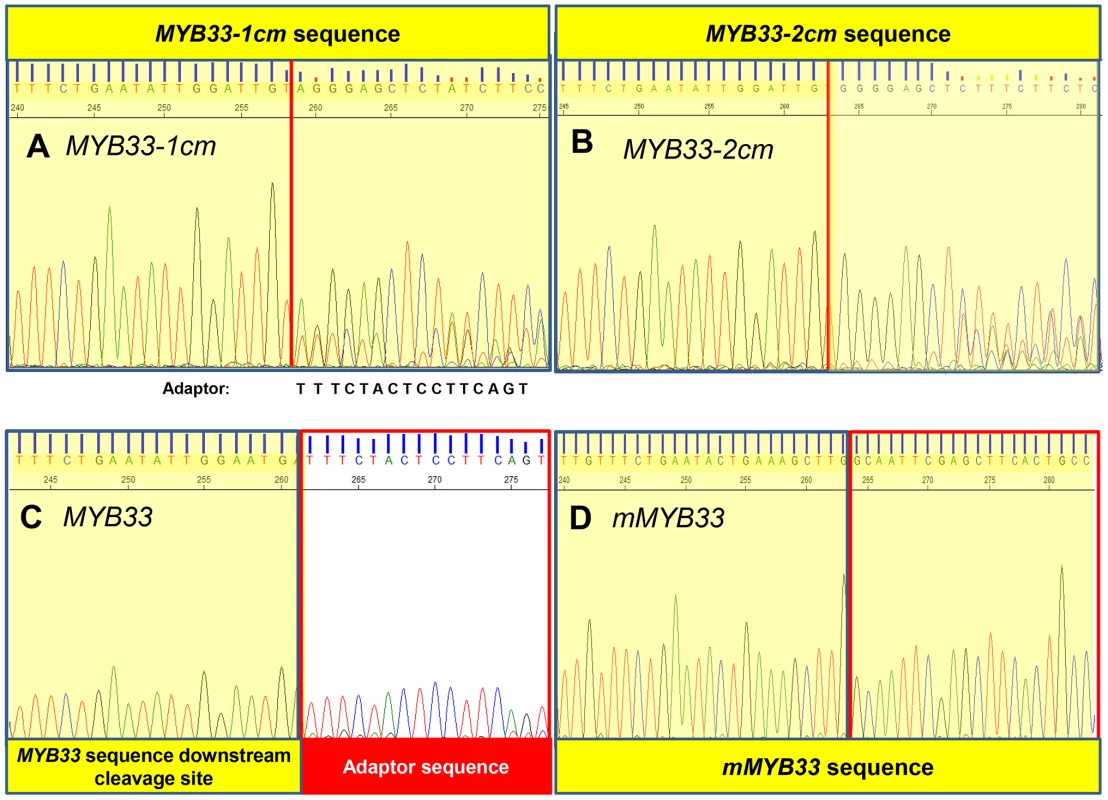
A fragment of the sequencing chromatography from 5′-RACE recovered 3′ MYB33 transcripts in (A) MYB33-1 cm plants. (B) MYB33-2 cm plants. (C) MYB33 plants. (D) mMYB33 plants. MYB33-1 cm, MYB33-2 cm, MYB33 and mMYB33 sequences are highlighted yellow, while the adaptor is left white. Red line: canonical miR159 cleavage site. MiRNA:target stoichiometry has a critical impact on the silencing outcome in MIR159a and MYB33 variant plants
It appears that central mismatches attenuate cleavage in both the miR159a variant-MYB33/MYB65 and miR159-MYB33 variant relationships, therefore additional factors must be impacting the differential silencing outcomes observed. Firstly, although the duplex pairs have identical mismatch positions, the nucleotides changes are not identical and hence will result in duplexes with different thermodynamics stabilities (Table S1), possibly impacting the silencing outcome. Secondly, although MYB33 and MYB65 were being transcribed at wild type levels in MIR159a1 and MIR159a2 plants, it is clear that mRNA from MYB33, MYB33-1 cm or MYB33-2 cm transgenes accumulated to higher levels than endogenous MYB33 (Figure 5D, 7D). This would be predicted to alter the stoichiometric ratio of MYB33 transcripts to miR159. Since target mRNA: miRNA stoichiometry is important for the silencing outcome in animal miRNA-target genes relationships [33]–[35], an altered miR159: MYB33 stoichiometry may explain the observed differential silencing outcomes.
As it is impossible to introduce identical central mismatch nucleotides in both the miR159a variant-MYB and miR159-MYB33 variant duplexes, we aimed to alter the stoichiometric ratio between MYB33 and miR159 in the MIR159a and MYB33 variant plants instead. This will determine whether stoichiometry can alter the silencing outcome, or whether the different thermodynamic stabilities of two different duplexes override any stoichiometric alteration.
Firstly, the wild type MYB33 transgene was transformed into individual MIR159a1 and MIR159a2 T3 transgenic lines (mir159ab genotype, wild-type phenotype) to see whether higher levels of MYB33 transcript could override the complementation by the miR159a variants. In contrast to the introduction of MYB33 into wild-type plants that resulted in no phenotypic abnormalities (Figure 5), the introduction of the MYB33 transgene into multiple lines of MIR159a1 and MIR159a2 plants resulted in a large proportion of transgenic plants displaying the characteristic phenotypic abnormalities of MYB33 expression, including leaf curl and smaller rosette size (Figure 9A). QRT-PCR analysis confirmed strong increases of MYB33 transcript levels in these MYB33 [MIR159a1 (mir159ab)] and MYB33 [MIR159a2 (mir159ab)] plants, whilst the mature miR159a1 and miR159a2 levels remained similar (Figure 9C). Consistent with the morphological defects, CP1 mRNA levels were elevated, especially in the MYB33 [MIR159a2 (mir159ab)] plants. This clearly demonstrates that the MIR159a variants are poor silencers of MYB33 when MYB33 is highly transcribed, resulting in an unfavourable stoichiometric miR159:MYB ratio for silencing.
Fig. 9. MYB33:miR159a stoichiometry has a critical impact on the silencing outcome in MIR159a and MYB33 variants. 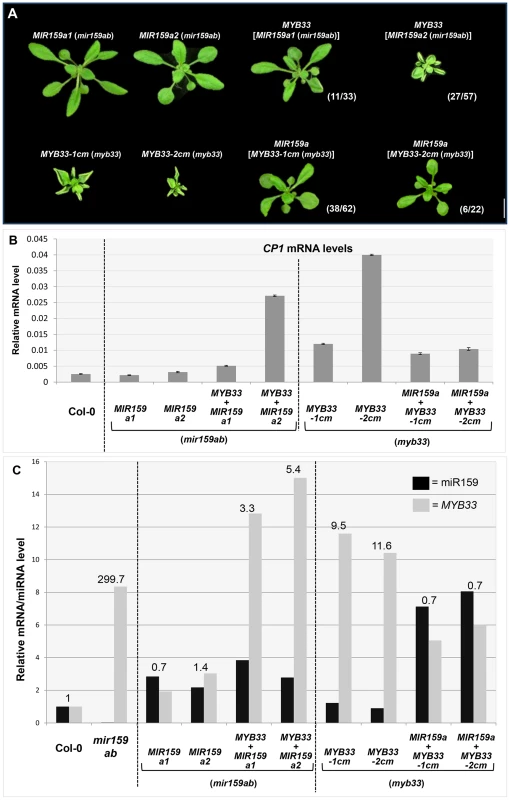
(A) Aerial view of rosettes of 27-day-old of MIR159a1 (mir159ab) and MIR159a2 (mir159ab) plants transformed with the MYB33 construct; and MYB33-1 cm (myb33) and MYB33-2 cm (myb33) plants transformed with MIR159a. All plants were grown side by side with the original MIR159a1, MIR159a2, MYB33-1 cm and MYB33-2 cm parental lines used for transformation. Scale bar = 10 mm. (B) The mRNA levels of CP1 in transgenic lines shown in (A). (C) The relative un-cleaved MYB33 mRNA level (grey bars) and mature miR159a/variant levels (black bars) in Col-0, mir159ab and transgenic lines shown in (A). All original mRNA levels were normalized with CYCLOPHILIN 5. All original miR159a levels were normalized with snoR101. All fold changes in (C) are relative to the levels in Col-0, which is set as 1. Measurements are the average of three technical replicates. Error bars represent the SEM. Secondly, the wild-type MIR159a construct was transformed into MYB33-1 cm and MYB33-2 cm T3 transgenic lines. The severe leaf curl and small rosette size observed in MYB33-1 cm and MYB33-2 cm plants were largely suppressed in many MIR159a (MYB33-1 cm) and MIR159a (MYB33-2 cm) transformants (Figure 9A). The introduction of MIR159a resulted in elevated levels of mature miR159a by approximately five-fold and a simultaneous reduction of the MYB33 transcript levels to approximately 50% of that found in parental MYB33-1 cm and MYB33-2 cm lines (Figure 9C). Consistent with these observations, the levels of CP1 were suppressed in these MIR159a (MYB33-1 cm) and MIR159a (MYB33-2 cm) transformants (Figure 9B).
