A Combination of Activation and Repression by a Colinear Hox Code Controls Forelimb-Restricted Expression of and Reveals Hox Protein Specificity
The acquisition of limbs during vertebrate evolution was a very successful innovation that enabled this group of species to diversify and colonise land. It has become clear recently that the primary driver behind the evolution of new structures, such as limbs, is the acquisition of novel regulatory elements that control when and where genes are activated rather than the proteins encoded by the genes themselves acquiring novel functions. We have identified the regulatory element from a gene, Tbx5. Activation of Tbx5 in the forelimb-forming region of the developing embryos is essential for forelimbs to form and disruption of human TBX5 causes limb abnormalities. We show that activation of Tbx5 in a restricted territory is achieved through a combination of activation inputs that are present broadly throughout the embryo flank and dominant, repressive inputs present only in more caudal regions of the flank. The sum of these inputs yields restricted activation in the rostral, forelimb-forming flank. Our results explain how the regulatory switches that were harnessed for the acquisition of limbs during evolution operate and how they can be turned off during the evolution of limblessness in species such as the snake.
Published in the journal:
. PLoS Genet 10(3): e32767. doi:10.1371/journal.pgen.1004245
Category:
Research Article
doi:
https://doi.org/10.1371/journal.pgen.1004245
Summary
The acquisition of limbs during vertebrate evolution was a very successful innovation that enabled this group of species to diversify and colonise land. It has become clear recently that the primary driver behind the evolution of new structures, such as limbs, is the acquisition of novel regulatory elements that control when and where genes are activated rather than the proteins encoded by the genes themselves acquiring novel functions. We have identified the regulatory element from a gene, Tbx5. Activation of Tbx5 in the forelimb-forming region of the developing embryos is essential for forelimbs to form and disruption of human TBX5 causes limb abnormalities. We show that activation of Tbx5 in a restricted territory is achieved through a combination of activation inputs that are present broadly throughout the embryo flank and dominant, repressive inputs present only in more caudal regions of the flank. The sum of these inputs yields restricted activation in the rostral, forelimb-forming flank. Our results explain how the regulatory switches that were harnessed for the acquisition of limbs during evolution operate and how they can be turned off during the evolution of limblessness in species such as the snake.
Introduction
Forelimbs and hindlimbs are derivatives of the lateral plate mesoderm (LPM) that arise at fixed positions along the vertebrate body axis. Limb formation is initiated by limb induction signals from axial tissues [1]. The presumptive limb-forming regions initially express two T-box genes prior to overt limb bud formation, Tbx5 in nascent forelimbs and Tbx4 in hindlimbs [2]–[5]. Genetic studies in the mouse have shown that both genes are crucial for normal limb outgrowth by activating Fgf10 in the limb mesenchyme [6]–[8]. Fgf10 subsequently induces Fgf8 expression in the apical ectodermal ridge (AER) and Fgf8 produced from the AER, in turn, maintains Fgf10 expression in mesenchyme to establish a positive feedback loop of Fgf signalling that maintains limb growth. Mutations in human TBX5 cause Holt-Oram Syndrome (HOS OMIM142900), a disorder characterised by upper limb and heart abnormalities [9], [10] and mutations in TBX4 cause Small Patella Syndrome (SPS OMIM 147891), a disorder characterised by knee, pelvis and toe defects [11]. Tbx5 is the earliest marker of presumptive forelimb mesenchyme and because activation of this factor within a defined region of the LPM ultimately dictates the position at which the forelimbs will arise, identifying the factors that control activation of this Tbx5 expression domain will reveal the mechanisms employed that allowed the acquisition of limbs in vertebrates and that dictate forelimb position in the embryo.
Tbx5 is initially expressed in the forelimb-forming region of LPM prior to the emergence of a bud and it is subsequently restricted to the forelimb region as development proceeds. Tbx5 is essential for forelimb formation and this exclusive requirement is limited to a short time window when limb bud initiation occurs [12]. Tbx4, the paralog of Tbx5, is able to rescue forelimb formation following conditional deletion of Tbx5 [13]. Furthermore, the ancestral Tbx4/5 gene represented by AmphiTbx4/5 of the limbless cephalochordate, amphioxus, can fully compensate for the loss of Tbx5 in the mouse [14]. This indicates the ancestral protein from a limbless organism has limb-inducing potential and supports a model in which evolution of a regulatory element sufficient to activate Tbx5 expression in the LPM was a critical step in the acquisition of limbs during vertebrate evolution.
Hox genes are conserved homeodomain-containing transcription factors that are arranged in clusters in the genome. The chromosomal organization of the genes in the complex reflects their expression pattern along the rostro-caudal body axis to determine positional identity [15], [16]. As relative positions of limbs, axial vertebrae and Hox expression domains are conserved among vertebrates in spite of the variable numbers of each type of vertebrae (e.g. cervical, thoracic, lumbar and sacral vertebrae), Hox genes have been good candidates as determinants of limb position [17], [18]. Despite the unquestionable importance of Hox genes in patterning the developing embryo, very little is known about their direct targets and mechanisms of action.
We have previously identified a Tbx5 regulatory element sufficient for early forelimb expression [19]. This element contains Hox binding sites that are required for the enhancer activity, thus implicating Hox genes in direct, positive regulation of Tbx5. However, since the ability to activate Tbx5 is not strictly restricted to Hox genes expressed only at forelimb level, the mechanism by which a rostro-caudal Hox code establishes forelimb-restriction of Tbx5 remained unknown. Here, we demonstrate how Hox paralogous group members act cooperatively to restrict expression of Tbx5 in the LPM, which ultimately determines the positions the forelimbs will emerge from the flank of the embryo. We show that mutations of a single Hox binding site in the Tbx5 forelimb regulatory element cause expanded reporter gene expression in caudal LPM. Rostral restriction in Tbx5 expression through repression in the caudal LPM is mediated by Hoxc8/9/10 genes and this repressive function is limited to Hox genes that are expressed in Tbx5-negative caudal LPM. We further map the Hoxc9 protein domains required to confer transcriptional repression that distinguishes these paralogs from other Hox proteins expressed throughout the flank of the embryo. Our results demonstrate how a nested, combinatorial code of Hox protein transcriptional activation and repression along the rostro-caudal embryo axis restricts Tbx5 expression to the forelimb and ultimately determines forelimb position.
Results
Hox binding sites are required for forelimb-restricted Tbx5 expression
Previously, we identified a short regulatory element within intron 2 of the mouse Tbx5 gene that recapitulates the dramatic forelimb-restricted expression of this gene [19]. This 361 base pair (bp) sequence contains six Hox binding sites (Hbs) (Fig. 1A). To analyze which sites within this minimal element are required for Tbx5 expression, we generated a series of constructs in which each individual Hbs site1-6 is mutated and tested their ability to activate a LacZ reporter gene in transgenic mice. While the Tbx5 int2(361) reporter construct drove forelimb-restricted expression of LacZ (Fig. 1B, H and [19]), mutation of either individual Hbs1, or Hbs3, or Hbs4, or Hbs5, or Hbs6 resulted in reduced reporter gene expression (Fig. 1C–G). Interestingly, in most cases residual expression was consistently detected in the anterior forelimb bud, however, mutation of Hbs5 produced mosaic expression throughout the limb. The six bp sequence (TGAGAG, bottom strand) situated 3′ of Hbs2 (6bp3′) is similar but not identical to both Pbx (TGAT) and Meis (TGACAG) canonical binding sequences [20]. Pbx and Meis are Hox cofactors that can bind DNA as heterodimers. Mutation of Hbs2 and 6bp3′ in the 361 bp core fragment produced a strikingly different result. Reporter expression was detected in the forelimb but was now also expanded throughout the interlimb and hindlimb-forming region (Fig. 1I). Since the number of transgenic embryos that showed expression with this construct was low, to study the effect of mutating the individual sites further, we used a 565 bp fragment (Tbx5 int2(565)) (Fig. 1J) that contains an additional 204 bp sequence 5′ to the 361 bp core element. The extra 204 bp sequence contains three putative Hox binding sites. However, these sites are not required to control the spatial restriction of expression since the 361 bp fragment produces forelimb-restricted expression equivalent to that observed with the 565 bp fragment (Fig. 1K and [19]). We have also previously shown that the fragment containing this 204 bp sequence and Hbs1 and Hbs2 cannot activate reporter gene expression indicating these sites are not sufficient for the enhancer activity [19]. As observed with the smaller fragment, mutations of Hbs2 and 6bp3′ caused caudally expanded LacZ reporter activity to include the LPM of interlimb and hindlimb-forming regions, which never normally express Tbx5 (Fig. 1L). These results suggest that these sites are required to restrict Tbx5 expression to the forelimb-forming region. The activity of the Hbs2+6bp3′-mutated construct is dependent on the presence of the other Hox sites since mutation of Hbs2 and 6bp3′ together with Hbs3-6 did not drive reporter expression at all (n = 0/6, data not shown). To distinguish the requirement for Hbs2 and 6bp3′, we next mutated either of these sites (Fig. 1M–N). Mutation of 6bp3′ did not affect the expression domain of the LacZ reporter (Fig. 1M), while mutation of Hbs2 caused caudal expansion (Fig. 1N) equivalent to that seen after mutating both Hbs2 and 6bp3′ (Fig. 1L). These results suggest that Hbs2 plays the predominant role restricting Tbx5 expression to the forelimb-forming region.
