-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaThe Phospholipid Flippase dATP8B Is Required for Odorant Receptor Function
The olfactory systems of insects are fundamental to critical behaviours such as finding mates, food and host plants. Insects can detect a wide range of environmental cues using three different families of olfactory receptor proteins. Why insects have three different families of receptor genes, and how they function together, is not fully understood. Here we identified a new gene, dATP8B, which is critically and specifically required for the function of only one of these receptor families in Drosophila. dATP8B is a member of the P4-type ATPases, or phospholipid flippases; these enzymes function in establishing a difference or asymmetry in lipid composition between the outer and inner leaflets of plasma membranes. This is thought to be important for many cellular membrane processes; however, specific functions of individual flippase proteins are not well described. We find that dATP8B is required for the function of the odorant receptor family, but not the ionotropic-like and gustatory receptor families. This further highlights the functional differences between these receptor families and suggests a role for phospholipids in the signalling of a specific family of receptor proteins.
Published in the journal: . PLoS Genet 10(3): e32767. doi:10.1371/journal.pgen.1004209
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004209Summary
The olfactory systems of insects are fundamental to critical behaviours such as finding mates, food and host plants. Insects can detect a wide range of environmental cues using three different families of olfactory receptor proteins. Why insects have three different families of receptor genes, and how they function together, is not fully understood. Here we identified a new gene, dATP8B, which is critically and specifically required for the function of only one of these receptor families in Drosophila. dATP8B is a member of the P4-type ATPases, or phospholipid flippases; these enzymes function in establishing a difference or asymmetry in lipid composition between the outer and inner leaflets of plasma membranes. This is thought to be important for many cellular membrane processes; however, specific functions of individual flippase proteins are not well described. We find that dATP8B is required for the function of the odorant receptor family, but not the ionotropic-like and gustatory receptor families. This further highlights the functional differences between these receptor families and suggests a role for phospholipids in the signalling of a specific family of receptor proteins.
Introduction
In insects such as Drosophila melanogaster the detection of environmental odours is achieved by arrays of olfactory receptor neurons (ORNs) housed in different types of chemosensory hairs (sensilla) on two olfactory organs, the antenna and the maxillary palp. Each class of ORN is tuned to specific chemical signals by expression of different olfactory receptor genes. The responses of most insect ORNs are reliant on members of two large and divergent families of olfactory receptor proteins, the odorant receptor (Or) and Ionotropic glutamate-like receptor (IR) families. Or proteins are seven trans-membrane domain proteins that are topologically inverted in comparison to G-protein-coupled receptors [1], [2], raising the question as to whether they do interact with G proteins. Indeed, several studies have concluded that Or-signalling is rather ionotropic, and that the functional receptor is a ligand-gated cation channel composed of a variable Or odorant-binding subunit and a co-receptor subunit called Orco [1]–[4]. Orco is required for Or proteins to be transported to the dendrites [5], and heterologous expression studies suggest Orco is also part of the functional receptor and is essential for the initial fast inward current upon ligand binding [6], [7]. However, there is also genetic and pharmacological evidence for a slower metabotropic transduction cascade (for an excellent recent review see [8]). For example, heterologous expression experiments demonstrated a slower metabotropic current after Or stimulation, as well as an increase in cAMP [4]. Orco has been suggested to be activated by cAMP [4], and in addition it has been suggested that phosphorylation of Orco by protein kinase C (PKC) is required for its activation [9]. However, several studies of loss of function of Gα–encoding genes in Drosophila have yielded conflicting results [10]–[12]. A recent study suggests that metabotropic regulation of Orco regulates Or sensitivity [13]. Overall, the mechanism of Or signalling appears to be complex, with both ionotropic and metabotropic pathway involvement, and despite much investigation is not fully understood.
IR genes encode a very different family of receptors, they are three trans-membrane domain ligand-gated ion channels that are related to ionotropic glutamate receptors [14]. IR proteins form heteromers but are not reliant on Orco [15], [16]. A third family of receptors, the gustatory receptors (Grs), are also seven trans-membrane proteins and are evolutionarily related to the Ors [17], [18]. Where the Or and IR families both detect a range of structurally diverse odorants and function in many ORNs [19], [20], only one functional class of ORN, specialized for detection of carbon dioxide (CO2), expresses Gr genes [21], [22]. Most other Gr genes are expressed in chemosensory neurons in taste sensilla on appendages of the fly that detect non-volatiles such as sugars and alkaloids [23], [24]. Although the Grs are evolutionarily related to Or genes [18], they are not reliant on Orco for function [5]. Their signalling properties have been much less extensively studied and it is not clear whether they utilize similar signalling mechanisms to Ors.
Relatively few other genes involved in the function of the peripheral olfactory system have been identified. Accordingly, to identify novel genes important for Drosophila peripheral olfaction, we conducted a screen for mutants defective in ORN responses. We identified a recessive mutation that specifically affects Or-expressing ORNs, dramatically reducing their sensitivity to ligands, but has no effect on IR or Gr-expressing sensory neurons. The causative gene, dATP8B, is a member of the phospholipid flippase family of P4-ATPases, which function to maintain the asymmetry in natural lipid composition between the outer and inner leaflets of cell membranes. Our results demonstrate that dATP8B is critically and specifically required for the function of the Or receptor family.
Results
To identify new genes involved in olfactory neuron function we screened 482 lines carrying homozygous viable EMS-induced mutations on chromosome III (from the Zuker Collection [25]) for defects in olfactory responses. Electroantennograms (EAGs) were employed to measure voltage changes across the antennal epithelium in response to a set of odorants known to excite a variety of different ORN classes. We identified one line (ll2) in which homozygous mutant flies showed reduced EAG responses to most tested odorants compared to heterozygous controls (p<0.05, Fig. 1A). Neurons on the second olfactory organ, the maxillary palp, showed an equivalent reduction in response to odorants in the mutant line (p<0.05, Fig. 1B). In contrast to the general odorants, the EAG response to carbon dioxide, which is generated by one specific antennal ORN class (ab1C) and mediated by two gustatory receptor genes, was unaffected in the mutant (Fig. 1A).
