-
Články
Top novinky
Reklama- Vzdělávání
- Časopisy
Top články
Nové číslo
- Témata
Top novinky
Reklama- Videa
- Podcasty
Nové podcasty
Reklama- Kariéra
Doporučené pozice
Reklama- Praxe
Top novinky
ReklamaGenetic Dissection of Photoreceptor Subtype Specification by the Zinc Finger Proteins Elbow and No ocelli
The eyes of many animals contain groups of photoreceptor cells with different chromatic sensitivities that can be arranged in complex patterns. It is of great interest to identify the genes and pathways shaping these ‘retinal mosaics’ which include stochastically distributed groups of cells, versus highly localized ones. In many insect eyes, which are composed of large numbers of unit eyes, or ommatidia, specialized photoreceptors are found only in the dorsal periphery, where they face the sky. These ommatidia are responsible for detecting linearly polarized skylight, which serves as an important navigational cue for these animals. Here we describe how two closely related proteins called Elbow and No ocelli interact with the transcription factors Homothorax and Orthodenticle to correctly specify the polarization detectors at the dorsal rim of the retina of Drosophila melanogaster. Interestingly, all four proteins are evolutionarily conserved from worms to humans, and they appear to be involved in similar developmental processes across species. Furthermore, human homologs of Elbow and No ocelli have been identified as promoters of luminal breast cancer. The newly identified role of these two proteins within a regulatory network might therefore enable new approaches in a number of important processes.
Published in the journal: . PLoS Genet 10(3): e32767. doi:10.1371/journal.pgen.1004210
Category: Research Article
doi: https://doi.org/10.1371/journal.pgen.1004210Summary
The eyes of many animals contain groups of photoreceptor cells with different chromatic sensitivities that can be arranged in complex patterns. It is of great interest to identify the genes and pathways shaping these ‘retinal mosaics’ which include stochastically distributed groups of cells, versus highly localized ones. In many insect eyes, which are composed of large numbers of unit eyes, or ommatidia, specialized photoreceptors are found only in the dorsal periphery, where they face the sky. These ommatidia are responsible for detecting linearly polarized skylight, which serves as an important navigational cue for these animals. Here we describe how two closely related proteins called Elbow and No ocelli interact with the transcription factors Homothorax and Orthodenticle to correctly specify the polarization detectors at the dorsal rim of the retina of Drosophila melanogaster. Interestingly, all four proteins are evolutionarily conserved from worms to humans, and they appear to be involved in similar developmental processes across species. Furthermore, human homologs of Elbow and No ocelli have been identified as promoters of luminal breast cancer. The newly identified role of these two proteins within a regulatory network might therefore enable new approaches in a number of important processes.
Introduction
The developing retina of Drosophila melanogaster is a powerful model for studying the specification of cell fates within a retinal mosaic. One important aspect is the localized specification of photoreceptor cell types in response to Wg/Wnt signaling. Wg emanating from the head cuticle specifies specialized ommatidia at the dorsal rim of the developing retina [1]–[3]. Here we show that the Drosophila NET family zinc finger proteins Elbow (Elb) and No ocelli (Noc) play a crucial role in this process. In fly wing imaginal discs, as well as in a mouse breast cancer model, NET proteins inhibit Wingless (Wg)/Wnt signaling [4]–[6], while expression of the C. elegans homolog TLP-1 is regulated by Wg/Wnt signaling [7]. Hence, the patterning of the ommatidial mosaic in the dorsal periphery of the retina serves as an attractive model to characterize the different roles of NET proteins.
The elbow/no ocelli complex of Drosophila is a ∼200 kb locus encoding two closely related proteins belonging to the NET family of zinc finger proteins [8]–[10] (Figure 1A). These proteins are related to Sp1-like transcription factors, but contain only one atypical C2H2 zinc finger with 8 amino acids between the two crucial cysteine residues, making these proteins unlikely to bind DNA directly (for review: [11]). They appear to function as repressors of transcription [12], [13] and contain several conserved amino acid motifs (Figure 1C). Elb and Noc bind the co-repressor Groucho through a conserved FKPY motif [10], while other NET family members use different domains for this interaction [6], [14]. The third highly conserved domain of NET proteins is an N-terminal ‘Sp motif’, a SPLALLA amino acid sequence shared with the vertebrate Sp1 family of transcription factors, but not with Drosophila Sp1 or Buttonhead (Btd) [7], [11]. Roles for this motif in protein degradation or transcriptional repression have been suggested [15], [16]. In vertebrates, NET family factors play an important role in the specification of motorneurons [17], and in the developing hindbrain [18], [19] and striatum [20], [21]. Loss of one NET family member, ZNF703, in humans promotes luminal breast cancer [22], [23]. In flies, Elb and Noc are important for proximo-distal patterning of the legs and for morphogenesis of tracheal branches [10], [24].
Fig. 1. Expression of elbow (elb) and No ocelli (noc) in photoreceptors. 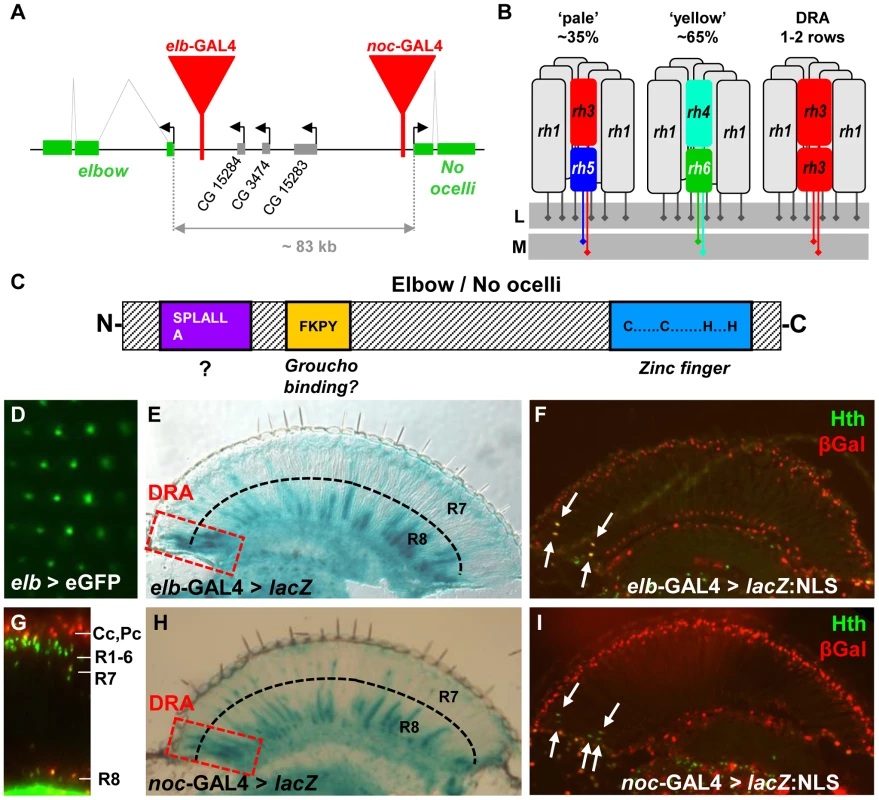
A. Schematic of the elb/noc locus with GAL4 enhancer traps shown as red triangles. B. Schematic summarizing Rhodopsin expression in the three main ommatididal subtypes of Drosophila. L = lamina; M = medulla. C. Domain structure of Elb/Noc proteins: like most members of the NET family of transcriptional repressors they contain a single C2H2 zinc finger domain at the C-terminus (light blue), as well as a conserved SPLALLA motif of unknown function at the N-terminus (purple). In between the two lies a Groucho-binding motif (yellow). D. GFP fluorescence of elb > UAS-eGFP observed under water immersion. Fluorescence localizes to the central photoreceptors of each ommatidium (R7 or R8), as well as non-neuronal cells (green blur). E. Cryostat cross section through an adult eye expressing βGalactosidase (elb > lacZ). Strong expression was observed in R7 and R8 of the Dorsal Rim Area (red dashed box). Additional expression exists in the brain, in many R8 cells, as well as few R7 cells throughout the retina. F. Elbow expression (elb > lacZ:NLS; red) co-localizes with DRA marker Homothorax (Anti-Hth; green; white arrows). G. Expression of elb-GAL4 driving UAS-lacZ:NLS in the adult retina (visualized on a Cryostat cross section): expression is restricted to R7 and R8 nuclei, as well as non-neuronal cone cell nuclei above the R1-6 level. H. Adult expression of noc-GAL4 is virtually indistinguishable from elb. Expression is strongest in R7 and R8 in the DRA (red dashed box), as well as subsets of R7 and R8 cells outside the DRA. I. No ocelli expression (noc > lacZ:NLS; red) also co-localizes with DRA marker Homothorax (Anti-Hth; green; white arrows). The Drosophila compound eye is composed of approximately 800 ommatidia (unit eyes), each containing 8 photoreceptor neurons (called R1 to R8), as well as pigment, cone, and bristle cells [25]. The outer photoreceptors (R1-6) have short axon fibers that terminate in the first optical ganglion, the lamina. The two remaining inner photoreceptors R7 and R8 have light-gathering structures (rhabdomeres) that are located in the center of the ommatidium, R7 situated on top of R8. Their long axonal fibers terminate in a deeper layer of the optic lobe, the medulla [26]. Specification of R7 and R8 depends on the Spalt complex (Sal), encoding two transcription factors, Spalt major (Salm) and Spalt related (Salr) [27]. The R7 cell type is then induced by Prospero (Pros) [28], while Senseless (Sens) determines R8 [29], [30].
A retinal mosaic arises from the molecular differences between R7/R8 of different ommatidia, creating functional heterogeneity [31] (Figure 1B). At least four ommatidial subtypes can be distinguished in flies. Two subtypes named ‘pale’ (p) and ‘yellow’ (y) are distributed randomly throughout the eye, with a ratio of 35% (p) to 65% (y) [32], [33]. R7 cells in p ommatidia always express UV-sensitive rhodopsin Rh3, while the underlying R8 express a blue-sensitive rhodopsin Rh5 [34], [35]. In y ommatidia, R7 express another UV-sensitive rhodopsin, encoded by the rh4 gene, while R8 cells contain a green-sensitive Rhodopsin, Rh6 [36]–[38]. This mosaic of chromatic sensitivities created by p/y ommatidia provides the substrate for Drosophila color vision [39], [40]. In the dorsal-most third of the adult eye, y ommatidia show an additional specialization by co-expressing both UV Rhodopsins Rh3 and Rh4 in R7 cells [41]. Finally, a narrow band of ommatidia along the dorsal head tissue, called the ‘dorsal rim area’ (DRA), manifests morphological and molecular specializations making these ommatidia ideal detectors for polarized light [42]. Inner photoreceptors R7 and R8 of DRA ommatidia are monochromatic since they both contain the UV Rhodopsin Rh3 [43]. Furthermore, the diameter of their rhabdomeres is enlarged, and the absence of rhabdomeric twist in DRA ommatidia results in high polarization sensitivity [42], [44], [45]. As a consequence, DRA ommatidia are both necessary and sufficient for detecting linearly polarized light emanating from the sky [45], [46].
We have shown that the development of DRA ommatidia in Drosophila depends on Homothorax (Hth) a homeodomain transcription factor homologous to vertebrate Meis factors [2],[47]. Hth is both necessary and sufficient for the specification of DRA ommatidia where it is expressed in both R7 and R8 [2]. Hth always co-localizes in nuclei with its ubiquitous cofactor Extradenticle (Exd), whose nuclear localization depends on Hth (for review: [48]). We have shown that the transcription factor Orthodenticle (Otd) is required as an activator of rh3 expression in DRA ommatidia [49]. Otd is expressed in all adult Drosophila photoreceptors, where it acts as a general activator of rh3 and rh5 expression [49], [50]. Otd also induces repression of rh6 through induction of another homeodomain transcription factor, the repressor ‘Defective proventriculus’ (Dve). In outer photoreceptors (R1-6), Otd and Dve act in a feedforward loop, resulting in repression of rh3 rh5 and rh6 by Dve in these cells [50]. In inner photoreceptors, Dve is repressed by Spalt (Sal) factors, allowing activation of rh3 and rh5 by Otd in ‘pale’ ommatidia. As a consequence, rh3 and rh5 are lost in otd mutants, while expression of rh6 is de-repressed into outer photoreceptors, due to the loss of Dve in these cells and the presence of the rh6-specific activator Pph13 [51].