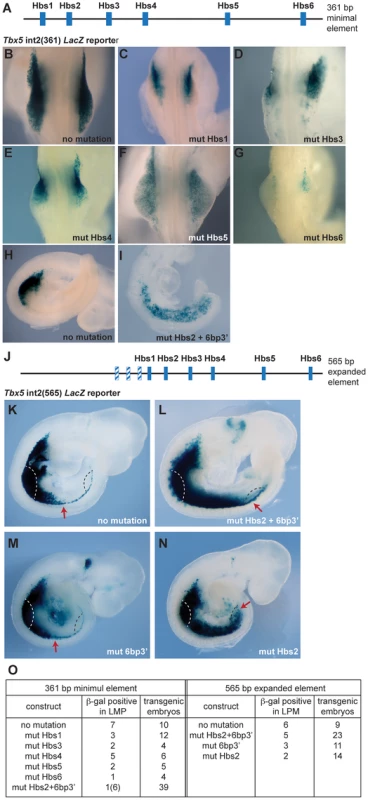
These results demonstrate that the binding sites within this regulatory element can be divided into 2 distinct functional groups. Hbs1 and 3-6 act as ‘on’ switches important for the amplitude of activation, whereas Hbs2 determines spatial resolution by hosting repressive complexes that restrict the domain of activation. This element can therefore have a binary function, serving as a site for the formation of transcriptional activation or repression complexes.
Hoxc8, Hoxc9 and Hoxc10 genes are expressed in Tbx5-negative, caudal lateral plate mesoderm
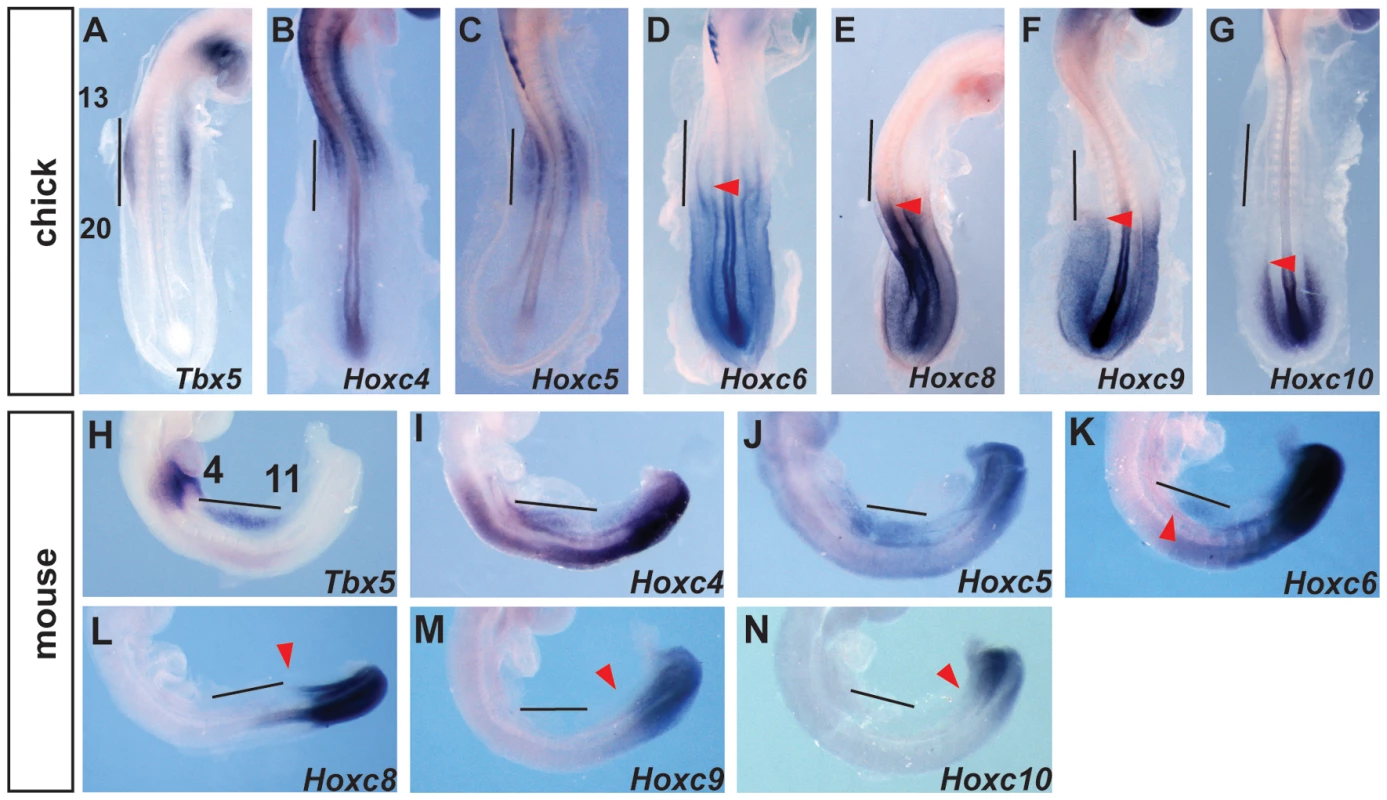
The presence of Hox binding sites in this element prompted us to search for candidate Hox genes that could be acting on this element as either positive or negative regulators of transcription. Previously, we have shown that PG 4 and 5 Hox genes can activate this regulatory element [19]. We now focused on Hox factors that could be mediating spatial resolution of this regulatory element by forming repressive complexes. We analysed the expression of Hox genes in chick and mouse embryos at stages when Tbx5 is first expressed in the forelimb-forming region. Tbx5 is first expressed at the level of somites 13–20 in chick and somites 4–11 in mouse embryos. As previously reported [19] the expression domains of Hox4 and Hox5 paralogs overlap with that of Tbx5 in both mouse and chick embryos (Fig. 2A–C, H–J). Since the expression patterns of HoxA, HoxB, HoxC and HoxD cluster genes are broadly similar, we show here the results of the HoxC cluster genes, as a representative example for simplicity. Hoxc6 is expressed within the caudal-most domain of Tbx5 as well as in more caudal LPM (Fig. 2D and K). Conversely, Hoxc8, Hoxc9 and Hoxc10 are exclusively expressed in caudal domains of the LPM that do not express Tbx5 (Fig. 2E–G and L–N) and are therefore candidates to repress Tbx5 expression.