Fig. 1. Mutations in dATP8B cause severe olfactory defects. 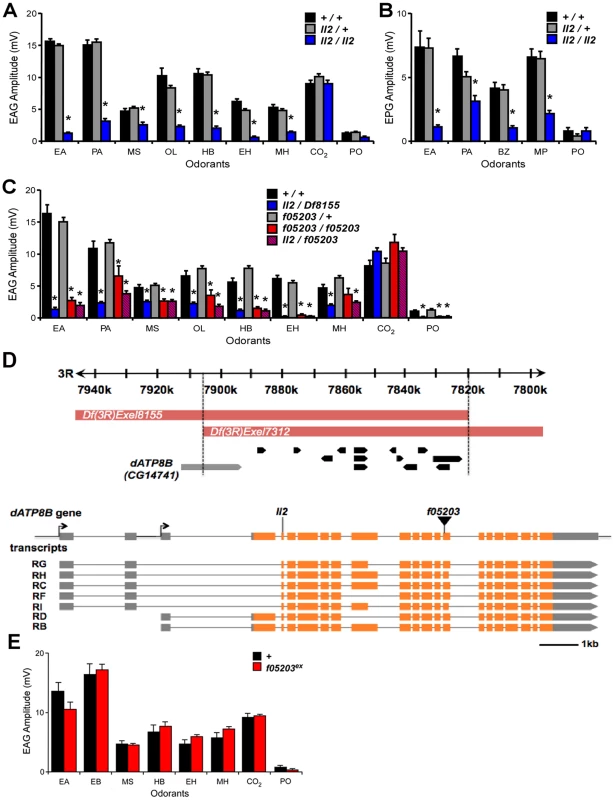
(A) Electroantennogram (EAG) and (B) electropalpogram (EPG) responses to a panel of odorants. Bars represent mean response ± SEM (n = 10), asterisks are significant differences. Responses of homozygous ll2 mutant flies (blue bars) are severely reduced when compared with controls (black bars) for all the tested odorants except CO2 (t-test, Bonferroni, p<0.05). Heterozygote flies (grey bars) are not affected. (C) A deletion that removes dATP8B and a piggyBac element inserted in dATP8B both fail to complement the ll2 phenotype. Bars represent mean EAG responses ± SEM (n = 6–10). Trans-heterozygotes for ll2 and deficiency Df(3R)Exel8155 (blue bars) have reduced EAG response compared to controls (black bars, t-tests, Bonferroni, p<0.05), and homozygotes for the piggyBac insertion dATP8Bf05203 (red bars) and trans-heterozygotes for ll2 and dATP8Bf05203 (red/blue hatch) show similar reductions when compared to heterozygote controls (grey bars; t-tests, Bonferroni, p<0.05). (D) The mapped genomic interval for the ll2 mutant and the gene model of dATP8B (CG14741). The candidate region contained 14 annotated genes. The identified nonsense mutation in dATP8B and the insertion site of the piggyBac line dATP8Bf05203 are in the 1st and the 10th common coding exon respectively, affecting all the annotated transcripts. For isoforms RC, RF, RG, RH and RI the EMS mutation causes R18-X and for isoforms RB and RD the mutation causes R197-X. Coding exons are colored in orange and the 3′ and the 5′ UTR are in grey. (E) The olfactory defect in the dATP8Bf05203 line is reverted when the piggyBac insertion is precisely excised. EAG responses of homozygous dATP8Bf05203-Ex flies (red bars) to a panel of odorants were not different from controls (black bars). Bars represent mean response ± SEM (n = 5, t-test, Bonferroni). Odorants are: EA, ethyl acetate, PA, pentyl acetate, MS, methyl salicylate, OL, 1-octen-3-ol, HB, ethyl 3-hydroxybutanoate, EH, ethyl hexanoate, MH, 6-methyl-5-hepten-2-one, BZ, benzaldehyde, MP, 4-methylphenol, PO, paraffin oil (solvent blank). We next used genomic deficiencies to map the mutation. We found that flies trans-heterozygous for the ll2 chromosome and either Df(3R)Exel7312 or Df(3R)Exel8155 showed the same mutant phenotype as homozygous ll2 flies (Fig. 1C). This localised the mutation to an 86.5kb genomic region that contains 14 annotated genes (Fig. 1D). Whole genome sequencing experiments identified a nonsense mutation in the CG14741 gene within this region (genomic location 7,902,447, mutation C7902447T). CG14741 has seven predicted isoforms; the nonsense mutation is in the first coding exon common to all predicted isoforms and thus is predicted to severely truncate all proteins encoded by the locus. The CG14741 gene has not previously been functionally characterised. However, sequence comparisons reveal it belongs to the type 4 P-type ATPase family of integral membrane transporter proteins [26]. This group of proteins includes four human members named ATPB1-4 and CG14741 as the sole Drosophila representative. Henceforth we refer to the gene CG14741 as dATP8B.
To confirm that the nonsense mutation in dATP8B was the cause of the olfactory defects observed in the ll2 line we examined an independent mutant allele, a line containing a piggyBac element inserted in the coding region and predicted to affect all the isoforms (dATP8B f05203, Fig. 1D). Flies homozygous for the dATP8B f05203 allele showed the same EAG defect as homozygotes for the original EMS allele (p<0.05, Fig. 1C). This phenotype was reverted when the piggyBac insertion was precisely excised (Fig. 1E), confirming that the olfactory defect is due to the insertion in dATP8B. We also showed that the EMS and piggyBac insertion alleles failed to complement; trans-heterozygotes for the two mutant alleles also exhibited the olfactory defect (p<0.05, Fig. 1C). Together, these data confirm that mutations in dATP8B cause a severe reduction in ORN responses.
In our initial EAG recordings we noted that the dATP8Bll2 mutant flies had normal responses to carbon dioxide (CO2). Unlike the other odorants tested, which are detected by ORNs that express members of the Or receptor family, CO2 is detected by the ab1C ORN which expresses two members of the Gr receptor family, Gr21a and Gr63a [21], [22]. These data suggested that the mutation might be specifically affecting Or-expressing ORNs rather than all ORNs. To determine if this was the case we characterised mutant responses from selected ORN classes on the antenna whose responses are determined by members of all three different receptor gene families. In each neuron type we examined that expresses Or genes we found that the responses to ligands were substantially reduced over a range of concentrations (Fig. 2). This was the case regardless of the morphological type of sensillum, as we found greatly reduced responses to 2-heptanone from the ab3B neuron in basiconic sensilla (Fig. 2A), to cis-vaccenyl acetate from the at1A neuron in trichoid sensilla (Fig. 2B), and to Z3-hexenol from the ac3B neuron in coeloconic sensilla (Fig. 2C).