Alleles of the gene encoding Otd are named ocelliless (oc) due to the function of otd in patterning the dorsal head cuticle where ocelli form. Therefore, oc and noc are both required for ocellar development [9], [52]. Furthermore, both genes show specific, overlapping expression at the anterior pole of the fly embryo [53], yet their regulatory relationship is not known. The vertebrate homologs of Otd are involved in retinal development, including OTX1, OTX2 and CRX (‘cone rod homeobox’), whose mutations cause retinal degeneration [54], [55]. Furthermore, OTX1 and OTX2 are required for the specification and regionalization of the forebrain and midbrain, while Otd is required for the development of the anterior brain in flies (for review: [56]). Several aspects of the otd mutant phenotype in flies can be rescued by either CRX or OTX2 in the retina [57], as well as the developing brain [58]. Furthermore, many aspects of the OTX1 phenotype in mice can be rescued by substitution with the Drosophila Otd protein [59], demonstrating that the molecular function of these factors is conserved.
Here we show that Elb and Noc play an important role in the specification of DRA ommatidia. Loss of both genes together leads to a phenotype identical to the loss of hth: the enlarged rhabdomere diameter of DRA inner photoreceptors is lost, and expression of Rh3 in DRA R8 cells is replaced with Rh6. Furthermore, the specific R8 marker Senseless (Sens) becomes de-repressed in R8 cells of DRA ommatidia. Since Hth expression in the DRA is normal in elb,noc double mutants, this indicates that Hth is unable to execute its DRA-inducing potential. Gain-of-function experiments in combination with site-directed mutagenesis of the three evolutionarily conserved domains in Elb and Noc reveal that an N-terminal SPLALLA motif as well as the unique zinc finger are crucial for the function of Elb and Noc in DRA ommatidia. Furthermore, Elb and Noc can genetically antagonize the activator or repressor functions of Otd in the retina through distinct protein domains. We therefore propose that NET family proteins might interact with Meis as well as Otx family genes, depending on the transcriptional context.
Results
elbow and no ocelli expression in photoreceptors
In a GAL4 enhancer trap screen for genes expressed in adult photoreceptors, we obtained two independent insertions in the elbow/no ocelli complex [8], [9], one localized 950 bp upstream of the transcription start of elbow (also referred to as ‘elbow B’, elB, or el; [10], and the other 285 bp upstream of no ocelli (noc; [24]) (Figure 1A). We previously showed that these two enhancer traps faithfully reproduce the expression patterns of the genes in which they are inserted, in both tracheae and leg imaginal discs [10], [24]. When crossed to UAS-GFP, the GFP signal under water immersion could be localized to the adult inner photoreceptors, with additional signal coming from non-neuronal cells, most likely cone cells (Figure 1D,G). GAL4 expression was strikingly similar between the two lines (Figure 1E,F,H,I). Expression was strongest in R7 and R8 cells in the ‘dorsal rim area’ (DRA), as well as in non-DRA R8 cells (Figure 1E,H), while expression in non-DRA R7 cells was much weaker (Supplemental Figure S1D,E). Different expression levels in R7 and R8 did not correlate with p/y-specific rhodopsin subtypes (Supplemental Figure S1F,G). In DRA ommatidia, strong GAL4 expression always co-localized with Homothorax (Hth) (Figure 1F,I). Although no expression was detected in adult outer photoreceptors R1-6 (Figure 1G), both GAL4 lines were expressed in larval R3 and R4 photoreceptors in eye-imaginal discs, with an onset 3–4 rows posterior to the morphogenetic furrow (Supplemental Figure S1A,B). No expression in R7 and R8 was detected at that stage. Co-expression with the inner photoreceptor marker Spalt (Sal) started around 50% pupation (Supplemental Figure S1C). Thus, elb/noc expression starts in R3 and R4 early in development, but becomes restricted to inner photoreceptor types during mid-pupal development, and is expressed most strongly in adult R7/R8 of DRA ommatidia, as well as non-DRA R8 cells.
Loss of typical DRA rhodopsin expression in elbow, no ocelli double mutants
We first tested single mutants for either elbow or no ocelli, for changes in the Rhodopsin pattern [38], [39]. Neither homozygous viable elb3.3.1 null mutants [24], nor whole mutant eyes generated using the null allele nocΔ64 (-/-) [10], generated with ey-Flip/GMR-hid [60], showed a Rhodopsin phenotype affecting R7 cells or R8 cells (Supplemental Figure S2A-F). We then used the same technique to generate elb3.3.1,noc Δ64 (-/-) double mutant eyes (Figure 2C,D,G). Although Rhodopsin expression appeared unaffected in most inner photoreceptors (Figure 2A–D, Supplemental Figure S2G,H), Rh3 expression was no longer detected in R8 cells in the DRA (Figure 2C). Instead, the R8 rhodopsin Rh6 was now expressed in the dorsal-most R8 cells (Figure 2D). Mutants lacking Hth function exhibit the same phenotype of ‘odd-coupled’ expression of Rh3/Rh6 in DRA ommatidia R7/R8 [2]. We stained elb3.3.1, noc Δ64 (-/-) double mutant eyes for Hth along with Rhodopsins (Figure 2E–G). In these double mutants, Hth expression was unaffected. DRA R7 co-expressed Hth and Rh3 while DRA R8 co-expressed Hth and Rh6 (Figure 2G). This situation was also identical to flies expressing dominant-negative Homothorax (HthHM) in all photoreceptors (Figure 2F). Hence, in the absence of elb and noc, DRA ommatidia are mis-specified into ‘odd-coupled’ Rh3/Rh6 ommatidia, despite the presence of Hth (Figure 2H). We therefore concluded that Elb and Noc act downstream of, or in parallel to Hth in the specification of DRA photoreceptor cell fates.
Fig. 2. The opsin phenotype of elb,noc double mutants is restricted to DRA ommatidia. 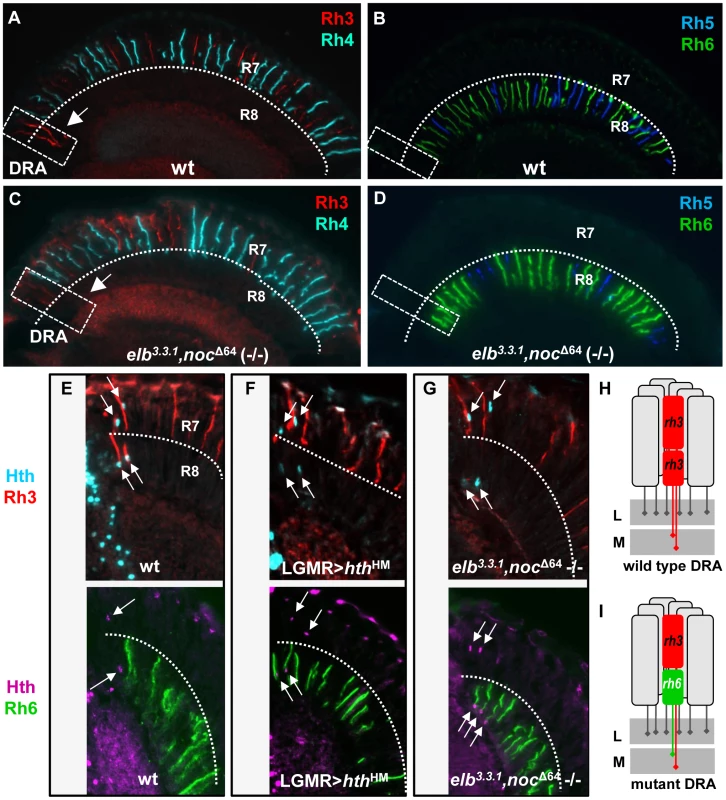
A. Random expression of R7 opsins Rh3 (red) and Rh4 (cyan) in wild type flies. Note expansion of Rh3 into the R8 layer in DRA ommatidia (white dashed box, arrow). B. Expression of R8 opsins Rh5 (blue) and Rh6 (green) in wild type flies. Both opsins are excluded from the DRA. C. Expression of R7 opsin in elb,noc double mutants appears largely normal, but Rh3 expression is lacking in DRA R8 cells (white dashed box, arrow). D. In elb,noc double mutants, expression of Rh6 (green) appears to extend all the way to the DRA. E–H. elb,noc double mutants phenocopy the opsin phenotype of homothorax loss-of-function in DRA. E. Top: wild type opsin expression in the DRA, marked with Hth (cyan) and Rh3 (red). Bottom: same genotype marked with Hth (purple) and Rh6 (green). F. Upon dominant negative loss of hth function (LGMR > hthHM), Rh6 (green, bottom) expands into DRA R8 cells, whereas R7 cells retain expression of Rh3 (red, top). G. Identical phenotype observed in elb,noc double mutants: co-expression of Rh3/Hth in R7DRA cells (top), and Hth/Rh6 in R8DRA cells (bottom). H,I. Summary of DRA phenotypes observed: typical monochromatic Rh3/Rh3 (red) coupling in wild type DRA ommatidia (H) is replaced with ‘odd’ coupled' ommatidia expressing Rh3 in R7 and Rh6 (green) in R8 at the dorsal rim (I), both in hth loss-of-function, as well as in elb,noc loss-of-function. L = lamina; M = medulla. Loss of all DRA markers in elb, no ocelli double mutants
DRA R8 cells express an R7 Rhodopsin and therefore lack features of R8 cells, like expression of the R8 transcription factor Sens [61]. Indeed, Sens becomes specifically de-repressed in hth mutant DRA R8 cells [2]. This phenotype correlates with the gain of Rh6 expression in hth mutant DRA R8 cells, since Sens has an inductive effect on rh6 expression [29], [62]. We stained elb3.3.1,noc Δ64 (-/-) double mutant retinas for Exd and Sens (Figure 3A–F). Like Hth, Exd was localized to the nuclei of DRA R7 and R8 in wild type (Figure 3A,B), as well as in elb,noc double mutants (Figure 3D; Supplemental Figure S3A,C). However, Sens was now co-expressed with Exd in R8 cells, similar to flies over-expressing dominant-negative HthHM (Figure 3C). Co-expression of Exd and Sens in DRA R8 cells was already visible in pupal retinas (50% APF; Figure 3E,F), arguing that R8 cells in DRA ommatidia lacking elb and noc became mis-specified before Rhodopsin expression begins. Outside the DRA, expression of inner photoreceptor markers Spalt, Prospero, and Senseless were indistinguishable from wild type (Supplemental Figure S3B,D–F). Unlike the R8 marker Sens, the R7 marker Prospero is not repressed in DRA R7 cells by Hth/Exd [2]. Hence, there was no immuno-histochemical way to tell whether DRA R7 cells had actually changed their fate in elb,noc (-/-) mutants. We therefore assessed the morphology of the eye tissue in tangential plastic sections in elb3.3.1,noc Δ64 (-/-) double mutant eyes (Figure 3G). Wild type DRA ommatidia display an enlarged R7 and R8 rhabdomere diameter [2], [42], [45], [63]. In mutant clones touching the dorsal eye rim, the typical DRA morphology was lost and resembled ommatidia from the main region of the eye. We have previously described the same morphological DRA phenotype in HthB2 (-/-) mutant clones [2]. We therefore concluded that all markers of DRA ommatidia were lost in elb3.3.1,noc Δ64 double mutants, despite persisting expression of Hth.