A ratio between the relative abundance of MYB33 and miR159a was calculated (levels of both MYB33 and miR159a in Col-0 were normalized to 1, Figure 9C). The higher this ratio was, the poorer the silencing outcome became; and vice versa. These data clearly demonstrate that the miRNA: target mRNA stoichiometric ratio has a critical role in determining the silencing outcome when central complementarity is compromised. Although we are unable to address whether thermodynamic differences between the duplexes may in part contribute to the differential silencing outcome, it, or any other factor, is clearly not strong enough to override stoichiometry.
Conserved nucleotides flanking the miR159 binding site of MYB33 are critical for efficient silencing
Interestingly, the stoichiometric ratio of endogenous miR159 to MYB33 appears less important, as even in transgenic plants transcribing MYB33 many fold higher than in wild-type, MYB33 appears to be strongly silenced (Figure 5, 6). This argues that the sensitivity of MYB33 to miR159 regulation is strong enough to override an unfavourable stoichiometric ratio. In order to understand this sensitivity, we initiated a characterization of the MYB33 region containing the miR159 binding site by aligning genes encoding MYB33 homologs from different monocotyledonous and dicotyledonous plant species. Alignment of the nucleotide sequences of 15 of these genes revealed multiple conserved nucleotides flanking the miR159 binding site, many of which are located in the third codon position (Figure 10A), suggesting that conservation is not at the protein but RNA level. Moreover, these nucleotides are among the strongest stretches of nucleotide conservation outside of the R2R3 MYB domain or the miR159 binding site (Figure S6), suggesting the conservation has functional significance.
Fig. 10. Nucleotides flanking the MYB33 miR159 target site are critical for conferring efficient silencing. 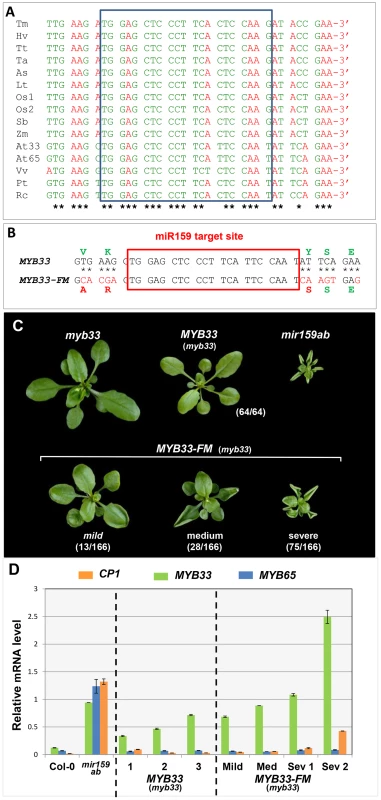
(A) ClustalW2 sequence alignment of MYB33 homologues. Nucleotides are grouped into codons of the reading frame. Conserved nucleotides are marked with *. The miR159 binding site is indicated with a pale blue box. Sequences include genes from monocotyledonous species (Tm: Triticum monococcum, Hv: Hordeum vulgare, Tt: Triticum turqidum, Ta: Triticum aestivum, As: Avena sativa, Lt: Lolium tremulentum, Os: Oryza sativa, Sb: Sorghum bicolor, Zm: Zea mays) and dicotyledonous species (At: Arabidopsis thaliana, Vv: Vitis vinifera, Pt: Populus trichocarpa, Rc: Ricinus communis). (B) Mutations made in the regions flanking the miR159 target site in the MYB33-FM construct. Corresponding nucleotide and protein changes are denoted red. (C) Aerial view of 4-week-old MYB33-FM transgenic plants in comparison with MYB33, myb33 and mir159ab plants grown side by side under long day conditions. The number of primary transformants showing the arbitrary phenotypic classification described in the text is listed in the bracket. Scale bar = 10 mm. (D) The mRNA levels of MYB33, MYB65 and CP1 in multiple biological samples of independent MYB33 and MYB33-FM transformants. Mild, Med (medium) and Sev (severe) denote the strength of phenotypic abnormalities observed in MYB33-FM transformants sampled. All mRNA levels were normalized to CYCLOPHILIN 5. Measurements are the average of three technical replicates with error bars representing the SEM. To test their role in miR159 regulation of MYB33, eleven nucleotides flanking the miR159 binding site were mutated in the genomic MYB33 construct to generate the MYB33-Flanking site Mutant construct (MYB33-FM, Figure 10B). These alterations result in sequence changes in the MYB33 protein, however all three resulting amino acid substitutions were conservative, minimizing possible changes to the biochemical properties of the protein. Moreover, these amino acids do not appear to be critical for MYB33 function, as a previous study has shown that a 33 bp deletion in the MYB33 coding region, including the miR159 binding site and these flanking nucleotides, still results in the production of a functional protein [30].
MYB33-FM and the positive control, MYB33, were individually transformed into myb33 plants, and multiple primary transformants were selected and analyzed. Consistent with previous results, none of the MYB33 plants showed any phenotypic abnormalities, indicating that MYB33 is fully repressed by miR159 (Figure 10C). In contrast, 116 out of 126 MYB33-FM plants showed strong developmental defects characterized by curly leaves and stunted growth (Figure 10C). As these are phenotypic characteristics of mir159ab, it appears that miR159 regulation of MYB33 has been compromised. Molecular analysis was carried out on RNA extracted from primary transformants that had been categorized according to phenotypic severities. As before, MYB33 transcript levels were elevated in MYB33 plants (Figure 10D), but remained strongly silenced as these plants were indistinguishable from wild-type (Figure 10D). Elevated MYB33 levels were also observed in all MYB33-FM plants examined, however, in contrast to MYB33 plants, the levels positively correlated with the phenotypic severity observed, as well as the CP1 mRNA levels (Figure 10D). By contrast, MYB65 mRNA levels were unchanged in all transgenic plants compared to wild type, indicating that miR159 activity has not been perturbed in any of the transgenic lines, therefore MYB33-FM is not acting as a decoy (Figure 10D). In conclusion, this data clearly demonstrates that in transgenic plants where MYB33-FM is highly transcribed, the silencing efficacy of miR159 against MYB33-FM is strongly perturbed.