Hoxc9 can repress Tbx5 expression via Hbs2
To determine whether caudally-expressed Hox genes can repress the Tbx5 forelimb-regulatory element, we compared the activities of Hoxc9 and Hoxc5 expression constructs when co-electroporated with the wild type Tbx5 int2(361) (Fig. 3A) LacZ reporter into the forelimb-forming region of HH stage 14–15 chick embryos. As expected, following electroporation of the Tbx5 int2(361) construct (with a dsRed reporter to assess electroporation efficiency (Fig. 3B)), β-gal activity is detected in successfully targeted forelimb LPM (Fig. 3B′) indicating that this mouse Tbx5 regulatory element can also function in chick. Following co-electroporation of a Hoxc9 expression construct with the Tbx5 int2(361) reporter, LacZ expression is repressed in the forelimb region (Fig. 3C′ white arrow). In contrast, performing the equivalent experiment with Hoxc5, which is expressed in the rostral, Tbx5-expressing LPM, does not negatively effect LacZ expression from the reporter (Fig. 3D′) demonstrating that the repressive activity is restricted to caudally-restricted Hox genes, such as Hoxc9.
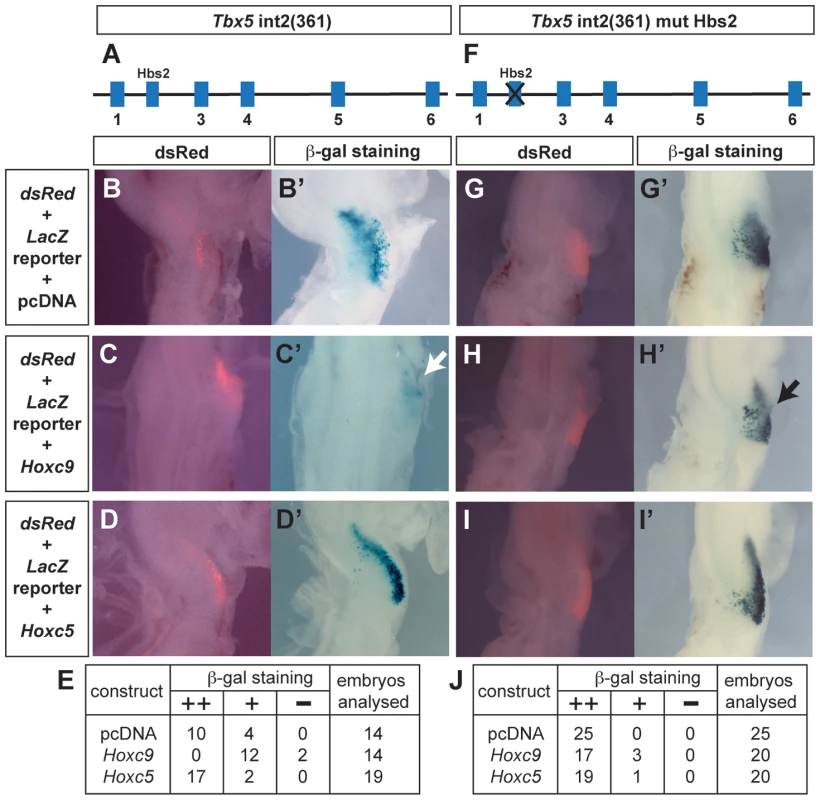
To determine whether Hoxc9 functions via Hbs2 to repress Tbx5 expression, we co-electroporated Hoxc9 with a Tbx5 int2(361) reporter in which Hbs2 is mutated (Fig. 3F). In transgenic mice, this mutation caused LacZ expression throughout the forelimb, interlimb and hindlimb regions (Fig. 1N) and electroporation of this reporter alone in the forelimb-forming region produced LacZ expression (Fig. 3G′) where cells have been successfully targeted as shown by the dsRed reporter (Fig. 3G). Co-electroporation of Hoxc9 with the Hbs2 mutated reporter did not repress LacZ expression (Fig. 3H′ black arrow). As expected, no effect was observed following co-electroporation with the Hoxc5 construct (Fig. 3I′). Since the expression of Hoxc8 and Hoxc10 are also restricted in caudal LPM, we tested if they can also repress the Tbx5 reporter activity similar to Hoxc9. Ectopic expression of either Hoxc8 (Fig. S1B′) or Hoxc10 (Fig. S1C′) reduced LacZ expression. Together, these results demonstrate that Hoxc8/9/10, which are normally expressed in the caudal LPM, have the ability to repress the Tbx5 regulatory element and that this repression is mediated via the Hbs2. In contrast, Hoxc5 does not exhibit equivalent repressive activity.
Ectopic Hoxc9 can repress Tbx5 expression
We next tested whether ectopic expression of Hoxc9 could repress endogenous Tbx5 expression in the forelimb-forming region. Electroporation of the right forelimb-forming region (Fig. 4A) with a Hoxc9 expression construct can repress the endogenous domain of Tbx5 (Fig. 4A′–A″). The electroporation protocol targets the proximal LPM most successfully and this is where the most profound repression of Tbx5 is observed consistent with Hoxc9 acting cell-autonomously.
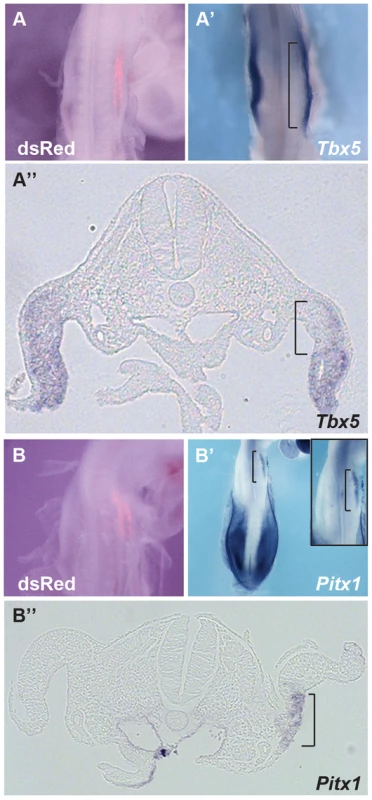
Although Tbx5 does not determine forelimb morphologies [13], its forelimb-restricted expression serves as a marker of forelimb identity. Since Hoxc9 is expressed in caudal LPM including the hindlimb region, we examined if, following ectopic activation of Hoxc9, hindlimb markers were activated in the forelimb region concomitant with down-regulation of Tbx5. Pitx1 is expressed in hindlimb, but not in forelimb, and determines some hindlimb morphologies [21]–[24]. Indeed, ectopic Pitx1 transcripts are detected in the forelimb (Fig. 4B′–B″) following electroporation of a Hoxc9 expression vector (Fig. 4B). The domain of ectopic Pitx1 is apparent in the proximal forelimb LPM consistent with the proximal bias in cells successfully targeted by electroporation and again consistent with a cell-autonomous mechanism of action.
Hox proteins can directly bind to Hbs2
To understand the molecular mechanisms of caudal Hox-specific repressive activity on Tbx5 expression, we compared the DNA binding abilities of Hoxc5 and Hoxc9 since paralogous-specific functions of Hox can be explained by different DNA binding specificities [25]. We performed electrophoretic mobility shift assays (EMSA) with an oligonucleotide probe that contains Hbs2 (Fig. 5E). in vitro translated Hoxc5 can bind to the probe (Fig. 5A lane 2). Addition of non-labelled oligo as a competitor abolished the DNA-protein complexes showing their specificity (Fig. 5A lane 3–4). Non-labelled oligo in which Hbs2 is mutated (mut Hbs2) did not affect the complexes, confirming that the protein occupies Hbs2 (Fig. 5A lane 5–6). Similar to Hoxc5, Hoxc9 makes a complex with this probe (Fig. 5B lane 2) and the specificity was confirmed by a competition assay (Fig. 5B lane 3–6). We then performed a super-shift assay using an antibody against a flag epitope present in the C- terminal of our recombinant Hox proteins (Fig. 5C). Addition of this antibody resulted in super-shifts of DNA-protein complexes (Fig. 5C lane 3 and 5), indicating these complexes contain Hoxc5 or Hoxc9 proteins. These results suggest that both Hoxc5 and Hoxc9 can bind Hbs2 in vitro.