Fig. 3. The DRA fate is lost in elb,noc double mutants. 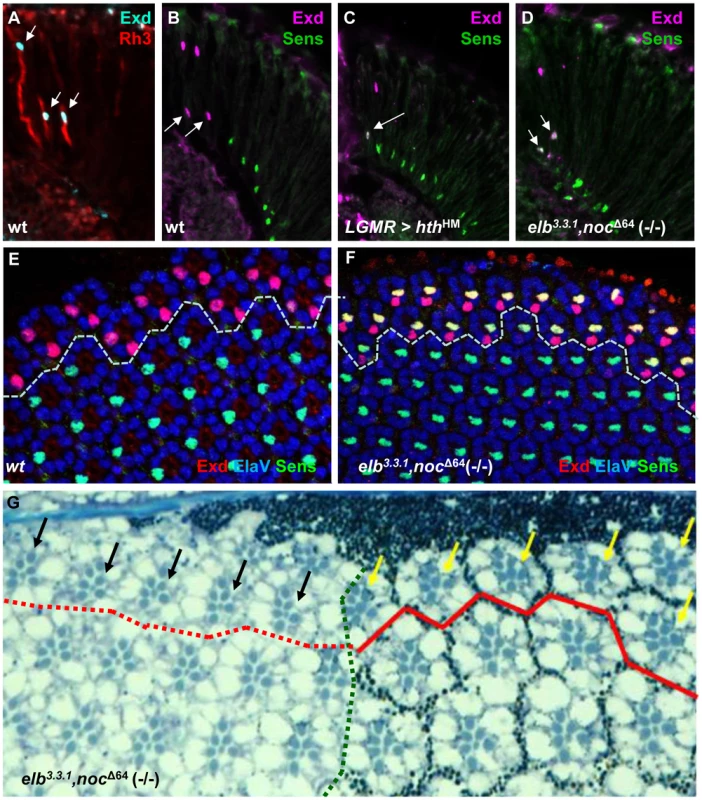
A. Nuclear localization of Exd (cyan) in R7 and R8 of wild type DRA ommatidia, marked with Rh3 (red). B. Exclusion of R8 marker Senseless (green) from wild type R8DRA cells (arrows), positively marked with Exd (purple). C–F. Exclusion between Extradenticle (Exd) and Senseless (Sens) is lost in R8DRA cells, in elb,noc double mutants (phenocopying homothorax loss-of-function). C. Co-expression of Exd (indirectly labeled using hth-lacZ:NLZ), and Sens (green) in homothorax LOF (LGMR > hthHM). D. Identical co-expression of Exd (purple) and Sens (green) is observed in elb,noc double mutants. E. Exclusion of Exd (red) in the DRA (dashed line) and Sens (green) in a whole mounted pupal retina from wild type flies (labeled with ElaV, blue). Weak co-staining is sometimes observed, at that developmental stage. F. Strong co-expression of Exd (red) and Sens (green) in R8 cells of all DRA ommatidia (dashed line), phenocopying hth loss-of-function. G. Morphological defects of elb,noc double mutant DRA ommatidia: typical enlarged rhabdomere diameter of central photoreceptors R7 and R8 (yellow arrows) is lost specifically (black arrows) in elb,noc double mutants clones touching the dorsal eye margin (marked by absence of photoreceptor pigment granules to the left of the green dotted line). The same phenotype was described for mutant clones of hthB2 [2]. Note that due to the absence of enlarged inner photoreceptor rhabdomeres the extent of the DRA could not be marked inside elb,noc mutant tissue (hence the dotted red line). Homothorax requires Elb and Noc for inducing DRA fates
Over-expression of Hth in all photoreceptors leads to a transformation of the entire retina into DRA ommatidia, with Rh3 expression expanding into all inner photoreceptors, while expression of Rh4, Rh5, Rh6, and Sens is lost [2]. We tested whether Elb and Noc were required for this function of Hth, focusing on expression of Rh6 and Sens as the most reliable markers (Figure 4A–C). First, we confirmed that expression of Rh6 was always lost in transgenic flies expressing a GFP:hth fusion protein directly attached to LGMR (Figure 4B; Supplemental Figure S4A–C; see Materials and Methods). However, when GFP:hth was over-expressed in elb3.3.1,noc Δ64 (-/-) double mutant eyes, Rh6 was found in R8 in the entire retina (Figure 4C). Rh3 was also detected, while Rh4 and Rh5 were absent (data not shown). Thus, the entire retina was transformed into ‘odd coupled’ Rh3/Rh6 ommatidia, like the ones we had described in the DRA of HthHM mutants. Interestingly, Elb and Noc remained restricted to inner photoreceptors when GFP:hth was over-expressed, but became expressed at high levels in all R7 and R8 (Figure 4D). Taken together, these data suggested that Elb and Noc act downstream of Hth in the specification of DRA ommatidia.
Fig. 4. Elb and Noc are crucial for Homothorax function in DRA specification. 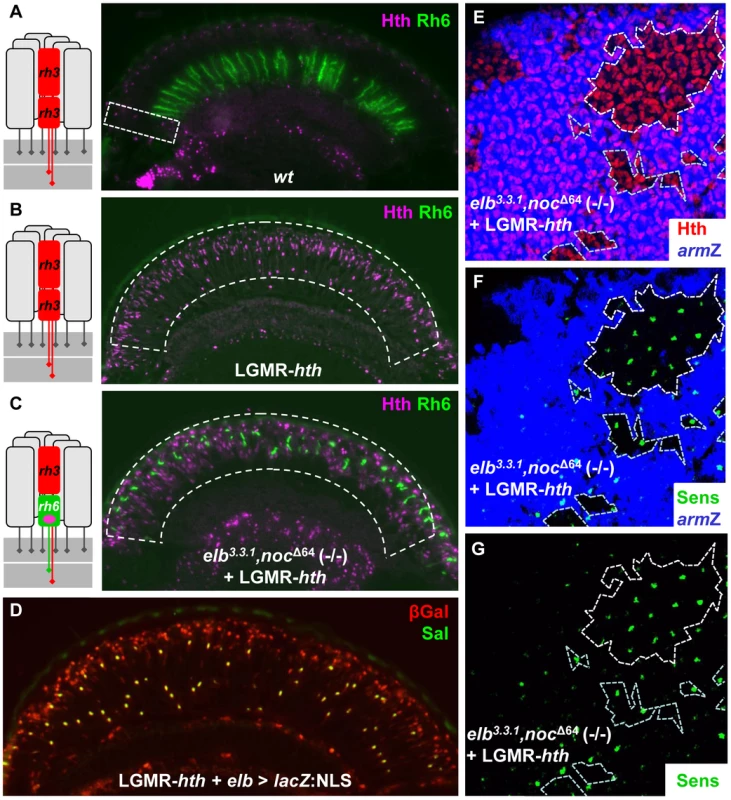
A.–C. Homothorax fails to ectopically induce the DRA fate in absence of Elb and Noc. (Ommatidial schematic to the left summarizes the only ommatidial subtype found inside the dashed white boxes; pink dot: nuclear Sens expression in R8). A. Exclusion of Rh6 (green) from the DRA (labeled with Hth, purple), in wild type flies (dashed white box). B. Complete loss of Rh6 upon over-expression of Hth (purple), under LGMR-GAL4 control. C. Co-expression of Rh6 (green) and Hth (purple) in elb,noc double mutant flies ectopically expressing Hth (green) in all photoreceptors. D. Expression of elb-GAL4 (visualized using UAS-lacZ:NLS) is never expanded into R1-6 by over-expression of Hth (green). Instead, strong expression of βGal (red) is observed in all inner photoreceptors R7 and R8 (co-labeled with Spalt, green). E–G. Homothorax fails to repress Senseless in absence of elb and noc. Whole mounted pupal retina expressing Hth in all photoreceptors (LGMR-hth; red) with homozygous clones lacking both elb and noc marked by absence of arm-lacZ (blue, E). The vast majority of strong Sens expression (green) is observed in R8 cells inside homozygous clones (F), as well as in close vicinity to mutant clones (G). As a second test of this epistatic relationship, we took advantage of the fact that the Wg pathway induces DRA ommatidia [1], [3]. Ectopic activation of the Wg pathway with constitutively active forms of Armadillo (armS10, or arm*; [64]) induces Hth in the entire dorsal eye and transforms it into DRA ommatidia, repressing Rh6 in all dorsal R8 cells [2). We therefore tested the requirement of elb and noc in the dorsal eye using direct GMR-arm* fusions [65]. While Hth was ectopically expressed in all inner photoreceptors of the dorsal eye in GMR-arm* flies, leading to the loss of Rh6 expression from the expanded DRA (Supplemental Figure S4D), we observed co-expression of Rh6 and Hth in R8 throughout the dorsal eye in an elb3.3.1,noc Δ64 (-/-) mutant background containing GMR-arm* (Supplemental Figure S4E). Rh4 and Rh5 were always excluded from the dorsal eye, while Rh3 expression remained (not shown). Hence, activating the Wg pathway in elb3.3.1,noc Δ64 (-/-) mutants induced Hth expression throughout the dorsal half of the eye, transforming it into ‘odd coupled’ Rh3/Rh6 ommatidia. Elb and Noc are therefore necessary for the ability of Hth to induce DRA fate in response to activating the Wingless pathway.
We also tested whether Hth required Elb and Noc for repression of Sens, when ectopically expressed by generating clones of elb3.3.1,noc Δ64 (-/-) mutant tissue in pupal retinas expressing LGMR - hth (Figure 4E). In these retinas, the vast majority of R8 cells (though not all) strongly expressing Sens were located within the elb3.3.1,noc Δ64 (-/-) double mutant tissue (Figure 4E–G). Therefore, Hth had lost the ability to repress Sens in the absence of Elb and Noc, which is consistent with a loss of DRA fate [2]. This requirement of Elb/Noc appeared not to be strictly cell autonomous since Sens-positive R8 cells could be observed outside elb,noc mutant clones, although almost always in their direct vicinity (Figure 4G) Taken together, we concluded that Hth function in the specification of DRA ommatidia in response Wg pathway activation was dependent on the presence of Elb and Noc.
Mutations in the Sp/SPLALLA motif and zinc finger affect DRA development
Homothorax is sufficient to induce the DRA fate in all ommatidia when over-expressed [2]. In contrast, over-expression of either Elb or Noc, or both proteins together never resulted in the induction of DRA markers (Supplemental Figure S5), and the gain-of-function phenotypes will be discussed in more detail below. All NET family zinc finger proteins share several conserved protein domains whose functional significance remains incompletely understood [11], [14]. We investigated the role of three domains in DRA specification: the conserved Sp/SPLALLA motif at the N-terminus, a more centrally located FKPY motif that was shown to interact with the transcriptional repressor Groucho, and the unusual zinc finger, located C-terminally (Figure 1C). We altered the amino acid sequence of each of these sequences individually using site-directed mutagenesis and placed the resulting cDNA for elb and noc in UAS-vectors. Rescue experiments using these point-mutated versions of elb or noc were not possible since the necessary driver lines elb-GAL4 and noc-GAL4 were (non-mutant) enhancer traps inserted within the elb.noc locus. We therefore tested whether these constructs affected DRA specification in a dominant-negative fashion when over-expressed, similar to HthHM.
We altered the Sp/SPLALLA motif of both Elb and Noc to TGIVIIV (Figure 5A; see Materials and Methods). Over-expression of UAS-elb[SPLALLA*] in all photoreceptors using LGMR-GAL4 indeed had a dominant negative effect on DRA development. Expression of Rh3 in DRA R8 cells was lost (Figure 5B) and, instead, Rh6 was expanded to the dorsal rim of the retina (Figure 5C). In these DRA R8 cells, we detected co-expression of Sens and Exd, which is an indication of DRA fate loss (Figure 5D). Expression of Rh3, Rh4 and Rh5 was normal outside the DRA in LGMR>elb[SPLALLA*] flies (Figure 5F). Interestingly, Rh6 was expanded into all outer photoreceptors R1-6, for reasons we will discuss below.
Fig. 5. Mutagenesis of Sp/SPLALLA motif or zinc finger transform Elb into a dominant-negative. 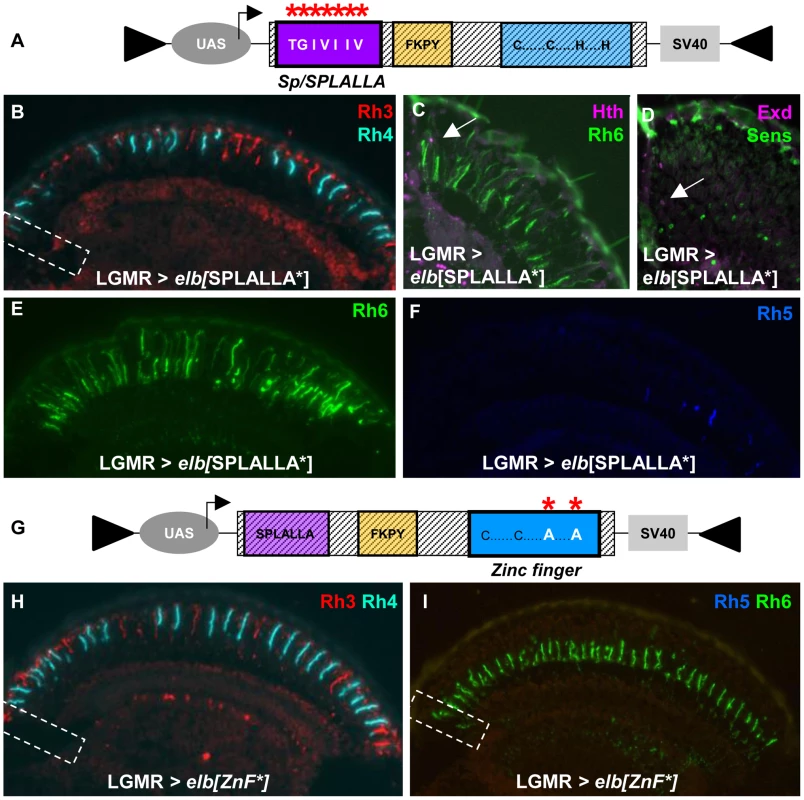
A. Schematic illustrating the UAS-construct for over-expression of an Elb protein with an altered Sp/SPLALLA motif (mutated to TGIVIIV, using site-directed mutagenesis; see Materials and Methods). B. Ectopic expression of mutated elb[SPLALLA*] using LGMR-GAL4 leads to a loss of typical Rh3 expression in R8DRA cells (white dashed box), while R7 expression appears normal in the rest of the retina. C. As a result, Rh6 (green) and Hth (purple) are co-expressed (white arrow) at the dorsal rim of the retina. D. Weak co-expression of Sens (green) and Exd (purple) was sometimes observed (white arrow), although not with 100% penetrance. E. Expression of Rh6 (green) is strongly expanded into outer photoreceptors R1-6, in LGMR > elb[SPLALLA*] flies. F. Expression of Rh5 (blue) appears normal in LGMR > elb[SPLALLA*] flies. G. Schematic illustrating the UAS-construct for over-expression of an Elb protein with a mutated zinc finger (both His were altered to Ala, using site-directed mutagenesis; see Materials and Methods). H. Over-expression of mutated elb[ZnF*] in all photoreceptors leads to a loss of DRA-specific Rh3 (red) expression in R8DRA cells (white dashed box). I. In the R8 cell layer, expression of Rh5 (blue) is specifically lost, and Rh6 (green) is the only remaining R8 rhodopsin, including the DRA (white dashed box). To test the involvement of the unique zinc finger motif for DRA specification, we mutated the two crucial Histidine residues in the zinc finger of both Elb and Noc proteins, replacing them with Alanine (Figure 5G; see Materials and Methods), thereby disrupting its ability to chelate zinc [66], [67]. Over-expression of elb[ZnF*] also resulted in a dominant-negative loss of the DRA: Rh3 was lost in R8 DRA cells and was replaced by Rh6, resulting in Rh3/Rh6 ‘odd-coupled’ ommatidia (Figure 5H,I).