Flanking or central nucleotide mutations perturb miR159 efficacy to similar extents
Our data has shown that under a poor stoichiometric ratio, both the central and flanking nucleotides are critical for strong miR159 efficacy against endogenous MYB33. In an attempt to determine the degree to which MYB33 silencing is perturbed by these different mutations, a GUS-reporter system was used to quantitatively measure silencing at both the mRNA and protein level. The MYB33, MYB33-1 cm, MYB33-2 cm and MYB33-FM transgenes were each translationally fused with a GUS reporter and corresponding transgenic lines were generated in myb33 plants [30]. For each sample, more than 50 primary transformants were bulked together from which both RNA and protein samples were prepared, so that both transcript (qRT-PCR) and GUS activity (MUG assays) quantification could be then performed, enabling direct comparison of measurements.
Un-cleaved MYB33 transcript levels in MYB33-1 cm:GUS and MYB33-FM:GUS were similar to the level in MYB33:GUS plants, whereas the MYB33-2 cm:GUS mRNA level was approximately three-fold higher (Figure 11F), again supporting that central mismatches attenuate cleavage. Despite MYB33-1 cm:GUS and MYB33-FM:GUS mRNA levels being similar to MYB33:GUS, these MYB33-1 cm:GUS and MYB33-FM:GUS mRNA must be translated more efficiently, as GUS activity in these lines are approximately three fold higher than in MYB33:GUS lines (Figure 11G). By contrast, the much higher MYB33-2 cm:GUS mRNA levels did not translate into dramatically higher GUS activity when compared to GUS activity in the MYB33-1 cm:GUS and MYB33-FM:GUS lines (Figure 11G), which again supports the existence of a strong non-cleavage silencing mechanism (Figure 2A).
Fig. 11. Quantification of MYB33 expression using the GUS reporter system. 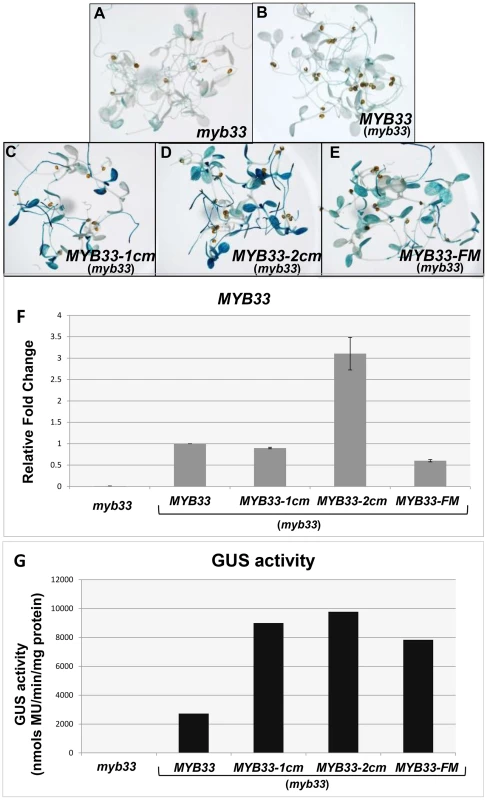
In situ GUS staining of 8-day-old seedlings of the control genotype, (A) myb33, and independent transformants of (B) MYB33:GUS, (C) MYB33-1 cm:GUS, (D) MYB33-2 cm:GUS, (E) MYB33-FM:GUS; all in the myb33 background. (F) The relative fold changes of MYB33 mRNA levels. All values were averaged from two individual biological replicates and the level in MYB33:GUS was normalized to 1. (G) β-Glucuronidase (GUS) activity as measured by MUG assays. Measurements represent three technical replicates and error bars are too small to be visible. Separately, in situ GUS staining was performed on over a hundred of primary transformants for each construct. The results were consistent with the MUG assays, with MYB33-1 cm:GUS MYB33-2 cm:GUS and MYB33-FM-GUS all having stronger staining than that of MYB33:GUS (Figure 11A–E). Therefore, the nucleotides that flank the miR159 binding site appear just as necessary for strong silencing of MYB33 as the central nucleotides of its miR159 binding site. Together, these data argues that a miRNA “target” site encompasses not only the miRNA binding site, but the sequence context with which it is in.
Discussion
Despite the extensive analysis of miRNAs in plants, there has been little investigation into the principles that govern their efficacy. Here, taking advantage of the miR159-MYB33/MYB65 module as a model system, we have investigated the principles that control the efficacy of silencing by a highly conserved plant miRNA.
Perfect central complementarity is not essential for strong miR159 efficacy
It is generally assumed that central matches are required for cleavage, and this is a prerequisite for a strong silencing outcome [36]. However, we functionally demonstrate in planta that miRNAs can potently silence target genes with two central mismatches. This directly challenges the empirical parameters of sequence requirements for miRNA-mediated gene silencing, which state there should be no mismatches at positions 10 and 11 of the miRNA-target gene pair [5]. There have been reports of single central mismatches at these positions in naturally occurring miRNA-target pairs. Firstly, degradome signatures have found such miRNAs that can cleave target mRNAs; however the strength of the silencing imposed or the functional outcome of these central mismatches is unknown [37]. Secondly, miR390 that can trigger tasiRNA production has a binding site which includes a single central mismatch, but it is unknown whether the miRNA can cleave its target in vivo [38].
However, it is clear that for strong regulation to occur under the scenario of central mismatches, the miRNA:target stoichiometric ratio must strongly favour the miRNA. Although we show that perfect central complementarity is not mandatory for miR159-guided cleavage, the introduction of central mismatches does attenuate cleavage. Firstly, the average steady state mRNA levels of MYB33 and MYB65 were elevated in MIR159a1 and MIR159a2 plants (Figure 3), where the ratio of un-cleaved MYB33 transcripts to 3′ end cleavage products increased with the number of central mismatches (Figure 4). Secondly, the sequencing profiles of the 5′-RACE products from MYB33-1 cm and MYB33-2 cm plants imply that the cleavage of these centrally mismatched MYB33 transgenes is attenuated (Figure 8). This is consistent with a large body of data that perfect central complementary promotes cleavage, whereas central mismatches compromise cleavage [3]. In fact, the Arabidopsis transcriptome has many mRNAs with potential miRNA binding sites with mismatches at positions 10 and 11, most of which have not been annotated as miRNA targets [32].
A potent non-cleavage mechanism silences MYB33
As one of the best-studied canonical miRNA targets being cleaved by miR159 and characterised by a strong degradome signature [8], [9], here we show that MYB33 is also efficiently silenced by a non-cleavage mechanism. In the case of MYB33 (Figure 5) or MIR159a2 (Figure 1–2) transgenic plants where MYB33 mRNA levels are high, this mechanism becomes more apparent, where it suppresses MYB33 expression to a phenotypically inconsequential level. This argues that cleavage is limiting to some degree, and this non-cleavage mechanism acts in combination to ensure silencing of un-cleaved transcripts. It is probable that this non-cleavage mechanism corresponds to mechanism commonly referred to as translational repression, the phenomenon where target protein accumulation is inconsistent with its transcript level due to miRNA regulation. The repression of MYB33 at both the transcript and translational level is consistent with the proposal that most known plant miRNAs regulate their targets using both mechanisms [3], [18]. Indeed, most targets which are claimed to be regulated primarily at the translational level also undergo cleavage, where the compositions of the degradome signatures for these “translationally repressed” targets appear indistinguishable from miRNA targets that appear to be predominantly regulated by cleavage [8], [9]
It has been hypothesised that cleavage could be a mechanism that results in the fast targeted and irreversible clearing of regulatory mRNAs [14], which in terms of expression would result in a switch outcome [19]. Although miR159 totally silences MYB33/MYB65, it is obvious that miR159 does not clear MYB33 transcripts, especially in the MYB33 transgenic plants. However, miR159 repression of MYB33 via this non-cleavage “translational repression” mechanism appears potent, where it seems to be having a “switch” effect. This is in contrast to “translational repression” in animals, which has been proposed to dampen or tune translation to obtain the desired level of protein synthesis [19].