Fig. 2. dATP8B is required for responses of Or but not IR or Gr- expressing sensory neurons. 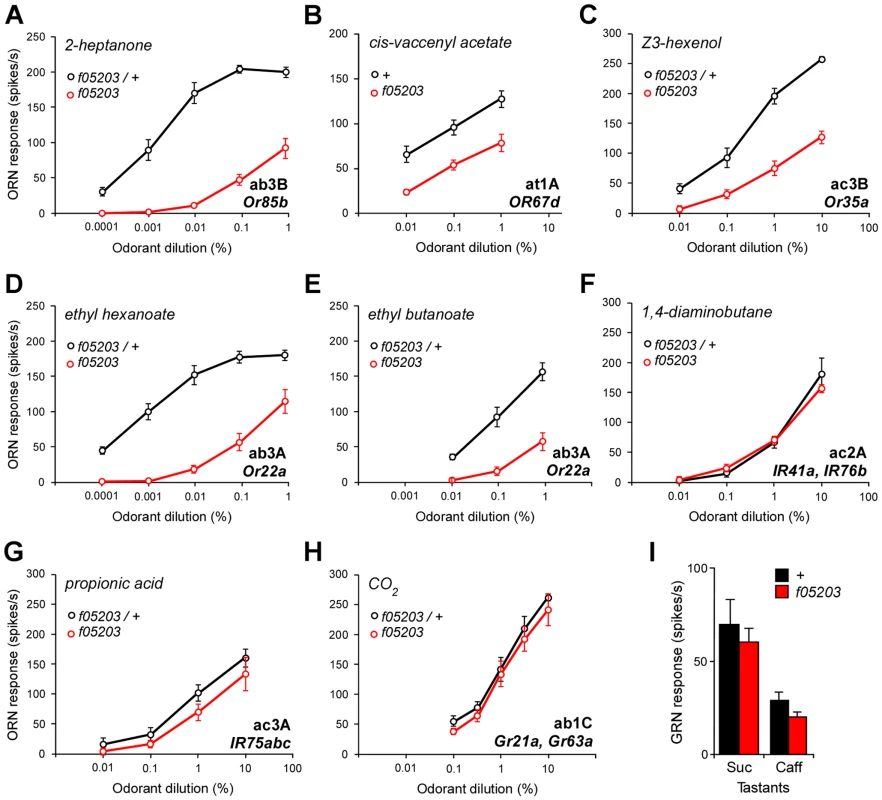
(A–C) Mutations in dATP8B reduce the sensitivity of Or-expressing neurons to their major ligands. Dose-response curves for neurons located in three different morphological types of sensilla are shown; (A) ab3B neurons in basiconic sensilla (Or85b) to 2-heptanone, (B) at1A neurons in trichoid sensilla (Or67d) to cis-vaccenyl acetate, (C) ac3B neurons in coeloconic sensilla (Or35a) to Z3-hexenol. In all cases sensitivity is significantly lowered for all doses in homozygous dATP8Bf05203 flies (red) compared to controls (black, t-test, Bonferroni, n = 6–9, recorded from 4–8 flies). (D–E) Mutations in dATP8B differentially alter responses of ab3A neurons (Or22a) to a major ligand ethyl hexanoate (D) and a minor ligand ethyl butanoate (E). (n = 7 sensilla, recorded from 5–6 flies). (F–I) Neurons expressing IR or Gr receptors are not affected by dATP8B mutations. Four different types of neurons are shown; (F) responses of ac2A neurons in coeloconic sensilla (IR41a and IR76b) to 1,4-diaminobutane, (G) responses of ac3A neurons in coeloconic sensilla (IR75a,b and c) to propionic acid, (H) responses of ab1C neurons in basiconic sensilla (Grs21a and 63a) to CO2, (I) responses from single neurons in labellar taste sensilla expressing Gr genes to 100 mM sucrose (Suc) or 10 mM caffeine (Caff) (mean ± SEM). In all cases control and mutant responses are not significantly different (n = 6–10 sensilla from 3–5 flies, t-tests, Bonferroni). Controls are either wild type or heterozygous dATP8Bf05203 mutants. These experiments also confirmed a finding we had noted from our EAG recordings, namely that the responses of dATP8B mutant flies, while greatly reduced, are not completely abolished. We found that at high odorant concentrations mutant neurons were still able to fire at relatively high rates (an average of 100 spikes per second; Figs. 2A–C). We also noted that the effect of the mutation can be different for different odorants activating the same receptor. For the ab3A neuron, which expresses Or22a, we found that the sensitivity to its high affinity ligand ethyl hexanoate was reduced by three log steps (Fig. 2D). However, when we recorded responses from ab3A neurons for a lower affinity ligand, ethyl butanoate, we found that the curve is shifted much less (Fig. 2E). This suggests that sensitivity to different ligands of the Or22a receptor is affected by the dATP8B mutation to differing extents.
In contrast, we found that mutations in dATP8B have no effect on responses of ORNs that express IR or Gr genes. This was the case for two ORN types that express IR genes; the responses to 1,4-diaminobutane of the ac2A neuron (Fig. 2F) and to propionic acid of the ac3A neuron (Fig. 2G) were unaffected in the mutant. Consistent with the initial EAG data, in dATP8B mutants the CO2 response from the ab1C neuron, which is determined by Gr genes, is not significantly reduced for any of the concentrations that evoke a wide range of excitation levels (Fig. 2H). We also performed recordings from gustatory neurons on the labellum expressing members of the Gr family, and showed that in dATP8B mutants the response to both sucrose and caffeine, mediated by different Gr receptors [23], was unaffected (Fig. 2I). Taken together, these data strongly suggest that dATP8B is specifically required for the function of Or-expressing neurons, and not for chemosensory neurons in general.
dATP8B was identified in a previous study as being present in the antennal proteome [27]. To confirm its expression in antennae and to determine the cell type in which dATP8B is expressed we performed immunohistochemistry using an anti-dATP8B antibody. Strong staining was seen within the shafts of the sensilla, the location of the outer dendrites of the ORNs (Fig. 3A). Staining was observed in both basiconic and trichoid sensilla, which both house neurons expressing Or genes. We could not easily visualise the sensilla shafts of the coeloconic sensilla. Staining was absent from the outer dendrites in the sensilla shafts in mutants for dATP8B (Fig. 3B). Signal was also observed in the inner dendrites and the cell bodies, however this signal was also observed in mutants for dATP8B, albeit more weakly, (compare Figs. 3A and B), and thus may be largely background staining.
Fig. 3. dATP8B is expressed and required in Or-expressing olfactory receptor neurons. 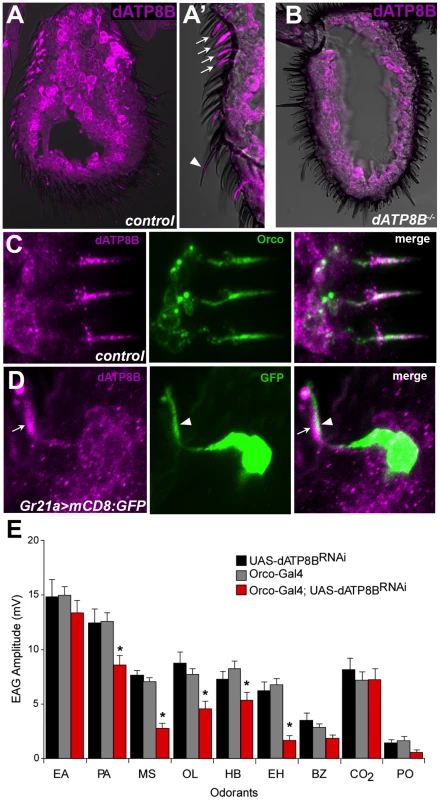
(A–A′) dATP8B protein localises to the dendrites of ORNs. 14 µm thick antennal sections from wild type flies (CS-5) were stained for anti-dATP8B (magenta). Strong anti-dATP8B staining is seen in the shafts of the sensilla (the location of the outer dendrites) of basiconic (arrows) and trichoid (arrowhead) sensilla. Staining is also seen in the inner dendrites and cell bodies, however this staining is also present in dATP8B mutants. (B) The strong outer dendrite staining of anti-dATP8B is absent in dATP8B mutants (ll2/Df(3R)Exel8155), indicating it is specific for dATP8B. (C) dATP8B and Orco co-localise in the outer dendrites. 14 µm thick antennal sections from wild type flies were stained for anti-dATP8B (magenta) and Orco (green). (D) dATP8B is absent from the outer dendrites of Gr21a-expressing (ab1C) neurons. 14 µm thick antennal sections from Gr21a>mCD8:GFP flies were stained for anti-dATP8B (magenta) and anti-GFP (green). (E) In Orco-GAL4: UAS-dATP8BRNAi flies (red bars) the EAG response is significantly reduced compared to controls (black and grey bars, t-test, Bonferroni, p<0.05) for some of the same odorants that are affected by the two dATP8B mutant alleles. Bars represent mean EAG responses ± SEM (n = 6–10). Odorants are: EA, ethyl acetate, PA, pentyl acetate, MS, methyl salicylate, OL, 1-octen-3-ol, HB, ethyl 3-hydroxybutanoate, EH, ethyl hexanoate, BZ, benzaldehyde, PO, paraffin oil (solvent blank). Given dATP8B function seems to only be required in the Or-expressing neurons we next asked if it is specifically expressed in Or-expressing, but not IR or Gr-expressing, neurons. We confirmed that dATP8B is expressed in the Or-expressing neurons by showing that it co-localises with Orco (Fig. 3C). We then asked if dATP8B is expressed in the ab1C neurons that express Gr21a. These neurons are housed in ab1 basiconic sensilla together with three other neurons, which all express Or genes. We visualised the ab1C neurons by using Gr21a-Gal4 to drive the membrane-localised mCD8:GFP and staining with anti-GFP. Strong GFP signal was observed in the outer and inner dendrites and cell bodies (Fig. 3D). Double staining with anti-GFP and anti-dATP8B showed that dATP8B is absent from the outer dendrites of Gr21a-expressing (ab1C) neurons (Fig. 3D). We saw many examples where the GFP-positive dendrites of the ab1C neurons ran parallel to but did not overlap dATP8B-positive dendrites within the sensilla shafts. We conclude that dATP8B localises to the outer dendrites in Or-expressing neurons, and likely does not in Gr-expressing neurons.