Finally, we altered the conserved, generic Groucho-binding motif present in both Elb and Noc (Figure 6A; FKPY → IEGS; see Materials and Methods). Over-expression of elb[Gro*] had no dominant-negative effect on DRA development (Figure 6B,C), and instead resulted in an unrelated rhodopsin phenotype (see below). We concluded from these experiments that point mutation of either Sp/SPLALLA-motif or zinc finger in Elb resulted in dominant-negative function, possibly by sequestering factors present in the DRA (possibly Hth), through the unaltered parts of the over-expressed inactivated protein. The dominant-negative function of over-expressed, point-mutated forms of Elb might therefore result in inactive protein complexes, similar to what has been described for HthHM [68]. Hence, these results suggested that Sp/SPLALLA domain and zinc finger were crucial for the in vivo function of Elb. Interestingly, the identical point-mutations in the Sp/SPLALLA or zinc finger motifs of the Noc protein did not result in a dominant-negative effect on DRA specification (Supplemental Figure S6A–F).
Fig. 6. Mutagenesis of conserved Elb domains interferes with different Otd functions. 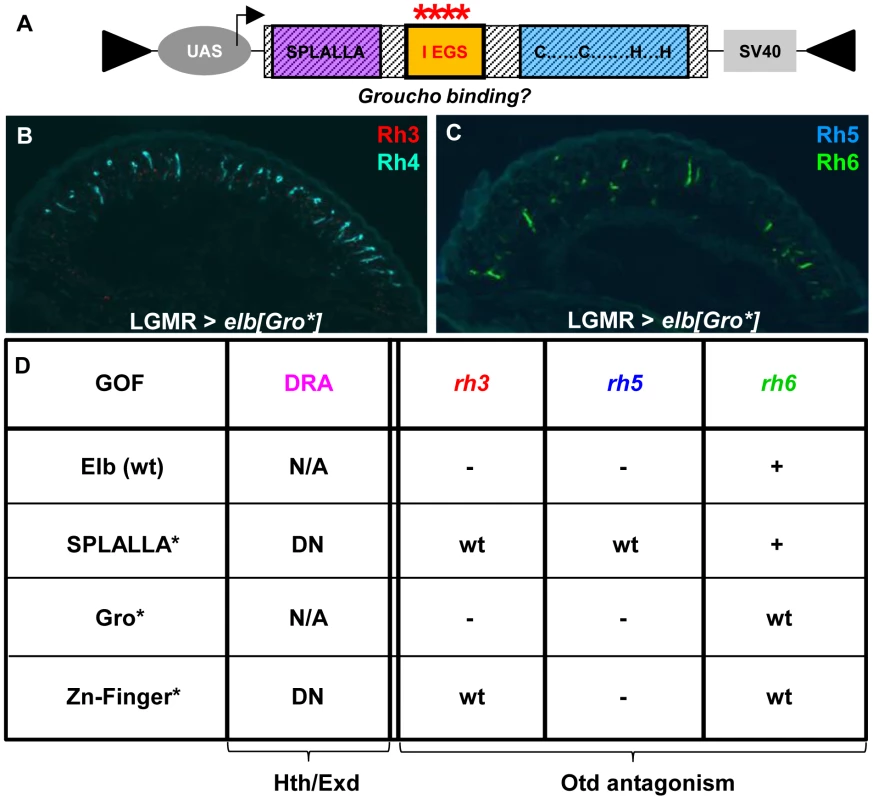
A. Schematic illustrating the UAS-construct for over-expression of an Elb protein with an altered Groucho-binding motif (FKPY → IEGS, see Materials and Methods). B. Over-expression of mutated elb[Gro*] in all photoreceptors using LGMR-GAL4 leads to a specific loss of Rh3 (red), while Rh4 expression (cyan) is normal. C. In R8 cells, Rh5 (blue) is specifically lost, while Rh6 expression (green) is normal. D. Table summarizing gain-of-function phenotypes observed using different point-mutated forms of Elb. Every mutant produces a specific phenotype that can be broken down into four aspects: DRA specification, rh3 expression, rh5 expression, and rh6 expression. Abbreviations: DN, dominant-negative; wt = expression like wild type; ‘-’ = loss of expression; ‘+’ = de-repression of expression into outer photoreceptors R1-6; ‘N/A’ = DRA specification could not be assessed due to ectopic loss of Rh3 expression. Mutagenesis of conserved Elb domains interferes with different functions of Orthodenticle (Otd)
Outside the DRA, over-expression of wild type Elb led to the loss of rh3 and rh5 expression, while rh6 was expanded into outer photoreceptors (Supplemental Figure S5A–G). This phenotype, which was observed both in wild type and in elb,noc double mutants backgrounds was identical to the loss of Otd function in otdUVI mutants [49], [50]. Gain-of-function experiments using UAS-noc resulted in a similar, but much milder phenotype, with only some outer photoreceptors de-repressing Rh6 (Supplemental Figure S5K,L). Importantly, over-expression of Elb did not repress larval or adult otd expression (Supplemental Figure S5I) and expression of elb/noc was unaltered in otdUVI mutants (Supplemental Figure S4F,G). Hence, since elb3.3.1,nocΔ64 mutant eyes showed no phenotype outside the DRA, we concluded that these gain-of-function phenotypes might be due to an antagonistic genetic interaction between Elb/Noc and Otd, which is expressed in all photoreceptors [69] (although it affects Rhodopsin expression in specific photoreceptor subtypes, see discussion) [49], [50].
Interestingly, the phenotypes observed outside the DRA when over-expressing different point-mutated versions of Elb and Noc separated different aspects of the otdUVI phenotype:
-
Over-expression of elb[Gro*] with LGMR-GAL4 resulted in the loss of Rh3 and Rh5 in the entire retina, while expression of Rh4 and Rh6 was normal (Figure 6A–C), leading to ‘empty’ pR7 and pR8 cells. Therefore, while elb[Gro*] was still able to antagonize the transcriptional activation of rh3 and rh5 by Otd in ‘pale’ ommatidia, Rh6 was not de-repressed in outer photoreceptors. It therefore appears that the putative Groucho-binding motif in Elb is specifically required to genetically antagonize the repressor function of Otd in R1-6, potentially by interfering with its ability to activate expression of the repressor Dve [50].
-
Over-expression of elb[SPLALLA*] led to a specific loss of Otd's ability to activate both rh3 and rh5 expression, but not to repress rh6 in outer photoreceptors. Therefore, the Sp/SPLALLA motif is specifically required for Elb to antagonize Otd's activator function on rh3 and rh5. Hence, the Gro-binding and Sp/SPLALLA domains therefore specifically affect opposite arms of the interlocked feedforward loops though which Otd and Dve activate or repress rhodopsin transcription [50].
-
The activator function of Otd could even further be separated though mutation of the Elb zinc finger, since Rh3 expression was normal outside the DRA in LGMR > elb[ZnF*] flies, while Rh5 remained lost (See Figure 6D for a table summarizing the effects of wild type, SPLALLA*, Gro*, and ZnF* gain-of-function experiments on DRA specification, as well as the different aspects of otdUVI loss-of-function). Like for the wild type Noc protein, the phenotypes obtained with point-mutated versions of Noc were extremely mild, only affecting the de-repression of Rh6 into outer photoreceptors for noc[ZnF*] (Supplemental Figure S6D-F), and noc[Gro*] (Supplemental Figure S6G-I). In conclusion, the different aspects of the otdUVI-like phenotype obtained in an Elb(wt) gain-of-function could be dissociated, by mutating either of the three conserved domains: the Sp/SPLALLA motif, the putative Groucho-binding domain, and the zinc finger.
VP16 and Engrailed[R] fusions of Noc have a strong effect on R8 Rhodopsin expression
To further investigate Elb/Noc's role as activators or repressors of transcription, and how they antagonize Otd, we generated fusions with the VP16 transcriptional activation domain [70], as well as the repressor domain of Engrailed [71] (Figure 7). Both kinds of fusion proteins were placed under UAS control for gain-of-function experiments (see Materials and Methods). Over-expression of VP16:elb with LGMR-GAL4 led to a severe disruption of retinal morphology (not shown) that was not observed with UAS-VP16:noc. We therefore focused our analysis on Noc fusion proteins (Figure 7A–E). Over-expression VP16:noc had no effect on R7 Rhodopsins, although Rh3 expression was weak in the DRA (Figure 7B). We confirmed that the DRA markers Hth and Rh3 were correctly expressed in DRA R8 in sevenless (sev) mutants (Figure 7C), showing that DRA ommatidia were indeed correctly specified. Rhodopsin expression in R8 outside the DRA was severely disrupted: Rh5 was found expanded to most R8 cells (figure 7D), while Rh6 expression appeared normal (figure 7E). The consequence was co-expression of Rh5 and Rh6 in many R8 cells (∼51%). In wild type flies, such co-expression is prevented by mutually exclusive genes in p and y R8 [72], [73]. We therefore concluded that VP16:noc had a direct activating effect on rh5, without affecting any other rhodopsin.
Fig. 7. VP16- and en[R]-fusions of Noc specifically affect R8 rhodopsin expression. ![VP16- and <i>en</i>[R]-fusions of Noc specifically affect R8 rhodopsin expression.](https://www.prolekarniky.cz/media/cache/resolve/media_object_image_small/media/image/d5e3823d92742601189bd1ca78ea29df.png)
A. Schematic of VP16:noc fusion cDNA generated in UAS-constructs for over-expression (see Materials and Methods). B,C. Mild effect of VP16:noc over-expression on DRA development: Rh3 expression (red) in the DRA is weak (arrows), yet detectable (B), and Hth expression (green) is normal (C, arrows). D,E. VP16:noc has a strong activating effect on Rh5 expression (blue), resulting in a high ratio of pR8 cells with Rh5. Since Rh6 expression appears normal (green), many R8 cells now co-express the two R8 rhodopsins (white arrows). F. Schematic of Engrailed fusion cDNAs generated for Noc (see Materials and Methods). G. Ectopic over-expression of en[R]:noc has no effect on R7 rhodopsin expression (Rh3 in red; Rh4 in cyan) and DRA specification (arrows). H. Expression of Rh5 (blue) is lost upon over-expression of en[R]:noc, and Rh6 (green) is the only R8 rhodopsin remaining. Over-expression of the repressor fusion en[R]:noc (Figure 7F) also had no effect on DRA specification (Figure 7G). However, expression of Rh5 was lost, while Rh3, Rh4, and Rh6 appeared normal in the rest of the retina (Figure 7H). This R8 phenotype observed with en[R]:noc was therefore the opposite of that observed with the activator fusion VP16:noc, with both Noc fusion proteins specifically affecting rh5 expression in opposite manner. Since Otd directly activates rh3 and rh5 transcription [49], [50], Noc might function as a direct antagonist of Otd's function in R8 cells, but not in R7. Interestingly, we found strong Elb/Noc expression outside the DRA only in R8 cells (Figure 1E–I; Supplemental Figure S1D–G).
Mutations in the human Elb/Noc homolog ZNF703 promote metastasis in luminal breast cancer [22], [23], [74]. To investigate if NET family protein functions are evolutionarily conserved, we generated UAS-constructs for over-expression of both human NET family proteins in Drosophila (UAS-ZNF503, UAS-ZNF703; see Materials & Methods). Gain-of-function phenotypes in the retina were very weak, but Rh5 was expanded in R8 cells, leading, in both cases, to co-expression with Rh6 (Supplemental Figure S7A–E). The C. elegans homolog TLP-1 was not active in this assay (Supplemental Figure S7F-H). This co-expression phenotype therefore resembles most closely what we had observed for the over-expression of a VP16:noc, suggesting that genetic interaction of NET family proteins with Otd/Otx proteins is a conserved feature of these factors.