Factors beyond sequence complementarity: miRNA:target mRNA stoichiometry
Although sequence complementarity is generally the sole factor considered when predicting plant miRNA-mediated silencing, here we show there are other strong contributing factors for the silencing outcome. Firstly, we demonstrate that the silencing outcome can be strongly influenced by the stoichiometric ratio of miRNA to target, a factor that has been largely ignored in plants to date. By transgenically altering the relative abundance of either MYB33 target or miR159 in the variant plants, we could change the silencing outcome independent of complementarity. It appears that the more inefficient a miRNA–target interaction is, the more important stoichiometry becomes. For instance, a combination of high target concentration together with mismatches at the cleavage site resulted in a poor silencing efficacy. This is consistent with previous findings in animals, where a target with central mismatches can saturate the miRNA at a lower concentration than a perfect matched target [33], [34]. This is because the miRISC catalytic rate is slowed down by central mismatches, therefore sequestering the miRNA from further rounds of silencing [35]. As miRISC recycling would make the silencing process more energy efficient, this could be a strong selective driver for perfect central matches in miRNA-target duplexes, which the vast majority of bona fide miRNA-target pairs contain [5].
Factors beyond sequence complementarity: the sequence context of the miR159 binding site
We have clearly demonstrated that nucleotides flanking a miRNA binding site can impact the efficacy of miRNA-mediated silencing. Based on the facts that the vast majority of MYB33-FM plants had developmental defects and the GUS expression level of MYB33-FM:GUS was similar to MYB33-2 cm:GUS, these flanking nucleotides clearly have a large impact on the efficacy of miR159-mediated silencing. This provides the first evidence in plants that the sequence in which a miRNA binding site is embedded plays a critical role in miRNA-target interactions, possibly affecting the silencing outcome comparably to nucleotides within the binding site itself.
We speculate that these flanking nucleotides provide a favourable context for recognition of the MYB33 mRNA by miR159, with important consequences for the efficacy and specificity of their interaction. We have previously shown that miR159 is functionally specific for MYB33 and MYB65 despite bioinformatics predicting approximately 20 genes to be miR159 regulated in Arabidopsis [21]. It is possible that the context of the miR159 binding site in MYB33/MYB65 denotes them as sensitive targets to miR159 regulation, and thereby is a strong contributing factor to this narrow miR159 functional specificity. This could have broad implications for miRNA target predictions and artificial miRNA design, both of which are solely based on sequence complementarity, but in which contextual features appear to have a strong impact on the silencing outcome [39], [40].
Furthermore, we have recently shown that the efficacy of miR159 in silencing MYB33 is tissue specific, where it has much weaker efficacy in the seed relative to the rosette [41]. As the miR159-MYB33 relationship has been conserved for many millions of years, it is conceivable that higher orders of regulation have arisen during this time. Possible factors, such as RNA secondary structures and RNA binding proteins have proven to play a critical role in both specificity and regulation of the silencing outcome in animals [42]–[44]. However, to date these features have been virtually ignored in plants. We believe our findings warrant attention to these possibilities.
A molecular model for plant miRNA mediated gene silencing
A molecular model for plant miRNA-mediated target recognition and subsequent silencing is proposed to explain our results (Figure 12). The nascent target transcripts are recognized and bound by the miRNA-loaded RISC (miRISCs). The ability/efficiency of the miRISCs to recognize target mRNA is influenced by at least three factors: the miRNA binding site complementarity, the target mRNA structure/accessibility (which together make the miRNA target site), and the relative concentration of target mRNA to the miRNA, which becomes increasingly important when cleavage, or recognition is attenuated. Inefficient target recognition can be caused by negative changes to any of the above factors solely, or combinatorially, and lead to a prolonged duration before the target gets bound by the miRISCs enabling translation. If the target mRNA gets recognized and bound by the miRISC, it becomes immediately non-translatable and hence silenced. Cleavage occurs from this pool of miRISC-mRNA complexes, the half-life of which is determined by the cleavage efficiency, which is strongly affected by the complementarity of the miRNA–target duplex, especially at positions 10 and 11. As it has been shown that miRISCs can direct multiple rounds of mRNA cleavage in vitro [45], [46], a major outcome of this event is not only to irreversibly destroy the target transcript but to recycle the miRISC so that it may participate in further target silencing. Of course additional experiments will be needed to determine how accurate this model is.
Fig. 12. A molecular model for plant miRNA-mediated gene silencing. 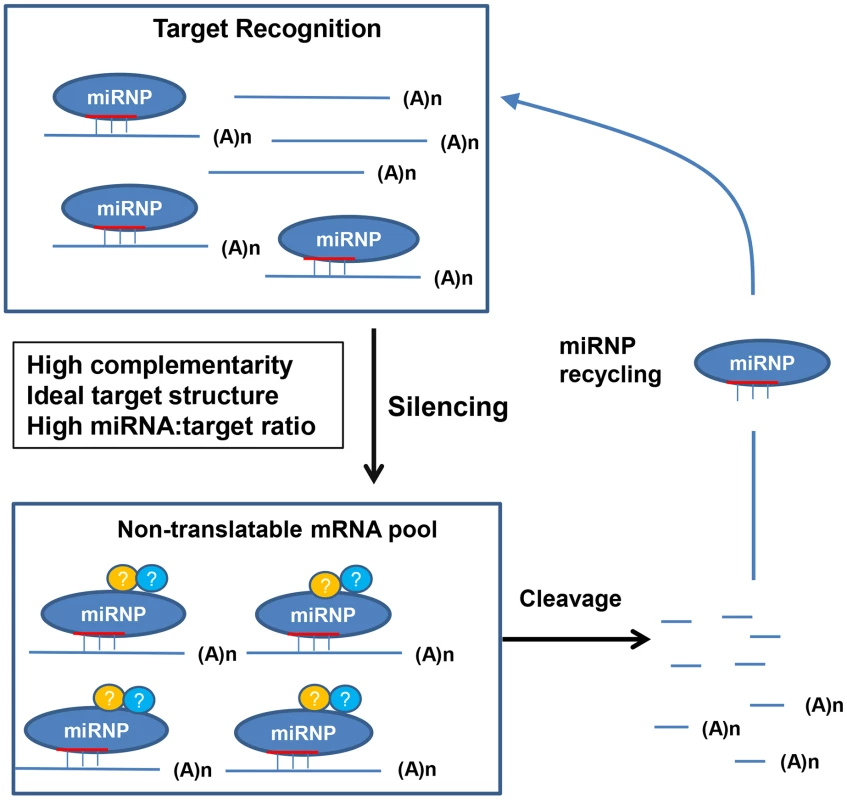
For MIR159a1 or MIR159a2 plants, introduction of central mismatches will increases miRISC-mRNA complex half-life due to an attenuated cleavage efficiency, and so leading to an increased target transcript abundance (Figure 1–4). However, as miR159-guided cleavage fails to clear MYB33/MYB65 transcripts (Figure 2), especially when MYB33 is highly transcribed (Figure 5), a non-cleavage (possibly translational repression) mechanism may be the initial or default state of miR159-mediated silencing, with cleavage being a secondary but inherently linked mechanism completing the silencing process, and simultaneously enabling miRISC recycling. When miRISC recycling does become limiting, the miRNA:target mRNA stoichiometry become an increasingly important factor. For instance, attenuated miR159 cleavage of the highly abundant MYB33-1 cm and MYB33-2 cm transcripts (Figure 7), may result in the miR159-RISC complex becoming limiting, leading to the inefficient silencing of these transgenes (Figure 7, 11). Finally, the non-ideal target structure of the MYB33-FM transgene, results in its poor recognition by miR159, enabling this transgene to be expressed (Figure 10, 11). Materials and Methods
Plant materials and growth conditions
Arabidopsis thaliana ecotype Columbia-0 (Col-0) was used in all experiments and is referred to as wild type. The myb33 mutant used is in ecotype Col-6 with a glaborous1 background mutation which has no trichomes [30] and the mir159ab mutant is as previously described [20]. Plants were grown on soil (Debco Plugger soil mixed with Osmocote Extra Mini fertilizer at 3.5 g/L) either under long day conditions (16 hr light/8 hr dark, 150 µmol/m2/sec at 22°C), or under short-day conditions (12 hr light/12 hr dark at 150 µmol/m2/sec at 22°C).