We also showed that dATP8B function is required in the ORNs, rather than another cell type, by using RNA interference (RNAi) to knock down dATP8B in the Or-expressing ORNs using an Orco driver. In Orco-GAL4:UAS-dATP8BRNAi flies the EAG response was significantly reduced for some of the odorants for which we also saw reduced responses in the two dATP8B mutant alleles, for example methyl salicylate and ethyl hexanoate (Fig. 3E). Although this defect is much less severe than seen in the loss of function mutants (Fig. 1A), taken together with the immunohistochemistry data these results suggest that dATP8B has a functional role in ORNs, rather than in another cell type such as support cells. As Orco has a relatively late onset of expression in pupal development [5], this result also suggests that dATP8B plays a role in these neurons after the initial development of olfactory sensilla.
The phenotype of dATP8B mutants bears a strong resemblance to that of mutations in the Orco gene, which is required for the localisation and function of the Or receptors, but not for the IR or Gr receptors [5], [15], [16]. We thus asked if loss of dATP8B caused Orco itself and/or the other Or proteins to be incorrectly localized. We tested this by using antibodies to examine the localization of both Orco and Or22a. In wild type flies anti-Orco staining is seen in both the outer dendrites and the cell bodies, for anti-Or22a there is strong staining in the outer dendrites only (Fig. 4). No difference in localization of either Orco or Or22a was observed in dATP8B mutants (Fig. 4). In addition, no noticeable difference was observed in the length or shape of the dendrites in the mutant. This confirmed an initial finding that the overall appearance of the olfactory sensilla in the mutant is normal (Fig. S1).
Fig. 4. Orco and Or22a localize normally to the dendrites in dATP8B mutants. 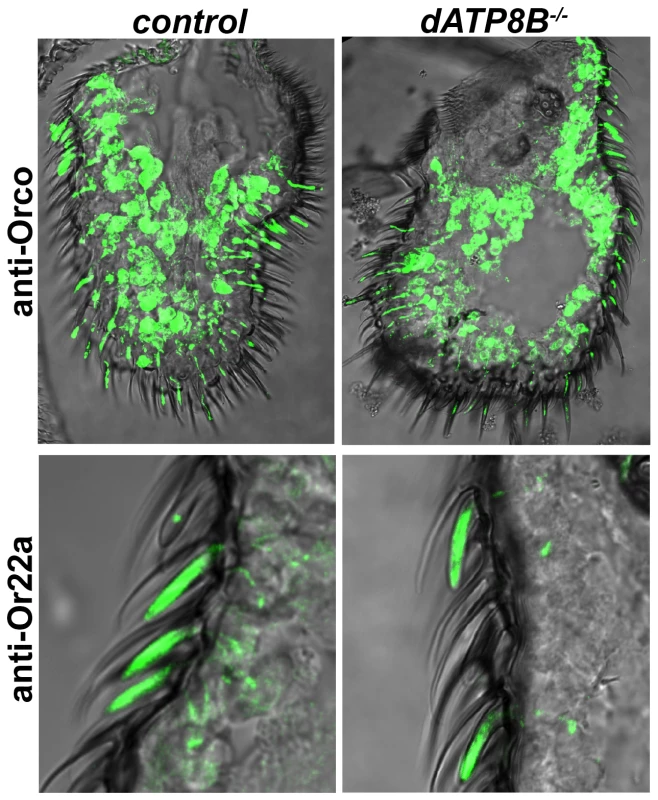
14 µm thick antennal sections from wild type flies (CS-5) were stained for anti-Orco or for anti-Or22a. No difference in either Orco or Or22a localisation to the outer dendrites was observed in dATP8B mutants (dATP8Bf05203) compared to control flies. Discussion
dATP8B is a member of the type 4 subfamily (P4-ATPases) of P-type ATPases. Unlike the other subfamilies, most of which encode ion transporters, the P4-ATPases are believed to share a distinct function as phospholipid translocases or “flippases” [26]. Eukaryotic plasma membranes have an asymmetrical distribution of phospholipids across the bilayer, with sphingolipids and phosphatidylcholine enriched in the exoplasmic leaflet, and more polar lipids such as phosphatidylserine and phosphatidylethanolamine enriched in the cytoplasmic leaflet. Phospholipid flippases contribute to asymmetry by “flipping” phospholipids from the exoplasmic to the cytoplasmic leaflet [28]. The physiological significance of this asymmetry is not well understood but it seems to be important for critical membrane processes such as vesicle trafficking and intracellular signalling [26], [29]. Disruption of the asymmetry may affect the conformation, membrane insertion, or trafficking, of integral membrane proteins. Alternatively it could affect lipid-signalling molecules, or membrane fluidity or bending. There are six members of the phospholipid flippase family in Drosophila and C.elegans and 14 in humans [26]. Genetic studies in C.elegans have suggested that the different members have different functions [30]. One of the six Drosophila flippases (CG33298) has been implicated in secretory vesicle formation and cholesterol homeostasis [31], the others have been uncharacterized to date.
dATP8B is the Drosophila homologue of four mammalian ATP8B proteins [26]. Of these only ATP8B1 has been substantially studied. Mutations in human ATP8B1 are associated with intrahepatic cholestasis [32] and also cause hearing loss [33]. The protein localizes to the apical plasma membrane of hepatocytes [32], [34], where it is thought to play a role in protection from the detergent effects of bile salts [35]. In hair cells, the sensory cells of the inner ear, ATP8B1 has been shown to localize to the stereocilia that transduce the mechanical vibrations in the cochlea [33]. ATP8B1 deficient mice exhibit a progressive degeneration of cochlea hair cells, possibly due to changes in the mechanical properties of stereocilia or the disruption of a Ca2+ transporter crucial for sensory transduction [33].