Discussion
The transcription factors Homothorax (Hth) and Extradenticle (Exd) have been well characterized as co-factors for Hox genes [48]. Hth/Exd can also act as co-factors for non-Hox transcription factors, like for Engrailed [75], [76]. Here we showed that loss of both Elb and Noc phenocopies the loss of Hth at the dorsal rim of the retina. All markers of DRA ommatidia are lost in elb,noc double mutants: Rh3 expression and Sens repression in DRA R8, as well as the DRA-specific inner photoreceptor rhabdomere morphology in DRA R7 and DRA R8. Our data shows that Elb/noc act downstream of Hth in the specification of DRA cell fates. Elb and Noc are expressed strongly in DRA R7 and R8. This expression is expanded to all R7 and R8 by ectopic Hth (but never into outer photoreceptors R1-6), while Hth expression is not affected in elb,noc double mutants. One possibility is that Elb/Noc serve as cofactors for Hth/Exd (Figure 8A), since Hth loses its potential to induce the DRA fate in a double mutant retina. The vertebrate homologs of Elb and Noc function as repressors of transcription [13]. Therefore, aspects of the Hth/Exd and Elb/Noc loss-of-function phenotypes could be due to a direct failure of their complex to repress common target genes. For instance, the de-repression of the R8 marker Sens by dominant-negative hthHM, as well as in elb,noc double mutants could be explained by loss of a repressor complex containing all four proteins. Interestingly, functional antagonism between the Hox/Hth/Exd complex and Sens have been described in the Drosophila embryo [77]. However, in this case the factors were shown to compete for overlapping binding sites in the promoter of the common target gene rhomboid. Gene expression profiling data revealed that the Hox gene Abd-B also directly represses Sens in the embryo using Hth/Exd as cofactors [78]. Elb and Noc might therefore provide a missing link for transcriptional repression of Sens by Hth/Exd.
Fig. 8. Model summarizing the roles of Elb/Noc action in photoreceptor cell fate specification. 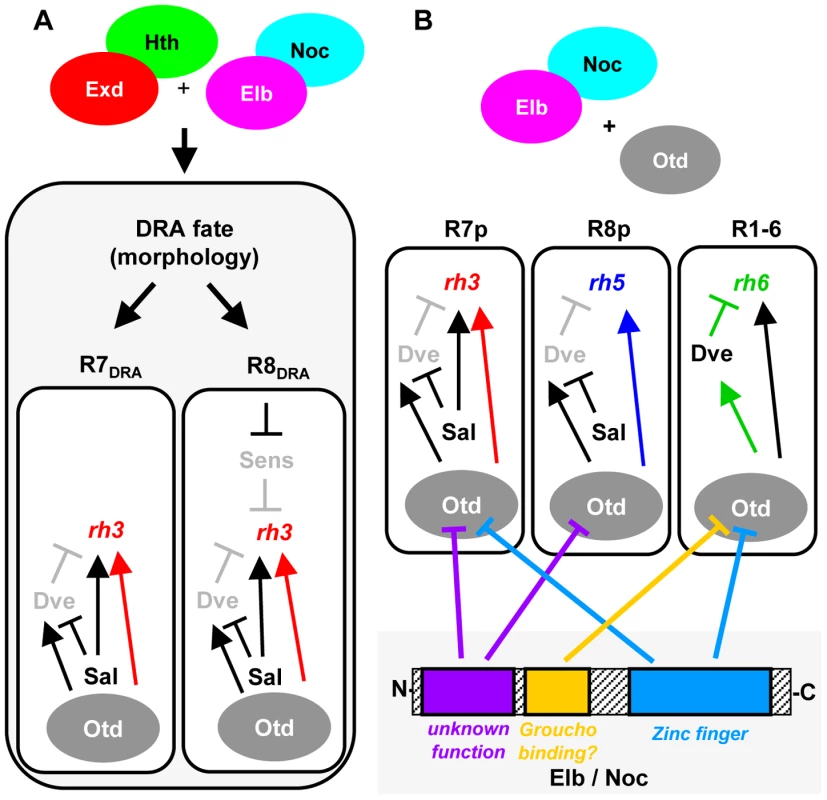
Model summarizing the role of Elb and Noc proteins in terminal specification of photoreceptor cell fates. A. Role of Elb and Noc in DRA ommatidia. Loss of function data suggests Elb & Noc function with Hth in specifying the DRA cell fates, both morphologically (rhabdomere diameter) and molecularly: repression of Sens in R8DRA by Hth/Exd and Elb/Noc is crucial for these cells to express the DRA rhodopsin marker Rh3. B. Dissection of Otd functions through antagonism with Elb/Noc protein domains. The similarity of elb/noc gain-of-function phenotypes to the previously published ocelliless (otd) loss-of-function suggests Otd/Elb/Noc proteins interact genetically. The different transcriptional effects of Otd on rh3 and rh5 expression (activation in ‘pale’ ommatidia), as well as rh6 (repression via Dve in outer photoreceptors) can be neutralized independently, using different mutated forms of Elb/Noc. The specific role of three conserved Elb/Noc domains (Sp/SPLALA, Gro-binding, Zn finger) in genetically antagonizing Otd function are indicated by inhibition arrows. Much work on NET family proteins has focused on functional characterization of their evolutionarily conserved domains. The C-terminus of NET proteins is required for nuclear localization [14], [79], as well as for self-association of the zebrafish ortholog Nlz1, although neither self-association nor heterodimerization with Nlz2 was found to be necessary for wild type function [79]. The ‘buttonhead box’ [80], a conserved 7–10 amino acid motif which we have not investigated in this study, may be required for transcriptional activation [81]. Deletion of the ‘buttonhead box’ in zebrafish Nlz proteins transformed them into dominant-negatives, an effect that was proposed to be due to reduced affinity to co-repressor Groucho and histone de-acetylases [12], [79]. Interestingly, deletion of N-terminal sequences, including the Sp/SPLALLA motif also leads to dominant negative proteins [79]. These data are consistent with our findings that a protein with a mutated Sp/SPLALLA motif has a dominant-negative effect on DRA specification. The Sp motif was proposed to mediate transcriptional repression by directly binding to cofactors [15]. It should be noted that both N-terminal Sp/SPLALLA deletion and the VP16 fusions have the same dominant-negative effect for zebrafish Nlz1 [12]. While this is consistent with a pure repressor function of the zebrafish protein, the differences between Sp/SPLALLA mutation and VP16-fusion (as well as the observation of a phenotype for the Engrailed fusion) reported in this study hint towards a more complex role of Elb and Noc in transcriptional regulation.
We showed that mutation of the conserved zinc finger of Elbow also transforms this protein into a dominant-negative. Usually, multiple zinc fingers are required for DNA binding, suggesting that the NET family zinc finger is a protein-protein interaction domain [11], [67]. Deletion of the zinc finger from zebrafish Nlz proteins leads to a loss of nuclear localization [79], and the Nlz1 zinc finger is necessary for transcriptional repression [13]. Although we cannot exclude the possibility that Elb and Noc bind DNA through their zinc finger, it is likely that mutation of the zinc finger either leads to an inactive complex by sequestration of another co-repressor, or that such complex could be trapped in the cytoplasm. Given that mutation of either Sp/SPLALLA motif or zinc finger both lead to a dominant-negative effect raises the possibility that protein binding to both motifs could be necessary for in vivo function, possibly through the formation of higher order transcriptional complexes.
Loss of both elb and noc does not result in Rhodopsin phenotypes outside the DRA. However, over-expression of different forms of Elb or Noc recapitulates all Rhodopsin phenotypes observed in otdUVI mutants [49], [50]. This phenotype might therefore arise from forcing a direct interaction between over-expressed Elb protein and Otd. Little is known about the regulatory relationship between Elb/Noc and Otd. However, the overlapping expression patterns and similar phenotypes for certain alleles of otd named ocelliless, and for no ocelli (noc) at the anterior pole of the fly embryo, as well as their common requirement in the morphogenesis of ocelli suggests that these proteins also interact positively outside of the retina. The antagonism we observed might therefore be a dominant-negative effect resulting from sequestration of the Otd protein by over-expressed Elb. Alternatively, different combinations of transcriptional cofactors present between tissues (for instance DRA versus non-DRA R8 cells) might decide whether Elb and Noc act in concert with Otd, or as antagonists.
In the retina, Otd acts in a ‘coherent feedforward loop’ with Spalt to directly activate transcription of rh3 and rh5 [50]. As a consequence, Rh3 and Rh5 are lost in otd mutants. Furthermore, Otd activates transcription of the repressor Dve, forming an ‘incoherent feedforward loop’, resulting in repression of rh3 and rh5 in outer photoreceptors. Since rh6 is activated by a distinct factor, Pph13 [51], loss of Otd leads to a specific de-repression of rh6 into outer photoreceptors [50]. We show that different domains of Elb specifically interfere with different aspects of Otd function in these feedforward loops (Figure 8B). Mutation of the Groucho-binding motif FKPY only abolishes the ability of over-expressed Elbow protein to antagonize Otd function in repressing rh6 in outer photoreceptors, while mutation of the Sp/SPLALLA motif specifically antagonizes Otd function in activating both rh3 and rh5, without affecting repression of rh6 in outer photoreceptors (mediated by induction of Dve). Furthermore, while the Elb zinc finger is also required for antagonizing the function of Otd in outer photoreceptors, it is also necessary for antagonizing activation of rh3 by Otd, but not rh5. Hence, these two activator functions of Otd could be separated by mutating the zinc finger.
The different Rhodopsin phenotypes caused by loss of Otd can be mapped to different protein domains [51]. Our data therefore reveal specific genetic interactions between the protein domains of Elb/Noc and Otd. Such interactions could be direct or be mediated through additional proteins. For instance, the Otd C-terminus mediates the repression of rh6 in outer photoreceptors [82], making it a possible interaction domain for Groucho binding to the Elb/Noc FKPY motif. The N-terminus of Otd is necessary for most activation potential on rh3, while activation of rh5 predominantly maps to the C-terminus [82]. This correlates well with the Rhodopsin-specific phenotypes we see after mutation of Sp/SPLALLA (affecting rh3 and rh5), or the zinc finger (affecting rh3 and rh6) motifs. Finally, our results using VP16 - and en[R]-fusions of Noc show that potentially direct transcriptional effects on rhodopsin genes can only be induced in R8 cells. Both fusion proteins specifically regulate expression of rh5, while all other rhodopsins remain unaffected. Elb and Noc are both expressed strongly in R8 cells outside of the DRA where they may contribute the repression of Rh5. The absence of a non-DRA R8 rhodopsin phenotype in elb,noc double mutants, as well as the R8-specific action of VP16:noc could therefore be due to the existence of redundant, R8-specific factors required for Elb/Noc function there, but not for DRA specification. These factors remain unknown, since we found that expression of elb and noc is not altered in homozygous mutants affecting p/y cell fate decisions in R8 cells (melt and wts, [72]; Supplemental Figure S8, A–D), like in R7 cells (Supplemental Figure S8E,F).
Mutations in the human Elb/Noc homolog ZNF703 promote metastasis [6]. We have shown that over-expression of both human NET family proteins UAS-ZNF503 and UAS-ZNF703 in the Drosophila retina result in weak co-expression of Rh5 and Rh6, resembling over-expression of a VP16:noc protein. It is therefore possible that the genetic interaction of NET family proteins with Otd/Otx proteins is evolutionarily conserved, especially since a central domain of Otd was previously shown to mediate mutual exclusion of Rh5 and Rh6 [82]. Here we present a new role for Drosophila NET proteins in retinal patterning. Both zebrafish homologs of Elb/Noc, Nlz1 and Nlz2 are also required for optic fissure closure during eye development [83]. Furthermore, expression of the Elb/Noc mouse homologue znf503 suggests that NET family genes are involved in the development of mammalian limbs [84]. Given previous reports from Drosophila on the proximo-distal specification of leg segments [24], it appears that NET family members act in similar processes across species. This raises the possibility that NET proteins serve as evolutionarily conserved modules that have been re-utilized for analogous processes during evolution. Based on our data, their conserved domain structure might be crucial for interacting with transcription factor networks involving conserved families of factors like Otx or Meis. Given their medical relevance in breast cancer, a better understanding of the role NET proteins play in the transcriptional control of tissue patterning will be of great importance.
Materials and Methods
Fly stocks
-
GAL4 drivers: elb-GAL4 and noc-GAL4 [24], LGMR-GAL4 [2], [85].