The generation of binary vectors and transgenic plants
For complementation of mir159ab, a 3642 bp genomic MIR159a (AT1G73687) fragment including the miR159 stem loop region and its extensive 5′ and 3′ flanking sequences, was PCR amplified from Arabidopsis genomic DNA with primers containing attB sites, and sub-cloned into the Gateway donor vector pDONOR/ZEO (Invitrogen) by performing BP reaction using BP Clonase II enzyme mix (Invitrogen). A mutagenesis approach based on Liu and Naismith [47] was then used to generate entry vectors for miR159a0, miR159a1, miR159a2, miR159a3 and miR159a4 variants. Two pairs of primers were designed for each variant to mutate the corresponding miR159a* and miR159a sequences, by performing PCR on 100 ng of MIR159a entry vector, following the setting of 1 cycle of 98°C for 2 min, 20 cycles of 98°C/10 sec, 55°C/30 sec and 72°C for 30 sec/kb extension time, finished with 1 cycle of 55°C/5 min, 72°C/10 min. Each pair of forward and reverse primers contained non-overlapping sequence at 3′ end and primer-primer complementary sequences at the 5′ end, to minimize primer dimerization and enable primers to use the PCR product as template. The non-overlapping sequences were larger than the complementary sequence and had a 5–10°C higher melting temperature. The mutations were placed in both the complementary region and non-overlapping region. The subsequent PCR product was digested with 2 µl DpnI enzyme at 37°C for five hours and purified using Wizard SV Gel and PCR Clean-Up System (Promega), and transformed into E. coli Alpha-Select Gold Efficiency competent cells (Bioline). All entry vectors were confirmed by diagnostic restriction enzyme digestion and sequencing, and were subjected to LR reaction with pMDC100 vector [48] to generate the corresponding binary vectors.
For MYB33 variants, a 4356 bp genomic fragment of MYB33 (AT5G06100), containing identical genomic elements to MYB33:GUS [30], was amplified by PCR from genomic DNA and sub-cloned into pDONOR/ZEO (Invitrogen) by BP reaction. This contained 1991 bp of genomic sequence upstream of the MYB33 start codon, the whole MYB33 coding region, and 585 bp of sequences 3′ of the MYB33 stop codon. The subsequent MYB33 entry vector was mutated using the same strategy described above, to generate the MYB33-0 cm, MYB33-1 cm, MYB33-2 cm, and the MYB33-FM entry vectors respectively. For the mMYB33 construct, the mutagenesis was performed on a MYB33 entry vector with a 24 bp Strep-II tag coding sequence inserted in front of the stop codon. After confirmation by diagnostic restriction enzyme digestion and sequencing, all these entry vectors were subjected to LR reaction with pMDC123 vector [48] to generate the corresponding binary vectors.
The MYB33:GUS and mMYB33:GUS constructs were generated previously [30]. For the generation of MYB33-1 cm:GUS, MYB33-2 cm:GUS and MYB33-FM:GUS translational fusions, the GUS gene was cleaved out of the MYB33:GUS construct with NcoI and ligated into the NcoI site of MYB33-1 cm, MYB33-2 cm and MYB33-FM, respectively. This resulted in the GUS gene being fused in frame to the coding region of MYB33-1 cm, MYB33-2 cm and MYB33-FM, respectively, 55 amino acids from the end of the gene. A clone containing the GUS gene in the right orientation and with the correct sequence was identified for each construct.
All expression vectors were transformed into Agrobacterium tumefaciens strain GV3101 by electroporation [49]. Using the floral dip method [50], MIR159a and its five variant constructs were transformed into the mir159ab mutant; while all MYB33-related constructs were transformed into myb33.
Phusion High Fidelity DNA Polymerase (Finnzymes) was used in all PCR reactions following the standard protocol provided by the manufacturer unless stated elsewhere. All primers used were listed in supplementary file, Table S2.
Expression analysis
TRIzol (Invitrogen) was used for RNA extraction of tissues from plants at different growth stages. The extraction procedure was carried out as per manufacturer's instructions except the following modifications: (1) Approximately 500 mg of plant material was used with 1 mL of Trizol reagent for each extraction; (2) Homogenization of tissues was carried out using a mortar and pestle; (3) The chloroform extraction step was repeated once; (4) Precipitation of RNA was carried out overnight at −20°C to maximize the recovery of small RNAs. RQ1 RNase-Free DNase (Promega) was used to treat RNA samples for qRT-PCR, except those for Taqman sRNA assays. 30–50 µg of total RNA was treated for each sample in a 100 µL reaction volume following the protocol provided, with the addition of RNaseOutRecobinant RNase Inhibitor (Invitrogen) at a concentration of 1 µL/10 µg RNA. Treated RNA was then purified using Spectrum Plant Total RNA Kit (Sigma Aldrich) following instructions from the manufacturer's manual. cDNA synthesis was carried out using SuperScript III Reverse Transcriptase (Invitrogen) and an oligo dT primer according to manufacturer's protocol. For each sample, 250 ng - 5 µg of total RNA was used. The 20 µL reaction was then diluted 50 times in nuclease free distilled water and used for subsequent qRT-PCR. For qRT-PCR, Platinum Taq DNA Polymerase (Invitrogen) with SYB Green (Sigma) and dNTPs (Fisher Biotec) added was used as a master mix. 10 µL of each cDNA sample was added to 9.6 µL of SYB/Taq master mix with 0.4 µL of forward and reverse primers at 10 µmol each. All qRT-PCR reactions (for both reference and genes of interests) were carried out on a Rotor-Gene Q real time PCR machine (QIAGEN) in triplicate, under the following cycling conditions: 1 cycle of 95°C/5 min, 45 cycles of 95°C/15 sec, 60°C/15 sec, 72°C/20 sec. Fluorescence was acquired at the 72°C step. A 55°C to 99°C melting cycle was then carried out. CYCLOPHILIN 5 (At2g29960) was used to normalise mRNA levels using the comparative quantitation program in the Rotor-Gene Q software package provided by QIAGEN. The value for each gene represents the average of triplicate assays.
QRT-PCR assays for mature miRNAs
Customized Taqman sRNA assays (Applied Biosystem) were used to quantitate the mature miR159a variants following protocols described by Allen et al., (2010). Each cDNA sample was assayed in triplicate using a Rotor-Gene Q real time PCR machine (QIAGEN) under the following cycling conditions: 1 cycle of 95°C/5 min, 45 cycles of 95°C/15 sec, 60°C/15 sec, 72°C/20 sec. Fluorescence was acquired at the 72°C step. Expression of all miR159a variants were normalized with sno101 using the comparative concentration analysis program from Rotor-Gene Q software (QIAGEN). The specificity of the assay was tested by performing miR159a2 assays on RNA from mir159ab, MIR159a0 and MIR159a1 plants, and only background signals were detected (data not shown).