Here we have found that dATP8B is essential specifically for the responses of Or-expressing neurons, and not for IR or Gr-expressing neurons in general. Our expression studies suggest that dATP8B is not expressed in the latter, although further studies are needed to confirm the generality of this. Several lines of evidence suggest that mutation in dATP8B is not disrupting the development or morphology of the Or-expressing neurons and that its function is required in adult ORNs. First, in spite of the dramatic effects on sensitivity to odorants, the affected ORNs are still functional neurons, as evidenced by some response at high odorant doses, and have normal morphological appearance. Second, we found two examples where Gr or IR-expressing unaffected neurons (ac3A and ab1C) are housed in the same sensilla as affected Or-expressing neurons. This indicates that the defect is intrinsic to the Or-expressing neurons, and is not due to altered sensillum morphology or perireceptor processes.
Our data suggest that dATP8B is not required for the localization of the Or proteins to the dendrites, although we note that we have only examined Orco and one ligand-binding Or protein (Or22a) and thus cannot completely rule out effects on localisation of other Or proteins. Nonetheless, it seems most likely that dATP8B is necessary instead for the function of the Or receptor complex and for primary receptor signal transduction processes. This is further supported by our finding that dATP8B mutations differentially affect sensitivity of an individual receptor to different odorants. At present there is no biochemical evidence that dATP8B functions as a phospholipid flippase, however all members of this family of P4-ATPases for which biochemical assays have been performed do function to translocate phospholipids [26], [28], [29]. If dATP8B functions as expected there are several ways it could affect Or signal transduction. An altered phospholipid composition of the plasma membrane in dATP8B mutants may affect Or-Orco interactions, or interfere with binding of odorants to the receptor complex. Alternatively, reduced availability or activity of membrane-localised signalling molecules may affect Or signalling. For example, phosphatidylserine (PS) is required for activation of PKC once it translocates to the plasma membrane in response to increases in diacylglycerol [36]. In dATP8B mutants reduced availability of PS in the inner leaflet could thus lead to loss of PKC activation. PKC has functions in many signalling pathways, and has been suggested to be important for Orco activation [8]. Another possibility is that mutations in dATP8B affect the minor plasma membrane phospholipid phosphatidylinositol 4,5-bisphosphate (PIP2). PIP2 is found primarily in the cytoplasmic leaflet of the membrane and does not flip between leaflets, but levels could potentially be disrupted by flippase disfunction. PIP2 has many signalling roles. Its cleavage products inositol 1,4,5-trisphosphate and diacylglycerol are key components of G protein-activated signalling pathways. In addition, a number of families of ion channel and ion transporter proteins, for example the transient receptor potential channels, are dependent on PIP2 for their activation [37].
In conclusion, we have identified a new olfactory gene, dATP8B, which is specifically required for odour responses of Or-expressing, but not Gr or IR-expressing, sensory neurons in Drosophila. Given the very high level of homology of dATP8B to known phospholipid flippases, our findings suggest a specific role for cell membrane phospholipids in Or receptor signalling, as well as providing further evidence for fundamental differences between the signalling mechanisms of the different families of insect olfactory receptors. Further studies of this interesting gene may provide insight into the potentially complex mechanisms of Or signalling.
Materials and Methods
Drosophila stocks
Drosophila stocks were reared on yeasted semolina/syrup medium in 30 ml vials at 22°C under a natural daylight cycle. All crosses were performed at 22°C. Flies carrying the dATP8Bll2 mutation were part of a collection of mutagenized stocks obtained from Charles Zuker's laboratory [25]. The piggyBac insertion line dATP8Bf05203 (BL18847) and deficiency lines Df(3R)Exel7312 (BL7966) and Df(3R)Exel8155 (BL7967) were obtained from Bloomington Stock Center. The RNAi line for dATP8B was obtained from the Vienna Drosophila RNAi Center (v102648). The Orco-Gal4 line was obtained from Leslie Vosshall (Rockefeller University) and the Gr21a-Gal4 line from Kristin Scott at University of California Berkeley.
Electrophysiological recordings and data analysis
Recordings from whole olfactory organs
We recorded electrical signals from whole antennae (electroantennogram, EAG) and maxillary palps (electropalpogram, EPG) as described in Tom et al. 2011 [38]. A single fly was immobilized and a reference electrode inserted in the eye. For EAGs the recording electrode was placed on the surface of the antenna and for EPGs on the palp. Changes in voltage (mV) in response to 1 s stimulation with odorants were amplified using an active probe and a serial-IDAC amplifier (Syntech, Hilversum, the Netherlands). EAGs and EPGs represent the summed activity of a population of ORNs.
Recordings from single olfactory sensilla
Activity of individual olfactory receptor neurons was studied using the single sensillum recording (SSR) technique as described elsewhere [39]. Action potentials were recorded by a glass electrode inserted at the base of an olfactory sensillum and amplified via an IDAC-4 amplifier (Syntech). Action potential firing rates during 500 ms stimulations were analysed by subtracting firing rates during the 2 seconds before stimulation. Action potentials from the different neurons in a single sensillum were separated as in de Bruyne et al. [39]. A two-tailed Student's t-test with a Bonferroni correction for multiple comparisons was used to compare firing rates. Odor stimulation for SSR, EAG and EPG recordings was by injecting volatiles from 5 ml disposable syringes into an airstream blown over the preparation. All odorants were at highest available purity (>98%, Sigma-Aldrich) and dissolved in paraffin oil at different dilution from 0.0001 to 10% v/v. Because of its low volatility, the Drosophila pheromone cis-vaccenyl acetate (>98%, Cayman Chemicals) was dissolved in hexane and delivered from a pasteur pipette that was briefly heated prior to use. Male flies of age 3–7 days were used for all electrophysiological recordings, except for the RNAi experiment where newly emerged male flies were incubated at 25°C for exactly 7 days before recordings were performed.
Recordings from taste sensilla
Single sensillum tip recordings were performed from large (L-type) and intermediate (Ib-type) sensilla in the labellum as described earlier [20]. Male flies were aged 3–7 days and prepared for recordings by insertion of a glass micropipette reference electrode filled with Ringer's solution. Tastants were dissolved in 30 mM tricholine citrate, which was used as the electrolyte for the recording electrode. Action potentials obtained by using a TasteProbe and IDAC-4 amplifier (Syntech), were counted during the 500-ms period after initial contact with the stimulus solution in the recording electrode, and multiplied by two to obtain firing rates in spikes per second. A two-tailed Student's t-test was used to compare firing rates between mutant and wild-type flies. Sucrose and caffeine were purchased from Sigma Aldrich.