-
UAS-constructs: UAS-elb (U. Weihe & S.M. Cohen), UAS-noc (U. Weihe & S.M. Cohen), UAS-VP16:elb (this study), UAS-VP16:noc (this study), UAS-en[R]:elb (this study), UAS-en[R]:noc (this study), UAS-elb[SPLALLA*] (this study), UAS-noc[SPLALLA*] (this study), UAS-elb[Gro*] (this study), UAS-noc[Gro*] (this study), UAS-elb[ZnF*] (this study), UAS-noc[ZnF*] (this study), UAS-TLP-1 (this study), UAS-ZNF703 (this study), UAS-ZNF503 (this study), UAS-GFP:hth (R. Mann), UAS-myc:hth (R. Mann), UAS-GFP:hthHM (R. Mann), UAS-armS10 [64], UAS-armΔN (F. Pichaud), UAS-lacZ (J. Treisman), UAS-lacZ:NLS (Bloomington Stock Center), UAS-eGFP (M. Wernet), UAS-GFP:NLS (Bloomington Stock Center).
-
p{PZ} enhancer traps: hthl(3)06762-PZ/TM3 (Bloomington stock center), svp-PZ/TM3 (U. Gaul).
-
clonal analysis: ey-Flip (B. Dickson), FRT40-elb,noc[64-1-4][24], FRT40 - nocΔ64 [10], FRT40-arm-lacZ (J. Treisman), FRT40-p[w+] (J. Treisman), FRT82B-arm-lacZ (F. Pichaud), FRT82B-ssD115.7 (I. Duncan), FRT82B-wtsP1/TM2 (T. Xu), melt32.1a (S. Cohen).
-
Viable mutants: elb3.3.1 (U. Weihe), elb3.3.4 (U. Weihe), otdUVI (R. Reinke).
-
other: LGMR-GFP:hth (this study), rh1-lacZ, rh3-lacZ, rh4-lacZ, rh5-lacZ, rh6-lacZ (Bloomington stock center), GMR-arm* [65]
Molecular biology
Generation of LongGMR-GFP:hth transgenes
The ORF of the GFP:hth fusion protein was excised from pUAST-GFP:hth (gift from R. Mann, Columbia University) using Xba1, and ligated into a pre-existing pCasper4-LongGMR-mcs-SV40 vector (B. Mollereau, M. Wernet, and C. Desplan, unpublished). Sequence available upon request.
Generation of VP16 and En[R] fusions
A ∼500 bp 5′ fragment of elb was amplified from the full-length cDNA [10] replacing the start ATG with an EcoR1 site (altering the amino acid sequence from MLQ to EFLG). This fragment was digested EcoR1/Xho1(489) and triple ligated into pVP16 (Clontech Laboratories Inc.) digested (EcoR1/Hind3), using an Xho1/Hind3 fragment providing the rest of the elb cDNA. The same strategy was used for VP16:noc: the start ATG of noc was replaced with an EcoR1 site (MVV → EFVV). The resulting ∼650 bp fragment was digested (EcoR1/Bgl2) and triple-ligated into pVP16 (EcoR1/Xho1), using a (Bgl2/Xho1) fragment from the full-length noc cDNA [10]. The VP16 fusions were then removed from pBSK (Bgl2/Xba1) and ligated into pUAST [86].
A 892 bp 5′ fragment from engrailed (en) containing its repressor domain as PCR-amplified from a full length clone (gift from T. Cook), introducing an EcoR1 site 5′ of the ATG. In parallel, the ATG's of both elb and noc [10] were then replaced with BamH1 sites (GGATCC). The En repressor domain was then digested EcoR1/BamH1 and ligated into pBSK (EcoR1/Xho1), together with a (BamH1/Xho1) fragment providing the rest of the elb or noc coding sequence, respectively. Fusion cDNAs were sequenced and subcloned from pSK into pUAST (EcoR1/Xho1).
Site–directed mutagenesis of SPLALLA, Gro, and ZnF motifs in elb and noc
a) Site-directed mutagenesis of conserved SPLALLA motifs in both Elb and Noc: The amino acid sequence of this motif was altered to TGIVIIV in both proteins, using overlapping PCR primers with an altered nucleotide sequence (AGT CCG TTG GCG CTA TTG GCC → ACT GGG ATT GTG ATA ATC GTC for elb, and AGT CCC TTG GCT CTG CTC CTA → ACT GGC ATT GTT ATT ATC GTA for noc). The resulting ∼500 bp mutant fragment was digested Spe1/Xho1 (elb), or Hind3/Bgl2 (noc), and ligated into pBSK together with a second fragment providing the rest of the respective cDNA (Xho1/Hind3 for elb, and Bgl2/Xho1 for noc).
b) Site-directed mutagenesis of conserved Groucho-binding motifs in both Elb and Noc: The amino acid sequence was altered to IEGS in both proteins, using overlapping PCR primers (TTT AAG CCC TAC → ATT GAG GGC TCC for elb, and TTC AAG CCC TAC → ATC GAG GGC TCC for noc). The resulting mutant fragment was digested Nco1 (elb, ∼600 bp), or Bgl2/Mlu1 (noc, ∼210 bp), and ligated into pBSK-elb (digested Nco1), or pBSK-noc (digested Bgl2/Mlu1), respectively.
c) Site-directed mutagenesis of conserved zinc fingers in both Elb and Noc: The two Histidines in both proteins were altered to Alanines, using overlapping PCR primers (CAT CTG CGC ACC CAT → GCT CTG CGC ACC GCT for elb, and CAT CTG CGC ACC CAT → GCT CTG CGC ACC GCT for noc). The resulting mutant fragment was digested BsiW1 (elb, resulting fragment size: ∼220 bp), or Nco1/Sac2 (noc, ∼420 bp), and ligated into pBSK-elb (digested BsiW1), or pBSK-noc (digested Nco1/Sac2), respectively.
Finally, all mutated cDNAs were sequenced and then subcloned (EcoR1/Xho1) from pBSK into pUAST [86].
Generation of UAS-TLP-1, UAS-ZNF503, and UAS-ZNF703
The entire ORF of C. elegans TLP-1 was PCR amplified from a full-length cDNA clone (gift from D. Fitch, NYU), using restriction sites attached to the primers: EcoR1 (5′), and Xho1 (3′). The product was digested EcoR1/Xho1 and ligated into pUAST [86]. The ORF's of human homologues ZNF503 and ZNF703 were excised EcoR1/Xho1 from full-length clones MGC2555 (image:3604473) and FLJ14299 (image:5527569), respectively (from ATCC Inc.) and ligated into pUAST (digested EcoR1/Xho1).
Immunohistochemistry
Primary antibodies used were anti-βGal rabbit polyclonal 1/5000 (Cappel), anti-βGal mouse monoclonal 1/500 (Promega), anti-Homothorax guinea pig polyclonal 1/500 (R. Mann, Columbia University), anti-ElaV mouse or rat monoclonals 1/10 (Iowa University Hybridoma bank), anti-24B10 mouse monoclonal 1/10 (Iowa University Hybridoma bank), anti-Prospero mouse monoclonal 1/4 (Iowa University Hybridoma bank), anti-Senseless guinea pig polyclonal 1/10 (H. Bellen, Baylor College), anti-Rh3 mouse monoclonal 1/100 (S. Britt, University of Colorado), anti-Rh3 chicken polyclonal 1/20 (T. Cook, University of Cincinnati), anti-Rh4 mouse monoclonal 1/100 S. Britt), anti-Rh5 mouse 1/100 (S. Britt, anti-Rh6 rabbit polyclonal 1/1000 [49].
Secondary antibodies were a) AlexaFluor488 coupled made in goat or donkey, anti-rabbit, mouse, rat or guinea pig (Molecular Probes), b) Cy3 or TxRed-coupled made in goat or donkey, anti-rabbit, mouse, rat, guinea pig or chicken (Jackson Immunochemicals) and c) Cy5 coupled made in goat or donkey, anti-mouse or rat (Jackson Immunochemicals).
Pupal dissections
Pupal retinas were staged and dissected essentially as previously described [2], were fixed with 4% formaldehyde for 25 min and washed with PBS+0.3% Triton X-100. Incubation with primary antibodies was performed at 4°C overnight in BNT [PBS(1x), 250 mM NaCl, 1% BSA, 1% Tween 20], and secondary antibodies (1/200 in BNT) were applied for at least 2 hours at RT.
Adult cryostat sections
All used transgenic constructs were crossed into a cn bw background [38] to eliminate eye pigmentation. Frozen sections of adult heads were performed using a cryostat microtome (Zeiss) and deposited on superfrost Plus slides (Fisher), as previously described [43]. For immuno Histochemistry conditions were the same as above. For X-Gal reactions, Horizontal eye sections were fixed 5±10 min in PBS 0.25% gluteraldehyde. They were stained in a solution of 7.2 mM Na2HPO4, 2.8 mM NaH2PO4, 150 mM NaCl, 1 mM MgCl2, 3 mM K3[Fe(CN)6], 3 mM K4[Fe(CN)6], containing a 1/30 dilution of X-Gal (30 mg/ml in dimethyl formamide). After washing in PBS, slides were mounted in aquamount (Lerner Laboratories, Fisher).
Adult plastic sections
For the morphological examinations with transmission light microscopy, the eyes were fixed with 2% glutaraldehyde (sometimes plus 1% OsO4) in 0.05 M Na-cacodylate buffer (pH 7.2) for 2 h at 4°C. Following post-fixation with 2% OsO4 in 0.05 M Na-cacodylate buffer (pH 7.2) for 2 h at 4°C, the tissue was dehydrated with 2,2-dimethoxypropane and embedded in Epon 812. 1 µm sections for light microscopy were stained with methylene blue.
Imaging software
All fluorescent microscopy was performed using a Nikon Microphot-SA and super high pressure mercury lamps (Hg 100 watts, Ushio Electric). Confocal microscopy was performed using a Leica TCS S2 system. Digital images were produced using SPOT software.
Supporting Information
Zdroje
1. TomlinsonA (2003) Patterning the peripheral retina of the fly: decoding a gradient. Dev Cell 5(5): 799–809.
2. Wernet M.F., Labhart T., Baumann F., Mazzoni E.O., Pichaud F., et al. (2003). Homothorax switches function of Drosophila photoreceptors from color to polarized light sensors. Cell 115 : 267–79.
3. XinN, BenchabaneH, TianA, NguyenK, KlofasL, et al. (2011) Erect Wing facilitates context-dependent Wnt/Wingless signaling by recruiting the cell-specific Armadillo-TCF adaptor Earthbound to chromatin. Development 138(22): 4955–67.
4. LuqueCM, MilánM (2007) Growth control in the proliferative region of the Drosophila eye-head primordium: the elbow-noc gene complex. Dev Bio 301 ((2)): 327–39.
5. PereaD, TerrienteJ, Díaz-BenjumeaFJ (2009) Temporal and spatial windows delimit activation of the outer ring of wingless in the Drosophila wing. Dev Biol 328(2): 445–55.
6. SlorachEM, ChouJ, WerbZ (2011) Zeppo1 is a novel metastasis promoter that represses E-cadherin expression and regulates p120-catenin isoform expression and localization. Genes Dev. 2011 Mar 1 25(5): 471–84.
7. ZhaoX, YangY, FitchDH, HermanMA (2002) TLP-1 is an asymmetric cell fate determinant that responds to Wnt signals and controls male tail tip morphogenesis in C. elegans. Development 129(6): 1497–508.
8. DavisT, TrenearJ, AshburnerM (1990) The molecular analysis of the el-noc complex of Drosophila melanogaster. Genetics 126(1): 105–19.
9. DavisT, AshburnerM, JohnsonG, GubbD, RooteJ (1997) Genetic and phenotypic analysis of the genes of the elbow-no-ocelli region of chromosome 2L of Drosophila melanogaster. Hereditas 126(1): 67–75.
10. DorfmanR, GlazerL, WeiheU, WernetMF, ShiloBZ (2002) Elbow and Noc define a family of zinc finger proteins controlling morphogenesis of specific tracheal branches. Development 129(15): 3585–96.
11. NakamuraM, RunkoAP, SagerströmCG (2004) A novel subfamily of zinc finger genes involved in embryonic development. J Cell Biochem 93(5): 887–95.
12. RunkoAP, SagerströmCG (2003) Nlz belongs to a family of zinc-finger-containing repressors and controls segmental gene expression in the zebrafish hindbrain. Dev Biol 262(2): 254–67.
13. NakamuraM, ChoeSK, RunkoAP, GardnerPD, SagerströmCG (2008) Nlz1/Znf703 acts as a repressor of transcription. BMC Dev Biol 8 : 108.
14. Pereira-CastroI, CostaAM, OliveiraMJ, BarbosaI, RochaAS, et al. (2013) Characterization of human NLZ1/ZNF703 identifies conserved domains essential for proper subcellular localization and transcriptional repression. J Cell Biochem 114(1): 120–33.
15. MurataY, KimHG, RogersKT, UdvadiaAJ, HorowitzJM (1994) Negative regulation of Sp1 trans-activation is correlated with the binding of cellular proteins to the amino terminus of the Sp1 trans-activation domain. J Biol Chem 269(32): 20674–81.