GUS staining
In situ GUS staining was performed on 8-day-old seedlings using the method previously described [30] with the following modifications: seedlings were collected and fixed with 90% acetone for 20 minutes at room temperature, followed by a wash with GUS staining buffer containing 50 mM Na phosphate buffer, pH 7.2, 0.2% Triton X-100, 2 mM potassium ferricyanide and 2 mM potassium ferrocyanide. Histochemical reactions were performed with 2 mM X-Gluc (5-bromo-4-chloro-3-indolyl-β-D-glucuronide) in GUS staining buffer at 37°C overnight. All fixative and substrate solutions were introduced into the plants with a 10–15 min vacuum infiltration. Plants were cleared with 70% ethanol for easy GUS observation.
MUG assays
MUG assays were performed using 12-day-old seedlings. 50–100 seedlings were ground into a fine powder in liquid N2 using a mortar and pestle and homogenized with 450 µl of GUS extraction buffer (0.5 M NAPO4, 0.5 M EDTA, 10% SDS, 10% Triton X-100, 1 M DTT) followed by centrifugation at 4°C for 10 min at 12,000 rpm. Subsequent supernatant containing total protein was transferred into a new microfuge tube and the protein concentration was determined by a standard Bradford assay. A GUS fluorometric assay was prepared in a microtitre plate using 20 µg of protein, 90 µl of GUS Assay buffer (1 mM 4-methylumbelliferyl-β-d-glucuronic acid (MUG) in GUS Extraction buffer) and GUS Extraction buffer to a final volume of 150 µl. GUS activity was determined by measuring fluorescence of 4-methylumbelliferone (4-MU) using a Fluostar Fluorometer. Successive fluorescence readings were determined at a wavelength of 355/460 nm over a two hours interval. GUS activity was determined from a set of 4-MU standards and expressed in nmols 4-MU/min/mg protein.
Modified 5′-Rapid Amplification of cDNA Ends (5′-RACE) of degraded MYB33 transcripts
RNA was extracted from either inflorescences or rosettes, and treated with DNaseI and purified as described above. A GeneRacer Kit (Invitrogen) was used to ligate 5 µg of the total purified RNA directly to 1 µg of the RNA oligo adapter provided, without carrying out the de-capping procedure described in the manual. After a one hour incubation at 37°C, the ligation mixture was diluted with 90 µL nuclease free distilled water, and 100 µL of phenol∶chloroform was added and was then vortexed vigorously. The aqueous phase was recovered by centrifugation at 14,680 rpm at room temperature for 5 min, and precipitated with 2 µL 10 mg/mL mussel glycogen, 10 µl 3 M sodium acetate pH 5.2 and 220 µL 95% ethanol overnight at −20°C. The RNA was pelleted by centrifugation at 14,680 rpm for 20 min at 4°C. After one wash with 70% ethanol, the pellet was dried and resuspended in 11 µL water. 1 µL of ligated RNA was analysed on a 1% agarose gel by electrophoresis. The remaining 10 µL was retro-transcribed in a 20 µL reaction. The cDNA synthesized (20 µL) was diluted 25 times, and 25 µL of this diluted cDNA was used in a 50 µL PCR reaction with nested GeneRacer oligo-specific and MYB33-specific primer. PCR was carried out using Platinum Taq DNA Polymerase (Invitrogen) using the setting of 1 cycle of 94°C/2 min; 30 cycles of 95°C/30 sec, 60°C/30 sec, 72°C/1–2 min; 1 cycle of 72°C for 5 min. The PCR products obtained were purified using Wizard SV Gel and PCR Clean-Up System (Promega) and sequenced using a MYB33 specific primer downstream the miR159 cleavage site.
Supporting Information
Zdroje
1. KozomaraA, Griffiths-JonesS (2010) miRBase: integrating microRNA annotation and deep-sequencing data. Nucl Acids Res 39: D152–157.
2. ChenX (2009) Small RNAs and Their Roles in Plant Development. Ann Rev Cell Dev Biol 25 : 21–44.
3. BrodersenP, VoinnetO (2009) Revisiting the principles of microRNA target recognition and mode of action. Nat Rev Mol Cell Biol 10 : 141–148.
4. Jones-RhoadesMW, BartelDP (2004) Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol Cell 14 : 787–799.
5. SchwabR, PalatnikJF, RiesterM, SchommerC, SchmidM, et al. (2005) Specific effects of microRNAs on the plant transcriptome. Dev Cell 8 : 517–527.
6. MalloryAC, ReinhartBJ, Jones-RhoadesMW, TangG, ZamorePD, et al. (2004) MicroRNA control of PHABULOSA in leaf development: importance of pairing to the microRNA 5′ region. EMBO J 23 : 3356–3364.
7. VaucheretH, VazquezF, CrétéP, BartelDP (2004) The action of ARGONAUTE1 in the miRNA pathway and its regulation by the miRNA pathway are crucial for plant development. Genes Dev 18 : 1187–1197.
8. Addo-QuayeC, EshooTW, BartelDP, AxtellMJ (2008) Endogenous siRNA and miRNA targets identified by sequencing of the Arabidopsis degradome. Curr Biol 18 : 758–762.
9. GermanMA, PillayM, JeongDH, HetawalA, LuoS, et al. (2008) Global identification of microRNA-target RNA pairs by parallel analysis of RNA ends. Nat Biotech 26 : 941–946.
10. DaiX, ZhuangZ, ZhaoPX (2011) Computational analysis of miRNA targets in plants: current status and challenges. Brief Bioinform 12 : 115–121.
11. AxtellMJ (2013) Classification and comparison of small RNAs from plants. Annu Rev Plant Biol 64 DOI:10.1146/annurev-arplant-050312-120043
12. DebernardiJM, RodriguezRE, MecchiaMA, PalatnikJF (2012) Functional Specialization of the Plant miR396 Regulatory Network through Distinct MicroRNA–Target Interactions. PLoS Genetics 8 (1) e1002419.
13. TodescoM, BalasubramanianS, CaoJ, OttF, SureshkumarS, et al. (2012) Natural variation in biogenesis efficiency of individual Arabidopsis thaliana microRNAs. Curr Biol 22 : 166–70.
14. RhoadesMW, ReinhartBJ, LimLP, BurgeCB, BartelB, et al. (2002) Prediction of plant microRNA targets. Cell 110 : 513–520.
15. AukermanMJ, SakaiH (2003) Regulation of flowering time and floral organ identity by a MicroRNA and its APETALA2-like target genes. Plant Cell 15 : 2730–2741.
16. ChenX (2004) A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science 303 : 2022–2025.
17. GandikotaM, BirkenbihlRP, HohmannS, CardonGH, SaedlerH, et al. (2007) The miRNA156/157 recognition element in the 3′ UTR of the Arabidopsis SBP box gene SPL3 prevents early flowering by translational inhibition in seedlings. Plant J 49 : 683–693.
18. BrodersenP, Sakvarelidze-AchardL, Bruun-RasmussenM, DunoyerP, YamamotoYY, et al. (2008) Widespread translational inhibition by plant miRNAs and siRNAs. Science 320 : 1185–1190.
19. BartelDP, ChenCZ (2004) Micromanagers of gene expression: the potentially widespread influence of metazoan microRNAs. Nat Rev Genet 5 : 396–400.
20. AllenRS, LiJ, StahleMI, DubroueA, GublerF, et al. (2007) Genetic analysis reveals functional redundancy and the major target genes of the Arabidopsis miR159 family. Proc Natl Acad Sci USA 104 : 16371–16376.