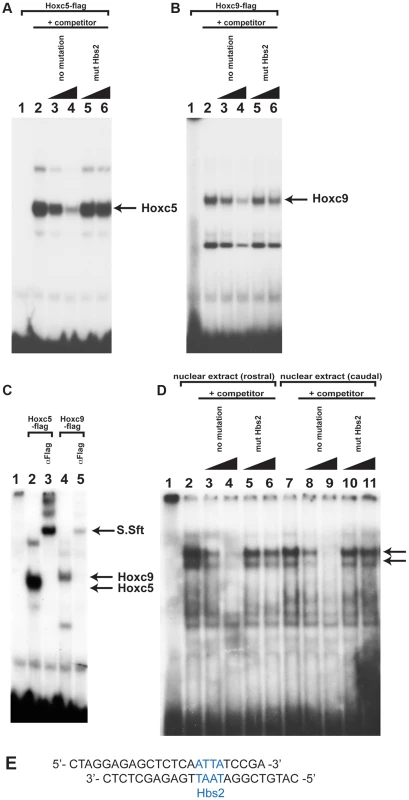
To examine whether the occupancy of Hbs2 in forelimb forming, Tbx5-positive LPM and Tbx5-negative caudal LPM is different, we carried out EMSA analysis using nuclear extracts from rostral or caudal LPM. We observed two bands of the same size using both rostral and caudal extracts (Fig. 5D lane 2 and 7 arrows). We confirmed the specificity of Hox binding by competition assay. While the no mutation oligo disrupts both of the two bands (Fig. 5D lane 3–4 and lane 8–9) the mut Hbs2 oligo can only very weakly compete the complexes (Fig. 5D lane 5–6 and lane 10–11), suggesting that Hbs2 is required for these DNA-protein complexes.
These results suggest that in vitro translated Hoxc9 and Hoxc5 can bind equivalently to Hbs2 and that the protein-DNA complexes from both rostral and caudal nuclear extract occupy Hbs2 specifically. Since the electroporation experiments demonstrate that the repression of the Tbx5 enhancer by Hoxc9 requires Hbs2 (Fig. 3), one of the Hox proteins forming a complex on Hbs2 using caudal nuclear extract as input is likely to be Hoxc9. In rostral LPM, since the repressive Hox genes, such as Hoxc8/9/10, are not expressed, the Hox proteins on Hbs2 using rostral nuclear extract as input are either activating Hox proteins, such as Hox PG4 and PG5 or Hox proteins with neutral function on Tbx5 expression. Thus, we propose a model in which HoxPG4 and PG5 protein complexes occupy Hbs2 in rostral forelimb forming LPM, while in Tbx5-negative caudal LPM the same site is occupied by Hoxc9 and/or Hoxc8/Hoxc10 containing-complexes that repress Tbx5 expression. Therefore, we conclude that a combination of restricted expression of Hox genes and the distinct activities of Hox proteins of different paralogous groups, which we demonstrate here, are harnessed to enable restricted expression of Tbx5 via the Hbs2.
The N-terminal region of Hoxc9 is sufficient to confer the ability to transcriptionally repress Tbx5
To further analyse the functional differences between Hoxc5 and Hoxc9, we generated chimeric forms of Hoxc5 and Hoxc9 proteins (Fig. 6A) and assayed their ability to repress the Tbx5 intron2 reporter construct (Fig. 6B–I). In both Hoxc5 and Hoxc9 the homeodomain is located in the C-terminus of the proteins. Paralog-specific DNA-binding properties have been reported to be determined by a specificity module spanning a Pbx-binding hexapeptide motif (W) present N-terminal to the homeodomain and the N-terminal arm of the homeodomain (NHD) [26] Fig. 6A). As would be predicted, a construct containing only the C-terminal half of Hoxc5 (Hoxc5C) cannot repress reporter gene expression (Fig. 6B–B′). Strikingly, addition of the N-terminal domain of Hoxc9 (N1N2) to the C-terminal half of Hoxc5 converts Hoxc5C into a chimeric protein (Hoxc9N5C) with Hoxc9-like repressor activity (Fig. 6C–C′). This supports our model that the opposing transcriptional activities of Hoxc5 and Hoxc9 do not lie in their distinct ability to bind Hox binding sites. To attempt to further refine the domain(s) responsible for transcriptional repression of Tbx5, we divided the N-terminus of Hoxc9 into two smaller domains, Hoxc9N1 and Hoxc9N2, and tested their function. Neither chimeric protein (Hoxc9N15C or Hoxc9N25C) showed clear repression of the reporter demonstrating that within the limits of this assay the entire N-terminus or the domain overlapping the junction between N1 and N2 is required for repressive activity (Fig. 6D–E).
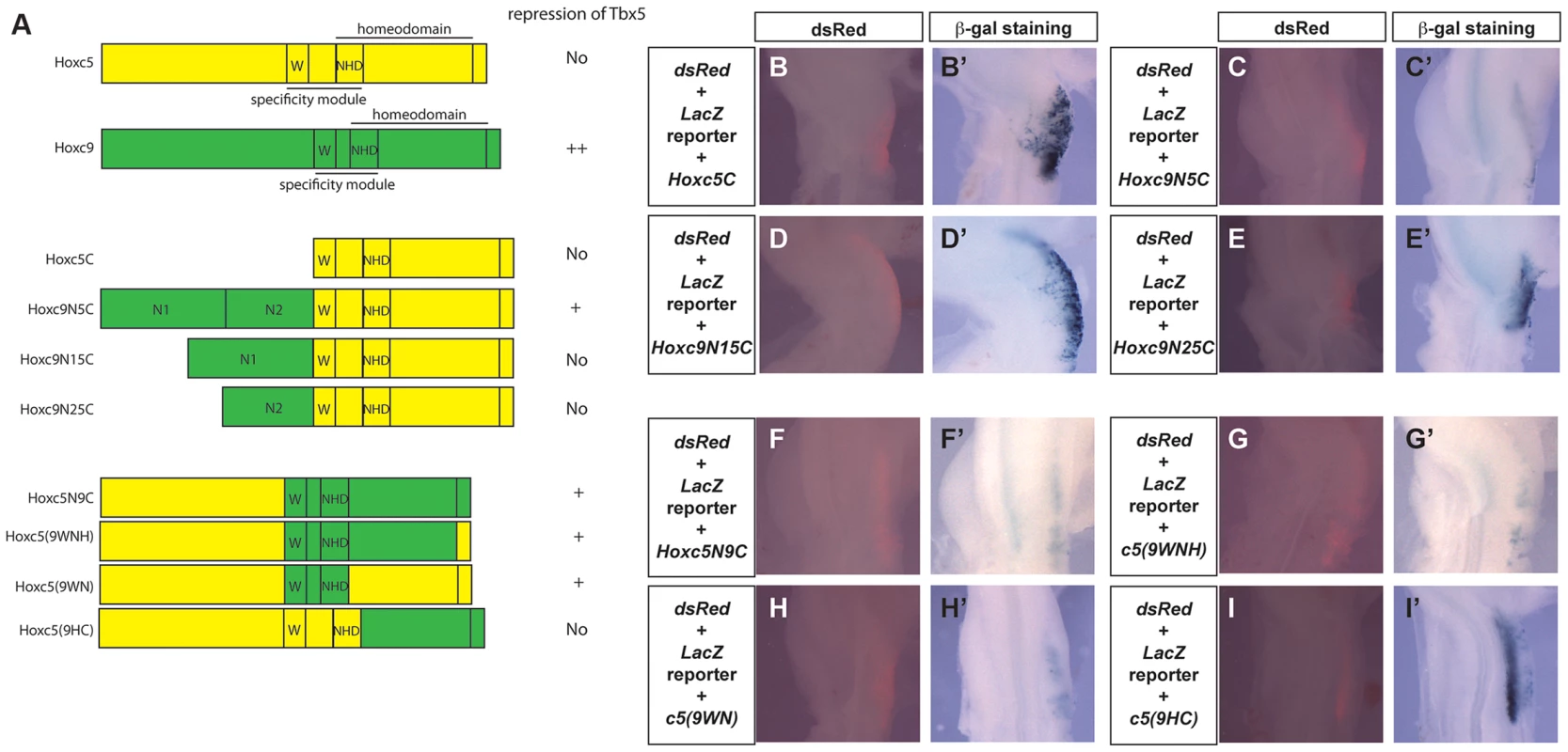
The specificity module of Hoxc9 can change the transcriptional properties of Hoxc5
Although Hoxc9N5C reduced a reporter gene expression, this repression was weaker than that seen with full length Hoxc9. We, therefore, examined if there are other domains in the C-terminal half of Hoxc9 that can contribute to transcriptional repression. A chimeric protein that contains the N-terminal half of Hoxc5 and C-terminal half of Hoxc9 (Hoxc5N9C) can reduce LacZ expression (Fig. 6F–F′), suggesting that there is an additional repression domain(s) in the C-terminal region of Hoxc9. Replacement of a short C-terminal tail (Hoxc5(9WNH)) with equivalent regions of Hoxc5 did not affect its ability to repress the reporter (Fig. 6G–G′). Strikingly insertion of 18 amino acids spanning the hexapeptides and the homeodomain N-terminal arm from Hoxc9 (Hoxc5(9WN)) is sufficient to convert Hoxc5 to a transcriptional repressor (Fig. 6H–H′). To further test the requirement of these domains, we generated another chimeric protein in which all of the regions upstream from homeodomain N-terminal arm were replaced (Hoxc5(9HC)). This protein did not suppress LacZ expression (Fig. 6I–I′). To confirm that the loss of repressive activity is not caused by the disruption of the 3D-structure of the chimeric protein, we performed EMSA to demonstrate that this protein (Hoxc5(9HC) and the chimeric proteins Hoxc5N9C and Hoxc5(9WNH) can all bind a DNA probe containing Hbs2 (data not shown). These results suggest that repression of Tbx5 by Hoxc9 is mediated by two domains: one N-terminal and the other in the specificity module that contains Pbx-binding hexapeptides and the N-terminal arm of the homeodomain.