16. SuK, RoosMD, YangX, HanI, PatersonAJ, et al. (1999) An N-terminal region of Sp1 targets its proteasome-dependent degradation in vitro. J Biol Chem 274(21): 15194–202.
17. JiSJ, PerizG, SockanathanS (2009) Nolz1 is induced by retinoid signals and controls motoneuron subtype identity through distinct repressor activities. Development 136(2): 231–40.
18. HoyleJ, TangYP, WielletteEL, WardleFC, SiveH (2004) nlz gene family is required for hindbrain patterning in the zebrafish. Dev Dyn 229(4): 835–46.
19. ChangSL, YanYT, ShiYL, LiuYC, TakahashiH, et al. (2011) Region - and cell type-selective expression of the evolutionarily conserved Nolz-1/zfp503 gene in the developing mouse hindbrain. Gene Expr Patterns 11(8): 525–32.
20. ChangCW, TsaiCW, WangHF, TsaiHC, ChenHY, et al. (2004) Identification of a developmentally regulated striatum-enriched zinc-finger gene, Nolz-1, in the mammalian brain. Proc Natl Acad Sci U S A 101(8): 2613–8.
21. UrbánN, Martín-IbáñezR, HerranzC, EsgleasM, CrespoE, et al. (2010) Nolz1 promotes striatal neurogenesis through the regulation of retinoic acid signaling. Neural Dev 24;5 : 21.
22. HollandDG, BurleighA, GitA, GoldgrabenMA, Perez-ManceraPA, et al. (2011) ZNF703 is a common Luminal B breast cancer oncogene that differentially regulates luminal and basal progenitors in human mammary epithelium. EMBO Mol Med 3(3): 167–80.
23. SircoulombF, NicolasN, FerrariA, FinettiP, BekhoucheI, et al. (2011) ZNF703 gene amplification at 8p12 specifies luminal B breast cancer. EMBO Mol Med 3(3): 153–66.
24. WeiheU, DorfmanR, WernetMF, CohenSM, MilánM (2003) Proximodistal subdivision of Drosophila legs and wings: the elbow-no ocelli gene complex. Development 131(4): 767–74.
25. Wolff, T R., Ready D.F. (1993). Pattern formation in the Drosophila Retina. In The Development of Drosophila melanogaster, M. M.-A. BateA, ed. (Cold Spring Harbor, Cold Spring Harbor Laboratory Press) pp., 1277–1325.
26. Meinertzhagen I.A. and Hanson,T.E. (1993). The development of the optic lobe. In The Development of Drosophila melanogaster (eds. Bate M. andMartinez Arias, A.).Plainview, NY: Cold Spring Harbor Laboratory Press, pp. 1363–1491.
27. Mollereau B., Dominguez M., Webel R., Colley, N J., Keung B., et al. (2001). Two-step process for photoreceptor formation in Drosophila. Nature 412 : 911–913.
28. CookT, PichaudF, SonnevilleR, PapatsenkoD, DesplanC (2003) Distinction between color photoreceptor cell fates is controlled by Prospero in Drosophila. Dev Cell 4(6): 853–64.
29. XieB, Charlton-PerkinsM, McDonaldE, GebeleinB, CookT (2007) Senseless functions as a molecular switch for color photoreceptor differentiation in Drosophila. Development 134(23): 4243–53.
30. MoreyM, YeeSK, HermanT, NernA, Blanco, etal (2008) Coordinate control of synaptic-layer specificity and rhodopsins in photoreceptor neurons. Nature 456(7223): 795–9.
31. Hardie R.C. (1985). Functional organization of the fly retina. In Progress in sensory physiology, H Autrum, D Ottoson, E. R Perl, R. FSchmidt, H Shimazu and W.D Willis, eds ( Berlin: Springer), pp 1–79.
32. BellML, EarlJB, BrittSG (2007) Two types of Drosophila R7 photoreceptor cells are arranged randomly: a model for stochastic cell-fate determination. J Comp Neurol 502(1): 75–85.
33. ThanawalaSU, RisterJ, GoldbergGW, ZuskovA, OlesnickyEC, et al. (2013) Regional modulation of a stochastically expressed factor determines photoreceptor subtypes in the Drosophila retina. Dev Cell 25(1): 93–105.
34. FeilerR, BjornsonR, KirschfeldK, MismerD, RubinGM, et al. (1992) Ectopic expression of ultraviolet-rhodopsins in the blue photoreceptor cells of Drosophila: visual physiology and photochemistry of transgenic animals. J Neurosci 12(10): 3862–8.
35. ChouWH, HallKJ, WilsonDB, WidemanCL, TownsonSM, et al. (1996) Identification of a novel Drosophila opsin reveals specific patterning of the R7 and R8 photoreceptor cells. Neuron 17(6): 1101–15.
36. PapatsenkoD, ShengG, DesplanC (1997) A new rhodopsin in R8 photoreceptors of Drosophila: evidence for coordinate expression with Rh3 in R7 cells. Development 124(9): 1665–73.
37. HuberA, SchulzS, BentropJ, GroellC, WolfrumU, et al. (1997) Molecular cloning of Drosophila Rh6 rhodopsin: the visual pigment of a subset of R8 photoreceptor cells. FEBS Lett 7 406(1-2): 6–10.
38. Chou, W H., Huber A., Bentrop J., Schulz S., Schwab K., et al. (1999). Patterning of the R7 and R8 photoreceptor cells of Drosophila: evidence for induced and default cell-fate specification. Development 126 : 607–616.
39. Wernet M.F., Desplan C. (2004). Building a retinal mosaic: cell fate decisions in the fly eye. Trends Cell Biol. 14 : 576–584.
40. YamaguchiS, Desplan, HeisenbergC (2010) M (2010) Contributions of photoreceptor subtypes to spectral wavelength preference in Drosophila. Proc Natl Acad Sci U S A 107(12): 5634–9.
41. MazzoniEO, CelikA, WernetMF, VasiliauskasD, JohnstonRJ, et al. (2008) Iroquois complex genes induce co-expression of rhodopsins in Drosophila. PLoS Biol 6(4): e97.
42. Labhart T., Meyer E.P. (1999). Detectors for polarized skylight in insects: a survey of ommatidial specializations in the dorsal rim area of the compound eye. Microsc. Res. Tech. 47 : 368–379.
43. Fortini, M.ERubin (1990) G.M (1990) Analysis of cis-acting requirements of the Rh3 and Rh4 genes reveals a bipartite organization to rhodopsin promoters in Drosophila melanogaster. Genes Dev 4(3): 444–63.
44. Hardie R.C. (1984). Properties of photoreceptors R7 and R8 in dorsal marginal ommatidia in the compound eyes of Musca and Calliphora. J. Comp. Physiol. A 154 : 157–165.
45. WernetMF, VelezMM, ClarkDA, Baumann-KlausenerF, BrownJR, et al. (2012) Genetic dissection reveals two separate retinal substrates for polarization vision in Drosophila. Curr Biol 22(1): 12–20.
46. WeirPT, DickinsonMH (2012) Flying Drosophila orient to sky polarization. Curr Biol 22(1): 21–7.
47. RieckhofGE, CasaresF, RyooHD, Abu-ShaarM, MannRS (1997) Nuclear translocation of extradenticle requires homothorax, which encodes an extradenticle-related homeodomain protein. 91(2): 171–83.
48. Ladam F., Sagerström C.G. (2013). Hox regulation of transcription - more complex(es). Dev Dyn. 2013 Jun 13. doi:10.1002/dvdy.23997.
49. TahayatoA, SonnevilleR, PichaudF, WernetMF, PapatsenkoD, et al. (2003) Otd/Crx, a dual regulator for the specification of ommatidial subtypes in the Drosophila retina. Dev. Cell 5(3): 391–402.
50. JohnstonRJ, OtakeY, SoodP, VogtN, BehniaR, et al. (2011) Interlocked feedforward loops control cell-type-specific Rhodopsin expression in the Drosophila eye. 145(6): 956–68.
51. MishraM, OkeA, LebelC, McDonaldEC, PlummerZ, et al. (2010) Pph13 and orthodenticle define a dual regulatory pathway for photoreceptor cell morphogenesis and function. Development 137(17): 2895–904.
52. FinkelsteinR, SmouseD, CapaciTM, SpradlingAC, PerrimonN (1990) The orthodenticle gene encodes a novel homeo domain protein involved in the development of the Drosophila nervous system and ocellar visual structures. Genes Dev 4(9): 1516–27.
53. HouHY, HefferA, AndersonWR, LiuJ, BowlerT, et al. (2009) Stripy Ftz target genes are coordinately regulated by Ftz-F1. Dev Biol 335(2): 442–53.
54. FreundCL, Gregory-EvansCY, FurukawaT, PapaioannouM, LooserJ, et al. (1997) Cone-rod dystrophy due to mutations in a novel photoreceptor-specific homeobox gene (CRX) essential for maintenance of the photoreceptor. Cell 91(4): 543–53.
55. SwainPK, ChenS, WangQL, AffatigatoLM, CoatsCL, et al. (1997) Mutations in the cone-rod homeobox gene are associated with the cone-rod dystrophy photoreceptor degeneration. Neuron 19(6): 1329–36.
56. ReichertH (2005) A tripartite organization of the urbilaterian brain: developmental genetic evidence from Drosophila. Brain Res Bull 66(4-6): 491–4.
57. TerrellD, XieB, WorkmanM, MahatoS, ZelhofA, et al. (2012) OTX2 and CRX rescue overlapping and photoreceptor-specific functions in the Drosophila eye. Dev Dyn 241(1): 215–28.
58. LeuzingerS, HirthF, GerlichD, AcamporaD, SimeoneA, et al. (1998) Equivalence of the fly orthodenticle gene and the human OTX genes in embryonic brain development of Drosophila. Development 125(9): 1703–10.
59. AcamporaD, AvantaggiatoV, TuortoF, BaroneP, ReichertH, et al. (1998) Murine Otx1 and Drosophila otd genes share conserved genetic functions required in invertebrate and vertebrate brain development. Development 125(9): 1691–702.
60. StowersRS, SchwarzTL (1999) A genetic method for generating Drosophila eyes composed exclusively of mitotic clones of a single genotype. Genetics 152(4): 1631–9.
61. FrankfortBJ, NoloR, ZhangZ, BellenH, MardonG (2001) senseless repression of rough is required for R8 photoreceptor differentiation in the developing Drosophila eye. Neuron 8 32(3): 403–14.
62. DomingosPM, BrownS, BarrioR, RatnakumarK, FrankfortBJ, et al. (2004) Regulation of R7 and R8 differentiation by the spalt genes. Dev Biol 273(1): 121–33.
63. Wada S. (1974). Spezielle randzonale Ommatidien der Fliegen (Diptera: Brachycera): Architektur und Verteilung in den Komplexaugen. Z. Morph. Tiere 77 : 87–125.
64. van deWetering, CavalloM, DooijesR, van BeestD, van EsMJ, et al. (1997) Armadillo coactivates transcription driven by the product of the Drosophila segment polarity gene dTCF. Cell 88(6): 789–99.
65. FreemanM, BienzM (2000) EGF receptor/Rolled MAP kinase signalling protects cells against activated Armadillo in the Drosophila eye. EMBO Rep 2(2): 157–62.
66. Wolfe, S A., Nekludova L., & Pabo, C O. (2000). DNA recognition by Cys2His2 zinc finger proteins. Annual Review of Biophysics and Biomolecular Structure, 29 : 183–212.
67. BrayerKJ, SegalDJ (2008) Keep your fingers off my DNA: protein-protein interactions mediated by C2H2 zinc finger domains. Cell Biochem Biophys 50(3): 111–31.
68. Ryoo, H D., Marty T., Casares F., Affolter M., and Mann, R S. (1999). Regulation of Hox target genes by a DNA bound Homothorax/Hox/Extradenticle complex. Development 126 : 5137–5148.
69. VandendriesER, JohnsonD, ReinkeR (1996) orthodenticle is required for photoreceptor cell development in the Drosophila eye. Dev Biol 173(1): 243–55.
70. HiraiH, TaniT, KikyoN (2010) Structure and functions of powerful transactivators: VP16, MyoD and FoxA. Int J Dev Biol 54(11-12): 1589–96.
71. MorganR (2006) Engrailed: complexity and economy of a multi-functional transcription factor. FEBS Lett 15 580(11): 2531–3.
72. Mikeladze-DvaliT, WernetMF, PistilloD, TelemanA, ChenYW, et al. (2005) The growth regulators Dlats and Melted interact in a bistable loop to specify opposite fates in R8 photoreceptors. Cell 122(5): 775–87.
73. JukamD, DesplanC (2011) Binary regulation of Hippo pathway by Merlin/NF2, Kibra, Lgl, and Melted specifies and maintains postmitotic neuronal fate. Dev Cell. 2011 Nov 15 21(5): 874–87.
74. BazarovAV, YaswenP (2011) Who is in the driver's seat in 8p12 amplifications? ZNF703 in luminal B breast tumors. Breast Cancer Res 25 13(3): 308.