21. AllenRS, LiJ, Alonso-PeralMM, WhiteRG, GublerF, et al. (2010) MicroR159 regulation of most conserved targets in Arabidopsis has negligible phenotypic effects. Silence 1 : 18.
22. Alonso-PeralMM, LiJ, LiY, AllenRS, SchnippenkoetterW, et al. (2010) The microRNA159-regulated GAMYB-like genes inhibit growth and promote programmed cell death in Arabidopsis. Plant Physiol 154 : 757–771.
23. FahlgrenN, HowellMD, KasschauKD, ChapmanEJ, SullivanCM, et al. (2007) High-throughput sequencing of Arabidopsis microRNAs: evidence for frequent birth and death of MIRNA genes. PLoS ONE 2: e219.
24. MartinezJ, TuschlT (2004) RISC is a 5′ phosphomonoester-producing RNA endonuclease. Genes Dev 18 : 975–980.
25. ParizottoEA, DunoyerP, Rahm, HimberC, VoinnetO (2004) In vivo investigation of the transcription, processing, endonucleolytic activity, and functional relevance of the spatial distribution of a plant miRNA. Genes Dev 18 : 2237–2242.
26. LlaveC, XieZ, KasschauKD, CarringtonJC (2002) Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science 297 : 2053–2056.
27. PalatnikJF, WollmannH, SchommerC, SchwabR, BoisbouvierJ, et al. (2007) Sequence and expression differences underlie functional specialization of Arabidopsis microRNAs miR159 and miR319. Dev Cell 13 : 115–125.
28. LlaveC, KasschauKD, RectorMA, CarringtonJC (2002) Endogenous and silencing-associated small RNAs in plants. Plant Cell 14 : 1605–1619.
29. SouretFF, KastenmayerJP, GreenPJ (2004) AtXRN4 degrades mRNA in Arabidopsis and its substrates include selected miRNA targets. Mol Cell 15 : 173–183.
30. MillarAA, GublerF (2005) The Arabidopsis GAMYB-like genes, MYB33 and MYB65, are microRNA-regulated genes that redundantly facilitate anther development. Plant Cell 17 : 705–721.
31. PalatnikJF, AllenE, WuX, SchommerC, SchwabR, et al. (2003) Control of leaf morphogenesis by microRNAs. Nature 425 : 257–263.
32. IvashutaS, BanksIR, WigginsBE, ZhangY, ZieglerTE, et al. (2011) Regulation of gene expression in plants through miRNA inactivation. PLoS ONE 6: e21330.
33. DoenchJG, SharpPA (2004) Specificity of microRNA target selection in translational repression. Genes Dev 18 : 504–511.
34. BrownBD, GentnerB, CantoreA, ColleoniS, AmendolaM, et al. (2007) Endogenous microRNA can be broadly exploited to regulate transgene expression according to tissue, lineage and differentiation state. Nat Biotech 25 : 1457–1467.
35. BaccariniA, ChauhanH, GardnerTJ, JayaprakashAD, SachidanandamR, et al. (2011) Kinetic analysis reveals the fate of a microRNA following target regulation in mammalian cells. Curr Biol CB 21 : 369–376.
36. Jones-RhoadesMW, BartelDP, BartelB (2006) MicroRNAs and their regulatory roles in plants. Annu Rev Plant Biol 57 : 19–53.
37. ZhengY, LiY-F, SunkarR, ZhangW (2012) SeqTar: an effective method for identifying microRNA guided cleavage sites from degradome of polyadenylated transcripts in plants. Nucl Acids Res 40: e28.
38. AxtellMJ, JanC, RajagopalanR, BartelDP (2006) A two-hit trigger for siRNA biogenesis in plants. Cell 127 : 565–577.
39. DevesonI, LiJ, MillarAA (2013) MicroRNAs with analogous target complementarities perform with highly variable efficacies in Arabidopsis. FEBS Lett 587 : 3703–3708.
40. LiJ-F, ChungHS, NiuY, BushJ, McCormackM, et al. (2013) Comprehensive protein-based artificial microRNA screens for effective gene silencing in plants. The Plant Cell 25 : 1507–1522.
41. Alonso-PeralMM, SunC, MillarAA (2012) MircoRNA159 can act as a switch or tuning microRNA independently of its abundance in Arabidopsis. PLoS ONE 7 (4) e34751.
42. AmeresSL, MartinezJ, SchroederR (2007) Molecular Basis for Target RNA Recognition and Cleavage by Human RISC. Cell 130 : 101–112.
43. KertesM, IovinoN, UnnerstallU, GaulU, SegalE (2007) The role of site accessibility in microRNA target recognition. Nat Genet 39 : 1278–1284.
44. LongD, LeeR, WilliamsP, ChanCY, AmbrosV, et al. (2007) Potent effect of target structure on microRNA function. Nat Struct Mol Biol 14 : 287–294.
45. HutvagnerG, ZamorePD (2002) A microRNA in a multiple-turnover RNAi enzyme complex. Science 297 : 2056–2060.
46. TangG, ReinhartBJ, BartelDP, ZamorePD (2003) A biochemical framework for RNA silencing in plants. Genes Dev 17 : 49–63.
47. LiuH, NaismithJ (2008) An efficient one-step site-directed deletion, insertion, single and multiple-site plasmid mutagenesis protocol. BMC Biotech 8 : 91.
48. CurtisMD, GrossniklausU (2003) A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133 : 462–469.
49. HellensR, MullineauxP, KleeH (2000) Technical Focus: a guide to Agrobacterium binary Ti vectors. Trends Plant Sci 5 : 446–451.