Discussion
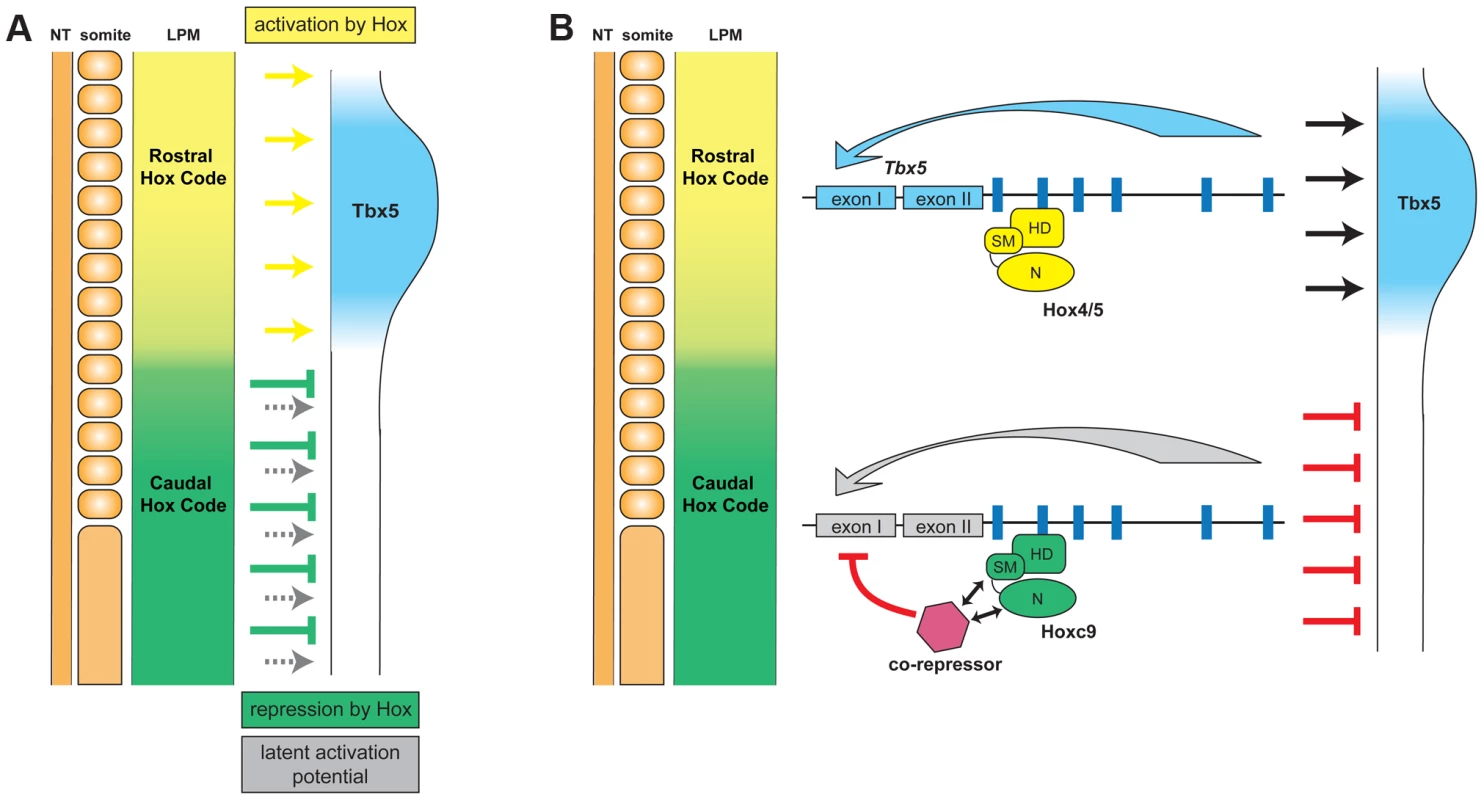
Using the forelimb regulatory element of Tbx5 as an assay, we have been able to distinguish the opposing transcriptional activities of different Hox paralogous group proteins. Hoxc9, as well as Hoxc8 and Hoxc10, that are normally expressed in the LPM caudal to the forelimb, can repress Tbx5 to restrict its expression to the forelimb level region of the LPM (Fig. 7). A single Hox binding site (Hbs2) in the Tbx5 forelimb enhancer is required for this restriction through repression, as mutation of this site causes caudal expansion of expression. Hoxc9 can suppress Tbx5 transcription through this site and this repressive activity is restricted to caudal Hox proteins. Previously, we showed that Hox4 and Hox5 paralogs positively regulate Tbx5 expression [19]. We reveal the combinatorial regulation of Tbx5 by distinct paralogous Hox gene inputs. Hox PG4 and PG5 genes expressed in forelimb-forming LPM form a transcriptional activation complex to positively regulate Tbx5, while Hoxc9, as well as Hoxc8 and Hoxc10 genes expressed in LPM at more caudal levels form a repressive complex to restrict Tbx5 expression. Together, our results reveal that the forelimb-restricted expression of Tbx5 is achieved through co-option of two characteristics of Hox genes, their colinear expression pattern along the rostro-caudal body axis and the functional specificity of Hox proteins from different paralogous groups.
Hox binding site specificity
Our results demonstrate that five of the six Hox binding sites (namely Hbs1 and Hbs3-6) within the Tbx5 forelimb regulatory element are required for the positive regulation of Tbx5 expression while a single site (Hbs2) is required for its repression in caudal LPM (Fig. 1). One possible mechanism to explain how these opposing transcriptional effects are mediated is that different Hox proteins have distinct binding preferences for these sites. For example Hox proteins that act as activators, such as Hox PG4 and PG5, have greater affinity for Hbs1 and 3-6 while repressive Hox proteins, such as Hoxc8/9/10, preferentially bind Hbs2. Our results do not support such a model. In this study, we show that both Hoxc5 and Hoxc9 proteins can bind repressive Hbs2 sites (Fig. 5), suggesting the repressive activity of Hbs2 is not mediated by preferential binding of repressive Hox proteins.