Genome sequencing and data analysis
Genomic DNA was extracted from adult heterozygous males using a QIAGEN Genomic-tip 20/G. A paired-end library with ∼300 bp insert size was prepared and sequenced by the Australian Genome Research Facility. In total ∼19 million 100 bp paired-end reads were generated using 0.5 lane on the Illumina HiSeq system. Sequencing reads were mapped to the Drosophila reference genome (Release 5 assembly) using BWA (Version 0.5.9) with default settings [40]. Integrative Genomics Viewer (Version 1.5.64) was used to visually inspect overall mapping quality of the candidate region [41]. After quality validation, consensus was generated using SAMtools (Version 0.1.13) with default settings [42]. Sequence variations were annotated using ANNOVAR's gene-based annotation option [43] with FlyBase Release 5.36 annotation [44]. SNPs from the Drosophila melanogaster Genetic Reference Panel [45] were used to filter naturally occurring variations. All computations were performed on the Monash Sun Grid. The nonsense mutation in dATP8B was verified with Sanger Sequencing using an independently prepared genomic DNA sample. The following primers were used to amplify a 655 bp region flanking the mutation site: forward primer 5′CATACGCATCCTTAACAGCC3′, reverse primer 5′ACCCAACAAATCCGATGACC3′.
Antibody production
cDNAs encoding six different regions of the CG14741-PB isoform (PE1, a.a. 2–236; PE2, 261–450; PE3, 527–630; PE4, 655–1115; PE5, 967–1359; PE6, 1562–1726) that are not part of predicted transmembrane domains were cloned into a pET100/D-TOPO vector (Invitrogen, Carlsbad, CA) such that the expressed peptide was tagged N-terminally with 6×His. The 6×His:PE1-6 peptides were expressed in E. coli and purified using a Ni-NTA column (Invitrogen). Peptides PE3, 5, and 6 were soluble and were individually injected into two guinea pigs by Cocalico Biologicals (Reamstown, PA). The obtained antisera (90 day protocol) were screened for antibodies against dATP8B using Western blots of both fly head protein extracts and the bacterially expressed peptides. The antisera obtained from injecting PE5 and PE6 showed positive signals and PE6 was used for immunostaining.
Immunohistochemistry
Antibodies and dilutions used were as follows: guinea pig anti-dATP8B (1∶10,000); rabbit anti-Orco (1∶5,000; Vosshall lab); rabbit anti-Or22a (1∶1,000; Vosshall lab); rabbit anti-GFP (1∶1,000; Life Technologies). Secondary antibodies raised in mouse, rabbit and guinea pig were Alexa-conjugated (Alexa Fluro 488 at 1∶250–500, Alexa Fluro 568 at 1∶500) (Molecular Probes). 14 µM cryo-sectioned adult heads were mounted on SuperFrost Plus slides (Thermo Scientific), dried for up to 3 hours, and then fixed in 4% paraformaldehyde/PBS for 30 mins at room temperature. Samples were washed for 10 minutes three times with PBST (PBS, 0.3% Triton-X-100), incubated in Block (5% normal goat serum in PBST) for 2 hours at room temperature and then incubated with the primary antibodies diluted in Block overnight at 4°C. After three 10 minutes washes with PBST, samples were incubated with secondary antibodies diluted in Block for 2 hours at room temperature. Sections were washed for 10 minutes three times with PBST before being mounted in Vectashield (Vector Labs). Samples were viewed and images acquired using a Nikon C1 confocal microscope.
Supporting Information
Zdroje
1. BentonR, SachseS, MichnickSW, VosshallLB (2006) Atypical membrane topology and heteromeric function of Drosophila odorant receptors in vivo. PLoS Biol 4 (2) e20.
2. SmartR, KielyA, BealeM, VargasE, CarraherC, et al. (2008) Drosophila odorant receptors are novel seven transmembrane domain proteins that can signal independently of heterotrimeric G proteins. Insect Biochem Mol Biol 38 : 770–780.
3. SatoK, PellegrinoM, NakagawaT, NakagawaT, VosshallLB, et al. (2008) Insect olfactory receptors are heteromeric ligand-gated ion channels. Nature 452 : 1002–1006.
4. WicherD, SchäferR, BauernfeindR, StensmyrMC, HellerR, et al. (2008) Drosophila odorant receptors are both ligand-gated and cyclic-nucleotide-activated cation channels. Nature 452 : 1007–1011.
5. LarssonMC, DomingosAI, JonesWD, ChiappeE, AmreinH, et al. (2004) Or83b encodes a broadly expressed odorant receptor essential for Drosophila olfaction. Neuron 43 : 703–714.
6. JonesPL, PaskGM, RinkerDC, ZwiebelLJ (2011) Functional agonism of insect odorant receptor ion channels. Proc Natl Acad Sci USA 108 : 8821–8825.
7. NakagawaT, PellegrinoM, SatoK, VosshallLB, TouharaK (2012) Amino acid residues contributing to function of the heteromeric insect olfactory receptor complex. PLoS ONE 7: e32372.
8. StenglM, FunkNW (2013) The role of the coreceptor Orco in insect olfactory transduction. J Comp Physiol A 199 : 897–909.
9. SargsyanV, GetahunMN, LlanosSL, OlssonSB, HanssonBS, et al. (2011) Phosphorylation via PKC regulates the function of the Drosophila odorant co-receptor. Front Cell Neurosci 5 : 5.
10. KainP, ChakrabortyTS, SundaramS, SiddiqiO, RodriguesV, et al. (2008) Reduced odor responses from antennal neurons of Gqα, phospholipase Cβ, and rdgA mutants in Drosophila support a role for a phospholipid intermediate in insect olfactory transduction. J Neurosci 28 : 4745–55.
11. YaoCA, CarlsonJR (2010) Role of G-proteins in odor-sensing and CO2-sensing neurons in Drosophila. J Neurosci 30 : 4562–72.
12. DengY, ZhangW, FarhatK, OberlandS, GisselmannG, et al. (2011) The stimulatory Gαs protein is involved in olfactory signal transduction in Drosophila. PLoS One 6 (4) e18605.
13. GetahunMN, OlssonSB, Lavista-LlanosS, HanssonBS, WicherD (2013) Insect odorant response sensitivity is tuned by metabotropically autoregulated olfactory receptors. PLoS One 8 (3) e58889.
14. BentonR, VanniceKS, Gomez-DiazC, VosshallLB (2009) Variant ionotropic glutamate receptors as chemosensory receptors in Drosophila. Cell 136 : 149–162.
15. AbuinL, BargetonB, UlbrichMH, IsacoffEY, KellenbergerS, et al. (2011) Functional architecture of olfactory ionotropic glutamate receptors. Neuron 69 : 44–60.
16. AiM, BlaisS, ParkJ-Y, MinS, NeubertTA, et al. (2011) Ionotropic glutamate receptors IR64a and IR8a form a functional odorant receptor complex in vivo in Drosophila. J Neurosci 33 : 10741–10749.
17. ClynePJ, WarrCG, CarlsonJR (2000) Candidate taste receptors in Drosophila. Science 287 : 1830–1834.
18. RobertsonHM, WarrCG, CarlsonJR (2003) Molecular evolution of the insect chemoreceptor gene superfamily in Drosophila melanogaster. Proc Natl Acad Sci USA 100 (Suppl 2): 14537–14542.
19. HallemEA, CarlsonJR (2006) Coding of odors by a receptor repertoire. Cell 125 : 143–160.
20. SilberingAF, RytzR, GrosjeanY, AbuinL, RamdyaP, et al. (2011) Complementary function and integrated wiring of the evolutionarily distinct Drosophila olfactory subsystems. J Neurosci 31 : 13357–13375.