75. KobayashiM, FujiokaM, TolkunovaEN, DekaD, Abu-ShaarM, et al. (2003) Engrailed cooperates with extradenticle and homothorax to repress target genes in Drosophila. Development 130(4): 741–51.
76. FujiokaM, GebeleinB, CoferZC, MannRS, JaynesJB (2012) Engrailed cooperates directly with Extradenticle and Homothorax on a distinct class of homeodomain binding sites to repress sloppy paired. Dev Biol 366(2): 382–92.
77. Li-KroegerD, WittLM, GrimesHL, CookTA, GebeleinB (2008) Hox and senseless antagonism functions as a molecular switch to regulate EGF secretion in the Drosophila PNS. Dev Cell 15(2): 298–308.
78. ZhaiZ, FuchsAL, LohmannI (2010) Cellular analysis of newly identified Hox downstream genes in Drosophila. Eur J Cell Biol 89(2-3): 273–8.
79. RunkoAP, SagerströmCG (2004) Isolation of nlz2 and characterization of essential domains in Nlz family proteins. J Biol Chem 279(12): 11917–25.
80. WimmerEA, JackleH, PfeifleC, CohenSM (1993) A Drosophila homologue of human Sp1 is a head-specific segmentation gene. Nature 66 : 690–694.
81. AthanikarJN, SanchezHB, OsborneTF (1997) Promoter selective transcriptional synergy mediated by sterol regulatory element binding protein and Sp1: A critical role for the Btd domain of Sp1. Mol Cell Biol 17 : 5193–5200.
82. McDonaldEC, XieB, WorkmanM, Charlton-PerkinsM, TerrellDA, et al. (2010) Separable transcriptional regulatory domains within Otd control photoreceptor terminal differentiation events. Dev Biol 347(1): 122–32.
83. BrownJD, DuttaS, BhartiK, BonnerRF, MunsonPJ, et al. (2009) Expression profiling during ocular development identifies 2 Nlz genes with a critical role in optic fissure closure. Proc Natl Acad Sci USA 106(5): 1462–7.
84. McGlinnE, RichmanJM, MetzisV, TownL, ButterfieldNC, et al. (2008) Expression of the NET family member Zfp503 is regulated by hedgehog and BMP signaling in the limb. Dev Dyn 237(4): 1172–82.
85. MosesK, RubinGM (1991). Glass encodes a site-specific DNA-binding protein that is regulated in response to positional signals in the developing Drosophila eye. Genes Dev 5 : 583–593.
86. BrandAH, PerrimonN (1993). Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118 : 401–415.
Štítky
Genetika Reprodukční medicína
Článek Sex Chromosome Turnover Contributes to Genomic Divergence between Incipient Stickleback SpeciesČlánek Final Pre-40S Maturation Depends on the Functional Integrity of the 60S Subunit Ribosomal Protein L3
Článek vyšel v časopisePLOS Genetics
Nejčtenější tento týden
2014 Číslo 3
-
Všechny články tohoto čísla
- , a Gene That Influences the Anterior Chamber Depth, Is Associated with Primary Angle Closure Glaucoma
- Genomic View of Bipolar Disorder Revealed by Whole Genome Sequencing in a Genetic Isolate
- The Rate of Nonallelic Homologous Recombination in Males Is Highly Variable, Correlated between Monozygotic Twins and Independent of Age
- Genetic Determinants Influencing Human Serum Metabolome among African Americans
- Heterozygous and Inherited Mutations in the Smooth Muscle Actin () Gene Underlie Megacystis-Microcolon-Intestinal Hypoperistalsis Syndrome
- Genome-Wide Meta-Analysis of Homocysteine and Methionine Metabolism Identifies Five One Carbon Metabolism Loci and a Novel Association of with Ischemic Stroke
- Cancer Evolution Is Associated with Pervasive Positive Selection on Globally Expressed Genes
- Genetic Diversity in the Interference Selection Limit
- Integrating Multiple Genomic Data to Predict Disease-Causing Nonsynonymous Single Nucleotide Variants in Exome Sequencing Studies
- An Evolutionary Analysis of Antigen Processing and Presentation across Different Timescales Reveals Pervasive Selection
- Cleavage Factor I Links Transcription Termination to DNA Damage Response and Genome Integrity Maintenance in
- DNA Dynamics during Early Double-Strand Break Processing Revealed by Non-Intrusive Imaging of Living Cells
- Genetic Basis of Metabolome Variation in Yeast
- Modeling 3D Facial Shape from DNA
- Dysregulated Estrogen Receptor Signaling in the Hypothalamic-Pituitary-Ovarian Axis Leads to Ovarian Epithelial Tumorigenesis in Mice
- Photoactivated UVR8-COP1 Module Determines Photomorphogenic UV-B Signaling Output in
- Local Evolution of Seed Flotation in Arabidopsis
- Chromatin Targeting Signals, Nucleosome Positioning Mechanism and Non-Coding RNA-Mediated Regulation of the Chromatin Remodeling Complex NoRC
- Nucleosome Acidic Patch Promotes RNF168- and RING1B/BMI1-Dependent H2AX and H2A Ubiquitination and DNA Damage Signaling
- The -Induced Arabidopsis Transcription Factor Attenuates ABA Signaling and Renders Seedlings Sugar Insensitive when Present in the Nucleus
- Changes in Colorectal Carcinoma Genomes under Anti-EGFR Therapy Identified by Whole-Genome Plasma DNA Sequencing
- Selection of Orphan Rhs Toxin Expression in Evolved Serovar Typhimurium
- FAK Acts as a Suppressor of RTK-MAP Kinase Signalling in Epithelia and Human Cancer Cells
- Asymmetry in Family History Implicates Nonstandard Genetic Mechanisms: Application to the Genetics of Breast Cancer
- Co-translational Localization of an LTR-Retrotransposon RNA to the Endoplasmic Reticulum Nucleates Virus-Like Particle Assembly Sites
- Sex Chromosome Turnover Contributes to Genomic Divergence between Incipient Stickleback Species
- DWARF TILLER1, a WUSCHEL-Related Homeobox Transcription Factor, Is Required for Tiller Growth in Rice
- Functional Organization of a Multimodular Bacterial Chemosensory Apparatus
- Genome-Wide Analysis of SREBP1 Activity around the Clock Reveals Its Combined Dependency on Nutrient and Circadian Signals
- The Yeast Sks1p Kinase Signaling Network Regulates Pseudohyphal Growth and Glucose Response
- An Interspecific Fungal Hybrid Reveals Cross-Kingdom Rules for Allopolyploid Gene Expression Patterns
- Temperate Phages Acquire DNA from Defective Prophages by Relaxed Homologous Recombination: The Role of Rad52-Like Recombinases
- Dying Cells Protect Survivors from Radiation-Induced Cell Death in
- Determinants beyond Both Complementarity and Cleavage Govern MicroR159 Efficacy in
- The bHLH Factors Extramacrochaetae and Daughterless Control Cell Cycle in Imaginal Discs through the Transcriptional Regulation of the Phosphatase
- The First Steps of Adaptation of to the Gut Are Dominated by Soft Sweeps
- Bacterial Regulon Evolution: Distinct Responses and Roles for the Identical OmpR Proteins of Typhimurium and in the Acid Stress Response
- Final Pre-40S Maturation Depends on the Functional Integrity of the 60S Subunit Ribosomal Protein L3
- Mitogen-Activated Protein Kinase (MAPK) Pathway Regulates Branching by Remodeling Epithelial Cell Adhesion
- CDP-Diacylglycerol Synthetase Coordinates Cell Growth and Fat Storage through Phosphatidylinositol Metabolism and the Insulin Pathway
- Coronary Heart Disease-Associated Variation in Disrupts a miR-224 Binding Site and miRNA-Mediated Regulation
- TBX3 Regulates Splicing : A Novel Molecular Mechanism for Ulnar-Mammary Syndrome
- Identification of Interphase Functions for the NIMA Kinase Involving Microtubules and the ESCRT Pathway
- Is a Cancer-Specific Fusion Gene Recurrent in High-Grade Serous Ovarian Carcinoma
- LILRB2 Interaction with HLA Class I Correlates with Control of HIV-1 Infection
- Worldwide Patterns of Ancestry, Divergence, and Admixture in Domesticated Cattle
- Parent-of-Origin Effects Implicate Epigenetic Regulation of Experimental Autoimmune Encephalomyelitis and Identify Imprinted as a Novel Risk Gene
- The Phospholipid Flippase dATP8B Is Required for Odorant Receptor Function
- Noise Genetics: Inferring Protein Function by Correlating Phenotype with Protein Levels and Localization in Individual Human Cells
- DUF1220 Dosage Is Linearly Associated with Increasing Severity of the Three Primary Symptoms of Autism
- Sugar and Chromosome Stability: Clastogenic Effects of Sugars in Vitamin B6-Deficient Cells
- Pheromone-Sensing Neurons Expressing the Ion Channel Subunit Stimulate Male Courtship and Female Receptivity
- Gene-Based Sequencing Identifies Lipid-Influencing Variants with Ethnicity-Specific Effects in African Americans
- Telomere Shortening Unrelated to Smoking, Body Weight, Physical Activity, and Alcohol Intake: 4,576 General Population Individuals with Repeat Measurements 10 Years Apart
- A Combination of Activation and Repression by a Colinear Hox Code Controls Forelimb-Restricted Expression of and Reveals Hox Protein Specificity
- An ER Complex of ODR-4 and ODR-8/Ufm1 Specific Protease 2 Promotes GPCR Maturation by a Ufm1-Independent Mechanism
- Epigenetic Control of Effector Gene Expression in the Plant Pathogenic Fungus
- Genetic Dissection of Photoreceptor Subtype Specification by the Zinc Finger Proteins Elbow and No ocelli
- Validation and Genotyping of Multiple Human Polymorphic Inversions Mediated by Inverted Repeats Reveals a High Degree of Recurrence
- CYP6 P450 Enzymes and Duplication Produce Extreme and Multiple Insecticide Resistance in the Malaria Mosquito
- GC-Rich DNA Elements Enable Replication Origin Activity in the Methylotrophic Yeast
- An Epigenetic Signature in Peripheral Blood Associated with the Haplotype on 17q21.31, a Risk Factor for Neurodegenerative Tauopathy
- Lsd1 Restricts the Number of Germline Stem Cells by Regulating Multiple Targets in Escort Cells
- RBPJ, the Major Transcriptional Effector of Notch Signaling, Remains Associated with Chromatin throughout Mitosis, Suggesting a Role in Mitotic Bookmarking
- The Membrane-Associated Transcription Factor NAC089 Controls ER-Stress-Induced Programmed Cell Death in Plants
- The Functional Consequences of Variation in Transcription Factor Binding
- Comparative Genomic Analysis of N-Fixing and Non-N-Fixing spp.: Organization, Evolution and Expression of the Nitrogen Fixation Genes
- An Insulin-to-Insulin Regulatory Network Orchestrates Phenotypic Specificity in Development and Physiology
- Suicidal Autointegration of and Transposons in Eukaryotic Cells
- A Multi-Trait, Meta-analysis for Detecting Pleiotropic Polymorphisms for Stature, Fatness and Reproduction in Beef Cattle
- Genome-Wide DNA Methylation Analysis Predicts an Epigenetic Switch for GATA Factor Expression in Endometriosis
- Genome-Wide DNA Methylation Analysis of Human Pancreatic Islets from Type 2 Diabetic and Non-Diabetic Donors Identifies Candidate Genes That Influence Insulin Secretion
- The and Hybrid Incompatibility Genes Suppress a Broad Range of Heterochromatic Repeats
- The Kil Peptide of Bacteriophage λ Blocks Cytokinesis via ZipA-Dependent Inhibition of FtsZ Assembly
- PLOS Genetics
- Archiv čísel
- Aktuální číslo
- Informace o časopisu
Nejčtenější v tomto čísle- Worldwide Patterns of Ancestry, Divergence, and Admixture in Domesticated Cattle
- Genome-Wide DNA Methylation Analysis of Human Pancreatic Islets from Type 2 Diabetic and Non-Diabetic Donors Identifies Candidate Genes That Influence Insulin Secretion
- Genetic Dissection of Photoreceptor Subtype Specification by the Zinc Finger Proteins Elbow and No ocelli
- GC-Rich DNA Elements Enable Replication Origin Activity in the Methylotrophic Yeast
Kurzy
Zvyšte si kvalifikaci online z pohodlí domova
Současné možnosti léčby obezity
nový kurzAutoři: MUDr. Martin Hrubý
Všechny kurzyPřihlášení#ADS_BOTTOM_SCRIPTS#Zapomenuté hesloZadejte e-mailovou adresu, se kterou jste vytvářel(a) účet, budou Vám na ni zaslány informace k nastavení nového hesla.
- Vzdělávání