An alternative model is that the transcriptional activity of the Hox complex bound at Hbs2 is determined by a co-factor(s). The sequence of Hbs2 is identical to the sequences of Hbs1 and Hbs3, therefore we compared the sequences surrounding these Hox binding sites. One distinguishing feature of Hbs2 identified using Mat Inspector (http://www.genomatix.de) is the presence of a 6-bp sequence named Pbx1-Meis1 complexes site located 3′ of Hbs2 (6bp3′). Pbx is a Hox co-factor that can attenuate Hox-mediated gene transcription by recruiting histone deacetylases (HDACs) [27]. Therefore, a possible mechanism of the transcriptional repression through Hbs2 is the recruitment of HDACs to Hbs2/6bp3′ by Pbx. To examine this model, we mutated this 6 bp sequence while leaving Hbs2 intact. This mutation did not cause expansion of the reporter gene unlike mutation of Hbs2 or mutations of both Hbs2 and 6bp3′ (Fig. 1), suggesting that the repression is independent of 6bp3′. Thus, our results do not support a role for Pbx determining the transcriptional activities of Hox proteins bound to the Tbx5 forelimb regulatory element.
Specificity of Hox function
The specificities of Hox proteins from different paralogous groups must be tightly regulated. One mechanism by which this is achieved is through distinct DNA binding specificity, for example homeodomains of Hoxc5 and Hoxc9 have different sequence preference in protein binding microarrays [25]. We found, however, that both Hoxc5 and Hoxc9 can bind Hbs2 (Fig. 5), suggesting specificity is not determined by distinct DNA binding abilities of Hox proteins. In addition, we also demonstrate that Hoxc9N5C chimeric protein, which contains the N-terminal repression domain of Hoxc9 fused to the homeodomain –containing C-terminus of Hoxc5, can repress Tbx5. Thus, the transcriptional repression specific to Hoxc9 is not mediated by DNA-binding specificity but rather achieved by transcriptional repression activities restricted to Hoxc9, which are mediated by two domains; the specificity module including the Pbx-binding hexapeptide and homeodomain N-terminal arm and a region N-terminal to the specificity module (Fig. 7).
The mechanism by which these domains confer repressive activity remains to be elucidated. One possible model is by interacting with other transcriptional regulatory domain(s) in the protein. The hexapeptide of AbdA represses dpp expression by inhibiting the function of a glutamine (Q)-rich C-terminal activation domain [28]. Mutations in the hexapeptide converts AbdA from a repressor to an activator without affecting DNA-binding site selection. Although Hoxc9 lacks this Q-rich domain, the hexapeptide of Hoxc9 may block the activity of an unidentified activation domain. Another possibility is that the length of the linker region between the hexapeptide and homeodomain determines transcriptional activity. Several Antp isoforms are produced that have different linker sizes. Synthetic Antp protein with a long linker behaves as an activator, while the short-linker construct acts as a repressor, suggesting the importance of linker size [29]. As Hoxc9 has a shorter linker than Hoxc5, this may favour its function as a repressor.
As it is unlikely that Hox protein itself directly represses Tbx5 transcription, we suggest the model that Hoxc9 supresses Tbx5 expression by interaction with co-repressor(s) (Fig. 7). One candidate is histone deacetylase (HDAC), which can bind Hox proteins directly [30], however, in EMSA we were unable to detect a HDAC/Hoxc9 complex on Hbs2, with in vitro translated proteins or nuclear extract from LPM (data not shown). Other potential collaborators are Smad proteins. In the Drosophila haltere, a Mad/Med/Shn complex works in combination with Ubx to repress Sal expression [31]. There is a potential Smad binding site proximal to Hbs2, however, we mutated this site and did not observe expansion in expression, rather it caused reduced expression in the distal limb bud, suggesting this Smad binding site may have a positive input on Tbx5 expression (). Other candidate repressors are engrailed (En) and sloppy paired (Slp) since, in Drosophila, they form a complex with Hox, Exd and Hth to repress transcription [32]–[34]. Neither of the two mouse En genes, Engrailed1 and Engrailed2, are expressed in LPM at pre-limb bud stages [35], [36]. The mammalian homolog of Slp, fork head box G1 (FoxG1)/brain factor 1 (BF-1) is also not expressed in LPM [37]. Therefore, the putative co-repressors enabling unique Hoxc9 repressive activity remain to be determined.
We have shown that Hoxc8 and Hoxc10 have transcriptional repression ability similar to Hoxc9 (Fig. S1). To gain an insight of the mechanisms of their function, we compared the amino acid sequences of Hoxc8, Hoxc9 and Hoxc10 (data not shown). We could not, however, find any obvious conserved domains outside of homeodomains. It is possible that they use different mechanisms to repress Tbx5 expression or that they share similar 3D structure domains in spite of their distinct amino acid sequences.
Patterning of LPM
Our analysis of the Tbx5 forelimb regulatory element reveals a direct link between patterning of the rostro-caudal axis of the embryo by Hox genes and the programme that controls positioning of the forelimb forming territory. A clear correlation between Hox expression and establishment of the forelimb territory of the LPM has previously been suggested [17], [18], [38]. Application of Fgf to the interlimb flank adjacent to the normal wing induces a wing-like extra limb that expresses Tbx5 [5], [39]. Prior to the emergence of the ectopic wing the endogenous expression of Hoxc9 is reduced [38] consistent with downregulation of Hoxc9 as a repressor of Tbx5 (and the subsequent forelimb programme) being essential for emergence of an ectopic wing bud from this region. In the limbless python, Hoxc8 expression is rostrally expanded to the anterior limit of the trunk [18]. Hoxc8 is expressed exclusively in Tbx5-negative caudal LPM at pre-limb bud stages in chick and mouse (Fig. 2) and it can, like Hoxc9, repress Tbx5 (Fig. S1). Our results therefore, explain the mechanisms that lead to loss of forelimbs in snake through the repression of Tbx5 following expansion of Hoxc8 expression throughout the trunk.
A previous study has demonstrated the presence of and a function for Hox9 genes in anterior-posterior patterning of the forelimb [40]. The complete loss of Hox9 paralogous group leads to the loss of Hand2 expression in posterior forelimb and a consequent reduction in Shh expression, while no effect on Tbx5 expression was reported. Failure to observe any caudal expansion of Tbx5 in this mutant can be simply explained by the redundant function of Hoxc8 and Hoxc10. The same study reported that Hoxc9 is expressed in the forelimb bud at E9.5, but it is undetectable by E10.5. Tbx5 expression is first initiated in the forelimb-forming region at E8.5. We therefore examined the expression of Hoxc9 at stages E8.5–E9.5 (data not shown), however, we did not detect expression of Hoxc9 in the forelimb-forming region, in contrast to the strong staining in caudal tissues. We therefore conclude that Hoxc9 is not present in the forelimb-forming region at stages when Tbx5 expression is first initiated. Later expression of Hoxc9 is not sufficient to cause detectable repression of the domain of Tbx5 already activated by Hox4/5 paralogous genes.
While we have shown that Hoxc8, Hoxc9 and Hoxc10 can repress Tbx5 expression, our study does not exclude the possibility that other caudally-expressed Hox genes have a similar repressive ability. We favour a model in which other caudally-expressed Hox paralogs have redundant functions in repression of Tbx5. Hoxc cluster null mice have no defects in the limb skeleton [41], however, the expression of Tbx5 in these mutants have not been reported and we predict that the ectopic expansion of Tbx5 in caudal LPM would not cause any skeletal defects. Further analysis will be required to uncover the requirement of caudally–restricted Hox paralogs, such as Hox8, Hox9 and Hox10 for Tbx5 repression in caudal LPM.
In addition, while our results clearly demonstrate the importance of specific Hox inputs to generate the restricted expression of Tbx5 in the LPM, a similar Hox protein code is present in axial tissues (neural tube and somites) that do not express Tbx5. The activity of the forelimb regulatory element of Tbx5 is restricted to LPM and this LPM restriction is maintained following mutation of Hbs2 that leads to caudal expansion in expression. One possible explanation for LPM restriction is the presence of unknown repressors in axial tissues or alternatively additional factors, which are active exclusively in LPM, are required for Tbx5 expression. Odd-skipped related (Osr) genes are candidates as they are expressed in LPM, but excluded from axial tissues such as neural tube and somites [42]. We mutated a putative Osr binding site within the Tbx5 forelimb regulatory element to test if reporter activity was lost. The activity of the element was unaffected, however, suggesting Osr genes are not required for Tbx5 LPM expression (Fig. S3).