21. JonesWD, CayirliogluP, KadowIG, VosshallLB (2007) Two chemosensory receptors together mediate carbon dioxide detection in Drosophila. Nature 445 : 86–90.
22. KwonJY, DahanukarA, WeissLA, CarlsonJR (2007) The molecular basis of CO2 reception in Drosophila. Proc Natl Acad Sci USA 104 : 3574–3578.
23. MontellC (2009) A taste of the Drosophila gustatory receptors. Curr Opin Neurobiol 19 : 345–353.
24. WeissLA, DahanukarA, KwonJY, BanerjeeD, CarlsonJR (2011) The molecular and cellular basis of bitter taste in Drosophila. Neuron 69 : 258–272.
25. KoundakjianEJ, CowanDM, HardyRW, BeckerAH (2004) The Zuker collection: a resource for the analysis of autosomal gene function in Drosophila melanogaster. Genetics 167 : 203–206.
26. TanakaK, Fujimura-KamadaK, YamamotoT (2011) Functions of phospholipid flippases. J Biochem 149 : 131–143.
27. AnholtRRH, WilliamsTI (2010) The soluble proteome of the Drosophila antenna. Chem Senses 35 : 21–30.
28. Van der VeldenLM, Van de GraafSFJ, KlompLWJ (2010) Biochemical and cellular functions of P4 ATPases. Biochem J 431 : 1–11.
29. PaulusmaCC, Oude ElferinkRPJ (2010) P4 ATPases – The physiological relevance of lipid flipping transporters. FEBS Letters 584 : 2708–2716.
30. LyssenkoNN, MitevaY, GilroyS, Hanna-RoseW, SchlegelRA (2008) An unexpectedly high degree of specialization and a widespread involvement in sterol metabolism among the C. elegans putative aminophospholipid translocases. BMC Dev Biol 8 : 96.
31. MaZ, LiuZ, HuangX (2012) Membrane phospholipid asymmetry counters the adverse effects of sterol overloading in the Golgi membrane of Drosophila. Genetics 190 : 1299–1308.
32. UjhazyP, OrtizD, MisraS, LiS, MoseleyJ, et al. (2001) Familial intrahepatic cholestasis 1: studies of localization and function. Hepatology 34 : 768–775.
33. StapelbroekJM, PetersTA, van BeurdenDHA, CurfsJHAJ, JoostenA, et al. (2009) ATP8B1 is essential for maintaining normal hearing. Proc Natl Acad Sci USA 106 : 9709–9714.
34. CaiSY, GautamS, NguyenT, SorokaCJ, RahnerC, et al. (2009) ATP8B1 deficiency disrupts the bile canalicular membrane bilayer structure in hepatocytes, but FXR expression and activity are maintained. Gastroenterology 136 : 1060–1069.
35. FolmerDE, van der MarkVA, Ho-MokKS, Oude ElferinkRPJ, PaulusmaCC (2009) Differential effects of progressive familial intrahepatic cholestasis type 1 and benign recurrent intrahepatic cholestasis type 1 mutations on canalicular localization of ATP8B1. Hepatology 50 : 1597–1605.
36. OrrJW, NewtonAC (1992) Interaction of Protein Kinase C with Phosphatidylserine. 2. Specificity and Regulation. Biochemistry 31 : 4667–73.
37. SuhB-C, HilleB (2008) PIP2 is a necessary factor for ion channel function: How and why? Annu Rev Biophys 37 : 175–95.
38. TomW, de BruyneM, HaehnelM, CarlsonJR, RayA (2011) Disruption of olfactory receptor neuron patterning in Scutoid mutant Drosophila. Mol Cell Neurosci 46 : 252–261.
39. de BruyneM, ClynePJ, CarlsonJR (1999) Odor coding in a model olfactory organ: the Drosophila maxillary palp. J Neurosci 19 : 4520–4532.
40. LiH, DurbinR (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25 : 1754–1760.
41. RobinsonJT, ThorvaldsdóttirH, WincklerW, GuttmanM, LanderES, et al. (2011) Integrative genomics viewer. Nat Biotechnol 29 : 24–26.
42. LiH, HandsakerB, WysokerA, FennellT, RuanJ, et al. (2009) The Sequence Alignment/Map format and SAMtools. Bioinformatics 25 : 2078–2079.
43. WangK, LiM, HakonarsonH (2010) ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 38: e164.
44. TweedieS, AshburnerM, FallsK, LeylandP, McQuiltonP, et al. (2009) FlyBase: enhancing Drosophila Gene Ontology annotations. Nucleic Acids Res 37: D555–9.
45. MackayTFC, RichardsS, StoneEA, BarbadillaA, AyrolesJF, et al. (2012) The Drosophila melanogaster Genetic Reference Panel. Nature 482 : 173–178.
Štítky
Genetika Reprodukční medicína
Článek Sex Chromosome Turnover Contributes to Genomic Divergence between Incipient Stickleback SpeciesČlánek Final Pre-40S Maturation Depends on the Functional Integrity of the 60S Subunit Ribosomal Protein L3
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2014 Číslo 3
-
Všechny články tohoto čísla
- , a Gene That Influences the Anterior Chamber Depth, Is Associated with Primary Angle Closure Glaucoma
- Genomic View of Bipolar Disorder Revealed by Whole Genome Sequencing in a Genetic Isolate
- The Rate of Nonallelic Homologous Recombination in Males Is Highly Variable, Correlated between Monozygotic Twins and Independent of Age
- Genetic Determinants Influencing Human Serum Metabolome among African Americans
- Heterozygous and Inherited Mutations in the Smooth Muscle Actin () Gene Underlie Megacystis-Microcolon-Intestinal Hypoperistalsis Syndrome
- Genome-Wide Meta-Analysis of Homocysteine and Methionine Metabolism Identifies Five One Carbon Metabolism Loci and a Novel Association of with Ischemic Stroke
- Cancer Evolution Is Associated with Pervasive Positive Selection on Globally Expressed Genes
- Genetic Diversity in the Interference Selection Limit
- Integrating Multiple Genomic Data to Predict Disease-Causing Nonsynonymous Single Nucleotide Variants in Exome Sequencing Studies
- An Evolutionary Analysis of Antigen Processing and Presentation across Different Timescales Reveals Pervasive Selection
- Cleavage Factor I Links Transcription Termination to DNA Damage Response and Genome Integrity Maintenance in
- DNA Dynamics during Early Double-Strand Break Processing Revealed by Non-Intrusive Imaging of Living Cells
- Genetic Basis of Metabolome Variation in Yeast
- Modeling 3D Facial Shape from DNA
- Dysregulated Estrogen Receptor Signaling in the Hypothalamic-Pituitary-Ovarian Axis Leads to Ovarian Epithelial Tumorigenesis in Mice
- Photoactivated UVR8-COP1 Module Determines Photomorphogenic UV-B Signaling Output in
- Local Evolution of Seed Flotation in Arabidopsis
- Chromatin Targeting Signals, Nucleosome Positioning Mechanism and Non-Coding RNA-Mediated Regulation of the Chromatin Remodeling Complex NoRC
- Nucleosome Acidic Patch Promotes RNF168- and RING1B/BMI1-Dependent H2AX and H2A Ubiquitination and DNA Damage Signaling
- The -Induced Arabidopsis Transcription Factor Attenuates ABA Signaling and Renders Seedlings Sugar Insensitive when Present in the Nucleus
- Changes in Colorectal Carcinoma Genomes under Anti-EGFR Therapy Identified by Whole-Genome Plasma DNA Sequencing