50. CloughSJ, BentAF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16 : 735–743.
Štítky
Genetika Reprodukční medicína
Článek Sex Chromosome Turnover Contributes to Genomic Divergence between Incipient Stickleback SpeciesČlánek Final Pre-40S Maturation Depends on the Functional Integrity of the 60S Subunit Ribosomal Protein L3
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2014 Číslo 3
-
Všechny články tohoto čísla
- , a Gene That Influences the Anterior Chamber Depth, Is Associated with Primary Angle Closure Glaucoma
- Genomic View of Bipolar Disorder Revealed by Whole Genome Sequencing in a Genetic Isolate
- The Rate of Nonallelic Homologous Recombination in Males Is Highly Variable, Correlated between Monozygotic Twins and Independent of Age
- Genetic Determinants Influencing Human Serum Metabolome among African Americans
- Heterozygous and Inherited Mutations in the Smooth Muscle Actin () Gene Underlie Megacystis-Microcolon-Intestinal Hypoperistalsis Syndrome
- Genome-Wide Meta-Analysis of Homocysteine and Methionine Metabolism Identifies Five One Carbon Metabolism Loci and a Novel Association of with Ischemic Stroke
- Cancer Evolution Is Associated with Pervasive Positive Selection on Globally Expressed Genes
- Genetic Diversity in the Interference Selection Limit
- Integrating Multiple Genomic Data to Predict Disease-Causing Nonsynonymous Single Nucleotide Variants in Exome Sequencing Studies
- An Evolutionary Analysis of Antigen Processing and Presentation across Different Timescales Reveals Pervasive Selection
- Cleavage Factor I Links Transcription Termination to DNA Damage Response and Genome Integrity Maintenance in
- DNA Dynamics during Early Double-Strand Break Processing Revealed by Non-Intrusive Imaging of Living Cells
- Genetic Basis of Metabolome Variation in Yeast
- Modeling 3D Facial Shape from DNA
- Dysregulated Estrogen Receptor Signaling in the Hypothalamic-Pituitary-Ovarian Axis Leads to Ovarian Epithelial Tumorigenesis in Mice
- Photoactivated UVR8-COP1 Module Determines Photomorphogenic UV-B Signaling Output in
- Local Evolution of Seed Flotation in Arabidopsis
- Chromatin Targeting Signals, Nucleosome Positioning Mechanism and Non-Coding RNA-Mediated Regulation of the Chromatin Remodeling Complex NoRC
- Nucleosome Acidic Patch Promotes RNF168- and RING1B/BMI1-Dependent H2AX and H2A Ubiquitination and DNA Damage Signaling
- The -Induced Arabidopsis Transcription Factor Attenuates ABA Signaling and Renders Seedlings Sugar Insensitive when Present in the Nucleus
- Changes in Colorectal Carcinoma Genomes under Anti-EGFR Therapy Identified by Whole-Genome Plasma DNA Sequencing
- Selection of Orphan Rhs Toxin Expression in Evolved Serovar Typhimurium
- FAK Acts as a Suppressor of RTK-MAP Kinase Signalling in Epithelia and Human Cancer Cells
- Asymmetry in Family History Implicates Nonstandard Genetic Mechanisms: Application to the Genetics of Breast Cancer
- Co-translational Localization of an LTR-Retrotransposon RNA to the Endoplasmic Reticulum Nucleates Virus-Like Particle Assembly Sites
- Sex Chromosome Turnover Contributes to Genomic Divergence between Incipient Stickleback Species
- DWARF TILLER1, a WUSCHEL-Related Homeobox Transcription Factor, Is Required for Tiller Growth in Rice
- Functional Organization of a Multimodular Bacterial Chemosensory Apparatus
- Genome-Wide Analysis of SREBP1 Activity around the Clock Reveals Its Combined Dependency on Nutrient and Circadian Signals
- The Yeast Sks1p Kinase Signaling Network Regulates Pseudohyphal Growth and Glucose Response
- An Interspecific Fungal Hybrid Reveals Cross-Kingdom Rules for Allopolyploid Gene Expression Patterns
- Temperate Phages Acquire DNA from Defective Prophages by Relaxed Homologous Recombination: The Role of Rad52-Like Recombinases
- Dying Cells Protect Survivors from Radiation-Induced Cell Death in
- Determinants beyond Both Complementarity and Cleavage Govern MicroR159 Efficacy in
- The bHLH Factors Extramacrochaetae and Daughterless Control Cell Cycle in Imaginal Discs through the Transcriptional Regulation of the Phosphatase
- The First Steps of Adaptation of to the Gut Are Dominated by Soft Sweeps
- Bacterial Regulon Evolution: Distinct Responses and Roles for the Identical OmpR Proteins of Typhimurium and in the Acid Stress Response
- Final Pre-40S Maturation Depends on the Functional Integrity of the 60S Subunit Ribosomal Protein L3
- Mitogen-Activated Protein Kinase (MAPK) Pathway Regulates Branching by Remodeling Epithelial Cell Adhesion
- CDP-Diacylglycerol Synthetase Coordinates Cell Growth and Fat Storage through Phosphatidylinositol Metabolism and the Insulin Pathway
- Coronary Heart Disease-Associated Variation in Disrupts a miR-224 Binding Site and miRNA-Mediated Regulation
- TBX3 Regulates Splicing : A Novel Molecular Mechanism for Ulnar-Mammary Syndrome
- Identification of Interphase Functions for the NIMA Kinase Involving Microtubules and the ESCRT Pathway
- Is a Cancer-Specific Fusion Gene Recurrent in High-Grade Serous Ovarian Carcinoma
- LILRB2 Interaction with HLA Class I Correlates with Control of HIV-1 Infection
- Worldwide Patterns of Ancestry, Divergence, and Admixture in Domesticated Cattle
- Parent-of-Origin Effects Implicate Epigenetic Regulation of Experimental Autoimmune Encephalomyelitis and Identify Imprinted as a Novel Risk Gene
- The Phospholipid Flippase dATP8B Is Required for Odorant Receptor Function
- Noise Genetics: Inferring Protein Function by Correlating Phenotype with Protein Levels and Localization in Individual Human Cells
- DUF1220 Dosage Is Linearly Associated with Increasing Severity of the Three Primary Symptoms of Autism
- Sugar and Chromosome Stability: Clastogenic Effects of Sugars in Vitamin B6-Deficient Cells
- Pheromone-Sensing Neurons Expressing the Ion Channel Subunit Stimulate Male Courtship and Female Receptivity
- Gene-Based Sequencing Identifies Lipid-Influencing Variants with Ethnicity-Specific Effects in African Americans
- Telomere Shortening Unrelated to Smoking, Body Weight, Physical Activity, and Alcohol Intake: 4,576 General Population Individuals with Repeat Measurements 10 Years Apart
- A Combination of Activation and Repression by a Colinear Hox Code Controls Forelimb-Restricted Expression of and Reveals Hox Protein Specificity
- An ER Complex of ODR-4 and ODR-8/Ufm1 Specific Protease 2 Promotes GPCR Maturation by a Ufm1-Independent Mechanism
- Epigenetic Control of Effector Gene Expression in the Plant Pathogenic Fungus
- Genetic Dissection of Photoreceptor Subtype Specification by the Zinc Finger Proteins Elbow and No ocelli
- Validation and Genotyping of Multiple Human Polymorphic Inversions Mediated by Inverted Repeats Reveals a High Degree of Recurrence
- CYP6 P450 Enzymes and Duplication Produce Extreme and Multiple Insecticide Resistance in the Malaria Mosquito
- GC-Rich DNA Elements Enable Replication Origin Activity in the Methylotrophic Yeast
- An Epigenetic Signature in Peripheral Blood Associated with the Haplotype on 17q21.31, a Risk Factor for Neurodegenerative Tauopathy
- Lsd1 Restricts the Number of Germline Stem Cells by Regulating Multiple Targets in Escort Cells
- RBPJ, the Major Transcriptional Effector of Notch Signaling, Remains Associated with Chromatin throughout Mitosis, Suggesting a Role in Mitotic Bookmarking
- The Membrane-Associated Transcription Factor NAC089 Controls ER-Stress-Induced Programmed Cell Death in Plants
- The Functional Consequences of Variation in Transcription Factor Binding
- Comparative Genomic Analysis of N-Fixing and Non-N-Fixing spp.: Organization, Evolution and Expression of the Nitrogen Fixation Genes
- An Insulin-to-Insulin Regulatory Network Orchestrates Phenotypic Specificity in Development and Physiology
- Suicidal Autointegration of and Transposons in Eukaryotic Cells
- A Multi-Trait, Meta-analysis for Detecting Pleiotropic Polymorphisms for Stature, Fatness and Reproduction in Beef Cattle
- Genome-Wide DNA Methylation Analysis Predicts an Epigenetic Switch for GATA Factor Expression in Endometriosis
- Genome-Wide DNA Methylation Analysis of Human Pancreatic Islets from Type 2 Diabetic and Non-Diabetic Donors Identifies Candidate Genes That Influence Insulin Secretion
- The and Hybrid Incompatibility Genes Suppress a Broad Range of Heterochromatic Repeats
- The Kil Peptide of Bacteriophage λ Blocks Cytokinesis via ZipA-Dependent Inhibition of FtsZ Assembly
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Worldwide Patterns of Ancestry, Divergence, and Admixture in Domesticated Cattle
- Genome-Wide DNA Methylation Analysis of Human Pancreatic Islets from Type 2 Diabetic and Non-Diabetic Donors Identifies Candidate Genes That Influence Insulin Secretion
- Genetic Dissection of Photoreceptor Subtype Specification by the Zinc Finger Proteins Elbow and No ocelli
- GC-Rich DNA Elements Enable Replication Origin Activity in the Methylotrophic Yeast
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