Conclusions
Our analysis of the Tbx5 forelimb regulatory element has revealed a mechanism by which Hox genes regulate embryonic patterning and how recruitment of regulatory elements allow for the acquisition of novel structures and independent modulation of their morphology. Mechanisms that control PG-specific Hox functions have been described in Drosophila [26], [43]–[48]. Vertebrates, however, have a minimum of 2–4 Hox genes from the same PG and functional redundancy between Hox proteins from the same PG makes it difficult to examine their specific functions experimentally. Here we used a direct target of Hox activity, a regulatory element of Tbx5, to analyse the mechanism of Hox functional specificity and distinguished DNA binding specificity and transcriptional activity. Interestingly, the Tbx5 forelimb regulatory element contains both activating sites and a repressive site in a relatively short fragment of 361 bp. Active complexes are not spatially restricted and can be formed by a range of Hox PG proteins present throughout the rostral-caudal LPM. Instead, restriction of Tbx5 expression is achieved by superimposing a dominant repressive (Hoxc8, c9 and c10) complex that ultimately determines the caudal boundary of Tbx5 expression. Thus, the regulation of Tbx5 expression in the LPM represents an excellent system to understand the interactions between neighbouring Hox binding sites and how the consequent output is integrated.
Materials and Methods
DNA constructs and transient transgenic analysis
For reporter analysis in chick and mouse, we used the BGZA reporter vector [49]. Putative DNA binding sites were searched by MatInspector (http://www.genomatix.de). Transgenic embryos were generated by the Procedural Service section, NIMR by standard pronuclear microinjection techniques. Mouse embryos were staged according to [50]. Noon on the day a vaginal plug was observed was taken to be E0.5 days of development. Mice carrying the LacZ transgene were identified by PCR using specific primers (LacZfwd, 5′GGTCGGCTTACGGCGGTGATTT3′; LacZrev, 5′AGCGGCGTCAGCAGTTGTTTTT3′). Sequences surrounding putative Hox binding sites and the mutations induced are as followings, binding sites are shown in bold; Hbs1, ACATTATTGGA; mut Hbs1, ACATGCTTGGA; Hbs2, GACTCTCAATTATC; mut Hbs2, GACTCTCAACGATC; mut 6bp3′, GACTGCAAATTATC; mut Hbs2+6bp3′, GACGCTTAACGATC; Hbs3, AGATAATTC; mut Hbs3, AGATCGTTC; Hbs4, CCTTATTAAGG; mut Hbs4, CCTTGGCAAGG; Hbs5, CCATTTATCTTG; mut Hbs5, CCATTCGTCTTG; Hbs6, TGTTATTT; mut Hbs6, TGTCGTTT.
Whole mount in situ hybridization
Whole mount in situ hybridizations were carried out essentially as previously described [51]. Probe templates for chick Hox genes, Pitx1, Tbx5 and mouse Hox genes have been described previously [4], [17], [19], [52], [53] Embryos were sectioned by the Histology service, NIMR.
In ovo electroporation of chick embryos
Fertilized chick embryos (Henry Stewart Ltd, Winter Egg Farm) were incubated at 38°C and staged according to Hamburger Hamilton (HH) [54]. Reporter constructs and/or Hox expression constructs were mixed with fast green dye tracer and injected into the coelom located between the somatic and splanchnic LPM. Electric pulses (three pulses 30 v, 50 ms, with 200 ms intervals for tungsten electrodes or three pulses 20 v, 50 ms, with 200 ms intervals for platinum electrodes) were then immediately applied. Only those embryos showing robust expression of dsRed reporter (pCAβ-dsRed-Express) were processed for further analysis.
In vitro translated protein and nuclear extracts from mouse embryos
In vitro translated proteins were produced using a TnT Coupled Reticulocyte Lysate System (Promega). Proteins were labelled with 35S-Methionine (PerkinElmer) to verify and quantify translation. LPM strips adjacent to somites 5–10 (rostral LPM nuclear extract) and lateral to somite 14 to its caudal extreme (caudal LPM nuclear extract) were dissected from E9 mouse embryos. Nuclear extracts were prepared using the NE-PER Nuclear and Cytoplasmic Extraction Kit (Pierce) following manufacturers instructions.
Electrophoretic mobility shift assays
Double-strand oligonucleotides were labelled with 32P by incubating with T4 polynucleotide kinase (NEB) for 30 minutes. 2 µl of in vitro translated protein or nuclear extract were blocked with 200 ng poly-dIdC, 2 µg of poly-dGdC or 2 µg of poly-dAdT in binding buffer (6.7 mM Tris-HCl pH 7.5, 50 mM NaCl, 0.67 mM EDTA, 0.67 mM DTT, 2 µg BSA, 4% glycerol) in a total volume of 22 µl for 15 minutes on ice. For super-shift, 2 µl of the antibody recognising flag epitope (Sigma, F3165) was added to the binding reaction and incubated for a further 15 minutes. Then, 1 µl of 32P -labelled double-stranded oligonucleotides were mixed and incubated for 30 minutes. The protein∶DNA hybrids were resolved on 6% PAGE in 0.5xTBE.
Supporting Information
Zdroje
1. DubocV, LoganMP (2011) Regulation of limb bud initiation and limb-type morphology. Dev Dyn 240: 1017–1027.
2. Gibson-BrownJJ, SIA, SilverLM, PapaioannouVE (1998) Expression of T-box genes Tbx2-Tbx5 during chick organogenesis. Mech Dev 74: 165–169.
3. IsaacA, Rodriguez-EstebanC, RyanA, AltabefM, TsukuiT, et al. (1998) Tbx genes and limb identity in chick embryo development. Development 125: 1867–1875.
4. LoganM, SimonHG, TabinC (1998) Differential regulation of T-box and homeobox transcription factors suggests roles in controlling chick limb-type identity. Development 125: 2825–2835.
5. OhuchiH, TakeuchiJ, YoshiokaH, IshimaruY, OguraK, et al. (1998) Correlation of wing-leg identity in ectopic FGF-induced chimeric limbs with the differential expression of chick Tbx5 and Tbx4. Development 125: 51–60.
6. RallisC, BruneauBG, Del BuonoJ, SeidmanCE, SeidmanJG, et al. (2003) Tbx5 is required for forelimb bud formation and continued outgrowth. Development 130: 2741–2751.
7. AgarwalP, WylieJN, GalceranJ, ArkhitkoO, LiC, et al. (2003) Tbx5 is essential for forelimb bud initiation following patterning of the limb field in the mouse embryo. Development 130: 623–633.
8. NaicheLA, PapaioannouVE (2003) Loss of Tbx4 blocks hindlimb development and affects vascularization and fusion of the allantois. Development 130: 2681–2693.
9. LiQY, Newbury-EcobRA, TerrettJA, WilsonDI, CurtisAR, et al. (1997) Holt-Oram syndrome is caused by mutations in TBX5, a member of the Brachyury (T) gene family. Nat Genet 15: 21–29.
10. BassonCT, BachinskyDR, LinRC, LeviT, ElkinsJA, et al. (1997) Mutations in human TBX5 cause limb and cardiac malformation in Holt-Oram syndrome. Nat Genet 15: 30–35.
11. BongersEM, DuijfPH, van BeersumSE, SchootsJ, Van KampenA, et al. (2004) Mutations in the human TBX4 gene cause small patella syndrome. Am J Hum Genet 74: 1239–1248.
12. HassonP, Del BuonoJ, LoganMP (2007) Tbx5 is dispensable for forelimb outgrowth. Development 134: 85–92.
13. MinguillonC, Del BuonoJ, LoganMP (2005) Tbx5 and Tbx4 are not sufficient to determine limb-specific morphologies but have common roles in initiating limb outgrowth. Dev Cell 8: 75–84.
14. MinguillonC, Gibson-BrownJJ, LoganMP (2009) Tbx4/5 gene duplication and the origin of vertebrate paired appendages. Proc Natl Acad Sci U S A 106: 21726–21730.