- Selection of Orphan Rhs Toxin Expression in Evolved Serovar Typhimurium
- FAK Acts as a Suppressor of RTK-MAP Kinase Signalling in Epithelia and Human Cancer Cells
- Asymmetry in Family History Implicates Nonstandard Genetic Mechanisms: Application to the Genetics of Breast Cancer
- Co-translational Localization of an LTR-Retrotransposon RNA to the Endoplasmic Reticulum Nucleates Virus-Like Particle Assembly Sites
- Sex Chromosome Turnover Contributes to Genomic Divergence between Incipient Stickleback Species
- DWARF TILLER1, a WUSCHEL-Related Homeobox Transcription Factor, Is Required for Tiller Growth in Rice
- Functional Organization of a Multimodular Bacterial Chemosensory Apparatus
- Genome-Wide Analysis of SREBP1 Activity around the Clock Reveals Its Combined Dependency on Nutrient and Circadian Signals
- The Yeast Sks1p Kinase Signaling Network Regulates Pseudohyphal Growth and Glucose Response
- An Interspecific Fungal Hybrid Reveals Cross-Kingdom Rules for Allopolyploid Gene Expression Patterns
- Temperate Phages Acquire DNA from Defective Prophages by Relaxed Homologous Recombination: The Role of Rad52-Like Recombinases
- Dying Cells Protect Survivors from Radiation-Induced Cell Death in
- Determinants beyond Both Complementarity and Cleavage Govern MicroR159 Efficacy in
- The bHLH Factors Extramacrochaetae and Daughterless Control Cell Cycle in Imaginal Discs through the Transcriptional Regulation of the Phosphatase
- The First Steps of Adaptation of to the Gut Are Dominated by Soft Sweeps
- Bacterial Regulon Evolution: Distinct Responses and Roles for the Identical OmpR Proteins of Typhimurium and in the Acid Stress Response
- Final Pre-40S Maturation Depends on the Functional Integrity of the 60S Subunit Ribosomal Protein L3
- Mitogen-Activated Protein Kinase (MAPK) Pathway Regulates Branching by Remodeling Epithelial Cell Adhesion
- CDP-Diacylglycerol Synthetase Coordinates Cell Growth and Fat Storage through Phosphatidylinositol Metabolism and the Insulin Pathway
- Coronary Heart Disease-Associated Variation in Disrupts a miR-224 Binding Site and miRNA-Mediated Regulation
- TBX3 Regulates Splicing : A Novel Molecular Mechanism for Ulnar-Mammary Syndrome
- Identification of Interphase Functions for the NIMA Kinase Involving Microtubules and the ESCRT Pathway
- Is a Cancer-Specific Fusion Gene Recurrent in High-Grade Serous Ovarian Carcinoma
- LILRB2 Interaction with HLA Class I Correlates with Control of HIV-1 Infection
- Worldwide Patterns of Ancestry, Divergence, and Admixture in Domesticated Cattle
- Parent-of-Origin Effects Implicate Epigenetic Regulation of Experimental Autoimmune Encephalomyelitis and Identify Imprinted as a Novel Risk Gene
- The Phospholipid Flippase dATP8B Is Required for Odorant Receptor Function
- Noise Genetics: Inferring Protein Function by Correlating Phenotype with Protein Levels and Localization in Individual Human Cells
- DUF1220 Dosage Is Linearly Associated with Increasing Severity of the Three Primary Symptoms of Autism
- Sugar and Chromosome Stability: Clastogenic Effects of Sugars in Vitamin B6-Deficient Cells
- Pheromone-Sensing Neurons Expressing the Ion Channel Subunit Stimulate Male Courtship and Female Receptivity
- Gene-Based Sequencing Identifies Lipid-Influencing Variants with Ethnicity-Specific Effects in African Americans
- Telomere Shortening Unrelated to Smoking, Body Weight, Physical Activity, and Alcohol Intake: 4,576 General Population Individuals with Repeat Measurements 10 Years Apart
- A Combination of Activation and Repression by a Colinear Hox Code Controls Forelimb-Restricted Expression of and Reveals Hox Protein Specificity
- An ER Complex of ODR-4 and ODR-8/Ufm1 Specific Protease 2 Promotes GPCR Maturation by a Ufm1-Independent Mechanism
- Epigenetic Control of Effector Gene Expression in the Plant Pathogenic Fungus
- Genetic Dissection of Photoreceptor Subtype Specification by the Zinc Finger Proteins Elbow and No ocelli
- Validation and Genotyping of Multiple Human Polymorphic Inversions Mediated by Inverted Repeats Reveals a High Degree of Recurrence
- CYP6 P450 Enzymes and Duplication Produce Extreme and Multiple Insecticide Resistance in the Malaria Mosquito
- GC-Rich DNA Elements Enable Replication Origin Activity in the Methylotrophic Yeast
- An Epigenetic Signature in Peripheral Blood Associated with the Haplotype on 17q21.31, a Risk Factor for Neurodegenerative Tauopathy
- Lsd1 Restricts the Number of Germline Stem Cells by Regulating Multiple Targets in Escort Cells
- RBPJ, the Major Transcriptional Effector of Notch Signaling, Remains Associated with Chromatin throughout Mitosis, Suggesting a Role in Mitotic Bookmarking
- The Membrane-Associated Transcription Factor NAC089 Controls ER-Stress-Induced Programmed Cell Death in Plants
- The Functional Consequences of Variation in Transcription Factor Binding
- Comparative Genomic Analysis of N-Fixing and Non-N-Fixing spp.: Organization, Evolution and Expression of the Nitrogen Fixation Genes
- An Insulin-to-Insulin Regulatory Network Orchestrates Phenotypic Specificity in Development and Physiology
- Suicidal Autointegration of and Transposons in Eukaryotic Cells
- A Multi-Trait, Meta-analysis for Detecting Pleiotropic Polymorphisms for Stature, Fatness and Reproduction in Beef Cattle
- Genome-Wide DNA Methylation Analysis Predicts an Epigenetic Switch for GATA Factor Expression in Endometriosis
- Genome-Wide DNA Methylation Analysis of Human Pancreatic Islets from Type 2 Diabetic and Non-Diabetic Donors Identifies Candidate Genes That Influence Insulin Secretion
- The and Hybrid Incompatibility Genes Suppress a Broad Range of Heterochromatic Repeats
- The Kil Peptide of Bacteriophage λ Blocks Cytokinesis via ZipA-Dependent Inhibition of FtsZ Assembly
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Worldwide Patterns of Ancestry, Divergence, and Admixture in Domesticated Cattle
- Genome-Wide DNA Methylation Analysis of Human Pancreatic Islets from Type 2 Diabetic and Non-Diabetic Donors Identifies Candidate Genes That Influence Insulin Secretion
- Genetic Dissection of Photoreceptor Subtype Specification by the Zinc Finger Proteins Elbow and No ocelli
- GC-Rich DNA Elements Enable Replication Origin Activity in the Methylotrophic Yeast
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