15. WellikDM (2009) Hox genes and vertebrate axial pattern. Curr Top Dev Biol 88: 257–278.
16. DubouleD (2007) The rise and fall of Hox gene clusters. Development 134: 2549–2560.
17. BurkeAC, NelsonCE, MorganBA, TabinC (1995) Hox genes and the evolution of vertebrate axial morphology. Development 121: 333–346.
18. CohnMJ, TickleC (1999) Developmental basis of limblessness and axial patterning in snakes. Nature 399: 474–479.
19. MinguillonC, NishimotoS, WoodS, VendrellE, Gibson-BrownJJ, et al. (2012) Hox genes regulate the onset of Tbx5 expression in the forelimb. Development 139: 3180–3188.
20. MannRS, AffolterM (1998) Hox proteins meet more partners. Curr Opin Genet Dev 8: 423–429.
21. LoganM, TabinCJ (1999) Role of Pitx1 upstream of Tbx4 in specification of hindlimb identity. Science 283: 1736–1739.
22. TakeuchiJK, Koshiba-TakeuchiK, MatsumotoK, Vogel-HopkerA, Naitoh-MatsuoM, et al. (1999) Tbx5 and Tbx4 genes determine the wing/leg identity of limb buds. Nature 398: 810–814.
23. DeLaurierA, SchweitzerR, LoganM (2006) Pitx1 determines the morphology of muscle, tendon, and bones of the hindlimb. Dev Biol 299: 22–34.
24. DubocV, LoganMP (2011) Pitx1 is necessary for normal initiation of hindlimb outgrowth through regulation of Tbx4 expression and shapes hindlimb morphologies via targeted growth control. Development 138: 5301–5309.
25. BergerMF, BadisG, GehrkeAR, TalukderS, PhilippakisAA, et al. (2008) Variation in homeodomain DNA binding revealed by high-resolution analysis of sequence preferences. Cell 133: 1266–1276.
26. JoshiR, SunL, MannR (2010) Dissecting the functional specificities of two Hox proteins. Genes Dev 24: 1533–1545.
27. GordonJA, HassanMQ, SainiS, MontecinoM, van WijnenAJ, et al. (2010) Pbx1 represses osteoblastogenesis by blocking Hoxa10-mediated recruitment of chromatin remodeling factors. Mol Cell Biol 30: 3531–3541.
28. MerabetS, KambrisZ, CapovillaM, BerengerH, PradelJ, et al. (2003) The hexapeptide and linker regions of the AbdA Hox protein regulate its activating and repressive functions. Dev Cell 4: 761–768.
29. PapadopoulosDK, Resendez-PerezD, Cardenas-ChavezDL, Villanueva-SeguraK, Canales-del-CastilloR, et al. (2011) Functional synthetic Antennapedia genes and the dual roles of YPWM motif and linker size in transcriptional activation and repression. Proc Natl Acad Sci U S A 108: 11959–11964.
30. LuY, GoldenbergI, BeiL, AndrejicJ, EklundEA (2003) HoxA10 represses gene transcription in undifferentiated myeloid cells by interaction with histone deacetylase 2. J Biol Chem 278: 47792–47802.
31. WalshCM, CarrollSB (2007) Collaboration between Smads and a Hox protein in target gene repression. Development 134: 3585–3592.
32. LelliKM, NoroB, MannRS (2011) Variable motif utilization in homeotic selector (Hox)-cofactor complex formation controls specificity. Proc Natl Acad Sci U S A 108: 21122–21127.
33. FujiokaM, GebeleinB, CoferZC, MannRS, JaynesJB (2012) Engrailed cooperates directly with Extradenticle and Homothorax on a distinct class of homeodomain binding sites to repress sloppy paired. Dev Biol 366: 382–392.
34. GebeleinB, McKayDJ, MannRS (2004) Direct integration of Hox and segmentation gene inputs during Drosophila development. Nature 431: 653–659.
35. WurstW, AuerbachAB, JoynerAL (1994) Multiple developmental defects in Engrailed-1 mutant mice: an early mid-hindbrain deletion and patterning defects in forelimbs and sternum. Development 120: 2065–2075.
36. BroccoliV, ColomboE, CossuG (2002) Dmbx1 is a paired-box containing gene specifically expressed in the caudal most brain structures. Mech Dev 114: 219–223.
37. FilosaS, Rivera-PerezJA, GomezAP, GansmullerA, SasakiH, et al. (1997) Goosecoid and HNF-3beta genetically interact to regulate neural tube patterning during mouse embryogenesis. Development 124: 2843–2854.
38. CohnMJ, PatelK, KrumlaufR, WilkinsonDG, ClarkeJD, et al. (1997) Hox9 genes and vertebrate limb specification. Nature 387: 97–101.
39. CohnMJ, Izpisua-BelmonteJC, AbudH, HeathJK, TickleC (1995) Fibroblast growth factors induce additional limb development from the flank of chick embryos. Cell 80: 739–746.
40. XuB, WellikDM (2011) Axial Hox9 activity establishes the posterior field in the developing forelimb. Proc Natl Acad Sci U S A 108: 4888–4891.
41. SuemoriH, NoguchiS (2000) Hox C cluster genes are dispensable for overall body plan of mouse embryonic development. Dev Biol 220: 333–342.
42. StrickerS, BrieskeN, HauptJ, MundlosS (2006) Comparative expression pattern of Odd-skipped related genes Osr1 and Osr2 in chick embryonic development. Gene Expr Patterns 6: 826–834.
43. ChanSK, MannRS (1993) The segment identity functions of Ultrabithorax are contained within its homeo domain and carboxy-terminal sequences. Genes Dev 7: 796–811.
44. GebeleinB, CuliJ, RyooHD, ZhangW, MannRS (2002) Specificity of Distalless repression and limb primordia development by abdominal Hox proteins. Dev Cell 3: 487–498.
45. NoroB, LelliK, SunL, MannRS (2011) Competition for cofactor-dependent DNA binding underlies Hox phenotypic suppression. Genes Dev 25: 2327–2332.
46. Furukubo-TokunagaK, FlisterS, GehringWJ (1993) Functional specificity of the Antennapedia homeodomain. Proc Natl Acad Sci U S A 90: 6360–6364.
47. LinL, McGinnisW (1992) Mapping functional specificity in the Dfd and Ubx homeo domains. Genes Dev 6: 1071–1081.
48. RyooHD, MannRS (1999) The control of trunk Hox specificity and activity by Extradenticle. Genes Dev 13: 1704–1716.
49. SummerbellD, AshbyPR, CoutelleO, CoxD, YeeS, et al. (2000) The expression of Myf5 in the developing mouse embryo is controlled by discrete and dispersed enhancers specific for particular populations of skeletal muscle precursors. Development 127: 3745–3757.
50. Kaufman MH (2001) The Atlas of Mouse Development, 2nd edn. Cambridge, UK: Academic Press.
51. RiddleRD, JohnsonRL, LauferE, TabinC (1993) Sonic hedgehog mediates the polarizing activity of the ZPA. Cell 75: 1401–1416.
52. NelsonCE, MorganBA, BurkeAC, LauferE, DiMambroE, et al. (1996) Analysis of Hox gene expression in the chick limb bud. Development 122: 1449–1466.
53. PetersonRL, PapenbrockT, DavdaMM, AwgulewitschA (1994) The murine Hoxc cluster contains five neighboring AbdB-related Hox genes that show unique spatially coordinated expression in posterior embryonic subregions. Mech Dev 47: 253–260.
54. HamburgerV, HamiltonHL (1992) A series of normal stages in the development of the chick embryo. 1951. Dev Dyn 195: 231–272.
Štítky
Genetika Reprodukční medicínaČlánek vyšel v časopise
PLOS Genetics
2014 Číslo 3
Nejčtenější v tomto čísle
- Worldwide Patterns of Ancestry, Divergence, and Admixture in Domesticated Cattle
- Genome-Wide DNA Methylation Analysis of Human Pancreatic Islets from Type 2 Diabetic and Non-Diabetic Donors Identifies Candidate Genes That Influence Insulin Secretion
- Genetic Dissection of Photoreceptor Subtype Specification by the Zinc Finger Proteins Elbow and No ocelli
- GC-Rich DNA Elements Enable Replication Origin Activity in the Methylotrophic Yeast
